- Department of Integrated Biological Science, Pusan National University, Busan, South Korea
The partner switching system (PSS) of the SigF regulatory pathway in Mycobacterium smegmatis has been previously demonstrated to include the anti-sigma factor RsbW (MSMEG_1803) and two anti-sigma factor antagonists RsfA and RsfB. In this study, we further characterized two additional RsbW homologs and revealed the distinct roles of three RsbW homologs [RsbW1 (MSMEG_1803), RsbW2 (MSMEG_6129), and RsbW3 (MSMEG_1787)] in the SigF PSS. RsbW1 and RsbW2 serve as the anti-sigma factor of SigF and the protein kinase phosphorylating RsfB, respectively, while RsbW3 functions as an anti-SigF antagonist through its protein interaction with RsbW1. Using relevant mutant strains, RsfB was demonstrated to be the major anti-SigF antagonist in M. smegmatis. The phosphorylation state of Ser-63 was shown to determine the functionality of RsfB as an anti-SigF antagonist. RsbW2 was demonstrated to be the only protein kinase that phosphorylates RsfB in M. smegmatis. Phosphorylation of Ser-63 inactivates RsfB to render it unable to interact with RsbW1. Our comparative RNA sequencing analysis of the wild-type strain of M. smegmatis and its isogenic Δaa3 mutant strain lacking the aa3 cytochrome c oxidase of the respiratory electron transport chain revealed that expression of the SigF regulon is strongly induced under respiration-inhibitory conditions in an RsfB-dependent way.
Introduction
Sigma factors reversibly associate with the core RNA polymerase and function as specific factors that direct transcription of a specific subset of genes. 28 sigma factor genes were found to occur in Mycobacterium smegmatis in contrast to 13 genes in Mycobacterium tuberculosis (Cole et al., 1998; Manganelli et al., 2004; Waagmeester et al., 2005; Rodrigue et al., 2006). SigF belongs to group III sigma factors and is dispensable for growth of M. tuberculosis and M. smegmatis (Chen et al., 2000; Williams et al., 2007; Provvedi et al., 2008; Singh et al., 2015). SigF is phylogenetically and functionally in close relation to the well-studied stress sigma factor SigB in Bacillus species (DeMaio et al., 1996, 1997; Hecker and Volker, 2001; Hecker et al., 2007; Bouillet et al., 2018).
The sigF gene is widely conserved in mycobacterial species (Singh and Singh, 2008; Sachdeva et al., 2010). In M. tuberculosis, SigF (Rv3286c) was shown to be involved in virulence, biofilm formation, and diverse stress responses (Chen et al., 2000; Geiman et al., 2004; Karls et al., 2006; Williams et al., 2007; Hartkoorn et al., 2012; Manganelli, 2014). SigF (MSMEG_1804) in M. smegmatis was suggested to play roles in adaptation to stationary phase and conditions of heat and oxidative stress (Gebhard et al., 2008; Humpel et al., 2010; Singh et al., 2015). Overexpression or deletion of sigF was reported to alter cell wall architectures in M. tuberculosis, Mycobacterium bovis, and M. smegmatis (Forrellad et al., 2013; Singh et al., 2015; Dutta et al., 2019). The inactivation of sigF in M. smegmatis was shown to result in the loss of carotenoid (isorenieratene) pigmentation, accompanying increased susceptibility to hydrogen peroxide (Provvedi et al., 2008; Humpel et al., 2010; Singh et al., 2015). The consensus sequence (GGWWT-N16–17-GGGTAY) was suggested for the mycobacterial SigF-recognizing promoters (Gebhard et al., 2008; Provvedi et al., 2008; Humpel et al., 2010). The sigF genes of M. tuberculosis and M. smegmatis were demonstrated to be cotranscribed with their cognate anti-sigma factor genes usfX (Rv3287c) and rsbW (MSMEG_1803), respectively (DeMaio et al., 1997; Gebhard et al., 2008). Transcription of the usfX-sigF operon in M. tuberculosis is driven from a SigF-dependent promoter, whereas the rsbW-sigF operon in M. smegmatis is transcribed from two promoters, a SigF-independent promoter located immediately upstream of rsbW and a SigF-dependent promoter upstream of the chaB (MSMEG_1802) gene that is located 103 bp upstream of rsbW (Gebhard et al., 2008).
M. tuberculosis sigF was found to be strongly induced within cultured human macrophages, during stationary phase of growth, and upon exposure to cold shock, nutrient depletion, oxidative stress, and several antibiotics (rifampicin, ethambutol, streptomycin, and cycloserine), as well as in persister cells (DeMaio et al., 1996; Graham and Clark-Curtiss, 1999; Michele et al., 1999; Mariani et al., 2000; Betts et al., 2002; Keren et al., 2011; Forrellad et al., 2013), while M. smegmatis sigF was shown to be expressed at similar levels throughout its growth phase and only marginally increased under SigF-activating conditions (Gebhard et al., 2008; Singh and Singh, 2008).
The functionality of SigF is regulated by the so-called partner switching system (PSS) including its cognate anti-sigma factor (anti-SigF) and anti-anti-sigma factors (anti-SigF antagonists) (Figure 1; DeMaio et al., 1997; Singh et al., 2015; Bouillet et al., 2018). Under non-stress (SigF-non-activating) conditions, SigF is held in an inactive state in complex with the anti-SigF (RsbW or UsfX). Under stress (SigF-activating) conditions, the release of SigF from its anti-SigF is accomplished by two anti-SigF antagonists, RsfA (Rv1365c in M. tuberculosis and MSMEG_1786 in M. smegmatis) and RsfB (Rv3687c in M. tuberculosis and MSMEG_6127 in M. smegmatis), which sequester the anti-SigF (Beaucher et al., 2002; Parida et al., 2005; Singh et al., 2015). RsfA is inactivated when a disulfide bond is formed between its two redox-responsive cysteine residues, while RsfB was suggested to be inactivated by phosphorylation (Beaucher et al., 2002; Manganelli et al., 2004). MSMEG_6129 was identified to be a protein kinase that phosphorylates RsfB in M. smegmatis (Bowman and Ghosh, 2014), while the kinase that is responsible for RsfB phosphorylation remains unknown in M. tuberculosis. Since an MSMEG_6129 mutant strain of M. smegmatis paradoxically displayed decreased expression of the SigF regulon relative to the wild-type (WT) strain (Bowman and Ghosh, 2014), it was uncertain whether MSMEG_6129 is the kinase that inactivates RsfB by phosphorylation. Dephosphorylation of RsfB homologs in Bacillus species was demonstrated to be catalyzed by the PP2C family of phosphatases (Voelker et al., 1996; Vijay et al., 2000; Chen et al., 2003). However, no study has been published regarding which gene product is responsible for dephosphorylation of phosphorylated RsfB in mycobacteria. The Rv1364c gene in M. tuberculosis was found to encode a multi-domain protein consisting of the sensor, PP2C phosphatase, GHKL (gyrase, Hsp90, histidine kinase, MutL) kinase, and anti-sigma antagonist domains (Sachdeva et al., 2008; Greenstein et al., 2009; Misra et al., 2019). Rv1364c was shown to interact with SigF in vitro, suggesting the possibility that it might serve as an anti-SigF along with the major anti-SigF UsfX (Misra et al., 2019). Although Rv1364c was demonstrated to possess both kinase and phosphatase activities that autophosphorylate and autodephosphorylate its anti-sigma antagonist domain (Greenstein et al., 2009), the question remains unanswered regarding whether it can phosphorylate and dephosphorylate RsfB to modulate the anti-SigF antagonist activity of RsfB.
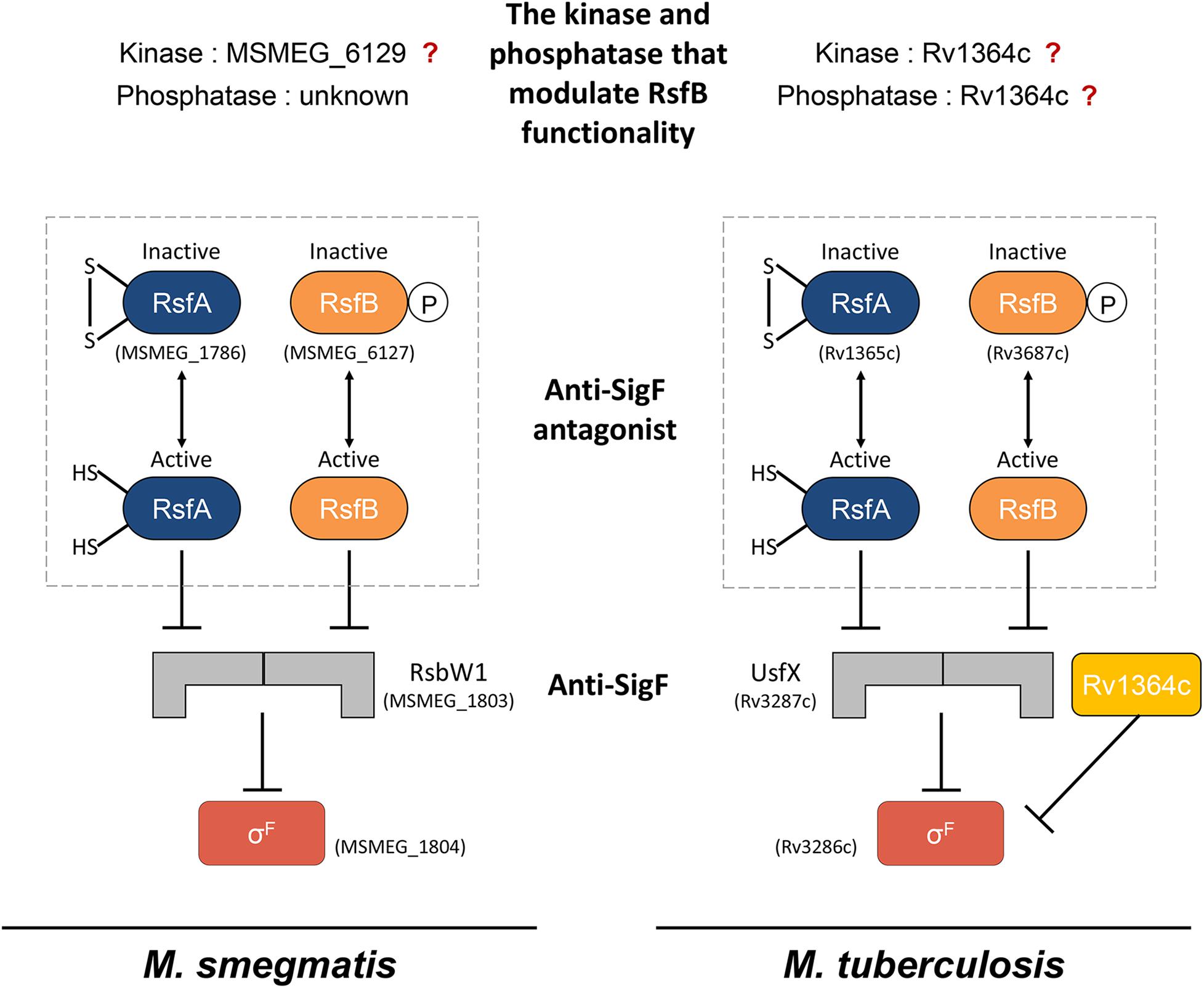
Figure 1. The schematic illustration outlining the current knowledge of the SigF PSS in M. smegmatis and M. tuberculosis. The “?” mark indicates the ambiguity of the roles of the corresponding proteins in the SigF PSS.
Using relevant mutant strains and protein interaction analyses, we here reveal the distinct roles of three RsbW homologs and two anti-SigF antagonists (RsfA and RsfB) in the SigF PSS of M. smegmatis. This study provides several lines of evidence showing that MSMEG_6129 is the only kinase in M. smegmatis that phosphorylates RsfB on Ser-63 to inactivate the functionality of RsfB as an anti-SigF antagonist. This study also presents the novel finding that expression of the SigF regulon in M. smegmatis is strongly induced under respiration-inhibitory conditions in an RsfB-dependent way.
Materials and Methods
Bacterial and Yeast Strains, Plasmids, and Culture Conditions
The bacterial and yeast strains and plasmids used in this study are listed in Table S1 in the supplementary material. Escherichia coli strains were cultivated in Luria-Bertani (LB) medium on a gyratory shaker (200 rpm) at 37°C. M. smegmatis strains were grown aerobically in Middlebrook 7H9 medium (Difco, Sparks, MD, United States) supplemented with 0.2% (w/v) glucose (7H9-glucose) and 0.02% (v/v) Tween 80 as an anti-clumping agent on a gyratory shaker at 37°C. Ampicillin (100 or 200 μg/ml for E. coli), kanamycin (50 μg/ml for E. coli and 15 or 30 μg/ml for M. smegmatis), chloramphenicol (34 μg/ml for E. coli) and hygromycin (200 μg/ml for E. coli and 50 μg/ml for M. smegmatis) were added to the growth medium when required. For treatment of M. smegmatis cultures with KCN, the cultures were grown to an optical density at 600 nm (OD600) of 0.45–0.5 and further incubated for 15 min following the addition of KCN to the cultures to a final concentration of 0.5 mM.
Saccharomyces cerevisiae strains were cultivated in YPD medium (Difco) or synthetic defined dropout (SD) medium (Clontech, Palo Alto, CA, United States) on a gyratory shaker at 30°C.
DNA Manipulation and Transformation
Standard protocols and manufacturers’ instructions were followed for recombinant DNA manipulations (Green and Sambrook, 2012). Transformation of M. smegmatis and S. cerevisiae with plasmids was conducted by electroporation and the lithium acetate (LiAc)-mediated method, respectively, as previously described (Snapper et al., 1990; Guthrie and Fink, 1991).
Site-Directed Mutagenesis
To introduce point mutations into the rsfB genes, PCR-based mutagenesis was performed using the Quick Change site-directed mutagenesis procedure (Stratagene, La Jolla, CA, United States). Synthetic oligonucleotides containing a mutated codon in the middle of their sequences were used to mutagenize the original codons. The primers used for mutagenesis are listed in Table S2. Mutations were verified by DNA sequencing.
