- Reference Center for Gender Specific Medicine, Istituto Superiore di Sanità, Rome, Italy
Coronaviruses are enveloped, single-stranded, positive-sense RNA viruses that can infect animal and human hosts. The infection induces mild or sometimes severe acute respiratory diseases. Nowadays, the appearance of a new, highly pathogenic and lethal coronavirus variant, SARS-CoV-2, responsible for a pandemic (COVID-19), represents a global problem for human health. Unfortunately, only limited approaches are available to treat coronavirus infections and a vaccine against this new coronavirus variant is not yet available. The plasma membrane microdomain lipid rafts have been found by researchers to be involved in the replication cycle of numerous viruses, including coronaviruses. Indeed, some pathogen recognition receptors for coronaviruses as for other viruses cluster into lipid rafts, and it is therefore conceivable that the first contact between virus and host cells occurs into these specialized regions, representing a port of cell entry for viruses. Recent data highlighted the peculiar pro-viral or anti-viral role played by autophagy in the host immune responses to viral infections. Coronaviruses, like other viruses, were reported to be able to exploit the autophagic machinery to increase their replication or to inhibit the degradation of viral products. Agents known to disrupt lipid rafts, such as metil-β-cyclodextrins or statins, as well as autophagy inhibitor agents, were shown to have an anti-viral role. In this review, we briefly describe the involvement of lipid rafts and autophagy in coronavirus infection and replication. We also hint how lipid rafts and autophagy may represent a potential therapeutic target to be investigated for the treatment of coronavirus infections.
Introduction
Coronaviruses (CoVs) are a large group of enveloped animal and human viruses, with a single-stranded positive-sense RNA genome. The Coronaviridae family, to which they belong, includes four genera of CoVs, indicated as Alphacoronavirus (αCoV), Betacoronavirus (βCoV), Gammacoronavirus (γCoV), and Deltacoronavirus (δCoV) (Cui et al., 2019; Chen et al., 2020). αCoV and βCoV have evolutionary evolved from bats and rodents, and have been responsible for human infections; whereas δCoV and γCoV derive from avian species (Woo et al., 2012). αCoV and βCoV are further divided into subgroups, 1a-1b and 2a-2d, respectively, (Graham et al., 2013; Drexler et al., 2014; Cui et al., 2019; Fung and Liu, 2019) (Supplementary Figure S1). Coronaviruses have been regarded as etiologic agents of both relatively mild and severe respiratory infections/diseases in humans (Zhao et al., 2012; Paules et al., 2020). HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1 cause the common cold and mild upper respiratory diseases in immunocompetent hosts, although some of them can be potentially virulent in infants, young children, and elder individuals. Highly pathogenic human coronaviruses, responsible for more severe respiratory diseases, include SARS-CoV that caused the 2003 pandemic outbreak of severe respiratory tract infection (Ksiazek et al., 2003), MERS-CoV, responsible for the 2012 outbreak in Middle Eastern countries (Zaki et al., 2012), and the novel SARS-CoV-2, actually causing a pandemic of severe pneumonia that started in China in December 2019 (Benvenuto et al., 2019; Zhu et al., 2020).
Coronavirus and coronavirus-like infections have been described also in domestic and wild animals, such as swine [porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus], cattle (BCoV), horses, camels, cats, dogs, rodents, birds, bats, rabbits, ferrets, mink, and various wildlife species (Fehr and Perlman, 2015; Lin et al., 2016; Banerjee et al., 2019; Cui et al., 2019; Ye et al., 2020). Although many coronavirus infections are subclinical in animals, however, they cause a range of diseases that can have serious consequences for animal health as well as for the economic losses in livestock. In addition, domestic animals may have been important intermediate hosts for virus transmission from natural hosts to humans (Fehr and Perlman, 2015; Ye et al., 2020).
Since only limited approaches are available to treat or prevent coronavirus infections (Zhang et al., 2020), the importance of identifying novel therapeutic targets is evident.
Viruses have evolved a close interplay with the host cell and the first encounter between the virus and target cell occurs at the plasma membrane. Lipid rafts are specialized plasma membrane microdomains involved in important processes of the virus infections and of the host target cells (Rosenberger et al., 2000).
Viruses exploit lipid rafts to gain infection of the target cells; therefore, pharmacologic depletion or disruption of lipid rafts may provide a tool to reduce or inhibit viral replication (Khandia et al., 2019).
Furthermore, it has been widely demonstrated that lipid rafts play a fundamental role in autophagic machinery: they are associated with autophagosome morphogenesis, either in the initiation or in the maturation phases (Matarrese et al., 2014).
Autophagy, a cell process involved in cellular homeostasis, stress, and immune responses to viral infections, is a two-edged process in virus infections, since it may have pro- or anti-viral roles (Kudchodkar and Levine, 2009; Khandia et al., 2019). Viruses, on the other side, during co-evolution with the host cell, developed mechanisms to usurp/exploit the host autophagic system (Jheng et al., 2014).
This minireview reports on the available knowledge about the interplay between coronaviruses, including the SARS-CoV-2, with lipid rafts and autophagic pathways, in order to focus the attention to novel potential targets to inhibit coronavirus infections.
