
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 October 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.573916
This article is part of the Research Topic Plant Microbiome: Interactions, Mechanisms of Action, and Applications View all 26 articles
Root-knot nematode, Meloidogyne incognita, is a devastating sedentary endoparasite that causes considerable damage to agricultural crops worldwide. Modern approaches targeting the physiological processes have confirmed the potential of FMRFamide like peptide (FLPs) family of neuromotor genes for nematode management. Here, we assessed the knock down effect of Mi-flp1, Mi-flp12, and Mi-flp18 of M. incognita and their combinatorial fusion cassette on infection and reproduction. Comparative developmental profiling revealed higher expression of all three FLPs in the infective 2nd stage juveniles (J2s). Further, Mi-flp1 expression in J2s could be localized in the ventral pharyngeal nerves near to metacarpal bulb of the central nervous system. In vitro RNAi silencing of three FLPs and their fusion cassette in M. incognita J2s showed that combinatorial silencing is the most effective and affected nematode host recognition followed by reduced penetration ability and subsequent infection into tomato and adzuki bean roots. Northern blot analysis of J2s soaked in fusion dsRNA revealed the presence of siRNA of all three target FLPs establishing successful processing of fusion gene dsRNA in the J2s. Further, evaluation of the fusion gene cassette is done through host-delivered RNAi in tobacco. Transgenic plants with fusion gene RNA-expressing vector were generated in which transgene integration was confirmed by PCR, qRT-PCR, and Southern blot analysis. Transcript accumulation of three FLPs constituting the fusion gene was reduced in the M. incognita females collected from the transgenic plants that provided additional evidence for successful gene silencing. Evaluation of positive T1 transgenic lines against M. incognita brought down the disease burden as indicated by various disease parameters that ultimately reduced the nematode multiplication factor (MF) by 85% compared to the wild-type plants. The study establishes the possibility of simultaneous silencing of more than one FLPs gene for effective management of M. incognita.
Plant-parasitic nematodes (PPNs) have proven to be one of the most challenging to control and stubborn pests causing an estimated yield loss amounting to $US 173 billion globally (Elling, 2013). Meloidogyne incognita, is the most widespread and damaging root-knot nematode (RKN) species in the world causing serious consequence for food security and economy of the farming community (Trudgill and Blok, 2001; Jones et al., 2013). Meloidogyne species are soil borne root feeders that cause severe root galling, stunted plant growth due to impaired nutrient absorption and inclination of roots to other pathogen invasion (Jones et al., 2013).
Considering the management strategies available to mitigate the RKNs, it is very challenging to select any single promising tool which is effective, eco-friendly and harmless to the non-target organisms (Bridge et al., 2005; De Waele et al., 2013). Genetic improvement through traditional breeding program faces various challenges due to non-availability of appropriate resistant source for a given crop. Management of PPNs currently relies greatly on resistant plants developed through conventional breeding methods. Further, evolution of races and pathotypes within the nematode population restricted the effectiveness of existing resistant genotypes, and selection pressure often leads to resistance-breaking biotypes of the pests. Chemical nematicides have widely been used to manage these PPNs. The recent introduction of nematicidal chemicals, viz., fluensulfone, fluazaindolizine, tioxazafen, and fluopyrum although have been claimed to be environmentally friendly and well-targeted against RKNs (Kearn et al., 2014; Slomczynska et al., 2014; Faske and Hurd, 2015; Lahm et al., 2017), the adverse effects of chemical pesticides cannot be overruled (Starr et al., 2002). Therefore, there has been a continuing demand for the progression of target-specific, environmentally sound and biodegradable pest management approaches and genetic engineering based techniques have emerged as a precious alternative and corresponding approach in this regard (Fairbairn et al., 2007; Danchin et al., 2013; Roderick et al., 2018; Chaudhary et al., 2019; Hada et al., 2020).
Targeting genes critical for the survival of nematodes provide environmentally sound strategy. Availability of M. incognita genome and several ESTs offers a repository for identification of candidate genes responsible for nematodes parasitism (McCarter et al., 2005; Abad et al., 2008). The knowledge of nematode genes responsible for successful infection and parasitism inside a host can give clue for arresting some vital physiological processes required for completing its life cycle.
The nematodes neuropeptides play many vital roles in modulating motor and sensory system and, regulate various physiological and behavioral processes including host recognition, infection, alimentation and reproduction. Neuropeptide signaling system has been identified as potential drug target due to its established effect in host recognition and infection inside the host (Maule et al., 2002; Kimber et al., 2007; Papolu et al., 2013).
FMRFamide-like peptides (FLPs) establish the most diverse and largest group of neuropeptides, leading to behavioral changes of nematodes by binding through G-protein coupled receptors (GPCRs) or peptide-gated ion channels (Husson et al., 2007; Atkinson L. E. et al., 2013; Rao et al., 2013). These FLPs transcripts are synthesized from flp genes on large propeptides, containing individual peptides in single or multiple copy number flanked by the cleavage sites having mono (R/K) or dibasic (KK/KR/RK/RR) amino acids. The propeptide molecules originated from the N-terminal signal sequences of peptide leads to synaptic drop of the mature peptide by secretory pathway (McVeigh et al., 2006). Fundamental physiological roles of a few FLPs are identified in Caenorhabditis elegans and Ascaris suum in their biology. However, most of the biological evidences on FLPs are lacking in PPNs, and M. incognita most likely due to their obligate nature which confines the use of conventional physiological procedures. Nineteen FLPs are reported from M. incognita out of which six have been established to have transcriptional confirmation viz., Mi-flp1, Mi-flp7, Mi-flp12, Mi-flp14, Mi-flp16, and Mi-flp18 (Magrane and UniProt Consortium, 2011). Multiple Sequence alignment of these known FLPs in RKNs exhibited less sequence similarity among them despite they share a typical RF-amide sequence at C-terminus. Their uniqueness could consequently be advantageous at generating sequence specific knockdown module by dsRNA approach to avoid any off target effects.
Increasing evidence for the development of RNAi based transgenics suggests that expression of M. incognita specific dsRNA molecule in the host plant may provide alternative management strategy. The simplicity and targeted knock down effect associated with RNAi has immense value for deciphering the gene function in many organisms including the obligate PPNs (Urwin et al., 2002; Rosso et al., 2005; Kimber et al., 2007; Shingles et al., 2007; Dalzell et al., 2009; Ibrahim et al., 2011; Niu et al., 2012; Chaudhary et al., 2019). Rosso et al. (2005) explored the RNAi for the first time in M. incognita J2s for the silencing of two genes, Mi-crt and Mi-pg1 which are expressed in the sub-ventral esophageal glands of the nematodes and probably involved in early parasitism.
Investigation on the FLPs genes function through RNAi has shown reduced migration, penetration and aberrant behavioral phenotypes in M. incognita, Meloidogyne graminicola, and Globodera pallida (Urwin et al., 2002; Kimber et al., 2007; Dalzell et al., 2010; Atkinson L. E. et al., 2013; Papolu et al., 2013; Dong et al., 2014; Banakar et al., 2015; Kumari et al., 2017). Further, effectiveness of in vitro RNAi of FLP genes (Gp-flp1, Gp-flp12, and Gp-flp18) in G. pallida (Kimber et al., 2007) and host-delivered RNAi of Mi-flp14, Mi-flp18 in M. incognita (Papolu et al., 2013) indicate that it would be intriguing to investigate the synergistic or additive impacts, if any, of concomitant knockdown of more than one flp gene. Multiple genes silencing concurrently has been successfully employed in C. elegans (Tischler et al., 2006; Gouda et al., 2010; Min et al., 2010). Tischler et al. (2006) simultaneously silenced four genes viz., lin-31, sma-4, unc-22, and lon-2 having different biological functions through RNAi by feeding modified bacteria to C. elegans that produced comparatively weaker phenotypes compared to the double genes. Gene pyramiding in plants has been proven to be a better approach for resistance against many pathogens which cause substantial yield losses (Urwin et al., 1998; Zhao et al., 2003; Abdeen et al., 2005; Turra et al., 2009; Roderick et al., 2012; de Souza Júnior et al., 2013; Walawage et al., 2013; Narusaka et al., 2014; Quilis et al., 2014; Tripathi et al., 2014; Chan et al., 2015). Targeting multiple genes has also been utilized for the development of nematode resistant transgenic plants. Urwin et al. (1998) developed transgenic Arabidopsis plants with a stacked gene construct, having a CpTI and Oc-I_D86 genes. The transgenic plants exhibited superior resistance to Heterodera schachtii. Similarly, Chan et al. (2015) delivered a dual gene construct, which included CeCPI and a PjCHI-1 genes in transgenic tomato plants with synthetic promoter, pMSPOA, having NOS-like and SP8a elements. These transgenic tomato plants having dual genes significantly reduced the RKN infection and reproduction compared with the plants transformed with a single gene.
In view of the above, we have selected three M. incognita FLP coding genes, Mi-flp1, Mi-flp12, and Mi-flp18 and their fusion sequence prepared synthetically for evaluating RNAi silencing effect on host finding, invasion and reproduction of M. incognita. Further, functional validation through host-delivered RNAi of FLPs-fusion gene in Nicotiana tabacum L. using Agrobacterium tumefaciens-mediated transformation and evaluation of transgenic plants against M. incognita has been undertaken. In addition, site of expression of a gene helps in better understanding of its function and hence expression site of Mi-flp1 which is not known is deciphered in the present study. Thus, the present investigation strengthens our knowledge on the effect of M. incognita Mi-flp1, Mi-flp12, Mi-flp18 and their fusion gene on nematode reproduction and plant parasitism potential when applied through host-delivered RNAi.
A pure population of M. incognita was maintained on eggplant (Solanum melongena cv. Pusa Purple Long) in the greenhouse. Further, Adzuki bean plants growing in CYG growth pouches1 infected with M. incognita were used for collecting the different developmental stages [eggs, pre parasitic juveniles (J2s), 3rd and 4th stage juveniles (J3s/J4s), and young adult females] of the nematode (Atkinson and Harris, 1989; Tyagi et al., 2013). Additionally, fresh egg masses from the infected plants were also collected for hatching the pre-parasitic J2s required for further experiments.
Freshly hatched J2s of M. incognita were collected and total RNA was isolated using NucleoSpin RNA extraction kit (Macherey-Nagel, Germany). RNA quality and quantity was analyzed using NanoDrop-2000 (Thermo Fisher Scientific, United States) and about 500 ng of total RNA was reverse transcribed to complementary DNA (cDNA) with cDNA synthesis Kit (Superscript VILO, Invitrogen, United States).
To design the primers for FLPs, sequences of Mi-flp1 (KC517344.1), Mi-flp12 (AY804187.1), and Mi-flp18 (AY729022.1) of M. incognita available in GenBank were used and aligned in the ClustalX version 1.81 (Thompson et al., 1997). Conserved nucleotide sequences were used and primers were designed using IDT OligoAnalyzer, and primer sequences are given in Supplementary Table S1. Selected genes were PCR amplified from cDNA and cloned into pGEM-T easy vector (Promega, United States) and confirmed the target gene inserts by sequencing. Selected sequences of Mi-flp1, Mi-flp12, and Mi-flp18 were further used for designing the fusion gene cassette by placing the three sequences continuously and synthesized artificially by Biomatik Custom Gene Synthesis (Biomatik Technologies, United States) and sub-cloned into pGEM-T easy vector (Promega, United States).
Different developmental stages of M. incognita (eggs, pre parasitic J2s, J3s/J4s, and young adult females) were collected and used for RNA extraction and cDNA preparation as elaborated above. Quantitative Real-time PCR (qRT-PCR) primers (Supplementary Table S1) were also designed using IDT OligoAnalyzer. qRT-PCR was carried out with SYBR Green PCR Master Mix (Eurogentec, Liège, Belgium) in realplex2 thermal cycler (Eppendorf, Germany). The conditions and melt curve program for qRT-PCR reaction was similar as previously depicted by Papolu et al. (2013). Relative fold-change in target gene expression was analyzed using comparative Ct method (Livak and Schmittgen, 2001) and log2- transformed. Egg stage gene expression was used as base level expression for comparing with other developmental stages. M. incognita 18S rRNA (Accessions-HE667742) was taken as a reference gene for the gene expression normalization. Three biological and three technical replicates were used for each of the samples during qRT-PCR analysis.
The plasmids harboring the target cDNA sequence of flp1 was used as a template to amplify the gene. Subsequently, PCR product was used for preparing the DIG- labeled sense and antisense DNA probes, separately (Roche, Switzerland). J2s fixation, permeabilization, hybridization of probes and signal detection and staining procedures were followed as described by Banakar et al. (2015). Processed nematodes were placed on microscope slides, covered with 0.28 mm glass coverslips and sealed with nail varnish. The observations were recorded in the Zeiss Imager M2m compound microscope. Images were recorded with Axion camera MRc5 through Axion vision Rel. 4.8 software (Zeiss Imager M2m, Carl Zeiss, Germany).
