- State Key Laboratory for Conservation and Utilization of Bio-Resources and Key Laboratory for Microbial Resources of the Ministry of Education, School of Life Sciences, Yunnan University, Kunming, China
Autophagy plays an important role in cell growth and development. The autophagy-related gene atg4 encodes a cysteine protease, which can cleave the carboxyl terminus of Atg8, thus plays a role in autophagosome formation in yeast and filamentous fungi. Arthrobotrys oligospora is well known for producing special trapping-devices (traps) and capturing nematodes. In this study, two ΔAolatg4 mutants were generated using targeted gene replacement and were used to investigate the biological functions of autophagy in A. oligospora. Autophagic process was observed using the AoAtg8-GFP fusion protein. The mutants showed a defective in hyphal growth and sporulation and were sensitive to chemical stressors, including menadione and Congo red. The spore yield of the ΔAolatg4 mutants was decreased by 88.5% compared to the wild type (WT), and the transcript levels of six sporulation-related genes, such as abaA, fluG, brlA, and wetA, were significantly downregulated during the conidiation stage. Deletion of Aolatg4 also affected the cell nuclei and mycelial septal development in A. oligospora. Importantly, autophagosome formation and the autophagic process were impaired in the ΔAolatg4 mutant. Moreover, the ΔAolatg4 mutant lost its ability to form mature traps. Our results provide novel insights into the roles of autophagy in A. oligospora.
Introduction
Autophagy is a conserved intracellular recycling process in all eukaryotic cells, in which autophagosomes are delivered to lysosomes or vacuoles for degradation (Reggiori and Klionsky, 2002; Nakatogawa et al., 2012; Ryter et al., 2013). The autophagic process is governed by autophagy-related (Atg) proteins, which orchestrate the different steps of autophagy, and about 19 Atg proteins are necessary for autophagosome formation (Farré and Subramani, 2016). Previous studies have revealed that the ubiquitin-like protein Atg8 localizes to the autophagosome and autophagy-related structures, thus it can be used a reliable marker for autophagy (Hirata et al., 2017). Atg4 is a cysteine protease, which plays a role in the Atg8 conjugation system of autophagy (Ichimura et al., 2000; Kirisako et al., 2000; Maruyama and Noda, 2018). In Saccharomyces cerevisiae, Atg8 is C-terminally conjugated to the phospholipid phosphatidylethanolamine (PE) to generate Atg8-PE, and Atg4 cleaves the amide bond of Atg8-PE to release the protein from PE in membranes (Yoshimoto et al., 2004; Nakatogawa et al., 2007, 2012). Therefore, Atg4 plays a vital role in autophagy by cleaving the Atg8-PE, which will help to recycle Atg8 for the next round of the conjugation reaction (Nakatogawa et al., 2007, 2012), and promote the elongation step of the isolation membrane directly (Hirata et al., 2017).
Recently, increasing knowledge of atg genes has been acquired from several filamentous fungi (Ying and Feng, 2019). Previous studies showed that Atg proteins play important roles in appressorium formation and virulence in the plant pathogenic fungi, such as Magnaporthe oryzae. For example, deletion of Moatg8 prevented starvation-induced autophagy, appressorium development, and conidium death in M. oryzae (Veneault-Fourrey et al., 2006; Liu et al., 2010). Subsequently, deletion of three atg genes (Mgatg1, Mgatg5, and Mgatg9), which are required for autophagy, blocked plant infection in M. oryzae (Liu et al., 2007, 2010; Lu et al., 2009; Yin et al., 2019), and deletion of Moatg4 resulted a significant defective in hyphal growth, conidiation, and appressorium formation in M. oryzae (Liu et al., 2010, 2012). Moreover, disruption of Bbatg1 and Bbatg8 impaired conidial germination and virulence in entomopathogenic fungus Beauveria bassiana (Ying et al., 2016). Recently, deletion of Bbatg11 caused a significantly defective in conidial germination, stress response, and virulence in B. bassiana (Ding et al., 2018). Therefore, atg genes play important roles in fungal growth, development, and differentiation.
Nematode-trapping (NT) fungi are natural enemies of nematodes since they may attack live nematodes and utilize them as a source of nutrients (Dijksterhuis et al., 1994). Arthrobotrys oligospora is a typical species of the NT fungi that can develop adhesive networks (traps) for nematode predation (Yang et al., 2011). In previous study, a knockout mutant of the gene Aoatg8 was constructed in A. oligospora and found that Aoatg8 is required for conidiation and trap formation (Chen et al., 2013). Similar to S. cerevisiae, M. oryzae, and B. bassiana, most atg genes, involved in macroautophagy and selective autophagy, are also conserved in A. oligospora (Ying and Feng, 2019). However, little is known about the functions of atg genes in A. oligospora and other NT fungi. In this study, we elucidated the effect of gene Aolatg4 on vegetative growth, spore production, and pathogenicity in A. oligospora by comparing the biochemical and phenotypical traits between the wild type (WT) strain and ΔAolatg4 mutants. Moreover, we observed autophagosome formation and autophagic process in the WT and ΔAolatg4 mutants using microscopic analysis of GFP-Atg8. Our results suggest that Aolatg4 plays important roles in autophagic process, and also regulates hyphal growth, conidiation, stress response, and trap formation in A. oligospora.
Materials and Methods
Strains and Culture Conditions
The fungus A. oligospora (strain no. ATCC 24927) and its derived strains were cultivated on potato dextrose agar (PDA), tryptone glucose (TG), and Corn-maizena Yeast extract (CMY) media as described previously (Yang et al., 2018). The fungal strains were incubated on CM and MM-N media for nutrient stress induced by nitrogen starvation (Talbot et al., 1993). Saccharomyces cerevisiae FY834 (strain no. ATCC 90845) was inoculated on SC-Ura medium for screening the recombinational clones (Park et al., 2011). Caenorhabditis elegans (strain N2) was incubated on oatmeal medium at 26°C for inducing the trap formation of A. oligospora and bioassay.
Sequence and Phylogenetic Analyses of AolAtg4
The autophagy gene Aolatg4 was retrieved from A. oligospora according to the orthologous gene atg4 in S. cerevisiae, and the corresponding protein AolAtg4 (AOL_s00083g501) was identified. The conserved domains of AolAtg4 were predicted using InterProScan.1 Similarities of orthologous Atg4 proteins from different fungi were predicted using the DNAman software package (Lynnon Biosoft, San Ramon, United States; Ma et al., 2020). A neighbor-joining tree for Atg4 from various fungi was constructed using the Mega 7.0 software (Kumar et al., 2016).
Construction of Aolatg4 Gene Deletion in A. oligospora
Deletion of the Aolatg4 gene was performed using homologous recombination, as described previously (Tunlid et al., 1999; Chen et al., 2013; Zhen et al., 2019). The upstream (2,150 bp) and downstream (2,098 bp) sequences corresponding to the Aolatg4 ORF (5' and 3' flanking regions) in A. oligospora were amplified using paired primers (Supplementary Table S1). Subsequently, the hph cassette was amplified using primers Hph-f and Hph-r (Supplementary Table S1), and the plasmid pSCN44 was used as the template (Bernhards and Pöggeler, 2011). Finally, three PCR fragments and a linearized pRS426 vector were co-transformed into yeast strain FY834 via electroporation (Bernhards and Pöggeler, 2011; Park et al., 2011). The complete fragment for gene disruption was amplified from the recombinant plasmid pRS426-ATG4-hph using primers AolAtg4-5f and AolAtg4-3r (Supplementary Table S1), then it was transformed into A. oligospora using protoplast transformation method (Tunlid et al., 1999; Liu et al., 2020). Hygromycin-resistant transformants were selected on PDAS medium containing 200 mg/ml hygromycin B (Amresco, Solon, United States; Zhen et al., 2019; Liu et al., 2020). Two deletion mutants of Aolatg4 gene were verified using PCR amplification and Southern blotting analyses as described previously (Xie et al., 2019, 2020).
