
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 November 2020
Sec. Virology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.591019
 Micaela Ziraldo1
Micaela Ziraldo1 Juan E. Bidart2
Juan E. Bidart2 Cecilia A. Prato3
Cecilia A. Prato3 María V. Tribulatti3
María V. Tribulatti3 Patricia Zamorano2
Patricia Zamorano2 Nora Mattion1
Nora Mattion1 Alejandra L. D’Antuono1*
Alejandra L. D’Antuono1*Although replication-defective human adenovirus type 5 (Ad5) vectors that express in situ the capsid-encoding region of foot-and-mouth disease virus (FMDV) have been proven to be effective as vaccines in relevant species for several viral strains, the same result was not consistently achieved for the O1/Campos/Brazil/58 strain. In the present study, an optimization of the Ad5 system was explored and was proven to enhance the expression of FMDV capsid proteins and their association into virus-like particles (VLPs). Particularly, we engineered a novel Ad5 vector (Ad5[PVP2]OP) which harbors the foreign transcription unit in a leftward orientation relative to the Ad5 genome, and drives the expression of the FMDV sequences from an optimized cytomegalovirus (CMV) enhancer-promoter as well. The Ad5[PVP2]OP vaccine candidate also contains the amino acid substitutions S93F/Y98F in the VP2 protein coding sequence, predicted to stabilize FMD virus particles. Cells infected with the optimized vector showed an ∼14-fold increase in protein expression as compared to cells infected with an unmodified Ad5 vector tested in previous works. Furthermore, amino acid substitutions in VP2 protein allowed the assembly of FMDV O1/Campos/Brazil/58 VLPs. Evaluation of several serological parameters in inoculated mice with the optimized Ad5[PVP2]OP candidate revealed an enhanced vaccine performance, characterized by significant higher titers of neutralizing antibodies, as compared to our previous unmodified Ad5 vector. Moreover, 94% of the mice vaccinated with the Ad5[PVP2]OP candidate were protected from homologous challenge. These results indicate that both the optimized protein expression and the stabilization of the in situ generated VLPs improved the performance of Ad5-vectored vaccines against the FMDV O1/Campos/Brazil/58 strain and open optimistic expectations to be tested in target animals.
Included in the A list of infectious diseases of animals of the Office International des Épizooties (OIE), foot-and-mouth disease (FMD) is one of the most contagious animal illnesses that affects several species of wild and domestic cloven-hoofed ruminants (Knight-Jones et al., 2016, 2017). Although the disease has been circulating for a long time, it remains of major concern to livestock farmers and official health authorities since it causes a huge socioeconomic impact, particularly in the agricultural sector. Due to the financial losses incurred, FMD is still considered the major disease concerning production and international trade in foodstuffs of animal origin, food safety, and economic development, affecting both small- and large-scale production (Knight-Jones and Rushton, 2013).
The causative agent, FMD virus (FMDV), is a positive-sense single-stranded RNA virus that belongs to the genus Aphthovirus of the Picornaviridae family. The World Reference Laboratory for FMD lists seven FMDV serotypes [Euroasiatic serotypes O, A, Asia1, and C and South African territories (SAT) serotypes SAT1, SAT2, and SAT3] in seven regional endemic pools, among which no cross-serotype protection is expected (FAO, 2020). Within the seven serotypes described, serotype O is the most prevalent and is distributed across six out of the seven described endemic pools (Brito et al., 2017). Furthermore, serotype O was responsible for the recent FMD outbreaks in Colombia (2017–2018), Ecuador (2011), and Venezuela (2013) (Centro Panamericano de Fiebre Aftosa, OPS, and OMS., 2019).
In FMD endemic regions, disease control is mainly achieved by regular vaccination (Knight-Jones and Rushton, 2013; Naranjo and Cosivi, 2013). In this regard, the currently employed vaccines are reliant on the use of highly purified, highly concentrated, inactivated FMDV antigen (Grubman and Baxt, 2004). Despite their effectiveness, these vaccine formulations have several drawbacks, including the difficulty to differentiate infected from vaccinated animals, the need for expensive high-containment Biosafety Level 4-OIE (BSL4-OIE) manufacturing facilities, and the generation of large amounts of infectious virus required for vaccine production. Another problem associated with the traditional FMD vaccine is the risk of live virus release during the manufacturing process or the unintentional use of incompletely inactivated products. These disadvantages highlight the need for new-generation genetically engineered vaccines.
At the moment, the most successful strategy in the development of new FMDV vaccines has been the production of a replication-defective human serotype 5 adenovirus (Ad5) that delivers the capsid coding region of FMDV along with the viral protease 3C (3Cpro), required for its processing (P12A3C cassette). Initial studies reported by Mayr et al. (1999) have shown that an Ad5 vector that delivers the capsid and 3Cpro coding regions of FMDV A12/119/Kent/UK/32 (Ad5-A12) was effective at protecting swine from clinical disease after direct contact challenge. Subsequently, it was demonstrated that one dose of Ad5-A24, which contains the P12A3C cassette from FMDV A24/Cruzeiro/BRA/55, confers early protection against homologous FMDV challenge in swine (Moraes et al., 2002) and cattle (Pacheco et al., 2005; Schutta et al., 2016). More recently, it has been shown that an Ad5 expressing FMDV O/Manisa/TUR/69 antigen (Ad5-O1Man) could fully protect swine at 7 days post-inoculation (dpi) (Fernández-Sainz et al., 2017). Furthermore, the Adt-O1Manisa vector in combination with Ad5-boIFNλ3, which drives the expression of bovine IFN-λ3, completely protects cattle against homologous challenge as early as 3 dpi (Diaz-San Segundo et al., 2016). On the contrary, in initial efficacy studies, the Ad5-O1/Campos/Brazil/58 (Ad5-O1C)-vectored vaccine has shown limited performance in target species such as swine (Caron et al., 2005). Consequently, several strategies have been implemented to improve Ad5-O1C vaccine performance. In this regard, the incorporation of the full-length coding sequence of the non-structural protein 2B (Ad5.O1C.2B) enhanced the FMDV-specific neutralizing antibody response in swine (Pena et al., 2008) and specific vaccine-induced T cell response in cattle (Moraes et al., 2011). On the other hand, the insertion of the RGD motif (Arg–Gly–Asp) into the adenovirus fiber protein (Adt.O1C.2B.RGD) did not significantly enhance vaccine efficacy in cattle (Medina et al., 2015). However, a similar adenovirus-vectored O1/Campos/Brazil/58 vaccine [Adt.O1C.2B.F(RGD)], but in combination with ENABL® adjuvant, afforded complete protection of cattle against clinical FMD (Barrera et al., 2018). Given that naturally occurring empty capsids in infected cells are less abundant for most strains of serotype O than for serotype A (Rweyemamu et al., 1979), the lack of assembled empty capsid proteins might explain why adenovirus-vectored subunit vaccines for the O1/Campos/Brazil/58 strain are not as effective as the equivalents from serotype A or even other O subtypes.
Foot-and-mouth disease virus empty capsids are safe and as immunogenic as the intact viral particles (Rweyemamu et al., 1979). In fact, empty capsid particles are much more immunogenic than the individual capsid proteins or even partially assembled sub-viral particles (e.g., pentamers) or disrupted products (Rweyemamu et al., 1979; Doel and Chong, 1982). FMDV empty capsids comprise identical pentameric protein subunits held together by tenuous non-covalent interactions, which are unstable in mildly acidic pH conditions (Curry et al., 1995) or at elevated temperatures (Curry et al., 1997). In this respect, attempts to increase capsid acid or thermal stability by replacement of some residues by others that could strengthen inter-pentamer interactions have demonstrated to be an effective tool for the development of improved engineered vaccines against FMDV (Porta et al., 2013; Kotecha et al., 2015; Scott et al., 2017).
In the present study, we generated an optimized Ad5-vectored FMDV subunit vaccine against the O1/Campos/Brazil/58 strain by favoring the expression of FMDV capsid proteins and their self-association into virus-like particles. We analyzed several serological parameters after intramuscular inoculation with the optimized Ad5[PVP2]OP candidate in a two-dose regimen formulated without adjuvants. Our results showed an enhanced vaccine performance, as demonstrated by the higher total and neutralizing antibody responses and an improved protection upon homologous challenge in a mouse model.
Human embryonic kidney 293A (HEK 293A), Madin–Darby bovine kidney (MDBK), and baby hamster kidney 21 (BHK-21) cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml of penicillin G, and 100 μg/ml of streptomycin (Thermo Fisher Scientific). The recombinant Ad5 viruses used in this study included: Ad5[G], which expresses the enhanced green fluorescent protein (GFP) reporter gene (Romanutti et al., 2013); Ad5[P], which contains the P12A polyprotein capsid precursor and the 3Cpro coding regions (P12A3C) of the FMDV O1/Campos/Brazil/58 strain under the control of the cytomegalovirus enhancer-promoter (pCMV) (D’Antuono et al., 2010; Romanutti et al., 2013); and two novel Ad5 constructions denominated Ad5[P]OP and Ad5[PVP2]OP, which are described below. The infectious FMDV strain O1/Campos/Brazil/58 used for challenge experiments was supplied by the Argentine FMD Reference Laboratory, Servicio Nacional de Sanidad y Calidad Agroalimentaria (SENASA). All experiments involving infectious virus were performed in the BSL4-OIE facilities at the Institute of Virology, CICVyA, INTA.
Non-replicating (ΔE1/E3) optimized (OP) Ad5 vectors expressing O1/Campos/Brazil/58 (O1/Campos) FMDV capsid proteins were constructed through Gateway recombination following the manufacturer’s instructions (Gateway System, Thermo Fisher Scientific). Firstly, the pENTR[P]OP shuttle plasmid was generated by inserting the pCMV, the chimeric β-globin/IgG intron, the P12A3C sequence, and the SV40 poly(A) signal from the pCI-P12A3C plasmid (Romanutti, 2012) between the attL1–attL2 recombination sites in pENTRTM4 (Thermo Fisher Scientific). Subsequently, amino acid substitutions were introduced into the FMDV VP2 full-length sequence using a QuikChange site-directed mutagenesis kit (Agilent Technologies) using pENTR[P]OP as the template and primers containing the mutated sequences (S93Ff 5′-CAAAGGTGTCTACGGCTTCCTGACTG ACTCGTATGC-3′, S93Fr 5′-GCATACGAGTCAGTCAGGAA GCCGTAGACACCTTTG-3′, Y98Ff 5′-CCTGACTGACTCGTT TGCATATATGAGAAACGG-3′, and Y98Fr 5′-CCGTTTCT CATATATGCAAACGAGTCAGTCAGG-3′). The resulting plasmid, designated as pENTR[PVP2]OP, contains nucleotide changes (agc>ttc and tat>ttt) at positions corresponding to residues S93 and Y98 of the VP2 protein sequence. Finally, the pENTR[PVP2]OP and pENTR[P]OP shuttle vectors were recombined in vitro into the pAd/PL/V5-DESTTM to obtain the pAd5[PVP2]OP and pAd5[P]OP genomes, respectively (Figures 1B,C). The correct insertion of the foreign sequences was confirmed by restriction digestion analysis and DNA sequencing. The final plasmids (pAd5[PVP2]OP and pAd5[P]OP) were linearized with PacI and transfected into HEK 293A cells using Lipofectamine 2000 (Thermo Fisher Scientific). The recombinant Ad5[PVP2]OP and Ad5[P]OP viruses were harvested 4–10 days post-transfection when approximately 80% cytopathic effect was observed. The Ad5 vectors were subsequently propagated and titrated into HEK 293A cells. Titration of Ad5 stocks was performed by the plaque-forming unit (PFU) method according to the manufacturer’s instructions (ViraPowerTM Adenoviral Expression System, Thermo Fisher). Vector titers were expressed as PFUs per milliliter. The average titers of the adenoviral stocks used in the experiments were as follows: Ad5[P], 7.5 × 109 PFU/ml; Ad5[P]OP, 5.8 × 109 PFU/ml; and Ad5[PVP2]OP, 5.2 × 109 PFU/ml.
