- 1Guangdong Provincial Key Laboratory for Plant Epigenetics, College of Life Science and Oceanography, Shenzhen University, Shenzhen, China
- 2Shenzhen Key Laboratory of Laser Engineering, College of Optoelectronic Engineering, Shenzhen University, Shenzhen, China
- 3Formerly, Department of Botany, Goa University, Taleigão, India
- 4No. 128/1J, Azad Co-Op Housing Society, Curca, India
- 5Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
A survey of bambusicolous fungi in Bijiashan Mountain Park, Shenzhen, Guangdong Province, China, revealed several Arthrinium-like taxa from dead sheaths, twigs, and clumps of Bambusa species. Phylogenetic relationships were investigated based on morphology and combined analyses of the internal transcribed spacer region (ITS), large subunit nuclear ribosomal DNA (LSU), beta tubulin (β-tubulin), and translation elongation factor 1-alpha (tef 1-α) gene sequences. Based on morphological characteristics and phylogenetic data, Arthrinium acutiapicum sp. nov. and Arthrinium pseudorasikravindrae sp. nov. are introduced herein with descriptions and illustrations. Additionally, two new locality records of Arthrinium bambusae and Arthrinium guizhouense are described and illustrated.
Introduction
Arthrinium Kunze is accommodated in Apiosporaceae, Xylariales, which is morphologically different from other xylariaceous genera by the presence of basauxic conidiophores and dark, aseptate, globose to lenticular conidia with a hyaline rim or germ slit (Minter, 1985; Petrini and Müller, 1986; Singh et al., 2012; Jiang et al., 2018; Pintos et al., 2019). Basauxic conidiophores simply mean elongation of conidiogenous cells from the basal growing point after formation of a single, terminal blastic conidium at its apex (Cole, 1986).
Arthrinium species are distributed worldwide in various hosts as endophytes, epiphytes, or saprobes, as well as plant pathogens on some commercial crops and ornamentals (Agut and Calvo, 2004; Ramos et al., 2010; Crous and Groenewald, 2013; Sharma et al., 2014; Senanayake et al., 2015; Wijayawardene et al., 2020). Also, species of Arthrinium (Schmidt and Kunze, 1817) inhabit a wide range of substrates, i.e., air, soil debris, lichens, marine algae (Agut and Calvo, 2004; Senanayake et al., 2015; Dai et al., 2016; Luo et al., 2019), and even human tissues (Sharma et al., 2014). The genus Arthrinium morphologically differs from other xylariaceous anamorphic genera by the presence of basauxic conidiogenous cells which arise from conidiophore mother cells (Schmidt and Kunze, 1817; Minter, 1985).
The commonly reported diseases by Arthrinium species are kernel blight of barley and brown culm streak of Phyllostachys praecox by A. arundinis, damping-off of wheat by A. sacchari, and culm rot of bamboos and Phyllostachys viridis by A. phaeospermum (Martínez-Cano et al., 1992; Mavragani et al., 2007; Chen et al., 2014; Li et al., 2016). Additionally, the causative agent of cutaneous infections of humans has been reported as A. phaeospermum (Rai, 1989; Zhao et al., 1990; De Hoog et al., 2000; Crous and Groenewald, 2013). Some Arthrinium species produce bioactive compounds with pharmacological and medicinal properties (Hong et al., 2015), while some produce industrially important enzymes (Shrestha et al., 2015). Crous and Groenewald (2013) reviewed the genus Arthrinium based on morphology and multigene phylogeny. There are several subsequent studies providing additions to the genus (Singh et al., 2012; Sharma et al., 2014; Crous et al., 2015; Senanayake et al., 2015; Dai et al., 2016, 2017; Hyde et al., 2016; Réblová et al., 2016; Jiang et al., 2018, 2020; Wang et al., 2018; Pintos et al., 2019).
Several studies revealed bambusicolous Arthrinium species from Poaceae and Cyperaceae host plants in China (Dai et al., 2016, 2017; Jiang et al., 2018, 2020; Wang et al., 2018), and several Arthrinium-like taxa on dead leaves, clumps, and twigs of bamboo were collected from Shenzhen (China) during this study. The aims of this study are identifying these Arthrinium-like taxa based on morphology and phylogeny and describe and illustrate them. Phylogenetic relationships were investigated based on DNA sequence data of the internal transcribed spacer region (ITS), large subunit nuclear ribosomal DNA (LSU), beta tubulin (β-tubulin), and translation elongation factor 1-alpha (tef 1-α), and two new Arthrinium species from bamboo are introduced as Arthrinium pseudorasikravindrae and A. acutiapicum and two locality records, Arthrinium bambusae and Arthrinium guizhouense, are described and illustrated.
Materials and Methods
Sample Collection and Fungal Isolation
Fresh specimens of Arthrinium-like taxa were collected from Bijiashan Mountain Park, Shenzhen, Guangdong Province, China, in September–October 2018, and the collection site has a tropical climate with abundant sunshine and rainfall all year round. The yearly average temperature is 22°C and vegetative type is tropical evergreen monsoon forests (Li et al., 2015). Specimens were brought to the laboratory in paper bags and they were examined under a stereomicroscope (Carl Zeiss Discovery V8), and blackish conidial mass and fruit bodies were observed. The fruit bodies were studied and photographed using a stereomicroscope fitted with a camera (ZEISS Axiocam 208). The micromorphological characters were studied and photographed using a compound microscope (Nikon Eclipse 80i) fitted with a digital camera (Canon 450D). All microscopic measurements such as the length of the conidiophores, conidiogenous cells, and conidia were made with Tarosoft image framework (v. 0.9.0.7).
Single conidial isolation was carried out following the method described by Senanayake et al. (2020a). Germinated conidia were aseptically transferred into fresh potato dextrose agar (PDA) plates, incubated at 20°C to obtain pure cultures, and later transferred to PDA slants and stored at 4°C for further study. Colony characteristics were recorded from cultures grown on PDA. Additionally, pure cultures were inoculated in 2% PDA together with sterilized bamboo leaves and incubated at 25°C for sporulation.
