
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 27 October 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.601352
This article is part of the Research TopicInterview with the Translational Apparatus: Stories of Intriguing Circuits and Mechanisms to Regulate Translation in BacteriaView all 8 articles
The carbon storage regulator (Csr) or repressor of stationary phase metabolites (Rsm) system of Gammaproteobacteria is among the most complex and best-studied posttranscriptional regulatory systems. Based on a small RNA-binding protein, CsrA and homologs, it controls metabolism, physiology, and bacterial lifestyle decisions by regulating gene expression on a vast scale. Binding of CsrA to sequences containing conserved GGA motifs in mRNAs can regulate translation, RNA stability, riboswitch function, and transcript elongation. CsrA governs the expression of dozens of transcription factors and other regulators, further expanding its influence on cellular physiology, and these factors can participate in feedback to the Csr system. Expression of csrA itself is subject to autoregulation via translational inhibition and indirect transcriptional activation. CsrA activity is controlled by small noncoding RNAs (sRNAs), CsrB and CsrC in Escherichia coli, which contain multiple high affinity CsrA binding sites that compete with those of mRNA targets. Transcription of CsrB/C is induced by certain nutrient limitations, cellular stresses, and metabolites, while these RNAs are targeted for degradation by the presence of a preferred carbon source. Consistent with these findings, CsrA tends to activate pathways and processes that are associated with robust growth and repress stationary phase metabolism and stress responses. Regulatory loops between Csr components affect the signaling dynamics of the Csr system. Recently, systems-based approaches have greatly expanded our understanding of the roles played by CsrA, while reinforcing the notion that much remains to be learned about the Csr system.
The csrA gene was discovered in studies aimed at finding regulators of gene expression in the stationary phase of bacterial growth. These studies used random transposon mutagenesis to identify trans-acting factors affecting glycogen biosynthesis and expression of a glgC’-‘lacZ translational reporter (Romeo and Gong, 1993; Romeo et al., 1993). At that time, transcript initiation was understood to be a focal point of genetic regulation, while translational regulation and posttranscriptional regulation were limited to a few examples, such as feedback inhibition of ribosomal protein synthesis (discussed in Babitzke et al., 2009, 2019; Vakulskas et al., 2015). Nevertheless, early observations on the regulation of glgC by CsrA began to associate this protein with posttranscriptional control, which involves CsrA-mediated repression associated with binding to the glgC untranslated mRNA leader, inhibition of ribosome loading, and mRNA destabilization (Liu et al., 1995, 1997; Liu and Romeo, 1997; Baker et al., 2002). The global regulatory roles of CsrA and its orthologs, e.g., RsmA, that began to emerge soon after its discovery included widespread effects on carbon metabolism, virulence of plant and animal pathogens, motility, and surface properties that mediate biofilm formation (Romeo et al., 1993; Sabnis et al., 1995; Mukherjee et al., 1996; Yang et al., 1996; Altier et al., 2000; Wei et al., 2001; Jackson et al., 2002). These and other roles of CsrA rely on the RNA-binding properties of this protein. The varied regulatory mechanisms mediated by CsrA depend on the positioning of its binding sites in target RNAs relative to other cis-acting elements and the ability of CsrA binding to alter RNA structure.
Purification of CsrA protein revealed its tight association with RNA, a large fraction of which consisted of a noncoding sRNA designated CsrB (Liu et al., 1997). The functions of this sRNA and a second similar one, CsrC (Weilbacher et al., 2003), defined a new paradigm of genetic regulation, in which a noncoding RNA molecule employs multiple high-affinity binding sites to sequester an mRNA-binding regulatory protein away from its target mRNAs. Predictably, genes and conditions that influence the biosynthesis and turnover of CsrB/C sRNAs affect the expression of genes in the CsrA regulon, although feedback in the system can sometimes obscure findings from single gene disruptions. The regulatory factors that control carbon storage regulator (Csr) sRNAs inform our understanding of how metabolic status and physiological conditions can impact translation and posttranscriptional processes to guide bacterial lifestyle decisions. This review focuses on regulatory mechanisms and circuitry involving CsrA and the Csr system of Escherichia coli, with occasional reference to homologous systems in other species.
CsrA is a small, highly conserved protein that is 61 amino acids in length in E. coli. Although early observations suggested a role for CsrA in posttranscriptional gene regulation, its amino acid sequence was originally found to be unrelated to known proteins and its structure and regulatory mechanism were unclear (Romeo et al., 1993; Liu et al., 1995). A major advancement in understanding its structure occurred when the 3D structures of three CsrA/RsmA family proteins were independently solved (Gutiérrez et al., 2005; Rife et al., 2005; Heeb et al., 2006). These structures revealed a novel RNA-binding topology and confirmed previous studies, indicating that E. coli CsrA functions as a homodimer (Dubey et al., 2003). In the active dimer, the CsrA monomer polypeptides exhibit a β-β-β-β-β-α secondary structure containing five consecutive antiparallel β-strands (β1–β5) and an α-helix, followed by a flexible C-terminus. The β-strands of each monomer interdigitate with those of the other monomer so that the dimer contains two interlocking five-stranded antiparallel β-sheets. The three central strands (β2–β4) of each β-sheet originate from one subunit while the two peripheral strands (β1, β5) come from the other subunit. The interlocking β-strands form a hydrophobic core from which the two α-helices extend.
Alanine scanning mutagenesis of E. coli CsrA revealed two identical surfaces, located on opposite sides of the protein, which are essential for RNA binding and regulation (Mercante et al., 2006). These two surfaces are comprised of the parallel β1 and β5 strands of opposing monomers, which form two positively charged subdomains and can allow the binding of two RNA molecules simultaneously. (Schubert et al., 2007; Mercante et al., 2009; Figure 1). Highly conserved residues L4 in β1 and R44 in β5 are particularly critical for CsrA-RNA interaction. R44A substitution in E. coli CsrA resulted in defective RNA binding and eliminated regulation of target genes (Mercante et al., 2006). R44 is also important for RNA binding by the Yersinia enterocolitica CsrA (Heeb et al., 2006) and Pseudomonas fluorescens RsmE regulation (Schubert et al., 2007). L4A substitution resulted in a partial loss of regulation by RsmE in P. fluorescens as well (Schubert et al., 2007). These structural and functional studies established CsrA and its homologs as a novel class of RNA-binding protein, containing a new RNA-binding fold that mediates the posttranscriptional regulation of gene expression.
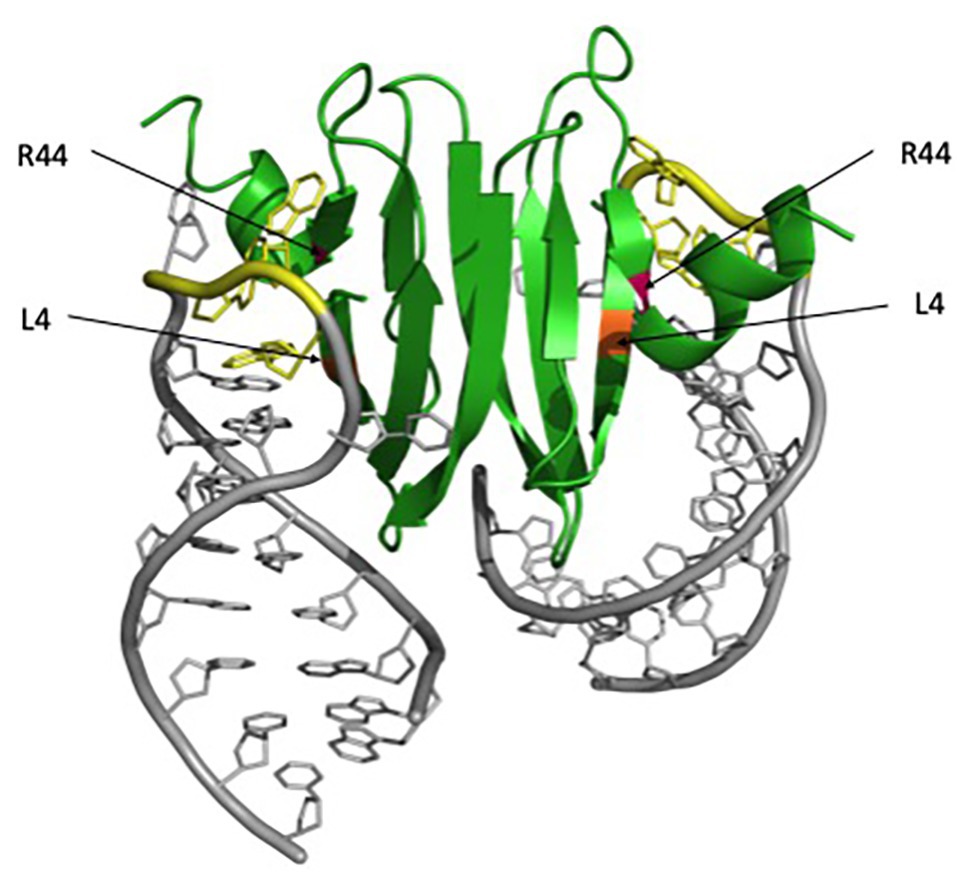
Figure 1. RsmA/CsrA binding to hcnA mRNA. The RsmA/CsrA protein is shown in green, critical RNA-binding residues are marked with asterisks, L4 is highlighted in orange, and R44 in magenta. The hcnA mRNA is shown in gray, with GGA motifs associated with binding highlighted in yellow. The figure was generated in PyMol using the Protein Data Bank (PDB) file 2JPP.
The first insights into the features of RNA that facilitate CsrA binding came with the identification of the noncoding sRNA CsrB, which interacts with CsrA and antagonizes its regulatory activity. The stoichiometry of the CsrA:CsrB complex indicated that each CsrB molecule binds ~18 CsrA subunits, forming a large globular ribonucleoprotein complex (Liu et al., 1997). The CsrB sRNA contained about the same number of repeated CAGGA(U/A/C)G sequence elements, suggestive of their function as targets for CsrA binding. These repeated sequence motifs were located within the loops of predicted hairpin structures or other single-stranded regions (Romeo, 1998). These early studies strongly suggested that a single-stranded GGA motif located within the loop of a hairpin structure serves as an important part of the CsrA binding site.
A more detailed exploration of the role of RNA sequence and secondary structure in CsrA binding was carried out using systematic evolution of ligands by exponential enrichment (SELEX). SELEX allowed the identification of high-affinity CsrA:RNA interactions from a large combinatorial RNA library. The SELEX-derived consensus sequence for a CsrA binding site was deduced to be RUACARGGAUGU. The ACA and GGA motifs were 100% conserved and the GU sequence was present in all but one ligand. Of the 55 RNAs identified, a majority (51) contained a GGA motif in the loop of a hairpin that was the most stable predicted structure, similar to the repeated elements found in CsrB. Most of the SELEX hairpins contained four or fewer contiguous base pairs, suggesting that very stable secondary structure might be disadvantageous for CsrA binding. Additionally, a majority (44) of the hairpin loops consisted of six nucleotides, with the upstream AC and downstream GU residues base-pairing together (Dubey et al., 2005). The studies also established that CsrA binding affinity is influences by both RNA primary sequence information and its structural context.
Solution nuclear magnetic resonance (NMR) of the CsrA homolog RsmE confirmed that differences in RNA sequence and structure can alter RsmE binding affinity and revealed that interaction of RNA with this protein introduces RNA curvature (Duss et al., 2014a,b). NMR structures of RsmE bound to high-affinity RNA targets revealed contacts involving a conserved hairpin structure containing an ANGGAN hexanucleotide loop. Hydrogen bond formation between the conserved A and GGA RNA bases and the RsmE backbone carbonyl and amino groups also occurs (Schubert et al., 2007). The N nucleotides of the sequence are variable, yet they affect binding affinity. The presence of additional nucleotides within the loop can decrease RsmE binding affinity, as can alterations in secondary structure, such as the absence of a hairpin structure or the involvement of the GGA motif in base pairing (Duss et al., 2014a).