Generation of AoAtg8-GFP Fusion Cassette
The promoter (1,345 bp) of the Aoatg8 gene in A. oligospora was amplified with the primers AoAtg8p-f and AoAtg8p-r, and the gene Aoatg8 was amplified with the primers AoAtg8-f and AoAtg8-r (Supplementary Table S1; Ding et al., 2018). The Aoatg8 gene was integrated into the BsrGΙ sites of the Ppk2 vector, and the corresponding promoter sequence was inserted into XhoΙ; the GFP gene was cloned at the BsrGΙ and XhoΙ sites. The plasmid ppk-GFP-atg8 was transformed into the WT and ΔAolatg4 mutant strains, as described previously (Liu et al., 2008).
Analyses of Mycelial Growth Under Different Media and Stress Conditions
For vegetative growth, A. oligospora WT strain and its derived mutants (ΔAolatg4) were propagated on PDA plate for 5 days at 28°C and then placed onto fresh media (CMY, TG, and PDA) to assess mycelial growth as described previously (Zhen et al., 2018). For stress tolerance analysis, WT and mutants were inoculated on the center of plates with stress conditions (TG containing 0.04, 0.06, and 0.08 mM menadione and Congo red, as well as 0.3, 0.5, and 0.7 M sorbitol), and cultured at 28°C for 7 days (Zhen et al., 2019). Relative growth inhibition (RGI) value of fungal strain was determined as previously described (Chen et al., 2014; Ma et al., 2020). Each experiment was performed three times.
To determine the transcription of genes Aolatg4 at the different growth stages, the fungus was incubated on CMY at 28°C, and the mycelia were collected at 3, 4, 5, 7, and 9 days, respectively (Zhen et al., 2019). Moreover, the WT and mutant were inoculated on CM medium for 48 h, and then the mycelia were transferred into MM-N and MM-C, as well CM medium containing 0.05 mg/ml rapamycin for 6 h at 28°C, respectively, and mycelial samples were collected to analyze the transcription of gene Aolatg4 under chemical and nutrient stresses.
Analysis of Conidia Yields and Transcription of Sporulation-Related Genes
Conidia yields were determined after the fungal colonies were cultivated on CMY plate at 28°C for 7 days. To restore conidiation, the fungal colonies were cultivated on CMY medium supplemented with 10 or 20 mM glucose. The spores of the colonies was scraped harvested in micro-centrifuge tubes (Biosharp Life Sciences, Hefei, China), the conidia yields of the WT and ΔAolatg4 mutants were calculated as described previously (Deng et al., 2012). To determine the transcription of sporulation-related genes at the different growth stages, the WT and mutant were incubated on CMY at 28°C, and their mycelia were collected at 3, 5, and 7 days, respectively.
Confocal Microscopy and Transmission Electron Microscopy Assays
Hyphae expressing the fusion gene of GFP-atg8 were cultured in CM medium at 28°C and 180 rpm for 48 h, and then transferred into MM-N medium for 6 h at 28°C in a 180 rpm shaker (Lv et al., 2017). For detection of the expression of GFP-Atg8 in trap cells, 50 μl conidial suspensions (105 conidia per ml) of the WT and mutants were inoculated on MM-N plate contained cellophane. After 24-h incubation at 28°C, ~100 nematodes (C. elegans) were added to induce trap formation. Finally, the localization of GFP-Atg8 in hyphae and traps was observed using a confocal laser scanning microscope (Leica, Mannheim, Germany). FM4-64 (Invitrogen, Carlsbad, United States) was used for vacuole staining as described previously (Ma et al., 2020). DAPI (Invitrogen, Carlsbad, United States) was used for nuclear staining as described previously (Xie et al., 2020).
Trap Formation of A. oligospora and Bioassay
For analysis of trap formation induced by nematodes, conidia harvested from 7-day-old CMY cultures were resuspended to 1 × 105 conidia per ml in sterile water. A fifty microliter conidial suspension of the WT and mutants was inoculated on separate WA plates and incubated at 28°C for 2 days (Xie et al., 2019). Arthrobotrys oligospora mycelia and ΔAolatg4 mutants were added to ~200 nematodes on each plate. Trap formation was observed at the time intervals, 12, 24, and 36 h under a light microscopy (Olympus, Tokyo, Japan). Staining with 0.1% Calcofluor White (CFW, Sigma-Aldrich, United States) was carried out to observe trap formation induced by nematodes (Xie et al., 2020). Trap production and nematode death rate were quantified as the total number of traps and nematodes present per unit area of the plates in WT and ΔAolatg4 mutants.
Quantitative Real-Time PCR Analysis
Total RNAs from WT and mutants were extracted using a RNA miniprep Kit (Axygen, Jiangsu, China), and reverse-transcribed into cDNAs. The cDNA samples were used as the template to determine the transcription of sporulation-related genes with paired primers (Supplementary Table S2; Xie et al., 2020). Real-Time PCR (RT-PCR) was performed to analyze the transcription of genes as described previously (Yang et al., 2013). β-Tubulin gene (Aotub) was used as an internal reference and transcript levels were calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Statistical Analysis
Data are presented as the mean ± standard deviation (SD). Prism 5 (GraphPad, San Diego, CA, United States) was used for the photographs and statistical analyses (one-way ANOVA). Tukey’s honestly significant difference (HSD), p < 0.05, was considered to indicate significant differences (Ma et al., 2020).
Results
Sequence and Phylogenetic Analyses of AolAtg4
The gene Aolatg4 encodes a 444-amino acid polypeptide with a molecular mass of 49.8 kDa and an isoelectric point of 4.8, which contains a conserved peptidase C54 (IPR005078) domain. AolAtg4 shares a high degree of similarity with orthologs from various fungi. It has 89.6 and 93.2% identity with the homologous Atg4 from two species of NT fungi Duddingtonia flagrans and Dactylellina haptotyla, respectively; 42.7–56.8% sequence identity with the orthologs from other filamentous fungi; and the lowest identity (32.3%) with its S. cerevisiae ortholog. Atg4 orthologs from diverse fungi was divided into two clades (A and B); Atg4 orthologs from three species of NT fungi clustered into clade B, and Atg4 from other filamentous fungi clustered into clade A (Supplementary Figure S1).
AolAtg4 Regulates Mycelial Growth, Cell Nuclei, and Septal Development
Two ∆Aolatg4 mutants (a and b) were verified using PCR amplification and Southern blot methods (Supplementary Figure S2). Mycelial growth showed a statistically significant difference between the WT strain and ΔAolatg4 mutants on different media (CMY, TYGA, and TG; Figure 1A and Supplementary Figure S3). The colony diameter of the WT and ∆Aolatg4 mutants was 7.75 ± 0.23 and 6.24 ± 0.12 cm, respectively on CMY plate in 6 days (Figure 1A). In TG, the colony diameter of the ∆Aolatg4 mutants was 5.45 ± 0.13 cm after incubation for 6 days, which was significantly smaller than the 7.92 ± 0.16 cm colony diameter of the WT strain (Figure 1A). In addition, the ΔAolatg4 mutants also showed different hyphal growth rate compared to the WT strain on CM and MM-N. The ΔAolatg4 mutants displayed decreased growth and sparse aerial hyphae on CM medium compared to the WT strain. The ΔAolatg4 mutants also grew slower than the WT strain on MM-N (Figures 1B,C).
