- 1College of Plant Protection, Henan Agricultural University, Zhengzhou, China
- 2State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
Fusarium wilt caused by Fusarium oxysporum f. sp. momordicae (FoM) is an important fungal disease that affects the production of bitter gourd. Hypovirulence-associated mycoviruses have great potential and application prospects for controlling the fungal disease. In this study, a novel ourmia-like virus, named Fusarium oxysporum ourmia-like virus 1 (FoOuLV1), was isolated from FoM strain HuN8. The viral genomic RNA is 2,712 nucleotides (nt) in length and contains an open reading frame (ORF) encoding a putative RNA-dependent RNA polymerase (RdRp) using either standard or mitochondrial codes. In strain HuN8, there was also a FoOuLV1-associated RNA segment with 1,173 nt in length with no sequence homology. Phylogenetic analysis showed that FoOuLV1 is a member of the genus Magoulivirus of the family Botourmiaviridae. FoOuLV1 was found to be associated with hypovirulence in FoM. Moreover, FoOuLV1 and its hypovirulence trait can be transmitted horizontally to other FoM strains and also to other formae speciale strains of F. oxysporum. In addition, FoOuLV1 showed significant biological control effect against the bitter gourd Fusarium wilt. To our knowledge, this study reveals the first description of a hypovirulence-associated ourmia-like mycovirus, which has the potential to the biological control of Fusarium wilt.
Introduction
Mycoviruses are widespread in all major filamentous fungi, yeasts, and oomycetes (Xie and Jiang, 2014; Ghabrial et al., 2015). According to a recent study, more than 300 mycoviral sequences have been logged in the National Center for Biotechnology Information (NCBI) database (Zhou et al., 2020), which are grouped into at least 19 families by the International Committee on Taxonomy of Viruses (ICTV). Most mycoviruses have double-stranded RNA (dsRNA); however, some mycoviruses have positive single-stranded RNA (+ssRNA), and a few mycoviruses have single-stranded DNA (ssDNA) or negative single-stranded RNA (−ssRNA; Ghabrial and Suzuki, 2009; Xie and Jiang, 2014; Ghabrial et al., 2015). The ssRNA mycoviruses are grouped into seven families: Alphaflexiviridae, Barnaviridae, Botourmiaviridae, Gammaflexiviridae, Hypoviridae, Mymonaviridae, and Narnaviridae (Amarasinghe et al., 2018). There are also unclassified mycoviruses (Li H. et al., 2019).
Ourmiavirus is a genus of viruses isolated from various plants. The genome of Ourmiavirus contains three linear + ssRNAs, each encoding a single protein: RNA-dependent RNA polymerase (RdRp), movement protein (MP), or capsid protein (CP; Lisa et al., 1988; Turina et al., 2017). The Ourmiavirus has a unique bacilliform virion structure (Avgelis et al., 1989). Viruses in the family Narnaviridae have a 2.5- to 3.0-kb genome that encodes only an RdRp with no capsid; they have been widely discovered in fungi, oomycetes, invertebrates, and plants (Cai et al., 2012; Bruenn et al., 2015; Shi et al., 2016). Recently, many mycoviruses containing only one RNA segment encoding RdRp have been identified to be phylogenetically related to Ourmiavirus genus (Xu et al., 2015; Shi et al., 2016; Amarasinghe et al., 2018; Zheng et al., 2019; Li C.X. et al., 2019). Therefore, the ICTV created a new family (Botourmiaviridae) to incorporate these ourmia-like viruses (Turina et al., 2017; Wang et al., 2020). Botourmiaviridae comprising five genera: Magoulivirus, Ourmiavirus, Botoulivirus, Scleroulivirus, and Penoulivirus (Zhou et al., 2020). Whether these ourmia-like mycoviruses have other RNA segments remains unknown (Wang et al., 2020).
The mycoviruses infection usually causes no associated symptoms and sometimes even has beneficial effects on their fungal hosts (Zheng et al., 2019). Some mycoviruses in plant pathogenic fungi can reduce the ability of their fungal hosts to cause disease in plants. This property, named hypovirulence, has explored for biological control of crop fungal diseases. (Nuss, 2005; Xie and Jiang, 2014). Cryphonectria hypovirus 1 (CHV1) and Sclerotiorum sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1) have been successfully used to control diseases caused by Cryphonectria parasitica and Sclerotiorum sclerotiorum (Anagnostakis, 1982; Yu et al., 2013). Besides these examples, many hypovirulence-associated mycoviruses, belonging to various genera, have been identified from diverse plant pathogenic fungi, such as Hubei sclerotinia RNA virus 1 (HuSRV1), Botryosphaeria dothidea botourmiavirus 1 (BdBOV-1), Alternaria alternata hypovirus 1 (AaHV1), and Rhizoctonia solani endornavirus 1 (RsEV1), etc. (Xie et al., 2011; Azhar et al., 2019; Zheng et al., 2019; Li H. et al., 2019). However, to our knowledge, no hypovirulence-associated ourmia-like mycovirus has been reported so far.
Fusarium is widely distributed in soil, associated with plants worldwide. It includes many important plant pathogenic fungi (Li P. et al., 2019). Many mycoviruses have been reported in different species of Fusarium genus, and a few of them have hypovirulent effects on their hosts, including: Fusarium graminearum virus 1 (FgV1), Fusarium graminearum virus-ch9 (FgV-ch9), Fusarium graminearum hypovirus 2 (FgHV2), and so on (Kwon et al., 2007; Darissa et al., 2012; Li et al., 2015). F. oxysporum is an important plant pathogenic fungus and causes vascular wilt in a wide variety of agricultural crop species. F. oxysporum is classified into different host-specific forms (formae speciales) based on the types of host plants (Di Pietro et al., 2003). Although a number of mycoviruses have been identified in the genus Fusarium, only three mycoviruses have been reported in F. oxysporum. Fusarium oxysporum chrysovirus 1 (FoCV1) was found in F. oxysporum f. sp. melonis and assigned to the family Chrysoviridae, but its complete genomic sequence has not been determined (Sharzei et al., 2007). Fusarium oxysporum f. sp. dianthi mycovirus 1 (FodV1), a new member of the family Chrysoviridae, has been isolated from F. oxysporum f. sp. dianthi and exerts a hypovirulent effect (Lemus-Minor et al., 2015). In addition, Fusarium oxysporum f. sp. dianthi mitovirus 1 (FodMV1) has been identified from F. oxysporum f. sp. dianthi, but it has no hypovirulence trait (Torres-Trenas and Pérez-Artés, 2020).
In this study, we identified and characterized a novel hypovirulence-inducing ourmia-like mycovirus from F. oxysporum f. sp. momordicae named Fusarium oxysporum ourmia-like virus 1 (FoOuLV1) and verified its hypovirulence trait and its horizontal transmission ability. In addition, we evaluated the biocontrol potential of FoOuLV1 by the pot and field experiments.
Materials and Methods
Fungal Isolates and Plant Materials
Fusarium oxysporum f. sp. momordicae strains HuN8 and SD-1 were isolated from bitter gourd, showing symptoms of Fusarium wilt disease in Hunan and Shandong Province, China, in 2017. F. oxysporum f. sp. cucumerinum (FoC) strain HK3 was kindly gifted by Dr. Xuehong Wu (China Agricultural University, Beijing, China). The strains were cultured on potato dextrose agar (PDA) medium at 28°C. Fungal DNA was isolated using standard phenol-chloroform extraction and ethanol precipitation, then used for polymerase chain reaction (PCR) amplification with universal primers (ITS1 and ITS4) and specific primers (fp7318 and fp7335) to perform formae speciales identification of F. oxysporum (van Dam et al., 2018). Bitter gourd seeds were purchased from Fujian Agricultural Science Agricultural Seed Development Co. Ltd. All primers used in this manuscript are listed in Supplementary Table 1.
RNA Extraction and RT-PCR Detection
The extraction of ssRNA and dsRNA was performed following the procedure described previously (Xie et al., 2006). Strains grew for 4–5 days on cellophane membranes of the PDA medium. Fresh mycelia (1–2 g) were harvested to isolate dsRNA. 2 × GPS (glycine 15.0 g/L, Na2HPO4 14.2 g/L, NaCl 35.1 g/L, and pH = 9.6) 400 μl, phenol (pH 8.0) 400 μl, chloroform-isoamyl alcohol (24:1) 400 μl, and 10% SDS 92 μl were added to a sterile centrifuge tube each 0.2 g sample and were shaken for 10 min at room temperature, then centrifuged at 12,000 revolutions per minute (rpm) for 10 min. The supernatant was transferred to another clean centrifuge tube and mixed with 114 μl anhydrous ethanol for every 600 μl of the supernatant. Then 0.04 g cellulose powder CF-11 (Sigma–Aldrich, St. Louis, MO, United States) was added. The supernatant was shaken for 30 min in an ice bath. After centrifugation at 12,000 rpm for 1 min, the supernatant was discarded, 600 μl of eluent buffer [10 × STE (0.5 M Tris; 1 M NaCl, 10 mM EDTA, and pH = 7.0) 10 ml, 95% ethanol 17 ml, DEPC-H2O 73 ml] was added to elute for two to three times. After centrifugation at 12,000 rpm for 2 min, the supernatant was discarded and added with 600 μl 1 × STE, mixed and shaken for 5 min, and centrifuged at 12,000 rpm for 5 min. Then the supernatant was transferred to a new centrifuge tube, and the same volume isopropyl alcohol, with acryl carrier nucleic acid co-precipitator (item No. RP2001, ®Bioteke Corporation, Beijing, China) were added to improve the yield of dsRNA. After centrifugation at 12,000 rpm at 4°C for 25 min, the precipitation was washed with 75% ethanol, dried at 37°C for 10 min, and dissolved in 25 μl DEPC-H2O. The dsRNA was treated with RNase-free DNase I (0.6 U/μl) and S1 nuclease (3 U/μl). The segments were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and visualized with gel documentation and image analysis system (InGenius LhR, Syngene, United Kingdom).
