
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 November 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.586764
This article is part of the Research Topic New Insights into the Transmission Dynamics and Control of Antimicrobial Resistance to Last-resort Antibiotics View all 22 articles
 Dairong Wang1,2†
Dairong Wang1,2† Xinli Mu1,3†
Xinli Mu1,3† Ying Chen1,3
Ying Chen1,3 Dongdong Zhao1,3
Dongdong Zhao1,3 Ying Fu3,4
Ying Fu3,4 Yan Jiang1,3
Yan Jiang1,3 Yiwei Zhu1,3
Yiwei Zhu1,3 Jingjing Quan1,3
Jingjing Quan1,3 Xiaoting Hua1,3
Xiaoting Hua1,3 Guofeng Mao5
Guofeng Mao5 Xi Li6*
Xi Li6* Yunsong Yu1,3*
Yunsong Yu1,3*Objectives: Bacteria carrying the Klebsiella pneumoniae carbapenemase genes have rapidly spread worldwide and have become a great threat to public health. The blaKPC–2 gene has been primarily located on plasmids cocirculating in various strains. However, chromosomal integration of the blaKPC–2 gene in Escherichia coli has not been reported. In the present study, we report the detection of the first clinical strain of E. coli ST131 with a blaKPC–2 gene, which integrated in the chromosome. E. coli strain EC3385 was identified and subjected to susceptibility testing and genotyping. The complete genome sequences of this strain and four Proteus mirabilis strains were obtained. Chromosomal integration of the blaKPC–2 gene was confirmed using a combination of short- and long-read sequencing. Comparative genetic analyses were performed and the origin of the chromosomal location of the blaKPC–2 gene was further analyzed. Whole-genome sequencing revealed that strain EC3385 belonged to the ST131 type and possessed various resistance and virulence genes. Sequence analysis showed that the blaKPC–2 gene was carried in a 24-kb insertion sequence on the chromosome. This insertion sequence possessed high sequence similarity to previously reported blaKPC–2-habouring plasmids of P. mirabilis in China. To the best of our knowledge, this is the first report of a clinical ST131 E. coli strain carrying blaKPC–2 on the chromosome. The blaKPC–2 gene was probably horizontally transferred from the P. mirabilis plasmid to the E. coli chromosome by the IS26 element, indicating that P. mirabilis might be an important reservoir of blaKPC–2 gene for E. coli. Furthermore, the E. coli ST131 strain carrying the chromosomal blaKPC–2 gene could be further spread due to its carbapenem resistance and high virulence. It is imperative to perform active surveillance to prevent further dissemination of KPC-2 type carbapenemase-producing isolates.
Bacteria carrying the Klebsiella pneumoniae carbapenemase genes (blaKPC) have rapidly spread worldwide and have become a great threat to public health because these bacteria are often associated with high morbidity and mortality (Wang et al., 2016; An et al., 2018). KPC-2 is the main type of KPC carbapenemase and is most common in K. pneumoniae bacteria. In China, clonal spreading is a main mode of transfer of KPC-2 type carbapenemase-producing K. pneumonia. Our previous research demonstrated that multilocus sequence type 11 (ST11) originated from a successful lineage of KPC-2 type carbapenemase-producing K. pneumonia in China (Qi et al., 2011).
In contrast to K. pneumoniae, E. coli strains have rarely been reported to carry the blaKPC–2 gene. However, recent reports found that the number of E. coli strains carrying the blaKPC–2 gene has increased. In addition, unlike K. pneumoniae, clonal spread has not been found for the blaKPC–2 gene of E. coli (Chen et al., 2014). These strains also have different clone types, such as ST131, ST410, ST2281, ST43, ST721, ST4385, and ST8 (Kim et al., 2012; Mavroidi et al., 2012; Tian et al., 2020). Notably, among these clone types, E. coli ST131, an international multidrug-resistant high-risk clone, has gained a further selective advantage as a result of acquiring carbapenem resistance (Rogers et al., 2011; Kim et al., 2012) and E. coli ST131 may become a successful lineage of KPC-2 type carbapenemase-producing E. coli.