Because each CsrA dimer contains two positively charged RNA-binding surfaces, and because many RNA targets of CsrA contain two or more binding sites, it was proposed that a CsrA protein might be able to interact simultaneously with two sites in a single transcript (Mercante et al., 2009). To explore this hypothesis, CsrA dimers with mutations (R44A) in one or both polypeptides, which alter one or both RNA-binding surfaces of the protein, were tested for interaction with RNAs containing two or more CsrA binding sites. Gel shift assays and other studies confirmed that CsrA is capable of “bridging” two sites on a single RNA that are separated by ≥10 to ≤63 nucleotides (nt), with an optimal intersite distance of >18 nt. Additionally, in vitro coupled transcription-translation reactions demonstrated that full repression of glgC requires a CsrA dimer to contain both of the functional RNA-binding pockets. Disruption of CsrA dual-site binding by RNA sequence alterations also interfered with regulation (Mercante et al., 2009). However, the requirement of two CsrA binding sites for full regulatory capability is apparently not essential, as one known CsrA target mRNA contains a single CsrA binding site (Baker et al., 2007). Structural studies using NMR and electron paramagnetic resonance spectroscopy (EPR) confirmed dual-site binding by the CsrA homolog RsmE. Furthermore, this protein bound to the individual sites within a multi-site sRNA target in a sequential order and with cooperativity (Duss et al., 2014b).
The identification of CsrA as a regulator of glgC expression in E. coli led to the first insights regarding CsrA regulatory mechanisms. glgC mRNA is destabilized by CsrA, and deletion studies of a glgC’-‘lacZ translational fusion revealed that CsrA-mediated regulation was dependent on the region surrounding the glgC initiation codon and did not require a glgC promoter (Liu et al., 1995). In vitro experiments with purified recombinant protein confirmed that CsrA binds specifically to glgC mRNA and inhibits expression posttranscriptionally (Liu and Romeo, 1997). Further exploration demonstrated that CsrA binds four sites in the untranslated glgC leader (Baker et al., 2002). The downstream binding site overlaps the Shine-Dalgarno (SD) sequence, while a second binding site lies within the single stranded loop of an RNA hairpin that is located just upstream of the SD sequence. High-affinity binding to the glgC hairpin tethers the CsrA protein to this mRNA and permits the CsrA dimer to bridge to the SD sequence, where it prevents translation (Mercante et al., 2009; Figure 2A). Because translation and mRNA stability are often subject to coordinate regulation, translational inhibition likely contributes to CsrA-mediated instability of the glgC transcript (Liu et al., 1995; Vytvytska et al., 2000).
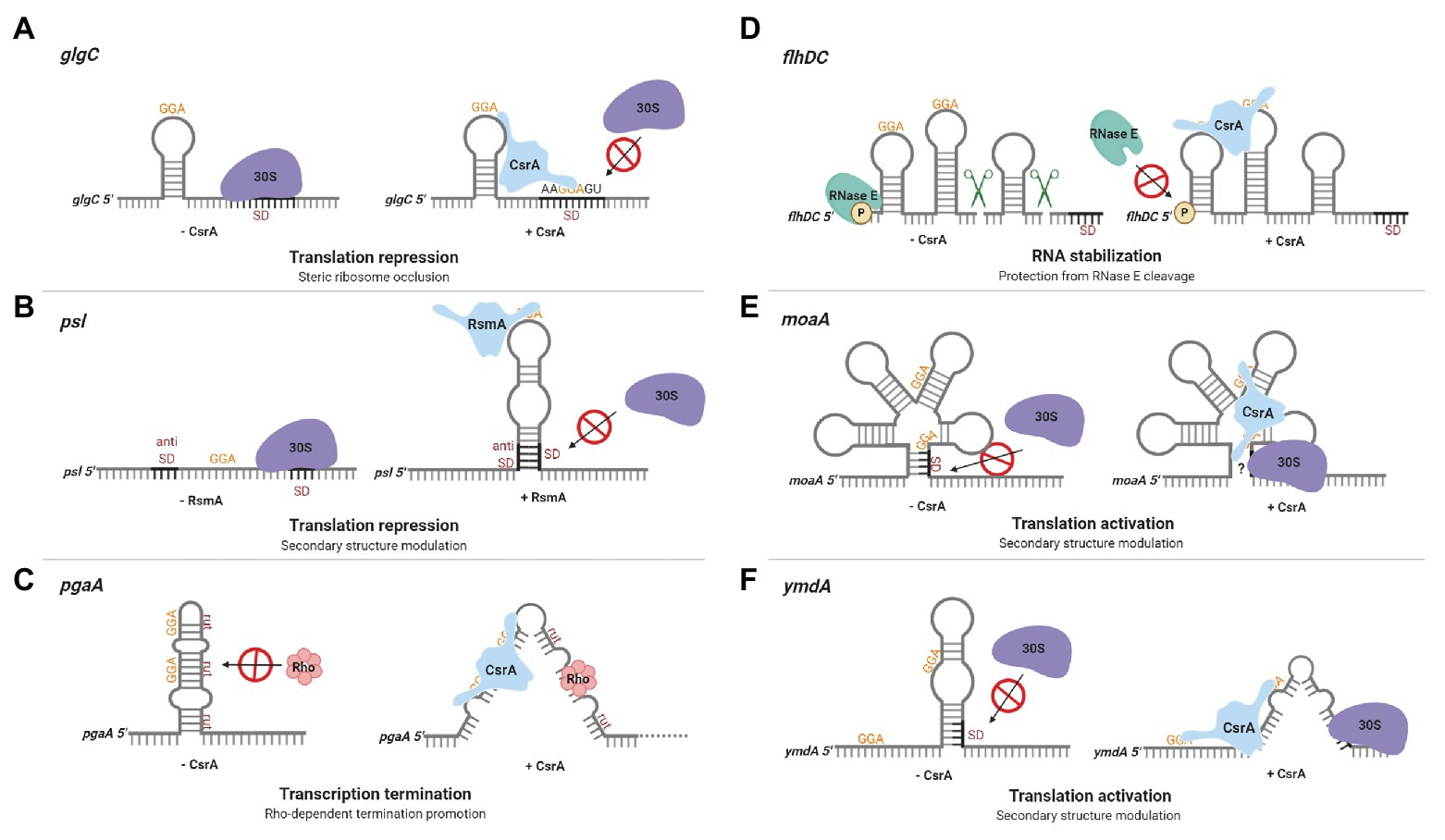
Figure 2. Regulation of gene expression by CsrA-family proteins. (A) CsrA repression of glgC translation in Escherichia coli. A CsrA homodimer binds to two sites in the glgC leader, a high-affinity site in a hairpin loop and a lower-affinity region overlapping the Shine-Dalgarno (SD) sequence, competing with ribosome (30S) binding and repressing translation. (B) Repression of psl translation in Pseudomonas aeruginosa by RsmA. The psl leader contains an unstable RNA hairpin formed by the SD and anti-SD regions. RsmA binding stabilizes the hairpin, prevents ribosome binding, and represses translation. (C) CsrA promotes Rho-dependent pgaA transcription termination in E. coli. In the absence of CsrA, the pgaA transcript forms a hairpin that sequesters Rho utilization (rut) sites. CsrA binding to two sites prevents this hairpin from forming, exposes the rut sites, and facilitates termination. (D) CsrA stabilization of the flhDC transcript in E. coli. CsrA binds to two sites located near the 5' end of the flhDC transcript, thus protecting this mRNA against RNase E cleavage and turnover. (E) CsrA activates moaA translation in E. coli. CsrA binds to two sites within an apparent Moco-responsive riboswitch aptamer in the moaA transcript. Binding at this location is proposed to expose the SD for ribosome binding, permitting translation. (F) CsrA activates ymdA translation in E. coli. In the absence of CsrA, ymdA mRNA forms a hairpin that sequesters the SD sequence. CsrA binding to a site within the hairpin and another site just upstream destabilizes the hairpin and permits ribosome access. These regulatory mechanisms are discussed in the text. This figure was created with BioRender.com.
Subsequent investigation of other CsrA targets revealed that translation inhibition by ribosome exclusion is a common mechanism of CsrA-mediated gene regulation. cstA, which encodes a protein involved in peptide transport during carbon starvation in E. coli, was identified as a putative CsrA target based on sequence identity to the glgC leader. Much like glgC, the cstA leader contains up to four CsrA binding sites, with one overlapping the SD sequence. Toeprinting analysis of CsrA and 30S ribosomal subunit association with cstA RNA demonstrated that bound CsrA sterically inhibits ribosome binding and represses the expression of this gene (Dubey et al., 2003). The same mechanism has been reported in gram-positive bacteria. In Bacillus subtilis, CsrA represses translation of the gene encoding the flagellin protein (hag) by binding to two sites in the hag leader mRNA. One of these sites overlaps the SD sequence and CsrA bound at this location blocks ribosome binding (Yakhnin et al., 2007). Similar mechanisms of translational repression occur in many other mRNA targets, with CsrA and its homologs binding to regions overlapping the SD sequence, start codon, and/or initially translated region (Wang et al., 2005; Martinez et al., 2011; Yakhnin et al., 2011a; Pannuri et al., 2012; Abbott et al., 2015; Park et al., 2015; Pourciau et al., 2019). A unique example of CsrA-dependent ribosome exclusion is the repression of sdiA, which encodes the N-acyl-L-homoserine lactone receptor in E. coli (Dyszel et al., 2010). Unlike other known CsrA targets, the two CsrA binding sites in the sdiA transcript are located entirely within the coding region, although the upstream CsrA binding site overlaps the ribosome-binding segment of this transcript (Yakhnin et al., 2011a).
In Pseudomonas aeruginosa, the CsrA homolog RsmA represses translation of the pslA mRNA by sequestering the SD sequence via an alternative mechanism (Irie et al., 2010). The psl locus is responsible for the production of a biofilm supporting exopolysaccharide and the corresponding transcript has an extensive untranslated leader. RsmA binds to a single stranded site within the loop of a hairpin in the psl leader. Base pairing in this imperfect hairpin occurs between the SD and anti-SD sequences and is unstable in the absence of RsmA. RsmA binding to the transcript stabilizes the hairpin, which prevents ribosome binding and represses translation (Irie et al., 2010; Figure 2B). A more recently discovered mechanism of CsrA-mediated repression involves translational coupling, wherein the translation of a gene depends on the translation of a closely located upstream gene (Babitzke et al., 2009). CsrA regulates expression of iraD, which encodes an anti-adapter protein that inhibits RssB-mediated RpoS proteolysis in E. coli. The iraD leader contains four CsrA binding sites, but CsrA binding does not directly inhibit ribosome access to the iraD translation initation region. Instead, CsrA represses translation of a short open reading frame upstream from and translationally coupled to iraD, thereby repressing iraD expression (Park et al., 2017).
There are six CsrA binding sites within the E. coli pgaA noncoding leader, which are involved in multiple regulatory mechanisms (Wang et al., 2005). The pgaABCD operon encodes proteins responsible for synthesis and secretion of a biofilm polysaccharide adhesin consisting of a partially N-deacetylated polymer of β-1,6-N-acetyl-D-glucosamine (Wang et al., 2004, 2016; Itoh et al., 2008). CsrA interacts with the two sites overlapping the SD sequence and start codon, which inhibits ribosome binding and initiation of translation. Mutation of these binding sites relieved ~60% of CsrA-dependent repression (Wang et al., 2005). Studies of the remaining sites revealed a mechanism whereby CsrA binding promotes Rho-dependent transcription termination. In the absence of CsrA, the pgaA leader forms a hairpin that sequesters Rho utilization (rut) sites. CsrA binding to two sites in this region prevents hairpin formation, exposes the rut sites, and facilitates Rho-dependent termination (Figueroa-Bossi et al., 2014; Figure 2C). The exceedingly sophisticated direct regulation of pgaABCD expression by CsrA is striking, as it affects transcript elongation, translation, and transcript stability (Wang et al., 2005). However, given that many other CsrA RNA targets apparently possess multiple CsrA binding sites, CsrA may regulate expression of other genes using similarly complex mechanisms.
