- 1Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, United States
- 2Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 3Earth and Environmental Sciences, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 4Molecular Biosciences and Integrated Bioimaging, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 5Department of Bioengineering, University of California, Berkeley, Berkeley, CA, United States
- 6Department of Civil and Environmental Engineering, The University of Tennessee, Knoxville, Knoxville, TN, United States
A nitrate- and metal-contaminated site at the Oak Ridge Reservation (ORR) was previously shown to contain the metal molybdenum (Mo) at picomolar concentrations. This potentially limits microbial nitrate reduction, as Mo is required by the enzyme nitrate reductase, which catalyzes the first step of nitrate removal. Enrichment for anaerobic nitrate-reducing microbes from contaminated sediment at the ORR yielded Bacillus strain EB106-08-02-XG196. This bacterium grows in the presence of multiple metals (Cd, Ni, Cu, Co, Mn, and U) but also exhibits better growth compared to control strains, including Pseudomonas fluorescens N2E2 isolated from a pristine ORR environment under low molybdate concentrations (<1 nM). Molybdate is taken up by the molybdate binding protein, ModA, of the molybdate ATP-binding cassette transporter. ModA of XG196 is phylogenetically distinct from those of other characterized ModA proteins. The genes encoding ModA from XG196, P. fluorescens N2E2 and Escherichia coli K12 were expressed in E. coli and the recombinant proteins were purified. Isothermal titration calorimetry analysis showed that XG196 ModA has a higher affinity for molybdate than other ModA proteins with a molybdate binding constant (KD) of 2.2 nM, about one order of magnitude lower than those of P. fluorescens N2E2 (27.0 nM) and E. coli K12 (25.0 nM). XG196 ModA also showed a fivefold higher affinity for molybdate than for tungstate (11 nM), whereas the ModA proteins from P. fluorescens N2E2 [KD (Mo) 27.0 nM, KD (W) 26.7 nM] and E. coli K12[(KD (Mo) 25.0 nM, KD (W) 23.8 nM] had similar affinities for the two oxyanions. We propose that high molybdate affinity coupled with resistance to multiple metals gives strain XG196 a competitive advantage in Mo-limited environments contaminated with high concentrations of metals and nitrate, as found at ORR.
Introduction
Molybdenum (Mo) is an essential metal for the growth of virtually all known life forms, including humans, plants and microorganisms, as it is required for the function of several key enzymes involved in the cycling of N, C, and S (Hamlin, 2016; Schwarz, 2016; Maia et al., 2017). Tungsten (W), an antagonist of Mo, is more uncommon in nature but required in some enzymes, most notably in archaea. Physiologically-relevant oxidation states of Mo and W are + 4, + 5, and + 6 (Maia et al., 2017). There are five distinct enzyme families that use Mo and/or W, represented by nitrogenase (Mo only, although some use vanadium), xanthine oxidase (Mo only), the sulfite oxidase (Mo only), DMSO reductase (most family members use Mo, a few use W) and tungsten-containing oxidoreductase (WOR, W only) (Hille et al., 2014; Maia et al., 2017). In most microorganisms, molybdate is taken up into the cell by the molybdate ATP-binding cassette or Mod transporter (ModABC), which can also take up tungstate (Grunden and Shanmugam, 1997; Self et al., 2001).
In the nitrogen cycle, Mo is utilized in three key steps, N2-fixation (by nitrogenase), nitrite oxidation (by nitrite oxidoreductase) and nitrate reduction (by nitrate reductase) (Zhang and Gladyshev, 2008). Hence Mo is required for the biological removal of nitrate from contaminated environments as the reductase is a key enzyme in both the denitrification (yielding N2) and dissimilatory nitrate reduction to ammonium (DNRA) pathways (Zhang and Gladyshev, 2008). Consequently, in natural environments, the availability of Mo can limit nitrate removal (Barron et al., 2009; Glass et al., 2012). Mo limitation can also negatively impact nitrate removal in contaminated environments, which can be caused by the extensive use of nitrate-containing fertilizers, the release of nitrate-containing industrial wastes, as well as mining and other anthropogenic activities leading to problems for human health and natural environments (Spalding and Exner, 1993; Kellman and Hillaire-Marcel, 2003; Diaz and Rosenberg, 2008; Gruber and Galloway, 2008; Powlson et al., 2008; Thorgersen et al., 2015; Zhang et al., 2015).
The Oak Ridge Reservation (ORR) in Tennessee, United States contains a nitrate-contaminated waste site – the S-3 ponds. These are four adjacent (∼9.5 million liters each) earthen reservoirs used for the disposal of waste liquids that had been produced from the Y-12 nuclear plant for more than 30 years (Brooks, 2001). The waste liquids contained high, and potentially toxic, concentrations of nitrate (up to 1.2 M) and a wide variety of metals, such as iron (up to 21 mM), aluminum (up to 180 mM), magnesium (up to 28 mM), and uranium (up 1.3 mM) (Brooks, 2001). In 1983, the waste liquids in the S-3 ponds were adjusted to about pH 9, and the precipitates formed were allowed to settle before the liquid was removed (Brooks, 2001; Revil et al., 2013). In 1988, the S-3 ponds were filled and capped and now serve as a parking lot (Revil et al., 2013). However, the area is still heavily contaminated and groundwater in the contamination plume emanating from the former S-3 ponds is at low pH (as low as 3.0) and contains high concentrations of nitrate (up to 230 mM), much higher than the surrounding pristine groundwater (less than 32 μM) (Nolan et al., 1998; Ge et al., 2019). In addition, the contaminating plume has elevated concentrations of over 20 metals, including uranium (up to 580 μM) (Smith et al., 2015; Thorgersen et al., 2015). In stark contrast, extremely low concentrations of Mo (in the picomolar range) were measured in this highly contaminated groundwater. It was demonstrated experimentally that the pM concentrations of Mo in ORR contaminated groundwater were likely a result of molybdate adsorption and incorporation into Fe- and Al-based minerals that are formed as the groundwater from the highly contaminated area (pH < 1) mixes with the surrounding groundwater (Moura et al., 2004; Ge et al., 2019).
Hence, a fundamental question is whether microorganisms that thrive in the unique ORR environment contaminated with high concentrations of metals and nitrate, yet containing only picomolar levels of Mo, have unique features that enhance Mo utilization. Herein, we describe the characterization of a novel nitrate-reducing Bacillus, designated strain EB106-08-02-XG196 (hereafter XG196), that was isolated from a sample of nitrate- and metal-contaminated ORR sediment (EB-106) located 21 m downstream of the S-3 ponds area (Ge et al., 2019). XG196 is resistant to high concentrations of a metal mixture that was designed to mimic the ORR contaminated groundwater. More importantly, it is also much less sensitive to Mo-limitation than other ORR isolates, including four other EB-106 strains and a microbe obtained from a non-contaminated ORR environment. The molecular basis for the ability of XG196 to thrive under Mo-limited conditions was investigated.
Materials and Methods
Sampling and Isolation of Strains
An 8-meter-deep borehole of 8.9 cm diameter (designated EB-106) located 21.1 meters downstream from the S-3 ponds area was drilled at ORR. The sediment was collected and cut into 22 cm segments under anaerobic conditions, as reported elsewhere (Ge et al., 2019; Moon et al., 2020). For microbial enrichment, sediment samples (1 g) were incubated anaerobically in 5 ml of a defined medium (pH 7.0) containing 1.3 mM KCl, 2 mM MgSO4, 0.1 mM CaCl2, 0.3 mM NaCl, 30 mM NaHCO3, 5 mM NaH2PO4 and 20 mM NaNO3, with added vitamins and minerals as described (Widdel and Bak, 1992). A mixture of 2 mM of different carbon sources (formate, acetate, ethanol, lactate, succinate, and glucose) and 0.1 g/L yeast extract was used as carbon sources. Metal mix (MM) was used to mimic the metal contamination in the groundwater near the ORR S-3 ponds. MM (1x) resulted in final concentrations in the media of 5 μM cadmium acetate (Cd(CH3COO)2⋅2H2O), 100 μM manganous chloride (MnCl2⋅2H2O), 30 μM cobalt chloride (CoCl2⋅6H2O), 100 μM nickel chloride (NiCl2⋅6H2O), 10 μM cupric chloride (CuCl2⋅2H2O), 10 μM ferrous ammonium sulfate (Fe(NH4)2(SO4)2 ⋅6H2O) and 100 μM uranyl acetate (UO2(CH3COO)2⋅2H2O) (Supplementary Table S1). For enrichment, all cultures were incubated anaerobically in anaerobic chamber (filled with 95% argon and 5% hydrogen) at room temperature for 2 to 7 days in media containing either 1 ×, 0.5 × or no MM of final concentrations. Cultures with turbidity were streaked out on plates (1.5% agar) using the same medium and were incubated at room temperature for single colony isolation. For purified microbial strains, the sequences of their 16S rRNA genes were determined as described below and compared with those in the BLASTN suite1 using default parameters (Altschul et al., 1990).
Nitrate Reductase Activity
Nitrate reductase activities of the EB-106 isolates were determined using whole cell suspensions (Filiatrault et al., 2013). Strains were grown anaerobically in Hungate tubes and cells were collected between mid-log phase and early stationary phase, then 15 μL of 5 mg/ml chloramphenicol was added to 1.5 ml of culture to inhibit protein synthesis. Cells were washed twice and re-suspended in buffer (50 mM phosphate buffer, pH 7.2) and the OD660 was determined. 200 μL of cells were mixed with 25 μL of methyl viologen (0.5 mg/ml) in an anaerobic sealed cuvette at 25°C. 100 μL of reaction solution (4 mg/ml Na2S2O4, 4 mg/ml NaHCO3 and 100 mM KNO3) was added to start the reaction. In control reaction buffer, Na2S2O4 was replaced with water. After incubation at room temperature for 5 min, the mixtures were vortexed in air to stop the reaction by oxidizing the electron donors (Na2S2O4 and reduced methyl viologen). The amount of nitrite produced was measured by adding 100 μL of sulfanilic acid (1% w/v in 20% HCl) to 30 μL of each reaction mixture followed by 100 μL of N-(1-naphthyl)ethylenediamine-HCl (1.3 mg/ml). The OD540 of each sample supernatant was measured and the amount of nitrite was calculated according to nitrite standards. The OD420 of the samples was also measured to account for light scattering by residual cells and cell fragments. Nitrate reductase specific activity is expressed as units/OD660, in which units are calculated using the formula 100 × [OD540 - (0.72 × OD420)]/(T × V), T is time in minutes and V is reaction volume in milliliters (Filiatrault et al., 2013; Thorgersen et al., 2019).
Carbon Sources Utilization for Anaerobic Growth Analysis
Growth on various carbon sources was determined at 25°C under anaerobic conditions using the standard medium lacking yeast extract and the organic mixture but containing either formate, acetate, ethanol, lactate, succinic acid, fumarate, xylose, xylitol, glucose, fructose, maltose, sodium benzoate, sodium 4-hydroxybenzoate, potassium sodium tartrate, proline, phenylalanine, arginine, threonine, leucine, glutamate, or glutamine (all at 2 mM) with and without nitrate (KNO3, 20 mM). Growth was measured in 400 μl wells on a 100-well plate (Bioscreen sterile plates HONEYCOMB, Thermo Fisher Scientific, Waltham, MA, United States) using a Bioscreen C (Thermo Labsystems, Thermo Fisher Scientific, Waltham, MA, United States) placed in a PLAS LABS anaerobic chamber under a 5% H2 and 95% Ar atmosphere. Optical density (OD600) of cultures in each wells were measured every 5 min, after the plate was shaken using the Bioscreen C to resuspend cells.
