
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 October 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.509380
 Paula J. Uriza1
Paula J. Uriza1 Cynthia Trautman1
Cynthia Trautman1 María M. Palomino2,3
María M. Palomino2,3 Joaquina Fina Martin2,3
Joaquina Fina Martin2,3 Sandra M. Ruzal2,3
Sandra M. Ruzal2,3 Mara S. Roset1*
Mara S. Roset1* Gabriel Briones1*
Gabriel Briones1*S-layers are bacterial structures present on the surface of several Gram-positive and Gram-negative bacteria that play a role in bacterial protection. In Lactobacillus acidophilus (L. acidophilus ATCC 4356), the S-layer is mainly composed of the protein SlpA. A tandem of two copies of the protein domain SLP-A (pfam: 03217) was identified at the C-terminal of SlpA, being this double SLP-A protein domain (in short dSLP-A) necessary and sufficient for the association of the protein to the L. acidophilus cell wall. A variety of proteins fused to the dSLP-A domain were able to spontaneously associate with high affinity to the cell wall of L. acidophilus and Bacillus subtilis var. natto, in a process that we termed decoration. Binding of dSLP-A-containing-proteins to L. acidophilus was stable at conditions that mimic the gastrointestinal transit in terms of pH, proteases, and bile salts. To evaluate if protein decoration of L. acidophilus can be adapted to generate an oral vaccine platform, a chimeric antigen derived from the bacterial pathogen Shiga-toxin-producing Escherichia coli (STEC) was constructed by fusing the sequences encoding the polypeptides EspA36–192, Intimin653–953, Tir240–378, and H7 flagellin352–374 (EITH7) to the dSLP-A domain (EITH7-dSLP-A). Recombinantly expressed EITH7-dSLP-A protein was affinity purified and combined with L. acidophilus cultures to allow the association of the chimeric antigen to the bacterial surface. EITH7-decorated L. acidophilus was orally administered to BALB/c mice and the induction of anti-EITH7 specific antibodies in sera and feces determined by ELISA. Mice presenting significantly higher anti-EITH7 antibodies titers were able to control more efficiently an experimental STEC infection than mice that received the non-decorated L. acidophilus carrier, indicating that antigen-decorated L. acidophilus can be adapted as a mucosal immunization delivery platform to elicit a protective immune response for vaccine purposes.
Vaccines have been the most successful medical therapy in Public Health preventing mortality and morbidity caused by several infectious diseases and saving millions of lives worldwide (Orenstein and Ahmed, 2017). Although most microbial pathogens enter the body through a particular mucosal barrier (gastrointestinal, respiratory, or genital mucosa) only a few vaccines using this route of administration have been approved, probably due to certain limitations in their protection performance (Lycke, 2012). It has been described that some oral vaccines performed poorly in developing countries compared with industrialized ones and this has been attributed to a syndrome named chronic environmental enteropathy (or tropical enteropathy). This syndrome is characterized by impairment in intestinal absorption that might lead to a reduction of bioavailability of zinc or vitamin A affecting the efficiency of adjuvant function and, therefore, impairing the induction of an effective immune response (Lycke, 2012). Another limitation related to oral administration of antigens is the natural proteolytic degradation that proteins suffer in the intestinal tract. Despite these limitations, oral immunization remains an ideal vaccine route to be explored, especially in global campaigns of vaccination; because its administration is simple (needleless administration) and non-medical trained personnel can perform it. Live vaccines based on attenuated microorganisms have been shown to be a very effective way to elicit a protective immunity as a consequence of the ability of the vaccine strain to replicate inside the host resembling the real infection, therefore, providing an effective antigenic stimulation that leads to the development of a protective memory immune response. The selection of attenuated microorganisms has been historically a serendipitous process based on the sequential replication of a pathogen either in laboratory in vitro conditions or within a non-natural host in order to isolate non-virulent strains (Plotkin, 2014; Minor, 2015). By this method, a few protective vaccines have been developed that have been widely used such as Oral Polio Virus (Sabin), BCG for tuberculosis, and the Yellow Fever vaccine YF17D (Galler et al., 1997; Plotkin, 2014). Despite a better performance of live-attenuated vaccines to induce individual and population protection, there are certain limitations such as the latent possibility of reversion to a full virulence or the risk that the residual virulence of a vaccine strain might be still dangerous for immune-compromised individuals. In addition, not all the microbial pathogens can be cultured in vitro and, in some cases, special biosafety facilities are also required, increasing production costs. To sort out these limitations, it has been proposed the use of vaccine strains as carriers for expressing heterologous antigens. Recently, a recombinant tetravalent vaccine against Dengue virus using the Yellow fever vaccine as a carrier was approved for humans, although, as a consequence of some complications that appeared during Phase III, the vaccine has had a limited use (Larson et al., 2019). In the case of vaccines based on genetically modified organisms (GMOs), they are subjected to strict regulations to allow its administration to animals or humans increasing production costs considerably (Seib et al., 2017).
Members of the Lactobacillus genus are considered as generally recognized as safe (GRAS) bacteria, since they are natural commensal bacteria that inhabit the gastrointestinal tract and genital mucosa and have been used for centuries in the preparation of fermented foods like yogurt, kefir, or sour cream. Given their mucosal localization, Lactobacillus species have been recently proposed as a recombinant carrier for oral vaccine development. For example, the human immunodeficiency virus (HIV) Gag, heat labile enterotoxin, rotavirus VP4, and the Helicobacter Adhesin Hp0410, have been expressed in Lactobacillus strains for vaccine purposes (Lecureux and Dean, 2018). However, these Lactobacillus strains are DNA-recombinant vaccines, which affect their GRAS status.
When residing in the gut, some Lactobacillus species interact with host cells exerting a strain-specific immunomodulatory effect, that contributes to its probiotic properties (Hymes et al., 2016; Johnson et al., 2016; Douillard and de Vos, 2019). This immune-modulation has been attributed to the recognition of different microbial-associated molecular patterns (MAMPs) mediated by the pattern recognition receptors (PRRs), expressed in immune and non-immune host cells (Abou-Saleh et al., 2016; Hymes et al., 2016; Douillard and de Vos, 2019). MAMPs-PRPs interaction triggers an inflammatory response, which is the main mechanism of action of adjuvants, such as the observed in the bacterium-like particles or Gram-positive enhancer matrixes used as antigen delivery platforms (Arce et al., 2019). In this report, we adapted Lactobacillus acidophilus as an oral vaccine platform to deliver heterologous antigens from a bacterial intestinal pathogen, the Shiga-toxin-producing Escherichia coli (STEC). STEC is a bacterial pathogen that causes hemorrhagic enteritis and the life-threatening hemolytic uremic syndrome (HUS) in humans (Goldwater, 2007). Immunization with STEC surface proteins (such as EspA, Tir, Intimin, or flagellin) induces an antibody humoral response that interferes with the effective STEC-host cell interaction. Based on this observation, two vaccine candidates have been developed for the prevention or reduction of STEC intestinal colonization in cattle (Potter et al., 2004; Babiuk et al., 2008). STEC deploys protein secretion machinery, the type three secretion system (T3SS), critical for host invasion and intestinal colonization. STEC T3SS is a “nanomachine” that the bacteria utilize to translocate effector proteins to the host cell cytosol to promote its tight association to the host cell surface. Initially, the effector protein Tir is “injected” by the STEC T3SS within the host cells and exposed on the host membrane, where it will be recognized by Intimin, a STEC outer membrane protein. When Tir interacts with Intimin, a host-signaling cascade is triggered that leads to the rearrangement of the intestinal epithelial cellular architecture, inducing the formation of actin “pedestals” beneath the attached bacteria. In addition, the T3SS effector protein EspA polymerizes forming a filamentous tip on the bacterial membrane which is essential for bacterial attachment and T3SS activity. Here, we constructed a STEC chimeric antigen composed by the peptides EspA36–192, Intimin653–935 (containing the Tir-binding domain), Tir258–361 (containing the Intimin-binding domain), and H7 flagellin352–374 (EITH7), which is protective against an E. coli O157:H7 challenge infection in the mouse model (Iannino et al., 2015), and fused in frame with the peptide SlpA284–444 of L. acidophilus. The resulting EITH7-SlpA284–444 was recombinantly expressed and mixed with a L. acidophilus culture to allow the external coating of the bacteria with the STEC antigen, in a process that we termed “decoration.”
The L. acidophilus S-layer is composed mainly of SlpA, although there are less-represented proteins such as SlpX (Palomino et al., 2016). After being secreted, the S-layer proteins can self-assemble forming a bi-dimensional symmetrical structure that completely covers the bacterial cell wall. In L. acidophilus, SlpA, in addition to its function as a protective coat for the bacteria, also participates in host cell adhesion (Ashida et al., 2011). It has been described that the S-layer can be removed from the bacterial surface by cation substitution solutions (LiCl) or hydrogen bond-breaking agents such as guanidine hydrochloride (GHCl) or urea (Sleytr et al., 2014). Interestingly, after dialysis, the isolated S-layer protein can associate back to the Lactobacillus cell wall or adhere to inert surfaces that can be adapted for biotechnological purposes (Sleytr et al., 2014; Fina Martin et al., 2019). SlpA encodes in its carboxy-terminal (SlpA284–444) a tandem of two copies of the protein domain denominated SLP-A (dSLP-A), a region that was shown to be necessary and sufficient for the association of SlpA to the bacterial cell wall (Fina Martin et al., 2019). Teichoic acid and wall teichoic acid present on the lactobacilli bacterial cell wall have been characterized as natural ligands for the dSLP-A domain (Fina Martin et al., 2019). The dSLP-A domain (pfam: 03217) is found only in Lactobacillus species, limited to S-layer forming lactobacilli of the L. acidophilus group, including Lactobacillus helveticus, Lactobacillus crispatus, Lactobacillus gallinarum, and Lactobacillus amylovorus (Avall-Jaaskelainen and Palva, 2005). This domain is also found in S-layer associated proteins (Johnson et al., 2013) related to probiotic activity, adhesion (Hymes et al., 2016), immunomodulation (Johnson et al., 2017), and cell division (Johnson et al., 2016).
