
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 21 October 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.593215
This article is part of the Research Topic Alternatives to Combat Bacterial Infections View all 31 articles
 Lucas Assoni
Lucas Assoni Barbara Milani
Barbara Milani Marianna Ribeiro Carvalho
Marianna Ribeiro Carvalho Lucas Natanael Nepomuceno
Lucas Natanael Nepomuceno Natalha Tedeschi Waz
Natalha Tedeschi Waz Maria Eduarda Souza Guerra
Maria Eduarda Souza Guerra Thiago Rojas Converso
Thiago Rojas Converso Michelle Darrieux*
Michelle Darrieux*With the alarming increase of infections caused by pathogenic multidrug-resistant bacteria over the last decades, antimicrobial peptides (AMPs) have been investigated as a potential treatment for those infections, directly through their lytic effect or indirectly, due to their ability to modulate the immune system. There are still concerns regarding the use of such molecules in the treatment of infections, such as cell toxicity and host factors that lead to peptide inhibition. To overcome these limitations, different approaches like peptide modification to reduce toxicity and peptide combinations to improve therapeutic efficacy are being tested. Human defense peptides consist of an important part of the innate immune system, against a myriad of potential aggressors, which have in turn developed different ways to overcome the AMPs microbicidal activities. Since the antimicrobial activity of AMPs vary between Gram-positive and Gram-negative species, so do the bacterial resistance arsenal. This review discusses the mechanisms exploited by Gram-positive bacteria to circumvent killing by antimicrobial peptides. Specifically, the most clinically relevant genera, Streptococcus spp., Staphylococcus spp., Enterococcus spp. and Gram-positive bacilli, have been explored.
Antimicrobial peptides, also known as host defense peptides (HDPs), are found in most life forms, being part of the innate immune system against pathogenic bacteria, fungi, parasites and viruses (Zasloff, 2002; Torrent et al., 2012). Due to the alarming increase in antimicrobial resistance to the commonly used drugs around the world and the lack in discovery of new drugs and alternative treatments, there is a growing concern among the scientific community that in a near future, the current clinical approaches might not be able to deal effectively with microbial infections. Therefore, AMPs have been suggested as an alternative therapeutic strategy, in combination or as a replacement for traditional antibiotics.
The development of antimicrobial resistance against AMPs is not as prevalent when compared to antibiotics, since AMPs targets are diverse and changes can interfere with the functionality of the cell, especially since the cell membrane is the main point of attack (Mahlapuu et al., 2016). However, bacteria can evolve quickly and grow resistant against AMPs in vitro (Andersson et al., 2016). Another approach in the use of AMPs is combination with traditional antibiotics, since both have shown to synergize, reducing microbial resistance (Moravej et al., 2018). A few AMPs have been translated into the clinic; polymyxins B, bacitracin, gramicidin S, daptomycin and vancomycin have been used for treatment of several types of bacteria. However, a number of questions are yet to be answered, such as the toxicity and stability in vivo of many peptides, as thoroughly reviewed (Jenssen et al., 2006; Vaara, 2009; Yount and Yeaman, 2012). The contact with human cells, such as erythrocytes, was shown to inhibit the activity of AMPs (Starr et al., 2016). Furthermore, physiological conditions of the host can interfere with the effectiveness of these molecules, along with the peptide’s pharmacokinetics (Jenssen et al., 2006). Though these are significant challenges, AMPs remain an interesting strategy and still expanding field, as many studies have tried molecular engineering as an approach to solve the concerns cited above. One such example is the production of synthetic D-enantiomeric peptides to avoid proteolytic degradation (de la Fuente-Nunez et al., 2015). So far, over three thousand different peptides have been identified, distributed among six different kingdoms (animalia, archaea, bacteria, fungi, plantae, protist), according to the Antimicrobial Peptide Database (APD) (aps.unmc.edu/AP/) (Wang et al., 2016). In humans, over 130 peptides have been described, and while the vast majority has been tested as potential antimicrobial drugs, AMPs have a larger impact than just direct antimicrobial effects, actively engaging with the host immune system, modulating its activity, promoting chemotaxis and cell recruitment, meddling with the inflammatory and wound healing pathways, among many different functions (Hancock et al., 2016; Mahlapuu et al., 2016; Haney et al., 2017). AMPs were also shown to have an anticarcinogenic effect, as extensively reviewed (Wang, 2014; Hancock et al., 2016; Haney et al., 2017; Yavari et al., 2018; Wang et al., 2019; Kunda, 2020).
An important group of antimicrobial peptides is the cathelicidins. The human representant of this group is LL-37, a cationic, amphipathic peptide, composed by 37 amino acid residues. Its precursor, hCAP18, was first isolated in neutrophils (Cowland et al., 1995; Sørensen et al., 1997) but can also be found in other cells, such as keratinocytes and mast cells (Frohm et al., 1997; Di Nardo et al., 2003). After its cleavage by neutrophil proteases, the peptide acquires its functional form (Sørensen et al., 2001). LL-37 effects have been extensively investigated, and include direct antimicrobial activity and immune modulation (Fabisiak et al., 2016; Mahlapuu et al., 2016; Xhindoli et al., 2016; Haney et al., 2017; Chen et al., 2018; Moravej et al., 2018). Cathelicidins are also found in many vertebrates, including farm animals, birds, reptiles and fish (Kościuczuk et al., 2012). Indolicidin, a 13 amino acid peptide expressed in bovine neutrophils, has antimicrobial activity against Gram-positive and Gram-negative bacteria (van Harten et al., 2018).
Another class of cationic and amphipathic antimicrobial peptides is the defensins, which can be divided in three main groups: α-defensins, β-defensins and θ-defensins. In humans, only α- and β-defensins can be found, while θ-defensins are present exclusively in Old World primates (Nguyen et al., 2003). Among human α-defensins, there are six peptides expressed: Human Neutrophil Peptide (HNP) 1 through 4 and Human Defensins (HD) 5 and 6. α-defensins can be found in many different tissues such as the gastrointestinal and respiratory epithelia, female reproductive tract and blood cells (Hancock et al., 2016). These peptides display direct antimicrobial activities and immunomodulatory effects, including chemotaxis (Wang, 2014; Moravej et al., 2018; Xu and Lu, 2020). β-defensins are expressed mainly in epithelial cells but also in monocytes, macrophages and dendritic cells (Hancock et al., 2016) and have an important role regulating the host microbiome (Meade and O’Farrelly, 2018; Xu and Lu, 2020).
Human Lactoferrin (hLF) is an 80 kDa bilobal glycoprotein, present in bodily fluids and neutrophils, which acts in the transport of metal ions, especially ferric iron (Fe3+) (Vogel, 2012). hLF displays a bacteriostatic effect through iron chelation, decreasing the extracellular concentration of this ion available to the microorganism. Furthermore, the iron-free molecule, Apolactoferrin (ApoLF), is able to interact with microbial cellular membranes, undergoing subsequent proteolysis which results in release of smaller and more potent cationic peptides, especially those found in the N-terminal lobe: Lactoferricin (LFcin), Lactoferrampin (LFampin) and LF1-11 (Sinha et al., 2013).
Human lysozyme, also named N-acetylmuramide glycanhydrolase, is often cited as the first antimicrobial protein discovered and is extensively used in industry (Ercan and Demirci, 2016; Wu T. et al., 2019). Lysozyme is a 14 kDa enzyme that binds to cell wall peptidoglycans, cleaving the links between different sugars, thus inducing cell rupture (Nawrocki et al., 2014; Wang, 2014). Similarly, to lactoferrin, peptides derived from the cleavage of lysozyme exhibit antimicrobial activity against Gram-positive and Gram-negative bacteria (Ibrahim et al., 2001, 2011; Mine et al., 2004; Hunter et al., 2005; Carrillo et al., 2018).
With an array of antimicrobial peptides being produced by different human cells, bacteria have developed a number of strategies to prevent AMP binding, to avoid their lytic effects or to degrade the peptides, in order to thrive in the human host. In the next sections, the different mechanisms employed by Gram-positive bacteria to circumvent AMP action will be explored. Table 1 and Figure 1 summarize the resistance mechanisms employed by these bacteria.
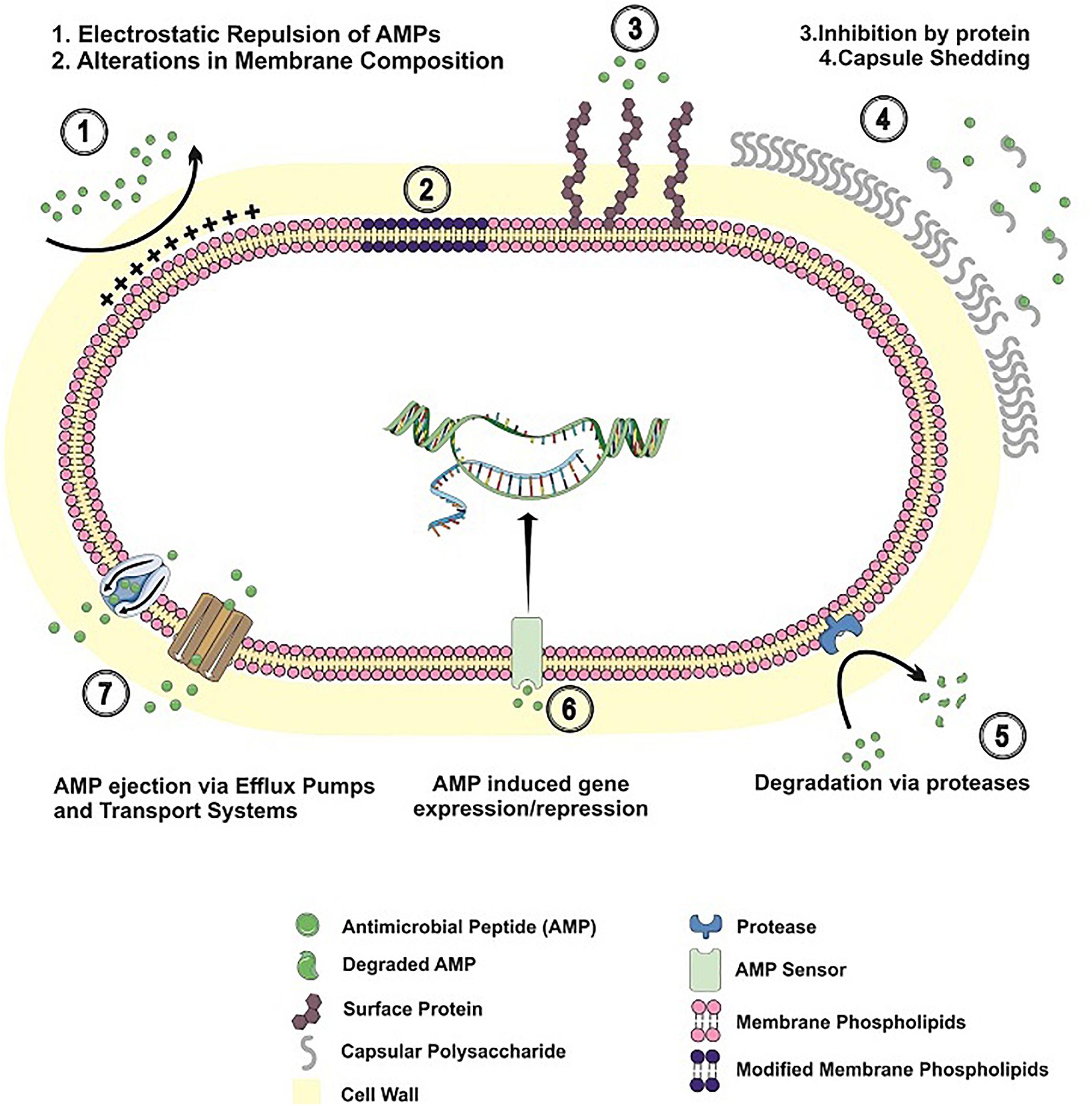
Figure 1. Resistance mechanisms to antimicrobial peptides in Gram-positive bacteria. AMP resistance mechanisms employed by Gram-positive bacteria are shown, including alterations in cell envelope charge/composition; AMP inhibition by binding to surface proteins/released capsular polysaccharide; AMP degradation by bacterial proteases; bacterial adaptation to AMP challenge; AMP extrusion by efflux pumps and transport systems.
Gram-positive bacilli include some pathogenic, anaerobic spore-forming species, such as Clostridium spp., Listeria monocytogenes, Bacillus anthracis, and Bacillus cereus (Chukwu et al., 2016; Schlech, 2019).
The genus Clostridium is composed of about 15 pathogenic species, of which the most common are Clostridium difficile, Clostridium perfringens, Clostridium tetani, and Clostridium botulinum. Although these species are similar, the pathologies caused by them are diverse (Fisher et al., 2005). C. tetani produces the tetanus neurotoxin (TeNT) that causes neurological disease (tetanus), characterized by muscle spasms and spastic paralysis of the limb muscles (Chapeton-Montes et al., 2019). C. botulinum produces the potent botulinum neurotoxin that causes a serious and fatal neuro-paralytic disease in humans and animals (botulism) (Brunt et al., 2018). C. difficile is the main causative agent of nosocomial diarrhea and gastroenteritis, which can lead to the development of asymptomatic or symptomatic diseases. Infection by C. difficile (ICD) has been increasingly reported in the United States (Lessa et al., 2015; Crobach et al., 2020). C. perfringens can also cause acute diarrhea, with an estimated death toll of 200,000 each year in Nigeria according to The World Health Organization (WHO) (Fisher et al., 2005; Chukwu et al., 2016).
Listeria monocytogenes is a foodborne pathogen that causes gastroenteritis in immunocompromised individuals, children, pregnant women and the elderly (Schlech, 2019). L. monocytogenes outbreaks in South Africa have reported around 1000 confirmed cases and 200 deaths in 2017–2018; in the United States, the bacterium was the causative agent in 147 confirmed cases and 33 deaths, making it the third most expensive foodborne pathogen in 2010, after C. botulinum (de Noordhout et al., 2014; Desai et al., 2019).
B. anthracis is the causative agent of anthrax and can manifest in four ways, namely: cutaneous, inhalation, gastrointestinal or injectable (Hagan et al., 2018; Chen et al., 2020). B. cereus causes foodborne diseases, such as gastrointestinal, diarrhea and emesis (Yu et al., 2019; Huang et al., 2020). In 2016, the European Union (EU) reported about 413 food-borne outbreaks caused by Bacillus toxins that affected 6657 people, ranking it the second most common cause of food-borne outbreaks in that year (Fiedler et al., 2019).
Since one of the most important mechanisms of AMP-based killing is the interaction with the negatively charged membrane, changing the membrane composition is a strategy used by many bacteria to survive AMPs action. Among those changes, the insertion of D-alanine in the lipoteichoic acids, a process named D-alanylation, is used to reduce the negative membrane charge, thus inhibiting interaction with AMPs. This resistance mechanism is regulated by the dlt operon, and it has been described in several Bacillus species, such as B. cereus (Abi Khattar et al., 2009), B. anthracis (Fisher et al., 2006), B. thuringiensis (Kamar et al., 2017) and B. subtilis (Cao and Helmann, 2004; May et al., 2005), L. monocytogenes (Abachin et al., 2002; Carvalho et al., 2015), C. difficile (McBride and Sonenshein, 2011a), C. butyricum (Wydau-Dematteis et al., 2015), Lactobacillus plantarum (Palumbo et al., 2006), and Lactobacillus reuteri (Walter et al., 2007).