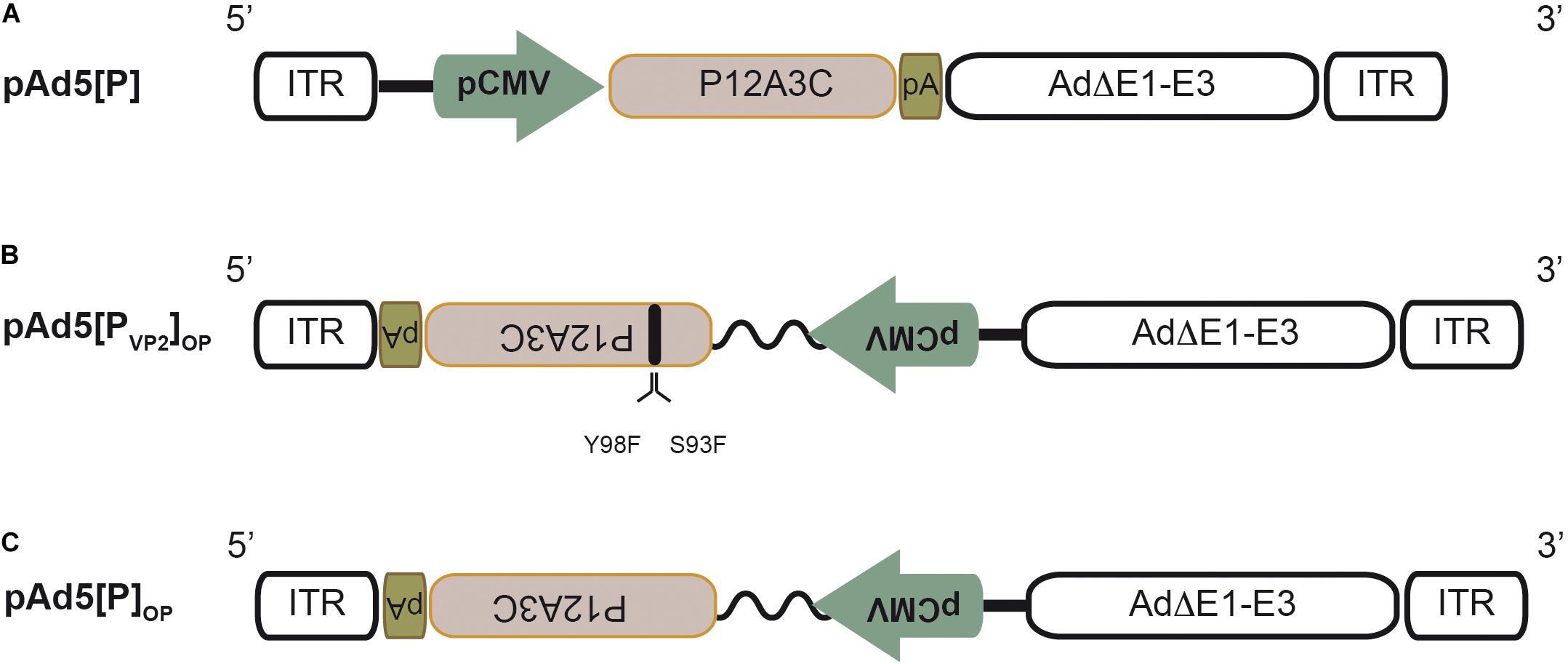
Figure 1. Schematic structure of the recombinant adenoviral genomes. (A) Recombinant pAd5[P] carrying the foot-and-mouth disease virus (FMDV) P12A3C cassette from O1/Campos strain (brown box) under the control of the cytomegalovirus enhancer-promoter (pCMV) in an E1/E3-deleted Ad5 backbone (Ad5ΔE1–E3). pA, SV40 poly(A) signal; ITR, inverted terminal repeats. (B) Recombinant pAd5[PVP2]OP. This Ad5 genome contains the expression unit at the E1 insertion site in a leftward orientation (3′→5′) in the ΔE1–E3 Ad5 backbone. The expression unit contains the FMDV P12A3C cassette under the control of the optimized pCMV (pCMV and human β-globin/IgG chimeric intron, zigzag line). Additionally, it harbors two non-synonymous substitutions in the VP2 coding sequence to express a S93F/Y98F VP2 mutant protein. (C) Recombinant pAd5[P]OP is identical to pAd5[PVP2]OP, but expresses a native VP2 capsid protein.
HEK 293A cells were infected with recombinant adenoviral particles at a multiplicity of infection (MOI) of 1. After 48 h incubation, the cells were washed with phosphate-buffered saline (PBS), lysed with 1× Laemmli buffer, and separated on 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The fractionated proteins were transferred to nitrocellulose membranes and probed with the corresponding primary antibodies. Anti-FMDV O1/Campos polyclonal guinea pig serum (Seki et al., 2009) was used as the primary antibody to detect the production of recombinant capsid proteins. Anti-β-tubulin monoclonal antibody (mAb; AC-15, Sigma-Aldrich) was used as an internal control. Horseradish peroxidase (HRP)-conjugated anti-guinea pig or anti-mouse IgG were used as secondary antibodies (Cappel Laboratories Inc.). The blots were developed by using SupersignaL® West Pico Chemiluminescent Substrate (Pierce), according to the manufacturer’s instructions. Protein band intensity was quantified by densitometry using ImageJ software (Abramoff et al., 2004).
HEK 293A and MDBK cells infected with Ad5 vectors at MOI 50 or 500, respectively, were lysed in NTE isotonic buffer (100 mM NaCl, 10 mM Tris–HCl, 1 mM EDTA, pH 7.4) containing 0.1% (v/v) Triton X-100 (Sigma-Aldrich). After 10 min incubation, the nuclei were removed by centrifugation and the supernatants were loaded onto 10–45% sucrose gradients in NTE and centrifuged at 22,000 rpm for 6 h in an SW28 rotor at 4°C. Density gradient fractions were collected and tested by enzyme-linked immunosorbent assay (ELISA) using non-cross-reactive anti-FMDV serotype O mAbs, as previously described (Seki et al., 2009). A stock of A/Argentina/2001 (A/Arg/01) inactivated FMD virus was also analyzed following the same protocol, but using an anti-A/Arg/01 polyclonal rabbit antiserum and specific mAbs against serotype A (Seki et al., 2009). Inactivated A/Arg/01 FMDV preparation was used as a marker of intact virions (146S), empty particles devoid of RNA (75S), and capsomer (12S) position in the gradient given the fact that the A/Arg/01 FMDV strain is able to generate a detectable amount of empty particles in cell cultures (Rowlands et al., 1975; Rweyemamu et al., 1979; Curry et al., 1995). For the detection of FMDV capsid proteins, eight fractions from the Ad5[PVP2]OP gradient were selected and concentrated by trichloroacetic acid (TCA) precipitation (Link and Labaer, 2011). The resultant pellets were resuspended in 1× Laemmli buffer, applied to standard 12% SDS-PAGE gels, and then transferred onto nitrocellulose membranes. The membranes were probed with an anti-FMDV O1/Campos polyclonal guinea pig serum (Seki et al., 2009) followed by the corresponding HRP-conjugated secondary antibody.
For negative-staining transmission electron microscopy (TEM), fractions containing FMDV subunits were adsorbed to carbon-coated Parlodion films and mounted on 300-mesh/in. copper grids (EMS, Fort Washington, PA, United States) for 10 min, washed once with water, and stained for 1 min with 2% phosphotungstic acid, pH 7.0 (Sigma-Aldrich). The specimens were analyzed in a transmission electron microscope (Zeiss EM 109T) equipped with a CCD camera (Ultrascan ES1000W, Gatan, Pleasanton, CA, United States) at an acceleration voltage of 100 kV.
To examine the quantity of FMDV messenger RNAs (mRNAs) accumulated in vitro, HEK 293A cells were infected with Ad5-FMD vectors at a MOI of 5 in 12-well plates. Twenty-four hours post-infection (hpi), the cells were harvested in TRIzol (Thermo Fisher Scientific).
RNA was isolated following the manufacturer’s instructions and quantified using a NanoDrop spectrophotometer. Aliquots of 200 ng of purified total RNA were treated with RQ1 DNase, RNase-free (Promega) and then reverse-transcribed with SuperScript II reverse transcriptase (Thermo Fisher Scientific) using specific primers for the 3Cpro coding sequence (3Crt 5′-CTCGTGGTGTGGTTCG-3′) and human actin (Act_r, 5′-CGTCACCGGAGTCCATCACGA-3′) housekeeping gene. The primers for the amplification of the 3Cpro or actin 110-bp DNA fragment were designed by Primer Express® software v3.0.1 (Applied Biosystems). Quantitative reverse transcription polymerase chain reactions (RT-qPCRs) were run in triplicate using 1/10th of the diluted complementary DNA (cDNA), 300 nmol of each primer (3Cf 5′-AAGATGGTCATGGGCA ACAC-3′ and 3Cr 5′-CGAGGTAAGCAGTGCCAAAC-3′ or Act_f 5′-GAGACCTTCAACACCCCAGCC-3′ and Act_r), and 10 μl of 2× FastStart Universal SYBR Green Master (Rox; Roche) in a final volume of 20 μl. PCR cycle conditions were set as follows: pre-incubation for 10 min at 95°C followed by 40 cycles, each including 15 s at 95°C and 60 s at 60°C. Dissociation curves were generated at the end of the run to verify the specificity of the reaction product. Relative quantification was performed by Applied Biosystems 7500 Real-Time PCR software v2.0.6 (Thermo Fisher Scientific) based on “cycle threshold” (Ct) values. Average 3Cpro Ct values were normalized to the average Ct values for actin, and the relative level of 3Cpro (R) was estimated as ΔΔCt-based fold change using the 2–ΔΔCt equation.
For transcript accumulation in vivo analysis, BALB/cJ mice were inoculated by the intramuscular (i.m.) route with 5 × 108 PFU of either the prior Ad5[P] or the optimized Ad5[PVP2]OP viruses (three animals per group). For transgene transcript accumulation analysis by RT-qPCR, draining popliteal lymph nodes (dPLNs) were isolated at 2 and 8 dpi, and cell suspensions were obtained in TRIzol (Thermo Fisher Scientific). cDNA synthesis was performed as described above, but using specific primers for 3Cpro (3Crt) and glyceraldehyde-3-phosphate dehydrogenase (GAPDHqr, 5′-CAGAAGGTGCGGAGATGATGA-3′) included as the housekeeping gene. RT-qPCR reactions were also run in triplicate using 1/10th of the diluted cDNA, 300 nmol of each primer (3Cf and 3Cr or GAPDHqf 5′-TGCTGGTGCCGAGTATGTTG-3′ and GAPDHqr). Average 3Cpro Ct values were normalized to the average Ct values for GAPDH, and the relative level of 3Cpro (R) was estimated as ΔΔCt-based fold change using the 2–ΔΔCt equation.
In the present study, a murine model was employed as a predictor of the immunogenicity and protection induced by Ad5-based FMD vaccines (Bidart et al., 2020; Gnazzo et al., 2020). The experiments carried out in mice reported in this manuscript have been performed following internationally recognized guidelines with the approval of the Institutional Committee for Care and Use of Experimental Animals, CICUAE (approval reference CICUAE CEVAN 01.1/2020, 02.1/2020, and INTA-CICVyA 10/2020).
Groups of 16 mice (4- to 6-week-old male BALB/cJ mice) were injected i.m. with 50 μl of the Ad5[PVP2]OP vaccine candidate (5 × 108 PFU per animal). For comparison purposes, three additional groups of mice were inoculated with Ad5[P]OP (n = 16), Ad5[P] (n = 8), or a commercial oil-adjuvanted inactivated O1/Campos strain vaccine (IV; n = 12). Animals inoculated with a recombinant Ad5 expressing GFP (Ad5[G]), considered as an unrelated antigen, were also included (n = 8). According to the 3R’s (replacement, reduction, and refinement), and due to ethical limitations, the number of animals used in each experimental group was reduced when possible, as in the case of groups inoculated with Ad5[P], Ad5[G], and IV. These groups were included only as control groups since they were already evaluated in previous works (D’Antuono et al., 2010; Romanutti et al., 2013). Table 1 summarizes the treatments given to the different groups of mice. Individual serum samples were collected from all mice at −2, 21, 35, and 45 dpi. Animal experiments were conducted in duplicate.
Anti-FMDV antibody titers were estimated by ELISA, following a modified protocol from Seki et al. (2009). Briefly, ELISA microplates (MaxiSorpTM, Nunc) were coated with a rabbit antiserum against the FMDV O1/Campos strain in carbonate–bicarbonate buffer (pH 9.6) and incubated overnight (ON) at 4°C. The inactivated O1/Campos 146S particles were then added to the wells and incubated for 1 h at 37°C. Subsequent steps were performed using a blocking buffer (PBS containing 3% horse serum and 0.05% Tween-20). Murine sera to be tested were serially diluted in blocking buffer and subsequently added to the coated ELISA plate and incubated for 1 h at 37°C. Peroxidase-labeled anti-mouse IgG, IgG1, or IgG2a antibodies were used to develop the reactions. Antibody (Ab) titers were expressed as the log10 of the reciprocal of serum dilutions giving at least twice the absorbance at 405 nm estimated in sera from animals inoculated with Ad5[G], which represents the negative control condition. Isotyping of the Ab response was carried out in sera at 45 dpi.
To examine the antigen avidity index, the plates were coated ON at 4°C with purified O1/Campos FMDV 146S particles in 50 mM carbonate/bicarbonate buffer (pH 9.6). After five washes with PBS-T (PBS with 0.05% Tween-20), 50 μl of a single dilution of serum samples (1/50) was added and incubated for 1 h at 37°C. The plates were washed twice with PBS-T and subsequently incubated with PBS–4 M urea for 15 min at room temperature, followed by two PBS-T washing steps. FMDV-specific antibodies were detected with the HRP-labeled anti-mouse conjugate incubated for 1 h at 37°C. The colorimetric reaction was revealed with the chromogen/substrate mixture ABTS/H2O2 at room temperature. The reaction was stopped after 30 min by the addition of 2% sodium fluoride and the absorbance was read at 405 nm. The optical density (OD) values of the samples were corrected by subtracting the mean blank OD values (cOD). The avidity index (AI) was calculated as the percentage of residual activity of each serum relative to the cOD of the untreated (not washed with urea) sample: AI% = (cOD sample with urea/cOD sample without urea) × 100 (Lavoria et al., 2012; Romanutti et al., 2013; Bidart et al., 2020).