Mo Accumulation Analysis
EB-106 isolates were grown in 500 ml of defined media with 1 μM Mo ((NH4)2MoO4) and harvested at mid log phase, washed three times with 10 ml of Tris buffer (Tris 50 mM, pH 8.0, containing 100 mM NaCl) and then resuspended in Tris buffer. Cells were lysed by sonication, then were spun down at 10,000 × G for 15 min and the supernatants were used for further centrifugation. The cytoplasmic extract (S100) was obtained after centrifugation at 100,000 × G for 1 h in a Beckman Coulter Optima L-90 ultracentrifuge. The membrane fraction was resuspended in 2 ml of Tris buffer. Both S100 and membrane fractions were diluted (1:40) with trace grade 2% nitric acid (VWR, Radnor, PA, United States) and incubated overnight prior to analysis by inductively coupled plasma mass spectrometry (ICP-MS) analysis to quantify Mo (Lancaster et al., 2014; Scott et al., 2015). Protein concentrations were measured using the Bradford assay (Bio-Rad protein assay kit, Bio-Rad, Berkeley, CA, United States). The amount of Mo accumulated is expressed as nmoles per gram of protein (nmol/g).
Molybdenum-Limited Growth
For the Mo-depleted medium, a solution was prepared that contained 1.3 mM KCl, 2 mM MgSO4, 0.1 mM CaCl2, and 0.3 mM NaCl together with the vitamins and minerals described above except that molybdenum and tungsten were not added (Widdel and Bak, 1992). Fe(NO3)3 (20 mM) was then added, which acidifies the solution to pH ∼ 2.5. The pH was then adjusted to pH 6.7 using trace grade NaOH (2.0 M) to induce precipitation of ferric hydroxide. As previously described (Ge et al., 2019), the Fe precipitates any contaminating Mo present in the medium components. The Mo-depleted growth medium was prepared by adding trace grade Fe(NO3)3 (7.4 μM), Na2SO4 (2 mM), NaHCO3 (30 mM), and NaH2PO4 (5 mM) and inoculated with 1% (vol/vol) washed XG77, XG146, XG95, XG201, or XG196 cells grown in media with no Mo added. Growth in this medium with and without added Mo (0.1, 0.5, 1, 5, 10, 50, or 500 nM Na2MoO4) was measured in quadruplet using the Bioscreen C described above. Mo and W competition analysis of XG196 and Pseudomonas fluorescens N2E2 under nitrate reducing conditions were performed using the same media with added Mo (0, 5, 50, 500, 5000, or 50000 nM Na2MoO4) and added W (0, 50 nM, 5 or 500 μM Na2WO4).
Metal Tolerance Assay
Each EB-106 isolate (XG77, XG146, XG95, XG201, and XG196) was incubated with individual metals at multiple concentrations, including manganous chloride (MnCl2⋅2H2O, 0 ∼ 6 mM), cobalt chloride (CoCl2⋅6H2O, 0∼600 μM), nickel chloride (NiCl2⋅6H2O, 0∼250 μM), cupric chloride (CuCl2⋅2H2O, 0∼250 μM), cadmium acetate Cd(CH3COO)2⋅2H2O, 0∼200 μM) or uranyl acetate (UO2(CH3COO)2⋅2H2O, 0∼2 mM), or with the MM (0∼2×) described above (Supplementary Table S1). Cultures were grown in defined medium described above containing 0.1 g/L yeast extract using the Bioscreen C described above and growth data were analyzed using R with Grofit package (Kahm et al., 2010). The average half maximal inhibitory concentration (IC50) was used to reflect the tolerance of each EB-106 isolate to each individual metal and the MM.
Genome Sequencing
The ZymoBead Genomic DNA kit was used to extract genomic DNA from strain XG196 and XG77. More than 1 μg of purified gnomic DNA from each strain was used for Illumina sequencing. The Illumina sequencing reads were trimmed using Trimmomatic 0.36, with parameters “-phred33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 ILLUMINACLIP:TruSeq3-PE.fa” (Bolger et al., 2014). The trimmed reads were assembled de novo using SPAdes v3.12.0 with parameters “-k 21,33,55,77” (Bankevich et al., 2012). Genes were identified using Prokka v1.12, with default parameters (Seemann, 2014). This pipeline was executed using the Department of Energy KnowledgeBase software platform (KBase2; Arkin et al., 2018). The genome of XG196 was submitted to GenBank (accession number: JABWSY000000000).
16S rDNA Sequencing and Phylogenetic Analysis
The 16S rDNA of isolate XG196 was amplified by PCR using universal bacterial primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-ACG GCT ACC TTG TTA CGA CTT-3′) from Integrated DNA Technologies, Coralville, IA, United States. DNA sequencing was carried out by GENEWIZ, South Plainfield, NJ, United States. The sequence was first analyzed by BLAST3 (Altschul et al., 1990), which indicated that XG196 is a Bacillus strain. The sequence was uploaded to the Ribosomal Database Project (RDP4) (Cole et al., 2013). The RDP tool Seqmatch was run to find the closest relatives of isolate XG196 and the Bacillus type strains with high quality 16S rDNA sequences (>1200 bp). The two closest relatives of XG196 and a total of 187 Bacillus type strains with one out group strain were selected to build the 16S rRNA phylogenetic tree by IQ-TREE using maximum likelihood (5 Nguyen et al., 2014). GTR + F + R6 model was selected by ModelFinder (Kalyaanamoorthy et al., 2017) and 1000 times of bootstrapping was run using UFBoot (Hoang et al., 2017).
Phylogenetic Analysis of Molybdate and Tungstate Binding Proteins
Accession numbers of ModA (family IPR005950) and WtpA (family IPR022498) were downloaded from the InterPro database6 (Mitchell et al., 2018). Information, such as sequence, mass, protein name, gene name, taxonomic lineage, cross-reference in PDB, cross-reference in KEGG, PubMed ID, etc. were all downloaded together. Proteins with candidadus/candidate organisms, uncultured organisms, fragment proteins, wrong/poorly-labeled organisms, and duplicates were removed from the list. Two lists (A and B) of strains were selected from downloaded candidates for ModA phylogenetic analysis. List A uses strains with ATCC (American Type Culture Collection) or DSM (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) reference IDs. For list B, all downloaded protein sequences were clustered by CD-HIT7 at 60% sequence identity (Huang et al., 2010). In both lists, all archaea, eukaryote sequences and sequences with KEGG cross-reference or 3D structures were kept. Several Bacillus type-strains and the top two closest isolate XG196 ModA relatives and ORR isolate P. fluorescens N2E2 from non-contaminated area were also kept. ModA/WtpA sequences from list A (617 sequences, Supplementary Material list A) and B (4623 sequences, Supplementary Material list B) were all used for tree building. Multiple sequence alignment was done by Clustal Omega8 (Madeira et al., 2019). IQ-tree were used to build the phylogenetic tree by maximum likelihood (Nguyen et al., 2014). LG + F + R10 model and WAG + R9 model were selected for list A and B ModA tree building by ModelFinder (Kalyaanamoorthy et al., 2017). 2000 and 3000 times of bootstrapping was run for list A and B ModA tree using UFBoot (Hoang et al., 2017). Signal peptide prediction analysis was performed for all list A sequences by SignalP5.09 (Armenteros et al., 2019).
Multi-Alignment and Structural Modeling Analysis of ModA
Multiple sequence alignments of XG196 ModA and selected proteins were first run by Clustal Omega10 (Madeira et al., 2019) and further analyzed with selected ModA proteins with structural data from the PDB11 using ESPript 3.012 (Robert and Gouet, 2014). The structures of the ModA proteins from Pyrococcus furiosus ATCC 43587 ModA (PDB: 3CG1) and Escherichia coli K12 ModA (PDB: 1AMF) were used for comparison with XG196 ModA. Mean identity and mean similarity of protein sequences were also calculated by ESPript 3.0. SWISS-MODEL13 (Brooks, 2001) was used to predict the model of XG196 using template ModA (PDB: 2H5Y) from Xanthomonas axonopodis pv. citri 306. UCSF Chimera14 (Pettersen et al., 2004) was used to visualize the model.
Expression and Purification of Recombinant ModA Proteins
ModA genes were amplified by PCR from the genomes of XG196, P. fluorescens N2E2 and E. coli K12. The primers are listed in Supplementary Table S2. The forward primer for the ModA gene of XG196 was designed to omit the N-terminal 20 amino acids, which include a signal peptide and a putative lipoprotein-attachment sight (Cys20). The forward primers for ModA genes of P. fluorescens N2E2 and E. coli K12 were designed to omit the N-terminal signal peptides, the first 23 and 25 amino acids, respectively. Signal sequences and lipoprotein-attachment site were predicted by SignalP-5.015 (Armenteros et al., 2019). The PCR amplicons were cloned into the pET24a (+) plasmid (Novagen). ModA proteins were expressed in E. coli Rosetta 2 (DE3)pLysS (Novagen) cells in LB media supplemented with kanamycin (50 μg/ml). Recombinant gene expression was induced at an OD600 ∼ 0.6 with 0.5 mM IPTG and the growth temperature was reduced from 37 to 25°C. Cells were harvested after 16 h and resuspended in start buffer (Tris 20 mM, pH 7.6, 100 mM NaCl, 5 mM imidazole). Cells were lysed by sonication and centrifuged to remove unlysed cells. The supernatant fractions were loaded onto a HisTrap FF crude column (GE health care) pre-equilibrated with start buffer and washed with two column volumes of wash buffer (Tris 20 mM, pH 7.6, 100 mM NaCl, 30 mM imidazole) and the recombinant ModA proteins were then eluted with elution buffer (Tris 20 mM, pH 7.6, 100 mM NaCl, 300 mM imidazole). ModA proteins were further purified by gel filtration using a Superdex 200 HiLoad 16/60 prep grade column (GE health care) equilibrated with Tris 20 mM, pH 7.6, containing 250 mM NaCl. Fractions containing the purified ModA protein as determined by SDS-PAGE were buffer exchanged to a low salt buffer (Tris 20 mM, pH 7.6, 90 mM NaCl) using an Amicon Ultra-15 10K centrifugal filter device at 4°C for 16 h for further ITC analysis. Mo in 40 μM of protein samples before and after dialysis were measured by ICP-MS. Trace grade of Tris (MilliporeSigma, St. Louis, MO, United States) and NaCl (MilliporeSigma, St. Louis, MO, United States) were used in protein purification and dialysis.
Isothermal Titration Calorimetry (ITC) Analysis
Molybdate (100 mM Na2MoO4) and tungstate (100 mM Na2WO4) stock solutions were prepared in trace grade ITC buffer (Tris 20 mM, pH 7.6, 90 mM NaCl) and then diluted to a final concentration of 0.3 or 0.4 mM using ITC buffer. ITC analysis was performed using a Malvern MicroCal PEAQ-ITC (Malvern Panalytical, Malvern, United Kingdom) at 25°C. Molybdate or tungstate were injected into the sample chamber (300 μL) containing 30 or 40 μM ModA to give a final molar ratio of oxyanion to ModA of 2:1. Displacement titrations were carried out by titrating molybdate or tungstate with chromate-saturated ModA (containing twofold of chromate from Na2CrO4) (Sigurskjold, 2000). Data were analyzed by Malvern MicroCal PEAQ-ITC analysis software (Malvern Panalytical, Malvern, United Kingdom). Each test was done twice and the average data were used.