Lipid Rafts and Autophagy: Focus on Coronavirus
Lipid rafts are plasma membrane microdomains (10–200 nm) enriched in cholesterol, glycosphingolipids, and phospholipids. They are also found in the endoplasmic reticulum (ER) (Browman et al., 2006), in the Golgi complex (Eberle et al., 2002), and on the membrane of endosomes (Sobo et al., 2007) and phagosomes (Dermine et al., 2001). There are two main types of lipid rafts based on their protein composition: “planar lipid rafts” and “caveolae” enriched by the proteins flotillin and caveolin, respectively (McGuinn and Mahoney, 2014). Both possess a similar lipid composition that confers resistance to solubilization by non-ionic detergents at low temperatures (Brown and Rose, 1992), and the characteristic of being able to be isolated in sucrose gradient (Sargiacomo et al., 1993). Because of their dynamic and heterogeneous structure, they can rapidly assemble and disassemble, changing their composition in response to intra- and extracellular stimuli (Simons and Toomre, 2000). A key component of lipid rafts is cholesterol that is the glue that maintains raft architecture, as demonstrated by the disorganization and disruption of lipid rafts upon depletion of plasma membrane cholesterol by the cholesterol depleting agent methyl-β-cyclodextrin (MβCD) (Simons and Ehehalt, 2002; Zidovetzki and Levitan, 2007). These membrane regions play an important role in a variety of cellular functions, but principally they recruit and concentrate several signaling molecules (Song et al., 1997; Galbiati et al., 1999; Razani et al., 1999; Patel and Insel, 2009) that, by interacting with caveolin and flotillin, form a sort of signal transduction platform. Therefore, lipid rafts are involved in many biological functions including endocytosis, signal transduction, cell communication, and regulation of autophagy (Nabi and Le, 2003; Shi et al., 2015).
Because of their capacity to cluster into a “phagocytic synapse” several pathogen recognition receptors (Toll-like receptors, C-type lectin receptors), lipid rafts are the focus of intense research in the field of infection. In particular, they are involved in several steps along a viral infection, such as virus entry into the host cell (fusion and internalization), viral protein transport, viral assembly, and budding processes (Hogue et al., 2012). Several enveloped (HIV-1, influenza viruses, coronavirus, flavivirus) and non-enveloped (SV40 and Rrotavirus) viruses exploit the raft platform to bind to their specific receptors (Ono and Freed, 2001; Pelkmans, 2005; Li et al., 2007; Takahashi and Suzuki, 2011).
Recent studies have implicated lipid rafts in coronavirus entry and egress through a multistep endocytic process, although the detailed mechanism remains to be disclosed (Li et al., 2007). Figure 1 depicts a schematic view of the role of lipid rafts in two paradigmatic human coronaviruses, HCoV-229E and SARS-CoV, infections. Briefly, the receptors for the two coronaviruses, aminopeptidase N (APN/CD13) and angiotensin converting enzyme 2 (ACE2), respectively, are located into lipid rafts and play a fundamental role in the initial step of the virus infection. APN/CD13, a zinc-binding aminopeptidase, is expressed in several cell types (endothelial, granulocytic, and monocytic cells; epithelial cells of the kidney; respiratory tract and intestine) and is required also for porcine, canine, and feline coronavirus recognition. ACE2, encoded on the X chromosome, is a metalloprotease long known to be a key player in the renin–angiotensin system that co-localizes with caveolin-1 and GM1 and is expressed on type I and II pneumocytes, enterocytes, endothelial cells of the heart and kidney, epithelial cells of the kidney, and the testis (Hamming et al., 2004). SARS-CoV-2, the etiological agent of the COVID-19 pandemic, also binds ACE2 to infect host cells, by similarity to SARS-CoV, from which it differentiates for point mutations on the spike protein, essential for receptor binding (Wang et al., 2020; Yan et al., 2020). By analogy to SARS-CoV, it can be hypothesized that SARS-CoV-2 may also use lipid rafts for its entry into the host cells.
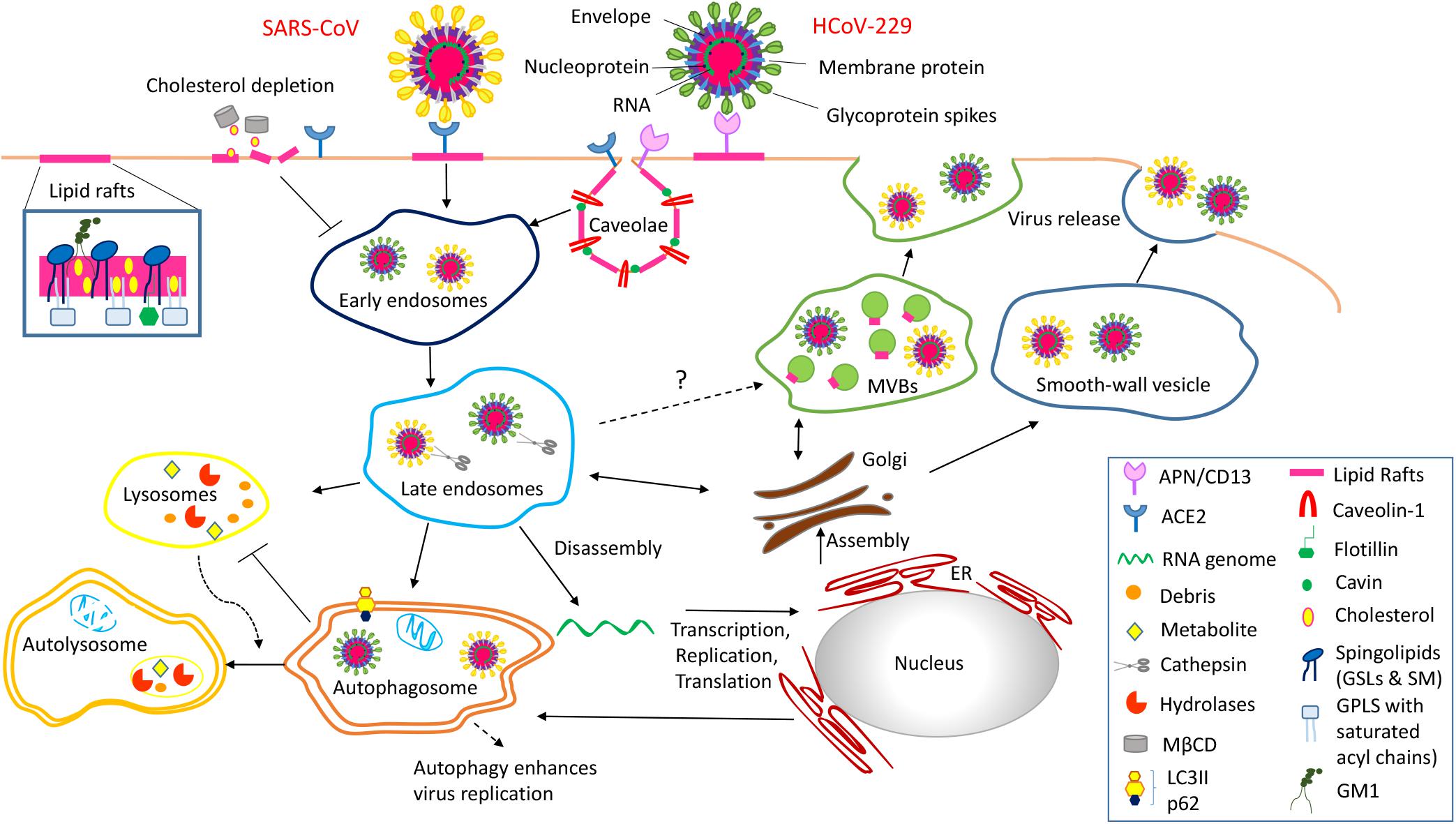
Figure 1. Schematic representation of the role of lipid rafts in Coronavirus infection of the host cells is a multistep endocytic process characterized by a series of complex events tightly regulated in space and time. Step 1 Entry process of coronavirus into the host cells is initiated by the binding of the spike glycoprotein with the specific receptor (ACE2, APN/CD13) located into lipid rafts/caveolae. This interaction causes conformational changes of the viral particle, which trigger specific signaling events necessary for the viral entry mechanism. Step 2 Lipid rafts/caveolae-mediated endocytosis is followed by intracellular trafficking of virus particles in transport vesicles (early and late endosomes). The low pH in late endosomes induces a conformational change in coronavirus that mediates fusion of the viral envelope with the endosomal membrane. Step 3 Viral genomes are translated in two polyproteins, pp1a