
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 October 2020
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.592496
The purpose of this research was to characterize the antibiotic resistance profiles of Campylobacter spp. derived from chicken and pig feces collected from farms in Jiangsu Province, China, and to analyze the relevant resistance mechanisms among antimicrobial-resistant Campylobacter spp. isolates. Antibiotic susceptibility to nine antibiotic agents was tested with the microdilution method in 93 Campylobacter spp. (45 C. jejuni and 25 C. coli from chickens; 23 C. coli from pigs). High rates of resistance were observed to nalidixic acid (79.6%), erythromycin (75.3%), tetracycline (68.8%), azithromycin (66.7%), ciprofloxacin (64.5%), and gentamicin (35.5%), with a lower resistance rate to florfenicol (8.6%). The prevalence of the tested antibiotic resistance in C. coli was higher than in C. jejuni from chickens. The rate of antimicrobial resistance to ciprofloxacin in C. coli isolates from chickens was 100.0%, and the C. coli isolates from pigs were all resistant to erythromycin (100%). Most of C. jejuni (64.4%) and C. coli (64.5%) isolates displayed multi-drug resistance. All the Campylobacter spp. isolates resistant to fluoroquinolones had the C257T mutation in the gyrA gene. All 64 tetracycline-resistant Campylobacter spp. isolates were positive for the tetO gene. The tetA gene was also amplified in 6.5% of Campylobacter spp. isolates, whereas tetB was not detected among the isolates. The A2075G point mutation in the 23S rRNA gene occurred in 86.1% (62/72) of the macrolides-resistant Campylobacter spp. isolates, and the ermB gene was identified in 49 Campylobacter spp. isolates (30 C. jejuni and 19 C. coli). Amino acid insertions or mutations in the L4 and L22 ribosomal proteins were not linked to macrolide resistance. These results highlight the high prevalence of resistance to multiple antibiotics, particular macrolides, among Campylobacter spp. from chickens and pigs in Jiangsu Province, China, which is probably attributable to the overuse of antimicrobials in chicken and pig production. These findings recommend the more cautious use of critical antimicrobial agents in swine and poultry production. Stringent and continuous surveillance is required to reduce the drug-resistant campylobacteriosis in food animals and humans.
Campylobacteriosis is an essentially foodborne zoonotic disease worldwide (Allos, 2001). Most of these human illnesses involve C. jejuni (81%) or C. coli (8.4%) (Sifré et al., 2015). Enteric diseases caused by the thermophilic species C. jejuni, C. coli, C. lari, and C. upsaliensis range from asymptomatic infections to severe inflammatory bloody diarrhea (Ketley, 1997). In addition, C. jejuni is often associated with the Guillain-Barre syndrome (Dingle et al., 2001). Campylobacter spp. widely and asymptomatically colonize the intestinal tracts of food producing animals (Altekruse et al., 1999; Harvey et al., 1999; Stanley and Jones, 2003), and C. jejuni is the main species affecting chickens, whereas C. coli is mainly found in pigs (Sáenz et al., 2000; Haruna et al., 2013). Poultry with no obvious clinical symptoms are the main reservoirs of these pathogens (Luangtongkum et al., 2006; Humphrey et al., 2007). Pigs are also usually subclinically infected with Campylobacter spp. During the slaughter process, intestinal content spillage could contaminate the meat and meat products (Weijtens et al., 1997; Gill et al., 1999).
Although Campylobacter infection is usually self-limiting, severe, long-lasting or systemic Campylobacter infections can be treated with antibacterial drugs (Allos, 2001). Tetracyclines, macrolides, and fluoroquinolones are used to control Campylobacter-mediated infections (Sifré et al., 2015). In food-producing animal industries, ciprofloxacin (a fluoroquinolone) is frequently used because it has a broad antibacterial spectrum. In recent decades, fluoroquinolone resistance in animal and human Campylobacter isolates have increased worldwide (Engberg et al., 2001; Moore et al., 2006; Wang et al., 2016). With this increase in fluoroquinolone-resistant Campylobacter, macrolides have become common drugs in treatment of campylobacteriosis (Luangtongkum et al., 2009). Macrolides, including tylosin, tilmicosin, tulathromycin, and tildipirosin, have been widely used in animal production in many countries (Poehlsgaard et al., 2012; McEwen and Collignon, 2018), and macrolide-resistant Campylobacter strains have been isolated in many regions (Kaakoush et al., 2015; Zhang et al., 2016) and from animals (Wang et al., 2016; Lim et al., 2017; Hafez and Attia, 2020). Tetracycline has also been extremely widely used in animal farming because it has broad-spectrum antibacterial activity, low cost, and excellent efficacy (Chopra and Roberts, 2001), and it may have cause high tetracycline resistance in Campylobacter from food-producing animals (Premarathne et al., 2017; Woźniak-Biel et al., 2018). The proportion of Campylobacter spp. strains resistant to ciprofloxacin, tetracycline, and erythromycin is increasing (García-Fernández et al., 2018). A tendency to detect increasing multidrug-resistant (MDR) strains has been reported in both humans (Feizabadi et al., 2007) and food-producing animals (Li et al., 2016; Neogi et al., 2020). The potential risk of MDR Campylobacter spread from animals to human is of great concern.
In Campylobacter, the C257T mutation in the gyrase gene (gyrA) is the most frequent mechanism causing quinolone and fluoroquinolone resistance (Iovine, 2013). Efflux pumps and ribosomal protection genes are two major mechanisms of tetracycline resistance in various bacteria (Roberts, 2005). The tetO gene encodes a ribosomal protection protein that is primarily responsible for tetracycline-resistance. It is carried on the chromosome (Chopra and Roberts, 2001) or transferred on plasmids (Gibreel et al., 2004). The efflux genes, tetA and tetB, encode membrane-bound efflux proteins that export tetracycline from the cell (Chopra and Roberts, 2001), and the presence of tetA in Campylobacter isolates was first detected by Abdi-Hachesoo et al. (2014). Campylobacter has three main mechanisms that confer resistance to macrolides: modifications to ribosome-associated genes (V region of 23S rRNA and the rplD and rplV ribosomal-protein-encoding genes) (Luangtongkum et al., 2009), multidrug efflux pumps mediated by CmeABC (Lin et al., 2002), and a ribosomal methylase encoded by ermB, which was first reported in China in 2014 (Qin et al., 2014). Since then, ermB-positive Campylobacter has been isolated from broilers in Spain and the United States (Florez-Cuadrado et al., 2016).
Several studies have been published in China that report relatively high levels of antimicrobial resistance in Campylobacter from animals in different areas (Ma et al., 2014, 2017). Furthermore, in a campylobacteriosis outbreak in China in 2018, and the domain isolate was resistant to nalidixic acid, tetracycline and ciprofloxacin (Qu et al., 2019). In China, a national annual antimicrobial-resistance surveillance system for Campylobacter in food-producing animals has been in place from 2017 to 2020 to strengthen the monitoring of antimicrobial resistance and ensure the safety of feed animals and public health. To obtain epidemiological data on Campylobacter from food-producing animals in Jiangsu Province, China, the resistance rates of Campylobacter strains in chicken and pig feces on farms and the relevant molecular mechanisms of their resistance to quinolones, tetracycline, and macrolides have been investigated.
