- 1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Research Unit Tropical Mycology and Plant-Soil Fungi Interactions, Faculty of Agronomy, University of Parakou, Parakou, Benin
- 4Department of Organismal Biology, Uppsala University, Uppsala, Sweden
Many species of Amanita sect. Phalloideae (Fr.) Quél. cause death of people after consumption around the world. Amanita albolimbata, a new species of A. sect. Phalloideae from Benin, is described here. The taxon represents the first lethal species of A. sect. Phalloideae known from Benin. Morphology and molecular phylogenetic analyses based on five genes (ITS, nrLSU, rpb2, tef1-α, and β-tubulin) revealed that A. albolimbata is a distinct species. The species is characterized by its smooth, white pileus sometimes covered by a patchy volval remnant, a bulbous stipe with a white limbate volva, broadly ellipsoid to ellipsoid, amyloid basidiospores, and abundant inflated cells in the volva. Screening for the most notorious toxins by liquid chromatography–high-resolution mass spectrometry revealed the presence of α-amanitin, β-amanitin, and phallacidin in A. albolimbata.
Introduction
Some species of Amanita Pers. are edible, whereas others are poisonous. All lethally poisonous Amanita species are in A. sect. Phalloideae that is characterized by a non-striate and non-appendiculate pileus and attenuate lamellulae, the presence of persistent annulus, a limbate volva on the bulbous stipe base, and amyloid basidiospores (Corner and Bas, 1962; Bas, 1969; Tulloss and Bhandary, 1992; Yang, 1997, 2005, 2015). Members of the section are responsible for greater than 90% of mushroom fatalities reported worldwide (Bresinsky and Besl, 1990; Unluoglu and Tayfur, 2003; Cai et al., 2014, 2016; Li et al., 2015). In Central Europe and North America, lethal amanitas such as A. bisporigera G.F. Atk., A. phalloides (Vaill.: Fr.) Link, A. suballiacea (Murrill) Murrill, A. verna (Bull.) Lam., and A. virosa Bertill. have caused dramatic human and animal poisoning cases (Wieland, 1973, 1986; Bresinsky and Besl, 1985). It has been reported that A. exitialis Zhu L. Yang and T. H. Li, A. fuliginea Hongo, A. fuligineoides P. Zhang and Zhu L. Yang, A. pallidorosea P. Zhang and Zhu L. Yang, A. rimosa P. Zhang and Zhu L. Yang, and A. subjunquillea S. Imai cause a lot of fungal poisoning cases in East Asia (Zhang et al., 2010; Deng et al., 2011; Chen et al., 2014; Li et al., 2014, 2020; Cai et al., 2016). The toxins in lethal amanitas are mainly amatoxins, phallotoxins, and virotoxins (Wieland et al., 1983; Wieland, 1986; Cai et al., 2016). Among them, amatoxins are 10–20 times more toxic than the other ones and represent the major toxins responsible for human poisoning (Li and Oberlies, 2005). These cyclopeptide toxins are able to resist high temperatures, and their consumption can cause severe liver and renal failure (Wieland, 1973, 1986; Chen et al., 2014).
Lethal amanitas have been extensively studied in Asia, Europe, and America, where more than 50 toxic taxa have been described (Zhang et al., 2010; Cai et al., 2014, 2016; Li et al., 2015; Yang, 2015; Thongbai et al., 2017; Cui et al., 2018). Although the genus Amanita is known worldwide, few taxa of the genus have been reported from tropical Africa (Walleyn and Verbeken, 1998; Eyi-Ndong et al., 2011; Yorou et al., 2014; Härkönen et al., 2015; De Kesel et al., 2017; Fraiture et al., 2019; Tulloss et al., 2020). Six species belonging to A. sect. Phalloideae are known from tropical Africa, of which three, including A. alliodora Pat., A. murinacea Pat., and A. thejoleuca Pat., were described from Madagascar (Patrouillard, 1924; Fraiture et al., 2019). The other three species, A. bweyeyensis Fraiture, Raspé and Degreef, A. harkoneniana Fraiture and Saarimäki, and A. strophiolata Beeli were described from DR Congo (Beeli, 1927, 1935; Fraiture et al., 2019).
In this study, a new member of A. sect. Phalloideae from tropical West Africa is described. Its macromorphological and micromorphological characteristics, as well as its phylogenetic relationships with other Amanita species are discussed. In addition, the screening of the species for the known toxins occurring in Amanita is reported.
Materials and Methods
Collections and Preservation
Specimens were opportunistically collected in Benin, West Africa (Figure 1), during the rainy season from June to September (2018-2019), especially in the forest dominated by Fabaceae/Leguminosae (Isoberlinia Craib and Stapf ex Holland, Anthonotha P. Beauv., Berlinia Sol. ex Hook. f.), Phyllanthaceae (Uapaca Baill.), and Dipterocarpaceae (Monotes A. DC.). Specimens were photographed in situ using a digital camera–type Canon EOS 60D. Macromorphological characteristics were recorded on fresh materials, according to Tulloss and Yang (2011). Color codes recorded from fresh materials follow Kornerup and Wanscher (1981). The fresh basidiomata were dried using an electric dryer Stockli Dorrex at 45°C for 1 day and stored thereafter as exsiccates with their label in sealable plastic bag–type minigrip. The dried specimens along with the holotype of the newly described species are deposited in the Mycological Herbarium of the University of Parakou (UNIPAR). Duplicates of dried specimens and the isotype of the new species are conserved at the Herbarium of Cryptogams of Kunming Institute of Botany, Chinese Academy of Sciences (KUN-HKAS). Small pieces of fresh basidiomata were also stored in CTAB lysis buffer (2% cetyl trimethylammonium bromide, 100 mM Tris–HCl, 20 mM EDTA, 1.4 M NaCl) and dried with silica gel for molecular investigations. Nomenclature aspects, as well as authorities for scientific names, have been double checked against Index Fungorum1 and in Tulloss et al. (2020).
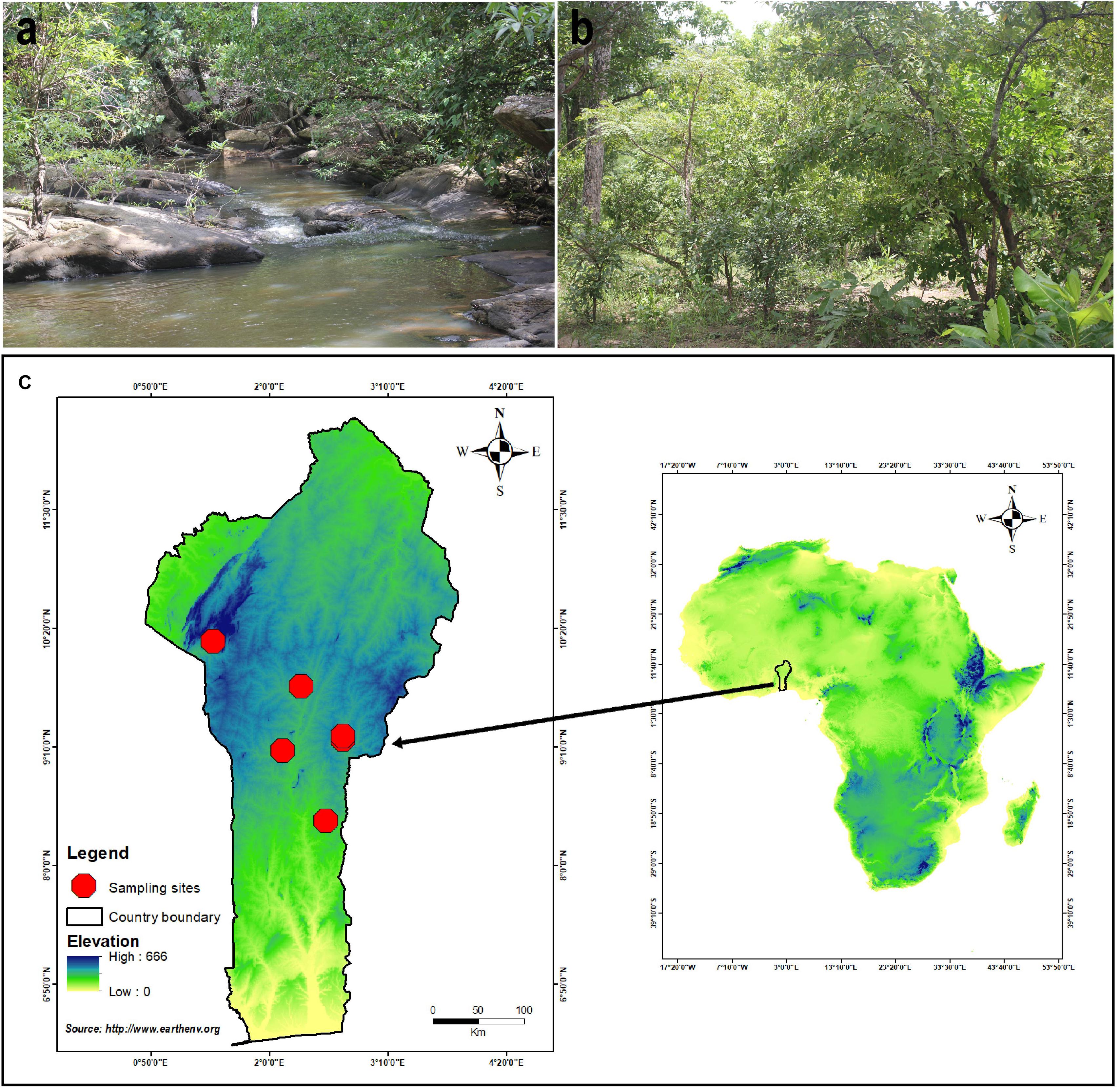
Figure 1. Distribution of A. albolimbata J.E.I. Codjia, N.S. Yorou and Zhu L. Yang: (a) Gallery forest associated with Uapaca guineensis. (b) Woodland associated with Uapaca togoensis and Isoberlinia doka. (c) Location of the sampling sites.
