
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 09 October 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.563944
This article is part of the Research Topic (p)ppGpp and Its Homologs: Enzymatic and Mechanistic Diversity Among the Microbes View all 19 articles
Single cell microorganisms including pathogens relentlessly face myriads of physicochemical stresses in their living environment. In order to survive and multiply under such unfavorable conditions, microbes have evolved with complex genetic networks, which allow them to sense and respond against these stresses. Stringent response is one such adaptive mechanism where bacteria can survive under nutrient starvation and other related stresses. The effector molecules for the stringent response are guanosine-5'-triphosphate 3'-diphosphate (pppGpp) and guanosine-3', 5'-bis(diphosphate) (ppGpp), together called (p)ppGpp. These effector molecules are now emerging as master regulators for several physiological processes of bacteria including virulence, persistence, and antimicrobial resistance. (p)ppGpp may work independently or along with its cofactor DksA to modulate the activities of its prime target RNA polymerase and other metabolic enzymes, which are involved in different biosynthetic pathways. Enzymes involved in (p)ppGpp metabolisms are ubiquitously present in bacteria and categorized them into three classes, i.e., canonical (p)ppGpp synthetase (RelA), (p)ppGpp hydrolase/synthetase (SpoT/Rel/RSH), and small alarmone synthetases (SAS). While RelA gets activated in response to amino acid starvation, enzymes belonging to SpoT/Rel/RSH and SAS family can synthesize (p)ppGpp in response to glucose starvation and several other stress conditions. In this review, we will discuss about the current status of the following aspects: (i) diversity of (p)ppGpp biosynthetic enzymes among different bacterial species including enteropathogens, (ii) signals that modulate the activity of (p)ppGpp synthetase and hydrolase, (iii) effect of (p)ppGpp in the production of antibiotics, and (iv) role of (p)ppGpp in the emergence of antibiotic resistant pathogens. Emphasis has been given to the cholera pathogen Vibrio cholerae due to its sophisticated and complex (p)ppGpp metabolic pathways, rapid mutational rate, and acquisition of antimicrobial resistance determinants through horizontal gene transfer. Finally, we discuss the prospect of (p)ppGpp metabolic enzymes as potential targets for developing antibiotic adjuvants and tackling persistence of infections.
Living organisms from three domains of life (bacteria, archaea, and eukarya) use a number of purine derivatives like guanosine pentaphosphate (pppGpp), guanosine tetraphosphate (ppGpp), cyclic di-GMP (c-di-GMP), etc., as intracellular signaling molecules. Bacteria use these small molecules to monitor intra- and extracellular environmental conditions and modulate their growth and multiplications in response to the availability of nutrients and related local cues (Potrykus and Cashel, 2008; Tozawa and Nomura, 2011; Hauryliuk et al., 2015). Over 50 years ago, Cashel identified the small molecule alarmones, pppGpp, and ppGpp (Figure 1), collectively known as (p)ppGpp, as the key players for the bacterial stringent response under nutrient limitations and other stressful conditions (Cashel, 1969). Later several studies revealed that the effector molecules of stringent response (p)ppGpp modulates bacterial multiplication rate and survival during nutrient limitations, exposure to antimicrobial compounds, xenobiotics, and osmotic stress (Hauryliuk et al., 2015; Irving and Corrigan, 2018). The alarmone (p)ppGpp is the derivatives of guanosine nucleosides, where guanosine triphosphate (GTP) pyrophosphokinases (RelA/SpoT/RSH/RelV/RelP/RelQ) transfer a pyrophosphate moiety from ATP to the 3'-OH position of GTP/guanosine diphosphate (GDP; Cashel, 1969; Gaca et al., 2015; Ronneau and Hallez, 2019). It has been shown that the intracellular level of (p)ppGpp is critical for modulation of different bacterial physiological processes mainly by regulating the activities of RNA polymerase (RNAP), DNA primase (DnaG), growth rate, and several other metabolic enzymes in Escherichia coli (Potrykus and Cashel, 2008; Potrykus et al., 2011; Hauryliuk et al., 2015; Zhang et al., 2018; Wang et al., 2019). In addition, (p)ppGpp also modulates bacterial growth and viability indirectly through depletion of cellular level of guanosine and adenosine nucleotides or by repressing transcription of genes required for active growth (Kriel et al., 2012). In nutrient rich growth condition, the basal cellular level of (p)ppGpp in E. coli is less than 0.2 mM (Mechold et al., 2013). Upon induction of stress, the level of (p)ppGpp may increase from 10 to 100-fold depending upon the type of stress and the enzymes involved in the biosynthesis of different biomolecules (Kalia et al., 2013). Elevated level of (p)ppGpp may work independently or synergistically with the transcriptional factor DksA, an RNAP binding small transcriptional factor (Paul et al., 2004). It was discovered earlier that the dksA gene product suppresses temperature-sensitive growth and filamentation of a dnaK deletion mutant of E. coli (Kang and Craig, 1990). Later, it has been established that both DksA and (p)ppGpp biosynthetic enzymes are crucial for the stringent response in Gram-negative bacteria since, ΔdksA and ΔrelAΔspoT mutants exhibit similar phenotypes (Gourse et al., 2018). In addition, overexpression of DksA can compensate the loss of (p)ppGpp in regulating uspA, livJ, and rrnBP1 (Magnusson et al., 2007). However, synergistic functions are not universal. DksA and (p)ppGpp can work independently or may have opposite effects on one another. For example, E. coli ΔdksA cells aggregate more efficiently compared to its isogenic wild-type strain. Similarly, overexpression of DksA decreases the adhesion of wild-type cells. In contrast, E. coli ΔrelAΔspoT mutant called (p)ppGpp0 cells failed to sediment in a similar experimental condition and the adhesion phenotype is not affected upon overexpression of DksA (Magnusson et al., 2007). In addition, transcription of the argX operon containing argX, hisR, leuT, and proM genes is activated by DksA but inhibited in the presence of (p)ppGpp and DksA (Lyzen et al., 2016).
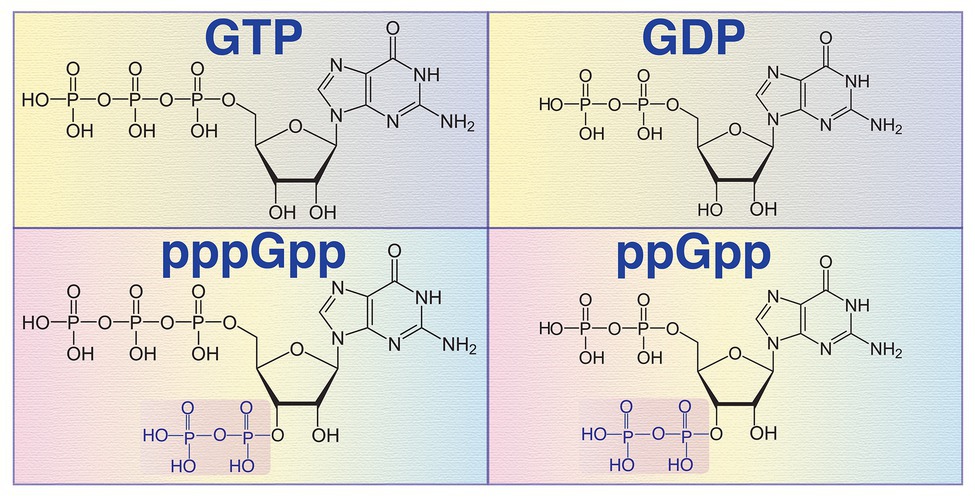
Figure 1. Chemical structures of guanosine triphosphate (GTP), guanosine diphosphate (GDP), guanosine pentaphosphate (pppGpp), and guanosine tetraphosphate (ppGpp) molecules. The pyrophosphate group of pppGpp and ppGpp at the 3' hydroxyl (OH) position is transferred by the (p)ppGpp synthetase from another purine nucleotide ATP.
