
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 November 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.597515
The BipA (BPI-inducible protein A) protein is ubiquitously conserved in various bacterial species and belongs to the translational GTPase family. Interestingly, the function of Escherichia coli BipA is not essential for cell growth under normal growth conditions. However, cultivation of bipA-deleted cells at 20°C leads to cold-sensitive growth defect and several phenotypic changes in ribosome assembly, capsule production, and motility, suggesting its global regulatory roles. Previously, our genomic library screening revealed that the overexpressed ribosomal protein (r-protein) L20 partially suppressed cold-sensitive growth defect by resolving the ribosomal abnormality in bipA-deleted cells at low temperature. Here, we explored another genomic library clone containing yebC, which encodes a predicted transcriptional factor that is not directly associated with ribosome biogenesis. Interestingly, overexpression of yebC in bipA-deleted cells diminished capsule synthesis and partially restored lipopolysaccharide (LPS) core maturation at a low temperature without resolving defects in ribosome assembly or motility, indicating that YebC may be specifically involved in the regulation of exopolysaccharide and LPS core synthesis. In this study, we collectively investigated the impacts of bipA-deletion on E. coli capsule, LPS, biofilm formation, and motility and revealed novel roles of YebC in extracellular polysaccharide production and LPS core synthesis at low temperature using this mutant strain. Furthermore, our findings suggest that ribosomal defects as well as increased capsule synthesis, and changes in LPS composition may contribute independently to the cold-sensitivity of bipA-deleted cells, implying multiple regulatory roles of BipA.
BipA (also known as TypA) is a GTPase belonging to translation/elongation factors (TRAFAC) family and shares structural similarity with translational GTPases (trGTPases), such as IF2, EF-Tu, EF-G, EF4, and RF3 (Ero et al., 2016). It is widely distributed in bacteria and plants (Margus et al., 2007; Wang et al., 2008). Like other ribosome-associated trGTPases, BipA binds to the ribosome only in the GTP-bound form, which in turn stimulates its GTP hydrolysis activity (deLivron and Robinson, 2008; deLivron et al., 2009; Kumar et al., 2015; Choi and Hwang, 2018).
While most trGTPases function as translation factors, BipA has been implicated as a ribosome assembly factor. In Escherichia coli, the deletion of bipA causes defects in growth and 50S ribosomal subunit assembly at low temperature (Choi and Hwang, 2018). E. coli bipA mutant exhibits the accumulation of 44S particles, precursors of 50S ribosomal subunits lacking L6 ribosomal protein (r-protein) and containing immature 16S and 23S rRNAs. Furthermore, the cold-sensitivity exhibited by bipA-deleted cells is suppressed by deletion of rluC, which encodes a pseudouridine synthase that introduces pseudouridine into U955, U2504, and U2580 of 23S rRNA located near the binding site of BipA (Conrad et al., 1998; Krishnan and Flower, 2008; Kumar et al., 2015). In addition, it was recently revealed that overexpressed r-protein L20 partially restores growth and ribosome maturation in a bipA-deleted strain at low temperature (Choi et al., 2019). These genetic links between BipA and RluC or L20 clearly support the notion that BipA plays a pivotal role in 50S ribosomal subunit maturation.
Interestingly, the function of BipA has also been implicated in various stress responses as its expression is significantly increased in response to stresses, such as low temperature, oxidative stress, antibiotics, and antimicrobial peptides (Qi et al., 1995; Farris et al., 1998; Barker et al., 2000; Pfennig and Flower, 2001; Wang et al., 2008; Neidig et al., 2013). In contrast, bipA is not essential for survival under normal growth condition (Pfennig and Flower, 2001). In Pseudomonas aeruginosa, deletion of bipA results in increased sensitivity to antibiotics such as meropenem, ceftazidime, tetracycline, polymyxin B, and colistin (Neidig et al., 2013). In Salmonella typhimurium, expression of bipA is induced by BPI (Bactericidal/permeability-increasing protein), which is a cationic antimicrobial protein produced by human neutrophils (Weiss and Olsson, 1987; Qi et al., 1995). Recently, we have shown that in non-pathogenic E. coli K-12 strain, cAMP-receptor protein (CRP) promotes the expression of bipA at low temperature (Choi and Hwang, 2018).
Unlike BipA in non-pathogenic E. coli, BipA in enteropathogenic E. coli (EPEC) is post-translationally regulated by phosphorylation of tyrosine residues enhancing GTPase activity. This BipA promotes the expression of two pathogenicity islands, espC and locus of enterocyte effacement (LEE), through positive regulation of LEE-encoded regulator (Ler) transcription (Farris et al., 1998; Freestone et al., 1998; Grant et al., 2003). In addition, a bipA-deleted EPEC mutant exhibited hypermotility phenotypes at 37°C (Grant et al., 2003). As a consequence, bipA-deleted EPEC strains lose the ability to induce rearrangement of the actin cytoskeleton, which is a target of bacterial virulence factors (Farris et al., 1998; Sal-Man et al., 2009). The EPEC bipA mutant exhibits increased transcription of group 2 capsule gene clusters at 20°C, suggesting that BipA represses K5 capsule production at 20°C (Rowe et al., 2000). Paradoxically, at 37°C, the level of these transcripts decreases in this bipA mutant, indicating that BipA is required for their maximum transcription at 37°C (Rowe et al., 2000). More peculiarly, however, the bipA-deleted K-12 strain presents decreased motility and overproduction of colanic acid (CA)-containing capsule at low temperature (Choi and Hwang, 2018), which is contrary to phenotypes in EPEC, implying that BipA may function differentially in EPEC and K-12 strains. Regarding lipopolysaccharide (LPS), an insertional mutation at the 3′ end of bipA was screened as a suppressor mutant for the E. coli K-12 waaQ mutation, which inactivates the entire waa operon and confers hypersensitivity to sodium dodecyl sulfate (SDS), bile salts, and the hydrophobic antibiotic novobiocin through impairment of LPS inner core biosynthesis (Moller et al., 2003). This insertional bipA mutant was partially functional and somehow restored LPS core biosynthesis by expressing the waa operon downstream of waaQ (Moller et al., 2003). In P. aeruginosa PAO1, transposon insertion in bipA leads to reduction in swarming motility and impairment of biofilm development (Overhage et al., 2007). In addition, deletion of bipA in P. aeruginosa PA14 causes a reduction in virulence in a Dictyostelium discoideum amoeba model (Rahme et al., 1995; Choi et al., 2002; Neidig et al., 2013). This PA14 mutant also exhibits reduction in the expression of Type III secretion system (T3SS) genes and impairment of biofilm formation (Neidig et al., 2013). Thus, it is likely that BipA is a crucial virulence factor regulating capsule, LPS, motility and biofilm formation.
In our previous study, we conducted genomic library screening in an E. coli K-12 bipA mutant to further comprehend its physiological role. The yebC gene was screened as a suppressor of cold-sensitivity (Choi et al., 2019). The YebC/PmpR family proteins are widespread and conserved in many bacteria (Wei et al., 2018). Even though its function could not be deduced from the X-ray structure of Aquifex aeolicus YebC (Shin et al., 2002), recent evidence supports the notion that in various bacteria, YebC is a potential transcriptional regulator with either activator or repressor function. For example, in Alishewanella sp. WH16-1, YebC induced by Cr(VI) positively regulates the transcription of ruvRCAB, contributing to Cr(VI), As(III), Sb(III), and Cd(II) resistance (Wu et al., 2019). In Lactobacillus delbrueckii subsp. lactis CRL 581, transcription of prtL, oppA, and optS is repressed by binding of YebC to the respective promoter regions (Brown et al., 2017). Interestingly, YebC of Edwardsiella piscicida binds to edwR or ETEA_0873 promoter regions resulting in the repression of quorum-sensing (QS) response regulator or activation of T3SS expression, respectively (Wei et al., 2018). In P. aeruginosa, YebC inhibits the transcription of pqsR, which is a Pseudomonas quinolone signal QS response regulator (Liang et al., 2008). These accumulated data raise the possibility that YebC may control the transcription of specific genes.
Despite the possible roles of YebC as a transcription factor in various bacteria, the function of YebC in E. coli has not been evaluated yet. Therefore, in the present study, considering pleiotropic phenotypes such as capsule production, LPS synthesis, biofilm formation, and motility altered by bipA-deletion, we explored the phonotypic impacts of overexpression of yebC on a bipA-deletion genetic background to elucidate the role of YebC.
The E. coli strains used in this study are listed in Table 1. The MG1655 yebC::kan (ESC48) and rcsF::kan (ESC53) were constructed by bacteriophage P1 transduction using JW1853 (yebC::kan) and JW0192 (rcsF::kan), respectively (Baba et al., 2006). The P1 lysate obtained from JW0192 was also employed to construct a double mutant (ESC54; ΔbipA, rcsF::kan). Kanamycin resistant cells were isolated and the disruption of each gene was confirmed by PCR using the primer sets listed in Supplementary Table S1. The removal of the kanamycin cassette at the bipA locus was carried out as described previously (Datsenko and Wanner, 2000). All E. coli strains were cultivated at either 37°C or 20°C in Luria–Bertani (LB) broth with chloramphenicol (50 μg/ml), kanamycin (50 μg/ml), or ampicillin (100 μg/ml) as needed. Bile salts was purchased from Sigma-Aldrich (catalog number: 48305).
Genomic library construction and suppressor screening were performed as described in our previous study (Choi et al., 2019). In brief, genomic DNA from bipA-deleted cells (ESC19) was purified and partially digested DNA fragments were ligated into pACYC184 yielding the ESC19 genomic library. After transforming ESC19 cells with the genomic library, suppressors of cold-sensitive growth were screened by selecting larger colonies than those harboring empty vector (pACYC184). To confirm their suppressing activities, library clones extracted from those larger colonies were retransformed into ESC19 cells. A total of 26 isolated library clones were sequenced from both ends to identify the inserted genomic DNA fragments.
To construct truncated library clones of pBIS05, DNA fragments of pBIS05 were amplified by PCR using pBIS05 as a template and the primer sets presented in Supplementary Table S1. The PCR fragments were ligated into the SmaI site of pUC19 followed by digestion with BamHI. Each digested DNA insert was ligated into the BamHI site of pACYC184, yielding pBIS05-1, pBIS05-2, and pBIS05-3. To replace the initiation codon AUG of yebC with stop codon UAG, site-directed mutagenesis PCR was carried out using pBIS05 as the template and the primer set in Supplementary Table S1.
