
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 29 October 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.578425
This article is part of the Research Topic XXXIII SIMGBM Congress 2019 - Environmental And Industrial Microbiology View all 11 articles
In the industry of fermented food and beverages, yeast cultures are often selected and standardized in order to ensure a better control of fermentation and a more stable product over time. Several studies have shown that the organoleptic characteristics of fermented products reflect geographic variations of the microbial community composition. Despite investigations of the worldwide distribution and genetic diversity of Saccharomyces cerevisiae, it is still unclear how and to what extent human intervention has shaped the brewer’s yeast population structure. The genotypic and phenotypic characterization of environmental yeast populations and their potential application in the fermentative processes can significantly enrich the industrial fermentation products. Social insects have proven to be closely associated to the yeasts ecology. The relationships between yeasts and insects represent a fundamental aspect for understanding the ecological and evolutionary forces shaping their adaptation to different niches. Studies on phylogenetic relationships of S. cerevisiae populations showed genetic differences among strains isolated from gut and non-gut environments (i.e., natural sources and fermentation). Recent evidences showed that insect’s gut is a reservoir and an evolutionary niche for Saccharomyces, contributing to its survival and evolution, favoring its dispersion, mating and improving the inter-specific hybrids production during hibernation. Here, we discuss the potential use of social insects for production of a wide range of hybrid yeasts from environmental Saccharomyces isolates suitable for industrial and biotechnological applications.
Many species of Saccharomycetes and non-Saccharomycetes are essential components of human production of fermented food and beverages. Saccharomyces cerevisiae, known as the brewer’s or bakery’s yeast, has been widely used for its fermentative capacity for thousands of years (Legras et al., 2007) (Figure 1). The fermentation process has allowed preservation of perishable food and made bioavailable nutrients and microelements, improving the quality of foods (Hatoum et al., 2012). The oldest traces of yeast and fermented foods have been found in the tomb of U-j of King/Jiahu in Henan in China (7,000 B.C.) (McGovern et al., 2004), in Iran (6,000 B.C.) (Fatahi et al., 2003), in wine jars of tomb of Scorpion King in Egypt (3,150 B.C.) (Cavalieri et al., 2003), and in Mesopotamia (about 4,000 to 3,100 B.C.) (McGovern, 2003), demonstrating that fermentation of cereals, honey and fruit, has been carried out since the dawn of civilization (McGovern et al., 2004; Kupfer, 2013, 2015). Successively, viticulture spread in Asia Minor and northern Africa, and around 1,000 B.C. arrived in Mediterranean countries (Legras et al., 2007). Beer production is supposed to be almost as ancient as wine and came from the Middle East, subsequently acquired by Germanic and Celtic populations around 1st century A.C. (Legras et al., 2007).
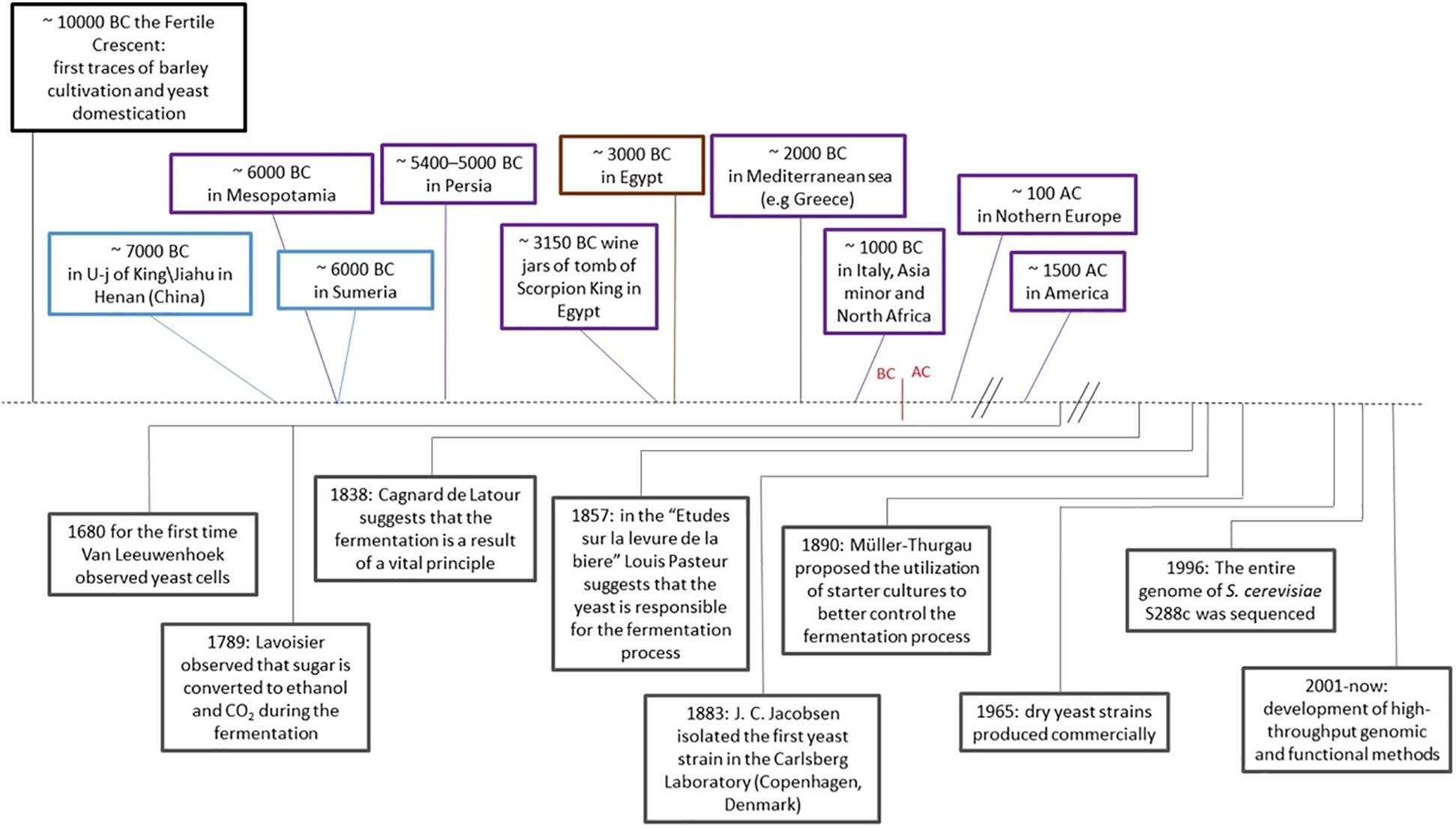
Figure 1. History of fermentation and yeast domestication. S. cerevisiae and human activities related to the production of fermented products. In the timeline: on the top of the dashed line, the oldest traces of S. cerevisiae across the human history and geography. Blue: fermented products; purple: wine; brown: bakery products. Below the dashed line, events related to S. cerevisiae in the history of fermentation and scientific advancements.
