
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 18 January 2021
Sec. Evolutionary and Genomic Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.605896
This article is part of the Research TopicPlasmid Transfer: Mechanisms, Ecology, Evolution, and ApplicationsView all 34 articles
 Ian S. Barton1†
Ian S. Barton1† Justin L. Eagan1†
Justin L. Eagan1† Priscila A. Nieves-Otero2
Priscila A. Nieves-Otero2 Ian P. Reynolds1
Ian P. Reynolds1 Thomas G. Platt2
Thomas G. Platt2 Clay Fuqua1*
Clay Fuqua1*Members of the Rhizobiaceae, often carry multiple secondary replicons in addition to the primary chromosome with compatible repABC-based replication systems. Unlike secondary chromosomes and chromids, repABC-based megaplasmids and plasmids can undergo copy number fluctuations and are capable of conjugative transfer in response to environmental signals. Several Agrobacterium tumefaciens lineages harbor three secondary repABC-based replicons, including a secondary chromosome (often linear), the Ti (tumor-inducing) plasmid and the At megaplasmid. The Ti plasmid is required for virulence and encodes a conjugative transfer (tra) system that is strictly regulated by a subset of plant-tumor released opines and a well-described acyl-homoserine lactone (AHL)-based quorum-sensing mechanism. The At plasmids are generally not required for virulence, but carry genes that enhance rhizosphere survival, and these plasmids are often conjugatively proficient. We report that the At megaplasmid of the octopine-type strain A. tumefaciens 15955 encodes a quorum-controlled conjugation system that directly interacts with the paralogous quorum sensing system on the co-resident Ti plasmid. Both the pAt15955 and pTi15955 plasmids carry homologs of a TraI-type AHL synthase, a TraR-type AHL-responsive transcription activator, and a TraM-type anti-activator. The traI genes from both pTi15955 and pAt15955 can direct production of the inducing AHL (3-octanoyl-L-homoserine lactone) and together contribute to the overall AHL pool. The TraR protein encoded on each plasmid activates AHL-responsive transcription of target tra gene promoters. The pAt15955 TraR can cross-activate tra genes on the Ti plasmid as strongly as its cognate tra genes, whereas the pTi15955 TraR is preferentially biased toward its own tra genes. Putative tra box elements are located upstream of target promoters, and comparing between plasmids, they are in similar locations and share an inverted repeat structure, but have distinct consensus sequences. The two AHL quorum sensing systems have a combinatorial effect on conjugative transfer of both plasmids. Overall, the interactions described here have implications for the horizontal transfer and evolutionary stability of both plasmids and, in a broad sense, are consistent with other repABC systems that often have multiple quorum-sensing controlled secondary replicons.
Many members of the Alphaproteobacteria (APB) group contain large secondary replicons, in addition to the primary chromosomal replicon with the majority of core genes. These secondary replicons which include megaplasmids, chromids (large replicons, >350 kb, that carry essential genes but are thought to have arisen by horizontal gene transfer), and secondary chromosomes often have repABC-based replication origins (Pinto et al., 2012; Fournes et al., 2018). Several APB, notably within the Family Rhizobiaceae, harbor multiple large secondary replicons that are also stably maintained by compatible repABC systems. These plasmids have extensive systems for their retention and proliferation, including copy number control, efficient partitioning with the chromosomes, and conjugative transfer (Pinto et al., 2012).
The Ti plasmid from the plant pathogen Agrobacterium tumefaciens is one of the most well-studied repABC replicons (Gordon and Christie, 2014), in which control of plasmid copy number and horizontal transfer is directly linked to bacterial population dynamics resulting from interactions with host plants. Octopine-type and nopaline-type Ti plasmids of A. tumefaciens have been most extensively studied for this regulation, but multiple other agrobacterial and rhizobial plasmids exhibit similar regulatory patterns (He and Fuqua, 2006; Wetzel et al., 2015). A. tumefaciens causes the plant neoplastic disease crown gall, by driving cross-kingdom horizontal gene transfer to host plants, delivering a segment of the Ti plasmid called the T-DNA, into host cells (Chilton et al., 1977). This process is strictly regulated by plant-released signals via the VirA-VirG two-component system (Winans, 1991) which activates virulence (vir) gene expression including the VirB Type IV secretion system (T4SS) that delivers the T-DNA to plant cells (Christie, 2001). In the plant, the T-DNA is ushered into the nucleus and inserted into the host genome (Gelvin, 2003). Stable integration of the T-DNA and expression of the transferred genes results in neoplastic growth of the infected tissue, and visible galls. These genes also encode proteins which drive production of unusual metabolites called opines that are utilized as semi-exclusive nutrients by agrobacteria in the rhizosphere via catabolic functions primarily encoded on the Ti plasmids (Dessaux et al., 1998). A subset of these opines, called conjugal opines, stimulate interbacterial conjugative transfer of the Ti plasmid, and increased Ti plasmid copy number (Klapwijk et al., 1978; Ellis et al., 1982). In most cases, the conjugal opines act indirectly, by stimulating expression of an acyl-homoserine lactone (AHL) quorum sensing (QS) mechanism. The Ti plasmid encodes a LuxI-type AHL synthase called TraI that synthesizes the AHL N-3-oxo-octanoyl-L-homoserine lactone (3-oxo-C8-HSL) (Fuqua and Winans, 1994; Hwang et al., 1994; Moré et al., 1996). As with many other AHL quorum sensing systems, the signal can diffuse across the membrane and out of the cell. However, as the population density of AHL-producing agrobacteria increases, the relative AHL concentration is also elevated, eventually signaling a bacterial quorum. The AHL-dependent, LuxR-type protein TraR, also encoded on the Ti plasmid, interacts with the AHL to activate transcription of conjugative transfer (tra and trb) genes and the repABC replication genes of the Ti plasmid, driving conjugation and increased copy number (Fuqua and Winans, 1996; Li and Farrand, 2000; Pappas and Winans, 2003). In addition, TraR activates traI expression, resulting in a positive feedback loop on AHL production. Expression of the Ti plasmid traR gene is under the tight transcriptional control of the conjugal opine, through an opine-responsive regulator (Fuqua and Winans, 1994). Therefore, conjugative transfer and copy number of the Ti plasmid is strictly regulated by the plant-tumor-released conjugal opine via traR expression, and AHL-producing bacterial population density via the QS mechanism. Other Ti plasmid-encoded functions modulate these processes, most notably the antiactivator TraM which directly inhibits TraR, and maintains the minimum threshold of the protein required to activate tra/trb and repABC expression (Fuqua et al., 1995; Hwang et al., 1995).
In the best studied A. tumefaciens strains, conjugative transfer and copy number of the co-resident At megaplasmid is not recognized to be strongly affected by the opine-responsive and quorum sensing control systems which regulate these processes for the Ti plasmid. Rather, At plasmid conjugative transfer is commonly regulated through the rctA and rctB repressor cascade, which seems to provide largely homeostatic control of At plasmid conjugation (Perez-Mendoza et al., 2005). However, in certain nopaline-type A. tumefaciens such as strain C58, the conjugal opines that activate quorum sensing can also stimulate transcription of the rctA and rctB genes (Lang et al., 2013), thereby providing a regulatory linkage between these plasmids.
Here, we show that the octopine-inducible, quorum sensing-dependent conjugation system of the Ti plasmid from A. tumefaciens 15955 interacts directly with the conjugation system of the co-resident At megaplasmid. We find that both plasmids contribute to the same AHL pool through independent traI AHL synthases, and that they each also encode AHL-responsive TraR transcription factors that can cross-regulate genes on both replicons, impacting the overall quorum-sensing response as well as conjugative transfer of both the Ti plasmid and the At megaplasmid. The co-regulation of conjugative transfer functions observed here suggests direct coordination between both replicons that may affect their population-level stability and increase overall horizontal transfer. Our observations provide another alternate example of complex interactions between conjugative transfer regulatory elements of co-resident repABC plasmids in the rhizobia (He et al., 2003; Castellani et al., 2019; Cervantes et al., 2019). Similar interactions are likely to occur within other systems with multiple repABC-based replicons, several examples of which exhibit quorum-sensing controlled replication and horizontal gene transfer systems.
All strains, plasmids, and oligonucleotides used in this study are described in Supplementary Tables 1, 2. Chemicals, antibiotics, and culture media were obtained from Fisher Scientific (Pittsburgh, PA), Sigma Aldrich (St. Louis, MO), and Goldbio (St. Louis, MO) unless otherwise noted. Plasmid design and verification as well as strain creation was performed as described previously (Morton and Fuqua, 2012a) with specific details in the text. Oligonucleotide primers were ordered from Integrated DNA Technologies (Coralville, IA). Single primer extension DNA sequencing was performed by ACGT, Inc. (Wheeling, IL). Plasmids were introduced into Escherichia coli via transformation with standard chemically competent preparations and into A. tumefaciens via electroporation or conjugation. In-frame deletions of traI and traR were constructed using a previously described allelic replacement method (Morton and Fuqua, 2012a).