Another mechanism of envelope modification is called lysinylation of the membrane; it consists of addition of L-lysine to the phosphatidylglycerol. A protein called MprF is essential for membrane lysinylation. In B. anthracis, a strain deficient in MprF was more susceptible to LL-37 and HNP-1 when compared with the wild type strain (Samant et al., 2009). In L. monocytogenes, MprF was shown to be essential in protection against gallidermin, HNP-1 and HNP-2 (Thedieck et al., 2006).
Bacteria can modify cell wall components, such as the peptidoglycan. O-acetylation of the peptidoglycan is able to reduce killing by lysozyme in L. monocytogenes and B. anthracis (Aubry et al., 2011; Laaberki et al., 2011; Rae et al., 2011). Modifications on cell wall constituents also include the N-deacetylation of the peptidoglycan and the glycosylation of the wall teichoic acids, which in L. monocytogenes and B. cereus (Psylinakis et al., 2005) is crucial to protect against lysozyme (Boneca et al., 2007), LL-37 and CRAMP, a cathelicidins found in mice (Meireles et al., 2020).
In B. subtilis, deacetylation of the N-acetylmuramic acid by the protein PdaC confers resistance against lysozyme attack (Fukushima et al., 2005; Kobayashi et al., 2012; Grifoll-Romero et al., 2019). Another protein, PdgA described in L. monocytogenes is responsible for a similar resistance mechanism inducing N-acetylation of the peptidoglycan; a study by Popowska et al. (2009) showed that a L. monocytogenes strain lacking PdgA was more susceptible to lysozyme and mutanolysin.
Changes in lipid composition are able to interfere with AMPs action, as shown for L. monocytogenes, where different proportions of lipids are found in bacteriocin resistant strains (Ming and Daeschel, 1993; Mazzotta and Montville, 1997; Verheul et al., 1997; Crandall and Montville, 1998; Naghmouchi et al., 2006, 2007). In Listeria innocua, changes in the proton motive force, via FoF1 ATPase, which altered the membrane potential were related with resistance to nisin (Maisnier-Patin and Richard, 1996; Bonnet et al., 2006).
A B. subtilis mutant resistant to daptomycin presents an irregular and more cationic membrane than the wild type, due to mutations in pgsA gene. The PgsA protein is responsible for the addition of phosphatidylglycerol to the membrane; in that sense, the diminished phosphatidylglycerol synthase function in the mutant strain was responsible for the increased resistance to daptomycin (Hachmann et al., 2011).
A strategy employed by many bacterial species to evade antimicrobial host defense is by expelling the molecules using efflux pumps or ABC transporters. The same mechanism has been implicated in AMP expulsion (Bernard et al., 2007; Collins et al., 2010; McBride and Sonenshein, 2011b).
Subtilin is an antibiotic produced by B. subtilis; to avoid self-destruction, the bacterium possess an ABC transporter called SpaIFEG, this transporter ejects the subtilin to the extracellular environment (Stein et al., 2005). B. licheniformis is capable of producing bacitracin, an antibacterial peptide also produced by other Bacilli; similarly, to B. subtilis, the bacterium is immune to the antimicrobial due to the action of the BcrABC transporter, which ejects the AMP before it affects the producer cell (Podlesek et al., 1995; Ohki et al., 2003b).
In C. difficile, the cpr operon is responsible for the extracellular transport of peptides (McBride and Sonenshein, 2011b; Suárez et al., 2013). A similar ABC transporter, AnrAB, is also found in L. monocytogenes, able to export AMPs and antimicrobials, hence hindering their efficiency (Collins et al., 2010). B. subtilis also has similar detoxification systems, BceAB-RS, PsdRS-AB (also named Yvc-PQ-RS) and YxdJK-LM. These transporters are important for resistance and cell wall stress signaling against AMPs and antimicrobial drugs, such as bacitracin and lantibiotics (Mascher et al., 2003; Ohki et al., 2003a; Bernard et al., 2007; Rietkotter et al., 2008; Dintner et al., 2011; Staron et al., 2011; Kallenberg et al., 2013). The YtsCD ABC transporter, independently or in association with YwoA, is responsible for bacitracin resistance in B. subtilis (Bernard et al., 2003).
Cell wall signaling can trigger the expression of many resistance-related genes such as sigma (σ) factors and global regulators in bacteria. In L. monocytogenes, sigma factors σB and σL and regulators such as VirR and LiaR regulate the expression of many virulence genes, such as the dlt operon, MprF – a protein responsible for adding L-lysine to membrane phospholipids–and ABC transporters, contributing to antimicrobial resistance (Mandin et al., 2005; Palmer et al., 2009; Samant et al., 2009; Bergholz et al., 2013). Lia-related regulators are also present in B. subtilis; in the presence of peptides that target the cell envelope, the stress sensor is activated and induces the expression of resistance genes such as LiaRS and other membrane modification genes (Mascher et al., 2004; Jordan et al., 2006; Hyyryläinen et al., 2007; Hachmann et al., 2009).
Clostridium difficile gene expression is also altered in the presence of AMPs. LL-37 induces overexpression of genes related to crucial functions; including those involved with cell wall and envelope homeostasis, ABC transporters and lysine metabolism (McQuade et al., 2012); similarly, bacitracin and lysozyme can alter the expression of extracellular σ factors (Ho and Ellermeier, 2011).
The mannose phosphotransferase (Man-PTS) pathway is an important resistance mechanism against bacteriocins. In L. monocytogenes, the activation of the Man-PTS pathway led to changes in metabolism, alteration of the membrane charge and addition of alanine in teichoic acids in strains resistant to class IIa bacteriocin (Ramnath et al., 2000; Gravesen et al., 2002a,b; Vadyvaloo et al., 2004a,b; Tessema et al., 2009; Wu X. et al., 2019). Although the Man-PTS pathway is a target for bacteriocins, in Lactococcus lactis and Lactococcus garvieae, it was shown to participate in resistance mechanisms, specifically in combination with LciA (Diep et al., 2007; Kjos et al., 2011; Daba et al., 2018; Tymoszewska et al., 2018). This pathway is also involved in bacteriocin resistance in L. plantarum, Leuconostoc mesenteroides, Lactobacillus salivarius, and Lactobacillus acidophilus, in combination with PedB, a protein that provides protection against the bacteriocin pediocin PA-1, in a complex, being able to avoid cell lysis by this AMP (Zhou et al., 2016). Similarly, in Listeria innocua, overexpression of pedB generated a more resistant phenotype (Monniot et al., 2012). In L. innocua, Man-PTS is regulated by a transcriptional activator (lin0142); inactivation of lin0142 is related to resistance to pediocin (Xue et al., 2005).
The B. subtilis sigma factor V (sigV) is activated in presence of Lysozyme, regulating important resistance genes such as oatA, dltABCD, and pbpX, promoting protection by virtue of membrane alterations (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011). The alternative sigma factor 54 (rpoN) is relevant in mesentericin Y105 resistance in Listeria monocytogenes; strains lacking the monocistronic unit of rpoN showed a higher susceptibility to this AMP - a phenotype reverted after complementation - indicating that the resistance genes are under regulation of rpoN (Robichon et al., 1997).
The genus Staphylococcus is responsible for various infections in humans like impetigo, scalded skin syndrome, toxic shock syndrome, pneumonia, endocarditis, urinary tract infections, and many others. The most clinically relevant members of this genus are Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus saprophyticus. They are grape-shaped, catalase producing Gram-positive spherical cocci. S. aureus are classified as coagulase positive, while S. epidermidis and S. saprophyticus do not show coagulase activity. Another trait shared by many staphylococci species is the presence of a carotenoid pigment called staphyloxanthin, which gives the colonies a golden color and has an inhibitory role against microbicide molecules and reactive oxygen species (ROS). Among their antigenic structures are the protein A, which binds to Fc region of immunoglobulin G (IgG) and prevents complement activation; the teichoic acids, which modulate mucosal adhesion and induce toxic shock through release of interleukin 1 (IL-1) and tumor necrosis factor (TNF); and polysaccharide capsule, with 11 different serotypes. Defense against AMPs in Staphylococcus spp.
Staphylococcus aureus is able to prevent AMP-mediated killing through modifications of the phosphatidylglycerol in the bacterial membrane by the multiple peptide resistance factor protein (MprF). The protein promotes the reaction of phosphatidylglycerol with lysin, generating lysylphosphatidylglycerol (Lys-PG), which is then translocated to the outer leaflet of the membrane (Oku et al., 2004; Staubitz et al., 2004; Ernst et al., 2009; Nasser et al., 2019). This results in a shift in membrane charge, and a subsequent repulsion of cationic AMPs.
The enhanced synthesis of the cationic phospholipid Lys-PG promotes changes in membrane fluidity also associated with increased resistance against different classes of AMPs in staphylococci. A study investigating the development of bacterial resistance to antimicrobial peptides demonstrated that exposure of S. aureus cultures to sub-lethal concentrations of magainin 2 and gramicidin D over several passages in vitro promoted resistance to these AMPs. The bacterial membrane adaptations induced by AMP exposure included an increase in net charge and altered membrane rigidity (Shireen et al., 2013).
Similarly, resistance to platelet microbicidal proteins (PMPs) in S. aureus has been linked with adaptations affecting membrane fluidity. A study investigating the mechanisms underlying S. aureus susceptibility to thrombin-induced PMP (tPMP-1) demonstrated that mutant strains with increased resistance to this AMP (either naturally occurring or artificially generated) displayed a high content of unsaturated lipids with longer chains (Bayer et al., 2000), which led to an enhanced membrane fluidity. Interestingly, tPMP resistance in S. aureus correlated with an increased virulence in both human and experimental endocarditis (Dhawan et al., 1998), highlighting the importance of this AMP in controlling S. aureus infection.
Resistance to cationic AMPs has also been associated with modifications in the cell wall teichoic acid by esterification with D-alanine, through the dlt operon, which reduces the net negative charge of the molecule. In S. aureus and S. xylosus, deletions of parts of the dlt operon induced a higher sensitivity to a variety of AMPs when compared to wild type strains. Interestingly, the increased susceptibility of the mutant strains were limited to cationic peptides, suggesting that electrostatic repulsion may be involved in resistance to cAMPs in S. aureus (Peschel et al., 1999; Jann et al., 2009; Simanski et al., 2013).
The presence of carotenoid pigments is another described mechanism of AMP resistance in staphylococci. These molecules are depicted as virulence factors, for their protective role against oxidative host defense mechanisms (Clauditz et al., 2006). Evidence suggests staphylococcal carotenoids can also provide protection against different antimicrobial peptides, through their effect on cell membrane stability (Mishra et al., 2011). In that work, a mutant strain with a defect in staphyloxanthin synthesis was compared with its supplemented counterpart in terms of susceptibility to a range of antimicrobial agents, including human HNP-1, PMPs, and polymyxin B. The supplemented strain showed a reduced susceptibility to the AMPs, which in this case was linked to a higher rigidity in the cell membrane. This apparent contrast with previous work showing a positive correlation between membrane fluidity and AMP resistance (Bayer et al., 2000) evidences the intricate balance driving peptide-cell membrane interactions. In that sense, extremes in rigidity or fluidity may hinder AMP insertion in the bacterial membrane.
Another mechanism of S. aureus evasion from AMPs is trapping them by surface or secreted proteins and polysaccharides. S. aureus secrets a plasminogen activating protein, staphylokinase (SK), which converts it into plasmin. High concentrations of plasmin on the bacterial surface promote fibrinolysis, favoring tissue invasion and dissemination (Braff et al., 2007). It has been shown that SK can bind to and inactivate mCRAMPs (cathelicidin murine antimicrobial peptides) and α-defensins secreted by neutrophils, including HNP 1-3 (Jin et al., 2004; Braff et al., 2007) reducing the activity of AMPs in 80%. In an in vivo trial with mice, S. aureus strains expressing SK were more resistant to α-defensin. Similarly, addition of purified SK was able to increase survival of strains that did not produce this protein in presence of α-defensin, in vitro (Jin et al., 2004).
Staphylococcus epidermidis synthesizes the exopolysaccharide intercellular adhesin (PIA), a positively charged polymer of the extracellular matrix in biofilms, which can promote hemagglutination. Studies using mutant strains lacking this polysaccharide have shown a role for PIA in resistance to LL-37 and β-defensin (HBD-3) (Vuong et al., 2004a,b; Kocianova et al., 2005). The mechanism responsible for PIA-mediated protection against AMPs seems to involve electrostatic repulsion, since the lytic activities of these antimicrobial peptides are dependent on the salt concentrations (Vuong et al., 2004b). Besides the protective effect against AMPs produced in the skin, PIA can also limit destruction of Staphylococcus by neutrophils, by forming a mechanical barrier that prevents bacterial uptake by phagocytes (Vuong et al., 2004b).
Additionally, S. aureus is able to sequester iron from the heme site of hemoglobin through the Iron-regulated surface determinant (Isd), which is then released into the cytoplasm for metabolization (Foster et al., 2014). This ability is responsible for the bacterial resistance to the bacteriostatic effects of lactoferrin and other iron-binding peptides. Furthermore, resistance to the bactericidal action of lactoferricin can be induced in S. aureus by growing the bacterium in increasing peptide concentrations–which also promoted cross-resistance to other antimicrobials, like indolicidin and penicillin G (Samuelsen et al., 2005).
Antimicrobial peptides are relatively resistant to bacterial surface or secreted proteases, yet some proteases can cleave a broad spectrum of AMPs; one such example is aureolysin, which inactivates LL-37 by cleaving peptide bonds in its C-terminus, between residues Arg19-Ile20, Arg23-Ile24, and Leu31-Val32 (Sieprawska-Lupa et al., 2004). Thus, Aureolysin expression allows a higher survival in environments with high concentrations of LL-37 such as the phagolysosomes in macrophages and neutrophils.
Staphylococcus aureus displays a phenotype known as small colony variant (SCV), which has been associated with persistent skin infections (Glaser et al., 2014). This phenotypic change allows S. aureus strains to evade innate immune responses, one of those being AMPs, since in SCVs, a higher MIC was observed (von Eiff et al., 2006; Garcia et al., 2013).
Analysis of four different AMPs found on the skin (beta-defensin – hBD-2 and -3, RNase 7, and LL-37) showed that SCV were more resistant to AMPs when compared with the wild type strains (Glaser et al., 2014). Similarly, a mutant strain with a hemin biosynthesis gene deletion, hemB, displaying a SCV phenotype, was less susceptible to three of the four AMPs tested, when compared with its complemented mutant exhibiting normal phenotype (Glaser et al., 2014). These results suggest that phase variation may be a mechanism of bacterial resistance to AMPs. This effect could be attributed to differences in membrane charge in the SCV strains, as suggested by Sadowska et al. (2002), however, in that work, SCVs showed an increased resistance to only a fraction of the AMPs tested.
Staphylococcus aureus expresses an AMP recognition system named Antimicrobial Peptide Sensor; this system comprises a sensor histidine kinase (ApsS), a DNA-binding response regulator (ApsR) and ApsX, responsible for interacting with the AMP. The ApsRSX regulators are responsible for the regulation of important genes related to AMP resistance, such as mprF, vraFG and the dlt operon (Li et al., 2007a,b; Martínez-García et al., 2019). Aps from S. epidermidis has shown the ability to interact with a broad variety of AMPs; in contrast to S. aureus in which Aps are active over a more limited spectrum of peptides (Joo and Otto, 2015). The ApsS is a transmembrane protein with an extracellular group sensitive to AMPs, composed by nine amino acids with a negative charge, which binds to AMPs and rapidly inactivates them (Li et al., 2007b).