Micromorphological Investigations
Microscopic structures were studied from dried materials mounted in 5% KOH and stained with Congo red to depict all tissues. The Melzer’s reagent was used to test the amyloidity of basidiospores. All measurements and line drawings were performed at 1,000× magnification, and a minimum of 20–30 basidiospores from each basidioma were measured in side view. Micromorphological investigations were performed by mean of a microscope-type Nikon Eclipse 50i. The abbreviation (n/m/p) is used to describe basidiospores where n is the number of basidiospores from m basidiomata of p collections. The basidiospores dimensions are provided using the notation (a)b – c(d) with the range b–c containing a minimum of 95% of the measured values, a and d in the brackets showing the two extreme values. Q is used for the ratio length/width of a spore in side view; Qm is the average Q of all basidiospores ± sample standard deviation. The measurements for the basidiospores were analyzed with Piximetre v5.10 (Henriot and Cheype, 2020). The descriptive terms are in accordance with Bas (1969), Yang (2005), Yang (2015), Cai et al. (2016), and Cui et al. (2018).
DNA Extraction, Amplification, and Sequencing
The total genomic DNA was obtained from materials preserved in CTAB or dried with silica gel following the modified CTAB procedure (Doyle and Doyle, 1987). Polymerase chain reaction (PCR) amplification and sequencing were performed in accordance with those described in Cai et al. (2014) and Cui et al. (2018). The following primer pairs were used for PCR amplification and sequencing: ITS1F and ITS4 to amplify ITS region (White et al., 1990; Gardes and Bruns, 1993); LROR and LR5 (Vilgalys and Hester, 1990) for nrLSU region; EF1-983F and EF1-1567R (Rehner and Buckley, 2005) for the translation elongation factor 1-α (tef1-α) region; ARPB2-6F and ARPB2-7R for RNA polymerase II second largest subunit (rpb2) region; and Am-b-tubulin F and Am-b-tubulin R (Cai et al., 2014) for beta-tubulin (β-tubulin) region.
Sequence Alignment and Phylogenetic Analyses
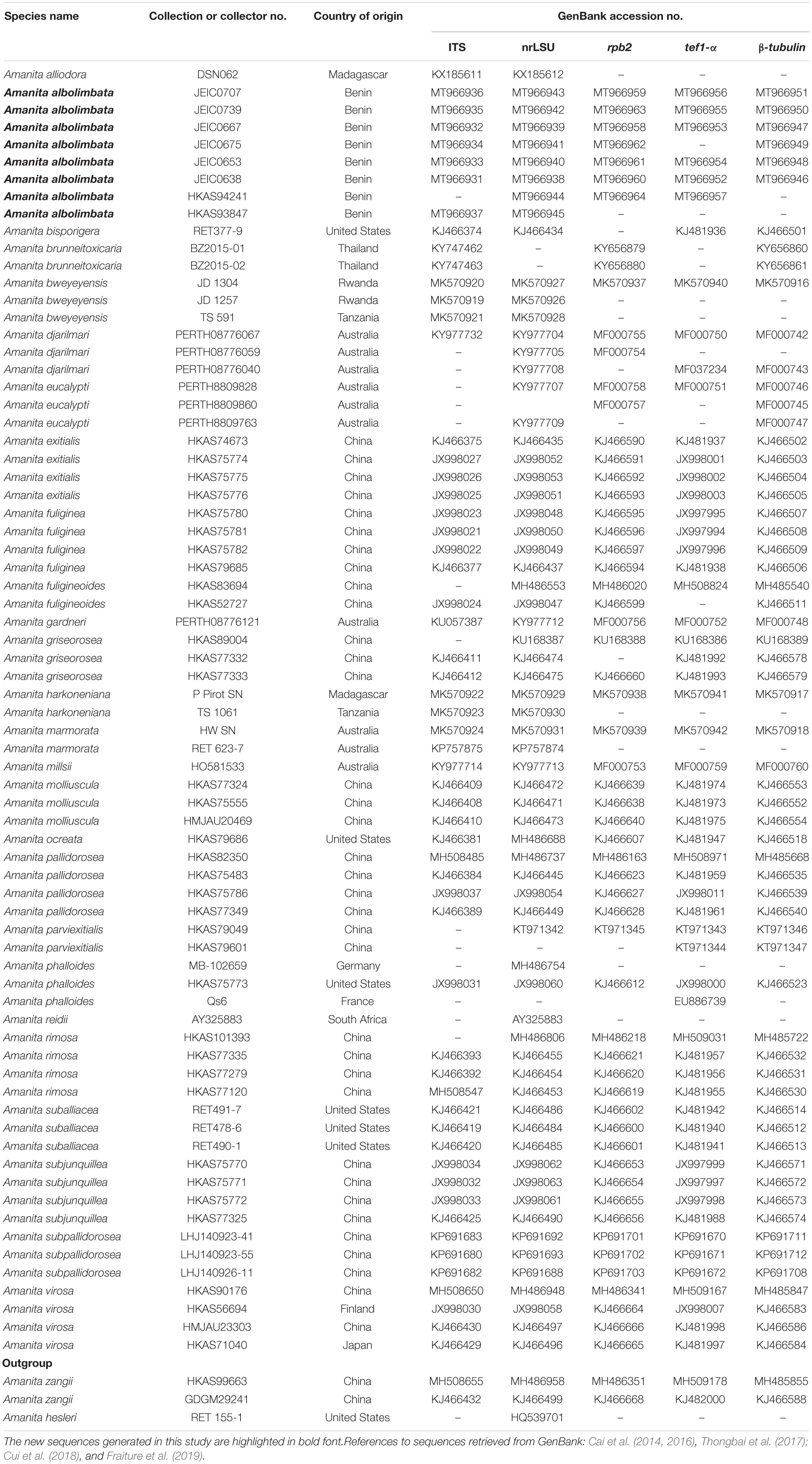
Thirty-four sequences (seven for ITS, eight for nrLSU, seven for rpb2, six for tef1-α, and six for β-tubulin) were newly generated for this study and deposited in GenBank (Table 1)2. Additional sequences were retrieved from previously published articles and GenBank (Table 1). The sequences were aligned using MAFFT v7.310 (Katoh and Standley, 2013) and edited manually when necessary using BioEdit v7.0.9 (Hall, 1999). The poorly aligned portions and divergent regions were eliminated using Gblocks v0.91b (Castresana, 2000; Talavera and Castresana, 2007). A concatenated dataset (including ITS, nrLSU, rpb2, tef1-α, β-tubulin) comprising 312 sequences was constructed using Phyutility v2.2 (Smith and Dunn, 2008) and used for phylogenetic analyses. Before using the concatenated dataset for phylogenetic analyses, the Incongruence Length Difference test in PAUP v4.0a168 (Swofford, 2002) was performed to detect any conflicts between the gene regions. As no incongruence (P = 0.363000) was detected, the maximum likelihood (ML) and Bayesian inference (BI) were used on the concatenated alignment for phylogenetic tree inference. The ML analysis was performed using RAxML v7.9.1 (Stamatakis, 2006) under the GTR + GAMMA + I nucleotide substitution model and performing non-parametric bootstrapping with 1,000 replicates. The BI was performed in MrBayes v3. 2 (Ronquist et al., 2012). The best substitution model was determined using the Akaike Information Criterion implemented in jModeltest v.2 on the CIPRES Science Gateway v3.1 (Miller et al., 2010). The BI was conducted with the following parameters: two runs, each with four simultaneous Markov chains, and trees were summarized every 1,000 generations. The analyses were completed after 20,000,000 generations when the average standard deviation of split frequencies was 0.002200 for the five-gene analysis, and the first 25% generations were discarded as burn-in. The phylograms from ML and BI analyses were visualized with FigTree v1.4.3 (Rambaut, 2009) and then edited in Adobe Illustrator CS6.
