- College of Biological and Pharmaceutical Engineering, Wuhan Polytechnic University, Wuhan, China
Pseudomonas is one of the most diverse bacterial genera identified in the environment. Genome sequence analysis has indicated that this genus can be clustered into three lineages and ten groups. Each group can adopt different mechanisms to thrive under zinc-depleted or high-zinc conditions, two environments that are frequently encountered during their environmental propagation. The response of three prominent Pseudomonas strains (Pseudomonas aeruginosa PAO1, Pseudomonas putida KT2440, and Pseudomonas fluorescens ATCC 13525T) to minimal inhibitory concentrations of zinc were compared using RNA-seq and ultra-performance liquid chromatography–tandem mass spectrometry analysis. Results demonstrated that the three strains shared only minimal similarity at the transcriptional level. Only four genes responsible for zinc efflux were commonly upregulated. P. aeruginosa PAO1 specifically downregulated the operons involved in siderophore synthesis and the genes that encode ribosomal protein, while upregulated the genes associated with antibiotic efflux and cell envelope biosynthesis. The membrane transporters in P. putida KT2440 were globally downregulated, indicating changes in cell permeability. Compared with P. aeruginosa PAO1 and P. putida KT2440, the most remarkable transcriptional variation in P. fluorescens ATCC 13525T is the significant downregulation of the type VI secretion system. Metabolite quantitative analysis showed that low concentrations of the metabolites involved in central carbon metabolism and amino acid synthesis were detected in the three strains. In summary, the cellular responses of the three strains under high-zinc condition is quite divergent. Although similar metal efflux systems were upregulated, the three strains employed different pathways to reduce zinc intrusion. In addition, zinc treatment can increase the difficulties of scavenging P. aeruginosa from its colonization area, and reduce the competitiveness of P. fluorescens in microbiota.
Introduction
Zn2+ is the second most abundant essential metal ion after Fe2+/3+ in all three kingdoms of life, which directly interacts with RNA polymerase, zinc-finger proteins and superoxide dismutase to maintain their functionality (Hobman et al., 2007; Watły et al., 2016; Gonzalez et al., 2018). Free Zn2+ in the cytoplasm is normally maintained at the femtomolar to picomolar level (Outten and O'Halloran, 2001). Once this threshold is surpassed, excess Zn2+ can replace other metal ions from proteins and alter the stability of biomolecules (Krezel and Maret, 2016; Peng et al., 2018). Although zinc is a redox-inactive metal under normal conditions, high levels of intracellular zinc clearly induce the generation of reactive oxygen species (ROS) as evidenced by the increased amount of oxidized protein (Alhasawi et al., 2014). In nature, the geological background level of zinc is low (Cheng, 2003). However, anthropogenic industries and domestic activities directly or indirectly release vast amounts of Zn2+ into the environment. In developing countries, mining activity coupled with poor effluent disposal and management often caused severe zinc pollution in soils or sediments. For instance, Vasile et al. (2008) analyzed the zinc partition in sediments taken from a river subjected of intense mining activities, and found that the easily soluble zinc ranged from 80 to 300 mg Kg−1. Klimek (2012) reported that the averaged concentration of water soluble zinc in the forest soils surrounded by industrial area achieved 82.30 mg Kg−1. With the progress of urbanization, large numbers of municipal wastewater treatment plants have been built. The sewage sludge is another material that accumulates zinc ions. Tu et al. (2012) used a four-step extraction method to extract heavy metals in municipal sludges from seven individual wastewater treatment plants. The results showed that zinc in some sludges is highly bioavailable (the water soluble zinc concentration reached 61.42±7.0 mg Kg−1). Fertilizing use of these sludges may cause severe zinc pollution in soils.
Prokaryotes are single cell organisms that have direct contact with their surrounding environment. Thus, these organisms are extremely vulnerable to environmental stresses (Blencowe and Morby, 2003). In response to metal stress, many bacteria have evolved numerous mechanisms, including mobilization, precipitation, biosorption and biogeochemical cycling, to respond to the adverse effects of metal ions (Singh et al., 2018). Pseudomonas is a ubiquitous bacterial genus with high adaptability to various environments (Santos et al., 2004). Many Pseudomonas species exhibit high capability to maintain the equilibrium of cytoplasmic metal ions, which directly affects metal availability and distribution (Haritha et al., 2008; Ramos and Filloux, 2010). In our previous study, the transcriptional responses of P. putida KT2440 to low, intermediate, and high levels of extracellular zinc stresses were thoroughly analyzed (Peng et al., 2018). The results showed that genes associated with metal transport and membrane homeostasis were significantly induced at the lowest zinc level. As zinc concentrations increased, the respiration chains, amino acids and carbon source metabolisms, as well as the biogenesis of Fe-S clusters were adjusted. However, Pseudomonas comprises ~200 species (Kohlstedt and Wittmann, 2019), which exhibit considerable diversity in their ecological habitat and physiology (Peix et al., 2017; Singh et al., 2018). Recent phylogenetic analysis has clustered Pseudomonas into three lineages and ten groups (Peix et al., 2017). Each Pseudomonas group probably uses different way to alleviate metal-induced intracellular disorder. A comparison of our transcriptome data with the results obtained by Alhasawi et al. (2014) showed partial differences between the response of P. putida and P. fluorescens to zinc. For example, the expression of NADPH-generating enzymes, including isocitrate dehydrogenase-NADP+, malic enzyme (ME), and glucose-6-phosphate dehydrogenase (G6PDH), was markedly increased in P. fluorescens. Meanwhile, the most upregulated NADPH-generating enzyme in P. putida KT2440 was ferredoxin-NADP+ reductase and the transcription of ME and G6PDH remained unchanged. In addition to P. fluorescens and P. putida, P. aeruginosa represents another important phylogenetic group in the Pseudomonas genus (Jensen et al., 2004; Peix et al., 2017). As a widespread environmental bacterium and an opportunistic animal pathogen, P. aeruginosa also encounters changing levels of zinc concentrations (Pederick et al., 2015). To date, studies on zinc homeostasis in P. aeruginosa mainly concentrated on zinc efflux and zinc associated virulence or antibiotic resistance (Braymer and Giedroc, 2014; Pederick et al., 2015; Gonzalez et al., 2018), other cellular responses remain poorly understood.
In the current study, three prominent Pseudomonas strains (P. putida KT2440, P. aeruginosa PAO1, and P. fluorescens ATCC 13525T) were challenged with external zinc at minimal inhibitory concentrations (MICs). Their responses were compared to provide a comprehensive understanding of the genes, the cellular processes and the cellular molecules that play crucial roles in zinc adaptation.