ICP-MS Analysis
Samples were vortexed and then diluted (at various concentrations depending on sample type) into 2% (vol/wt) trace-grade nitric acid (VWR, Radnor, PA, United States) in acid-washed 15 mL polypropylene tubes. Samples were analyzed by an Agilent 7900 ICP-MS fitted with MicroMist nebulizer, UHMI-spray chamber, Pt cones, x-lens and an octopole reaction system (ORS) collision cell with He-mode (Agilent Technologies, Santa Clara, CA, United States) as described (Ge et al., 2019).
Metagenome Annotation and Analysis of Nitrate-Reducing Bacteria in ORR
Previously published metagenome sequence reads of samples from ORR groundwater were obtained from the NCBI database under BioProject PRJNA513876 (Tian et al., 2020). Metagenomic reads were preprocessed using BBtools version 38.60 (no references known16) to remove Illumina adapters, perform quality filtering and trimming, and remove PhiX174 spike-ins. The script bbduk.sh was run with parameters ktrim = r k = 23 mink = 11 hdist = 1 ref = adapters.fa tbo tpe 2 to remove any remaining standard Illumina adapters given in adapters.fa. The script was run again with parameters bf1 k = 27 hdist = 1 qtrim = rl trimq = 17 cardinality = t ref = phix174_Illumina.fa to perform quality filtering and trimming, and to remove Illumina PhiX174 spike ins given in the file phix174_Illumina.fa. We assembled the reads using SPAdes version 3.13.0 (Bankevich et al., 2012) with parameters –meta -k 21,33,55,77,99,127. We predicted protein-coding genes using Prodigal v 2.6.3 (Hyatt et al., 2010) with parameters -n -p single. Predicted protein-coding genes were annotated on the contigs using eggNOG mapper (v2) with default parameters (Huerta-Cepas et al., 2017). The number of predicted genes for each protein of interest was normalized by the number of raw reads obtained from metagenome sequencing.
Results
Isolation and Physiological Characterization of Bacillus Strain XG196
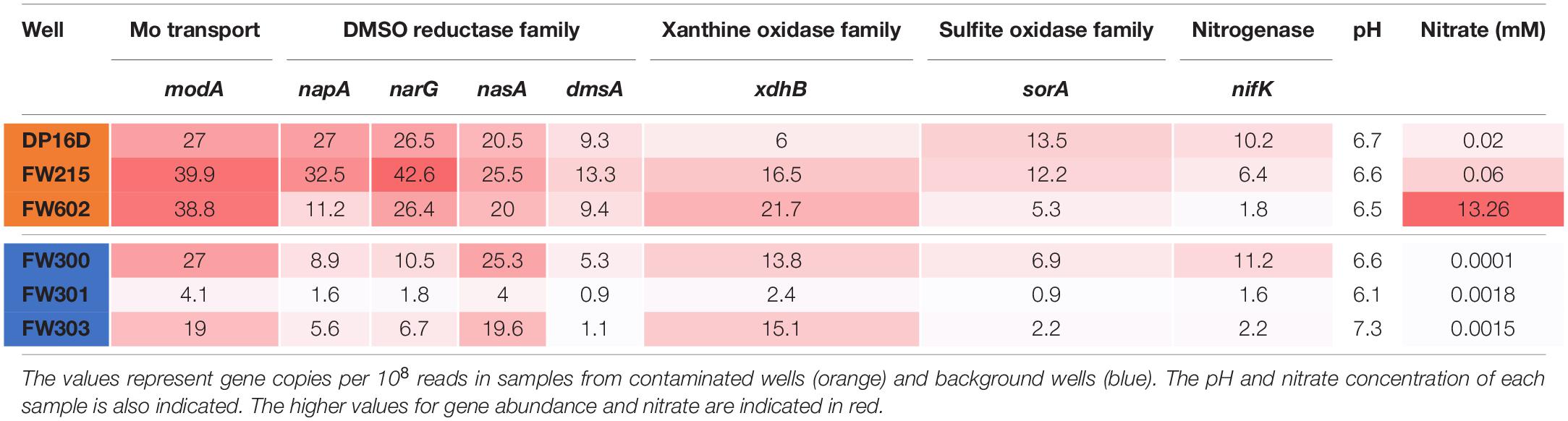
In order to isolate nitrate-reducing microbes with a high affinity for molybdate from the metal- and nitrate-contaminated ORR site, sediments from the contaminated EB-106 vertical core were used for enrichment and isolation. This 8-m core, taken about 21 m downstream of the contamination source (the S-3 ponds), was cut into 22 cm segments under anaerobic conditions (Ge et al., 2019; Moon et al., 2020). The EB-106 core covered the vadose zone (0–300 cm, the area between the land surface and water table), the capillary fringe (300–350 cm, the subsurface layer between vadose zone and the water table), and saturated zone (350–800 cm, the region below the water table) of the soil (Figure 1A). The groundwater passing through the saturated zone of the EB-106 core flows from the contamination site and is considered to be highly contaminated. A total of 88 unique nitrate-reducing bacteria were isolated from EB-106 sediment samples under nitrate-reducing conditions in a medium containing a combination of carbon sources (2 mM of formate, acetate, ethanol, lactate, succinate and glucose, together with 0.1 g/L yeast extract) and various levels of metal contaminants (no, 0.5 × MM or 1.0 × MM). Five strains, XG77, XG95, XG146, XG196 and XG201, were selected for further characterization based on their ability to grow anaerobically on nitrate, their nitrate reductase activities and metal resistance properties. XG77, XG95, and XG196 were identified as Bacillus strains, while XG146 and XG201 were identified as Ensifer and Enterobacter strains, respectively, by 16S rDNA sequences (Figure 1B). All five were isolated from the contaminated saturated zone (below 350 cm) (Figures 1A,B).
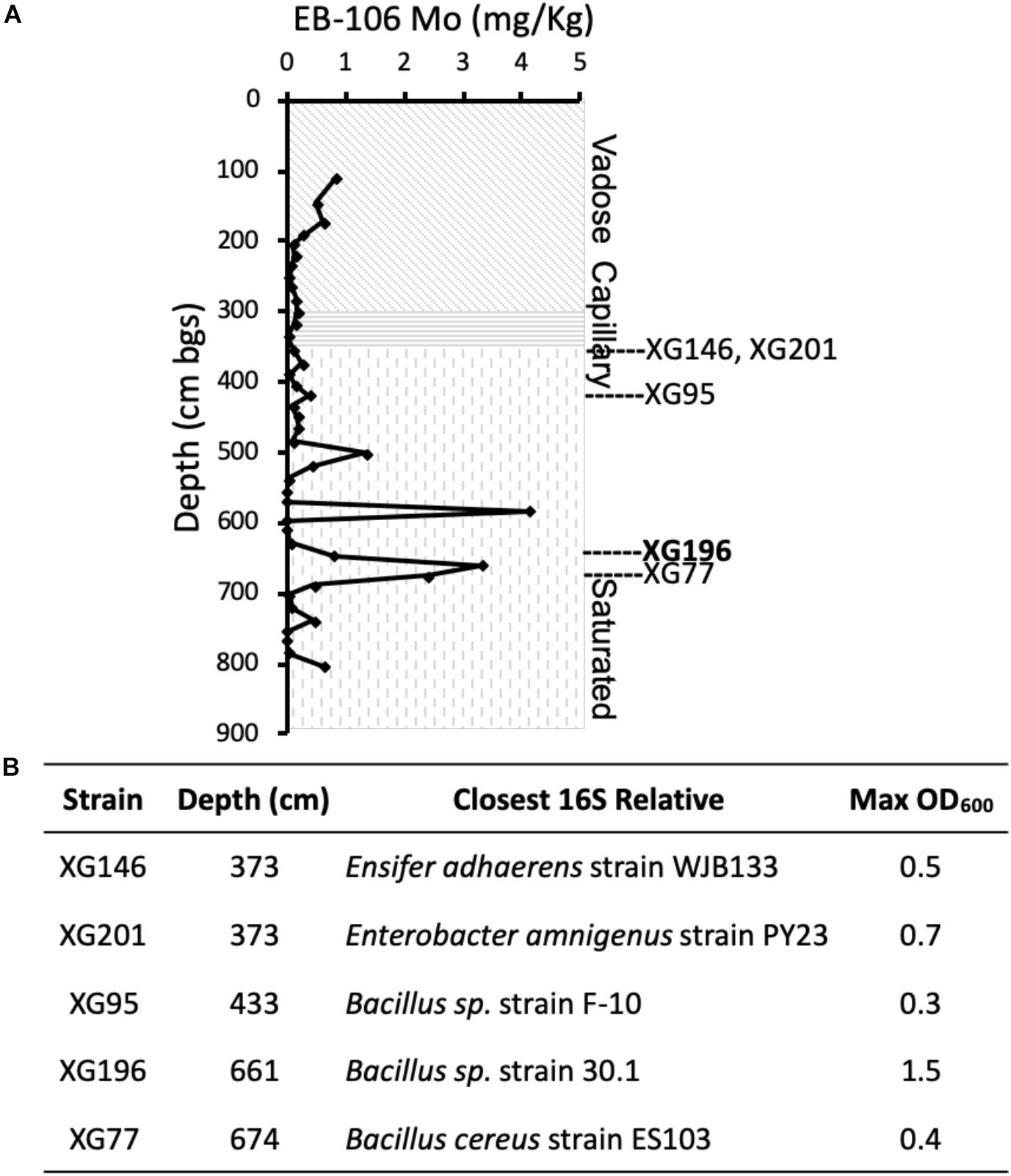
Figure 1. Five EB-106 sediment isolates used in this study. (A) Sketch of core EB-106. Subsurface layers: vadose zone (diagonal), capillary zone (horizontal), and saturated zone (vertical). Each point represents average Mo concentration at different depths. Depths of EB-106 sediments where XG146, XG201, XG95, XG196, and XG77 were isolated are indicated. (B) Properties of the ORR strains. Max OD600 was determined under nitrate reducing conditions with 20 mM carbon sources (lactate for XG196 and xylose for the other strains).
As shown in Supplementary Table S3, all five isolates use various carbon sources (2 mM) for nitrate-reducing growth. Isolate XG196 exhibited more robust growth on maltose (OD600max = 0.95), glucose (0.63), xylose (0.48), fructose (0.48), proline (0.47), glutamate (0.47), lactate (0.33), arginine (0.29) and fumarate (0.25), than other tested carbon sources (formate, acetate, ethanol, succinic acid, xylitol, benzoate, 4-hydeoxybenzoate, tartrate, phenylalanine, threonine, leucine and glutamine, OD600max ≤ 0.11). Xylose, glucose, fructose, maltose, proline, and glutamine also support the growth of isolate XG196 by fermentation. Lactate was selected as the carbon source for further characterization of strain XG196, as it supported robust nitrate-reducing growth and did not support fermentative growth. Xylose was selected for the other four EB-106 isolates as they exhibited good growth (XG77, OD600max 0.22; XG95, 0.22; XG146, 0.20; and XG201, 0.18) on xylose under nitrate-reducing conditions but did not use xylose for fermentation. Higher biomass yields were obtained with 20 mM of the preferred carbon source (lactate for isolate XG196 or xylose for strains XG146, XG201, XG95, and XG77). The maximum OD600 values for XG146, XG201, XG95, and XG77 increased from 0.20, 0.18, 0.22 and 0.22 to 0.5, 0.7, 0.25, and 0.4 respectively. XG196 reached the highest cell density under nitrate-reducing conditions, with OD600 reaching 1.5 when lactate was increased to 20 mM (Figure 1B). Nitrate reductase activities of XG146, XG201, XG95, XG77 and XG196 were determined using whole cell suspensions from cultures collected under anaerobic nitrate-reducing conditions using 20 mM nitrate (MacGregor et al., 1974; Filiatrault et al., 2013). Strains XG77 and XG196 showed higher nitrate reductase activities than the other three isolates (Supplementary Figure S1).
