
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 November 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.573056
This article is part of the Research Topic Plant Microbiome: Interactions, Mechanisms of Action, and Applications View all 26 articles
The conventional definition of endophytes is that they do not cause disease, whereas pathogens do. Complicating this convention, however, is the poorly explored phenomenon that some microbes are endophytes in some plants but pathogens in others. Black cottonwood or poplar (Populus trichocarpa) and wheat (Triticum aestivum) are common wild and crop plants, respectively, in the Pacific Northwest USA. The former anchors wild, riparian communities, whereas the latter is an introduced domesticate of commercial importance in the region. We isolated Fusarium culmorum – a well-known pathogen of wheat causing both blight and rot – from the leaf of a black cottonwood tree in western Washington. The pathogenicity of this cottonwood isolate and of a wheat isolate of F. culmorum were compared by inoculating both cottonwood and wheat in a greenhouse experiment. We found that both the cottonwood and wheat isolates of F. culmorum significantly reduced the growth of wheat, whereas they had no impact on cottonwood growth. Our results demonstrate that the cottonwood isolate of F. culmorum is endophytic in one plant species but pathogenic in another. Using sequence-based methods, we found an additional 56 taxa in the foliar microbiome of cottonwood that matched the sequences of pathogens of other plants of the region. These sequence-based findings suggest, though they do not prove, that P. trichocarpa may host many additional pathogens of other plants.
The plant microbiome is thought to aid plants under stressful conditions to enable them to adapt to new habitats. This is, in essence, the habitat-adapted symbiosis hypothesis (Rodriguez et al., 2004, 2008). This hypothesis is at the heart of current attempts to engineer microbiome-based adaptation to climate change, and to other stresses of a rapidly changing world (Busby et al., 2017). One of the challenges in this effort is that plants can asymptomatically host pathogens of other plants (Porras-Alfaro and Bayman, 2011). A concern is thus that successful bio-engineering of one plant could lead to undesirable, non-target, spillover effects in another, via augmentation of a pathogen of the latter.
In plant microbiome studies currently, and in earlier studies, pathogens of other plants are regularly reported on the basis of the taxa that they represent. Pathogenic function is seldom, however, confirmed experimentally. For example, in a study of the endophytes of Centaurea stoebe (Shipunov et al., 2008), the following pathogens were reported without proof of function: Botrytis cinerea, the cause of gray mold of many other plants (though never of C. stoebe itself), Diaporthe helianthi, the cause of Phomopsis stem canker of sunflower, Gibberella avenacea, the cause of many rots, blights and declines of many crops (though again, not of C. stoebe itself), and many other taxa of Fusarium and Alternaria that are pathogens of plants other than C. stoebe. Sequence-based evidence of the pathogen that causes the “mal secco” disease of citrus was even found in achenes of C. stoebe, although inoculations were never performed to confirm function (Shipunov et al., 2008; Migheli et al., 2009). In a study of endophytes of Bromus tectorum, Fusarium oxysporum was reported (Baynes et al., 2012). Fusarium oxysporum causes vascular wilts of a very wide range of plants, as well as blights, rots and damping off (Farr and Rossman, 2020), but the endophytic isolate in B. tectorum might possibly have been non-pathogenic. Similarly, species of Geniculosporium and Xylaria, pathogenic to various plants, were found as endophytes in Abies, or fir, trees (Carroll and Carroll, 1978). Further examples from other woody plants have been summarized in a study of white pine endophytes that often resembled pathogens of other plants (Ganley et al., 2004). In all of these studies, pathogenic function was suggested by the identification of isolates and/or sequences representative of pathogenic taxa of other plants; function was not, however, proven via inoculation assay in these studies.
In contrast, in studies focused on invaded plant communities, spillover of a pathogen from one host to another has been functionally demonstrated. For example, Alternaria spp., isolated from the reservoir host, Schedonorus arundinaceus (tall fescue, a highly invasive grass species in North America), could be inoculated into co-occurring grasses in which these fungi caused disease symptoms and decreased biomass (Wilson et al., 2014). Pathogen spillover from a reservoir host to other plants can also operate within seed banks in plant communities invaded by Bromus tectorum (Beckstead et al., 2010). Pathogen spillover could be ecologically common and important in natural plant communities, invaded or not, but a shortage of studies hampers generalization.
