- 1Department of Preventive Veterinary Medicine, College of Veterinary Medicine, Northwest A&F University, Yangling, China
- 2Key Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 3Department of Infection Biology, Institute of Infection and Global Health, University of Liverpool, Liverpool, United Kingdom
Porcine reproductive and respiratory syndrome (PRRS), caused by PRRS virus (PRRSV), is a widespread viral disease that has led to huge economic losses for the global swine industry. Non-structural protein 9 (Nsp9) of PRRSV possesses essential RNA-dependent RNA polymerase (RdRp) activity for viral RNA replication. Our previous report showed that Nsp9-specific nanobody, Nb6, was able to inhibit PRRSV replication. In this study, recombinant Nsp9 and Nsp9-Nb6 complex were prepared then characterized using bio-layer interferometry (BLI) and dynamic light scattering (DLS) analyses that demonstrated high-affinity binding of Nb6 to Nsp9 to form a homogeneous complex. Small-angle X-ray scattering (SAXS) characterization analyses revealed that spatial interactions differed between Nsp9 and Nsp9-Nb6 complex molecular envelopes. Enzyme-linked immunosorbent assays (ELISAs) revealed key involvement of Nsp9 residues Ile588, Asp590, and Leu643 and Nb6 residues Tyr62, Trp105, and Pro107 in the Nsp9-Nb6 interaction. After reverse genetics-based techniques were employed to generate recombinant Nsp9 mutant viruses, virus replication efficiencies were assessed in MARC-145 cells. The results revealed impaired viral replication of recombinant viruses bearing I588A and L643A mutations as compared with replication of wild type virus, as evidenced by reduced negative-strand genomic RNA [(−) gRNA] synthesis and attenuated viral infection. Moreover, the isoleucine at position 588 of Nsp9 was conserved across PRRSV genotypes. In conclusion, structural analysis of the Nsp9-Nb6 complex revealed novel amino acid interactions involved in viral RNA replication that will be useful for guiding development of structure-based anti-PRRSV agents.
Introduction
Porcine reproductive and respiratory syndrome (PRRS), first reported in 1987 in the United States, is one of the most widespread and severe viral disease threats to the global swine industry (Wensvoort et al., 1991; Nan et al., 2017). This disease is characterized by reproductive failures in sows and respiratory diseases in pigs of all ages (Lunney et al., 2010). The etiological agent, PRRS virus (PRRSV), belongs to the genus Betaarterivirus of the family Arteriviridae within the order Nidovirales. PRRSV isolates are divided into two major genotypes: Betaarterivirus suid 1 (PRRSV-1) and Betaarterivirus suid 2 (PRRSV-2) (Lin et al., 2020). These two genotypes share less than 70% identity across their entire genomes (Guo et al., 2018). In 2006, an atypical PRRS outbreak caused by a highly pathogenic PRRSV strain (HP-PRRSV) occurred in China, leading to a country-wide epidemic (Tian et al., 2007; Tong et al., 2007).
Porcine reproductive and respiratory syndrome virus is an enveloped RNA virus with a single positive-strand genome of approximately 15 kb. It contains at least ten open reading frames (ORFs), including ORF1a, ORF1ab, ORF2a, ORF2b, ORF3, ORF4, ORF5a, ORF5, ORF6, and ORF7 (Fang and Snijder, 2010; Snijder et al., 2013). ORF1a and ORF1ab occupy three quarters of the genome and encode two large polyproteins. Translation products of ORF1a and ORF1ab, pp1a and pp1ab, respectively, are processed by host and viral proteases into 16 non-structural proteins (Nsps) including Nsp1α/β, Nsp2-6, Nsp7α/β, Nsp8-12, Nsp2TF, and Nsp2N (Fang and Snijder, 2010; Fang et al., 2012). Among these Nsps, Nsp9 is highly conserved (Darwich et al., 2011) and possesses RNA-dependent RNA polymerase (RdRp) activity that is crucial for viral replication and transcription complex (RTC) formation (Snijder et al., 2013), while also contributing to HP-PRRSV virulence (Li et al., 2014). Meanwhile, Nsp9 also interacts with other cellular factors required for PRRSV infection, including annexin A2, retinoblastoma protein and DEAD-box RNA helicase 5 (Dong et al., 2014; Zhao et al., 2015). Thus, the apparent indispensable role played by Nsp9 in PRRSV functions warrants its evaluation as a promising anti-PRRSV target for development of antiviral agents. Indeed, RNA interference-based interventions targeting Nsp9 have already been shown to block PRRSV replication in permissive cell lines (Xie et al., 2014).
Nanobodies (Nbs), also known as camel single-domain antibodies (sdAbs), are derived from the variable region of Camellidae heavy chain-only antibodies (VHH) (Hamers-Casterman et al., 1993). Nbs possess various advantages compared to conventional monoclonal antibodies (Hassanzadeh-Ghassabeh et al., 2013), such as small size, ease of genetic manipulation, high specificity and solubility. Moreover, Nbs can be easily produced in prokaryotic or eukaryotic hosts, making them ideal candidates for drug development (Marasco, 2001). Our previous report demonstrated that a PRRSV Nsp9-specific Nb, designated Nb6, inhibited PRRSV replication in vitro when expressed intracellularly in PRRSV-permissive cells (Liu et al., 2015). Meanwhile, we have also demonstrated that expressed recombinant fusion proteins incorporating Nb6 fused with cell-penetrating peptides (CPP) exhibited broad antiviral activities against various PRRSV isolates of both genotypes (Wang et al., 2019). However, the antiviral mechanism whereby Nb6 targets Nsp9 to disrupt PRRSV function is unknown. In this study, recombinant PRRSV-Nsp9 and Nsp9-Nb6 complex were prepared then subjected to bio-layer interferometry (BLI) and structure-based biophysical analyses to visualize structural changes associated with Nsp9-Nb6 complex formation. Next, enzyme-linked immunosorbent assays (ELISAs) were conducted to identify key amino acid residues involved in the Nsp9-Nb6 interaction. In addition, we generated recombinant Nsp9-mutant viruses using a reverse genetics-based approach then assessed mutant virus replication efficiencies in vitro to determine key residues involved in viral replication.
Materials and Methods
Cells and Viruses
Porcine reproductive and respiratory syndrome virus-permissive MARC-145 cells (derived from African green monkey kidney cells) and CRL-2843-CD163 (a cell line stably expressing CD163 in CRL-2843) constructed in our laboratory were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, United States) and routinely maintained in Roswell Park Memorial Institute 1640 medium (RPMI 1640, Thermo Fisher Scientific), respectively, supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, Carlsbad, CA, United States) at 37°C in 5% CO2. The PRRSV strain used in this study was HP-PRRSV SD16 (GenBank #: JX087437).
Recombinant Nb6 and Nsp9 Expression and Purification
The sequence of Nb6 was amplified by PCR from template plasmid pEGFP-Nb6 (Liu et al., 2015) using primers listed in Table 1. The resulting fragment was isolated and digested with XbaI and XhoI (New England Biolabs, Ipswich, MA, United States), then was ligated to pPICZαA expression vector using DNA ligase (New England Biolabs), yielding the construct pPICZαA-Nb6. The construct was transformed into competent Escherichia coli DH5α then transformants were screened for growth on low salt LB (LLB) agar plates (0.5% yeast extract, 1% tryptone, 0.5% NaCl, 2% agar) containing 30 μg/ml Zeocin (Thermo Fisher Scientific). Positive colonies were verified by DNA sequencing (Sangon Biotech, Shanghai, China).
Competent Pichia pastoris strain X-33 cells (Thermo Fisher Scientific) were prepared for DNA transformation according to the manufacturer’s instructions. The plasmid pPICZαA-Nb6 was linearized using SacI (TaKaRa, Dalian, China) then transformed into X-33 via electroporation (1500 V, 25 μF, 200 Ω for 6 ms). After incubation for 1 h at 30°C in 1 M sorbitol without agitation, transformed X-33 cells were cultured on YPD agar plates (1% yeast extract, 2% peptone, 2% glucose, 2% agar) containing 100 μg/ml Zeocin at 30°C for 3 days. Colonies were screened to detect transformants harboring the gene encoding Nb6 integrated within host chromosomal DNA. Positive recombinant X-33 transformants for chromosomally integrated Nb6 were grown in BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4 × 10–5% biotin, 1% v/v glycerol) to an OD600 reading of 2-6 then were cultured in BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4 × 10–5% biotin, 1% v/v methanol). Protein expression was induced by daily additions of 1% (v/v) methanol for 5 days. After 5 days of induction, the cell culture supernatant was collected and recombinant proteins were purified using a Novagen Ni-NTA column (Merck, Darmstadt, Germany) pre-equilibrated with 20 mM Tris–HCl, pH 8.0, 150 mM NaCl. The target protein was then eluted with 20 mM Tris–HCl, pH 8.0, 150 mM NaCl and 150 mM imidazole and further purified using GE Superdex 75 10/300 GL on GE AKTATM Pure Chromatography System (GE Healthcare, Pittsburgh, PA, United States) with 20 mM Tris–HCl, pH 8.0 and 150 mM NaCl as elution buffer. Eluted proteins were concentrated using Millipore ultracentrifugation filter tubes (Merck) to 10 mg/ml in 20 mM Tris–HCl, pH 8.0, 150 mM NaCl.
Expression and purification of Nsp9 were performed as reported previously (Liu et al., 2015). Briefly, Nsp9-coding DNA amplified from template plasmid pBAC-SD16FL (Wang C. et al., 2013) was inserted between NcoI and XhoI sites of pET-28a then the resulting plasmid was transformed into E. Coli Transetta strain (Transgen, Beijing, China). Expressed Nsp9 protein was purified using metal affinity chromatography (Ni-NTA column; Novagen) and size-exclusion chromatography (Superdex 200 increase 10/300 GL; GE Healthcare).
Preparation of Nsp9-Nb6 Complex
Nsp9-Nb6 complex was generated by incubating excess purified Nb6 with purified Nsp9 in buffer containing 20 mM Tris–HCl, pH 8.0, 150 mM NaCl at 4°C overnight. The complex was then isolated via gel filtration chromatography (Superdex 75 10/300 GL; GE healthcare) to remove excess Nb6.
Bio-Layer Interferometry (BLI) and Dynamic Light Scattering (DLS) Analyses
In order to determine constants for equilibrium dissociation (KD), association rate (kon) and dissociation rate (koff) to understand binding of Nb6 to Nsp9, BLI analyses were carried out using ForteBio’s Octet optical biosensor system (Sartorius, Fremont, CA, United States). Briefly, biotinylated Nsp9 was generated using EZ-Link NHS-LC-LC-Biotin (Thermo Fisher Scientific) then was loaded onto the streptavidin biosensor (Sartorius). Various concentrations of Nb6 (1 μM, 500, 250, 125, and 62.5 nM) were tested.
Dynamic Light Scattering analyses were performed using a DLS measurement instrument (Malvern, Malvern, United Kingdom) equipped with a 2-ml micro-sampling cell maintained at 25°C. The Nsp9-Nb6 complex was diluted to 1.5 μM in buffer containing 20 mM Tris–HCl, pH 8.0, 150 mM NaCl. All samples were filtered through a 0.22-μm Millipore filter membrane to remove any dust particles prior to DLS analyses. Each sample was transferred to a cuvette that was placed into the unit and equilibrated for 2 min at 4°C before measurements were taken.