Materials and Methods
Bacterial Strains, Growth Conditions, and MIC Value Determination
P. putida KT2440 (ATCC 47054), P. aeruginosa PAO1 (ATCC 47085), and P. fluorescens ATCC 13525T were obtained from American Type Culture Collection (ATCC, Manassa, Virginia, USA). These strains were routinely grown in a semi-synthetic cation-defined medium (CDM) (Pederick et al., 2015) at their optimum growth temperatures (30°C for P. putida KT2440 and P. fluorescens ATCC 13525T, 37 °C for P. aeruginosa PAO1). The MIC values of zinc ions were determined as described by Mergeay et al. (1985). The exponential cells of the three strains were serially diluted and then spread onto CDM agar plates containing zinc sulfate from 0.2 mmol L−1 to 2.0 mmol L−1 (0.1 mmol L−1 interval). The MIC values were defined as the lowest concentrations of zinc sulfate at which no colonies were observed after 48 h of incubation at optimum temperature. For zinc treatment, the exponential cells grown in the CDM culture were harvested via centrifugation and inoculated into new CDM containing zinc sulfate at MIC to achieve a cell density of 107 CFU mL−1. After zinc treatment for 1 h, the cells were stabilized using RNA protect bacterial reagent (Qiagen, Valencia, CA, USA) at a ratio of 2:1 (2 mL of reagent per 1 mL of culture). The control groups were grown in CDM without zinc. Three biological replicates were performed for each treatment.
RNA Extraction and RNA-seq Analysis
The total RNAs was extracted separately from each replicate using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Residual genomic DNA contamination was removed from the total RNA samples via DNAase I digestion. The quality and quantity of total RNA was measured using an Agilent 2100 Bioanalyser. RNA-seq libraries were prepared and sequenced at the Beijing Genomics Institute (Beijing, China). 150 bp pair-end reads were sequenced using an Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA).
Raw data generated from Illumina sequencing platform were trimmed using Skewer version 0.2.2 (Jiang et al., 2014). After Quality control (Peng et al., 2018), Bowtie 2 version 2.2.3 (Langmead and Salzberg, 2012) was used to align the remaining reads to the Pseudomonas genomes (Winsor et al., 2015). The RPKM (reads per kilobase of transcript per million mapped reads) values were used for calculating gene expression level using DESeq2 version 2.2.2 (Love et al., 2014). DESeq generated all pairwise comparisons of treatments and associated adjusted P-values (Padj) controlling for the false discovery rate. Genes with average absolute fold changes higher than 2.0 and Padj values <0.01 were classified as differentially expressed genes (DEGs). The functional clusters of orthologous groups classification of all DEGs was performed automatically using the Integrated Microbial Genomes portal (Markowitz et al., 2013).
Validation of RNA-seq Results via RT-qPCR
To verify the validity of the RNA-seq data, RT-qPCR was performed with the same RNA samples used for RNA-seq analysis. Six genes were selected from each Pseudomonas strain. Therefore, a total of 18 genes were used for validation. The gene-specific primer pairs were designed using Beacon designer (Supplementary Table 1). The expression levels of the selected genes were calculated through the 2−ΔΔCt method (Livak and Schmittgen, 2001). For cDNA analysis, 1 μg of RNA was reverse-transcribed using PrimeScript RT reagent kit (Takara, Dalian, China) and random hexamer primers following the manufacture's protocol. The quantities of cDNA were analyzed using the qPCR-SYBR Premix ExTaq (Takara, Dalian, China) in a Bio-Rad iCycler (Bio-Rad, Berkeley, CA, USA). PCR conditions were 95°C for 1 min, followed by 40 cycles of 95°C for 15 s, annealing temperature for 15 s, and 72°C for 15 s. A melting curve analysis was then performed at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. Shapiro-Wilk test (SPSS 20.0, Chicago, IL, USA) was conducted for the fold-change values obtained from the RNA-seq and RT-qPCR (Gómez-Sagasti et al., 2015). Given that all data followed a normal distribution, Pearson's coefficients were calculated to illustrate the correlation level between two data groups.
Metabolite Extraction and UPLC-MS/MS Analysis
The bacterial samples used for the metabolome analysis were prepared as described by Chavez-Dozal et al. (2015). Zinc-treated Pseudomonas cells were collected from each biological replicate via centrifugation at 10,000 × g for 5 min at 4°C. After washing two times with ice-cold phosphate-buffered saline (pH 7.4), the cell pellets were frozen with liquid nitrogen. To extract the metabolites, 50 mg of the frozen samples were dispersed in 1 mL of liquid containing methanol/acetonitrile/water (2:2:1), sonicated for 10 min at 0°C, and then centrifuged at 14,000 × g for 20 min at 4°C. The resulting supernatants were lyophilised and reconstituted in 100 μL of acetonitrile/water (1:1) before UPLC-MS/MS analysis.
An Agilent 1290 HPLC system (Agilent, Santa Clara, CA, USA) equipped with an HILIC analytical column (amide 1.7 μm, 2.1 × 100 mm, Waters, Milford, MA, USA) was used to analyse the extract of each sample. The mobil phase was water (containing 25 mmol L−1 CH3COONH4 and 25 mmol L−1 H2O, Solvent A), and acetonitrile (Solvent B). The dilution programme was operated for 0.5 min 95:5 (B:A) then switched to 40:60 for 8.5 min before returning to 95:5 for 9 min further. The mass spectra were recorded by an Agilent 6540 Q-TOF mass spectrometer operating in the positive and negative ion modes using an Agilent electrospray ionization source. After each spectrum was normalized to the total spectral intensity, the profile data were imported into SIMCA-P+ software (Umetrics AB, Umeå, Sweden) and pareto-scaled to perform orthogonal partial least squares-discriminate analysis (OPLS-DA). The significance of the metabolites was ranked using the variable importance in projection score from the OPLS-DA model. For the univariate analysis of a specific metabolite, statistical significance was determined using Student's two-sample t-test. The metabolites with average absolute fold changes higher than 2.0 and Padj values <0.05 were defined as statistically different.
Results
MICs of Zinc Sulfate
Given that metal ions exert does-dependent effects on bacterial cells, comparing the cellular responses of different strains at the same zinc stress level is important. In this study, the MIC of ZnSO4 was used in the treatment. P. putida KT2440 was highly tolerant to zinc and exhibited a zinc MIC value of up to 1.1 mmol L−1. P. fluorescens was the least zinc-tolerant strain with an MIC value of approximately 0.4 mmol L−1. P. aeruginosa PAO1 had intermediate levels of zinc tolerance. A cation-defined medium (CDM) plate that contained more than 0.7 mmol L−1 zinc completely inhibited its colony formation. Therefore, in the following RNA-seq and UPLC-MS/MS analysis, 0.7, 1.1, and 0.4 mmol L−1 zinc sulfate were used to compare the zinc-induced cellular responses in P. aeruginosa PAO1, P. putida KT2440, and P. fluorescens ATCC 13525T.