Selected FLP genes were amplified from the pGEM-T clones using M13 universal primers. Double-stranded RNAs (dsRNAs) were synthesized for each of the target genes as described earlier by Shivakumara et al. (2016), with minor modifications as 2 μl of labeled dUTP with Alexa Flore 488 (10 mM) was used. Additionally, dsRNA of GFP (green fluorescent protein, GenBank: HF675000) was prepared to be used as negative control. Synthesis of dsRNA was confirmed by resolving on 1% (w/v) agarose gel and visualized in gel documentation system (AlphaImager® analyzer, United States).
About 2000 Freshly hatched J2s were soaked in 100 μl soaking solution having 50 μl of 50 mM octopamine (DL-octopamine hydrochloride, Sigma, United Kingdom), 2 μl labeled dsRNA (1 μg/μl) and 48 μl nuclease free water (Urwin et al., 2002) for 18 h in dark on a rotator (10 rpm) at room temperature. dsRNA soaking was done for all the target FLPs singly and combinatorial fusion gene in triplicates. J2s incubated separately in dsRNA of GFP and water were used as negative controls. Fluorescence was measured using EnSpire® Multimode Plate Reader (PerkinElmer, United States) with default settings and excitation wavelength 488 λ and emission wavelength was 520 λ. The dsRNA soaked worms were placed in the 96 well black OptiPlateTM-96 F (PerkinElmer, United States). About 100 nematodes were used in each well with 12 replications for both control and dsRNA soaked worms. dsRNA uptake was also confirmed by microscopic observation using a ZEISS SteREO Discovery V20 microscope.
Juveniles soaked overnight in dsRNA solutions as aforesaid for different FLPs genes were washed with nuclease free water and frozen immediately in liquid nitrogen. Total RNA was isolated, quantified, and reverse transcribed. qRT-PCR has been carried out to quantify the transcript accumulation of all the target genes so as to confirm target-specific knockdown as described above. Three biological and three technical replicates were kept for each of the samples and gene expression was normalized using 18S rRNA. Relative fold-change in target gene expression was analyzed using comparative Ct method (Livak and Schmittgen, 2001) and log2- transformed. Statistical analysis was carried out to assess the significance difference between treatments and water control worms.
Northern hybridization of the fusion dsRNA soaked J2s was done to confirm the processing of the dsRNA into siRNAs using a standard protocol (Sambrook et al., 1989). Nematodes were incubated in the fusion cassette dsRNA solution as mentioned above. Total small RNA was isolated from 100 mg of nematodes tissue using mirVanaTM miRNA Isolation Kit (Thermo Fisher Scientific, United States) and resolved on 30% denaturing PAGE (Polyacrylamide Gel Electrophoresis) gel and transferred onto a nitrocellulose membrane (Bio-Rad, United States) using iBlot® gel transfer device (Invitrogen, United States). Probes of Mi-flp1, Mi-flp12, and Mi-flp18 were separately prepared, hybridized, and detected as described earlier by Papolu et al. (2013).
To assess the knockdown effect of candidate genes on M. incognita infectivity, we evaluated penetration/invasion of dsRNA-soaked J2s in tomato roots on a soil less pluronic PF-127 medium (Wang et al., 2009; Chaudhary et al., 2019). Tomato seedlings were placed on pluronic gel medium in Petri plate and inoculated with dsRNA soaked J2s at a distance of 1.5 cm from the root tip. Sixteen replications were taken for each treatment. There were six treatments viz., Mi-flp1 dsRNA, Mi-flp12 dsRNA, Mi-flp18 dsRNA, fusion cassette dsRNA, GFP dsRNA, and water control. Number of J2s that moved toward the root tips and present in the vicinity up to 1 mm diameter at different time intervals viz., 4, 8, 24, and 72 h were counted. Additionally, number of J2s penetrated at 24 and 72 h after inoculation was observed after roots staining with acid fuchsin (Byrd et al., 1983) and documented.
Tomato plants were grown in 18 cells seedling trays (JPP54, Jain Plasto Pack, India) for 25 days under greenhouse conditions. dsRNA feeding and washing steps were followed as mentioned above. For all the treatments and controls, 200 J2s were inoculated per plant (2 nematodes / g of soil) and provided 72 h for nematode penetration and were harvested. Roots were washed to remove the adhering soil and stained with acid fuchsin (Byrd et al., 1983) to calculate the number of invaded J2s in roots. There were six treatments as listed above and ten replications for each treatment.
Silencing effect of Mi-flp1, Mi-flp12, Mi-flp18 and their synthetic fusion gene on M. incognita development and reproduction was carried out by inoculating dsRNA-soaked nematodes on 7-days old adzuki bean (Vigna angularis var. angularis) roots in CYG seed growth pouches (Banakar et al., 2015 and Shivakumara et al., 2016). Six root tips of each plant were inoculated with about 15–20 J2s, and incubated in a growth chamber at 27°C with 70% humidity and a light intensity of 300 lux (Jeio Tech Company Ltd., South Korea). Plants were supplemented with nutrient solution (Hoagland solution) after 7 days of inoculation at 3 days interval until harvest. At 35 DPI, plants were harvested, and disease burden per plant was scored using total number of galls, total number of endoparasites/females, number of egg masses, and number of eggs per egg mass, which were taken to calculate the multiplication factor (Papolu et al., 2013). Nematodes treated with GFP dsRNA and water were taken as controls. The experiments have three biological and ten technical replicates.
On the basis of in-vitro RNAi outcome, the fusion gene cassette of FLPs containing Mi-flp1, Mi-flp12, and Mi-flp18 was used for preparing hpRNA vector to examine host induced gene silencing effects. RNAi Gateway vector pB7GWIWG2(II) was procured from VIB-UGent Center for Plant Systems Biology, Ghent University, Ghent, Belgium. The commercially synthesized fusion gene cassette was PCR amplified from corresponding recombinant pGEMT clones and sub-cloned into the pDONR 221 (an entry vector). Subsequently, cloned into the destination vector “pB7GWIWG2(II)” using LR recombination in sense and antisense orientation intervening with an intron by GATEWAY recombination based cloning kit (Invitrogen, United States). The recombinant vector containing the hpRNA construct was transformed into E. coli (DH5α) cells. The positive clones were thereafter mobilized into A. tumefaciens strain EHA105, and confirmed the correct orientation of target gene by PCR using target gene specific primers, CaMV 35S promoter and attB2, CaMV 35S terminator and attB2, bar gene specific primers (Supplementary Table S1). Further, A. tumefaciens having fusion gene cassette was used for tobacco (N. tabacum L. cv. Petit Havana) transformation for functional validation as illustrated earlier by Papolu et al. (2013) and Shivakumara et al. (2019) with minor modifications.
Putative transgenic plants expressing hairpin fusion gene cassette were subjected to molecular analysis to confirm the presence of transgene. For this, genomic DNA was isolated from young leaf tissues of various putative transgenic events and wild-type (WT) plants following NucleoSpin Plant II DNA kit (Macherey-Nagel, Germany). Primary confirmation of the transgene was done by performing PCR using different sets of primers (Supplementary Table S1). The amplified product was resolved on 1.0% (w/v) agarose gel.
To confirm the T-DNA integration and copy number, genomic DNA (∼12 μg) from the PCR-positive T1 plants was digested with SacI (20U/μl) restriction enzyme (New England Biolabs, United Kingdom) by incubating at 37°C for 16 h. Digested DNA samples were separated on 0.8% agarose gel, transferred onto a nitrocellulose membrane (Bio-Rad, United States). Fusion gene specific probe was synthesized and labeled with digoxigenin (DIG) probe labeling kit (Roche, Switzerland). Hybridization, washing, detection and blot development was done as earlier illustrated by Papolu et al. (2013).
The confirmed transgenic T1 lines were further subjected to qRT-PCR to interpret the FLPs transcripts abundance. Total RNA was isolated from the T1 plants leaves using NucleoSpin plant II RNA kit (Macherey-Nagel, Germany), about 500 ng of total RNA was converted to cDNA using Superscript VILO cDNA synthesis kit (Invitrogen, United States), and qRT-PCR was performed with the target genes specific primers in the realplex2 thermal cycler (Eppendorf, Germany). N. tabacum 18S rRNA (Accession: HQ384692.1) was used for gene expression normalization. Three biological and three technical replicates were taken for the study and data were analyzed following Livak and Schmittgen (2001).
Transgenic lines (T1) harboring hpRNA construct of fusion gene were subjected to nematode parasitism assays. 30-days old plants were grown in 4-inch diameter pots containing 200 g of soil and sand (5:1), each plant inoculated @ 2 M. incognita J2s per g of soil and maintained in a growth chamber at 28°C, 70% relative humidity, 14:10 h light:dark. Plants were harvested at 35 DPI, and the nematode parasitic success was documented as interprets earlier by Papolu et al. (2013).
Additionally, about 20 mature females were dissected out from the transgenics and WT plants, total RNA was extracted and reverse-transcribed (300 ng RNA) as mentioned above. Further transcripts accumulation was quantified using qRT-PCR with Mi-flp1, Mi-flp12, and Mi-flp18 and M. incognita 18S rRNA was used as reference gene. Three biological and three technical replicates were considered for the study. Fold change in expression was analyzed using 2–Δ Δ CT (Livak and Schmittgen, 2001).
The experimental data were analyzed using one-way analysis of variance (ANOVA) with completely randomized design (CRD), and significance was decisive at P = 0.05 and P = 0.01. The mean values of treatments were subjected to Duncan Multiple Range Significant Difference Test (DMRT).
Polymerase chain reaction (PCR) amplification, cloning and sequencing of 232, 349, and 407 bp sequences of Mi-flp1, Mi-flp12, and Mi-flp18 amplified from the cDNA revealed 100% similarity with the previously reported sequences in NCBI database (Figure 1A). Differential expression of FLP genes in different developmental stages of M. incognita was performed by qRT-PCR. Based on expression of all three FLPs in eggs as reference, we observed that all three FLPs were significantly (P < 0.05) upregulated in the pre-parasitic J2s in relation to consistent down-regulation in J3/J4 and adult females (Figure 1B). Reference gene expression was constant in the different developmental stages. The expression stability was confirmed in three biological and three technical replications.
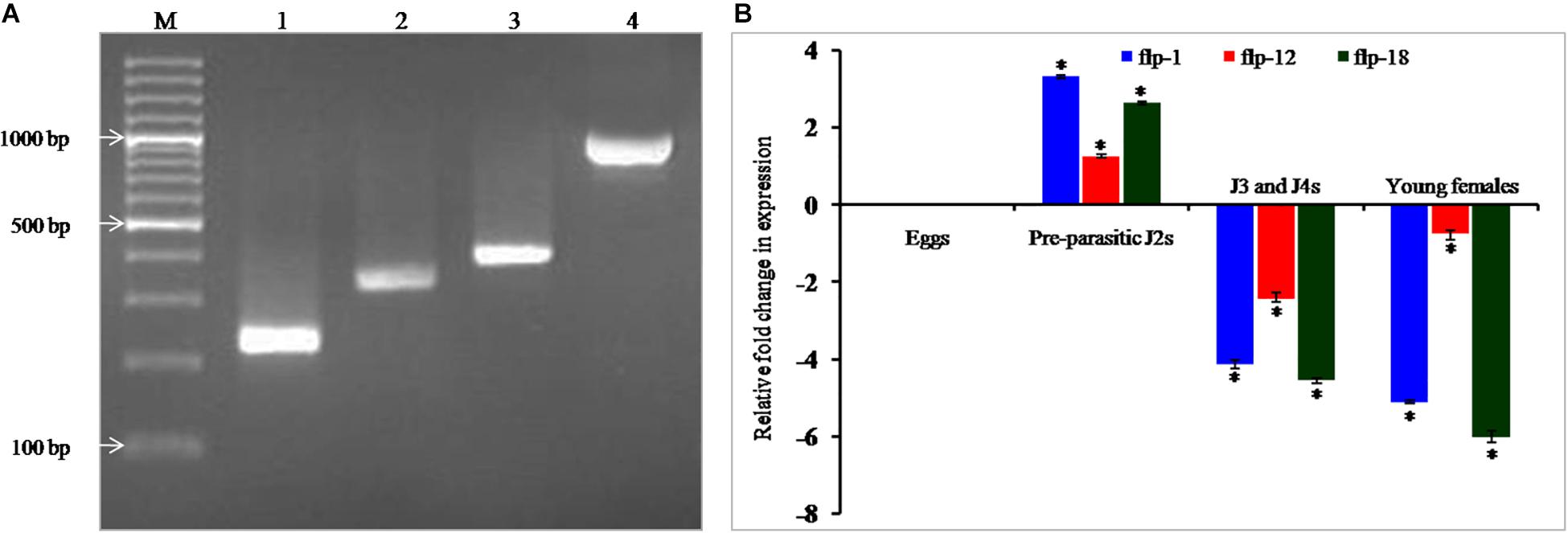
Figure 1. (A) PCR amplification of FLPs of M. incognita. Lanes – 1: Mi-flp1, 2: Mi-flp12, 3: Mi-flp18, 4: FLPs-fusion cassette, M: 100 bp marker, (B) Relative fold-change in the target genes expression in different developmental stages of M. incognita. Each bar represents the mean ± SE of n = 3, and asterisks indicate significant difference at P < 0.05.