Small-Angle X-ray Scattering (SAXS) Assay and Structural Modeling
Small-angle X-ray Scattering measurements of Nsp9 and Nsp9-Nb6 complex molecular envelopes in the buffer containing 20 mM Tris–HCl, pH 8.0, 150 mM NaCl were conducted on the SIBYLS Beamline 12.3.1 at the Shanghai Synchrotron Radiation Facility (SSRF) BL19U2. All data sets were measured with three exposure times (0.5, 1, and 6 s) at 283 K. Three concentrations (2, 3, and 5 mg/ml) of Nsp9 and the complex were used for measurements. Data of buffers were collected between every two protein samples. Scattering data for triplicate solutions were then scaled and average values were subtracted. All curves obtained for the three different solution concentrations were also scaled and merged to generate an average scattering curve. Quality scores of scattering curves were analyzed using the program PRIMUS (Konarev et al., 2003) to assure that no radiation damage and no obvious protein aggregation had occurred before selection of the best sample for use in further analyses. Initial radius of gyration (Rg) values were calculated from Guinier plots and the P(r) distribution function was calculated using the GNOM program (Petoukhov et al., 2012). Low-resolution shapes of Nsp9 and the Nsp9-Nb6 complex in solution were modeled using DAMMIF server (Franke and Svergun, 2009) based on P1 symmetry and an asymmetric unit. Continuous and meaningful shapes were extracted and averaged using DAMAVER server (Volkov and Svergun, 2003). Protein structure models of Nsp9 and Nb6 were predicted using the I-TASSER program1 as previously described (Liu et al., 2016) then models were built onto the envelopes using the Chimera program (Pettersen et al., 2004). Figures were generated using PyMOL (Delano, 2002).
Verification of the Interaction Between Nsp9 and Nb6 in vitro
The Nsp9-Nb6 interaction was evaluated by ELISA using 96-well microtiter plates coated with Nsp9 mutant protein (200 ng/well). After incubation with biotinylated Nb6, Nb-bound proteins were detected using horseradish peroxidase (HRP)-conjugated streptavidin (Proteintech, Wuhan, China) with wild type Nsp9 included as the control. Similarly, biotinylated wild type (WT) Nsp9 was utilized to test its binding to the Nb6 mutants.
Reverse Genetics-Based Mutagenesis of PRRSV
Reverse genetics-based mutagenesis of PRRSV was conducted after DNA from a known infectious viral strain (SD16) was cloned to generate pBAC-SD16 (Wang C. et al., 2013). Amino acid (aa) residues at positions 588, 590, and 643 of Nsp9 were mutated via site-directed mutagenesis using primers listed in Table 1. After cDNA fragments containing desired mutations were digested with BmtI and AscI, they were ligated to pBAC-SD16 and constructs were further validated by DNA sequencing. To rescue mutated viruses, pBAC-SD16 and constructs containing mutated Nsp9 sites were introduced into MARC-145 cells using Attractene Transfection Reagent according to the manufacturer’s protocol (Qiagen, Hilden, Germany). Cell culture supernatants were harvested 3 days post-transfection and indirect immunofluorescence assay (IFA) and western blotting (WB) analyses were conducted to confirm replication of recombinant viruses. Then the cell culture supernatants serially passaged in MARC-145 cells three times; virus rescue was confirmed using whole-genome sequencing. Rescued viruses were designated rSD16-I588A, rSD16-D590A, and rSD16-L643A.
Indirect Immunofluorescence Assay (IFA)
Virus-transfected (infected) or un-transfected (un-infected) cells were fixed with 4% paraformaldehyde at room temperature (RT) for 20 min then washed three times with PBS. Fixed cells were immediately incubated with primary antibody 6D10 diluted in PBS (pH 7.4, containing 1% BSA and 0.1% Triton X-100) at RT for 2 h. Interaction of primary antibody with corresponding target was visualized using FITC-conjugated goat anti-mouse IgG (Jackson, West Grove, PA, United States) at RT for 1 h. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) and observed via fluorescence microscopy (Zeiss, Oberkochen, Germany).
Western Blotting
Cells were lysed using ice-cold NP-40 lysis buffer (Sorlarbio, Beijing, China) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) (Beyotime, Beijing, China). Cellular proteins were separated by SDS-PAGE then proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). Next, membranes were blocked by incubation in PBS buffer containing 5% skim milk supplemented with 0.1% Tween-20 then membrane-bound proteins were probed with anti-PRRSV nucleocapsid (N)-specific monoclonal antibody (Clone No. 6D10; made in-house) for 1 h at 37°C. Antibody-bound PRRSV-N protein on membranes was detected after incubation with HRP-conjugated secondary antibodies followed by development using ECL chemiluminescence substrate. Housekeeping protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control.
Growth Kinetics of Mutant Viruses in MARC-145 Cells
Viral growth kinetics for wild type (WT) and mutant viruses were analyzed as previously described (Li et al., 2015). Briefly, MARC-145 cells grown in 24-well plates were infected with either WT or mutant viruses at 0.01 MOI. After a 1-h inoculation at 37°C, cell culture supernatants were replaced with fresh medium containing 3% FBS at 37°C. Culture supernatants were collected at 12, 24, 36, 48, 72, and 96 h post-infection (hpi). Viral titers of serial dilutions of harvested supernatants were calculated using the Reed-Muench method (Reed and Muench, 1938).
Real-Time Quantitative PCR (qPCR)
MARC-145 cells infected with mutated or control viruses were harvested using TRizol Reagent (Thermo Fisher Scientific) according to the manufacturer’s instruction. Total RNA was reverse transcribed using a PrimeScript RT Kit (TaKaRa) following the manufacturer’s instructions then qPCR was performed using reactions containing SYBR Green PCR Master Mix (Roche, Mannheim, Germany) using an ABI 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) with condition was: 95°C for 60 s; 40 cycles of 95°C for 15 s and 55°C for 34 s. Analysis of β-actin transcription was included in order to normalize relative expression levels of target genes as described previously (Zhang et al., 2017a,b) and calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Detection of Viral Negative-Strand Genomic RNA [(−) gRNA] by Reverse Transcription PCR (RT-PCR)
MARC-145 cells were infected with mutant or wild type viruses (0.01 MOI) for 24 h prior to RNA isolation. To remove contaminating DNA, RNA preparations were further processed using a DNA-free Kit (Thermo Fisher Scientific). RT-PCR was employed to detect (−) gRNAs as previously described (Lu et al., 2011; Zhao et al., 2018). Briefly, first-strand cDNA was synthesized using forward primer F-6 (Table 1) using reverse transcriptase Superscript III (Thermo Fisher Scientific) and 2 μg of total RNA. After removing remaining RNA with RNase A (Thermo Fisher Scientific), nested PCR was performed to detect (−) gRNA using primer pairs F-6/R-683 and F-12/R-343 (Table 1) via amplification with PrimeSTAR® Max DNA Polymerase (TaKaRa) with reaction program was: 30 cycles of 98°C for 30 s, 55°C for 15 s and 72°C for 10 s. Gray values for PCR products were measured using Image J software.
Statistical Analysis
All experiments were conducted for ≥ 3 replicate samples then were independently repeated. Statistical analysis was conducted using GraphPad Prism 6, with significance assessed using Student t-test. A p-value < 0.05 was considered statistically significant.
Results
Preparation of Nsp9-Nb6 Complex
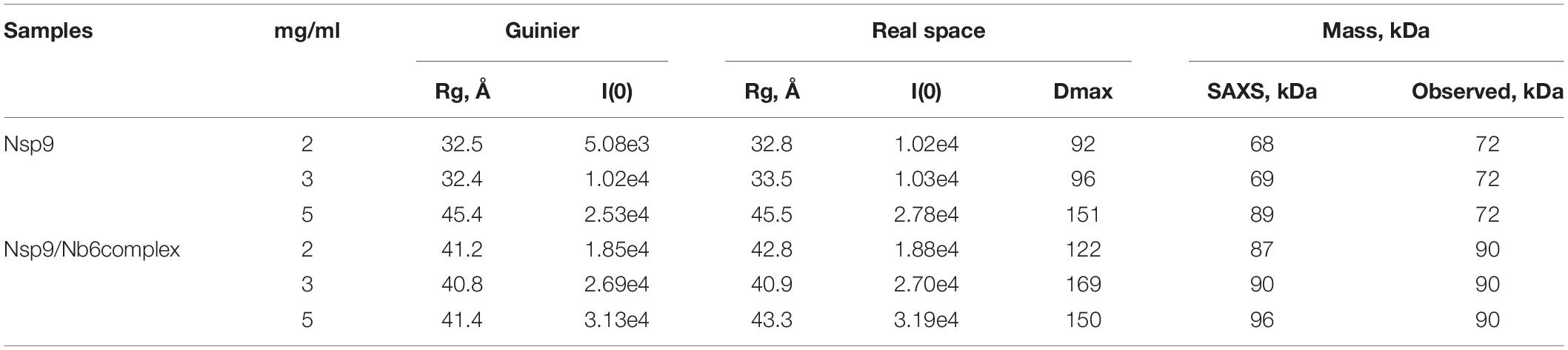
To discover the molecular structure of PRRSV-Nsp9, soluble Nsp9 was recombinantly expressed in a prokaryotic expression system and Nb6 was produced using a Pichia pastoris expression system. Both proteins were purified using metal-affinity and size-exclusion chromatography then were detected using SDS-PAGE (Figures 1A,B). Next, BLI analyses were carried out to study the interaction between recombinant Nsp9 and Nb6. As shown in Figure 1C, Nb6 bound to Nsp9 with high affinity, with a KD value of approximately 10 nM. Kinetic data analysis (kon: 1.44 × 102 M–1 s–1 and Koff: 2.56 × 10–5 s–1) further showed that the interaction involved a quick association process and a slow dissociation process.