To extract total RNA, strains were grown on the cellophane membrane overlying a PDA plate for 4–5 days, and 100–200 mg fresh mycelia were harvested and ground to powder in liquid nitrogen. Total RNA was prepared using an RNA reagent (Newbio Industry, Wuhan, China), according to the manufacturer’s instructions.
For RT-PCR detection, first-strand cDNAs were synthesized using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (®TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. Then the PCR amplification was carried out using the specific primers (FV-L-S and FV-L-A for L-segment; FV-S-S and FV-S-A for S-segment). All primers are listed in Supplementary Table 1.
cDNA Cloning and Sequencing
The cDNA library was constructed using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech) according to the manufacturer’s instructions.
To obtain initial sequence clones, a random primer (RACE3RT) was used for RT-PCR amplification. The PCR products were cloned into a pMD18-T vector (Takara, Dalian, China) for Sanger sequencing, with the internal gaps between the initial sequences filled by RT-PCR. The terminal sequence cloning was performed according to the method previously described (Potgieter et al., 2009). All primers used for cDNA cloning and sequencing are listed in Supplementary Table 1.
Sequence and Phylogenetic Analysis
Open reading frames (ORFs) and conserved domains were predicted using ORF finder and CD-search on the website of NCBI1 and motifs scan website2. Multiple sequence alignments were performed, and the phylogenetic tree was constructed using MEGA7 software (Kumar et al., 2016). Potential secondary structures were predicted by Mfold version 2.3 (Zuker, 2003).
Viral Transmission Assay
Hyphal fusion was conducted to investigate viral horizontal transmission between Fusarium oxysporum f. sp. momordicae (FoM) strains and FoC strains, following the procedure described previously (Li H. et al., 2019). The SD-1HygR and HK3 HygR strains which obtained hygromycin resistance by agrobacterium-mediated transformation from the strains SD-1 and HK3. The agrobacterium-mediated transformation was following the procedure described previously (Mullins et al., 2001). The agrobacterium (EHA105) and the plasmid (pTFCM) were kindly gifted by Dr. Daohong Jiang (Huazhong Agricultural University, Wuhan, China), which were described previously (Li et al., 2005). The biological phenotype of strains SD-1HygR and HK3 HygR were the same as the strains SD-1 and HK3.
A schematic diagram of the co-culturing performed using the viral transmission assay is shown in Supplementary Figure 1. Strains HuN8, SD-1HygR, and HK3HygR were grown for 4–5 days on PDA medium. Then, mycelia blocks of strains HuN8 and the SD-1HygR (or HK3HygR) were inoculated on the same PDA plate, growing for 4–5 days until the colony of HuN8 and the SD-1HygR (or HK3HygR) covered each other. Mycelia blocks of the common colony were transferred to new PDA plates with hygromycin (50 mg/ml), growing for 2–3 days. The mycelia could grow on the hygromycin-resistant PDA plates and became the derivative strains. The viral transmission was evaluated based on dsRNA extraction or RT-PCR detection.
Virulence Assay of in vitro Inoculation and Pot and Field Experiments
The virulence of FoM strains was assessed by in vitro inoculation and the pot and field experiments on bitter gourd plants. For in vitro experiment, young leaves and petioles of bitter gourd were inoculated with FoM strains, moisturized for 4–5 days, observed, and photographed. The leaves inoculated with nothing were used as control. Four leaves were used for a group in each experiment, and all the experiments were repeated three times.
For the pot experiment, bitter gourd seedlings with two or three leaves grown in sterile soil were inoculated with 10 ml of F. oxysporum spore (107 ml–1) suspension into the root, cultivated in the growth chamber, observed, and photographed. The plants inoculated with water were used as control. Four plants were used for a group in each experiment, and all the experiments were repeated three times.
The virulence assay of FoC strains was similar with the FoM strains. Cucumber seedlings with two leaves grown in sterile soil were inoculated with 10 ml of F. oxysporum spores (107 ml–1) suspension, cultivated, observed, and photographed. Plants inoculated with water were used as control. Four plants were used for a group in each experiment, and all the experiments were repeated three times.
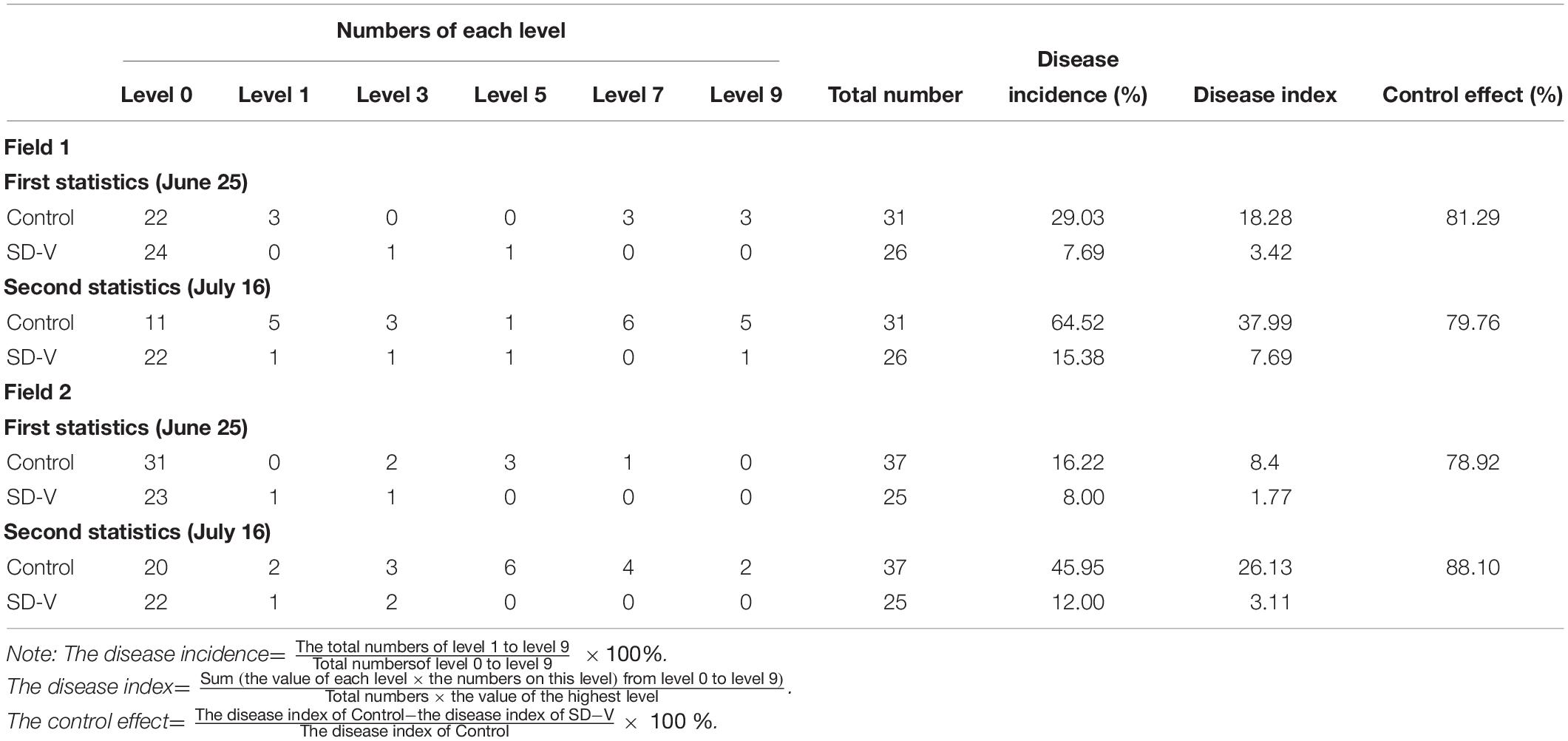
The field experiment was conducted from 21 May 2019 to 16 July 2019 in two fields where Fusarium wilt had been present for many years in Yuanyang, Henan Province, China. The bitter gourd seedlings (about 30 seedlings for each group in each field) were planted according to the experimental design (a schematic representation of the experiment distribution is shown in Supplementary Figure 2). In the treatment group, 10 ml of spores (107 ml–1) of the SD-V strains were inoculated into the area surrounding the roots on 21 May 2019; at the same time, the control group was instead inoculated with water. The disease incidence, severity, and index were assessed twice, on 25 June and on 16 July. Disease incidence was defined as the percentage of infected plants, and disease severity was rated on a scale of 0–9 as follows: level 0, no symptoms; level 1, <30% of leaves showing leaf veins with yellowing; level 3, 30–70% of leaves showing leaf veins with yellowing; level 5, leaf veins on the whole plant are yellowed, but the growth and development of the plant are not affected; level 7, the whole plant is yellowed and wilted, and the vascular bundle has turned brown and stopped growing and developing; and level 9, the plant is dead. The different levels of symptoms are shown in Supplementary Figure 3.
