- 1Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
- 2Department of Infectious Diseases, Central Clinical School, Monash University, Melbourne, VIC, Australia
- 3Department of Basic Health Sciences, State University of Maringá, Maringá, Brazil
- 4Department of Clinical Analysis, Health Sciences Center, Federal University of Santa Catarina, Florianópolis, Brazil
- 5Department of Clinical Analysis, University of São Paulo, São Paulo, Brazil
Integrative conjugative elements (ICEs) are widespread in many bacterial species, often carrying antibiotic resistance determinants. In the present work, we screened a collection of Proteus mirabilis clinical isolates for the presence of type 1 SXT/R391 ICEs. Among the 76 isolates analyzed, 5 of them carry such elements. The complete sequences of these elements were obtained. One of the isolates carried the CMY-2 beta-lactamase gene in a transposon and is nearly identical to the element ICEPmiJpn1 previously described in Japan, and later shown to be present in other parts of the world, indicating global spread of this element. Nevertheless, the Brazilian isolate carrying ICEPmiJpn1 is not clonally related to the other lineages carrying the same element around the world. The other ICEs identified in this work do not carry known antibiotic resistance markers and are diverse in variable gene content and size, suggesting that these elements may be responsible for the acquisition of other advantageous traits by bacteria. Some sequences carried by these elements in Brazilian strains were not previously found in other SXT/R391 variants.
Introduction
Proteus mirabilis is one of the most common causes of urinary tract infections (Armbruster and Mobley, 2012). This bacterium also has the potential to cause other diseases, including a number of nosocomial infections (O’Hara et al., 2000). Proteus is one of the genera of gram-negative bacteria in which ICEs (Integrative Conjugative Elements) of the SXT/R391 family are usually found, similarly to Morganella, Providencia, Vibrio and others (Wozniak et al., 2009). These mobile elements so far constitute the most commonly found family of conjugative transposons according to the ICEberg database (Liu et al., 2019). They have the ability to integrate in a specific locus in the chromosome. Type 1 SXT/R391 ICEs integrate into the 5’ portion of the prfC gene, while types 2, 3, and 4 integrate into the 3’ portion of the tRNA-Ser gene. (Hochhut and Waldor, 1999; Bioteau et al., 2018). Excision from the chromosome and conjugation are stimulated by DNA damaging conditions. A LexA-like repressor, SetR, is cleaved in a RecA-dependent manner (Beaber et al., 2004), leading to expression of the SetCD activators, which in turn increase the expression of genes related to transfer of the ICE (Poulin-Laprade et al., 2015). Once inside a new host, integrase (encoded by int) mediates site specific integration of the ICE in prfC in a way that a functional allele of this gene is maintained in the chromosome (Hochhut and Waldor, 1999). Notably, these elements are also able to mobilize other genomic islands in trans (Daccord et al., 2010), therefore playing an important role in microbial genomic plasticity.
The basic structure of these elements is more or less conserved among variants from different species, with a conserved core of genes involved in the basic functions of excision, conjugation and integration (Bioteau et al., 2018). Variable regions and insertion hotspots are points of new DNA insertion, in which genes related to antimicrobial and heavy metal resistance can be acquired. These elements are large and the insertion points often contain other types of genes unrelated to resistance, many of which with unknown function (Wozniak et al., 2009; Rodríguez-Blanco et al., 2016).
These elements have attracted a great deal of interest, due to their potential to spread resistance determinants. In V. cholerae, these elements were originally identified as conferring resistance to sulfamethoxazole, trimethoprim, streptomycin and chloramphenicol (Waldor et al., 1996). In P. mirabilis, many instances of occurrence of such elements have been reported. Interestingly, some P. mirabilis strains harbor ICE SXT/R391 variants which do not carry known antibiotic resistance determinants, including the type strain HI4320 (Pearson et al., 2008; Wozniak et al., 2009). This observation raises the question of which selective advantage would be conferred by such large elements not carrying resistance genes. Nevertheless, SXT/R391 elements also mediate resistance dissemination in P. mirabilis. Of particular interest and prevalence, the variant ICEPmiJpn1, originally identified in Japan, carries a CMY-2 beta-lactamase gene (Harada et al., 2010). This particular variant of SXT/R391 ICEs was subsequently identified in P. mirabilis isolates from human and animal origins in Spain, India, France, China, and Ireland (Mata et al., 2011; Aberkane et al., 2016; Lei et al., 2016; Mac Aogáin et al., 2016; Bie et al., 2017).
Other variants of this element were recently identified in different P. mirabilis strains, carrying resistance to fluoroquinolones, beta-lactams, fosfomycin, and a rRNA methyltransferase conferring resistance to oxazolidinones and lincosamides (Bie et al., 2017; Lei et al., 2018). In other Proteus species, SXT/R391 elements also carry resistance to multiple drugs (He et al., 2020). In order to gain further insight into the biology, evolution and role of these elements in antimicrobial resistance spread, we investigated the presence and genetic background of SXT/R391 ICEs in Brazilian P. mirabilis clinical isolates.
Materials and Methods
Screening of SXT/R391 Elements in P. mirabilis Isolates
Screening of SXT/R391 elements was performed using previously described primers to detect int (sxtintF: 5′ TCGATGATGGTCTCTAGCTG 3′ and sxtintR: 5′ TCAGTTAGCTGGCTCGATGC 3′) (Mata et al., 2011) and rumA (rumAF: 5′ TGGTGACCACACCAAATATCTC 3′ and rumAR: 5′ AAGCCAAGCGCCTTCGTATT 3′). rumA primers were designed in this study to amplify a conserved region in this gene, spanning nucleotides 47 to 418 of the rumA coding region in ICEPmiUSA1 from strain HI4320, used in this study as the prototypic SXT/R391 element from P. mirabilis. While rumA primers could detect genes from all types of SXT/R391 ICEs (1, 2, 3 and 4), primers for int are specific for type 1 elements, which are the focus of this study. Isolates were screened by colony PCR, using rpoB amplification as a positive control for the reaction by using primers rpoBF: 5′ GAATGTCAGATCCGTGGTGT 3′, and rpoBR: 5′ CCAACCGCAGAGAGATCATA 3′. A total of 76 P. mirabilis clinical isolates were investigated, which were collected between 2014 and 2015 from patients in hospitals located in the cities of São Paulo and Maringá, in Southeast and Southern Brazil, respectively (SisGen number A9D703D). PCR was performed as follows. 1 U of Taq DNA polymerase (Sinapse Inc., Brazil) was used in 25 μL reactions containing 20 ng of genomic DNA and 0.25 μM of each primer. The following cycling conditions were used: initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 1 min, annealing for 30 s and extension at 72°C (calculated for 1 min/kbp), followed by final extension at 72°C for 10 min. The annealing temperature of each reaction is: int - 51°C, rumA - 60°C, and rpoB - 54°C.
