- Institute of Oceanic Research and Development, Tokai University, Shizuoka, Japan
Glucose is the most favorable carbon source for many bacteria, and these bacteria have several glucose-responsive networks. We proposed new glucose responsive system, which includes protein acetylation and probable translation control through TsaEBD, which is a tRNA modification enzyme required for the synthesis of threonylcarbamoyl adenosine (t6A)-tRNA. The system also includes nucleoid-associated protein YlxR, regulating more than 400 genes including many metabolic genes and the ylxR-containing operon driven by the PylxS promoter is induced by glucose. Thus, transposon mutagenesis was performed for searching regulatory factors for PylxS expression. As a result, ywlE was identified. The McsB kinase phosphorylates arginine (Arg) residues of proteins and the YwlE phosphatase counteracts against McsB through Arg-dephosphorylation. Phosphorylated Arg has been known to function as a tag for ClpCP-dependent protein degradation. The previous analysis identified TsaD as an Arg-phosphorylated protein. Our results showed that the McsB/YwlE system regulates PylxS expression through ClpCP-mediated protein degradation of TsaD. In addition, we observed that glucose induced ywlE expression and repressed mcsB expression. It was concluded that these phenomena would cause glucose induction (GI) of PylxS, based on the Western blot analyses of TsaD-FLAG. These observations and the previous those that many glycolytic enzymes are Arg-phosphorylated suggested that the McsB/YwlE system might be involved in cell growth in glucose-containing medium. We observed that the disruption of mcsB and ywlE resulted in an increase of cell mass and delayed growth, respectively, in semi-synthetic medium. These results provide us broader insights to the physiological roles of the McsB/YwlE system and protein Arg-phosphorylation.
Introduction
Glucose is the most favorable carbon source for many bacteria, so bacteria have developed several glucose-responsive networks (Deutscher, 2008). In Gram-positive bacteria, such as Bacillus subtilis, the transcription factor, catabolite control protein A (CcpA), is the master regulator for carbon catabolite regulation (Deutscher, 2008; Fujita, 2009). Incorporating glucose into the bacterial cells increases the metabolite, fructose-1,6-bisphosphate, which triggers HPr phosphorylation at Ser46. HPr is a phosphocarrier protein in the sugar phosphotransferase system and P-Ser-HPr activates CcpA, causing widespread transcriptional changes. Moreover, there are several additional glucose-responsive transcription factors, such as CcpC, CcpN, CggR, and GlcT (Fujita, 2009).
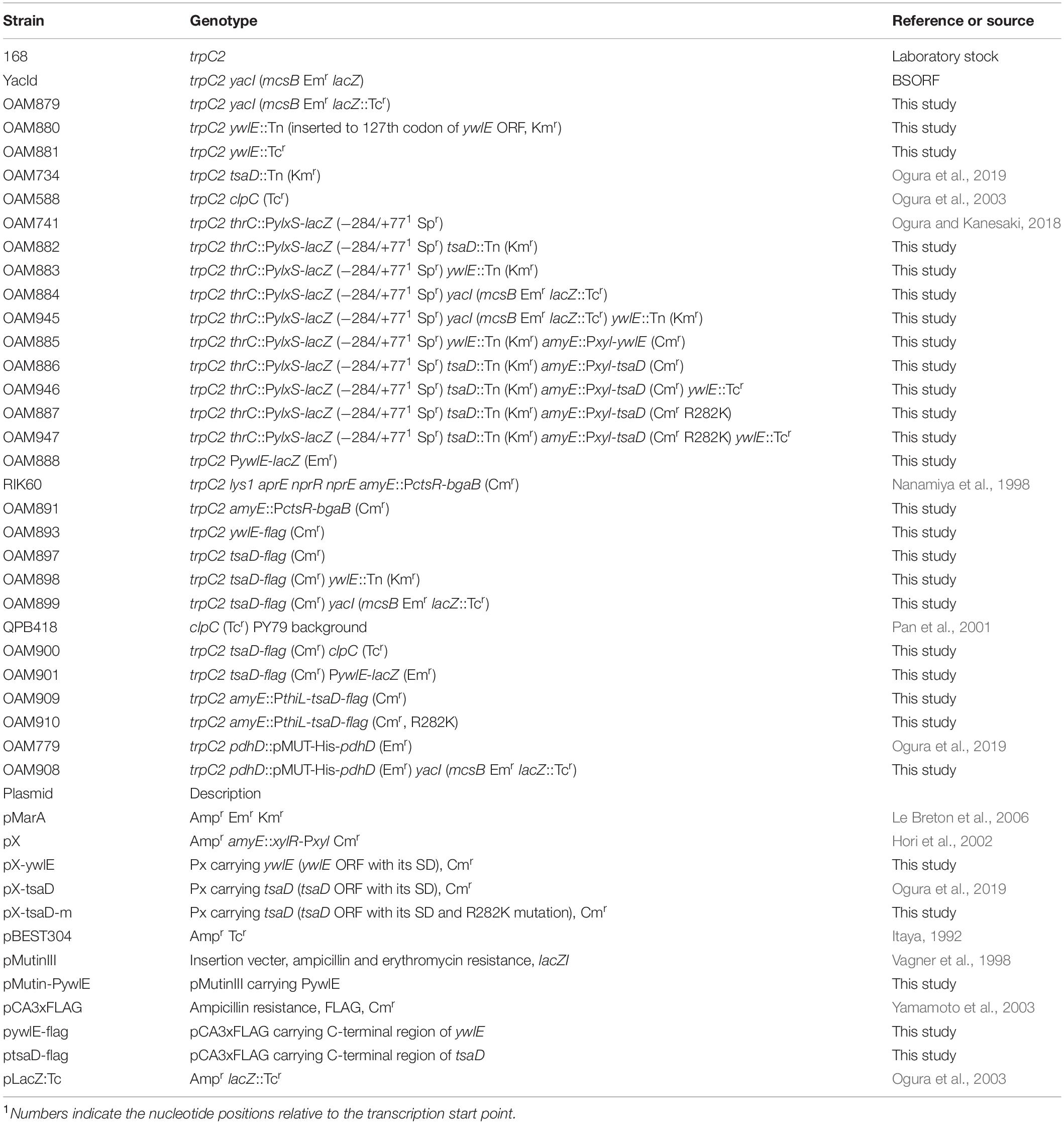
Several lines of evidence suggest another glucose responsive system (GRS) which includes protein acetylation and probable translational control (Figure 1; Ogura and Asai, 2016; Ogura and Kanesaki, 2018; Ogura et al., 2019, 2020). Glucose addition to culture medium often induces protein acetylation in E. coli and B. subtilis (Kosono et al., 2015; Schilling et al., 2015). Proteomic analysis of B. subtilis revealed that CshA, a DEAD-box helicase, is acetylated (Lehnik-Habrink et al., 2013; Kosono et al., 2015). We recently found that glucose stimulates CshA lysine acetylation (Ogura and Asai, 2016) and CshA associates with RNA polymerase (RNAP) (Delumeau et al., 2011). The association between acetylated CshA and RNAP enhances its SigX affinity, leading to glucose induction (GI) of sigX (Shiwa et al., 2015; Helmann, 2016; Ogura and Asai, 2016). GI of sigX caused by CshA acetylation is susceptible to pyruvate dehydrogenase (PDH) mutations in pdhABCD (Gao et al., 2002; Ogura and Asai, 2016). pdh gene disruption would reduce the intracellular acetyl-CoA pool and flux resulting from loss of PDH activity, that is, pyruvate conversion to acetyl-CoA (Gao et al., 2002). ylxR, a regulator of GRS, is another gene subject to GI caused by CshA (Ogura and Kanesaki, 2018). YlxR has characteristics specific to nucleoid-associated proteins (NAPs) and regulates the transcription of more than 400 genes (Dillon and Dorman, 2010; Ogura and Kanesaki, 2018). Further, YlxR is involved in tsaEBD-containing operon expression (Ogura et al., 2019). TsaEBD is a tRNA modification enzyme that is required for the synthesis of threonylcarbamoyl adenosine (t6A) (Thiaville et al., 2015; Zhang et al., 2015). t6A-modified tRNA is conserved in three domains of life and its deficiency sometimes causes severe dysfunctions (Thiaville et al., 2016; Arrondel et al., 2019; Ogura et al., 2019). In B. subtilis, several lines of evidence suggest a relationship between low t6A and protein quality control, including PDH (Ogura et al., 2019). Thus, t6A is required for stable acetyl-CoA supply through control of PDH activity. In other words, GRS constitutes feedback regulatory networks (Ogura et al., 2020).