Fungarium materials are deposited in the Herbarium of Cryptogams, Kunming Institute of Botany, Academia Sinica (HKAS), and all the ex-type living cultures are deposited at the Culture Collection of Kunming Institute of Botany (KUMCC). Index Fungorum numbers1 were obtained for the new strains.
DNA Extraction, PCR Amplification, and Sequencing
Fresh fungal mycelium grown on PDA for 2 weeks at 20°C in the dark was used for DNA extraction using fungal genomic DNA extraction kit (Biospin DNA Extraction Kit, Bioer Technology, Co. Ltd., Hangzhou, China) following the manufacturer’s protocols. Polymerase chain reactions (PCR) and sequencing were carried out for the following loci: the complete ITS region with the primer pair of ITS1/ITS4 (White et al., 1990); the LSU ribosomal DNA, amplified and sequenced as a single fragment with primers LR0R/LR5 (Vilgalys and Hester, 1990); the tef 1-α gene with primers EF1-728F/EF2 (Carbone and Kohn, 1999; Rehner, 2001); and the β-tubulin gene with primers bt2a and bt2b (Glass and Donaldson, 1995).
The PCR amplification reactions were carried out with the following protocol. The total volume of the PCR reaction was 25 μl reaction volume containing 1 μl of DNA template, 1 μl of each forward and reverse primer, 12.5 μl of 2 × PCR Master Mix, and 9.5 μl of double-distilled sterilized water (ddH2O). The reaction was conducted by running for 35 cycles following the condition of Senanayake et al. (2020b). The PCR products were observed on 1% agarose electrophoresis gel stained with ethidium bromide. Purification and sequencing of PCR products were carried out at Sunbiotech Company, Beijing, China. Sequence quality was checked and sequences were condensed with DNASTAR Lasergene v.7.1. Sequences derived in this study were deposited in GenBank and accession numbers were obtained (Table 1).
Sequence Alignments and Phylogenetic Analyses
BLASTn searches were made using the newly generated sequences to assist in taxon sampling for phylogenetic analyses. Jiang et al. (2018, 2020), Wang et al. (2018), and Pintos et al. (2019) were followed to obtain sequences from GenBank that are listed in Table 1. The concatenated ITS, LSU, β-tubulin, and tef 1-α sequence dataset comprised 101 strains of Arthrinium, while the outgroup taxon was Pestalotiopsis chamaeropis (CBS 237.38). DNA sequences of the ITS, LSU, β-tubulin, and tef 1-α were aligned using the online version of MAFFT v. 7.0362 (Katoh et al., 2020) with default settings and manually adjusted using BioEdit 7.1.3 (Hall, 1999) to allow maximum alignment and minimum gaps. Further, single gene alignments were combined to obtain the final multiloci alignment that was containing 2,817 nucleotide characters, viz. 681 of ITS, 875 of LSU, 434 of β-tubulin, and 827 of tef 1-α. Both single and concatenated alignments were used for the analyses.
Maximum likelihood analyses were performed by RAxML (Stamatakis and Alachiotis, 2010) implemented in raxmlGUIv.1.5 (Silvestro and Michalak, 2012) using the ML + rapid bootstrap setting and the GTR + I + G model of nucleotide substitution with 1,000 replicates. The matrix was partitioned for the different gene regions included in the combined multilocus analyses.
For the Bayesian inference (BI) analyses, the optimal substitution model for the combined dataset was determined to be GTR + I + G using the MrModeltest software v. 2.2 (Nylander, 2004). The BI analyses was computed with MrBayes v. 3.2.6 (Ronquist et al., 2012) with four simultaneous Markov chain Monte Carlo chains from random trees over 10 M generations (trees were sampled every 500th generation).
The distribution of log-likelihood scores was observed to check whether sampling is in stationary phase or not, and Tracer v1.5 was used to check if further runs were required to reach convergence or not (Rambaut and Drummond, 2007). The Bayesian analyses lasted until the average standard deviation of split frequencies has a value less than 0.01, and the consensus tree and posterior probabilities were calculated after discarding the first 20% of the sampled trees as burn-in. The phylogram was visualized in FigTree v. 1.4 (Rambaut, 2009). All the phylogenetic trees derived from this study were deposited in TreeBase3 under accession number S27147.
Results
Phylogenetic Inferences
All individual trees generated under different criteria and from single gene datasets were essentially similar in topology and not significantly different from the tree generated from the concatenated dataset (not discussed herein). Additionally, this tree topology is similar to previous studies on Arthrinium (Dai et al., 2016; Jiang et al., 2018, 2020; Wang et al., 2018; Pintos et al., 2019).
Maximum likelihood analysis of Arthrinium species in this study with 1,000 bootstrap replicates yielded the best ML tree (Figure 1) with the likelihood value of –29,933.362493 and the following model parameters: estimated base frequencies—A = 0.239654, C = 0.250345, G = 0.255054, and T = 0.254948; substitution rates—AC = 1.275584, AG = 2.530572, AT = 1.397969, CG = 1.184045, CT = 4.063803, and GT = 1.0; proportion of invariable sites—I = 0.203121; gamma distribution shape parameter—α = 0.54383. The alignment contained a total of 1,756 distinct alignment patterns and 28.72% of undetermined characters. After discarding the first 20% of generations, 36,000 trees remained from which 50% consensus trees and posterior probabilities (PP) were calculated (Figure 1). Maximum likelihood bootstrap values ≥ 60% and BI ≥ 0.95 are given at each node. Tree topologies of the ML and Bayesian analyses were similar to each other and there are no significant differences.
