- 1Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation Base of Ministry of Science and Technology, Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2The State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
- 3College of Plant Protection, Nanjing Agricultural University, Nanjing, China
Rice stripe virus (RSV) causes rice stripe disease, which is one of the most serious rice diseases in eastern Asian countries. It has been shown that overexpression of RSV coat protein (CP) in rice plants enhances resistance against virus infection. However, the detailed mechanism underlying RSV CP-mediated virus resistance remains to be determined. In this study, we show that both translatable and non-translatable RSV CP transgenic Arabidopsis plants exhibited immunity to virus infection. By using deep sequencing analysis, transgene-derived small interfering RNAs (t-siRNAs) from non-translatable CP transgenic plants and virus-derived small interfering RNAs (vsiRNAs) mapping in the CP region from RSV-infected wild-type plants showed similar sequence distribution patterns, except for a significant increase in the abundance of t-siRNA reads compared with that of CP-derived vsiRNAs. To further test the correlation of t-siRNAs with RSV immunity, we developed RSV CP transgenic Arabidopsis plants in an siRNA-deficient dcl2/3/4 mutant background, and these CP transgenic plants showed the same sensitivity to RSV infection as non-transgenic plants. Together, our data indicate that the expression of RSV CP protein from a transgene is not a prerequisite for virus resistance and RSV CP-mediated resistance is mostly associated with the RNA silencing mechanism in Arabidopsis plants.
Introduction
RNA silencing is a conserved antiviral defense mechanism that has been used to increase resistance to plant virus infections (Pumplin and Voinnet, 2013; Guo et al., 2019). In antiviral RNA silencing, host Dicer-like ribonucleases (DCLs) cleave viral double-stranded RNAs (dsRNAs) that are formed during virus replication and transcription into 21-24-nucleotide (nt) viral small interfering RNAs (vsiRNAs) (Pumplin and Voinnet, 2013; Guo et al., 2019). Secondary vsiRNAs can be produced by dicing secondary viral dsRNA synthesized by host-encoded RNA-dependent RNA polymerases (RdRps) (Wassenegger and Krczal, 2006; Zhang et al., 2015). These vsiRNAs are mainly loaded into the Argonaute (Ago) protein-containing RNA-induced silencing complex (RISC) to posttranscriptionally repress target viral and host RNAs by sequence complementarity (Carbonell and Carrington, 2015; Fang and Qi, 2016). The Arabidopsis thaliana genome encodes four DCLs, among which DCL1 produces microRNA, whereas DCL2, DCL3, and DCL4 produces 22-, 24-, and 21-nt vsiRNAs, respectively (Guo et al., 2019). DCL4 and DCL2 are the primary components of antiviral defense in plants and exhibit redundant or cooperative functions (Diaz-Pendon et al., 2007; Guo et al., 2019). The 21-nt vsiRNA produced by DCL4 is the most dominant species of vsiRNA in RNA virus-infected plants (Deleris et al., 2006; Wang et al., 2010). DCL2-dependent 22-nt vsiRNAs also accumulate abundantly when DCL4 is absent or suppressed by viruses, and these 22-nt vsiRNAs seem to be less effective at mediating antiviral RNA silencing (Wang et al., 2011). DCL3 produces 24-nt siRNAs that are associated with transcriptional repression of transposons and repeat sequences in plants (Blevins et al., 2015; Singh et al., 2019) and may enhance antiviral defense against DNA viruses (Raja et al., 2008; Jackel et al., 2016). In Arabidopsis plants, AGO1 and AGO2 are the two major plant antiviral argonautes against RNA viruses by associating with vsiRNAs based on the identity of the 5′ terminal nucleotide (Mi et al., 2008; Carbonell and Carrington, 2015).
Rice stripe virus (RSV) is the type species in the genus Tenuivirus and causes rice stripe disease, which is one of the most serious rice diseases in eastern Asia (Falk and Tsai, 1998; Xiong et al., 2008). It is transmitted by the small brown planthopper (SBPH) (Laodelphax striatellus Fallén) in a circulative and propagative manner (Falk and Tsai, 1998; Li et al., 2015). Under experimental control, RSV also infects Nicotiana benthamiana by mechanical sap inoculation and Arabidopsis plants by viruliferous SBPH inoculation (Xiong et al., 2008; Sun et al., 2011, 2016). The genome of RSV contains four single-stranded RNA segments, RNA1, RNA2, RNA3, and RNA4, in order of their decreasing molecular weights (Toriyama and Watanabe, 1989). RNA1 is negative sense and encodes a putative viral RdRp (Barbier et al., 1992). The other small three RNA segments encode two proteins using an ambisense coding strategy. RNA2 encodes NS2, a viral RNA silencing suppressor (Du et al., 2011), and NSvc2, a putative membrane glycoprotein (Yao et al., 2014). RNA3 encodes NS3, another RNA silencing suppressor (Xiong et al., 2009) and a coat protein (CP). SP (a disease-specific protein) (Kong et al., 2014) and NSvc4 (a movement protein) (Xiong et al., 2008) are encoded by RNA4.
To manage rice stripe disease caused by RSV, the use of insecticides against SBPH vectors and the exploitation of genetically resistant cultivars are the most effective approaches; however, they all have concerning features, such as high costs, environmental pollution, insecticide resistance and limited resistance resources (Otuka, 2013). Recently, RNA silencing-based resistance has been shown to be an effective and powerful method to enhance plant resistance against viruses in transgenic plants (Galvez et al., 2014; Lindbo and Falk, 2017). Plants transformed with a hairpin construct consisting of an inverted repeat virus-specific sequence are able to induce RNA silencing and exhibit immunity to virus infection (Lindbo and Falk, 2017). For RSV, transgenic rice plants with the RSV CP or NSvc4 hairpin constructs exhibit near immunity against RSV infection (Shimizu et al., 2011; Sasaya et al., 2014). Chimeric CP/SP RNA silencing construct transgenic rice plants show strong resistance against two different RSV isolates (Ma et al., 2011). In addition to hairpin constructs, plant expression of the viral CP gene also confers plant viral resistance, which is mostly but not completely due to an RNA silencing-mediated resistance mechanism (Lindbo and Falk, 2017). It has been shown that overexpression of the RSV CP gene in transgenic rice plants enhances resistance against the virus (Hayakawa et al., 1992). Recent research has also shown that RSV CP mediates virus resistance through jasmonate-AGO18 signaling in rice plants (Han et al., 2020; Yang et al., 2020). However, considering that AGO18 is the key component of RNA silencing in rice plants, whether RSV CP-mediated virus resistance involves the RNA silencing pathway remains to be addressed.
In this study, using the RSV-Arabidopsis pathosystem, we showed that both translatable and non-translatable RSV CP transgenic Arabidopsis plants exhibit immunity to RSV infection. By using deep sequencing analysis and comparing transgenic-derived siRNAs (t-siRNAs) from non-translatable versions of CP transgenic plants and CP-derived virus-derived siRNAs (vsiRNAs) from RSV-infected wild-type plants, we found that siRNA mapped to the CP sequence segment showed a similar pattern in both transgenic plants and virus-infected wild-type plants, except that greater accumulation of siRNAs was seen in transgenic immune plants. In the siRNA-deficient dcl2/3/4 mutant background, CP transgenic plants showed the same RSV susceptibility as non-transgenic plants. Together, our collective results indicated that RSV CP-mediated virus resistance depends on the function of DCLs and mostly involved in RNA silencing in Arabidopsis plants.
Materials and Methods
Sources of Virus, Vectors, and Plant Materials
Rice stripe virus-infected rice plants were collected from Jiangsu Province in China, and the virus was confirmed by using a western blot assay (Sun et al., 2016). Non-viruliferous instar nymphs of SBPHs reared on rice seedlings (Oryza sativa L. cv. Wuyujing No. 3) were collected and fed on RSV-infected rice plants for 3 days to acquire the virus. The virus was maintained by successive transovarial infections in SBPHs on rice seedlings in an insect-rearing room at 25°C.
Arabidopsis thaliana (Col-0) and dcl2/3/4 mutant seeds were obtained from Dr. Xiuren Zhang (Texas A&M University, College Station, United States) and grown in potting soil in a growth chamber at 23°C under 200 μmol m–2 s–1 illumination and 16-h light/8-h dark cycle conditions.
RSV Inoculation Assay
Transgenic and wild-type Arabidopsis plants at 2 weeks were inoculated with 10 viruliferous SBPHs per plant and were kept in a glass incubator containing ten plants. SBPHs were sprayed with insecticide after a 4-day inoculation access period. Plants were kept in a growth chamber for symptom development, and non-viruliferous insects were used for mock inoculation. To evaluate of resistance to RSV, thirty Arabidopsis plants of each line were inoculated with RSV. The seedlings were harvested at different time points for RNA and protein extractions. Each RSV inoculation assay was repeated three times.
DNA Constructs and Transgenic Plants
The RSV CP gene was amplified from RSV-infected rice plant leaf tissue using RT-PCR performed with CP-specific primers (Supplementary Table S1). The CP PCR products were cloned into the pDONR-zero vector (Invitrogen, Carlsbad, CA, United States) and subsequently transferred into the binary vector pBA-Flag-Myc4-DC (Zhang et al., 2005) using the Gateway cloning system following the manufacturer’s instructions (Invitrogen Corporation). The constructed binary vector pBA-Flag-Myc4-CP was transformed into Arabidopsis plants using Agrobacterium tumefaciens strain ABI by the floral dip method (Clough and Bent, 1998; Zhang et al., 2006). The CP overexpression transgenic Arabidopsis plants were selected on standard MS medium containing 10 mg/L glufosinate ammonium (Sigma-Aldrich, St. Louis, MO, United States).
