
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 November 2020
Sec. Microbe and Virus Interactions with Plants
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.584767
 Jin-Wei Zhou1
Jin-Wei Zhou1 Ai-Qun Jia2*
Ai-Qun Jia2* Xiao-Juan Tan3*
Xiao-Juan Tan3* Hong Chen4
Hong Chen4 Bing Sun5
Bing Sun5 Tian-Zi Huang1
Tian-Zi Huang1 Yu He1
Yu He1 Pei-Li Li5
Pei-Li Li5 En-Qi Liu1*
En-Qi Liu1*The impact of 1-(4-amino-2-hydroxyphenyl)ethanone (AHPE) from the metabolites of endophytic fungus Phomopsis liquidambari on quorum sensing (QS) of Agrobacterium tumefaciens was evaluated for the first time in this study. Exposure to AHPE at concentrations ranging from 12.5 to 50 μg/mL, the β-galactosidase activity, acyl-homoserine lactone level, swimming motility, chemotaxis, and flagella formation were significantly inhibited. qRT-PCR quantification combined with the docking analysis demonstrated that AHPE affected the QS system of A. tumefaciens by repressing the transcriptional levels of traI and traR rather than signal mimicry. 1H NMR-based metabolic analysis indicated that the metabolism of A. tumefaciens was notably disturbed with AHPE treatment. AHPE treatment also resulted in the enhanced oxidative stress in A. tumefaciens. The enhanced oxidative stress lead to the disorder of energy supply, protein synthesis, and nucleotide metabolism, and ultimately attenuated the pathogenicity of A. tumefaciens. Our study indicated that AHPE can serve as a potential pesticide to defend against A. tumefaciens.
Agrobacterium tumefaciens is a soil-borne phytopathogen that is responsible for crown gall disease in plant (Gelvin, 2003). It can affect more than 1000 various species of dicotyledonous plants and some monocotyledonous plants including many agronomically important crops (DeCleene and DeLey, 1976). A. tumefaciens causes crown gall disease by transferring and integrating its T-DNA into the plant genome and then inducing the biosynthesis of auxin and cytokinin (Gohlke and Deeken, 2014). Increased auxin and cytokinin levels result in uncontrolled development of crown galls due to the changed balance between these phytohormones. Crown gall disease caused by A. tumefaciens leads to substantial economic losses worldwide, mostly pertaining to perennial horticulture plants, by causing reduced growth, vigor, and yield, and severe diseases may cause partial or complete death of infected plants. An effective way to control crown gall disease is biological control by Agrobacterium radiobacter strain K84, a bacterium that can excrete agrocin 84 that is toxic to A. tumefaciens (Moore and Warren, 1979). However, this protection is limited because it is not suitable for all A. tumefaciens strains. Alternative measures depending on RNA interference to repress the expressions of genes involved in auxin synthesis were advocated for crown gall controlling (Lee et al., 2003). However, the methods were also limited for practical application due to the use of transgenic techniques. Therefore, developing durable disease control approaches for combating A. tumefaciens disease is needed.
T-DNA also encodes for synthesis of opines (Schell et al., 1979), which are specific growth substrates and signals used by the virulent Agrobacterium spp. Some opines, called conjugative opines, are required for synthesis of a quorum sensing (QS) signal, acylated homoserine lactones (AHLs) 3-oxo-octanoyl-homoserine (3-oxo-C8-HSL). The QS signal 3-oxo-C8-HSL was synthesized by the TraI AHL synthase (Zhang et al., 1993). The TraR protein is an AHL-responsive LuxR-type transcription factor that activates genes controlling Ti plasmid replication (rep) and conjugal transfer (Chen et al., 2004). At inducing concentrations, the AHL is bound by TraR, causing the formation of a stable, active homodimer with one AHL bound per TraR protomer. Dimerized TraR-AHL has an increased affinity for the control (tra-box) sequences upstream of target rep and tra operons, resulting in binding of the DNA and, in turn, transcriptional activation (Fuqua et al., 1996; Zhu and Winans, 1999; Chen et al., 2004). A link between a high copy number of Ti plasmid and severity of tumor symptoms was established (Pappas and Winans, 2003) and, consequently, anti-virulence strategies targeting QS have been proposed to attenuate Agrobacterium-induced symptoms on plants (Molina et al., 2003; Chevrot et al., 2006).
The chemical 1-(4-amino-2-hydroxyphenyl)ethanone (AHPE) (Supplementary Figure 1) was firstly purified from the metabolites of Phomopsis liquidambari S47, one endophytic fungus isolated from the leaves of Punica granatum. To date, literature about the effect of AHPE on QS and virulence of A. tumefaciens has not been reported. In this research, the impact of AHPE on QS was evaluated by using the reporter strain A. tumefaciens A136 (pCF218/PCF372) (traI: lacZ) (McLean et al., 1997) and the metabolic response of A. tumefaciens to AHPE was determined by using the 1H NMR-based global metabolomics.
Phomopsis liquidambari S47 was preserved in China Center for Type Culture Collection with the ID of M2018476. The AHL-responsive reporter strain A. tumefaciens A136 was kindly donated by Prof. Jun Zhu from University of Pennsylvania. The wild-type strain A. tumefaciens C58 was provided by Prof. Min-Liang Guo from Yangzhou University. All strains were cultivated in A. tumefaciens (AT) medium (K2HPO4 10.5 g, KH2PO4 4.5 g, MgSO4 0.2 g, FeSO4 5 mg, CaCl2 10 mg, MnCl2 2 mg, (NH4)2SO4 2 g, mannitol 2 g, H2O 1000 mL, pH 7.0) at 28°C unless otherwise specified. The chemical 1-(4-amino-2-hydroxyphenyl)ethanone (AHPE) was purified from the metabolites of P. liquidambari S47 by forward and reversed silica gel chromatography, HPLC, and gel permeation chromatography.
The minimum inhibitory concentration (MIC) of AHPE was determined according to the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute (CLSI), 2011) with a bacterial concentration of 1–5 × 105 CFU/mL. AHPE was serially two-fold diluted in AT medium. For growth profile analysis, overnight cultures of A. tumefaciens C58 was inoculated into 30 mL of AT medium to obtain OD620 = 0.05. The cultures were added with various concentrations of AHPE and then cultured at 28°C for 24 h. Salicylic acid (15 μg/mL) and DMSO served as the positive and negative control, respectively. Growth was analyzed by reading OD value at 620 nm (Biotek Elx800, United States).
Overnight cultures of A. tumefaciens A136 (OD620 = 0.5) were 0.1% inoculated into 100 mL of AT medium with or without AHPE. Aliquots (300 μL) of 3-oxo-C8-HSL (1 mM, Sigma-Aldrich, United States) were added into the cultures and then incubated at 28°C for 17 h. Salicylic acid (15 μg/mL) and DMSO were used as positive and negative control, respectively. After incubation, the OD600 was determined. And then, 200 μL of cultures were added with 800 μL of Z-buffer, 100 μL of 0.1% SDS, 150 μL chloroform, and 100 μL of ONPG (4 mg/mL). The mixture was incubated at water bath at 28°C until the mixture turned yellow, and then 600 μL of 1 M sodium carbonate solution was added for reaction termination. The reaction time (T, min) was recorded. After centrifugation, the OD420 value was determined. The β-galactosidase activity was determined using the following formula as described previously (Stachel et al., 1985):
The potential impact of AHPE on QS was evaluated by measuring 3-oxo-C8-HSL level produced by A. tumefaciens C58 (Morin et al., 2003). Briefly, 50 μL cultures of A. tumefaciens C58 were inoculated into 50 mL of AT medium and then incubated with AHPE (12.5, 25, and 50 μg/mL) at 28°C for 17 h. DMSO served as the negative control. After centrifugation at 12000 rpm, the supernatant was extracted with the same volume of acidified ethyl acetate. The solvent was evaporated and residues were redissolved with methanol. The level of 3-oxo-C8-HSL was quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Zhou et al., 2018b).
The swimming motility assay was performed according to Merritt (Merritt et al., 2007) with minor modifications. Briefly, 2 μL overnight cultures of A. tumefaciens C58 (OD620 = 0.5) were inoculated in the ATGN medium (K2HPO4 10.5 g, KH2PO4 4.5 g, MgSO4 0.2 g, FeSO4 5 mg, CaCl2 10 mg, MnCl2 2 mg, (NH4)2SO4 15 mM, glucose 5 g, agar 3 g, H2O 1000 mL, pH 7.0) supplemented with various concentrations of AHPE (25 and 50 μg/mL). Salicylic acid (15 μg/mL) and DMSO served as the positive and negative control, respectively. After 24-h incubation, the swimming diameter was recorded.
The chemotaxis assay was evaluated as described by Ding (Ding and Christie, 2003). Briefly, 2 μL overnight cultures of A. tumefaciens C58 (OD620 = 0.5) were inoculated in one side of the ATGN medium (without glucose) supplemented with various concentrations of AHPE (25 and 50 μg/mL). One strip filter paper with glucose was placed 4 cm away from the bacterial solution and then incubated at 28oC for 24 h. Salicylic acid (15 μg/mL) and DMSO served as the positive and negative control, respectively. After incubation, the chemotaxis diameter was determined.