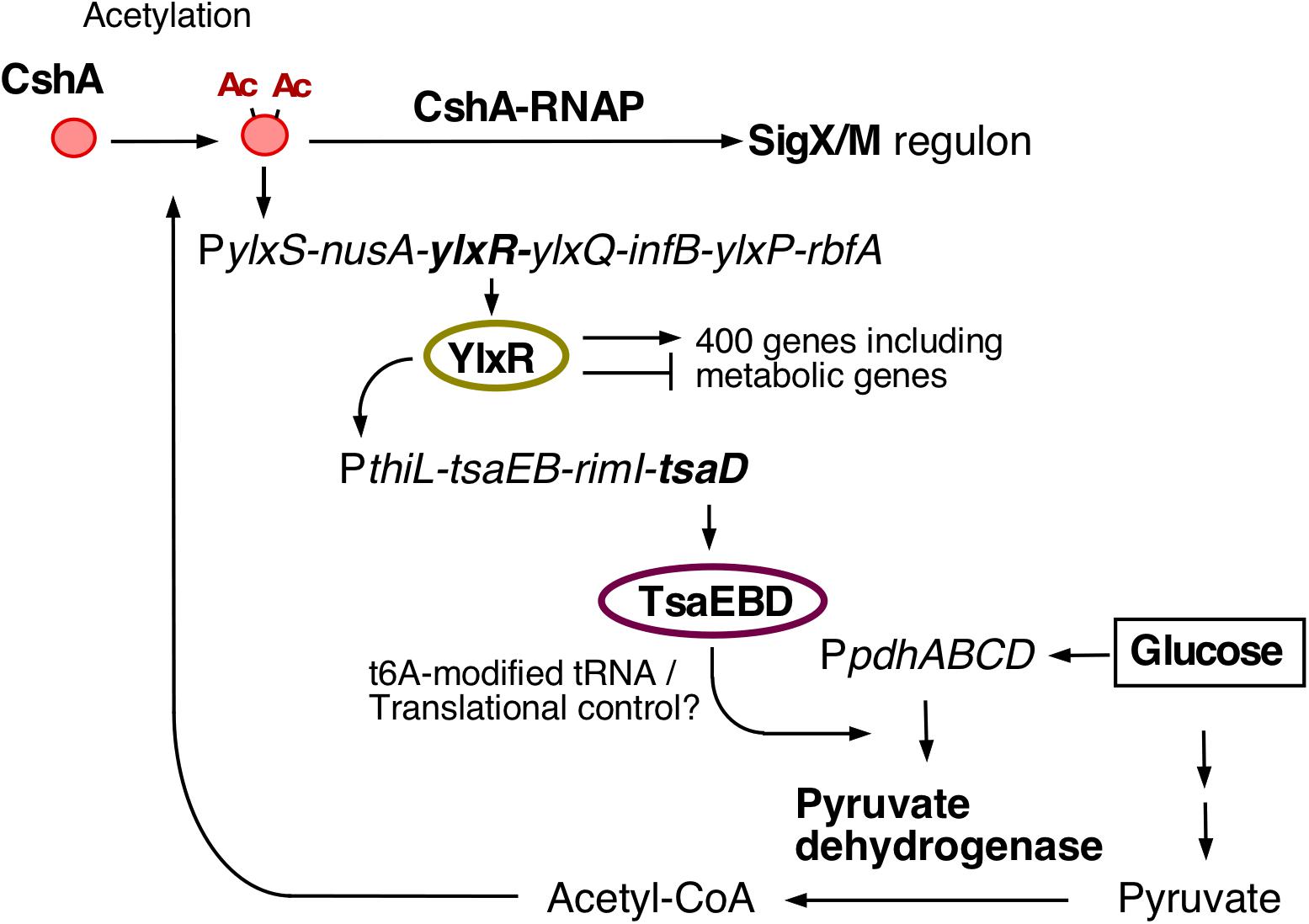
Figure 1. The current model for glucose-responsive system (GRS). The indicated pathways were previously identified: in vivo association of CshA with RNAP (Delumeau et al., 2011), glucose-stimulated CshA acetylation and sigX regulation (Kosono et al., 2015; Ogura and Asai, 2016), CshA-dependent PylxS regulation driving YlxR expression, which regulates metabolic genes (Ogura and Kanesaki, 2018), and transcriptional regulation of tsaEBD through the functional YlxR-binding to the promoter of tsaEBD, whose products are assembled and regulate pyruvate dehydrogenase translation (Ogura et al., 2019). Pyruvate dehydrogenase provides acetyl-CoA, which would be the acetyl moiety source for CshA acetylation. In the tsaD disruptant grown with glucose cellular acetyl-CoA pool was reduced (Ogura et al., 2019). Each pathway is supported by experimental evidence, however, considering the whole regulatory cascade there is a room for further verification. Arrows indicate transcription, translation, acetylation, enzymatic reaction, transcriptional activation or metabolic reaction. T-bars indicate transcriptional repression. Ac, acetyl moiety; RNAP, RNA polymerase.
Several bacterial protein modifications include Lys-acetylation, His-Asp phosphorylation for signal transduction by two-component regulatory systems, and Ser/Thr/Tyr phosphorylation (Dworkin, 2015; Mijakovic et al., 2016; Carabetta and Cristea, 2017). Emerging evidence suggests that Arg-phosphorylation is another protein modification in bacteria (Mijakovic et al., 2016). This modification was first identified in the transcriptional repressor CtsR-involving heat-shock response, and is regulated by McsA, McsB kinases, and YwlE phosphatase. Inactivated CtsR is degraded by ClpCP ATP-dependent protease (Krueger et al., 2001; Fuhrmann et al., 2009, 2013; Elsholz et al., 2012). Later, Arg-phosphorylation was identified as a tag for protein degradation by ClpCP (Trentini et al., 2016). Several efforts to identify Arg-phosphorylated proteins in B. subtilis and Staphylococcus aureus revealed many targets, including metabolic enzymes, translation-related proteins, and some transcription factors (Elsholz et al., 2012; Schmidt et al., 2014; Trentini et al., 2014; Junker et al., 2019).
Here, we report that ywlE is a regulatory factor for ylxR expression, which is driven by PylxS. Previous analysis identified TsaD as an Arg-phosphorylated protein (Trentini et al., 2014). McsB kinase and YwlE phosphatase regulate PylxS via ClpCP-mediated protein degradation of TsaD. In addition, we observed that glucose represses mcsB and clpC expression (Ishii et al., 2013) and induces ywlE expression. Many glycolytic and TCA cycle enzymes are Arg-phosphorylated, suggesting that the McsB/YwlE system might be involved in cell growth in glucose-containing medium (Elsholz et al., 2012; Schmidt et al., 2014; Trentini et al., 2014; Junker et al., 2019; Zhou et al., 2019). mcsB and ywlE disruption resulted in increased cell mass and delayed growth, respectively. These results provide broad insights to the physiological roles of the McsB/YwlE system and protein Arg-phosphorylation.
Materials and Methods
Strains, Media, and PCR
All B. subtilis strains used in this study are shown in Table 1 and Supplementary Figure 1. One-step competence medium (MC) (Kunst et al., 1994), Schaeffer’s sporulation medium (SM) (Schaeffer et al., 1965), Luria-Bertani (LB) medium (Lennox, Difco, MI, United States), and Antibiotic medium 3 (Difco) were used. Antibiotic concentrations were described previously (Ogura and Tanaka, 1996; Ogura et al., 1997). Synthetic oligonucleotides were commercially prepared by Tsukuba Oligo Service (Ibaraki, Japan) and are listed in Supplementary Table 1. For PCR-mediated construction of strains and plasmids, PrimeSTAR MAX DNA polymerase (Takara Co., Shiga, Japan) was used. For screening of recombinant DNA during plasmid construction, LA PCR DNA polymerase (Takara Co.) was used.
Strain Construction
The ywlE::Tcr unit in OAM881 was constructed using PCR. Briefly, Tcr from pBEST304 (Itaya, 1992) and the upstream and downstream ywlE regions with overlapping Tcr regions were amplified using primers listed in Supplementary Table 1, and then combined by PCR. The unit was directly transformed into B. subtilis 168. Total DNA was taken using DNeasy kit (Qiagen, Venlo, Netherlands) from the resultant Tcr strain for PCR-based confirmation of the expected chromosomal structure. The wild and mutant strains bearing tag-added tsaD at the amyE locus (OAM909 and OAM 910) were constructed by the PCR-based method shown in Supplementary Figure 1. The ORF associated with its own promoter was sequenced.
Plasmid Construction
The plasmids used in this study are listed in Table 1. For PCR, B. subtilis 168 chromosomal DNA was used as template. To construct pMutin-PywlE, PCR products were amplified using the oligonucleotides pair pMut-PywlE-F(H)/pMut-PywlE-R(B), digested with HindIII/BamHI, and cloned into pMutin3 treated with the same enzymes (Vagner et al., 1998). To construct pX-ywlE, PCR products were amplified using the oligonucleotides pair pX-ywlE-Spe/pX-ywlE-Bam, digested with SpeI/BamHI, and cloned into a pX plasmid treated with the same enzymes (Hori et al., 2002). To construct pX-tsaD-m, PCR products were amplified using the oligonucleotide pairs pX-gcp-Spe/tsaD-M1 and pX-gcp-Bam/tsaD-M2. Both PCR products were combined by PCR using the oligonucleotide pair pX-gcp-Spe/pX-gcp-Bam. The final PCR product was digested with SpeI/BamHI and cloned into pX treated with the same enzymes. To construct pywlE-flag and ptsaD-flag, PCR products were amplified using the oligonucleotides pairs Pflag-ywlE-F-H/Pflag-ywlE-R-Xb and Pflag-tsaD-F-H/Pflag-tsaD-R-Xb and digested with HindIII/XbaI. Each product was cloned into pCA3xFLAG treated with the same enzymes (Yamamoto et al., 2003).
Transposon Mutagenesis
The transposon delivery vector pMarA was introduced into the strain OAM741 (Le Breton et al., 2006). The resultant strain was incubated in LB medium containing kanamycin at 30°C overnight. The cells were diluted and plated onto sporulation medium with 1.5% agar plates containing X-gal (100 μg/mL), kanamycin, spectinomycin and 2% glucose. The plates were incubated at 42°C. White colonies were selected. The insertion mutations were backcrossed into the parental strain and used with the Lac assay. Total DNA was isolated from the candidate strain, SauIIIA1-digested, ligated, and amplified with inverse PCR using oligonucleotides 695 and 696, as described previously (Chan et al., 2014; Ogura and Asai, 2016; Supplementary Table 1). The PCR products were sequenced using the oligonucleotide 696.