Other than the function in stringent response, (p)ppGpp also plays important roles in modulating bacterial virulence gene expression (Dalebroux et al., 2010; and the references therein), sporulation (Crawford and Shimkets, 2000), biofilm formation (He et al., 2012), antibiotic resistance (Wu et al., 2010; Strugeon et al., 2016), tolerance (Kim et al., 2018), and persistence (Hauryliuk et al., 2015; Harms et al., 2016). In order to access host cell nutrients, colonization on the cell surface and detachment from mucosal surface, pathogenic bacteria use (p)ppGpp signaling networks to modulate expression of genes those are part of secretion systems, flagellar components, adhesins, and serine/metallo proteases (Dalebroux et al., 2010; Pal et al., 2012; and reference therein). Regulation of spore formation in certain bacteria mediated by (p)ppGpp through complex array of regulatory circuits that sense the environmental signals through altered levels of intracellular (p)ppGpp leading to rapid change in the expression of relevant genes involved in spore formation (Crawford and Shimkets, 2000). It has been shown that the stringent response positively modulates biofilm formation in E. coli, Vibrio cholerae, and Streptococcus mutans (He et al., 2012; Teschler et al., 2015; Strugeon et al., 2016). In Pseudomonas aeruginosa, the antibiotic tolerance of nutrient-limited and biofilm dwelling cells is mediated by active responses to starvation where stringent response plays a crucial role (Nguyen et al., 2011). This starvation mediated protective mechanism in P. aeruginosa has been shown to be linked with tolerance under reduced level of oxidative stress in bacterial cells and, therefore, inactivating this protective mechanism sensitized biofilms by several orders of magnitude to different classes of antibiotics allowing enhanced efficacy of antibiotic treatment in experimental infection in an animal model (Nguyen et al., 2011).
From various studies, it appears that emergence of instant antibiotic resistant clones in a susceptible bacterial population solely depends on: (i) target modifications, (ii) reduced accessibility of antibiotics to the target, (iii) decreased effective concentration of antibiotic by reducing the membrane permeability or by increasing efflux activity, and (iv) acquisition of antibiotic resistance genes from other microbial species (Gaca et al., 2015; Verma et al., 2019; Das et al., 2020; Jung et al., 2020; Pant et al., 2020). A recent study has shown that the expression levels of ~300 and ~400 genes (total ~700 genes) are upregulated and downregulated, respectively, within 5 min upon induction of (p)ppGpp (Sanchez-Vazquez, 2018). In Enterococcus faecalis (p)ppGpp0 cells, it has been found that the genes and pathways involved in pyruvate production and heterolactic fermentation are induced (Gaca et al., 2013). More importantly, (p)ppGpp induced RpoS expression, the stress response sigma factor, may lead to overproduction of error prone DNA polymerase IV (Pol IV; Storvik and Foster, 2010). In addition to antibiotic resistance, (p)ppGpp also reduces efficacy of antibiotics by inducing antibiotic tolerant persister cell formation in Gram-negative and Gram-positive bacterial populations (Korch et al., 2003; Kaspy et al., 2013).
As of 5th August 2020, more than 27,936 articles and reports are available in the National Center for Biotechnology Information1 on the stringent response in bacteria. Considering this vast literature, however, the present review will focus on: (i) metabolisms, biosynthetic enzymes, signals, interaction partners, and targets of (p)ppGpp; (ii) effect of (p)ppGpp in antibiotic production; (iii) role of (p)ppGpp in antibiotic resistance in bacterial pathogens; and (iv) (p)ppGpp biosynthetic enzymes as potential targets for developing antibiotic adjuvants.
Principally, (p)ppGpp homeostasis in bacterial cells depends on the availability and activity of the four classes of enzymes: (i) multi-domain bifunctional (p)ppGpp synthetase-hydrolase, (ii) multi-domain monofunctional (p)ppGpp synthetase, (iii) short alarmone synthetase (SAS), and (iv) short alarmone hydrolase (SAH), also called RelH (Figure 2). Bifunctional (p)ppGpp synthetase-hydrolase enzymes (Rel/RSH/SpoT) can modulate their conformation depending on the environmental conditions with the help of their regulatory domains and stimulate their GTP/GDP pyrophosphokinase or pyrophosphohydrolase activities to synthesize or hydrolyze (p)ppGpp, respectively (Ronneau and Hallez, 2019). In the presence of (p)ppGpp synthetase, hydrolase activity of the bifunctional enzyme is essential for viability (Xiao et al., 1991; Das and Bhadra, 2008). However, bacterial cells can survive in the absence of (p)ppGpp synthetase activity in nutrient rich environments but it is essential for viability in nutrient limited medium (Xiao et al., 1991). Enzymes with (p)ppGpp synthetase-hydrolase activities are widely conserved across bacterial phyla (Atkinson et al., 2011). Nevertheless, presence of (p)ppGpp synthetase-hydrolase domain containing proteins has also been reported in eukarya including Arabidopsis thaliana, Nicotiana tabacum, and Chlamydomonas reinhardtii (Sun et al., 2010; Ito et al., 2012). In Firmicutes, (p)ppGpp synthetase-hydrolase domain containing enzymes are the major source of stringent response effector molecules during amino acid, glucose, and fatty acid starvations (Wolz et al., 2010). In contrast, several Proteobacteria use the bifunctional (p)ppGpp synthetase-hydrolase enzyme for alarmone synthesis during glucose and fatty acid starvations but not during amino acid starvation (Xiao et al., 1991; Cashel, 1969). Upon sensing nutrient depletions or other stress conditions, Rel/RSH/SpoT catalyze pyrophosphorylation of GDP or GTP at the 3' hydroxyl (OH) position using ATP as pyrophosphate donor (Figure 3). Once reprogramming of cellular functions from vegetative to survival mode is accomplished with the help of elevated intracellular levels of (p)ppGpp, the bifunctional Rel/RSH enzyme then may change its conformation from (p)ppGpp synthetase-ON/hydrolase-OFF to (p)ppGpp hydrolase-ON/synthetase-OFF state to reduce the (p)ppGpp level to restore again the gene expression and metabolic functions of enzymes associated with growth and multiplication under favorable conditions (Hogg et al., 2004). Although the exact molecular mechanism is unknown, but it has been proposed that the regulatory domains located at the carboxy terminal domain of Rel/RSH/SpoT play important roles in the modulation of synthetase and hydrolase functions of the bifunctional (p)ppGpp synthetase-hydrolase enzymes (Angelini et al., 2012).
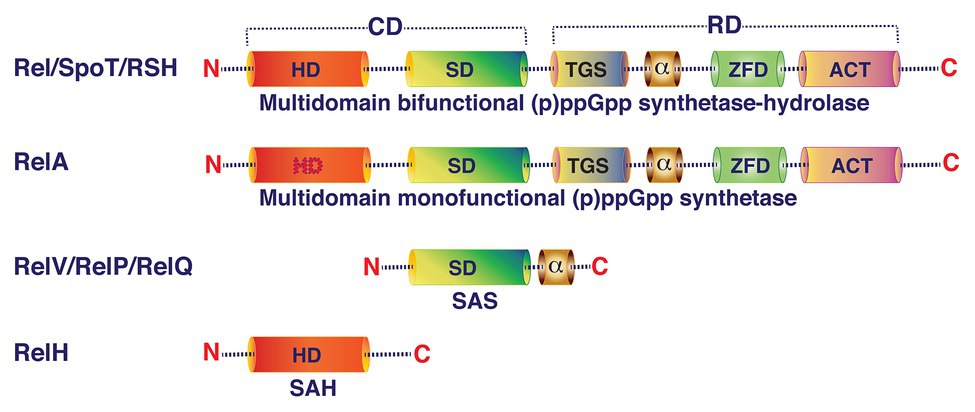
Figure 2. Schematic representation of the domain organization of (p)ppGpp synthetase/hydrolase family of proteins. Bifunctional long multidomain (p)ppGpp synthetase-hydrolase and monofunctional long multidomain (p)ppGpp synthetase enzymes are composed of two regions: (i) N-terminal catalytic domain (CD) and (ii) C-terminal regulatory domain (RD). Both synthetase and hydrolase domains (SD and HD, respectively) are functional in bifunctional enzymes (Rel/RSH/SpoT). In monofunctional multidomain, (p)ppGpp synthetase enzyme (RelA) hydrolase activity is lost due to accumulation of spontaneous mutations in the catalytic residues. The C-terminal regulatory region of the multidomain protein contains TGS domain (ThrRS, GTPase, and SpoT), conserved α helical domain, ZFD domain (zinc-finger or conserved cysteine domain), and ACT (aspartate kinase, chorismate, and TyrA) RNA recognition motif. The small alarmone synthetase (SAS) enzyme (RelV/RelP/RelQ) contains SD and a small oligomeric α-domain. The RelH/small alarmone hydrolase (SAH) contains only the HD.
