
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 October 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.587750
Antibiotic resistance currently presents one of the biggest threats to humans. The development and implementation of strategies against the spread of superbugs is a priority for public health. In addition to raising social awareness, approaches such as the discovery of new antibiotic molecules and the elucidation of resistance mechanisms are common measures. Accordingly, the two-component system (TCS) of Streptomyces coelicolor AbrB1/B2, offer amenable ways to study both antibiotic production and resistance. Global transcriptomic comparisons between the wild-type strain S. coelicolor M145 and the mutant ΔabrB, using RNA-Seq, showed that the AbrB1/B2 TCS is implicated in the regulation of different biological processes associated with stress responses, primary and secondary metabolism, and development and differentiation. The ΔabrB mutant showed the up-regulation of antibiotic biosynthetic gene clusters and the down-regulation of the vancomycin resistance gene cluster, according to the phenotypic observations of increased antibiotic production of actinorhodin and undecylprodigiosin, and greater susceptibility to vancomycin. The role of AbrB1/B2 in vancomycin resistance has also been shown by an in silico analysis, which strongly indicates that AbrB1/B2 is a homolog of VraR/S from Staphylococcus aureus and LiaR/S from Enterococcus faecium/Enterococcus faecalis, both of which are implied in vancomycin resistance in these pathogenic organisms that present a serious threat to public health. The results obtained are interesting from a biotechnological perspective since, on one hand, this TCS is a negative regulator of antibiotic production and its high degree of conservation throughout Streptomyces spp. makes it a valuable tool for improving antibiotic production and the discovery of cryptic metabolites with antibiotic action. On the other hand, AbrB1/B2 contributes to vancomycin resistance and is a homolog of VraR/S and LiaR/S, important regulators in clinically relevant antibiotic-resistant bacteria. Therefore, the study of AbrB1/B2 could provide new insight into the mechanism of this type of resistance.
The emergence and spread of antibiotic resistant bacteria currently present great challenges to humans as highlighted by the World Health Organization (WHO) and the American and European Centers for Disease Control and Prevention, CDC and ECDC, respectively. Antibiotic resistance can affect any person at any time of life and poses many risks, especially with respect to advancements in modern health care such as organ transplants and cancer therapy. The latest global antimicrobial resistance and use surveillance system (GLASS) report published by the WHO stated that over two million people are infected with antibiotic resistant bacteria (published in 2020, data collected until July 2019). However, if we consider the lack of data from countries like China (data only submitted from 78 countries/territories), and the partial implementation of surveillance systems in the countries enrolled in GLASS, the magnitude of the problem could even be considered substantially worse than the data suggests. The negative impact of this threat not only affects human health, but also animal health and world economies (CDC, 2019; GLASS, 2020).
The development and implementation of strategies against the spread of superbugs is a priority. Although raising social awareness and preventative measures can be effective, the discovery of new antibiotics that improve patient treatment options and outcomes still remain essential. In relation to this, the ubiquitous genus Streptomyces, with more than 600 species and several sequenced genomes, has a great capacity to produce secondary metabolites with a myriad of antimicrobial, antitumoral, and antifungal activities. Many of these metabolites are considered cryptic, owing to the lack of or the reduced ability to be produced in laboratory conditions (Hwang et al., 2014; Barka et al., 2016; Gómez-Escribano et al., 2016; Blaskovich et al., 2017; van der Meij et al., 2017; van der Heul et al., 2018; Mouncey et al., 2019).
The use of two-component systems (TCSs) are among the several strategies used to fight antibiotic resistance and are global regulators that coordinate biological processes in bacteria in response to internal and external stimuli. Many TCSs are involved in the regulation of resistance mechanisms through the activation of antibiotic response systems and changes in cell physiology that increase antibiotic resistance, making them a promising therapeutic target (Bhagirath et al., 2019; Tierney and Rather, 2019). For example, GraR/S is involved in resistance to cationic antimicrobial peptides in Staphylococcus aureus through the modification of teichoic acids that changes the bacterial surface charge (Yang et al., 2012), or CopR/S and CzcR/S which increase carbapenem resistance through the repression of a porin in Pseudomonas aeruginosa (Perron et al., 2004; Caille et al., 2007; Raavi et al., 2017). In addition, TCSs are also involved in the control of antibiotic production and have been extensively used in yield enhancement and metabolite mining (Liu et al., 2013; Rodríguez et al., 2013; Baral et al., 2018; Xia et al., 2020). For instance, MtrA/B acts as a positive regulator of chloramphenicol biosynthesis in Streptomyces venezuelae (Som et al., 2017) and PhoR/P is implied in the control of antimicrobial production at least in three Streptomyces species (Santos-Beneit, 2015): Streptomyces lividans (Sola-Landa et al., 2003), Streptomyces coelicolor (Santos-Beneit et al., 2009; Fernández-Martínez et al., 2012), and Streptomyces natalensis (Mendes et al., 2007).
Canonical TCSs are composed of a histidine kinase (HK) and a response regulator (RR). HK detects changes outside or inside the cells and, through the activation of the corresponding RR, trigger cellular responses, usually by regulating gene expression. This regulation is based on a histidine-aspartic phosphorylation cascade from the HK to the RR. The number of TCSs in an organism is dependent on ecologic niche, and members of the genus Streptomyces have approximately 90 TCSs (Álvarez et al., 2016).
In S. coelicolor, 23 out of the 100 TCSs in its genome have been studied, of which many have been linked to antibiotic regulation (Rodríguez et al., 2013). Some of these TCSs are global regulators of several antibiotic pathways, such as AbsA1/A2 (Lewis et al., 2019), DraK/R (Yeo et al., 2014), and RapA1/A2 (Lu et al., 2007), while others only regulate single secondary metabolite gene clusters such as CutR/S (Chang et al., 1996) and EcrA1/A2 (Li et al., 2004).
In our laboratory, the search for TCSs in S. coelicolor implicated in antibiotic production has led to the identification of three systems partially similar to the antibiotic regulatory system AbsA1/A2 and a related orphan RR. These systems are: AbrA1/A2 (SCO1744/45), AbrB1/B2 (SCO2165/66), AbrC1/C2/C3 (SCO4598/97/96), and Aor1 (SCO2281). The AbrA1/A2 system plays a negative role in both antibiotic production and differentiation (Yepes et al., 2011; Rico et al., 2014b). The AbrC1/C2/C3 system, an atypical TCS with two HK and one RR, acts as a positive regulator of antibiotic production and differentiation (Yepes et al., 2011; Rico et al., 2014a; Rodríguez et al., 2015). And Aor1 seems to be crucial in the antibiotic production linked to stress responses (Antoraz et al., 2017). Moreover, the biotechnological application of these systems has been demonstrated, where the overexpression of the RR AbrC3 in different species of Streptomyces enhanced antibiotic and antitumoral production (Rico et al., 2014a; Becerril et al., 2018). The use of a deletion mutant of the negative regulator system AbrA1/A2, as a heterologous host for antibiotic biosynthetic gene clusters, has also been successful (Rico et al., 2014b).
In this work we describe the role of AbrB1/B2 in S. coelicolor. The phenotypic and transcriptomic analyses of this TCS show that AbrB1/B2 is a negative regulator of antibiotic production and a positive regulator of vancomycin resistance. Also, an in silico analysis indicates that this system is a homolog of VraR/S and LiaR/S from S. aureus and Enterococcus faecium/Enterococcus faecalis, respectively, which plays an important role in the mechanism of vancomycin resistance in these pathogenic organisms (Kuroda et al., 2000; Qureshi et al., 2014; Tierney and Rather, 2019).
Information on the bacterial strains is shown in Supplementary Table 1. Escherichia coli DH5α and ET12567 (non-methylating strain) were cultivated at 37°C in LB medium and were used to manipulate DNA and to transform S. coelicolor respectively (this organism does no admit methylated DNA). S. coelicolor strains were cultivated at 30°C, R2YE was used for the transformations, MSA for sporulation (Kieser et al., 2000) and YEPD (Rose et al., 1990) for spore quantification.
For the phenotypic assay on solid media, S. coelicolor strains were grown on NMMP, YEPD, NA, LB, R2YE, and R5. The phenotypic assay as well as the cultures for the remaining experiments were performed using NMMP liquid media.
In the antagonistic assay, patches of S. coelicolor strains were grown during 10 days at 30°C on NMMP plates. Then, circular sections of the patches were collected and placed on BHI plates inoculated with a lawn of Staphylococcus epidermidis or E. faecalis. The plates were first incubated at 4°C for 5 h (to allow diffusion of any produced molecules) and then at 37°C for 24 h.
When necessary, the media were supplemented with the following antibiotics: E. coli media – ampicillin (100 μg mL–1), apramycin (15 μg mL–1), kanamycin (50 μg mL–1), chloramphenicol (25 μg mL–1), or nalidixic acid (25 μg mL–1), and S. coelicolor media – neomycin (20 μg mL–1), or thiostrepton (10 μg mL–1).
Photographs of colonies were obtained using an OLYMPUS E-620 camera. Photomicrographs were taken with a RT Monochrome (SPOT) in an Axiophot (ZEISS) microscope.
The plasmids and primers used in this work are listed in Supplementary Tables 2, 3, respectively. Plasmid isolations, restriction enzyme digests, ligations and E. coli and S. coelicolor transformations were carried out using the methods by Green and Sambrook (2012) and Kieser et al. (2000), following the manufacturer’s instructions.
The complementation plasmid pKCAbrB was generated in several steps. The DNA fragment including the abrB1/B2 operon and its promoter was amplified from the S. coelicolor M145 genome by PCR using DSM-002 and DSM-004 primers and cloned into the EcoRI/HindIII sites of pXHis1 to yield pXAbrB. Afterward, the BglII fragment from this plasmid containing the genes and promoter was cloned into the same site of pKC796.
The multicopy plasmid pNBAbrB was generated by cloning the DSM-002/DSM-004 DNA fragment previously indicated into the EcoRI/HindIII sites of pN702GEM3.
The pCRISPR-Cas9 plasmid provided by Tong et al. (2015) was used to generate the tools needed for creating the AbrB1/B2 system deletion mutant. The single guide RNA (sgRNA) used to target abrB1/B2 genes (GCAGGACACGGATCGTCATA) was designed using the web resource http://staff.biosustain.dtu.dk/laeb/crispy_scoeli/ and the whole sgRNA guide was amplified by PCR using primers SAM-051 and SAM-067. pCRISPR-Cas9 was used as template and the amplified product was cut with NcoI/SnaBI enzymes and introduced into pCRISPR-Cas9 digested with the same enzymes, yielding the pCRISPR-Cas9-sgB plasmid. In parallel, a DNA fragment containing the flanking regions of the AbrB1/B2 system was introduced into this new plasmid to provide a homologous recombination template to repair the cut produced by Cas9 and to delete the abrB1/B2 operon in Streptomyces. First, two PCRs were performed to generate fragments containing either the 1-Kb upstream abrB1/B2 operon (oligonucleotides SAM-069/SAM-070) or the 1-Kb downstream operon (oligonucleotides SAM-071/SAM-072). Afterward, the final DNA fragment used as a template was obtained by an overlapping PCR using primers SAM-069/SAM-072. This fragment was introduced into the XbaI site of pCRISPR-Cas9-sgB plasmid to generate the final plasmid pCRISPR-Cas9-AbrB. All of the resulting plasmids were sequenced for verification.
All the three plasmids, the empty one (pCRISPR-Cas9), the one harboring only the sgRNA (pCRISPR-Cas9-sgB) and the final plasmid with the guide and the template (pCRISPR-Cas9-AbrB), were introduced into E. coli ET12567/pUZ8002 by transformation, and subsequently into S. coelicolor by interspecific conjugation; apramycin-resistant colonies were then selected. To induce the loss of the plasmids (which were thermosensitive), the colonies obtained were grown at 37°C for 48 h in R2YE medium; apramycin-sensitive colonies were then selected. Nalidixic acid was used to eliminate any remaining E. coli colonies.
