
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 18 September 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.556227
This article is part of the Research TopicPlasmid Transfer: Mechanisms, Ecology, Evolution, and ApplicationsView all 34 articles
Resistance to ciprofloxacin, a treatment choice for Salmonella infections, has increased dramatically in recent years in particular in serotype Salmonella Derby with most of strains carrying chromosome-encoded multiple plasmid-mediated quinolone resistance (PMQR) genes. In this work, we discovered a conjugative plasmid, pSa64-96kb, in a Salmonella Derby isolate, namely Sa64, which could extract and fuse to a multiple drug resistance (MDR) DNA fragment containing two PMQR genes, aac(6’)-Ib-cr, and qnrS2 located on the chromosome of the Salmonella strain. This process led to the formation of a new 188 kb fusion plasmid, which could be then subsequently transmitted to recipient strain Escherichia coli J53. The chromosomal MDR DNA fragment was shown to be flanked by one copy of IS26 element at each end and could be excised from the chromosome to form circular intermediate, which was then fused to pSa64-96kb and form a single plasmid through IS26 mediated homologous recombination. The role of IS26 on enhancing the efficacy of fusion and transmission of this chromosomal MDR DNA fragment was further proven in other Salmonella strains. These findings showed that dynamic interaction between specific chromosomal fragment and plasmids may significantly enhance resistance development and transferability of mobile resistance-encoding elements among bacterial pathogens.
Non-typhoidal Salmonella represents the primary bacterial causes of food-borne gastroenteritis throughout the world (Gomez et al., 1997). Antibiotics are unessential for treating salmonellosis, yet it can be lifesaving in cases of serious or systemic infections (Hohmann, 2001). Ciprofloxacin has been the agent of choice in treatment of Salmonella infection in adults. However, mounting evidences indicated the increasing prevalence of non-typhoidal Salmonella isolates exhibiting multidrug resistance phenotypes has significantly compromised the efficacy of present strategies applied to prevent and administer diseases related to food-borne infections (Molbak, 2005). Quinolone and fluoroquinolone resistance in Salmonella have been low and are commonly associated with a single or double mutations in the gyrA and/or parC gene (Hooper, 2001; Chen et al., 2004). Plasmid-mediated quinolone resistance (PMQR) genes have been described in recent years, which contribute to the extension of low-level quinolone resistance in Enterobacteriaceae. Three patterns of PMQR determinates have been described to date: (1) the Qnr types, which are mainly classified into seven different families and encoded pentapeptide duplicate proteins binding to DNA gyrase through imitating double stranded DNA, avoiding the binding of fluoroquinolones to gyrase; (2) the aac(6’)-Ib-cr, a modified aminoglycoside acetyltransferase that could hydrolyze fluoroquinolones; and (3) the efflux pumps QepA and OqxAB. However, detection of these PMQR genes in Salmonella remains rare before 2009, with the first detection of qnrA and qnrS genes being reported in Europe (Gunell et al., 2009).
In recent years, the percentage of resistance to ciprofloxacin has grown drastically in both environmental and clinical collections throughout the world, particularly in China and surrounding regions (Wong et al., 2014b). An interesting phenomenon is that the majority of ciprofloxacin-resistant Salmonella isolates displayed relatively low resistance levels (MIC < 8 μg/ml) to ciprofloxacin, with the proportion of strains exhibiting high resistance (MIC > 32 μg/ml) being very small and mostly belonging to the serotype S. Indiana. Strikingly, a significant proportion of these ciprofloxacin-resistant strains did not carry any mutations in the target genes. Instead, various types of PMQR genes were detectable in these strains. The oqxAB and aac(6’)-Ib-cr genes are usually carried by the same isolate, which might be related to the increased frequency of ciprofloxacin resistance in nosocomial Salmonella isolates recently (Wong et al., 2014a). Other PMQR genes such as qnrS was also increasingly reported in Salmonella (Lin et al., 2015). These PMQR determinates are commonly detected in the chromosomes and non-conjugative plasmids in S. Derby. Recently, conjugative ciprofloxacin resistance has been reported, including (1) conjugative plasmids encoding PMQR genes, and (2) helper plasmids mediating transmission of non-conjugative plasmids harboring multiple PMQR determinants, which might accelerate the further dissemination of resistance to ciprofloxacin in Salmonella (Chen et al., 2018, 2019a,b).
Here, we report the characterization of an IncI1 type of plasmid, which can excise the chromosomal DNA fragment containing multiple PMQR genes encoding ciprofloxacin resistance and transfer it to other bacteria through conjugation. Such recurrent genetic events could significantly increase the transferability of ciprofloxacin resistance genes among Salmonella and other Enterobacteriaceae species, potentially leading to a sharp increase in ciprofloxacin resistance in a range of bacterial pathogens in the near future.
Salmonella strains Sa64 and Sa79 were isolated from a pork sample in supermarket in Shenzhen, China, in 2014 as described in our previous study on Salmonella surveillance in Shenzhen, China (Chen et al., 2018). The strains were confirmed to be Salmonella by MALDI-TOF MS and serotyped with commercial antiserum (Difco, Detroit, MI, United States) through the Kauffmann–White scheme. Antimicrobial-resistance phenotypes of these strains against a variety of antimicrobial agents (Table 1) were conducted with agar dilution and explained in agreement with the CLSI guidelines (CLSI, 2016). Quality control strains were Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922.