The localization of Mi-flp1 expression was performed by in situ hybridization in the M. incognita J2s. A 232 bp probe generated for Mi-flp1 hybridized to the ventral pharyngeal nerves of the metacorporeal bulb in the nervous system. Intense staining was observed in the ventral pharyngeal nerves of the metacorporeal bulb (Figure 2B). However, precise identification of neurons was difficult in this tightly packed region of the nervous system. So, the neural map of C. elegans was used for interpreting the site of expression of the Mi-flp1 in M. incognita J2s (White et al., 1986). Accordingly, Mi-flp1 gene expression appeared to be localized in the AVK, AVA, AVE, RIG, RMG, AIY, and AIA pharyngeal MN neurons of M. incognita J2s (Figure 2D). There was absence of a hybridization signal in the control juveniles with DIG labeled sense probes Figures 2A,C).
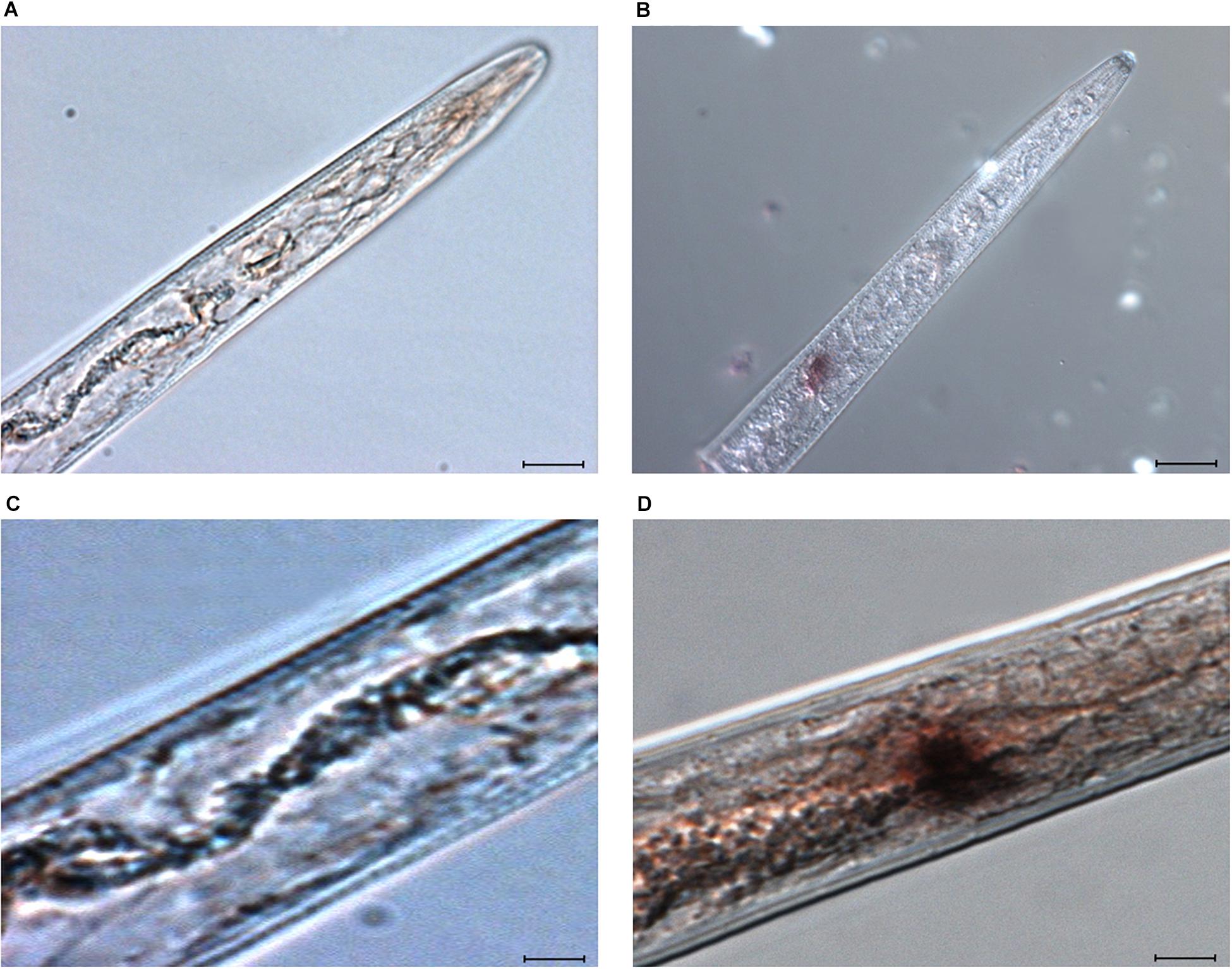
Figure 2. In situ hybridisation of DIG-labeled cDNA of Mi-flp1 probe in the anterior of M. incognita J2s. (A,C) No hybridization signal in the control juveniles with DIG labeled sense probes, (B,D) Intense staining was observed in the nematode nerve ring region, revealing the Mi-flp1 gene expression in ventral pharyngeal nerves near to metacarpal bulb of the central nervous system. Mi-flp1 expression site is indicated by brown/black color due to the enzymatic breakdown of a chromogenic substrate by alkaline phosphatase conjugated to anti-DIG antibody. Scale bar = 10 and 20 μm.
The comparative expression levels of target mRNAs were quantified by qRT-PCR in nematodes silenced for Mi-flp1, Mi-flp12, Mi-flp18, and fusion gene compared to water control nematodes. Silencing of Mi-flp1 and Mi-flp18 singly led to down-regulation (P < 0.05) of the target genes compared to water control (Figure 3A). However, silencing of Mi-flp12 singly resulted in significant (P < 0.05) up-regulation of the target gene over control. Silencing of the fusion gene having all three FLP genes (Mi-flp1, Mi-flp12, and Mi-flp18) resulted in significant (P < 0.05) down-regulation of Mi-flp1 and Mi-flp12 transcripts and up-regulation of the Mi-flp18 (Figure 3B). Here, target gene specific knockdown could be confirmed with suitable controls and at first, none of the target genes was silenced in the nematodes incubated in GFP dsRNA. Reference gene expression was equal in both the dsRNA treated and water control worms. The expression stability was confirmed in three biological and three technical replications.
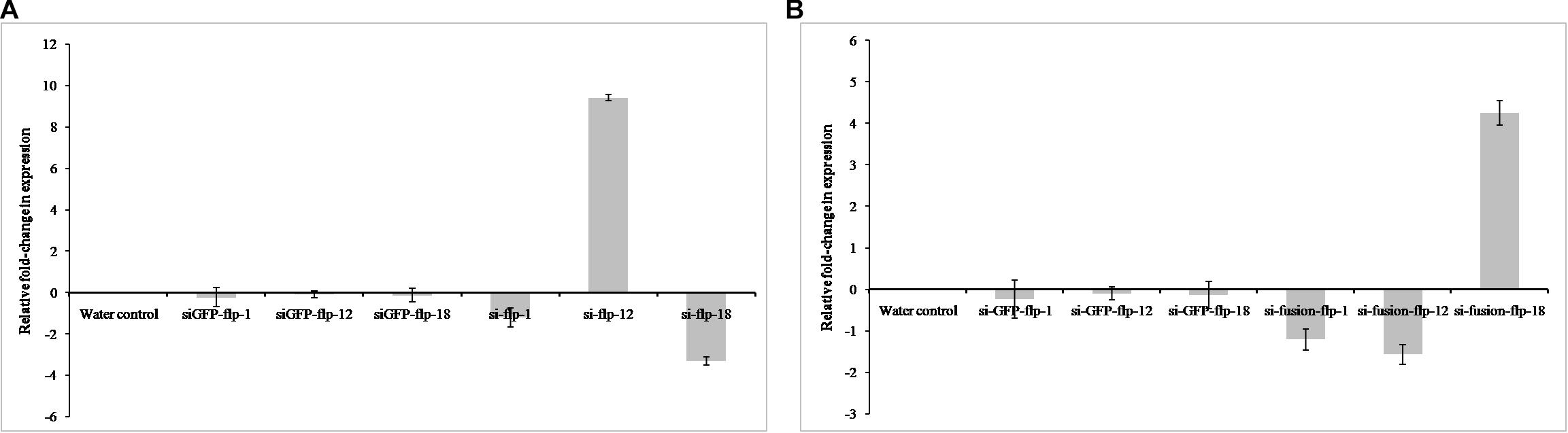
Figure 3. Quantification of target gene transcripts in dsRNA treated J2s of M. incognita. The relative expression levels of target mRNA were quantified by qRT-PCR in M. incognita silenced for (A) Mi-flp1, Mi-flp12, and Mi-flp18, (B) FLPs-fusion gene, compared to non-native GFP control and water control. 18s rRNA was used as a reference gene and fold-change was calculated using 2– Δ Δ CT method. Each bar represents the mean ± SE of n = 3 (P < 0.05).
As a key segment of successful RNAi, production of siRNAs of all three FLP genes was demonstrated by northern blot analysis in M. incognita J2s incubated in fusion dsRNA. The presence of siRNAs of all the target genes was abundant in fusion dsRNA soaked nematodes (Figure 4A). However, no expression was detected in water control nematodes.
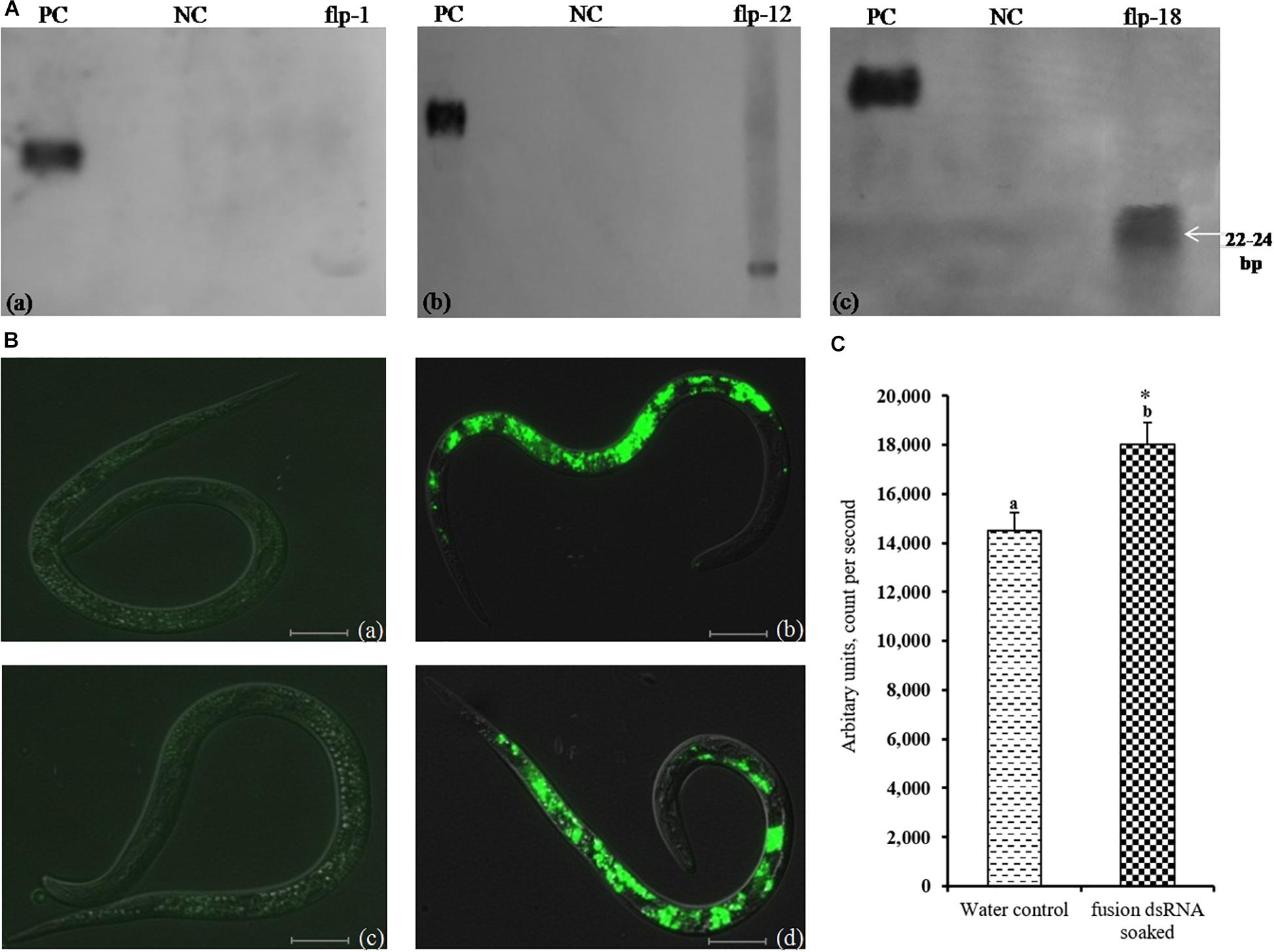
Figure 4. (A) Northern blot analysis for detection of siRNA in M. incognita J2s incubated in FLPs-fusion dsRNA. (a) Mi-flp1, (b) Mi-flp12, (c) Mi-flp18. PC – positive control (gene specific labeled probe: 25 pg); NC – negative control (total RNA from control healthy worms), (B) dsRNA uptake confirmation of M. incognita J2s incubated in FLPs-fusion dsRNA labeled with Alexa Flore 488 dUTPs using florescent microscope, (a,c) J2s without dsRNA, (b,d) dsRNA ingested J2s, fluorescence was localized in the stylet, esophageal gland cells, and intestine. Scale bar = 10 and 20 μm. (C) Difference in the fluorescence count between the water control and FLPs-fusion dsRNA soaked worms. Maximum fluorescence was quantified in 5000 flashes per minute using EnSpire® Multimode Plate Reader (t = 2.306, p > 0000001). a,b*Represents the values are statistical significant (t = 2.306, p > 0000001).