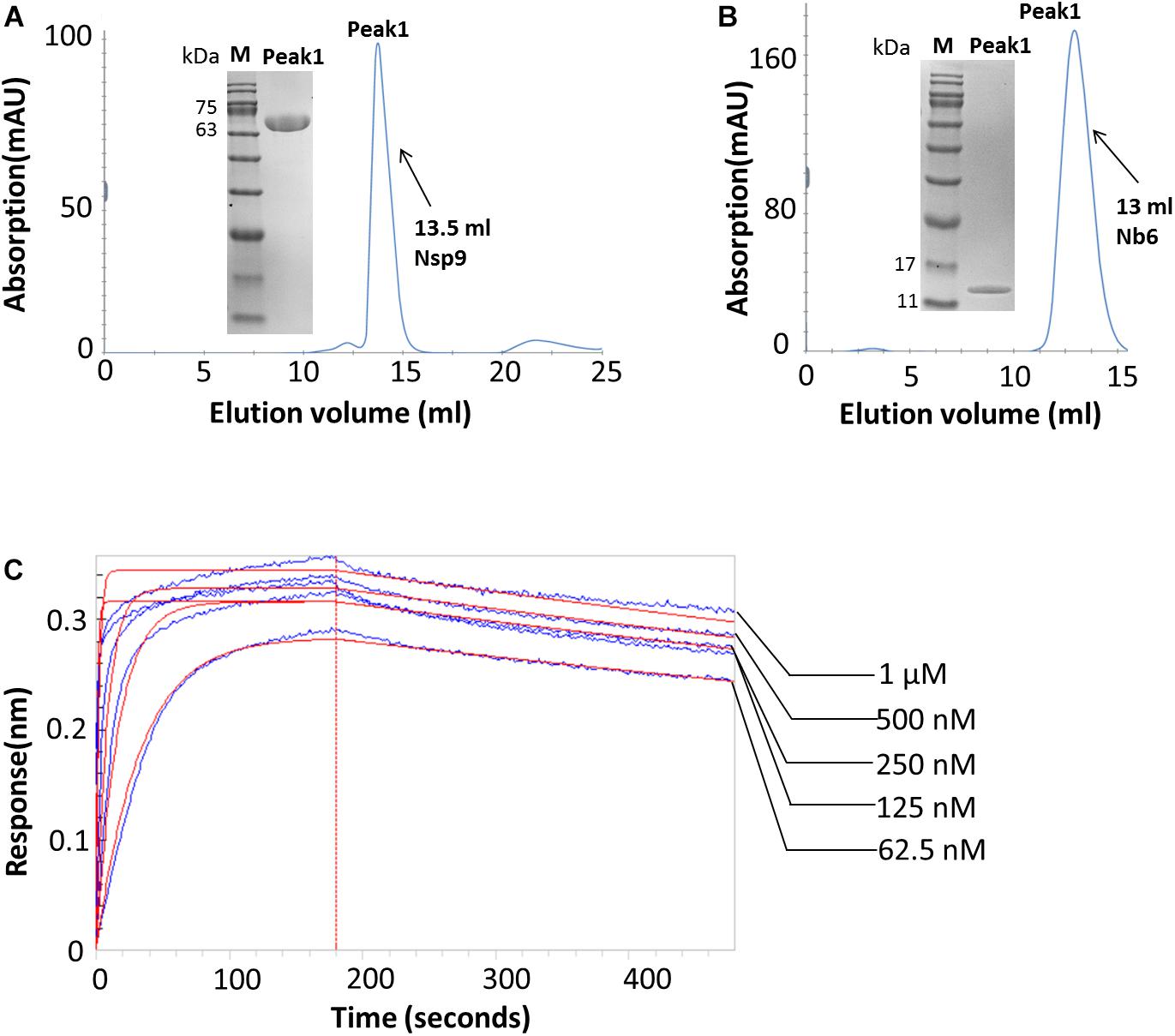
Figure 1. Expression and analysis of recombinant Nsp9 and Nb6. (A) Size-exclusion chromatography of recombinant Nsp9 on calibrated Superdex 200 increase 10/300 GL column. The target protein was eluted at a retention volume of 13.5 ml, corresponding to approximately 72 kDa as monomer. Characterization of recombinant Nsp9 after two-step purification was analyzed by SDS–PAGE (Inset), M: protein ladder (10–180 kDa, Sorlarbio, Beijing, China), Gel: 12% precast polyacrylamide gel (Item No. NP0341BOX, Thermo Fisher Scientific). (B) Size-exclusion chromatography of recombinant Nb6 on calibrated Superdex 75 10/300 GL column. The target protein was eluted at a retention volume of 13 ml, corresponding to approximately 15 kDa as monomer. Characterization of recombinant Nb6 after two-step purification was analyzed by SDS–PAGE (Inset), M: protein ladder (10–180 kDa, Sorlarbio, Beijing, China), Gel: 12% precast polyacrylamide gel (Item No. NP0341BOX, Thermo Fisher Scientific). (C) Analysis of purified Nb6 binding to Nsp9. Gradient concentrations (62.5 nM–1 μM) of Nb6 in PBS buffer (pH7.4), containing 0.02% tween-20 and 0.1% BSA were passing through the streptavidin biosensor loaded with biotinylated Nsp9, and the signals were recorded.
To generate the Nsp9-Nb6 complex, Nsp9 was mixed with an excess of Nb6 and the mixture was incubated at 4°C overnight. The complex was purified using gel filtration chromatography and detected via SDS–PAGE. As demonstrated in Figure 2A, two elution peaks were observed: Peak1 represented the complexed form of the two proteins and Peak2 represented unbound Nb6. Additionally, DLS analysis suggested that the complex that formed was homogeneous (Figure 2B).
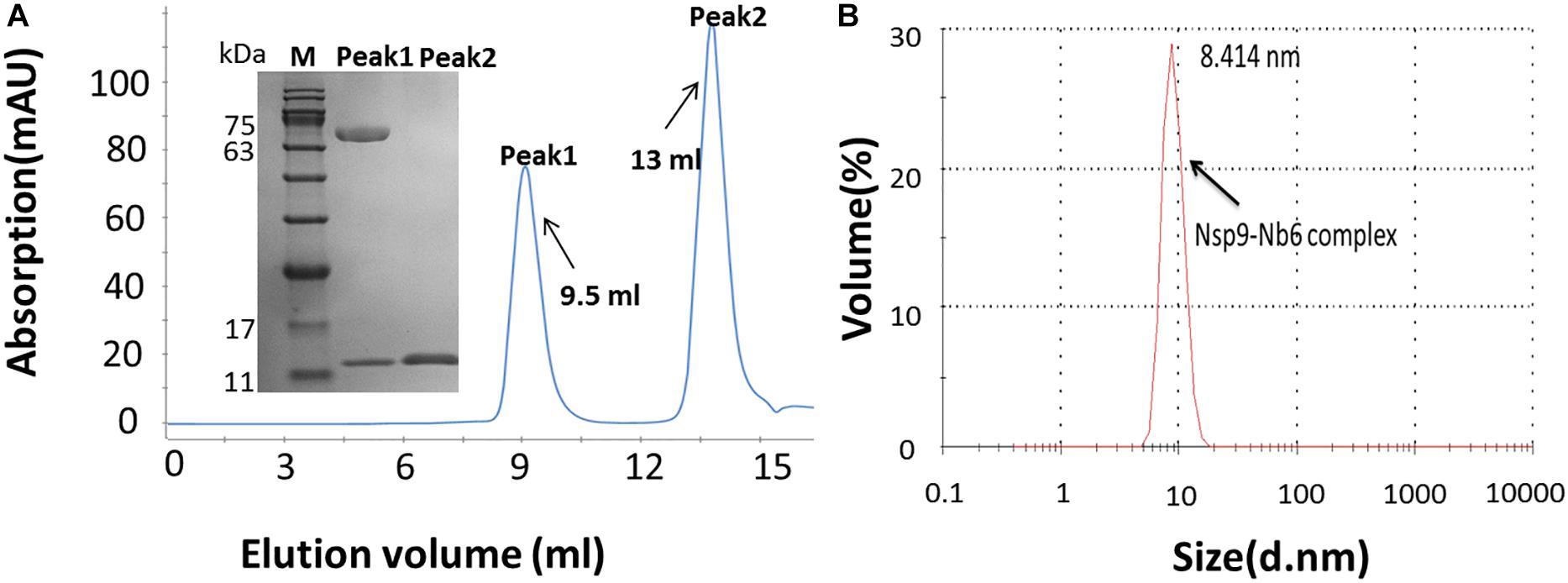
Figure 2. The Nsp9-Nb6 complex exists as a heterodimer. (A) The Nsp9-Nb6 complex was purified with high purity by calibrated Superdex 75 10/300 GL column. Two peaks were observed, with the retention volume of the Peak1 was 9.5 ml, corresponding to the heterodimer of the Nsp9-Nb6 complex, and the retention volume of the Peak 2 was about 13 ml, corresponding to the monomer of Nb6. Characterization of the complex after purification was analyzed by SDS–PAGE (Inset) M: protein ladder (10–180 kDa, Sorlarbio, Beijing, China), Gel: 12% precast polyacrylamide gel (Item No. NP0341BOX, Thermo Fisher Scientific). (B) DLS of Nsp9-Nb6 complex at 1.5 μM. The hydrodynamic molecular radius of the complex is 8.414 nm.
Structural Characterization of Nsp9 and Nsp9-Nb6 Complex by SAXS
After a tremendous effort to directly generate the crystal structure of Nsp9-Nb6 complex had failed (data not shown), we next used SAXS to characterize molecular envelope structures of Nsp9 and the Nsp9-Nb6 complex. Three concentrations (2, 3 and 5 mg/ml) of Nsp9 alone were prepared and analyzed, with results showing Nsp9 protein masses of 68, 69, and 89 kDa, respectively. However, no evidence for the existence of higher order oligomers was found at any of the three protein concentrations. Considering the inherent error of the SAXS technique (Fischer et al., 2010), these results suggest that Nsp9 may exist as a monomer in solution (Table 2). Based on its highest scattering curve signal-to-noise ratio and lowest observed level of radiation damage (Figure 3A), the 2-mg/ml sample was selected for further analysis. The clear linear performance of the Guinier plot (inset image, Figure 3A) derived from this curve supported a monodispersed protein sample and yielded an Rg value of 32.4 Å, a value consistent with that obtained by scattering analysis (Rg of 33.5 Å, Table 2). The P(r) function suggested that Nsp9 was not present as a globular assembly in solution (Figure 3B), due to its maximum dimension approaching 96 Å. Kratky analysis was used to evaluate the degree of folding (Mertens and Svergun, 2010) and demonstrated (Figure 3C) a peak value around 11.5 at low q values that reverted to zero at high q values, suggesting that recombinant Nsp9 obtained here was correctly folded.