Antimicrobial Susceptibility Assays
The antibiotic resistance profiles of ICE-harboring strains were determined by disk-diffusion method following guidelines from CLSIs (CLSI, 2016). The antibiotics tested were: amoxicillin-clavulanate (AMC) 20/10 μg, piperacillin-tazobactam (PPT) 100/10 μg, ampicillin (AMP) 10 μg, cephalothin (CFL) 30 μg, cefotaxime (CTX) 30 μg, ceftriaxone (CRO) 30 μg, cefepime (CPM) 30 μg, ceftazidime (CAZ) 30 μg, ertapenem (ETP) 10 μg, imipenem (IPM) 10 μg, meropenem (MER) 10 μg, nalidixic acid (NAL) 30 μg, ciprofloxacin (CIP) 5 μg, levofloxacin (LEV) 5 μg, norfloxacin (NOR) 10 μg, amikacin (AMI) 30 μg, gentamicin (GEN) 10 μg, tobramycin (TOB) 10 μg, and sulfamethoxazole-trimethoprim (SUT) 1.25/23.75 μg. The test was performed using Müeller-Hinton agar (MHA) plates and as a control we used the reference strain Escherichia coli ATCC 25922.
Genome Sequencing and Assembly
For all strains, total genomic DNA was extracted from saturated cultures using the Wizard Genomic DNA Kit (Promega) and the genome sequencing was performed by MicrobesNG1 with Illumina HiSeq using a 250 bp paired end protocol. Only for strain PmBR607, the Oxford Nanopore Sequencing (ONT) was also performed. For this purpose, total DNA was extracted using the Purelink genomic DNA mini kit (Invitrogen), and libraries were prepared using the Rapid Barcoding Kit (SQK-RBK004), and sequencing was performed with FLO-MIN106D flowcell.
Illumina reads were filtered by quality and adapters were removed using trimomatic v.0.392 with minimum quality threshold PHRED < 20 for passed reads. de novo assembly was achieved using SPAdes v. 3.11 (Bankevich et al., 2012). For ONT reads the quality filter was performed using guppy 2.1.3 and the hybrid assembly was obtained with Unicycler (Wick et al., 2017). The annotation was performed by NCBI Prokaryotic Genome Annotation Pipeline (PGAP).
For PmBR19, Sanger sequencing was also used to complete the gap between two contigs containing the ICE sequence. For this purpose, the gap region was amplified with primers designed based on PmBR19 genome sequence aligned to ICEPmiJpn1 (KT894734): tn10F (5′ TTCGTTGCTTGTGAGGTGAG 3′) and tn10R (5′ AAACAACGGCTGGAATGTGC 3′). Then the PCR product was purified with NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel) and Sanger sequencing was performed with primers tn10F, tn10R and 3 more primers designed for the gap region based on ICEPmiJpn1 sequence Tn10.1 (5′ GCCACGAGTAAGTCTTCCCT 3′), Tn10.2 (5′ GTCAGCCTCTTATAGCCTAAAGT 3′) and Tn10.3 (5′ GCCACGCATTACTTGACTGT 3′). The consensus sequence obtained from this Sanger sequencing was used to obtain the full sequence of ICEPmiJpn1 from PmBR19. The genome assemblies were deposited in Genbank BioProject database under accession code PRJNA576511.
Conjugation Assays
Filter mating assays were performed to test ICEPmiJpn1 (PmBR19) mobility. Overnight cultures of the donor strain PmBR19 (carrying ICEPmiJpn1 conferring resistance to Ampicillin) and recipient strain E. coli MG1655 RifR, were harvested by centrifugation and mixed at 1:1 ratio. The mixture was plated onto a sterile 0.22 μm pore size membrane filter placed on a non-selective MacConkey agar, and incubated overnight at 37°C. Then the filter was washed with LB broth and the non-diluted mixture was plated onto MacConkey agar plates containing rifampicin 100 μg/mL and ampicillin 100 μg/mL, in order to count transconjugants (i.e., Lac+, RifR, AmpR). Additionally, serial dilutions were plated onto MacConkey agar containing rifampicin to count recipient cells (i.e., Lac+, RifR). The plates were incubated overnight at 37°C for 24 hours, and CFU counts used to obtain the frequency of transconjugants per recipient cells. The reported frequency is the average of 4 independent experiments.
Phylogenetic Analysis