Polysome and subunit profiling experiments were carried out as described previously (Choi and Hwang, 2018). In brief, MG1655, ESC48, or ESC19 transformants harboring plasmid were cultured to the logarithmic phase at 37°C or 20°C in LB medium supplemented with the appropriate antibiotics. To trap polysomes, chloramphenicol was added to cultures at a final concentration of 100 μg/ml (for the strains MG1655 and ESC48) or 150 μg/ml (for the ESC19 transformants). After an additional 3 min of incubation, the cells were harvested by centrifugation. The cell pellets were resuspended in Buffer BP [20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 100 mM NH4Cl, and 5 mM β-mercaptoethanol (BME)]. Cells were then lysed by freeze–thaw cycle, and the cell lysates were obtained by centrifugation. Then, cleared lysates were loaded into 5–40% sucrose gradients in Buffer BP and resolved by ultracentrifugation at 4°C using a Beckman SW41 rotor at 170,000 × g for 2.5 h. For ribosomal subunit profiling experiments, the cells were lysed in Buffer BS [20 mM Tris–HCl (pH 7.5), 1 mM MgCl2, 100 mM NH4Cl, and 5 mM BME], and cleared cell lysates were prepared similarly as described for polysome profiling experiments. Then, lysates were subjected to 5–25% sucrose gradients in Buffer BS followed by centrifugation at 4°C in the same rotor for 3.5 h at 170,000 × g.
The morphology of colonies grown for 18 h at 37°C or for 96 h at 20°C on LB agar plates was observed using Azure C200 photo documentation (Azure, United States). For capsule staining, 5 μl of the resuspended colonies in 0.4% saline solution were dropped on a clean glass microscope slide and mixed with 10 μl of 1% crystal violet solution. Then, the mixture was spread using the edge of a clean cover glass to form a thin smear by placing the end of cover glass at an angle to the end of the slide containing the cells. After air drying for 10 min, crystal violet was washed with 20% copper sulfate solution. Then, to stain the background around E. coli cells, 1% Congo red solution was added to the extent that the smear was sufficiently covered. After staining for 10 min, Congo red was gently rinsed away with distilled water followed by an additional air drying. Then, capsule was observed under oil immersion with an Axio Observer (ZEISS, Germany) light microscope at 1,000 × magnification.
Colonies that formed on LB agar plates were scraped using 10 ml of fresh LB medium, and the collected cells were centrifuged to remove supernatant. Total RNA extraction and cDNA synthesis were carried out as we previously described (Choi and Hwang, 2018). We performed qRT-PCR using 1 μl of cDNA and 2 pmole of each gene-specific primer (Supplementary Table S1) in a 20 μl volume with 2× SYBR Green master mix (Qiagen). The reactions were carried out on a QuantStudio 3 Real-Time PCR system (Applied Biosystems) using the following cycling parameters: 95°C for 15 min and 40 cycles of denaturation at 95°C for 15 s, primer annealing at 55°C for 30 s, and extension at 72°C for 30 s. The gapA gene served as an endogenous reference (Navasa et al., 2011), and relative quantification (RQ) values were calculated using the comparative Ct method (Livak and Schmittgen, 2001).
Lipopolysaccharide was extracted from E. coli strains using the hot-phenol and diethyl ether method as previously described (Davis and Goldberg, 2012). Cells grown overnight in 3 ml of LB containing appropriate antibiotics were spread on LB agar plates. After incubation at 37°C or 20°C, colonies were scraped using 10 ml of fresh LB medium. The pelleted cells were resuspended in 200 μl of 1× SDS buffer [50 mM Tris–HCl (pH 6.8), 2% BME, 2% sodium dodecyl sulfate (SDS), and 10% glycerol] and boiled in a water bath for 15 min. Samples were cooled at room temperature for 15 min. Five microliters each of DNase I (10 mg/ml) and RNase A (10 mg/ml) solutions were added to the sample, and these mixtures were incubated at 37°C for 30 min. Next, 10 μl of Proteinase K (10 mg/ml) was added, followed by another incubation at 59°C for 3 h. 200 μl of ice-cold Tris–saturated phenol was added to each sample, and the samples were briefly vortexed for 5 to 10 s. The phenol mixtures were incubated at 65°C for 15 min, vortexing occasionally. After cooling at room temperature, 1 ml of diethyl ether was added to each sample, and the mixture was vortexed for 5 to 10 s. The samples were centrifuged at 17,000 × g for 10 min, and LPS was carefully extracted from the bottom layer. LPS samples were separated using 12.5% Tricine SDS-PAGE and visualized by silver staining as previously described (Morrissey, 1981).
Quantification of biofilm formation in glass tubes was performed as described previously (Naves et al., 2008). Twenty-five microliters of overnight culture were inoculated into 2.5 ml of fresh LB in glass tubes (16 × 100 mm). The cells were incubated for 24 h at 37°C or 48 h at 20°C, followed by removal of the culture medium. The tubes were rinsed once with 2.7 ml of 0.85% saline solution. After drying at 37°C for 30 min, 2.7 ml of 0.1% crystal violet dye was added to each glass tube for 5 min at room temperature. After this, the tubes were washed three times with 3 ml of distilled water, and the excess water was removed from the glass tubes by inverting the tubes and tapping them on paper towels. The glass tubes were allowed to dry and then photographed. The crystal violet was solubilized by adding 3 ml of 95% ethanol to the glass tube. Absorbance at 540 nm was then measured and recorded using a MultiskanTM GO Microplate Spectrophotometer (Thermo Fisher ScientificTM). Tubes containing only LB were used as controls, and their OD540 values were subtracted from all samples.
Results are presented as the mean ± SD of three independent experiments. The data were analyzed by unpaired two-tailed t-test. The statistical analyses were performed with the following significance levels: NS, non-significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Previously, we constructed a genomic library using genomic DNA from the strain ESC19 (bipA-deletion) and searched for an element(s) in the E. coli genome that can revert cold-sensitivity in the strain ESC19. A total of 26 library clones were isolated as positive suppressor clones. We have shown that among them, overexpression of rplT, whose gene product is r-protein L20, partially rescued the ribosomal defects caused by the deletion of bipA at low temperature. Among the remaining clones that were screened, we attempted to investigate the suppressor clone, pBIS05, which contains the nudB-yebC-ruvC genomic DNA fragment. Using this clone, three truncated clones, pBIS05-1, pBIS05-2, and pBIS05-3, were constructed to identify the gene that was responsible for suppression (Figure 1A). Then, these clones were transformed into the ESC19 cells, and the transformants were tested for their abilities to grow normally at a non-permissive temperature (20°C) on LB medium containing chloramphenicol and kanamycin. As shown in Figure 1B, cells harboring pACYC184 had a longer lag phase compared to ESC19 cells with pACYC184BipA. Nevertheless, the growth rates of these two transformants in the mid-exponential phase appeared to be similar. The lag phase of ESC19 cells harboring pBIS05, pBIS05-2, or pBIS05-3 was shorter than that of ESC19 cells with pACYC184, while their growth rates were in a similar range to those of the two control strains. Notably, the ESC19 cells transformed with pBIS05-1 grew as poorly as the cells harboring pACYC184 (Figure 1B). The replacement of yebC start codon in pBIS05 with a stop codon led to a loss of its suppressive activity, indicating that YebC is a responsible suppressor protein (Supplementary Figure S1). These suppressive phenotypes were further confirmed in solid medium at 20°C. In Figure 1C, the pattern of colony formation ability of ESC19 cells transformed with pBIS05 variants indicated very similar suppressive effects as in Figure 1B, indicating that the yebC gene is indeed a suppressor gene. Despite this suppression activity, it should be noted that the suppression was partial.
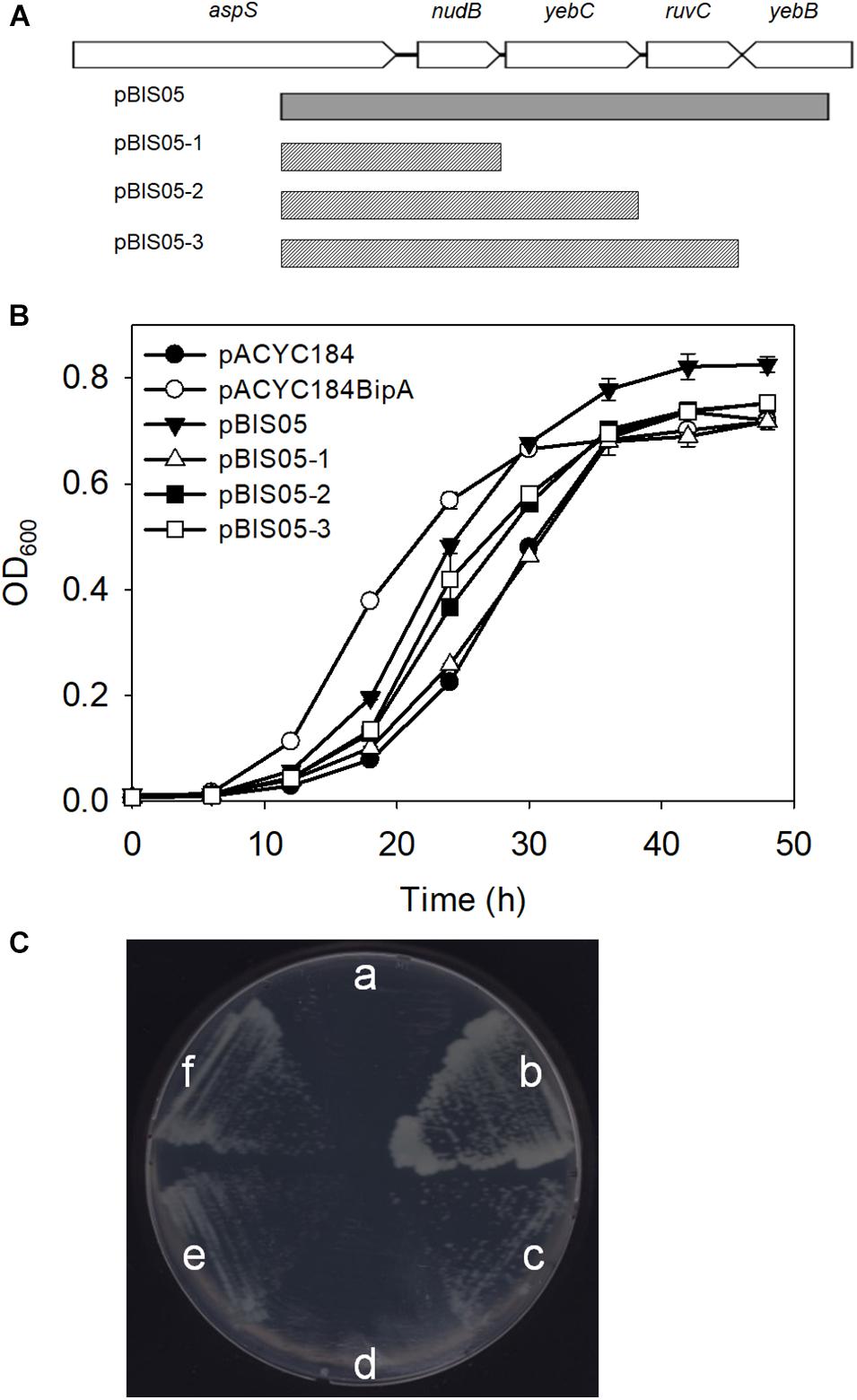
Figure 1. Genomic library clone that suppresses cold-sensitive growth defects of the ESC19 strain at 20°C. (A) Schematic diagram of the genomic locus containing the suppressor gene. The gray square box represents the suppressor library clone. The scratched boxes represent the truncated clones constructed from pBIS05. (B,C) Recovered growth phenotype of the ESC19 strain caused by the isolated suppressor. ESC19 cells were transformed with each plasmid as presented above. Each transformant was incubated in LB medium containing chloramphenicol and kanamycin at 20°C. Cultures were diluted five times before measurement of the optical density at 600 nm. Experiments were performed in three independent replicates and error bars represent SD. Overnight cultures were diluted 100-fold with LB medium containing chloramphenicol and kanamycin. LB agar plates supplemented with same antibiotics were streaked with 2.5 μl of diluted culture. (a), pACYC184; (b), pACYC184BipA; (c), pBIS05; (d), pBIS05-1; (e), pBIS05-2; and (f), pBIS05-3.
