- 1Department of Oral Biology, College of Dentistry, University of Florida, Gainesville, FL, United States
- 2Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL, United States
Lack of LrgAB renders cariogenic Streptococcus mutans more sensitive to oxidative stress, as well as limits the capacity of this organism to re-uptake pyruvate upon starvation. This study was aimed at investigating the ecological and metabolic contribution of LrgAB to competitive fitness, using S. mutans strains, that either lack or overexpress lrgAB. These experiments revealed that impaired aerobic growth of the ΔlrgAB mutant can be effectively restored by supplementation of pyruvate, and that perturbated expression of lrgAB significantly affects pyruvate flux and the conversion of pyruvate to acetyl-CoA by the Pdh pathway, verifying that LrgAB is closely linked to pyruvate catabolism. In vitro competition assays revealed that LrgAB plays an important role in S. mutans competition with H2O2-producing S. gordonii, an interaction which can also be modulated by external pyruvate. However, no obvious competitive disadvantage was observed against S. gordonii by either the S. mutans lrgAB mutant or lrgAB overexpression strain in vivo using a mouse caries model. Organic acid analysis of mouse dental biofilms revealed that metabolites produced by the host and/or dental plaque microbiota could complement the deficiency of a lrgAB mutant, and favored S. mutans establishment compared to S. gordonii. Collectively, these results reinforce the importance of the oral microbiota and the metabolic environment in the oral cavity battleground, and highlight that pyruvate uptake through LrgAB may be crucial for interspecies competition that drives niche occupancy.
Introduction
Cariogenic bacteria such as Streptococcus mutans are very effective at metabolizing a wide range of carbon sources, producing strong organic acids, and surviving in acidic environments, collectively allowing these organisms to thrive in the oral cavity of humans consuming a variety of diets (Burne, 1998; Marsh, 2003; Burne et al., 2009). In particular, the ability of S. mutans to efficiently acquire carbohydrates and coordinate its metabolism in response to environmental fluctuations is critical for outcompeting non-cariogenic indigenous oral microbiota in caries-conducive conditions. These abilities are also closely linked to the capacity of S. mutans to withstand a variety of stressors encountered in the oral cavity. All these processes are complex and cross-regulated with multiple metabolic pathways and virulence properties (Lemos and Burne, 2008; Smith and Spatafora, 2012).
Over the past decade, we have studied the S. mutans LrgAB system, originally hypothesized to induce cell death and lysis with its partner operon CidAB, and subsequently shown to play a significant role in modulating a variety of key S. mutans virulence traits, including autolysis, biofilm formation, oxidative and heat stresses, and genetic competence (Ahn et al., 2010, 2012, 2017; Rice et al., 2017). Interestingly, the lrgAB mutation was also shown to affect major metabolic pathways associated with carbohydrate, amino acid, fatty acid/lipid, nucleotide metabolism and transport (Ahn et al., 2017; Rice et al., 2017). Recently, we reported that LrgAB functions as a stationary phase pyruvate uptake system (Ahn et al., 2019) and its function is regulated by two global regulators, CcpA (Kim et al., 2019) and CodY (Ahn et al., 2020c), that are important for cellular responses to altered carbohydrate availability and amino acid limitations (Henkin, 1996; Sonenshein, 2005, 2007; Deutscher, 2008; Gorke and Stulke, 2008). More interestingly, we demonstrated that excess pyruvate produced during growth can be subsequently utilized as a carbon source during stationary phase (Ahn et al., 2019, 2020a), presumably promoting long-term survival of S. mutans when in a metabolically competitive environment with other oral microbiota. Given that pyruvate is a central carbon metabolite directly linked to key metabolic pathways and has a relatively fast-turnover during stationary phase, environmental pyruvate (or other metabolites) may therefore modulate interspecies competition during caries development. Although the role and regulation of LrgAB have been relatively well characterized, its ecological importance has not yet been fully investigated. In this study, we characterize how LrgAB contributes to the competitive fitness of S. mutans in vitro and in vivo, using a dual-species model with S. gordonii, a H2O2-producing oral commensal. The experimental outcomes described herein provide guidance for further defining the cariological and ecological significance of LrgAB.
Materials and Methods
Growth Conditions
All S. mutans and S. gordonii strains were grown in BHI (brain heart infusion) medium (Difco) as overnight cultures at 37°C in a 5% CO2 atmosphere. When necessary, antibiotics were added to cultures as follows: kanamycin (1 mg/ml) and spectinomycin (1 mg/ml). Overnight cultures were diluted 1:50 into fresh BHI broth, grown to mid-exponential phase (OD600∼0.4–0.5), and then used as seed cultures for the assays described below. The media used for these assays included BHI, chemically defined FMC (Terleckyj and Shockman, 1975; Terleckyj et al., 1975), and TV (Tryptone/Vitamin) (Burne et al., 1999), the latter two supplemented with 11 mM glucose (named FMC11 and TV11, respectively), as BHI already contains 11 mM glucose. Each medium was supplemented with α-ketoglutaric acid (αKG; Sigma-Aldrich), oxaloacetic acid (OA; Sigma-Aldrich), or sodium pyruvate (pyr; Fisher Scientific), at concentrations as indicated for each experiment. To achieve anaerobic conditions, sterile mineral oil was placed on top of the cultures.
Bacterial Strains and Mutant Construction
S. mutans strains used in this study were UA159 (wildtype) and isogenic lrgAB-deficient mutant (ΔlrgAB) (Ahn et al., 2010), lrgAB-overexpression strain (SAB161), and lytST-overexpression strain (SAB163) (Ishkov et al., 2020). The SAB161 strain, constitutively expressing lrgAB, was constructed as previously described (Ahn and Rice, 2016). Briefly, the promoter region of lrgA (PlrgA) was replaced by a fragment (ΩKm-Pldh) containing a polar kanamycin resistance gene (ΩKm) and a ldh promoter region (Pldh). For this, two ∼0.5 kb fragments flanking the −35 and −10 sequences of the lrgA promoter were PCR-amplified, ligated into the ΩKm-Pldh cassette, previously generated by PCR amplification (Ahn and Rice, 2016), and transformed into S. mutans. Transformants were selected on BHI agar containing kanamycin, and double-crossover recombination was confirmed by PCR and sequencing. A GFP reporter assay to monitor PlrgA activity consisted of the previously created PlrgA-gfp construct (Kim et al., 2019) in the shuttle vector pDL278 (LeBlanc et al., 1992), transformed into SAB161. As a control strain, the empty pDL278 plasmid was also transformed in this mutant. The GFP reporter and control constructs in wildtype, ΔlrgAB, and SAB163 backgrounds were previously generated (Kim et al., 2019; Ahn et al., 2020b; Ishkov et al., 2020). For a dual-species competition assay with S. mutans, S. gordonii wildtype DL1 (generating H2O2 in a SpxB-dependent manner) and ΔspxB (spxB-deficient) strains (Huang et al., 2016) were used.
Microplate Assays
To monitor growth patterns over time, seed cultures of each strain were diluted 1:100 into 350 μl fresh medium in individual wells of a 100-well honeycomb plate and the optical density at 600 nm (OD600) measured at 37°C at 30 min intervals using a Bioscreen C growth curve analysis system. At least three independent experiments, each in quadruplicate, were performed. A representative result is presented in each relevant figure. To monitor lrgAB expression (PlrgA activity) over growth, S. mutans strains harboring a PlrgA-gfp reporter fusion or pDL278 were used, as described above. Seed cultures of each reporter strain were diluted 1:50 into 175 μl fresh medium in individual wells of a 96-well plate (black walls, clear bottoms; Corning), which was subsequently loaded into a Synergy microplate reader (BioTek) controlled by Gen5 software (Ahn et al., 2019, 2020a; Kim et al., 2019). The optical density for growth curves and green fluorescence intensity for lrgAB expression were monitored at 600 and 485 nm/520 nm (excitation/emission), respectively, at 30 min intervals for 18–24 h. The fluorescence of wildtype harboring the plasmid without the reporter gene fusion was subtracted from fluorescence readings of each S. mutans strain harboring the PlrgA-gfp gene fusion. At least three independent replicates, each in triplicate, were performed. A representative result is presented in each relevant figure.
Measurement of Extracellular Pyruvate Levels
S. mutans UA159 wildtype, SAB161, and SAB163 were grown in FMC medium containing 11 mM glucose. Measurements of extracellular pyruvate during growth were taken from 250 μl samples at 1–2 h intervals and 100 μl was used to monitor growth (OD600). Extracellular pyruvate levels during early-exponential growth and stationary phase were predicted based on previously published data (Ahn et al., 2019). Thus, pyruvate quantification in this study was focused on samples taken at late-exponential and early-stationary phases, time points at which lrgAB induction and LrgAB-dependent pyruvate uptake occur. The remaining 150 μl of each sample was centrifuged for 2 min at 18,000 × g to remove cells, and pyruvate concentrations of supernatants quantified with an EnzyChromTM pyruvate assay kit (BioAssay Systems, Hayward, CA, United States), according to the manufacturer’s instructions.
Quantitative Real-Time PCR (qPCR) Assay
To measure growth-dependent gene expression of lrgA, lytS, pdhC, and pfl, using qPCR, S. mutans wild-type and isogenic mutant strains were grown in BHI, at 37°C in a 5% (vol/vol) CO2 atmosphere, and cells were harvested at early-exponential (EE; OD600∼0.2) and -stationary (ES; OD600∼1.0) phase. RNA extraction, reverse-transcriptase (RT) reaction, qPCR, and data analysis were performed as described elsewhere (Ahn et al., 2012; Ahn and Rice, 2016; Rice et al., 2017). Expression values were normalized against an internal standard (gyrA) and presented as relative copy number of mRNA (copies/μl) for comparison with each gene. Statistical analyses were performed on data generated from n = 3 independent experiments using an unpaired t-test.
In vitro Competition Assays
For dual-species competition assays between each S. mutans strain (UA159, ΔlrgAB, SAB161, or SAB163) and S. gordonii strain (DL1 or ΔspxB), 200 μl of each competing seed culture (one from S. mutans and the other from S. gordonii), were each adjusted to OD600 = 0.5 in BHI medium, centrifuged, the pellet washed with PBS (phosphate buffer saline), resuspended in 200 μl PBS, and then diluted 1:100 into 200 μl fresh medium in individual wells of a 96-well plate (CostarTM3595, Corning). After 24 h incubation at 37°C in an aerobic atmosphere, 5 μl culture from each well was inoculated on Prussian blue agar plates (Saito et al., 2007), indicating the production of H2O2. The plates were incubated for an additional 24 h at 37°C in an aerobic atmosphere. Growth and blue precipitation zones were documented and analyzed with VisionWorks® software (Analytik Jena, Upland, CA, United States). Original grayscale .tif images of each plate were each converted to RGB color, and mid-tone color balanced (−50, 0, +31) in Adobe Photoshop 21.2.1 to improve the contrast and clarity of each image.