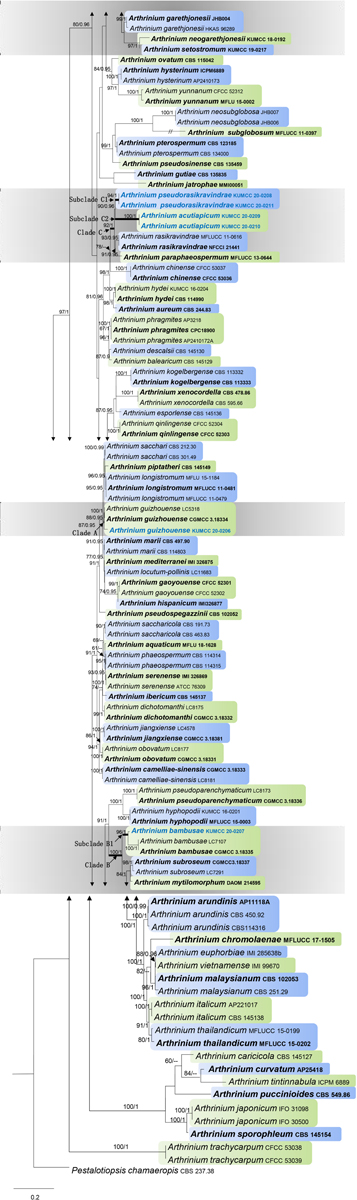
Figure 1. Phylogram generated from maximum likelihood analysis based on combined internal transcribed spacer region (ITS), large subunit nuclear ribosomal DNA (LSU), β-tubulin, and tef 1-α sequence data. Bootstrap support values greater than 50% and Bayesian posterior probabilities greater than 0.90 are given at the nodes. The tree is rooted with Pestalotiopsis chamaeropis (CBS 237.38). Ex-type strains are in bold and the newly obtained sequences are indicated in blue bold.
There are 101 Arthrinium strains in this study together with a new isolate that is introduced here. All the ex-type strains of Arthrinium species were included if available, and other authentic strains were selected when sequences from ex-type strains are unavailable. Our new isolate KUMCC 20-0206 clustered with the type strain of A. guizhouense (CGMCC 3.18334) and another representative strain (LC5318) with 87% ML and 0.95 PP support. This clade (clade A) has a close phylogenetic affinity to Arthrinium longistromum, A. piptatheri, and A. sacchari with 95% ML and 0.95 PP support. Two strains of A. bambusae (CGMCC 3.18335 and LC7107) and the new isolate KUMCC 20-0207 were grouped in a separate clade with 96% ML and 1.00 PP support. This clade (subclade B1) shares a monophyletic relationship to Arthrinium garethgonesii, A. mytilomotphum, A. neogarethjonesii, A. setostromun, and A. subroseum with strong bootstrap supports (100% ML, 1.00 PP, clade B, Figure 1). Two new isolates, KUMCC 20-0208 and KUMCC 20-0211, were monophyletic in subclade C1 (Figure 1) with 90% ML and 0.96 PP support. Subclade C2 is also monophyletic with two novel strains, viz. KUMCC 20–0209 and KUMCC 20–0210, which are sisters to subclade C1 with 90% ML and 0.96 PP support (clade C, Figure 1). With these four new strains, clade C shares a close phylogenetic affiliation to A. paraphaeospermum and A. rasikravindrae.
Taxonomy
Arthrinium acutiapicum Senan. and Cheew. sp. nov. Figure 2

Figure 2. Arthrinium acutiapicum (HKAS 107673). (A) Host. (B) Fungarium specimen. (C) Conidiomata on substrate. (D) Surface view of culture on potato dextrose agar (PDA). (E) Reverse view of culture on PDA. (F) Conidia and conidiogenous cells. (G,H) Conidia. Scale bars: (C) = 500 μm, (F–H) = 10 μm.
Index Fungorum number: IF557868
Etymology: Species epithet “acuti” refers to pointed and “apicum” refers to apex of conidiogenous cells.
Holotype: HKAS 107673
Saprobic on dead twigs of Bambusa bambos (L.) Voss. Hyphae 1.5–2.5 μm diam., hyaline, branched, septate, sparse. Sexual morph: undetermined. Asexual morph: Conidiomata 350–450 μm, pycnidial, immersed, aggregated, scattered, subglobose, ostiolate, black, coriaceous. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–7 × 2–3 μm ( = 6.3 × 2.1 μm, n = 30), holoblastic, develop from conidiophore mother cells, erect, basauxic, cylindrical to ampulliform, apex pointed and hyaline, smooth and thick-walled, pale brown. Conidia 7.5–10 × 8.5–12 μm ( = 9.3 × 11.9 μm, n = 30), globose in surface view, subglobose to oval in side view, apex and base blunted, smooth-walled, brown to dark brown, with a dark equatorial slit.
Culture Characteristics
Colonies grew on PDA at 20°C in the dark attenuated 2 cm diam., within 7 days, flat, circular, entire margin, wooly, with abundant aerial mycelia, white in surface view and off-white to yellow in reverse. Sporulation occurred after 10 days on PDA incubated at 20°C in the dark without any host substrate. Conidia seem black mass and well spread on culture.
Specimen Examined
China, Guangdong Province, Shenzhen City, Futian District, northwest of Futian, Bijiashan Park, on dead twigs of B. bambos (L.) Voss (Poaceae), 23 September 2018, IS, SI 86 (HKAS 107673, holotype), ex-type culture, KUMCC 20-0210; ibid 15 October 2018, IS, SI 86-1 (HKAS 107674, paratype), ex-paratype culture KUMCC 20-0211.
Notes
Arthrinium acutiapicum forms a distinct subclade (subclade C2, Figure 1) with strong bootstrap support values (ML/PP = 90/0.96) in our phylogenetic analysis, which is a sister to the newly introduced species A. pseudorasikravindrae. Additionally, A. acutiapicum shows close phylogenetic affinities to A. paraphaeospermum, A. pseudorasikravindrae, and A. rasikravindrae in clade C (Figure 1) and A. chinense. Blast results of ITS, LSU, β-tubulin, and tef 1-α sequences of A. acutiapicum show high similarity to A. hydei, A. paraphaeospermum, and A. rasikravindrae. Morphologically, A. acutiapicum is distinct from A. pseudorasikravindrae by the reduction of conidiophores to conidiogenous cells, cylindrical to ampulliform, pale brown conidiogenous cells with pointed, hyaline apex and brown to dark brown, smooth-walled conidia with dark equatorial slit. Additionally, A. acutiapicum is distinct from A. rasikravindrae by the reduction of conidiophores to pale brown conidiogenous cells and dimorphous, acropleurogenously arising conidia.