A total of 150 chicken cloacal swabs were collected on 15 chicken farms (seven layer farms and eight broiler farms), and 100 pig feces samples were collected on 10 pig farms, all located in five cities of Jiangsu Province, China. These cities (Changzhou, Suqian, Nantong, Yancheng, and Yangzhou) are the major food animal production areas for chickens and pigs in Jiangsu Province. Ten cloacal swabs or fresh feces samples were taken from randomly selected animals on each farm. After the samples were collected, they were transported to the laboratory under refrigerated conditions within 24 h. The collected samples were diluted with phosphate-buffered saline (PBS). After dilution, 50 μL of the appropriate concentration diluent was streaked onto campylobacter blood-free selective agar base (modified CCDA-preston; Oxoid CM1183, United Kingdom) containing cefoperazone (LKT, C1630) and amphotericin B (Wako, 011-1363). The agar plates were incubated at 42°C for 36 h under a microaerophilic atmosphere containing 85% N2, 10% CO2 and 5% O2. The suspected colonies were subcultured on Mueller-Hinton agar (MH, Difco MD) containing 5% defibrinated sheep blood under microaerophilic conditions at 42°C for 36 h, and confirmed with multiplex PCR, as described previously (Huang et al., 2009). The primers used are listed in Table 1. All the positive isolates were stored in brain-heart infusion (BHI) broth with 20% glycerol at −80°C.
The minimum inhibitory concentrations (MICs) of antimicrobial agents for Campylobacter were assessed with the broth microdilution method, using commercially available Sensititre® susceptibility plates for Campylobacter (TREK Diagnostic Systems, East Grinstead, United Kingdom), according to the manufacturer’s instructions. Nine antimicrobial agents belonging to seven classes were tested: fluoroquinolones (ciprofloxacin [CIP] and nalidixic acid [NAL]), macrolides (erythromycin [ERY] and azithromycin [AZM]), ketolides (telithromycin [TEL]), tetracyclines (tetracycline [TET]), aminoglycosides (gentamicin [GEN]), phenicols (florfenicol [FFN]), and lincosamides (clindamycin [CLI]). The MIC breakpoints for resistance were interpreted according to the NARMS-2014 recommendations for Campylobacter. C. jejuni strain ATCC 33560 and Mueller-Hinton broth with TES buffer and lysed horse blood were used as the positive control and the negative control, respectively. The results can be considered satisfactory if QC results are within range. To ensure the reproducibility of the MIC data, assays were repeated twice, each in duplicate. Multi-drug resistance was defined as resistance to three or more antimicrobial classes.
All genomic DNA of the Campylobacter isolates were extracted with the TIANamp Bacterial DNA Kit (Tiangen, Beijing, China), according to the manufacturer’s protocol, and stored at −20°C.
Ciprofloxacin resistance: The fluoroquinolone-resistance-determining region (QRDR) of the gyrA gene was amplified with PCR, as previously reported by Zirnstein et al. (1999). The PCR product of 673 bp was sequenced and compared with the sequences of the gyrA gene of Campylobacter obtained from GenBank.
Tetracycline resistance: Three tetracycline resistance genes (tetA, tetB, and tetO) were investigated in the Campylobacter isolates with PCR, as previously described (Abdi-Hachesoo et al., 2014).
Erythromycin resistance: region V of 23S rRNA (Jensen and Aarestrup, 2001), the ribosomal protein L4 gene rplD, and the ribosomal protein L22 gene rplV were amplified with PCR (Cagliero et al., 2006). The PCR products were then directly sequenced at Sangon Biotech (Shanghai, China). The DNA sequences obtained were compared with the sequence of the C. jejuni NCTC 11168 genome to identify specific mutations that had occurred in the ribosome. To test the ribosomal RNA methylase gene, ermB, it was amplified as reported by Qin et al. (2014). The oligonucleotide sequences of the primers and the sizes of the PCR products are given in Table 1.
The percentage differences in resistance to the nine tested antimicrobial agents between the C. jejuni and C. coli isolates were analyzed with the χ2 test in SAS 9.2 (SAS Institute Inc., Cary, NC, United States). P < 0.05 was considered statistically significant.
This study was performed according to the guidelines of the Animal Welfare and Ethical Censor Committee at Jiangsu Institute of Poultry Science. No chickens or pigs were sacrificed for the present study. When collecting cloacal swabs, well-trained farm workers held the chickens. Fresh feces from pigs were collected without any manipulation of the pigs.
A total 93 Campylobacter strains were isolated and identified from the 250 samples collected, with a total isolation rate of 37.2%. The occurrence of Campylobacter in the food animals ranged from 0 to 57.5%. Among them, 45 C. jejuni and 25 C. coli were isolated from chickens. The occurrence of Campylobacter was higher in the broilers (57.5%) than in the layers (34.3%, P < 0.0001). Twenty-three of the Campylobacter isolates from pigs were identified as C. coli (Table 2).
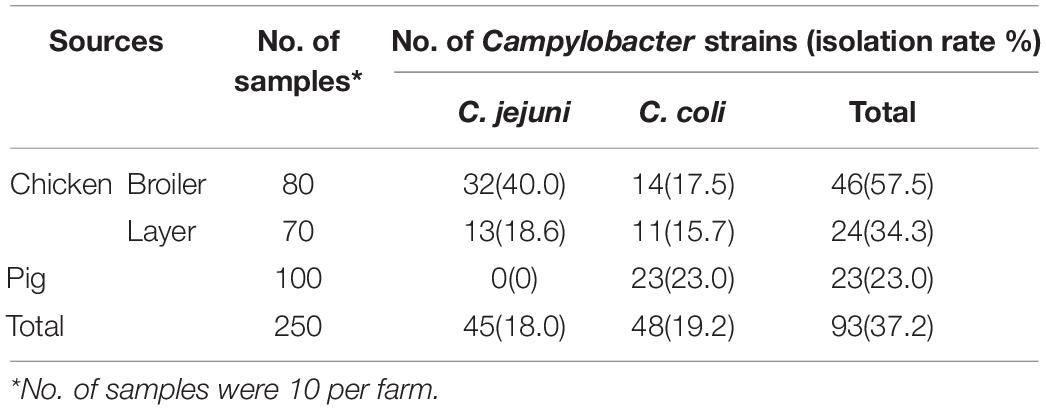
Table 2. Prevalence of Campylobacter strains isolated from chicken and pig in Jiangsu Province, China.