Results
Viruses in F. oxysporum f. sp. momordicae Strain HuN8
Total RNA was extracted from the FoM strain to perform NGS. The assembled sequences were used for homology searches against the NCBI virus amino acid sequence database using BLASTX. Eight contigs could be assembled into one long contig from the database. The long contig was similar to the sequence of Magnaporthe oryzae ourmia-like virus (SBQ28480.1), thus was named FoOuLV1.
Based on the RT-PCR analysis, we found that FoOuLV1 was harbored in FoM strain HuN8, which was isolated from the stem of a diseased bitter gourd plant collected in Hunan Province, China. Strain HuN8 displayed a similar colony morphology to strain SD-1 (virus free) on PDA medium (Figure 1A) and induced slight leaf yellowing, while the virus-free strain SD-1 induced obvious leaf yellowing (Figure 1B). The dsRNA was extracted from strain HuN8 and treated with DNase I to digest the genomic DNA. We found that two segments, the L-segment and the S-segment, were harbored in strain HuN8 (Figure 1C).
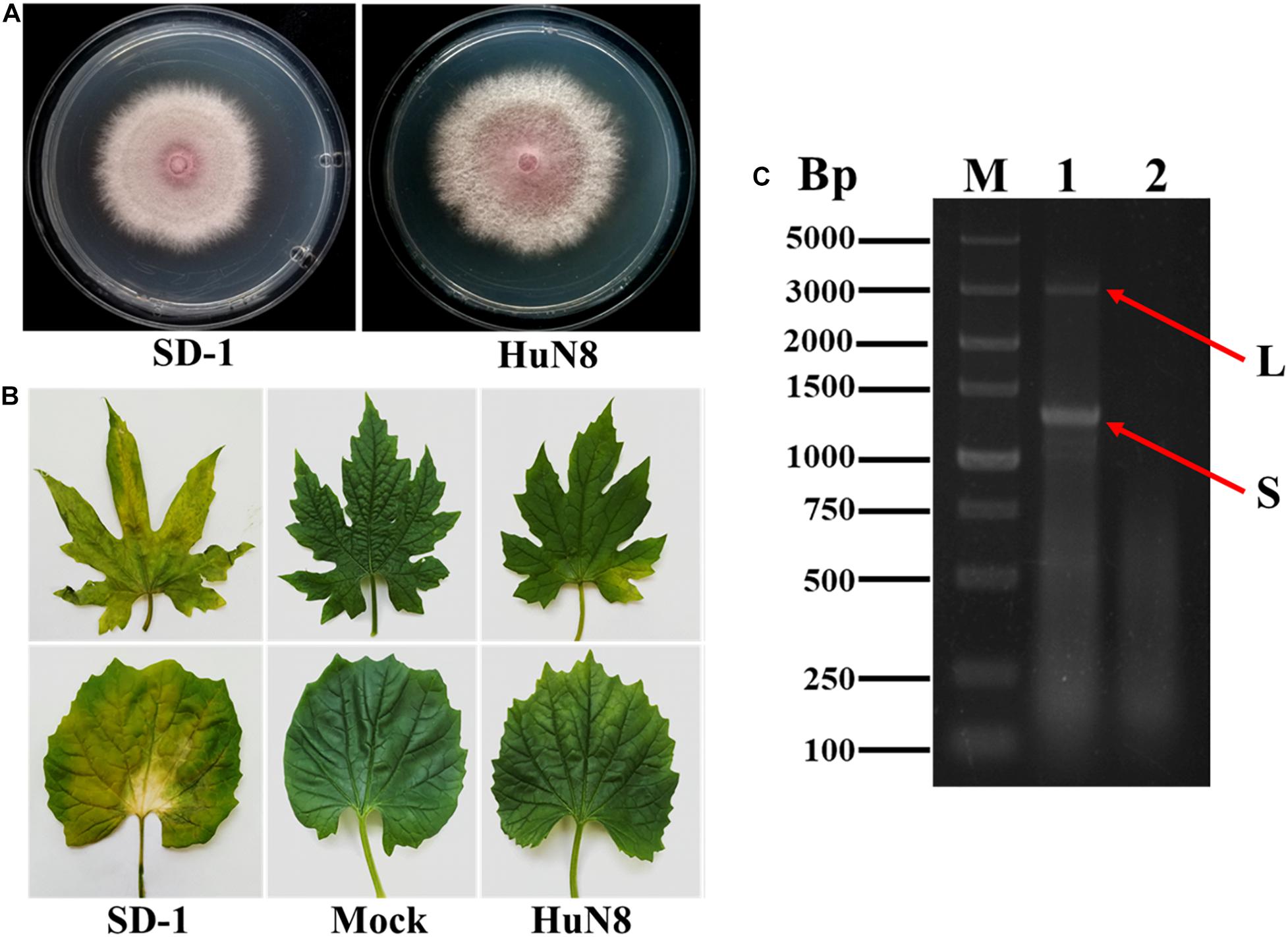
Figure 1. The biological characteristics and dsRNA pattern of Fusarium oxysporum f. sp. momordicae strain HuN8. (A) Colony morphology of strain HuN8 and virulent strain SD-1 (cultured on PDA for 2 days at 28°C). (B) Pathogenicity of strain HuN8 and SD-1 on the detached bitter gourd leaves (72 h post-inoculation at 28°C). Mock, blank control. (C) Agarose gel electrophoretic analysis of the dsRNA extracted from strain HuN8. The dsRNA was treated with DNase I. M, molecular weight marker; 1, dsRNA extracted from strain HuN8; and 2, dsRNA extracted from strain SD-1.
Molecular Characterization of FoOuLV1
The full-length sequence of the L-segment (GenBank Accession No. MT551010) in FoOuLV1 was 2,712 nt with a GC content of 55.5%. The genome contained only one large ORF that initiated at position 56 and terminated at position 2,612 based on the universal or mitochondrial genetic codes, potentially encoding 701 amino acid residues with a calculated molecular mass of 78.9 kDa from an AUG triplet to a UAG triplet (Figure 2A). Using the Mfold RNA structure software, the complex secondary structures of the L-segment of FoOuLV1 were predicted. The results indicated that the first 50 nt at the 5′ terminus were folded into two stable stem–loop structures, while the last 32 nt at the 3′ terminus formed a stable stem–loop structure (Figure 2B). Stem–loop structures are typical features of members of Narnaviridae (including mitovirus) and Ourmiavirus.
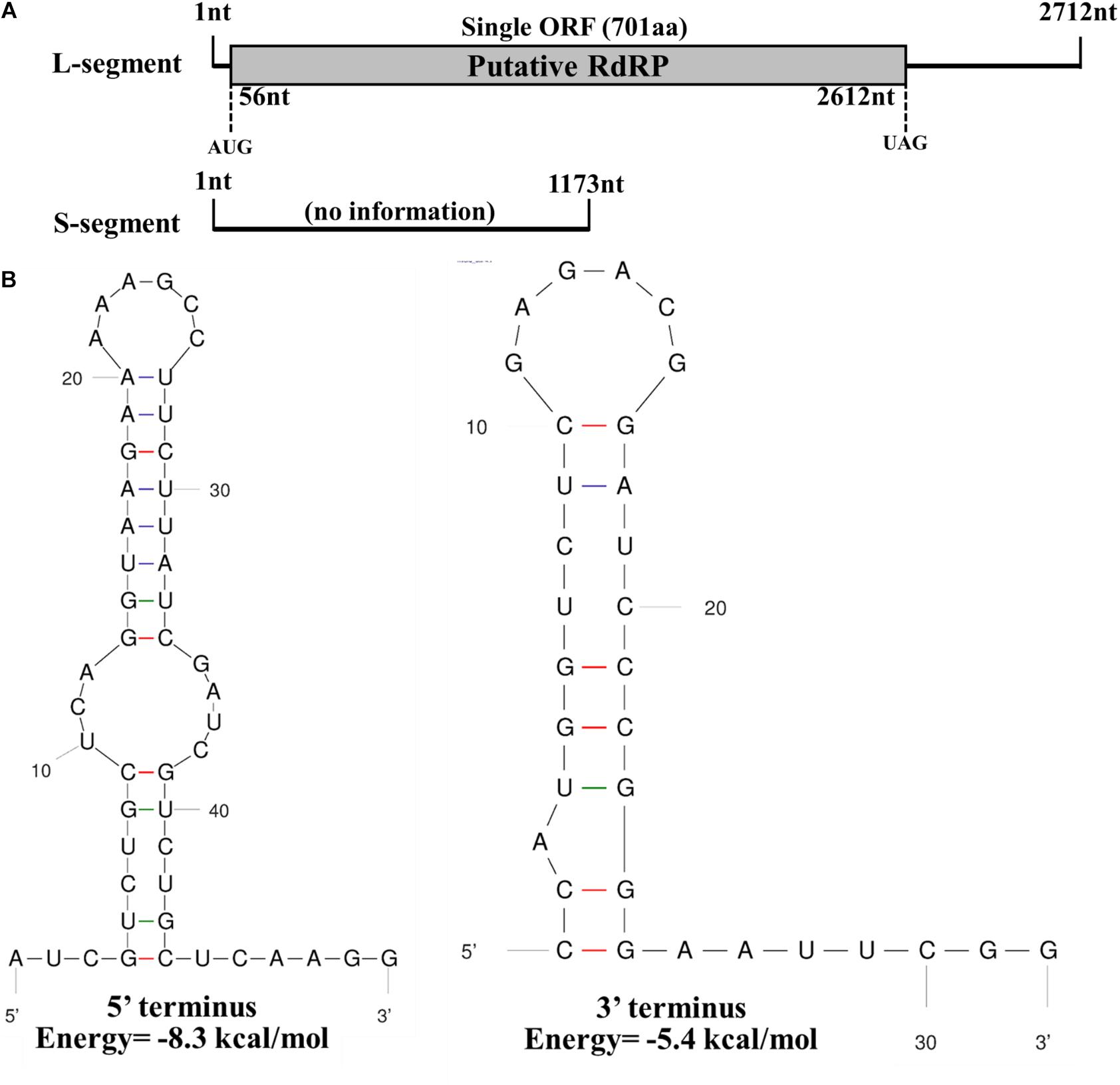
Figure 2. Genomic organization and terminal structure of Fusarium oxysporum ourmia-like virus 1 (FoOuLV1). (A) Schematic representation of the putative genomic organization of FoOuLV1. The open reading frame (ORF) shown as a gray box (56–2,162 nt) was putatively encoding the RNA-dependent-RNA polymerase (RdRp) domain. (B) Potential secondary structures of the 5′- (left) and 3′-termini (right) of the L fragment of FoOuLV1.
