- 1Department of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC, United States
- 2Department of Horticultural Science, North Carolina State University, Raleigh, NC, United States
Host resistance is one of the few strategies available to combat the soil borne pathogenic fungus Verticillium dahliae. Understanding pathogen diversity in populations is key to successfully deploying host resistance. In this study the genomes of 18 V. dahliae isolates of races 1 (n = 2), 2 (n = 4), and 3 (n = 12) from Japan, California, and North Carolina were sequenced and mapped to the reference genome of JR2 (from tomato). The genomes were analyzed for phylogenetic and pathogen specific signatures to classify specific strains or genes for future research. Four highly clonal lineages/groups were discovered, including a lineage unique to North Carolina isolates, which had the rare MAT1-1 mating type. No evidence for recombination between isolates of different mating types was observed, even in isolates of different mating types discovered in the same field. By mapping these 18 isolates genomes to the JR2 reference genome, 193 unique candidate effectors were found using SignalP and EffectorP. Within these effectors, 144 highly conserved effectors, 42 mutable effectors (truncated or present in some isolates but absent in others), and 7 effectors present in highly variable regions of the chromosomes were discovered. Of the 144 core effectors, 21 were highly conserved in V. alfalfae and V. longisporum, 7 of which have no known function. Within the non-core effectors 30 contained large numbers of non-synonymous mutations, while 15 of them contained indels, frameshift mutations, or were present on highly variable regions of the chromosome. Two of these highly variable region effectors (HVREs) were only present in race 2 isolates, but not in race 3 isolates. The race 1 effector Ave1 was also present in a highly variable region. These data may suggest that these highly variable regions are enriched in race determinant genes, consistent with the two-speed genome hypothesis.
Introduction
Verticillium dahliae is a destructive soil-borne pathogen that infects hundreds of plant species (Bhat and Subbarao, 1999; Pegg and Brady, 2002; Klosterman et al., 2009). The pathogen can survive in soil for over a decade without a viable host present (Green, 1980; Xiao et al., 1998). Selected soil fumigants are widely used in vegetable fields as one of the few chemical-based strategies to suppress this pathogen (Gullino et al., 2002). Host resistance is a key management strategy for tomato growers (Klosterman et al., 2009) where race 1 predominates. Race 1 V. dahliae is characterized by the presence of a functional copy of the Ave1 effector which is recognized by the host resistance gene Ve1 (Diwan et al., 1999; de Jonge et al., 2012). Non-race 1 isolates (frequently described as “race 2”) have been discovered to be widely distributed in North Carolina since the 1980s (Bender and Shoemaker, 1984). Until recently the population of V. dahliae isolates infecting tomato were separated into race 1 and non-race 1 strains. Usami et al. (2017) revealed that new races could be distinguished amongst non-race 1 strains. Isolates that were pathogenic on tomato lines harboring the Ve1 gene only, but found non-pathogenic on the tomato rootstock “Aibou” (aka “Aiboh” or V2) and “Ganbarune-Karis,” which contain an additional gene, were designated as race 2. Isolates pathogenic on “Aibou” and “Ganbarune-Karis” were described as race 3 (“non-race1, race2”). It was also shown that selfed “Aibou” resulted in progeny with a resistant to susceptible ratio of 3:1, indicating the race 2 resistance is conferred by a single dominant locus present in “Aibou” (Usami et al., 2017). In a more recent study, it was shown that “race 3” could be derived from the race 1 strain Vdp4 by knocking out the race 1 effector Ave1 (Kano and Usami, 2019). These findings suggest that races 2 and 3 isolates may have emerged from different phylogenetic origins. Understanding diversity within pathogen populations is vital for successful breeding programs, as well as providing fundamental knowledge of strain evolution. There are currently no genomic sequences available of V. dahliae isolates from tomato that are also from the United States. The absence of genomic data from the United States presents a serious information gap for breeders and researchers.
The first layer of biochemical plant defense is achieved through the detection of pathogen associated molecular patterns (PAMPs) and microbe-associated molecular patterns (MAMPs) such as flagella and chitin (Jones and Dangl, 2006; Akamatsu et al., 2013; Newman et al., 2013: Hayafune et al., 2014). Upon detection of pathogen signal molecules, plants activate their immune systems through pattern triggered immunity pathways (PTI) via salicylic acid, reactive oxygen species, MAP kinase pathways, phytoalexin production, and other biochemical processes (Ebel, 1986; Howe et al., 1992; Raskin, 1992; Peumans and Van Damme, 1995; Kombrink and Somssich, 1997; Sharma et al., 2011). In V. dahliae, chitin-binding lysin motif (LysM) effectors sequester chitin and thus increase host susceptibility by preventing the activation of PTI (Kombrink et al., 2017). The LysM effector VdPDA1 (polysaccharide deacetylase) recently characterized in V. dahliae is one of several effectors responsible for scavenging chitin pre-resistance gene recognition, which aids in hiding chitin from the plant’s PAMP recognition genes. A PDA1 homolog was also discovered in Fusarium spp. and other Verticillium spp. (Gao et al., 2019).
Effector triggered immunity (ETI) is typically a much more aggressive defense response than PTI (Tsuda and Katagiri, 2010). Host resistance genes typically encode cytoplasmic proteins that contain nucleotide binding site leucine rich repeats (NBS-LRR), which can directly or indirectly recognize the presence of pathogen effectors (Belkhadir et al., 2004). In foliar tissue, ETI often results in the hypersensitive response (HR), which leads to localized cell death and eventually systemic acquired resistance (SAR) (Kombrink and Schmelzer, 2001). While ETI generally results in HR in plant foliage, very little is known about ETI in roots. The Ve1 locus in tomatoes results in nearly complete immunity to strains containing the Ave1 effector (de Jonge et al., 2012). Co-expression of the Ave1 gene and Ve1 gene in foliar tissue of Nicotiana glutinosa results in the hypersensitive response (HR) (Song et al., 2017). However, there is only fragmentary evidence that HR occurs in tomato roots (Sutherland, 1991). A comparison of Ve1+ and Ve1- plants infected with V. dahliae strains Le1087 (race 1) and Le1811 (non-race 1) indicates that many defense reactions such as phenylalanine ammonia-lyase (PAL) and other enzymes are differentially expressed in incompatible interactions in roots (Hu et al., 2019). It is unclear whether race 1 or 2 resistant plants are eliciting an HR response or a more complex resistance response more akin to PTI or the post-HR SAR (Hu et al., 2019).
Currently, there are two publicly available annotated V. dahliae reference genomes, JR2 (GCA_000400815.2) and VdLs17 (GCF_000150675.1). JR2 was isolated from a tomato plant in Canada, and sequenced using PacBio at 250x coverage, and the scaffolds have been assembled into 8 distinct chromosomes. The JR2 genome was annotated using a combination of in silico gene prediction and 35 fungal proteomes, which resulted in the prediction of 11,426 genes (de Jonge et al., 2012). VdLs17 was isolated from lettuce in California and was sequenced using Illumina next generation sequencing at 7× coverage and assembled to 55 scaffolds. The VdLs17 genome was annotated using a combination of manual curation, BLAST prediction, and ab initio gene prediction uncovering 10,535 genes (Klosterman et al., 2011). Genome annotation typically involves ab initio gene discovery through the in silico recognition of open reading frames (ORFs), and the input of messenger RNA data from RNA-sequencing projects (Campbell et al., 2014a,b). Both methods have their limitations as ab initio annotation has difficulties accounting for introns within coding sequences, and mRNA data relies on specific genes being expressed in large quantities at the time of sampling (Yandell and Ence, 2012). In contrast, whole genome re-sequencing relies on mapping reads from next generation sequencing (NGS) of different isolates to reference genomes and subsequent extraction of gene information, observing indels and nucleotide polymorphisms. However, mapping to a reference genome relies on the reference genome and the re-sequenced isolate genome being closely related. Gene prediction algorithms such as EffectorP and SignalP can be used to filter annotated genes. SignalP detects signal peptide motifs on the N-terminus of protein sequences that is required for secretion through the canonical secretion pathway to outside the cell. As the vast majority of effectors must be secreted from fungal cells to interact with their hosts, this is a powerful tool for recognizing candidate effectors (Melhem et al., 2013; Armenteros et al., 2019). However, many other non-effector proteins are secreted. To refine predictions of candidate effectors other prediction algorithms such as EffectorP have been developed. EffectorP was trained on 94 experimentally confirmed effector genes from a diverse set of fungal pathogens (Sperschneider et al., 2016). In 2011, 127 effectors were discovered (using SignalP 3.0 and EffectorP 1.0) on the V. dahliae VdLs17 genome and 112 on the V. alfalfae VaMs.102 genome (Klosterman et al., 2011). V. dahliae is an asexually reproducing ascomycete, with no known sexual stage (Usami et al., 2009; Short et al., 2014). Chromosomal rearrangement via transposable elements (TEs), random mutation, and horizontal gene transfer all contribute to V. dahliae diversity (Chen et al., 2018; Shi-Kunne et al., 2018). VdLs17 (non-race 1) and JR2 (race 1) have vastly different chromosomal arrangements despite having few nucleotide differences (de Jonge et al., 2013). More research is needed to elucidate diversity within V. dahliae populations to understand how evolutionary processes are affecting pathogenesis.
The main objectives of this study were to: (i) illuminate the phylogenetic structure, and characterize genomic features, such as mating type and recombination, of V. dahliae tomato isolates from Japan, California, and North Carolina using whole genome re-sequencing and (ii) identify core and divergent effectors, as well as those associated with specific races. By combining whole genome resequencing, and mining for candidate effector genes with macroscopic phenotypic data, we provide new insights into this pathogen’s diversity and pathogenicity factors.
Materials and Methods
Fungal Isolation
North Carolina isolates were obtained from V. dahliae infested tomato fields from Henderson, Jackson, Haywood, and Buncombe counties, all in the temperate, high elevation, growing region of western North Carolina. V. dahliae was isolated from infected tomato by surface sterilizing stem segments and placing them on Sorenson’s NP-10 media for 2 weeks at 26°C (Kabir et al., 2004). Spore suspensions were diluted to 1 × 102 conidia per mL and streaked on potato dextrose agar (PDA, Difco Lab., Detroit, MI, United States). Single spore isolates were obtained by hyphal tip isolation from 3-day old single spore colonies. California isolates Le1087 (race 1) and Le1811 (race 2) were supplied by Dr. Krishna V. Subbarao at UC Davis. California isolates Ca70 and Ca36 were supplied by Suraj Gurung from Sakata Seed America, Inc., Salinas, CA. DNA from all Japanese isolates [GFCa2, To22, Vdp4, Vd141 (aka Ud-141), GFCB5, and HoMCF] was kindly provided by Dr. Toshiyuki Usami at Chiba University in Japan (Usami et al., 2017).