In addition, K. pneumoniae carbapenemase genes have been primarily located on plasmids cocirculating with various strains (Nordmann et al., 2011). They are considered a major mechanism responsible for the dramatic increase in the prevalence of carbapenem-resistant Enterobacteriaceae isolates. Plasmid DNA can act as a temporary “lending library” allowing vital genes to survive various selective pressures (Harrison et al., 2015). Notably, in vitro data demonstrated that once a gene is incorporated into a chromosome, it is maintained through replication without being subject to selective pressures, and gene loss from bacterial populations is rare (Bergstrom et al., 2000; Bahl et al., 2009; Carraro et al., 2015). Interestingly, the earliest observed chromosomal blaKPC gene integration events have been sporadic in gram-negative bacteria, such as Pseudomonas aeruginosa in 2006 (Villegas et al., 2007), Raoultella spp. in 2008 (Castanheira et al., 2009) and Acinetobacter baumannii in 2009 (Martínez et al., 2014). Recently, chromosomal integration has been described in four K. pneumoniae ST258 isolates (Conlan et al., 2014; Chen et al., 2015; Mathers et al., 2017). However, blaKPC–2 gene chromosomal integration events in E. coli have not been reported.
In the present study, we report the detection of the first clinical strain of E. coli ST131 with a chromosomal blaKPC–2 gene integrated in the chromosome. In addition, the genetic origin of this gene was further analyzed using whole-genome sequencing.
A patient was admitted to the hospital for a craniocerebral infarction in 2017. A carbapenem-resistant strain of E. coli EC3385 was isolated from sputum because the patient developed hospital-acquired pneumonia (HAP) secondary to postoperative intubation during the hospitalization. In addition, four P. mirabilis strains isolated at the same period (Table 1) as E. coli EC3385 in the ICU department were analyzed retrospectively. These strains were preliminary identified by the VITEK 2 system (Sysmex-bioMérieux, Marcy l’Etoile, France) and further confirmed by 16S rRNA sequencing.
Antibiotic susceptibility was determined using the VITEK 2 system and broth microdilution method and the results were interpreted according to the Clinical and Laboratory Standard Institute (CLSI) guidelines (CLSI, 2017) except for tigecycline and colistin, which were interpreted according to the European Committee on Antimicrobial Susceptibility Testing breakpoints for Enterobacteriaceae1.
Total genomic DNA extraction and analysis were performed as previously described (Li et al., 2018). Briefly, E. coli strain EC3385 and four P. mirabilis strains were cultured to mid-logarithmic phase in 50 ml of MH medium at 37°C. The genomic DNA of these strains was extracted using a QIAamp DNA MiniKit (Qiagen, Valencia, CA, United States) following the manufacturer’s recommendations. The DNA library was prepared using a Nextera XT DNA library preparation kit (Illumina, Inc., Cambridge, United Kingdom), and genomic DNA was sequenced on an Illumina HiSeq 4000 instrument with a 150-bp paired-end approach at a depth of approximately 200 ×. The raw reads of the strains were assembled into draft genomes using the CLC Genomics Workbench 10.0.
In addition, E. coli EC3385 strain sequencing was further performed via a single molecule real-time (SMRT) technique using a PacBio RS II platform and the resulting sequences were assembled de novo using the hierarchical genome assembly process (HGAP) with the default settings of the SMRT Analysis v2.3.0 software package (Shen et al., 2017).
The Rapid Annotation using Subsystems Technology (RAST) annotation website server2 was used to annotate the genomes. Multi-locus sequence typing (MLST) of resistance genes and the Inc-type plasmid of the strain were performed using the MLST 1.8 server, ResFinder 3.0, Virulence Finder 1.5, and Plasmid Finder 1.3, which are available at the Center for Genomic Epidemiology3. Graphical maps were generated by the CGView server4. A comparison of the insert sequence of this strain and its related plasmids was performed with EasyFig 2.2.2 (Sullivan et al., 2011).
Phylogenetic analysis of these P. mirabilis strains was performed. Genome sequences of other P. mirabilis strains were downloaded from the RefSeq database. Our strains were annotated by Prokka (Seemann, 2014) using the P. mirabilis proteins from the RefSeq database as a prior reference. The core genome was determined by Roary (Page et al., 2015) using Mafft for multiple sequence alignment. A maximum-likelihood phylogenetic tree was inferred by RAxML (Stamatakis, 2014) using the GTRGAMMA model for nucleotide substitution and running with 100 bootstraps. The phylogenetic tree was visualized by iTOL (Letunic and Bork, 2019).
The complete nucleotide sequences of the chromosome and three plasmids of E. coli strain EC3385 reported in the present study were deposited in the GenBank nucleotide database under accession numbers CP029420, CP029421, and CP029422, respectively.
Sequence data from four P. mirabilis strains were also deposited in GenBank as follows:
CAV1042, CP018671.1; CAV1392, CP011578.1; CAV1453, CP018356.1.