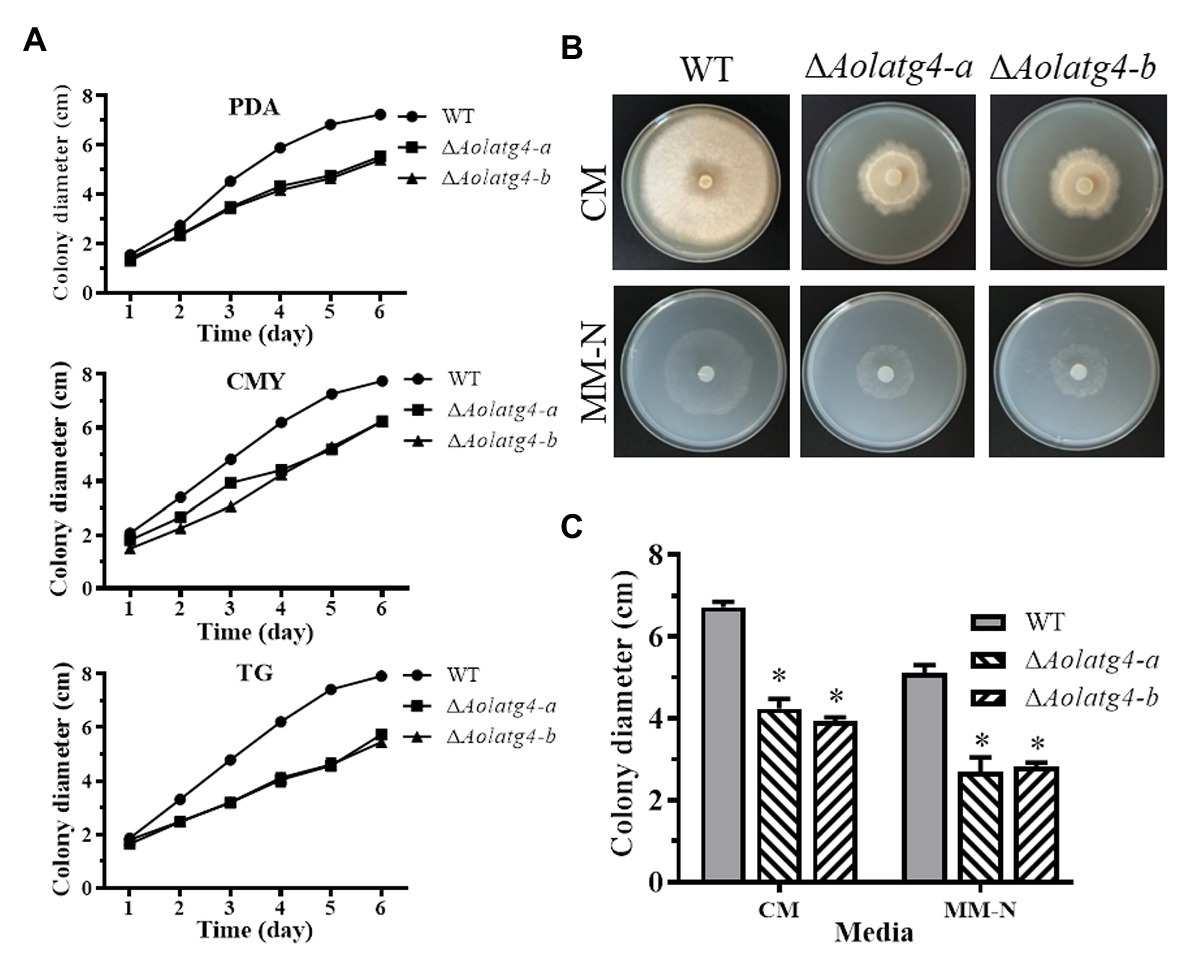
Figure 1. Comparison of mycelial growth of the wild type (WT) and ΔAolatg4 mutants on different meida. (A) Colony diameters of fungal strains incubated on potato dextrose agar (PDA), CMY, and tryptone glucose (TG) media. Each experiment was performed three times. (B) Colony morphology of fungal stains incubated on CM and MM-N media at 28°C for 7 days. (C) Colony diameters of fungal strains incubated on CM and MM-N plates for 7 days. Error bars: standard deviation, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
The transcription of gene Aolatg4 was increased in A. oligospora during the conidiation stage (7–9 days; Supplementary Figure S4A). Moreover, the transcript level of gene Aolatg4 was upregulated under chemical and nutrient stresses, including MM-N, MM-C, and CM containing 0.05 mg/ml rapamycin (Supplementary Figure S4B).
The cell nuclei were observed in WT and ΔAolatg4 mutants using DAPI staining. The hyphal cells of the WT contained 11–22 nuclei, whereas only 5–9 nuclei were observed in ΔAolatg4 mutants (Supplementary Figure S5A). Moreover, more septa were observed in the hyphae of the ΔAoatg4 mutants than in the WT strain using CFW staining (Supplementary Figure S5B).
AolAtg4 Plays a Critical Role in Conidiation
The wild type strain had luxuriant aerial hyphae when cultured on CMY plates, in contrast to the sparse aerial hyphae in the ∆Aolatg4 mutants (Figure 2A). Spore production in the ∆Aolatg4 mutants was significantly decreased. The mutants produced (1.2 ± 0.08) × 106 conidia/cm2, whereas the WT strain produced (10 ± 0.24) × 106 conidia/cm2; the conidial yield of the mutants was decreased by 88.4% (Figure 2B). Meanwhile, the conidiation of the ∆Aolatg4 mutant was partially restored when supplemented with 10 or 20 mM glucose (Supplementary Figure S6). Furtherly, we determined conidial germination on MM medium, at 4 h, 27.9% of the conidia of ∆Aolatg4 mutants germinated, compared to 40.8% of those of the WT strain, and 12 h later, the spore germination rates of the mutants and WT strain were 62.2 and 83.3%, respectively (Figure 2D). Moreover, 15.9% of the spores of the mutants were deformed compared to the WT strain (Figure 2C).