As discussed above, BipA may play global regulatory roles in capsule formation, motility, and pathogenicity in cells. Among these, the most apparent phenotype conferred by the bipA-deletion is cold-sensitivity caused by failure of 50S ribosomal subunit assembly (Choi and Hwang, 2018). We have shown that the exogenous expression of rplT restored the growth of the ESC19 strain at low temperature by restoring aberrant 50S ribosomal subunit assembly (Choi et al., 2019). Thus, to investigate whether the ribosomal defects of the ESC19 strain could be suppressed by overexpression of yebC, the polysomes and ribosomal subunits of the suppressed cells were analyzed by sucrose density gradient sedimentation experiments. ESC19 cells harboring pACYC184, pACYC184BipA, and pBIS05-2 were cultivated at 37°C or 20°C until mid-exponential phase and collected by centrifugation. Then, the cell pellets were subjected to sucrose density gradient sedimentation as described in section “Materials and Methods.” As expected, ESC19 cells harboring any of the plasmids exerted normal polysome and subunit profiles without any aberrant or accumulated particles at 37°C (left panels in Figures 2A,B). However, at low temperature, ESC19 cells harboring pACYC184 accumulated a significant level of free 30S ribosomal subunits in the polysome profile. Notably, the peak of 50S ribosomal subunits was almost half of that of 30S ribosomal subunits, and shoulder peak appeared to the right of 50S ribosomal subunits (top right panel in Figure 2A). Consistently, two abnormal particles appeared between the 50S and 30S ribosomal subunit peaks with a concomitant reduction of 50S ribosomal subunits in the subunit profile results for this strain (top right panel in Figure 2B). These results suggest that the absence of BipA at low temperature caused accumulation of unprocessed precursors and/or destabilized 50S ribosomal subunits. ESC19 cells expressing bipA showed a normal accumulation of 50S and 30S ribosomal subunit particles in polysomes at both temperatures (Figure 2A, center). Likewise, unprocessed precursors and/or destabilized 50S ribosomal subunits did not appear between 50S and 30S ribosomal subunit peaks at either temperature (Figure 2B, center), indicating complementation of pACYC184BipA. Interestingly, ESC19 cells transformed with pBIS05-2 exerted very similar polysome profile to those of ESC19 cells transformed with pACYC184 at 20°C (bottom right panel in Figure 2A). Consistently, abnormal 50S ribosomal subunit particles appeared between 50S and 30S peaks in the subunit profiles at 20°C (bottom right panel in Figure 2B). These results suggest that the yebC suppressor gene did not contribute significantly to 50S ribosomal subunit biogenesis, although it alleviated cold-sensitivity. Notably, both normal and abnormal 50S subunit particles overlapped at similar positions in polysome profiles because of a high sucrose gradient concentration and short sedimentation distance (right panel in Figure 2A).
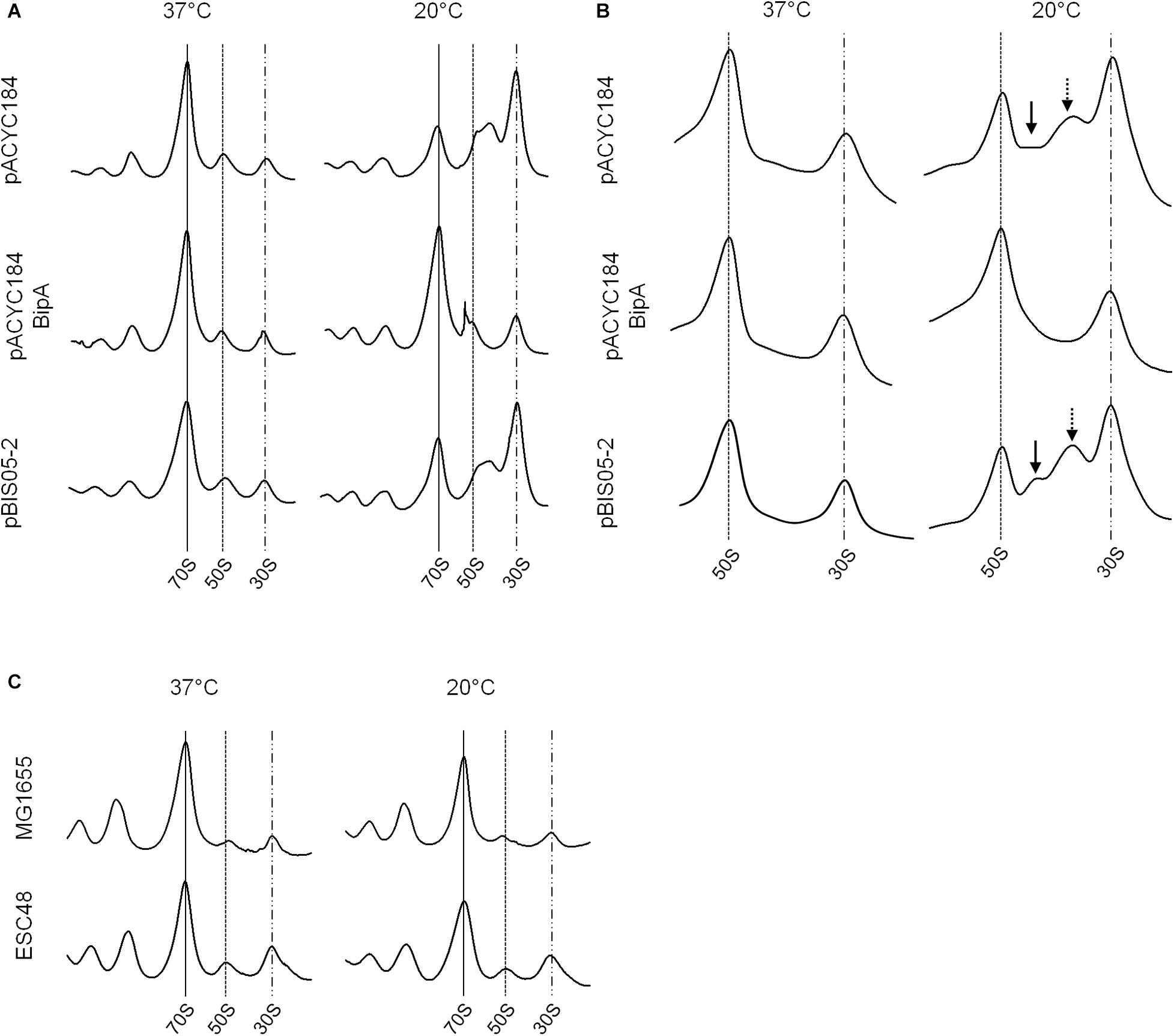
Figure 2. Ribosomal abnormality retained in the suppressor strain. Polysome profiles (A) and subunit profiles (B) of ESC19 cells harboring pACYC184, pACYC184BipA, and pBIS05-2. Transformants were grown at 37°C or 20°C in LB medium containing chloramphenicol until they reached an OD600 of ∼0.6. Then, cell lysates were prepared and analyzed as described in section “Materials and Methods.” Arrows indicate abnormal ribosomal particles. (C) Analyses of polysome profiles of the MG1655 and ESC48 strains. The polysomes were analyzed using the lysate of cells grown at 37°C or 20°C in LB medium until the exponential phase as in (A).
In order to exclude a supportive role of YebC in 50S ribosome biosynthesis, the polysome profiles of wild-type (MG1655) and yebC-deleted (ESC48) strains were explored. The yebC mutant showed no significant ribosomal defects compared to wild-type at either temperature, suggesting that the function of YebC is not associated with ribosome assembly under the given conditions (Figure 2C). This raises the question of whether the cold-sensitivity of the bipA-deleted mutant is not solely due to ribosomal defects.
Interestingly, when we initially confirmed the suppression activities of pBIS02 (a library clone expressing r-protein L20) and pBIS05, we noticed that transformants with pBIS05 were non-mucoid at low temperature unlike those with pBIS02. Therefore, to elucidate a possible role of YebC in suppression at low temperature, we observed the colony morphologies of the suppressed cells. First, we confirmed capsule production in ESC19 at low temperature as shown in Supplementary Figure S2A, and a clear zone around ESC19 cells was visualized using crystal violet and Congo red staining methods (Supplementary Figure S2B). Next, pACYC184, pACYC184BipA, pBIS02-2 (a subclone of pBIS02), or pBIS05-2 was transformed to the ESC19 strain, and the transformants were streaked onto LB agar plates containing chloramphenicol and kanamycin followed by incubation at 37°C or 20°C. As shown in Figure 3A, all transformants were non-mucoid at 37°C. However, at low temperature, the ESC19 strain with pACYC184 showed a substantially mucoid phenotype, which was reversed to non-mucoid when pACYC184BipA was introduced. Likewise, transformants with pBIS05-2 showed the same morphological phenotype at 20°C without being mucoid. Notably, pBIS02-2 and pACYC184BipAN128D did not restore the mucoid phenotype (Figure 3A and Supplementary Figure S3A).
