
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 September 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.580081
This article is part of the Research Topic Plant Microbiome: Interactions, Mechanisms of Action, and Applications View all 26 articles
 Dao-Jun Guo1,2,3,4
Dao-Jun Guo1,2,3,4 Rajesh Kumar Singh2,3,4
Rajesh Kumar Singh2,3,4 Pratiksha Singh2,3,4
Pratiksha Singh2,3,4 Dong-Ping Li5
Dong-Ping Li5 Anjney Sharma2,3,4
Anjney Sharma2,3,4 Yong-Xiu Xing1*
Yong-Xiu Xing1* Xiu-Peng Song2,3
Xiu-Peng Song2,3 Li-Tao Yang1
Li-Tao Yang1 Yang-Rui Li1,2,3,4*
Yang-Rui Li1,2,3,4*Sugarcane is the leading economic crop in China, requires huge quantities of nitrogen in the preliminary plant growth stages. However, the use of an enormous amount of nitrogen fertilizer increases the production price, and have detrimental results on the environment, causes severe soil and water pollution. In this study, a total of 175 endophytic strains were obtained from the sugarcane roots, belonging to five different species, i.e., Saccharum officinarum, Saccharum barberi, Saccharum robustum, Saccharum spontaneum, and Saccharum sinense. Among these, only 23 Enterobacter strains were chosen based on nitrogen fixation, PGP traits, hydrolytic enzymes production, and antifungal activities. Also, all selected strains were showed diverse growth range under different stress conditions, i.e., pH (5–10), temperature (20–45°C), and NaCl (7–12%) and 14 strains confirmed positive nifH, and 12 strains for acdS gene amplification, suggested that these strains could fix nitrogen along with stress tolerance properties. Out of 23 selected strains, Enterobacter roggenkampii ED5 was the most potent strain. Hence, this strain was further selected for comprehensive genome analysis, which includes a genome size of 4,702,851 bp and 56.05% of the average G + C content. Genome annotations estimated 4349 protein-coding with 83 tRNA and 25 rRNA genes. The CDSs number allocated to the KEGG, COG, and GO database were 2839, 4028, and 2949. We recognized a total set of genes that are possibly concerned with ACC deaminase activity, siderophores and plant hormones production, nitrogen and phosphate metabolism, symbiosis, root colonization, biofilm formation, sulfur assimilation and metabolism, along with resistance response toward a range of biotic and abiotic stresses. E. roggenkampii ED5 strain was also a proficient colonizer in sugarcane (variety GT11) and enhanced growth of sugarcane under the greenhouse. To the best of our knowledge, this is the first information on the whole-genome sequence study of endophytic E. roggenkampii ED5 bacterium associated with sugarcane root. And, our findings proposed that identification of predicted genes and metabolic pathways might describe this strain an eco-friendly bioresource to promote sugarcane growth by several mechanisms of actions under multi-stresses.
Agricultural extension in the 20th era has been deeply managed by the application of farm technologies, high-quality varieties, strong tillage, irrigation, chemical fertilizers, and pesticides (Foley et al., 2005). Sugarcane is the main energy and sugar crop that is consumed in several industries as a raw material. China positions the third major sugarcane growing country and produces approximately ten million tons of sugar every year (FAO, 2018) and Guangxi is the leading sugar-producing region of China (Li et al., 2016). The nitrogen fertilizer use is very high for commercial sugarcane production in China, extremely greater as compared to Brazil and other nations (Li et al., 2015). Whereas, constant exploit of nitrogen (N) fertilizers for an extended time increases the production cost as well as causes harmful results on the soil and environment health (Li and Yang, 2015).
Environmentally protected approaches such as bio-fertilizers are seriously required to improve crop/sugarcane growth, nitrogen fixation, and reduce yield loss in different stress conditions to retain sustainable crop production. The utilization of plant growth-promoting (PGP) endophytic bacteria is an efficient approach to stabilizing and improving crop yield due to these bacteria may have ecological benefits more than epiphytic and rhizospheric bacteria as they directly contact with the plants (James, 2000). Endophytic microbes, inhabit and survive inside plant tissue are widely investigated in several plants (Hardoim et al., 2015; Kumar et al., 2017), can support plant growth by several ways such as improving the soil nutrient uptake and germination rate, altering the phytohormone levels and improving plant biotic and abiotic stresses. In addition, secondary aids consist of the biological control of plant pathogens and the induction of induced systemic resistance (ISR) in plants (Rosenblueth and Martínez-Romero, 2006; Ryan et al., 2008; Mei and Flinn, 2010).
Biological nitrogen fixation (BNF) has been confirmed to give 30–80% of the total N for the sugarcane (Boddey et al., 1991; Döbereiner, 1997; Taulé et al., 2012; Urquiaga et al., 2012; Santi et al., 2013). Several nitrogen-fixing bacteria have been reported from inside and rhizosphere of sugarcane plants can fix N related to sugarcane plants (Gillis et al., 1989; Sevilla et al., 2001; Oliveira et al., 2002; Baldani and Baldani, 2005; Li et al., 2017). BNF decreases the sugarcane production cost and sugarcane is cultivated by an extremely less quantity of N inputs as a result of BNF in Brazil (Yong-Xiu et al., 2015). Diazotrophic endophytes are adaptable microorganisms, able to supply nutrients even in absence of nodules in plants, and a method named associative N-fixation (Carvalho et al., 2014). Previous studies showed that a few nitrogen-fixing genera of Enterobacteriaceae family have been enhanced nitrogenase activity and N-fixation in sugarcane (Mirza et al., 2001; Loiret et al., 2004; Govindarajan et al., 2007; Magnani et al., 2010; Lin et al., 2012; Taulé et al., 2012). Hence, there is a need to explore the endophytic diazotrophs belongs to Enterobacter genera in the major non- leguminous crops like sugarcane to improve nitrogen fixation, minimize the production cost, decrease the use of chemical fertilizer, and reduce environmental pollution.
Sugarcane production is usually affected by many pathogens and that can accrue in germplasm of sugarcane and cause major crop harm constraining the growth, dropping the stalk weight, and interrupt the sugar recovery. At present, above 120 diseases have been accounted for worldwide (Chen, 1982; Rott et al., 2000), whereas above 60 have been accounted for in China (Lu et al., 1997; Huang and Li, 2014, 2016). Out of these, pokkah-boeng, pineapple, red rot, smut, and wilt diseases cause significant yield damage (Viswanathan and Rao, 2011). Sugarcane production is also influenced by several abiotic stresses like drought, heavy metal, pH, temperature, and salt. NaCl is the major leading salt causing soil salinity, which affects plant growth and yield. Enormous effects of elevated salinity in plants consist of enzyme inactivation; reduction in K and Ca uptake by plants, protein synthesis inhibition, premature leaves senescence, development of burn-like lesions, a decline in respiration, and photosynthesis rate, and loss of cellular integrity, etc. (Munns, 2002). Whereas, heavy metals accretion in soils directly influences the pH and texture of the soil and finally may decrease the plant’s growth by exerting harmful results on a variety of biological processes in plants (Moftah, 2000). Additionally, drought stress stimulates cellular reactive oxygen species (ROS) production, which can oxidize many cellular components, lastly triggering cell death (Barrera, 2012).
The complete-genome study can be used to categorize genes implicated in the positive effects of plant growth-promoting bacteria (PGPB), offer the perception of the molecular and functional mechanisms (Kang et al., 2016; Qin et al., 2017; Oh et al., 2018). Earlier, complete genome analysis of some other Enterobacter stains is accessible (Ren et al., 2010; Taghavi et al., 2010; Liu et al., 2012; Andrés-Barrao et al., 2017) excluding E. roggenkampii. Therefore, the complete genome sequence accessibility of endophytic E. roggenkampii isolated from sugarcane root will help in full understanding of the diverse biological mechanisms and determining the characteristics of this bacteria, plus gene identification that is contributing to the positive activity of PGPB, improve sugarcane growth under abiotic and biotic stresses.
The objectives of this research are (i) to isolate Enterobacter strains from the roots of five different sugarcane species grown in the field of Guangxi, China (ii) to study their plant growth-promoting (PGP) and nitrogenase activities, as well as biocontrol potential against sugarcane and other plant pathogens (iii) to detect the nifH and acdS genes amplification (iv) to study their hydrolytic enzymes (chitinase, glucanase, cellulase, and protease) production (v) to investigate their capacity to tolerate several abiotic stresses (pH, temperature, and NaCl), (vi) to examine the colonization pattern of selected most prominent E. roggenkampii ED5 strain in sugarcane plant through confocal laser scanning microscopy (CLSM) and scanning electron microscopy (SEM), and (vii) to sequence the E. roggenkampii ED5 genome, a prospect to create the allocation of nitrogen-fixing, PGP, and stress-related genes. Here, we report the first statement of the forthcoming application of E. roggenkampii ED5 endophytic bacteria, isolated from sugarcane root, as a potential agent to improve growth and nitrogen fixation in sugarcane, stress alleviation, and biocontrol against pathogens.
Five different sugarcane species were selected in this study, i.e., Saccharum officinarum, Saccharum barberi, Saccharum robustum, Saccharum spontaneum, and Saccharum sinense. All these five sugarcane plant samples were obtained from the nursery of Sugarcane Germplasm Resources, Guangxi Academy of Agricultural Sciences, Sugarcane Research Centre, Nanning, China. Only root samples were selected for the isolation of different endophytic bacteria at the elongation stage. For each sample, five plants were selected, and five different root samples were composed of each sugarcane species. The roots samples with white tips, indicated the active growth, were used for the isolation.
One gram of fresh roots pieces was squashed in one mL of sterile 5% sucrose solution after sterilization (Dobereiner et al., 1993). Roots were cleaned with tap water, disinfected superficially by 70% ethanol for 5 min, once more rinsed and disinfected with 3% sodium hypochlorite for 5 min. For sterilization, roots were cleaned by sterilized double-distilled water, and then samples were dried with sterile filter paper. To check the disinfection method accomplishment, the former washing double distilled water was spread on the nutrient agar (NA) and potato dextrose agar (PDA) plates, kept at 30 ± 2 and 26 ± 2°C in an incubator for 3–5 days. The results were utilized as a sterilization control, and no fungal and bacterial colonies were capable to develop on the plates (Slama et al., 2019). Six different media were chosen for the nitrogen-fixing endophytic bacteria isolation, i.e., Ashby’s glucose, Ashby’s mannitol, Burk medium, Jensen medium, NA, and Yeast mannitol agar. The composition of all used different media is provided in Supplementary Table S1. All root samples were crushed with 5% of the sucrose solution. And, ten-fold serial dilutions from 10–2 to 10–5 of the 100 μL aliquots suspensions were spread into all different mediums in triplicates. After morphologically different strains of emerging spots or layers from the root, pieces were selected after 5–7 days at 30 ± 2°C, and individually bacterial colonies were further successive purification. The endophytic bacterial strains were stored in 25% glycerol at −20°C.
All endophytic strains were assessed for their in vitro antifungal activities against Fusarium moniliforme, Fusarium cubense, Botrytis cinereal, Ceratocystis paradoxa, and Sporisorium scitamineum with the slight modification of Singh et al. (2020) method on NA plus PDA (1:1) medium. These all fungal pathogens were obtained from Agriculture College, Guangxi University, Nanning. A 5 mm diameter of actively growing pathogen culture disk was cut from the PDA plate and put in the middle of PDA: NA plates. All bacterial strains (106 cell mL–1) were streaked on the plate around 3 cm from the pathogen disk and kept at 28 ± 2°C, till the mycelia of fungal pathogens were completely grown in the control plate (without bacterial strains). The antifungal activity was evaluated by determining the growth inhibition in response to selected pathogens. The inhibition percentage was observed by Singh et al. (2013) and strains displaying ≥50% inhibition of mycelial growth were measured as potential biocontrol agents.
The hydrolytic enzymes production is a common mechanism used by bacteria to prevent the growth of pathogenic microorganisms. In this study, production of four hydrolytic enzymes, i.e., chitinase (catalog no. MM1062O1), protease (MM1206O1), glucanase (MM91504O1), and cellulase (MM91502O1) was measured by enzyme-linked immune sorbent assays (ELISA) kits (Wuhan Colorful Gene Biological Technology Co. Ltd, China). A pure colony was transferred into 10 mL of LB broth medium and placed at 180 rpm for 36 h at 32°C in incubator shaker. Bacterial culture was centrifuged at 12,000 rpm for 5 min to acquire a supernatant. The supernatant for all strains was used for different enzyme activities assay by ELISA kits. The complete extraction method was performed at 4°C. The ELISA was done in 96-well microtiter plates coated with the antigen against the selected enzymes, according to Singh et al. (2018); Singh P. et al. (2019) procedure.
Growth of all selected endophytic bacterial strains was examined for their capacity to tolerate several abiotic stress conditions, i.e., temperature (20–45°C), pH (5–10), and NaCl (7–12%) in LB broth by spectrophotometer at 600 nm and the uninoculated medium was used as a blank.
0.1 mL bacterial suspension was transferred in LB broth medium (5 mL) of and tubes were incubated at 20, 25, 30, 35, 40, and 45°C for 36 h in a shaker incubator at 120 rpm and O.D. was recorded at 600 nm.
The pH of the LB broth medium was attuned to 5, 6, 7, 8, 9, and 10 with sterile buffers. 0.1 mL fresh cultures were transferred in 5 ml of LB broth medium comprising different pH and kept at 37°C; 120 rpm in incubator shaker and after 36 h growth was measured at 600 nm.
Five mL of LB broth medium supplemented with 7, 8, 9, 10, 11, and 12% NaCl was distributed in 30 mL tubes and autoclaved. 0.1 mL bacterial suspension was inoculated in LB broth tubes and incubated at 37°C/120 rpm in shaker incubator and growth was calculated at 600 nm after 36 h.
All endophytic strains were examined for different PGP traits, i.e., Indole acetic acid (IAA), Phosphate (P) solubilization, siderophore, hydrogen cyanide (HCN), and ammonia production, following the standard protocol of Lorck (1948), Schwyn and Neilands (1987), Brick et al. (1991), Glickmann and Dessaux (1995), and Dey et al. (2004), respectively. Each analysis was completed in three biological repeats.
Indole acetic acid production was estimated by the colorimetric method in the presence of tryptophan in the medium at different concentration levels. The potential of bacterial isolates to solubilize P was qualitatively evaluated by the Pikovskaya medium supplemented with tri-calcium phosphate. The strains were transferred on a plate and kept at 30 ± 2°C for 5–7 days and the development of a clear hallow zone around the bacterial isolates indicated P-solubilizing capacity. All selected endophytic strains were screened for siderophores production and development of halo zone on the chrome azurol S medium confirmed siderophore production. The HCN production capacity of all strains was evaluated on PDA medium with 4.4 g L–1 glycine to produce hydrocyanic acid. A filter paper soddens with 0.5% picric acid and 2% Na2CO3 was put on a cover plate, after that sealed by Parafilm and kept at 28°C, and change in color of filter paper confirmed the HCN production. All strains were incubated in 10% sterile peptone H2O at 30 ± 2°C for 72 h and change in yellow color by the addition of Nessler’s reagent (0.5 mL) confirmed the ammonia production.