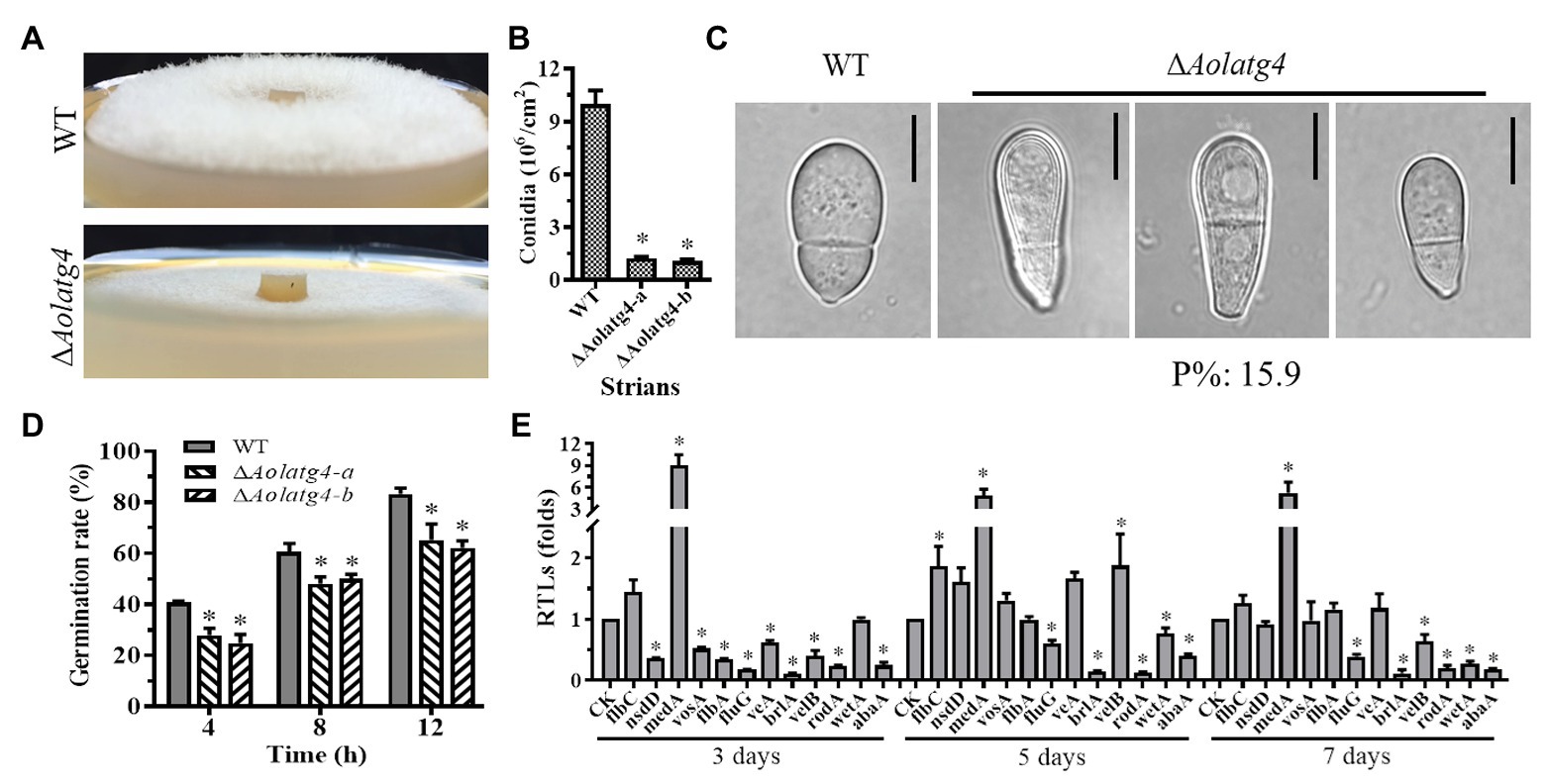
Figure 2. Comparison of aerial hyphae, conidiation, and the transcript of sporulation-related genes between the WT and ΔAolatg4 mutants. (A) Comparison of aerial hyphae between the WT and ΔAolatg4 mutants; the fungal strains were incubated on CMY for 7 days at 28°C. (B) The conidia yields collected from strains grown in CMY for 7 days. (C) The conidia of the WT and ΔAolatg4 mutants. P%: the percentage of the deformed spores in the mutant. (D) Spores of fungal strains germinate; the spores of the WT and ΔAolatg4 mutants were incubated in MM-N liquid medium for 4, 8, and 12 h. (E) Comparison of sporulation-related genes between the WT and ΔAolatg4 mutants. CK was denoted as a standard for statistical analysis of the RTL. Each experiment was performed three times. Error bars: standard deviation, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
The transcript levels of 12 sporulation-related genes changed in the ∆Aolatg4 mutant compared to those of the WT strain. Among them, nine genes (abaA, brlA, flbA, fluG, flbC, nsdD, rodA, veA, and vosA) were remarkably downregulated on day 3, and six genes including abaA, fluG, brlA, rodA, velB, and wetA were all downregulated on day 3, 5, and 7. However, medA was remarkably upregulated at three time points (Figure 2E).
AolAtg4 Regulates Stress Resistance in A. oligospora
The mycelial growth of WT was affected by TG plate-supplemented chemical stress (menadione, Congo red, and sorbitol), whereas the growth of the mutants was promoted by 0.04–0.06 mM and inhibited by 0.08 mM menadione; the growth of the mutants showed no change with 0.04–0.06 mM Congo red but was inhibited by 0.08 mM Congo red; in contrast, the growth of the mutants was promoted by 0.3–0.7 M sorbitol (Figure 3A). The RGI value of the ∆Aolatg4 mutants (83%) was higher than that of the WT strain (63%) on TG plates containing 0.08 mM Congo red or menadione (Figures 3B,C), whereas the RGI values of the mutant (5.6, 17.0, and 44%) were lower than that of the WT (14.9, 26.3, and 54.3%) on TG plates containing 0.3, 0.5, and 0.7 M sorbitol (Figure 3D), respectively.
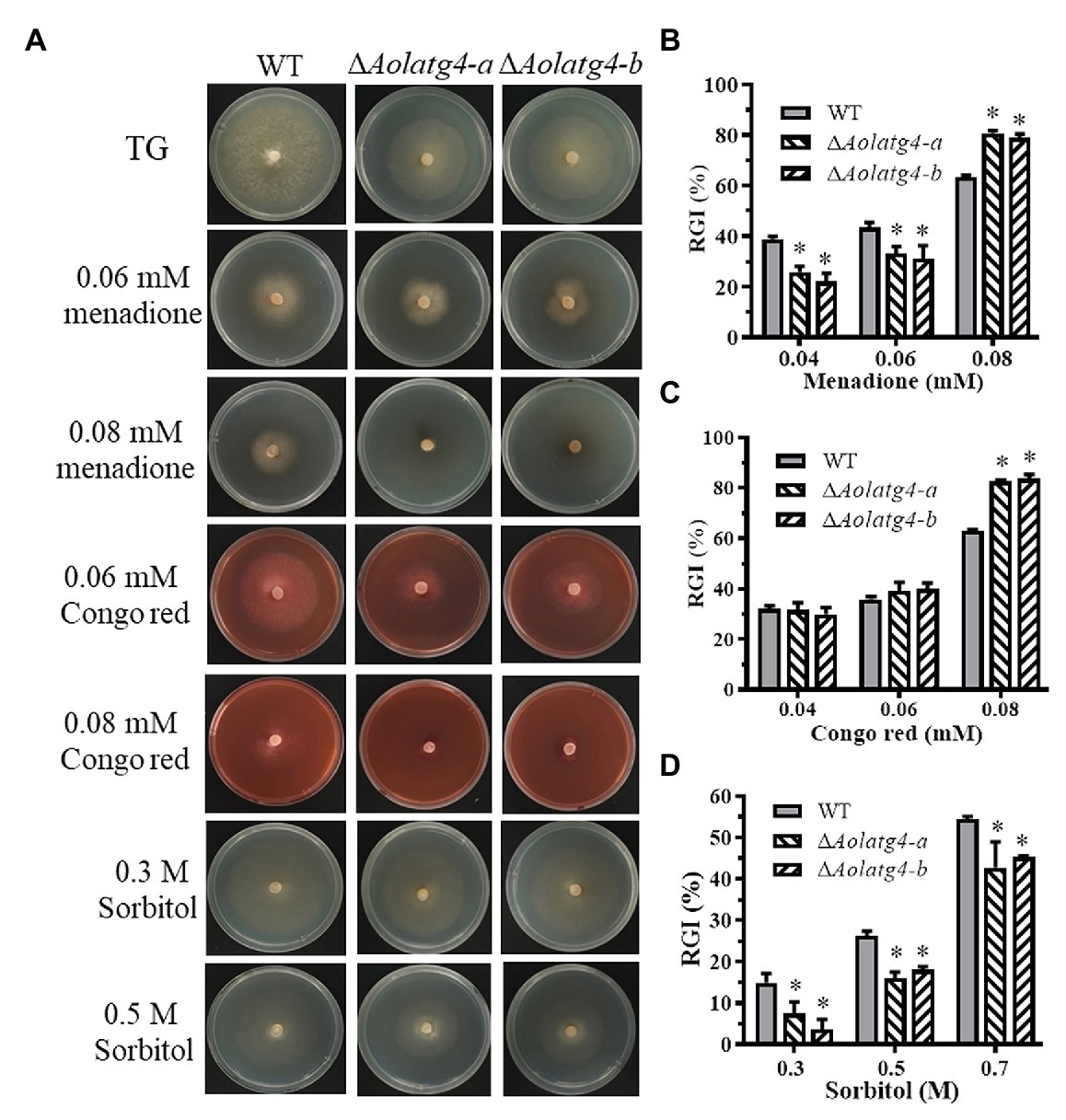
Figure 3. Comparison of stress resistance between WT and ΔAolatg4 mutants. (A) The WT strain and ΔAolatg4 mutant were incubated on TG medium containing menadione, sorbitol, and Congo red. (B) Relative growth inhibition (RGI) values of the fungal strains incubated on TG medium containing 0.04–0.08 mM menadione for 7 days. (C) RGI values of the fungal strains incubated on TG medium containing 0.04–0.08 mM Congo red for 7 days. (D) RGI values of the fungal strains incubated on TG medium containing 0.3–0.7 M sorbitol for 7 days. Each experiment was performed three times. Error bars: standard deviation, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
AolAtg4 Regulates Autophagic Process
To determine the blockage of the autophagic pathway in the ∆Aolatg4 mutants, we constructed the GFP-Atg8 fusion protein and expressed it in the WT and ∆Aolatg4 mutants. These strains were cultured in CM medium (rich-nutrient condition) for 48 h, few autophagosomes were observed in peripheral of vacuole in the hyphae of the WT and ∆Aolatg4 mutants, whereas no GFP signal was accumulated in vacuoles (Figure 4A). When fungal strains were cultured in CM medium for 24 h, and then transferred to MM-N medium (nitrogen starvation condition) and incubated for 6 h. Under starvation stress, the GFP signals were migrated into the mycelial vacuoles of the WT strain, whereas no GFP signal was accumulated in vacuoles of the ∆Aolatg4 mutants but instead was distributed dispersedly in hyphal cells (Figure 4B). Moreover, the vacuoles were significantly increased in numbers in the mutants compared to the WT strain (Figure 4B). To confirm whether autophagy was affected by deletion of the gene Aolatg4 in A. oligospora, the autophagosomes in vacuoles were observed in the WT and ∆Aolatg4 mutants using TEM. When fungal strains were cultured in MM-N medium for 6 h, autophagosomes were clearly observed in the vacuoles of the WT strain, while few autophagosomes or autophagosome-like structures (22.6% vs. to the WT strain) were observed in vacuoles of ∆Aolatg4 mutants (Figure 4C). Moreover, we further determine the localization of GFP-Atg8 in traps of the WT and mutants. GFP signals accumulated in vacuoles of the traps and adjacent cells of the WT strain, and no GFP signal was observed in vacuoles of the traps in the ∆Aolatg4 mutants (Figure 5).