Despite the prevalence of CsrA-mediated repression mechanisms, CsrA was recognized early as an activator of glycolytic gene expression via undetermined mechanisms (Sabnis et al., 1995). In addition, CsrA was found to activate the expression of flhDC, the master operon responsible for eliciting the motility and chemotaxis gene expression cascade in E. coli (Wei et al., 2001). The flhDC operon encodes the subunits of a DNA-binding protein (FlhD4C2) that recognizes class II flagellar promoters. In the absence of CsrA, the 5' monophosphorylated end of the flhDC transcript is accessible to RNase E, which binds to and cleaves this mRNA, facilitating its turnover. CsrA binding to two sites located near the 5' end of the flhDC transcript protects it against RNase E attack and stabilizes this mRNA (Yakhnin et al., 2013; Figure 2D).
CsrA also activates mRNA translation. In Pseudomonas aeruginosa, the CsrA homolog RsmA differentially regulates the expression of two genes involved in the biosynthesis of phenazine, phz1 and phz2. RsmA represses the translation of phz1 and activates the translation of phz2. RsmA-mediated activation of phz2 seems to occur by a mechanism, which is opposite to that of psl repression in some respects. RsmA binding to the phz2 leader is hypothesized to prevent the formation of secondary structure that sequesters the SD sequence, thus activating phz2 translation (Ren et al., 2014).
CsrA also activates translation of moaA in E. coli by altering RNA structure. The moaABCDE operon encodes proteins required for molybdenum cofactor (Moco) biosynthesis. Translation of moaA is apparently inhibited via a Moco-responsive riboswitch aptamer, which is thought to sequester the SD sequence (Regulski et al., 2008). CsrA binding to two sites within the aptamer seemingly alters the structure to expose the SD to ribosome binding (Patterson-Fortin et al., 2013; Figure 2E). To our knowledge, these studies of moaA regulation by CsrA brought to light the first example of dual posttranscriptional regulation mediated by the binding of a cofactor (Moco) and an RNA-binding protein to the aptamer domain of a riboswitch.
The first detailed molecular mechanism for CsrA-mediated translational activation was recently established in a study exploring the expression of the uncharacterized E. coli protein YmdA. The ymdA gene was identified as the most strongly activated direct mRNA target of CsrA in the E. coli genome in an integrated transcriptomics study (Potts et al., 2017) and may play a role in biofilm formation (Kim and Kim, 2017). Reporter assays and mRNA half-life studies confirmed that CsrA posttranscriptionally activates ymdA expression and stabilizes the ymdA transcript (Renda et al., 2020). In the absence of CsrA, ymdA mRNA forms a hairpin that sequesters the SD sequence, inhibiting ribosome binding. CsrA-ymdA RNA footprint experiments revealed two CsrA binding sites in the ymdA leader, one located within the ymdA hairpin and the other just upstream of it. Disruption of CsrA binding by a mutation at either position eliminated CsrA-dependent activation in vivo. Toeprinting analysis of CsrA and 30S ribosomal subunit association with the ymdA transcript further revealed that CsrA binding destabilizes the SD-sequestering hairpin, permitting ribosome access and translation (Figure 2F).
sRNAs participate in multiple regulatory networks, allowing bacteria to rapidly alter gene expression in response to environmental cues in order to make lifestyle decisions (Mehta et al., 2008; Gottesman and Storz, 2011; Wagner and Romby, 2015; Nitzan et al., 2017). While most of the well-characterized bacterial sRNAs function by base pairing with their mRNA targets, the discovery of CsrB and CsrC RNAs of E. coli introduced sRNAs that act by binding to and inhibiting the activities of an RNA-binding regulatory protein (Liu et al., 1997, 1998; Romeo, 1998; Heeb et al., 2002; Valverde et al., 2003; Weilbacher et al., 2003; Kay et al., 2005; Babitzke and Romeo, 2007). CsrB from E. coli contains 22 GGA sequences, most of which serve as CsrA binding sites (Liu et al., 1997; Vakulskas et al., 2015, 2016). Another E. coli sRNA, CsrC, is structurally and functionally related to CsrB (Weilbacher et al., 2003). Phylogenetic analyses and other studies suggest that sRNAs in the CsrB family are widespread in Gammaproteobacteria and control genes responsible for a myriad of metabolic pathways and physiological functions (Zere et al., 2015; Potts et al., 2018), as well as complex traits such as biofilm formation (Jackson et al., 2002; Parker et al., 2017), host-microbe interactions, and pathogenesis (reviewed in Vakulskas et al., 2015).
Synthesis of the E. coli CsrB and CsrC sRNAs depends on the two-component signal transduction system (TCS) BarA-UvrY (Suzuki et al., 2002). Orthologs of BarA-UvrY are known by a variety of names and activate the synthesis of CsrA-inhibitory sRNAs in many Gammaproteobacteria (Pernestig et al., 2001; Vakulskas et al., 2015; Zere et al., 2015). Metabolic end products, namely short chain carboxylate compounds such as formate and acetate (R-COOH), stimulate the kinase activity of BarA, which phosphorylates the DNA binding response regulator, UvrY. BarA also possesses phosphatase activity, as deduced by genetics experiments (Camacho et al., 2015). The phosphorylated form of UvrY binds to DNA and activates transcription from the csrB and csrC promoters (Pernestig et al., 2001; Chavez et al., 2010; Camacho et al., 2015; Zere et al., 2015). Other activators of CsrB/C synthesis include the stringent response factors, ppGpp and DksA, and two DEAD-box RNA helicases, DeaD (CsdA) and SrmB (Edwards et al., 2011; Vakulskas et al., 2014; Zere et al., 2015). DeaD helicase functions by overcoming inhibitory basepairing within uvrY mRNA to activate translation, while SrmB stimulates csrB/C expression by unknown mechanisms.
Turnover of CsrB/C sRNAs in E. coli is tightly regulated. In addition to the housekeeping endonuclease RNase E and the 3'-5' exonuclease polynucleotide phosphorylase (PNPase), it requires a non-nucleolytic EAL-GGDEF domain-containing protein, CsrD or MshH (Suzuki et al., 2006; Leng et al., 2016; Vijayakumar et al., 2018). Unlike classical EAL-GGDEF domain proteins, CsrD neither synthesizes nor degrades the signaling molecule (3'-5')-cyclic-diguanylate (c-di-GMP). Instead, the availability of a preferred carbon source, such as glucose, results in dephosphorylation of EIIAGlc of the PTS pathway, which can then bind to the EAL domain of CsrD and trigger CsrB/C decay (Figure 3). This binding interaction is part of a complex membrane-localized mechanism that is required for turnover of these sRNAs in the presence of CsrA (Suzuki et al., 2006; Leng et al. 2016; Vakulskas et al., 2016; Hadjeras et al., 2019). CsrA binding to CsrB RNA blocks cleavage by RNase E at an unstructured site just upstream of the intrinsic terminator, which is necessary for initiation of turnover. The CsrD-EIIAGlc interaction overcomes the inhibitory activity of CsrA (Leng et al., 2016; Vakulskas et al., 2016). While it seems that most or perhaps all Gammaproteobacteria express sRNAs that antagonize CsrA activity, CsrD orthologs appear to be restricted to the Enterobacteriaceae, Shewanellaceae, and Vibrionaceae. Interestingly, in contrast to short half-lives of CsrB/C in E. coli, Csr family sRNAs in species that lack a CsrD homolog are relatively stable, with half-lives reported from ∼20 min to >60 min (Valverde et al. 2004; Sonnleitner et al., 2006; Vakulskas et al., 2016). Moreover, the stability of the latter sRNAs is decreased in the absence of a CsrA homolog, while disruption of csrA has little or no effect on CsrB/C decay in csrD wild type (WT) strains of E. coli (Gudapaty et al., 2001). Thus, the evolution of CsrD was proposed to have provided a mechanism that allows the turnover of Csr sRNAs in Enterobacteriaceae and close relatives to respond to nutritional status. Viewed comprehensively, CsrB/C decay in E. coli is enhanced when preferred carbon sources are available, while their synthesis is activated by end products of metabolism (Chavez et al., 2010; Leng et al., 2016; Vakulskas et al., 2016).
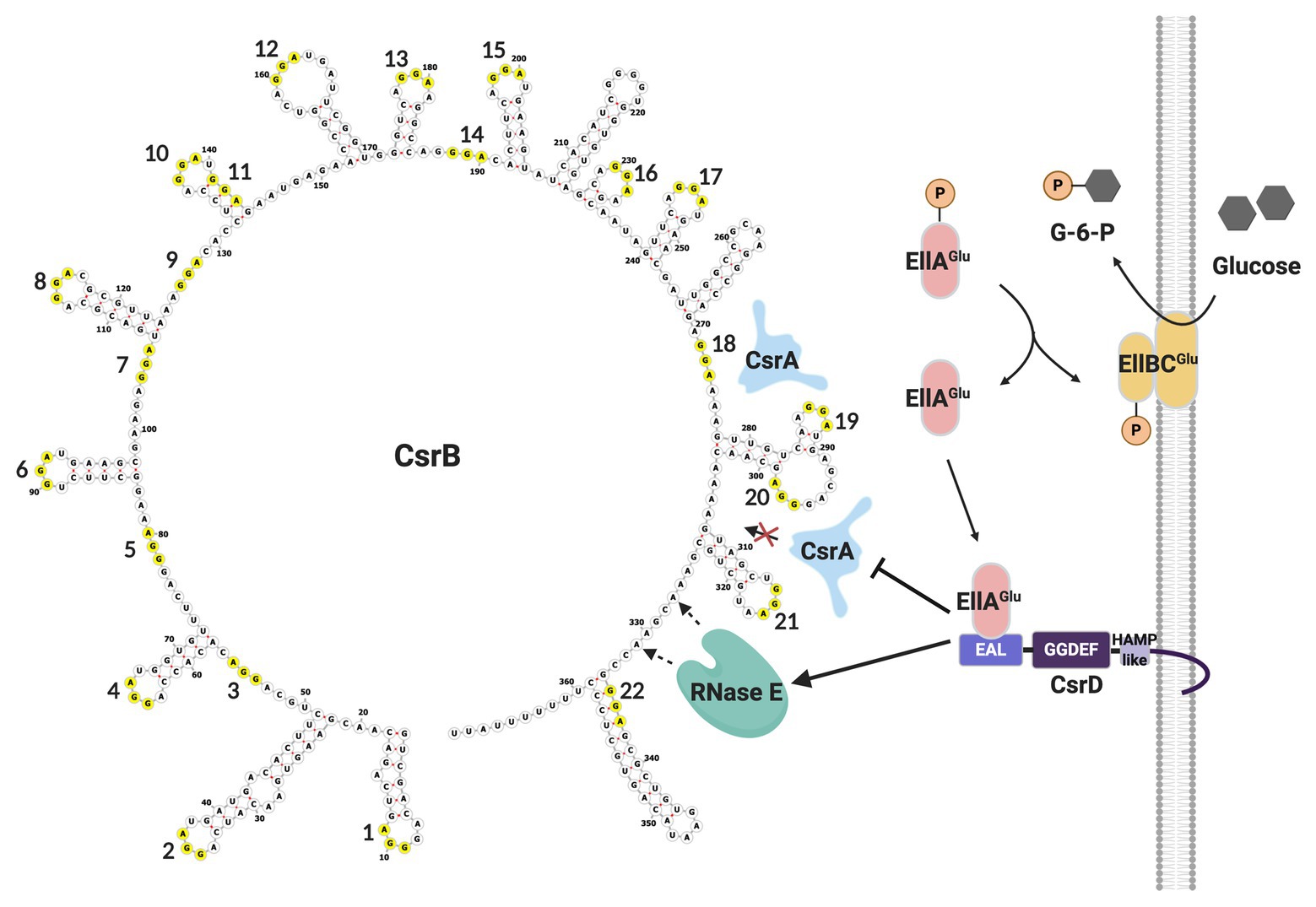
Figure 3. CsrB sRNA secondary structure and decay pathway. GGA motifs are numbered and circled in yellow. As glucose is transported into the cell, EllAGlu protein becomes dephosphorylated and able to bind to the EAL domain of CsrD, potentiating CsrB decay. In the absence of CsrD-EIIAGlc, CsrA binding to CsrB protects it from RNase E cleavage and turnover. This figure was created with BioRender.com.