Arthrinium bambusae M. Wang and L. Cai, in Wang et al. (2018) Figure 3
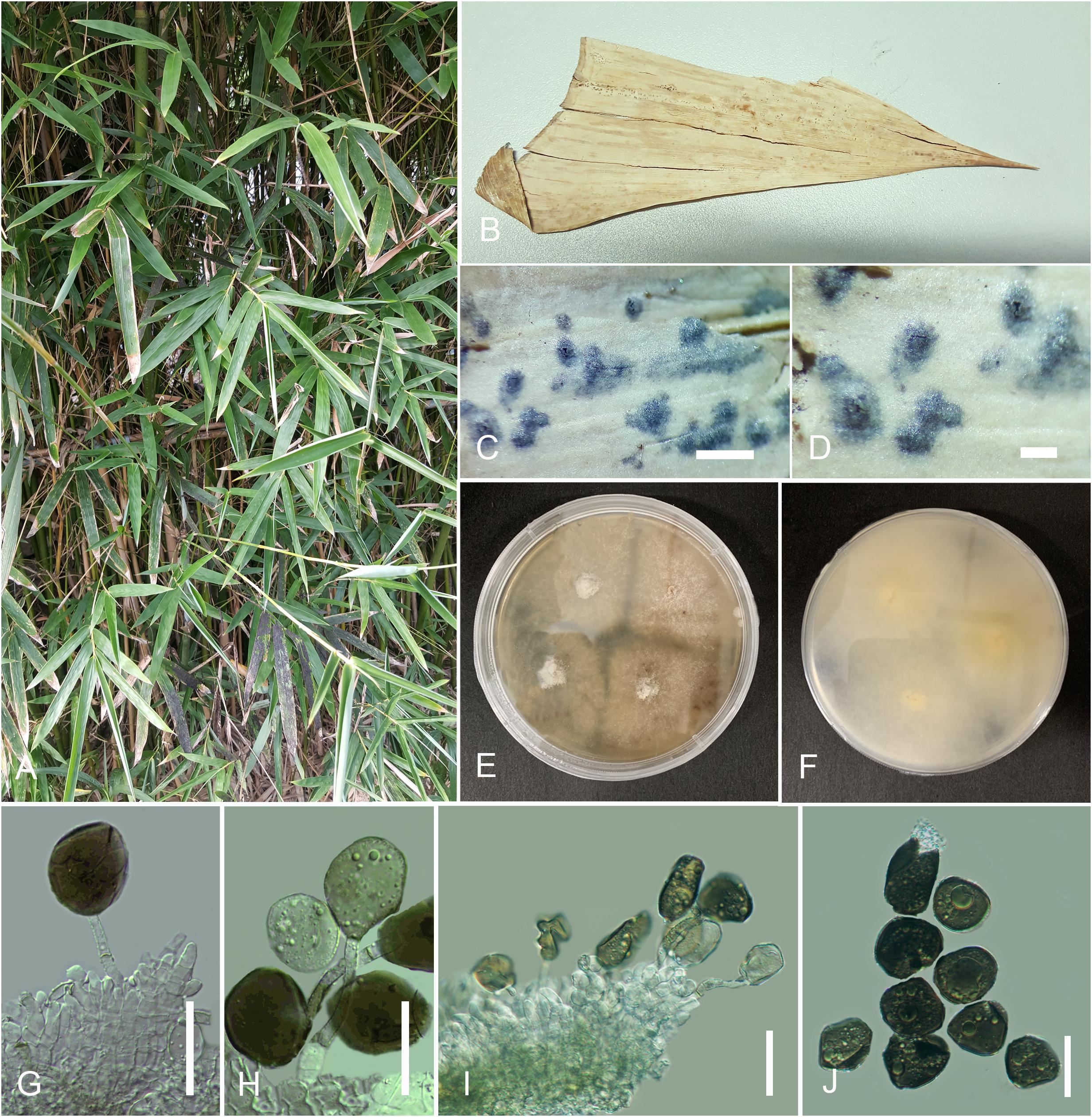
Figure 3. Arthrinium bambusae (HKAS 107671). (A) Host. (B) Fungarium specimen. (C,D) Conidiomata on substrate. (E,F) Culture on potato dextrose agar (PDA; surface and reverse view). (G–I) Conidia and conidiogenous cells. (J) Conidia. Scale bars: (C,D) = 100 μm, (G–J) = 15 μm.
Index Fungorum number: IF 824906
Saprobic on sheath of Bambusa dolichoclada Hayata. Hyphae 1–3 μm diam., hyaline, branched, septate, sparse. Sexual morph: undetermined. Asexual morph: appears as black, spotty patches on host surface. Conidiomata immersed, pycnidial, scattered, globose to slightly conical, ostiolate, black, coriaceous. Conidiomatal wall thin, comprising several layers of black, large cells of textura angularis. Conidiophores reduced to conidiogenous cells or rarely inconspicuous with cylindrical, thick-walled, hyaline. Conidiogenous cells 5–12 × 3–10 μm ( = 8.6 × 4 μm, n = 30), basauxic, holoblastic, develop from conidiophore mother cells, with periclinal thickening, doliiform to ampulliform or lageniform, erect, aggregated in clusters on hyphae, hyaline to pale brown, smooth. Conidia 10.5–17.5 × 8–15 μm ( = 15 × 12 μm, n = 30), subglobose to ellipsoid, guttulate, smooth to finely roughened, olivaceous to dark brown.
