- 1Biology Department, Faculty of Science, Taibah University, Medina, Saudi Arabia
- 2Bacteriology Section, Botany and Microbiology Department, Faculty of Science, Assiut University, Assiut, Egypt
Initial colonization of human gut by bacteria is an important step in controlling its microbiota and health status. This study followed the initial colonization by lactic acid bacteria (LAB) in colon of new born babies through following its occurrence in their stool at first week of their life. The LAB occurrence in the neonates’ stool was followed on MRS agar medium. The isolated LAB from male and female newborn babies of normal birth and cesarean section surgical delivery were molecular biologically identified by phylogenetic analysis of 16S rRNA gene sequence. From the 24 investigated newborn babies, three LAB taxa, Lactobacillaceae, Enterococcus, and Streptococcus, were detected in their stool at first week of their life. Lactobacillaceae represented 20.8% of total colonized LAB in newborn babies in the culture-dependent approach used in this study and included three species namely Limosilactobacillus reuteri (previously known as Lactobacillus reuteri), Lacticaseibacillus rhamnosus (previously known as Lactobacillus rhamnosus) and Ligilactobacillus agilis (previously known as Lactobacillus agilis). Enterococcus faecalis and E. faecium were detected where E. faecalis was the highest dominant, representing 62.5% of total LAB colonizing newborn babies. This result suggests that this bacterium has high potency for colonization and might be important for controlling the initial settlement of microbiota in healthy newborn babies. Only one species of Streptococcus namely Streptococcus agalactiae was detected in 8.33% total of the investigated newborn babies indicating high competency by other LAB for colonization and that this bacteria, in spite of its pathogenicity, is commensal in its low existence in healthy babies. The explored potency of natural initial colonization of the LAB species E. faecalis, E. faecium, L. reuteri, L. rhamnosus, and L. agilis of which many health beneficial strains were previously reported, would be important for future applications. Despite the controversy in evaluating its health benefits, E. faecalis as a potent competitor to other LAB refers to its importance in initial colonization of healthy babies colon at first week of their life. Further future studies, with more number of samples and characterization, would be of importance for evaluating the potential use of beneficial Enterococcus strains which could improve intestinal ecosystem.
Highlights
- Initial colonization of healthy 24 newborn babies by various lactic acid bacteria was followed.
- Three genus of lactic acid bacteria Lactobacillus, Enterococcus, and Streptococcus were detected.
- The species Lactobacillus reuteri, Lactobacillus rhamnosus, and Lactobacillus agilis were found.
- All Lactobacillus species represented as a whole 20.8% of total colonized lactic acid bacteria in newborn babies.
- Enterococcus faecalis represented 62.5% of total lactic acid bacteria colonized newborn babies.
Introduction
The initial bacterial colonization of human gut after birth is of importance for the control of microbiota settlement and for many health benefits. Microbiota colonizing the gut helps in driving post-natal maturation of the developing infant gut and plays also a role in of the mucosal immune system development (Gronlund et al., 2000; Hooper and Gordon, 2001; Hooper et al., 2002; Macpherson and Harris, 2004; Sjögren et al., 2009; Livingston et al., 2010; Donovan et al., 2012; Brugman et al., 2015; Timmerman et al., 2017). Some strains of LAB are able to promote intestinal health of their hosts (Lionetti et al., 2006; Ma et al., 2009; Jones et al., 2012; Kechagia et al., 2013; Mackos et al., 2016; Linares et al., 2017). Some strains of Limosilactobacillus reuteri (previously known as Lactobacillus reuteri; Zheng et al., 2020) is known for its beneficial roles in health (Mackos et al., 2013; Keller et al., 2014; Hou et al., 2015; Lee et al., 2016; Hsu et al., 2017; Mu et al., 2018), colonizing the human and animal intestine and producing the antimicrobial compound reuterin (Talarico and Dobrogosz, 1989; Cadieux et al., 2008; Morita et al., 2008; Spinler et al., 2008; Schaefer et al., 2010; Mishra et al., 2012; Mu et al., 2018; Asare et al., 2020; Senatore et al., 2020) which can inhibit the growth of several Gram-positive and Gram-negative bacteria (Cleusix et al., 2007). It has also been shown that some strains of Lacticaseibacillus rhamnosus (previously known as Lactobacillus rhamnosus; Zheng et al., 2020) have beneficial roles in health promotion (Martinez et al., 2009; Allonsius et al., 2017; George-Kerry et al., 2018; Westerik et al., 2018a,b).
Application of molecular identification tests along with metagenomic analysis for profiling the human colon microbiota (Qin et al., 2010; Maccaferri et al., 2011; Wang et al., 2015; Almeida et al., 2019; Franco-Duarte et al., 2019) would be of importance for diagnosing the health development of newborn babies. This would help in better understanding the colonization and various roles of colon beneficial bacteria for human health. The cost of molecular identification tests of microbes is continuously ongoing to be the cheapest. Along with its accuracy and fastness (Franco-Duarte et al., 2019), molecular identification tests would be the best choice not only for research purposes but for applicable gut health status diagnosis in near future. Thus, profiling the normal colon flora including the health beneficial bacterial species though molecular identification tests along with metagenomic analysis is a hot topic that would enrich the data base for following up the health development in newborn babies.
The health beneficial roles and use of Enterococcus strains is a controversial topic. The health benefits of specific strains of Enterococcus faecalis and Enterococcus faecium were characterized (Hlivak et al., 2005; Franz et al., 2011; Cebrián et al., 2012; Hanchi et al., 2018; Zommiti et al., 2018). Some strains of E. faecalis and E. faecium and some others strains belonging to the species Enterococcus durans, Enterococcus hirae, Enterococcus lactis, and Enterococcus munditii had the potential to promote health to their hosts (Nami et al., 2014; Pieniz et al., 2014; Nami et al., 2015; Gupta and Tiwari, 2015; Haghshenas et al., 2016; van Zyl et al., 2016). However, extensive studies for safety regulation is required upon application. Following the occurrence of these species of Enterococcus and other LAB in the human gut in newborn healthy babies, would explore its potency for colonization and possible roles at the early stage of life and initial establishment of gut microbiome. In this study, the occurrence of cultivable LAB in newborn infants’ stool was followed at the first week of their life for exploring its potency for initial colonization of healthy newborn babies colon.
Materials and Methods
Colonization of LAB at first week of life of newborn babies was followed by following the occurrence of theses bacteria in newborn infants’ stool in Almadinah Almunawarah, Kingdom of Saudi Arabia (KSA).
Sampling and LAB Isolation
LAB occurrence in stool of healthy newborn babies in the first week of their life (age 2–6 days old) was investigated. The stool of 24 newborn babies were sampled, 12 from who had normal delivery and 12 from who had cesarean birth. In each of these groups, six newborn babies were girls and six were boys. The healthy newborn babies were selected randomly. LAB occurrence in stool of healthy newborn babies was investigated using de Man, Rogosa, and Sharpe agar (MRS agar, HiMedia Laboratories Limited, Mumbai, India)]. Serial dilution of stool samples was conducted and subsequently LAB were detected on the MRS medium after 48 h of inoculation and incubation in CO2 incubator (5% CO2) at 35°C and colony forming units (CFU) was followed by most probable number (MPN) techniques (MacFaddin, 1985). The MRS plates with low detectable number of colonies were used for determining the CFU of dominating colonies with similar morphological and bacterial biochemical characteristics of which an isolated strain was subcultured in MRS agar to obtain a purified strain.
Specie-Level Identification of LAB Strains
The specie-level identification of LAB strains was conducted by phylogenetic analysis of 16S rRNA encoding gene sequence. The bacterial strains were cultured in MRS broth medium for 48 h for genomic DNA extraction.
DNA Isolation and 16S rRNA Encoding Gene Amplification
Bacterial cells from 1 mL culture of each LAB strain were collected by centrifugation and used for genomic DNA extraction. The DNA extraction was conducted using the Promega Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, Wisconsin, United States) following the kit manufacturer instructions. The extracted genomic DNA of each LAB strain was subsequently used as a template for PCR amplification of 16S rRNA encoding gene.
The PCR amplification of 16S rRNA encoding gene was conducted using the universal 27F forward and 1492R reverse primers where the sequence of the 27F forward primer was (5′-AGAGTTTGATC[A/C]TGGCTCAG-3′) and the sequence of the 1492R reverse primer was (5′-G[C/T]TACCTTGTTACGACTT-3′) (Lane, 1991). A near-full length amplification by PCR of the 16S rRNA encoding gene was performed with a reaction mixture (25 μl) composed by 10× Taq buffer (100 mM Tris-HCl, pH 8) 2.5 μl; MgCl2 (1.25 mM); dNTPs (100 μM) (Invitrogen, Carlsbad, CA, United States); forward and reverse primer (1.2 μM); Taq DNA polymerase (Invitrogen, United States) (0.5 U) and the bacterial genomic DNA as a template of about 5 ng. PCR was performed using a Thermal Cycler (Model 2720; Applied Biosystems, Foster City, California, United States). PCR program was implemented as follows: initial denaturation step at 95°C for 5 min followed by 35 amplification cycles of [94°C for 1 min (denaturation); 56°C for 1 min (annealing); 72°C for 1 min (extension)] and a subsequent final extension step at 72°C for 10 min. Analyzing the PCR amplification products was conducted using agarose electrophoresis on agarose (1%) gels containing 5 μg/mL ethidium bromide with using a DNA size marker [(Invitrogen, United States) 1 kb Plus DNA ladder].