Overnight cultures of A. tumefaciens C58 (OD620 = 0.5) were 0.1% inoculated into fresh AT medium supplemented with various concentrations of AHPE (25 and 50 μg/mL) and incubated at 28°C for 17 h. Salicylic acid (15 μg/mL) and DMSO served as the positive and negative control, respectively. After incubation, 1 mL of bacterial cultures were mixed with 50 μL of 37% methanol and then centrifugated at 5000 rpm for 2 min. Cells were washed with distilled water and then resuspended with 200 μL of distilled water. The resuspension (5 μL) was dropped on a slide, dried naturally, and then stained with the flagella staining solution (Beijing Solarbio Science and Technology Co., Ltd., China) for 5 min. After staining, the excess dye was rinsed off and flagella were observed under light microscope (Nikon 80i, Japan).
Docking simulation was performed using the AutoDock 4.2 program. The structures of AHPE and 3-oxo-C8-HSL were drew with ChemBioDraw 12.0. All water molecules were removed in the protein files (TraR, PDB: 1H0M), polar hydrogen atoms were added, and non-polar hydrogen atoms were merged using the Hydrogen module in the AutoDock Tools (ADT). Gasteiger charges were assigned and the Lamarckian genetic algorithm was used to obtain the conformation of compounds in the binding pocket. The RMSD difference within 2 Å to give the best scored poses. Protein ligand interactions were obtained by the online tool PLIP1 and analyzed using PyMOL. A docking grid with a size of 50∗50∗50 was used. For each PDB structure, the center coordinate was obtained from active site. The default settings for docking in AutoDock 4.2 software were used except for num_modes (50). ADT shows the docking scores as affinity of binding, and the scores were expressed as kilocalories per mole (Zhou et al., 2018a).
Agrobacterium tumefaciens C58 was cultivated as described above with DMSO served as the negative control. Cells were harvested by centrifugation at 4°C and 12000 rpm, and then washed with precooled PBS. Cell pellets were added with methanol/H2O (3.8 mL, 1/0.9) and homogenized by 4-min intermittent sonication. After homogenization, 4 mL of chloroform was supplemented. The mixtures were centrifugated and the upper layer was collected. Methanol was removed with an organomation and then lyophilized. The dried metabolites were resolved in D2O and 0.05% (w/v) sodium 3-(trimethylsilyl) propionate-2,2,3,3-d4 (TSP) for referencing purposes. 1H NMR analysis and data processing were performed according to Chen (Chen et al., 2017). The fold changes in metabolites and associated p-values were presented in fold change plots.
Agrobacterium tumefaciens C58 was cultivated as described above with DMSO served as the negative control. Cells were harvested by centrifugation at 4°C and 12000 rpm, and then washed with precooled sterile PBS. RNA was extracted using RNA extraction kit (Tiangen Biotech, Beijing, China) and the quantitative real-time PCR (qRT-PCR) reaction was performed according to Zhou (Zhou et al., 2019) with the reference gene lepA (atu0241) set as the internal control (Lang et al., 2016). The primers were listed in Supplementary Table 1.
Tobacco infection assay was performed as described by Liu (Liu et al., 2012) and Dandurishvili (Dandurishvili et al., 2011). Briefly, A. tumefaciens C58 was 0.1% inoculated into AT medium containing 25 and 50 μg/mL of AHPE, and incubation at 28°C and 180 rpm for 17 h. Salicylic acid (15 μg/mL) and DMSO were used as the positive and negative control, respectively. Cells were harvested by centrifugation, resuspended in PBS buffer to obtain an OD620 = 0.1 (approximately 1 × 108 cfu/mL). Stems of four-week-old tobaccos were inoculated by slight injury to the stem using a needle dipped in A. tumefaciens C58 suspension. The blank control was sprayed with water. Pictures and tumor weight were recorded after four-week of infection.
The MIC of AHPE for A. tumefaciens strains was determined to be 100 μg/mL. The growth analysis was evaluated by employing AHPE at sub-MIC concentrations. Results indicated that exposure to AHPE at 12.5, 25, and 50 μg/mL exhibited no inhibitory impact on bacterial growth (Figure 1).
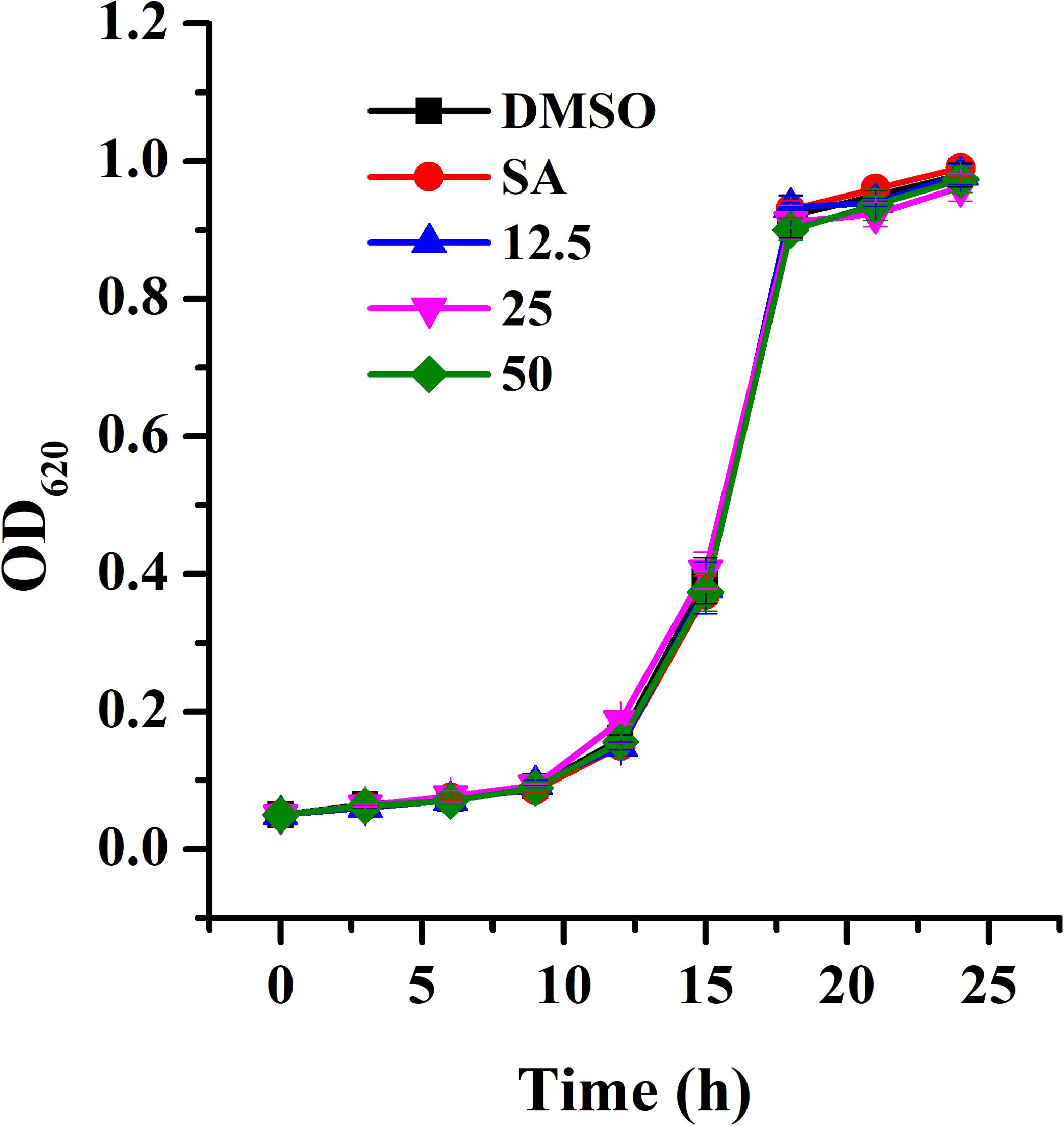
Figure 1. Growth profile of A. tumefaciens treated with AHPE. Growth was evaluated at various concentration of AHPE (12.5, 25, and 50 μg/mL) for 24 h. DMSO and salicylic acid (15 μg/mL) were used as the negative and positive controls, respectively. Error bars represent standard deviations of three measurements.
The effect of AHPE on QS system of A. tumefaciens was evaluated by β-galactosidase assay by using the AHL-responsive reporter strain A. tumefaciens A136 which contained a traI:lacZ fusion (McLean et al., 1997). Exposure to AHPE at 25 and 50 μg/mL resulted in approximately 53 and 72% inhibition in β-galactosidase activity when compared with the DMSO control (Figure 2), which was more efficient than that of 15 μg/mL of salicylic acid (Figure 2).