Culture Characteristics
Colonies grew on PDA at 20°C in the dark attenuated 2 cm diam., within 7 days, flat, spreading, margin circular, with abundant aerial mycelia, surface and reverse white to off-white.
Specimen Examined
China, Guangdong Province, Shenzhen City, Futian District, northwest of Futian, Bijiashan Park, on sheath of B. dolichoclada Hayata (Poaceae), 23 September 2018, IS, SI 80 (HKAS 107671), living culture, KUMCC 20-0207.
Notes
Arthrinium bambusae was introduced by Wang et al. (2018) from Guangdong Province, China, where our collection also was obtained. However, the exact locality is not mentioned in the original description there. The morphology of our collection was obtained from fungal structures on the host specimen, while Wang et al. (2018) had described the fungus from sporulated cultures. However, the morphology of our collection is similar to the holotype. Phylogenetically, A. bambusae clusters with A. garethjonesii, A. neogarethjonesii, A. mytilomorphum, A. setostromum, and A. subroseum with strong bootstrap value (ML/PP = 100/1), and the A. bambusae isolate (KUMCC 20-0207) clustered well with the ex-type culture (ML/PP = 96/1).
Arthrinium guizhouense M. Wang and L. Cai, in Wang et al. (2018)Figure 4

Figure 4. Arthrinium guizhouense (HKAS 107672). (A) Host. (B) Fungarium specimen. (C) Conidiomata on substrate. (D) Surface view of culture on potato dextrose agar (PDA). (E) Reverse view of culture on PDA. (F–H) Conidia and conidiogenous cells. (I) Conidia. Scale bars: (C) = 1,000 μm, (F–I) = 5 μm.
Index Fungorum number: IF824909
Saprobic on dead twigs of Bambusa multiplex (Lour.) Raeusch. ex Schult. f., appear as black mass coming out from ruptured bark. Hyphae hyaline, branched, septate, 1.5–6 μm diam. Sexual morph: undetermined. Asexual morph: Conidiomata 450–350 μm, picnidial, semi-immersed, aggregated, scattered, globose, ostiolate, black, coriaceous. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–8 × 3–5 μm ( = 5.3 × 4 μm, n = 30), develop from conidiophore mother cells, erect, subglobose, basauxic, ampulliform or doliiform, hyaline to pale brown, smooth. Conidia 5–7 × 4–7 μm ( = 6.3 × 5.7 μm, n = 30), globose or subglobose, smooth to finely roughened, dark brown to black, with a longitudinal, hyaline, thin, germ slit.
Culture Characteristics
Colonies grew on PDA at 20°C in the dark attenuated 2 cm diam., within 5 days, flat, wooly, margin circular, with slight aerial mycelia, surface initially white, becoming grayish white and reverse yellowish white.
Specimen Examined
China, Guangdong Province, Shenzhen City, Futian District, northwest of Futian, Bijiashan Park, on twigs of B. multiplex (Lour.) Raeusch. ex Schult. f. (Poaceae), 23 September 2018, I.C. Senanayake, SI 84 (HKAS 107672), living culture, KUMCC 20-0206.
Notes
NCBI blast result for β-tubulin sequences of this isolate gives high sequence similarities to A. guizhouense (99.55%), A. sacchari (98.25%), A. arundinis (98.25%), and A. marii (95.62%) while A. guizhouense (93.41%) and A. marii (92.94%) for tef 1-α. Additionally, high blast similarities for ITS loci are A. marii (99.54%), A. sacchari (99.22%), A. phaeospermum (99.20%), A. pseudospegazzinii (98.13%), A. longistromum (98.72%), and A. guizhouense (99.83%) while A. marii (100%), A. sacchari (100%), A. guizhouense (100%), and Apiospora montagnei (100%) for LSU. In the phylogenetic analysis, this isolate (KUMCC 20-0206) clusters with the ex-holotype strain of A. guizhouense (CGMCC3.18334) with moderate support value (ML/PP = 87/0.95). Morphologically, this collection is closely similar to the holotype specimen of A. guizhouense having brown to black, smooth to finely roughened, globose or subglobose, conidia with pale brown, subglobose, ampulliform or doliiform conidiogenous cells. However, the holotype of A. guizhouense has been collected from the air in karst cave in Guizhou Province, China, and this collection was obtained from bamboo twigs in Guangdong Province. Hence, HKAS 107672 is identified as A. guizhouense based on morphology and phylogeny. This is the first record of A. guizhouense in Guangdong Province and on bamboo.
Arthrinium pseudorasikravindrae Senan., and Cheew. sp. nov. Figure 5

Figure 5. Arthrinium pseudorasikravindrae (HKAS 107669). (A) Host. (B) Fungarium specimen. (C) Surface view of culture on potato dextrose agar (PDA). (D) Reverse view of culture on PDA. (E) Conidial mass on cultures. (F–J) Conidia, conidiogenous cells, and conidiophores. (K–N) Conidia (concentric pale rings are arrowed in I). Scale bars: (F–N) = 10 μm.
Index Fungorum number: IF 557870
Etymology: Species epithet the morphological similarity of this collection to Arthrinium rasikravindrae.
Holotype: HKAS 107669
Saprobic on sheaths of B. dolichoclada Hayata. Mycelium 1.5–3 μm in diam., consisting of smooth, hyaline, septate, branched, hyphae. Sexual morph: undetermined. Asexual morph: Conidiophores 10–15 × 3–7 μm ( = 12.3 × 5.2 μm, n = 30), basauxic, straight or flexuous, cylindrical, hyaline, thick, smooth-walled, aseptate. Conidiogenous cells 4–10 × 1.2–5 μm ( = 8.6 × 4.2 μm, n = 30), holoblastic, develop from conidiophore mother cells, ampulliform, cylindrical or doliiform, hyaline to olivaceous. Conidia 5–10 × 5.5–11 μm ( = 9.3 × 10.1 μm, n = 30), globose in face view, lenticular in side view, with a pale longitudinal slit, dark brown, thick-walled, finely roughened with one or two concentric pale rings.