1-Aminocyclopropane-1-carboxylate deaminase activity of all strains was studied based on the capability to utilize ACC as a nitrogen source on nitrogen-free Dworkin and Foster (DF) medium (Jacobson et al., 1994). DF medium deprived of ACC was used as the negative control, whereas DF medium with ACC (3 mM) or (NH4)2SO4 (0.2% w/v) was used as a positive control. The plates were kept at 30 ± 2°C for 3–5 days and ACC deaminase activity was confirmed by the strain growth on ACC plates. Quantitative ACC deaminase activity estimation was estimated by the procedure of Honma and Shimomura (1978).
The nitrogen-fixing capacity of each strain was examined by the ARA method (Hardy et al., 1968), and the procedure was followed by Li et al. (2017) with some modification.
Genomic DNA was isolated for all selected endophytic strains with DNA isolation kit (CWBIO, Beijing, China) and DNA was confirmed by gel electrophoresis (0.8% w/v) and quantified by Nanophotometer spectrophotometer (Pearl, Implen-3780). The 16S rRNA gene was amplified by using a pair of pA-F and pH-R universal primer through PCR and PCR condition was followed as Li et al. (2017) (Supplementary Table S2), and the purified PCR product was sequenced (Sangon Biotech, Shanghai, China).
Phylogenetic analysis and evolutionary relationship of the selected Enterobacter strains were studied through the comparison of 16S rRNA gene sequences with reference sequences of the National Center for Biotechnology Information (NCBI) GenBank database. The alignment of sequences was completed with ClustalW (Saitou and Nei, 1987). The phylogenetic tree was created by molecular evolutionary genetics analysis (MEGA) software (version 7.0) (Kumar et al., 2016) and unweighted pair group process through arithmetic mean (UPGMA) (Sneath and Sokal, 1973) in a Kimura two-parameter model (Tamura et al., 2004). The bootstrap examination was finished via Felsenstein procedure with 1000 pseudoreplication (Felsenstein, 1985).
The nifH and acdS genes amplification of all the selected strains was achieved with degenerate sets of primer following the PCR conditions of Li et al. (2011, 2017), as presented in Supplementary Table S2. All amplified products of PCR were purified and cloned according to the manufacturer’s instructions (TaKaRa, Japan) and then sequenced (Sangon Biotech, Shanghai, China). All sequences obtained for both genes were checked through the blastn suite search engine in the NCBI GenBank database.
The root colonization inside the sugarcane plant was confirmed through Green Fluorescent Protein (GFP) and Scanning Electron Microscopy (SEM) techniques. The pPROBE-pTetr-TT plasmid having the GFP gene was obtained from the Agriculture College, Guangxi University, Sugarcane laboratory, Nanning, China. Strain ED5 was mixed with plasmid vector (1:2 ratio) in LB broth and incubated at 30 ± 2°C for 36 h in an orbital shaker for 120 rpm. Sugarcane plantlets were shifted in the glass bottle inside the bacterial suspension and kept in the growth chamber. After 72 h plantlets were taken away and washed with autoclaved water. Micro-propagated cultivated sugarcane plantlets were cut into a small section and observed by a CLSM (Leica DMI 6000, Germany) (Singh et al., 2020). Sugarcane plant samples (stem and root tissues) were selected for the SEM analysis, both samples were cut into small pieces by knife and fixed in glutaraldehyde solution (Catalog G1102, Servicebio) overnight at 4°C. The samples were washed three times with distilled water and dehydrated in ethanol 30, 50, 70, 90, 95, and 100% for 15 min and finally isoamyl acetate for 15 min. After drying the samples with critical point dryer, colonization of E. roggenkampii ED5 was observed in sugarcane by using the SEM (Hitachi model SU8100), according to the protocol of Singh et al. (2013).
The different plant growth parameters such as chlorophyll content, leaf area, plant height, root weight, shoot weight, photosynthesis, and transpiration rate were observed in sugarcane variety GT11 at 30 and 60 days after inoculation of strain ED5.
Genomic DNA was isolated from the overnight liquid cell suspension of E. roggenkampii strain by Wizard Genomic DNA Kit (Promega). DNA quality and concentration were estimated by TBS-380 fluorometer (Turner BioSystems Inc., Sunnyvale, CA, United States) and DNA with high quality (OD260/280 = 1.8 ∼ 2.0 > 20 μg) was employed for additional experiment. The genome was sequenced by a fusion of Nanopore and Illumina sequencing platforms. The Illumina data were employed to assess the complexity of the genome. For Illumina sequencing, as a minimum 1 μg genomic DNA was utilized for every isolate in the assembly of the sequencing library. DNA fragments were incised into 400–500 bp by a Covaris M220 Focused Acoustic Shearer. Illumina sequencing libraries were prepared by NEXTflex Rapid DNA-Seq Kit. Briefly, 5′ prime ends were first end-repaired and phosphorylated. Next, the 3′ ends were A- tailed and ligated to sequencing adapters. The third step was to enrich adapters-ligated products using PCR. The organized libraries were used for paired-end Illumina sequencing (2 × 150 bp) on an Illumina HiSeq X Ten. For Nanopore sequencing, 15 μg of genomic DNA was spin in a Covaris G-TUBE (Covaris, MA) to cut the genomic DNA into ∼10 kb fragments, then performed magnetic bead purification and connect the sequencing adapters to both ends.
The data obtained by Nanopore and Illumina platform were used for bioinformatics analysis and all the analyses were done with the free online platform of Majorbio Cloud Platform1 from Shanghai Majorbio Bio-pharm Technology Co., Ltd. The whole-genome sequence was assembled using both Nanopore reads and Illumina reads. A statistic of quality information was applied for quality trimming, by which the low-quality data can be removed to form clean data. The reads then assembled into a contig using a hierarchical genome assembly process (HGAP) and canu (Koren et al., 2017), and the circular step was checked and completed, generating a complete genome with seamless chromosomes and plasmids. Finally, error correction of the Nanopore assembly results was performed using the Illumina reads using Pilon.
The Glimmer version 3.02 was used for coding sequence (CDS) prediction and predicted CDSs were annotated from NR, Swiss-Prot, Pfam, Gene Ontology (GO), Clusters of Orthologus Groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Delcher et al., 2007) by sequence alignment tools, i.e., Basic Local Alignment Search Tool (BLAST), Diamond and HMMER. The tRNA-scan-SE (v1.2.1) (Borodovsky and Mcininch, 1993) and Barrnap were used for tRNA prediction and rRNA prediction, as well as antismash software was used for the secondary metabolite genes prediction. In short, every protein query was aligned, and annotations of accurately matched subjects (e-value < 10–5) were completed for gene annotation.
Complete genome similarity was calculated with ANI. The Enterobacter strains gene sequences were obtained from the NCBI database. Based on the selected E. roggenkampii ED5 16S rRNA gene and 10 house-keeping genes (dnaG, frr, rpoB, pgk, rplB, infC, pyrG, rpmA, smpB, and rpsB) online NCBI Blast search program2 was used to compare the ED5 strain with closely related eight strains. ANI results were analyzed using R version 3.5.1 gplots 3.0.4 software and presented as heat map and vegan 2.5–6 software was used for hierarchical cluster analysis.
All genome analysis process was completed by the manufacturer’s instructions. All PGP and biocontrol tests were done in three replicates and data were considered through analysis of variance followed by Duncan’s multiple range test. Data were showed as the mean plus the standard error of the mean and evaluated by the Student t-test with p-value < 0.05 was indicated significant.
A total of 175 endophytic bacterial strains were isolated by using six different selective mediums from the roots of five sugarcane species (S. officinarum, S. barberi, S. robustum, S. spontaneum, and S. sinense). Among these, only 90 strains were selected which exhibited various nitrogenase and PGP activities, as well as biocontrol potential against sugarcane and other crops pathogens. After 16S rRNA gene sequencing, we preferred only 23 Enterobacter strains for further study (Supplementary Figure S1). PGP activities of all 23 selected strains are presented in Table 1.
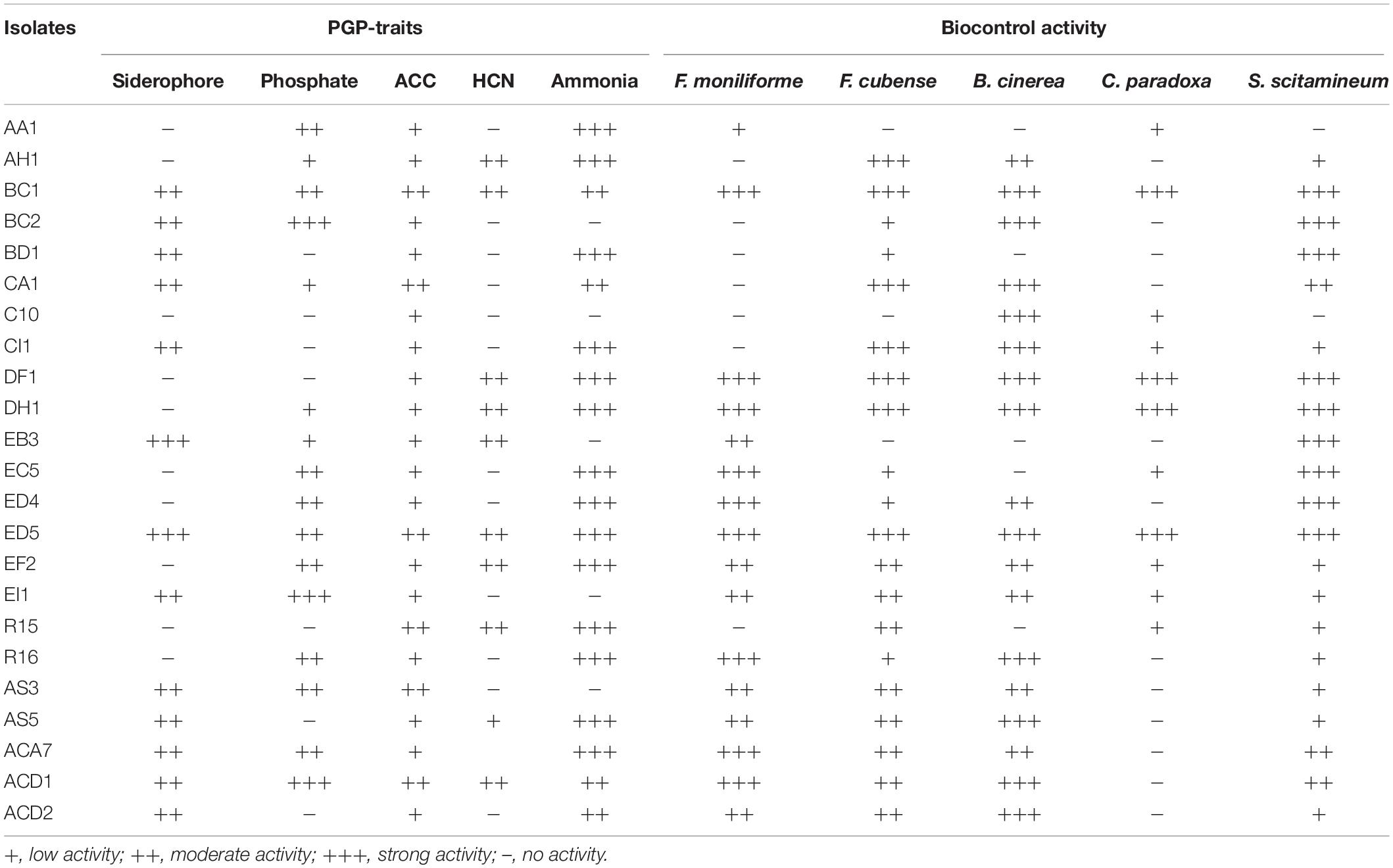
Table 1. Plant growth-promoting (PGP) and biocontrol activities of selected endophytic strains from the roots of different sugarcane species.
Out of all 23 strains, in vitro siderophore production results showed that 13 (56.52%) strains confirmed positive response by producing halo orange zone in CAS agar medium and two strains (EB3 and ED5) were showed strong activity. For P- solubilization, only 16 (69.56%) strains have the potential to produce a zone of inhibition to solubilize tricalcium phosphate on Pikovskaya’s media and three strains (BC2, EI1, and ACD1) displayed strong activity (Table 1). Both assays were performed by measuring 3 mm or larger zone of inhibition on specific medium following incubation at 30 ± 2°C for 3–5 days. Further, Table 1 indicated that 10 (43.47%) and 18 (78.26%) strains were proficient for HCN, and ammonia production with more strains established positive ammonia production test than that of HCN.
Biocontrol activity of all these endophytic bacteria was also analyzed in response to five different plant pathogens. The results presented in Table 1, designated that 21 (91.3%), 11 (47.82%), 16 (69.56%), 20 (86.95%), and 18 (78.26%) isolates were antagonistic against S. scitamineum, C. paradoxa, F. moniliforme, F. cubense, and B. cinerea correspondingly, with ED5, DH1, and DF1 strains possessed strong biocontrol activity against all pathogens.
ACCD activity was measured by all the strains which showed the potential to use ACC as a solitary source of nitrogen in DF minimal medium and the result illustrated the growth of all strains on plate medium. In addition, further screened for quantitative ACCD activity and varying ranged of activity was observed by all strains from 212.73 to 1192.74 nmol α-ketobutyrate mg–1h–1. The highest ACCD activity was examined by strain EB3 followed by ED5 and ED4 (Table 2). The nitrogen-fixing capacity of all isolates was measured through the ARA method which varied from 8.23 to 29.60 nmoL C2H4 mg protein h–1. Strain ED5 recorded the maximum, whereas BC1 showed the minimum nitrogenase activity (Table 2).