Murine in vivo Caries Model
A mouse caries protocol was performed as previously described (Culp et al., 2015; Shields et al., 2018) with modifications. All procedures with solutions and samples were performed under BSL2 conditions and mice were kept under ABSL2 conditions. Briefly, inbred 3-weeks-old female SPF BALB/cJ mice (The Jackson Laboratory, Bar Harbor, ME, United States) were placed in pairs in sterile cages and grouped into four groups of 20 mice. Two days later, mice were given drinking water containing 0.8 mg/ml sulphamethoxazole/0.16 mg/ml trimethoprim for a total of 10 days to suppress indigenous oral bacteria, followed by a 3-day washout period with sterile drinking water. On the following day (designated experimental day 0) mice were placed on a modification of powdered AIN-93G purified diet containing 37.5% (wt/vol) total sucrose (Cat. # TD.160810, Envigo, Madison, WI, United States). Mice were also given 4% (wt/vol) sterile sucrose water. In initial colonization experiments, mice were inoculated on experimental day 0 and each successive 4 days with 50 μl of 1.5% (wt/vol) carboxymethylcellulose containing approximately 1 × 109 cells of either wild type S. mutans UA159 (WT), ΔlrgAB (lrgAB-deficient), SAB161 (lrgAB-overexpressing) or with carboxymethylcellulose alone (mock inoculations). In competition experiments, mice were first inoculated over five consecutive days starting on experimental day 0 with 50 μl 1.5% (wt/vol) carboxymethylcellulose containing approximately 1 × 109 cells of wild type S. gordonii DL1 followed on experimental day 7 with five consecutive daily inoculations with 50 μl 1.5% (wt/vol) carboxymethylcellulose containing approximately 1 × 109 cells of either wild type S. mutans UA159 (WT), ΔlrgAB (lrgAB-deficient), SAB161 (lrgAB-overexpressing) or with carboxymethylcellulose alone (mock S. mutans). Oral swabs were taken at intervals to monitor colonization using HydraFlock® 6” Sterile Micro Ultrafine Flock swabs (Puritan Medical Products, Guilford, ME). At the end of each experiment mice were euthanized by CO2 asphyxiation followed by cervical dislocation. The protocol was reviewed and approved by the Institutional Animal Care and Use Committee at University of Florida (IACUC protocol #201810470).
Swab tips were vortexed (3 times for 5 s) in 1 ml sterile PBS, the tips removed and 200 μl added of ice-cold PBS containing approximately 5 × 108 depurinated cells of laboratory strain S. mitis UF2. The tube was then vortexed 5 s and centrifuged (10,000 × g, 10 min at 4°C) to pellet recovered cells. Cell pellets were then processed for DNA isolation using the DNeasy UltraClean Microbial kit (Qiagen Inc., Germantown, MD, United States) as per manufacturer’s instructions. In preliminary experiments, employment of a high concentration of depurinated cells was found to greatly enhance quantitative pelleting and recovery of low cell numbers and subsequent DNA, thus increasing the sensitivity of qPCR assays. The cells walls of depurinated cells remain intact and therefore at high concentrations likely act as a carrier to help limit non-specific binding and promote pelleting of recovered bacteria. Purine bases in genomic DNA are lost by depurination, producing apurinic sites and rendering DNA undetectable in all three qPCR assays. Cells from a 200 ml culture (OD600 = 0.5) of laboratory strain S. mitis UF2 in BHI were pelleted (4 × 50 ml at 10,000g × 7 min at 4°C) and each pellet resuspended in 11 ml sterile ice-cold PBS. Cell were pooled and again centrifuged. The pellet was resuspended in 35 ml of 0.2 N HCl and placed in a 70°C water bath for 90 min with vortexing (5 × 2 s) every 15 min. Cells were then pelleted as before and the 90 min incubation in fresh 0.2 N HCl repeated. Cells were washed 3 times with 30 ml sterile ice-cold PBS. Before the third centrifugation the cell concentration was estimated from OD600 and the subsequent cell pellet resuspended in sterile ice-cold PBS to a concentration of approximately 2.5 × 109 cells/ml, then aliquoted and stored at −75°C.
To assess dental colonization, the left and right halves of each mandible were aseptically extracted by first breaking the fibrous symphysis at the rostral midline, then gripping one incisor and pulling the left or right half of mandibular bone away from the temporal mandibular joint and nearly all associated soft tissue. Then, under a dissecting microscope, any remaining extraneous soft tissue near the molar teeth was removed by scraping with a scalpel followed by removal of bone approximately 2 mm anterior and posterior to the three molar teeth. Molar teeth with remaining underlying bone were sonicated on ice in 1 ml sterile PBS, pH 7.4, in siliconized 2 mL microcentrifuge tubes. Molar teeth with remaining bone were then aseptically removed using sterile forceps. Approximately 5 × 108 depurinated cells of laboratory strain S. mitis UF2 were then added, the tube vortexed 5 s and centrifuged (10,000 × g, 10 min at 4°C) to pellet recovered cells. Cell pellets were then processed for DNA isolation as described above for swabs. Supernatants were then immediately used for analyses of organic acids as described below.
Quantitative PCR was used to estimate total recovered bacterial genomes and recovered genomes of inoculated strains in each DNA sample. DNA isolation using the DNeasy UltraClean Microbial kit (Qiagen) resulted in 50 μl of DNA that was diluted to 125 μl with nuclease free water resulting in DNA in 4 mM Tris-HCl, pH 8.0. Samples from swabs were stored at −75°C in aliquots. Samples from molars were treated similarly, but further diluted 10-fold with 4 mM Tris-HCl, pH 8.0 to eliminate interference in qPCR assays caused by unknown components in the samples, then aliquoted and stored at −75°C. Each qPCR assay included 9 μl of diluted DNA and resultant genome numbers from the average of triplicates then multiplied by either 13.89 (for swab DNAs) or 138.89 (for mandibular DNAs) to calculated total recovered genomes in each sample. To estimate total recovered bacteria, degenerate primers were used to PCR amplify conserved regions of the ubiquitous single-copy gene, rpsL (30S ribosomal protein S12) (Lang et al., 2013; Shields et al., 2018). Recovery of mouse commensals was then estimated by subtracting recovered genomes for inoculated strains from total recovered bacterial genomes. Primers and qPCR conditions used are given in Supplementary Table S1. Standard curves were derived from DNA samples isolated from each strain grown to mid-exponential phase in BHI. S. mutans UA159 was used as standard for rpsL assays. Efficiencies, slopes and r2 values for standard curves were greater than 90%, −3.205 and 0.978, respectively. Statistical comparisons of colonization between groups were by one-way ANOVA with Tukey’s multiple comparisons test after conversion of genomes recovered to their log values in order to stabilize variances.
Smooth surface and sulcal caries of mandibular and maxillary molars were scored by a single calibrated examiner using Larson’s modification of the Keyes’ scoring system, as described previously (Culp et al., 2015). The linear evaluations of carious enamel involvement are expressed as E, while severities of carious lesions, based on degree of dentin involvement, are expressed as Ds (dentin exposed) and Dm (3/4 of the dentin affected). To stabilize variances, caries scores were expressed as proportions of their maximum possible values (124 for smooth surface caries and 56 for sulcal surface caries) and then the arcsine of the square root of the proportions calculated, as described previously (Culp et al., 2005). Transformed scores were compared by analysis of variance with the Tukey-Kramer post hoc test using Prism v8.1 (GraphPad Software, San Diego, CA, United States).
Organic Acid Analysis of Mouse Plaque Samples
The supernatants from sonicates of mandibles from four mice were pooled, yielding five samples of pooled supernatants for each group of twenty mice. Each pooled supernatant was then aspirated into a sterile 5 ml Luer-LokTM syringe using a 16-gauge needle (BD). The needle was removed and a 0.2 μm, 13 mm PVDF disposable filter was fastened to the 5 mL syringe (GE Whatman). Each sample was then filtered into a sterile 15 ml tube (Corning). Any sample remaining in the filter was collected by passing 5 ml of air through the filter. Samples were centrifuged at 1,000 × g for 1 min at 4°C to collect supernatants to the bottom of each 15 ml tube. The tubes were then frozen at −75°C and lyophilized. Metabolomics experiments and analyses were performed in the Metabolomics Core of CVI (Cardiovascular Institute) at University of Pennsylvania. Briefly, each lyophilized sample was dissolved in 150 μl of 50% MeOH and vortexed for 3 min. Thereafter, centrifugation was carried out at 10,000 rpm for 10 min, and 100 μl of the supernatant was obtained. The resuspended metabolite samples (10 μl) were injected onto Luna NH2 column (Phenomenex, CA, United States). LC-MS/MS analysis was performed using Agilent 6420 triple quadrupole (Agilent, CA, United States). Data Acquisition software was used to monitor and control LC-MS. Metabolites were separated on the analytical column at a flow rate of 150 μl/min and the LC gradient method was set as follows: t = 0 min, 85% B; t = 15 min, 0% B; t = 38 min, 0% B; t = 40 min, 85% B; t = 50 min, 85%. Solvent A was 20 mM ammonium acetate + 20 mM ammonium hydroxide in 95:5 water: acetonitrile and solvent B was 100% acetonitrile. The separated metabolites were identified by dynamic multiple reaction monitoring (MRM). Dynamic MRM involves sorting for the retention time of a specific parent molecular ion, the parent molecular weight, the optimized collision energy to produce a specific fragment ion and the fragment ion. Before sample analysis, authentic samples of carboxylic acids were used to verify dynamic MRM transition for quantitative dynamic MRM analysis. Using dynamic MRM, metabolites were identified, and relative quantification was determined by area of the peak in the chromatogram of each metabolite. Data was normalized by protein quantification.
Results
Either Lack of Expression or Overexpression of lrgAB Causes Induction of lrgAB Expression at Stationary Phase
To further characterize the role and regulation of LrgAB, a lrgAB constitutive overexpression strain of S. mutans UA159 was created (SAB161), whereby lrgAB expression was placed under control of the S. mutans ldh promoter and ribosome binding site (RBS). For comparison, we also included SAB163 (Ahn et al., 2020a), recently created to overexpress lytST in a similar manner to SAB161. Given that expression of lrgAB is under tight positive regulation by LytST (Ahn et al., 2010, 2012), lrgAB is also overexpressed in SAB163, which was recently observed by GFP quantification assays using a PlrgA-gfp reporter strain in SAB163 (Ahn et al., 2020a). We compared expression of lrgAB and lytST transcripts in wildtype (UA159), SAB161 and SAB163 at early-exponential (EE) and early-stationary (ES) growth phases by qPCR (Figure 1). We first confirmed that lrgA was dramatically upregulated at ES (> about 4-logs), compared to EE in the wild-type strain, while such a remarkable stationary-phase induction was not observed during the transition from EE to ES in either SAB161 or SAB163 (Figure 1A), although a slight increase was observed at ES in SAB163. This observation confirmed that lrgAB expression is constitutively over-expressed in SAB161 and is independent of growth phase. However, SAB163 still maintained a modest induction of lrgAB expression in stationary phase, and overexpressed lytST was responsible for increased lrgAB expression at EE (relative to wildtype), as recently observed (Ahn et al., 2020a). Consistent with our previous observation (Ahn et al., 2012), lytS expression levels in the wildtype were approximately 7.7-fold elevated during the transition to stationary phase (Figure 1B). This increment was moderately alleviated in SAB161, showing only a 2.6-fold increase, suggesting that lytST may be subject to possible feedback regulation though the lytST-lrgAB circuit. As expected, lytS was overexpressed in SAB163 throughout growth and its expression was at least a log higher than that observed in wildtype and SAB161 (Figure 1B). Thus, these two derivative strains, SAB161 and SAB163, overexpressing lrgAB during growth, were further investigated in this study, in conjunction with a ΔlrgAB mutant strain (referred to below as “lrgAB deficient”).
