- 1Academy of Agricultural and Forestry Sciences, Qinghai University, Xining, China
- 2Qinghai Key Laboratory of Hulless Barley Genetics and Breeding, Xining, China
- 3Qinghai Subcenter of National Hulless Barley Improvement, Xining, China
Long-term continuous cropping influences the nutrient of soil and microbiome of the rhizosphere, resulting in the yield decrease of crops. Tibetan barley is a dominant cereal crop cultivated at high altitudes in Tibet. Its growth and yield are negatively affected by continuous cropping; however, the response of the rhizosphere microbial community to continuous cropping remains poorly understood. To address this question, we investigated the bacterial community structure and conducted predictive functional profiling on rhizosphere soil from Tibetan barley monocropped for 2–6 years. The results revealed that long-term continuous cropping markedly decreased total nitrogen and available nitrogen in rhizosphere soil. Illumina high-throughput sequencing of 16S rRNA genes indicated that the bacterial community was altered by continuous cropping; operational taxonomic units (OTUs), Shannon index, and Faith Phylogenetic Diversity decreased with increasing monocropping duration. Relative abundances of family Pseudomonadaceae, Cytophagaceae, and Nocardioidaceae were significantly increased, while those of Chitinophagaceae and Sphingomonadaceae were significantly decreased (all p < 0.05). Besides, continuous cropping significantly increased the abundance of bacteria associated with chemoheterotrophy, aromatic compound degradation, and nitrate reduction (p < 0.05). Generalized boosted regression model analysis indicated that total nitrogen was the most important contributor to the bacterial community diversity, indicating their roles in shaping the rhizosphere bacterial community during continuous cropping. Overall, continuous cropping had a significant impact on the structure of bacterial communities in rhizosphere soil of Tibetan barley, and these results will improve our understanding of soil bacterial community regulation and soil health maintenance in Tibetan barley farm systems.
Introduction
High altitude ecosystems are generally characterized by low temperature, variable rainfall, reduced atmospheric pressure, and soil nutritional stress. Cold in high altitude region is the major factor that has confounded effects on both microbial biodiversity and soil physicochemical properties (Kumar et al., 2019). Plants and microbes are co-evolved and interact with each other in the environment. Thus, high altitude regions are the research center in understanding the interactions between certain microbes and the plants cultivated in the cold environment (Rawat et al., 2020). Lower species richness and diversity were observed in soil with higher elevations (Bryant et al., 2008; Wang et al., 2015). In a study of three vertical climate zones on the Qinghai-Tibetan Plateau, China (av. 4,000 m above sea level), soil bacteria exhibited more apparent elevational zonation features and decreased diversity patterns with increasing elevation (Wang et al., 2015). However, the microbial community structure of rhizosphere soil in high altitude region is less understood (Praeg et al., 2019).
Continuous cropping, defined as a single crop being cultivated in the same field year after year, is commonly practiced in China due to limited arable lands and lower associated agronomic management costs compared to rotation cropping (Zhang and He, 2004; Chen et al., 2009). However, long-term monoculture cropping can lead to serious soil sickness. Previous studies reported that continuous cropping often leads to restricted growth and crop yields with different time-scales (Ventura and Watanabe, 1978; Lua et al., 2017; Zhao et al., 2017). Furthermore, Changes in the diversity and composition of soil microorganisms by continuous cropping can disrupt ecosystem function, balance, and health; in turn, such phenomena negatively affect soil productivity and ultimately lead to reduced plant biomass and yield (Chen et al., 2012). Dynamic successions of the rhizosphere microbial community during continuous cropping have been identified in soybeans (Bai et al., 2015), cotton (Wei and Yu, 2018), peanut (Chen et al., 2014), and cucumber (Ali et al., 2019). However, these crops are usually cultivated in normal altitude regions, and the role of continuous cropping in shaping the microbiome structure of plants cultivated at high altitudes remains poorly investigated. Moreover, previous studies have proven that continuous cropping disturbed the microbial community structure in the rhizosphere soil, and loss of biodiversity has been identified as a common phenomenon in soil with long-term continuous cropping (Li et al., 2010, 2018; Chen et al., 2014). But, the effects of continuous cropping on the functional groups of rhizosphere microbe are less explored.
Tibetan hulless barley (Hordeum vulgare L., qingke) has been completely adapted to the extreme plateau climate after thousands of years of domestication and cultivation on the Qinghai-Tibetan Plateau, China (av. 4,000 m above sea level) (Zeng et al., 2018). Tibetan barley accounts for approximately 70% of the farming land in Tibet and serves as a food staple, animal fodder, and ingredient in various foods (Choo, 2002). However, the sustainability and development of Tibetan barley production are impeded by conventional farming practices, such as continuous cropping (Zhang et al., 2012). We hypothesized that the long-term continuous cropping of Tibetan barley alters the bacterial community structure and function in rhizosphere soil, resulting in specific responses of rhizosphere soil microbes to continuous cropping. Thus, understanding the influence of continuous cropping on the microbial community in the soil rhizosphere is pivotal for designing effective farm management practices to relieve soil depletion associated with Tibetan barley cultivation.
In this study, Tibetan barley fields cultivated for various durations were used to investigate the effect of continuous cropping on the rhizosphere bacterial community. Dynamic succession of the bacterial community structure in the rhizosphere was explored by 16S rRNA gene high-throughput sequencing. Changes in the potential ecological functions of the rhizosphere bacterial community during continuous cropping were also investigated. These results provide a comprehensive understanding of the rhizosphere microbial response to continuous Tibetan barley cropping and may help improve farming strategies by regulating the structure and function of rhizosphere soil flora to reduce the negative impacts of continuous cropping.
Materials and Methods
Field Description and Yield Investigation
The study field was located in Qinghai province, China (37°21′N, 101°44′E) at an altitude of 2,870 m. The Qinghai-Tibet plateau features a cold, warm, and humid climate, with an average annual sunshine duration of 2,575 h, average annual temperature of 1.3°C, annual rainfall of 530–560 mm, and annual evaporation of 1,128–1,343 mm (Zhang et al., 2000). The soil in the field is Kastanozems according to the FAO classification (FAO, 1990), with 4.5 g kg–1 total nitrogen, 1.0 g kg–1 total phosphorus, 22.0 g kg–1 total potassium, 60.5 g kg–1 organic matter, 245 mg kg–1 available nitrogen, 20 mg kg–1 available phosphorus, and 200 mg kg–1 available potassium. The experimental field contained four plots, each 10.0 × 10.0 m in size. Starting in June 2011, Tibetan barley (Hordeum vulgare L., qingke) is an annual crop. In the period of 2011–2016, the plants were cultivated in April and harvested in August, yearly. Blended fertilizer with 22.5 g m–2 (NH4)2SO4 and 11.5 g m–2 urea was applied annually before seeding. About 60,000 seeds were sowed at each plot, and approximately 250 plant m–2 successfully matured. All other field management activities were performed according to local farming habits. Harvesting was performed annually when plants reached full maturity, and seeds were manually separated to measure yield.
Rhizosphere Soil Collection and Physiochemical Analyses
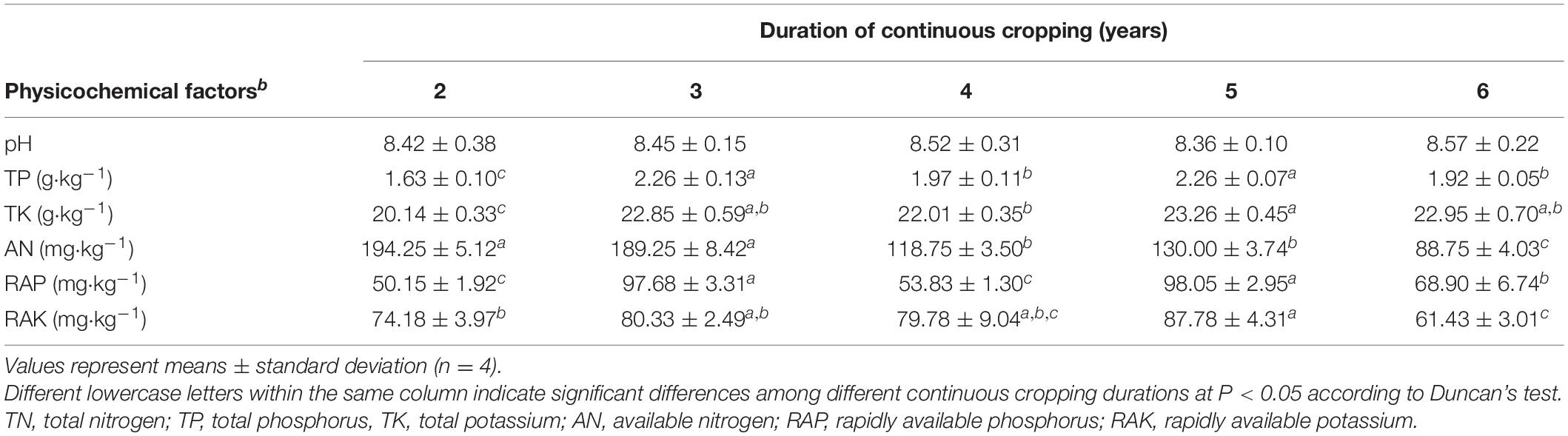
Rhizosphere soil samples were collected annually during the flowering period. Approximately 15 plants sample were collected from five different sites using a “Z” pattern in each experimental plot. Then, the composite sample was constituted from the 15 plants. Rhizosphere soils were collected for analysis as reported (Li et al., 2018). Briefly, plants were carefully pulled from the ground and the conglutinating soil from the roots was mildly crushed and shaken to collect the soil located within the plant root surfaces. Quadruple soil and plant samples were collected from the four experimental fields. The rhizosphere soil was immediately transported on ice to the laboratory and stored at −20°C for DNA extraction and chemical analysis. Half of each combined soil sample was air-dried and filtered through a 0.149 mm sieve. The sample pH was determined using a pH meter with a glass electrode (FE20-Five Easy PlusTM, Mettler Toledo, Greifensee, Switzerland). Total nitrogen (TN), available nitrogen (AN), total phosphorus (TP), total potassium (TK), rapidly available phosphorus (RAP), and rapidly available potassium (RAK) were measured as previously described (Van Raij et al., 1986; Nichols and Wright, 2006).