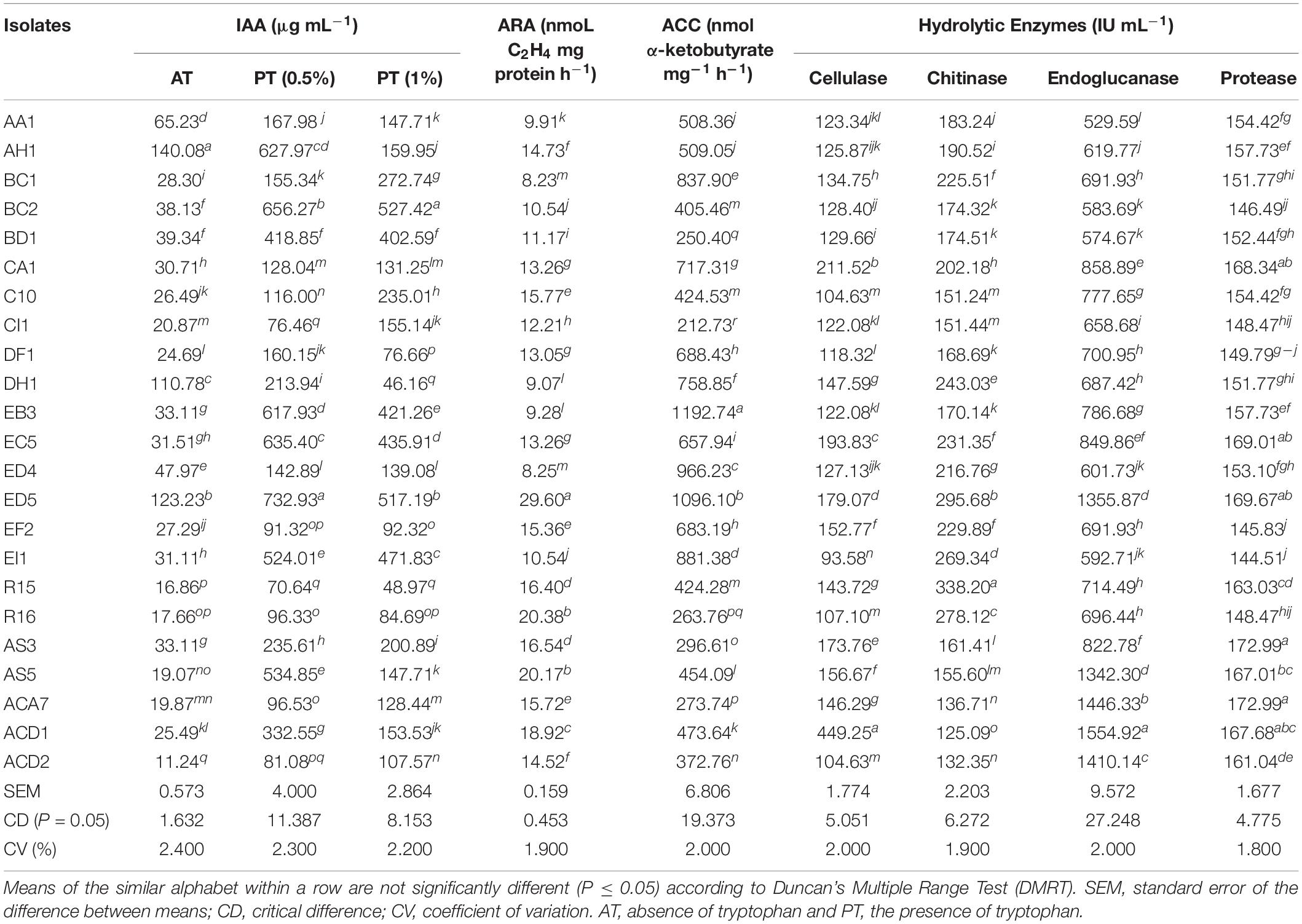
Table 2. In vitro quantitative assays for ARA, ACC, and hydrolytic enzymes of isolated endophytic strains.
Indole acetic acid biosynthesis is an essential trait of PGPR strains and results elucidated that all these isolates had a diverse ability to synthesize IAA, which are presented in Table 2. The quantitative IAA synthesis ranged from 70.64 to 732.93 μg mL–1 and 46.16 to 527.42 μg mL–1 in medium supplemented with 0.5 and 1% tryptophan and from 11.24 to 140.08 μg mL–1 in medium deprived of tryptophan. In the presence of 0.5% tryptophan, the minimum and maximum IAA production were recorded R15 and ED5 strains. While strains BC2 and DH1 confirmed the highest and lowest IAA production in medium supplemented with 1% of tryptophan. For medium devoid of tryptophan, the greatest IAA production was observed in AH1 and the least for ACD2 strains, respectively.
The quantitative estimation of four hydrolytic enzymes, i.e., cellulase, chitinase, endoglucanase, and protease was also measured for all the selected strains using the ELISA kit. All strains showed activity ranged between 93.58–449.25, 125.09–338.2, 529.59–1554.92, and 144.51–172.99 IU mL–1 for cellulase, chitinase, endoglucanase, and protease enzymes, respectively (Table 2). The strains R15 and AS3 showed maximum chitinase and protease activities; with ACD1 strain confirmed maximum cellulase and endoglucanase activities. Whereas, ACD1 and AA1 strains presented minimum chitinase and endoglucanase activities, and EI1 strain displayed minimum cellulase and protease activities (Table 2).
The growth of all selected strains was measured at 600 nm in different abiotic stress conditions, i.e., temperature (20–45°C), pH (5–10), and salt (7–12%), as displayed in Figure 1. Strain ACD1 established the greatest growth followed by CA1, AA1, and ED5 strains in LB broth medium supplemented with 7–12% NaCl, whereas the lowest growth was observed by the DH1 strain (Figure 1A). For pH, strains BC1, ED5, ED4, and CI1 showed maximum ability to grow in an extensive pH varying from 5 to 10. Alternatively, stains R16 and BD1 were least pH tolerant (Figure 1B). In the case of temperature, strain ED5 exhibited the highest and AS3 confirmed lowest temperature tolerance up to 45°C (Figure 1C).
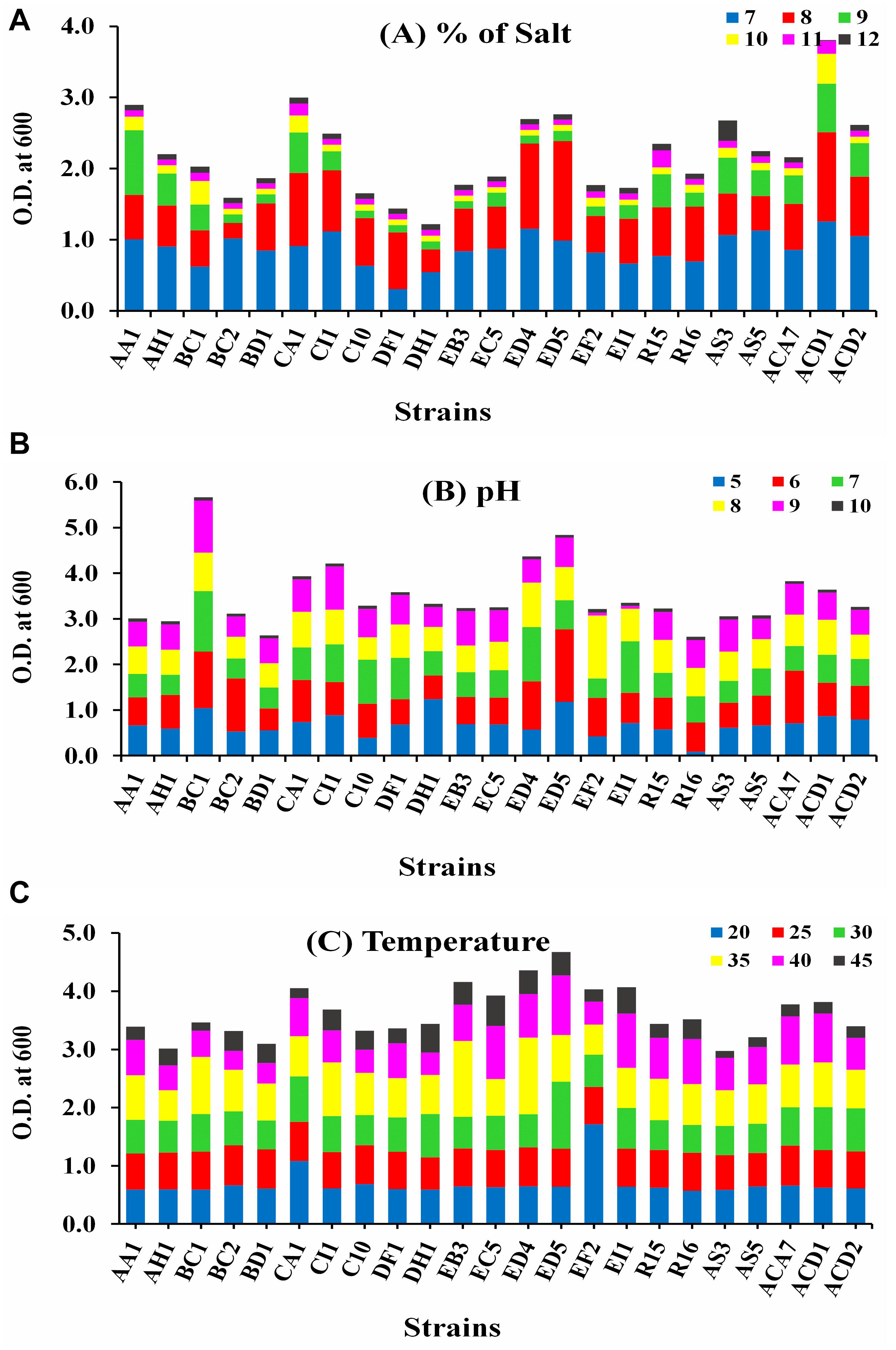
Figure 1. Intrinsic result of various abiotic stresses of selected endophytic Enterobacter strains growth (A) salt (7–12%), (B) pH (5–10), and (C) temperature (20–45°C).
Endophytic strains were recognized through 16S rRNA gene sequencing and all achieved sequences were matched with nucleotide sequences of the national center for biotechnology information (NCBI) GenBank database by basic local alignment search tool (BlastN) program. We alienated 23 strains into 10 different species of Enterobacter i.e., Enterobacter ludwigii (2), Enterobacter cloacae (5), Enterobacter tabaci (2), Enterobacter sp. (5), Enterobacter asburiae (3), Enterobacter cancerogenus (1), Enterobacter oryzae (2), Enterobacter aerogenes (1), Enterobacter roggenkampii (1), and Enterobacter mori (1), based on ≥97% score similarity value. And all sequences were deposited in the NCBI GenBank from accession numbers MT613360-MT613382.
The phylogenetic tree was formed by a comparison of 16S rRNA gene partial sequences of the selected 23 isolates with the reference strains sequences of the NCBI GenBank public database. The phylogenetic tree which was created by 1000 bootstrap sampling showed two major sets and Pseudomonas putida strain was employed as the reference strain to divide Enterobacter strains (Figure 2).
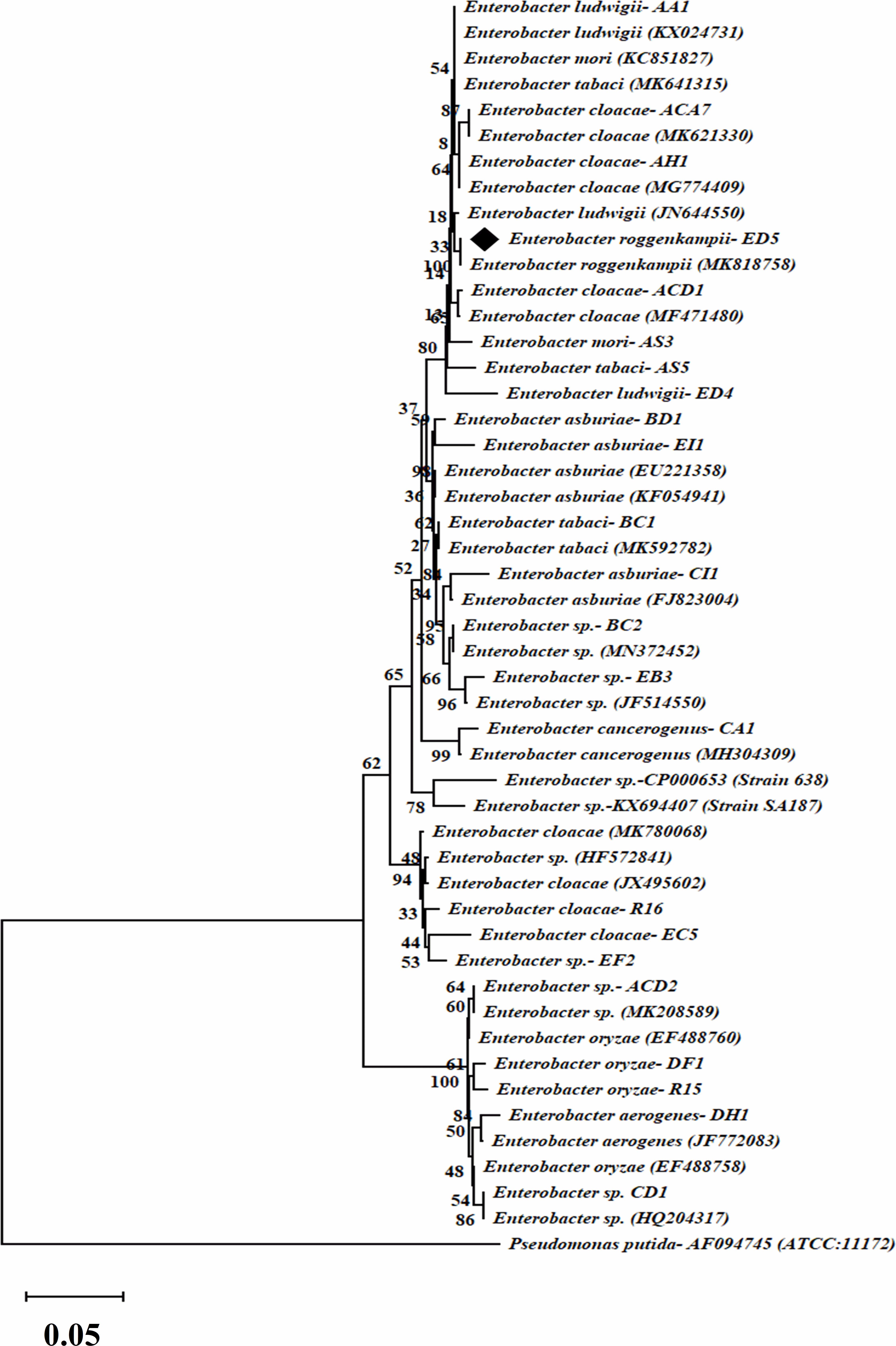
Figure 2. Dendrogram of 16S rRNA gene sequences of selected twenty-three endophytic Enterobacter isolates. The evolutionary distance was calculated by the UPGMA technique. Bootstrap analysis of 1,000 replications is specified as % confidence values for specific branching. Bar indicates % similarity and P. putida, as an outgroup.
Genomic DNA of all selected 23 endophytic strains was used to amplify nifH and acdS genes. Only 14 out of the 23 strains were confirmed positive nifH gene amplification, with a band size of 360 bp (Supplementary Figure S2) and a dendrogram was also created (Figure 3), whereas 12 confirmed positive acdS genes amplification with a band size of ∼ 755 bp (Supplementary Figure S3). All positive nifH and acdS strains were cloned and sequenced. After sequencing a BlastN search was finished and found all the sequenced clones were similar to the nifH gene sequences of NCBI GenBank database. In the case of acdS gene, only some sequenced clones showed similarity with acdS gene of the NCBI GenBank database and sequences not submitted. The identified nifH sequences were deposited in NCBI GenBank with accession numbers (MT649070-MT649083).