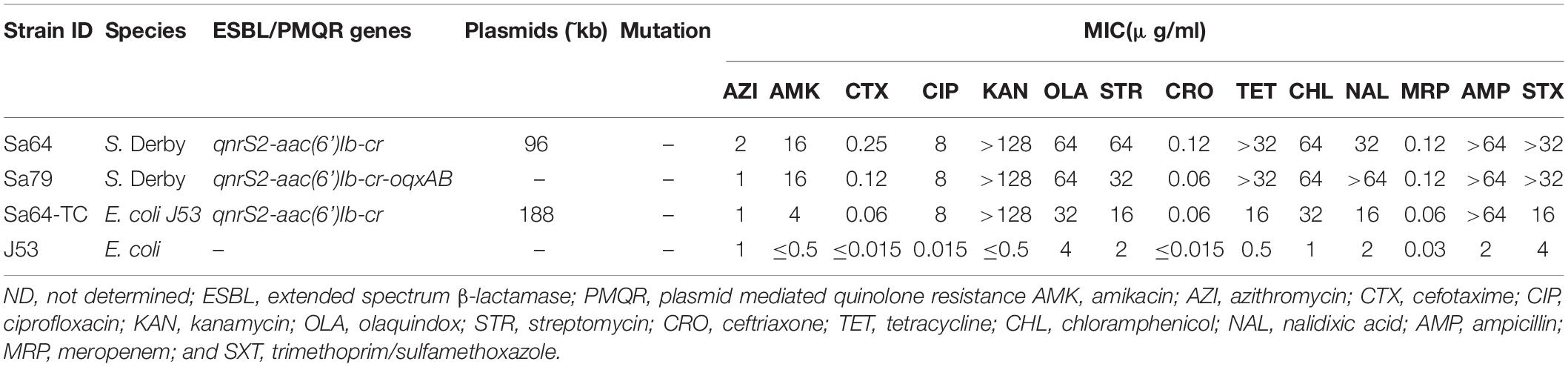
Table 1. Genetic and phenotypic characteristic of ciprofloxacin-resistant Salmonella isolates Sa64, Sa79, and its corresponding transconjugants.
Conjugation experiments were performed following a previous study (Li et al., 2015a) to test the transferability of the aac(6’)-Ib-cr- qnrS2 bearing plasmid, using a sodium-azide-resistance E. coli J53 as the recipient. Briefly, overnight incubation of Sa64 and strain E. coli J53 were blended in a rate of 1:4 in Luria Bertani (LB) nutrient broth, which was then exposed to overnight culture on an LB agar media. The admixture was then disseminated on a selective Eosin Methylene Blue agar plate supplemented with ciprofloxacin (0.5 mg/L) and sodium azide (100 mg/L) to pick out transconjugants that had obtained the PMQR-carrying plasmid. And other conjugation experiments were performed in the similar procedure.
Salmonella Derby strains Sa64 and its corresponding transconjugants were subjected to S1-PFGE to obtain the length of the plasmids. Briefly, S1-nuclease was used to digest agarose embedded DNA at 37°C for 15 min. The restriction bands were dispersed by using a Chef Mapper electrophoresis system (Bio-Rad, Hercules, CA, United States) with 2.16 to 63.8 S pulse times in 0.5 Tris–borate-EDTA buffer at 14°C. DNA fingerprinting of H9812 was used as DNA dimension marker. The gels were subjected to GelRed staining, and DNA bands were imaged with UV transillumination (Bio-Rad). Southern bolt hybridization was conducted according to the manufacturer’s directions of the Detection Starter Kit II (Roche Diagnostics), using the digoxigenin-labeled qnrS gene probe.
Whole genome sequencing was conducted to acquire comprehensive understanding of the ciprofloxacin PMQR genes harbored by Sa64, Sa79, and Sa64-TC with the Illumina HiSeq 2500 sequencing, Nanopore MinION long-read sequencing. Genome sequence was assembled with SPAdes 3.12.1 (Bankevich et al., 2012). Long contigs assembled from Nanopore was used to align and join contigs obtained from Illumina assembly with the CLC Genomics Workbench v10 (CLC bio, Denmark). The complete genome sequence was annotated using the RAST server (Overbeek et al., 2013). Multilocus sequence typing (MLST) service of the Center for Genomic Epidemiology was used to test Sequence type (ST) typing and screen PMQR-genes as described previously (Chen et al., 2007). EasyFig and BRIG were used to compare genome contents, and plot linear and circular plasmid maps, respectively. A polymerase chain reaction (PCR) assays was performed as described previously (Li et al., 2015b) to check the circular intermediate status of chromosomally encoded multiple drug resistance (MDR) fragment using primer sets listed in Supplementary Table 1.
Our surveillance project on ciprofloxacin resistance in Salmonella in food samples collected from the wet markets and supermarkets in Shenzhen, China, allowed us to identify a Salmonella isolate that could transfer its phenotype of ciprofloxacin resistance via conjugation. S1-PFGE analysis was then conducted on this strain and its transconjugant showing that there was only one plasmid with the size of 96 kb in strain Sa64, whereas a larger plasmid of 188 kb was detectable in the transconjugant Sa64-TC, suggesting that the plasmid in strain Sa64 acquired an extra DNA fragment during the conjugation process. This finding prompted us to investigate the molecular mechanism underlying the conjugative transmission of ciprofloxacin resistance in Salmonella. Strain Sa64 was found to belong to ST40 and exhibited resistance to a wide range of antimicrobial agents, containing ciprofloxacin, kanamycin, tetracycline, and Sulfamethoxazole-methoxazole, but remained sensitive to cephalosporin and meropenem. The ciprofloxacin-resistance (CipR) phenotype of this strain was able to be transferred through conjugation. Mutations were not detected in the four target genes, parC, gyrA, gyrB, and parE. PCR screening of known PMRQ genes showed that this strain harbored two PMQR determinants, aac(6’)Ib-cr, and qnrS2, which were also detectable in the transconjugants (Table 1). Southern hybridization using the qnrS2 probe suggested that the qnrS2 gene was located in the chromosome of strain Sa64 and the 188 kb plasmid in the transconjugant Sa64-TC, respectively, indicating that the ciprofloxacin-resistant genes were originally existed on the chromosome of strain Sa64 and then captured by the 96 kb plasmid and transmitted to E. coli J53 through conjugation (Figure 1). Interestingly, another CipR isogenic Salmonella strain, namely S. Derby Sa79, sharing the similar structure in both chromosome (Supplementary Figure 1) and chromosomal MDR DNA fragment (Supplementary Figure 2A) but not plasmid, does not show the potential ability to transfer its ciprofloxacin resistance to the recipient strain E. coli J53 in this study.