β-Galactosidase Assay and Determination of the Protein Concentration
β-Galactosidase activity in M. smegmatis was assayed spectrophotometrically as described elsewhere (Oh and Kaplan, 1999). Protein concentration was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, United States) with bovine serum albumin as a standard protein.
Reverse Transcription-PCR and Quantitative Real-Time PCR
RNA isolation from M. smegmatis strains, preparation of cDNA, reverse transcription PCR (RT-PCR), and quantitative real-time PCR (qRT-PCR) were conducted as described previously (Kim et al., 2010). The primers used for cDNA synthesis, RT-PCR, and qRT-PCR are listed in Table S2.
Protein Purification
The C-terminally His6-tagged WT and mutant forms of RsfB were expressed in E. coli BL21 (DE3) strains harboring the pT7-7 derivative plasmids (pT7-7rsfB, pT7-7rsfBS63A, and pT7-7rsfBS63E). The strains harboring the pT7-7 derivatives were cultivated aerobically at 37°C in LB medium containing 100 μg/ml ampicillin to an OD600 of 0.4–0.6. Expression of the rsfB gene was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, and then cells were further grown for 4 h at 30°C. For purification of RsfB from M. smegmatis, the M. smegmatis strains containing pMHRsfB or pMHRsfBS63E were grown aerobically to an OD600 of 0.5–0.8 at 37°C in 7H9-glucose medium supplemented with 15 μg/ml kanamycin. Expression of the rsfB gene was induced by the addition of acetamide to a final concentration of 0.2% (w/v), and then cells were further grown for 7 h at 37°C. After 400 ml of E. coli or M. smegmatis cultures were harvested, cells were resuspended in 10 ml of buffer A [(20 mM Tris–HCl pH 8.0), 100 mM NaCl] containing 10 U/ml DNase I and 10 mM MgCl2. The resuspended cells were disrupted twice for E. coli or five times for M. smegmatis using a French pressure cell, and cell-free crude extracts were obtained by centrifugation twice at 14,000 × g for 15 min. 500 μl of 50% (v/v) slurry (bed volume: 250 μl) of Ni-Sepharose high performance resin (GE Healthcare, Piscataway, NJ, United States) was packed into a column. After equilibration of the resin with 10 bed volumes of buffer A, cell-free crude extracts were loaded into the column. The resin was washed with 40 bed volumes of buffer A containing 10 mM imidazole, 20 bed volumes of buffer A containing 30 mM imidazole, and then His6-tagged RsfB was eluted with 10 bed volumes of buffer A containing 100 mM imidazole. The eluted His6-tagged RsfB was diluted with buffer A to 10 mM imidazole and subjected to affinity chromatography again. The resin was washed with 20 bed volumes of buffer A containing 30 mM imidazole, and then His6-tagged RsfB was finally eluted with 10 bed volumes of buffer A containing 100 mM imidazole. Imidazole and NaCl were removed from purified RsfB by means of a PD-10 desalting column (GE Healthcare) equilibrated with 20 mM Tris–HCl (pH 8.0).
Purification of RsbW1 was conducted using E. coli BL21 (DE3) strains carrying pT7-7rsbW1. Cell-free crude extracts were loaded into the column packed with Ni-Sepharose resin. The resin was washed with 40 bed volumes of buffer A containing 10 mM imidazole, 20 bed volumes of buffer A containing 30 mM imidazole, and then His6-tagged RsfB was eluted with 10 bed volumes of buffer A containing 100 mM imidazole. RsbW2 and RsbW3 were purified from the E. coli BL21(DE3) strain carrying pT7-7rsbW2 and the E. coli Rosetta-gami 2 (DE3) strain with pT7-7rsbW3, respectively, in the same way as that of RsbW1 except for the modified wash and elution steps (40 bed volumes of buffer A containing 5 mM imidazole and 30 bed volumes of buffer A containing 50 mM imidazole for the wash step; 10 bed volumes of buffer A containing 250 mM imidazole for the elution step). SigF was purified from the E. coli BL21(DE3) strain carrying pT7-7sigF in the same way as RsbW2 except for the modified wash step (40 bed volumes of buffer A containing 10 mM imidazole and then 40 bed volumes of buffer A containing 25 mM imidazole).
Western Blotting Analysis
To detect expressed SigF, RsbW1, and RsfB in cells, Western blotting analyses using rabbit polyclonal antibodies against the corresponding proteins were performed as described previously (Mouncey and Kaplan, 1998). For detection of His6-tagged proteins a mouse monoclonal antibody against His-3 (Santa Cruz Biotechnology, Santa Cruz, CA, United States; sc8036) was employed. The rabbit polyclonal antibodies and His-3 monoclonal antibody were used at a 1:20,000 and 1:2,000 dilution, respectively. To detect GroEL, a mouse monoclonal antibody against HSP65 (Santa Cruz Biotechnology; sc58170) was used at a 1:2,000 dilution. Alkaline phosphatase-conjugated anti-rabbit IgG produced in goat (Sigma, St. Louis, CA, United States; A0545) or alkaline phosphatase-conjugated anti-mouse IgG produced in rabbit (Sigma; A4312) was used at a 1:10,000 dilution for the detection of the primary antibodies.
Analysis of in vitro Protein–Protein Interactions Using Non-denaturing PAGE
The mixture of two purified proteins in 20 mM Tris–HCl (pH 8.0) solution containing 20 mM β-mercaptoethanol was mixed with the same volume of 2× Binding buffer [40 mM Tris–HCl (pH 8.0), 0.01 mM EDTA (pH 8.0), 10 mM MgCl2, 20% (v/v) glycerol] and incubated for 25 min at room temperature. After the addition of 10× sample buffer [50 mM Tris–HCl (pH 6.8), 40% (w/v) sucrose, 0.05% (w/v) bromophenol blue], the mixtures were subjected to non-denaturing PAGE [7.5% (w/v) acrylamide] using electrophoresis buffer [2.5 mM Tris–HCl (pH 8.3), 19.2 mM glycine], which was initially run at 80 V for 1 h and subsequently at 100 V for 4 h. Non-denaturing PAGE was conducted at 4°C.
Analysis of in vivo Protein–Protein Interactions Using Copurification Assay
To examine protein interactions of RsbW1 with RsbW2 and RsbW3 in M. smegmatis, copurification assay using Ni-Sepharose resin was performed. The C-terminally His6-tagged RsbW2 and RsbW3 were expressed in the WT strains of M. smegmatis harboring pMHRsbW2 and pMHRsbW3, respectively. The strains were grown aerobically to an OD600 of 0.45–0.5 at 37°C in 7H9-glucose medium supplemented with 15 μg/ml kanamycin and 0.1% (w/v) acetamide. After 200 ml of M. smegmatis cultures were harvested, cells were resuspended in 8 ml of buffer A containing 10 U/ml DNase I and 10 mM MgCl2. The resuspended cells were disrupted five times using a French pressure cell, and cell-free crude extracts were obtained by centrifugation twice at 14,000 × g for 15 min. 500 μl of the 50% (v/v) slurry (bed volume 250 μl) of Ni-Sepharose resin was packed into a column. After equilibration of the resin with 10 bed volumes of buffer A, cell-free crude extracts were loaded into the column. The resin was washed with 125 bed volumes of buffer A containing 10 mM imidazole and then His6-tagged RsbW2 and RsbW3 were eluted with 10 bed volumes of buffer A containing 250 mM imidazole. RsbW2 and RsbW3 in the eluents were detected by Western blotting analysis with a His-3 monoclonal antibody. The presence of RsbW1 in the eluents was determined using Western blotting analysis with RsbW1 polyclonal antibodies.
Analysis of in vivo Protein–Protein Interactions Using Yeast Two-Hybrid Assay
S. cerevisiae AH109 strains cotransformed with both pGADT7linker and pGBKT7 derivatives were grown in SD medium (Clontech, Palo Alto, CA, United States) lacking leucine and tryptophan (SD/-Leu/-Trp). The overnight cultures were diluted with distilled water to an OD600 of 0.6 and spotted onto both solid SD/-Leu/-Trp plates and histidine-deficient SD/-Leu/-Trp/-His plates containing various concentrations of 3-amino-1,2,4-triazole (3-AT) for spotting assays. The plates were incubated at 30°C for 3–5 days.
RNA Sequencing and Gene Expression Profiling
Three biological replicate cultures of the WT and Δaa3 strains were grown aerobically to an OD600 of 0.45–0.5. Total RNA of each culture was isolated as described previously (Kim et al., 2010). rRNA was removed from the isolated total RNA using a Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina, San Diego, CA, United States). The RNA sequencing libraries were created using a TruSeq RNA Sample Prep Kit v2 (Illumina) with the standard low-throughput protocol. Sequencing of the six libraries was conducted on an Illumina HiSeq 4000 platform at Macrogen Inc. (Seoul, South Korea) using the HiSeq 3000–4000 sequencing protocol and TruSeq 3000–4000 SBS Kit v3 reagent (Illumina). Paired-end reads (101 bp) were then mapped to the reference genome sequence of M. smegmatis mc2155 (GCF_000015005.1_ASM1500v1) with the program Bowtie 1.1.2 using default settings. Summarized statistics of RNA sequencing alignment are listed in Table S3. The differentially expressed genes (DEGs) were subsequently identified pair-wise by the edgeR package in R language (Robinson et al., 2010). In this analysis, the genes with P-value < 0.05 and | log2 fold change of gene expression (FC)| > 2 were regarded as DEGs. The data described in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE155251.
In vitro Kinase Assay
Purified RsfB was mixed with purified RsbW1, RsbW2, or RsbW3 in 30 μl of reaction buffer [20 mM Tris–Cl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, and 10 mM MnCl2]. The reactions were started by adding 100 μM ATP and incubated for 30 min at 30°C. The reactions were terminated by adding 7.5 μl of gel-loading buffer [250 mM Tris–Cl (pH 6.8), 50% (w/v) glycerol, 500 mM dithiothreitol (DTT), 10% (w/v) SDS, 5% (v/v) β-mercaptoethanol, and 0.5% (w/v) bromophenol blue]. Proteins were resolved by Phos-tag SDS-PAGE prepared as described elsewhere (Barbieri and Stock, 2008). The duplicated reaction mixtures were subjected to normal SDS-PAGE. The gels were stained with Coomassie brilliant blue (CBB).
Results
Induction of the SigF Regulon Under Respiration-Inhibitory Conditions and the Genetic Organization of the Genes Involved in the SigF PSS
Comparative RNA sequencing analysis of the WT strain of M. smegmatis and its isogenic Δaa3 mutant strain with a deletion in ctaC encoding subunit III of the aa3 cytochrome c oxidase led us to identify 103 DEGs whose expression is upregulated in the Δaa3 mutant by more than four-fold with a P-value less than 0.05 relative to the WT strain. As shown in Figure 2A and Table S4, 61 genes of the 103 DEGs were found to overlap with the genes belonging to the known SigF regulon (Singh et al., 2015), suggesting that the genes of the SigF regulon are strongly upregulated in M. smegmatis, when the major terminal oxidase of the electron transport chain (ETC) is inactivated. Among the 61 identified genes, we selected two genes (MSMEG_1777 and MSMEG_1782) with the large FC and RPKM (reads per kilo base pair per million mapped reads) values in the Δaa3 mutant, and examined the expression levels of the genes in the WT (control) and Δaa3 mutant strains grown aerobically, as well as in the WT strain treated with KCN, an inhibitor of the aa3 cytochrome c oxidase (Figure 2B). Consistent with the RNA sequencing result, expression of MSMEG_1777 and MSMEG_1782 was significantly increased in the Δaa3 mutant and the WT strain treated with KCN as compared to that in the control WT strain. We also included the Δf1f2f3 mutant strain of M. smegmatis with deletions in three furA paralogous genes in this experiment, since it had been reported that the genes of the SigF regulon are strongly downregulated in the Δf1f2f3 mutant relative to the WT strain (Lee et al., 2018). As expected, expression of MSMEG_1777 and MSMEG_1782 was significantly decreased in the mutant, confirming the MSMEG_1777 and MSMEG_1782 genes belong to the SigF regulon. Based on this result, we hereafter used the MSMEG_1777 gene as a marker gene of the SigF regulon to determine the functionality of SigF.