β-Galactosidase Analysis
Growth conditions and β-galactosidase analysis procedures were previously described (Ogura and Asai, 2016; Ogura and Kanesaki, 2018). β-galactosidase activity from BgaB was determined at 54°C. β-galactosidase analysis using chlorophenol red β-D-galactopyranoside (CPRG, Roche, Germany) was performed as described previously (Ogura et al., 2019).
Western Blot Analysis
To determine the amounts of each FLAG protein and SigA, cells were grown in 50 mL sporulation medium with or without 2% glucose in 200 mL flasks. At the appropriate growth phase, cells were harvested and washed with 1 mL TBS buffer (10 mM Tris-HCl pH 7.5 and 150 mM NaCl) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). To determine protein stability, cells were grown in 100 mL sporulation medium with or without 2% glucose in 500 mL flasks. At the appropriate growth phase, chloramphenicol was added at a final concentration of 150 μg/mL. Then, 25 mL culture was sequentially harvested and washed with 1 mL TBS buffer containing 1 mM PMSF. Cells were disrupted with a French Pressure cell to obtain whole cell extracts. Western blot analysis was performed as previously described (Hata et al., 2001). Monoclonal mouse anti-FLAG M2 antibody (F3165) was purchased from Sigma-Aldrich (Darmstadt, Germany). Polyclonal rabbit anti-SigA antibody was previously described (Ogura, 2016). Monoclonal mouse anti-His tag antibody was purchased from Medical and Biological Laboratories (Nagoya, Japan). These antibodies were diluted (1/1000) in Can Get Signal solution 1 (ToYoBo, Tokyo, Japan). Can Get Signal solution 2 (ToYoBo) was used for POD-conjugated Anti-rabbit/mouse IgG secondary antibody (Roche, Mannheim, Germany). Band intensities were analyzed using Adobe Photoshop version 2.0.
Results
Screening for Deficient PylxS-lacZ Expression in Transposon-Mediated Gene Disruptants
We previously identified the genes responsible for GI of sigX, including ylxR, and characterized these genes as a part of a GRS (Figure 1; Ogura and Asai, 2016; Ogura and Kanesaki, 2018; Ogura et al., 2019, 2020). The ylxR-containing operon is driven by PylxS, and is composed of the transcription and translation genes, NusA transcription elongation factor and translation initiation factor B (infB), respectively (Mondal et al., 2016). This operon is subject to CshA-regulated GI (Ogura and Kanesaki, 2018). To identify additional genes involved in CshA-regulated GI, we screened for transposon (Tn)-insertion mutations that reduced the expression of ylxS operon in the presence of glucose. Several candidate genes were obtained from approximately 12,000 colonies. Of these, we identified that ywlE encoding an Arg-phosphatase. Tn insertion into the ywlE gene reduced PylxS-lacZ expression on solid Schaeffer’s sporulation medium (Figure 2E), although the same strain showed moderately decreased PylxS-lacZ expression compared to wild type in liquid medium irrespective of the presence of glucose (Figures 2A1,2). This difference may be due to different growth conditions. It should be noted that in the ywlE disruptant significant GI was observed (see below).
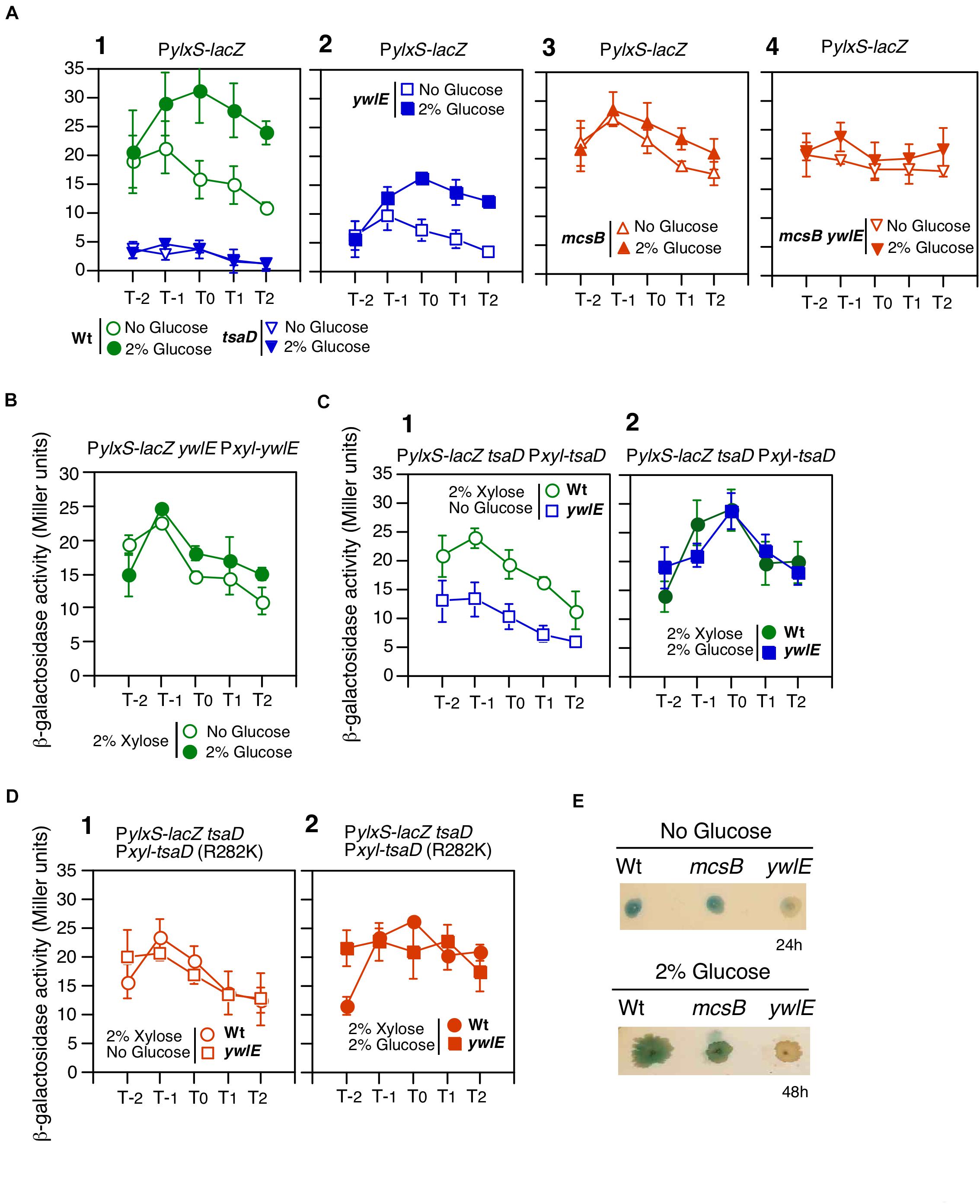
Figure 2. Effects of ywlE and mcsB disruption and phosphorylated-Arg residue mutation of tsaD on PylxS-lacZ expression. (A–D) β-Gal activity in sporulation medium. Data represent means and SD from three independent experiments. The x-axis represents growth time in hours relative to the end of vegetative growth (T0). The relevant genotype and the presence of glucose or xylose are indicated. The strain lacking lacZ showed less than 1 Miller units under the condition with or without glucose. (A) PylxS-lacZ expression in mutant strains. 1, OAM741[wild] and OAM882[tsaD]; 2, OAM883[ywlE]; 3, OAM884[mcsB]; 4, OAM945[ywlE mcsB]. (B) Complementation test of the ywlE mutation using OAM885. (C) Complementation test of tsaD using OAM886[wild] and OAM946[ywlE]. (D) Complementation test of the tsaDR282K mutation using OAM887[Wt] and OAM947[ywlE]. (E) PylxS-lacZ expression on sporulation medium agar plates. Each strain (OAM741[wild], OAM884[mcsB], and OAM883[ywlE]) was inoculated onto 1.5% agar sporulation medium plates containing 100 μg/mL X-gal and spectinomycin, and incubated at 37°C. Images were taken at the indicated time.
Involvement of the McsB/YwlE System in PylxS Expression
Next, we performed a complementation test of ywlE disruption by artificial overexpression of xylose-inducible ywlE at amyE. Under xylose and no glucose condition, PylxS expression was higher than that of the ywlE disruptant (Figures 2A2,B). This indicated that ywlE disruption indeed decreased PylxS expression. Moreover, addition of glucose did not cause GI, suggesting that overproduction of ywlE abolished the GI of PylxS-lacZ.
YwlE phosphatase dephosphorylates Arg-phosphorylated proteins, which are generated by McsB kinase. Thus, we examined how mcsB disruption affects PylxS-lacZ expression. We observed that in the mcsB mutant, basal PylxS-lacZ expression was increased, similar to levels observed under glucose-containing culture, though glucose was not present (Figure 2A3). Thus, mcsB disruption abolished GI of this operon. It should be noted that the results in the mcsB disruptant were quite similar to those in the ywlE-overexpressing strain (Figures 2A3,B). This is reasonable, because ywlE-overexpression is essentially equivalent to mcsB disruption. When cells had mcsB disruption, further disruption of ywlE would not affect PylxS expression, because dephosphorylation has no effect if phosphorylation does not occur. This was the case (Figure 2A4). Arg-phosphorylated protein is known to be a target for ClpCP-dependent protein degradation (Trentini et al., 2016). Therefore, GI of PylxS-lacZ expression should be abolished in the clpC mutant. We constructed a clpC-deficient PylxS-lacZ strain and observed expression with and without glucose. We observed no GI during log-phase but did slight increase of expression with glucose in the early stationary phase due to an unknown reason (Supplementary Figure 2A). Taken together, we concluded that the McsB/YwlE pair is a newly-identified PylxS regulatory factor. Apparently, McsB is involved in GI of PylxS (see section “Discussion”). Moreover, YwlE is required for sufficient expression of PylxS, irrespective of the presence of glucose, under the condition when McsB is functional.