Genomic DNA was extracted from the putative S. coelicolor M145 wild-type strain and the corresponding ΔabrB mutant, and the correct deletion of abrB1/B2 genes was verified by PCR (external primers EP: RCD-005/RCD-006; and internal primers IP: RCD-007/RCD-008) (Supplementary Figure 1).
Antibiotic production was assayed on different solid media plates inoculated with 105 spores added in a 5 μL drop, incubated for several days at 30°C, and monitored for any changes in phenotype (two drops of each strain per plate). RED production is detected as red-colored colonies and ACT production as a blue halo around the colonies.
Antibiotic production was quantified from cultures in liquid NMMP medium inoculated with 4 × 106 spores mL–1. ACT and RED antibiotic production were quantified using the spectrophotometric method described in Yepes et al. (2011).
These experiments were all performed in quadruplicate.
Streptomyces coelicolor M145 wild-type strain and its mutant ΔabrB were grown in liquid NMMP medium inoculated with 4 × 106 spores mL–1 at 30°C for 24 and 36 h in triplicate biological samples. The cells were collected and treated with RNAprotect Bacterial Reagent (QIAGEN), and RNA was extracted using the RNeasy Mini Kit (QIAGEN) and treated with RNase-free DNaseI (Thermo Fisher Scientific). The quantity and quality of the RNA samples were checked using a bioanalyzer 2200 TapeStation (Agilent).
Macrogen Inc. (South Korea), provided the RNA-Seq data. The rRNA was removed using the Ribo-Zero rRNA Removal Kit and one library per sample was obtained using TruSeq Stranded Total RNA (NEB Microbe). Sequencing was performed in a NovaSeq6000 (Illumina), generating PE (paired-end) reads of 150 bases. The sequencing data were converted into FASTQ raw data for the RNA-Seq analysis using Illumina package bcl2fastq.
RaNA-Seq software was used to perform the FASTQ pre-processing (fastp 0.19.4) and quantification (salmon 0.9.1) of the samples (Prieto and Barrios, 2019). Total reads obtained for each library ranged between 36 and 52 million, and around 99% passed the quality filter; between 89 and 94% of the cleaned-read set was successfully mapped to the reference genome (Streptomyces coelicolor A3(2) chromosome; GenBank NC_003888). The comparative analysis was performed with DEseq2 (Wald’s test and parametric fit type applied). STRING-Database1 was used for the detailed study of the results obtained.
The RNA-Seq data have been deposited in the Sequence Read Archives (BioProject ID: PRJNA637401; BioSample Accessions: SAMN15105054, SAMN15105055, SAMN15105056, SAMN15105057).
Oligonucleotides were designed using the Primer Quest Tool (IDT) (Supplementary Table 3). These experiments were performed in quadruplicate. RNA was obtained using the RNeasy Mini Kit (QIAGEN) and treated with RNase-free DNaseI (Thermo Fisher Scientific). cDNA was synthesized using iScript Reverse Transcription Supermix (Bio-Rad). Quantitative reverse transcription PCR (RT-qPCR) was carried out using the TB GreenTM Premix Ex TaqTM (TAKARA). The manufacturer’s instructions were followed in all of the previous steps. Technical triplicates were performed for each sample and the RT and NTC controls were included to check the absence of DNA or environmental contamination. The control rpsL (SCO4659) was used as the housekeeping gene. The amplification profile was monitored on a CFX96 Touch Real Time PCR Detection System (Bio-Rad). Absolute quantification was performed using decimal serial dilutions of S. coelicolor genomic DNA with a known number of copies as the standard reference.
Plates containing NMMP solid medium were inoculated with 2 × 105 spores (and the serial dilutions) of the S. coelicolor M145 wild-type strain and the deletion mutant ΔabrB. Increasing vancomycin or penicillin G concentrations were used (1, 10, 50, 100, and 500 μg mL–1) to test the susceptibility of the strains to these two antibiotics. Survival rates were calculated as the number of growing colonies at each antibiotic concentration in comparison to the number of colonies growing on NMMP without it. The experiments were performed in quadruplicate.
Comparison of amino acid sequences were performed in EMBOSS Needle (EMBL-EBI), which creates an optimal global alignment of two sequences using the Needleman–Wunsch algorithm, applying a BLOSUM62 substitution matrix (Madeira et al., 2019).
The relation among the RRs was inferred using the maximum likelihood method and the Le Gascuel model (Le and Gascuel, 2008). The tree with the highest log likelihood (−2758.14) is shown in Supplementary Figure 2. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The initial tree for the heuristic search was obtained automatically by applying the neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then by selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model the evolutionary rate differences among sites (five categories, +G parameter = 21.3358). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved five amino acid sequences. There was a total of 242 positions in the final dataset. The analysis was conducted in MEGA X (Kumar et al., 2018).
The homology modeling of the AbrB1 RR was performed in SWISS-MODEL (Waterhouse et al., 2018) using the VraR structure (4GVP.1.A) as a template. The model obtained shows a GMQE of 0.69 and a QMEAN of −1.08. High local quality was obtained in the REC domain and in the HTH-LuxR domain, while the local quality of the linker region was lower due to the presence of gaps. The structure assessment of the model was performed using MolProbity v4.4 (Chen et al., 2010) and the score obtained was 1.17.
AbrB1/B2 is a canonical TCS comprised by a RR (SCO2165, named as abrB1) and a HK (SCO2166, named as abrB2) coupled in the genome (Supplementary Figure 3A). Both genes seem to form an operon and their coding regions overlap by four nucleotides.
The RR AbrB1 belongs to the NarL family which is characterized by a REC receiver domain (position 3–119; contains the putative phosphorylation site: Asp55) and an HTH-LuxR DNA-binding domain (position 162–218) (Supplementary Figure 3B). The HK AbrB2 shows a classic architecture domain, containing three putative transmembrane regions (positions 21–36, 46–68, and 120–142), an extracellular input domain (position 69–119), a phosphoacceptor/dimerization HiskA-3 domain (position 193–261; contains the putative phosphorylation site: His203), and a HATPase-c domain (position 286–399) (Supplementary Figure 3C) [SMART database2 (Letunic and Bork, 2018), P2CS database3 (Barakat et al., 2011)].
AbrB1/B2 is highly conserved in Streptomyces spp. This TCS is present with a high degree of amino acid sequence identity (over 75% in the case of the RR) to a wide range of Streptomyces species: Streptomyces griseus, Streptomyces scabiei, Streptomyces venezuelae, Streptomyces avermitilis, Streptomyces albus, and Streptomyces hygroscopicus among others (Supplementary Figure 4).
The complete abrB1/B2 TCS was deleted in S. coelicolor M145 using the CRISPR-Cas9 system (Tong et al., 2015; Supplementary Figure 1).
The phenotype of the deletion strain, ΔabrB, was analyzed on several solid culture media: NMMP, YEPD, NA, LB, R2YE and R5. There were no observable differences related to morphological differentiation (appearance of aerial mycelium and sporulation) between the wild-type and mutant strains on any of the media tested. Nevertheless, enhanced actinorhodin (ACT) production was observed as a more intense or wider blue halo around the colonies on NMMP, YEPD, and NA media (Figure 1A). According to these observations, a wider inhibitory halo was produced in the mutant strain, compared to the wild-type, when the Gram+ bacteria S. epidermidis or E. faecalis were added to the media (actinorhodin is an antibiotic active against Gram+ bacteria) (Figure 1B). We chose to use the minimal medium NMMP for further assays.
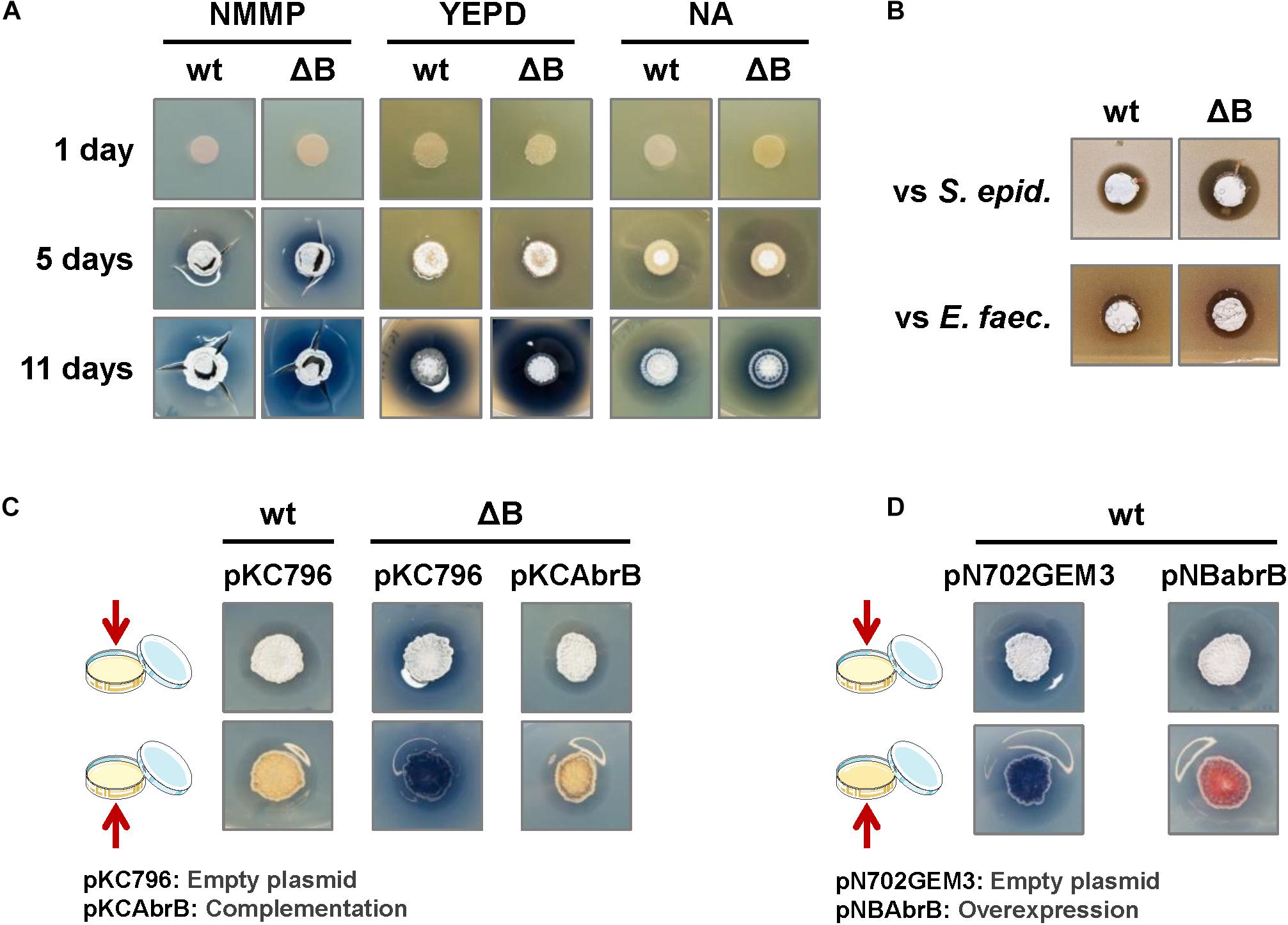
Figure 1. Phenotypic analysis of the S. coelicolor mutant ΔabrB strain. (A) Comparison of ACT production (blue halo around the colonies) and the differentiation of S. coelicolor M145 wild-type strain (wt) and its deletion mutant ΔabrB (ΔB) growing during 1, 5, and 11 days on several types of media (NMMP, YEPD, and NA). Photos of the top view of the plates. (B) Antagonistic assay: inhibitory effect of S. coelicolor M145 wild-type strain (wt) and its mutant ΔabrB (ΔB) growing on a lawn of the Gram+ bacteria Staphylococcus epidermidis (S. epid.) and Enterococcus faecalis (E. faec.). Photos of the top view of the plates. (C) Complementation of ACT production in S. coelicolor M145 ΔabrB mutant (ΔB) with the integrative plasmid pKCAbrB after 5 days on NMMP medium. The wt and ΔB strains carrying the empty plasmid (pKC796) were used as controls to compare the phenotypes. Photos of the top and bottom views of the plates. (D) Overexpression of the AbrB1/B2 system (pNBabrB) compared with the empty plasmid (pN703GEM3), used as control, in the wild-type strain (wt) on NMMP medium after 10 days. Photos of the top and bottom views of the plates.