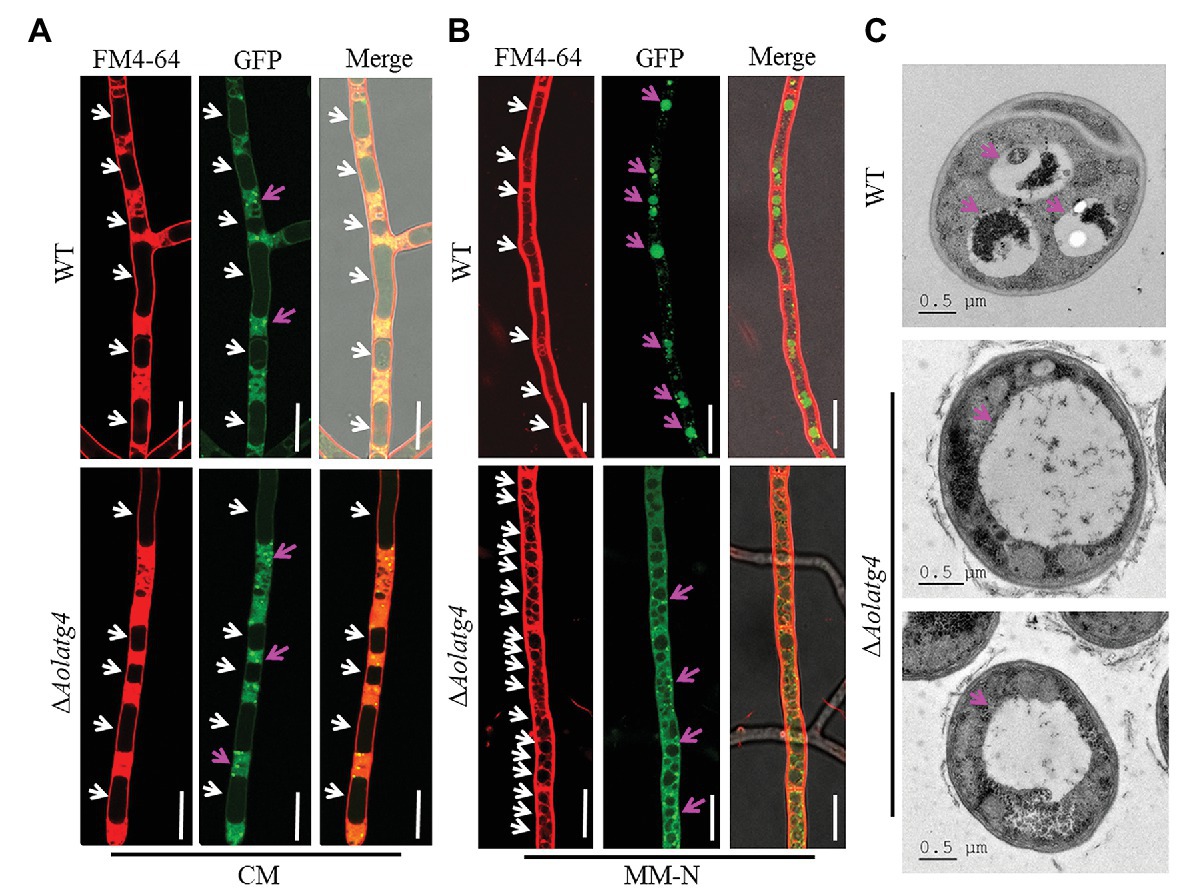
Figure 4. Observation of autophagosomes in the hyphal vacuoles of the WT and ∆Aolatg4 mutants. (A) Observation of the WT and ∆Aolatg4 mutants expressing GFP-Atg8; the strains were cultivated in liquid CM medium at 28°C for 48 h. (B) Observation of the WT and ∆Aolatg4 mutants expressing GFP-Atg8; the strains were grown in liquid CM medium at 28°C for 24 h, and then transferred to liquid MM-N medium for 6 h. The vacuoles were stained by FM4-64 and examined by fluorescence microscopy. White arrow: vacuoles; red arrow: autophagosomes. Scale bars = 10 μm. (C) The vacuoles of hyphal cells were observed using transmission electron microscopy. Arrows indicate the vacuole. Scale bars = 0.5 μm.
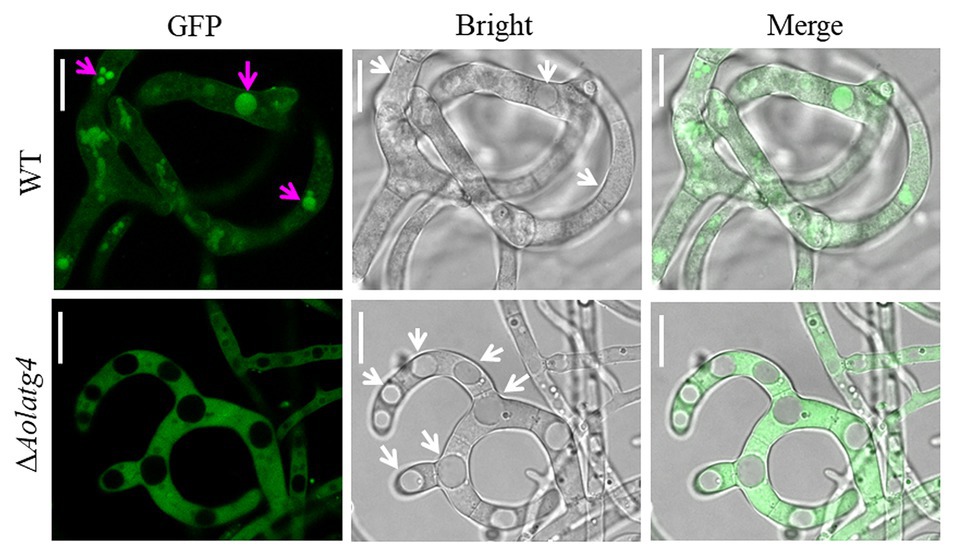
Figure 5. Detection of autophagosomes in the traps of the WT and ∆Aolatg4 mutants. Conidial suspensions of the indicated strains were inoculated on MM-N plate contained cellophane. After 24-h incubation at 28°C, about 100 nematodes were added on each plate. Autophagosomes in traps were observed using fluorescence microscopy. White arrow: vacuoles; red arrow: autophagosomes. Scale bars = 10 μm.