Phylogenetic analysis of 74 different ICEs from both our study and public databases3 was performed by SNP calling and phylogeny inference using CSI Phylogeny 1.44 (Kaas et al., 2014). The entire sequences of these elements were used as inputs. All P. mirabilis ICEs analyzed in previous publications were included, the only exception were very similar ICEs from the same study, in which case only one representative was used. Also, representative ICEs from other species were included in the analysis, as well as previously uncharacterized elements from P. mirabilis found in whole genome sequences deposited in the NCBI database. Accession numbers are provided in Supplementary Table S1. The same pipeline was used to infer phylogenetic relationships of P. mirabilis strains containing the ICEs for which whole genome sequence is available, as well as a similar number of strains devoid of this element. Accession numbers of these genome sequences are provided in Supplementary Table S2. This analysis uses FastTree to build the approximately-maximum-likelihood trees (Price et al., 2010).
Results
Occurrence of SXT/R391 Elements in P. mirabilis
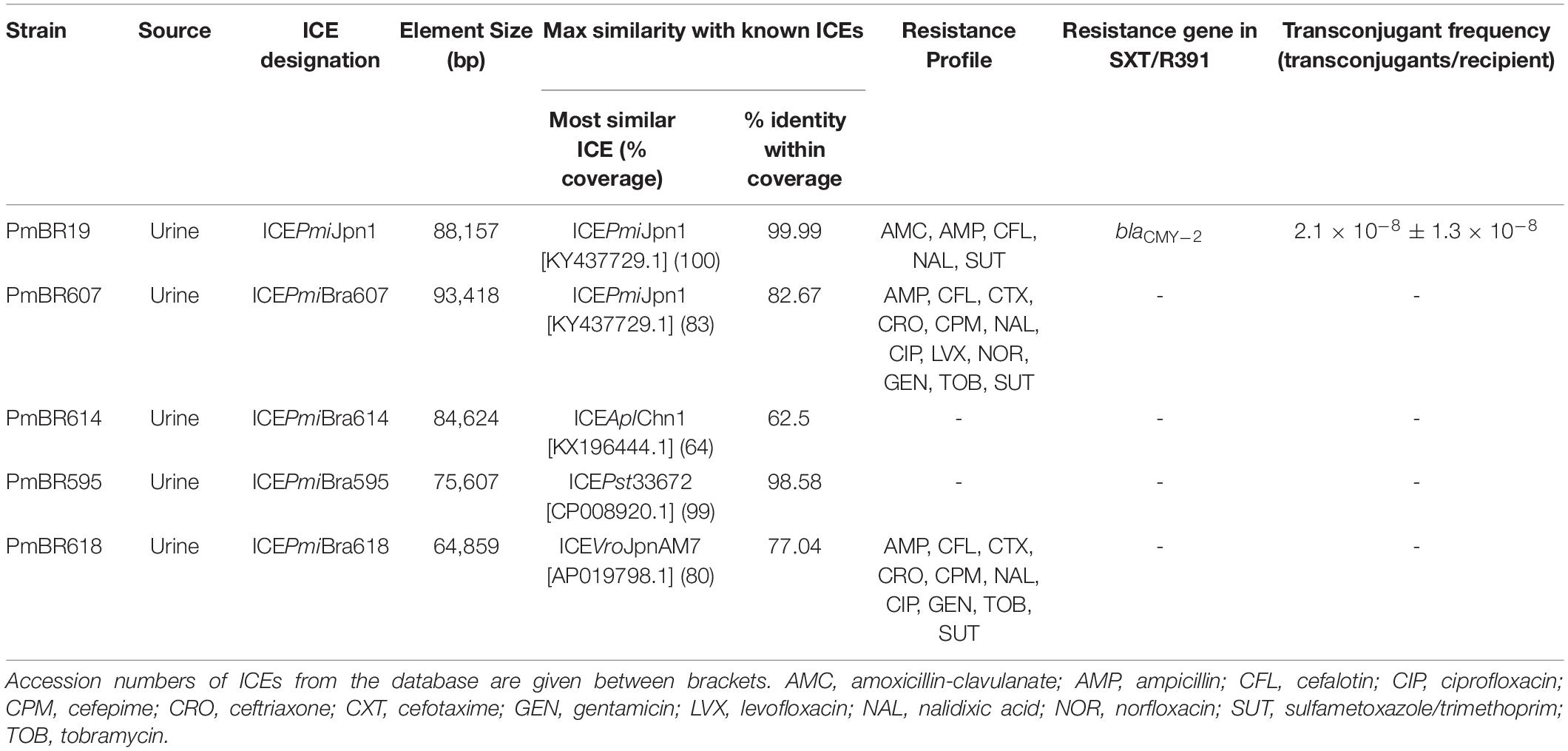
The distribution of SXT/R391 ICEs in P. mirabilis clinical isolates from Brazil was determined by screening a collection of isolates using a PCR strategy to detect the integrase (int) and rumA genes, two conserved core genes. Among 76 isolates, 5 carried int and rumA, indicating the presence of a type 1 SXT/R391 ICE. This represents a prevalence of 6.5% of this genetic element among these clinical isolates. All 5 SXT/R391 ICE-harboring isolates originated from urine samples. Antimicrobial resistance profiles of the ICE-harboring isolates, determined by disk diffusion method (according to CLSI guidelines) (CLSI, 2016), are shown in Table 1. Isolates PmBR19, PmBR607 and PmBR618 are multidrug-resistant (MDR) (Magiorakos et al., 2012). Nevertheless, isolates PmBR614 and PmBR595 are not MDR, despite carrying SXT/R391 ICEs.
Comparative Genomic Analysis of SXT/R391 ICEs
Isolates bearing SXT/R391 elements were subject to genome sequencing with the Illumina platform. The resulting assemblies produced the entire sequence of the ICEs contained in one single contig for isolates PmBR595, PmBR614, and PmBR618. In the assembly of Illumina data from PmBR19, the ICE was split in two different contigs. Nevertheless, we detected that its structure is identical to ICEPmiJpn1, and Sanger sequencing was used to complete the gap between the two contigs. For isolate PmBR607, the ICE was also fragmented in two or more contigs, with a structure similar to ICEPmiJpn1, but with a significant variation, since the sequence inserted in variable region V, which includes the CMY-2 beta-lactamase, was not present in any contig. To obtain the structure of this ICE, we used Nanopore sequencing. Illumina and Nanopore reads were assembled in a complete genome. A summary of the most important aspects of the ICEs, their assigned names according to the proposal of Burrus and co-workers (Burrus et al., 2006) and the characteristics of the respective bacterial strains are shown in Table 1. The structures of the ICEs are depicted in Figure 1 in comparison to the structure of the ICEPmiUSA1 from the type strain HI4320. The precise locations of each ICE in our genome assemblies are described in Supplementary Table S3.
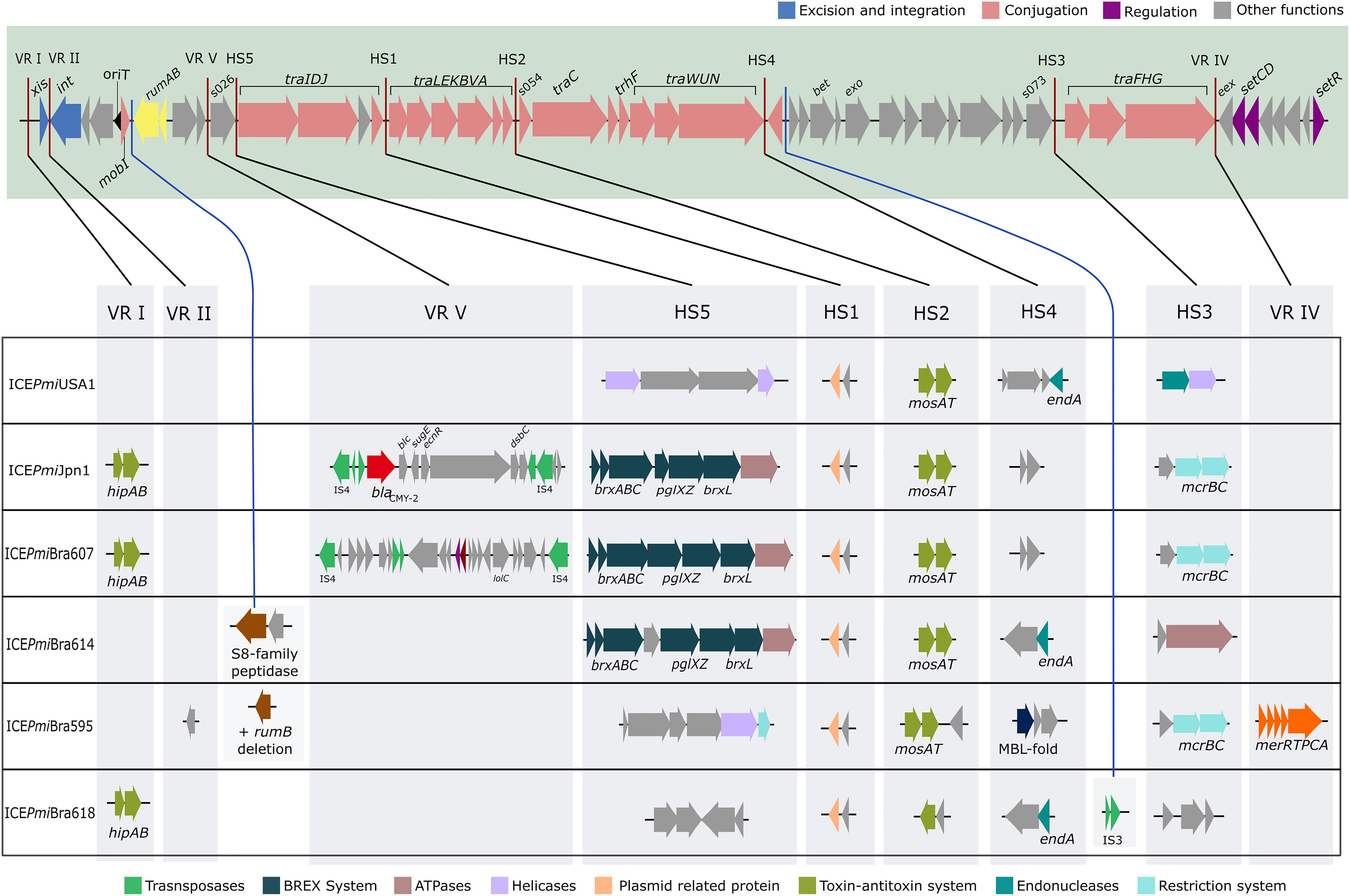
Figure 1. Schematic view of SXT/R391 ICEs from P. mirabilis from Brazil, and ICEPmiUSA1 (HI4320). In the shadowed box the backbone of this family of ICEs with conserved core genes is represented. Among the backbone genes, those colored blue are associated with excision and integration, genes colored pink are associated with transfer machinery, and genes colored purple are associated with regulation. Other genes with unknown function or with functions apparently not associated with transfer process are represented in gray, and the operon rumAB involved in error-prone DNA repair is colored yellow. Red lines indicate the position of hotspots (HS) and variable regions (VR), and genes associated with these regions in each strain in this study are represented in the scheme beneath. Blue lines represent the novel insertion points identified in this study. Similar variable structure or genes with similar putative functions in HS or VR in different ICEs were colored in the same color (as indicated below the scheme). Only the strain PmBR19 carries an antibiotic resistance gene (blaCMY–2 in red arrow) and its ICE is identical to ICEPmiJpn1. The remainders of the ICEs were named according to the convention of the field. ICEPmiBra595 has a mercury resistance gene in VR IV (in orange color).