DNA Extraction
Single spore isolates were grown on PDA for up to 10 days. Plugs from those plates were used to inoculate autoclaved 150 mL conical flasks filled with 50 mL of potato dextrose broth (PDB, Difco Lab., Detroit). After 4 days on PDB, the resulting mycelia was decanted into a 50 mL Falcon centrifuge tube and spun at 6000 rpm for 10 min. The supernatant was discarded, and the pelleted mycelia was dried under a laminar flow hood on autoclaved filter paper for 5–10 min. Pellets were frozen with liquid nitrogen and ground with a mortar and pestle to a fine powder. DNA was extracted using a phenol-chloroform extraction (Usami et al., 2007). High molecular weight and overall DNA quality was confirmed by North Carolina State University (NC State) Genome Sciences Laboratory (GSL) in Raleigh NC using an Agilent 2200 TapeStation and the Agilent 2100 Bioanalyzer (Santa Clara, CA, United States).
Whole Genome Sequencing
Library preparation and sequencing took place at the NC State GSL. Genomic DNA libraries of Vdp4 and GFCa2 were prepared using Nextera DNA Flex Library Prep Kit from Illumina (San Diego, CA, United States). Vdp4 and GFCa2 genomes were sequenced at 813x and 167x coverage using MiSeq v.2 150 bp PE flow cell. Genome libraries from the 16 other isolates was prepared using TruSeq Nano LT DNA kit by Illumina (San Diego, CA, United States) and were sequenced at 20–30x coverage using MiSeq v3 300 bp PE flow cell.
Mapping to Reference Genomes and Coding Sequence Extraction
Adapters were removed, paired-end reads were merged, and QC scores <10 were trimmed in Geneious (Biomatters Ltd., Auckland, New Zealand). The paired reads were mapped to the 8 chromosomes of the JR2 reference genome (BioProject Accession PRJNA175765) using the Geneious mapper (Biomatters, Ltd., Auckland, New Zealand) with the “only non-default” option turned on being “Find structural variants, short insertions, and deletions of any size”. Annotations were transferred to the consensus sequence at 75% similarity and a cost matrix of 65% similarity (5.0/−4.0). Coding sequences were extracted from the consensus sequence for each individual isolate sequence with a minimum coverage of 2×. Each coding sequence was translated in Geneious (Biomatters, Ltd., Auckland, New Zealand). This process was repeated for the same 18 isolates mapped to the 55 contigs of the VdLs17 reference genome BioProject PRJNA28529 NCBI.
Secreted Effector Discovery
Coding sequences from the consensus sequence of each isolate were filtered through SignalP 5.0 to determine if a signal peptide was present on the N-terminus of the gene coding sequence (Armenteros et al., 2019). During SignalP filtering the signal peptide, a ∼25 amino acid long sequence, was removed. Secreted sequences were then filtered through EffectorP (Sperschneider et al., 2016). Candidate effectors with transmembrane helices were identified using TMHMM Server v. 2.0 and were removed (Moller et al., 2001).
Phylogenetic Analysis
The consensus sequences of all 18 isolates and the reference genome JR2’s (PRJNA175765) chromosomes were individually aligned using Mauve (Darling et al., 2004). Gaps and ambiguous sequences were removed using Geneious (Biomatters, Ltd., Auckland, New Zealand). Segregating sites and recombining sites determined using DnaSP v5 (Rozas et al., 2003). MAT1-2 were identified if they mapped to the JR2 region where the gene is present. MAT1-1 were identified by mapping the sequences isolates to the AB505215 contig.
Eight chromosome consensus sequences were concatenated from the 18 isolates from this study, as well as 8 outgroup genomes from Gibriel et al. (2019) and the reference genome JR2 and were aligned using Mauve (Darling et al., 2004). Genetic distances were calculated using the Jukes-Cantor model (Jukes and Cantor, 1969). Resampling was completed using the bootstrap method with 1000 replicates. Pathogenicity on tomato confirmed in this study or in other studies using universally susceptible standard “Bonny Best” (Faino et al., 2015; Gibriel et al., 2019).
Unassembled Reads Effector Discovery
The unassembled reads that did not map to the VdLs17 reference genome were assembled using the Geneious de novo assembler at the default settings in version 2020.1.0. Open reading frames (ORFs) were annotated to the assembled contigs. ORF settings were set a minimum nucleotide length of 100. All ORFs annotations were extracted and translated to amino acids in Geneious and those that did not start with an ATG were removed. Amino acid sequences were processed by SignalP v. 5.0, and EffectorP v. 2.0. Candidate effectors with transmembrane helices were identified using TMHMM Server v. 2.0 and were removed. Sequences were analyzed for secreted effectors specific to race 2 isolates.
Pathogenicity Assay
Pathogenic race designations of 12 isolates were confirmed by inoculating 3-week-old differential tomato cultivars in the greenhouse. Three tomato cultivars tested were “Bonny Best” (universal susceptible), “Red Defender” (Ve1+/race 1 resistant, race 2 susceptible), and “Aibou” (Ve1+/race 1 resistant, race 2 resistant). Inoculum was prepared by growing each isolate on PDA for 1–2 weeks. Distilled de-ionized autoclaved water at ∼22°C was used to wash spores from PDA plates. Spores were filtered through a double layer of autoclaved cheesecloth and diluted to 1 × 107 conidia mL–1. Ten mL of this spore suspension were injected into the soil ∼1 cm from the base of the plant stem using a 10 mL sterile pipette. Disease ratings from 0 to 5 (0 = 0; 1 = 1–20; 2 = 21–40; 3 = 41–60; 4 = 61–80; 5 = 81–100) for V-shaped foliar chlorosis/necrosis symptoms were recorded every 7 days for 45 days post inoculation. Wilting was rated on an identical scale. Each experiment contained 4 reps. Wilting and chlorosis/necrosis scores were used to calculate area under the disease progress curve (AUDPC) scores for each isolate/cultivar combination. Plants were planted in 530 mL plastic cups with 2-ply Sungro (70–80% Canadian sphagnum peat moss; 5–10% vermiculite plus dolomite limestone) potting soil. Plants were top watered for 2-weeks post inoculation. Two weeks post inoculation, holes were cut in the bottom of the cups, which were placed in trays 12 plants per tray with 2–5 cm of tap water. A 100 mL aliquot of fertilizer “Miracle-Gro Water-Soluble All-Purpose Plant Food” at a concentration of 1g/1000 mL was added to each tray once a week. Photosynthetic photon flux density was 251 μmol photon m–2 s–1 with a blue, green, and red percentages being 37, 37.1, and 25.9% respectively, and were on for 12 h each day. Temperatures ranged between 23 and 27°C, and relative humidity ranged between 30 and 55%. Additional pathogenicity assays were conducted with “Aibou” for six key isolates (Le1087, Ca36, NC86, JL5c, KJ14a, and VdLs17) and a water control. To determine if “Bowman” and or “Ganbarune-Karis” most likely contained the race 2 resistance gene they were inoculated in a complementary experiment with water, Ca36, NC86, and KJ14a using identical parameters.
Gene Ontology (GO) Analysis
Full amino acid coding sequences of all potential effectors from the isolates mapped to the JR2 reference genome were analyzed using Blast2GO 5.2.5 (Conesa et al., 2005). Annotations were determined by blasting sequences against the BLASTP database with default settings. Sequences were scanned using EMBL-EBI InterPro. GO annotations were determined using default settings. GO descriptions were trimmed using GO slim generic. Effectors with GO annotations indicating the proteins were involved in eukaryotic transmembrane functions were removed.
Results
Race Designation of V. dahliae Isolates
Four experiments were conducted to determine race designations of isolates. In experiment 1, isolates tested were pathogenic on “Bonny Best,” and only Le1087 was non-pathogenic on “Red Defender” (race 1 resistant) (Supplementary Table 1). Le1087, Ca36, and TC18a were non-pathogenic on “Aibou” (race 1 and 2 resistant). NC86, FL10b, NC85, KJ14a, FF5a, FL7a, JL5c, FL9b, and VdLs17 were all pathogenic on “Aibou” (Supplementary Table 1). The water control showed no symptoms in all cultivars. In experiment 2, race typing on “Aibou” confirmed that Le1087 and Ca36 were non-pathogenic on “Aibou,” while NC86, JL5c, KJ14a, and VdLs17 were pathogenic on “Aibou” indicating they can be classified as race 3 (Supplementary Table 2). Experiment 3 demonstrated that Ca70, Le1811, and FL10b were also race 3 (Supplementary Table 3). Experiment 4 confirmed that the race 2 isolate Ca36 was non-pathogenic on Bowman and Ganbarune-Karis, and race 3 isolates were pathogenic on those cultivars (Supplementary Table 4). In sum, the data showed Le1087 is race 1, that Ca36, and TC18a are race 2, and NC86, FL10b, NC85, KJ14a, FF5a, FL7a, JL5c, Le1811, Ca70, and FL9b are race 3. VdLs17 is also pathogenic on “Aibou” (Supplementary Table 2). The race typing of Japanese isolates included in this study were Vdp4 (race 1, Vdp4ΔAve1 = race 3), GFCa2 (race 2), Vd141 (race 2), To22 (Race 2), GFCB5 (Race 3), HoMCF (Race 3) as documented in two separate publications (Usami et al., 2017; Kano and Usami, 2019).