Analysis of Toxins by Liquid Chromatography–High-Resolution Mass Spectrometry
Dried basidiomata of the target taxon have been used for toxin analyses using the method of Lüli et al. (2019). Toxins were extracted from basidiomata, using methanol–water–0.01 M hydrochloric acid (5:4:1, vol/vol) as the extraction buffer. Dried material (0.05 g) was crushed into fine powder in a mortar and pestle with liquid nitrogen. Then, 1.5 mL aforementioned buffer was added, and the suspension transferred into 1.5-mL centrifuge tubes. The tubes were kept at room temperature for 30 min, followed by centrifugation (12,000 rpm) for 3 min. Finally, the supernatant was transferred into new centrifuge tubes for mass spectrometry analysis.
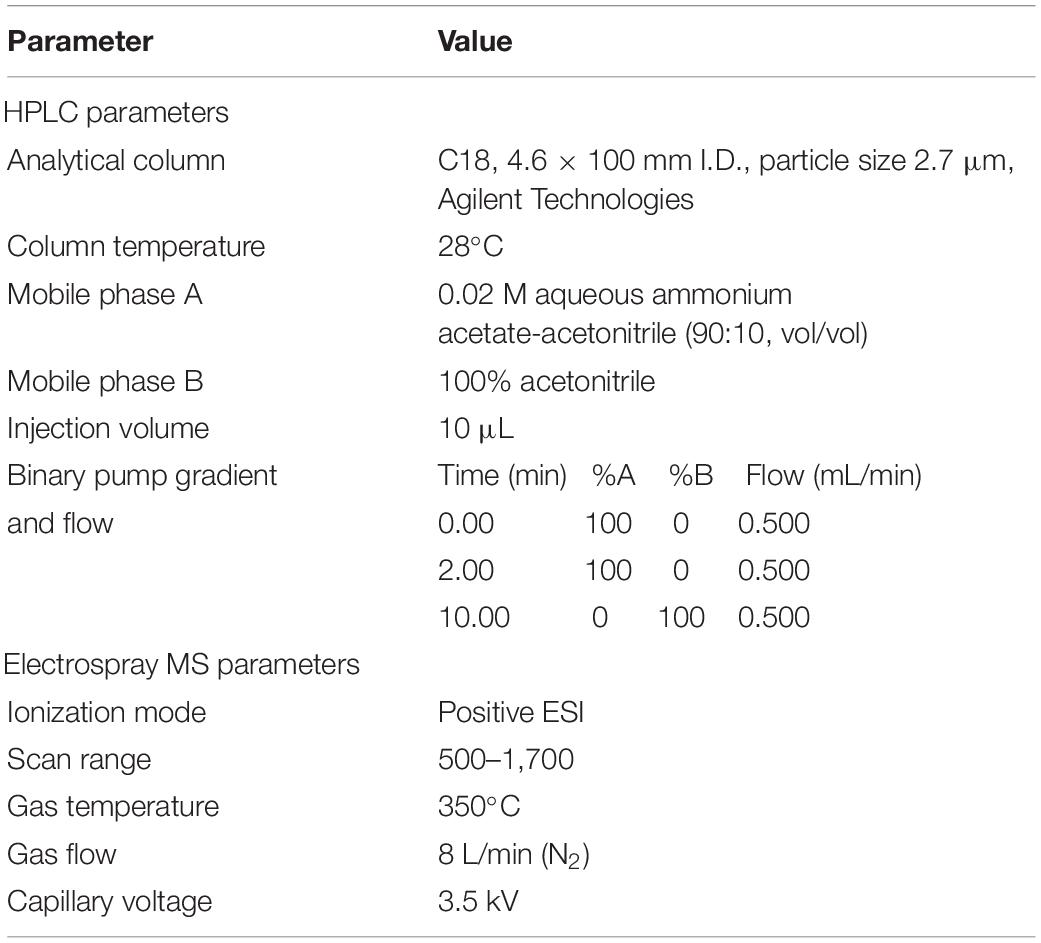
The presence of cyclic peptides, especially α-amanitin, β-amanitin, phalloidin, and phallacidin (standards provided by Sigma Chemical Co, United States), was evaluated through the liquid chromatography–high-resolution mass spectrometry (LC-HRMS) using 1290 Infinity II HPLC systems coupled with 6540 UHD precision mass Q-TOF instruments under the conditions listed in Table 2.
Results
Phylogenetic Data
The topologies of ML and BI phylogenetic trees obtained in this study are practically the same (Supplementary Figures S1, S2). In the combined dataset (ITS, nrLSU, rpb2, tef1-α, and β-tubulin), 312 sequences were included. The combined dataset contained 3,024 total characters, including 2,070 constant (proportion = 0.684524), 93 variable and parsimony-uninformative, and 861 parsimony-informative. The target species, A. albolimbata, forms a well-supported distinct lineage (MLB = 100%, BPP = 1.0) and is a close sister to the Asian species (A. parviexitialis Qing Cai, Zhu L. Yang and Yang-Yang Cui) (Figure 2 and see also Supplementary Figures S1–S7). Within the section, A. albolimbata is genetically distant from other African species such as A. alliodora, A. bweyeyensis, and A. harkoneniana. In the phylogenetic tree, the African and Australian species hold the basal positions.
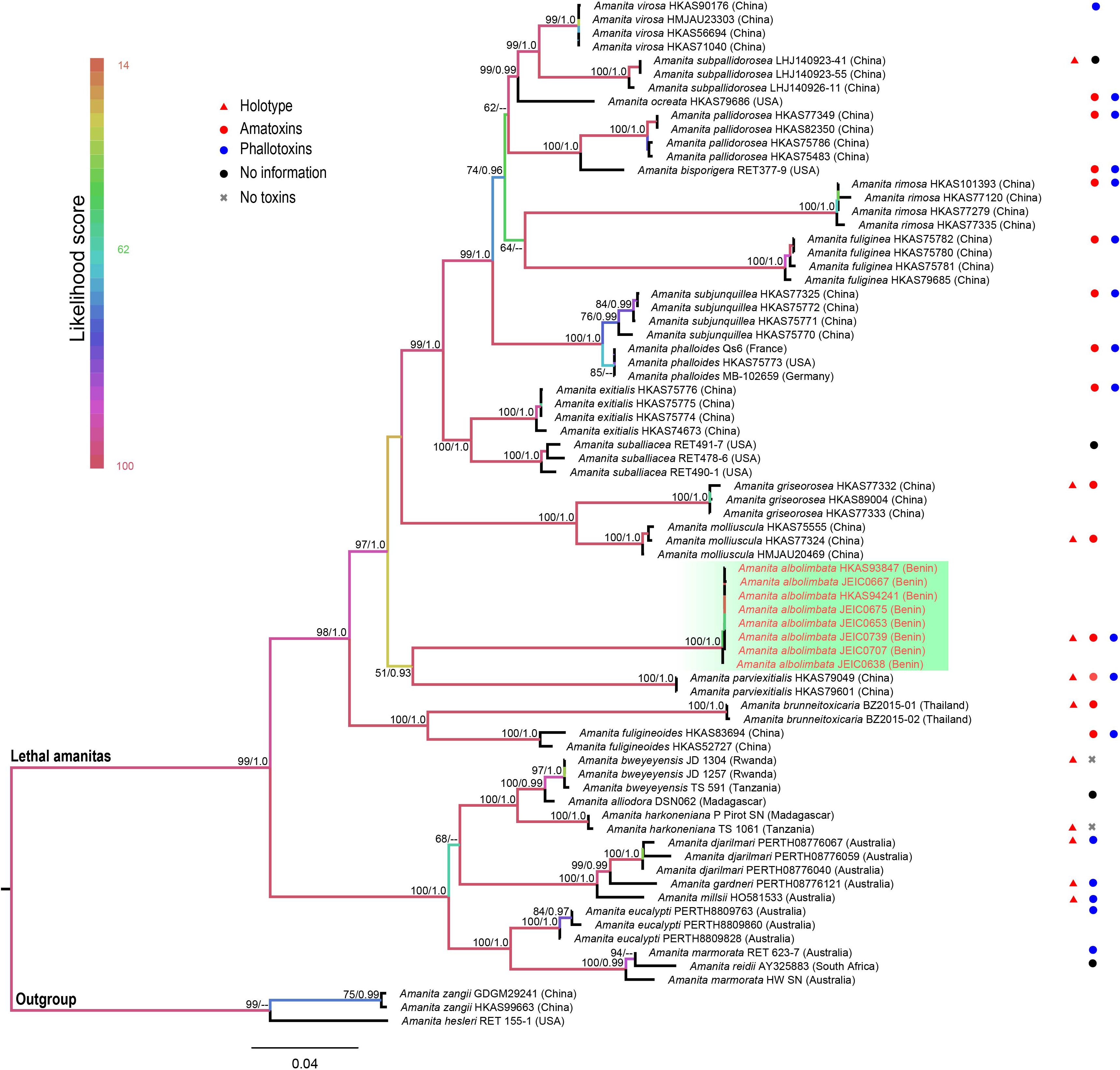
Figure 2. Phylogenetic relationship and placement of Amanita albolimbata within A. sect. Phalloideae inferred from the combined dataset (ITS, nrLSU, rpb2, tef1-α, and β-tubulin) using maximum likelihood (ML). Bootstrap values ≥50% and Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Taxonomy
Amanita albolimbata J.E.I. Codjia, N.S. Yorou and Zhu L. Yang, sp. nov.