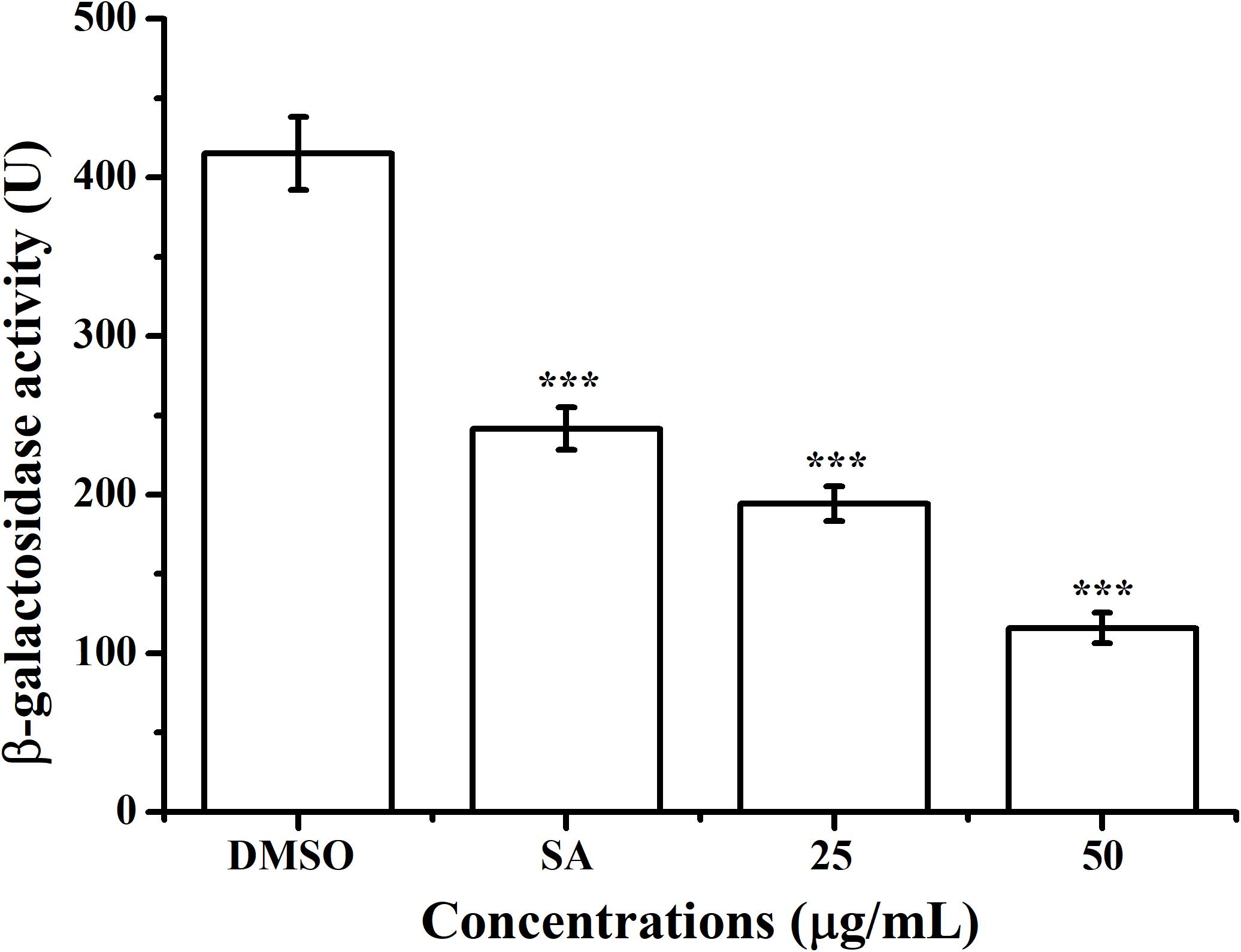
Figure 2. Effect of AHPE (25 and 50 μg/mL) on β-galactosidase activity using A. tumefaciens A136 as the AHL-responsive reporter strain. Salicylic acid (15 μg/mL) and DMSO were used as positive and negative control, respectively. Statistical differences were determined by ANOVA followed by Tukey-Kramer test. ***p < 0.001 versus the DMSO control.
The putative impact of AHPE on QS of A. tumefaciens C58 was further evaluated by investigating the AHL level secreted by this organism. LS-MS/MS confirmed the presence of the signal molecule 3-oxo-C8-HSL in the supernatant of A. tumefaciens C58 (Figures 3C–G). Exposure to AHPE at 12.5, 25, and 50 μg/mL significantly decreased the peaks and areas of 3-oxo-C8-HSL, which was evidenced by the HPLC chromatograms (Figures 3D–F). Relative quantification indicated that AHPE treatment resulted in the reduced production of 3-oxo-C8-HSL by about 50, 65, and 70%, respectively, related to the DMSO-treated control (Figure 3H).
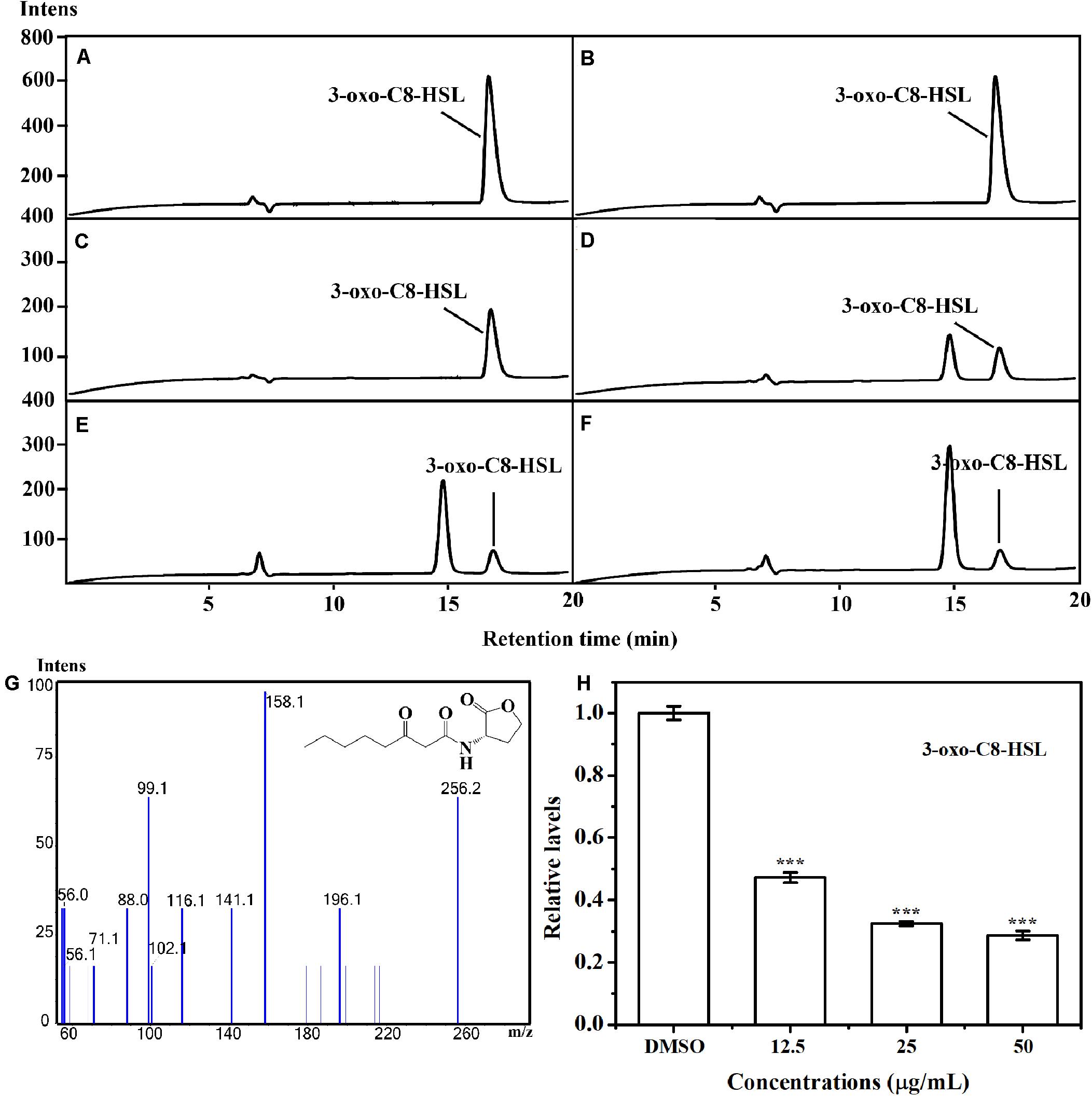
Figure 3. Relative quantification of 3-oxo-C8-HSL by LC-MS/MS chromatograms. HPLC chromatograms of 3-oxo-C8-HSL produced by A. tumefaciens C58 treated with (C) DMSO and (D–F) AHPE (12.5, 25, and 50 μg/mL, respectively). (A) and (B) represented the standard chemicals of 3-oxo-C8-HSL. (G) MS/MS spectra of 3-oxo-C8-HSL. (H) Quantitative analysis of 3-oxo-C8-HSL treated with 12.5, 25, and 50 μg/mL of AHPE, respectively. Statistical differences were determined by ANOVA followed by Tukey-Kramer test. ***p < 0.001 versus the DMSO control.