In a third area of research, on pathogens shared by crop and weedy plants, there are examples of pathogen movement from weed to crop, and its epidemiological significance (Wisler and Norris, 2005). Crop rotation to reduce pathogen inoculum can even fail because weedy plants host the pathogen when the crop is absent. For instance, weedy Chenopodium album can host Verticillium albo-atrum that can then re-infect alfalfa when it is again grown in a given field (Busch and Smith, 1982). Rust fungi can be hosted by wild oats and then infect domesticated oats (Burdon et al., 1983). Movement of pathogens in the other direction (i.e., from crop plants to weeds or to wild plants) has been little studied (Blitzer et al., 2012), which is attributed to little study generally of natural plant pathosystems (Power and Mitchell, 2004).
A recent, molecular field study of foliar fungi of Populus trichocarpa revealed taxa that are putative pathogens of other plants (Barge et al., 2019). P. trichocarpa is the wild black cottonwood tree of riparian communities in the Pacific Northwest. It is frequently found in close proximity to wheat and other crops of the region. Here, we summarize the pathogens of other plants that were found in the sequence-based study (Barge et al., 2019), and report on an experiment testing the pathogenicity of Fusarium culmorum isolated from P. trichocarpa on both wheat and cottonwood. Fusarium culmorum is a well-known pathogen of wheat, that has not been reported as a pathogen of P. trichocarpa. However, F. culmorum has been reported as a mutualist in dunegrass of the region (Rodriguez et al., 2008). We therefore expected that our assay with a cottonwood isolate could reveal positive, negative, or neutral effects on growth of cottonwood and wheat.
We selected one putative pathogen for a pathogenicity experiment: Fusarium culmorum. An isolate of F. culmorum collected from asymptomatic leaves of P. trichocarpa in a previous study (SNO-11, Busby et al., 2016) produced a colony resembling a wheat isolate of Fusarium culmorum, a known pathogen of wheat. We identified the isolate on the basis of micro-morphology as Fusarium. To obtain a species-level identification we extracted DNA from the isolate, then amplified and sequenced the full ITS region. The Sanger sequence of the full ITS region for this isolate is archived in the NCBI genbank (accession number MN154167). This particular cottonwood, or poplar isolate, P. trichocarpa (FCP) was then used in the inoculation experiment below to test the hypothesis of endophytes as functional pathogens of other plants. For comparative purposes we used a wheat isolate of F. culmorum (FCW) from a previous study (Ridout and Newcombe, 2016).
Cultures of each strain were grown on 4% potato dextrose agar (PDA) for 2 weeks. At 2 weeks plates were flushed with sterile distilled water (SDW) and conidia were loosened with a sterile, bent glass rod. The solution was homogenized with a tissue macerator (Tissue Tearor™, BioSpec Products, Bartlesville, OK, USA) and brought to volume with SDW. Conidial concentrations of the FCW and FCP suspensions were adjusted to 106 conidia per milliliter using a Neubauer hemacytometer.
Seeds of P. trichocarpa (collected from a wild tree growing in Moscow, ID) and T. aestivum (University of Idaho line 306 UI-SRG, Lot: 1209 Moscow HRWW5, class hard red winter) were sown into soilless mix (Sunshine Professional Growing Mix #1, Sun Gro Horticulture, Sacramento, CA) in four-inch pots. Seeds were germinated and seedlings grown at a diurnal temperature cycle of 18°C/15°C with a 16-h day length. Ninety pots were sown for each species. Seedlings were grown in the greenhouse for 12 days prior to inoculation.