AolAtg4 Regulates Trap Formation and Pathogenicity
The vegetative hyphae of A. oligospora and other NT fungi can develop diverse traps for nematode predation. After addition of nematodes for 12 h, fresh traps containing one or two hyphal loops (immature traps) were observed on the plates culturing the WT strain, whereas few traps were observed on the plates culturing the ∆Aolatg4 mutants. Mature traps containing multiple hyphal loops were observed at 24 and 36 h in the WT strain (Figures 6A,B); 812 and 1,150 traps were observed in the WT strain at 24 and 36 h, respectively, whereas 96 and 98 traps per plate were produced by the ∆Aolatg4 mutants at 24 and 36 h, respectively (Figure 6C). We further observed the traps produced by the WT and ∆Aolatg4 mutants; the traps of the WT strain consisted of six or more hyphal loops, whereas traps developed by the ∆Aolatg4 mutants contained only 2–3 hyphal loops (Figure 6B) even at 48 h or longer.
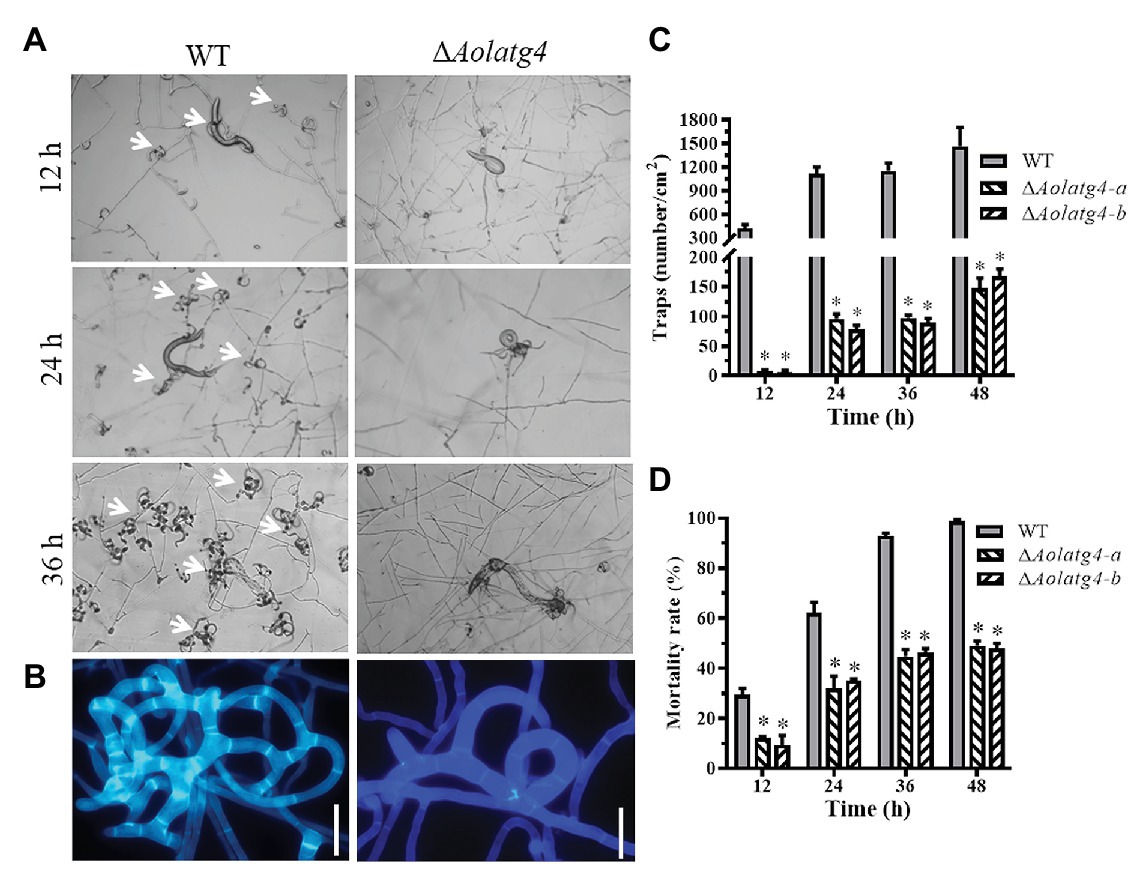
Figure 6. Analysis of trap formation and nematicidal activity of the WT and ΔAolatg4 mutants. (A) Traps induced by nematodes in WT and mutants at 12, 24, and 36 h. White arrow: traps. (B) The enlarged traps produced by the WT and mutants at 48 h; the traps were stained with Calcofluor White (CFW; 10 μg/ml). Scale bars = 10 μm. (C) The traps produced by the WT and mutants at different time points. (D) The mortality rate (%) of nematodes at different time points. Each experiment was performed three times. Error bars: standard deviation, asterisk: significant difference between mutant and WT (Tukey’s HSD, p < 0.05).
The fungal strains began to capture nematodes with trap formation; 29.6% of nematodes were captured by the WT strain and 10.8% of nematodes were captured by the ∆Aolatg4 mutants at 12 h. Subsequently, 62.2, 92.8, and 98.6% nematodes were captured by the WT strain at 24, 36, and 48 h (Figure 6D), respectively. In contrast, 33.5, 45.5, and 48.5% of nematodes were captured by the ΔAolatg4 mutants at the corresponding time points (Figure 6D).
Discussion
Autophagy is a conserved biological process in eukaryotes and contributes to maintaining cellular homeostasis, which is strictly regulated by Atg proteins (Mizushima et al., 2011; Maruyama and Noda, 2018). Recent years, increasing studies have shown that Atg proteins involve in the regulation of vegetative growth, conidiation, infection structure, and virulence of fungi (Liu et al., 2012; Ying and Feng, 2019). Previous research suggested that Aoatg8 gene is necessary for hyphal growth, sporulation, and trap formation in A. oligospora (Chen et al., 2013). In this study, we characterized the gene Aolatg4 in A. oligospora, and our analysis showed that deletion of Aolatg4 impaired autophagic process, thus affecting diverse phenotypic traits, such as hyphal growth, conidiation, and nematicidal activity in A. oligospora.
Deletion of the gene Aolatg4 caused defective growth on the PDA, CMY, and TG media compared to the WT strain, and mycelial growth of the mutants was also reduced on CM and MM-N media (Figures 1B,C). Meanwhile, the aerial mycelia of the ∆Aolatg4 mutants became very sparse (Figure 2A). Similarly, deletion of the gene Moatg4 in M. oryzae caused a significant reduction in hyphal growth (Liu et al., 2010), and colonies of the ∆Fgatg4 mutants were significantly smaller than those of the WT strain under nutrient-rich conditions (PDA plates) in Fusarium graminearum (Lv et al., 2017). These analyses show that atg4 plays pivotal roles in mycelium growth under routine or nitrogen starvation conditions.