Western Blot Assay
Western blot was performed following the protocol described by Sun et al. (2016). In brief, Arabidopsis total proteins were separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked and inoculated with an anti-RSV SP (our laboratory), anti-Myc (Sigma-Aldrich, St. Louis, MO, United States), anti-YFP (Genscript, Nanjing, China) or anti-actin antibody (Enogene, Nanjing, China) overnight at 4°C. Signals were developed in ECL buffer (Transgen Biotech, Beijing, China) and recorded with a Tanon 5200 Luminescent Imaging Workstation (Tanon, Shanghai, China).
RT-PCR
Total RNA was isolated from the plant samples using RNAiso Plus reagent (Takara, Dalian, China), and first-strand cDNA was synthesized from 1 μg of total RNA using an iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA, United States). PCR amplification was performed using 2 × Taq Master Mix (Vazyme Biotech, Nanjing, China) with RSV CP-specific primers (Supplementary Table S1). EF1a was used as a loading control.
Quantitative Reverse-Transcription PCR
Total RNA was isolated from the plant samples using RNAiso Plus reagent (Takara, Dalian, China), and cDNA synthesis and PCR were performed as described previously (Sun et al., 2016). Briefly, first-strand cDNA was synthesized from 1 μg of total RNA using an iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA, United States). qRT-PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, United States) with a Bio-Rad iQ5 Real-Time PCR system with gene-specific primers (Supplementary Table S1). EF1a was used as an internal standard, and all qRT-PCR experiments were performed at least three times.
Deep Sequencing and Analysis of Small RNA Sequences
Total RNA was extracted from RSV-infected and mock-treated NOP-CP transgenic or non-transgenic plants each with triplicate biological replicates, using RNAiso Plus reagent (Takara, Dalian, China), and used for small RNA library construction. A total of 12 small RNA libraries were sequenced using an Illumina HiSeq 4000 sequencer to generate single-end 50-bp sequences at the Beijing Genome Institute (BGI, Shenzhen, China). Raw data were obtained after removing the adapter sequences using cutadapt (v1.16) (Martin, 2011). The clean data were then mapped to the RSV genome (NCBI accession numbers: NC_003755.1, NC_003754.1, NC_003776.1, NC_003753.1), allowing up to one mismatch. Reads showing matches with the RSV reference sequences were retained and further analyzed using the CLC Genomics Workbench program (QIAGEN, Hilden, Germany), Perl scripts and Excel tools. Small RNA-seq data was deposited in the NCBI BioProject database with accession code PRJNA649376.
Results
RSV CP Transgenic Arabidopsis Plants Show Immunity to RSV Infection
Previous studies demonstrated that transgenic rice plants expressing RSV CP exhibited a significant level of resistance to virus infection (Hayakawa et al., 1992; Yang et al., 2020). Our previous studies also showed that RSV infected Arabidopsis plants and caused significant symptoms, including stunted growth and vein chlorosis on leaves (Sun et al., 2011, 2016). To test whether RSV CP-mediated resistance can be extended to Arabidopsis plants, we first generated CP transgenic Arabidopsis plants. Full-length RSV CP was cloned from virus-infected rice plants and transferred into the binary vector pBA-Flag-Myc4-DC (Zhang et al., 2005) using the Gateway cloning system (Figure 1B). This recombinant vector was transformed into Arabidopsis plants (Col-0 ecotype) by the floral dip method (Clough and Bent, 1998; Zhang et al., 2006). These T2 generation transgenic plants overexpressing RSV CP with Flag-Myc4 epitopes were examined by western blot assay. The expression of Flag-Myc4-CP was detected only in transgenic plants with an anti-Myc antibody (Figure 1B).
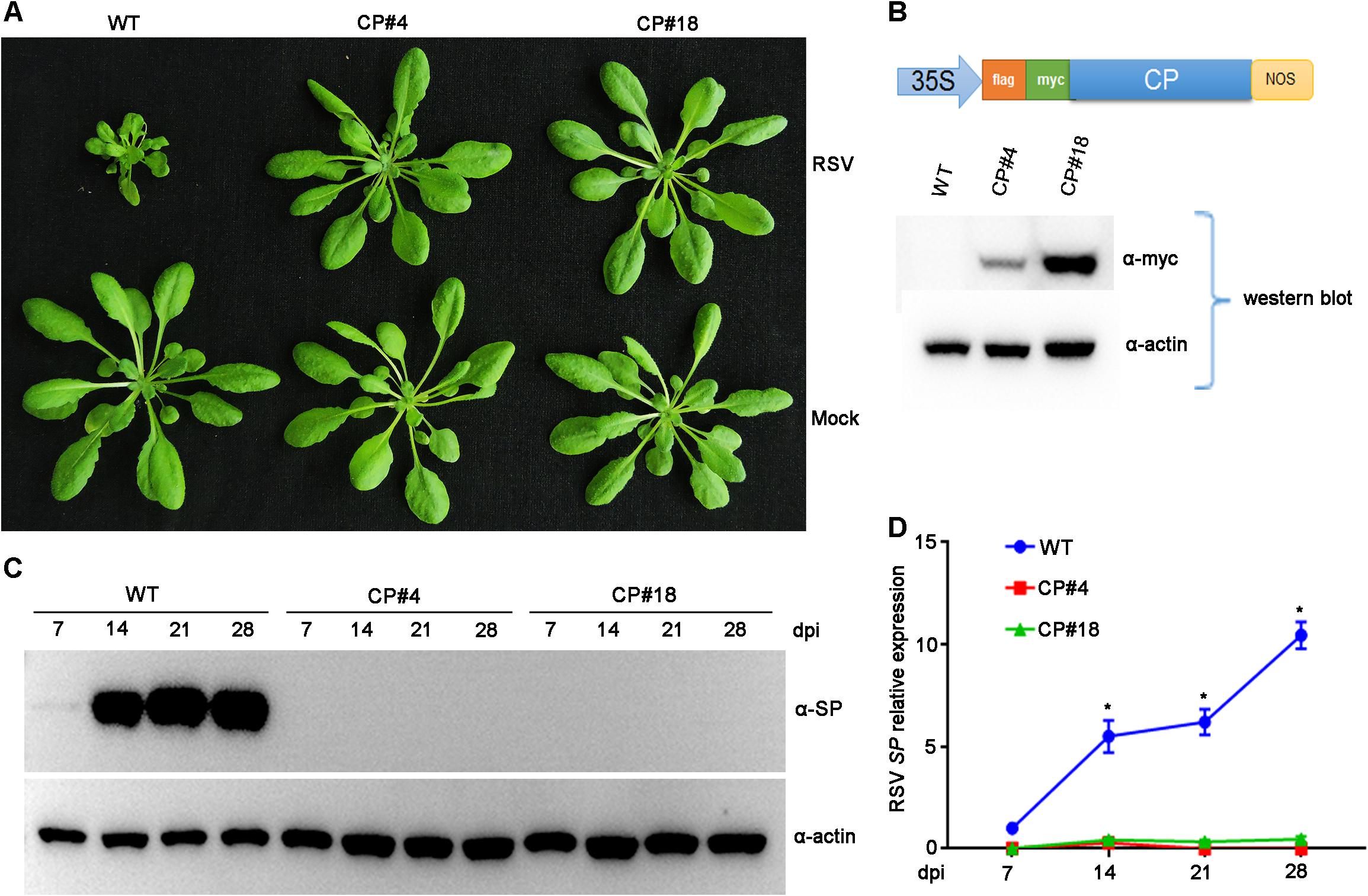
Figure 1. Examination of the Rice stripe virus (RSV) resistance of CP transgenic Arabidopsis plants. (A) Symptoms of mock-inoculated and RSV-infected CP transgenic (CP#4, CP#18) and wild-type (WT) Arabidopsis plants. Photographs were taken at 4 weeks post-inoculation. (B) The upper panel shows the schematic representation of the constructed binary vector pBA-Flag-Myc4-CP, and the lower panel shows western blot analysis of CP transgenic lines with an anti-Myc antibody. The actin protein level served as a loading control. (C) Western blot analysis of the time course of RSV-encoded SP protein accumulation in RSV-infected CP transgenic (CP#4, CP#18) and WT Arabidopsis plants using an SP-specific antibody. The actin protein level served as a loading control. (D) qRT-PCR analysis of the time course of RSV SP mRNA transcription levels in RSV-infected CP transgenic and WT Arabidopsis plants. Signal intensities for each transcript were normalized with those for EF1-α. Values are means ± SD (n = 3). *p ≤ 0.05 (Student’s t-test).
To test RSV resistance, both CP transgenic (CP#4, CP#18) and wild-type (Col-0 ecotype) Arabidopsis plants were challenged with RSV by viruliferous SBPHs at 2-week-old stages. In three independent experiments, all wild-type plants showed typical RSV symptoms, including severe stunting at 4 weeks post-inoculation. In contrast, all the CP transgenic Arabidopsis plants did not show any symptoms and displayed resistance to RSV infection (Figure 1A). Moreover, RSV-encoded SP protein was detected in wild-type Arabidopsis plants by western blot assay at 14 to 28 days post-inoculation (dpi) but not in CP transgenic plants (Figure 1C). qRT-PCR assays showed that RSV SP mRNA accumulated to significantly higher levels in wild-type plants than in CP transgenic plants (Figure 1D). These results therefore show that RSV CP induces virus resistance in Arabidopsis plants.