Sequencing and Sequence Analysis
The PCR products were purified and subsequently cycle sequenced at Macrogen Korea sequencing facility (Seoul, Korea). Direct cycle sequencing using same forward and reverse primers in both directions was performed for sequencing the PCR purified product using automated florescent dye terminator sequencing method (Sanger et al., 1977) with DNA Analyzer 3730XL (Applied Biosystems, CA, United States) at Macrogen Korea sequencing facility (Seoul, Korea). The sequence reads of the 16S rRNA gene sequence with forward and the reverse primers were assembled and compared with closest matches of DNA sequences found in GenBank using the search tools of nucleotide-nucleotide BLAST at www.ncbi.nlm.nih.gov/blast/Blast.cgi in the NCBI (National Center for Biotechnology and Information) website. The alignments of the 16S rRNA gene sequences were done according to Thompson et al. (1997) by Clustal W1.83 XP. Using neighbor-joining method (Saitou and Nei, 1987), the phylogenetic trees of 16S rRNA encoding gene sequences were constructed with MEGAX software (Kumar et al., 2018). An outgroup [Bacillus subtilis strain JCM 1465; accession number (NR_113265)] was used.
Nucleotide Sequences Accession Numbers
The 16S rRNA encoding gene partial nucleotide sequence of each LAB strain was deposited under an accession number for each in the GenBank nucleotide sequence database. The accession numbers of the deposited 16S gene sequence of all LAB strains are outlined in Table 1.
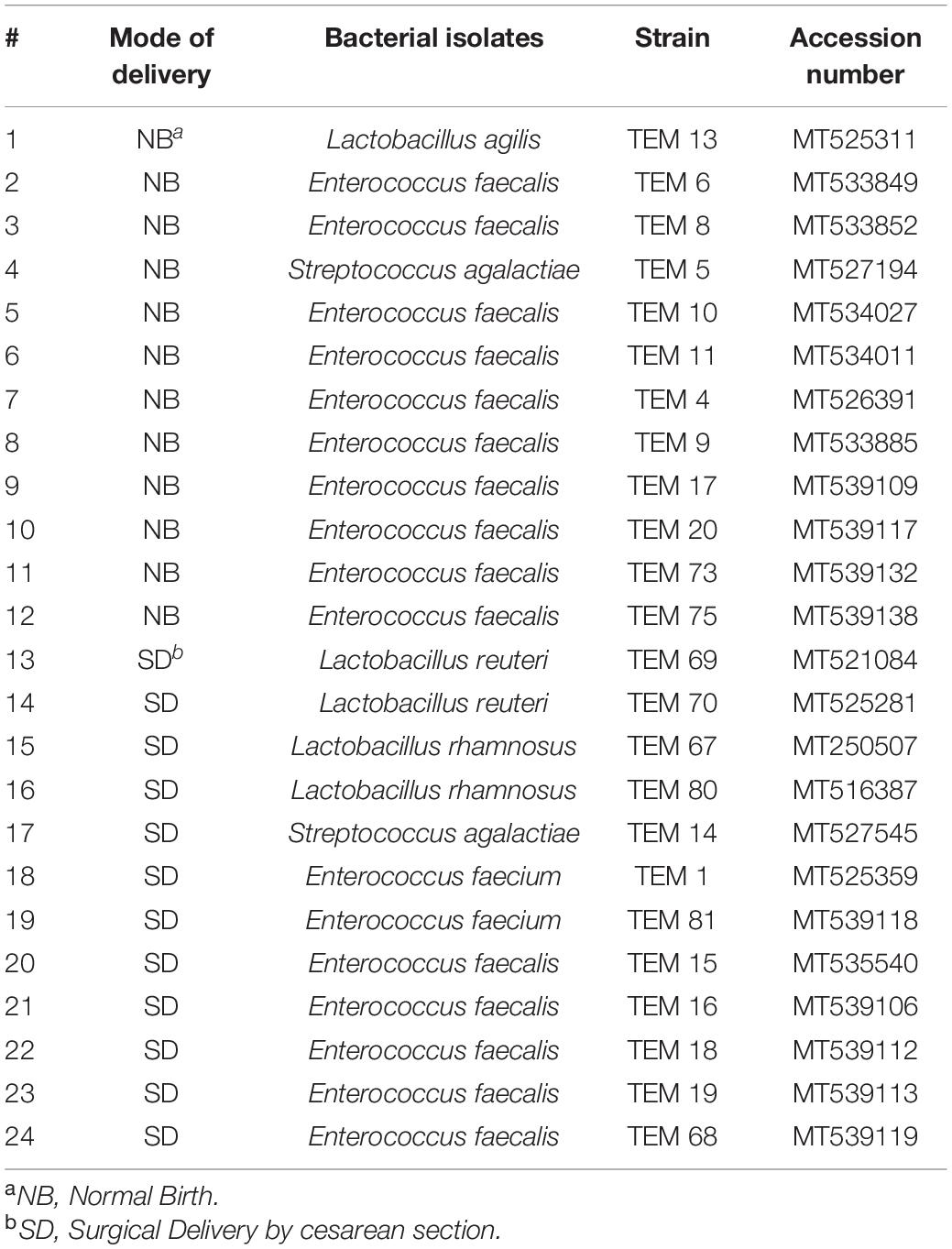
Table 1. Lactic acid bacterial strains detected in stool of newborn babies from various delivery modes (normal birth and surgical delivery by cesarean section) at first week of life (age 2–6 days old).
Results
In this study, the occurrence of LAB in newborn infants’ stool was followed to explore the colonization of various LAB in colon of newborn babies at first week of their life. All of the bacterial strains isolated from the stool of the 24 newborn babies were identified by phylogenetic analysis of 16S gene sequence. From all of the newborn babies subjects investigated for colonization by LAB, three LAB genus namely Enterococcus, Lactobacillus, and Streptococcus were detected (Table 1) in stool of newborn subjects at this stage of initial colonization in this start of babies life. The isolated LAB species was dominant in each case of the 24 healthy newborn babies which is probably because the colonization is initial at the first week of the babies life.
From the 24 babies, two subjects retained L. rhamnosus which represent 8.33% of the total LAB occurrence in all of the 24 investigated newborn subjects. The 16S rRNA encoding gene of the isolated L. rhamnosus strain TEM 67 and strain TEM 80 was sequenced and submitted to gene bank for accession number (Table 1). Phylogenetic analysis of the 16S rRNA encoding gene sequence was conducted for identification of L. rhamnosus (Figure 1). Two other species of Lactobacillus were detected namely L. reuteri and L. agilis. Limosilactobacillus reuteri was detected in stool samples of two newborn subjects representing a similar percent of occurrence to L. rhamnosus of the total LAB in all investigated newborn subjects. Phylogenetic analysis of L. reuteri of the 16S rRNA encoding gene sequence was conducted (Figure 2). Ligilactobacillus agilis was the most rare of all LAB detected out the 24 investigated newborn subjects where it represented 4.16% of the total LAB occurrence. This bacterial strain was found in stool a newborn baby of normal birth vaginal delivery, suggesting that vaginal delivery normal birth process might be the source of the inoculation by this bacterium to newborn baby through vaginal birth canal. The molecular biological identification by phylogenetic analysis for the isolated L. agilis strain TEM 13 is shown (Figure 3). The colonization of newborn babies colon by the three species of Lactobacillaceae (L. rhamnosus, L. reuteri, and L. agilis) at this early stage of life indicate its importance for the healthy babies where several strains of theses species was previously reported as health beneficial ones. The two bacterial species with controversial hypothesized health benefits of Enterococcus namely E. faecalis and E. faecium were detected where E. faecalis was the most abundant bacterium detected in babies subjects. Phylogenetic analysis tree of 16S rRNA gene sequence for molecular biological identification of E. faecalis and E. faecium is shown in Figures 4, 5, respectively. The occurrence and molecular characterization of LAB in newborn infants’ stool in Al-Madinah, KSA reveals higher potency of the facultative anaerobes E. faecalis for earlier colonization of healthy babies colon than other detected LAB. Enterococcus appeared in newborn infants’ stool whatever the mode of delivery or gender (Table 1) indicating a high potency of this facultative anaerobic bacterium to colonize babies gut at this early stage of life. The results in this study on healthy newborn babies might supports the hypothesis that E. faecalis, of which some health beneficial strains were reported, can be used with potential initial colonization as this specie naturally and potentially colonizes healthy babies colon as appeared in their stool at the first week of their life with no disorder syndrome suggesting its compatibility for a healthy conditions in human gut and for the colon microbiome initial settlement at the start of human life. Streptococcus genus existence was low compared to total LAB and only one species namely S. agalactiae was found (Table 1) and identified by 16S phylogenetic analysis (Figure 6). The low occurrence of S. agalactiae in stool of healthy newborn babies subjects (Figure 7) compared to total LAB indicates high competition by other LAB Enterococcus and Lactobacillus against colonization by this S. agalactiae pathogenic bacterium. In spite of its rare existence, the detection of the pathogenic bacterium S. agalactiae in healthy newborn babies might also indicate its commensal status.
