
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 November 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.570269
A correction has been applied to this article in:
Corrigendum: Ribosomal Protein L40e Fused With a Ubiquitin Moiety Is Essential for the Vegetative Growth, Morphological Homeostasis, Cell Cycle Progression, and Pathogenicity of Cryptococcus neoformans
 Jingyu Zhao1,2†
Jingyu Zhao1,2† Yali Yang1,3†
Yali Yang1,3† Yibin Fan4†
Yibin Fan4† Jiu Yi1
Jiu Yi1 Chao Zhang1
Chao Zhang1 Zhongkai Gu5
Zhongkai Gu5 Weihua Pan1
Weihua Pan1 Julin Gu2*
Julin Gu2* Wanqing Liao1*
Wanqing Liao1* Wei Fang1*
Wei Fang1*Ubiquitin is a highly conserved protein required for various fundamental cellular processes in eukaryotes. Herein, we first report the contribution of the ubiquitin fusion protein Ubi1 (a ubiquitin monomer fused with the ribosome protein L40e, Rpl40e) in the growth and pathogenicity of Cryptococcus neoformans. UBI1 deletion resulted in severe growth restriction of C. neoformans, whose growth rate was positively correlated with UBI1 expression level. The growth defect of the ubi1Δ strain could be closely associated with its morphological abnormalities, such as its reduced ribosome particles. In addition, the ubi1Δ mutant also displayed increased cell ploidy, cell cycle arrest, and decreased intracellular survival inside macrophages. All these phenotypes were reversed by the reconstitution of the full-length UBI1 gene or RPL40a domain. Mouse survival and fungal burden assays further revealed a severely attenuated pathogenicity for the ubi1Δ mutant, which is probably associated with its reduced stress tolerance and the induction of T-helper 1-type immune response. Taken together, Ubi1 is required for maintaining the vegetative growth, morphological homeostasis, cell cycle progression, and pathogenicity in vivo of C. neoformans. The pleiotropic roles of Ubi1 are dependent on the presence of Rpl40e and associated with its regulation of cryptococcal ribosome biogenesis.
Cryptococcus neoformans is an important invasive fungal pathogen, which could cause life-threatening meningoencephalitis in both the immunocompetent and immunocompromised population (Fang et al., 2015; Maziarz and Perfect, 2016). Globally, it is estimated that there are approximately 278,000 new cases of cryptococcal infections each year, which cause 181,000 deaths annually (Rajasingham et al., 2017). In mammalian hosts, phagocytic cell-based immunity is critical to controlling fungal infections (Casadevall et al., 2018). Reactive oxygen and nitrogen species generated by macrophages, neutrophils, and other phagocytic cells are implicated in killing C. neoformans and other fungal pathogens (Seider et al., 2010; May et al., 2016). However, C. neoformans can effectively sense and overcome the hostile living environment in vivo (such as limited nutrition, high temperature, oxidative stress, etc.) via a variety of virulence factors (Kronstad et al., 2012; Alspaugh, 2015; Zaragoza, 2019). For example, yeast cells escape phagocytosis by immune cells by producing a capsule, which can also promote the intracellular proliferation and extracellular dissemination of C. neoformans (O’Meara and Alspaugh, 2012). Melanin can effectively remove free radicals in the host and interfere with macrophage polarization, thereby enhancing the intracellular survival of C. neoformans (Zaragoza, 2019).
Ubiquitination is a reversible post-translational modification mechanism in eukaryotes, involved in a variety of biological processes such as ribosome biogenesis, transcriptional regulation, protein quality control, and signal transduction (Harper and Schulman, 2006). Over the last decade, people have gradually realized the potential roles of the ubiquitin system in the growth and pathogenicity of C. neoformans. The ubiquitin-proteasome inhibitor bortezomib significantly inhibits the production of the most critical pathogenic factor, the capsule, in C. neoformans (Geddes et al., 2016). Our previous studies revealed that the deubiquitinase Ubp5 is essential for the production of various pathogenic factors (such as high thermotolerance, melanin, and the capsule), stress responses, sexual reproduction, and pathogenicity in both C. neoformans and Cryptococcus gattii (Fang et al., 2012; Meng et al., 2016). Similarly, deletion of the ubiquitin ligase Fbp1 causes sexual reproduction defects, decreased stress tolerance, and attenuated virulence in vivo in C. neoformans (Liu et al., 2011; Liu and Xue, 2014). These studies suggest that the ubiquitin system is crucial for C. neoformans to adapt to the environmental niche of a mammalian host.
Ubiquitin is a core element in the ubiquitin-proteasome system, and its functions and molecular mechanisms in the growth and pathogenicity of fungal pathogens remain unclear. Ubiquitin usually exists as a polyubiquitin precursor (Ubi4) or a hybrid protein with a fused ribosomal protein, such as Rpl40e (Ubi1/2 precursors) or Rps31e (Ubi3 precursor) (Clague and Urbé, 2010). In Saccharomyces cerevisiae, the ribosomal protein moiety is required for 60S or 40S subunit production, subunit assembly, and subsequent translation (Finley et al., 1989; Fernández-Pevida et al., 2012). The ubiquitin moiety acts as a chaperone by facilitating efficient ribosome biogenesis (Lacombe et al., 2009; Martín-Villanueva et al., 2019, 2020). The ubi1Δ or ubi2Δ mutant shows a slow-growth phenotype while UBI3 deletion or double deletions of UBI1 and UBI2 cause a lethal phenotype in S. cerevisiae (Finley et al., 1989). In C. neoformans, ubiquitin is mainly composed of two encoding genes: the UBI1 gene (encoding a fusion protein of a ubiquitin monomer and Rpl40e) and the UBI4 gene (encoding a polyubiquitin containing five ubiquitin repeats) (Spitzer and Spitzer, 1995). Here we first report the identification and characterization of the pleiotropic roles of UBI1 in the growth and pathogenicity in vivo of C. neoformans.
All the cryptococcal strains and plasmids used in this study are shown in Supplementary Table S1. The wild-type (WT) H99 strain was cultured on YPD agar medium (1% yeast extract, 2% peptone, and 2% dextrose) and different transformants were cultured on selective media (YPD plus 200 mg L–1 G418 or 100 mg L–1 nourseothricin). Condition media (YPD plus different concentrations of bathocuproinedisulfonic acid or cupric sulfate) were used to induce or suppress the expression of UBI1 in promoter-reconstituted strains. Several media for phenotypic assays (such as capsule-inducing medium, DOPA/caffeic-acid medium, and Titan Cells Medium) and DMEM medium for the J774 macrophage killing assay were prepared as previously reported (Fang et al., 2012; Trevijano-Contador et al., 2018).
The Ubi1 homolog of C. neoformans was identified by BLAST search of the serotype A (H99) genome database1. The UBI1 gene (CNAG_00370) was disrupted by biolistic transformation, with the disruption cassette generated by overlapping PCR. Specifically, the upstream flanking region (982-bp) and downstream flanking region (859-bp) of the UBI1 gene were amplified from the genomic DNA of H99 with the primer pairs ZJY001/ZJY002 and ZJY003/ZJY004, respectively. ZJY005 and M13F were used to amplify the neomycin phosphotransferase II (NEO, 2098-bp) cassette from the plasmid pJAF1. These PCR products were gel-purified and used as templates to generate a 3.9-kb ubi1:NEO deletion construct. The linearized construct was transformed into H99 using a gene gun (Toffaletti et al., 1993). Stable transformants were selected by G418 resistance and colony PCR, using appropriate primers. Deletion of UBI1 was further confirmed by Southern blot hybridization; the NEO probe was generated by PCR with the primers NEO-F and NEO-R (data not shown).
To obtain the complete sequence of the UBI1 gene, a 3.8-kb DNA fragment, containing an open reading frame (ORF), promoter, and terminator region, was PCR amplified from H99 genomic DNA, sequenced, and then cloned into the plasmid pCH233 using the In-Fusion® EcoDryTM Cloning System (Clontech, Palo Alto, CA, United States). The reconstructed plasmid pUBI1-NAT was linearized with BamHI and transformed into the H99 ubi1Δ strain by biolistic transformation (Toffaletti et al., 1993). Positive colonies were screened on YPD agar plus nourseothricin. The reconstitution of UBI1 was confirmed by diagnostic PCR (primers ZJY010 and ZJY011) and Southern blot hybridization using a NAT probe (primers NAT-F and NAT-R).
A copper-regulated mutant strain was constructed using the overlap PCR technique. A 982-bp fragment of the 5′ region of UBI1 was identical to the upstream flanking region of the ubi1:NEO deletion construct. A 956-bp fragment, starting from the initiation codon of UBI1, was amplified with the primers ZJY012 and ZJY013. The 2664-bp NeoR-PCTR4 fragment was amplified from the plasmid pNEO-CTR4 with the primers ZJY014 and ZJY015. All the PCR products were gel-purified and then used as templates to generate a 4.6-kb overlap fragment with the primers ZJY001 and ZJY013, which was then transformed into H99 using a biolistic delivery system. Stable transformants were screened on YPD agar containing G418 and BCS. The copper-regulated UBI1 mutants (PCTR4-UBI1) were selected using colony PCR and confirmed by Southern blotting and real-time PCR.
To determine the contributions of the different Ubi1 domains to its biological functions, truncated UBI1 gene fragments, the N-terminal domain (monoubiquitin, UB, 1–76) and the C-terminal domain (Ribosomal unit protein L40e, Rpl40e, 77-129), were cloned into the pCH233 vector. For the construction of the pUB-NAT vector, a 1624-bp fragment containing the UBI1 promoter and monoubiquitin ORF was amplified from the H99 genomic DNA, using the primers ZJY008 and ZJY016, and infused with the UBI1 terminator fragment (941-bp, primers ZJY017 and ZJY009) by overlap PCR. The truncated monoubiquitin gene fragment was inserted into digested plasmid pCH233 by XbaI using an infusion kit, as described above. The vector pRPL40a-NAT was constructed in a similar way: a 1478-bp fragment containing the RPL40a ORF and UBI1 terminator was amplified using the primers ZJY019 and ZJY009 and then fused with the UBI1 promoter fragment. Then, the linearized vector (pRPL40a-NAT or pUB-NAT) was integrated into the H99 ubi1Δ strain by biolistic transformation. Stable transformants were selected and confirmed by diagnostic PCR and quantitative real-time PCR. All the primers used in this study are listed in Supplementary Table S2.
Genomic DNA (20 μg) from each strain was extracted, using the CTAB method (Pitkin et al., 1996), and digested by the appropriate restriction endonucleases overnight. The DNA were separated on a 0.8% agarose gel, and then transferred to positively charged nylon membranes (Roche Applied Science, Indianapolis, IN, United States). The membranes were hybridized with the NEO or NAT probes overnight, washed, and then analyzed. The hybridized DNA bands were visualized on film after 3–10 min of exposure.
Cryptococcus neoformans was cultured in liquid YPD media until it reached mid-log phase. It was then pelleted, washed twice, frozen at −80°C, and lyophilized overnight. The cell pellets were broken with glass beads (2 mm diameter) and then the total RNA was isolated using the Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA, United States) following the manufacturer-provided protocol. Library preparation and RNA sequencing were performed by Genergy Biotech (Shanghai) Co., Ltd. The total RNA samples were purified and prepared as previously described (Trapnell et al., 2010). High-throughput sequencing was performed on a Hiseq3000 Genome Analyzer (Illumina, United States). To achieve sufficient sequence coverage, both the WT and mutant strains were sequenced with 100-bp paired-end reads. All the reads were mapped to the reference genome of C. neoformans from the Broad Institute website, using TopHat v1.3.0 (Trapnell et al., 2009). Less than 1% of the reads in all samples were excluded for poor quality. Among the aligned reads, the fragments per kilobase of transcript per million fragments mapped (FPKM) were counted to evaluate the transcriptional level, using Cufflinks v.1.0.3. The transcriptomic differences between the strains were analyzed and determined using CuffDiff (Trapnell et al., 2010). A gene was considered differently expressed if the transcriptional change was greater than 2.0-fold and the P-value less than 0.05 after Benjamini–Hochberg correction.
