
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 November 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.584789
This article is part of the Research Topic Genomic Basis of Antibiotic Resistance and Virulence in Acinetobacter View all 17 articles
 Yi-Tzu Lee1,2†
Yi-Tzu Lee1,2† Hsing-Yu Chen3†
Hsing-Yu Chen3† Ya-Sung Yang4
Ya-Sung Yang4 Yu-Ching Chou5
Yu-Ching Chou5 Tein-Yao Chang6
Tein-Yao Chang6 Wei-Jane Hsu3
Wei-Jane Hsu3 I-Chieh Lin6 ACTION Study Group
I-Chieh Lin6 ACTION Study Group Jun-Ren Sun6*
Jun-Ren Sun6*Carbapenem-resistant Acinetobacter nosocomialis (CRAn) is a significant public health concern. Tigecycline non-susceptible CRAn (Tn-CRAn) isolates have emerged worldwide. Tigecycline resistance is mainly related to the overexpression of AdeABC efflux pump controlled by AdeRS two-component system (TCS). Two novel tetracycline derivatives, omadacycline and eravacycline, may present a treatment option for CRAn. This study investigated the in vitro antimicrobial activity of tigecycline, omadacycline and eravacycline against clinical CRAn isolates and the contribution of efflux pumps in their resistance. Eighty-nine clinical CRAn isolates, including 57 Tn-CRAn isolates were evaluated for minimum inhibitory concentrations (MICs) by the broth microdilution. The relationship between the antimicrobial resistance and efflux pump expression was assessed by their responses to the efflux pump inhibitor 1-(1-naphthylmethyl)-piperazine (NMP). The contribution of the AdeABC efflux pump in their resistance was determined by the complementation of the AdeRS two-component system in wild-type, adeRS operon and adeB gene knockout strains. Among the 89 isolates, omadacycline and eravacycline MICs were correlated closely with those of tigecycline. They demonstrated improved potency, based on MIC90 values, by showing a 4 to 8-fold greater potency than tigecycline. The synergetic effects of tigecycline, omadacycline and eravacycline with NMP were observed in 57 (100%), 13 (22.8%), and 51 (89.5%) of Tn-CRAn isolates, respectively. Further analysis showed that the laboratory strain carrying the Type 1 adeRS operon increased the tigecycline, omadacycline and eravacycline MICs by 4–8-folds, respectively. Eravacycline demonstrated improved potency over tigecycline against populations of CRAn, including Tn-CRAn isolates. The over-expression of AdeABC efflux pumps was directly activated by the AdeRS two-component system and simultaneously reduced the susceptibilities of tigecycline, eravacycline, and omadacycline. Omadacycline and eravacycline MICs were correlated closely with those of eravacycline.
Acinetobacter species has become a major nosocomial pathogen associated with high mortality in immunocompromised patients on account of its rapid acquisition of resistance (Ayoub Moubareck and Hammoudi Halat, 2020). When Acinetobacter spp. develops extended drug resistance to sulbactam, tigecycline, and colistin, the antimicrobial choices become scarce and difficult (Montana et al., 2015). New and novel antimicrobial agents are therefore needed.
Acinetobacter nosocomialis is an emerging opportunistic pathogen that is usually grouped into the Acinetobacter calcoaceticus-baumannii complex (Acb complex) (Vijayakumar et al., 2019). In Asia, the carbapenem resistant rate among infections caused by A. nosocomialis is as high as 30% (Chen et al., 2018; Singkham-In and Chatsuwan, 2018; Chen et al., 2019). Tigecycline has been regarded as one of the final armamentaria against carbapenem resistant A. nosocomialis (CRAn). Unfortunately, tigecycline non-susceptible A. nosocomialis has also increasingly emerged in recent years (Yang et al., 2019).
Resistance-nodulation-cell division (RND) efflux pumps are ubiquitous in gram-negative bacteria and have been shown to play an important role in antimicrobial resistance (Coyne et al., 2011; Li et al., 2015b). It has been shown that three efflux pumps, AdeABC, AdeFGH, and AdeIJK, are associated with tigecycline resistance in Acinetobacter species (Coyne et al., 2011). The expression of the AdeABC is controlled by the AdeRS two-component system (TCS). The expression of AdeFGH and AdeIJK is controlled by AdeL, a LysR-type transcriptional regulator and AdeN, a TetR-like transcriptional regulator, respectively (Xu et al., 2019). Our previous study has demonstrated that the overexpression of the AdeABC efflux pump plays a major role in exporting tigecycline in tigecycline non-susceptible A. nosocomialis (Yang et al., 2019). Type 1 AdeRS TCS was found to be associated with tigecycline non-susceptible in clinical Acinetobacter nosocomialis in isolates (Yang et al., 2019). However, the correlation between the Type 1 AdeRS TCS pattern and tigecycline resistance is still uncertain and requires further investigation.
Omadacycline and eravacycline are novel tetracycline derivatives similar to tigecycline, and they were found to have broad spectrum activity against various multi-drug resistant pathogens (Zhanel et al., 2018; Karlowsky et al., 2019; Kaushik et al., 2019). Because of the structural resemblance to tigecycline, omadacycline and eravacycline are believed to have good activity against clinical CRAn. In addition, there were no reports indicating omadacycline and eravacycline susceptibilities among tigecycline non-susceptible CRAn (Tn-CRAn). The aim of this study was to examine the antimicrobial susceptibilities of omadacycline and eravacycline against the emerging CRAn, in comparison with tigecycline. Furthermore, we also investigated the role of efflux pump-mediated resistance to omadacycline and eravacycline.