Growth of the EB-106 isolates was determined under nitrate-reducing growth conditions in the presence of increasing concentrations of a single metal (Cd, Ni, Cu, Co, Mn, or U) or the MM metal mixture containing all six metals, which mimics the concentrations of metals found in the ORR contaminated groundwater (Supplementary Table S1). The effects of the metals on growth was determined by calculating the IC50 values. Generally, strain XG196 had the highest metal tolerance of the five stains to the metal contaminants in the EB-106 sediments. Specifically, isolate XG196 had the highest IC50 values when grown with Ni2+ (119 μM), Co2+ (220 μM), Mn2+ (>900 μM), U6+ (2,000 μM) and the metal mixture (1.2×) and the second highest IC50 value when grown with Cu2+ (94 μM) (Supplementary Figure S2). Strain XG196 also grew in the presence of very high concentrations of nitrate and nitrite, with IC50 values of 299 and 99 mM, respectively (Supplementary Figure S2).
To analyze the dependence of growth under nitrate-reducing conditions on Mo, the five EB-106 strains and one strain previously isolated from non-contaminated ORR groundwater (P. fluorescens N2E2) were grown with increasing concentrations of molybdate in Mo depleted media prepared with trace metal grade chemicals in order to lower the amount of contaminating Mo to picomolar concentrations in cultures (∼400 pM, Ge et al., 2019). As shown in Figure 2A, strain XG196 showed the highest percentage of maximum growth (84% of highest OD600) even when no Mo was added to the medium, while the other EB-106 sediment strains tested required at least 1 nM Mo to reach ≥ 74% of maximum growth. P. fluorescens N2E2, which has been used as a reference strain in other ORR contamination studies (Thorgersen et al., 2015; Ge et al., 2019), had the lowest percentage (as low as 43%) of maximum growth when less than 1 nM Mo was added.
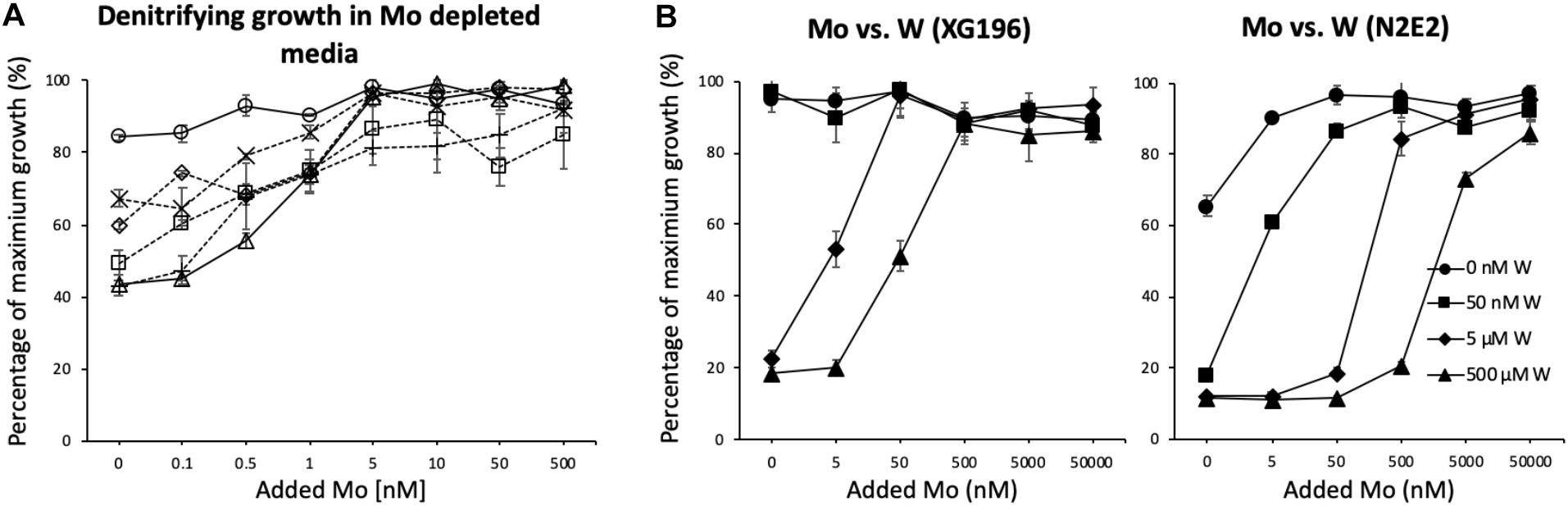
Figure 2. Strain XG196 is resistant to Mo limiting growth conditions. (A) Percentage of maximum growth (OD600) of ORR strains XG196 (Bacillus, circle), XG77 (Bacillus, cross), XG201 (Enterobacter, diamond), XG95 (Bacillus, square), XG146 (Ensifer, plus sign), and Pseudomonas fluorescens N2E2 (triangle) under nitrate-reducing conditions in Mo depleted media with the indicated concentration of added Mo. (B) Percentage of maximum growth of XG196 (left) and P. fluorescens N2E2 (right) under nitrate reducing conditions with tungstate at the indicated concentration competing with molybdate.
Tungstate is a competitive inhibitor of molybdate transport (Grunden and Shanmugam, 1997; Hu et al., 1997; Self et al., 2001). A Mo/W competitive growth analysis of isolate XG196 and P. fluorescens N2E2 under nitrate reducing conditions showed that low concentrations of W (up to 50 nM) do not affect the nitrate-dependent growth of XG196 but limits the growth of P. fluorescens N2E2 to only 20% of the maximum (Figure 2B). At higher W concentrations, W inhibits nitrate-dependent growth of both isolate XG196 and P. fluorescens N2E2. However, strain XG196 requires the addition of less Mo to resume maximal growth. For example, when 5 μM W was added to their media, XG196 only required 50 nM Mo to reach maximum growth, but P. fluorescens N2E2 required at least two orders of magnitude more Mo (5,000 nM; Figure 2B). Our hypothesis is that XG196 has a much higher affinity for molybdate than the other strains tested, especially that of strain N2E2. The environment from which strain N2E2 was isolated has much higher molybdate concentrations (approximately 10 nM) than the contaminated groundwater (Mo < 1 nM) (Smith et al., 2015; Thorgersen et al., 2015). A higher affinity for molybdate could give a growth advantage to XG196 by nitrate reduction under Mo-limited conditions.
Genomic and 16S rDNA Analysis of XG196
The draft genome of strain XG196 contained 6,010,169 bp in 55 contigs longer than 500 bp with a 38.35% G + C content. A total of 5721 coding sequences were predicted. The genome sequencing information from strain XG196 was submitted to the National Center for Biotechnology Information (NCBI) genome database and the accession number is JABWSY000000000. Nitrate reduction-related genes were annotated in the XG196 genome, including for nitrate reductase (napA and napB), copper-containing nitrite reductase (aniA) and nitrous-oxide reductase (nosZ), while the gene encoding nitric oxide reductase (nor) was missing. Some assimilatory nitrate reduction-related genes (nasC, nasD, and nasE) were also present in the genome. Genes encoding the molybdate ABC transport system (modA and modB) were also identified. The 16S rDNA sequence (1487 bp) of strain XG196 identified the organism as a member of the Bacillus genus. In order to characterize it at the species level, a total of 190 16S rDNA sequences, which include those of two XG196 close relatives, 186 Bacillus type strains and one out group strain, were used to build a phylogenetic tree using maximum likelihood by IQ-TREE (Supplementary Figure S3). Strain XG196 is closely related to B. niacini RB-113 (non-type, 99.176%), B. sp. LMG20241 (non-type, 99.663%), B. niacini IFO15566 (type, 99.193%), and B. drentensis LMG 21831 (type, 99.084%).
Phylogenetic Analysis of the Molybdate Binding Protein (ModA) of XG196
ModA is the molybdate-binding protein component of the molybdate ModABC transporter. We hypothesize that the ability of XG196 to grow by nitrate reduction using an extremely low concentration of Mo [<1 nM in contaminated groundwater close to S-3 ponds area (Ge et al., 2019)] is because its ModA has an unusually high affinity for molybdate. Phylogenetic analysis of XG196 ModA based on protein sequences of about 600 ATCC and DSM strains, including those from Archaea, Bacteria and Eukaryota, showed that it is, indeed, distinct from those of the ORR isolate P. fluorescens N2E2 (N2E2) and of E. coli K12 (Figure 3). The same conclusion was reached by a similar analysis using over 4,000 strains (Supplementary Figure S4). Most ModA proteins in proximity to XG196 ModA on the phylogenetic tree originate from other Bacillus strains, most of which were also isolated from soil, but their sequence identities are only about 50% (Supplementary Table S4). The two closest relatives of XG196 ModA are from Rhodococcus qingshengii (entry: A0A4R6A6K9, 85.9% identity) and Bacillus sp. 7884-1 (entry: A0A268JZS1, 85.9% identity) by UniProt BLAST (Figure 4 and Supplementary Table S4).
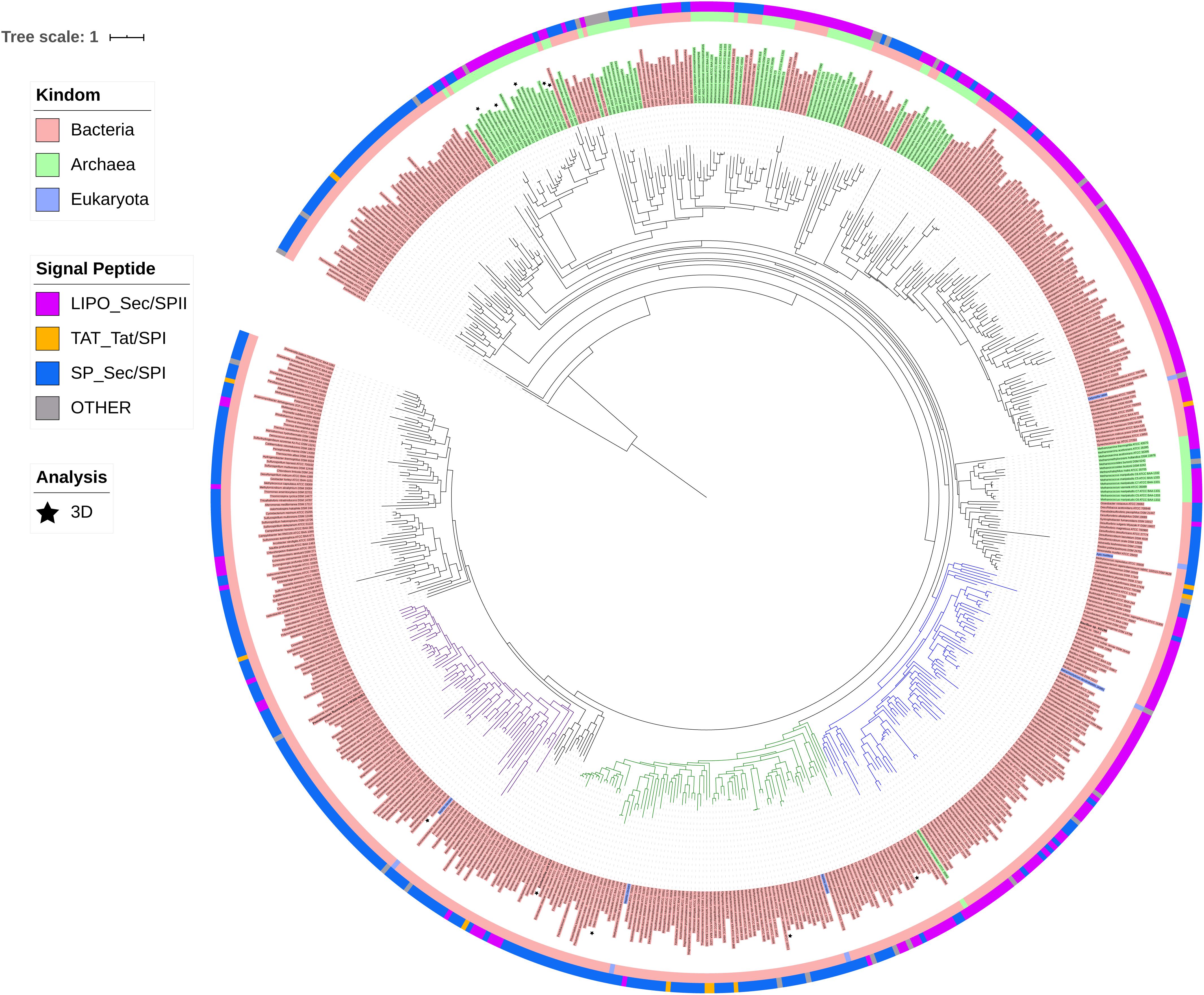
Figure 3. Phylogenetic analysis of ModA. Rooted phylogenetic tree of ModA and WtpA from Bacteria (pink, inner circle), Archaea (green, inner circle), and Eukaryota (blue, inner circle). Outer circle indicates different signal peptide type (LIPO_Sec/SPII in purple, TAT-Tat/SPI in yellow, SP_Sec/SPI in blue and other in gray). ModA proteins with PDB 3D structure data were indicated as black stars. Clades where XG196 ModA (blue clade), N2E2 ModA (purple clade), and E. coli ModA (green clade) belong to were also labeled in different colors.