E. coli was cultured in standard LB broth with or without 1.5% (w v−1) agar. Unless noted otherwise, A. tumefaciens strains were grown on AT minimal medium containing 0.5% (w v−1) glucose and 15 mM ammonium sulfate without added FeS04 (ATGN) (Morton and Fuqua, 2012b) or AT with 5% (w v−1) sucrose as the sole carbon source (ATSN). When required, appropriate antibiotics were added to the medium as follows: for E. coli, 100 μg ml−1 ampicillin (Ap), 50 μg ml−1 gentamicin (Gm), and 50 μg ml−1 kanamycin (Km); and for A. tumefaciens, 300 μg ml−1 Gm, 300 μg ml−1 Km, and 300 μg ml−1 spectinomycin (Sp). When indicated, A. tumefaciens 15955 strains were grown in ATGN media supplemented with 3.25 mM octopine (Octopine, + Oct.) and/or 0.1 μM synthetic 3-oxo-octanoyl-L-homoserine lactone (+AHL, 3-oxo-C8-HSL).
The genome assembly for A. tumefaciens 15955 was deposited into GenBank by Academia Sinica (Genbank Access. No. 7601828). Multiple and pairwise sequence alignments were performed with Clustal Omega (Sievers et al., 2011) using default parameters. Promoter sequence motifs were generated with WebLogo (Crooks et al., 2004).
Plasmid-borne expression constructs were created using the LacIQ encoding, IPTG-inducible series of pSRK vectors (Khan et al., 2008). Coding sequences of traR and traI from pAt15955 (At15955_50350 and At15955_49060, respectively) and pTi15955 (At15955_53990 and At15955_54620, respectively) were PCR amplified from A. tumefaciens 15955 gDNA using primers for the corresponding gene (Supplementary Table 3) and Phusion DNA polymerase (New England Biolabs, Beverly, MA). PCR products were ligated into pGEM-T easy (Promega, Madison, WI), excised by restriction enzyme digest, and ligated into either pSRKKm or pSRKGm. Final pSRK constructs were confirmed with sequencing and restriction enzyme digestion prior to electroporation into A. tumefaciens.
Promoter expression reporter plasmids were created in pRA301 (Akakura and Winans, 2002a). Promoter regions for traA (At15955_53920 for pTi15955 and At15955_50400 for pAt15955) and traI (~500 bp) were amplified with PCR (as above) using A. tumefaciens 15955 gDNA and the appropriate primers (Supplementary Table 3). All reporter gene fusions have lacZ translationally fused in frame at the start codon of each gene. PCR products were then assembled into restriction enzyme digests of pRA301 in a two-part NEBuilder isothermal assembly reaction (New England Biolabs, Beverly, MA), verified by PCR and sequencing, and introduced into A. tumefaciens via electroporation.
For octopine and AHL induction of traA and traI promoters, A. tumefaciens 15955 carrying the appropriate pRA301-based reporter constructs were grown to an optical density at 600 nm (OD600) of 0.4–0.6 in ATGN supplemented with appropriate antibiotics and spotted onto 0.2 μm sterile filter disks (0.45 μm pore size) placed on small ATGN plates (35 × 10 mm) supplemented with octopine (3.25 mM) and/or AHL (0.1 μM 3-oxo-C8-HSL) and incubated for 48 h at 28°C. A. tumefaciens 15955 strains were then removed from the filter discs, diluted to an OD600 ~0.6 in sterile water and used as in standard β-galactosidase assays, as previously described (Miller, 1972).
To determine the effect of traR expression on promoter activities of PtraA and PtraI from pAt15955 and pTi15955, A. tumefaciens 15955 and A. tumefaciens NTL4 derivatives carrying pSRK-based traR expression constructs and pRA301-based lacZ reporter constructs (Supplementary Table 1) were grown in ATGN liquid cultures with appropriate antibiotics and 400 μM IPTG in the presence or absence of AHL to an OD600 ~0.6 and used in standard β-galactosidase assays.
For AHL detection on solid media, we utilized the AHL biosensor strain [A. tumefaciens NTL4 (pCF218)(pCF372)] (Fuqua and Winans, 1996). This biosensor relies on a high expression, plasmid-borne copy of the AHL-responsive TraR protein from A. tumefaciens R10 (pCF218), and a second plasmid carrying a TraR-dependent promoter fused to lacZ (traI-lacZ, pCF372), both harbored in A. tumefaciens NTL4, a Ti-plasmidless strain which lacks an AHL synthase, and therefore does not produce endogenous AHLs. The biosensor is a sensitive detection system for exogenous AHLs, expressing lacZ only when inducing AHLs are present (McLean et al., 1997). Strain derivatives to be tested for AHLs were grown overnight to an OD600 0.4–0.6 in ATGN supplemented with appropriate antibiotics and then each strain to be tested was struck onto ATGN agar supplemented with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) ~1 cm from the AHL biosensor strain. Media used in assays to test for AHL production with A. tumefaciens NTL4-derivatives also had 400 μM IPTG unless otherwise noted. The AHL biosensor was also used to detect AHL in cell-free supernatant from A. tumefaciens strains. Strains to be tested were grown overnight in ATGN supplemented with appropriate antibiotics to an OD600 of 0.4–0.6 and 1 mL of culture was then pelleted, supernatant removed and re-centrifuged to remove additional cell debris, and filter sterilized (0.2 μm) to create cell-free supernatant (CFS). One hundred microliter of CFS was then added to cultures of the AHL biosensor, grown to an OD600 0.4–0.6, and assayed for promoter activity using standard β-galactosidase assays. Relative AHL levels for CFS were then reported as Miller units normalized to the OD600 of the tested culture.
Conjugation assays were performed as described previously (Heckel et al., 2014) with alterations. Briefly, liquid cultures of donors and recipients were grown to mid-logarithmic phase in ATGN at 30°C and normalized to OD600 ~0.6, mixed 1:1, and 100 μL was spotted onto 0.2 μm pore size cellulose acetate filter disks placed on small ATGN plates (35 × 10 mm). Conjugations were performed on ATGN with A. tumefaciens 15955 ectopically expressing plasmid-borne Plac-traR fusions with or without 400 μM IPTG and appropriate antibiotics. For AHL rescue of conjugation in the traI double mutant, varying amounts of 3-oxo-C8-HSL were also added. Following incubation (18 h, 28°C), cells were suspended in liquid ATGN, serially diluted, and spotted onto media supplemented with appropriate antibiotics to enumerate donors and transconjugants. Conjugation efficiencies were determined as the ratio of transconjugants per output donor.
Assays were performed with at least three biological replicates. Standard deviation was determined and presented as error bars. Where appropriate, significance was conservatively evaluated using 2-sided, paired t-tests.
A. tumefaciens 15955 is a member of the Genomospecies Group 1 along with similar derivatives A. tumefaciens Ach5 and A6 (Lassalle et al., 2011). Sequence analysis of pAt15955 identified conjugation genes homologous to the quorum-sensing regulated tra/trb system encoded on pTi15955 and other Ti plasmids (Alt-Mörbe et al., 1996; Farrand et al., 1996), including tra and trb structural genes and homologs of the quorum sensing regulators traI, traR, and traM (Supplementary Figure 1A). The conjugation system encoded on pAt15955 is distinct from those found on many other At plasmids, including those of the type strain A. tumefaciens C58, a nopaline-type strain (Genomospecies 8), in which pAtC58 conjugates via a virB-like type IV secretion system (avhB) (Chen et al., 2002) regulated by rctA/rctB-like regulators commonly found for other rhizobial plasmids (Perez-Mendoza et al., 2005; Nogales et al., 2013) (Supplementary Figure 1). Given the presence of TraI, TraR and TraM homologs on pAt15955 (Supplementary Figures 1B,C), it was conceivable that the conjugative transfer of this plasmid might be regulated by quorum sensing and that pAt15955 and pTi15955 could contribute and respond to a common AHL pool. We previously showed that pAt15955 encodes a functional conjugation system and observed that conjugation of pAt15955Δ270 (a derivative with a 270 kb deletion on pAt) which lacks cis-encoded trb conjugation functions (Supplementary Figure 1A), could be stimulated by octopine-induction in a pTi15955-dependent manner (Barton et al., 2019) suggesting a direct interaction between the conjugation systems of both plasmids.
To compare the impact of octopine and AHL cues on the expression of pAt15955 and pTi15955 conjugative transfer (tra) genes, we constructed independent translational lacZ fusions to the upstream regions and start codons of the predicted traA nickase/helicase homologs and the traI AHL synthase homologs, carried on the broad host range plasmid pRA301 (Akakura and Winans, 2002b) (Supplementary Table 2, Methods). These fusion plasmids were introduced into wild type A. tumefaciens 15955, and used in β-galactosidase assays to measure gene expression in the presence or absence of 0.1 μM 3-oxo-C8-HSL and/or 3.25 mM octopine (Figure 1, Materials and Methods). The β-galactosidase activity imparted by the traA-lacZ fusions from pTi15955 (green bars) and pAt15955 (blue bars) was low in the absence of octopine or AHL addition (Figure 1A). Addition of octopine alone (medium-fill bars) or octopine plus AHL (dark-fill bars) significantly increased activity from the traAAt-lacZ and traATi-lacZ fusions, although overall activity of each was relatively low. The same pattern of regulation was observed for 15955 harboring the traI-lacZ fusions, although the level of activity from both traITi-lacZ fusions was considerably higher than the traA-lacZ fusions, and addition of octopine and AHL together had a stronger effect than octopine alone (Figure 1B). The traIAt-lacZ fusion (blue bars) showed a similar pattern although its overall expression was lower than the traITi fusion (green bars). Taken together with our prior observation of pTi15955-dependent octopine-induction of pAt15955 conjugation (Barton et al., 2019), this data suggests that pTi15955 is directly involved in activation of pAt15955 tra gene expression in response to octopine (Fuqua and Winans, 1994, 1996). Additionally, octopine-induction had differential effects on pAt15955 and pTi15955 gene activation, where octopine addition stimulated comparable modest expression from both traA genes (Figure 1A), but had a more dramatic effect on traITi expression than that of traIAt (Figure 1B), suggesting complex cross-regulation between both plasmids and supporting our previous observations for pAt15955 conjugation control, as well as previous work on the activation of octopine-type Ti plasmid conjugation (Fuqua and Winans, 1996, Fuqua and Winans, 1994).