Culture Characteristics
Colonies grew on PDA at 20°C in the dark attenuated 2 cm diam., within 5 days, flat, spreading, circular, margin filiform with abundant aerial mycelia, surface white to off-white and reverse pale yellow, sporulation occurs on 2% PDA incubated at 25°C after 2 weeks, black, conidial mass concentrated at colony margins. Sporulation occurred after 10 days on PDA incubated at 20°C in the dark without any host substrate. Conidia seem black mass and spread mostly in colony margins.
Specimen Examined
China, Guangdong Province, Shenzhen City, Futian District, northwest of Futian, Bijiashan Park, on sheath of B. dolichoclada Hayata (Poaceae), 23 September 2018, IS, SI 73 (HKAS 107669, holotype), ex-type culture, KUMCC 20-0208; ibid October 15, 2018, IS, SI 73-1 (HKAS 107670, paratype), ex-paratype culture KUMCC 20-0211.
Notes
Blast results of ITS, LSU, β-tubulin, and tef 1-α sequences of A. pseudorasikravindrae (KUMCC 20-0208, KUMCC 20-0211) show high similarity to A. hydei, A. paraphaeospermum, and A. rasikravindrae. In our phylogenetic analysis, A. pseudorasikravindrae forms a subclade (subclade C1, Figure 1) with strong bootstrap support values (ML/PP = 90/0.96), which is a sister to the newly introduced species A. acutiapicum. Additionally, A. pseudorasikravindrae shows close phylogenetic affinities to A. chinense, A. paraphaeospermum, and A. rasikravindrae (Figure 1).
Arthrinium pseudorasikravindrae is morphologically distinct from the above species (Table 2) by its thick-walled, finely roughened conidia with pale, equatorial slit and ampulliform, cylindrical or doliiform, basauxic conidiogenous cells. The morphology of A. pseudorasikravindrae is compared with other closely related species (Table 2). Therefore, considering morphological and molecular uniqueness, these isolates are introduced here as belonging to a new species, A. pseudorasikravindrae. HKAS 107669 and HKAS 107670 represent a distinct clade (clade A, Figure 1) which not known before in the phylogenetic analysis, and hence, these collections are introduced here as a new species based on their morphology and phylogeny.

Table 2. Morphological comparisons of species which are phylogenetically closely related to Arthrinium acutiapicum and Arthrinium pseudorasikravindrae.
Discussion
Bamboo is an important group of flowering plants that helps to conserve and manage forest ecosystems and reduce soil erosion and it is also important for panda conservation and many more commercial applications such as making fishing rod, flute, paper, flooring material, etc. and as food for humans and livestock (Chapman and Peat, 1992). Members of bamboo belong to the family Poaceae comprising more than 115 genera with approximately 1,450 species (Gratani et al., 2008; Kelchner and Group, 2013), and bamboo occurs in all tropical, subtropical, and temperate regions as herbaceous or woody plants. Microfungi associate with bamboo in many ways and phytopathogenic or endophytic microfungi form diseases while saprobic microfungi help to decompose plant debris (Zhang and Wang, 1999; Hyde et al., 2002a,b).
The first monograph on bambusicolous fungi was published with 258 fungal species by Hino and Katumoto (1960), and 63 new species were introduced by Petrini et al. (1989). Eriksson and Yue (1998) provided a checklist of the ascomycetes on bamboo, while Zhang and Wang (1999) recorded 213 species described from bamboo in China. Kuai (1996) listed phytopathogenic bambusicolous fungi in China and Taiwan. Hyde et al. (2002a) reviewed bambusicolous fungi that grow on all bamboo substrates including the leaves, culms, branches, rhizomes, and roots and enlisted more than 1,100 species, which belong to 228 genera. Dai et al. (2018) have reviewed the taxonomy of bambusicolous fungi. This study is one of the articles in the series on bambusicolous microfungi in Guangdong Province. Herein, we collected Arthrinium-like taxa from bamboo plant samples from Shenzhen, Guangdong Province, China. Currently, there are 81 species in the Arthrinium (Species Fungorum, 2020) and only 61 have molecular data. More than 30% of holotypes of Arthrinium species have been collected in China (Table 1). Therefore, the aims of this paper were to study Arthrinium-like fungi in Guangdong Province and to introduce several putative new species by comparing them morphologically and genetically with existing taxa.
According to morphology and phylogeny, two novel Arthrinium species were obtained with two new locality records. Most phylogenetic studies on Arthrinium used ITS, β-tubulin, and tef 1-α; however, LSU has been added to the analyses here. Negligible variations occur in tree topology in spite of adding LSU. A. guizhouense (HKAS 107672) is the first record in Guangdong Province and also from bamboo. The holotype of A. guizhouense was collected from the air in kart caves in Guizhou Province, China (Wang et al., 2018). This suggests that fungal conidioma in plant hosts release the conidia and conidia can survive in the air for a sufficiently long time. Our strain of A. bambusae is identical to the holotype which was collected from Guangdong Province on bamboo (Wang et al., 2018). Hence, this specimen can be used as an epitype if the holotype cannot be used for taxonomic purpose. The morphological differences between these two Arthrinium species are listed in Table 2. However, the life mode, host, and colony characters of these two species are not significantly different.
Data Availability Statement
The datasets generated in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
IS designed the study, performed the morphological study and phylogenetic analyses, and wrote the manuscript. JB, NX, and RC reviewed and edited the manuscript. All authors approved the final manuscript.