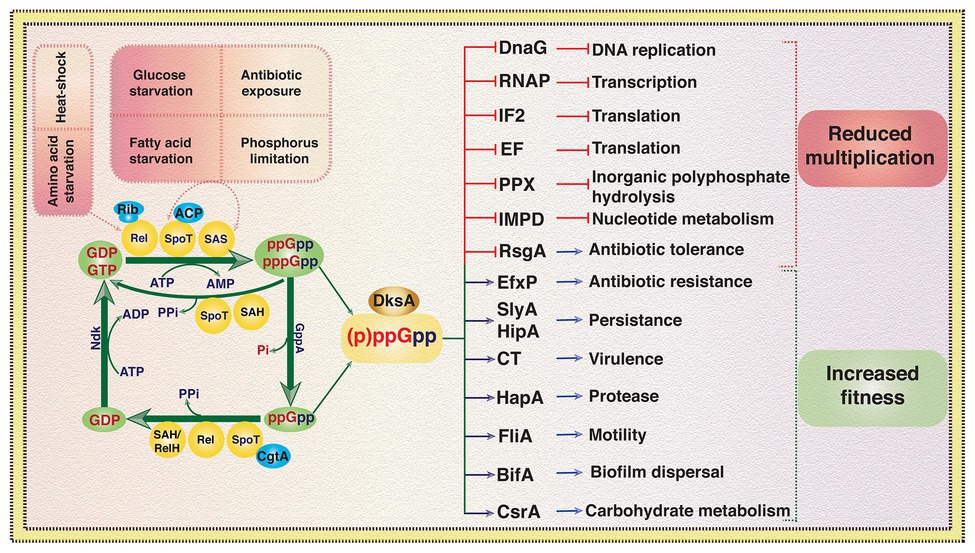
Figure 3. An overview of signaling, metabolism, and effect of (p)ppGpp in virulence, antibiotic resistance, and other cellular processes in bacteria. GDP and GTP are converted to (p)ppGpp during stress conditions or upon exposure to xenobiotics due to transfer of a pyrophosphate moiety from ATP to the 3'-OH position of ribosugar of guanosine nucleotide. GTP pyrophospho kinase activity of Rel, SpoT, and SAS can mediate the transfer of pyrophosphate to GTP or GDP depending on the stress signaling and activity of other cellular proteins like acyl carrier protein (ACP) and functional state of protein synthesis machinery, i.e., ribosome (Rib). pppGpp can be converted to ppGpp due to phosphatase activity of GppA. SpoT, Rel, and SAH can hydrolyze the pyrophosphate moiety from 3'-OH position of pppGpp and ppGpp and generate GTP or GDP, respectively. Guanine nucleotide binding protein of the Obg/GTP1 subfamily CgtA induces (p)ppGpp hydrolase activity of SpoT. Nucleoside diphosphate kinase (Ndk) converts GDP to GTP using ATP as phosphate donor. Elevated intracellular level of (p)ppGpp independently or in conjunction with transcriptional factor DksA regulates several cellular processes including DNA replication, transcription, protein biosynthesis, carbohydrate metabolism, virulence, motility, antibiotic resistance, and biofilms formation. (p)ppGpp regulates several proteins involved in nucleotide metabolism, persistence, antibiotic resistance, etc., however, for simplicity, a representative protein for each case is depicted here and more details can be found in the references cited. SAS, small alarmone synthase; SAH, small alarmone hydrolase; GppA, guanosine-5'-triphosphate, 3'-diphosphate pyrophosphatase; DnaG, DNA primase; RNAP, RNA polymerase; IF2, initiation factor 2; EF, elongation factor; PPX, exopolyphosphatase; IMPD, inosine monophosphate dehydrogenase; RsgA, small ribosomal subunit biogenesis GTPase; EfxP, efflux pumps; SlyA, transcriptional regulator of Salmonella enterica; HipA, toxin component of Escherichia coli TA module; CT, cholera toxin; HapA, hemagglutinin protease; FliA, flagella regulatory sigma factor; BifA, biofilm formation protein; CsrA, carbon storage regulator (Dalebroux et al., 2010; Ronneau and Hallez, 2019; and the references therein).
Monofunctional (p)ppGpp synthetase, the enzyme that synthesizes (p)ppGpp only, further subdivided into two major sub-classes based on their size and domain organization: (i) long multi-domain monofunctional (p)ppGpp synthetase and (ii) short monodomain monofunctional (p)ppGpp synthetase also known as SAS (Figure 2). Presence of multi-domain monofunctional (p)ppGpp synthetase (for example, RelA) is common in Proteobacteria. RelA is a ribosome-associated protein, which recognizes stalled ribosomes due to presence of uncharged tRNA in the A-site of ribosome during amino acid starvation. It has been reported that the large subunit ribosomal protein L11 is crucial for the activation of (p)ppGpp synthetase activity of RelA (Parker et al., 1976; Cashel et al., 1996). Wendrich et al. (2002) initially proposed a “hopping model” for RelA mediated (p)ppGpp synthesis during amino acid starved condition. According to this model the RelA molecule hops from ribosome to ribosome to synthesize one (p)ppGpp molecule per dissociation event. Later, English et al. (2011) using single-molecule tracking methodology proposed an “extended hopping” model for RelA mediated (p)ppGpp synthesis during amino acid starvation. According to the extended hopping model several molecules of (p)ppGpp are synthesized by the free but enzymatically active RelA upon its dissociation from ribosomes. However, findings of Li et al. (2016) contradict the extended hopping model and they proposed a “short hopping time” model for RelA mediated (p)ppGpp synthesis during amino acid starvation. According to this model, RelA synthesizes (p)ppGpp while bound to the 70S ribosomes. Currently, however, the reasons behind these differences are not clear. Although amino acid starvation is the major cause of activation of (p)ppGpp synthetase function of RelA, heat shock can also induce its synthetase activity in E. coli (Gallant et al., 1977; English et al., 2011). Short monodomain monofunctional (p)ppGpp synthetases are widely distributed among Firmicutes (Bacillus subtilis, S. mutans, E. faecalis, etc.), Actinobacteria (Mycobacterium smegmatis), Proteobacteria (V. cholerae), Archaea (Methanosarcina acetivorans), and Eukarya (Dictyostelium discoideum) (Lemos et al., 2007; Nanamiya et al., 2008; Das et al., 2009; Atkinson et al., 2011; Murdeshwar and Chatterji, 2012; and the references therein). Unlike RelA, the SAS sub-class can recognize glucose, fatty acids, and other starvations and catalyze (p)ppGpp synthesis by transferring a pyrophosphate moiety from ATP to the 3'-OH position of GTP or GDP probably following same mechanistic pathways like RelA/RSH/Rel enzymes (Figure 3). Since SAS enzymes are devoid of any additional regulatory domain, their regulation for (p)ppGpp synthesis activity may primarily be dependent at the gene expression level (Dasgupta et al., 2014; Ronneau and Hallez, 2019) and needs further investigation.
The monodomain monofunctional (p)ppGpp hydrolase, also known as small alarmone hydrolase (SAH), was initially identified and functionally characterized in the metazoa as an ortholog of bacterial SpoT (Sun et al., 2010). Like multi-domain bacterial (p)ppGpp hydrolases, the metazoan ortholog, called Mesh1, contains an active site for (p)ppGpp hydrolysis and it carries the conserved His-Asp box motif for binding with Mn2+. Later in silico analyses of more than 1,000 genome sequences identified seven subgroups of SAH (Atkinson et al., 2011). Recently, an SAH from Corynebacterium glutamicum has functionally been characterized and designated RelH (Ruwe et al., 2018). However, the regulatory signals, interaction partners, and importance of SAH in either kingdom are not clear. Thus, more intense research on these aspects of SAH is needed.