Expression of CP Protein From a Transgene Is Not a Prerequisite for Resistance in Arabidopsis Plants
Previous studies demonstrated that the mechanism of CP-mediated resistance depended on the particular virus system and transgenic plants being analyzed (Lindbo and Falk, 2017). To demonstrate whether expression of RSV CP protein is necessary for resistance, we generated Arabidopsis plants overexpressing a non-translatable protein mutant of RSV CP (NOP-CP) in which the first codon ATG of the CP gene was replaced with TAG (Figure 2B), and the NOP-CP sequence was inserted into the pBA-Flag-Myc4-DC vector and tagged with Flag-Myc4 epitopes. These T2 generation transgenic plants were confirmed by RT-PCR and western blot assays. As expected, transcripts of NOP-CP mRNA were detected with primers specific to the CP sequence only in transgenic Arabidopsis plants (Figure 2B). However, signals of NOP-CP protein were not visible in either transgenic or wild-type plants with an anti-Myc antibody. These results indicate that NOP-CP transgenic Arabidopsis plants expressed only CP mRNA but not protein. To further analyze whether there was still a truncated RSV CP protein encoding C-terminal truncation in the corresponding region of CP nucleotides in NOP-CP gene. YFP-tagged CP or NOP-CP in C-terminal were transiently expressed in Nicotiana benthamiana. The transcripts of NOP-CP-YFP mRNA were detected in Nicotiana benthamiana (Supplementary Figure S1B). However, signals of NOP-CP-YFP protein were not visible in tobacco plants using confocal imaging and western blot assays (Supplementary Figures S1A,C). These data indicated that NOP-CP-YFP gene expressed only CP mRNA but not protein in Nicotiana benthamiana.
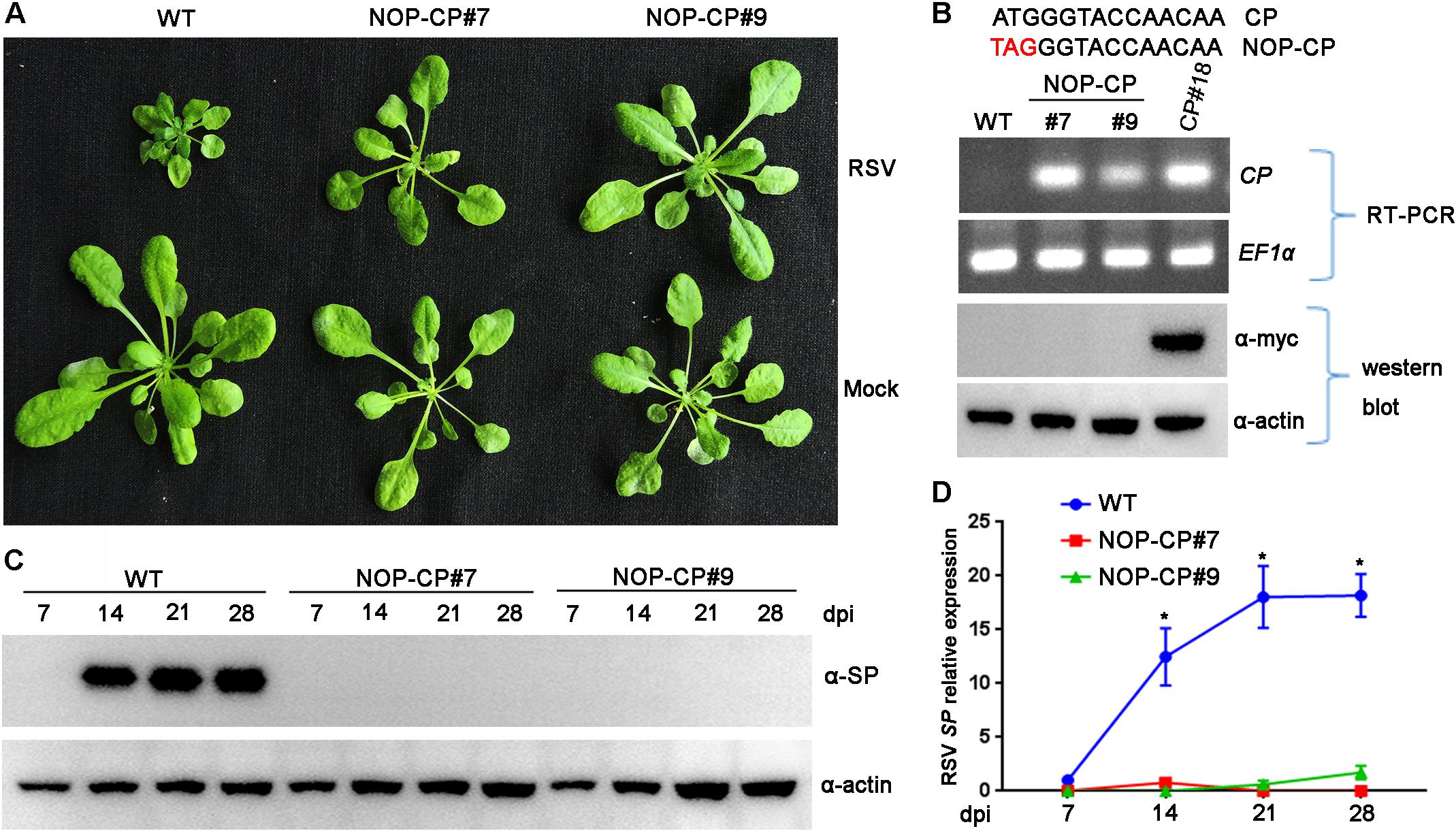
Figure 2. Evaluation of NOP-CP transgenic Arabidopsis plants for resistance to Rice stripe virus (RSV). (A) Symptoms of mock-inoculated and RSV-infected NOP-CP transgenic (NOP-CP#7, NOP-CP#9) and wild-type (WT) Arabidopsis plants. Photographs were taken at 3 weeks post-inoculation. (B) The upper panel shows the schematic representation of the NOP-CP nucleotide sequence. The middle panel shows RT-PCR analysis of NOP-CP mRNA transcription levels of NOP-CP transgenic lines, and the EF1-α mRNA level served as a loading control. The lower panel shows western blot analysis of NOP-CP transgenic lines with an anti-Myc antibody, and the actin protein level served as a loading control. (C) Western blot analysis of the time course of RSV-encoded SP protein accumulation in RSV-infected NOP-CP transgenic (NOP-CP#7, NOP-CP#9) and WT Arabidopsis plants using an SP-specific antibody. The actin protein level served as a loading control. (D) qRT-PCR analysis of the time course of RSV SP mRNA transcription levels in RSV-infected NOP-CP transgenic and Col-0 Arabidopsis plants. Signal intensities for each transcript were normalized with those for EF1-α. Values are means ± SD (n = 3). *p ≤ 0.05 (Student’s t-test).
To test RSV resistance, NOP-CP transgenic (NOP-CP#7, NOP-CP#9) and wild-type Arabidopsis plants were inoculated with RSV viruliferous SBPHs. In three independent trials, no RSV symptoms were observed in the NOP-CP transgenic Arabidopsis plants (Figure 2A), in contrast with the severe stunting symptoms in wild-type plants. Consistently, the accumulation of viral SP protein was not detected in the NOP-CP transgenic plants by western blot (Figure 2C), and fewer SP mRNA transcripts were detected in NOP-CP transgenic plants than in wild-type plants (Figure 2D). These results show that Arabidopsis transgenic plants with a non-translatable RSV CP sequence are immune to virus infection and that RSV CP-mediated resistance is independent of CP protein expression.
Deep Sequencing of Small RNA From NOP-CP Transgenic and Wild-Type Arabidopsis Plants
To better resolve whether the small RNA population is associated with RSV CP-mediated resistance and minimize the impact of CP protein on the immune response, 12 libraries were constructed with small RNA isolated from mock- and RSV-infected NOP-CP transgenic and wild-type Arabidopsis plants. Each library generated 7–15 million clean reads ranging from 18 to 30 nt (Table 1). Three biological replicates of each group were analyzed and showed very similar patterns. The results described below were derived from the average generated from three biological replicates. The size distributions of total small RNAs within these different treatments were similar; the dominant size was 22 nt, followed by 21 nt and 23 nt (Figure 3A). We mapped clean small RNA (18–30 nt) reads to the RSV genome and CP transgenic sequence and obtained RSV-derived, CP-derived siRNAs. In RSV-infected wild-type Arabidopsis plants, we identified 160,436 vsiRNAs, accounting for 1.55% of the total, whereas only 1,481 reads matched to the RSV genome in mock control plants. In NOP-CP transgenic Arabidopsis plants, almost all reads matched to the RSV genome were generated from the NOP-CP segment sequence, accounting for 0.09% of the total reads in RSV-infected plants and 0.27% in mock plants (Table 1). Unexpectedly, in NOP-CP transgenic Arabidopsis plants, RSV infection resulted in less accumulation of NOP-CP-derived small RNA than in mock plants despite generating more small RNA reads mapped to other RSV genomic regions (Table 1). The sequencing results confirmed that the accumulation of NOP-CP-derived siRNAs from transgenic sequences was associated with the immunity of these plants to RSV infection.