We inoculated T. aestivum and P. trichocarpa with the F. culmorum isolated from P. trichocarpa (FCP) and a pathogenic strain of F. culmorum (FCW) known to cause crown rot in T. aestivum (Washington State University/ USDA ARS, Pullman WA: cereal pathogen collections). Twelve days following sowing, 30 seedlings each of P. trichocarpa and T. aestivum were inoculated at the crown with either 10 mL of FCP suspension, 10 mL of FCW suspension, or 10 mL of SDW (sterile distilled water) for a negative control. The plants were kept in the greenhouse for 47 days while disease developed. Visual analyses were made to determine disease presence or absence in inoculated seedlings. T. aestivum seedlings were then harvested by removing the above-ground vegetation at the crown; F. culmorum causes crown rot and thus affects above-ground biomass. This biomass was then dried in an oven at 60°C for roughly 48 h, although drying time varied somewhat. Above-ground biomass of P. trichocarpa seedlings was harvested 4 days later and dried at 60°C for 70 h. Dry biomass was determined for all the seedlings.
We used ITS sequence data from a previously published molecular field survey of Populus trichocarpa foliar fungi across ten watersheds in the Pacific Northwest, USA (Busby et al., 2016) to identify fungal taxa likely to cause disease in other plants. A complete description of this molecular field survey can be found in Busby et al. (2016). In brief, we sampled leaves from six trees in 10 populations. Leaves were surface sterilized and lyophilized prior to DNA extraction. We used a modified version of the primer set ITS1F and ITS2 for sequencing on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) (Smith and Peay, 2014). PCR products were cleaned using the Agencourt Ampure XP kit (Beckman Coulter, Brea, CA, USA), quantified using the Qubit hs-DS-DNA kit (Invitrogen, Carlsbad, CA, USA) on a Tecan Infinite F200 Pro plate reader (Tecan, Morrisville, NC, USA) (285 nm excitation and 530 nm emission), then pooled at equimolar concentrations prior to 250-bp paired-end sequencing using Illumina MiSeq. Raw sequence data are deposited in NCBI's Short Read Archive (accession no. SRP064132, http://www.ncbi.nlm.nih.gov/books/NBK47529/).
Both QIIME (Caporaso et al., 2010) and UPARSE (Edgar, 2013) were used to process the sequence data. Forward and reverse reads were paired with USEARCH v.7.0.1001, and discarded if they contained > 0.25 expected errors. High-quality sequences were grouped into operational taxonomic units (OTUs) in USEARCH using UPARSE-OTU and UPARSE OTU algorithms at 97% similarity. Taxonomy was assigned using the BLAST algorithm in QIIME, which uses the UNITE fungal ITS database. Additionally, the 500 most abundant OTUs were checked using BLAST searches against the NCBI GenBank. In total, 968 fungal taxa were identified. For this study, we identified putative pathogens in the dataset by searching for all taxa in the USDA SMML databases (Farr and Rossman, 2020). We calculated the proportional abundance of each putative pathogen within a sample by dividing the number of pathogen reads in the sample by the number of total reads in the sample. We additionally calculated the mean proportional abundance of each putative pathogen for each tree population.
All analyses were performed in RStudio v.3.6.3 (RStudio Team 2015). We used a one-way analysis of variance to compare dry weights among the treatment groups in both T. aestivum and P. trichocarpa.
Both isolates of F. culmorum – from cottonwood and wheat – caused visual symptoms of disease (Figure 1) and a reduction in above-ground biomass for wheat; neither symptoms nor a reduction in biomass were seen in cottonwood (Figure 2). Ten days after the inoculation, wheat leaves became symptomatic. The first necrotic lesions appeared on the twelfth day post-inoculation. The uninoculated wheat controls remained asymptomatic throughout the experiment (Figure 1). After destructive sampling, the mean mass of the control plants was 7.70 grams, compared to 6.22 and 6.35 g, respectively (F = 11.13, p = 4.84 × 10−5; Figure 2). Thus, both isolates were associated with an ~20% reduction in wheat biomass. In contrast, both isolates of F. culmorum – from cottonwood and wheat – caused no visible symptoms of disease (Figure 1) nor impacted cottonwood biomass (F = 0.33, p = 0.72; Figure 2).