Accumulation of (p)ppGpp in the cytosol due to nutrient limitations or other stress conditions leads to change in bacterial cellular physiology by: (i) reprogramming of transcription of rRNA operons, ribosomal protein encoding genes, and others by regulating RNAP activity, (ii) stalling DNA replication by inhibiting DnaG, and (iii) modulating metabolic pathways by regulating activity of the associated enzymes (Figure 3). Single or multiple molecules of (p)ppGpp binds to its targets and modulate their activity independently or synergistically with its functional partner DksA. In E. coli, (p)ppGpp binds to at least two sites of RNAP enzyme (Ross et al., 2013). Crystal structure of RNAP holoenzyme showed that (p)ppGpp binds to the cleft surrounded by the α, β', and ω subunits (site 1) and also at an interface of RNAP and DksA (site 2). Binding of (p)ppGpp induces allosteric changes in RNAP, which either affects its catalytic activity by modulating the Mg+2 nucleotidyl transfer efficacy or by reducing stability of RNAP-promoter complex (Kanjee et al., 2012). DksA binds to the secondary channel of RNAP and can potentiate the effects (p)ppGpp by modulating RNAP-promoter complex stability (Perederina et al., 2004). In B. subtilis, (p)ppGpp regulates rRNA and ribosomal protein encoding genes transcription by interfering (p)ppGpp homeostasis. Elevated levels of (p)ppGpp inhibit hypoxanthine phosphoribosyl transferase (HprT) and GMP kinase (Gmk) activity in Firmicutes and reduce the intracellular GTP pool (Figure 3). HprT is essential for the conversion of hypoxanthine to IMP and guanine to GMP, while Gmk catalyzes synthesis of GDP from GMP. Both the enzymes are crucial for GTP biosynthesis in B. subtilis and other Firmicutes (Denapoli et al., 2013). It has been shown recently that in Gram-positive bacteria (p)ppGpp may bind with several ribosome-associated GTPases like RsgA, RbgA, Era, and HflX leading to strong inhibition of their activities (Corrigan et al., 2016). More recently, Zhang et al. (2018) have also reported similar target proteins, namely, RsgA, Era, HflX, etc., of (p)ppGpp in E. coli. However, the exact biochemical mechanism by which (p)ppGpp can inhibit GTPase activity is yet to be fully elucidated. Nonetheless, the findings clearly point toward the critical roles of (p)ppGpp in controlling ribosomal assembly/biogenesis in bacterial stress survival.
It is well established that (p)ppGpp drastically reduces bacterial multiplication by inhibiting initiation and elongation of DNA replication (Wang et al., 2007). It has been shown that (p)ppGpp binds to the RNA primer biosynthesis enzyme primase (DnaG) and directly inhibits its primer biosynthesis activity, which is essential for initiation of DNA replication (Figure 3). Initial finding hypothesized that the effect of (p)ppGpp in DNA replication is mostly restricted at the oriC region during initiation of DNA replication (Gourse and Keck, 2007). However, subsequent studies demonstrated that the arrest of DNA replication occurs throughout the chromosome during stringent response. Although elevated levels (>10–20-fold) of (p)ppGpp is detrimental to bacterial growth, complete lack of (p)ppGpp, called (p)ppGpp0 phenotype, render several bacterial species auxotrophic to different amino acids, defective in cell division, deficient in protease production, and poor survival upon exposure to xenobiotics (Hauryliuk et al., 2015). More importantly, increasing numbers of reports suggest that elevated level of (p)ppGpp reduces efficacy of several clinically important antibiotics, while (p)ppGpp0 strains are susceptible to numerous antibiotics with reduced minimum inhibitory concentration (MIC; Gaca et al., 2013; Kamarthapu et al., 2016; Hobbs and Boraston, 2019 and the references therein).
Production of antibiotics in Actinobacteria, one of the most diverse groups of Gram-positive bacteria that produce most of the clinically used antibiotics, is influenced by the availability of nutrients and external signals. Such factors work either by modulating the expression level of antibiotic biosynthetic gene clusters or through pleiotropic regulators, which play important roles in bacterial intracellular signaling (van der Heul et al., 2018). Role of (p)ppGpp in antibiotic production has been demonstrated both in environmental isolates as well as in clinically important bacterial pathogens (Ochi, 1987; Jin et al., 2004; Gomez-Escribano et al., 2008; van der Heul et al., 2018). Different soil living Gram-positive bacteria including B. subtilis, Streptomyces clavuligerus, produce a variety of antibiotics to inhibit the growth of other bacteria living in the same environment (van der Meij et al., 2017; and the references therein). Various reports suggest that (p)ppGpp may either positively or negatively regulate antibiotic production in Actinobacteria. For example, Streptomyces coelicolor synthesizes two well-known antibiotics, the polyketide actinorhodin, and the tripyrolle undecylprodigiosin and the production of each them was decreased in the ΔrelA mutant of S. coelicolor under a continuous culture condition (Kang et al., 1998). On the other hand, it has been shown in S. clavuligerus that (p)ppGpp negatively regulates the production of clavulanic acid and cephamycin C antibiotics in the absence of (p)ppGpp biosynthetic gene relA (Gomez-Escribano et al., 2008). Role of (p)ppGpp in the antibiotic production has also been demonstrated in other species of Streptomyces including Streptomyces antibioticus and Streptomyces griseus (Sivapragasam and Grove, 2019). (p)ppGpp may promote antibiotic production in these bacteria by inducing transcription of antibiotic biosynthetic gene clusters. In fact, it has been shown that the elevated level of intracellular (p)ppGpp in S. coelicolor leads to reduction in the expression of vegetative sigma factor and induction of expression of alternative sigma factor, which play important role in the transcription of antibiotic biosynthesis gene cluster (Hesketh et al., 2007).
Antibiotics, natural or synthetic chemical compounds that interfere with microbial growth or eliminate microbes from their niche, are one of the most important drugs in the medical history that has ever been discovered and deployed to prevent or cure microbial infections (Fischbach and Walsh, 2009). However, emergence of antibiotic resistance (AMR) and rapid dissemination of resistant traits in the clinically important bacterial species has drastically reduced the efficacy of several therapeutic agents commonly used to prevent or treat microbial infection (Das et al., 2017). AMR could be intrinsic or acquired and a wide range of metabolic pathways can confer resistance phenotype in bacteria (Figure 4; Das et al., 2020). Horizontal gene transfer (HGT), de novo mutation and changes in the expression profile of secreted proteins determine the rate of emergence of resistant variants (Davies and Davies, 2010). Alarmone molecule (p)ppGpp induces HGT while bacterial species are living in biofilms (Strugeon et al., 2016). Elevated level of (p)ppGpp derepressed the intI1 promoter (P intI1 ) by inducing autoproteolytic activity of SOS response master regulator LexA, possibly through reducing the activity of exopolyphosphatase enzyme PPX (Strugeon et al., 2016). In normal physiological conditions, LexA dimer reduces the expression of IntI1 integrase by inhibiting P intI1 . Increased expression of integrin integrase due to (p)ppGpp mediated moderate increase of SOS response in biofilm environment helps integron mediated acquisition/exchange of antibiotic resistance gene cassettes through HGT (Figure 4).