The bacterial strains and plasmids used in this study are listed in Table 1. Between 2012 and 2018, clinical CRAn isolates were collected from AntimiCrobial studies in Taiwan Operating Network (ACTION), which included six medical centers located in different parts of Taiwan, including (alphabetically) the Changhua Christian Hospital (CCH) in Central Taiwan, Kaohsiung Medical University Hospital (KMUH) in southern Taiwan, Mackay Memorial Hospital (MMH) in Northern Taiwan, National Taiwan University Hospital (NTUH) in Northern Taiwan, Taipei Veterans General Hospital (TVGH) in Northern Taiwan, Tri-Service General Hospital (TSGH) of the National Defense Medical Center (NDMC) in Northern Taiwan and National Institute of Infectious Diseases and Vaccinology, National Health Research Institute (NHRI) in Northern Taiwan. All isolates were identified by MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) and confirmed by a sequence analysis of the rpoB gene (Bartual et al., 2005). The isolates were subjected to multi-locus sequence typing (MLST) according to the Pasteur scheme (Diancourt et al., 2010). The genetic relationship among the MLST types of Tn-CRAn isolates was reconstructed using the Phyloviz program.1
Antibiotic susceptibility testing was performed using Sensititre GNX2F (Thermo Fisher Scientific, Waltham, MA, United States) following the manufacturer’s protocols. Minimum inhibitory concentrations (MICs) of tigecycline, omadacycline and eravacycline were determined by broth microdilution. The U.S. Food and Drug Administration (FDA) recommendation for tigecycline susceptibility breakpoints of Enterobacteriaceae (susceptible MIC≤2 mg/L; intermediate >2 and <8 mg/L; resistant ≥8 mg/L) was used as the MIC interpretation criteria (Yang et al., 2019). Omadacycline and eravacycline were currently without breakpoints. Escherichia coli ATCC 25922 was utilized as a quality control for each series of isolates tested. The effect of 1-(1-naphthylmethyl)-piperazine (NMP) on antibiotic activity was determined by the Mueller-Hinton broth (MHB) in the presence and absence of NMP (50 mg/L), respectively. The NMP effects were interpreted and reported as follows: good effect for a 4-fold or more reduction and poor effect for a less than 2-fold reduction in the MIC value (Yang et al., 2019).
ATCC17903ΔadeRS (AnΔadeRS) and ATCC17903ΔadeB (AnΔadeB) were unmarked deletion mutants created by a previously described method for generating markerless deletions in A. nosocomialis strain ATCC17903 (An wt) as suggested in a previous study (Sun et al., 2017). Primers are shown in Table 2. Suicide plasmid pMo130-TelR was used for deleting the adeRS and adeB operons. Approximately 1-kb fragments upstream and downstream from the target genes were amplified and cloned in pMo130-TelR. Successful ATCC17903 transformants were first selected on LB agar plates containing 10 mg/L kanamycin (first crossovers). Kanamycin-resistant clones were isolated, and single homologous recombination events were screened for by PCR. Kanamycin -resistant clones were then plated on an LB agar containing 10% sucrose to be selected for the deletion of the adeRS operon or adeB gene by a second crossover and allelic replacement.
The plasmid pS01 was digested with SmaI and BamHI to delete its lacIq gene and T7 early gene promoters. The digested plasmids were ligated with the respective gene fragments to generate a series of recombinant pRS clones, including pRS-AnadeRS (wt) and pRS-AnadeRS (Type1). Recombinant plasmids were introduced by electroporation into AnΔadeRS and AnΔadeB and selected on LB agar plates containing 5 mg/L tetracycline.
The transcription levels of adeB, adeJ, and adeG genes were measured by quantitative real-time PCR assays as suggested in a previous study (Yang et al., 2019). ATCC 17903 transforms were grown in Luria-Bertani (LB) broth until mid-log phase. Total RNA was extracted using an RNeasy Mini Kit (Qiagen Sciences, Germantown, MD, United States). Reverse transcription was performed using a QuantiNova Reverse Transcription Kit (Qiagen). Real-time PCR was performed with the QuantiNova SYBR Green PCR Kit (Qiagen). The mRNA of rpoB gene was used as a control and ATCC 17903 as a reference to the standard expression level.
Statistical analysis was carried out using the SPSS version 20.0 software package. The calculation of statistical differences between various groups was based on the Fisher’s exact test. Pairs of outcomes (Correlation of MICs between two antibiotics) were examined for evidence of monotonic associations using Spearman’s correlation coefficients. Differences were considered statistically significant when p < 0.05.