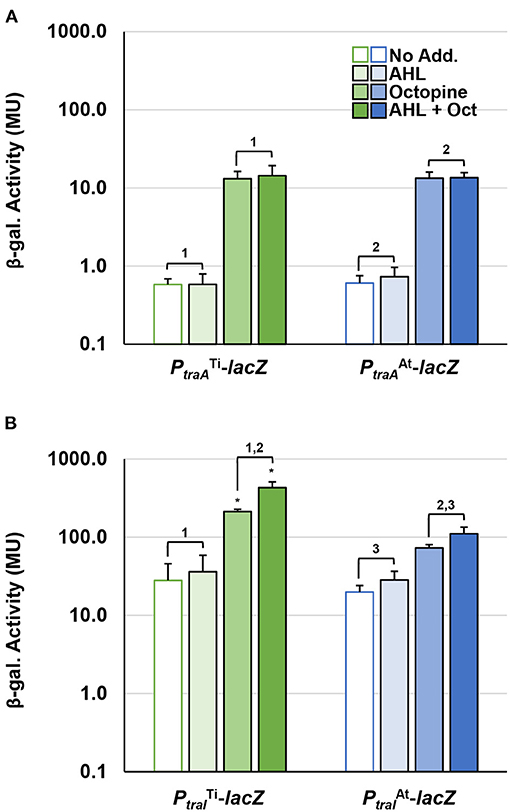
Figure 1. Octopine-induced activation of PtraA and PtraI from pAt and pTi in WT A. tumefaciens 15955. WT A. tumefaciens 15955 carrying a plasmid-borne copy of either PtraA (top) or PtraI (bottom) from pTi15955 (green) or pAt15955 (blue) translationally fused at their start codons to lacZ were spotted on 0.2 μm cellulose acetate filter discs placed on ATGN with or without 3.25 mM octopine (Oct.) and/or 0.1 μM N-3-oxo-C8-HSL (AHL) and incubated at 28°C for 48 h. Bars with no, light, medium, or dark fill indicate no addition, addition of AHL, addition of octpine, or addition of both octopine and AHL, respectively. Cells were resuspended and promoter activities were calculated as Miller Units (Methods). Bars under the same bracket are not significantly different, unless they are marked with an asterisk (p < 0.05). Bars under brackets that have the same number above them (1, 2, 3) in each panel (A,B) denote a significant difference in averages (p < 0.05) between the bars under the similarly numbered brackets.
The pAt15955 plasmid has two annotated traI homologs, one of which is encoded as the predicted first gene of a traI-trb operon (traIAt_1, At15955_49060) in a position similar to pTi-encoded systems, and another ~20 Kbp distant from trb in an uncharacterized locus (traIAt_2, At15955_RS25205) (Supplementary Figures 1A,C). Both are within the large segment of pAt15955 that is deleted upon curing of pTi15955 (blue bar in Supplementary Figure 1), and a pAt15955 derivative with this deletion is unable to conjugate independently (Barton et al., 2019). The annotated coding sequence of traIAt_2 specifies a protein that has a 25 amino acid C-terminal extension relative to TraITi and TraIAt_1 (Supplementary Figure 2). TraIAt_1 is 100% identical to a TraI homolog we identified earlier from the related Genomospecies 1 strain A. tumefaciens A6, that we found to specify production of 3-oxo-C8-HSL (Wang et al., 2014). To evaluate the functionality of the three traI homologs in A. tumefaciens 15955, each coding sequence was fused in-frame with the lacZ start codon under control of Plac in the broad host range vector, pSRKGm, to create the following constructs: pSRKGm Plac-traIAt_1 (pIB302), pSRKGm Plac-traIAt_2 (pIB305) and pSRKGm Plac-traITi (pIB303) (Supplementary Table 2). These plasmids were introduced individually into A. tumefaciens NTL4, a strain that lacks the Ti plasmid and does not produce AHL (Supplementary Table 1) to create derivatives that were used in agar plate-based AHL crossfeeding assays (McLean et al., 1997) with the A. tumefaciens NTL4(pCF218)(pCF372) AHL biosensor (Figure 2A). With this biosensor, diffusible AHLs in the producing strain cause induction of the AHL-responsive biosensor resulting in blue X-Gal pigmentation via increased β-galactosidase activity. The biosensor responds optimally to 3-oxo-C8-HSL, but will also detectably respond at diminishing levels to a broad range of non-cognate AHLs, from acyl chain lengths of C6-C14, and either 3-oxo-, 3-OH or fully reduced at the β-carbon (Zhu et al., 1998). Expression of either normal length traI from pAt15955 or pTi15955 grown in the presence of 400 μM IPTG to induce expression of the Plac-traI fusion was able to generate detectable AHL via the plate-based assay, validating that both genes are functional (Figure 2A). However, no AHL production was detected from the strain harboring the traIAt_2 plasmid, suggesting it is non-functional or produces a product not recognized by the AHL biosensor strain.
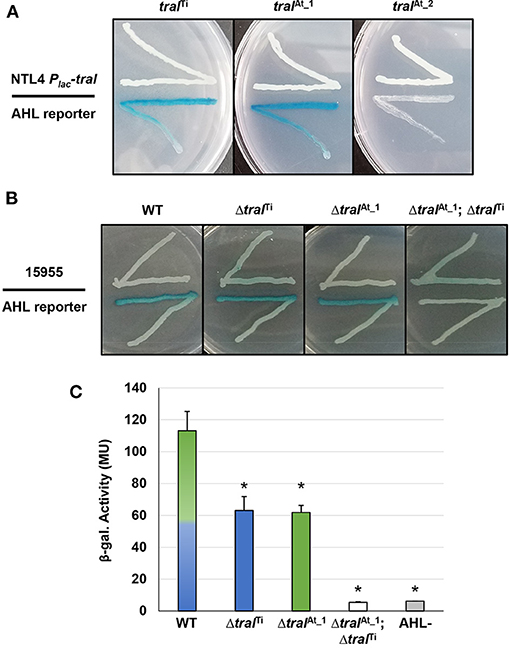
Figure 2. Qualitative and quantitative AHL assays of A. tumefaciens strains ectopically expressing traI genes, and traI mutant derivatives. (A) AHL production by A. tumefaciens NTL4 (Ti plasmidless, AHL−) ectopically expressing traI genes from pAt15955 and pTi15955 expressed from Plac (top streaks). These strains were used in a plate-based AHL production diffusion assays (McLean et al., 1997) with NTL4(pCF218)(pCF372), (AHL reporter, bottom streak). The traI designations are listed above. Media was ATGN agar supplemented with IPTG (400 μM) and X-Gal (40 ug/ml). (B) A. tumefaciens 15955 or derivatives with clean deletions in traI from pTi15955 (ΔtraITi), pAt15955 (ΔtraIAt_1), or both (ΔtraITi; ΔtraIAt_1) (top streak) were used in plate-based AHL production assays with A. tumefaciens NTL4(pCF218)(pCF372) (AHL reporter, bottom streak). (C) Cell-free supernatants from A. tumefaciens 15955 (WT, mixed green/blue bar) and derivatives mutated for traI on pTi15955 (ΔtraITi, blue bar), pAt15955 (ΔtraIAt_1, green bar), or both (ΔtraITi; ΔtraIAt_1, open bar) were added to subcultures of the AHL biosensor strain, A. tumefaciens NTL4(pCF218)(pCF372), and allowed to grow until mid-exponential phase. Standard β-gal assays were then conducted on reporter strain cultures to determine relative AHL levels in the cell-free supernatant, calculated as β-gal activity (Methods). AHL- indicates cell-free supernatant from the biosensor strain A. tumefaciens NTL4(pCF218)(pCF372) (gray bar). Assays performed as three biological replicates, and bars marked with asterisks are significantly different from wild type (p < 0.05).
