- 1Institute for Frontier Life and Medical Sciences, Kyoto University, Kyoto, Japan
- 2Hakubi Center for Advanced Research, Kyoto University, Kyoto, Japan
- 3Department of Virology, Tohoku University Graduate School of Medicine, Sendai, Japan
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a worldwide pandemic. Many projections concerning the outbreak, such as the estimated number of cases and deaths in upcoming months, have been made available. However, what happens to the virus after the pandemic subsides has not been fully explored. In this article, we discuss the ways that past and present human viruses have emerged via zoonotic transmission, the mechanisms that they have acquired the ability for effective transmission among humans, the process to sustain a chain of transmission to coexist with humans, and the factors important for complete containment leading to eradication of viruses. These aspects of viral disease may provide clues for the future path that SARS-CoV-2 might take in relation to human infection.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported from China in December 2019. As of August 2020, the world is gripped by a pandemic of the virus, with numbers of cases and deaths increasing globally. People are trying to reduce new infections by non-pharmaceutical intervention (Flaxman et al., 2020; Hsiang et al., 2020; Lai et al., 2020). A global research effort is underway for the development of specific remedies and vaccines for the virus (European Centre for Disease Prevention and Control, 2020; Wiersinga et al., 2020). Many projections concerning the outbreak, such as the estimated number of cases and deaths in upcoming months, have been made available (Holmdahl and Buckee, 2020; Jewell et al., 2020; Metcalf et al., 2020). However, what happens to the virus after the pandemic subsides has not been fully explored. What can we expect in the future, and will the virus coexist with us or go extinct? (Figure 1).
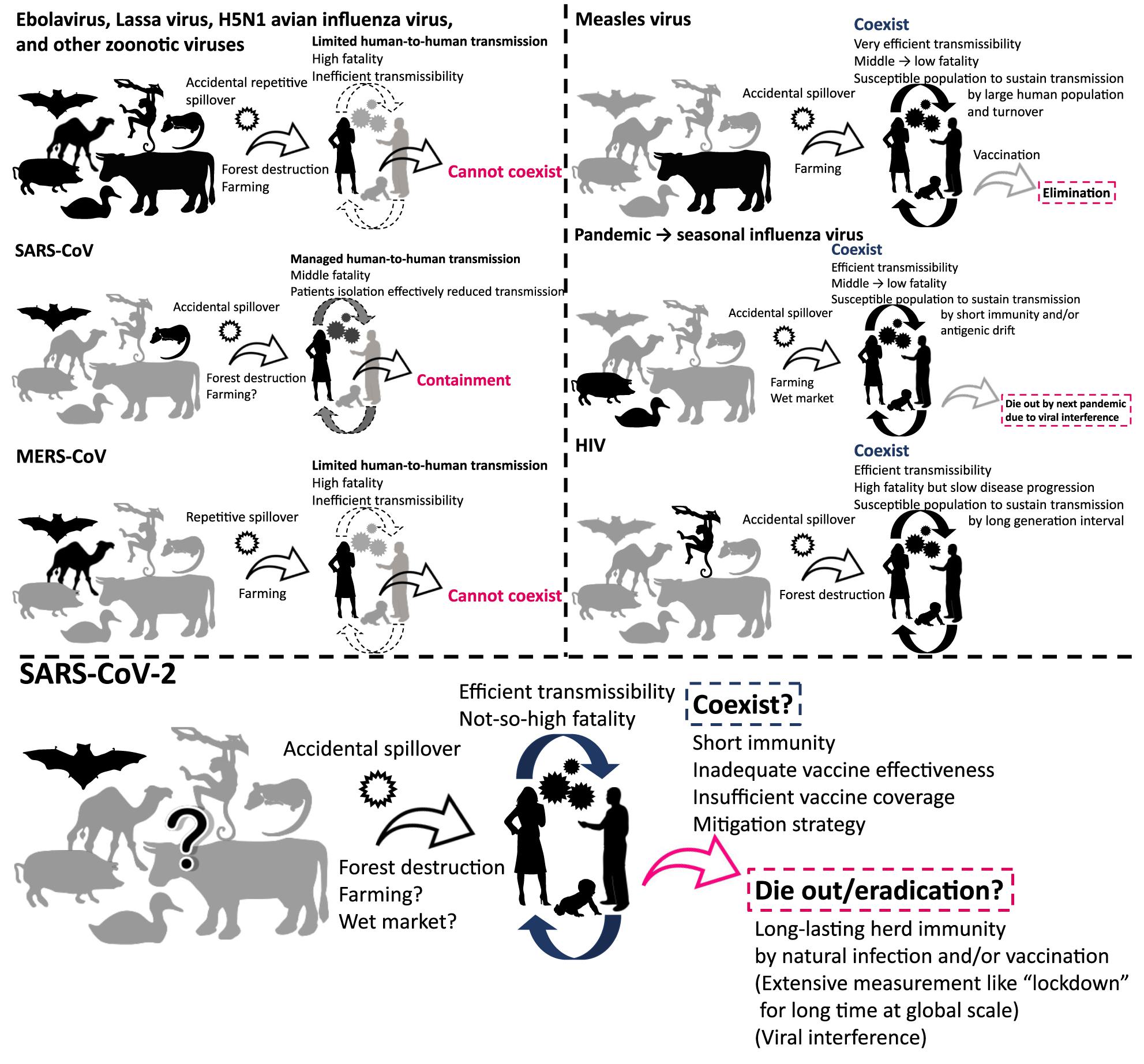
Figure 1. Viruses that cannot coexist and that have coexisted with humans. The figure illustrates schematic overview of the history of emergence, adaptation, transmission, and consequence of viruses.
SARS-CoV-2 is believed to have been introduced by zoonotic transmission (Lu et al., 2020; Wu et al., 2020; Zhou et al., 2020). Zoonosis, which is identified as a disease transmitted from animals to humans, is a major route of introduction of new viruses to humans. Zoonotic viruses were repeatedly introduced to people as a spillover from natural hosts. However, many of them cannot sustain a chain of transmission in human populations for extended periods. On the other hand, some viruses, such as pandemic influenza virus and human immunodeficiency virus (HIV), have adapted to humans after zoonotic introduction, acquiring the ability to sustain human-to-human transmissions (Wasik et al., 2019). In this article, we provide an overview and discuss how past and present human viruses emerged, spread, and persisted or disappeared after zoonotic introduction. These aspects of viral disease may provide clues for the future path that SARS-CoV-2 might take in relation to human infection.
Zoonotic Transmission, Adaptation to Humans, and Eradication of Viruses
The chance of zoonotic transmission of virus increases by frequent contact between humans and animals. This phenomenon could be partially attributed to forest destruction for urban development and exploitation of natural resources (Figure 1; Kessler et al., 2018; Gibb et al., 2020). The emergence of HIV and repetitive spillover of Ebolavirus might be caused by such activities (Whiteside and Zebryk, 2015). The transmission of simian immunodeficiency virus to humans and its subsequent adaptation led to the generation of HIV (Sharp and Hahn, 2011). Farming is an obvious occupation where human-animal contact is likely to occur on a regular basis. Zoonotic transmission of animal viruses from farm animals to humans may have resulted in the emergence of pandemic influenza and measles viruses (Webster et al., 1992; Düx et al., 2020). In the past ∼100 years, we witnessed the emergence of pandemic influenza caused by introductions of novel viruses from avian or swine species (Webster et al., 1992): Spanish flu in 1918, Asian flu in 1957, Hong Kong flu in 1968, and “Swine flu (H1N1pdm09 virus)” in 2009. The descendant viruses of Hong Kong flu and Swine flu now coexist in human communities as seasonal influenza (Morens and Taubenberger, 2011; Taubenberger et al., 2019). Measles virus was introduced by (relatively) ancient zoonotic transmission that has acquired its exclusive circulation in humans; phylogenetic analyses suggest that a bovine virus was transmitted to humans 1,000–2,500 years ago, leading to its adaptation as measles virus (Furuse et al., 2010; Düx et al., 2020). In addition, wet market selling of live animals is often recognized as the source of zoonotic virus transmission to humans (Cowling et al., 2013). Modern-day extensive forest destruction and large-scale farming likely increase the risk of zoonotic transmission of novel viruses to humans (Dobson et al., 2020; Gibb et al., 2020). Further, urbanization and globalization of today’s world make outbreaks that could have been contained in a local area easily and rapidly spared to other parts of world (Neiderud, 2015; Wu et al., 2017). One health approach including surveillance system to detect outbreaks of unknown disease in humans and animals at a global level must be important for better preparedness in future (Watsa, 2020).