A total of 89 clinical CRAn isolates were collected, including 57 Tn-CRAn from ACTION medical centers. The antimicrobial susceptibilities of Tn-CRAn isolates are shown in Supplementary Table S1. Multilocus sequence typing revealed the Tn-CRAn isolates belonged to three sequence types (ST): ST410 (55 isolates), ST68 (1 isolate) and ST1272 (1 isolate) (Supplementary Figure S1). Tigecycline susceptible CRAn (Ts-CRAn) isolates were randomly selected with the following STs: ST1272 (14 isolates), ST433 (6 isolates), ST410 (6 isolates), ST68 (4 isolates) and ST217 (2 isolates). The overall tigecycline MICs of CRAn isolates ranged from 0.25 to 32 mg/L; omadacycline, 0.25 to 16 mg/L; and eravacycline, 0.03 to 8 mg/L (Table 3). Among the 57 isolates, 34 displayed lower omadacycline MICs than tigecycline MICs (Figure 1A) and all displayed lower eravacycline MICs than tigecycline MICs (Figure 1B). Correlation analysis was carried out to test the correlation between levels of resistance to the three tetracycline derivatives (Table 4). There was a strong correlation among MICs for the three antibiotics (Spearman’s correlation coefficients = 0.829 to 0.852, both P < 0.05). Taken together, these data indicate that omadacycline and eravacycline showed a positive correlation with tigecycline activity, but they were not associated with the MLST profile. Eravacycline demonstrated improved potency over tigecycline, based on MIC90 values, showing a 4-fold and 4- to 8-fold greater potency against populations of Tn-CRAn and Ts-CRAn, respectively (Table 5).
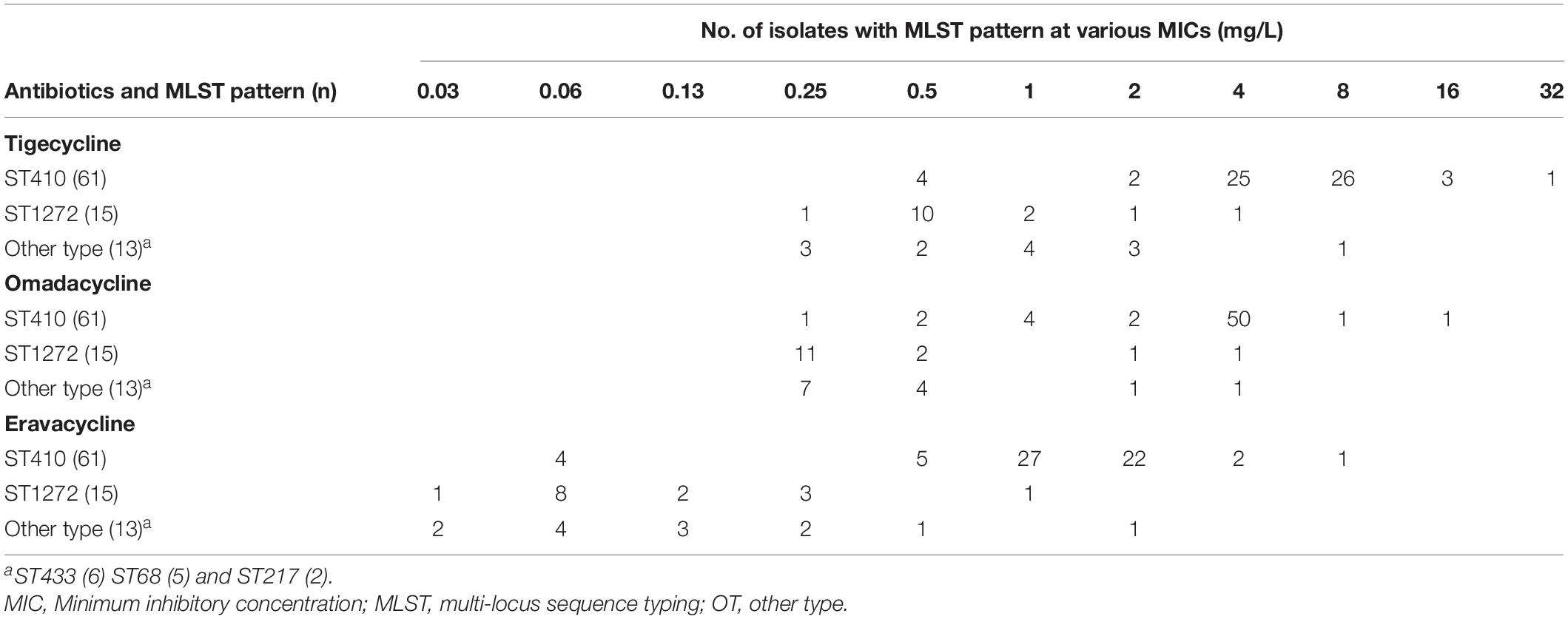
Table 3. Minimum inhibitory concentration distributions of tigecycline, omadacycline, and eravacycline in relation to multi-locus sequence typing types.