RNA was converted to cDNA with the SuperScript III First-Strand Synthesis Kit for RT-PCR (Invitrogen Corp., Carlsbad, CA, United States), according to the manufacturer’s instructions. Quantitative real-time PCR was performed to verify the RNA sequencing results using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, United States). The ACT1 gene was utilized as an internal control to normalize gene amplification for each sample. The real-time PCR conditions were as follows: an initial denaturing cycle of 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 s, and annealing/extension at 60°C for 20 s. All primers used for the RT-PCR assays are listed in Supplementary Table S2.
To test the effect of UBI1 deletion on growth rate, the WT, mutant (ubi1Δ), reconstituted (ubi1Δ:UBI1) and partially reconstituted (ubi1Δ:RPL40a) strains were grown overnight at 30°C in liquid YPD medium. The cell numbers were counted by hemocytometer and 106 colony-forming units (CFUs) from each culture were transferred to flasks containing 30 mL fresh YPD medium and incubated at 30°C. Optical density at 600 nm (OD600) was measured for each culture at 4-h intervals, and growth curves plotted in Microsoft Excel. To detect the relationship between cryptococcal UBI1 expression and its growth rate, 106 CFUs of the WT and the PCTR4-UBI1 mutant strain were incubated under different conditions (YPD containing 25 μM CuSO4, YPD, and YPD containing 50 μM BCS) at 30°C. OD600 was recorded at each time point and growth curves plotted. Doubling time was used to evaluate the growth rate of the cryptococcal strains under distinct conditions, and calculated according to the formula: Td = t × Lg2/(LgNt − LgN0). Td, doubling time; t, culture time; Nt, OD600 value after t hours of culture; N0, initial OD600 value at the time of inoculation.
For colony size measurement, different strains were grown in YPD liquid medium overnight at 30°C, washed twice with PBS buffer, and resuspended at a concentration of 103 CFU per mL. -Each strain was spread (100 μL) on a YPD solid medium and grown at 30°C for 2 weeks. Photographs were taken on day 9 and day 14.
For the budding rate assay, 106 CFU of each strain were inoculated into 30 mL YPD liquid medium and incubated to the mid-log (OD600 = 1.5) or stationary phase (OD600 = 4.0). Each culture was diluted to appropriate concentrations, and then stained with India ink. The budding yeast cells were then counted using an hemocytometer. Daughter cells were not considered budding cells if their size was larger than 50% of their parent cell size. The budding rate (%) was calculated with the formula: budding cells/total cells × 100. The total number of cells for each strain were not less than 500, and each test was repeated three times.
For cell size measurement, 50 μL culture from each strain (WT, ubi1Δ, ubi1Δ:UBI1, and ubi1Δ:RPL40a) at mid-log phase was washed three times in 500 μL 1 × PBS, then resuspended in 100 μL 1 × PBS, and visualized with 25 μL India Ink. Images were collected using the 63× objective of a Zeiss LSM inverted confocal microscope (Jena, Germany), and the diameters of at least 100 cells were measured using Adobe Photoshop.
Cells (2 × 104 cells for each strain) were harvested, fixed, and stained with propidium iodide (Sigma-Aldrich, St. Louis, MO, United States), and then analyzed using a BD FACSCalibur flow cytometer (Becton Dickinson Biosciences, Sparks, MD, United States). CellQuest Pro software was used for cell collection, and data analysis was performed using ModFit and FlowJo. The G1, S, and G2/M phases were identified using the Dean-Jett-Fox mathematical model.
For capsule analysis, the WT, mutant, and reconstituted strains of C. neoformans were incubated in 1/10 Sabouraud medium at 37°C and 5% CO2 for 3 days (Zaragoza and Casadevall, 2004). Capsule measurements were performed as previously described (Maxson et al., 2007). Melanin production was visualized in cells spotted on L-DOPA agar and caffeic-acid medium at 30°C for 7 days (Fang et al., 2012). For titan-like cell induction, cryptococcal strains were incubated in Titan Cells Medium as previously described (Trevijano-Contador et al., 2018), which contains 5% Sabouraud and 5% fetal calf serum diluted in 50 mM MOPS (Ph7.3) plus 15 μM sodium azide (Sigma-Aldrich). For stress assays, each strain was cultivated to saturation in YPD medium, serially diluted (1–106 dilutions), and spotted onto YPD or YNB agar media containing different stress-inducing agents, as previously reported (Fang et al., 2012).
The impact of UBI1 deletion on the morphology of C. neoformans was evaluated with transmission electron microscopy (TEM). Cells of the WT, ubi1Δ, and ubi1Δ:UBI1 strains were cultured to log phase in YPD liquid medium. The cells were harvested and fixed using the procedures developed by Reese et al. (2007). Ultrathin sections were cut using a Leica ultramicrotome, collected on copper grids, and stained with uranyl acetate and Satos lead. The samples were examined under a JEM-1230 transmission electron microscope (JOEL, Japan) using an acceleration voltage of 80 kV.
The survival rate of the WT, ubi1Δ, ubi1Δ:UBI1, and ubi1Δ:RPL40a strains of C. neoformans within macrophage-like J744A.1 cells was assessed as previously described (Aslanyan et al., 2017). Macrophage cells (50 μL; 2 × 106/mL) were aliquotted into the wells of a 96-well plate and activated by gamma interferon-c and lipopolysaccharide. The activated macrophages were co-incubated with 106 CFU of a cryptococcal strain opsonized by a monoclonal antibody (C66441M, bought from Meridian Life Science, Inc.) for 2 h at 37°C in 5% CO2. The extracellular yeast cells were removed using PBS buffer and spread onto YPD agar for phagocytosis efficacy assessment. The monolayers were incubated in fresh DMEM media overnight, and then disrupted with 0.5% SDS to lyse the macrophages. The lysates were diluted and plated on YPD agar to calculate the viable fungal cells after incubation at 30°C for 3–5 days.
The animal protocol was approved by the Committee on Ethics of Biomedicine Research, Second Military Medical University. The protocol was carried out in strict accordance with the Guide for Care and Use of Laboratory Animals issued by the Chinese Ministry of Science and Technology. All efforts were made to minimize animal suffering and to reduce the number of mice used.
Female BALB/c mice (5 weeks old) were infected with the WT, ubi1Δ, or ubi1Δ:UBI1 strains of C. neoformans by intranasal inoculation, according to an established protocol (Fang et al., 2012). For the survival assay, ten mice per group were infected with 105 yeast cells suspended in 50 mL PBS. The infected animals were monitored and sacrificed based on predetermined endpoints, as previously described (Rittershaus et al., 2006). For the fungal burden assay, the lungs and brains were removed from the sacrificed mice (four mice for each group) after 3, 7, 14, and 21 days. Organ samples were weighed and homogenized. The homogenate (200 mL) was plated onto YPD agar with appropriate dilution, and incubated at 30°C for 3–5 days. The number of CFUs was calculated and expressed as CFU/g of tissue. For histological analysis, the lungs were removed from the infected mice at the indicated time points and fixed in 10% neutral buffered formalin. Paraffin sections (5 μm) of the lungs were stained with PAS (for C. neoformans) and counterstained with hematoxylin and eosin.
All values are expressed at mean ± S.D. unless otherwise indicated. Statistical analyses were performed using SPSS 18.0 (IBM, Armonk, NY, United States). Differences between paired groups were evaluated using analysis of variance and student’s t-test (two-tailed). Data from the survival assay in the murine infection model were analyzed using the Mantel–Cox test. P-values less than 0.05 were considered statistically significant.
A search of the C. neoformans genome database revealed that the UBI1 (accession number CNAG_00370) coding region was 787 bp long with five exons and encodes a 129-amino-acid protein. The ubiquitin hybrid protein UBI1 consisted of a monoubiquitin domain fused to a 53-aa C-terminal extension protein (Ribosomal unit protein L40a, Rpl40a) (Figure 1A). Other putative eukaryotic UBI1 orthologs displayed an identical composition of domain structures, including the single ubiquitin moiety and the ribosomal protein L40e or S27e (Figure 1A). The putative sequence of Cn-Ubi1 was highly similar to its counterparts in other fungi (Schizosaccharomyces pombe, 96% identity; S. cerevisiae, 95% identity) and other eukaryotic species (Caenorhabditis elegans, 89% identity; Drosophila melanogaster, 88% identity) (Figure 1B), indicating that the Ubi1 protein is highly conserved across eukaryotic species.
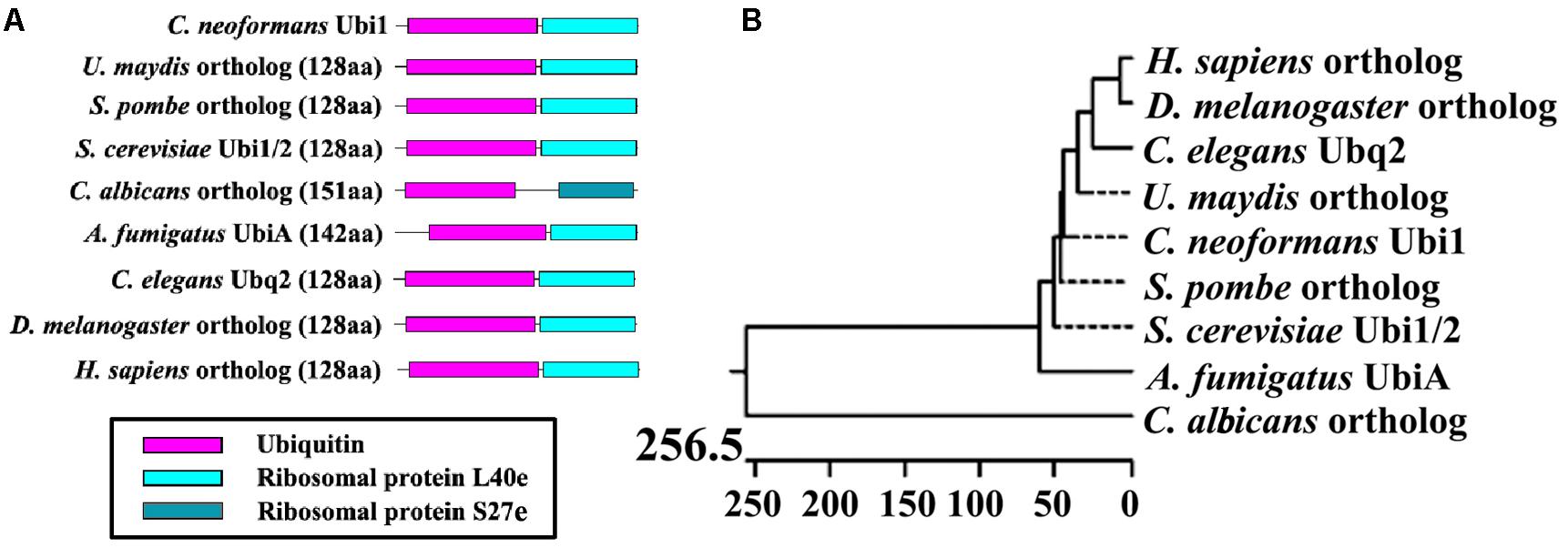
Figure 1. Bioinformatic analysis of putative Ubiquitin hybrid protein Ubi1. (A) Comparison of Ubi1 orthologs between Cryptococcus neoformans and other species. Ubi1 ortholog diagrams show functional protein domains, including a monoubiquitin and a ribosomal protein (L40e or S27e), which were identified by the Pfam database. (B) Phylogenetic analysis was depicted by Jotun Hein alignment using DNASTAR software. The protein sequences of putative Ubi1 orthologs were retrieved from the following databases: C. neoformans Ubi1 (CNAG_00370) from the C. neoformans H99 database, and its orthologs among other eukaryotic species from the protein databank of the NCBI website. XP_011388728, Ustilago maydis (Um); NP_593923, Schizosaccharomyces pombe (Sp); CAA86130 and CAA51949, S. cerevisiae (Sc); XP_715397, Candida albicans (Ca); XP_001481702, Aspergillus fumigatus (Af); NP_499695, Caenorhabditis elegans (Ce); NP_001260018, Drosophila melanogaster (Dm); NP_001307950, Homo sapiens (Hs). The bar marker indicates genetic distance.