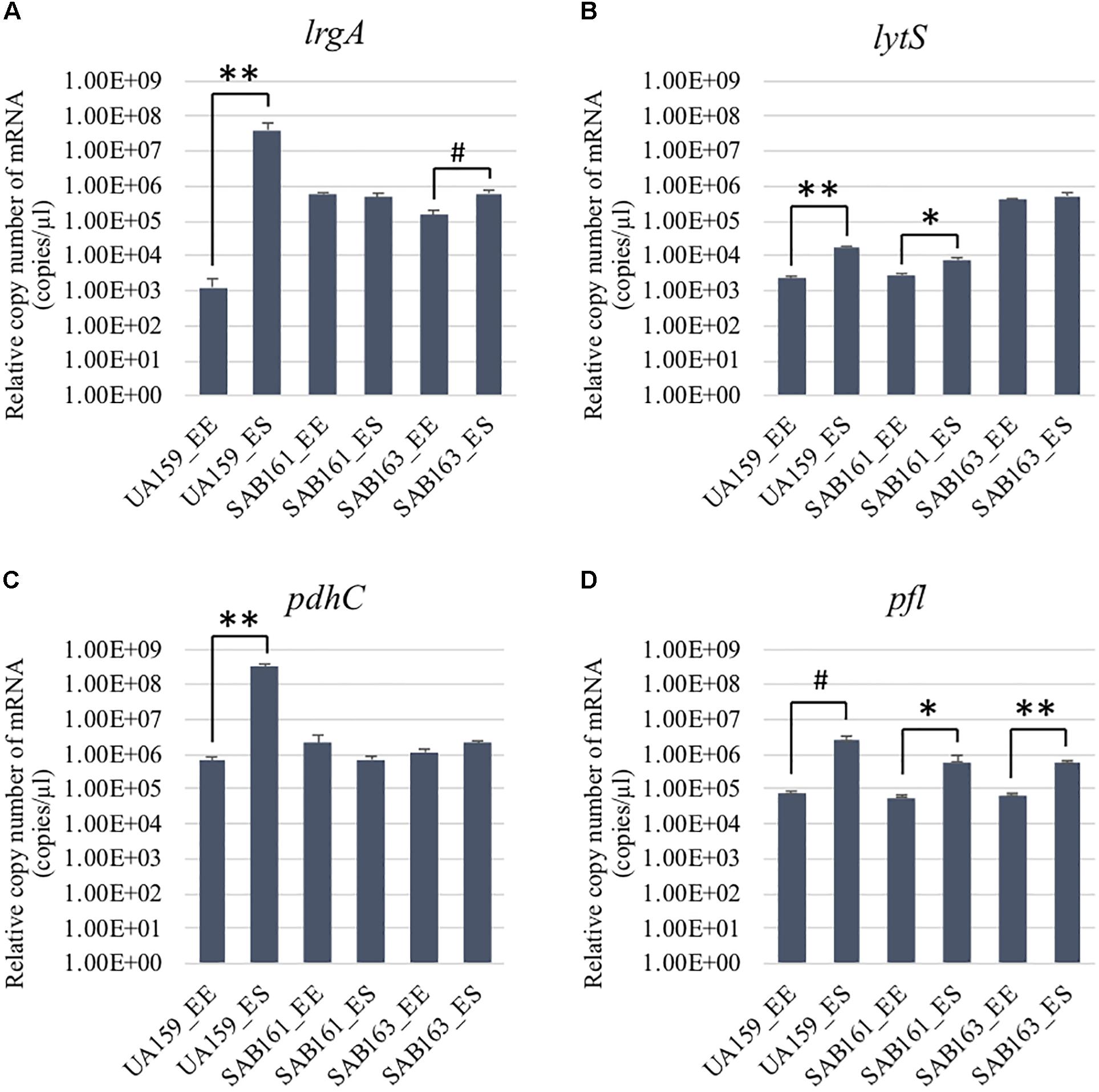
Figure 1. Growth-dependent expression determined by qRT-PCR in the lrg derivative strains. The expression of lrgA (A), lytS, (B) pdhC (C), and pfl (D) was measured at early-exponential (EE; OD600∼0.15–0.2) and -stationary (ES; OD600∼1.0) growth phases of S. mutans UA159 (WT), SAB161 (lrgAB-overexpressing), and SAB163 (lytST-overexpressing) (see section “Materials and Methods” for details). Data are averages of three independent biological replicates. Differences in relative gene expression between strains were evaluated for statistical significance by Student’s t-test (unpaired; one-tailed). **P < 0.001; *P < 0.005; #P < 0.01.
We recently reported that stationary phase PlrgA activation was further increased in both the ΔlrgAB and SAB163 strains, relative to wildtype (Ahn et al., 2020a, b; Ishkov et al., 2020). For measurement of PlrgA activity in SAB161 during growth, the PlrgA-gfp construct was introduced into SAB161 and PlrgA activity compared between the wildtype, ΔlrgAB, SAB161 and SAB163 backgrounds. Interestingly, PlrgA activity was increased about threefold in SAB161 at stationary growth (Supplementary Figure S1C), compared to that in wildtype (Supplementary Figure S1A). This induction was approximately 2.5-fold higher than in the ΔlrgAB background (Supplementary Figure S1B), and about 40% lower than in the SAB163 background (Supplementary Figure S1D). Another interesting finding was that the uncontrolled overexpression of lrgAB (in SAB 161) had no effect on PlrgA activity during exponential growth, confirming that PlrgA is specifically activated upon entry into stationary phase. This is in line with the sharp induction of high-level PlrgA activity in SAB163 between late-exponential and stationary growth (Supplementary Figure S1D), consistent with our previously published results (Ahn et al., 2020a). However, overexpression of lytST (SAB163) remarkably relieved the repression of lrgAB during exponential growth (Supplementary Figure S1D). Therefore, these observations demonstrate that either lack of expression or overexpression of lrgAB triggers the ability of S. mutans to induce lrgAB expression at stationary phase, although these effects could be exerted through different cellular and metabolic routes.
Either Lack of Expression or Overexpression of lrgAB Promotes Pyruvate Excretion During Growth
We previously demonstrated that the maximum level of pyruvate excreted during growth of the ΔlrgAB strain was about 50% elevated compared to wildtype (Ahn et al., 2019), implying that stationary phase lrgAB induction may be positively correlated with excreted pyruvate levels. Thus, we cultivated wildtype, SAB161 and SAB163 under the same conditions used previously to assess extracellular pyruvate levels during growth (FMC11 medium, at 37°C in a 5% CO2 atmosphere) (Ahn et al., 2019). As shown Figures 2B,C, the maximum level (∼1,000 μM) of pyruvate excreted during growth of the SAB161 and SAB163 strains was about threefold higher than wildtype (∼350 μM, Figure 2A), which may contribute to the observed increase in PlrgA activity observed in these strains (Supplementary Figures S1C,D). Stationary phase pyruvate re-uptake in SAB161 was somewhat decelerated (Figure 2B) relative to the wildtype (Figure 2A) and SAB163 (Figure 2C) strains. These results suggest that lack of expression or overexpression of lrgAB can each increase pyruvate excretion compared to wildtype S. mutans, although its underlying mechanism remains unknown. Thus, we next evaluated how either lack of expression or overexpression of lrgAB impacts the capacity of this organism to respond to and take up external pyruvate, particularly during aerobic growth, as oxygen availability was shown previously to influence the efficacy of LrgAB to function as a pyruvate uptake system (Ahn et al., 2019), consequently contributing to phenotypes associated with lrgAB expression. We aerobically cultivated S. mutans wildtype and derivative strains in BHI, supplemented by different concentrations (0, 1, 10, and 40 mM) of exogenous pyruvate. All strains efficiently utilized external pyruvate to sustain growth at stationary phase (Figure 3), in accordance with increased lrgAB promoter activity at stationary phase (Supplementary Figure S1). As expected, the ΔlrgAB strain experienced delayed exponential growth and entered stationary phase at a lower overall OD600 (Ahn et al., 2010; Ahn and Rice, 2016) relative to wildtype (Figures 3A,B). However, the impaired aerobic growth phenotype of the ΔlrgAB mutant was considerably restored by addition of either 10 mM or 80 mM pyruvate (Figure 3B). This growth restoration effect was not previously recognized when the cell was tested in FMC medium supplemented by pyruvate (Ahn et al., 2019), suggesting that the effect of LrgAB deficiency may depend on specific external metabolic/nutritional conditions. It is also noteworthy that exogenous pyruvate promoted growth of all strains to a similar extent, despite considerably different stationary phase PlrgA activities (Supplementary Figure S1), suggesting that the LrgAB-mediated pyruvate uptake and its cellular consequence are not proportional to the level of lrgAB expression, possibly due to post-transcriptional effects or metabolic adjustments.
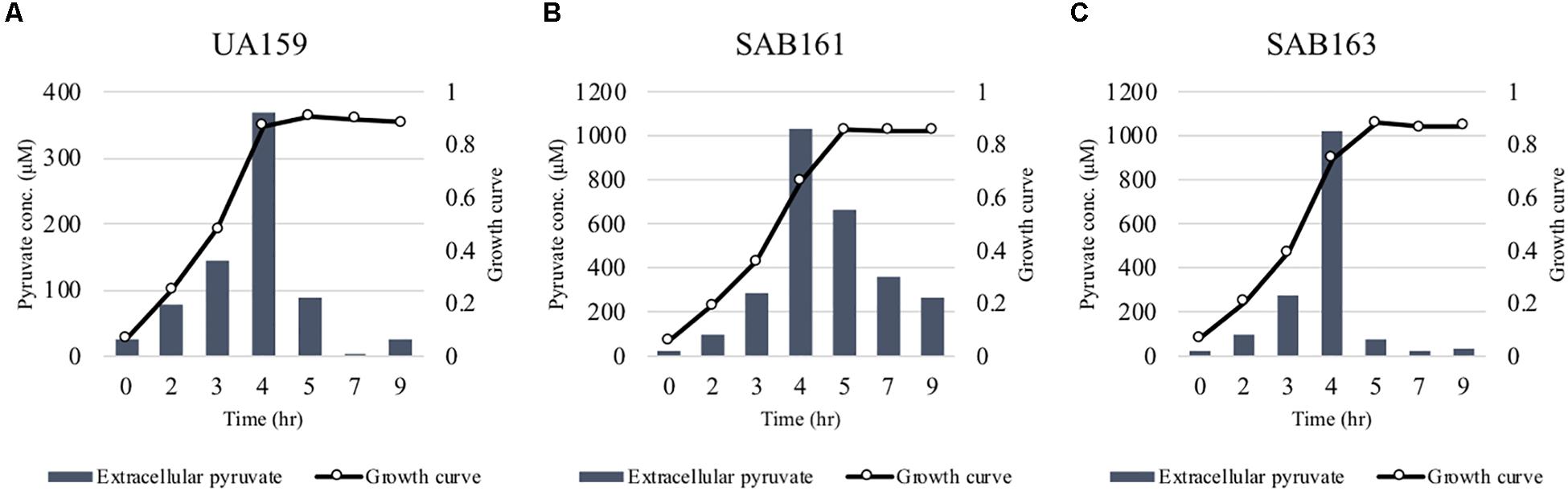
Figure 2. Measurement of extracellular pyruvate during growth of the lrg derivative strains. The strains, including S. mutans UA159 (WT, A), SAB161 (lrgAB-overexpressing, B), and SAB163 (lytST-overexpressing, C) were cultivated in FMC medium supplemented by 11 mM glucose. For time course measurements of extracellular pyruvate and growth, samples were taken at 1 or 2 h intervals (see section “Materials and Methods” for details). The concentration of pyruvate was determined using an EnzyChromTM pyruvate assay kit, and growth was measured by the optical density at 600 nm (OD600). Bars indicates the average concentration of extracellular pyruvate; solid line with circles indicates the corresponding growth curve. The results are representative of two independent experiments.