The antimicrobial resistance to the nine tested antimicrobial agents in these Campylobacter isolates are given in Table 3. High rates of resistance were observed to nalidixic acid (79.6%), erythromycin (75.3%), tetracycline (68.8%), azithromycin (66.7%), ciprofloxacin (64.5%), and gentamicin (35.5%) in the Campylobacter spp. strains, with a lower resistance rate to florfenicol (8.6%). Overall, the prevalence of resistance to the antibiotics tested was higher in C. coli than in C. jejuni from chickens. Resistance to fluoroquinolones was highest (80%–100%) in Campylobacter from chickens, except for ciprofloxacin in C. jejuni. The second highest rate of resistance was to tetracyclines, followed resistance to macrolides. The resistance rates of the C. jejuni (66.7%) and C. coli isolates (100.0%) from chickens to ciprofloxacin differed significantly (P = 0.0011). However, there was no significant difference between the resistance rates of the C. jejuni and C. coli isolates from chicken to the other antimicrobial agents. Resistance to erythromycin (100%) and azithromycin (82.6%) was much higher in the C. coli isolates from pigs than in C. coli (76.0%, P = 0.012 and 64.0%, P = 0.1472, respectively) or in C. jejuni (62.2%, P = 0.0007 and 60.0%, P = 0.05942, respectively) from chickens. The rates of resistance to ciprofloxacin (21.7%), nalidixic acid (65.2%), and tetracycline (43.5%) in the C. coli isolates from pigs were lower than in the C. coli isolates from chickens (100.0%, P < 0.0001, and 92.0%, P = 0.0225, and 88.0%, P = 0.0011, respectively).
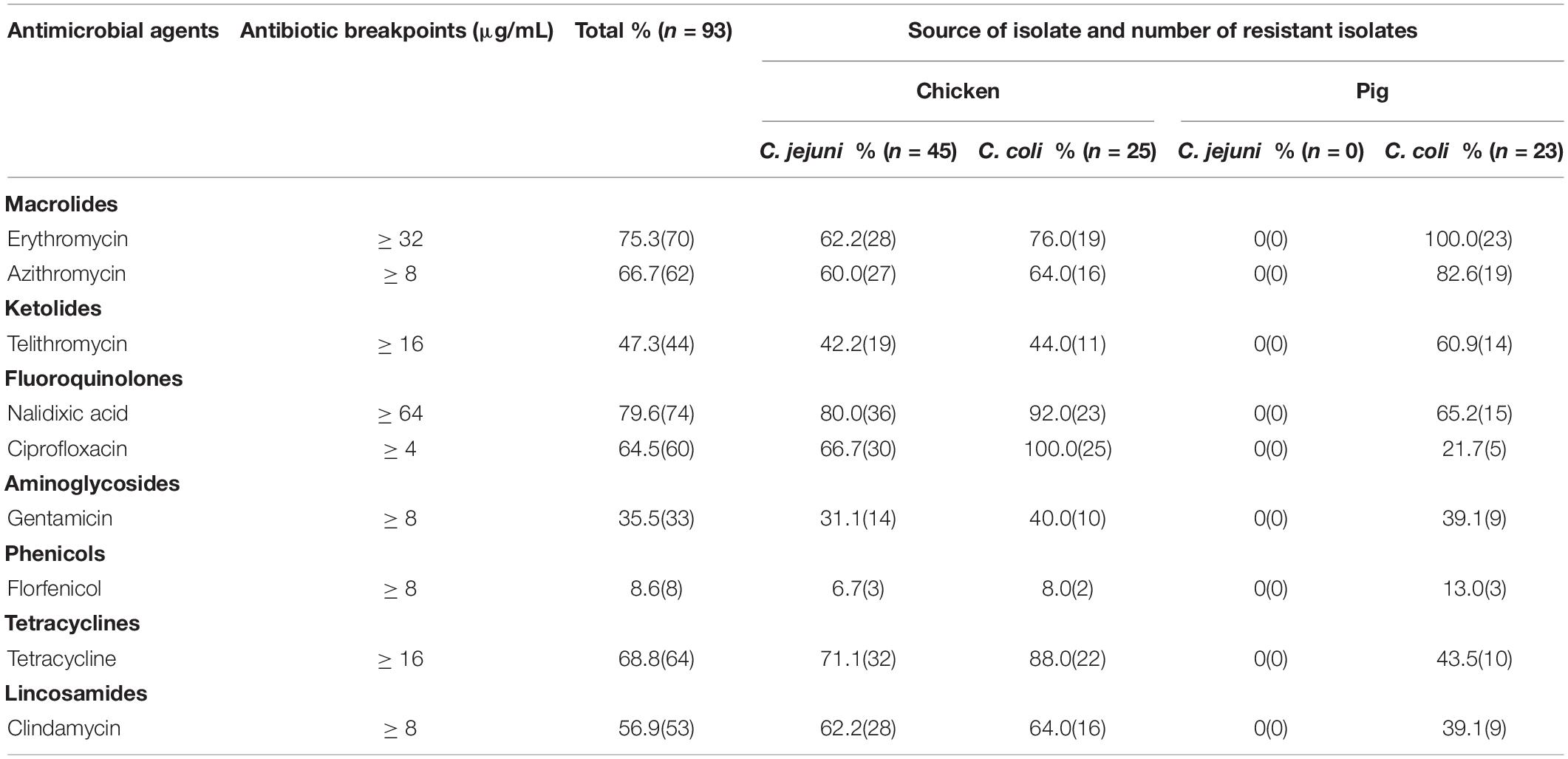
Table 3. The rate of antimicrobial resistance in C. jejuni and C. coli isolated from chicken and pig samples.
In this study, 88 out of the examined 93 isolates (94.6%) were resistant to at least one antimicrobial agent, whereas five C. jejuni isolates (5.4%) were susceptible to all the antimicrobial agents tested. Resistance to one antimicrobial class was identified in two (4.4%) out of the 45 C. jejuni isolates and five (10.4%) of the 48 C. coli isolates. Eighteen isolates (19.4%; nine C. jejuni and nine C. coli) were resistant to two antimicrobial classes. Sixty-three (67.7%) of all the isolates (29 C. jejuni and 34 C. coli) were classified as MDR, and the rates of resistance profiles that included 3, 4, 5, 6, and 7 antimicrobial classes were 7.5%, 17.2%, 20.4%, 18.3%, and 4.3%, respectively. In terms of the antimicrobial resistance pattern, 23 antimicrobial resistance (AMR) patterns were observed in the C. jejuni isolates, whereas 32 AMR patterns were detected in the C. coli isolates. The main AMR in Campylobacter spp. isolates was the combination of fluoroquinolones, tetracyclines, and macrolides (Table 4).
In this study, all phenotypically ciprofloxacin-and/or nalidixic-acid-resistant strains carried the C257T transition in gyrA, which resulted in a T86I amino acid substitution. No mutation at other position was detected in gyrA gene.
When we screened for tetracycline resistance genes, the tetO gene was detected in all 64 tetracycline-resistant Campylobacter spp. strains. The tetA gene was present in six (6.45%) isolates, including two C. coli and four C. jejuni. The tetB resistance gene was not detected in any Campylobacter isolate.
The A2075G point mutation within domain V of the 23S rRNA gene was observed in 62 highly macrolide-resistant Campylobacter isolates, including 28 C. jejuni and 34 C. coli. No mutation at other position associated with macrolide resistance was detected in this region in any isolate tested.
Two amino acid changes (V121A and M192I) in ribosomal protein L4 (encoded by the rplD gene) were detected in both macrolide-resistant and macrolide-susceptible Campylobacter isolates (Tables 5, 6). Eight macrolide-resistant and four macrolide-susceptible Campylobacter isolates carried a V121A substitution in the L4 protein. Nine macrolide-resistant and three macrolide-susceptible Campylobacter isolates showed the variant M192I in the L4 protein. Five Campylobacter isolates contained both substitutions in the L4 protein.