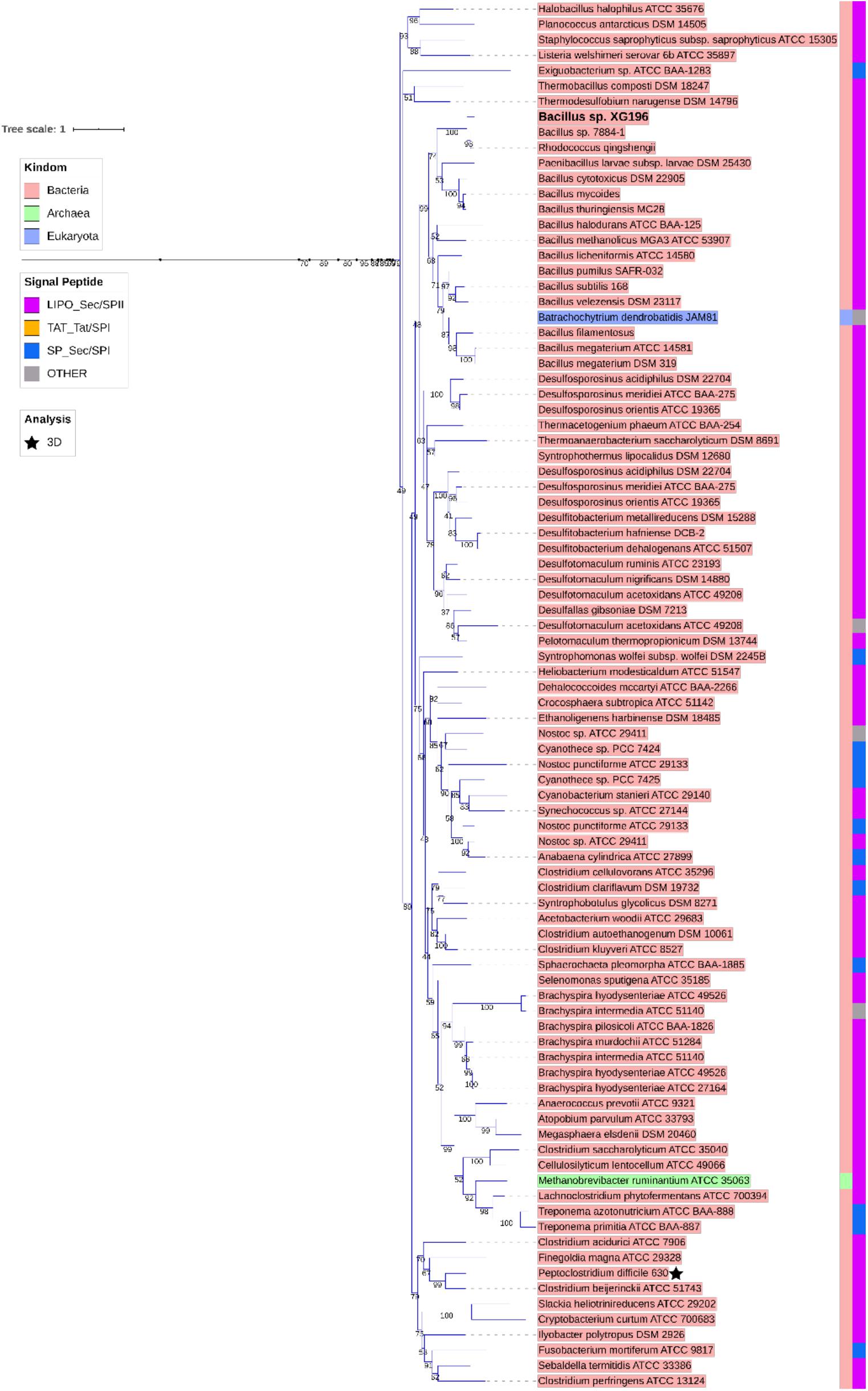
Figure 4. Zoomed-in clade of XG196 ModA and its close relatives. Bootstrap data are shown in the middle of each branch.
The structures of the ModA proteins of Peptoclostridium difficile 630, also known as Clostridium difficile 630 (PDB: 4KD5), X. axonopodis pv. citri 306 (PDB: 2H5Y, ), E. coli K12 (PDB: 1AMF, ), Vibrio cholerae serotype O1 ATCC 39315 (PDB:4RXL, ), and Azotobacter vinelandii (PDB: 1ATG, ) have been determined (Figures 3, 5) (Hu et al., 1997; Lawson et al., 1998; Santacruz et al., 2006). Each binds a single molybdate (or tungstate) ion. In addition, some archaea are able to utilize tungsten, a metal seldom used in biology, in their pyranopterin-containing enzymes (other than Mo-dependent nitrate reductase) (Cabello et al., 2004; Bevers et al., 2006). These tungsten-utilizing microorganisms take up tungstate using a transporter (WtpA) that is highly homologous to ModA (Supplementary Figure S5), and the structures of WtpA from Methanosarcina acetivorans ATCC 35395 (PDB: 3CFX, ), Methanocaldococcus jannaschii ATCC 43067 (PDB: 3CFZ, ), Pyrococcus furiosus ATCC 43587 (PDB: 3CG1, ), Archaeoglobus fulgidus ATCC 49558 (PDB: 3CIJ, ) and P. horikoshii ATCC 700860 (PDB: 3CG3, ) are known, all of which bind one tungstate ion (Supplementary Figure S5) (Hollenstein et al., 2009).
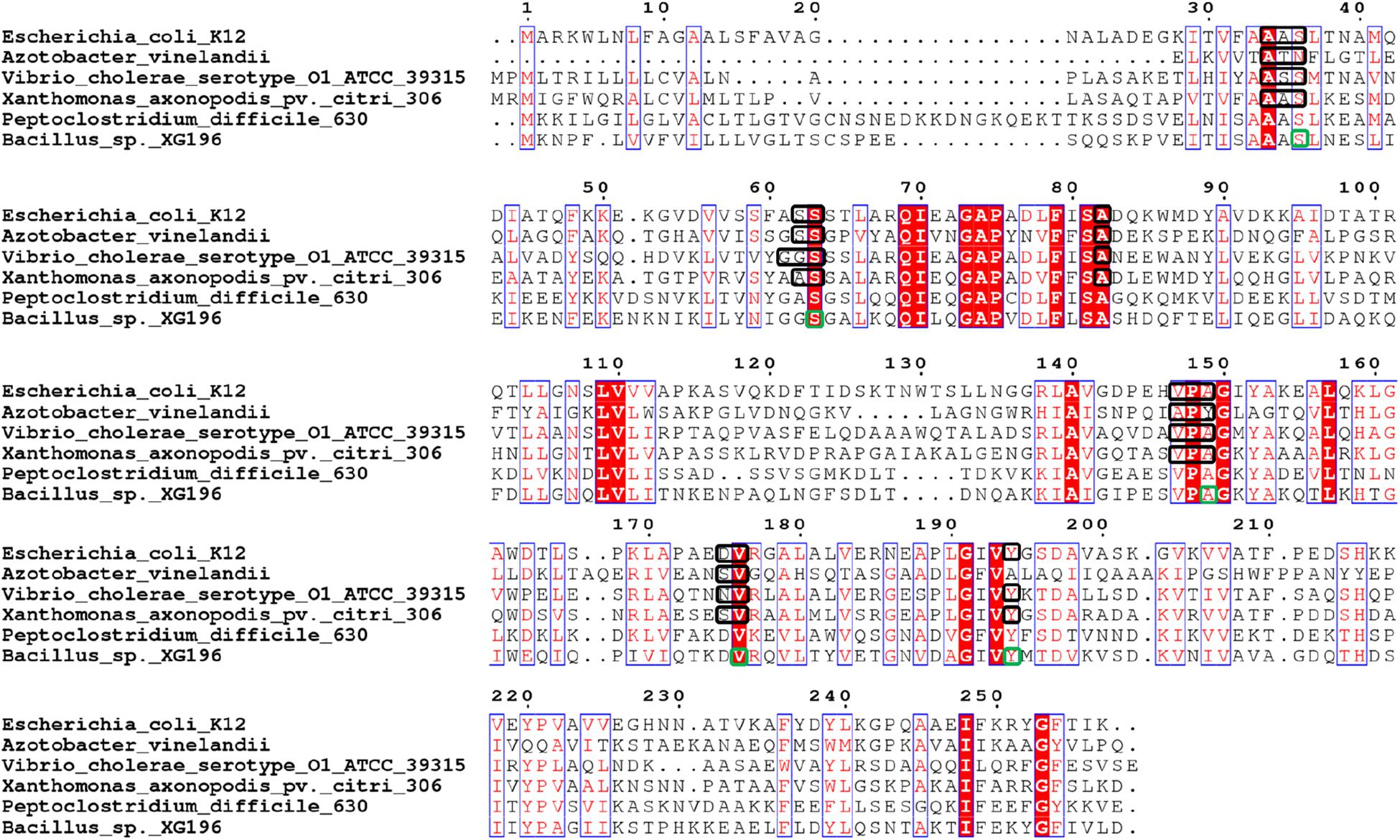
Figure 5. Multi-alignment analysis of Peptoclostridium difficile 630 (ModA, UniProt Entry: Q18A64, PDB: 4KD5), Xanthomonas axonopodis pv. citri 306 (ModA, UniProt Entry: Q8PHA1, PDB: 2H5Y, ), Escherichia coli K12 (ModA, UniProt Entry: P37329, PDB: 1AMF, ), Vibrio cholerae serotype O1 ATCC 39315 (ModA, UniProt Entry: Q9KLL7, PDB:4RXL, ), Azotobacter vinelandii (ModA, UniProt Entry: Q7SIH2, PDB: 1ATG, ) and Bacillus sp. XG196. Identical residues are in white and boxed in red, while similar residues are in red and boxed in blue. Molybdate/tungstate binding residues of ModA proteins resolved by crystal structures were indicated in black squares. Molybdate binding residues of XG196 ModA identified by protein modeling using X. axonopodis pv. citri 306 ModA (PDB: 2H5Y) are indicated in green squares.