The CshA-regulated GRS regulates ylxS and sigX promoters. To determine whether McsB/YwlE affects the GRS or specifically PylxS, we examined the effects of the mutations introduced into the sigX-lacZ strain. We observed significantly decreased GI of sigX-lacZ expression in the ywlE disruptant, and only very slightly increased basal expression without glucose in the mcsB disruptant (Supplementary Figure 2B). These results show that YwlE affects sigX expression similar to PylxS, but to a lesser extent, suggesting that the McsB/YwlE pair is more important for the expression of PylxS than of PsigX.
TsaD as an McsB/YwlE Target Protein in PylxS Expression
We tried to identify the potential McsB/YwlE target in GRS using previous global analyses of Arg-phosphate proteins (Figure 1). From one of the analyses, TsaD was identified as a phosphorylated protein at Arg282 (Trentini et al., 2014). TsaD is a component of the TsaEBD complex, which catalyzes t6A-modified tRNA production and plays role in glucose-mediated sigX induction via translation of pyruvate dehydrogenase subunits in the presence of glucose (Zhang et al., 2015; Thiaville et al., 2015, 2016; Ogura et al., 2019; Figure 1). In fact, tsaD disruption caused severely decreased PylxS-lacZ expression (Figure 2A1). First, we observed that artificial and ectopic expression of xylose-inducible tsaD at amyE complemented tsaD disruption without glucose (compare the expression observed in tsaD in Figure 2A1 to wt in Figure 2C1). Next, we observed the decreased PylxS-lacZ expression in the ywlE disruptant without glucose (ywlE in Figure 2C1). These results demonstrated the negative effect of ywlE disruption on PylxS in this tsaD-induction system. Under the tsaD-overproducing condition with glucose, the similar expression levels of PylxS-lacZ to that in the wild type strain was observed, indicating that tsaD-overexpression suppressed the effect of the ywlE disruption on PylxS expression (Figure 2C2).
Since the above control experiments worked well, we next examined the possible effect of mutant TsaDR282K on PylxS-lacZ expression. Under the condition where tsaDR282K was artificially induced without glucose, PylxS-lacZ expression in the ywlE disruptant was similar to that in the strain with functional ywlE (Figure 2D1). Thus, the R282K mutation suppressed the negative effect of ywlE disruption on PylxS expression. These data indicate that the decreased expression of PylxS-lacZ in the ywlE disruptant is dependent on Arg-phosphorylation of TsaD. In the presence of glucose, the mutant strain showed similar phenotypes to the wild-type strain, as expected (Figure 2D2). Moreover, PylxS-lacZ expression was significantly high even without xylose compared to that in the tsaD disruptant, probably due to leaky production of TsaD (Supplementary Figure 2C). The results without xylose were similar to those with xylose, indicating that the TsaD protein amounts to sufficiently activate PylxS-lacZ are very small.
Glucose-Dependent ywlE Induction
Previously, we reported that ctsR/mcsAB/clpC operon expression is repressed in glucose-containing sporulation medium (Ishii et al., 2013). Glucose addition also represses ClpC protein expression, which was observed in Western blot analysis (Ishii et al., 2013). The previously observed β-Gal activity was, however, very low, and therefore, we used a more sensitive β-Gal substrate, CPRG. As shown in Figure 3A, glucose addition clearly decreased the PctsR expression. Thus, we examined the possible effect of glucose on ywlE expression, because YwlE functionally counteracts McsB. Since ywlE expression is driven by two upstream promoters, the lacZ reporter gene was inserted into the immediate downstream region of the ywlE promoter (Figure 3B). We observed that ywlE expression was fourfold induced by glucose after entry into the stationary phase (Figure 3B). PywlE-lacZ showed a glucose-dependent response at more than 0.5% glucose, which was also induced by glycerol but not acetate or succinate (data not shown). These carbon source reactivities were similar to those of PylxS-lacZ (Ogura and Kanesaki, 2018). To determine which of the two promoters are responsible for GI of ywlE, we constructed ectopic lacZ-fusions with the 0.4 and 1.0 Kb upstream regions of ywlE at amyE. The short and long fusions contain PywlE and both promoters, respectively. The two fusion expression was only 3- to 2.5-fold induced by glucose (Supplementary Figure 2D). These results indicated that PywlE is responsible for GI and that GI of ywlE was fully observed only in the original chromosome region. This may be due to chromosomal position effect (Bryant et al., 2014).
Figure 3. Glucose-induction of ywlE and -repression of mcsB. (A,B) β-Gal activity in sporulation medium using CPRG (A) and ONPG (B) is shown. Data represent means and SD from three independent experiments. The x-axis is the same as in Figure 2. The presence of glucose is indicated. The strain lacking lacZ/bgaB showed less than 1 Miller units under the condition with or without glucose. (A) PctsR-bgaB expression, OAM891. The chromosomal structure of the mcsB-containing operon is shown alongside the panel. (B) PywlE-lacZ expression, OAM888. The chromosomal structures of the ywlE-containing operon and the region around the PywlE-lacZ fusion are shown alongside the panel. (C) Western blot analysis of YwlE-FLAG. The growth phase is indicated in hours relative to the end of vegetative growth (T0) in sporulation medium. Equal protein amounts of whole cell extracts were analyzed in 15% polyacrylamide gels for Western blot using anti-FLAG-tag monoclonal antibody. SigA was used as a control. (D) Model of TsaD Arg-phosphorylation control. Arrows and T-bars indicate transcriptional activation or phosphorylation/dephosphorylation, and transcriptional repression or protein degradation, respectively. R, arginine residue to be phosphorylated; circled P, phosphate residue.
The GI of ywlE was also observed at the protein level as expected (Figure 3C). Notably, ywlE-FLAG is functionally equivalent to the wild type protein because in the tag-carrying strain, similar GI and basal expression of PylxS-lacZ were observed (left, Supplementary Figure 2E). These findings strongly suggested that the TsaD phosphorylation state is glucose-dependent, leading to altered protein levels from ClpCP-dependent degradation of Arg-phosphorylated proteins (Figure 3D).
Glucose-Mediated Control of TsaD Degradation
Since Arg-phosphorylated TsaD may be subjected to ClpCP-dependent degradation, we examined TsaD stability using Western blot analysis. The FLAG-tagged TsaD is functionally equivalent to the wild type protein, because similar GI and basal expression of PylxS-lacZ was observed to that in the wild-type strain (right, Supplementary Figure 2E). To determine protein stability, which we defined as protein degradation rate after inhibiting protein synthesis, chloramphenicol was added to the culture medium. TsaD-FLAG protein was quantified in sequentially-sampled cells. First, we examined wild type strain without glucose and observed fast protein degradation, however, in cells grown with glucose, TsaD-FLAG was stabilized (left, Figure 4A). In the ywlE disruptant, TsaD-FLAG was more unstable compared to the wild-type strain irrespective of the presence of glucose (mid-left, Figure 4A). The observation of glucose-mediated stabilization even in the ywlE disruptant is reasoned by catabolite repression of mcsB and clpC. Based on these results, it is possible that mcsB or clpC disruption stabilizes TsaD-FLAG. Similarly, ywlE overproduction, which should be equivalent to mcsB disruptant, would stabilize TsaD-FLAG. Thus, we examined protein stability in these strains and observed the expected TsaD-FLAG stabilization irrespective of the presence of glucose (Figure 4A). Next, to confirm the significance of R282, we constructed an ectopic TsaD-FLAG expression system driven by its own promoter at amyE (Figure 4B). Subsequently, the Arg residue to be phosphorylated was changed to Lys in the TsaD protein at amyE (TsaDR282K). Using the expression system, we observed similar protein degradation profiles of wild-type protein as in the cases of the strain bearing TsaD-FLAG in its original location (left, Figures 4A,B). As expected, the mutant TsaDR282K-FLAG protein was significantly stabilized in the absence of glucose compared to the wild type (Figure 4B). Similar degradation rates were observed for wild and mutant TsaD with glucose. These were consistent with the observations for the wild and mcsB strains bearing TsaD-FLAG in original location. These results indicate that the McsB/YwlE system, including ClpCP, is involved in the control of TsaD stability. These results are consistent with the results shown in Figure 2, and strongly suggest that control of TsaD stability through Arg-phosphorylation regulates PylxS-lacZ expression.