Illumina MiSeq Sequencing of Bacterial 16S rRNA Genes
Total genomic DNA was extracted from 0.25 g of rhizosphere soil using the MoBio Soil DNA isolation kit (MO BIO Laboratories Inc., Carlsbad, CA, United States) according to the manufacturer’s protocol. The bacterial V4–V5 region of the 16S rRNA gene was amplified using the universal prokaryotic primers 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 907R (5′-CCG TCA ATT CMT TTR AGT TT-3′) (Zhou et al., 2011) with the addition of a 12 bp unique barcode and the required Illumina adapters. PCRs were performed in a 30-μL mixture containing 10 μL TaKaRa Ex Taq® PCR premix (TaKaRa Bio Inc., Kusatsu, Japan), 0.5 μL forward primer (10 μM), 0.5 μL reverse primer (10 μM), 1 μL DNA template (20 ng), and 18 μL PCR-grade water under the following PCR conditions: initial denaturation at 95°C for 3 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 30 s, extension at 72°C for 1 min, and then a final extension at 72°C for 10 min. Samples were amplified in triplicate, and replicate PCRs for each sample were pooled and purified using a QIAquick Gel Extraction Kit (Qiagen Inc., Chatsworth, CA, United States). PCR products were then sequenced using an Illumina MiSeq system at Sangon Biotech Co., Ltd. (Shanghai, China).
16S rRNA Sequence Analysis
The raw paired-end reads were merged using FLASH Version 1.2.11 (Magoč and Salzberg, 2011). Merged sequences were subsequently analyzed using the bioinformatics pipeline of Quantitative Insights Into Microbial Ecology (QIIME) version 2018.6 (Caporaso et al., 2010). First, demultiplexing was summarized using demux1 to classify sequences from each sample. Then, primer sequences were removed using “cutadapt” in QIIME2. After that, chimeric sequences were removed and sequences were clustered using QIIME 2’s q2-feature-classifier plugin2 (Bokulich et al., 2018). The clean data were then clustered into OTUs at 97% similarity using UCLUST (Edgar, 2010), and the longest sequence within each OTU was selected as the representative sequence. Taxonomic assignment of each OTU was conducted using QIIME 2’s q2-feature-classifier plugin3 and the SILVA version 132 database (Quast et al., 2012). The OTU table was rarefied to the minimum sample count (36,836 sequences) for subsequent analysis, including alpha and beta diversities and species composition. Alpha-diversity was described using OTUs, the Shannon index, and Faith Phylogenetic Diversity (PD).
Predictive Functional Profiling of Bacterial Communities Using 16S rRNA Data
The predicted ecological functions of the prokaryotic communities were analyzed by mapping the taxonomy into certain metabolic pathways using FaProTax software (Louca et al., 2016). The OTU table generated from 16S rRNA sequence analysis was matched to annotated OTU information with species information in the FaProTax database to determine the predicted ecological function.
Statistical Analysis
ANOVA and Spearman’s correlations were analyzed using SPSS Statistical Software Package ver. 20.0 (SPSS Inc., Chicago, IL, United States). Correlations between the community composition and environmental parameters were also analyzed by RDA (redundancy analysis) using the “vegan” package in R software (version 3.5.1). Linear regression models in R software were used to analyze the dynamic tendency of geochemical parameters, taxonomic abundance, and functional profiles against years of continuous cropping. Pearson relationships between physicochemical properties of rhizosphere soils and predictive functional profiles of bacterial communities were explored by SPSS 20.0. A correlation between two items was considered statistically robust if the Spearman’s correlation coefficient (ρ) was >0.8 and the P-value was <0.05. Co-occurrence network visualization was conducted on the interactive platform of Gephi (Bastian et al., 2009). To accurately predict and explain the relationships between bacterial data and chemical variables, generalized boosted regression modeling (GBM) analysis (5,000 trees used for boosting, 10-fold cross-validation, and three-way interactions) was performed to quantitatively evaluate the relative influence of individual chemical factors on bacterial community diversities using the R package “gbm” function (De’ath, 2007).
Results
Tibetan Barley Yields and Physicochemical Properties of Rhizosphere Soils
Yields consistently decreased during 6 years of Tibetan barley monoculture, from 19.42 ± 0.74 kg ha–1 to 12.40 ± 0.45 kg ha–1 (Supplementary Figure S1). Environmental parameters of the rhizosphere soil were also influenced by continuous cropping, although rhizosphere soil pH exhibited no obvious changes throughout the study and was slightly alkaline (pH ranged from 8.06 to 8.91) (Table 1). The concentration of TN and AN decreased with continuous cropping. However, phosphorus (TP and RAP) and potassium (TK and RAK) fluctuated and did not exhibit increasing or decreasing trends throughout the experimental period (Table 1).
Dynamic Succession of the Rhizosphere Bacterial Community
A total of 799,745 qualified 16S rRNA gene sequences were obtained with an average of 58,670 sequences per sample. Each sample was rarified to 36,836 sequences to reduce the effect of sequence numbers on bacterial diversity and taxonomic composition analysis. The richness of the rhizosphere bacterial community declined during 6 years of continuous cropping (Figure 1). The number of OTUs decreased from 1,226 ± 7 at 2 years to 1,066 ± 22 at 6 years of continuous cropping, and the Faith PD decreased from 86.73 ± 0.71 to 73.04 ± 2.1. The Shannon index decreased from 9.75 ± 0.02 to 9.52 ± 0.05 with successive years of continuous cropping (p > 0.05, ANOVA) (Figure 1). RDA was performed to summarize the relationships between bacterial communities and soil variables. The results showed that the first two axes of the RDA accounted for 46.13% of the total community variance. The RDA plot showed a clear separation in the space among the samples at different years of continuous cropping. Bacterial communities from the second and third years (CC2Y and CC3Y) were separated from those from later years (CC4Y, CC5Y, and CC6Y) along the first RDA axis. This separation was influenced mainly by AN, TN, pH. The structure of CC2Y and CC3Y communities are related to lower pH and higher AN and TN values in contrast to CC4Y, CC5Y, and CC6Y communities. Communities from the last three years were separated along the second RDA axis, and especially those from the fourth and fifth years were related to higher TP and TK. CC3Y and CC5Y are related to higher RAK and RAP (Figure 2). By contrast, the impact of pH on bacterial community composition was minimal.
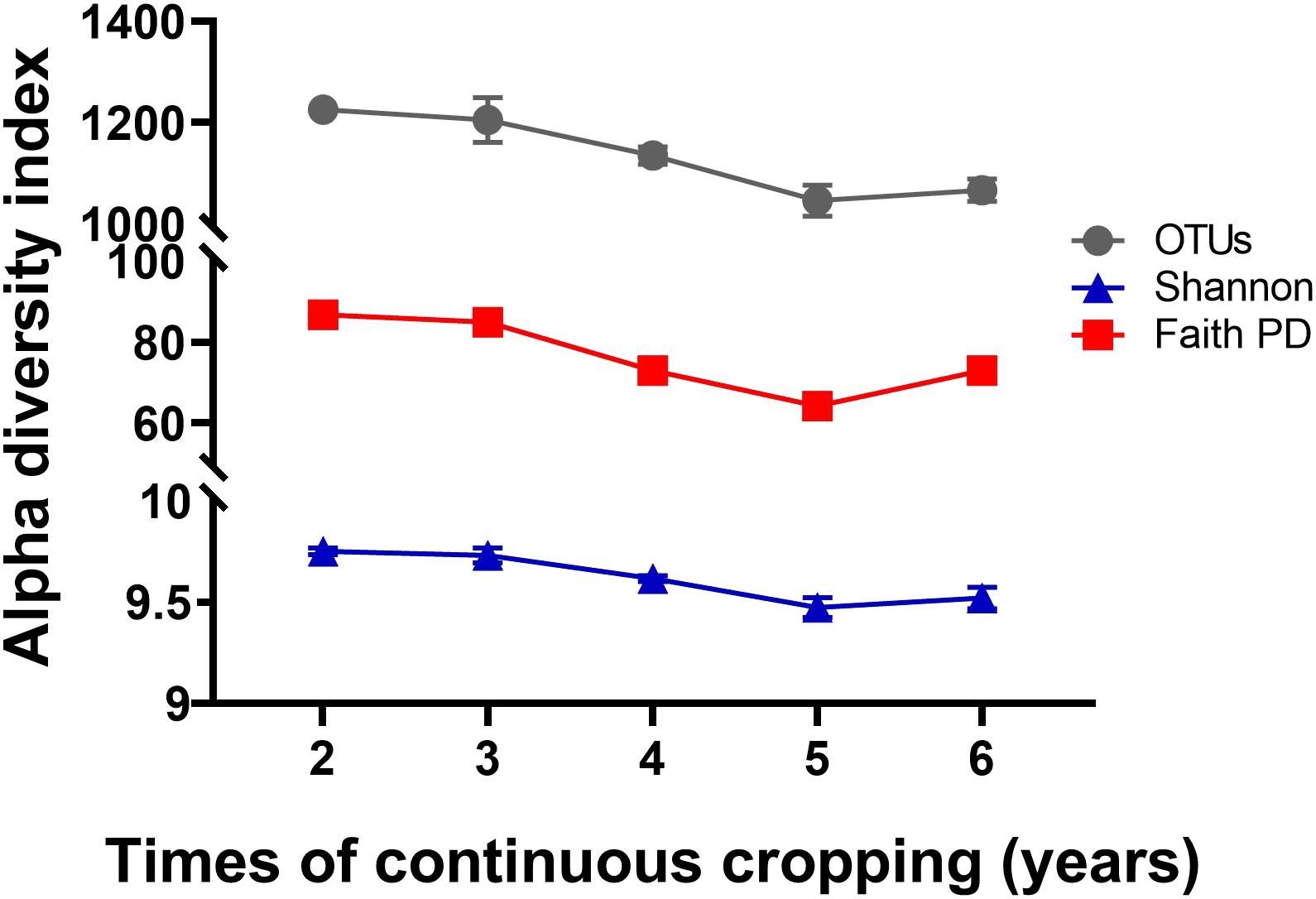
Figure 1. Effects of continuous cropping on bacterial alpha diversities for different continuous cropping durations. OTUs, Shannon index, and Faith PD in rhizosphere soils continuously cropped for 2 to 6 years; Symbols represent means ± SDs (n = 4).