The GraRS regulators induce the expression of mprF and dltABCD, when activated together with vraFG, as a response to AMPs and glycopeptides, whereas mutant strains negative for graRS or vraFG were more susceptible to the peptides as the surface alterations generated as protection mechanisms were reduced (Meehl et al., 2007; Neoh et al., 2008; Yang et al., 2012).
The agr global transcriptional regulator induced a super expression of dltD, a member of the dlt operon (Dunman et al., 2001). Another regulator is the LytSR, a transmembrane electrical potential sensor (Patel and Golemi-Kotra, 2015). AMP cell wall damage is partially due to changes in membrane polarization; therefore, deletion of LytSR increased susceptibility to HNP-1 and tPMPs, Interestingly, no conformational changes were found in mutant cells membrane, indicating an alternative resistance pathway (Yang et al., 2013).
The VraTSR is a bacterial sensor which responds to stress; it is involved in S. aureus resistance do methicillin (MRSA) and other antimicrobials that target the cell wall (Boyle-Vavra et al., 2013; Lee et al., 2019). Exposure to AMPs activates operons VraSR e VraDE, leading to a change in the transcriptional profile with the repression of virulence and metabolism genes, and an induction of genes that regulate envelope homeostasis (Pietiäinen et al., 2009).
Enterococci are a group of Gram-positive cocci comprising more than 30 species, of which E. faecalis and Enterococcus faecium are the most clinically relevant (Fiore et al., 2019). They can be found in several environments such as water, soil and food, and are able to colonize the gastrointestinal tract of different animals. Enterococci are a leading cause of nosocomial infections – including endocarditis, urinary tract infections and bacteremia–being responsible for 14% of hospital infections in the United States (Weiner et al., 2016). The problem is aggravated by the increased intrinsic resistance and tolerance exhibited by these bacteria against several commercial antimicrobial agents, including β–lactams such as cephalosporins, and vancomycin (Kristich et al., 2014). In addition, enterococci rapidly acquire resistance to many classes of antibiotics upon treatment, thus posing a great public health threat.
Similarly, to many other species previously cited, enterococci reshape their cell envelope composition in response to AMPs (Cremniter et al., 2006; Mehla and Sood, 2011; Mishra et al., 2012; Kandaswamy et al., 2013), with lysinated phosphatidylglycerol (Kraus and Peschel, 2006; Bao et al., 2012; Kumariya et al., 2015), addition of D-alanine to teichoic acids via dlt (Fabretti et al., 2006; Hirt et al., 2018) or MprF (Bao et al., 2012), also, the N-acetylglucosamine deacetylase PdgA (EF1843) contributes to lysozyme resistance in E. faecalis, by promoting peptidoglycan deacetylation (Benachour et al., 2012).
Many Enterococci species are able to perform O-acetylation of the cell wall peptidoglycan (Pfeffer et al., 2006) a mechanism related with resistance to lysozyme (Hebert et al., 2007).
Proteases and inhibitors are found in E. faecalis, either degrading or binding to the peptide, preventing their lytic effects. Among the proteases, GelE and SerE, a gelatinase and a serine protease, respectively, are able to degrade LL-37, HYL-20 – an α-helical amphipathic analog of a natural AMP present in bees – and GL13K, a peptide found in human saliva (Schmidtchen et al., 2002; Sieprawska-Lupa et al., 2004; Sedgley et al., 2009; Nesuta et al., 2017; Hirt et al., 2018). In addition, extracellular dermatan sulfate – a product released from proteoglycans after the activity of extracellular proteinases – was able to inhibit the activity of HNP-1 on E. faecalis (Schmidtchen et al., 2001), representing an important virulence mechanism for this bacterium.
The Bcr transporter family is related to bacitracin resistance and is found in many enterococci species (Matos et al., 2009). In E. faecalis, BcrABD is an ABC transporter expressed in the presence of bacitracin. It is regulated by BcrR, which is responsible for the extracellular pumping of the polypeptide (Manson et al., 2004). However, the BcrAB is not the only mechanism of bacitracin resistance in E. faecalis; other two-component regulatory systems and ABC transporters were also described (Gebhard et al., 2014). In S. aureus, LtnIFE is responsible for protection against lacticin. E. faecium possess homologs with similar function (Draper et al., 2009).
In E. faecalis and E. faecium, the Man-PTS pathway is also related to resistance against bacteriocins, however, there are several implications in metabolic pathways which could hinder the host colonization (Héchard et al., 2001; Opsata et al., 2010; Geldart and Kaznessis, 2017). Undecaprenyl pyrophosphate phosphatase (UppP) is also related to bacitracin resistance in E. faecium by reducing the amount of substrate for bacitracin-mediated cell death (Shaaly et al., 2013). Another regulator crucial for successful host colonization is the sigma factor SigV, which is involved in resistance to lysozyme, but not to nisin (Le Jeune et al., 2010). rpoN is responsible for encoding the sigma factor 54 in E. faecalis, an important factor for bacteriocin resistance. Interestingly, sensibility to other AMPs did not change in absence of this sigma factor (Dalet et al., 2000).
Both E. faecalis and E. faecium share the LiaFSR stress-induced regulatory pathway. LiaFS is the homolog of VraTS from S. aureus. Strains lacking liaR showed higher sensitivity against daptomycin, and LL-37, HBD-3, nisin, gallidermin–a type A lantibiotic, the synthetic antimicrobial peptide RP-1, mersacidin–a type B lantibiotic and friulimicin, a cationic lipopeptide in E. faecalis (Reyes et al., 2015; Wang et al., 2017). The deletion of liaF, along with gdpD, promoted a similar increase in resistance against daptomycin (Arias et al., 2011). The liaFSR and related genes, such as liaX, a sensor that inhibits LiaFSR, are directly related to the cell envelope alterations in response to antimicrobials (Tran et al., 2013; Khan et al., 2019).
Group A Streptococci (GAS) includes bacterial species such as Streptococcus pyogenes and Streptococcus mutans (Gold et al., 1973; Bessen et al., 1996). These bacteria are beta-hemolytic cocci and known to cause several diseases in humans, including mild conditions like scarlet fever, impetigo, strep throat, caries and cellulitis, and more severe illnesses like necrotizing fasciitis (flesh eating disease) and toxic shock syndrome (TSS) (Kristian et al., 2005).
Streptococcus pyogenes comprises the considerable majority of Group A Streptococci (GAS); it is a pathogen responsible for several human diseases such as pharyngitis, scarlet fever, toxic shock syndrome, pneumonia and others (Lauth et al., 2009). Recent studies have shown that GAS was able to resist the action of several human antimicrobial peptides such as cathelicidin, LL-37 and the α-defensin (HNP-1) (Kristian et al., 2005; Lauth et al., 2009; Rafei et al., 2020). The surface exposed M-protein is used to classify the bacterium into different serotypes (Bessen et al., 1996; Lauth et al., 2009). Lauth et al. (2009) have shown that the N-terminal portion of M-protein can interact with LL-37, preventing its action on the bacterium membrane.
Streptococcus mutans is an important pathogen that colonizes the human oral cavity being the most important caries agent (Gold et al., 1973). Interestingly, several S. mutans strains have been described as resistant to salivary AMPs and bacitracin (Tsuda et al., 2002; Kitagawa et al., 2011; Tian et al., 2018).
A study by Phattarataratip et al. (2011) compared S. mutans strains isolated from 60 children divided into two groups (caries-free and caries-active) and they found that strains isolated from the caries-active group were significantly more resistant to salivary AMPs such as LL-37, α-defensins and β-defensins, in comparison to caries-free strains. Their analysis also correlates this resistance to an ecological advantage over the less resistant strains, which reinforces the importance of AMPs in controlling S. mutans colonization (Phattarataratip et al., 2011).
Since most AMPs present cationic nature, the negative charge of the bacterial surface is important for the bactericidal activity of these molecules. Kristian et al. (2005) showed that the D-alanylation (regulated by the operon dlt (DltABCD)) of S. pyogenes lipoteichoic acid is related with resistance to cationic AMPs, lysozyme and low pH, and it was also associated with an increased survival against neutrophil killing; this phenomenon is due to the increase of positive surface charge caused by the D-alanylation on the cell membrane. In another study, (Cox et al., 2009) using a knockout strain for the dltABCD operon found that the DltA mutant displayed a drastic reduction in the expression of M protein and SIC (Serum Inhibitor of Complement) (Frick et al., 2003), showing that the operon dlt (DltABCD), specifically the dltA gene regulates the expression of genes involved in AMP resistance.
A study published by Tsuda et al. (2002) investigated the mechanisms that allow S. mutans to resist bacitracin; they found that mutant strains lacking the rgp locus (a six gene operon) presented up to five times more sensibility to bacitracin than the wild type counterpart. A possible mechanism to explain this sensitiveness is the fact that the rgp locus is involved in the synthesis of rhamnose-glucose polysaccharide (RGP), a cell wall component; mutations affecting this process render the bacterium more sensitive to bacitracin (Yamashita et al., 1998).
Streptococcus pyogenes is able to limit LL-37 action through degradation by the cysteine proteinase, SpeB. In presence of the inhibitor E64 (which inhibited the cysteine proteinase) the bacterium’s ability to degrade LL-37 was hampered, making it more susceptible to this CAMP. This effect highlights the importance of proteinase SpeB in LL-37 degradation (Schmidtchen et al., 2002).
Similarly, to previously described for Enterococci, S. pyogenes secretes proteases that are able to cleave proteoglycans containing dermatan sulfate, releasing it to the extracellular space. The extracellular dermatan sulfate was able to neutralize neutrophil-derived alpha-defensin, protecting the bacteria from its bactericidal activity (Schmidtchen et al., 2002).
M-protein is the most studied protein in S. pyogenes; variations in M-protein sequence are used to classify the bacterium into different serotypes (Lauth et al., 2009). A study by Lauth et al. (2009), showed that the M protein type 1 protects the bacterium from killing by cathelicidins LL-37 (human) and mCRAMP (mouse). The proposed mechanism involves M1 binding to and trapping the cathelicidin before it can reach the cell wall. They also showed that this protection is type specific once M protein type 49 did not protects the bacterium the same extension of M1, moreover, they found that strains isolated from invasive diseases patients were more resistant to LL-37 action than the strains isolated from asymptomatic patients (Lauth et al., 2009).
Another strategy employed by S. pyogenes to resist AMP attack is the Serum Inhibitor of Complement (SIC). This protein was initially identified as a virulence factor protecting the bacterium against killing by the complement system membrane attack complex (Akesson et al., 1996). Further studies from the same group showed that SIC is important for bacterium full virulence, once it is able to bind to defensins and LL-37, protecting the bacterium against these molecules (Frick et al., 2003).
Streptococcus mutans express the ABC transporter, mbr, an operon composed by 4 genes. Mutant strains that do not express the full transporter were 100 to 120-fold more sensitive to bacitracin than the wild type strain (Tsuda et al., 2002). A more recent study from the same group, analyzed the transcriptome of the bacterium after exposure to bacitracin. They found 8 genes (SMU.302, SMU.862, SMU.863, SMU.864, mbrA, mbrB, SMU.1479, SMU.1856c) that were upregulated upon AMP challenge; of those, the MbrC protein acts as a transcriptional regulator for MbrA and MbrB–which are part of the ABC transporter and are required for bacitracin resistance–and it also controls the expression of SMU.863 and SMU.864, also described as ABC transporters involved in bacitracin resistance by S. mutans (Kitagawa et al., 2011).
The S. mutans bceABRS operon encodes an ABC transporter (BceAB) and a two-component system BceRS. The entire four-component system was shown to be important for protection against bacitracin, defensins (α and β), LL-37 and histatin (Tian et al., 2018). In contrast with wild type S. mutans, mutant strains lacking each bceABRS gene failed to form biofilms in response to a sub-inhibitory concentration of β-defensin. This data suggest that BceABRS also acts as a sensor, promoting a switch to an AMP resistant phenotype upon challenge (Tian et al., 2018).
Streptococcus agalactiae, also referred to as Group B Streptococci (GBS), is an opportunistic pathogen that colonizes the gastrointestinal, genitourinary tracts and, in women, the vaginal mucosa. The biggest concern regarding infections with GBS is in pregnant women, because it can be transmitted vertically and results in serious neonatal consequences, causing several diseases to the newborn, such as meningitis, sepsis and pneumonia (Shabayek and Spellerberg, 2017).
The incidence of infections by S. agalactiae is twice as high in pregnant women when compared to non-pregnant women. Most GBS infections occur during labor, but there is also a chance of infection after delivery. In the United States, GBS infection rates range from 0.1 to 0.8 per 1,000 childbirths. Worldwide, the rates in pregnant women are 0.38 per 1,000 childbirths, with 0.2 in 1000 mortality rate (Raabe and Shane, 2019). GBS infection is also associated with an increased chance of premature delivery. Around the world, premature birth is an important contribution to the death of newborns; approximately 10% of deaths in neonates are caused by GBS infection (Vornhagen et al., 2017).
In S. agalactiae, the dlt operon is essential for resistance against AMPs. Deletion of dltA hinders bacterial survival ability in vivo and reduces the resistance to AMPs, possibly due to an increased interaction with the peptide. Interestingly, the D-alanylation of the membrane seems to induce resistance by enhancing cell envelope strength rather than the interference with the ionic charge of the membrane (Poyart et al., 2001, 2003; Saar-Dover et al., 2012).
Streptococcus agalactiae is intrinsically resistant to nisin via NSR or SaNSR, a nisin-specific enzyme that cleaves and hinders the activity of the peptide. It is expressed by the nsr operon with other lantibiotic resistance genes, such as nsrFP and nsrRK (Khosa et al., 2013, 2015, 2016). However, modified nisin molecules were able to maintain activity against strains possessing SaNSR (Hayes et al., 2019; Zaschke-Kriesche et al., 2019). Another mode of escaping the degrading activity of AMPs is via inhibitory molecules capable of binding to the nisin site of SaNSR (Porta et al., 2019). A phosphoglycerate kinase of GBS was also identified to participate in AMP resistance. Though the mechanism is unknown, it is supposed to include direct binding of the peptides (Boone and Tyrrell, 2012).
NsrFP is an ABC transporter which exports nisin to the extracellular medium. The transporter binds to the N-terminal portion of the peptide and releases it, preventing cell death, even in absence of the two-component regulator NsrRK (Reiners et al., 2017).
In S. sanguinis, a study involving multiple gene screening reported a role for sag1003 in AMP resistance against nisin and bacitracin. The gene is predicted to be an efflux pump against AMPs and a transposon-induced mutagenesis caused a higher sensitivity against both AMPs in a plate-based minimum inhibitory concentration (MIC) assay (Boone and Tyrrell, 2012).
Streptococcus agalactiae pili are important against host defense mechanisms, such as AMPs. The sequestration of AMPs by pili prevents the interaction with the membrane targets. Strains lacking pilB, one of the pilus subunit proteins, were more sensitive to AMPs and less virulent overall, supposedly by virtue of resistance against LL-37, mCRAMP and polymyxin B. Heterologous overexpression of PilB from S. agalactiae in L. lactis showed similar results (Maisey et al., 2008).