We evaluated the efficiency of L. acidophilus decorated with STEC antigens as a vaccine platform. Mice were orally immunized with EITH7-decorated L. acidophilus to evaluate its ability to elicit a protective immune response against STEC. Our results suggest that the antigen decoration strategy of lactobacilli can be an interesting alternative as a vaccine platform.
Bacterial strains and plasmids used are summarized in Table 1. Escherichia coli strains were cultured at 37°C on Luria Bertani (LB) medium (Sigma, St. Louis, MO, United States) or Sorbitol MacConkey agar (SMAC; Britania, Argentina). Lactobacillus acidophilus ATCC 4356 was cultured at 37°C on DeMan, Ragosa, and Sharpe medium (MRS; Britania, Argentina). Bacillus subtilis var. natto (ATCC 15245) was cultured in LB or LB agar. When necessary, culture media were supplemented with antibiotics at the following concentrations: ampicillin 50 μg/ml, kanamycin 50 μg/ml, and nalidixic acid 20 μg/ml. All experiments involving STEC were conducted in a BSL3 facility.
A DNA fragment encoding for the last 160 amino acids of SlpA (SlpA284–444) of L. acidophilus that contains a tandem of two SLP-A protein domains (dSLP-A) was synthesized by Integrated DNA Technologies, Inc. (IDT, United States). A synthetic gene encoding EspA36–192, Intimin653–953, Tir240–378, and flagellin H7domain (EITH7) from STEC EDL933 strain was synthesized by GenScript Company (United States; Iannino et al., 2015). The peptide (EAAAK)4 was intervened between individual proteins to promote their proper folding (Iannino et al., 2015). The FLAG epitope (DYKDDDDK) was fused to the C-terminal of EITH7 to monitor protein expression.
The DNA sequence encoding the L. acidophilus dSLP-A domain (SlpA284–444) was subcloned into pGEX-2T vector (GE-Healthcare; Table 1), and the resultant protein GST-dSLP-A was expressed and purified by Glutathione Sepharose Fast Flow (GE-Healthcare) affinity chromatography as described by manufacturer instruction. The encoding sequence of EITH7 was subcloned into pLC3 expression vector (Table 1) and afterward the dSLP-A encoding sequence was placed at the C-terminal as shown in Table 1. The resulting plasmid pLC3-EITH7-dSLP-A was transformed into E. coli BL21. Expression of EITH7-dSLP-A protein was induced by a final concentration of 0.25 mM of Isopropyl β-D-1-thiogalactopyranoside (Sigma-Aldrich) and purified by amylose affinity chromatography (New England BioLabs) as manufacturer instruction.
Sequences of fliC from Salmonella enterica serovar Typhimurium LT2, Brucella abortus omp19, and the mouse lectin gal8 were cloned into pLC3-EITH7- dSLP-A, replacing EITH7 from the construction (Table 1). Resulting M-FliC-dSLP-A, M-Omp19-dSLP-A, and M-Gal8-dSLP-A proteins was recombinantly expressed as described above and purified at the same experimental conditions used for M-EITH7-dSLP-A. For amplification of the sequences of Gal8 and Omp19 by PCR, the following primers were used: pFwGal8 5'-tgaCATATGgctagcatgttgtccttaaa-3', pRvGal8, 5'-gtaGTCGACccagctccttacatccag-3', pFwOmp19 5'-tgaCATATGatgcagagctcccggctt-3', and pRvOmp19 5'-tgaGTCGACgcgcgacagcgtcacgg-3'.
Eight-week-old BALB/c mice were immunized intraperitoneally with 10 μg of purified recombinant GST-dSLP-A protein or M-EITH7 protein using aluminum hydroxide (Imject Alum ThermoFisher) as an adjuvant and boosted at 2–4 weeks with 5 μg of each protein. A week after the last immunization, mice were bled, and sera were stored at −20°C for further use.
Protein samples were treated with cracking buffer (50 mM Tris-HCl pH 6.8, 2% wt/vol sodium dodecyl sulfate, 0.1% wt/vol bromophenol blue, 10% wt/vol glycerol, and 50 mM dithiothreitol) and incubated at 100°C for 5 min. Protein electrophoresis separation was performed at 120 V for 1 h on a 10% SDS-PAGE gel. Subsequently, proteins were transferred to a nitrocellulose membrane using a semi-dry electroblotting transfer unit (Bio-Rad, Hercules, CA, United States) for 45 min at 15 V. After 1 h blocking buffer [0.1% Tween 20 and 1% dry skim milk in phosphate-buffered saline (PBS) buffer], membranes were incubated for 1 h with the corresponding primary antibody. Membranes were washed four times during 5 min with washing buffer (0.1% Tween 20 in PBS buffer) and incubated for 1 h with a 1:20.000 blocking buffer dilution of IRDye fluorophore-labeled secondary antibodies (LI-COR, Lincoln, NE, United States). After washing four times, membranes were scanned using the Odyssey Imaging System (LI-COR).
Overnight cultures of lactobacilli were harvested by centrifugation. After centrifugation, the bacterial pellet was washed three times with cold PBS. An equal number of lactobacilli (5 × 108 CFU) were resuspended and incubated for 30 min with different concentrations of dSLP-A-fusion proteins in a final volume of 100 μl. Each fraction of decorated lactobacilli was washed three times with cold PBS. L. acidophilus was pre-treated for 15 min with a 5 M LiCl solution to remove the wild type SlpA. The same protocol was used also for decoration of B. subtilis.
For studies of binding stability, decorated bacteria were subjected to different conditions that mimic the gastrointestinal environment (pH, bile acids, and protease). After being collected by centrifugation, L. acidophilus was washed twice with PBS and processed for SDS-PAGE and western blot analysis. GST-dSLP-A decorated bacteria were incubated 30 min at pH2 (stomach-like conditions), 60 min at pH6.5 (proximal small bowel-like), 120 min at pH7.5 (mid-small bowel-like), and 240 min at pH7.5 (distal small bowel-like; Evans et al., 1988). For protease stability assay, a pancreatin (SIGMA-ALDRICH) concentration of 10 μg/ml was used. For bile acid stability were used different concentrations of bile acids (Britania; 4–40 mg/ml) at different incubation times (10, 15, 30, and 60 min; Pfeiler et al., 2007).
Eight-week-old BALB/c mice (female) were orally immunized with 4 × 108 UFC of lactobacilli or 4 × 108 CFU of lactobacilli carrying 20 μg of M-EITH7-dSLP-A or with M-EITH7-dSLP-A alone in a three doses scheme (0, 2, and 4 weeks). Prior oral immunization mice were fasted for 6 h and food was placed back 1 h after oral immunization.
Ten days after the last immunization, mice were challenged by intragastric inoculation of 1010 CFU of STEC in 100 μl of PBS. Previously, food was removed from mouse cages 6 h before the oral infection challenge. Bacterial fecal shedding was monitored daily for over 25 days. Briefly, mouse fecal pellets were collected, suspended in 500 μl of PBS, serially diluted, and plated onto SMAC agar with 20 μg/ml of nalidixic acid. Culture plates were incubated at 37°C for 24 h, and colony-forming units (CFUs) were determined.
The 96-well plate was coated with purified M-EITH7 protein (Table 1; 125 ng/well) in carbonate-bicarbonate buffer pH 9.6 for the iELISA test. To determine fecal antibodies, feces were resuspended in buffer PBS containing 1% of BSA (Sigma, St. Louis, MO, United States), 25 mM of EDTA, 2 mM of phenylmethylsulfonyl fluoride (PMSF; Sigma, St. Louis, MO, Unites States). The suspension was clarified by centrifugation and its supernatant diluted (1:2) and then used for an ELISA test during the same day. Sera samples were collected and stored at −20°C until use. Sera were 1:200 diluted in the blocking buffer (0.1% Tween 20 in PBS buffer). To reveal antibody presence, HRP-conjugated anti-mouse IgA (1:500) or HRP-anti-mouse-IgG (1:1000) were added and incubated for 1 h at 25°C. HRP activity was determined with 3, 3',5,5' Tetramethylbenzidine (Sigma), and optical density was measured at 450 nm (OD450). A positive serum was generated against the EITH7 antigen by ip immunization that was included in all the ELISA test plates as an internal reference value.
Lactobacillus acidophilus or B. subtilis previously decorated with dSLP-A-tagged proteins were incubated during 30 min on coverslips pretreated with poly-L-Lysine (Sigma, St. Louis, MO, United States) to allow the bacterial adhesion. After washing twice with PBS, samples were fixed with PFA (4% in PBS) for 15 min at room temperature. After fixation, coverslips were washed twice with PBS, and the remaining PFA was quenched by incubation with 50 mM NH4Cl (Sigma, St. Louis, MO, United States) for 1 h or overnight. Coverslips were washed with PBS and blocked during 1 h with 10% Bovine Serum Albumin (Sigma, St. Louis, MO, United States) in PBS. Later, covers were incubated 1 h with anti-EITH7 mouse sera diluted (1:500) in blocking buffer, washed with PBS, and incubated 1 h more with an ALEXA488- anti-IgG conjugated (ThermoFisher), diluted (1:10000) in blocking buffer. Finally, samples were examined with a confocal laser-scanning microscope Olympus FV1000 using a PlanApo N (60 × 1.42 NA) oil objective.
One-way ANOVA with Bonferroni post hoc test and Student’s t-test (GraphPad Software) was used for statistical analysis.
Experimental procedure of this study (permit number 15/2018) was approved by the Committee on the Ethics of Animal Experiments of the Universidad Nacional de San Martín (UNSAM), under the recommendations for animal experimentation (Helsinki Declaration and its amendments, Amsterdam Protocol of welfare and animal protection and National Institutes of Health, United States NIH, guidelines: Guide for the Care and Use of Laboratory Animals).