Nematodes soaked in Alexa Flore 488 labeled florescent dsRNA showed fluorescence after 18 h of incubation and fluorescence was localized in the stylet, esophageal gland cells, and intestine (Figure 4B). Maximum fluorescence was quantified in 5000 flashes per minute in fluorescence multimode plate reader. The data were subjected to the Student’s t-test, and there was a significant difference between the water control and dsRNA soaked worms (Figure 4C) (t = 2.306, p > 0000001).
Freshly hatched J2s of M. incognita were soaked in respective dsRNAs of the target genes for 18 h in the presence of 50 mM octopamine. In general, target genes silencing reduced M. incognita attraction to tomato roots compared to water control nematodes, and the effect was noticeable promptly after 4 h. Least number of nematodes were observed in fusion dsRNA soaked nematodes followed by Mi-flp1 (P = 0.05), and a significant difference was observed between the fusion gene and single gene silenced nematodes at 8 h (Figure 5A). Silencing of each of the target genes decreased the M. incognita penetration/invasion at 24 and 72 h compared to the water control and GFP treatments (Figure 5B). However, simultaneous silencing of all the three FLPs through fusion gene provided the highest reduction in penetration (P = 0.001) (Figure 5C). Similarly, attraction and penetration of dsRNA soaked worms into tomato roots in seedling trays resulted in the significant reduction of nematode penetration after 72 h of inoculation. The findings demonstrated that the silencing of fusion gene significantly reduced the nematode attraction and penetration more than other treatments (P = 0.001) (Supplementary Figure S1).
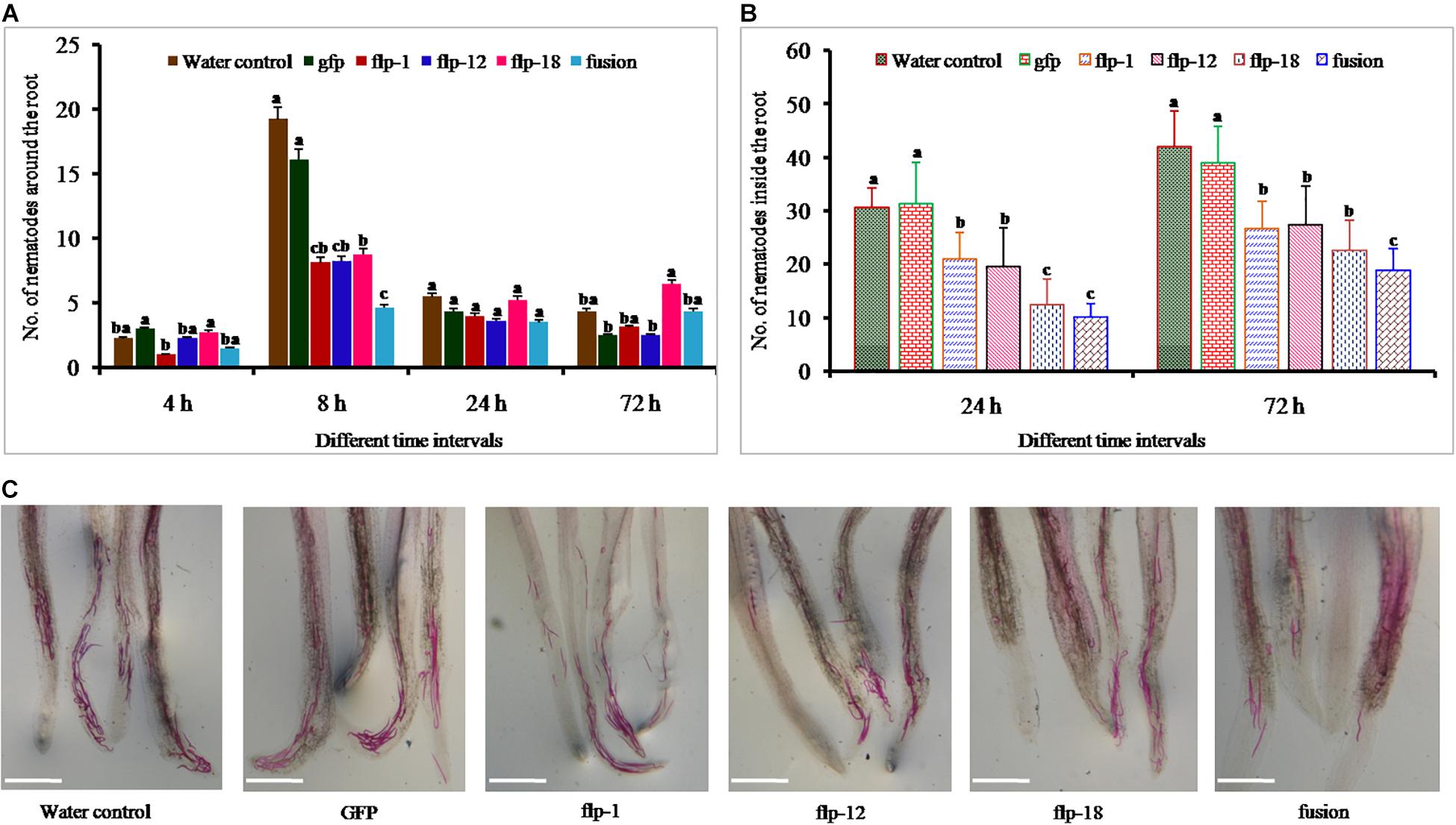
Figure 5. (A) Penetration ability of Mi-flp1, Mi-flp12, Mi-flp18, and FLPs-fusion dsRNA-soaked M. incognita J2s in tomato roots on PF-127 medium at 4, 8, 24, and 72 h. Each bar represents the mean standard error (n = 16); bars with letters stand for a significant difference at P > 0.05, (B) Number of nematodes inside tomato roots at 24 and 72 h of FLPs and GFP treated J2s and control worms in water, (C) Stained worms in infected tomato roots at 72 h post inoculation. Worms treated with non-native dsRNA (GFP) and worms in water were used as controls. Scale bar = 500 μm. a,b,cRepresents the values are statistical significant (P = 0.001).
Root-knot nematode bioassay was carried out on adzuki beans in the CYG seed growth pouch system. After 35 DPI, each plant was scored for disease parameters in terms of total number of galls, endoparasites/females, egg masses and eggs per egg mass. Target gene silencing through soaking in dsRNA of three FLP genes singly and fusion gene resulted in reduction in average galling by 54% compared to the control. This also led to reduction in total endoparasites/females and egg masses by 63% and 54%, respectively (P = 0.01). A significant difference was observed between the fusion dsRNA soaked J2s compared to all other treatments (P = 0.01). However, within single gene silenced nematodes, Mi-flp18 was significantly different from Mi-flp1 and Mi-flp12 (P = 0.01). All the treatments demonstrated considerable differences from each other and showed reduction in nematode reproduction up to 58% (P = 0.01) (Figure 6).
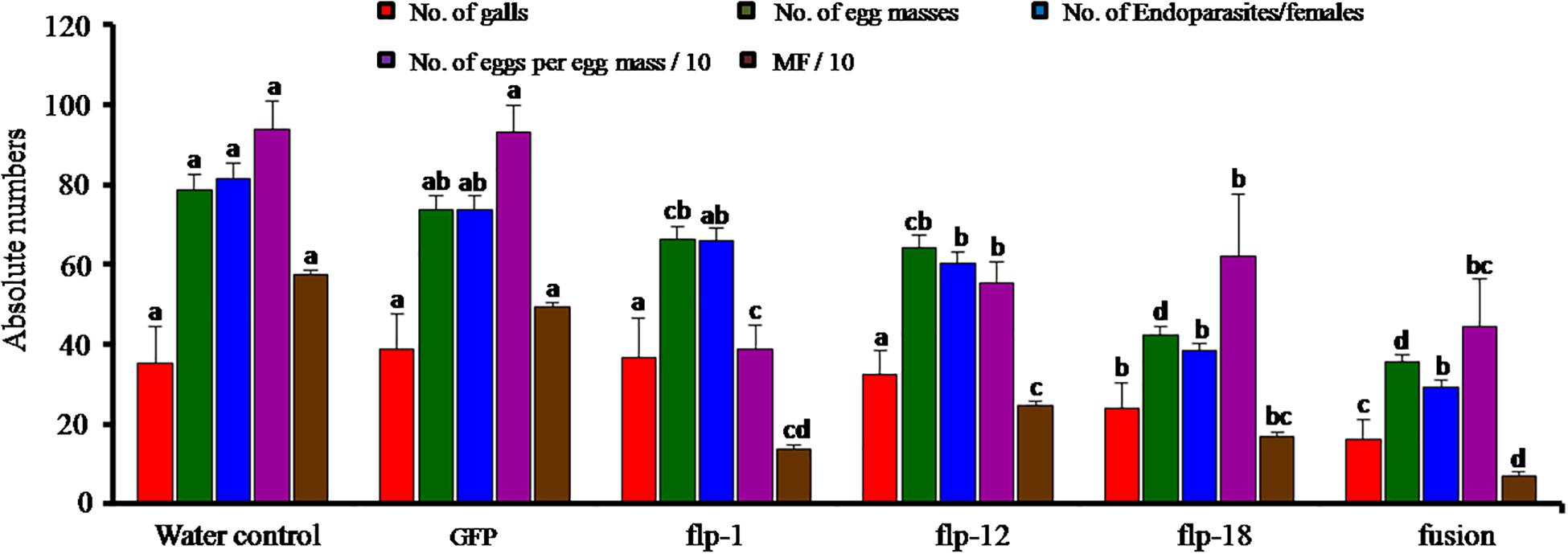
Figure 6. Effect of in vitro silencing of FLPs gene and their fusion gene on developmental and reproduction of M. incognita in adzuki bean. Total numbers of galls, total endoparasites, egg masses, eggs per egg mass and the respective multiplication factor (MF) of M. incognita in adzuki bean infected with dsRNA-treated J2s at 35 DPI. Values are means ± standard errors (n = 9) and letters for a given variable differed significantly amongst treatments at P < 0.05. J2s treated with GFP dsRNA and worms in water are used as controls.
Based on the outcome of in-vitro RNAi experiments, in-planta functional validation analyses have been carried out with FLPs-fusion gene. A commercially synthesized fusion gene using coding nucleotide sequences of Mi-flp1, Mi-flp12, and Mi-flp18 was cloned and confirmed by sequencing. FLPs-fusion gene cassette cloned into RNAi Gateway vector pB7GWIWG2(II) is shown in Supplementary Figure S2A. PCR with different sets of primers confirmed the orientation of the sense and antisense strands of the fusion gene in the hpRNA construct.
Tobacco cv. Petit Havana was transformed with hpRNA construct harboring the fusion gene using a standardized leaf-based regeneration and transformation method and T0 plants were generated (Supplementary Figure S2B). To confirm the complete T-DNA integration into the T0 plants, initial analysis was carried out using PCR analyses with different sets of primers. Presence of the fusion gene, promoter, terminator and the selectable marker gene was determined in all the independent events and no amplification was detected in WT plant (Supplementary Figure S3).
The transgenic T1 plants were identified and set up by selfing the positive T0 plants and re-confirmed via PCR with the same set of primers as stated above, which amplifies the expected target fragments in the tested events, signifying integration of the fusion gene construct in the progeny plants (Supplementary Figure S4).