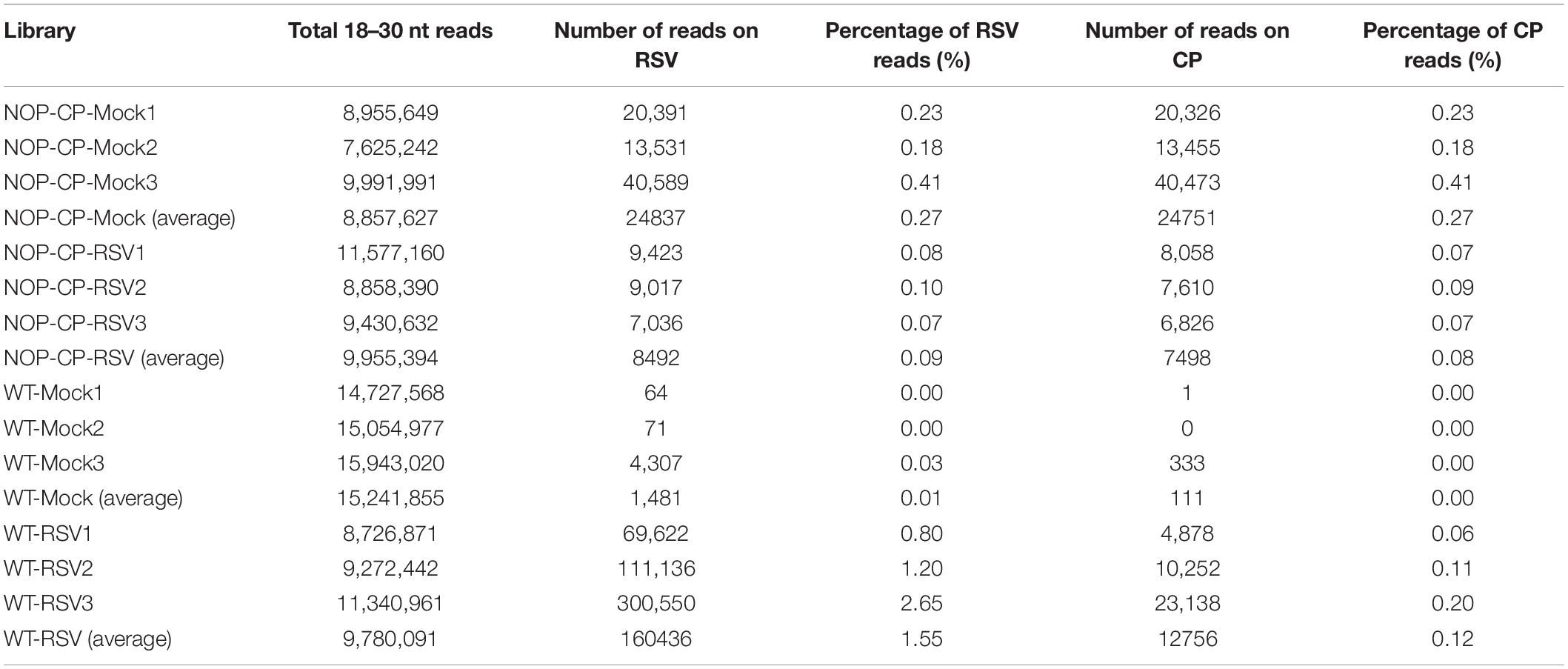
Table 1. Twelve libraries constructed with small RNAs from wild-type (WT) and NOP-CP transgenic (NOP-CP) Arabidopsis plants after mock control (Mock) or RSV inoculation (RSV), including three biological replicates each.
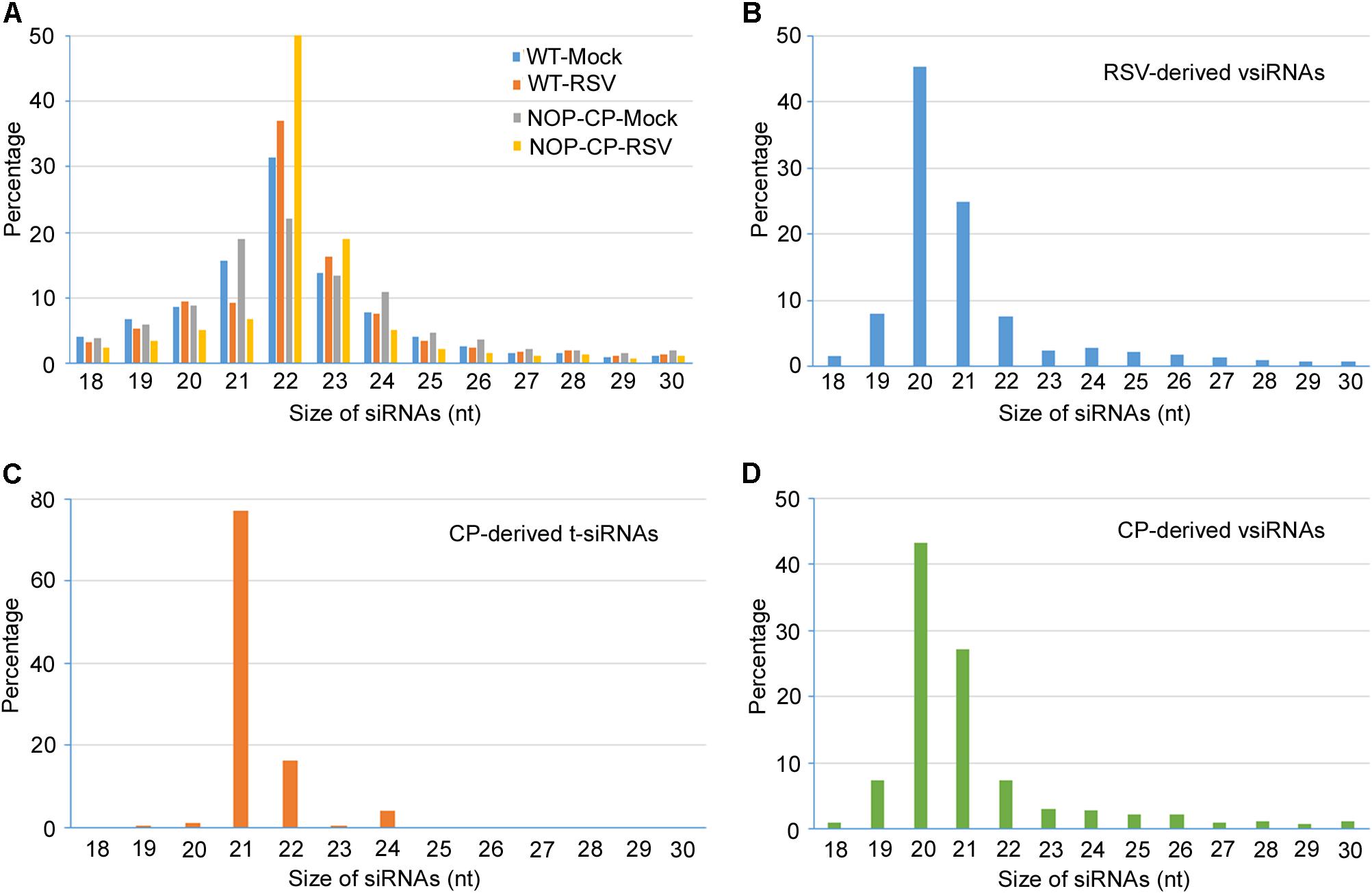
Figure 3. Size distribution of 18–30 nucleotide small RNAs. (A) Size distribution of total small RNA from mock-inoculated and RSV-infected NOP-CP transgenic and Col-0 Arabidopsis plants. (B) Size distribution of small RNA from vsiRNAs from RSV-infected Col-0 Arabidopsis plants. (C) Size distribution of CP-derived t-siRNAs from NOP-CP transgenic plants. (D) Size distribution of CP-derived vsiRNAs from RSV-infected Col-0 plants.
Characterization of vsiRNA in RSV-Infected Wild-Type Arabidopsis Plants
To further decipher the regulatory functions of vsiRNAs, we studied the profiles of vsiRNA in RSV-infected Arabidopsis plants. Size distribution analysis showed that RSV-derived vsiRNAs were predominantly 20 and 21 nt, accounting for 45% and 25% of the total, respectively (Figure 3B). For CP-derived t-siRNAs, the dominant sizes were 21 nt and 22 nt (Figure 3C), whereas the most dominant sizes of CP-derived vsiRNAs were 20 nt and 21 nt (Figure 3D), suggesting that DCL4 is the major producer of CP-derived t-siRNA and that RSV vsiRNA production involves multiple DCL functions.
The canonical 21–24-nt small RNAs from the viral sequences were further analyzed. The 21–24-nt vsiRNA reads were mapped to the RSV genomic sequence to explore their origin. As shown in Figure 4A and Supplementary Figures S2, S3, the proportions of vsiRNAs from both polarities were almost continuous, but some genomic regions showed higher mapping frequencies. In the RSV RNA1 genome, the complementary strand of the 3′ terminus produced significantly more vsiRNAs, and other RNA segments also exhibited some hotspots for vsiRNA generation (Figure 4A and Supplementary Figures S2, S3). The sense and antisense RSV RNA strands generated almost equivalent amounts of sense (49.5%) and antisense (50.5%) vsiRNA (Figure 4B). In addition, preferential occurrence of A/U at the 5′ terminus was identified in vsiRNAs (Figure 4C). These results suggest that RSV vsiRNAs are potentially loaded into AGO1/AGO2-containing RISCs.