V. dahliae Genome Statistics
Genomes were sequenced using NGS by Illumina MiSeq. Vdp4 had a coverage of 813x (based on an estimated genome size of 32 Mbp) with 98.3% of the total 86,689,970 paired reads (150 bp paired end MiSeq) assembled to the JR2 reference genome. GFCa2 had a coverage of 167x, with 98.59% of reads mapping to the JR2 reference genome. The other 16 genomes had coverages ranging from 20× to 25× with 94.3–95.9% of the totals of 1,046,827 and 1,329,386 paired reads (300 bp paired end MiSeq) (Supplementary Table 5). It is noteworthy that the additional depth of sequence coverage of Vdp4 and GFCa2 added little improvement to the genome assemblies mapped to JR2. Coding sequences extracted from the 18 sequenced genomes based on presence in the JR2 reference genome ranged from 10896 to 11404 (Supplementary Table 6). Coding sequences with signal peptides ranged from 1025 to 1055, and effectors from 162 to 174 (Supplementary Table 6). Genome statistics based on comparison to the VdLs17 reference genome were similar and are shown in Supplementary Tables 7, 8.
Genetic Diversity of V. dahliae
Based on whole genome phylogenetic analysis, isolates in this study were placed in to 4 distinct lineages/groups. Group 1 contained Vdp4 and HoMCF; Group 2: JL5c, FL9b, Le1811, CA70, and GFCB5; Group 3: FL7a, FF5a, KJ14a, and NC85; and Group 4: FL10b, NC86, GFCa2, Le1087, To22, Ca36, and Vd141 (Figure 1). VdLs17 is also in group 1 but was not included in Figure 1. Among the isolates in this study, strains classified as race 3 predominated Groups 2 and 3 and were also present in Groups 1 and 4. Within this study, Group 3 isolates were found exclusively in North Carolina, United States and were all MAT1-1. The only other MAT1-1 strain in our study was Vd39, an isolate from sunflower. Searching GenBank revealed the only other publicly available genome similar to Group 3 isolates is DVD-s29 from Canada, which is also a MAT1-1 strain.
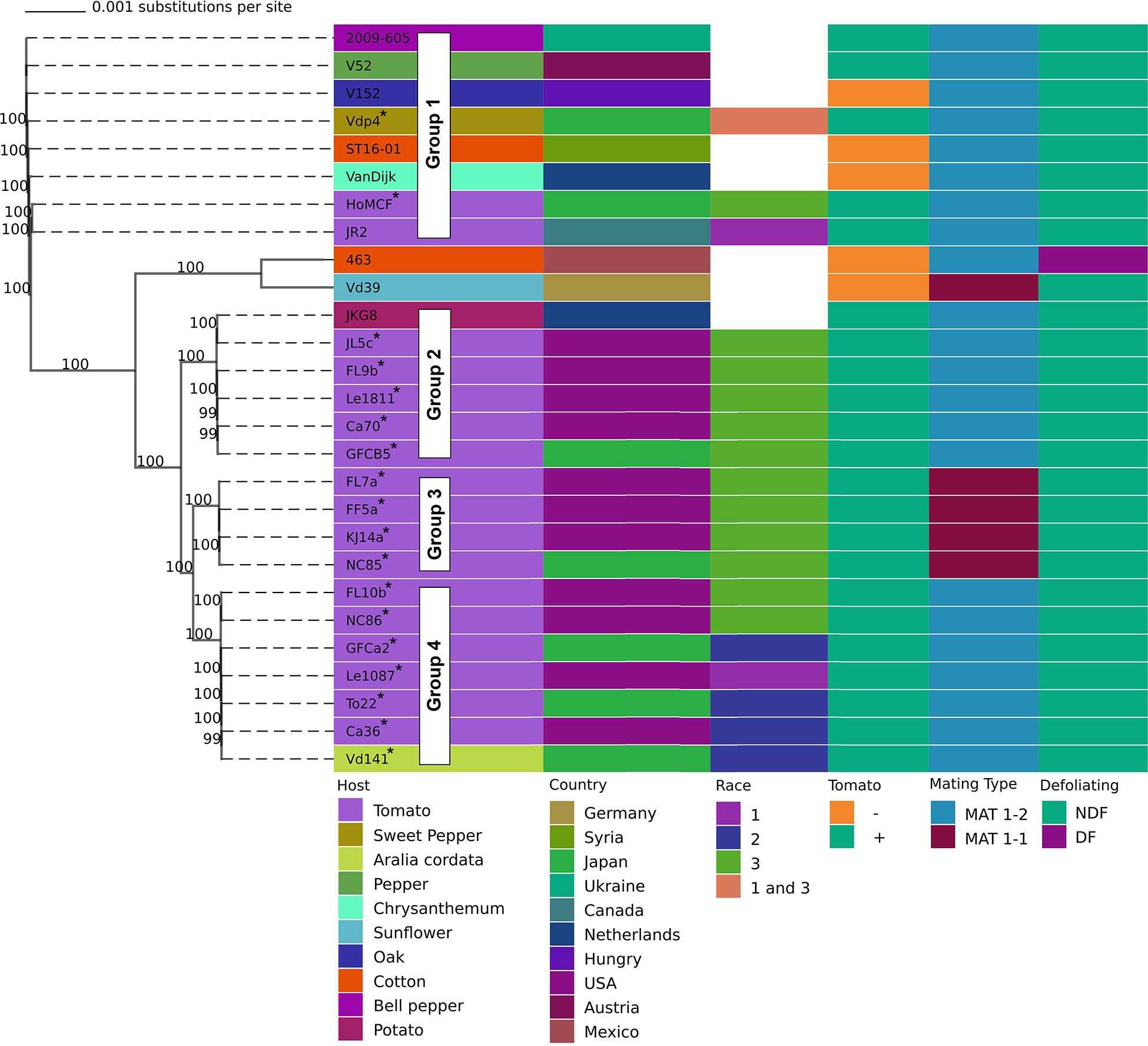
Figure 1. Phylogenetic tree developed from whole genome alignment of V. dahliae isolates from multiple hosts, countries, and mating types. Bootstrap values determined using 1000 replications. Isolates marked with * were sequenced for this study. Pathogenicity on tomato confirmed using universally susceptible tomato lines. Four phylogenetically distinct groups among isolates infecting tomato are indicated.
Genome and Effector Analyses
Across the 18 re-sequenced V. dahliae genomes, and the JR2 reference genome, 193 unique secreted effector regions were discovered using SignalP and EffectorP, and after sequences with transmembrane helices were removed (Table 1). Contained in all 19 genomes were 144 core effectors. There were 42 mutable effectors (MEs) that contained large truncations, large numbers of non-synonymous mutations, insertions, deletions, that caused them to fail being recognized as coding sequences in general, secreted proteins, or effectors. There were 7 effectors that were found in highly variable regions (HVREs), where locally collinear blocks (LCBs) were inconsistently maintained across all genomes (Table 1). Effectors were present on all chromosomes. Chromosome 1 contained the largest number of effectors at 37, while chromosome 3 had the fewest at 17. A total of 141,726 segregating sites containing nucleotide polymorphic sites were present on all genomes (Table 1). Across all chromosomes there were 481 sites of recombination, representing 0.33% of all segregating sites.
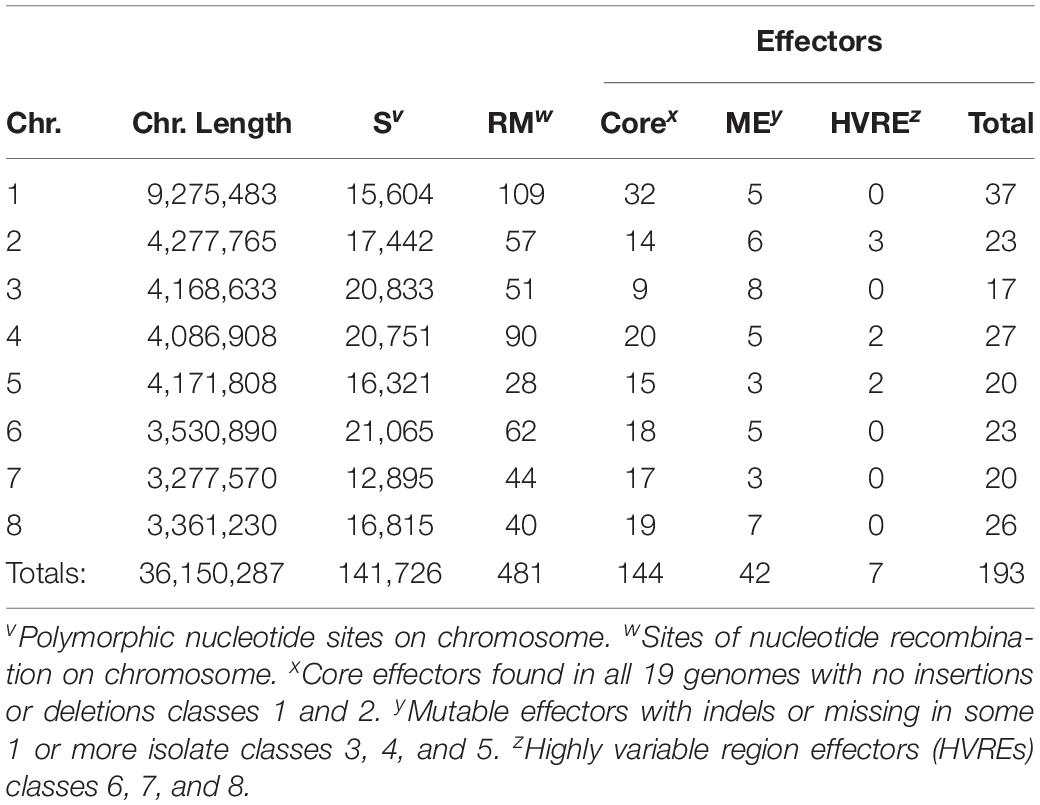
Table 1. Chromosome statistics of 18 isolates mapped to the JR2 reference genome. Segregating, recombining sites, and core/mutable/HVR effectors are displayed.
Gene Ontology (GO) Analysis of All Putative Effectors
A GO analysis of all 193 effectors revealed a wide range of biological, molecular, and cellular component functions (Figures 2, 3 and Supplementary Figures 1–3). Simplified versions of the GO analysis are shown in Figures 2, 3, with complete versions in Supplementary Figures 1–3. The most common biological processes were metabolic processes and cellular processes representing 40 and 18 effectors, respectively (Figure 2 and Supplementary Figure 1) with at least 40 effectors involved in carbohydrate metabolic processes, and 13 involved in cellular macromolecule metabolic processes. In the molecular processes category, 65 effectors had catalytic activity, of those 33 had hydrolase activity, and 24 having lyase activity (Figure 3 and Supplementary Figure 2). There were 12 effectors that had binding activity, with 8 of those involved in nucleic acid binding (Figure 3 and Supplementary Figure 2). Sixty-three effectors had cellular component functions, with 45 possibly being cellular anatomical entities, and 34 of those having extracellular region functions (Supplementary Figure 3). These data revealed we have little current knowledge of the possible function of the majority of effectors while many of those with a possible function bind targets and have hydrolyzing ability.