and pp1ab, which encode the non-structural viral proteins that form the replication transcription complex. This complex produces genomic RNA as well as multiple subgenomic mRNAs encoding structural proteins. Translation of mRNA encoding for the nucleocapsid proteins occurs in the cytoplasm where the newly synthesized proteins interact with new genomes to form ribonucleoprotein particles. In contrast, matrix, envelope and spike proteins translation occurs into the ER. Coronavirus uses also the autophagy machinery for replication and has evolved strategies to avoid autophagy-induced lysosomal degradation. Step 4 After assembly the progeny viral particles, virus-containing vesicles (smooth-wall vesicles) are budded and released into the extracellular environment through fusion with the plasma membrane (exocytosis). Alternatively, we speculate that coronavirus might utilize multivescicular bodies (MVBs) and take advantage of the exosomal pathway for egress.
The steps following coronavirus entry are not clearly identified. A recent study demonstrated that SARS-CoV exploits the activity of cathepsin L, an endosomal cysteine protease, to initiate proteolysis and activation of membrane fusion within endosomes (Simmons et al., 2013). In addition, coronavirus mouse hepatitis virus (MHV) infection is inhibited when early endosome-associated proteins, RAB5 and EEA1, are down-regulated by siRNA (Burkard et al., 2014) and infectious bronchitis virus (IBV) release of the nucleocapsid into the cytoplasm (Wang et al., 2018) occurs through the fusion with endosome–lysosome membranes. These data suggest that coronaviruses exploit the early and late endosome compartment after viral entry, as several other viruses (adenovirus, human papillomavirus, polyomavirus, African swine fever virus, and influenza virus) (Lakadamyali et al., 2004; Sánchez et al., 2017; Spriggs et al., 2019).
Recent evidence shows that there is a close interplay between lipid rafts and autophagy during physiological cell function, as lipid rafts can regulate autophagy by interacting with autophagosomes and autophagy-related proteins, such as ATG5 and ATG12 (Chen et al., 2014; Garofalo et al., 2016). In addition a functional and structural link between lipid rafts and autophagy comes from the evidence that chemical (e.g., MβCD, fumonisin B1) or biological (e.g., siRNA inhibition of key enzymes involved in the sphingolipids metabolism) molecules capable of altering the chemical composition or molecular organization of lipid rafts have also a strong impact on autophagy (Khandia et al., 2019).
Autophagy is an evolutionarily conserved (from yeast to mammals), selective, and finely regulated process that generates ATP and precursors for the synthesis of macromolecules (Ktistakis and Tooze, 2016) and is considered as a survival process put in place by cells under stressful conditions, such as pathogen microbial infections. It plays a key role in cellular homeostasis and is responsible for the turnover of cellular organelles (Yu et al., 2018), as its main function is to remove and recycle non-essential, damaged, or obsolete cellular components, such as whole organelles or macromolecules (Mizushima and Komatsu, 2011). During autophagy, cytoplasmic portions are enclosed into double membrane bound vesicles (autophagosomes) that then fuse with late endosomes/lysosomes, whose contents are degraded by lysosomal proteases. Autophagy has been shown to have additional function in innate immunity, by degradation of viruses, or intracellular pathogens, as well as by presenting pathogen components to the immune system (Mao et al., 2019).
Autophagy can have a pro-viral role, promoting virus infection and propagation, as well as an antiviral role, essentially depending on the virus strain, on the phase of infection, on the infected cell type, but also on the cellular microenvironment (Lennemann and Coyne, 2015). During co-evolution with their natural hosts, viruses developed the ability to hijack autophagic mechanisms to their advantage by using them for immune escaping, or using autophagosomes as a replicative niche. However, data on autophagy interplay with coronaviruses are scant so far, related on no more than 50 works published between 2004 and 2015. In general, it is suggested that CoVs can interact with some components of the autophagic pathway in an opposite way with a dual effect: utilizing autophagy components to promote viral replication and/or to inhibit degradation of viral products through the autophagic pathway (Cong et al., 2017). In some coronaviruses, infection results are still contradictory, as in the case of MERS-CoV (Corman et al., 2016), which has been reported to induce phosphorylation changes in key kinases, such as AKT1 and mTOR, that regulate the early steps of autophagic process (Kindrachuk et al., 2015), thus stimulating autophagy in infected cells. In contrast, Gassen et al. reported that MERS-CoV activates the SKP2 kinase, consequently reducing Beclin1 (BECN1) activity and inhibiting the fusion of autophagosomes with lysosomes, which results in autophagy inhibition (Gassen et al., 2019).