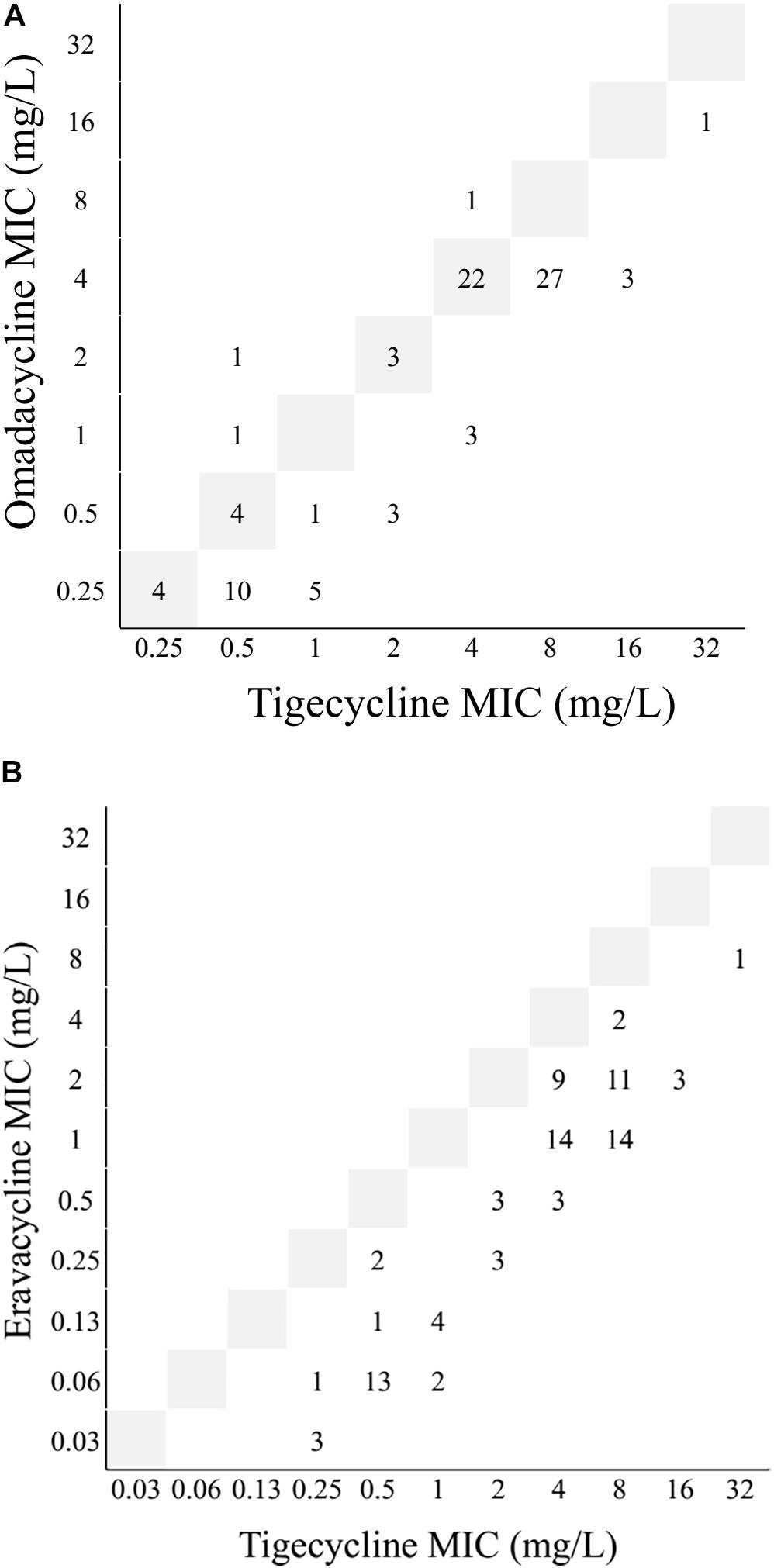
Figure 1. Interrelationship between minimum inhibitory concentrations (MICs) of omadacycline, eravacycline and tigecycline against 89 Acinetobacter nosocomialis isolates. (A) omadacycline vs. tigecycline. (B) eravacycline vs. tigecycline. Gray boxes represented the line of equivalence. Numbers below this line indicated omadacycline or eravacycline was more active, and numbers above indicate tigecycline was more active.
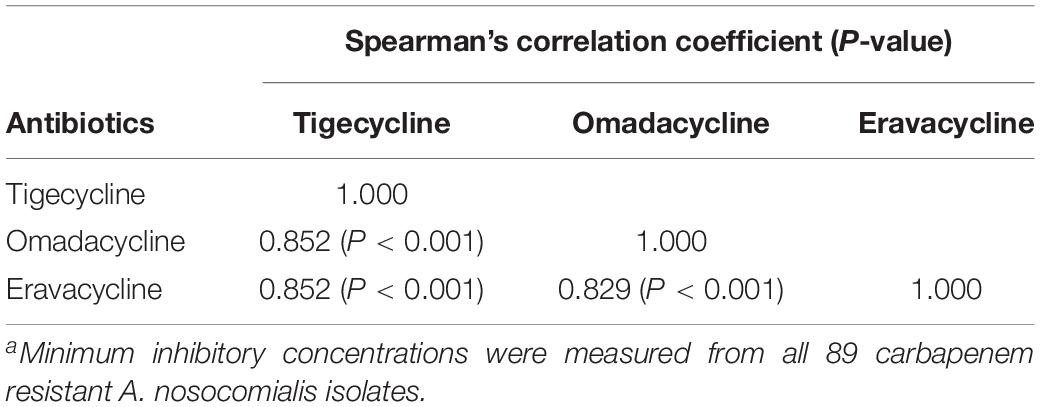
Table 4. Correlations between minimum inhibitory concentrations of tigecycline, omadacycline, and eravacycline against carbapenem resistant Acinetobacter nosocomialisa.