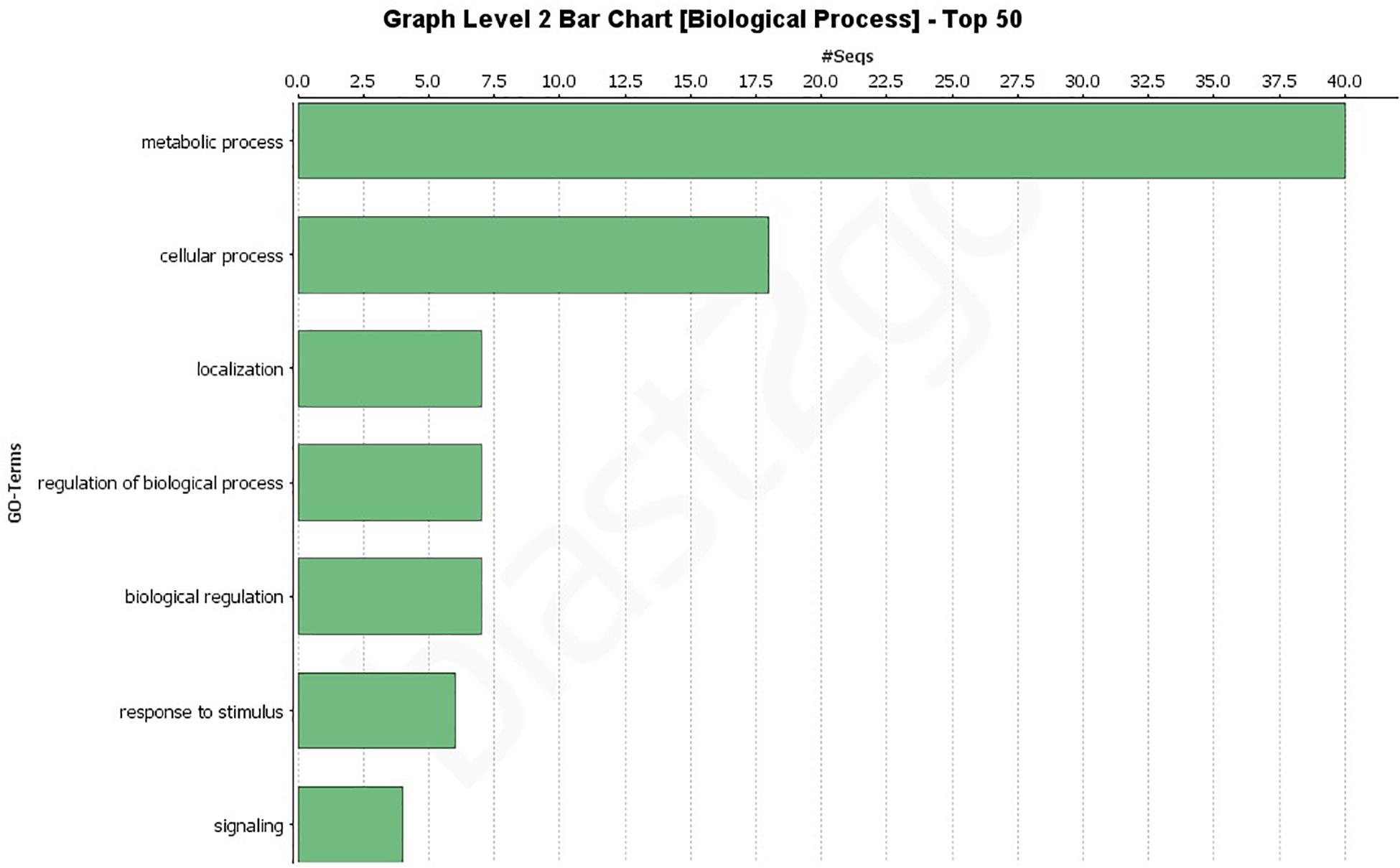
Figure 2. Gene Ontology (GO) level 2 bar chart of the biological processes of all 193 effectors found across 18 genomes and the JR2 reference genome. See Supplementary Figure 1 for full network analysis of all 193 effectors.
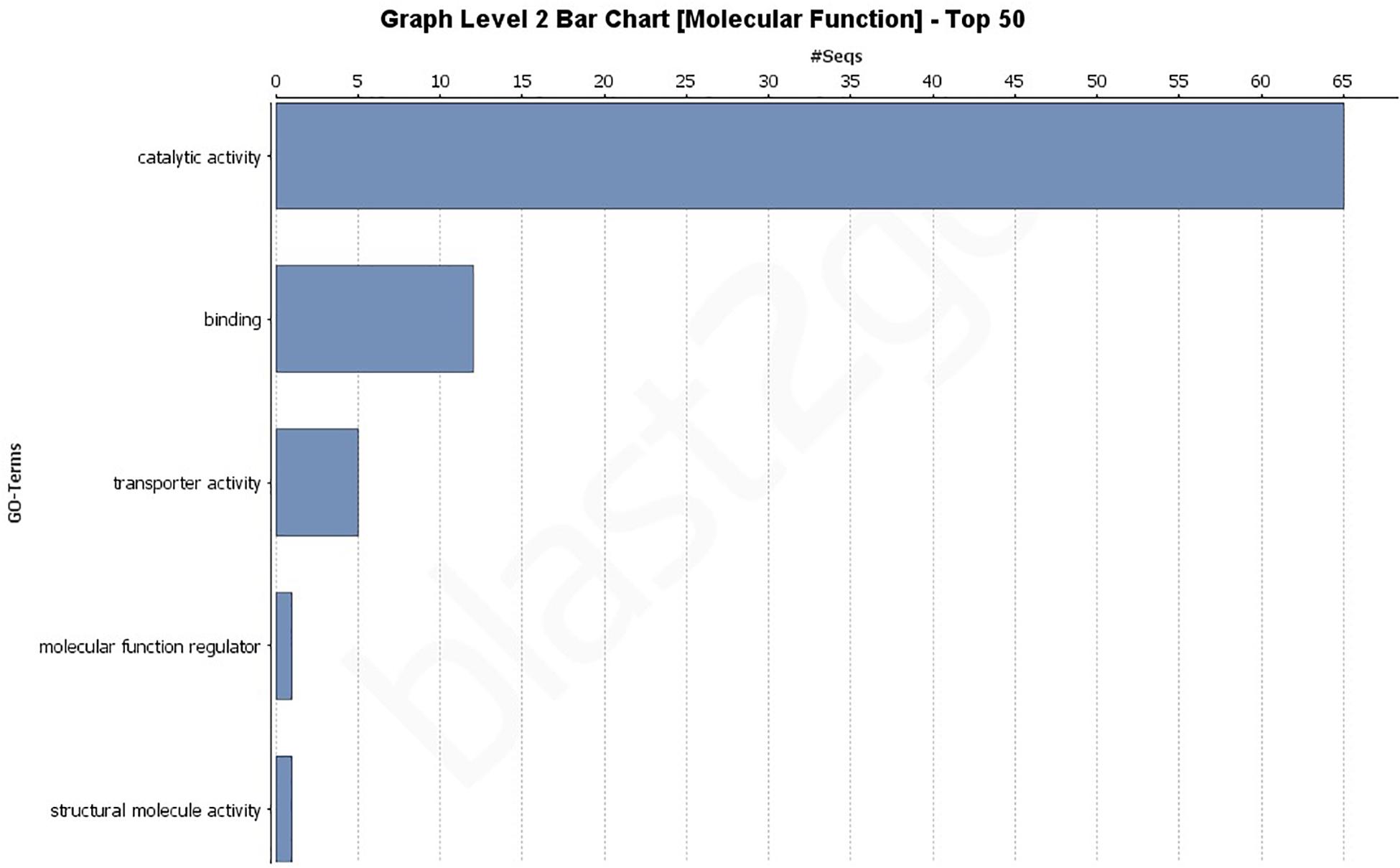
Figure 3. Gene Ontology (GO) level 2 bar chart of the molecular function of all 193 effectors found across 18 genomes and the JR2 reference genome. See Supplementary Figure 2 for full network analysis of all 193 effectors.
Classification and Genome Distribution of Effectors
The 193 effectors were broken down into 8 classes based on specific features (Table 2). Class 1 contained 75 core effectors (found in all 19 V. dahliae genomes) that had no non-synonymous mutations (NSM). There were 69 effectors classified as class 2 core effectors with 1 or more NSM. Class 3 contained 4 core effectors with truncation on the C-terminus in one or more isolates. There were 30 effectors classified as class 4 which contained non-core effectors that failed to pass through the SignalP or EffectorP pipelines in one or more isolates. Class 5 contained 8 effectors which had coding sequences missing in at least one genome. There were 4 effectors placed in Class 6 which were present in locally collinear blocks (LCBs) which were missing from one or more isolates. Class 7 contained a single effector (Ave1) present only in a highly variable region in an LCB only present in race 1 isolates JR2, Le1087, and Vdp4. The remaining 2 effectors were classified as Class 8 effectors and were present only race 2 isolates, and the race 1 isolates JR2, and Le1087 (Table 2). A full list of all unique effectors extracted from the 18 genomes and the JR2 reference genome, with their gene names and classification, is available in Supplementary Table 9.