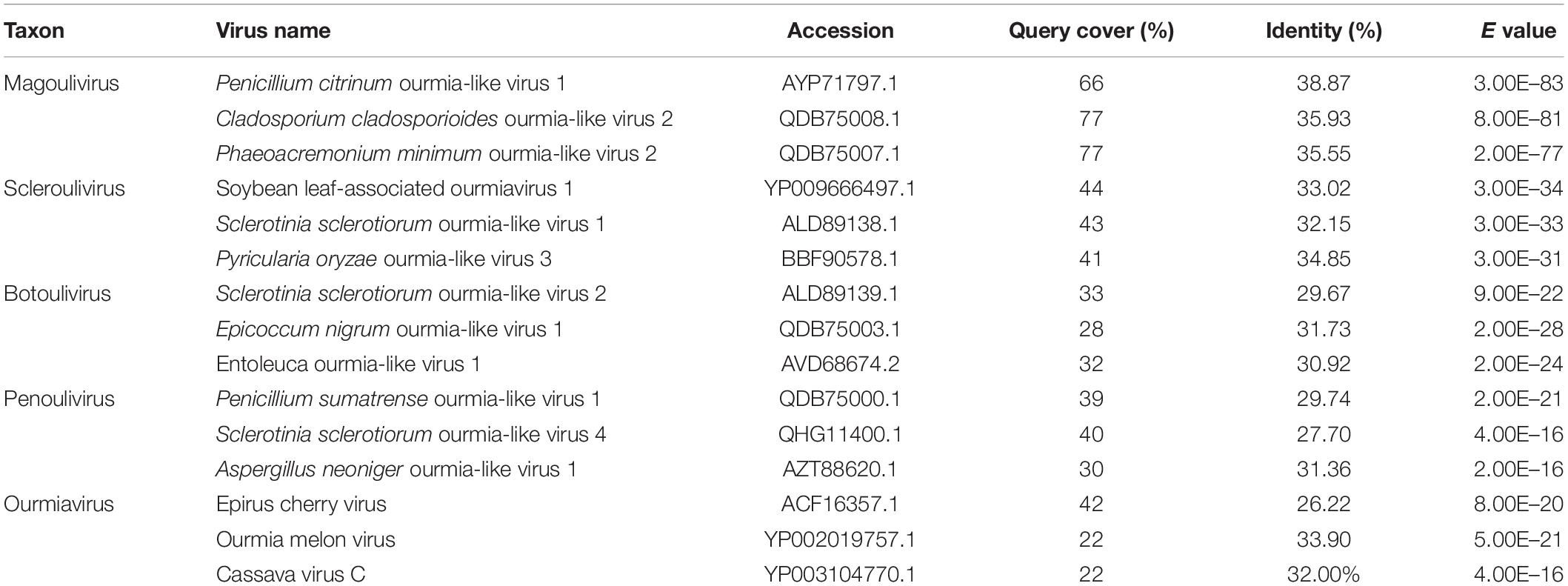
Using the homology search on BLASTP, the ORF encoded a protein with one conserved domain, which was closely related to RdRps of Penicillium citrinum ourmia-like virus 1, Cladosporium cladosporioides ourmia-like virus 2, and Phaeoacremonium minimum ourmia-like virus 2 (Table 1). Furthermore, a conserved domain database (CDD) search and multiple protein alignment suggested that the predicted RdRp domain contained seven conserved motifs (Figure 3).

Figure 3. Multiple alignment of the amino acid sequences of RNA-dependent RNA polymerase (RdRp) proteins encoded by FoOuLV1 and other selected ourmia-like viruses. Horizontal lines above the alignment indicate seven motifs (Roman numerals I to VII). Abbreviations: CcOLV1, Cladosporium cladosporioides ourmia-like virus 1; RsOLV1, Rhizoctonia solani ourmia-like virus 1; PcOLV1, Penicillium citrinum ourmia-like virus 1; AsOLV1, Acremonium sclerotigenum ourmia-like virus 1; MoOLV, Magnaporthe oryzae ourmia-like virus; CcOLV2, Cladosporium cladosporioides ourmia-like virus 1; and PmOLV2, Phaeoacremonium minimum ourmia-like virus 2.
The full-length sequence of the S-segment (GenBank Accession No. MT551011) associated with FoOuLV1 was composed of 1,173 nt with a GC content of 43.8%, and the segment contained no ORFs (Figure 2A). There was no significant similar information from NCBI database using BLASTX.
Phylogenetic Analysis of FoOuLV1
To examine the phylogenetic relationship between FoOuLV1 and other mycoviruses, the phylogenetic tree was constructed using a maximum likelihood method based on the RdRp amino acid sequences of FoOuLV1 and other related viruses from Narnaviridae (including Narnarvirus, Chrysovirus, Tetramycovirus, and Mitovirus) and Botourmiaviridae (including Penoulivirus, Botoulivirus, Scleroulivirus, Magoulivirus, and Ourmiavirus). The results indicated that FoOuLV1 was closely related to the genus Magoulivirus and clustered with viruses such as Cladosporium cladosporioides ourmia-like virus, Phaeoacremonium minimum ourmia-like virus 2, and Magnaporthe oryzae ourmia-like virus. Thus, FoOuLV1 was determined to be a new member of Magoulivirus within the family Botourmiaviridae (Figure 4).
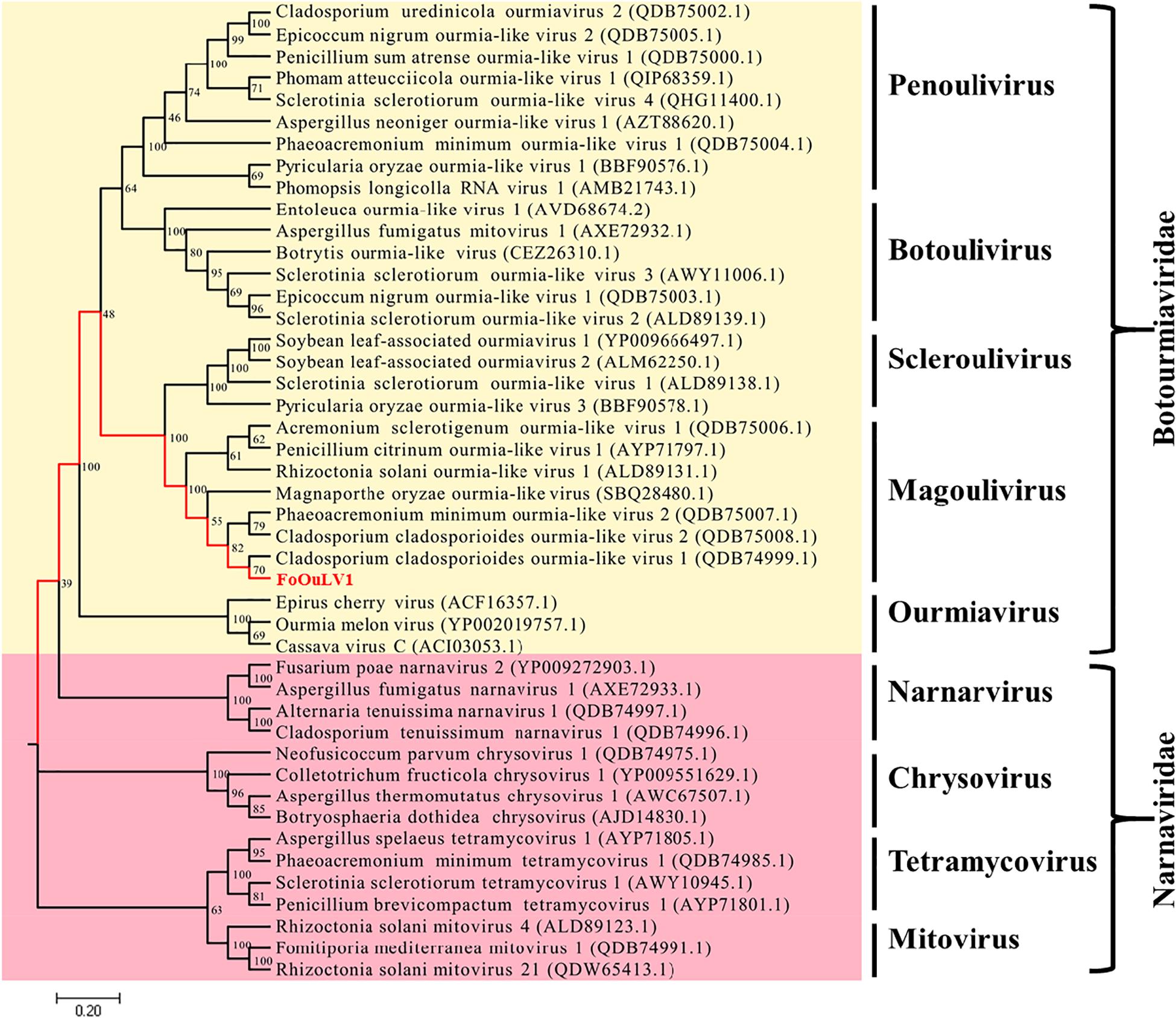
Figure 4. Maximum likelihood phylogenetic tree based on the RdRps of FoOuLV1 and other related viruses. The phylogenetic analysis was generated using the MEGA7 software with 1,000 bootstrap replicates. The scale bar represents a genetic distance of 0.2.