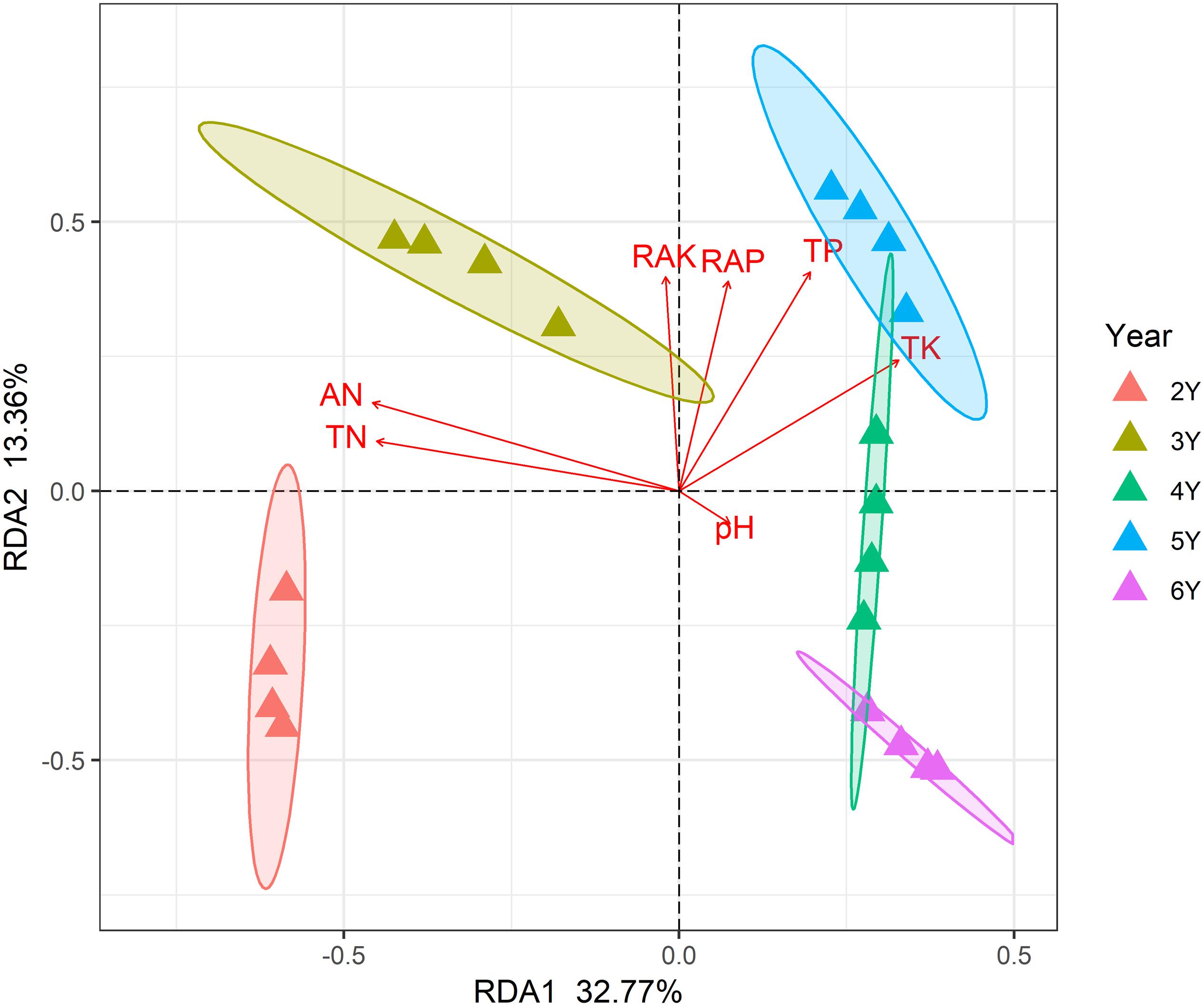
Figure 2. Redundancy analysis (RDA) biplot of bacterial 16S rRNA genes in the rhizosphere soils showing geochemical factors and samples.
The predominant phyla in the rhizosphere soil were Proteobacteria (42.72% ± 1.69% relative abundance), Bacteroidetes (12.33% ± 1.95%), and Actinobacteria (11.2% ± 3.12%) (Supplementary Figure S2A). At the family level, the bacterial community was dominated by Sphingomonadaceae and Xanthomonadaceae (Supplementary Figure S2B). The relative abundance of Sphingomonadaceae fluctuated and tended to decrease from 9.47% ± 0.05% to 7.36% ± 0.30% throughout the experiment. The relative abundance of Xanthomonadaceae increased from 2.00% ± 0.09% to 5.64% ± 0.75% at 4 years of monocropping and was restored to 2.37% ± 0.14% after 6 years of continuous cropping (Supplementary Figure S2B). We also explore the changes of 20 most abundant OTUs during continuous cropping. The abundance of OTU1 (classified as Unclassified Exiguobacterium), OTU5 (sugarcane phytoplasma), OTU9 (Unclassified Pseudonocardiaceae), OTU11 (Unclassified Xanthomonadaceae), and OTU12 (Unclassified Nocardioides) were significantly increased after 6 years of continuous cropping. By contrast, the OTUs displayed significantly decreased, including OTU2 (Sphingomonas jaspsi), OTU14 (Unclassified Brevundimonas), OTU17 (Unclassified Sphingomonadaceae), and OTU20 (Burkholderiales bacterium X4) (Supplementary Table S1). Significant linear correlations between taxonomic abundance and cropping duration were only identified in the less abundant families (Supplementary Table S2). The relative abundances of Pseudomonadaceae (r2 = 0.844, p < 0.05, Supplementary Figure S3A), Cytophagaceae (r2 = 0.696, p < 0.05, Supplementary Figure S3B), and Nocardioidaceae (r2 = 0.656, p < 0.05, Supplementary Figure S3C) were positively related to the duration of continuous cropping. Chitinophagaceae (r2 = 0.695, p < 0.05, Supplementary Figure S3D) exhibited a significantly negative relationship with cropping duration. The dynamic succession of the above families suggested that functionalities carried out by affiliated bacteria were critical in the rhizosphere biotope during continuous cropping.
Changes in Bacterial Community Functionality With Continuous Cropping Duration
Fifty-one ecological functions were annotated by FaProTax software, in which chemoheterotrophy (average abundance 33.42% ± 4.36%), aerobic chemoheterotrophy (31.79% ± 4.19%), animal parasites or symbionts (2.53% ± 0.28%), aromatic compound degradation (2.49% ± 0.73%), and nitrate reduction (1.69% ± 0.28%) were the predominant functional groups in the rhizosphere soil of Tibetan barley (Figure 3 and Supplementary Table S3). The results of linear correlation analysis revealed that the relative abundances of the taxa associated with chemoheterotrophy (r2 = 0.51), aerobic chemoheterotrophy (r2 = 0.53), aromatic compound degradation (r2 = 0.73), and nitrate reduction (r2 = 0.41) increased significantly with continuous cropping duration (all p < 0.05; Supplementary Figure S4, Table S4). Furthermore, significantly positive relationships were identified between abundant families and functions. However, dark hydrogen oxidation was only positively related to Sphingomonadaceae, Comamonadaceae, and Chitinophagaceae (all p < 0.05; Figure 4). Interestingly, the three families mentioned above were negatively related to the functional groups, which were positively correlated with other bacterial families, suggesting that the functionalities of Sphingomonadaceae, Comamonadaceae, and Chitinophagaceae were different from other bacterial families in the rhizosphere biotope of Tibetan barley.