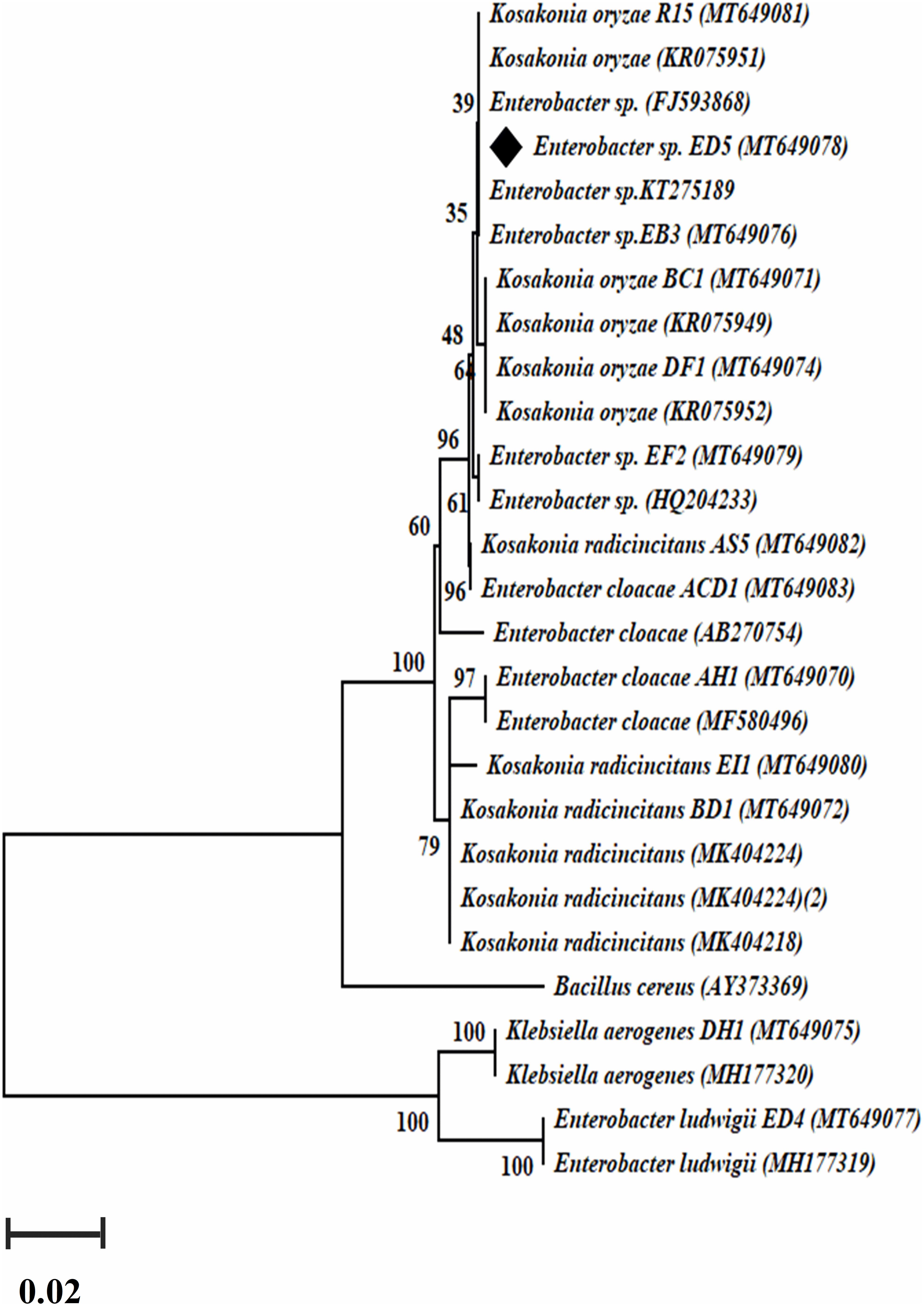
Figure 3. The dendrogram was created by a nifH gene sequences of the amplified Enterobacter strains by the neighbor-joining method.
The root colonization and colony morphology of ED5 strain was examined by SEM and CLSM (Figure 4), as this bacterium confirmed many PGP traits, excellent nitrogen-fixing potential, antifungal activity against plant pathogens, as well as survived in various abiotic stress circumstances. These techniques helped to study the interaction mechanism of the potential strains. In this study, E. roggenkampii ED5 strain was chosen for localization assessment in sugarcane cultivar with SEM and CLSM. Figures 4C,D, SEM results confirmed the colonization of E. roggenkampii in both stem and root tissues of sugarcane.
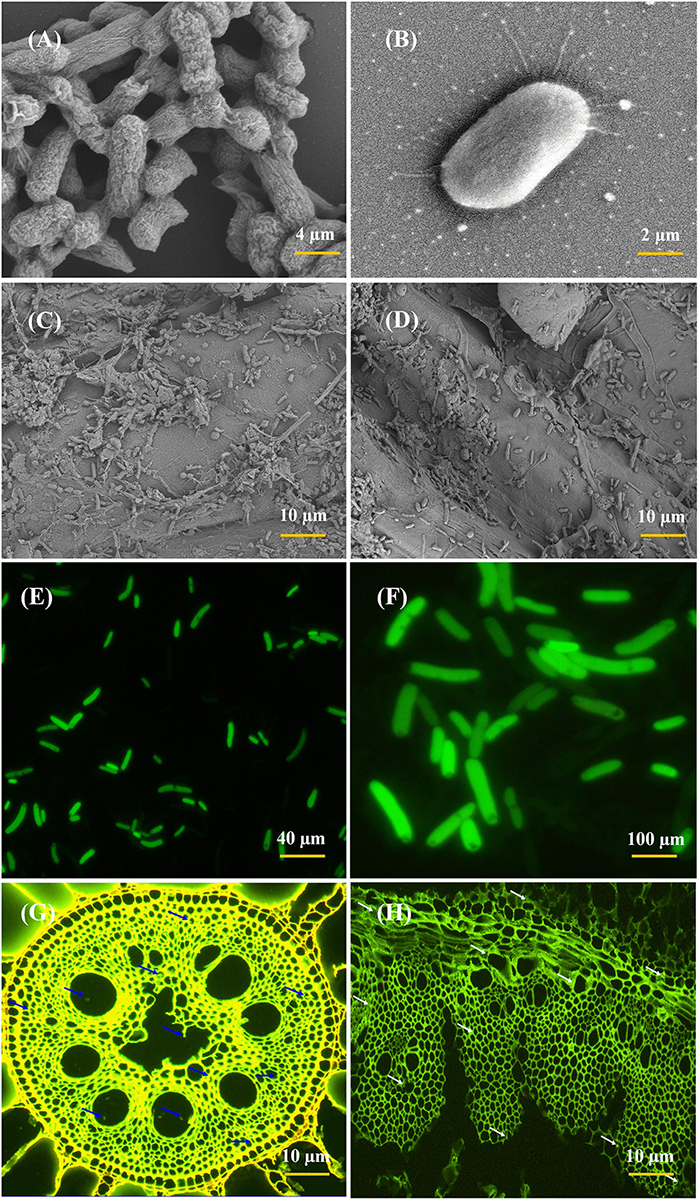
Figure 4. Scanning electron microscopy (SEM) and CLSM micrographs of most efficient endophytic E. roggenkampii ED5 strain and its colonization in sugarcane plant parts at the root and stem regions. Panels (A,B) is the SEM images showing the morphology of ED5 strain and, (C,D) is the colonization images obtained after the inoculation of ED5 strain in root and stem tissues of sugarcane. Panels (E,F) showing the CLSM micrographs of GFP-tagged endophytic ED5 strain, and (G,H), showing the colonization in the roots and stems of sugarcane by GFP-tagged E. roggenkampii ED5. CLSM images showing the selected strain ED5 in green dots of auto-fluorescence in both root and stem tissues, respectively, and bacterial cells are specified by blue and white arrowheads. Both micrographs confirmed the colonization of inoculated endophytic E. roggenkampii ED5 strain in sugarcane.
Whereas the GFP-tagged ED5 isolate transferred in sugarcane plants was also observed after 3 days of incubation and bacteria colonization was spotted as a green circle in all over of the plant stem and root tissues (Figures 4G,H). The density of ED5 strain had increased after incubation, and colonization of GFP-tagged strain was detected through the green auto-fluorescence produced as small dots in both roots and stems plant parts (Figure 4).
All physiological parameters (chlorophyll content, leaf area, plant height, root weight, shoot weight, photosynthesis, and transpiration rate) were significantly increased by inoculation of strain ED5 compared to control in GT11sugarcane cultivar at 30 and 60 days (Table 3).
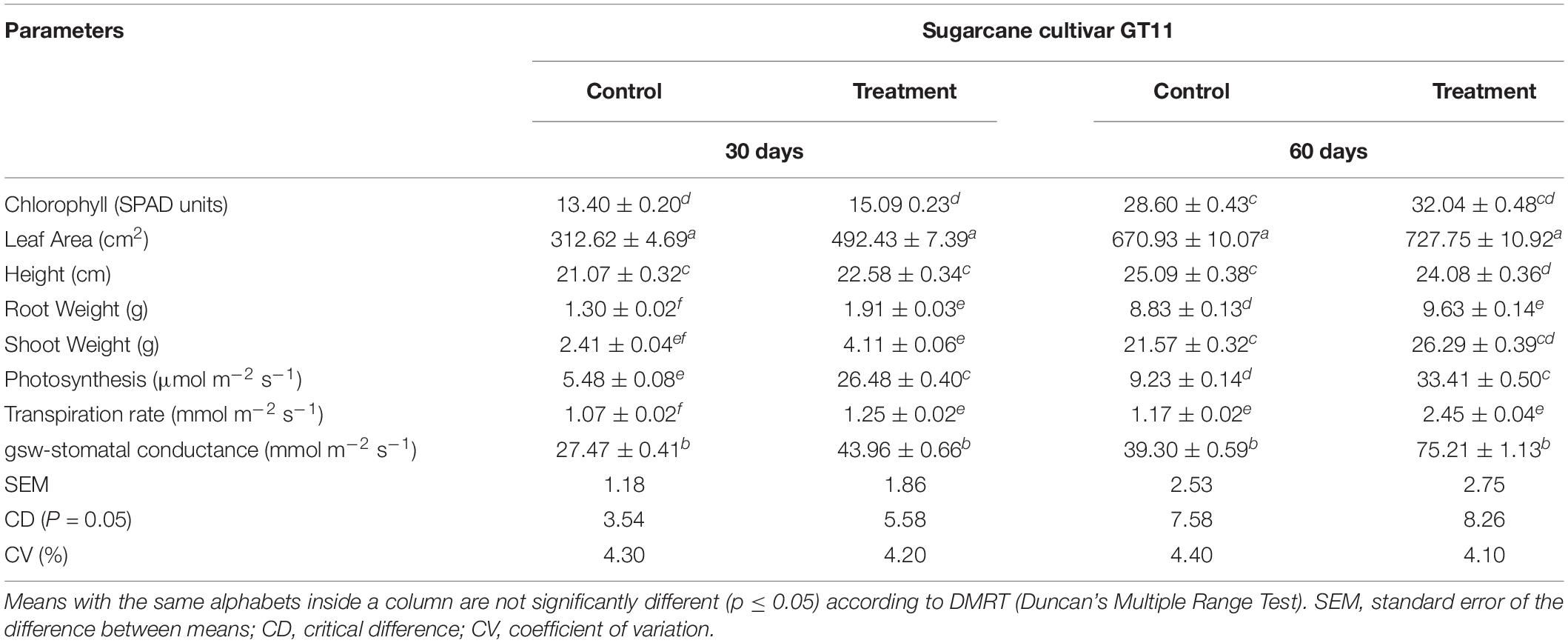
Table 3. Evaluation of E. roggenkampii ED5 strain on the plant growth parameters of sugarcane under greenhouse conditions after inoculation at 30 and 60 days.
The general properties of the endophytic ED5 strain genome are presented in Table 4, which comprised 4,698,609 base pairs of a circular chromosome with an average 56.05% G + C content. There were about 4349 predicted CDSs (Figure 5A). In addition, the E. roggenkampii genome included 83 tRNA and 25 rRNA (9, 5S; 8, 16S, and 8, 23S) genes. The CDSs number allocated to the KEGG, COG, and GO database were 2839, 4028, and 2949 (Supplementary Figures S4–S6). And a circular plasmid with 4242 base pairs of DNA and the G + C content of 45.66% (Table 4). Plasmid genome annotations estimated protein-coding with 6 genes, and results involved mRNA-degrading endonuclease, hypothetical protein, a transcriptional regulator, and RNA polymerase (Figure 5B). Here, we used Island Path-DIMOB, PHAST, and Minced software to predict the presence of 7 gene islands, 5 CRISPR, and 2 prophages in the ED5 genome. Clustered regularly interspaced short palindromic repeats (CRISPR) are hypervariable loci extensively dispersed in bacteria which offer acquired resistance toward foreign genetic elements. It is composed of many short and conserved repeat regions and spacers. A total of 5 CRISPRs were predicted from the genome of strain ED5 with 25 bp shortest and 43 bp longest direct repeat sequences. Prophages are repeatedly confined in sequenced bacterial genomes through a simple semantic script and contain 90 CDS genes, mainly related to hypothetical protein, cold shock-like protein, phage tail protein, DNA polymerase V subunit UmuC, etc. Whereas, gene islands contain 160 CDS genes, mainly related to pyrimidine utilization protein, hypothetical protein, Type VI secretion protein, etc. (Table 4). A complete genome sequence of this strain has been submitted at Gen-Bank/EMBL/DDBJ with accession numbers CP058253–CP058254.
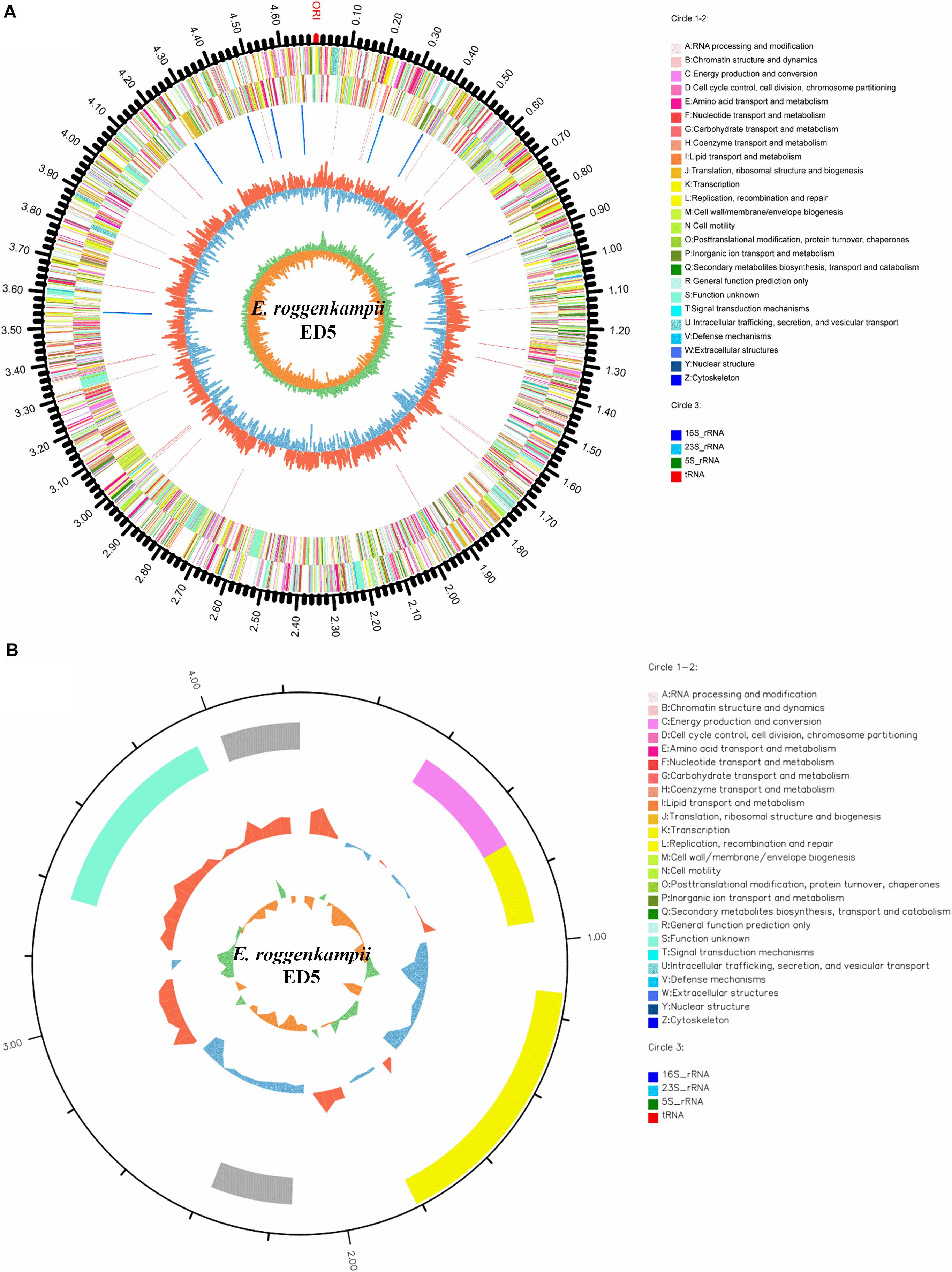
Figure 5. Circular representation of chromosome and plasmid of endophytic nitrogen-fixing E. roggenkampii ED5 strain isolated from sugarcane root. The inner and innermost rings display the GC content and skew. ORI; the origin of replication in chromosome map. (A,B): A–Z, respectively, show the functional classification of the CDS genes in the chromosome and plasmid with the colors of the COG database and circle 3; different colors show different RNA types.