Sera were examined for anti-FMDV O1/Campos neutralizing antibodies (nAbs) as described before (Quattrocchi et al., 2014). Briefly, serial dilutions of complement inactivated sera were incubated for 1 h at 37°C with 100 fifty percent tissue culture infective dose (TCID50) of the infective FMDV O1/Campos. Then, virus–serum mixtures were seeded on BHK-21 monolayers. After 40 min at 37°C, fresh DMEM 2% fetal calf serum (FCS) was added to the monolayers, which were incubated at 37°C under 5% CO2. Cytopathic effects were observed after 48 h. Titers are expressed as log10 of the reciprocal of the serum dilution that neutralizes 50% of 100 TCID50 of the infective FMDV O1/Campos using the fixed virus–variable serum method.
Mice were challenged by intraperitoneal (i.p.) inoculation of 104.5 TCID50/ml of infective FMDV O1/Campos strain per mouse. The groups of mice described in Table 1 were challenged at 49 dpi in BSL4-OIE facilities and analyzed for the presence of viremia 24 h post-challenge. Protection against FMDV was assessed as previously described, with minor modifications (Carrillo et al., 1998; Quattrocchi et al., 2011). Twenty-four hours after challenge (50 dpi), the animals were anesthetized and bled by the retro-orbital route. Heparinized blood was spread undiluted on BHK-21 cell monolayers; after virus adsorption, the monolayers were washed with sterile PBS. Fresh DMEM with 2% FCS was added and the cells were kept for 48 h at 37°C in a 5% CO2 incubator. An animal was considered protected if the cell monolayer did not present a cytopathic effect after a blind passage. Protection percentages were calculated as P% = (number of protected mice/number of challenged mice) × 100.
Bone marrow was obtained from the femurs and tibias of 8-week-old C57BL/6J males by flushing RPMI 1640 medium through the bone interior. Then, red blood cells were lysed and the cells were cultured at 37°C and 5% CO2 in six-well plates (1.5 × 106 cells per well) in 2 ml of RPMI 1640 containing 10% FBS, 50 μg/ml gentamycin (Sigma), and 20 ng/ml of recombinant GM-CSF (GenScript). The media was renovated on days 3 and 5, and on day 8 the cells were harvested and the percentage of bone marrow dendritic cells (BMDCs) was determined by the co-expression of CD11c and major histocompatibility complex (MHC) class II surface markers.
Bone marrow dendritic cells (5 × 105 cells/well) were stimulated with 500 ng/ml lipopolysaccharide (LPS) as a positive control condition or infected with Ad5[P] or Ad5[PVP2]OP at different MOIs (50 and 250) for 1 h at 37°C and 5% CO2 in RPMI 1640. Then, the supernatant was removed and RPMI 1640 containing 10% FBS was added and the cells were incubated at 37°C and 5% CO2. After 24 h, the cells were washed and resuspended in 100 μl of ice-cold PBS–azide plus anti-FcγR mAb (CD16/32; clone 93) and incubated for 30 min on ice. Then, APC anti-mouse CD11c (clone M1/70), FITC anti-mouse MHC-II (clone M5/114.15.2), PE anti-mouse CD80 (clone 16-10AI), and PerCP anti-mouse CD86 (clone PO3) mAbs were added in the recommended concentrations and incubated for additional 40 min on ice. Next, the cells were washed, fixed with 1% p-formaldehyde in PBS, and analyzed by flow cytometry. FlowMax cytometer PASIII (Partec, Münster, Germany) and Flowjo software (FlowJo, Ashland, OR, United States) were used for the analysis. All mAbs and their isotype controls were from Biolegend.
All sequences were retrieved by BLAST using FMDV O1/Campos VP2 (Uniprot accession no. Q8UZC1) as the query in the NCBI non-redundant database. Multiple sequence alignments were done by t_coffee (Di Tommaso et al., 2011) on a set of 116 non-redundant VP2 sequences and on a subset of 22 representative VP2 sequences. Amino acid conservation scores were retrieved from The ConSurf Server1 (Berezin et al., 2004; Ashkenazy et al., 2016). Both alignments exhibited comparable conservation scores at the analyzed positions. Data are available upon request. A three-dimensional (3D) model of the O1/Campos VP2 protein was created with MODELLER (Marti-Renom et al., 2000; Eswar et al., 2006) based on the full-length FMDV O1 crystal structure [Protein Data Bank (PDB) code 1QQP.3] (Fry et al., 1993). Validation of the model was carried out by ProSA (Wiederstein and Sippl, 2007) and Verify3d (Eisenberg et al., 1997). Also, the stereochemical quality of the model was assessed by the Ramachandran plot using PROCHECK (Laskowski et al., 1993). Superimposition (Maiti et al., 2004) with 1QQP.3 gave a global root mean square deviation (RMSD) of 0.32 Å across 216 equivalent positions. The interfacing region at the inter-pentamer contact of FMDV O1/Campos VP2 was modeled using the PDB coordinates 1BBT and 5DDJ as templates. All the evaluations supported the correctness of the models.
Statistical analysis was performed using the GraphPad Prism 6 software (CA, United States). Comparison between mean values of the two groups (i.e., mean Ab titers) was assessed by unpaired non-parametric Mann–Whitney test. Comparison of the RNA levels in infected cells and dPLNs as well as the activation marker levels among the different Ad5-infected BMDCs was performed using two-way analysis of variance (ANOVA). Differences were considered significant for p < 0.05.
Previously, we have described an adenovirus vector (Ad5[P]) that contains the FMDV O1/Campos P12A polyprotein and 3Cpro coding regions placed under the control of the pCMV into the E1 locus in the rightward orientation (5′→3′) relative to the Ad5 genome (D’Antuono et al., 2010; Romanutti et al., 2013). Ad5[P] (Figure 1A) was able to express and properly process the FMDV capsid proteins, but it was unable to induce protective immunity in a murine experimental model (D’Antuono et al., 2010; Romanutti et al., 2013). Therefore, to improve the Ad5-vectored O1/Campos subunit vaccine performance, we explored the impact of increasing the expression levels of FMDV capsid proteins as well as improving their ability to self-associate into VLPs on vaccine immunogenicity and efficacy. Accordingly, we engineered an optimized novel Ad5 vector that contains modifications in the FMDV autonomous expression unit.
Firstly, to enhance the expression of the O1/Campos capsid proteins, we placed the P12A3C transgene under the transcriptional control of an optimized pCMV, which comprises the pCMV and the human chimeric β-globin/IgG intron regulatory element, which is well known to produce strong intron-mediated enhancement-related effects on mRNA expression (Buchman and Berg, 1988; Wu et al., 2008; Choi et al., 2014). Additionally, in order to avoid aberrant splicing between the transgene and the adenoviral genes, the FMDV expression unit was embedded into the E1 locus in a leftward orientation (3′→5′) (Rubinchik et al., 2001; Nakai et al., 2007; Suzuki et al., 2015). Secondly, to favor the self-assembly of the FMDV O1/Campos capsid proteins into VLPs, we introduced substitutions in amino acids located in the N-terminal of the VP2 capsid protein, whose side chains are engaged in inter-pentamer interactions. In particular, the replacements S93F and Y98F were described as essential to increase the pH and thermal stability of the less stable FMD O1/Manisa and SAT2 virus particles and recombinant empty capsids (Kotecha et al., 2015; Rincon et al., 2015; Ganji et al., 2018). We surmised that introducing these mutations in both residues would favor O1/Campos empty capsid formation in situ. Therefore, we defined the presence and disposition of these relevant residues involved in VP2–VP2 contacts at the capsid inter-pentamer interfaces. To that end, we generated a molecular model of the VP2 monomer employing the 3D structure of FMDV O1BFS (99% sequence identity) as a template (PDB 1BBT). Based on the location in this model, as well as on the conservation score of the individual positions across seven representative FMDV O subtypes in multiple sequence alignments (data not shown), strictly conserved amino acids S93 and Y98 were corroborated as candidates for mutagenesis (Figure 2). By this rationale, we generated the Ad5[PVP2]OP vaccine candidate that contains the leftward-directed FMDV expression unit under the control of the optimized pCMV. Additionally, it carries two non-synonymous substitutions in the VP2 coding sequence to express a S93F/Y98F VP2 mutant protein (Figure 1B).
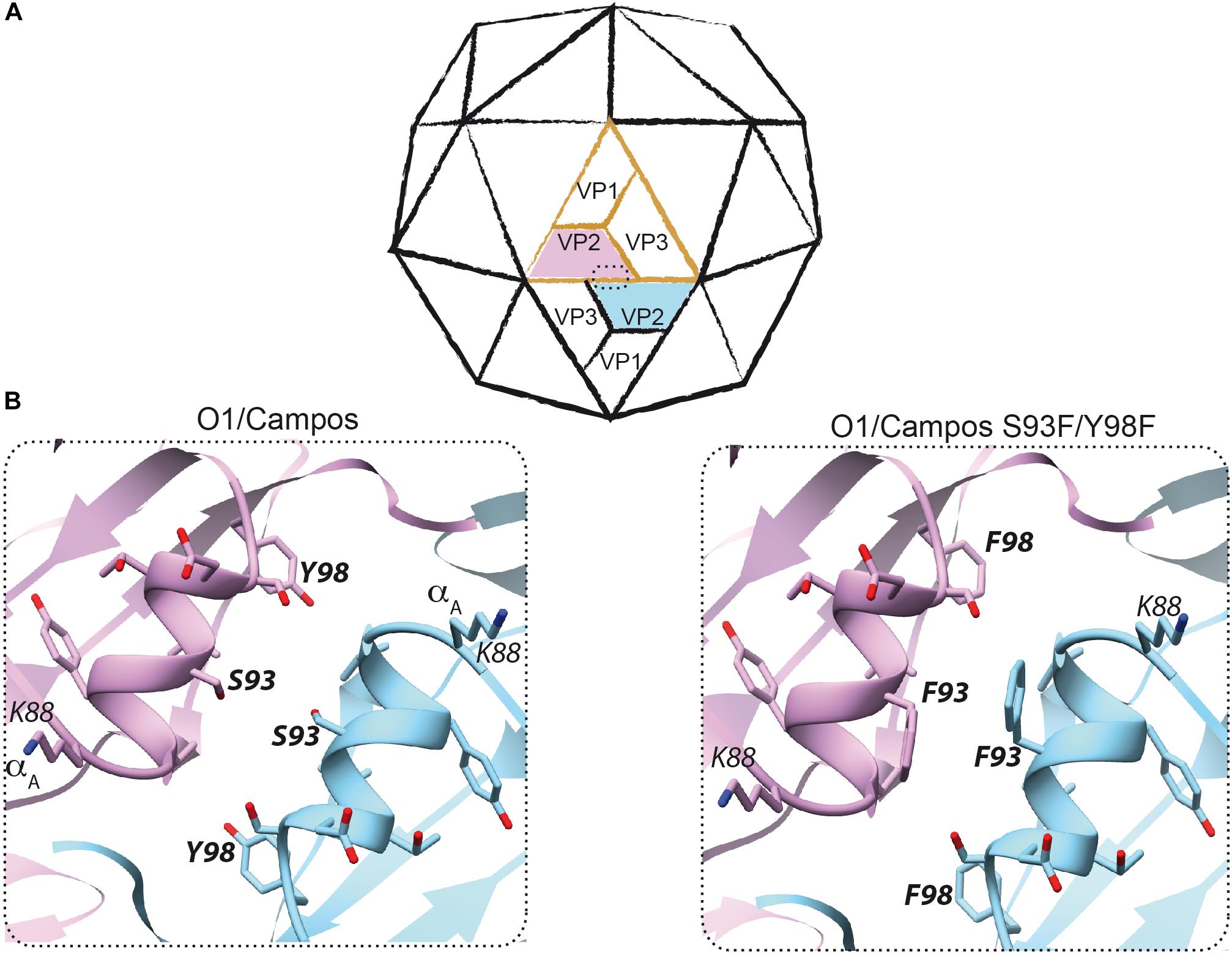
Figure 2. Rational design to produce empty capsids of foot-and-mouth disease virus (FMDV) O1/Campos. (A) Scheme of the icosahedral FMDV capsid. A protomer consisting of VP1, VP2, VP3, and the internal polypeptide VP4 (not visible) is shown in orange. The interface between two pentamers at the two-fold symmetry axis, where two neighboring VP2–VP2 interact, is enclosed within a dashed rectangle. (B) The VP2 structure of FMDV O1/Campos was modeled as described in the section “Materials and Methods”. This model led to the prediction and potential localization of residues S93 and Y98, located on the α-helix A (amino acids K88–Y98) at the two-fold symmetry axis.
Complementary, to evaluate specifically the contribution of the introduced double mutation upon the in vitro VLP assembly as well as its influence on the induction of adaptive immune response in mice, we also generated the Ad5[P]OP vector to be used as a control. Regarding expression unit structure and orientation, this vector is identical to Ad5[PVP2]OP, but expresses a wild-type VP2 capsid protein (Figure 1C).