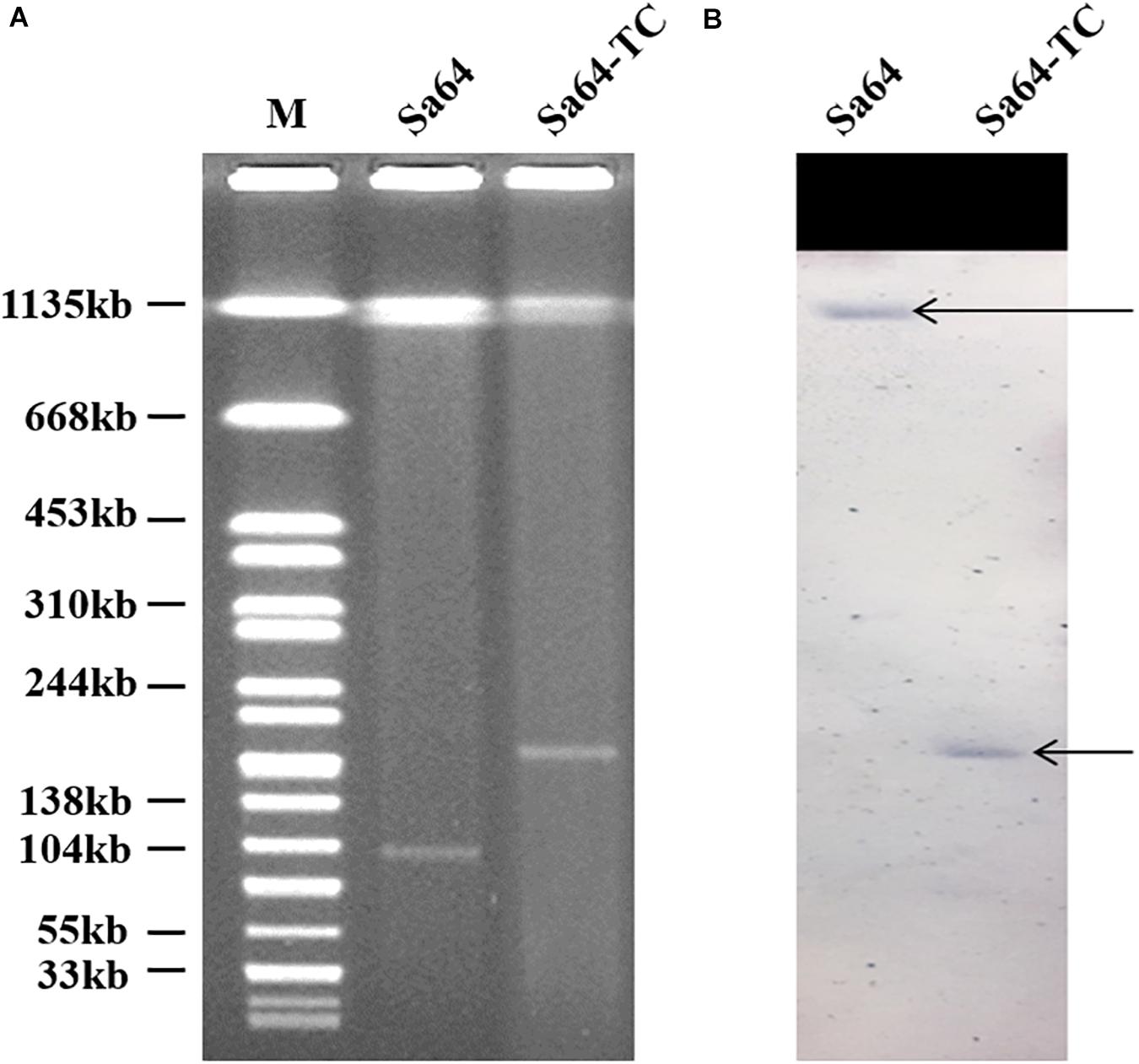
Figure 1. S1-PFGE of ciprofloxacin-resistant Salmonella strain Sa64, and its corresponding transconjugants. (A) S1-PFGE patterns of Salmonella isolate Sa64 and its corresponding transconjugant; (B) Southern blot hybridization results of Salmonella isolate Sa64 and its relevant transconjugant. The arrow depicts the position of chromosomal DNA and plasmid band in which positive hybridization semaphore of the qnrS gene was detectable.