Mycobank: MB836777
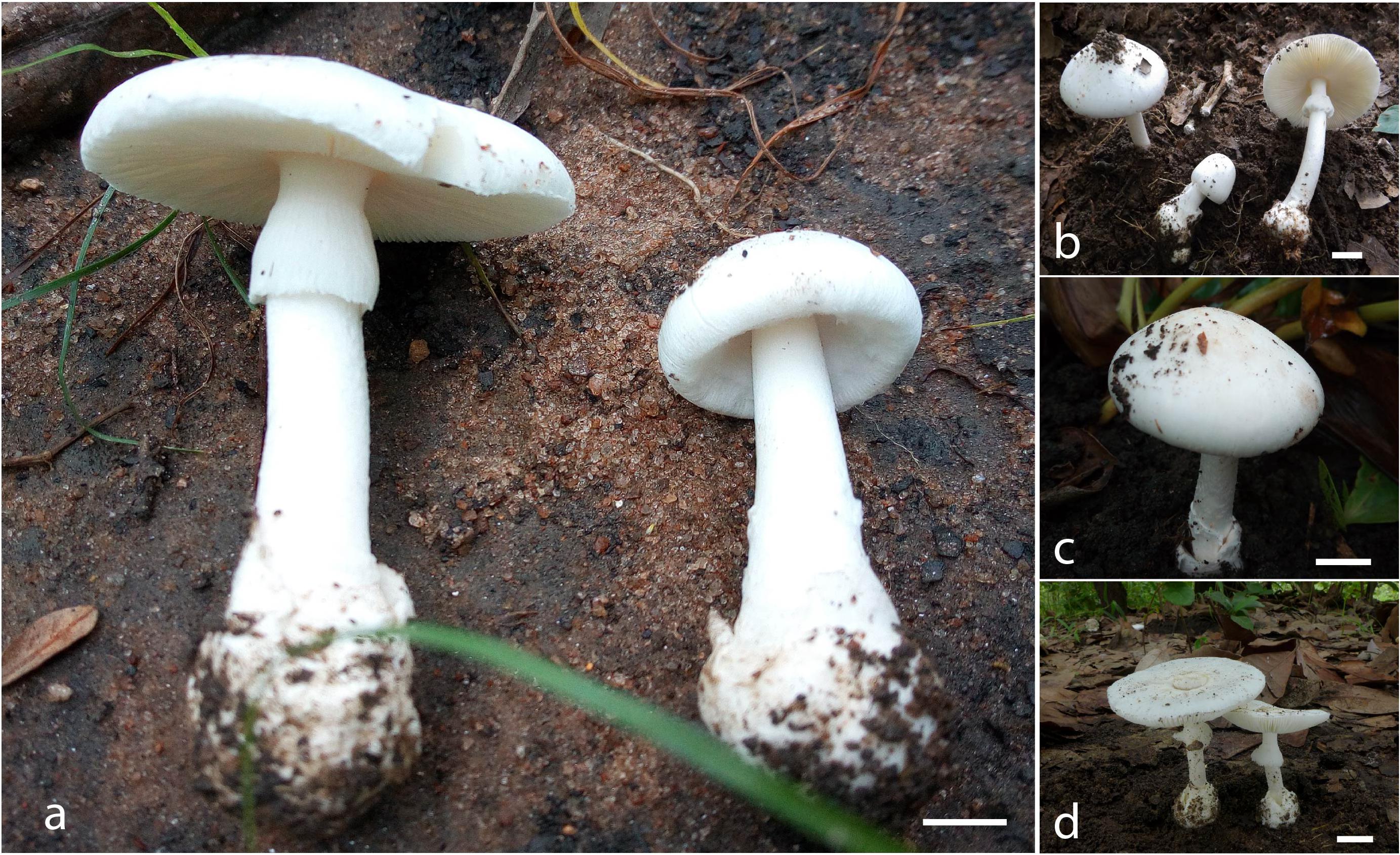
Figure 3. Basidiomata of A. albolimbata [(a) holotype JEIC0739; (b) JEIC0653; (c) JEIC0667; (d) HKAS93847]. Scale bar: 1 cm (photos by Codjia J.E.I. and Gang Wu).
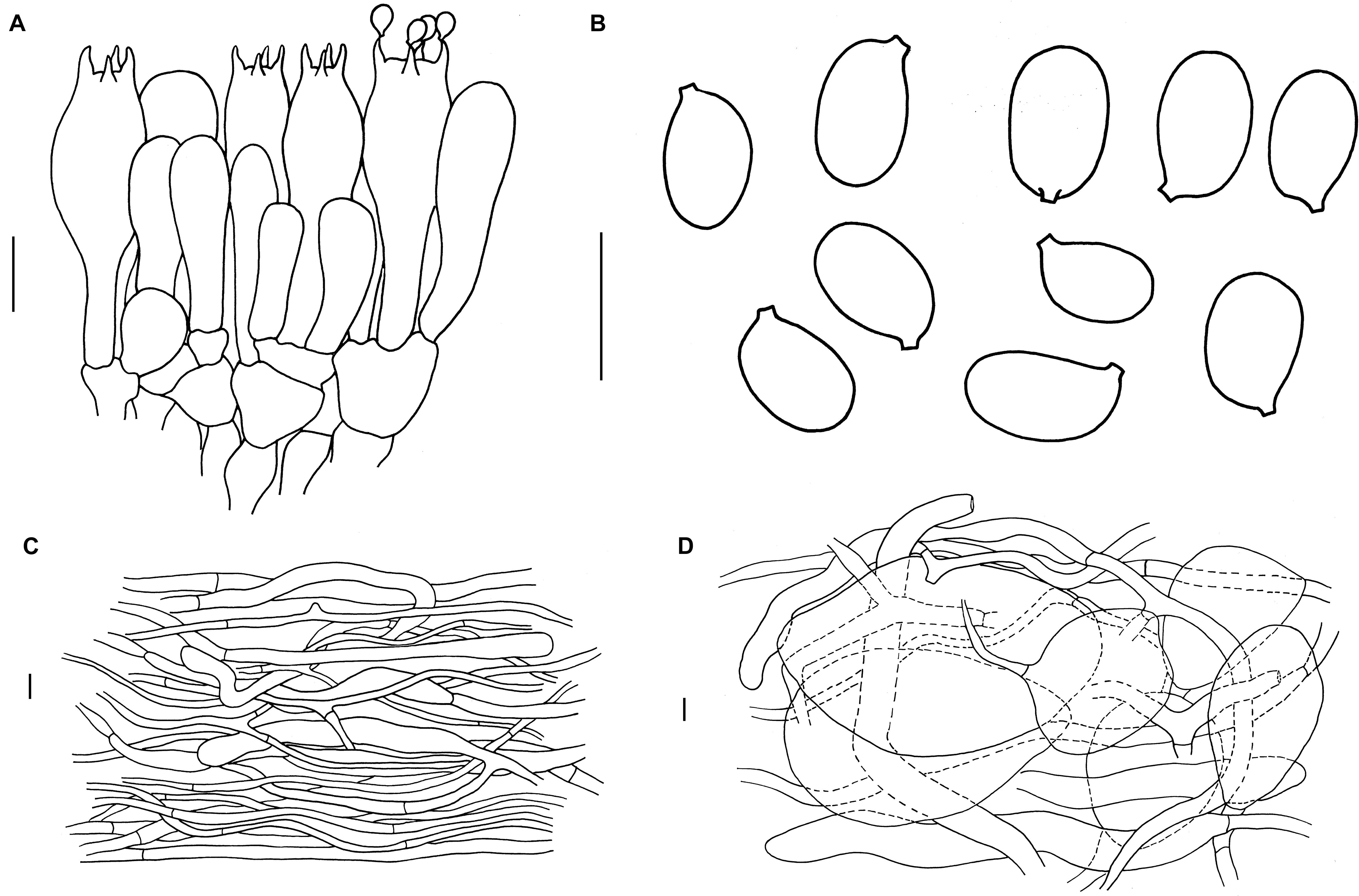
Figure 4. Microscopic features of A. albolimbata. Holotype JEIC0739 [(A) hymenium and subhymenium; (B) basidiospores]. HKAS93847 [(C) pileipellis; (D) elements of inner part of volva]. Scale bars: a–d = 10 μm.