In the first place, we evaluated the impact of the optimization approach in FMDV capsid protein expression. To that end, HEK 293A cells were infected with Ad5[PVP2]OP or Ad5[P]OP, and FMDV capsid protein accumulation was analyzed by Western blot using a polyclonal Ab against O1/Campos FMDV (Figure 3A). As expected, neither VP0, VP3, nor VP1 was detected upon infection with Ad5[G] (lane 1). A discrete band corresponding to the mature capsid proteins VP0, VP1, and VP3 was identified in lysates from Ad5[P]-, Ad5[P]OP-, and Ad5[PVP2]OP-infected cells (lane 2, 3, and 4, respectively), indicating that these vectors are able to deliver the expression of FMDV polyprotein, which is properly processed. Interestingly, cells infected with either the Ad5[PVP2]OP or Ad5[P]OP vector also yielded a quantitative stronger signal than those cells infected with Ad5[P], indicative of an increased capsid protein accumulation. Quantification by ELISA (data not shown) and densitometry (samples infected at a MOI of 1) indicated an approximately 14-fold increase in VP0 accumulation in cells infected with Ad5[PVP2]OP or Ad5[P]OP vectors as compared to Ad5[P] (Figure 3A). Moreover, no remarkable differences on the FMDV protein expression levels were observed between the cells infected with Ad5[P]OP or Ad5[PVP2]OP (lanes 3 and 4, respectively). Consistently, HEK 293A cells infected with the Ad5[P]OP or Ad5[PVP2]OP vectors accumulated significantly higher levels of 3Cpro mRNA (two-fold increase) when compared to those infected with the Ad5[P] vector (Figure 3B).
Figure 3. Analysis of foot-and-mouth disease virus (FMDV) capsid protein expression. Lysates of human embryonic kidney (HEK) 293A cells infected with Ad5[P], Ad5[P]OP, or Ad5[PVP2]OP were analyzed by SDS-PAGE and Western blot using FMDV anti-O1/Campos polyclonal serum (A). An Ad5 encoding an irrelevant antigen (green fluorescent protein), Ad5[G], was also included. To estimate FMDV structural protein accumulation, the ratios of VP0 to β-tubulin bands were calculated and normalized to the Ad5[P] VP0/β-tubulin ratio, set at 1.0. Mean values from two independent experiments are presented. Standard deviations ranged from 0% to 2% (not shown). VP0, VP1/VP3, and β-tubulin are indicated with an arrow. (B) Analysis of 3C mRNA expression in HEK 293A cells infected with Ad5[G], Ad5[P], Ad5[P]OP, or Ad5[PVP2]OP by quantitative real-time PCR. HEK 293A cells were infected with the corresponding Ad5 vector at multiplicity of infection (MOI) 5. Twenty-four hours post-infection, total RNA was isolated and used to amplify a fragment of the FMDV 3Cpro open reading frame (ORF) and, as a control, a fragment of human Actin ORF by RT-qPCR. For each experimental group, the FMDV 3Cpro transcript level relative to the Ad5[P] group was calculated by using the 2− ΔΔCt equation. Data correspond to the average and standard deviation from two independent experiments. *p < 0.01 (ANOVA).
In brief, these results showed that the insertion of the optimized leftward-directed FMDV expression unit boosts FMDV capsid protein expression through enhanced FMDV transcript levels delivered by the Ad5 vector. This phenomenon does not correlate with the introduction of S93F and Y98F substitutions within the VP2 capsid protein.
Subsequently, the effect of the targeted amino acid mutations on the FMDV VP2 capsid protein in the self-assembly of empty capsids was investigated. In preliminary experiments, we found that ultracentrifugation in sucrose gradients of lysates derived from non-permissive MDBK cells infected with Ad5[PVP2]OP showed a sedimentation pattern consistent with the generation of 75S FMDV subunits (Supplementary Figure S1).
To confirm whether this finding was specific for the VP2-introduced mutations, lysates from HEK 293A cells infected with Ad5[PVP2]OP, Ad5[P], or Ad5[P]OP were analyzed by sucrose gradient ultracentrifugation and the resulting fractions were examined using serotype-specific antigen ELISAs (Seki et al., 2009). Considering that FMDV O1/Campos does not produce easily detectable native empty particles in cell cultures, inactivated FMDV A/Argentina/2001 (A/Arg/01) was analyzed in parallel as a marker for 146S, 75S, and 12S particles. In lysates from cells infected with Ad5[P], Ad5[P]OP, or Ad5[PVP2]OP, FMDV capsid proteins predominantly associated in macromolecular complexes that sedimented at a lower rate than did the empty capsids (fractions 23–36) (Figure 4A). However, in Ad5[PVP2]OP-infected cell lysates, a fraction of the self-assembled subunits was detected at the same rate as the native empty FMDV strain A/Arg/01 capsids (75S). The absence of the 75S fraction in the lysates from cells infected with Ad5[P]OP is indicative that amino acid substitutions S93F and Y98F (introduced into the VP2 sequence) favor FMDV empty capsid particles assembly in situ. Distribution analysis of the FMDV capsid proteins throughout the sucrose gradient from HEK 293A cells infected with Ad5[PVP2]OP by Western blot revealed specific bands corresponding to VP0 (37 kDa) and VP1/VP3 (23 kDa) (Figure 4B). We roughly quantified the amount of VLPs produced in cells infected with the Ad5[PVP2]OP vector by estimating the area under the curve. According to this method, VLPs represented approximately 12 ± 5% of the total antigenic material produced in infected cells. These results were confirmed by indirect ELISA using a linear standard curve derived from a reference 146S O1/Campos FMDV (data not shown).
Figure 4. Assembly of recombinant capsid proteins into subviral particles. (A) Lysates of HEK 293A cells infected with Ad5[P], Ad5[P]OP, or Ad5[PVP2]OP were loaded onto 10–45% sucrose gradients. Inactivated native A/Argentina/2001 foot-and-mouth disease virus (iFMDV A/Arg/01) was also included as a marker (black dotted line). Fractions were collected after centrifugation at high speed and analyzed by ELISA. The positions of the FMDV virions (146S), empty capsids (75S), and capsomers (12S) are indicated. (B) Eight fractions (5, 10, 15, 20, 25, 30, 35, and 40) were analyzed by Western blot with specific anti-FMDV O1/Campos polyclonal antibody. VP0 and VP1/VP3 are indicated with an arrow. (C) Negative-staining TEM of in situ generated FMDV empty capsid particles (right) and iFMDV 146S and 75S particles (left). Scale bar, 50 nm. Virus-like particles are indicated (filled inverted triangle).
Finally, fractions were obtained from the middle of the gradients (fractions 15–20) and were analyzed by negative-staining TEM. VLPs of approximately 30 nm in diameter could be observed in the fractions derived from HEK 293A cells infected with the Ad5[PVP2]OP candidate (Figure 4C, right). These structures were indistinguishable from the native empty viral particles generated by A/Arg/01 (Figure 4C, left). Thus, the Ad5[PVP2]OP vaccine candidate favors the production of uniform round O1/Campos VLPs with a diameter of about 30 nm, which is similar to the average diameter of the authentic FMDV particles.
Our in vitro results indicated that the Ad5[PVP2]OP vaccine candidate is more efficient in supporting FMDV capsid protein expression than the initial Ad5[P] vector. Thereafter, we tested whether the use of the leftward-directed FMDV expression unit containing the optimized pCMV would enhance FMDV mRNA accumulation in vivo. To that end, mice were inoculated with Ad5[G], Ad5[P], or Ad5[PVP2]OP and FMDV 3Cpro transcript accumulation was assessed at 2 and 8 dpi in mouse dPLNs by RT-qPCR (Figure 5). At 2 dpi, animals inoculated with the Ad5[PVP2]OP candidate accumulated approximately twofold increased levels of 3Cpro transcripts compared with those inoculated with the previous Ad5[P] (p < 0.01). Furthermore, despite a decline, 3Cpro transcripts were still detected in mice inoculated with the Ad5[PVP2]OP candidate at 8 dpi. These data indicate that the Ad5[PVP2]OP vaccine candidate also drives the expression of higher levels of FMDV mRNA than the Ad5[P] vector in vivo.
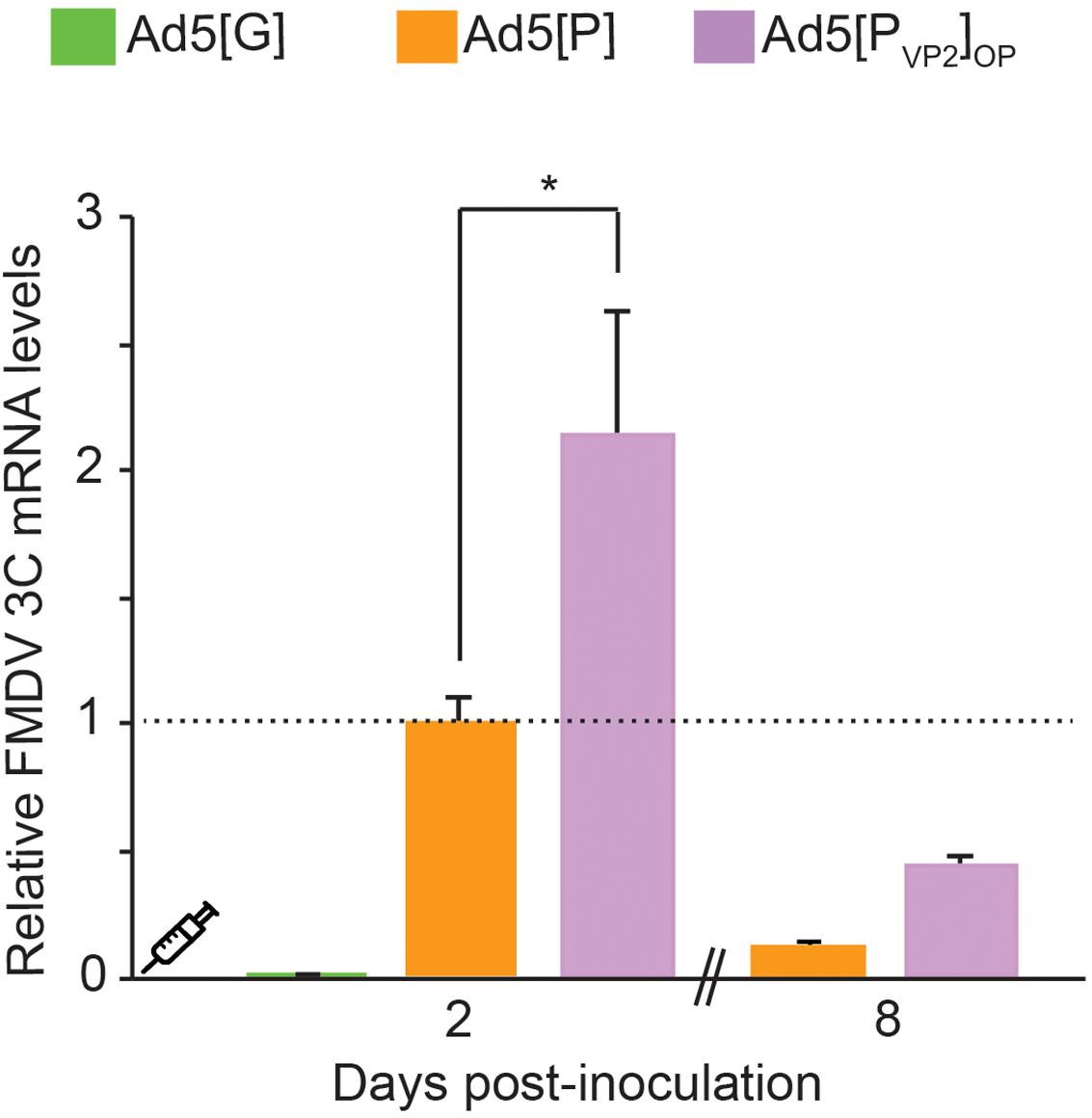
Figure 5. Comparative detection of foot-and-mouth disease virus (FMDV) transcripts in draining popliteal lymph nodes (dPLNs) of inoculated mice. Comparison of transcript accumulation levels in dPLNs after inoculation of BALB/cJ mice. Animals (n = 3) were inoculated i.m. with 5 × 108 PFU of Ad5[G], Ad5[P], or Ad5[PVP2]OP. Total RNA was extracted from dPLNs at 2 and 8 dpi, respectively, and a fragment of the FMDV 3Cpro open reading frame (ORF) and the GAPDH ORF were amplified by RT-qPCR. For each experimental group, the FMDV 3Cpro transcript level relative to the Ad5[P] group was calculated by using the 2− ΔΔCt equation. Data correspond to the average and standard deviation from two independent experiments. *p < 0.01 (ANOVA).
Immunogenicity experiments comparing the prior Ad5[P] vector and the newly optimized Ad5[PVP2]OP vaccine candidate were assessed in mice following the inoculation schedule shown in Table 1. In order to understand the nature of the immune response afforded by Ad5[PVP2]OP, we also analyzed the immunogenicity of the assembly deficient Ad5[P]OP vector. Anti-FMDV endpoint ELISA was performed with serum from individual animals bled at −2, 21, 35, and 45 dpi. As was previously reported (D’Antuono et al., 2010; Romanutti et al., 2013), after the first dose (21 dpi), significant differences in specific Ab titers could not be established between the different experimental groups of mice (data not shown). The Ab titers significantly increased in all groups after a booster immunization (35 dpi), with no further increase at 45 dpi (Figure 6A). Animals primed and boosted with the optimized vector Ad5[PVP2]OP or Ad5[P]OP developed specific FMDV Ab titers that were significantly higher than the ones elicited by the animals inoculated with Ad5[P] and similar to those by the animals primed with the oil-adjuvanted inactivated O1/Campos FMDV vaccine (p < 0.05). As expected, the administration of a vector encoding an unrelated antigen, Ad5[G], did not elicit measurable anti-FMDV antibodies (not shown).