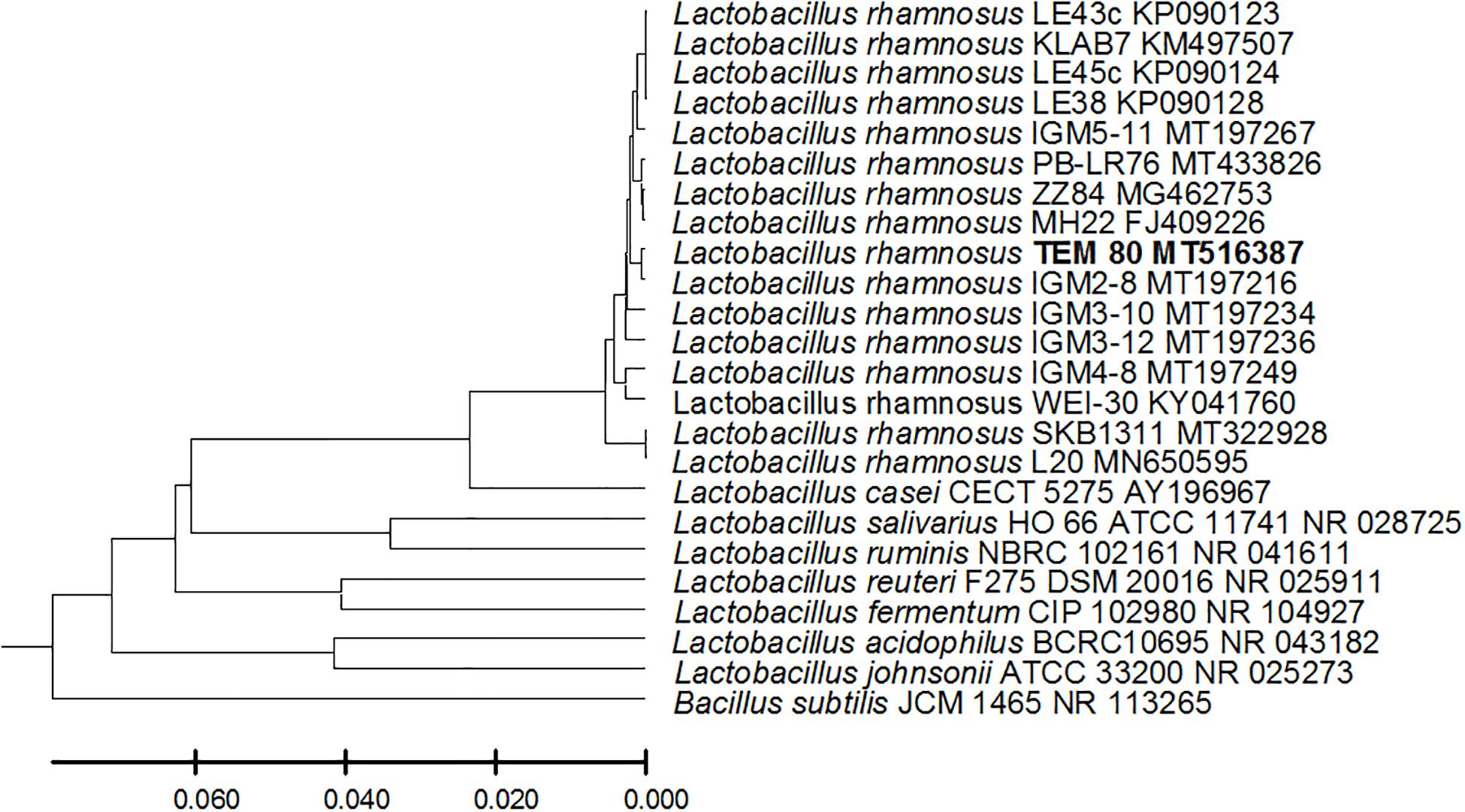
Figure 1. Lactobacillus rhamnosus strain TEM 80 16S rRNA encoding gene phylogenetic analysis. Phylogenetic tree of Lactobacillus rhamnosus strain TEM 80 isolated from newborn infants’ stool showing relationship with its closest bacterial neighbors strains from NCBI. The Lactobacillus rhamnosus strain TEM 80 is shown in bold. The neighbor-joining tree of isolated Lactobacillus rhamnosus strain TEM 80 and other bacterial strains was determined using 16S rRNA encoding gene sequences and the frequency filter in MEGA-X analysis package software. An out group Bacillus subtilis strain JCM 1465 (accession number; NR_113265) was used. A scale bar segment indicates 2% estimated difference in sequence. Each strain accession number in the NCBI database is shown.
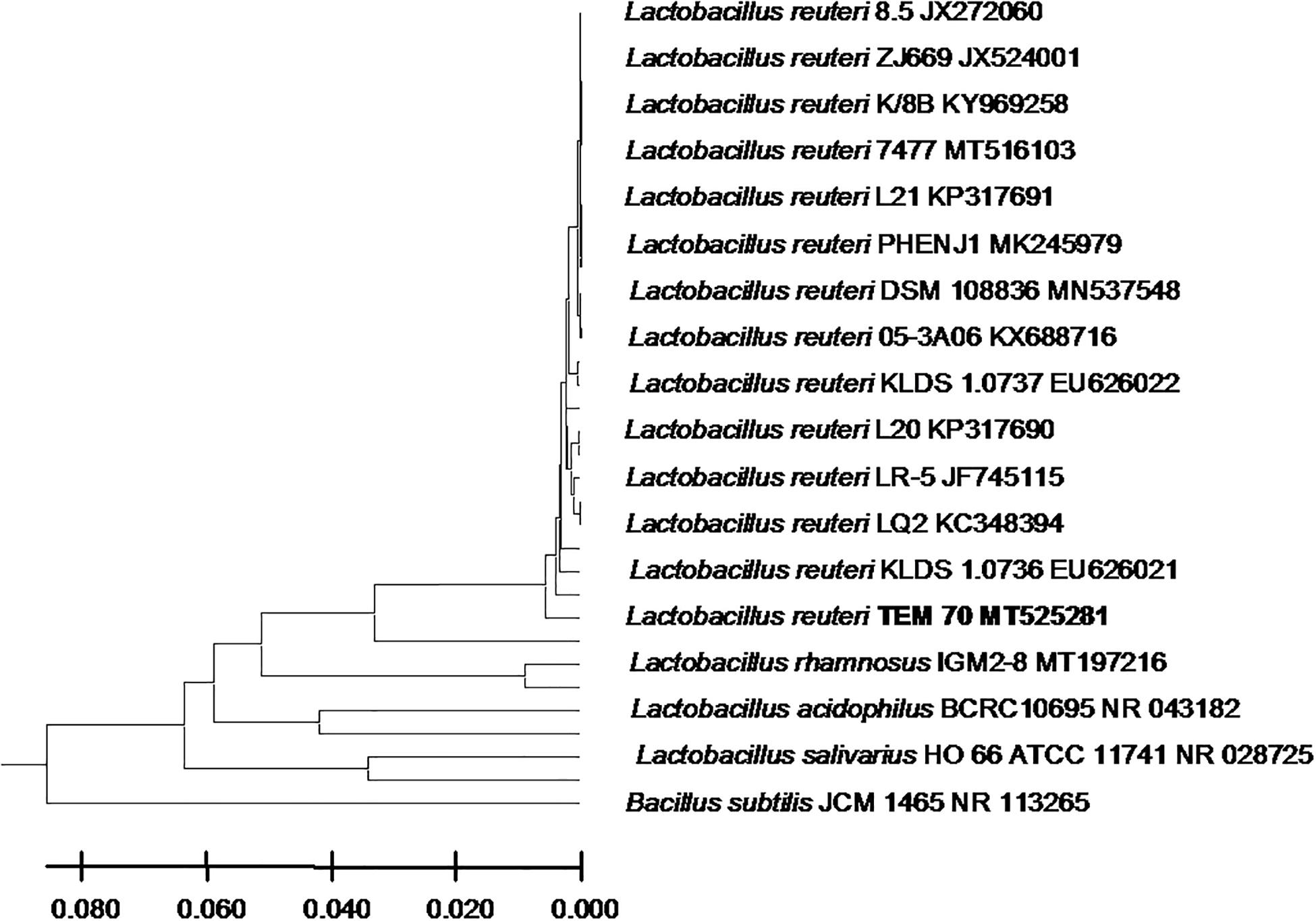
Figure 2. Lactobacillus reuteri strain TEM 70 16S rRNA encoding gene phylogenetic analysis. Phylogenetic tree of Lactobacillus reuteri strain TEM 70 isolated from newborn infants’ stool showing relationship with its closest bacterial neighbors strains from NCBI. The Lactobacillus reuteri strain TEM 70 is shown in bold. The neighbor-joining tree of isolated Lactobacillus reuteri strain TEM 70 and other bacterial strains was determined using 16S rRNA encoding gene sequences and the frequency filter in MEGA-X analysis package software. An out group Bacillus subtilis strain JCM 1465 (accession number; NR_113265) was used. A scale bar segment indicates 2% estimated difference in sequence. Each strain accession number in the NCBI database is shown.