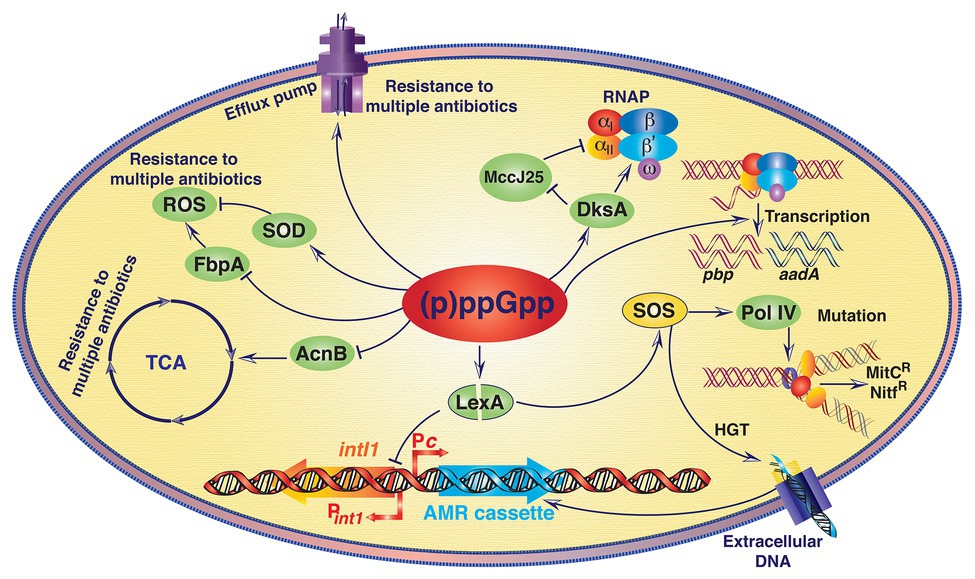
Figure 4. A schematic representation showing the (p)ppGpp mediated antimicrobial resistance (AMR) in bacteria. Except AMR cassette acquisition at the integron integrase locus all other processes that are directly or indirectly associated with antibiotic resistance due to increased level of (p)ppGpp are non-inheritable. (p)ppGpp induces antibiotic resistance in bacteria via distinct mechanisms (see details in text), which are as follows. (i) (p)ppGpp increases the efficiency of horizontal gene transfer (HGT) and AMR cassette acquisition by derepressing the integron integrase 1 (intI1) gene promoter (P intI1 ). Elevated level of (p)ppGpp induces autoproteolytic activity of SOS response master regulator LexA, and thus increases expression of the intI1 gene. In addition, SOS induction also derepressed the expression of error-prone DNA polymerase IV (Pol IV) and upturns accumulation of spontaneous point mutations via DNA replication, which in turn escalates development of resistant variants. (ii) (p)ppGpp upregulates the expression of the aminoglycoside adenylyl transferase gene (aadA), penicillin binding protein coding gene (pbp), and different components of the efflux pumps. (iii) (p)ppGpp helps DksA to compete with microcin J25 (MccJ25) peptide antibiotic for binding with RNA polymerase (RNAP) and confers resistance against this peptide antibiotic. (iv) (p)ppGpp represses the expression of aconitase B (AcnB) and suppresses tricarboxylic acid (TCA) cycle and increases non-inheritable resistance to antibiotics. (v) (p)ppGpp represses the expression of Fe(III) ABC transporter substrate-binding protein (FbpA) and activates superoxide dismutase (SOD) activity. Both the modulations reduce the level of reactive oxygen species (ROS) leading to increase in resistance against tetracycline and other antibiotics. Solid arrows indicate activating interactions and T-arrows indicate inhibiting interactions. Bent arrows at the integron locus indicate promoter for AMR cassette (Pc) and P intI1 . MitCR, Mitomycin C resistance; NitfR, Nitrofurantoin resistance.
It has been observed that bacterial (p)ppGpp0 strains are usually sensitive to diverse xenobiotics, antibiotics, and ultraviolet radiation (Hobbs and Boraston, 2019; and the references therein). Bacterial population that does not carry any specific antibiotic resistance trait or altered target, a small fraction of this population of cells may develop persistence to tolerate lethal doses of antibiotics, which seems to be mediated by the elevated level of intracellular (p)ppGpp. From several recent studies, it is gradually becoming clear that the increased intracellular levels of (p)ppGpp most likely activates the bacterial toxin-antitoxin module, also called TA module, through a complex regulatory mechanism, and the released toxin moiety then helps in maintaining the high intracellular levels of (p)ppGpp and thus, leading to the genesis of persister cells (Korch et al., 2003; Kaspy et al., 2013; Harms et al., 2016). It has been demonstrated recently that (p)ppGpp may play an important role in the nucleotide excision DNA repair process. Absence of (p)ppGpp or its functional partner DksA render bacterial cells highly sensitive to mitomycin C, 4-nitroquinoline 1-oxide and nitrofurantoin, the antibiotics that kill bacterial species by interfering DNA or RNA synthesis (Kamarthapu and Nudler, 2015). Bacterial cells with higher basal level of (p)ppGpp are more resistant to antibiotics that work through interfering nucleic acid biosynthesis or damage. (p)ppGpp directly modulates the activity of RNAP, therefore, it could support UvrD for the repair of damaged DNA through transcription coupled DNA repair pathway. In addition, induction of SOS during stress conditions including exposure to sub-lethal concentration of antibiotics also derepressed the expression of error-prone DNA polymerase IV and increases accumulation of spontaneous mutations, which in turn escalate development of resistant variants (Layton and Foster, 2005). When Salmonella enterica cells are treated with aminoglycoside antibiotics, they showed (p)ppGpp mediated upregulation of expression of the aminoglycoside adenylyl transferase gene (aadA) and thus, resulting in resistance to streptomycin and spectinomycin antibiotics (Koskiniemi et al., 2011).
Increased basal level of (p)ppGpp also contributes in the emergence of antibiotic resistant bacterial cells by directly modulating the expression profile of genes encoding penicillin binding proteins (PBPs) or components of the efflux pumps (Aedo and Tomasz, 2016). Rodionov and Ishiguro (1995) have reported about the importance of (p)ppGpp in inducing resistance against beta-lactam antibiotics namely penicillin. They showed that controlled expression of the relA gene resulted in inhibition of peptidoglycan and phospholipid biosynthesis subsequently resulting in penicillin tolerance. Oxacillin resistance in Staphylococcus aureus is directly associated with increased (p)ppGpp synthetase activity of RelQ or decreasing (p)ppGpp hydrolase activity of the Rel enzyme (Mwangi et al., 2013). Similarly, increase in intracellular (p)ppGpp concentration in E. faecalis has been found to be responsible for the development of non-inheritable resistance against the antibiotic vancomycin (Abranches et al., 2009). Very recently it has been shown that the rel gene deleted Mycobacterium tuberculosis cells become more susceptible to isoniazid killing under nutrient starved condition and also in the lungs of infected mice (Dutta et al., 2019). Thus, it appears that inhibition of Rel may be a useful approach to eliminate M. tuberculosis persister cells. On the other hand, a direct correlation has been found between intracellular (p)ppGpp accumulation and increase in resistance against the peptide antibiotic microcin in E. coli (Pomares et al., 2008). It was hypothesized that (p)ppGpp helps DksA to compete with microcin J25 peptide antibiotic for binding with RNAP (Figure 4). A very recent study showed that (p)ppGpp induces the expression of efflux pump related genes in Acinetobacter baumannii (Jung et al., 2020). MIC of several antibiotics drastically reduced in (p)ppGpp0 A. baumannii strain compared to its wild-type ancestor, possibly due to reduction in expression of antibiotics expelling efflux pumps (Jung et al., 2020). In E. coli, homeostasis of purine nucleosides is regulated by the nucleosidase PpnN by cleaving nucleoside monophosphates. (p)ppGpp binds to the PpnN and stimulates its catalytic activity, which helps cells to tolerate increased concentration of antibiotics (Zhang et al., 2019). The most common antibiotics for the treatment of cholera patients are tetracycline, erythromycin, and chloramphenicol. A recent study has shown that the increased level of intracellular (p)ppGpp helps V. cholerae cells to reduce the efficacy of all these antibiotics, at least under the laboratory testing conditions (Kim et al., 2018). (p)ppGpp metabolic cycle in V. cholerae is fairly unique in comparison to other Gram-negative bacteria (Das et al., 2009). The alarmone helps cholera pathogens for survival, disease development, and antibiotic resistance. Wild-type V. cholerae strain and its (p)ppGpp overproducing isogenic mutant ΔrelAΔspoT can survive better in the presence of different antibiotics compared to (p)ppGpp° ΔrelAΔspoTΔrelV strain. It was hypothesized that (p)ppGpp suppresses tricarboxylic acid cycle by repressing the aconitase B encoding gene acnB and increases non-inheritable resistance to antibiotics (Kim et al., 2018). In addition, (p)ppGpp also represses the expression of iron (III) ABC transporter substrate-binding protein in the wild-type V. cholerae strain, which is important for the generation of reactive oxygen species (ROS) and increased susceptibility to tetracycline (Figure 4). Nevertheless, it has also been reported that the ΔdksA V. cholerae mutants are also highly sensitive to different antibiotics compared to isogenic wild-type strain (Kim et al., 2018). In a murine infection model, substantial increase in the efficacy of several fluoroquinolones against the (p)ppGpp0 P. aeruginosa mutant strain has been noticed compared to that of isogenic wild-type strain (Nguyen et al., 2011). In fact, it has been observed that increased intracellular concentration of (p)ppGpp may modulate non-inheritable resistance to different antibiotics possibly through regulation of bacterial ROS production. Several studies have demonstrated that the high intracellular levels of (p)ppGpp is associated with the antioxidant defense mechanisms in P. aeruginosa (Nguyen et al., 2011; Martins et al., 2018). Superoxide dismutase (SOD), an enzyme that catalyzes the dismutation of the superoxide (O2 −) radical into ordinary molecular oxygen (O2) and hydrogen peroxide (H2O2), is a key enzyme involved in stringent response mediated drug resistance in stationary phase cells of P. aeruginosa (Martins et al., 2018). It has been shown that deletions of (p)ppGpp biosynthetic genes reduce the SOD activity and diminish drug tolerance phenotypes in stationary phase cells of P. aeruginosa. Complementation of SOD activity in the (p)ppGpp0 P. aeruginosa was able to restore the non-inheritable resistance against several antibiotics almost similar to that of isogenic wild-type strain. The authors have also found that in the absence of SOD activity, the membrane permeability of P. aeruginosa cells is high, which leads to increase in drug internalization and thus raising the efficacy of antibiotics.