In general, it has been hypothesized that coronaviruses would induce accumulation of autophagic vacuoles to obtain a larger availability of the membrane structure necessary for their replication (Gassen et al., 2019), which is consistent to the observed co-localization of the rodent coronavirus, MHV, replication complex, with the autophagic proteins LC3 and Apg12 (Prentice et al., 2004). Likewise, the SARS-CoV takes advantage of autophagic pathway to replicate and transcribe its own genome, as suggested by inhibition of its replication through inactivation of GSK-3 (glycogen synthase kinase-3), a serine/threonine kinase that inhibits autophagy, through the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) (Wu et al., 2009; Zúñiga et al., 2010). A very recent study showed that SARS-CoV-2, similar to MERS-CoV, strongly reduced the autophagic flux in infected cell lines downregulating the AMPK/mTORC1 pathway, altered autophagy-relevant signaling, and also reduced autophagosome/lysosome fusion efficiency (Gassen et al., 2020). As a result, the SARS-CoV-2 virus could take advantage of the autophagy reduction, thus preventing viral product degradation and enhancing double membrane vesicles (DMV) availability, indispensable for their replication.
The cell organelle most involved in the dynamic membrane changes is the ER. It is therefore not surprising that some coronaviruses, through the insertion of their trans-membrane glycoproteins at the ER level, are able to induce ER stress. This is the case, for example, of the ORF3 protein produced by porcine epidemic diarrhea virus (PEDV), which induced ER stress-dependent autophagy in different porcine and human cell types (Zou et al., 2019). In the same vein, MHV was observed to use the host machinery to export vesicular ER to originate membranes for the genesis of DMV. As regards the non-structural protein (NSP) 6 of the avian infectious bronchitis virus (IBV), a gamma-coronavirus, in addition to providing membranes useful for the replication of the virus, it could prevent its degradation within the lysosomes, effectively escaping the autophagy-based cellular defensive mechanisms. Similar properties have also been observed for MHV and SARS NSP6 proteins and PRRSV arterivirus NSP5, NSP6, and NSP7 (Cottam et al., 2014).
Discussion
Coronaviruses may represent a threat for human and animal health for their potential to cross the species barrier, thus acquiring human virulence, as evidenced by the previous SARS-CoV and MERS-CoV outbreaks and by the ongoing COVID-19 pandemic. Notwithstanding, treatment and prophylaxis measures to control coronavirus infections are lacking so far, either in humans or in livestock animals.
As outlined in this review, lipid rafts and autophagic pathways play a pivotal role in coronavirus infection, being critical for viral entry and replication, as well as for viral release from the host cells. Actually, lipid rafts are the focus of intense research in the field of infection, and it is conceivable to consider targeting some lipid raft components in order to inhibit virus infection at the cell level.
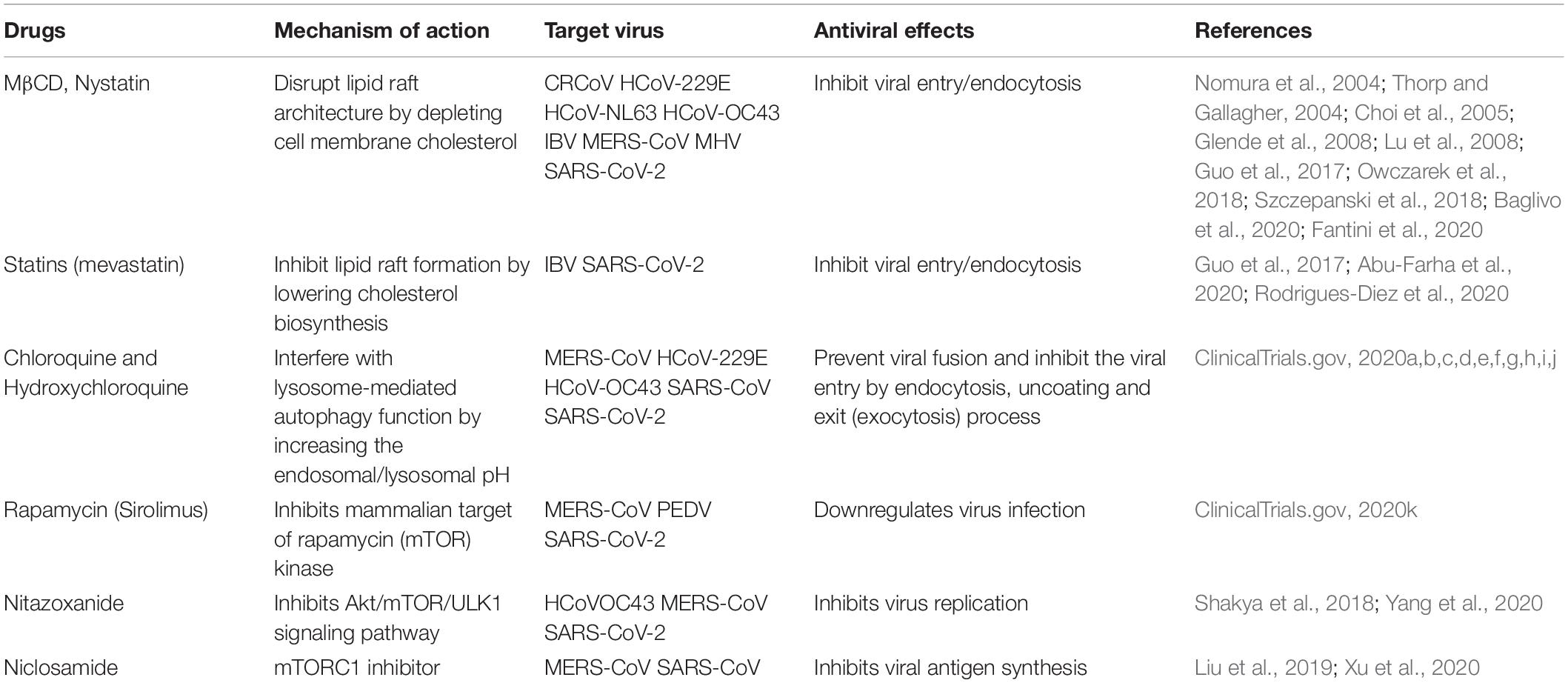
In particular, lipid raft disruption by cholesterol-depleting agents has been shown to inhibit infection of several microbes by blocking their entry into the host cells (Nomura et al., 2004; Thorp and Gallagher, 2004; Choi et al., 2005; Riethmüller et al., 2006; Glende et al., 2008; Lu et al., 2008; Guo et al., 2017; Owczarek et al., 2018; Szczepanski et al., 2018; Abu-Farha et al., 2020; Baglivo et al., 2020; Fantini et al., 2020; Rodrigues-Diez et al., 2020; Table 1). In human immunodeficiency virus (Ono and Freed, 2001), herpes simplex virus, and rotavirus infections, MβCD, a widely used raft-disrupting agent, has been shown to affect virus entry, thereby reducing their infectivity (Dou et al., 2018; Wudiri et al., 2017). In addition, in Japanese encephalitis virus and dengue virus infection, disruption of lipid rafts by MβCD has been shown to decrease viral infection acting at both viral entry and intracellular replication step (Lee et al., 2008). β-cyclodextrins are extensively utilized in pharmaceutical formulations as excipients to enhance solubility, bioavailability, and stability of many drugs (Loftsson and Brewster, 2012). In the light of the above-described antiviral activity, further studies on pharmacokinetic and safety of MβCD could foster its clinical application as an antimicrobial agent in humans. Statins, used in human therapy for their ability to inhibit cellular synthesis of cholesterol, have also been reported to have an anti-viral effect, inhibiting infection of flavivirus, such as dengue virus (DENV), hepatitis C virus (HCV), West Nile virus (WNV), and Zika virus (Mackenzie et al., 2007; Andrus and East, 2010; Martínez-Gutierrez et al., 2011; Españo et al., 2019), and their application to inhibition of the coronavirus infection would be worth considering.