Establishing infection in new hosts often requires numerous adaptive changes, such as receptor-specificity adjustments (Li et al., 2005; Yamada et al., 2006; Taubenberger and Kash, 2010), optimizing the compatibility with host’s factors (Long et al., 2016), and overcoming host antiviral defenses (Sawyer et al., 2004; Rajsbaum et al., 2012; Sauter and Kirchhoff, 2019). For example, when avian influenza virus adapted to humans, mutations in the HA gene altered avidity for cellular receptor from avian-type to human-type, mutations in the PB2 gene increased viral polymerase activity in human cells by interaction with a host’s factor, ANP32A, and mutations in the NS1 gene regulated innate immunity in a species-specific manner (Long et al., 2019). Detailed virological and physiological mechanisms of genetic mutations that affect adaptation to the new hosts are reviewed in Wasik et al. (2019); Letko et al. (2020). Also, we explore another aspect of farming that might contribute to zoonotic transmission. During viral replication in host cells, genetic mutations are randomly induced into viral genomes (Bordería et al., 2011). Yet, generated genetic diversity cannot be effectively transmitted from infector to infectee due to the so-called bottleneck effect (Domingo et al., 1996). However, when farm animals are raised in densely crowded conditions, an inter-host genetic bottleneck may be loosened (Domingo and Holland, 1997). Virus with mutations that enhance infectivity in humans may be induced and transmitted under such conditions, increasing the chances that a mutant virus will establish infection in a person.
After introduction to humans, viruses must sustain a chain of transmission to coexist as “human viruses” (Figures 1, 2). First, transmission should be efficient enough that the reproduction number, the average number of secondary cases per infectious case, is >1 (Delamater et al., 2019). This ability might be closely tied to achieving efficient replication ability in new hosts via genetic mutations. Second, viruses should not consume susceptible hosts; a certain fraction of susceptible hosts is needed to sustain transmission. Measles virus is known for great transmission efficiency due to air-borne transmission (Guerra et al., 2017; National Center for Immunization and Respiratory Diseases, 2017). When a virus is highly transmissible, shows a short generation interval, and induces life-long immunity; size of host population and turnover are important for sustaining viral transmission. Mathematical models show that a population of 250,000–500,000 is needed to maintain viral transmission (Black, 1966). For influenza virus, short-lived acquired immunity and viral evolution called antigenic drift are considered to allow escape from host immunity (Ferguson et al., 2003; Smith et al., 2004) although the duration of protective immunity against the virus is controversial (Couch and Kasel, 1983; Xia et al., 2005; Kucharski et al., 2015). These properties encourage sustained transmissions by creating an adequate proportion of susceptible hosts. Long generation interval due to chronic infection is considered another way that viruses maintain a population of susceptible hosts, as exemplified by HIV (Gran et al., 2008). The virus is known for its long latent phase followed by slow disease progression to acquired immunodeficiency syndrome, which takes more than a decade. Third, viruses should not kill too many infected individuals to maintain an ecologically fit life cycle. Notably, mortality of newly introduced viruses can decrease after a certain period of circulation as the pandemic influenza transforms into a seasonal influenza, possibly due to immune response to reinfection and viral adaptation in humans in post-pandemic epidemics (Barry et al., 2008; Dawood et al., 2012; Lin et al., 2012). Similarly, mortalities of measles and smallpox are reportedly high in naïve populations, but less in populations where such viruses coexist (Elwood, 1989; Shanks et al., 2011).
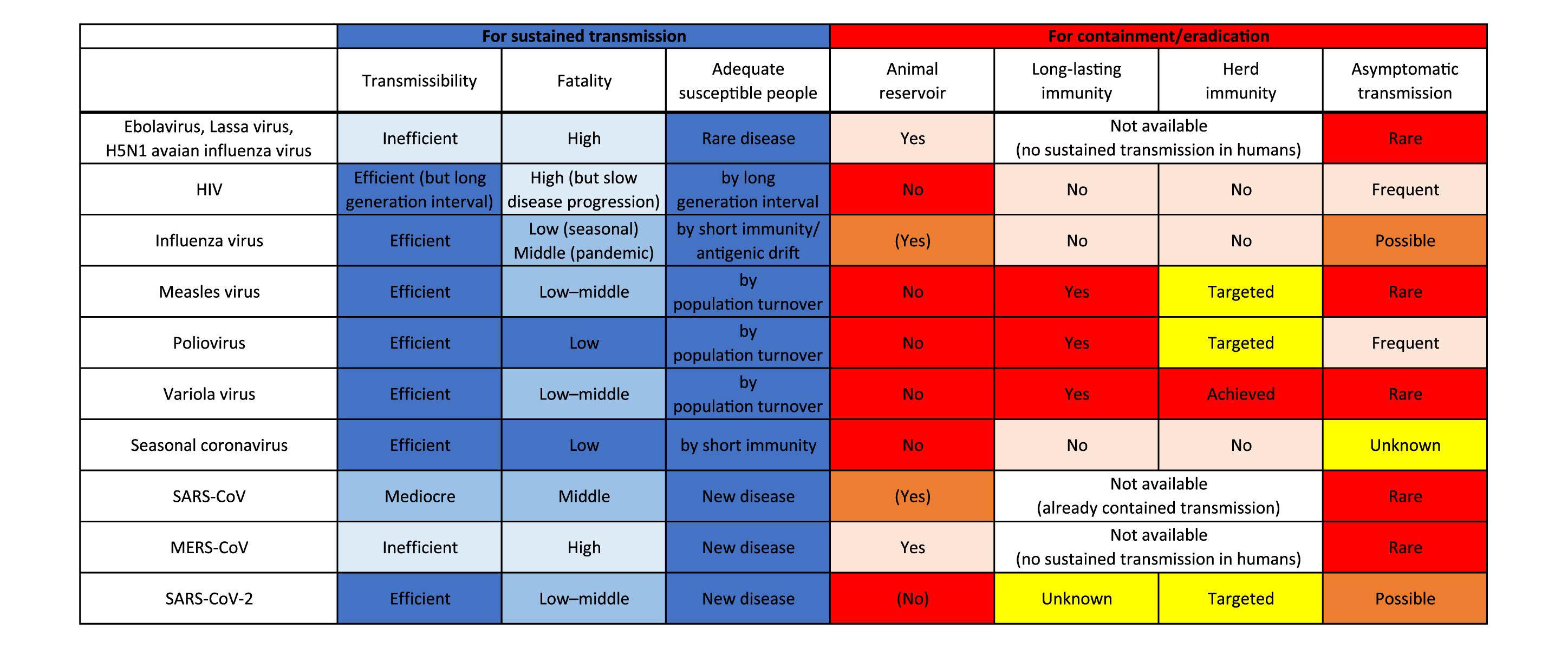
Figure 2. Factors responsible for sustained transmission and containment/eradication of viruses. Deeper blue colors indicate more adapted characteristics for sustained transmission in humans, and deeper red colors indicate more favorable features for containment/eradication of viruses.