Global Transcription Features of the Three Pseudomonas Strains
The numbers of differentially expressed genes (DEGs) during the zinc treatment of P. aeruginosa PAO1, P. putida KT2440, and P. fluorescens ATCC 13525T are provided in Table 1. The comparison of the zinc-treated P. aeruginosa PAO1 with their control samples identified 529 genes that were differently transcribed. Amongst which, 406 genes, 7.1% of the total coding DNA sequence (CDS) in the genome, were upregulated, whereas, 123 (2.2%) genes were downregulated. The comparison of the zinc-treated P. putida KT2440 with their control bioreplicates showed that, the expression of 607 genes was significantly altered, with the majority (344 genes, 6.0%) displaying significant induction in transcription compared with those (263 genes, 4.6%) demonstrating decreased transcription. The lowest number of DEGs was observed in P. fluorescens ATCC 13525T, with only 48 induced genes and 19 repressed genes (Table 1). In general, KEGG analysis showed that large quantities of DEGs identified in P. aeruginosa PAO1 and P. putida KT2440 can be clustered into the metabolism class (Figure 1A), particularly the carbohydrate and energy metabolism subclasses. As an opportunistic animal pathogen, P. aeruginosa PAO1 also regulated many genes belonging to the human disease class. The cellular response of P. fluorescens ATCC 13525T differed from those of P. aeruginosa and P. putida, in which ~50% of DEGs with clear KEGG classifications were categorized into the environmental information processing group. To better compared the cellular responses occurred in the three Pseudomonas strains, the DNA sequence of the DEGs was extracted from each genome, and then blast against the DEGs identified in the other two genomes with a match cutoff of 70% and an E-value exponent cutoff of 1-e5. As shown in Figure 1B, only four orthologs were commonly regulated by the three Pseudomonas strains, and 1044 genes were strain-specifically regulated. The transcriptional patterns of P. aeruginosa and P. putida presented high similarities with 58 commonly regulated DEGs. In accordance with the KEGG analysis, P. fluorescens stands out in in all pair-wise blast analysis. Only 19 orthologs were commonly regulated by P. fluorescens and P. aeruginosa, and this value was reduced to 7 in P. fluorescens vs. P. putida analysis.
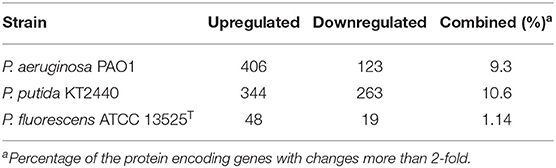
Table 1. The quantity of DEGs in P. aeruginosa PAO1, P. putida KT2440, and P. fluorescens ATCC 13525T.
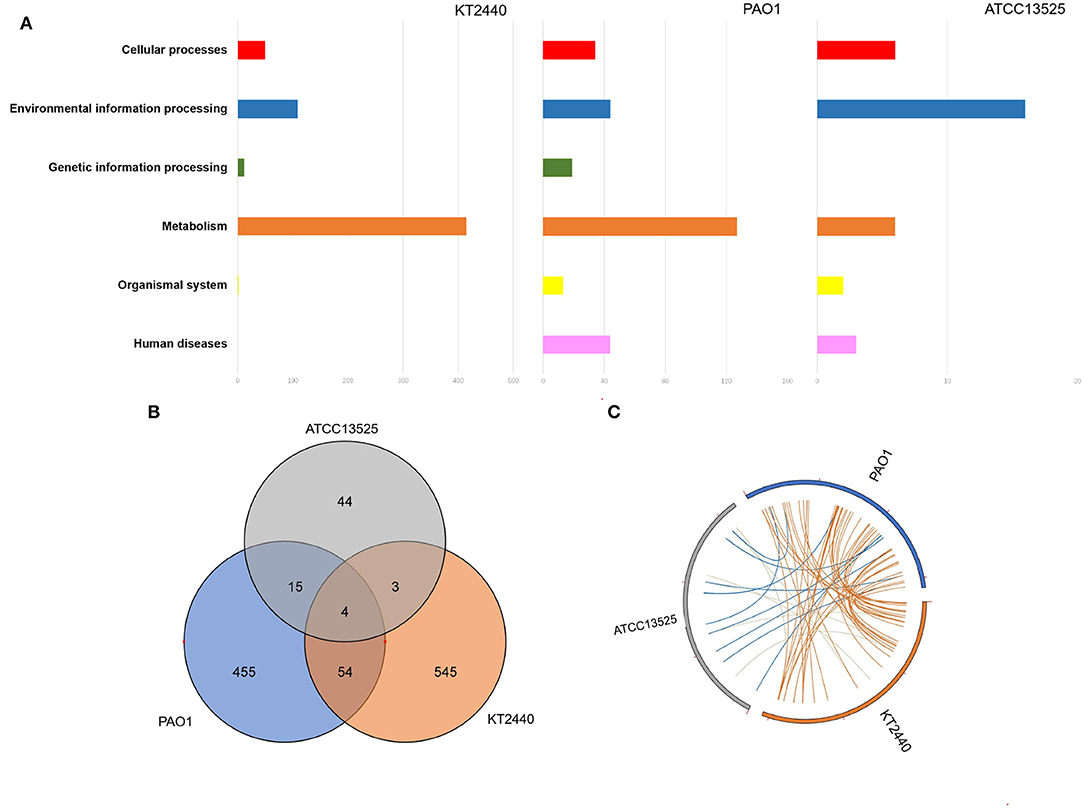
Figure 1. Global transcription features of three Pseudomonas strains. (A) KEGG analysis of DEGs identified in P. aeruginosa PAO1, P. putida KT2440, and P. fluorescence ATCC 13525T. (B) Venn diagram showing the DEGs commonly or strain-specifically regulated by these three Pseudomonas strains. (C) Genome location of these genes regulated by more than one strain.