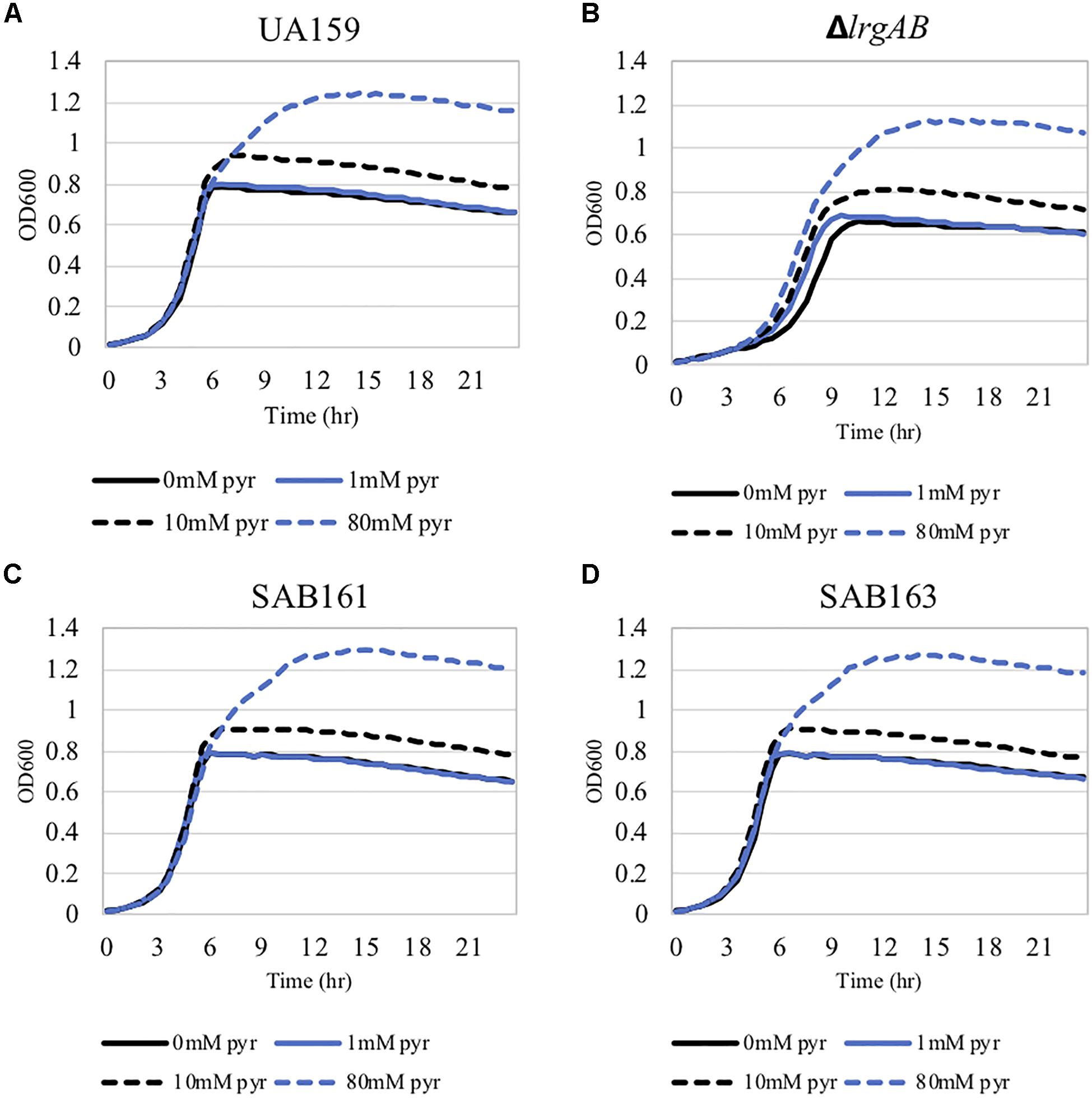
Figure 3. The effect of exogenously added pyruvate on the aerobic growth of the lrg-derivative strains. The strains, including S. mutans UA159 (WT, A), ΔlrgAB (lrgAB-deficient, B), SAB161 (lrgAB-overexpressing, C), and SAB163 (lytST-overexpressing, D) in BHI medium supplemented by different concentrations of exogenous pyruvate (0, 1, 10, and 40 mM). OD600 was monitored every 30 min at 37°C using the Bioscreen C lab system. The results are representative of three independent experiments.
Lack or Overexpression of lrgAB Alleviates the Conversion of Pyruvate to Acetyl-CoA
The finding that exogenous pyruvate restores the impaired aerobic growth phenotype of the ΔlrgAB strain (Figure 3B), further supports our previous hypothesis that external pyruvate may be internalized into the cell during exponential growth, independently of LrgAB (Ahn et al., 2019). We reported previously that stationary phase lrgAB induction and pyruvate uptake are efficiently inhibited by low concentrations (∼0.01 mM) of the toxic pyruvate analog, 3FP (3-fluoropyruvate) (Ahn et al., 2019). This analog competes with pyruvate and inhibits cell growth by binding to cellular pyruvate dehydrogenase (Pdh) complexes (Lang et al., 1987; Flournoy and Frey, 1989). S. mutans was unable to normally grow in the presence of 10 mM 3FP (Ahn et al., 2019). In this study, when we cultivated S. mutans wildtype and derivative strains in BHI, containing different levels (0, 1, and 10 mM) of 3FP, the ΔlrgAB strain was more resistant to 3FP than wildtype, particularly in the presence of 10 mM 3FP (Figure 4B). In contrast, SAB161 and SAB163 displayed elevated sensitivity even at low (1 mM) concentrations of 3FP (Figures 4C,D, respectively), compared to the wildtype and ΔlrgAB strains (Figures 4A,B, respectively), and their growth was even more severely impaired in the presence of 10 mM 3FP (Figures 4C,D) compared to wildtype. When this experiment was repeated in TV medium, a growth condition in which induction of lrgAB is not observed (Ahn et al., 2020a), growth trends in the presence of 3FP were similar to those observed in BHI, except that growth impairments were more severe in all strains tested (Supplementary Figure S2). Therefore, these results suggest that external pyruvate may be taken up by the cell through LrgAB during growth, which may subsequently affect metabolic enzymes such as Pdh. To investigate whether either lack of expression or overexpression of lrgAB actually affects expression of pyruvate metabolic pathways, we used qPCR to measure growth-dependent expression of pdhC transcripts, encoding one of the components of the Pdh complex, in wildtype, SAB161 and SAB163 strains. As shown in Figure 1C, the pdhC transcript levels were about 500-fold elevated during the transition from EE (early-exponential) to ES (early-stationary) in the wildtype, which is consistent with our previous microarray data (Ahn et al., 2012, 2019). However, this induction was completely abolished in both the SAB161 and SAB163 strains (Figure 1C). Given that the Pdh enzymes are responsible for the conversion of pyruvate to acetyl-CoA, this result suggests that pyruvate utilization via Pdh may be interrupted when lrgAB is overexpressed. In the previous study with the ΔlytS mutant all pdhDABC genes were still considerably upregulated at ES (approximately 45–212-fold) compared to EE, although upregulation was smaller than in the wildtype (approximately 284–366-fold). Collectively, these results suggest that overexpression of lrgAB, and to a lesser extent the absence of lrgAB, interferes with the catabolism of pyruvate through the Pdh pathway. Pyruvate is also converted by pyruvate formate lyase (Pfl) to acetyl-CoA and formate, particularly in the absence of oxygen. Interestingly, expression of pfl was increased about 32-fold as wildtype cells approached stationary phase when grown in a 95% air – 5% CO2 atmosphere (Figure 1D). This expression pattern is comparable to the approximately 38-fold increase in wildtype pfl expression observed in our previous microarray experiment (Ahn et al., 2012). The magnitude of pfl induction was less alleviated in SAB161 and SAB163, approximately 10- and 8.5-fold, respectively (Figure 1D). In our previous microarray experiment, expression of pfl in the ΔlytS strain was still about 35-fold increased during the transition to stationary phase (Ahn et al., 2012). Although the Pfl enzyme is unlikely to be active in this culture condition (5% CO2), it is possible that regulation of pfl gene expression is affected by both lack of expression and overexpression of lrgAB. These data also suggest that LrgAB, responsible for stationary phase pyruvate uptake, is closely linked to pyruvate catabolism.
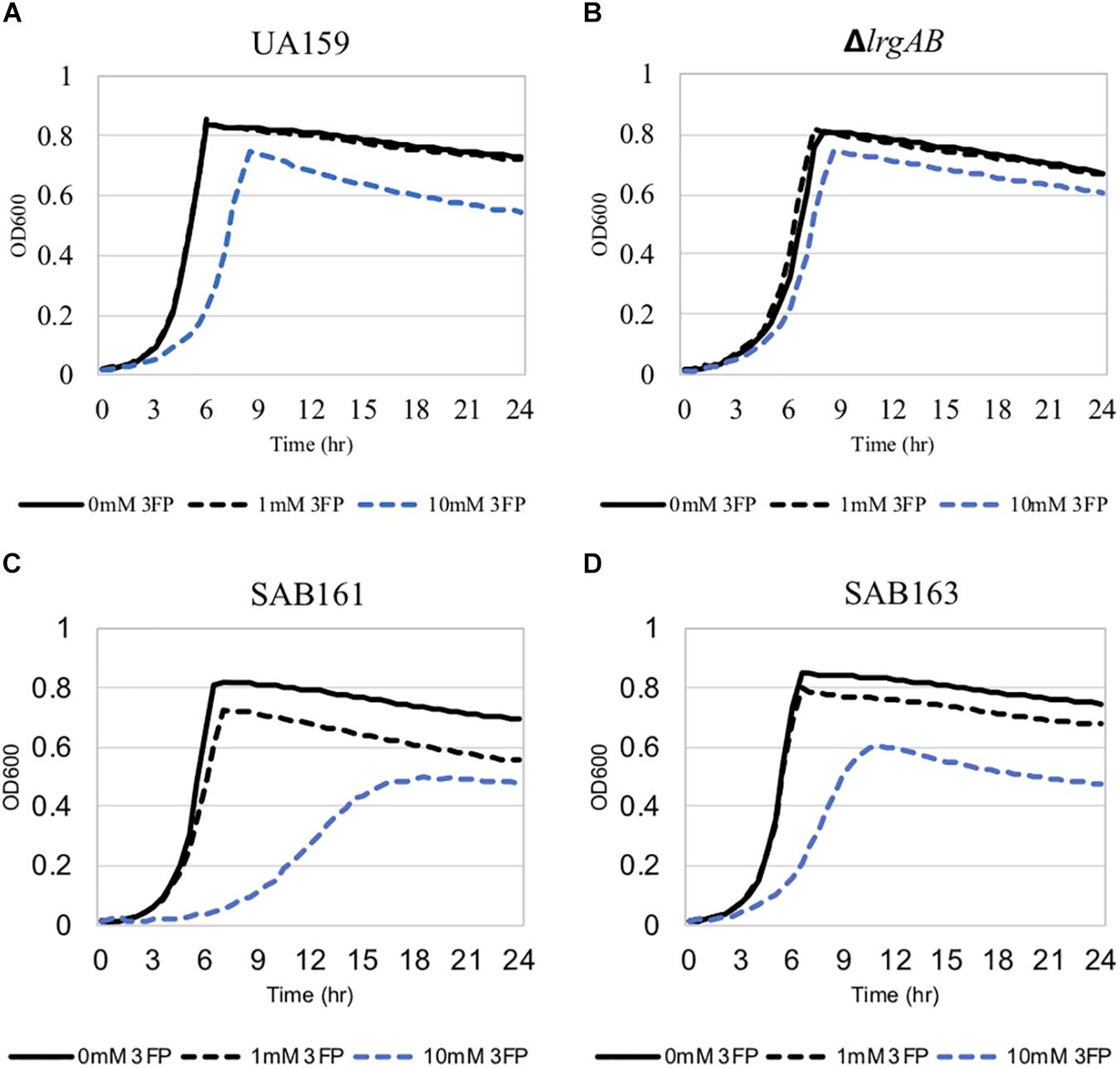
Figure 4. The effect of 3-fluoropyruvate (3FP; pyruvate analog) on the growth of the lrg-derivative strains. The strains, including S. mutans UA159 (WT, A), ΔlrgAB (lrgAB-deficient, B), SAB161 (lrgAB-overexpressing, C), and SAB163 (lytST-overexpressing, D), were cultivated in BHI medium supplemented by different concentrations of 3FP (0, 1, and 10 mM). Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C lab system. The results are representative of three independent experiments.
