
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 December 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.592146
This article is part of the Research Topic New Insights into the Transmission Dynamics and Control of Antimicrobial Resistance to Last-resort Antibiotics View all 22 articles
To investigate the prevalence, probable mechanisms and serotype correlation of colistin resistance in clinical isolates of Salmonella from patients in China, Salmonella isolates were collected from fecal and blood samples of patients. In this study, 42.8% (136/318) clinical isolated Salmonella were resistant to colistin. MIC distribution for colistin at serotype level among the two most prevalent serotypes originating from humans in China indicated that Salmonella Enteritidis (83.9% resistance, 125/149) were significantly less susceptible than Salmonella Typhimurium (15.3% resistance, 9/59, P < 0.01). mcr genes and mutations in PmrAB confer little for rate of colistin resistant Salmonella isolated from human patients. Phylogenetic tree based on core-genome single nucleotide polymorphisms (SNPs) was separately by the serotypes and implied a diffused distribution of MICs in the same serotype isolates. Relatvie expression levels of colistin resistant related pmr genes were significantly higher in non-mcr colistin resistant S. Typhimurium than in colistin sensitive S. Typhimurium, but no discernable differences between colistin resistant and sensitive S. Enteritidis, indicating a different mechanism between colistin resistant S. Typhimurium and S. Enteritidis. In conclusion, colistin susceptibility and colistin resistant mechanism of clinical isolated Salmonella were closely associated with specific serotypes, at least in the two most prevalent serotype Enteritidis and Typhimurium. We suggest clinical microbiology laboratory interpreting Salmonella colistin MIC results in the serotype level.
Polymyxins, which include polymyxin B and colistin, are cyclic polypeptide antibiotics that are synthesized by members of the Paenibacillus genus (Olaitan et al., 2014). As an old class of antimicrobials, polymyxins have been increasingly revitalized as a last resort drug to combat infections caused by multidrug-resistant (MDR) and carbapenem-resistant bacteria (Poirel et al., 2017). The target of polymyxins is the outer membrane lipopolysaccharide (LPS) of Gram-negative bacteria. Binding of polymyxin to the LPS increases the permeability of the bacterial membrane, leading to leakage of the cytoplasmic content and ultimately cell death (Olaitan et al., 2014). Resistance to polymyxins can be conferred via mutations in chromosomal genes, such as genes involved in the PmrAB/PhoPQ two-component systems, which promote the expression of LPS modification related genes, such as pmrC, pmrE, and pmrHFJKLM operon (Olaitan et al., 2014; Poirel et al., 2017). Plasmid-mediated colistin resistance genes (mcr) have since discovery in 2015 been frequently observed, and their continued widespread dissemination has became a challenge to public health worldwide (Liu et al., 2016; Luo et al., 2020).
Salmonella is one of the most important causes of foodborne diarrheal disease, even bloodstream infection (Ashton et al., 2017; Mohan et al., 2019). Although colistin is not a standardized option for treating infections caused by Salmonella in humans, the increasing reports of MDR and carbapenem-resistant Salmonella, in particular strains harboring mcr-genes isolated from patients (Kock et al., 2018; Lu et al., 2019; Sun et al., 2020), indicate that these pathogens constitute a public health concern which calls more attention to the epidemic characteristics of colistin resistance. Colistin resistance in Salmonella spp. can be conferred by mutations in pmrAB, or more rarely, by the colistin resistance gene mcr (Cui et al., 2017). Interestingly, our daily experimental results indicated the rate of colistin resistance among clinical isolates of Salmonella spp. are considerably higher compared to other Enterobacteriaceae, such as Escherichia coli and Klebsiella pneumoniae (Zhang et al., 2017). The objective of this study was to investigate the prevalence and probable mechanisms of colistin resistance in clinical isolates of Salmonella in China.
A total of 318 isolates of Salmonella spp. were collected from clinical samples nationwide in China from 2014 to 2018, including 218 isolates from bloodstream infection and 100 isolates from fecal samples that were derived from diarrhea patients (Supplementary Table 1). Salmonella serotyping was conducted according to the White–Kauffmann–Le Minor scheme (9th Edition) by performing a slide agglutination test (State Serum Institute (SSI), Copenhagen, Denmark).
Cation-adjusted Mueller-Hinton Broth (Oxoid, Basingstoke, United Kingdom) dilutions were used for colistin MIC determination according to the CLSI-EUCAST joint recommendations. E. coli ATCC 25922 was used as a control. The experiment was conducted in duplicate on at least two separate occasions. The higher MIC was accepted for analysis in the duplicative test within one double dilution difference, or a third replicate would be measured (Kulengowski et al., 2019). The results were interpreted according to the EUCAST colistin breakpoint for Enterobacteriaceae (MIC > 2 mg/L, resistant).
The isolates were screened by using primers targeting mcr-1 to mcr-9 from previous studies (Luo et al., 2017; Borowiak et al., 2020). For the colistin-resistant isolates, pmrA and pmrB were amplified by primers pmrAB-F and pmrAB-R (Supplementary Table 2). The positive PCR products were sequenced with Sanger sequencing for verification. The sequences of pmrA and pmrB were compared to that of S. Typhimurium LT2 (GenBank: GCA_000006945.2), a colistin-susceptible Salmonella reference strain.