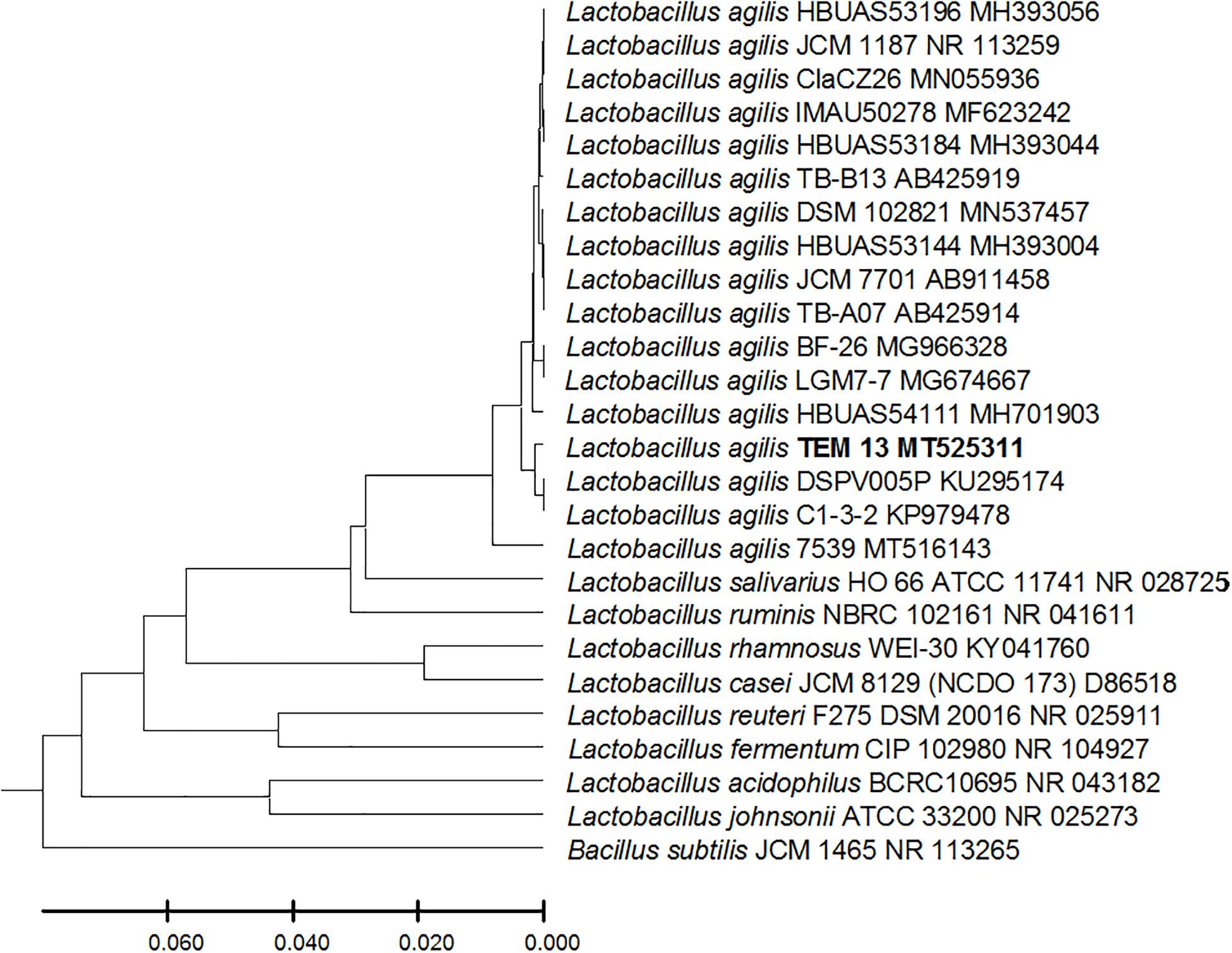
Figure 3. Lactobacillus agilis strain TEM 13 16S rRNA encoding gene phylogenetic analysis. Phylogenetic tree of Lactobacillus agilis strain TEM 13 isolated from newborn infants’ stool showing relationship with its closest bacterial neighbors strains from NCBI. The Lactobacillus agilis strain TEM 13 is shown in bold. The neighbor-joining tree of isolated Lactobacillus agilis strain TEM 13 and other bacterial strains was determined using 16S rRNA encoding gene sequences and the frequency filter in MEGA-X analysis package software. An out group Bacillus subtilis strain JCM 1465 (accession number; NR_113265) was used. A scale bar segment indicates 2% estimated difference in sequence. Each strain accession number in the NCBI database is shown.
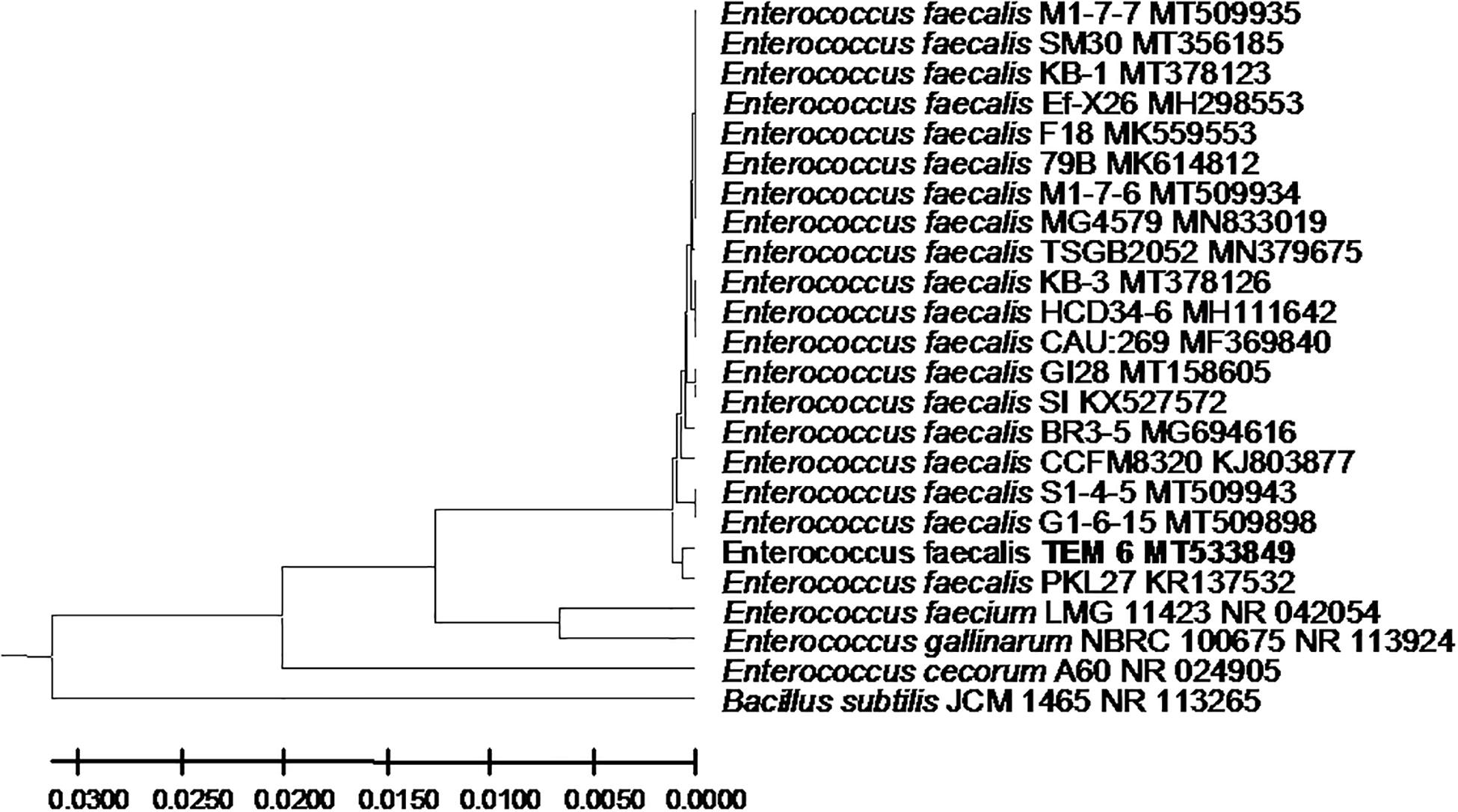
Figure 4. Enterococcus faecalis strain TEM 6 16S rRNA encoding gene phylogenetic analysis. Phylogenetic tree of Enterococcus faecalis strain TEM 6 isolated from newborn infants’ stool showing relationship with its closest bacterial neighbors strains from NCBI. The Enterococcus faecalis strain TEM 6 is shown in bold. The neighbor-joining tree of isolated Enterococcus faecalis strain TEM 6 and other bacterial strains was determined using 16S rRNA encoding gene sequences and the frequency filter in MEGA-X analysis package software. An out group Bacillus subtilis strain JCM 1465 (accession number; NR_113265) was used. A scale bar segment indicates 2% estimated difference in sequence. Each strain accession number in the NCBI database is shown.