Etymology
“Albo” (white), “limbata” (limb bearing volva), meaning white limbate volva.
Type
BENIN. Donga Province: Bassila, 09°07′58″N, 2°07′43″E, Forest Reserve of Bassila, woodland of Uapaca togoensis Pax (Phyllanthaceae), date: 06 August 2019, leg. and det. Jean Evans I. CODJIA, Holotype JEIC0739 (UNIPAR), Isotype (KUN-HKAS 107736), GenBank Acc. No.: (ITS = MT966935, nrLSU = MT966942, rpb2 = MT966963, tef1-α = MT966955, β-tubulin = MT966950).
Basidiomata Small-Sized
Pileus 38–58 mm in diameter, hemispherical when young and then expanding to regularly convex or applanate at the maturity, smooth, without umbo, subviscid when moist, sometimes the presence of patchy volval remnants, white (1A1); margin non-striate, non-appendiculate, white (1A1). Lamellae free, white (1A1), lamellulae attenuate. Stipe 55–87 mm long × 6–13 mm diameter, white, nearly cylindrical, covered with white (1A1) squamules. Annulus present, superior, white (1A1), membranous. Basal bulb of the stipe globose, surrounded by a white (1A1) limbate membranous volva (also white inside). Context stuffed, white (1A1). Odor and taste not recorded.
Lamellar Trama Bilateral
Mediostratum 36–50 μm wide, composed of abundant subfusiform to ellipsoidal inflated cells (30–90 × 15–30 μm), and abundant filamentous hyphae, 3–4 μm wide; vascular hyphae scarce. Lateral stratum composed of ellipsoid to subglobose inflated cells (25–40 × 20–30 μm), diverging at an angle of ca. 30°–45° to mediostratum; filamentous hyphae abundant, 3–9 μm wide. Subhymenium 35–45 μm thick, with 2–3 layers of ovoid to subglobose or irregular cells, 9–12 × 8–10 μm. Basidia (Figure 3a) [40/4/4] 30–48 × 9.5–11 (–14) μm, clavate, 4-spored; basal septa lacking clamps. Basidiospores (Figure 3b) [90/4/4] (7.5–) 9 (–11) × (5–) 6–7 (–7.5) μm, Q = (1.2) 1.4–1.5 (1.7), Qm = 1.4 ± 0.05, broadly ellipsoid to ellipsoid, smooth, colorless, amyloid. Lamellar edge sterile, composed of subglobose to ellipsoid, inflated cells (15–40 × 10–25 μm), filamentous hyphae, 3–9 μm wide, irregularly arranged or running parallel to lamellar edge. Pileipellis 250–370 μm thick, 2-layered; suprapellis up to 100–170 μm thick, slightly gelatinized, composed of arranged, thin-walled, colorless to nearly colorless, ellipsoid to clavate terminal cells 80–180 × 10–20 μm, mixed with filamentous hyphae 3–5 μm wide; subpellis up to 150–200 μm thick, composed of undifferentiated, filamentous hyphae 2–5 μm wide; vascular hyphae scarce. Inner part of volva composed of longitudinally arranged elements: filamentous hyphae abundant 9- to 15-μm-wide, colorless, thin to slightly thick-walled branching; inflated cells abundant, subglobose to ellipsoid, 60–155 × 35–90 μm, colorless, thin to slightly thick-walled, terminal, mixed with long clavate terminal cells 130–210 × 15–20 μm; vascular hyphae rare. Outer part of volva similar to the inner part of volva but with less abundant inflated cells. Stipe trama composed of longitudinally arranged, long clavate terminal cells 130–540 × 15–25 μm; filamentous hyphae scattered to abundant, 3–10 μm wide; vascular hyphae scarce. Clamps absent in all tissues.
Habitat
Solitary, rarely or in small group of 2 or 3 individuals, on the ground in woodland and gallery forests, associated with Uapaca guineensis Müll. Arg. or U. togoensis (Phyllanthaceae) and Isoberlinia doka Craib and Stapf (Fabaceae/Leguminosae).
Distribution
Currently known from Benin, but likely occurs more widely in the region in similar vegetation.
Additional specimens examined were BENIN. Colline Province: Ouèssè, 08°26′34.4.0″ N, 02°33′09.0″ E, open forest, date: 02 July 2015, leg. B. Feng 1854 (HKAS94241), date: 03 July 2015, leg. G. Wu 1470 (HKAS93847). Borgou Province: Ndali, 09°14′31.93″N, 02°43′22.7″E, Forest Reserve of Ndali, date: 31 July 2019, leg. and det. Jean Evans I. CODJIA, JEIC0638. Donga Province: Bassila, 09°07′58″N, 2°07′43″E, Forest Reserve of Bassila, date: 02 August 2019, leg and det. Jean Evans I. CODJIA, JEIC0707. Atacora Province: Kota, 10°12′39″ N, 01°26′45.8″ E, gallery forest of Kota, date: 08 August 2019, leg. and det. Jean Evans I. CODJIA, JEIC0653. Borgou Province: Okpara, 09°16′34.8″N, 02°43’12.8″E, Forest Reserve of Okpara, date: 22 August 2019, leg. and det. Jean Evans I. CODJIA, JEIC0667, date: 11 September 2019, leg. and det. Jean Evans I. CODJIA, JEIC0675.
Analysis of Toxins by LC-HRMS
Amanita albolimbata contains three cyclic peptides: α-amanitin, β-amanitin, and phallacidin (Figure 5). The formula of α-amanitin is C39H54N10O14S with a monoisotopic mass of 918.3541. The calculated mass of the [M + H]+ ion is 919.3614, and the measured mass was 919.3609 with mass discrepancy of 0.59 ppm. The formula of β-amanitin is C39H53N9O15S with a monoisotopic mass of 919.3381. The calculated mass of the [M + H]+ ion is 920.3455, and the measured mass was 920.3461 with mass discrepancy of 0.7 ppm. The formula of phallacidin is C37H50N8O13S with a monoisotopic mass of 846.3218. The calculated mass of the [M + H]+ ion is 847.3291, and the measured mass was 847.3293 with mass discrepancy of 0.26 ppm. The measured masses of the two other adduct ions for above cyclic peptides, [M + Na]+ and [M + K]+, are also included in Figure 5. No corresponding mass was identified for phalloidin.