Other than mycelial growth, conidiation was also seriously affected in the ΔAolatg4 mutants. The spore yield of the ΔAolatg4 mutants was decreased by 88.4%. Further, 15.4% conidia of the mutants became spindly from obovoid, as seen in the WT strain (Figure 2B). The transcript of six sporulation-related genes, such as abaA, brlA, fluG, rodA, and wetA, was significantly downregulated at the conidiation stage, whereas the medA gene was remarkably upregulated at all time points in the ΔAolatg4 mutant compared to the WT strain. These genes play a crucial role in conidiation in filamentous fungi. For example, abaA, brlA, and wetA are the key genes encoding central regulators of conidiation in species of Aspergillus (Park and Yu, 2012), F. graminearum (Son et al., 2013), and B. bassiana (Zhang et al., 2019). The rodA gene is required for the rodlet layer formation and hydrophobicity in conidia of A. fumigatus (Park and Yu, 2016); and gene velB is required for conidiation in A. oligospora; ΔAovelB mutants displayed serious sporulation defects (Zhang et al., 2019). Interestingly, the conidiation defect in the ΔAolatg4 mutants could be restored by glucose, suggested that it was directly related to autophagy defect. Similarly, deletion of Moatg4 caused a significant defective in conidiation and conidial germination in M. oryzae (Liu et al., 2010). Moreover, the gene Moatg8 was significantly induced during asexual development in M. oryzae, and the conidial yield was dramatically reduced in the ΔMoatg4 mutant (Deng et al., 2009). Therefore, atg4 plays a conserved and pivotal role in conidiation in A. oligospora and M. oryzae.
Moreover, deletion of the gene Aolatg4 resulted in an increase in sensitivity to chemical stressors, such as menadione and Congo red (Figure 3). Similarly, deletion of Hoatg5 caused defective in hyphal growth on CM plates containing cell wall-perturbing agents, including CFW, SDS, and Congo red, in the endophytic fungus Harpophora oryzae (Liu et al., 2016). Moreover, the vegetative growth of ΔBbatg1 and ΔBbatg8 mutants was inhibited by menadione in B. bassiana (Ying et al., 2016). These results indicate that autophagy is involved in the regulation of fungal sensitivity to cell wall-disturbing agents and antioxidants.
The ∆atg4 mutants were almost completely blockage in autophagy and produced few small autophagosomes in S. cerevisiae under starvation conditions (Hirata et al., 2017). In filamentous fungi such as Sordaria macrospora and M. oryzae, orthologs of the atg4 gene have been characterized in autophagy. In S. macrospora, Smatg4 is required for nonselective macroautophagy and selective macropexophagy (Voigt and Pöggeler, 2013). No autophagosomes were accumulated in the cytoplasm or autophagic bodies in the lumen of vacuoles in ΔMoatg4 mutants (Liu et al., 2010). Moreover, the ΔAoatg4 mutant in Aspergillus oryzae indicated a defect in autophagy according to observation of the behavior of GFP-AoAtg8 (Kikuma et al., 2006). In this study, we found that autophagosomes were obviously decreased in the ∆Aolatg4 mutants under starvation conditions by TEM observation; we further confirmed using the GFP-Atg8 fusion protein that autophagic process was impaired in the hyphae and traps of ΔAolatg4 mutant. Our results showed that autophagosome formation was impaired in ∆Aolatg4 mutants. Therefore, Atg4 plays a vital role in autophagosome formation and autophagic processes in yeast and filamentous fungi.
Arthrobotrys oligospora is a common species of NT fungi, and trap formation is an important indicator for the lifestyle switch of NT fungi (Su et al., 2017). In previous study, deletion of the Aoatg8 gene suppressed nematode-induced autophagy and trap formation, suggested that autophagy plays an important role in trap formation of A. oligospora (Chen et al., 2013). In this study, we further characterized the role of Aolatg4 in trap formation and pathogenicity. Our results showed that Aolatg4 also contributes to A. oligospora autophagy; meanwhile, trap formation and nematicidal activity was remarkably decreased in the ∆Aolatg4 mutants, and the mutant only produced immature traps containing 2–3 hyphal loops. Similarly, deletion of the gene Moatg4 causes loss of appressorium ability to penetrate rice and barley (Liu et al., 2010). These results showed that atg4 regulates the development of infection structure, thus playing a vital role in pathogenicity in pathogenic fungi.
In summary, our results demonstrate that AolAtg4 plays an important role in hyphal growth, sporulation, stress resistance, trap formation, and regulation of autophagic process in A. oligospora. Our results provide a basis for investigating the roles of atg genes in A. oligospora and other NT fungi, which will help to explore the regulation mechanisms of vegetative growth, development, and differentiation of NT fungi.
Conclusion
We identified and characterized an autophagy gene, Aolatg4, from the fungus A. oligospora. Aolatg4 plays a crucial role in autophagic process, and is important for conidiation, trap formation, and resistance to oxidants and cell-wall-perturbing agents in this fungus. Except for above phenotypic traits, Aolatg4 also regulates the development of cell nuclei and mycelial septa in A. oligospora. Our findings provide a novel understanding of atg genes that regulate hyphal growth, conidiation, trap formation, and pathogenicity of NT fungi.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Author Contributions
JY conceived and designed the study. DZ and JY wrote the manuscript. DZ conducted the experiments. MX, NB, and LY analyzed the data. JY and K-QZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was jointly supported by the National Natural Science Foundation of China (nos. 31960556 and U1402265), the Applied Basic Research Foundation of Yunnan Province (202001BB050004), and the National Basic Research Program of China (2013CB127503).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.592524/full#supplementary-material
Footnotes
References
Bernhards, Y., and Pöggeler, S. (2011). The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr. Genet. 57, 133–149. doi: 10.1007/s00294-010-0333-z
Chen, Y. L., Gao, Y., Zhang, K. Q., and Zou, C. G. (2013). Autophagy is required for trap formation in the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. Rep. 5, 511–517. doi: 10.1111/1758-2229.12054
Chen, Y., Zhu, J., Ying, S. H., and Feng, M. G. (2014). Three mitogen-activated protein kinases required for cell wall integrity contribute greatly to biocontrol potential of a fungal entomopathogen. PLoS One 9:e87948. doi: 10.1371/journal.pone.0087948
Deng, Y. Z., Qu, Z., He, Y., and Naqvi, N. I. (2012). Sorting nexin Snx41 is essential for conidiation and mediates glutathione-based antioxidant defense during invasive growth in Magnaporthe oryzae. Autophagy 8, 1058–1070. doi: 10.4161/auto.20217
Deng, Y. Z., Ramos, P. M., and Naqvi, N. I. (2009). Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy 5, 33–43. doi: 10.4161/auto.5.1.7175
Dijksterhuis, J., Veenhuis, M., Harder, W., and Nordbring-Hertz, B. (1994). Nematophagous fungi: physiological aspects and structure–function relationships. Adv. Microb. Physiol. 36, 111–143. doi: 10.1016/S0065-2911(08)60178-2
Ding, J. L., Peng, Y. J., Chu, X. L., Feng, M. G., and Ying, S. H. (2018). Autophagy-related gene BbATG11 is indispensable for pexophagy and mitophagy, and contributes to stress response, conidiation and virulence in the insect mycopathogen Beauveria bassiana. Environ. Microbiol. 20, 3309–3324. doi: 10.1111/1462-2920.14329
Farré, J. C., and Subramani, S. (2016). Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat. Rev. Mol. Cell Biol. 17, 537–552. doi: 10.1038/nrm.2016.74