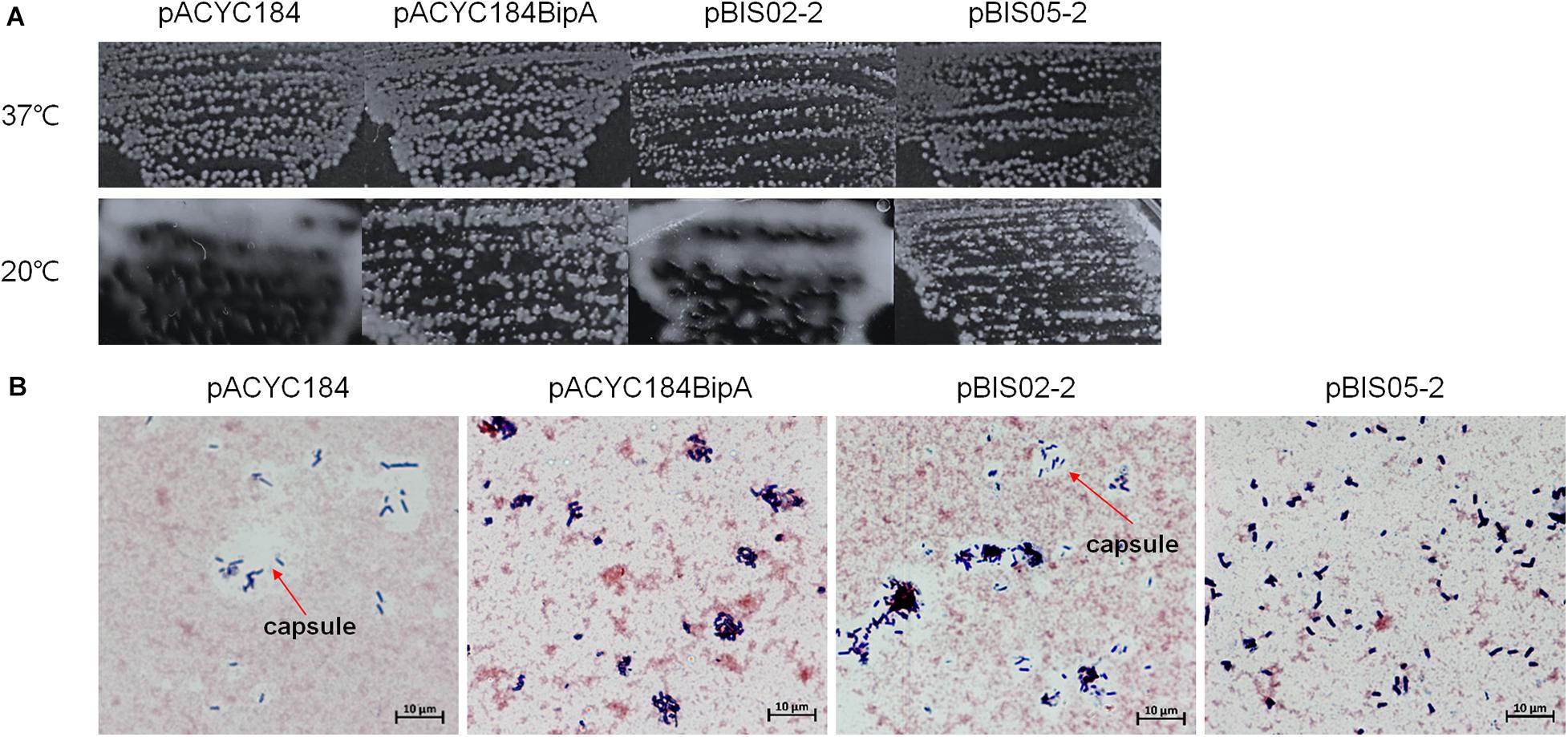
Figure 3. Colony morphologies of the ESC19 transformants. (A) ESC19 cells transformed with pACYC184, pACYC184BipA, pBIS02-2, or pBIS05-2 were grown on LB agar plates supplemented with the appropriate antibiotics overnight at 37°C or 4 days at 20°C. The suppressor clone (pBIS02-2) expressing r-protein L20 was tested as a mucoid suppressor. (B) Capsule staining by crystal violet and Congo red. Colonies from LB plates incubated at 20°C for 4 days were stained and observed as described in section “Materials and Methods.” Arrows indicate capsule.
Colonies of these transformants were stained with crystal violet and Congo red to microscopically observe individual cells as described in section “Materials and Methods.” In Figure 3B, a large clear zone was observed surrounding ESC19 cells harboring pACYC184, which was complemented by expression of bipA from pACYC184BipA. However, the ESC19 strain harboring pBIS05-2 did not form the clear zone, which is formed by excessive production of exopolysaccharide. These results suggest that the deletion of bipA at low temperature led to capsule overproduction and that overexpressed YebC in ESC19 cells may repress production of capsule. Consistently, ESC19 transformants with pBIS02-2 had a clear exopolysaccharide layer around them, indicating that overexpressed L20 in ESC19 cells was unable to repress capsule synthesis. The ESC48 strain did not produce excessive capsular exopolysaccharide at 20°C (Supplementary Figure S2). These results imply that the function of BipA in capsule production repression at low temperature may prevail over that of YebC.
To investigate the role of YebC in suppressing capsule production, transcriptional changes in genes that are involved in capsule synthesis were analyzed using qRT-PCR. We chose six genes in the cps operon, a gene cluster functioning in CA biosynthesis (Stevenson et al., 1996; Rahn et al., 1999). The gmd and ugd genes are involved in CA precursor synthesis (Stevenson et al., 1996; Sturla et al., 1997), and wcaA and wcaK encode CA modification enzymes (Scott et al., 2019). WcaD polymerizes CA units, and Wzc exports colonic acid across the inner membrane (Stevenson et al., 1996; Paulsen et al., 1997).
First, we compared the mRNA levels of those genes in strains MG1655 and ESC19 grown at 37°C or 20°C. Cells grown on LB agar plates were collected by scraping, total RNAs were extracted from the collected cells, and qRT-PCR analysis was conducted. As shown in Figure 4A, both strains grown at 37°C did not have significant changes in transcript levels of any of the six genes. However, at 20°C, the transcript levels of gmd (11.1-fold), ugd (2.7-fold), and wcaA (4.3-fold) were markedly higher in the ESC19 strain than in MG1655, demonstrating capsule overproduction in ESC19 at 20°C.
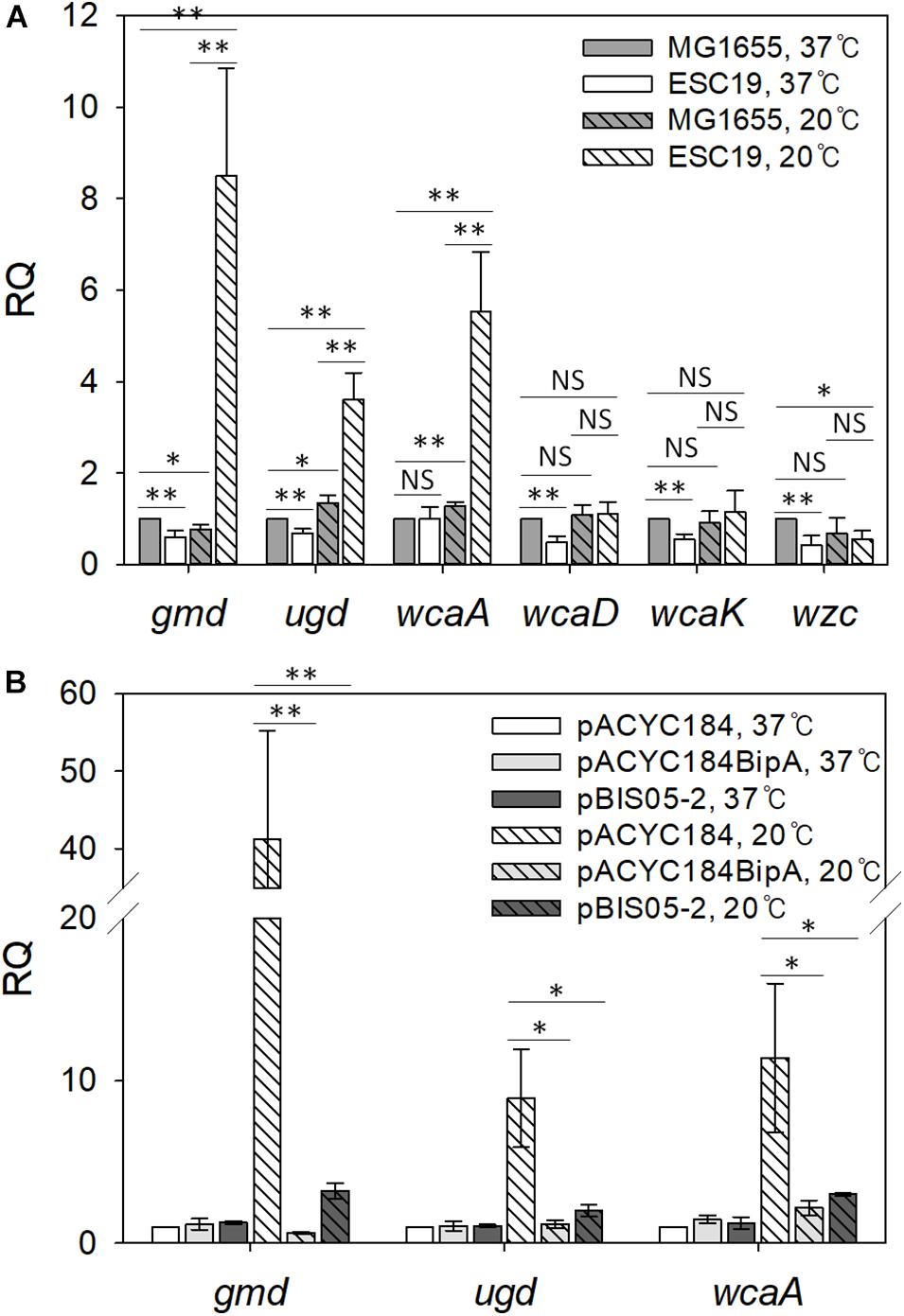
Figure 4. Analysis of transcript levels of genes involved in colanic acid synthesis. (A) Quantification of transcript levels of the cps genes in MG1655 and ESC19 strains at 37°C or 20°C. Cells were grown on LB agar plates at 37°C for 18 h or at 20°C for 5–6 days, and total RNAs were extracted and analyzed using qRT-PCR for gmd, ugd, wcaA, wcaD, wcaK, and wzc transcripts. The expression levels were calculated relative to the expression level of MG1655 grown at 37°C. (B) qRT-PCR analysis of gmd, ugd, and wcaA in ESC19 transformants with pACYC184, pACYC184BipA, or pBIS05-2 at 37°C or 20°C. The expression levels were calculated relative to the expression level of ESC19 with pACYC184 grown at 37°C. The expression levels were normalized to that of the endogenous control gene gapA (glyceraldehyde-3-phosphate dehydrogenase A). Error bars denote SD. NS, non-significant; ∗p < 0.05; ∗∗p < 0.01.