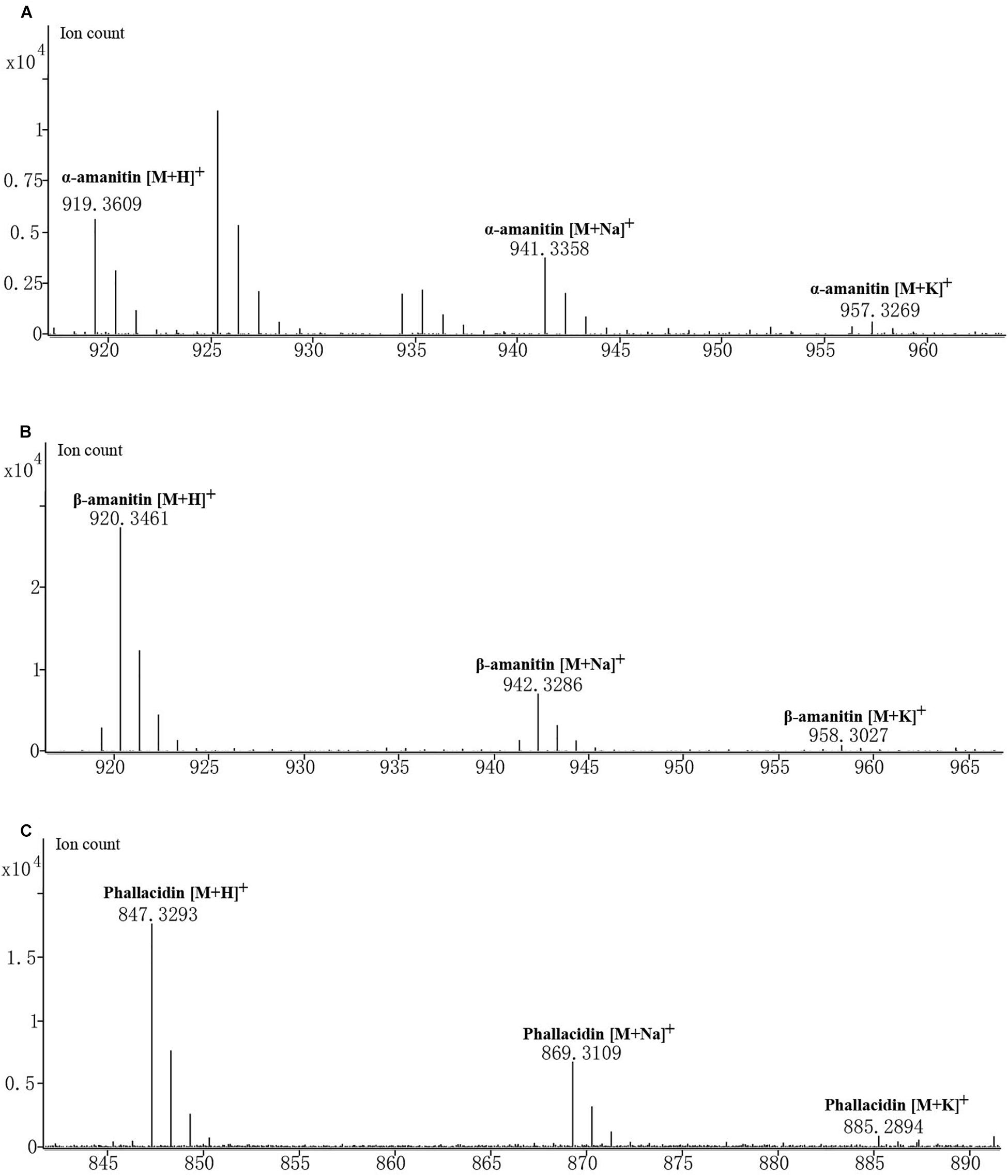
Figure 5. Mass spectrogram of cyclic peptides produced in A. albolimbata. (A) α-Amanitin. (B) β-Amanitin. (C) Phallacidin.
Discussion
Species Delimitation
Most of the fatal mushroom poisonings are caused by lethal amanitas belonging to A. sect. Phalloideae (Bresinsky and Besl, 1990; Unluoglu and Tayfur, 2003; Cai et al., 2014, 2016; Li et al., 2015; Cui et al., 2018). In tropical Africa, very few lethal amanitas have been reported (Walleyn and Verbeken, 1998; Fraiture et al., 2019; Tulloss et al., 2020). Only six species are known from tropical Africa including A. alliodora, A. murinacea, and A. thejoleuca described from Madagascar and A. bweyeyensis, A. harkoneniana, and A. strophiolata described from DR Congo (central Africa). Amanita albolimbata represents a new lethal Amanita from tropical Africa and can be recognized by its white basidiomata with a convex or applanate pileus without umbo, a limbate volva with inner part composed of abundant inflated cells, and broadly ellipsoid to ellipsoid basidiospores.
The multigene phylogenetic analyses revealed that A. albolimbata is an independent lineage in A. sect. Phalloideae. Surprisingly, the species is genetically distant from other African species such as A. alliodora, A. bweyeyensis, and A. harkoneniana that form a clade. Among lethal amanitas from tropical Africa, A. albolimbata is similar to A. strophiolata because of the white basidiomata, but unfortunately, we do not have any material of the latter species to test its phylogenetic relationship to other species. However, A. strophiolata presents some distinct morphological characteristics that clearly separate it from A. albolimbata. Amanita strophiolata was described by Beeli (1927, 1935) from DR Congo and is distinguished by its umbonate pileus, often yellowish at center, the absence of volval remnants on pileus, ellipsoid to elongate basidiospores (10–11 × 6–7 μm), a distinctive annulus in the form of a funnel, and larger basidiomata.
Amanita albolimbata also shows some similarities with Asian, European, and American species including A. exitialis, A. bisporigera, A. molliuscula, A. parviexitialis, and A. virosa, based on the white basidiomata. Those species have never been reported in tropical Africa and differ from the African species by morphological characteristics. Amanita exitialis was described from China (Yang and Li, 2001) and subsequently has been collected from India (Bhatt et al., 2003). The species is distinguished from A. albolimbata by the absence of voval remnants on pileus, globose-to-subglobose basidiospores (9.5–12 × 9–11.5 μm), 2-spored basidia, scarce inflated cells in the inner part of the volva, and larger basidiomata (Yang and Li, 2001; Cui et al., 2018). Amanita bisporigera was described from North America and characterized by the absence of voval remnants on pileus, a skirt-like annulus, globose-to-subglobose basidiospores (7.8–9.6 × 7–9 μm), 2-spored basidia and sometimes 4-spored, and larger basidiomata (Jenkins, 1986; Tulloss et al., 1995; Yang and Li, 2001; Yang, 2015). The species mainly has 2-spored basidia, in late spring and early summer; later in year, it may sometimes mainly or entirely have 4-spored basidia (Tulloss and Possiel, 2005). Amanita molliuscula was described from China and characterized by the absence of voval remnants on pileus, globose-to-subglobose basidiospores (7.5–9 × 7–8 μm), and larger basidiomata (Cai et al., 2016; Cui et al., 2018). Amanita parviexitialis was described from China and characterized by the absence of voval remnants on pileus sometimes slightly brownish at center, subglobose, rarely globose to broadly ellipsoidal basidiospores (7.5–9.5 × 7–9 μm), 2-spored basidia, and smaller basidiomata (Cai et al., 2016; Cui et al., 2018). Amanita virosa is widely distributed across Europe and temperate to subtropical Asia (Neville and Poumarat, 2004; Zhang et al., 2010; Li et al., 2015; Yang, 2015; Cai et al., 2016; Cui et al., 2018). It has an umbonate pileus, white, often cream at the center, the absence of volval remnants on pileus, the presence of globose-to-subglobose basidiospores (8–11 × 8–10 μm), scarce inflated cells in the inner part of the volva, and larger basidiomata (Cai et al., 2016; Cui et al., 2018). Generally, A. albolimbata sometimes has a patchy volval remnant on the pileus, which is typically absent for A. exitialis, A. bisporigera, A. molliuscula, A. strophiolata, and A. virosa (Beeli, 1927, 1935; Tulloss and Possiel, 2005; Cai et al., 2016; Cui et al., 2018).
Amanita albolimbata is also distinct from other white species in Amanita sect. Phalloideae by its ecology. It occurs in woodland and gallery forests, associated with U. guineensis or U. togoensis (Phyllanthaceae) and I. doka (Fabaceae/Leguminosae), whereas A. strophiolota grows in swampy forests (Beeli, 1927, 1935). Amanita exitialis, A. bisporigera, A. molliuscula, A. parviexitialis, and A. virosa occur in forests of Pinaceae and Fagaceae (Tulloss et al., 1995; Cai et al., 2016; Cui et al., 2018).
In the multigene phylogenetic tree, the African and Australian taxa are basal. This suggests that the lethal amanitas originated from the palaeotropical areas. Cai et al. (2014) also suggested a possible palaeotropical origin of lethal amanitas and highlighted the need for more molecular–phylogenetic studies on collections from the tropics and the Southern Hemisphere.
Toxicity in Amanita
For centuries, wild mushrooms have been consumed massively and popular in the human diet because of their matchless taste, protein content, and medicinal properties (de Román et al., 2006; Cheung, 2010). However, the high interest on wild mushroom collections and consumption could increase the risk of poisoning by lethal mushrooms. During picking, confusions could easily be made between edible and poisonous mushrooms because of their morphological similarities. Many mushroom poisoning cases have been reported worldwide and have mainly been caused by members of A. sect. Phalloideae (Zhang et al., 2010; Cai et al., 2014, 2016; Yang, 2015; Li et al., 2015, 2020; Thongbai et al., 2017). Consequently, much attention has been devoted to the species producing toxins within A. sect. Phalloideae (Chen et al., 2014; Li et al., 2014; Garcia et al., 2015; Cai et al., 2016).