Funding
This study was funded by Chiang Mai University postdoctoral research fund, Science and technology Project of Shenzhen City, Shenzhen Bureau of Science, Technology and Information (JCYJ20180305123659726), and National Natural Science Foundation of China (No. 31601014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the kind support given by the laboratory staff of the College of Life Science and Oceanography, ShenzhenUniversity, Shenzhen, China, for the support in doing molecularwork. IS thanks Chiang Mai University and NX thanks the National Natural Science Foundation of China for funding this research. Our special thanks go to staff members of the Herbarium of Cryptogams, Kunming Institute of Botany, Academia Sinica (HKAS) and the Culture Collection of Kunming Institute of Botany (KUMCC) for depositing all the authentic cultures and fungarium specimens. IS thanks Dr. Shaun Pennycook for proposing Latin names of the new species.
Footnotes
- ^ http://www.indexfungorum.org
- ^ http://mafft.cbrc.jp/alignment/server/large.html
- ^ www.treebase.org
References
Agut, M., and Calvo, M. A. (2004). In vitro conidial germination in Arthrinium aureum and Arthrinium phaeospermum. Mycopathologia 157, 363–367. doi: 10.1023/B:MYCO.0000030432.08860.f3
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Chapman, G. P., and Peat, W. E. (1992). An Introduction to Grasses (including Bamboos and Cereals). London: Redwood Press Ltd.
Chen, K., Wu, X. Q., Huang, M. X., and Han, Y. Y. (2014). First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 98, 1274. doi: 10.1094/PDIS-02-14-0165-PDN
Cole, G. T. (1986). Models of cell differentiation in conidial fungi. Microbiol. Rev. 50, 95–132. doi: 10.1128/MMBR.50.2.95-132.1986
Crous, P. W., and Groenewald, J. Z. (2013). A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4, 133–154. doi: 10.5598/imafungus.2013.04.01.13
Crous, P. W., Wingfield, M. J., Le Roux, J. J., Richardson, D. M., Strasberg, D., Shivas, R. G., et al. (2015). Fungal planet description sheets: 371-399. Persoonia 35, 264–327. doi: 10.3767/003158515X690269
Dai, D. Q., Jiang, H. B., Tang, L. Z., and Bhat, D. J. (2016). Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7, 1332–1345. doi: 10.5943/mycosphere/7/9/7
Dai, D. Q., Phookamsak, R., Wijayawardene, N. N., Li, W. J., Bhat, D. J., Xu, J. C., et al. (2017). Bambusicolous fungi. Fungal Divers. 82, 1–105. doi: 10.1007/s13225-016-0367-8
Dai, D. Q., Tang, L. Z., and Wang, H. B. (2018). “A review of bambusicolous ascomycetes,” in Bamboo: Current and Future Prospects, ed. H. P. S., Abdul Khalil (IntechOpen), 210.
De Hoog, G. S., Guarro, J., Gene, J., and Figueras, M. J. (2000). Atlas of Clinical Fungi, 2nd Edn, Utrecht: Centraalbureau voor Schimmelcultures.
Eriksson, O., and Yue, J. Z. (1998). Bambusicolous pyrenomycetes, an annotated check-list. Myconet 1, 25–78.
Glass, N. L., and Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/AEM.61.4.1323-1330.1995
Gratani, L., Crescente, M. F., Varone, L., Fabrini, G., and Digiulio, E. (2008). Growth pattern and photosynthetic activity of different bamboo species growing in the Botanical Garden of Rome. Flora Morphol. Distrib. Funct. Ecol. Plants 203, 77–84. doi: 10.1016/j.flora.2007.11.002
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hino, I., and Katumoto, K. (1960). Icones Fungorum Bambusicolorum Japonicorum. Kobe: The Fuji Bamboo Garden.
Hong, J. H., Jang, S., Heo, Y. M., and Min, M. (2015). Investigation of marine-derived fungal diversity and their exploitable biological activities. Mar. Drugs 13, 4137–4155. doi: 10.3390/md13074137
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Jones, E. B. G., et al. (2016). Fungal diversity notes 367-490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Hyde, K. D., Zhou, D., and Dalisayl, T. (2002a). Bambusicolous fungi: a review. Fungal Divers. 9, 1–14.
Hyde, K. D., Zhou, D., McKenzie, E., Ho, W., and Dalisay, T. (2002b). Vertical distribution of saprobic fungi on bamboo culms. Fungal Divers. 11, 109–118.
Jiang, N., Li, J., and Tian, C. M. (2018). Arthrinium species associated with bamboo and reed plants in China. Fungal Syst. Evol. 2, 1–9. doi: 10.3114/fuse.2018.02.01
Jiang, N., Liang, Y. M., and Tian, C. M. (2020). A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia 72, 77–83.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2020). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kelchner, S. A., and Group, B. P. (2013). Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 67, 404–413. doi: 10.1016/j.ympev.2013.02.005
Kuai, S. Y. (1996). A checklist of pathogenic Bambusicolous fungi of mainland China and Taiwan. J. For. Sci. Technol. 4, 64–71.
Li, B. J., Liu, P. Q., Jiang, Y., Weng, Q. Y., and Chen, Q. H. (2016). First report of culm rot caused by Arthrinium phaeospermum on Phyllostachys viridis in China. Plant Dis. 100, 1013–1013. doi: 10.1094/PDIS-08-15-0901-PDN
Li, L., Chan, P. W., Wang, D., and Tan, M. (2015). Rapid urbanization effect on local climate: intercomparison of climate trends in Shenzhen and Hong Kong, 1968-2013. Clim. Res. 63, 145–155. doi: 10.3354/cr01293
Luo, Z.-L., Hyde, K. D., Liu, J.-K., Maharachchikumbura, S. S. N., Jeewon, R., Bao, D.-F., et al. (2019). Freshwater sordariomycetes. Fungal Divers. 99, 451–660. doi: 10.1007/s13225-019-00438-1
Martínez-Cano, C., Grey, W. E., and Sands, D. C. (1992). First report of Arthrinium arundinis causing kernel blight on barley. Plant Dis. 76:1077. doi: 10.1094/PD-76-1077B
Mavragani, D. C., Abdellatif, L., McConkey, B., Hamel, C., and Vujanovic, V. (2007). First report of damping-off of durum wheat caused by Arthrinium sacchari in the semi-arid Saskatchewan fields. Plant Dis. 91:469. doi: 10.1094/PDIS-91-4-0469A
Minter, D. W. (1985). A re-appraisal of the relationships between Arthrinium and other hyphomycetes. Proc. Indian Acad. Sci. 94, 281–308.