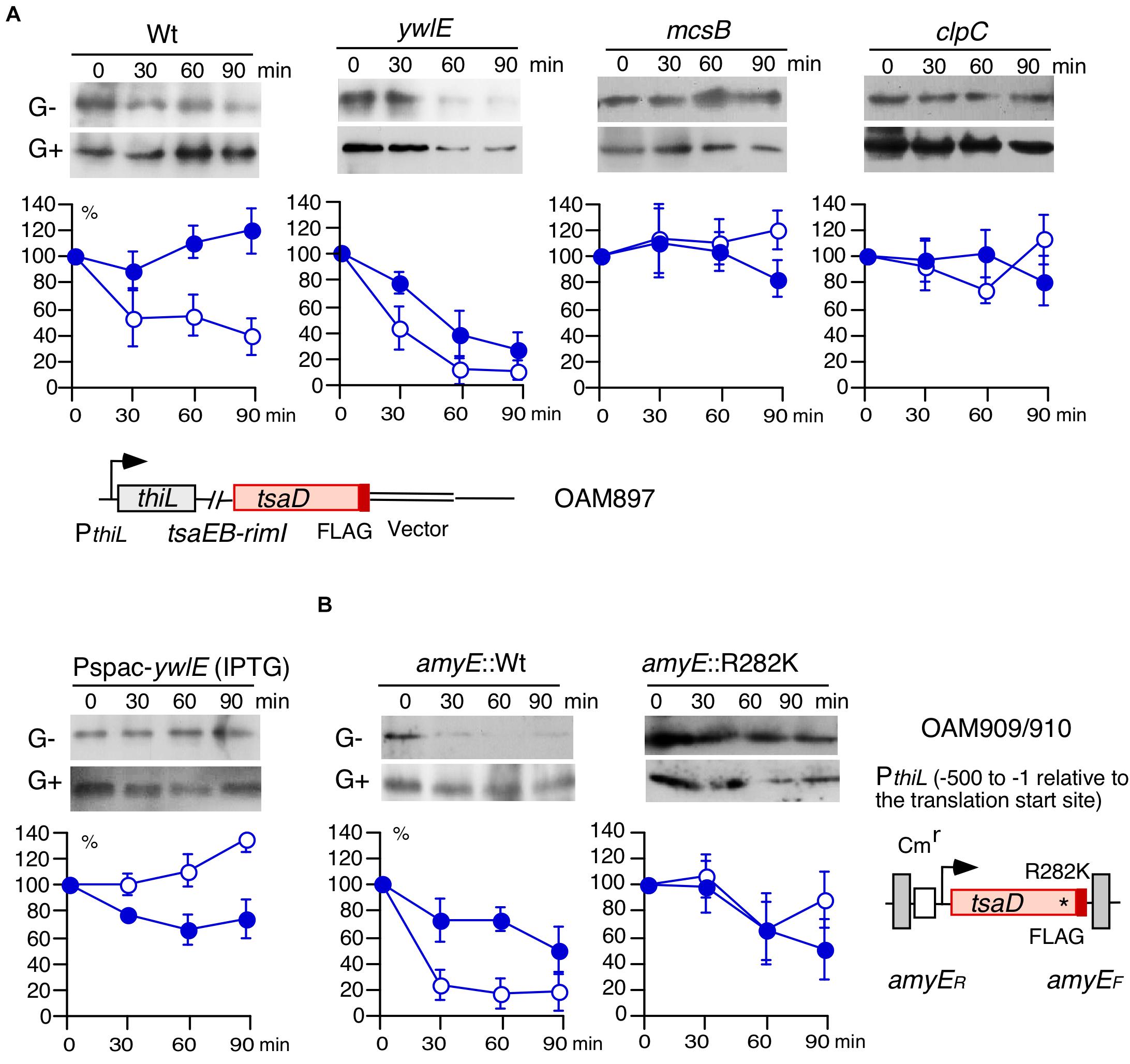
Figure 4. Western blot analysis of TsaD-FLAG. Equal protein amounts of whole cell extracts were analyzed in 12.5% polyacrylamide gels for Western blots using anti-FLAG-tag monoclonal antibody. The chromosomal structures of OAM897 and OAM 909/910 are shown. Boxes and bent arrows show open-reading frames and promoters, respectively. The promoter region (−500/−1 relative to the translation start site) has full promoter activity (Ogura et al., 2019). (A,B) Protein stability analysis. Band intensities are shown in the graphs. After inhibiting protein synthesis, more than 100% of Arg-phosphorylated TsaD was sometimes observed. It is likely that apparent ratio of TsaD, if not degraded, to the total protein amounts increased due to other protein degradation systems. Time indicates culture sampling interval after chloramphenicol addition, which was added at T1 in sporulation medium culture. G+ and G− indicate the presence or absence of 2% glucose. Closed and open symbols indicate results from the medium containing glucose or no glucose, respectively. Means and SD (error bars) are shown from three to five biologically-independent samples. A, OAM897[wild], OAM898[ywlE], OAM899[mcsB], OAM900[clpC], and OAM901[Pspac-ywlE]. For OAM901, 1 mM IPTG (final concentration) was added. B, OAM909[wild] and OAM910[R282K mutant]. * indicates the introduced nucleotide change.
Glucose-Mediated PdhD-His Protein Stabilization and Effect of ywlE/mcsB Mutation on Cell Growth
Many glycolytic and TCA-cycle enzymes, including PdhD, which is involved in PylxS regulation, were previously identified in the analysis of Arg-phosphorylated proteins (Elsholz et al., 2012; Schmidt et al., 2014; Trentini et al., 2014; Figures 1, 5A). We observed that PdhD-His was degraded after quenching protein synthesis (Figure 5B). Moreover, in the mcsB disruptant, PdhD-His was more stabilized than the case for the wild-type, strongly suggesting that this degradation is mediated by PdhD Arg-phosphorylation. Based on these observations and the model shown in Figure 3D, we expected that glucose addition to the culture medium would result in lower protein degradation because of glucose-mediated ywlE induction and mcsB repression. Our results confirmed this notion (Figure 5B). Notably, glucose addition results in an increase of pdhABCD operon transcription (Blencke et al., 2003). This effect would reflect enhanced protein expression by glucose. To further examine the Arg-phosphorylation effects on PdhD protein stability, we constructed an ectopic PdhD-His expression system driven by the pdhABCD operon promoter at amyE. The PdhD-His from this strain, however, was highly unstable and addition of glucose did not significantly stabilize the protein (data not shown). Thus, we did not perform this mutational analysis. Based on these results, it was concluded that the protein degradation of PdhD is mediated by Arg-phosphorylation.
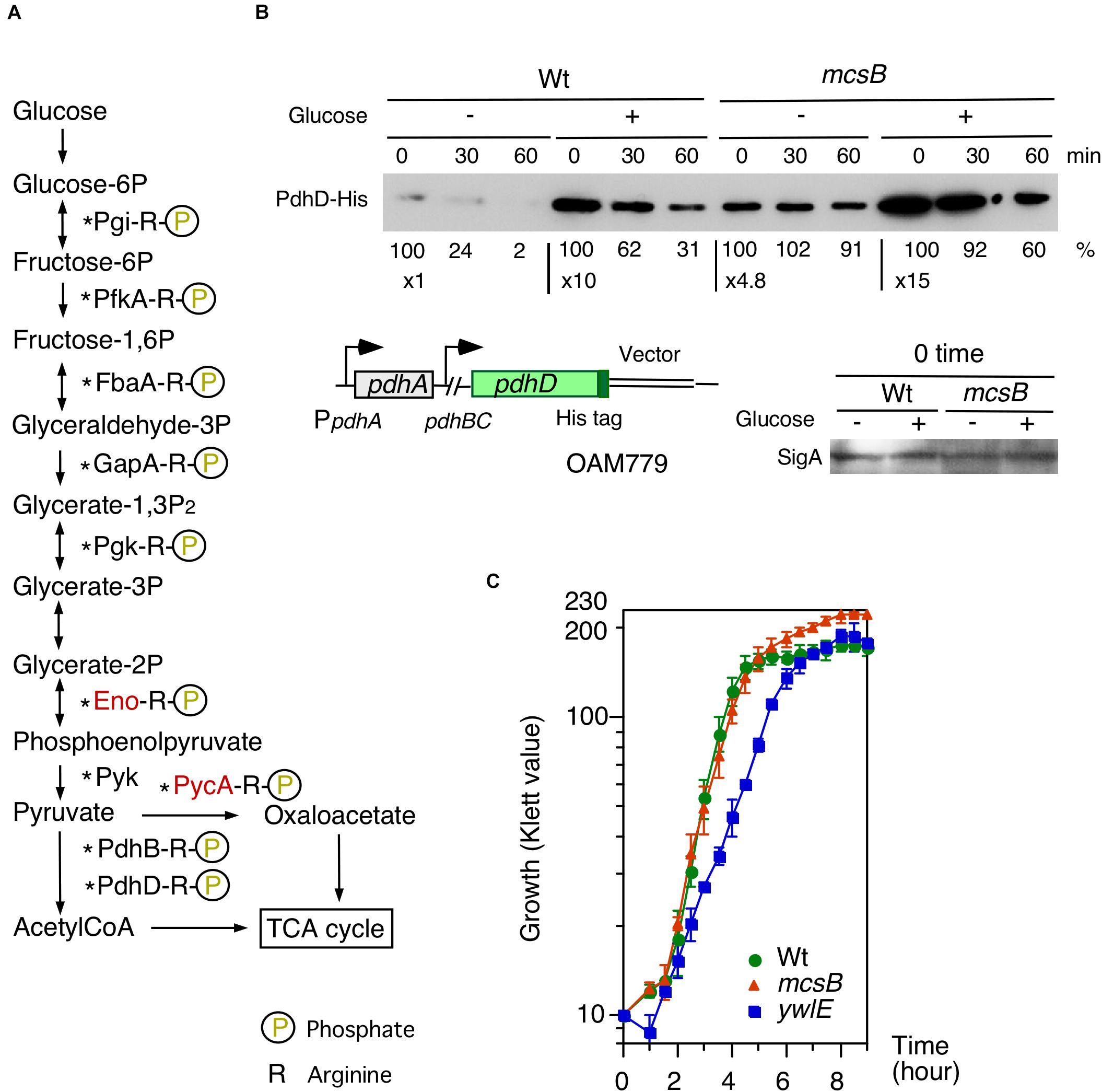
Figure 5. Effects of glucose on PdhD-His stability and cell growth profiles in mcsB and ywlE disruptants. (A) Glycolytic pathways and enzymes detected in Arg-phosphorylated forms. Enzymes in red letters are likely to be regulated by ClpCP-dependent degradation (Gerth et al., 2008). Asterisks show the enzymes that have been reported to be acetylated (Kosono et al., 2015; Carabetta et al., 2016). (B) PdhD-His western analysis. Cells were grown in sporulation medium with and without 2% glucose. Equal protein amounts of whole cell extracts were analyzed in 12.5% polyacrylamide gel for Western blots using anti-His-tag monoclonal antibody. Time indicates sampling interval after chloramphenicol addition, which was added at T1. Band intensities are indicated below the panels. The chromosomal structure of OAM779 is shown. Boxes and bent arrows show open-reading frames and promoters, respectively. OAM779 [Wt] and OAM908 [mcsB]. As a control, SigA is shown for time 0 samples. (C) Cell growth profiles of each mutant. Overnight culture grown in A3 medium was inoculated to 4 mL semisynthetic MC (modified competence) medium in an L-tube. Growth was monitored with a Klett calorimeter (Thermo Fisher Scientific, Waltham, MA, United States). Means and SD from three independent experiments are shown. 168[wild], OAM879[mcsB], and OAM881[ywlE].