Next, to examine whether YebC could repress the expression of gmd, ugd, and wca genes, we carried out the same experiment using the suppressed strain. As we expected, in the ESC19 cells with pBIS05-2 at 20°C, RQ values for gmd, ugd, and wcaA transcripts decreased to 3.2, 2.0, and 3.8, respectively, indicating similar expression levels to the complemented strain (Figure 4B). At 37°C, there were no significant differences in the expression levels of these three genes among transformants harboring each plasmid. These results suggest that overexpressed YebC led to a reduction in transcript levels of gmd, ugd, and wcaA, and subsequently restored capsule production in the ESC19 strain at 20°C. Considering that Gmd, Ugd, and WcaA precede WcaD, WcaK, and Wzc in CA biosynthesis (Ranjit and Young, 2016; Scott et al., 2019), it is likely that YebC may be involved in regulation of earlier steps of CA synthesis at low temperature.
Recently, it was reported that CA synthesis is activated in response to a defect in the core oligosaccharide of LPS through the regulator of capsule synthesis (Rcs) phosphorelay system, resulting in secretion of large amounts of exopolysaccharide and mucoid colony morphology (Ren et al., 2016). Furthermore, as mentioned above, BipA seems to be functionally associated with LPS core synthesis (Moller et al., 2003). Therefore, we examined the sensitivity of the suppressor strains to bile salts, to which E. coli cells with defective LPS core are known to be hypersensitive (Picken and Beacham, 1977). Overnight cultures of MG1655, ESC19, or ESC48 strains were streaked onto LB agar plates supplemented with 2–5 to 26 g/L bile salts and incubated at 37°C or 20°C. As shown in Figure 5A, the ESC19 strain did not form colonies on the LB plate containing 26 g/L bile salts, whereas MG1655 and ESC48 strains grew normally at 37°C in the presence of bile salts at concentrations up to 26 g/L. In contrast, the growth of MG1655 and ESC48 strains were inhibited by 25 g/L bile salts at 20°C. Interestingly, ESC19 cells did not form colonies at a much lower concentration of bile salts (20 g/L). The hypersensitivity of ESC19 to bile salts at low temperature implies that E. coli may go through changes in composition or structure of LPS core at 20°C and that BipA is functionally implicated in LPS synthesis at 20°C. Next, the suppressor clones were transformed into ESC19 cells to examine whether YebC could recover the sensitivity of ESC19 cells to bile salts at 20°C. Each transformant was streaked on LB plates containing bile salts and incubated at 37°C or 20°C. The complemented ESC19 strain with pACYC184BipA grew well at 37°C on LB plates containing bile salts without any abnormality, while the growth of the other ESC19 transformants were inhibited by 26 g/L bile salts (Figure 5B). However, at 20°C, the ESC19 transformant with pACYC184 became sensitive at ≥23 g/L of bile salts. Notably, ESC19 cells with pBIS05-2 were insensitive to 23 g/L bile salts. The growth of ESC19 cells harboring pACYC184 was inhibited at this concentration of bile salts, suggesting that partial suppression by YebC may be mediated by the recovery of LPS biosynthesis.
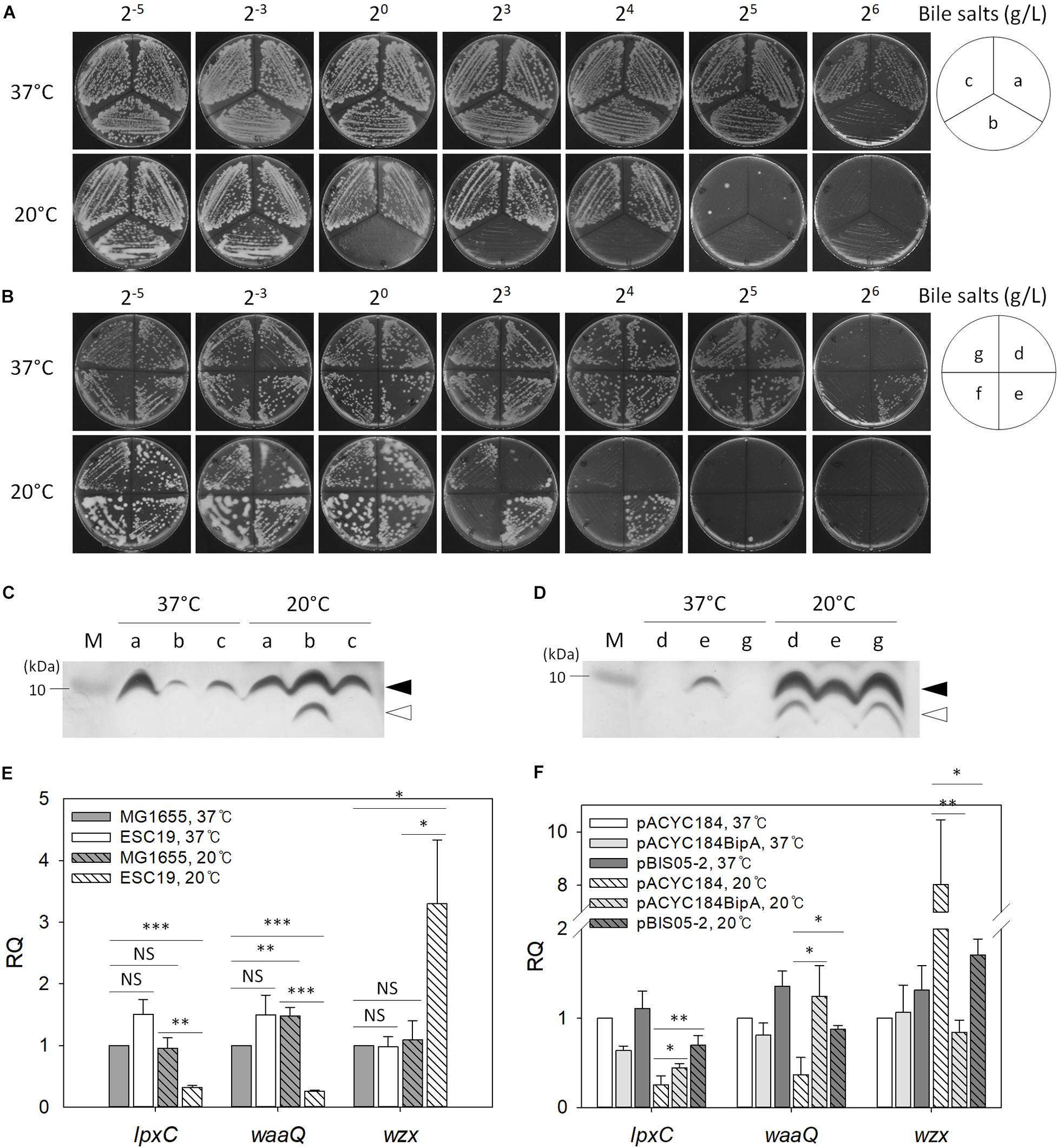
Figure 5. The effect of bipA-deletion on LPS biosynthesis. Overnight cultures of the MG1655, ESC19, and ESC48 strains (A) or the ESC19 transformants with pACYC184, pACYC184BipA, pBIS02-2, or pBIS05-2 (B) were diluted with LB medium to an OD600 of 0.2. Then, 3 μl of the diluted cultures were streaked onto LB plates supplemented with different concentrations of bile salts as presented above, after which the plates were incubated at 37°C or 20°C. The media for transformants contained chloramphenicol and kanamycin. (a), MG1655; (b), ESC19; (c), ESC48; (d), ESC19 with pACYC184; (e), ESC19 with pACYC184BipA; (f), ESC19 with pBIS02-2, and (g), ESC19 with pBIS05-2. (C,D) Tricine SDS-PAGE analysis of LPS. LPS was extracted as described in section “Materials and Methods” and subjected to 12.5% Tricine-polyacrylamide gel electrophoresis followed by silver staining. M, PageRulerTM Unstained Protein Ladder (Thermo Fisher ScientificTM). Alphabetical labels are the same as in (A,B). Filled and open arrows represent mature and precursor LPS core, respectively. (E,F) Relative quantification of mRNA levels of genes involved in LPS biosynthesis. Total RNA was extracted and subjected to qRT-PCR analysis. The relative quantities of lpxC, waaQ, and wzx were normalized to that of the endogenous control gene gapA. Error bars denote the SD using three experimental replicates. NS, non-significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Next, to investigate any LPS core defect in the ESC19 strain, we analyzed the LPS extracted from the various strains. The LPS isolated from MG1655, ESC19, and ESC48 strains were examined using Tricine SDS-PAGE followed by silver staining. As shown in Figure 5C, ESC19 cells showed a normal LPS core, although accumulation was reduced at 37°C. Interestingly, LPS of the ESC19 strain exhibited increased amounts of mature LPS core with a concomitant accumulation of LPS core precursor at 20°C. The upper LPS core band was almost undetectable in the ESC19 transformants with pACYC184 or pBIS05-2, probably because the very small amount of LPS core in ESC19 cells was rapidly consumed as a substrate for O-antigen polymerization at 37°C. However, at 20°C, cells harboring pACYC184 or pBIS05-2 accumulated LPS core precursor, which was not observed in ESC19 with pACYC184BipA. Our results suggest that maturation of LPS core at low temperature is affected by the deletion of bipA, eventually making this mutant sensitive to bile salts.
To further verify the impairment of LPS core synthesis, qRT-PCR was carried out targeting genes involved in lipid A synthesis (lpxC), core oligosaccharide synthesis (waaQ), and polysaccharide synthesis (wzx) (Anderson et al., 1985, 1988, 1993; Liu et al., 1996; Yethon et al., 1998). As shown in Figure 5E, significant changes were not observed with any of the three transcripts in both strains at 37°C, while the ESC19 strain showed substantial reductions in the RQ values for lpxC (0.32) and waaQ (0.26) at 20°C. In contrast, wzx transcript levels increased 3.0-fold compared to the MG1655 grown at 20°C, suggesting that the initial steps of LPS biosynthesis may be hampered in ESC19 cells at low temperature.
Interestingly, when ESC19 cells with pBIS05-2 were grown at 20°C, these transcriptional changes were reversed. As shown in Figure 5F, qRT-PCR analysis revealed that the expression levels of lpxC and waaQ were significantly reduced (4.0- and 2.7-fold, respectively) in ESC19 cells with pACYC184 grown at 20°C compared to those grown at 37°C. Transcript levels of lpxC and waaQ were restored by overexpression of yebC to similar levels as complemented ESC19 cells. These results suggest that partial restoration of bile salt sensitivity by YebC resulted from modulation of the transcriptional profile of genes involved in LPS synthesis.