Motilities and chemotaxis are important indexes to evaluate the survival and infection abilities of A. tumefaciens. Excellent motilities and chemotaxis are beneficial for bacteria to strive for more nutrients and host infection in the environment (Erhardt, 2016). In addition, A. tumefaciens moves forward for survival and infection was governed by clockwise rotation of flagella (Guo et al., 2017). Therefore, inhibition of motilities, chemotaxis, and flagella are essential for suppressing the pathogenicity of A. tumefaciens. For the DMSO-treated group, the swimming diameter was about 6.5 ± 0.3 cm (Figure 4A). However, the swimming diameter was reduced to 2.0 ± 0.3 cm (Figure 4C) and 0.7 ± 0.1 cm (Figure 4D), respectively, when exposed to 25 and 50 μg/mL of AHPE. The inhibitory efficiency of AHPE was superior to that of salicylic acid (Figure 4B). AHPE also showed dose-dependently suppressed effect on chemotaxis, which was evidenced by the chemotaxis diameter (Figures 4E–H). After AHPE treatment (25 and 50 μg/mL), the diameter of chemotaxis was obviously decreased from 3.0 ± 0.2 cm (Figure 4E) to 1.4 ± 0.2 cm (Figure 4G) and 0.4 ± 0.2 cm (Figure 4H), respectively. Similarly, a notable suppression on flagella formation was also detected after treatment with AHPE (Figures 4K,L) when compared with the DMSO-treated control (Figure 4I). The inhibited efficiency of AHPE was superior to that of salicylic acid (Figure 4J).
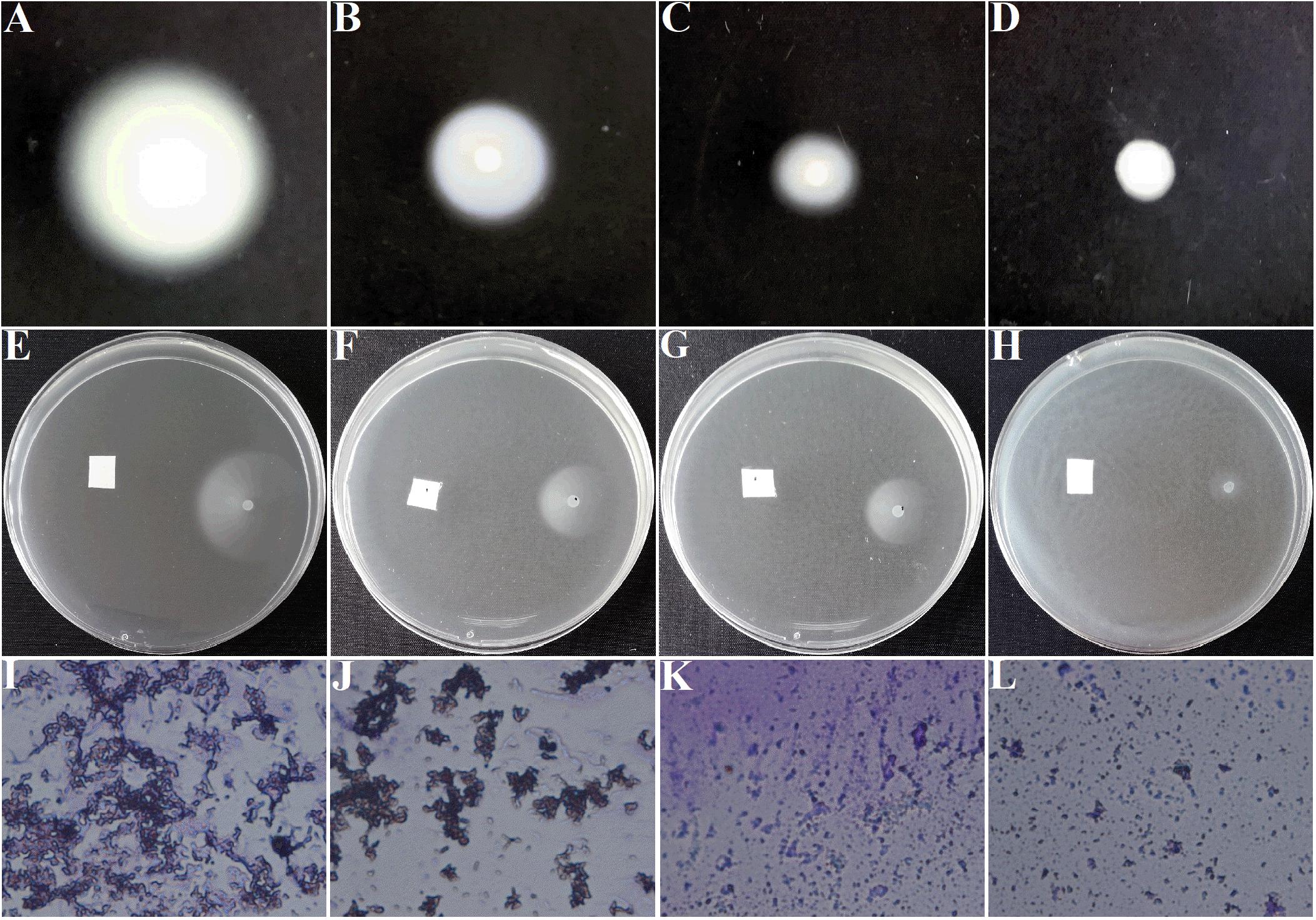
Figure 4. Effect of AHPE on swimming motility (A–D), chemotaxis (E–H), and flagella formation (I–L). Images of (A,E,I) represented DMSO-treated groups; (B,F,J) represented 15 μg/mL of salicylic acid-treated groups; (C,G,K) represented 25 μg/mL of AHPE-treated groups; and (D,H,L) represented 50 μg/mL of AHPE-treated groups.
As presented in Figure 5A, the amino residue of 3-oxo-C8-HSL showed H-binding interaction with Asp70 at a distance of 2.81 Å and the carbonyl group demonstrated H-binding interaction with Thr129 at a distance of 3.26 Å. The hydroxyl group and carbonyl group of AHPE showed H-interactions with Trp57, respectively (Figure 5B). AHPE binds to TraR with the energy of −6.24 kcal/mol, whereas 3-oxo-C8-HSL was −8.09, indicating that 3-oxo-C8-HSL has stronger binding affinity to TraR compared with AHPE.
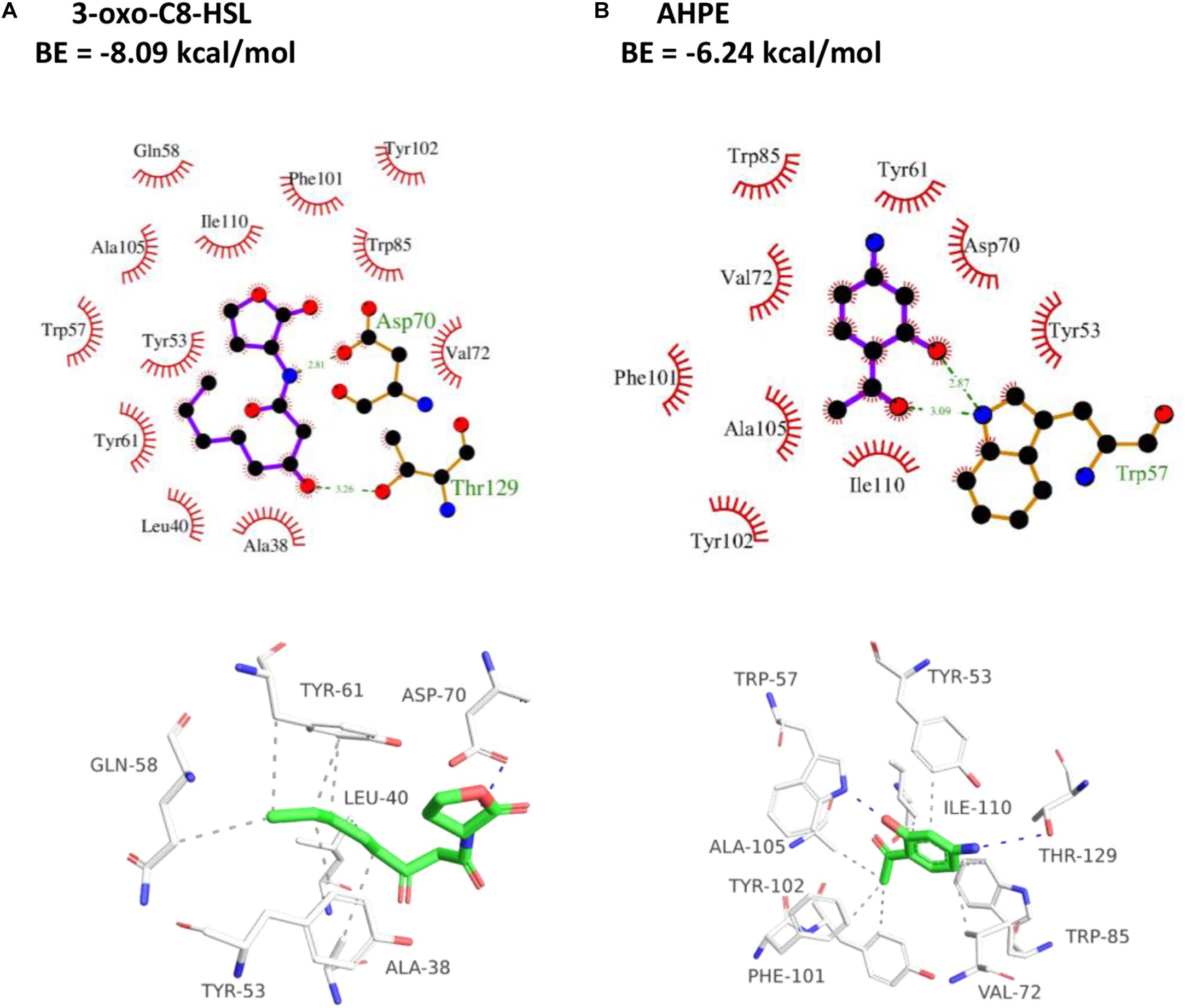
Figure 5. 2D (top) and 3D (bottom) schematics of receptor-ligand interactions of TraR with 3-oxo-C8-HSL (A) and AHPE (B), respectively.