Genomic DNA of 136 randomly selected Salmonella spp. isolates (including 64 colistin resistant and 72 susceptible isolates) were extracted using Gentra Puregene Yeast/Bact. Kit (Qiaqen, Hilden, Germany). Genomes were sequenced using the Illumina HiSeq 2500-PE150 platform (Illumina, San Diego, CA, United States). Quality-trimmed raw sequence data was assembled by using SPAdes 3.13.0. Annotation was performed by uploading the data to the RAST server (rast.nmpdr.org). All the assembled genomes were deposited in NCBI, the genome accession numbers were listed in Supplementary Table 1.
A phylogenetic tree based on SNPs in the core-genome was constructed via kSNP version 3.0 (Gardner et al., 2015). iTOL (V4) was used for the display, manipulation and annotation of phylogenetic trees (Letunic and Bork, 2019). Multi-sequence typing (MLST) were identified using BacWGSTdb (Ruan and Feng, 2016). All whole-genome sequenced Salmonella genomes were analyzed by using the SeqSero 1.2 software (Zhang et al., 2015)1 for serotype prediction. The results from SeqSero were compared to the traditional Kauffman-White serotyping. When the serotype of one isolate from SeqSero 1.2 was inconsistent with the serotyping from the slide agglutination test, the serotype was determined according to the phylogenetic clusters using core-genome SNPs.
Overnight cultures of the target Salmonella isolates were diluted 1:100 and subcultured in MH medium for ∼4 h at 37°C (OD600 ∼0.6). Cells were collected at 4°C by centrifuging at 10,000 rpm for 1 min, and RNA was extracted using TRIzol Reagent (Invitrogen). DNase I-treated RNA was obtained using an RNeasy Mini Kit (QIAGEN, No. 75142), and mRNA expression levels of the target genes were examined using real-time PCR primers listed in Supplementary Table 2. qRT-PCR was performed using an ABI 7300 96-well system (Applied Biosystems) with SYBR Premix Ex Taq II (cat. no. RR820A; TaKaRa). Expression levels of target genes were normalized against the 16S rRNA gene of S. enterica using the standard curve method.
Hypothesis testing was performed by stratifying the analyses for isolates of Salmonella Enteritidis and Salmonella Typhimurium. Due to the low prevalence of isolates with other serotypes, these were excluded from the analyses. Fisher’s exact tests were used to determine if resistant and susceptible phenotypes were equally distributed in S. Enteritidis and S. Typhimurium, and among isolates from blood and intestinal samples. A Mann-Whitney test was performed to determine if MICs were equally distributed among isolates of S. Enteritidis and S. Typhimurium. For the statistical analysis of relative expression levels of pmr genes, values are presented as means ± standard deviation (SD). Rank-sum tests were performed for pair-wise comparisons of groups, and P < 0.05 (two-tailed) was considered significant.
The colistin MICs of the tested Salmonella spp. isolates ranged from 0.25 mg/L to 16 mg/L (Figure 1). The two most prevalent MICs were 8 mg/L (38.4%, 122/318) and 1 mg/L (33.3%, 106/318). 42.8% (136/318) of colistin MICs of the isolates were measured as > 2 mg/L. MIC50 and MIC90 of the isolates were 2 and 8 mg/L, respectively. 48.6% (106/218) of the bloodstream infection isolates were identified as colistin-resistant, whereas 30% (30/100) of the isolates from intestinal samples were determined as colistin-resistant.
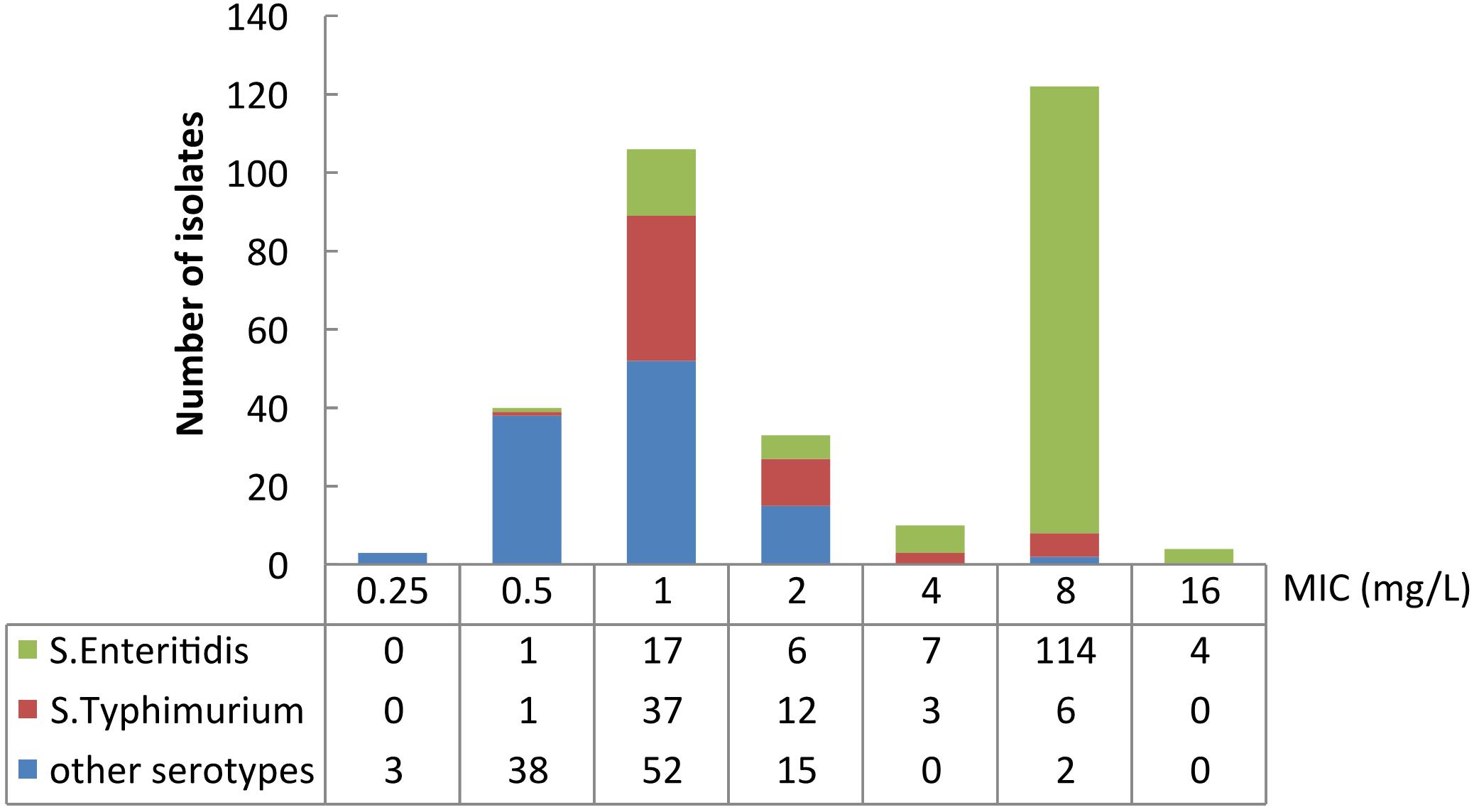
Figure 1. Number of isolates and MIC distribution of different serotypes of Salmonella. “Other serotypes” presents serotypes except for Enteritidis and Typhimurium.