Figure 1. Fusarium culmorum isolates caused disease in wheat (A) but not in poplars (B). Wheat and poplars were inoculated with an isolate of F. culmorum from wheat (FCW) and from P. trichocarpa (FCP), or with sterile water for the control (CTRL).
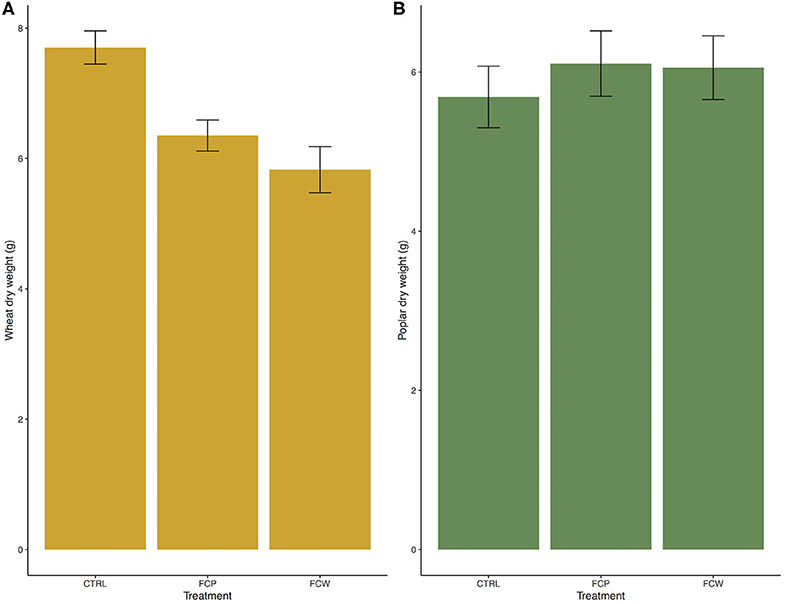
Figure 2. Fusarium culmorum isolates reduced biomass in wheat (A) but not in poplars (B). FCW: isolate of F. culmorum from wheat. FCP: isolate of F. culmorum from P. trichocarpa. CTRL: sterile water control. Bars are standard error.
The USDA SMML Fungal Database shows that F. culmorum has previously been reported from 134 plant species, in 22 families (Farr and Rossman, 2020). It is most common as a pathogen of plants of the grass family, Poaceae, but it is common in plants of other families as an endophyte. This report is the first to mention any species of Populus and even its family, Salicaceae, as hosts of endophytic F. culmorum. The 23 families in which F. culmorum has now been reported are the following: Amaranthaceae, Anacardiaceae, Apiaceae, Asparagaceae, Asteraceae, Brassicaceae, Caryophyllaceae, Chenopodiaceae, Convolvulaceae, Cucurbitaceae, Cyperaceae, Ericaceae, Fabaceae, Fagaceae, Lamiaceae, Liliaceae, Linaceae, Malvaceae, Pinaceae, Poaceae, Polygonaceae, Salicaceae, and Solanaceae.
Many pathogens like F. culmorum are pathogenic in only some of the plant species from which they have been reported. In others they are present as endophytes or even as mutualists (Rodriguez and Redman, 2008; Rodriguez et al., 2008). In some, such as Washington's coastal dunegrass (Leymus mollis), F. culmorum is a mutualist that improves both salt and drought tolerance. With unequal effects on competing species of plants, microbes like F. culmorum could be important drivers of “apparent competition” (Orrock and Witter, 2010; Cobey and Lipsitch, 2013). This occurs when one competitor is favored by hosting a pathogen of the other.
We identified 56 sequence-based taxa in the foliar microbiome of cottonwood that are pathogens of other plants of the region (Table 1), but that are not known to cause disease in P. trichocarpa (Newcombe, 1996). Cryptodiaporthe pulchella, which is a pathogen of a few species of Populus and Salix, has not been recorded as a pathogen of P. trichocarpa (Farr and Rossman, 2020). Similarly, Knufia cryptophialidica affects P. tremuloides but not P. trichocarpa (Farr and Rossman, 2020). Some pathogens of P. trichocarpa were found as foliar endophytes in our earlier studies, and those have been reported and discussed elsewhere (Busby et al., 2016; Barge et al., 2019).