To determine whether both functional traI genes are expressed in WT A. tumefaciens 15955 and contribute to the overall AHL pool, plate-based AHL detection assays were conducted (as above) with WT A. tumefaciens 15955 and strains isogenic for deletion of traI gene from the Ti plasmid (ΔtraITi), the At plasmid (ΔtraIAt_1), or both (ΔtraITiΔtraIAt_1) (Supplementary Table 1, Figure 2B). AHL production was detected for WT A. tumefaciens 15955 and the individual deletion mutants, but the double mutant did not produce inducing levels of AHL (Figure 2B), indicating that both traI genes are expressed in A. tumefaciens 15955 and each contribute to detectable levels of AHL. To assess the relative amount of AHL produced by each TraI protein in standard growth media, quantitative β-gal assays were conducted with the AHL reporter (NTL4 pCF218 pCF372) using cell-free supernatant from the isogenic traI mutant strains (Figure 2C). Cell-free supernatant from turbid cultures from either traI mutant strain provided half of the B-gal activity relative to WT 15955, which indicates roughly equal contribution from each traI under standard growth conditions in the absence of octopine. The double deletion mutant resulted in background activity equivalent to an AHL- control strain (Figure 2C).
One explanation for octopine-induced activation of pAt15955 conjugation is direct regulation of pAt15955 tra genes by traRTi. To determine the effect of traRTi on conjugation in A. tumefaciens 15955, we generated Plac-traR constructs fused in-frame at the lacZ start codon and carried on pSRKKm (pIB309) or pSRKGm (pIB307). These plasmids were independently introduced into WT A. tumefaciens 15955 derivatives that carry antibiotic resistance marked derivatives of either pTi15955 (IB123, GmR) or pAt15955 (IB125, KmR), respectively, and assayed for conjugation efficiencies of pTi15955 or pAt15955 upon induction of traRTi with 400 μM IPTG (Figure 3, green and blue bars, respectively). As with other Ti plasmids, pTi15955 plasmid does not conjugate at detectable levels in the absence of octopine, but this requirement can be bypassed with ectopic expression of traR (Fuqua and Winans, 1994). Expression of traRTi stimulated conjugation of pTi15955 to a level consistent with our previous findings, and also strongly stimulated pAt15955 conjugation (Figure 3).
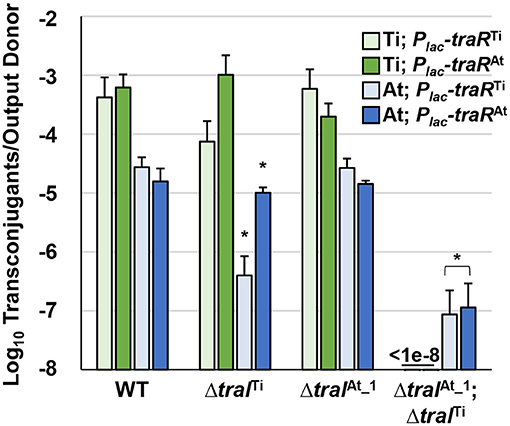
Figure 3. Stimulation of pTi15955 and pAt15955 conjugation by traR expression is dependent on pAt15955- and pTi15955-encoded traI. A tumefaciens 15955 derivatives with a GmR cassette integrated at a neutral location on pTi15955 (green bars) or a KmR cassette similarly integrated on pAt15955 (blue bars) harboring either a plasmid-borne copy of Plac-traR from pTi15955 (light green and light blue bars) or pAt15955 (dark green and dark blue bars) were mixed 1:1 with a plasmidless recipient (A. tumefaciens ERM52, SpR) and spotted on ATGN agar containing 400 μM IPTG. After 24 h incubation, transconjugants and donors were enumerated and conjugation frequencies were calculated as transconjugants per output donor (Methods). Green and blue bars depict pTi15955 and pAt15955 conjugation frequencies, respectively. Asterisks denote significant difference between averages relative to wild type (p < 0.05).
As with many quorum sensing systems, stimulation of conjugation by traRTi in previously studied A. tumefaciens strains requires elevated levels of AHL, augmented by positive feedback on AHL production, to fully activate TraR by stabilizing functional dimerization and DNA binding (Hwang et al., 1999; Qin et al., 2000). To test how the two traI genes contribute to AHL-mediated induction of pTi15955 and pAt15955 conjugation, experiments were performed as above in derivatives deleted for traITi or traIAt_1, or both. Interestingly, conjugation of pTi15955 and of pAt15955 was activated by expression of traRTi in the presence of either version of traI, but was extremely low or undetectable (<10−7 conjugation efficiency) when both were mutated (Figure 3 light green and light blue bars). However, while expression of traRTi was able to stimulate equivalent levels of conjugation in the wild type or in the ΔtraIAt_1 mutant, deletion of the traITi decreased conjugation frequency of both plasmids by 10-fold or greater, although it was still detectable over the ΔtraITiΔtraIAt_1 double mutant. Conjugation of pAt15955 from the ΔtraITiΔtraIAt mutant could be rescued with exogenous addition of 3-oxo-C8-HSL, confirming that the extremely low conjugation from this mutant was due to differences in AHL levels (Supplementary Figure 3).
LuxR-type proteins are defined by several well-conserved amino acid positions in the N-terminal domain that mediate AHL recognition, and in their C-terminal domains that usually coordinate DNA binding. TraRAt is very similar to TraRTi (63.2% identity) and both share the majority of these defining conserved residues (Supplementary Figure 4). However, they differ at position 194 where TraRTi has a well-conserved isoleucine (I194) residue whereas at this position TraRAt has an alanine (A194) residue, an amino acid residue not found in most other LuxR homologs (Whitehead et al., 2001). To determine whether the traR homolog from pAt15955 could stimulate conjugation of either pAt15955 or pTi15955, conjugation experiments were performed as above with ectopic expression of Plac-traRAt (Figure 3, dark green and dark blue bars). The traRAt plasmid was able to stimulate conjugation of pTi15955 and pAt15955 to a magnitude similar for traRTi in a traI-dependent manner (Figure 3). However, in contrast to traRTi, traRAt was able to effectively stimulate conjugation of pTi15955 and pAt15955 when either traI gene, traITi or traIAt_1 was present (Figure 3, dark green and dark blue bars), suggesting that traRAt can potentiate AHL production from either traI gene.
Our findings have revealed that the traI genes from pAt15955 and pTi15955 are activated by octopine and AHL, presumably by one or both TraR proteins (TraRTi and TraRAt), but that the traITi gene is more strongly expressed than traIAt_1 (Figure 1B). As such, one explanation for the reduced conjugation frequencies in the ΔtraITi mutant is that although TraIAt_1 generates similar basal levels of AHL in the absence of octopine induction (Figures 2B,C), its expression is less strongly activated by TraRTi and thus does not create the same level of positive feedback to drive maximal AHL production. We performed plate-based AHL detection assays with the isogenic traI mutants harboring the Plac-traRTi plasmid (Figure 4A). Ectopic expression of traRTi clearly drives strong AHL synthesis in wild type, consistent with positive autoregulation of AHL production by activated TraR. This elevated AHL production was not observed in the ΔtraITi mutant or in the ΔtraITiΔtraIAt_1 double mutant, but was clear for the ΔtraIAt_1 mutant (Figure 4A). This observation suggested that the weak TraRTi activation of traIAt_1 expression resulting in less pronounced increases in AHL levels might underlie the diminished conjugation observed in the traITi mutant. Conversely, we explored how traRAt impacted AHL production with the isogenic traI derivatives expressing traRAt (Figure 4B). In contrast to traRTi, AHL production was enhanced by expression of traRAt when either traI gene was present (Figure 4B).
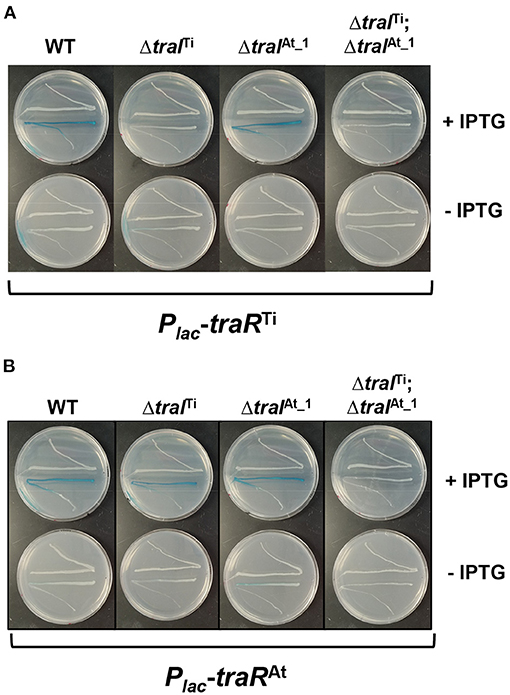
Figure 4. AHL production induced by ectopic expression of traRTi and traRAt in A. tumefaciens 15955. A. tumefaciens 15955 expressing a plasmid-borne copy of (A) traRTi or (B) traRAt with or without in-frame deletions in traITi, traIAt_1, or both (top streaks) were used as AHL donors for A. tumefaciens NTL4(pCF218)(pCF372) (bottom streaks) on ATGN media supplemented with XGal, with or without 400 μM IPTG (top and bottom plates, respectively). Genotypes of A. tumefaciens 15955 AHL donor strain relative to traI indicated above.