The bceRSAB is a detoxification system in GBS, regulating the gene expression against AMPs, such as dltA, promoting resistance. Strains lacking the regulator BceR showed an increased susceptibility against bacitracin and LL-37 and reduced overall virulence (Yang et al., 2019).
The insertion of an inactivation transposon in sag1003 induced a reduction of phosphoglycerate kinase in the cell wall (Boone and Tyrrell, 2012).
The two-component regulator NsrRK is responsible for the transcriptional control of the NSR pathway (nsr and nsrFP) in L. lactis strains capable of synthesizing nisin. In GBS, a very similar nsr operon was described, indicating the possibility of an analogous system (Khosa et al., 2013, 2016).
Hamilton et al. (2006) identified a surface-associated penicillin-binding protein called PBP1a, which is encoded by the ponA gene. A mutant ΔponA strain was more susceptible to AMPs from cathelicidin and defensin families, but the exact mechanism involved in this protection is still unknown (Hamilton et al., 2006; Jones et al., 2007).
Streptococcus pneumoniae (pneumococcus) is responsible for around 1 million deaths worldwide every year, and an increasing drug resistance case reporting (Tramper-Stranders, 2018). It is the main causative agent in community acquired bacterial pneumonia, and it can also cause otitis media, conjunctivitis, sinusitis and more severe diseases like meningitis and bacteremia.
Pneumococci are frequent colonizers of the upper respiratory tract, and a single person may be colonized with multiple strains concomitantly for months. Asymptomatic carriers are also the main source of pneumococcal transmission (Khan and Pichichero, 2014). In this highly colonized niche, AMP resistance confers an important competitive advantage both inter and intra species. Pneumococci display a vast number of adaptations that promote increased AMP resistance, from envelope modifications to AMP sequestration, as described next.
A vast majority of clinically relevant pneumococcal isolates are covered by a thick polysaccharide capsule with variable structure, which protects the bacterium from host immune defenses. Based on their high immunogenicity and protective efficacy, capsular polysaccharides comprise the basis of the current pneumococcal vaccines, alone or in fusion with carrier proteins (Darrieux et al., 2015; Geno et al., 2015; Converso et al., 2020).
Variations in capsule polysaccharide () locus determine the classification of pneumococci in over 95 different serotypes. These include mainly negative structures, with a few being neutral or positive. Negatively charged free capsular polysaccharides (but not neutral or positive ones) have displayed a role in preventing AMP attack. These purified anionic CPS were able to increase the resistance of non-encapsulated mutant pneumococci to HNP-1 and polymyxin B, an effect that was abrogated when the CPSs lost their negative charge through reaction with polycations. One proposed mechanism is that exposure to antimicrobial peptides triggers CPS release, which trap the AMPs and shield the bacterium (Llobet et al., 2008). This capsule shedding has been demonstrated to occur in vivo, thus comprising a potential strategy to prevent AMP-mediated killing. Capsule shedding can be triggered by autolysin (LytA) activity, promoting bacterial resistance to LL-37 and favoring colonization (Kietzman et al., 2016).
Surface-attached capsular polysaccharides, on the other hand, have shown the opposite effect, rendering the bacteria more susceptible to AMP action, in comparison with non-encapsulated isogenic mutants (Beiter et al., 2008). This effect was observed with different capsular types, including CPS 2, 4, 9V and 19F, and the zwitterionic serotype 1. As shown for other Gram-positive bacteria, D-alanylation of teichoic acids in non-encapsulated pneumococci results in increased resistance against killing by neutrophil extracellular trap (NET)-derived components (Beiter et al., 2008). This effect is aided by surface proteins, like the choline binding protein LytA and PgdE, which contribute to reduce the surface negative charge (discussed further). In that sense, the presence of capsule could mask the underlying protective mechanisms against AMPs. This apparent detrimental effect of capsule production over pneumococcal sensitivity to AMPs is possibly overcome by the capsule shedding as previously discussed, and also by its ability to protect the bacterium against mucus and phagocytic cell repulsion (Geno et al., 2015). Furthermore, the effect may not be applicable to all capsular serotypes; great variations in carriage, invasiveness and prevalence exist among capsule types, which have been associated with variations in surface net charge (Li et al., 2013). In that sense, the investigation of AMP resistance in a higher number of pneumococcal serotypes may provide new insights into the role of surface CPS on AMP resistance. For instance, type 4 TIGR4 and its isogenic capsule-negative mutant have shown increased sensibility to CXCL10, LL-37, and nisin, when compared with the type 2 strain, D39 (Bruce et al., 2018).
Another study has shown that non-encapsulated pneumococci are more resistant to neutrophil proteases, elastase and cathepsin G–a feature that also contributes to the ability to colonize the nasopharynx (van der Windt et al., 2012).
Cell wall modifications by the dlt operon have also been shown to promote resistance against nisin and gallidermin in pneumococci, an effect that was consistent with an increased release of D-alanine upon hydrolysis in wild type versus dltA-negative mutant stains (Kovacs et al., 2006).
Pneumococci express two enzymes, PgdA and Adr, that modify peptidoglycans on the bacterial cell wall. PgdA is a N-acetylglucosamine deacetylase (Vollmer and Tomasz, 2000), while Adr is an O-acetyl transferase that acetylates muramic acid residues on the peptidoglycan backbone (Crisostomo et al., 2006). Double mutant strains unable to perform these modifications displayed lower ability to colonize lysozyme-sufficient mice, but behaved similarly, to wild type pneumococci in mice lacking lysozyme production. In contrast, mutants in only one of the molecules colonized mice more efficiently than the wild type strain, in both Lys-producing and Lys-deficient mice (Davis et al., 2008). Taken together, these results indicate that the ability to limit lysozyme attack by modifying the cell wall contributes to successful colonization of the host.
Studies from our group and others have reported a role for pneumococcal surface protein A (PspA) in bacterial resistance to AMPs. PspA is an exposed virulence factor with structural and serological variability (Goulart et al., 2013; Converso et al., 2017b) that has been successfully evaluated as a vaccine candidate in different infection models (Darrieux et al., 2007; Goulart et al., 2013; Converso et al., 2017a, 2020). It prevents complement activation/deposition on the pneumococcal surface, limiting bacterial uptake by phagocytes (Ren et al., 2004; Mukerji et al., 2012).
Pneumococcal surface protein A can bind to and prevent the lytic action of lactoferrin (Hakansson et al., 2001; Shaper et al., 2004). Furthermore, anti-PspA antibodies induced by vaccination were able to enhance the bactericidal effect of apolactoferrin (the iron-free form of the molecule) by blocking PspA interaction with that protein (Shaper et al., 2004; Andre et al., 2015). This protective effect of PspA over pneumococci was diminished when lactoferrin was combined with lysozyme (Andre et al., 2015). This set of data suggests PspA is able to prevent the lytic action of cationic peptides against pneumococci, possibly by binding to these molecules through their active sites. This interaction has been demonstrated for lactoferrin (Senkovich et al., 2007).
Pneumococcal surface protein A has also been shown to interfere with the bactericidal activity of NETs (Martinez et al., 2019). Mutants lacking PspA were more susceptible to trapping by NETs, an effect that was dependent on PspA type. In addition, incubation with anti-PspA antibodies promoted NET formation (Martinez et al., 2019). Taken together, the data indicates that PspA is able to directly prevent killing by AMPs, and also to limit the bactericidal mechanisms of neutrophils.
Pneumococci express and efflux pump, MefE/Mel, which confers resistance to macrolides. mfE expression is induced upon bacterial incubation in presence of LL-37. In consequence, pneumococci develop resistance to LL-37 and erythromycin in vitro (Zahner et al., 2010).
A second, MacAB-like efflux pump described in S. pneumoniae, comprised by the spr0693-spr0694-spr0695 operon, is also involved in resistance against antimicrobial peptides and antibiotics, like LL-37, nisin and bacitracin (Majchrzykiewicz et al., 2010; Yang et al., 2018).
The oligopeptide import ABC transport system Opp (AmiACDEF) has been implicated in resistance against CXCL10, a chemokine with antimicrobial activity against several pathogens (Yung and Murphy, 2012). In that work, mutant strains lacking the permease were less susceptible to CXCL10 and nisin, when compared with the parent D39 strain. Although the precise mechanism responsible for this effect is not fully understood, it is known that AmiA-F has additional pleiotropic roles in pneumococcal physiology, quorum sensing, and virulence (Bruce et al., 2018).
Cell wall modifications in pneumococci can be triggered by AMPs. Treatment with lysozyme leads to upregulation of the dlt locus through the CiaRH sensoring system, resulting in lipoteichoic acid (LTA) modifications and increased inflammatory responses, which in that case contributed to bacterial shedding and transmission (Zafar et al., 2019). Thus, D-alanylation of the cell wall – a mechanism of AMP resistance shared among different Gram-positive microbes–can be induced, in pneumococci, by treatment with antimicrobial proteins that target the bacterial cell wall.
Incubation with LL-37 can also trigger an adaptive response in pneumococci. The transcriptome analysis of pneumococci treated with LL-37 revealed a profound effect on the bacterial genome, with 10% of the genes displaying an altered expression upon challenge (Majchrzykiewicz et al., 2010). The up-regulated genes included those involved in cell wall biosynthesis (dlt), bacteriocin production, virulence (such as the proteases HtrA e PrtA) and bacteriocin production, as well as transcriptional regulators and putative ABC transporters. Interestingly, the serino-protease HtrA is also involved in resistance against other environmental stressors, like high temperature and oxidative stress (Dawid et al., 2009). The choline binding protein PspA and LysM protein (SP 0107)–predicted to be involved in cell wall metabolism–were down regulated in presence of LL-37. Interestingly, LL-37 had a much more dramatic effect on pneumococcal gene expression patterns, when compared with bacterial-derived AMPs that act on the same bacterial targets (nisin and bacitracin). Furthermore, mutant strains lacking these genes revealed an increased susceptibility to treatment with LL-37, confirming the employment of multiple defense strategies against AMPs in pneumococci (Majchrzykiewicz et al., 2010). A more recent study evaluating the proteome of pneumococci treated with LL-37 has also reported a large number of proteins with altered abundance, including transporters, proteins involved in gene regulation and cell wall modification, virulence factors (such as Pht family) and the protease HtrA (Mucke et al., 2020). This result suggests that multiple mechanisms cooperate in pneumococcal response to AMPs.
A study investigating the susceptibility of multiple pneumococcal isolates – both clinical and from carriage–to LL-37 and HNP-1 found great variations in AMP resistance, with no correlation with AMP or capsule type, although clinical isolates were, in general, more susceptible than were carriage isolates (Habets et al., 2012). Furthermore, the study reported that AMP challenge could affect bacterial fitness in competitive assays. This result suggests a role for AMPs in driving intraspecific competition among pneumococci in the nasopharynx, contributing the bacterial genetic diversity in this niche.
Antimicrobial peptides are central players in the innate immune defense against pathogenic bacteria. Unsurprisingly, microbes have developed several strategies to overcome AMP activity, which allow them to efficiently colonize/invade the host. The present review summarizes the strategies adopted by Gram-positive pathogenic bacteria to resist AMP action. Some of these mechanisms, like cell wall modifications, are shared by several pathogens, highlighting their pivotal contribution to bacterial survival within the host. Other factors such as surface proteins and virulence factors are microbe-specific, revealing a myriad of adaptations that comprise the bacterial arsenal against AMPs.
The alarming increase in antibiotic resistance has prompted the search for alternative treatment options. In this scenario, AMPs emerge as a promising strategy to control bacterial infections. This rationale is reinforced by the demonstration that antibiotic resistance in bacteria usually correlates with a collateral sensitivity to AMPs (Lazar et al., 2018).
Several approaches employing AMPs have been tested with encouraging results. The use of AMP combinations is of particular interest, since these molecules can potentiate each other’s action, and also improve the therapeutic efficacy of conventional antibiotics, through synergistic interactions (Reffuveille et al., 2014), This represents an excellent strategy to slow down or minimize bacterial resistance development. In that sense, a better comprehension of the mechanisms employed by bacteria to resist AMP action is pivotal for the development of more effective therapeutic strategies. Furthermore, since many bacterial molecules involved in AMP resistance are important virulence factors, the present review presents numerous potential targets for vaccine development, and also contributes to elucidate the mechanisms driving intra-and interspecies competition within the host.