Mutations in LPS synthesis have been shown to affect the ability of E. coli to adhere to abiotic surfaces, leading to significantly reduced biofilm formation (Genevaux et al., 1999). In the case of CA capsule, it has been known to play an inhibitory role in biofilm formation by shielding bacterial surface adhesin and releasing capsular polysaccharide (Roberts, 1996; Whitfield, 2006). Therefore, weakened cell-surface contacts and reduced cell-cell interactions can antagonize biofilm maturation (Beloin et al., 2008). Lastly, since the deletion of bipA was also reported to impair biofilm formation in P. aeruginosa (Overhage et al., 2007; Neidig et al., 2013), we investigated whether the reduced capsule production and restoration of LPS core maturation in suppressed cells can affect biofilm formation under the given conditions. To address this, biofilms were quantified using the crystal violet staining method. First, to assess the effect of bipA or yebC deletion on biofilm-forming ability, the strains MG1655, ESC19, and ESC48 were grown in LB medium at 37°C for 18 h or 20°C for 48 h. The biofilms formed on the inner surface of glass tubes were quantified as described in section “Materials and Methods.” As shown in Figure 6A, the MG1655 strain cultivated at 20°C showed a noticeable increase in biofilm formation (38-fold) compared to cells grown at 37°C and to ESC48 cells. Interestingly, the amount of biofilm produced was greatly reduced in the ESC19 strain (4.3-fold) compared to MG1655 at 20°C.
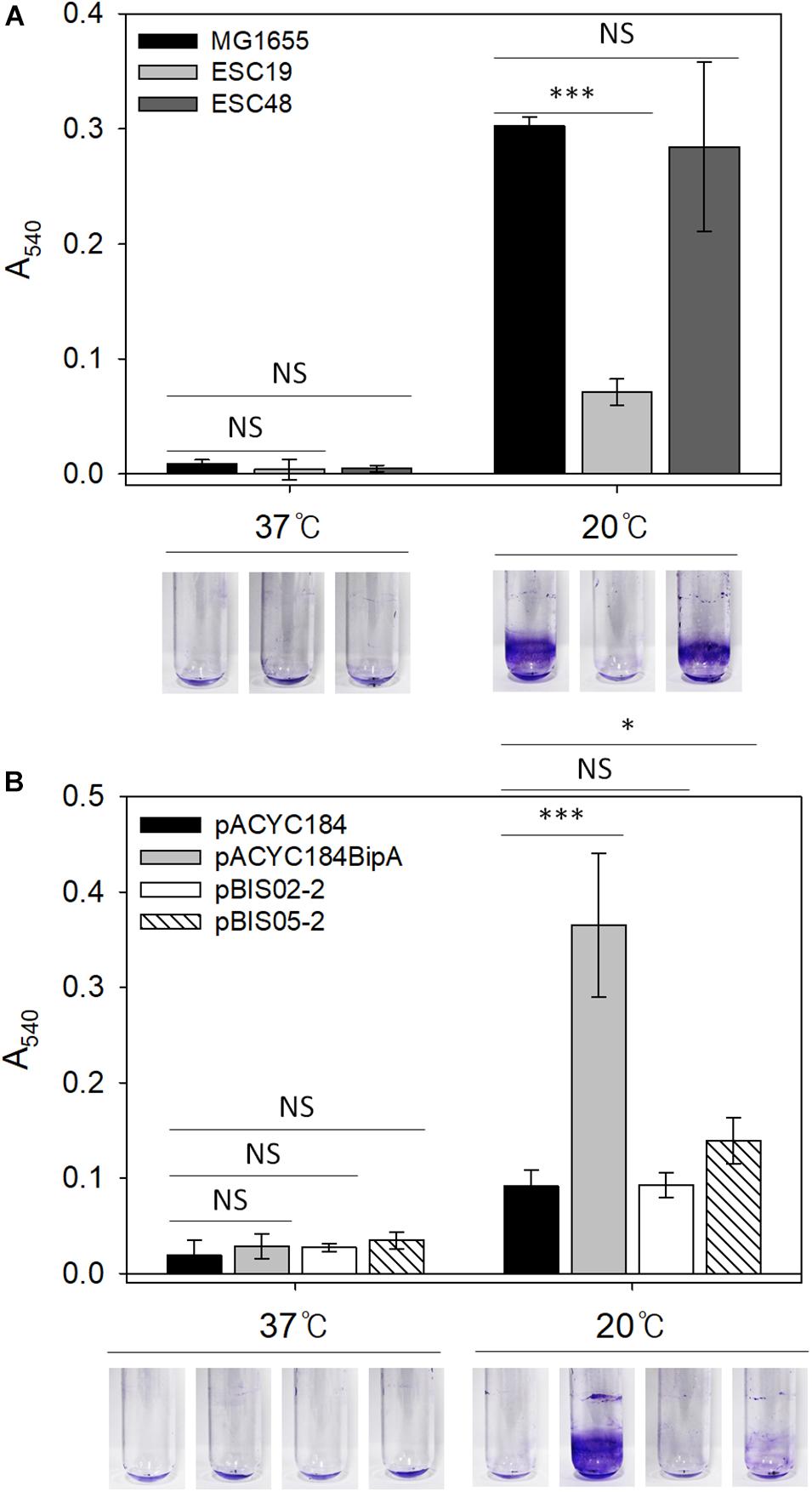
Figure 6. Biofilm formation assay of MG1655, ESC19, and ESC48 strains (A) and ESC19 transformants with pACYC184, pACYC184BipA, pBIS02-2, or pBIS05-2 (B). Cells were grown in 2.5 ml of LB broth with appropriate antibiotics at 37°C for 24 h or 20°C for 48 h. Then, biofilm biomass on inner surface of glass tube was stained with 0.1% crystal violet and dissolved in 95% ethanol. The absorbance of the resulting dissolved dye was measured at a wavelength of 540 nm. A representative photograph is shown from three independent experiments. Error bars represent SD using three replicates. NS, non-significant; ∗p < 0.05; ∗∗∗p < 0.001.
Next, to examine whether YebC could restore impaired biofilm-forming ability in the ESC19 strain, the biofilms produced by ESC19 transformants with pACYC184, pACYC184BipA, pBIS02-2, or pBIS05-2 were quantified. The biofilms produced by the complemented ESC19 cells harboring pACYC184BipA were significantly increased at 20°C compared to those of ESC19 cells with pACYC184 (Figure 6B). ESC19 cells expressing YebC showed slightly increased biofilm production, suggesting that expression of yebC partially restored the biofilm-forming ability of ESC19 at 20°C. The defective biofilm production of pBIS02-2 was not recovered. These findings suggest that biofilm formation was prevented by overproduced capsule in ESC19 cells at 20°C and that overexpressed YebC resulted in partial restoration of biofilm production by repressing capsule synthesis.
In this study, we have shown that the global regulator BipA is functionally associated with ribosome biosynthesis, capsule production, LPS core synthesis, and biofilm formation. Among the pleiotropic phenotypes caused by bipA-deletion, the most conspicuous was defective ribosome assembly at low temperature, although BipA was not essential for this function at 37°C (Choi and Hwang, 2018). Defective rRNA processing and ribosome assembly causes many ribosome assembly factor mutants to be cold-sensitive (Connolly and Culver, 2009). Interestingly, overexpression of yebC in the bipA mutant did not rescue ribosomal abnormalities, however, confers an ability to grow at low temperature (Figure 2). This prompts us to speculate that BipA is involved in other cellular functions as well as ribosome biogenesis. As a ribosome-associated GTPase, BipA shares several characteristic features with other trGTPases, such as LepA and EF-G (Ero et al., 2016). The primary and tertiary structures of BipA are highly similar with those of LepA and EF-G, and the enzymatic behaviors of BipA are the same as these proteins regarding GTP-dependent ribosome binding and ribosome-induced GTP hydrolysis. Furthermore, the ribosomal binding sites of these three GTPases overlap (Ero et al., 2016).
This leads us to assume that BipA is very likely to function as a translational GTPase. From this point of view, we can hypothesize three possibilities regarding the roles of BipA. First, BipA may engage in assembly of subgroups of complete ribosomes at low temperature. The de novo synthesis of ribosomes is regulated in response to nutritional starvation and various stress conditions, and recent reports demonstrate that in E. coli, r-proteins L31 and L36 are encoded by rpmE/ykgM (or rpmE2) and rpmJ/ykgO (or rpmJ2), respectively (Lilleorg et al., 2019; Ghulam et al., 2020). In E. coli K-12, there are seven redundant rRNA genes having heterogeneous nucleotide sequences, and the most variable regions are located at helices 63 and 98 (Anton et al., 1999). Furthermore, the promoters of these rDNA operons were differentially responsive to various stimuli, causing an accumulation of distinct ribosome species under various environmental conditions (Condon et al., 1992; Maeda et al., 2015; Kurylo et al., 2018). Therefore, expression of rRNAs or r-protein paralogs ultimately forms a dynamic population of ribosomes with heterogeneous protein composition depending on growth conditions. Ribosomes act as thermosensors (Vanbogelen and Neidhardt, 1990), and upon cold-shock, assembly factors such as CsdA and RbfA are promptly induced, indicating that remodeling by rRNA folding and r-protein incorporation may be undertaken to facilitate proper ribosome biosynthesis (Jones and Inouye, 1996; Jones et al., 1996). During this process, BipA may recognize those heterogeneous ribosome populations to promote effective ribosome assembly at low temperature. Recently, these divergent ribosomes were revealed to contribute to differential translation of a body of transcripts in Vibrio vulnificus (Song et al., 2019).
Second, whole transcription profiles vary between 37 and 20°C, especially during the acclimation phase (Phadtare et al., 1999), and the effective translation of cold-shock related genes is required for cells to adapt to low temperature. The absence of BipA in cold-exposed cells may cause a cold-sensitive phenotype with deficient expression of a factor(s) that are required for normal growth under cold-stress conditions. The cold-shock inducible protein, initiation factor IF3, was shown to preferentially translate cold-shock mRNAs (Giuliodori et al., 2004; Bruijn, 2016). Therefore, it is possible that BipA itself may act as a translational factor with bias.
Lastly, in concert with the two hypothetical roles discussed above, BipA may regulate the transcription of capsule or LPS-related genes either directly or indirectly. As mentioned earlier, premature transcription terminator sequences were inserted into waaQ, a promoter-adjacent gene in the waa operon, which shuts down the transcription of the entire operon. However, an additional bipA mutation in the waaQ mutant relieved the failure in LPS core synthesis by an uncharacterized mechanism, implying that genes downstream of waaQ are somehow transcribed by mutant BipA (Moller et al., 2003).