Functional Analysis of DEGs Identified in More Than One Strain
All the four DEGs (cadA, cadR, czcR, and czcS) commonly upregulated by the three strains were involved in metal efflux. cadA has been recognized as an efficient P-type ATPase that moves several divalent heavy metal ions out of the cellular membrane (Leedjärv et al., 2008), whilst czcRS has been proven as an important two-component system that connects metal efflux and porins. In P. aeruginosa, unphosphorylated CzcR decreased the expression of outer membrane porins, OprD. By contrast, the presence of phosphorylated CzcR induced the most effective metal efflux system, czcCBA (Perron et al., 2004). DEGs mutually regulated by P. aeruginosa PAO1 and P. putida KT2440 are listed in Supplementary Table 2. As expected, czcCBA operons, which are known to be induced by several divalent metal ions, were found to be highly responsive. The analysis of other DEGs with known functions showed that most of these DEGs can be manually clustered into three categories. The genes involved in membrane structure and channels constituted the largest functional category. In this group, seven genes, including desA, ompB, dgkA, warB, warA, amgR and PA4819 (PP_0034), were strongly upregulated (>5 fold), whereas oprD and kguT were mutually downregulated. In the other two groups, genes associated with central metabolism and those responsible for protein folding and degradation were generally upregulated. For P. fluorescens, the most prominent response mutually identified in P. fluorescens and P. aeruginosa was the upregulation of acyl-CoA dehydrogenase (RS07420, RS06160) and protein-tyrosine-phosphatase (RS10450). A ton-dependent receptor (RS23050) and a non-ribosomal peptide synthatase were markedly down-regulated (RS27315) (Supplementary Table 3). In addition to the four DEGs commonly regulated by the three strains, P. fluorescens and P. putida shared three more transcripts (Supplementary Table 4). Two of them were responsible for active transmembrane transport (RS21740, RS19005). The induction of asparaginase (RS29965) was also identified. The genome locations of these genes that are regulated by more than one strain are spread throughout the genomes, no obvious genomic island was observed (Figure 1C).
DEGs Specifically Identified in P. aeruginosa PAO1
The DEGs specifically identified in P. aeruginosa PAO1 are shown in Supplementary Table 5. The fold-change data indicated that more energy were required for P. aeruginosa to alleviate zinc-induced toxicity. The proton-translocating NADH:ubiquinone oxidoreductase complex (nuoGHIJLMN) and the pyruvate dehydrogenase complex (PA3416-3417), which transforms pyruvate into acetyl-CoA were upregulated by ~2.3-fold, whilst the synthesis of ribosomal protein was significantly downregulated (rpsB, rpsS, rpsT, rpsR, rpsF, rplS, rplW, rplJ, and rplU). A distinct group of operons involved in lipid A synthesis/modification was highly induced (arnBCADEF), suggesting that the P. aeruginosa cells were in a cell-envelope-stressed state. In support of this, there was also significant upregulation of other genes previously linked to bacterial cell envelope homeostasis, such as glmM (phosphoglucosamine mutase) and pagL (lipid A 3-O-deacylase). The direct replacement of iron from their binding sites has been recognized as an important mechanism for zinc to exhibit cytotoxicity. Therefore, it is reasonable that the transcription of pvdQAPNOFEDJ that responsible for pyoverdine biosynthesis was downregulated. Using UV-visible spectrophotometry analysis (Hoegy et al., 2014), the concentration of pyoverdine presenced in the cultures was also determined. The biosynthesis of pyoverdine in zinc treated P. aeruginosa PAO1 (40.1 μmol L−1) reduced about 66.8% compared with control samples (121.4 μmol L−1) (Supplementary Figure 1).
Given that P. aeruginosa is an opportunistic pathogen, monitoring the transcriptional variation of pathogenicity related genes is important. The MIC of zinc strongly enhanced the transcription of mexRAB-oprM (Poole et al., 1996) and mexXY (Morita et al., 2012), two important multidrug efflux systems responsible for antibiotic resistance. Moreover, one operon involved in alginate synthesis (algU-mucABC) was significantly activated. The overproduction of exopolysaccharide alginate caused mucoid conversion in P. aeruginosa (Mathee et al., 1999; Rao et al., 2011), which increased bacterial metal tolerance via metal chelation (Gómez-Sagasti et al., 2015). Amongst all the DEGs specifically identified in P. aeruginosa, the most upregulated gene was ptrA. The function of PtrA was studied by two groups, but contrasting conclusions were reported. One group showed that PtrA directly binds to ExsA, which in turn, suppresses the expression of the type III secretion system (T3SS) (Ha et al., 2004). By contrast, the other group demonstrated that PtrA is a periplasmic protein, the expression of which increases the Cu tolerance of P. aeruginosa without affecting basal ExsA (Elsen et al., 2011). Our data supported that PtrA is not a T3SS repressor because the transcription of the T3SS gene clusters remained unchanged when ptrA was considerably induced by zinc.
DEGs Specifically Identified in P. putida KT2440
In our previous study, the transcriptional response of P. putida KT2440 to stress-inducing concentrations of zinc was analyzed. The results showed that different zinc stress levels strongly influenced (>4 fold change) the transcription of genes from four functional groups, including metal transporting genes, genes associated with membrane homeostasis, antioxidant-encoding genes and genes involved in basic cellular metabolism (Peng et al., 2018). In this study, a twofold change was used as a criterion, and a total of 545 genes were identified as DEGs specifically regulated in P. putida KT2440. Approximately 64% of the downregulated operons encode nutrient uptake transporters (Supplementary Table 6), which indicated that P. putida KT2440 decreased the permeability of the cell envelope under zinc treatment. Only the transporters responsible for methoine, sulfate, and sulfonates uptake were upregulated, suggesting that, unlike P. aeruginosa PAO1, P. putida KT2440 did not suffer from a severe iron metabolism perturbation, but the sulfur-containing molecules were considerably disrupted. The synthesis of ribosomal protein was another remarkable difference between P. aeruginosa PAO1 and P. putida KT2440. In P. aeruginosa PAO1, the transcription of ribosomal protein encoding genes was generally downregulated. Meanwhile, their orthologs in P. putida KT2440 were highly stable, and even the transcription of rpmH was increased. In addition, the MIC of zinc significantly increased the transcription of the nickel (nikABCDE) and arsenic (arsBR) efflux systems in P. putida KT2440. Such phenomenon was not observed in the other two strains.
DEGs Specifically Identified in P. fluorescens ATCC 13525T
Compared with the transcriptional responses of P. aeruginosa PAO1 and P. putida KT2440, the downregulation of type VI secretion system (T6SS) was the most remarkable feature of P. fluorescens ATCC 13525T under zinc stresses (Supplementary Table 7). T6SS is widely distributed across diverse bacterial species; around one-third of the sequenced Gram-negative bacteria possess T6SS-associated genes (Alteri and Mobley, 2016). Bacterial T6SS functions as a contractile nanomachine that delivers effectors upon direct contact with a target cell (Drebes Dörr and Blokesch, 2018). Given that these effectors have different functions but frequently disturb the cellular structure, such as the cell wall, nucleic acid, or membrane compartment, T6SS is mainly perceived as an antibacterial weapon (Alteri and Mobley, 2016). In recent studies, the T6SSs in Burkholderia thailandensis and Yersinia pseudotuberculosis were found to secrete metal binding proteins which facilitate the intracellular accumulation of Zn2+ (Wang et al., 2015; Si et al., 2017). Similar capability of ion transport by T6SS was also found in P. aeruginosa. Its T6SS effector, TseF, facilities the delivery of outer membrane vesicles associated iron to bacterial cells by engaging the Fe-pyochelin receptor FptA and the porin OprF (Lin et al., 2017). Based on these published results and the RNA-seq results obtained in this study, we infer that T6SS also plays an important role in zinc transport in P. fluorescens, the downregulation of which reduced the intrusion of zinc across the outer membrane.