The antimicrobial susceptibility test results showed that E. coli strain EC3385 was resistant to multiple antimicrobial agents, including cephalosporins, carbapenems and fluoroquinolones, but it was susceptible to aminoglycosides, ceftazidime-avibactam, colistin, and tigecycline (Table 1).
Multi-locus sequence typing analysis showed that this strain belonged to the ST131 type. The ST131-type E. coli clonal group emerged in the mid-2000s and has since spread extensively throughout the world (Can et al., 2015). Currently, the ST131 type is a very successful pandemic clone associated with community- and hospital-acquired infections. Many studies have demonstrated that this clone has high virulence potential and is associated with treatment failure (Can et al., 2015). In this study, VirulenceFinder analysis showed the presence of multiple potential virulence factors, such as iss (increased serum survival), lpfA (long polar fimbriae), and gad (glutamate decarboxylase) (Table 2). In addition, this clone is responsible for the rapid increase in β-lactam resistance among E. coli, mainly due to the production of CTX-M type extended spectrum β-lactamase enzymes (ESBLs) (Nicolas-Chanoine et al., 2014).
Interestingly, the isolate in this study did not carry additional genes encoding the CTX-M enzyme. A recent study reported that ESBL-negative ST131 strains have also been isolated worldwide (Ripabelli et al., 2020). In this study, no ESBL-encoding gene was detected in E. coli strain EC3385; instead, the blaKPC–2 gene, which encodes the KPC-2 type β-lactamase was identified by PCR amplification and sequencing.
Escherichia coli strain EC3385 carried the blaKPC–2 gene, which is primarily located on plasmids. However, further plasmid transfer and location experiments were not successful (data not shown), suggesting that the blaKPC–2 gene was located on the chromosome. Notably, a CTX-M type β-lactamase gene was found to be integrated in the chromosome of a high-risk E. coli ST131 clone by vertical transmission (Cerquetti et al., 2010; Stoesser et al., 2016), indicating that the ST131 type E. coli strain might have the ability to integrate resistance genes into chromosomes.