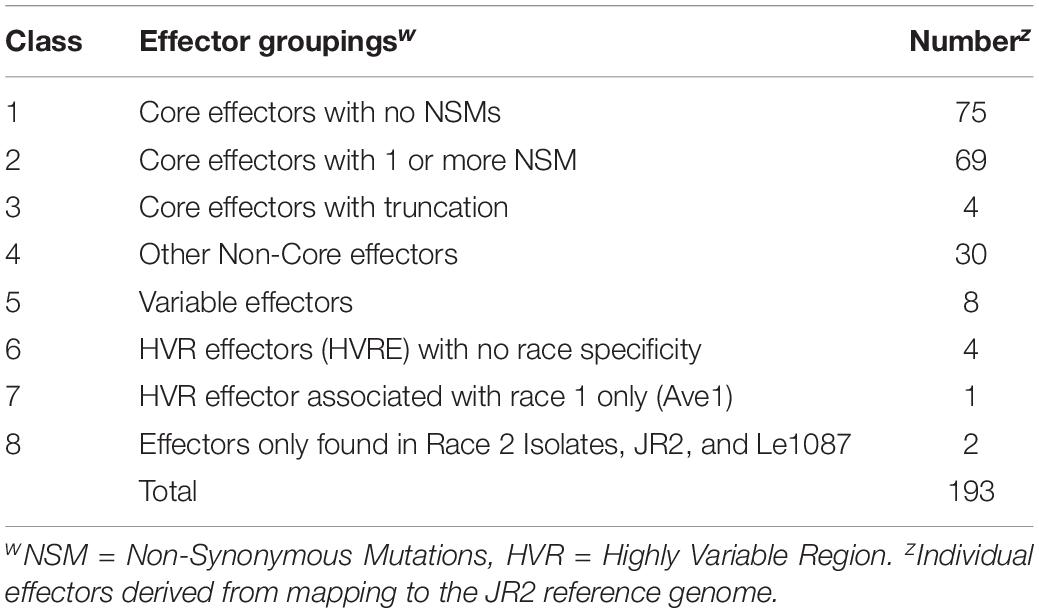
Table 2. Unique effector-like regions discovered through filtering coding sequences through SignalP and EffectorP. Coding sequences were extracted from mapping reads from 18 V. dahliae genomes to the JR2 reference genome (ASM15067v2).
Within the class 1 effectors, 21 effectors were highly conserved both in sequence and sequence lengths in all 19 genomes and V. alfalfae VaMs.102 (GCA_000150825) and V. longisporum (GCA_001268165). These cross-species core effectors display a diverse set of characteristics, 17 of which had GO descriptions (Table 3). In contrast, of the 15 genes assigned to classes 5–8 considered as non-core, only 8 had a specified GO description (Table 4). One of these non-core effectors has cysteine-rich domains found in CFEM-containing proteins (Kulkarni et al., 2003). Another is the race 1 effector Ave1 (de Jonge et al., 2012).
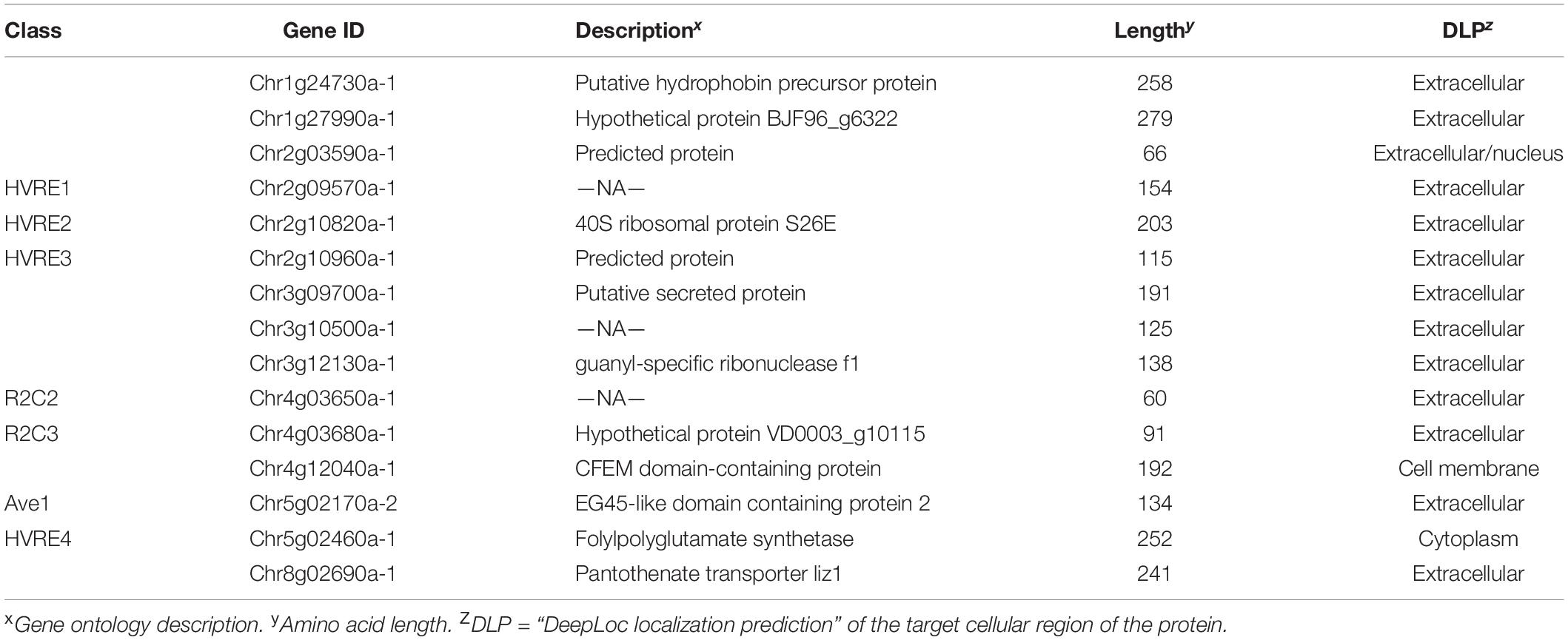
Table 4. Gene ontology and cellular localization analysis of effectors present in HVRs, or with missing coding sequences due to indels, of 18 V. dahliae genomes.
Predicted Effector Sequences Present in Race 2 Genomes
Two race 2 candidate effectors were discovered using the JR2 reference genome (R2C2 and R2C3) (Table 5). In Mauve alignments different colors are used to differentiate locally collinear blocks (LCBs). LCBs are regions which are non-rearranged homologous regions present in more than one isolates genome. Both these effectors were found in highly variable regions of chromosome 4 (Figures 4, 5). The region containing these effectors was missing in the near isogenic group 4 race 3 isolates FL10b and NC86, but were present in race 2 isolates, JR2 (race 1, group 1), and Le1087 (race 1, group 4) (Figure 5). Within the analysis of effectors from the VdLs17 reference genome (a lettuce isolate pathogenic on race 2 resistant tomatoes), there was a single gene which contained a non-synonymous mutation found only in race 2 isolates, but not in race 3 isolates (Table 5 and Figure 6). This race 2 candidate (R2C1) is a LysM effector, VdPDA1, which binds to chitin to prevent the recognition of chitin by host defenses (Gao et al., 2019). Notably, VdPDA1 was not recognized as a secreted effector in the JR2 reference genome because the provided annotation starts with a TTG start codon that adds 18 amino acids to the beginning of the gene sequence. JR2 contains the race 3 haplotype of VdPDA1, although its phenotype is unknown.
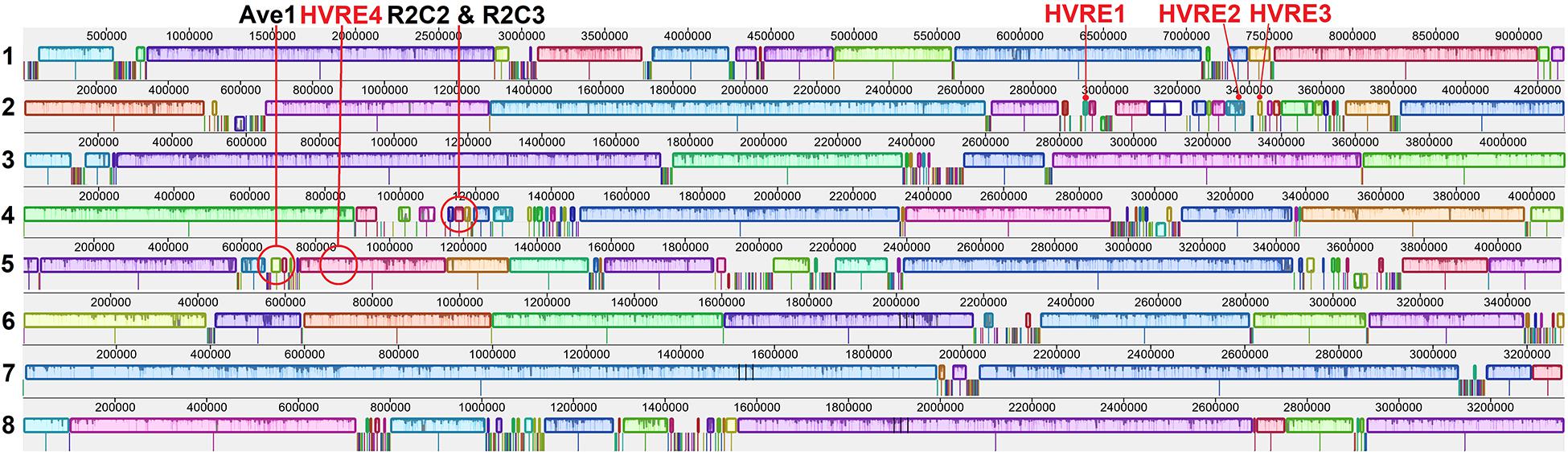
Figure 4. Mauve alignments of chromosomes 1–8 of JR2 with 18 genomes included in this study. Locally collinear blocks (LCBs) and highly variable regions (HVRs) are displayed. Shown here are the 3 race specific (black text) and 4 other effectors (red text) present in these HVRs on chromosomes 2, 4, and 5. Different colors are used to differentiate LCBs.

Figure 5. Mauve alignment of the specific region of JR2 chromosome 4 containing 2 race 2 candidate effectors (R2C2 and R2C3). Ca36 (race 2) and NC86 (race 3) are near-isogenic isolates belonging to group 4. Both candidate effectors are absent from NC86 and other race 3 isolates (not shown).