In frame deletions were created in the traRTi (ΔtraRTi) and traRAt (ΔtraRAt) genes of A. tumefaciens 15955 to generate single mutants as well as a double mutant (ΔtraRTiΔtraRAt). The plasmids carrying lacZ fusions to the traITi and traIAt genes were independently introduced into these mutants, and β-galatosidase activity was measured following growth on solid medium under a range of different conditions (Figures 5A,B). In wild type A. tumefaciens 15955, the traITi-lacZ fusion was strongly induced on media with octopine (3.25 mM) with or without the exogenous AHL 3-oxo-C8-HSL (0.1 μM) (Figure 5A). Octopine-dependent induction was abolished in the ΔtraRTi mutant, but retained in the ΔtraRAt mutant. Basal level activity was also decreased in the ΔtraRTi mutant but a modest increase was observed in the presence of AHL (Figure 5A). In contrast, basal activity from this lacZ fusion was slightly increased in the ΔtraRAt mutant. The double mutant lacking both traR genes had very low activity that was unaffected by any of the different conditions.
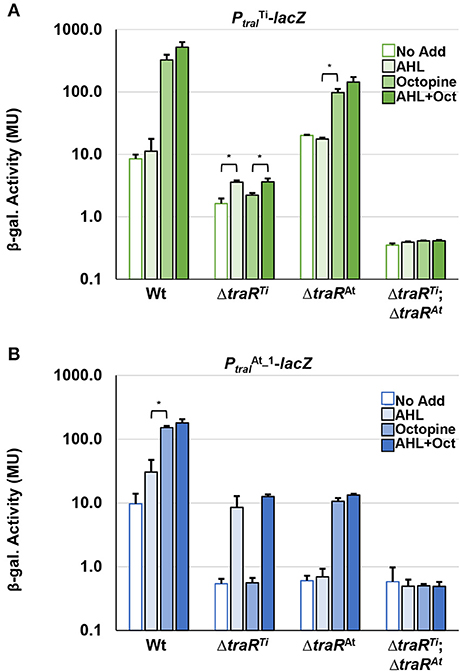
Figure 5. Mutational analysis of traR gene with traI reporter fusions. WT A. tumefaciens 15955 and mutant derivatives (ΔtraRTi and ΔtraRAt), and (ΔtraRTiΔtraRAt) carrying a plasmid-borne copy of either PtraI from pTi15955 (A, green bars) or from pAt15955 (B, blue bars) translationally fused to lacZ were spotted on 0.2 μm cellulose acetate filter discs placed on ATGN on its own (open bars), with 0.1 μM N-3-oxo-C8-HSL (AHL, light-fill bars), with 3.25 mM octopine (Oct., medium-fill bars), and with both (dark-fill bars) Cells were resuspended and promoter activities were calculated as Miller Units (Methods). Bars are standard deviation. All bars that differ by 10-fold or more are significant (p < 0.05). Bars that differ by <10-fold, that are however significantly different (p < 0.05) are designated with a bracketed asterisk.
Expression of the traIAt_1-lacZ fusion showed a different pattern of regulation in the mutant backgrounds. In the wild type background AHL alone was able to significantly increase β-galactosidase activity roughly 3-fold, and addition of octopine with and without exogenous AHL increased this even further (Figure 5B). In the ΔtraRTi mutant the overall level of β-galactosidase decreased about 10-fold relative to wild type, but addition of AHL conversely increased the activity 10-fold from that amount. Octopine addition had no effect in the ΔtraRTi mutant. The activity imparted from the traIAt_1-lacZ fusion also decreased 10-fold in the traRAt mutant relative to wild type, but in contrast to the traRTi mutant exogenous addition of AHL alone had no effect, whereas octopine or AHL plus octopine both resulted in a 10-fold induction. As with the traRTi-lacZ fusion, low level activity from the traIAt_1-lacZ fusion was unaffected by either octopine or AHL alone or in combination in the ΔtraITiΔtraIAt_1 mutant (Figure 5B).
Introduction of compatible plasmids expressing either the traRTi or traRAt gene from the Plac promoter into wild type 15955 and all three different traR mutants resulted in strong IPTG-inducible β-galactosidase activity from both the traITi-lacZ and traRAt-lacZ fusions, in the absence of exogenous AHL or octopine (Supplementary Figure 5). This is consistent with other A. tumefaciens strains where ectopic expression of the traR protein, and its positive impact on AHL production, can bypass the need for conjugal opine addition or the effect of exogenous AHL (Fuqua and Winans, 1994). Ectopic expression of the traRTi gene however stimulated notably lower β-galactosidase activity from the traIAt_1-lacZ fusion than it did for traITi-lacZ, and this decreased even further in the ΔtraR mutant backgrounds, with the lowest activity in the ΔtraRTiΔtraRAt mutant. Ectopic expression of traRAt seemed to have an equivalent impact on both promoters, and this was largely unaffected in the traR deletion mutants.
Our observations on plasmid conjugation, AHL production, and tra gene expression suggested that TraRTi and TraRAt might have different specificities for target promoters. Well-studied TraR proteins are known to bind upstream of regulated promoters at conserved tra box elements (Fuqua and Winans, 1996; Zhu and Winans, 1999), sequence motifs that can readily be identified on pTi15955 upstream of tra genes. Sequence analysis of the presumptive promoter regions of conjugative transfer genes on pAt15955 identified similar inverted repeat, tra box-type elements (Supplementary Figures 6A,B), suggesting direct regulation by one or both TraR proteins. However, the tra box consensus motif for pAt15955-encoded genes is slightly different from that of the pTi15955 elements (Supplementary Figure 6B).
The traI-lacZ and traA-lacZ fusions from pAt15955 and pTi15955 were introduced into the AHL− Ti plasmid-cured A. tumefaciens NTL4 strain harboring either the traRTi or traRAt expression plasmids, that provide IPTG-inducible expression. Very low-β-galactosidase activity was imparted by all of the fusions in the absence of exogenous AHL, even with high level expression of either traR gene in the presence of 800 μM IPTG (Figure 6). In the presence of AHL, the traAAt-lacZ was strongly activated by traRAt, but about 4-fold more weakly activated by traRTi, whereas the difference for activation of the traATi-lacZ fusion was less pronounced (Figure 6A, compare blue-filled bars relative to green-filled bars). Activation patterns for the traI fusions were similar to the traA fusions, though the magnitude of β-galactosidase activity was many fold greater (Figure 6B). The traIAt_1-lacZ fusion was again more weakly activated by traRTi than by traRAt (blue-filled bars), with the activation of the traITi-lacZ closer to equivalency for the two regulators (compare green-filled bars). Thus, for both gene fusions derived from pAt15955 the Ti-plasmid encoded traR leads to weaker induction, whereas the equivalent gene fusions from pTi15955 are strongly activated by both the TraRTi and TraRAt.
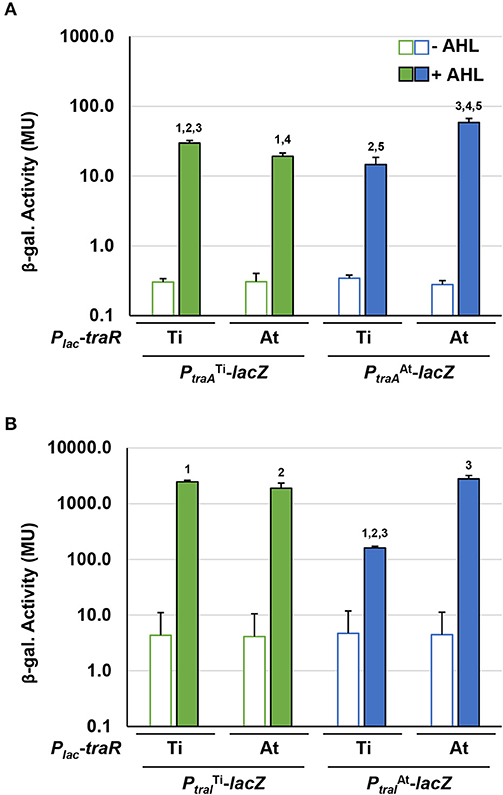
Figure 6. Differential activation of pAt15955 and pTi15955 PtraA and PtraI by traR. Plasmid-borne copies of PtraA-lacZ or PtraI-lacZ from pTi15955 (green) or pAt15955 (blue) and Plac-traR with traR genes from either pAt15955 (At) or pTi15955 (Ti) were moved into A. tumefaciens NTL4 and β-galactosidase activity (MU) of PtraA (A) or PtraI (B) were determined in the presence or absence of 0.1 μM 3-oxo-C8-HSL (+ AHL and – AHL, respectively). Bars with the same numbers possess significant difference in averages (p < 0.05).
Members of the Family Rhizobiaceae often harbor multiple repABC-based secondary replicons that are compatible with each other and are replicated and segregated faithfully during the cell cycle (Cevallos et al., 2008; Castillo-Ramirez et al., 2009). Unlike secondary chromosomes and chromids, megaplasmids, and plasmids within this group often encode conjugation machinery and undergo copy number fluctuations tied with functions such as virulence, symbiosis, and horizontal transfer. The Ti plasmid from A. tumefaciens is one of the most well-studied repABC-controlled replicons, where virulence and horizontal transfer are tightly controlled by environmental signals. In addition to proper coordination with the cell cycle, tight regulation of copy number and horizontal gene transfer of large secondary replicons serves to limit the costs associated with DNA replication and horizontal transfer.