In contrast, zoonotic viruses, such as Ebolavirus, Lassa virus, and H5N1 avian influenza virus, have not adapted well enough to humans to achieve effective transmission. Host susceptibility is inadequate, viral replication ability is inefficient, and/or fatality is too high. Case fatality rates of these viral zoonoses in the accidental host, humans, were extraordinarily high, ranging over 10–90% (Heeney, 2006). Such severe illness reduces the chance of transmission by death, hospitalization, and decreased mobility of patients. Inefficient transmission can also be attributed to the mode of transmission such as blood- (and other body fluids-) borne transmission for some viruses including Ebolavirus (Chowell and Nishiura, 2014). These factors result in failure to establish a sustained chain of transmission (Figures 1, 2).
Viruses that once established sustained human-to-human transmission can disappear. A good example is a replacement of previously circulating seasonal influenza virus by newly emerged pandemic influenza virus. When a human-adapted influenza virus emerged after zoonotic transmission of avian or swine influenza virus as a pandemic virus, the seasonal human influenza virus that had been circulating before will disappear. Then, the pandemic virus becomes the “new” seasonal human influenza virus (Webster et al., 1992). This phenomenon is likely due to interference between the viruses (Palese and Wang, 2011; Furuse and Oshitani, 2016). Vaccination also can eradicate viruses. Smallpox variola virus has been identified as the first and currently the only human virus eradicated by vaccination. Meanwhile, measles virus and poliovirus remain to be the target for eradication by vaccination. Several important factors are required for successful eradication: (1) virus should circulate only among humans; that is, no animal reservoir can exist; (2) vaccine should produce a long-lasting immunity; (3) vaccination coverage should be high enough to achieve herd immunity at a global scale; and (4) most infections should cause symptomatic illness. Variola virus was able to meet all the four conditions, but unmet factors (3) and (4) for poliovirus and unmet factor (3) for measles virus make the ongoing effort to eradicate them challenging (Figure 2; Kew et al., 2005; Moss and Strebel, 2011; Furuse and Oshitani, 2017; Chard et al., 2020).
Coronaviruses Including SARS-CoV-2
Seasonal human coronaviruses, including 229E, OC43, NL63, and HKU1, can cause common cold-like respiratory symptoms, circulating exclusively in humans. Members of the family Coronaviridae, however, can be found in both humans and other animals (Vijaykrishna et al., 2007). Hence, an ancient zoonotic origin of seasonal human coronaviruses is likely (Forni et al., 2017). Severe acute respiratory syndrome coronavirus (SARS-CoV) caused a multi-country outbreak in 2003. Its closely related virus was found in bats, and civets were considered as an intermediate host for transmission of the virus from bats to humans (Shi and Hu, 2008). Viral infection was sustained by sequential human-to-human transmissions (Anderson et al., 2004). The virus is often found to cause severe illness, and transmissibility becomes high at the time when patients develop severe symptoms (Anderson et al., 2004). Thanks to these characteristics, sensitive detection and timely isolation of patients, along with appropriate standard precautions in nosocomial settings, can effectively reduce new transmission (Fraser et al., 2004). As a result, the outbreak was contained within a year (Figures 1, 2). Middle East respiratory syndrome coronavirus (MERS-CoV) was introduced by zoonotic transmission from camels; the camel virus was reported to be likely originated from a bat virus (Mohd et al., 2016). Whereas some instances of human-to-human transmission of the virus did occur within households and in nosocomial settings, overall transmission was not sufficient to sustain transmission in humans (Figures 1, 2; Raj et al., 2014). Still, the high prevalence of the virus in domesticated camels caused repetitive spillover of the virus (Azhar et al., 2014).
SARS-CoV-2 is also believed to have come from a bat virus (Boni et al., 2020; Lu et al., 2020; Wu et al., 2020; Zhou et al., 2020). Although viruses close to SARS-CoV-2 were discovered in pangolins (Lam et al., 2020; Xiao K. et al., 2020; Zhang et al., 2020), determining the intermediate host between bats and humans remains elusive (Boni et al., 2020; Han, 2020; Li et al., 2020; Liu P. et al., 2020). The secondary attack rate and therefore reproduction number of SARS-CoV-2 in humans are found to be much higher than those of SARS-CoV and MERS-CoV (Jing et al., 2020; Kucharski et al., 2020; Liu Y. et al., 2020; Madewell et al., 2020). Even transmission via aerosol of SARS-CoV-2 is possible (Morawska and Milton, 2020; Van Doremalen et al., 2020). The case-fatality is relatively low for SARS-CoV-2 (∼1%) compared with those of SARS-CoV (∼15%) and MERS-CoV (∼30%) (Mahase, 2020a; Peeri et al., 2020). These characteristics of SARS-CoV-2 appear to be typical features of human viruses (Figure 2).
Unfortunately, it seems unlikely that SARS-CoV-2 will disappear naturally or by current intervention strategies, including isolation of patients, quarantine of contacts, physical distancing, and standard precautions including wearing a mask and hand washing. It is possible to contain the outbreak at local level; some countries or regions were able to reduce their new cases to almost zero by extensive measures such as “lockdown” and strict “border control” (Baker M. et al., 2020; Erkhembayar et al., 2020; Wang C. J. et al., 2020; Wu and McGoogan, 2020). However, these interventions cannot be sustained for months or years worldwide (Chowdhury et al., 2020; Weill et al., 2020). Existence of asymptomatic and presymptomatic transmission makes public health intervention difficult (Fraser et al., 2004). Given high transmissibility (Kucharski et al., 2020; Liu Y. et al., 2020) and a high proportion of asymptomatic infections (Day, 2020; Oran and Topol, 2020), a subsequent wave of infections seems inevitable (Baker R. E. et al., 2020; Ferguson et al., 2020; Kissler et al., 2020; López and Rodó, 2020). As we discussed, a virus should avoid consumption of a susceptible population for sustained transmission. Ironically, mitigation strategies (i.e., “flattening the curve”) can ensure a susceptible uninfected population for a longer time, perhaps years (Kissler et al., 2020; Stewart et al., 2020). Seroprevalence against the virus is still low even in heavily affected areas by July 2020 (Eckerle and Meyer, 2020; Havers et al., 2020; Pollán et al., 2020; Stringhini et al., 2020). Further, regardless of mitigation, the virus can sustain transmission when immunity against it does not last long or virus keeps evolving to evade host immunity. Short immunity periods are the case for seasonal human coronaviruses (Callow et al., 1990; Galanti and Shaman, 2020; Kissler et al., 2020). Although natural infection with SARS-CoV-2 can induce neutralizing antibodies in humans (Brouwer et al., 2020; Chi et al., 2020; Ju et al., 2020; Shi R. et al., 2020), we still do not know if the infection confers immunity to prevent reinfection and, if so, for what duration (Altmann and Boyton, 2020; Ibarrondo et al., 2020; Isho et al., 2020; Kirkcaldy et al., 2020; Long et al., 2020; Robbiani et al., 2020; Staines et al., 2020). Many cases that tested positive after once turning negative are currently considered as reactivation, long-term shedding, or detection of residual genetic material (An et al., 2020; Mahase, 2020b; Xiao A. T. et al., 2020) although a few cases of possible reinfection have been reported (Bloomberg, 2020). There is also concern that preexisting immunity might worsen the disease severity by mechanisms called antibody-dependent enhancement (Arvin et al., 2020; Eroshenko et al., 2020).