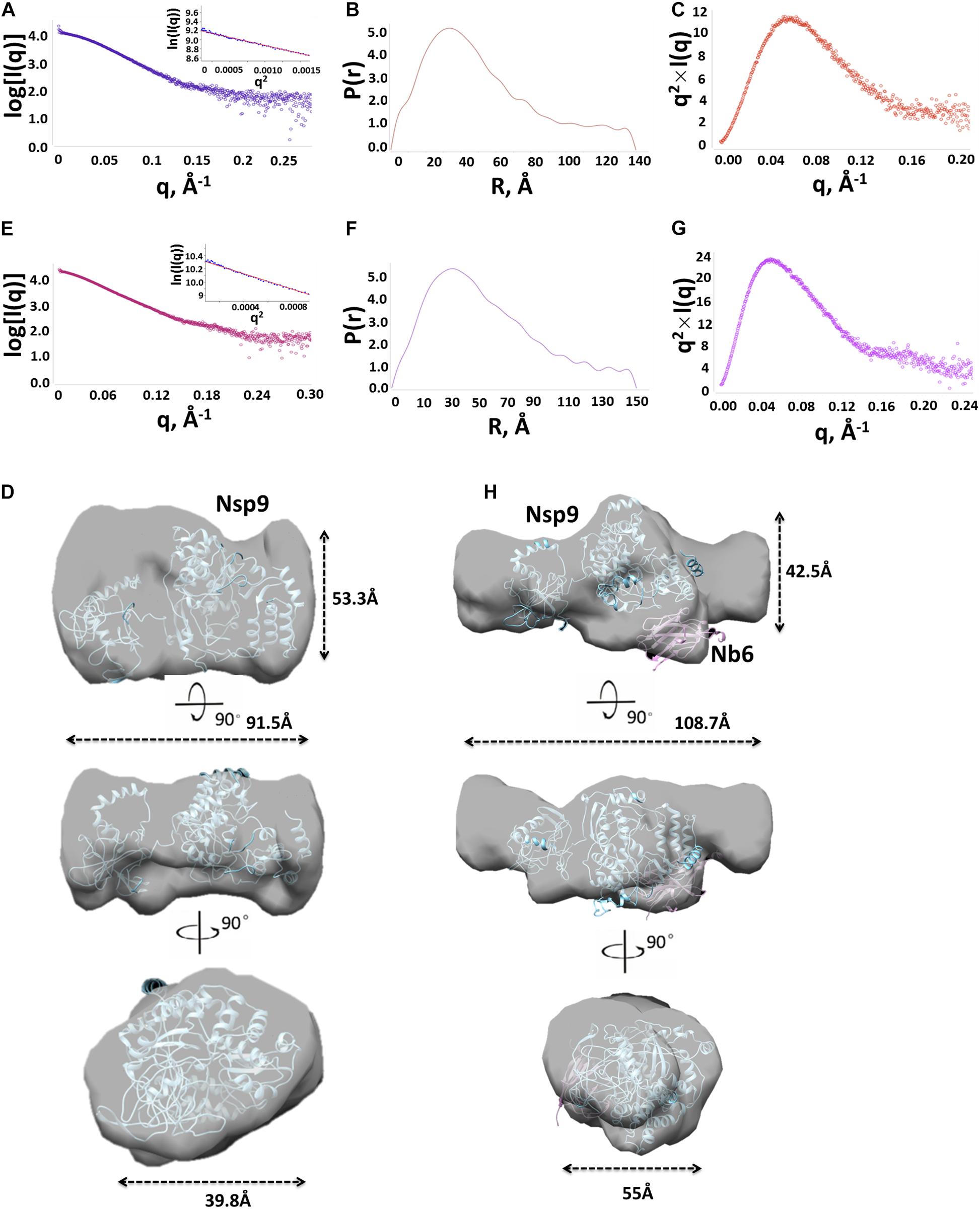
Figure 3. Structural characterization of Nsp9 and Nsp9-Nb6 complex by SAXS. (A) Experimental SAXS curve (open circles) of Nsp9 in solution. The Guinier region and the corresponding linear fitting were shown in the inset. (B) Pair-distance distribution function derived from the experimental curve. (C) Kratky plot calculated from the experimental data. Bell shaped curves indicate compact structures. (D) Superimposition of the experimental envelope (as shown in surface) onto the model of Nsp9 represented in cartoon. (E) Experimental SAXS curve (open circles) of Nsp9-Nb6 complex in solution. The Guinier region and the corresponding linear fitting were shown in the inset. (F) Pair-distance distribution function derived from the experimental curve. (G) Kratky plot calculated from the experimental data. Bell shaped curves indicate compact structures. (H) Superimposition of the experimental envelope (as shown in surface) onto the heterogeneous model of Nsp9 and Nb6 heterodimer represented in cartoon.
To build the molecular envelope of Nsp9 from the scattering curve, it is important to know whether molecular symmetry is present in the particle, since this knowledge is essential for assessing the reliability of a molecular shape determined by SAXS. We first built the monomer molecular envelope using DAMMIF server (Franke and Svergun, 2009) by imposing a one-fold symmetry constraint onto Nsp9 then the DAMAVER program was used to generate an average molecular envelope. The model of Nsp9 was predicted using the I-TASSER server and built onto the averaged molecular envelope obtained using CHIMERA (Pettersen et al., 2004). Using this method, the molecular envelope was determined to be asymmetric, with a barrel shape of overall dimensions of approximately 91.5 Å × 53.3 Å × 39.8 Å (Figure 3D).
Meanwhile, the molecular envelope of the Nsp9-Nb6 complex was built from the scattering curve in parallel. First, SAXS results of the complexes at three concentrations yielded molecular masses of 87, 90, and 96 kDa, respectively, indicating that the Nsp9-Nb6 complex was a heterodimer in solution. Next, the 5 mg/ml sample was chosen for further analysis, due to its higher scattering curve signal-to-noise ratio and absence of radiation damage; the resulting Guinier plot (Figure 3E) exhibited a clearly linear presentation that indicated complexed protein was present in a monodispersed state (Figure 3E). The Rg value was 41.4 Å which was consistent with the value obtained using Real Space analysis (Rg of 43.3 Å, Table 2). The P(r) function suggested that the Nsp9-Nb6 complex was not present as a globular assembly in solution, due to its maximum dimension approaching 150 Å (Figure 3F). Kratky analysis give a peak value of about 24 at low q values and zero at high q values, suggesting that the complex was correctly folded (Figure 3G). The average molecular envelope was obtained using DAMMIF server results based on a one-fold symmetry constraint followed by analysis using the DAMAVER program. Meanwhile, the Nsp9-Nb6 complex structure was predicted using the online I-TASSER server then the predicted structural models were built onto the averaged molecular envelope using CHIMERA. Ultimately, the molecular envelope of the complex was also shown to be asymmetric, with a Z-shape with overall dimensions of approximately 108.7 Å × 55 Å × 42.5 Å (Figure 3H).
As demonstrated above, it was notable that a molecular envelope shape change occurred when Nb6 bound to Nsp9, with the shape becoming longer and a protrusion forming in the middle of the shape that were consistent with Nb6 represented as the protrusion of the complex. Ultimately, the final secondary structure models fitted tightly together with the SAXS molecular envelope, with complex formation shown to involve Nsp9 aa residues Ile588, Asp590, Leu643 and Asp646 and Nb6 aa residues Trp105, Pro107, Tyr62 and Asp64 (Figure 4).
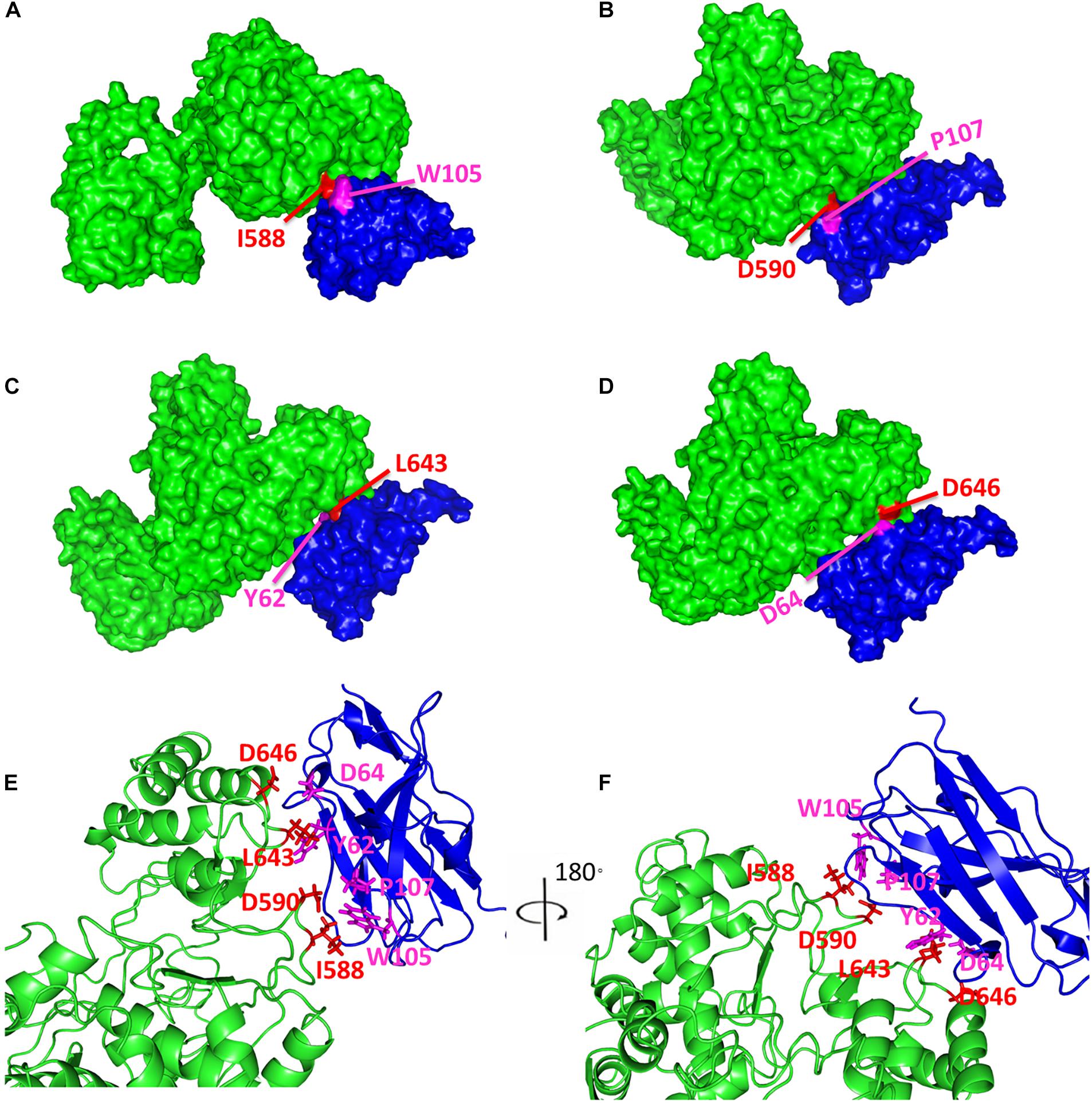
Figure 4. Predicted model of Nsp9-Nb6 complex. The surface representations of Nsp9-Nb6 complex are shown in (A–D). Nsp9 is colored in green, Nb6 is in blue, Ile588 (A), Asp590 (B), Leu643 (C) and Asp646 (D) in Nsp9 are highlighted in red, Trp105 (A), Pro107 (B), Tyr62 (C) and Asp64 (D) in Nb6 are highlighted in magentas. (E,F) The predicted complex of Nsp9 and Nb6 are shown in cartoon diagrams.
Screening for Amino Acid Residues Involved in the Binding Interaction Between Nsp9 and Nb6
After the abovementioned structure model was used to predict putative key Nsp9 residues (aa 588, 590, 643, 646) required for interacting with Nb6, site-directed mutagenesis was carried out to generate a panel of recombinant single-site Nsp9 mutants with aa substitutions of those key residues. Next, mutant Nsp9 proteins were expressed, isolated, and then tested for binding to biotinylated Nb6 via ELISA. Based on ELISA results, the interaction of mutant protein Nsp9-D646A with Nb6 was similar to that of WT Nsp9 with Nb6. However, the Nsp9 I588A mutant exhibited a complete loss of binding to Nb6, while D590A and L643A mutants exhibited only impaired binding to Nb6 (Figure 5A). Conversely, single-site Nb6 mutants were also studied. Although the binding of Nb6-D64A was similar to the WT control, Nb6-W105A and Nb6-P107A mutants did not bind to Nsp9 at all, while Nb6-Y62A showed partial binding (Figure 5B). When considered together, these results suggest that Nsp9 aa residues 588, 590, and 643 interacted with Nb6 aa residues 105, 107, and 62 in a reciprocal fashion to form the Nsp9-Nb6 complex.