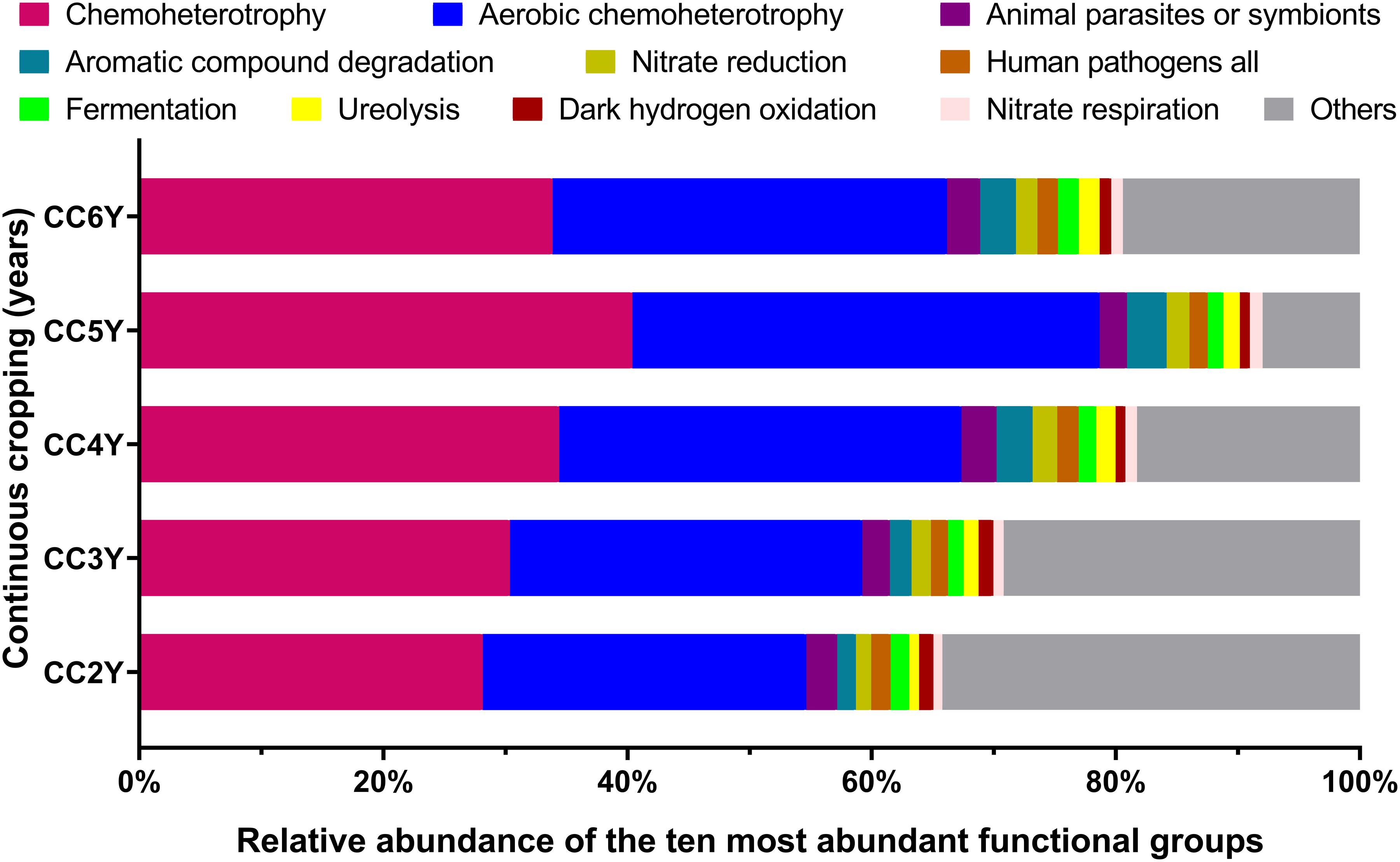
Figure 3. Distribution of bacterial ecological function in the FaProTax database for different continuous cropping durations. CC2Y, CC3Y, CC4Y, CC5Y, and CC6Y represent continuous cropping for 2, 3, 4, 5, and 6 years, respectively. Values represent means (n = 4). The 10 most abundant bacterial functional groups are shown.
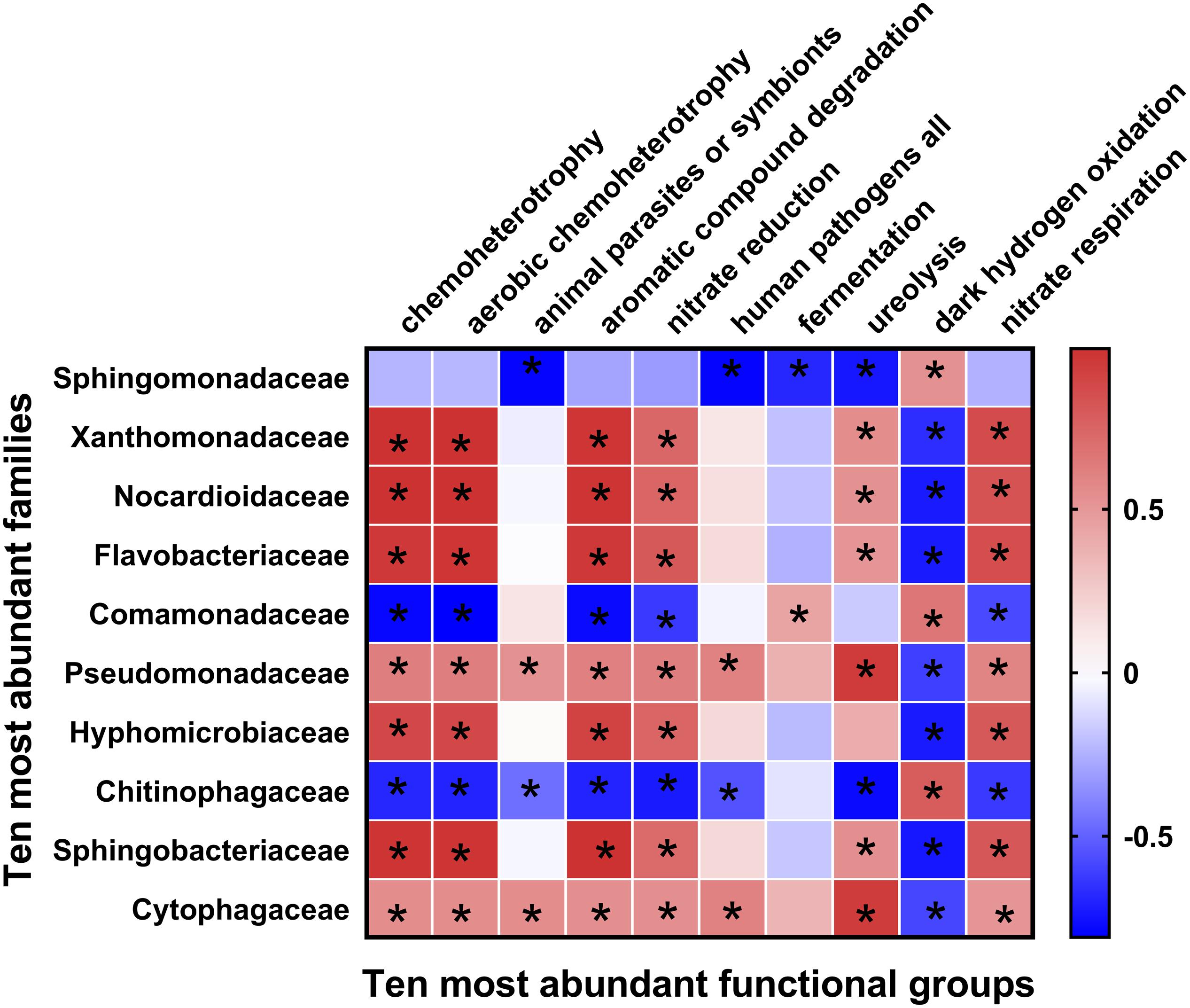
Figure 4. Heatmap of correlations between bacterial families with ecological functions in rhizosphere soil. Heatmap values ranged from +0.5 to −0.5. Values above/below zero represent positive/negative correlations between bacterial families and parameters analyzed. *P < 0.05 for the indicated comparisons.
Linking Bacterial Community Structure and Functionality to Rhizosphere Soil Properties
We hypothesized that bacterial communities in monocropped rhizosphere soil are shaped by innate environmental parameters. GBM analysis indicated that TN had the greatest relative influence on OTU numbers (45.55%), Shannon index (44.64%), and Faith PD (32.02%) of the bacterial communities studied (Figure 5), implying that nitrogen biotransformation was most influenced by changes in bacterial community diversity during continuous cropping. Pearson’s correlation coefficients between relative abundances of bacterial families and environmental parameters revealed that TN was positively correlated with the abundance of Comamonadaceae but negatively related to Hyphomicrobiaceae (all p < 0.01; Supplementary Figure S5). Furthermore, Comamonadaceae abundance was significantly negatively related to RAK (p < 0.01; Supplementary Figure S5). The correlation-based network analysis revealed that AN was positively related to dark hydrogen oxidation, and TK was positively related to nitrate respiration, but negative relationships were observed between other geochemical properties and the relative abundance of functional groups (all p < 0.05, Figure 6). Because pH remained constant throughout the experimental period, no remarkable relationship was observed between pH and ecological function. Some functional groups were significantly correlated with phosphorus and potassium concentrations in rhizosphere soil; for example, TK was significantly positively related to nitrate respiration, aromatic compound degradation, chemoheterotrophy, and aerobic chemoheterotrophy (Figure 6).