Still, the virus could go extinct. A key for containment lies in how we reduce the pool of susceptible people. Reduction can be achieved by natural infection of most people and/or by the introduction of world-scale mass vaccination. We pointed out previously in this article that four factors are important for such eradication: (1) no animal reservoir, (2) a long-lasting immunity, (3) an adequate proportion of immunized people, and (4) a small proportion of asymptomatic infections (Figure 2). For SARS-CoV-2, (1) no known non-human animal reservoir exists, but the virus can infect other animals suggesting possible animal-to-human transmission (Newman et al., 2020; Oreshkova et al., 2020; Shi J. et al., 2020; Sit et al., 2020). (2) The titer of antibody against SARS-CoV-2 in patients has been observed to decline during the early convalescent phase (Ibarrondo et al., 2020; Long et al., 2020). Still, vaccine candidates for SARS-CoV-2 showed the efficacy to induce neutralizing antibodies in humans (Folegatti et al., 2020; Jackson et al., 2020; Mulligan et al., 2020; Zhu et al., 2020) and it is possible that not only humoral immunity but cellular immunity counteracts SARS-CoV-2 infections (Altmann and Boyton, 2020; Grifoni et al., 2020; Le Bert et al., 2020; Ni et al., 2020; Weiskopf et al., 2020). Protective immunity by infection or vaccination in animal models lasted for at least a short period (Chandrashekar et al., 2020; Corbett et al., 2020; Deng et al., 2020; Gao et al., 2020; van Doremalen et al., 2020; Wang H. et al., 2020; Yu et al., 2020). Further, cross-reactivity of immune response between SARS-CoV-2 and seasonal human coronaviruses has been reported; however, it is uncertain whether viral interference occurs between SARS-CoV-2 and other viruses (Baker R. E. et al., 2020; Braun et al., 2020; Grifoni et al., 2020; Kissler et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Ng et al., 2020; Sette and Crotty, 2020; Wec et al., 2020; Weiskopf et al., 2020). (3) We must produce large quantities of vaccines and distribute them even to remote areas and conflict zones. We are now struggling to meet this challenge for measles and polio eradication (Peck et al., 2019; Chard et al., 2020). Meanwhile, a recent analysis that considered the heterogeneity in infectivity among individuals showed herd immunity required to contain this outbreak could be considerably smaller than previously thought (Britton et al., 2020; Gomes et al., 2020). (4) Unfortunately, a substantial proportion of infections with the virus do not cause notable symptoms (Day, 2020; Oran and Topol, 2020), and asymptomatic/presymptomatic people can transmit infection (Bai et al., 2020; Furukawa et al., 2020; Furuse et al., 2020; He et al., 2020). Assessing outbreak containment and determining when and how the virus dies out are thus difficult.
Concluding Remarks
Zoonotic viruses and human viruses have different characteristics, even though many human viruses originally come from animal viruses. The animal-human interface brought SARS-CoV-2 into human populations, and the virus has already acquired the ability for effective transmission among humans. Projecting the future of the current outbreak remains to be difficult. The virus could become a “human virus” and coexist in the world population. For this scenario, we can still hope that its morbidity and mortality lower as they do for post-pandemic influenza. Even if natural infection or vaccination cannot provide a long-lasting immunity to prevent reinfection, it could reduce the severity of the disease (Gao et al., 2020; Ng et al., 2020; Wang H. et al., 2020; Yu et al., 2020). Future development of antiviral agents could contribute to that as well (European Centre for Disease Prevention and Control, 2020; Siemieniuk et al., 2020; Wiersinga et al., 2020). It is less likely but still possible that the virus will go extinct. However, many obstacles exist for the latter scenario to materialize.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
HO conceived the study. YF reviewed the literatures and wrote the first draft of the manuscript. Both authors reviewed the draft and approved the final version of the manuscript.
Funding
The work was supported in part by the Leading Initiative for Excellent Young Researchers (grant number 16809810) from the Ministry of Education, Culture, Sports, Science, and Technology in Japan. The funder had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Altmann, D. M., and Boyton, R. J. (2020). SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci. Immunol. 5:eabd6160. doi: 10.1126/sciimmunol.abd6160
An, J., Liao, X., Xiao, T., Qian, S., Yuan, J., Ye, H., et al. (2020). Clinical characteristics of the recovered COVID-19 patients with re-detectable positive RNA test. medRxiv [Preprint]. doi: 10.1101/2020.03.26.20044222
Anderson, R. M., Fraser, C., Ghani, A. C., Donnelly, C. A., Riley, S., Ferguson, N. M., et al. (2004). Epidemiology, transmission dynamics and control of SARS: the 2002-2003 epidemic. Philos. Trans. R. Soc. B Biol. Sci. 359, 1091–1105. doi: 10.1098/rstb.2004.1490
Arvin, A. M., Fink, K., Schmid, M. A., Cathcart, A., Spreafico, R., Havenar-Daughton, C., et al. (2020). A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 584, 353–363. doi: 10.1038/s41586-020-2538-8
Azhar, E. I., El-Kafrawy, S. A., Farraj, S. A., Hassan, A. M., Al-Saeed, M. S., Hashem, A. M., et al. (2014). Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 370, 2499–2505. doi: 10.1056/NEJMoa1401505
Bai, Y., Yao, L., Wei, T., Tian, F., Jin, D. Y., Chen, L., et al. (2020). Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA J. Am. Med. Assoc. 323, 1406–1407. doi: 10.1001/jama.2020.2565
Baker, M., Kvalsvig, A., Verrall, A. J., Telfar-Barnard, L., and Wilson, N. (2020). New Zealand’s elimination strategy for the COVID-19 pandemic and what is required to make it work. N. Z. Med. J. 133, 10–14.
Baker, R. E., Yang, W., Vecchi, G. A., Metcalf, C. J. E., and Grenfell, B. T. (2020). Susceptible supply limits the role of climate in the early SARS-CoV-2 pandemic. Science 369, 315–319. doi: 10.1126/science.abc2535
Barry, J. M., Viboud, C., and Simonsen, L. (2008). Cross-protection between successive waves of the 1918–1919 influenza pandemic: epidemiological evidence from US army camps and from Britain. J. Infect. Dis. 198, 1427–1434. doi: 10.1086/592454
Black, F. L. (1966). Measles endemicity in insular populations: critical community size and its evolutionary implication. J. Theor. Biol. 11, 207–211. doi: 10.1016/0022-5193(66)90161-5
Bloomberg (2020). Two Chinese Patients Test Positive Months After Virus Recovery. Available at: https://www.bloomberg.com/news/articles/2020-08-13/two-chinese-patients-test-positive-months-after-virus-recovery (accessed August 14, 2020).