According to the experimental results on glucose-mediated stability control of PdhD, we expected that in glucose-containing medium, YwlE/McsB may be involved in cell growth. To test this, we used semi-synthetic medium, because more stable cell growth profiles are obtained compared to complex medium containing natural components. The semi-synthetic MC (modified competence) medium contains 2% glucose, 0.1% citrate, 0.2% glutamate, 0.1% casamino acids, tryptophan, salts, and minerals (Kunst et al., 1994). It should be noted that in this medium the PctsR-bgaB and PywlE-lacZ expression was similar to those in glucose-added sporulation medium (Supplementary Figure 2F). The ywlE disruptant showed delayed cell growth in the log-phase, but overall cell mass was similar to the wild-type (Figure 5C). However, while the mcsB disruptant showed a similar cell growth profile in the log-phase, the final cell mass was significantly larger and about 40% increased. These results show that McsB and YwlE affect cell growth in MC medium.
Discussion
In this study, we identified the McsB/YwlE system controlling Arg-phosphorylation of target proteins in GRS, particularly as a regulatory factor for TsaD. Since Arg-phosphorylated proteins are degraded by ClpCP, the McsB/YwlE system regulates the fate of such proteins. In fact, we observed changes of degradation rates of TsaD after mutations to the McsB/YwlE system or substitution of the Arg-to-Lys mutation in TsaD. TsaD would function via efficient translation through modification of tRNA decoding ANN codons. Thus, glucose-mediated TsaD enhancement may have global effects on cellular physiology, because many proteins have ANN codons.
Overproduction of tsaD resulted in abolishment of GI of PylxS-lacZ, whereas simultaneous overproduction of tsaD and ywlE disruption, which decreased protein stability of TsaD, lowered PylxS expression in the absence of glucose and rescued GI. This GI would be attributed to catabolite repression of mcsB because YwlE is abcent. These observations indicated that glucose-mediated increase from the low levels of cellular amount of TsaD is critical for GI of PylxS. The fact that the overproduction of more stable TsaDR282K in the strain with ywlE disruption did not show GI of PylxS is consistent with the above notion. In the tsaD-overexpressing strain, there was no further increase in PylxS expression from that in the wild type strain even with glucose, suggesting that these levels of TsaD might also exceed the saturation levels required for PylxS expression. Otherwise, too much amount of TsaD might inhibit further enhancement of PylxS expression. Disruption of mcsB or overproduction of ywlE, which is equivalent to mcsB disruption, also resulted in abolishment of GI of PylxS-lacZ. Since the mcsB disruptant lacks the glucose target and requirement of another target, ywlE, is canceled because of absence of McsB, it is reasonable that addition of glucose has no effect on PylxS expression. These results support the notion that PylxS-lacZ induction is caused by glucose-mediated control of Arg-phosphorylation state of TsaD. On the contrary, YwlE is a positive factor for PylxS expression, when McsB is present. Collectively, these results indicate a central role for McsB/YwlE in GI of PylxS-lacZ (Figure 6). When glucose was added, TsaD levels increased, probably leading to efficient translation of the pdhABCD operon. In addition, glucose increased pdhABCD transcription and stabilized PdhD. Perhaps PdhB is also stabilized through decreased Arg-phosphorylation. Then, enhanced PDH levels would stably supply acetyl-CoA as a source of acetylated CshA, which is a positive regulator for PylxS. Then PylxS-driven ylxR activates tsaD-containing operon expression (Figure 6).
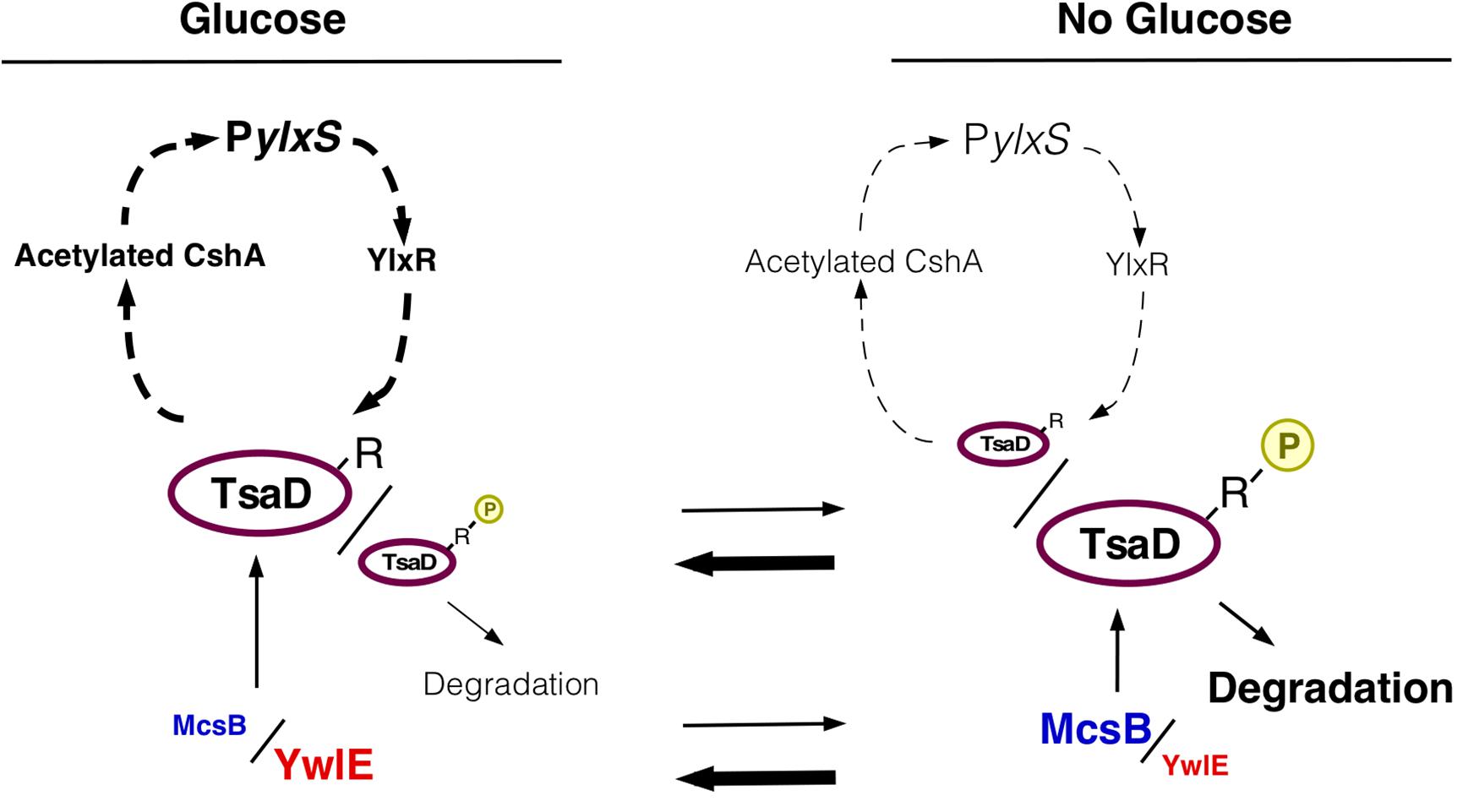
Figure 6. Working hypothesis of glucose-mediated PylxS regulation through TsaD in GRS. Description of figures is the same as in Figures 1, 3D. The arrowheads with dotted line indicate indirect positive regulation (see Figure 1).
Signals controlling the McsB/YwlE system occur in two environmental states–heat-shock and oxidative stress (Krueger et al., 2001; Fuhrmann et al., 2016). Heat-shock induces the ctsR/mcsAB/clpC operon through an intrinsic CtsR thermosensor function. McsA and McsB are required for CtsR degradation. Oxidative stress is sensed by cysteine residue of YwlE, leading to YwlE inactivation (Fuhrmann et al., 2016). The global analyses of Arg-phosphorylated proteins revealed many genes not involved in heat-shock and oxidative stress responses. Indeed, presence of several other unknown signals for the McsB/YwlE system has been pointed out (Suskiewicz and Clausen, 2016). Our study raised the possibility that glucose availability regulates the McsB/YwlE system, because glucose induced PywlE-lacZ and repressed PctsR-bgaB. The expression of PctsR-bgaB is activated by Spx and repressed by CtsR (Elsholz et al., 2017; Rojas-Tapias and Helmann, 2019). We note that in the ctsR-inactivation background, elevated PctsR-bgaB expression was still repressed by glucose (Ishii et al., 2013). In the spx inactivation background, PctsR-bgaB expression was further repressed by glucose (Ogura M, unpublished results).