Resistance mechanisms of all the colistin-resistant isolates were investigated by screening for mcr genes and sequencing of pmrA and pmrB. Out of 136 colistin-resistant isolates, two were as mcr-1 positive (S039-44712 and S040-44862), none had other mcr genes, and two had pmrAB mutations which affected the amino acid sequence (S132-54197 and S133-54550). The isolates with pmrAB mutations carried a missense point mutation in pmrA (T89S), and five missense mutations in pmrB (M15T, G73S, V74I, I83V, A111T). However, none of the mutations were predicted to affect the protein function.
18 serotypes were identified among the 318 isolates. S. Enteritidis (n = 149) and S. Typhimurium (n = 59) were found to be the overall most prevalent. S. Enteritidis (122/218) was the most common among isolates from bloodstream infection samples, followed by S. Typhimurium (12/218), whereas the inverse was true among isolates from patients with intestinal infections, with 27.0% (27/100) S. Enteritidis and 47.0% (47/100) S. Typhimurium (P < 0.01).
The rate of colistin-resistant S. Enteritidis and S. Typhimurium were 83.9% (125/149) and 15.3% (9/59), respectively. Isolates of S. Enteritidis were significantly less susceptible than isolates of S. Typhimurium (P < 0.01) and had significantly higher MICs (P < 0.01). S. Enteritidis was most prevalent among the colistin-resistant isolates, representing 91.9% (125/136) of all with the resistance phenotype, followed by S. Typhimurium (6.6%, 9/136). All colistin-resistant isolates were S. Enteritidis and S. Typhimurium, except for the two isolates (S132-54197 and S133-54550) which were identified as Salmonella Gallinarum (1/2) and Salmonella Goldcoast (1/1), respectively.
In order to discover the relation between Salmonella serotypes, phylogenetic evolution and MICs, Salmonella serotypes were confirmed and phylogenetic trees based on core-genome SNPs were generated. The whole-genome sequenced isolates were predicted serotypes using SeqSero 1.2 and were compared to the traditional Kauffman-White serotyping. Phylogenetic analysis was employed to confirm the serotyping, by using core-genome SNPs as described above. 63 S. Enteritidis, 45 S. Typhimurium and 28 isolates of other serotypes were identified from the 136 whole-genome sequenced isolates, and were 98.4% (63/64), 97.8% (45/46), and 92.9% (28/30) identical to the traditional Kauffman-White serotyping, respectively.
Phylogenetic trees based on core-genome SNPs were generated for WGS isolates of all serotypes (Figure 2), for all S. Enteritidis isolates (Figure 3) and for all S. Typhimurium isolates (Figure 4). The relation between serotypes, sequence types and MICs was inferred in the phylogenetic tree (Figure 2). The isolates clustered by serotypes and sequence types in the phylogenetic tree but not by MICs. The S. Enteritidis isolates showed little variation in their core genomes. The numbers of SNPs of serotype Enteritidis isolates ranged from 1 to 236, which was very closely in evolution, but did not constitute a single clone. Sequence type of all serotype Enteritidis were ST11 (Figure 3). Five colistin-susceptible isolates (S049, S064, S145, S146, S147) clustered together while the remaining five colistin-susceptible isolates (S148, S149, S150, S151, S152) were more diffusely distributed (Figure 3). In contrast, isolates of S. Typhimurium showed a considerable diversity in their core genomes (Figure 4). The two mcr-1 positive S. Typhimurium isolates were both monophasic variant of Typhimurium and clustered together, while the other colistin-resistant S. Typhimurium isolates were more dispersed.