Nylander, J. A. A. (2004). MrModeltest 2.0. Program Distributed by the Author. Sweden: Evolutionary Biology Centre, Uppsala University.
Petrini, L. E., and Müller, E. (1986). Haupt-und nebenfruchtformen europäischer Hypoxylon-Arten (Xylariaceae, Sphaeriales) und verwandter Pilze. Mycol. Helv. 1, 501–627.
Petrini, O., Candoussau, F., and Petrini, L. (1989). Bambusicolous fungi collected in southwestern France 1982–1989. Mycologia Helvetica 3, 263–279.
Pintos, A., Alvarado, P., Planas, J., and Jarling, R. (2019). Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. Hosts. Mycokeys 49, 15–48. doi: 10.3897/mycokeys.49.32115
Rai, M. K. (1989). Mycosis in man due to Arthrinium phaeospermum var. indicum. First case report of mycosis due to Arthrinium phaeospermum var. indicum in humans. Mycoses 32, 472–475. doi: 10.1111/j.1439-0507.1989.tb02285.x
Rambaut, A. (2009). FigTree v1.4: Tree Figure Drawing Tool. Available online at: http://tree.bio.ed.ac.uk/software/figtre?e/ (accessed July 25, 2020).
Rambaut, A., and Drummond, A. J. (2007). Tracer v1, 4. Available online at: http://beast.bio.ed.ac.uk/Tracer (accessed July 1, 2020).
Ramos, H. P., Braun, G. H., Pupo, M. T., and Said, S. (2010). Antimicrobial activity from endophytic fungi Arthrinium state of Apiospora montagnei Sacc. and Papulaspora immersa. Braz. Arch. Biol. Technol. 53, 629–632. doi: 10.1590/S1516-89132010000300017
Réblová, M., Miller, A. N., Rossman, A. Y., Seifert, K., Crous, P., Hawksworth, D., et al. (2016). Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7, 131–153. doi: 10.5598/imafungus.2016.07.01.08
Rehner, S. (2001). Primers for Elongation Factor 1-α (EF1-α). Washington, DC: Insect Biocontrol Laboratory: USDA, ARS, PSI.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Senanayake, I. C., Jeewon, R., Hyde, K. D., Bhat, J. D., and Cheewangkoon, R. (2020a). Taxonomy and phylogeny of Leptosillia cordylinea sp. nov. from China. Phytotaxa 435, 213–226. doi: 10.11646/phytotaxa.435.3.1
Senanayake, I. C., Rathnayake, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020b). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere (in press).
Senanayake, I. C., Maharachchikumbura, S. S., Hyde, K. D., Bhat, J. D., Jones, E. G., Wendt, L., et al. (2015). Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 73, 73–144. doi: 10.1007/s13225-015-0340-y
Sharma, R., Kulkarni, G., Sonawane, M. S., and Shouche, Y. S. (2014). A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience 55, 118–123. doi: 10.1016/j.myc.2013.06.004
Shrestha, P., Ibáñez, A. B., Bauer, S., Glassman, S. I., Szaro, T. M., Bruns, T. D., et al. (2015). Fungi isolated from Miscanthus and sugarcane: biomass conversion, fungal enzymes, and hydrolysis of plant cell wall polymers. Biotechnol. Biofuels 8:1. doi: 10.1186/s13068-015-0221-3
Silvestro, D., and Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337. doi: 10.1007/s13127-011-0056-0
Singh, S. M., Yadav, L. S., Singh, P. N., Hepat, R., Sharma, R., and Singh, S. K. (2012). Arthrinium rasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon 122, 449–460. doi: 10.5248/122.449
Species Fungorum (2020). Available online at: http://www.speciesfungorum.org/Names/Names.asp (accessed July 10, 2020).
Stamatakis, A., and Alachiotis, N. (2010). Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. J. Bioinform. 26, 132–139. doi: 10.1093/bioinformatics/btq205
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990
Wang, M., Tan, X. M., Liu, F., and Cai, L. (2018). Eight new Arthrinium species from China. Mycokeys 34, 1–24. doi: 10.3897/mycokeys.34.24221
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Al-Ani, L. K. T., Tedersoo, L., Haelewaters, D., Fiuza, P. O., et al. (2020). Outline of Fungi and fungus-like taxa. Mycosphere 11, 1060–1456. doi: 10.5943/mycosphere/11/1/8
Zhang, L. Q., and Wang, X. G. (1999). Fungus resource of bamboos in China. J. Bamboo Res. 18, 66–72.
Keywords: Apiosporaceae, bamboo, fungal taxonomy, new locality records, novel species
Citation: Senanayake IC, Bhat JD, Cheewangkoon R and Xie N (2020) Bambusicolous Arthrinium Species in Guangdong Province, China. Front. Microbiol. 11:602773. doi: 10.3389/fmicb.2020.602773
Received: 11 September 2020; Accepted: 02 November 2020;
Published: 14 December 2020.
Edited by:
Peter Edward Mortimer, Kunming Institute of Botany, Chinese Academy of Sciences, ChinaReviewed by:
Dong-Qin Dai, Qujing Normal University, ChinaSaowaluck Tibpromma, Kunming Institute of Botany, Chinese Academy of Sciences, China
Copyright © 2020 Senanayake, Bhat, Cheewangkoon and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ratchadawan Cheewangkoon, cmF0Y2hhLjIyMkBnbWFpbC5jb20=; Ning Xie, U2hhaW5pbkBtc24uY24=
 Indunil C. Senanayake
Indunil C. Senanayake Jayarama D. Bhat3,4
Jayarama D. Bhat3,4 Ratchadawan Cheewangkoon
Ratchadawan Cheewangkoon