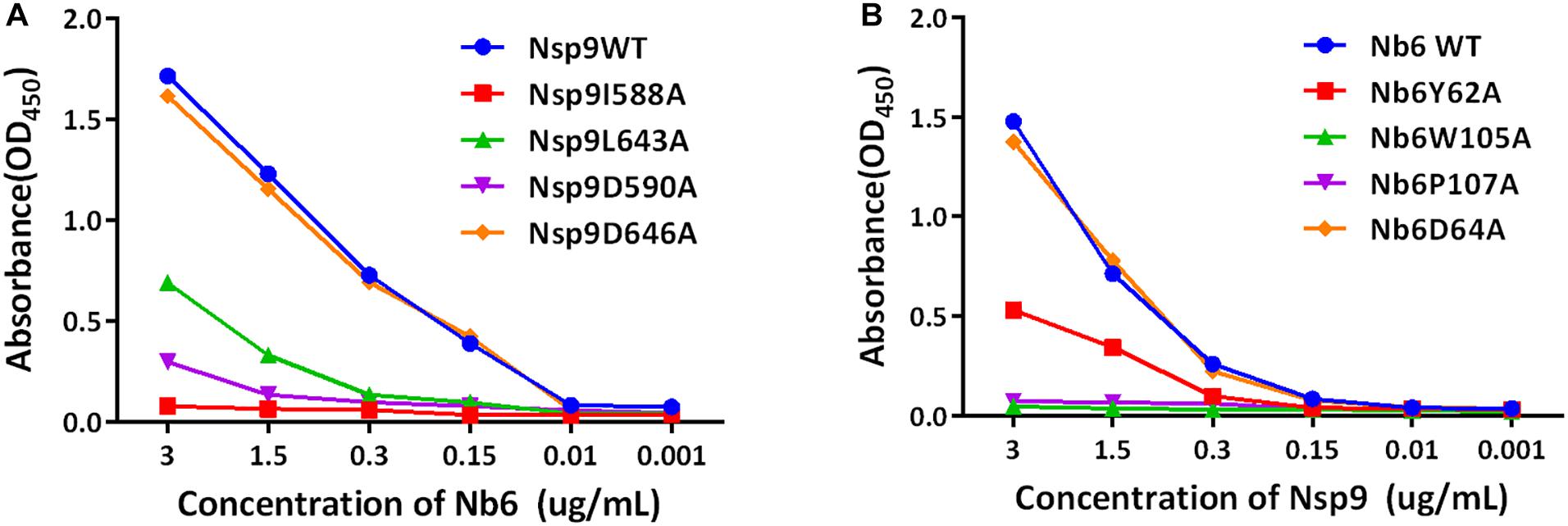
Figure 5. Identification of residues involved in the interaction between Nsp9 and Nb6 by ELISAs. (A) Binding efficiency of the WT and mutant Nsp9 to Nb6. 96-well microtiter plates were coated with the WT and mutant Nsp9 (200 ng/well). After incubation with biotinylated Nb6, bound proteins were detected by HRP-conjugated streptavidin. (B) Binding efficiency of the WT and mutant Nb6 to Nsp9. 96-well microtiter plates were coated with the WT and mutant Nb6 (200 ng/well). After incubation with biotinylated Nsp9, bound proteins were detected by HRP-conjugated streptavidin. The binding efficiency was calculated on the basis of optical density (OD450). Assays were performed in triplicate and data are presented as means ± SD.
Mutations of Residues 588 and 643 of PRRSV-Nsp9 Affect Viral Replication in vitro
To characterize the involvement and biological significance of the abovementioned residues in PRRSV replication, a reverse genetics-based system was employed using an infectious clone of HP-PRRSV strain SD16 (pBAC-SD16) as the backbone for mutagenesis. However, only reverse-genetically engineered virus with Nsp9 substitutions I588A, D590A or L643A could be rescued, with replication of recombinant viruses confirmed based on PRRSV-N protein expression in MARC-145 cells using IFA and WB (Figure 6).
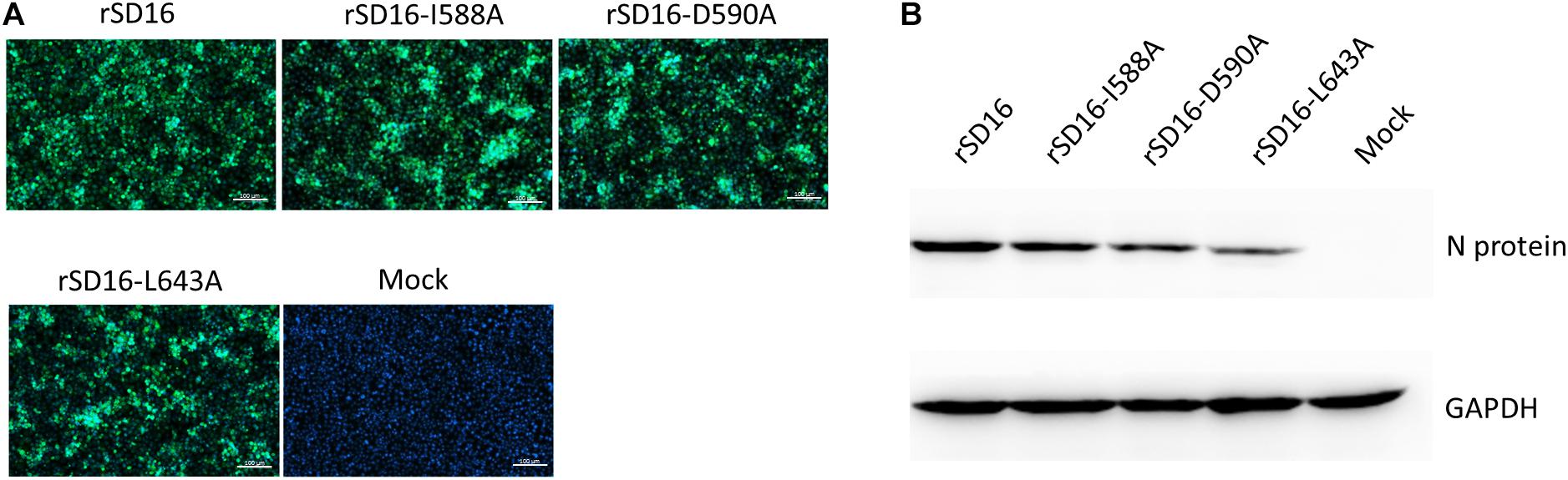
Figure 6. Rescue of recombinant PRRSV bearing Nsp9 mutation. (A) MARC-145 cells transfected with DNA-launched infectious clones of wild type PRRSV-SD16 strain (rSD16) or infectious clones bearing indicated mutations in the putative interaction sites (I588A, D590A, L643A) between Nsp9 and Nb6 on rSD16 backbone. IFA were performed to detect PRRSV-N protein expression and visualized by FITC-conjugated goat anti-mouse IgG. Overlay images were visualized for cell nuclei (blue) and N protein (green) with mock-transfected cells were included as control. The scale bars are 100 μm. (B) WB analysis of PRRSV-N protein expression in MARC-145 cells transfected with DNA-launched infectious clones of indicated mutants or wild type virus. Mock-transfected cells were used as control.
To compare these mutants to wild type virus, mutant virus growth kinetics were characterized in MARC-145 cells. As shown in Figure 7A, titers of all viruses reached peak levels at 48 hpi then decreased from 48 hpi to 96 hpi, with mutant virus rSD16-D590A and rSD16 exhibiting similar growth kinetics. At 24 hpi, average rSD16-I588A and rSD16-L643A virus titers reached 4.208 log10TCID50/ml and 4.417 log10TCID50/ml, respectively, that were both significantly lower than the rSD16 titer. At 36 hpi, mutant viruses rSD16-I588A and rSD16-L643A titers were 5.71 log10TCID50/ml and 5.5 log10TCID50/ml, respectively, which were also markedly lower than the rSD16 titer. Virus titers of mutants rSD16-I588A and rSD16-L643A, measured at 48 h, 72 h and 96, attained similar levels at 24 h and 36 h as compared to wild type virus or rSD16-D590A (Figure 7A). Meanwhile, detailed analysis of viral RNA copy numbers at early time points after MARC-145 cell infected with mutant viruses and wild type virus demonstrated similar trends as well (Figure 7B), implying that mutation of Nsp9 residues at positions 588 and 643 (I588A and L643A) could potentially impair PRRSV RNA replication. Therefore, to further investigate viral genome replication, intracellular RNAs were isolated from transfected MARC-145 cells and analyzed via RT-PCR analysis for (−) gRNA. Based on the semi-quantification method, rSD16-I588A and rSD16-L643A (−) gRNA synthesis levels were approximately 12-fold and 10-fold lower than the wild type rSD-16 level, respectively (Figures 7C,D), thus supporting our speculation that I588A and L643A Nsp9 mutations may reduce viral RdRp activity. Meanwhile, further evidence was obtained using fresh MARC-145 cells and CRL-2843-CD163 cells inoculated with recombinant viruses (at a MOI of 0.1) via monitoring of viral replication by immunofluorescence after incubating inoculated cells with monoclonal antibody specific for PRRSV-N protein (Figures 8A,C). A quick enumeration of immunofluorescent cell numbers revealed reduced replication levels of recombinant viruses bearing I588A or L643A mutations relative to wild type virus levels after inoculation of cells with the same initial dose of virus (Figures 8B,D). Taken together, the abovementioned results indicate that the mutations of Nsp9 residues 588 and 643 decreased replication of SD16 strain PRRSV in vitro.
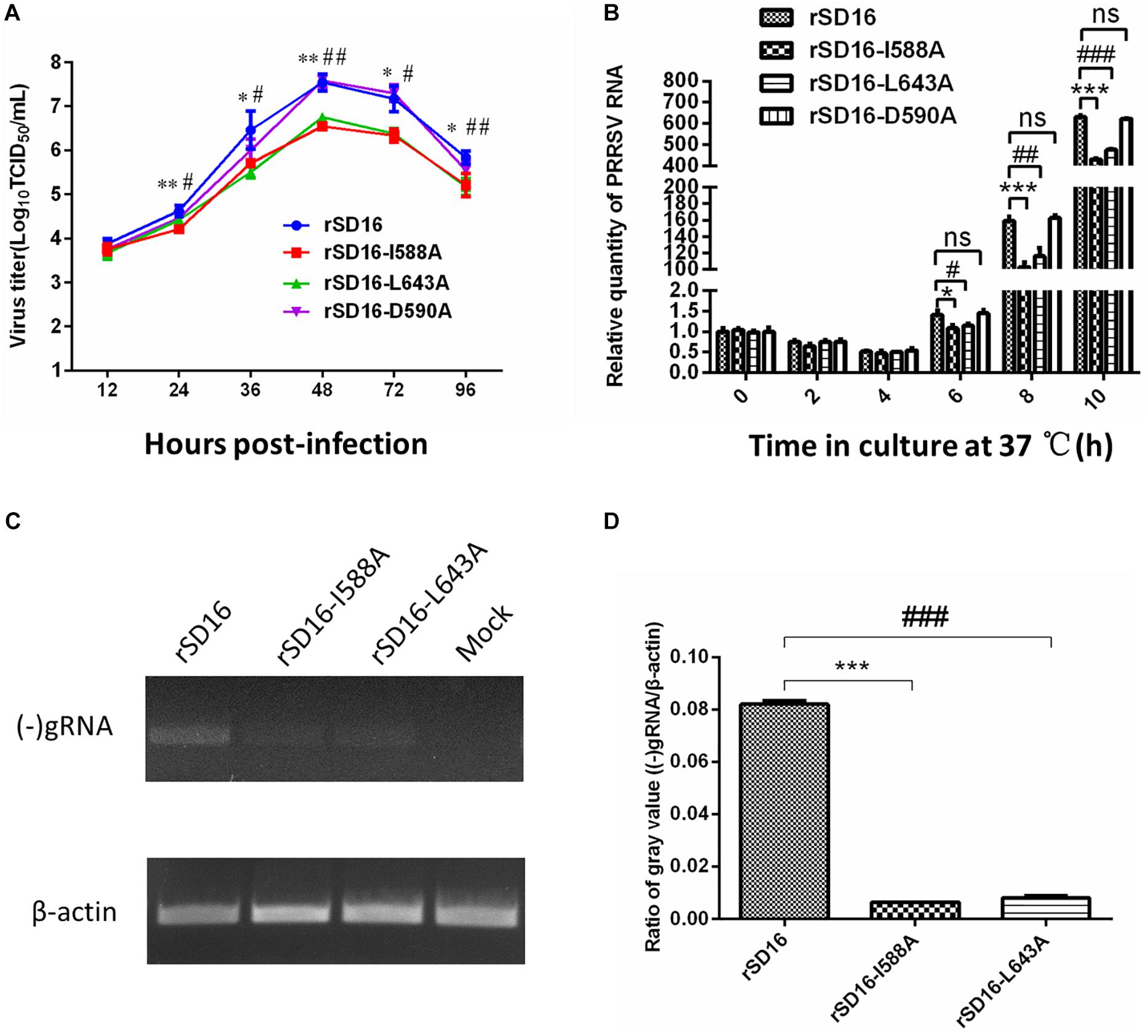
Figure 7. The effect of Nsp9 mutants on PRRSV replication. (A) The growth kinetics of the rescued recombinant viruses bearing different mutations in Nsp9 were analyzed. (B) Relative levels of PRRSV RNA of recombinant viruses were further analyzed for early time points. Data represent means ± SD from three independent experiments. Asterisk (*) indicates a significant difference between rSD16 and rSD16-I588A (*p < 0.05; **p < 0.01; ***p < 0.001). Pound (#) shows a significant difference between rSD16 and rSD16-L643A (#p < 0.05; ##p < 0.01; ###p < 0.001); ns, not significant. (C) Total cellular RNAs were extracted from recombinant virus-transfected MARC-145 cells at 24 hpi, and then subjected to reverse transcribed-PCR (RT-PCR) and agarose gel electrophoresis. The mRNA level of β-actin was included as reference for total RNA input. (D) The ratio of the gray value for PCR products of (−) gRNA/β-actin calculated using Image J software. Asterisk (*) indicates a significant difference between rSD16 and rSD16-I588A (***p < 0.001). Pound (#) shows a significant difference between rSD16 and rSD16-L643A (###p < 0.001).