The long lasting association of S. cerevisiae with fermentative processes has led to propose the idea that its wide use caused its domestication as adaptation to different fermented products (Legras et al., 2007; Sicard and Legras, 2011). Yet the effect of domestication on trait selection has been proven only in the yeast strains used for the beer production (Piskur et al., 2006; Berlowska et al., 2015; Gallone et al., 2016, 2018).
The art of fermentation had been developed empirically from generation to generation. Scientific awareness related to biochemical transformations during fermentation started around the end of the 18th and in 19th century (Gay-Lussac, 1815; Kützing, 1837; Cagniard-Latour, 1838). In 1883, the first pure yeast culture was created by Emil Christian Hansen for beer production (Figure 1). Hansen had been brought to work from Copenhagen University to the Carlsberg’s Laboratories by Jacobsen, the founder of the Carlsberg Empire, to standardize the quality of beer by isolating and stabilizing the microorganism used in the brewing process. Subsequently in 1890, Hermann Mueller-Thurgau planned the process for a better control and repeatability of wine fermentations with starter cultures (Marsit and Dequin, 2015) (Figure 1). The rise of the industry of fermented products and the application of innovative biotechnological methods for selection of enhanced yeast strains or production of hybrids suitable for different types of fermentable substrates enables a much more defined control of the fermentation process than the generally used strains.
The ability of yeasts to metabolize sugars by producing ethanol through anaerobic fermentation even when oxygen is available (the Crabtree effect) (Chambers and Pretorius, 2010) and in presence of high glucose concentrations (Otterstedt et al., 2004) has enabled S. cerevisiae to gain an evolutionary advantage over other microorganisms (Albergaria and Arneborg, 2016). Ethanol production and acidification of the growth medium have always kept bacteria and other aerobic molds under control. The microbial communities, including yeasts, can play a key role in triggering fermentation processes conferring a typical bouquet, thanks to volatile compounds (El-Sayed et al., 2005; Christiaens et al., 2014). S. cerevisiae produces several aromatic esters: ethyl acetate (varnish, nail polish, and fruity aroma), isoamyl acetate (banana and pear), isobutyl acetate (banana), phenylethyl acetate (fruity and flowery), ethyl hexanoate (apple, banana, and violets), ethyl octanoate (pineapple and pear), and ethyl decanoate (floral) (Ruiz et al., 2019).
The question of the natural environmental niche for S. cerevisiae was debated for a long time (Goddard and Greig, 2015). Environmental S. cerevisiae strains are subjected to harsh conditions and they developed survival strategies, which are not retained when laboratory or industrial strains are cultured under most favorable conditions. S. cerevisiae has been isolated from different natural environments, such as oak trees (Sampaio and Goncalves, 2008; Zhang et al., 2010; Hyma and Fay, 2013), maize (Hayford and Jespersen, 1999; Halm et al., 2004), various fermentations and other substrates, including soil (van der Aa Kuhle et al., 2001; Oba et al., 2011; Dunn et al., 2012; Kubo et al., 2014).
Saccharomyces cerevisiae has adapted itself and evolved to different environmental niches (Goddard and Greig, 2015; Peter et al., 2018). Evidences indicated that, although S. cerevisiae is found in abundance in environments, such as wineries, it does not originate from grapevines or grape berries. Mortimer and Polsinelli (Mortimer and Polsinelli, 1999) demonstrated that S. cerevisiae was found at very low frequency on unripe and intact grape berries (0.05% on average), while during grape maturation it was present on average with a frequency of 25% on broken berries (Mortimer et al., 1994; Polsinelli et al., 1996; Mortimer and Polsinelli, 1999). Thus, ripe and crushed grape berries (with high concentration of fermentable sugars), represent a suitable environment for S. cerevisiae. In a recent study (Taylor et al., 2014) the rare presence of S. cerevisiae in intact grapes was confirmed: one S. cerevisiae cell was found among 20,000 cells of other fungi by metagenomic approach.