CsrB, CsrC, as well as two basepairing sRNAs, McaS and GadY, have been reported to activate biofilm formation when they are overexpressed. This effect on biofilm was proposed to occur because these sRNAs bind to and antagonize CsrA activity, thus increasing pgaA expression (Wang et al., 2005; Itoh et al., 2008; Pannuri et al., 2012; Jørgensen et al., 2013; Parker et al., 2017). The basepairing sRNAs contain at least two GGA motifs and bind to CsrA with high affinity in vitro (Jørgensen et al., 2013; Potts et al., 2017). UV-crosslinking immunoprecipitation and sequencing (CLIP-seq) revealed that CsrA also interacts directly with other sRNAs in vivo in both Salmonella and E. coli (Holmqvist et al., 2016; Potts et al., 2017). The physiological functions associated with these interactions remain to be elucidated. Interestingly, CsrA was recently reported to interact with and promote pairing of the sRNA SR1 with the mRNA ahrC in B. subtilis (Müller et al., 2019). In addition, the CsrA homolog RsmA in P. aeruginosa has been shown to sometimes bind to nascent mRNAs in conjunction with the sRNA-mRNA chaperone protein Hfq (Gebhardt et al., 2020). These findings warrant additional studies and hint that CsrA may play important roles in sRNA-mRNA or other RNA-RNA interactions.
While CsrA binding to Csr-family sRNAs plays a regulatory role in many Gammaproteobacteria, some organisms employ a protein that antagonizes CsrA activity. FliW was the first example of this kind of protein, which is present in the gram-positive firmicute Bacillus subtilis (Mukherjee et al., 2011). B. subtilis CsrA inhibits translation of hag mRNA that encodes the flagellar filament protein. Free FliW directly inhibits CsrA from binding to and occluding the SD sequence of the hag transcript and, thus, releaves CsrA-mediated repression of hag translation (Yakhnin et al., 2007; Mukherjee et al., 2011). The Hag flagellin, together with FliW and CsrA, are involved in a partner-switching mechanism promoting Hag homeostasis (Mukherjee et al., 2011; Figure 4). Unlike CsrB/C sRNAs, FliW does not interact with the RNA-binding amino acid residues in the RNA-binding pocket of CsrA. Moreover, structural analysis of FliW-CsrA complex revealed that FliW interacts with the extended C-terminus of CsrA (Altegoer et al., 2016; Mukherjee et al., 2016). As such, FliW has been proposed to allosterically antagonize CsrA via a noncompetitive mechanism (Altegoer et al., 2016; Mukherjee et al., 2016). A recent study revealed that the Hag-FliW-CsrAdimer system functions at a nearly equimolar ratio. The components participate in a three-node negative-feedback loop that maintains Hag homeostasis in the cytoplasm, with similarities to toxin-antitoxin systems (Oshiro et al., 2019). Interestingly, FliW appears to be widely dispersed among motile bacteria and has been found to interact with CsrA in the gram-negative bacterium Campylobacterium jejuni (Dugar et al., 2016; Radomska et al., 2016; Li et al., 2018). However, fliW is notably absent from Gammaproteobacteria, and the presence of this gene is anti-correlated with genes encoding BarA-UvrY and their homologs, which are required for transcription of Csr-family sRNAs in many species (Zere et al., 2015). Whether FliW and sRNAs function together to modulate CsrA activity in any bacterial species remains to be determined.
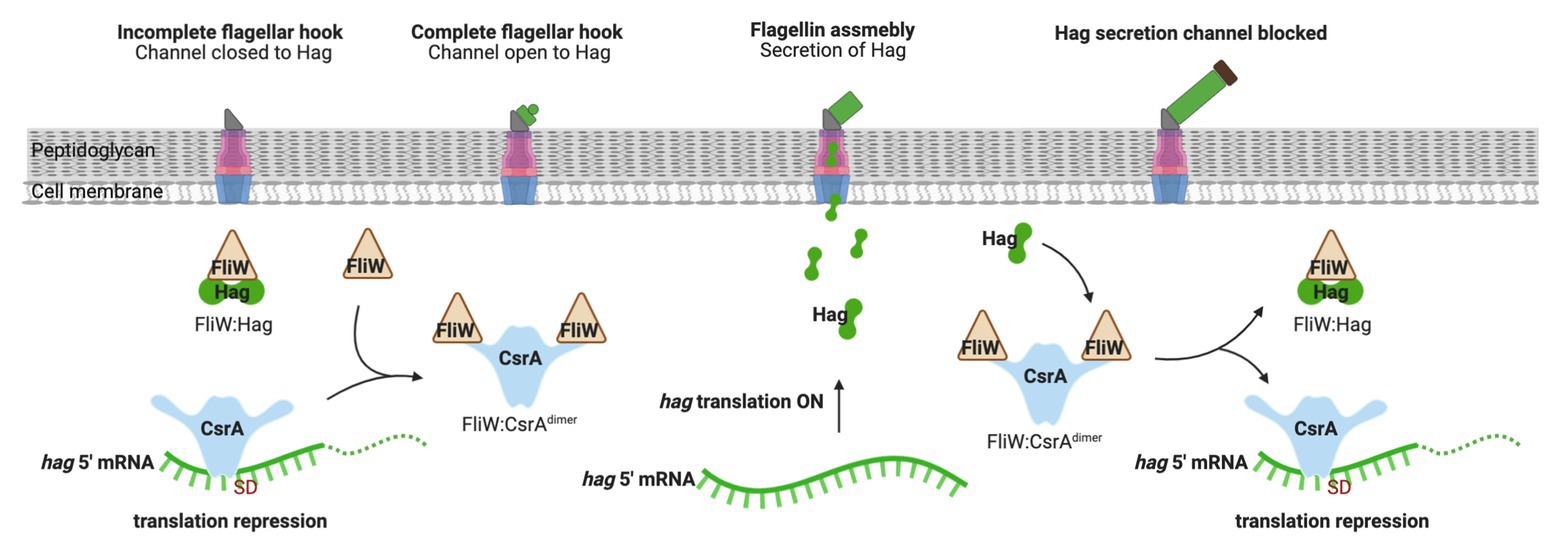
Figure 4. Regulation of flagellum biosynthesis in Bacillus subtilis by a partner-switching mechanism involving CsrA, Hag, and FliW. Prior to flagellar hook assembly, FliW is bound in a complex with cytoplasmic Hag (flagellin) protein and free CsrA represses hag translation by occluding ribosome access. Once assembly of the flagellar hook structure is completed, Hag is actively secreted to assemble the flagellum filament and FliW is released. Free FliW binds to CsrA, derepessing hag translation to allow flagellin synthesis. Upon capping of the channel, intracellular levels of Hag increase. Hag then binds to FliW, which releases CsrA to inhibit hag translation. This figure was created with BioRender.com.
In enteropathogenic E. coli (EPEC), CsrA regulates genes of the locus of enterocyte effacement, the Lee locus, which are involved in the Type III secretion system (T3SS) of this bacterium (Bhatt et al., 2009). Furthermore, CsrA binds to CesT in this bacterium, a chaperone protein needed for colonization of host intestinal epithelial cells via the T3SS (Katsowich et al., 2017). Although FliW and CesT are both protein antagonists of CsrA, they employ distinct mechanisms for performing this role. The CesT binding region in CsrA extensively overlaps the RNA-binding pockets, which is indicative of a competitive antagonism, similar to that of the Csr sRNAs (Ye et al., 2018). Furthermore, the CsrA-CesT interaction was apparent only at high local concentrations of CesT, suggesting that it enables bacteria to redirect gene expression after the effector proteins with which CesT interacts, especially Tir, have been injected into the host (Ye et al., 2018; Elbaz et al., 2019). Because the regulatory interactions of Tir-CesT-CsrA are responsible for the remodeling of virulence and metabolic gene expression, this regulation is likely required for EPEC survival at the host intestinal epithelium.
As described above, CsrA activity in E. coli is regulated by sequestration by its sRNA antagonists, CsrB and CsrC. The short half-lives of these RNAs (Gudapaty et al., 2001; Weilbacher et al., 2003; Leng et al., 2016) allow for rapid adjustments of CsrA activity in response to factors affecting sRNA transcription and stability. Antagonism of the stable CsrA protein by these RNAs occurs even during growth arrest, making this regulation robust to variations in growth rate (Adamson and Lim, 2013). Additionally, the presence of two negative regulators of CsrA activity implies a functional redundancy, which is known to decrease nongenetic noise and increase regulatory precision (Kafri et al., 2006). In both E. coli and S. enterica serovar Typhimurium, transcription of csrB (but not csrC) is activated by integration host factor (IHF), suggesting that the sRNAs may also have distinct regulatory roles in some species (Martínez et al., 2014; Zere et al., 2015).
The Escherichia coli Csr system is comprised of multiple positive and negative feedback loops that tightly control the levels of each molecular component (Figure 5). For example, CsrA indirectly activates transcription of its sRNA antagonists through its effects on the BarA-UvrY TCS (Gudapaty et al., 2001; Suzuki et al., 2002; Weilbacher et al., 2003). Phosphorylated UvrY activates expression of CsrB/C, and CsrA has positive effects on uvrY expression at both the transcriptional and translational levels (Camacho et al., 2015; Zere et al., 2015). CsrA is also necessary for proper switching of the membrane-bound BarA protein from its phosphatase to kinase activity (Camacho et al., 2015). Because CsrB and CsrC depend on free CsrA for their synthesis while also antagonizing its activity, disruption of the expression of either regulatory RNA results in a compensatory increase in the level of the other (Weilbacher et al., 2003; Suzuki et al., 2006). Additionally, CsrA in both E. coli and Salmonella indirectly stabilizes CsrB/C by repressing the expression of CsrD, which targets these antagonists for RNase E-dependent degradation (Suzuki et al., 2006; Jonas et al., 2008). These negative feedback loops support rapid signal propagation and demonstrably reduce the time required for the system to achieve a steady state (Adamson and Lim, 2013).
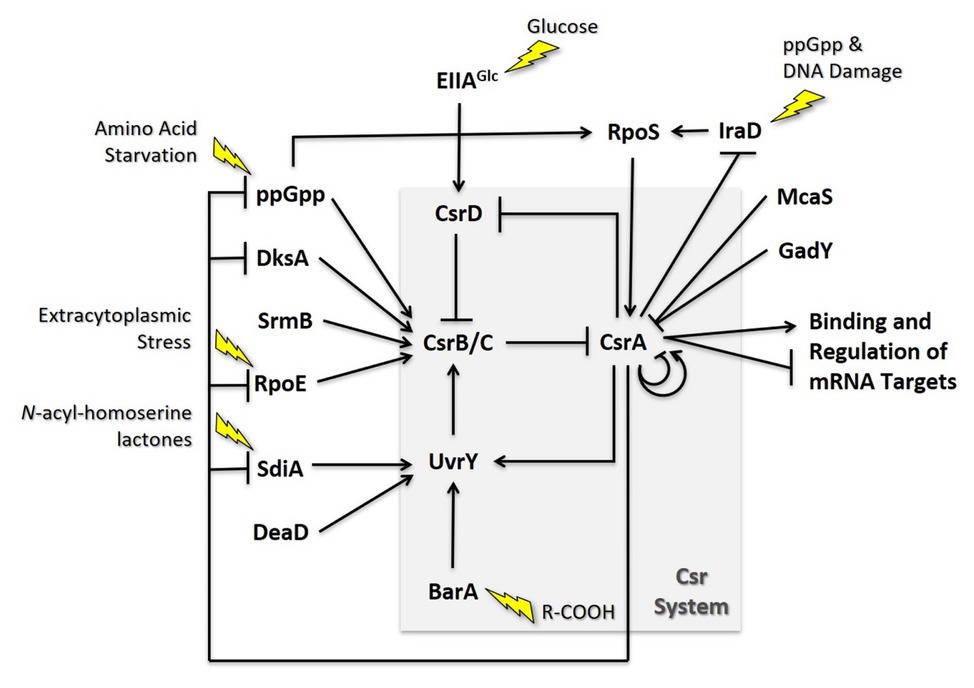
Figure 5. Regulatory circuitry impacting the Csr system of E. coli. These interactions are discussed in the text.