To analyze the presence of conserved protein domains within the SlpA amino acid sequence, a BLAST search was performed. As shown in Figure 1A, two copies of a 60-amino acids protein domain denominated SLP-A (pfam03217; Figure 1A; Boot et al., 1995) were identified within de carboxy- terminal of SlpA. Both copies of the SLP-A domain (dSLP-A) are required for an efficient cell wall binding of SlpA to the surface of L. acidophilus (Fina Martin et al., 2019). In addition, as shown in Figure 1A, the amino-terminal of SlpA (SlpA1–31) encodes a predicted signal peptide (analyzed by a deep neural network-based method SignalP-5.0) and the peptide region SlpA31–284 for a Herpes BLLF1 protein domain (pfam05109), a protein region that has been shown to be required for self-assembly of SlpA on the bacterial surface to form the S-layer (Smit et al., 2002). As shown in the scheme presented in Figure 1A, an expression vector was constructed for fusing in frame any heterologous proteins (h-proteins) of interest with the dSLP-A domain. Also, the sequence of the FLAG epitope was inserted between the heterologous protein of interest and the dSLP-A domain as a reporter tag. As shown in Figure 1B and Table 1, two different plasmids backbones were used for cloning proteins of interest, fused to the dSLP-A domain: the pLC3 (Savinov et al., 2012) and the plasmid pGEX-2T that allows the fusion of polypeptides with the maltose-binding protein (MBP) or the glutathione S-tranferase (GST), respectively, both for affinity purification purposes. Consequently, a set of dSLP-A-fused proteins were constructed and were used here (Figure 1B). As shown in Figure 1C, three different examples of heterologous proteins such as the Brucella protease inhibitor Omp19 (Ibañez et al., 2015), the Salmonella flagellin FliC (Mcclelland et al., 2001), and the mouse lectin GAL8 (Tribulatti et al., 2007) fused to the dSLP-A domain were able to efficiently bind to 108 CFU of L. acidophilus.
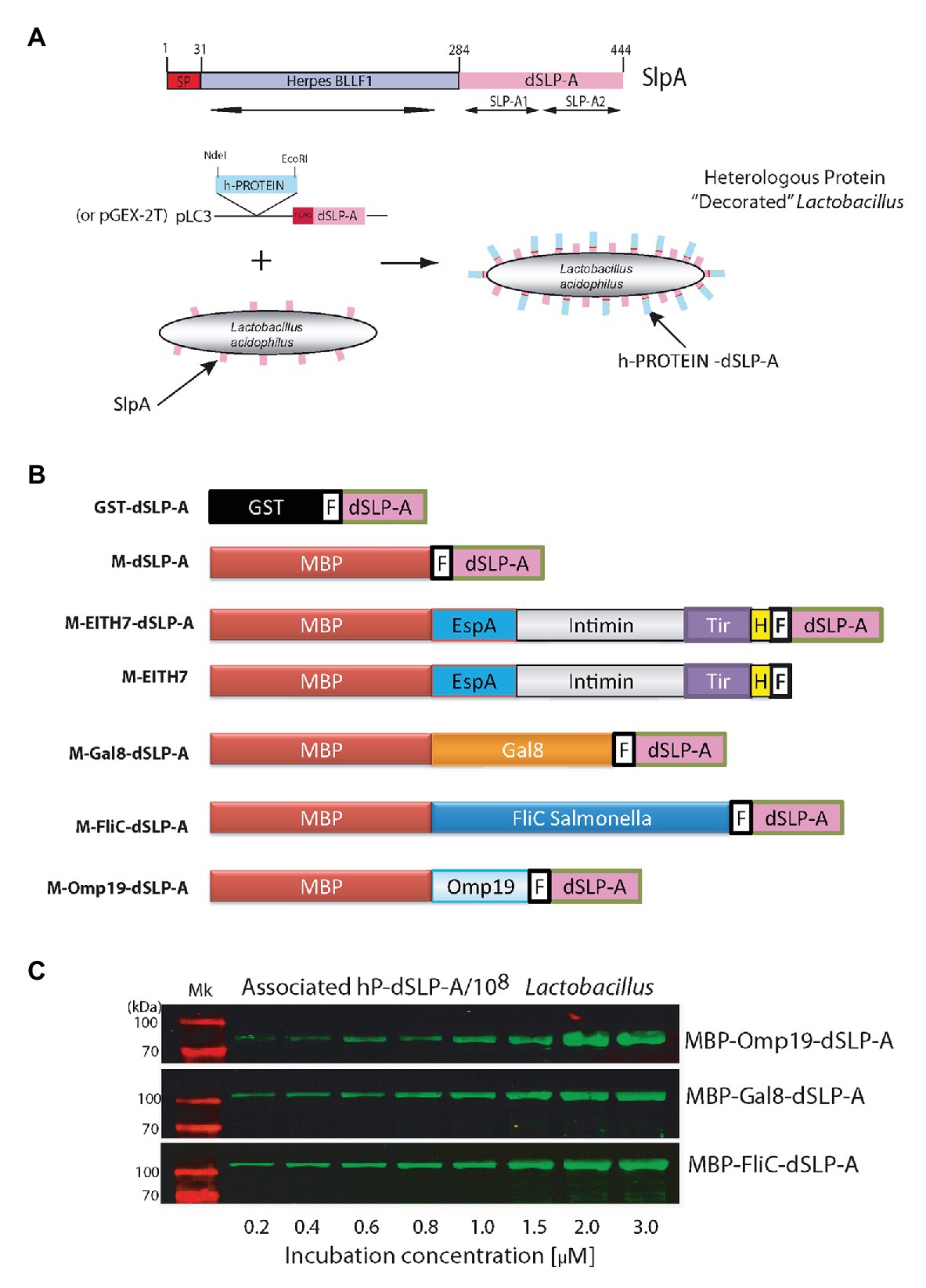
Figure 1. (A) The scheme shows all the protein domains detected in the sequence of the protein L. acidophilus SlpA predicted by the NCBI BLAST program. The binding domain named dSLP-A is formed by a tandem of two copies of the SLP-A domain (pfam03217): SLP-A1 and SLP-A2. SlpA1–31 encodes for a secretion signal sequence predicted by SignalP-5.0 (SP). In SlpA 31–284 region, a Herpes BLLF1 domain was detected (pfam05109). A cartoon showing the decoration process is presented which includes the expression vector used for tagging different heterologous proteins (h-PROTEIN) with the dSLP-A domain. (B) Different constructions used in this report. (C) Western blot analysis shows the increasing association of dSLP-A-tagged proteins to the Lactobacillus cell wall that correlates with the increasing concentration of these proteins in the incubation solution. TransGen Biotech Pre-stained markers blue-Plus Pre-stained markers 120, 100, 70, 50, 40, 30, 25, and 14 kDa. GST, glutathione S-transferase; MBP, maltose-binding protein; F, FLAG epitope; H, Escherichia coli H7 epitope.
As described in Table 1, the sequence of the dSLP-A domain was inserted in pGEX-2T to construct the reporter protein GST-dSLP-A (Figure 1B). Lactobacillus acidophilus decorated with GST-dSLP-A were subjected to environmental conditions that bacteria encounter during the transit through the intestinal tract (pH variations, bile salts, or degradation by intestinal proteases) to evaluate the stability of dSLP-A binding in these conditions (Figure 2).
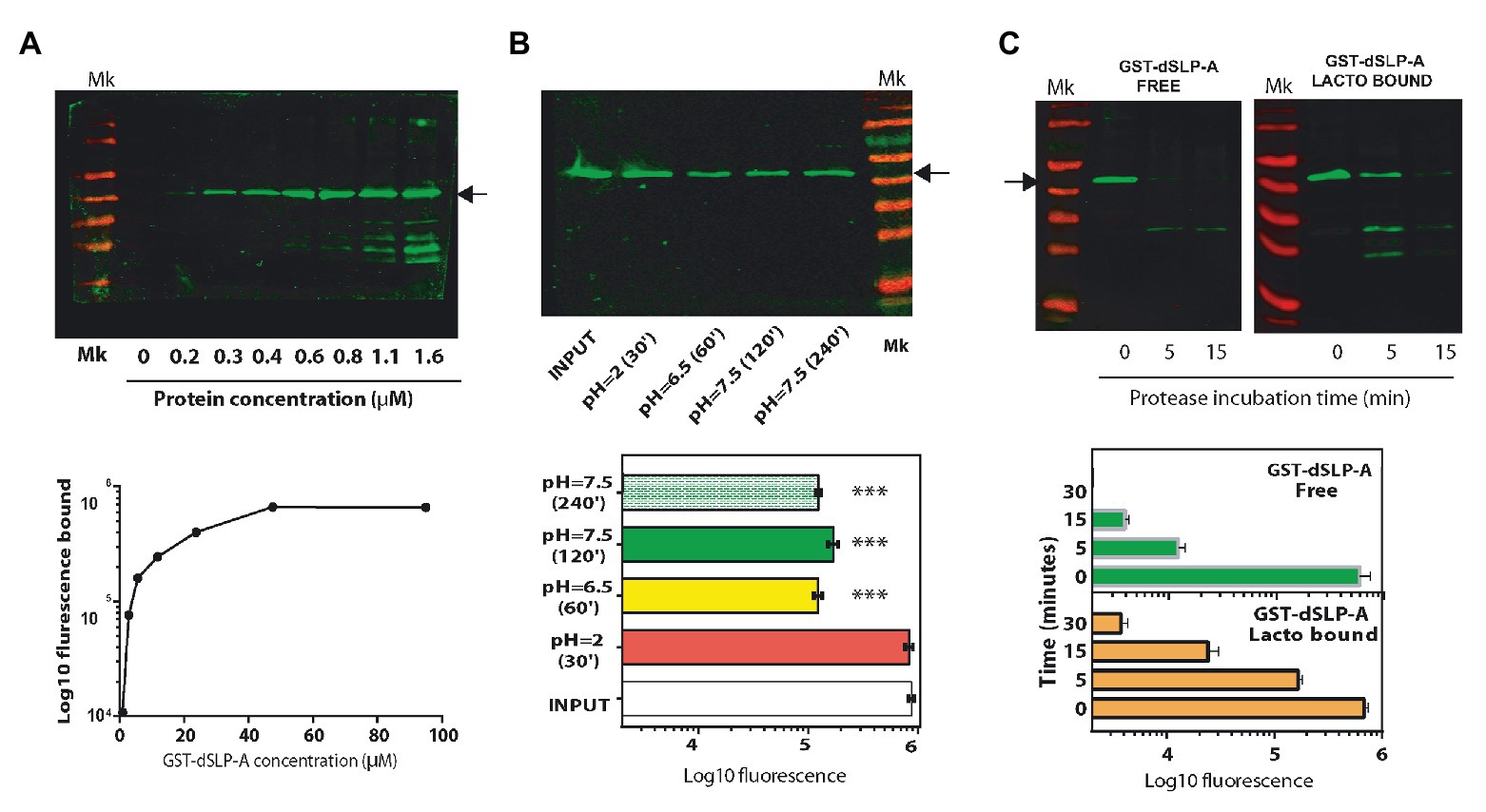
Figure 2. (A) Western blot analysis revealed with the Odyssey system (up panel), and its quantification (down panel) using the Image Studio software showing the association of the protein GST-dSLP-A to the surface of 108 cells of L. acidophilus. Increasing concentrations of GST-dSLP-A (at μM concentration) correlates with the increasing amount of this protein associated with the bacterial cell wall that follows a typical saturation curve. (B) Western blot showing the amount of retained GST-dSLP-A on the bacterial cell wall after the incubation for 30 min at pH2 (stomach-like conditions), 60 min at pH6.5 (proximal small bowel-like), 120 min at pH7.5 (mid-small bowel-like), and 240 min at pH7.5 (distal small bowel-like) compared with the control (INPUT; up panel). Quantification is shown in the down panel. One way ANOVA test and Bonferroni post hoc test were performed to analyze the statistical significance of these results (***p < 0.001). (C) Western blot analysis comparing the speed of degradation of GST-dSLP-A bound to L. acidophilus or GST-dSLP-A free in solution incubated with pancreatin (free vs. bound). Arrow indicates the migration of GST-dSLP-A (47.2 kDa). Thermo Scientific Pre-stained markers 180, 130, 100, 70, 55, 40, 35, 25, 15, and 10 kDa. Results shown are representative of two independent experiments.