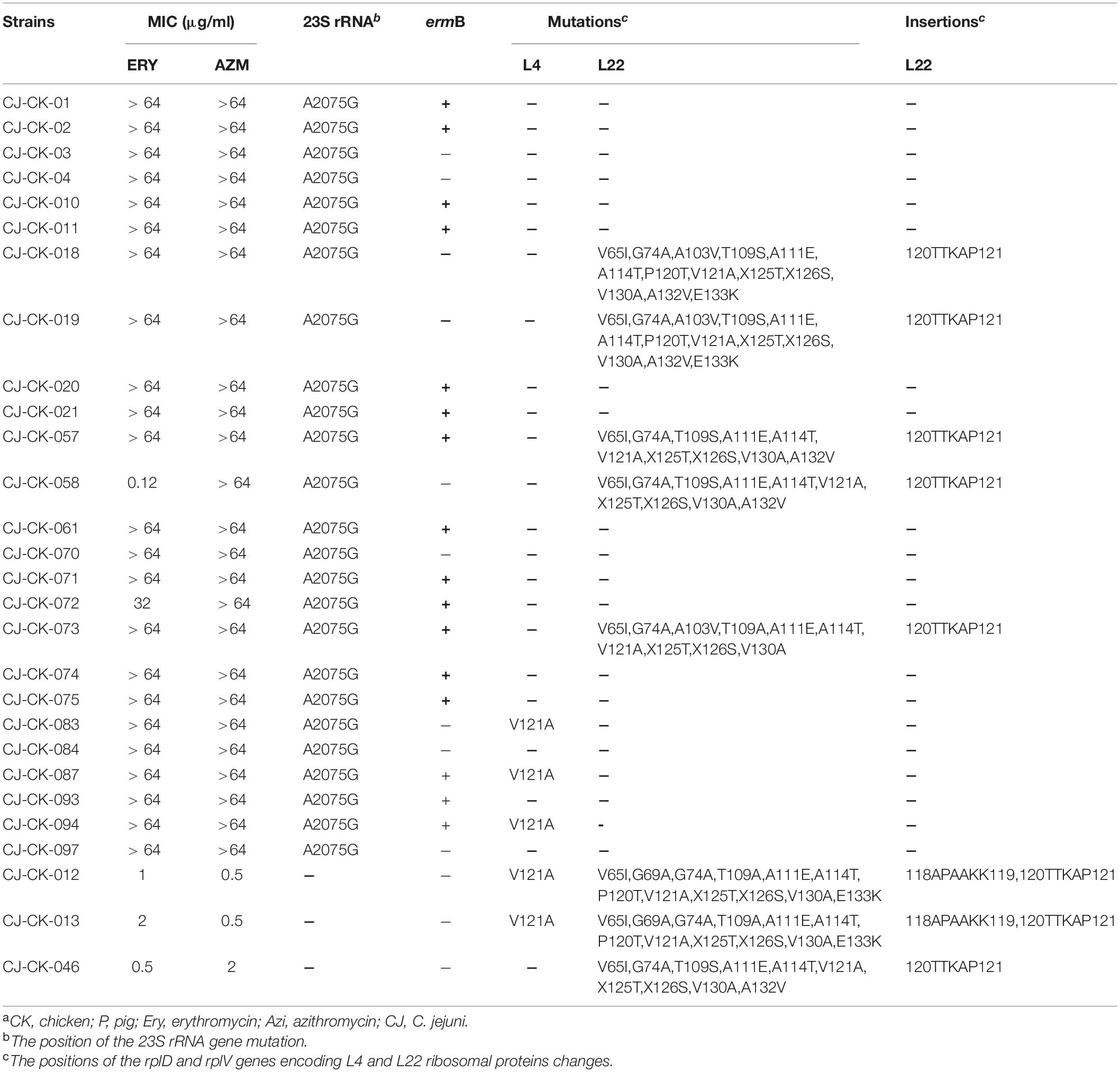
Table 5. MIC, presence of ermB gene, mutation and insertion of 23S rRNA and L4 and L22 ribosomal protein in C. jejuni strains having different resistance to azithromycin and erythromycina.
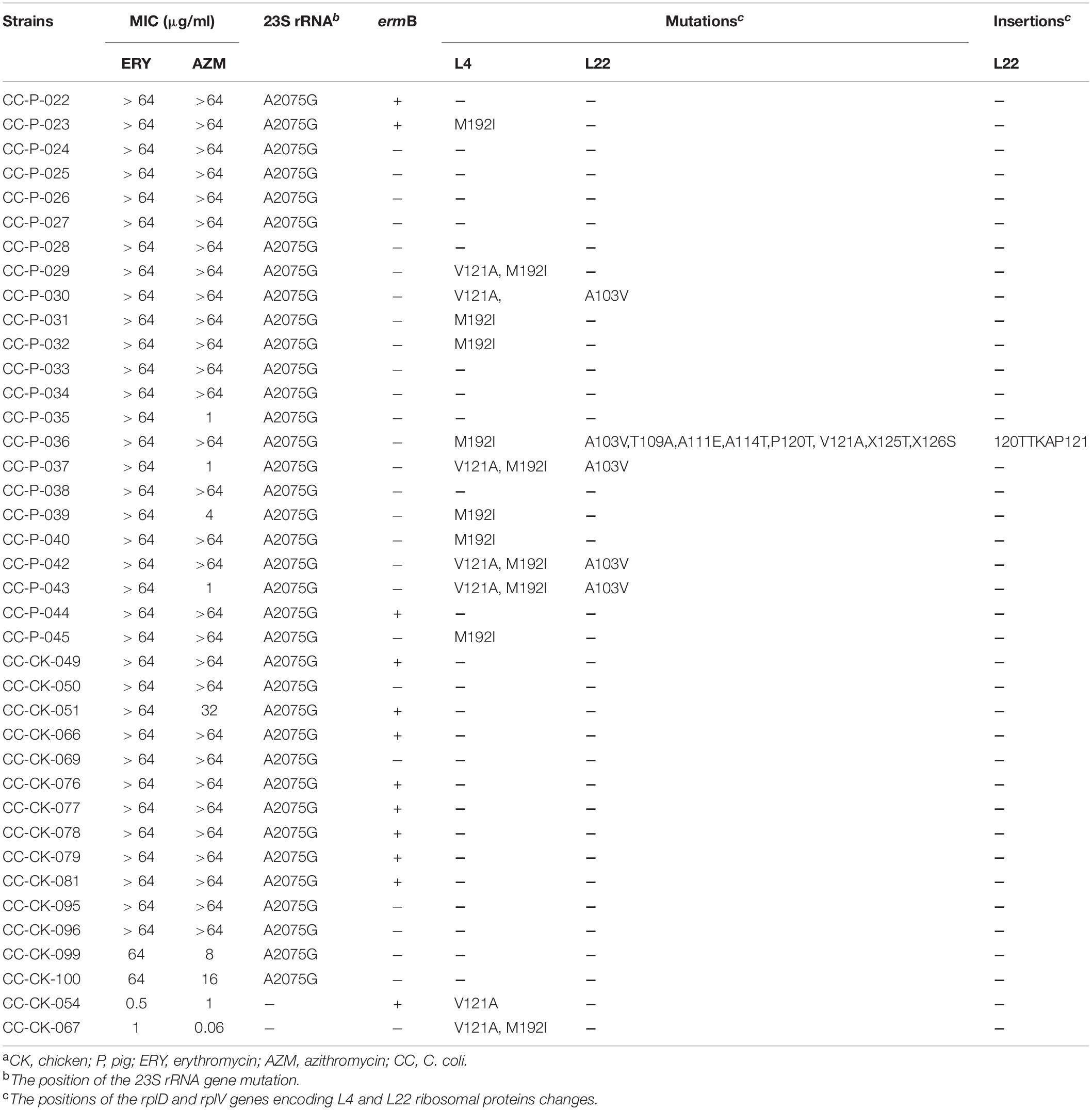
Table 6. MIC, presence of ermB gene, mutation and insertion of 23S rRNA and L4 and L22 ribosomal protein in C. coli strains having different resistance to azithromycin and erythromycina.