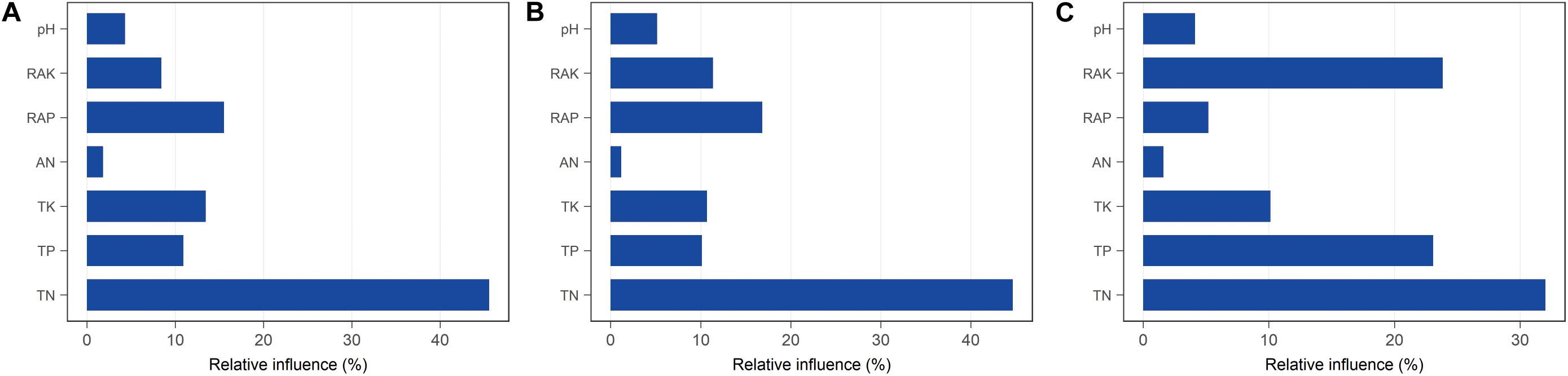
Figure 5. Relative influence (%) of individual physicochemical parameters on bacterial diversity in rhizosphere soil with different continuous cropping durations. (A) OTUs; (B) Shannon index; (C) Faith PD.
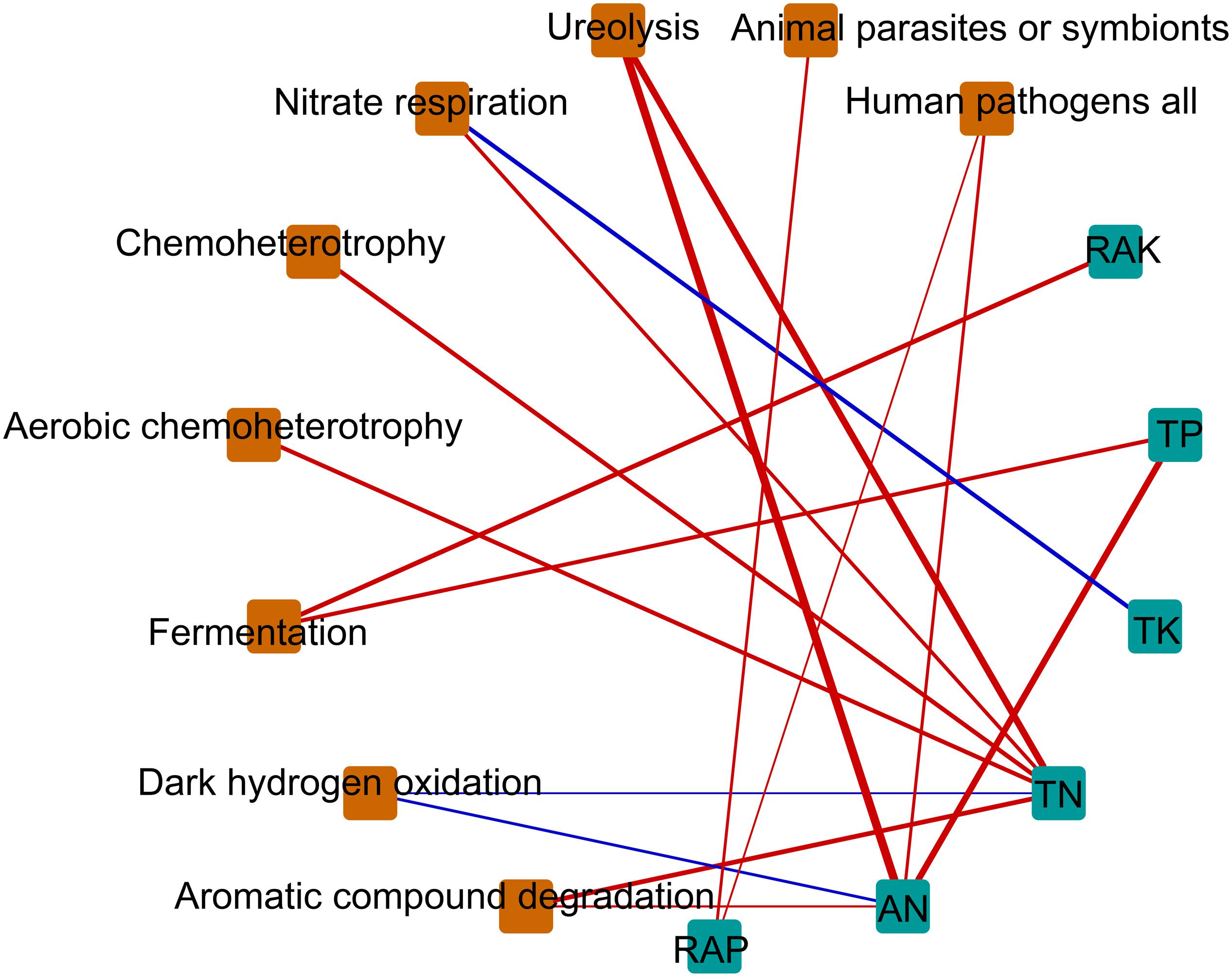
Figure 6. Correlation-based network analysis showing potential interactions between physicochemical parameters and ecological function. Node size is proportional to a functional group’s average relative abundance (log transformation) across all the samples. Lines connecting nodes (edges) represent positive (blue) or negative (red) co-occurrence relationships. TN, total nitrogen; TP, total phosphorus, TK, total potassium; AN, available nitrogen; RAP, rapidly available phosphorus; RAK, rapidly available potassium.
Discussion
It has been suggested that continuous monoculture systems have negative impacts on soil function and sustainability (Liu et al., 2006; Chen et al., 2009, 2015; Nayyar et al., 2009; Bado et al., 2010; Li et al., 2010; Zhao et al., 2018). However, little is known about how soil geochemical properties are affected by continuous cropping in Tibetan barley. In this study, we found TN and AN were decreased with continuous Tibetan barley cropping (Table 1). Continuous cropping may alter soil structure and lead to the hardening of soil (Rezapour and Samadi, 2012) and hollowness of the tillable layer (Mikha and Rice, 2004), producing obstacles for nutrition transport from bulk soil to rhizosphere soil. The TN and AN tapering may be derived from the change of soil structure after long-term Tibetan barley continuous cropping. The ability of the rhizosphere soil to transform nitrogen elements decreases during continuous cropping, and the decreased TN and AN may attribute to the low yields of Tibetan barley in the fields. The grain yields of Tibetan barley ranged from 1.5 to 6.1 t ha–2 with different concentrations of urea N fertilization (Cui et al., 2016). According to the Standard of Qinghai Province (DB63/T1536-2020), the high, intermediate, or low yields of Tibetan barley are 5.25 to 6.00 t ha–2, 4.50 to 5.25 t ha–2, and 4.05 to 4.5 t ha–2, respectively. In this study, yields of Tibetan barley consistently decreased during 6 years of monoculture, from 4.37 ± 0.17 t ha–2 to 2.79 ± 0.10 t ha–2 (Supplementary Figure S1), and were all classified as low levels of crop productivity. In accordance with our results, previous studies reported that continuous cropping often leads to restricted growth and crop yields with different time-scales (Ventura and Watanabe, 1978; Lua et al., 2017; Zhao et al., 2018), which could result from the increase in autotoxicity of root exudates (Gao and Zhang, 1998), the accumulation of pathogens (Li et al., 2014), and the disturbance of the microbial community structure in the soil (Hamid et al., 2016). In our study, bacterial community structures and functions in rhizosphere soil were significantly altered by the continuous cropping of Tibetan barley, which may influence element transformation in the soil and nutrition uptake up by the plant.
Bacterial community diversity significantly decreased with the increased continuous cropping duration of Tibetan barley (Figure 1). Decreased microbial diversity was also observed in rhizosphere soil with continuous cropping of cucumbers, rice, ginseng, and peas (Reichardt et al., 1997; Yao et al., 2006; Nayyar et al., 2009; Tan et al., 2017). These findings indicated that diversity reduction in soil microbial communities with continuous cropping could be common. The results of the present study provided evidence that diversity, taxonomic richness, and evenness of rhizosphere soil bacteria were strongly influenced by continuous cropping (Figure 1). OTU numbers and Faith PD (phylogenetic diversity) for rhizosphere soil decreased significantly with an increased duration of continuous cropping. In addition, changes in soil chemical properties were likely to drive bacterial community changes in rhizosphere soil. We found that TN the most important factor explaining the alpha diversity reduction of the rhizosphere bacterial community during continuous cropping (Figure 5). Soil nitrogen availability is known to drive rhizosphere bacterial community assembly during plant growth (Bell et al., 2015), and available N, NO3–-N, and NH4+-N were the main environmental factors affecting the distribution of the community structure in intensive plant monoculture (Li G. et al., 2016).