As shown in Figure 2A, GST-dSLP-A coating of L. acidophilus follows a saturation curve reaching a plateau at micromolar concentrations. The stability of the GST-dSLP-A association with L. acidophilus wall was tested in the environmental conditions mimicking different portions of the human intestinal tract (stomach, duodenum, jejunum, and ileum) in terms of pH and time of transit through the intestinal tract (Evans et al., 1988). As shown in Figure 2B, GST-dSLP-A (INPUT) binding to L. acidophilus was stable for 30 min at pH 2 and 37°C, a condition that resembles the stomach-like environment. In addition, the stability of GST-dSLP-A bacterial association at proximal small bowel-like conditions (1 h, pH 6.5–37°C), mid-small bowel-like conditions (2 h, pH 7.5–37°C), and distal small bowel-like conditions (4 h, pH 7.5–37°C) was partially affected compared against the input control (p < 0.001; Figure 2B). No significant difference was detected between treatments at pH 6.5/37°C, 2 h or 4 h at pH7.5/37°C. Interestingly, GST-dSLP-A association with L. acidophilus was not affected by increasing concentration of bile salts even when the concentration was five times higher than physiological concentrations (Supplementary Figure S1; Pfeiler et al., 2007).
As shown in Figure 2C, binding of GST-dSLP-A to L. acidophilus protected the degradation of this protein when incubated with the digestive enzymes cocktail named Pancreatin (containing trypsin, chymotrypsin, pepsin, pancreatin, and elastase) compared with an equivalent amount of non-associated GST-dSLP-A. Free GST-dSLP-A was degraded close to 10 times faster than GST-dSLP-A bound to L. acidophilus.
To evaluate the possibility of using L. acidophilus as an antigen delivery system, the chimeric antigen EspA36–192-Intimin653–953-Tir240–378 H7-flagellin352–374 (EITH7; from E. coli O157:H7 EDL933) was constructed by gene synthesis as described in the section Materials and Methods, fused in frame with the dSLP-A domain and subcloned into the pLC3 expression vector (MBP-EspA36–192-Intimin653–953-Tir240–378- H7-flagellin352–374 SlpA284–444 or M-EITH7-dSLP-A; Figure 1B). As shown in Figure 3A, the protein M-EITH7-dSLP-A strongly associated with the cell wall of L. acidophilus with a similar saturation curve as the one observed with GST-dSLP-A (Figure 2A).
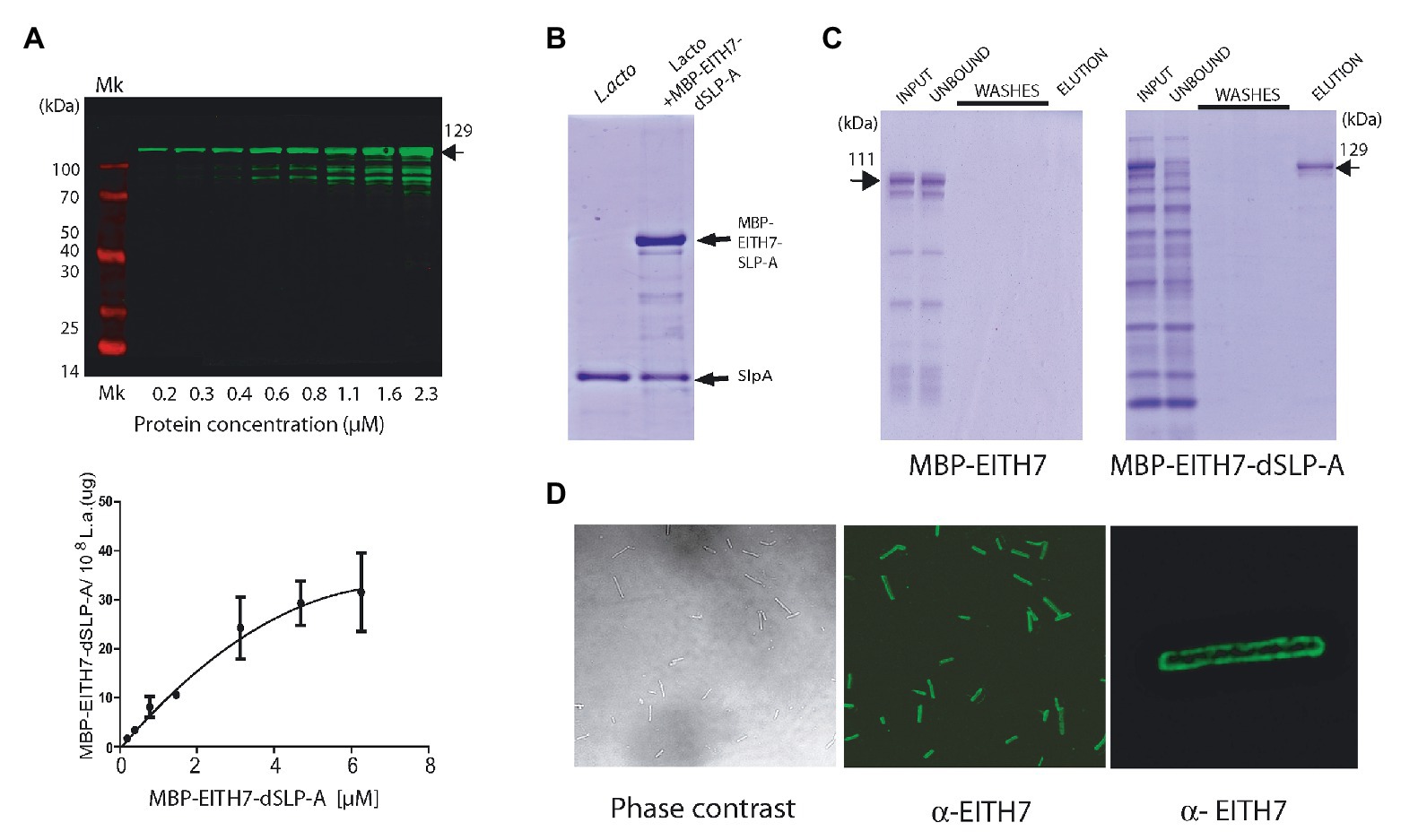
Figure 3. (A) Western blot (up panel) and its quantification (down panel) showing the association of the MBP-EITH7-dSLP-A to 108 cells of L. acidophilus. Incubation with increasing concentration of MBP-EITH7-dSLP-A correlates with the increment in the association of this protein to the bacterial cell wall that follows a typical saturation curve. As described, by comparison with a standard curve that correlates micrograms of proteins with the log10 of the fluorescence is possible to convert fluorescence to micrograms of MBP-EITH7-dSLP-A bound to Lactobacillus (down panel). (B) SDS-PAGE analysis was performed to evaluate the binding of MBP-EITH7-dSLP-A to Lactobacillus. Purified MBP-EITH7-dSLP-A was incubated with 108 bacteria for 30 min at 4°C and afterward centrifuged to separate bacterial pellet from the protein input solution. After two washes the pellet was subjected to 5 M LiCl to elute bacterium-associated proteins. (C) To control binding specificity of MBP-EITH7-dSLAP to Lactobacillus, proteins MBP-EITH7 (as a negative control) and MBP-EITH7-dSLP-A were incubated with 108 bacteria for 30 min at 37°C, centrifuged, and the bacterial pellet were washed twice and resuspended in 5 M LiCl to elute protein associated to Lactobacillus. (D) Immune-fluorescence using an anti-EITH7 antibody. Confocal microscopy shows the homogenous distribution of MBP-EITH7 on the L. acidophilus surface after decoration. TransGen Biotech Pre-stained markers blue-Plus Pre-stained markers 120, 100, 70, 50, 40, 30, 25, and 14 kDa.