The integrative plasmid pKCAbrB carrying the abrB1/B2 system under its own promoter was able to complement the ΔabrB phenotype, abolishing the increment of ACT production observed in the deletion mutant (Figure 1C). On the contrary, the overexpression of the abrB1/B2 system with the multicopy plasmid pNBAbrB (abrB genes under the control of its own promoter) produced a phenotype where S. coelicolor M145 exhibited reduced antibiotic production (Figure 1D).
The increment in antibiotic production was also observed in NMMP liquid medium. Quantification of undecylprodigiosin (RED) production allowed us to detect an increase of more than 10-fold of RED in the ΔabrB strain from 36 to 68 h (Figure 2A). Quantification of ACT also showed a 20-fold increment in the mutant strain compared to the wild-type at 60 h and a 1.5-fold increase at 68 h (Figure 2A). No differences were detected between both strains with respect to growth rate nor to the germination time (8 h) under this culture condition (Figure 2B).
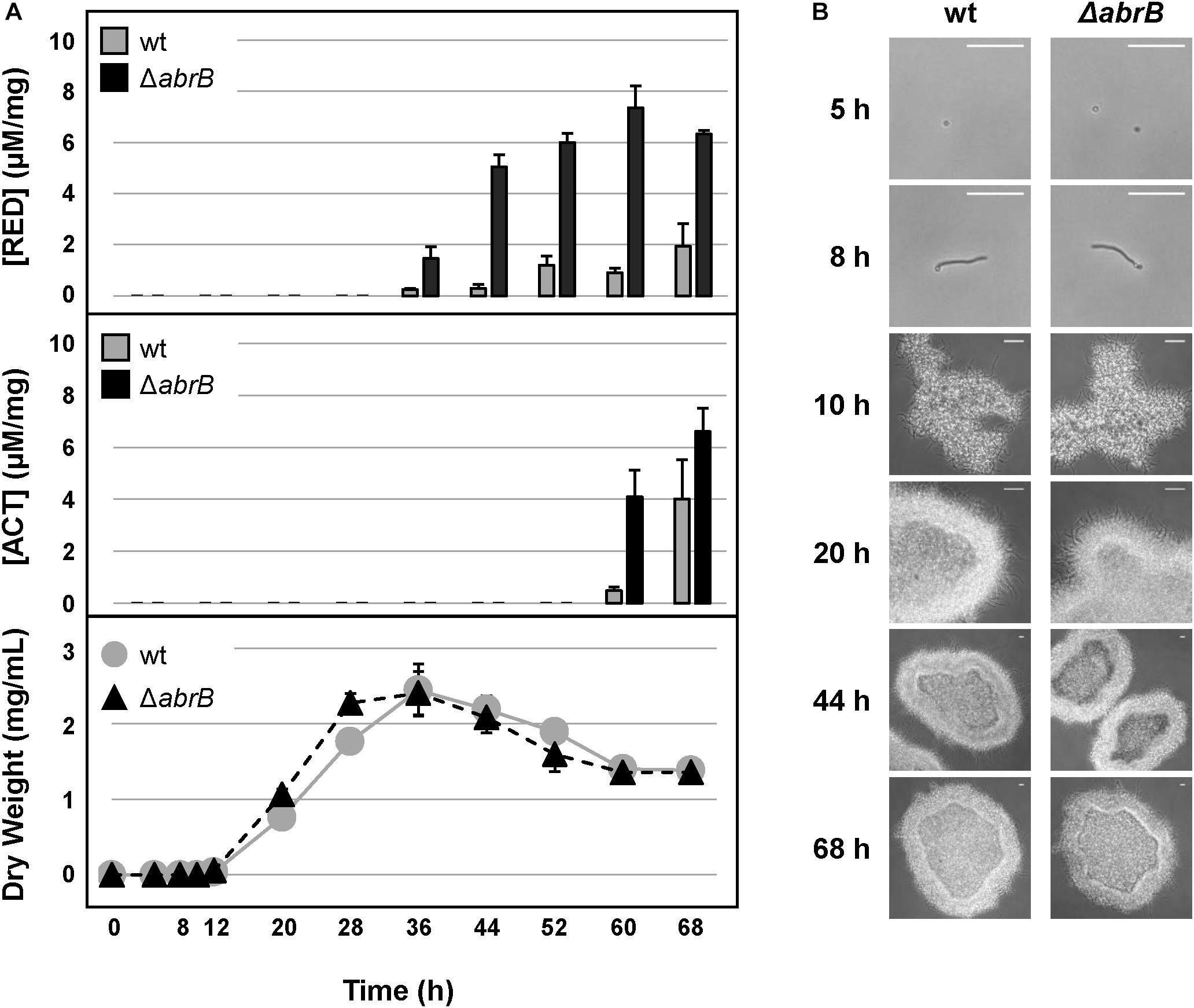
Figure 2. Antibiotic production and growth in liquid medium. (A) RED (top) and ACT (middle) production (μM/mg mycelium) along the growth curves of S. coelicolor M145 wild-type strain (wt, gray circles) and the mutant ΔabrB (black triangles) in NMMP liquid medium (bottom). Error bars show the standard deviation of the quadruplicate assays. (B) Germination and growth progression of the wt and ΔabrB strains (photomicrographs). White-bar (up-right): 50 μm.
These results highlight the role of the TCS AbrB1/B2 as a negative regulator of antibiotic production under these conditions with no apparent effect on growth or morphological differentiation.
To examine the regulation cascade triggered by the AbrB1/B2 system more closely, a global comparative transcriptomic assay between the mutant strain and the wild-type was performed.
The expression pattern of both genes (abrB1 and abrB2) was determined by RT-qPCR in the wild-type M145 strain at 24, 36, 48, and 60 h in NMMP liquid medium. Both genes were expressed over the time course studied, although an expression peak was observed at the earlier time of 24 h (mid exponential growth phase) which decreased at 36 h (early stationary growth phase) until reaching a relative constant expression at 48 and 60 h (Supplementary Figure 5). Consequently, an RNA-Seq analysis was performed in NMMP medium at 24 and 36 h, the time points where expression of the abrB1/B2 system was the highest.
The samples clearly showed a differential expression clustering for each strain at each of the time points using principal component analysis (PCA) (Figure 3A). To perform the global analysis, the normalized data were filtered according to the following criteria: padj ≤ 0.05 and fold change of two (log2FC ≤ -1; log2FC ≥ 1). At 24 h, 15 genes were differentially expressed between both strains: 6 up-regulated and 9 down-regulated in the mutant ΔabrB relative to the wild-type (Figure 3B and Supplementary Table 4). At 36 h, 88 genes were differentially expressed between both strains: 51 up-regulated and 37 down-regulated in the mutant ΔabrB relative to the wild-type (Figure 3B and Supplementary Table 5).
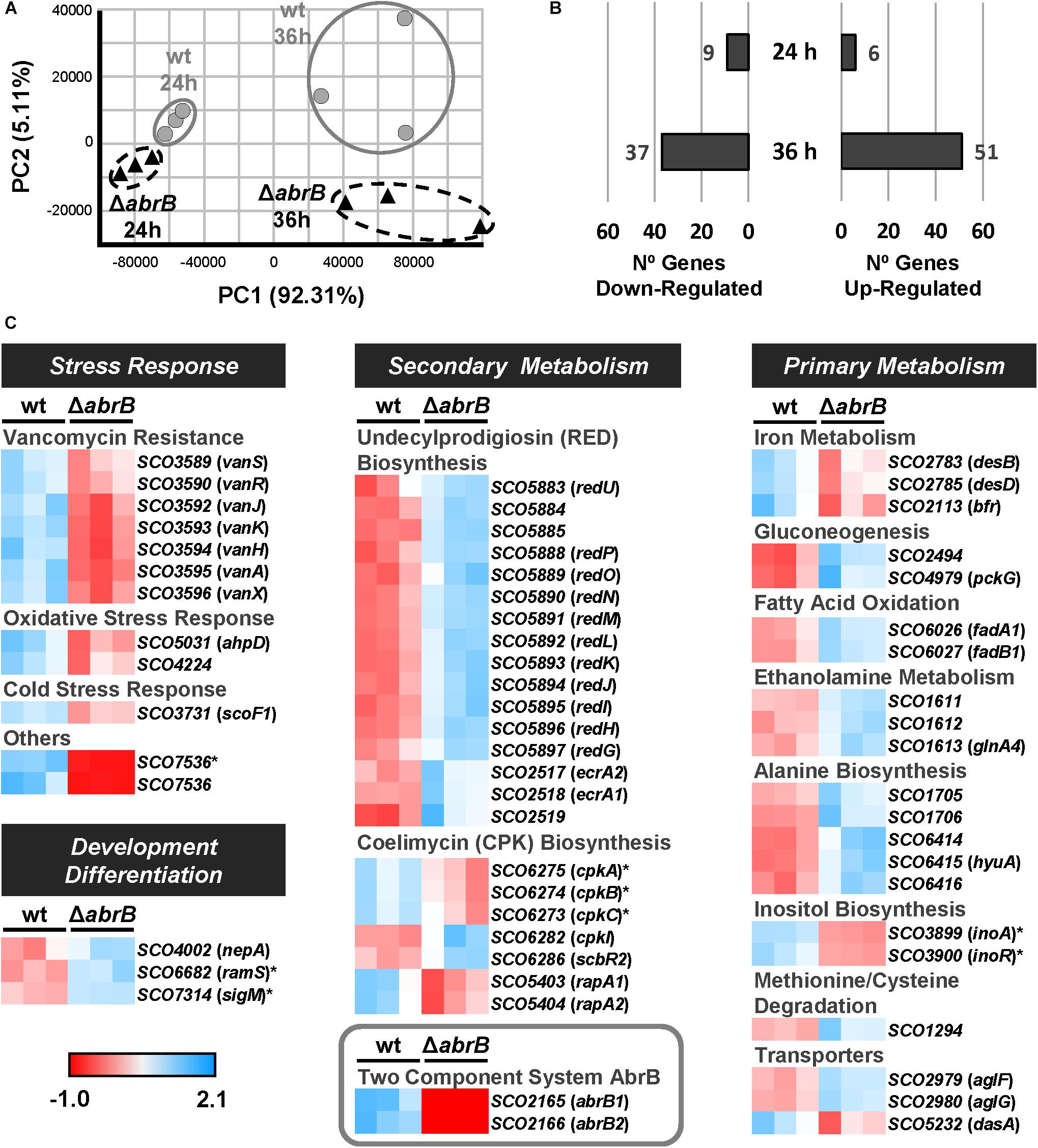
Figure 3. Overview of RNA-Seq assay. (A) Principal component analysis (PCA) of the RNA-Seq samples: triplicates of the wt S. coelicolor M145 (wt; gray circles) and the mutant ΔabrB (black triangles) at 24 and 36 h. (B) Number of up-regulated (FC > 2) and down-regulated (FC < –2) genes in the mutant strain relative to the wt at 24 and 36 h. The selected genes passed the filter of padj ≤ 0.05. (C) Heatmaps of standardized TPMs (transcripts per million) from the differentially expressed genes between S. coelicolor M145 wt and mutant ΔabrB strains at 24 h (marked with asterisks) and 36 h. The genes with unknown functions or biological roles have not been included.