CsrA also directly and indirectly regulates its own expression. In Escherichia coli, csrA expression is complex, involving five promoters, two sigma factors, and four CsrA binding sites. Transcription from σs-dependent promoter 3 is indirectly activated by CsrA and is largely responsible for the substantial increase in csrA expression when cells transition to stationary-phase growth. Furthermore, CsrA binds to four sites in its own leader, one of which overlaps the SD sequence, and thereby inhibits its own translation (Yakhnin et al., 2011b). This incoherent feedforward loop likely enhances CsrA expression in response to σs-inducing stressors while providing another mechanism for rapid repression when free CsrA levels are high. The structure of the Csr system and its interrelated regulatory pathways suggest a multifaceted arrangement of autoregulation, a common characteristic of systems that maintain homeostasis. Its complex regulatory circuitry involves multiple network motifs that tightly control the levels and/or activity of each molecular component. This arrangement likely reduces cell-to-cell variability under a given growth condition, supports rapid intracellular signaling, and may limit stochastic fluctuations in a system responsible for both responding to and coordinating the influence other signaling networks (Alon, 2007; Adamson and Lim, 2013; Potts et al., 2017).
Among the most striking findings to come to light from transcriptomics and other systems-based studies are that CsrA regulates the expression of dozens of transcription factors and other regulatory genes (Edwards et al., 2011; Potts et al., 2017, 2018, 2019; Sowa et al., 2017). These effects expand the regulatory influence of CsrA via indirect effects on the structural genes that respond to the CsrA-controlled regulators. These findings illustrate the importance of CsrA, and posttranscriptional regulation in general, in determining the complex structure of bacterial regulatory networks. Although the biology of many of these regulatory interactions has not been elucidated, a few examples of how the Csr system interacts with other regulatory factors and systems have been studied. These include direct roles of CsrA in controlling expression of hfq, pnp, sdiA, and nhaR (Baker et al., 2007; Yakhnin et al., 2011a; Pannuri et al., 2012; Park et al., 2015), as well as examples in which Csr system components have been shown to both control and be controlled by other regulators.
CsrA was one of the first regulators of bacterial biofilm formation to be identified (Romeo et al., 1993; Jackson et al., 2002). Since that time, it has been found to play a complex regulatory role in biofilm formation, accomplished by interactions at several levels (Figure 6A). The major influence of CsrA is to act as a posttranscriptional repressor of the pgaABCD operon, which is needed for the biosynthesis and secretion of an adhesive glycosaminoglycan, variously referred to as PGA, PNAG, dPNAG, or PIA (Wang et al., 2004, 2005, 2016). As described above, binding to pgaA leader mRNA leads to repression of its translation, destabilization, and transcription termination. Besides interacting directly with pgaABCD mRNA, CsrA represses the translation of a LysR-family transcription factor, NhaR, which binds upstream of the single pgaABCD promoter and activates transcription in response to high pH and [Na+] (Goller et al., 2006; Pannuri et al., 2012). Finally, CsrA represses the expression of genes needed for production of c-di-GMP, an alarmone that allosterically activates the polymerization of PGA by the PgaCD protein and, thus, enhances biofilm formation (Jonas et al., 2008; Steiner et al., 2013). These findings demonstrate that CsrA effects multi-tier regulation of PGA biosynthesis and biofilm circuitry (Romeo et al., 2013; Figure 6A).
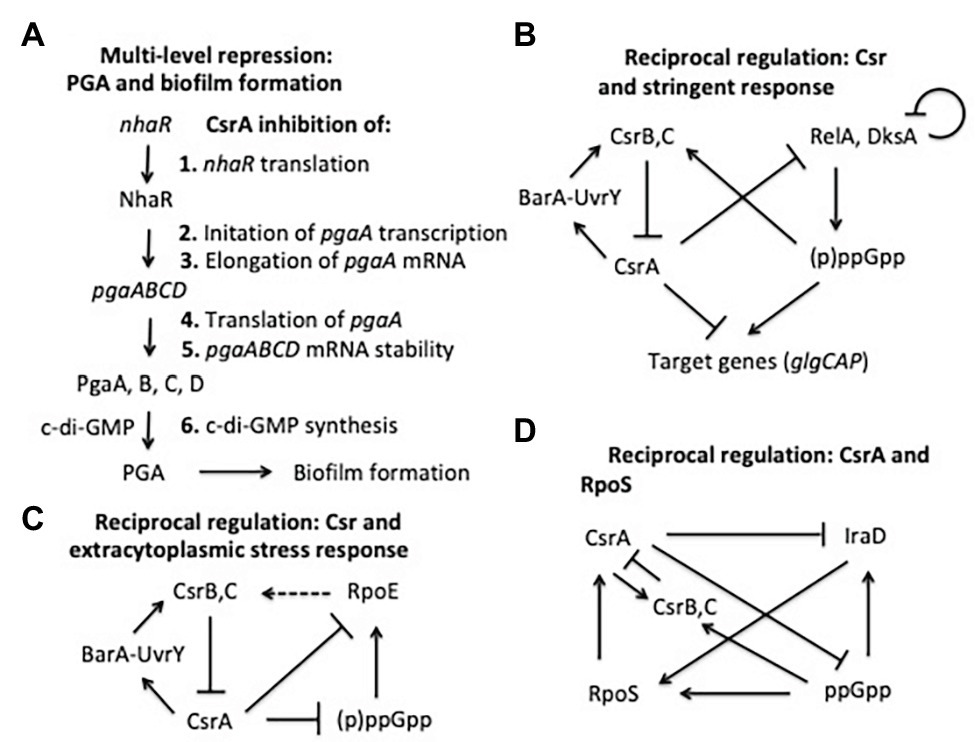
Figure 6. Regulatory circuits involving CsrA interactions with other regulators. The models shown here are described in the text.
As previously discussed, the regulation of flhDC expression in E. coli represented the first mechanism of genetic activation by CsrA to be elucidated. The flhDC operon encodes the two subunits of a transcription factor, FlhD4C2 that initiates the three-tier cascade of gene expression needed for temporal development of the flagellum and chemotaxis in E. coli and its relatives (Lee et al., 2011; Fitzgerald et al., 2014). A csrA::kan transposon mutation was found to eliminate flagellum production and motility by reducing the expression of flhDC below the threshold necessary for acquiring motility (Wei et al., 2001). Importantly, this defect could be corrected by ectopic expression of csrA, as well as flhDC, suggesting that the main role of CsrA in motility is to activate flhDC expression. Nevertheless, transcriptomics studies revealed that flhDC mRNA is not the only target of CsrA that is involved in E. coli motility; CsrA activates the expression of multiple motility genes (Potts et al., 2017). FlhD4C2 itself directly activates transcription of second tier genes of the motility cascade of gene expression, including fliA, which encodes σ28. This alternative sigma factor is required for transcription of the third tier of motility genes. The direct effects of CsrA on FlhD4C2 may impact the expression of all motility genes. However, CLIP-seq analyses and CsrA:RNA co-purification followed by RNA-seq also identified transcripts of a variety of flagellar genes in addition to flhDC as probable direct targets of CsrA binding (Edwards et al., 2011; Potts et al., 2017). These include the regulatory genes fliA (σ28) and flgK, a morphogene involved in controlling the switch from secretion of hook proteins to filament proteins during flagellum assembly. Together, these findings indicate that CsrA plays a complex role as a multi-tier regulator within the motility cascade, with biological implications remaining to be determined.
As mentioned above, the stringent response system, based on the nucleotide alarmone (p)ppGpp, stimulates transcription of CsrB and CsrC sRNAs. In addition, CsrA represses relA and dksA expression, and (p)ppGpp levels are elevated during the stringent response in a csrA mutant relative to the WT background (Edwards et al., 2011). These regulatory effects define reciprocal regulatory circuitry, through which the Csr system reinforces the effects of the stringent response system. Expression of CsrB/C is activated by ppGpp when cells are starved for amino acids or other nutrients, thus reducing CsrA activity, increasing relA expression and the potential for additional (p)ppGpp production (Jonas and Melefors, 2009; Edwards et al., 2011; Figure 6B). Similar to ppGpp, DksA acts by binding to RNA polymerase, and often potentiates the effects of ppGpp (Paul et al., 2004; Aberg et al., 2009). CsrA also posttranscriptionally represses expression of dksA, although the DksA protein feedback inhibits its own transcription, which can mask the effects of CsrA (Chandrangsu et al., 2011; Edwards et al., 2011).
Another example in which the Csr system interacts reciprocally with a stress response system is found in the extracytoplasmic stress response system (Figure 6C). The extracytoplasmic stress system detects misfolded proteins in the periplasm, relying on the sigma factor σE (RpoE) for transcription of stress response genes when this occurs (Ades, 2008; Yakhnin et al., 2017). In the absence of stress, σE is largely sequestered at the cytoplasmic membrane by its interaction with the membrane-bound anti-sigma factor RseA. Upon stress, proteolytic cleavage of RseA leads to release of σE, making it available for interaction with the RNA polymerase core enzyme. RNA-seq studies first suggested that CsrA binds to the rpoE transcript (Edwards et al., 2011). Biochemical studies revealed that CsrA binds to three sites in rpoE mRNA, two of which overlap the SD sequence and the initiation codon, leading to repression of translation (Yakhnin et al., 2017). In addition, RpoE indirectly activates transcription of CsrB and CsrC sRNAs, although the mechanisms responsible for this remain to be determined. This circuitry was seen as increasing CsrB/C synthesis during extracytoplasmic stress, thus limiting CsrA activity and leading to an increase in rpoE translation. When the stress has been resolved, CsrB/C levels should decline and CsrA activity should increase, helping to reestablish σE levels to a pre-stress state. Because (p)ppGpp and DksA activate expression of rpoE independently of RseA under nutrient limitation (Costanzo and Ades, 2006; Costanzo et al., 2008), this pathway via σE may also contribute to the increase in CsrB/C sRNA levels during the stringent response (Edwards et al., 2011; Zere et al., 2015). Restoration of nutrients should decrease (p)ppGpp levels, helping to reset the system.
Among the most global of the E. coli regulatory factors is the alternative sigma factor σs (RpoS), which activates gene expression upon entry into the stationary phase of growth and exposure to a variety of physicochemical stresses (Battesti et al., 2011). Regulation of σs levels is complex and multifactorial, including inhibition of its turnover by anti-adapter proteins that prevent the RssB adapter protein from triggering its cleavage by ClpXP protease under a variety of stresses. Expression of the anti-adapter protein IraD is activated by ppGpp accumulation and DNA damage (Merrikh et al., 2009a,b). CsrA was found to interact in vivo with the iraD transcript by using CLIP-seq analysis and was seen to inhibit its expression (Potts et al., 2017). This regulation involved a new mechanism, in which CsrA directly repressed the translation of a short open reading frame, ORF27 (idlP), located immediately upstream and slightly overlapping the iraD coding region (Park et al., 2017). Repression of iraD translation by CsrA was mediated entirely through translational coupling of iraD to idlP. RpoS acts at two of the five csrA promoters, activating CsrA transcription upon entry into stationary phase (Yakhnin et al., 2011b). CsrB and CsrC sRNAs also accumulate at this time, each of which can sequester multiple CsrA proteins (Gudapaty et al., 2001; Weilbacher et al., 2003). The latter effect seemingly dominates, such that overall CsrA activity decreases while CsrA protein accumulates. Thus, RpoS should be stabilized in the stationary phase via effects of the Csr system on IraD. Similarly, under stress-related induction of ppGpp production, CsrA expression should be activated via RpoS. Coordinately, CsrA activity will be decreased by the ppGpp-dependent increase in the accumulation of CsrB/C (Edwards et al., 2011), which should override the increase in CsrA protein. Activation of CsrA transcription via RpoS as cultures enter stationary phase would seem to ensure that sufficient CsrA will be available to rapidly restore gene expression needed for growth resumption upon replenishment of nutrients or relief of stress, although this remains to be demonstrated experimentally (Figure 6D).
A final example of reciprocal regulatory interactions of Csr with other regulatory systems involves the E. coli N-acyl-homoserine lactone (HSL) receptor SdiA (Dyszel et al., 2010). As described above, CsrA represses translation of sdiA, while genetic studies indicate that SdiA activates transcription of the gene encoding UvrY, the key transcriptional activator of csrB/C (Suzuki et al., 2002). The resulting feedback activation loop is consistent with autoinduction, a well-described feature of HSLs that mediate quorum sensing. However, SdiA is an orphan HSL receptor in E. coli and closely related species, which cannot synthesize HSLs. While SdiA maintains weak activity in the absence of HSL, binding to cognate HSLs of other species improves its function and permits sensing of the microbial environment (Smith and Ahmer, 2003; Dyszel et al., 2010; Venturi and Ahmer, 2015). The finding that HSLs produced by other species act as a signal to E. coli via SdiA, with important effects on physiology (Hughes et al., 2010; Sperandio, 2010; Lu et al., 2017), suggests that the Csr system may participate in the regulation of bacterial interspecies communications, in addition to its well-studied roles in virulence and host-microbe interactions (Vakulskas et al., 2015).