To determine the gene location, whole-genome sequencing was performed. The whole-genome sequencing data were assembled, and a circular chromosome and two plasmids were generated (Table 2). The size of the genome was 4,910,422 bp, with a GC content of 50.9%, 66 rRNA operons, 267 tRNAs, and 4749 predicted protein-coding sequences (Table 2). Two plasmids approximately 89 to 101 kb in size and having a GC content between 50.5 and 46.3% were grouped into identifiable replicon types (Table 2 and Figure 1). Notably, the chromosomal location of the blaKPC–2 gene was determined using PacBio sequencing. Furthermore, the resequencing results further confirmed that the blaKPC–2 gene was located on the chromosome.
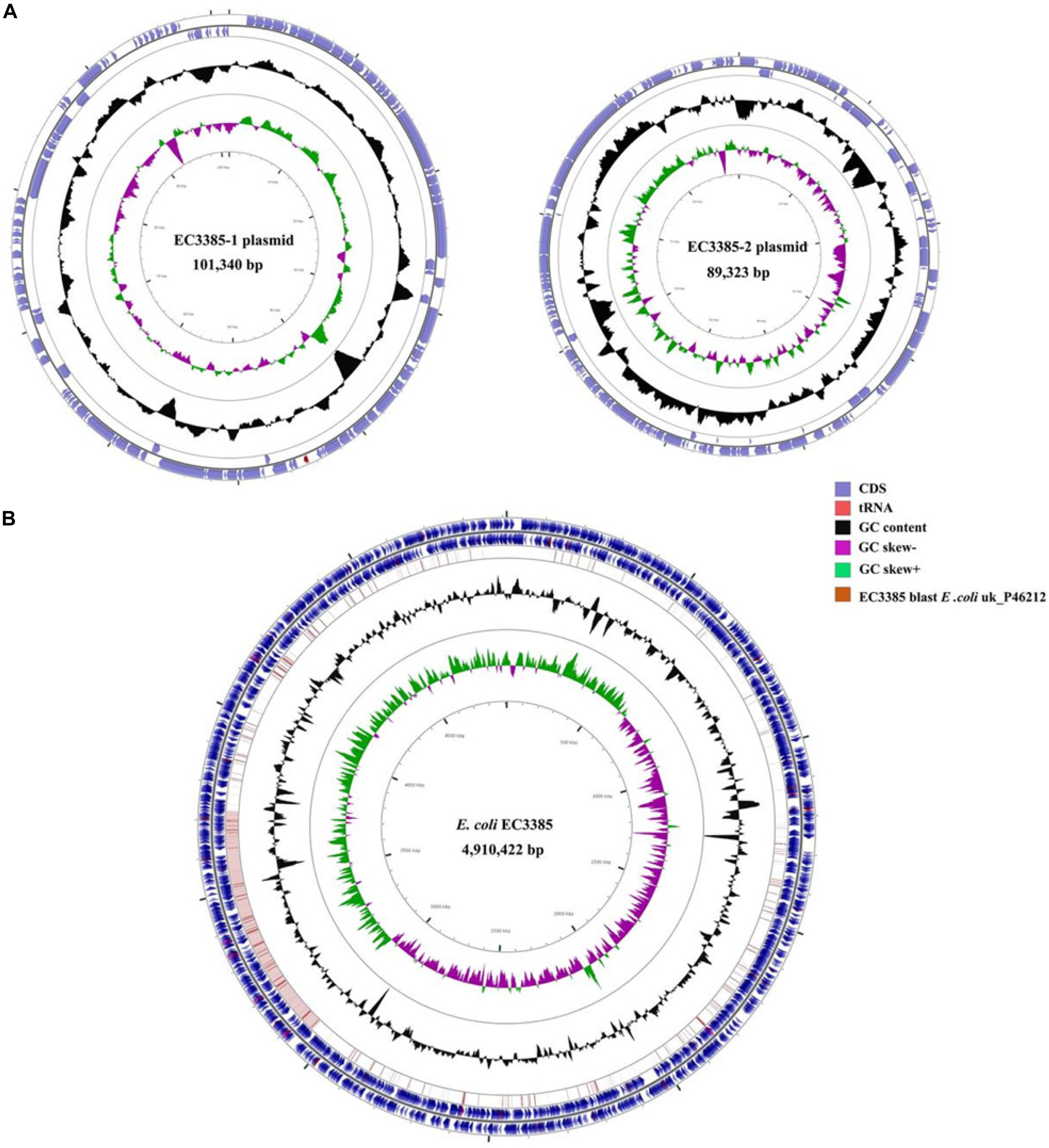
Figure 1. Circular maps of the E. coli EC3385 genome and its plasmids. (A) Circular graphs of two plasmids. (B) Circular graph of the EC3385 genome sequence and genome alignment. Blue arrows denote coding sequences, red arrows denote tRNA genes, and replication genes are denoted by green arrows. Genome alignment between EC3385 and E. coli uk_P46212 is shown in the outer circle in pink, and the GC content is shown in the inner circle in black. The region surrounding the blaKPC–2 gene is highlighted with a red frame.
To evaluate the molecular basis of chromosomal integration, the chromosomal region encompassing the blaKPC–2 gene in the closed PacBio assembly of the ST131 type EC3385 isolate was aligned to reference the strain E. coli uk_P46212 (GenBank accession number CP013658), which belongs to the ST131 clone type. Relative to the reference, the EC3385 strain had a 24-kb insertion sequence in the chromosome, which included Tn1722 and several ISs (Figure 2).
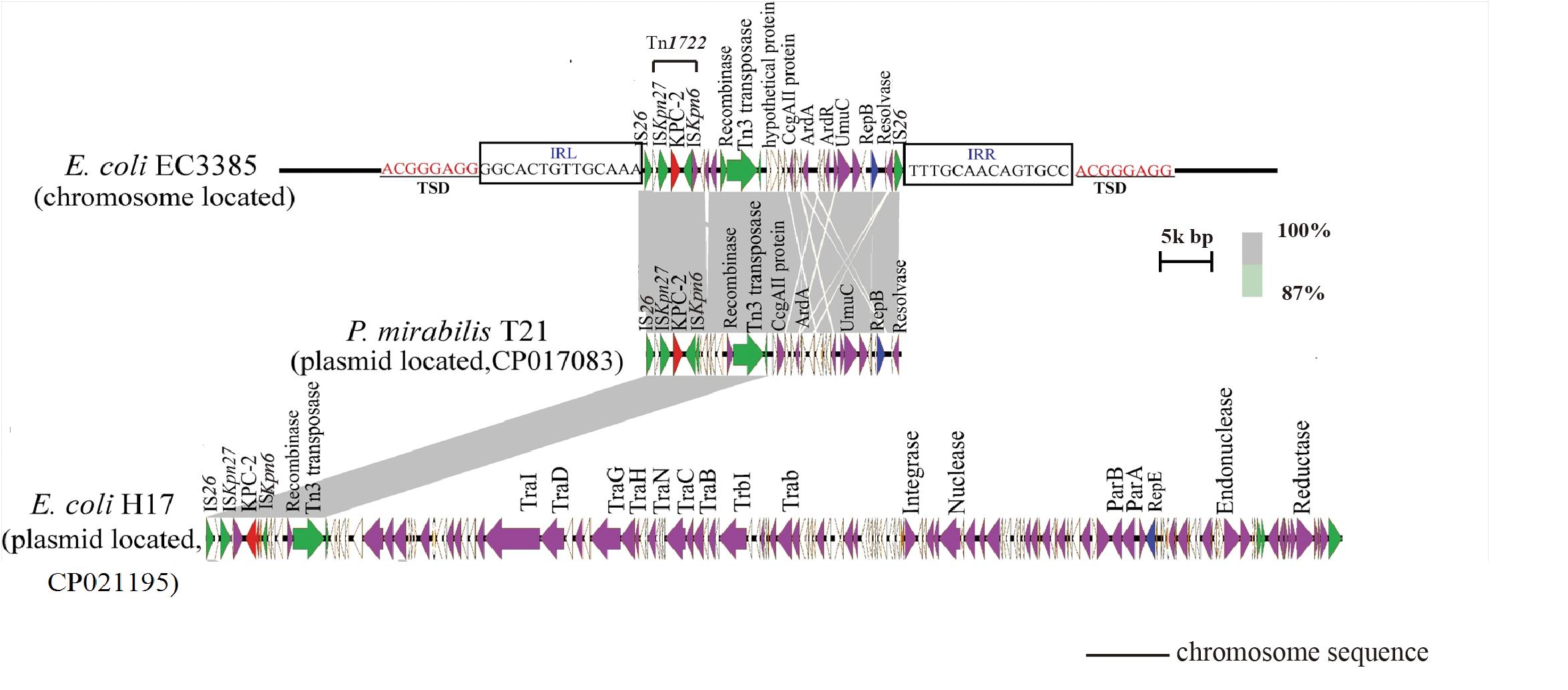