The differentially expressed genes belonged to several biological processes: stress response, primary and secondary metabolism, and development/differentiation (Figure 3C).
The mutant ΔabrB strain exhibited a down-regulation of several response systems to different types of stress: antibiotic resistance, oxidative stress and cold shock.
The complete gene cluster of vancomycin resistance, vanSRJKHAX (SCO3589/90/92/93/94/95/96), was downregulated in the mutant at 36 h. This cluster is divided into four transcriptional units: vanSR (TCS which regulates cluster expression in response to the presence of vancomycin), vanJ, vanK, and vanHAX (enzymes that lead to vancomycin resistance through the modification of the D-Ala-D-Ala terminus of Lipid II, a cell wall precursor, which makes this compound unrecognizable to the antibiotic) (Hong et al., 2004; Hutchings et al., 2006; Koteva et al., 2010).
In the mutant strain various genes related to stress response were down-regulated at 36 h: ahpD (SCO5031) and SCO4224. AhpD belongs to the alkyl-hydroperoxide-reductase system, one of the most relevant antioxidant systems of S. coelicolor (Hahn et al., 2002). In the case of SCO4224, although its product has not yet been characterized, its deletion restricted the growth of the closely specie S. lividans in the presence of oxidative stress (Darbon et al., 2012).
The gene scoF1 (SCO3731), which encodes a potential cold-shock protein quite similar to CspA (Kormanec and Sevcikova, 2000), was down-regulated at 36 h in the ΔabrB strain.
Finally, the differential expression exhibited by SCO7536 was particularly noteworthy, with the highest fold-changes at both 24 h (FC -17) and 36 h (FC -27). This gene seems to encode a potential drug exporter, although at present there are no studies regarding this gene.
According to the phenotypic analysis previously described, most of the genes of the red gene cluster, which manage RED production (Stankovic et al., 2014) were up-regulated at 36 h in the mutant strain. These genes were redUPONMLKJIHG (SCO5883/88/89/90/91/92/93/94/95/96/97) and SCO5884/85, whose role in the biosynthetic pathway are unknown. The cluster-specific regulator redD (SCO5877) did not pass the filters threshold of differential expression. However, the operon SCO2517-19 which contains ecrA1/A2 (SCO2518/17), a TCS that positively regulates RED biosynthesis (Li et al., 2004), was up-regulated in the mutant strain at 36 h.
CPK production was also affected in the ΔabrB mutant strain, where several related genes were differentially expressed: cpkABC (SCO6275/74/73), cpkI (SCO6282), scbR2 (SCO6286) and rapA1/A2 (SCO5403/04). The core genes of the CPK biosynthetic gene cluster, cpkABC, were down-regulated in the mutant strain at 24 h, while cpkI, involved in post-polyketide tailoring (Pawlik et al., 2007; Bednarz et al., 2019), was up-regulated in the mutant strain at 36 h. The regulator scbR2 was also up-regulated in the mutant strain at 36 h. The main role of this gene is to coordinate the production of coelimycin, undecylprodigiosin, actinorhodin and calcium-dependent antibiotics, acting as a repressor of coelimycin biosynthesis and an activator of the remaining compounds (Xu et al., 2010; Bednarz et al., 2019), which was in accordance with the results obtained in this work. The TCS rapA1/A2 was down-regulated in the mutant strain at 36 h; this system acts as a positive regulator of coelimycin and actinorhodin production (Lu et al., 2007).
Although increased ACT production was phenotypically observed in the mutant, few genes related to this process passed the filters established, most probably due to the early time points at which RNA-Seq was performed. However, the previously mentioned regulator ScbR2 and the TCS RapA are known to be related to ACT production. ScbR2 acts as a positive regulator of ACT biosynthesis, and its over-expression in the mutant strain agreed with this phenotype. On the contrary, RapA (also a positive regulator of this process) was under-expressed in the mutant, which could mean ScbR2 has a dominant effect over RapA. Aside from these regulatory systems, there were several genes that did not pass through the filters (0.05 < padj < 0.08) but were still relevant to the analysis: actI-ORF3 (SCO5089), actVII (SCO5090), actIV (SCO5091), and actVB (SCO5092) (Supplementary Table 6). These genes (up-regulated in the mutant strain) belong to the act gene cluster and encode enzymes related to ACT production (Okamoto et al., 2009).
Systems for iron uptake (desBD) and iron storage (bfr) were down-regulated at 36 h in the mutant strain. The genes desB (SCO2783) and desD (SCO2785) participate in deferoxamine biosynthesis, siderophores that mediate iron uptake (Barona-Gómez et al., 2004). The bfr gene encodes a bacterioferritin, an intracellular iron-storage protein.
The mutant ΔabrB exhibited an over-expression of genes SCO2494 and pckG (SCO4979), implicated in the early steps of gluconeogenesis at 36 h. Both genes form part of the phosphoenolpyruvate-pyruvate-oxaloacetate node (PEP-PYR-OXA), a central point in carbon metabolism. The gene SCO2494 encodes a pyruvate phosphate dikinase, while the gene pckG encodes a phosphoenolpyruvate carboxykinase (Llamas-Ramírez et al., 2020).
The fadA1/B1 (SCO6026/27) system was up-regulated at 36 h in the mutant strain. This system is involved in the final steps of fatty acid β-oxidation (Menéndez-Bravo et al., 2017).
The operon SCO1611-13 was up-regulated in the mutant strain at 36 h. Although the entire operon is related with ethanolamine metabolism, only glnA4 (SCO1613) has been studied more in depth. GlnA4 is a γ-glutamyl-ethanolamine synthetase, an enzyme that allows S. coelicolor to use ethanolamine as an alternative nitrogen/carbon source (Rexer et al., 2006; Krysenko et al., 2019).
The mutant ΔabrB showed an over-expression at 36 h of genes implicated in the alanine biosynthesis: SCO1705/06 and SCO6414/15/16, according to several databases (KEGG4, BioCyc5, etc.).
The operon inoAR (SCO3899/3900) was down-regulated at 24 h in the mutant ΔabrB. This operon which leads inositol biosynthesis is formed by the regulator InoR and the myo-inositol phosphate synthase InoA. Although inositol is usually related to the process of differentiation as a phospholipid precursor (Zhang et al., 2010, 2012), this molecule also has a role as a mycothiol precursor, a compound that protects S. coelicolor from oxidative stress, like glutathione in other organisms (Park and Roe, 2008).
The gen SCO1294 was up-regulated in S. coelicolor at 36 h. Even though this gene has not been studied in S. coelicolor, its homolog in S. avermitilis (SAV7062) has been described as a L-methionine-γ-liase that degrades methionine, cysteine, and derivatives (Kudou et al., 2015).
Several transporter systems were differentially expressed between the wild-type and the mutant strains. The operon aglFG (SCO2979/2980) was up-regulated in the mutant at 36 h, and in silico studies suggest the operon constitutes an α-glucosides permease (Bertram et al., 2004). The gene dasA (SCO5232) was down-regulated in the mutant strain at 36 h, where DasA takes part in the uptake system of N,N′-diacetyl-chitobiose and similar molecules such as N-acetyl-glucosamine (Saito et al., 2007).
The genes nepA (SCO4002), ramS (SCO6682), and sigM (SCO7314) were also over-expressed in the mutant strain. NepA is a structural protein of the cell wall and is related to spore dormancy (Dalton et al., 2007; de Jong et al., 2009). RamS is the SapB precursor, a morphogenetic peptide in aerial mycelium development (Kodani et al., 2004). SigM, or Sigma Factor M, is the last step in the regulation cascade of Sigma Factor B and participates in differentiation and in the protection against oxidative and osmotic stress (Lee et al., 2005).
In order to validate the RNA-Seq data by RT-qPCR, three up-regulated genes, ecrA1 (SCO2518), redG (SCO5897), and mfnB (SCO6440) and three down-regulated genes, bfr (SCO2113), vanJ (SCO3592), and SCO7536, were selected. The fold-changes obtained were similar to those obtained by RNA-Seq for the down-regulated genes, but there were more differences in the fold change obtained by RT-qPCR than by RNA-Seq (Supplementary Table 7) for the up-regulated genes.
Finally, no potential motifs were found implied in the regulation of this set of genes by AbrB1/B2, which could mean that the DNA recognition motif of the RR AbrB1 was highly degenerated.
Although AbrB1/B2 seemed to control the basal levels of the vancomycin resistance gene cluster (vanSRJKHAX), as can be seen in the RNA-Seq results, this did not necessarily be affecting the resistance potential of S. coelicolor in the presence of vancomycin, because this resistance operon (van) mainly responds to the presence of the antibiotic through the VanRS TCS.
Therefore, to determine the role of AbrB1/B2 in this process, the survival rates of specific spore concentrations of the M145 wild-type strain and the ΔabrB mutant were examined on NMMP solid medium with increasing concentrations of vancomycin. The deletion of the AbrB1/B2 system triggered a higher susceptibility of S. coelicolor to vancomycin (Figure 4). This result confirmed, according to the alteration of the basal levels of the vancomycin resistance gene cluster (van), that AbrB1/B2 also contributes to vancomycin resistance in S. coelicolor.