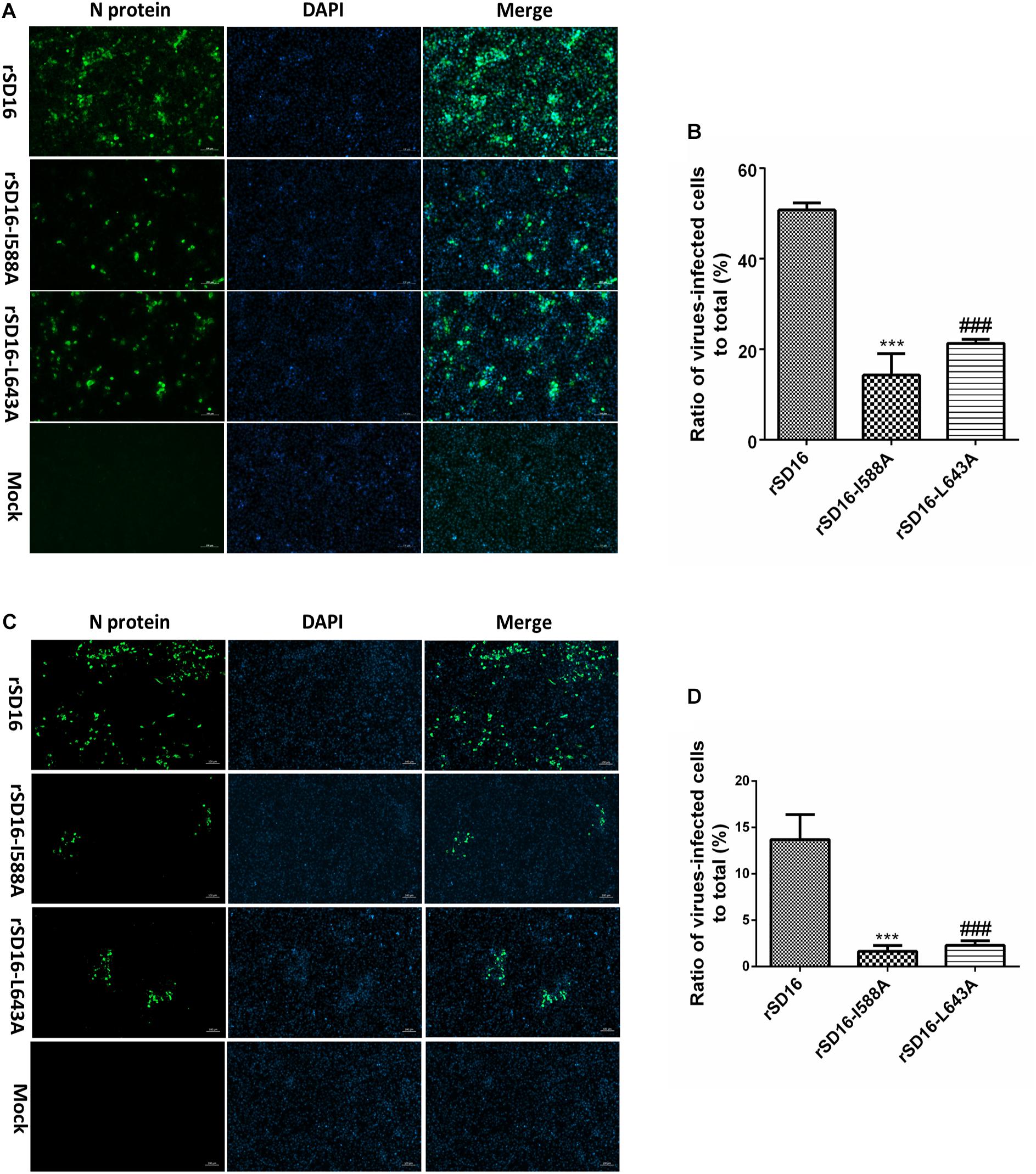
Figure 8. Recombinant PRRSV viruses bearing mutations of residues 588 and 643 of Nsp9 impaired PRRSV replication in vitro. MARC-145 Cells (A) and CRL-2843-CD163 cells (C) were infected with mutated viruses or wild type rSD16 (0.1 MOI). IFA was performed at 48 hpi with the monoclonal antibody recognized PRRSV-N protein (Clone No. 6D10) and visualized by FITC-conjugated goat anti-mouse IgG. Overlay images were visualized for cell nuclei (blue) and N protein (green). The scale bars are 100 μm. (B,D) The ratio of virus-infected cells/total cells calculated using Image J software. Asterisk (*) indicates a significant difference between rSD16 and rSD16-I588A (***p < 0.001). Pound (#) shows a significant difference between rSD16 and rSD16-L643A (###p < 0.001).
Amino Acid Residue 588 of Nsp9 Is Conserved in Both PRRSV Genotypes
Due to the fact that our data suggested that Nsp9 amino acid resides 588 and 643 were crucial for PRRSV replication in cells, Nsp9 amino acid sequences for the aa561-689 region from 32 PRRSV strains (12 Betaarterivirus suid 1 isolates and 20 Betaarterivirus suid 2 isolates) were obtained from GenBank and aligned using the Clustal W module of Lasergene 7.1 (DNASTAR, Inc.). The results showed that residue Ile at position 588 was conserved among strains across both PRRSV genotypes, while the Leu residue at 643 was only conserved in the 20 Betaarterivirus suid 2 sequences analyzed here, but not among Betaarterivirus suid 1 isolates analyzed (Figure 9). Although our reverse genetics-based system was based on the Betaarterivirus suid 2 SD-16 strain, our data suggest that these two highly conserved Nsp9 residues (588 and 643) may be crucial for PRRSV replication and thus may serve as novel targets of anti-PRRSV therapies to achieve viral attenuation in vivo.
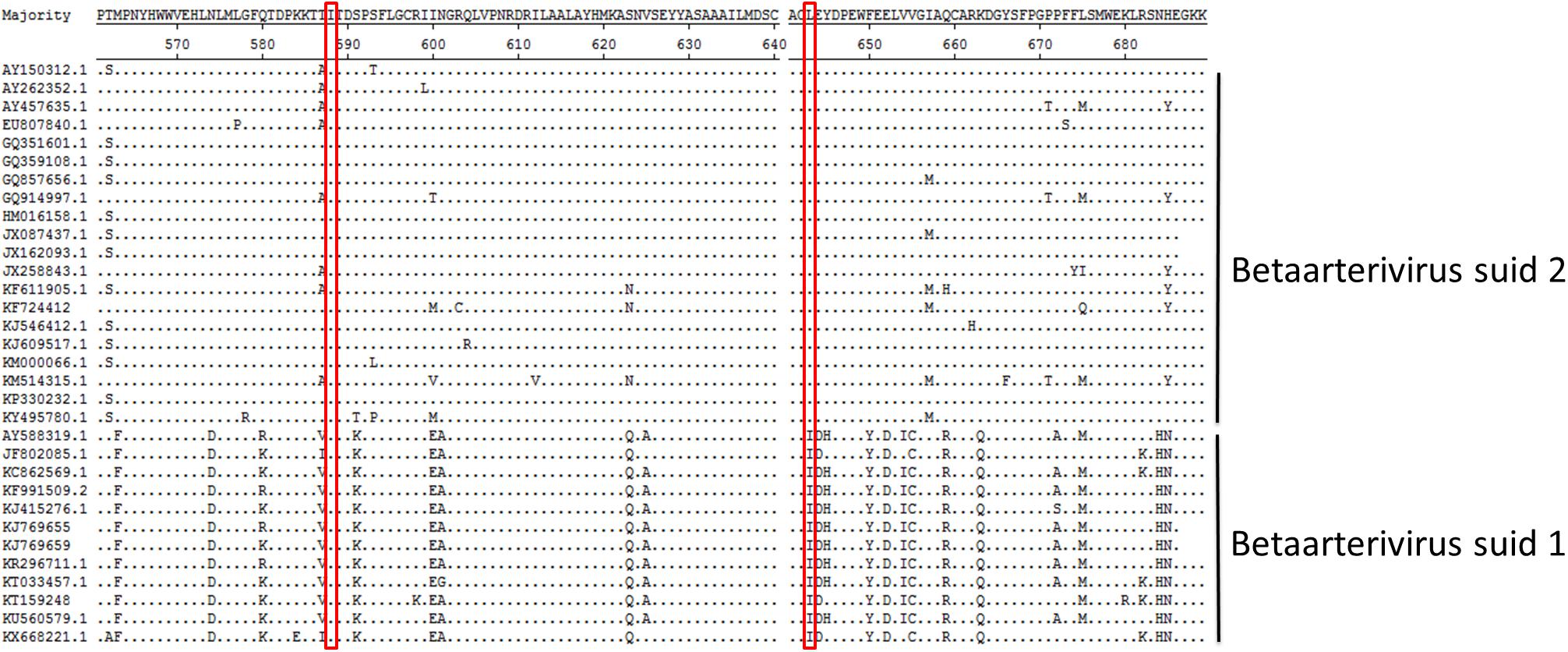
Figure 9. The sequence alignment of aa residues 561-689 in PRRSV Nsp9. The sequences of Nsp9 from the 32 PRRSV strains were pulled from Genbank and then analyzed using Lasergene. The residues at 588 and 643 of Nsp9 are shown in red frame with dots indicating identical amino acid residues.