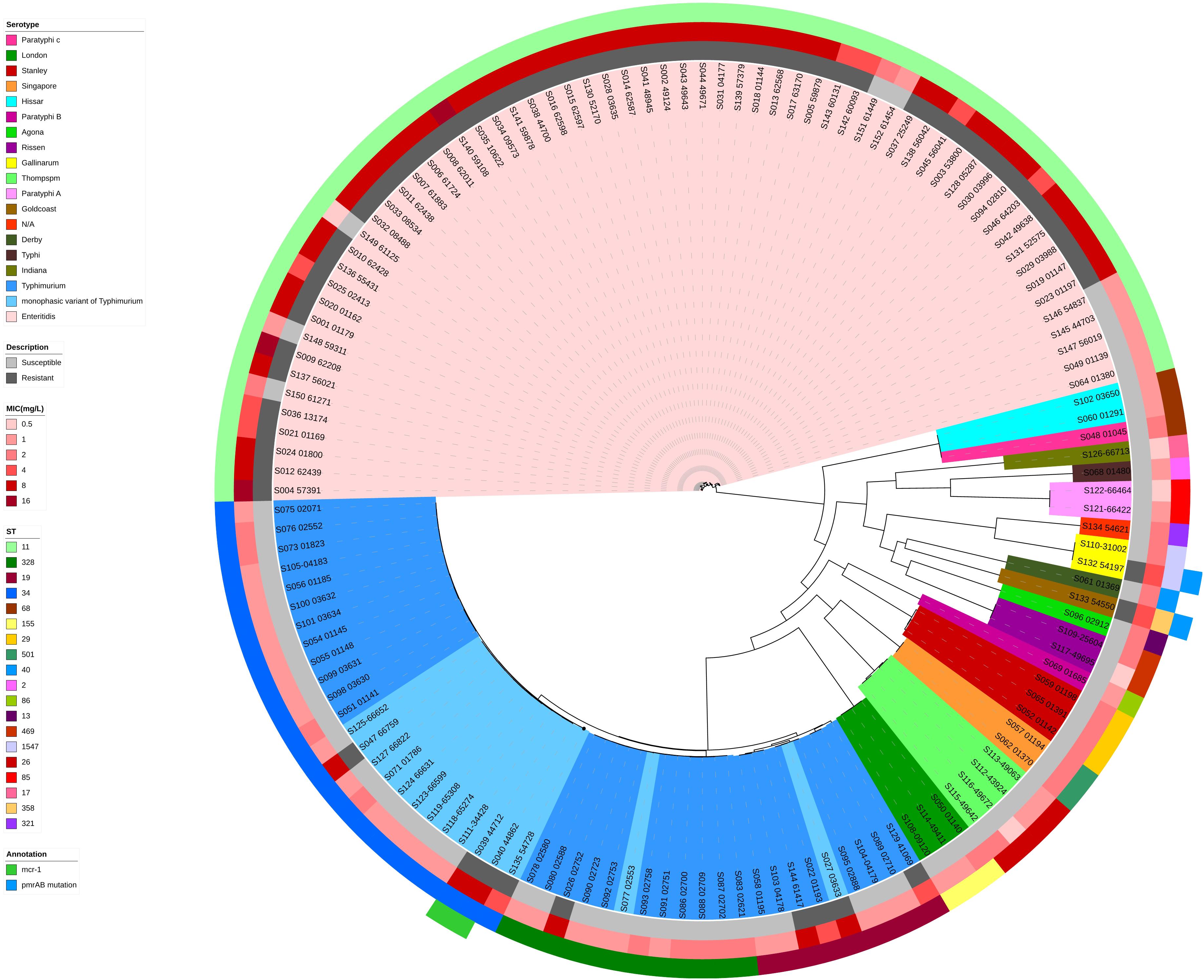
Figure 2. Maximum parsimony tree of the whole-genome sequenced 136 clinical Salmonella isolates. MICs (mg/L) and sequence types of colistin among different serotype Salmonella isolates were shown. Maximum parsimony tree were generated based on core-genome SNPs by kSNP 3.0. Isolates with different serotypes are indicated in the inner ring with different colors. Colistin MICs in mg/L are shown, along with corresponding mcr-1 gene and pmrAB mutations known to be associated with phenotypic resistance. For the description of colistin susceptibility, resistance corresponds to MIC > 2 mg/L and susceptible corresponds to MIC ≤ 2 mg/L.