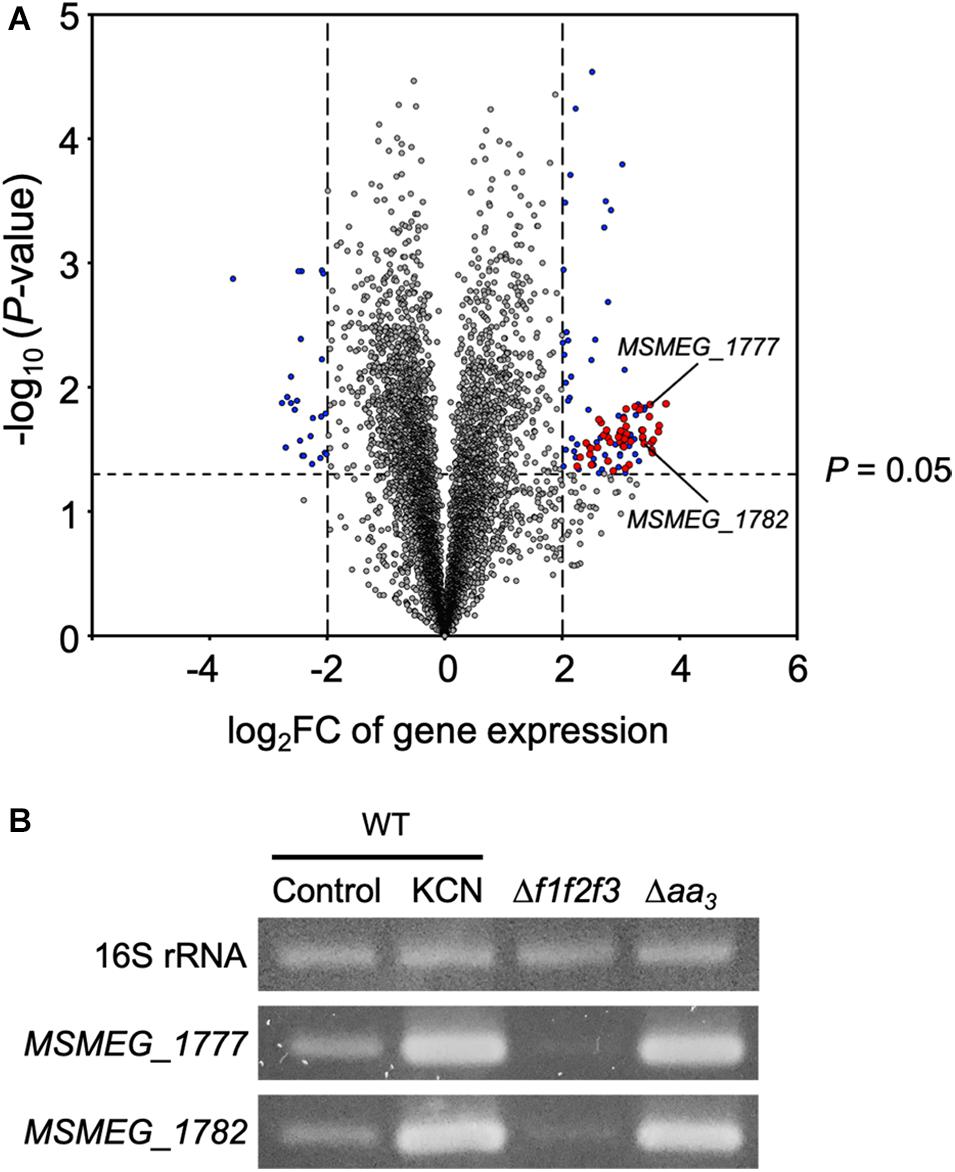
Figure 2. Overlap between the SigF regulon and the genes induced in the Δaa3 mutant strain of M. smegmatis. (A) Volcano plot showing the DEGs in the Δaa3 mutant strain relative to the WT strain. RNA sequencing was carried out using RNA extracted from three biological replicate cultures of the WT and Δaa3 strains aerobically grown to an OD600 of 0.45–0.5 in 7H9-glucose medium. The x-axis represents log2 fold change of gene expression (log2FC) in the Δaa3 mutant strain relative to the WT strain, and the y-axis represents –log10 (P-value). The vertical dashed lines on the graph mark the border lines indicating the log2FC values of –2 and 2. The genes, which are differently regulated by more than | log2FC| > 2 with P-value < 0.05, are depicted by blue-filled circles. Among the identified DEGs, the genes belonging to the SigF regulon are denoted by red-filled circles. The reported downregulated genes (log2FC < –2) in a sigF mutant strain relative to the WT strain were regarded as the genes belonging to the SigF regulon (Singh et al., 2015). Two representative genes (MSMEG_1777 and MSMEG_1782), whose expression was confirmed by RT-PCR, are indicated by the arrows. (B) RT-PCR analysis showing the expression levels of the MSMEG_1777 and MSMEG_1782 in the WT, Δf1f2f3, and Δaa3 mutant strains. The M. smegmatis strains were grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium. For treatment of M. smegmatis cultures with KCN, the WT strain was grown to an OD600 of 0.45–0.5, followed by the addition of KCN to the final concentration of 0.5 mM. The WT cultures were further grown under KCN-untreated (control) or treated (0.5 mM KCN) conditions for 15 min. RT-PCR for 16S rRNA was performed to assure that equal amounts of total RNA were employed for RT-PCR.
As a first step to understand the mechanism underlying the strong induction of the SigF regulon under respiration-inhibitory conditions, we decided to investigate the SigF PSS in detail. The rsbW-sigF (MSMEG_1803-MSMEG_1804) operon has been previously identified, and the role of RsbW (MSMEG_1803) as an anti-SigF in M. smegmatis has been suggested on the basis of its overexpression phenotype and its protein interaction with SigF (Singh et al., 2015). The genes encoding the proposed anti-SigF antagonists RsfA and RsfB have been also previously identified (Singh et al., 2015). To identify the additional genes whose products are likely to be involved in the SigF PSS, a BLAST search using the RsbW (MSMEG_1803) sequence as a query sequence was performed against the M. smegmatis mc2155 genome, which revealed two additional genes (MSMEG_1787 and MSMEG_6129) that encode the RsbW homologs. As shown in Figure 3, The MSMEG_1787 and MSMEG_6129 genes were found to be located in the vicinity of the rsfA and rsfB genes, respectively. The RsbW homologs, whose genes are adjacent to sigF, rsfB, and rsfA, were named as RsbW1, RsbW2, and RsbW3, respectively. Among the three RsbW homologs, RsbW1 composed of 138 amino acids shows the highest homology (66.2% identity) to UsfX of M. tuberculosis. The rsbW2 gene, whose product consists of 148 amino acids, appears to form an operon with the upstream gene rsfB. Downstream of rsbW2 occurs a putative operon that contains the gene (MSMEG_6130) encoding a histidine kinase and two adjacent genes (MSMEG_6128 and MSMEG_6131) encoding N-terminally receiver domain-containing proteins. Sequence analysis revealed that MSMEG_6128 and MSMEG_6131 contain a DNA-binding domain and a PP2C phosphatase domain at their C-termini, respectively. RsbW2 was previously reported to encode the protein kinase that phosphorylates the anti-SigF antagonist RsfB (Bowman and Ghosh, 2014). However, its inactivation by mutation was reported not to produce the anticipated phenotype like an increase in expression of the SigF regulon (Bowman and Ghosh, 2014), casting doubt as to whether RsbW2 is the kinase that inactivates RsfB by phosphorylation. The rsbW3 gene was found to code for the largest RsbW homolog composed of 194 amino acids. When the amino acid sequence of RsbW3 was aligned with that of RsbW1, RsbW3 was found to have an N-terminal extension of 30 amino acids which is not present in RsbW1 (Supplementary Figure S1).
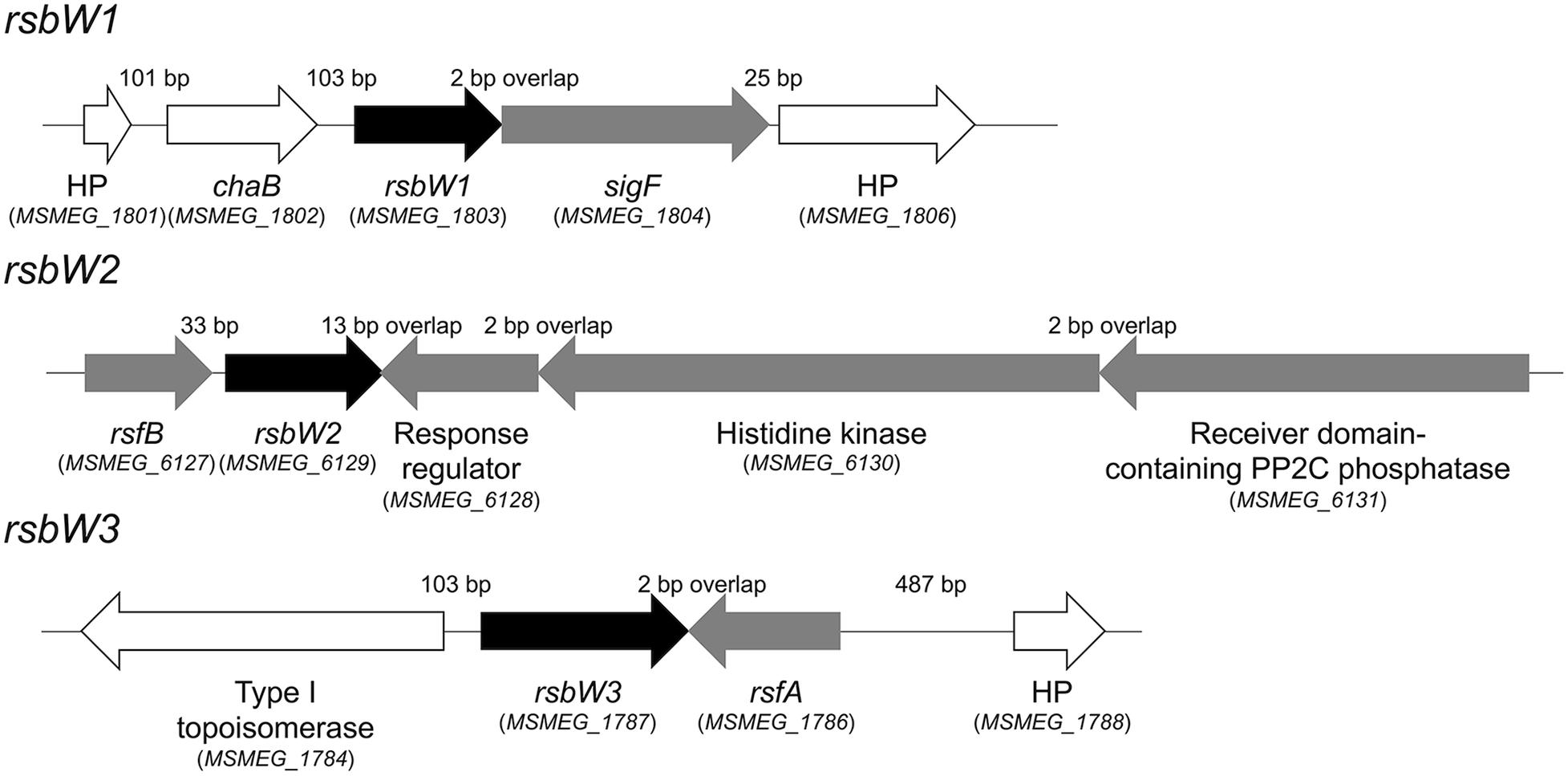
Figure 3. Genetic organization of the rsbW1, rsbW2, and rsbW3 loci of M. smegmatis mc2 155. The arrows indicate the open reading frames and their transcriptional direction. The locus tag numbers of the genes are presented in parentheses below the gene names. The lengths of the intergenic and overlapping regions are given as the nucleotide numbers above the schematic diagrams. The genes encoding hypothetical proteins are denoted by HP.
The Roles of Three RsbW Homologs in the Regulation of SigF Functionality
To examine whether the identified RsbW homologs function as anti-SigF, we individually inactivated the rsbW1, rsbW2, and rsbW3 genes by deleting the corresponding genes, yielding the ΔrsbW1, ΔrsbW2, and ΔrsbW3 mutant strains of M. smegmatis (Supplementary Figure S2). As shown in Supplementary Figure S3, both ΔrsbW1 and ΔrsbW2 mutants formed yellow-colored colonies on solid agar plates unlike the WT, ΔrsbW3, and ΔsigF strains of M. smegmatis, implying that biosynthesis of the carotenoid isorenieratene is increased in the ΔrsbW1 and ΔrsbW2 mutant strains. Given the previous report that the genes involved in biosynthesis of isorenieratene belong to the SigF regulon in M. smegmatis (Provvedi et al., 2008; Humpel et al., 2010), it is likely that expression of the SigF regulon is increased in the ΔrsbW1 and ΔrsbW2 mutants. Unfortunately, the ΔrsbW1 and ΔrsbW2 mutants were found to be instable in terms of yellow pigmentation. They lost the yellow color during cultivation in liquid growth medium, and the altered strains did not restore the yellow pigmentation in solid growth plates. For this reason, we did not examine expression of the SigF-dependent genes in the mutants. Instead, overexpression effects of rsbW1, rsbW2, and rsbW3 on MSMEG_1777 expression were examined to assess the anti-SigF activity of the three RsbW homologs. The genes of the RsbW homologs were overexpressed from an acetamide-inducible promoter on pMHRsbW1, pMHRsbW2, and pMHRsbW3 that are derivatives of the pMH201 integration vector. The expression level of MSMEG_1777 in M. smegmatis was determined using an MSMEG_1777:lacZ translational fusion, pNCII1777. As shown in Figure 4, overexpression of rsbW1 or rsbW2 led to a significant decrease in MSMEG_1777 expression in M. smegmatis compared to the control strain with pNCII1777 and pMH201, while overexpression of rsbW3 rather strongly increased the expression of MSMEG_1777. Overexpression of rsbW1, rsbW2, and rsbW3 in the M. smegmatis strains was verified by Western blotting analysis using a His-tag antibody. Altogether, the results indicate that overexpression of rsbW1 and rsbW2 inhibits the transcriptional activity of SigF, while that of rsbW3 increases the functionality of SigF.
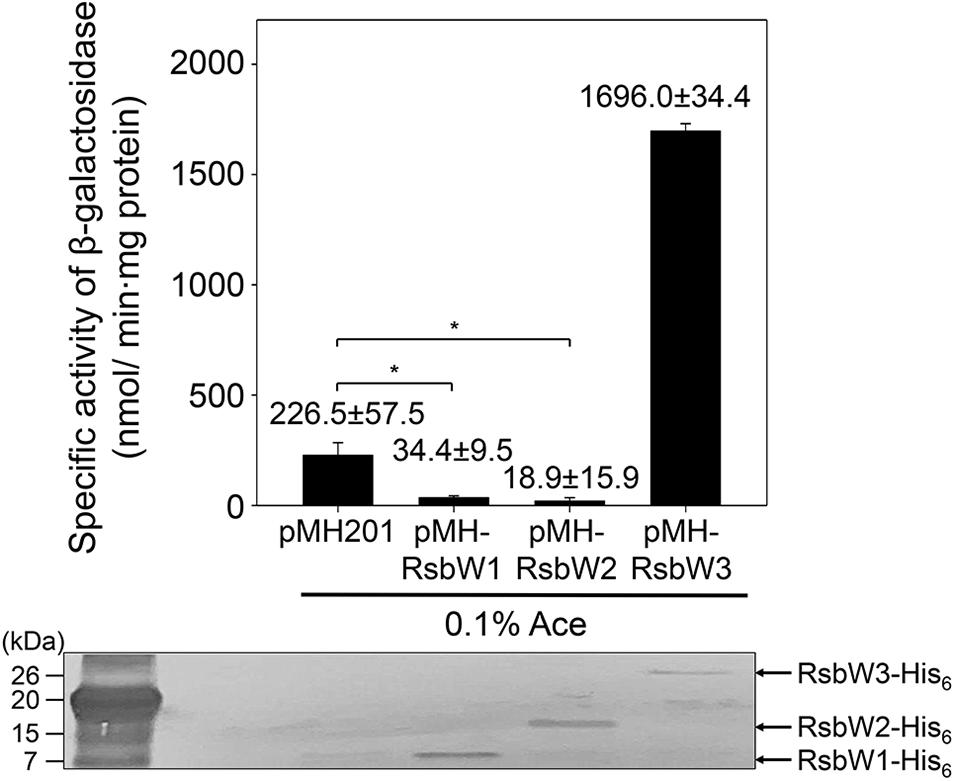
Figure 4. Overexpression effects of rsbW1, rsbW2, and rsbW3 on MSMEG_1777 expression. The rsbW1, rsbW2, and rsbW3 genes were overexpressed from the pMH201 derivatives pMHRsbW1, pMHRsbW2, and pMHRsbW3, respectively. The overexpression effects of rsbW1, rsbW2, and rsbW3 on the expression level of MSMEG_1777 were determined in the M. smegmatis strains harboring the MSMEG_1777:lacZ translational fusion plasmid (pNCII1777) and the pMH201 derivatives. As a control, the M. smegmatis strain harboring both pNCII1777 and the empty pMH201 vector was employed in the experiment. The promotor activity of MSMEG_1777 was measured by determining β-galactosidase activity in the strains grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium in the presence of 0.1% (w/v) acetamide (Ace). All values are the means of the results from three biological replicates. The error bars indicate the standard deviations. Western blotting analysis was conducted for the detection of expressed His6-tagged RsbW1, RsbW2, and RsbW3. Cell-free crude extracts (30 μg) were separated on SDS-PAGE, followed by Western blotting analysis with a His-tag antibody. *P < 0.05.