The conserved distribution of stringent response across the bacterial phyla and its important roles in developing persistent bacterial infections made it a promising drug target. Subpopulation of bacteria can survive upon exposure to lethal concentration of antibiotic through growth arrest, while others are not (Balaban et al., 2004). Persister bacteria exhibit non-inheritable resistance to the bactericidal antibiotics (Dutta et al., 2019). It is well known that the stringent response effector molecules (p)ppGpp plays an important role in fine-tuning bacterial growth during nutritional stress and exposure to antibiotics. It helps bacterial pathogens to survive during growth-limiting conditions, to increase non-inheritable resistance and to develop persister cells for prolong infection (Abranches et al., 2009; Dutta et al., 2019). The major bacterial persistent infections are usually caused by M. tuberculosis, Helicobacter pylori, Salmonella Typhi, and several others, where the pathogens can overcome the antimicrobial effect of routinely used antimicrobials (Kester and Fortune, 2014). As discussed above, several studies have indicated that the stringent response plays a pivotal role in developing non-inheritable resistance and persister cell formation. Therefore, in recent years’ attempts have been made to develop stringent response inhibitors to block long-term persistence, sporulation, and biofilm formation with a hope that blocking such important cellular functions may help in increasing the efficacy of antimicrobial drugs (Wexselblatt et al., 2012, 2013; Syal et al., 2017a,b). Thus, a number of potential stringent response inhibitory molecules, namely, 2'-deoxyguanosine-3'-5'-di(methylene bisphosphonate), relacin, relacin analog 2d, vitamin C, etc., have been developed and shown to be effective against B. subtilis, Bacillus anthracis, M. smegmatis, and E. coli both under in vitro and in vivo growth conditions (Wexselblatt et al., 2012, 2013; Syal et al., 2017a,b). Relacin, a well-studied ppGpp analog in which 3'-pyrophosphate moieties were substituted with glycyl-glycine dipeptide, inhibits (p)ppGpp synthetic activity of Rel/RelA proteins of both Gram-positive and Gram-negative bacteria under in vitro conditions. It has been demonstrated that relacin reduces bacterial viability by preventing the cells entering into stationary growth phase (Wexselblatt et al., 2012). Thus, it seems that inhibiting the (p)ppGpp synthesis activity in pathogens by stringent response inhibitor is a potential approach to reduce viability of persister and shortening the duration of antibiotic treatment of a patient. Development of such promising inhibitors against RelA/SpoT/RSH enzymes and its use as antibiotic adjuvants can potentially help in eradicating several chronic bacterial infections.
Although the (p)ppGpp metabolic pathway was originally discovered in bacteria about 50 years ago, continued research on this important molecule has made it clear that these alarmones are widely synthesized by the living organisms from all three domains of life. Enzymes involved in the (p)ppGpp metabolism and signals that activate or repress stringent response share substantial similarity between different organisms. Overwhelming evidences established that other than nutrient limitations, antibiotics, and non-antibiotic xenobiotics activate stringent response and help microbes to survive even in the absence of specific resistance genes. In addition to the regulation of DNA replication, RNA transcription and protein synthesis, (p)ppGpp molecules have a wide range of heterogeneous targets depending on the organisms and their habitats. It is becoming clear that the modulations of the activities of several such targets/proteins are linked with the non-heritable resistance to antibiotics. The genetic program linked with the bacterial antibiotic resistance in the absence of specific resistance genes appears to have multiple components. Activity of several such molecules intricately depends on the alarmone (p)ppGpp and other small signaling molecules, which warrants further in depth research for effective management of infectious diseases.
BD and RKB designed the review, done literature search, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study received financial support from the Dept. of Biotechnology (DBT), Govt. of India (no. BT/PR9983/MED/97/194/2013), Translational Health Science and Technology Institute core funds (2019-2020) and partial support from MLP118 laboratory project of CSIR-IICB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors acknowledge Prof. G. Balakrish Nair and Dr. Amit Ghosh for critical reading and valuable suggestions.
Abranches, J., Martinez, A. R., Kajfasz, J. K., Chavez, V., Garsin, D. A., and Lemos, J. A. (2009). The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191, 2248–2256. doi: 10.1128/JB.01726-08
Aedo, S., and Tomasz, A. (2016). Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 2311–2317. doi: 10.1128/AAC.02697-15
Angelini, S., My, L., and Bouveret, E. (2012). Disrupting the acyl carrier protein/SpoT interaction in vivo: identification of ACP residues involved in the interaction and consequence on growth. PLoS One 7:e36111. doi: 10.1371/journal.pone.0036111
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L., and Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. doi: 10.1126/science.1099390
Cashel, M. (1969). The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244, 3133–3141.
Cashel, M., Gentry, D. R., Hernandez, V. J., and Vinella, D. (1996). “The stringent response” in Escherichia coli and Salmonella: Cellular and molecular biology. ed. F. C. Neidhardt (Washington, DC: ASM Press), 1458–1496.
Corrigan, R. M., Bellows, L. E., Wood, A., and Grundling, A. (2016). ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 113, E1710–E1719. doi: 10.1073/pnas.1522179113
Crawford, E. W. Jr., and Shimkets, L. J. (2000). The stringent response in Myxococcus xanthus is regulated by SocE and the CsgA C-signaling protein. Genes Dev. 14, 483–492.