Researchers wondered how S. cerevisiae could be carried on the grapes. The agents proposed to play a role in spreading of microorganisms in the environment are animal vectors (e.g., insects and birds) (Mortimer and Polsinelli, 1999; Goddard et al., 2010; Stefanini et al., 2012). Unlike bacteria and fungal spores that can easily be dispersed by other means (e.g., air/wind) (Madden et al., 2018), yeast spores are not adapted for wind-borne transmission. Many decades ago (Grace and Collins, 1976), testing the dispersion rate of bacteria and yeasts spores using a wind tunnel, demonstrated that the wind is unable to disperse S. cerevisiae cells when these are adhered to the leaf surface.
Evidences showed that insects have a mutualistic relationship with yeasts (Belisle et al., 2012; Stefanini et al., 2012, 2016; Madden et al., 2018) and play a key role for yeast dispersion in natural environments. Saccharomyces spp., in particular S. cerevisiae, were detected in different insects worldwide (Stefanini et al., 2012; Buser et al., 2014; Jimenez et al., 2017; Meriggi et al., 2020; YeastFinder1; Table 1).
Social insects represent a fulcrum in the yeast ecology and evolution. At the same time, volatile compounds produced by yeasts attract insects that preferentially foraged nectar sources, and influence their behavior and physiology (Becher et al., 2012; Babcock et al., 2017; Stefanini, 2018).
In our previous studies (Stefanini et al., 2012, 2016), we demonstrated not only that wasps contribute to dispersion of yeast strains into the environment, but also that they can host yeasts in their gut, contributing to their survival and biodiversity. Polistes dominula allows the transmission of yeasts to the progeny (Stefanini et al., 2012), ensuring the presence of yeast in the colony and in the foraging area. This make a flow of yeast cells in the environments (in the vineyard, on grapes) from autumn to spring. These observations allow to explain where yeasts can reside during the winter and how reappear during the spring and summer, an unsolved question up until few years ago.
In spite these evidences, specific factors that select natural yeasts in the insect gut have not yet been found. Our recent findings showed that yeast has evolved strategies to adapt to the gastrointestinal tract of insects (Stefanini et al., 2012, 2016; Ramazzotti et al., 2018). The wall of the ascospore is able to resist to the insects gastric digestion, allowing survival within the host gut (Coluccio et al., 2008; Stefanini et al., 2012, 2016). More recently, we demonstrated that host’s gut, not only of insect but also human, is a potential reservoir for yeasts (Ramazzotti et al., 2018). Genetic and phenotypic differences, including peculiar cell wall composition, different ability to sporulation and to induce host immune response were discovered between strains isolated from human and insect’s gut and non-gut environment, suggesting the existence of gut-specific features that could represent a selective advantage for survival and expansion in the gut environment (Ramazzotti et al., 2018).
Based on our previous studies (Stefanini et al., 2012, 2016), the insect’s gut is an advantageous ecological niche for S. cerevisiae, favoring the intra− and inter−species mating of yeast cells, and allowing increased fitness of hybrids, thus representing an environment favoring the generation of yeast genetic biodiversity.
The current used industrial strains represent a small fraction of the natural biodiversity (Liti et al., 2009). The nature could provide unknown strains with relevant characteristics that may enhance the industrial fermentations. Specific strategies could help to transfer these properties to industrial strains or create novel strains with best performance for fermentation processes. Here, we propose and discuss the use of wasp’s gut for accelerated selection and production of hybrids suitable for different types of fermentable substrates.
Recent genomic studies provided a comprehensive overview of the biodiversity of wild and industrial Saccharomyces strains (Liti et al., 2009; Schacherer et al., 2009). Diversity within S. cerevisiae population structure was at least in part associated to its industrial application. According to the domestication hypothesis, this long-term process has resulted in different strains with specific characteristics suited for industrial fermentation, clustering differently from wild populations (Fay and Benavides, 2005; Liti et al., 2009; Schacherer et al., 2009; Sicard and Legras, 2011). Several studies reported genome-wide signatures of clonal expansion of yeast strains, as well as convergent evolution of industrially relevant traits in separate lineages (Liti et al., 2009; Gallone et al., 2016; Parapouli et al., 2020). Yet the only bona fide evidence for selection on genomic regions associated to domestication was reported by Gallone et al. (2019) that studied the whole-genome of more than 200 industrial yeasts showing that about 25% consisted of interspecific hybrids derived from S. cerevisiae, S. kudriavzevii, S. eubayanus, and S. uvarum. Langdon et al. (2019) analyzed the genomes of 122 interspecies hybrids and introgressed strains in Saccharomyces genus revealing three domesticated lineages, including wild lineages from Europe and Northern continents of the world. These evidences show that industrial yeasts are the result of selection following clonal expansion and adaptation of specific strains, shaped by genetic drift caused by bottlenecking.
Beer’s yeasts present the strongest and maybe only genetic and phenotypic signatures of domestication (Gallone et al., 2019; Langdon et al., 2019). The strong selective pressure imposed over many generations allowed to obtain desirable phenotypes, but has also dramatically affected the genomic structure and stability of domesticated yeasts (Legras et al., 2007; Gallone et al., 2016, 2018). Hybrid strains are preferred in the industrial fermentation because they show phenomena, such as “vigor of the hybrid” that confer better fermentation capacity in terms of speed, use of alternative sugar source and an enriched pattern of aromatic compounds (Bellon et al., 2011; Piotrowski et al., 2012; Bellon et al., 2013; Gamero et al., 2013; Snoek et al., 2015).