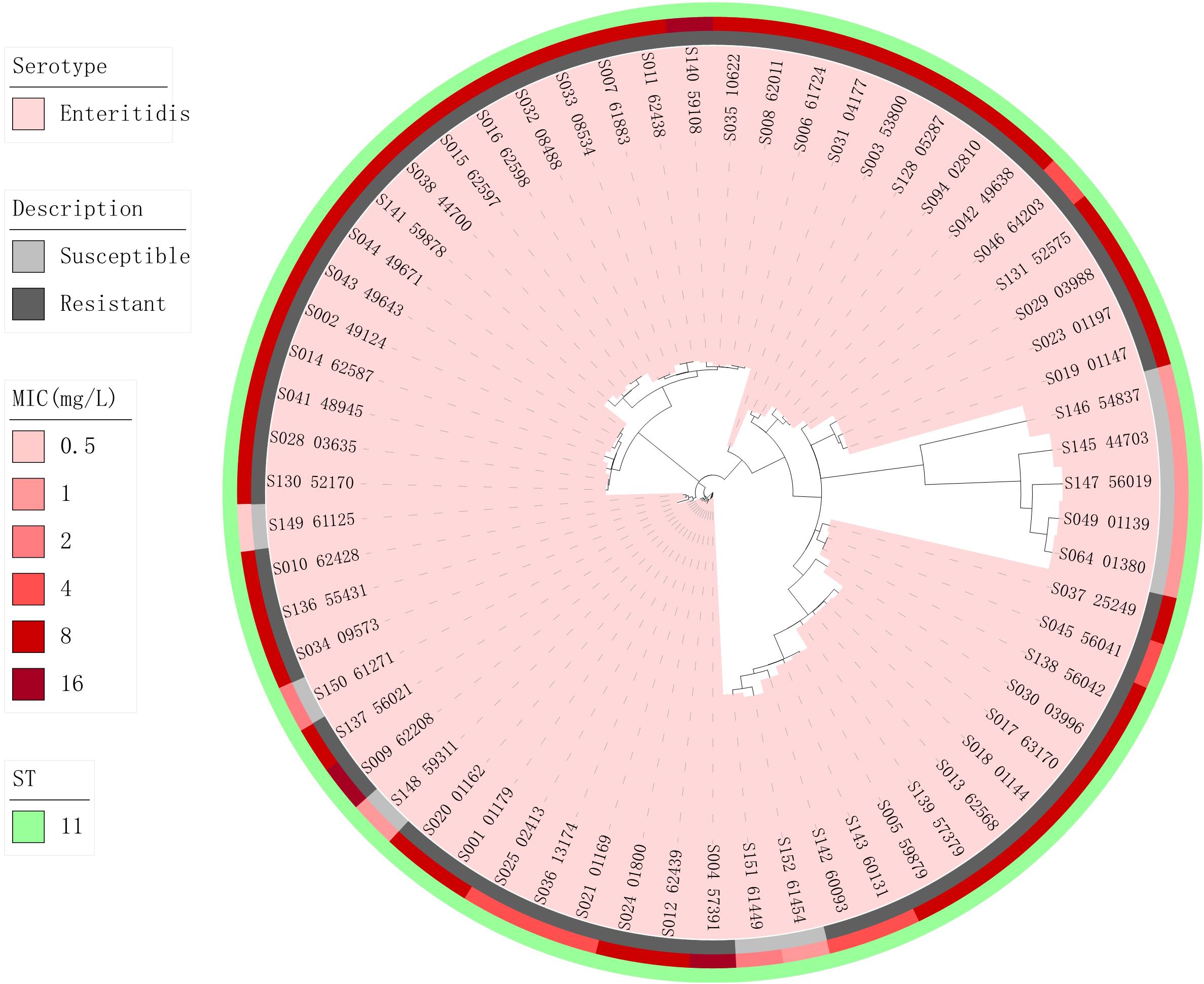
Figure 3. Zoomed-in maximum parsimony tree showing the Enteritidis serotype sub-clade of the clinical Salmonella isolates. Colistin MICs in mg/L and sequence types are shown. For the description of colistin susceptibility, resistance corresponds to MIC > 2 mg/L and susceptible corresponds to MIC ≤ 2 mg/L.