Influence of FoOuLV1 to the Virulence of FoM
Pathogenicity detection of FoOuLV1-containing strain HuN8 and virus-free strain SD-1 on the detached bitter gourd leaves showed that FoOuLV1 reduced the virulence of the FoM strain as described above (Figure 1B). Fusarium wilt caused by F. oxysporum is a systemic infection disease characterized by a typical symptom of whole plant wilting. In order to further determine whether the presence of FoOuLV1 reduced the virulence of FoM strain in terms of infecting living bitter gourd plants, the FoM strains HuN8 and SD-1 were inoculated onto bitter gourd seedlings at the stage of two or three leaves. Pathogenicity data were investigated before, during, and after the presentation of typical wilt symptoms.
At the early stage (15 dpi), the basal leaves of bitter gourd inoculated with SD-1 strain showed the symptoms of vein fading and yellowing on leaves, while the leaves of bitter gourd inoculated with HuN8 strain showed no symptoms (Figures 5A,D). At the middle stage (19 dpi), most of the leaves of bitter gourd inoculated with SD-1 strain had shown the typical symptoms of Fusarium wilt (leaf vein fading and yellowing), while those leaves inoculated with HuN8 strain still showed no symptoms (Figures 5B,E). At the later stage (23 dpi), the whole bitter gourd plant inoculated with SD-1 strain exhibited dryness and wilting, while the bitter gourd inoculated with HuN8 strain remained no symptoms during the entire observation period (Figures 5C,F).
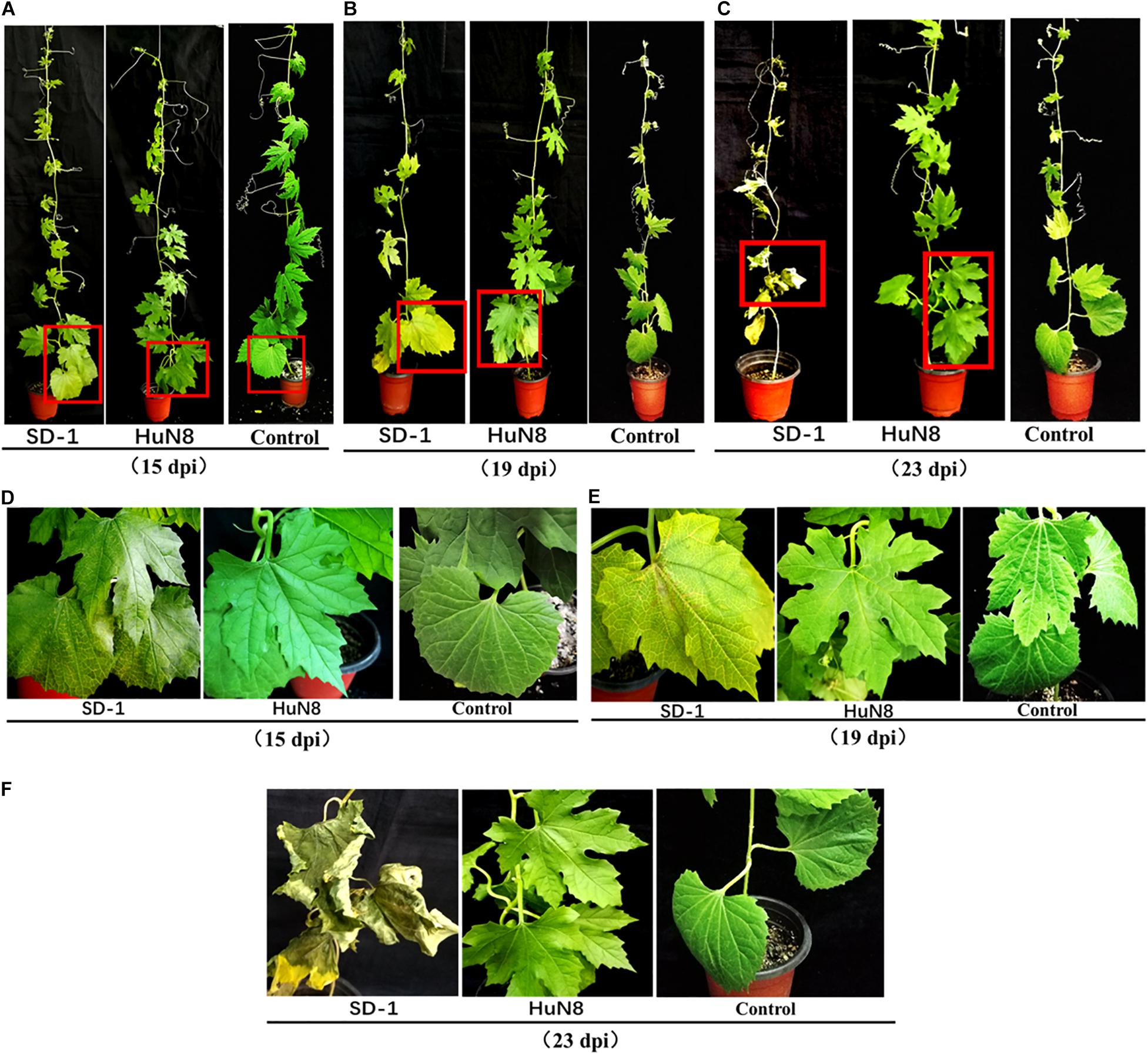
Figure 5. Pathogenicity detection of strains HuN8 and SD-1 in living bitter gourd seedlings. (A–C) Symptoms at the early (15 dpi), middle (19 dpi), and later (23 dpi) stages. (D–F) The enlarged images of the red boxes in (A–C), respectively.
Horizontal Transmission of FoOuLV1 Among FoM Strains
The dual-culture technique was used to determine whether the hypovirulence traits and RNA elements of FoOuLV1 could be transmitted horizontally. The results indicated that the L- and S-elements were both successfully transmitted from the hypovirulent strain HuN8 to the mycovirus-free strain SD-1HygR, yielding the derivative strain SD-V. The colony morphology of strain SD-V was similar to that of the strain SD-1 (Figure 6A). The presence of L- and S-segments was confirmed in strain SD-V by dsRNA extraction (Supplementary Figure 4A) and RT-PCR with specific primers (Figure 6B). The hypovirulence traits were also successfully transmitted, as indicated by the pathogenicity detection results: the whole bitter gourd plants inoculated with strain SD-1 exhibited dryness and wilting, while plants inoculated with strain SD-V showed no symptoms, appearing in the same way as the strain HuN8 (Figure 6C).