Likewise, the pharmacological modulation of autophagic processes can represent an attractive therapeutic strategy for elimination of the viral pathogen or containment of the infection (Rubinsztein et al., 2009; Clark et al., 2018).
In fact, different drugs described as inhibitors or inducers of the autophagy that control host cell pathways process involved in coronavirus infection, have sparked interest for their potential antiviral activity (Shakya et al., 2018; Liu et al., 2019; Xu et al., 2020; Yang et al., 2020; Table 1). One of this, chloroquine (CQ), and its derivative hydroxychloroquine (HCQ), known as anti-malarial drugs (O’Neill et al., 1998), which are able to inhibit autophagy by raising the lysosomal pH (Golden et al., 2015), have also been evaluated for HIV infection (Romanelli et al., 2004). Very recently, clinicians have paid attention to CQ and HCQ as a possible treatment of patients infected by the novel emerged SARS-CoV-2 (ClinicalTrials.gov, 2020a,b,c,d,e,f,g,h,i,j). This insight is also supported by some recent works, including a recent publication by Gao et al. that indicates some positive effects of CQ on the course of pneumonia associated with the infection and on the reduction in healing (Cortegiani et al., 2020; Gao et al., 2020; Geleris et al., 2020; Yu et al., 2020).
On the contrary, other studies have highlighted the ineffectiveness of these drugs both in the viremic phase and in respiratory complications (Boulware et al., 2020; Mahévas et al., 2020; Rosenberg et al., 2020). Although some data, both referring to COVID-19 and SARS, have highlighted some positive effects of CQ and HCQ in the evolution of the disease, thus showing their therapeutic potential, at the moment, there are no data that can support the use of these drugs to control the entire pathological process related to SARS-CoV-2 (Gies et al., 2020).
In the same vein, rapamycin, also known as sirolimus, an autophagy inducer already used as an immunosuppressant, has been tested, with some success, in the treatment of COVID-19 (NCT04341675) (ClinicalTrials.gov, 2020k). These cases may represent a repositioning of the drugs with clinical success in treatment areas beyond their original approved use. According to this, the antiviral in vitro activity of spermidine, niclosamide, and nitazoxanide (known autophagy inducers) vs. SARS-CoV-2 was recently reported (Shakya et al., 2018; Liu et al., 2019; Xu et al., 2020; Yang et al., 2020). Thus, a prophylactic approach to COVID-19 using these drugs, which are well tolerated, clinically applied, or FDA-approved compounds, would be rational (Gassen et al., 2020). Importantly, treatments for emerging infections by targeting host cell pathways, rather than the infectious agent directly, or to complement antivirals with drugs that enhance host cell resistance mechanisms have thus become an active and promising therapeutic strategy. This strategy is even more important and urgent to be explored in the case of such potentially and suddenly pandemic virus family, as coronaviruses are.
Author Contributions
KF contributed to the design of this study, drafted the manuscript, and drew figures relative to lipid rafts. SA contributed to the conception of the idea and drafted the manuscript and figure relative to coronavirus description. DP conducted a wide literature search and drafted the manuscript on the lipid rafts section. EI drafted the manuscript on the autophagy section. PM provided autophagy expertise, contributed to draft and revision of the manuscript, and concurred with the final version of the review. MG provided lipid rafts expertise, and contributed to draft and review the manuscript and to the organization of the review. AR conceived the idea, drafted and revised the manuscript, coordinated the activity, and concurred with the final version of the review. All authors contributed to the article and approved the submitted version.