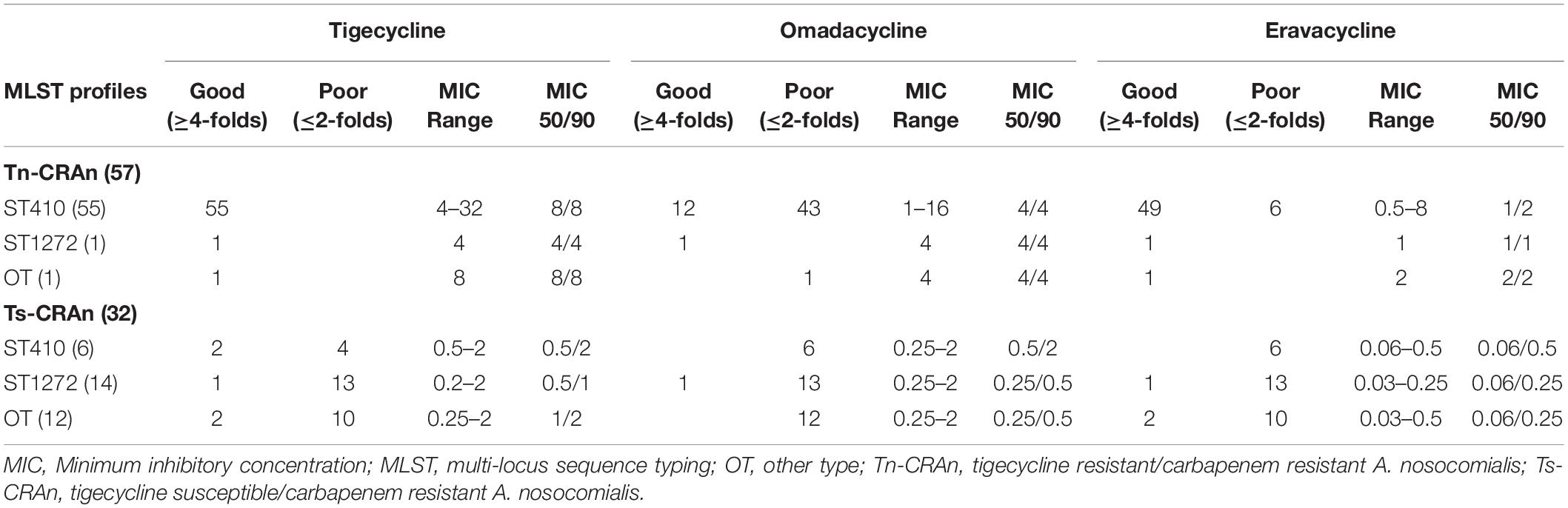
Table 5. Minimum inhibitory concentration reduction effects of 1-(1-naphthylmethyl)-piperazine (NMP) in relation to multi-locus sequence typing types between tigecycline resistant/carbapenem resistant and tigecycline susceptible/carbapenem resistant Acinetobacter nosocomialis.
The effects of the NMP on MICs of the three antibiotics against CRAn isolates are reported in Table 5. Among those, Tn-CRAn isolates showed better response to NMP in the three antibiotics than Ts-CRAn isolates (Fisher’s exact test; P < 0.05). All Tn-CRAn isolates showed a more than 4-fold decrease in eravacycline MICs with NMP. However, only seven Tn-CRAn isolates reached such reduction in omadacycline MICs with NMP. Among the NMP effects in the three antibiotics against Tn-CRAn isolates, Tigecycline and eravacycline with NMP both showed significantly more MIC reductions than omadacycline with NMP (Fisher’s exact test; P < 0.05).
To confirm the roles of AdeABC efflux pumps in the resistance to the three antibiotics, AdeRS operon and adeB gene deletion mutants were generated and tested. The sequence alignment of AdeR and AdeS sequences from Type 1 AdeRS TCS (GenBank accession number MH321430) and ATCC 17903 AdeRS TCS (wt) were shown in Supplementary Figure S2. The two amino acid differences were found between the AdeRS patterns, which contained S16N in AdeS and T137S in AdeR. The AnΔadeRS transformed with pRS-AnadeRS (Type1) showed higher MICs of the three antibiotics in comparison with pRS-adeRS (wt) [tigecycline (1 vs. 0.13 mg/L); omadacycline (2 vs. 0.25 mg/L); eravacycline (0.25 vs. 0.06 mg/L)] (Table 6). The transcription levels of adeB genes in AnΔadeRS with pRS-AnadeRS (Type1) were higher than that with pRS-adeRS (wt) (27.4-folds vs. 1.4-folds). The increase of omadacycline and eravacycline MIC was similar to tigecycline MIC and correlated with the expression of AdeABC efflux pump in the transformant. The transcription levels of adeJ and adeG in the two transforms were not significantly different. The MICs of the three antibiotics did not increase while being transformed with those recombinant plasmids into the AnΔadeB strain. These findings suggested that AdeABC efflux pumps medicated by AdeRS TCS played a role in the resistance to the three antibiotics in A. nosocomialis.