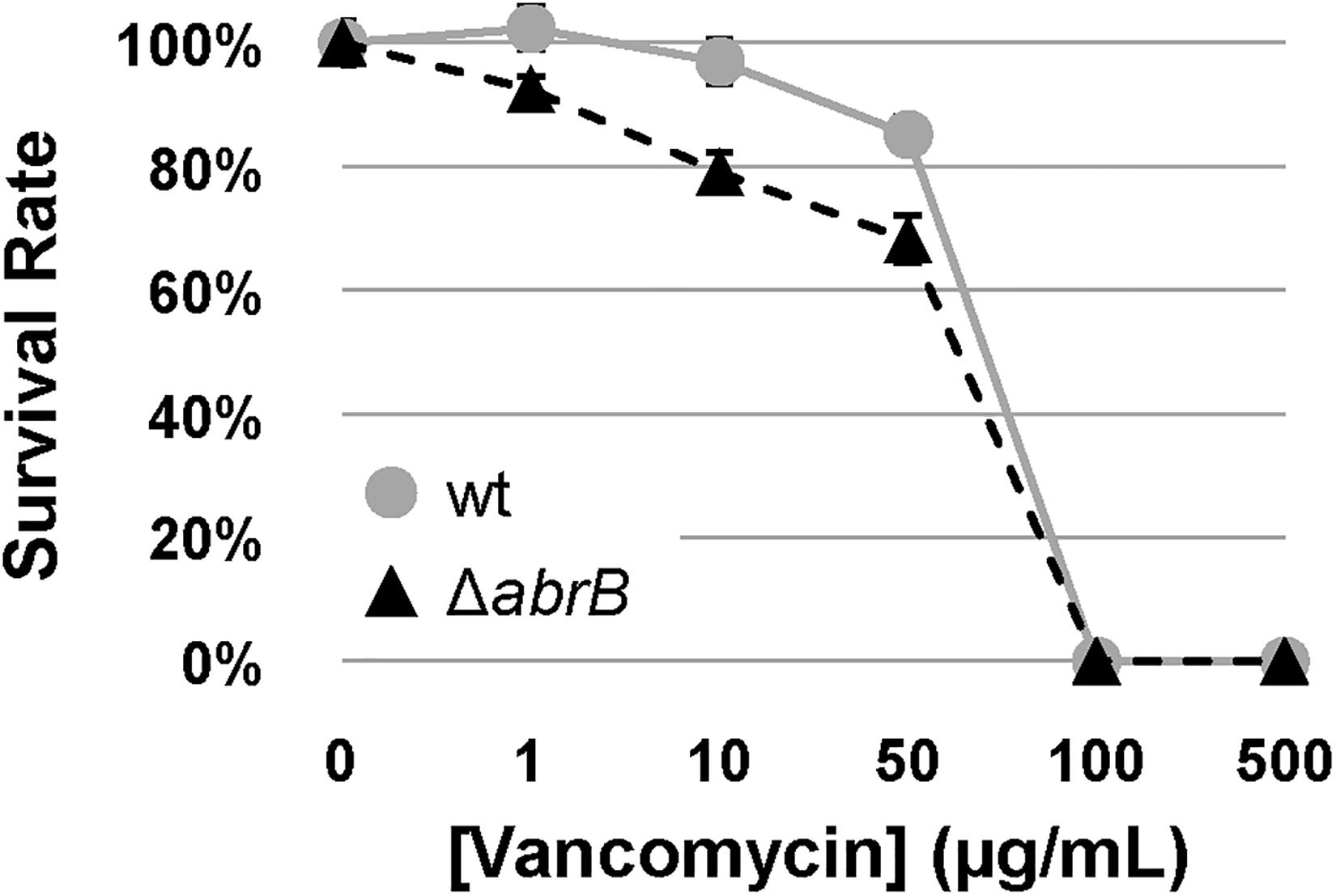
Figure 4. Vancomycin survival assay. Survival assay of spores from S. coelicolor M145 wt (wt; gray circles) and mutant ΔabrB strains (black triangles) against different concentrations of vancomycin in NMMP plates. Error bars show the standard deviation of quadruplicate assays.
The role of AbrB1/B2 in vancomycin resistance led us to look for their homologs in clinically relevant bacteria, such as enterococci, whose vancomycin-resistance strains (VREs) constitute a serious threat to public health. The search and analysis were made based on the RRs, which are much better characterized than HKs.
In the literature, the only system described to regulate VanR/S (like AbrB1/B2 in S. coelicolor, as observed in this study) was VraR/S in S. aureus (Qureshi et al., 2014). The pairwise alignment of the amino acid sequences of AbrB1 and VraR showed an identity and similarity over 30 and 50%, respectively, similar to that described for other homologs found in both species such as VanR (Supplementary Table 8). Furthermore, a maximum likelihood analysis of AbrB1 (from S. coelicolor) and VraR (from S. aureus), plus other RRs involved in vancomycin resistance in S. aureus, like VanR, WalR, and GraR, revealed that VraR had a closer relationship with AbrB1 than with the other staphylococci RRs (Supplementary Figure 2). Similar results were obtained when AbrB1 was compared with LiaR from E. faecium and E. faecalis, a closely related homolog of VraR (despite the sequence differences, both VraR and LiaR share exactly the same tertiary structure, experimentally obtained, and fulfill the same biological function).
Relevant structural data have been published on VraR and LiaR (Donaldson, 2008; Leonard et al., 2013; Davlieva et al., 2015, 2016). All the relevant residues identified in both regulators were conserved in AbrB1, both at the amino acid sequence level and at the level of the three-dimensional structure [homology modeling of AbrB1 was performed in SWISS-MODEL using the structure of VraR (4GVP.1.A) as a template] (Figure 5):
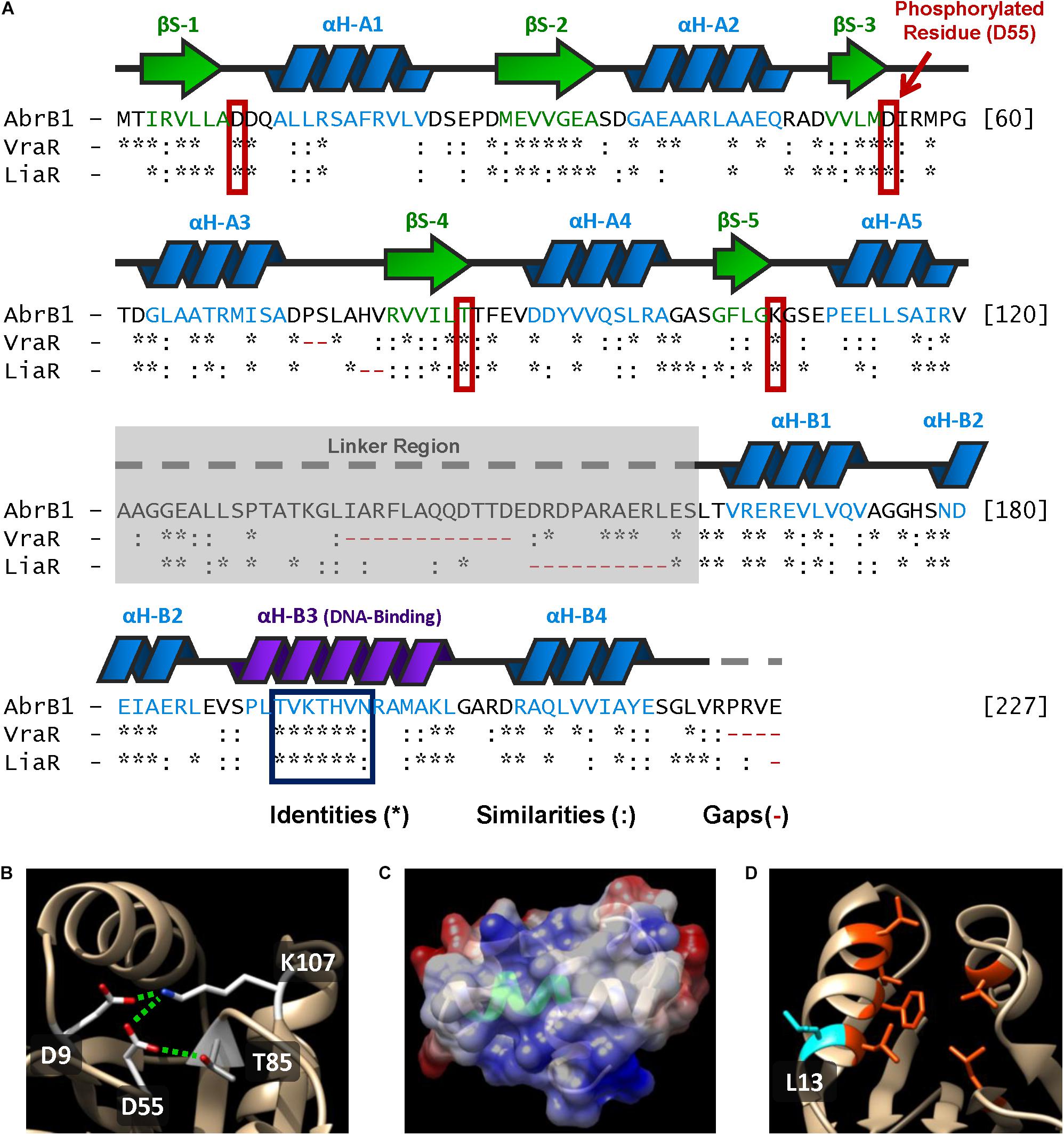
Figure 5. Structural features of AbrB1 and its comparison with VraR and LiaR. (A) Sequence of the RR AbrB1. Secondary structures are shown above the amino acid sequence (beta-strands in green, and alpha-helices in blue). Comparison with VraR from S. aureus and LiaR from E. faecium is shown below the AbrB1 sequence; identities (*), similarities (:), and gaps (-) are indicated (pairwise alignment was performed in EMBOSS Needle applying a BLOSUM62 substitution matrix). Active site residues are squared in red, while conserved DNA-recognition site is squared in blue. (B) Phosphorylation site in AbrB1 structural model: D55 (phosphorylated residue), D9, K107, and T85. Interactions between residues (inactive form) are shown as dashed green lines. (C) DNA-binding helix in AbrB1 structural model. Conserved recognition site is shown in green. Electrostatic surface is represented (positive charge as blue, and negative charge as red). (D) Hydrophobic “pocket” in REC domain for the dimerization process. The residues that conform this “pocket” are shown in orange (Leu14, Phe18, Leu 21, Val22, Leu84, and Leu114); protuberant L13 (which interacts with the “pocket” of the other protomer) is shown in blue.
1. Phosphorylation site residues: Asp55 (putative phosphorylated residue), Asp9, Thr85, and Lys107 (residues with which Asp55 interacts in the inactive form, non-phosphorylated) (Figure 5B).
2. DNA recognition site: from Thr192 to Asn198 (Figure 5C). Additionally, the residues that stabilize DNA binding were also conserved in AbrB1: Arg165, Asn179, and Arg209.
3. Hydrophobic “pocket” in REC domain for the dimerization process (Figure 5D). Although in this feature the residues were not completely conserved, the hydrophobicity of this region was maintained: Leu14, Phe18, Leu 21, Val22, Leu84, and Leu114. The protuberant methionine that interacts with the hydrophobic “pocket” of the other protomer in VraR was substituted by leucine (Leu13) in AbrB1, which shows an almost identical hydrophobicity (Finney et al., 1980; Guy, 1985).
4. Hydrophobic nucleus of the alpha-helix α4 in the HTH-LuxR domain for the dimerization process: from Leu212 to Ala216. Like the previous feature, hydrophobicity was conserved although the residues were not identical.
5. The core of REC domain (beta-strands) and the core of HTH-LuxR domain (alpha-helices) show a high degree of conservation.
A summary of AbrB1/B2 regulatory cascade is shown in Figure 6.