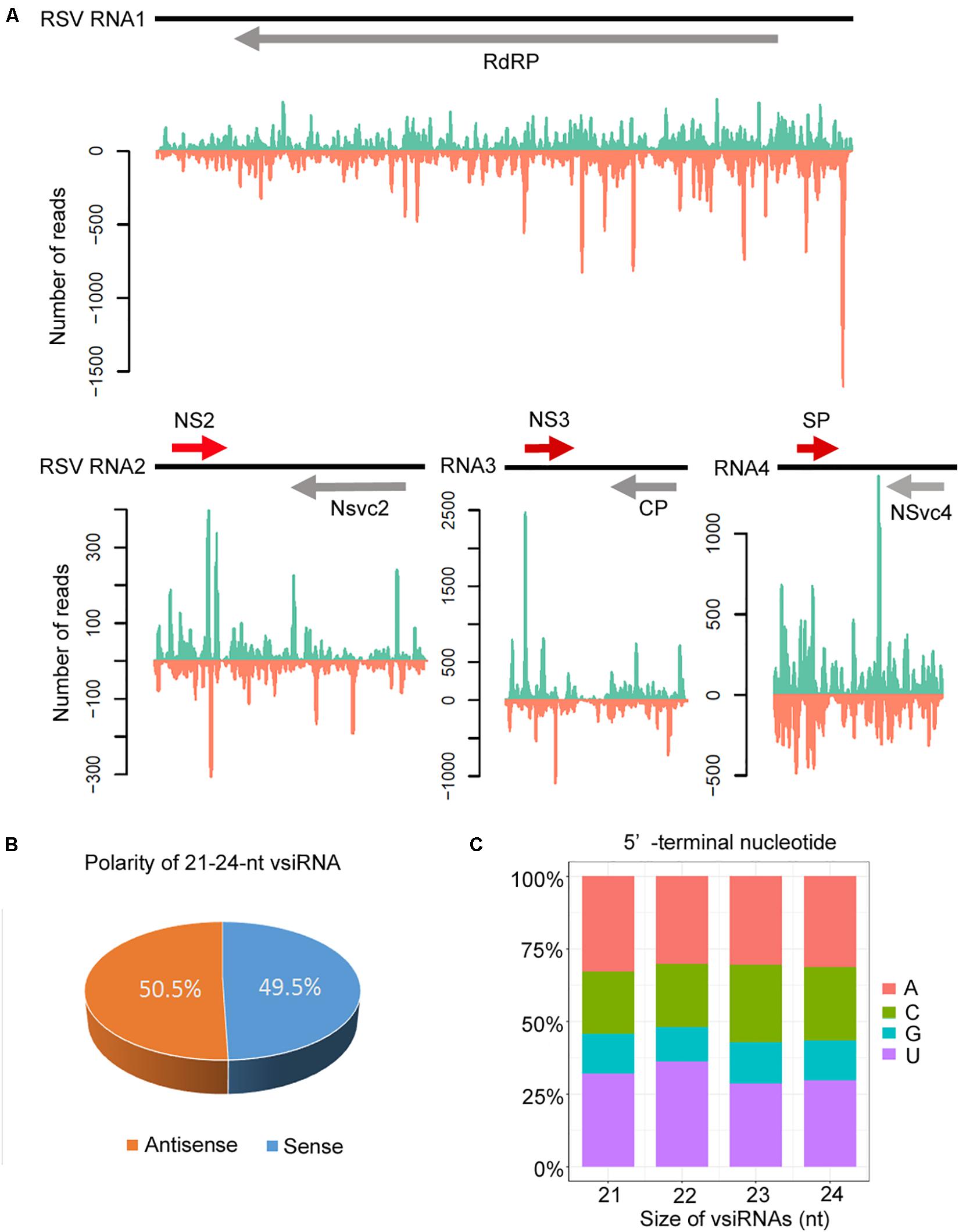
Figure 4. Profile of 21–24 nucleotide (nt) virus-derived small interfering (vsiRNA) derived from RSV-infected Col-0 plants. (A) Distribution of vsiRNAs along the RSV genome in both positive (blue) and negative (red) polarity. Genome organization of RSV positioned proportionally above. CP, coat protein; RdRp, RNA-dependent RNA polymerase; SP, disease-specific protein. (B) Accumulation of sense and antisense vsiRNAs. (C) 5′-terminal nucleotide frequency of 21–24 nt vsiRNAs.
Characterization of t-siRNA in NOP-CP Transgenic Arabidopsis Plants
To understand the significance of t-siRNA in CP-mediated RSV resistance, we characterized and compared t-siRNAs generated from the NOP-CP transgene of transgenic Arabidopsis plants and vsiRNAs mapping to the same region (CP-derived vsiRNA) in RSV-infected wild-type plants. The sequence distribution profile of 21–24-nt t-siRNA from the NOP-CP transgene region showed that t-siRNA accumulated in several hotspot regions throughout the transgenic sequence. Interestingly, CP vsiRNAs mapping to the CP region also showed a very similar profile, with peaks in the same position, although the t-siRNA peak read number was much higher than that for vsiRNAs (Figure 5A and Supplementary Figure S4). The similar t-siRNA and vsiRNA distribution profiles indicated that a sequence bias in the t-siRNA population depends on a nucleotide sequence preference and that there is a similar mechanism for t-siRNA and vsiRNA generation.
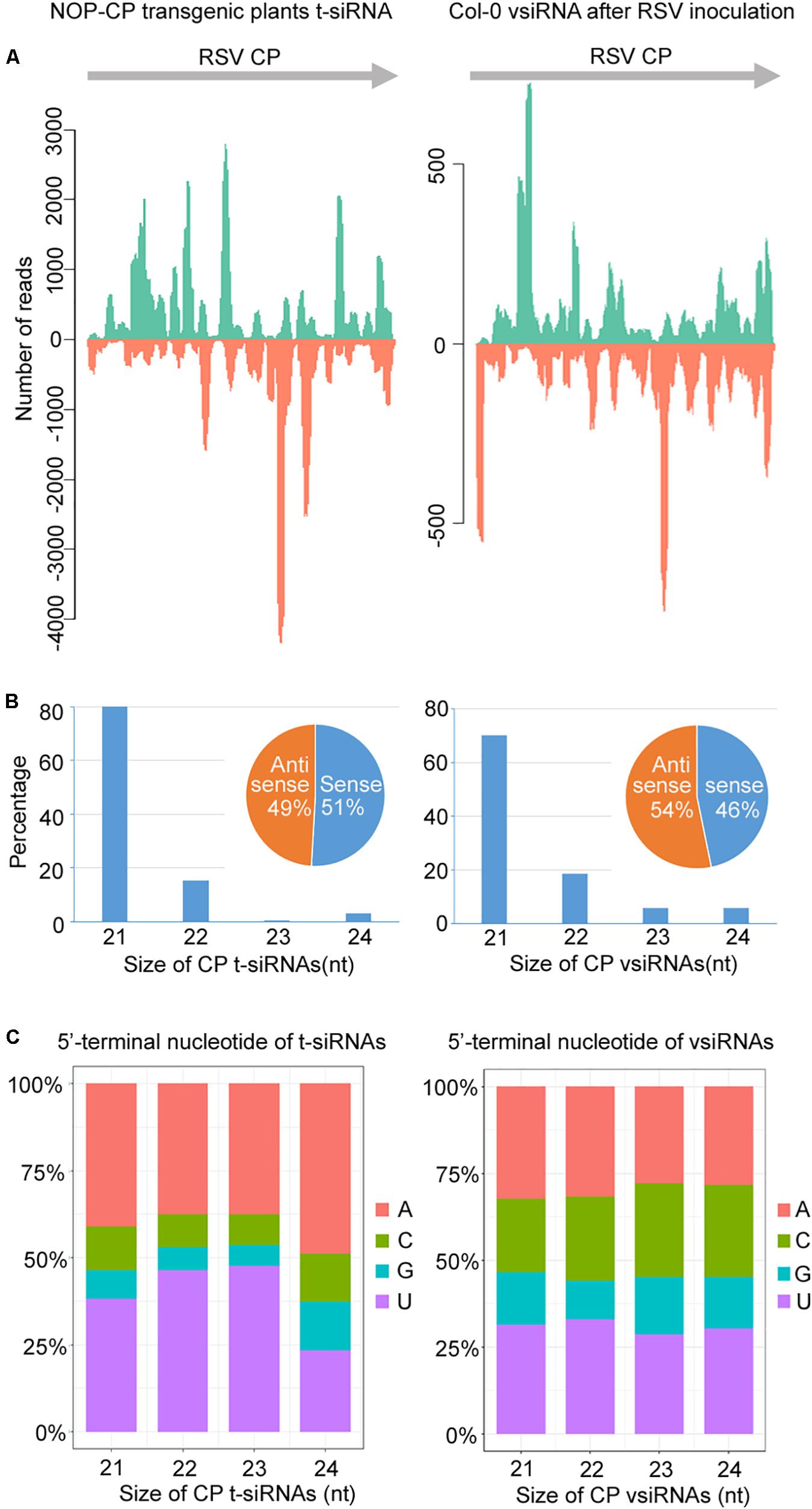
Figure 5. Characterization and comparison of 21–24 nt transgenic-derived small interfering RNA (t-siRNA) mapped to the CP sequence in NOP-CP transgenic plants (left) and virus-derived small interfering RNA (vsiRNA) mapped to the RSV CP sequence in RSV-infected Col-0 plants (right). (A) Distribution of t-siRNAs and vsiRNAs along the CP sequence in both positive (blue) and negative (red) polarity. Note that the scale used for the NOP-CP transgenic plants is different from that used for the RSV-infected Col-0 plants. (B) Size distribution of t-siRNAs and vsiRNAs. Pie graph showing the percentage of the sense and antisense t-siRNAs and vsiRNAs. (C) 5′-terminal nucleotide frequency of 21–24 nt t-siRNAs and vsiRNAs.
The size distributions of t-siRNAs showed that 21 nt was the most dominant size, representing 80% of the total t-siRNAs (Figure 5B). Similarly, the 21-nt classes accounted for the majority of the CP vsiRNAs, representing 70% of the total CP vsiRNAs (Figure 5B), indicating similar sRNA processing properties in transgene- and virus-derived sRNA. To explore the origin of t-siRNAs, the strand specificity and locations of t-siRNAs were analyzed. As shown in Figures 5A,B, almost equivalent proportions of sense and antisense sRNAs were generated in both t-siRNAs and CP vsiRNAs, although the sense (51%) t-siRNA amounts were slightly greater than the antisense orientation amounts (49%), whereas for CP vsiRNAs, more reads were counted in the antisense (54%) orientation. The 5′ terminal nucleotides were characterized for 21–24 nt t-siRNAs and CP-vsiRNAs. The U/A were the most abundant 5′ nucleotides in all four sizes of both t-siRNAs and CP-vsiRNAs (Figure 5C).