The McsB/YwlE system has profound effects on gene transcription, including the ComK-regulon (Elsholz et al., 2012). Moreover, genome-wide analysis of Arg-phosphorylated proteins indicated DegU, a transcription factor controlling multiple physiological phenomena including biofilm formation, genetic competence, motility, and exoprotease production, is Arg-phosphorylated (Kobayashi, 2007; Tsukahara and Ogura, 2008; Ogura and Tsukahara, 2010; Chan et al., 2014; Schmidt et al., 2014; Trentini et al., 2014, 2016; Ogura, 2016). We performed in vitro degradation assays using Asp-phosphorylated DegU and showed ClpCP-dependent DegU-P degradation (Ogura and Tsukahara, 2010). In this system, the role of McsB/YwlE should be clarified by future study. Recent studies revealed that the McsB/YwlE system plays critical roles for spore germination (Zhou et al., 2019). Immediately after receiving trigger nutrients, activated YwlE dephosphorylates Arg-phosphorylated SigA and Tig, important factors for transcription and translation, respectively. As a result, the ywlE disruptant showed delayed germination. This is a distinct physiological phenotype of the ywlE disruptant.
In the glucose-based semi-synthetic medium, the ywlE disruptant showed decreased growth rate (Figure 5C). This raised the possibility that Arg-phosphorylated glycolytic enzyme dephosphorylation plays a role in normal cell growth. Indeed, degradation of Eno and Pyc by ClpCP have been identified in 2D gel analysis of the B. subtilis proteome after culture in minimal medium with glutamate and without citrate (Gerth et al., 2008). Moreover, these results are consistent with previous observations that Eno and Pyc are Arg-phosphorylated proteins (Schmidt et al., 2014; Trentini et al., 2014). However, the study addressing the role of Arg-phosphorylation of GapA revealed that GapA was stable after change of glucose to malate as carbon source (Gerth et al., 2017). The recent study showed that pyruvate kinase significantly affects Z-ring formation and showed that pyruvate is a key metabolite that coordinates bacterial growth and cell division (Monahan et al., 2014). These metabolic activities, which may be regulated by YwlE, would be related to cell growth. Moreover, many glycolytic enzymes are acetylated, such as Pgi, GapA, Pgk, PdhD, and PycA (Kosono et al., 2015). These proteins have been reported to be Arg-phosphorylated, perhaps leading to degradation (Schmidt et al., 2014; Trentini et al., 2014). In B. subtilis, acetylated Eno is inhibited and FbaA and Pyk are likely to be acetylated at critical lysine residues for enzyme activities (Nakayasu et al., 2017). Considering glucose-mediated ywlE induction, glucose has positive (dephosphorylation of Arg-phosphorylated enzyme leading to protein stabilization, which is the case for PdhD) and negative effects (acetylation and inhibition) on glycolytic enzymes through these two protein modification systems. Perhaps the overall balance between both effects might be likely to be positive, because we observed that in the presence of glucose, wild-type cells showed significantly higher cell mass at the early stationary phase compared to cells without glucose (Ogura et al., 2019). Moreover, a recent study showed that wild type, mcsB, and ywlE strain growth was unchanged in LB medium, which hardly contains glucose (Zhou et al., 2019), confirming our observation that growth changes are dependent on glucose. The mcsB disruptant showed increased cell mass at the early stationary phase. In the mcsB disruptant, glycolytic enzymes are not Arg-phosphorylated, suggesting that glycolytic enzyme levels should be high, leading to more available energy. This would result in increased cell mass. Together, the cell growth phenotypes of the mcsB and ywlE disruptants probably involve Arg-phosphorylation-mediated protein degradation of glycolytic enzymes, such as PdhD. Arg-phosphorylation-mediated and ClpCP-dependent in vivo protein degradation of specific proteins under normal conditions is likely, but thus far remain unidentified (Schmidt et al., 2014).
Glucose has profound effects, including CcpA-mediated repression and activation of more than a hundred genes. Further, glucose likely activates and inhibits several unidentified transcription factors (for example, the transcription factor responsible for glucose-mediated pdhABCD induction is not known), and participates in transcriptome alterations (Blencke et al., 2003; Deutscher, 2008; Fujita, 2009). CcpA is indirectly involved in catabolite repression of ctsR/mcsAB/clpC and the previous transcriptome analyses revealed that CcpA is involved in regulation of ctsR/clpC expression in a synthetic medium (Moreno et al., 2001; Ishii et al., 2013). Our preliminary RNA-seq analysis to assess the effect of glucose on the transcriptome (cells grown in sporulation medium at T1) showed decreases of mRNA levels of ctsR, mcsA, mcsB and clpC to 44, 26, 30, and 31% of respective levels in the wild type strain (Ogura and Kanesaki, unpublished results). These are consistent with catabolite repression of PctsR. Taken together, our findings indicate that (1) TsaD is a target protein of McsB/YwlE, (2) glucose induces ywlE and represses mcsB/clpC, and (3) McsB/YwlE affects cell growth in glucose-containing semi-synthetic medium. The overall results provide profound insights on understanding B. subtilis cell physiology responses to environmental cues, including glucose.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
MO performed the experiments and wrote the manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Number 18K05415 and the Research Program of the Institute of Oceanic Research and Development.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank R. Losick (Harvard University), H. Yamamoto (Shinshu University), and F. Kawamura (Rikkyo University) for kindly supplying the bacterial strains and plasmids used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.590828/full#supplementary-material
References
Arrondel, C., Missoury, S., Snoek, R., Patat, J., Menara, G., Collinet, B., et al. (2019). Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 10:3967.
Blencke, H. M., Homuth, G., Ludwig, H., Mäder, U., Hecker, M., and Stülke, J. (2003). Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5, 133–149. doi: 10.1016/s1096-7176(03)00009-0
Bryant, J. A., Sellars, L. E., Busby, S. J. W., and Lee, J. (2014). Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 42, 11383–11392. doi: 10.1093/nar/gku828
Carabetta, V. J., and Cristea, I. M. (2017). Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol. 199, 107–117.
Carabetta, V. J., Greco, T. M., Tanner, A. W., Cristea, I. M., and Dubnau, D. (2016). Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems 1:e00005-16.
Chan, J. M., Guttenplan, S. B., and Kearns, D. B. (2014). Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis. J. Bacteriol. 196, 740–753. doi: 10.1128/jb.01217-13
Delumeau, O., Lecointe, F., Muntel, J., Guillot, A., Guédon, E., Monnet, V., et al. (2011). The dynamic protein partnership of RNA polymerase in Bacillus subtilis. Proteomics 11, 2992–3001. doi: 10.1002/pmic.201000790
Deutscher, J. (2008). The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11, 87–93. doi: 10.1016/j.mib.2008.02.007
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Dworkin, J. (2015). Ser/Thr phosphorylation as a regulatory mechanism in bacteria. Curr. Opin. Microbiol. 24, 47–52. doi: 10.1016/j.mib.2015.01.005
Elsholz, A. K., Turgay, K., Michalik, S., Hessling, B., Gronau, K., Oertel, D., et al. (2012). Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 109, 7451–7456. doi: 10.1073/pnas.1117483109
Elsholz, A. K. W., Birk, M. S., Charpentier, E., and Turgay, K. (2017). Functional Diversity of AAA+ protease complexes in Bacillus subtilis. Front. Mol. Biosci. 4:44. doi: 10.3389/fmolb.2017.00044
Fuhrmann, J., Mierzwa, B., Trentini, D. B., Spiess, S., Lehner, A., Charpentier, E., et al. (2013). Structural basis for recognizing phosphoarginine and evolving residue-specific protein phosphatases in gram-positive bacteria. Cell. Rep. 3, 1832–1839. doi: 10.1016/j.celrep.2013.05.023
Fuhrmann, J., Schmidt, A., Spiess, S., Lehner, A., Turgay, K., Mechtler, K., et al. (2009). McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science 324, 1323–1327. doi: 10.1126/science.1170088
Fuhrmann, J., Subramanian, V., Kojetin, D. J., and Thompson, P. R. (2016). Activity-based profiling reveals a regulatory link between oxidative stress and protein arginine phosphorylation. Cell Chem. Biol. 23, 967–977. doi: 10.1016/j.chembiol.2016.07.008
Fujita, Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259. doi: 10.1271/bbb.80479
Gao, H., Jiang, X., Pogliano, K., and Aronson, A. I. (2002). The E1beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 184, 2780–2788. doi: 10.1128/jb.184.10.2780-2788.2002
Gerth, U., Kock, H., Kusters, I., Michalik, S., Switzer, R. L., and Hecker, M. (2008). Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 190, 321–331. doi: 10.1128/jb.01233-07
Gerth, U., Krieger, E., Zühlke, D., Reder, A., Völker, U., and Hecker, M. (2017). Stability of proteins out of service - The GapB case of Bacillus subtilis. J. Bacteriol. 199:e00148-17.