As described in the section Material and Methods, we estimated that 108 cells of L. acidophilus were able to load up to 30 μg of protein M-EITH7-dSLP-A (Figure 3A, lower panel). Interestingly, to remove the recombinant S-layer attached to L. acidophilus cell wall formed by M-EITH7-dSLP-A, it was necessary to treat the antigen decorated bacteria with chaotropic solutions (5 M LiCl), with the same stringent conditions required for removing the wild type S-layer (Figure 3B).
To determine the strict requirement of dSLP-A domain for the binding specificity of M-EITH7-dSLP-A to L. acidophilus, cultures of these bacteria were incubated with M-EITH7 (without dSLP-A, see Figure 1B) or M-EITH7-dSLP-A (Figure 3C). Figure 3C shows that incubation with L. acidophilus retained proteins with high specificity from a protein lysate input only when dSLP-A was present in the construct (see input vs. unbound fraction). Accordingly, elution with 5 M LiCl only released proteins in the case of M-EITH7-dSLP-A indicating that the binding was dependent on the dSLP-A domain. When observed by immune-fluorescence confocal microscopy, it was possible to determine that M-EITH7-dSLP-A completely coated the surface of the bacteria L. acidophilus with a homogeneous distribution (Figure 3D).
Teichoic acid and wall teichoic acid which are present on the lactobacilli bacterial cell wall have been characterized as natural ligands for the dSLP-A domain (Fina Martin et al., 2019). Therefore, it is conceivable to expect that, in addition to L. acidophilus, dSLP-A-tagged proteins could bind also to any bacteria that display teichoic acids on its cell wall. Thus, dSLP-A-tagged proteins (GST-dSLP-A and M-EITH7-SLP-A) were also able to associate with B. subtilis var. natto (also a GRAS microorganism; Figure 4A). Association of the chimeric antigen M-EITH7-SLP-A to the surface of B. subtilis natto was also confirmed by confocal microscopy (Figure 4B).
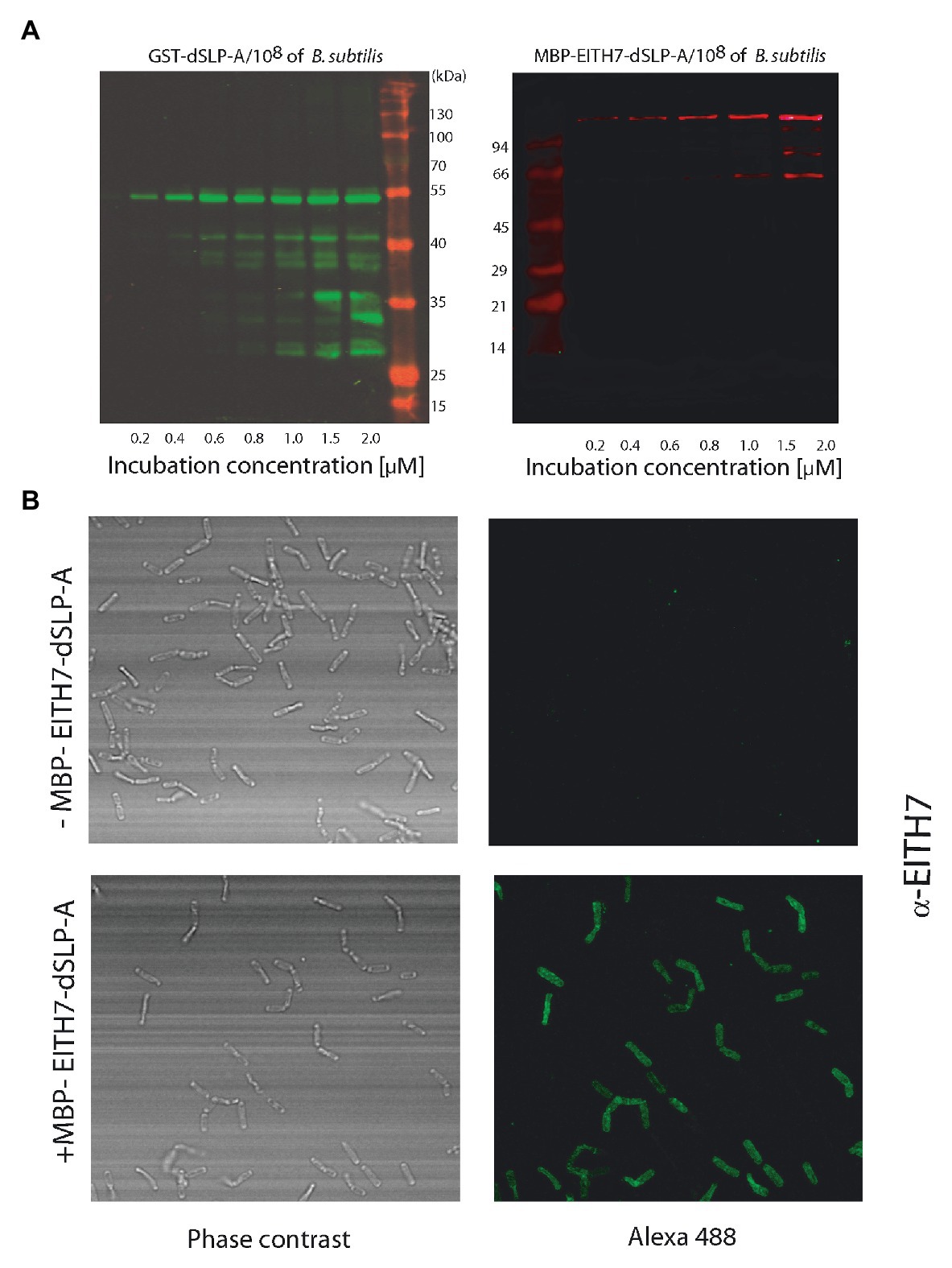
Figure 4. (A) Western blot analysis shows the increased binding of GST-dSLP-A (left) or MBP-EITH7-dSLP-A (right) to Bacillus subtilis surface that correlates with the increasing concentration of these proteins in the incubation solution. (Mk) Pre-stained molecular markers. (B) Confocal immunofluorescence microscopy of B. subtilis (upper panel) or B. subtilis decorated with MBP-EITH7-dSLP-A (lower panel) revealed with anti-EITH7 as the primary antibody.
Figure 5A shows a diagram of the immunization protocol used. BALB/c mice were immunized with three oral doses of different antigenic preparations (at 0, 2, and 4 weeks) and afterward (6 weeks post-immunization), all the immunized groups were experimentally challenged with 1010 CFU of E. coli O157:H7. Interestingly, after the first dose of M-EITH7-dSLP-A associated to L. acidophilus (10 days post-immunization), a significant increase in fecal EITH7-specific antibodies (IgA) was detected compared with the oral administration of the free M-EITH7-dSLP-A antigen or L. acidophilus alone used as a negative control. Similar results were also observed at 20 days after the first immunization (10 days after the second immunization; Figure 5B). In addition, the presence of EITH7-specific antibodies (IgG and IgA) in mouse sera was also determined by ELISA (Figure 6). A significant increase of anti-EITH7 titers in sera, IgG (Figure 6A), and IgA antibodies (Figure 6B) was detected 10 days post-immunization although at 20 days post-immunization only anti-EITH7 IgG was significantly increased. Of relevance, oral administration of wild type L. acidophilus that naturally carries a full-length SlpA (our negative control) or the administration of the free antigen M-EITH7-dSLP-A (Figure 6B, M-EITH7-dSLP-Afree) elicited no significant increase in IgA titers in feces (Figure 5B) or in sera (Figure 6). Altogether, these results indicate that only oral administration of M-EITH7-dSLP-A carried by L. acidophilus was able to induce a significant mucosal and humoral immune response against the EITH7 antigen. These results suggest that the association of EITH7 to L. acidophilus allows the effective delivery of the antigen to the competent sites for an effective antigen presentation to induce a specific immune response.