To understand the function of the ubiquitin hybrid protein Ubi1 in C. neoformans, both deletion mutant (ubi1Δ) and reconstituted (ubi1Δ:UBI1) strains were constructed using a biolistic system and confirmed via diagnostic PCR and Southern Hybridization (Supplementary Figure S1). Intriguingly, the ubi1Δ mutant displayed severe growth restriction. Therefore, we first compared its growth rate with the WT (H99) and reconstituted strains in rich medium (YPD) at 30°C. After a short lag phase of 4 h, the H99 and ubi1Δ:UBI1 strains rapidly entered the log phase, and they reached the stationary phase within 36 h. In contrast, the ubi1Δ mutant remained in the lag phase for 48 h and only reached the stationary phase after 108 h (Figure 2A). The generation times of the H99 and ubi1Δ mutant strains were 3.7 and 12.2 h, respectively, suggesting that Ubi1 is critical for maintaining growth rate in C. neoformans. In addition, the ubi1Δ mutant strain displayed significantly smaller colonies than either the H99 or ubi1Δ:UBI1 strains after 9 or 14 days incubation on YPD agar at 30°C (Figure 2B). We next evaluated the budding rate of each strain (Figure 2C). At mid-log phase, the ubi1Δ mutant strain displayed about a 55.5% budding rate, significantly lower than those of the H99 (66.8%) or reconstituted (67.3%) strains (P < 0.001). At stationary phase, however, there were no statistical difference in the budding rate of each strain. Our data reveal that the ubiquitin hybrid protein Ubi1 is important for cryptococcal vegetative growth.
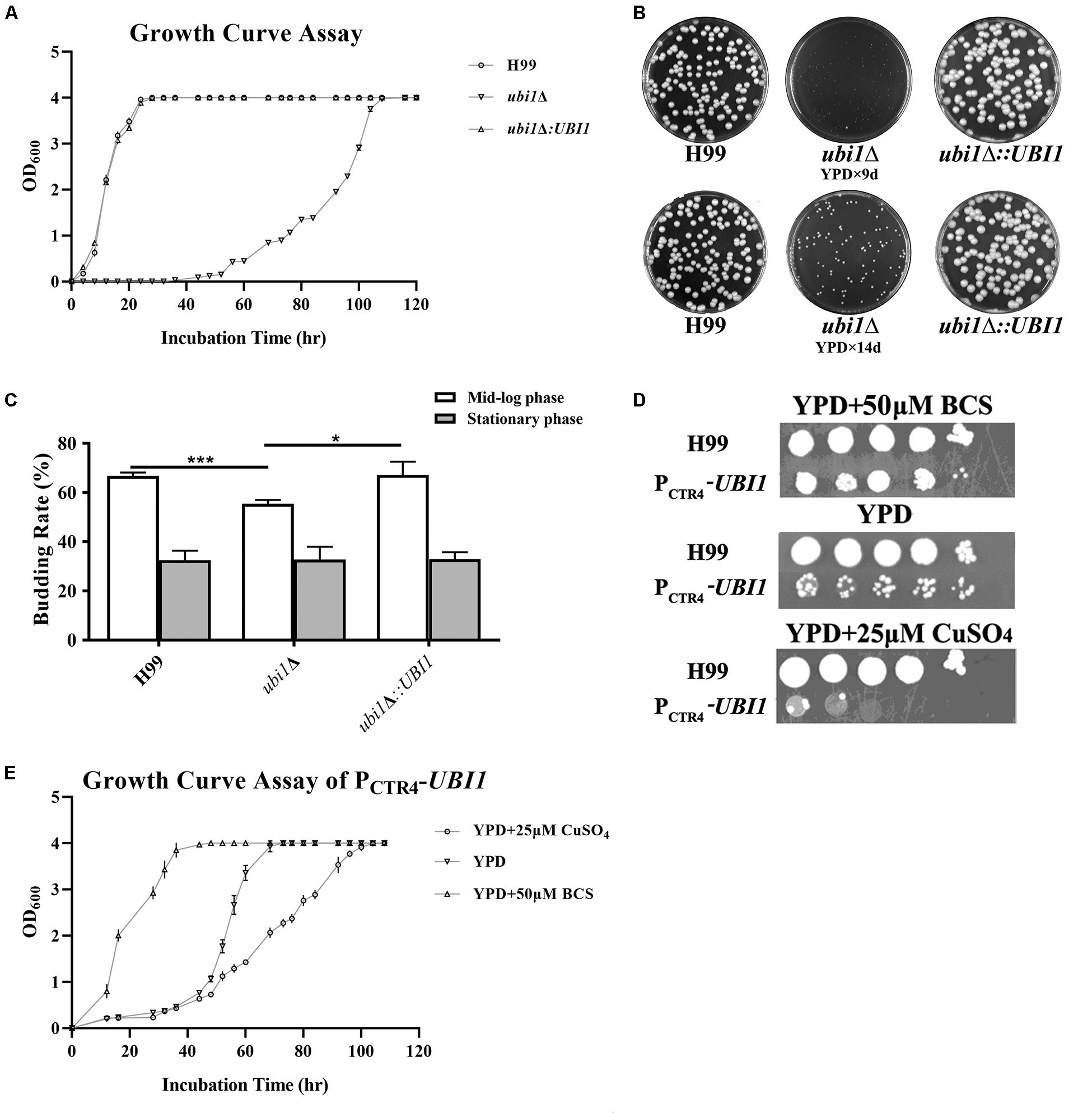
Figure 2. Ubi1 is essential for growth and proliferation in C. neoformans. (A) Growth rate assay. Samples (106 CFU) of the wild-type (WT, H99), mutant (ubi1Δ), and reconstituted (ubi1Δ:UBI1) strains were transferred to 30 mL fresh YPD medium in flasks and incubated at 30°C. OD600 was measured for each culture at 4-h intervals. All assays were performed in triplicate. (B) Colony size measurement. Samples (100 CFU) of each strain were spread on YPD agar medium and then incubated at 30°C for 2 weeks. Photographs of each petri dish were taken on day 9 and day 14. (C) Budding rate assay. Each strain was incubated to the mid-log or stationary phase in YPD liquid medium at 30°C. Each culture was diluted and stained with India ink. The budding rates (%) were calculated with a hemocytometer using the formula: budding cells/total cells × 100 for each strain. The total number of cells for each strain was not less than 500, and each test was repeated three times. ***P < 0.001, *P < 0.05. (D) The growth performance of the condition-regulated strain (PCTR4-UBI1) under YPD agar with/without CuSO4 or BCS. The WT and PCTR4-UBI1 mutants were grown to saturation in YPD liquid medium, 10-fold serially diluted, and 5 μL cells spotted on different YPD solid media. The samples were incubated at 30°C for 1 week and then photographed. (E) The growth rate of the condition-regulated strain (PCTR4-UBI1) was inversely correlated with copper concentration and, thus, positively correlated with UBI1 expression level.
To further clarify the role of the ubiquitin hybrid protein Ubi1 inyeast growth, the copper-repressible promoter PCTR4 was inserted upstream of the UBI1 gene to enable its expression in C. neoformans to be regulated by copper. Quantitative real-time PCR was performed to examine UBI1 mRNA levels under the control of varying levels of copper to correlate with growth rate. Compared to H99 grown in regular YPD medium, the UBI1 transcript showed a 66-fold reduction when grown in YPD containing CuSO4 but an 80-fold increase when grown in YPD containing BCS (Supplementary Figure S2A). At 30°C, H99 displayed normal growth in YPD medium and in YPD containing BCS or CuSO4, with a doubling time of about 4 h (3.7–4.2 h) in the different media (Supplementary Figure S2B). In contrast, the PCTR4 reconstituted strain (PCTR4-UBI1) showed significant growth arrest in the YPD agar containing CuSO4, partial growth arrest in YPD, and normal growth in the YPD containing BCS, with doubling times of 11.6, 7.1, and 4.2 h for those three media, respectively (Figure 2E). The growth performance of PCTR4-UBI1 on YPD agar, with or without copper ions, was highly consistent with the results obtained from the liquid media (Figure 2D), further indicating that the cryptococcal proliferation rate is positively correlated with UBI1 expression level.
Next, we studied the effects of Ubi1 on the cell morphology and intracellular structure of C. neoformans using TEM. Compared with the WT strain, deletion of UBI1 caused several significant morphological alterations in C. neoformans cells, including irregularly shaped and larger cryptococcal cells (Figures 3A,D), uneven cell wall thickness, more projections or pseudopodia-like structures on the cell membrane (Figures 3D,E), fewer free ribosome particles (Figures 3C,F), more irregularly shaped mitochondria, and fewer cristae (Figure 3F). Moreover, ubi1Δ mutant cells had more sac-like or tubular cristae, whereas WT cells had more lamellar cristae (Figures 3C,F). When the UBI1 gene was reconstituted, the cellular morphology and intracellular structure was completely restored to WT status (Figures 3G–I), indicating that the hybrid protein Ubi1 is essential for proper morphological maintenance and organelle functioning in C. neoformans.