Reverse Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) Validation
To confirm the RNA-seq data, six genes were selected from each strain, and their transcription was analyzed via RT-qPCR with three biological replicates. As shown in Figure 2, the dynamic transcription patterns of all the genes were consistent with the RNA-seq data (Figure 2A), and the observed fold changes for each gene were moderately correlated (R2 ranged from 80.1 to 89.1%) (Figure 2B). Therefore, the RT-qPCR results validated the accuracy of the RNA-seq data. Moreover, previous studies on the transcriptomic profiles of P. putida KT2440 were useful in further confirming the results obtained in this study.
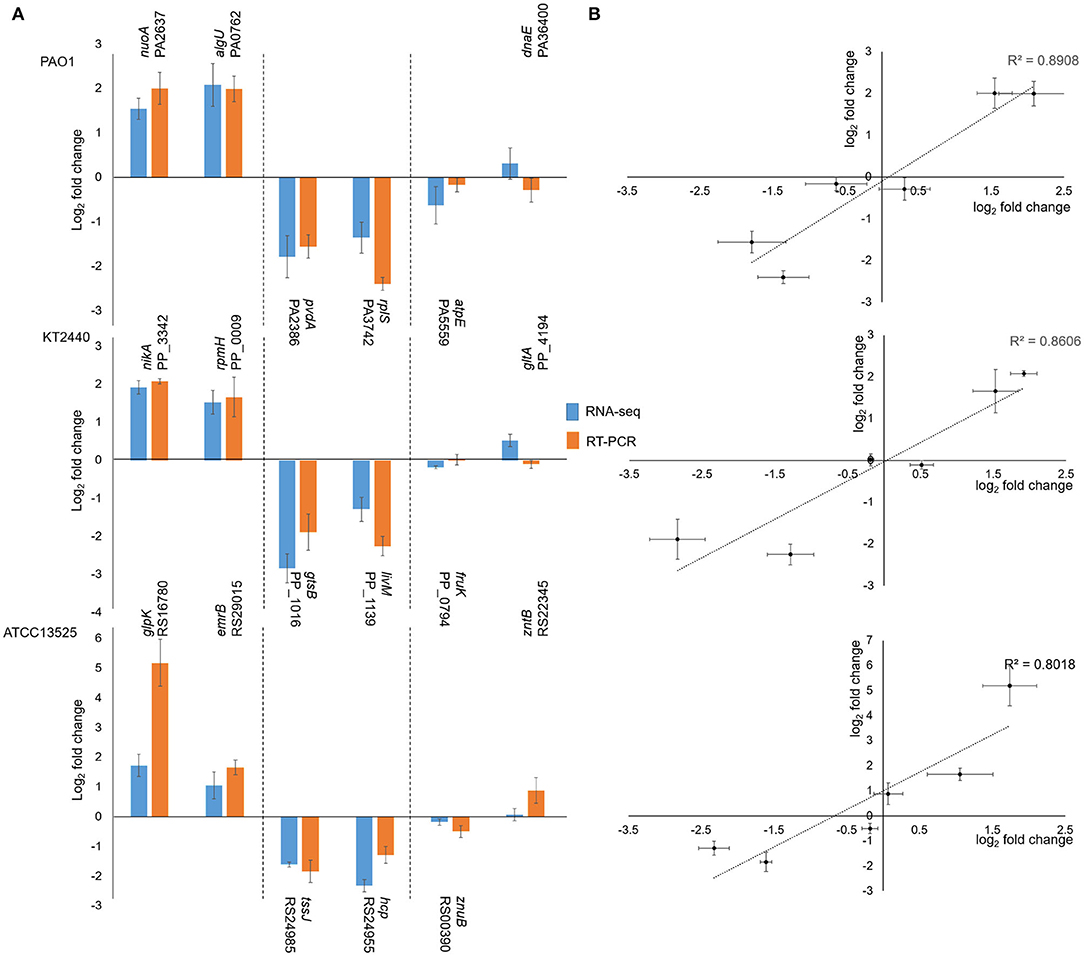
Figure 2. Correlation between the results obtained by RNA-seq and RT-PCR. (A) Rectangles represent fold changes of selected genes. Bars illustrate standard deviations of three replicates. (B) Correlation plot between fold change values of selected genes observed by RNA-seq and RT-PCR. Horizontal bars represent standard deviations for RNA-seq data, and vertical bars illustrated standard deviations of RT-PCR results. As the fold change values followed the normal distribution, Pearson' coefficients were calculated.
Metabolite Changes in the Three Pseudomonas Strain
Considering that large quantities of DEGs identified in the transcriptome analysis were genes associated with metabolism, we detected the concentrations of metabolites in these bacterial cells. Metabolic profiles of whole-cell extracts were obtained from zinc-treated and control samples exponentially cultured in CDM. Six replicates were performed for each sample. A total of 83 annotated metabolites were significantly changed, and approximately 76% of which were down-regulated. Only two metabolites were commonly regulated by the three Pseudomonas strains (pyroglutamic, 2-phosphoglycerate). The metabolite profiles of P. aeruginosa and P. putida shared more similarity, 18 metabolites were commonly regulated (Figure 3). However, after referring to the KEGG database, the metabolic pathways affected by the MICs of zinc imposed to the three Pseudomonas strains exhibited higher similarity (Supplementary Tables 8–10). Several metabolic intermediates associated with central carbon metabolisms (the Entner-Doudoroff pathway, tricarboxylic cycle, and pentose phosphate pathway) were significantly downregulated. The concentrations of citrate, phosphoenolpyruvate, and 2-phosphoglycerate in the zinc-treated P. aeruginosa PAO1 was only approximately 20% of that detected in the control samples. Similar phenomena were also observed in P. putida KT2440 and P. fluorescens ATCC 13525T. Compared with the P. putida cells without zinc treatment, the concentrations of citrate, succinate, and 2-phosphoglycerate were reduced by approximately 60.4, 72.6, and 78.2%, respectively. These observations are consistent with the RNA-seq data. The RPKM values of mdh (malate dehydrogenase) and idh (isocitrate dehydrogenase) in P. putida were only about 70% of that calculated in the cells without zinc treatment, and the transcription of eda (2-keto-3-deoxy-6-phosphogluconate aldolase) were downregulated about 1.7 and 1.5-fold in P. putida and P. fluorescens, respectively. The metabolites involved in amino acids metabolism were also reduced. For example, a wide range of amino acids (ornithine, Lys, Pro, Asp, Trp, and His) or intermediates involved in their biosynthesis were decreased in P. putida. And lower concentrations of metabolites involved in Arg, Glu, Lys, Cys, and Met metabolisms were observed in P. aeruginosa.