Furthermore, we also characterized and compared the t-siRNA populations of NOP-CP transgenic Arabidopsis plants with mock treatment and RSV inoculation. The results showed that the t-siRNAs of NOP-CP transgenic plants after mock and RSV inoculation shared highly similar distribution profiles, although the read number of total t-siRNAs in RSV inoculation plants was significantly less than that in the mock treatment plants (Supplementary Figure S5 and Table 1). Characterization of t-siRNAs in NOP-CP transgenic Arabidopsis plants indicates that NOP-CP-derived t-siRNAs may be associated with CP-mediated RSV resistance in Arabidopsis.
Plant DCLs 2, 3, and 4 Are Indispensable for CP-Mediated RSV Resistance in Arabidopsis
Previous studies have demonstrated that plant DCLs are responsible for generating transgene-derived t-siRNAs and virus-derived vsiRNAs (Guo et al., 2019). To test our hypothesis that the accumulation of t-siRNAs is associated with RSV resistance in CP transgenic Arabidopsis plants, we generated CP transgene plants in the Arabidopsis dcl2/3/4 mutant background. These T2 generation transgenic plants were confirmed by western blot using an anti-Myc antibody (Figure 6B). An RSV infection assay showed that CP transgenic Arabidopsis plants in the dcl2/3/4 background (dcl2/3/4;CP) developed severe stunting symptoms similar to those of non-transgenic dcl2/3/4 and Col-0 plants (Figure 6A). Western blot and qRT-PCR results both showed that both RSV SP mRNA and proteins accumulated to similar levels in dcl2/3/4;CP plants and non-transgenic dcl2/3/4 and Col-0 plants (Figures 6C,D and Supplementary Figure S6). These results indicate that CP-mediated RSV resistance depends on the function of DCLs in Arabidopsis.
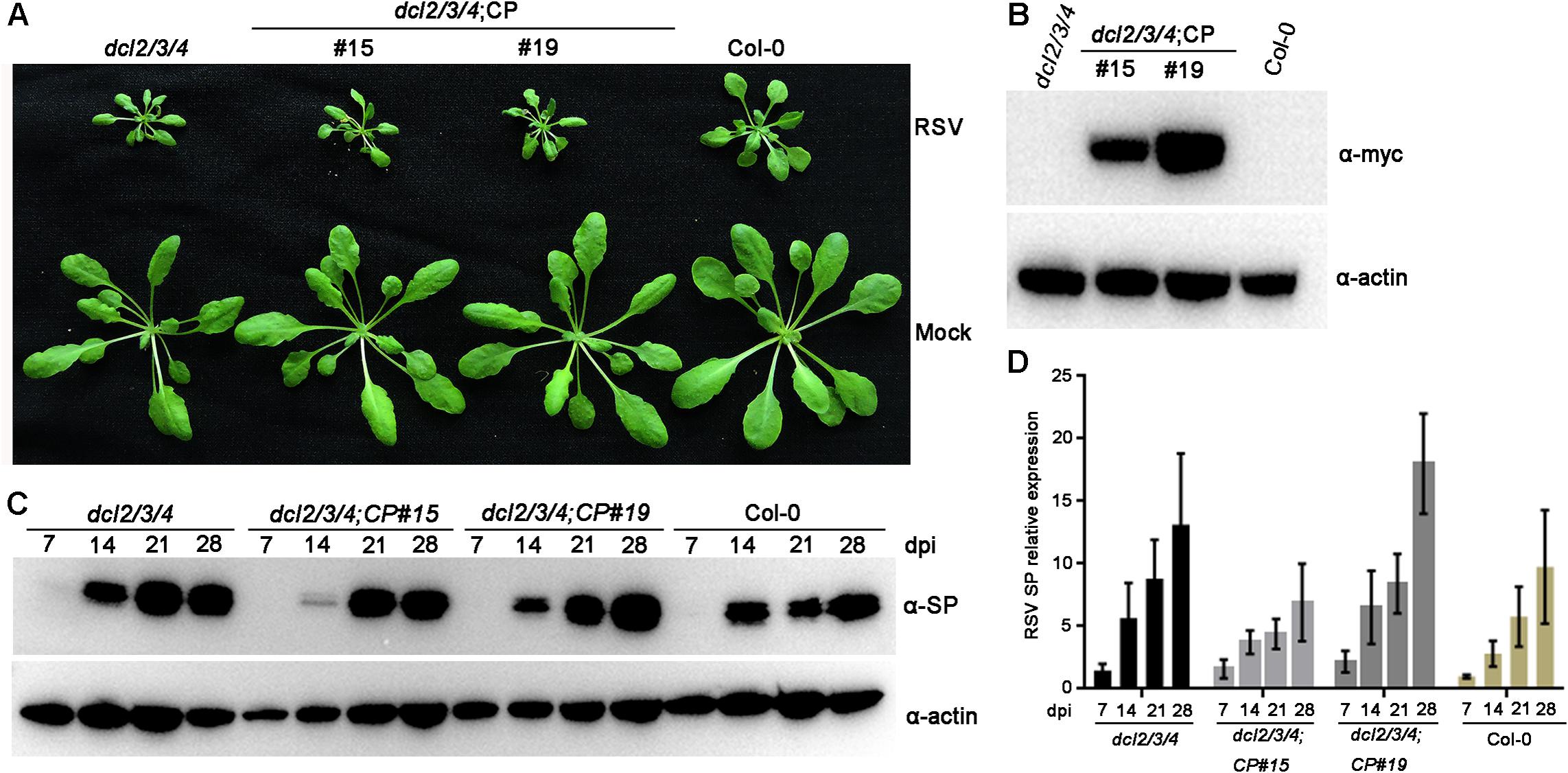
Figure 6. Rice stripe virus resistance examination of CP transgenic plants in the dcl2/3/4 background. (A) Symptoms of mock-inoculated and RSV-infected CP transgenic plants in the dcl2/3/4 background (dcl2/3/4;CP#15, dcl2/3/4;CP#19) and dcl2/3/4 and Col-0 Arabidopsis plants. Photographs were taken at 4 weeks post-inoculation. (B) Western blot analysis of CP transgenic lines in the dcl2/3/4 background with an anti-Myc antibody. The actin protein level served as a loading control. (C) Western blot analysis of the time course of RSV-encoded SP protein accumulation in RSV-infected CP transgenic plants in the dcl2/3/4 background and dcl2/3/4 (dcl2/3/4;CP#15, dcl2/3/4;CP#19) and Col-0 Arabidopsis plants using an SP-specific antibody. The actin protein level served as a loading control. (D) qRT-PCR analysis for the time course of RSV SP mRNA transcription levels in RSV-infected CP transgenic plants in the dcl2/3/4 background (dcl2/3/4;CP#15, dcl2/3/4;CP#19) and dcl2/3/4 and Col-0 Arabidopsis plants. Signal intensities for each transcript were normalized with those for EF1-α. Values are means ± SD (n = 3).
Discussion
Since the concept of pathogen-derived resistance (PDR) was proposed (Sanford and Johnston, 1985) and confirmed by expressing tobacco mosaic virus (TMV) CP in transgenic tobacco plants, resulting in TMV resistance (Abel et al., 1986), CP-mediated resistance has been widely used to protect various plant species against virus infection (Galvez et al., 2014). In the case of RSV, the overexpression of CP in rice plants resulted in partial resistance to virus infection, but no related mechanism was determined (Hayakawa et al., 1992). Recent studies have also demonstrated that the RSV CP triggers jasmonate accumulation and subsequently induces the plant antiviral immune response (Han et al., 2020; Yang et al., 2020) and that RSV CP-mediated resistance requires AGO18 protein expression in rice plants (Yang et al., 2020). Thus, RSV CP-mediated resistance appears to be a protein-mediated phenomenon in rice plants, similar to TMV CP-mediated resistance in tobacco plants (Lindbo and Falk, 2017). However, the effectiveness of transgene-induced silencing could not be ruled out in these previous studies, as some resistant lines containing translatable transgenes also generated RNA-mediated resistance correlating with t-siRNAs. In this study, we provide evidence that the expression of RSV CP in transgenic Arabidopsis plants is not a prerequisite for CP-mediated resistance. First, both translatable and non-translatable versions of CP transgenic Arabidopsis plants all showed immunity to RSV infection. Second, by using deep sequencing analysis, CP-mediated resistance in transgenic Arabidopsis plants was associated with high accumulation of 21–24-nt t-siRNAs. Third, CP overexpression in the dcl2/3/4 mutant background resulted in similar sensitivity to RSV infection in non-transgenic dcl2/3/4 plants. Our results strongly indicate that RSV CP-mediated resistance is due to an RNA silencing-mediated resistance mechanism in Arabidopsis, similar to that indicated in tobacco etch virus (TEV) CP-mediated resistance (Lindbo et al., 1993) and tomato spotted wilt virus (TSWV) N (nucleocapsid)-mediated resistance (Catoni et al., 2013).
RNA silencing-directed resistance is mediated by 21–24 nt siRNAs (Hamilton et al., 2002; Ruiz-Ferrer and Voinnet, 2009) generated through the cleavage of a double-stranded or an imperfect stem-loop RNA molecule by plant DCLs (Moazed, 2009). In this paper, Illumina deep sequencing was performed to characterize the small RNA population and gain insights into the RNA-based virus resistance mechanism during RSV-Arabidopsis interactions. Bioinformatic analysis of the deep sequencing data indicated that RSV infection triggered the generation of relatively large amounts of vsiRNA, accounting for 1.55% of the total 18–30-nt reads in Arabidopsis plants (Table 1), in contrast with an average of 0.29% of the total small RNA reads mapped to the RSV genome in RSV-infected natural host rice plants (Yang et al., 2018). This difference is probably a result of the different virus titers in different RSV-host pathosystems. In general, DCL4-generated 21-nt vsiRNAs are the most abundant class, following DCL-dependent 22-nt vsiRNAs (Guo et al., 2019). In RSV-infected Arabidopsis, 20-nt vsiRNAs was the most abundant species (Figure 3B). This result is similar to the rice ragged stunt virus (RRSV) vsiRNAs in rice plants (Li et al., 2018). In RSV-infected rice and tobacco plants, 20 nt vsiRNAs also account for the high proportion (Yan et al., 2010; Jiang et al., 2012; Xu et al., 2012). These results indicate that 20 nt vsiRNAs may play a role in RSV-plants interaction. The mechanism of biogenesis and function of 20 nt visRNA remains elusive, though the biogenesis of 20 nt miRNAs is dependent on DCL1 (Lee et al., 2015). Further studies are needed to elucidate the role of Arabidopsis DCL1 in the production RSV vsiRNAs.