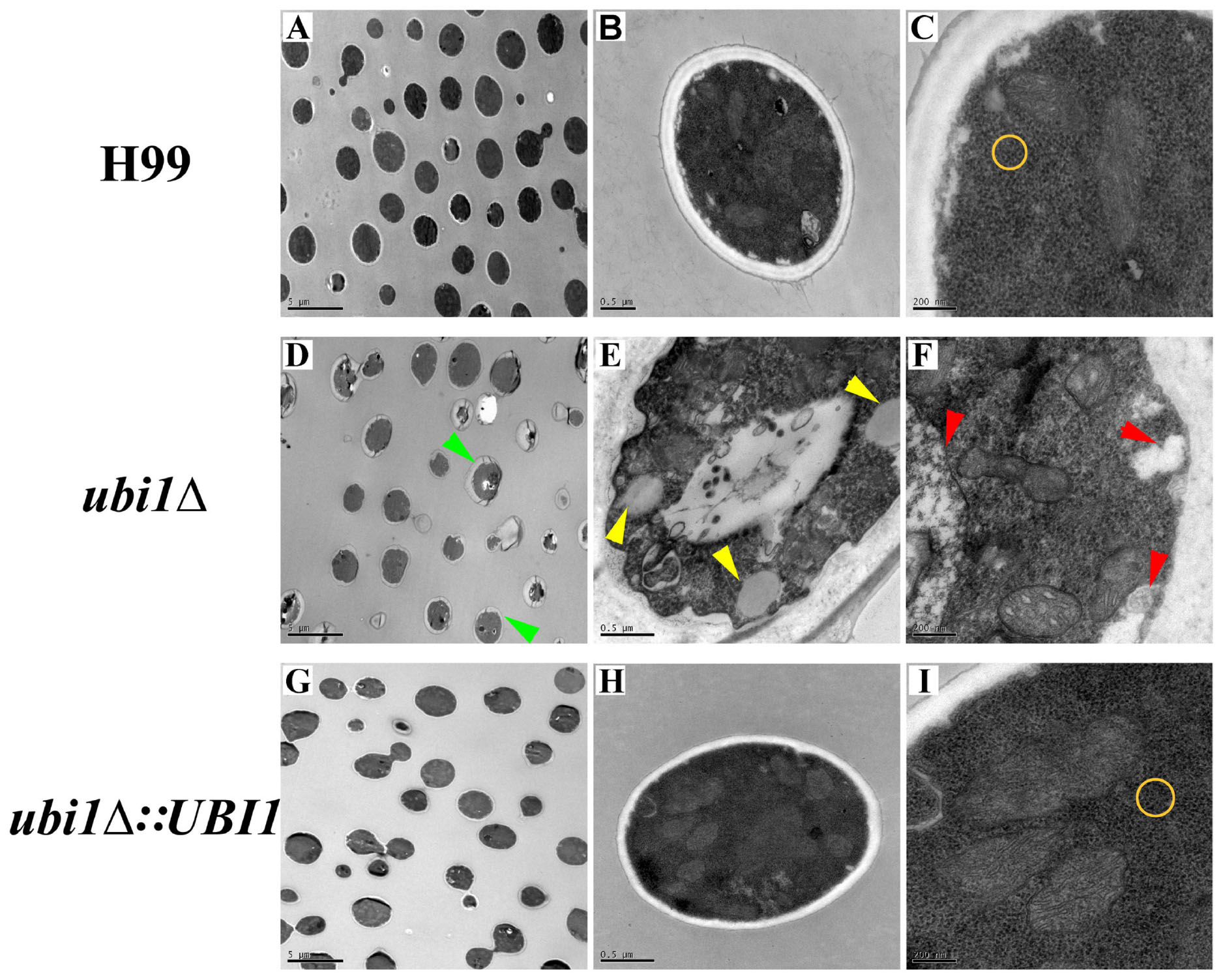
Figure 3. Impact of UBI1 deletion on the cell morphology and intracellular structure of C. neoformans. The TEM images represent different strains as follows, H99 (A–C), ubi1Δ (D–F), and ubi1Δ:UBI1 (G,H). Sizes of the scale bar: 5 μm for (A,D,G); 0.5 μm for (B,E,H); and 200 nm for (C,F,I). Green arrow, irregular cell shape and uneven cell wall thickness; red arrow, swelling mitochondria with dissoluted ridge; yellow arrow, intracellular vacuoles; yellow circle, comparison of ribosomal density.
Ribosomes are key organelles that mediate protein translation, and thus determine the growth and proliferation of eukaryotic cells (Cheng et al., 2019). In S. cerevisiae, Rpl40e deletion induced a slow-growth phenotype, characterized by impaired 60S ribosomal subunit biogenesis (Finley et al., 1989; Fernández-Pevida et al., 2012). To investigate the roles of Rpl40e in regulating cryptococcal growth, we generated a truncated UBI1-targeting vector containing only the C-terminal RPL40a domain (Figure 4A) and transformed it into a ubi1Δ mutant strain. The reconstituted ubi1Δ:RPL40a strain was confirmed by diagnostic PCR and real-time PCR. The transcriptional levels of RPL40a were essentially the same between ubi1Δ:RPL40a and the WT or reconstituted (ubi1Δ:UBI1) strains (Figure 4B). Compared with the knockout strain ubi1Δ, the growth rate of the partially reconstituted strain ubi1Δ:RPL40a was significantly increased, and was similar to those of the WT and fully reconstituted ubi1Δ:UBI1 strains (Figure 4C), indicating that the C-terminal RPL40a domain of Ubi1 is responsible for its function in regulating cryptococcal growth.
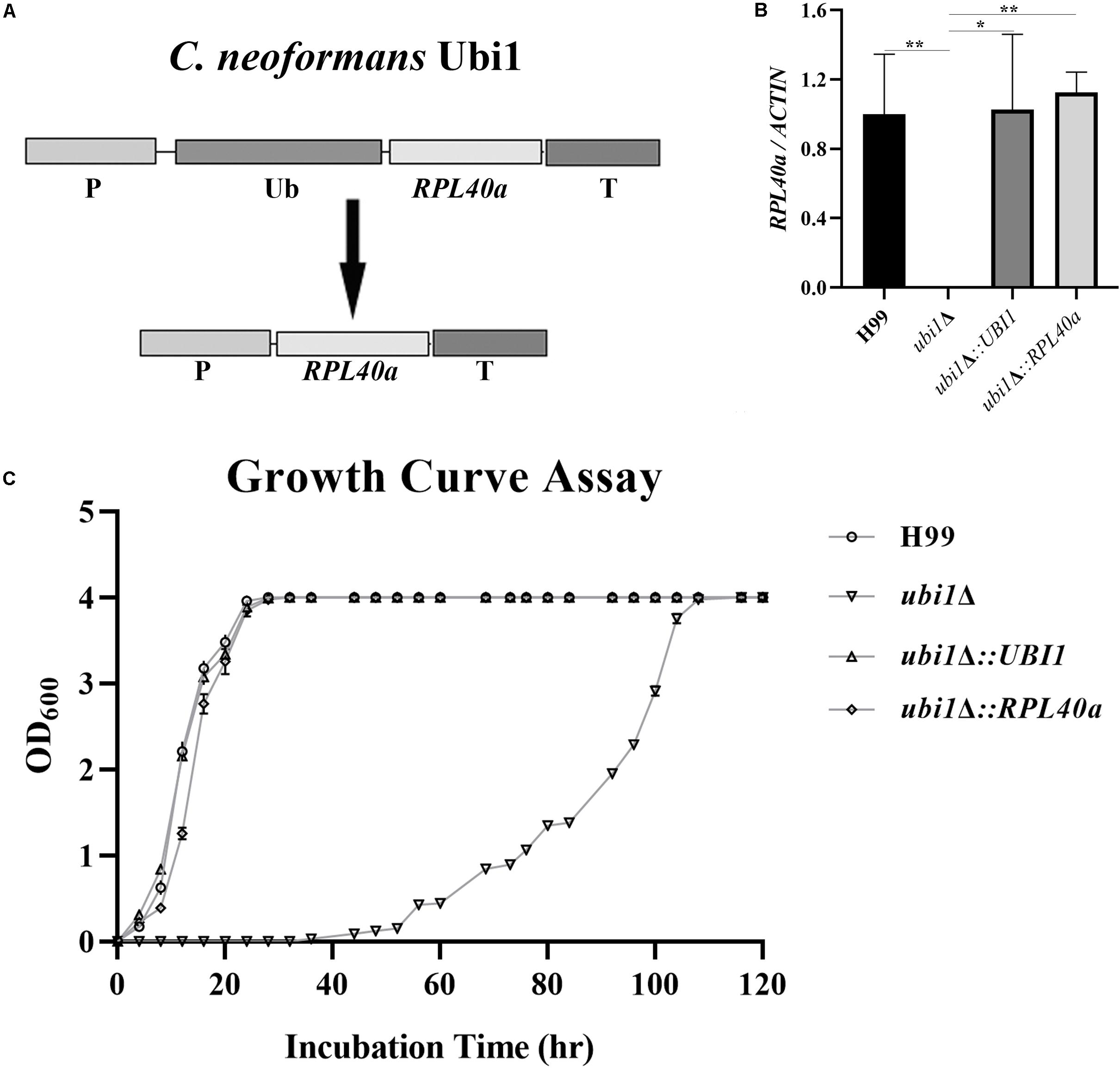
Figure 4. The C-terminal domain Rpl40e is responsible for the function of Ubi1 in regulating cryptococcal growth. (A) Construction of the partially reconstituted strain ubi1Δ:RPL40a. The truncated gene, RPL40a, containing the UBI1 promoter, RPL40a ORF, and UBI1 terminator, was cloned into the vector pCH233. The generated vector, pRPL40a-NAT, was linearized and integrated into the mutant strain ubi1Δ. (B) Confirmation of the partially reconstituted strain ubi1Δ:RPL40a via real-time PCR. The strain ubi1Δ:RPL40a displayed a similar transcriptional level to that of the WT (H99) or the fully reconstituted (ubi1Δ:UBI1) strain. The test was repeated three times for each strain. *P < 0.05, **P < 0.01. (C) After introduction of the RPL40a domain, the ubi1Δ mutant restored its growth rate, similar to that of WT or the fully reconstituted strain. Each experiment was performed in triplicate.
Cell size is a key factor for proliferative capacity and growth rate regulation in most eukaryotic cells. Cell size homeostasis is controlled at the G1/S phase boundary, primarily by preventing cell division until a “critical cell size” is attained (Aldea et al., 2017). The cell size of the ubi1Δ mutant strain of C. neoformans was therefore examined. When cultured to mid-log phase in rich medium (YPD) at 30°C, the average diameters of H99, ubi1Δ, ubi1Δ:UBI1, and ubi1Δ:RPL40a cells were 5.30 ± 0.30, 6.43 ± 0.76, 5.63 ± 0.36, and 5.17 ± 0.41 μm, respectively (P < 0.0001) (Figure 5A). Propidium iodide staining, followed by a fluorescence flow cytometry assay, showed that the deletion of Ubi1 resulted in irregular DNA content in C. neoformans, which ranged from 1N to more than 4N (49%). Reconstitution of UBI1 or RPL40a reversed the ploidy of the ubi1Δ cells back to a regular 1N/2N mode (Figure 5B). Moreover, normal cell cycle progression was blocked by UBI1 deletion (manifested by the depletion of S phase cells), and was rescued by the introduction of either full-length Ubi1 or only RPL40a (Figure 5C).
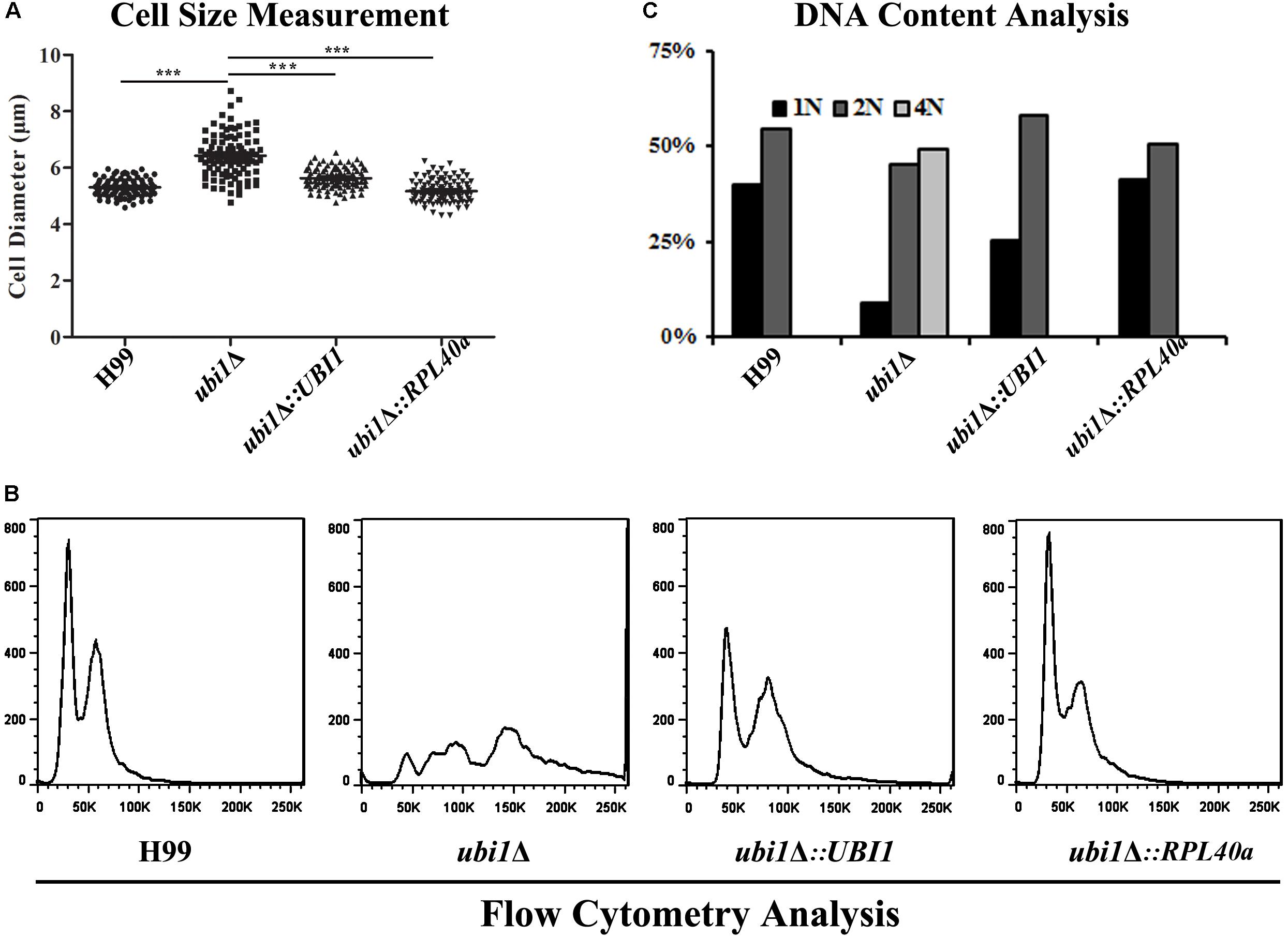
Figure 5. The C-terminal domain Rpl40e is responsible for the function of Ubi1 in regulating cryptococcal growth. (A) Deletion of Ubi1 led to enlargement of cell size in C. neoformans. Each strain was incubated to mid-log phase at 30°C, washed, and then visualized using the 63× objective of a Zeiss LSM inverted confocal microscope (Jena, Germany). ***P < 0.001. For each culture, the cell sizes of at least 100 cells were measured by using Adobe Photoshop Software. Flow cytometry analysis was performed to assess the effect of UBI1 deletion on the ploidy (B) and cell cycle progression (C) of C. neoformans. Each experiment was performed in duplicate.