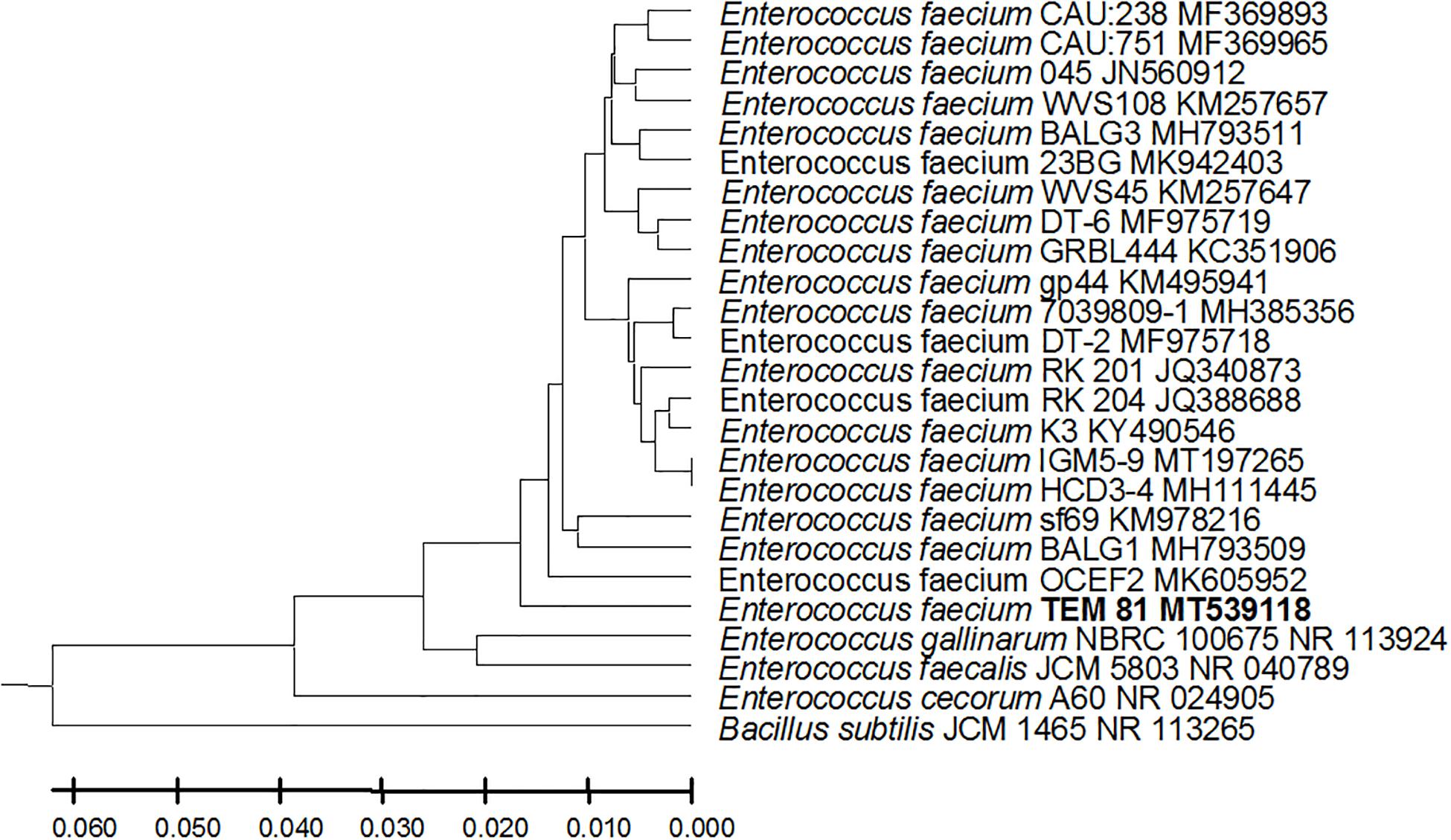
Figure 5. Enterococcus faecium strain TEM 81 16S rRNA encoding gene phylogenetic analysis. Phylogenetic tree of Enterococcus faecium strain TEM 81 isolated from newborn infants’ stool showing relationship with its closest bacterial neighbors strains from NCBI. The Enterococcus faecium strain TEM 81 is shown in bold. The neighbor-joining tree of isolated Enterococcus faecium strain TEM 81 and other bacterial strains was determined using 16S rRNA encoding gene sequences and the frequency filter in MEGA-X analysis package software. An out group Bacillus subtilis strain JCM 1465 (accession number; NR_113265) was used. A scale bar segment indicates 2% estimated difference in sequence. Each strain accession number in the NCBI database is shown.

Figure 6. Streptococcus agalactiae strains TEM 5 16S rRNA encoding gene phylogenetic analysis. Phylogenetic tree of Streptococcus agalactiae strains TEM 5 isolated from newborn infants’ stool showing relationship with its closest bacterial neighbors strains from NCBI. The Streptococcus agalactiae strains TEM 5 is shown in bold. The neighbor-joining tree of isolated Streptococcus agalactiae strains TEM 5 and other bacterial strains was determined using 16S rRNA encoding gene sequences and the frequency filter in MEGA-X analysis package software. An out group Bacillus subtilis strain JCM 1465 (accession number; NR_113265) was used. A scale bar segment indicates 2% estimated difference in sequence. Each strain accession number in the NCBI database is shown.
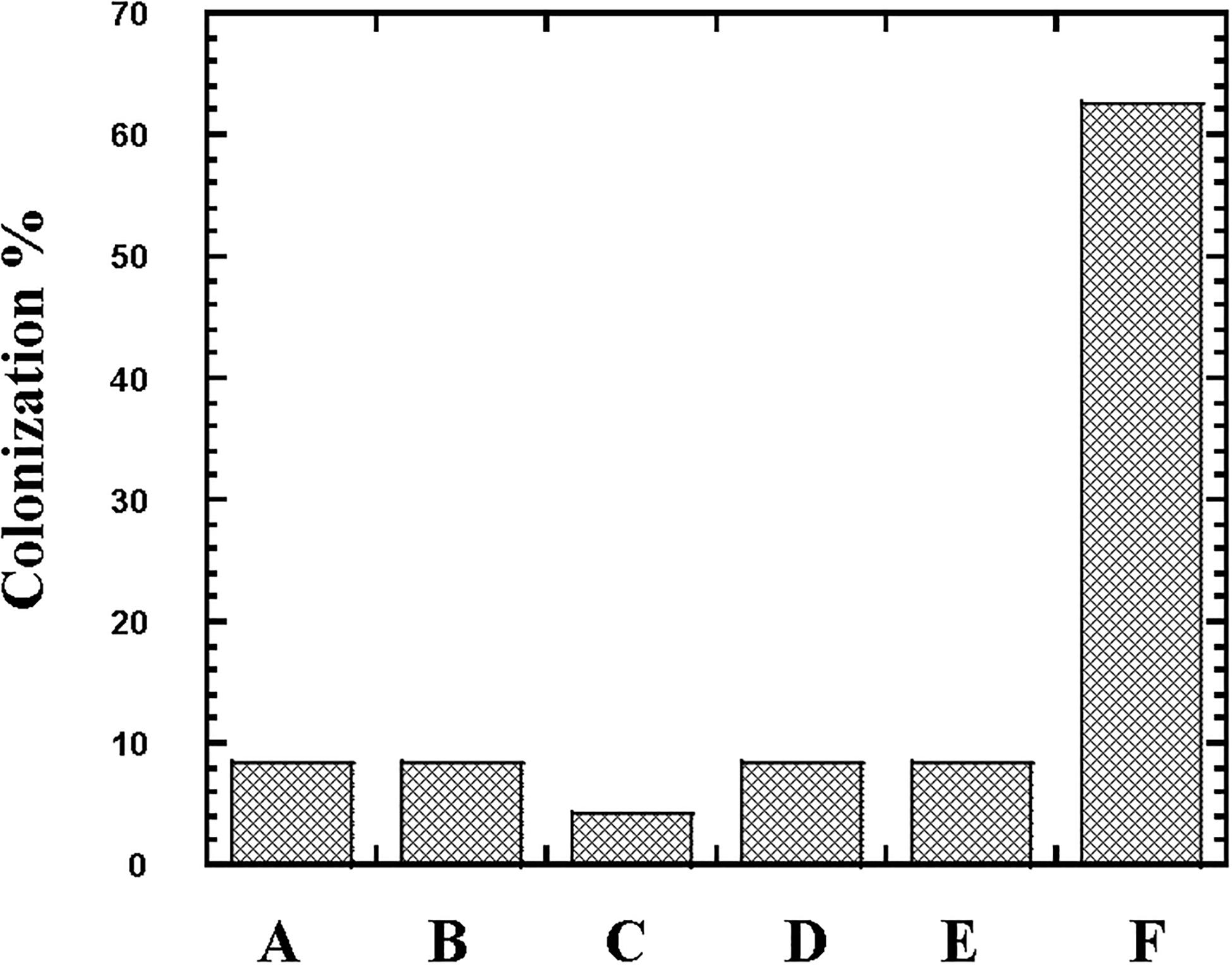
Figure 7. Colonization (%) of the lactic acid bacteria Lactobacillus, Streptococcus and Enterococcus in newborn babies subjects. The occurrence of Lactobacillus reuteri (Column A), Lactobacillus rhamnosus (Column B), Lactobacillus agilis (Column C), Streptococcus agalactiae (Column D), Enterococcus faecium (Column E), and Enterococcus faecalis (Column F) in stool of 24 newborn babies subjects was followed at first week of their life.
Discussion
The health beneficial properties of some strains of the LAB E. faecalis, L. reuteri, L. rhamnosus, L. agilis, and others were previously reported. This study was devoted for investigating the initial colonization of healthy newborn babies colon at the first week of their life by various LAB through following its occurrence in the neonates’ stool. The study targeted to follow up the potency of colonization by these LAB species of which many strains health beneficial properties were previously reported where investigating the initial colonization potency of these LAB species explored in this study is important to know for assessment of the usage efficiency of these LAB species in applications.