Metabolic changes of A. tumefaciens C58 treated with AHPE were presented in Figure 6. A total of 35 metabolites including organic acid, organic amine, amino acids, and energy-related compounds were assigned according to their hydrogen chemical shift and peaks’ shape (Table 1). The PCA score plot showed a significant separation between the untreated and AHPE-treated groups (Figure 7A), which indicated that AHPE has a marked impact on metabolism in A. tumefaciens C58. From the S-plot (Figure 7B) and loading plots (Figures 7C,D), the levels of lysine, putrescine, beta-alanine, sarcosine, N, N-dimethylglycine, choline, glycine, sucrose, uracil, and NADP+ were significantly decreased, while lactate, glutamate, dimethylamine, and UDP-galactose were significantly increased after exposure to AHPE. The assignments and fold changes between groups were displayed in Table 1.
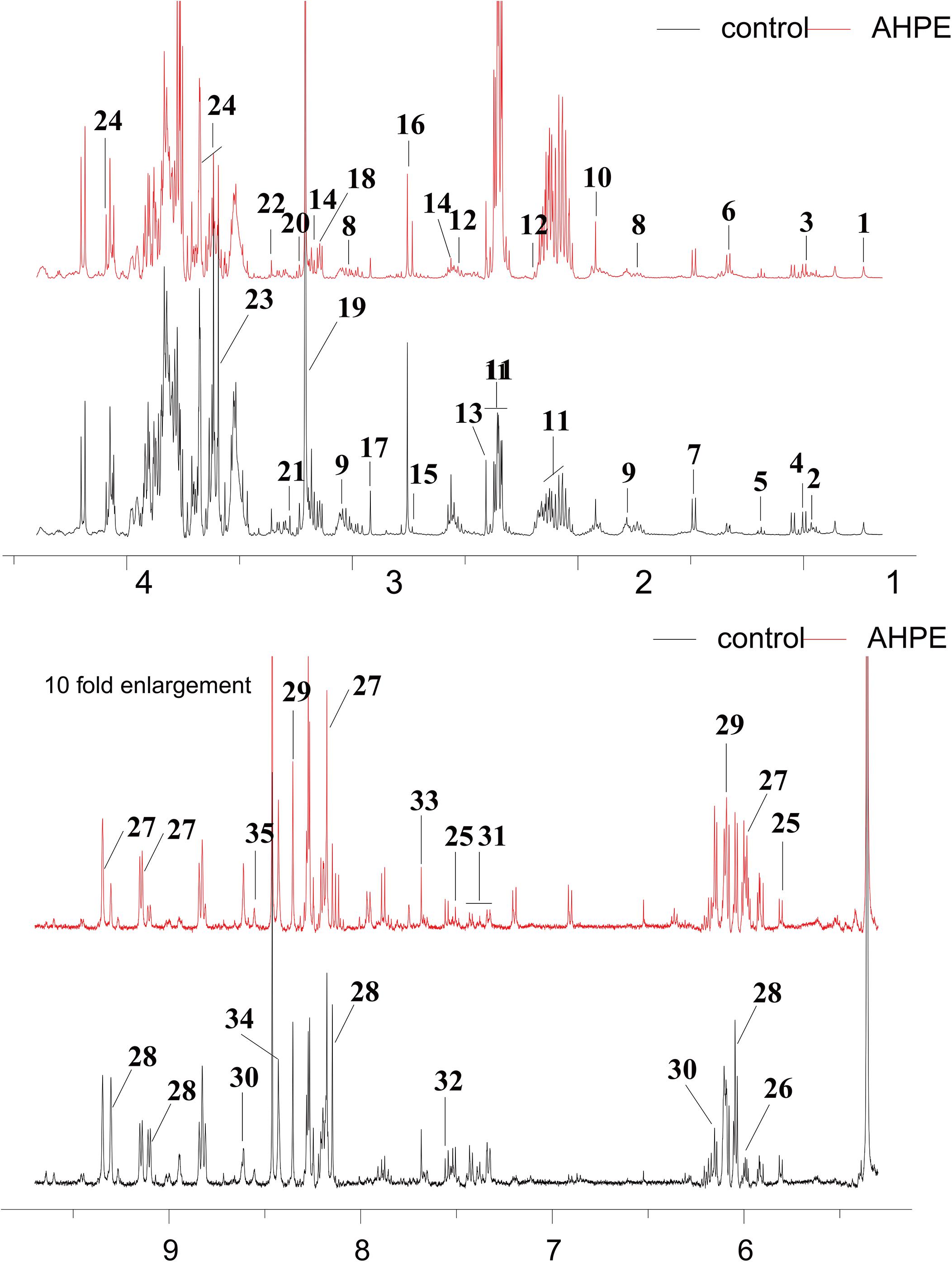
Figure 6. 1H NMR spectra of A. tumefaciens extracts from AHPE-treated (red line) and control group (black line). Labeled metabolites were assigned in Table 1.
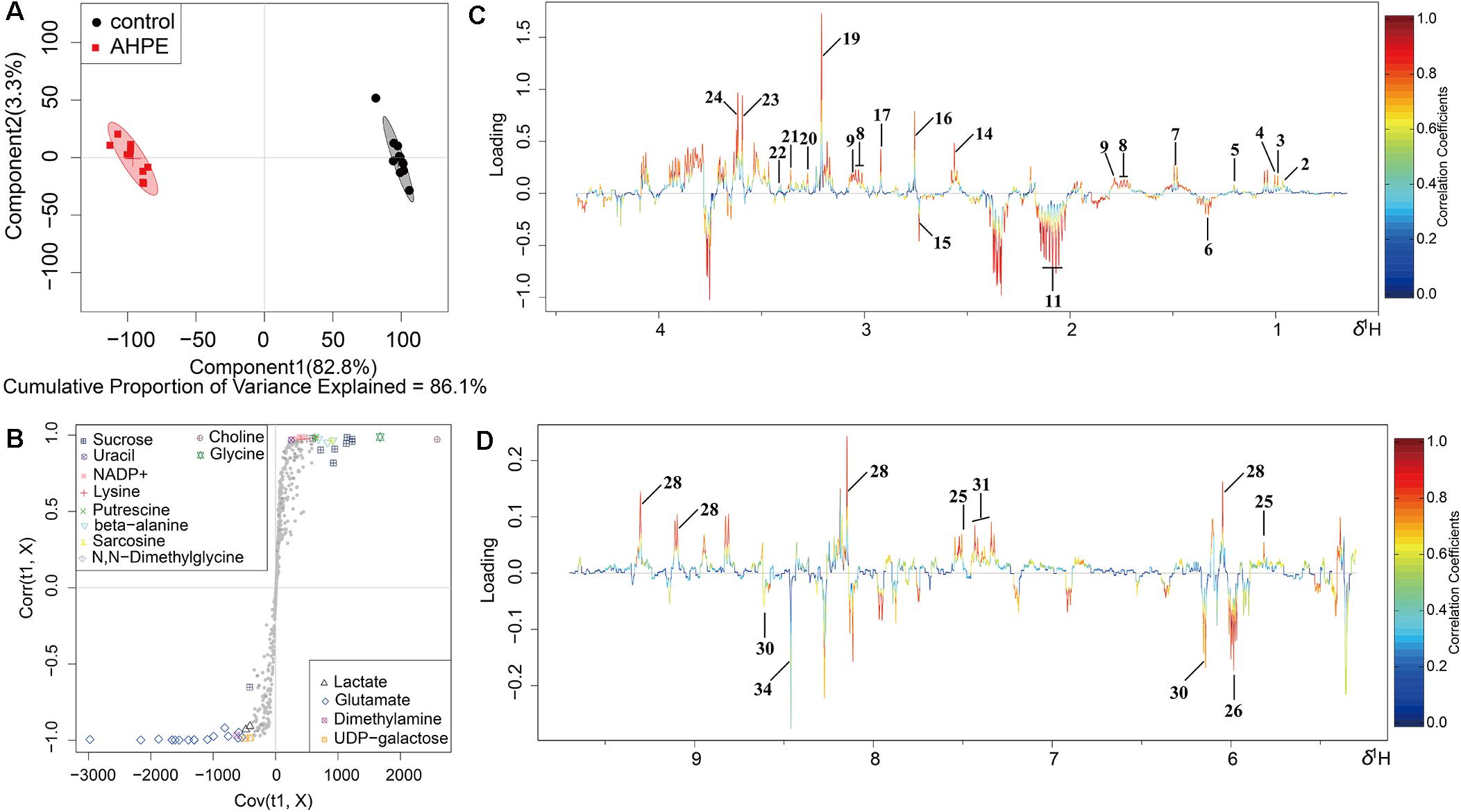
Figure 7. OSC-PLS-DA of metabolomics profiles from AHPE-treated and control groups. (A) PCA score plot. (B) S-plot points represent different variables (metabolites). (C,D) Color-coded loading plot after removal of water signals and affected regions.
qRT-PCR was employed to evaluate the effect of AHPE on transcriptional levels of five genes involved in QS, virulence, and antioxidase, that is traR, traI, virA, virG, and sodB. Results indicated that AHPE exposure lead to a significant downregulation in the expressions of traR and traI by about 53 and 40%, respectively. Similarly, the expressions of virA, virG, and sodB, three genes involved in the synthesis of virulence factors and superoxide dismutase, were also notably reduced after exposure to AHPE (Figure 8). Therefore, we speculated that the downregulated expressions of these QS and virulence-related genes would inevitably lead to the reduced pathogenicity of A. tumefaciens C58.