Funding
The authors gratefully acknowledge the financial support obtained from the Italian Ministry of Health (to MG), grant PE-2013-02354961, and Peretti Foundation (to PM), grant 3603.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Massimo Delle Femmine for his support with the figures.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01821/full#supplementary-material
FIGURE S1 | Taxonomy of CoVs Classification scheme of some representative human (in red) and animal (in black) coronaviruses.Abbreviations: FCoV, feline coronavirus; TGEV, transmissible gastroenteritis virus; PRCV, porcine respiratory coronavirus; HCoV, human coronavirus; PEDV, porcine epidemic diarrhea virus; Ro-BatCoV, Rousettus bat CoV; PHEV, porcine hemagglutinating encephalomyelitis virus; MHV, mouse hepatitis virus; BCV, bovine coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; Bat SARSr-CoV, Bat SARS-like coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; BuCoV, bulbul coronavirus; PDCoV, porcine deltacoronavirus; BWCoV, beluga whale coronavirus; IBV, avian infectious bronchitis virus; TCoV, Turkey coronavirus.
References
Abu-Farha, M., Thanaraj, T. A., Qaddoumi, M. G., Hashem, A., Abubaker, J., and Al-Mulla, F. (2020). The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci. 21:3544. doi: 10.3390/ijms21103544
Andrus, M. R., and East, J. (2010). Use of statins in patients with chronic Hepatitis C. S. Med. J. 103, 1018–1024. doi: 10.1097/SMJ.0b013e3181f0c6b4
Baglivo, M., Baronio, M., Natalini, G., Beccari, T., Chiurazzi, P., Fulcheri, E., et al. (2020). Natural small molecules as inhibitors of coronavirus lipiddependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Biom. 91, 161–164. doi: 10.23750/abm.v91i1.9402
Banerjee, A., Kulcsar, K., Misra, V., Frieman, M., and Mossman, K. (2019). Bats and coronaviruses. Viruses 11:E41. doi: 10.3390/v11010041
Benvenuto, D., Giovanetti, M., Ciccozzi, A., Spoto, S., Angeletti, S., and Ciccozzi, M. (2019). The2019-new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 92, 455–459. doi: 10.1002/jmv.25688
Boulware, D. R., Pullen, M. F., Bangdiwala, A. S., Pastick, K. A., Lofgren, S. M., Okafor, E. C., et al. (2020). A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. doi: 10.1056/NEJMoa2016638 [Epub ahead of print].
Browman, D. T., Resek, M. E., Zajchowski, L. D., and Robbins, S. M. (2006). Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119, 3149–3160. doi: 10.1242/jcs.03060
Brown, D. A., and Rose, J. K. (1992). Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544. doi: 10.1016/0092-8674(92)90189-j
Burkard, C., Verheije, M. H., Wicht, O., van Kasteren, S. I., van Kuppeveld, F. J., Haagmans, B. L., et al. (2014). Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 10:e1004502. doi: 10.1371/journal.ppat.1004502
Chen, Y., Liu, Q., and Guo, D. (2020). Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423. doi: 10.1002/jmv.25681
Chen, Z. H., Cao, J. F., Zhou, J. S., Liu, H., Che, L. Q., Mizumura, K., et al. (2014). Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am. J. Physiol. Lung. Cell Mol. Physiol. 306, 1016–1025. doi: 10.1152/ajplung.00268.2013
Choi, K. S., Aizaki, H., and Lai, M. M. (2005). Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release. J. Virol. 79, 9862–9871. doi: 10.1128/JVI.79.15.9862-9871.2005
Clark, A. E., Sabalza, M., Gordts, P. L. S. M., and Spector, D. H. (2018). Human cytomegalovirus replication is inhibited by the autophagy-inducing compounds trehalose and SMER28 through distinctively different mechanisms. J. Virol. 92, e2015–e2017. doi: 10.1128/JVI.02015-17
ClinicalTrials.gov (2020a). Chemoprophylaxis With Hydroxychloroquine in Healthcare Personnel in Contact With COVID-19 Patients: A Randomized Controlled Trial (PHYDRA Trial) (NCT04318015). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020b). Chloroquine/Hydroxychloroquine Prevention of Coronavirus Disease (COVID-19) in the Healthcare Setting (COPCOV) (NCT04303507). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020c). Effectiveness of Hydroxychloroquine in Covid-19 Patients. Bethesda, MD: ClinicalTrials.gov (NCT04328272).
ClinicalTrials.gov (2020d). Efficacy and Safety of Hydroxychloroquine for Treatment of COVID-19 (NCT04261517). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020e). Hydroxychloroquine for the Treatment of Patients With Mild to Moderate COVID-19 to Prevent 106. Progression to Severe Infection or Death (NCT04323631). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020f). Hydroxychloroquine Post Exposure Prophylaxis (PEP) for Household Contacts of COVID-19 Patients: A NYC Community-Based Randomized Clinical Trial (NCT04318444). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020g). Hydroxychloroquine Treatment for Severe COVID-19 Pulmonary Infection (HYDRA Trial) (HYDRA) (NCT04315896). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020h). Hydroxychloroquine Versus Placebo in COVID-19 Patients at Risk for Severe Disease (HYCOVID) (NCT04325893). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020i). Safety and Efficacy of Hydroxychloroquine Associated With Azithromycin in SARS-Cov-2 Virus (COVID-19) (Coalition-I) (NCT04322123). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020j). Safety and Efficacy of Hydroxychloroquine Associated With Azithromycin in SARS-CoV2 Virus (Coalition Covid-19 Brasil II) (NCT04321278). Bethesda, MD: ClinicalTrials.gov.