Figure 2. Linear comparison of the 24-kb insertion sequence with closely related plasmids. The gray regions between plasmids indicate nucleotide identity (87–100%) determined by BLASTn. Arrows indicate predicted open reading frames (ORFs). The primary structural characteristics of the 24-kb insertion sequence are compared to plasmids pT21 (CP017083) and pH17 (CP021195). Colored arrows represent ORFs, with red, purple, yellow, green, and white representing antibiotic resistance genes; replication, recombination and repair genes; plasmid stability genes; mobile elements and plasmid transfer related-genes; and genes with unknown function genes, respectively.
The blaKPC–2 gene in the E. coli EC3385 strain was carried on this 24-kb composite transposon-like element flanked by two IS26 elements, which undergo replicative transposition with 8-bp target site duplication (TSD) (ACGGGAGG). This finding suggests the mobilization of this blaKPC–2 gene by the composite transposon formed by IS26 (Figure 2). IS26 has been demonstrated to undergo frequent intramolecular transposition. The structure of the insert sequence leads to the speculation that the IS26 element may facilitate recombination between the plasmid and chromosome (He et al., 2015).
A further BLAST search of the 24-kb insertion sequence against the GenBank database5 revealed that this sequence is highly similar to plasmid pT21 (GenBank accession no.CP017083), which was described in a KPC-2 type carbapenemase- producing P. mirabilis strain isolated in Zhejiang, China (Hua et al., 2020), with 99.9% query coverage and a maximum of 100% identity (Figure 2). In contrast, this 24-kb insertion sequence is only partly similar (47% query coverage and a maximum of 100% identity) to plasmid pH17-2 (GenBank accession no. CP021195) of a KPC-2 type carbapenemase-producing E. coli strain isolated in China (Figure 2; Zhao et al., 2018), indicating that capture of the chromosomal blaKPC–2 gene from P. mirabilis by plasmids is possible.