LA, TC, BM, MC, NW, LN, MG, and MD drafted the manuscript. LA, TC, and MD revised the manuscript. LA and MC drafted the table. LA, MC, and MD produced the Figure. All authors read and approved the final manuscript. None of the authors have any competing interests.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abachin, E., Poyart, C., Pellegrini, E., Milohanic, E., Fiedler, F., Berche, P., et al. (2002). Formation of D-alanyl-lipoteichoic acid is required foradhesion and virulence of Listeria monocytogenes. Mol. Microbiol 43, 1–14. doi: 10.1046/j.1365-2958.2002.02723.x
Abi Khattar, Z., Rejasse, A., Destoumieux-Garzón, D., Escoubas, J. M., Sanchis, V., Lereclus, D., et al. (2009). The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J. Bacteriol. 191, 7063–7073. doi: 10.1128/jb.00892-09
Akesson, P., Sjoholm, A. G., and Bjorck, L. (1996). Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 271, 1081–1088. doi: 10.1074/jbc.271.2.1081
Andersson, D. I., Hughes, D., and Kubicek-Sutherland, J. Z. (2016). Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 26, 43–57. doi: 10.1016/j.drup.2016.04.002
Andre, G. O., Politano, W. R., Mirza, S., Converso, T. R., Ferraz, L. F., Leite, L. C., et al. (2015). Combined effects of lactoferrin and lysozyme on Streptococcus pneumoniae killing. Microb. Pathog. 89, 7–17. doi: 10.1016/j.micpath.2015.08.008
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138
Aubry, C., Goulard, C., Nahori, M. A., Cayet, N., Decalf, J., Sachse, M., et al. (2011). OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204, 731–740. doi: 10.1093/infdis/jir396
Bao, Y., Sakinc, T., Laverde, D., Wobser, D., Benachour, A., Theilacker, C., et al. (2012). Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One 7:e38458. doi: 10.1371/journal.pone.0038458
Bayer, A. S., Prasad, R., Chandra, J., Koul, A., Smriti, M., Varma, A., et al. (2000). In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect. Immun. 68, 3548–3553. doi: 10.1128/iai.68.6.3548-3553.2000
Beiter, K., Wartha, F., Hurwitz, R., Normark, S., Zychlinsky, A., and Henriques-Normark, B. (2008). The capsule sensitizes Streptococcus pneumoniae to alpha-defensins human neutrophil proteins 1 to 3. Infect. Immun. 76, 3710–3716. doi: 10.1128/IAI.01748-07
Benachour, A., Ladjouzi, R., Le Jeune, A., Hebert, L., Thorpe, S., Courtin, P., et al. (2012). The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. J. Bacteriol. 194, 6066–6073. doi: 10.1128/JB.00981-12
Bergholz, T. M., Tang, S., Wiedmann, M., and Boor, K. J. (2013). Nisin Resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl. Environ. Microbiol. 79, 5682–5688. doi: 10.1128/aem.01797-13
Bernard, R., Guiseppi, A., Chippaux, M., Foglino, M., and Denizot, F. (2007). Resistance to bacitracin in Bacillus subtilis: unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189, 8636–8642. doi: 10.1128/JB.01132-07
Bernard, R., Joseph, P., Guiseppi, A., Chippaux, M., and Denizot, F. (2003). YtsCD and YwoA, two independent systems that confer bacitracin resistance to Bacillus subtilis. FEMS Microbiol. Lett. 228, 93–97. doi: 10.1016/S0378-1097100300738-00739
Bessen, D. E., Sotir, C. M., Readdy, T. L., and Hollingshead, S. K. (1996). Genetic correlates of throat and skin isolates of group A streptococci. J. Infect. Dis. 173, 896–900. doi: 10.1093/infdis/173.4.896
Boneca, I. G., Dussurget, O., Cabanes, D., Nahori, M. A., Sousa, S., Lecuit, M., et al. (2007). A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U.S.A. 104, 997–1002. doi: 10.1073/pnas.0609672104
Bonnet, M., Rafi, M. M., Chikindas, M. L., and Montville, T. J. (2006). Bioenergetic mechanism for nisin resistance, induced by the acid tolerance response of Listeria monocytogenes. Appl. Environ. Microbiol. 72, 2556–2563. doi: 10.1128/AEM.72.4.2556-2563.2006
Boone, T. J., and Tyrrell, G. J. (2012). Identification of genes affecting expression of phosphoglycerate kinase on the surface of group B streptococcus. Can. J. Microbiol. 58, 433–441. doi: 10.1139/w2012-015
Boyle-Vavra, S., Yin, S., Jo, D. S., Montgomery, C. P., and Daum, R. S. (2013). VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 83–95. doi: 10.1128/AAC.01651-01612
Braff, M. H., Jones, A. L., Skerrett, S. J., and Rubens, C. E. (2007). Staphylococcus aureus exploits cathelicidin antimicrobial peptides produced during early pneumonia to promote staphylokinase-dependent fibrinolysis. J. Infect. Dis. 195, 1365–1372. doi: 10.1086/513277
Bruce, K. E., Rued, B. E., Tsui, H. T., and Winkler, M. E. (2018). The Opp (AmiACDEF) oligopeptide transporter mediates resistance of Serotype 2 Streptococcus pneumoniae D39 to killing by chemokine CXCL10 and other antimicrobial peptides. J. Bacteriol. 200:e00745-17. doi: 10.1128/JB.00745-17
Brunt, J., Carter, A. T., Pye, H. V., and Peck, M. W. (2018). The orphan germinant receptor protein GerXAO (but not GerX3b) is essential for L-alanine induced germination in Clostridium botulinum Group II. Sci. Rep. 8:7060. doi: 10.1038/s41598-018-25411-x
Cao, M., and Helmann, J. D. (2004). The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186, 1136–1146. doi: 10.1128/jb.186.4.1136-1146.2004
Carrillo, W., Lucio, A., Gaibor, J., Morales, D., and Vasquez, G. (2018). Isolation of antibacterial hydrolysates from hen egg white lysozyme and identification of antibacterial peptides. J. Med. Food 21, 808–818. doi: 10.1089/jmf.2017.0134
Carvalho, F., Atilano, M. L., Pombinho, R., Covas, G., Gallo, R. L., Filipe, S. R., et al. (2015). L-rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog. 11:e1004919. doi: 10.1371/journal.ppat.1004919
Chapeton-Montes, D., Plourde, L., Bouchier, C., Ma, L., Diancourt, L., Criscuolo, A., et al. (2019). The population structure of Clostridium tetani deduced from its pan-genome. Sci. Rep. 9:11220. doi: 10.1038/s41598-019-47551-4
Chen, M., Lyu, Y., Feng, E., Zhu, L., Pan, C., Wang, D., et al. (2020). SpoVG is necessary for sporulation in Bacillus anthracis. Microorganisms 8:548. doi: 10.3390/microorganisms8040548
Chen, X., Zou, X., Qi, G., Tang, Y., Guo, Y., Si, J., et al. (2018). Roles and mechanisms of human cathelicidin LL-37 in cancer. Cell Physiol. Biochem. 47, 1060–1073. doi: 10.1159/000490183
Chukwu, E. E., Nwaokorie, F. O., Coker, A. O., Avila-Campos, M. J., Solis, R. L., Llanco, L. A., et al. (2016). Detection of toxigenic Clostridium perfringens and Clostridium botulinum from food sold in Lagos, Nigeria. Anaerobe 42, 176–181. doi: 10.1016/j.anaerobe.2016.10.009
Clauditz, A., Resch, A., Wieland, K. P., Peschel, A., and Gotz, F. (2006). Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953. doi: 10.1128/IAI.00204-06
Collins, B., Curtis, N., Cotter, P. D., Hill, C., and Ross, R. P. (2010). The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4416–4423. doi: 10.1128/AAC.00503-10
Converso, T. R., Assoni, L., Andre, G. O., Darrieux, M., and Leite, L. C. C. (2020). The long search for a serotype independent pneumococcal vaccine. Expert. Rev. Vaccines 19, 57–70. doi: 10.1080/14760584.2020.1711055
Converso, T. R., Goulart, C., Darrieux, M., and Leite, L. C. C. (2017a). A protein chimera including PspA in fusion with PotD is protective against invasive pneumococcal infection and reduces nasopharyngeal colonization in mice. Vaccine 35, 5140–5147. doi: 10.1016/j.vaccine.2017.08.010
Converso, T. R., Goulart, C., Rodriguez, D., Darrieux, M., and Leite, L. C. C. (2017b). Rational selection of broadly cross-reactive family 2 PspA molecules for inclusion in chimeric pneumococcal vaccines. Microb. Pathog. 109, 233–238. doi: 10.1016/j.micpath.2017.06.004
Cowland, J. B., Johnsen, A. H., and Borregaard, N. (1995). hCAP-18, a cathelin-pro-bactenecin-like protein of human neutrophil specific granules. FEBS Lett. 368, 173–176. doi: 10.1016/0014-5793(95)00634-l
Cox, K. H., Ruiz-Bustos, E., Courtney, H. S., Dale, J. B., Pence, M. A., Nizet, V., et al. (2009). Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes. PLoS One 4:e5366. doi: 10.1371/journal.pone.0005366
Crandall, A. D., and Montville, T. J. (1998). Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl. Environ. Microbiol. 64, 231–237. doi: 10.1128/aem.64.1.231-237.1998
Cremniter, J., Mainardi, J. L., Josseaume, N., Quincampoix, J. C., Dubost, L., Hugonnet, J. E., et al. (2006). Novel mechanism of resistance to glycopeptide antibiotics in Enterococcus faecium. J. Biol. Chem. 281, 32254–32262. doi: 10.1074/jbc.M606920200
Crisostomo, M. I., Vollmer, W., Kharat, A. S., Inhulsen, S., Gehre, F., Buckenmaier, S., et al. (2006). Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol. Microbiol. 61, 1497–1509. doi: 10.1111/j.1365-2958.2006.05340.x
Crobach, M. J. T., Ducarmon, Q. R., Terveer, E. M., Harmanus, C., Sanders, I., Verduin, K. M., et al. (2020). The bacterial gut microbiota of adult patients infected, colonized or noncolonized by Clostridioides difficile. Microorganisms 8:677. doi: 10.3390/microorganisms8050677
Daba, G. M., Ishibashi, N., Gong, X., Taki, H., Yamashiro, K., Lim, Y. Y., et al. (2018). Characterisation of the action mechanism of a Lactococcus-specific bacteriocin, lactococcin Z. J. Biosci. Bioeng. 126, 603–610. doi: 10.1016/j.jbiosc.2018.05.018
Dalet, K., Briand, C., Cenatiempo, Y., and Hechard, Y. (2000). The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr. Microbiol. 41, 441–443. doi: 10.1007/s002840010164
Darrieux, M., Goulart, C., Briles, D., and Leite, L. C. (2015). Current status and perspectives on protein-based pneumococcal vaccines. Crit. Rev. Microbiol. 41, 190–200. doi: 10.3109/1040841X.2013.813902
Darrieux, M., Miyaji, E. N., Ferreira, D. M., Lopes, L. M., Lopes, A. P., Ren, B., et al. (2007). Fusion proteins containing family 1 and family 2 PspA fragments elicit protection against Streptococcus pneumoniae that correlates with antibody-mediated enhancement of complement deposition. Infect. Immun. 75, 5930–5938. doi: 10.1128/IAI.00940-07
Davis, K. M., Akinbi, H. T., Standish, A. J., and Weiser, J. N. (2008). Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 4:e1000241. doi: 10.1371/journal.ppat.1000241
Dawid, S., Sebert, M. E., and Weiser, J. N. (2009). Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191, 1509–1518. doi: 10.1128/JB.01213-08
de la Fuente-Nunez, C., Reffuveille, F., Mansour, S. C., Reckseidler-Zenteno, S. L., Hernandez, D., Brackman, G., et al. (2015). D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205. doi: 10.1016/j.chembiol.2015.01.002
de Noordhout, C. M., Devleesschauwer, B., Angulo, F. J., Verbeke, G., Haagsma, J., Kirk, M., et al. (2014). The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect. Dis. 14, 1073–1082. doi: 10.1016/S1473-3099(14)70870-9
Desai, A. N., Anyoha, A., Madoff, L. C., and Lassmann, B. (2019). Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: a review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 84, 48–53. doi: 10.1016/j.ijid.2019.04.021
Dhawan, V. K., Bayer, A. S., and Yeaman, M. R. (1998). In vitro resistance to thrombin-induced platelet microbicidal protein is associated with enhanced progression and hematogenous dissemination in experimental Staphylococcus aureus infective endocarditis. Infect. Immun. 66, 3476–3479. doi: 10.1128/iai.66.7.3476-3479.1998
Di Nardo, A., Vitiello, A., and Gallo, R. L. (2003). Cutting edge: mast cell antimicrobial activity is mediated by expression of cathelicidin antimicrobial peptide. J. Immunol. 170, 2274–2278. doi: 10.4049/jimmunol.170.5.2274
Diep, D. B., Skaugen, M., Salehian, Z., Holo, H., and Nes, I. F. (2007). Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U.S.A. 104, 2384–2389. doi: 10.1073/pnas.0608775104
Dintner, S., Staron, A., Berchtold, E., Petri, T., Mascher, T., and Gebhard, S. (2011). Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes Bacteria. J. Bacteriol. 193, 3851–3862. doi: 10.1128/JB.05175-11
Draper, L. A., Grainger, K., Deegan, L. H., Cotter, P. D., Hill, C., and Ross, R. P. (2009). Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol. Microbiol. 71, 1043–1054. doi: 10.1111/j.1365-2958.2008.06590.x
Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., et al. (2001). Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353. doi: 10.1128/jb.183.24.7341-7353.2001
Ercan, D., and Demirci, A. (2016). Recent advances for the production and recovery methods of lysozyme. Crit. Rev. Biotechnol. 36, 1078–1088. doi: 10.3109/07388551.2015.1084263
Ernst, C. M., Staubitz, P., Mishra, N. N., Yang, S. J., Hornig, G., Kalbacher, H., et al. (2009). The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. doi: 10.1371/journal.ppat.1000660
Fabisiak, A., Murawska, N., and Fichna, J. (2016). LL-37: cathelicidin-related antimicrobial peptide with pleiotropic activity. Pharmacol. Rep. 68, 802–808. doi: 10.1016/j.pharep.2016.03.015
Fabretti, F., Theilacker, C., Baldassarri, L., Kaczynski, Z., Kropec, A., Holst, O., et al. (2006). Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74, 4164–4171. doi: 10.1128/IAI.00111-06
Fiedler, G., Schneider, C., Igbinosa, E. O., Kabisch, J., Brinks, E., Becker, B., et al. (2019). Antibiotics resistance and toxin profiles of Bacillus cereus-group isolates from fresh vegetables from German retail markets. BMC Microbiol. 19:250. doi: 10.1186/s12866-019-1632-2
Fiore, E., Van Tyne, D., and Gilmore, M. S. (2019). Pathogenicity of enterococci. Microbiol. Spectr. 7, 1–23. doi: 10.1128/microbiolspec.GPP3-0053-2018
Fisher, D. J., Miyamoto, K., Harrison, B., Akimoto, S., Sarker, M. R., and McClane, B. A. (2005). Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56, 747–762. doi: 10.1111/j.1365-2958.2005.04573.x
Fisher, N., Shetron-Rama, L., Herring-Palmer, A., Heffernan, B., Bergman, N., and Hanna, P. (2006). The dltABCD operon of Bacillus anthracis sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J. Bacteriol. 188, 1301–1309. doi: 10.1128/jb.188.4.1301-1309.2006
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Hook, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Frick, I. M., Akesson, P., Rasmussen, M., Schmidtchen, A., and Bjorck, L. (2003). SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J. Biol. Chem. 278, 16561–16566. doi: 10.1074/jbc.M301995200