In order to confirm the pattern of transgene integration and copy number in T1 plants, PCR positive plants were validated by Southern blot analysis. It was observed that the transgenic lines, viz., 25–6, 28–9, 38–5, and 47–2 showed single copy insertions while 27–4 and 36–12 exhibited double copies of the transgene. There was absence of any hybridization in the WT plants (Figure 7A).
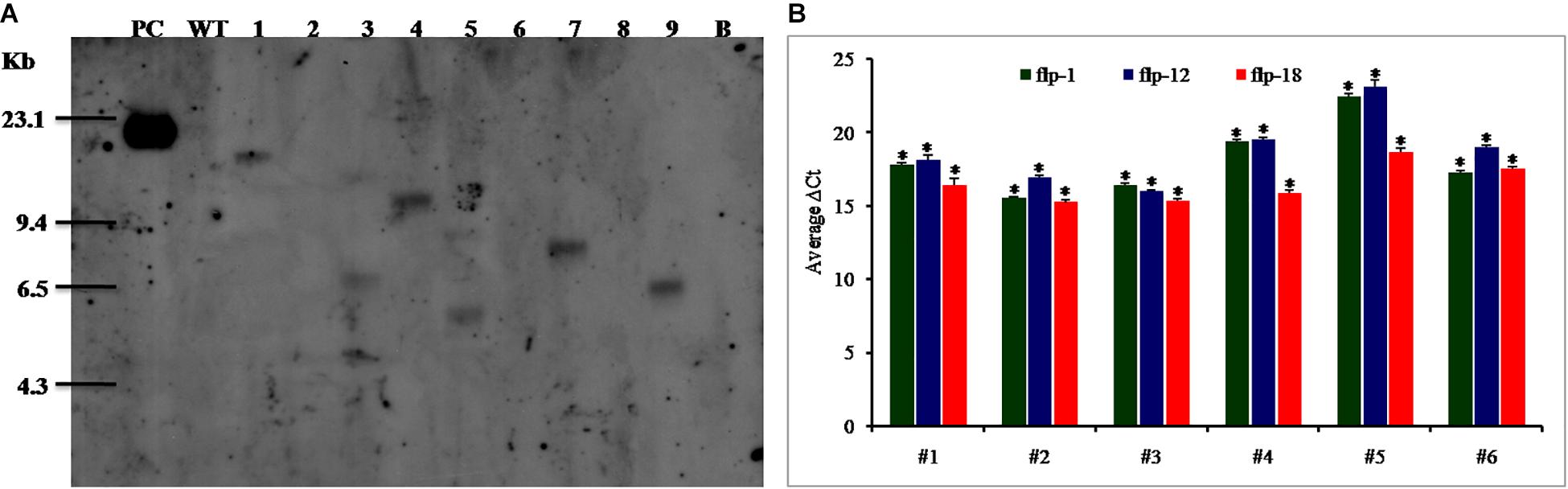
Figure 7. (A) Southern blot analysis of transgenic lines harboring dsRNA construct of FLPs-fusion gene. Lanes 1–9 shows transgenic lines 25-6, 26-2, 27-4, 28-9, 36-12, 37-1, 38-5, 40-5, and 47-2. PC – positive control (linearized recombinant vector pB7GWIWG2(II) (25 pg); WT – negative control (wild-type plant); B – blank, (B) Expression analysis of target genes in Southern positive T1 plants. Mi-flp1, Mi-flp12, and Mi-flp18 expressing lines (#1 to #6 represent lines– 25-6, 27-4, 28-9, 36-12, 38-5, and 47-2). ΔCt values were calculated using difference in the Ct mean of target gene and reference gene (tobacco 18S rRNA). Each bar represents the mean ± SE of n = 3, and asterisks shows significant difference at P < 0.05. Higher ΔCt values specify the lower expression of transgene in the corresponding events.
Further, transgenic plants expressing fusion gene construct were confirmed by qRT-PCR. The results indicated significant expression of a transgene in all the selected events, and among them events 27–4 and 28–9 had the highest expression level with all three tested genes and the event 38–5 showed the least in terms of average ΔCT values (Figure 7B). There was no transcript detected in WT plants.
T1 generation plants of positive transgenic events were evaluated against M. incognita. The nematode infection was determined after 35 DPI in terms of total number of galls, endoparasites, egg masses, and eggs per egg masses. The results showed average galling in the range of 41–70 in different T1 lines compared to WT plants which was observed to be about 90 that led to a reduction of 22–54% in average galling in transgenic events. This eventually resulted in significant lowering in total endoparasites in the range of 65–95 in T1 lines compared to about 120 in WT plants. As a result, 21–46% reduction was observed in total endoparasites in the transgenic events. Similarly, the average number of egg masses and eggs per egg mass was present in the range of 28–58 and 214–320 in T1 plants of different transgenic events whereas WT plants showed about 80 and 482, respectively. This resulted in significant reduction in fecundity in the transgenics events by 25–65% and 32–56%, respectively. Finally, derived MF was in range of 13.3–41.3 in T1 lines while WT plants documented about 86.02 which demonstrated a reduction of 85% in transgenic events compared to the WT plants (Figures 8A,B).
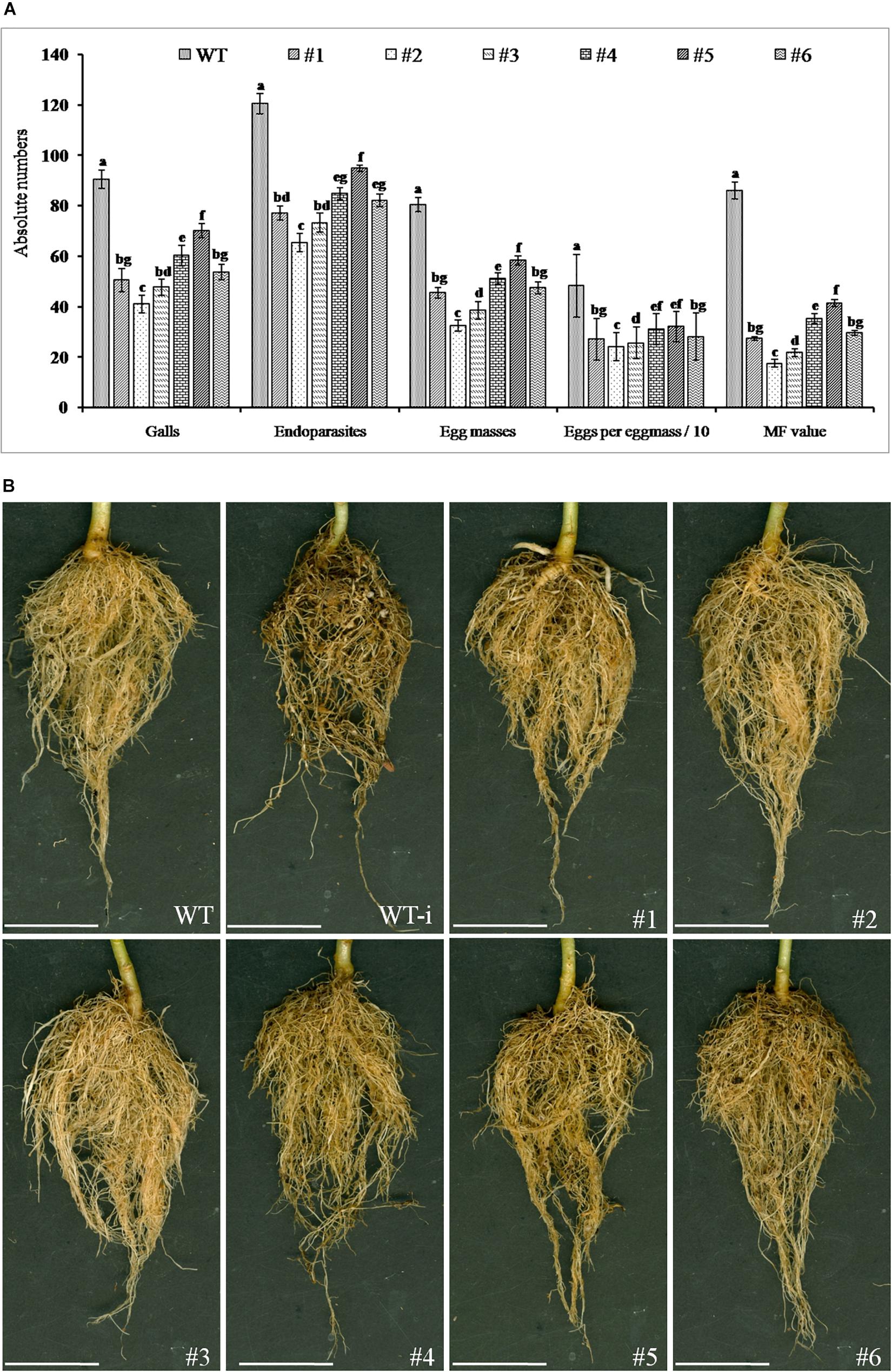
Figure 8. Effect of host-delivered RNAi of FLPs-fusion gene on development and reproduction of M. incognita. (A) Performance of transgenic plants was assessed in terms of number of galls, total endoparasites, egg masses, eggs per egg mass, and multiplication factor (MF) in different transgenic lines (#1- 25-6, #2- 27-4, #3- 28-9, #4- 36-12, #5- 38-5, and #6- 47-2) and WT plants at 35 DPI in soil. Each bar represents the mean ± SE of n = 5, and bars with different letters (within each parameter) indicate significant difference at P < 0.05. (B) Comparison of M. incognita infection in roots of transgenic and WT plants at 35 DPI; representative roots of (WT) healthy wild-type plants, (WT-i) nematode infected wild-type plants, transgenic events #1 to #6: 25-6, 27-4, 28-9, 36-12, 38-5, and 47-2 infected with M. incognita, scale bar = 5 cm.
In order to examine the effect of host-delivered RNAi on suppressing the target transcripts in the worms, qRT-PCR was carried out with adult females of M. incognita dissect out from the transgenic plants. The expression of Mi-flp1, Mi-flp12, and Mi-flp18 in M. incognita females was down-regulated significantly (P < 0.05) by 0.7 ± 0.2 to 1.5 ± 0.1 fold in case of Mi-flp1, 0.9 ± 0.1 to 1.8 ± 0.2 fold in case of Mi-flp12, and 1.2 ± 0.2 to 2.2 ± 0.3 fold in case of Mi-flp18, respectively, compared to females from WT plants (Figure 9).
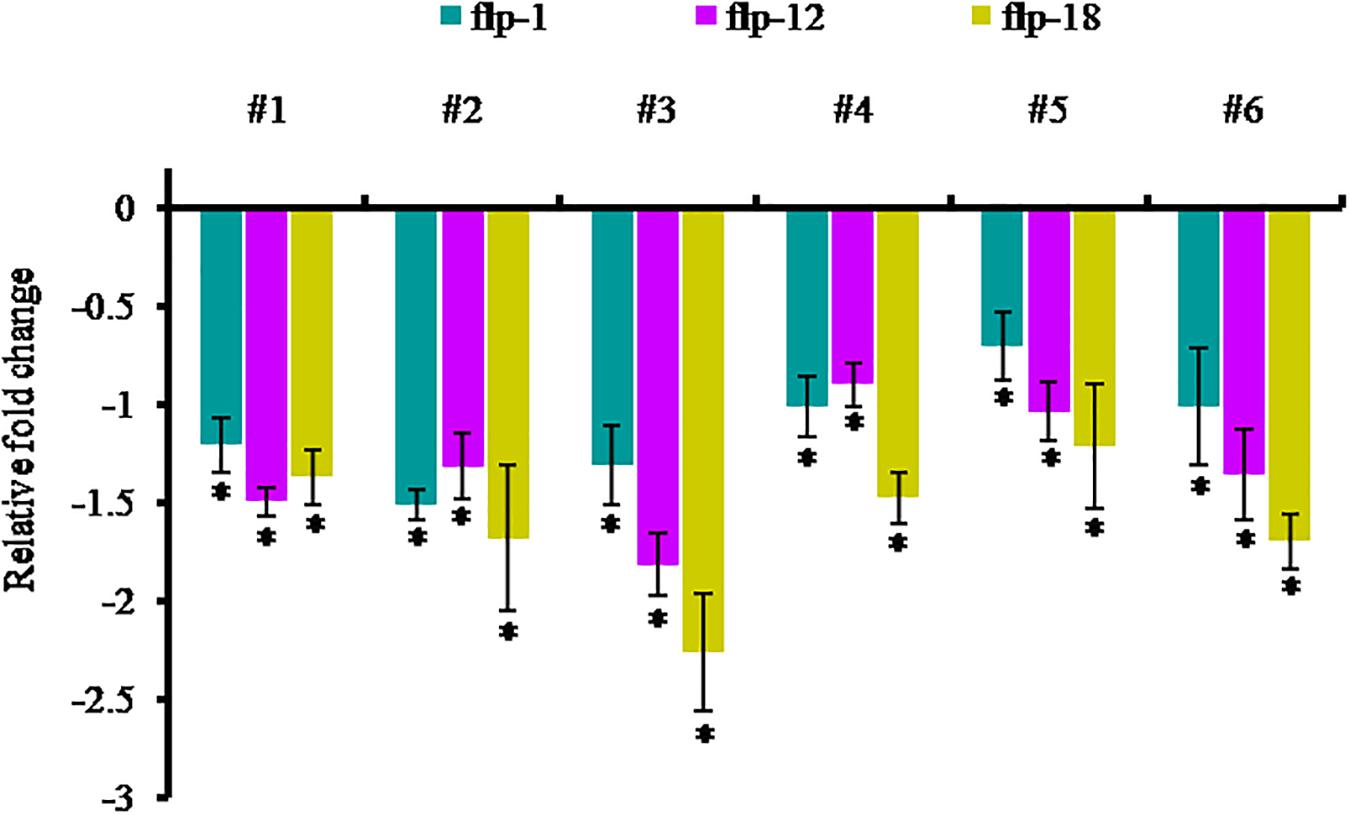
Figure 9. Transcript levels of Mi-flp1, Mi-flp12, and Mi-flp18 in M. incognita females extracted from dsRNA expressing transgenic events (#1 to #6: 25-6, 27-4, 28-9, 36-12, 38-5, and 47-2). Expression was measured as relative fold-change, analyzed using 2– Δ Δ CT method. Each bar represents the mean ± SE of n = 3, and asterisks specify the significant difference at P < 0.05.