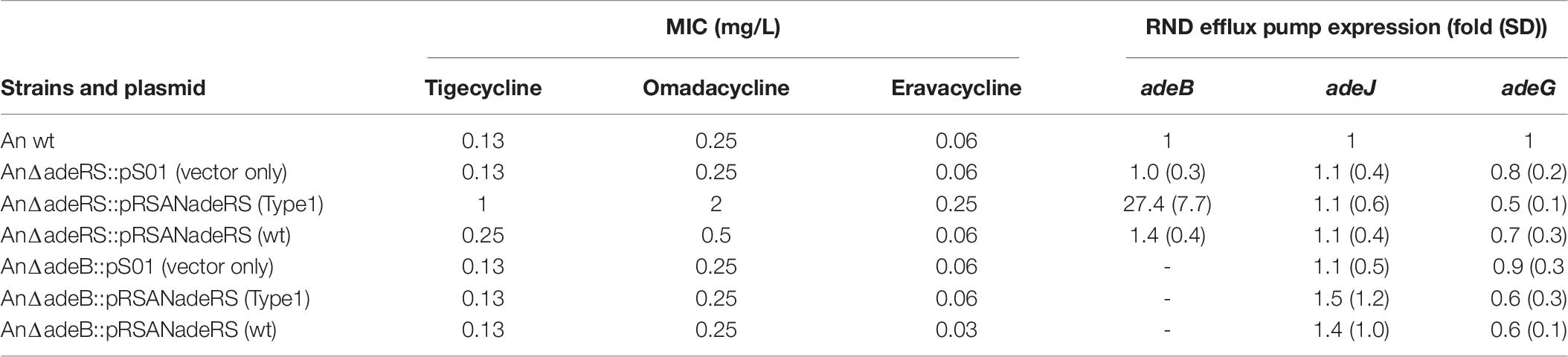
Table 6. Minimum inhibitory concentrations of tigecycline, omadacycline and eravacycline against various Acinetobacter nosocomialis transformants.
This is the first study elucidating the contribution of efflux pumps in the resistance to novel tetracycline derivative antibiotics, eravacycline and omadacycline in clinical carbapenem resistant A nosocomialis isolates. We found that there was a strong correlation among the MICs of the two antibiotics and tigecycline. NMP pump inhibitors reduced the MICs of tigecycline and eravacycline against Tn-CRAn isolates. Since the synergistic effect with NMP for omadacycline susceptibility was poor, it suggests that other mechanisms are contributing to omadacycline resistance. Further studies confirmed that AdeRS TCS played a role in the resistance to the three antibiotics in A. nosocomialis.
ST410 was found to be the most dominant type in clinical Tn-CRAn isolates in Taiwan, which is consistent with our previous studies (Chen et al., 2018; Yang et al., 2019). This may indicate that there is an unknown mechanism in ST410 that provides fitness advantage in a hospital setting, which could easily transform to carbapenem and/or tigecycline resistance. Further evaluation is needed to elucidate the relationship between ST410 and tigecycline resistance, such as whole-genome sequence analysis by next generation sequencing. MICs of omadacycline and eravacycline for the ST410 Tn-CRAn isolates were unimodally distributed, with clustering at 4 vs. 1 to 2 mg/L. In previous studies, the A. baumannii with higher MICs of tigecycline also had a strong positive correlation with higher eravacycline MICs, and the eravacycline MICs were generally 2-fold lower than that of tigecycline (Livermore et al., 2016; Seifert et al., 2018). In this study, we observed a strong correlation among the MICs of the three antibiotics against A. nosocomialis. The results suggested that the resistant mechanisms of A. nosocomialis against these three antibiotics might be similar due to the similar tetracycline-like structures and related to the AdeABC efflux pump.
1-(1-naphthylmethyl)-piperazine is a type of naphthyl derivative and is often used as an efflux pump inhibitor (EPI) in research (Sonnet et al., 2012; Yang et al., 2019). EPI has the ability to reverse antibiotic resistance, but is partly related to the ability to inhibit the AdeABC efflux pump (Pannek et al., 2006). In the current study, we found that omadacycline with NMP has weaker synergistic activity compared to the other two antibiotics. Moreover, we found that this was not related to the overexpression of the AdeABC efflux pump system. However, other findings about NMP under sub-inhibitory concentrations have also been reported in recent years, including reduction of the mass of preformed biofilm, inhibition of virulence factors and membrane destabilization (Bina et al., 2009; Anes et al., 2018; Anes et al., 2019). During survival in an environmental stress containing NMP, Klebsiella pneumoniae would initiate many genes involved in repairing and maintaining membrane homeostasis, up-regulation of other efflux systems and lipopolysaccharide modification genes (Anes et al., 2019). These findings may also suggest that sub-inhibitory concentrations of NMP not only inhibit the AdeABC efflux pump, but also drive other specific mechanisms of resistance to omadacycline.
The resistant mechanisms against tigecycline in Acinetobacter spp. included RND efflux pump overexpression, modifications at the ribosomal binding site, and enzymatic inactivation by tetracycline monooxygenase TetX (Chen et al., 2014; Beabout et al., 2015; Li et al., 2015a; Wang et al., 2019; Yang et al., 2019). Although the plasmid mediated TetX gene has been found to confer resistance to tigecycline, the overexpression of chromosomal RND efflux pumps, especially the AdeABC efflux pump is still a widespread tigecycline resistance determinant (Wang et al., 2019; Yang et al., 2019). The AdeABC efflux pump has been demonstrated to be present in most clinical A. nosocomialis and A. baumannii isolates (Yang et al., 2019; Yoon et al., 2013). The AdeABC efflux pump was confirmed to export not only tigecycline but also beta-lactam, fluoroquinolone, aminoglycoside and chloramphenicol (Coyne et al., 2011; Xu et al., 2019). As mentioned in previous studies, the overexpression of AdeABC efflux pump has been shown to be related to amino acid substitutions in AdeRS TCS (Gerson et al., 2018; Yoon et al., 2013). Our results indicated that the two amino acid substitutions in AdeRS TCS may be associated to the overexpression of AdeABC efflux pump. S16N is in the transmembrane domain of AdeS, while T137S is in the receiver domain of AdeR. Further structural and biochemical studies are needed to unravel how the two amino acid substitutions affect AdeRS TCS signal transmission.