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C., and Swanson, M. S. (2010). ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199. doi: 10.1128/MMBR.00046-09
Das, B., and Bhadra, R. K. (2008). Molecular characterization of Vibrio cholerae ΔrelA ΔspoT double mutants. Arch. Microbiol. 189, 227–238. doi: 10.1007/s00203-007-0312-z
Das, B., Chaudhuri, S., Srivastava, R., Nair, G. B., and Ramamurthy, T. (2017). Fostering research into antimicrobial resistance in India. BMJ 358:j3535. doi: 10.1136/bmj.j3535
Das, B., Pal, R. R., Bag, S., and Bhadra, R. K. (2009). Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol. Microbiol. 72, 380–398. doi: 10.1111/j.1365-2958.2009.06653.x
Das, B., Verma, J., Kumar, P., Ghosh, A., and Ramamurthy, T. (2020). Antibiotic resistance in Vibrio cholerae: understanding the ecology of resistance genes and mechanisms. Vaccine 38(Suppl. 1), A83–A92. doi: 10.1016/j.vaccine.2019.06.031
Dasgupta, S., Basu, P., Pal, R. R., Bag, S., and Bhadra, R. K. (2014). Genetic and mutational characterization of the small alarmone synthetase gene relV of Vibrio cholerae. Microbiology 160, 1855–1866. doi: 10.1099/mic.0.079319-0
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Denapoli, J., Tehranchi, A. K., and Wang, J. D. (2013). Dose-dependent reduction of replication elongation rate by (p)ppGpp in Escherichia coli and Bacillus subtilis. Mol. Microbiol. 88, 93–104. doi: 10.1111/mmi.12172
Dutta, N. K., Klinkenberg, L. G., Vazquez, M. J., Segura-Carro, D., Colmenarejo, G., Ramon, F., et al. (2019). Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 5:eaav2104. doi: 10.1126/sciadv.aav2104
English, B. P., Hauryliuk, V., Sanamrad, A., Tankov, S., Dekker, N. H., and Elf, J. (2011). Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl. Acad. Sci. U. S. A. 108, E365–E373. doi: 10.1073/pnas.1102255108
Fischbach, M. A., and Walsh, C. T. (2009). Antibiotics for emerging pathogens. Science 325, 1089–1093. doi: 10.1126/science.1176667
Gaca, A. O., Colomer-Winter, C., and Lemos, J. A. (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. doi: 10.1128/JB.02577-14
Gaca, A. O., Kajfasz, J. K., Miller, J. H., Liu, K., Wang, J. D., Abranches, J., et al. (2013). Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. mBio 4, e00646–e00713. doi: 10.1128/mBio.00646-13
Gallant, J., Palmer, L., and Pao, C. C. (1977). Anomalous synthesis of ppGpp in growing cells. Cell 11, 181–185. doi: 10.1016/0092-8674(77)90329-4
Gomez-Escribano, J. P., Martin, J. F., Hesketh, A., Bibb, M. J., and Liras, P. (2008). Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154, 744–755. doi: 10.1099/mic.0.2007/011890-0
Gourse, R. L., Chen, A. Y., Gopalkrishnan, S., Sanchez-Vazquez, P., Myers, A., and Ross, W. (2018). Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184. doi: 10.1146/annurev-micro-090817-062444
Gourse, R. L., and Keck, J. L. (2007). Magic spots cast a spell on DNA primase. Cell 128, 823–824. doi: 10.1016/j.cell.2007.02.020
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
He, H., Cooper, J. N., Mishra, A., and Raskin, D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972. doi: 10.1128/JB.00014-12
Hesketh, A., Chen, W. J., Ryding, J., Chang, S., and Bibb, M. (2007). The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. doi: 10.1186/gb-2007-8-8-r161
Hobbs, J. K., and Boraston, A. B. (2019). (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect. Dis. 5, 1505–1517. doi: 10.1021/acsinfecdis.9b00204
Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004). Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117, 57–68. doi: 10.1016/s0092-8674(04)00260-0
Irving, S. E., and Corrigan, R. M. (2018). Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology 164, 268–276. doi: 10.1099/mic.0.000621
Ito, D., Kato, T., Maruta, T., Tamoi, M., Yoshimura, K., and Shigeoka, S. (2012). Enzymatic and molecular characterization of arabidopsis ppGpp pyrophosphohydrolase, AtNUDX26. Biosci. Biotechnol. Biochem. 76, 2236–2241. doi: 10.1271/bbb.120523
Jin, W., Kim, H. K., Kim, J. Y., Kang, S. G., Lee, S. H., and Lee, K. J. (2004). Cephamycin C production is regulated by relA and rsh genes in Streptomyces clavuligerus ATCC27064. J. Biotechnol. 114, 81–87. doi: 10.1016/j.jbiotec.2004.06.010
Jung, H. W., Kim, K., Islam, M. M., Lee, J. C., and Shin, M. (2020). Role of ppGpp-regulated efflux genes in Acinetobacter baumannii. J. Antimicrob. Chemother. 75, 1130–1134. doi: 10.1093/jac/dkaa014
Kalia, D., Merey, G., Nakayama, S., Zheng, Y., Zhou, J., Luo, Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. doi: 10.1039/c2cs35206k
Kamarthapu, V., Epshtein, V., Benjamin, B., Proshkin, S., Mironov, A., Cashel, M., et al. (2016). ppGpp couples transcription to DNA repair in E. coli. Science 352, 993–996. doi: 10.1126/science.aad6945
Kamarthapu, V., and Nudler, E. (2015). Rethinking transcription coupled DNA repair. Curr. Opin. Microbiol. 24, 15–20. doi: 10.1016/j.mib.2014.12.005
Kang, P. J., and Craig, E. A. (1990). Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 172, 2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990
Kang, S. G., Jin, W., Bibb, M., and Lee, K. J. (1998). Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3(2) grown in continuous culture. FEMS Microbiol. Lett. 168, 221–226. doi: 10.1111/j.1574-6968.1998.tb13277.x
Kanjee, U., Ogata, K., and Houry, W. A. (2012). Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85, 1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x
Kaspy, I., Rotem, E., Weiss, N., Ronin, I., Balaban, N. Q., and Glaser, G. (2013). HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4:3001. doi: 10.1038/ncomms4001
Kester, J. C., and Fortune, S. M. (2014). Persisters and beyond: mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit. Rev. Biochem. Mol. Biol. 49, 91–101. doi: 10.3109/10409238.2013.869543
Kim, H. Y., Go, J., Lee, K. M., Oh, Y. T., and Yoon, S. S. (2018). Guanosine tetra- and pentaphosphate increase antibiotic tolerance by reducing reactive oxygen species production in Vibrio cholerae. J. Biol. Chem. 293, 5679–5694. doi: 10.1074/jbc.RA117.000383
Korch, S. B., Henderson, T. A., and Hill, T. M. (2003). Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x
Koskiniemi, S., Pränting, M., Gullberg, E., Näsvall, J., and Andersson, D. I. (2011). Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol. Microbiol. 80, 1464–1478. doi: 10.1111/j.1365-2958.2011.07657.x
Kriel, A., Bittner, A. N., Kim, S. H., Liu, K., Tehranchi, A. K., Zou, W. Y., et al. (2012). Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48, 231–241. doi: 10.1016/j.molcel.2012.08.009
Layton, J. C., and Foster, P. L. (2005). Error-prone DNA polymerase IV is regulated by the heat shock chaperone GroE in Escherichia coli. J. Bacteriol. 187, 449–457. doi: 10.1128/JB.187.2.449-457.2005
Lemos, J. A., Lin, V. K., Nascimento, M. M., Abranches, J., and Burne, R. A. (2007). Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65, 1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x
Li, W., Bouveret, E., Zhang, Y., Liu, K., Wang, J. D., and Weisshaar, J. C. (2016). Effects of amino acid starvation on RelA diffusive behavior in live Escherichia coli. Mol. Microbiol. 99, 571–585. doi: 10.1111/mmi.13252
Lyzen, R., Maitra, A., Milewska, K., Kochanowska-Lyzen, M., Hernandez, V. J., and Szalewska-Palasz, A. (2016). The dual role of DksA protein in the regulation of Escherichia coli pArgX promoter. Nucleic Acids Res. 44, 10316–10325. doi: 10.1093/nar/gkw912
Magnusson, L. U., Gummesson, B., Joksimovic, P., Farewell, A., and Nystrom, T. (2007). Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 189, 5193–5202. doi: 10.1128/JB.00330-07