To investigate the mechanism underlying the transfer of ciprofloxacin resistance from Sa64 to E. coli strain J53, the complete sequences of the chromosome and the 96 kb plasmid in strain Sa64, as well as the ∼188 kb plasmid from the transconjugant Sa64-TC were obtained by using both Nanopore and Illumina sequencing platforms. The chromosome of Sa64 was found to be 4,824,198 bp in length with a Guanine and Cytosine (GC) content of 52.1%, and comprise 5,253 predicted coding sequences. BLANTN analysis showed that it exhibited high homology (92% coverage and 99% identity) to the chromosome of Salmonella strain ST350 (CP019407, recovered from a Salmonella enterica subsp. enterica serovar Borreze strain) isolated several decades ago from pig feces. Comparative genomic analysis showed that the chromosome of Sa64 had acquired an extra DNA fragment when compared to strain ST350 (Supplementary Figure 1). This extra DNA fragment was shown to be 92,690 bp in size and exhibit a GC content of 51.3%. BLAST of this DNA fragment showed that it displayed high homology (99% identity with 75% and 81% coverage, respectively) to other MDR-encoding plasmids such as pSTM6-275 (CP019647) and pbl10-220 (CP025340; Figure 2A), both of which were isolated from Salmonella strains in pig feces. These data suggest that this ∼92 kb DNA fragment in the chromosome of Sa64 might be acquired through integration of an MDR plasmid into the chromosome. Analysis of Nanopore reads of strain Sa64 has identified ten reads covering two fusion regions between chromosome and the ∼92 kb DNA fragment, which confirmed the presence of MDR plasmid in chromosome in strain Sa64 (Supplementary Figure 3). Sequence analysis showed that this DNA fragment contained a variety of antimicrobial resistance genes surrounded by different IS elements, such as blaOXA, qnrS2, floR, aac(6’)-Ib-cr, tet(A), and aph genes, and some other resistance gene cassettes surrounded by diverse mobile elements. In this fragment, resistance genes that might contribute to the ciprofloxacin resistance phenotype were found to be located in a complex mobile element comprising various genes, with a structure of IS26-aac(6’)Ib-cr-carB-arr3-emrE-IS6-qnrS2-IS26, suggesting that IS26 played a critical role in the formation of this mobile element and subsequent integration into the chromosome of strain Sa64, with the potential of being reacquired by other plasmids harbored by the host strain.
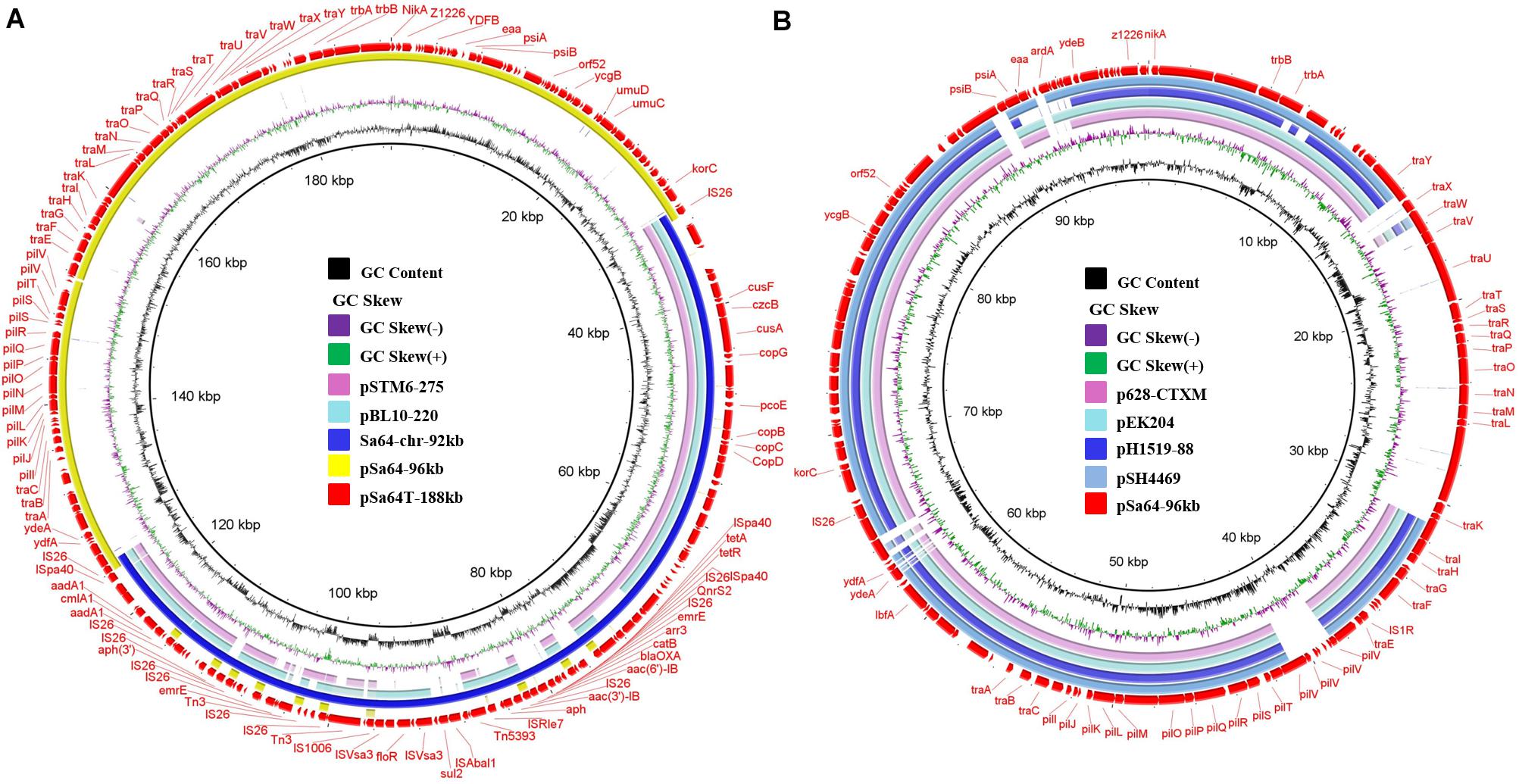
Figure 2. Circular alignment of plasmid pSa64-188kb and the pSa64-96kb. (A) The red circle depicts plasmid pSa64-188kb, which is used as reference; basic genetic loci in this plasmid are tagged. The light pink and green circle, respectively, indicate pSTM6-275 (275,801 bp, CP019647.1) and pBL10-220k (220,231 bp, CP025340.1), respectively; the circle in deep blue color represents DNA fragament Sa64-chr-92k from chromosomal of Sa64 and yellow circle represents pSa64-96kb in this study; (B) The light pink and blue circle represent plasmid p628-CTXM (85,338 bp: KP987217.1) and pEK204 (93,732 bp: EU935740.1), respectively; The blue circle with deep and light color, respectively, depicts pH1519-88 (88,678 bp: KJ484630.1) and pSH4469 (91,109 bp: KJ406378.1) in the NCBI database. The outmost circle in red color represents pSa64-96kb. Sequence was produced via combination of Illumina and PacBio sequencing data.