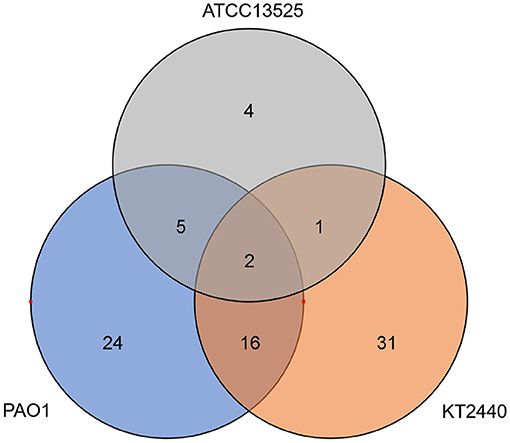
Figure 3. The quantity of metabolites commonly or strain-specifically regulated by P. aeruginosa PAO1, P. putida KT2440, and P. fluorescence ATCC 13525T.
Discussion
The genus Pseudomonas is one of the most diverse bacterial genera, comprising ~200 species isolated from sources ranging from vegetation to water and contaminated soil to animal clinical samples (Singh et al., 2018). With relatively large genomes (typically between 6 and 7 Mbp), many of these species have evolved delicate gene regulation systems that allow them to thrive under various environmental stresses (Jensen et al., 2004; Balasubramanian and Mathee, 2009). By combining the transcriptomic and metabolic data obtained in the present study, an overview of the global effects of zinc on three representative Pseudomonas strains was provided.
P. aeruginosa PAO1 was originally isolated from a human wound and widely studied as a model opportunistic human pathogen (Stover et al., 2000). Soil-originating P. putida KT2440 is the best-characterized strain from this species. It is used worldwide as the primary strain for genetic and physiological studies (Regenhardt et al., 2002). High genetic diversity was observed among bacteria classified as P. fluorescens, which can be further clustered into three smaller taxonomic subclades (Scales et al., 2015). Strains classified into one subclade shared a higher amount of conserved genes than the species-complex as a whole. Comparative genomic analysis showed a high level of genome similarity between P. putida KT2440 and P. aeruginosa PAO1, about 85% of the CDS are shared (Jensen et al., 2004). Although P. fluorescens ATCC 13525T behaved as an out-group amongst the three strains, our bioinformatic analysis indicated that there is still 29% CDS are shared between P. fluorescens ATCC 13525T and P. putida KT2440, and 33% CDS are shared between P. fluorescens ATCC 13525T and P. aeruginosa PAO1. By contrast, the transcriptomic features of the three strains in response to the MICs of zinc exhibited significant divergence. Only 4 orthologs were commonly regulated, and all these orthologs were involved in metal efflux. The exclusion of cytoplasmic ions by membrane efflux pumps has been recognized as an important strategy for prokaryotic cells to withstand metal toxicity (Nies, 2007). However, this process is not energy-efficient, and damages can happen before these efflux systems are fully expressed. Reducing the entry of excess metal ions can more effectively prevent the generation of metal-induced cytotoxicity. Based on our data, the three strains may use different way to preclude the entry of excess zinc ions under high zinc conditions. P. aeruginosa PAO1 significantly decreased the secretion of siderophore, which was also found to complex with zinc, nickel, cobalt and magnesium as well as iron (Sasirekha and Srividya, 2016; Shi et al., 2017). P. putida universally reduced the expression of membrane transporters. P. fluorescens ATCC 13525T has the lowest MIC value of zinc, and a relatively small number of genes were transcriptionally changed. Among these genes, T6SS gene cluster was remarkably downregulated, whose analogs in B. thailandensis play an important role in zinc uptake under oxidative stresses (Si et al., 2017). In addition to the change in zinc transport, it interesting to note the side effect caused by zinc ions. The increased alginate synthesis can facilitate bacterial adherence, biofilm formation, neutralize oxygen radicals, and act as a barrier to phagocytosis (Sautter et al., 2012). Coupled with the upregulated antibiotic efflux systems, the scavenging of zinc-exposed P. aeruginosa can be more difficult. P. fluorescens has been studied extensively as a plant growth promoter that synthesizes toxic metabolites against phytopathogenic microorganisms, and enhancing nutrient availability in soil (Loper et al., 2012). The downregulated transcription of T6SS can reduce their competence to occupy phyllosphere and rhizosphere, which in turn, decreases the effectiveness of P. fluorescens to be used as a biocontrol agent.
As transcriptome analysis showed that large quantities of DEGs identified in P. aeruginosa, P. putida, and P. fluorescens can be categorized into the metabolism KEGG group, Hence, different metabolic intermediates were measured using UPLC-MS/MS analysis to further explore the mechanisms underlying the effects of zinc exposure on the metabolic pathway. Two classes of metabolites were significantly decreased, including central carbon metabolism and amino acids synthesis, which may be one of the important reason why the growth of these bacterial cells stopped. Dipeptide is another group of metabolites that identified at lower concentrations after zinc treatment, especially in P. aeruginosa (Pro-Arg, and Ile-Leu) and P. putida (Pro-Val, Val-Ala, and Ile-Ala). High percentages of branched amino acids were observed, indicating that these dipeptides were not generated by protein degradation. The changes of Ile/Leu/Val containing short peptides were also observed by Jousse et al. (2017). Their metabolomics data revealed that the concentrations of Ala-Leu, Leu-Leu, Ala-Arg, Pro-Phe, Pro-Ile, Leu-Phe, and Val-Val were significantly decreased in cold-shocked P. syringae. Beside the downregulated compounds discussed above, some metabolites were observed at high concentrations, such as nucleoside/base (adenosine, deoxycytidine, uracil, cytosine) and proline. Proline has been reported to play important roles in protecting protein denaturation and stabilizing protein synthesis, the upregulation of which commonly occurs in cells under heavy metal stresses (Alia and Saradhi, 1991; Choudhary et al., 2007). Because energy generation and central carbon metabolism were severely affected and the upregulation of exonuclease in P. aeruginosa PAO1 (PA4937) and P. putida KT2440 (PP_0034, PP_0353) was observed (<2 fold), the presence of high concentrations of nucleotide and base may be produced by the recycling of denatured nucleic acid.
In this study, we compared the zinc-induced cellular responses of three prominent Pseudomonas strains from different phylogenetic groups. The analysis of the obtained transcriptomic profiles showed that 1,120 genes transcriptionally changed by more than twofold. The exclusion of cytoplasmic ions and reducing the unexpected intrusion of excess zinc are important for P. aeruginosa, P. putida, and P. fluorescens to withstand external zinc stress. Metabolic analysis showed that low concentrations of the metabolic intermediates involved in central carbon metabolism and amino acid biosynthesis were identified. Dipeptides that contained branched-chain amino acids may play important roles during their zinc adaptation.