Fifteen amino acid substitutions in the L22 ribosomal protein were observed: V65I (n = 8), G69A (n = 2), G74A (n = 8), A103V (n = 8), T109A (n = 4), T109S (n = 5), A111E (n = 9), A114T (n = 9), P120T (n = 5), V121A (n = 9), X125T (n = 9), X126S (n = 9), V130A (n = 8), E133K (n = 4), and A132V (n = 5). The L22 protein of the tested isolates also contained two amino acid insertions (118APAAKK119 and 120TTKAP121) relative to the L22 protein of C. jejuni NCTC 11168 (Tables 5, 6).
The ermB gene was identified in 49 isolates (52.7%, 49/93): 30 C. jejuni (66.7%) and 19 C. coli (39.6%). The ermB-positive rate in the chicken-origin Campylobacter spp. isolates (65.7%, 46/70) was significantly higher than that in the Campylobacter isolates with a pig origin (13.0%, 3/23). Notably, only one C. jejuni isolate showing high level macrolide resistance did not contain the 23S rRNA mutation, but did carry the ermB gene. Twenty-seven ermB-positive isolates also contained the A2075G 23S rRNA mutation (Tables 5, 6).
Thermophilic Campylobacter are zoonotic pathogens, and antimicrobial resistance in Campylobacter has gradually become a major public health problem in both developed and developing countries (Wieczorek and Osek, 2013). In the present study, 45 C. jejuni and 25 C. coli were isolated from chickens, and 23 C. coli were isolated from pigs. The C. jejuni was the predominant Campylobacter species in chicken isolates (64.3%), followed by C. coli (35.7%), and C. coli was the dominant Campylobacter species in pigs, accounting for 100% of the pig isolates. The isolation rates of Campylobacter on pig farms was 23.0%, which was within the range of 13.4%–26.1% reported by Wang et al. (2016). In China, laying hens are used as stewed chicken. Hence, Campylobacter in layers can also pose a risk for consumers. The isolation rates of Campylobacter on layer farms (34.3%) was lower than the rate on broiler farms (57.5%). The total isolation rates of Campylobacter on chicken farms in Jiangsu Province was 46.7%, which was higher than the isolation rates in other regions of China (Wang et al., 2016). The isolation rate of Campylobacter differed in different regions, which may be attributable to the different prevalence of Campylobacter in different regions or to different animal breeding conditions, breeding scales, sample numbers, and the isolation methods used by various laboratories.
Telithromycin belongs to a class of semisynthetic macrolides, the ketolides, and has been designed to treat respiratory infections. In this study, resistance to telithromycin was highly prevalent among the Campylobacter isolates: 42.2% of C. jejuni isolates from chickens, 44.0% of C. coli isolates from chickens, and 60.9% of C. coli isolates from pigs were resistant to telithromycin. We noted a difference in telithromycin resistance between C. coli from chickens and those from pigs, but the relationship between the emergence of ketolide resistance among Campylobacter and the macrolides used in pig production is not yet clear. The resistance rate to gentamicin in the C. coli isolates from chickens (40.0%) was similar to that for C. jejuni isolates (31.1%). These findings are inconsistent with the results of Li et al. (2016), who reported that the prevalence of gentamicin resistance among C. coli isolates was 93.6%, whereas that in C. jejuni isolates was 17.9%. Overall, our results indicate that the drug resistance rates in C. coli for the antimicrobial classes tested were higher than those in C. jejuni from chickens. In recent years, MDR Campylobacter strains have been a growing global public health problem. In this study, a high prevalence of MDR Campylobacter isolates was detected (up to 64.5%). However, the MDR rate was lower than in other countries, such as Italy (100%) (Fraqueza et al., 2014). The prevalence of MDR isolates of C. coli (68%) was similar to that of C. jejuni (64.4%) in chickens, which is inconsistent with the results of Wang et al. (2016). The reason for this discrepancy may be the small number of samples examined in the present study.
High rates of fluoroquinolone resistance were observed among the 93 Campylobacter isolates. We found that the rates of fluoroquinolone resistance in C. coli (92.0% and 100%) and C. jejuni isolates (80.0% and 66.7%) from chickens were higher than the rates of resistance in C. coli isolates from pigs (65.2% and 21.7%). This was attributable to the use of fluoroquinolones as the preferred drugs against bacterial infections in poultry production, and has been caused by the unreasonable and uncontrolled use of antimicrobial agents in poultry production in China. These results are consistent with previous reports in China, in which the Campylobacter strains isolated from chickens were resistant to quinolones (Ma et al., 2014; Li et al., 2016). Therefore, the high prevalence of fluoroquinolone-resistant Campylobacter isolates in chickens may cause serious public health problems.
Quinolone resistance in Campylobacter spp. is usually caused by the point mutation T86I in the gyrase protein, and is the most frequently detected mechanism (Hormeño et al., 2016). In this study, 79 Campylobacter isolates (39 C. jejuni and 40 C. coli) with the quinolone (ciprofloxacin or nalidixic acid) resistance phenotype were positive for the T86I mutation. Other authors have reported similar results, where all ciprofloxacin-resistant Campylobacter strains carried the same mutation (Woźniak-Biel et al., 2018; Elhadidy et al., 2020).
Given the long-term use of tetracyclines in feed additives for livestock and poultry, large numbers of tetracycline-resistant isolates are found in animal reservoirs (Premarathne et al., 2017; Hafez and Attia, 2020). It has previously been reported that 95.6% of C. jejuni isolates from chickens, 97.5% of C. coli isolates from chickens and 97.5% of C. coli isolates from pigs in China were resistant to tetracycline (Wang et al., 2016). In the present study, a relatively high level of resistance to tetracycline was observed in the C. jejuni (71.1%) and C. coli (88.0%) isolates from chickens.