Contribution of LrgAB to Competition Between S. mutans and S. gordonii, in vitro
Given the fast-turnover nature of pyruvate upon nutrient limitation, the re-uptake of pyruvate through LrgAB may represent a competitive advantage for S. mutans against other endogenous species of the oral microbiome. To this end, we further explored how LrgAB contributes to S. mutans competition with other oral streptococci. We utilized a dual-species competition model, in vitro, with H2O2-generating S. gordonii, because LrgAB has been previously shown to be especially important for coping with oxidative stress, such as H2O2 challenge (Rice et al., 2017). Each S. mutans derivative strain and S. gordonii wildtype DL1 was cultured on Prussian blue agar medium, widely used to detect the presence of H2O2 (Saito et al., 2007). As shown in Figure 5A, S. gordonii DL1 produced a blue precipitation zone indicating H2O2 production, while S. mutans produced no distinct precipitation zone. Thus, when S. mutans cells outcompete S. gordonii cells, the blue precipitation zone should be reduced. However, the S. gordonii strain ΔspxB, lacking SpxB (pyruvate oxidase) that mediates production of H2O2 in the presence of oxygen, also produced a blue precipitation zone which was slightly smaller than wildtype DL1 yet still distinct from S. mutans (Figure 5A), suggesting that ΔspxB is still capable of producing H2O2 or another oxidant that reacts with this stain (presumably by an SpxB-independent mechanism). When each S. mutans strain was co-cultured with wildtype DL1 for 24 h and spotted onto Prussian blue agar medium, the sizes of blue zones among S. mutans strains were not qualitatively different (Figure 5B, left panel) and were also similar to DL1 alone as seen in Figure 5A. Thus, S. gordonii DL1 outcompeted all S. mutans strains. In contrast, when co-cultured with ΔspxB, all S. mutans strains, except for ΔlrgAB, appeared to be more competitive against ΔspxB, as manifested by generation of less blue precipitate by the spxB mutant (Figure 5B, right panel). These results suggest that LrgAB contributes to the ability of S. mutans to cope with H2O2 challenge, but this contribution is overshadowed by the higher levels of H2O2 generated by S. gordonii wildtype DL1.
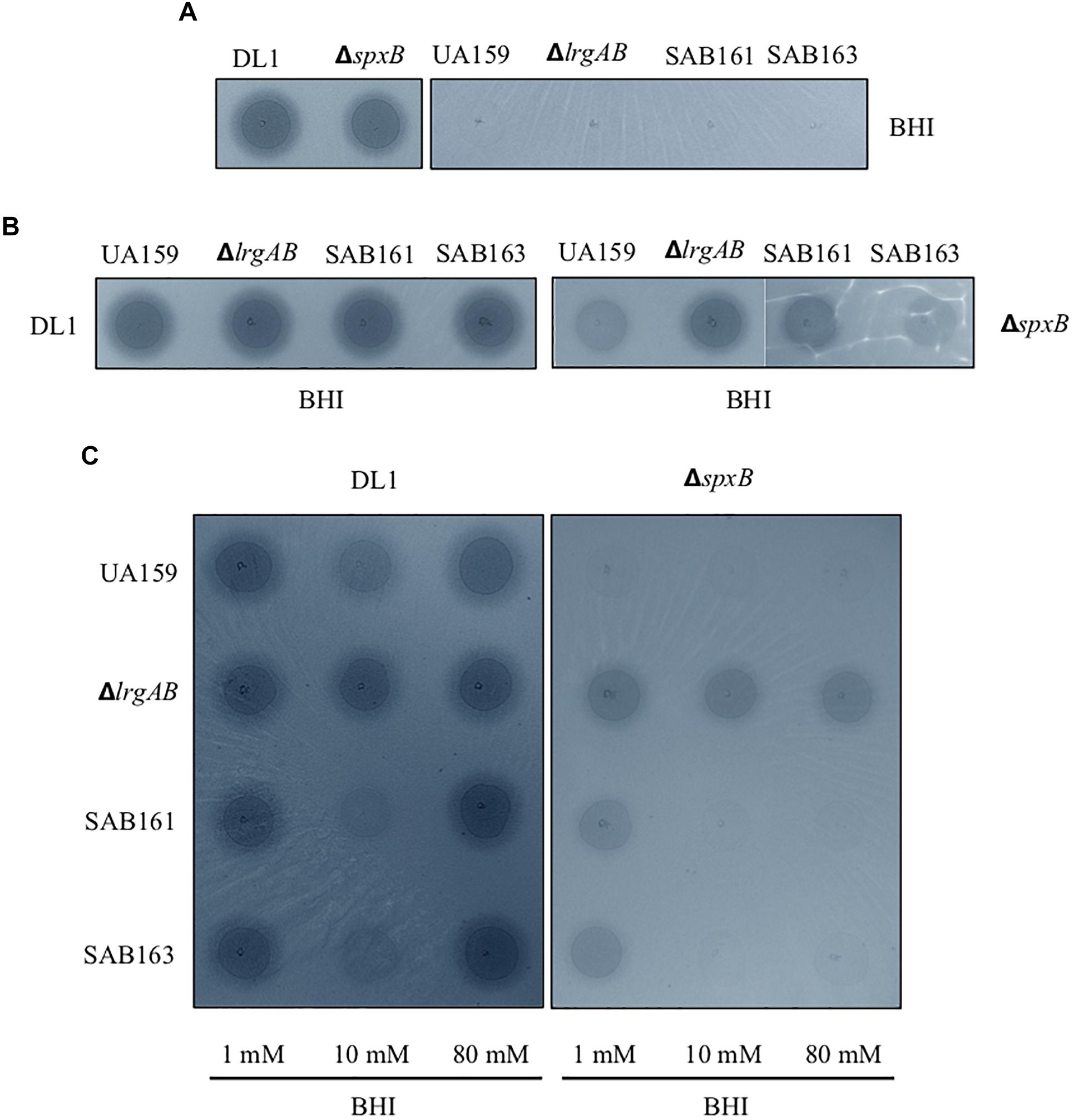
Figure 5. Dual-species competition assays between S. mutans lrg-derivative strains and S. gordonii DL1, or ΔspxB. The lrg-derivative strains include S. mutans UA159 (WT), ΔlrgAB (lrgAB-deficient), SAB161 (lrgAB-overexpressing), and SAB163 (lytST-overexpressing). Each S. mutans strains was cultured singly (A) or together in competition with either S. gordonii DL1 (left panels of B and C) or ΔspxB (right panels of B,C) in plain BHI medium (A,B) or in BHI medium supplemented by 1, 10, and 80 mM exogenous pyruvate (C). All cultures were for 24 h at 37°C in an aerobic atmosphere, and then was inoculated on Prussian blue agar plates. After an additional 24 h, the growth and blue precipitation zone were observed and analyzed with VisionWorks® software. All images were subjected to identical color balancing settings in Adobe Photoshop as described in section “Materials and Methods.”
To investigate whether external pyruvate influences the interaction between S. mutans and S. gordonii, we added pyruvate at concentrations of 1, 10, and 80 mM to the co-cultures of S. mutans and S. gordonii strains. Supplementation with 1 mM pyruvate resulted in no obvious difference in competition between any of the S. mutans strains and DL1 (Figure 5C, leftmost column of left panel). However, in the presence of 10 mM pyruvate, S. mutans SAB161 and SB163 were more competitive against DL1 (Figure 5C, middle column of left panel), indicating LrgAB in the presence of high concentrations of external pyruvate can function to enhance the competitiveness of S. mutans against S. gordonii. Interestingly, this competitive advantage for S. mutans growth was not observed in the presence of 80 mM pyruvate, due possibly to negative feedback regulation for stationary phase lrgAB induction and pyruvate uptake in response to high concentrations of external pyruvate (≥40 mM) (Ahn et al., 2019). In competition with ΔspxB, a much clearer competitive advantage for S. mutans strains was observed at all concentration of pyruvate, except for ΔlrgAB (Figure 5C, right panel), further suggesting LrgAB in the presence of external pyruvate confers a competitive fitness to S. mutans when coping with oxidative stress.
Contribution of LrgAB to S. mutans Competition With S. gordonii, in vivo
To interrogate whether LrgAB mediates a competitive advantage for S. mutans against S. gordonii in vivo, as well as to evaluate the cariological potential of S. mutans when LrgAB is absent or overexpressed, we used a well-defined mouse caries model (Culp et al., 2005, 2011). First, we used qPCR assays to compare oral colonization (estimated from oral swabs at days 10, 29, and 48, following the initial inoculation, Supplementary Figure S3A) and dental colonization (from sonicates of mandibular molars) by each of the three S. mutans strains (wildtype, ΔlrgAB or SAB161) plus a fourth uninoculated group as a negative control (mock). In addition, colonization by the total indigenous mouse commensal bacteria was assessed (Supplementary Figure S3B). All three S. mutans strains colonized the oral cavity at similar levels, gradually displaying an increase at day 48 versus day 10. Dental colonization by all three strains were also similar (Supplementary Figure S3B). Interestingly, recovery of mouse commensal bacteria increased with time when mice were inoculated with either of the S. mutans strains, whereas in the absence of S. mutans (mock group), the number of recovered commensal bacteria was unchanged. However, recovered commensals at day 10 in all three S. mutans groups were significantly less than in the mock group, indicating all S. mutans strains had a negative initial impact on oral commensal bacteria. However, as a whole, the commensal bacterial community was able to eventually grow. Recoveries of S. mutans strains and mouse commensal bacteria from molar biofilms were, in all cases, not statistically different, although the average values were consistently higher for each S. mutans strain compared to commensals. Caries scores reflected this colonization status (Supplementary Table S2), showing that the absence or overexpression of lrgAB had no obvious effect on the cariogenicity of S. mutans in this model.
In a second in vivo experiment, we further examined the contribution of LrgAB to competitiveness of S. mutans against S. gordonii DL1. The experimental design is depicted in Figure 6A. All mice were first inoculated with a single pure culture of S. gordonii DL1. One week later, three groups of 20 mice were inoculated with either S. mutans wildtype, ΔlrgAB or SAB161, while the fourth group (Mock S. mutans) underwent inoculations without added S. mutans. Oral colonization by S. gordonii DL1 was consistent over time in the mock group, whereas oral colonization by DL1 in the presence of all three strains of S. mutans was negatively impacted at day 35, but were able to recover to levels seen in the mock group by day 54, except in the ΔlrgAB group, in which DL1 levels remained below the mock group (Figure 6B). The establishment of DL1 on molars was hindered by subsequent inoculation with all of the S. mutans strains (Figure 6B), which is in contrast to the in vitro data, in which S. gordonii DL1 was dominant against S. mutans on Prussian Blue agar medium plates (Figure 5B, left panel). Both oral colonization and incorporation of S. mutans into dental biofilms were similar between all three strains of S. mutans, although oral colonization, but not dental colonization, appeared lower in the presence of DL1 (Figure 6B) versus in its absence (Supplementary Figure S3B). With respect to mouse commensal bacteria, DL1 had no apparent impact on either oral or dental colonization (compare upper left panels in Figure 6B with Supplementary Figure S3B). Recoveries of mouse commensals from oral swabs after challenge with each of the three strains of S. mutans showed some minor variations but remained mostly persistent. The presence within the dental biofilm of mouse commensals was unimpeded when challenged with either of these three S. mutans strains. Again, caries scores reflected colonization status (Supplementary Table S3), suggesting that either lack or overexpression of lrgAB had no major impact on competition with S. gordonii.