The plasmid harbored by Salmonella strain Sa64, designated as pSa64-96kb, was 96,571 bp in length, with GC content of 49.7%. BLASTN analysis indicated that this plasmid was 99% homologous to plasmid pS68 (Accession number: KU130396) of which was acquired from E. coli at 99% coverage. Plasmid pSa64-96kb carried IncI1 replicon together with the transfer proteins (tra locus), the pilus formation protein (pil locus), the shufflon-specific DNA recombinase (rci), and the nikAB-trbABC region encoding genes. It also showed homology (98.9% similarity, 72–76% coverage) to some other plasmids, including E. coli plasmid pEK204 (EU935740.1) and Klebsiella pneumoniae plasmids p628-CTXM (KP987217.1) and pH1519-88 (KJ484630.1), and a Shigella sonnei plasmid, pSH4469 (KJ406378.1), all of which contained a blaCTX–M gene belonging to group 1 variant and contributed to worldwide transmission of ceftriaxone resistance in Salmonella (Figure 2B). The insertion sites of the blaCTX–M variants were very similar on these plasmids (Supplementary Figure 2B). However, no antimicrobial resistance gene was carried by pSa64-96kb, and thus it could be considered as the prototype of such IncI1 plasmids. Additional tra genes responsible for conjugation were carried by pSa64-96kb but were replaced by diverse antimicrobial resistance genes in other IncI plasmids during plasmid evolution (Figure 2B and Supplementary Figure 2).
The plasmid recovered from the transconjugant S64-TC was a circular IncI1 plasmid of 188,966 bp, with 229 predicated coding sequences and, a GC content of 50.5%. It was designated as pSa64T-188kb. Sequence analysis showed that a ∼92 kb DNA fragment, (Sa64-chr-92kb) and the plasmid pSa64-96kb, which existed in the parental strain as separate entity, have merged and formed the fusion plasmid pSa64T-188kb though IS26 (Figures 2B, 3A), which was subsequently transferred to the recipient strain J53 through conjugation. Taken together, our data could help explain the conjugative transmission of ciprofloxacin resistance in Salmonella Sa64 strain. The carriage of multiple PMQR genes [aac(6’)Ib-cr and qnrS2] in the MDR fragment located in the chromosome of Sa64 could confer resistance to ciprofloxacin (CIP MIC = 8 μg/ml), which was further confirmed by the increase of CIP MIC from 0.01 to 8 μg/ml after acquisition of conjugative plasmid pSa64T-188kb bearing this MDR fragment carrying aac(6’)Ib-cr and qnrS2 in E. coli J53.