In this study, the high tendency for colonization of newborn gut by E. faecalis at this early stage of life and initial formation of the gut microbiome refers to its efficiency in colonization. In each case of the 24 healthy newborn babies, the isolated LAB species was dominant. This is probably because the colonization is initial and hence E. faecalis, representing 62.5% of total LAB colonizing healthy newborn babies, is shown as a better competitor than other LAB in the initial colonization. Because of their tolerance to wide range of temperature and pH, many Enterococcus species are highly competitive (Hanchi et al., 2018). The only Enterococci yet suggested to be used for health benefits are some specific strains of E. faecalis and E. faecium (Franz et al., 2011; Cebrián et al., 2012). Such high competitiveness E. faecalis for colonization can be also attributed to their ability to produce bacteriocins (Cebrián et al., 2012; Hanchi et al., 2018) that would support the potency of colonization of Enterococcus against other bacteria. This ability to effectively colonize the gut of newborn babies by E. faecalis at this early stage of life might be related to other factors including its facultative anaerobiosis metabolism where a successful colonization by Lactobacillaceae and other microaerobes or anaerobes in the human gut would require installation of microaerobic and anaerobic conditions in the lumen of the colon. Such installation of microaerobic conditions might require time and varies from a newborn subject to the other at this initial stage of colonization. The facultative anaerobes including Enterococcus and the widely spread Escherichia coli would play important role in consuming molecular oxygen for installing microaerobic and anaerobic conditions suitable for vitality and possible later enriched colonization by Lactobacilli and other microaerobic and anaerobic bacteria where aerobic and facultative anaerobic bacterial consumption of oxygen along with other mechanisms of O2 consumption was suggested to maintain the lumen of the gut in a deeply anaerobic state that is important for obligate anaerobes (Friedman et al., 2018). Colonization by L. rhamnosus, L. reuteri, and L. agilis detected in only around 20% of total of the newborn babies subjects indicates the possibility of variation in ability to install the microaerobic and anaerobic conditions suitable for viability of these bacteria and its potential of colonization. The high potency for colonization of E. faecalis in healthy newborn babies might prevent colonization by virulent pathogenic bacteria possibly through its ability to produce antimicrobial agents bacteriocins (Franz et al., 2011; Hanchi et al., 2018). In addition to L. reuteri which is known to produce the antimicrobial reuterin (Talarico et al., 1988; Spinler et al., 2008; Jones and Versalovic, 2009; Greifova et al., 2017; Mu et al., 2018; Asare et al., 2020; Senatore et al., 2020) these bacteria in healthy newborn babies might act as defenders against colonization by virulent pathogenic bacteria in this early stage of life. While the antimicrobial reuterin produced by L. reuteri can inhibit other microorganisms (Cleusix et al., 2007) that can help in controlling the initial microbial colonization in newborn babies to reduce the possible colonization by pathogenic bacteria. Many other benefits of L. reuteri includes reduction of infections by harmful bacteria, decreasing bacterial translocation, promoting health, increasing the absorption of minerals, vitamins and nutrients, modulating the immune responses of the host, and promoting the integrity of gut mucosa (Wang et al., 1995; Adawi et al., 1997; Nikawa et al., 2004; Niv et al., 2005; Saggioro et al., 2005; Tubelius et al., 2005; Imase et al., 2007; McFall-Ngai, 2007; Francavilla et al., 2008; Indrio et al., 2008; Spinler et al., 2008; Hou et al., 2015; Mu et al., 2018). Controlling the initial microbiotal colonization in newborn babies can also be aided by L. rhamnosus which appeared in stool of newborn babies in similar detection percent to L. reuteri and is also well-known for its healthy effects (Szajewska et al., 2015; Urbańska et al., 2016; Allonsius et al., 2017; Westerik et al., 2018a,b). The colonization with S. agalactiae (Group B Streptococcus) was low in stool of healthy newborn babies subjects. Streptococcus agalactiae is commensal bacterium in the human intestinal and genitourinary tracts (Kaambo et al., 2018) where in healthy adults, this Group B Streptococcus rarely cause infections; however, it may occasionally cause morbidity in pregnant women (Sørensen et al., 2010; Brigtsen et al., 2015; Kaambo et al., 2018) and is a major risk factor in newborns for Group B Streptococcus early onset invasive disease (GBS EOD) (Kwatra et al., 2014; Brigtsen et al., 2015; Kaambo et al., 2018). The rare colonization by this bacterium indicates that other dominant LAB might hinders its colonization. Such rare existence of this bacterium in healthy newborn babies might also indicate that this commensal bacterium is not harmful except in case of overgrowth which would be hindered by other LAB such as L. reuteri and E. faecalis producing antimicrobial agents. The antagonistic effect of Lactobacilli against S. agalactiae is well-documented (Ho et al., 2016; Sharpe et al., 2019). Streptococcus agalactiae is well-known as a commensal member of the vaginal microflora (El Aila et al., 2009; Meyn et al., 2009; Sangeetha et al., 2015; Kaambo et al., 2018; Shabayek and Spellerberg, 2018) suggesting that vaginal canal is the source of inoculation in newborn babies of vaginal delivery normal birth. However, the bacterium colonized not only newborn babies of vaginal delivery normal birth but also babies of cesarean section surgical delivery, indicating that inoculation might also occur from surrounding environments or during feeding process. The third species of Lactobacilli appeared in stool of newborn baby of normal birth vaginal delivery was L. agilis. This Lactobacilli was previously isolated from human vagina (Ocana et al., 1999) suggesting that inoculation of newborn babies by this bacterium might occur during normal delivery through the vagina. Some strains of L. agilis was suggested as health beneficial strains that can be applied as a feed additive for poultry (Lan et al., 2003; Ren et al., 2019) and was considered as one of the vaginal Lactobacilli potential antagonistic against Candida spp. (Gil et al., 2010). Thus, this bacterium can also act for minimizing the colonization of newborn babies colon by pathogenic bacteria through competing for the colonization space in addition to other antimicrobial agents producing L. reuteri, E. faecalis, and E. faecium.
In near future, the use of molecular identification tests with the broadening gene bank data base would be of importance for applicable diagnosis of bacterial flora in newborn babies for following up their health development status. In addition, the metagenomic analysis for profiling the whole microbiota in colon would be very helpful for achieving this applicable future diagnosis target of health development assessments including the initial colonization of the colon by beneficial bacterial species.
In conclusion, the results in this study showed that E. faecalis was most dominant representing more than 60% of the total LAB in stool of healthy newborn babies and is hence a better competitor to other LAB including L. reuteri and L. rhamnosus in the initial colonization of healthy babies colon at first week of their life. These results in healthy babies suggest possible beneficial role of this bacterium in addition to the detected Lactobacilli for minimizing colonization by pathogenic LAB such as S. agalactiae which showed minimum initial colonization of newborn babies at the first week of their life.
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board (IRB) in Medina [Head of IRB Committee (Abduhameed Alsubhi)]. Informed consent was obtained from parents for the collection of infant stools.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adawi, D., Kasravi, F. B., Molin, G., and Jeppsson, B. (1997). Effect of Lactobacillus supplementation with and without arginine on liver damage and bacterial translocation in an acute liver injury model in the rat. Hepatology 25, 642–647. doi: 10.1002/hep.510250325
Allonsius, C. N., van den Broek, M., De Boeck, I., Kiekens, S., Oerlemans, E., Kiekens, F., et al. (2017). Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb. Biotechnol. 1, 1753–1763. doi: 10.1111/1751-7915.12799
Almeida, A., Mitchell, A. L., Boland, M., Forster, S., Gloor, G. B., Tarkowska, A., et al. (2019). A new genomic blueprint of the human gut microbiota. Nature 568, 499–504. doi: 10.1038/s41586-019-0965-1
Asare, P. T., Zurfluh, K., Greppi, A., Lynch, D., Schwab, C., Stephan, R., et al. (2020). Reuterin demonstrates potent antimicrobial activity against a broad panel of human and poultry meat Campylobacter spp. isolates. Microorganisms 8:78. doi: 10.3390/microorganisms8010078
Brigtsen, A. K., Dedi, L., Melby, K. K., Holberg-Petersen, M., Radtke, A., Lyng, R. V., et al. (2015). Comparison of PCR and serotyping of group B Streptococcus in pregnant women: the Oslo GBS-study. J. Microbiol. Methods 108, 31–35. doi: 10.1016/j.mimet.2014.11.001
Brugman, S., Perdijk, O., van Neerven, R. J., and Savelkoul, H. F. (2015). Mucosal immune development in early life: setting the stage. Arch. Immunol. Ther. Exp. 63, 251–268. doi: 10.1007/s00005-015-0329-y
Cadieux, P., Wind, A., Sommer, P., Schaefer, L., Crowley, K., Britton, R. A., et al. (2008). Evaluation of reuterin production in urogenital probiotic Lactobacillus reuteri RC-14. Appl. Environ. Microbiol. 74, 4645–4649. doi: 10.1128/aem.00139-08
Cebrián, R., Baños, A., Valdivia, E., Pérez-Pulido, R., Martínez-Bueno, M., and Maqueda, M. (2012). Characterization of functional, safety, and probiotic properties of Enterococcus faecalis UGRA10, a new AS-48-producer strain. Food Microbiol. 30, 59–67. doi: 10.1016/j.fm.2011.12.002
Cleusix, V., Lacroix, C., Vollenweider, S., Duboux, M., and Le Blay, G. (2007). Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol. 7:101.
Donovan, S. M., Wang, M., Li, M., Friedberg, I., Schwartz, S. L., and Chapkin, R. S. (2012). Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv. Nutr. 3, 450S–455S.