Data Availability Statement
The RNA-seq datasets for this study can be found in the NCBI short read archive database under the bioproject accession number PRJNA606809.
Author Contributions
LL and PL conceived the experiment. LL, WL, and JC performed the experiment. LL and PL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Natural Science Foundation of China (Grant no. 31700100) and the Hubei Provincial Natural Science Foundation of China (Grant no. 2017CFB207).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Xi Chen at Wuhan Institute of Virology for her technical help during bioinformatic analysis. A preprint has been uploaded to Research Square, Lei et al. (2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.573857/full#supplementary-material
References
Alhasawi, A., Auger, C., Appanna, V. P., Chahma, M., and Appanna, V. D. (2014). Zinc toxicity and ATP production in Pseudomonas fluorescens. J. Appl. Microbiol. 117, 65–73. doi: 10.1111/jam.12497
Alia, and Saradhi, P. P. (1991). Proline accumulation under heavy metal stress. J. Plant Physiol. 138, 554–558. doi: 10.1016/S0176-1617(11)80240-3
Alteri, C. J., and Mobley, H. L. T. (2016). The versatile type VI secretion system. Microbiol. Spectr. 4:2. doi: 10.1128/microbiolspec.VMBF-0026-2015
Balasubramanian, D., and Mathee, K. (2009). Comparative transcriptome analyses of Pseudomonas aeruginosa. Hum. Genom. 3, 349–361. doi: 10.1186/1479-7364-3-4-361
Blencowe, D. K., and Morby, A. P. (2003). Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27, 291–311. doi: 10.1016/S0168-6445(03)00041-X
Braymer, J. J., and Giedroc, D. P. (2014). Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr. Opin. Chem. Biol. 19, 59–66. doi: 10.1016/j.cbpa.2013.12.021
Chavez-Dozal, A., Gorman, C., and Nishiguchi, M. K. (2015). Proteomic and metabolomic profiles demonstrate variation among free-living and symbiotic Vibrio fischeri biofilms. BMC Microbiol. 15:226. doi: 10.1186/s12866-015-0560-z
Cheng, S. (2003). Heavy metal pollution in China: origin, pattern and control. Environ. Sci. Pollut. Res. 10, 192–198. doi: 10.1065/espr2002.11.141.1
Choudhary, M., Jetley, U. K., Abash Khan, M., Zutshi, S., and Fatma, T. (2007). Effect of heavy metal stress on proline, malondialdehyde, and superoxide dismutase activity in the cyanobacterium Spirulina platensis-S5. Ecotoxicol. Environ. Saf. 66, 204–209. doi: 10.1016/j.ecoenv.2006.02.002
Drebes Dörr, N. C., and Blokesch, M. (2018). Bacterial type VI secretion system facilitates niche domination. Proc. Nat. Acad. Sci. U.S.A. 115, 8855–8857. doi: 10.1073/pnas.1812776115
Elsen, S., Ragno, M., and Attree, I. (2011). PtrA is a periplasmic protein involved in Cu tolerance in Pseudomonas aeruginosa. J. Bacteriol. 193, 3376–3378. doi: 10.1128/JB.00159-11
Gómez-Sagasti, M. T., Becerril, J. M., Epelde, L., Alkorta, I., and Garbisu, C. (2015). Early gene expression in Pseudomonas fluorescens exposed to a polymetallic solution. Cell Biol. Toxicol. 31, 39–81. doi: 10.1007/s10565-015-9294-9
Gonzalez, M. R., Ducret, V., Leoni, S., and Perron, K. (2018). Pseudomonas aeruginosa zinc homeostasis: key issues for an opportunistic pathogen. Biochim. Biophys Acta Gene Regul Mech. 1862, 722–733. doi: 10.1016/j.bbagrm.2018.01.018
Ha, U.-H., Kim, J., Badrane, H., Jia, J., Baker, H. V., Wu, D., et al. (2004). An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol. Microbiol. 54, 307–320. doi: 10.1111/j.1365-2958.2004.04282.x
Haritha, A., Rodrigue, A., and Mohan, P. M. (2008). A comparative analysis of metal transportomes from metabolically versatile Pseudomonas. BMC Res. Notes 1:88. doi: 10.1186/1756-0500-1-88
Hobman, J. L., Yamamoto, K., and Oshima, T. (2007). “Transcriptomic responses of bacterial cells to sublethal metal ion stress,” in Molecular Microbiology of Heavy Metals, D. H. Nies, and S. Silver, eds. (Heidelberg: Springer), 375–405.
Hoegy, F., Mislin, G. L. A., and Schalk, I. J. (2014). “Pyoverdine and pyochelin measurements,” in Pseudomonas Methods and Protocols, A. Filloux, and J. Ramos, eds, (Heidelberg: Springer), 293–301. doi: 10.1007/978-1-4939-0473-0_24
Jensen, L. J., Skovgaard, M., Sicheritz-Pontén, T., Hansen, N. T., Johansson, H., Jørgensen, M. K., et al. (2004). “Comparative genomics of four Pseudomonas species,” in Pseudomonas, J. L. Ramos, ed. (Boston: Springer), 139–164. doi: 10.1007/978-1-4419-9086-0_5
Jiang, H., Lei, R., Ding, S.-W., and Zhu, S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 15:182. doi: 10.1186/1471-2105-15-182
Jousse, C., Dalle, C., Canet, I., Lagrée, M., Traïkia, M., Lyan, B., et al. (2017). Metabolomic study of the response to cold shock in a strain of Pseudomonas syringae isolated from cloud water. Metabolomics 14:11. doi: 10.1007/s11306-017-1295-7
Klimek, B. (2012). Effect of long-term zinc pollution on soil microbial community resistance to repeated contamination. Bull. Environ. Contam. Toxicol. 88, 617–622. doi: 10.1007/s00128-012-0523-0
Kohlstedt, M., and Wittmann, C. (2019). GC-MS-based (13)C metabolic flux analysis resolves the parallel and cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. Metab. Eng. 54, 35–53. doi: 10.1016/j.ymben.2019.01.008
Krezel, A., and Maret, W. (2016). The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 611, 3–19. doi: 10.1016/j.abb.2016.04.010
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leedjärv, A., Ivask, A., and Virta, M. (2008). Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 190, 2680–2689. doi: 10.1128/JB.01494-07
Lei, L., Chen, J., Liao, W., and Liu, P. (2020). Determining the different mechanisms used by Pseudomonas species to cope with minimal inhibitory concentrations of zinc via comparative transcriptomic analyses. Res. Square [Preprint]. Available online at: https://researchsquare.com/article/rs-17097/v1 (accessed March 13, 2020).
