
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 22 December 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.605229
Increasing concerns about nosocomial infection, food and environmental safety have prompted the development of rapid, accurate, specific and ultrasensitive methods for the early detection of critical pathogens. Pseudomonas aeruginosa is one of the most common pathogens that cause infection. It is ubiquitous in nature, being found in water, soil, and food, and poses a great threat to public health. The conventional detection technologies are either time consuming or readily produce false positive/negative results, which makes them unsuitable for early diagnosis and spot detection of P. aeruginosa. To circumvent these drawbacks, many efforts have been made to develop biosensors using aptamers as bio-recognition elements. Various aptamer-based biosensors for clinical diagnostics, food, and environmental monitoring of P. aeruginosa have been developed in recent years. In this review, we focus on the latest advances in aptamer-based biosensors for detection of P. aeruginosa. Representative biosensors are outlined according to their sensing mechanisms, which include optical, electrochemical and other signal transduction methods. Possible future trends in aptamer biosensors for pathogen detection are also outlined.
Pseudomonas aeruginosa, an opportunistic Gram-negative bacterium, is one of the most intractable multidrug-resistant bacteria causing severe nosocomial infections and poses an increasing threat to human health (Curran et al., 2018; Horcajada et al., 2019; Jean et al., 2020). In 2017, P. aeruginosa was listed as a critical pathogen by the World Health Organization. It is widely distributed in soil, water, air, animals and humans. Its widespread habitats make it difficult to control. Immunocompromised patients or people with damaged tissue barriers are often susceptible to serious P. aeruginosa infection, which causes diseases such as keratitis, endophthalmitis, pneumonia, urinary tract infections, lung diseases, cystic fibrosis, neutropenia, and sepsis (Winstanley et al., 2016; Paz-Zarza et al., 2019; Tummler, 2019; Kowalski et al., 2020). Moreover, P. aeruginosa tends to form biofilm, which is difficult to eradicate and has 10- to 1,000-fold higher resistance to antibiotic concentrations compared with equivalent floating bacteria (Lewis, 2001; Pang et al., 2019). Therefore, rapid, accurate, and ultrasensitive identification of P. aeruginosa is critical for both empirical antipseudomonal therapy and the guarantee of food security.
Diagnostic methods for P. aeruginosa presently include conventional culture-dependent colony counting, immunodetection, polymerase chain reaction (PCR), and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Rajapaksha et al., 2019). The traditional culture-based approach, regarded as the golden standard for pathogenic microorganism detection, is the most commonly used method (Perry, 2017). However, it is labor-intensive and time consuming (48–120 h), which makes it unsuitable for rapid detection of P. aeruginosa (Varadi et al., 2017). Immunological tests are also commonly used for clinical diagnoses. However, they are dependent on antibodies, which are sensitive to temperature and pH changes, and prone to false negatives/positives (Mauch and Levy, 2014). Another approach for P. aeruginosa detection is PCR-based method, which requires strict protocols (Deschaght et al., 2011). MALDI-TOF-MS requires sophisticated instrumentation and highly trained personnel. Bacterial enrichment is also needed before analysis, resulting in a long turnaround time (Perry, 2017). Therefore, the development of accurate, sensitive, specific, and rapid methods for identifying P. aeruginosa remains a challenging research goal. Biosensors are promising alternatives for pathogen detection (Khater et al., 2017; Nasseri et al., 2018; Silva et al., 2018; Furst and Francis, 2019; Cesewski and Johnson, 2020). In recent years, biosensors have been widely used in the microbial field, because of their rapidity, sensitivity, high specificity, and low cost (Farooq et al., 2018; Zhang et al., 2019a). In general, specific interactions between molecular recognition elements and pathogens can be converted into electrical, optical, or other signal outputs for analysis and detection. Therefore, the bio-recognition element is the most critical part in biosensors (Morales and Halpern, 2018; Zhang and Tian, 2018). Antibodies have been used as recognition elements in biosensors for decades. However, antibody development requires a tedious and complicated in vivo screening process, which leads to long development time and high cost. Moreover, antibodies are sensitive to temperature and pH changes and prone to denaturation (Alahi and Mukhopadhyay, 2017). Thus, researchers have sought to identify new molecular recognition elements that can retain the high affinity and specificity of antibodies without their shortcomings.
Aptamer, a promising alternative to antibodies as molecular recognition tools, has spurred a great interest since 1990 (Ellington and Szostak, 1990; Tuerk and Gold, 1990; Tan et al., 2020). They are short single-stranded DNA or RNA, selected in vitro from a synthesized random library using systematic evolution of ligands by exponential enrichment (SELEX) (Wang C. H. et al., 2019a; Wang T. et al., 2019; Yan et al., 2019). By folding into specific tertiary structures, aptamers bind to their targets with high affinity and specificity by complementary shape interactions (Gao et al., 2019). Compared with antibodies, aptamer-based biosensors have many advantages, including lower cost, less batch-to-batch variation, automated chemical synthesis, and more flexible modification strategies (Radom et al., 2013; Groff et al., 2015; Moutsiopoulou et al., 2019; Chen et al., 2020). Furthermore, targets such as toxins or haptens cannot be used to produce antibodies with high affinity, and in these cases aptamers possess a significant advantage. These excellent characteristics provide an opportunity for aptamers to replace antibodies in all analytical formats. To date, a myriad of aptamer-based biosensors have been developed to detect various bacteria, such as Escherichia coli (Hua et al., 2018; Dong et al., 2020; Kaur et al., 2020; Wang et al., 2020), Salmonella enterica (Fang et al., 2014; Chinnappan et al., 2017; Dao et al., 2018; Luo et al., 2020), and Staphylococcus aureus (Shrivastava et al., 2018; Cai et al., 2020; Fu et al., 2020; Pla et al., 2020). For P. aeruginosa, multiple aptamer-based sensors (aptasensors) have also been developed using optical, electrochemical and other signal sensing approaches. In this review, we first briefly introduce aptamers that have been selected against P. aeruginosa. Subsequently, we focus on the latest advances in aptamer-based biosensors detection of P. aeruginosa. Finally, the current challenges and future trends of aptamer-based biosensors in P. aeruginosa detection are further discussed.
For the detection of P. aeruginosa, several specific aptamers have so far been isolated. Wang et al. (2011) selected an ssDNA aptamer specifically binding to P. aeruginosa (ATCC 27853) using a whole-bacterium SELEX strategy. Aptamers obtained through such kind of derived SELEX strategy (for example Figure 1), can bind to targets on bacteria in their native and most stable conformation, without changes before selection (Liu et al., 2020). The most specific aptamer, named F23, can identify the target bacteria with a low nanomolar dissociation constant (Kd = 17.27 ± 5.00 nM). F23 laid the foundation for many sensitive and rapid detection methods for P. aeruginosa. Similarly, another specific aptamer against P. aeruginosa (ATCC 27853) with a Kd of 15.16 ± 3.62 nM was obtained by other researchers (Wang et al., 2018). Moreover, Soundy and Day (2017) obtained three chimeric DNA aptamers, with Kd values in the nanomolar range, against live biofilm-derived PA692 P. aeruginosa cells. Although these aptamers were derived from biofilms, they can bind to both planktonic- and biofilm-grown cells. Another interesting example is provided by Davydova et al. (2017) who developed a 2′-fluoro RNA aptamer capable of internalizing into P. aeruginosa cells. As specific bio-recognition molecules, these above aptamers have provided effective tools for the rapid detection, early diagnosis, and therapeutic targeting of P. aeruginosa.
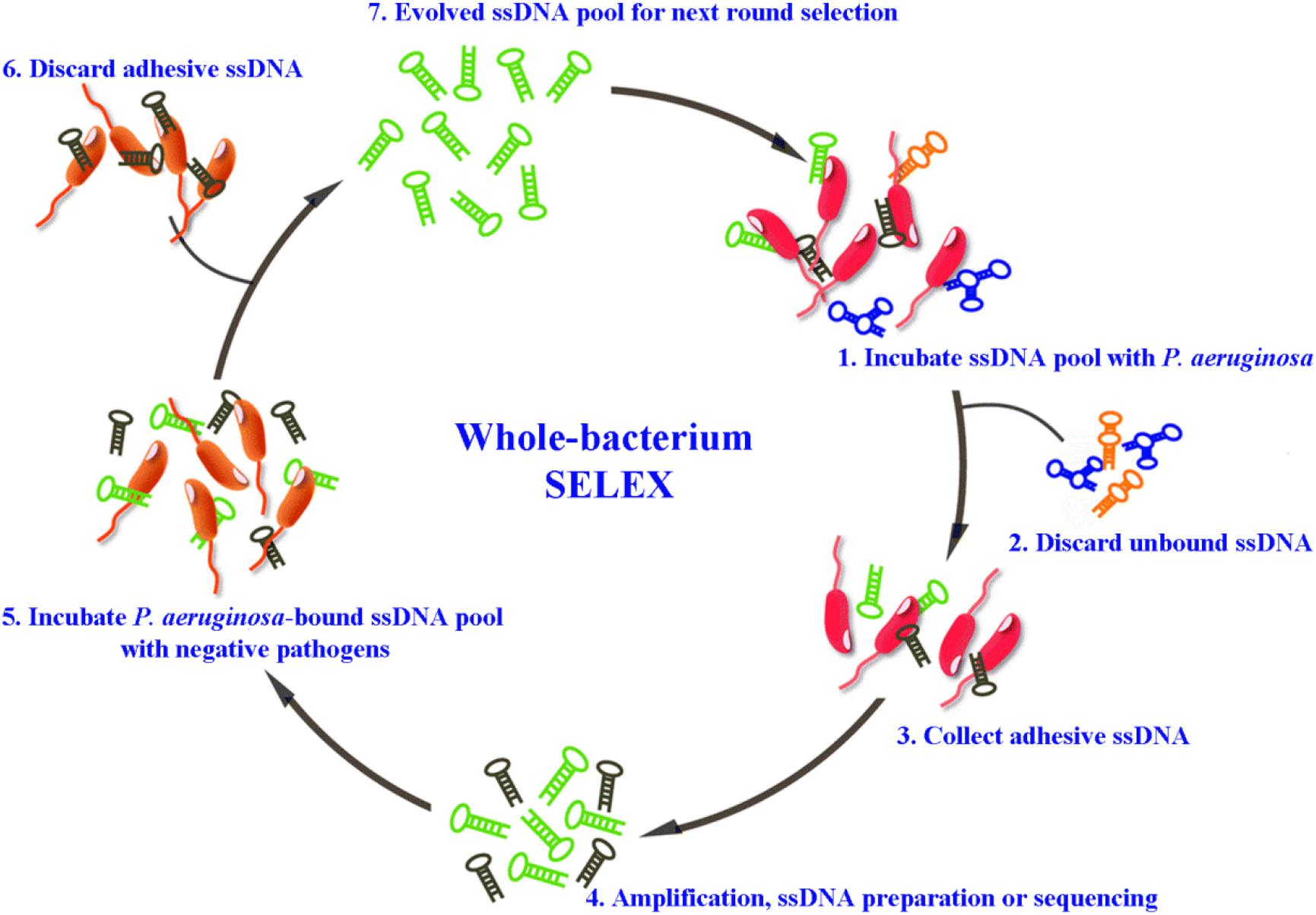
Figure 1. Schematic representation of whole-bacterium aptamer screening in P. aeruginosa using systematic evolution of ligands by exponential enrichment (SELEX) (Liu et al., 2020).