The prerequisite for a protein to act as an anti-sigma factor is protein–protein interactions between the protein and its cognate sigma factor. To determine protein–protein interactions between the RsbW homologs and SigF, we performed yeast two-hybrid assay (Y2H). For the Y2H assay, the rsbW1, rsbW2, and rsbW3 genes were cloned into the prey vector pGADT7linker, whereas the sigF gene was cloned into the bait vector pGBKT7. As shown in Figure 5A, the yeast strain coexpressing RsbW1 and SigF grew well on solid growth medium without histidine (−His) in the presence of up to 5 mM 3-AT. In contrast, coexpression of either RsbW2 or RsbW3 with SigF did not lead to growth of yeast on −His medium in the presence of 3-AT. As expected, the yeast strains coexpressing either RsbW1 or SigF alone did not grow on −His medium in the presence of 3-AT. We next examined in vitro protein–protein interactions between SigF and RsbW homologs by means of non-denaturing PAGE analysis using purified SigF and RsbW homologs. Since the RsbW3 protein was not purified to homogeneity, the partially purified protein was used in the experiment. As shown in Figure 5B, RsbW1 was shown to interact with SigF as judged by the formation of a new band representing the SigF-RsbW1 complex and disappearance of the SigF band in the non-denaturing PAGE gel. In contrast, the presence of RsbW2 and RsbW3 in the binding mixtures did not result in a decrease in the SigF band intensity in the non-denaturing PAGE gel, indicating that RsbW2 and RsbW3 do not interact with SigF. Taken together, the Y2H and non-denaturing PAGE results suggest that SigF interacts only with RsbW1 among the three RsbW homologs.
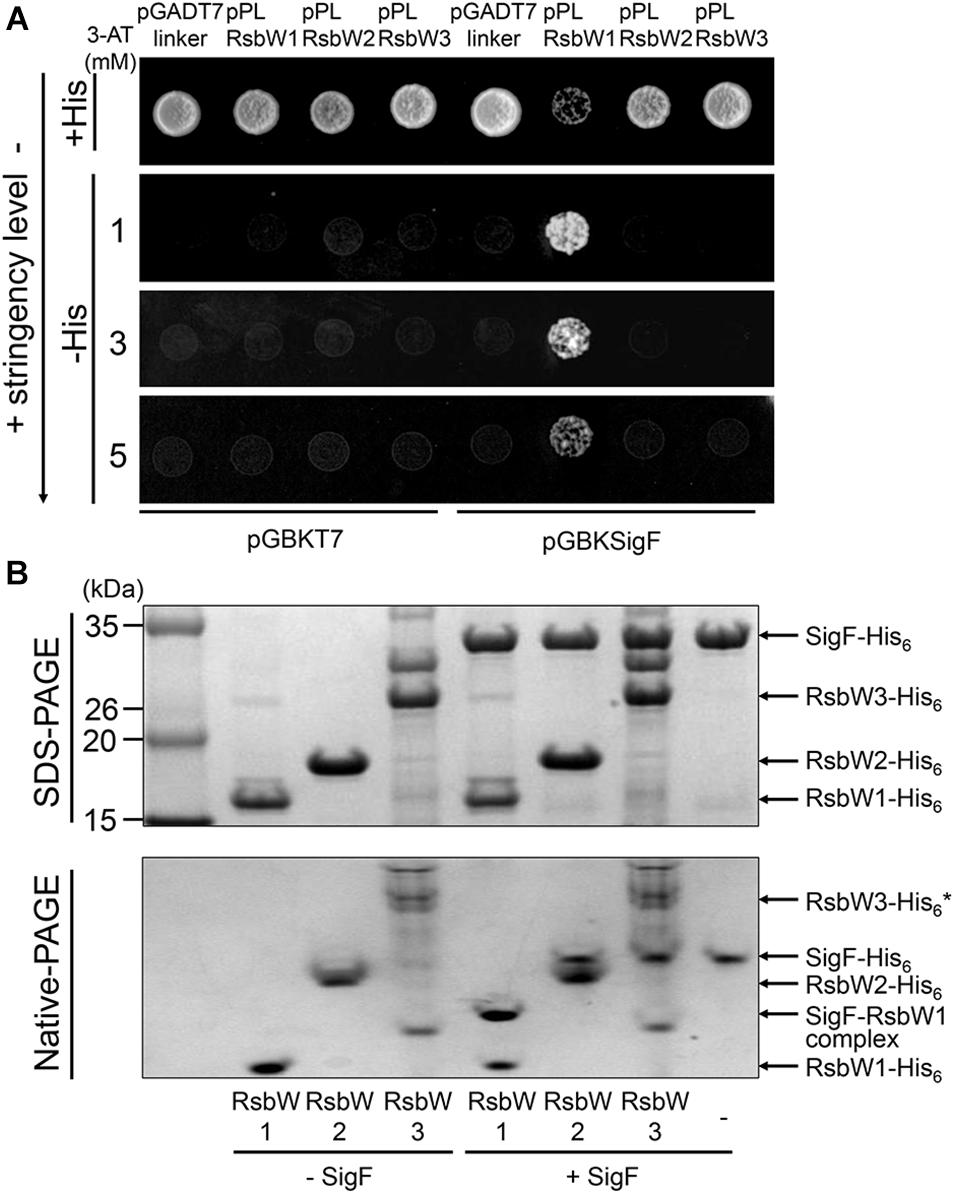
Figure 5. Determination of protein–protein interactions between SigF and RsbW homologs using Y2H assay and non-denaturing PAGE. (A) Y2H assay. The sigF gene is cloned into pGBKT7 (encoding the GAL4 DNA-binding domain), yielding pGBKSigF. The rsbW1, rsbW2, rsbW3 genes were cloned into the pGADT7linker (encoding the GAL4 activation domain), resulting in pPLRsbW1, pPLRsbW2, and pPLRsbW3, respectively. The yeast strains cotransformed with both pGBKSigF and pGADT7linker derivative plasmids were employed for Y2H assay. To discriminate false positive interactions, the yeast strains with empty pGBKT7 and either pGADT7linker or the pGADT7linker derivatives were included in Y2H assay as negative controls. All yeast strains were spotted onto SD/-Leu/-Trp plates (+His) and histidine-deficient SD/-Leu/-Trp/-His plates (–His) containing different concentrations of 3-AT. The numbers to the left indicate the concentration of 3-AT. (B) Non-denaturing PAGE analysis. 100 pmol of purified SigF was mixed with purified RsbW1 (200 pmol), RsbW2 (200 pmol), or RsbW3 in binding buffer and incubated for 30 min at 25°C (+SigF). The mixtures were subjected to both SDS-PAGE (upper panel) and native PAGE (lower panel). As controls, the binding mixtures without the addition of SigF, which contain the RsbW homologs alone, were included in the experiment (–SigF). The gels were stained with CBB. The bands representing the RsbW homologs, SigF, and the RsbW1-SigF complex are indicated by the arrows. * Since RsbW3 was not purified to homogeneity, the RsbW3 band in the native PAGE gel is uncertain.
The observed overexpression effect of rsbW3 on MSMEG_1777 led us to assume that RsbW3 might serve as an anti-SigF antagonist. To examine this assumption, protein–protein interactions between RsbW3 and three RsbW homologs were assessed using Y2H analysis (Figure 6A). For the Y2H assay, the rsbW1, rsbW2, and rsbW3 genes were cloned into pGADT7linker, and the rsbW3 gene was cloned into pGBKT7. Only the yeast strain coexpressing RsbW1 and RsbW3 grew on −His medium containing 0.5 mM 3-AT, indicating a possible protein interaction between RsbW1 and RsbW3. Protein–protein interactions between RsbW1 and RsbW3 were also assessed by copurification analysis using affinity chromatography (Figure 6B). RsbW1 was copurified with His6-tagged RsbW3 from crude extracts of the WT strain of M. smegmatis expressing His6-tagged RsbW3, whereas RsbW1 was not copurified with His6-tagged RsbW2 from the M. smegmatis strain expressing His6-tagged RsbW2, confirming protein–protein interactions between RsbW1 and RsbW3.
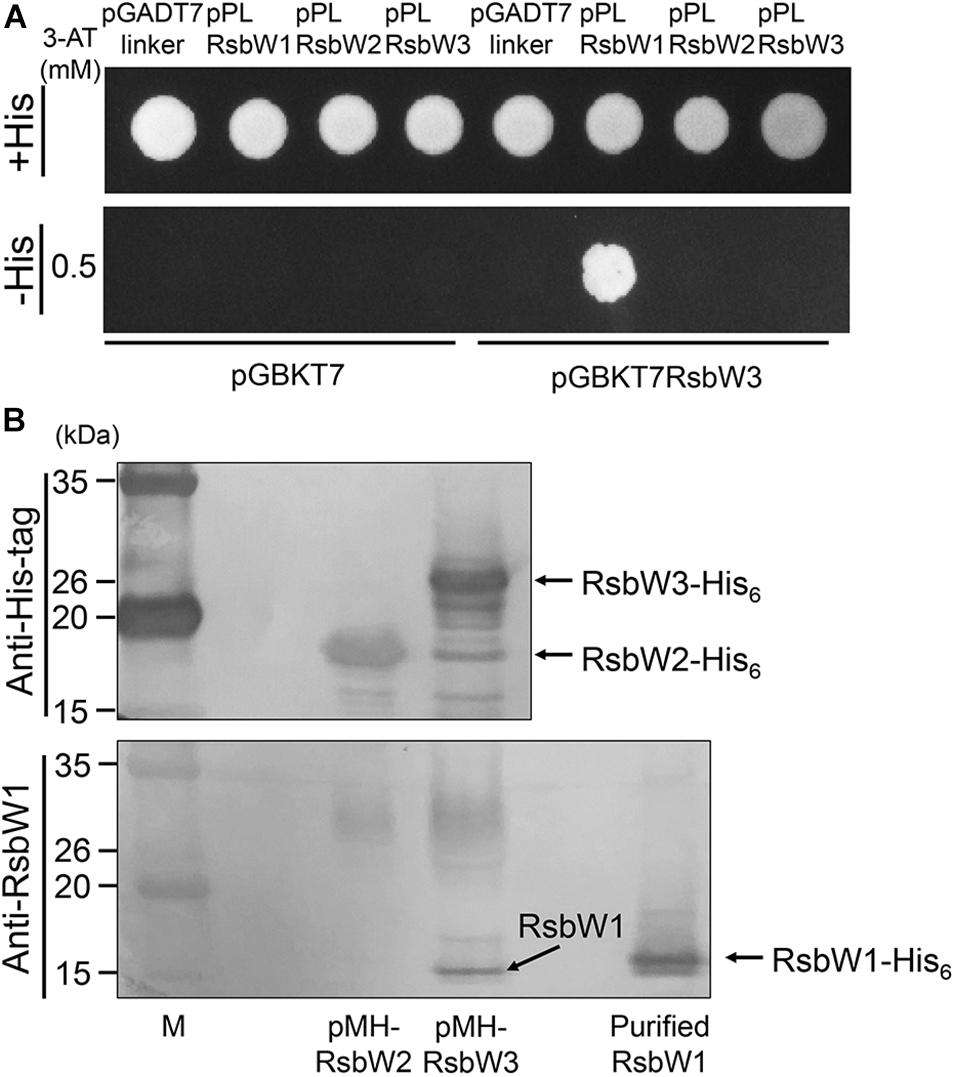
Figure 6. Determination of protein–protein interactions between RsbW3 and RsbW homologs using Y2H and copurification assays. (A) Y2H assay. The yeast strains carrying both pGBKT7-derived pGBKRsbW3 and pGADT7linker-derived plasmids (pPLRsbW1, pPLRsbW2, and pPLRsbW3) were employed for Y2H assay. The yeast strains harboring the empty pGBKT7 or pGADT7linker vector were used as negative controls. All yeast strains were spotted onto SD/-Leu/-Trp plates (+His) and histidine-deficient SD/-Leu/-Trp/-His plates (–His) containing 0.5 mM 3-AT. (B) Copurification assay. His6-tagged RsbW2 and RsbW3 were expressed in the WT strains of M. smegmatis with pMHRsbW2 and pMHRsbW3, respectively, and partially purified from their crude extracts by means of affinity chromatography using Ni-Sepharose resin. The purified proteins were subjected to Western blotting analyses using a His-3 monoclonal antibody for detection of His6-tagged RsbW2 and RsbW3 (upper panel), as well as RsbW1 polyclonal antibodies for detection of RsbW1 (lower panel). The bands representing RsbW1 and His6-tagged RsbW1, RsbW2, and RsbW3 are indicated by the arrows. M, molecular weight marker lanes.