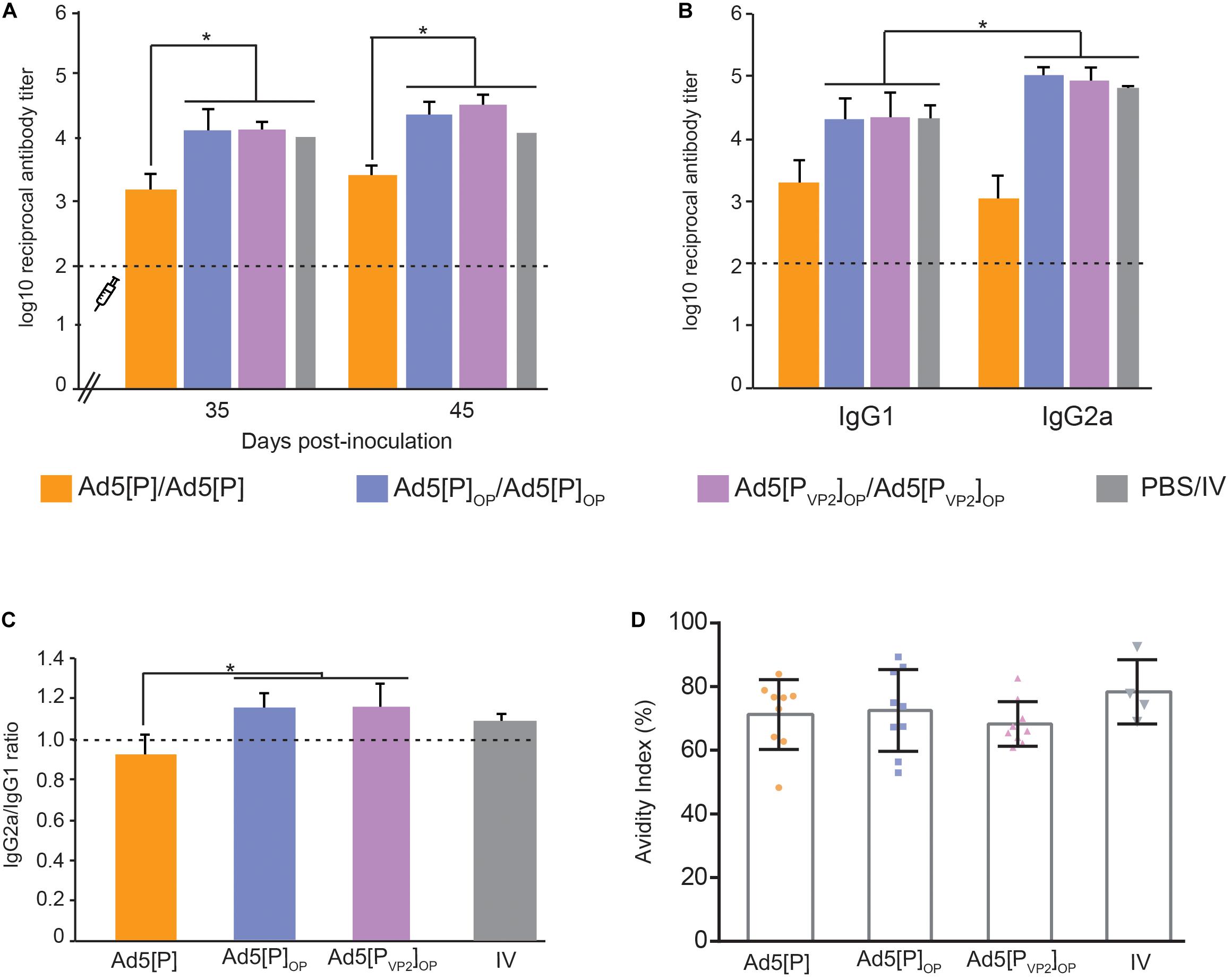
Figure 6. Immune responses in mice after the administration of Ad5[P]OP and Ad5[PVP2]OP particles. (A) Kinetics of foot-and-mouth disease (FMD) antigen-specific antibodies in BALB/cJ mice. The syringe indicates the booster inoculation (at 28 dpi). Asterisks indicate statistical significance in antibody titers (p < 0.05). FMD virus (FMDV)-specific antibody titers were determined by enzyme-linked immunosorbent assay (ELISA) in sera from animals bled at 35 and 45 days post-inoculation. (B) Isotype antibody profile induced by different Ad5 candidates. ELISA titers of anti-FMDV-specific IgG1 or IgG2a determined at 45 dpi are shown. Asterisk indicates the statistically significant difference between the IgG1 and IgG2a levels (p < 0.05). (C) IgG2a/IgG1 ratio measured in each experimental group. The consistency of the IgG ratios was assessed through the determination of the significant differences in the titers of both isotypes in each group (shown at the top of the bars). (D) Avidity of the antibodies induced by different immunogens. Serum samples from inoculated mice at 45 dpi were used to estimate the avidity index as a percentage. The average index corresponded to the optical density (OD) 405 nm of the urea-treated samples divided by the OD 405 nm of those PBS-treated. Dots correspond to values from individual animals of the indicated group; the mean value is indicated (solid line).
The Ab isotype analysis at 45 dpi revealed that the sera from animals vaccinated with both optimized vectors induced higher FMDV-specific IgG1 and IgG2a titers than Ad5[P], but interestingly, Ad5[PVP2]OP and Ad5[P]OP evoked IgG2a as the predominant isotype (p < 0.05; Figures 6B,C), similar to the animals vaccinated with the oil-adjuvanted inactivated vaccine. Therefore, Ad5[PVP2]OP and Ad5[P]OP seemed likely to induce a Th1-dependent humoral immune response and showed the highest IgG2a-to-IgG1 ratio of all the experimental groups.
To extend this analysis, we compared the avidity of specific FMDV antibodies in each experimental group. Specific antibodies elicited at 45 dpi by the inoculation of Ad5[PVP2]OP, Ad5[P]OP, or Ad5[P] had similar high avidity indexes (>50%), which did not differ significantly from those promoted by the inactivated conventional FMDV vaccine (Figure 6D).
As a whole, these data indicate that the expression levels of FMDV antigens driven by the Ad5[PVP2]OP and Ad5[P]OP vectors were sufficient to elicit a strong immune response comparable to the oil-adjuvanted inactivated O1/Campos FMDV commercial vaccine.
According to the immunization scheme shown in Table 1, animals were challenged at 49 dpi with the infective O1/Campos virus strain and were analyzed for the presence of viremia 24 h post-challenge. As shown in Figure 7A, protection was found in 62.5% (10 out of 16) of the Ad5[P]OP group and 94% (15 out of 16) of the Ad5[PVP2]OP group. No protection (0%) was found in the groups inoculated with Ad5[P] or Ad5[G], and 100% protection was observed in animals inoculated with one dose of the inactivated commercial FMDV vaccine (12 out of 12).
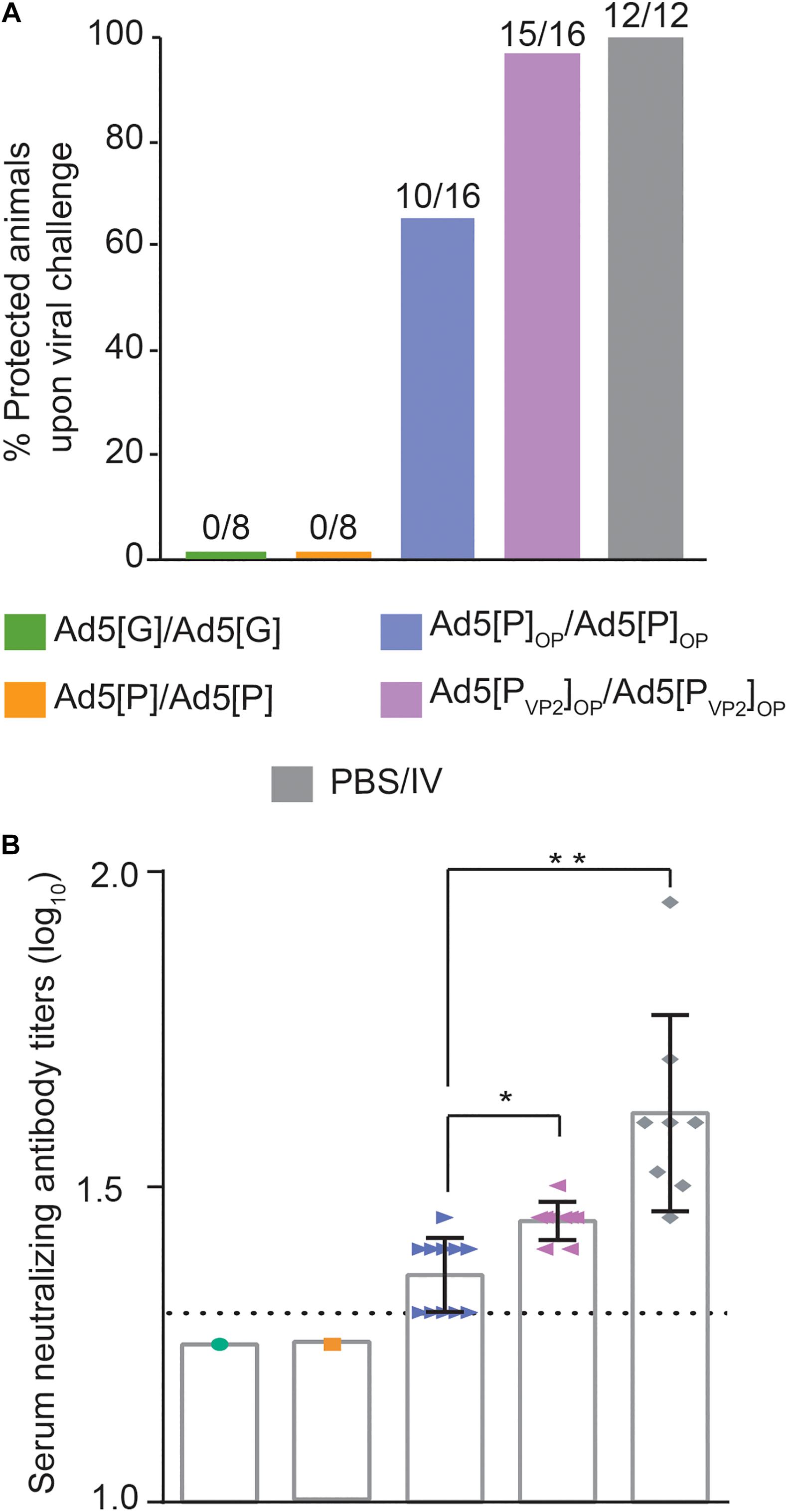
Figure 7. Protection against challenge with infectious foot-and-mouth disease virus (FMDV) O1/Campos. (A) Protection from viral challenge elicited by the different Ad5-FMD vectors. Groups of mice were inoculated with Ad5[P], Ad5[P]OP, Ad5[PVP2]OP, Ad5[G], or a commercial FMD vaccine and challenged with infective FMDV at 49 dpi. Animals were considered protected if viremia was absent after a blind passage. Protection percentages were calculated as 100 × (number of vaccinated animals without viremia/number of vaccinated animals). (B) Virus neutralization test (VNT) at day 45 post-inoculation with 5 × 108 PFU, expressed as log10 of the reciprocal of the serum dilution that neutralizes 50% of 100 TCID50 of infective FMDV O1/Campos (fixed virus–variable serum method). *p < 0.05, **p < 0.01 with respect to the Ad5[P] group. A serum pool was made every two animals of a group in order to perform the serum neutralization test.
Animals vaccinated with either the Ad5[PVP2]OP or Ad5[P]OP vector developed specific FMDV-neutralizing Ab responses (Figure 7B). As expected, the highest nAb titers were found in mice inoculated with the inactivated commercial FMDV vaccine (1.60 ± 0.15). However, the mean of the nAb titer of the group inoculated with Ad5[P]OP remained near the test detection limit (1.36 ± 0.02), while the mean titer of the group inoculated with the Ad5[PVP2]OP vaccine candidate (1.45 ± 0.03) was significantly higher than that of Ad5[P]OP (p = 0.0175) and was consistent with the afforded protection (Figure 7A). For animals of the non-protected groups (Ad5[P] and Ad5[G]), the nAb titers were found to be below the assay detection limit (<1.3).
Dendritic cells are potent antigen-presenting cells that initiate and modulate the host immune response. Our in vivo data showed that Ad5[PVP2]OP is highly immunogenic and is the most efficient in conferring protection after homologous challenge. Since Ad5[P] and Ad5[PVP2]OP are antigenically equivalent and only differ in their ability to deliver FMDV subunits, we sought to determine the effect of these gene transfer vectors on the activation phenotype of mouse BMDCs. To that end, BMDCs were infected with Ad5[P] or Ad5[PVP2]OP at different MOIs (50 or 250) or mock-infected. Twenty-four hours later, the BMDC phenotype was assessed by using flow cytometry analysis for CD80, CD86, and MHC-II surface expression. As expected, treatment of mouse BMDCs with LPS, included as a maturation control, resulted in a significant upregulation of MHC-II, CD80, and CD86 expression (Figure 8). Infection of mouse BMDCs with Ad5[P] or Ad5[PVP2]OP at MOI 50 did not induce the upregulation of any of the studied surface markers. In contrast, infection with Ad5[P] or Ad5[PVP2]OP at MOI 250 led to a twofold increase of the MHC-II, CD80, and CD86 expression levels as compared to non-infected BMDCs. Overall, these results suggest that BMDC activation depended on a threshold dose since no effect on maturation was apparent at a MOI of 50, and the maturation rate at a MOI of 250 was similar to that induced by LPS. Notably, both Ad5[P] and Ad5[PVP2]OP stimulated a strong activation of mouse BMDCs.