Systems-based methodologies provide powerful approaches for investigating the roles of global regulatory factors, such as CsrA. Integrated systems approaches, which combine two or more complementary techniques applied to isogenic strains, grown under uniform conditions, have proven especially useful for generating hypotheses to guide the discovery of new regulatory roles, mechanisms, and circuitry. Integrated transcriptomics allowed global regulatory effects of CsrA on transcript levels, translation, and/or RNA stability to be analyzed in the context of the RNA binding interactions that may underlie the effects. While most of the vast information gathered in such studies needs to be confirmed and extended before the causal mechanisms and circuitry are understood, the implications of such findings for the regulatory landscape of CsrA are profound.
A study using RNA-seq to identify cellular RNAs that copurify with CsrA, coupled with proteomics analyses to determine the global effects of a csrA mutation on protein levels provided the impetus for investigating the interconnecting roles of the Csr and stringent response systems, described above (Edwards et al., 2011). A more recent integrated transcriptomics study combined CLIP-Seq for monitoring in vivo RNA binding interactions of CsrA with ribosome profiling to quantitatively monitor the positioning of translating ribosomes and RNA-Seq to reveal both steady state transcript levels and transcript stability in WT and csrA mutant strains (Potts et al., 2017). This study uncovered many new potential roles of CsrA, e.g., in regulating iron metabolism, toxin-antitoxin systems, membrane homeostasis, and expression of many regulatory factors such as global transcription factors and sRNAs. Evidence that CsrA binds in vivo to mRNA encoding iraD and affects the level of this transcript compelled the iraD studies described above (Park et al., 2017). CsrA affected the expression of several genes involved in iron homeostasis (Potts et al., 2017). A follow-up study used in vivo and in vitro approaches to demonstrate that CsrA binding interactions directly repress the translation of genes for three iron storage proteins, ftnB, bfr, and dps, which are expressed in the stationary phase of growth and affect oxidative stress. In addition, repression of ftnB and bfr by CsrA was found to be essential for normal cell growth under iron-limiting conditions (Pourciau et al., 2019).
The integrated transcriptomics study also provided evidence that CsrA regulates diverse new targets including transcriptional regulators, RNases, and sRNAs (Potts et al., 2017). Transcriptional factors identified (e.g., fliA, lrp, cra, and soxS) regulate distinct physiological properties ranging from motility, amino acid metabolism, and central carbon metabolism, to resistance to a variety of stresses. Lrp alone regulates at least 10% of E. coli’s genome (Tani et al., 2002), suggestive of a major new route of global regulation by CsrA. The finding that CsrA regulated genes for ribonucleases (including rng, rnb, and orn), which along with its known role in regulating pnp (Park et al., 2015), may help to explain the contrasting negative effects of CsrA on overall RNA translation and abundance vs. its action as a stabilizer of total RNA decay (Esquerré et al., 2016; Potts et al., 2017, 2018). Of the new sRNA interactions identified, CsrA bound with high affinity to GadY, Spot42, GcvB, and MicL (Potts et al., 2017). These findings call for research into the physiological and mechanistic implications and the potential CsrA-dependent regulatory circuits that they create.
Another recent integrated transcriptomics approach demonstrated the flexibility of CsrA as a condition-specific regulator in Salmonella. The study employed ribosome profiling and RNA-seq to examine the effects of CsrA on ribosome occupancy, transcript levels, and transcript stability in the rich medium Luria Broth (LB) and the acidic and nutrient limiting medium mLPM (Potts et al., 2019). These media favor the expression of genes in Salmonella pathogenicity islands SPI-1 and SPI-2, respectively, which are primarily associated with growth in the intestine vs. the Salmonella-containing vacuole of the macrophage. While certain genes were regulated under both growth conditions, others were subject to condition-specific regulation by CsrA, including key regulators involved in stress and virulence such as rpoS, slyA, and spvR. The results from this study illustrated how the Csr system may act as a physiological switch in responding to external stimuli, with potential regulatory consequences for virulence, metabolism, and stress responses.
CLIP-seq studies from both E. coli and Salmonella revealed the unexpected finding that CsrA binds not only to mRNA leader regions but also binds extensively within the coding regions of mRNAs (Holmqvist et al., 2016; Potts et al., 2017). Furthermore, ribosome pause sites were significantly enriched near CsrA crosslinking sites (Potts et al., 2017). Because the ribosome profiling pattern was unaltered in a csrA mutant strain, CsrA does not grossly affect ribosome pausing. However, ribosome pausing may facilitate CsrA binding to sites that would otherwise be protected by active translation. Whether CsrA binding near ribosome pause sites has regulatory consequences is of considerable interest, as ribosome pausing occurs during nutrient limitation and other stresses (Subramaniam et al., 2014), and might provide a means of altering the availability of free CsrA protein under such conditions.
An approach combining RNA-seq with epistasis analysis (Epi-seq) was used to distinguish two alternative models for how the CsrD protein exerts global effects on gene expression: by altering the stability of CsrB/C RNAs and, thus, CsrA activity (described above), or by acting as a transcription factor that regulates numerous transcripts, as proposed based on observations from a transcriptomics study (Esquerré et al., 2016). Because global effects of CsrA on transcripts remained mostly unaltered by deletion of csrD, while disruption of csrA eliminated all effects of csrD and deletion of csrB and csrC eliminated almost all effects, it was concluded that the CsrD acts as a global regulator through its role in CsrB/C sRNA decay.
Metabolomics studies of CsrA have revealed striking changes in metabolite levels upon csrA disruption, some of which were predictable from earlier investigations, but many of which were unanticipated. Studies in E. coli K-12 showed that metabolites in glycolysis upstream of phosphofructokinase (PfkA) accumulate (Morin et al., 2016), in agreement with CsrA effects on this enzyme and its transcript (Sabnis et al., 1995; Potts et al., 2017). Studies in E. coli Nissle 1917 revealed that the influence of CsrA on metabolite levels and metabolic flux were conditional, differing dependent upon the metabolic fate of the carbon source that was used for growth (Revelles et al., 2013). A study conducted in EPEC showed that deletion of csrA resulted in a significant change in over half of the metabolites observed (Berndt et al., 2019). In line with previous work conducted in CsrA transposon insertion mutants, the deletion led to an accumulation of glycogen and fructose-6-phosphate (Berndt et al., 2019).
An unanticipated finding from the EPEC metabolomics study was that nucleotides were depleted, while nucleosides and nucleobases accumulated in the csrA mutant. These changes were associated with a corresponding drop in expression of nucleotide synthesis genes such as cdd, rihB, and deoA (Berndt et al., 2019). These findings might help to explain why csrA deletion mutants often have severe growth defects or are unable to grow (Altier et al., 2000; Timmermans and Van Melderen, 2009; Mey et al., 2015). Furthermore, while aromatic amino acids were depleted, an intermediate in their synthesis, shikimate was observed to have accumulated drastically and the expression of shikimate kinase (AroL) dropped significantly. Finally, colanic acid accumulation was accompanied by a large increase in the expression of genes in the colanic biosynthesis pathway in the csrA mutant (Berndt et al., 2019). The precise mechanisms for these metabolic alterations are not fully understood.
Posttranscriptional regulation by the RNA-binding protein CsrA in E. coli and related bacterial Csr systems is crucial for maintenance of robust growth and for orchestrating major lifestyle decisions in response to nutritional and stress conditions. Binding interactions of CsrA-family proteins with RNA targets are known in atomic detail and give rise to diverse regulatory mechanisms. Interactions of CsrA with Csr-family sRNAs have long been known to sequester this protein and, thus, modulate its activity and the mechanisms and physiology behind Csr sRNA synthesis and turnover are partially understood. In contrast, CsrA interactions with multiple basepairing sRNAs, recently identified by transcriptomics analyses, are poorly understood, requiring investigation of both mechanisms and biological functions. The recent discovery of extensive binding of CsrA deep within coding regions of mRNAs in proximity to ribosome pause sites also requires further examination. CsrA regulates dozens of regulators and the Csr system has been found to participate in complex circuitry with some of these regulators. These interactions extend the regulatory reach of the Csr system and allow the CsrA regulon to respond to broader aspects of physiology and metabolism. While Csr systems are among the best-known bacterial posttranscriptional regulators, many new discoveries will be required for a full appreciation of their pervasive roles.
TR conceived and outlined this review. All authors contributed to the article and approved the submitted version.
Support was provided by NIH R01 NIGMS 059969.
The authors declare that this review was written in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We wish to thank our students, postdocs, and collaborators who contributed to many of the findings discussed in this review.