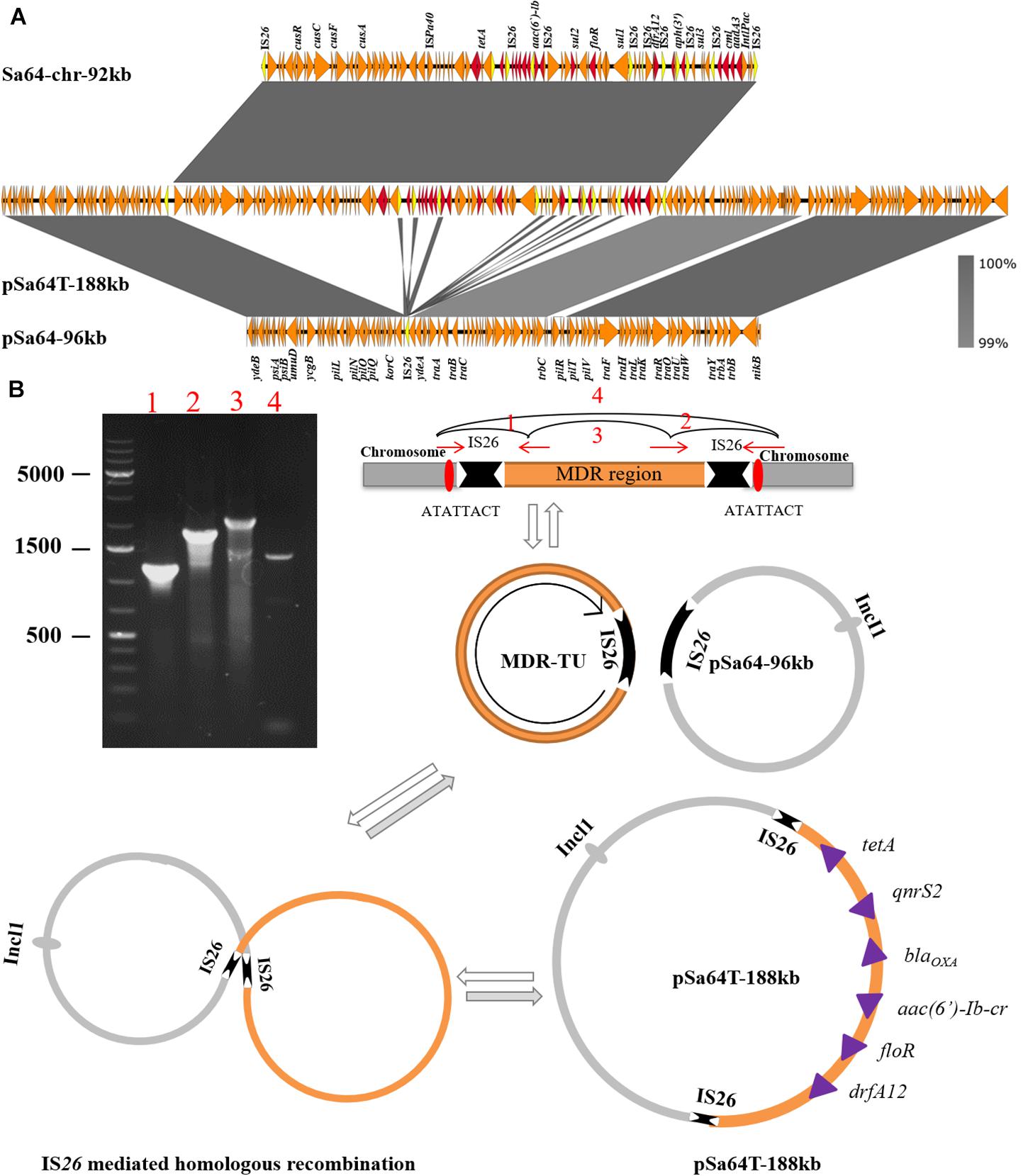
Figure 3. Mechanisms of plasmid recombination. (A) Structure alignment of two plasmids and the MDR region in the chromosome. Duplicated IS26 elements are highlighted in yellow color and drug-resistance genes are depicted in red arrows; (B) Proposed IS element-mediated plasmid and drug-resistant chromosomal fragament fusion in Sa64, PCR products 3 and 4 confirm that TU could be dynamic existed in a circular intermediate form and also could be assembled in chromosome via replicative transposition event by the hot spot (ATATTACT). While the plasmid pSa64T-188kb is the cointegrate generated by homologous recombination bewteen TU and plasmid pSa64-96kb.
The molecular mechanisms underlying this DNA transmission and plasmid fusion were further investigated. Detailed analysis of the aligned sequences of these two plasmids and the chromosomal DNA fragments revealed that two copies of IS26 were found to be flanked at both ends of chromosomal MDR fragment, Sa64-chr-92kb, and one copy of IS26 on plasmid pSa64-96kb. These data suggested that the insertion of Sa64-chr-92kb to plasmid pSa64-96kb could be due to homologous recombination mediated by IS26. We then ask how the chromosomal DNA fragment could be integrated into plasmid by recombination. It has been reported that DNA fragment flanked by IS26 could form circular intermediate known as a translocatable unit (TU; Harmer and Hall, 2015, 2016). To test if MDR Sa64-chr-92kb fragment could also form TU, four PCR assays using primers listed in Supplementary Table 1 were performed. PCR products 1 and 2 were successfully amplified with corrected sizes and sequences suggesting that these four primers were valid (Figure 3B). The amplification of product 3 as shown in Figure 3B was also obtained with right size and sequence, which aligned to the IS26 sequence confirming that a TU has been formed in S. Derby Sa64 strain. In addition, PCR product 4 with size and sequence covering one copy of IS26 and adjacent sequences suggested that some population of Sa64 carried one copy of IS26 with the whole MDR fragment being excised out (Figure 3B). This data also indicated that the dynamic presence of TU and chromosomal encoded MDR fragment in a total population of Sa64. Analysis of product 4 sequence and sequences flanking MDR fragment on the chromosome allowed us to identify a hot spot (ATATTACT) located right outside of IS26, which implied that Sa64 obtained this MDR fragment through being attacked by IS26 flanking the MDR region from other sources most likely from other plasmid at this hot spot on the chromosome. After confirmation of the presence of TU in Sa64, it is further speculated that this TU could undergo cointegration with pSa64-96kb to form pSa64T-188kb through homologous recombination at IS26 region (Figure 3B).