To further clarify the origin of the blaKPC–2 gene, four blaKPC–2-producing P. mirabilis strains isolated during the same period (approximately 2 months, Table 1) as E. coli EC3385 in the ICU department were analyzed retrospectively. These four P. mirabilis strains were all isolated from the sputum of different patients. Notably, a maximum-likelihood phylogenetic analysis between the four P. mirabilis strains and P. mirabilis T21 carrying the pT21 plasmid revealed that these strains were clustered together and belonged to the same clone (Figure 3A). Moreover, the whole-genome sequence analysis revealed that the four P. mirabilis strains all possessed a 24-kb insertion sequence (Figure 3B), indicating that this 24-kb insertion sequence that integrated into the chromosome of the E. coli EC3385 strain may have been acquired from P. mirabilis strains. In addition, two P. mirabilis strains were isolated before the E. coli EC3385 strain was identified, indicating that KPC-2 type carbapenemase-producing P. mirabilis strains may have spread in this ICU department. A limitation of this study is the lack of the direct links regarding the transmission between KPC-2 type carbapenemase- producing P. mirabilis and E. coli EC3385 strains. However, because the patients had stayed in the same department, it is most likely they were exposed to a common source.
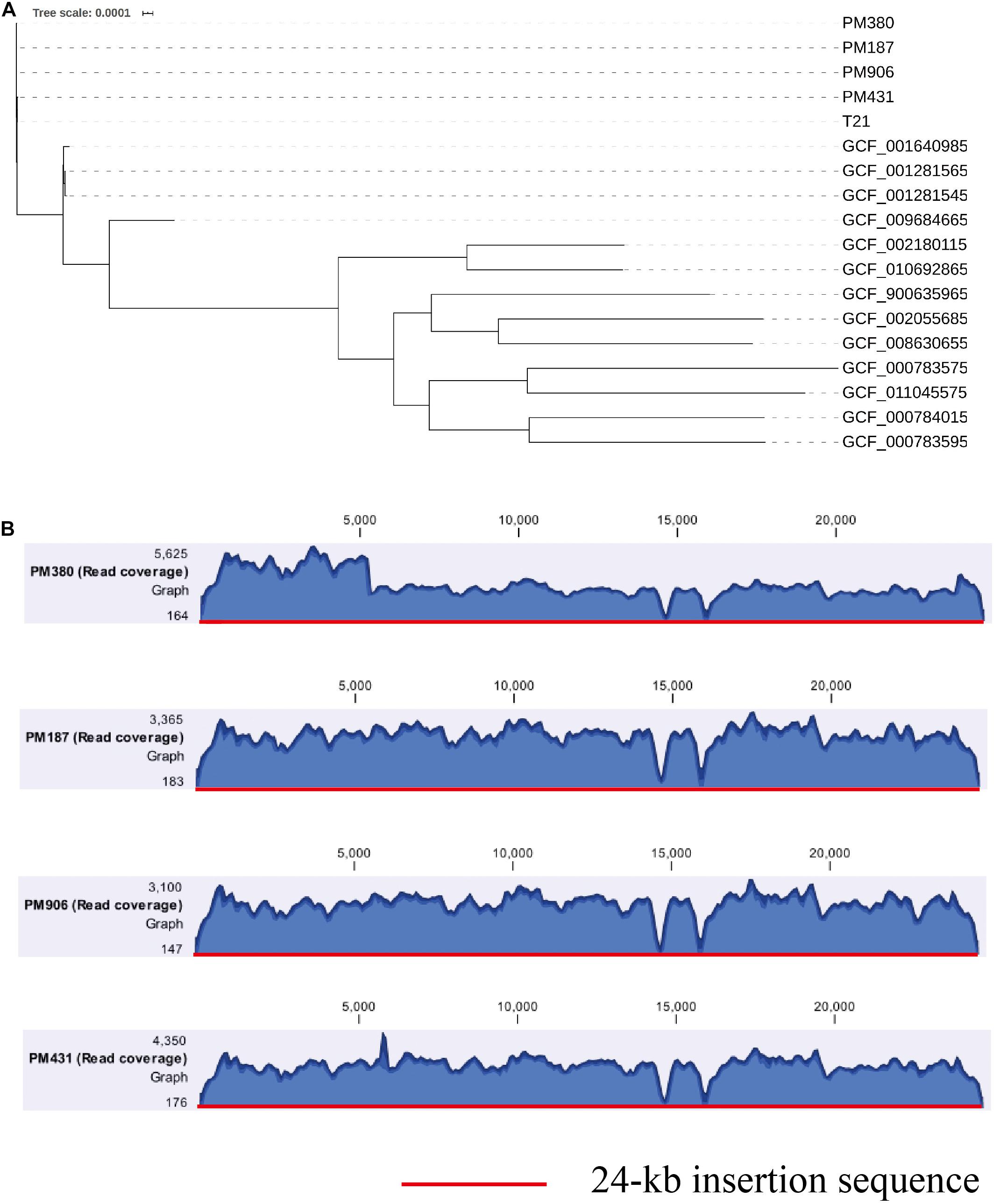
Figure 3. (A) A maximum-likelihood phylogenetic analysis of P. mirabilis strains. (B) Coverage of Illumina reads for four P. mirabilis strains (PM380, PM187, PM906, and PM431) mapped to the 24-kb insertion shown in Figure 2. The mean coverage for each strain is denoted by a curve (blue).