In Arabidopsis, AGO1 and AGO2 play crucial roles in antivirus defense (Carbonell and Carrington, 2015) and associate with small RNAs that exhibit uridine and adenosine at their 5′ ends, respectively (Mi et al., 2008). Our study also revealed preferential occurrence of uridine or adenosine residues at the 5′ terminus in RSV vsiRNA, showing the conserved AGO complexes binding vsiRNA for antivirus defense. Compared with previous studies, the 5′-terminal nucleotides of RSV visRNAs from Arabidopsis, rice and tobacco plants were similar (Yan et al., 2010; Jiang et al., 2012; Xu et al., 2012; Yang et al., 2018), suggesting that AGO1 and AGO2 play an important role in virus defense against RSV in plants.
Regarding the genomic regions from which RSV vsiRNAs were generated, fewer vsiRNAs were produced from RNA2 than from the other three RNA segments in Arabidopsis (Figure 4); however, in RSV-infected rice plants, RNA1 generated fewer vsiRNAs than the other three RNA segments (Xu et al., 2012; Yang et al., 2018). These distinct RSV vsiRNA origins in plant hosts might be explained by the evolutionary adaptation of RSV, which escapes RNA silencing antivirus machinery, thus facilitating successful virus infection in rice plants. In this study, RSV-derived vsiRNAs equally originate from the sense and antisense strands (Figure 4B), consistent with previous reports in RSV-rice interaction (Yan et al., 2010; Yang et al., 2018). These results indicate that the RSV vsiRNAs may originated from double-stranded replication intermediates.
To characterize CP t-siRNAs that confer resistance to RSV infection, the origin, composition and abundance of CP sequence-derived t-siRNAs were analyzed and compared with those of CP-derived vsiRNAs. The sequence distribution, 5′ terminal nucleotides and strand specificity exhibited very similar patterns between CP-derived t-siRNAs and CP-derived vsiRNAs, except that the amount of t-siRNAs was nearly twice that of vsiRNAs, indicating that the activation of t-siRNAs mediates antiviral defense derived from the transgene. A prerequisite step for t-siRNA biogenesis is a sufficient quantity of transgene RNA that has a dsRNA nature. Multiple studies have demonstrated that transgenes can often integrate into the host genome in complex structures, such as multiple copies inverted to each other, and transcription of transgenic DNA from these complex integrations could lead to dsRNA structures (Lindbo and Falk, 2017). Another possibility is that the single-stranded RNA transcript from transgene DNA is converted to dsRNAs by SUPPRESSOR OF GENE SILENCING 3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) (Guo et al., 2019). Further genetic and biochemical studies will be necessary to address which pathway is responsible for t-siRNA biogenesis in CP transgenic Arabidopsis plants.
In Arabidopsis plants, DCL4, DCL2 and DCL3 process transgene-derived dsRNAs to 21–24 nt siRNAs in a redundant and hierarchical manner (Deleris et al., 2006), which are incorporated into AGO1 or AGO2 effectors to guide cleavage of target RNAs (Pumplin and Voinnet, 2013). To further confirm our hypothesis that the accumulation of t-siRNAs was associated with RSV immunity in CP transgenic Arabidopsis plants, CP transgenic plants in the dcl2/3/4 triple mutant background were developed. As expected, these transgenic plants exhibited susceptibility to RSV infection similar to that of non-transgenic plants, thus supporting our hypothesis. Our results also showed that dcl2/3/4 triple mutant plants exhibited increased severity of disease symptoms and virus titers compared with those of Col-0 plants in RSV challenge assays, and similar results were obtained with other RNA viruses (Guo et al., 2019). Further investigation is needed to elucidate the special role of DCLs in producing t-siRNAs in CP transgenic Arabidopsis plants. The mechanism of RSV CP-mediated virus resistance revealed by this study provides an in-depth understanding of RNA-based antiviral immunity in RSV-Arabidopsis interactions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FS and TZ conceived and designed the experiments. FS, PH, and WW conducted the experiments. YL, LD, FS, and TZ analyzed the data. FS and TZ wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by the National Key R&D Program of China (grant no. 2017YFD0100400), the Natural Science Foundation of Jiangsu Province (grant no. BK20171322), the National Natural Science Foundation of China (grant no. 31761143012), and TA CR-JSTD Bilateral Co-funding R&D Project (grant no. BZ2020024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful Dr. Xiuren Zhang at Texas A&M University for providing pBA-Flag-Myc4-DC vector and Arabidopsis Col-0 and dcl2/3/4 mutant seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.591619/full#supplementary-material
Supplementary Figure S1 | Characterization and expression of CP-YFP and NOP-CP-YFP in Nicotiana benthamiana. (A) Confocal imaging assays show the expression of CP-YFP but not NOP-CP-YFP in Nicotiana benthamiana. (B) RT-PCR analysis of CP mRNA transcription levels of CP-YFP and NOP-CP-YFP in Nicotiana benthamiana. The ubc mRNA level served as a loading control. (C) Western blot analysis of the expression of CP-YFP and NOP-CP-YFP in Nicotiana benthamiana using an YFP-specific antibody. The actin protein level served as a loading control.
Supplementary Figure S2 | Profile of 21–24 nucleotide (nt) virus-derived small interfering RNA (vsiRNA) derived from RSV-infected Col-0 plants, biological replicate 2.
Supplementary Figure S3 | Profile of 21–24 nucleotide (nt) virus-derived small interfering (vsiRNA) derived from RSV-infected Col-0 plants biological replicate 3.
Supplementary Figure S4 | Characterization and comparison of 21–24 nt transgenic-derived small interfering RNA (t-siRNA) mapped to the CP sequence in NOP-CP transgenic plants (left) and virus-derived small interfering RNA (vsiRNA) mapped to the RSV CP sequence in RSV-infected Col-0 plants (right) from two other biological replicates.
Supplementary Figure S5 | Characterization and comparison of 21–24 nt transgenic-derived small interfering RNA (t-siRNA) mapped to the CP sequence in NOP-CP transgenic plants after mock control (left) and RSV inoculation (right). (A) Distribution of t-siRNAs along the CP sequence in both positive (blue) and negative (red) polarity. Note that the scale used for the mock control is different from that used for the RSV inoculation. (B) Size distribution of t-siRNAs. Pie graph showing the percentage of the sense and antisense t-siRNAs. (C) 5′-terminal nucleotide frequency of 21–24 nt t-siRNAs.
Supplementary Figure S6 | Comparison of RSV resistance in CP transgenic plants in dcl2/3/4 and Col-0 background. (A) Western blot analysis of RSV-encoded SP protein accumulation in RSV-infected CP transgenic Arabidopsis plants in dcl2/3/4 and Col-0 background at 28 dpi. The actin protein level served as a loading control. (B) qRT-PCR analysis of RSV SP mRNA transcription levels in RSV-infected CP transgenic plants in dcl2/3/4 and Col-0 background at 28 dpi. Signal intensities for each transcript were normalized with those for EF1-α. Values are means ± SD (n = 3). ∗p ≤ 0.05 (Student’s t-test).
Supplementary Table S1 | Primer sequence used for the cloning of transgenes and testing the viral infection.
References
Abel, P. P., Nelson, R. S., De, B., Hoffmann, N., Rogers, S. G., Fraley, R. T., et al. (1986). Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232, 738–743. doi: 10.1126/science.3457472
Barbier, P., Takahashi, M., Nakamura, I., Toriyama, S., and Ishihama, A. (1992). Solubilization and promoter analysis of RNA polymerase from rice stripe virus. J. Virol. 66, 6171–6174. doi: 10.1016/0166-0934(92)90013-4
Blevins, T., Podicheti, R., Mishra, V., Marasco, M., Wang, J., Rusch, D., et al. (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4:e09591. doi: 10.7554/eLife.09591
Carbonell, A., and Carrington, J. C. (2015). Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 27, 111–117. doi: 10.1016/j.pbi.2015.06.013
Catoni, M., Lucioli, A., Doblas-Ibáňez, P., Accotto, G. P., and Vaira, A. M. (2013). From immunity to susceptibility: virus resistance induced in tomato by a silenced transgene is lost as TGS overcomes PTGS. Plant J. 75, 941–953. doi: 10.1111/tpj.12253
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Deleris, A., Gallego-Bartolome, J., Bao, J. S., Kasschau, K. D., Carrington, J. C., and Voinnet, O. (2006). Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science 313, 68–71. doi: 10.1126/science.1128214
Diaz-Pendon, J. A., Li, F., Li, W. X., and Ding, S. W. (2007). Suppression of antiviral silencing by Cucumber Mosaic Virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19, 2053–2063. doi: 10.1105/tpc.106.047449
Du, Z., Xiao, D., Wu, J., Jia, D., Yuan, Z., Liu, Y., et al. (2011). p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol. Plant Pathol. 12, 808–814. doi: 10.1111/j.1364-3703.2011.00716.x
Falk, B. W., and Tsai, J. H. (1998). Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 36, 139–163. doi: 10.1146/annurev.phyto.36.1.139
Fang, X., and Qi, Y. (2016). RNAi in plants: an argonaute-centered view. Plant Cell 28, 272–285. doi: 10.1105/tpc.15.00920
Galvez, L. C., Banerjee, J., Pinar, H., and Mitra, A. (2014). Engineered plant virus resistance. Plant Sci. 228, 11–25. doi: 10.1016/j.plantsci.2014.07.006
Guo, Z., Li, Y., and Ding, S. W. (2019). Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 19, 31–44. doi: 10.1038/s41577-018-0071-x
Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. doi: 10.1093/emboj/cdf464
Han, K., Huang, H., Zheng, H., Ji, M., Yuan, Q., Cui, W., et al. (2020). Rice stripe virus coat protein induces the accumulation of jasmonic acid, activating plant defence against the virus while also attracting its vector to feed. Mol. Plant Pathol. doi: 10.1111/mpp.12995 [Epub ahead of print].