Here, we have evaluated the role of three FLP genes of M. incognita, Mi-flp1, Mi-flp12, and Mi-flp18 singly and in combination for their effect on nematode reproduction and parasitism. Neuropeptides, mainly FLPs, are associated with neurosensory motor function, feeding, secretion, locomotion and reproduction necessary for parasitism (Maule et al., 2002; Kimber et al., 2007; Johnston et al., 2010). Thus, FLPs interference can be directed to disruption of several functions. The conservation of FLPs genes among nematodes and its probable role in locomotion and reproduction has attracted attention for exploitation of these neuromodulator genes as possible targets for nematode management which is environmentally friendly and target specific (Piggott et al., 2011; Peymen et al., 2014; Morris et al., 2017). Quite a few neuropeptides have been identified in PPNs, and FMRFamide-like immune reactivity has also been recorded in the nervous systems of Heterodera glycines, Globodera rostochiensis, G. pallida and M. incognita and M. graminicola (Atkinson et al., 1988; Maule et al., 2002; McVeigh et al., 2005; Kimber et al., 2007; Dalzell et al., 2010; Holden-Dye and Walker, 2011; Atkinson L. E. et al., 2013; Papolu et al., 2013; McCoy et al., 2014; Kumari et al., 2017; Warnock et al., 2017). So far, nineteen FLPs have been identified in the M. incognita and out of which six have been established to have transcriptional evidence (Abad et al., 2008) and host-delivered RNAi of Mi-flp14 and Mi-flp18 in N. tabacum provided excellent reduction in nematode parasitism and reproductive potential (Papolu et al., 2013).
In this direction, present work is focused on initial functional characterization of three flp genes, Mi-flp1, Mi-flp12, and Mi-flp18 of M. incognita singly and in combination employing a well established in vitro RNAi strategy to assess the comparative effect on nematode parasitism. Subsequently, three FLPs integrated into a fusion gene is used for silencing through host-delivered RNAi in tobacco to establish any synergistic or additive effects of the simultaneous silencing of three genes on M. incognita. The target dsRNA sequences were analyzed in dsCheck database to find out any off-target sites in the existing database, and no similarity was detected for the processed siRNAs. Thus, silencing of these genes possibly will not produce any off-target effects on another organisms and signifies the rational design of dsRNA molecule in turn to decrease the feasible risk.
The three FLPs transcripts were differentially expressed in different developmental stages of M. incognita and attain maximum expression in infective J2s, in contrast to basal level expression in eggs. Further, expression was down-regulated in J3/J4 and young females. Similarly, Papolu et al. (2013) observed higher expression of Mi-flp14 and Mi-flp18 in pre-parasitic J2s. These results re-emphasize the importance of FLPergic system in the initial parasitic process of M. incognita.
In situ hybridization assay in the present study revealed Mi-flp1 expression in ventral pharyngeal nerve cord near the metacorporeal bulb of M. incognita J2s. Expression of Mi-flp1 in some common nerve cells connecting the amphidial nerves to the central nervous system revealed its function similar to that of Mi-flp18. In G. pallida, Gp-flp1 expression was shown to be present in different nerve cell bodies’ viz., PHA, PVQ, LUA, PHB, ALN, and PVC compared to C. elegans. Two nerves (PHA and PHB) are ciliated and innervate the phasmids in the caudal region. ALN runs laterally from the tail to the nerve ring and has central synapses with motor-neurons innervating musculature in the head of C. elegans (Rogers et al., 2003). Our findings are in line with earlier reports in G. pallida by Kimber et al. (2002). Hence, it is possible that Mi-flp1 could have similar functional role in M. incognita. Previously, we had reported the expression site of Mi-flp12 and Mi-flp18 in the nerve cells, involved in the various biological and physiological process of M. incognita J2s (Banakar et al., 2015; Banakar and Rao, 2016). These findings support the involvement of the three FLPs in locomotion, host finding, reproduction and other physiological roles in the nematodes (White et al., 1986; Kimber et al., 2002, 2007; Rogers et al., 2003; Johnston et al., 2010; Kumari et al., 2017).
RNAi has been the mainstay tool for functional validation of genes in phytonematodes (Lilley et al., 2007; Dutta et al., 2015a, b). However, reports of gene silencing to explore the function of FLPergic molecules in plant nematodes are finite (Kimber et al., 2007; Dalzell et al., 2010; Atkinson N. J. et al., 2013; Papolu et al., 2013; Dong et al., 2014; Banakar et al., 2015). Here, we disrupted three individual FLPs and their combinatorial fusion gene through in vitro RNAi (dsRNA soaking) in M. incognita J2s which resulted in perturbed the expression of target transcripts of each of the FLPs. These transient natures of gene silencing effects detected in our study are in line with the earlier report of Kimber et al. (2007). Similarly, de Souza Júnior et al. (2013) reported the concurrent silencing of three proteases using a single dsRNA expression cassette that resulted in unexpected up-regulation of Mi-asp1 during combinatorial silencing of three genes (de Souza Júnior et al., 2013). Likewise, Bakhetia et al. (2008) found an increase in the dg13 expression during the combinatorial RNAi evaluation of dg13 and dg14 genes of H. glycines. The possible reason underlying an expected upregulated expression during both single and combinatorial silencing of three flps in the present work could be due to interaction between FLPs in different biological process. In-silico analysis using string database revealed that flp1, flp12, and flp18 are co-expressed in C. elegans, and flp18 together with flp1 plays a homeostatic role by acting on the GABAergic neural transmission at neuromuscular junctions to inhibit over excitation of the locomotor circuit. Further, we have demonstrated the successful processing of combinatorial dsRNA by northern blot analysis in the fusion dsRNA soaked worms that showed the presence of siRNA of each of the three flps in M. incognita J2s. This is the first established report in PPNs showing positive gene silencing evidence by northern blot hybridization and combinatorial dsRNA processing into siRNA.
The effect of in vitro silencing of Mi-flp1, Mi-flp12, Mi-flp18 and the fusion genes on attraction and penetration of J2s approaching tomato roots was examined on PF-127 and also in soil. Worms treated with dsRNA corresponding to both individual genes and fusion cassette markedly defer in level of attraction and penetration of J2s indicating that the target genes are important in early plant-nematode interactions. Fusion gene silencing is the most effective in terms of reduction of nematode attraction and penetration compared to single gene silenced and GFP dsRNA treated controls. This establishes the synergistic effect due to simultaneous knockdown of three FLPs particularly on chemo-sensation and host finding ability of the nematode. Similar phenotypic effects were observed due to RNAi silencing of gp-flp1, gp-flp12, and gp-flp18 in G. pallida that completely inhibited the migratory behavior after 24 h of incubation (Kimber et al., 2007). The significance of Mi-flp18 in M. incognita host finding (chemotaxis), migration and infection was also established through in vitro RNAi previously from our lab by Papolu et al. (2013), and Banakar et al. (2015) and the results are similar to the present finding. In C. elegans, analysis of flp18 and the related nerve cell mutants were defective in chemo-sensation, dauer formation, foraging and fat accumulation and also showed reduced consumption of oxygen (Albertson and Thompson, 1976; Avery and Horvitz, 1990; Avery, 1993; Tsalik and Hobert, 2003; Wakabayashi et al., 2004; Gray et al., 2005; Cohen et al., 2009). Here, the observed decrease in nematode migration and penetration could be attributed to the gene silencing effect of FLPs that interrupted neuromotor actions involved in olfaction, chemotaxis and infectivity.
The reduced infection after knockdown of individual FLPs and their fusion gene also resulted in considerable retardation of infection, development and multiplication of M. incognita in adzuki bean. Amongst treatments, the fusion gene was the most effective in reducing the infection as indicated by least number of total endoparasites in the roots and significantly affected the fecundity in reduction of eggs per egg mass. The result was comparable to the penetration assays on tomato roots. Our studies could be supported by Moffett et al. (2003) in C. elegans where flp1 inhibits rhythmic contractions of the ovijector, the organ that controls egg-laying when flp1 was tested in different concentrations on ovijector. These reports strongly suggest that Mi-flp1, Mi-flp12, and Mi-flp18 are involved in nematode perception of host plants, feeding and reproduction. Therefore, all three FLPs could be potential targets for future management of M. incognita.
On confirming the in vitro RNAi silencing of Mi-flp1, Mi-flp12, Mi-flp18 and fusion gene, additional proof for the possibility of simultaneous silencing of three genes and consequent effects on M. incognita was undertaken by in planta validation with a highly effective tobacco system. Using Agrobacterium-mediated genetic transformation, transgenic plants were generated expressing FLPs-fusion dsRNA. Molecular characterization confirmed the presence, integration and inheritance of the T-DNA harboring the hpRNA construct independently in six transformed lines. Though, the transcripts abundance at mRNA level of transgene measured by qRT-PCR was variable amongst events, signifying that the transgene was integrated at random sites in the plant genome. Additionally, resistance offered due to the expression of transgene was confirmed by bioefficacy analysis. Bioefficacy analysis indicated a pronounced decrease in M. incognita development and reproduction in most of the T1 lines compared to WT plants with up to 85% reductions in derived MF. The significant reduction of MF in transgenic lines has proved efficient RNAi silencing of FLPs-fusion gene. Earlier study by Papolu et al. (2013) demonstrated that tobacco plants expressing RNAi construct of the single gene Mi-flp18 had reduced the numbers of galls and final MF compared to WT plants. The present findings are better than the earlier reports in reducing the MF in transgenic lines and on par with other parameters observed in M. incognita. In the present study, silencing of more than one gene at a time with multiple roles had a higher impact on reducing the nematode parasitism in plants. The pyramiding expression of three FLP genes involved in more than one function produced synergistic effects by a detrimental effect on development of M. incognita. Our results are supported by the finding of Urwin et al. (1998), where transgenic Arabidopsis expressing a stacked gene construct having a cowpea trypsin inhibitor and a cystatin displayed improved resistance to H. schachtii. Similarly, a dsFusion RNAi construct having three proteases gene viz., Mi-ser1, Mi-cpl1, and Mi-asp1 interfere the nematode fecundity and successfully reduced disease burden in tobacco. These studies demonstrated the gene pyramiding in transgenic plants expressing more than one gene not only offer two defense systems across worms but also widen the spectrum of resistance to different nematode species (de Souza Júnior et al., 2013). Transgenic plants were morphologically similar and comparable to WT-plants, indicating the specificity of gene silencing.
In order to assess the host delivered silencing effect of Mi-flp1, Mi-flp12, and Mi-flp18 on nematode development, adult females were dissect out from T1 transgenic plants and WT-plants and subjected to qRT-PCR analysis. A considerable reduction (up to 2.2-fold) in gene expression was observed in the young females from transgenic plants compared to that from WT-plants. The current findings are is in confirmation with the previous reports showing inhibitory effect of flp1 and other 15 flp genes in modulating ovijector of A. suum by cessation of contractile activity of oviduct, affecting egg laying (Moffett et al., 2003). Loss of flp1 gene in C. elegans resulted in reduced locomotion and egg laying rate (Chang et al., 2015) that supports the findings of present work.