Since capsules and melanin are important pathogenic factors for cryptococcal invasion into the host, we further examined their expression in each strain. As shown in Figure 6, UBI1 deletion induced a slight increase in the capsule production of C. neoformans (Figures 6A,B, P < 0.001), but fully blocked its melanin secretion. Reconstitution of the UBI1 or RPL40a gene restored the normal production of both capsules and melanin in the ubi1Δ mutant. Besides, we also evaluated the effect of UBI1 deletion on the formation of titan cells, which confers distinct advantages to C. neoformans against the host during infection (Zaragoza and Nielsen, 2013). After incubation in Titan Cells Medium, the ubi1Δ mutant exhibited a significant enlargement of total cell size and cell body size compared to WT or reconstituted strain (Figure 6D; P < 0.001), indicating that Ubi1 is involved in titan-like cell formation in vitro.
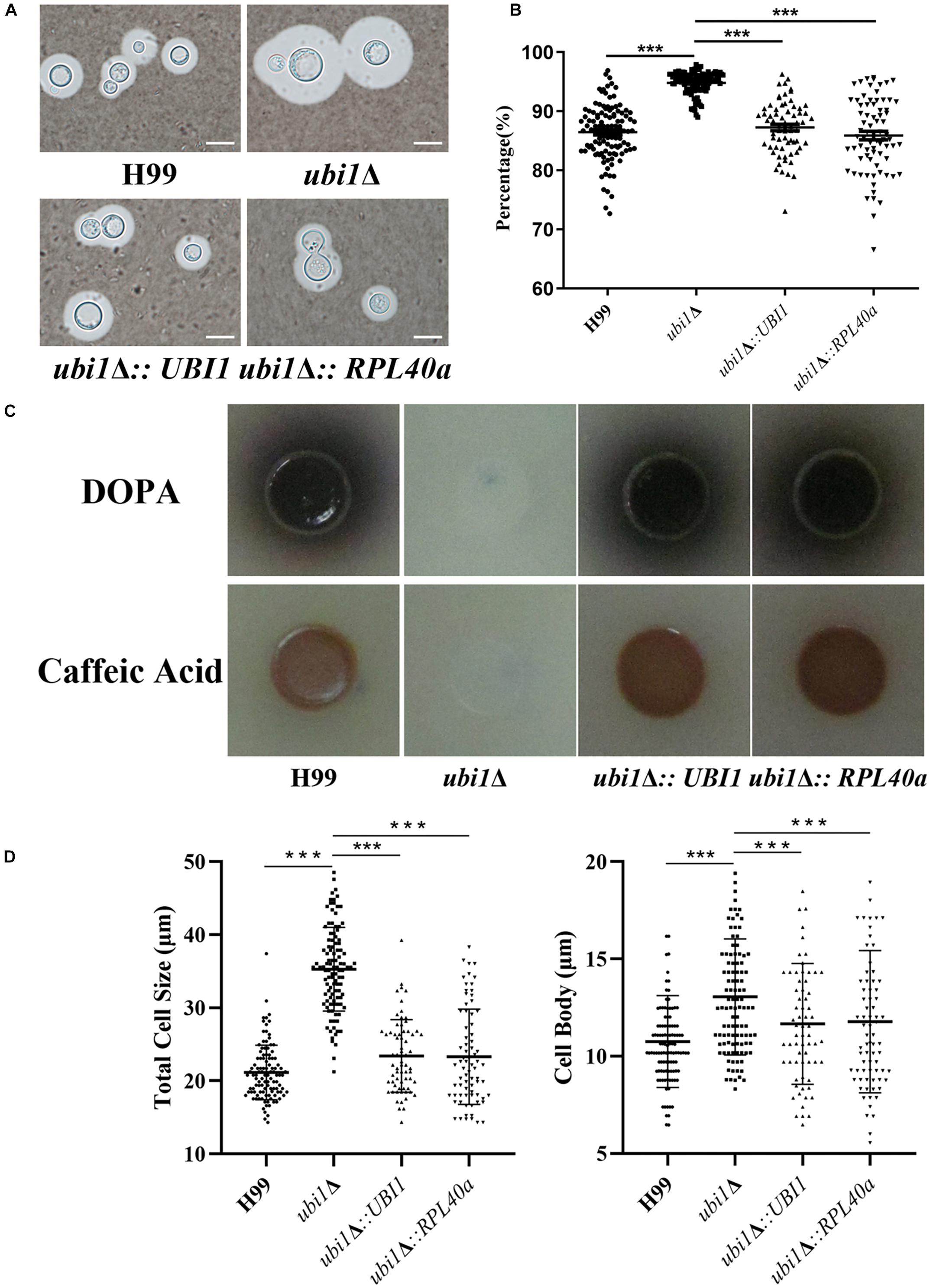
Figure 6. Ubi1 is involved in the production of capsule and melanin in C. neoformans. (A) Capsule induction assay. All the strains were cultured on DME medium at 37°C for 5 days. The capsules were examined by staining them with India ink and visualizing them at 100× magnification (scale bar = 5 μm). (B) Relative capsule volume detection. Total (cell and capsule) and cell-only diameter were measured via Adobe Photoshop Software for more than 50 cells of each strain. Then, the relative ratio of the capsule was calculated with the formula: (Total Volume-Packed Volume)/Total Volume. (C) Melanin production assay. Strains were grown on L-DOPA or caffeic-acid medium for 5 days at 30°C. (D) Titan-like cell induction assay. Strains were grown on Titan Cells Medium at 37°C and 5% CO2 for 3 days. Measurements of total cell sizes (left) and cell body sizes (right). ***P < 0.001.
To examine the roles of Ubi1 in stress response, different C. neoformans strains were exposed to various stresses. High temperature, oxidative stress (H2O2 and NaNO2), sorbitol, high salt (NaCl and KCl), Congo red, and SDS all exacerbated the growth restriction or growth defect of the ubi1Δ strain (Supplementary Figure S3). Both the ubi1Δ:UBI1 and ubi1Δ:RPL40a strains exhibited growth similar to the WT (H99) in the presence of the above-mentioned stressors (Supplementary Figure S3). Together, these phenotypic assays in vitro suggest that Ubi1 may be essential for cryptococcal adaptability to various environmental niches.
Next, we examined the effect of UBI1 deletion on cryptococcal virulence via macrophage killing and mouse survival assays. WT H99, ubi1Δ, and reconstituted strains (ubi1Δ:UBI1 and ubi1Δ:RPL40a) were co-cultured with IFN-γ and LPS-activated J774A.1 macrophages. There were no differences among the strains in the initial uptake into the macrophages (data not shown). However, deletion of the Ubi1 gene reduced the intracellular survival of C. neoformans by about 97% (P < 0.0001 vs. H99), while reconstituting UBI1 or RPL40a partially restored its intracellular survival rate (Figure 7A).
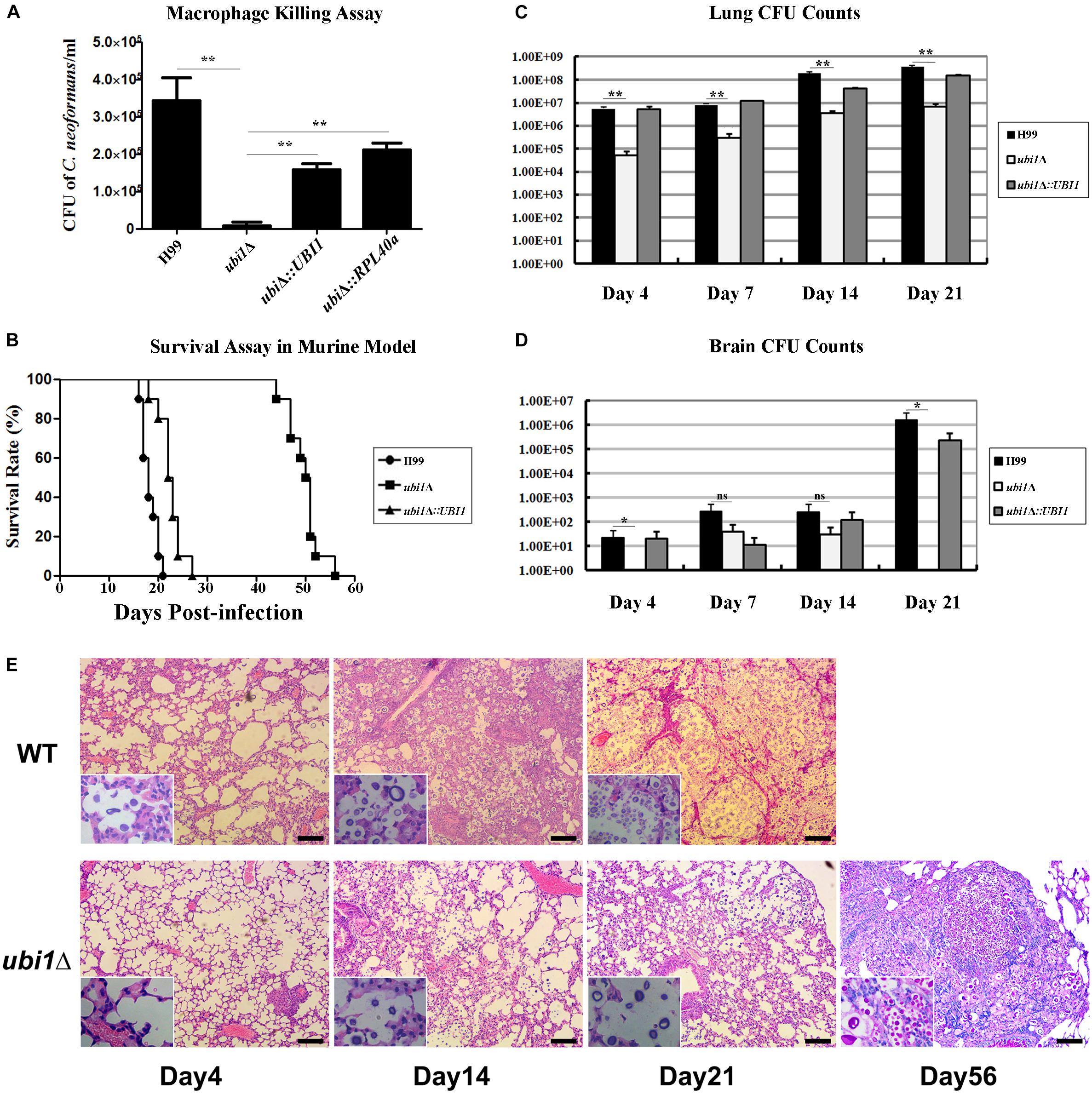
Figure 7. Ubi1 is required for C. neoformans virulence. (A) C. neoformans colonies were counted after uptake and killing by activated macrophages overnight and macrophage lysate was diluted and cultured on YPD plates. **P < 0.01. (B) The mouse survival assay showed that ubi1Δ C. neoformans-infected mice had significantly longer survival times (P < 0.01). Mouse lung (C) and brain (D) fungal burdens were evaluated by counting fungal colonies after culturing serially diluted tissue lysate on YPD plates. *P < 0.05, **P < 0.01; ns, no significant difference. (E) Ubi1 deletion altered host pulmonary inflammation responses to C. neoformans. Representative photographs of hematoxylin and eosin-stained mouse lungs at different times post-infection. Scale bar = 100 μm.