XG196 ModA was modeled using ModA (PDB: 2H5Y) from X. axonopodis pv. citri 306 as the template, which has 37% sequence identity with XG196 ModA and contains molybdate as the ligand in the crystal structure. Based on the residues invoved in molybdate binding in X. axonopodis pv. citri 306 ModA, Ser36, Ser63, Ala149, Val176, and Tyr194 of XG196 ModA are predicted to directly bind molybdate via hydrogen bonds (Figure 5 and Supplementary Figure S6). However, from the modeling it is not clear why the XG196 protein has increased affinity for the metal. Although archaeal and bacterial WtpA/ModA proteins are evolutionally distant, the residues involved in metal binding are partially conserved, suggesting a similar ligand binding mechanism (Figure 3 and Supplementary Figure S5). Multi-alignment analysis of the ModA proteins from E. coli K12, XG196, two close relatives of XG196 ModA (from Rhodococcus qingshengii and Bacillus sp. 7884-1), other Bacillus ModA proteins from phylogenetic analysis (Figure 3 and Supplementary Table S4) and of EB-106 isolate XG77, isolated from sediments of similar depth with isolate XG196 (Figures 1A,B), are shown in Supplementary Figure S7. The mean sequence identity and similarity of these ModA sequences is about 12.2 and 65.4%, respectively. The sizes of these ModA proteins are similar (about 250 residues) and their sequences are conserved at 11 out of 12 the molybdate binding residues found in E. coli K12 ModA (Ala34, Ala35, Ser36, Ser63, Ala82, Val147, Pro148, Ala149, Asp175, Val176, and Tyr/Phe194), the exception being position Ser/Gly/Ala62. It seems that ModA proteins are quite similar, particularly XG196 ModA and other Bacillus ModA proteins, and novel attributes of the XG196 protein are not obvious, especially in the deduced oxyanion binding site.
ITC Analysis of ModA Proteins
To determine their molybdate-binding properties, the genes encoding the ModA proteins from XG196, N2E2 and E. coli were expressed in, and the recombinant proteins were purified from, E. coli. ICP-MS analysis showed that XG196 ModA(40 μM) can naturally bind about 67 nM of Mo even when trace grade chemicals were used, higher than what N2E2 ModA (15 nM) and E. coli ModA (9 nM) can bind (Supplementary Figure S8). After dialysis in low salt ITC buffer, all ModA proteins can pick up a little bit more Mo from the ITC buffer (XG196 ModA to 82 nM, N2E2 ModA 19 nM, and E. coli ModA 10 nM). On average, XG196 ModA, N2E2 ModA and E. coli ModA bound 0.002, 0.0005, and 0.0003 of molybdate per protein, respectively, which are far away from being saturated. ITC analysis showed that these proteins contain a single binding site for molybdate (values were 1.10 ± 0.01, 0.95 ± 0.08, and 0.92 ± 0.01, respectively). However, the molybdate binding curves showed that XG196 ModA had a KD value for molybdate of 2.21 ± 1.03 nM, which is about one order of magnitude lower than those of N2E2 (27.0 ± 6.2 nM) and E. coli (25.01 ± 3.7 nM) ModA (Table 1 and Supplementary Figure S9). Hence, XG196 ModA has a much higher affinity for molybdate, consistent with results from the physiological study showing that XG196 is able to grow by nitrate reduction using Mo concentrations (<1 nM) that limit the growth of other bacteria, including N2E2. The tungstate-binding affinity of XG196 ModA was about fivefold higher than that for molybdate (KD 11.15 ± 1.34 nM), and about half of the tungstate dissociation constent values for the ModA proteins of N2E2 and E. coli (26.6 ± 2.0 and 23.7 ± 0.6 nM, respectively; see Table 1 and Supplementary Figure S9). The stoichiometry of tungstate binding to each of these proteins was also 1:1, as found for molybdate. Hence, the lower binding affinity for molybdate than tungstate of XG196 ModA is consistent with the better growth of the organism under nitrate-reducing conditions than N2E2 when tungstate is present (Figure 2B).
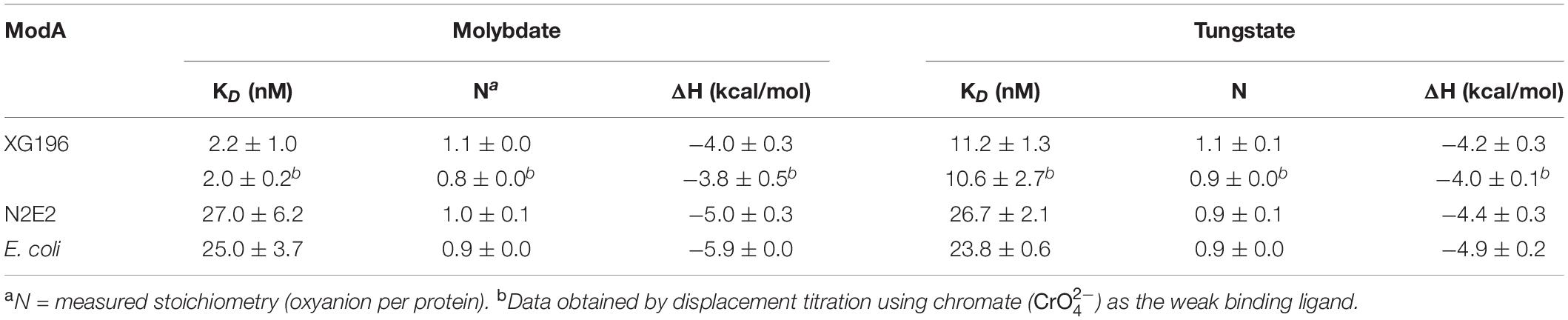
Table 1. Molybdate and tungstate binding properties of ModA proteins determined by isothermal titration calorimetry and displacement titration.
The KD value of XG196 ModA for molybdate was extremely low (≤2 nM), which is only just within the confidence range of the direct ITC approach. Hence, another approach known as displacement titration analysis was used (Sigurskjold, 2000; Krainer and Keller, 2015). Chromate () was use as the weak binding ligand. ITC analysis of XG196 ModA binding chromate gave the following results: KD = 1.56 ± 0.05 μM, N = 0.83 ± 0.09, and ΔH = −1.71 ± 0.26 kcal/mol. XG196 ModA saturated with chromate was then titrated with molybdate or tungstate. The results for molybdate (KD = 2.04 ± 0.19, N = 0.84 ± 0.02, ΔH = −3.76 ± 0.49 kcal/mol) and tungstate (KD = 10.6 ± 2.6, N = 0.87 ± 0.02, ΔH = −4.01 ± 0.49 kcal/mol), are similar to those obtained using ITC analysis of direct molybdate or tungstate titrations (Table 1).
Gene Abundance of Mo-Related Proteins in ORR Groundwater
To better understand the utilization of Mo in the ORR environment, the abundances of ModA genes and genes encoding representative proteins from the four families of Mo proteins were analyzed in ORR groundwater samples from both contaminated and background wells. As shown in Table 2, the abundance of Mo-related genes are generally higher in ORR contaminated groundwater samples. In particular, the abundance of modA (encoding ModA) and napA/narG (encoding dissimilatory nitrate reductase Mo-containing subunit) are significantly higher in contaminated well FW021, FW104 and FW106 (modA 27 to 39.9 copies per 108 reads, napA 11.2 to 32.5 copies per 108 reads, and narG 26.4 to 42.6 copies per 108 reads) than in background well FW300, FW301, and FW305 (modA 4.1 to 27 copies per 108 reads, napA 1.6 to 8.9 copies per 108 reads, and narG 1.8 to 10.5 copies per 108 reads). In contrast, the abundance of nasA (encoding assimilatory nitrate reductase Mo-containing subunit), dmsA (encoding DMSO reductase Mo-containing subunit), xdhB (xanthine oxidase/dehydrogenase) and sorA (encoding sulfite oxidoreductase Mo-containing subunit) were only slightly higher in contaminated wells, while the abundance of nifK (encoding nitrogenase) is similar in both contaminated and background wells (Table 2). The higher abundance of modA, napA, and narG relative to other Mo-related protein genes in the contaminated wells is likely an adaptive advantage given the high nitrate concentrations (0.02–13.3 mM), which are about 1000-fold higher than in the background wells (0.1–1.8 μM).
Discussion
The ORR S-3 ponds contamination plume is unique as it contains high concentrations of nitrate (up to 230 mM in groundwater) and various metals (Cd, Ni, Cu, Co, Mn, U, etc.) at low pH (∼3) (Brooks, 2001; Revil et al., 2013). Yet, we previously showed that in this unique environment, Mo is generally limiting for microbial nitrate reduction (Thorgersen et al., 2015). Previous studies have revealed that complex microbial communities survive in this contaminated site (Abulencia et al., 2006; Vishnivetskaya et al., 2011). The overall goal of this research was to elucidate the molecular mechanisms that give certain microorganisms competitive advantages in these extreme habitats. Bacillus strain XG196 was isolated from contaminated core EB-106 that was drilled adjacent to the origin of the contamination (the S-3 ponds). XG196 was shown to grow by nitrate reduction in the presence of an exceedingly low concentration of Mo that contaminated its defined medium from the inoculum and the chemicals that make the media (to which no Mo was added). The ability to grow with limited Mo appears to be due to its molybdate-binding protein, ModA, which has a very high affinity for molybdate (KD ∼ 2 nM). This is the lowest KD value yet reported for any ModA to date and it is also the first ModA characterized from a Bacillus strain. Previous studies have typically reported molybdate affinities with ModA proteins that are more than an order of magnitude lower (Corcuera et al., 1993; Bevers et al., 2006; Smart et al., 2009; Aryal et al., 2012). A similarly high affinity but for tungstate was reported for the Wtp protein of W-dependent P. furiosus, a member of the archaea domain. Its binding affinity of molybdate is about fivefold lower (KD = 11 ± 5 nM) than that found here for XG196 ModA (Bevers et al., 2006).
The molybdate binding affinity of E. coli ModA measured in this study (∼25 nM) is consistent with what has been reported by others (KD = 20–26 nM; Corcuera et al., 1993; Imperial et al., 1998). The KD value for molybdate of N2E2 ModA is about 27 nM, consistent with the poor nitrate-reducing growth observed in Mo-limited media compared to XG196. In addition, the ModA proteins of E. coli and N2E2 have very similar KD values for both molybdate and tungstate, hence, neither protein is able to distinguish between these two oxyanions, consistent with what has been reported for E coli ModA (Rech et al., 1996; Imperial et al., 1998). In contrast, XG196 ModA has a fivefold higher affinity for molybdate compared to tungstate, which could give the organism a selective advantage in scavenging molybdate for growth in the presence of tungstate as seen in the Mo/W competition growth studies herein (Figure 2).