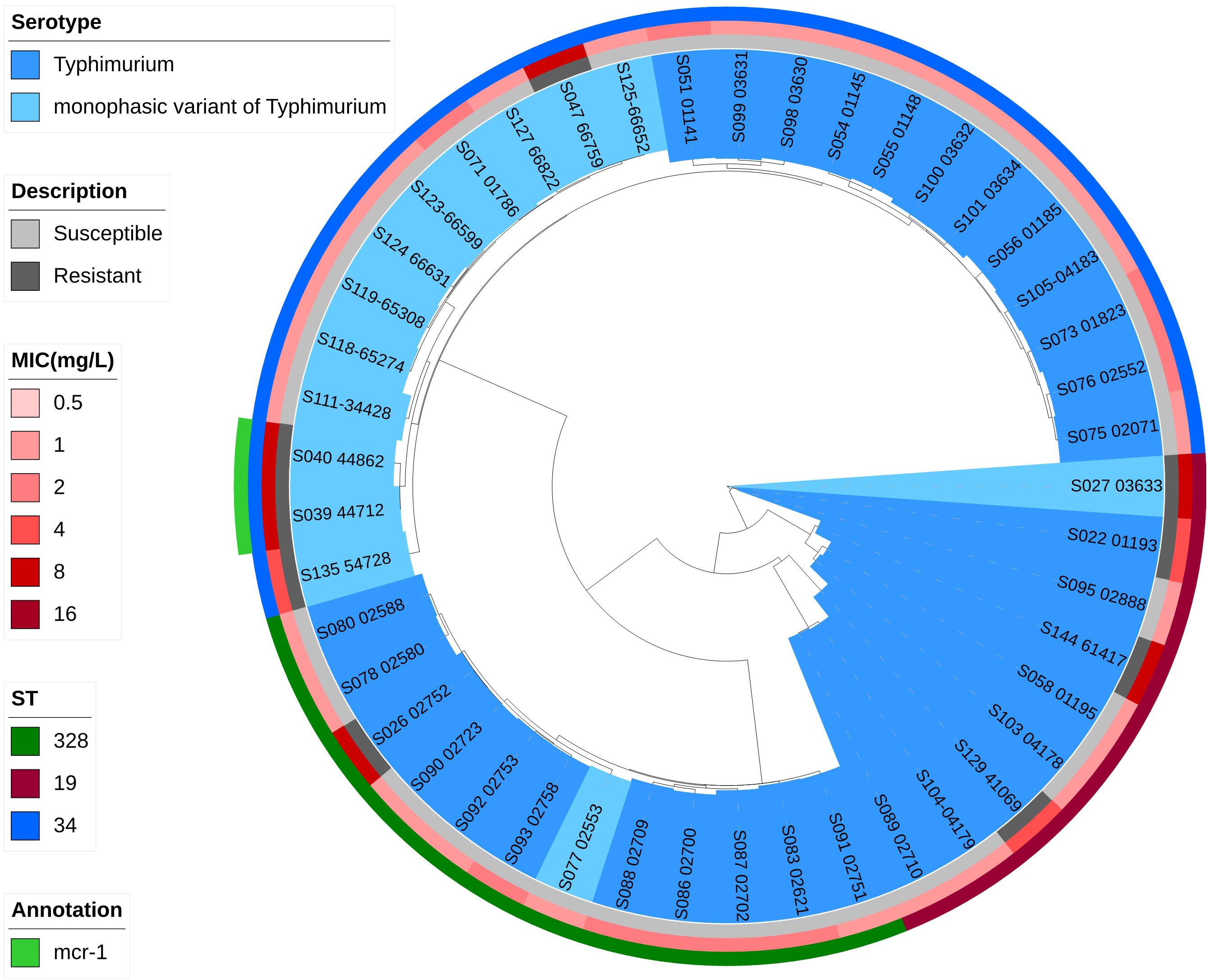
Figure 4. Zoomed-in maximum parsimony tree showing the Typhimurium and monophasic variant of Typhimurium (Salmonella 4,[5],12:i:-) serotypes sub-clade of the clinical Salmonella isolates. Colistin MICs in mg/L and sequence types are shown. For the description of colistin susceptibility, resistance corresponds to MIC > 2 mg/L and susceptible corresponds to MIC ≤ 2 mg/L.
In order to discover if there are different mechanisms between colistin resistant S. Typhimurium and S. Enteritidis, expression levels of LPS modification related pmr genes (pmrC, pmrD, pmrE, and pmrHFJKLM operon) in colistin sensitive S. Typhimurium or S. Enteritidis were compared with non-mcr colistin resistant S. Typhimurium or S. Enteritidis, respectively. Seven isolates of each serotype were colistin resistance (Supplementary Table 1) and seven were colistin sensitive (Supplementary Table 1). Relatvie expression levels of all pmr genes were enhanced in colistin resistant S. Typhimurium than in colistin sensitive S. Typhimurium (P < 0.05), but no discernable differences between colistin resistant and sensitive S. Enteritidis (Figure 5), indicating a different mechanism between colistin resistant S. Typhimurium and S. Enteritidis.