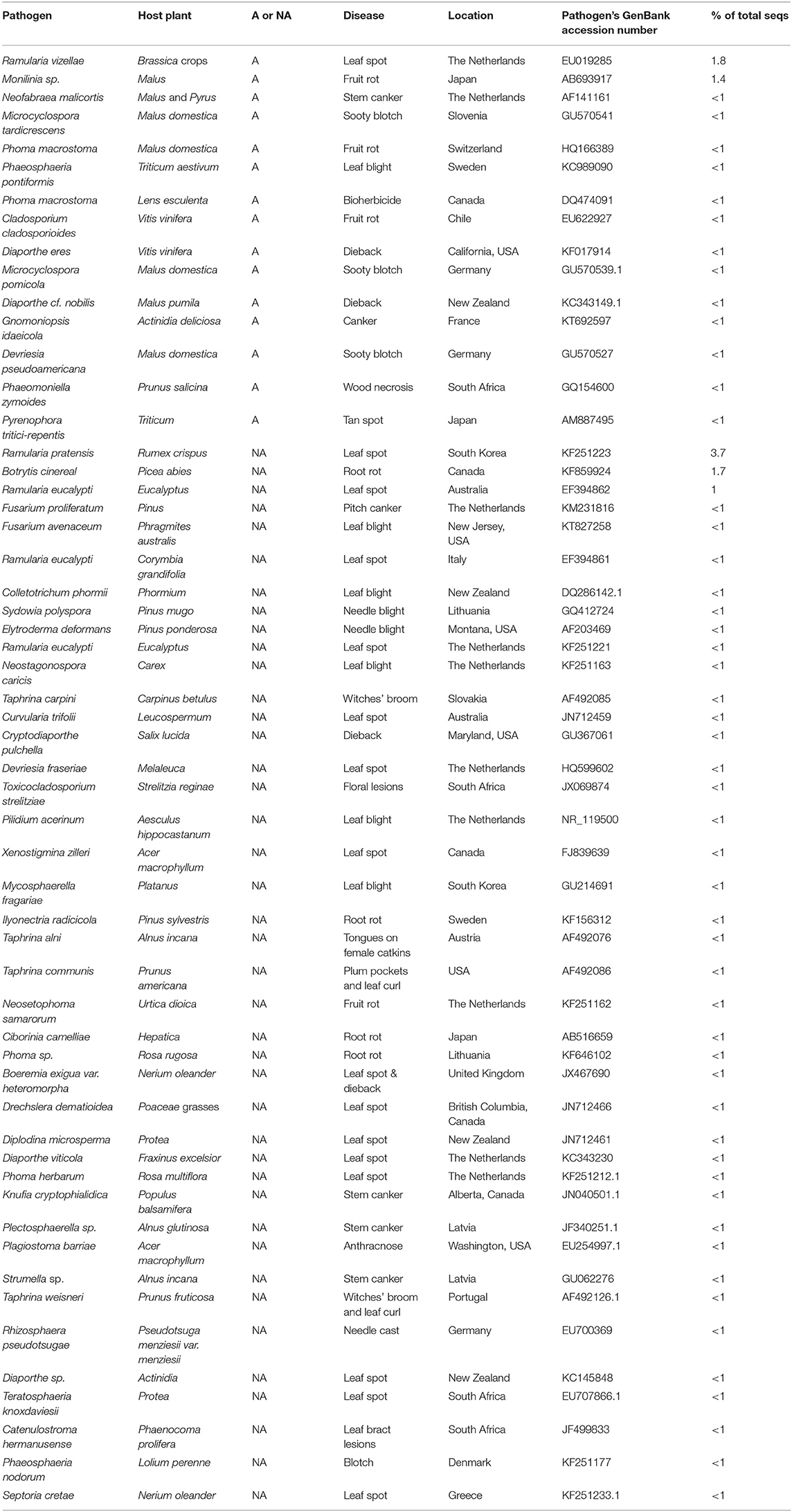
Table 1. High-throughput sequencing of leaves of Populus trichocarpa indicated GenBank- and USDA SMML-reported pathogens of agricultural (A) or non-agricultural plants (NA), their diseases, locations, accession numbers, and % of total sequences in the dataset.