Frohm, M., Agerberth, B., Ahangari, G., Stâhle-Bäckdahl, M., Lidén, S., Wigzell, H., et al. (1997). The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 272, 15258–15263. doi: 10.1074/jbc.272.24.15258
Fukushima, T., Kitajima, T., and Sekiguchi, J. (2005). A polysaccharide deacetylase homologue, PdaA, in Bacillus subtilis acts as an N-acetylmuramic acid deacetylase in vitro. J. Bacteriol. 187, 1287–1292. doi: 10.1128/jb.187.4.1287-1292.2005
Garcia, L. G., Lemaire, S., Kahl, B. C., Becker, K., Proctor, R. A., Denis, O., et al. (2013). Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 68, 1455–1464. doi: 10.1093/jac/dkt072
Gebhard, S., Fang, C., Shaaly, A., Leslie, D. J., Weimar, M. R., Kalamorz, F., et al. (2014). Identification and characterization of a bacitracin resistance network in Enterococcus faecalis. Antimicrob. Agents Chemother. 58, 1425–1433. doi: 10.1128/AAC.02111-13
Geldart, K., and Kaznessis, Y. N. (2017). Characterization of class IIa bacteriocin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 61:e02033-16. doi: 10.1128/AAC.02033-16
Geno, K. A., Gilbert, G. L., Song, J. Y., Skovsted, I. C., Klugman, K. P., Jones, C., et al. (2015). Pneumococcal capsules and their types: past, present, and future. Clin. Microbiol. Rev. 28, 871–899. doi: 10.1128/CMR.00024-15
Glaser, R., Becker, K., von Eiff, C., Meyer-Hoffert, U., and Harder, J. (2014). Decreased susceptibility of Staphylococcus aureus small-colony variants toward human antimicrobial peptides. J. Invest. Dermatol. 134, 2347–2350. doi: 10.1038/jid.2014.176
Gold, O. G., Jordan, H. V., and Van Houte, J. (1973). A selective medium for Streptococcus mutans. Arch. Oral Biol. 18, 1357–1364. doi: 10.1016/0003-9969(73)90109-x
Goulart, C., da Silva, T. R., Rodriguez, D., Politano, W. R., Leite, L. C., and Darrieux, M. (2013). Characterization of protective immune responses induced by pneumococcal surface protein A in fusion with pneumolysin derivatives. PLoS One 8:e59605. doi: 10.1371/journal.pone.0059605
Gravesen, A., Jydegaard Axelsen, A. M., Mendes da Silva, J., Hansen, T. B., and Knochel, S. (2002a). Frequency of bacteriocin resistance development and associated fitness costs in Listeria monocytogenes. Appl. Environ. Microbiol. 68, 756–764. doi: 10.1128/aem.68.2.756-764.2002
Gravesen, A., Ramnath, M., Rechinger, K. B., Andersen, N., Jänsch, L., Héchard, Y., et al. (2002b). High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148(Pt 8), 2361–2369. doi: 10.1099/00221287-148-8-2361
Grifoll-Romero, L., Sainz-Polo, M. A., Albesa-Jove, D., Guerin, M. E., Biarnes, X., and Planas, A. (2019). Structure-function relationships underlying the dual N-acetylmuramic and N-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC. J. Biol. Chem. 294, 19066–19080. doi: 10.1074/jbc.RA119.009510
Guariglia-Oropeza, V., and Helmann, J. D. (2011). Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 193, 6223–6232. doi: 10.1128/JB.06023-11
Habets, M. G., Rozen, D. E., and Brockhurst, M. A. (2012). Variation in Streptococcus pneumoniae susceptibility to human antimicrobial peptides may mediate intraspecific competition. Proc. Biol. Sci. 279, 3803–3811. doi: 10.1098/rspb.2012.1118
Hachmann, A. B., Angert, E. R., and Helmann, J. D. (2009). Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53, 1598–1609. doi: 10.1128/AAC.01329-08
Hachmann, A. B., Sevim, E., Gaballa, A., Popham, D. L., Antelmann, H., and Helmann, J. D. (2011). Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55, 4326–4337. doi: 10.1128/AAC.01819-10
Hagan, A. K., Plotnick, Y. M., Dingle, R. E., Mendel, Z. I., Cendrowski, S. R., Sherman, D. H., et al. (2018). Petrobactin protects against oxidative stress and enhances sporulation efficiency in Bacillus anthracis sterne. mBio 9:e02079-18. doi: 10.1128/mBio.02079-18
Hakansson, A., Roche, H., Mirza, S., McDaniel, L. S., Brooks-Walter, A., and Briles, D. E. (2001). Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69, 3372–3381. doi: 10.1128/IAI.69.5.3372-3381.2001
Hamilton, A., Popham, D. L., Carl, D. J., Lauth, X., Nizet, V., and Jones, A. L. (2006). Penicillin-binding protein 1a promotes resistance of group B streptococcus to antimicrobial peptides. Infect. Immun. 74, 6179–6187. doi: 10.1128/IAI.00895-06
Hancock, R. E., Haney, E. F., and Gill, E. E. (2016). The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334. doi: 10.1038/nri.2016.29
Haney, E. F., Mansour, S. C., and Hancock, R. E. (2017). Antimicrobial peptides: an introduction. Methods Mol. Biol. 1548, 3–22. doi: 10.1007/978-1-4939-6737-7_1
Hayes, K., Field, D., Hill, C., O’Halloran, F., and Cotter, L. (2019). A novel bioengineered derivative of nisin displays enhanced antimicrobial activity against clinical Streptococcus agalactiae isolates. J. Glob. Antimicrob. Resist. 19, 14–21. doi: 10.1016/j.jgar.2019.04.010
Hebert, L., Courtin, P., Torelli, R., Sanguinetti, M., Chapot-Chartier, M. P., Auffray, Y., et al. (2007). Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75, 5390–5398. doi: 10.1128/IAI.00571-07
Héchard, Y., Pelletier, C., Cenatiempo, Y., and Frère, J. (2001). Analysis of sigma(54)-dependent genes in Enterococcus faecalis: a mannose PTS permease (EII(Man)) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology. 147(Pt 6), 1575–1580. doi: 10.1099/00221287-147-6-1575
Hirt, H., Hall, J. W., Larson, E., and Gorr, S. U. (2018). A D-enantiomer of the antimicrobial peptide GL13K evades antimicrobial resistance in the Gram positive bacteria Enterococcus faecalis and Streptococcus gordonii. PLoS One 13:e0194900. doi: 10.1371/journal.pone.0194900
Ho, T. D., and Ellermeier, C. D. (2011). PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect. Immun. 79, 3229–3238. doi: 10.1128/IAI.00019-11
Ho, T. D., Hastie, J. L., Intile, P. J., and Ellermeier, C. D. (2011). The Bacillus subtilis extracytoplasmic function σ factor σ(V) is induced by lysozyme and provides resistance to lysozyme. J. Bacteriol. 193, 6215–6222. doi: 10.1128/jb.05467-11
Huang, Y., Flint, S. H., and Palmer, J. S. (2020). Bacillus cereus spores and toxins – The potential role of biofilms. Food Microbiol. 90:103493. doi: 10.1016/j.fm.2020.103493
Hunter, H. N., Jing, W., Schibli, D. J., Trinh, T., Park, I. Y., Kim, S. C., et al. (2005). The interactions of antimicrobial peptides derived from lysozyme with model membrane systems. Biochim. Biophys. Acta 1668, 175–189. doi: 10.1016/j.bbamem.2004.12.004
Hyyryläinen, H. L., Pietiäinen, M., Lundén, T., Ekman, A., Gardemeister, M., Murtomäki-Repo, S., et al. (2007). The density of negative charge in the cell wall influences two-component signal transduction in Bacillus subtilis. Microbiology 153(Pt 7), 2126–2136. doi: 10.1099/mic.0.2007/008680-0
Ibrahim, H. R., Imazato, K., and Ono, H. (2011). Human lysozyme possesses novel antimicrobial peptides within its N-terminal domain that target bacterial respiration. J. Agric. Food Chem. 59, 10336–10345. doi: 10.1021/jf2020396
Ibrahim, H. R., Thomas, U., and Pellegrini, A. (2001). A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J. Biol. Chem. 276, 43767–43774. doi: 10.1074/jbc.M106317200
Jann, N. J., Schmaler, M., Kristian, S. A., Radek, K. A., Gallo, R. L., Nizet, V., et al. (2009). Neutrophil antimicrobial defense against Staphylococcus aureus is mediated by phagolysosomal but not extracellular trap-associated cathelicidin. J. Leukoc. Biol. 86, 1159–1169. doi: 10.1189/jlb.0209053
Jenssen, H., Hamill, P., and Hancock, R. E. (2006). Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511. doi: 10.1128/CMR.00056-05
Jin, T., Bokarewa, M., Foster, T., Mitchell, J., Higgins, J., and Tarkowski, A. (2004). Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172, 1169–1176. doi: 10.4049/jimmunol.172.2.1169
Jones, A. L., Mertz, R. H., Carl, D. J., and Rubens, C. E. (2007). A streptococcal penicillin-binding protein is critical for resisting innate airway defenses in the neonatal lung. J. Immunol. 179, 3196–3202. doi: 10.4049/jimmunol.179.5.3196
Joo, H. S., and Otto, M. (2015). Mechanisms of resistance to antimicrobial peptides in staphylococci. Biochim. Biophys. Acta 1848(Pt B), 3055–3061. doi: 10.1016/j.bbamem.2015.02.009
Jordan, S., Junker, A., Helmann, J. D., and Mascher, T. (2006). Regulation of LiaRS-dependent gene expression in bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188, 5153–5166. doi: 10.1128/JB.00310-06
Kallenberg, F., Dintner, S., Schmitz, R., and Gebhard, S. (2013). Identification of regions important for resistance and signalling within the antimicrobial peptide transporter BceAB of Bacillus subtilis. J. Bacteriol. 195, 3287–3297. doi: 10.1128/JB.00419-13
Kamar, R., Rejasse, A., Jehanno, I., Attieh, Z., Courtin, P., Chapot-Chartier, M. P., et al. (2017). DltX of Bacillus thuringiensis is essential for D-alanylation of teichoic acids and resistance to antimicrobial response in insects. Front. Microbiol. 8:1437. doi: 10.3389/fmicb.2017.01437
Kandaswamy, K., Liew, T. H., Wang, C. Y., Huston-Warren, E., Meyer-Hoffert, U., Hultenby, K., et al. (2013). Focal targeting by human beta-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 110, 20230–20235. doi: 10.1073/pnas.1319066110
Khan, A., Davlieva, M., Panesso, D., Rincon, S., Miller, W. R., Diaz, L., et al. (2019). Antimicrobial sensing coupled with cell membrane remodeling mediates antibiotic resistance and virulence in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 116, 26925–26932. doi: 10.1073/pnas.1916037116
Khan, M. N., and Pichichero, M. E. (2014). The host immune dynamics of pneumococcal colonization: implications for novel vaccine development. Hum. Vaccin. Immunother. 10, 3688–3699. doi: 10.4161/21645515.2014.979631
Khosa, S., AlKhatib, Z., and Smits, S. H. (2013). NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 394, 1543–1549. doi: 10.1515/hsz-2013-0167
Khosa, S., Hoeppner, A., Kleinschrodt, D., and Smits, S. H. (2015). Overexpression, purification, crystallization and preliminary X-ray diffraction of the nisin resistance protein from Streptococcus agalactiae. Acta Crystallogr. F Struct. Biol. Commun. 71(Pt 6), 671–675. doi: 10.1107/S2053230X15006226
Khosa, S., Lagedroste, M., and Smits, S. H. (2016). Protein defense systems against the lantibiotic nisin: function of the immunity protein NisI and the resistance protein NSR. Front. Microbiol. 7:504. doi: 10.3389/fmicb.2016.00504
Kietzman, C. C., Gao, G., Mann, B., Myers, L., and Tuomanen, E. I. (2016). Dynamic capsule restructuring by the main pneumococcal autolysin LytA in response to the epithelium. Nat. Commun. 7:10859. doi: 10.1038/ncomms10859
Kitagawa, N., Shiota, S., Shibata, Y., Takeshita, T., and Yamashita, Y. (2011). Characterization of MbrC involved in bacitracin resistance in Streptococcus mutans. FEMS Microbiol. Lett. 318, 61–67. doi: 10.1111/j.1574-6968.2011.02238.x
Kjos, M., Nes, I. F., and Diep, D. B. (2011). Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl. Environ. Microbiol. 77, 3335–3342. doi: 10.1128/AEM.02602-10
Kobayashi, K., Sudiarta, I. P., Kodama, T., Fukushima, T., Ara, K., Ozaki, K., et al. (2012). Identification and characterization of a novel polysaccharide deacetylase C (PdaC) from Bacillus subtilis. J. Biol. Chem. 287, 9765–9776. doi: 10.1074/jbc.M111.329490
Kocianova, S., Vuong, C., Yao, Y., Voyich, J. M., Fischer, E. R., DeLeo, F. R., et al. (2005). Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Invest. 115, 688–694. doi: 10.1172/JCI23523
Kościuczuk, E. M., Lisowski, P., Jarczak, J., Strzałkowska, N., Jóźwik, A., Horbańczuk, J., et al. (2012). Cathelicidins: family of antimicrobial peptides: a review. Mol. Biol. Rep. 39, 10957–10970.
Kovacs, M., Halfmann, A., Fedtke, I., Heintz, M., Peschel, A., Vollmer, W., et al. (2006). A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188, 5797–5805. doi: 10.1128/JB.00336-06
Kraus, D., and Peschel, A. (2006). Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr. Top. Microbiol. Immunol. 306, 231–250. doi: 10.1007/3-540-29916-5_9
Kristian, S. A., Datta, V., Weidenmaier, C., Kansal, R., Fedtke, I., Peschel, A., et al. (2005). D-alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187, 6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005
Kristich, C. J., Rice, L. B., and Arias, C. A. (2014). “Enterococcal infection—treatment and antibiotic resistance,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
Kumariya, R., Sood, S. K., Rajput, Y. S., Saini, N., and Garsa, A. K. (2015). Increased membrane surface positive charge and altered membrane fluidity leads to cationic antimicrobial peptide resistance in Enterococcus faecalis. Biochim. Biophys. Acta 1848, 1367–1375. doi: 10.1016/j.bbamem.2015.03.007
Kunda, N. K. (2020). Antimicrobial peptides as novel therapeutics for non-small cell lung cancer. Drug Discov. Today 25, 238–247. doi: 10.1016/j.drudis.2019.11.012
Laaberki, M. H., Pfeffer, J., Clarke, A. J., and Dworkin, J. (2011). O-Acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J. Biol. Chem. 286, 5278–5288. doi: 10.1074/jbc.M110.183236
Lauth, X., von Kockritz-Blickwede, M., McNamara, C. W., Myskowski, S., Zinkernagel, A. S., Beall, B., et al. (2009). M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J. Innate Immun. 1, 202–214. doi: 10.1159/000203645
Lazar, V., Martins, A., Spohn, R., Daruka, L., Grezal, G., Fekete, G., et al. (2018). Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731. doi: 10.1038/s41564-018-0164-0
Le Jeune, A., Torelli, R., Sanguinetti, M., Giard, J. C., Hartke, A., Auffray, Y., et al. (2010). The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One 5:e9658. doi: 10.1371/journal.pone.0009658
Lee, H., Boyle-Vavra, S., Ren, J., Jarusiewicz, J. A., Sharma, L. K., Hoagland, D. T., et al. (2019). Identification of small molecules exhibiting oxacillin synergy through a novel assay for inhibition of vraTSR expression in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 63:e02593-18.