An example of yeast’s domestication trait is the ability to ferment maltotriose. This trait evolved independently and through different genetic pathways in the two main beer lineages, such as ale (by the top-fermenting S. cerevisiae) and lager (by the bottom – fermenter S. pastorianus, a interspecific hybrid S. cerevisiae × S. eubayanus), suggesting strong selection pressure (Gallone et al., 2016, 2018). S. pastorianus can ferment at lower temperatures than S. cerevisiae (Dunn and Sherlock, 2008; Libkind et al., 2011). This hybridization process has combined the efficiency of S. cerevisiae in the sugar metabolism and the cryoprotective capacities of S. eubayanus (Hebly et al., 2015; Krogerus et al., 2015). The cryoprotective capacity was demonstrated to be related with the mitochondrial genome inheritance. S. eubayanus (Baker et al., 2019) and S. uvarum (Li et al., 2019) mitochondrial genome provided with low-temperature tolerance to the interspecies hybrids compared when the same hybrids inherited the mitochondrial genome of S. cerevisiae. In addition, hybrids between natural strains of S. kudriavzevii × S. cerevisiae, selected to confer diverse flavors, are used for the production of Trappist beers (a subgroup of ale) (Gonzalez et al., 2008; Peris et al., 2018; Gallone et al., 2019; Langdon et al., 2019).
On the other hand, several authors suggested that during centuries of wine production, S. cerevisiae acquired remarkable resistance/tolerance to high sugar concentrations and nitrogen metabolic activity, through adaptive horizontal gene transfer and copy number variations (Fay and Benavides, 2005; Aa et al., 2006; Ezov et al., 2006; Ruderfer et al., 2006; Legras et al., 2007; Stefanini and Cavalieri, 2018). These events potentially conferred competitive advantages during must fermentation (Almeida et al., 2017), and production of a wide spectrum of aromatic profiles (González et al., 2007). Triple hybrid S. cerevisiae × S. kudriavzevii × S. uvarum has shown to be able to use fructose more efficiently than S. uvarum strain and to restart fermentation (Christ et al., 2015), avoiding the upper hand of bacteria and the consequent spoilage of the must. Inactive or stuck fermentations are detrimental to wine/beer production. These events have been reduced by the commercial availability of selected yeast strains used as starter. However, this practice limits the developing of wild yeasts during fermentation (Parapouli et al., 2020). Camarasa et al. (2011) investigated the fermentative efficiency of a set of S. cerevisiae strains, observing that strains isolated from sugar-rich environments were able to complete the fermentation process, while the laboratory or environmental strains were unable. The extensive genetic diversity of environmental S. cerevisiae isolates, in particular of interspecific hybrids, could be an extremely significant source of innovation for biotechnological applications (Parapouli et al., 2020).
The quest for increased and improved productivity and adaptability to changing consumer preferences lead to the study and development of industrial strains with novel and desired properties.
Non-genetically modified organisms (non-GMO) and GMO techniques can be used for selection of yeast strains with suitable traits and industrially relevant phenotypes. Jan Steensels et al. (2014) extensively described these approaches, indicating their advantages and limitations. Non-GMO techniques have been developed to create performant yeast variants, that can be freely used in industrial fermentations, without encounter any problems with legislation and/or consumer acceptance.
Currently there are four main approaches for generate artificial diversity in yeast strain using sexual hybridization: (i) direct mating – crossing of two haploid cells or spores of opposite mating types; (ii) rare mating – crossing of strains without sporulation by occasional and rare homothallic mating-type switch; (iii) mass mating – crossing of multiple parental strains or a heterogeneous population of the same parental strain; and (iv) genome shuffling of multiple strains. In addition, asexual hybridization includes cytoduction (a method that transfer cytoplasmically inherited traits) and protoplast fusion (asexually merging of cells after cell wall distruption in osmotic medium). Overall, the above mentioned approaches could have important impact in the optimization of industrial processes, at technical and economic level. Our previous studies demonstrated that the insects’ gut represents the environment where S. cerevisiae mates and interspecific hybrids arise spontaneously. After 2 and 4 months of hibernation in the wasp’s gut, S. paradoxus can survive only in hybrid shape with S. cerevisiae. The rate of inbreeding in S. cerevisiae spores increases up to ten times when inoculated in the insect gut. The high frequency of outbreeding coincides with the rates of mosaicism and genetic diversity in yeast strains (Reuter et al., 2007).
In last years, the interest for insects for food and applicative purposes is growing. The insects breeding is a sustainable method with low energy impact and represents a great opportunity for large-scale industrial applications. Social insects could play a role in the evolution and genetic recombination of yeasts. They seems to be a perfect niche in which the formation of hybrids occurs naturally and much more efficiently than in any other wild place, without employing artificial strategies of genetic manipulation, as recently showed by Peris et al. (2020) making synthetic hybrids by six yeast species. Wasps could provide yeast communities with the level of genetic variation required in time to adapt to a changing environment. Given the fast pace at which climate change is affecting the man made, “ersatz” environment where fermented products are produced, maintaining the genetic variation force driven by the presence of insects could be very important to support the adaptability of yeast to these changes. Considering that many yeasts used in the fermentation industry are interspecific hybrid strains, we could speculate that insects have been the breeders of the past, having likely provided a great deal to the evolution of brewing and wine making. The need of product diversification present in the fermented beverages field could look with great interest at the potential of using wasps as breeding places to produce yeasts of biotechnological interest, including yeasts producing beverages with a reduced ethanol content.