Optical methods have been widely applied in various fields, owing to their convenience, low-cost, rapidity, and resistance to electrical noise. They can be divided into several subclasses, which are based on absorption, reflection, refraction, dispersion, infrared, chemiluminescence, and fluorescence (Paniel et al., 2013). Among them, fluorescence spectroscopy, surface plasmon resonance (SPR), and surface-enhanced Raman scattering (SERS) are the most employed optical techniques because of their enhanced sensitivity.
After an electron is promoted to an excited state by absorbing a high-energy photon, fluorescence emission of a lower-energy photon occurs when the electron returns to its original ground state (Chen et al., 2018). Aptamer-based fluorescent biosensors mainly detect their targets through changes in fluorescence polarization or intensity that result from the interaction between the fluorescently labeled aptamer and its corresponding target.
Traditional organic fluorescent dyes, which have long been used in sensors, include carboxyfluorescein (FAM), fluorescein isothiocyanate (FITC), and acridine orange (Paniel et al., 2013). A fluorescence biosensor based on an FITC-aptamer was developed by Kim et al. (2013) for P. aeruginosa detection. The aptasensor has been used to analyze drinking water, with a lower limit of detection (LOD) of 5.07 colony-forming units (CFU)/mL. However, its linear range (5.64 to 102 CFU/mL) is suboptimal. Another fluorescence aptasensor was reported, based on aptamer hybridization with carboxyfluorescein-labeled complementary DNA (FAM-cDNA) in combination with magnetic separation (Figure 2A) (Zhong et al., 2018). In the absence of P. aeruginosa, FAM-cDNA hybridizes with the aptamer, which is covalently bound to the surface of magnetic nanoparticles (MNPs). In the presence of P. aeruginosa, FAM-cDNA will be displaced from the aptamer by the bacterium and therefore released from the MNPs. The quantification of P. aeruginosa can then be determined from the fluorescence intensity of the FAM-cDNA in the supernatant after magnetic separation. The assay has an outstanding linear response to the logarithm of P. aeruginosa concentrations ranging from 10 to 108 CFU/mL, with a LOD as low as 1 CFU/mL. The whole detection process can be finished within 1.5 h. Gao et al. (2018) likewise engineered a fluorescent aptasensor based on the same DNA hybridization principle, with graphene oxide quantum dots (GOQDs) serving as the quencher. As shown in Figure 2B, upon addition of P. aeruginosa, the aptamer specifically binds to it as a bio-recognition element. FAM-cDNA prefers to hybridize with the aptamer, resulting in the desorption of FAM-cDNA from GOQDs and recovery of FAM fluorescence. The aptasensor shows a linear response to P. aeruginosa concentration in the range from 1.28 × 103 to 2.00 × 107 CFU/mL, with a LOD of 100 CFU/mL. The system can complete all detection steps within 2 h and has been used to detect P. aeruginosa in drinking water, orange juice, and popsicle samples. Although organic fluorescent dyes are still the most widely used fluorescent dyes in biosensors currently, there exist some well-known disadvantages, such as interference from background fluorescence, short lifetime as well as poor stability against photo-bleaching. It is crucial to solve these problems for their further application in rapid detection of pathogens.
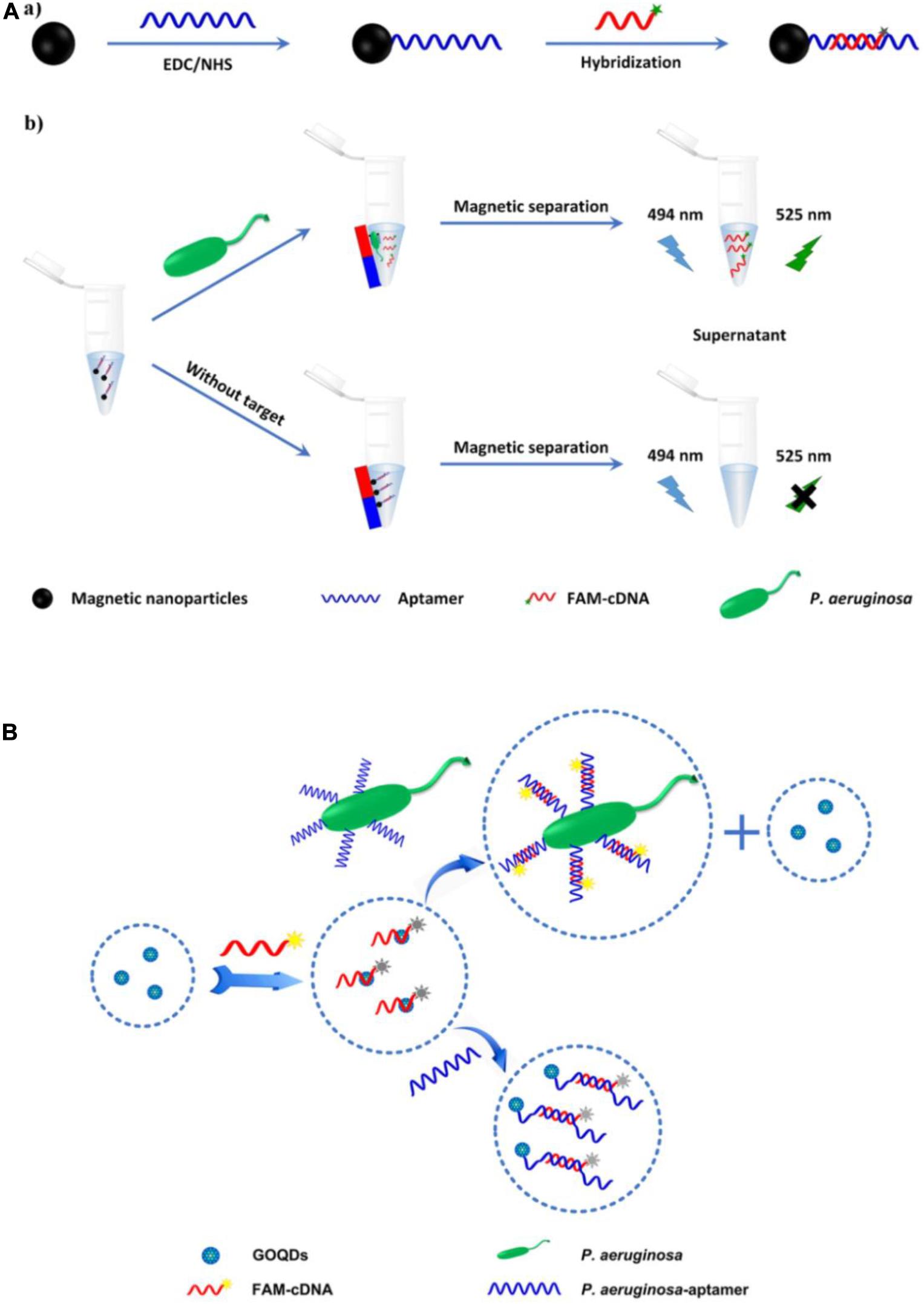
Figure 2. (A) Schematic illustration of the fabrication of magnetic nanoparticles modified by the hybrid of aptamer and carboxyfluorescein-labeled complementary DNA (a), and the principle of P. aeruginosa detection (b) (Zhong et al., 2018). (B) Schematic illustration of the principle of fluorescent aptasensor detection of P. aeruginosa (Gao et al., 2018).
Compared with organic dyes, fluorescent nanomaterials possess numerous advantageous characteristics, such as a broader absorption spectrum, a narrower and size-tunable emission spectrum, and better stability against photobleaching (Yao et al., 2014). Owing to their attractive properties, fluorescent nanomaterials, such as carbon dots (CDs), quantum dots (QDs), and upconversion nanoparticles (UCNPs), have been widely applied in biosensor fabrication (Chen et al., 2018). Wang et al. (2018) constructed an aptamer-based fluorescence assay, in which the specific aptamer was conjugated with photoluminescent CDs as the fluorescent probe and graphene oxide (GO) served as the anchor and quencher. The use of GO can reduce the background signals of CDs and improve sensitivity. This aptasensor displayed excellent specificity toward P. aeruginosa, with a lower LOD of 9 CFU/mL, and was free from interference of many ubiquitous bacteria, including Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Enterococcus faecalis, and Clostridium perfringens. For the simultaneous detection of four types of bacteria (Listeria monocytogenes, Staphylococcus aureus, Salmonella typhimurium, and P. aeruginosa), an aptasensor exploiting Stokes and anti-Stokes photoluminescence of two QDs and two UCNPs was developed (Figure 3A) (Yüce et al., 2018). The system was composed of aptamer-modified QD and UCNP probes and partially complementary DNA-modified magnetic beads (MBs). The fluorescent signals before and after incubation with targets were detected after magnetic separation using sequential excitation at 335 nm for the QDs and 980 nm for the UCNPs. By complementing QDs with UCNPs, the problems associated with fluorescence signal overlap of QDs and the inherently poor quantum yields or limited color options of UCNPs were overcome using this multiplex sensing method.