Both RsfA and RsfB Are Functional as Anti-SigF Antagonists, and RsfB Is the Major Anti-SigF Antagonist in M. smegmatis
After having established the distinct roles of three RsbW homologs in the SigF PSS, we next examined the roles of the suggested anti-SigF antagonists RsfA and RsfB in vivo. We constructed deletion mutants of rsfA and rsfB in the background of both WT and Δaa3 strains, and the expression level of MSMEG_1777 in the WT and mutant strains of M. smegmatis was comparatively determined (Figures 7A,B). The inactivation of the aa3 cytochrome c oxidase was used for an induction condition of the SigF regulon. The expression level of MSMEG_1777 was shown to be increased by 4.9-fold in the Δaa3 strain of M. smegmatis relative to that in the WT strain grown under the same conditions. Expression of MSMEG_1777 was abolished in the ΔsigF and Δaa3ΔsigF mutant strains. Both the results confirmed that transcription of MSMEG_1777 depends on SigF, and that expression of the SigF regulon is induced under respiration-inhibitory conditions. Expression of MSMEG_1777 was decreased by 26% in the ΔrsfA mutant compared to the WT strain. The Δaa3ΔrsfA mutant also showed a 31% decrease in MSMEG_1777 expression relative to the Δaa3 mutant strain. It is noteworthy that inactivation of rsfB almost abolished expression of MSMEG_1777 in both WT and Δaa3 mutant strains. These results suggest that both RsfA and RsfB serve as anti-SigF antagonists, and that RsfB is the major anti-SigF antagonist in M. smegmatis under both SigF-activating and SigF-non-activating conditions. To confirm the roles of RsfA and RsfB as anti-SigF antagonists, we examined whether MSMEG_1777 expression correlates with the expression levels of RsfA and RsfB. The rsfA and rsfB genes were expressed from an acetamide-inducible promoter on pMHRsfA and pMHRsfB, respectively. The expression level of MSMEG_1777 was determined in the ΔrsfA mutant with pMHRsfA and the ΔrsfB mutant with pMHRsfB with increasing concentrations of acetamide in growth medium. As shown in Figures 7C,D, the expression level of MSMEG_1777 in the M. smegmatis strains with either pMHRsfA or pMHRsfB were gradually increased with increasing concentrations of acetamide, and lower concentrations of acetamide were required for similar levels of MSMEG_1777 induction in M. smegmatis with pMHRsfA compared to M. smegmatis with pMHRsfB. Western blotting analysis showed that the amount of expressed RsfB was proportional to the concentration of acetamide. We did not detect the expressed RsfA by Western blotting analysis in the concentration range of acetamide used in the experiment. Taken together, these results provide the strong evidence that both RsfA and RsfB function as anti-SigF antagonists in M. smegmatis.
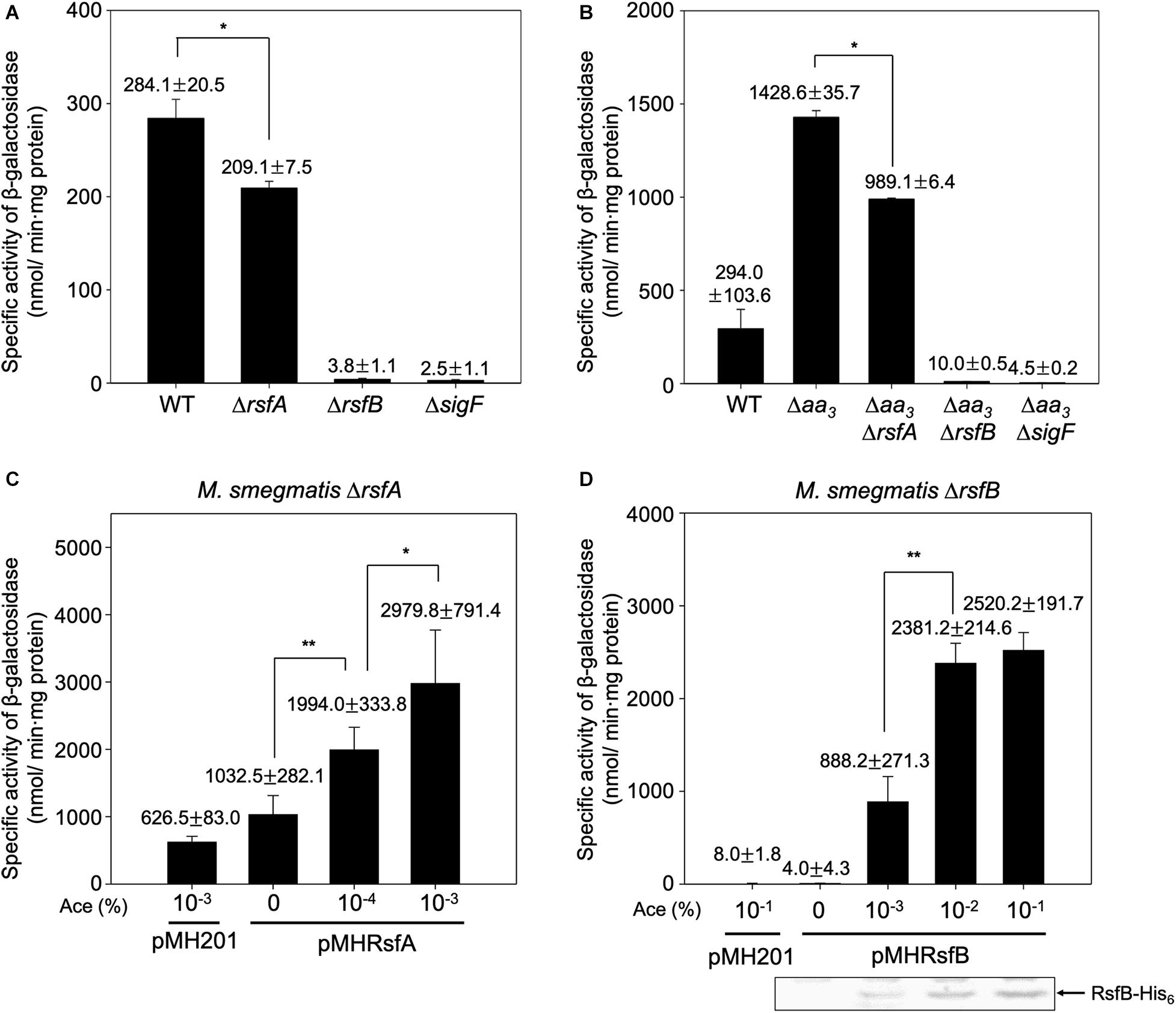
Figure 7. Effects of inactivation and overexpression of rsfA and rsfB on expression of MSMEG_1777 in M. smegmatis. The expression level of MSMRG_1777 was determined in the WT, ΔrsfA, ΔrsfB, ΔsigF, Δaa3, Δaa3ΔrsfA, Δaa3ΔrsfB, and Δaa3ΔsigF strains of M. smegmatis harboring pNCII1777 (A,B). The M. smegmatis strains were grown aerobically to an OD600 of 0.45–0.5 in 7H9 medium. Cell-free crude extracts were used to measure β-galactosidase activity. All values are the means of the results from three biological replicates. The error bars indicate the standard deviations. *P < 0.05. Effects of increasing expression of rsfA and rsfB on MSMEG_1777 expression were examined in the ΔrsfA and ΔrsfB strains harboring pNCII1777, respectively (C,D). The pMH201-derived pMHRsfA plasmid was used for the controllable expression of rsfA in the ΔrsfA strain, and pMHRsfB was used for the controllable expression of rsfB in the ΔrsfB strain. The strains were grown aerobically to an OD600 of 0.45–0.5 in 7H9 medium supplemented with the indicated concentrations of acetamide (Ace). Cell-free crude extracts were used to measure β-galactosidase activity. All values are the means of the results from three biological replicates. The error bars indicate the standard deviations. Western blotting analysis was performed for the detection of expressed His6-tagged RsfB. Cell-free crude extracts (30 μg) were separated on SDS-PAGE, followed by Western blotting analysis with a His-tag antibody. **P < 0.01 and *P < 0.05.
We further examined whether the significantly reduced expression of MSMEG_1777 in the ΔrsfB mutant is caused by the decreased expression of sigF or the increased expression of rsbW1. Using Western blotting analysis, the protein levels of expressed SigF and RsbW1 were determined in the WT, ΔrsfA, and ΔrsfB strains that were grown aerobically to an OD600 of 0.45–0.5 (Supplementary Figure S4). The Western blotting result showed that the protein levels of SigF and RsbW1 in the ΔrsfA and ΔrsfB mutant strains are not different from those in the WT strain, indicating that the cellular levels of SigF and RsbW1 in M. smegmatis are not decreased under SigF-non-activating conditions. This finding can be explained by the presence of a SigF-independent promoter immediately upstream of the rsbW1-sigF operon (Gebhard et al., 2008).
The Functionality of RsfB Is Controlled Through Phosphorylation of Ser-63 by RsbW2
A previous study has reported that purified RsbW2 (MSMEG_6129) phosphorylates RsfB (MSMEG_6127) in vitro (Bowman and Ghosh, 2014). Using LC-tandem mass spectrometry, eight Ser/Thr residues (Ser-3, Thr-10, Thr-20, Thr-25, Thr-27, Thr-32, Ser-42, and Ser-63) in RsfB have been identified to be the phosphorylation sites by RsbW2 (Bowman and Ghosh, 2014). To specify the functionally important residue(s) among the identified phosphorylation sites in RsfB, we examined the functionality of the mutant forms of RsfB (T10A, T220A, T25A, T27A, T32A, S42A, S63A, and S63E) by determining MSMEG_1777 expression in the ΔrsfB mutant expressing the mutant forms of RsfB. The Ser-3 was excluded from the experiment since the residue is not present in Bacillus subtilis RsbV (Figure 8A). The ΔrsfB mutant of M. smegmatis was complemented by introducing the pMV306 derivatives that carry the WT or mutant rsfB genes. The WT and ΔrsfB strains carrying the pMV306 empty vector were included in the experiment as positive and negative controls, respectively. As shown in Figure 8B, the S63A mutation led to a drastic increase in MSMEG_1777 expression in M. smegmatis, whereas the T32A mutation resulted in abolishment of MSMEG_1777 expression. The expression level of MSMEG_1777 was shown to be decreased by about 50% in the M. smegmatis strain expressing the phosphomimetic S63E form of RsfB compared to the control M. smegmatis strain expressing WT RsfB. Western blotting analysis revealed that the WT and mutant forms of RsfB were expressed at similar levels in the M. smegmatis strains used in the experiment. These results suggest the followings: (i) unphosphorylated RsfB is the active form of RsfB as an anti-SigF antagonist, (ii) phosphorylation of RsfB on Ser-63 decreases the functionality of RsfB as in the RsfB homologs such as RsbV and SpoIIAA of B. subtilis (Bouillet et al., 2018), (iii) Thr-32 is important for anti-SigF antagonist activity of RsfB.
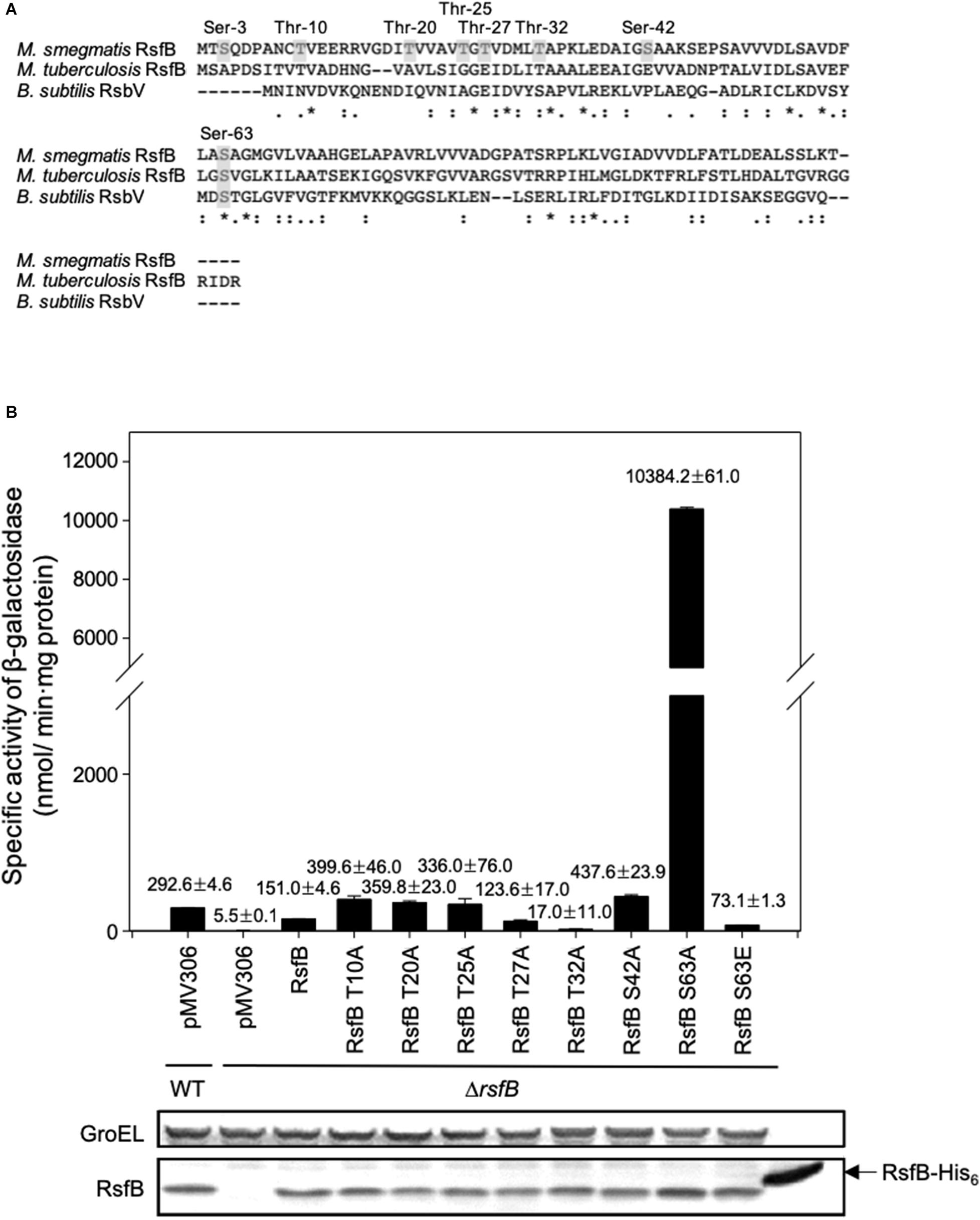
Figure 8. Identification of the amino acid residues that are responsible for the inactivation of RsfB by phosphorylation. (A) Multiple sequence alignment of the RsfB homologs of M. smegmatis, M. tuberculosis, and B. subtilis was generated using ClustalW. The asterisks and colons denote the conserved and conservatively substituted amino acid residues, respectively. The residues of M. smegmatis RsfB, which were identified to be phosphorylated by MSMEG_6129 in vitro (Bowman and Ghosh, 2014), are shown in the gray background. (B) Effects of T10A, T20A, T25A, T27A, S42A, S63A, and S63E mutations on the functionality of RsfB in vivo. The ΔrsfB strain harboring pNCII1777 was complemented with pMV306RsfB and its derivatives carrying the mutated rsfB gene (pMVRsfBT10A, pMVRsfBT20A, pMVRsfBT25A, pMVRsfBT27A, pMVRsfBT32A, pMVRsfBS42A, pMVRsfBS63A, pMVRsfBS63E). The complementation test was performed by determining the expression level of MSMEG_1777 in the M. smegmatis strains. As controls, the M. smegmatis WT and ΔrsfB mutant strains with both pNCII1777 and the empty vector pMV306 were included in the experiment. The M. smegmatis strains were grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium. Cell-free crude extracts were used to measure β-galactosidase activity. All values are the means of the results from three biological replicates. The error bars indicate the standard deviations. Protein levels of the WT and mutant forms of RsfB expressed in the strains were detected by Western blotting analysis with RsfB polyclonal antibodies. As a loading control, GroEL was detected by a GroEL monoclonal antibody.