Boni, M. F., Lemey, P., Jiang, X., Lam, T. T.-Y., Perry, B. W., Castoe, T. A., et al. (2020). Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. [Epub ahead of print]. doi: 10.1038/s41564-020-0771-4
Bordería, A. V., Stapleford, K. A., and Vignuzzi, M. (2011). RNA virus population diversity: implications for inter-species transmission. Curr. Opin. Virol. 1, 643–648. doi: 10.1016/j.coviro.2011.09.012
Braun, J., Loyal, L., Frentsch, M., Wendisch, D., Georg, P., Kurth, F., et al. (2020). SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 20:353. doi: 10.1038/s41586-020-2598-9
Britton, T., Ball, F., and Trapman, P. (2020). A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science 369, 846–849. doi: 10.1126/science.abc6810
Brouwer, P. J. M., Caniels, T. G., van der Straten, K., Snitselaar, J. L., Aldon, Y., Bangaru, S., et al. (2020). Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369:eabc5902. doi: 10.1126/science.abc5902
Callow, K. A., Parry, H. F., Sergeant, M., and Tyrrell, D. A. J. (1990). The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 105, 435–446. doi: 10.1017/S0950268800048019
Chandrashekar, A., Liu, J., Martinot, A. J., McMahan, K., Mercado, N. B., Peter, L., et al. (2020). SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369, 812–817. doi: 10.1126/science.abc4776
Chard, A. N., Datta, S. D., Tallis, G., Burns, C. C., Wassilak, S. G. F., Vertefeuille, J. F., et al. (2020). Progress Toward Polio Eradication — Worldwide, January 2018–March 2020. MMWR Morb. Mortal. Wkly. Rep. 69, 784–789. doi: 10.15585/mmwr.mm6925a4
Chi, X., Yan, R., Zhang, J., Zhang, G., Zhang, Y., Hao, M., et al. (2020). A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655. doi: 10.1126/science.abc6952
Chowdhury, R., Luhar, S., Khan, N., Choudhury, S. R., Matin, I., and Franco, O. H. (2020). Long-term strategies to control COVID-19 in low and middle-income countries: an options overview of community-based, non-pharmacological interventions. Eur. J. Epidemiol. [Epub ahead of print]. doi: 10.1007/s10654-020-00660-1
Chowell, G., and Nishiura, H. (2014). Transmission dynamics and control of Ebola virus disease (EVD): a review. BMC Med. 12:196. doi: 10.1186/s12916-014-0196-0
Corbett, K. S., Flynn, B., Foulds, K. E., Francica, J. R., Boyoglu-Barnum, S., Werner, A. P., et al. (2020). Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. [Epub ahead of print]. doi: 10.1056/NEJMoa2024671
Couch, R. B., and Kasel, J. A. (1983). Immunity to influenza in man. Annu. Rev. Microbiol. 37, 529–549. doi: 10.1146/annurev.mi.37.100183.002525
Cowling, B. J., Jin, L., Lau, E. H. Y., Liao, Q., Wu, P., Jiang, H., et al. (2013). Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382, 129–137. doi: 10.1016/S0140-6736(13)61171-X
Dawood, F. S., Iuliano, A. D., Reed, C., Meltzer, M. I., Shay, D. K., Cheng, P. Y., et al. (2012). Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 12, 687–695. doi: 10.1016/S1473-3099(12)70121-4
Day, M. (2020). Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ 369:m1375. doi: 10.1136/bmj.m1375
Delamater, P. L., Street, E. J., Leslie, T. F., Yang, Y. T., and Jacobsen, K. H. (2019). Complexity of the basic reproduction number (R0). Emerg. Infect. Dis. 25, 1–4. doi: 10.3201/eid2501.171901
Deng, W., Bao, L., Liu, J., Xiao, C., Liu, J., Xue, J., et al. (2020). Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 369, 818–823. doi: 10.1126/science.abc5343
Dobson, A. P., Pimm, S. L., Hannah, L., Kaufman, L., Ahumada, J. A., Ando, A. W., et al. (2020). Ecology and economics for pandemic prevention. Science 369, 379–381. doi: 10.1126/science.abc3189
Domingo, E., Escarmís, C., Sevilla, N., Moya, A., Elena, S. F., Quer, J., et al. (1996). Basic concepts in RNA virus evolution. FASEB J. 10, 859–864. doi: 10.1096/fasebj.10.8.8666162
Domingo, E., and Holland, J. J. (1997). RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51, 151–178. doi: 10.1146/annurev.micro.51.1.151
Düx, A., Lequime, S., Patrono, L. V., Vrancken, B., Boral, S., Gogarten, J. F., et al. (2020). Measles virus and rinderpest virus divergence dated to the sixth century BCE. Science 368, 1367–1370. doi: 10.1126/science.aba9411
Eckerle, I., and Meyer, B. (2020). SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet 396, 514–515. doi: 10.1016/S0140-6736(20)31482-3
Erkhembayar, R., Dickinson, E., Badarch, D., Narula, I., Thomas, G. N., Ochir, C., et al. (2020). Early policy actions and emergency response to the COVID-19 pandemic in Mongolia: experiences and challenges. Lancet Glob. Heal. [Epub ahead of print]. doi: 10.1016/s2214-109x(20)30295-3
Eroshenko, N., Gill, T., Keaveney, M. K., Church, G. M., Trevejo, J. M., and Rajaniemi, H. (2020). Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat. Biotechnol. 38, 789–791. doi: 10.1038/s41587-020-0577-1
European Centre for Disease Prevention and Control (2020). Vaccines and Treatment of COVID-19. Available at: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/vaccines-and-treatment (accessed August 11, 2020).
Ferguson, N. M., Galvani, A. P., Bush, R. M., Galvanl, A. P., and Bush, R. M. (2003). Ecological and immunological determinants of influenza evolution. Nature 422, 428–433. doi: 10.1038/nature01509
Ferguson, N. M., Laydon, D., Nedjati-Gilani, G., Imai, N., Ainslie, K., Baguelin, M., et al. (2020). Impact of Non-Pharmaceutical Interventions (NPIs) to Reduce COVID-19 Mortality and Healthcare Demand. London: Imperial College, 3–20.