Optimization of current strategies and novel technologies such as next-generation sequencing, together with a better understanding of complex phenotypes allows to create yeasts with more variants and better adapted to the industrial goals. In the near future, the fermented beverages industry could benefit significantly from the possibility to breed insects massively and use them as a forge for accelerated selection and production of hybrids suitable for different types of fermentable substrates, further modeling these according to biotechnological requirements.
We can conclude that the yeasts–insect association certainly goes beyond the simple link between vectors and transported. In a future perspective, a better understanding of the ecology and relationships between insects and yeasts can play a key role in producing fermented beverages meeting the needs of tomorrow’s consumers.
MDP, DC, and NM ideated the review. MDP and NM gathered the data. MDP, NM, and DC wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the University of Florence and Regione Toscana POR FSE 2014–2020, VESPATER project, and the H2020-EU project 863059 – 2019-2024 Food and Nutrition Security Cloud FNS-Cloud-https://cordis.europa.eu/project/id/863059/it.
Aa, E., Townsend, J. P., Adams, R. I., Nielsen, K. M., and Taylor, J. W. (2006). Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 702–715. doi: 10.1111/j.1567-1364.2006.00059.x
Albergaria, H., and Arneborg, N. (2016). Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100, 2035–2046. doi: 10.1007/s00253-015-7255-0
Almeida, P., Barbosa, R., Bensasson, D., Goncalves, P., and Sampaio, J. P. (2017). Adaptive divergence in wine yeasts and their wild relatives suggests a prominent role for introgressions and rapid evolution at noncoding sites. Mol. Ecol. 26, 2167–2182. doi: 10.1111/mec.14071
Babcock, T., Gries, R., Borden, J., Palmero, L., Mattiacci, A., Masciocchi, M., et al. (2017). Brewer’s yeast, Saccharomyces cerevisiae, enhances attraction of two invasive yellowjackets (hymenoptera: vespidae) to dried fruit and fruit powder. J. Insect. Sci. 17:91. doi: 10.1093/jisesa/iex065
Baker, E. P., Peris, D., Moriarty, R. V., Li, X. C., Fay, J. C., and Hittinger, C. T. (2019). Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5:eaav1869. doi: 10.1126/sciadv.aav1869
Batra, L. R., Batra, S. W. T., and Bohart, G. E. (1973). The mycoflora of domesticated and wild bees (Apoidea). Mycopathol. Mycol. Appl. 49, 13–44. doi: 10.1007/BF02057445
Becher, P. G., Flick, G., Rozpedowska, E., Schmidt, A., Hagman, A., Lebreton, S., et al. (2012). Yeast, not fruit volatiles mediate Drosophila melanogaster attraction, oviposition and development. Funct. Ecol. 26, 822–828.
Belisle, M., Peay, K. G., and Fukami, T. (2012). Flowers as islands: spatial distribution of nectar-inhabiting microfungi among plants of Mimulus aurantiacus, a hummingbird-pollinated shrub. Microb. Ecol. 63, 711–718. doi: 10.1007/s00248-011-9975-8
Bellon, J. R., Eglinton, J. M., Siebert, T. E., Pollnitz, A. P., Rose, L., de Barros Lopes, M., et al. (2011). Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl. Microbiol. Biotechnol. 91, 603–612. doi: 10.1007/s00253-011-3294-3
Bellon, J. R., Schmid, F., Capone, D. L., Dunn, B. L., and Chambers, P. J. (2013). Introducing a new breed of wine yeast: interspecific hybridisation between a commercial Saccharomyces cerevisiae wine yeast and Saccharomyces mikatae. PLoS One 8:e62053. doi: 10.1371/journal.pone.0062053
Berlowska, J., Kregiel, D., and Rajkowska, K. (2015). Biodiversity of brewery yeast strains and their fermentative activities. Yeast 32, 289–300. doi: 10.1002/yea.3041
Broderick, N. A., and Lemaitre, B. (2012). Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307–321. doi: 10.4161/gmic.19896
Buser, C. C., Newcomb, R. D., Gaskett, A. C., and Goddard, M. R. (2014). Niche construction initiates the evolution of mutualistic interactions. Ecol. Lett. 17, 1257–1264. doi: 10.1111/ele.12331
Camarasa, C., Sanchez, I., Brial, P., Bigey, F., and Dequin, S. (2011). Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147. doi: 10.1371/journal.pone.0025147
Cavalieri, D., McGovern, P. E., Hartl, D. L., Mortimer, R., and Polsinelli, M. (2003). Evidence for s. cerevisiae fermentation in ancient wine. J. Mol. Evol. 57, (Suppl. 1), S226–S232. doi: 10.1007/s00239-003-0031-2
Chambers, P. J., and Pretorius, I. S. (2010). Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep. 11, 914–920. doi: 10.1038/embor.2010.179
Christ, E., Kowalczyk, M., Zuchowska, M., Claus, H., Löwenstein, R., Szopinska-Morawska, A., et al. (2015). An exemplary model study for overcoming stuck fermentation during spontaneous fermentation with the aid of a saccharomyces triple hybrid. J. Agric. Sci. 7, 18–34. doi: 10.5539/jas.v7n8p18
Christiaens, J. F., Franco, L. M., Cools, T. L., De Meester, L., Michiels, J., Wenseleers, T., et al. (2014). The fungal aroma gene ATF1 promotes dispersal of yeast cells through insect vectors. Cell Rep. 9, 425–432. doi: 10.1016/j.celrep.2014.09.009
Coluccio, A. E., Rodriguez, R. K., Kernan, M. J., and Neiman, A. M. (2008). The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One 3:e2873. doi: 10.1371/journal.pone.0002873
Dunn, B., Richter, C., Kvitek, D. J., Pugh, T., and Sherlock, G. (2012). Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22, 908–924. doi: 10.1101/gr.130310.111
Dunn, B., and Sherlock, G. (2008). Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 18, 1610–1623. doi: 10.1101/gr.076075.108
El-Sayed, A. M., Heppelthwaite, V. J., Manning, L. M., Gibb, A. R., and Suckling, D. M. (2005). Volatile constituents of fermented sugar baits and their attraction to lepidopteran species. J. Agric. Food Chem. 53, 953–958. doi: 10.1021/jf048521j
Ezov, T. K., Boger-Nadjar, E., Frenkel, Z., Katsperovski, I., Kemeny, S., Nevo, E., et al. (2006). Molecular-genetic biodiversity in a natural population of the yeast Saccharomyces cerevisiae from “Evolution Canyon”: microsatellite polymorphism, ploidy and controversial sexual status. Genetics 174, 1455–1468. doi: 10.1534/genetics.106.062745
Fatahi, R., Badi, A. E., Assil, N. B., Ehlenbacher, S. A. M., and Amani, Z. Z. (2003). Characterization of Iranian grapevine cultivars using microsatellite markers. Vitis 42, 185–192.