Phylogenetic analysis showed that XG196 ModA is distinct from previously described ModA proteins, including that of E. coli K12 (Corcuera et al., 1993; Aryal et al., 2012), the WtpA/ModA proteins from the bacterium Azotobacter vinelandii (Lawson et al., 1998), and the archaea P. horikoshii (Hollenstein et al., 2009) and P. furiosus (Bevers et al., 2006). Multi-alignment analysis indicates that XG196 ModA is quite similar to those of other Bacillus species based on their sequence (65.4% mean similarity) and their deduced oxyanion binding sites (Supplementary Figure S7). However, it is hard to conclude that all of these Bacillus ModA proteins have molybdate affinities as high as that of XG196 ModA since the molybdate-binding residues are highly conserved. Unfortunately, modeling of XG196 ModA (Supplementary Figure S6) did not shed light on why it has a much higher affinity for molybdate than structurally-characterized proteins. ModA proteins contain a signal peptide at the N terminus that enables the protein to be transported across the membrane. ModA signal peptides fall into one of four different groups: Sec/SPI, Sec/SPII, Tat/SPI, and other (Nielsen et al., 2019). Surprisingly, the ModA from XG196 grouped with the ModA proteins from archaea, and these are all predicted to be lipopeptides and belong to the Sec/SPII group, while N2E2 and E. coli ModA proteins belong to the Sec/SPI group with non-lipopeptides (Figure 3). Substrate-binding lipoproteins are widely observed in gram-positive bacteria (Sutcliffe and Russell, 1995; Hutchings et al., 2009). It is believed that the lipopeptides can tether substrate-binding proteins in order to prevent their loss into the growth environment because of the absence of the retentive outer membrane in gram-positive bacteria (Sutcliffe and Russell, 1995). At present, not enough information is available to distinguish “high” affinity molybdate transporters (like XG196 ModA) from “low” affinity ones (like those of N2E2 and E. coli ModA) based only on sequence similarity or the deduced molybdate-binding residues. Structural determinations of high affinity ModA proteins in addition to that of XG196 will be required to elucidate the molecular basis as to why these particular proteins bind molybdate so tightly.
Strain XG196 exhibited higher nitrate reductase specific activity than the other EB-106 strains XG95, XG146, and XG201 (Supplementary Figure S1) and accumulated more Mo in its cytoplasm than XG77, XG95, XG146, and XG201 (Supplementary Figure S10). This could be the result of the higher molybdate affinity of its ModA, which must provide more than sufficient Mo for the biosynthesis of functional pyranopterin cofactor in nitrate reductase (Schwarz and Mendel, 2006) when Mo is limited in the environment. XG196 also accumulated the second highest concentration of Mo in the membrane fractions compared to the other EB-106 strains, which might be the result of a high nitrate reductase concentration in the membrane because of more than sufficient Mo taken up from environment. However, these results might not be directly related to the high affinity of ModA for molybdate. Nitrate reductases with high specific activities or high affinity molybdate storage proteins described in previous studies (Pienkos and Brill, 1981; Grunden and Shanmugam, 1997) could also contribute to XG196 being able to grow robustly under nitrate reducing conditions with limited Mo. Further study is required to clarify this issue.
Mo is removed from groundwater in the ORR contaminated area but not from the non-contaminated area as a result of Fe and Al precipitation (Ge et al., 2019). The low Mo concentrations (picomolar range) in the ORR contaminated environment is unusual but not unique. Low Mo concentrations (5–70 nM) occur in naturally-acidic groundwater (pH 2.4 to 2.9) (Nordstrom, 2015), in an acid mine drainage (<10 nM) (Sánchez-España et al., 2016), in harbors (<20 nM) as a result of sedimentary processes (Morford et al., 2007), and in various aquifers, including the Yorkshire Chalk aquifer (<10 nM) due to co-precipitation with or adsorption to sulfide minerals under strong reducing conditions (Smedley et al., 2014). These environments have significantly lower Mo concentrations than most freshwater and open seawater systems, which are typically > 300 nM (Smedley and Kinniburgh, 2017). Limiting Mo concentrations in natural water systems could lead to other environmental problems, for example, by affecting critical steps in the nitrogen cycle, such as nitrate reduction, leading to nitrate accumulation or to slowing down of nitrate removal from contaminated water or soil systems.
There are several factors that affect nitrate reduction in the ORR contaminated environment besides lack of the essential metal Mo. These include the acidic conditions, high nitrate concentrations, the presence of heavy metal contaminants, and limited availability of carbon sources to serve as electron donors for nitrate reduction (Smith et al., 2015; Thorgersen et al., 2015; Ge et al., 2019). Other factors, such as O2 concentrations in the soil and groundwater (Zumft, 1997; Qu et al., 2016), temperature and denitrifier community composition (Wallenstein et al., 2006), can also affect the efficiency of nitrate reduction. Meanwhile, the higher abundance of genes encoding the molybdate transport protein (modA) and assimilatory nitrate reductase Mo-containing subunits (napA/narG) in nitrate-contaminated wells indicates enhanced nitrate reduction in the ORR contaminated groundwater. The high abundance of modA could result in a greater uptake of molybdate into cells for the biosynthesis of dissimilatory nitrate reductase, enabling microorganisms to survive in the nitrate-contaminated and Mo-limited ORR environment. These numerous complex environmental factors make it difficult to study the relationships between nitrate reduction and natural microbial communities. There are therefore many unanswered questions at present that can be addressed in part by characterizing novel microbial stains with unique molecular mechanisms, as reported here for XG196 and its ModA protein. Such microorganisms could also be instrumental in developing novel methods to remove contaminating nitrate in complex waste environments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: “https://www.ncbi.nlm.nih.gov/nuccore/JABWSY000000000.”
Author Contributions
XG, MT, and FP designed this study, performed the experiments, analyzed and interpreted the data. MA directed the research. AD, J-MC, and PN carried out the genome sequencing. SG-D, LL, and TN performed ORR metagenome annotation and analysis. TH contributed to environmental sampling. XG wrote the manuscript. MT, FP, AD, J-MC, PN, SG-D, PA, AA, TH, and MA contributed to its revision. All authors contributed to the article and approved the submitted version.
Funding
This material by ENIGMA (Ecosystems and Networks Integrated with Genes and Molecular Assemblies) (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory, is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract number DE-AC02-05CH11231.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Kenneth Lowe and Dominique Joyner for their invaluable help in collecting sediment samples, John Rose for protein modeling analysis, Astrid Terry for project management, and Scott Pegan and Brendan Freitas for their assistance in ITC analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.587127/full#supplementary-material
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov
- ^ http://www.kbase.us
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://rdp.cme.msu.edu/
- ^ http://www.iqtree.org/
- ^ https://www.ebi.ac.uk/interpro/
- ^ http://weizhongli-lab.org/cd-hit/
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://www.rcsb.org/
- ^ http://espript.ibcp.fr/ESPript/ESPript/
- ^ https://swissmodel.expasy.org/
- ^ http://www.cgl.ucsf.edu/chimera
- ^ http://www.cbs.dtu.dk/services/SignalP-5.0/
- ^ https://jgi.doe.gov/data-and-tools/bbtools/
References
Abulencia, C. B., Wyborski, D. L., Garcia, J. A., Podar, M., Chen, W., Chang, S. H., et al. (2006). Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72, 3291–3301. doi: 10.1128/aem.72.5.3291-3301.2006
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569.
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Aryal, B. P., Brugarolas, P., and He, C. (2012). Binding of ReO4- with an engineered MoO42–binding protein: towards a new approach in radiopharmaceutical applications. J. Biol. Inorg. Chem. 17, 97–106. doi: 10.1007/s00775-011-0833-4
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barron, A. R., Wurzburger, N., Bellenger, J. P., Wright, S. J., Kraepiel, A. M., and Hedin, L. O. (2009). Molybdenum limitation of asymbiotic nitrogen fixation in tropical forest soils. Nat. Geosci. 2, 42–45. doi: 10.1038/ngeo366
Bevers, L. E., Hagedoorn, P.-L., Krijger, G. C., and Hagen, W. R. (2006). Tungsten transport protein A (WtpA) in Pyrococcus furiosus: the first member of a new class of tungstate and molybdate transporters. J. Bacteriol. 188, 6498–6505. doi: 10.1128/jb.00548-06
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brooks, S. C. (2001). Waste Characteristics of the Former S-3 Ponds and Outline of Uranium Chemistry Relevant to NABIR Field Research Center Studies. Oak Ridge, TN: Oak Ridge National Laboratory.
Cabello, P., Roldan, M. D., and Moreno-Vivian, C. (2004). Nitrate reduction and the nitrogen cycle in archaea. Microbiology 150, 3527–3546. doi: 10.1099/mic.0.27303-0
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., Mcgarrell, D. M., Sun, Y., et al. (2013). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642.
Corcuera, G. L., Bastidas, M., and Dubourdieu, M. (1993). Molybdenum uptake in Escherichia coli K12. Microbiology 139, 1869–1875. doi: 10.1099/00221287-139-8-1869
Diaz, R. J., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Filiatrault, M. J., Tombline, G., Wagner, V. E., Van Alst, N., Rumbaugh, K., Sokol, P., et al. (2013). Pseudomonas aeruginosa PA1006, which plays a role in molybdenum homeostasis, is required for nitrate utilization, biofilm formation, and virulence. PLoS One 8:e55594. doi: 10.1371/journal.pone.0055594
Ge, X., Vaccaro, B. J., Thorgersen, M. P., Poole, F. L., Majumder, E. L., Zane, G. M., et al. (2019). Iron- and aluminium-induced depletion of molybdenum in acidic environments impedes the nitrogen cycle. Environ. Microbiol. 21, 152–163. doi: 10.1111/1462-2920.14435
Glass, J. B., Axler, R. P., Chandra, S., and Goldman, C. R. (2012). Molybdenum limitation of microbial nitrogen assimilation in aquatic ecosystems and pure cultures. Front. Microbiol. 3:331. doi: 10.3389/fmicb.2012.00331
Gruber, N., and Galloway, J. N. (2008). An Earth-system perspective of the global nitrogen cycle. Nature 451, 293–296. doi: 10.1038/nature06592
Grunden, A. M., and Shanmugam, K. (1997). Molybdate transport and regulation in bacteria. Arch. Microbiol. 168, 345–354. doi: 10.1007/s002030050508
Hamlin, R. L. (2016). “Molybdenum,” in Handbook of Plant Nutrition, eds A. V. Barker and D. J. Pilbeam (Boca Raton, FL: CRC Press), 391–410.