BLAST analysis confirmed that the element identified in strain PmBR19 is 99.9% identical to the world-wide spread ICEPmiJpn1 (Table 1), therefore we maintained its original name. This variant carries the CMY-2 beta-lactamase gene flanked by insertion sequences in a composite transposon, inserted in variable region V (Figure 1). The only major difference between the ICE from the Brazilian strain and the one originally identified in Japan (Harada et al., 2010) is a 2478 bp deletion in the 3’-end of pglX gene and 456 bp deletion in the 5’-end of pglZ gene in the former. These genes are part of the BREX system involved in phage resistance (Goldfarb et al., 2015; Slattery et al., 2020).
Conjugation of the ICEPmiJpn1 element from P. mirabilis PmBr19 to E. coli MG1655 was achieved, albeit at a low frequency (Table 1), in the order of 2 × 10–8 transconjugants/recipient cell. Conjugation of the original ICEPmiJpn1 originally detected in Japan was compatible with the observed in our assays (10–9 transconjugants/donor) (Harada et al., 2010). Nevertheless, the same element present in strains of avian origin was transferred at much higher rates, in the order of 10–5 transconjugants/recipient (Aberkane et al., 2016). The conjugation frequency of other SXT/R391 ICEs vary widely (from 10–4 to 10–8), depending on the element and strain (Lei et al., 2016, 2018; Li et al., 2016; Bie et al., 2017; Siebor et al., 2018; Slattery et al., 2020). Therefore, our results are in agreement with the previously identified range of conjugation frequencies for these elements in P. mirabilis.
ICEPmiBra607 (PmBR607) is very similar to ICEPmiJpn1. Variable regions I, and IV, and hotspots 1-5 carry the same genes inserted in both variants (Figure 1). Nevertheless, a different composite transposon (∼10 kbp long) is located inside variable region V. This transposon is also flanked by IS4-like repeats, similarly to the one present in ICEPmiJpn1, but contains a number of small ORFs, none of them encoding for known resistance determinants (Figure 1). The strain bearing ICEPmiBra607 also has other noteworthy genomic features. It carries two plasmids, one of them encoding quinolone resistance (qnrD1), identical to plasmids previously identified among Enterobacterales (NZ_CP049755.1). The other plasmid (NZ_CP049754.1) has partial similarity to other enterobacterial plasmids, with a unique structure. This plasmid does not carry any resistance genes. Some pseudogenes are found in this plasmid, indicating gene decay and a lack of selective pressure for its functions in this strain, but the presence of one toxin-antitoxin pair is probably preventing its loss. Finally, the chromosome of this strain (NZ_CP049753.1) contains three integrons bearing resistance determinants, one copy of In293 and two copies of In2-4 (Moura et al., 2009). After the initial submission of this work, two whole genome sequences were added to the NCBI database, which contain SXT/R391 elements similar to ICEPmiJpn1 and ICEPmiBra607. We nominated these elements as ICEPmiHN2p and ICEPmiL901, in reference to their respective strain names (Supplementary Table S1). ICEPmiL901 is highly similar to ICEPmiBra607, containing the same insertion in variable region 5. ICEPmiHN2p contains both composite transposons in variable region 5: the one containing the CMY-2 beta-lactamase gene found in ICEPmiJpn1, and the one found in ICEPmiBra607, in a tandem array (data not shown).
ICEPmiBra595 (PmBR595) is highly similar to the one found in the genome of Providencia stuartii ATCC33672 (Frey et al., 2014). This ICE, to the best of our knowledge, does not have an official assigned designation in the literature, and has not been analyzed in depth. We hereafter refer to the P. stuartii element as ICEPst33672 (Table 1). It contains a heavy metal resistance cassette in variable region IV (Figure 1) similar as those found in R391 and pMERPH (Böltner et al., 2002; Ryan et al., 2019), and encodes a small protein containing a metallo-beta-lactamase fold in hotspot 4, but does not carry known antimicrobial resistance determinants. Interestingly, a nonsense mutation is present in traG in the Brazilian isolate (data not shown), probably impairing the conjugation function of this element, given its importance to the mating-pair stabilization (Firth and Skurray, 1992).
No known antimicrobial or heavy metal resistance genes were found in ICEPmiBra614 (PmBR614) and ICEPmiBra618 (PmBR618), which have unique structures and therefore consist of new elements (Table 1, Figure 1). ICEPmiBra618 carry homologs of the hipAB genes in VRI, similarly to ICEPmiJpn1 and ICEPmiBra607. This toxin-antitoxin system is implicated in the formation of persisters cells (Moyed and Bertrand, 1983; Black et al., 1991; Schumacher et al., 2009), a trait that could provide selective advantage in the clinical setting for strains carrying these elements. Another toxin-antitoxin system known as mosAT is present in HS2 from all ICEs, except ICEPmiBra618, which carries another type of toxin without a known antitoxin. The mosAT (for maintenance of SXT) toxin-antitoxin system was previously shown to prevent ICE-loss from the host chromosome (Wozniak and Waldor, 2009).
Novel Features of SXT/R391 Elements in P. mirabilis From Brazil
ICEPmiBra607 encodes a novel composite transposon not related to any known element in sequence databases, present in variable region V, as described above. Interestingly, one of the genes present in this novel region encodes a putative transporter of the lolC family, albeit the other components of this ABC transport system are not present in the transposon. Other genes encoded by this transposon may be involved in bacterial virulence, such as genes encoding for a lysozyme inhibitor (G9C79_07940) and putative adhesion proteins (G9C79_07900 and G9C79_07905). A complete list of all the genes encoded in the variable and hotspot regions of all elements is present in Supplementary Table S4.