Fay, J. C., and Benavides, J. A. (2005). Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1:e5. doi: 10.1371/journal.pgen.0010005
Gallone, B., Mertens, S., Gordon, J. L., Maere, S., Verstrepen, K. J., and Steensels, J. (2018). Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr. Opin. Biotechnol. 49, 148–155. doi: 10.1016/j.copbio.2017.08.005
Gallone, B., Steensels, J., Mertens, S., Dzialo, M. C., Gordon, J. L., Wauters, R., et al. (2019). Interspecific hybridization facilitates niche adaptation in beer yeast. Nat. Ecol. Evol. 3, 1562–1575. doi: 10.1038/s41559-019-0997-9
Gallone, B., Steensels, J., Prahl, T., Soriaga, L., Saels, V., Herrera-Malaver, B., et al. (2016). Domestication and Divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397–1410. doi: 10.1016/j.cell.2016.08.020
Gamero, A., Tronchoni, J., Querol, A., and Belloch, C. (2013). Production of aroma compounds by cryotolerant Saccharomyces species and hybrids at low and moderate fermentation temperatures. J. Appl. Microbiol. 114, 1405–1414. doi: 10.1111/jam.12126
Gay-Lussac, J. L. (1815). Sur l’analyse de l’alcohol et de l’ether sulfurique et sur les produits de la fermentation. Ann. Chim. Phys. 95, 311.
Goddard, M. R., Anfang, N., Tang, R., Gardner, R. C., and Jun, C. (2010). A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ. Microbiol. 12, 63–73. doi: 10.1111/j.1462-2920.2009.02035.x
Goddard, M. R., and Greig, D. (2015). Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 15:fov009. doi: 10.1093/femsyr/fov009
Gonzalez, S. S., Barrio, E., and Querol, A. (2008). Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 74, 2314–2320. doi: 10.1128/AEM.01867-07
González, S. S., Gallo, L., Climent, M. D., Barrio, E., and Querol, A. (2007). Caratterizzazione enologica di ibridi naturali da S. cerevisiae e S. Kudriavzevii. Int. J. Food Microbiol. 116, 11–1.
Grace, J., and Collins, M. A. (1976). “Spore liberation from leaves by wind,” in Microbiology of Aerial Plant Surfaces, eds C. H. Dickinson and T. F. Preece (New York, NY, London: Academic Press), 185–198.
Halm, M., Hornbaek, T., Arneborg, N., Sefa-Dedeh, S., and Jespersen, L. (2004). Lactic acid tolerance determined by measurement of intracellular pH of single cells of Candida krusei and Saccharomyces cerevisiae isolated from fermented maize dough. Int. J. Food Microbiol. 94, 97–103. doi: 10.1016/j.ijfoodmicro.2003.12.019
Hatoum, R., Labrie, S., and Fliss, I. (2012). Antimicrobial and probiotic properties of yeasts: from fundamental to novel applications. Front. Microbiol. 3:421. doi: 10.3389/fmicb.2012.00421
Hayford, A. E., and Jespersen, L. (1999). Characterization of Saccharomyces cerevisiae strains from spontaneously fermented maize dough by profiles of assimilation, chromosome polymorphism. PCR and MAL genotyping. J. Appl. Microbiol. 86, 284–294. doi: 10.1046/j.1365-2672.1999.00653.x
Hebly, M., Brickwedde, A., Bolat, I., Driessen, M. R., de Hulster, E. A., van den Broek, M., et al. (2015). S. cerevisiae x S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 15:fov005. doi: 10.1093/femsyr/fov005
Hyma, K. E., and Fay, J. C. (2013). Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in north American vineyards. Mol. Ecol. 22, 2917–2930. doi: 10.1111/mec.12155
Jimenez, S. I., Carroll, C., Babcock, T., Derstine, N., Hadwin, A., Moore, M., et al. (2017). Yeasts harbored by vespine wasps in the Pacific Northwest. Environ. Entomol. 46, 217–225. doi: 10.1093/ee/nvw173
Kircher, H. W., Phariss, R. L., Rosenstein, F. U., Baldwin, D., and Fogleman, J. C. (1982). Response of Drosophila to cis- and trans-22-dehydrocholesterol: I. A survey. Lipids 17, 209–214. doi: 10.1007/BF02535105
Krogerus, K., Magalhaes, F., Vidgren, V., and Gibson, B. (2015). New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 42, 769–778. doi: 10.1007/s10295-015-1597-6
Kubo, R., Ohta, K., Funakawa, S., Kitabatake, N., Araki, S., and Izawa, S. (2014). Isolation of lactic acid-tolerant Saccharomyces cerevisiae from Cameroonian alcoholic beverage. J. Biosci. Bioeng. 118, 657–660. doi: 10.1016/j.jbiosc.2014.05.007
Kupfer, P. (2013). “Weinstraße vor der Seidenstraße—Weinkulturen zwischen Georgien und China,” in Kulturgut Rebe und Wein, eds H. König and H. Decker (Germany: Springer Spektrum), 3–17.