The ANI results showed that the genome of ED5 presents 98.5259% ANI to E. roggenkampii FDAARGOS and 98.507% ANI to E. roggenkampii ECY546, respectively. Cluster analysis showed that they were closely related. The ANI value of strain ED5 and other strains were less than 95%, the highest value was 93.1342% for E. asburiae CAV1043 and the lowest was 79.69% for Citrobacter werkmanii MGYG-HGUT-02535. These ANI results indicated that strain ED5 belongs to E. roggenkampii (Figure 6).
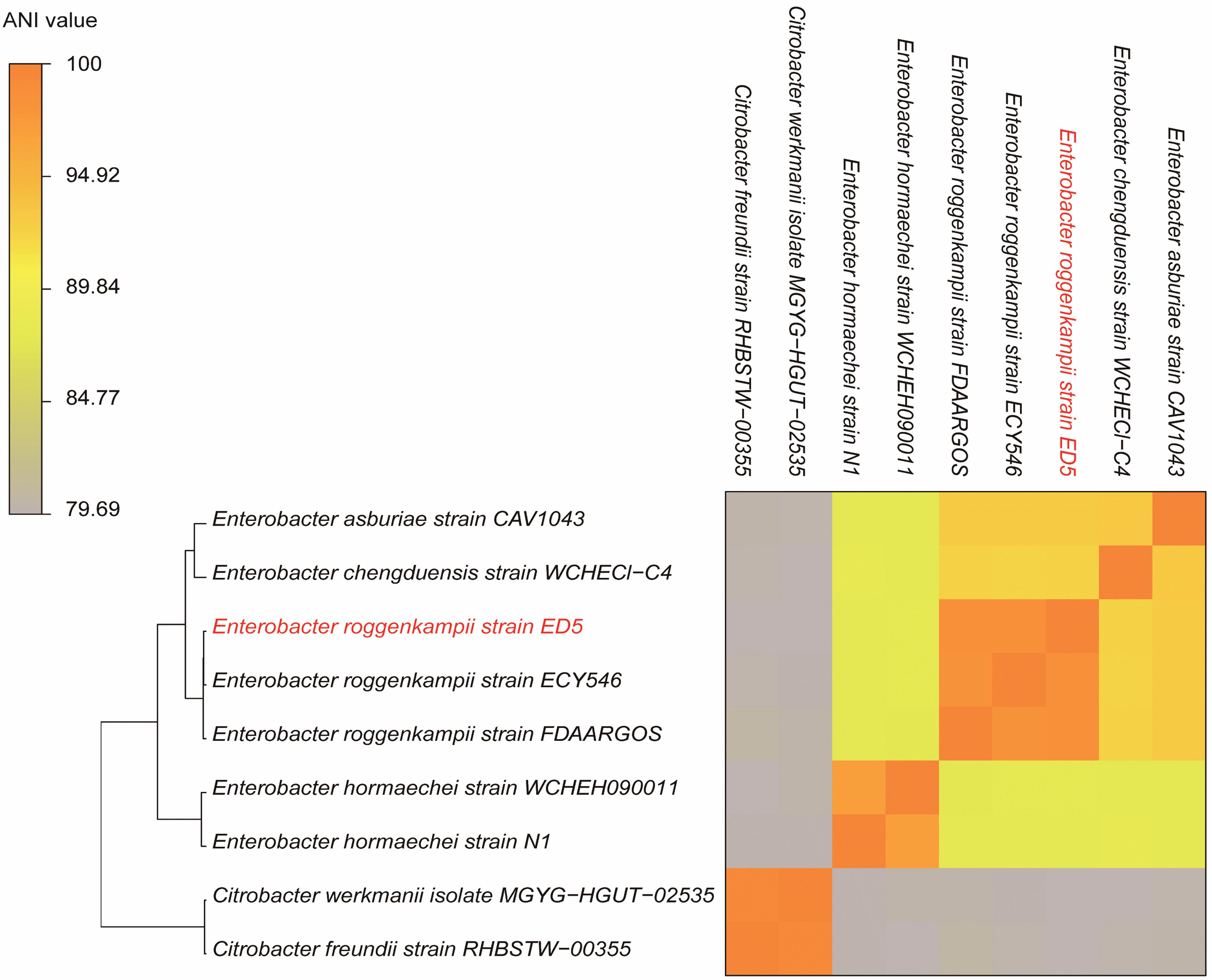
Figure 6. The heat maps of ANI (average nucleotide identity) between strain ED5 and phylogenetically eight closely related species. The ANI value among strain ED5 and E. roggenkampii FDAARGOS was 98.529% and E. roggenkampii ECY546 was 98.507%.
Examination of the recognized CDSs exposed the genome includes genes that encode proteins, related with nitrogen metabolism (iscU, norRV, and gltBD), ACC deaminase (dcyD), siderophores (fes, entFS, and fepA) plant hormones, phosphate metabolism, biofilm formation, root colonization, sulfur assimilation and metabolism, which are contributing in plant growth enhancement, were spotted (Table 5). The number of predicted gene clusters for secondary metabolite production such as NRPS, thiopeptide, Hserlactone, siderophore, and aryl-polyene are shown in Figure 7.
Also, genes involved in plant resistance response, i.e., antimicrobial peptide, synthesis of resistance inducers, hydrolase genes such as chitinase, cellulase, α- amylase, GTP cyclohydrolase, glutamate dehydrogenase, xylan 1,4beta-xylosidase, and glucosidase, whereas, oxidoreductases genes such as superoxide dismutase (SOD), glutathione peroxidase (GPX) and peroxiredoxin (PRXS) were also categorized (Table 5). Strain ED5 genome predicted some key genes of volatile substances such as 2,3-butanediol (alsD and ilvABCDEHMY), methanethiol (metH and idi) and isoprene (gcpE and ispE) and might be involved in biocontrol mechanism of strain ED5 (Table 5). Some symbiosis-related genes were also observed in strain ED5 genome, which might play a role in the establishment of symbiosis with the sugarcane plant (Table 5).
Enterobacter roggenkampii genome study also confirmed the existence of numerous genes involved in different abiotic stresses tolerance, mainly, the cold shock (cspA), heat shock (smpB hslR, ibpA, ibpB, and hspQ), drought resistance (nhaA, chaABC, proABPQSVWX, betABT, gabD, trkAH, kup, and kdpABCDEF), and heavy metals (cobalt, zinc, cadmium, magnesium, copper, mercury, lead, and manganese) resistance were identified (Table 6). Pathogenic and non-pathogenic bacteria secrete some protein-like virulence factors to adapt and survive in their living host. In this study, strain ED5 genome showed five types of secretion systems such as Type I, Type II, Type VI, Sec-SRP, and twin-arginine translocase (Tat), involving 49 genes by using Diamond Version 0.8.35 software (Figure 8).
In China, farmer’s applying higher doses of chemical fertilizers especially N-fertilizers to enhance growth and yields of sugarcane, but the use of the higher amount of chemical fertilizers increases the production cost as well as have unfavorable results on the environment, causes severe soil and water pollution, the decline in beneficial microbial flora associated with PGP, and nitrogen mineralization, etc. (Herridge et al., 2008; Li and Yang, 2015; Singh et al., 2020). The main objective of this research work is lookup an endophytic microbe that fixes nitrogen for prolong periods in sugarcane as well as another crop. Therefore, here we have focused to isolate and identify only on root endophytic strains of Enterobacter genus, as this is an important genus of nitrogen fixation. A total of 23 endophytic Enterobacter strains were designated and identified with 16S rRNA gene sequencing with E. roggenkampii was the most prominent strain. Endophytic bacterial strains interact with the plant extra efficiently than rhizospheric bacteria and increasingly provide several benefits to the host plant, generally growth promotion, and tolerance to biotic and abiotic stresses, also carry the genes essential for BNF, to change dinitrogen gas (N2) into usable forms of nitrogen, ACC deaminase activity, P- solubilization, and produce plant hormones, for example, IAA (Gaiero et al., 2013; Beltran-Garcia et al., 2014; Lebeis, 2014; Santoyo et al., 2016; Maksimov et al., 2018; White et al., 2018).
The Enterobacter strains are well-known nitrogen fixers, plant colonizers, and highly resistant to biotic and abiotic stresses (English et al., 2010; Zhu et al., 2013; Sarkar et al., 2018; Macedo-Raygoza et al., 2019). All selected strains showed significant biocontrol activities against several pathogens used by dual culture method, and E. roggenkampii (ED5) showed highest antagonistic activity against F. moniliforme, F. cubense, B. cinerea, C. paradoxa, and S. scitamineum which indicates the potential application for management of diseases caused by various pathogens. Whereas, E. roggenkampii strain is unknown for the ability to produce secondary metabolites, various PGP traits including colonization ability, and environmental stresses. Greenhouse experiment confirmed that selected strain ED5 improve the growth of physical parameters in sugarcane. Because previously no information was reported to compare this strain, we need to go for complete genome sequencing and annotation of this endophytic strain, which offers a useful platform to study all nitrogen-fixing, PGP, and stress tolerance mechanisms. Here, in this study, a complete genomic analysis of ED5 strain identified several genes clusters related to antimicrobial peptide, synthesis of resistance inducers, and hydrolases, including pagP, sapB, alsD, ilvABCDEHMY, metH, idi, gcpE, ispE, sacA, yxeP, ycjT, ribA, folE, gdhA, bglAFX, malZ, xynB, amyA, and some unknown gene name. Identification of the genes associated with the production of antimicrobial compounds especially to stimulate the antibiotic production recommends the biocontrol ability of strain ED5 as well as its function as a different PGP trait and nitrogenase activity genes that can indirectly stimulate plant health by defeating the pathogens (Shariati et al., 2017). We identified several genes that are known to support the production of antimicrobial compounds and they additionally contained genes for chitinase, cellulase, and beta-glucosidase enzyme that damage the pathogenic fungi cell walls, and similar genes are also reported earlier in other strains (Cho et al., 2015; Shariati et al., 2017; Luo et al., 2018; Yang et al., 2019) as well as growth-stimulating volatile compounds, are produced by some of the most efficient PGPR strains, including Enterobacter spp. (Weilharter et al., 2011).
The endophytic PGP Enterobacter strains were used as microbial inoculants in many crops globally, to decrease the application of chemical fertilizers and increase the yield of the crops, in addition to maintaining soil fertility (Singh et al., 2017; Daur et al., 2018; de Zélicourt et al., 2018). Therefore, this study explored almost all PGP traits like nitrogen fixation, IAA, siderophore, phosphate, ACC, HCN, and ammonia production of the selected strains isolated from sugarcane root. Several other studies also showed that all PGP traits comprising bacterial strains from sugarcane used as bio-inoculants and increased sugarcane yield (Li et al., 2017; Singh et al., 2020). The genome of E. roggenkampii covers several genes contributing to plant-beneficial roles, such as ACC, siderophore, ammonia, IAA production, phosphate metabolism, and nitrogen fixation. The presence of important genes encoding for PGP mechanisms was also determined previously, and some related genes were informed by Asaf et al. (2018) and Kang et al. (2020).
Nitrogen is one of the essential micronutrients for plant growth, while nitrogen metabolism is the main metabolic activity of bacterial cells. Earlier, several Enterobacteriaceae for example E. oryzae Ola51T, Enterobacter agglomerans, and E. cloacae were accounted as nitrogen-fixers (Kreutzer et al., 1991; Peng et al., 2009; Laili et al., 2017). The nifH is a well-recognized functional gene and its amplification via degenerate primers is a convenient method to confirm the nitrogen-fixation capability of the strains (Zehr and Capone, 1996; Rosado et al., 1998). In this study, all endophytic bacteria established nitrogen-fixing potential through the ARA method in an N-free medium. However, only 14 strains confirmed nifH gene amplification at around 360 bp of band size. Most prominent strain E. roggenkampii genome encloses six nitrogen metabolism associated genes, i.e., iscU, norRV, and gltBD with one unknown gene name, which proved that the strain is directly connected with nitrogen metabolisms such as nitrogen fixation, cyanate hydrolysis, nitrosative stress, and ammonia assimilation. Gene iscU is responsible for nitrogen fixation protein nifU and related proteins; nifU protein contributes a major role in the Fe-S cluster congregation, which is necessary for nitrogen fixation (Smith et al., 2005). In contrast, Andrés-Barrao et al. (2017) reported Enterobacter sp. SA187 genome includes dissimilatory nitrate reduction genes apart from genes coding for the nitrogenase enzyme (nifDHK). Klebsiella variicola GN02 and K. variicola DX120E genome hold numerous genes associated with nitrogen fixation, for example, nif gene cluster (nifHDK and nifLA), nitrogen metabolism-regulatory genes (ntrBC and glnD), and ammonium carrier gene (amtB) (Lin et al., 2012, 2015; Biaosheng et al., 2019).
Phosphorus is another vital and limiting macronutrient for the plant’s production, along with nitrogen. Specific bacteria play an important part in supplying accessible inorganic phosphorous in the form of orthophosphate (PO43–) to the plant, owing to phosphate is generally existing in the soil in the form of insoluble compounds and plants are only proficient to receive free orthophosphate (PO43–) (Bergkemper et al., 2016). In the present study, 16 Enterobacter strains showed phosphate solubilization traits. Similar to our results, other Enterobacter strains such as E. asburiae (Gyaneshwar et al., 1999), Enterobacter sp. EnB1 (Delgado et al., 2014), E. cloacae SBP-8 (Singh et al., 2017), and Enterobacter sp. SA187 (Andrés-Barrao et al., 2017) have been also reported as phosphate solubilizers. The genome of ED5 includes 14 genes (pit, pstABCS, phoUAEBRH, and ugpBE, with one unknown gene name) coding for phosphate metabolism. The Pit system is constitutive, whereas Pst transporter is inhibited by phosphate and induced under phosphate limitation (Jansson, 1988). Andrés-Barrao et al. (2017) reported that the Enterobacter sp. SA187 genome comprises genes coding for phosphate uptake, low-affinity inorganic phosphate transporter, and phosphate starvation response.