Loss of biodiversity has been identified as a common phenomenon in soil with long-term continuous cropping. In this study, alpha diversities (OTU numbers and Faith PD indices) declined during 5 years of continuous cropping but increased at 6 years (Figure 1). Similar results have also been observed in soil with continuous cropping of cotton and tea (Li Y.C. et al., 2016; Wei and Yu, 2018; Xi et al., 2019; Liang et al., 2020). We inferred that changes in soil chemical properties could have contributed to the distinct variations in bacterial community structures, as soil chemical properties play critical roles in microbial community structures (Kuramae et al., 2012). It is also worth noting that changes in bacterial community diversity and structure in Tibetan barley monoculture might also be caused by root exudate effects. Root exudates directly influence the assemblage and activities of the rhizosphere microbiota. Plants not only provide nutrients for microbial communities but also contain a series of antimicrobial metabolites in their root exudates (Bais et al., 2006; Berg and Smalla, 2009). It has been confirmed that continuous cropping accumulates root exudates, which results in a microbial community structure shift (Reichardt et al., 1997; Strickland et al., 2015; Wu et al., 2019). Monocropping results in plant roots repeatedly releasing the same exudates in the same fields long term, which can result in significant colonization by certain pathogenic or beneficial microorganisms that utilize these substrates (Brimecombe et al., 2001). Thus, the continuous cropping system was speculated as a dominant factor disrupting the balance of the bacterial community structure in Tibetan barley rhizosphere soil.
Continuous cropping not only affects bacterial diversity but also bacterial community composition in rhizosphere soil. The abundances of Pseudomonadaceae, Cytophagaceae, and Nocardioidaceae increased significantly throughout the experiment (Supplementary Figure S3), indicating that the bacteria in these lineages were more adaptable to the continuous cropping environment. The relative abundance of Pseudomonas (in the family of Pseudomonadaceae) increased from 0 to 10-yr in soil with continuous cropping of tea (Li Y.C. et al., 2016). A previous study demonstrated that Pseudomonas density was closely related to plant growth (Rumberger et al., 2007). Furthermore, we found the relative abundance of Pseudomonas was positively related to most of the predicted functionalities of bacterial communities in rhizosphere soil (Figure 4). The above results indicated that bacteria affiliated with the family Pseudomonadaceae positively contributed to the effects of Tibetan barley continuous cropping. The relative abundance of Actinobacteria increased during continuous cropping, and Xanthomonadaceae increased at 4 years of monocropping and was restored after 6 years of continuous cropping (Supplementary Figure S3). Similarly, Actinobacteria was strongly enriched in the soil after long-term cotton continuous cropping (Xi et al., 2019). Actinobacteria take part in the global carbon cycle and break down soil organic matter (Upchurch et al., 2008). Thus, members of the phylum Actinobacteria in continuous Tibetan barley cropping soils may have a role in carbon transformation in rhizosphere soil. Moreover, high-throughput sequencing technology reveals that continuous cropping of American ginseng results in an increase of Xanthomonadaceae (Dong et al., 2017). However, in a study of consecutive monoculture of sweet potato, the abundance of Xanthomonadaceae in soil decreased considerably as the number of continuous cropping years increased (Li et al., 2018). A decrease in Nocardioidaceae abundance was observed in rhizosphere soil during continuous cropping of Tibetan barley (Supplementary Figure S3). The abundances of Nocardioidaceae decreased with the increasing years of consecutive monoculture in rhizosphere soils of Achyranthes bidentata (Wang et al., 2019). However, the functions carried out by Xanthomonadaceae and Nocardioidaceae in the soil are less understood, and their relationship with plant continuous cropping should be explored in future studies. Chitinophagaceae was the only family detected whose abundance significantly decreased with increasing continuous cropping duration (Supplementary Figure S3). However, the functions carried out by Chitinophagaceae warrant further exploration. Members of the genus Sphingomonas have raised increasing attention due to their ability for organic carbon degradation and their ubiquity in the environment (Bastiaens et al., 2000; Leys et al., 2004). OTUs classified as Sphingomonas (OTU2, OTU4, OTU6, and OTU8; Supplementary Table S1) dominated in the rhizosphere bacterial communities, suggesting their roles in utilizing organic carbon or root exudates in the rhizosphere soil of Tibetan barley during continuous cropping. It was also observed that phytoplasma (OTU5), exhibited an increasing pattern during continuous cropping. Phytoplasma is associated with the yellow leaf syndrome disease in sugarcane (Parmessur et al., 2002), however, the role of phytoplasma in the rhizosphere environment is less understood, and its response to continuous cropping should be illustrated in the future. In summary, we speculate that the functional capabilities of the microbiome can greatly affect the structural and functional diversity of the bacterial community during continuous cropping of Tibetan barley.
Overwhelming scientific evidence has demonstrated that the rhizosphere soil microbial community diversity and structure are strongly influenced by continuous cropping systems. However, the impact of continuous cropping on the functions of rhizosphere microbial communities of plants cultivated at high altitudes remains poorly understood. In this study, FaProTax software (Louca et al., 2016) was used to predict the ecological functions of the rhizosphere soil bacterial community during continuous cropping of Tibetan barley. The results indicated that chemoheterotrophy and aerobic chemoheterotrophy-related species were dominant in rhizosphere soil (Figure 3), which concurred with previous soil findings (Legrand et al., 2018; Che et al., 2019; Liang et al., 2020). We observed that the relative abundances of 16S rRNA gene sequences assigned to chemoheterotrophy and aerobic chemoheterotrophy increased significantly with continuous cropping duration (Supplementary Figure S4). In continuous cropping systems, root exudates, including the secretion of a diverse array of carbon-containing metabolites, such as sugars, amino acids, and phenolic acids, are easily decomposable and act mainly as carbon sources for soil microbes (Bais et al., 2006). Increasing evidence suggests that the continuous cropping of plants can enrich the soil in root exudates (Guo et al., 2011). Thus, the accumulation of root exudates may be a major reason for the increased chemoheterotrophs during continuous cropping of Tibetan barley.
Plant–soil feedback occurs in the duration of continuous cropping, with consequent effects on plants grown and soil microbial communities. Many secondary metabolites secreted by the plant can inhibit microbial growth by autotoxicity or allelopathy effects (Bennett and Klironomos, 2019). Thus, secondary metabolites should be another important factor in determining the bacterial community structure of rhizosphere soil under continuous cropping conditions. Long-term continuous cropping causes substantial enrichment of harmful secondary metabolites secreted by plants, which eventually leads to the changes in the structure and function of rhizosphere microbial communities (Shen et al., 2020). Long-term continuous cropping in soybean causes substantial changes to the types and amounts of secondary metabolites secreted by the roots, leading to an enrichment of harmful secondary metabolites (phenolic acids, benzene, and esters), which eventually results in autotoxicity (Cui et al., 2018). It was also showed that secondary metabolites (ferulic and p-hydroxybenzoic acids) influenced the microbial composition of the cucumber rhizosphere (Jin et al., 2020), and phenolic acids could change soil microbial composition over continuous cropping seasons in tobacco (Shen et al., 2020). Secondary metabolism is involved in the tolerance to drought and salinity in Tibetan barley (Ahmed et al., 2015; Yuan et al., 2018). However, the changes of secondary metabolites in the rhizosphere of Tibetan barley during continuous cropping have not been reported, and their effects on rhizosphere soil microbes should be explored in the future.
Conclusion
In summary, our study demonstrated TN was the most important contributor to bacterial community diversity during continuous cropping, of which Pseudomonadaceae, Cytophagaceae, and Nocardioidaceae were key bacteria. Moreover, continuous cropping significantly increased the relative abundances of bacterial groups capable of chemoheterotrophy, aromatic compound degradation, and nitrate reduction. However, we only predicted bacterial function from a taxonomy assignment in this study. Further research should focus on obtaining direct evidence of changes in soil microbial functions in our continuous cropping system through approaches such as metagenomics or metatranscriptomic sequencing. The effects of continuous cropping on bacterial ecology in the rhizosphere of Tibetan barley should be given more attention to developing improved practices for continuous soil management and accelerated soil resource recycling in areas of high altitude.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA622628.
Author Contributions
YY and XY carried out the molecular experiments, analyzed data, and wrote the manuscript. YY and LA carried out data analyses. YB and DX contributed to the field experiments and collected samples. KW conceived the study, contributed to the design, and interpreted the research. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Key R&D Program of China (Grant nos. 2019YFD1001705 and 2019YFD1001700), the National Science Foundation of China (Grant nos. 32060447, 31660388, and 31960427), the China Agriculture Research System (Grant no. CARS-05), and the Project of Application and Fundamental Research in Qinghai Science and Technology Department (Grant no. 2019-ZJ-7075).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.551444/full#supplementary-material
Supplementary Figure 1 | Effects of continuous cropping on Tibetan Barley yield. Values are means ± standard deviation (n = 4).
Supplementary Figure 2 | Relative abundance of bacterial taxa in rhizosphere soil of Tibetan barley with different continuous cropping durations. (A) Relative abundances of the 10 most abundant bacterial phyla. (B) Relative abundances of the 10 most abundant bacterial families. Symbols represent means (n = 4).
Supplementary Figure 3 | Relative abundances of Pseudomonadaceae (A), Cytophagaceae (B), Nocardioidaceae (C), and Chitinophagaceae (D) plotted against years of continuous cropping of Tibetan barley.
Supplementary Figure 4 | Relative abundance of ecological function plotted against years of continuous cropping of Tibetan barley. (A) chemoheterotrophy; (B) aerobic chemoheterotrophy; (C) aromatic compound degradation; (D) nitrate reduction.
Supplementary Figure 5 | Spearman’s correlation heatmap based on population abundance and environmental variables for families with abundance >1%. AN, available nitrogen; TN, total nitrogen; RAK, rapidly available potassium; RAP, rapidly available phosphorus; TP, total phosphorus, TK, total potassium. Positive correlations (red), negative correlations (blue). *P < 0.05 and **P < 0.01 for the indicated comparisons.