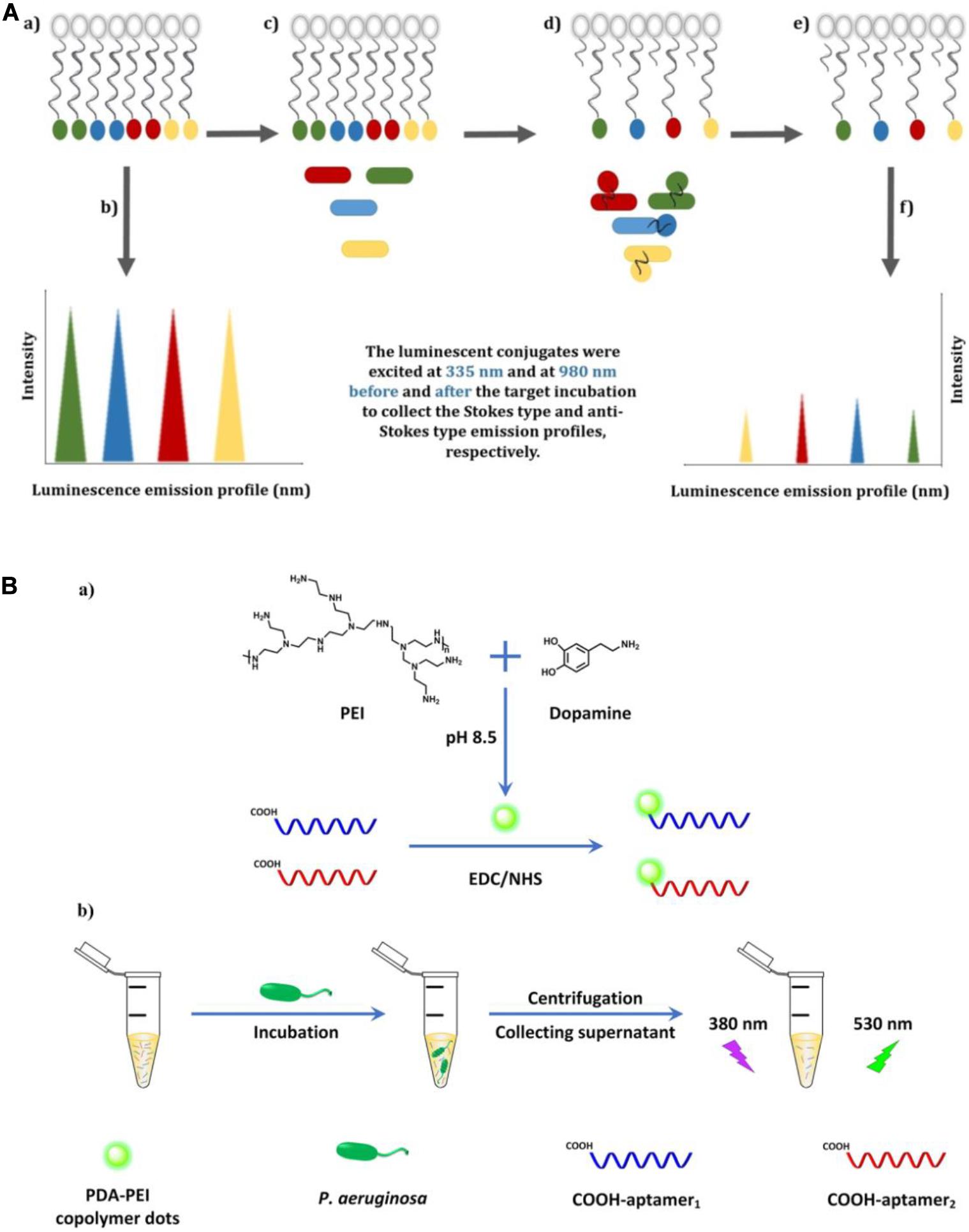
Figure 3. (A) Illustration of the multiplex-detection of conjugates of luminescent upconversion nanoparticles and quantum dots (a), luminescence emission profiles of the conjugates before interaction with the targets (b), addition of the targets into the conjugate solution (c), binding of the luminescent aptamer–nanoparticle probes to the targets (d), separation of the unbound conjugates (e), and luminescence emission profiles of the conjugates after interaction with the targets (f) (Yüce et al., 2018). (B) Schematic illustration of the fabrication of dual-aptamer-labeled polydopamine–polyethyleneimine (PDA-PEI) probe (a), and the detection procedure for P. aeruginosa (b) (Zhong et al., 2020).
In contrast to general fluorescent nanomaterials, polymer-based fluorescent nanomaterials display excellent biocompatibility and biodegradability, and are easy to prepare via self-polymerization (Zhang et al., 2015). Zhong et al. (2020) engineered a fluorescent biosensor based on dual-aptamer-labeled polydopamine–polyethyleneimine (PDA-PEI) copolymer dots (Figure 3B). The PDA-PEI copolymer dots have good aqueous dispersibility and are very stable against photobleaching, extreme pH, and high ionic strength. In the presence of P. aeruginosa, the aptamer-modified PDA-PEI dots can specifically capture the bacteria. After centrifugation, the bacterial conjugates were isolated from the solution, with the unbound aptamer-modified PDA-PEI dots remaining in the supernatant. By determining the decrease of fluorescence intensity in the supernatant, the amount of P. aeruginosa can be quantified, with the whole detection process being finished within 1.5 h. The LOD was determined to be as low as 1 CFU/mL.
These aptasensors based on fluorescent nanomaterials exhibit excellent performance in rapid detection of P. aeruginosa with high sensitivity and good selectivity. Although the preparation of fluorescent nanomaterials is complicated, with the further improvement and development of fabrication technologies, fluorescent nanomaterials will probably take place of organic dyes in biosensors in the future.
SPR is an ultrasensitive, real-time, and label-free analytical technique (Gorodkiewicz and Lukaszewski, 2018). When light is incident at a critical angle on the interface of two media with different refractive indices, resonant absorption of free electrons in the metal occurs (Zhou et al., 2019), which leads to a diminution of the reflected light. When an analyte binds to the interface, an intensity change and/or a shift of the absorbance peak is observed, with the magnitude and position of the displacement being related to the characteristics of the surface interaction (Prabowo et al., 2018). The surface-constrained collective oscillation of free electrons produced by the interaction of light with metal nanoparticles (e.g., Au, Ag, and Cu) is known as localized surface plasmon resonance (LSPR) (Csaki et al., 2018). Compared with traditional SPR biosensors, LSPR biosensors have numerous advantages, such as low cost, miniaturization and portability, and have been widely used to detect various analytes, including small molecules, proteins and clinical biomarkers (Unser et al., 2015; Csaki et al., 2018).
Hu et al. (2018) reported an aptamer-based LSPR sensing platform to detect whole-cell P. aeruginosa strain PAO1 (Figure 4A). They used nanosphere lithography to manufacture a sensor surface containing a hexagonal array of Au nanotriangles. By immobilizing aptamers on the sensor, P. aeruginosa cells were captured from solution onto the sensor surface, resulting in a red shift of the LSPR extinction maximum (Hu et al., 2018). The approach is extraordinarily sensitive, with a lower LOD of 10 CFU/mL. Yoo et al. (2015) used three specific aptamers as recognition elements and introduced another LSPR sensor array for the multiplexed detection of L. acidophilus, S. typhimurium, and P. aeruginosa. As shown in Figure 4B, a multispot gold-capped nanoparticle array chip was used in this system, which was composed of a dielectric layer comprising a thin gold (Au) layer on silica (Si) nanoparticles (NPs)-absorbed glass slide. The presence of targets manifest as a change in the LSPR peak intensity upon incubation of the chip with a sample volume of only 3 μL.
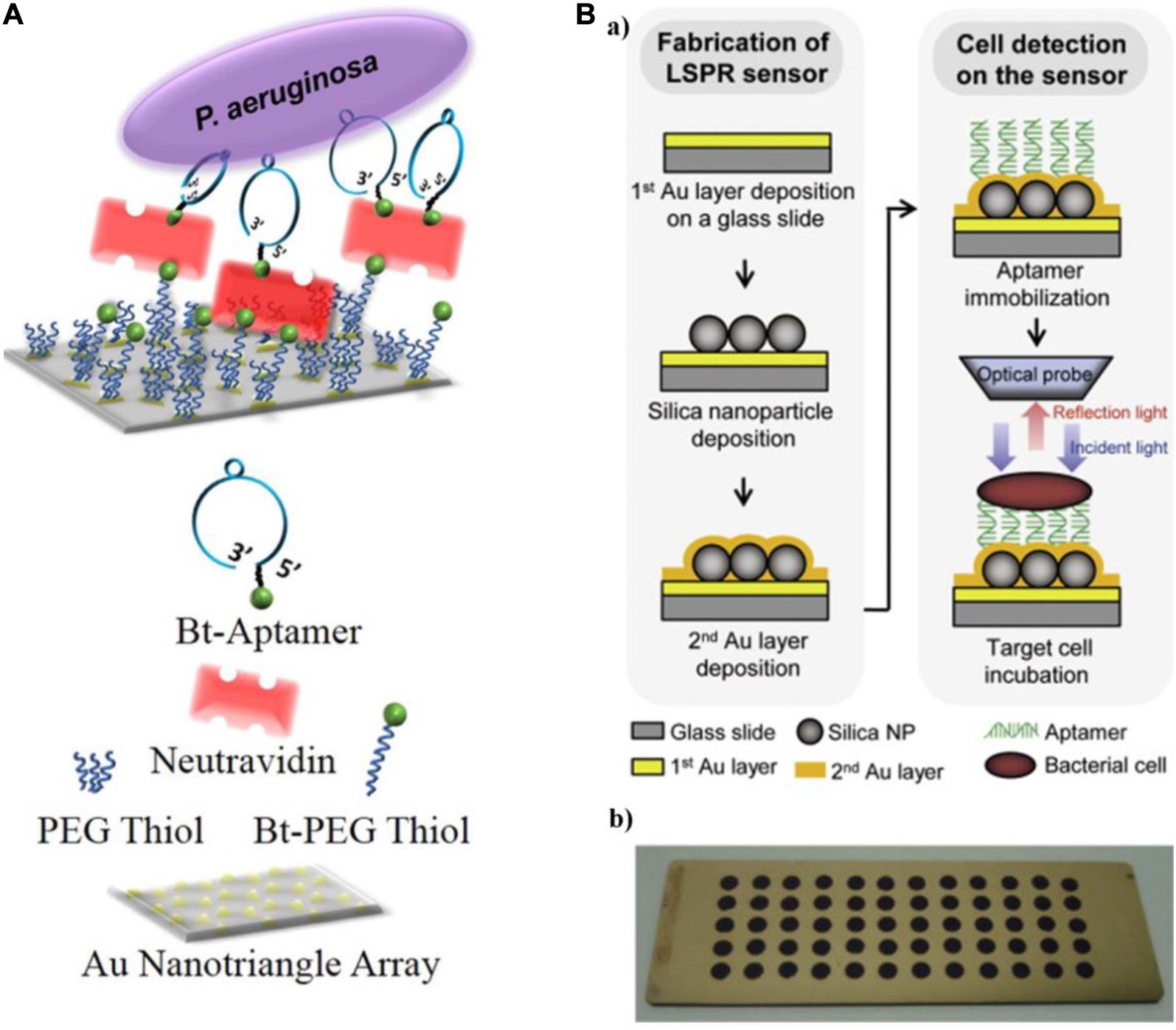
Figure 4. (A) Schematic diagram of a localized surface plasmon resonance (LSPR) sensor chip (top) and legend (bottom) (Hu et al., 2018). (B) Schematic representing the detection of bacterial cells using the aptamer-functionalized LSPR-based sensor (a), and multispot gold-capped nanoparticle array chip (b). Each spot (diameter of 3 mm) on the chip consists of Si nanoparticles (NPs) with a dielectric structure formed by the deposition of a gold layer (Yoo et al., 2015).
Despite the fact that LSPR-based aptasensors can accomplish label-free detection of P. aeruginosa with high sensitivity and good specificity, they need costly optical system as well as long preparation time.
The detection of pathogens using SERS has attracted much attention, owing to its ultra-sensitive, non-destructive, fingerprint identification (Xu et al., 2019). Raman spectroscopy measures the inelastic scattering of light by analytes in the sample medium. SERS is produced by analytes adsorbed on rough metal surfaces (Zong et al., 2018).