Hille, R., Hall, J., and Basu, P. (2014). The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038. doi: 10.1021/cr400443z
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2017). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hollenstein, K., Comellas-Bigler, M., Bevers, L. E., Feiters, M. C., Meyer-Klaucke, W., Hagedoorn, P.-L., et al. (2009). Distorted octahedral coordination of tungstate in a subfamily of specific binding proteins. J. Biol. Inorg. Chem. 14, 663–672. doi: 10.1007/s00775-009-0479-7
Hu, Y., Rech, S., Gunsalus, R. P., and Rees, D. C. (1997). Crystal structure of the molybdate binding protein ModA. Nat. Struct. Biol. 4, 703–707. doi: 10.1038/nsb0997-703
Huang, Y., Niu, B., Gao, Y., Fu, L., and Li, W. (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. doi: 10.1093/bioinformatics/btq003
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., Von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Hutchings, M. I., Palmer, T., Harrington, D. J., and Sutcliffe, I. C. (2009). Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol. 17, 13–21. doi: 10.1016/j.tim.2008.10.001
Hyatt, D., Chen, G.-L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Imperial, J., Hadi, M., and Amy, N. K. (1998). Molybdate binding by ModA, the periplasmic component of the Escherichia coli mod molybdate transport system. Biochim. Biophys. Acta 1370, 337–346. doi: 10.1016/s0005-2736(98)00003-0
Kahm, M., Hasenbrink, G., Lichtenberg-Fraté, H., Ludwig, J., and Kschischo, M. (2010). grofit: fitting biological growth curves with R. J. Stat. Softw. 33:37740.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kellman, L., and Hillaire-Marcel, C. (2003). Evaluation of nitrogen isotopes as indicators of nitrate contamination sources in an agricultural watershed. Agric. Ecosyst. Environ. 95, 87–102. doi: 10.1016/s0167-8809(02)00168-8
Krainer, G., and Keller, S. (2015). Single-experiment displacement assay for quantifying high-affinity binding by isothermal titration calorimetry. Methods 76, 116–123. doi: 10.1016/j.ymeth.2014.10.034
Lancaster, W. A., Menon, A. L., Scott, I., Poole, F. L., Vaccaro, B. J., Thorgersen, M. P., et al. (2014). Metallomics of two microorganisms relevant to heavy metal bioremediation reveal fundamental differences in metal assimilation and utilization. Metallomics 6, 1004–1013. doi: 10.1039/c4mt00050a
Lawson, D. M., Williams, C. E., Mitchenall, L. A., and Pau, R. N. (1998). Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 Å resolution crystal structure of Azotobacter vinelandii ModA. Structure 6, 1529–1539. doi: 10.1016/s0969-2126(98)00151-8
MacGregor, C. H., Schnaitman, C. A., Normansell, D. E., and Hodgins, M. G. (1974). Purification and properties of nitrate reductase from Escherichia coli K12. J. Biol. Chem. 249, 5321–5327.
Madeira, F., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., Basutkar, P., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641.
Maia, L. B., Moura, I., and Moura, J. J. (2017). “Molybdenum and tungsten-containing enzymes: an overview,” in Molybdenum and Tungsten Enzymes, eds R. Hille, C. Schulzke, and M. L. Kirk (London: The Royal Society of Chemistry), 1–80. doi: 10.1039/9781782628828-00001
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2018). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360.
Moon, J.-W., Paradis, C. J., Joyner, D. C., Von Netzer, F., Majumder, E. L., Dixon, E. R., et al. (2020). Characterization of subsurface media from locations up-and down-gradient of a uranium-contaminated aquifer. Chemosphere 255:126951. doi: 10.1016/j.chemosphere.2020.126951
Morford, J. L., Martin, W. R., Kalnejais, L. H., François, R., Bothner, M., and Karle, I.-M. (2007). Insights on geochemical cycling of U, Re and Mo from seasonal sampling in Boston Harbor, Massachusetts, USA. Geochim. Cosmochim. Acta 71, 895–917. doi: 10.1016/j.gca.2006.10.016
Moura, J. J., Brondino, C. D., Trincão, J., and Romao, M. J. (2004). Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J. Biol. Inorg. Chem. 9, 791–799. doi: 10.1007/s00775-004-0573-9
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nielsen, H., Tsirigos, K. D., Brunak, S., and Von Heijne, G. (2019). A brief history of protein sorting prediction. Protein J. 38, 200–216. doi: 10.1007/s10930-019-09838-3
Nolan, B. T., Ruddy, B. C., Hitt, K. J., and Helsel, D. R. (1998). A national look at nitrate contamination of ground water. Water Cond. Purif. 39, 76–79.
Nordstrom, D. K. (2015). Baseline and premining geochemical characterization of mined sites. Appl. Geochem. 57, 17–34. doi: 10.1016/j.apgeochem.2014.12.010
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Pienkos, P. T., and Brill, W. (1981). Molybdenum accumulation and storage in Klebsiella pneumoniae and Azotobacter vinelandii. J. Bacteriol. 145, 743–751. doi: 10.1128/jb.145.2.743-751.1981
Powlson, D. S., Addiscott, T. M., Benjamin, N., Cassman, K. G., De Kok, T. M., Van Grinsven, H., et al. (2008). When does nitrate become a risk for humans? J. Environ. Qual. 37, 291–295. doi: 10.2134/jeq2007.0177
Qu, Z., Bakken, L. R., Molstad, L., Frostegård, Å., and Bergaust, L. L. (2016). Transcriptional and metabolic regulation of denitrification in Paracoccus denitrificans allows low but significant activity of nitrous oxide reductase under oxic conditions. Environ. Microbiol. 18, 2951–2963. doi: 10.1111/1462-2920.13128
Rech, S., Wolin, C., and Gunsalus, R. P. (1996). Properties of the periplasmic ModA molybdate-binding protein of Escherichia coli. J. Biol. Chem. 271, 2557–2562. doi: 10.1074/jbc.271.5.2557
Revil, A., Skold, M., Karaoulis, M., Schmutz, M., Hubbard, S. S., Mehlhorn, T. L., et al. (2013). Hydrogeophysical investigations of the former S-3 ponds contaminant plumes, oak ridge integrated field research challenge site, Tennessee. Geophysics 78, 29–41.
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324.
Sánchez-España, J., Yusta, I., Gray, J., and Burgos, W. D. (2016). Geochemistry of dissolved aluminum at low pH: extent and significance of Al–Fe (III) coprecipitation below pH 4.0. Geochim. Cosmochim. Acta 175, 128–149. doi: 10.1016/j.gca.2015.10.035
Santacruz, C., Balan, A., Ferreira, L. C. D. S., and Barbosa, J. (2006). Crystallization, data collection and phasing of the molybdate-binding protein of the phytopathogen Xanthomonas axonopodis pv. citri. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commoun. 62, 289–291.
Schwarz, G. (2016). Molybdenum cofactor and human disease. Curr. Opin. Chem. Biol. 31, 179–187. doi: 10.1016/j.cbpa.2016.03.016
Schwarz, G., and Mendel, R. R. (2006). Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 57, 623–647. doi: 10.1146/annurev.arplant.57.032905.105437
Scott, I. M., Rubinstein, G. M., Lipscomb, G. L., Basen, M., Schut, G. J., Rhaesa, A. M., et al. (2015). A new class of tungsten-containing oxidoreductase in Caldicellulosiruptor, a genus of plant biomass-degrading thermophilic bacteria. Appl. Environ. Microbiol. 81, 7339–7347. doi: 10.1128/aem.01634-15
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Self, W. T., Grunden, A. M., Hasona, A., and Shanmugam, K. T. (2001). Molybdate transport. Res. Microbiol. 152, 311–321.
Sigurskjold, B. W. (2000). Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal. Biochem. 277, 260–266. doi: 10.1006/abio.1999.4402
Smart, J. P., Cliff, M. J., and Kelly, D. J. (2009). A role for tungsten in the biology of Campylobacter jejuni: tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter. Mol. Microbiol. 74, 742–757. doi: 10.1111/j.1365-2958.2009.06902.x
Smedley, P., Cooper, D., Ander, E., Milne, C., and Lapworth, D. (2014). Occurrence of molybdenum in British surface water and groundwater: distributions, controls and implications for water supply. Appl. Geochem. 40, 144–154. doi: 10.1016/j.apgeochem.2013.03.014
Smedley, P. L., and Kinniburgh, D. G. (2017). Molybdenum in natural waters: a review of occurrence, distributions and controls. Appl. Geochem. 84, 387–432. doi: 10.1016/j.apgeochem.2017.05.008
Smith, M. B., Rocha, A. M., Smillie, C. S., Olesen, S. W., Paradis, C., Wu, L., et al. (2015). Natural bacterial communities serve as quantitative geochemical biosensors. mBio 6:e00326-15.
Spalding, R. F., and Exner, M. E. (1993). Occurrence of nitrate in groundwater—a review. J. Environ. Qual. 22, 392–402. doi: 10.2134/jeq1993.00472425002200030002x
Sutcliffe, I. C., and Russell, R. (1995). Lipoproteins of gram-positive bacteria. J. Bacteriol. 177, 1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995
Thorgersen, M. P., Ge, X., Poole, F. L., Price, M. N., Arkin, A. P., and Adams, M. W. (2019). Nitrate-utilizing microorganisms resistant to multiple metals from the heavily contaminated Oak Ridge Reservation. Appl. Environ. Microbiol. 85:e00896-19.
Thorgersen, M. P., Lancaster, W. A., Vaccaro, B. J., Poole, F. L., Rocha, A. M., Mehlhorn, T., et al. (2015). Molybdenum availability is key to nitrate removal in contaminated groundwater environments. Appl. Environ. Microbiol. 81, 4976–4983. doi: 10.1128/aem.00917-15
Tian, R., Ning, D., He, Z., Zhang, P., Spencer, S. J., Gao, S., et al. (2020). Small and mighty: adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 8:51.
Vishnivetskaya, T. A., Mosher, J. J., Palumbo, A. V., Yang, Z. K., Podar, M., Brown, S. D., et al. (2011). Mercury and other heavy metals influence bacterial community structure in contaminated Tennessee streams. Appl. Environ. Microbiol. 77, 302–311. doi: 10.1128/aem.01715-10
Wallenstein, M. D., Myrold, D. D., Firestone, M., and Voytek, M. (2006). Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16, 2143–2152. doi: 10.1890/1051-0761(2006)016[2143:ecodca]2.0.co;2
Widdel, F., and Bak, F. (1992). “Gram-negative mesophilic sulfate-reducing bacteria,” in The Prokaryotes, eds A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (New York, NY: Springer), 3352–3378. doi: 10.1007/978-1-4757-2191-1_21
Zhang, Q., Sun, J., Liu, J., Huang, G., Lu, C., and Zhang, Y. (2015). Driving mechanism and sources of groundwater nitrate contamination in the rapidly urbanized region of south China. J. Contam. Hydrol. 182, 221–230. doi: 10.1016/j.jconhyd.2015.09.009
Zhang, Y., and Gladyshev, V. N. (2008). Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 379, 881–899. doi: 10.1016/j.jmb.2008.03.051
Keywords: Bacillus sp. XG196, nitrate, nitrate reductase, molybdenum limitation, molybdate transport
Citation: Ge X, Thorgersen MP, Poole FL II, Deutschbauer AM, Chandonia J-M, Novichkov PS, Gushgari-Doyle S, Lui LM, Nielsen T, Chakraborty R, Adams PD, Arkin AP, Hazen TC and Adams MWW (2020) Characterization of a Metal-Resistant Bacillus Strain With a High Molybdate Affinity ModA From Contaminated Sediments at the Oak Ridge Reservation. Front. Microbiol. 11:587127. doi: 10.3389/fmicb.2020.587127
Received: 24 July 2020; Accepted: 22 September 2020;
Published: 19 October 2020.
Edited by:
Partha Basu, Indiana University, Purdue University Indianapolis, United StatesReviewed by:
Ulrike Kappler, The University of Queensland, AustraliaOded Lewinson, Technion Israel Institute of Technology, Israel
Copyright © 2020 Ge, Thorgersen, Poole, Deutschbauer, Chandonia, Novichkov, Gushgari-Doyle, Lui, Nielsen, Chakraborty, Adams, Arkin, Hazen and Adams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael W. W. Adams, YWRhbXNtQHVnYS5lZHU=
 Xiaoxuan Ge
Xiaoxuan Ge Michael P. Thorgersen
Michael P. Thorgersen Farris L. Poole II1
Farris L. Poole II1 Adam M. Deutschbauer
Adam M. Deutschbauer Sara Gushgari-Doyle
Sara Gushgari-Doyle Lauren M. Lui
Lauren M. Lui Romy Chakraborty
Romy Chakraborty Adam P. Arkin
Adam P. Arkin Terry C. Hazen
Terry C. Hazen Michael W. W. Adams
Michael W. W. Adams