In summary, to the best of our knowledge, this is the first report of a clinical ST131 E. coli strain carrying the blaKPC–2 gene in the chromosome. The blaKPC–2 gene was probably horizontally transferred from the P. mirabilis plasmid to the E. coli chromosome by the IS26 mobile element, indicating that P. mirabilis might be an important reservoir of the blaKPC–2 gene for E. coli. Furthermore, the discovery of a chromosomal the blaKPC–2 gene in an E. coli strain is alarming. This gene will be maintained through replication without being subject to selective pressures, as the loss of chromosomal elements from bacterial populations is rare. Therefore, the E. coli ST131 strain carrying the blaKPC–2 gene in the chromosome would be further spread due to its own carbapenem resistance and high virulence. It is imperative to perform active surveillance to prevent further dissemination of KPC-2 type carbapenemase-producing isolates.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
YY and XL conceived and designed the experiments. DW, XM, and YC performed the experiments. DZ, YZ, XH, GM, JQ, and YF analyzed the data. DW, XL, and YJ wrote the manuscript. All authors read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (31700125) and The Project Supported by Zhejiang Provincial Natural Science Foundation of China (Grant no. LY20H200007 and LGF20H200004) and the Medical and Health Research Project of Zhejiang Province, China (2018KY229 and 2020KY420). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
An, J., Lai, K., Ma, Y., Guo, L., Ye, L., Luo, Y., et al. (2018). Emergence of multiple carbapenemase-producing organisms in single patients: an increasing threat to treatment of infection. J. Antimicrob. Chemother. 73, 544–546. doi: 10.1093/jac/dkx411
Bahl, M. I., Hansen, L. H., and Sørensen, S. J. (2009). Persistence mechanisms of conjugative plasmids. Methods Mol. Biol. 532, 73–102. doi: 10.1007/978-1-60327-853-9_5
Bergstrom, C. T., Lipsitch, M., and Levin, B. R. (2000). Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505–1519.
Can, F., Azap, O. K., Seref, C., Ispir, P., Arslan, H., and Ergonul, O. (2015). Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin. Infect. Dis. 60, 523–527. doi: 10.1093/cid/ciu864
Carraro, N., Poulin, D., and Burrus, V. (2015). Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet. 11:e1005298. doi: 10.1371/journal.pgen.1005298
Castanheira, M., Deshpande, L. M., DiPersio, J. R., Kang, J., Weinstein, M. P., and Jones, R. N. (2009). First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the sentry antimicrobial surveillance program. J. Clin. Microbiol. 47, 4129–4130. doi: 10.1128/jcm.01502-09
Cerquetti, M., Giufrè, M., García-Fernández, A., Accogli, M., Fortini, D., Luzzi, I., et al. (2010). Ciprofloxacin-resistant, CTX-M-15-producing Escherichia coli ST131 clone in extraintestinal infections in Italy. Clin. Microbiol. Infect. 16, 1555–1558. doi: 10.1111/j.1469-0691.2010.03162.x
Chen, L., Chavda, K. D., DeLeo, F. R., Bryant, K. A., Jacobs, M. R., Bonomo, R. A., et al. (2015). Genome sequence of a Klebsiella pneumoniae sequence type 258 isolate with prophage-encoded K. pneumoniae carbapenemase. Genome Announc. 3:e659-15. doi: 10.1128/genomeA.00659-15
Chen, L., Hu, H., Chavda, K. D., Zhao, S., Liu, R., Liang, H., et al. (2014). Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob. Agents Chemother. 58, 2422–2425. doi: 10.1128/aac.02587-13
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Conlan, S., Deming, C., Tsai, Y. C., Lau, A. F., Dekker, J. P., Korlach, J., et al. (2014). Complete genome sequence of a Klebsiella pneumoniae isolate with chromosomally encoded carbapenem resistance and colibactin synthesis loci. Genome Announc. 2, e1332–e1314. doi: 10.1128/genomeA.01332-14
Harrison, E., Guymer, D., Spiers, A. J., Paterson, S., and Brockhurst, M. A. (2015). Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25, 2034–2039. doi: 10.1016/j.cub.2015.06.024
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15