Wu et al. (2018) fabricated a bimodal (SERS and colorimetric) aptasensor for quantitation of P. aeruginosa (Figure 5). A horseradish peroxidase (HRP)-linked aptamer against P. aeruginosa was conjugated to 30-nm gold nanoparticles (AuNPs) as a colorimetric probe, while aptamer’s cDNA fragment was coupled with 15-nm AuNPs as a SERS probe. In the absence of targets, the two probes automatically assemble into a duplex. However, upon exposure to P. aeruginosa, the aptamer dissociates from the cDNA and binds to its target. The previous asymmetric dimers, in which the small gaps between two differently sized AuNPs contribute to intense Raman signal enhancement, become isolated AuNPs. After centrifugation, the SERS signal from the supernatant is decreased significantly, while a colored signal is generated the from HRP-linked aptamer in the resuspended pellet after addition of 3,3′,5,5′-tetramethylbenzidine (TMB) and hydrogen peroxide. The LOD of the aptasensor is calculated to be 20 CFU/mL using SERS detection and 50 CFU/mL using colorimetric detection. The accuracy of the system is ensured by the positive colorimetric response and a concomitant negative SERS response.
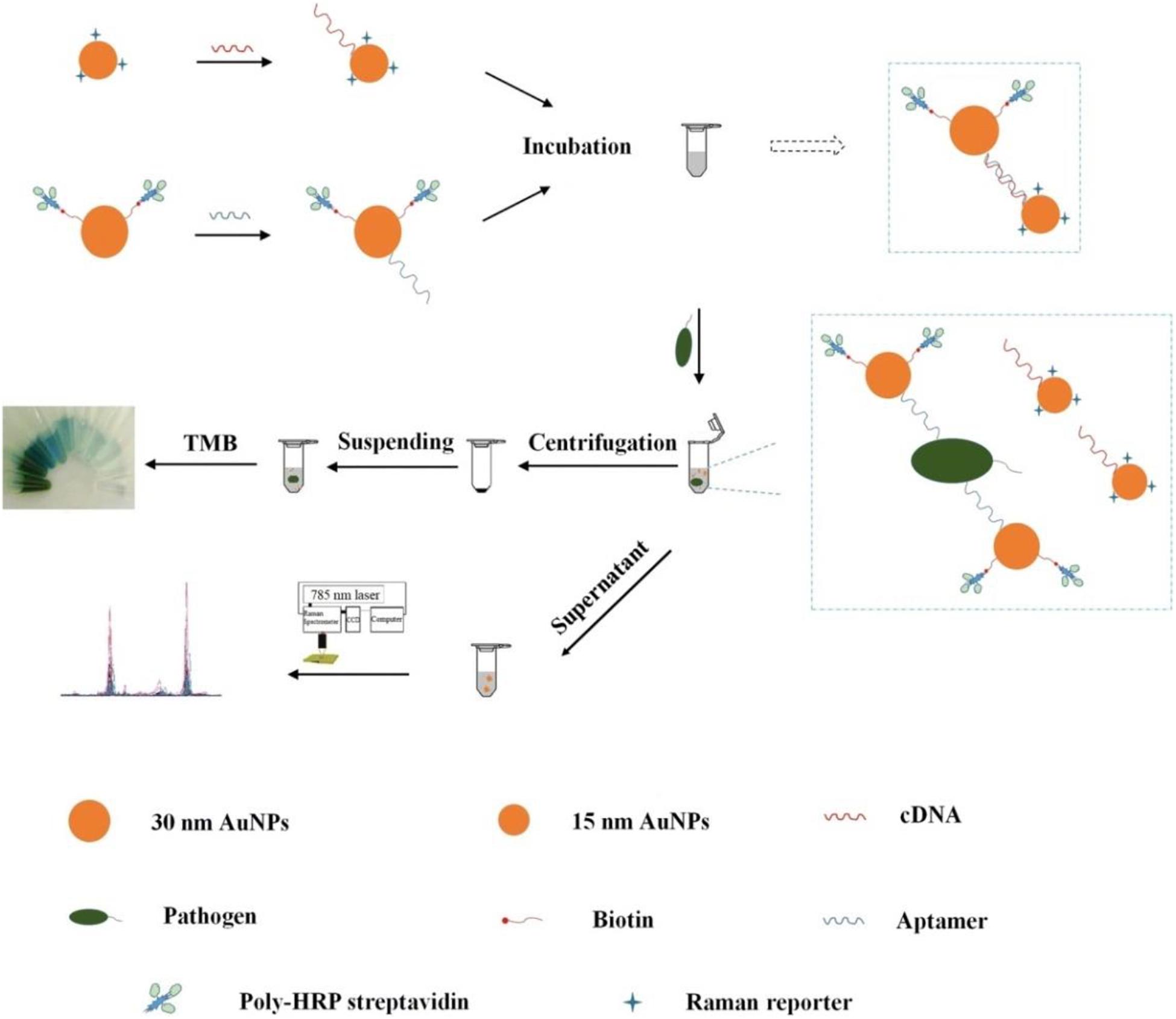
Figure 5. Schematic illustration of the fabrication procedure of the dual mode aptasensor for sensitive detection of P. aeruginosa (Wu et al., 2018).
In spite of the ultra-sensitivity and high accuracy of SERS-based aptasensors, the high cost of instrument and need of professional operation may restrict their common application.
Electrochemical biosensors have a long history and remain attractive tools for identification and quantification of analytes. They demonstrate several advantages, being highly sensitive, rapid, economical, amenable to miniaturization, and easy in signal translation and operation (Maduraiveeran et al., 2018). An electrochemical biosensor is typically composed of an electrode fixed with a molecular identification element and an electrochemically active element. The presence of the target analyte will result in changes on the surface of the electrode, which are converted to electrochemical signals (current, impedance, potential, or conductance) (Li et al., 2019). Correspondingly, these biosensors can be classified into amperometric/voltammetric, impedimetric, potentiometric, and conductometric sensors (Paniel et al., 2013). The combination of these techniques with aptamers has promoted the development of aptamer-based biosensors for pathogen identification in real complex samples. Amperometry/voltammetry and impedimetry are the most frequently used electrochemical techniques for P. aeruginosa detection.
Amperometry/voltammetry is the most common and successful electrochemical method in research and practical applications to environment monitoring, medical analysis, and food sanitation. Several groups have constructed numerous electrochemical aptasensors of this type and used them to quantify P. aeruginosa. Das et al. (2019) exploited the inherent peroxidase-like nanozyme activity of gold nanoparticles (GNPs) to develop an aptamer-mediated sensor of live P. aeruginosa (Figure 6A). The peroxidase-mimetic activity of GNPs is inhibited by the simple adsorption of the aptamer onto their surface. However, in the presence of P. aeruginosa, the aptamer binds with high affinity to bacterium, restoring the peroxidase-like activity of the GNPs, which catalyzes the oxidation of TMB. With TMB as an electrochemically active material, the aptasensor was constructed using a disposable screen-printed carbon electrode (Das et al., 2019). This approach is ultrasensitive, with a lower LOD of 60 CFU/mL in water and a detection time of 10 min. Zhang et al. (2019b) constructed another novel aptasensor for rapid and sensitive detection of P. aeruginosa. As shown in Figure 6B, a Cu-ZrMOF (zirconium series metal-organic framework) was synthesized to provide a large surface area for aptamer/DNA adsorption and high catalytic activity. The Cu-ZrMOF/aptamer/DNA nanocomposite was used to identify P. aeruginosa captured on an antibody-modified electrode surface, and served as an electrochemical probe by catalyzing the decomposition of H2O2. To increase the electron transfer for satisfactory detection sensitivity, the gold electrode was coated with Super P® and AuNPs. The biosensor is capable of quantifying P. aeruginosa with a lower LOD of 2 CFU/mL within 120 min. More importantly, this biosensor has been validated using urine samples spiked with P. aeruginosa, which indicates its promising potential for clinical applications. Shahrokhian and Ranjbar (2019) fabricated an electrochemical biosensor with aptamers immobilized in an engineered zeolitic imidazolate framework-8 (ZIF-8) (Figure 6C). The hollow porous ZIF-8 provided a large surface area containing abundant carboxylic acid groups in low density with a favorable open structure to immobilize a large amount of aptamers via amino-reactive coupling. Because of the unique π-π stacking interactions between the aptamer and GO, differential pulse voltammetric detection was used with ferrocene-graphene oxide (Fc-GO) as an electroactive indicator. Upon binding with P. aeruginosa, the aptamer undergoes conformational changes, resulting in the release of Fc-GO from the electrode surface and a change in the effective electron transfer to the electrode. Using this “signal-off” strategy, the biosensor displays a broad linear dynamic range with a lower LOD of 1 CFU/mL. The reported amperometric/voltammetric aptasensors exhibit excellent performance for quantitative determination of P. aeruginosa with high sensitivity, good specificity and ultralow LOD. However, the poor stability of electrode will have unfavorable effects on their application in pathogen detection.
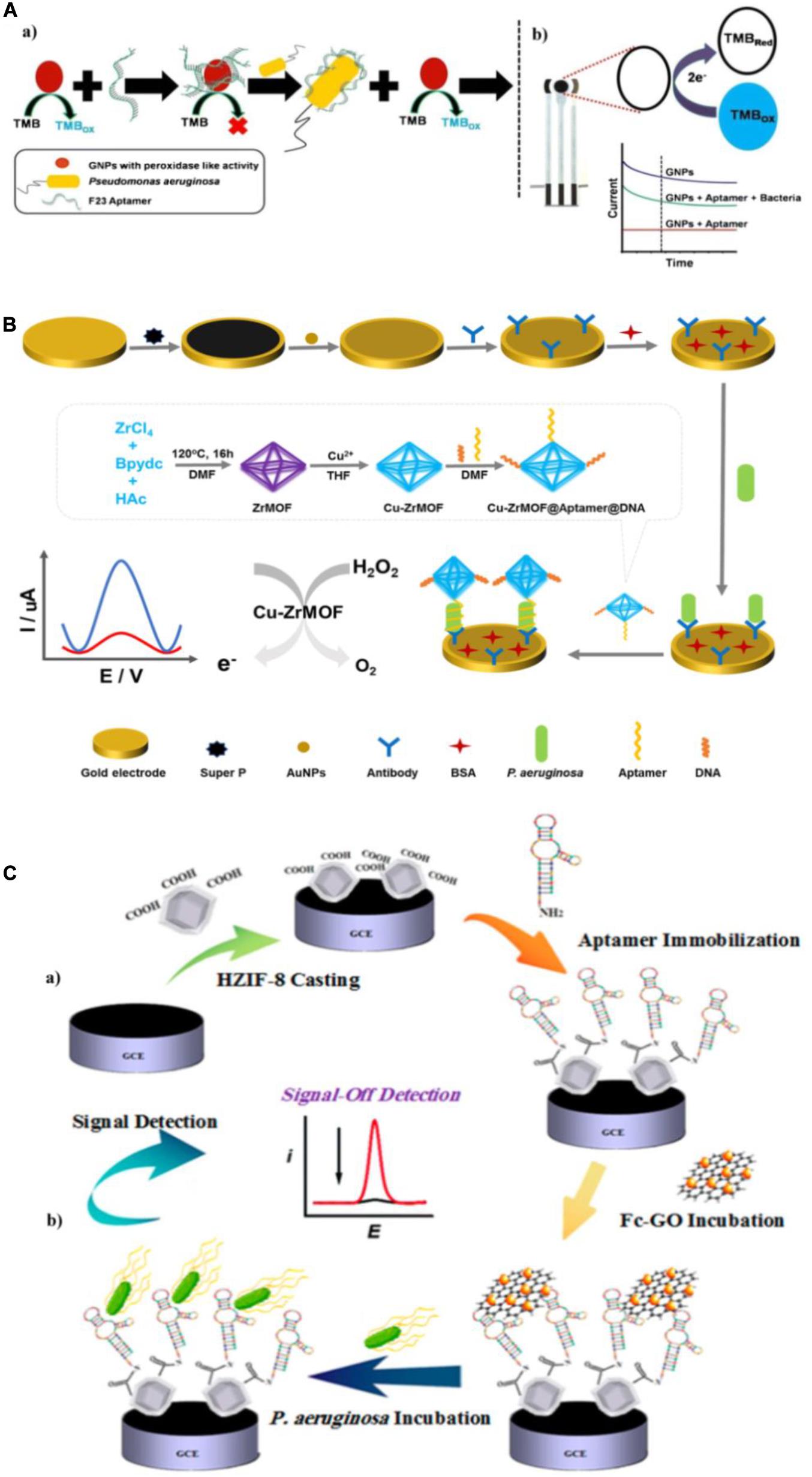
Figure 6. (A) Schematic representation of the detection of P. aeruginosa using the nanozyme activity of gold nanoparticles (GNPs) (a), and the electrochemical detection of tetramethylbenzidine (TMB) reduction on a screen-printed carbon electrode in the presence of P. aeruginosa (b) (Das et al., 2019). (B) Schematic of the electrochemical detection of P. aeruginosa (Zhang et al., 2019b). (C) Schematic illustration of aptasensor fabrication (a) and “signal-off” detection of P. aeruginosa (b) (Shahrokhian and Ranjbar, 2019).