The tetO gene is the most commonly reported tetracycline resistance gene in Campylobacter spp. (Luangtongkum et al., 2009), and is located on the chromosome or transmissible plasmids (Abdi-Hachesoo et al., 2014; Narvaez-Bravo et al., 2017). In this study, all 64 tetracycline-resistant Campylobacter isolates (32 C. jejuni and 32 C. coli) carried the tetO gene. The high prevalence of the tetO gene reflects the high rate of tetracycline resistance in Campylobacter spp. isolated from chickens and pigs in China.
Chopra and Roberts (2001) reported that the efflux proteins encoded by tetA and tetB can export tetracycline from the cell. In this study, the tetA detection rate in the Campylobacter isolates was 6.5%, but there was no evidence of tetB. Abdi-Hachesoo et al. (2014) reported detecting the tetA resistance gene in 18% of Campylobacter spp. isolated from poultry carcasses in Iran. Nguyen et al. (2016) reported that the detection rates of tetA gene in C. jejuni and C. coli isolates from chickens in Kenya were 90.3% and 100%, respectively. Previous studies have proved that the tetA gene can be located on mobile elements, such as plasmids, and can be horizontally transferred among bacterial strains (Szczepanowski et al., 2004). Campylobacter spp. with the tetA gene may be involved in the distribution of this resistance gene to other food-borne bacteria in the animal industry.
The prevalence of Campylobacter resistance to macrolides varies according to geographic region (Rożynek et al., 2013). In the present study, erythromycin and azithromycin resistance were detected in 62.2%–100.0% and 60.0%–82.6% of Campylobacter isolates from pigs and chickens. In Korea in 2010, Lim et al. reported that 15.5% of Campylobacter isolates from chicken and swine feces or carcasses showed phenotypic resistance to erythromycin (Lim et al., 2017). Rozynek et al. reported a low rate of erythromycin resistance (4.7%), which was similar to that reported in chicken-origin isolates in other European countries (Rożynek et al., 2013). In the present study, the rates of erythromycin and azithromycin resistance were much higher in C. coli from chickens and pigs than in C. jejuni from chickens, as has been observed in previous studies (Chen et al., 2010; Wang et al., 2016). The rate of erythromycin-resistant C. coli isolates in pigs (100%) was higher than the rate in chickens (72.0%), which was much higher than has been reported in previous studies in China (Qin et al., 2011). Notably, in the present study, the erythromycin resistance rate in C. jejuni from chickens (62.2%) was higher than those reported in most previous studies undertaken in China (Chen et al., 2010; Li et al., 2016; Wang et al., 2016; Zhang et al., 2016). High levels of macrolide resistance were attributable to the widespread and increasing use of macrolides in animal production, including tylosin, tilmicosin, erythromycin, and kitasamycin.
When the macrolide-resistant isolates were screened, a range of different molecular resistance mechanisms were identified, including mutations in the 23S rRNA, rplD, and rplV genes and the presence of ermB. Our results show that the A2075G mutation in the 23S rRNA gene in the Campylobacter isolates might be the cause of their high resistance to azithromycin and erythromycin. The rate of the A2075G mutation in the present study was 98.4% (62/63) in the Campylobacter isolates with high level resistance to erythromycin, which is similar to the frequencies reported in a previous study (Luangtongkum et al., 2009). The mutation was not present in Campylobacter isolates that were susceptible to azithromycin or erythromycin or that displayed low level or intermediate resistance to these agents. No other mutations in the 23S rRNA gene were detected in this study.
Several studies have reported that ribosomal protein L4 mutations at amino acid positions 192 and 121, which conferred no significant difference between macrolide-resistant and -susceptible strains (Lehtopolku et al., 2011; Qin et al., 2011). In the present study, several mutations, including V65I, G74A, A103V, T109A, T109S, A111E, A114T, V130A, and A132V, in ribosomal protein L22 were detected in macrolide-susceptible and macrolide-resistant strains. This result is similar to a previous report by Lim et al. (2017). However, in the present study, several mutations are reported for the first time, including G69A, P120T, V121A, X125T, X126S, and E133K, which were found in both macrolide-susceptible and -resistant isolates. We noted one amino acid insertion (118APAAKK119) in the L22 ribosomal protein in nine Campylobacter isolates and another amino acid insertion (120TTKAP121) in L22 in two macrolide-susceptible C. jejuni isolates. The L22 protein of C. jejuni 81-176 also contains amino acid insertion 118APAAKK119 (Hao et al., 2013). Our results suggested that these changes were unlikely to contribute directly to macrolide resistance.
The mechanism of resistance mediated by ermB is particularly noteworthy because this gene can transfer macrolide resistance horizontally among Campylobacter strains and confer high-level resistance (Qin et al., 2014). In the present study, the detection rate of ermB was 52.7%, which was higher than those reported in other areas of China (Wang et al., 2014; Li et al., 2016). We observed that the rate of ermB-positive C. jejuni isolates (30/45) was significantly higher than that of ermB-positive C. coli isolates (19/48, P = 0.0089), which is inconsistent with previous reports (Zhang et al., 2016; Liu et al., 2017). The prevalence of the ermB gene in the Campylobacter from chickens (65.7%, n = 46) was significantly higher than in those from pigs (13.0%, n = 3, P < 0.0001). Notably, 27 (43.5%, 27/62) ermB-positive isolates also carried the A2075G mutation in 23S rRNA, which is a higher rate than in previous reports by Wang et al. (38%, 22/58) (Wang et al., 2014) and Zhang et al. (1.9%, 3/157) (Zhang et al., 2016). These results confirmed that ermB was markedly more prevalent among Campylobacter strains isolated from chickens than in those from pigs. The ermB gene must be monitored in Campylobacter because it is so highly transmittable. In general, the A2075G point mutation in domain V of the 23S rRNA and the ermB gene were the main causes of macrolide resistance in the Campylobacter isolates from Jiangsu Province, China.
In conclusion, this study has demonstrated that Campylobacter spp. isolated from chickens and pigs had high drug resistance rates to fluoroquinolones, tetracyclines and macrolides. The emergence of MDR Campylobacter strains is attributable to the widespread use of antibiotics in poultry and pig production. Monitoring antimicrobial resistance in Campylobacter and the prudent use of antimicrobials in animal-food production are essential to reducing antimicrobial resistance in bacterial pathogens.
All datasets generated for this study are included in the article/supplementary material.
The animal study was reviewed and approved by Animal Welfare and Ethical Censor Committee at Jiangsu Institute of Poultry Science. Written informed consent was obtained from the owners for the participation of their animals in this study.
QZ, JZ, XZ, and MT performed antibiotic susceptibility tests. XT and JL collected the samples. XT, MT, and SZ did PCR amplification and DNA sequence analysis. QZ conducted the statistical analysis of data. MT completed the manuscript. YG directed the study and assisted in the completion of the manuscript. All authors read and agreed to submit the manuscript.