The opine-dependent quorum sensing system is well-characterized in Agrobacterium species harboring the wide host range octopine-type and nopaline-type Ti plasmids, with prototypes pTiR10 and pTiC58, respectively (Gordon and Christie, 2014). Based on extensive genomic sequencing the Agrobacterium genus has been phylogenetically subdivided into over 10 different genomospecies (Lassalle et al., 2011), with pTiR10 found in Genomospecies 4 and pTiC58 found in Genomospecies 8. In these systems there is a single quorum sensing system comprised of the TraR transcription factor, the TraI AHL synthase, and the TraM antiactivator, which together regulate Ti plasmid conjugation and copy number. However, in several members of the Agrobacterium G1 genomospecies with octopine-type Ti plasmids, such as A. tumefaciens 15955, A. tumefaciens A6, and A. tumefaciens Ach5, the At plasmid carries a second TraR-TraI-TraM quorum sensing system, that is closely related to the Ti-encoded regulators. We have demonstrated that this second At-encoded system controls conjugative transfer through regulation of tra genes on the At plasmid, but that it can also have a profound impact on Ti plasmid functions. Likewise, the Ti-encoded system can impact At plasmid conjugative transfer, most notably imparting octopine-inducible control over pAt15955 conjugation (Barton et al., 2019). These co-resident quorum sensing systems interdigitate at multiple levels to regulate horizontal transfer of both plasmids (Figure 7). Both TraI gene products are highly similar to each other, and drive AHL synthesis. Although the chemical identity of the AHL is not formally shown here for A. tumefaciens 15955, it has been reported for the closely related A. tumefaciens A6 that both the Ti and At traI genes specify production of 3-oxo-C8-HSL (Wang et al., 2014). The TraI proteins between A6 and 15955 show 100% amino acid identity. Furthermore, we show here that either traI gene can function to facilitate conjugation of both plasmids, but loss of both traI genes together abolishes conjugation of each plasmid. Therefore, we predict that the two TraI proteins in A. tumefaciens 15955 primarily contribute to the same pool of 3-oxo-C8-HSL (Figure 7). This pool of AHL is required for TraR-mediated activation of tra genes and plasmid conjugation, through protein-ligand interactions with their amino-terminal ligand binding domains.
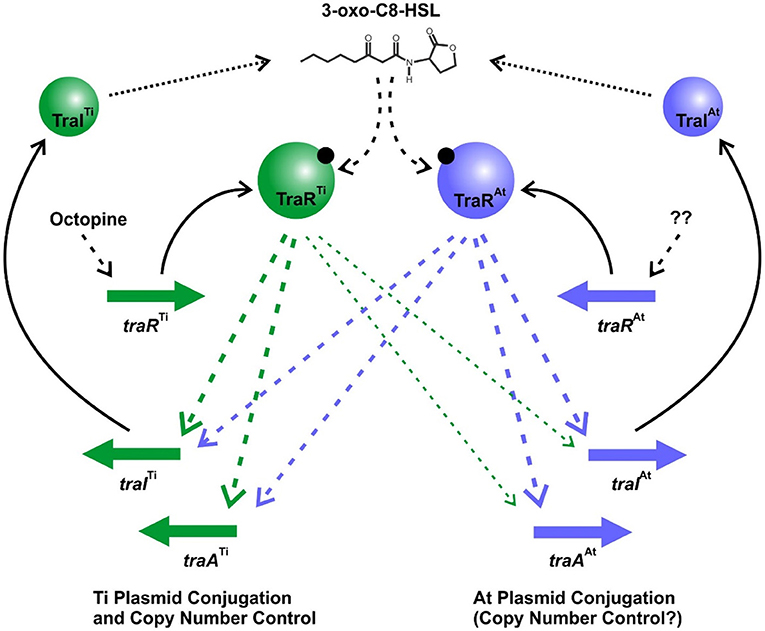
Figure 7. Model of Dual Quorum Sensing Systems in A. tumefaciens 15955. Green shaded symbols representing proteins and genes from pTi15955, and blue shaded symbols represent proteins and genes from pAt15955. Solid arrows indicate translation of specific genes, dotted arrows represent enzymatic synthesis of AHL, and dashed arrows are regulatory impacts of each system. The thinner arrows from TraRTi to the pAt15955 target genes reflect the observed weaker effect of this protein on their expression.
In the presence of inducing levels of AHL the TraR proteins can bind to DNA and activate expression of target promoters, and stimulate plasmid conjugative transfer. The presence of octopine, naturally produced by plant tissue transformed with T-DNA encoding the octopine synthase genes, has been shown to activate expression of the Ti plasmid traR gene in other octopine-type plasmids (Fuqua and Winans, 1994). The operon structure upstream of the pTi15955 traR is identical to that on these better studied octopine-type Ti plasmids (Supplementary Figure 1B). We posit that octopine increases expression of traRTi on pTi15955 as well, and that this increase stimulates expression of the traITi gene, which in turn elevates AHL levels, creating a positive feedback loop as observed for many LuxR-LuxI-type systems (Fuqua and Greenberg, 2002). Ectopically increasing expression of traRTi bypasses the need for octopine. However, traRTi only weakly stimulates the traI gene on the At plasmid, which in turn limits its impact on AHL synthesis, thereby explaining the weaker stimulation of both Ti plasmid and At plasmid conjugation in the ΔtraITi mutant (Figure 3). The traRAt gene product appears to activate expression of both the traIAt_1 and traITi with rough equivalence. However, there is no evidence that its expression is affected by octopine, and consistent with this, in the absence of the traRTi gene octopine has no impact on the tra gene expression or conjugation of either plasmid. We hypothesize that under laboratory conditions, traRAt expression is low, and insufficient to stimulate significant positive feedback on AHL production, and thus pTi15955 conjugative transfer is extremely infrequent or undetectable in the absence of octopine. The basal expression of the traITi gene is in fact modestly elevated in the ΔtraRAt mutant raising the possibility that TraRAt plays a mildly inhibitory role in the absence of octopine. In contrast to pTi15955, low level conjugation of pAt15955 is detectable in the absence of octopine (Barton et al., 2019). The observation that even this minimal conjugation is largely abolished in the ΔtraITiΔtraIAt double mutants suggests that quorum sensing is functioning to regulate the process even at its lowest levels. It also is plausible that expression of the traRAt gene is regulated by a condition that we have not yet identified such as has been observed in other rhizobia, and under certain conditions might elevate pAt15955 and thereby indirectly pTi15955 conjugation (Banuelos-Vazquez et al., 2020).
The molecular mechanisms by which the two TraR proteins function in the same cell remains an area for future studies. There are abundant examples of multiple LuxR-type regulators functioning in a single bacterial species, often in some form of hierarchal network. For example, in Pseudomonas aeruginosa there are three distinct LuxR-type regulators, LasR, RhlR, and QscR with overlapping regulons (Ding et al., 2018). In other Alphaproteobacteria multiple LuxR proteins are also common. For example, in the marine bacterium Ruegeria sp. KLH11 the SsaR LuxR-type protein activates motility in response to long chain AHLs generated by the SsaI AHL synthase, and also regulates the activity of a second LuxR-type protein SsbR and two AHL synthases SsbI and SscI (Zan et al., 2012, 2015). Multiple, co-resident LuxR-LuxI-type proteins are well-documented in the rhizobia, and their regulatory interactions are notably complex and convoluted. Rhizobium leguminosarum biovar viciae has a cascade of multiple LuxR and LuxI-type proteins including several that control plasmid conjugation (Lithgow et al., 2002; Cervantes et al., 2019). The biocontrol agent known as Rhizobium rhizogenes K84 encodes two TraR homologs (TraRacc and TraRnoc) on the single pAtK84b plasmid that combine to control conjugative transfer of this plasmid (Oger et al., 1998). Remarkably, in K84 expression of each traR gene is regulated by a different conjugal opine, nopaline and agrocinopine A/B, and consequently both opines independently stimulate pAtK84b conjugation through a single conjugative machinery. Recently, two of the repABC plasmids of Sinorhizobium fredii GR64, pSfr64a, and pSfr64b have been reported to encode two separate TraR-TraM-TraI quorum sensing systems, in addition to a quorum sensing regulatory system encoded by the chromosome, NgrR-NgrI (Cervantes et al., 2019). All three of these quorum sensing systems impact each other and contribute to control of pSfr64a and pSfr64b conjugation. Similar to S. fredii GR64, A. tumefaciens 15955 and its immediate relatives have dual, highly conserved TraR-TraI-TraM systems on two separate plasmids that each encode their own conjugative transfer machinery. Although there is no chromosomal LuxR-LuxI system in A. tumefaciens 15955, our findings reveal that the plasmid encoded quorum sensing systems are clearly interdependent.
Dimerization is known to be a requirement for DNA binding among diverse LuxR homologs including TraR. Given the similarity of the TraRTi and TraRAt proteins (63.2% identity), heterodimer formation is plausible, potentially altering the activity of each TraR protein. Interestingly, A. tumefaciens 15955 is known to encode a truncated copy of the traRTi gene, named TrlR that is 88% identical, but ablated for the DNA binding domain, expression of which is inducible by the non-conjugative opine mannopine (Oger et al., 1998; Zhu et al., 1998). TrlR is a dominant negative homolog of TraR, that forms an inactive heterodimer with TraRTi and prevents DNA binding (Chai et al., 2001). In the absence of mannopine, TrlR is not expressed. Our genetic analysis revealed that mutation of traRAt modestly elevated expression of the traITi-lacZ fusion in response to octopine, suggesting that TraRAt may also have a mild dominant negative effect on TraRTi.