Supplementary Table 1 | The changes of the 20 most abundant OTUs during continuous cropping.
Supplementary Table 2 | Relative abundance of families against years of continuous cropping of Tibetan barley.
Supplementary Table 3 | Dynamics of ecological function during continuous cropping of Tibetan barley.
Supplementary Table 4 | Relative abundance of ecological function against years of continuous cropping of Tibetan barley.
Footnotes
- ^ https://github.com/qiime2/q2-demux
- ^ https://github.com/qiime2/q2-dada2
- ^ https://github.com/qiime2/q2-feature-classifier
References
Ahmed, I. M., Nadira, U. A., Bibi, N., Cao, F. B., He, X. Y., Zhang, G. P., et al. (2015). Secondary metabolism and antioxidants are involved in the tolerance to drought and salinity, separately and combined, in Tibetan wild barley. Environ. Exp. Bot. 111, 1–12. doi: 10.1016/j.envexpbot.2014.10.003
Ali, A., Imran Ghani, M., Li, Y. H., Ding, H. Y., Meng, H. W., and Cheng, Z. H. (2019). Hiseq base molecular characterization of soil microbial community, diversity structure, and predictive functional profiling in continuous cucumber planted soil affected by diverse cropping systems in an intensive greenhouse region of Northern China. Int. J. Mol. Sci. 20:2619. doi: 10.3390/ijms20112619
Bado, B. V., Aw, A., and Ndiaye, M. (2010). Long-term effect of continuous cropping of irrigated rice on soil and yield trends in the Sahel of West Africa. Nutr. Cycl. Agroecosyst. 88, 133–141. doi: 10.1007/s10705-010-9355-7
Bai, L., Cui, J. Q., Jie, W. G., and Cai, B. Y. (2015). Analysis of the community compositions of rhizosphere fungi in soybeans continuous cropping fields. Microbiol. Res. 180, 49–56. doi: 10.1016/j.micres.2015.07.007
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bastiaens, L., Springael, D., Wattiau, P., Harms, H., Dewachter, R., Verachtert, H., et al. (2000). Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66, 1834–1843. doi: 10.1128/aem.66.5.1834-1843.2000
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: An Open Source Software for Exploring and Manipulating Networks. Menlo Park, CA: AAAI.
Bell, C. W., Asao, S., Calderon, F., Wolk, B., and Wallenstein, M. D. (2015). Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol. Biochem. 85, 170–182. doi: 10.1016/j.soilbio.2015.03.006
Bennett, J. A., and Klironomos, J. (2019). Mechanisms of plant–soil feedback: interactions among biotic and abiotic drivers. New Phytol. 222, 91–96. doi: 10.1111/nph.15603
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Brimecombe, M. J., De Leij, F. A., and Lynch, J. M. (2001). “The effect of root exudates on rhizosphere microbial populations,” in The Rhizosphere: Biochemistry and Organic Substances at the Soil–Plant Interface, eds R. Pinton, Z. Varaninin, and P. Nannipieri (New York: Marcel Dekker), 95–140.
Bryant, J. A., Lamanna, C., Morlon, H., Kerkhoff, A. J., Enquist, B. J., and Green, J. L. (2008). Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U.S.A. 105, 11505–11511. doi: 10.1073/pnas.0801920105
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Che, R. X., Wang, Y. F., Li, K. X., Xu, Z. H., Hu, J. M., Wang, F., et al. (2019). Degraded patch formation significantly changed microbial community composition in alpine meadow soils. Soil Tillage Res. 195:104426. doi: 10.1016/j.still.2019.104426
Chen, H. Q., Hou, R. X., Gong, Y. S., Li, H. W., Fan, M. S., and Kuzyakov, Y. (2009). Effects of 11 years of conservation tillage on soil organic matter fractions in wheat monoculture in Loess Plateau of China. Soil Tillage Res. 106, 85–94. doi: 10.1016/j.still.2009.09.009
Chen, M. N., Li, X., Yang, Q. L., Chi, X. Y., Pan, L. J., Chen, N., et al. (2012). Soil eukaryotic microorganism succession as affected by continuous cropping of peanut - Pathogenic and beneficial fungi were selected. PLoS One 7:e0040659. doi: 10.1371/journal.pone.0040659
Chen, M. N., Li, X., Yang, Q. L., Chi, X. Y., Pan, L. J., Chen, N., et al. (2014). Dynamic succession of soil bacterial community during continuous cropping of peanut (Arachis hypogaea L.). PLoS One 9:e101355. doi: 10.1371/journal.pone.0101355
Chen, S. L., Zhou, B. L., Lin, S. S., Li, X., and Ye, X. L. (2015). Accumulation of cinnamic acid and vanillin in eggplant root exudates and the relationship with continuous cropping obstacle. Afr. J. Biotechnol. 10:1338. doi: 10.5897/ajb10.1338
Choo, T. M. (2002). Genetic resources of Tibetan barley in China. Crop Sci. 42, 1759–1761. doi: 10.2135/cropsci2002.1759
Cui, J. H., Yang, H. J., Yu, C. Q., Bai, S., Wu, T. T., Song, S. S., et al. (2016). Effect of urea fertilization on biomass yield, chemical composition, in vitro rumen digestibility and fermentation characteristics of straw of highland barley planted in Tibet. J. Agric. Sci. 154, 151–164. doi: 10.1017/s0021859615000805
Cui, J. Q., Sun, H. B., Sun, M. B., Liang, R. T., Jie, W. G., and Cai, B. Y. (2018). Effects of Funneliformis mosseae on root metabolites and rhizosphere soil properties to continuously-cropped soybean in the potted-experiments. Int. J. Mol. Sci. 19:2160. doi: 10.3390/ijms19082160
De’ath, G. (2007). Boosted trees for ecological modeling and prediction. Ecology 88, 243–251. doi: 10.1890/0012-9658(2007)88[243:btfema]2.0.co;2
Dong, L. L., Xu, J., Zhang, L. J., Yang, J., Liao, B. S., Li, X. W., et al. (2017). High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chinese Med. 12:18. doi: 10.1186/s13020-017-0139-8
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Gao, Z. Q., and Zhang, S. X. (1998). Continuous cropping obstacle and rhizospheric rnicroecology 1. Root exrxlates and their ecological effects. J. Appl. Ecol. 9, 549–554.
Guo, Z. Y., Kong, C. H., Wang, J. G., and Wang, Y. F. (2011). Rhizosphere isoflavones (daidzein and genistein) levels and their relation to the microbial community structure of mono-cropped soybean soil in field and controlled conditions. Soil Biol. Biochem. 43, 2257–2264. doi: 10.1016/j.soilbio.2011.07.022
Hamid, M. I., Hussain, M., Wu, Y. P., Zhang, X. L., Xiang, M. C., and Liu, X. Z. (2016). Successive soybean-monoculture cropping assembles rhizosphere microbial communities for the soil suppression of soybean cyst nematode. FEMS Microbiol. Ecol. 93:fiw222. doi: 10.1093/femsec/fiw222
Jin, X., Wu, F. Z., and Zhou, X. G. (2020). Different toxic effects of ferulic and p-hydroxybenzoic acids on cucumber seedling growth were related to their different influences on rhizosphere microbial composition. Biol. Fertil. Soils 56, 125–136. doi: 10.1007/s00374-019-01408-0
Kumar, S., Suyal, D. C., Yadav, A., Shouche, Y., and Goel, R. (2019). Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS One 14:e0213844. doi: 10.1371/journal.pone.0213844
Kuramae, E. E., Yergeau, E., Wong, L. C., Pijl, A. S., Van Veen, J. A., and Kowalchuk, G. A. (2012). Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 79, 12–24. doi: 10.1111/j.1574-6941.2011.01192.x
Legrand, F., Picot, A., Cobo-Díaz, J. F., Carof, M., Chen, W., and Le Floch, G. (2018). Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 132, 135–145. doi: 10.1016/j.apsoil.2018.08.016
Leys, N. M. E. J., Ryngaert, A., Bastiaens, L., Verstraete, W., Top, E. M., and Springael, D. (2004). Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70, 1944–1955. doi: 10.1128/aem.70.4.1944-1955.2004
Li, C. G., Li, X. M., Kong, W. D., Wu, Y., and Wang, J. G. (2010). Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil 330, 423–433. doi: 10.1007/s11104-009-0216-6
Li, J. G., Shen, M. C., Hou, J. F., Li, L., Wu, J. X., and Dong, Y. H. (2016). Effect of different levels of nitrogen on rhizosphere bacterial community structure in intensive monoculture of greenhouse lettuce. Sci. Rep. 6:25305. doi: 10.1038/srep25305
Li, W. H., Liu, Q. Z., and Chen, P. (2018). Effect of long-term continuous cropping of strawberry on soil bacterial community structure and diversity. J. Integr. Agric. 17, 2570–2582. doi: 10.1016/S2095-3119(18)61944-6
Li, X. G., Ding, C. F., Zhang, T. L., and Wang, X. X. (2014). Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 72, 11–18. doi: 10.1016/j.soilbio.2014.01.019
Li, Y. C., Li, Z., Li, Z. W., Jiang, Y. H., Weng, B. Q., and Lin, W. X. (2016). Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 121, 787–799. doi: 10.1111/jam.13225