Hayakawa, T., Zhu, Y., Itoh, K., Kimura, Y., Izawa, T., Shimamoto, K., et al. (1992). Genetically engineered rice resistant to rice stripe virus, an insect-transmitted virus. Proc. Natl. Acad. Sci. U.S.A. 89, 9865–9869. doi: 10.1073/pnas.89.20.9865
Jackel, J. N., Storer, J. M., Coursey, T., and Bisaro, D. M. (2016). Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on geminivirus chromatin. J. Virol. 90, 7529–7540. doi: 10.1128/JVI.00656-16
Jiang, L., Qian, D., Zheng, H., Meng, L. Y., Chen, J., Le, W. J., et al. (2012). RNA-dependent RNA polymerase 6 of rice (oryza sativa) plays role in host defense against negative-strand RNA virus, rice stripe virus. Virus Res. 163, 512–519. doi: 10.1016/j.virusres.2011.11.016
Kong, L., Wu, J., Lu, L., Xu, Y., and Zhou, X. (2014). Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol. Plant 7, 691–708. doi: 10.1093/mp/sst158
Lee, W. C., Lu, S. H., Lu, M. H., Yang, C. J., Wu, S. H., and Chen, H. M. (2015). Asymmetric bulges and mismatches determine 20-nt microRNA formation in plants. RNA Biol. 12, 1054–1066. doi: 10.1080/15476286.2015.1079682
Li, S., Wang, S., Wang, X., Li, X., Zi, J., Ge, S., et al. (2015). Rice stripe virus affects the viability of its vector offspring by changing developmental gene expression in embryos. Sci. Rep. 5:7883. doi: 10.1038/srep07883
Li, Z., Zhang, T., Huang, X., and Zhou, G. (2018). Impact of two reoviruses and their coinfection on the rice RNAi system and vsiRNA production. Viruses 10:594. doi: 10.3390/v10110594
Lindbo, J. A., and Falk, B. W. (2017). The impact of “Coat Protein-Mediated Virus Resistance” in applied plant pathology and basic research. Phytopathology 107, 624–634. doi: 10.1094/PHYTO-12-16-0442-RVW
Lindbo, J. A., Silva-Rosales, L., Proebsting, W. M., and Dougherty, W. G. (1993). lnduction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5, 1749–1759. doi: 10.1105/tpc.5.12.1749
Ma, J., Song, Y., Wu, B., Jiang, M., Li, K., Zhu, C., et al. (2011). Production of transgenic rice new germplasm with strong resistance against two isolations of rice stripe virus by RNA interference. Transgenic Res. 20, 1367–1377. doi: 10.1007/s11248-011-9502-1
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Moazed, D. (2009). Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420. doi: 10.1038/nature07756
Otuka, A. (2013). Migration of rice planthoppers and their vectored re-emerging and novel rice viruses in East Asia. Fron.t Microbiol. 4:309. doi: 10.3389/fmicb.2013.00309
Pumplin, N., and Voinnet, O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11, 745–760. doi: 10.1038/nrmicro3120
Raja, P., Sanville, B. C., Buchmann, R. C., and Bisaro, D. M. (2008). Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007. doi: 10.1128/JVI.00719-08
Ruiz-Ferrer, V., and Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. doi: 10.1146/annurev.arplant.043008.092111
Sanford, J. C., and Johnston, S. A. (1985). The concept of parasite-derived resistance-deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 113, 395–405. doi: 10.1016/s0022-5193(85)80234-4
Sasaya, T., Nakazono-Nagaoka, E., Saika, H., Aoki, H., Hiraguri, A., Netsu, O., et al. (2014). Transgenic strategies to confer resistance against viruses in rice plants. Front. Microbiol. 4:409. doi: 10.3389/fmicb.2013.00409
Shimizu, T., Nakazono-Nagaoka, E., Uehara-Ichiki, T., Sasaya, T., and Omura, T. (2011). Targeting specific genes for RNA interference is crucial to the development of strong resistance to rice stripe virus. Plant Biotechnol. J. 9, 503–512. doi: 10.1111/j.1467-7652.2010.00571.x
Singh, J., Mishra, V., Wang, F., Huang, H. Y., and Pikaard, C. S. (2019). Reaction mechanisms of Pol IV, RDR2, and DCL3 drive RNA channeling in the siRNA-directed DNA methylation pathway. Mol. Cell 75, 576.e5–589.e5. doi: 10.1016/j.molcel.2019.07.008
Sun, F., Fang, P., Li, J., Du, L., Lan, Y., Zhou, T., et al. (2016). RNA-seq-based digital gene expression analysis reveals modification of host defense responses by rice stripe virus during disease symptom development in Arabidopsis. Virol. J. 13:202. doi: 10.1186/s12985-016-0663-7
Sun, F., Yuan, X., Zhou, T., Fan, Y., and Zhou, Y. (2011). Arabidopsis is susceptible to rice stripe virus infections. J. Phytopathol. 159, 767–772. doi: 10.1111/j.1439-0434.2011.01840.x
Toriyama, S., and Watanabe, Y. (1989). Characterization of single- and double-stranded RNAs in particles of rice stripe virus. J. Gen. Virol. 70, 505–511. doi: 10.1099/0022-1317-70-3-505
Wang, X. B., Jovel, J., Udomporn, P., Wang, Y., Wu, Q., Li, W. X., et al. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23, 1625–1638. doi: 10.1105/tpc.110.082305
Wang, X. B., Wu, Q., Ito, T., Cillo, F., Li, W. X., Chen, X., et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A. 107, 484–489. doi: 10.1073/pnas.0904086107
Wassenegger, M., and Krczal, G. (2006). Nomenclature and functions of RNA directed RNA polymerases. Trends Plant Sci. 11, 142–151. doi: 10.1016/j.tplants.2006.01.003
Xiong, R., Wu, J., Zhou, Y., and Zhou, X. (2008). Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82, 12304–12311. doi: 10.1128/JVI.01696-08
Xiong, R., Wu, J., Zhou, Y., and Zhou, X. (2009). Characterization and subcellular localization of an RNA silencing suppressor encoded by rice stripe tenuivirus. Virology 387, 29–40. doi: 10.1016/j.virol.2009.01.045
Xu, Y., Huang, L., Fu, S., Wu, J., and Zhou, X. (2012). Population diversity of rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphgax striatellus. PLoS One 7:e46238. doi: 10.1371/journal.pone.0046238
Yan, F., Zhang, H., Adams, M. J., Yang, J., Peng, J., Antoniw, J. F., et al. (2010). Characterization of siRNAs derived from rice stripe virus in infected rice plants by deep sequencing. Arch. Virol. 155, 935–940. doi: 10.1007/s00705-010-0670-8
Yang, M., Xu, Z., Zhao, W., Liu, Q., Li, Q., Lu, L., et al. (2018). Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus. BMC Plant Biol. 18:219. doi: 10.1186/s12870-018-1438-7
Yang, Z., Huang, Y., Yang, J., Yao, S., Zhao, K., Wang, D., et al. (2020). Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28, 89–103. doi: 10.1016/j.chom.2020.05.001
Yao, M., Liu, X., Li, S., Xu, Y., Zhou, Y., Zhou, X., et al. (2014). Rice stripe tenuivirus NSvc2 glycoproteins targeted to the golgi body by the N-terminal transmembrane domain and adjacent cytosolic 24 amino acids via the COP I- and COP II-dependent secretion pathway. J. Virol. 88, 3223–3234. doi: 10.1128/JVI.03006-13
Zhang, C., Wu, Z., Li, Y., and Wu, J. (2015). Biogenesis, function, and applications of virus-derived small RNAs in plants. Front. Microbiol. 6:1237. doi: 10.3389/fmicb.2015.01237
Zhang, X., Garreton, V., and Chua, N. H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543. doi: 10.1101/gad.1318705
Keywords: Rice stripe virus, CP-mediated resistance, RNA silencing, deep sequence, dcl2/3/4
Citation: Sun F, Hu P, Wang W, Lan Y, Du L, Zhou Y and Zhou T (2020) Rice Stripe Virus Coat Protein-Mediated Virus Resistance Is Associated With RNA Silencing in Arabidopsis. Front. Microbiol. 11:591619. doi: 10.3389/fmicb.2020.591619
Received: 05 August 2020; Accepted: 26 October 2020;
Published: 13 November 2020.
Edited by:
Xifeng Wang, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection (CAAS), ChinaReviewed by:
Won Kyong Cho, Seoul National University, South KoreaYongliang Zhang, China Agricultural University, China
Hongying Zheng, Ningbo University, China
Fei Yan, Ningbo University, China
Copyright © 2020 Sun, Hu, Wang, Lan, Du, Zhou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Sun, c3VuZmVuZzEyMDFAMTI2LmNvbQ==; Tong Zhou, emhvdXRvbmdAamFhcy5hYy5jbg==
 Feng Sun
Feng Sun Peng Hu1,2
Peng Hu1,2 Yijun Zhou
Yijun Zhou Tong Zhou
Tong Zhou