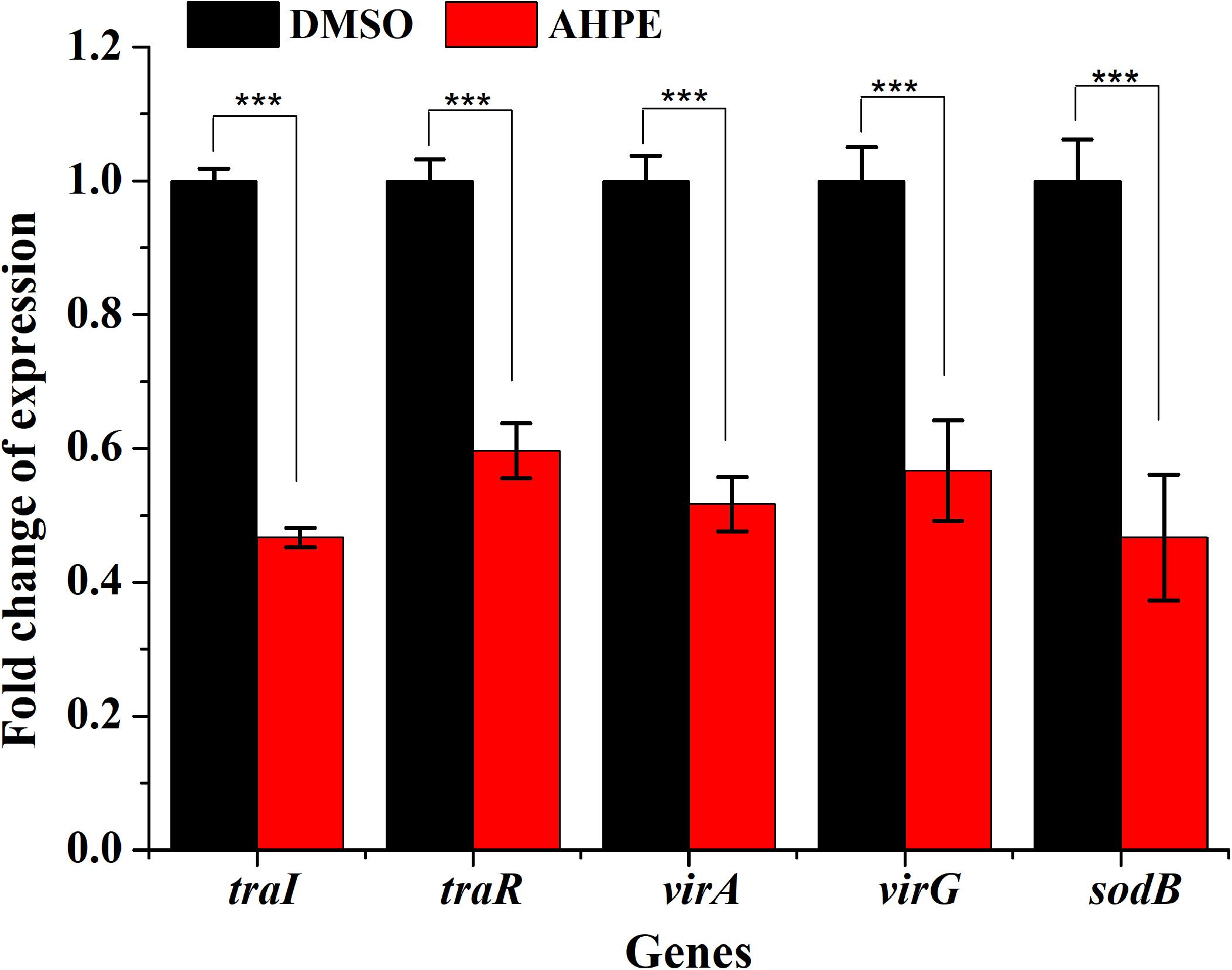
Figure 8. Effect of AHPE on expressions of genes involved in QS, virulence and antioxidant enzyme. ***p < 0.001 versus the DMSO control.
The effect of AHPE on tobacco stem infection was investigated. As shown in Figure 9A, exposure to AHPE at 25 and 50 μg/mL significantly inhibited the formation of crown galls caused by A. tumefaciens C58. Quantification analysis indicated that the weight of crown galls was remarkably decreased after treatment with AHPE compared to the salicylic acid and DMSO-treated groups (Figure 9B). The result indicated that AHPE has the potential to function as a pesticide in preventing A. tumefaciens from infecting tobacco plants at seedling stage.
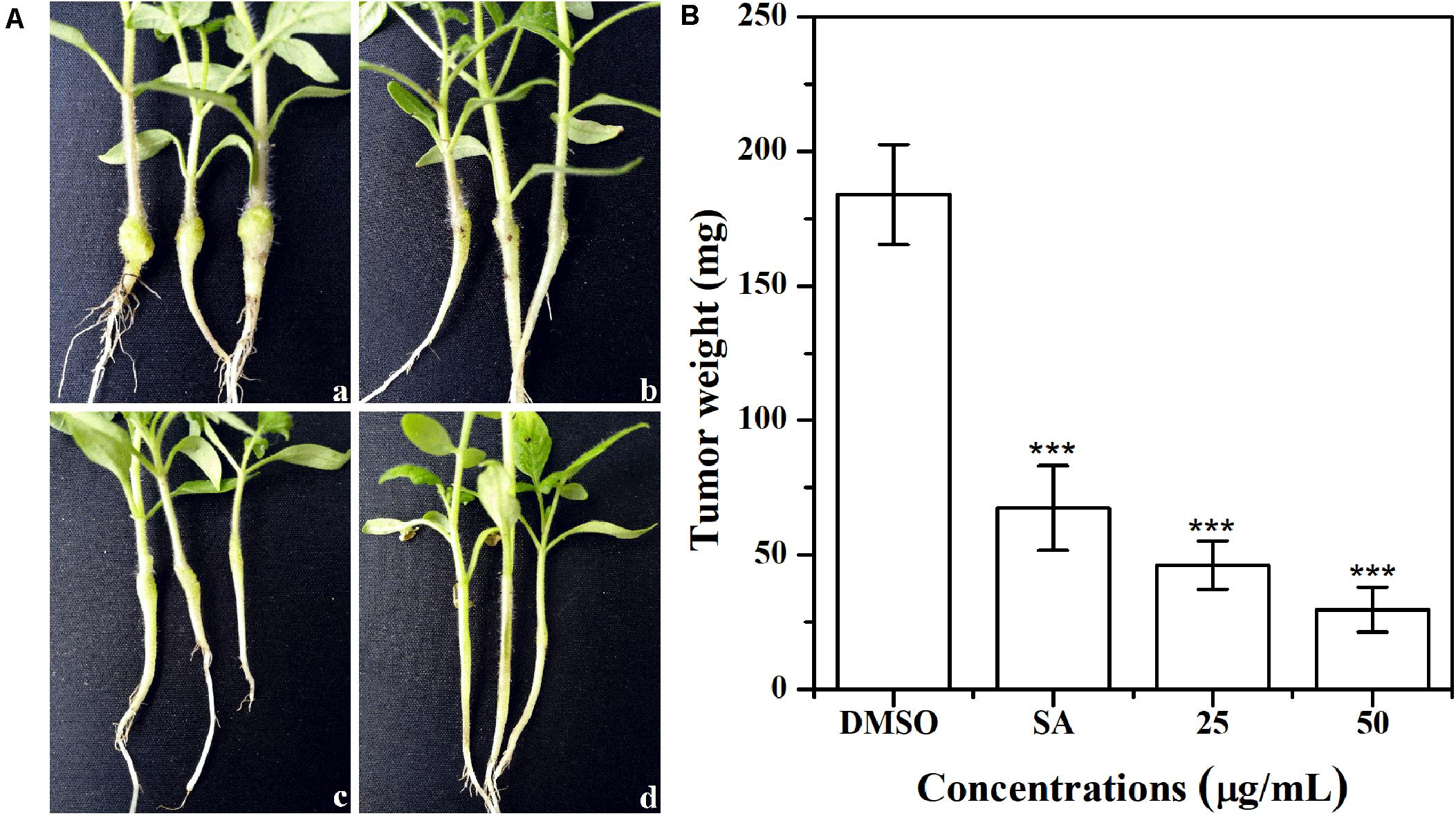
Figure 9. Effect of AHPE on tobacco plant infections. (A) Inoculation with A. tumefaciens C58 treated with (a) DMSO, (b) salicylic acid (15 μg/mL), (c) 25 μg/mL of AHPE, and (d) 50 μg/mL of AHPE, respectively. (B) Quantification of crown gall weight treated with or without AHPE. ***p < 0.001 versus DMSO or resveratrol-treated control.