Kupfer, P. (2015). “Der älteste Wein der Menschheit in China: Jiahu und die Suche nach den Ursprüngen der eurasischenWeinkultur,” in Wo aber der Wein fehlt, stirbt der Reiz des Lebens eds H. Decker, H. König, and W. Zwickel (Germany: Nünnerich-Asmus Verlag), 12–25.
Kützing, F. T. (1837). Mikroskopische Untersuchungen über die Hefe und Esssigmutter, nebst mehreren anderen dazu gehörigen vegetabilischen Gebilden. J. Prakt. Chem. 11, 385–409.
Langdon, Q. K., Peris, D., Baker, E. P., Opulente, D. A., Nguyen, H. V., Bond, U., et al. (2019). Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 3, 1576–1586. doi: 10.1038/s41559-019-0998-8
Legras, J. L., Merdinoglu, D., Cornuet, J. M., and Karst, F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16, 2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x
Li, X. C., Peris, D., Hittinger, C. T., Sia, E. A., and Fay, J. C. (2019). Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci. Adv. 5:eaav1848. doi: 10.1126/sciadv.aav1848
Libkind, D., Hittinger, C. T., Valerio, E., Goncalves, C., Dover, J., Johnston, M., et al. (2011). Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 14539–14544. doi: 10.1073/pnas.1105430108
Liti, G., Carter, D. M., Moses, A. M., Warringer, J., Parts, L., James, S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. doi: 10.1038/nature07743
Madden, A. A., Epps, M. J., Fukami, T., Irwin, R. E., Sheppard, J., Sorger, D. M., et al. (2018). The ecology of insect-yeast relationships and its relevance to human industry. Proc. Biol. Sci. 285:20172733. doi: 10.1098/rspb.2017.2733
Marsit, S., and Dequin, S. (2015). Diversity and adaptive evolution of Saccharomyces wine yeast: a review. FEMS Yeast Res. 15:fov067. doi: 10.1093/femsyr/fov067
McGovern, P. E. (2003). “Ancient wine,” in The Search for the Origins of Viniculture (Princeton, NJ, USA: Princeton University Press), 464.
McGovern, P. E., Zhang, J., Tang, J., Zhang, Z., Hall, G. R., Moreau, R. A., et al. (2004). Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. U.S.A. 101, 17593–17598. doi: 10.1073/pnas.0407921102
Meriggi, N., Di Paola, M., Vitali, F., Rivero, D., Cappa, F., Turillazzi, F., et al. (2019). Saccharomyces cerevisiae induces immune enhancing and shapes gut microbiota in social wasps. Front. Microbiol. 10:2320. doi: 10.3389/fmicb.2019.02320
Meriggi, N., Di Paola, M., Cavalieri, D., and Stefanini, I. (2020). Saccharomyces cerevisiae– insects association: impacts, biogeography, and extent. Front. Microbiol. 11:1629.
Mortimer, R., and Polsinelli, M. (1999). On the origins of wine yeast. Res. Microbiol. 150, 199–204. doi: 10.1016/S0923-2508(99)80036-9
Mortimer, R. K., Romano, P., Suzzi, G., and Polsinelli, M. (1994). Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10, 1543–1552. doi: 10.1002/yea.320101203
Oba, T., Suenaga, H., Nakayama, S., Mitsuiki, S., Kitagaki, H., Tashiro, K., et al. (2011). Properties of a high malic acid-producing strains of Saccharomyces cerevisiae isolated from sake mash. Biosci. Biotechnol. Biochem. 75, 2025–2029. doi: 10.1271/bbb.110262
Otterstedt, K., Larsson, C., Bill, R. M., Stahlberg, A., Boles, E., Hohmann, S., et al. (2004). Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 5, 532–537. doi: 10.1038/sj.embor.7400132
Parapouli, M., Vasileiadis, A., Afendra, A. S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Peris, D., Alexander, W. G., Fisher, K. J., Moriarty, R. V., Basuino, M. G., Ubbelohde, E. J., et al. (2020). Synthetic hybrids of six yeast species. Nat. Commun. 11:2085. doi: 10.1038/s41467-020-15559-4
Peris, D., Perez-Torrado, R., Hittinger, C. T., Barrio, E., and Querol, A. (2018). On the origins and industrial applications of Saccharomyces cerevisiae x Saccharomyces kudriavzevii hybrids. Yeast 35, 51–69. doi: 10.1002/yea.3283
Peter, J., De Chiara, M., Friedrich, A., Yue, J. X., Pflieger, D., Bergstrom, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Phaff, H. J., and Knapp, E. P. (1956). The taxonomy of yeasts found in exudates of certain trees and other natural breeding sites of some species of Drosophila. Antonie Van Leeuwenhoek 22, 117–130. doi: 10.1007/BF02538319
Piotrowski, J. S., Nagarajan, S., Kroll, E., Stanbery, A., Chiotti, K. E., Kruckeberg, A. L., et al. (2012). Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve. BMC Evol. Biol. 12:46. doi: 10.1186/1471-2148-12-46