Impedimetric biosensors have drawn much interest in recent years, owing to their reduced matrix interference, relatively simple electrical measurements, and the possibility of automation (Paniel et al., 2013). Roushani et al. (2019) constructed an impedimetric aptasensor using a glassy carbon electrode (GCE) modified with silver nanoparticles (AgNPs) (Figure 7A). Free amino groups of the aptamer were covalently coupled to the AgNP/GCE surface. The interaction between P. aeruginosa and aptamers significantly reduced the charge transfer from the hexacyanoferrate redox system to the surface of the electrode. The quantification of P. aeruginosa is carried out by monitoring the change in charge transfer resistance using the hexacyanoferrate redox system as an electrochemical probe. This assay was used to measure P. aeruginosa in spiked serum samples, with a lower LOD of 33 CFU/mL. Sarabaegi and Roushani (2019) designed an electrochemical aptasensor (Figure 7B) with a specific aptamer covalently linked to a GCE surface, and the hexacyanoferrate redox system as an electrochemical probe. They used nanosized chitosan (NC) particles to modify the GCE, form a sensitive layer, and improve performance. The aptasensor has a lower LOD of 3 CFU/mL and has been used to detect P. aeruginosa in serum samples. These impedimetric aptasensors have been recognized as promising analytical tools for P. aeruginosa detection. Nevertheless, they require time-consuming and costly processes to model the electrodes. More suitable materials are in demand for fabrication of impedimetric aptasensors.
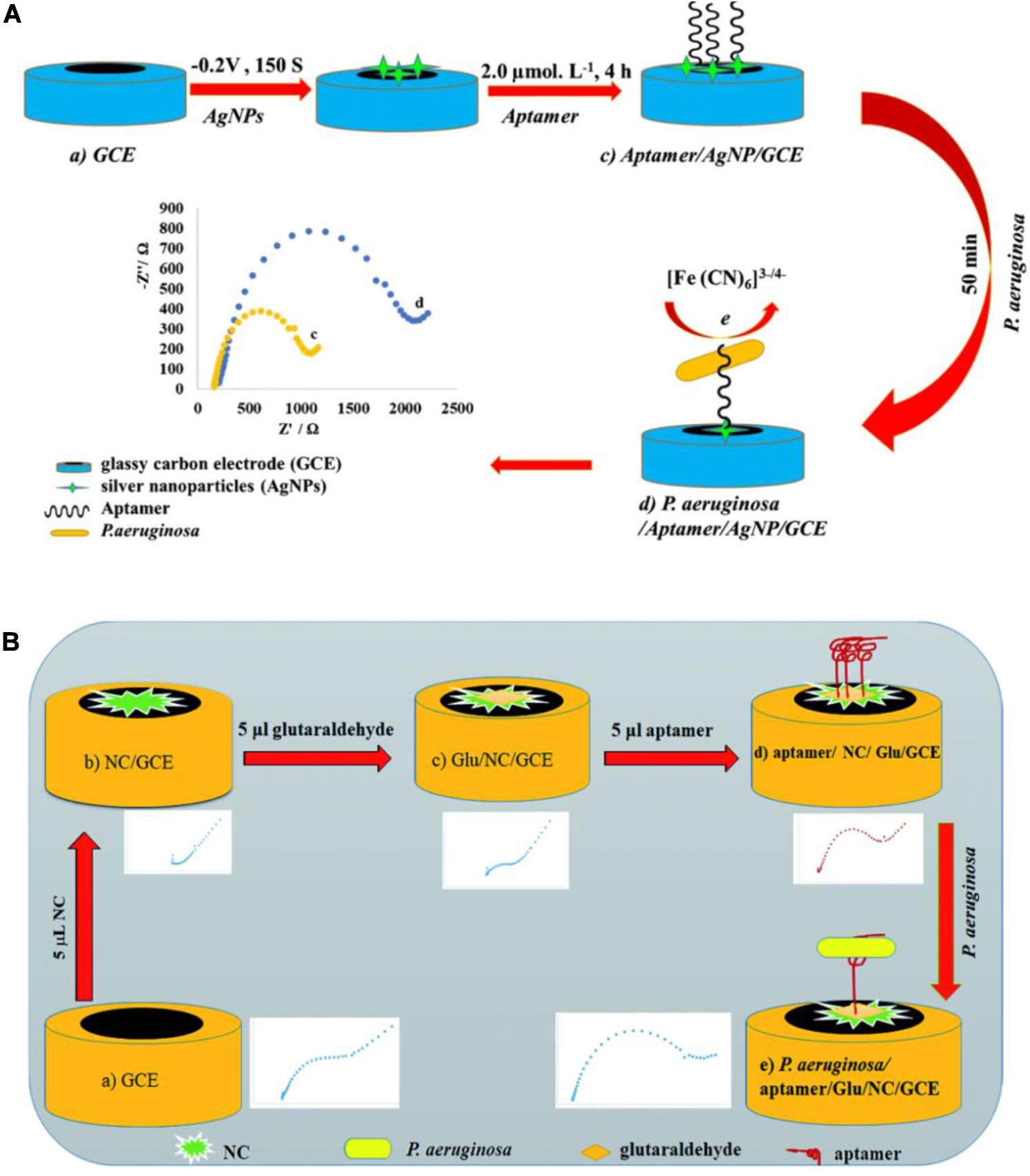
Figure 7. (A) Schematic view of the development of a P. aeruginosa/aptamer/silver nanoparticle/glassy carbon electrode complex for P. aeruginosa detection (Roushani et al., 2019). (B) Schematic view of the development of a P. aeruginosa/aptamer/glutaraldehyde (Glu)/nanosized chitosan (NC)/glassy carbon electrode complex for P. aeruginosa detection (Sarabaegi and Roushani, 2019).
The electrode material plays a key role in electrochemical biosensor construction because it directly influences the reaction speed, stability, sensitivity, electron transfer efficiency, and active site capacity. Since the 1970s, various chemical modifications to the electrode surface have been introduced to control the electrochemical process. In recent years, the use of nanotechnology to improve the sensitivity, electrochemical activity, and conductivity of electrochemical sensors has become prevalent. Compared with macroscopic materials, nanomaterials exhibit superior optical and electrical properties because of their special structures. Various nanomaterials, such as gold nanoparticles, graphene, and carbon nanotubes, have been commonly used to construct electrochemical aptasensors, owing to their large specific surface area and biocompatibility. Moreover, the emergence of new nanocomposites, such as graphene–metal nanoparticles and carbon nanotubes modified with metal nanoparticles, has advanced the development of electrochemical aptasensors. In the future, the identification of new nanomaterials and composite nanomaterials will further improve the sensitivity of electrochemical biosensors.
Piezoelectric crystal biosensors, which are based on mass-sensitive transduction, can offer direct label-free analysis with excellent sensitivity and specificity. They accomplish quantification of targets by measuring changes in their oscillation frequency induced by small mass variations on surface, which can be caused by specific interactions between functional molecules, such as analyte and aptamer. Shi et al. (2019) developed an aptamer/polyadenylated DNA interdigitated gold electrode piezoelectric sensor. As shown in Figure 8A, the sandwich-type MB/aptamer/polyadenylated DNA complex was combined with a gold interdigital electrode connected to a multichannel series piezoelectric quartz crystal (Au IDE-MSPQC). In the absence of P. aeruginosa, polyadenylated DNA is bound to the aptamer through partially complementary interaction. In the presence of P. aeruginosa, the bacterium binds to the aptamer, displacing the polyadenylated DNA. After magnetic separation, the strong affinity between adenine and Au causes the adsorption of the polyadenylated DNA on the Au IDE surface, resulting in frequency shift of the MSPQC sensor. The LOD of this method is as low as 9 CFU/mL in buffer and 52 CFU/mL in simulated blood samples.