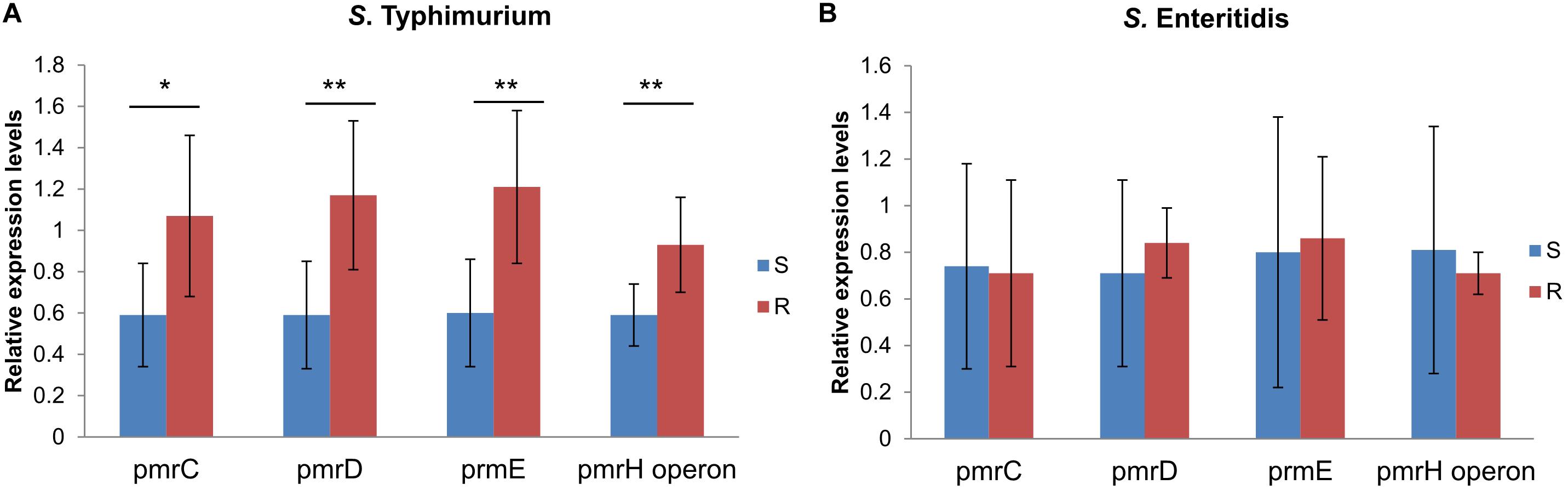
Figure 5. Relative expression levels of pmrC, pmrD, pmrE and pmrHFJKLM operon in colistin resistant and sensitive isolates of serotype Typhimurium (A) and Enteritidis (B). Gene expression levels of each target gene were identified and compared with 16s rRNA expression levels of each isolates. Relative gene expression levels of colistin sensitive (S) group and colistin resistant (R) group in serotype Typhimurium and Enteritidis were compared using p-values calculated by Statistical Package for the Social Sciences (SPSS) using rank-sum tests, respectively (*P < 0.05, **P < 0.01).
Previous studies on colistin susceptibility of Salmonella spp. utilizing surveillance data from poultry, food and human clinical sources in European countries and the United States, have indicated a correlation between Salmonella serotype and colistin MICs (Tyson et al., 2018; Alvarez et al., 2019). Though the occurrence of Salmonella serotypes vary from different countries and sources, the inhomogeneity was confirmed in the current study in which the high rate of colistin resistance of Salmonella spp. isolated from human patients from China was clearly linked to specific serotypes.
S. Enteritidis and S. Typhimurium were the predominant serotypes overall (with S. Enteritidis being the most prevalent), however, there was a sizeable and significant difference in colistin susceptibility and MICs between isolates of the two serotypes. Interestingly, the colistin resistance rate of bloodstream infection isolates was significantly higher compared to that of isolates from intestinal samples (P < 0.01). This difference was attributable to that S. Enteritidis were significantly more common among bloodstream isolates compared to the S. Typhimurium (P < 0.01). This indicates that not only is S. Enteritidis more likely to be colistin-resistant compared to S. Typhimurium, but also more frequently cause severe infections in China.
Carriage of mcr genes and missense mutations causing amino acid substitutions in PmrAB are the most commonly reported colistin resistance mechanisms in Salmonella spp. Surprisingly, in this study, only four isolates were observed with either of these resistance mechanisms, including two S. Typhimurium isolates carried mcr-1 and two isolates had missense mutations in pmrAB. According to previous studies in China, S. Typhimurium appear to more easily acquire mcr genes compared to S. Enteritidis (Li et al., 2016; Cui et al., 2017; Luo et al., 2020). This may be because S. Typhimurium in general is more susceptible to colistin than S. Enteritidis, and so tend to acquire mcr genes in order to adapt to colistin selection pressure. No pmrAB mutations were found in colistin resistant S. Enteritidis, though the high rate of colistin resistance were observed in this serotype. Relatvie expression levels of pmr genes were enhanced in colistin resistant S. Typhimurium than in colistin sensitive S. Typhimurium, but no discernable differences between colistin resistant and sensitive S. Enteritidis, which indicating a pmr related mechanism that confers to colistin resistance to non-mcr S. Typhimurium, but a pmr unrelated mechanism in colistin resistant S. Enteritidis. The large proportion of isolates with unknown resistance mechanisms in the current study indicate that other, as of yet uncharacterized, resistance mechanisms may be more important for Salmonella spp.
Aside from that unknown colistin resistance mechanisms could account for the difference in colistin resistance rates of S. Enteritidis and S. Typhimurium in this study, there could also be intrinsic differences between the serotypes which play an important role. The target of polymyxins is the outer membrane LPS of Gram-negative bacteria. These suggests that the O-antigen (surface LPS of the cell) might play a role in colistin susceptibility. Whether and how different O-antigen confer a different colistin susceptibility phenotype of Salmonella needs to be further investigation.
An epidemiological cut-off value for colistin regarding Salmonella spp. is lacking in the data presented by EUCAST; currently, only the MIC distribution of S. Dublin is presented. The results in this study indicate a very high rate of colistin resistance among S. Enteritidis in clinical isolates in China, and this could be associated to elevated wild type MICs compared to other serotypes. Further studies including isolates of S. Enteritidis from other countries and other sources are warranted to explore this possibility.
The original contributions presented in the study are publicly available. This data can be found in NCBI under accession Nos. WPIS00000000-WPNX00000000.
The studies involving human participants were reviewed and approved by the Ethics Committee of the First Affilated Hospital of Zhejiang University. The patients/participants provided their written informed consent to participate in this study.