The different toxins documented in those species are mainly amatoxins, phallotoxins, and virotoxins, which can cause severe damages, like liver and renal failure (Wieland, 1973, 1986; Chen et al., 2014). Amanita exitialis, A. bisporigera, A. brunneitoxicaria, A. djarilmari, A. eucalypti, A. fuliginea, A. fuligineoides, A. gardeneri, A. marmorata, A. millsii, A. molliuscula, A. ocreata, A. pallidorosea, A. parviexitialis, A. phalloides, A. rimosa, A. suballiacea, A. subjunquillea, A. verna, and A. virosa are known to contain those toxins and are distributed across Asia, America, Australia, and Europe (Wieland, 1973, 1986; Bresinsky and Besl, 1985; Chen et al., 2014; Li et al., 2014; Garcia et al., 2015; Cai et al., 2016).
Until now, no lethal amanitas had been reported from West Africa. However, lethal amanitas have been documented from Central Africa and Madagascar (Fraiture et al., 2019). Amanita albolimbata represents the first lethal species of A. sect. Phalloideae known from West Africa. The most notorious toxins, α-amanitin, β-amanitin, and phallacidin, have also been detected in the species.
Numerous amanitoid taxa are harvested and consumed by local people in tropical Africa (Codjia and Yorou, 2014; Yorou et al., 2014; Boni and Yorou, 2015; De Kesel et al., 2017; Fadeyi et al., 2017; Milenge et al., 2018; Soro et al., 2019). Because of the whitish color of the basidiomata, A. albolimbata can be confused with A. subviscosa Beeli. Amanita subviscosa is commonly harvested and used as food by local people in Benin (Yorou et al., 2014; Boni and Yorou, 2015; Fadeyi et al., 2017, 2019). Still, A. subviscosa displays contrasting morphological characteristics with A. albolimbata by a slightly squamulose and viscous pileus, slightly striated margin, slightly bulbous, and slightly furfuraceous and hollow stipe, with a distinctive membranous volva (Beeli, 1935). The lack of A. albolimbata in various ethnomycological investigations (Yorou et al., 2014; Boni and Yorou, 2015; Fadeyi et al., 2017; Soro et al., 2019) attests that either local people are aware about its toxicity, or some fatal but unrecorded cases did occur within rural communities. However, it is important to educate local people on the best ways to discriminate morphologically close taxa in order to avoid the consumption of lethal Amanita species for an effective prevention of future poisoning incidents.
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZLY, JEIC, and NSY conceived and designed the research. JEIC collected the species, performed the molecular phylogenetic analyses and the taxonomic studies, and wrote the first draft of the manuscript. JEIC and QC generated the DNA sequences. JEIC and SWZ carried out the cyclic peptide toxins analyses. QC, HL, MR, NSY, and ZLY critically revised and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the International Partnership Program of Chinese Academy of Sciences (No. 151853KYSB201 70026), Yunnan Ten-Thousand-Talents Plan – Yunling Scholar Project, and the FORMAS Grant (No. 226-20141109).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Gang Wu and Bang Feng from Kunming Institute of Botany, CAS, for providing additional materials and images for this study. We thank Yang-Yang Cui, Pan Meng Wang, Si-Peng Jian, Xin Xu, and Kui Wu (Kunming Institute of Botany, CAS) for their kind assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.599047/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree inferred by Maximum Likelihood analysis based on combined dataset (ITS, nrLSU, rpb2, tef1-α, and β-tubulin). Bootstrap values ≥50% are reported on branches. Sequences generated in this study are highlighted in red.
Supplementary Figure 2 | Phylogenetic tree inferred by Bayesian Inference analysis based on combined dataset (ITS, nrLSU, rpb2, tef1-α, and β-tubulin). Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Supplementary Figure 3 | Phylogenetic tree inferred by Maximum Likelihood analysis based on ITS sequences. Bootstrap values ≥50% and Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Supplementary Figure 4 | Phylogenetic tree inferred by Maximum Likelihood analysis based on nrLSU sequences. Bootstrap values ≥50% and Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Supplementary Figure 5 | Phylogenetic tree inferred by Maximum Likelihood analysis based on rpb2 sequences. Bootstrap values ≥50% and Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Supplementary Figure 6 | Phylogenetic tree inferred by Maximum Likelihood analysis based on tef1-α sequences. Bootstrap values ≥50% and Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Supplementary Figure 7 | Phylogenetic tree inferred by Maximum Likelihood analysis based on β-tubulin sequences. Bootstrap values ≥50% and Bayesian posterior probabilities ≥0.90 are reported on branches. Sequences generated in this study are highlighted in red.
Footnotes
References
Bas, C. (1969). Morphology and subdivision of Amanita and a monograph on its section Lepidella. Persoonia 5, 285–573.
Beeli, M. (1927). Contribution à l’étude de la flore mycologique du Congo. II. Bull. Soc. R. Bot. Belg. 59, 101–112.
Beeli, M. (1935). Flore iconographique des champignons du Congo. Amanita. Fasc. I. Bruxelles: Office de Publicité, 1–27.
Bhatt, R. P., Tulloss, R. E., Semwal, K. C., Bhatt, V. K., Moncalvo, J. M., and Stephenson, S. L. (2003). Amanitaceae reported from India. A critically annotated checklist. Mycotaxon 88, 249–270.
Boni, S., and Yorou, N. S. (2015). Diversité et Variabilité inter-ethniques dans la consommation de champignons sauvages de la région de N’Dali au Bénin. Tropicultura 33, 266–276.
Bresinsky, A., and Besl, H. (1985). Giftpilze, mit einer Einführung in die Pilzbestimmung. Stuttgart: Wissenschaftl. Verlagsgesellschaft.
Bresinsky, A., and Besl, H. (1990). A Colour Atlas of Poisonous Fungi: A Handbook for Pharmacist, Doctor and Biologist. London: Wolf Publishing Ltd.
Cai, Q., Cui, Y. Y., and Yang, Z. L. (2016). Lethal Amanita species in China. Mycologia 108, 993–1009. doi: 10.3852/16-008
Cai, Q., Tulloss, R. E., Tang, L., Tolgor, B., Zhang, P., Chen, Z. H., et al. (2014). Multi-locus phylogeny of lethal Amanitas: implications for species diversity and historical biogeography. BMC Evol. Biol. 14:143. doi: 10.1186/1471-2148-14-143
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen, Z. H., Zhang, P., and Zhang, Z. G. (2014). Investigation and analysis of 102 mushroom poisoning cases in Southern China from 1994 to 2012. Fungal Divers. 64, 123–131. doi: 10.1007/s13225-013-0260-7
Cheung, P. C. K. (2010). The nutritional and health benefits of mushrooms. Nutr. Bull. 35, 292–299. doi: 10.1111/j.1467-3010.2010.01859.x
Codjia, J. E. I., and Yorou, N. S. (2014). Ethnicity and gender variability in the diversity, recognition and exploitation of Wild Useful Fungi in Pobè region (Benin, West Africa). J. Appl. Biosci. 78, 6729–6742. doi: 10.4314/jab.v78i0.14
Corner, E. J. H., and Bas, C. (1962). The genus Amanita in Singapore and Malaya. Persoonia 2, 241–304.
Cui, Y. Y., Cai, Q., Tang, L. P., Liu, J. W., and Yang, Z. L. (2018). The family Amanitaceae: molecular phylogeny, higher-rank taxonomy and the species in China. Fungal Divers. 91, 5–230. doi: 10.1007/s13225-018-0405-9
De Kesel, A., Degreef, J., and Kasongo, B. (2017). Champignons Comestibles du Haut-Katanga (R.D. Congo). Bruxelles: ABC taxa, 17.
de Román, M., Boa, E., and Woodward, S. (2006). Wild-gathered fungi for health and rural livelihoods. Proc. Nutr. Soc. 65, 190–197. doi: 10.1079/pns2006491
Deng, W. Q., Li, T. H., Xi, P. G., Gan, L. X., Xiao, Z. D., and Jiang, Z. D. (2011). Peptide toxin components of Amanita exitialis basidiocarps. Mycologia 103, 946–949. doi: 10.3852/10-319
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Eyi-Ndong, H., Degreef, J., and De Kesel, A. (2011). Champignons Comestibles des Forêts Denses d’Afrique Centrale: Taxonomie et Identification, Vol. 10. Bruxelles: ABC taxa.