Figure 8. Bone marrow dendritic cell (BMDC) activation induced by Ad5[P] or Ad5[PVP2]OP infection. BMDCs were infected with Ad5[P] or Ad5[PVP2]OP at multiplicity of infection (MOI) of 50 and 250, as described in the section “Materials and Methods.” Then, the cells were stained with specific monoclonal antibodies (mAbs) for phenotypic and activation markers and analyzed by flow cytometry. (A) Representative histograms depict the fluorescence intensities of CD80, CD86, and major histocompatibility complex (MHC) class II molecules on MHCII+CD11c+ gated population. (B) Bars represent the geometric mean of fluorescence intensity (MFI) ± SD. Isotype, cells were stained with control isotype mAbs; LPS-stimulated, cells were treated with 500 ng/ml lipopolysaccharide (LPS). *p < 0.05, **p < 0.01, ***p < 0.001 compared to non-stimulated cells (ANOVA).
Although recombinant adenovirus vectors have demonstrated promising capabilities as vaccine candidates for several FMDV strains (Grubman et al., 2009), it has been more difficult to achieve the same results with some strains of serotype O, specifically for the epidemiologically relevant O1/Campos/Brazil/58 strain (Caron et al., 2005; D’Antuono et al., 2010; Moraes et al., 2011; Romanutti et al., 2013; Medina et al., 2015).
It is known that the performance of FMDV vaccines is dependent on the amount of intact viral particles (146S) (Terry et al., 1982) or natural empty capsids (75S) (Rweyemamu et al., 1989) in the vaccine’s preparation. In this study, we explored the hypothesis that enhanced expression of the FMDV capsid proteins and their self-association into VLPs would improve the immunogenicity and efficacy of the Ad5-FMD O1/Campos vaccine. Thereby, a novel recombinant Ad5-vectored vaccine (Ad5[PVP2]OP) was engineered by modifying the sense of transcription of the foreign expression unit and by expressing the FMDV proteins from an optimized pCMV. The hypothesis was further co-implemented with the introduction of specific amino acid replacements (S93F and Y98F) within the N-terminal of VP2, reported to stabilize the generated empty capsids (Kotecha et al., 2015).
The in vitro data reported in this work support the working hypothesis and revealed a substantial increase (∼14-fold) in FMDV capsid protein accumulation in cells transduced with the optimized Ad5[PVP2]OP (and also Ad5[P]OP) vector compared with those cells transduced by our group’s first-generation vector, Ad5[P] (Figure 3A). Moreover, the Ad5[PVP2]OP vaccine candidate was able to deliver an increased FMDV transcript accumulation (Figure 3B). Therefore, these results showed that the insertion of the heterologous transcription unit in the leftward orientation relative to the Ad5 genome, and the expression of FMDV sequences from an optimized pCMV, led to an increase in mRNA and capsid protein accumulation levels in transduced cells. The data was very relevant since Pena et al. (2008) reported a 1.5- to 1.7-fold increase when using an identical CMV enhancer/promoter and chimeric intron combination to drive the expression of FMDV A24 Cruzeiro capsid proteins, indicating that the positioning of the optimized P12A3C expression unit in an opposite transcriptional orientation relative to the Ad5 genome is a suitable additional strategy for optimizing FMDV gene expression in vitro (Rubinchik et al., 2001; Nakai et al., 2007; Suzuki et al., 2015).
As predicted, the Ad5[PVP2]OP vaccine candidate was able to drive the assembly of FMDV O1/Campos VLPs in infected permissive and non-permissive cells, while Ad5[P] and Ad5[P]OP were not. Sucrose gradients of Ad5[PVP2]OP-infected cells showed a FMDV protein fraction sedimenting at the same rate as the native empty particles (75S) of the A/Arg/01 strain produced in culture (Figure 4A and Supplementary Figure S1). Indeed, examination by negative-staining TEM revealed that the O1/Campos VLPs closely resemble the native FMD empty capsid particles in size and overall particle morphology (Figure 4C). Thus, the introduced double-phenylalanine substitution at positions S93 and Y98 increased the yields of assembly of FMDV O1/Campos empty capsids. The phenotype exhibited by this mutated VP2 confirmed previously reported results indicating that a 10-residue region spanning α-helix A (amino acids 88–98 for the O serotype) of VP2 protein is relevant for the infectivity and stability of FMDV (Ellard et al., 1999; Porta et al., 2013; Kotecha et al., 2015; Ganji et al., 2018). Furthermore, these studies have shown that residues within this region are responsible for non-covalent interactions between adjacent pentamers at the icosahedral two-fold axis, where neighboring VP2–VP2 proteins interact. As shown in the O1/Campos VP2 model, these residues are likely committed to local inter-pentameric subunit associations (Figure 2).
The serological data reinforce the initial hypothesis since the amount of FMDV transgene expressed by Ad5[PVP2]OP (and Ad5[P]OP) is sufficient to elicit a strong immune response in mice, comparable to the oil-adjuvanted inactivated O1/Campos FMDV vaccine. It is of note that the humoral and protective responses against FMDV detected in our murine model have a good established correlation with cattle (Habiela et al., 2014; Bidart et al., 2020; Gnazzo et al., 2020). We demonstrated that the Ad5[PVP2]OP vaccine candidate elicited an enhanced antibody response as compared to our first-generation vector, Ad5[P] (Figures 6A, 7B). Notably, 94% of the animals inoculated with Ad5[PVP2]OP were completely protected from a severe FMD virus challenge (Figure 7A), which suggests that, at a similar dose of the tried Ad5-vectored vaccines, Ad5[PVP2]OP provides additional expression of the FMDV proteins and/or ability in antigen presentation to immune cells sufficient to protect the vaccinated animals. It is known that a higher antigenic mass of the O1/Campos strain is required to achieve equivalent potency in inactivated vaccines as compared to other serotype antigens (Doel et al., 1994; Doel, 2003; Pena et al., 2008). It appears that the efficacy of the Ad5-vectored O1/Campos vaccine is also dependent on the amount of transgene expressed in transduced cells. In this regard, significant differences in transgene expression were detected in the dPLN populations isolated from mice inoculated with Ad5[PVP2]OP or Ad5[P] (Figure 5). While the levels of RNA do not necessarily correlate with the protein expression levels determined in vitro (Figure 3), these differences in FMDV RNA levels could explain the improved efficacy and the higher potency elicited by the Ad5[PVP2]OP candidate in vivo as compared to the Ad5[P] vector.
The levels of neutralizing antibodies were markedly higher in mice inoculated with Ad5[PVP2]OP than in those inoculated with Ad5[P] (Figure 7B). Although serum-neutralizing antibodies have been considered the most relevant parameter that correlates with protection against FMDV (Doel, 2003; Paton et al., 2005; World Organisation for Animal Health, 2012), the contribution of non-neutralizing antibodies in the protection against infection and disease has also been described (McCullough et al., 1992; Pay and Hingley, 1992; Dunn et al., 1998). In this sense, higher levels of isotype IgG1 and IgG2a specific antibodies were quantified in the groups inoculated with the optimized vectors than in those mice inoculated with Ad5[P] (Figures 6B,C). These increased IgG1 and IgG2a specific antibodies may have played a relevant role in FMDV opsonization and its subsequent clearance by phagocytic cells of the immune system (McCullough et al., 1988). Further studies to explore the contribution of other mechanisms involved in the protection induced by Ad5[PVP2]OP in the mouse model, as the induction of a detectable T cell response, are needed.
Dendritic cells (DCs) play an important role in both the initiation of innate immunity and the development of adaptive immunity (Hochrein and O’Keeffe, 2008). Previous reports have demonstrated that recombinant Ad5 induces the maturation of dendritic cells by upregulating the expression of major histocompatibility complex class I and II antigens, co-stimulatory molecules (CD40, CD80, and CD86), and the adhesion molecule CD54 (ICAM-1) (Morelli et al., 2000). The preliminary shown in this work suggested that BMDCs infected with Ad5[P] or Ad5[PVP2]OP upregulated the expression of MHC class II and co-stimulatory molecules (CD80 and CD86) to a similar extent (Figure 8). This data adds evidence to previous reports that demonstrated that an unequal relationship exists between the rate of transgene expression and the extent of DC maturation (Morelli et al., 2000; Miller et al., 2003; Philpott et al., 2004). FMDV structural and non-structural proteins have been shown to antagonize the host innate response in mammalian cells by preventing the translational (Guzylack-Piriou et al., 2006) or transcriptional (de Los Santos et al., 2006) regulation of host proteins. Although Ad5 vectors expressing FMDV proteins seem to induce BMDCs to adopt an activated phenotype, it remains unclear whether other biological functions of these cells, which include antigen uptake, migration to regional lymph nodes, and antigen-specific T-lymphocyte activation, are affected.
When the optimized vectors were compared in mouse challenge experiments, a remarkable finding was the ability of the Ad5[PVP2]OP candidate to confer higher protection than Ad5[P]OP (94% and 60%, respectively). Evaluation of specific parameters of the humoral immune response such as total specific antibody titers, isotype balance, and specific antibody avidity indicated no significant difference in these attributes (Figure 6). In this scenario, differences in other responses such as innate and cellular immunity cannot be discarded. For example, an increased antibody-dependent phagocytosis (Lannes et al., 2012; Quattrocchi et al., 2011) and T CD8-mediated cytotoxicity (Guzman et al., 2010; Patch et al., 2013) may exert a synergistic effect, explaining the higher protection afforded by Ad5[PVP2]OP. Furthermore, our results may indicate that elements in addition to transgene expression levels are likely to have the potential to significantly influence the enhanced resistance to challenge in Ad5[PVP2]OP-vaccinated animals. It is then tempting to suggest that the superior ability of Ad5[PVP2]OP in the assembly of FMDV VLPs shown in vitro may have accounted for its enhanced efficacy in vivo.
One of the foremost factors reported to influence the potency of vaccine preparations is the structural integrity of the whole (146S) FMD viral particles, mainly due to the capacity of this antigen to elicit antibodies against exposed epitopes that can drive to viral clearance in the case of a natural infection (Cartwright et al., 1982; Doel and Chong, 1982; Rao et al., 1994). In particular, neutralizing antibody production is associated mainly with 146S particles (Bahnemann, 1975; Pay and Hingley, 1992), whereas disrupted viral particles (12S pentamers) are less immunogenic and elicit a diminished neutralizing response (Doel and Chong, 1982; Rao et al., 1994). It has recently been demonstrated that FMDV-specific production of IFN-γ is strongly affected by the integrity of the viral capsids (Bucafusco et al., 2015). Additionally, FMDV-specific IFN-γ production in vaccinated cattle has been associated with the activation of antigen-specific CD4+ T cells (Oh et al., 2012), which are responsible for supporting antibody-neutralizing responses (Carr et al., 2013). In this regard, a comparative analysis of the antigenic reactivity with a panel of 13 mAbs specific for FMDV serotype O (Seki et al., 2009) revealed that the FMDV subunits launched by Ad5[PVP2]OP exhibit an improved reactivity with neutralizing and non-neutralizing mAbs as compared to those antigens generated by the assembly deficient Ad5[P]OP (Supplementary Figure S2). Moreover, we demonstrated that the Ad5[PVP2]OP vaccine candidate elicited moderate but significantly higher titers of nAb than did Ad5[P]OP (Figure 7B). Therefore, it is tempting to speculate that FMDV subunits generated by the Ad5[PVP2]OP vaccine candidate form an oligomeric protein scaffold capable of presenting immunogenic epitopes and proteins in a more efficient manner, similar to the native FMD virion.
Traditional inactivated FMDV vaccines have played and still play a preponderant role in preventing and controlling the disease worldwide (Caridi et al., 2017; Khalifa et al., 2017). However, the drawbacks associated with its production, the duration of immunity, the necessity of a strict cold chain to preserve particle stability, among others, have led to continuous attempts to develop a large variety of alternative FMD vaccines. In this respect, we have generated the optimized Ad5[PVP2]OP candidate which launches the expression of high levels of FMDV O1/Campos subunits that partially self-associate into VLPs. The results herein demonstrated the remarkable antigenicity and efficacy of this platform in a very well-supported murine model. Notwithstanding, the potency of the Ad5[PVP2]OP vaccine candidate should be further studied in target species, such as pigs or cattle, whenever possible.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by 1. Institutional Committee for Care and Use of Experimental Animals (CICUAE) Affiliation: Instituto de Virología, Centro de Investigaciones en Ciencias Veterinarias, Instituto Nacional de Tecnología Agropecuaria, (INTA-CICVyA), N. Repetto y De Los Reseros s/n, Hurlingham (1686), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Buenos Aires, Argentina. 2. Institutional Committee for Care and Use of Experimental Animals (CIEMAE) Affiliation: Centro de Virología Animal, Consejo Nacional de Investigaciones Científicas y Técnicas (CEVAN-CONICET), Saladillo 2468, C1440FFX, Buenos Aires, Argentina.