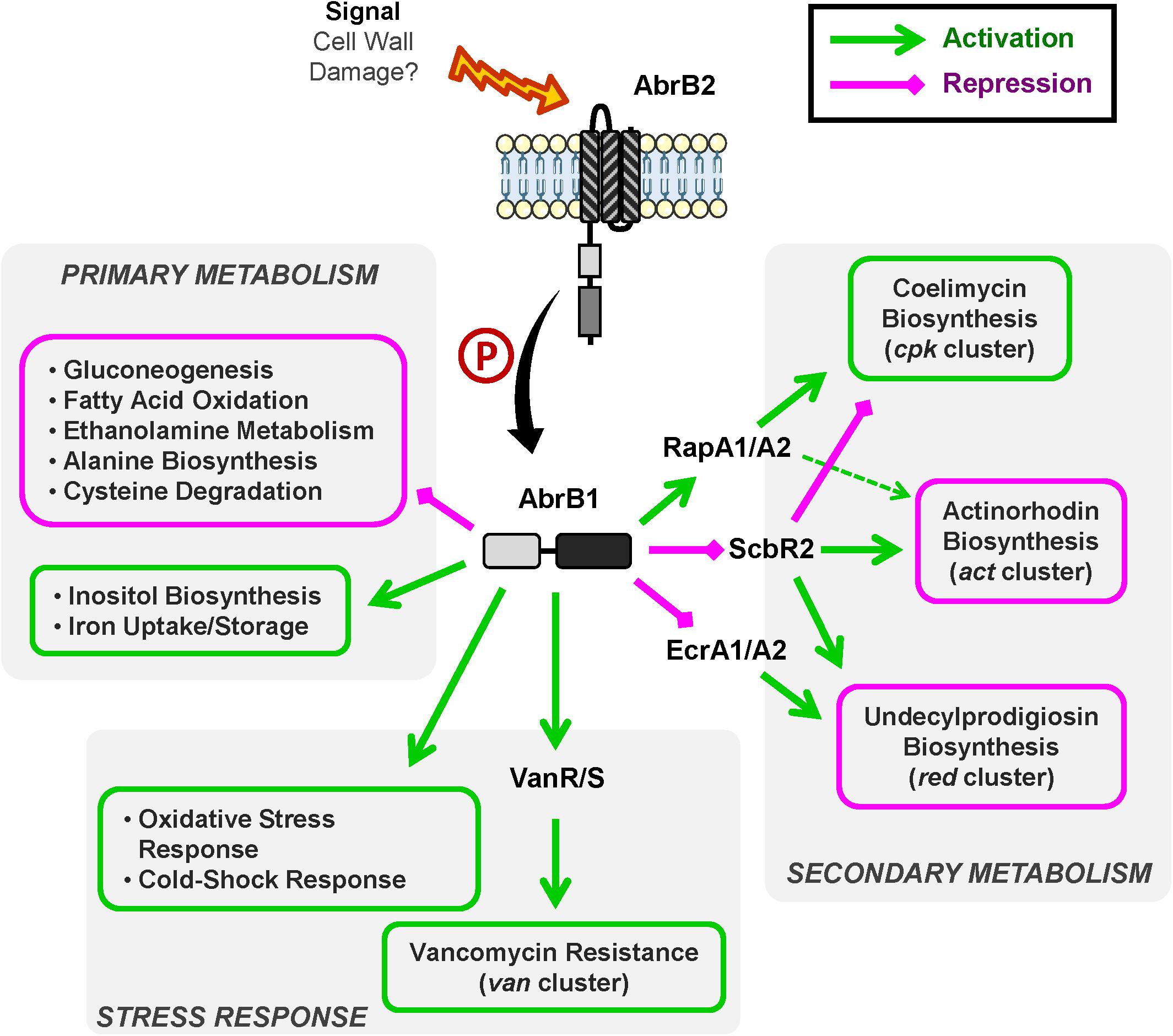
Figure 6. Overview of AbrB1/B2 regulatory cascade. Summary of positively regulated processes (green) and negatively regulated processes (magenta) by the TCS AbrB1/B2.
The role of AbrB1/B2 as negative regulator of antibiotic biosynthesis has been shown at the phenotypic and transcriptional levels. This TCS is involved in RED and ACT production in S. coelicolor. The regulatory cascade controlling these pathways involves the TCS EcrA1/A2 in the case of RED and the regulator ScbR2 in the case of the regulation of both RED and ACT synthesis.
In sum, AbrB1/B2 could be a repressor of EcrA1/A2, a positive regulator of RED biosynthesis (Li et al., 2004), which is linked to the negative regulation of RED production. This regulation could be in part due to cross-talk events, due to the high similarity present between EcrA1/A2 and AbrB1/B2 (the RRs have an identity/similarity in their amino acid sequences of 53 and 66%, respectively). ScbR2 could also be negatively regulated by AbrB1/B2, which would equate to a repression of ACT and RED production and an activation of CPK, because ScbR2 coordinates antibiotic production in S. coelicolor as a positive regulator of ACT and RED and a negative regulator of CPK (Xu et al., 2010; Bednarz et al., 2019). Furthermore, the action of ScbR2 seems to be dominant over the TCS RapA1/A2 in the control of ACT production, since RapA1/A2 is a positive regulator of ACT and CPK synthesis (Lu et al., 2007) but is positively regulated by AbrB1/B2.
The negative regulation of antibiotic production by AbrB1/B2 may seem contradictory, as CPK is down-regulated in the absence of this TCS (a regulatory cascade that seems to imply ScbR2 and RapA1 regulators). However, although the classical role of CPK has been attributed to antibiotic activity, this role is unclear from a physiological point of view. Despite the antibiotic properties of coelimycin A, this molecule is highly unstable and spontaneously reacts with compounds present in the medium, such as N-acetylcysteine and glutamate, generating coelimycin P1 and P2 (compounds without antibiotic properties), respectively (Gottelt et al., 2010; Gómez-Escribano et al., 2012).
The down-regulation of antibiotic production by AbrB1/B2 could be, at the physiological level, a strategy to maintain cellular energetics, since both antibiotic synthesis and cell wall remodeling in the vancomycin resistance are processes of high energetic cost.
Vancomycin is a glycopeptide antibiotic that inhibits cell wall formation by joining peptidoglycan precursors (Lipid II) and blocking crosslink formation. Vancomycin only affects Gram+ bacteria (this antibiotic is not able to cross the external membrane present in Gram− bacteria), and its clinical importance is key since it is the unique effective treatment against S. aureus MRSA. The vancomycin resistance gene cluster (van) is present in multiple human pathogens (S. aureus, E. faecalis, and E. faecium) and actinomycetes (glycopeptide antibiotic producers such as Amycolatopsis orientalis and Streptomyces toyocaensis and non-producers such as S. coelicolor). However, despite the central role of the van cluster, other systems like VraR/S and GraR/S also contribute to vancomycin resistance in different ways (Hong et al., 2008).
In this work, the role of AbrB1/B2 as a positive regulator of vancomycin resistance has been shown at the phenotypic and transcriptional levels. This TCS acts as a positive regulator of the van cluster and as a result increases vancomycin resistance. The regulation of this resistance mechanism could explain a great part of RNA-Seq results, as will be detailed below:
1. Antibiotic responses, oxidative stress and iron metabolism can be integrated as follows. The presence of antibiotics increases ROS (reactive-oxygen species) levels, which contribute to antibiotic toxicity to bacteria, although the molecular mechanism for this is still under debate. One of the proposed theories suggests that antibiotic action in essential cellular processes (like cell wall synthesis in the case of vancomycin) could generate some dysfunctions in the respiratory chain. This would provoke an increase in the concentration of ROS which would destabilize Fe-S clusters, releasing iron and giving rise to Fenton’s reaction. This would enhance, as positive feedback, the toxic effect of the ROS, and, subsequently, the antibiotic (Ezraty and Barras, 2016). All of this could explain the differential expression of the genes related to oxidative stress (both directly like ahpD and indirectly like inoAR) and the iron uptake/storage detected in the RNA-Seq analysis.
2. The link between antibiotic resistance and thermal shock is not new. It has been described that ribosomes affecting antibiotics induce heat-shock proteins and cold-shock proteins depending on its mechanism of action (VanBogelen and Neidhardt, 1990). Furthermore, it has been proposed a report system that precisely use thermal-shock response promoters (like the cspA promoter) to discover new antibiotics (Bianchi and Baneyx, 1999).
3. Changes in primary metabolism (gluconeogenesis, fatty acids oxidation, ethanolamine metabolism, etc.) could be also attributed to vancomycin resistance and cell wall remodeling. Alterations in carbon and fatty acids metabolism has been previously reported in a transcriptomic assay related to VraR (homolog of AbrB1, as it was shown in this study) in vancomycin-resistance S. aureus strains (Kuroda et al., 2000). For example, vancomycin-resistance is based on Lipid II modification (D-Ala-D-Ala terminus is substituted by D-Ala-D-Lac), therefore, the overexpression of genes related to alanine biosynthesis in the mutant ΔabrB is probably a consequence of the change in alanine requirements.
4. Despite the absence of significant changes in development and differentiation (wild-type and mutant strain show a similar growth, germination time, etc.), nepA, ramS, and sigM were differentially expressed in the RNA-Seq assay. Changes in the expression of nepA could be a side effect of cell wall remodeling due to vancomycin resistance, since this gene encodes a cell wall structural protein. In the case of ramS, the absence of phenotypic changes in minimal medium has been previously described, since only chaplins are needed for aerial mycelium formation in this medium (Kodani et al., 2004; Capstick et al., 2007) and its differential expression could also be provoked by cell-wall remodeling. Finally, sigM, a sigma factor related to differentiation and protection against osmotic and oxidative stress (Lee et al., 2005; Gaskell et al., 2007), could only be manifesting its role in oxidative stress in these conditions, coordinating the alternative action of alkyl-hydroperoxide-reductase and catalase systems.
The homology of AbrB1/B2 to VraR/S and LiaR/S also supports its role in the mechanism of vancomycin resistance. Both VraR/S (from S. aureus) and LiaR/S (present in a multitude of bacteria such as E. faecium, Bacillus subtilis, Streptococcus mutans, etc.) are involved in cell envelope stress responses. These systems are not only induced by vancomycin, but also other cell wall active antibiotics (like β-lactams in the case of VraR/S) and other perturbations in cell wall integrity. This suggest that these TCSs detect cell wall damage rather than the antibiotics themselves. Thus, they have been cataloged as “sentinel” systems capable of sensing cell wall perturbations, although the specific activation signal is not clear. Both VraR/S and LiaR/S respond to cell envelope stress through the activation of systems involved in the maintenance of cell wall integrity (such as the peptidoglycan biosynthetic pathway). Mutations and overexpression of these systems have been related to VREs (for example, VISA strains of S. aureus show high expression levels of VraR/S that provoke cell wall thickening) (Kuroda et al., 2003; Mascher et al., 2004; Gardete et al., 2006; Yin et al., 2006; Belcheva and Golemi-Kotra, 2008; Suntharalingam et al., 2009; Klinzing et al., 2013; Miller et al., 2014; Qureshi et al., 2014; Draper et al., 2015; Chen et al., 2016; Tierney and Rather, 2019).
The connection between cell wall integrity and vancomycin resistance has been previously evidenced in S. coelicolor. In this organism the resistance to vancomycin is phosphate-dependent: high concentrations of inorganic phosphate in the media increase the susceptibility to this antibiotic. Surprisingly, this process is not mediated by the main regulator of phosphate metabolism, the TCS PhoRP (Santos-Beneit and Martín, 2013), but it seems to be related with the cell wall metabolism, since the deletion or mutation of genes implied in this process (SCO2594 and SCO1213; this last one is a gatD homolog) overcomes the phosphate effect on vancomycin resistance (Santos-Beneit et al., 2014; Santos-Beneit, 2018).
Our working hypothesis for future work is that the role of AbrB1/B2 could be to detect cell wall damage (similar to VraR/S and LiaR/S) and regulate systems that contribute to the maintain of cell wall integrity such as the van cluster whose TCS (VanR/S) directly detects vancomycin molecules (Koteva et al., 2010; Novotna et al., 2015; Lockey et al., 2020) and promotes cell wall remodeling. Thus, a preliminary experiment testing the susceptibility to penicillin G agrees with the working hypothesis, being the ΔabrB mutant more sensitive to this ß-lactam (Supplementary Figure 6).
The results obtained in this work are interesting from a biotechnological perspective since, on one hand, this TCS is a negative regulator of antibiotic production, and its high degree of conservation throughout Streptomyces spp. makes it a valuable tool for improving antibiotic production and the discovery of cryptic metabolites with antibiotic action. On the other hand, AbrB1/B2 contributes to vancomycin resistance and is a homolog of VraR/S and LiaR/S, important regulators in clinically relevant antibiotic-resistant bacteria (S. aureus MRSA and vancomycin-resistance enterococci, VRE, are considered serious threats to public health that require prompt and sustained action according to the CDC in its latest report on antibiotic resistance). Therefore, the study of AbrB1/B2 could provide new insight into the mechanism of this type of resistance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA637401.
RSN and SA conducted the experiments. RSN, JA, and MD analyzed the results. RIS, MD, SA, and RSN designed the experiments and wrote the manuscript. All authors have read and approved the final manuscript.