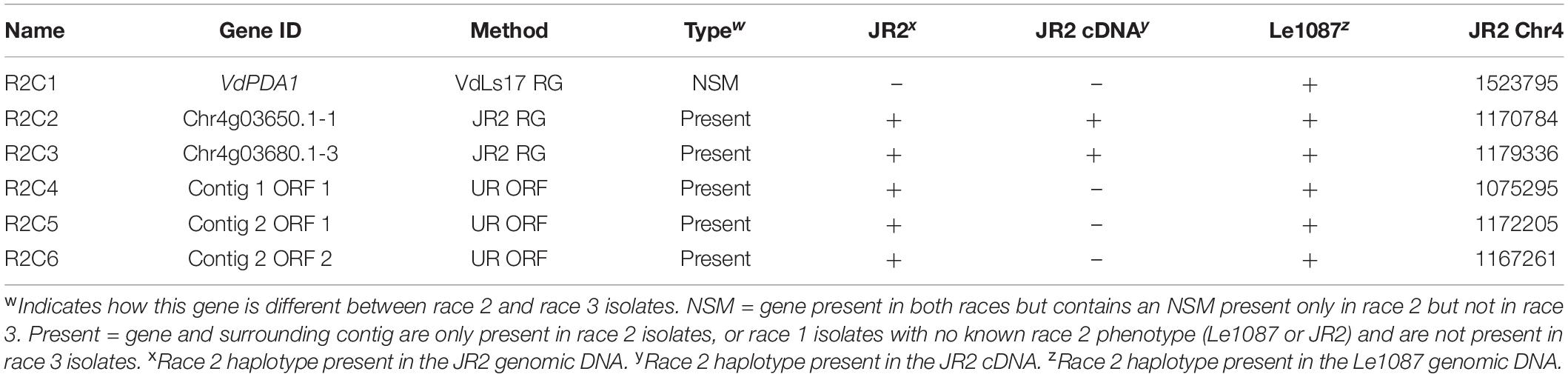
Table 5. Race 2 candidate effectors (R2C1-6) from three different discovery methods: VdLs17 reference genome; JR2 reference genome; and an ab initio gene discovery of the unassembled reads of the race 2 isolates mapped to the VdLs17 reference genome.
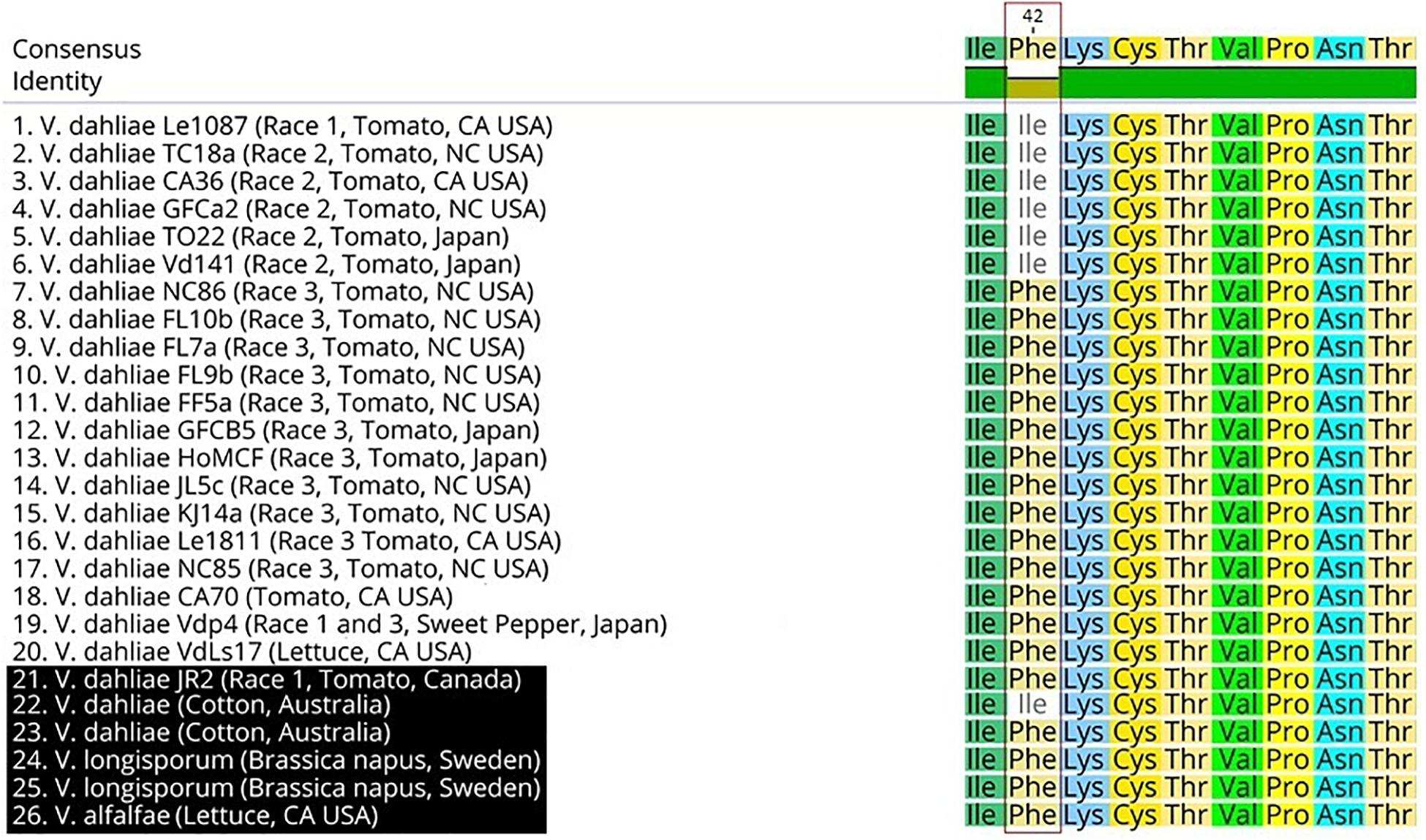
Figure 6. Protein sequence alignment of the R2C1 VdPDA1 gene. Sequences highlighted in black (21–26) have not been tested for the race2/3 phenotype. A single nucleotide polymorphism of A to T results in an isoleucine (ATC) to phenylalanine (TTC) conversion.
De novo Assembly of Unmapped Reads (Not Mapped to the VdLs17 Genome), Open Reading Frame (ORF) and Effector Analysis
Using an ORF analysis of a de novo assembly of reads that did not map to the race 3 reference genome VdLs17, three race 2 specific putative effectors (R2C4-6) were discovered (Table 5). The contigs containing these putative effectors were 29.4 Kb (contig 1) and 14.6 Kb (contig 2) in length. These putative effectors bore no sequence homology with known genes in BLAST and were not found in the JR2 cDNA library. Both contigs 1 and 2 resided in the highly variable region on JR2 chromosome 4 where R2C2 and R2C3 are located but were not annotated on that reference genome.
Discussion
In this study, we present a first report of race 2 and 3 isolates in North America, as well as genome sequence and analyses of 18 V. dahliae of isolates from various races pathogenic on tomato from a wide range of geographic regions to reveal specific phylogenetic and race specific signatures. Isolates infecting tomato were grouped into four phylogenetically distinct lineages. Group 3 contained only mating type MAT1-1, while all other groups were MAT1-2. No significant evidence of recombination was observed on any of the chromosomes (Table 1). Of the 193 effectors identified using the JR2 reference genome, over 75% had few to no changes to their amino acid sequences among genomes, indicating their function may be necessary or they are involved in fitness of V. dahliae (Table 2). At least 21 effectors were highly conserved between V. dahliae, V. longisporum, and V. alfalfae (Table 3). Fifteen effectors had coding sequences missing from a number of isolates, with 7 of those effectors found in highly variable regions, and 2 of which were specific to race 2 (Tables 4, 5). We also found additional race 2 specific ORFs with effector signatures in assembled reads that did not align to the VdLs17 (race 3) reference genome.
Phylogenetic Groupings
We found group 1 isolates to be the most phylogenetically distinct group, while the other groups were clustered much closer together (Figure 1). Group 1 also contains both reference genomes VdLs17 and JR2. According to other studies, VdLs17 and JR2 also have radically different chromosome structures, despite being phylogenetically similar to each other, having undergone chromosomal rearrangement in the not so distant past (de Jonge et al., 2013; Shi-Kunne et al., 2018). These data raise productive avenues of further research concerning the comparative chromosomal structure of groups 2, 3, and 4. Interestingly group 3 was only found in North Carolina, United States, and the only other publicly available genome comparable to group 3 was found from Canada (DVD-s29) which is also MAT1-1. An expanded analysis of a world-wide collection could ascertain the distribution of groups 1–4, potentially uncover additional groups, and provide a broader phylogenetic framework to offer additional insights regarding V. dahliae biology, race, and host range attributes. Further research is also needed into mating type genes, specifically group 3. While there is very little evidence of recombination, this may be due in part to the low number of MAT1-1, which make up only 1% of isolates, according to an analysis of 1120 isolates (Short et al., 2014). Group 3, which is unique to North Carolina and one location in Canada, has this rare MAT1-1 locus and is race 3. Interestingly several isolates of opposite mating types were found in the same fields, such as FL7a and FL9b, and NC85 and NC86 (Figure 1), although there is no evidence these isolates recombined in the recent past (Table 1).
Secreted Effector Genes
In this study V. dahliae isolates pathogenic on tomato were shown to have between 162 and 174 secreted effectors using the JR2 reference genome (Supplementary Table 6). A study of the VdLs17 genome in 2011 showed only 127 effectors, however, the method by which they were classified as effectors was by sequence length (<400 bp) and being rich in cysteine (Klosterman et al., 2011). A more recent study of the VdLs17 (using SignalP v4.1 and EffectorP v1.0) reported 179 (Gibriel et al., 2019). In this study, additional secreted effectors were found. It is unclear if this increase in secreted effectors is due to the discovery of more true effector genes or more false positives. Phenotyping secreted effectors should be a priority to increase our understanding of these crucial genes. Both core and highly variable effectors may play vital functions that could be exploited by researchers to influence pathogenicity. At least 65 effectors were annotated as having some form of catalytic activity, 24 lyase, 33 hydrolase, and 15 oxidoreductase activity. This may indicate that a large number of these secreted effectors are involved in the metabolism or recognition of carbohydrates (Figure 3 and Supplementary Figure 2). Further analysis of these genes should be conducted to determine how many genes recognized as effectors are also carbohydrate active enzymes (CAZymes). In general, more in depth analysis of each individual secreted effector is warranted.