Abbott, Z. D., Yakhnin, H., Babitzke, P., and Swanson, M. S. (2015). csrR, a paralog and direct target of CsrA, promotes Legionella pneumophila resilience in water. mBio 6:e00595. doi: 10.1128/mBio.00595-15
Aberg, A., Fernández-Vázquez, J., Cabrer-Panes, J. D., Sánchez, A., and Balsalobre, C. (2009). Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J. Bacteriol. 191, 3226–3236. doi: 10.1128/JB.01410-08
Adamson, D. N., and Lim, H. N. (2013). Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation. Proc. Natl. Acad. Sci. U. S. A. 110, 13120–13125. doi: 10.1073/pnas.1308476110
Ades, S. E. (2008). Regulation by destruction: design of the sigmaE envelope stress response. Curr. Opin. Microbiol. 11, 535–540. doi: 10.1016/j.mib.2008.10.004
Alon, U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461. doi: 10.1038/nrg2102
Altegoer, F., Rensing, S. A., and Bange, G. (2016). Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein. Proc. Natl. Acad. Sci. U. S. A. 113, 10168–10173. doi: 10.1073/pnas.1602425113
Altier, C., Suyemoto, M., and Lawhon, S. D. (2000). Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect. Immun. 68, 6790–67977. doi: 10.1128/iai.68.12.6790-6797.2000
Babitzke, P., Baker, C. S., and Romeo, T. (2009). Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol. 63, 27–44. doi: 10.1146/annurev.micro.091208.073514
Babitzke, P., Lai, Y. -J., Renda, A. J., and Romeo, T. (2019). Posttranscription initiation control of gene expression mediated by bacterial RNA-binding proteins. Annu. Rev. Microbiol. 73, 43–67. doi: 10.1146/annurev-micro-020518-115907
Babitzke, P., and Romeo, T. (2007). CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10, 156–163. doi: 10.1016/j.mib.2007.03.007
Baker, C. S., Eöry, L. A., Yakhnin, H., Mercante, J., Romeo, T., and Babitzke, P. (2007). CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 189, 5472–5481. doi: 10.1128/JB.00529-07
Baker, C. S., Morozov, I., Suzuki, K., Romeo, T., and Babitzke, P. (2002). CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44, 1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Berndt, V., Beckstette, M., Volk, M., Dersch, M., and Brönstrup, M. (2019). Metabolome and transcriptome-wide effects of the carbon storage regulator A in enteropathogenic Escherichia coli. Sci. Rep. 9:138. doi: 10.1038/s41598-018-36932-w
Bhatt, S., Edwards, A. N., Nguyen, H. T., Merlin, D., Romeo, T., and Kalman, D. (2009). The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect. Immun. 77, 3552–3568. doi: 10.1128/IAI.00418-09
Camacho, M. I., Alvarez, A. F., Chavez, R. G., Romeo, T., Merino, E., and Georgellis, D. (2015). Effects of the global regulator CsrA on the BarA/UvrY two-component signaling system. J. Bacteriol. 197, 983–991. doi: 10.1128/JB.02325-14
Chandrangsu, P., Lemke, J. J., and Gourse, R. L. (2011). The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol. Microbiol. 80, 1337–1348. doi: 10.1111/j.1365-2958.2011.07649.x
Chavez, R. G., Alvarez, A. F., Romeo, T., and Georgellis, D. (2010). The physiological stimulus for the BarA sensor kinase. J. Bacteriol. 192, 2009–2012. doi: 10.1128/JB.01685-09
Costanzo, A., and Ades, S. E. (2006). Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3',5'-bispyrophosphate (ppGpp). J. Bacteriol. 188, 4627–4634. doi: 10.1128/JB.01981-05
Costanzo, A., Nicoloff, H., Barchinger, S. E., Banta, A. B., Gourse, R. L., and Ades, S. E. (2008). ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor σE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67, 619–632. doi: 10.1111/j.1365-2958.2007.06072.x
Dubey, A. K., Baker, C. S., Romeo, T., and Babitzke, P. (2005). RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11, 1579–1587. doi: 10.1261/rna.2990205
Dubey, A. K., Baker, C. S., Suzuki, K., Jones, A. D., Pandit, P., Romeo, T., et al. (2003). CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185, 4450–4460. doi: 10.1128/jb.185.15.4450-4460.2003
Dugar, G., Svensson, S. L., Bischler, T., Wäldchen, S., Reinhardt, R., Sauer, M., et al. (2016). The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun. 7:11667. doi: 10.1038/ncomms11667
Duss, O., Michel, E., Diarra dit Konté, N., Schubert, M., and Allain, F. H. (2014a). Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition. Nucleic Acids Res. 42, 5332–5346. doi: 10.1093/nar/gku141
Duss, O., Michel, E., Yulikov, M., Schubert, M., Jeschke, G., and Allain, F. H. (2014b). Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 509, 588–592. doi: 10.1038/nature13271
Dyszel, J. L., Soares, J. A., Swearingen, M. C., Lindsay, A., Smith, J. N., and Ahmer, B. M. (2010). E. coli K-12 and EHEC genes regulated by SdiA. PLoS One 5:e8946. doi: 10.1371/journal.pone.0008946
Edwards, A. N., Patterson-Fortin, L. M., Vakulskas, C. A., Mercante, J. W., Potrykus, K., Vinella, D., et al. (2011). Circuitry linking the Csr and stringent resonse global regulatory systems. Mol. Microbiol. 80, 1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x
Elbaz, N., Socol, Y., Katsowich, N., and Rosenshine, I. (2019). Control of Type III secretion system effector/chaperone ratio fosters pathogen adaptation to host-adherent lifestyle. mBio 10:e02074–19. doi: 10.1128/mBio.02074-19
Esquerré, T., Bouvier, M., Turlan, C., Carpousis, A. J., Girbal, L., and Cocaign-Bousquet, M. (2016). The Csr system regulates genome-wide mRNA stability and transcription and thus gene expression in Escherichia coli. Sci. Rep. 6:25057. doi: 10.1038/srep25057
Figueroa-Bossi, N., Schwartz, A., Guillemardet, B., D’Heygère, F., Bossi, L., and Boudvillain, M. (2014). RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 28, 1239–1251. doi: 10.1101/gad.240192.114
Fitzgerald, D. M., Bonocora, R. P., and Wade, J. T. (2014). Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 10:e1004649. doi: 10.1371/journal.pgen.1004649
Gebhardt, M. J., Kambara, T. K., Ramsey, K. M., and Dove, S. L. (2020). Widespread targeting of nascent transcripts by RsmA in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 117, 10520–10529. doi: 10.1073/pnas.1917587117
Goller, C., Wang, X., Itoh, Y., and Romeo, T. (2006). The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 188, 8022–8032. doi: 10.1128/JB.01106-06
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Gudapaty, S., Suzuki, K., Wang, X., Babitzke, P., and Romeo, T. (2001). Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 183, 6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001
Gutiérrez, P., Li, Y., Osborne, M. J., Pomerantseva, E., Liu, Q., and Gehring, K. (2005). Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J. Bacteriol. 187, 3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005
Hadjeras, L., Poljak, L., Bouvier, M., Morin-Ogier, Q., Canal, I., Cocaign-Bousquet, M., et al. (2019). Detachment of the RNA degradosome from the inner membrane of Escherichia coli results in a global slowdown of mRNA degradation, proteolysis of RNase E and increased turnover of ribosome-free transcripts. Mol. Microbiol. 111, 1715–1731. doi: 10.1111/mmi.14248
Heeb, S., Blumer, C., and Haas, D. (2002). Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184, 1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002
Heeb, S., Kuehne, S. A., Bycroft, M., Crivii, S., Allen, M. D., Haas, D., et al. (2006). Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 355, 1026–1036. doi: 10.1016/j.jmb.2005.11.045
Holmqvist, E., Wright, P. R., Li, L., Bischler, T., Barquist, L., Reinhardt, R., et al. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011. doi: 10.15252/embj.201593360
Hughes, D. T., Terekhova, D. A., Liou, L., Hovde, C. J., Sahl, J. W., Patankar, A. V., et al. (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U. S. A. 107, 9831–9836. doi: 10.1073/pnas.1002551107
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. doi: 10.1111/j.1365-2958.2010.07320.x
Itoh, Y., Rice, J. D., Goller, C., Pannuri, A., Taylor, J., Meisner, J., et al. (2008). Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 190, 3670–3680. doi: 10.1128/JB.01920-07
Jackson, D. W., Suzuki, K., Oakford, L., Simecka, J. W., Hart, M. E., and Romeo, T. (2002). Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184, 290–301. doi: 10.1128/jb.184.1.290-301.2002
Jonas, K., Edwards, A. N., Simm, R., Romeo, T., Römling, U., and Melefors, O. (2008). The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70, 236–257. doi: 10.1111/j.1365-2958.2008.06411.x
Jonas, K., and Melefors, O. (2009). The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiol. Lett. 297, 80–86. doi: 10.1111/j.1574-6968.2009.01661.x
Jørgensen, M. G., Thomason, M. K., Havelund, J., Valentin-Hansen, P., and Storz, G. (2013). Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 27, 1132–1145. doi: 10.1101/gad.214734.113
Kafri, R., Levy, L., and Pilpel, Y. (2006). The regulatory utilization of genetic redundancy through responsive backup circuits. Proc. Natl. Acad. Sci. U. S. A. 103, 11653–11658. doi: 10.1073/pnas.0604883103
Katsowich, N., Elbaz, N., Pal, R. R., Mills, E., Kobi, S., Kahan, T., et al. (2017). Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria. Science 355, 735–739. doi: 10.1126/science.aah4886
Kay, E., Dubuis, C., and Haas, D. (2005). Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. U. S. A. 102, 17136–17141. doi: 10.1073/pnas.0505673102
Kim, M., and Kim, K. -S. (2017). Stress-responsively modulated ymdAB-clsC operon plays a role in biofilm formation and apramycin susceptibility in Escherichia coli. FEMS Microbiol. Lett. 364, 1–9. doi: 10.1093/femsle/fnx114
Lee, Y. Y., Barker, C. S., Matsumura, P., and Belas, R. (2011). Refining the binding of the Escherichia coli flagellar master regulator, FlhD4C2, on a base-specific level. J. Bacteriol. 193, 4057–4068. doi: 10.1128/JB.00442-11
Leng, Y., Vakulskas, C. A., Zere, T. R., Pickering, B. S., Watnick, P. I., Babitzke, P., et al. (2016). Regulation of CsrB/C sRNA decay by EIIA(Glc) of the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol. 99, 627–639. doi: 10.1111/mmi.13259
Li, J., Gulbronson, C. J., Bogacz, M., Hendrixson, D. R., and Thompson, S. A. (2018). FliW controls growth-phase expression of Campylobacter jejuni flagellar and non-flagellar proteins via the post-transcriptional regulator CsrA. Microbiology 164, 1308–1319. doi: 10.1099/mic.0.000704
Liu, Y., Cui, Y., Mukherjee, A., and Chatterjee, A. K. (1998). Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol. Microbiol. 29, 219–234. doi: 10.1046/j.1365-2958.1998.00924.x
Liu, M. Y., Gui, G., Wei, B., Preston, J. F. 3rd, Oakford, L., Yüksel, U., et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. doi: 10.1074/jbc.272.28.17502
Liu, M. Y., and Romeo, T. (1997). The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179, 4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997
Liu, M. Y., Yang, H., and Romeo, T. (1995). The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177, 2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995
Lu, Y., Zeng, J., Wu, B., E, S., Wang, L., Cai, R., et al. (2017). Quorum sensing N-acyl homoserine lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front. Cell. Infect. Microbiol. 7:7. doi: 10.3389/fcimb.2017.00007
Martínez, L. C., Martínez-Flores, I., Salgado, H., Fernández-Mora, M., Medina-Rivera, A., Puente, J. L., et al. (2014). In silico identification and experimental characterization of regulatory elements controlling the expression of the Salmonella csrB and csrC genes. J. Bacteriol. 196, 325–336. doi: 10.1128/JB.00806-13
Martínez, L. C., Yakhnin, H., Camacho, M. I., Georgellis, D., Babitzke, P., Puente, J. L., et al. (2011). Integration of a complex regulatory cascade involving the SirA/BarA and Cssr global regulatory systems that controls expression of the Salmonella SP-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol 80, 1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x
Mehta, P., Goyal, S., and Wingreen, N. S. (2008). A quantitative comparison of sRNA-based and protein-based gene regulation. Mol. Syst. Biol. 4:221. doi: 10.1038/msb.2008.58
Mercante, J., Edwards, A. N., Dubey, A. K., Babitzke, P., and Romeo, T. (2009). Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J. Mol. Biol. 392, 511–528. doi: 10.1016/j.jmb.2009.07.034
Mercante, J., Suzuki, K., Cheng, X., Babitzke, P., and Romeo, T. (2006). Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 281, 31832–31842. doi: 10.1074/jbc.M606057200
Merrikh, H., Ferrazzoli, A. E., Bougdour, A., Olivier-Mason, A., and Lovett, S. T. (2009a). A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc. Natl. Acad. Sci. U. S. A. 106, 611–616. doi: 10.1073/pnas.0803665106
Merrikh, H., Ferrazzoli, A. E., and Lovett, S. T. (2009b). Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli. J. Bacteriol. 191, 7436–7446. doi: 10.1128/JB.00412-09
Mey, A. R., Butz, H. A., and Payne, S. M. (2015). Vibrio cholerae CsrA regulates ToxR levels in response to amino acids and is essential for virulence. mBio 6:e01064. doi: 10.1128/mBio.01064-15