Agrobacterium tumefaciens is a soil-born phytopathogen that can cause crown gall disease in more than 1000 species of dicots and certain monocots. Crown gall disease sometimes will result in significant economic losses by bringing disastrous crop failure. During the infection, T-DNA is transferred from A. tumefaciens plasmid into plants and integrated into plant genome (Hao et al., 2007). And then, the integrated genes induce the synthesis of auxin and cytokinin, disrupt the balance of plant hormones, lead to the formation of tumor, and ultimately result in the death of plants. Studies have shown that conjugal transfer (tra) genes were mediated by QS (Fuqua and Winans, 1994). Therefore, interfering with the QS would be an efficient strategy for attenuating the virulence of A. tumefaciens. In this study, a natural compound, 1-(4-amino-2-hydroxyphenyl)ethanone (AHPE), was firstly isolated from the metabolites of P. liquidambari S47. However, the anti-QS and anti-virulence activities of AHPE against A. tumefaciens have not been documented. Here, AHPE was evaluated for its potential to inhibit the QS and relative virulence of A. tumefaciens. AHPE showed a concentration-dependent reduction in β-galactosidase activity and AHL production, thus indicating the breakdown of QS in A. tumefaciens. AHPE also disturbed the metabolic profiles of A. tumefaciens as evidenced by the altered metabolites involved in oxidative stress, energy metabolism, protein synthesis and nucleotide metabolism.
The effect of AHPE on QS was firstly evaluated by determining β-galactosidase activity using A. tumefaciens A136. Results showed significantly reduced β-galactosidase activity after treatment with AHPE, thus indicating the breakdown of QS in A. tumefaciens. As AHL binds to the receptor TraR and subsequently activates the expressions of the lacZ reporter gene (McLean et al., 1997), we then quantified the production of AHL secreted by A. tumefaciens. Results indicated that AHPE exposure resulted in remarkable repression on 3-oxo-C8-HSL secretion. We subsequently used qRT-PCR to determine the transcriptional levels of QS-correlated genes. The expressions of traI and traR were significantly repressed compared with the untreated control. The results mentioned above combined with the docking analysis demonstrated that the breakdown of QS might be obtained by inhibiting the transcriptional levels of traI and traR rather than signal mimicry.
Previous study has shown that the virulence factors of A. tumefaciens are encoded by the conjugative Ti plasmid, the dissemination of which is controlled by a QS signaling based on the synthesis and perception of 3-oxo-C8-HSL (Lang and Faure, 2014). The suppression of 3-oxo-C8-HSL production would inevitably result in the reduced virulence in A. tumefaciens. Results from the present study showed that the swimming motility, chemotaxis, and flagella formation were significantly inhibited after exposure to AHPE. The results were in line with the decreased AHL level and repressed gene expressions, which further confirmed the dysfunctional QS of A. tumefaciens. It has been proven that the AHL signal 3-oxo-C8-HSL can bind to the TraR protein and the formed TraR-AHL then activates genes controlling Ti plasmid replication and conjugal transfer (Chen et al., 2004). The suppression of TraR and 3-oxo-C8-HSL secretion with AHPE treatment would result in the inhibition on Ti plasmid replication and conjugal transfer, and consequently lead to the weakened pathogenicity of A. tumefaciens. This has been evidence by the reduced crown gall disease after inoculation with A. tumefaciens. The pathogenicity of A. tumefaciens might be also attenuated by the inhibited motility and flagella formation. Additionally, the mediation of plasmid copy number is a complex process that might be influenced by different physiological states (Cho and Winans, 2005; Pappas, 2008; Lang and Faure, 2014).
1H NMR-based analysis demonstrated that the metabolism of A. tumefaciens was prominently disturbed with AHPE exposure. Among these changed metabolites, some were correlated with oxidative stress, quorum sensing, and protein synthesis including putrescine, arginine, isoleucine, leucine, valine, choline, and dimethylamine. Putrescine can be synthesized from arginine via arginine decarboxylase (Nakada and Itoh, 2003). The decreased arginine would inevitably result in reduced level of putrescine. It has been proven that putrescine production was significantly enhanced after supplementation with QS signals (Zhu et al., 2016). The decreased putrescine was in accordance with the decreased 3-oxo-C8-HSL level. Isoleucine, leucine, and valine are branched-chain amino acids which play important roles in maintaining cell physiological function (Zhou et al., 2019). The significant reduction of these metabolites indicated that the protein synthesis might be disordered due to the breakdown of QS in A. tumefaciens. Choline is one of the components of phospholipids and is involved in maintaining membrane integrity (Zhou et al., 2019). The decreased choline indicated the increased oxidative stress after AHPE treatment. To rebalance the redox level inside the cell and repair the damaged membrane caused by oxidative stress, the choline was heavily consumed. The decomposition of choline resulted in the enhanced level of dimethylamine as dimethylamine was the downstream metabolite of choline and used as carbon and nitrogen sources for cell growth (Dumas et al., 2006). The increased dimethylamine implied the disorder of protein synthesis and energy supply caused by oxidative stress.
Glutamate is crucial for glutathione (GSH) synthesis and GSH is an important redox substance for maintaining the normal reduction state of cells. The increased glutamate level indicated the intensified oxidative stress caused by AHPE. To counteract oxidative stress and reduce cell damage, more glutamate needs to be produced for GSH synthesis. In addition, betaine, one important antioxidant in organism, is essential for eliminate free radicals and maintaining membrane integrity (Schmidley, 1990). The decrease of betaine indicated the increase of free radicals. To repair the damage caused by free radicals and maintain the normal function of cells, betaine would be consumed in large quantities. In addition, the transcriptional level of sodB encoding SOD was significantly repressed. The decreased activity of SOD would further aggravate the oxidative stress, resulting in the disorder of physiological function of A. tumefaciens.
AHPE also induced disorder of energy metabolism in A. tumefaciens, which was evidenced by the marked increase of succinate. Succinate is an important intermediate of the tricarboxylic acid (TCA) cycle (Chen et al., 2017). The increase of succinate would inevitably result in the disorder of TCA cycle. As the most important energy source, the disorder of TCA cycle would inevitably lead to the disturbance of energy supply and eventually change the physiological function of cells. Because the energy supply was disturbed, other pathways such as anaerobic respiration would be enhanced for compensation, which could be confirmed by the significant increase in acetate. This result was in line with the research by Ringø (Ringo et al., 1984). It has been proven that the acceleration of anaerobic respiration could result in the reduced pathogenicity in organism (Lee et al., 2011). Such an effect was also confirmed by the motility assay and tobacco infection assay in this study.
In comparison with the untreated control, the levels of adenosine and AMP were notably increased. Adenosine was transformed from AMP by dephosphorylation, while AMP was derived from cyclic AMP (cAMP) by cAMP-specific phosphodiesterase and cyclic 3′,5′-phosphodiesterase (Zhou et al., 2019). The increase of AMP may be attributed to the excessive decomposition of cAMP. NADP+ was involved in redox reaction, reductants synthesis, and energy supply (Ying, 2008). The decrease of NADP+ indicated that the oxidative stress was enhanced. To counteract the oxidative stress, NADP+ had to be overconsumed.
In conclusion, AHPE treatment resulted in the dysfunction of QS in A. tumefaciens most likely by repressing the transcriptional levels of traI and traR. AHPE exposure also inhibited swimming motility, chemotaxis, and flagella formation, and lead to enhanced oxidative stress. The enhanced oxidative stress resulted in the disorder of energy metabolism, amino acid metabolism, and nuclear acid metabolism, and ultimately weakened the virulence of A. tumefaciens. Therefore, AHPE may act as a potential pesticide in preventing A. tumefaciens from infecting tobacco plants or other plant species at seedling stage.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
J-WZ, A-QJ, X-JT, and E-QL conceived and designed the experiments. J-WZ, HC, and BS performed the experiments. J-WZ, T-ZH, YH, and P-LL analyzed the data. J-WZ, A-QJ, X-JT, and E-QL wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grants from the National Natural Science Foundation of China (32000091 and 41766006), National Key Research and Development Program of China (2017YFD0201401), Research Starting Fund of Xuzhou University of Technology (02900142), and Cultivation Fund of Xuzhou University of Technology (00700001).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.584767/full#supplementary-material
Supplementary Figure 1 | Chemical structure of AHPE.
Supplementary Table 1 | PCR Primers for qRT-PCR.