Fifteen pathogens of agricultural or cultivated plants were found to varying extent in leaves of P. trichocarpa sampled in the Pacific Northwest (Table 1). These 15 fungal taxa identified by sequence homology with GenBank accessions were associated with many regionally important plants. Malus (apple) stood out, as 7 of the 15 pathogens were associated with this leading orchard crop of the region. Three wheat pathogens (i.e., Pyrenophora tritici-repentis, Phaeosphaeria pontiformis, Phaeosphaeria nodorum) were found in addition to the cultured and pathogenicity-tested isolate of F. culmorum. Another three taxa of fungal pathogens were found that have been associated with damage to cherry and grape, which are also regionally important crops. It is important to note that the agricultural host plants listed in Table 1 represent only a fraction of the broad host ranges of this group of 15 pathogen taxa. For example, Phoma macrostoma is potentially pathogenic to all dicots (Bailey et al., 2011), and as such this fungus has been of interest as a bioherbicide that could be used with monocot crops.
Among the 41 pathogens of non-agricultural plants, there were some with very broad host ranges (e.g., Botrytis cinerea) and others that were very host-specific (Table 1). Elytroderma deformans is an example of the latter as it only causes disease of species of Pinus subgenus Pinus, otherwise known as the “hard” or diploxylon pines. Elytroderma deformans is also limited in its distribution in North America, and its presence as an endophyte has been reported only in Pinus subgenus Strobus (Ganley et al., 2004). Thus, this report is the first for E. deformans in an angiosperm tree. A second example was found in the four taxa of Taphrina that are similarly host-specific pathogens. They parasitize species of Prunus, Alnus, and Carpinus. A third example, Ciborinia camelliae, specifically affects flowers of species of Camellia (Farr and Rossman, 2020). The most abundant pathogen was one that most closely matched a GenBank accession of Ramularia pratensis found on Rumex crispus in Korea. It is a specialized pathogen of leaves of species in only three genera, including Rumex, of the Polygonaceae. These genera and Ramularia pratensis are now found around the world where these plants have been introduced (Farr and Rossman, 2020).
In some cases the mean proportional abundance of a putative pathogen in the ITS dataset was consistent with expectations based on the abundance and distribution of the pathogen's preferred host. For example, F. proliferatum was more abundant in the two Idaho sites where its host, Pinus ponderosa (Ocamb et al., 2002), is dominant. Similarly, R. vizellae was more abundant in the western Washington sites where Brassica seed production occurs (Du Toit, 2004).
The putative pathogens of other plants, revealed here by ITS metabarcoding, would need to be tested for pathogenicity, as we have done with F. culmorum. If confirmed as pathogens, they could be either unspecialized and broad-range like F. culmorum, or highly host-specific. Host specificity varies considerably among the fungal pathogens of Populus (Newcombe, 1996) and among plant pathogens generally. Host ranges tend to be broader when they are compiled from identifications of pathogens and narrower when investigated via functional assay (Newcombe, 2003; Benítez et al., 2013; Sarmiento et al., 2017). However, we have not explored the transmissibility of the 56 pathogens, nor even that of F. culmorum, since we used artificial inoculation to prove the latter's pathogenicity toward wheat. Would cottonwood seedlings infected with Ramularia pratensis transmit their infection in nature to competing species of Rumex, Rheum or Oxyria? Would the competitive balance be tipped due to cottonwood's tolerance and the susceptibility of the others? Further studies are needed to explore how hosting pathogens of neighboring plants contributes to apparent competition.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MN154167, in NCBI's Short Read Archive (accession no. SRP064132, http://www.ncbi.nlm.nih.gov/books/NBK47529/).
GN and PB conceived of the idea for the study and wrote the manuscript with input from SF and MR. SF and MR conducted experiments. PB analyzed the data. All authors contributed to the article and approved the submitted version.