Lessa, F. C., Mu, Y., Bamberg, W. M., Beldavs, Z. G., Dumyati, G. K., Dunn, J. R., et al. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834. doi: 10.1056/NEJMoa1408913
Li, M., Cha, D. J., Lai, Y., Villaruz, A. E., Sturdevant, D. E., and Otto, M. (2007a). The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66, 1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x
Li, M., Lai, Y., Villaruz, A. E., Cha, D. J., Sturdevant, D. E., and Otto, M. (2007b). Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U.S.A. 104, 9469–9474. doi: 10.1073/pnas.0702159104
Li, Y., Weinberger, D. M., Thompson, C. M., Trzcinski, K., and Lipsitch, M. (2013). Surface charge of Streptococcus pneumoniae predicts serotype distribution. Infect. Immun. 81, 4519–4524. doi: 10.1128/IAI.00724-13
Llobet, E., Tomas, J. M., and Bengoechea, J. A. (2008). Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology (Reading) 154(Pt 12), 3877–3886. doi: 10.1099/mic.0.2008/022301-0
Mahlapuu, M., Hakansson, J., Ringstad, L., and Bjorn, C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell Infect. Microbiol. 6:194. doi: 10.3389/fcimb.2016.00194
Maisey, H. C., Quach, D., Hensler, M. E., Liu, G. Y., Gallo, R. L., Nizet, V., et al. (2008). A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 22, 1715–1724. doi: 10.1096/fj.07-093963
Maisnier-Patin, S., and Richard, J. (1996). Cell wall changes in nisin-resistant variants of Listeria innocua grown in the presence of high nisin concentrations. FEMS Microbiol. Lett. 140, 29–35. doi: 10.1111/j.1574-6968.1996.tb08310.x
Majchrzykiewicz, J. A., Kuipers, O. P., and Bijlsma, J. J. (2010). Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob. Agents Chemother. 54, 440–451. doi: 10.1128/AAC.00769-09
Mandin, P., Fsihi, H., Dussurget, O., Vergassola, M., Milohanic, E., Toledo-Arana, A., et al. (2005). VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57, 1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x
Manson, J. M., Keis, S., Smith, J. M., and Cook, G. M. (2004). Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 48, 3743–3748. doi: 10.1128/AAC.48.10.3743-3748.2004
Martinez, P. J., Farhan, A., Mustafa, M., Javaid, N., Darkoh, C., Garrido-Sanabria, E., et al. (2019). PspA facilitates evasion of pneumococci from bactericidal activity of neutrophil extracellular traps (NETs). Microb. Pathog. 136:103653. doi: 10.1016/j.micpath.2019.103653
Martínez-García, S., Chávez-Cabrera, C., Quintana, E. T., Marsch-Moreno, R., Ibáñez-Hernández, M. A., Zenteno, J. C., et al. (2019). Differential expression of the apsXRS system by antimicrobial peptide LL-37 in commensal and clinical Staphylococcus epidermidis isolates. Indian J. Microbiol. 59, 295–303. doi: 10.1007/s12088-019-00800-6
Mascher, T., Margulis, N. G., Wang, T., Ye, R. W., and Helmann, J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50, 1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x
Mascher, T., Zimmer, S. L., Smith, T. A., and Helmann, J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004
Matos, R., Pinto, V. V., Ruivo, M., and Lopes Mde, F. (2009). Study on the dissemination of the bcrABDR cluster in Enterococcus spp. reveals that the BcrAB transporter is sufficient to confer high-level bacitracin resistance. Int. J. Antimicrob. Agents 34, 142–147. doi: 10.1016/j.ijantimicag.2009.02.008
May, J. J., Finking, R., Wiegeshoff, F., Weber, T. T., Bandur, N., Koert, U., et al. (2005). Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium’s susceptibility to antibiotics that target the cell wall. FEBS J. 272, 2993–3003. doi: 10.1111/j.1742-4658.2005.04700.x
Mazzotta, A. S., and Montville, T. J. (1997). Nisin induces changes in membrane fatty acid composition of Listeria monocytogenes nisin-resistant strains at 10 degrees C and 30 degrees C. J. Appl. Microbiol. 82, 32–38. doi: 10.1111/j.1365-2672.1997.tb03294.x
McBride, S. M., and Sonenshein, A. L. (2011a). The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157(Pt 5), 1457–1465. doi: 10.1099/mic.0.045997-0
McBride, S. M., and Sonenshein, A. L. (2011b). Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect. Immun. 79, 167–176. doi: 10.1128/IAI.00731-10
McQuade, R., Roxas, B., Viswanathan, V. K., and Vedantam, G. (2012). Clostridium difficile clinical isolates exhibit variable susceptibility and proteome alterations upon exposure to mammalian cationic antimicrobial peptides. Anaerobe 18, 614–620. doi: 10.1016/j.anaerobe.2012.09.004
Meade, K. G., and O’Farrelly, C. (2018). beta-defensins: farming the microbiome for homeostasis and health. Front. Immunol. 9:3072. doi: 10.3389/fimmu.2018.03072
Meehl, M., Herbert, S., Gotz, F., and Cheung, A. (2007). Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 2679–2689. doi: 10.1128/AAC.00209-07
Mehla, J., and Sood, S. K. (2011). Substantiation in Enterococcus faecalis of dose-dependent resistance and cross-resistance to pore-forming antimicrobial peptides by use of a polydiacetylene-based colorimetric assay. Appl. Environ. Microbiol. 77, 786–793. doi: 10.1128/AEM.01496-10
Meireles, D., Pombinho, R., Carvalho, F., Sousa, S., and Cabanes, D. (2020). Listeria monocytogenes wall teichoic acid glycosylation promotes surface anchoring of virulence factors, resistance to antimicrobial peptides, and decreased susceptibility to antibiotics. Pathogens 9:290. doi: 10.3390/pathogens9040290
Mine, Y., Ma, F., and Lauriau, S. (2004). Antimicrobial peptides released by enzymatic hydrolysis of hen egg white lysozyme. J. Agric. Food Chem. 52, 1088–1094. doi: 10.1021/jf0345752
Ming, X., and Daeschel, M. A. (1993). Nisin resistance of foodborne bacteria and the specific resistance responses of Listeria monocytogenes scott A. J. Food Protoc. 56, 944–948. doi: 10.4315/0362-028X-56.11.944
Mishra, N. N., Bayer, A. S., Tran, T. T., Shamoo, Y., Mileykovskaya, E., Dowhan, W., et al. (2012). Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 7:e43958. doi: 10.1371/journal.pone.0043958
Mishra, N. N., Liu, G. Y., Yeaman, M. R., Nast, C. C., Proctor, R. A., McKinnell, J., et al. (2011). Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55, 526–531. doi: 10.1128/AAC.00680-10
Monniot, C., Zebre, A. C., Ake, F. M., Deutscher, J., and Milohanic, E. (2012). Novel listerial glycerol dehydrogenase- and phosphoenolpyruvate-dependent dihydroxyacetone kinase system connected to the pentose phosphate pathway. J. Bacteriol. 194, 4972–4982. doi: 10.1128/JB.00801-12
Moravej, H., Moravej, Z., Yazdanparast, M., Heiat, M., Mirhosseini, A., Moosazadeh Moghaddam, M., et al. (2018). Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 24, 747–767. doi: 10.1089/mdr.2017.0392
Mucke, P. A., Maass, S., Kohler, T. P., Hammerschmidt, S., and Becher, D. (2020). Proteomic adaptation of Streptococcus pneumoniae to the human antimicrobial peptide LL-37. Microorganisms 8:413. doi: 10.3390/microorganisms8030413
Mukerji, R., Mirza, S., Roche, A. M., Widener, R. W., Croney, C. M., Rhee, D. K., et al. (2012). Pneumococcal surface protein A inhibits complement deposition on the pneumococcal surface by competing with the binding of C-reactive protein to cell-surface phosphocholine. J. Immunol. 189, 5327–5335. doi: 10.4049/jimmunol.1201967
Naghmouchi, K., Drider, D., Kheadr, E., Lacroix, C., Prevost, H., and Fliss, I. (2006). Multiple characterizations of Listeria monocytogenes sensitive and insensitive variants to divergicin M35, a new pediocin-like bacteriocin. J. Appl. Microbiol. 100, 29–39. doi: 10.1111/j.1365-2672.2005.02771.x
Naghmouchi, K., Kheadr, E., Lacroix, C., and Fliss, I. (2007). Class I/Class IIa bacteriocin cross-resistance phenomenon in Listeria monocytogenes. Food Microbiol. 24, 718–727. doi: 10.1016/j.fm.2007.03.012
Nasser, A., Moradi, M., Jazireian, P., Safari, H., Alizadeh-Sani, M., Pourmand, M. R., et al. (2019). Staphylococcus aureus versus neutrophil: scrutiny of ancient combat. Microb Pathog 131, 259–269. doi: 10.1016/j.micpath.2019.04.026
Nawrocki, K. L., Crispell, E. K., and McBride, S. M. (2014). Antimicrobial peptide resistance mechanisms of gram-positive bacteria. Antibiotics (Basel) 3, 461–492. doi: 10.3390/antibiotics3040461
Neoh, H. M., Cui, L., Yuzawa, H., Takeuchi, F., Matsuo, M., and Hiramatsu, K. (2008). Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52, 45–53. doi: 10.1128/AAC.00534-00507
Nesuta, O., Budesinsky, M., Hadravova, R., Monincova, L., Humpolickova, J., and Cerovsky, V. (2017). How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides. Pathog. Dis. 75:ftx091. doi: 10.1093/femspd/ftx091
Nguyen, T. X., Cole, A. M., and Lehrer, R. I. (2003). Evolution of primate theta-defensins: a serpentine path to a sweet tooth. Peptides 24, 1647–1654. doi: 10.1016/j.peptides.2003.07.023
Ohki, R., Giyanto, Tateno, K., Masuyama, W., Moriya, S., Kobayashi, K., et al. (2003a). The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49, 1135–1144. doi: 10.1046/j.1365-2958.2003.03653.x
Ohki, R., Tateno, K., Okada, Y., Okajima, H., Asai, K., Sadaie, Y., et al. (2003b). A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185, 51–59. doi: 10.1128/jb.185.1.51-59.2003
Oku, Y., Kurokawa, K., Ichihashi, N., and Sekimizu, K. (2004). Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology (Reading) 150(Pt 1), 45–51. doi: 10.1099/mic.0.26706-0
Opsata, M., Nes, I. F., and Holo, H. (2010). Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 10:224. doi: 10.1186/1471-2180-10-224
Palmer, M. E., Wiedmann, M., and Boor, K. J. (2009). sigma(B) and sigma(L) contribute to Listeria monocytogenes 10403S response to the antimicrobial peptides SdpC and nisin. Foodborne Pathog. Dis. 6, 1057–1065. doi: 10.1089/fpd.2009.0292
Palumbo, E., Deghorain, M., Cocconcelli, P. S., Kleerebezem, M., Geyer, A., Hartung, T., et al. (2006). D-alanyl ester depletion of teichoic acids in Lactobacillus plantarum results in a major modification of lipoteichoic acid composition and cell wall perforations at the septum mediated by the Acm2 autolysin. J. Bacteriol. 188, 3709–3715. doi: 10.1128/jb.188.10.3709-3715.2006
Patel, K., and Golemi-Kotra, D. (2015). Signaling mechanism by the Staphylococcus aureus two-component system LytSR: role of acetyl phosphate in bypassing the cell membrane electrical potential sensor LytS. F1000Res. 4:79. doi: 10.12688/f1000research.6213.2
Peschel, A., Otto, M., Jack, R. W., Kalbacher, H., Jung, G., and Gotz, F. (1999). Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410. doi: 10.1074/jbc.274.13.8405
Pfeffer, J. M., Strating, H., Weadge, J. T., and Clarke, A. J. (2006). Peptidoglycan O acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J. Bacteriol. 188, 902–908. doi: 10.1128/JB.188.3.902-908.2006
Phattarataratip, E., Olson, B., Broffitt, B., Qian, F., Brogden, K. A., Drake, D. R., et al. (2011). Streptococcus mutans strains recovered from caries-active or caries-free individuals differ in sensitivity to host antimicrobial peptides. Mol. Oral Microbiol. 26, 187–199. doi: 10.1111/j.2041-1014.2011.00607.x
Pietiäinen, M., François, P., Hyyryläinen, H. L., Tangomo, M., Sass, V., Sahl, H. G., et al. (2009). Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:1471–2164. doi: 10.1186/1471-2164-10-429
Podlesek, Z., Comino, A., Herzog-Velikonja, B., Zgur-Bertok, D., Komel, R., and Grabnar, M. (1995). Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16, 969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x
Popowska, M., Kusio, M., Szymanska, P., and Markiewicz, Z. (2009). Inactivation of the wall-associated de-N-acetylase (PgdA) of Listeria monocytogenes results in greater susceptibility of the cells to induced autolysis. J. Microbiol. Biotechnol. 19, 932–945. doi: 10.4014/jmb.0810.557
Porta, N., Zaschke-Kriesche, J., Frieg, B., Gopalswamy, M., Zivkovic, A., Etzkorn, M., et al. (2019). Small-molecule inhibitors of nisin resistance protein NSR from the human pathogen Streptococcus agalactiae. Bioorg. Med. Chem. 27:115079. doi: 10.1016/j.bmc.2019.115079
Poyart, C., Lamy, M. C., Boumaila, C., Fiedler, F., and Trieu-Cuot, P. (2001). Regulation of D-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183, 6324–6334. doi: 10.1128/JB.183.21.6324-6334.2001
Poyart, C., Pellegrini, E., Marceau, M., Baptista, M., Jaubert, F., Lamy, M. C., et al. (2003). Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 49, 1615–1625. doi: 10.1046/j.1365-2958.2003.03655.x
Psylinakis, E., Boneca, I. G., Mavromatis, K., Deli, A., Hayhurst, E., Foster, S. J., et al. (2005). Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J. Biol. Chem. 280, 30856–30863. doi: 10.1074/jbc.M407426200
Raabe, V. N., and Shane, A. L. (2019). Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 7, 1–13. doi: 10.1128/microbiolspec.GPP3-0007-2018
Rae, C. S., Geissler, A., Adamson, P. C., and Portnoy, D. A. (2011). Mutations of the Listeria monocytogenes peptidoglycan N-deacetylase and O-acetylase result in enhanced lysozyme sensitivity, bacteriolysis, and hyperinduction of innate immune pathways. Infect. Immun. 79, 3596–3606. doi: 10.1128/IAI.00077-11
Rafei, R., Hawli, M., Osman, M., Dabboussi, F., and Hamze, M. (2020). Distribution of emm types and macrolide resistance determinants among group A streptococci in the Middle East and North Africa region. J. Glob. Antimicrob. Resist. 22, 334–348. doi: 10.1016/j.jgar.2020.02.005
Ramnath, M., Beukes, M., Tamura, K., and Hastings, J. W. (2000). Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin A-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 66, 3098–3101. doi: 10.1128/aem.66.7.3098-3101.2000
Reffuveille, F., de la Fuente-Nunez, C., Mansour, S., and Hancock, R. E. (2014). A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371. doi: 10.1128/AAC.03163-14
Reiners, J., Lagedroste, M., Ehlen, K., Leusch, S., Zaschke-Kriesche, J., and Smits, S. H. J. (2017). The N-terminal region of nisin is important for the BceAB-Type ABC transporter NsrFP from Streptococcus agalactiae COH1. Front. Microbiol. 8:1643. doi: 10.3389/fmicb.2017.01643
Ren, B., McCrory, M. A., Pass, C., Bullard, D. C., Ballantyne, C. M., Xu, Y., et al. (2004). The virulence function of Streptococcus pneumoniae surface protein A involves inhibition of complement activation and impairment of complement receptor-mediated protection. J. Immunol. 173, 7506–7512. doi: 10.4049/jimmunol.173.12.7506
Reyes, J., Panesso, D., Tran, T. T., Mishra, N. N., Cruz, M. R., Munita, J. M., et al. (2015). A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J. Infect. Dis. 211, 1317–1325. doi: 10.1093/infdis/jiu602
Rietkotter, E., Hoyer, D., and Mascher, T. (2008). Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68, 768–785. doi: 10.1111/j.1365-2958.2008.06194.x
Robichon, D., Gouin, E., Débarbouillé, M., Cossart, P., Cenatiempo, Y., and Héchard, Y. (1997). The rpoN (sigma54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179, 7591–7594. doi: 10.1128/jb.179.23.7591-7594.1997