Hata, M., Ogura, M., and Tanaka, T. (2001). Involvement of stringent factor RelA in expression of the alkaline protease gene aprE in Bacillus subtilis. J. Bacteriol. 183, 4648–4651. doi: 10.1128/jb.183.15.4648-4651.2001
Helmann, J. D. (2016). Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr. Opin. Microbiol. 30, 122–132. doi: 10.1016/j.mib.2016.02.002
Hori, K., Kaneko, M., Tanji, Y., Xing, X. H., and Unno, H. (2002). Construction of self-disruptive Bacillus megaterium in response to substrate exhaustion for polyhydroxybutyrate production. Appl. Microbiol. Biotechnol. 59, 211–216. doi: 10.1007/s00253-002-0986-8
Ishii, H., Tanaka, T., and Ogura, M. (2013). The Bacillus subtilis response regulator gene degU is positively regulated by CcpA and by catabolite-repressed synthesis of ClpC. J. Bacteriol. 195, 193–201. doi: 10.1128/jb.01881-12
Itaya, M. (1992). Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci. Biotechnol. Biochem. 56, 685–686. doi: 10.1271/bbb.56.685
Junker, S., Maaß, S., Otto, A., Hecker, M., and Becher, D. (2019). Toward the quantitative characterization of arginine phosphorylations in Staphylococcus aureus. J. Proteome Res. 18, 265–279.
Kobayashi, K. (2007). Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 66, 395–407.
Kosono, S., Tamura, M., Suzuki, S., Kawamura, Y., Yoshida, A., Nishiyama, M., et al. (2015). Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 10:e0131169. doi: 10.1371/journal.pone.0131169
Krueger, E., Zuehlke, D., Witt, E., Ludwig, H., and Hecker, M. (2001). Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20, 852–863. doi: 10.1093/emboj/20.4.852
Kunst, F., Msadek, T., Rapoport, G. (1994). “Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis,” in Regulation of Bacterial Differentiation, eds Piggot, P. J., Moran, C. P. Jr., and Youngman, P. (Washington, DC: ASM Press), 1–20.
Le Breton, Y., Mohapatra, N. P., and Haldenwang, W. G. (2006). In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72, 327–333. doi: 10.1128/aem.72.1.327-333.2006
Lehnik-Habrink, M., Rempeters, L., Kovács, ÁT., Wrede, C., Baierlein, C., Krebber, H., et al. (2013). DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J. Bacteriol. 195, 534–544. doi: 10.1128/jb.01475-12
Mijakovic, I., Grangeasse, C., and Turgay, K. (2016). Exploring the diversity of protein modifications: special bacterial phosphorylation systems. FEMS Microbiol. Rev. 40, 398–417. doi: 10.1093/femsre/fuw003
Monahan, L. G., Hajduk, I. V., Blaber, S. P., Charles, I. G., and Harry, E. J. (2014). Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5:e00935-14.
Mondal, S., Yakhnin, A. V., Sebastian, A., Albert, I., and Babitzk, E. P. (2016). NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 1:15007.
Moreno, M. S., Schneider, B. L., Maile, R. R., Weyler, W., and Saier, M. H. Jr. (2001). Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39, 1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x
Nakayasu, E. S., Burnet, M. C., Walukiewicz, H. E., Wilkins, C. S., Shukla, A. K., Brooks, S., et al. (2017). Ancient regulatory role of lysine acetylation in central metabolism. mBio 8:e01894–17. doi: 10.1128/mBio.01894-17
Nanamiya, H., Ohashi, Y., Asai, K., Moriya, S., Ogasawara, N., Fujita, M., et al. (1998). ClpC regulates the fate of a sporulation initiation sigma factor, sigmaH protein, in Bacillus subtilis at elevated temperatures. Mol. Microbiol. 29, 505–513. doi: 10.1046/j.1365-2958.1998.00943.x
Ogura, M. (2016). Post-transcriptionally generated cell heterogeneity regulates biofilm formation in Bacillus subtilis. Genes Cells 21, 335–349. doi: 10.1111/gtc.12343
Ogura, M., and Asai, K. (2016). Glucose induces ECF sigma factor genes, sigX and sigM, independent of cognate anti-sigma factors through acetylation of CshA in Bacillus subtilis. Front. Microbiol. 7:1918. doi: 10.3389/fmicb.2016.01918
Ogura, M., and Kanesaki, Y. (2018). Newly identified nucleoid-associated-like protein YlxR regulates metabolic gene expression in Bacillus subtilis. mSphere 3:e00501-18.
Ogura, M., Ohshiro, Y., Hirao, S., and Tanaka, T. (1997). A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol. 179, 6244–6253. doi: 10.1128/jb.179.20.6244-6253.1997
Ogura, M., Sato, T., and Abe, K. (2019). Bacillus subtilis YlxR, which is involved in glucose-responsive metabolic changes, regulates expression of tsaD for protein quality control of pyruvate dehydrogenase. Front. Microbiol. 10:923. doi: 10.3389/fmicb.2019.00923
Ogura, M., Shimane, K., Asai, K., Ogasawara, N., and Tanaka, T. (2003). Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol. Microbiol. 49, 1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x
Ogura, M., Shindo, K., and Kanesaki, Y. (2020). Bacillus subtilis nucleoid-associated protein YlxR is involved in bimodal expression of the fructoselysine utilization operon (furBONMD-yurJ) promoter. Front. Microbiol. 11:2024. doi: 10.3389/fmicb.2020.02024
Ogura, M., and Tanaka, T. (1996). Transcription of Bacillus subtilis degR is σD-dependent and suppressed by multicopy proB through σD. J. Bacteriol. 178, 216–222. doi: 10.1128/jb.178.1.216-222.1996
Ogura, M., and Tsukahara, K. (2010). Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol. Microbiol. 75, 1244–1259. doi: 10.1111/j.1365-2958.2010.07047.x
Pan, Q., Garsin, D. A., and Losick, R. (2001). Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol. Cell. 8, 873–883. doi: 10.1016/s1097-2765(01)00362-8
Rojas-Tapias, D. F., and Helmann, J. D. (2019). Identification of novel Spx regulatory pathways in Bacillus subtilis uncovers a close relationship between the CtsR and Spx regulons. J. Bacteriol. 201:e00151-19.
Schaeffer, P., Millet, J., and Aubert, J. (1965). Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. U.S.A. 54, 704–711.
Schilling, B., Christensen, D., Davis, R., Sahu, A. K., Hu, L. I., Walker-Peddakotla, A., et al. (2015). Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 98, 847–863. doi: 10.1111/mmi.13161
Schmidt, A., Trentini, D. B., Spiess, S., Fuhrmann, J., Ammerer, G., Mechtler, K., et al. (2014). Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response. Mol. Cell. Proteomics 13, 537–550. doi: 10.1074/mcp.m113.032292
Shiwa, Y., Yoshikawa, H., Tanaka, T., and Ogura, M. (2015). Bacillus subtilis degSU operon is regulated by the ClpXP-Spx regulated proteolysis system. J. Biochem. 157, 321–330. doi: 10.1093/jb/mvu076
Suskiewicz, M. J., and Clausen, T. (2016). Chemical biology interrogates protein arginine phosphorylation. Cell. Chem. Biol. 23, 888–890. doi: 10.1016/j.chembiol.2016.08.003
Thiaville, P. C., El Yacoubi, B., Köhrer, C., Thiaville, J. J., Deutsch, C., Iwata-Reuyl, D., et al. (2015). Essentiality of threonylcarbamoyladenosine (t(6)A), a universal tRNA modification, in bacteria. Mol. Microbiol. 98, 1199–1221. doi: 10.1111/mmi.13209
Thiaville, P. C., Legendre, R., Rojas-Benítez, D., Baudin-Baillieu, A., Hatin, I., Chalancon, G., et al. (2016). Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell. 3, 29–45. doi: 10.15698/mic2016.01.473
Trentini, D. B., Fuhrmann, J., Mechtler, K., and Clausen, T. (2014). Chasing phosphoarginine proteins: development of a selective enrichment method using a phosphatase trap. Mol. Cell. Proteomics 13, 1953–1964. doi: 10.1074/mcp.o113.035790
Trentini, D. B., Suskiewicz, M. J., Heuck, A., Kurzbauer, R., Deszcz, L., Mechtler, K., et al. (2016). Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature 539, 48–53. doi: 10.1038/nature20122
Tsukahara, K., and Ogura, M. (2008). Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 8:8. doi: 10.1186/1471-2180-8-8
Vagner, V., Dervyn, E., and Ehrlich, S. D. (1998). A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144, 3097–3104. doi: 10.1099/00221287-144-11-3097
Yamamoto, H., Kurosawa, S., and Sekiguchi, J. (2003). Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 185, 6666–6677. doi: 10.1128/jb.185.22.6666-6677.2003
Zhang, W., Collinet, B., Perrochia, L., Durand, D., and van Tilbeurgh, H. (2015). The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli. Nucleic Acids Res. 43, 1804–1817. doi: 10.1093/nar/gku1397
Keywords: glucose response, YwlE phosphatase, ClpCP protease, protein acetylation, glycolysis
Citation: Ogura M (2020) Glucose-Mediated Protein Arginine Phosphorylation/Dephosphorylation Regulates ylxR Encoding Nucleoid-Associated Protein and Cell Growth in Bacillus subtilis. Front. Microbiol. 11:590828. doi: 10.3389/fmicb.2020.590828
Received: 03 August 2020; Accepted: 07 September 2020;
Published: 25 September 2020.
Edited by:
Ilana Kolodkin-Gal, Weizmann Institute of Science, IsraelReviewed by:
Kazuo Kobayashi, Nara Institute of Science and Technology (NAIST), JapanLei Shi, Chalmers University of Technology, Sweden
Copyright © 2020 Ogura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mitsuo Ogura, b2d1cmFtQHNjYy51LXRva2FpLmFjLmpw
 Mitsuo Ogura
Mitsuo Ogura