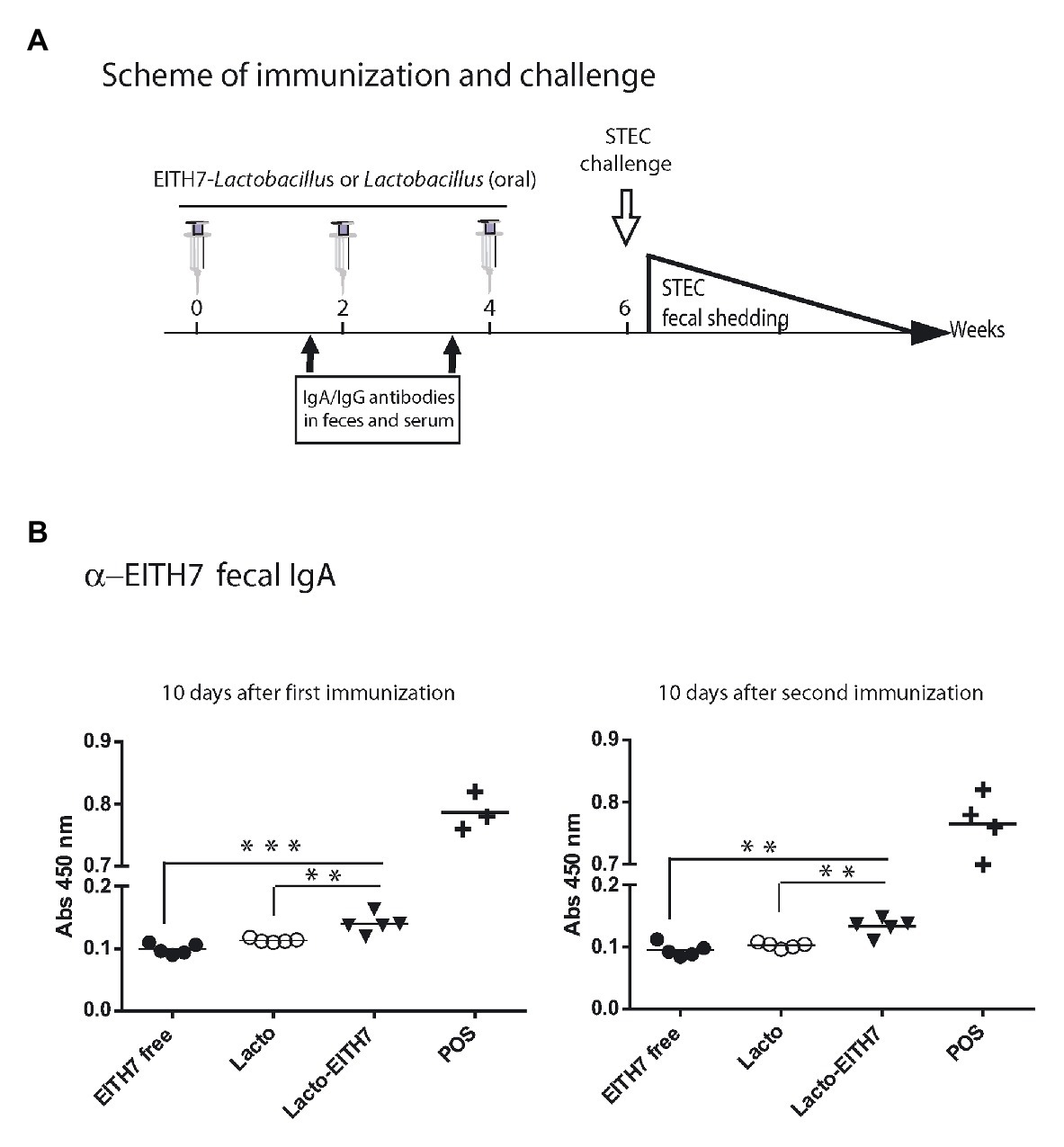
Figure 5. (A) Scheme of immunization and study of vaccine protection against a challenging infection in a mouse model. (B) BALB/c mice were orally immunized with free 20 μg of MBP-EITH7-dSLP-A, 4 × 108 UFC of lactobacilli, or 4 × 108 CFU of lactobacilli decorated with 20 μg of MBP-EITH7-dSLP-A in a scheme of three doses. Indirect ELISA for detection of mucosal IgA in feces (n = 6) was performed at 10 and 20 days post-immunization. A reference positive serum against EITH7 generated by i.p. immunization was used as a positive control (POS). One way ANOVA and Bonferroni’s Multiple Comparison post hoc Test (*p < 0.05), (***p < 0.01), and (***p < 0.001) was performed to analyze the statistical significance of results. Results shown are representative of three independent experiments.
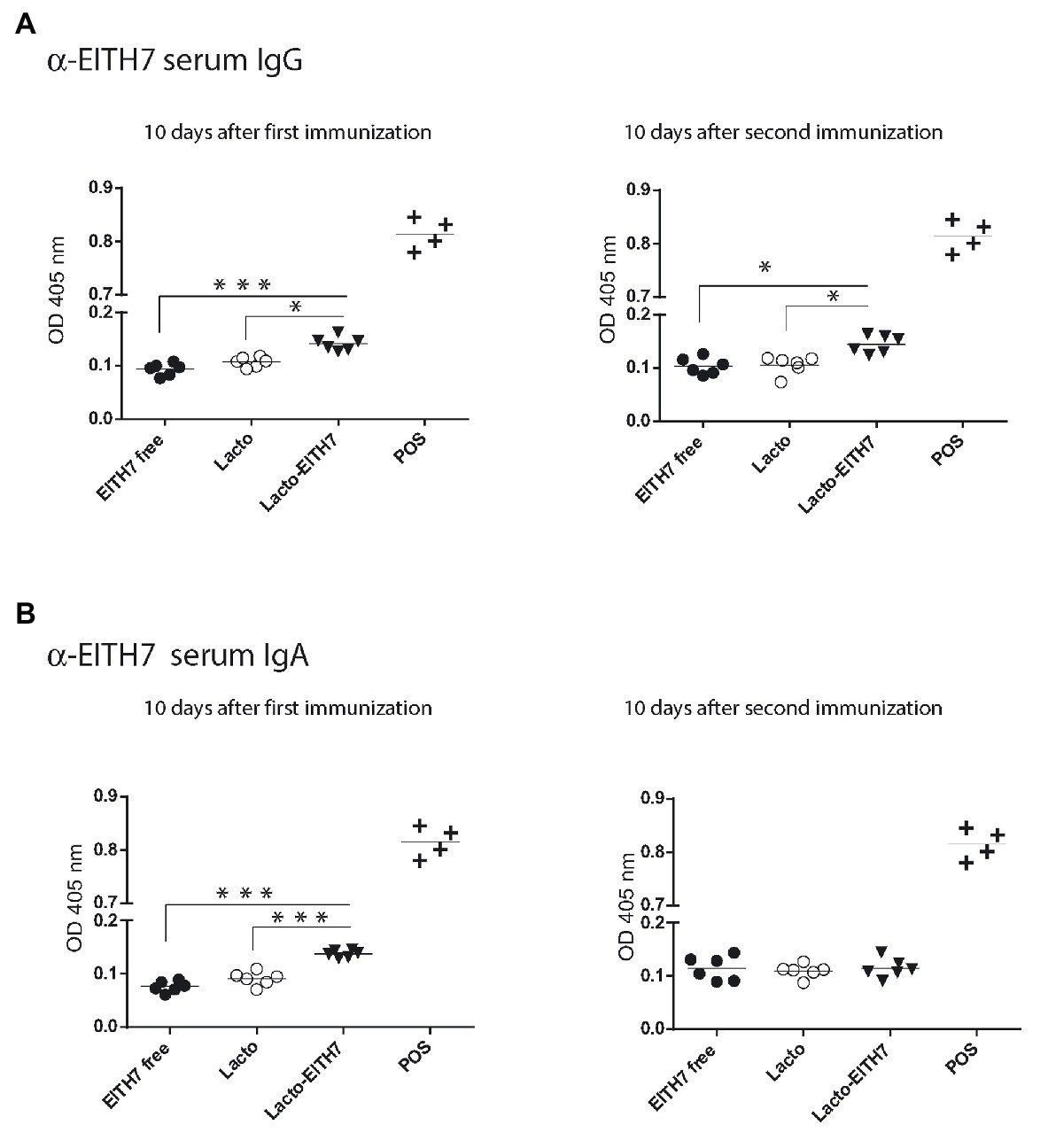
Figure 6. Indirect ELISA for the detection of serum anti-EITH7 specific IgG (A) and IgA (B) antibodies (n = 6). A reference positive serum against EITH7 generated by i.p. immunization, as described in the section Materials and Methods, was used as a POS. One way ANOVA and Bonferroni’s Multiple Comparison Test was used to analyze the statistical significance of results. Results shown are representative of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001.
As shown in Figures 5, 6, induction of a mucosal and humoral immune response in mice required the administration of the antigen M-EITH7-dSLP-A bound to L. acidophilus while either the free M-EITH7-dSLP-A or L. acidophilus were not sufficient to elicit an EITH7-specific immune response. To determine if this antibody immune response correlated with an effective protection against an experimental infection with E. coli O157:H7, mice were immunized with different preparations and orally challenged with 1010 CFU of STEC.
Bacterial fecal shedding was determined to estimate the efficiency of protection elicited by the antigen preparation. As shown in Figure 7, mice immunized with either the antigen decorated L. acidophilus-(M-EITH7-dSLP-A; white dots) or with L. acidophilus (black dots) shed equivalent levels of STEC in mouse feces during the first-week post-challenge. Interestingly, after 11 days post-challenge, a significant reduction in fecal shedding was observed in the group that received L. acidophilus-(M-EITH7-dSLP-A) compared with the control group. Mice that received the carrier L. acidophilus alone kept shedding more than 104 STEC/g in feces for almost 3 weeks post-challenge while the group that received antigen-decorated L. acidophilus-(M-EITH7-dSLP-A) shed less than 102 STEC/g at the same time post-challenge. These results indicate that the EITH7 antigen delivered by Lactobacillus as a vaccine carrier was able to induce a protective immune response that correlates with the differential immune response observed.
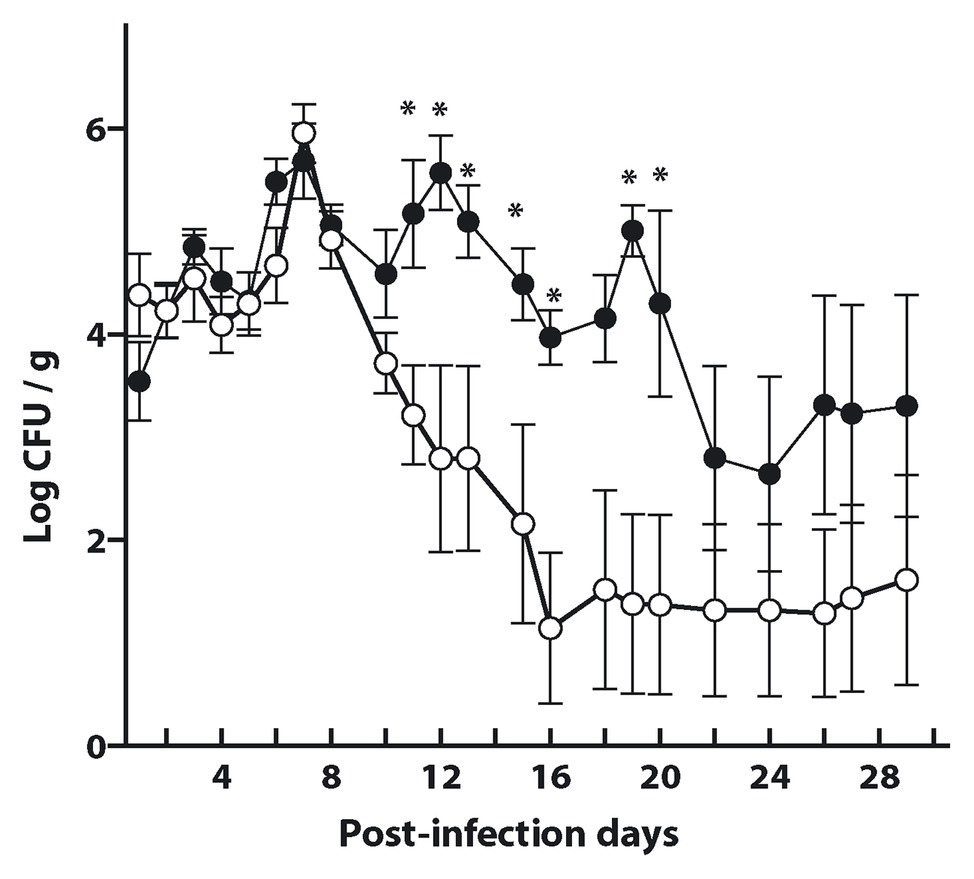
Figure 7. L. acidophilus and L. acidophilus decorated with EITH7-dSLP-A were administrated three times (0, 2, and 4 weeks) to mice. Ten days after the last immunization, mice were experimentally infected with 1010 CFU of E. coli O157:H7 and bacterial shedding in feces was monitored daily and the CFU per gram of feces was determined (n = 6). Results shown are representative of three independent experiments. Student’s t-test was used to analyze the statistical significance of results. *p < 0.05.