El Aila, N. A., Tency, I., Claeys, G., Saerens, B., De Backer, E., Temmerman, M., et al. (2009). Genotyping of Streptococcus agalactiae (group B streptococci) isolated from vaginal and rectal swabs of women at 35-37 weeks of pregnancy. BMC Infect. Dis. 9:153. doi: 10.1186/1471-2334-9-153
Francavilla, R., Lionetti, E., Castellaneta, S. P., Magistà, A. M., Maurogiovanni, G., Bucci, N., et al. (2008). Inhibition of Helicobacter pylori infection in humans by Lactobacillus reuteri ATCC 55730 and effect on eradication therapy: a pilot study. Helicobacter 13, 127–134. doi: 10.1111/j.1523-5378.2008.00593.x
Franco-Duarte, R., Èernáková, L., Kadam, S., Kaushik, K. S., Salehi, B., Bevilacqua, A., et al. (2019). Advances in chemical and biological methods to identify microorganisms - From past to present. Microorganisms 7:130. doi: 10.3390/microorganisms7050130
Franz, C. M. A. P., Huch, M., Abriouel, H., Holzapfel, W., and Gálvez, A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Friedman, E. S., Bittinger, K., Esipova, T. V., Hou, L., Chau, L., Jiang, J., et al. (2018). Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc. Natl. Acad. Sci. U S A. 115, 4170–4175. doi: 10.1073/pnas.1718635115
George-Kerry, R., Patra, J. K., Gouda, S., Park, Y., Shin, H.-S., and Das, G. (2018). Benefaction of probiotics for human health: A review. J. Food Drug Anal. 26, 927–939. doi: 10.1016/j.jfda.2018.01.002
Gil, N. F., Martinez, R. C. R., Gomes, B. C., Nomizo, A., and De Martinis, E. C. P. (2010). Vaginal lactobacilli as potential probiotics against Candida spp. Braz. J. Microbiol. 41, 6–14. doi: 10.1590/s1517-83822010000100002
Greifova, G., Majekova, H., Greif, G., Body, P., Greifova, M., and Dubnickova, M. (2017). Analysis of antimicrobial and immunomodulatory substances produced by heterofermentative Lactobacillus reuteri. Folia Microbiol. 62, 515–524. doi: 10.1007/s12223-017-0524-9
Gronlund, M. M., Arvilommi, H., Kero, P., Lehtonen, O. P., and Isolauri, E. (2000). Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch. Dis. Child. Fetal. Neonatal. Ed. 83, F186–F192.
Gupta, A., and Tiwari, S. K. (2015). Probiotic potential of bacteriocin-producing Enterococcus hirae strain LD3 isolated from dosa batter. Ann. Microbiol. 65, 2333–2342. doi: 10.1007/s13213-015-1075-4
Haghshenas, B., Haghshenas, M., Nami, Y., Yari Khosroushahi, A., Abdullah, N., Barzegari, A., et al. (2016). Probiotic assessment of Lactobacillus plantarum 15HN and Enterococcus mundtii 50H isolated from traditional dairies microbiota. Adv. Pharm. Bull. 6, 37–47. doi: 10.15171/apb.2016.07
Hanchi, H., Mottawea, W., Sebei, K., and Hammami, R. (2018). The genus Enterococcus: between probiotic potential and safety concerns - an update. Front. Microbiol. 9:1791. doi: 10.3389/fmicb.2018.01791
Hlivak, P., Odraska, J., Ferencik, M., Ebringer, L., Jahnova, E., and Mikes, Z. (2005). One-year application of probiotic strain Enterococcus faecium M-74 decreases serum cholesterol levels. Bratisl. Lek. Listy 106, 67–72.
Ho, M., Chang, Y. Y., Chang, W. C., Lin, H. C., Wang, M. H., Lin, W. C., et al. (2016). Oral Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 to reduce Group B Streptococcus colonization in pregnant women: A randomized controlled trial. Taiwan J. Obstet. Gynecol. 55, 515–518. doi: 10.1016/j.tjog.2016.06.003
Hooper, L. V., and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. doi: 10.1126/science.1058709
Hooper, L. V., Midtvedt, T., and Gordon, J. I. (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307.
Hou, C., Zeng, X., Yang, F., Liu, H., and Qiao, S. (2015). Study and use of the probiotic Lactobacillus reuteri in pigs: a review. J. Anim. Sci. Biotechnol. 6:14.
Hsu, T. C., Huang, C. Y., Liu, C. H., Hsu, K. C., Chen, Y. H., and Tzang, B. S. (2017). Lactobacillus paracasei GMNL-32. Lactobacillus reuteri GMNL-89 and L. reuteri GMNL-263 ameliorate hepatic injuries in lupus-prone mice. Br. J. Nutr. 117, 1066–1074. doi: 10.1017/s0007114517001039
Imase, K., Tanaka, A., Tokunaga, K., Sugano, H., Ishida, H., and Takahashi, S. (2007). Lactobacillus reuteri tablets suppress Helicobacter pylori infection-a double-blind randomised placebo-controlled cross-over clinical study. Kansenshogaku Zasshi 81, 387–393. doi: 10.11150/kansenshogakuzasshi1970.81.387
Indrio, F., Riezzo, G., Raimondi, F., Bisceglia, M., Cavallo, L., and Francavilla, R. (2008). The effects of probiotics on feeding tolerance, bowel habits, and gastrointestinal motility in preterm newborns. J. Pediatr. 152, 801–806. doi: 10.1016/j.jpeds.2007.11.005
Jones, M. L., Martoni, C. J., and Prakash, S. (2012). Cholesterol lowering and inhibition of sterol absorption by Lactobacillus reuteri NCIMB 30242: a randomized controlled trial. Eur. J. Clin. Nutr. 66, 1234–1241. doi: 10.1038/ejcn.2012.126
Jones, S. E., and Versalovic, J. (2009). Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 9:35. doi: 10.1186/1471-2180-9-35
Kaambo, E., Africa, C., Chambuso, R., and Passmore, J. S. (2018). Vaginal microbiomes associated with aerobic vaginitis and bacterial vaginosis. Front. public Health 6:78. doi: 10.3389/fpubh.2018.00078
Kechagia, M., Basoulis, D., Konstantopoulou, S., Dimitriadi, D., Gyftopoulou, K., Skarmoutsou, N., et al. (2013). Health benefits of probiotics: a review. ISRN Nutr. 2013:481651.
Keller, M. K., Nohr Larsen, I., Karlsson, I., and Twetman, S. (2014). Effect of tablets containing probiotic bacteria (Lactobacillus reuteri) on early caries lesions in adolescents: a pilot study. Benef. Microbes 5, 403–407. doi: 10.3920/bm2013.0089
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwatra, G., Adrian, P. V., Shiri, T., Buchmann, E. J., Cutland, C. L., and Madhi, S. A. (2014). Serotype-specific acquisition and loss of group B Streptococcus recto-vaginal colonization in late pregnancy. PLoS One 9:e98778. doi: 10.1371/journal.pone.0098778
Lan, P. T., Binh, L.-T., and Benno, Y. (2003). Impact of two probiotic Lactobacillus strains feeding on fecal lactobacilli and weight gains in chicken. J. Gen. Appl. Microbiol. 49, 29–36. doi: 10.2323/jgam.49.29
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic acid Technique sin Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (NewYork, NY: John Wiley and Sons), 115–175.
Lee, J., Yang, W., Hostetler, A., Schultz, N., Suckow, M. A., Stewart, K. L., et al. (2016). Characterization of the anti-inflammatory Lactobacillus reuteri BM36301 and its probiotic benefits on aged mice. BMC Microbiol. 16:69. doi: 10.1186/s12866-016-0686-7
Linares, D. M., Gómez, C., Renes, E., Fresno, J. M., Tornadijo, M. E., Ross, R. P., et al. (2017). Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front. Microbiol. 8:846. doi: 10.3389/fmicb.2017.00846
Lionetti, E., Miniello, V. L., Castellaneta, S. P., Magistá, A. M., de Canio, A., Maurogiovanni, G., et al. (2006). Lactobacillus reuteri therapy to reduce side-effects during anti-Helicobacter pylori treatment in children: a randomized placebo controlled trial. Aliment. Pharmacol. Ther. 24, 1461–1468. doi: 10.1111/j.1365-2036.2006.03145.x
Livingston, M., Loach, D., Wilson, M., Tannock, G. W., and Baird, M. (2010). Gut commensal Lactobacillus reuteri 100-23 stimulates an immunoregulatory response. Immunol. Cell Biol 88, 99–102. doi: 10.1038/icb.2009.71
Ma, X., Mao, Y. K., Wang, B., Huizinga, J. D., Bienenstock, J., and Kunze, W. (2009). Lactobacillus reuteri ingestion prevents hyperexcitability of colonic DRG neurons induced by noxious stimuli. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G868–G875.
Maccaferri, S., Biagi, E., and Brigidi, P. (2011). Metagenomics: key to human gut microbiota. Dig. Dis. 29, 525–530. doi: 10.1159/000332966
MacFaddin, J. (1985). Media for isolation-cultivation-identification-maintenance of medical bacteria, Baltimore. Maryland: Williams and Wilkins.
Mackos, A. R., Eubank, T. D., Parry, N. M., and Bailey, M. T. (2013). Probiotic Lactobacillus reuteri attenuates the stressor-enhanced severity of Citrobacter rodentium infection. Infect. Immun. 81, 3253–3263. doi: 10.1128/iai.00278-13
Mackos, A. R., Galley, J. D., Eubank, T. D., Easterling, R. S., Parry, N. M., Fox, J. G., et al. (2016). Social stress-enhanced severity of Citrobacter rodentium-induced colitis is CCL2-dependent and attenuated by probiotic Lactobacillus reuteri. Mucosal Immunol. 9, 515–526. doi: 10.1038/mi.2015.81
Macpherson, A. J., and Harris, N. L. (2004). Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485. doi: 10.1038/nri1373
Martinez, R. C., Franceschini, S. A., Patta, M. C., Quintana, S. M., Candido, R. C., Ferreira, J. C., et al. (2009). Improved treatment of vulvovaginal candidiasis with fluconazole plus probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14. Lett Appl Microbiol. 48, 269–274.