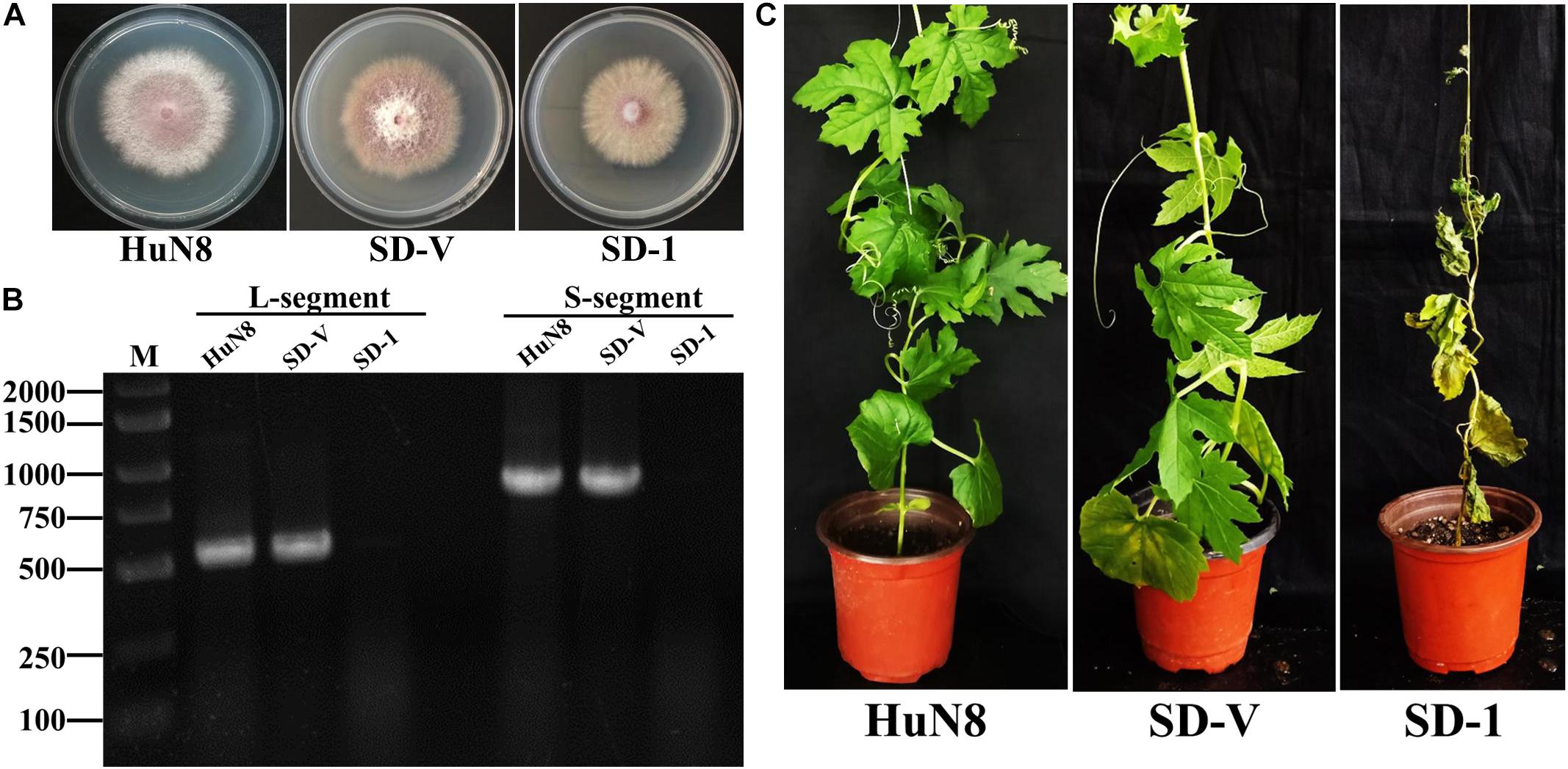
Figure 6. FoOuLV1 infectivity and pathogenicity in FoM strains. (A) Colony morphology of the HuN8 strain, the virulent strain SD-1, and the derivative strain SD-V (cultured on PDA for 2 days at 28°C). (B) Detection of the L- and S-segments by RT-PCR. M, DNA marker. (C) Pathogenicity detection in a living bitter gourd seedling.
Horizontal Transmission of FoOuLV1 Between Different Special Form Strains of F. oxysporum
The specialization of F. oxysporum is closely related to its pathogenicity. In order to determine whether the hypovirulent traits could be transmitted from FoM strain to FoC strain, dual culture was performed. The results showed the L- and S-segments were successfully transmitted from FoM strain HuN8 to virus-free FoC strain HK3HygR, yielding the derivative strains HK3-V-1 and HK3-V-2. The L- and S-segments were detected by dsRNA extraction (Supplementary Figure 4B) and RT-PCR (Figure 7A). The hypovirulent traits were also successfully transmitted: whole cucumber plants inoculated with the strain HK3 exhibited dryness and wilting, while plants inoculated with strains HK3-V-1 and HK3-V-2 showed no symptoms (Figure 7B).
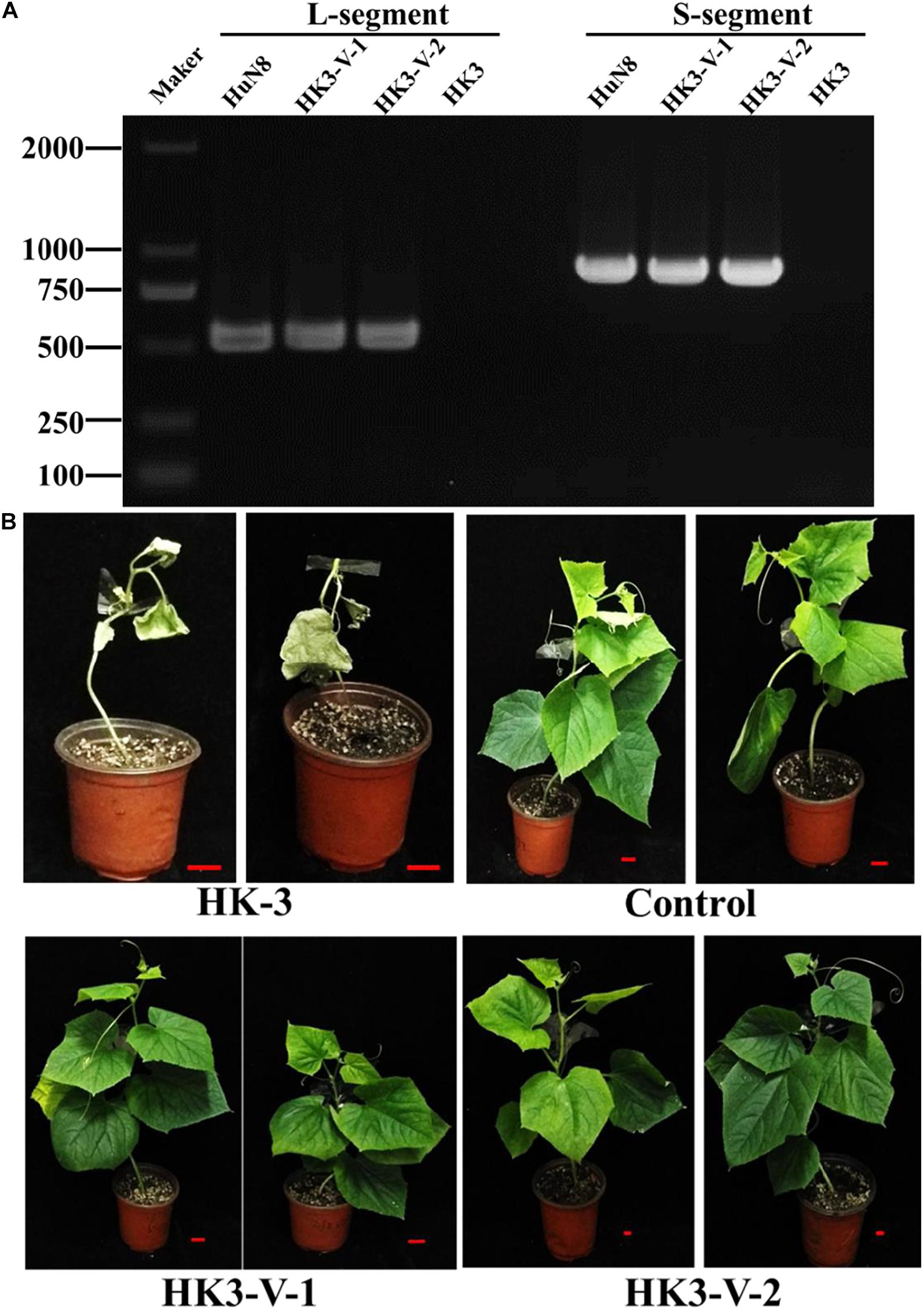
Figure 7. FoOuLV1 infectivity and pathogenicity in FoC strains. (A) Detection of L- and S-segments by RT-PCR. (B) Pathogenicity detection in living cucumber seedlings. HuN8 is the FoM strain (with FoOuLV1), HK3 is the mycovirus-free FoC strain, HK3-V-1, and HK3-V-2 are the FoC derivative strains obtained from the dual-culture technique with strains HuN8 and HK3.
Biological Control Effects of FoOuLV1 Against Fusarium Wilt in Bitter Gourd
To verify the biological control effect of FoOuLV1 against Fusarium wilt in bitter gourd plants, field experiment was conducted from 21 May to 16 July 2019 in two fields. The results are shown in Table 2. According to the first results obtained from Field 1 on 25th June, the disease incidence of bitter gourd Fusarium wilt in the control group was 29.03% and the disease index was 18.28. In contrast, in the mycovirus FoOuLV1-treated group, the disease incidence was only 7.69%, the disease index was only 3.42, and the control effect reached 81.29%. A similar result was observed in Field 2, where the control effect reached 78.92%. The second results collected on 16th July showed a similar control effect (79.76% in Field 1 and 88.10% in Field 2; Table 1).
Discussion
Genomic Differences Between FoOuLV1 and Other Ourmia-Like Viruses
Recently, an increasing number of ourmia-like viruses have been reported in diverse fungi, including two ourmia-like viruses from Magnaporthe oryzae (Illana et al., 2017; Li C.X. et al., 2019), three ourmia viruses from Pyricularia oryzae (Ohkita et al., 2019), two ourmia viruses from Sclerotinia sclerotiorum (Marzano et al., 2016; Wang et al., 2020), and other ourmia viruses from Botrytis (Donaire et al., 2016), Phomopsis longicolla (Hrabáková et al., 2017), and Phoma matteucciicola (Zhou et al., 2020). All of them are phylogenetically related to those in the newly established family Botourmiaviridae, which currently comprised five genera, according to current taxonomic information from ICTV and research report. Members of the genus Ourmiavirus are found only in plant-infecting viruses and usually harbor three RNA segments encoding RdRp, MP, and CP protein. Viruses in the genera Botoulivirus, Penoulivirus, and Magoulivirus are only isolated from fungi, while Scleroulivirus viruses are isolated from both fungi and plants. The mycoviruses in these four genera contain only one RNA segment encoding RdRp (Crivelli et al., 2011; Wang et al., 2020).
In this study, we described a novel ourmia-like mycovirus FoOuLV1, which is identified in F. oxysporum for the first time and a new member of the genus Magoulivirus within the family Botourmiaviridae. The genome of FoOuLV1 only has one ORF encoding RdRp in the L-segment similar to the other ourmia-like mycoviruses (Guo et al., 2019; Wang et al., 2020; Zhou et al., 2020). In addition, it contains a shorter segment named S-segment, which has no blast information. In this regard, the FoOuLV1 is significantly different from other ourmia-like mycoviruses.