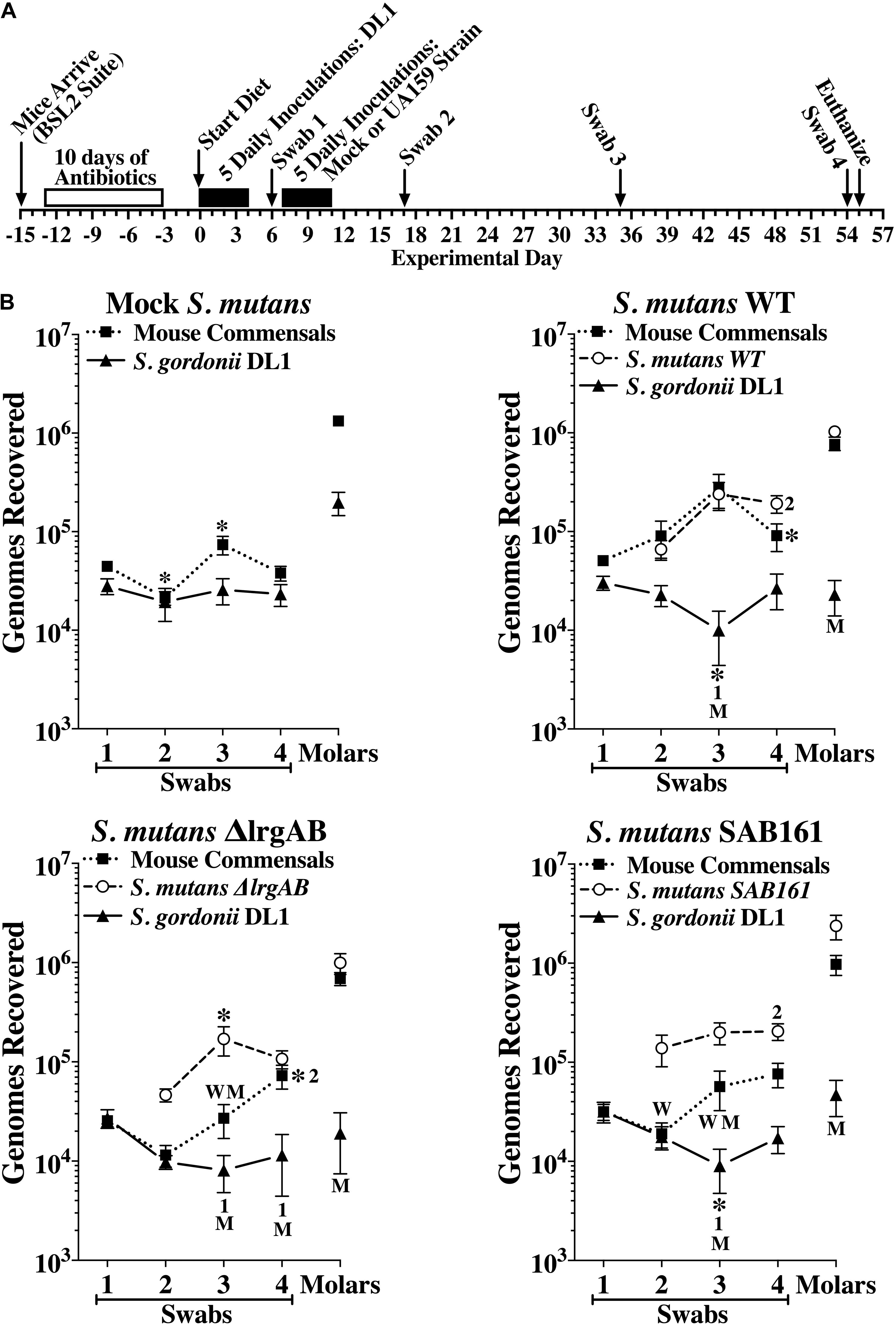
Figure 6. Colonization of the oral cavity and mandibular molars of mice by wild type S. mutans UA159 (WT), ΔlrgAB (lrgAB-deficient) and SAB161 (lrgAB-overexpressing) strains with S. gordonii DL1. (A) Timeline of key events in the experiment (see section “Materials and Methods” for details). (B) Colonization results for each indicated inoculated strain and mouse oral commensals (Commensals) from oral swabs 1–4 and from sonicates of mandibular molars taken at experimental days indicated in the timeline. Results are mean ± SE (n = 20 mice per group) of recovered genomes estimated by qPCR. Statistical comparisons by one-way ANOVA with Tukey-Kramer multiple comparisons test. *p ≤ 0.05 versus the previous swab or an earlier swab as indicated by the swab number, or versus the same point in either the mock (M) or wild type (W) group.
Deficiency of lrgAB May Be Complemented by Other External Metabolites
The in vivo competition data above was somewhat surprising, because even the ΔlrgAB strain displayed successful colonization of molar biofilms, in which the surrounding micro-environment is likely aerobic. In fact, when the ΔlrgAB strain is aerobically cultured on an agar plate, growth of the mutant is almost completely inhibited (Ahn et al., 2010). However, as shown above in Figure 3B, the impaired aerobic growth phenotype of the ΔlrgAB mutant was restored in vitro by external pyruvate (Figure 3B). S. mutans and other members of the oral microbiota can excrete various metabolic organic acids, particularly α-keto acids, including pyruvate (pyr), α-ketoglutarate (αKG) and oxaloacetate (OA), which are known to possess H2O2-scavenging activity (Nath et al., 1995; Giandomenico et al., 1997; Long and Halliwell, 2011, 2012). Both αKG and OA restored S. mutans’ growth in the presence of a H2O2 challenge (Supplementary Figure S4), similar to pyruvate (Ahn et al., 2019). However, unlike pyruvate, αKG and OA did not promote enhanced growth at stationary phase when challenged with H2O2, suggesting that these organic acids may not be utilized as a secondary carbon source in this growth condition. However, αKG and to a great extent OA restored the aerobic growth defect of the ΔlrgAB mutant strain (Figure 7). These observations suggest that common metabolites, such as α-keto acids, within the oral microflora may have impacted the outcomes of our in vivo S. gordonii DL1 – S. mutans competition experiment. Efforts were thus directed to analyze samples from this experiment by mass spectrometry for a profile of organic acids in supernatants after pelleting cells from mandibular sonicates, an acellular extract of molar biofilms. Results between each of the four groups of mice are shown in Supplementary Table S4 and demonstrate remarkably high concentrations of lactate in all four groups, likely due to carbohydrate fermentation by oral resident bacteria, even in the absence of S. mutans (mock group), suggesting that a high proportion of acidogenic and aciduric species could already reside in the mouse oral microbiota. Pyruvate concentrations in all groups were relatively low compared to most other organic acids, possibly due to its fast-turnover. Interestingly, there were no statistically significant differences between groups for each of the assayed organic acids, suggesting extracellular metabolites may be dynamically balanced by microbiota within dental biofilms in response to ecological fluctuations, even within the more acidic environment imparted by S. mutans.
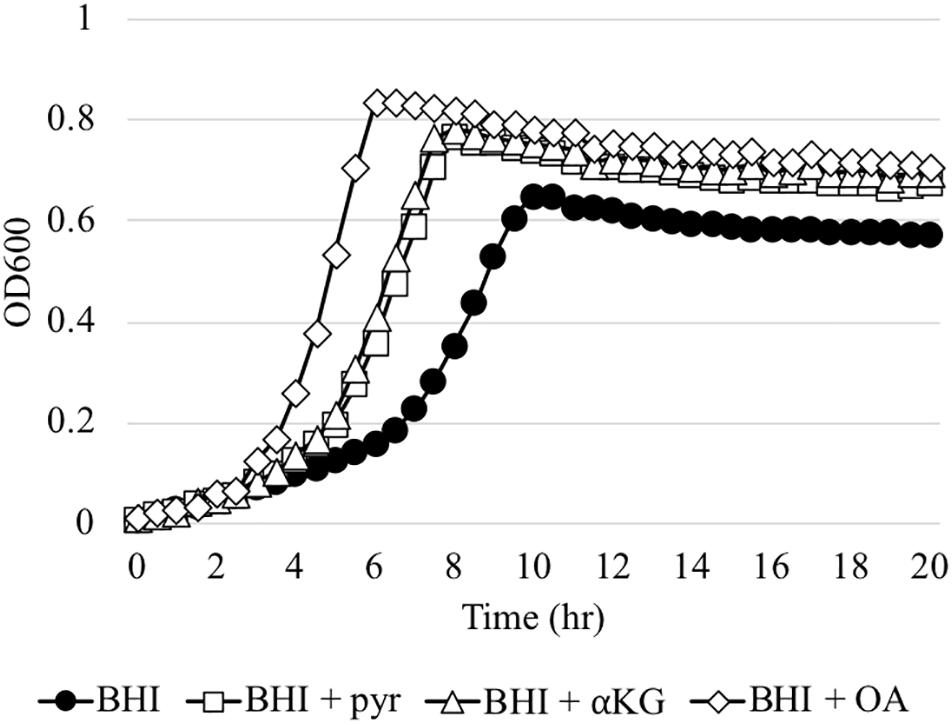
Figure 7. The effect of α-keto-acids on the aerobic growth impairment of the ΔlrgAB mutant strain. The ΔlrgAB strain was aerobically cultivated in plain BHI medium and BHI supplemented by 1 mM exogenous pyruvate (pyr), α-ketoglutarate (αKG), and oxaloacetate (OA). Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C lab system. The results are representative of three independent experiments.
Discussion
In this present study, we further characterize the role and regulation of LrgAB in S. mutans focusing on its cariological and ecological significance to better understand its function in a condition mimicking the in vivo oral milieu. One important finding is that LrgAB is closely linked to the conversion of pyruvate to acetyl-CoA through the Pdh pathway, likely modulating both the intracellular and overflow (excreted) levels of pyruvate. Although the underlying mechanisms remain to be elucidated, overexpression of lrgAB almost completely inhibits induction of the pdh genes in S. mutans that are normally dramatically upregulated during the transition to stationary growth phase (Ahn et al., 2012). Lack of lrgAB, which observed in the ΔlytS mutant, also appeared to modestly alleviate stationary phase pdh induction, relative to that observed in the wildtype. Thus, metabolic changes in response to perturbated expression of lrgAB may have a negative effect on the activity of Pdh. In line with this supposition, either lack or overexpression of lrgAB promoted pyruvate excretion during S. mutans growth. Intimate linkage between LrgAB and pyruvate catabolism was anticipated from our previous results (Ahn et al., 2020b) in which stationary-phase lrgAB induction and pyruvate uptake were also completely inhibited by disruption of the Pta-AckA pathway, responsible for the conversion of acetyl-CoA to acetate with the production of one molecule of ATP. Collectively, these findings suggest the catabolic rate of pyruvate is an important trigger to take up external pyruvate through LrgAB, and vice versa. Results, moreover, are consistent with previous results demonstrating both lrgAB and pta-ackA expression are directly under the control of two global regulators CcpA and CodY (Kim and Burne, 2017; Kim et al., 2019; Ahn et al., 2020c). Considering Pdh is highly regulated by various metabolic intermediates, including pyruvate (Hansen and Henning, 1966; Schwartz and Reed, 1970; Shen and Atkinson, 1970; Datta, 1991), and that pdh genes are commonly upregulated in response to three environmental stressors (i.e., aerobic, heat and vancomycin challenge) (Rice et al., 2017), it is reasonable to posit that pyruvate catabolism and lrg regulation work together and are modulated by the cell’s metabolic status and environmental condition.
Another notable finding is that lrgAB expression was correlated with the magnitude of growth inhibition by 3FP, known to interfere with the cellular Pdh complex (Lang et al., 1987; Flournoy and Frey, 1989), thus suggesting external pyruvate is taken up through LrgAB during exponential growth, in addition to the stationary phase (Ahn et al., 2019). This possibility was also suggested by our recent observation, showing that the addition of pyruvate to tryptone medium elicited PlrgA activation during exponential growth and an enhanced growth rate (Ahn et al., 2020a). It therefore is plausible that pyruvate is dynamically fluxed through LrgAB in response to external pyruvate levels, or other unknown metabolic/nutritional signals, whereas uptake and utilization of external pyruvate for re-growth may require activation of the LytST-LrgAB circuit by a metabolic cue.