ICEPmiBra618 has a novel insertion at HS5, not previously identified in any other ICE. This DNA sequence produces no significant hits in BLAST analysis (Supplementary Table S5). In spite of the lack of sequence conservation at the DNA level, it encodes 4 predicted proteins, which are conserved among different gammproteobacteria: two hypothetical proteins, one site-specific DNA-methyltransferase and one helix-turn-helix domain-containing protein (Supplementary Table S4).
Other interesting finding regards novel insertion points in the elements identified. VRs and HSs have been identified early in the first comparative genomic analyses of the ICEs, and later expanded to include newly observed points of DNA insertion (Beaber et al., 2002; Wozniak et al., 2009; Bie et al., 2017). In the elements identified in our study, two new points of DNA insertion have been observed. In ICEPmiBra614 and ICEPmiBra595 there is a point of DNA insertion between the 3‘ends of mobI and rumB. In ICEPmiBra618 there is a point of insertion close to HS4, where an IS3-like element is located (Figure 1).
Analysis of sequence similarity in DNA inserted in VRs and HSs is suggestive of a complex evolution pattern of these elements, in particular for ICEPmiBra614 and ICEPmiBra618 (Supplementary Table S5). These elements seem to be mosaics of other previously reported ICEs from several species. For instance, ICEPmiBra614 HS5 content is more similar to the insertion seen in the same position in ICEPmiChn2 (98% sequence identity), while HS4 has the same insertion as ICEAplChn1 from Actinobacillus pleuropneumoniae (99.9% identity). In HS3, this element contains a sequence with high similarity to ICESupCHN110003 from Shewanella upenei (99.9% identity). A similar convoluted pattern is seen in ICEPmiBra618, for which each insertion point is more similar to a sequence from a different ICE, from a different organism.
Phylogenetic Relationships Between P. mirabilis SXT/R391 ICEs
We analyzed the relationship between the ICEs identified in our study and other representative ICEs from the literature and databases. For this purpose, first we analyzed their phylogenetic relationships based on SNPs in their conserved regions using the CSI phylogeny pipeline (Kaas et al., 2014; Figure 2). As suggested by their common structure and similar content in hotspots and variable regions, ICEPmiBra607 and ICEPmiJpn1 are highly similar. ICEPmiBra595 is more similar to ICEPmiBra618, although they carry markedly different contents in variable and hotspot regions (Figure 1, Supplementary Tables S4,S5). ICEPmiBra614 is more distantly related to the other elements found in this work in P. mirabilis originated from Brazil. In another analysis, using a tree constructed based on the concatenated sequence of int, setR, and setCD, conserved genes located in both ends of the element, the close relationship of ICEPmiBra607 and ICEPmiJpn1 is confirmed. Nevertheless, the relationships among the other ICEs are changed, ICEPmiBra595 being more distantly related to the other elements from our study, and ICEPmiBra618 and ICEPmiBra614 more closely related (Supplementary Figure S1). Taken together with the seemingly mosaic nature of the ICEs described above (Supplementary Table S5), this is suggestive of recombination and shuffling of these elements.
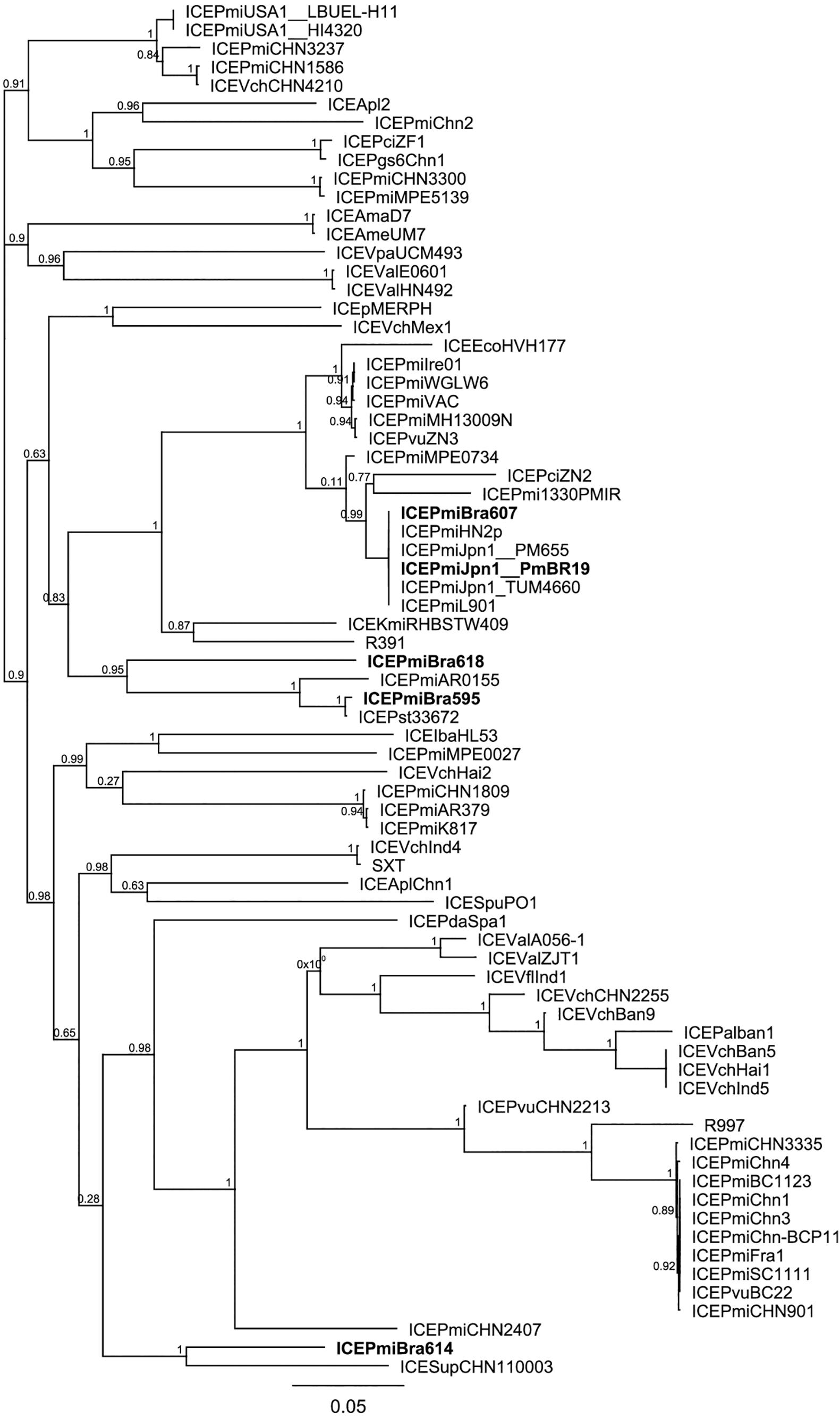
Figure 2. Phylogenetic relationships of SXT/R391 elements. The phylogenetic tree of SXT/R391 ICEs was built based on SNPs in their conserved regions (CSI Phylogeny pipeline) and using the approximate maximum-likelihood method in FastTree. Support values (ranging from 0 to 1) are shown next to the nodes. ICEs SXT/R391 from this study are indicated by bold letters, and strain names are shown after underscore for some ICEs found in multiple strains.