Hirata, E., Ohya, Y., and Suzuki, K. (2017). Atg4 plays an important role in efficient expansion of autophagic isolation membranes by cleaving lipidated Atg8 in Saccharomyces cerevisiae. PLoS One 12:e0181047. doi: 10.1371/journal.pone.0181047
Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. doi: 10.1038/35044114
Kikuma, T., Ohneda, M., Arioka, M., and Kitamoto, K. (2006). Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot. Cell 5, 1328–1336. doi: 10.1128/EC.00024-06
Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., et al. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276. doi: 10.1083/jcb.151.2.263
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, X. H., Gao, H. M., Xu, F., Lu, J. P., Devenish, R. J., and Lin, F. C. (2012). Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy 8, 1415–1425. doi: 10.4161/auto.21274
Liu, T., Huang, Y., Chen, X. X., Long, X., Yang, Y. H., Zhu, M. L., et al. (2020). Comparative transcriptomics reveals features and possible mechanisms of glucose-mediated soil fungistasis relief in Arthrobotrys oligospora. Front. Microbiol. 10:3143. doi: 10.3389/fmicb.2019.03143
Liu, X. H., Liu, T. B., and Lin, F. C. (2008). Monitoring autophagy in Magnaporthe oryzae. Methods Enzymol. 451, 271–294. doi: 10.1016/S0076-6879(08)03219-9
Liu, T. B., Liu, X. H., Lu, J. P., Zhang, L., Min, H., and Lin, F. C. (2010). The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 6, 74–85. doi: 10.4161/auto.6.1.10438
Liu, X. H., Lu, J. P., Zhang, L., Dong, B., Min, H., and Lin, F. C. (2007). Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 6, 997–1005. doi: 10.1128/EC.00011-07
Liu, N., Ning, G. A., Liu, X. H., Feng, X. X., Lu, J. P., Mao, L. J., et al. (2016). An autophagy gene, HoATG5, is involved in sporulation, cell wall integrity and infection of wounded barley leaves. Microbiol. Res. 192, 326–335. doi: 10.1016/j.micres.2016.08.008
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J. P., Liu, X. H., and Feng, X. X. (2009). An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae. Curr. Genet. 55, 461–473. doi: 10.1007/s00294-009-0259-5
Lv, W. Y., Wang, C. Y., Yang, N., Que, Y. W., Talbot, N. J., and Wang, Z. Y. (2017). Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 7:11062. doi: 10.1038/s41598-017-11640-z
Ma, Y. X., Yang, X. W., Xie, M. H., Zhang, G. S., Yang, L., Bai, N., et al. (2020). The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet. Biol. 138:103352. doi: 10.1016/j.fgb.2020.103352
Maruyama, T., and Noda, N. (2018). Autophagy-regulating protease Atg4: structure, function, regulation and inhibition. J. Antibiot. 71, 72–78. doi: 10.1038/ja.2017.104
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Nakatogawa, H., Ishii, J., Asai, E., and Ohsumi, Y. (2012). Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy 8, 177–186. doi: 10.4161/auto.8.2.18373
Park, G., Colot, H. V., Collopy, P. D., Krystofova, S., Crew, C., Ringelberg, C., et al. (2011). High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol. Biol. 722, 179–189. doi: 10.1007/978-1-61779-040-9_13
Park, H. S., and Yu, J. H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Park, H. S., and Yu, J. H. (2016). Developmental regulators in Aspergillus fumigatus. J. Microbiol. 54, 223–231. doi: 10.1007/s12275-016-5619-5
Reggiori, F., and Klionsky, D. J. (2002). Autophagy in the eukaryotic cell. Eukaryot. Cell 1, 11–21. doi: 10.1128/EC.01.1.11-21.2002
Ryter, S. W., Cloonan, S. M., and Choi, A. M. (2013). Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cell 36, 7–16. doi: 10.1007/s10059-013-0140-8
Son, H., Kim, M. G., Min, K., Seo, Y. S., Lim, J. Y., Choi, G. J., et al. (2013). AbaA regulates conidiogenesis in the ascomycete fungus Fusarium graminearum. PLoS One 8:e72915. doi: 10.1371/journal.pone.0072915
Su, H., Zhao, Y., Zhou, J., Feng, H., Jiang, D., Zhang, K. Q., et al. (2017). Trapping devices of nematode-trapping fungi: formation, evolution, and genomic perspectives. Biol. Rev. 92, 357–368. doi: 10.1111/brv.12233
Talbot, N. J., Ebbole, D. J., and Hamer, J. E. (1993). Identification and characterization of Mpg1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. doi: 10.1105/tpc.5.11.1575
Tunlid, A., Ahman, J., and Oliver, R. P. (1999). Transformation of the nematode trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 173, 111–116. doi: 10.1111/j.1574-6968.1999.tb13491.x
Veneault-Fourrey, C., Barooah, M., Egan, M., Wakley, G., and Talbot, N. J. (2006). Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312, 580–583. doi: 10.1126/science.1124550
Voigt, O., and Pöggeler, S. (2013). Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy 9, 33–49. doi: 10.4161/auto.22398
Xie, M. H., Bai, N., Yang, J. L., Jiang, K. X., Zhou, D. X., Zhao, Y. N., et al. (2020). Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 10:3065. doi: 10.3389/fmicb.2019.03065
Xie, M. H., Wang, Y., Tang, L., Yang, L., Zhou, D. X., Li, Q., et al. (2019). AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 21, 4648–4661. doi: 10.1111/1462-2920.14785
Yang, X. W., Ma, N., Yang, L., Zheng, Y., Zhen, Z., Li, Q., et al. (2018). Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 102, 4601–4613. doi: 10.1007/s00253-018-8929-1
Yang, J. K., Wang, L., Ji, X., Feng, Y., Li, X., Zou, C., et al. (2011). Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7:e1002179. doi: 10.1371/journal.ppat.1002179
Yang, J. K., Yu, Y., Li, J., Zhu, W., Geng, Z. Y., Jiang, D. W., et al. (2013). Characterization and functional analyses of the chitinase-encoding genes in the nematode-trapping fungus Arthrobotrys oligospora. Arch. Microbiol. 195, 453–462. doi: 10.1007/s00203-013-0894-6
Yin, Z. Y., Chen, C., Yang, J., Feng, W. Z., Liu, X. Y., Zuo, R. F., et al. (2019). Histone acetyltransferase MoHat1 acetylates autophagy-related proteins MoAtg3 and MoAtg9 to orchestrate functional appressorium formation and pathogenicity in Magnaporthe oryzae. Autophagy 15, 1234–1257. doi: 10.1080/15548627.2019.1580104
Ying, S. H., and Feng, M. G. (2019). Insight into vital role of autophagy in sustaining biological control potential of fungal pathogens against pest insects and nematodes. Virulence 10, 429–437. doi: 10.1080/21505594.2018.1518089
Ying, S. H., Liu, J., Chu, X. L., Xie, X. Q., and Feng, M. G. (2016). The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana. Sci. Rep. 6:26376. doi: 10.1038/srep26376
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Zhang, G. S., Zheng, Y. Q., Ma, Y. X., Yang, L., Niu, X. M., Zhou, D. X., et al. (2019). The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 10:1917. doi: 10.3389/fmicb.2019.01917
Zhen, Z. Y., Xing, X. J., Xie, M. H., Yang, L., Yang, X. W., Zheng, Y. Q., et al. (2018). MAP kinase Slt2 orthologs play similar roles in conidiation, trapformation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 116, 42–50. doi: 10.1016/j.fgb.2018.04.011
Zhen, Z. Y., Zhang, G. S., Yang, L., Ma, N., Li, Q., Ma, Y. X., et al. (2019). Characterization and functional analysis of calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 103, 819–832. doi: 10.1007/s00253-018-9504-5
Keywords: Arthrobotrys oligospora, autophagy-related gene Aolatg4, conidiation, trap formation, autophagosome, stress resistance
Citation: Zhou D, Xie M, Bai N, Yang L, Zhang K-Q and Yang J (2020) The Autophagy-Related Gene Aolatg4 Regulates Hyphal Growth, Sporulation, Autophagosome Formation, and Pathogenicity in Arthrobotrys oligospora . Front. Microbiol. 11:592524. doi: 10.3389/fmicb.2020.592524
Edited by:
Chengshu Wang, Chinese Academy of Sciences, ChinaCopyright © 2020 Zhou, Xie, Bai, Yang, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinkui Yang, amlua3VpOTYwQHludS5lZHUuY24=
 Duanxu Zhou
Duanxu Zhou Meihua Xie
Meihua Xie Na Bai
Na Bai Le Yang
Le Yang Ke-Qin Zhang
Ke-Qin Zhang Jinkui Yang
Jinkui Yang