This study was supported by the Agriculture and Food Research Initiative Competitive Grant no. 2011- 68005-30407 from the USDA National Institute of Food and Agriculture (GN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Josh Miller for help with the inoculation experiment, and Gillian Bergmann and Lucas Nebert for help with manuscript preparation.
Bailey, K. L., Pitt, W. M., Falk, S., and Derby, J. (2011). The effects of Phoma macrostoma on nontarget plant and target weed species. Biol. Control. 58, 379–386. doi: 10.1016/j.biocontrol.2011.06.001
Barge, E. G., Leopold, D. R., Peay, K. G., Newcombe, G., and Busby, P. E. (2019). Differentiating spatial from environmental effects on foliar fungal communities of Populus trichocarpa. J. Biogeogr. 46, 2001–2011. doi: 10.1111/jbi.13641
Baynes, M., Newcombe, G., Dixon, L., Castlebury, L., and O'Donnell, K. (2012). A novel plant–fungal mutualism associated with fire. Fungal Biol. 116, 133–144. doi: 10.1016/j.funbio.2011.10.008
Beckstead, J., Meyer, S. E., Connolly, B. M., Huck, M. B., and Street, L. E. (2010). Cheatgrass facilitates spillover of a seed bank pathogen onto native grass species. J Ecol. 98, 168–177. doi: 10.1111/j.1365-2745.2009.01599.x
Benítez, M. S., Hersh, M. H., Vilgalys, R., and Clark, J. S. (2013) Pathogen regulation of plant diversity via effective specialization. Trends Ecol. Evol. 28, 705–711. doi: 10.1016/j.tree.2013.09.005
Blitzer, E. J., Dormann, C. F., Holzschuh, A., Klein, A. M., Rand, T. A., and Tscharntke, T. (2012). Spillover of functionally important organisms between managed and natural habitats. Agric. Ecosyst. Environ. 146, 34–43. doi: 10.1016/j.agee.2011.09.005
Burdon, J. J., Oates, J. D., and Marshall, D. R. (1983). Interactions between avena and puccinia species. I. The Wild Hosts: Avena barbata pott ex link, A. fatua L., and A. ludoviciana Durieu. J Appl Ecol. 20, 571–584. doi: 10.2307/2403527
Busby, P. E., Peay, K. G., and Newcombe, G. (2016). Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytolo. 209, 1681–1692. doi: 10.1111/nph.13742
Busby, P. E., Soman, C., Wagner, M. R., Friesen, M. L., Kremer, J., Bennett, A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15:e2001793. doi: 10.1371/journal.pbio.2001793
Busch, L. V., and Smith, E. A. (1982). Reaction of a number of cultivated plants and weed species to an alfalfa isolate of Verticillium albo-atrum in Ontario, Canada. Can. J. Plant Pathol. 4, 266–268. doi: 10.1080/07060668209501294
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carroll, G. C., and Carroll, F. E. (1978). Studies on the incidence of coniferous needle endophytes in the Pacific Northwest. Can. J. Bot. 56, 3034–3043. doi: 10.1139/b78-367
Cobey, S., and Lipsitch, M. (2013). Pathogen diversity and hidden regimes of apparent competition. Am. Nat. 181, 12–24. doi: 10.1086/668598
Du Toit, L. J. (2004). Management of diseases in seed crops. Encycl. Plant. Crop. Sci. 675–677. doi: 10.1081/E-EPCS-120019947
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Farr, D. F., and Rossman, A. Y. Fungal Databases U.S. National Fungus Collections, ARS, USDA. (2020). Available online at: https://nt.ars-grin.gov/fungaldatabases/ (accessed March 25, 2020).
Ganley, R. J., Brunsfeld, S. J., and Newcombe, G. (2004). A community of unknown, endophytic fungi in western white pine Proc. Natl Acad Sci. U.S.A. 101, 10107–10112. doi: 10.1073/pnas.0401513101
Migheli, Q., Cacciola, S. O., Balmas, V., Pane, A., Ezra, D., and di San Lio, G. M. (2009). Mal secco disease caused by Phoma tracheiphila: a potential threat to lemon production worldwide. Plant Dis. 93, 852–867. doi: 10.1094/PDIS-93-9-0852
Newcombe, G. (1996). “The specificity of fungal pathogens of Populus,” in Biology of Populus and its Implications for Management and Conservation. Part I, Chapter 10, eds R. F. Stettler, H. D. Bradshaw, Jr., P. E. Heilman, and T. M. Hinckley (Ottawa, ON: NRC Research Press), 223–246.