Although, the CRAn isolates were collected from six major medical centers in Taiwan, one of the limitations in our study is that such small sample sizes may not represent the genotype distribution of CRAn in the world. Another limitation is that the contribution of potential mechanism other than efflux pump was not examined. On the basis of our current data, we suggest that Type 1 AdeRS TCS could directly stimulate the expression of the AdeABC efflux pump and is associated with eravacycline and omadacycline resistance. Thus, Type 1 AdeRS TCS could be a potential target to screen inhibitors as tetracycline derivative adjuvants for future combination therapy.
In conclusion, eravacycline and omadacycline MICs of CRAn were correlated closely with those of tigecycline and eravacycline. They demonstrated improved potency over tigecycline against populations of CRAn, including Tn-CRAn isolates. The synergistic activity of NMP on eravacycline and tigecycline was observed, but not on omadacycline. The type 1 AdeRS TCS pattern was able to reduce the susceptibilities of the three antibiotics against A. nosocomialis by the regulation of AdeABC efflux pumps medicated by AdeRS TCS.
The members of the ACTION study group include Yea-Yuan Chang (National Yang-Ming University Hospital, Yilan, Taiwan), Y-SY (Division of Infectious Diseases and Tropical Medicine, Department of Internal Medicine, Tri-Service General Hospital, National Defense Medical Centre, Taipei, Taiwan), Chung-Ting Chen (Department of Emergency Medicine, Taipei Veterans General Hospital, Taipei, Taiwan), Yuag-Meng Liu (Changhua Christian Hospital, Changhua, Taiwan), Shu-Chen Kuo (National Institute of Infectious Diseases and Vaccinology, National Health Research Institute, Maoli County, Taiwan), Chang-Pan Liu (MacKay Memorial Hospital, Taipei, Taiwan), Te-Li Chen (Graduate Institute of Life Sciences, National Defense Medical Centre, Taipei, Taiwan), and Y-TL (Taipei Veterans General Hospital, Taipei, Taiwan).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Y-TL, Y-SY, Y-CC, T-YC, W-JH, and I-CL conceptualized the study, performed the experiments, and analyzed the data. J-RS, H-YC and Y-TL was responsible for funding acquisition, aided in conceptualization of the study, and writing of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Yang-Ming University Hospital (RD2019-011), Taipei City Hospital (TPCH-109-20), and the Ministry of Science and Technology (MOST 108-2320-B-016-009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by the ACTION study group.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.584789/full#supplementary-material
Anes, J., Martins, M., and Fanning, S. (2018). Reversing antimicrobial resistance in multidrug-resistant Klebsiella pneumoniae of clinical origin using 1-(1-Naphthylmethyl)-piperazine. Microb. Drug Resist. Online ahead of print.
Anes, J., Sivasankaran, S. K., Muthappa, D. M., Fanning, S., and Srikumar, S. (2019). Exposure to sub-inhibitory concentrations of the chemosensitizer 1-(1-naphthylmethyl)-piperazine creates membrane destabilization in multi-drug resistant Klebsiella pneumoniae. Front. Microbiol. 10:92. doi: 10.3389/fmicb.2019.00092
Ayoub Moubareck, C., and Hammoudi Halat, D. (2020). Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics (Basel) 9:119. doi: 10.3390/antibiotics9030119
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A., Wisplinghoff, H., and Rodriguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/jcm.43.9.4382-4390.2005
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhaes, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother 59, 5561–5566. doi: 10.1128/aac.00547-15
Bina, X. R., Philippart, J. A., and Bina, J. E. (2009). Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J. Antimicrob Chemother 63, 103–108. doi: 10.1093/jac/dkn466
Chen, F. J., Huang, W. C., Liao, Y. C., Wang, H. Y., Lai, J. F., Kuo, S. C., et al. (2019). Molecular epidemiology of emerging carbapenem resistance in Acinetobacter nosocomialis and Acinetobacter pittii in Taiwan, 2010 to 2014. Antimicrob Agents Chemother 63, e2007–e2018.
Chen, H. Y., Yang, Y. S., Hsu, W. J., Chou, Y. C., Huang, L. S., Wang, Y. C., et al. (2018). Emergence of carbapenem-resistant Acinetobacter nosocomialis strain ST410 harbouring plasmid-borne blaOXA-72 gene in Taiwan. Clin. Microbiol. Infect. 24, 1023–1024. doi: 10.1016/j.cmi.2018.04.009
Chen, Q., Li, X., Zhou, H., Jiang, Y., Chen, Y., Hua, X., et al. (2014). Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J Antimicrob Chemother 69, 72–76. doi: 10.1093/jac/dkt319
Coyne, S., Courvalin, P., and Perichon, B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55, 947–953. doi: 10.1128/aac.01388-10
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 7:e10034. doi: 10.1371/journal.pone.0010034
Gerson, S., Nowak, J., Zander, E., Ertel, J., Wen, Y., Krut, O., et al. (2018). Diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J. Antimicrob Chemother 73, 1501–1508. doi: 10.1093/jac/dky083
Karlowsky, J. A., Steenbergen, J., and Zhanel, G. G. (2019). Microbiology and preclinical review of omadacycline. Clin. Infect. Dis. 69, S6–S15.