The in vivo virulence of the WT, ubi1Δ, and ubi1Δ:UBI1 strains was evaluated using a mouse survival assay. Mice infected with the ubi1Δ strain exhibited prolonged survival (49.8 ± 3.3 days, P < 0.001 compared to WT) while the average survival times of WT- and ubi1Δ:UBI1-infected mice were 18.3 ± 1.6 and 22.5 ± 2.4 days, respectively (Figure 7B), indicating a significant role of Ubi1 in the pathogenesis of C. neoformans. Since pneumonia and meningoencephalitis were the primary clinical manifestations of cryptococcosis, the fungal burdens of the lungs and brain of C. neoformans-infected mice were assessed. As shown in Figure 7C, the cryptococcal burden gradually increased in the lungs of mice infected by the ubi1Δ mutant, but was significantly lower (about 30–110 fold) than those of WT- and ubi1Δ:UBI1-infected mice (Figure 7C). The brain fungal burden of WT- and ubi1Δ:UBI1-infected mice remained low until 21 days post-infection (pi). However, ubi1Δ infection produced a low brain fungal burden 7 and 14 days pi and almost no detectable living yeast cells 4 days or 21 days pi (Figure 7D). These results indicate that UBI1 deletion might lead to a chronic pulmonary infection, with delayed or reduced brain invasion by C. neoformans.
As the mice infected by the ubi1Δ strain displayed significantly prolonged survival, but finally succumbed to chronic pulmonary infection, we next assessed the effects of UBI1 deletion on the pathological changes in mouse lung and immune responses to C. neoformans. In BALB/c mice infected with the WT strain, a few yeast cells were confined to the alveolar space or septum, along with mild infiltration of the neutrophils, at the early stage of infection (day 4 pi; Figure 7E). On day 14 pi, mild destruction of the alveolar architecture owing to propagated cryptococci and severe inflammation were observed. The inflammatory cells were primarily mononuclear cells with a few eosinophils. Widespread pulmonary alveolar destruction and a large quantity of fungi with much fewer inflammatory cells were seen 21 days pi-.
Mouse lungs infected with the ubi1Δ mutant strain exhibited a different histological pattern. On day 4 pi, limited yeast cells were present in the alveolar septum, accompanied by increased inflammatory cell infiltration. Intriguingly, on days 14 and 21 pi, there was a significant increase of fungal cells present in the alveolar space but without any exacerbation of inflammation. By day 56 pi, however, a large proportion of the pulmonary alveoli were destroyed and filled with a multitude of yeast cells in the presence of mononuclear and lymphoid cells. It was also noted that a proportion of titan cells, an important C. neoformans morphotype characterized by enlarged cells with extensive capsules, were observed at the late stage of infection (day 56 pi) which occurred around 14 days pi in the lungs of mice infected with the WT strain (Figure 7E).
In order to characterize the effect of cryptococcal UBI1 deletion on pulmonary inflammatory response, we assessed changes in the immune cell population using flow cytometry with total lung cells on days 7, 14, and 21 pi. Mouse lungs infected with the ubi1Δ strain had significantly fewer total leukocytes, neutrophils, and dendritic cells than the WT strain-infected mouse lungs at all time points. Monocyte numbers were substantially higher in the WT-infected mouse lungs only on 14 days pi, while the number of eosinophils was not different at any time point. The total numbers of T cells, CD4+ T cells, and CD8+ T cells were markedly higher in the ubi1Δ strain-infected mouse lungs than in the WT-infected ones at all infection stages. The B cells showed a biphasic change pattern; the ubi1Δ strain-infected mouse lungs had fewer B cells 14 days pi but significantly more B cells 21 days pi than the WT-infected mouse lungs (Figure 8). Accordingly, mice produced different patterns of cytokines upon infection with WT and ubi1Δ C. neoformans. The levels of TNF-α, IL-20p40, and IL-17A in the ubi1Δ strain-infected mice were much higher than those in the WT-infected mice at all examined time points. However, the IL-4 level was drastically higher in the WT-infected mice at every stage of infection. The IFN-γ level was higher in the WT-infected mice 7 days pi but in the ubi1Δ C. neoformans-infected mice 14 days pi (Figure 9).
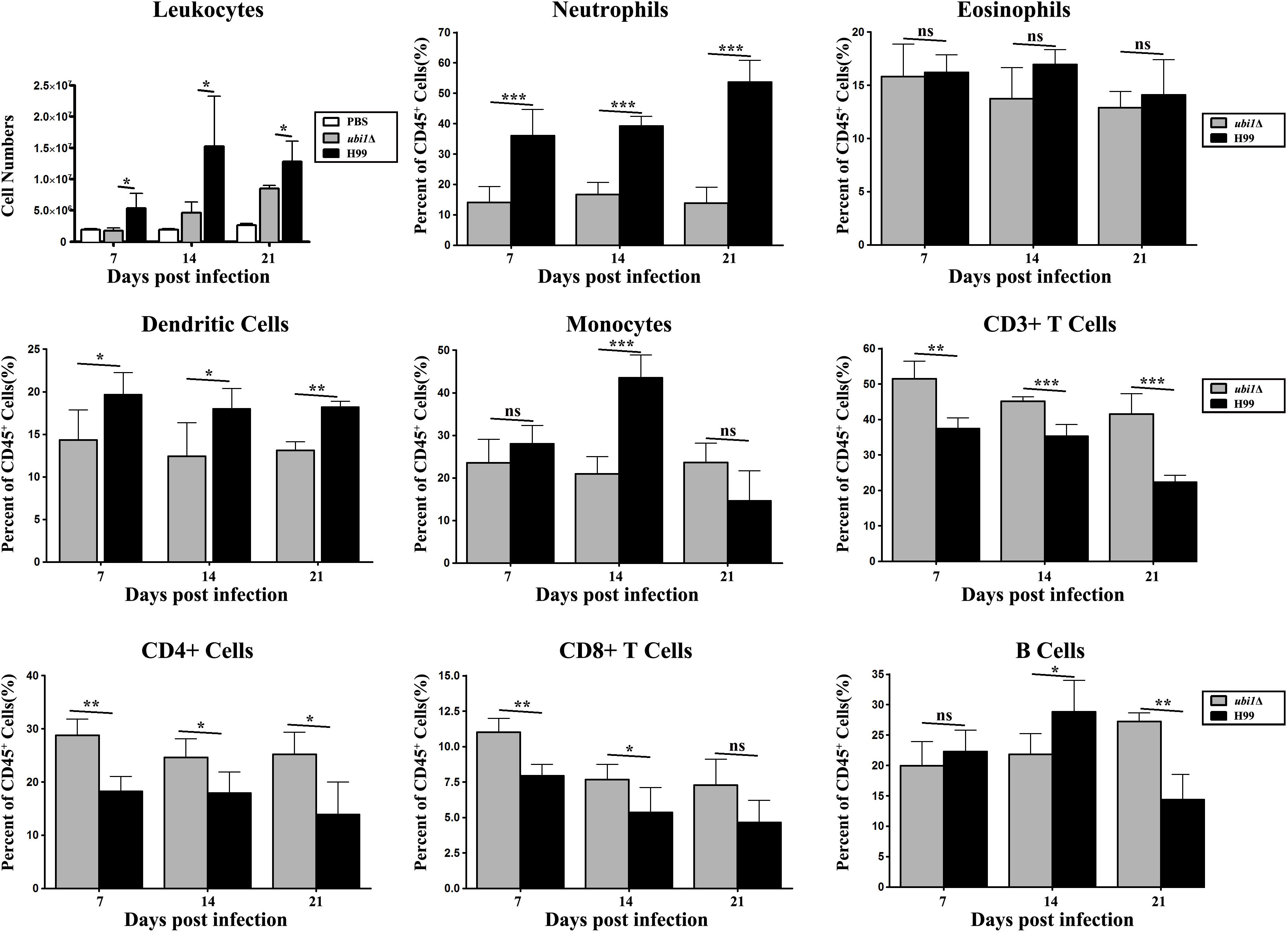
Figure 8. Ubi1 regulated pulmonary recruitment of leukocyte subsets during cryptococcal infection. Different leukocytes and lymphocytes from mice infected by C. neoformans were assessed by flow cytometry. Results represent mean ± SEM, N = 4 or more mice per group. *P < 0.05, **P < 0.01, ***P < 0.001; ns, no significant difference.