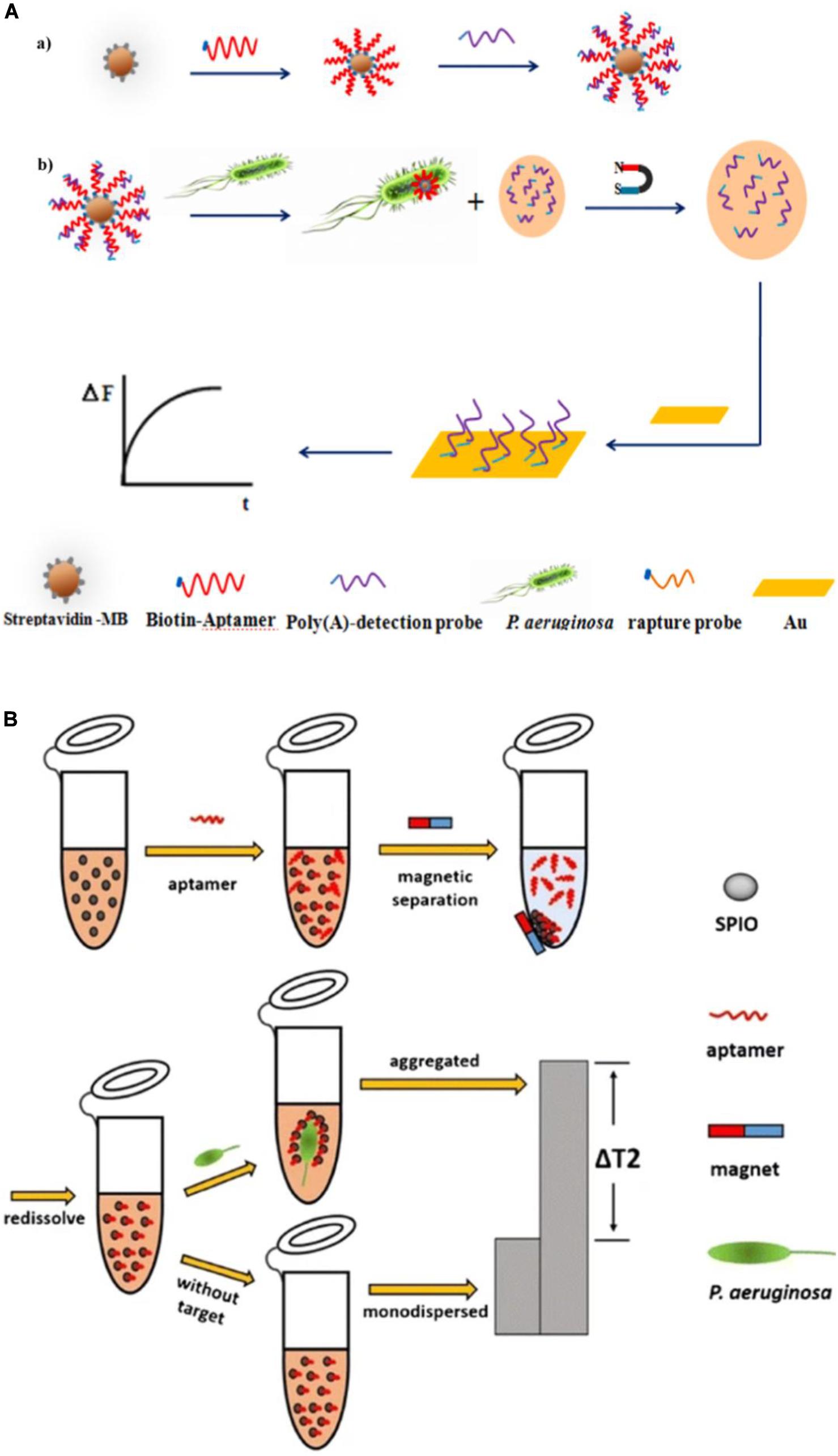
Figure 8. (A) P. aeruginosa detection strategy using a magnetic bead/aptamer/polyadenylated DNA complex and a gold interdigital electrode connected to a multichannel series piezoelectric quartz crystal. The preparation of magnetic bead/aptamer/polyadenylated DNA (a) and the detection of P. aeruginosa by the adsorption of released polyadenylated DNA onto the gold interdigital electrode surface (b) (Shi et al., 2019). (B) Schematic illustration of the magnetic relaxation switch aptasensor for P. aeruginosa detection (Jia et al., 2017).
The magnetic relaxation switch (MRSw) has drawn increasing attention since 2002 (Perez et al., 2002). Its high sensitivity results from the magnetic amplification principle, and enables a wide variety of targets, including proteins, cancer cells, bacteria, and viruses to be measured (Bamrungsap et al., 2011; Ghazani et al., 2014; Chen et al., 2015; Wang et al., 2015). Aggregation of magnetic nanoparticles affects billions of surrounding water molecules, dephasing the precession of nuclear spins of water protons. The resulting change in the spin–spin relaxation time (T2) permits sensitive target detection. Jia et al. (2017) reported an MRSw-based aptasensor for P. aeruginosa detection (Figure 8B), with the aptamer covalently bound to superparamagnetic iron oxide (SPIO) nanoparticles. P. aeruginosa is captured by the aptamer, which leads to aggregation of the SPIO and a substantial increase in the T2 value of the neighboring water molecules, measured using low-field nuclear magnetic resonance. The assay has a lower LOD of 50 CFU/mL.
These aptasensors above also exhibit great potential for P. aeruginosa detection. They offer alternative tools for pathogen analysis in water, food, and clinical samples.
Aptasensor is a powerful and promising sensing method for analysis of P. aeruginosa in food, water, and clinical samples. In this review, recent advances in the development of aptasensors for P. aeruginosa detection are comprehensively summarized. According to the different sensing mechanisms, representative aptasensors are classified into optical, electrochemical and other signal transduction methods, which are listed in Table 1. Optical aptasensors, including fluorescence, SPR and SERS methods, are advantageous for their properties of simplicity, rapid response and high sensitivity. Among the optical methods, the fluorescence is extensively used in quantitative determination of P. aeruginosa, and exhibits high sensitivity and good specificity. However, fluorescence are easy to be interfered by auto-fluorescence or background fluorescence. Compared with fluorescent methods, novel optical methods, such as SPR and SERS, are more stable, rapid and sensitive in detection of P. aeruginosa. Nevertheless, the needs of costly equipment and tedious sample preparation to some extent restrict their further application. Electrochemical aptasensors, including amperometric/voltammetric and impedimetric methods, exhibit higher sensitivity and accuracy, in comparison with optical methods. Even though they require time-consuming and costly processes to model the electrodes, electrochemical aptasensors are still recognized as the most promising analytical tools for P. aeruginosa detection. Apart from optical and electrochemical approaches, piezoelectric crystal aptasensors and MRSw-based aptasensors also exhibit great potential for P. aeruginosa quantitation.
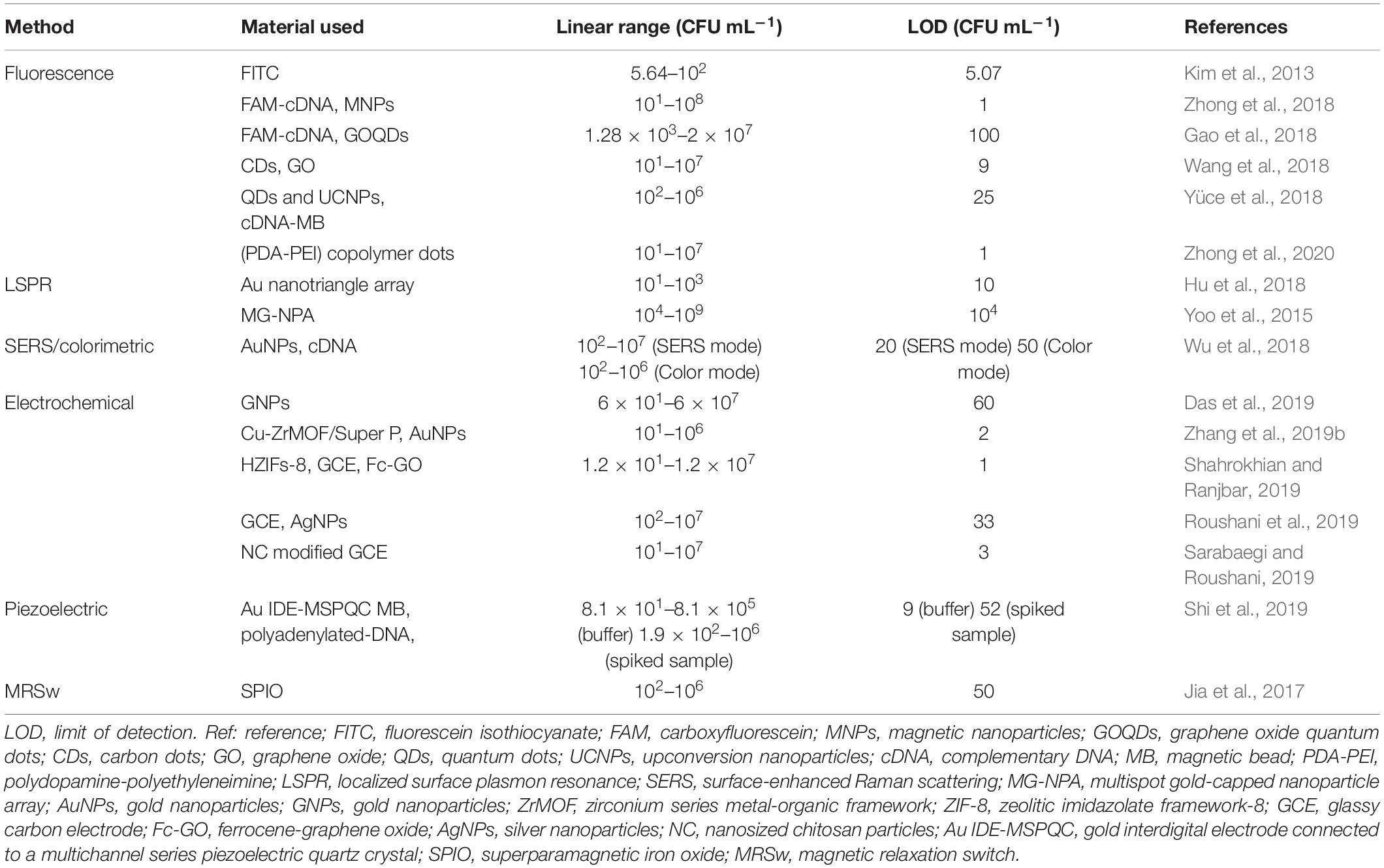
Table 1. Comparison of analytical performance of the presented aptamer-based detection methods for P. aeruginosa.
In the past decade, the development of aptasensors is one of the most active research fields. However, detection of P. aeruginosa by aptasensors is still under development. Possible reasons are analyzed as follows. First, relatively few specific aptamers for P. aeruginosa have been obtained, which hinders the further development of aptasensors in pathogen detection. More efficient screening methods are required to identify pathogen-specific aptamers. Second, pathogens are more complicated than other targets, because multiple different components exist on their surfaces, such as proteins, polysaccharides and endotoxins. Therefore, compared with one single aptamer, a group of specific aptamers will provide better selectivity and reliability in practical aptasensor detection. Third, the stability of aptasensor requires to be further improved. Because of the flexibility in structure and charged property, aptamers are more susceptible to interferences from practical samples. Therefore, aptamers need to be further chemically optimized before aptasensor fabrication.
In the future, miniaturization, high throughput and multi-item joint testing are expected to be the focus of research. Aptasensors may someday find its place in the market to compete with the expensive antibody-based methods that are still most commonly used rapid methods at the present time.
XZ surveyed the literature and wrote the manuscript. SG conceived the topic of the manuscript and synopsis, and helped in shaping the manuscript. JW and XH provided the critical feedback and reviewed the final manuscript. All authors have read and finalized the manuscript.