Flaxman, S., Mishra, S., Gandy, A., Unwin, H. J. T., Mellan, T. A., Coupland, H., et al. (2020). Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 584, 257–261. doi: 10.1038/s41586-020-2405-7
Folegatti, P. M., Ewer, K. J., Aley, P. K., Angus, B., Becker, S., Belij-Rammerstorfer, S., et al. (2020). Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478. doi: 10.1016/s0140-6736(20)31604-4
Forni, D., Cagliani, R., Clerici, M., and Sironi, M. (2017). Molecular evolution of human coronavirus genomes. Trends Microbiol. 25, 35–48. doi: 10.1016/j.tim.2016.09.001
Fraser, C., Riley, S., Anderson, R. M., and Ferguson, N. M. (2004). Factors that make an infectious disease outbreak controllable. Proc. Natl. Acad. Sci. U.S.A. 101, 6146–6151. doi: 10.1073/pnas.0307506101
Furukawa, N. W., Brooks, J. T., and Sobel, J. (2020). Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerg. Infect. Dis. 26:e201595. doi: 10.3201/eid2607.201595
Furuse, Y., and Oshitani, H. (2016). Mechanisms of replacement of circulating viruses by seasonal and pandemic influenza A viruses. Int. J. Infect. Dis. 51, 6–14. doi: 10.1016/j.ijid.2016.08.012
Furuse, Y., and Oshitani, H. (2017). Global transmission dynamics of measles in the measles elimination era. Viruses 9:82. doi: 10.3390/v9040082
Furuse, Y., Sando, E., Tsuchiya, N., Miyahara, R., Yasuda, I., Ko, Y. K., et al. (2020). Clusters of coronavirus disease in communities, Japan, January–April 2020. Emerg. Infect. Dis. 26:202272. doi: 10.3201/eid2609.202272
Furuse, Y., Suzuki, A., and Oshitani, H. (2010). Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virol. J. 7:52. doi: 10.1186/1743-422X-7-52
Galanti, M., and Shaman, J. (2020). Direct observation of repeated infections with endemic coronaviruses. J. Infect. Dis. [Epub ahead of print]. doi: 10.1093/infdis/jiaa392
Gao, Q., Bao, L., Mao, H., Wang, L., Xu, K., Yang, M., et al. (2020). Development of an inactivated vaccine candidate for SARS-CoV-2. Science [Epub ahead of print]. doi: 10.1126/science.abc1932
Gibb, R., Redding, D. W., Chin, K. Q., Donnelly, C. A., Blackburn, T. M., Newbold, T., et al. (2020). Zoonotic host diversity increases in human-dominated ecosystems. Nature 584, 398–402. doi: 10.1038/s41586-020-2562-8
Gomes, M. G. M., Aguas, R., Corder, R. M., King, J. G., Langwig, K. E., Souto-Maior, C., et al. (2020). Individual variation in susceptibility or exposure to SARS-CoV-2 lowers the herd immunity threshold. MedRxiv [Preprint]. doi: 10.1101/2020.04.27.20081893
Gran, J. M., Wasmuth, L., Amundsen, E. J., Lindqvist, B. H., and Aalen, O. O. (2008). Growth rates in epidemic models: application to a model for HIV/AIDS progression. Stat. Med. 27, 4817–4834. doi: 10.1002/sim.3219
Grifoni, A., Weiskopf, D., Ramirez, S. I., Mateus, J., Dan, J. M., Moderbacher, C. R., et al. (2020). Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489.e15–1501.e15. doi: 10.1016/j.cell.2020.05.015
Guerra, F. M., Bolotin, S., Lim, G., Heffernan, J., Deeks, S. L., Li, Y., et al. (2017). The basic reproduction number (R0) of measles: a systematic review. Lancet Infect. Dis. 17, e420–e428. doi: 10.1016/S1473-3099(17)30307-9
Han, G. Z. (2020). Pangolins harbor SARS-CoV-2-related coronaviruses. Trends Microbiol. 28, 515–517. doi: 10.1016/j.tim.2020.04.001
Havers, F. P., Reed, C., Lim, T., Montgomery, J. M., Klena, J. D., Hall, A. J., et al. (2020). Seroprevalence of Antibodies to SARS-CoV-2 in 10 Sites in the United States, March 23-May 12, 2020. JAMA Intern. Med. [Epub ahead of print]. doi: 10.1001/jamainternmed.2020.4130
He, X., Lau, E. H. Y., Wu, P., Deng, X., Wang, J., Hao, X., et al. (2020). Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 26, 672–675. doi: 10.1038/s41591-020-0869-5
Heeney, J. L. (2006). Zoonotic viral diseases and the frontier of early diagnosis, control and prevention. J. Intern. Med. 260, 399–408. doi: 10.1111/j.1365-2796.2006.01711.x
Holmdahl, I., and Buckee, C. (2020). Wrong but useful — what Covid-19 epidemiologic models can and cannot tell us. N. Engl. J. Med. 383, 303–305. doi: 10.1056/nejmp2016822
Hsiang, S., Allen, D., Annan-Phan, S., Bell, K., Bolliger, I., Chong, T., et al. (2020). The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature 584, 262–267. doi: 10.1038/s41586-020-2404-8
Ibarrondo, F. J., Fulcher, J. A., Goodman-Meza, D., Elliott, J., Hofmann, C., Hausner, M. A., et al. (2020). Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. [Epub ahead of print]. doi: 10.1056/nejmc2025179
Isho, B., Abe, K. T., Zuo, M., Jamal, A. J., Rathod, B., Wang, J. H., et al. (2020). Evidence for sustained mucosal and systemic antibody responses to SARS-CoV-2 antigens in COVID-19 patients. MedRxiv [Preprint]. doi: 10.1101/2020.08.01.20166553
Jackson, L. A., Anderson, E. J., Rouphael, N. G., Roberts, P. C., Makhene, M., Coler, R. N., et al. (2020). An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. [Epub ahead of print]. doi: 10.1056/nejmoa2022483
Jewell, N. P., Lewnard, J. A., and Jewell, B. L. (2020). Predictive mathematical models of the Covid-19 pandemic: underlying principles and value of projections. JAMA J. Am. Med. Assoc. 323, 1893–1894. doi: 10.1001/jama.2020.6585
Jing, Q.-L., Liu, M.-J., Zhang, Z.-B., Fang, L.-Q., Yuan, J., Zhang, A.-R., et al. (2020). Articles Household secondary attack rate of COVID-19 and associated determinants in Guangzhou, China: a retrospective cohort study. Lancet [Epub ahead of print]. doi: 10.1016/S1473-3099(20)30471-0
Ju, B., Zhang, Q., Ge, J., Wang, R., Sun, J., Ge, X., et al. (2020). Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119. doi: 10.1038/s41586-020-2380-z
Kessler, M. K., Becker, D. J., Peel, A. J., Justice, N. V., Lunn, T., Crowley, D. E., et al. (2018). Changing resource landscapes and spillover of henipaviruses. Ann. N. Y. Acad. Sci. 1429, 78–99. doi: 10.1111/nyas.13910
Kew, O. M., Sutter, R. W., De Gourville, E. M., Dowdle, W. R., and Pallansch, M. A. (2005). Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 59, 587–635. doi: 10.1146/annurev.micro.58.030603.123625
Kirkcaldy, R. D., King, B. A., and Brooks, J. T. (2020). COVID-19 and postinfection immunity: limited evidence, many remaining questions. JAMA J. Am. Med. Assoc. 323, 2245–2246. doi: 10.1001/jama.2020.7869
Kissler, S. M., Tedijanto, C., Goldstein, E., Grad, Y. H., and Lipsitch, M. (2020). Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 368, 860–868. doi: 10.1126/science.abb5793
Kucharski, A. J., Lessler, J., Read, J. M., Zhu, H., Jiang, C. Q., Guan, Y., et al. (2015). Estimating the life course of influenza A(H3N2) antibody responses from cross-sectional data. PLoS Biol. 13:e1002082. doi: 10.1371/journal.pbio.1002082
Kucharski, A. J., Russell, T. W., Diamond, C., Liu, Y., Edmunds, J., Funk, S., et al. (2020). Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect. Dis. [Epub ahead of print]. doi: 10.1016/S1473-3099(20)30144-4
Lai, S., Ruktanonchai, N. W., Zhou, L., Prosper, O., Luo, W., Floyd, J. R., et al. (2020). Effect of non-pharmaceutical interventions to contain COVID-19 in China. Nature [Epub ahead of print]. doi: 10.1038/s41586-020-2293-x
Lam, T. T. Y., Jia, N., Zhang, Y. W., Shum, M. H. H., Jiang, J. F., Zhu, H. C., et al. (2020). Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature [Epub ahead of print]. doi: 10.1038/s41586-020-2169-0