Cross Species Core Effectors
Within the core effectors, 21 effectors were found not only in the 18 V. dahliae genomes in this study and the JR2 reference genome, but also V. alfalfae and V. longisporum (Table 3). Some of these highly conserved effectors have been shown to be important pathogenicity factors. Chr1g21900a-1 is a metalloprotease which is vital for V. dahliae virulence (Keykhasaber and Thomma, 2011). Chr6g08770a-1 is a necrosis- and ethylene-inducing protein and ethylene inducing peptide (VdNEP) which has been linked to wilting, and this gene is homologous to a similar gene in Fusarium oxysporum (Wang et al., 2004; Bae et al., 2006; Zhou et al., 2012). There are also six putative proteins with no known function (Table 3). More research is needed to identify the function of these predicted protein sequences. Some of these highly conserved core effectors may also be good targets for targeted host resistance research, such as host induced gene silencing.
Highly Variable Region Effectors Found on the JR2 Reference Genome
While 15 effectors were completely absent from one or more isolates genome, seven of those were present in highly variable regions; one of which is the race 1 effector Ave1, and two of which were present only in race 2 strains. There were also three putative effectors found in the ORF analysis in these regions. Further research is needed into these race 2 candidate genes, specifically the 5 found in the highly variable regions. If any of the race 2 candidates found in highly variable regions are shown to be the true race 2 effector, this would add significant support the hypothesis that V. dahliae is operating under the two-speed genome hypothesis whereby “effector genes reside in genomic compartments that are considerably more plastic than the core genome” (Faino et al., 2016).
VdPDA1 and Other LysM Effectors
The only candidate effector with identified function is VdPDA1, which contains a chitin binding domain. Effector triggered immunity against LysM effectors is unknown in tomatoes. The non-LysM effector Avr4 in Cladosporium fulvum protects chitin from hydrolysis by plant chitinases and can be recognized by host defenses in tomato (van den Burg et al., 2006). The VdPDA1 gene may have chitin binding properties that are also found in other virulence factors of V. dahliae (Miya et al., 2007; Kombrink et al., 2017; Gao et al., 2019). A homologous VdPDA1 (and functional) locus also exists in Fusarium oxysporum f. sp. vasinfectum which may function similarly, to the V. dahliae gene (Gao et al., 2019).
Race Designations of V. dahliae Isolates
With the discovery of a new source of host resistance in tomato against V. dahliae race 2, new questions emerge about the nomenclature surrounding this pathosystem. Except for Vdp4 (races 1 and 3) and JR2 (race 1), all isolates in groups 1–3, as circumscribed in this study, are race 3 (i.e., can infect “Aibou”). Ave1 is the effector responsible for the race 1 phenotype (de Jonge et al., 2012). Interestingly, in 2019 it was shown that deletion of Ave1 from Vdp4 resulted in the ability to be pathogenic on “Aibou,” i.e., become race 3 (Kano and Usami, 2019). In contrast the race 1 isolate Le1087 is in the phylogenetically distinct group 4. Le1087 also contains all the race 2 candidate secreted effectors. Group 4 is the only group with race 2 isolates, which suggests that within this group race 2 evolved from race 1 by loss of the Ave1 effector. Indeed, with the exception of Le1087, all isolates in group 4 lack Ave1. Further evidence for this hypothesis could be provided by deleting Ave1 from Le1087 and evaluating pathogenicity on Aibou. Our results also call into the question the race nomenclature. Race 3 may simply represent isolates that lack races 1 and 2 effectors, which could mean that race 3 actually comprises many races. Race 3 and further race designations will require the identification of additional differential germplasm. Before the Usami et al., 2017 race 2 publication, many scientific publications referred to non-race 1 isolates as race 2.
Conclusion and Perspectives
Whole genome analysis revealed 4 highly clonal lineages/groups of V. dahliae. Group 3 was unique to western North Carolina and has the rare MAT1-1 lineage. The recently described races 2 and 3 are both present in North Carolina and California in major tomato growing areas. Group 4 contains near-isogenic isolates of races 1, 2, and 3, indicating recent and rapid evolution. Many highly conserved cross-species effectors were observed. Using the JR2 reference genome there were seven effectors located in highly variable regions of the genome, the race 1 effector Ave1, and two race 2 specific effectors. In addition to those effectors, 4 other candidate race 2 effectors were identified using other methods.
To determine the nature of race 2 resistance in tomato, “Aibou,” and “Ganbarune-Karis,” were crossed and selfed, and their progeny were evaluated for resistance to race 2. The ratio of resistant to susceptible plants was 3:1 (Usami et al., 2017). This indicates that the resistance to race 2 is controlled by a single dominant locus. To maximize the genetic gain in this population, focus must be given to elucidate the plant-pathogen interactions at the molecular level. It appears that race 3 is widely prevalent in the region, thus, rapid and precision breeding is necessary by strategically integrating modern genomics approaches with advanced breeding techniques. Cost-effective third-generation sequencing technologies are now available to capture the SNPs via high-throughput genotyping. The identification and functional characterization of genes associated with disease resistance to all known races would be useful to ensure broad-spectrum and durable resistance to V. dahliae.
Data Availability Statement
Raw data for this project is available through Sequence Read Archive (SRA) bioproject PRJNA640002, https://www.ncbi.nlm.nih.gov/bioproject/640002. Biosamples SAMN15297318–SAMN15297335.
Author Contributions
TI conceived and designed the study with the guidance of YO, FL, and RD. TI performed the laboratory and greenhouse experiments. TI analyzed and interpreted the data with the aid of RD and YO. All authors contributed to the writing the manuscript. Grant writing and funding was provided by FL, TA, and RD. All authors have read and approved the final version of the manuscript.
Funding
Funding for this research was provided by a USDA-NIFA specialty crop research initiative under award number 2016-51181-25404. Addition funding and resources for this project were supplied by the NC Agricultural Foundation, Inc. and the North Carolina Tomato Growers Association.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Toshiyuki Usami at the Graduate School of Horticulture, Chiba University Japan for his contribution of DNA from V. dahliae isolates from Japan. We would also like to thank Dr. Krishna Subbarao at University of California, Davis for his contribution of V. dahliae isolates from California and Sakata Seed America, Inc. for providing United States isolates and seed for this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.573755/full#supplementary-material
Supplementary Figure 1 | Combined GO graph of the biological process of all 193 effectors.
Supplementary Figure 2 | Combined GO graph of the molecular functions of all 193 effectors.
Supplementary Figure 3 | Combined GO graph of the cellular component functions of all 193 effectors.
Supplementary Figure 4 | Water control “Bowman” seedling 45 days post inoculation.
Supplementary Figure 5 | Full amino acid alignment of VdPDA1.
Supplementary Figure 6 | “Bowman” seedling 45 days post inoculation with race 2 isolate Ca36.
Supplementary Figure 7 | “Bowman” seedling 45 days post inoculation with the race 3 isolate KJ14a.
Supplementary Figure 8 | “Bowman” seedling 45 days post inoculation with the race 3 isolate NC86.
Supplementary Table 1 | Experiment 1 of screening sequenced isolates against differential tomato lines. Bonny Best = universal susceptible; Red Defender = Ve1+ V2-; Aibou = Ve1+ V2+. Wilt and chlorosis/necrosis AUDPC scores displayed with Tukey’s HSD letters indicating significance groupings.
Supplementary Table 2 | Experiment 2 of screening sequenced isolates against differential tomato lines. Bonny Best = universal susceptible; Red Defender = Ve1+ V2-; Aibou = Ve1+ V2+. Wilt and chlorosis/necrosis AUDPC scores displayed with Tukey’s HSD letters indicating significance groupings.
Supplementary Table 3 | Experiment 3 of screening sequenced isolates against differential tomato lines. Bonny Best = universal susceptible; Red Defender = Ve1+ V2-; Aibou = Ve1+ V2+. Wilt and chlorosis/necrosis AUDPC scores displayed with Tukey’s HSD letters indicating significance groupings.
Supplementary Table 4 | Experiment 4 of screening sequenced isolates against differential tomato lines. Bonny Best = universal susceptible; Red Defender = Ve1+ V2-; Bowman = Ve1+ V2+, Ganbarune-Karis (GBK) = Ve1+ V2+. Wilt and chlorosis/necrosis AUDPC scores displayed with Tukey’s HSD letters indicating significance groupings.
Supplementary Table 5 | Total, assembled, and unassembled paired reads to the JR2 reference genome.
Supplementary Table 6 | Total number of secreted coding sequence regions and secreted effectors present on the consensus sequence of reads mapped to the JR2 reference genome.
Supplementary Table 7 | Total, assembled, and unassembled paired reads to the VdLs17 reference genome.
Supplementary Table 8 | Total number of secreted coding sequence regions and secreted effectors present on the consensus sequence of reads mapped to the VdLs17 reference genome.
Supplementary Table 9 | Spreadsheet of secreted effectors from JR2 RG analysis, and GO function definition.
Supplementary Table 10 | Meta data of V. dahliae isolates sequenced in this study.
References
Akamatsu, A., Wong, H. L., Fujiwara, M., Okuda, J., Nishide, K., and Uno, K. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476. doi: 10.1016/j.chom.2013.03.007
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Bae, H., Kim, M. S., Sicher, R. C., Bae, H. J., and Bailey, B. A. (2006). Necrosis-and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiol. 141, 1056–1067. doi: 10.1104/pp.106.076869
Belkhadir, Y., Subramaniam, R., and Dangl, J. L. (2004). Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. doi: 10.1016/j.pbi.2004.05.009
Bender, C. G., and Shoemaker, P. B. (1984). Prevalence of Verticillium wilt of tomato and virulence of Verticillium dahliae race 1 and race 2 isolates in western North Carolina. Plant Dis. 68, 305–309. doi: 10.1094/pd-69-305
Bhat, R. G., and Subbarao, K. V. (1999). Host range specificity in Verticillium dahliae. Phytopathology 89, 1218–1225. doi: 10.1094/phyto.1999.89.12.1218
Campbell, M. S., Holt, C., Moore, B., and Yandell, M. (2014a). Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinform. 48, 4–11.