When compared to the species tree (Figure 3), the relationships between the ICEs show interesting features. Although clonally related and with similar resistance profiles (Figure 3, Table 1), strains PmBR607 (ICEPmiBra607) and PmBR618 (ICEPmiBra618) share ICEs with markedly different content in VRs and HSs (Figure 1, Supplementary Table S5). The Brazilian isolate PmBR19,the Japanese isolate TUM4660 and PM655 from Ireland all share the same element ICEPmiJpn1, but are not identical, suggesting horizontal transmission of this ICE among different P. mirabilis strains. In fact all three strains are phylogenetically closer to other P. mirabilis strains either devoid of SXT/R391 elements, or bearing different versions of such elements. Strain PmBR595 (bearing ICEPmiBra595) is closely related to strain FDAARGOS_81, although the later does not possess a SXT/R391 element in its genome, again indicating loss or horizontal acquisition of this element in the evolutionary history of P. mirabilis lineages. The same is true for strains PmBr614 and WGLW4, closely related, but the former carrying ICEPmiBra614, while the latter is devoid of any conjugative element.
Figure 3. Phylogenetic tree comparing Brazilian Proteus mirabilis isolates with previously sequenced strains. Whole genome SNPs alignment from CSI Phylogeny analysis was used to build the tree. An approximate maximum-likelihood tree was obtained using FastTree. Brazilian strains from this study are indicated by bold letters. Strains containing the same ICEs are indicated along with corresponding ICE names after the underscore.
Identification of Exclusion Groups
As previously described, SXT/R391 ICEs are divided in two different exclusion groups, S, related to the SXT element, and R, related to the R391 element (Marrero and Waldor, 2007). Both Eex and TraG determine the exclusion groups, which can be differentiated based on the presence of specific aminoacids in their primary sequences, in particular the 56 C-terminal aminoacids in Eex, and three specific positions in TraG. We therefore sought to determine to which exclusion group each of the five ICEs identified in this study belong to. The comparison of the relevant aminoacid residues with the ones present in SXT and R391 revealed that ICEPmiBra595 and ICEPmiBra618 belong to the R group, while ICEPmiBra614, ICEPmiBra607, and ICEPmiJpn1 belong to the S group (Figure 4).

Figure 4. Exclusion group analysis. Comparison of the sequences of Eex (A) and TraG (B) of the five ICEs from this study to SXT and R391. The relevant variable residues are colored. In blue, residues identical to SXT and in red, residues identical to R391.
Discussion
SXT/R391 ICEs have been recognized in the last two decades as important agents in the dissemination of antimicrobial resistance (Burrus et al., 2006; Bioteau et al., 2018). In our work, we identified 5 of these elements in P. mirabilis isolated in the clinical setting in Brazil. One of these elements is the ICEPmiJpn1, which carries the CMY-2 beta-lactamase and is present in several different isolates from different origins around the globe (Harada et al., 2010; Mata et al., 2011; Aberkane et al., 2016; Lei et al., 2016; Mac Aogáin et al., 2016; Bie et al., 2017). To the best of our knowledge, this is the first description of P. mirabilis carrying this element in the Americas. The global spread of P. mirabilis strains carrying this particular variant indicates a strong selective advantage provided by this version of the ICE.
On the other hand, most of the elements identified in this work do not carry resistance determinants. Recently it has been proposed that type 1 SXT/R391 elements (i.e., those in which the ICE is inserted in the 5‘ end of prfC, such as the ones identified in this work) are mostly associated with the dissemination of antibiotic resistance, in the case of clinical isolates, and with heavy metal resistance, in the case of environmental isolates (Bioteau et al., 2018). This does not seem to be the case of the ICEs identified in P. mirabilis clinical isolates in our work, since most of them do not carry any known resistance determinants. Many of the studies searching for SXT/R391 elements were focused in multi-resistant clinical strains (Mata et al., 2011; Mac Aogáin et al., 2016; Lei et al., 2018), and this fact may have introduced a bias in the current database of known SXT/R391 elements5 (Liu et al., 2019). Our study aimed at the identification of such elements regardless of the antimicrobial resistance status of the strains. On the other hand, the relatively small dataset obtained in our work does not dismiss the idea that most of these elements are associated with antimicrobial resistance. It is an interesting question for further examination whether most of the type 1 elements from the SXT/R391 family are associated or not with antimicrobial resistance, and if this pattern varies in the different species in which they usually occur. More recently, another example of an element devoid of resistance genes was identified in a P. mirabilis isolated from wastewater (Slattery et al., 2020).
Another possibility would be that these elements carry yet uncharacterized resistance determinants. Nevertheless, for the strains characterized in this work, this is not likely to be the case. The antimicrobial resistance pattern correlates with resistance genes identified outside the ICE in the whole genome sequence of these strains, according to analysis performed on the CARD database (data not shown). These elements probably influence other aspects of cellular physiology. In one example, one SXT/R391 element from a V. cholerae strain has been shown to increase c-di-GMP levels in the cell, affecting motility and biofilm formation (Bordeleau et al., 2010). Given that many of the genes present in VRs and HSs encode proteins of unknown function, it is a pressing matter to understand which effects SXT/R391 elements may have in the host cell.
Genes involved in DNA repair, and toxin-antitoxin systems are other elements frequently present in SXT/R391 ICEs (Wozniak et al., 2009). These may also provide selective advantage to the host cell under certain environmental conditions. Of particular interest, many ICEs carry homologs of the hipAB system, involved in the formation of persisters (Moyed and Bertrand, 1983; Black et al., 1994; Schumacher et al., 2009). Interestingly, hipAB have been implicated in stability of R391 like mosAT have been demonstrated for SXT, indicating a role as addiction modules for the ICE (Wozniak and Waldor, 2009; Carraro et al., 2015). Whether these toxin-antitoxin systems could contribute to the formation of persisters is still unknown.
All ICEs from our study have the same genes inserted in HS1, suggesting a common ancestry. ICEPmiJpn1, ICEPmiBra607, and ICEPmiBra618 all have the above-mentioned hipAB system inserted in VRI. Nevertheless, it is interesting to note that the ICEs identified in this work share the same genes inserted in VRs and HSs with many different SXT/R391 elements. Furthermore, particularly for ICEPmiBra614 and ICEPmiBra618, DNA in each insertion point bears more similarity with a different ICE (Supplementary Table S5), which is suggestive of an intricate evolutionary path of these elements.
It is unlikely that the pattern observed in Supplementary Table S5 indicates that the same DNA sequence has entered in the same insertion point of many different ICEs in independent events in the course of evolution. A more parsimonious hypothesis would be that each DNA sequence entered at one given insertion point in one element, and these elements are being constantly shuffled by recombination. Such recombination would be possible if two different ICEs could coexist in the cell. It has already been demonstrated that there are two independent exclusion groups, which permit that two different variants to be inserted in tandem in the cell (Hochhut et al., 2001; Burrus and Waldor, 2004; Marrero and Waldor, 2007). ICE recombination has already been demonstrated experimentally, and is facilitated by the Bet/Exo recombination machinery encoded by these elements (Garriss et al., 2009, 2013). For the ICEs described in this study, we identified that ICEPmiBra618 and ICEPmiBra595 belong to the R group, and that ICEPmiJpn1, ICEPmiBra607 and ICEPmiBra614 are from the S group (Figure 4).