To examine the phosphorylation state of RsfB in M. smegmatis grown under SigF-non-activating conditions and whether Ser-63 is the only residue that is phosphorylated, we expressed the WT and S63E mutant forms of His6-tagged RsfB in both M. smegmatis and E. coli, purified the proteins, and determined their phosphorylation state using Phos-tag SDS-PAGE analysis. As shown in Figure 9, most fractions of WT RsfB purified from M. smegmatis were found to be phosphorylated, while S63E RsfB purified from M. smegmatis was not phosphorylated at all. Both WT and S63E mutant forms of RsfB purified from E. coli were found to be unphosphorylated. The results indicate that Ser-63 in RsfB is the residue that is phosphorylated in M. smegmatis, and that E. coli does not have the protein kinase that can phosphorylate RsfB.
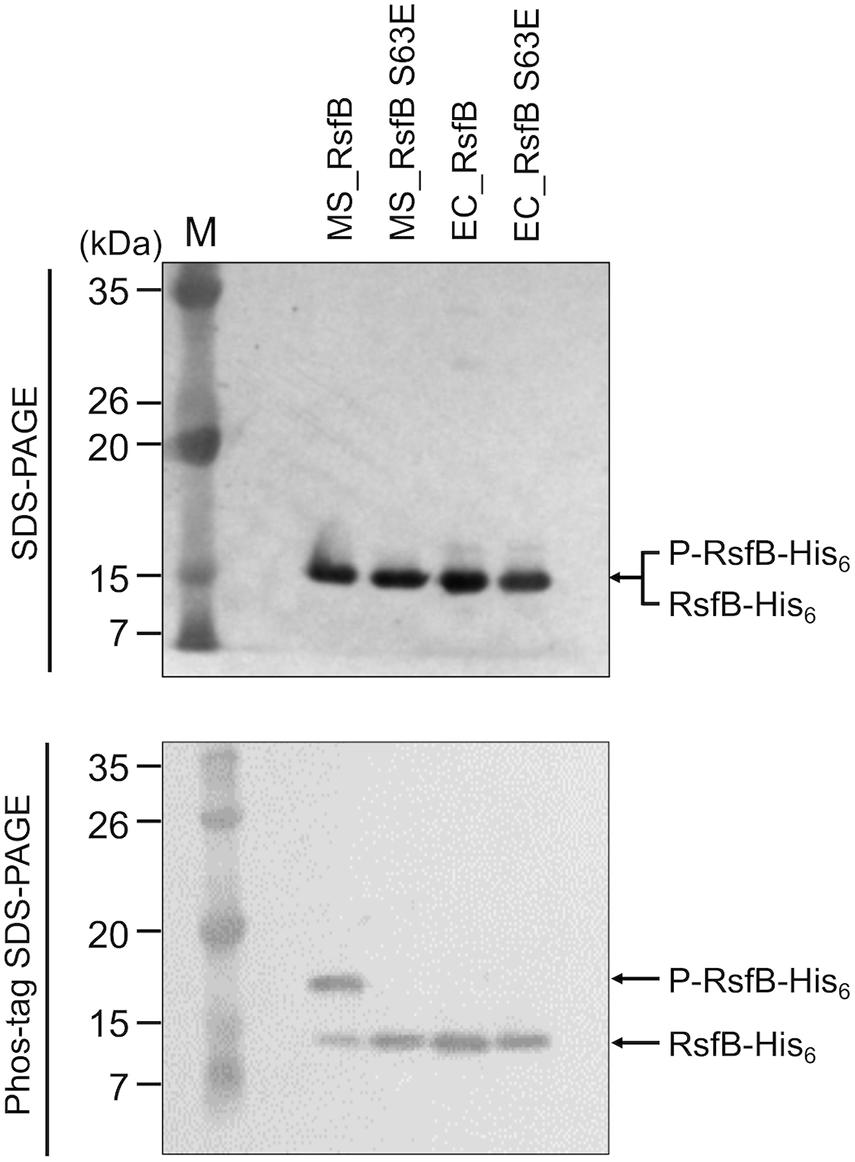
Figure 9. Phosphorylation state of the WT and S63E mutant forms of RsfB in M. smegmatis and E. coli. The WT and S63E mutant forms of His6-tagged RsfB were purified from E. coli and M. smegmatis that were aerobically grown to an OD600 of 0.4–0.5. 2 μg each of the WT and S63E mutant forms of RsfB were subjected to SDS-PAGE and Phos-tag SDS-PAGE, followed by Western blotting analysis with RsfB polyclonal antibodies. The bands representing unphosphorylated RsfB (RsfB-His6) and phosphorylated RsfB (P-RsfB-His6) are indicated by the arrows. MS_RsfB, RsfB purified from M. smegmatis. EC_RsfB, RsfB purified from E. coli M, molecular weight marker lanes.
We next examined the effect of Ser-63 phosphorylation on protein–protein interactions between RsfB and RsbW1. In place of phosphorylated RsfB, the phosphomimetic S63E mutant form of RsfB was employed for non-denaturing PAGE analysis. The WT and S63A RsfB proteins purified from E. coli were used as unphosphorylated RsfB. As shown in Figure 10, both WT RsfB and S63A RsfB interacted with purified RsbW1 and formed the retarded bands representing the RsfB-RsbW1 complex in non-denaturing PAGE. The intensity of the RsfB-RsbW1 complex bands was increased up to the ratio of RsbW1 to RsfB to be 1:1 with increasing amounts of WT RsfB and S63A RsfB. In contrast, the S63E mutant form of RsfB did not give rise to the RsfB-RsbW1 complex band even at high concentrations of S63E RsfB. The results suggest that phosphorylation of Ser-63 inactivates RsfB to render it unable to interact with RsbW1.
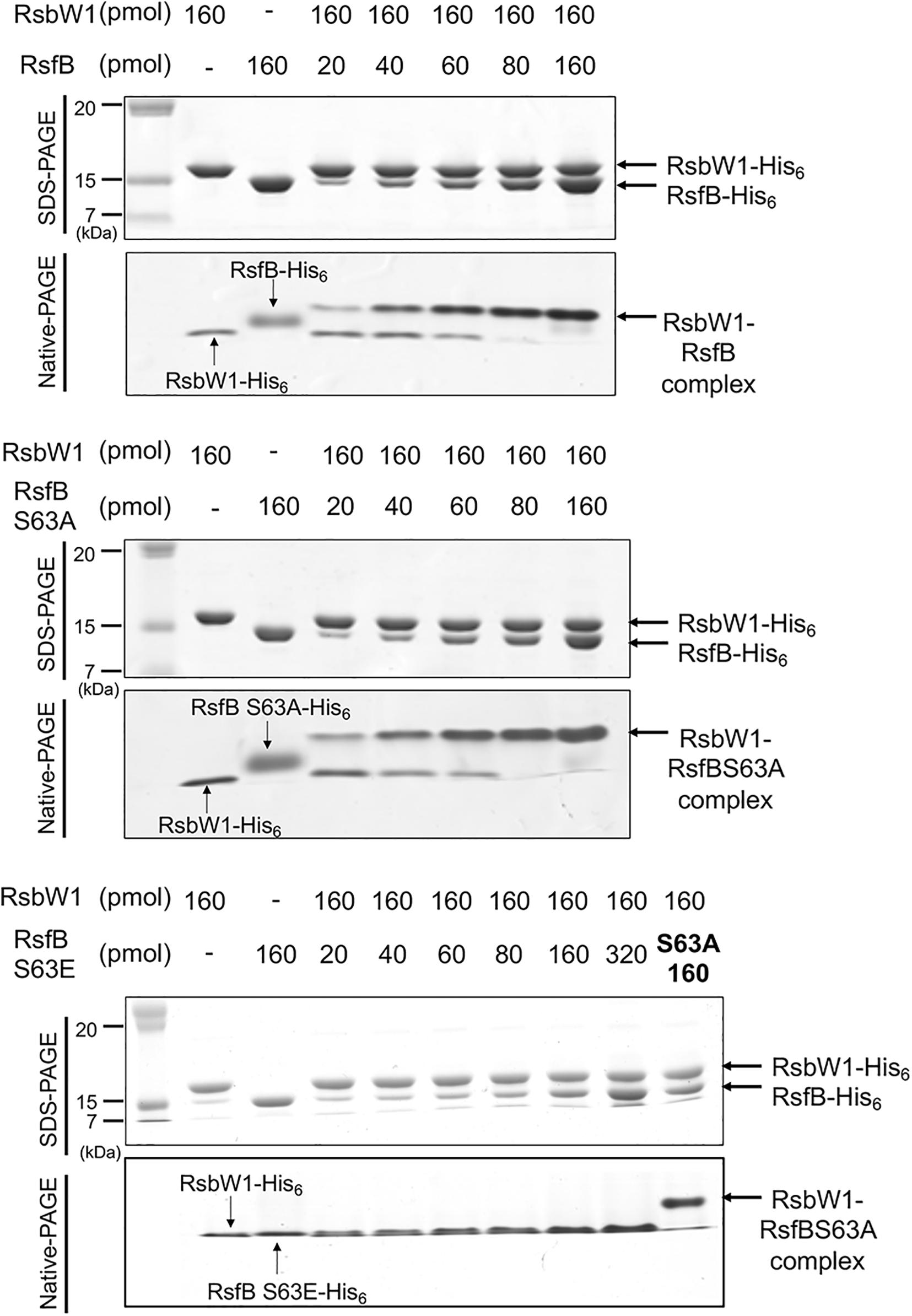
Figure 10. Determination of protein–protein interactions between RsbW1 and several forms of RsfB (WT, S63A, and S63E) by non-denaturing PAGE. 160 pmol of purified RsbW1 was mixed with increasing amounts of the purified WT and mutant forms (S63A and S63E) of RsfB in binding buffer [40 mM Tris–HCl (pH 8.0), 0.01 mM EDTA, 10 mM MgCl2, 20% (v/v) glycerol] and incubated for 30 min at 25°C. The mixtures were subjected to both SDS-PAGE (upper panel) and native PAGE (lower panel). The gels were stained with CBB. The bands representing RsbW1, RsfB (WT and mutant forms), and RsbW1-RsfB complex are indicated by the arrows.
Since the RsbW homologs of Bacillus SigB had been demonstrated to function as both anti-SigB and the protein kinase phosphorylating the anti-SigB antagonist (Benson and Haldenwang, 1993; Dufour and Haldenwang, 1994), we wondered whether in addition to RsbW2, RsbW1, and RsbW3 have the protein kinase activity phosphorylating RsfB. To examine this possibility, we performed in vitro kinase assay using purified RsbW homologs and RsfB. As shown in Figure 11A, only RsbW2 could phosphorylate unphosphorylated RsfB purified from E. coli, which is in good agreement with the fact that RsbW1 and RsbW3 are closely clustered with anti-sigma factors lacking the kinase activity, while RsbW2 is clustered with kinase-positive anti-sigma factors (Supplementary Figure S5). We also examined the phosphorylation state of RsfB in the WT and ΔrsbW2 mutant strains grown under SigF-non-activating conditions using Phos-tag SDS-PAGE and Western blotting analysis (Figure 11B). The Δ5437 mutant of M. smegmatis was included in the experiment, since it had been suggested that MSMEG_5437 is a Ser/Thr protein kinase that might modulate RsbW2 activity by phosphorylation (Bowman and Ghosh, 2014). Phos-tag SDS-PAGE showed that RsfB in the ΔrsbW2 mutant was not phosphorylated in contrast to RsfB in the WT and Δ5437 strains of M. smegmatis, indicating that RsbW2 is the only protein kinase that phosphorylates RsfB in M. smegmatis, and that RsbW2 is still active in the Δ5437 mutant.