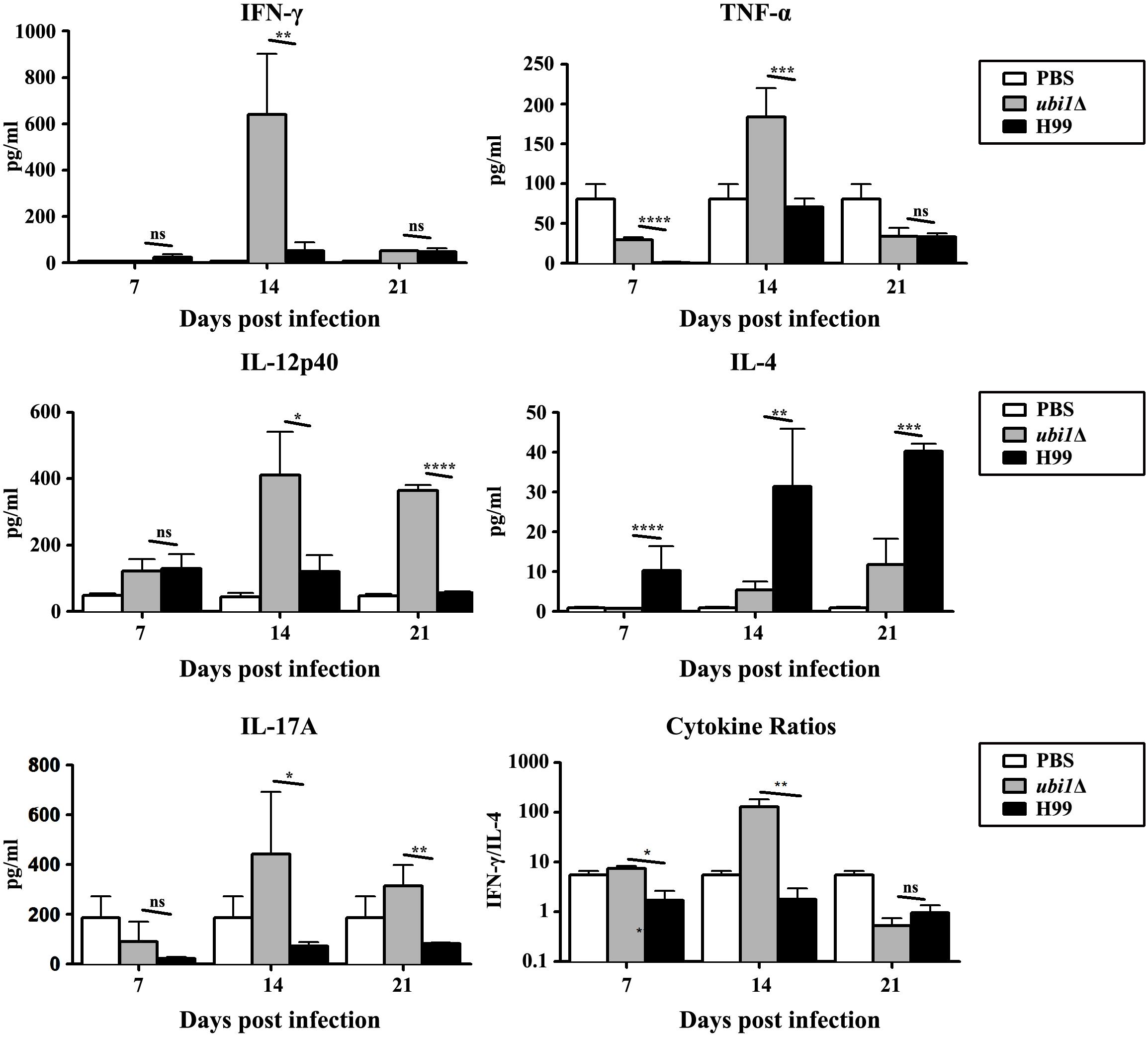
Figure 9. UBI1 deletion induced the robust development of pulmonary Th1 cytokine bias during cryptococcal infection. Specified inflammatory cytokines of C. neoformans-infected mice were analyzed using ELISA. Results represent mean ± SEM, N = 4 or more mice per group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, no significant difference.
To explore the molecular basis for the defect of in vitro growth and in vivo invasion caused by UBI1 deletion, RNA sequencing was performed to compare the transcriptional profiles of WT and ubi1Δ cells. The data have been submitted to the GEO database under accession number GSE1423742. A total of 236 genes were differentially expressed (141 upregulated and 95 downregulated) by at least twofold (P < 0.05) between these two strains (Table 1). The genes regulated by UBI1 deletion covered a variety of functional categories. First, a group of genes involved in ribosome biogenesis and RNA processing and transport were significantly upregulated in the ubi1Δ mutant compared to the WT. Consistent with the reduced ribosome particles in the TEM assay (Figure 3), these results suggest that UBI1 deletion might induce ribosome stress and dysfunction in C. neoformans. Furthermore, several genes involved in nutritional metabolism were differentially expressed by the UBI1 mutation. Several enzymes in amino acid metabolism (except the sulfur-containing amino acids), steroid biosynthesis, and oxidative phosphorylation were repressed, suggesting that UBI1 deletion might alter metabolic flux in C. neoformans. Ubi1 inactivation also resulted in the downregulation of genes associated with cell cycle progression (such as TEM1 and CDC31) and nucleotide metabolism, which might partially explain the cell cycle arrest in the ubi1Δ mutant (Figure 5). Finally, UBI1 deletion also led to the differential expression of several genes involved in other basic cellular processes, such as DNA repair, protein processing, and signaling pathways.
Ubiquitin is a highly conserved post-translational modifier, involved in a variety of cellular processes (Clague and Urbé, 2010). In most eukaryotes, ubiquitin is expressed as two classes of proteins: (i) a hybrid protein between a ubiquitin monomer and a ribosomal protein (such as Rpl40e or Rps31e), and (ii) a polyubiquitin precursor including a polymer of several tandem ubiquitin monomers. The current study characterized the roles of the ubiquitin hybrid protein, Ubi1, in the growth and virulence of C. neoformans. Deletion of UBI1 resulted in growth inhibition, cell ploidy increase, morphological abnormalities, cell cycle arrest, and attenuated virulence, which were all corrected by reconstituting ubi1Δ cells with either the full-length UBI1 gene or the C-terminal RPL40a domain.
The most striking phenotypic observation in the cryptococcal ubi1Δ strain was the dramatic growth restriction, even under rich growth conditions. Several lines of evidence strongly support that the growth rate defect caused by UBI1 deletion is closely associated with abnormal ribosomal biosynthesis. First, the TEM assay showed a significant reduction of ribosomes with sparse distribution in the ubi1Δ cells, compared with the WT or ubi1Δ:UBI1 strains of C. neoformans. A deficiency of ribosomes, the conserved molecular machines that mediate protein translation, is closely associated with decreased bulk protein synthesis and thus a defective cell growth rate (Steffen et al., 2012; Cheng et al., 2019). Furthermore, reconstitution of the UBI1 or RPL40a domain restored the exponential growth rate in cryptococcal strains with UBI1 deletion. Rpl40e is a highly conserved large subunit ribosomal protein in eukaryotes (Finley et al., 1989; Kobayashi et al., 2016). In S. cerevisiae, Rpl40e is generated by the proteolytic cleavage of ubiquitin precursor proteins (Ubi1 and Ubi2) and is required for 60S subunit production, subunit assembly, and the translocation process during translation elongation (Finley et al., 1989; Fernández-Pevida et al., 2012). Deletion of either RPL40-encoding gene was characterized by a slow-growth phenotype, while double deletions caused a lethal phenotype in S. cerevisiae (Finley et al., 1989). Our transcriptome analysis further revealed that UBI1 deletion in C. neoformans induced the significant upregulation of several genes involved in ribosome biogenesis and RNA processing and transport. Therefore, we speculate that Ubi1 might play similar roles in the ribosomal biogenesis and homeostasis of C. neoformans. The detailed mechanism requires further investigation.
In addition to growth rate, we found that Ubi1 also participated in regulating cell ploidy variation, morphological homeostasis, and cell cycle progression in C. neoformans. UBI1 deletion induced a substantial increase of cryptococcal cells with 4N DNA content. Ploidy changes usually influence basic cellular properties, including cellular morphology. For example, cryptococcal Titan cells (highly polyploid cells) have a gigantic cell size (>10 μm), an enlarged capsule, and a dramatically thicker cell wall (Zaragoza et al., 2010; Dambuza et al., 2018). Consistent with the ploidy increase, the ubi1Δ strain also exhibited significant morphological alterations, such as enlargement of the cell size and extracellular capsule, uneven cell wall thickness, and more pseudopodia-like structures on the cell membrane. The ploidy alterations might be associated with the cell cycle arrest observed in ubi1Δ cells. Previous studies revealed that abrogation of ribosome biogenesis generated cell cycle arrest (Gómez-Herreros et al., 2013; Thapa et al., 2013; Polymenis and Aramayo, 2015). In S. cerevisiae, the repression of nine 60S ribosomal protein genes, including RPL40a, leads to cell cycle arrest in the G2/M phase (Thapa et al., 2013). Similarly, we observed that cell clusters with >2N DNA content accounted for 49% of ubi1Δ cells, suggestive of a G2/M cell cycle phenotype in C. neoformans. The phenotype is consistent with the transcriptomic profile of the ubi1Δ strain, in which expression of some proteins required for the G2/M phase of progression, such as Tem1 and Cdc31, were significantly downregulated (Shirayama et al., 1994; Ivanovska and Rose, 2001). Furthermore, the depletion of S phase cells was another cell cycle phenotype of the ubi1Δ strain. It also exhibited remarkable inhibition of the transcription of several genes involved in nucleotide anabolism of C. neoformans. Together, these data highlight the importance of Ubi1 (especially Rpl40e) in regulating cell ploidy regularity, morphological homeostasis, and cell cycle progression.
Finally, UBI1 deletion induced a dramatic attenuation in the survival and infectivity of C. neoformans in mammalian hosts. Our macrophage and mouse experiments exhibited significantly reduced intracellular survival and fungal burdens for the ubi1Δ strain, which is consistent with the results of the stress assay. The phenotype is probably attributable to the growth rate defect of cryptococcal mutant cells owing to ribosomal deficiency. Intriguingly, growth in vivo of the ubi1Δ strain seemingly outpaces the WT strain in the early stage of pulmonary infection (Figure 7C). The difference of growth rates in vivo and in vitro is confounding, which requires further exploration. In addition, the attenuated virulence of the ubi1Δ strain was closely associated with the induction of a T-helper (Th)1-type response, which could promote the elimination of intracellular pathogens (Arora et al., 2011; Davis et al., 2013). Histology sections demonstrated more localized and mild inflammation in the lungs of ubi1Δ-infected animals, and presented infiltration with fewer total leukocytes, neutrophils, and dendritic cells but a higher proportion of T lymphocytes than mice infected with the WT strain. Furthermore, ubi1Δ-infected mice showed a significant increase of Th1-type cytokines (such as IFN-γ, TNF-α, IL-12, and IL-17A) while mice in the WT group showed an accumulation of the anti-inflammatory cytokine IL-4. This distinct immune response profile might be attributed to cryptococcal morphological alterations owing to UBI1 deletion. The irregular shapes and uneven cell walls might significantly alter pathogen-associated molecular pattern exposure in ubi1Δ cells. For example, the ubi1Δ mutants showed a drastic defect in melanin secretion, and cryptococcal laccase (melanin synthase) induces immune modulation from Th1/Th17 responses toward Th2 responses (Qiu et al., 2012). Despite the attenuated virulence, however, the ubi1Δ strain eventually caused delayed lethality in mice, which probably died of chronic pulmonary infection. Previous reports revealed that the deletion of genes encoding 60S subunit proteins, including Rpl40e, leads to lifespan extension in S. cerevisiae (Steffen et al., 2008, 2012). UBI1 deletion might also enhance longevity in C. neoformans, which would ultimately alter its survival and pathogenic patterns in the mammalian host.
In eukaryotes, ubiquitin is usually expressed as a hybrid protein with a fused ribosomal protein, such as Rpl40e or Rps31e (Clague and Urbé, 2010). The ubiquitin moiety could act as a molecular chaperone to the ribosomal proteins by facilitating their efficient production, folding, and ribosome assembly (Lacombe et al., 2009; Martín-Villanueva et al., 2019, 2020). Expression of ubi1Δub-HA, as the only source of Rpl40e, caused a severe slow-growth phenotype and aggregation of ribosomal proteins in S. cerevisiae (Martín-Villanueva et al., 2019). In the current study, however, partial reconstitution of the Rpl40e domain could rescue various abnormal phenotypes of the cryptococcal ubi1Δ strain, such as growth rate defect, cell ploidy variation, cell cycle arrest, and attenuated intracellular survival. Our work reveals the evolutionary convergence and divergence of the ubiquitin hybrid protein in different fungal species. The role of the ubiquitin moiety in the ribosome biogenesis, growth, and pathogenicity of C. neoformans requires further investigation.
In summary, we demonstrated that the ubiquitin hybrid protein is required for maintaining vegetative growth, morphological homeostasis, cell cycle progression, and pathogenicity in vivo of C. neoformans. The Rpl40e component could abrogate the effects of UBI1 deletion on cryptococcal growth and virulence, strongly supporting the importance of the ubiquitin hybrid protein in the ribosome biogenesis of C. neoformans.
The datasets generated in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Committee on Ethics of Biomedicine Research, Second Military Medical University.
JZ, YY, YF, JG, and WF conceived, designed, and performed the experiments. JY, CZ, ZG, and WF analyzed the data. JG, WL, and WF drafted the manuscript. All authors approved the final version of the manuscript.