Our analysis also suggests many events of ICE transfer or loss among Proteae and with other bacteria. Strain PmBR595 harbors the exact same element found in P. stuartii (ICEPmiBra595). On the other hand, this strain is markedly genetically close to P. mirabilis FDAARGOS_81, which does not carry any ICE. At least one event of ICE gain or loss is clear from this observation, and the same is true for strains WGLW4 and PmBr614. Another interesting scenario is presented by strains PmBR607 (ICEPmiBra607) and PmBR618 (ICEPmiBra618). Those strains are very closely related but carry different ICEs. ICEPmiBra607 and ICEPmiBra618 carry different DNA content in VRs and HSs, and show a considerable phylogenetic distance. In this case, either multiple events of ICE gain and loss occurred in these strains, or extensive inter-ICE recombination produced new variants containing different DNA insertions in VRs and HSs.
ICEPmiJpn1 is another interesting case, since it has been detected in P. mirabilis isolates from many parts of the world (Mata et al., 2011; Aberkane et al., 2016; Lei et al., 2016; Mac Aogáin et al., 2016; Bie et al., 2017). In our study, we found another example of P. mirabilis carrying this element. Two whole genome sequences of P. mirabilis strains containing this element are present in the database: strains TUM4660 (the one from which this element was first isolated in Japan) and PM655 isolated in Ireland. The Brazilian strain carrying this element (PmBR19) is not closely related to either TUM4660 or PM655, according to our phylogenetic analysis. Therefore, this particular SXT/R391 variant is likely being transmitted to and maintained in different strains, which indicates a strong selective advantage, probably linked to the world-wide massive use of beta-lactams.
Conclusion
In conclusion, our study described for the first time the genetic structure of SXT/R391 ICEs present in P. mirabilis in Brazil, which include three previously unknown elements. Future studies are needed to understand the prevalence of these elements, in particular the ones not carrying antimicrobial resistance genes.
Data Availability Statement
The datasets generated for this study can be found in the NCBI database, under BioProject PRJNA576511 and genome assemblies GCA_011149675.1, GCA_009184605.1, GCA_009183735.1, GCA_009183705.1, and GCA_009183685.1.
Ethics Statement
This study used strains obtained in the cities of São Paulo and Maringá, in Brazil. Committee for Ethics in Research (C.E.P.) from ICB, University of São Paulo, issued certificate of exemption 706/14 approving the study, which collected no patient-related information. Committee for Ethics in Research Involving Human Beings (COPEP) from State University of Maringá also approved the study (CAAE 318.0.093.000-11/COPEP UEM).
Author Contributions
RG, JS, and MF conceived the study. JS, MF, LC, MT, MA, TS, NL, and RG conducted experimental work and data analysis. JS and RG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001, and by grants 2015/11348-3, 2018/23872-7, and 2019/19435-3 from FAPESP. Genome sequencing was provided by Microbes NG (http://www.microbesng.uk) which is supported by the BBSRC (grant number BB/L024209/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all the members of our research groups for helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.571472/full#supplementary-material
Footnotes
- ^ http://www.microbesng.uk
- ^ http://www.usadellab.org/cms/?page=trimmomatic
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ https://cge.cbs.dtu.dk/services/CSIPhylogeny/
- ^ https://db-mml.sjtu.edu.cn/ICEberg
References
Aberkane, S., Compain, F., Decré, D., Dupont, C., Laurens, C., Vittecoq, M., et al. (2016). High Prevalence of Sxt / R391-Related Integrative and Conjugative Elements Carrying bla Cmy-2 in Proteus mirabilis Isolates from Gulls in the South of France. Antimicrob. Agents Chemother. 60, 1148–1152. doi: 10.1128/Aac.01654-15
Armbruster, C. E., and Mobley, H. L. T. (2012). Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat. Rev. Microbiol. 10, 743–754. doi: 10.1038/nrmicro2890
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beaber, J. W., Burrus, V., Hochhut, B., and Waldor, M. K. (2002). Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 59, 2065–2070. doi: 10.1007/s000180200006
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Bie, L., Wu, H., Wang, X.-H., Wang, M., and Xu, H. (2017). Identification and characterization of new members of the SXT/R391 family of integrative and conjugative elements (ICEs) in Proteus mirabilis. Int. J. Antimicrob. Agents 50, 242–246. doi: 10.1016/j.ijantimicag.2017.01.045
Bioteau, A., Durand, R., and Burrus, V. (2018). Redefinition and Unification of the SXT/R391 Family of Integrative and Conjugative Elements. Appl. Environ. Microbiol. 84, 418–485 e. doi: 10.1128/AEM.00485-18
Black, D. S., Irwin, B., and Moyed, H. S. (1994). Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176, 4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994
Black, D. S., Kelly, A. J., Mardis, M. J., and Moyed, H. S. (1991). Structure and Organization of hip, an Operon That Affects Lethality Due to Inhibition of Peptidoglycan or DNA Synthesis. J. Bacteriol. 173, 5732–5739.
Böltner, D., MacMahon, C., Pembroke, J. T., Strike, P., and Osborn, A. M. (2002). R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184, 5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002
Bordeleau, E., Brouillette, E., Robichaud, N., and Burrus, V. (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ. Microbiol. 12, 510–523. doi: 10.1111/j.1462-2920.2009.02094.x
Burrus, V., and Waldor, M. K. (2004). Formation of SXT Tandem Arrays and SXT-R391 Hybrids. J. Bacteriol. 186, 2636–2645. doi: 10.1128/JB.186.9.2636-2645.2004
Burrus, V., Marrero, J., and Waldor, M. K. (2006). The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55, 173–183. doi: 10.1016/j.plasmid.2006.01.001
Carraro, N., Poulin, D., and Burrus, V. (2015). Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner. PLoS Genet. 11:e1005298. doi: 10.1371/journal.pgen.1005298
CLSI (2016). Performance standards for antimicrobial susceptibility testing, 26th Edn, Wayne, PA: Clinical and Laboratory Standards Institute.
Daccord, A., Ceccarelli, D., and Burrus, V. (2010). Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 78, 576–588. doi: 10.1111/j.1365-2958.2010.07364.x
Firth, N., and Skurray, R. (1992). Characterization of the F plasmid bifunctional conjugation gene, traG. MGG Mol. Gen. Genet. 232, 145–153. doi: 10.1007/BF00299147
Frey, K. G., Bishop-Lilly, K. A., Daligault, H. E., Davenport, K. W., Bruce, D. C., Chain, P. S., et al. (2014). Full-genome assembly of reference strain Providencia stuartii ATCC 33672. Genome Announc. 2, 1014–1082 e. doi: 10.1128/genomeA.01082-14
Garriss, G., Poulin-Laprade, D., and Burrus, V. (2013). DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R391 family. J. Bacteriol. 195, 1991–2003. doi: 10.1128/JB.02090-12
Garriss, G., Waldor, M. K., and Burrus, V. (2009). Mobile Antibiotic Resistance Encoding Elements Promote Their Own Diversity. PLoS Genet. 5:e1000775. doi: 10.1371/journal.pgen.1000775
Goldfarb, T., Sberro, H., Weinstock, E., Cohen, O., Doron, S., Charpak−Amikam, Y., et al. (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34, 169–183. doi: 10.15252/embj.201489455
Harada, S., Ishii, Y., Saga, T., Tateda, K., and Yamaguchi, K. (2010). Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 54, 3545–3550. doi: 10.1128/AAC.00111-10
He, D., Wang, L., Zhao, S., Liu, L., Liu, J., Hu, G., et al. (2020). A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J. Antimicrob. Chemother. 75, 1159–1164. doi: 10.1093/jac/dkaa012
Hochhut, B., and Waldor, M. K. (1999). Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32, 99–110.