It has been described that strains of Lactobacillus can stimulate the specific and the innate immune responses and, consequently, have been proposed as vaccine carriers (Pardo et al., 2016; Szatraj et al., 2017; Lecureux and Dean, 2018). Activation of the innate immune response is mediated by the PRRs expressed on immune cells that recognize conserved molecular structures known as MAMPs that activate the production of cytokines, chemokines, and other innate effectors. Host recognition of Lactobacillus is predominantly mediated by TLR2 receptor that recognizes teichoic and lipoteichoich acids which are present in the Lactobacillus cell wall leading to maturation of dendritic cells (Konstantinov et al., 2008) and the differentiation of monocytic precursors to Langerhans-like cells (Song et al., 2018). Also, it was shown that the L. acidophilus S-layer protein is able to interact with the DC-SIGN receptor (Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin; CD209) a major dendritic cell receptor, interfering with the association of several human viruses such as HIV (Petrova et al., 2013) and JUNV (Martinez et al., 2012).
Here, we report the adaptation of L. acidophilus to deliver antigens to the gastrointestinal mucosa. Traditionally, Lactobacillus species have been engineered to express viral and bacterial antigens to elicit protective immune responses (Lecureux and Dean, 2018). This approach has a regulatory problem as it generates GMOs that are strictly regulated by several government offices (Directive 2001/18/EC of the European Parliament) if they will be released to the environment. In our system, antigens are passively attached to the surface of L. acidophilus with no alteration of the genotype and, therefore, with no generation of GMO microorganisms. For this reason, vaccine candidates based on this antigen decoration strategy are expected to have an easier, faster, and less-expensive regulatory approval process. Taking this into consideration, we studied if this strategy is suitable for the development of a vaccine candidate against the STEC. STEC infections can induce the development of the HUS, particularly in children below 5-year old that are the most susceptible population group (Fernandez-Brando et al., 2017). In addition to the severe consequences of HUS in children, it is also important to mention the economical burden that this disease represents for Public Health (e.g., STEC infection is a recurrent cause for kidney transplant in children and young people). Despite all these problems, there are no approved therapies or licensed vaccines against this bacterial infection for human use. The development of a safe oral vaccine could be of critical relevance for preventing HUS in children. Recently, two papers have proposed the adaptation of Lactobacillus for STEC vaccine development (Ferreira et al., 2011; Lin et al., 2017). In one of these reports, Lin et al. (2017) engineered L. acidophilus to express a chimeric STEC antigen composed by EspA and Tir (named E-T) with a signal peptide for the secretion to the bacterial supernatant. Interestingly, these authors have shown that E-T-vaccinated mice were protected against an experimental infection (1010 E. coli O157), similar to what we have shown here. Interestingly, although the fecal immune response (determined by ELISA signal) was similar to our results, immunization with E-T secreting-Lactobacillus induced an antibody immune response in sera than was relatively much stronger than the one we observed (Figure 6). It is interesting to speculate if these discrepancies between results could be reflecting differences in antigen dose since a recombinant strain of Lactobacillus has the capacity to express the heterologous antigens after it replicates. In our case, antigen-decorated bacteria are limited to a one-shot opportunity to deliver its cargo to the mucosal tissues, since the de novo expression of the recombinant antigen is not possible. This limitation could be sorted out either by optimizing the amount of antigen to be delivered increasing the number of decorated bacteria or by augmenting the number of doses. Since the amount of decorating antigen can be finely tuned and monitored it is possible to optimize the immunization protocol to induce a more effective immune response. Additionally, the system can be improved since it allows the co-decoration of L. acidophilus with proteins with different activities like immune modulators such as the TLR5-agonist of Salmonella flagellin, the lectin GAL8, or with the protease inhibitor B. abortus Omp19. Interestingly, all these molecules can modulate the induction of the immune response by altering the way that antigens are presented to the immune system, enhancing its half-life, or providing a particular environment of cytokines for a more efficient antigen presentation. As shown here, the fusion of the dSLP-A domains to all protein tested allows the adhesion of the resulting chimera protein to the surface of L. acidophilus. As shown in Figure 4, in addition to L. acidophilus, dSLP-A-tagged proteins can also bind to different bacteria that display teichoic acids on its cell wall such as B. subtilis. This could be particularly useful to optimize a prime/boost immunization strategy by simply switching the bacterial carrier when a second dose is required. BLAST sequence analysis showed that L. acidophilus dSLP-A domain displays more than 75% amino acid identity to SlpA sequences present in L. helveticus, L. crispatus, L. amyolovorus, and L. gallinarum (Supplementary Table S1). Since L. helveticus and L. crispatus are vaginal mucosa resident bacteria (Nunn and Forney, 2016), it could be interesting to explore if they can be adapted as vaccine carriers for the induction of protective immune responses at genital mucosa against sexually transmitted diseases like HBV, HPV, or HIV.
In this report, and as a proof of concept, we demonstrated that L. acidophilus could be adapted as a vaccine carrier by coating the bacteria with antigens from a microbial pathogen mimicking its surface, to induce a protective immune response for vaccination purposes.
Here, we have explored the adaptation of a GRAS bacterium such as L. acidophilus as an antigen delivery system. Decoration of the L. acidophilus with heterologous proteins might allow the development of a universal platform for intestinal delivery of proteins or enzymes with therapeutic value. Of interest, all these biotechnological applications will be achieved without the generation of genetically modified bacteria or the introduction of antibiotic resistances.
All datasets generated for this study are included in the article/Supplementary Material.
The animal study was reviewed and approved by Experimental procedure of this study (permit number 15/2018) was approved by the Committee on the Ethics of Animal Experiments of the Universidad Nacional de San Martín (UNSAM), under the recommendations for animal experimentation (Helsinki Declaration and its amendments, Amsterdam Protocol of welfare and animal protection and National Institutes of Health, USA NIH, guidelines: Guide for the Care and Use of Laboratory Animals).
SR, MR, and GB conceived and design the experiments. PU, CT, MP, and JM performed experiments. PU, MR, and GB analyzed data. SR and MP contribute with reagent, material. MR and GB wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica, Buenos Aires, Argentina (PICT 2013-0662, PICT 2016-0412, and PICT 2018-00778), CONICET (P-UE 0086), and the Universidad Nacional de San Martín, Buenos Aires, Argentina (Puente-006SM). MP, SR, MR, and GB are members of the Research Career of CONICET. PU and JM are doctoral fellow from CONICET.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Juan E. Ugalde for his careful and critical reading of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.509380/full#supplementary-material
Abou-Saleh, R. H., Peyman, S. A., Johnson, B. R., Marston, G., Ingram, N., Bushby, R., et al. (2016). The influence of intercalating perfluorohexane into lipid shells on nano and microbubble stability. Soft Matter 12, 7223–7230. doi: 10.1039/c6sm00956e
Arce, L. P., Raya Tonetti, M. F., Raimondo, M. P., Muller, M. F., Salva, S., Alvarez, S., et al. (2019). Oral vaccination with hepatitis E virus capsid protein and immunobiotic bacterium-like particles induce intestinal and systemic immunity in mice. Probiotics Antimicrob. Proteins 12, 961–972. doi: 10.1007/s12602-019-09598-7
Ashida, N., Yanagihara, S., Shinoda, T., and Yamamoto, N. (2011). Characterization of adhesive molecule with affinity to Caco-2 cells in Lactobacillus acidophilus by proteome analysis. J. Biosci. Bioeng. 112, 333–337. doi: 10.1016/j.jbiosc.2011.06.001
Avall-Jaaskelainen, S., and Palva, A. (2005). Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29, 511–529. doi: 10.1016/j.femsre.2005.04.003
Babiuk, S., Asper, D. J., Rogan, D., Mutwiri, G. K., and Potter, A. A. (2008). Subcutaneous and intranasal immunization with type III secreted proteins can prevent colonization and shedding of Escherichia coli O157:H7 in mice. Microb. Pathog. 45, 7–11. doi: 10.1016/j.micpath.2008.01.005
Boot, H. J., Kolen, C. P., and Pouwels, P. H. (1995). Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 177, 7222–7230. doi: 10.1128/jb.177.24.7222-7230.1995
Douillard, F. P., and de Vos, W. M. (2019). Biotechnology of health-promoting bacteria. Biotechnol. Adv. 37:107369. doi: 10.1016/j.biotechadv.2019.03.008
Evans, D. F., Pye, G., Bramley, R., Clark, A. G., Dyson, T. J., and Hardcastle, J. D. (1988). Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29, 1035–1041. doi: 10.1136/gut.29.8.1035
Fernandez-Brando, R. J., Amaral, M. M., Ciocchini, A. E., Bentancor, L. V., Trelles, J. A., Da Rocha, M., et al. (2017). Microbiological and serological control of Escherichia coli O157: H7 in kindergarten staff in Buenos Aires city and suburban areas. Medicina 77, 185–190.