This study was supported by the National Natural Science Foundation of China (81772159, 81501728, 81401651, and 81720108026), Shanghai Science and Technology of Committee (17DZ2270900), and Zhejiang Provincial Natural Science Foundation of China (LY20H110002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.570269/full#supplementary-material
Aldea, M., Jenkins, K., and Csikász-Nagy, A. (2017). Growth rate as a direct regulator of the start network to set cell size. Front. Cell Dev. Biol. 5:57. doi: 10.3389/fcell.2017.00057
Alspaugh, J. A. (2015). Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol. 78, 55–58. doi: 10.1016/j.fgb.2014.09.004
Arora, S., Olszewski, M. A., Tsang, T. M., McDonald, R. A., Toews, G. B., and Huffnagle, G. B. (2011). Effect of cytokine interplay on macrophage polarization during chronic pulmonary infection with Cryptococcus neoformans. Infect. Immun. 79, 1915–1926. doi: 10.1128/IAI.01270-10
Aslanyan, L., Ekhar, V. V., DeLeon-Rodriguez, C. M., and Martinez, L. R. (2017). Capsular specific IgM enhances complement-mediated phagocytosis and killing of Cryptococcus neoformans by methamphetamine-treated J774.16 macrophage-like cells. Int. Immunopharmacol. 49, 77–84. doi: 10.1016/j.intimp.2017.05.024
Casadevall, A., Coelho, C., and Alanio, A. (2018). Mechanisms of Cryptococcus neoformans-mediated host damage. Front. Immunol. 9:855. doi: 10.3389/fimmu.2018.00855
Cheng, Z., Mugler, C. F., Keskin, A., Hodapp, S., Chan, L. Y., Weis, K., et al. (2019). Small and large ribosomal subunit deficiencies lead to distinct gene expression signatures that reflect cellular growth rate. Mol. Cell 73, 36.e10–47.e10. doi: 10.1016/j.molcel.2018.10.032
Clague, M. J., and Urbé, S. (2010). Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685. doi: 10.1016/j.cell.2010.11.012
Dambuza, I. M., Drake, T., Chapuis, A., Zhou, X., Correia, J., Taylor-Smith, L., et al. (2018). The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog. 14:e1006978. doi: 10.1371/journal.ppat.1006978
Davis, M. J., Tsang, T. M., Qiu, Y., Dayrit, J. K., Freij, J. B., Huffnagle, G. B., et al. (2013). Macrophage M1/M2 polarization dynamically adapts to changes in cytokine microenvironments in Cryptococcus neoformans infection. mBio 4:e00264-13. doi: 10.1128/mBio.00264-13
Fang, W., Fa, Z., and Liao, W. (2015). Epidemiology of Cryptococcus and Cryptococcosis in China. Fungal Genet. Biol. 78, 7–15. doi: 10.1016/j.fgb.2014.10.017
Fang, W., Price, M. S., Toffaletti, D. L., Tenor, J., Betancourt-Quiroz, M., Price, J. L., et al. (2012). Pleiotropic effects of deubiquitinating enzyme Ubp5 on growth and pathogenesis of Cryptococcus neoformans. PLoS One 7:e38326. doi: 10.1371/journal.pone.0038326
Fernández-Pevida, A., Rodríguez-Galán, O., Díaz-Quintana, A., Kressler, D., and de la Cruz, J. (2012). Yeast ribosomal protein L40 assembles late into precursor 60 S ribosomes and is required for their cytoplasmic maturation. J. Biol. Chem. 287, 38390–38407. doi: 10.1074/jbc.M112.400564
Finley, D., Bartel, B., and Varshavsky, A. (1989). The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401. doi: 10.1038/338394a0
Geddes, J. M., Caza, M., Croll, D., Stoynov, N., Foster, L. J., and Kronstad, J. W. (2016). Analysis of the protein kinase A-Regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin-proteasome pathway in capsule formation. mBio 7:e01862-15. doi: 10.1128/mBio.01862-15
Gómez-Herreros, F., Rodríguez-Galán, O., Morillo-Huesca, M., Maya, D., Arista-Romero, M., de la Cruz, J., et al. (2013). Balanced production of ribosome components is required for proper G1/S transition in Saccharomyces cerevisiae. J. Biol. Chem. 288, 31689–31700. doi: 10.1074/jbc.M113.500488
Harper, J. W., and Schulman, B. A. (2006). Structural complexity in ubiquitin recognition. Cell 124, 1133–1136. doi: 10.1016/j.cell.2006.03.009
Ivanovska, I., and Rose, M. D. (2001). Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics 157, 503–518.
Kobayashi, M., Oshima, S., Maeyashiki, C., Nibe, Y., Otsubo, K., Matsuzawa, Y., et al. (2016). The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci. Rep. 6:36780. doi: 10.1038/srep36780
Kronstad, J., Saikia, S., Nielson, E. D., Kretschmer, M., Jung, W., Hu, G., et al. (2012). Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryotic Cell 11, 109–118. doi: 10.1128/ec.05273-11
Lacombe, T., Garcia-Gomez, J. J., de la Cruz, J., Roser, D., Hurt, E., and Linder, P. (2009). Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol. Microbiol. 72, 69–84. doi: 10.1111/j.1365-2958.2009.06622.x
Liu, T. B., Wang, Y., Stukes, S., Chen, Q., Casadevall, A., and Xue, C. (2011). The F-box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryotic Cell 10, 791–802. doi: 10.1128/ec.00004-11
Liu, T. B., and Xue, C. (2014). Fbp1-mediated ubiquitin-proteasome pathway controls Cryptococcus neoformans virulence by regulating fungal intracellular growth in macrophages. Infect. Immun. 82, 557–568. doi: 10.1128/IAI.00994-13
Martín-Villanueva, S., Fernández-Pevida, A., Fernández-Fernández, J., Kressler, D., and de la Cruz, J. (2020). Ubiquitin release from eL40 is required for cytoplasmic maturation and function of 60S ribosomal subunits in Saccharomyces cerevisiae. FEBS J. 287, 345–360. doi: 10.1111/febs.14999
Martín-Villanueva, S., Fernández-Pevida, A., Kressler, D., and de la Cruz, J. (2019). The ubiquitin moiety of Ubi1 is required for productive expression of ribosomal protein eL40 in Saccharomyces cerevisiae. Cells 8:850. doi: 10.3390/cells8080850
Maxson, M. E., Cook, E., Casadevall, A., and Zaragoza, O. (2007). The volume and hydration of the Cryptococcus neoformans polysaccharide capsule. Fungal Genet. Biol. 44, 180–186. doi: 10.1016/j.fgb.2006.07.010
May, R. C., Stone, N. R., Wiesner, D. L., Bicanic, T., and Nielsen, K. (2016). Cryptococcus: from environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 14, 106–117. doi: 10.1038/nrmicro.2015.6
Maziarz, E. K., and Perfect, J. R. (2016). Cryptococcosis. Infect. Dis. Clin. North Am. 30, 179–206. doi: 10.1016/j.idc.2015.10.006
Meng, Y., Zhang, C., Yi, J., Zhou, Z., Fa, Z., Zhao, J., et al. (2016). Deubiquitinase Ubp5 Is Required for the Growth and Pathogenicity of Cryptococcus gattii. PLoS One 11:e0153219. doi: 10.1371/journal.pone.0153219
O’Meara, T. R., and Alspaugh, J. A. (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408. doi: 10.1128/cmr.00001-12
Pitkin, J. W., Panaccione, D. G., and Walton, J. D. (1996). A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142(Pt 6), 1557–1565. doi: 10.1099/13500872-142-6-1557
Polymenis, M., and Aramayo, R. (2015). Translate to divide: control of the cell cycle by protein synthesis. Microb Cell 2, 94–104. doi: 10.15698/mic2015.04.198
Qiu, Y., Davis, M. J., Dayrit, J. K., Hadd, Z., Meister, D. L., Osterholzer, J. J., et al. (2012). Immune modulation mediated by cryptococcal laccase promotes pulmonary growth and brain dissemination of virulent Cryptococcus neoformans in mice. PLoS One 7:e47853. doi: 10.1371/journal.pone.0047853
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17, 873–881. doi: 10.1016/S1473-3099(17)30243-8
Reese, A. J., Yoneda, A., Breger, J. A., Beauvais, A., Liu, H., Griffith, C. L., et al. (2007). Loss of cell wall alpha(1-3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 63, 1385–1398. doi: 10.1111/j.1365-2958.2006.05551.x
Rittershaus, P. C., Kechichian, T. B., Allegood, J. C., Merrill, A. H., Hennig, M., Luberto, C., et al. (2006). Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 116, 1651–1659. doi: 10.1172/JCI27890
Seider, K., Heyken, A., Luttich, A., Miramon, P., and Hube, B. (2010). Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr. Opin. Microbiol. 13, 392–400. doi: 10.1016/j.mib.2010.05.001
Shirayama, M., Matsui, Y., and Toh-E, A. (1994). The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14, 7476–7482. doi: 10.1128/mcb.14.11.7476
Spitzer, E. D., and Spitzer, S. G. (1995). Structure of the ubiquitin-encoding genes of Cryptococcus neoformans. Gene 161, 113–117. doi: 10.1016/0378-1119(95)00231-t
Steffen, K. K., MacKay, V. L., Kerr, E. O., Tsuchiya, M., Hu, D., Fox, L. A., et al. (2008). Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302. doi: 10.1016/j.cell.2008.02.037
Steffen, K. K., McCormick, M. A., Pham, K. M., MacKay, V. L., Delaney, J. R., Murakami, C. J., et al. (2012). Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191, 107–118. doi: 10.1534/genetics.111.136549
Thapa, M., Bommakanti, A., Shamsuzzaman, M., Gregory, B., Samsel, L., Zengel, J. M., et al. (2013). Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes. Mol. Biol. Cell 24, 3620–3633. doi: 10.1091/mbc.E13-02-0097
Toffaletti, D. L., Rude, T. H., Johnston, S. A., Durack, D. T., and Perfect, J. R. (1993). Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175, 1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Trevijano-Contador, N., de Oliveira, H. C., García-Rodas, R., Rossi, S. A., Llorente, I., and Zaballos, Á (2018). Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 14:e1007007. doi: 10.1371/journal.ppat.1007007
Zaragoza, O. (2019). Basic principles of the virulence of Cryptococcus. Virulence 10, 490–501. doi: 10.1080/21505594.2019.1614383
Zaragoza, O., and Casadevall, A. (2004). Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 6, 10–15. doi: 10.1251/bpo68
Zaragoza, O., Garcia-Rodas, R., Nosanchuk, J. D., Cuenca-Estrella, M., Rodriguez-Tudela, J. L., and Casadevall, A. (2010). Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. doi: 10.1371/journal.ppat.1000945
Keywords: Cryptococcus neoformans, ubiquitin, growth restriction, cellular morphology, virulence, immune evasion
Citation: Zhao J, Yang Y, Fan Y, Yi J, Zhang C, Gu Z, Pan W, Gu J, Liao W and Fang W (2020) Ribosomal Protein L40e Fused With a Ubiquitin Moiety Is Essential for the Vegetative Growth, Morphological Homeostasis, Cell Cycle Progression, and Pathogenicity of Cryptococcus neoformans. Front. Microbiol. 11:570269. doi: 10.3389/fmicb.2020.570269
Received: 07 June 2020; Accepted: 20 August 2020;
Published: 05 November 2020.
Edited by:
Carolina Coelho, University of Exeter, United KingdomReviewed by:
Michael S. Price, Liberty University, United StatesCopyright © 2020 Zhao, Yang, Fan, Yi, Zhang, Gu, Pan, Gu, Liao and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julin Gu, d3VqZ2psQDEyNi5jb20=; Wanqing Liao, bGlhb3dhbnFpbmdAc29odS5jb20=; Wei Fang, d2VpZmFuZzA4MTc4MkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.