Fadeyi, O. G., Assogba, F. M., Chabi, D. D. C. B., Yorou, N. S., and Gbenou, J. D. (2019). Ethnomycology, myco-chemical analyzes and antioxidant activity of eleven species of the genus Amanita (Basidiomycota, fungi) from Benin (West Africa). J. Pharm. Phytochem. 8, 335–341.
Fadeyi, O. G., Badou, A. S., Aignon, L. H., Codjia, J. E. I., Moutouama, J. K., and Yorou, N. S. (2017). Études ethnomycologiques et identification des champignons sauvages comestibles les plus consommés dans la région des Monts-Kouffè au Bénin (Afrique de l’ouest). Agron. Afr. 29, 93–109.
Fraiture, A., Amalfi, M., Raspé, O., Kaya, E., Akata, I., and Degreef, J. (2019). Two new species of Amanita sect. Phalloideae from Africa, one of which is devoid of amatoxins and phallotoxins. MycoKeys 53, 93–125. doi: 10.3897/mycokeys.53.34560
Garcia, J., Costa, V. M., Carvalho, A., Baptista, P., de Pinho, P. G., de Lourdes Bastos, M., et al. (2015). Amanita phalloides poisoning: mechanisms of toxicity and treatment. Food Chem. Toxicol. 86, 41–55. doi: 10.1016/j.fct.2015.09.008
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41, 95–98.
Härkönen, M., Niemelä, T., Kotiranta, H., and Pierce, G. (2015). Zambian Mushrooms and Mycology. Helsinki, FL: Finnish Museum of Natural History, 29.
Katoh, K., and Standley, D. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kornerup, A., and Wanscher, J. H. (1981). Taschenlexikon der Farben, 3rd Edn. Zürich: Muster-Schmidt Verlag.
Li, C., and Oberlies, N. H. (2005). The most widely recognized mushroom: chemistry of the genus Amanita. Life Sci. 78, 532–538. doi: 10.1016/j.lfs.2005.09.003
Li, H. J., Xie, J. W., Zhang, S., Zhou, Y. J., Ma, P. B., Zhou, J., et al. (2015). Amanita subpallidorosea, a new lethal fungus from China. Mycol. Prog. 14:43. doi: 10.1007/s11557-015-1055-x
Li, H. J., Zhang, H. S., Zhang, Y. Z., Zhang, K. P., Zhou, J., Yin, Y., et al. (2020). Mushroom poisoning outbreaks — China, 2019. China CDC Weekly 2, 19–27. doi: 10.46234/ccdcw2020.005
Li, P., Deng, W. Q., and Li, T. H. (2014). The molecular diversity of toxin gene families in lethal Amanita mushrooms. Toxicon 83, 59–68. doi: 10.1016/j.toxicon.2014.02.020
Lüli, Y., Cai, Q., Chen, Z. H., Sun, H., Zhu, X., Li, X., et al. (2019). Genome of lethal Lepiota venenata and insights into the evolution of toxin-biosynthetic genes. BMC Genomics 20:198. doi: 10.1186/s12864-019-5575-7
Milenge, K. H., Nshimba, S. M. H., Masumbuko, N. C., Degreef, J., and De Kesel, A. (2018). Uses and importance of wild fungi: traditional knowledge from the Tshopo province in the Democratic Republic of the Congo. J. Ethnobiol. Ethnomed. 14:13. doi: 10.1186/s13002-017-0203-6
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop, New Orleans, LA, 1–8.
Neville, P., and Poumarat, S. (2004). Amaniteae: Amanita, Limacella & Torrendia. Alassio: Edizioni Candusso.
Patrouillard, N. (1924). Basidiomycètes nouveaux de Madagascar. Bull. Mus. Natl. Hist. Nat. 30, 526–532.
Rambaut, A. (2009). FigTree Version 1.3.1. Avalaibale at: http://tree.bio.ed.ac.uk.
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 12–20. doi: 10.1093/sysbio/sys029
Smith, S. A., and Dunn, C. W. (2008). Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24, 715–716. doi: 10.1093/bioinformatics/btm619
Soro, B., Koné, N. A., Vanié-Léabo, L. P. L., Konaté, S., Bakayoko, A., and Koné, D. (2019). Phytogeographical and sociolinguistical patterns of the diversity, distribution, and uses of wild mushrooms in Côte d’Ivoire, West Africa. J. Ethnobiol. Ethnomed. 15:5. doi: 10.1186/s13002-019-0284-5
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Swofford, D. L. (2002). PAUP∗. Phylogenetic Analysis Using Parsimony (∗and Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates.
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Thongbai, B., Miller, S. L., Stadler, M., Wittstein, K., Hyde, K. D., Lumyong, S., et al. (2017). Study of three interesting Amanita species from Thailand: morphology, multiple-gene phylogeny and toxin analysis. PLoS One 12:e018213. doi: 10.1371/journal.pone.0182131
Tulloss, R. E., and Bhandary, H. R. (1992). Amanita chepangiana —a new species from Nepal. Mycotaxon 43, 25–31.
Tulloss, R. E., Bhatt, R., Stephenson, S., and Kumar, A. (1995). Key to Species of Amanita Section Phalloideae from North and Central America. Available online at: http://www.amanitaceae.org/?Phalloideae+of+North+and+Central+America (accessed August 2, 2020).
Tulloss, R. E., and Possiel, L. (2005). Key to Species of Amanita Section Phalloideae From North and Central America. Avaliable at: http://www.Amanitaceae.org/?Phalloideae+of+North+and+Central+America.
Tulloss, R. E., Possiel, L., and Mazzella, E. (2020). Checklist of Amanita taxa found in sub-Saharan Africa. Avaliable at: http://www.Amanitaceae.org/?sub-Saharan+Africa (accessed April 16, 2020).
Tulloss, R. E., and Yang, Z. L. (2011). “Morphological study of Amanita (Fungi: Agaricales)—notes on methodology,” in Amanita Workshop, 6th Edn, eds R. E. Tulloss and C. Rodríguez Caycedo (Cook College: Rutgers University).
Unluoglu, I., and Tayfur, M. (2003). Mushroom poisoning: an analysis of the data between 1996 and 2000. Eur. J. Emerg. Med. 10, 23–26. doi: 10.1097/00063110-200303000-00007
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Walleyn, R., and Verbeken, A. (1998). Notes on the genus Amanita in Sub-Saharan Africa. Walleyn 131, 156–161.
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (California: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wieland, T., Goetzendoerfer, C., Dabrowski, J., Lipscomb, W. N., and Shoham, G. (1983). Unexpected similarity of the structures of the weakly toxic amanitin (S)-sulfoxide and the highly toxic (R)-sulfoxide and sulfone as revealed by proton nuclear magnetic resonance and x-ray analysis. Biochemistry 22, 1264–1271. doi: 10.1021/bi00274a043
Yang, Z. L., and Li, T. H. (2001). Notes on three white Amanitae of section Phalloideae (Amanitaceae) from China. Mycotaxon 78, 439–448.
Yorou, N. S., Koné, N. G. A., Guissou, M., Guelly, K. A., Maba, L. D., Ekué, M., et al. (2014). “Biodiversity and sustainable use of wild edible fungi in the sudanian centre of endemism: a plea for valorisation,” in Ectomycorrhizal Symbioses in Tropical and Neotropical Forests, eds A. M. Bâ, K. L. McGuire, and A. G. Diédhiou (Boca Raton, FL: CRC Press), 241–269. doi: 10.1201/b16536-14
Keywords: amatoxins, phallotoxins, poisoning risk, taxonomy, tropical Africa
Citation: Codjia JEI, Cai Q, Zhou SW, Luo H, Ryberg M, Yorou NS and Yang ZL (2020) Morphology, Multilocus Phylogeny, and Toxin Analysis Reveal Amanita albolimbata, the First Lethal Amanita Species From Benin, West Africa. Front. Microbiol. 11:599047. doi: 10.3389/fmicb.2020.599047
Received: 26 August 2020; Accepted: 21 October 2020;
Published: 20 November 2020.
Edited by:
Rajesh Jeewon, University of Mauritius, MauritiusReviewed by:
Jing Si, Beijing Forestry University, ChinaYusufjon Gafforov, Academy of Science of the Republic of Uzbekistan, Uzbekistan
Copyright © 2020 Codjia, Cai, Zhou, Luo, Ryberg, Yorou and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhu L. Yang, ZnVuZ2lAbWFpbC5raWIuYWMuY24=
 Jean Evans I. Codjia
Jean Evans I. Codjia Qing Cai
Qing Cai Sheng Wen Zhou
Sheng Wen Zhou Hong Luo
Hong Luo Martin Ryberg
Martin Ryberg Nourou S. Yorou
Nourou S. Yorou Zhu L. Yang1*
Zhu L. Yang1*