BPI interacts with the lipid A moiety of LPS and competitively replaces Ca2+ and Mg2+ divalent metal ions, which chelate the negative charge of phosphates in LPS. This interaction consequently perturbs the normal arrangement of LPS molecules, resulting in membrane rupture and eventually cytotoxicity (Levy, 2000). Upon exposure to this antimicrobial protein, Salmonella typhimurium induces the expression of bipA (Qi et al., 1995). Thus, it is likely that BipA may sense perturbations in the outer membrane and modulate the expression of capsule and/or LPS-related genes. We speculate that in the absence of BipA at low temperature, expression of LPS core production genes may be disturbed, eventually damaging outer membrane integrity. In addition to LPS-triggered damage, cold-shock stress induces a decrease in membrane fluidity, an increase in permeability, and a change in fatty acid composition of lipid A (Carty et al., 1999; Cao-Hoang et al., 2008, 2010). Mutant r-protein L6 also affects membrane stability (Bosl and Bock, 1981). It is noteworthy that the bipA mutant accumulates L6-deficient 50S ribosomal subunits at low temperature (Choi and Hwang, 2018). Thus, under the combinatorial effects of injuries or alternations of membrane and LPS, the Rcs phosphorelay system is likely to stimulate capsule production. In E. coli, CA capsule is synthesized and tightly controlled by a signal transduction system governed by the Rcs phosphorelay system (Trisler and Gottesman, 1984; Stout and Gottesman, 1990). Upon receiving signals, an outer membrane-integrated lipoprotein, RcsF, interacts with an inner membrane protein, IgaA and this interaction activates the downstream RcsB-RcsC two-component system (Cho et al., 2014). This activation phosphorylates the cytosolic domain of RcsC, a transmembrane sensor histidine kinase. Then, the phosphate group is transferred to RcsD, an intermediate inner membrane phosphorelay protein, which is eventually transferred to the effector protein, RcsB (Takeda et al., 2001; Hagiwara et al., 2003; Majdalani and Gottesman, 2005). Phosphorylated RcsB functions as a transcriptional activator for the expression of the cps operon for capsule formation (Stout and Gottesman, 1990). RcsB must form a heterodimer complex with another positive regulator, RcsA, in order to regulate the transcription of the cps operon (Stout et al., 1991). Furthermore, expression of rcsA is negatively regulated by H-NS and CRP, consequently repressing capsule production (Sledjeski and Gottesman, 1995; Lin et al., 2013). Intriguingly, however, the rcsF-bipA double deletion mutant was able to produce capsule at low temperature (Supplementary Figure S5). This may be inferred by the facts that RcsB can respond to another signal and that the repressed expression of igaA in ESC19 cells triggers the Rcs system, consequently producing exopolysaccharide capsule (Potrykus and Wegrzyn, 2004).
Unlike capsule production, the activated Rcs system shuts down the expression of flhDC for flagella production, leading to immobility (Francez-Charlot et al., 2003). We have confirmed that the ESC19 strain showed almost no motility at 20°C and that pBIS02-2 and pBIS05-2 did not contribute to cellular motility (Supplementary Figure S4), suggesting that YebC functions in capsule and LPS core synthesis but not motility.
Here, we demonstrated that overexpression of yebC ameliorated defects in growth and capsule synthesis of ESC19 at low temperature (Figures 1, 3). In addition, suppressed cells were insensitive to bile salts and displayed increased mRNA levels of lpxC and waaQ (Figure 5), suggesting that the role of YebC in the suppression is confined to the regulation of capsule production and LPS core synthesis. Thus, based on our findings and earlier reports on DNA binding activity of YebC, it is likely that YebC in the bipA mutant specifically represses the transcription of capsule and LPS core synthesis genes. Currently, we are carrying out SELEX (Systematic evolution of ligands by exponential enrichment) experiments to isolate target gene(s) that YebC may bind and modulate. This will help us to gain further insights into the physiological function of YebC. The most intriguing finding is that BipA controls the LPS core biosynthesis at low temperature, which may ultimately explain the pleiotropic phenotypes of bipA-deletion. This present study is the first report that investigates the role of YebC in E. coli and demonstrates the relationship of YebC with capsule and LPS core synthesis. Taken together, our results will provide a better understanding of the role of BipA in the cold-stress response, LPS core synthesis, and capsule synthesis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
EC designed the study, executed the experiments, analyzed the data, and wrote the manuscript. HJ and CO assisted the experiments. JH supervised EC and HJ. All authors discussed the results and approved the final manuscript.
This work was supported by a Two-Year Research Grant of Pusan National University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.597515/full#supplementary-material
Anderson, M. S., Bulawa, C. E., and Raetz, C. R. (1985). The biosynthesis of gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem. 260, 15536–15541.
Anderson, M. S., Bull, H. G., Galloway, S. M., Kelly, T. M., Mohan, S., Radika, K., et al. (1993). UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem. 268, 19858–19865.
Anderson, M. S., Robertson, A. D., Macher, I., and Raetz, C. R. (1988). Biosynthesis of lipid A in Escherichia coli: identification of UDP-3-O-[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine as a precursor of UDP-N2,O3-bis[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine. Biochemistry 27, 1908–1917. doi: 10.1021/bi00406a017
Anton, A. I., Martinez-Murcia, A. J., and Rodriguez-Valera, F. (1999). Intraspecific diversity of the 23S rRNA gene and the spacer region downstream in Escherichia coli. J. Bacteriol. 181, 2703–2709. doi: 10.1128/jb.181.9.2703-2709.1999
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050
Barker, H. C., Kinsella, N., Jaspe, A., Friedrich, T., and O’connor, C. D. (2000). Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol. Microbiol. 35, 1518–1529. doi: 10.1046/j.1365-2958.2000.01820.x
Beloin, C., Roux, A., and Ghigo, J. M. (2008). Escherichia coli biofilms. Curr. Top Microbiol. Immunol. 322, 249–289. doi: 10.1007/978-3-540-75418-3_12
Blattner, F. R., Plunkett, G. III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Bosl, A., and Bock, A. (1981). Ribosomal mutation in Escherichia coli affecting membrane stability. Mol. Gen. Genet. 182, 358–360. doi: 10.1007/bf00269684
Brown, L., Villegas, J. M., Elean, M., Fadda, S., Mozzi, F., Saavedra, L., et al. (2017). YebC, a putative transcriptional factor involved in the regulation of the proteolytic system of Lactobacillus. Sci. Rep. 7:8579. doi: 10.1038/s41598-017-09124-1
Bruijn, F. J. D. (2016). Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. Hoboken, NJ: John Wiley & Sons, Inc.
Cao-Hoang, L., Dumont, F., Marechal, P. A., and Gervais, P. (2010). Inactivation of Escherichia coli and Lactobacillus plantarum in relation to membrane permeabilization due to rapid chilling followed by cold storage. Arch. Microbiol. 192, 299–305. doi: 10.1007/s00203-010-0555-y
Cao-Hoang, L., Dumont, F., Marechal, P. A., Le-Thanh, M., and Gervais, P. (2008). Rates of chilling to 0 degrees C: implications for the survival of microorganisms and relationship with membrane fluidity modifications. Appl. Microbiol. Biotechnol. 77, 1379–1387. doi: 10.1007/s00253-007-1279-z
Carty, S. M., Sreekumar, K. R., and Raetz, C. R. (1999). Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction At 12 degrees C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 274, 9677–9685. doi: 10.1074/jbc.274.14.9677
Cho, S. H., Szewczyk, J., Pesavento, C., Zietek, M., Banzhaf, M., Roszczenko, P., et al. (2014). Detecting envelope stress by monitoring beta-barrel assembly. Cell 159, 1652–1664. doi: 10.1016/j.cell.2014.11.045
Choi, E., and Hwang, J. (2018). The GTPase BipA expressed at low temperature in Escherichia coli assists ribosome assembly and has chaperone-like activity. J. Biol. Chem. 293, 18404–18419. doi: 10.1074/jbc.ra118.002295
Choi, E., Jeon, H., Oh, J. I., and Hwang, J. (2019). Overexpressed L20 rescues 50S ribosomal subunit assembly defects of bipA-deletion in Escherichia coli. Front. Microbiol. 10:2982. doi: 10.3389/fmicb.2019.02982
Choi, J. Y., Sifri, C. D., Goumnerov, B. C., Rahme, L. G., Ausubel, F. M., and Calderwood, S. B. (2002). Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. J. Bacteriol. 184, 952–961. doi: 10.1128/jb.184.4.952-961.2002
Condon, C., Philips, J., Fu, Z. Y., Squires, C., and Squires, C. L. (1992). Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. Embo J. 11, 4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x
Connolly, K., and Culver, G. (2009). Deconstructing ribosome construction. Trends Biochem. Sci. 34, 256–263. doi: 10.1016/j.tibs.2009.01.011
Conrad, J., Sun, D. H., Englund, N., and Ofengand, J. (1998). The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23S ribosomal RNA. J. Biol. Chem. 273, 18562–18566. doi: 10.1074/jbc.273.29.18562
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Davis, M. R. Jr., and Goldberg, J. B. (2012). Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J. Vis. Exp. 63:e3916. doi: 10.3791/3916
deLivron, M. A., Makanji, H. S., Lane, M. C., and Robinson, V. L. (2009). A novel domain in translational GTPase BipA mediates interaction with the 70S ribosome and influences GTP hydrolysis. Biochemistry 48, 10533–10541. doi: 10.1021/bi901026z
deLivron, M. A., and Robinson, V. L. (2008). Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. J. Bacteriol. 190, 5944–5952. doi: 10.1128/jb.00763-08
Ero, R., Kumar, V., Chen, Y., and Gao, Y. G. (2016). Similarity and diversity of translational GTPase factors EF-G, EF4, and BipA: from structure to function. RNA Biol. 13, 1258–1273. doi: 10.1080/15476286.2016.1201627
Farris, M., Grant, A., Richardson, T. B., and O’connor, C. D. (1998). BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol. Microbiol. 28, 265–279. doi: 10.1046/j.1365-2958.1998.00793.x
Francez-Charlot, A., Laugel, B., Van Gemert, A., Dubarry, N., Wiorowski, F., Castanie-Cornet, M. P., et al. (2003). RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49, 823–832. doi: 10.1046/j.1365-2958.2003.03601.x
Freestone, P., Trinei, M., Clarke, S. C., Nystrom, T., and Norris, V. (1998). Tyrosine phosphorylation in Escherichia coli. J. Mol. Biol. 279, 1045–1051. doi: 10.1006/jmbi.1998.1836
Genevaux, P., Bauda, P., Dubow, M. S., and Oudega, B. (1999). Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172, 1–8. doi: 10.1007/s002030050732