Pathogenic bacteria such as Salmonella often contain target gene mutations and exhibit resistance to ciprofloxacin. Double and single mutations in gyrA and parC genes associated with ciprofloxacin resistance phenotypes in salmonella were shown to be consistent with previously reports (Chen et al., 2007). Salmonella isolates were rarely resistant to ciprofloxacin in the past decades mainly due to the low occurrence of gyrA double mutations. In 2005, quinolone resistance conferred by the acquisition of PMQR genes was first observed in Salmonella, while mutations in PMQR determinants only mediated quinolone resistance, but not resistance to fluoroquinolones such as ciprofloxacin (Ferrari et al., 2013; Wong et al., 2014a; Kim et al., 2016). The prevalence of ciprofloxacin resistance in Salmonella climbed sharply in several countries, especially China, with up to ∼ 30–40% resistance rate detectable among strains of specific serotypes (Lin et al., 2015; Pribul et al., 2017). These emerging ciprofloxacin-resistant Salmonella strains were found to harbor only a single gyrA mutation and an extrachromosomal PMQR gene, or several PMQR genes with no mutations. Apparently, the ciprofloxacin-resistance phenotypes of these isolates were attributable to the combined effects of the PMQR gene product, and/or target mutations because the PMQR gene products can drastically reduce the antimicrobial effect of fluoroquinolones via antibiotic efflux, competitive inhibition by binding of antibiotics, and inactivation with enzymes, resulting in emergence of high-level quinolone resistance strains without target gene mutations. However, ciprofloxacin resistance strains of Salmonella whose resistance phenotypes are transferable have rarely been reported, since the majority of PMQR genes were normally located in non-conjugative plasmid or the chromosome of Salmonella (Lin et al., 2015). However, the ciprofloxacin-resistance phenotype encoded by two types of conjugative plasmids has been reported in Salmonella previously (Chen et al., 2018), suggesting that ciprofloxacin resistance is now readily transferrable.
This study identified a chromosomal DNA fragment carrying PMQR and other drug-resistance genes that was assembled via plasmid-mediated integration into the Salmonella chromosome. Importantly, such chromosomal fragment may readily become transferable upon being captured by a conjugative plasmid in Salmonella. Emergence of such chromosome-derived, ciprofloxacin-encoding conjugative plasmids comprises a severe public health threat since ciprofloxacin is first option for curing life-threatening Salmonellosis. Comprehensive analysis of the genetic content among strains Sa64, Sa79, and ST350 showed that one insertion sequence, IS26, commonly co-occurred with antimicrobial resistance genes and were highly transferable between plasmids and the chromosome. The plasmid pSa64-96kb, which contains IncI1 replicon, was found to be an important resistance-encoding vector commonly harbored by various bacterial species. One part of this plasmid, the IncI1 replicon, may readily be fused with other drug-resistance genes at high frequency. Such fusion plasmids, such as pSa64T-188kb, exhibit a broader host spectrum and might help further expand the resistance profile of the host strain. It has been shown that plasmids are functionally dynamic, readily forming hybrid plasmids through recombination. Although the structure of Sa64-chr-92kb in the chromosome of the test strain was found to be stable via Nanopore long-reads sequencing. No TU of MDR fragment, Sa64-chr-92kb, could be detected with sequencing depth of 40 multiplicative, while it was shown that this type of TU was presented in some population of Sa64 by PCR amplification. This is probably due to the low number of this TU in total population of Sa64, which could not be picked up by Nanopore sequencing with low sequencing depth, which is consistent with the weak band of product 4 obtained by PCR. Plasmid cointegration is not rare among bacteria, and it has been reported to be related to spreading of antimicrobial resistance genes, such as cephalosporin resistance encoded by beta-lactamases genes (Leelaporn et al., 1996; He et al., 2015; Wong et al., 2016). For example, Harmer and Hall (2015, 2016) found that the insertion sequence IS26 played a role in reorganizing plasmids in clinically isolated MDR bacteria via replicative transposition (Kim et al., 2016). Our recent work also reported that dissemination of IncI1 plasmids can integrate with various blaCTX–M genes such as the blaCTX–M group 1 and group 2 elements, contributing to an increasing prevalence of ceftriaxone resistance (Wong et al., 2016). These plasmids ranged from 75 kb to 100 kb in size and contained blaCTX–M typical structures, ISEcp1-blaCTX–M–14-IS903-iroN or flanked by ISEcp1 element and a truncated orf477 in up- and downstream regions, respectively. Our study further extends the role of this type IncI1plasmid as helper plasmid to fuse with chromosomal MDR fragment or MDR plasmid to help conjugate these non-conjugative elements to other bacteria with even broader host spectrum.
In conclusion, this study showed that dynamic interaction between specific chromosomal fragment and plasmids may significantly enhance resistance development and transferability of mobile resistance-encoding elements in bacterial pathogens. Mobile elements including both IS26 and the IncI1 plasmid played important roles during the transmission process. IS26 mediated homologous recombination of antimicrobial resistance genes with the IncI1 conjugative helper plasmid, during which a recombined fusion plasmid was generated. The fusion plasmid was transferred to a new host with helper of conjugative elements encoded by the IncI plasmid. Fusion between IncI1 type plasmids and various antibiotic-resistance gene-bearing mobile elements and chromosomal fragments is expected to bring about a sharp increase in the number of mobile resistance elements circulating among populations of bacterial pathogens, thereby posing a severe threat to the effectiveness of traditional antibiotics in treatment of infections caused by Salmonella pathogens. Meanwhile, such fusion events explain that bacteria may acquire antibiotic resistance genes from a route other than conjugation. Furthermore, such events will continue to produce novel plasmids conferring enhanced resistance phenotypes with expanded bacterial hosts.
The sequencing data of the plasmids and chromosome have been deposited in GenBank under the accession numbers pSa64T-188kb (CP034251), pSa64-96kb (CP034252), Sa64-chr (CP034250), and Sa79-chr (SGWG00000000).
CY collected the strain and finished the wet experiments. KC performed the sequencing and bioinformatics analysis. EC, KC, and SC participated in research design and manuscript writing. SC and WY supervised the whole project. All authors contributed to the article and approved the submitted version.