Deciphering the competitive fitness between species within the oral microbiota is crucial for understanding the contribution of LrgAB to the pathogenic potential of S. mutans and changes that lead to dysbiosis. This present study therefore employed a dual-species model of S. mutans and S. gordonii to further characterize the ecological significance of LrgAB. In competition assays using Prussian blue agar medium, we found S. gordonii DL1 was relatively dominant over all tested S. mutans derivative strains when cultured aerobically, although this was not surprising given that DL1 generates H2O2 under ample oxygen in a SpxB-dependent manner (Kreth et al., 2005, 2008). However, LrgAB contribution to this competition was not evident. As anticipated, DL1 dominance was attenuated in an isogenic spxB-deficient mutant (ΔspxB), which only displayed dominance against the ΔlrgAB strain, reinforcing a role for LrgAB in S. mutans’ competition with H2O2-generating S. gordonii. Given that pyruvate is excreted as an overflow metabolite, it should be a common carbon source in microbiome environments such as the oral cavity (Takahashi et al., 2010; Gawron et al., 2019). More interestingly, external pyruvate modulated this interspecies competition, with 10 mM pyruvate conferring a great competitive advantage to S. mutans wildtype and SAB161 but not ΔlrgAB. This advantage may be due to the ability of pyruvate to scavenge H2O2, as evidenced by excretion of more pyruvate by S. mutans during growth in the presence of 10 mM pyruvate than in its absence, enabling more efficient and rapid scavenging of H2O2. Also, pyruvate at 10 mM may be more efficiently utilized by S. mutans compared to DL1 to promote its growth at stationary phase. The loss of this advantage by increasing exogenous pyruvate to 80 mM may be due to negative feedback regulation through LytST, as observed by ≥40 mM pyruvate (Ahn et al., 2019). Conversely, 80 mM pyruvate may have enhanced DL1’s capacity to generate H2O2, as its dominance was lost upon deletion of spxB. Although the molecular events behind these observations cannot be fully clarified under the present experimental approach, these results may have implications for therapeutic application of pyruvate to restrict the survival of S. mutans or enhance the fitness of H2O2-producing non-cariogenic bacteria.
The outcomes of competition between S. mutans and DL1 in vivo were rather unexpected, in that both the absence or overexpression of S. mutans lrgAB had little effect on competitiveness against DL1. Moreover, despite prior dental colonization by DL1, S. mutans consistently outnumbered DL1 leading to high caries scores in all experimental groups. Considering the ΔlrgAB mutant is highly sensitive to oxygen and H2O2 (Ahn et al., 2010; Turner et al., 2019), and given the superiority of DL1 in competition with all three strains of S. mutans in vitro, our in vivo results indicate this dual-species competition is affected considerably by in vivo environmental factors. Such factors include interactions with other oral microbiota, host factors and diet. The highly cariogenic mouse diet may have imparted a competitive advantage for S. mutans, although Tanzer et al. (2012) previously reported that S. mutans’ competitiveness against DL1 is independent of a high sucrose diet. In addition, in another in vivo study, we found strains of human commensal streptococci that are competitive with S. mutans under the same high sucrose diet (manuscript submitted). Another possibility is that the micro-environment within the dental biofilm is micro-aerobic, therefore better supporting growth of the ΔlrgAB mutant compared to a more aerobic condition. Dietary sucrose triggers production of extracellular glucans by S. mutans, creating an anaerobic microenvironment (Kleinberg, 2002; Paes Leme et al., 2006; Bowen and Koo, 2011; Koo et al., 2013) that may be disadvantageous to DL1, as it requires oxygen to generate competitive amounts of H2O2. This anoxic biofilm environment may also complement the aerobic growth impairment of the ΔlrgAB strain. S. mutans also utilizes dietary sucrose to produce acid, thus lowering the surrounding pH and leading to a shift in the oral microbiota composition toward a higher proportion of other acidogenic and aciduric species (Marsh, 2003; Takahashi and Nyvad, 2008; Bradshaw and Lynch, 2013; Rosier et al., 2014). The fact that a comparable amount of lactate was detected even in the mock group of mice not inoculated with S. mutans, suggests select mouse commensal species within dental biofilms also produce lactic acid during fermentation of sucrose, that may help foster S. mutans’ colonization. Moreover, the extremely low levels of caries in the mock group further suggests acids produced by mouse commensals are likely diffused away from tooth enamel surfaces, possibly due to significantly less production of a surrounding extracellular glucan-like matrix. Because total recovered mouse commensals from dental biofilms were similar between the mock group and the three S. mutans groups, one or more species of mouse commensals are likely also aciduric and thus flourish even when in competition with S. mutans. Future in vivo studies of dual-species competition may consider the use of gnotobiotic mice, a less extreme cariogenic diet or delineation of colonization by specific indigenous species and their phenotypes.
The observation that DL1 failed to compete with the oxygen-sensitive S. mutans ΔlrgAB strain in vivo, extended our interest in the ecological role of external metabolites in interspecies competition, which was initiated by our finding that external pyruvate restored impaired aerobic growth of the ΔlrgAB strain. Subsequently, we found the same effect when supplementing the culture with other α-keto acids, such as α-ketoglutarate and oxaloacetate, further supporting an ecological contribution of metabolic intermediates excreted presumably by indigenous microbiota. The ability of α-keto acids to buffer external sources of oxidative stress, such as H2O2, may have assisted the oxygen-sensitive ΔlrgAB strain to cope with oxidative stress in vivo. The reaction of α-keto acids, such as α-ketoglutarate, oxaloacetate, and pyruvate, with H2O2 is expected to generate CO2 and either acetate, succinate or malonate as major byproducts (Giandomenico et al., 1997; Oh et al., 2002; Berry and Toms, 2006; Satpute et al., 2014). Acetate, in particular, would also be taken up into the cell in parallel with pyruvate under nutrient-limited growth conditions (Jolkver et al., 2009; Paczia et al., 2012), and was recently reported to affect stationary phase lrgAB induction in S. mutans (Ahn et al., 2020a). Changes in levels of α-keto acids was also reported to coordinate catabolism of amino acids and glucose in Escherichia coli (Zampieri et al., 2019). These dynamically changing external metabolites could force bacteria to continuously modulate their uptake, which in turn may require continuous redirection of intracellular fluxes. These external metabolites may therefore have modulated the resources available for indigenous microbiota, and also affected the capacity of DL1 to produce competitive amounts of H2O2. Mouse host factors (e.g., salivary peroxidase) could also attenuate localized H2O2 concentrations (Tenovuo, 1976; Tenovuo and Pruitt, 1984; Carlsson, 1987; Ericson and Bratt, 1987). In fact, H2O2 itself is unstable and readily degraded in nature. These factors combined may have attenuated establishment of DL1 within the indigenous microbiota community.
We also compared organic acids from molar sonicates to explore correlations between the metabolic activity of oral microbiota in dental biofilms and the contribution of LrgAB to caries development. Surprisingly, the abundance of each organic acid was similar among each of the four groups, suggesting the exo-metabolome may be rapidly balanced through continuous adjustments of intercellular fluxes and cross-competition within the murine oral indigenous microbiota. Nevertheless, it is noteworthy that together with glucose, malate is the preferred carbon source for B. subtilis and is known to repress transcription of pftAB (previously, ysbAB), a lrgAB homolog in this organism (Charbonnier et al., 2017). Our previous transcriptomic analysis also showed that the mleS gene (SMU.137), encoding an enzyme catalyzing conversion of L-malate to lactic acid and CO2, was > 3-fold downregulated in the ΔlrgAB strain compared to the wildtype (Rice et al., 2017). This malolactic enzyme was further proposed to enhance the acid tolerance response by increasing the cytoplasmic pH through production of CO2 (Sheng and Marquis, 2007). However, given caries and recovered malate levels were similar between the wildtype and ΔlrgAB strains, the ΔlrgAB strain must use alternative and/or redundant mechanisms for acid tolerance, in addition to metabolism of malate. Nevertheless, it may be interesting to study how malate uptake and metabolism are linked to lrg and pyruvate uptake regulation.
In summary, the present study implemented the use of both in vitro and in vivo competition models to address how LrgAB contributes to niche competition and modulates the cariogenic potential of S. mutans. The results suggest that the role and regulation of LrgAB are coordinated in response to both external and internal metabolic fluxes and dynamics that are more variable in vivo, and consequently precluded detection of a specific cariological and ecological role for LrgAB. Although knowledge of niche competition with non-mutans streptococci in rodents with indigenous oral flora is limited, this in vivo study opens the possibility of examining the relationship between ecological fitness and metabolite fluxes that has been overlooked in interpreting the outcomes of in vivo interspecies competition experiments.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at University of Florida.
Author Contributions
S-JA contributed to conception, design, acquisition, analysis, and interpretation, and drafted the manuscript. WH and SD contributed to acquisition, analysis, and interpretation. KR contributed to conception and interpretation, and edited the manuscript. DC contributed to conception, design, acquisition, analysis, and interpretation, and edited the manuscript. All authors gave the final approval and agreed to be accountable for all aspects of the work.
Funding
This study was supported by NIDCR R01 DE025237.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Robert A. Burne (Department of Oral Biology, University of Florida) for providing all the resources needed.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02119/full#supplementary-material
References
Ahn, S. J., Deep, K., Turner, M. E., Ishkov, I., Waters, A., Hagen, S. J., et al. (2019). Characterization of LrgAB as a stationary phase-specific pyruvate uptake system in Streptococcus mutans. BMC Microbiol. 19:223. doi: 10.1186/s12866-019-1600-x
Ahn, S. J., Desai, S., Blanco, L., Lin, M., and Rice, K. C. (2020a). Acetate and potassium modulate the stationary-phase activation of lrgAB in Streptococcus mutans. Front. Microbiol. 11:401. doi: 10.3389/fmicb.2020.00401
Ahn, S. J., Desai, S., Lin, M., and Rice, K. C. (2020b). The Pta-AckA pathway regulates LrgAB-mediated pyruvate uptake in Streptococcus mutans. Microorganisms 8:846. doi: 10.3390/microorganisms8060846
Ahn, S. J., Kim, H. M., Desai, S., Deep, K., and Rice, K. C. (2020c). Regulation of cid and lrg expression by CodY in Streptococcus mutans. Microbiologyopen 9:e1040. doi: 10.1002/mbo3.1040
Ahn, S. J., Gu, T., Koh, J., and Rice, K. C. (2017). Remodeling of the Streptococcus mutans proteome in response to LrgAB and external stresses. Sci. Rep. 7:14063. doi: 10.1038/s41598-017-14324-w
Ahn, S. J., Qu, M. D., Roberts, E., Burne, R. A., and Rice, K. C. (2012). Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol. 12:187. doi: 10.1186/1471-2180-12-187
Ahn, S. J., and Rice, K. C. (2016). Understanding the Streptococcus mutans Cid/Lrg system through CidB function. Appl. Environ. Microbiol. 82, 6189–6203. doi: 10.1128/AEM.01499-16