Another component of the quorum sensing systems commonly encoded on Ti plasmids is the TraM anti-activator (Fuqua et al., 1995; Hwang et al., 1995). TraM forms a multimeric complex with TraR, abrogating its activity and targeting it for proteolysis (Swiderska et al., 2001; Chen et al., 2007; Costa et al., 2012). A. tumefaciens 15955 encodes TraM proteins on pAt15955 and pTi15955, that are 65% identical. Genetic and biochemical analysis of a second TraM homolog from the close relative A. tumefaciens A6 demonstrated that it can form a complex with TraR encoded by the Ti plasmid and adopts the same three-dimensional structure as TraMTi (Chen et al., 2006; Wang et al., 2014). The two TraM proteins seemed to be functionally redundant in A. tumefaciens A6, and mutation of both genes was required to disregulate the Ti plasmid TraR. Subsequent studies prior to genome sequencing also identified traR and traI genes linked to this second traM gene, all of which we now recognize to be carried on the pAtA6 plasmid (Wang et al., 2014). Although we did not examine the role for the traMTi and traMAt in the current study on A. tumefaciens 15955, it seems very likely that they play similar roles as those in A6. As such, the orchestration between the pTi15955 and pAt15955 quorum sensing systems to control plasmid transfer, very likely also includes these TraM anti-activators and their ability to inhibit their cognate and non-cognate TraR proteins.
We observed that TraRTi only weakly activated the pAt15955 gene fusions tested, while it strongly affected the homologous pTi15955 gene fusions. In contrast, TraRAt was largely equivalent in activation of both pTi15955 and pAt15955 targets, suggesting that it has a more permissive DNA binding specificity. Comparatively, the tra box elements identified upstream of these and other potential target genes, are similar 18 bp inverted repeats, but there are several differences at specific positions, that may distinguish the pTi15955 targets from the pAt15955 sequences. Amino acid sequence differences between TraRTi and TraRAt within the C-terminal DNA-binding domain are likely to impart the DNA binding site specificity. Although the proteins are quite similar overall in the DNA binding domain (69% identity), there are several differences including a conserved isoleucine residue (I194) in TraRTi (either an I or V in most LuxR-type proteins, Whitehead et al., 2001), that is an alanine (A194) in TraRAt (Supplementary Figure 6). The octopine-type A. tumefaciens R10 TraR protein which is encoded on pTiR10, was structurally characterized in association with the 3-oxo-C8-HSL ligand and bound to a synthetic tra box oligonucleotide (Zhang et al., 2002). Two of the primary contacts with DNA residues within the tra box were Arg206 (R206) and Arg210 (R210) which are conserved in the 15955 TraRTi protein (Supplementary Figure 4, asterisks). However, these positions are divergent in 15955 TraRAt and are Lys206 and Glu210 (Supplementary Figure 4). Thus, it seems the potential mechanism of tra box recognition may be different between these two regulators. Our findings suggest that sequence divergence between the two proteins imparts a less restrictive DNA binding site specificity to TraRAt than to TraRTi. However, testing the binding of each TraR paralogue to pAt15955 and pTi15955 promoters in vitro would provide an effective way to directly evaluate their specificity.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
IB, TP, and CF designed the experiments and wrote the paper. TP, IB, JE, IR, PN-O, and CF performed the experiments. All authors contributed to the article and approved the submitted version.
This work was supported by National Institutes of Health Grant (R01 GM092660 to CF and James D. Bever), the Indiana University Faculty Research Support Program (FRSP) (to CF), the National Science Foundation (NSF) Graduate Research Fellowship Program (GGVP004842 to PN-O), and the Kansas NSF EPSCoR First Award Program (OIA-1656006 to TP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would also like to thank Isam Madi and Reilly Jensen for help with strain construction.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.605896/full#supplementary-material
Akakura, R., and Winans, S. C. (2002a). Constitutive mutations of the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. J. Biol. Chem. 277, 5866–5874. doi: 10.1074/jbc.M110555200
Akakura, R., and Winans, S. C. (2002b). Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. J. Biol. Chem. 277, 15773–15780. doi: 10.1074/jbc.M200109200
Alt-Mörbe, J., Stryker, J. L., Fuqua, C., Li, P.-L., Farrand, S. K., and Winans, S. C. (1996). The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmids is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J. Bacteriol. 178, 4248–4257. doi: 10.1128/JB.178.14.4248-4257.1996
Banuelos-Vazquez, L. A., Castellani, L. G., Luchetti, A., Romero, D., Torres Tejerizo, G. A., and Brom, S. (2020). Role of plant compounds in the modulation of the conjugative transfer of pRet42a. PLoS ONE 15:e0238218. doi: 10.1371/journal.pone.0238218
Barton, I. S., Platt, T. G., Rusch, D. B., and Fuqua, C. (2019). Destabilization of the tumor-inducing plasmid from an octopine-type Agrobacterium tumefaciens lineage drives a large deletion in the co-resident At megaplasmid. G3. 9, 3489–3500. doi: 10.1534/g3.119.400554
Castellani, L. G., Nilsson, J. F., Wibberg, D., Schluter, A., Puhler, A., Brom, S., et al. (2019). Insight into the structure, function and conjugative transfer of pLPU83a, an accessory plasmid of Rhizobium favelukesii LPU83. Plasmid 103, 9–16. doi: 10.1016/j.plasmid.2019.03.004
Castillo-Ramirez, S., Vazquez-Castellanos, J. F., Gonzalez, V., and Cevallos, M. A. (2009). Horizontal gene transfer and diverse functional constrains within a common replication-partitioning system in Alphaproteobacteria: the repABC operon. BMC Genomics. 10:536. doi: 10.1186/1471-2164-10-536
Cervantes, L., Miranda-Sanchez, F., Torres Tejerizo, G., Romero, D., and Brom, S. (2019). Plasmid pSfr64a and the symbiotic plasmid pSfr64b of Sinorhizobium fredii GR64 control each other's conjugative transfer through quorum-sensing elements. Plasmid 106:102443. doi: 10.1016/j.plasmid.2019.102443
Cevallos, M. A., Cervantes-Rivera, R., and Gutierrez-Rios, R. M. (2008). The repABC plasmid family. Plasmid 60, 19–37. doi: 10.1016/j.plasmid.2008.03.001
Chai, Y., Zhu, J., and Winans, S. C. (2001). TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40, 414–421. doi: 10.1046/j.1365-2958.2001.02385.x
Chen, G., Jeffrey, P. D., Fuqua, C., Shi, Y., and Chen, L. (2007). Structural basis for antiactivation in bacterial quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 104, 16474–16479. doi: 10.1073/pnas.0704843104
Chen, G., Wang, C., Fuqua, C., Zhang, L. H., and Chen, L. (2006). Crystal structure and mechanism of TraM2, a second quorum-sensing antiactivator of Agrobacterium tumefaciens strain A6. J. Bacteriol. 188, 8244–8251. doi: 10.1128/JB.00954-06
Chen, L., Chen, Y., Wood, D. W., and Nester, E. W. (2002). A new Type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J. Bacteriol. 184, 4838–4845. doi: 10.1128/JB.184.17.4838-4845.2002
Chilton, M. D., Drummond, M. H., Merio, D. J., Sciaky, D., Montoya, A. L., Gordon, M. P., et al. (1977). Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11, 263–271. doi: 10.1016/0092-8674(77)90043-5
Christie, P. J. (2001). Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40, 294–305. doi: 10.1046/j.1365-2958.2001.02302.x
Costa, E. D., Chai, Y., and Winans, S. C. (2012). The quorum-sensing protein TraR of Agrobacterium tumefaciens is susceptible to intrinsic and TraM-mediated proteolytic instability. Mol. Microbiol. 84, 807–815. doi: 10.1111/j.1365-2958.2012.08037.x
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dessaux, Y., Petit, A., Farrand, S. K., and Murphy, P. J. (1998). “Opines and opine-like molecules involved in plant-Rhizobiaceae interactions,” in The Rhizobiaceae: Molecular Biology of Model Plant-Associated Bacteria eds Spaink H. P., Kondorosi A., and Hooykaas P. J. J. (Boston, MA: Kluwer Academic Press), 173–197. doi: 10.1007/978-94-011-5060-6_9
Ding, F., Oinuma, K. I., Smalley, N. E., Schaefer, A. L., Hamwy, O., Greenberg, E. P., et al. (2018). The Pseudomonas aeruginosa orphan quorum sensing signal receptor QscR regulates global quorum sensing gene expression by activating a single linked operon. MBio 9, e01274–e01218. doi: 10.1128/mBio.01274-18
Ellis, J. G., Kerr, A., Petit, A., and Tempé, J. (1982). Conjugal transfer of nopaline and agropine Ti-plasmids - the role of agrocinopines. Mol. Gen. Genet. 186, 269–273. doi: 10.1007/BF00331861