The FoOuLV1 Is Unique Compared With Other Ourmia-Like Mycoviruses
Some mycoviruses in pathogenic fungi can cause a decline in the pathogenicity of the host fungi. This is known as mycovirus-mediated hypovirulence and has attracted research attention, owing to its importance in controlling fungal diseases (Nuss, 2005; Suzuki et al., 2018). To date, many families of mycoviruses have been identified as being associated with fungal hypovirulence, including the following: Hypoviridae (e.g., CHV1), Megabirnaviridae [e.g., Rosellinia necatrix megabirnavirus 1 (RnMBV1)], Reoviridae [e.g., Rosellinia necatrix mycoreovirus 3 (RnMYRV-3)], and Narnaviridae [e.g., Sclerotinia sclerotiorum mitovirus 1 (SsMV1)] (Hillman et al., 1990; Kanematsu et al., 2004; Sasaki et al., 2007; Chiba et al., 2009; Xu et al., 2015). In addition, some unclassified hypoviruses (such as SsHADV-1 and Sclerotinia sclerotiorum hypovirus 1) have been recognized as inducing hypovirulence (Yu et al., 2010; Xie et al., 2011).
A large number of ourmia-like viruses have been reported in diverse fungi, but none of them exhibit hypovirulence. As reported in this study, FoOuLV1 is the first ourmia-like mycovirus to be reported that exhibits hypovirulence. The S-segment associated with FoOuLV1 is unique among ourmia-like mycoviruses. A previous study showed that SsHV1 and its satellite-like RNA were able to co-infect the hypovirulent strain SZ-150, with the satellite-like RNA in conferring hypovirulence on S. sclerotiorum (Xie et al., 2011). Therefore, S-segment associated with FoOuLV1 may play an important role in its hypovirulence.
The Transmission of FoOuLV1 Is Important for F. oxysporum
Mycoviruses are generally transmitted by hyphal anastomosis and during sporogenesis, with the hyphal anastomosis occurring naturally between individuals belonging to the same or closely related vegetative compatibility groups (Lee et al., 2014). Furthermore, transmission via protoplast fusion has been reported in many mycoviruses (van Diepeningen et al., 1998; Kanematsu et al., 2010). The efficient transmission observed among isolates under natural conditions is considered to be an important condition for the successful application of mycoviruses to the control of plant fungal disease.
Fusarium wilt caused by F. oxysporum affects a large number of economically important crops. The pathogenic species contain a diversity of host-plant–specific forms (Torres-Trenas and Pérez-Artés, 2020), with different formae speciales potentially existing together in the soil concurrently. The hypovirulent traits of FoOuLV1 reported here can be transmitted horizontally from FoM strains to virus-free FoM strains and to FoC strains. This indicates that FoOuLV1 could be potentially used to control Fusarium wilt in various crops.
Potential Use of FoOuLV1
A large number of hypovirulent mycoviruses have been identified in previous studies, and some have been explored as potential biocontrol agents against fungal diseases. CHV1 has successfully been used to control chestnut blight in Europe (Anagnostakis, 1982), while SsHADV1 was used to control the disease caused by S. sclerotiorum (Yu et al., 2013). Except for these two mycoviruses, attempts to control diseases in field using mycoviruses are rare.
Many mycoviruses have been identified from the genus Fusarium (Cho et al., 2013); but, to our knowledge, only three mycoviruses have been identified in F. oxysporum (FoCV1, FodV1, and FodMV1). Furthermore, among these, only FodV1 induces hypovirulence (Sharzei et al., 2007; Lemus-Minor et al., 2015; Torres-Trenas and Pérez-Artés, 2020). In this study, FoOuLV1 was not only able to induce hypovirulence in F. oxysporum but also exhibited significant biological control effects against bitter gourd Fusarium wilt in the pot and field experiments. This suggests that FoOuLV1 has a potential to be used in the future.
Conclusion
In this study, we characterized a novel mycovirus (FoOuLV1) related to members of the Magoulivirus genera from a phytopathogenic fungus, F. oxysporum f. sp. momordicae. Although some ourmia-like mycoviruses have been identified in diverse fungi, this is the first report of a mycovirus in F. oxysporum f. sp. momordicae and the first report of an ourmia-like mycovirus in F. oxysporum. Furthermore, FoOuLV1 is the first ourmia-like mycovirus harboring an associated RNA segment and possessing a hypovirulence-inducing trait. Therefore, this virus provides a valuable experimental system to study the interaction of ourmia-like mycovirus and its fungal host. In addition, FoOuLV1 is also a potential biocontrol agent that could be further studied to control Fusarium wilt.
Data Availability Statement
The sequence file of FoOuLV1 is available from the NCBI, GenBank Accession Nos. MT551010 and MT551011.
Author Contributions
YiZ designed the research. YuZ, YS, ML, XW, HX, and YiZ performed the experimental work. YiZ, JX, and CW analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (31901934) and the Special Fund for Agro-scientific Research in the Public Interest (201503110).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our deep gratitude to Dr. Jinlu Wu (National University of Singapore), Dr. Yanping Fu (Huazhong Agricultural University), and Yan Shi (Henan Agricultural University) for providing advice and guidance and to Dr. Xuehong Wu for providing research materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.569869/full#supplementary-material
Footnotes
References
Amarasinghe, G. K., Aréchiga Ceballos, N. G., Banyard, A. C., Basler, C. F., Bavari, S., Bennett, A. J., et al. (2018). Taxonomy of the order Mononegavirales: update 2018. Arch. Virol. 163, 2283–2294. doi: 10.1007/s00705-018-3814-x
Anagnostakis, S. L. (1982). Biological control of chestnut blight. Science 215, 466–471. doi: 10.1126/science.215.4532.466
Avgelis, A., Barba, M., and Rumbos, I. (1989). Epirus cherry virus, an unusual virus isolated from cherry with rasp−leaf symptoms in Greece. J. Phytopathol. 126, 51–58. doi: 10.1111/j.1439-0434.1989.tb01089.x
Azhar, A., Mu, F., Huang, H., Cheng, J., Fu, Y., Hamid, M. R., et al. (2019). A novel RNA virus related to Sobemoviruses confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Viruses 11:759. doi: 10.3390/v11080759
Bruenn, J. A., Warner, B. E., and Yerramsetty, P. (2015). Widespread Mitovirus sequences in plant genomes. PeerJ 3:e876. doi: 10.7717/peerj.876
Cai, G., Myers, K., Fry, W. E., and Hillman, B. I. (2012). A member of the virus family Narnaviridae from the plant pathogenic oomycete Phytophthora infestans. Arch. Virol. 157, 165–169. doi: 10.1007/s00705-011-1126-5
Chiba, S., Salaipeth, L., Lin, Y.-H., Sasaki, A., Kanematsu, S., and Suzuki, N. (2009). A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 83, 12801–12812. doi: 10.1128/jvi.01830-09
Cho, W. K., Lee, K. M., Yu, J., Son, M., and Kim, K. H. (2013). Insight into mycoviruses infecting Fusarium species. Adv. Virus Res. 86, 273–288. doi: 10.1016/B978-0-12-394315-6.00010-6
Crivelli, G., Ciuffo, M., Genre, A., Masenga, V., and Turina, M. (2011). Reverse genetic analysis of Ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation. J. Virol. 85, 5091–5104. doi: 10.1128/jvi.02565-10
Darissa, O., Adam, G., and Schäfer, W. (2012). A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 134, 181–189. doi: 10.1007/s10658-012-9977-5
Di Pietro, A., Madrid, M. P., Caracuel, Z., Delgado-Jarana, J., and Roncero, M. I. G. (2003). Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. doi: 10.1046/j.1364-3703.2003.00180.x
Donaire, L., Rozas, J., and Ayllón, M. A. (2016). Molecular characterization of Botrytis ourmia-like virus, a mycovirus close to the plant pathogenic genus Ourmiavirus. Virology 489, 158–164. doi: 10.1016/j.virol.2015.11.027
Ghabrial, S. A., Castón, J. R., Jiang, D., Nibert, M. L., and Suzuki, N. (2015). 50-plus years of fungal viruses. Virology 479–480, 356–368. doi: 10.1016/j.virol.2015.02.034
Ghabrial, S. A., and Suzuki, N. (2009). Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384. doi: 10.1146/annurev-phyto-080508-081932
Guo, J., Zhu, J. Z., Zhou, X. Y., Zhong, J., Li, C. H., Zhang, Z. G., et al. (2019). A novel ourmia-like mycovirus isolated from the plant pathogenic fungus Colletotrichum gloeosporioides. Arch. Virol. 164, 2631–2635. doi: 10.1007/s00705-019-04346-2
Hillman, B. I., Shapira, R., and Nuss, D. L. (1990). Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80, 950–956. doi: 10.1094/phyto-80-950
Hrabáková, L., Koloniuk, I., and Petrzik, K. (2017). Phomopsis longicolla RNA virus 1 – novel virus at the edge of myco- and plant viruses. Virology 506, 14–18. doi: 10.1016/j.virol.2017.03.003
Illana, A., Marconi, M., Rodríguez-Romero, J., Xu, P., Dalmay, T., Wilkinson, M. D., et al. (2017). Molecular characterization of a novel ssRNA ourmia-like virus from the rice blast fungus Magnaporthe oryzae. Arch. Virol. 162, 891–895. doi: 10.1007/s00705-016-3144-9
Kanematsu, S., Arakawa, M., Oikawa, Y., Onoue, M., Osaki, H., Nakamura, H., et al. (2004). A reovirus causes hypovirulence of Rosellinia necatrix. Phytopathology 94, 561–568. doi: 10.1094/PHYTO.2004.94.6.561
Kanematsu, S., Sasaki, A., Onoue, M., Oikawa, Y., and Ito, T. (2010). Extending the fungal host range of a partitivirus and a mycoreovirus from Rosellinia necatrix by inoculation of protoplasts with virus particles. Phytopathology 100, 922–930. doi: 10.1094/PHYTO-100-9-0922
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, S.-J., Lim, W.-S., Park, S.-H., Park, M.-R., and Kim, K.-H. (2007). Molecular characterization of a dsRNA mycovirus, Fusarium graminearum virus-DK21, which is phylogenetically related to hypoviruses but has a genome organization and gene expression strategy resembling those of plant potex-like viruses. Mol. Cells 23, 304–315. doi: 10.1007/s10059-009-0112-1
Lee, K. M., Cho, W. K., Yu, J., Son, M., Choi, H., Min, K., et al. (2014). A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS One 9:e100989. doi: 10.1371/journal.pone.0100989
Lemus-Minor, C. G., Cañizares, M. C., García-Pedrajas, M. D., and Pérez-Artés, E. (2015). Complete genome sequence of a novel dsRNA mycovirus isolated from the phytopathogenic fungus Fusarium oxysporum f. sp. dianthi. Arch. Virol. 160, 2375–2379. doi: 10.1007/s00705-015-2509-9
Li, C. X., Zhu, J. Z., Gao, B. D., Zhu, H. J., Zhou, Q., and Zhong, J. (2019). Characterization of a novel ourmia-like mycovirus infecting Magnaporthe oryzae and implications for viral diversity and evolution. Viruses 11:223. doi: 10.3390/v11030223
Li, H., Bian, R., Liu, Q., Yang, L., Pang, T., Salaipeth, L., et al. (2019). Identification of a novel hypovirulence-inducing hypovirus from Alternaria alternata. Front. Microbiol. 10:1076. doi: 10.3389/fmicb.2019.01076
Li, M., Gong, X., Zheng, J., Jiang, D., Fu, Y., and Hou, M. (2005). Transformation of Coniothyrium minitans, a parasite of Sclerotinia sclerotiorum, with Agrobacterium tumefaciens. FEMS Microbiol. Lett. 243, 323–329.