Liang, S. C., Deng, J. J., Jiang, Y., Wu, S. J., Zhou, Y. B., and Zhu, W. X. (2020). Functional distribution of bacterial community under different land use patterns based on FaProTax function prediction. Polish J. Environ. Stud. 29, 1245–1261. doi: 10.15244/pjoes/108510
Liu, X., Herbert, S., Hashemi, A., Zhang, X., and Ding, G. (2006). Effects of agricultural management on soil organic matter and carbon transformation - A review. Plant Soil Environ. 52:531. doi: 10.17221/3544-pse
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lua, Q., Zhanga, J. C., and Chena, L. S. (2017). Impact of monoculture of poplar on the rhizosphere microbial communities over time. Pedosphere 30, 487–495. doi: 10.1016/S1002-0160(17)60416-8
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mikha, M. M., and Rice, C. W. (2004). Tillage and manure effects on soil and aggregate-associated carbon and nitrogen. Soil Sci. Soc. Am. J. 68, 809–816. doi: 10.2136/sssaj2004.8090
Nayyar, A., Hamel, C., Lafond, G., Gossen, B. D., Hanson, K., and Germida, J. (2009). Soil microbial quality associated with yield reduction in continuous-pea. Appl. Soil Ecol. 43, 115–121. doi: 10.1016/j.apsoil.2009.06.008
Nichols, K. A., and Wright, S. F. (2006). Carbon and nitrogen in operationally defined soil organic matter pools. Biol. Fertil. Soils 43, 215–220. doi: 10.1007/s00374-006-0097-2
Parmessur, Y., Aljanabi, S., Saumtally, S., and Dookun-Saumtally, A. (2002). Sugarcane yellow leaf virus and sugarcane yellows phytoplasma: elimination by tissue culture. Plant Pathol. 51, 561–566. doi: 10.1046/j.1365-3059.2002.00747.x
Praeg, N., Pauli, H., and Illmer, P. (2019). Microbial diversity in bulk and rhizosphere soil of Ranunculus glacialis along a high-alpine altitudinal gradient. Front. Microbiol. 10:1429. doi: 10.3389/fmicb.2019.01429
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rawat, J., Yadav, N., and Pande, V. (2020). “Chapter 7 - Role of rhizospheric microbial diversity in plant growth promotion in maintaining the sustainable agrosystem at high altitude regions,” in Recent Advancements in Microbial Diversity, eds S. De Mandal and P. Bhatt (Cambridge, MA: Academic Press), 147–196. doi: 10.1016/b978-0-12-821265-3.00007-4
Reichardt, W., Mascarina, G., Padre, B., and Doll, J. (1997). Microbial communities of continuously cropped, irrigated rice fields. Appl. Environ. Microbiol. 63, 233–238. doi: 10.1128/aem.63.1.233-238.1997
Rezapour, S., and Samadi, A. (2012). Assessment of inceptisols soil quality following long-term cropping in a calcareous environment. Environ. Monit. Assess. 184, 1311–1323. doi: 10.1007/s10661-011-2042-6
Rumberger, A., Merwin, I. A., and Thies, J. E. (2007). Microbial community development in the rhizosphere of apple trees at a replant disease site. Soil Biol. Biochem. 39, 1645–1654. doi: 10.1016/j.soilbio.2007.01.023
Shen, H., Yan, W. H., Yang, X. Y., He, X. H., Wang, X., Zhang, Y. T., et al. (2020). Co-occurrence network analyses of rhizosphere soil microbial PLFAs and metabolites over continuous cropping seasons in tobacco. Plant Soil 425, 119–135. doi: 10.1007/s11104-020-04560-x
Strickland, M. S., Mcculley, R. L., Nelson, J. A., and Bradford, M. A. (2015). Compositional differences in simulated root exudates elicit a limited functional and compositional response in soil microbial communities. Front. Microbiol. 6:817. doi: 10.3389/fmicb.2015.00817
Tan, Y., Cui, Y. S., Li, H. Y., Kuang, A. X., Li, X. R., Wei, Y. L., et al. (2017). Diversity and composition of rhizospheric soil and root endogenous bacteria in Panax notoginseng during continuous cropping practices. J. Basic Microbiol. 57, 337–344. doi: 10.1002/jobm.201600464
Upchurch, R., Chiu, C. Y., Everett, K., Dyszynski, G., Coleman, D. C., and Whitman, W. B. (2008). Differences in the composition and diversity of bacterial communities from agricultural and forest soils. Soil Biol. Biochem. 40, 1294–1305. doi: 10.1016/j.soilbio.2007.06.027
Van Raij, B., Quaggio, J. A., and Da Silva, N. M. (1986). Extraction of phosphorus, potassium, calcium, and magnesium from soils by an ion-exchange resin procedure. Commun. Soil Sci. Plant Anal. 17, 547–566. doi: 10.1080/00103628609367733
Ventura, W., and Watanabe, I. (1978). Growth inhibition due to continuous cropping of dryland rice and other crops. Soil Sci. Plant Nutr. 24, 375–389. doi: 10.1080/00380768.1978.10433117
Wang, J. T., Cao, P., Hu, H. W., Li, J., Han, L. L., Zhang, L. M., et al. (2015). Altitudinal distribution patterns of soil bacterial and archaeal communities along mt. Shegyla on the Tibetan Plateau. Microb. Ecol. 69, 135–145. doi: 10.1007/s00248-014-0465-7
Wang, J. Y., Wu, L. K., Tantai, H. P., Khan, M. U., Letuma, P., Wu, H. M., et al. (2019). Properties of bacterial community in the rhizosphere soils of Achyranthes bidentata tolerant to consecutive monoculture. Plant Growth Regul. 89, 167–178. doi: 10.1007/s10725-019-00523-0
Wei, Z., and Yu, D. (2018). Analysis of the succession of structure of the bacteria community in soil from long-term continuous cotton cropping in Xinjiang using high-throughput sequencing. Arch. Microbiol. 200, 653–662. doi: 10.1007/s00203-018-1476-4
Wu, H. M., Qin, X. J., Wang, J. Y., Wu, L. K., Chen, J., Fan, J. K., et al. (2019). Rhizosphere responses to environmental conditions in Radix pseudostellariae under continuous monoculture regimes. Agric. Ecosyst. Environ. 270-271, 19–31. doi: 10.1016/j.agee.2018.10.014
Xi, H., Shen, J. L., Qu, Z., Yang, D. Y., Liu, S. M., Nie, X. H., et al. (2019). Effects of long-term cotton continuous cropping on soil microbiome. Sci. Rep. 9, 18297–18297. doi: 10.1038/s41598-019-54771-1
Yao, H. Y., Jiao, X. D., and Wu, F. Z. (2006). Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 284, 195–203. doi: 10.1007/s11104-006-0023-2
Yuan, H. J., Zeng, X. Q., Shi, J., Xu, Q. J., Wang, Y. L., Jabu, D. Z., et al. (2018). Time-course comparative metabolite profiling under osmotic stress in tolerant and sensitive Tibetan hulless barley. BioMed. Res. Int. 2018:9415409. doi: 10.1155/2018/9415409
Zeng, X. Q., Guo, Y., Xu, Q. J., Mascher, M., Guo, G. G., Li, S. C., et al. (2018). Origin and evolution of qingke barley in Tibet. Nat. Commun. 9:5433. doi: 10.1038/s41467-018-07920-5
Zhang, D., Fengquan, L., and Jianmin, B. (2000). Eco-environmental effects of the Qinghai-Tibet Plateau uplift during the Quaternary in China. Environ. Geol. 39, 1352–1358. doi: 10.1007/s002540000174
Zhang, D., Zhou, Z. H., Zhang, B., Du, S. H., and Liu, G. C. (2012). The effects of agricultural management on selected soil properties of the arable soils in Tibet, China. CATENA 93, 1–8. doi: 10.1016/j.catena.2012.01.004
Zhang, M. K., and He, Z. L. (2004). Long-term changes in organic carbon and nutrients of an Ultisol under rice cropping in southeast China. Geoderma 118, 167–179. doi: 10.1016/S0016-7061(03)00191-5
Zhao, H. T., Li, T. P., Zhang, Y., Hu, J., Bai, Y. C., Shan, Y. H., et al. (2017). Effects of vermicompost amendment as a basal fertilizer on soil properties and cucumber yield and quality under continuous cropping conditions in a greenhouse. J. Soils Sediments 17, 2718–2730. doi: 10.1007/s11368-017-1744-y
Zhao, Q. Y., Xiong, W., Xing, Y. Z., Sun, Y., Lin, X. J., and Dong, Y. P. (2018). Long-term coffee monoculture alters soil chemical properties and microbial communities. Sci. Rep. 8:6116. doi: 10.1038/s41598-018-24537-2
Keywords: Tibetan barley, continuous cropping, rhizosphere soil, bacterial community structure, predictive functional profiling
Citation: Yao Y, Yao X, An L, Bai Y, Xie D and Wu K (2020) Rhizosphere Bacterial Community Response to Continuous Cropping of Tibetan Barley. Front. Microbiol. 11:551444. doi: 10.3389/fmicb.2020.551444
Received: 13 April 2020; Accepted: 05 November 2020;
Published: 30 November 2020.
Edited by:
Paulo José Pereira Lima Teixeira, University of São Paulo, BrazilReviewed by:
Ederson Da Conceicao Jesus, Brazilian Agricultural Research Corporation (EMBRAPA), BrazilXingang Zhou, Northeast Agricultural University, China
Chunjie Tian, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, China
Copyright © 2020 Yao, Yao, An, Bai, Xie and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunlun Wu, d2tscWFhZkBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Youhua Yao1,2,3†
Youhua Yao1,2,3† Kunlun Wu
Kunlun Wu