Although, there are some reports showing efficacy of FLPs for disrupting the neuromotor functions, to the best of our knowledge, this is the first report to demonstrate effectiveness of Mi-flp1 and Mi-flp12 and their fusion gene along with Mi-flp18 for the management of M. incognita using in vitro gene silencing as well as host-induced gene silencing. Significant reduction in nematode MF, due to knockdown of FLPs would be of enormous value to bring down the resident population pressure in soil for the successive crops. Given the transient nature of RNAi effects, it is practically not possible to attain absolute nematode resistance (Rosso et al., 2009). As RKNs complete 3–4 or more generations during a cropping season, about 60% decline in multiplication is adequate to reduce nematode population below the economic threshold (Fuller et al., 2008). Additionally, sustainable management can be accomplished by pyramiding few flp genes for efficient interference of various physiological processes requisite for successful completion of nematode lifecycle.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
UR conceived, designed, and supervised the experiments. PB and AH performed all the experiments. PP carried out Northern blot analysis. AH analyzed the data and wrote the original draft of the manuscript. UR, AH, and PB finalized the draft of the manuscript. All authors read and approved the final manuscript.
This work partly carried out by the Ph.D. student PB acknowledges the Post Graduate School, Indian Council of Agricultural Research-Indian Agricultural Research Institute, New Delhi for financial assistance for his doctoral degree. Funding from the Department of Biotechnology, Government of India to UR through grant number BT/PR5908/AGR/36/727/2012 “Translational research on root-knot nematode tolerant RNAi transgenics based on vital parasite gene targets from validation to proof-of-concept to selection of event(s) in the field under confined conditions” is acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the Director and the Joint Director (Research), ICAR-Indian Agricultural Research Institute, New Delhi, India, for extending all the support and facilities to complete the study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.573916/full#supplementary-material
Supplementary Figure 1 | Effect of in vitro RNAi on host invasion of M. incognita in soil. Penetration of dsRNA soaked worms into roots of tomato seedlings after 72 h of inoculation. Each bar represents the mean standard error (n = 6); bars with letters indicate significant difference at P > 0.05. Nematodes treated with GFP dsRNA (non-native control) and worms in water were used as controls.
Supplementary Figure 2 | (A) T-DNA region of binary vector used for FLPs-fusion gene silencing in tobacco cv. Petit Havana. RNAi suppression construct – pB7GWIWG2(II). Inverted repeats of fusion gene fragment were cloned into the destination vector. RB – T-DNA right border, LB – left border, bar – selectable marker gene, P35S – Cauliflower mosaic virus (CaMV) 35S promoter, T35S – CaMV 35S terminator, fusion gene – M. incognita FMRFamide like peptides (FLPs) fusion gene, attB1 and attB2 – LR reaction sites, (B) Agrobacterium- mediated transformation and regeneration of tobacco cv. Petit Havana. (a) Leaf explants of 1 cm2 cut from young tobacco leaves (b) callusing of co-cultivated explants (c) selection of transformed calli in presence of glufosinate (d) regeneration of selected explants (e) appearance of roots in rooting medium (f) hardened plants in glasshouse (g) successful plant growth and maturation (h) fruiting and seed setting.
Supplementary Figure 3 | PCR analysis of genomic DNA extracted from the primary transgenic plants harboring RNAi–fusion gene cassette. (A) Amplification of the target gene using gene specific primers, (B) Amplification of sense strand using primers 35S promoter forward and attB2 reverse, (C) Amplification of the antisense strand using primers 35S terminator forward and attB2 reverse, (D) Amplification of bar gene to confirm the presence and inheritance of dsRNA into transformed plants. M – 100 bp molecular marker. WT – negative control (WT-plant); Lanes 1–13- independent T0 events (25, 26, 27, 28, 36, 37, 38, 40, 47, 48, 52, 53, and 84).
Supplementary Figure 4 | PCR analysis of genomic DNA extracted from the T1 transgenic lines harboring RNAi–fusion gene cassette. M – 100 bp molecular marker; WT – negative control (WT-plant); Lanes 1–12 – show independent progenies of T0 events (25-6, 26-2, 27-4, 28-9, 36-12, 37-1, 38-5, 40-5, 47-2, 48-3, 52-7, and 53-1). (A) Target gene amplification, (B) bar gene amplification.
Abad, P., Gouzy, J., Aury, J. M., Castagnone-Sereno, P., Danchin, E. G., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi: 10.1038/nbt.1482
Abdeen, A., Virgos, A., Olivella, E., Villanueva, J., Aviles, X., Gabarra, R., et al. (2005). Multiple insect resistance in transgenic tomato plants over-expressing two families of plant proteinase inhibitors. Plant Mol. Biol. 57, 189–202. doi: 10.1007/s11103-004-6959-9
Albertson, D. G., and Thompson, J. N. (1976). The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275, 299–325. doi: 10.1098/rstb.1976.0085
Atkinson, H. J., and Harris, P. D. (1989). Changes in nematode antigens recognized by monoclonal antibodies during early infections of soya beans with the cyst nematode Heterodera glycines. Parasitology 98, 479–487. doi: 10.1017/S0031182000061576
Atkinson, H. J., Isaac, R. E., Harris, P. D., and Sharpe, C. M. (1988). FMRFamide-like immunoreactivity within the nervous system of the nematodes Panagrellus redivius, Caenorhabditis elegans and Heterodera glycines. J. Zool. 216, 663–671. doi: 10.1111/j.1469-7998.1988.tb02464.x
Atkinson, L. E., Stevenson, M., McCoy, C. J., Marks, N. J., Fleming, C., Zamanian, M., et al. (2013). flp-32 Ligand/receptor silencing phenocopy faster plant pathogenic nematodes. PLoS Pathog. 9:e1003169. doi: 10.1371/journal.ppat.1003169
Atkinson, N. J., Lilley, C. J., and Urwin, P. E. (2013). Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 162, 2028–2041. doi: 10.1104/pp.113.222372
Avery, L. (1993). Motor neuron M3 controls pharyngeal muscle relaxation timing in Caenorhabditis elegans. J. Exp. Biol. 175, 283–297.
Avery, L., and Horvitz, H. R. (1990). Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J. Exp. Zool. 253, 263–270. doi: 10.1002/jez.1402530305
Bakhetia, M., Urwin, P. E., and Atkinson, H. J. (2008). Characterisation by RNAi of pioneer genes expressed in the dorsal pharyngeal gland cell of Heterodera glycines and the effects of combinatorial RNAi. Int. J. Parasitol. 38, 1589–1597. doi: 10.1016/j.ijpara.2008.05.003
Banakar, P., and Rao, U. (2016). In-Situ hybridization for localization of FMRF amide like peptide mi-flp-12 in Meloidogyne incognita. Indian J. Nematol. 46, 145–150.
Banakar, P., Sharma, A., Lilley, C. J., Gantasala, N. P., Kumar, M., and Rao, U. (2015). Combinatorial in vitro RNAi of two neuropeptide genes and a pharyngeal gland gene on Meloidogyne incognita. Nematology 17, 155–167. doi: 10.1163/15685411-00002859
Bridge, J., Plowright, R. A., and Peng, D. (2005). “Nematode parasites of rice,” in Plant-Parasitic Nematodes in Subtropical and Tropical Agriculture, eds M. Luc, R. A. Sikora, and J. Bridge (Wallingford: CABI International), 87–130. doi: 10.1079/9780851997278.0087
Byrd, D. W. Jr., Kirkpatrick, T., and Barker, K. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143.
Chan, Y. L., He, Y., Hsiao, T. T., Wang, C. J., Tian, Z., and Yeh, K. W. (2015). Pyramiding taro cystatin and fungal chitinase genes driven by a synthetic promoter enhances resistance in tomato to root-knot nematode Meloidogyne incognita. Plant Sci. 231, 74–81. doi: 10.1016/j.plantsci.2014.11.010
Chang, Y. J., Burton, T., Ha, L., Huang, Z., Olajubelo, A., and Li, C. (2015). Modulation of locomotion and reproduction by FLP neuropeptides in the nematode Caenorhabditis elegans. PLoS One 10:e0135164. doi: 10.1371/journal.pone.0135164
Chaudhary, S., Dutta, T. K., Tyagi, N., Shivakumara, T. N., Papolu, P. K., Chobhe, K. A., et al. (2019). Host-induced silencing of Mi-msp-1 confers resistance to root-knot nematode Meloidogyne incognita in eggplant. Transgenic Res. 28, 327–340. doi: 10.1007/s11248-019-00126-5
Cohen, M., Reale, V., Olofsson, B., Knights, A., Evans, P., and de Bono, M. (2009). Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 9, 375–385. doi: 10.1016/j.cmet.2009.02.003
Dalzell, J. J., McMaster, S., Fleming, C. C., and Maule, A. G. (2010). Short interfering RNA-mediated gene silencing in Globoderapallida and Meloidogyne incognita infective stage juveniles. Int. J. Parasitol. 40, 91–100. doi: 10.1016/j.ijpara.2009.07.003
Dalzell, J. J., McMaster, S., Johnston, M. J., Kerr, R., Fleming, C. C., and Maule, A. G. (2009). Non-nematode-derived double-stranded RNAs induce profound phenotypic changes in Meloidogyne incognita and Globoderapallida infective juveniles. Int. J. Parasitol. 39, 1503–1516. doi: 10.1016/j.ijpara.2009.05.006
Danchin, E. G., Arguel, M. J., Campan-Fournier, A., Perfus-Barbeoch, L., Magliano, M., Rosso, M. N., et al. (2013). Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PLoS Pathog. 9:e1003745. doi: 10.1371/journal.ppat.1003745
de Souza Júnior, J. D. A., Coelho, R. R., Lourenço, I. T., da Rocha Fragoso, R., Viana, A. A. B., de Macedo, L. L. P., et al. (2013). Knocking-down Meloidogyne incognita proteases by plant-delivered dsRNA has negative pleiotropic effect on nematode vigor. PLoS One 8:e85364. doi: 10.1371/journal.pone.0085364
De Waele, D., Das, K., Zhao, D., Tiwari, R. K. S., Shrivastava, D. K., Vera-Cruz, C., et al. (2013). Host response of rice genotypes to the rice root-knot nematode (Meloidogyne graminicola) under aerobic soil conditions. Arch. Phytopathol. Plant Protect. 46, 670–681. doi: 10.1080/03235408.2012.749702
Dong, L., Li, X., Huang, L., Gao, Y., Zhong, L., Zheng, Y., et al. (2014). Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J. Exp. Bot. 65, 131–141. doi: 10.1093/jxb/ert356
Dutta, T. K., Banakar, P., and Rao, U. (2015a). The status of RNAi-based transgenic research in plant nematology. Front. Microbiol. 5:760. doi: 10.3389/fmicb.2014.00760
Dutta, T. K., Papolu, P. K., Banakar, P., Choudhary, D., Sirohi, A., and Rao, U. (2015b). Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front. Microbiol. 6:260. doi: 10.3389/fmicb.2015.00260
Elling, A. A. (2013). Major emerging problems with minor Meloidogyne species. Phytopathology 103, 1092–1102. doi: 10.1094/PHYTO-01-13-0019-RVW
Fairbairn, D. J., Cavallaro, A. S., Bernard, M., Mahalinga-Iyer, J., Graham, M. W., and Botella, J. R. (2007). Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226, 1525–1533. doi: 10.1007/s00425-007-0588-x
Faske, T. R., and Hurd, K. (2015). Sensitivity of Meloidogyne incognita and Rotylenchulus reniformis to fluopyram. J. Nematol. 47, 316–321.