Several helpful bacteria comprise PGP activity that is occurred by various mechanisms, such as inactivation or production of ACCD enzyme activity. PGPB including ACCD decreased the ethylene content in plants and encouraged root elongation (Penrose and Glick, 2003). In this study, all strains showed ACCD enzyme activity whereas, only 12 strains confirmed acdS gene amplification of ∼750–755 bp. Interestingly, one dcyD gene, coding for ACC deaminase, was present in E. roggenkampii genome. ACCD activity has been reported in many Pseudomonas, Bacillus, and Mesorhizobium strains, along with members of Enterobacter genus such as E. cloacae UW4, E. cloacae CAL2, E. cancerogenus, and Enterobacter sp. EN-21 (Shah et al., 1998; Holguin and Glick, 2001; Glick, 2014; Li et al., 2017; Kruasuwan and Thamchaipenet, 2018; Singh et al., 2020). IAA production from tryptophan by indole pyruvate is another approach of PGPB to improve plant growth (Taghavi et al., 2009). We observed that all endophytic strains were capable to synthesize IAA, and E. roggenkampii holds trpBCEFS, and trpGD genes code for enzymes concerned in this pathway. Moreover, we identified one gene auxin efflux carrier (mdcF) related to auxin biosynthesis, confirm their potential to be used as growth regulators. In similar to our findings, previously also well- recognized that the existence of tryptophan associated genes in genomes of bacteria is related to IAA production (Tadra-Sfeir et al., 2011; Gupta et al., 2014). As reported in Enterobacter strain 638 (Taghavi et al., 2010) and E. cloacae UW5 (Coulson and Patten, 2015) improved IAA levels and stimulate root development. Asaf et al. (2018) found tryptophan biosynthesis genes (trpABD) involved in IAA production was found in Sphingomonas sp. LK11 genome (Asaf et al., 2018).
PGPB developed a particular method for iron absorption by siderophores production, which transfers this component into their cells (Arora et al., 2013). In this study, 13 isolates showed positive siderophore production. Siderophore production by these strains expects importance for iron nutrition of plants matures in iron-limited situations. ED5 strain demonstrated strong siderophore activity and siderophore enterobactin (fes, entFS, and fepA) biosynthesis pathway was also observed in its genome study. Consistent with this study, the siderophore enterobactin pathway (fepEGDC) was detected in E. cloacae SBP-8 (Singh et al., 2017) and Bacillus subtilis EA-CB0575 genomes (Franco-Sierra et al., 2020).
A biofilm is a surface-linked efficient microorganism confined by a polymeric matrix including self-making exopolysaccharides, extracellular DNA and proteins related through the biotic surface (Hall-Stoodley and Stoodley, 2002; Vlamakis et al., 2013; Teschler et al., 2015). A large amount of beneficial microbial community’s structure recommended a biofilm, and some of the most fascinating recommended helpful biofilms are used in agriculture (Kumar et al., 2019; Singh M. P. et al., 2019). In the endophytic and rhizospheric zone, several kinds of bacterial species involve plant roots and create a biofilm which offers benefits to each other. Nowadays, plant-associated microorganisms have concerned a lot of interest because of the considerable effects of plant health and productivity (Bogino et al., 2013). Some of the PGPR showed antagonistic activity in response to phytopathogens by starting biofilm-like assemblies that have been previously reported to Bacillus cereus (Xu et al., 2014), Paenibacillus (Timmusk et al., 2015), and Pseudomonas stutzeri (Wang et al., 2017). In the biofilm, a cell-to-cell communication raises the gene expression of both up-down regulation, for enhancing the adaptation of microorganisms in both biotic and abiotic environments. In the E. roggenkampii genome, 21 genes, tomb, luxS, efp, flgABCDEFGHIJKLNM, motAB, sacA, and hfq which are associated to exopolysaccharides biosynthesis, protein, and biofilm formation were also found (Gupta et al., 2014; Ju et al., 2018).
Beneficial microorganism generally colonizes on the surface and inside tissues of various sections in the plant, i.e., root, stem, and leaf, the place they stay either commensally or perform helpful features (Johnston and Raizada, 2011). The interactions between beneficial microbes of the host plant might play an essential part in the achievement of microbial bioinoculant for improving the production of crops, but there is no strong thought on the entire role of colonization on the plant microbiome. The GFP pictures showed a signal of the occurrence of E. roggenkampii in the intercellular regions also as cell aggregates or isolated single cells in the root of sugarcane (Li et al., 2017; Singh et al., 2020). The SEM images confirmed of selected inoculated strain E. roggenkampii in stem and roots specified that the forms of adherence in sugarcane, and similar observation were also found in other endophytic strains like Paenibacillus polymyxa, Rhizobium sp. and Burkholderia sp. (Timmusk et al., 2005; Singh et al., 2009). In this study, we also clearly confirmed the result of root colonization genes present in E. roggenkampii through genome analysis. We recognized a great number of root colonization genes present in E. roggenkampii at different stages: chemotaxis (cheABRVWYZ, tsr, trg, aer, tar, and mcp) for signal transduction, motility (flhEABCD, fliABCDEFGHIJKLMNOPQRSTYZ) for genes regulation, adhesive structure (pilDT, and hofC), play a significant function in host–microbes interactions (Dorr et al., 1998; Krause et al., 2006) and adhesin production (pgaABCD). A similar observation was also reported by Cho et al. (2015).
Hydrogen sulfide (H2S) production by PGPR has been described to improve plant growth, and root colonization (Dooley et al., 2013). H2S, as an important molecule with beneficial effects on P-solubilization, responds with ferric phosphate to crop ferrous with the release of phosphate (Shariati et al., 2017). Sulfur is a vital nutrient for plant increase and development, along with related to stress tolerance in plants (Gill and Tuteja, 2011), because lacking sulfur in plant cause severe losses in crop yield and production. In the genome of E. roggenkampii ED5 genes associated with sulfate transporters (cysACDJHIKMNPUWZ) were found. Earlier, the cysP gene function was verified with a strain Escherichia coli transformed by a plasmid expressing B. subtilis cysP gene through a mutated sulfate transport (Mansilla and de Mendoza, 2000). The operon determined by cysP gene in B. subtilis is accountable for sulfur metabolism, for example, the sulfate adenylyltransferase gene (Aguilar-Barajas et al., 2011). The strain E. roggenkampii genome encodes the set of genes that are responsible for H2S biosynthesis, including the cysACDEGHIJKMNPQSUWZ, and fdx genes presented in assimilated sulfate reduction. The existence of an ATP-binding transporter gene that contains periplasmic binding proteins cysP, cysW, and cysA were determined in the genome of ED5 that discovered these genes might be elaborated in the transportation of thiosulfate or inorganic sulfate to cells, earlier reported in Pseudomonas sp. UW4 (Duan et al., 2013). The occurrence of these genes in microorganisms has been associated with the oxidation of sulfur and sulfur-conjugated secondary metabolites (Kwak et al., 2014). Also, sulfur oxidation effects soil pH and successively recovers solubility of micronutrients, i.e., N, P, K, Mg, and Zn (Vidyalakshmi et al., 2009). Hence, this type of beneficial endophytic microbes can offer enhanced mineral achievement and distribution to the host plants (Kang et al., 2020).
Abiotic stresses are highly injurious for the plants, the most unfavorable influence from physiological to the molecular level of the plants. Drought and heavy metal stress greatly decrease crop yield (Khan et al., 2018; Kang et al., 2020). Drought is the leading reason for preventive crop production over massive areas of the Earth (Saleem et al., 2018), and estimated to cause severe plant growth difficulties for above 50% of the arable lands in 2050 (Vinocur and Altman, 2005; Kasim et al., 2013). However, heavy metal recognized as a principal hazard in many terrestrial ecosystems globally (Shahid et al., 2015). Currently, significant industrialization will directly increase the heavy metals in soils, that have damaging outcomes on plant growth and productiveness (Forstner and Wittmann, 2012). The numerous micro-organisms have an intrinsic ability to manage abiotic stresses and improve plant growth. Earlier, several PGPR genera have been described to succeed drought and heavy metal stresses through various mechanisms, and to improve plant tolerance especially abiotic stresses (Dary et al., 2010; Hassan et al., 2017), as well as to improve the production of crops (Sessitsch et al., 2013).
A number of mechanisms have been described earlier about the heavy metal stress resistance of many microbial species (Kunito et al., 1996). Investigation of the ED5 genome discovered the occurrence of various genes presented in the homeostasis of heavy metals like copper, zinc, manganese, cadmium, etc. A genes czcD, znuABC, zupT, zntAB are found and these proteins were linked to the role of generating tolerance to about divalent cations with cobalt, zinc, and cadmium, owing to its capability to automatically produce an efflux of metal ions (Mima et al., 2009). Additionally, the magnesium and cobalt transport genes corAC and cobA were found in the ED5 genome. These genes have been stated to be elaborate in manganese transport, mntR and mntH genes considered the primary manganese transporters in bacteria (Kang et al., 2020). The copper (Cu) is an essential component for biological progressions, and a similar metal cofactor of several enzymes like monooxygenases, dioxygenases, and SOD (Giner-Lamia et al., 2012). The genes found in ED5 genome like copCD, cusABCFRS, and copA, have similar operon has been existing in bacterial species of Pseudomonas psychrotolerans, Pseudomonas syringae, Xanthomonas campestris, and E. coli (Duncan et al., 1985; Lee et al., 1992; Silver et al., 1993; Voloudakis et al., 1993; Cooksey, 1994; Kang et al., 2020). To further verify the ability of drought tolerance, we mentioned extraordinary resistance mechanisms in ED5, inclusive of tolerance to heavy metal, pH, and temperature stresses. Genes related to drought stress were determined in the ED5 genome are nhaA, chaABC, proABPQSVWX, betABT, gabD, trkAH, kup, kdpABCDEF.
The present study selected the numbers of nitrogen-fixing endophytic strains of Enterobacter genus from the sugarcane roots. All strains exhibited many PGP traits, biocontrol activity, as well as tolerance to various environmental conditions, and E. roggenkampii ED5 was the most prominent strain among all. Therefore, the employ of efficient endophytic bacteria is an opportunity to improve crop yield and comprehensive genome sequencing of ED5 strain has revealed many prospects to study this potential endophytic strain in the future. Also, the ED5 genome carried a set of universal genes that contributed to PGP, nitrogen fixation, and response to several stresses. So, it can be summarized that ED5 strain may be used as a possible alternate for chemical fertilizers and play an important part in improving ecosystem quality. However, field trials are required to explain the usability of the E. roggenkampii ED5 in the field earlier than it can be established as a plant growth promoter for utilizing in sustainable agriculture.
The datasets generated in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
D-JG, RS, PS, and Y-RL planned the proposal and experiments. RS, PS, and D-JG accomplished the experiments. AS, D-PL, and X-PS performed the data examination. L-TY, Y-XX, and Y-RL contributed to study and resources. D-JG, RS, and PS wrote the original manuscript. Y-XX and Y-RL reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This study was sponsored by Guangxi Special Funds for Bagui Scholars and Distinguished Experts (2013-3), Guangxi Special Fund for Scientific Base, and Talent (GKAD17195100). The National Natural Science Foundation of China (31471449, 31171504, and 31101122), Fund for Guangxi Innovation Teams of Modern Agriculture Technology (GJNYTXGXCXTD-03-01), and Fund of Guangxi Academy of Agricultural Sciences (2015YT02, GNKB2017028, and GNKB2018034).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.580081/full#supplementary-material
Aguilar-Barajas, E., Díaz-Pérez, C., Ramírez-Díaz, M. I., Riveros-Rosas, H., and Cervantes, C. (2011). Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24, 687–707. doi: 10.1007/s10534-011-9421-x
Andrés-Barrao, C., Lafi, F. F., Alam, I., de Zélicourt, A., Eida, A. A., Bokhari, A., et al. (2017). Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 8:2023. doi: 10.3389/fmicb.2017.02023
Arora, N. K., Tewari, S., and Singh, R. (2013). ”Multifaceted plant-associated microbes and their mechanisms diminish the concept of direct and indirect PGPRs,” in Plant Microbe Symbiosis; Fundamentals and Advances, ed. N. K. Arora (Lucknow: Springer), 411–449. doi: 10.1007/978-81-322-1287-4_16
Asaf, S., Khan, A. L., Khan, M. A., Al-Harrasi, A., and Lee, I. J. (2018). Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3 Biotech 8:389. doi: 10.1007/s13205-018-1403-z
Baldani, J. I., and Baldani, V. L. D. (2005). History on the biological nitrogen fixation research in graminaceous plants, special emphasis on the Brazilian experience. An. Acad. Bras. Cienc. 77, 549–579. doi: 10.1590/S0001-37652005000300014
Barrera, G. (2012). Oxidative stress and lipid peroxidation products in cancer progression and therapy. ISRN Oncol. 2012:137289. doi: 10.5402/2012/137289
Beltran-Garcia, M. J., White, J. F., Padro, F. M., Prieto, K. R., Yamaguchi, L. F., Torres, M. S., et al. (2014). Nitrogen acquisition in Agave tequilana from degradation of endophytic bacteria. Sci. Rep. 4:6938. doi: 10.1038/srep06938
Bergkemper, F., Schöler, A., Engel, M., Lang, F., Krüger, J., Schloter, M., et al. (2016). Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ. Microbiol. 18, 1988–2000.
Biaosheng, L., Zhaozhao, S., Yulei, J., Yulong, Z., Lifang, W., Jinling, F., et al. (2019). Biological characteristics and genome-wide sequence analysis of endophytic nitrogen-fixing bacteria Klebsiella variicola GN02. Biotechnol. Biotechnol. Equip. 33, 108–117. doi: 10.1080/13102818.2018.1555010
Boddey, R. M., Urquiaga, S., Reis, V., and Döbereiner, J. (1991). Biological nitrogen fixation associated with sugarcane. Plant Soil 137, 111–117.
Bogino, P., Oliva, M., Sorroche, F., and Giordano, W. (2013). The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 14, 15838–15859. doi: 10.3390/ijms140815838
Borodovsky, M., and Mcininch, J. (1993). Gene mark, parallel gene recognition for both DNA strands. Comput. Chem. 17, 123–133. doi: 10.1016/0097-8485(93)85004-V
Brick, J. M., Bostock, R. M., and Silverstone, S. E. (1991). Rapid in situ assay for indole acetic acid production by bacteria immobilized on nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538. doi: 10.1128/AEM.57.2.535-538.1991
Carvalho, T. L. G., Balsemao-Pires, E., Saraiva, R. M., Ferreira, P. C. G., and Hemerly, A. S. (2014). Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J. Exp. Bot. 65, 5631–5642.