Hua, X., Zhang, L., Moran, R. A., Xu, Q., Sun, L., van Schaik, W., et al. (2020). Cointegration as a mechanism for the evolution of a KPC-producing multidrug resistance plasmid in Proteus mirabilis. Emerg. Microbes Infect. 9, 1206–1218. doi: 10.1080/22221751.2020.1773322
Kim, Y. A., Qureshi, Z. A., Adams-Haduch, J. M., Park, Y. S., Shutt, K. A., and Doi, Y. (2012). Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clin. Infect. Dis. 55, 224–231. doi: 10.1093/cid/cis387
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, X., Fu, Y., Shen, M., Huang, D., Du, X., Hu, Q., et al. (2018). Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7:59. doi: 10.1186/s13756-018-0349-6
Martínez, T., Vázquez, G. J., Aquino, E. E., Martínez, I., and Robledo, I. E. (2014). ISEcp1-mediated transposition of blaKPC into the chromosome of a clinical isolate of Acinetobacter baumannii from puerto rico. J. Med. Microbiol. 63(Pt 12), 1644–1648. doi: 10.1099/jmm.0.080721-0
Mathers, A. J., Stoesser, N., Chai, W., Carroll, J., Barry, K., Cherunvanky, A., et al. (2017). Chromosomal integration of the Klebsiella pneumoniae Carbapenemase Gene, bla(KPC), in Klebsiella species is elusive but not rare. Antimicrob. Agents Chemother. 61, e1823–e1816. doi: 10.1128/aac.01823-16
Mavroidi, A., Miriagou, V., Malli, E., Stefos, A., Dalekos, G. N., Tzouvelekis, L. S., et al. (2012). Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int. J. Antimicrob. Agents 39, 247–250. doi: 10.1016/j.ijantimicag.2011.11.003
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/cmr.00125-13
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., and Yu, Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Ripabelli, G., Sammarco, M. L., Scutellà, M., Felice, V., and Tamburro, M. (2020). Carbapenem-resistant KPC- and TEM-producing Escherichia coli ST131 isolated from a hospitalized patient with urinary tract infection: first isolation in molise region, Central Italy, July 2018. Microb. Drug Resist. 26, 38–45. doi: 10.1089/mdr.2019.0085
Rogers, B. A., Sidjabat, H. E., and Paterson, D. L. (2011). Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66, 1–14. doi: 10.1093/jac/dkq415
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, P., Yi, M., Fu, Y., Ruan, Z., Du, X., Yu, Y., et al. (2017). Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbiol. 55, 199–205. doi: 10.1128/jcm.01581-16
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stoesser, N., Sheppard, A. E., Pankhurst, L., De Maio, N., Moore, C. E., Sebra, R., et al. (2016). Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162. doi: 10.1128/mBio.02162-15
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tian, X., Zheng, X., Sun, Y., Fang, R., Zhang, S., Zhang, X., et al. (2020). Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002-2017. Infect. Drug. Resist. 13, 501–512. doi: 10.2147/idr.s232010
Villegas, M. V., Lolans, K., Correa, A., Kattan, J. N., Lopez, J. A., and Quinn, J. P. (2007). First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 51, 1553–1555. doi: 10.1128/aac.01405-06
Wang, Q., Zhang, Y., Yao, X., Xian, H., Liu, Y., Li, H., et al. (2016). Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1679–1689. doi: 10.1007/s10096-016-2710-0
Keywords: E. coli, KPC-2, cre, resistance mechanism, whole genome sequencing
Citation: Wang D, Mu X, Chen Y, Zhao D, Fu Y, Jiang Y, Zhu Y, Quan J, Hua X, Mao G, Li X and Yu Y (2020) Emergence of a Clinical Escherichia coli Sequence Type 131 Strain Carrying a Chromosomal blaKPC–2 Gene. Front. Microbiol. 11:586764. doi: 10.3389/fmicb.2020.586764
Received: 24 July 2020; Accepted: 26 October 2020;
Published: 13 November 2020.
Edited by:
Yi-Wei Tang, Cepheid, United StatesReviewed by:
Fupin Hu, Fudan University, ChinaCopyright © 2020 Wang, Mu, Chen, Zhao, Fu, Jiang, Zhu, Quan, Hua, Mao, Li and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Li, bGl4aV8wNjExQDE2My5jb20=; Yunsong Yu, eXZ5czExOUB6anUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.