Saar-Dover, R., Bitler, A., Nezer, R., Shmuel-Galia, L., Firon, A., Shimoni, E., et al. (2012). D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. PLoS Pathog 8:e1002891. doi: 10.1371/journal.ppat.1002891
Sadowska, B., Bonar, A., von Eiff, C., Proctor, R. A., Chmiela, M., Rudnicka, W., et al. (2002). Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol. Med. Microbiol. 32, 191–197. doi: 10.1111/j.1574-695x.2002.tb00553.x
Samant, S., Hsu, F. F., Neyfakh, A. A., and Lee, H. (2009). The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 191, 1311–1319. doi: 10.1128/jb.01345-08
Samuelsen, O., Haukland, H. H., Jenssen, H., Kramer, M., Sandvik, K., Ulvatne, H., et al. (2005). Induced resistance to the antimicrobial peptide lactoferricin B in Staphylococcus aureus. FEBS Lett. 579, 3421–3426. doi: 10.1016/j.febslet.2005.05.017
Schlech, W. F. (2019). Epidemiology and clinical manifestations of Listeria monocytogenes infection. Microbiol. Spectr. 7, 1–12. doi: 10.1128/microbiolspec.GPP3-0014-2018
Schmidtchen, A., Frick, I. M., Andersson, E., Tapper, H., and Bjorck, L. (2002). Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46, 157–168. doi: 10.1046/j.1365-2958.2002.03146.x
Schmidtchen, A., Frick, I. M., and Björck, L. (2001). Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial alpha-defensin. Mol. Microbiol. 39, 708–713. doi: 10.1046/j.1365-2958.2001.02251.x
Sedgley, C. M., Clewell, D. B., and Flannagan, S. E. (2009). Plasmid pAMS1-encoded, bacteriocin-related “Siblicide” in Enterococcus faecalis. J. Bacteriol. 191, 3183–3188. doi: 10.1128/JB.00147-09
Senkovich, O., Cook, W. J., Mirza, S., Hollingshead, S. K., Protasevich, I. I., Briles, D. E., et al. (2007). Structure of a complex of human lactoferrin N-lobe with pneumococcal surface protein a provides insight into microbial defense mechanism. J. Mol. Biol. 370, 701–713. doi: 10.1016/j.jmb.2007.04.075
Shaaly, A., Kalamorz, F., Gebhard, S., and Cook, G. M. (2013). Undecaprenyl pyrophosphate phosphatase confers low-level resistance to bacitracin in Enterococcus faecalis. J. Antimicrob. Chemother. 68, 1583–1593. doi: 10.1093/jac/dkt048
Shabayek, S., and Spellerberg, B. (2017). Acid stress response mechanisms of group B Streptococci. Front. Cell Infect. Microbiol. 7:395. doi: 10.3389/fcimb.2017.00395
Shaper, M., Hollingshead, S. K., Benjamin, W. H. Jr., and Briles, D. E. (2004). PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin [corrected]. Infect. Immun. 72, 5031–5040. doi: 10.1128/IAI.72.9.5031-5040.2004
Shireen, T., Singh, M., Das, T., and Mukhopadhyay, K. (2013). Differential adaptive responses of Staphylococcus aureus to in vitro selection with different antimicrobial peptides. Antimicrob. Agents Chemother. 57, 5134–5137. doi: 10.1128/AAC.00780-13
Sieprawska-Lupa, M., Mydel, P., Krawczyk, K., Wojcik, K., Puklo, M., Lupa, B., et al. (2004). Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48, 4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004
Simanski, M., Glaser, R., Koten, B., Meyer-Hoffert, U., Wanner, S., Weidenmaier, C., et al. (2013). Staphylococcus aureus subverts cutaneous defense by D-alanylation of teichoic acids. Exp. Dermatol. 22, 294–296. doi: 10.1111/exd.12114
Sinha, M., Kaushik, S., Kaur, P., Sharma, S., and Singh, T. P. (2013). Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int. J. Pept. 2013:390230. doi: 10.1155/2013/390230
Sørensen, O., Arnljots, K., Cowland, J. B., Bainton, D. F., and Borregaard, N. (1997). The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood 90, 2796–2803. doi: 10.1182/blood.v90.7.2796.2796_2796_2803
Sørensen, O. E., Follin, P., Johnsen, A. H., Calafat, J., Tjabringa, G. S., Hiemstra, P. S., et al. (2001). Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97, 3951–3959. doi: 10.1182/blood.v97.12.3951
Staron, A., Finkeisen, D. E., and Mascher, T. (2011). Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55, 515–525. doi: 10.1128/AAC.00352-10
Starr, C. G., He, J., and Wimley, W. C. (2016). Host cell interactions are a significant barrier to the clinical utility of peptide antibiotics. ACS Chem. Biol. 11, 3391–3399. doi: 10.1021/acschembio.6b00843
Staubitz, P., Neumann, H., Schneider, T., Wiedemann, I., and Peschel, A. (2004). MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 231, 67–71. doi: 10.1016/S0378-1097(03)00921-2
Stein, T., Heinzmann, S., Dusterhus, S., Borchert, S., and Entian, K. D. (2005). Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187, 822–828. doi: 10.1128/JB.187.3.822-828.2005
Suárez, J. M., Edwards, A. N., and McBride, S. M. (2013). The Clostridium difficile cpr Locus is regulated by a noncontiguous two-component system in response to type A and B. J. Bacteriol. 195, 2621–2631. doi: 10.1128/jb.00166-13
Tessema, G. T., Moretro, T., Kohler, A., Axelsson, L., and Naterstad, K. (2009). Complex phenotypic and genotypic responses of Listeria monocytogenes strains exposed to the class IIa bacteriocin sakacin P. Appl. Environ. Microbiol. 75, 6973–6980. doi: 10.1128/AEM.00608-09
Thedieck, K., Hain, T., Mohamed, W., Tindall, B. J., Nimtz, M., Chakraborty, T., et al. (2006). The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62, 1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x
Tian, X. L., Salim, H., Dong, G., Parcells, M., and Li, Y. H. (2018). The BceABRS four-component system that is essential for cell envelope stress response is involved in sensing and response to host defence peptides and is required for the biofilm formation and fitness of Streptococcus mutans. J. Med. Microbiol. 67, 874–883. doi: 10.1099/jmm.0.000733
Torrent, M., Pulido, D., Rivas, L., and Andreu, D. (2012). Antimicrobial peptide action on parasites. Curr. Drug Targets 13, 1138–1147. doi: 10.2174/138945012802002393
Tramper-Stranders, G. A. (2018). Childhood community-acquired pneumonia: a review of etiology- and antimicrobial treatment studies. Paediatr. Respir. Rev. 26, 41–48. doi: 10.1016/j.prrv.2017.06.013
Tran, T. T., Panesso, D., Mishra, N. N., Mileykovskaya, E., Guan, Z., Munita, J. M., et al. (2013). Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4:e00281-13. doi: 10.1128/mBio.00281-13
Tsuda, H., Yamashita, Y., Shibata, Y., Nakano, Y., and Koga, T. (2002). Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46, 3756–3764. doi: 10.1128/aac.46.12.3756-3764.2002
Tymoszewska, A., Diep, D. B., and Aleksandrzak-Piekarczyk, T. (2018). The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A, B and C. Sci. Rep. 8:15790.
Vaara, M. (2009). New approaches in peptide antibiotics. Curr. Opin. Pharmacol. 9, 571–576. doi: 10.1016/j.coph.2009.08.002
Vadyvaloo, V., Arous, S., Gravesen, A., Hechard, Y., Chauhan-Haubrock, R., Hastings, J. W., et al. (2004a). Cell-surface alterations in class IIa bacteriocin-resistant Listeria monocytogenes strains. Microbiology 150(Pt 9), 3025–3033. doi: 10.1099/mic.0.27059-0
Vadyvaloo, V., Snoep, J. L., Hastings, J. W., and Rautenbach, M. (2004b). Physiological implications of class IIa bacteriocin resistance in Listeria monocytogenes strains. Microbiology 150(Pt 2), 335–340. doi: 10.1099/mic.0.26731-0
van der Windt, D., Bootsma, H. J., Burghout, P., van der Gaast-de Jongh, C. E., Hermans, P. W., and van der Flier, M. (2012). Nonencapsulated Streptococcus pneumoniae resists extracellular human neutrophil elastase- and cathepsin G-mediated killing. FEMS Immunol. Med. Microbiol. 66, 445–448. doi: 10.1111/j.1574-695X.2012.01028.x
van Harten, R. M., van Woudenbergh, E., van Dijk, A., and Haagsman, H. P. (2018). Cathelicidins: immunomodulatory antimicrobials. Vaccines (Basel) 6:63. doi: 10.3390/vaccines6030063
Verheul, A., Russell, N. J., Van, T. H. R., Rombouts, F. M., and Abee, T. (1997). Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 63, 3451–3457. doi: 10.1128/AEM.63.9.3451-3457.1997
Vogel, H. J. (2012). Lactoferrin, a bird’s eye view. Biochem. Cell Biol. 90, 233–244. doi: 10.1139/o2012-016
Vollmer, W., and Tomasz, A. (2000). The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275, 20496–20501. doi: 10.1074/jbc.M910189199
von Eiff, C., Peters, G., and Becker, K. (2006). The small colony variant (SCV) concept – the role of staphylococcal SCVs in persistent infections. Injury 37(Suppl. 2), S26–S33. doi: 10.1016/j.injury.2006.04.006
Vornhagen, J., Adams Waldorf, K. M., and Rajagopal, L. (2017). Perinatal group B streptococcal infections: virulence factors, immunity, and prevention strategies. Trends Microbiol. 25, 919–931. doi: 10.1016/j.tim.2017.05.013
Vuong, C., Kocianova, S., Voyich, J. M., Yao, Y., Fischer, E. R., DeLeo, F. R., et al. (2004a). A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279, 54881–54886. doi: 10.1074/jbc.M411374200
Vuong, C., Voyich, J. M., Fischer, E. R., Braughton, K. R., Whitney, A. R., DeLeo, F. R., et al. (2004b). Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 6, 269–275. doi: 10.1046/j.1462-5822.2004.00367.x
Walter, J., Loach, D. M., Alqumber, M., Rockel, C., Hermann, C., Pfitzenmaier, M., et al. (2007). D-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 9, 1750–1760. doi: 10.1111/j.1462-2920.2007.01292.x
Wang, G. (2014). Human antimicrobial peptides and proteins. Pharmaceuticals (Basel) 7, 545–594. doi: 10.3390/ph7050545
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. doi: 10.1093/nar/gkv1278
Wang, J., Dou, X., Song, J., Lyu, Y., Zhu, X., Xu, L., et al. (2019). Antimicrobial peptides: promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 39, 831–859. doi: 10.1002/med.21542
Wang, X., Davlieva, M., Reyes, J., Panesso, D., Arias, C. A., and Shamoo, Y. (2017). A novel phosphodiesterase of the GdpP family modulates cyclic di-AMP levels in response to cell membrane stress in daptomycin-resistant enterococci. Antimicrob. Agents Chemother. 61:e01422-16. doi: 10.1128/AAC.01422-16
Weiner, L. M., Webb, A. K., Limbago, B., Dudeck, M. A., Patel, J., Kallen, A. J., et al. (2016). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011-2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. doi: 10.1017/ice.2016.174
Wu, T., Jiang, Q., Wu, D., Hu, Y., Chen, S., Ding, T., et al. (2019). What is new in lysozyme research and its application in food industry? A review. Food Chem. 274, 698–709. doi: 10.1016/j.foodchem.2018.09.017
Wu, X., Ju, X., Du, L., Wang, L., He, R., and Chen, Z. (2019). The Man-PTS subunit IIC is responsible for the sensitivity of Listeria monocytogenes to durancin GL. Food Sci. Nutr. 8, 150–161. doi: 10.1002/fsn3.1285
Wydau-Dematteis, S., Louis, M., Zahr, N., Lai-Kuen, R., Saubamea, B., Butel, M. J., et al. (2015). The functional dlt operon of Clostridium butyricum controls the D-alanylation of cell wall components and influences cell septation and vancomycin-induced lysis. Anaerobe 35(Pt B), 105–114. doi: 10.1016/j.anaerobe.2015.09.001
Xhindoli, D., Pacor, S., Benincasa, M., Scocchi, M., Gennaro, R., and Tossi, A. (2016). The human cathelicidin LL-37 — A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta 1858, 546–566. doi: 10.1016/j.bbamem.2015.11.003
Xu, D., and Lu, W. (2020). Defensins: a double-edged sword in host immunity. Front. Immunol. 11:764. doi: 10.3389/fimmu.2020.00764
Xue, J., Hunter, I., Steinmetz, T., Peters, A., Ray, B., and Miller, K. W. (2005). Novel activator of mannose-specific phosphotransferase system permease expression in Listeria innocua, identified by screening for pediocin AcH resistance. Appl. Environ. Microbiol. 71, 1283–1290. doi: 10.1128/AEM.71.3.1283-1290.2005
Yamashita, Y., Tsukioka, Y., Tomihisa, K., Nakano, Y., and Koga, T. (1998). Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180, 5803–5807. doi: 10.1128/JB.180.21.5803-5807.1998
Yang, H. B., Hou, W. T., Cheng, M. T., Jiang, Y. L., Chen, Y., and Zhou, C. Z. (2018). Structure of a MacAB-like efflux pump from Streptococcus pneumoniae. Nat. Commun. 9:196. doi: 10.1038/s41467-017-02741-4
Yang, S. J., Bayer, A. S., Mishra, N. N., Meehl, M., Ledala, N., Yeaman, M. R., et al. (2012). The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 80, 74–81. doi: 10.1128/IAI.05669-11
Yang, S. J., Xiong, Y. Q., Yeaman, M. R., Bayles, K. W., Abdelhady, W., and Bayer, A. S. (2013). Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 3875–3882. doi: 10.1128/AAC.00412-13
Yang, Y., Luo, M., Zhou, H., Li, C., Luk, A., Zhao, G., et al. (2019). Role of two-component system response regulator bceR in the antimicrobial resistance, virulence, biofilm formation, and stress response of group B Streptococcus. Front. Microbiol. 10:10. doi: 10.3389/fmicb.2019.00010
Yavari, B., Mahjub, R., Saidijam, M., Raigani, M., and Soleimani, M. (2018). The potential use of peptides in cancer treatment. Curr Protein Pept. Sci. 19, 759–770. doi: 10.2174/1389203719666180111150008
Yount, N. Y., and Yeaman, M. R. (2012). Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 52, 337–360. doi: 10.1146/annurev-pharmtox-010611-134535
Yu, S., Yu, P., Wang, J., Li, C., Guo, H., Liu, C., et al. (2019). A study on prevalence and characterization of Bacillus cereus in ready-to-eat foods in China. Front. Microbiol. 10:3043. doi: 10.3389/fmicb.2019.03043
Yung, S. C., and Murphy, P. M. (2012). Antimicrobial chemokines. Front. Immunol. 3:276. doi: 10.3389/fimmu.2012.00276
Zafar, M. A., Hammond, A. J., Hamaguchi, S., Wu, W., Kono, M., Zhao, L., et al. (2019). Identification of pneumococcal factors affecting pneumococcal shedding shows that the dlt locus promotes inflammation and transmission. mBio 10:e01032-19. doi: 10.1128/mBio.01032-19
Zahner, D., Zhou, X., Chancey, S. T., Pohl, J., Shafer, W. M., and Stephens, D. S. (2010). Human antimicrobial peptide LL-37 induces MefE/Mel-mediated macrolide resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 54, 3516–3519. doi: 10.1128/AAC.01756-09
Zaschke-Kriesche, J., Behrmann, L. V., Reiners, J., Lagedroste, M., Groner, Y., Kalscheuer, R., et al. (2019). Bypassing lantibiotic resistance by an effective nisin derivative. Bioorg. Med. Chem. 27, 3454–3462. doi: 10.1016/j.bmc.2019.06.031
Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. doi: 10.1038/415389a
Keywords: antimicrobial peptides, gram-positive, AMP, resistance, streptococci
Citation: Assoni L, Milani B, Carvalho MR, Nepomuceno LN, Waz NT, Guerra MES, Converso TR and Darrieux M (2020) Resistance Mechanisms to Antimicrobial Peptides in Gram-Positive Bacteria. Front. Microbiol. 11:593215. doi: 10.3389/fmicb.2020.593215
Received: 10 August 2020; Accepted: 03 September 2020;
Published: 21 October 2020.
Edited by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoReviewed by:
Cesar de la Fuente-Nunez, University of Pennsylvania, United StatesCopyright © 2020 Assoni, Milani, Carvalho, Nepomuceno, Waz, Guerra, Converso and Darrieux. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle Darrieux, c2FtcGFpb21pY2hlbGxlQHVvbC5jb20uYnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.