Meyn, L. A., Krohn, M. A., and Hillier, S. L. (2009). Rectal colonization by group B Streptococcus as a predictor of vaginal colonization. Am. J. Obstet. Gynecol 201, .e1–.e76.
Mishra, S. K., Malik, R. K., Manju, G., Pandey, N., Singroha, G., Behare, P., et al. (2012). Characterization of a reuterin-producing Lactobacillus reuteri BPL-36 strain isolated from human infant fecal sample. Probiot. Antimicrob. Prot. 4, 154–161. doi: 10.1007/s12602-012-9103-1
Morita, H., Toh, H., Fukuda, S., Horikawa, H., Oshima, K., Suzuki, T., et al. (2008). Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 15, 151–161. doi: 10.1093/dnares/dsn009
Mu, Q., Tavella, V. J., and Luo, X. M. (2018). Role of Lactobacillus reuteri in human health and diseases. Front Microbiol. 9:757. doi: 10.3389/fmicb.2018.00757
Nami, Y., Abdullah, N., Haghshenas, B., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2014). Probiotic assessment of Enterococcus durans 6HL and Lactococcus lactis 2HL isolated from vaginal microflora. J. Med. Microbiol. 63, 1044–1051. doi: 10.1099/jmm.0.074161-0
Nami, Y., Haghshenas, B., Haghshenas, M., Abdullah, N., and Yari Khosroushahi, A. (2015). The prophylactic effect of probiotic Enterococcus lactis IW5 against different human cancer cells. Front. Microbiol. 6:1317. doi: 10.3389/fmicb.2015.01317
Nikawa, H., Makihira, S., Fukushima, H., Nishimura, H., Ozaki, Y., Ishida, K., et al. (2004). Lactobacillus reuteri in bovine milk fermented decreases the oral carriage of mutans streptococci. Int. J. Food Microbiol. 95, 219–223. doi: 10.1016/j.ijfoodmicro.2004.03.006
Niv, E., Naftali, T., Hallak, R., and Vaisman, N. (2005). The efficacy of Lactobacillus reuteri ATCC 55730 in the treatment of patients with irritable bowel syndrome-A double blind, placebo-controlled, randomized study. Clin. Nutr. 24, 925–931. doi: 10.1016/j.clnu.2005.06.001
Ocana, V. S., Bru, E., De Ruiz Holgado, A. A., and Nader-Macias, M. E. (1999). Surface characteristics of lactobacilli isolated from human vagina. J. Gen. Appl. Microbiol. 45, 203–212. doi: 10.2323/jgam.45.203
Pieniz, S., Andreazza, R., Anghinoni, T., Camargo, F., and Brandelli, A. (2014). Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control. 37, 251–256. doi: 10.1016/j.foodcont.2013.09.055
Qin, J., Li, R., Raes, J., Arumugam, M., and Burgdorf, K. S. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 464, 59–65.
Ren, H., Zentek, J., and Vahjen, W. (2019). Optimization of production parameters for probiotic Lactobacillus strains as feed additive. Molecules 24:3286. doi: 10.3390/molecules24183286
Saggioro, A., Caroli, M., Pasini, M., Bortoluzzi, F., Girardi, L., and Pilone, G. (2005). Helicobacter pylori eradication with Lactobacillus reuteri. A double blind placebo-controlled study. Dig. Liver Dis. 37(Suppl. 1), 407–413. doi: 10.1097/mcg.0000000000000007
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sangeetha, K. T., Golia, S., and Vasudha, C. L. (2015). A study of aerobic bacterial pathogens associated with vaginitis in reproductive age group women (15-45 years) and their sensitivity pattern. Int. J. Res. Med. Sci. 3, 2268–2273. doi: 10.18203/2320-6012.ijrms20150615
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-termination inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467.
Schaefer, L., Auchtung, T. A., Hermans, K. E., Whitehead, D., Borhan, B., and Britton, R. A. (2010). The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 156(Pt 6), 1589–1599. doi: 10.1099/mic.0.035642-0
Senatore, G., Mastroleo, F., Leys, N., and Mauriello, G. (2020). Growth of Lactobacillus reuteri DSM17938 under two simulated microgravity systems: Changes in reuterin production, gastrointestinal passage resistance and stress genes expression response. Astrobiology 20, 1–14. doi: 10.1089/ast.2019.2082
Shabayek, S., and Spellerberg, B. (2018). Group B Streptococcal colonization, molecular characteristics, and epidemiology. Front. Microbial. 9:437. doi: 10.3389/fmicb.2018.00437
Sharpe, M., Shah, V., Freire-Lizama, T., Cates, E. C., McGrath, K., David, I., et al. (2019). Effectiveness of oral intake of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on Group B Streptococcus colonization during pregnancy: a midwifery-led double-blind randomized controlled pilot trial J. Matern Fetal Neonatal Med. 2019, 1–8. doi: 10.1080/14767058.2019.1650907
Sjögren, Y. M., Tomicic, S., Lundberg, A., Böttcher, M. F., Björkstén, B., Sverremark-Ekström, E., et al. (2009). Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 39, 1842–1851. doi: 10.1111/j.1365-2222.2009.03326.x
Sørensen, U. B. S., Poulsen, K., Ghezzo, C., Margarit, I., and Kilian, M. (2010). Emergence and global dissemination of host-specific Streptococcus agalactiae clones. MBio 1, e178–e110.
Spinler, J. K., Taweechotipatr, M., Rognerud, C. L., Ou, C. N., Tumwasorn, S., and Versalovic, J. (2008). Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 14, 166–171. doi: 10.1016/j.anaerobe.2008.02.001
Szajewska, H., Urbańska, M., Chmielewska, A., Weizman, Z., and Shamir, R. (2015). Meta-analysis: Lactobacillus reuteri strain DSM 17938 (and the original strain ATCC 55730) for treating acute gastroenteritis in children. Benef. Microbes 5, 285–293. doi: 10.3920/bm2013.0056
Talarico, T. L., Casas, I. A., Chung, T. C., and Dobrogosz, W. J. (1988). Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32, 1854–1858. doi: 10.1128/aac.32.12.1854
Talarico, T. L., and Dobrogosz, W. J. (1989). Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 33, 674–679. doi: 10.1128/aac.33.5.674
Thompson, D., Gibson, J., Plewinak, F., Jeanmougin, F., and Higgins, G. (1997). The ClastalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acid Res. 25, 4867–4887.
Timmerman, H. M., Rutten, N., Boekhorst, J., Saulnier, D. M., Kortman, G., Contractor, N., et al. (2017). Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci. Rep. 7:8327.
Tubelius, P., Stan, V., and Zachrisson, A. (2005). Increasing work-place healthiness with the probiotic Lactobacillus reuteri: a randomised, double-blind placebo-controlled study. Environ. Health 4:25.
Urbańska, M., Gieruszczak-Białek, D., and Szajewska, H. (2016). Systematic review with meta-analysis: Lactobacillus reuteri DSM 17938 for diarrhoeal diseases in children. Aliment. Pharmacol. Ther. 43, 1025–1034. doi: 10.1111/apt.13590
van Zyl, W., Deane, S., and Dicks, L. (2016). Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 excludes Listeria monocytogenes from the GIT, as shown by bioluminescent studies in mice. Benef. Microbes 7, 227–235. doi: 10.3920/bm2015.0082
Wang, W. L., Xu, S. Y., Ren, Z. G., Tao, L., Jiang, J. W., and Zheng, S. S. (2015). Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 21, 803–814. doi: 10.3748/wjg.v21.i3.803
Wang, X. D., Soltesz, V., Molin, G., and Andersson, R. (1995). The role of oral administration of oatmeal fermented by Lactobacillus reuteri R2LC on bacterial translocation after acute liver failure induced by subtotal liver resection in the rat. Scand. J. Gastroenterol. 30, 180–185. doi: 10.3109/00365529509093259
Westerik, N., Kort, R., Sybesma, W., and Reid, G. (2018a). Lactobacillus rhamnosus probiotic food as a tool for empowerment across the value chain in Africa. Front. Microbiol. 9:1501. doi: 10.3389/fmicb.2018.01501
Westerik, N., Reid, G., Sybesma, W., and Kort, R. (2018b). The probiotic Lactobacillus rhamnosus for alleviation of Helicobacter pylori-associated gastric pathology in east Africa. Front. Microbiol. 9:1873. doi: 10.3389/fmicb.2018.01873
Zheng, J., Wittouck, S., Salvetti, E., Franz, C., Harris, H., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: Enterococcus faecalis, Lacticaseibacillus rhamnosus, Limosilactobacillus reuteri, newborn babies, stool, initial colonization
Citation: Al-Balawi M and Morsy FM (2020) Enterococcus faecalis Is a Better Competitor Than Other Lactic Acid Bacteria in the Initial Colonization of Colon of Healthy Newborn Babies at First Week of Their Life. Front. Microbiol. 11:2017. doi: 10.3389/fmicb.2020.02017
Received: 07 June 2020; Accepted: 30 July 2020;
Published: 29 September 2020.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Babak Haghshenas, Kermanshah University of Medical Sciences, IranAmarela Terzić-Vidojević, University of Belgrade, Serbia
Stefania Silvi, University of Camerino, Italy
Copyright © 2020 Al-Balawi and Morsy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fatthy Mohamed Morsy, fmorsy@aun.edu.eg
†These authors have contributed equally to this work
 Mohammad Al-Balawi
Mohammad Al-Balawi Fatthy Mohamed Morsy
Fatthy Mohamed Morsy