This work was supported by the National Natural Science Foundation of China (82071011, 81770925, and 81790641) and Science and Technology Commission of Shanghai Municipality (19ZR1471900).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alahi, M. E. E., and Mukhopadhyay, S. C. (2017). Detection methodologies for pathogen and toxins: a review. Sensors 17:1885. doi: 10.3390/s17081885
Bamrungsap, S., Shukoor, M. I., Chen, T., Sefah, K., and Tan, W. (2011). Detection of lysozyme magnetic relaxation switches based on aptamer-functionalized superparamagnetic nanoparticles. Anal. Chem. 83, 7795–7799. doi: 10.1021/ac201442a
Cai, R., Yin, F., Chen, H., Tian, Y., and Zhou, N. (2020). A fluorescent aptasensor for Staphylococcus aureus based on strand displacement amplification and self-assembled DNA hexagonal structure. Mikrochim. Acta 187:304. doi: 10.1007/s00604-020-04293-9
Cesewski, E., and Johnson, B. N. (2020). Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 159:112214. doi: 10.1016/j.bios.2020.112214
Chen, K., Zhou, J., Shao, Z., Liu, J., Song, J., Wang, R., et al. (2020). Aptamers as versatile molecular tools for antibody production monitoring and quality control. J. Am. Chem. Soc. 142, 12079–12086. doi: 10.1021/jacs.9b13370
Chen, L., Hwang, E., and Zhang, J. (2018). Fluorescent nanobiosensors for sensing glucose. Sensors 18:1440. doi: 10.3390/s18051440
Chen, Y., Xianyu, Y., Wang, Y., Zhang, X., Cha, R., Sun, J., et al. (2015). One-step detection of pathogens and viruses: combining magnetic relaxation switching and magnetic separation. ACS Nano 9, 3184–3191. doi: 10.1021/acsnano.5b00240
Chinnappan, R., AlAmer, S., Eissa, S., Rahamn, A. A., Abu Salah, K. M., and Zourob, M. (2017). Fluorometric graphene oxide-based detection of Salmonella enteritis using a truncated DNA aptamer. Mikrochim. Acta 185:61. doi: 10.1007/s00604-017-2601-9
Csaki, A., Stranik, O., and Fritzsche, W. (2018). Localized surface plasmon resonance based biosensing. Expert. Rev. Mol. Diagn. 18, 279–296. doi: 10.1080/14737159.2018.1440208
Curran, C. S., Bolig, T., and Torabi-Parizi, P. (2018). Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 197, 708–727. doi: 10.1164/rccm.201705-1043SO
Dao, T. N. T., Yoon, J., Jin, C. E., Koo, B., Han, K., Shin, Y., et al. (2018). Rapid and sensitive detection of Salmonella based on microfluidic enrichment with a label-free nanobiosensing platform. Sens. Actuators B Chem. 262, 588–594. doi: 10.1016/j.snb.2017.12.190
Das, R., Dhiman, A., Kapil, A., Bansal, V., and Sharma, T. K. (2019). Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 411, 1229–1238. doi: 10.1007/s00216-018-1555-z
Davydova, A. S., Vorobyeva, M. A., Kabilov, M. R., Tikunova, N. V., Pyshnyi, D. V., and Venyaminova, A. G. (2017). In vitro selection of cell-internalizing 2’-modified RNA aptamers against Pseudomonas aeruginosa. Russ. J. Bioorg. Chem. 43, 58–63. doi: 10.1134/s1068162016060030
Deschaght, P., Van Daele, S., De Baets, F., and Vaneechoutte, M. (2011). PCR and the detection of Pseudomonas aeruginosa in respiratory samples of CF patients. A literature review. J. Cyst. Fibros. 10, 293–297. doi: 10.1016/j.jcf.2011.05.004
Dong, X., Shi, Z., Xu, C., Yang, C., Chen, F., Lei, M., et al. (2020). CdS quantum dots/Au nanoparticles/ZnO nanowire array for self-powered photoelectrochemical detection of Escherichia coli O157:H7. Biosens. Bioelectron. 149:111843. doi: 10.1016/j.bios.2019.111843
Ellington, A. D., and Szostak, J. W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. doi: 10.1038/346818a0
Fang, Z., Wu, W., Lu, X., and Zeng, L. (2014). Lateral flow biosensor for DNA extraction-free detection of Salmonella based on aptamer mediated strand displacement amplification. Biosens. Bioelectron. 56, 192–197. doi: 10.1016/j.bios.2014.01.015
Farooq, U., Yang, Q., Ullah, M. W., and Wang, S. (2018). Bacterial biosensing: recent advances in phage-based bioassays and biosensors. Biosens. Bioelectron. 118, 204–216. doi: 10.1016/j.bios.2018.07.058
Fu, F., Zhang, Y., Li, L., Wang, H., Li, Q., Tao, X., et al. (2020). Intracellular pathogen detection based on dual-recognition units constructed fluorescence resonance energy transfer nanoprobe. Anal. Chem. 92, 11462–11468. doi: 10.1021/acs.analchem.0c02695
Furst, A. L., and Francis, M. B. (2019). Impedance-Based Detection of Bacteria. Chem. Rev. 119, 700–726. doi: 10.1021/acs.chemrev.8b00381
Gao, R., Zhong, Z., Gao, X., and Jia, L. (2018). Graphene oxide quantum dots assisted construction of fluorescent aptasensor for rapid detection of Pseudomonas aeruginosa in food samples. J. Agric. Food Chem. 66, 10898–10905. doi: 10.1021/acs.jafc.8b02164
Gao, S., Hu, W., Zheng, X., Cai, S., and Wu, J. (2019). Functionalized aptamer with an antiparallel G-quadruplex: structural remodeling, recognition mechanism, and diagnostic applications targeting CTGF. Biosens. Bioelectron. 142:111475. doi: 10.1016/j.bios.2019.111475
Ghazani, A. A., Pectasides, M., Sharma, A., Castro, C. M., Mino-Kenudson, M., Lee, H., et al. (2014). Molecular characterization of scant lung tumor cells using iron-oxide nanoparticles and micro-nuclear magnetic resonance. Nanomedicine 10, 661–668. doi: 10.1016/j.nano.2013.10.008
Gorodkiewicz, E., and Lukaszewski, Z. (2018). Recent progress in surface plasmon resonance biosensors (2016 to Mid-2018). Biosensors 8:132. doi: 10.3390/bios8040132
Groff, K., Brown, J., and Clippinger, A. J. (2015). Modern affinity reagents: recombinant antibodies and aptamers. Biotechnol. Adv. 33, 1787–1798. doi: 10.1016/j.biotechadv.2015.10.004
Horcajada, J. P., Montero, M., Oliver, A., Sorli, L., Luque, S., Gomez-Zorrilla, S., et al. (2019). Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32:e31-19. doi: 10.1128/CMR.00031-19
Hu, J., Fu, K., and Bohn, P. W. (2018). Whole-cell Pseudomonas aeruginosa localized surface plasmon resonance aptasensor. Anal. Chem. 90, 2326–2332. doi: 10.1021/acs.analchem.7b04800
Hua, R., Hao, N., Lu, J., Qian, J., Liu, Q., Li, H., et al. (2018). A sensitive potentiometric resolved ratiometric photoelectrochemical aptasensor for Escherichia coli detection fabricated with non-metallic nanomaterials. Biosens. Bioelectron. 106, 57–63. doi: 10.1016/j.bios.2018.01.053
Jean, S. S., Chang, Y. C., Lin, W. C., Lee, W. S., Hsueh, P. R., and Hsu, C. W. (2020). Epidemiology, treatment, and prevention of nosocomial bacterial pneumonia. J. Clin. Med. 9:275. doi: 10.3390/jcm9010275
Jia, F., Xu, L., Yan, W., Wu, W., Yu, Q., Tian, X., et al. (2017). A magnetic relaxation switch aptasensor for the rapid detection of Pseudomonas aeruginosa using superparamagnetic nanoparticles. Microchim. Acta 184, 1539–1545. doi: 10.1007/s00604-017-2142-2
Kaur, H., Shorie, M., and Sabherwal, P. (2020). Electrochemical aptasensor using boron-carbon nanorods decorated by nickel nanoparticles for detection of E. coli O157:H7. Mikrochim. Acta 187:461. doi: 10.1007/s00604-020-04444-y
Khater, M., de la Escosura-Muniz, A., and Merkoci, A. (2017). Biosensors for plant pathogen detection. Biosens. Bioelectron. 93, 72–86. doi: 10.1016/j.bios.2016.09.091
Kim, L. H., Yu, H.-W., Kim, Y.-H., Kim, I. S., and Jang, A. (2013). Potential of fluorophore labeled aptamers for Pseudomonas aeruginosa detection in drinking water. J. Korean Soc. Appl. Biol. Chem. 56, 165–171. doi: 10.1007/s13765-013-3019-7
Kowalski, R. P., Nayyar, S. V., Romanowski, E. G., Shanks, R. M. Q., Mammen, A., Dhaliwal, D. K., et al. (2020). The prevalence of bacteria, fungi, viruses, and acanthamoeba from 3,004 cases of keratitis, endophthalmitis, and conjunctivitis. Eye Contact Lens 46, 265–268. doi: 10.1097/ICL.0000000000000642
Lewis, K. (2001). Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45, 999–1007. doi: 10.1128/AAC.45.4.999-1007.2001
Li, Z., Mohamed, M. A., Vinu Mohan, A. M., Zhu, Z., Sharma, V., Mishra, G. K., et al. (2019). Application of electrochemical aptasensors toward clinical diagnostics, food, and environmental monitoring: review. Sensors 19:5435. doi: 10.3390/s19245435
Liu, D. J., Hu, B., Peng, D. F., Lu, S., Gao, S. X., Li, Z. G., et al. (2020). Isolation ssDNA aptamers specific for both live and viable but nonculturable state Vibrio vulnificus using whole bacteria-SEILEX technology. RSC Adv. 10, 15997–16008. doi: 10.1039/c9ra10733a
Luo, F., Li, Z., Dai, G., Lu, Y., He, P., and Wang, Q. (2020). Simultaneous detection of different bacteria by microchip electrophoresis combined with universal primer-duplex polymerase chain reaction. J. Chromatogr. A 1615:460734. doi: 10.1016/j.chroma.2019.460734
Maduraiveeran, G., Sasidharan, M., and Ganesan, V. (2018). Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 103, 113–129. doi: 10.1016/j.bios.2017.12.031
Mauch, R. M., and Levy, C. E. (2014). Serum antibodies to Pseudomonas aeruginosa in cystic fibrosis as a diagnostic tool: a systematic review. J. Cyst. Fibros. 13, 499–507. doi: 10.1016/j.jcf.2014.01.005
Morales, M. A., and Halpern, J. M. (2018). Guide to selecting a biorecognition element for biosensors. Bioconjug. Chem. 29, 3231–3239. doi: 10.1021/acs.bioconjchem.8b00592
Moutsiopoulou, A., Broyles, D., Dikici, E., Daunert, S., and Deo, S. K. (2019). Molecular aptamer beacons and their applications in sensing, imaging, and diagnostics. Small 15:e1902248. doi: 10.1002/smll.201902248
Nasseri, B., Soleimani, N., Rabiee, N., Kalbasi, A., Karimi, M., and Hamblin, M. R. (2018). Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 117, 112–128. doi: 10.1016/j.bios.2018.05.050