QL and YX conceived and designed the experiments. YW, HF, XY, and YC performed the experiments. QL, BB, and BZ analyzed the data. QL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2017YFC1600100), the National Natural Science Foundation of China (81702040), the National Science Foundation of Zhejiang province, China (LY20H190002), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) (2016-00640), and the Swedish Foundation for International Cooperate in Research and Higher Education (STINT) (CH2016-6707).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Jinru Ji, Chaoqun Ying, and Yunhui Fang, for the support of technical support and quality control.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.592146/full#supplementary-material
Alvarez, J., Lopez, G., Muellner, P., de Frutos, C., Ahlstrom, C., Serrano, T., et al. (2019∗). Identifying emerging trends in antimicrobial resistance using Salmonella surveillance data in poultry in Spain. Transbound. Emerg. Dis. 67, doi: 10.1111/tbed.13346
Ashton, P. M., Owen, S. V., Kaindama, L., Rowe, W. P. M., Lane, C. R., Larkin, L., et al. (2017). Public health surveillance in the UK revolutionises our understanding of the invasive Salmonella typhimurium epidemic in Africa. Genome Med. 9:92. doi: 10.1186/s13073-017-0480-487
Borowiak, M., Baumann, B., Fischer, J., Thomas, K., Deneke, C., Hammerl, J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011-2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080
Cui, M., Zhang, J., Gu, Z., Li, R., Chan, E. W., Yan, M., et al. (2017). Prevalence and molecular characterization of mcr-1-positive Salmonella strains recovered from clinical specimens in China. Antimicrob Agents Chemother 61:e02471-16. doi: 10.1128/AAC.02471-2416
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Kock, R., Daniels-Haardt, I., Becker, K., Mellmann, A., Friedrich, A. W., Mevius, D., et al. (2018). Carbapenem-resistant enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin. Microbiol. Infect. 24, 1241–1250. doi: 10.1016/j.cmi.2018.04.004
Kulengowski, B., Ribes, J. A., and Burgess, D. S. (2019). Polymyxin B EtestR compared with gold-standard broth microdilution in carbapenem-resistant Enterobacteriaceae exhibiting a wide range of polymyxin B MICs. Clin. Microbiol. Infect. 25, 92–95. doi: 10.1016/j.cmi.2018.04.008
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, 256–259. doi: 10.1093/nar/gkz239
Li, X. P., Fang, L. X., Song, J. Q., Xia, J., Huo, W., Fang, J. T., et al. (2016). Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci. Rep. 6:38511. doi: 10.1038/srep38511
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-427
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai. China, 2006–2016. EBioMedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Luo, Q., Wan, F., Yu, X., Zheng, B., Chen, Y., Gong, C., et al. (2020). MDR Salmonella enterica serovar Typhimurium ST34 carrying mcr-1 isolated from cases of bloodstream and intestinal infection in children in China. J. Antimicrob. Chemotherapy 75, 92–95. doi: 10.1093/jac/dkz415
Luo, Q., Yu, W., Zhou, K., Guo, L., Shen, P., Lu, H., et al. (2017). Molecular epidemiology and colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front. Microbiol. 8:2262. doi: 10.3389/fmicb.2017.02262
Mohan, A., Munusamy, C., Tan, Y. C., Muthuvelu, S., Hashim, R., Chien, S. L., et al. (2019). Invasive Salmonella infections among children in Bintulu, Sarawak, Malaysian Borneo: a 6-year retrospective review. BMC Infect Dis. 19:330. doi: 10.1186/s12879-019-3963-x
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Ruan, Z., and Feng, Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44, 682–687. doi: 10.1093/nar/gkv1004
Sun, R. Y., Ke, B. X., Fang, L. X., Guo, W. Y., Li, X. P., Yu, Y., et al. (2020). Global clonal spread of mcr-3-carrying MDR ST34 Salmonella enterica serotype Typhimurium and monophasic 1,4,[5],12:i:- variants from clinical isolates. J. Antimicrob Chemother 75, 1756–1765. doi: 10.1093/jac/dkaa115
Tyson, G. H., Bodeis-Jones, S., Caidi, H., Cook, K., Dessai, U., Haro, J., et al. (2018). Proposed epidemiological cutoff values for ceftriaxone, cefepime, and colistin in Salmonella. Foodborne Pathog Dis. 15, 701–704. doi: 10.1089/fpd.2018.2490
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Keywords: Salmonella enterica, serotype, colistin susceptibility, clinical isolates, phylogenetic analysis, pmr genes
Citation: Luo Q, Wang Y, Fu H, Yu X, Zheng B, Chen Y, Berglund B and Xiao Y (2020) Serotype Is Associated With High Rate of Colistin Resistance Among Clinical Isolates of Salmonella. Front. Microbiol. 11:592146. doi: 10.3389/fmicb.2020.592146
Received: 06 August 2020; Accepted: 16 November 2020;
Published: 18 December 2020.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Xiaodong Xia, Northwest A&F University, ChinaCopyright © 2020 Luo, Wang, Fu, Yu, Zheng, Chen, Berglund and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Xiao, eGlhb3lvbmdob25nQHpqdS5lZHUuY24=; eGlhby15b25naG9uZ0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.