Li, P., Bhattacharjee, P., Wang, S., Zhang, L., Ahmed, I., and Guo, L. (2019). Mycoviruses in Fusarium species: an update. Front. Cell. Infect. Microbiol. 9:257. doi: 10.3389/fcimb.2019.00257
Li, P., Zhang, H., Chen, X., Qiu, D., and Guo, L. (2015). Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 481, 151–160. doi: 10.1016/j.virol.2015.02.047
Lisa, V., Milne, R. G., Accotto, G. P., Boccardo, G., Caciagli, P., and Parvizy, R. (1988). Ourmia melon virus, a virus from Iran with novel properties. Ann. Appl. Biol. 112, 291–302. doi: 10.1111/j.1744-7348.1988.tb02065.x
Marzano, S.-Y. L., Nelson, B. D., Ajayi-Oyetunde, O., Bradley, C. A., Hughes, T. J., Hartman, G. L., et al. (2016). Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 90, 6846–6863. doi: 10.1128/jvi.00357-16
Mullins, E. D., Chen, X., Romaine, P., Raina, R., Geiser, D. M., and Kang, S. (2001). Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91, 173–180. doi: 10.1094/PHYTO.2001.91.2.173
Nuss, D. L. (2005). Hypovirulence: mycoviruses at the fungal–plant interface. Nat. Rev. Microbiol. 3, 632–642. doi: 10.1038/nrmicro1206
Ohkita, S., Lee, Y., Nguyen, Q., Ikeda, K., Suzuki, N., and Nakayashiki, H. (2019). Three ourmia-like viruses and their associated RNAs in Pyricularia oryzae. Virology 534, 25–35. doi: 10.1016/j.virol.2019.05.015
Potgieter, A. C., Page, N. A., Liebenberg, J., Wright, I. M., Landt, O., and van Dijk, A. A. (2009). Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J. Gen. Virol. 90(Pt 6), 1423–1432. doi: 10.1099/vir.0.009381-0
Sasaki, A., Kanematsu, S., Onoue, M., Oikawa, Y., Nakamura, H., and Yoshida, K. (2007). Artificial infection of Rosellinia necatrix with purified viral particles of a member of the genus Mycoreovirus reveals its uneven distribution in single colonies. Phytopathology 97, 278–286. doi: 10.1094/PHYTO-97-3-0278
Sharzei, B., Banihashemi, Z. A. D., and Afsharifar, A. R. (2007). Detection and characterization of a double-stranded RNA mycovirus in Fusarium oxysporum f. sp. melonis. Iran J. Plant Pathol. 43, 9–26.
Shi, M., Lin, X. D., Tian, J. H., Chen, L. J., Chen, X., Li, C. X., et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. doi: 10.1038/nature20167
Suzuki, N., Ghabrial, S. A., Kim, K. H., Pearson, M., Marzano, S. Y. L., Yaegashi, H., et al. (2018). ICTV virus taxonomy profile: hypoviridae. J. Gen. Virol. 9, 615–616. doi: 10.1099/jgv.0.001055
Torres-Trenas, A., and Pérez-Artés, E. (2020). Characterization and incidence of the first member of the genus Mitovirus identified in the phytopathogenic species Fusarium oxysporum. Viruses 12:279. doi: 10.3390/v12030279
Turina, M., Hillman, B. I., Izadpanah, K., Rastgou, M., and Rosa, C. (2017). ICTV virus taxonomy profile: Ourmiavirus. J. Gen. Virol. 9, 129–130. doi: 10.1099/jgv.0.000725
van Dam, P., de Sain, M., ter Horst, A., van der Gragt, M., and Rep, M. (2018). Use of comparative genomics-based markers for discrimination of host specificity in Fusarium oxysporum. Appl. Environ. Microb. 84:e01868-17.
van Diepeningen, A. D., Debets, A. J. M., and Hoekstra, R. F. (1998). Intra- and interspecies virus transfer in Aspergilli via protoplast fusion. Fungal Genet. Biol. 25, 171–180. doi: 10.1006/fgbi.1998.1096
Wang, Q., Mu, F., Xie, J., Cheng, J., Fu, Y., and Jiang, D. (2020). A single ssRNA segment encoding RdRp is sufficient for replication, infection, and transmission of ourmia-like virus in fungi. Front. Microbiol. 11:379. doi: 10.3389/fmicb.2020.00379
Xie, J., and Jiang, D. (2014). New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52, 45–68. doi: 10.1146/annurev-phyto-102313-050222
Xie, J., Wei, D., Jiang, D., Fu, Y., Li, G., Ghabrial, S., et al. (2006). Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotina sclerotorium. J. Gen. Virol. 87(Pt 1), 241–249. doi: 10.1099/vir.0.81522-0
Xie, J., Xiao, X., Fu, Y., Liu, H., Cheng, J., Ghabrial, S. A., et al. (2011). A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 418, 49–56. doi: 10.1016/j.virol.2011.07.008
Xu, Z., Wu, S., Liu, L., Cheng, J., Fu, Y., Jiang, D., et al. (2015). A Mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res. 197, 127–136. doi: 10.1016/j.virusres.2014.12.023
Yu, X., Li, B., Fu, Y., Jiang, D., Ghabrial, S. A., Li, G., et al. (2010). A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U.S.A. 107, 8387–8392. doi: 10.1073/pnas.0913535107
Yu, X., Li, B., Fu, Y., Xie, J., Cheng, J., Ghabrial, S. A., et al. (2013). Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. U.S.A. 110, 1452–1457. doi: 10.1073/pnas.1213755110
Zheng, L., Shu, C., Zhang, M., Yang, M., and Zhou, E. (2019). Molecular characterization of a novel endornavirus conferring hypovirulence in rice sheath blight fungus Rhizoctonia solani AG-1 IA strain GD-2. Viruses 11:178. doi: 10.3390/v11020178
Zhou, J., Wang, Y., Liang, X., Xie, C., Liu, W., Miao, W., et al. (2020). Molecular characterization of a novel ourmia-like virus infecting Phoma matteucciicola. Viruses 12:231. doi: 10.3390/v12020231
Keywords: Fusarium oxysporum, ourmia-like virus, hypovirulence, mycovirus, transfection
Citation: Zhao Y, Zhang Y, Wan X, She Y, Li M, Xi H, Xie J and Wen C (2020) A Novel Ourmia-Like Mycovirus Confers Hypovirulence-Associated Traits on Fusarium oxysporum. Front. Microbiol. 11:569869. doi: 10.3389/fmicb.2020.569869
Received: 05 June 2020; Accepted: 12 November 2020;
Published: 09 December 2020.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Jeesun Chun, Jeonbuk National University, South KoreaDavid Turra, Università degli Studi di Napoli Federico II, Italy
Copyright © 2020 Zhao, Zhang, Wan, She, Li, Xi, Xie and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiyi Wen, d2VuY2FpeWlAaGVuYXUuZWR1LmNu
 Ying Zhao
Ying Zhao Yuanyan Zhang1
Yuanyan Zhang1 Jiatao Xie
Jiatao Xie