Cho, S. T., Chang, H. H., Egamberdieva, D., Kamilova, F., Lugtenberg, B., and Kuo, C. H. (2015). Genome analysis of Pseudomonas fluorescens PCL1751, a rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS One 10:e0140231. doi: 10.1371/journal.pone.0140231
Cooksey, D. A. (1994). Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 14, 381–386. doi: 10.1111/j.1574-6976.1994.tb00112.x
Coulson, T. J. D., and Patten, C. L. (2015). Complete genome sequence of Enterobacter cloacae UW5, a rhizobacterium capable of high levels of indole-3-acetic acid production. Genome Announc. 3:e00843-15.
Dary, M., Chamber-Pérez, M., Palomares, A., and Pajuelo, E. (2010). “In situ” phytostabilisation of heavy metal polluted soils using Lupinus luteus inoculated with metal resistant plant-growth promoting rhizobacteria. J. Hazard. Mater. 177, 323–330. doi: 10.1016/j.jhazmat.2009.12.035
Daur, I., Saad, M. M., Eida, A. A., Ahmad, S., Shah, Z. H., Ihsan, M. Z., et al. (2018). Boosting alfalfa (Medicago sativa L.) production with rhizobacteria from various plants in Saudi Arabia. Front. Microbiol. 9:477. doi: 10.3389/fmicb.2018.00477
de Zélicourt, A., Synek, L., Saad, M. M., Alzubaidy, H., Jalal, R., Xie, Y., et al. (2018). Ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2-keto-4-methylthiobutyric acid production. PLoS Genet. 14:e1007273. doi: 10.1371/journal.pgen.1007273
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Delgado, M., Mendez, J., Rodriìguez-Herrera, R., Aguilar, C. N., Cruz-Hernaìndez, M., and Balagurusamy, N. (2014). Characterization of phosphate solubilizing bacteria isolated from the arid soils of a semi-desert region of north-east Mexico. Biol. Agric. Hortic. 30, 211–217. doi: 10.1080/01448765.2014.909742
Dey, R., Pal, K. K., Bhatt, D. M., and Chauhan, S. M. (2004). Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth promoting rhizobacteria. Microbiol. Res. 159, 371–394.
Döbereiner, J. (1997). Biological nitrogen fixation in the tropics: social and economic contributions. Soil Biol. Biochem. 29, 771–774. doi: 10.1016/s0038-0717(96)00226-x
Dobereiner, J., Reis, V. M., Paula, M. A., and Olivares, F. L. (1993). “Endophytic diazotrophs in sugar cane, cereals and tuber plants,” in New Horizons in Nitrogen Fixation, eds R. Palacios, J. Mora, and W. E. Newton (Dordrecht: Kluwer Academic), 671–676. doi: 10.1007/978-94-017-2416-6_55
Dooley, F. D., Nair, S. P., and Ward, P. D. (2013). Increased growth and germination success in plants following hydrogen sulfide administration. Nitric Oxide 31:S24.
Dorr, J., Hurek, T., and Reinhold-Hurek, B. (1998). Type IV pili are involved in plant–microbe and fungus–microbe interactions. Mol. Microbiol. 30, 7–17. doi: 10.1046/j.1365-2958.1998.01010.x
Duan, J., Jiang, W., Cheng, Z. Y., Heikkila, J. J., and Glick, B. R. (2013). The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp UW4. PLoS One 8:e58640. doi: 10.1371/journal.pone.0058640
Duncan, R., Camakaris, J., Lee, B. T., and Luke, R. K. (1985). Inducible plasmid-mediated copper resistance in Escherichia coli. J. Gen. Microbiol. 131, 939–943. doi: 10.1099/00221287-131-4-939
English, M. M., Coulson, T. J. D., Horsman, S. R., and Patten, C. L. (2010). Over expression of hns in the plant growth-promoting bacterium Enterobacter cloacae UW5 increases root colonization. J. Appl. Microbiol. 108, 2180–2190.
FAO (2018). Top Sugarcane Production, Food and Agriculture Organization of the United Nations. Rome: FAO.
Felsenstein, J. (1985). Confidence limits on phylogenies, an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Foley, J. A., Defries, R., Asner, G. P., Barford, C., Bonan, G., Carpenter, S. R., et al. (2005). Global consequences of land use. Science 309, 570–574. doi: 10.1126/science.1111772
Forstner, U., and Wittmann, G. T. (2012). Metal Pollution in the Aquatic Environment. Berlin: Springer Science & Business Media.
Franco-Sierra, N. D., Posada, L. F., Santa-María, G., Romero-Tabarez, M., Villegas-Escobar, V., and Álvarez, J. C. (2020). Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct. Integr. Genomic 20, 575–589. doi: 10.1007/s10142-020-00736-x
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., and Dunfield, K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750. doi: 10.3732/ajb.1200572
Gill, S. S., and Tuteja, N. (2011). Cadmium stress tolerance in crop plants, probing the role of sulfur. Plant Signal. Behav. 6, 215–222. doi: 10.4161/psb.6.2.14880
Gillis, M., Kersters, K., Hoste, B., Janssens, D., Kroppenstedt, R. M., Stephan, M. P., et al. (1989). Acetobactev diazotrophicus sp. nov., a nitrogen-fixing acetic acid bacterium associated with sugarcane. Int. J. Syst. Bacteriol. 39, 361–364. doi: 10.1099/00207713-39-3-361
Giner-Lamia, J., Lopez-Maury, L., Reyes, J. C., and Florencio, F. J. (2012). The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 159, 1806–1818. doi: 10.1104/pp.112.200659
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Glickmann, E., and Dessaux, Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61, 793–796. doi: 10.1128/aem.61.2.793-796.1995
Govindarajan, M., Kwon, S. W., and Weon, H. Y. (2007). Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph Klebsiella sp. GR9. World J. Microbiol. Biotechnol. 23, 997–1006. doi: 10.1007/s11274-006-9326-y
Gupta, A., Gopal, M., Thomas, G. V., Manikandan, V., Gajewski, J., Thomas, G., et al. (2014). Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops Coconut, Cocoa and Arecanut. PLoS One 9:e104259. doi: 10.1371/journal.pone.0104259
Gyaneshwar, P., Parekh, L. J., Archana, G., Poole, P. S., Collins, M. D., Hutson, R. A., et al. (1999). Involvement of a phosphate starvation inducible glucose dehydrogenase in soil phosphate solubilization by Enterobacter asburiae. FEMS Microbiol. Lett. 171, 223–229. doi: 10.1111/j.1574-6968.1999.tb13436.x
Hall-Stoodley, L., and Stoodley, P. (2002). Developmental regulation of microbial biofilms. Curr. Opin. Biotechnol. 13, 228–233. doi: 10.1016/s0958-1669(02)00318-x
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttila, C. S., Campisano, A., Dorin, M., et al. (2015). The hidden world within plants, ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/mmbr.00050-14
Hardy, R. W. F., Holsten, R. D., Jackson, E. K., and Burns, R. C. (1968). The acetylene ethylene assay for N2 fixation, laboratory and field evaluation. Plant Physiol. 43, 1185–1207. doi: 10.1104/pp.43.8.1185
Hassan, T. U., Bano, A., and Naz, I. (2017). Alleviation of heavy metals toxicity by the application of plant growth promoting rhizobacteria and effects on wheat grown in saline sodic field. Int. J. Phytoremediation 19, 522–529. doi: 10.1080/15226514.2016.1267696
Herridge, D. F., Peoples, M. B., and Boddey, R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 3, 1–18. doi: 10.1007/s11104-008-9668-3
Holguin, G., and Glick, B. R. (2001). Expression of the ACC deaminase gene from Enterobacter cloacae UW4 in Azospirillum brasilense. Microb. Ecol. 41, 281–288. doi: 10.1007/s002480000040
Honma, M., and Shimomura, T. (1978). Metabolism of 1-aminocyclopropane-1- carboxylic acid. Agric. Biol. Chem. 42, 1825–1831.
Huang, Y. K., and Li, W. F. (2014). The List of Harmful Organism and Natural Enemies Resources in Modern Sugarcane. Beijing: China Agriculture Press.
Huang, Y. K., and Li, W. F. (2016). Colored Atlas of Control on Diseases, Insect Pests and Weeds of Modern Sugarcane. Beijing: China Agriculture Press.
Jacobson, C. B., Pasternak, J. J., and Glick, B. R. (1994). Partial purification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 40, 1019–1025. doi: 10.1139/m94-162
James, E. K. (2000). Nitrogen fixation in endophytic and associative symbiosis. Food Crop Res. 65, 197–209. doi: 10.1016/s0378-4290(99)00087-8
Jansson, M. (1988). Phosphate uptake and utilization by bacteria and algae. Hydrobiologia 170, 177–189. doi: 10.1007/978-94-009-3109-1_11
Johnston, M. D., and Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in zea across boundaries of evolution, ethnography and ecology. PLoS One 6:e20396. doi: 10.1371/journal.pone.0020396
Ju, X., Li, J., Zhu, M., Lu, Z., Lv, F., Zhu, X., et al. (2018). Effect of the luxS gene on biofilm formation and antibiotic resistance by Salmonella serovar Dublin. Food Res. Int. 107, 385–393. doi: 10.1016/j.foodres.2018.02.039
Kang, S. M., Asaf, S., Khan, A. L., Lubna, Khan, A., Mun, B. G., et al. (2020). Complete genome sequence of Pseudomonas psychrotolerans CS51, a plant growth-promoting bacterium, under heavy metal stress conditions. Microorganisms 8:382. doi: 10.3390/microorganisms8030382
Kang, S. M., Asaf, S., Kim, S. J., Yun, B. W., and Lee, I. J. (2016). Complete genome sequence of plant growth-promoting bacterium Leifsonia xyli SE134, a possible gibberellin and auxin producer. J. Biotechnol. 239, 34–38. doi: 10.1016/j.jbiotec.2016.10.004
Kasim, W. A., Osman, M. E., Omar, M. N., El-Daim, I. A., Bejai, S., and Meijer, J. (2013). Control of drought stress in wheat using plant-growth-promoting bacteria. J. Plant Growth Regul. 32, 122–130. doi: 10.1007/s00344-012-9283-7
Khan, N., Bano, A., Rahman, M. A., Rathinasabapathi, B., and Babar, M. A. (2018). UPLC-HRMS based untargeted metabolic profiling reveals changes in Chickpea (Cicer arietinum) Metabolome following long term drought stress. Plant Cell Environ. 42, 115–132. doi: 10.1111/pce.13195
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Krause, A., Ramakumar, A., Bartels, D., Battistoni, F., Bekel, T., Boch, J., et al. (2006). Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24, 1385–1391. doi: 10.1038/nbt1243
Kreutzer, R., Steibl, H. D., Dayananda, S., Dippe, R., Halda, L., Buck, M., et al. (1991). “Genetic characterization of nitrogen fixation in Enterobacter strains from the rhizosphere of cereals,” in Nitrogen Fixation. Developments in Plant and Soil Sciences, Vol. 48, eds M. Polsinelli, R. Materassi, and M. Vincenzini (Dordrecht: Springer).
Kruasuwan, W., and Thamchaipenet, A. (2018). 1-Aminocyclopropane-1-carboxylate (ACC) deaminase-producing endophytic diazotrophic Enterobacter sp. EN-21 modulates salt–stress response in sugarcane. J. Plant Growth Regul. 37, 849–858. doi: 10.1007/s00344-018-9780-4
Kumar, J., Sharma, V. K., Parmar, S., Singh, P., and Singh, R. K. (2019). “Biofilm, A microbial assemblage on the surface-A boon or bane?,” in New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms, eds M. K. Yadav and B. P. Singh (Amsterdam: Elsevier), 139–150. doi: 10.1016/b978-0-444-64279-0.00011-6
Kumar, M., Grader, G., Sessitsch, A., Maki, A., van Elsas, J. D., and Nissien, R. (2017). Plants assemble species specific bacterial communities from common core taxa in three arcto-alpine climate zones. Front. Microbiol. 8:12. doi: 10.3389/fmicb.2017.00012
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7, molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kunito, T., Kusano, T., Oyaizu, H., Senoo, K., Kanazawa, S., and Matsumoto, S. (1996). Cloning and sequence analysis of czc genes in Alcaligenes sp. strain CT14. Biosci. Biotechnol. Biochem. 60, 699–704. doi: 10.1271/bbb.60.699
Kwak, M. J., Jeong, H., Madhaiyan, M., Lee, Y., Sa, T. M., Oh, T. K., et al. (2014). Genome information of Methylobacterium oryzae, a plant-probiotic methylotroph in the phyllosphere. PLoS One 9:e106704. doi: 10.1371/journal.pone.0106704
Laili, N. S., Radziah, O., and Zaharah, S. S. (2017). Isolation and characterization of plant growth-promoting rhizobacteria (PGPR) and their effects on growth of strawberry (Fragaria ananassa Duch.) Bangladesh J. Bot. 46, 277–282.
Lebeis, S. L. (2014). The potential for give and take in plant-microbiome relationships. Front. Plant Sci. 5:287. doi: 10.3389/fpls.2014.00287
Lee, Y., Hendson, M., and Schroth, M. (1992). Cloning and characterization of copper-resistance genes from Xanthomonas campestris pv. juglandis. Phytopathology 82:1125.
Li, H. B., Singh, R. K., Singh, P., Song, Q. Q., Xing, Y. X., Yang, L. T., et al. (2017). Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front. Microbiol. 8:1268. doi: 10.3389/fmicb.2017.01268
Li, R. Y., and Yang, L. T. (2015). Sugarcane agriculture and sugar industry in China. Sugar Tech 17, 1–8. doi: 10.1007/s12355-014-0342-1
Li, Y. R., Song, X. P., Wu, J. M., Li, C. N., Liang, Q., Liu, X. H., et al. (2016). Sugar industry and improved sugarcane farming technologies in China. Sugar Tech 18, 603–611. doi: 10.1007/s12355-016-0480-8
Li, Y. R., Zhou, X. Z., and Yang, L. T. (2015). Biological nitrogen fixation in sugarcane and nitrogen transfer from sugarcane to cassava in an intercropping system. Inter. J. Sci. Nat. 6, 214–218.
Li, Z., Chang, S., Lin, L., Li, Y., and An, Q. (2011). A colorimetric assay of 1- aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett. Appl. Microbiol. 53, 178–185. doi: 10.1111/j.1472-765x.2011.03088.x
Lin, L., Li, Z., Hu, C., Zhang, X., Chang, S., Yang, L., et al. (2012). Plant growth-promoting nitrogen-fixing enterobacteria are in association with sugarcane plants growing in Guangxi. Microbes Environ. 27, 391–398. doi: 10.1264/jsme2.me11275
Lin, L., Wei, C., Chen, M., Wang, H., Li, Y., Li, Y., et al. (2015). Complete genome sequence of endophytic nitrogen-fixing Klebsiella variicola strain DX120E. Stand. Genomic Sci. 10:22.
Liu, W. Y., Chung, K. M. K., Wong, C. F., Jiang, J. W., Hui, R. K. H., and Leung, F. C. C. (2012). Complete genome sequence of the endophytic Enterobacter cloacae subsp. cloacae strain ENHKU01. J. Bacteriol. 194:5965. doi: 10.1128/jb.01394-12
Loiret, F. G., Ortega, E., Kleiner, D., Ortega-Rodés, P., Rodés, R., and Dong, Z. (2004). A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J. Appl. Microbiol. 97, 504–511. doi: 10.1111/j.1365-2672.2004.02329.x
Lorck, H. (1948). Production of hydrocyanic acid by bacteria. Physiol. Plant 1, 142–146. doi: 10.1111/j.1399-3054.1948.tb07118.x
Lu, G. D., Li, C. C., Pan, C. Z., and Zhang, X. B. (1997). Sugarcane diseases in China. Sugarcane 10, 9–12.