Our laboratory was funded by projects BIO2015-66958-R (MINECO/FEDER), PID2019-107716RB-I00, SA036G19 (Regional Government of Castile and Leon) and, from the Regional Government of Castile and Leon/FEDER program (“Escalera de Excelencia” CLU-2017-03/14-20) to the Institute of Functional Biology and Genomics (IBFG). SA was supported by a fellowship awarded by the Regional Government of Castile and Leon (grant CSI099A12-1) and, RSN has been supported by an FPU fellowship from Spanish Ministry of Education, Culture and Sport (FPU17/02862).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Ana M. Martínez-Carrasco for her excellent technical support and Emma Keck for revising the English.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.587750/full#supplementary-material
Álvarez, A. F., Barba-Ostria, C., Silva-Jiménez, H., and Georgellis, D. (2016). Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ. Microbiol. 18, 3210–3226. doi: 10.1111/1462-2920.13397
Antoraz, S., Rico, S., Rodríguez, H., Sevillano, L., Alzate, J. F., Santamaría, R. I., et al. (2017). The orphan response regulator Aor1 is a new relevant piece in the complex puzzle of Streptomyces coelicolor antibiotic regulatory network. Front. Microbiol. 8:2444. doi: 10.3389/fmicb.2017.02444
Barakat, M., Ortet, P., and Whitworth, D. E. (2011). P2CS: a database of prokaryotic two-component systems. Nucleic Acids Res. 39, D771–D776. doi: 10.1093/nar/gkq1023
Baral, B., Akhgari, A., and Metsa-Ketela, M. (2018). Activation of microbial secondary metabolic pathways: avenues and challenges. Synth. Syst. Biotechnol. 3, 163–178. doi: 10.1016/j.synbio.2018.09.001
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H. P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/mmbr.00019-15
Barona-Gómez, F., Wong, U., Giannakopulos, A. E., Derrick, P. J., and Challis, G. L. (2004). Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 126, 16282–16283. doi: 10.1021/ja045774k
Becerril, A., Álvarez, S., Braña, A. F., Rico, S., Díaz, M., Santamaría, R. I., et al. (2018). Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS One 13:e0198145. doi: 10.1371/journal.pone.0198145
Bednarz, B., Kotowska, M., and Pawlik, K. J. (2019). Multi-level regulation of coelimycin synthesis in Streptomyces coelicolor A3(2). Appl. Microbiol. Biotechnol. 103, 6423–6434. doi: 10.1007/s00253-019-09975-w
Belcheva, A., and Golemi-Kotra, D. (2008). A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283, 12354–12364. doi: 10.1074/jbc.M710010200
Bertram, R., Schlicht, M., Mahr, K., Nothaft, H., Saier, M. H. Jr., and Titgemeyer, F. (2004). In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186, 1362–1373. doi: 10.1128/jb.186.5.1362-1373.2004
Bhagirath, A. Y., Li, Y., Patidar, R., Yerex, K., Ma, X., Kumar, A., et al. (2019). Two component regulatory systems and antibiotic resistance in gram-negative pathogens. Int. J. Mol. Sci. 20:1781. doi: 10.3390/ijms20071781
Bianchi, A. A., and Baneyx, F. (1999). Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl. Environ. Microbiol. 65, 5023–5027. doi: 10.1128/aem.65.11.5023-5027.1999
Blaskovich, M. A., Butler, M. S., and Cooper, M. A. (2017). Polishing the tarnished silver bullet: the quest for new antibiotics. Essays Biochem. 61, 103–114. doi: 10.1042/EBC20160077
Caille, O., Rossier, C., and Perron, K. (2007). A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189, 4561–4568. doi: 10.1128/jb.00095-07
Capstick, D. S., Willey, J. M., Buttner, M. J., and Elliot, M. A. (2007). SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol. Microbiol. 64, 602–613. doi: 10.1111/j.1365-2958.2007.05674.x
CDC (2019). CDC. Antibiotic Resistance Threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services.
Chang, H. M., Chen, M. Y., Shieh, Y. T., Bibb, M. J., and Chen, C. W. (1996). The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol. Microbiol. 21, 1075–1085.
Chen, H., Xiong, Z., Liu, K., Li, S., Wang, R., Wang, X., et al. (2016). Transcriptional profiling of the two-component regulatory system VraSR in Staphylococcus aureus with low-level vancomycin resistance. Int. J. Antimicrob. Agents 47, 362–367. doi: 10.1016/j.ijantimicag.2016.02.003
Chen, V. B., Arendall, W. B. III, Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66(Pt 1), 12–21. doi: 10.1107/S0907444909042073
Dalton, K. A., Thibessard, A., Hunter, J. I., and Kelemen, G. H. (2007). A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 64, 719–737. doi: 10.1111/j.1365-2958.2007.05684.x
Darbon, E., Martel, C., Nowacka, A., Pegot, S., Moreau, P. L., and Virolle, M. J. (2012). Transcriptional and preliminary functional analysis of the six genes located in divergence of phoR/phoP in Streptomyces lividans. Appl. Microbiol. Biotechnol. 95, 1553–1566. doi: 10.1007/s00253-012-3995-2
Davlieva, M., Shi, Y., Leonard, P. G., Johnson, T. A., Zianni, M. R., Arias, C. A., et al. (2015). A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin. Nucleic Acids Res. 43, 4758–4773. doi: 10.1093/nar/gkv321
Davlieva, M., Tovar-Yanez, A., DeBruler, K., Leonard, P. G., Zianni, M. R., Arias, C. A., et al. (2016). An adaptive mutation in Enterococcus faecium LiaR associated with antimicrobial peptide resistance mimics phosphorylation and stabilizes LiaR in an activated state. J. Mol. Biol. 428, 4503–4519. doi: 10.1016/j.jmb.2016.09.016
de Jong, W., Manteca, A., Sanchez, J., Bucca, G., Smith, C. P., Dijkhuizen, L., et al. (2009). NepA is a structural cell wall protein involved in maintenance of spore dormancy in Streptomyces coelicolor. Mol. Microbiol. 71, 1591–1603. doi: 10.1111/j.1365-2958.2009.06633.x
Donaldson, L. W. (2008). The NMR structure of the Staphylococcus aureus response regulator VraR DNA binding domain reveals a dynamic relationship between it and its associated receiver domain. Biochemistry 47, 3379–3388. doi: 10.1021/bi701844q
Draper, L. A., Cotter, P. D., Hill, C., and Ross, R. P. (2015). Lantibiotic resistance. Microbiol. Mol. Biol. Rev. 79, 171–191.
Ezraty, B., and Barras, F. (2016). The ‘liaisons dangereuses’ between iron and antibiotics. FEMS Microbiol. Rev. 40, 418–435. doi: 10.1093/femsre/fuw004
Fernández-Martínez, L. T., Santos-Beneit, F., and Martín, J. F. (2012). Is PhoR-PhoP partner fidelity strict? PhoR is required for the activation of the pho regulon in Streptomyces coelicolor. Mol. Genet. Genomics 287, 565–573. doi: 10.1007/s00438-012-0698-4
Finney, J. L., Gellatly, B. J., Golton, I. C., and Goodfellow, J. (1980). Solvent effects and polar interactions in the structural stability and dynamics of globular proteins. Biophys. J. 32, 17–33. doi: 10.1016/s0006-3495(80)84913-7
Gardete, S., Wu, S. W., Gill, S., and Tomasz, A. (2006). Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 3424–3434. doi: 10.1128/aac.00356-06
Gaskell, A. A., Crack, J. C., Kelemen, G. H., Hutchings, M. I., and Le Brun, N. E. (2007). RsmA is an anti-sigma factor that modulates its activity through a [2Fe-2S] cluster cofactor. J. Biol. Chem. 282, 31812–31820. doi: 10.1074/jbc.M705160200
GLASS (2020). Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020. Geneva: World Health Organization.
Gómez-Escribano, J. P., Alt, S., and Bibb, M. J. (2016). Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar. Drugs 14:78. doi: 10.3390/md14040078
Gómez-Escribano, J. P., Song, L., Fox, D., Yeo, V., Bibb, M., and Challis, G. (2012). Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 3, 2716–2720. doi: 10.1039/c2sc20410j
Gottelt, M., Kol, S., Gómez-Escribano, J. P., Bibb, M., and Takano, E. (2010). Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156(Pt 8), 2343–2353. doi: 10.1099/mic.0.038281-0
Green, M. R., and Sambrook, J. (2012). Molecular Cloning: a Laboratory Manual, 4th Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Guy, H. R. (1985). Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys. J. 47, 61–70. doi: 10.1016/s0006-3495(85)83877-7
Hahn, J. S., Oh, S. Y., and Roe, J. H. (2002). Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184, 5214–5222. doi: 10.1128/jb.184.19.5214-5222.2002
Hong, H. J., Hutchings, M. I., and Buttner, M. J. (2008). Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 631, 200–213. doi: 10.1007/978-0-387-78885-2_14
Hong, H. J., Hutchings, M. I., Neu, J. M., Wright, G. D., Paget, M. S., and Buttner, M. J. (2004). Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52, 1107–1121. doi: 10.1111/j.1365-2958.2004.04032.x
Hutchings, M. I., Hong, H. J., and Buttner, M. J. (2006). The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 59, 923–935. doi: 10.1111/j.1365-2958.2005.04953.x
Hwang, K. S., Kim, H. U., Charusanti, P., Palsson, B. O., and Lee, S. Y. (2014). Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 32, 255–268. doi: 10.1016/j.biotechadv.2013.10.008
Kieser, T., Hopwood, D. A., Bibb, J. M., Chater, K. F., and Buttner, M. J. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Klinzing, D. C., Ishmael, N., Hotopp, J. C. D., Tettelin, H., Shields, K. R., Madoff, L. C., et al. (2013). The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology 159(Pt 7), 1521–1534. doi: 10.1099/mic.0.064444-0
Kodani, S., Hudson, M. E., Durrant, M. C., Buttner, M. J., Nodwell, J. R., and Willey, J. M. (2004). The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 101, 11448–11453. doi: 10.1073/pnas.0404220101
Kormanec, J., and Sevcikova, B. (2000). Identification and transcriptional analysis of a cold shock-inducible gene, cspA, in Streptomyces coelicolor A3(2). Mol. Genet. Genomics 264, 251–256. doi: 10.1007/s004380000298
Koteva, K., Hong, H. J., Wang, X. D., Nazi, I., Hughes, D., Naldrett, M. J., et al. (2010). A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 6, 327–329. doi: 10.1038/nchembio.350
Krysenko, S., Matthews, A., Okoniewski, N., Kulik, A., Girbas, M. G., Tsypik, O., et al. (2019). Initial metabolic step of a novel ethanolamine utilization pathway and its regulation in Streptomyces coelicolor M145. mBio 10:e00326-19.