Piskur, J., Rozpedowska, E., Polakova, S., Merico, A., and Compagno, C. (2006). How did Saccharomyces evolve to become a good brewer? Trends Genet. 22, 183–186. doi: 10.1016/j.tig.2006.02.002
Polsinelli, M., Romano, P., Suzzi, G., and Mortimer, R. (1996). Multiple strains of Saccharomyces cerevisiae on a single grape vine. Lett. Appl. Microbiol. 23, 110–114. doi: 10.1111/j.1472-765X.1996.tb00043.x
Ramazzotti, M., Stefanini, I., Di Paola, M., De Filippo, C., Rizzetto, L., Berna, L., et al. (2018). Population genomics reveals evolution and variation of Saccharomyces cerevisiae in the human and insects gut. Environ. Microbiol. 21, 50–71. doi: 10.1111/1462-2920.14422
Reuter, M., Bell, G., and Greig, D. (2007). Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 17, R81–R83. doi: 10.1016/j.cub.2006.11.059
Ruderfer, D. M., Pratt, S. C., Seidel, H. S., and Kruglyak, L. (2006). Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38, 1077–1081. doi: 10.1038/ng1859
Ruiz, J., Kiene, F., Belda, I., Fracassetti, D., Marquina, D., Navascues, E., et al. (2019). Effects on varietal aromas during wine making: a review of the impact of varietal aromas on the flavor of wine. Appl. Microbiol. Biotechnol. 103, 7425–7450. doi: 10.1007/s00253-019-10008-9
Sampaio, J. P., and Goncalves, P. (2008). Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 74, 2144–2152. doi: 10.1128/AEM.02396-07
Sandhu, D. K., and Waraich, M. K. (1985). Yeasts associated with pollinating bees and flower nectar. Microb. Ecol. 11, 51–58. doi: 10.1007/BF02015108
Schacherer, J., Shapiro, J. A., Ruderfer, D. M., and Kruglyak, L. (2009). Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458, 342–345. doi: 10.1038/nature07670
Sicard, D., and Legras, J. L. (2011). Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 334, 229–236. doi: 10.1016/j.crvi.2010.12.016
Snoek, T., Picca Nicolino, M., Van den Bremt, S., Mertens, S., Saels, V., Verplaetse, A., et al. (2015). Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol. Biofuels 8:32. doi: 10.1186/s13068-015-0216-0
Steensels, J., Snoek, T., Meersman, E., Picca Nicolino, M., Voordeckers, K., and Verstrepen, K. J. (2014). Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995. doi: 10.1111/1574-6976.12073
Stefanini, I. (2018). Yeast-insect associations: it takes guts. Yeast 35, 315–330. doi: 10.1002/yea.3309
Stefanini, I., and Cavalieri, D. (2018). Metagenomic approaches to investigate the contribution of the vineyard environment to the quality of wine fermentation: potentials and difficulties. Front. Microbiol. 9:991. doi: 10.3389/fmicb.2018.00991
Stefanini, I., Dapporto, L., Berna, L., Polsinelli, M., Turillazzi, S., and Cavalieri, D. (2016). Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. U.S.A. 113, 2247–2251. doi: 10.1073/pnas.1516453113
Stefanini, I., Dapporto, L., Legras, J. L., Calabretta, A., Di Paola, M., De Filippo, C., et al. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 13398–13403. doi: 10.1073/pnas.1208362109
Suh, S. O., McHugh, J. V., Pollock, D. D., and Blackwell, M. (2005). The beetle gut: a hyperdiverse source of novel yeasts. Mycol. Res. 109(Pt 3), 261–265. doi: 10.1017/S0953756205002388
Taylor, M. W., Tsai, P., Anfang, N., Ross, H. A., and Goddard, M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. doi: 10.1111/1462-2920.12456
van der Aa Kuhle, A., Jesperen, L., Glover, R. L., Diawara, B., and Jakobsen, M. (2001). Identification and characterization of Saccharomyces cerevisiae strains isolated from West African sorghum beer. Yeast 18, 1069–1079. doi: 10.1002/yea.756
Keywords: Saccharomyces cerevisiae, social insects, yeast–insect association, hybrids, wine, beer, biotechnological application, ecology
Citation: Di Paola M, Meriggi N and Cavalieri D (2020) Applications of Wild Isolates of Saccharomyces Yeast for Industrial Fermentation: The Gut of Social Insects as Niche for Yeast Hybrids’ Production. Front. Microbiol. 11:578425. doi: 10.3389/fmicb.2020.578425
Received: 30 June 2020; Accepted: 06 October 2020;
Published: 29 October 2020.
Edited by:
Sara Borin, University of Milan, ItalyReviewed by:
Rashmi Chandra, University of Waterloo, CanadaCopyright © 2020 Di Paola, Meriggi and Cavalieri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Duccio Cavalieri, ZHVjY2lvLmNhdmFsaWVyaUB1bmlmaS5pdA==; Y2F2YWxpZXJpLnVuaWZpQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.