Luo, Y., Cheng, Y., Yi, J., Zhang, Z., Luo, Q., Zhang, D., et al. (2018). Complete genome sequence of industrial biocontrol strain Paenibacillus polymyxa HY96-2 and further analysis of its biocontrol mechanism. Front. Microbiol. 9:1520. doi: 10.3389/fmicb.2018.01520
Macedo-Raygoza, G. M., Valdez-Salas, B., Prado, F. M., Prieto, K. R., Yamaguchi, L. F., Kato, M. J., et al. (2019). Enterobacter cloacae, an endophyte that establishes a nutrient-transfer symbiosis with Banana plants and protects against the Black sigatoka pathogen. Front. Microbiol. 10:804. doi: 10.3389/fmicb.2019.00804
Magnani, G. S., Didonet, C. M., Cruz, L. M., Picheth, C. F., Pedrosa, F. O., and Souza, E. M. (2010). Diversity of endophytic bacteria in Brazilian sugarcane. Genet. Mol. Res. 9, 250–258. doi: 10.4238/vol9-1gmr703
Maksimov, I. V., Maksimova, T. I., Sarvarova, E. R., and Blagova, D. K. (2018). Endophytic bacteria as effective agents of new-generation biopesticides (review). Appl. Biochem. 54, 128–140. doi: 10.1134/S0003683818020072
Mansilla, M. C., and de Mendoza, D. (2000). The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 146, 815–821. doi: 10.1099/00221287-146-4-815
Mei, C., and Flinn, B. S. (2010). The use of beneficial microbial endophytes for plant biomass and stress tolerance improvement. Recent Pat. Biotechnol. 4, 81–95. doi: 10.2174/187220810790069523
Mima, T., Kohira, N., Li, Y., Sekiya, H., Ogawa, W., Kuroda, T., et al. (2009). Gene cloning and characteristics of the RND-type multidrug efflux pump Mux ABC-OpmB possessing two RND components in Pseudomonas aeruginosa. Microbiology 155, 3509–3517. doi: 10.1099/mic.0.031260-0
Mirza, M. S., Ahmad, W., Latif, F., Haurat, J., Bally, R., Normand, P., et al. (2001). Isolation, partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant Soil 237, 47–54.
Moftah, A. E. (2000). Physiological responses of lead-polluted tomato and eggplant to the antioxidant ethylendiure. Minufiya J. Agric. Res. 25, 933–955.
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Oh, M., Han, J. W., Lee, C., Choi, G. J., and Kim, H. (2018). Nematicidal and plant growth-promoting activity of Enterobacter asburiae HK169, genome analysis provides insight into its biological activities. J. Microbiol. Biotechnol. 28, 968–975. doi: 10.4014/jmb.1801.01021
Oliveira, A. L. M., Urquiaga, S., Döbereiner, J., and Baldani, J. I. (2002). The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 242, 205–215.
Peng, G., Zhang, W., Luo, H., Xie, H., Lai, W., and Tan, Z. (2009). Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia. Int. J. Syst. Evol. Microbiol. 59, 1650–1655. doi: 10.1099/ijs.0.005967-0
Penrose, D. M., and Glick, B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 118, 10–15. doi: 10.1034/j.1399-3054.2003.00086.x
Qin, S., Feng, W. W., Wang, T. T., Ding, P., Xing, K., and Jiang, J. H. (2017). Plant growth-promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant Soil 416, 117–132. doi: 10.1007/s11104-017-3192-2
Ren, Y., Zhou, Z., Guo, X., Li, Y., Feng, L., and Wang, L. (2010). Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J. Bacteriol. 192, 2463–2464. doi: 10.1128/jb.00067-10
Rosado, A. S., Duarte, G. F., Seldin, L., and Elsas, J. D. V. (1998). Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR amplified gene fragments. Appl. Environ. Microbiol. 64, 2770–2779.
Rosenblueth, M., and Martínez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/mpmi-19-0827
Rott, P., Bailey, R. A., Comstock, J. C., Croft, B. J., and Saumtally, A. S. (2000). A Guide to Sugarcane Diseases. Montpellier: CIRAD and ISSCT.
Ryan, R. P. R., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes, recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Saitou, N., and Nei, M. (1987). The neighbor-joining method, a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Saleem, A. R., Brunetti, C., Khalid, A., Della, R. G., Raio, A., Emiliani, G., et al. (2018). Drought response of Mucuna pruriens (L.) DC. Inoculated with ACC deaminase and IAA producing rhizobacteria. PLoS One 13:e0191218. doi: 10.1371/journal.pone.0191218
Santi, C., Bogusz, D., and Franche, C. (2013). Biological nitrogen fixation in non-legume plants. Ann. Bot. 111, 743–767. doi: 10.1093/aob/mct048
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda, M. C., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Sarkar, A., Ghosh, P. K., Pramanik, K., Mitra, S., Soren, T., Pandey, S., et al. (2018). A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res. Microbiol. 169, 20–32. doi: 10.1016/j.resmic.2017.08.005
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sessitsch, A., Kuffner, M., Kidd, P., Vangronsveld, J., Wenzel, W. W., Fallmann, K., et al. (2013). The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 60, 182–194. doi: 10.1016/j.soilbio.2013.01.012
Sevilla, M., Burris, R. H., Gunapala, N., and Kennedy, C. (2001). Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and Nif-mutant strains. Mol. Plant Microbe Interact. 14, 358–366. doi: 10.1094/mpmi.2001.14.3.358
Shah, S., Li, J., Moffatt, B. A., and Glick, B. R. (1998). Isolation and characterization of ACC deaminase genes from two different plant growth promoting rhizobacteria. Can. J. Microbiol. 44, 833–843. doi: 10.1139/w98-074
Shahid, M., Khalid, S., Abbas, G., Shahid, N., Nadeem, M., Sabir, M., et al. (2015). “Heavy metal stress and crop productivity,” in Crop Production and Global Environmental Issues, ed. K. Hakeem (Berlin: Springer), 1–25. doi: 10.1007/978-3-319-23162-4_1
Shariati, J. V., Malboobi, M. A., Tabrizi, Z., Tavakol, E., Owilia, P., and Safari, M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 7:15610.
Silver, S., Lee, B. T., Brown, O. N. L., and Cooksey, D. A. (1993). “Bacterial plasmid resistances to copper, cadmium and zinc,” in The Chemistry of Copper and Zinc Triads, eds A. J. Welch and S. U. Chapman (London: The Royal Society of Chemistry), 38–53.
Singh, M. K., Kushwaha, C., and Singh, R. K. (2009). Studies on endophytic colonization ability of two upland rice endophytes, Rhizobium sp. and Burkholderia sp., using green ?uorescent protein reporter. Curr. Microbiol. 59, 240–243. doi: 10.1007/s00284-009-9419-6
Singh, M. P., Singh, P., Li, H. B., Song, Q. Q., and Singh, R. K. (2019). “Microbial biofilms, development, structure, and their social assemblage for beneficial applications,” in New and Future Developments in Microbial Biotechnology and Bioengineering, eds M. K. Yadav and B. P. Singh (Amsterdam: Elsevier), 125–138. doi: 10.1016/b978-0-444-64279-0.00010-4
Singh, P., Song, Q. Q., Singh, R. K., Li, H. B., Solanki, M. K., Malviya, M. K., et al. (2019). Proteomic analysis of the resistance mechanisms in sugarcane during Sporisorium scitamineum infection. Int. J. Mol. Sci. 20:569. doi: 10.3390/ijms20030569
Singh, P., Song, Q. Q., Singh, R. K., Li, H. B., Solanki, M. K., Yang, L. T., et al. (2018). Physiological and molecular analysis of sugarcane (varieties-F134 and NCo310) during Sporisorium scitamineum interaction. Sugar Tech 21, 631–644. doi: 10.1007/s12355-018-0671-6
Singh, R. K., Kumar, D. P., Solanki, M. K., Singh, P., Srivastava, S., Srivastva, A. K., et al. (2013). Multifarious plant growth promoting characteristics of chickpea rhizosphere associated Bacilli help to suppress soil-borne pathogens. Plant Growth Regul. 73, 91–101. doi: 10.1007/s10725-013-9870-z
Singh, R. K., Singh, P., Li, H. B., Song, Q. Q., Guo, D. J., Solanki, M. K., et al. (2020). Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: a comprehensive study of plant-microbe interactions for growth enhancement in Saccharum spp. BMC Plant Biol. 20:220. doi: 10.1186/s12870-020-02400-9
Singh, R. P., Nalwaya, S., and Jha, P. N. (2017). The draft genome sequence of the plant growth promoting rhizospheric bacterium Enterobacter cloacae SBP-8. Genom. Data 12, 81–83. doi: 10.1016/j.gdata.2017.03.006
Slama, H. B., Triki, M. A., Bouket, A. C., Mefteh, F. B., Alenezi, F. N., Luptakova, L., et al. (2019). Screening of the high-rhizosphere competent Limoniastrum monopetalum culturable endophyte microbiota allows the recovery of multifaceted and versatile biocontrol agents. Microorganisms 7:249. doi: 10.3390/microorganisms7080249
Smith, A. D., Jameson, G. N. L., Santos, P. C. D., Agar, J. N., Naik, S., Krebs, C., et al. (2005). NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44, 12955–12969. doi: 10.1021/bi051257i
Tadra-Sfeir, M. Z., Souza, E. M., Faoro, H., Muller-Santos, M., Baura, V. A., Tuleski, T. R., et al. (2011). Naringenin regulates Expression of genes involved in cell wall synthesis in Herbaspirillum seropedicae. Appl. Environ. Microbiol. 77, 2180–2183. doi: 10.1128/aem.02071-10
Taghavi, S., Garafola, C., Monchy, S., Newman, L., Hoffman, A., Weyens, N., et al. (2009). Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75, 748–757. doi: 10.1128/aem.02239-08
Taghavi, S., Van der Lelie, D., Hoffman, D. A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Taulé, C., Mareque, C., Barlocco, C., Hackembruch, F., Reis, V. M., Sicardi, M., et al. (2012). The contribution of nitrogen fixation to sugarcane (Saccharum officinarum L.), and the identification and characterization of part of the associated diazotrophic bacterial community. Plant Soil 356, 35–49. doi: 10.1007/s11104-011-1023-4
Teschler, J. K., Zamorano-Sánchez, D., Utada, A. S., and Warner, C. J. A. (2015). Living in the matrix, assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268. doi: 10.1038/nrmicro3433
Timmusk, S., Grantcharova, N., Gerhart, E., and Wagner, H. (2005). Paenibacillus polymyxa invades plant roots and forms bio?lms. Appl. Environ. Microbiol. 71, 7292–7300.
Timmusk, S., Kim, S. B., Nevo, E., Abd El Daim, I., Ek, B., Bergquist, J., et al. (2015). Sfp-type PPTase inactivation promotes bacterial biofilm formation and ability to enhance wheat drought tolerance. Front. Microbiol. 6:387. doi: 10.3389/fmicb.2015.00387
Urquiaga, S., Xavier, R. P., de Morais, R. F., Batista, R. B., Schultz, N., Leite, J. M., et al. (2012). Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant Soil 356, 5–21. doi: 10.1007/s11104-011-1016-3
Vidyalakshmi, R., Paranthaman, R., and Bhakyaraj, R. (2009). Sulphur oxidizing bacteria and pulse nutrition, a review world. J. Agric. Sci. 5, 270–278.
Vinocur, B., and Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress, achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Viswanathan, R., and Rao, G. P. (2011). Disease scenario and management of major sugarcane diseases in India. Sugar Tech 13, 336–353. doi: 10.1007/s12355-011-0102-4
Vlamakis, H., Chai, Y., Beauregard, P., and Losick, R. (2013). Sticking together, building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Voloudakis, A. E., Bender, C. L., and Cooksey, D. A. (1993). Similarity between copper resistance genes from Xanthomonas campestris and Pseudomonas syringae. Appl. Environ. Microbiol. 59, 1627–1634. doi: 10.1128/aem.59.5.1627-1634.1993
Wang, D., Xu, A., Elmerich, C., and Ma, L. Z. (2017). Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions. ISME J. 11, 1602–1613. doi: 10.1038/ismej.2017.30
Weilharter, A., Mitter, B., Shin, M. V., Chain, P. S., Nowak, J., and Sessitsch, A. (2011). Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J. Bacteriol. 193, 3383–3384. doi: 10.1128/jb.05055-11
White, J. F., Kingsley, K. L., Verma, S. K., and Kowalski, K. P. (2018). Rhizophagy cycle: an oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms 6:95. doi: 10.3390/microorganisms6030095
Xu, Y. B., Chen, M., Zhang, Y., Wang, M., Wang, Y., Huang, Q. B., et al. (2014). The phosphotransferase system gene ptsI in the endophytic bacterium Bacillus cereus is required for biofilm formation, colonization, and biocontrol against wheat sharp eyespot. FEMS Microbiol. Lett. 354, 142–152. doi: 10.1111/1574-6968.12438
Yang, E., Sun, L., Ding, X., Sun, D., Liu, J., and Wang, W. (2019). Complete genome sequence of Caulobacter flavus RHGG3T, a type species of the genus Caulobacter with plant growth-promoting traits and heavy metal resistance. 3 Biotech 9:42.
Yong-Xiu, X., Chun-Yan, W., Yao, M., Yang, L.-T., Si-Liang, H., and Yang-Rui, L. (2015). Nitrogen fixing and plant growth-promoting ability of two endophytic bacterial strains isolated from sugarcane stalks. Sugar Tech 18, 373–379. doi: 10.1007/s12355-015-0397-7
Zehr, J. P., and Capone, D. G. (1996). Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb. Ecol. 32, 263–281.
Keywords: endophyte, E. roggenkampii, nitrogen fixation, PGPB, root colonization, stress, sugarcane, whole-genome sequencing
Citation: Guo D-J, Singh RK, Singh P, Li D-P, Sharma A, Xing Y-X, Song X-P, Yang L-T and Li Y-R (2020) Complete Genome Sequence of Enterobacter roggenkampii ED5, a Nitrogen Fixing Plant Growth Promoting Endophytic Bacterium With Biocontrol and Stress Tolerance Properties, Isolated From Sugarcane Root. Front. Microbiol. 11:580081. doi: 10.3389/fmicb.2020.580081
Received: 04 July 2020; Accepted: 25 August 2020;
Published: 22 September 2020.
Edited by:
Prem Lal Kashyap, Indian Institute of Wheat and Barley Research (ICAR), IndiaReviewed by:
Worarat Kruasuwan, National Center for Genetic Engineering and Biotechnology (BIOTEC), ThailandCopyright © 2020 Guo, Singh, Singh, Li, Sharma, Xing, Song, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Xiu Xing, ZG9jdW1lbnQxMjZAMTI2LmNvbQ==; Yang-Rui Li, bGl5ckBneGFhcy5uZXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.