Ahn, S. J., Rice, K. C., Oleas, J., Bayles, K. W., and Burne, R. A. (2010). The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156(Pt 10), 3136–3147. doi: 10.1099/mic.0.039586-0
Berry, E. V., and Toms, N. J. (2006). Pyruvate and oxaloacetate limit zinc-induced oxidative HT-22 neuronal cell injury. Neurotoxicology 27, 1043–1051. doi: 10.1016/j.neuro.2006.05.011
Bowen, W. H., and Koo, H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45, 69–86. doi: 10.1159/000324598
Bradshaw, D. J., and Lynch, R. J. (2013). Diet and the microbial aetiology of dental caries: new paradigms. Int. Dent. J. 63(Suppl. 2), 64–72. doi: 10.1111/idj.12082
Burne, R. A. (1998). Oral streptococci. products of their environment. J. Dent. Res. 77, 445–452. doi: 10.1177/00220345980770030301
Burne, R. A., Ahn, S. J., Wen, Z. T., Zeng, L., Lemos, J. A., Abranches, J., et al. (2009). Opportunities for disrupting cariogenic biofilms. Adv. Dent. Res. 21, 17–20. doi: 10.1177/0895937409335593
Burne, R. A., Wen, Z. T., Chen, Y. Y., and Penders, J. E. (1999). Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181, 2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999
Carlsson, J. (1987). Salivary peroxidase: an important part of our defense against oxygen toxicity. J. Oral Pathol. 16, 412–416. doi: 10.1111/j.1600-0714.1987.tb02077.x
Charbonnier, T., Le Coq, D., McGovern, S., Calabre, M., Delumeau, O., Aymerich, S., et al. (2017). Molecular and Physiological Logics of the Pyruvate-Induced Response of a Novel Transporter in Bacillus subtilis. mBio 8:e00976-17. doi: 10.1128/mBio.00976-17
Culp, D. J., Quivey, R. Q., Bowen, W. H., Fallon, M. A., Pearson, S. K., and Faustoferri, R. (2005). A mouse caries model and evaluation of aqp5-/- knockout mice. Caries Res 39, 448–454. doi: 10.1159/000088179
Culp, D. J., Robinson, B., Cash, M. N., Bhattacharyya, I., Stewart, C., and Cuadra-Saenz, G. (2015). Salivary mucin 19 glycoproteins: innate immune functions in Streptococcus mutans-induced caries in mice and evidence for expression in human saliva. J. Biol. Chem. 290, 2993–3008. doi: 10.1074/jbc.M114.597906
Culp, D. J., Robinson, B., Parkkila, S., Pan, P. W., Cash, M. N., Truong, H. N., et al. (2011). Oral colonization by Streptococcus mutans and caries development is reduced upon deletion of carbonic anhydrase VI expression in saliva. Biochim. Biophys. Acta 1812, 1567–1576. doi: 10.1016/j.bbadis.2011.09.006
Datta, A. (1991). Characterization of the inhibition of Escherichia coli pyruvate dehydrogenase complex by pyruvate. Biochem. Biophys. Res. Commun. 176, 517–521. doi: 10.1016/0006-291x(91)90955-7
Deutscher, J. (2008). The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11, 87–93. doi: 10.1016/j.mib.2008.02.007
Ericson, T., and Bratt, P. (1987). Interactions between peroxide and salivary glycoprotein: protection by peroxidase. J. Oral Pathol. 16, 421–424. doi: 10.1111/j.1600-0714.1987.tb02079.x
Flournoy, D. S., and Frey, P. A. (1989). Inactivation of the pyruvate dehydrogenase complex of Escherichia coli by fluoropyruvate. Biochemistry 28, 9594–9602. doi: 10.1021/bi00451a007
Gawron, K., Wojtowicz, W., Lazarz-Bartyzel, K., Lamasz, A., Qasem, B., Mydel, P., et al. (2019). Metabolomic status of the oral cavity in chronic periodontitis. In Vivo 33, 1165–1174. doi: 10.21873/invivo.11587
Giandomenico, A. R., Cerniglia, G. E., Biaglow, J. E., Stevens, C. W., and Koch, C. J. (1997). The importance of sodium pyruvate in assessing damage produced by hydrogen peroxide. Free Radic. Biol. Med. 23, 426–434. doi: 10.1016/s0891-5849(97)00113-5
Gorke, B., and Stulke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624. doi: 10.1038/nrmicro1932
Hansen, H. G., and Henning, U. (1966). Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochim. Biophys. Acta 122, 355–358. doi: 10.1016/0926-6593(66)90076-2
Henkin, T. M. (1996). The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135, 9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x
Huang, X., Palmer, S. R., Ahn, S. J., Richards, V. P., Williams, M. L., Nascimento, M. M., et al. (2016). A highly arginolytic Streptococcus species that potently antagonizes Streptococcus mutans. Appl. Environ. Microbiol. 82, 2187–2201. doi: 10.1128/AEM.03887-15
Ishkov, I. P., Ahn, S. J., Rice, K. C., and Hagen, S. J. (2020). Environmental triggers of lrgA expression in Streptococcus mutans. Front. Microbiol. 11:18. doi: 10.3389/fmicb.2020.00018
Jolkver, E., Emer, D., Ballan, S., Kramer, R., Eikmanns, B. J., and Marin, K. (2009). Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 191, 940–948. doi: 10.1128/JB.01155-08
Kim, H. M., Waters, A., Turner, M. E., Rice, K. C., and Ahn, S. J. (2019). Regulation of cid and lrg expression by CcpA in Streptococcus mutans. Microbiology 165, 113–123. doi: 10.1099/mic.0.000744
Kim, J. N., and Burne, R. A. (2017). CcpA and CodY Coordinate Acetate Metabolism in Streptococcus mutans. Appl. Environ. Microbiol. 83:e03274-16. doi: 10.1128/AEM.03274-16
Kleinberg, I. (2002). A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit. Rev. Oral Biol. Med. 13, 108–125. doi: 10.1177/154411130201300202
Koo, H., Falsetta, M. L., and Klein, M. I. (2013). The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J. Dent. Res. 92, 1065–1073. doi: 10.1177/0022034513504218
Kreth, J., Merritt, J., Shi, W., and Qi, F. (2005). Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187, 7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005
Kreth, J., Zhang, Y., and Herzberg, M. C. (2008). Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190, 4632–4640. doi: 10.1128/JB.00276-08
Lang, J. M., Darling, A. E., and Eisen, J. A. (2013). Phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices. PLoS One 8:e62510. doi: 10.1371/journal.pone.0062510
Lang, V. J., Leystra-Lantz, C., and Cook, R. A. (1987). Characterization of the specific pyruvate transport system in Escherichia coli K-12. J. Bacteriol. 169, 380–385. doi: 10.1128/jb.169.1.380-385.1987
LeBlanc, D. J., Lee, L. N., and Abu-Al-Jaibat, A. (1992). Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28, 130–145. doi: 10.1016/0147-619x(92)90044-b
Lemos, J. A., and Burne, R. A. (2008). A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154(Pt 11), 3247–3255. doi: 10.1099/mic.0.2008/023770-0
Long, L. H., and Halliwell, B. (2011). Artefacts in cell culture: alpha-Ketoglutarate can scavenge hydrogen peroxide generated by ascorbate and epigallocatechin gallate in cell culture media. Biochem. Biophys. Res. Commun. 406, 20–24. doi: 10.1016/j.bbrc.2011.01.091
Long, L. H., and Halliwell, B. (2012). The effects of oxaloacetate on hydrogen peroxide generation from ascorbate and epigallocatechin gallate in cell culture media: potential for altering cell metabolism. Biochem. Biophys. Res. Commun. 417, 446–450. doi: 10.1016/j.bbrc.2011.11.136
Marsh, P. D. (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149(Pt 2), 279–294. doi: 10.1099/mic.0.26082-0
Nath, K. A., Ngo, E. O., Hebbel, R. P., Croatt, A. J., Zhou, B., and Nutter, L. M. (1995). alpha-Ketoacids scavenge H2O2 in vitro and in vivo and reduce menadione-induced DNA injury and cytotoxicity. Am. J. Physiol. 268(1 Pt 1), C227–C236. doi: 10.1152/ajpcell.1995.268.1.C227
Oh, T. J., Kim, I. G., Park, S. Y., Kim, K. C., and Shim, H. W. (2002). NAD-dependent malate dehydrogenase protects against oxidative damage in Escherichia coli K-12 through the action of oxaloacetate. Environ. Toxicol. Pharmacol. 11, 9–14. doi: 10.1016/s1382-6689(01)00093-x
Paczia, N., Nilgen, A., Lehmann, T., Gatgens, J., Wiechert, W., and Noack, S. (2012). Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 11:122. doi: 10.1186/1475-2859-11-122
Paes Leme, A. F., Koo, H., Bellato, C. M., Bedi, G., and Cury, J. A. (2006). The role of sucrose in cariogenic dental biofilm formation–new insight. J. Dent. Res. 85, 878–887. doi: 10.1177/154405910608501002
Rice, K. C., Turner, M. E., Carney, O. V., Gu, T., and Ahn, S. J. (2017). Modification of the Streptococcus mutans transcriptome by LrgAB and environmental stressors. Microb. Genom. 3:e000104. doi: 10.1099/mgen.0.000104
Rosier, B. T., De Jager, M., Zaura, E., and Krom, B. P. (2014). Historical and contemporary hypotheses on the development of oral diseases: are we there yet? Front. Cell. Infect. Microbiol. 4:92. doi: 10.3389/fcimb.2014.00092
Saito, M., Seki, M., Iida, K., Nakayama, H., and Yoshida, S. (2007). A novel agar medium to detect hydrogen peroxide-producing bacteria based on the prussian blue-forming reaction. Microbiol. Immunol. 51, 889–892. doi: 10.1111/j.1348-0421.2007.tb03971.x
Satpute, R., Lomash, V., Hariharakrishnan, J., Rao, P., Singh, P., Gujar, N., et al. (2014). Oxidative stress and tissue pathology caused by subacute exposure to ammonium acetate in rats and their response to treatments with alpha-ketoglutarate and N-acetyl cysteine. Toxicol. Ind. Health 30, 12–24. doi: 10.1177/0748233712448117
Schwartz, E. R., and Reed, L. J. (1970). Regulation of the activity of the pyruvate dehydrogenase complex of Escherichia coli. Biochemistry 9, 1434–1439. doi: 10.1021/bi00808a019
Shen, L. C., and Atkinson, D. E. (1970). Regulation of pyruvate dehydrogenase from Escherichia coli. Interactions of adenylate energy charge and other regulatory parameters. J. Biol. Chem. 245, 5974–5978.
Sheng, J., and Marquis, R. E. (2007). Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 272, 196–201. doi: 10.1111/j.1574-6968.2007.00744.x
Shields, R. C., Zeng, L., Culp, D. J., and Burne, R. A. (2018). Genomewide identification of essential genes and fitness determinants of Streptococcus mutans UA159. mSphere 3:e00031-18. doi: 10.1128/mSphere.00031-18
Smith, E. G., and Spatafora, G. A. (2012). Gene regulation in S. mutans: complex control in a complex environment. J. Dent. Res. 91, 133–141. doi: 10.1177/0022034511415415
Sonenshein, A. L. (2005). CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8, 203–207. doi: 10.1016/j.mib.2005.01.001
Sonenshein, A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927. doi: 10.1038/nrmicro1772
Takahashi, N., and Nyvad, B. (2008). Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 42, 409–418. doi: 10.1159/000159604
Takahashi, N., Washio, J., and Mayanagi, G. (2010). Metabolomics of supragingival plaque and oral bacteria. J. Dent. Res. 89, 1383–1388. doi: 10.1177/0022034510377792
Tanzer, J. M., Thompson, A., Sharma, K., Vickerman, M. M., Haase, E. M., and Scannapieco, F. A. (2012). Streptococcus mutans out-competes Streptococcus gordonii in vivo. J. Dent. Res. 91, 513–519. doi: 10.1177/0022034512442894
Tenovuo, J. (1976). The variation of salivary peroxidase activities in persons of different oral health. Acta Odontol. Scand. 34, 163–168. doi: 10.3109/00016357609002564
Tenovuo, J., and Pruitt, K. M. (1984). Relationship of the human salivary peroxidase system to oral health. J. Oral Pathol. 13, 573–584. doi: 10.1111/j.1600-0714.1984.tb01459.x
Terleckyj, B., and Shockman, G. D. (1975). Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11, 656–664. doi: 10.1128/iai.11.4.656-664.1975
Terleckyj, B., Willett, N. P., and Shockman, G. D. (1975). Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11, 649–655. doi: 10.1128/iai.11.4.649-655.1975
Turner, M. E., Huynh, K., Carney, O. V., Gross, D., Carroll, R. K., Ahn, S. J., et al. (2019). Genomic instability of TnSMU2 contributes to Streptococcus mutans biofilm development and competence in a cidB mutant. Microbiologyopen 8:e934. doi: 10.1002/mbo3.934
Keywords: Streptococcus mutans, pyruvate, LrgAB, dual-species, caries, mouse model
Citation: Ahn S-J, Hull W, Desai S, Rice KC and Culp D (2020) Understanding LrgAB Regulation of Streptococcus mutans Metabolism. Front. Microbiol. 11:2119. doi: 10.3389/fmicb.2020.02119
Received: 29 June 2020; Accepted: 11 August 2020;
Published: 03 September 2020.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Jens Kreth, Oregon Health and Science University, United StatesMarlise Inez Klein, Paulista State University, Brazil
Mingyun Li, Sichuan University, China
Copyright © 2020 Ahn, Hull, Desai, Rice and Culp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Joon Ahn, c2FobkBkZW50YWwudWZsLmVkdQ==
 Sang-Joon Ahn
Sang-Joon Ahn William Hull
William Hull Shailja Desai
Shailja Desai Kelly C. Rice
Kelly C. Rice David Culp
David Culp