AD and NM developed the idea and design of the study. MZ, JB, and CP performed the experiments. MZ and AD analyzed the data and interpreted the results. AD and NM wrote the original draft. MZ, JB, CP, PZ, MT, and NM wrote and critically revised the manuscript. All authors read and approved the final manuscript.
CEVAN-CONICET and INTA are members of the Inter-Institutional FMD Research and Development Network of Argentina (RIIDFA). This work was supported in part by FONCyT (PID2013-0022 and PICT2016-0810), CONICET, and the Fundación de Estudios en Virología Animal (FEVAN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank O. Periolo, C. Seki, B. Robiolo, C. De Vicenzo, M. Gonzalez, J. Acevedo, S. Rojana, and I. García Briones for their technical assistance. We also thank the reviewers of this manuscript for their constructive comments and suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.591019/full#supplementary-material
Supplementary Figure 1 | Assembly of recombinant structural proteins into subviral particles. Lysates of MDBK cells infected with Ad5[PVP2]OP were loaded onto 10–45% sucrose gradients. Fractions were collected and analyzed by ELISA. Inactivated native A/Argentina/2001 FMDV (iFMDV A/Arg/01) was included as a marker (black dotted line). The positions of the FMDV virions (146S), empty capsids (75S) and capsomers (12S) are indicated.
Supplementary Figure 2 | Antigenic reactivity of FMDV subunits launched by Ad5[P]OP and Ad5[PVP2]OP. Lysates of HEK 293A cells infected with Ad5[P]OP or Ad5[PVP2]OP were analyzed by using specific mAbs for O1/Campos FMDV by ELISA. The reactivities of FMDV subunits against several mAbs are shown as the absorbance values (OD 405 nm) detected for each specific mAb. The antigenic reactivity for inactivated O1/Campos virus (iFMDV O1/Campos) was included as reference sample.
Abramoff, M. D., Magalhaes, P. J., and Ram, S. J. (2004). Image processing with ImageJ. Biophoton. Int. 11, 36–42.
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. doi: 10.1093/nar/gkw408
Bahnemann, H. G. (1975). Binary ethylenimine as an inactivant for foot-and-mouth disease virus and its application for vaccine production. Arch. Virol. 47, 47–56. doi: 10.1007/BF01315592
Barrera, J., Brake, D. A., Schutta, C., Ettyreddy, D., Kamicker, B. J., Rasmussen, M. V., et al. (2018). Versatility of the adenovirus-vectored foot-and-mouth disease vaccine platform across multiple foot-and-mouth disease virus serotypes and topotypes using a vaccine dose representative of the AdtA24 conditionally licensed vaccine. Vaccine 36, 7345–7352. doi: 10.1016/j.vaccine.2018.10.031
Berezin, C., Glaser, F., Rosenberg, J., Paz, I., Pupko, T., Fariselli, P., et al. (2004). ConSeq: The identification of functionally and structurally important residues in protein sequences. Bioinformatics 20, 1322–1324. doi: 10.1093/bioinformatics/bth070
Bidart, J. E., Kornuta, C., Gammella, M., Gnazzo, V., Soria, I., Langellotti, C. A., et al. (2020). A new cage-like particle adjuvant enhances protection of foot and mouth disease vaccine. Front. Vet. Sci. 7:396. doi: 10.3389/fvets.2020.00396
Brito, B. P., Rodriguez, L. L., Hammond, J. M., Pinto, J., and Perez, A. M. (2017). Review of the Global Distribution of foot-and-mouth disease virus from 2007 to 2014. Transbound Emerg Dis. 64, 316–332. doi: 10.1111/tbed.12373
Bucafusco, D., Di Giacomo, S., Pega, J., Schammas, J. M., Cardoso, N., Capozzo, A. V., et al. (2015). Foot-and-mouth disease vaccination induces cross-reactive IFN-gamma responses in cattle that are dependent on the integrity of the 140S particles. Virology 476, 11–18. doi: 10.1016/j.virol.2014.11.023
Buchman, A. R., and Berg, P. (1988). Comparison of intron-dependent and intron-independent gene expression. Mol. Cell Biol. 8, 4395–4405. doi: 10.1128/mcb.8.10.4395
Caridi, F., Vázquez-Calvo, Á, Borrego, B., McCullough, K. C., Summerfield, A., Sobrino, F., et al. (2017). Preserved immunogenicity of an inactivated vaccine based on foot-and-mouth disease virus particles with improved stability. Vet. Microbiol. 203, 275–279. doi: 10.1016/j.vetmic.2017.03.031
Caron, L., Brum, M. C. S., Moraes, M. P., Goldei, W. T., and Grubman, M. J. (2005). Granulocyte-macrophage colony-stimulating factor does not increase the potency or efficacy of a foot-and-mouth disease virus subunit vaccine. Pesq. Vet. Bras. 25, 150–158.
Carr, B. V., Lefevre, E. A., Windsor, M. A., Inghese, C., Gubbins, S., Prentice, H., et al. (2013). CD4+ T-cell responses to foot-and-mouth disease virus in vaccinated cattle. J. Gen. Virol. 94, 97–107. doi: 10.1099/vir.0.045732-45730
Carrillo, C., Wigdorovitz, A., Oliveros, J. C., Zamorano, P. I., Sadir, A. M., Gomez, N., et al. (1998). Protective immune response to foot-and-mouth disease virus with VP1 expressed in transgenic plants. J. Virol. 72, 1688–1690.
Cartwright, B., Morrell, D. J., and Brown, F. (1982). Nature of the antibody response to the foot-and-mouth disease virus particle, its 12S protein subunit and the isolated immunizing polypeptide VP1. J. Gen. Virol. 63, 375–381. doi: 10.1099/0022-1317-63-2-375
Centro Panamericano de Fiebre Aftosa, OPS, and OMS. (2019). Informe de Situación de los Programas de Erradicación de la Fiebre Aftosa en Sudamérica y Panamá, año 2018. Washington, D.C: Organización Panamericana de la Salud.
Choi, J. H., Yu, N. K., Baek, G. C., Bakes, J., Seo, D., Nam, H. J., et al. (2014). Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain 7:17. doi: 10.1186/1756-6606-7-17
Curry, S., Abrams, C. C., Fry, E., Crowther, J. C., Belsham, G. J., Stuart, D. I., et al. (1995). Viral RNA modulates the acid sensitivity of foot-and-mouth disease virus capsids. J. Virol. 69, 430–438. doi: 10.1128/JVI.69.1.430-438.1995
Curry, S., Fry, E., Blakemore, W., Abu-Ghazaleh, R., Jackson, T., King, A., et al. (1997). Dissecting the roles of VP0 cleavage and RNA packaging in picornavirus capsid stabilization: the structure of empty capsids of foot-and-mouth disease virus. J. Virol. 71, 9743–9752. doi: 10.1128/JVI.71.12.9743-9752.1997
D’Antuono, A., Laimbacher, A. S., La Torre, J., Tribulatti, V., Romanutti, C., Zamorano, P., et al. (2010). HSV-1 amplicon vectors that direct the in situ production of foot-and-mouth disease virus antigens in mammalian cells can be used for genetic immunization. Vaccine 28, 7363–7372. doi: 10.1016/j.vaccine.2010.09.011
de Los Santos, T., De Avila Botton, S., Weiblen, R., and Grubman, M. J. (2006). The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 80, 1906–1914. doi: 10.1128/JVI.80.4.1906-1914.2006
Di Tommaso, P., Moretti, S., Xenarios, I., Orobitg, M., Montanyola, A., Chang, J. M., et al. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17. doi: 10.1093/nar/gkr245
Diaz-San Segundo, F., Montiel, N. A., Sturza, D. F., Perez-Martin, E., Hickman, D., Ramirez-Medina, E., et al. (2016). Combination of Adt-O1Manisa and Ad5-boIFNlambda3 induces early protective immunity against foot-and-mouth disease in cattle. Virology 499, 340–349. doi: 10.1016/j.virol.2016.09.027
Doel, T. R., and Chong, W. K. (1982). Comparative immunogenicity of 146S, 75S and 12S particles of foot-and-mouth disease virus. Arch. Virol. 73, 185–191. doi: 10.1007/BF01314726
Doel, T. R., Williams, L., and Barnett, P. V. (1994). Emergency vaccination against foot-and-mouth disease: rate of development of immunity and its implications for the carrier state. Vaccine 12, 592–600. doi: 10.1016/0264-410x(94)90262-90263
Dunn, C. S., Samuel, A. R., Pullen, L. A., and Anderson, J. (1998). The biological relevance of virus neutralisation sites for virulence and vaccine protection in the guinea pig model of foot-and-mouth disease. Virology 247, 51–61. doi: 10.1006/viro.1998.9175
Eisenberg, D., Lüthy, R., and Bowie, J. U. (1997). VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 277, 396–404. doi: 10.1016/s0076-6879(97)77022-77028
Ellard, F. M., Drew, J., Blakemore, W. E., Stuart, D. I., and King, A. M. Q. (1999). Evidence for the role of His-142 of protein 1C in the acid-induced disassembly of foot-and-mouth disease virus capsids. J. Gen. Virol. 80(Pt 8), 1911–1918. doi: 10.1099/0022-1317-80-8-1911
Eswar, N., Webb, B., Marti-Renom, M. A., Madhusudhan, M. S., Eramian, D., Shen, M. Y., et al. (2006). Comparative protein structure modeling using modeller. Curr. Protoc. Bioinform. Chapter 5:Unit–5.6. doi: 10.1002/0471250953.bi0506s15
FAO (2020). Food-and-mouth Disease, Quarterly Report (April-June 2020): FAST Reports: Foot-and-mouth and Similar Transboundary Animal Diseases. Available online at: http://www.fao.org/3/cb0479en/cb0479en.pdf
Fernández-Sainz, I., Medina, G. N., Ramirez-Medina, E., Koster, M. J., Grubman, M. J., and De Los Santos, T. (2017). Adenovirus-vectored foot-and-mouth disease vaccine confers early and full protection against FMDV O1 manisa in swine. Virology 502, 123–132. doi: 10.1016/j.virol.2016.12.021
Fry, E., Acharya, R., and Stuart, D. (1993). Methods used in the structure determination of foot-and-mouth disease virus. Acta Crystallogr A 49(Pt 1), 45–55. doi: 10.1107/s0108767392005737
Ganji, V. K., Biswal, J. K., Lalzampuia, H., Basagoudanavar, S. H., Saravanan, P., Tamil Selvan, R. P., et al. (2018). Mutation in the VP2 gene of P1-2A capsid protein increases the thermostability of virus-like particles of foot-and-mouth disease virus serotype O. Appl. Microbiol. Biotechnol. 102, 8883–8893. doi: 10.1007/s00253-018-9278-9279
Gnazzo, V., Quattrocchi, V., Soria, I., Pereyra, E., Langellotti, C., Pedemonte, A., et al. (2020). Mouse model as an efficacy test for foot-and-mouth disease vaccines. Transbound Emerg Dis. doi: 10.1111/tbed.13591 Online ahead of print.
Grubman, M. J., and Baxt, B. (2004). Foot-and-mouth disease. Clin. Microbiol. Rev. 17, 465–493. doi: 10.1128/cmr.17.2.465-493.2004
Grubman, M. J., Moraes, M. P., Schutta, C., Barrera, J., Neilan, J., Ettyreddy, D., et al. (2009). Adenovirus serotype 5-vectored foot-and-mouth disease subunit vaccines: the first decade. Fut. Virol. 5:51. doi: 10.2217/fvl.09.68
Guzman, E., Taylor, G., Charleston, B., and Ellis, S. A. (2010). Induction of a cross-reactive CD8+ T cell response following foot-and-mouth disease virus vaccination. J. Virol. 84, 12375–12384. doi: 10.1128/JVI.01545-1510
Guzylack-Piriou, L., Bergamin, F., Gerber, M., McCullough, K. C., and Summerfield, A. (2006). Plasmacytoid dendritic cell activation by foot-and-mouth disease virus requires immune complexes. Eur. J. Immunol. 36, 1674–1683. doi: 10.1002/eji.200635866
Habiela, M., Seago, J., Perez-Martin, E., Waters, R., Windsor, M., Salguero, F. J., et al. (2014). Laboratory animal models to study foot-and-mouth disease: a review with emphasis on natural and vaccine-induced immunity. J. Gen. Virol. 95, 2329–2345. doi: 10.1099/vir.0.068270-68270
Hochrein, H., and O’Keeffe, M. (2008). Dendritic Cell Subsets and Toll-Like Receptors. Berlin: Springer.