Kaushik, A., Ammerman, N. C., Martins, O., Parrish, N. M., and Nuermberger, E. L. (2019). In vitro activity of new tetracycline analogs omadacycline and eravacycline against drug-resistant clinical isolates of Mycobacterium abscessus. Antimicrob Agents Chemother 63:e00470-19.
Li, X., Liu, L., Ji, J., Chen, Q., Hua, X., Jiang, Y., et al. (2015a). Tigecycline resistance in Acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase. Eur. J. Clin. Microbiol. Infect. Dis. 34, 625–631. doi: 10.1007/s10096-014-2272-y
Li, X. Z., Plesiat, P., and Nikaido, H. (2015b). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/cmr.00117-14
Livermore, D. M., Mushtaq, S., Warner, M., and Woodford, N. (2016). In vitro activity of eravacycline against carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother 60, 3840–3844. doi: 10.1128/aac.00436-16
Montana, S., Vilacoba, E., Traglia, G. M., Almuzara, M., Pennini, M., Fernandez, A., et al. (2015). Genetic variability of aders two-component system associated with tigecycline resistance in XDR-Acinetobacter baumannii isolates. Curr. Microbiol. 71, 76–82. doi: 10.1007/s00284-015-0829-3
Pannek, S., Higgins, P. G., Steinke, P., Jonas, D., Akova, M., Bohnert, J. A., et al. (2006). Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide. J. Antimicrob Chemother 57, 970–974. doi: 10.1093/jac/dkl081
Seifert, H., Stefanik, D., Sutcliffe, J. A., and Higgins, P. G. (2018). In-vitro activity of the novel fluorocycline eravacycline against carbapenem non-susceptible Acinetobacter baumannii. Int. J. Antimicrob Agents 51, 62–64. doi: 10.1016/j.ijantimicag.2017.06.022
Singkham-In, U., and Chatsuwan, T. (2018). Mechanisms of carbapenem resistance in Acinetobacter pittii and Acinetobacter nosocomialis isolates from Thailand. J. Med. Microbiol. 67, 1667–1672. doi: 10.1099/jmm.0.000845
Sonnet, P., Izard, D., and Mullie, C. (2012). Prevalence of efflux-mediated ciprofloxacin and levofloxacin resistance in recent clinical isolates of Pseudomonas aeruginosa and its reversal by the efflux pump inhibitors 1-(1-naphthylmethyl)-piperazine and phenylalanine-arginine-beta-naphthylamide. Int. J. Antimicrob Agents 39, 77–80. doi: 10.1016/j.ijantimicag.2011.08.005
Sun, J. R., Chiang, Y. S., Shang, H. S., Perng, C. L., Yang, Y. S., and Chiueh, T. S. (2017). Phenotype microarray analysis of the AdeRS two-component system in Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2343–2353. doi: 10.1007/s10096-017-3066-9
Vijayakumar, S., Biswas, I., and Veeraraghavan, B. (2019). Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci. OA. 5:FSO395.
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline and omadacycline in clinical Acinetobacter baumannii. Antimicrob Agents Chemother. 64:e013 26-19.
Xu, C., Bilya, S. R., and Xu, W. (2019). adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 30:100549. doi: 10.1016/j.nmni.2019.100549
Yang, Y. S., Chen, H. Y., Hsu, W. J., Chou, Y. C., Perng, C. L., Shang, H. S., et al. (2019). Overexpression of AdeABC efflux pump associated with tigecycline resistance in clinical Acinetobacter nosocomialis isolates. Clin. Microbiol. Infect. 512, e511–e512.
Yoon, E. J., Courvalin, P., and Grillot-Courvalin, C. (2013). RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 57, 2989–2995. doi: 10.1128/aac.02556-12
Zhanel, G. G., Baxter, M. R., Adam, H. J., Sutcliffe, J., and Karlowsky, J. A. (2018). In vitro activity of eravacycline against 2213 Gram-negative and 2424 Gram-positive bacterial pathogens isolated in Canadian hospital laboratories: CANWARD surveillance study 2014-2015. Diagn. Microbiol. Infect. Dis. 91, 55–62. doi: 10.1016/j.diagmicrobio.2017.12.013
Keywords: tigecycline, Acinetobacter, omadacycline, eravacycline, efflux pump
Citation: Lee Y-T, Chen H-Y, Yang Y-S, Chou Y-C, Chang T-Y, Hsu W-J, Lin I-C, ACTION Study Group and Sun J-R (2020) AdeABC Efflux Pump Controlled by AdeRS Two Component System Conferring Resistance to Tigecycline, Omadacycline and Eravacycline in Clinical Carbapenem Resistant Acinetobacter nosocomialis. Front. Microbiol. 11:584789. doi: 10.3389/fmicb.2020.584789
Received: 18 July 2020; Accepted: 06 October 2020;
Published: 02 November 2020.
Edited by:
Ayush Kumar, University of Manitoba, CanadaCopyright © 2020 Lee, Chen, Yang, Chou, Chang, Hsu, Lin, ACTION Study Group and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Ren Sun, dHNnaHN1bkBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.