Martins, D., McKay, G., Sampathkumar, G., Khakimova, M., English, A. M., and Nguyen, D. (2018). Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 115, 9797–9802. doi: 10.1073/pnas.1804525115
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S., and Cashel, M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189. doi: 10.1093/nar/gkt302
Murdeshwar, M. S., and Chatterji, D. (2012). MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J. Bacteriol. 194, 4003–4014. doi: 10.1128/JB.00258-12
Mwangi, M. M., Kim, C., Chung, M., Tsai, J., Vijayadamodar, G., Benitez, M., et al. (2013). Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Resist. 19, 153–159. doi: 10.1089/mdr.2013.0053
Nanamiya, H., Kasai, K., Nozawa, A., Yun, C. S., Narisawa, T., Murakami, K., et al. (2008). Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67, 291–304. doi: 10.1111/j.1365-2958.2007.06018.x
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. doi: 10.1126/science.1211037
Ochi, K. (1987). A rel mutation abolishes the enzyme induction needed for actinomycin synthesis by Streptomyces antibioticus. Agric. Biol. Chem. 51, 829–835. doi: 10.1271/bbb1961.51.829
Pal, R. R., Bag, S., Dasgupta, S., Das, B., and Bhadra, R. K. (2012). Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J. Bacteriol. 194, 5638–5648. doi: 10.1128/JB.00518-12
Pant, A., Bag, S., Saha, B., Verma, J., Kumar, P., Banerjee, S., et al. (2020). Molecular insights into the genome dynamics and interactions between core and acquired genomes of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. doi: 10.1073/pnas.2006283117 [Epub ahead of print]
Parker, J., Watson, R. J., and Friesen, J. D. (1976). A relaxed mutant with an altered ribosomal protein L11. Mol. Gen. Genet. 144, 111–114. doi: 10.1007/BF00277313
Paul, B. J., Barker, M. M., Ross, W., Schneider, D. A., Webb, C., Foster, J. W., et al. (2004). DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322. doi: 10.1016/j.cell.2004.07.009
Perederina, A., Svetlov, V., Vassylyeva, M. N., Tahirov, T. H., Yokoyama, S., Artsimovitch, I., et al. (2004). Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118, 297–309. doi: 10.1016/j.cell.2004.06.030
Pomares, M. F., Vincent, P. A., Farias, R. N., and Salomon, R. A. (2008). Protective action of ppGpp in microcin J25-sensitive strains. J. Bacteriol. 190, 4328–4334. doi: 10.1128/JB.00183-08
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Potrykus, K., Murphy, H., Philippe, N., and Cashel, M. (2011). ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575. doi: 10.1111/j.1462-2920.2010.02357.x
Rodionov, D. G., and Ishiguro, E. E. (1995). Direct correlation between overproduction of guanosine 3', 5'-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J. Bacteriol. 177, 4224–4229. doi: 10.1128/jb.177.15.4224-4229.1995
Ronneau, S., and Hallez, R. (2019). Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol. Rev. 43, 389–400. doi: 10.1093/femsre/fuz009
Ross, W., Vrentas, C. E., Sanchez-Vazquez, P., Gaal, T., and Gourse, R. L. (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429. doi: 10.1016/j.molcel.2013.03.021
Ruwe, M., Ruckert, C., Kalinowski, J., and Persicke, M. (2018). Functional characterization of a small alarmone hydrolase in Corynebacterium glutamicum. Front. Microbiol. 9:916. doi: 10.3389/fmicb.2018.00916
Sanchez-Vazquez, P. (2018). Genome-wide effects of ppGpp binding to RNA polymerase on E. coli gene expression. [PhD Thesis]. USA: University of Wisconsin-Madison.
Sivapragasam, S., and Grove, A. (2019). The link between purine metabolism and production of antibiotics in Streptomyces. Antibiotics 8:76. doi: 10.3390/antibiotics8020076
Storvik, K. A., and Foster, P. L. (2010). RpoS, the stress response sigma factor, plays a dual role in the regulation of Escherichia coli’s error-prone DNA polymerase IV. J. Bacteriol. 192, 3639–3644. doi: 10.1128/JB.00358-10
Strugeon, E., Tilloy, V., Ploy, M. C., and Da Re, S. (2016). The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7, e00868–e00916. doi: 10.1128/mBio.00868-16
Sun, D., Lee, G., Lee, J. H., Kim, H. Y., Rhee, H. W., Park, S. Y., et al. (2010). A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 17, 1188–1194. doi: 10.1038/nsmb.1906
Syal, K., Bhardwaj, N., and Chatterji, D. (2017b). Vitamin C targets (p)ppGpp synthesis leading to stalling of long-term survival and biofilm formation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 364:fnw282. doi: 10.1093/femsle/fnw282
Syal, K., Flentie, K., Bhardwaj, N., Maiti, K., Jayaraman, N., Stallings, C. L., et al. (2017a). Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob. Agents Chemother. 61, e00443–e00517. doi: 10.1128/AAC.00443-17e00443-17
Teschler, J. K., Zamorano-Sanchez, D., Utada, A. S., Warner, C. J., Wong, G. C., Linington, R. G., et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268. doi: 10.1038/nrmicro3433
Tozawa, Y., and Nomura, Y. (2011). Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol. 13, 699–709. doi: 10.1111/j.1438-8677.2011.00484.x
van der Heul, H. U., Bilyk, B. L., McDowall, K. J., Seipke, R. F., and van Wezel, G. P. (2018). Regulation of antibiotic production in Actinobacteria: new perspectives from the post- genomic era. Nat. Prod. Rep. 35, 575–604. doi: 10.1039/c8np00012c
van der Meij, A., Worsley, S. F., Hutchings, M. I., and van Wezel, G. P. (2017). Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416. doi: 10.1093/femsre/fux0005
Verma, J., Bag, S., Saha, B., Kumar, P., Ghosh, T. S., Dayal, M., et al. (2019). Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 116, 6226–6231. doi: 10.1073/pnas.1900141116
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15:756. doi: 10.1038/s41589-019-0296-4
Wang, J. D., Sanders, G. M., and Grossman, A. D. (2007). Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–875. doi: 10.1016/j.cell.2006.12.043
Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A., and Nierhaus, K. H. (2002). Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788. doi: 10.1016/s1097-2765(02)00656-1
Wexselblatt, E., Kaspy, N., Glaser, G., Katzhendler, J., and Yavin, E. (2013). Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur. J. Med. Chem. 70, 497–504. doi: 10.1016/j.ejmech.2013.10.036
Wexselblatt, E., Oppenheimer-Shaanan, Y., Kaspy, I., London, N., Schueler-Furman, O., Yavin, E., et al. (2012). Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog. 8:e1002925. doi: 10.1371/journal.ppat.1002925
Wolz, C., Geiger, T., and Goerke, C. (2010). The synthesis and function of the alarmone (p)ppGpp in firmicutes. Int. J. Med. Microbiol. 300, 142–147. doi: 10.1016/j.ijmm.2009.08.017
Wu, J., Long, Q., and Xie, J. (2010). (p)ppGpp and drug resistance. J. Cell. Physiol. 224, 300–304. doi: 10.1002/jcp.22158
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990.
Zhang, Y. E., Baerentsen, R. L., Fuhrer, T., Sauer, U., Gerdes, K., and Brodersen, D. E. (2019). (p)ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Mol. Cell 74, 1239–1249. doi: 10.1016/j.molcel.2019.03.035
Keywords: bacterial pathogen, antibiotic resistance, spoT, relA, stringent response, (p)ppGpp, alarmone
Citation: Das B and Bhadra RK (2020) (p)ppGpp Metabolism and Antimicrobial Resistance in Bacterial Pathogens. Front. Microbiol. 11:563944. doi: 10.3389/fmicb.2020.563944
Received: 20 May 2020; Accepted: 09 September 2020;
Published: 09 October 2020.
Edited by:
Michael Cashel, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), United StatesReviewed by:
Agnieszka Szalewska-Pałasz, University of Gdansk, PolandCopyright © 2020 Das and Bhadra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhabatosh Das, YmhhYmF0b3NoQHRoc3RpLnJlcy5pbg==; Rupak K. Bhadra, cnVwYWtiaGFkcmFAaWljYi5yZXMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.