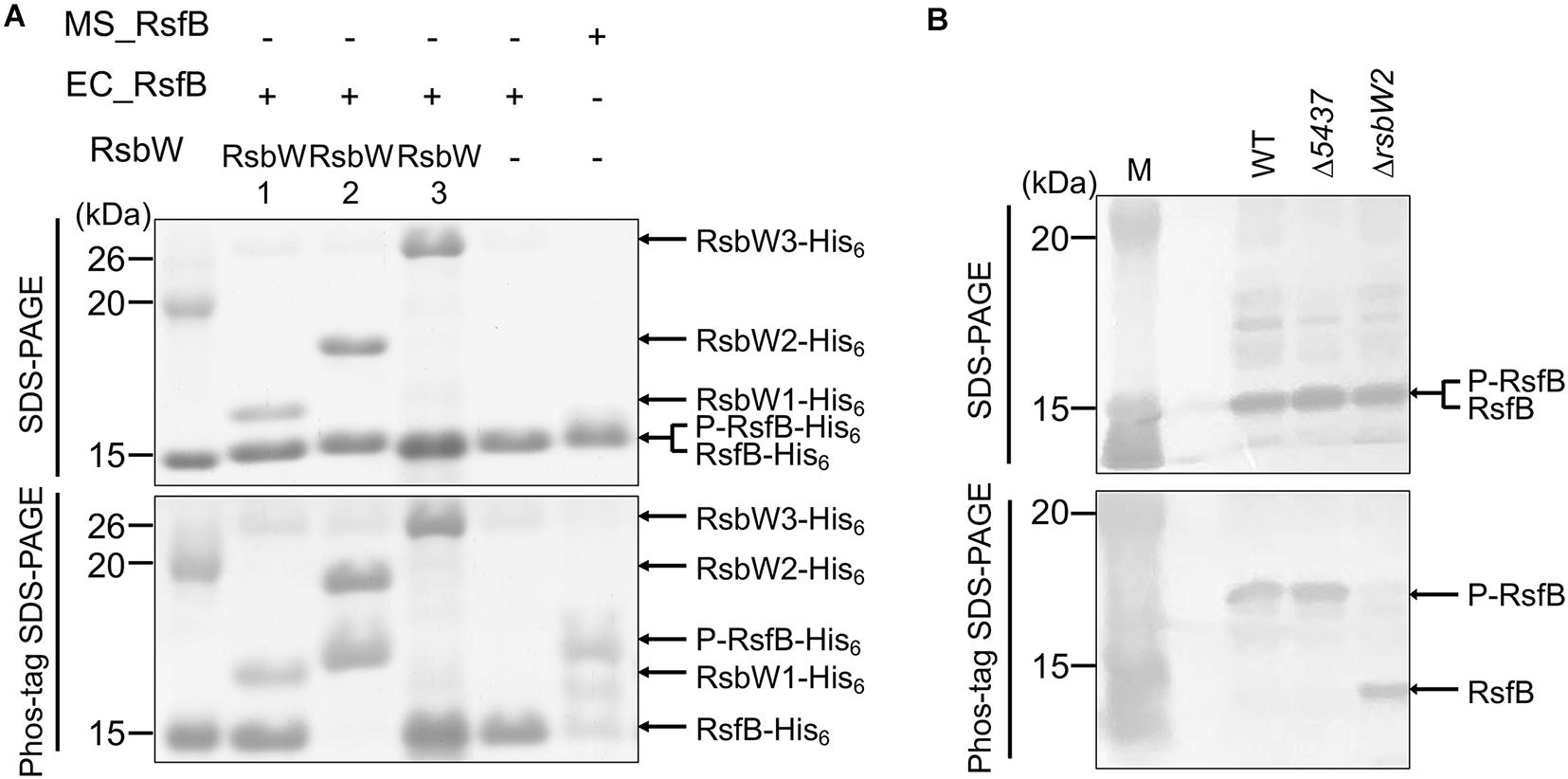
Figure 11. Determination of kinase activities of the RsbW homologs acting on RsfB. (A) RsfB purified from E. coli was mixed with partially purified RsbW1, purified RsbW2, or partially purified RsbW3 in reaction buffer [20 mM Tris–Cl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, and 10 mM MnCl2]. The phosphorylation reactions were initiated by the addition of 100 μM ATP and performed for 30 min at 30°C. Subsequently, the reaction mixtures were subjected to both SDS-PAGE (upper panel) and Phos-tag SDS-PAGE (lower panel). The gels were stained with CBB. RsfB partially purified from M. smegmatis was used as the phosphorylated control of RsfB. The bands of the His6-tagged RsbW homologs, unphosphorylated RsfB (RsfB-His6), and phosphorylated RsfB (P-RsfB-His6) are indicated by the arrows. MS_RsfB, RsfB purified from M. smegmatis. EC_RsfB, RsfB purified from E. coli. (B) The phosphorylation state of the RsfB protein in the WT, Δ5437, and ΔrsbW2 mutant strains of M. smegmatis. The strains were grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium. Cell-free crude extracts (60 μg) of the strains were separated simultaneously on both SDS-PAGE and Phos-tag SDS-PAGE, followed by Western blotting analysis with RsfB polyclonal antibodies. The bands representing unphosphorylated RsfB (RsfB) and phosphorylated RsfB (P-RsfB) are indicated by the arrows. M, molecular weight marker lanes.
Discussion
The genome of M. smegmatis contains three genes encoding RsbW homologs (RsbW1, RsbW2, and RsbW3). Among them, RsbW1 shows the highest degree of homology to UsfX that is a known anti-SigF in M. tuberculosis. Y2H and non-denaturing PAGE analysis revealed the interaction of RsbW1 with SigF, which is in good agreement with the previous result from bacterial two-hybrid assay (Singh et al., 2015). Overexpression of rsbW1 in M. smegmatis led to a significant reduction in expression of MSMEG_1777 that is under the control of SigF. Furthermore, disruption of the rsbW1 gene by deletion resulted in an increase in yellow pigmentation of M. smegmatis colonies, which appears to be the result of increased isorenieratene biosynthesis. All of these results indicate that RsbW1 is a bona fide anti-SigF in M. smegmatis.
RsbW2 is most deviated among the RsbW homologs with regard to the reciprocal sequence homology (Supplementary Figure S5). In contrast to RsbW1, RsbW2 was shown not to interact with SigF in Y2H and non-denaturing PAGE analysis, implying that RsbW2 does not play a direct role as an anti-SigF. However, overexpression and inactivation of rsbW2 gave rise to the same phenotype as those of rsbW1 in terms of both MSMEG_1777 expression and colony pigmentation. These results suggest that RsbW2 has an activity to decrease SigF functionality without direct binding to SigF. A clue about the anti-SigF activity of RsbW2 came from the kinase motifs (N, G1, and G2) that are conserved in RsbW2. Like B. subtilis RsbW that can inactivate the anti-SigB antagonist RsbV through phosphorylation (Dufour and Haldenwang, 1994), RsbW2 has the protein kinase activity that inhibits the functionality of RsfB by phosphorylation of Ser-63. RsfB was shown to exist in an unphosphorylated form in the ΔrsbW2 mutant in contrast to the isogenic WT strain in which most fractions of RsfB exist in a phosphorylated form, which indicates that RsbW2 is the kinase that can phosphorylate RsfB in M. smegmatis. It is worth noting that RsbW of the SigB PSS in Bacillus species and Streptomyces coelicolor acts as both protein kinase for its cognate anti-SigB antagonist (RsbV) and anti-sigma factor (Benson and Haldenwang, 1993; Dufour and Haldenwang, 1994; Voelker et al., 1996; Lee et al., 2004; van Schaik and Abee, 2005), while RsbW1 and RsbW2 of M. smegmatis are specialized to function as anti-SigF and protein kinase, respectively.
Interestingly, both ΔrsbW1 and ΔrsbW2 mutants of M. smegmatis showed phenotypic instability in terms of yellow pigmentation. When first obtained, both the mutants exhibited yellow pigmentation on solid 7H9-glucose medium. However, when the mutant strains were passed through successive subcultures in solid and especially liquid growth media, the mutant strains lost yellow pigmentation and showed only basal levels of MSMEG_1777 expression (data not shown). This observation implies that excessive expression of the SigF regulon is detrimental to M. smegmatis, leading to secondary mutations that mitigate the expression of the SigF regulon.
RsbW3 is closer to RsbW1 than RsbW2 in terms of sequence homology. In contrast to rsbW1, overexpression of rsbW3 in M. smegmatis resulted in a significant increase in MSMEG_1777 expression. Both the lack of protein–protein interactions between RsbW3 and SigF and the overexpression effect of rsbW3 suggest the role of RsbW3 as an anti-SigF antagonist. Given both the quaternary structure (homodimers) of RsbW (SpoIIAB)-like anti-sigma factors (Campbell et al., 2002; Masuda et al., 2004) and the results demonstrating protein–protein interactions between RsbW1 and RsbW3 (Figure 6), we assume that RsbW3 likely inactivates RsbW1 by forming a heterodimer when overexpressed. The presence of a SigF–recognizing promoter (GTTT-N17-GGGTAA) upstream of rsbW3 (Table S4) and abolishment of rsbW3 expression by the inactivation of the sigF gene (Supplementary Figure S6) indicate that rsbW3 belongs to the SigF regulon. Based on these findings, we suggest that RsbW3 serves as a booster for expression of the SigF regulon under SigF-activating conditions via the positive feedback loop.
So far, the roles of RsfA and RsfB as anti-SigF antagonists in mycobacteria have been predicted from both their protein interactions with anti-SigF (RsbW and UsfX) and the result from in vitro transcription analysis (Beaucher et al., 2002; Malik et al., 2008, 2009; Singh et al., 2015). Through both deletion and overexpression of rsfA and rsfB, we first demonstrated the physiological roles of RsfA and RsfB as anti-SigF antagonists in M. smegmatis in vivo. As judged by the RPKM values obtained from RNA sequencing analysis (Lee et al., 2018), the transcript level of rsfB was estimated to be ∼6-fold higher than that of rsfA in M. smegmatis grown aerobically to an OD600 of 0.4–0.5 in 7H9-glucose medium (Supplementary Figure S7). The difference in the expression levels of rsfA and rsfB might give a clue explaining the dominant role of RsfB as an anti-SigF antagonist. The observation that expression of MSMEG_1777 was nearly abolished in the ΔrsfB mutant despite the presence of RsfA implies that the cellular level of active RsfA might not be sufficient to quarantine RsbW1 to such an extent as to induce the SigF regulon in the absence of RsfB. The result of rsfA and rsfB overexpression using an acetamide-inducible promoter clearly showed that when RsfA is sufficiently expressed, it acts as an anti-SigF antagonist more efficiently than RsfB. This observation is in good agreement with the previous report demonstrating that RsfA interacts more strongly with RsbW1 (UsfX) than RsfB (Singh et al., 2015). When the ΔrsfA mutant was complemented by introducing pMHRsfA (pMH201:rsfA), the strain did not grow at 0.01% acetamide in contrast to the ΔrsfB mutant carrying pMHRsfB (pMH201:rsfB) that grew, albeit slowly, even in the presence of 0.1% acetamide (data not shown). The inability of the ΔrsfA mutant with pMHRsfA to grow in the presence of 0.01% acetamide reinforces our assumption that expression of the SigF regulon in excess is toxic to M. smegmatis.
We found that Ser-63 is the amino acid residue of RsfB that is phosphorylated by RsbW2. It was also demonstrated that the S63A mutant form of RsfB interacts with RsbW1 with a similar affinity as the unphosphorylated form of WT RsfB, while the phosphomimetic (S63E) form of RsfB does not interact with RsbW1. These results confirm that the phosphorylation state of Ser-63 determines the functionality of RsfB as an anti-SigF antagonist. The importance of the corresponding serine residue in the functionality of anti-sigma factor antagonists has been reported for several RsfB homologs (RsfB of M. tuberculosis, RsbV and SpoIIAB of B. subtilis) (Diederich et al., 1994; Najafi et al., 1995; Yang et al., 1996; Beaucher et al., 2002).
The Δaa3 mutant of M. smegmatis lacking the aa3 cytochrome c oxidase of the respiratory ETC has been reported to exhibit 53% of the oxygen consumption rate observed for the isogenic WT strain (Jeong et al., 2018), indicating that electron flow through the ETC is inhibited in the mutant by ∼50% relative to the WT strain. The finding that expression of the SigF regulon is significantly induced in the Δaa3 mutant relative to the WT stain implies that the availability of free active SigF is increased in response to inhibition of the respiratory ETC. This observation is in good agreement with the suggestion that SigF makes direct contributions to transcriptomic remodeling in M. smegmatis under hypoxic growth conditions (Martini et al., 2019). The activation of SigF under respiration-inhibitory conditions might result from energy limitation as in the case of the SigB PSS in Bacillus species (Hecker and Volker, 2001; Marles-Wright and Lewis, 2007; de Been et al., 2011; Paget, 2015), or from other factors associated with ETC functions such as changes in the redox state of electron carriers, membrane potential, and proton motive force, etc. The inactivation of the aa3 cytochrome c oxidase in the background of the ΔrsfB mutant was shown not to lead to induction of MSMEG_1777 expression. This result implies that RsfB mediates the induction of the SigF regulon under respiration-inhibitory conditions.
We demonstrated that the sole protein kinase that phosphorylates RsfB in M. smegmatis is RsbW2. RsbW2 has been suggested to be phosphorylated by a Ser/Thr protein kinase, MSMEG_5437 (Bowman and Ghosh, 2014), although the role of MSMEG_5437 in the SigF PSS remains elusive. In the vicinity of the rsfB-rsbW2 operon occur the genes encoding a histidine kinase (MSMEG_6130) and a receiver domain-containing PP2C phosphatase (MSMEG_6131). The PP2C-family phosphatases are known to be responsible for dephosphorylation of the anti-SigB antagonist RsbV in Bacillus species and S. coelicolor (de Been et al., 2011). Indeed, our preliminary result showed that MSMEG_6131 could dephosphorylate the phosphorylated RsfB protein (data not shown). It is conceivable that the phosphorylation state of RsfB might be modulated by the combined control of the kinase activity of RsbW2 and the phosphatase activity of MSMEG_6131 that might be regulated by MSMEG_5437 Ser/The protein kinase and MSMEG_6130 histidine kinase, respectively. Further study is required to reveal the mechanism by which inhibition of the respiratory ETC leads to the activation of SigF in M. smegmatis.
Data Availability Statement
The RNA sequencing data described in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE155251.