Newcombe, G. (2003). Puccinia tanaceti: specialist or generalist? Mycol Res. 107, 797–802. doi: 10.1017/S0953756203008025
Ocamb, C. M., Juzwik, J., and Martin, F. B. (2002). Fusarium spp., and Pinus strobus seedlings: root disease pathogens and taxa associated with seed. New Forests 24, 67–79. doi: 10.1023/A:1020589121134
Orrock, J. L., and Witter, M. S. (2010). Multiple drivers of apparent competition reduce re-establishment of a native plant in invaded habitats. Oikos 119, 101–108. doi: 10.1111/j.1600-0706.2009.17831.x
Porras-Alfaro, A., and Bayman, P. (2011). Hidden fungi, emergent properties: endophytes and microbiomes. Annu. Rev. Phytopathol. 49, 291–315. doi: 10.1146/annurev-phyto-080508-081831
Power, A. G., and Mitchell, C. E. (2004). Pathogen spillover in disease epidemics. Am. Nat. 164, S79–S89. doi: 10.1086/424610
Ridout, M., and Newcombe, G. (2016). Disease suppression in winter wheat from novel symbiosis with forest fungi. Fungal Ecol. 20, 40–48. doi: 10.1016/j.funeco.2015.10.005
Rodriguez, R., and Redman, R. (2008). More than 400 million years of evolution and some plants still can't make it on their own: plant stress tolerance via fungal symbiosis. J. Exp. Bot. 59, 1109–1114. doi: 10.1093/jxb/erm342
Rodriguez, R. J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. doi: 10.1038/ismej.2007.106
Rodriguez, R. J., Redman, R. S., and Henson, J. M. (2004). The role of fungal symbioses in the adaptation of plants to high stress environments. Mitig Adapt. Strategies Glob. Change 9, 261–272. doi: 10.1023/B:MITI.0000029922.31110.97
Sarmiento, C., Zalamea, P.C., Dalling, J. W., Davis, A. S., Stump, S. M., U'Ren, J. M. et al. (2017). Soilborne fungi have host affinity and host-specific effects on seed germination and survival in a lowland tropical forest. Proc. Natl. Acad. Sci.U.S.A. 114, 11458–11463. doi: 10.1073/pnas.1706324114
Shipunov, A., Newcombe, G., Raghavendra, A. K., and Anderson, C. L. (2008). Hidden diversity of endophytic fungi in an invasive plant. Am. J. Bot. 95, 1096–1108. doi: 10.3732/ajb.0800024
Smith, D. P., and Peay, K. G. (2014). Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE 9:e90234. doi: 10.1371/journal.pone.0090234
Wilson, H. E., Carroll, G. C., Roy, B. A., and Blaisdell, G. K. (2014). Tall fescue is a potential spillover reservoir host for Alternaria species. Mycologia 106, 22–31. doi: 10.3852/12-330
Keywords: fungal endophyte, plant pathogen, pathogen spillover, Fusarium, Populus, Triticum
Citation: Newcombe G, Fraser SJ, Ridout M and Busby PE (2020) Leaf Endophytes of Populus trichocarpa Act as Pathogens of Neighboring Plant Species. Front. Microbiol. 11:573056. doi: 10.3389/fmicb.2020.573056
Received: 15 June 2020; Accepted: 15 September 2020;
Published: 17 November 2020.
Edited by:
Nicole Marie Gerardo, Emory University, United StatesReviewed by:
Paul-Camilo Zalamea, University of South Florida, United StatesCopyright © 2020 Newcombe, Fraser, Ridout and Busby. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Posy E. Busby, cG9zeS5idXNieUBvcmVnb25zdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.