ClinicalTrials.gov (2020k). Sirolimus Treatment in Hospitalized Patients With COVID-19 Pneumonia (SCOPE) (NCT04341675). Bethesda, MD: ClinicalTrials.gov.
Cong, Y., Verlhac, P., and Reggiori, F. (2017). The interaction between nidovirales and autophagy components. Viruses 9:E182. doi: 10.3390/v9070182
Corman, V. M., Albarrak, A. M., Omrani, A. S., Albarrak, M. M., Farah, M. E., Almasri, M., et al. (2016). Viral shedding and antibody response in 37 patients with middle east respiratory syndrome coronavirus infection. Clin. Infect. Dis. 62, 477–483. doi: 10.1093/cid/civ951
Cortegiani, A., Ingoglia, G., Ippolito, M., Giarratano, A., and Einav, S. (2020). A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care 57, 279–283. doi: 10.1016/j.jcrc.2020.03.005
Cottam, E. M., Whelband, M. C., and Wileman, T. (2014). Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10, 1426–1441. doi: 10.4161/auto.29309
Cui, J., Li, F., and Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. NatRevMicrobiol. 17, 181–192. doi: 10.1038/s41579-018-0118-9
Dermine, J. F., Duclos, S., Garin, J., St-Louis, F., Rea, S., Parton, R. G., et al. (2001). Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J. Biol. Chem. 276, 18507–18512. doi: 10.1074/jbc.M101113200
Dou, X., Li, Y., Han, J., Zarlenga, D. S., Zhu, W., Ren, X., et al. (2018). Cholesterol of lipid rafts is a key determinant for entry and post-entry control of porcine rotavirus infection. BMC Vet. Res. 14:45. doi: 10.1186/s12917-018-1366-7
Drexler, J. F., Corman, V. M., and Drosten, C. (2014). Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 101, 45–56. doi: 10.1016/j.antiviral.2013.10.013
Eberle, H. B., Serrano, R. L., Füllekrug, J., Schlosser, A., Lehmann, W. D., Lottspeich, F., et al. (2002). Identification and characterization of a novel human plant pathogenesis-related protein that localizes to lipid-enriched microdomains in the Golgi complex. J. Cell Sci. 115, 827–838.
Españo, E., Nam, J. H., Song, E. J., Song, D., Lee, C. K., and Kim, J. K. (2019). Lipophilic statins inhibit Zika virus production in Vero cells. Sci. Rep.. 9, 11461. doi: 10.1038/s41598-019-47956-1
Fantini, J., Di Scala, C., Chahinian, H., and Yahi, N. (2020). Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 91, 161–164.
Fehr, A. R., and Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23. doi: 10.1007/978-1-4939-2438-7_1
Fung, T. S., and Liu, D. X. (2019). Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 73, 529–557. doi: 10.1146/annurev-micro-020518-115759
Galbiati, F., Volonte, D., Meani, D., Milligan, G., Lublin, D. M., Lisanti, M. P., et al. (1999). The dually acylated NH2-terminal domain of gi1alpha is sufficient to target a green fluorescent protein reporter to caveolin-enriched plasma membrane domains. Palmitoylation of caveolin-1 is required for the recognition of dually acylated g-protein alpha subunits in vivo. J. Biol. Chem. 274, 5843–5850. doi: 10.1074/jbc.274.9.5843
Gao, J., Tian, Z., and Yang, X. (2020). Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 14, 72–73. doi: 10.5582/bst.2020.01047
Garofalo, T., Matarrese, P., Manganelli, V., Marconi, M., Tinari, A., Gambardella, L., et al. (2016). Evidence for the involvement of lipid rafts localized at the ER-mitochondria associated membranes in autophagosome formation. Autophagy 12, 917–935. doi: 10.1080/15548627.2016.1160971
Gassen, N. C., Niemeyer, D., Muth, D., Corman, V. M., Martinelli, S., Gassen, A., et al. (2019). Rein T.SKP2 attenuates autophagy through beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 10:5770. doi: 10.1038/s41467-019-13659-4
Gassen, N. C., Papies, J., Bajaj, T., Dethlof, F., Emanuel, J., Weckmann, K., et al. (2020). Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. bioRxiv [Preprint] doi: 10.1101/2020.04.15.997254
Geleris, J., Sun, Y., Platt, J., Zucker, J., Baldwin, M., Hripcsak, G., et al. (2020). Observational study of hydroxychloroquine in hospitalized patients with COVID-19. N. Engl. J. Med. 382, 2411–2418. doi: 10.1056/NEJMoa2012410
Gies, V., Bekaddour, N., Dieudonné, Y., Guffroy, A., Frenger, Q., Gros, F., et al. (2020). Beyond anti-viral effects of chloroquine/hydroxychloroquine. Front. Immunol. doi: 10.3389/fimmu.2020.01409 [Epub ahead of print].