Campbell, M. S., Law, M., Holt, C., Stein, J. C., Moghe, G. D., Hufnagel, D. E., et al. (2014b). MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524. doi: 10.1104/pp.113.230144
Chen, J., Liu, C., Gui, Y., Si, K., Zhang, D., Wang, J., et al. (2018). Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 217, 756–770. doi: 10.1111/nph.14861
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
de Jonge, R., Bolton, M. D., Kombrink, A., van den Berg, G. C., Yadeta, K. A., and Thomma, B. P. (2013). Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282. doi: 10.1101/gr.152660.112
de Jonge, R., Esse, H. P. V., Maruthachalam, K., Bolton, M. D., Santhanam, P., Saber, M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A 109, 5110–5115. doi: 10.1073/pnas.1119623109
Diwan, N., Fluhr, R., Eshed, Y., Zamir, D., and Tanksley, S. D. (1999). Mapping of Ve in tomato: a gene conferring resistance to the broad-spectrum pathogen, Verticillium dahliae race 1. Theor. Appl. Genet. 98, 315–319. doi: 10.1007/s001220051075
Ebel, J. (1986). Phytoalexin synthesis: the biochemical analysis of the induction process. Annu. Rev. Phytopathol. 24, 235–264. doi: 10.1146/annurev.py.24.090186.001315
Faino, L., Seidl, M. F., Datema, E., van den Berg, G. C., Janssen, A., Wittenberg, A. H., et al. (2015). Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. mBio 6:e00936-15.
Faino, L., Seidl, M. F., Shi-Kunne, X., Pauper, M., van den Berg, G. C., Wittenberg, A. H., et al. (2016). Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 26, 1091–1100. doi: 10.1101/gr.204974.116
Gao, F., Zhang, B.-S., Zhao, J.-H., Huang, J.-F., Jia, P.-S., Wang, S., et al. (2019). Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 5, 1167–1176. doi: 10.1038/s41477-019-0527-4
Gibriel, H., Li, J., Zhu, L., Seidl, M., and Thomma, B. (2019). Verticillium dahliae strains that infect the same host plant display highly divergent effector catalogs. bioRxiv [Preprint]. doi: 10.1101/528729v1
Green, R. J. (1980). Soil factors affecting survival of microsclerotia of Verticillium dahliae. Phytopathology. 70, 353–355. doi: 10.1094/phyto-70-353
Gullino, M. L., Minuto, A., and Garibaldi, A. (2002). Soil fumigation with chloropicrin in Italy: experimental results on melon, eggplant and tomato. Meded. Rijksuniv. Te Gent Fak. Van Landbouwkd. En Toegepaste Biol. Wet. 67, 171–180.
Hayafune, M., Berisio, R., Marchetti, R., Silipo, A., Kayama, M., Desaki, Y., et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U.S.A. 111, E404–E413.
Howe, L. R., Leevers, S. J., Gómez, N., Nakielny, S., Cohen, P., and Marshall, C. J. (1992). Activation of the MAP kinase pathway by the protein kinase raf. Cell 71, 335–342. doi: 10.1016/0092-8674(92)90361-f
Hu, X., Puri, K. D., Gurung, S., Klosterman, S. J., Wallis, C. M., Britton, M., et al. (2019). Proteome and metabolome analyses reveal differential responses in tomato -Verticillium dahliae-interactions. J. Proteomics. 207:103449. doi: 10.1016/j.jprot.2019.103449
Jukes, T., and Cantor, C. (1969). “Evolution of protein molecules,” in Mammalian Protein Metabolism, ed. H. N. Munro (New York: Academic Press), 21–132. doi: 10.1016/b978-1-4832-3211-9.50009-7
Kabir, Z., Bhat, R., and Subbarao, K. (2004). Comparison of media for recovery of Verticillium dahliae from soil. Plant Dis. 88, 49–55. doi: 10.1094/pdis.2004.88.1.49
Kano, K., and Usami, T. (2019). Race 3 tomato Verticillium wilt pathogen potentially derives from race 1 isolate. Soil Microorg. 73, 71–78.
Keykhasaber, M., and Thomma, B. P. H. J. (2011). Secreted Serine and Metalloproteases are Required for Virulence in Verticillium dahliae. 41–42. EPS Ph. D. thesis, Wageningen University, Wageningen.
Klosterman, S. J., Atallah, Z. K., Vallad, G. E., and Subbarao, K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. doi: 10.1146/annurev-phyto-080508-081748
Klosterman, S. J., Subbarao, K. V., Kang, S., Veronese, P., Gold, S. E., Thomma, B. P., et al. (2011). Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7:e1002137. doi: 10.1371/journal.ppat.1002137
Kombrink, A., Rovenich, H., Shi-Kunne, X., Rojas-Padilla, E., van den Berg, G. C., Domazakis, E., et al. (2017). Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol. Plant Pathol. 18, 596–608. doi: 10.1111/mpp.12520
Kombrink, E., and Schmelzer, E. (2001). The hypersensitive response and its role in local and systemic disease resistance. Eur. J. Plant Pathol. 107, 69–78.
Kombrink, E., and Somssich, I. (1997). Pathogenesis-Related Proteins and Plant Defense. Cham: Springer, 107–128.
Kulkarni, R. D., Kelkar, H. S., and Dean, R. A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121. doi: 10.1016/s0968-0004(03)00025-2
Melhem, H., Min, X. J., and Butler, G. (2013). The Impact of SignalP 4.0 on the Prediction of Secreted Proteins. Piscataway, NJ: IEEE, 16–22.
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Moller, S., Croning, M. D. R., and Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653. doi: 10.1093/bioinformatics/17.7.646
Newman, M.-A., Sundelin, T., Nielsen, J. T., and Erbs, G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4:139. doi: 10.3389/fpls.2013.00139
Peumans, W. J., and Van Damme, E. (1995). Lectins as plant defense proteins. Plant Physiol. 109:347. doi: 10.1104/pp.109.2.347
Rozas, J., Sánchez-DelBarrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi: 10.1093/bioinformatics/btg359
Sharma, N., Sharma, K., Gaur, R., and Gupta, V. (2011). Role of chitinase in plant defense. Asian J. Biochem. 6, 29–37. doi: 10.3923/ajb.2011.29.37
Shi-Kunne, X., Faino, L., van den Berg, G. C., Thomma, B. P., and Seidl, M. F. (2018). Evolution within the fungal genus Verticillium is characterized by chromosomal rearrangement and gene loss. Environ. Microbiol. 20, 1362–1373. doi: 10.1111/1462-2920.14037
Short, D. P., Gurung, S., Hu, X., Inderbitzin, P., and Subbarao, K. V. (2014). Maintenance of sex-related genes and the co-occurrence of both mating types in Verticillium dahliae. PLoS One 9:e112145. doi: 10.1371/journal.pone.0112145
Song, Y., Zhang, Z., Seidl, M. F., Majer, A., Jakse, J., Javornik, B., et al. (2017). Broad taxonomic characterization of Verticillium wilt resistance genes reveals an ancient origin of the tomato Ve1 immune receptor. Mol. Plant Pathol. 18, 195–209. doi: 10.1111/mpp.12390
Sperschneider, J., Gardiner, D. M., Dodds, P. N., Tini, F., Covarelli, L., Singh, K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210, 743–761. doi: 10.1111/nph.13794
Sutherland, M. W. (1991). The generation of oxygen radicals during host plant responses to infection. Physiol. Mol. Plant Pathol. 39, 71–93.
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Usami, T., Ishigaki, S., Takashina, H., Matsubara, Y., and Amemiya, Y. (2007). Cloning of DNA fragments specific to the pathotype and race of Verticillium dahliae. J. Gen. Plant Pathol. 73, 89–95. doi: 10.1007/s10327-006-0334-4
Usami, T., Itoh, M., and Amemiya, Y. (2009). Asexual fungus Verticillium dahliae is potentially heterothallic. J. Gen. Plant Pathol. 75:422. doi: 10.1007/s10327-009-0197-6
Usami, T., Momma, N., Kikuchi, S., Watanabe, H., Hayashi, A., Mizukawa, M., et al. (2017). Race 2 of Verticillium dahliae infecting tomato in Japan can be split into two races with differential pathogenicity on resistant rootstocks. Plant Pathol. 66, 230–238. doi: 10.1111/ppa.12576
van den Burg, H. A., Harrison, S. J., Joosten, M. H., Vervoort, J., and de Wit, P. J. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant. Microbe Interact. 19, 1420–1430. doi: 10.1094/mpmi-19-1420
Wang, J. Y., Cai, Y., Gou, J. Y., Mao, Y. B., Xu, Y. H., Jiang, W. H., et al. (2004). VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl. Environ. Microbiol. 70, 4989–4995. doi: 10.1128/aem.70.8.4989-4995.2004
Xiao, C. L., Subbarao, K. V., Schulbach, K. F., and Koike, S. T. (1998). Effects of crop rotation and irrigation on Verticillium dahliae microsclerotia in soil and wilt in cauliflower. Phytopathology. 88, 1046–1055. doi: 10.1094/phyto.1998.88.10.1046
Yandell, M., and Ence, D. (2012). A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 13, 329–342. doi: 10.1038/nrg3174
Keywords: Solanum lycopersicum, secreted effectors, whole genome resequencing, soilborne fungus, Verticillium dahliae race 2
Citation: Ingram TW, Oh Y, Adhikari TB, Louws FJ and Dean RA (2020) Comparative Genome Analyses of 18 Verticillium dahliae Tomato Isolates Reveals Phylogenetic and Race Specific Signatures. Front. Microbiol. 11:573755. doi: 10.3389/fmicb.2020.573755
Received: 17 June 2020; Accepted: 11 November 2020;
Published: 30 November 2020.
Edited by:
Fred Asiegbu, University of Helsinki, FinlandReviewed by:
Georgios Tzelepis, Swedish University of Agricultural Sciences, SwedenStefan Kusch, RWTH Aachen University, Germany
Copyright © 2020 Ingram, Oh, Adhikari, Louws and Dean. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas W. Ingram, dHdpbmdyYTJAbmNzdS5lZHU=; Ralph A. Dean, cmFkZWFuMkBuY3N1LmVkdQ==
 Thomas W. Ingram
Thomas W. Ingram Yeonyee Oh
Yeonyee Oh Tika B. Adhikari
Tika B. Adhikari Frank J. Louws
Frank J. Louws Ralph A. Dean
Ralph A. Dean