This work was supported by the National Natural Science Foundation of China grant (31700005), the Independent Research Funding of Public Welfare Research Institutes in Jiangsu Province (BM 2018026), Yangzhou Modern Agriculture (YZ2020049).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdi-Hachesoo, B., Khoshbakht, R., Sharifiyazdi, H., Tabatabaei, M., Hosseinzadeh, S., and Asasi, K. (2014). Tetracycline resistance genes in Campylobacter jejuni and C. coli isolated from poultry carcasses. Jundishapur. J. Microbiol. 7:e12129. doi: 10.5812/jjm.12129
Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206. doi: 10.1086/319760
Altekruse, S. F., Stern, N. J., Fields, P. I., and Swerdlow, D. L. (1999). Campylobacter jejuni–an emerging foodborne pathogen. Emerg. Infect. Dis. 5, 28–35. doi: 10.3201/eid0501.990104
Cagliero, C., Mouline, C., Cloeckaert, A., and Payot, S. (2006). Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 50, 3893–3896. doi: 10.1128/AAC.00616-06
Chen, X., Naren, G. W., Wu, C. M., Wang, W., Dai, L., Xia, L. N., et al. (2010). Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet. Microbiol. 144, 133–139. doi: 10.1016/j.vetmic.2009.12.035
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Dingle, K. E., Colles, F. M., Wareing, D. R., Fox, A. J., Bolton, F. E., Bootsma, H. J., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39, 14–23. doi: 10.1128/JCM.39.1.14-23.2001
Elhadidy, M., Ali, M. M., El-Shibiny, A., Miller, W. G., Elkhatib, W. F., Botteldoorn, N., et al. (2020). Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS One 15:e0227833. doi: 10.1371/journal.pone.0227833
Engberg, J., Aarestrup, F. M., Taylor, D. E., Gerner-Smidt, P., and Nachamkin, I. (2001). Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7, 24–34. doi: 10.3201/eid0701.010104
Feizabadi, M. M., Dolatabadi, S., and Zali, M. R. (2007). Isolation and drug-resistant patterns of Campylobacter strains cultured from diarrheic children in Tehran. Jpn. J. Infect. Dis. 60, 217–219.
Florez-Cuadrado, D., Ugarte-Ruiz, M., Quesada, A., Palomo, G., Domínguez, L., and Porrero, M. C. (2016). Description of an erm(B)-carrying Campylobacter coli isolate in Europe. J. Antimicrob. Chemother. 71, 841–843. doi: 10.1093/jac/dkv383
Fraqueza, M. J., Martins, A., Borges, A. C., Fernandes, M. H., Fernandes, M. J., Vaz, Y., et al. (2014). Antimicrobial resistance among Campylobacter spp. strains isolated from different poultry production systems at slaughterhouse level. Poult. Sci. 93, 1578–1586. doi: 10.3382/ps.2013-03729
García-Fernández, A., Dionisi, A. M., Arena, S., Iglesias-Torrens, Y., Carattoli, A., et al. (2018). Human Campylobacteriosis in Italy: emergence of multi-drug resistance to ciprofloxacin, tetracycline, and erythromycin. Front. Microbiol. 9:1906. doi: 10.3389/fmicb.2018.01906
Gibreel, A., Tracz, D. M., Nonaka, L., Ngo, T. M., Connell, S. R., and Taylor, D. E. (2004). Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48, 3442–3450. doi: 10.1128/AAC.48.9.3442-3450.2004
Gill, C. O., Badoni, M., and McGinnis, J. C. (1999). Assessment of the adequacy of cleaning of equipment used for breaking beef carcasses. Int. J. Food Microbiol. 46, 1–8. doi: 10.1016/s0168-1605(98)00181-0
Hafez, M. H., and Attia, Y. A. (2020). Challenges to the poultry industry: current 453 perspectives and strategic future after the COVID-19 outbreak. Front. Vet. Sci. 7:516. doi: 10.3389/fvets.2020.00516
Hao, H., Yuan, Z., Shen, Z., Han, J., Sahin, O., Liu, P., et al. (2013). Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 57, 1369–1378. doi: 10.1128/AAC.01927-12
Haruna, M., Sasaki, Y., Murakami, M., Mori, T., Asai, T., Ito, K., et al. (2013). Prevalence and antimicrobial resistance of Campylobacter isolates from beef cattle and pigs in Japan. J. Vet. Med. Sci. 75, 625–628. doi: 10.1292/jvms.12-0432
Harvey, R. B., Young, C. R., Ziprin, R. L., Hume, M. E., Genovese, K. J., Anderson, R. C., et al. (1999). Prevalence of Campylobacter spp. isolated from the intestinal tract of pigs raised in an integrated swine production system. J. Am. Vet. Med. Assoc. 215, 1601–1604.
Hormeño, L., Palomo, G., Ugarte-Ruiz, M., Porrero, M. C., Borge, C., Vadillo, S., et al. (2016). Identification of the main quinolone resistance determinant in Campylobacter jejuni and Campylobacter coli by MAMA-DEG PCR. Diagn. Microbiol. Infect. Dis. 84, 236–239. doi: 10.1016/j.diagmicrobio.2015.11.002
Huang, J. L., Xu, H. Y., Bao, G. Y., Zhou, X. H., Ji, D. J., Zhang, G., et al. (2009). Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiol. Infect. 137, 1111–1120. doi: 10.1017/S0950268809002039
Humphrey, T., O’Brien, S., and Madsen, M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117, 237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006
Iovine, N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. doi: 10.4161/viru.23753
Jensen, L. B., and Aarestrup, F. M. (2001). Macrolide resistance in Campylobacter coli of animal origin in Denmark. Antimicrob. Agents Chemother. 45, 371–372. doi: 10.1128/aac.45.1.371-372.2001
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15
Ketley, J. M. (1997). Pathogenesis of enteric infection by Campylobacter. Microbiology 143, 5–21. doi: 10.1099/00221287-143-1-5
Lehtopolku, M., Kotilainen, P., Haanperä-Heikkinen, M., Nakari, U. M., Hänninen, M. L., Huovinen, P., et al. (2011). Ribosomal mutations as the main cause of macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 55, 5939–5941. doi: 10.1128/AAC.00314-11
Li, B., Ma, L., Li, Y., Jia, H., Wei, J., Shao, D., et al. (2016). Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in Shanghai, China. Foodborne Pathog. Dis. 14, 96–102. doi: 10.1089/fpd.2016.2186
Lim, S. K., Moon, D. C., Chae, M. H., Kim, H. J., Nam, H. M., Kim, S. R., et al. (2017). Macrolide resistance mechanisms and virulence factors in erythromycin-resistant Campylobacter species isolated from chicken and swine feces and carcasses. J. Vet. Med. Sci. 78, 1791–1795. doi: 10.1292/jvms.16-0307
Lin, J., Michel, L. O., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/aac.46.7.2124-2131.2002
Liu, D., Deng, F., Gao, Y., Yao, H., Shen, Z., Wu, C., et al. (2017). Dissemination of erm(B) and its associated multidrug-resistance genomic islands in Campylobacter from 2013 to 2015. Vet. Microbiol. 204, 20–24. doi: 10.1016/j.vetmic.2017.02.022