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., and Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Paniel, N., Baudart, J., Hayat, A., and Barthelmebs, L. (2013). Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 64, 229–240. doi: 10.1016/j.ymeth.2013.07.001
Paz-Zarza, V. M., Mangwani-Mordani, S., Martinez-Maldonado, A., Alvarez-Hernandez, D., Solano-Galvez, S. G., and Vazquez-Lopez, R. (2019). [Pseudomonas aeruginosa: pathogenicity and antimicrobial resistance in urinary tract infection]. Rev. Chilena Infectol. 36, 180–189. doi: 10.4067/S0716-10182019000200180
Perez, J. M., Josephson, L., O’Loughlin, T., Hogemann, D., and Weissleder, R. (2002). Magnetic relaxation switches capable of sensing molecular interactions. Nat. Biotechnol. 20, 816–820. doi: 10.1038/nbt720
Perry, J. D. (2017). A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin. Microbiol. Rev. 30, 449–479. doi: 10.1128/CMR.00097-16
Pla, L., Santiago-Felipe, S., Tormo-Mas, M. Á., Pemán, J., Sancenón, F., Aznar, E., et al. (2020). Aptamer-capped nanoporous anodic alumina for Staphylococcus aureus detection. Sens. Actuators B Chem. 320:128281. doi: 10.1016/j.snb.2020.128281
Prabowo, B. A., Purwidyantri, A., and Liu, K. C. (2018). Surface plasmon resonance optical sensor: a review on light source technology. Biosensors 8:80. doi: 10.3390/bios8030080
Radom, F., Jurek, P. M., Mazurek, M. P., Otlewski, J., and Jelen, F. (2013). Aptamers: molecules of great potential. Biotechnol. Adv. 31, 1260–1274. doi: 10.1016/j.biotechadv.2013.04.007
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., and Chapman, J. (2019). A review of methods for the detection of pathogenic microorganisms. Analyst 144, 396–411. doi: 10.1039/c8an01488d
Roushani, M., Sarabaegi, M., and Pourahmad, F. (2019). Impedimetric aptasensor for Pseudomonas aeruginosa by using a glassy carbon electrode modified with silver nanoparticles. Mikrochim. Acta 186:725. doi: 10.1007/s00604-019-3858-y
Sarabaegi, M., and Roushani, M. (2019). A nano-sized chitosan particle based electrochemical aptasensor for sensitive detection of P. aeruginosa. Anal. Methods 11, 5591–5597. doi: 10.1039/c9ay01509d
Shahrokhian, S., and Ranjbar, S. (2019). Development of a sensitive diagnostic device based on zeolitic imidazolate frameworks-8 using ferrocene–graphene oxide as electroactive indicator for Pseudomonas aeruginosa detection. ACS Sustain. Chem. Eng. 7, 12760–12769. doi: 10.1021/acssuschemeng.9b01314
Shi, X., Zhang, J., and He, F. (2019). A new aptamer/polyadenylated DNA interdigitated gold electrode piezoelectric sensor for rapid detection of Pseudomonas aeruginosa. Biosens. Bioelectron. 132, 224–229. doi: 10.1016/j.bios.2019.02.053
Shrivastava, S., Lee, W. I., and Lee, N. E. (2018). Culture-free, highly sensitive, quantitative detection of bacteria from minimally processed samples using fluorescence imaging by smartphone. Biosens. Bioelectron. 109, 90–97. doi: 10.1016/j.bios.2018.03.006
Silva, N. F. D., Magalhaes, J., Freire, C., and Delerue-Matos, C. (2018). Electrochemical biosensors for Salmonella: state of the art and challenges in food safety assessment. Biosens. Bioelectron. 99, 667–682. doi: 10.1016/j.bios.2017.08.019
Soundy, J., and Day, D. (2017). Selection of DNA aptamers specific for live Pseudomonas aeruginosa. PLoS One 12:e0185385. doi: 10.1371/journal.pone.0185385
Tan, W., Li, L., Xu, S., Yan, H., Li, X., Yazd, H. S., et al. (2020). Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angew. Chem. Int. Ed. Engl. doi: 10.1002/anie.202003563
CrossRef Full Text Online ahead of print | PubMed Abstract | Google Scholar
Tuerk, C., and Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. doi: 10.1126/science.2200121
Tummler, B. (2019). Emerging therapies against infections with Pseudomonas aeruginosa. F1000Res 8:F1000FacultyRev-1371. doi: 10.12688/f1000research.19509.1
Unser, S., Bruzas, I., He, J., and Sagle, L. (2015). Localized surface plasmon resonance biosensing: current challenges and approaches. Sensors 15, 15684–15716. doi: 10.3390/s150715684
Varadi, L., Luo, J. L., Hibbs, D. E., Perry, J. D., Anderson, R. J., Orenga, S., et al. (2017). Methods for the detection and identification of pathogenic bacteria: past, present, and future. Chem. Soc. Rev. 46, 4818–4832. doi: 10.1039/c6cs00693k
Wang, C. H., Wu, J. J., and Lee, G. B. (2019). Screening of highly-specific aptamers and their applications in paper-based microfluidic chips for rapid diagnosis of multiple bacteria. Sens. Actuators B Chem. 284, 395–402. doi: 10.1016/j.snb.2018.12.112
Wang, T., Chen, C., Larcher, L. M., Barrero, R. A., and Veedu, R. N. (2019). Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 37, 28–50. doi: 10.1016/j.biotechadv.2018.11.001
Wang, H., Chi, Z., Cong, Y., Wang, Z., Jiang, F., Geng, J., et al. (2018). Development of a fluorescence assay for highly sensitive detection of Pseudomonas aeruginosa based on an aptamer-carbon dots/graphene oxide system. RSC Adv. 8, 32454–32460. doi: 10.1039/c8ra04819c
Wang, K. Y., Zeng, Y. L., Yang, X. Y., Li, W. B., and Lan, X. P. (2011). Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 30, 273–278. doi: 10.1007/s10096-010-1074-0
Wang, P. Y., Wang, A. C., Hassan, M. M., Ouyang, Q., Li, H. H., and Chen, Q. S. (2020). A highly sensitive upconversion nanoparticles-WS2 nanosheet sensing platform for Escherichia coli detection. Sens. Actuators B Chem. 320:128434. doi: 10.1016/J.Snb.2020.128434
Wang, S. B., Zhang, Y., An, W. T., Wei, Y. L., Wang, Y., and Shuang, S. M. (2015). [Fluorescent and magnetic relaxation switch immunosensor for the detecting foodborne pathogen Salmonella enterica in water samples]. Guang Pu Xue Yu Guang Pu Fen Xi 35, 3105–3110.
Winstanley, C., O’Brien, S., and Brockhurst, M. A. (2016). Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 24, 327–337. doi: 10.1016/j.tim.2016.01.008
Wu, Z., He, D., Cui, B., and Jin, Z. (2018). A bimodal (SERS and colorimetric) aptasensor for the detection of Pseudomonas aeruginosa. Mikrochim. Acta 185:528. doi: 10.1007/s00604-018-3073-2
Xu, K., Zhou, R., Takei, K., and Hong, M. (2019). Toward flexible surface-enhanced raman scattering (SERS) sensors for point-of-care diagnostics. Adv. Sci. 6:1900925. doi: 10.1002/advs.201900925
Yan, J., Xiong, H., Cai, S., Wen, N., He, Q., Liu, Y., et al. (2019). Advances in aptamer screening technologies. Talanta 200, 124–144. doi: 10.1016/j.talanta.2019.03.015
Yao, J., Yang, M., and Duan, Y. (2014). Chemistry, biology, and medicine of fluorescent nanomaterials and related systems: new insights into biosensing, bioimaging, genomics, diagnostics, and therapy. Chem. Rev. 114, 6130–6178. doi: 10.1021/cr200359p
Yoo, S. M., Kim, D. K., and Lee, S. Y. (2015). Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 132, 112–117. doi: 10.1016/j.talanta.2014.09.003
Yüce, M., Kurt, H., Hussain, B., Ow-Yang, C. W., and Budak, H. (2018). Exploiting stokes and anti-stokes type emission profiles of aptamer-functionalized luminescent nanoprobes for multiplex sensing applications. ChemistrySelect 3, 5814–5823. doi: 10.1002/slct.201801008
Zhang, L., and Tian, Y. (2018). Designing recognition molecules and tailoring functional surfaces for in vivo monitoring of small molecules in the brain. Acc. Chem. Res. 51, 688–696. doi: 10.1021/acs.accounts.7b00543
Zhang, X., Jiang, X., Hao, Z., and Qu, K. (2019a). Advances in online methods for monitoring microbial growth. Biosens. Bioelectron. 126, 433–447. doi: 10.1016/j.bios.2018.10.035
Zhang, X., Xie, G., Gou, D., Luo, P., Yao, Y., and Chen, H. (2019b). A novel enzyme-free electrochemical biosensor for rapid detection of Pseudomonas aeruginosa based on high catalytic Cu-ZrMOF and conductive Super P. Biosens. Bioelectron. 142:111486. doi: 10.1016/j.bios.2019.111486
Zhang, X., Wang, K., Liu, M., Zhang, X., Tao, L., Chen, Y., et al. (2015). Polymeric AIE-based nanoprobes for biomedical applications: recent advances and perspectives. Nanoscale 7, 11486–11508. doi: 10.1039/c5nr01444a
Zhong, Z., Gao, R., Chen, Q., and Jia, L. (2020). Dual-aptamers labeled polydopamine-polyethyleneimine copolymer dots assisted engineering a fluorescence biosensor for sensitive detection of Pseudomonas aeruginosa in food samples. Spectrochim. Acta A Mol. Biomol. Spectrosc. 224:117417. doi: 10.1016/j.saa.2019.117417
Zhong, Z., Gao, X., Gao, R., and Jia, L. (2018). Selective capture and sensitive fluorometric determination of Pseudomonas aeruginosa by using aptamer modified magnetic nanoparticles. Mikrochim. Acta 185:377. doi: 10.1007/s00604-018-2914-3
Zhou, J., Qi, Q., Wang, C., Qian, Y., Liu, G., Wang, Y., et al. (2019). Surface plasmon resonance (SPR) biosensors for food allergen detection in food matrices. Biosens. Bioelectron. 142:111449. doi: 10.1016/j.bios.2019.111449
Keywords: Pseudomonas aeruginosa, aptamer, molecular recognition, biosensor, detection
Citation: Zheng X, Gao S, Wu J and Hu X (2020) Recent Advances in Aptamer-Based Biosensors for Detection of Pseudomonas aeruginosa. Front. Microbiol. 11:605229. doi: 10.3389/fmicb.2020.605229
Received: 11 September 2020; Accepted: 03 December 2020;
Published: 22 December 2020.
Edited by:
Yang-Chun Yong, Jiangsu University, ChinaReviewed by:
Koshy Philip, University of Malaya, MalaysiaCopyright © 2020 Zheng, Gao, Wu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihong Wu, amlob25nd3VAZnVkYW4uZWR1LmNu; Xiaobo Hu, aHV4aWFvYm9AdmlwLnNpbmEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.