Chen, G., Malenkos, J. W., Cha, M. R., Fuqua, C., and Chen, L. (2004). Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52, 1641–1651. doi: 10.1111/j.1365-2958.2004.04110.x
Chen, T., Sheng, J., Fu, Y., Li, M., Wang, J., and Jia, A. Q. (2017). 1H NMR-based global metabolic studies of Pseudomonas aeruginosa upon exposure of the quorum sensing inhibitor resveratrol. J. Proteome Res. 16, 824–830. doi: 10.1021/acs.jproteome.6b00800
Chevrot, R., Rosen, R., Haudecoeur, E., Cirou, A., Shelp, B. J., Ron, E., et al. (2006). GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 103, 7460–7464. doi: 10.1073/pnas.0600313103
Cho, H., and Winans, S. C. (2005). VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc. Natl. Acad. Sci. U.S.A. 102, 14843–14848. doi: 10.1073/pnas.0503458102
Clinical and Laboratory Standards Institute (CLSI) (2015). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard 10th Edn, M07-A10. Wayne, PA: CLSI.
Dandurishvili, N., Toklikishvili, N., Ovadis, M., Eliashvili, P., Giorgobiani, N., Keshelava, R., et al. (2011). Broad-range antagonistic rhizobacteria Pseudomonas fluorescens and Serratia plymuthica suppress Agrobacterium crown gall tumours on tomato plants. J. Appl. Microbiol. 110, 341–352. doi: 10.1111/j.1365-2672.2010.04891.x
DeCleene, M., and DeLey, J. (1976). The host range of crown gall. Bot. Rev. 42, 389–466. doi: 10.1007/bf02860827
Ding, Z., and Christie, P. J. (2003). Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185, 760–771. doi: 10.1128/jb.185.3.760-771.2003
Dumas, M. E., Barton, R. H., Toye, A., Cloarec, O., Blancher, C., Rothwell, A., et al. (2006). Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U.S.A. 103, 12511–12516. doi: 10.1073/pnas.0601056103
Erhardt, M. (2016). Strategies to block bacterial pathogenesis by interference with motility and chemotaxis. Curr. Top. Microbiol. Immunol. 398, 185–205. doi: 10.1007/82_2016_493
Fuqua, C., Winans, S. C., and Greenberg, E. P. (1996). Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751. doi: 10.1146/annurev.micro.50.1.727
Fuqua, W. C., and Winans, S. C. (1994). A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176, 2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/mmbr.67.1.16-37.2003
Gohlke, J., and Deeken, R. (2014). Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 5:155. doi: 10.3389/fpls.2014.00155
Guo, M., Huang, Z., and Yang, J. (2017). Is there any crosstalk between the chemotaxis and virulence induction signaling in Agrobacterium tumefaciens? Biotechnol. Adv. 35, 505–511. doi: 10.1016/j.biotechadv.2017.03.008
Hao, Y., Charles, T. C., and Glick, B. R. (2007). ACC deaminase from plant growth-promoting bacteria affects crown gall development. Can. J. Microbiol. 53, 1291–1299. doi: 10.1139/W07-099
Lang, J., and Faure, D. (2014). Functions and regulation of quorum-sensing in Agrobacterium tumefaciens. Front. Plant Sci. 5:14. doi: 10.3389/fpls.2014.00014
Lang, J., Gonzalez-Mula, A., Taconnat, L., Clement, G., and Faure, D. (2016). The plant GABA signaling downregulates horizontal transfer of the Agrobacterium tumefaciens virulence plasmid. N. Phytol. 210, 974–983. doi: 10.1111/nph.13813
Lee, H., Humann, J. L., Pitrak, J. S., Cuperus, J. T., Parks, T. D., Whistler, C. A., et al. (2003). Translation start sequences affect the efficiency of silencing of Agrobacterium tumefaciens T-DNA oncogenes. Plant Physiol. 133, 966–977. doi: 10.1104/pp.103.026534
Lee, K. M., Yoon, M. Y., Park, Y., Lee, J. H., and Yoon, S. S. (2011). Anaerobiosis-induced loss of cytotoxicity is due to inactivation of quorum sensing in Pseudomonas aeruginosa. Infect. Immun. 79, 2792–27800. doi: 10.1128/IAI.01361-10
Liu, J., Li, H., Miao, M., Tang, X., Giovannoni, J., Xiao, F., et al. (2012). The tomato UV-damaged DNA-binding protein-1 (DDB1) is implicated in pathogenesis-related (PR) gene expression and resistance to Agrobacterium tumefaciens. Mol. Plant Pathol. 13, 123–134. doi: 10.1111/j.1364-3703.2011.00735.x
McLean, R. J., Whiteley, M., Stickler, D. J., and Fuqua, W. C. (1997). Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154, 259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x
Merritt, P. M., Danhorn, T., and Fuqua, C. (2007). Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189, 8005–8014. doi: 10.1128/JB.00566-07
Molina, L., Constantinescu, F., Michel, L., Reimmann, C., Duffy, B., and Défago, G. (2003). Degradation of pathogen quorum-sensing molecules by soil bacteria: a preventive and curative biological control mechanism. FEMS Microbiol. Ecol. 45, 71–81. doi: 10.1016/S0168-6496(03)00125-9
Moore, L. W., and Warren, G. (1979). Agrobacterium radiobacter strain K84 and biological control of crown gall. Annu. Rev. Phytopathol. 17: 163–179. gall of grape: biology and disease management. Annu. Rev. Phytopathol. 37, 53–80.
Morin, D., Grasland, B., Vallee-Rehel, K., Dufau, C., and Haras, D. (2003). On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002, 79–92. doi: 10.1016/s0021-9673(03)00730-1
Nakada, Y., and Itoh, Y. (2003). Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 149(Pt 3), 707–714. doi: 10.1099/mic.0.26009-0
Pappas, K. M. (2008). Cell-cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid 60, 89–107. doi: 10.1016/j.plasmid.2008.05.003
Pappas, K. M., and Winans, S. C. (2003). A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plamsid replication genes. Mol. Microbiol. 48, 1059–1073. doi: 10.1046/j.1365-2958.2003.03488.x
Ringo, E., Stenberg, E., and Strom, A. R. (1984). Amino acid and lactate catabolism in trimethylamine oxide respiration of Alteromonas putrefaciens NCMB 1735. Appl. Environ. Microbiol. 47, 1084–1089. doi: 10.1128/aem.47.5.1084-1089.1984
Schell, J., Van Montagu, M., De Beuckeleer, M., De Block, M., Depicker, A., De Wilde, M., et al. (1979). Interactions and DNA transfer between Agrobacterium tumefaciens, the Ti-plasmid and the plant host. Proc. R. Soc. Lond. Biol. Sci. 204, 251–266. doi: 10.1098/rspb.1979.0026
Schmidley, J. W. (1990). Free radicals in central nervous system ischemia. Stroke 21, 1086–1090. doi: 10.1161/01.str.21.7.1086
Stachel, S. E., An, G., Flores, C., and Nester, E. W. (1985). A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. Embo J. 4, 891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x
Ying, W. (2008). NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 10, 179–206. doi: 10.1089/ars.2007.1672
Zhang, L., Murphy, P. J., Kerr, A., and Tate, M. E. (1993). Agrobacterium conjugation and gene regulation by N-acyl homoserine lactones. Nature 362, 446–448. doi: 10.1038/362446a0
Zhou, J. W., Hou, B., Liu, G. Y., Jiang, H., Sun, B., Wang, Z. N., et al. (2018a). Attenuation of Pseudomonas aeruginosa biofilm by hordenine: a combinatorial study with aminoglycoside antibiotics. Appl. Microbiol. Biotechnol. 102, 9745–9758. doi: 10.1007/s00253-018-9315-8
Zhou, J. W., Luo, H. Z., Jiang, H., Jian, T. K., Chen, Z. Q., and Jia, A. Q. (2018b). Hordenine: a novel quorum sensing inhibitor and antibiofilm agent against Pseudomonas aeruginosa. J. Agric. Food Chem. 66, 1600–1628. doi: 10.1021/acs.jafc.7b05035
Zhou, J. W., Muhammad, J., Sun, B., Yang, R., Wadood, A., Wang, J. S., et al. (2019). Metabolomic analysis of quorum sensing inhibitor hordenine on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 103, 6271–6285. doi: 10.1007/s00253-019-09878-w
Zhu, J., and Winans, S. C. (1999). Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. U.S.A. 96, 4832–4837. doi: 10.1073/pnas.96.9.4832
Keywords: Agrobacterium tumefaciens, quorum sensing, 1-(4-amino-2-hydroxyphenyl)ethanone, metabolomics, pathogenicity
Citation: Zhou J-W, Jia A-Q, Tan X-J, Chen H, Sun B, Huang T-Z, He Y, Li P-L and Liu E-Q (2020) 1-(4-Amino-2-Hydroxyphenyl)Ethenone Suppresses Agrobacterium tumefaciens Virulence and Metabolism. Front. Microbiol. 11:584767. doi: 10.3389/fmicb.2020.584767
Received: 20 July 2020; Accepted: 20 October 2020;
Published: 12 November 2020.
Edited by:
Marco Scortichini, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Randy Ortiz-Castro, National Council of Science and Technology (CONACYT), MexicoCopyright © 2020 Zhou, Jia, Tan, Chen, Sun, Huang, He, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-Qun Jia, amlhYWlxdW5AZ21haWwuY29t; Xiao-Juan Tan, eGp0YW5AYWhudS5lZHUuY24=; En-Qi Liu, bGVxQHh6aXQuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.