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Fut. Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Luangtongkum, T., Morishita, T. Y., Ison, A. J., Huang, S., McDermott, P. F., and Zhang, Q. (2006). Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72, 3600–3607. doi: 10.1128/AEM.72.5.3600-3607.2006
Ma, H., Su, Y., Ma, L., Ma, L., Li, P., Du, X., et al. (2017). Prevalence and characterization of Campylobacter jejuni isolated from retailed chicken in Tianjin, China. J. Food Prot. 80, 1032–1040. doi: 10.4315/0362-028X.JFP-16-561
Ma, L., Wang, Y., Shen, J., Zhang, Q., and Wu, C. (2014). Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181, 77–84. doi: 10.1016/j.ijfoodmicro.2014.04.023
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 6, 1–26. doi: 10.1128/microbiolspec.ARBA-0009-2017
Moore, J. E., Barton, M. D., Blair, I. S., Corcoran, D., Dooley, J. S., Fanning, S., et al. (2006). The epidemiology of antibiotic resistance in Campylobacter. Microb. Infect. 8, 1955–1966. doi: 10.1016/j.micinf.2005.12.030
Narvaez-Bravo, C., Taboada, E. N., Mutschall, S. K., and Aslam, M. (2017). Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada. Int. J. Food Microbiol. 253, 43–47. doi: 10.1016/j.ijfoodmicro.2017.04.019
Neogi, S. B., Islam, M. M., Islam, S., Akhter, A. H. M. T., Sikder, M. M. H., Yamasaki, S., et al. (2020). Risk of multi-drug resistant Campylobacter spp. and residual antimicrobials at poultry farms and live bird markets in Bangladesh. BMC Infect. Dis. 20:278. doi: 10.1186/s12879-020-05006-6
Nguyen, T. N., Hotzel, H., Njeru, J., Mwituria, J., El-Adawy, H., Tomaso, H., et al. (2016). Antimicrobial resistance of Campylobacter isolates from small scale and backyard chicken in Kenya. Gut Pathog. 8:39. doi: 10.1186/s13099-016-0121-5
Poehlsgaard, J., Andersen, N. M., Warrass, R., and Douthwaite, S. (2012). Visualizing the 16-membered ring macrolides tildipirosin and tilmicosin bound to their ribosomal site. ACS Chem. Biol. 7, 1351–1355. doi: 10.1021/cb300105p
Premarathne, J., Anuar, A. S., Thung, T. Y., Satharasinghe, D. A., Jambari, N. N., Abdul-Mutalib, N. A., et al. (2017). Prevalence and antibiotic resistance against tetracycline in Campylobacter jejuni and C. coli in cattle and beef meat from Selangor, Malaysia. Front. Microbiol. 8:2254. doi: 10.3389/fmicb.2017.02254
Qin, S., Wang, Y., Zhang, Q., Zhang, M., Deng, F., Shen, Z., et al. (2014). Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J. Antimicrob. Chemother. 69, 964–968. doi: 10.1093/jac/dkt492
Qin, S. S., Wu, C. M., Wang, Y., Jeon, B., Shen, Z. Q., Wang, Y., et al. (2011). Antimicrobial resistance in Campylobacter coli isolated from pigs in two provinces of China. Int. J. Food Microbiol. 146, 94–98. doi: 10.1016/j.ijfoodmicro.2011.01.035
Qu, M., Zhang, M., Zhang, X., Jia, L., Xu, J., Chu, Y., et al. (2019). Molecular and epidemiological analysis of a Campylobacter jejuni outbreak in China, 2018. J. Infect. Dev. Ctries 13, 1086–1094. doi: 10.3855/jidc.11408
Roberts, M. C. (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203. doi: 10.1016/j.femsle.2005.02.034
Rożynek, E., Maćkiw, E., Kamińska, W., Tomczuk, K., Antos-Bielska, M., Dzierżanowska-Fangrat, K., et al. (2013). Emergence of macrolide-resistant Campylobacter strains in chicken meat in Poland and the resistance mechanisms involved. Foodborne Pathog. Dis. 10, 655–660. doi: 10.1089/fpd.2012.1333
Sáenz, Y., Zarazaga, M., Lantero, M., Gastanares, M. J., Baquero, F., and Torres, C. (2000). Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob. Agents Chemother. 44, 267–271. doi: 10.1128/aac.44.2.267-271.2000
Sifré, E., Salha, B. A., Ducournau, A., Floch, P., Chardon, H., Mégraud, F., et al. (2015). EUCAST recommendations for antimicrobial susceptibility testing applied to the three main Campylobacter species isolated in humans. J. Microbiol. Methods 119, 206–213. doi: 10.1016/j.mimet.2015.10.018
Stanley, K., and Jones, K. (2003). Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94, (Suppl. 1), 104S–113S. doi: 10.1046/j.1365-2672.94.s1.12.x
Szczepanowski, R., Krahn, I., Linke, B., Goesmann, A., Pühler, A., and Schlüter, A. (2004). Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150(Pt 11), 3613–3630. doi: 10.1099/mic.0.27317-0
Wang, Y., Dong, Y., Deng, F., Liu, D., Yao, H., Zhang, Q., et al. (2016). Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008-14. J. Antimicrob. Chemother. 71, 666–669. doi: 10.1093/jac/dkv382
Wang, Y., Zhang, M., Deng, F., Shen, Z., Wu, C., Zhang, J., et al. (2014). Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents Chemother. 58, 5405–5412. doi: 10.1128/AAC.03039-14
Weijtens, M. J., van der Plas, J., Bijker, P. G., Urlings, H. A., Koster, D., van Logtestijn, J. G., et al. (1997). The transmission of Campylobacter in piggeries; an epidemiological study. J. Appl. Microbiol. 83, 693–698. doi: 10.1046/j.1365-2672.1997.00301.x
Wieczorek, K., and Osek, J. (2013). Antimicrobial resistance mechanisms among Campylobacter. Biomed. Res. Int. 2013:340605. doi: 10.1155/2013/340605
Woźniak-Biel, A., Bugla-Płoskońska, G., Kielsznia, A., Korzekwa, K., Tobiasz, A., Korzeniowska-Kowal, A., et al. (2018). High Prevalence of resistance to fluoroquinolones and tetracycline Campylobacter Spp. isolated from poultry in Poland. Microb. Drug Resist. 24, 314–322. doi: 10.1089/mdr.2016.0249
Zhang, A., Song, L., Liang, H., Gu, Y., Zhang, C., Liu, X., et al. (2016). Molecular subtyping and erythromycin resistance of Campylobacter in China. J. Appl. Microbiol. 121, 287–293. doi: 10.1111/jam.13135
Zirnstein, G., Li, Y., Swaminathan, B., and Angulo, F. (1999). Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37, 3276–3280. doi: 10.1128/JCM.37.10.3276-3280.1999
Keywords: Campylobacter, antimicrobial resistance, resistance mechanism, chicken, pig
Citation: Tang M, Zhou Q, Zhang X, Zhou S, Zhang J, Tang X, Lu J and Gao Y (2020) Antibiotic Resistance Profiles and Molecular Mechanisms of Campylobacter From Chicken and Pig in China. Front. Microbiol. 11:592496. doi: 10.3389/fmicb.2020.592496
Received: 07 August 2020; Accepted: 08 October 2020;
Published: 27 October 2020.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Hosny El-Adawy, Institut für bakterielle Infektionen und Zoonosen, Friedrich Loeffler Institut, GermanyCopyright © 2020 Tang, Zhou, Zhang, Zhou, Zhang, Tang, Lu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yushi Gao, Z2FveXMxMDBAc2luYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.