Farrand, S. K., Hwang, I., and Cook, D. M. (1996). The tra region of the nopaline-type Ti plasmid is chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 178, 4233–4247. doi: 10.1128/JB.178.14.4233-4247.1996
Fournes, F., Val, M. E., Skovgaard, O., and Mazel, D. (2018). Replicate once per cell cycle: replication control of secondary chromosomes. Front. Microbiol. 9:1833. doi: 10.3389/fmicb.2018.01833
Fuqua, C., Burbea, M., and Winans, S. C. (1995). Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J. Bacteriol. 177:7. doi: 10.1128/JB.177.5.1367-1373.1995
Fuqua, C., and Greenberg, E. P. (2002). Listening in on bacteria: acylhomoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695. doi: 10.1038/nrm907
Fuqua, C., and Winans, S. C. (1996). Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:6. doi: 10.1128/JB.178.2.435-440.1996
Fuqua, W. C., and Winans, S. C. (1994). A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:11. doi: 10.1128/JB.176.10.2796-2806.1994
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Gordon, J. E., and Christie, P. J. (2014). The Agrobacterium Ti plasmids. Microbiol Spectr. 2:10.1128/microbiolspec.PLAS-0010-2013. doi: 10.1128/microbiolspec.PLAS-0010-2013
He, X., Chang, W., Pierce, D. L., Seib, L. O., Wagner, J., and Fuqua, C. (2003). Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185, 809–822. doi: 10.1128/JB.185.3.809-822.2003
He, X., and Fuqua, C. (2006). Rhizosphere communication: quorum sensing by the rhizobia. J. Microbiol. Biotechnol. 16, 1661–1677. doi: 10.1098/rstb.2007.2041
Heckel, B. C., Tomlinson, A. D., Morton, E. R., Choi, J. H., and Fuqua, C. (2014). Agrobacterium tumefaciens exoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J. Bacteriol. 196, 3221–3233. doi: 10.1128/JB.01751-14
Hwang, I., Cook, D. M., and Farrand, S. K. (1995). A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177, 449–458. doi: 10.1128/JB.177.2.449-458.1995
Hwang, I., Li, P.-L., Zhang, L., Piper, K. R., Cook, D. M., Tate, M. E., et al. (1994). TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. U.S.A. 91:5. doi: 10.1073/pnas.91.11.4639
Hwang, I., Smyth, A. J., Luo, Z.-Q., and Farrand, S. K. (1999). Modulation quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34, 282–294 doi: 10.1046/j.1365-2958.1999.01595.x
Khan, S. R., Gaines, J., Roop, R. M. I. I, and Farrand, S. K. (2008). Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74, 5053–5062. doi: 10.1128/AEM.01098-08
Klapwijk, P. M., Scheulderman, T., and Schilperoort, R. (1978). Coordinated regulation of octopine degradation and conjugative transfer of Ti plasmids in Agrobacterium tumefaciens : evidence for a common regulatory gene and separate operons. J. Bacteriol. 136, 775–785. doi: 10.1128/JB.136.2.775-785.1978
Lang, J., Planamente, S., Mondy, S., Dessaux, Y., Morera, S., and Faure, D. (2013). Concerted transfer of the virulence Ti plasmid and companion At plasmid in the Agrobacterium tumefaciens-induced plant tumour. Mol. Microbiol. 90, 1178–1189. doi: 10.1111/mmi.12423
Lassalle, F., Campillo, T., Vial, L., Baude, J., Costechareyre, D., Chapulliot, D., et al. (2011). Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 3, 762–781. doi: 10.1093/gbe/evr070
Li, P. L., and Farrand, S. K. (2000). The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J. Bacteriol. 182, 179–188. doi: 10.1128/JB.182.1.179-188.2000
Lithgow, J. K., Wilkinson, A., Hardman, A., Rodelas, B., Wisniewski-Dye, F., Williams, P., et al. (2002). The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 37, 81–97. doi: 10.1046/j.1365-2958.2000.01960.x
McLean, R. J. C., Whiteley, M., Stickler, D. J., and Fuqua, W. C. (1997). Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154, 259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x
Moré, M. I, Finger, L. D., Stryker, J. L., Fuqua, C., Eberhard, A., and Winans, S. C. (1996). Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272, 1655–1658. doi: 10.1126/science.272.5268.1655
Morton, E. R., and Fuqua, C. (2012a). Genetic manipulation of Agrobacterium. Curr. Protoc. Microbiol. Unit 3D 2:1–15. doi: 10.1002/9780471729259.mc03d02s25
Morton, E. R., and Fuqua, C. (2012b). Laboratory maintenance of Agrobacterium. Curr. Protoc. Microbiol. Unit 3D 1:1–6. doi: 10.1002/9780471729259.mc03d01s24
Nogales, J., Blanca-Ordonez, H., Olivares, J., and Sanjuan, J. (2013). Conjugal transfer of the Sinorhizobium meliloti 1021 symbiotic plasmid is governed through the concerted action of one- and two-component signal transduction regulators. Environ. Microbiol. 15, 811–821. doi: 10.1111/1462-2920.12073
Oger, P., Kim, K.-S., Sackett, R. L., Piper, K. R., and Farrand, S. K. (1998). Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27, 277–288. doi: 10.1046/j.1365-2958.1998.00671.x
Pappas, K. M., and Winans, S. C. (2003). A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol. Microbiol. 48:15. doi: 10.1046/j.1365-2958.2003.03488.x
Perez-Mendoza, D., Sepulveda, E., Pando, V., Munoz, S., Nogales, J., Olivares, J., et al. (2005). Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids. J. Bacteriol. 187, 7341–7350. doi: 10.1128/JB.187.21.7341-7350.2005
Pinto, U. M., Pappas, K. M., and Winans, S. C. (2012). The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 10, 755–765. doi: 10.1038/nrmicro2882
Qin, Y., Luo, Z.-Q., Smyth, A. J., Gao, P., Beck von Bodman, S., and Farrand, S. K. (2000). Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19, 5212–5221. doi: 10.1093/emboj/19.19.5212
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 11:539. doi: 10.1038/msb.2011.75
Swiderska, A., Berndtson, A. K., Cha, M.-R., Li, L., Beaudoin, G. M. J. I., Zhu, J., et al. (2001). Inhibition of the Agrobacterium tumefaciens TraR quorum-sensing regulator: interactions with the TraM anti-activator. J. Biol. Chem. 276, 49449–49458. doi: 10.1074/jbc.M107881200
Wang, C., Yan, C., Fuqua, C., and Zhang, L. H. (2014). Identification and characterization of a second quorum-sensing system in Agrobacterium tumefaciens A6. J. Bacteriol. 196, 1403–1411. doi: 10.1128/JB.01351-13
Wetzel, M. E., Olsen, G. J., Chakravartty, V., and Farrand, S. K. (2015). The repABC plasmids with quorum-regulated transfer systems in members of the Rhizobiales divide into two structurally and separately evolving groups. Genome Biol. Evol. 7, 3337–3357. doi: 10.1093/gbe/evv227
Whitehead, N. A., Barnard, A. M. L., Slater, H., Simpson, N. J. L., and Salmond, G. P. C. (2001). Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x
Winans, S. C. (1991). An Agrobacterium two-component regulatory system for the detection of chemicals released from plant wounds. Mol. Microbiol. 5, 2345–2350. doi: 10.1111/j.1365-2958.1991.tb02080.x
Zan, J., Choi, O., Meharena, H., Uhlson, C. L., Churchill, M. E., Hill, R. T., et al. (2015). A solo luxI-type gene directs acylhomoserine lactone synthesis and contributes to motility control in the marine sponge symbiont Ruegeria sp. KLH11. Microbiology 161, 50–56. doi: 10.1099/mic.0.083956-0
Zan, J., Cicirelli, E. M., Mohamed, N. M., Sibhatu, H., Kroll, S., Choi, O., et al. (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85, 916–933. doi: 10.1111/j.1365-2958.2012.08149.x
Zhang, R. G., Pappas, K. M., Brace, J. L., Miller, P. C., Oulmassov, T., Molyneaux, J. M., et al. (2002). Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417, 971–974. doi: 10.1038/nature00833
Zhu, J., Beaber, J. W., More, M. I., Fuqua, C., Eberhard, A., and Winans, S. C. (1998). Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180, 5398–5405. doi: 10.1128/JB.180.20.5398-5405.1998
Keywords: plasmids, conjugative transfer, quorum sensing (QS), repABC system, gene regulation
Citation: Barton IS, Eagan JL, Nieves-Otero PA, Reynolds IP, Platt TG and Fuqua C (2021) Co-dependent and Interdigitated: Dual Quorum Sensing Systems Regulate Conjugative Transfer of the Ti Plasmid and the At Megaplasmid in Agrobacterium tumefaciens 15955. Front. Microbiol. 11:605896. doi: 10.3389/fmicb.2020.605896
Received: 13 September 2020; Accepted: 21 December 2020;
Published: 18 January 2021.
Edited by:
Daniel Yero, Autonomous University of Barcelona, SpainReviewed by:
Miguel Angel Cevallos, National Autonomous University of Mexico, MexicoCopyright © 2021 Barton, Eagan, Nieves-Otero, Reynolds, Platt and Fuqua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clay Fuqua, Y2Z1cXVhQGluZGlhbmEuZWR1
†Present address: Ian S. Barton, Department of Microbiology and Immunology, Brody School of Medicine, East Carolina University, Greenville, NC, United States
Justin L. Eagan, Department of Medical Microbiology and Immunology, University of Wisconsin-Madison, Madison, WI, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.