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loper, J. E., Hassan, K. A., Mavrodi, D. V., Davis, E. W., Lim, C. K., Shaffer, B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. doi: 10.1371/journal.pgen.1002784
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Markowitz, V. M., Chen, I. M., Palaniappan, K., Chu, K., Szeto, E., Pillay, M., et al. (2013). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42:D560. doi: 10.1093/nar/gkt963
Mathee, K., Ciofu, O., Sternberg, C., Lindum, P. W., Campbell, J. I. A., Jensen, P., et al. (1999). Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145, 1349–1357. doi: 10.1099/13500872-145-6-1349
Mergeay, M., Nies, D. H., Schlegel, H. G., Gerits, J., and Gijsegem, F. V. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162, 328–334. doi: 10.1128/JB.162.1.328-334.1985
Morita, Y., Tomida, J., and Kawamura, Y. (2012). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. microbiol. 3:408. doi: 10.3389/fmicb.2012.00408
Nies, D. H. (2007). “Transition Metal Homeostasis,” in Molecualr Microbiology of Heavy Metals, D. H. Nies, and S. Silver, ed. (Heidelberg: Springer), 117–142. doi: 10.1007/7171_2006_075
Outten, C. E., and O'Halloran, T. V. (2001). Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492. doi: 10.1126/science.1060331
Pederick, V. G., Eijkelkamp, B. A., Begg, S. L., Ween, M. P., McAllister, L. J., Paton, J. C., et al. (2015). ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci. Rep. 5:13139. doi: 10.1038/srep13139
Peix, A., Ramírez-Bahena, M.-H., and Velázquez, E. (2017). The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 57, 106–116. doi: 10.1016/j.meegid.2017.10.026
Peng, J., Miao, L., Chen, X., and Liu, P. (2018). Comparative transcriptome analysis of Pseudomonas putida KT2440 revealed its response mechanisms to elevated levels of zinc stress. Front. Microbiol. 9:1669. doi: 10.3389/fmicb.2018.01669
Perron, K., Caille, O., Rossier, C., Delden, C. V., and Köhler, T. (2004). CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279, 8761–8768. doi: 10.1074/jbc.M312080200
Poole, K., Tetro, K., Qx, Z., Neshat, S., Heinrichs, D., and Bianco, N. (1996). Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40, 2021–2028. doi: 10.1128/AAC.40.9.2021
Rao, J., Damron, F. H., Basler, M., Digiandomenico, A., Sherman, N. E., Fox, J. W., et al. (2011). Comparisons of two proteomic analyses of non-mucoid and mucoid Pseudomonas aeruginosa clinical isolates from a cystic fibrosis patient. Front. Microbiol. 2:162. doi: 10.3389/fmicb.2011.00162
Regenhardt, D., Heuer, H., Heim, S., Fernandez, D. U., Strompl, C., Moore, E. R., et al. (2002). Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4, 912–915. doi: 10.1046/j.1462-2920.2002.00368.x
Santos, V. A. P. M.D., Heim, S., Moore, E. R. B., Strätz, M., and Timmis, K. N. (2004). Insight into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6, 1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x
Sasirekha, B., and Srividya, S. (2016). Siderophore production by Pseudomonas aeruginosa FP6, a biocontrol strain for Rhizoctonia solani and Colletotrichum gloeosporioides causing diseases in chilli. Agric. Nat. Resour. 50, 250–256. doi: 10.1016/j.anres.2016.02.003
Sautter, R., Ramos, D., Schneper, L., Ciofu, O., Wassermann, T., Koh, C.-L., et al. (2012). A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and AlgT/U activity results in the loss of alginate production. Gene 498, 242–253. doi: 10.1016/j.gene.2011.11.005
Scales, B. S., Erb-Downward, J. R., Huffnagle, I. M., LiPuma, J. J., and Huffnagle, G. B. (2015). Comparative genomics of Pseudomonas fluorescens subclade III strains from human lungs. BMC Genom. 16:1032. doi: 10.1186/s12864-015-2261-2
Shi, P., Xing, Z., Zhang, Y., and Chai, T. (2017). Effect of heavy-metal on synthesis of siderophores by Pseudomonas aeruginosa ZGKD3. IOP Conference Series. Earth Environ. Sci. 52:012103. doi: 10.1088/1742-6596/52/1/012103
Si, M., Wang, Y., Zhang, B., Zhao, C., Kang, Y., Bai, H., et al. (2017). The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep. 20, 949–959. doi: 10.1016/j.celrep.2017.06.081
Singh, S., Singh, P., Gupta, A., Singh, A., and Keshri, J. (2018). “Tolerance of heavy metal toxicity using PGPR strains of Pseudomonas species,” in PGPR Amelioration in Sustainable Agriculture, A. K. Singh, A. Kumar, and P. K. Singh, ed. (Cambridge: Woodhead Publishing), 239–252. doi: 10.1016/B978-0-12-815879-1.00012-4
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Tu, J., Zhao, Q., Wei, L., and Yang, Q. (2012). Heavy metal concentration and speciation of seven representative municipal sludges from wastewater treatment plants in northeast China. Environ. Monit. Assess. 184, 1645–1655. doi: 10.1007/s10661-011-2067-x
Vasile, G. D., Nicolau, M., and Vlãdescu, L. (2008). Zinc speciation in sediments from a polluted river, as an estimate of its bioaccesibility. Environ. Monit. Assess. 160, 71–81. doi: 10.1007/s10661-008-0658-y
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. Plos Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Watły, J., Potocki, S., and Rowińska-Zyrek, M. (2016). Zinc homeostasis at the bacteria/host interface-from coordination chemistry to nutritional immunity. Chemistry 22, 15992–16010. doi: 10.1002/chem.201602376
Keywords: Pseudomonas, zinc, comparative analysis, transcriptomic, minimal inhibit concentration
Citation: Lei L, Chen J, Liao W and Liu P (2020) Determining the Different Mechanisms Used by Pseudomonas Species to Cope With Minimal Inhibitory Concentrations of Zinc via Comparative Transcriptomic Analyses. Front. Microbiol. 11:573857. doi: 10.3389/fmicb.2020.573857
Received: 18 June 2020; Accepted: 16 November 2020;
Published: 03 December 2020.
Edited by:
Hauke Smidt, Wageningen University and Research, NetherlandsReviewed by:
Patrick Kyle Taylor, Simon Fraser University, CanadaMatilde Fernandez, University of Granada, Spain
Copyright © 2020 Lei, Chen, Liao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pulin Liu, cGxsaXVAd2hwdS5lZHUuY24=
 Lei Lei
Lei Lei Jiahui Chen
Jiahui Chen Weifang Liao
Weifang Liao Pulin Liu
Pulin Liu