Ghulam, M. M., Catala, M., and Abou Elela, S. (2020). Differential expression of duplicated ribosomal protein genes modifies ribosome composition in response to stress. Nucleic Acids Res. 48, 1954–1968. doi: 10.1093/nar/gkz1183
Giuliodori, A. M., Brandi, A., Gualerzi, C. O., and Pon, C. L. (2004). Preferential translation of cold-shock mRNAs during cold adaptation. RNA 10, 265–276. doi: 10.1261/rna.5164904
Grant, A. J., Farris, M., Alefounder, P., Williams, P. H., Woodward, M. J., and O’connor, C. D. (2003). Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48, 507–521. doi: 10.1046/j.1365-2958.2003.t01-1-03447.x
Hagiwara, D., Sugiura, M., Oshima, T., Mori, H., Aiba, H., Yamashino, T., et al. (2003). Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185, 5735–5746. doi: 10.1128/jb.185.19.5735-5746.2003
Jones, P. G., and Inouye, M. (1996). RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol. Microbiol. 21, 1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x
Jones, P. G., Mitta, M., Kim, Y., Jiang, W. N., and Inouye, M. (1996). Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 76–80. doi: 10.1073/pnas.93.1.76
Krishnan, K., and Flower, A. M. (2008). Suppression of delta bipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. J. Bacteriol. 190, 7675–7683. doi: 10.1128/jb.00835-08
Kumar, V., Chen, Y., Ero, R., Ahmed, T., Tan, J., Li, Z., et al. (2015). Structure of BipA in GTP form bound to the ratcheted ribosome. Proc. Natl. Acad. Sci. U.S.A. 112, 10944–10949. doi: 10.1073/pnas.1513216112
Kurylo, C. M., Parks, M. M., Juette, M. F., Zinshteyn, B., Altman, R. B., Thibado, J. K., et al. (2018). Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 25, 236–248. doi: 10.1016/j.celrep.2018.08.093
Levy, O. (2000). A neutrophil-derived anti-infective molecule: bactericidal/permeability-increasing protein. Antimicrob. Agents Chemother. 44, 2925–2931. doi: 10.1128/aac.44.11.2925-2931.2000
Liang, H., Li, L., Dong, Z., Surette, M. G., and Duan, K. (2008). The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 190, 6217–6227. doi: 10.1128/jb.00428-08
Lilleorg, S., Reier, K., Pulk, A., Liiv, A., Tammsalu, T., Peil, L., et al. (2019). Bacterial ribosome heterogeneity: changes in ribosomal protein composition during transition into stationary growth phase. Biochimie 156, 169–180. doi: 10.1016/j.biochi.2018.10.013
Lin, C. T., Chen, Y. C., Jinn, T. R., Wu, C. C., Hong, Y. M., and Wu, W. H. (2013). Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One 8:e54430. doi: 10.1371/journal.pone.0054430
Liu, D., Cole, R. A., and Reeves, P. R. (1996). An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 178, 2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maeda, M., Shimada, T., and Ishihama, A. (2015). Strength and regulation of seven rRNA promoters in Escherichia coli. PLoS One 10:e0144697. doi: 10.1371/journal.pone.0144697
Majdalani, N., and Gottesman, S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405. doi: 10.1146/annurev.micro.59.050405.101230
Margus, T., Remm, M., and Tenson, T. (2007). Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics 8:15. doi: 10.1186/1471-2164-8-15
Moller, A. K., Leatham, M. P., Conway, T., Nuijten, P. J., De Haan, L. A., Krogfelt, K. A., et al. (2003). An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71, 2142–2152. doi: 10.1128/iai.71.4.2142-2152.2003
Morrissey, J. H. (1981). Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117, 307–310. doi: 10.1016/0003-2697(81)90783-1
Navasa, N., Rodriguez-Aparicio, L. B., Ferrero, M. A., Moteagudo-Mera, A., and Martinez-Blanco, H. (2011). Growth temperature regulation of some genes that define the superficial capsular carbohydrate composition of Escherichia coli K92. FEMS Microbiol. Lett. 320, 135–141. doi: 10.1111/j.1574-6968.2011.02300.x
Naves, P., Del Prado, G., Huelves, L., Gracia, M., Ruiz, V., Blanco, J., et al. (2008). Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J. Appl. Microbiol. 105, 585–590.
Neidig, A., Yeung, A. T. Y., Rosay, T., Tettmann, B., Strempel, N., Rueger, M., et al. (2013). TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol. 13:77. doi: 10.1186/1471-2180-13-77
Overhage, J., Lewenza, S., Marr, A. K., and Hancock, R. E. W. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189, 2164–2169. doi: 10.1128/jb.01623-06
Paulsen, I. T., Beness, A. M., and Saier, M. H. Jr. (1997). Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 143(Pt 8), 2685–2699. doi: 10.1099/00221287-143-8-2685
Pfennig, P. L., and Flower, A. M. (2001). BipA is required for growth of Escherichia coli K12 at low temperature. Mol. Genet. Genomics 266, 313–317. doi: 10.1007/s004380100559
Phadtare, S., Alsina, J., and Inouye, M. (1999). Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2, 175–180. doi: 10.1016/s1369-5274(99)80031-9
Picken, R. N., and Beacham, I. R. (1977). Bacteriophage-resistant mutants of Escherichia coli K12. Location of receptors within the lipopolysaccharide. J. Gen. Microbiol. 102, 305–318. doi: 10.1099/00221287-102-2-305
Potrykus, J., and Wegrzyn, G. (2004). The ypdI gene codes for a putative lipoprotein involved in the synthesis of colanic acid in Escherichia coli. FEMS Microbiol. Lett. 235, 265–271. doi: 10.1111/j.1574-6968.2004.tb09598.x
Qi, S. Y., Li, Y., Szyroki, A., Giles, I. G., Moir, A., and Oconnor, C. D. (1995). Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol. Microbiol. 17, 523–531. doi: 10.1111/j.1365-2958.1995.mmi_17030523.x
Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G., and Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. doi: 10.1126/science.7604262
Rahn, A., Drummelsmith, J., and Whitfield, C. (1999). Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 181, 2307–2313. doi: 10.1128/jb.181.7.2307-2313.1999
Ranjit, D. K., and Young, K. D. (2016). Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid. J. Bacteriol. 198, 1230–1240. doi: 10.1128/jb.01034-15
Ren, G., Wang, Z., Li, Y., Hu, X., and Wang, X. (2016). Effects of lipopolysaccharide core sugar deficiency on colanic acid biosynthesis in Escherichia coli. J. Bacteriol. 198, 1576–1584. doi: 10.1128/jb.00094-16
Roberts, I. S. (1996). The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50, 285–315. doi: 10.1146/annurev.micro.50.1.285
Rowe, S., Hodson, N., Griffiths, G., and Roberts, I. S. (2000). Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J. Bacteriol. 182, 2741–2745. doi: 10.1128/jb.182.10.2741-2745.2000
Sal-Man, N., Biemans-Oldehinkel, E., and Finlay, B. B. (2009). Structural microengineers: pathogenic Escherichia coli redesigns the actin cytoskeleton in host cells. Structure 17, 15–19. doi: 10.1016/j.str.2008.12.001
Scott, P. M., Erickson, K. M., and Troutman, J. M. (2019). Identification of the functional roles of six key proteins in the biosynthesis of enterobacteriaceae colanic acid. Biochemistry 58, 1818–1830. doi: 10.1021/acs.biochem.9b00040
Shin, D. H., Yokota, H., Kim, R., and Kim, S. H. (2002). Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus. Proc. Natl. Acad. Sci. U.S.A. 99, 7980–7985. doi: 10.1073/pnas.132241399
Sledjeski, D., and Gottesman, S. (1995). A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 2003–2007. doi: 10.1073/pnas.92.6.2003
Song, W., Joo, M., Yeom, J. H., Shin, E., Lee, M., Choi, H. K., et al. (2019). Divergent rRNAs as regulators of gene expression at the ribosome level. Nat. Microbiol. 4, 515–526. doi: 10.1038/s41564-018-0341-1
Stevenson, G., Andrianopoulos, K., Hobbs, M., and Reeves, P. R. (1996). Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178, 4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996
Stout, V., and Gottesman, S. (1990). RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172, 659–669. doi: 10.1128/jb.172.2.659-669.1990
Stout, V., Torres-Cabassa, A., Maurizi, M. R., Gutnick, D., and Gottesman, S. (1991). RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173, 1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991
Sturla, L., Bisso, A., Zanardi, D., Benatti, U., De Flora, A., and Tonetti, M. (1997). Expression, purification and characterization of GDP-D-mannose 4,6-dehydratase from Escherichia coli. FEBS Lett. 412, 126–130. doi: 10.1016/s0014-5793(97)00762-x
Takeda, S., Fujisawa, Y., Matsubara, M., Aiba, H., and Mizuno, T. (2001). A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC –> YojN –> RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40, 440–450. doi: 10.1046/j.1365-2958.2001.02393.x
Trisler, P., and Gottesman, S. (1984). lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160, 184–191. doi: 10.1128/jb.160.1.184-191.1984
Vanbogelen, R. A., and Neidhardt, F. C. (1990). Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 5589–5593. doi: 10.1073/pnas.87.15.5589
Wang, F., Zhong, N. Q., Gao, P., Wang, G. L., Wang, H. Y., and Xia, G. X. (2008). SsTypA1, a chloroplast-specific TypA/BipA-type GTPase from the halophytic plant Suaeda salsa, plays a role in oxidative stress tolerance. Plant Cell Environ. 31, 982–994. doi: 10.1111/j.1365-3040.2008.01810.x
Wei, L., Wu, Y., Qiao, H., Xu, W., Zhang, Y., Liu, X., et al. (2018). YebC controls virulence by activating T3SS gene expression in the pathogen Edwardsiella piscicida. FEMS Microbiol. Lett. 365. doi: 10.1093/femsle/fny137
Weiss, J., and Olsson, I. (1987). Cellular and subcellular localization of the bactericidal/permeability-increasing protein of neutrophils. Blood 69, 652–659. doi: 10.1182/blood.v69.2.652.bloodjournal692652
Whitfield, C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68. doi: 10.1146/annurev.biochem.75.103004.142545
Wu, S., Xia, X., Wang, D., Zhou, Z., and Wang, G. (2019). Gene function and expression regulation of RuvRCAB in bacterial Cr(VI), As(III), Sb(III), and Cd(II) resistance. Appl. Microbiol. Biotechnol. 103, 2701–2713. doi: 10.1007/s00253-019-09666-6
Yethon, J. A., Heinrichs, D. E., Monteiro, M. A., Perry, M. B., and Whitfield, C. (1998). Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273, 26310–26316. doi: 10.1074/jbc.273.41.26310
Keywords: GTPase, YebC, cold shock, capsule, BipA, lipopolysaccharide (LPS)
Citation: Choi E, Jeon H, Oh C and Hwang J (2020) Elucidation of a Novel Role of YebC in Surface Polysaccharides Regulation of Escherichia coli bipA-Deletion. Front. Microbiol. 11:597515. doi: 10.3389/fmicb.2020.597515
Received: 21 August 2020; Accepted: 23 October 2020;
Published: 09 November 2020.
Edited by:
Jai Justin Tree, University of New South Wales, AustraliaReviewed by:
Yong-Gui Gao, Nanyang Technological University, SingaporeCopyright © 2020 Choi, Jeon, Oh and Hwang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihwan Hwang, aHdhbmdqaEBwdXNhbi5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.