Khalifa, M., El-Deeb, A., Zeidan, S., Hussein, H., and Abu-El-Naga, H. (2017). Enhanced protection against FMDV in cattle after prime- boost vaccination based on mucosal and inactivated FMD vaccine. Vet. Microbiol. 210, 1–7. doi: 10.1016/j.vetmic.2017.08.014
Knight-Jones, T. J., Robinson, L., Charleston, B., Rodriguez, L. L., Gay, C. G., Sumption, K. J., et al. (2016). Global Foot-and-Mouth disease research update and gap analysis: 2 - epidemiology. wildlife and economics. Transbound Emerg Dis. 63(Suppl. 1), 14–29. doi: 10.1111/tbed.12522
Knight-Jones, T. J., and Rushton, J. (2013). The economic impacts of foot and mouth disease - what are they, how big are they and where do they occur? Prev. Vet. Med. 112, 161–173. doi: 10.1016/j.prevetmed.2013.07.013
Knight-Jones, T. J. D., Mclaws, M., and Rushton, J. (2017). Foot-and-Mouth disease impact on smallholders - what do we know, what don’t we know and how can we find out more? Transbound Emerg Dis. 64, 1079–1094. doi: 10.1111/tbed.12507
Kotecha, A., Seago, J., Scott, K., Burman, A., Loureiro, S., Ren, J., et al. (2015). Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design. Nat. Struct. Mol. Biol. 22, 788–794. doi: 10.1038/nsmb.3096
Lannes, N., Python, S., and Summerfield, A. (2012). Interplay of foot-and-mouth disease virus, antibodies and plasmacytoid dendritic cells: virus opsonization under non-neutralizing conditions results in enhanced interferon-alpha responses. Vet. Res. 43:64. doi: 10.1186/1297-9716-43-64
Laskowski, R. A., Moss, D. S., and Thornton, J. M. (1993). Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067. doi: 10.1006/jmbi.1993.1351
Lavoria, M. A., Di-Giacomo, S., Bucafusco, D., Franco-Mahecha, O. L., Perez-Filgueira, D. M., and Capozzo, A. V. (2012). Avidity and subtyping of specific antibodies applied to the indirect assessment of heterologous protection against Foot-and-Mouth Disease virus in cattle. Vaccine 30, 6845–6850. doi: 10.1016/j.vaccine.2012.09.011
Link, A. J., and Labaer, J. (2011). Trichloroacetic Acid (TCA) Precipitation of Proteins. Cold Spring Harbor Lab. Course Manual. 2011, 993–994. doi: 10.1101/pdb.prot5651
Maiti, R., Van Domselaar, G. H., Zhang, H., and Wishart, D. S. (2004). SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 32, W590–W594. doi: 10.1093/nar/gkh477
Marti-Renom, M. A., Stuart, A. C., Fiser, A., Sanchez, R., Melo, F., and Sali, A. (2000). Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325. doi: 10.1146/annurev.biophys.29.1.291
Mayr, G. A., Chinsangaram, J., and Grubman, M. J. (1999). Development of replication-defective adenovirus serotype 5 containing the capsid and 3C protease coding regions of foot-and-mouth disease virus as a vaccine candidate. Virology 263, 496–506. doi: 10.1006/viro.1999.9940
McCullough, K. C., Bruckner, L., Schaffner, R., Fraefel, W., Muller, H. K., and Kihm, U. (1992). Relationship between the anti-FMD virus antibody reaction as measured by different assays, and protection in vivo against challenge infection. Vet. Microbiol. 30, 99–112. doi: 10.1016/0378-1135(92)90106-90104
McCullough, K. C., Parkinson, D., and Crowther, J. R. (1988). Opsonization-enhanced phagocytosis of foot-and-mouth disease virus. Immunology 65, 187–191.
Medina, G. N., Montiel, N., Diaz-San Segundo, F., Sturza, D., Ramirez-Medina, E., Grubman, M. J., et al. (2015). Evaluation of a fiber-modified adenovirus vector vaccine against foot-and-mouth disease in cattle. Clin. Vaccine Immunol. 23, 125–136. doi: 10.1128/CVI.00426-415
Miller, G., Lahrs, S., Shah, A. B., and Dematteo, R. P. (2003). Optimization of dendritic cell maturation and gene transfer by recombinant adenovirus. Cancer Immunol. Immunother 52, 347–358. doi: 10.1007/s00262-003-0379-376
Moraes, M. P., Mayr, G. A., Mason, P. W., and Grubman, M. J. (2002). Early protection against homologous challenge after a single dose of replication-defective human adenovirus type 5 expressing capsid proteins of foot-and-mouth disease virus (FMDV) strain A24. Vaccine 20, 1631–1639. doi: 10.1016/s0264-410x(01)00483-482
Moraes, M. P., Segundo, F. D., Dias, C. C., Pena, L., and Grubman, M. J. (2011). Increased efficacy of an adenovirus-vectored foot-and-mouth disease capsid subunit vaccine expressing nonstructural protein 2B is associated with a specific T cell response. Vaccine 29, 9431–9440. doi: 10.1016/j.vaccine.2011.10.037
Morelli, A. E., Larregina, A. T., Ganster, R. W., Zahorchak, A. F., Plowey, J. M., Takayama, T., et al. (2000). Recombinant adenovirus induces maturation of dendritic cells via an NF-kappaB-dependent pathway. J. Virol. 74, 9617–9628. doi: 10.1128/jvi.74.20.9617-9628.2000
Nakai, M., Komiya, K., Murata, M., Kimura, T., Kanaoka, M., Kanegae, Y., et al. (2007). Expression of pIX gene induced by transgene promoter: possible cause of host immune response in first-generation adenoviral vectors. Hum. Gene. Ther. 18, 925–936. doi: 10.1089/hum.2007.085
Naranjo, J., and Cosivi, O. (2013). Elimination of foot-and-mouth disease in South America: lessons and challenges. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120381. doi: 10.1098/rstb.2012.0381
Oh, Y., Fleming, L., Statham, B., Hamblin, P., Barnett, P., Paton, D. J., et al. (2012). Interferon-gamma induced by in vitro re-stimulation of CD4+ T-cells correlates with in vivo FMD vaccine induced protection of cattle against disease and persistent infection. PLoS One 7:e44365. doi: 10.1371/journal.pone.0044365
Pacheco, J. M., Brum, M. C., Moraes, M. P., Golde, W. T., and Grubman, M. J. (2005). Rapid protection of cattle from direct challenge with foot-and-mouth disease virus (FMDV) by a single inoculation with an adenovirus-vectored FMDV subunit vaccine. Virology 337, 205–209. doi: 10.1016/j.virol.2005.04.014
Patch, J. R., Kenney, M., Pacheco, J. M., Grubman, M. J., and Golde, W. T. (2013). Characterization of cytotoxic T lymphocyte function after foot-and-mouth disease virus infection and vaccination. Viral. Immunol. 26, 239–249. doi: 10.1089/vim.2013.0011
Paton, D. J., Valarcher, J. F., Bergmann, I., Matlho, O. G., Zakharov, V. M., Palma, E. L., et al. (2005). Selection of foot and mouth disease vaccine strains–a review. Rev. Sci. Tech. 24, 981–993.
Pay, T. W., and Hingley, P. J. (1992). Foot and mouth disease vaccine potency tests in cattle: the interrelationship of antigen dose, serum neutralizing antibody response and protection from challenge. Vaccine 10, 699–706. doi: 10.1016/0264-410x(92)90092-x
Pena, L., Moraes, M. P., Koster, M., Burrage, T., Pacheco, J. M., Segundo, F. D., et al. (2008). Delivery of a foot-and-mouth disease virus empty capsid subunit antigen with nonstructural protein 2B improves protection of swine. Vaccine 26, 5689–5699. doi: 10.1016/j.vaccine.2008.08.022
Philpott, N. J., Nociari, M., Elkon, K. B., and Falck-Pedersen, E. (2004). Adenovirus-induced maturation of dendritic cells through a PI3 kinase-mediated TNF-alpha induction pathway. Proc. Natl. Acad. Sci. U.S.A. 101, 6200–6205. doi: 10.1073/pnas.0308368101
Porta, C., Kotecha, A., Burman, A., Jackson, T., Ren, J., Loureiro, S., et al. (2013). Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen. PLoS Pathog 9:e1003255. doi: 10.1371/journal.ppat.1003255
Quattrocchi, V., Langellotti, C., Pappalardo, J. S., Olivera, V., Di Giacomo, S., Van Rooijen, N., et al. (2011). Role of macrophages in early protective immune responses induced by two vaccines against foot and mouth disease. Antiviral Res. 92, 262–270. doi: 10.1016/j.antiviral.2011.08.007
Quattrocchi, V., Pappalardo, J. S., Langellotti, C., Smitsaart, E., Fondevila, N., and Zamorano, P. (2014). Early protection against foot-and-mouth disease virus in cattle using an inactivated vaccine formulated with Montanide ESSAI IMS D 12802 VG PR adjuvant. Vaccine 32, 2167–2172. doi: 10.1016/j.vaccine.2014.02.061
Rao, M. G., Butchaiah, G., and Sen, A. K. (1994). Antibody response to 146S particle, 12S protein subunit and isolated VP1 polypeptide of foot-and-mouth disease virus type Asia-1. Vet. Microbiol. 39, 135–143. doi: 10.1016/0378-1135(94)90094-90099
Rincon, V., Rodriguez-Huete, A., and Mateu, M. G. (2015). Different functional sensitivity to mutation at intersubunit interfaces involved in consecutive stages of foot-and-mouth disease virus assembly. J. Gen. Virol. 96, 2595–2606. doi: 10.1099/vir.0.000187
Romanutti, C., D’antuono, A., Palacios, C., Quattrocchi, V., Zamorano, P., La Torre, J., et al. (2013). Evaluation of the immune response elicited by vaccination with viral vectors encoding FMDV capsid proteins and boosted with inactivated virus. Vet. Microbiol. 165, 333–340. doi: 10.1016/j.vetmic.2013.04.017
Romanutti, C. (2012). Evaluación de la respuesta inmune inducida por distintos inmunógenos recombinantes dirigidos contra el Virus de la Fiebre Aftosa. Doctoral Thesis, University of Buenos Aires – Faculty of Exact and Natural Sciences. Available online at: http://hdl.handle.net/20.500.12110/tesis_n5056_Romanutti
Rowlands, D. J., Sangar, D. V., and Brown, F. (1975). A comparative chemical and serological study of the full and empty particles of foot-and mouth disease virus. J. Gen. Virol. 26, 227–238. doi: 10.1099/0022-1317-26-3-227
Rubinchik, S., Lowe, S., Jia, Z., Norris, J., and Dong, J. (2001). Creation of a new transgene cloning site near the right ITR of Ad5 results in reduced enhancer interference with tissue-specific and regulatable promoters. Gene. Ther. 8, 247–253. doi: 10.1038/sj.gt.3301364
Rweyemamu, M. M., Terry, G., and Pay, T. W. (1979). Stability and immunogenicity of empty particles of foot-and-mouth disease virus. Arch. Virol. 59, 69–79. doi: 10.1007/BF01317896
Rweyemamu, M. M., Unehara, O., Giorgi, W., Medeiros, R., Lucca, D., and Baltazar, M. (1989). Effect of formaldehyde and binary ethyleneimine (BEI) on the integrity of foot and mouth disease virus capsid. Rev. Sci. Tech. 8, 747–764. doi: 10.20506/rst.8.3.425
Schutta, C., Barrera, J., Pisano, M., Zsak, L., Grubman, M. J., Mayr, G. A., et al. (2016). Multiple efficacy studies of an adenovirus-vectored foot-and-mouth disease virus serotype A24 subunit vaccine in cattle using homologous challenge. Vaccine 34, 3214–3220. doi: 10.1016/j.vaccine.2015.12.018
Scott, K. A., Kotecha, A., Seago, J., Ren, J., Fry, E. E., Stuart, D. I., et al. (2017). SAT2 Foot-and-Mouth Disease Virus structurally modified for increased thermostability. J. Virol. 91, e2312–e2316. doi: 10.1128/JVI.02312-2316
Seki, C., Robiolo, B., Periolo, O., Iglesias, M., D’antuono, A., Maradei, E., et al. (2009). Rapid methodology for antigenic profiling of FMDV field strains and for the control of identity, purity and viral integrity in commercial virus vaccines using monoclonal antibodies. Vet. Microbiol. 133, 239–251. doi: 10.1016/j.vetmic.2008.07.011
Suzuki, M., Kondo, S., Pei, Z., Maekawa, A., Saito, I., and Kanegae, Y. (2015). Preferable sites and orientations of transgene inserted in the adenovirus vector genome: the E3 site may be unfavorable for transgene position. Gene. Ther. 22, 421–429. doi: 10.1038/gt.2014.124
Terry, G. M., Clark, R. P., and Rweyemamu, M. M. (1982). Variations in the buoyant density of foot-and-mouth disease virus strains. Arch. Virol. 71, 333–341. doi: 10.1007/BF01315063
Wiederstein, M., and Sippl, M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410. doi: 10.1093/nar/gkm290
World Organisation for Animal Health (2012). “Foot and Mouth Disease (infection with FMDV),” in OIE Terrestrial Manual, ed. OIE (Paris: OIE).
Keywords: foot-and-mouth disease virus, replication-defective human adenovirus type 5, genetic vaccine, mutated VP2 capsid protein, virus-like particles O1/Campos, neutralizing antibodies
Citation: Ziraldo M, Bidart JE, Prato CA, Tribulatti MV, Zamorano P, Mattion N and D’Antuono AL (2020) Optimized Adenoviral Vector That Enhances the Assembly of FMDV O1 Virus-Like Particles in situ Increases Its Potential as Vaccine for Serotype O Viruses. Front. Microbiol. 11:591019. doi: 10.3389/fmicb.2020.591019
Received: 03 August 2020; Accepted: 28 September 2020;
Published: 04 November 2020.
Edited by:
Douglas Paul Gladue, Agricultural Research Service, United States Department of Agriculture, United StatesReviewed by:
Teresa de los Santos, Agricultural Research Service, United States Department of Agriculture, United StatesCopyright © 2020 Ziraldo, Bidart, Prato, Tribulatti, Zamorano, Mattion and D’Antuono. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alejandra L. D’Antuono, adantuono@gmail.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.