Kudou, D., Yasuda, E., Hirai, Y., Tamura, T., and Inagaki, K. (2015). Molecular cloning and characterization of l-methionine gamma-lyase from Streptomyces avermitilis. J. Biosci. Bioeng. 120, 380–383. doi: 10.1016/j.jbiosc.2015.02.019
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuroda, M., Kuroda, H., Oshima, T., Takeuchi, F., Mori, H., and Hiramatsu, K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49, 807–821. doi: 10.1046/j.1365-2958.2003.03599.x
Kuroda, M., Kuwahara-Arai, K., and Hiramatsu, K. (2000). Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269, 485–490. doi: 10.1006/bbrc.2000.2277
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Lee, E. J., Karoonuthaisiri, N., Kim, H. S., Park, J. H., Cha, C. J., Kao, C. M., et al. (2005). A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 57, 1252–1264. doi: 10.1111/j.1365-2958.2005.04761.x
Leonard, P. G., Golemi-Kotra, D., and Stock, A. M. (2013). Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc. Natl. Acad. Sci. U.S.A. 110, 8525–8530. doi: 10.1073/pnas.1302819110
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Lewis, R. A., Wahab, A., Bucca, G., Laing, E. E., Moller-Levet, C. S., Kierzek, A., et al. (2019). Genome-wide analysis of the role of the antibiotic biosynthesis regulator AbsA2 in Streptomyces coelicolor A3(2). PLoS One 14:e0200673. doi: 10.1371/journal.pone.0200673
Li, Y. Q., Chen, P. L., Chen, S. F., Wu, D., and Zheng, J. (2004). A pair of two-component regulatory genes ecrA1/A2 in S. coelicolor. J. Zhejiang Univ. Sci. 5, 173–179. doi: 10.1631/jzus.2004.0173
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/mmbr.00054-12
Llamas-Ramírez, R., Takahashi-Iniguez, T., and Flores, M. E. (2020). The phosphoenolpyruvate-pyruvate-oxaloacetate node genes and enzymes in Streptomyces coelicolor M-145. Int. Microbiol. 23, 429–439. doi: 10.1007/s10123-019-00116-x
Lockey, C., Edwards, R. J., Roper, D. I., and Dixon, A. M. (2020). The extracellular domain of two-component system sensor kinase VanS from Streptomyces coelicolor binds vancomycin at a newly identified binding site. Sci. Rep. 10:5727. doi: 10.1038/s41598-020-62557-z
Lu, Y., Wang, W., Shu, D., Zhang, W., Chen, L., Qin, Z., et al. (2007). Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 77, 625–635. doi: 10.1007/s00253-007-1184-5
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mascher, T., Zimmer, S. L., Smith, T. A., and Helmann, J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004
Mendes, M. V., Tunca, S., Antón, N., Recio, E., Sola-Landa, A., Aparicio, J. F., et al. (2007). The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab. Eng. 9, 217–227. doi: 10.1016/j.ymben.2006.10.003
Menéndez-Bravo, S., Paganini, J., Avignone-Rossa, C., Gramajo, H., and Arabolaza, A. (2017). Identification of FadAB complexes involved in fatty acid beta-oxidation in Streptomyces coelicolor and construction of a triacylglycerol overproducing strain. Front. Microbiol. 8:1428. doi: 10.3389/fmicb.2017.01428
Miller, W. R., Munita, J. M., and Arias, C. A. (2014). Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 12, 1221–1236. doi: 10.1586/14787210.2014.956092
Mouncey, N. J., Otani, H., Udwary, D., and Yoshikuni, Y. (2019). New voyages to explore the natural product galaxy. J. Ind. Microbiol. Biotechnol. 46, 273–279. doi: 10.1007/s10295-018-02122-w
Novotna, G. B., Kwun, M. J., and Hong, H. J. (2015). In vivo characterization of the activation and interaction of the VanR-VanS two-component regulatory system controlling glycopeptide antibiotic resistance in two related Streptomyces species. Antimicrob. Agents Chemother. 60, 1627–1637. doi: 10.1128/AAC.01367-15
Okamoto, S., Taguchi, T., Ochi, K., and Ichinose, K. (2009). Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem. Biol. 16, 226–236. doi: 10.1016/j.chembiol.2009.01.015
Park, J. H., and Roe, J. H. (2008). Mycothiol regulates and is regulated by a thiol-specific antisigma factor RsrA and sigma(R) in Streptomyces coelicolor. Mol. Microbiol. 68, 861–870. doi: 10.1111/j.1365-2958.2008.06191.x
Pawlik, K., Kotowska, M., Chater, K. F., Kuczek, K., and Takano, E. (2007). A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch. Microbiol. 187, 87–99. doi: 10.1007/s00203-006-0176-7
Perron, K., Caille, O., Rossier, C., Van Delden, C., Dumas, J. L., and Kohler, T. (2004). CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279, 8761–8768. doi: 10.1074/jbc.M312080200
Prieto, C., and Barrios, D. (2019). RaNA-Seq: interactive RNA-Seq analysis from FASTQ files to functional analysis. Bioinformatics doi: 10.1093/bioinformatics/btz854 [Epub ahead of print].
Qureshi, N. K., Yin, S., and Boyle-Vavra, S. (2014). The role of the Staphylococcal VraTSR regulatory system on vancomycin resistance and vanA operon expression in vancomycin-resistant Staphylococcus aureus. PLoS One 9:e85873. doi: 10.1371/journal.pone.0085873
Raavi, Mishra, S., and Singh, S. (2017). Prevention of OprD regulated antibiotic resistance in Pseudomonas aeruginosa biofilm. Microb. Pathog. 112, 221–229. doi: 10.1016/j.micpath.2017.08.007
Rexer, H. U., Schaberle, T., Wohlleben, W., and Engels, A. (2006). Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 186, 447–458. doi: 10.1007/s00203-006-0159-8
Rico, S., Santamaría, R. I., Yepes, A., Rodríguez, H., Laing, E., Bucca, G., et al. (2014a). Deciphering the regulon of Streptomyces coelicolor AbrC3, a positive response regulator of antibiotic production. Appl. Environ. Microbiol. 80, 2417–2428. doi: 10.1128/aem.03378-13
Rico, S., Yepes, A., Rodríguez, H., Santamaría, J., Antoraz, S., Krause, E. M., et al. (2014b). Regulation of the AbrA1/A2 two-component system in Streptomyces coelicolor and the potential of its deletion strain as a heterologous host for antibiotic production. PLoS One 9:e109844. doi: 10.1371/journal.pone.0109844
Rodríguez, H., Rico, S., Díaz, M., and Santamaría, R. I. (2013). Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microb Cell Fact. 12:127. doi: 10.1186/1475-2859-12-127
Rodríguez, H., Rico, S., Yepes, A., Franco-Echevarría, E., Antoraz, S., Santamaría, R. I., et al. (2015). The two kinases, AbrC1 and AbrC2, of the atypical two-component system AbrC are needed to regulate antibiotic production and differentiation in Streptomyces coelicolor. Front. Microbiol. 6:450. doi: 10.3389/fmicb.2015.00450
Rose, M. D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: a Laboratory Course Manual. New York, NY: Cold Spring Harbour Laboratory Press.
Saito, A., Shinya, T., Miyamoto, K., Yokoyama, T., Kaku, H., Minami, E., et al. (2007). The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N’-diacetylchitobiose in Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 73, 3000–3008. doi: 10.1128/aem.02612-06
Santos-Beneit, F. (2015). The Pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6:402. doi: 10.3389/fmicb.2015.00402
Santos-Beneit, F. (2018). Genome sequencing analysis of Streptomyces coelicolor mutants that overcome the phosphate-depending vancomycin lethal effect. BMC Genomics 19:457. doi: 10.1186/s12864-018-4838-z
Santos-Beneit, F., Fernández-Martínez, L. T., Rodríguez-García, A., Martín-Martín, S., Ordoñez-Robles, M., Yagüe, P., et al. (2014). Transcriptional response to vancomycin in a highly vancomycin-resistant Streptomyces coelicolor mutant. Future Microbiol. 9, 603–622. doi: 10.2217/fmb.14.21
Santos-Beneit, F., and Martín, J. F. (2013). Vancomycin resistance in Streptomyces coelicolor is phosphate-dependent but is not mediated by the PhoP regulator. J. Glob. Antimicrob. Resist. 1, 109–113. doi: 10.1016/j.jgar.2013.03.003
Santos-Beneit, F., Rodriguez-Garcia, A., Sola-Landa, A., and Martin, J. F. (2009). Crosstalk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72, 53–68. doi: 10.1111/j.1365-2958.2009.06624.x
Sola-Landa, A., Moura, R. S., and Martín, J. F. (2003). The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U.S.A. 100, 6133–6138. doi: 10.1073/pnas.0931429100
Som, N. F., Heine, D., Holmes, N., Knowles, F., Chandra, G., Seipke, R. F., et al. (2017). The MtrAB two-component system controls antibiotic production in Streptomyces coelicolor A3(2). Microbiology 163, 1415–1419. doi: 10.1099/mic.0.000524
Stankovic, N., Senerovic, L., Ilic-Tomic, T., Vasiljevic, B., and Nikodinovic-Runic, J. (2014). Properties and applications of undecylprodigiosin and other bacterial prodigiosins. Appl. Microbiol. Biotechnol. 98, 3841–3858. doi: 10.1007/s00253-014-5590-1
Suntharalingam, P., Senadheera, M. D., Mair, R. W., Levesque, C. M., and Cvitkovitch, D. G. (2009). The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191, 2973–2984. doi: 10.1128/jb.01563-08
Tierney, A. R., and Rather, P. N. (2019). Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 14, 533–552. doi: 10.2217/fmb-2019-0002
Tong, Y., Charusanti, P., Zhang, L., Weber, T., and Lee, S. Y. (2015). CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 4, 1020–1029. doi: 10.1021/acssynbio.5b00038
van der Heul, H. U., Bilyk, B. L., McDowall, K. J., Seipke, R. F., and van Wezel, G. P. (2018). Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 35, 575–604. doi: 10.1039/c8np00012c
van der Meij, A., Worsley, S. F., Hutchings, M. I., and van Wezel, G. P. (2017). Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416. doi: 10.1093/femsre/fux005
VanBogelen, R. A., and Neidhardt, F. C. (1990). Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 5589–5593. doi: 10.1073/pnas.87.15.5589
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Xia, H., Zhan, X., Mao, X. M., and Li, Y. Q. (2020). The regulatory cascades of antibiotic production in Streptomyces. World J. Microbiol. Biotechnol. 36:13.
Xu, G., Wang, J., Wang, L., Tian, X., Yang, H., Fan, K., et al. (2010). “Pseudo” gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285, 27440–27448. doi: 10.1074/jbc.M110.143081
Yang, S. J., Bayer, A. S., Mishra, N. N., Meehl, M., Ledala, N., Yeaman, M. R., et al. (2012). The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 80, 74–81. doi: 10.1128/iai.05669-11
Yeo, K. J., Hong, Y. S., Jee, J. G., Lee, J. K., Kim, H. J., Park, J. W., et al. (2014). Mechanism of the pH-induced conformational change in the sensor domain of the DraK Histidine kinase via the E83, E105, and E107 residues. PLoS One 9:e107168. doi: 10.1371/journal.pone.0107168
Yepes, A., Rico, S., Rodríguez-García, A., Santamaría, R. I., and Díaz, M. (2011). Novel two-component systems implied in antibiotic production in Streptomyces coelicolor. PLoS One 6:e19980. doi: 10.1371/journal.pone.0019980
Yin, S., Daum, R. S., and Boyle-Vavra, S. (2006). VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 336–343. doi: 10.1128/AAC.50.1.336-343.2006
Zhang, G., Tian, Y., Hu, K., Feng, C., and Tan, H. (2010). SCO3900, co-transcripted with three downstream genes, is involved in the differentiation of Streptomyces coelicolor. Curr. Microbiol. 60, 268–273. doi: 10.1007/s00284-009-9536-2
Keywords: two-component system, Streptomyces, antibiotic production, antibiotic resistance, vancomycin, RNA-Seq
Citation: Sánchez de la Nieta R, Antoraz S, Alzate JF, Santamaría RI and Díaz M (2020) Antibiotic Production and Antibiotic Resistance: The Two Sides of AbrB1/B2, a Two-Component System of Streptomyces coelicolor. Front. Microbiol. 11:587750. doi: 10.3389/fmicb.2020.587750
Received: 27 July 2020; Accepted: 22 September 2020;
Published: 09 October 2020.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Yinhua Lu, Shanghai Normal University, ChinaCopyright © 2020 Sánchez de la Nieta, Antoraz, Alzate, Santamaría and Díaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margarita Díaz, bWFyZGlAdXNhbC5lcw==; Ramón I. Santamaría, c2FudGFAdXNhbC5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.