Discussion
Since its discovery, PRRS has caused significant economic losses to the global swine industry due to the ineffectiveness of vaccinations and the lack of efficient antiviral strategies. In China this situation has worsened since 2006, when HP-PRRSV emerged there. Although tremendous efforts have been dedicated to elucidating PRRSV pathogenesis, contributing factors underlying viral virulence remain elusive, including regulatory mechanisms involving host immunity, viral neutralizing targets and protective immune responses against PRRSV. In early reports, antagonism of host innate immunity was considered the major underlying factor for PRRSV virulence, as several PRRSV proteins, such as Nsp1α, Nsp1β, Nsp2, Nsp4, Nsp11 and PRRSV-N, were demonstrated to block IFNs induction and IFN-activated JAK/STAT signaling (Chen et al., 2010; Li et al., 2010; Patel et al., 2010; Sun et al., 2010; Wang et al., 2013a,b; Wang and Zhang, 2014; Yang et al., 2017) or the NF-κB signaling pathway (Huang et al., 2014). However, this explanation for viral virulence has been challenged by discoveries of PRRSV isolates that maintain a virulent phenotype in vivo but which also induce robust IFNs production and other innate immune response in vitro (Nan et al., 2012; Sun et al., 2016). Therefore, it appears that PRRSV proteins other than innate immunity antagonists also may be involved in PRRSV virulence and pathogenesis.
Since the emergence of HP-PRRSV in China, researchers have conducted several studies to determine viral factors responsible for increased virulence using reverse genetics-based approaches. Unexpectedly, swapping of genetic fragments between infectious clones of highly pathogenic and low pathogenic clones derived from HP-PRRSV isolates has indicated that Nsp9 and Nsp10 are contributing factors to HP-PRRSV virulence in vivo (Li et al., 2014). Moreover, a more recent report has demonstrated that two Nsp9 aa residues (aa519 and aa544) contribute to pathogenicity of HP-PRRSV in vivo by improving PRRSV replication efficiency (Zhao et al., 2018). Therefore, additional investigations of PRRSV-Nsp9 are required to enhance our understanding of PRRSV pathogenesis.
As a PRRSV-specific RdRp, Nsp9 is crucial for viral RNA replication and transcription (Beerens et al., 2007). It has been demonstrated that both viral and cellular factors associate with Nsp9 to regulate viral RNA synthesis, including PRRSV-N protein and host DHX9 RNA helicase and retinoblastoma protein (Dong et al., 2014; Liu et al., 2016). However, the crystal structure of Nsp9 remains unsolved; even our own persistent efforts have failed to produce a high resolution crystal structure of PRRSV-Nsp9. Therefore, we propose that a protein-interacting partner may be needed to stabilize Nsp9 in solution and act as a chaperone to promote suitable crystal formation. In our previous work, we developed the PRRSV-Nsp9-specific nanobody Nb6 to serve as such a chaperone. Nanobodies, unlike conventional antibodies, contain a long stable convex loop within the complementarity determining region 3 (CDR3) that permits them to penetrate deeply into interior pockets of antigens, such as active sites of enzymes (Hassanzadeh-Ghassabeh et al., 2013). Notably, Nbs can also stabilize proteins in certain conformations, with past reports demonstrating successful use of Nbs for inducing lattice formation during crystallization (Lam et al., 2009; Rasmussen et al., 2011; Park et al., 2012). Therefore, recombinant Nsp9 and corresponding Nb6 were produced here to generate the Nsp9-Nb6 complex. However, although crystallization of the Nsp9-Nb6 complex was demonstrated, we ultimately could not obtain suitable crystals for crystallographic analysis. Consequently, we used SAXS to determine the molecular envelope of Nsp9 and the Nsp9-Nb6 complex. SAXS is a well-established technique for generating low-resolution structures of macromolecules in solution (Svergun, 2010), especially for characterization of solution-based protein-to-protein or protein-to-nucleic acid complexes (Gabel, 2015).
Our SAXS data indicate that Nsp9 was monomeric in solution, with the monomer assuming an asymmetric barrel shape (Figure 3D). For Nsp9-Nb6 heterodimers, the molecular envelope was asymmetric and assumed a Z-shape (Figure 3H) and was not present as a globular assembly or fully folded protein in solution. Notably, a conformational change of the molecular envelope occurred when Nb6 bound to Nsp9 that generated a conformation that differed from that of Nsp9 alone. In the Nsp9-Nb6 complex, the molecular envelope was longer and a protrusion was present in the middle of the shape (Figures 3D,H). Therefore, we speculated that Nb6 was situated at the location of the protrusion. In our model, complex formation involved residues Ile588, Asp590, Leu643 and Asp646 of Nsp9 and Trp105, Pro107, Tyr62 and Asp64 of Nb6 (Figure 4). Next, we carried out site-directed mutagenesis of putative amino acid residues responsible for the Nsp9-Nb6 interaction then conducted ELISAs to detect interactions between Nsp9 mutants and Nb6 and interactions between Nb6 mutants and Nsp9. Our result demonstrated that residues at positions 588, 590, 643 of Nsp9 and at positions 62, 105, 107 of Nb6 were involved in the Nsp9-Nb6 interaction (Figure 5). These results are consistent with our previous results demonstrating that Nb6 interacting regions spanned two discontinuous regions located within the C-terminal region of Nsp9 (residues 454-551 and 599-646) (Wang et al., 2019).
Importantly, our previous study had demonstrated that Nb6 blocked PRRSV replication by targeting Nsp9 both in MARC-145 cells and porcine alveolar macrophages (PAMs). In the present study, we identified critical amino acid residues involved in the Nsp9-Nb6 interaction and hypothesized that Nsp9 residues 588, 590 and 643 may affect viral replication. To determine the importance of these residues to PRRSV function, reverse genetics-based mutagenesis was applied to generate mutated viruses bearing a single-mutation or double-mutations. Ultimately, we observed that double-mutations of Nsp9 prevented PRRSV replication rescue of recombinant virus in vitro, implying that these residues are crucial for PRRSV functions (data not shown). Meanwhile, viral replication levels and (−) gRNA synthesis levels for single-mutant viruses rSD16-I588A and rSD16-L643A were lower than corresponding wild type rSD16 levels, indicating that mutations altering Nsp9 amino acids 588 or 643 reduced PRRSV RNA replication.
Based on structural models, PRRSV Nsp9 contains an RdRp domain within its C-terminal sequence (Gorbalenya et al., 1989). Although both residues required for Nb6-Nsp9 interaction (Ile588 and Leu643) are located within the Nsp9 C terminus, their locations are far from the enzymatic center of the RdRp domain, implying that the interaction of Nb6 with Nsp9 may alter the 3-dimensional structure of Nsp9 to block RdRp activity. Alternatively, it is also possible that the interaction between Nb6 and Nsp9 interferes with associations of Nsp9 with other viral or cellular proteins required for viral RTC assembly, although further investigations are required to test these speculations. Nevertheless, sequence alignments generated here indicate that residue Ile588 of PRRSV-Nsp9 is conserved among all Betaarterivirus suid 1 and Betaarterivirus suid 2 isolates analyzed, while residue Leu643 is only conserved in Betaarterivirus suid 2 isolates. These observations suggest that these residues are crucial for PRRSV replication across and within PRRSV genotypes, respectively. As many viruses enhance their pathogenicity by increasing viral replication efficiency (Hanley et al., 2001; Watanabe et al., 2009), it would be interesting to know whether mutation of these residues would attenuate phenotypic PRRSV virulence in vivo. Indeed, a previous report has suggested that mutation of certain Nsp9 residues leads to an attenuated phenotype (Xu et al., 2018; Zhao et al., 2018). However, more evidence is needed than that which has been collected so far (including results of this work) to enhance our understanding of PRRSV Nsp9.
In conclusion, the application of SAXS to understand molecular envelopes of Nsp9 and the Nsp9-Nb6 complex has permitted us to identify two novel residues of Nsp9 required for the Nsp9-Nb6 interaction. Moreover, results obtained here have further elucidated the molecular mechanism whereby Nb6 acts as a novel PRRSV inhibitor while also revealing crucial residues conserved across PRRSV genotypes or within Betaarterivirus suid 2 that are essential for PRRSV replication in vitro. These findings enhance our understanding of PRRSV replication and will likely facilitate development of anti-PRRSV drugs and future vaccines.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YW and RL performed the experiment, analyzed the data, and drafted the manuscript. SQ, JW, HL, ZL, and HM contributed to protein expression. LY, HR, and MW contributed to PRRSV replication assay. JH and JS contributed to the immunofluorescence assay. YN, GZ, and E-MZ conceived the study, carried out additional analyses, and finalized the manuscript. All authors contributed to the revising manuscript.
Funding
This study is supported by the grants from National key R&D Program of China (Grant Nos. 2017YFD0501006 and 2016YFD05007) and National Natural Science Foundation of China (Grant No. 31430084).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the BL19U2 beamline at Shanghai Synchrotron Radiation Facility, Shanghai, People’s Republic of China, for assistance during data collection and Dr. Xiangjing Fu of Zhengzhou University for providing the Fortebio instrument.
Footnotes
References
Beerens, N., Selisko, B., Ricagno, S., Imbert, I., van der Zanden, L., Snijder, E. J., et al. (2007). De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J. Virol. 81, 8384–8395. doi: 10.1128/JVI.00564-07
Chen, Z., Lawson, S., Sun, Z., Zhou, X., Guan, X., Christopher-Hennings, J., et al. (2010). Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398, 87–97. doi: 10.1016/j.virol.2009.11.033
Darwich, L., Gimeno, M., Sibila, M., Diaz, I., de la Torre, E., Dotti, S., et al. (2011). Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet. Microbiol. 150, 49–62. doi: 10.1016/j.vetmic.2011.01.008
Delano, W. L. (2002). The PyMol molecular graphics system. Proteins Struct. Funct. Bioinform. 30, 442–454.
Dong, J., Zhang, N., Ge, X., Zhou, L., Guo, X., and Yang, H. (2014). The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology 464–465, 432–440. doi: 10.1016/j.virol.2014.07.036
Fang, Y., and Snijder, E. J. (2010). The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 154, 61–76. doi: 10.1016/j.virusres.2010.07.030
Fang, Y., Treffers, E. E., Li, Y., Tas, A., Sun, Z., Van, D. M. Y., et al. (2012). Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. U.S.A. 109, 17321– 17322.
Fischer, H., Neto, M. D. O., Napolitano, H. B., Polikarpov, I., and Craievich, A. F. (2010). Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43, 101–109. doi: 10.1107/s0021889809043076
Franke, D., and Svergun, D. I. (2009). DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42(Pt 2), 342–346. doi: 10.1107/S0021889809000338
Gabel, F. (2015). Small-angle neutron scattering for structural biology of protein-RNA complexes. Methods Enzymol. 558, 391–415. doi: 10.1016/bs.mie.2015.02.003
Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P., and Blinov, V. M. (1989). Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17, 4847–4861. doi: 10.1093/nar/17.12.4847
Guo, Z., Chen, X., Li, R., Qiao, S., and Zhang, G. (2018). The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol. J. 15, 1–14.
Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E. B., et al. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448. doi: 10.1038/363446a0
Hanley, K. A., Lee, J. J. Jr., Murphy, B. R., and Whitehead, S. S. (2001). Paired charge-to-alanine mutagenesis of dengue Virus Type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 76:525. doi: 10.1128/jvi.76.2.525-531.2002
Hassanzadeh-Ghassabeh, G., Devoogdt, N., De Pauw, P., Vincke, C., and Muyldermans, S. (2013). Nanobodies and their potential applications. Nanomedicine 8, 1013–1026. doi: 10.2217/nnm.13.86
Huang, C., Zhang, Q., Guo, X. K., Yu, Z. B., Xu, A. T., Tang, J., et al. (2014). Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-kappaB essential modulator. J. Virol. 88, 10934–10945. doi: 10.1128/JVI.01396-14
Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. J., and Svergun, D. I. (2003). PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282. doi: 10.1107/s0021889803012779
Lam, A. Y., Pardon, E., Korotkov, K. V., Hol, W. G. J., and Steyaert, J. (2009). Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus. J. Struct. Biol. 166, 0–15.
Li, H., Zheng, Z., Zhou, P., Zhang, B., Shi, Z., Hu, Q., et al. (2010). The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J. Gen. Virol. 91(Pt 12), 2947–2958. doi: 10.1099/vir.0.025205-0
Li, Y., Zhou, L., Zhang, J., Ge, X., Zhou, R., Zheng, H., et al. (2014). Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. 10:e1004216. doi: 10.1371/journal.ppat.1004216
Li, Z., Wang, G., Wang, Y., Zhang, C., Huang, B., Li, Q., et al. (2015). Immune responses of pigs immunized with a recombinant porcine reproductive and respiratory syndrome virus expressing porcine GM-CSF. Vet. Immunol. Immunopathol. 168, 40–48. doi: 10.1016/j.vetimm.2015.08.003
Lin, W., Kaewprom, K., Wang, S., Lin, C., Yang, C., Chiou, M., et al. (2020). Outbreak of porcine reproductive and respiratory syndrome Virus 1 in Taiwan. Viruses 12:316. doi: 10.3390/v12030316
Liu, H., Wang, Y., Duan, H., Zhang, A., Liang, C., Gao, J., et al. (2015). An intracellularly expressed Nsp9-specific nanobody in MARC-145 cells inhibits porcine reproductive and respiratory syndrome virus replication. Vet. Microbiol. 181, 252–260. doi: 10.1016/j.vetmic.2015.10.021
Liu, L., Tian, J., Nan, H., Tian, M., Li, Y., Xu, X., et al. (2016). Porcine reproductive and respiratory syndrome virus nucleocapsid protein interacts with Nsp9 and cellular DHX9 to regulate Viral RNA synthesis. J. Virol. 90, 5384–5398. doi: 10.1128/JVI.03216-15
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J., Gao, F., Wei, Z., Liu, P., Liu, C., Zheng, H., et al. (2011). A 5’-proximal Stem-loop Structure of 5’ untranslated region of porcine reproductive and respiratory syndrome virus genome is key for virus replication. Virol. J. 8:172. doi: 10.1186/1743-422x-8-172
Lunney, J. K., Benfield, D. A., and Rowland, R. R. R. (2010). Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 154, 1–6. doi: 10.1016/j.virusres.2010.10.009
Marasco, W. (2001). Intrabodies as antiviral agents. Curr. Top. Microbiol. Immunol. 260, 247–270. doi: 10.1007/978-3-662-05783-4_13
Mertens, H. D. T., and Svergun, D. I. (2010). Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 172, 128–141. doi: 10.1016/j.jsb.2010.06.012
Nan, Y., Wang, R., Shen, M., Faaberg, K. S., Samal, S. K., and Zhang, Y. J. (2012). Induction of type I interferons by a novel porcine reproductive and respiratory syndrome virus isolate. Virology 432, 261–270. doi: 10.1016/j.virol.2012.05.015
Nan, Y., Wu, C., Gu, G., Sun, W., Zhang, Y. J., and Zhou, E. M. (2017). Improved vaccine against PRRSV: current progress and future perspective. Front. Microbiol. 8:1635. doi: 10.3389/fmicb.2017.01635
Park, Y. J., Pardon, E., Wu, M., Steyaert, J., and Hol, W. G. J. (2012). Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody. Nucleic Acids Res. 40, 1828–1840. doi: 10.1093/nar/gkr867
Patel, D., Nan, Y., Shen, M., Ritthipichai, K., Zhu, X., and Zhang, Y. J. (2010). Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 84, 11045–11055. doi: 10.1128/JVI.00655-10
Petoukhov, M. V., Franke, D., Shkumatov, A. V., Tria, G., Kikhney, A. G., Gajda, M., et al. (2012). New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45(Pt 2), 342–350. doi: 10.1107/S0021889812007662
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Rasmussen, S. G., Choi, H. J., Fung, J. J., Pardon, E., Casarosa, P., Chae, P. S., et al. (2011). Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–180. doi: 10.1038/nature09648
Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 27, 493–497. doi: 10.1093/oxfordjournals.aje.a118408
Snijder, E. J., Kikkert, M., and Fang, Y. (2013). Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 94(Pt 10), 2141–2163. doi: 10.1099/vir.0.056341-0
Sun, H., Pattnaik, A. K., Osorio, F. A., and Vu, H. L. (2016). Identification of viral genes associated with the interferon-inducing phenotype of a synthetic porcine reproductive and respiratory syndrome virus strain. Virology 499, 313–321. doi: 10.1016/j.virol.2016.09.018
Sun, Z., Chen, Z., Lawson, S. R., and Fang, Y. (2010). The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 84, 7832–7846. doi: 10.1128/JVI.00217-10
Svergun, D. I. (2010). Small-angle X-ray and neutron scattering as a tool for structural systems biology. Biol. Chem. 391, 737–743. doi: 10.1515/BC.2010.093
Tian, K., Yu, X., Zhao, T., Feng, Y., Cao, Z., Wang, C., et al. (2007). Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526
Tong, G. Z., Zhou, Y. J., Hao, X. F., Tian, Z. J., An, T. Q., and Qiu, H. J. (2007). Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg. Infect. Dis. 13, 1434–1436.
Volkov, V. V., and Svergun, D. I. (2003). Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864.
Wang, C., Huang, B., Kong, N., Li, Q., Ma, Y., Li, Z., et al. (2013). A novel porcine reproductive and respiratory syndrome virus vector system that stably expresses enhanced green fluorescent protein as a separate transcription unit. Vet. Res. 44:104. doi: 10.1186/1297-9716-44-104
Wang, R., Nan, Y., Yu, Y., Yang, Z., and Zhang, Y. J. (2013a). Variable interference with interferon signal transduction by different strains of porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 166, 493–503. doi: 10.1016/j.vetmic.2013.07.022
Wang, R., Nan, Y., Yu, Y., and Zhang, Y. J. (2013b). Porcine reproductive and respiratory syndrome virus Nsp1beta inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-alpha1 degradation. J. Virol. 87, 5219–5228. doi: 10.1128/JVI.02643-12
Wang, L., Zhang, L., Huang, B., Li, K., Hou, G., Zhao, Q., et al. (2019). A nanobody targeting viral nonstructural protein 9 inhibits porcine reproductive and respiratory syndrome virus replication. J. Virol. 93:e01888-18. doi: 10.1128/JVI.01888-18
Wang, R., and Zhang, Y. J. (2014). Antagonizing interferon-mediated immune response by porcine reproductive and respiratory syndrome virus. Biomed. Res. Int. 2014:315470. doi: 10.1155/2014/315470
Watanabe, T., Shinji, S., Kyoko, J., Kim, J. H., Hatta, M., and Kawaoka, Y. (2009). Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc. Natl. Acad. Sci. U.S.A. 106, 588–592. doi: 10.1073/pnas.0806959106
Wensvoort, G., Terpstra, C., Pol, J. M. A., Ter Laak, E. A., Bloemraad, M., De Kluyver, E. P., et al. (1991). Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet. Q 13, 121–130. doi: 10.1080/01652176.1991.9694296
Xie, J., Zhou, H., Cui, J., Chen, Y., Zhang, M., Deng, S., et al. (2014). Inhibition of porcine reproductive and respiratory syndrome virus by specific siRNA targeting Nsp9 gene. Infect. Genet. Evol. 28, 64–70. doi: 10.1016/j.meegid.2014.08.008
Xu, L., Zhou, L., Sun, W., Zhang, P., Ge, X., Guo, X., et al. (2018). Nonstructural protein 9 residues 586 and 592 are critical sites in determining the replication efficiency and fatal virulence of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Virology 517, 135–147. doi: 10.1016/j.virol.2018.01.018
Yang, L., Wang, R., Ma, Z., Xiao, Y., Nan, Y., Wang, Y., et al. (2017). Porcine reproductive and respiratory syndrome virus antagonizes JAK/STAT3 signaling via nsp5, which induces STAT3 degradation. J. Virol. 91:e02087-16. doi: 10.1128/JVI.02087-16
Zhang, A., Duan, H., Li, N., Zhao, L., Haung, B., and Yani, S. (2017a). Heme oxygenase-1 metabolite biliverdin, not iron, inhibits porcine reproductive and respiratory syndrome virus replication. Free Radic. Biol. Med. 102, 149–161. doi: 10.1016/j.freeradbiomed.2016.11.044
Zhang, A., Zhao, L., Li, N., Duan, H., and Xiao, S. (2017b). Carbon monoxide inhibits porcine reproductive and respiratory syndrome virus replication by the cyclic GMP/Protein Kinase G and NF-κB signaling pathway. J. Virol. 91:e01866-16.
Zhao, K., Gao, J. C., Xiong, J. Y., Guo, J. C., Yang, Y. B., Jiang, C. G., et al. (2018). Two residues in NSP9 contribute to the enhanced replication and pathogenicity of highly pathogenic porcine reproductive and respiratory syndrome virus. J. Virol. 92:e02209-17. doi: 10.1128/JVI.02209-17
Keywords: PRRSV, Nsp9, RdRp, nanobody, SAXS
Citation: Wang Y, Li R, Qiao S, Wang J, Liu H, Li Z, Ma H, Yang L, Ruan H, Weng M, Hiscox JA, Stewart JP, Nan Y, Zhang G and Zhou E-M (2020) Structural Characterization of Non-structural Protein 9 Complexed With Specific Nanobody Pinpoints Two Important Residues Involved in Porcine Reproductive and Respiratory Syndrome Virus Replication. Front. Microbiol. 11:581856. doi: 10.3389/fmicb.2020.581856
Received: 10 July 2020; Accepted: 26 October 2020;
Published: 12 November 2020.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Yongming Sang, Tennessee State Museum, United StatesJesus Hernandez, Consejo Nacional de Ciencia y Tecnología (CONACYT), Mexico
Copyright © 2020 Wang, Li, Qiao, Wang, Liu, Li, Ma, Yang, Ruan, Weng, Hiscox, Stewart, Nan, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuchen Nan, bmFueXVjaGVuMjAxNUBud3N1YWYuZWR1LmNu; Gaiping Zhang, emhhbmdnYWlwQDEyNi5jb20=; emhhbmdnYWlwaW5nMjAwM0AxNjMuY29t; En-Min Zhou, emhvdWVtQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Yan Wang
Yan Wang Rui Li
Rui Li Songlin Qiao2
Songlin Qiao2 Zhijun Li
Zhijun Li Julian A. Hiscox
Julian A. Hiscox James P. Stewart
James P. Stewart Yuchen Nan
Yuchen Nan En-Min Zhou
En-Min Zhou