Ferreira, P. C., Da Silva, J. B., Piazza, R. M., Eckmann, L., Ho, P. L., and Oliveira, M. L. (2011). Immunization of mice with Lactobacillus casei expressing a beta-intimin fragment reduces intestinal colonization by Citrobacter rodentium. Clin. Vaccine Immunol. 18, 1823–1833. doi: 10.1128/CVI.05262-11
Fina Martin, J., Palomino, M. M., Cutine, A. M., Modenutti, C. P., Fernández Do Porto, D. A., Allievi, M. C., et al. (2019). Exploring lectin-like activity of the S-layer protein of Lactobacillus acidophilus ATCC 4356. Appl. Microbiol. Biotechnol. 103, 4839–4857. doi: 10.1007/s00253-019-09795-y
Galler, R., Freire, M. S., Jabor, A. V., and Mann, G. F. (1997). The yellow fever 17D vaccine virus: molecular basis of viral attenuation and its use as an expression vector. Braz. J. Med. Biol. Res. 30, 157–168. doi: 10.1590/s0100-879x1997000200002
Goldwater, P. N. (2007). Treatment and prevention of enterohemorrhagic Escherichia coli infection and hemolytic uremic syndrome. Expert Rev. Anti-Infect. Ther. 5, 653–663. doi: 10.1586/14787210.5.4.653
Hymes, J. P., Johnson, B. R., Barrangou, R., and Klaenhammer, T. R. (2016). Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 2676–2685. doi: 10.1128/AEM.00024-16
Iannino, F., Herrmann, C. K., Roset, M. S., and Briones, G. (2015). Development of a dual vaccine for prevention of Brucella abortus infection and Escherichia coli O157:H7 intestinal colonization. Vaccine 33, 2248–2253. doi: 10.1016/j.vaccine.2015.03.033
Ibañez, A. E., Coria, L. M., Carabajal, M. V., Delpino, M. V., Risso, G. S., Cobiello, P. G., et al. (2015). A bacterial protease inhibitor protects antigens delivered in oral vaccines from digestion while triggering specific mucosal immune responses. J. Control. Release 220, 18–28. doi: 10.1016/j.jconrel.2015.10.011
Johnson, B. R., Hymes, J., Sanozky-Dawes, R., Henriksen, E. D., Barrangou, R., and Klaenhammer, T. R. (2016). Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming Lactobacilli. Appl. Environ. Microbiol. 82, 134–145. doi: 10.1128/AEM.01968-15
Johnson, B. R., O’flaherty, S., Goh, Y. J., Carroll, I., Barrangou, R., and Klaenhammer, T. R. (2017). The S-layer associated serine protease homolog PrtX impacts cell surface-mediated microbe-host interactions of Lactobacillus acidophilus NCFM. Front. Microbiol. 8:1185. doi: 10.3389/fmicb.2017.01185
Johnson, B., Selle, K., O’flaherty, S., Goh, Y. J., and Klaenhammer, T. (2013). Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 159, 2269–2282. doi: 10.1099/mic.0.070755-0
Konstantinov, S. R., Smidt, H., de Vos, W. M., Bruijns, S. C., Singh, S. K., Valence, F., et al. (2008). S layer protein a of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U. S. A. 105, 19474–19479. doi: 10.1073/pnas.0810305105
Larson, H., Hartigan-Go, K., and de Figueiredo, A. (2019). Regarding response by Dans et. al. to our article, “vaccine confidence plummets in the Philippines following dengue vaccine scare: why it matters to pandemic preparedness.”. Hum. Vaccin. Immunother. 15:630. doi: 10.1080/21645515.2018.1556079
Lecureux, J. S., and Dean, G. A. (2018). Lactobacillus mucosal vaccine vectors: immune responses against bacterial and viral antigens. mSphere 3:e00061–18. doi: 10.1128/mSphere.00061-18
Lin, R., Zhang, Y., Long, B., Li, Y., Wu, Y., Duan, S., et al. (2017). Oral immunization with recombinant Lactobacillus acidophilus expressing espA-Tir-M confers protection against enterohemorrhagic Escherichia coli O157:H7 challenge in mice. Front. Microbiol. 8:417. doi: 10.3389/fmicb.2017.00417
Lycke, N. (2012). Recent progress in mucosal vaccine development: potential and limitations. Nat. Rev. Immunol. 12, 592–605. doi: 10.1038/nri3251
Martinez, M. G., Prado Acosta, M., Candurra, N. A., and Ruzal, S. M. (2012). S-layer proteins of Lactobacillus acidophilus inhibits JUNV infection. Biochem. Biophys. Res. Commun. 422, 590–595. doi: 10.1016/j.bbrc.2012.05.031
Mcclelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856. doi: 10.1038/35101614
Minor, P. D. (2015). Live attenuated vaccines: historical successes and current challenges. Virology 479-480, 379–392. doi: 10.1016/j.virol.2015.03.032
Nunn, K. L., and Forney, L. J. (2016). Unraveling the dynamics of the human vaginal microbiome. Yale J. Biol. Med. 89, 331–337.
Orenstein, W. A., and Ahmed, R. (2017). Simply put: vaccination saves lives. Proc. Natl. Acad. Sci. U. S. A. 114, 4031–4033. doi: 10.1073/pnas.1704507114
Palomino, M. M., Waehner, P. M., Fina Martin, J., Ojeda, P., Malone, L., Sánchez Rivas, C., et al. (2016). Influence of osmotic stress on the profile and gene expression of surface layer proteins in Lactobacillus acidophilus ATCC 4356. Appl. Microbiol. Biotechnol. 100, 8475–8484. doi: 10.1007/s00253-016-7698-y
Pardo, R., Blasco, N., Vila, M., Beiroa, D., Nogueiras, R., Canas, X., et al. (2016). EndoG knockout mice show increased brown adipocyte recruitment in white adipose tissue and improved glucose homeostasis. Endocrinology 157, 3873–3887. doi: 10.1210/en.2015-1334
Petrova, M. I., Mathys, L., Lebeer, S., Noppen, S., Van Damme, E. J., Tanaka, H., et al. (2013). Inhibition of infection and transmission of HIV-1 and lack of significant impact on the vaginal commensal lactobacilli by carbohydrate-binding agents. J. Antimicrob. Chemother. 68, 2026–2037. doi: 10.1093/jac/dkt152
Pfeiler, E. A., Azcarate-Peril, M. A., and Klaenhammer, T. R. (2007). Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189, 4624–4634. doi: 10.1128/JB.00337-07
Plotkin, S. (2014). History of vaccination. Proc. Natl. Acad. Sci. U. S. A. 111, 12283–12287. doi: 10.1073/pnas.1400472111
Potter, A. A., Klashinsky, S., Li, Y., Frey, E., Townsend, H., Rogan, D., et al. (2004). Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine 22, 362–369. doi: 10.1016/j.vaccine.2003.08.007
Savinov, A., Pan, J., Ghosh, P., and Hatfull, G. F. (2012). The Bxb1 gp47 recombination directionality factor is required not only for prophage excision, but also for phage DNA replication. Gene 495, 42–48. doi: 10.1016/j.gene.2011.12.003
Seib, K., Pollard, A. J., De Wals, P., Andrews, R. M., Zhou, F., Hatchett, R. J., et al. (2017). Policy making for vaccine use as a driver of vaccine innovation and development in the developed world. Vaccine 35, 1380–1389. doi: 10.1016/j.vaccine.2016.10.080
Sleytr, U. B., Schuster, B., Egelseer, E. M., and Pum, D. (2014). S-layers: principles and applications. FEMS Microbiol. Rev. 38, 823–864. doi: 10.1111/1574-6976.12063
Smit, E., Jager, D., Martinez, B., Tielen, F. J., and Pouwels, P. H. (2002). Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein-protein interaction of two subdomains. J. Mol. Biol. 324, 953–964. doi: 10.1016/s0022-2836(02)01135-x
Song, J., Lang, F., Zhao, N., Guo, Y., and Zhang, H. (2018). Vaginal Lactobacilli induce differentiation of monocytic precursors toward langerhans-like cells: in vitro evidence. Front. Immunol. 9:2437. doi: 10.3389/fimmu.2018.02437
Szatraj, K., Szczepankowska, A. K., and Chmielewska-Jeznach, M. (2017). Lactic acid bacteria—promising vaccine vectors: possibilities, limitations, doubts. J. Appl. Microbiol. 123, 325–339. doi: 10.1111/jam.13446
Keywords: Lactobacillus, shiga toxigenic Escherichia coli, S-layer (fusion) proteins, oral vaccination, antigen delivery system
Citation: Uriza PJ, Trautman C, Palomino MM, Fina Martin J, Ruzal SM, Roset MS and Briones G (2020) Development of an Antigen Delivery Platform Using Lactobacillus acidophilus Decorated With Heterologous Proteins: A Sheep in Wolf’s Clothing Story. Front. Microbiol. 11:509380. doi: 10.3389/fmicb.2020.509380
Received: 01 November 2019; Accepted: 01 October 2020;
Published: 27 October 2020.
Edited by:
Leonard Peruski, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Brant R. Johnson, AgBiome Inc, United StatesCopyright © 2020 Uriza, Trautman, Palomino, Fina Martin, Ruzal, Roset and Briones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mara S. Roset, bXJvc2V0QGlpYi51bnNhbS5lZHUuYXI=; Gabriel Briones, Z2JyaW9uZXNAaWliLnVuc2FtLmVkdS5hcg==; Z2JyaW9uZXNAaWliaW50ZWNoLmNvbS5hcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.