Le Bert, N., Tan, A. T., Kunasegaran, K., Tham, C. Y. L., Hafezi, M., Chia, A., et al. (2020). SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462. doi: 10.1038/s41586-020-2550-z
Letko, M., Seifert, S. N., Olival, K. J., Plowright, R. K., and Munster, V. J. (2020). Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 18, 461–471. doi: 10.1038/s41579-020-0394-z
Li, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., et al. (2005). Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24, 1634–1643. doi: 10.1038/sj.emboj.7600640
Li, X., Giorgi, E. E., Marichannegowda, M. H., Foley, B., Xiao, C., Kong, X.-P., et al. (2020). Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 27:eabb9153. doi: 10.1126/sciadv.abb9153
Lin, Y. P., Xiong, X., Wharton, S. A., Martin, S. R., Coombs, P. J., Vachieri, S. G., et al. (2012). Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 109, 21474–21479. doi: 10.1073/pnas.1218841110
Liu, P., Jiang, J. Z., Wan, X. F., Hua, Y., Li, L., Zhou, J., et al. (2020). Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 16:e1008421. doi: 10.1371/journal.ppat.1008421
Liu, Y., Gayle, A. A., Wilder-Smith, A., and Rocklöv, J. (2020). The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 27:taaa021. doi: 10.1093/jtm/taaa021
Long, J. S., Giotis, E. S., Moncorgé, O., Frise, R., Mistry, B., James, J., et al. (2016). Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529, 101–104. doi: 10.1038/nature16474
Long, J. S., Mistry, B., Haslam, S. M., and Barclay, W. S. (2019). Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 17, 67–81. doi: 10.1038/s41579-018-0115-z
Long, Q.-X., Tang, X.-J., Shi, Q.-L., Li, Q., Deng, H.-J., Yuan, J., et al. (2020). Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 26, 1200–1204. doi: 10.1038/s41591-020-0965-6
López, L., and Rodó, X. (2020). The end of social confinement and COVID-19 re-emergence risk. Nat. Hum. Behav. 4, 746–755. doi: 10.1038/s41562-020-0908-8
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Madewell, Z. J., Yang, Y., Longini, I. M., Halloran, M. E., and Dean, N. E. (2020). Household transmission of SARS-CoV-2: a systematic review and meta-analysis of secondary attack rate. medRxiv [Preprint]. doi: 10.1101/2020.07.29.20164590
Mahase, E. (2020a). Coronavirus covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ 368:m641. doi: 10.1136/bmj.m641
Mahase, E. (2020b). Covid-19: WHO and South Korea investigate reconfirmed cases. BMJ 369:m1498. doi: 10.1136/bmj.m1498
Mateus, J., Grifoni, A., Tarke, A., Sidney, J., Ramirez, S. I., Dan, J. M., et al. (2020). Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 4:eabd3871. doi: 10.1126/science.abd3871
Metcalf, C. J. E., Morris, D. H., and Park, S. W. (2020). Mathematical models to guide pandemic response. Science 369, 368–369. doi: 10.1126/science.abd1668
Mohd, H. A., Al-Tawfiq, J. A., and Memish, Z. A. (2016). Middle east respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir susanna lau. Virol. J. 13:87. doi: 10.1186/s12985-016-0544-0
Morawska, L., and Milton, D. K. (2020). It is time to address airborne transmission of COVID-19. Clin. Infect. Dis. [Epub ahead of print]. doi: 10.1093/cid/ciaa939
Morens, D. M., and Taubenberger, J. K. (2011). Pandemic influenza: certain uncertainties. Rev. Med. Virol. 21, 262–284. doi: 10.1002/rmv.689
Moss, W. J., and Strebel, P. (2011). Biological feasibility of measles eradication. J. Infect. Dis. 204(Suppl. 1), S47–S53. doi: 10.1093/infdis/jir065
Mulligan, M. J., Lyke, K. E., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al. (2020). Phase 1/2 study of COVID-19 RNA vaccine BNT162b1 in adults. Nature [Epub ahead of print]. doi: 10.1038/s41586-020-2639-4
National Center for Immunization, and Respiratory Diseases (2017). Measles | Transmission | CDC. Atlanta: NCIRD.
Neiderud, C. J. (2015). How urbanization affects the epidemiology of emerging infectious diseases. Afr. J. Disabil. 5:27060. doi: 10.3402/iee.v5.27060
Newman, A., Smith, D., Ghai, R. R., Wallace, R. M., Torchetti, M. K., Loiacono, C., et al. (2020). First reported cases of SARS-CoV-2 infection in companion animals — New York, March–April 2020. MMWR Morb. Mortal. Wkly. Rep. 69, 710–713. doi: 10.15585/mmwr.mm6923e3
Ng, K., Faulkner, N., Cornish, G., Rosa, A., Earl, C., Wrobel, A., et al. (2020). Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans. bioRxiv [Preprint]. doi: 10.1101/2020.05.14.095414
Ni, L., Ye, F., Cheng, M. L., Feng, Y., Deng, Y. Q., Zhao, H., et al. (2020). Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52, 971.e3–977.e3. doi: 10.1016/j.immuni.2020.04.023
Oran, D. P., and Topol, E. J. (2020). Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. [Epub ahead of print]. doi: 10.7326/m20-3012
Oreshkova, N., Molenaar, R. J., Vreman, S., Harders, F., Oude Munnink, B. B., Hakze-van der Honing, R. W., et al. (2020). SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 25:2001005. doi: 10.2807/1560-7917.es.2020.25.23.2001005
Palese, P., and Wang, T. T. (2011). Why do influenza virus subtypes die out? A hypothesis. mBio 2:e00150-11. doi: 10.1128/mBio.00150-11
Peck, M., Gacic-Dobo, M., Diallo, M. S., Nedelec, Y., Sodha, S. S., and Wallace, A. S. (2019). Global routine vaccination coverage, 2018. MMWR Morb. Mortal. Wkly. Rep. 68, 937–942. doi: 10.15585/mmwr.mm6842a1
Peeri, N. C., Shrestha, N., Rahman, M. S., Zaki, R., Tan, Z., Bibi, S., et al. (2020). The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 49, 717–726. doi: 10.1093/ije/dyaa033
Pollán, M., Pérez-Gómez, B., Pastor-Barriuso, R., Oteo, J., Hernán, M. A., Pérez-Olmeda, M., et al. (2020). Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet 396, 535–544. doi: 10.1016/S0140-6736(20)31483-5
Raj, V. S., Osterhaus, A. D. M. E., Fouchier, R. A. M., and Haagmans, B. L. (2014). MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 5, 58–62. doi: 10.1016/j.coviro.2014.01.010
Rajsbaum, R., Albrecht, R. A., Wang, M. K., Maharaj, N. P., Versteeg, G. A., Nistal-Villán, E., et al. (2012). Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza a virus NS1 protein. PLoS Pathog. 8:e1003059. doi: 10.1371/journal.ppat.1003059
Robbiani, D. F., Gaebler, C., Muecksch, F., Lorenzi, J. C. C. C., Wang, Z., Cho, A., et al. (2020). Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442. doi: 10.1038/s41586-020-2456-9
Sauter, D., and Kirchhoff, F. (2019). Key viral adaptations preceding the AIDS pandemic. Cell Host Microbe 25, 27–38. doi: 10.1016/j.chom.2018.12.002
Sawyer, S. L., Emerman, M., and Malik, H. S. (2004). Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:e275. doi: 10.1371/journal.pbio.0020275
Sette, A., and Crotty, S. (2020). Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 20, 457–458. doi: 10.1038/s41577-020-0389-z
Shanks, G. D., Lee, S. E., Howard, A., and Brundage, J. F. (2011). Extreme mortality after first introduction of measles virus to the Polynesian Island of Rotuma, 1911. Am. J. Epidemiol. 173, 1211–1222. doi: 10.1093/aje/kwq504
Sharp, P. M., and Hahn, B. H. (2011). Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1:a006841. doi: 10.1101/cshperspect.a006841