Morin, M., Ropers, D., Letisse, F., Laguerre, S., Portais, J. C., Cocaign-Bousquet, M., et al. (2016). The post-transcriptional regulatory system CSR controls the balance of metabolic pools in upper glycolysis of Escherichia coli. Mol. Microbiol. 100, 686–700. doi: 10.1111/mmi.13343
Mukherjee, A., Cui, Y., Liu, Y., Dumenyo, C. K., and Chatterjee, A. K. (1996). Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology 142, 427–434. doi: 10.1099/13500872-142-2-427
Mukherjee, S., Oshiro, R. T., Yakhnin, H., Babitzke, P., and Kearns, D. B. (2016). FliW antagonizes CsrA RNA binding by a noncompetitive allosteric mechanism. Proc. Natl. Acad. Sci. U. S. A. 113, 9870–9875. doi: 10.1073/pnas.1602455113
Mukherjee, S., Yakhnin, H., Kysela, D., Sokoloski, J., Babitzke, P., and Kearns, D. B. (2011). CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 82, 447–461. doi: 10.1111/j.1365-2958.2011.07822.x
Müller, P., Gimpel, M., Wildenhain, T., and Brantl, S. (2019). A new role for CsrA: promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 16, 972–987. doi: 10.1080/15476286.2019.1605811
Nitzan, M., Rehani, R., and Margalit, H. (2017). Integration of bacterial small RNAs in regulatory networks. Annu. Rev. Biophys. 46, 131–148. doi: 10.1146/annurev-biophys-070816-034058
Oshiro, R. T., Rajendren, S., Hundley, H. A., and Kearns, D. B. (2019). Robust stoichiometry of FliW-CsrA governs flagellin homeostasis and cytoplasmic organization in Bacillus subtilis. mBio 10:e00533–19. doi: 10.1128/mBio.00533-19
Pannuri, A., Yakhnin, H., Vakulskas, C. A., Edwards, A. N., Babitzke, P., and Romeo, T. (2012). Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J. Bacteriol. 194, 79–89. doi: 10.1128/JB.06209-11
Park, H., McGibbon, L. C., Potts, A. H., Yakhnin, H., Romeo, T., and Babitzke, P. (2017). Translational repression of the RpoS antiadapter IraD by CsrA is mediated via translational coupling to a short upstream open reading frame. mBio 8:e01355–17. doi: 10.1128/mBio.01355-17
Park, H., Yakhnin, H., Connolly, M., Romeo, T., and Babitzke, P. (2015). CsrA participates in a PNPase autoregulatory mechanism by selectively repressing translation of pnp transcripts that have been previously processed by RNase III and PNPase. J. Bacteriol. 197, 3751–3759. doi: 10.1128/JB.00721-15
Parker, A., Cureoglu, S., De Lay, N., Majdalani, N., and Gottesman, S. (2017). Alternative pathways for Escherichia coli biofilm formation revealed by sRNA overproduction. Mol. Microbiol. 105, 309–325. doi: 10.1111/mmi.13702
Patterson-Fortin, L. M., Vakulskas, C. A., Yakhnin, H., Babitzke, P., and Romeo, T. (2013). Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J. Mol. Biol. 425, 3662–3677. doi: 10.1016/j.jmb.2012.12.010
Paul, B. J., Barker, M. M., Ross, W., Schneider, D. A., Webb, C., Foster, J. W., et al. (2004). DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322. doi: 10.1016/j.cell.2004.07.009
Pernestig, A. K., Melefors, O., and Georgellis, D. (2001). Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276, 225–231. doi: 10.1074/jbc.M001550200
Potts, A. H., Guo, Y., Ahmer, B. M. M., and Romeo, T. (2019). Role of CsrA in stress responses and metabolism important for Salmonella virulence revealed by integrated transcriptomics. PLoS One 14:e0211430. doi: 10.1371/journal.pone.0211430
Potts, A. H., Leng, Y., Babitzke, P., and Romeo, T. (2018). Examination of Csr regulatory circuitry using epistasis analysis with RNA-seq (Epi-seq) confirms that CsrD affects gene expression via CsrA, CsrB and CsrC. Sci. Rep. 8:5373. doi: 10.1038/s41598-018-23713-8
Potts, A. H., Vakulskas, C. A., Pannuri, A., Yakhnin, H., Babitzke, P., and Romeo, T. (2017). Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 8:1596. doi: 10.1038/s41467-017-01613-1
Pourciau, C., Pannuri, A., Potts, A., Yakhnin, H., Babitzke, P., and Romeo, T. (2019). Regulation of iron storage by CsrA supports exponential growth of Escherichia coli. mBio 10:e01034–19. doi: 10.1128/mBio.01034-19
Radomska, K. A., Ordoñez, S. R., Wösten, M. M., Wagenaar, J. A., and van Putten, J. P. (2016). Feedback control of Campylobacter jejuni flagellin levels through reciprocal binding of FliW to flagellin and the global regulator CsrA. Mol. Microbiol. 102, 207–220. doi: 10.1111/mmi.13455
Regulski, E. E., Moy, R. H., Weinberg, Z., Barrick, J. E., Yao, Z., Ruzzo, W. L., et al. (2008). A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 68, 918–932. doi: 10.1111/j.1365-2958.2008.06208.x
Ren, B., Shen, H., Lu, Z. J., Liu, H., and Xu, Y. (2014). The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLoS One 9:e89653. doi: 10.1371/journal.pone.0089653
Renda, A. J., Poly, S., Lai, Y. -J., Pannuri, A., Yakhnin, H., Potts, A. H., et al. (2020). CsrA-mediated translational activation of ymdA expression in Escherichia coli. mBio 11:e00849–20. doi: 10.1128/mBio.00849-20
Revelles, O., Millard, P., Nougayrède, J. P., Dobrindt, U., Oswald, E., Létisse, F., et al. (2013). The carbon storage regulator (Csr) system exerts a nutrient-specific control over central metabolism in Escherichia coli strain Nissle 1917. PLoS One 8:e66386. doi: 10.1371/journal.pone.0066386
Rife, C., Schwarzenbacher, R., McMullan, D., Abdubek, P., Ambing, E., Axelrod, H., et al. (2005). Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Protein 61, 449–453. doi: 10.1002/prot.20502
Romeo, T. (1998). Global regulation by the small RNA binding protein CsrA and the noncoding RNA CsrB. Mol. Microbiol. 29, 1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x
Romeo, T., and Gong, M. (1993). Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J. Bacteriol. 175, 5740–5741. doi: 10.1128/jb.175.17.5740-5741.1993
Romeo, T., Gong, M., Liu, M. Y., and Brun-Zinkernagel, A. M. (1993). Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175, 4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993
Romeo, T., Vakulskas, C. A., and Babitzke, P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324. doi: 10.1111/j.1462-2920.2012.02794.x
Sabnis, N. A., Yang, H., and Romeo, T. (1995). Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270, 29096–29104. doi: 10.1074/jbc.270.49.29096
Schubert, M., Lapouge, K., Duss, O., Oberstrass, F. C., Jelesarov, I., Haas, D., et al. (2007). Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 14, 807–813. doi: 10.1038/nsmb1285
Smith, J. N., and Ahmer, B. M. M. (2003). Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185, 1357–1366. doi: 10.1128/jb.185.4.1357-1366.2003
Sonnleitner, E., Schuster, M., Sorger-Domenigg, T., Greenberg, E. P., and Bläsi, U. (2006). Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59, 1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x
Sowa, S. W., Gelderman, G., Leistra, A. N., Buvanendiran, A., Lipp, S., Pitaktong, A., et al. (2017). Integrative FourD omics approach profiles the target network of the carbon storage regulatory system. Nucleic Acids Res. 45, 1673–1686. doi: 10.1093/nar/gkx048
Sperandio, V. (2010). SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen. Gut Microbes 1, 432–435. doi: 10.4161/gmic.1.6.14177
Steiner, S., Lori, C., Boehm, A., and Jenal, U. (2013). Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 32, 354–368. doi: 10.1038/emboj.2012.315
Subramaniam, A. R., Zid, B. M., and O’Shea, E. K. (2014). An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159, 1200–1211. doi: 10.1016/j.cell.2014.10.043
Suzuki, K., Babitzke, P., Kushner, S. R., and Romeo, T. (2006). Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20, 2605–2017. doi: 10.1101/gad.1461606
Suzuki, K., Wang, X., Weilbacher, T., Pernestig, A. K., Melefors, O., Georgellis, D., et al. (2002). Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184, 5130–5140. doi: 10.1128/jb.184.18.5130-5140.2002
Tani, T. H., Khodursky, A., Blumenthal, R. M., Brown, P. O., and Matthews, R. G. (2002). Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99, 13471–13476. doi: 10.1073/pnas.212510999
Timmermans, J., and Van Melderen, L. (2009). Conditional essentiality of the csrA gene in Escherichia coli. J. Bacteriol. 191, 1722–1724. doi: 10.1128/JB.01573-08
Vakulskas, C. A., Leng, Y., Abe, H., Amaki, T., Okayama, A., Babitzke, P., et al. (2016). Antagonistic control of the turnover pathway for the global regulatory sRNA CsrB by the CsrA and CsrD proteins. Nucleic Acids Res. 44, 7896–7910. doi: 10.1093/nar/gkw484
Vakulskas, C. A., Pannuri, A., Cortés-Selva, D., Zere, T. R., Ahmer, B. M., Babitzke, P., et al. (2014). Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol. Microbiol. 92, 945–958. doi: 10.1111/mmi.12606
Vakulskas, C. A., Potts, A. H., Babitzke, P., Ahmer, B. M., and Romeo, T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224. doi: 10.1128/MMBR.00052-14
Valverde, C., Heeb, S., Keel, C., and Haas, D. (2003). RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50, 1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x
Valverde, C., Lindell, M., Wagner, E. G., and Haas, D. (2004). A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J. Biol. Chem. 279, 25066–25074. doi: 10.1074/jbc.M401870200
Venturi, V., and Ahmer, B. M. M. (2015). LuxR solos are becoming major players in cell-cell communication in bacteria. Front. Cell. Infect. Microbiol. 5:89. doi: 10.3389/fcimb.2015.00089
Vijayakumar, V., Vanhove, A. S., Pickering, B. S., Liao, J., Tierney, B. T., Asara, J. M., et al. (2018). Removal of a membrane anchor reveals the opposing regulatory functions of Vibrio cholerae glucose-specific enzyme IIA in biofilms and the mammalian intestine. mBio 9:e00858–18. doi: 10.1128/mBio.00858-18
Vytvytska, O., Moll, I., Kaberdin, V. R., von Gabain, A., and Bläsi, U. (2000). Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14, 1109–1118.
Wagner, E. G. H., and Romby, P. (2015). Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208. doi: 10.1016/bs.adgen.2015.05.001
Wang, X., Dubey, A. K., Suzuki, K., Baker, C. S., Babitzke, P., and Romeo, T. (2005). CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56, 1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x
Wang, Y., Pannuri, A., Ni, D., Zhou, H., Cao, X., Lu, X., et al. (2016). Structural basis for translocation of a biofilm-supporting exopolysaccharide across the bacterial outer membrane. J. Biol. Chem. 291, 10046–10057. doi: 10.1074/jbc.M115.711762
Wang, X., Preston, J. F. 3rd, and Romeo, T. (2004). The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734. doi: 10.1128/jb.186.9.2724-2734.2004
Wei, B. L., Brun-Zinkernagel, A. M., Simecka, J. W., Pruss, B. M., Babitzke, P., and Romeo, T. (2001). Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40, 245–256. doi: 10.1046/j.1365-2958.2001.02380.x
Weilbacher, T., Suzuki, K., Dubey, A. K., Wang, X., Gudapaty, R., Morozov, I., et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. doi: 10.1046/j.1365-2958.2003.03459.x
Yakhnin, H., Aichele, R., Ades, S. E., Romeo, T., and Babitzke, P. (2017). Circuitry linking the global Csr- and σE-dependent cell envelope stress response systems. J. Bacteriol. 199:e00484–17. doi: 10.1128/JB.00484-17
Yakhnin, H., Baker, C. S., Berezin, I., Evangelista, M. A., Rassin, A., Romeo, T., et al. (2011a). CsrA represses translation of sdiA, which encodes the N-acylhomoserine-L-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J. Bacteriol. 193, 6162–6170. doi: 10.1128/JB.05975-11
Yakhnin, A. V., Baker, C. S., Vakulskas, C. A., Yakhnin, H., Berezin, I., Romeo, T., et al. (2013). CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol. Microbiol. 87, 851–866. doi: 10.1111/mmi.12136
Yakhnin, H., Pandit, P., Petty, T. J., Baker, C. S., Romeo, T., and Babitzke, P. (2007). CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 64, 1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x
Yakhnin, H., Yakhnin, A. V., Baker, C. S., Sineva, E., Berezin, I., Romeo, T., et al. (2011b). Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Eσ70 and EσS, and indirect transcriptional activation by CsrA. Mol. Microbiol. 81, 689–704. doi: 10.1111/j.1365-2958.2011.07723.x
Yang, H., Liu, M. Y., and Romeo, T. (1996). Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178, 1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996
Ye, F., Yang, F., Yu, R., Lin, X., Qi, J., and Chen, Z. (2018). Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT. Nat. Commun. 9:1196. doi: 10.1038/s41467-018-03625-x
Keywords: RNA-binding proteins, sRNAs, posttranscriptional regulation, regulatory circuitry, translation control
Citation: Pourciau C, Lai Y-J, Gorelik M, Babitzke P and Romeo T (2020) Diverse Mechanisms and Circuitry for Global Regulation by the RNA-Binding Protein CsrA. Front. Microbiol. 11:601352. doi: 10.3389/fmicb.2020.601352
Received: 31 August 2020; Accepted: 07 October 2020;
Published: 27 October 2020.
Edited by:
Stefano Marzi, UPR9002 Architecture et Réactivité de l’ arN, FranceCopyright © 2020 Pourciau, Lai, Gorelik, Babitzke and Romeo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tony Romeo, dHJvbWVvQHVmbC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.