Author Contributions
J-IO, S-YS, and YO: conception or design of the study. YO, S-YS, H-JK, GH, and H-YK: acquisition of the data. YO, S-YS, JH, and J-IO: analysis or interpretation of the data and writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2020R1A2C1005305).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.588487/full#supplementary-material
References
Barbieri, C. M., and Stock, A. M. (2008). Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem. 376, 73–82. doi: 10.1016/j.ab.2008.02.004
Beaucher, J., Rodrigue, S., Jacques, P. E., Smith, I., Brzezinski, R., and Gaudreau, L. (2002). Novel Mycobacterium tuberculosis anti-σ factor antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 45, 1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x
Benson, A. K., and Haldenwang, W. G. (1993). Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 90, 2330–2334. doi: 10.1073/pnas.90.6.2330
Betts, J. C., Lukey, P. T., Robb, L. C., Mcadam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Bouillet, S., Arabet, D., Jourlin-Castelli, C., Mejean, V., and Iobbi-Nivol, C. (2018). Regulation of σ factors by conserved partner switches controlled by divergent signalling systems. Environ. Microbiol. Rep. 10, 127–139. doi: 10.1111/1758-2229.12620
Bowman, J., and Ghosh, P. (2014). A complex regulatory network controlling intrinsic multidrug resistance in Mycobacterium smegmatis. Mol. Microbiol. 91, 121–134. doi: 10.1111/mmi.12448
Campbell, E. A., Masuda, S., Sun, J. L., Muzzin, O., Olson, C. A., Wang, S., et al. (2002). Crystal structure of the Bacillus stearothermophilus anti-σ factor SpoIIAB with the sporulation σ factor σF. Cell 108, 795–807. doi: 10.1016/s0092-8674(02)00662-1
Chen, C. C., Lewis, R. J., Harris, R., Yudkin, M. D., and Delumeau, O. (2003). A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49, 1657–1669. doi: 10.1046/j.1365-2958.2003.03663.x
Chen, P., Ruiz, R. E., Li, Q., Silver, R. F., and Bishai, W. R. (2000). Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68, 5575–5580. doi: 10.1128/iai.68.10.5575-5580.2000
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
de Been, M., Francke, C., Siezen, R. J., and Abee, T. (2011). Novel σB regulation modules of Gram-positive bacteria involve the use of complex hybrid histidine kinases. Microbiology 157, 3–12. doi: 10.1099/mic.0.045740-0
DeMaio, J., Zhang, Y., Ko, C., and Bishai, W. R. (1997). Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Int. J. Tuberc. Lung Dis. 78, 3–12. doi: 10.1016/s0962-8479(97)90010-1
DeMaio, J., Zhang, Y., Ko, C., Young, D. B., and Bishai, W. R. (1996). A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 93, 2790–2794. doi: 10.1073/pnas.93.7.2790
Diederich, B., Wilkinson, J. F., Magnin, T., Najafi, M., Errington, J., and Yudkin, M. D. (1994). Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor σF of Bacillus subtilis. Genes Dev. 8, 2653–2663. doi: 10.1101/gad.8.21.2653
Dufour, A., and Haldenwang, W. G. (1994). Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176, 1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994
Dutta, D., Srivastava, V., Tripathi, A., Singh, V., Ansari, M. M., Pant, G., et al. (2019). Mycobacterium bovis sigF mutant exhibits altered surface phenotype and compromised pathogenesis. Tuberculosis 118, 1–13. doi: 10.1016/j.tube.2019.07.006
Forrellad, M. A., Klepp, L. I., Gioffre, A., Sabio, Y., Garcia, J., Morbidoni, H. R., et al. (2013). Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4, 3–66. doi: 10.4161/viru.22329
Gebhard, S., Humpel, A., Mclellan, A. D., and Cook, G. M. (2008). The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 154, 2786–2795. doi: 10.1099/mic.0.2008/018044-0
Geiman, D. E., Kaushal, D., Ko, C., Tyagi, S., Manabe, Y. C., Schroeder, B. G., et al. (2004). Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72, 1733–1745. doi: 10.1128/iai.72.3.1733-1745.2004
Graham, J. E., and Clark-Curtiss, J. E. (1999). Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U.S.A. 96, 11554–11559. doi: 10.1073/pnas.96.20.11554
Green, M., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual, 4th Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Greenstein, A. E., Hammel, M., Cavazos, A., and Alber, T. (2009). Interdomain communication in the Mycobacterium tuberculosis environmental phosphatase Rv1364c. J. Biol. Chem. 284, 29828–29835. doi: 10.1074/jbc.M109.056168
Guthrie, C., and Fink, G. R. (1991). Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–932.
Hartkoorn, R. C., Sala, C., Uplekar, S., Busso, P., Rougemont, J., and Cole, S. T. (2012). Genome-wide definition of the SigF regulon in Mycobacterium tuberculosis. J. Bacteriol. 194, 2001–2009. doi: 10.1128/JB.06692-11
Hecker, M., Pane-Farre, J., and Volker, U. (2007). SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236. doi: 10.1146/annurev.micro.61.080706.093445
Hecker, M., and Volker, U. (2001). General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44, 35–91. doi: 10.1016/s0065-2911(01)44011-2
Humpel, A., Gebhard, S., Cook, G. M., and Berney, M. (2010). The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J. Bacteriol. 192, 2491–2502. doi: 10.1128/JB.00035-10
Jeong, J. A., Park, S. W., Yoon, D., Kim, S., Kang, H. Y., and Oh, J. I. (2018). Roles of alanine dehydrogenase and Induction of Its gene in Mycobacterium smegmatis under respiration-inhibitory conditions. J. Bacteriol. 200:e00152-18. doi: 10.1128/JB.00152-18
Karls, R. K., Guarner, J., Mcmurray, D. N., Birkness, K. A., and Quinn, F. D. (2006). Examination of Mycobacterium tuberculosis sigma factor mutants using low-dose aerosol infection of guinea pigs suggests a role for SigC in pathogenesis. Microbiology 152, 1591–1600. doi: 10.1099/mic.0.28591-0
Keren, I., Minami, S., Rubin, E., and Lewis, K. (2011). Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11. doi: 10.1128/mBio.00100-11
Kim, M. J., Park, K. J., Ko, I. J., Kim, Y. M., and Oh, J. I. (2010). Different roles of DosS and DosT in the hypoxic adaptation of Mycobacteria. J. Bacteriol. 192, 4868–4875. doi: 10.1128/JB.00550-10
Lee, E. J., Cho, Y. H., Kim, H. S., Ahn, B. E., and Roe, J. H. (2004). Regulation of σB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 186, 8490–8498. doi: 10.1128/JB.186.24.8490-8498.2004
Lee, H. N., Ji, C. J., Lee, H. H., Park, J., Seo, Y. S., Lee, J. W., et al. (2018). Roles of three FurA paralogs in the regulation of genes pertaining to peroxide defense in Mycobacterium smegmatis mc2 155. Mol. Microbiol. 108, 661–682. doi: 10.1111/mmi.13956
Malik, S. S., Luthra, A., and Ramachandran, R. (2009). Interactions of the M. tuberculosis UsfX with the cognate sigma factor SigF and the anti-anti sigma factor RsfA. Biochim. Biophys. Acta 1794, 541–553. doi: 10.1016/j.bbapap.2008.11.007
Malik, S. S., Luthra, A., Srivastava, S. K., and Ramachandran, R. (2008). Mycobacterium tuberculosis UsfX (Rv3287c) exhibits novel nucleotide binding and hydrolysis properties. Biochem. Biophys. Res. Commun. 375, 465–470. doi: 10.1016/j.bbrc.2008.08.043
Manganelli, R. (2014). Sigma factors: key molecules in Mycobacterium tuberculosis physiology and virulence. Microbiol. Spectr. 2, 1–23. doi: 10.1128/microbiolspec.MGM2-0007-2013
Manganelli, R., Provvedi, R., Rodrigue, S., Beaucher, J., Gaudreau, L., and Smith, I. (2004). σ factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186, 895–902. doi: 10.1128/jb.186.4.895-902.2004
Mariani, F., Cappelli, G., Riccardi, G., and Colizzi, V. (2000). Mycobacterium tuberculosis H37Rv comparative gene-expression analysis in synthetic medium and human macrophage. Gene 253, 281–291. doi: 10.1016/s0378-1119(00)00249-3
Marles-Wright, J., and Lewis, R. J. (2007). Stress responses of bacteria. Curr. Opin. Struct. Biol. 17, 755–760. doi: 10.1016/j.sbi.2007.08.004
Martini, M. C., Zhou, Y., Sun, H., and Shell, S. S. (2019). Defining the Transcriptional and Post-transcriptional Landscapes of Mycobacterium smegmatis in Aerobic Growth and Hypoxia. Front. Microbiol. 10:591. doi: 10.3389/fmicb.2019.00591
Masuda, S., Murakami, K. S., Wang, S., Anders Olson, C., Donigian, J., Leon, F., et al. (2004). Crystal structures of the ADP and ATP bound forms of the Bacillus anti-σ factor SpoIIAB in complex with the anti-anti-σ SpoIIAA. J. Mol. Biol. 340, 941–956. doi: 10.1016/j.jmb.2004.05.040
Michele, T. M., Ko, C., and Bishai, W. R. (1999). Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43, 218–225. doi: 10.1128/aac.43.2.218
Misra, R., Menon, D., Arora, G., Virmani, R., Gaur, M., Naz, S., et al. (2019). Tuning the Mycobacterium tuberculosis alternative sigma factor sigf through the multidomain regulator Rv1364c and osmosensory kinase protein Kinase D. J. Bacteriol. 201, 1–16. doi: 10.1128/JB.00725-18
Mouncey, N. J., and Kaplan, S. (1998). Redox-dependent gene regulation in Rhodobacter sphaeroides 2.4.1T: effects on dimethyl sulfoxide reductase (dor) gene expression. J. Bacteriol. 180, 5612–5618. doi: 10.1128/jb.180.21.5612-5618.1998
Najafi, S. M., Willis, A. C., and Yudkin, M. D. (1995). Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific σF of Bacillus subtilis. J. Bacteriol. 177, 2912–2913. doi: 10.1128/jb.177.10.2912-2913.1995
Oh, J. I., and Kaplan, S. (1999). The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38, 2688–2696. doi: 10.1021/bi9825100
Paget, M. S. (2015). Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5, 1245–1265. doi: 10.3390/biom5031245
Parida, B. K., Douglas, T., Nino, C., and Dhandayuthapani, S. (2005). Interactions of anti-sigma factor antagonists of Mycobacterium tuberculosis in the yeast two-hybrid system. Tuberculosis 85, 347–355. doi: 10.1016/j.tube.2005.08.001
Provvedi, R., Kocincova, D., Dona, V., Euphrasie, D., Daffe, M., Etienne, G., et al. (2008). SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacteriol. 190, 7859–7863. doi: 10.1128/JB.00714-08
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodrigue, S., Provvedi, R., Jacques, P. E., Gaudreau, L., and Manganelli, R. (2006). The σ factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30, 926–941. doi: 10.1111/j.1574-6976.2006.00040.x
Sachdeva, P., Misra, R., Tyagi, A. K., and Singh, Y. (2010). The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J. 277, 605–626. doi: 10.1111/j.1742-4658.2009.07479.x
Sachdeva, P., Narayan, A., Misra, R., Brahmachari, V., and Singh, Y. (2008). Loss of kinase activity in Mycobacterium tuberculosis multidomain protein Rv1364c. FEBS J. 275, 6295–6308. doi: 10.1111/j.1742-4658.2008.06753.x
Singh, A. K., Dutta, D., Singh, V., Srivastava, V., Biswas, R. K., and Singh, B. N. (2015). Characterization of Mycobacterium smegmatis sigF mutant and its regulon: overexpression of SigF antagonist (MSMEG_1803) in M. smegmatis mimics sigF mutant phenotype, loss of pigmentation, and sensitivity to oxidative stress. Microbiology 4, 896–916. doi: 10.1002/mbo3.288
Singh, A. K., and Singh, B. N. (2008). Conservation of Sigma F in mycobacteria and its expression in Mycobacterium smegmatis. Curr. Microbiol. 56, 574–580. doi: 10.1007/s00284-008-9126-8
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T., and Jacobs, W. R. Jr. (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x
van Schaik, W., and Abee, T. (2005). The role of σB in the stress response of Gram-positive bacteria - targets for food preservation and safety. Curr. Opin. Biotechnol. 16, 218–224. doi: 10.1016/j.copbio.2005.01.008
Vijay, K., Brody, M. S., Fredlund, E., and Price, C. W. (2000). A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35, 180–188. doi: 10.1046/j.1365-2958.2000.01697.x
Voelker, U., Voelker, A., and Haldenwang, W. G. (1996). Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178, 5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996
Waagmeester, A., Thompson, J., and Reyrat, J. M. (2005). Identifying sigma factors in Mycobacterium smegmatis by comparative genomic analysis. Trends Microbiol. 13, 505–509. doi: 10.1016/j.tim.2005.08.009
Williams, E. P., Lee, J. H., Bishai, W. R., Colantuoni, C., and Karakousis, P. C. (2007). Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 189, 4234–4242. doi: 10.1128/JB.00201-07
Keywords: aa3 cytochrome c oxidase, anti-sigma factor, anti-anti-sigma factor, electron transport chain, Mycobacterium, partner switching system, protein kinase, SigF
Citation: Oh Y, Song S-Y, Kim H-J, Han G, Hwang J, Kang H-Y and Oh J-I (2020) The Partner Switching System of the SigF Sigma Factor in Mycobacterium smegmatis and Induction of the SigF Regulon Under Respiration-Inhibitory Conditions. Front. Microbiol. 11:588487. doi: 10.3389/fmicb.2020.588487
Received: 29 July 2020; Accepted: 22 October 2020;
Published: 11 November 2020.
Edited by:
Morigen Morigen, Inner Mongolia University, ChinaReviewed by:
Vinay Kumar Nandicoori, National Institute of Immunology (NII), IndiaKaixia Mi, Chinese Academy of Sciences, China
Copyright © 2020 Oh, Song, Kim, Han, Hwang, Kang and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeong-Il Oh, am9oQHB1c2FuLmFjLmty
†These authors have contributed equally to this work
 Yuna Oh
Yuna Oh Su-Yeon Song†
Su-Yeon Song† Jihwan Hwang
Jihwan Hwang Jeong-Il Oh
Jeong-Il Oh