Shi, J., Wen, Z., Zhong, G., Yang, H., Wang, C., Huang, B., et al. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368, 1016–1020. doi: 10.1126/science.abb7015
Shi, R., Shan, C., Duan, X., Chen, Z., Liu, P., Song, J., et al. (2020). A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature 584, 120–124. doi: 10.1038/s41586-020-2381-y
Shi, Z., and Hu, Z. (2008). A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 133, 74–87. doi: 10.1016/j.virusres.2007.03.012
Siemieniuk, R. A., Bartoszko, J. J., Ge, L., Zeraatkar, D., Izcovich, A., Pardo-Hernandez, H., et al. (2020). Drug treatments for covid-19: living systematic review and network meta-analysis. BMJ 370:2980. doi: 10.1136/bmj.m2980
Sit, T. H. C., Brackman, C. J., Ip, S. M., Tam, K. W. S., Law, P. Y. T., To, E. M. W., et al. (2020). Infection of dogs with SARS-CoV-2. Nature [Epub ahead of print]. doi: 10.1038/s41586-020-2334-5
Smith, D. J., Lapedes, A. S., de Jong, J. C., Bestebroer, T. M., Rimmelzwaan, G. F., Osterhaus, A. D. M. E., et al. (2004). Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376. doi: 10.1126/science.1097211
Staines, H. M., Kirwan, D. E., Clark, D. J., Adams, E. R., Augustin, Y., Byrne, R. L., et al. (2020). Dynamics of IgG seroconversion and pathophysiology of COVID-19 infections. medRxiv [Preprint]. doi: 10.1101/2020.06.07.20124636
Stewart, G., Heusden, K., and Dumont, G. A. (2020). How control theory can help us control Covid-19. IEEE Spectr. 57, 22–29. doi: 10.1109/MSPEC.2020.9099929
Stringhini, S., Wisniak, A., Piumatti, G., Azman, A. S., Lauer, S. A., Baysson, H., et al. (2020). Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet 396, 313–319. doi: 10.1016/S0140-6736(20)31304-0
Taubenberger, J. K., and Kash, J. C. (2010). Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7, 440–451. doi: 10.1016/j.chom.2010.05.009
Taubenberger, J. K., Kash, J. C., and Morens, D. M. (2019). The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci. Transl. Med. 11:eaau5485. doi: 10.1126/scitranslmed.aau5485
Van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., et al. (2020). Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 382, 1564–1567. doi: 10.1056/NEJMc2004973
van Doremalen, N., Lambe, T., Spencer, A., Belij-Rammerstorfer, S., Purushotham, J. N., Port, J. R., et al. (2020). ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature [Epub ahead of print]. doi: 10.1038/s41586-020-2608-y
Vijaykrishna, D., Smith, G. J. D., Zhang, J. X., Peiris, J. S. M., Chen, H., and Guan, Y. (2007). Evolutionary insights into the ecology of coronaviruses. J. Virol. 81, 4012–4020. doi: 10.1128/jvi.02605-06
Wang, C. J., Ng, C. Y., and Brook, R. H. (2020). Response to COVID-19 in Taiwan: big data analytics, new technology, and proactive testing. JAMA J. Am. Med. Assoc. 323, 1341–1342. doi: 10.1001/jama.2020.3151
Wang, H., Zhang, Y., Huang, B., Deng, W., Quan, Y., Wang, W., et al. (2020). Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 182, 713.e9–721.e9. doi: 10.1016/j.cell.2020.06.008
Wasik, B. R., De Wit, E., Munster, V., Lloyd-Smith, J. O., Martinez-Sobrido, L., and Parrish, C. R. (2019). Onward transmission of viruses: how do viruses emerge to cause epidemics after spillover? Philos. Trans. R. Soc. B Biol. Sci. 374:20190017. doi: 10.1098/rstb.2019.0017
Watsa, M. (2020). Rigorous wildlife disease surveillance. Science 369, 145–147. doi: 10.1126/science.abc0017
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M., and Kawaoka, Y. (1992). Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179. doi: 10.1128/mmbr.56.1.152-179.1992
Wec, A. Z., Wrapp, D., Herbert, A. S., Maurer, D. P., Haslwanter, D., Sakharkar, M., et al. (2020). Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369, 731–736. doi: 10.1126/science.abc7424
Weill, J. A., Stigler, M., Deschenes, O., and Springborn, M. R. (2020). Social distancing responses to COVID-19 emergency declarations strongly differentiated by income. Proc. Natl. Acad. Sci. U.S.A. 117, 19658–19660. doi: 10.1073/pnas.2009412117
Weiskopf, D., Schmitz, K. S., Raadsen, M. P., Grifoni, A., Okba, N. M. A., Endeman, H., et al. (2020). Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 5:eabd2071. doi: 10.1126/sciimmunol.abd2071
Whiteside, A., and Zebryk, N. (2015). Commentary/Commentaire Ebola and AIDS in Africa. Can. J. Afr. Stud. 49, 409–419. doi: 10.1080/00083968.2015.1014381
Wiersinga, W. J., Rhodes, A., Cheng, A. C., Peacock, S. J., and Prescott, H. C. (2020). Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA J. Am. Med. Assoc. [Epub ahead of print]. doi: 10.1001/jama.2020.12839
Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. doi: 10.1038/s41586-020-2008-3
Wu, T., Perrings, C., Kinzig, A., Collins, J. P., Minteer, B. A., and Daszak, P. (2017). Economic growth, urbanization, globalization, and the risks of emerging infectious diseases in China: a review. Ambio 46, 18–29. doi: 10.1007/s13280-016-0809-2
Wu, Z., and McGoogan, J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. JAMA [Epub ahead of print]. doi: 10.1001/jama.2020.2648
Xia, Y., Gog, J. R., and Grenfell, B. T. (2005). Semiparametric estimation of the duration of immunity from infectious disease time series: influenza as a case-study. J. R. Stat. Soc. Ser. C Appl. Stat. 54(Pt 3), 659–672. doi: 10.1111/j.1467-9876.2005.05383.x
Xiao, A. T., Tong, Y. X., and Zhang, S. (2020). False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J. Med. Virol. [Epub ahead of print]. doi: 10.1002/jmv.25855
Xiao, K., Zhai, J., Feng, Y., Zhou, N., Zhang, X., Zou, J. J., et al. (2020). Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583, 286–289. doi: 10.1038/s41586-020-2313-x
Yamada, S., Suzuki, Y., Suzuki, T., Le, M. Q., Nidom, C. A., Sakai-Tagawa, Y., et al. (2006). Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444, 378–382. doi: 10.1038/nature05264
Yu, J., Tostanoski, L. H., Peter, L., Mercado, N. B., McMahan, K., Mahrokhian, S. H., et al. (2020). DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811. doi: 10.1126/science.abc6284
Zhang, T., Wu, Q., and Zhang, Z. (2020). Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 30:1578. doi: 10.1016/j.cub.2020.03.022
Zhou, P., Yang, X. Lou, Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhu, F. C., Guan, X. H., Li, Y. H., Huang, J. Y., Jiang, T., Hou, L. H., et al. (2020). Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 396, 479–488. doi: 10.1016/S0140-6736(20)31605-6
Keywords: COVID-19, SARS-CoV-2, epidemiology, transmission, zoonosis, evolution
Citation: Furuse Y and Oshitani H (2020) Viruses That Can and Cannot Coexist With Humans and the Future of SARS-CoV-2. Front. Microbiol. 11:583252. doi: 10.3389/fmicb.2020.583252
Received: 16 July 2020; Accepted: 19 August 2020;
Published: 18 September 2020.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Yohei Kurosaki, Nagasaki University, JapanTakamasa Ueno, Kumamoto University, Japan
Makoto Takeda, National Institute of Infectious Diseases (NIID), Japan
Seiya Yamayoshi, University of Tokyo, Japan
Copyright © 2020 Furuse and Oshitani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuki Furuse, ZnVydXNleS5reW90b0BnbWFpbC5jb20=
 Yuki Furuse
Yuki Furuse Hitoshi Oshitani
Hitoshi Oshitani