Fuller, V. L., Lilley, C. J., and Urwin, P. E. (2008). Nematode resistance. New Phytol. 180, 27–44. doi: 10.1111/j.1469-8137.2008.02508.x
Gouda, K., Matsunaga, Y., Iwasaki, T., and Kawano, T. (2010). An altered method of feeding RNAi that knocks down multiple genes simultaneously in the nematode Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 74, 2361–2365. doi: 10.1271/bbb.100579
Gray, J. M., Hill, J. J., and Bargmann, C. I. (2005). A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 102, 3184–3191. doi: 10.1073/pnas.0409009101
Hada, A., Kumari, C., Phani, V., Singh, D., Chinnusamy, V., and Rao, U. (2020). Host-induced silencing of FMRFamide-like peptide genes, flp-1 and flp-12, in rice impairs reproductive fitness of the root-knot nematode Meloidogyne graminicola. Front. Plant Sci. 11:894. doi: 10.3389/fpls.2020.00894
Holden-Dye, L., and Walker, R. J. (2011). Neurobiology of plant parasitic nematodes. Invert. Neurosci. 11, 9–19. doi: 10.1007/s10158-011-0117-2
Husson, S. J., Mertens, I., Janssen, T., Lindemans, M., and Schoofs, L. (2007). Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog. Neurobiol. 82, 33–55. doi: 10.1016/j.pneurobio.2007.01.006
Ibrahim, H. M., Hosseini, P., Alkharouf, N. W., Hussein, E. H., Abd El Kader, Y., Aly, M. A., et al. (2011). Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics 12:220. doi: 10.1186/1471-2164-12-220
Johnston, M. J. G., McVeigh, P., McMaster, S., Fleming, C. C., and Maule, A. G. (2010). FMRFamide-like peptides in root knot nematodes and their potential role in nematode physiology. J. Helminthol. 84, 253–265. doi: 10.1017/S0022149X09990630
Jones, J. T., Haegeman, A., Danchin, E. G., Gaur, H. S., Helder, J., Jones, M. G., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Kearn, J., Ludlow, E., Dillon, J., O’Connor, V., and Holden-Dye, L. (2014). Fluensulfone is a nematicide with a mode of action distinct from anticholinesterases and macrocyclic lactones. Pestic. Biochem. Physiol. 109, 44–57. doi: 10.1016/j.pestbp.2014.01.004
Kimber, M. J., Fleming, C. C., Prior, A., Jones, J. T., Halton, D. W., and Maule, A. G. (2002). Localisation of Globodera pallida FMRFamide-related peptide encoding genes using in situ hybridisation. Int. J. Parasitol. 32, 1095–1105. doi: 10.1016/S0020-7519(02)00084-X
Kimber, M. J., McKinney, S., McMaster, S., Day, T. A., Fleming, C. C., and Maule, A. G. (2007). flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J. 21, 1233–1243. doi: 10.1096/fj.06-7343com
Kumari, C., Dutta, T. K., Chaudhary, S., Banakar, P., Papolu, P. K., and Rao, U. (2017). Molecular characterization of FMRFamide-like peptides in Meloidogyne graminicola and analysis of their knockdown effect on nematode infectivity. Gene 619, 50–60. doi: 10.1016/j.gene.2017.03.042
Lahm, G. P., Desaeger, J., Smith, B. K., Pahutski, T. F., Rivera, M. A., Meloro, T., et al. (2017). The discovery of fluazaindolizine: a new product for the control of plant parasitic nematodes. Bioorg. Med. Chem. Lett. 27, 1572–1575. doi: 10.1016/j.bmcl.2017.02.029
Lilley, C. J., Bakhetia, M., Charlton, W. L., and Urwin, P. E. (2007). Recent progress in the development of RNA interference for plant parasitic nematodes. Mol. Plant Pathol. 8, 701–711. doi: 10.1111/j.1364-3703.2007.00422.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Magrane, M., and UniProt Consortium (2011). UniProt knowledgebase: a hub of integrated protein data. Database 2011:bar009. doi: 10.1093/database/bar009
Maule, A. G., Mousley, A., Marks, N. J., Day, T. A., Thompson, D. P., Geary, T. G., et al. (2002). Neuropeptide signaling systems-potential drug targets for parasite and pest control. Curr. Top. Med. Chem. 2, 733–758. doi: 10.2174/1568026023393697
McCarter, J. P., Bird, D. M., and Mitreva, M. (2005). Nematode gene sequences: update for December 2005. J. Nematol. 37:417.
McCoy, C. J., Atkinson, L. E., Zamanian, M., McVeigh, P., Day, T. A., Kimber, M. J., et al. (2014). New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteom. 3, 262–272. doi: 10.1016/j.euprot.2014.04.002
McVeigh, P., Geary, T. G., Marks, N. J., and Maule, A. G. (2006). The FLP-side of nematodes. TRENDS Parasitol. 22, 385–396. doi: 10.1016/j.pt.2006.06.010
McVeigh, P., Leech, S., Mair, G. R., Marks, N. J., Geary, T. G., and Maule, A. G. (2005). Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int. J. Parasitol. 35, 1043–1060. doi: 10.1016/j.ijpara.2005.05.010
Min, K., Kang, J., and Lee, J. (2010). A modified feeding RNAi method for simultaneous knockdown of more than one gene in Caenorhabditis elegans. Biotechniques 48, 229–232. doi: 10.2144/000113365
Moffett, C. L., Beckett, A. M., Mousley, A., Geary, T. G., Marks, N. J., Halton, D. W., et al. (2003). The ovijector of Ascaris suum: multiple response types revealed by Caenorhabditis elegans FMRFamide-related peptides. Int. J. Parasitol. 33, 859–876. doi: 10.1016/S0020-7519(03)00109-7
Morris, R., Wilson, L., Sturrock, M., Warnock, N. D., Carrizo, D., Cox, D., et al. (2017). A neuropeptide modulates sensory perception in the entomopathogenic nematode Steinernema carpocapsae. PLoS Pathog. 13:e1006185. doi: 10.1371/journal.ppat.1006185
Narusaka, M., Hatakeyama, K., Shirasu, K., and Narusaka, Y. (2014). Arabidopsis dual resistance proteins, both RPS4 and RRS1, are required for resistance to bacterial wilt in transgenic Brassica crops. Plant Signal. Behav. 9:e29130. doi: 10.4161/psb.29130
Niu, J., Jian, H., Xu, J., Chen, C., Guo, Q., Liu, Q., et al. (2012). RNAi silencing of the Meloidogyne incognita Rpn7 gene reduces nematode parasitic success. Eur. J. Plant Pathol. 134, 131–144. doi: 10.1007/s10658-012-9971-y
Papolu, P. K., Gantasala, N. P., Kamaraju, D., Banakar, P., Sreevathsa, R., and Rao, U. (2013). Utility of host delivered RNAiof two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode, Meloidogyne incognita. PLoS One 8:e80603. doi: 10.1371/journal.pone.0080603
Peymen, K., Watteyne, J., Frooninckx, L., Schoofs, L., and Beets, I. (2014). The FMRFamide-like peptide family in nematodes. Front. Endocrinol. 5:90. doi: 10.3389/fendo.2014.00090
Piggott, B. J., Liu, J., Feng, Z., Wescott, S. A., and Xu, X. S. (2011). The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell 147, 922–933. doi: 10.1016/j.cell.2011.08.053
Quilis, J., López-García, B., Meynard, D., Guiderdoni, E., and San Segundo, B. (2014). Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant Biotechnol. J. 12, 367–377. doi: 10.1111/pbi.12143
Rao, U., Thakur, P. K., Prasad, N., Banakar, P., and Kumar, M. (2013). Identification of neuropeptides, flp-1 and flp-12 targeting neuromuscular system of rice root knot nematode (RRKN) Meloidogyne graminicola. Bioinformation 9, 182–186. doi: 10.6026/97320630009182
Roderick, H., Tripathi, L., Babirye, A., Wang, D., Tripathi, J., Urwin, P. E., et al. (2012). Generation of transgenic plantain (Musa spp.) with resistance to plant pathogenic nematodes. Mol. Plant Pathol. 13, 842–851. doi: 10.1111/j.1364-3703.2012.00792.x
Roderick, H., Urwin, P. E., and Atkinson, H. J. (2018). Rational design of biosafe crop resistance to a range of nematodes using RNA interference. Plant Biotechnol. J. 16, 520–529. doi: /10.1111/pbi.12792
Rogers, C., Reale, V., Kim, K., Chatwin, H., Li, C., Evans, P., et al. (2003). Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat. Neurosci. 6, 1178–1185. doi: 10.1038/nn1140
Rosso, M. N., Dubrana, M. P., Cimbolini, N., Jaubert, S., and Abad, P. (2005). Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microbe Interact. 18, 615–620. doi: 10.1094/MPMI-18-0615
Rosso, M. N., Jones, J. T., and Abad, P. (2009). RNAi and functional genomics in plant parasitic nematodes. Annu. Rev. Phytopathol. 47, 207–232. doi: 10.1146/annurev.phyto.112408.132605
Sambrook, J., Fristch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Shingles, J., Lilley, C. J., Atkinson, H. J., and Urwin, P. E. (2007). Meloidogyne incognita: molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp. Parasitol. 115, 114–120. doi: 10.1016/j.exppara.2006.07.008
Shivakumara, T. N., Papolu, P. K., Dutta, T. K., Kamaraju, D., Chaudhary, S., and Rao, U. (2016). RNAi-induced silencing of an effector confers transcriptional oscillation in another group of effectors in the root-knot nematode, Meloidogyne incognita. Nematology 18, 857–870. doi: 10.1163/15685411-00003003
Shivakumara, T. N., Somvanshi, V. S., Phani, V., Chaudhary, S., Hada, A., Budhwar, R., et al. (2019). Meloidogyne incognita (Nematoda: Meloidogynidae) sterol-binding protein Mi-SBP-1 as a target for its management. Int. J. Parasitol. 49, 1061–1073. doi: 10.1016/j.ijpara.2019.09.002
Slomczynska, U., South, M. S., Bunkers, G., Edgecomb, D., Wyse-Pester, D., Selness, S., et al. (2014). “Tioxazafen: A new broad-spectrum seed treatment nematicide,” in ACS Symposium Series, Vol. 1204, Discovery and Synthesis of Crop Protection Products, eds P. Maienfisch and T. M. Stevenson (Chesterfield, MO: Monsanto Corporation), 129–147. doi: 10.1021/bk-2015-1204.ch010
Starr, J. L., Bridge, J., and Cook, R. (2002). “Resistance to plant-parasitic nematodes: history, current use and future potential,” in Plant resistant to parasitic nematodes, eds J. L. Starr, R. Cook, and J. Bridge (Wallingford: CAB International), 1–22. doi: 10.1079/9780851994666.0001
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tischler, J., Lehner, B., Chen, N., and Fraser, A. G. (2006). Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 7:R69. doi: 10.1186/gb-2006-7-8-r69
Tripathi, J. N., Lorenzen, J., Bahar, O., Ronald, P., and Tripathi, L. (2014). Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotechnol. J. 12, 663–673. doi: 10.1111/pbi.12170
Trudgill, D. L., and Blok, V. C. (2001). Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39, 53–77. doi: 10.1146/annurev.phyto.39.1.53
Tsalik, E. L., and Hobert, O. (2003). Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J. Neurobiol. 56, 178–197. doi: 10.1002/neu.10245
Turra, D., Bellin, D., Lorito, M., and Gebhardt, C. (2009). Genotype-dependent expression of specific members of potato protease inhibitor gene families in different tissues and in response to wounding and nematode infection. J. Plant Physiol. 166, 762–774. doi: 10.1016/j.jplph.2008.10.005
Tyagi, N., Sharma, A., Kamaraju, D., Banakar, P., and Rao, S. B. (2013). Soil less medium for culturing and multiplication of root knot and cyst nematodes for basic and applied studies. Ann. Plant Protect. Sci. 21, 148–153.
Urwin, P. E., Lilley, C. J., and Atkinson, H. J. (2002). Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 15, 747–752. doi: 10.1094/MPMI.2002.15.8.747
Urwin, P. E., McPherson, M. J., and Atkinson, H. J. (1998). Enhanced transgenic plant resistance to nematodes by dual proteinase inhibitor constructs. Planta 204, 472–479. doi: 10.1007/s004250050281
Wakabayashi, T., Kitagawa, I., and Shingai, R. (2004). Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci. Res. 50, 103–111. doi: 10.1016/j.neures.2004.06.005
Walawage, S. L., Britton, M. T., Leslie, C. A., Uratsu, S. L., Li, Y., and Dandekar, A. M. (2013). Stacking resistance to crown gall and nematodes in walnut rootstocks. BMC Genomics 14:668. doi: 10.1186/1471-2164-14-668
Wang, C., Bruening, G., and Williamson, V. M. (2009). Determination of preferred pH for root-knot nematode aggregation using pluronic F-127 gel. J. Chem. Ecol. 35, 1242–1251. doi: 10.1007/s10886-009-9703-8
Warnock, N. D., Wilson, L., Patten, C., Fleming, C. C., Maule, A. G., and Dalzell, J. J. (2017). Nematode neuropeptides as transgenic nematicides. PLoS Pathog. 13:e1006237. doi: 10.1371/journal.ppat.1006237
White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340. doi: 10.1098/rstb.1986.0056
Keywords: root-knot nematodes, FMRFamide-like peptides, neuromotor functions, double-stranded RNA, combinatorial RNAi, transgenic plants
Citation: Banakar P, Hada A, Papolu PK and Rao U (2020) Simultaneous RNAi Knockdown of Three FMRFamide-Like Peptide Genes, Mi-flp1, Mi-flp12, and Mi-flp18 Provides Resistance to Root-Knot Nematode, Meloidogyne incognita. Front. Microbiol. 11:573916. doi: 10.3389/fmicb.2020.573916
Received: 18 June 2020; Accepted: 06 October 2020;
Published: 23 October 2020.
Edited by:
Gustavo Santoyo, Universidad Michoacana de San Nicolás de Hidalgo, MexicoReviewed by:
Akanksha Singh, Central Institute of Medicinal and Aromatic Plants, Council of Scientific and Industrial Research (CSIR), IndiaCopyright © 2020 Banakar, Hada, Papolu and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uma Rao, dW1hcmFvQGlhcmkucmVzLmlu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.