The research was supported by the National Key R&D Program (2018YFD0500300) and Collaborative Research Fund of the Hong Kong Research Grant Council (C5026-16G).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.556227/full#supplementary-material
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Chen, K., Chan, E. W. C., and Chen, S. (2019a). Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerg. Microb. Infect. 8, 396–403. doi: 10.1080/22221751.2019.1585965
Chen, K., Dong, N., Chan, E. W., and Chen, S. (2019b). Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microb. Infect. 8, 857–865. doi: 10.1080/22221751.2019.1626197
Chen, K., Dong, N., Zhao, S., Liu, L., Li, R., Xie, M., et al. (2018). Identification and characterization of conjugative plasmids that encode ciprofloxacin resistance in Salmonella. Antimicrob. Agents Chemother. 62:e00575-18.
Chen, S., Cui, S., McDermott, P. F., Zhao, S., White, D. G., Paulsen, I., et al. (2007). Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar Typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 51, 535–542. doi: 10.1128/aac.00600-06
Chen, S., Zhao, S., White, D. G., Schroeder, C. M., Lu, R., Yang, H., et al. (2004). Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70, 1–7. doi: 10.1128/aem.70.1.1-7.2004
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical Lab Standards Institute.
Ferrari, R., Galiana, A., Cremades, R., Rodriguez, J. C., Magnani, M., Tognim, M. C., et al. (2013). Plasmid-mediated quinolone resistance (PMQR) and mutations in the topoisomerase genes of Salmonella enterica strains from Brazil. Braz. J. Microbiol. 44, 651–656. doi: 10.1590/S1517-83822013000200046
Gomez, T. M., Motarjemi, Y., Miyagawa, S., Käferstein, F., and Stöhr, K. (1997). Foodborne salmonellosis. World health statistics quarterly. Rapport Trimestriel de Statistiques Sanitaires Mondiales 50, 81–89.
Gunell, M., Webber, M. A., Kotilainen, P., Lilly, A. J., Caddick, J. M., Jalava, J., et al. (2009). Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob. Agents Chemother. 53, 3832–3836. doi: 10.1128/AAC.00121-09
Harmer, C. J., and Hall, R. M. (2015). IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6:e01866-15. doi: 10.1128/mBio.01866-15
Harmer, C. J., and Hall, R. M. (2016). IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762-15.
Hohmann, E. L. (2001). Nontyphoidal salmonellosis. Clin. Infect. Dis. 32, 263–269. doi: 10.1086/318457
Hooper, D. C. (2001). Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7, 337–341. doi: 10.3201/eid0702.700337
Kim, J., Han, X., Bae, J., Chui, L., Louie, M., Finley, R., et al. (2016). Prevalence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with resistance and reduced susceptibility to fluoroquinolones from human clinical cases in Alberta, Canada, 2009-13. J. Antimicrob. Chemother. 71, 2988–2990. doi: 10.1093/jac/dkw232
Leelaporn, A., Firth, N., Paulsen, I. T., and Skurray, R. A. (1996). IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J. Bacteriol. 178, 6070–6073. doi: 10.1128/jb.178.20.6070-6073.1996
Li, R., Lin, D., Chen, K., Wong, M. H. Y., and Chen, S. (2015a). First detection of AmpC β-lactamase blaCMY-2 on a conjugative IncA/C plasmid in Vibrio parahaemolyticus of food origin. Antimicrob. Agents Chemother. 59, 4106–4111. doi: 10.1128/aac.05008-14
Li, R., Wong, M. H., Zhou, Y., Chan, E. W., and Chen, S. (2015b). Complete nucleotide sequence of a conjugative plasmid carrying bla(PER-1). Antimicrob. Agents Chemother. 59, 3582–3584. doi: 10.1128/AAC.00518-15
Lin, D., Chen, K., Chan, E. W.-C., and Chen, S. (2015). Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 5, 1–8.
Molbak, K. (2005). Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 41, 1613–1620. doi: 10.1086/497599
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2013). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214.
Pribul, B. R., Festivo, M. L., Rodrigues, M. S., Costa, R. G., Rodrigues, E. C., de Souza, M. M., et al. (2017). Characteristics of quinolone resistance in Salmonella spp. isolates from the food chain in Brazil. Front. Microbiol. 8:299. doi: 10.3389/fmicb.2017.00299
Wong, M. H., Chan, E. W., Liu, L. Z., and Chen, S. (2014a). PMQR genes oqxAB and aac(6’)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 5:521. doi: 10.3389/fmicb.2014.00521
Wong, M. H., Yan, M., Chan, E. W., Biao, K., and Chen, S. (2014b). Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob. Agents Chemother. 58, 3752–3756. doi: 10.1128/AAC.02770-13
Keywords: Salmonella, ciprofloxacin resistance, chromosomal fragment, PMQR genes, conjugative helper plasmid
Citation: Yang C, Chen K, Chan EW-C, Yao W and Chen S (2020) Transmission of Chromosomal MDR DNA Fragment Encoding Ciprofloxacin Resistance by a Conjugative Helper Plasmid in Salmonella. Front. Microbiol. 11:556227. doi: 10.3389/fmicb.2020.556227
Received: 27 April 2020; Accepted: 19 August 2020;
Published: 18 September 2020.
Edited by:
Charlene Renee Jackson, United States Department of Agriculture (USDA), United StatesReviewed by:
Xiang-Dang Du, Henan Agricultural University, ChinaCopyright © 2020 Yang, Chen, Chan, Yao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Yao, eWFvd2VuNjdqcEBuamF1LmVkdS5jbg==; Sheng Chen, c2hlY2hlbkBjaXR5dS5lZHUuaGs=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.