Hochhut, B., Beaber, J. W., Woodgate, R., and Waldor, M. K. (2001). Formation of Chromosomal Tandem Arrays of the SXT Element and R391, Two Conjugative Chromosomally Integrating Elements That Share an Attachment Site. J. Bacteriol. 183, 1124–1132. doi: 10.1128/JB.183.4.1124-1132.2001
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Lei, C.-W., Chen, Y.-P., Kang, Z.-Z., Kong, L.-H., and Wang, H.-N. (2018). Characterization of a Novel SXT/R391 Integrative and Conjugative Element Carrying cfr, bla CTX-M-65, fosA3, and aac(6’)-Ib-cr in Proteus mirabilis. Antimicrob. Agents Chemother. 62, 818–849 e. doi: 10.1128/AAC.00849-18
Lei, C.-W., Zhang, A.-Y., Wang, H.-N., Liu, B.-H., Yang, L.-Q., and Yang, Y.-Q. (2016). Characterization of SXT/R391 Integrative and Conjugative Elements in Proteus mirabilis Isolates from Food-Producing Animals in China. Antimicrob. Agents Chemother. 60, 1935–1938. doi: 10.1128/AAC.02852-15
Li, X., Du, Y., Du, P., Dai, H., Fang, Y., Li, Z., et al. (2016). SXT/R391 integrative and conjugative elements in Proteus species reveal abundant genetic diversity and multidrug resistance. Sci. Rep. 6:37372. doi: 10.1038/srep37372
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucl. Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
Mac Aogáin, M., Rogers, T. R., and Crowley, B. (2016). Identification of emergent bla CMY-2 -carrying Proteus mirabilis lineages by whole-genome sequencing. N. microb. N. Infect. 9, 58–62. doi: 10.1016/j.nmni.2015.11.012
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marrero, J., and Waldor, M. K. (2007). The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J. Bacteriol. 189, 3302–3305. doi: 10.1128/JB.01902-06
Mata, C., Navarro, F., Miró, E., Walsh, T. R., Mirelis, B., and Toleman, M. (2011). Prevalence of SXT/R391-like integrative and conjugative elements carrying bla CMY-2 in Proteus mirabilis. J. Antimicrob. Chemother. 66, 2266–2270. doi: 10.1093/jac/dkr286
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Moyed, H. S., and Bertrand, K. P. (1983). hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155, 768–775. doi: 10.1128/jb.155.2.768-775.1983
O’Hara, C. M., Brenner, F. W., and Miller, J. M. (2000). Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev. 13, 534–546. doi: 10.1128/CMR.13.4.534-546.2000
Pearson, M. M., Sebaihia, M., Churcher, C., Quail, M. A., Seshasayee, A. S., Luscombe, N. M., et al. (2008). Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190, 4027–4037. doi: 10.1128/JB.01981-07
Poulin-Laprade, D., Matteau, D., Jacques, P. E., Rodrigue, S., and Burrus, V. (2015). Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucl. Acids Res. 43, 2045–2056. doi: 10.1093/nar/gkv071
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 - Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Rodríguez-Blanco, A., Lemos, M. L. M. L., and Osorio, C. R. (2016). Unveiling the pan-genome of the SXT/R391 family of ICEs: molecular characterisation of new variable regions of SXT/R391-like ICEs detected in Pseudoalteromonas sp. and Vibrio scophthalmi. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 109, 1141–1152. doi: 10.1007/s10482-016-0716-3
Ryan, M. P., Slattery, S., and Pembroke, J. T. (2019). A novel arsenate−resistant determinant associated with ICEpMERPH, a member of the SXT/R391 group of mobile genetic elements. Genes 10:048. doi: 10.3390/genes10121048
Schumacher, M. A., Piro, K. M., Xu, W., Hansen, S., Lewis, K., and Brennan, R. G. (2009). Molecular Mechanisms of HipA-Mediated Multidrug Tolerance and its Neutralization by HipB. Science 323, 396–401. doi: 10.1126/science.1163806
Siebor, E., de Curraize, C., and Neuwirth, C. (2018). Genomic context of resistance genes within a French clinical MDR Proteus mirabilis: identification of the novel genomic resistance island GIPmi1. J. Antimicrob. Chemother. 73, 1808–1811. doi: 10.1093/jac/dky126
Slattery, S., Tony Pembroke, J., Murnane, J. G., and Ryan, M. P. (2020). Isolation, nucleotide sequencing and genomic comparison of a Novel SXT/R391 ICE mobile genetic element isolated from a municipal wastewater environment. Sci. Rep. 10:8716. doi: 10.1038/s41598-020-65216-5
Waldor, M. K., Tschäpe, H., and Mekalanos, J. J. (1996). A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178, 4157–4165. doi: 10.1128/JB.178.14.4157-4165.1996
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wozniak, R. A. F., and Waldor, M. K. (2009). A Toxin–Antitoxin System Promotes the Maintenance of an Integrative Conjugative Element. PLoS Genet. 5:e1000439. doi: 10.1371/journal.pgen.1000439
Keywords: ICE, SXT/R391, CMY-2, Proteus mirabilis, WGS, nanopore, resistance
Citation: Sato JL, Fonseca MRB, Cerdeira LT, Tognim MCB, Sincero TCM, Noronha do Amaral MC, Lincopan N and Galhardo RS (2020) Genomic Analysis of SXT/R391 Integrative Conjugative Elements From Proteus mirabilis Isolated in Brazil. Front. Microbiol. 11:571472. doi: 10.3389/fmicb.2020.571472
Received: 10 June 2020; Accepted: 29 September 2020;
Published: 20 October 2020.
Edited by:
John R. Battista, Louisiana State University, United StatesReviewed by:
Hong-Ning Wang, Sichuan University, ChinaMichael P. Ryan, University of Limerick, Ireland
Copyright © 2020 Sato, Fonseca, Cerdeira, Tognim, Sincero, Noronha do Amaral, Lincopan and Galhardo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodrigo S. Galhardo, cmdhbGhhcmRAdXNwLmJy
†These authors have contributed equally to this work
 Juliana L. Sato
Juliana L. Sato Marina R. B. Fonseca
Marina R. B. Fonseca Louise T. Cerdeira
Louise T. Cerdeira Maria C. B. Tognim3
Maria C. B. Tognim3 Thais C. M. Sincero
Thais C. M. Sincero Nilton Lincopan
Nilton Lincopan Rodrigo S. Galhardo
Rodrigo S. Galhardo