Glende, J., Schwegmann-Wessels, C., Al-Falah, M., Pfefferle, S., Qu, X., Deng, H., et al. (2008). Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology 381, 215–221. doi: 10.1016/j.virol.2008.08.026
Golden, E. B., Cho, H. Y., Hofman, F. M., Louie, S. G., Schönthal, A. H., and Chen, T. C. (2015). Quinoline-based antimalarial drugs: a novel class of autophagy inhibitors. Neurosurg. Focus. 2015:E12. doi: 10.3171/2014.12.FOCUS14748
Graham, R. L., Donaldson, E. F., and Baric, R. S. (2013). A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 11, 836–848. doi: 10.1038/nrmicro3143
Guo, H., Huang, M., Yuan, Q., Wei, Y., Gao, Y., Mao, L., et al. (2017). The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS ONE 12:e0170123. doi: 10.1371/journal.pone.0170123
Hamming, I., Timens, W., Bulthuis, M. L., Lely, A. T., Navis, G., and van Goor, H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus, A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637. doi: 10.1002/path.1570
Hogue, I. B., Llewellyn, G. N., and Ono, A. (2012). Dynamic association between HIV-1 gag and membrane domains. Mol. Biol. Int. 2012:979765. doi: 10.1155/2012/979765
Jheng, J. R., Ho, J. Y., and Horng, J. T. (2014). ER stress, autophagy, and RNA viruses. Front. Microbiol. 5:388. doi: 10.3389/fmicb.2014.00388
Khandia, R., Dadar, M., Munjal, A., Dhama, K., Karthik, K., Tiwari, R., et al. (2019). A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells 8:674. doi: 10.3390/cells8070674
Kindrachuk, J., Ork, B., Hart, B. J., Mazur, S., Holbrook, M. R., Frieman, M. B., et al. (2015). Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 59, 1088–1099. doi: 10.1128/aac.03659-14
Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966. doi: 10.1056/NEJMoa030781
Ktistakis, N. T., and Tooze, S. A. (2016). Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 26, 624–635. doi: 10.1016/j.tcb.2016.03.006
Kudchodkar, S. B., and Levine, B. (2009). Viruses and autophagy. Rev. Med. Virol. 19, 359–378. doi: 10.1002/rmv.630
Lakadamyali, M., Rust, M. J., and Zhuang, X. (2004). Endocytosis of influenza viruses. Microbes Infect. 6, 929–936. doi: 10.1016/j.micinf.2004.05.002
Lee, C. J., Lin, H. R., Liao, C. L., and Lin, Y. L. (2008). Cholesterol effectively blocks entry of flavivirus. J. Virol. 82, 6470–6480. doi: 10.1128/JVI.00117-08
Lennemann, N. J., and Coyne, C. B. (2015). Catch me if you can: the link between autophagy and viruses. PLoS Pathog. 11:e1004685. doi: 10.1371/journal.ppat.1004685
Li, G. M., Li, Y. G., Yamate, M., Li, S. M., and Ikuta, K. (2007). Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 9, 96–102. doi: 10.1016/j.micinf.2006.10.015
Lin, C. M., Saif, L. J., Marthaler, D., and Wang, Q. (2016). Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 226, 20–39. doi: 10.1016/j.virusres.2016.05.023
Liu, Y., Luo, X., Shan, H., Fu, Y., Gu, Q., Zheng, X., et al. (2019). Niclosamide triggers non-canonical LC3 lipidation. Cells 8:248. doi: 10.3390/cells8030248
Loftsson, T., and Brewster, M. E. (2012). Cyclodextrinsasfunctionalexcipients: methods to enhancecomplexationefficiency. J. Pharm. Sci. 101, 3019–3032. doi: 10.1002/jps.23077
Lu, Y., Liu, D. X., and Tam, J. P. (2008). Lipid rafts are involved in SARS-CoV entry into vero E6 cells. Biochem. Biophys. Res. Commun. 369, 344–349. doi: 10.1016/j.bbrc.2008.02.023
Mackenzie, J. M., Khromykh, A. A., and Parton, R. G. (2007). Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2, 229–239. doi: 10.1016/j.chom.2007.09.003
Mahévas, M., Tran, V. T., Roumier, M., Chabrol, A., Paule, R., Guillaud, C., et al. (2020). Clinical efficacy of hydroxychloroquine in patients with covid-19 pneumonia who require oxygen: observational comparative study using routine care data. BMJ 369:m1844. doi: 10.1136/bmj.m1844
Mao, J., Lin, E., He, L., Yu, J., Tan, P., and Zhou, Y. (2019). Autophagy and viral infection. Adv. Exp. Med. Biol. 1209, 55–78. doi: 10.1007/978-981-15-0606-2_5
Martínez-Gutierrez, M., Castellanos, J. E., and Gallego-Gómez, J. C. (2011). Statins reduce dengue virus production via decreased virion assembly. Intervirology 54, 202–216. doi: 10.1159/000321892
Matarrese, P., Garofalo, T., Manganelli, V., Gambardella, L., Marconi, M., Grasso, M., et al. (2014). Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy 10, 750–765. doi: 10.1080/15548627.2016.1160971
McGuinn, K. P., and Mahoney, M. G. (2014). Lipid rafts and detergent-resistant membranes in epithelial keratinocytes. Methods Mol. Biol. 1195, 133–144. doi: 10.1007/7651_2014_71
Mizushima, N., and Komatsu, M. (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. doi: 10.1016/j.cell.2011.10.026
Nabi, I. R., and Le, P. U. (2003). Caveolae/raft-dependent endocytosis. J. Cell Biol. 161, 673–677. doi: 10.1083/jcb.200302028
Nomura, R., Kiyota, A., Suzaki, E., Kataoka, K., Ohe, Y., Miyamoto, K., et al. (2004). Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 78, 8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004
O’Neill, P. M., Bray, P. G., Hawley, S. R., Ward, S. A., and Park, B. K. (1998). 4-Aminoquinolines-past, present, and future: a chemical perspective. Pharmacol. Ther. 77, 29–58. doi: 10.1016/s0163-7258(97)00084-3
Ono, A., and Freed, E. O. (2001). Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U.S.A. 98, 13925–13930. doi: 10.1073/pnas.241320298
Owczarek, K., Szczepanski, A., Milewska, A., Baster, Z., Rajfur, Z., Sarna, M., et al. (2018). Early events during human coronavirus OC43 entry to the cell. Sci. Rep. 8, 7124. doi: 10.1038/s41598-018-25640-0
Patel, H. H., and Insel, P. A. (2009). Lipid rafts and caveolae and their role in compartmentation of redox signaling. Antioxid. Redox. Signal. 11, 1357–1372. doi: 10.1089/ars.2008.2365
Paules, C. I., Marston, H. D., and Fauci, A. S. (2020). Coronavirus infections-more than just the common cold. JAMA 323, 707–708.
Pelkmans, L. (2005). Secrets of caveolae- and lipid raft-mediated endocytosis revealed by mammalian viruses. Biochim. Biophys. Acta 1746, 295–304. doi: 10.1016/j.bbamcr.2005.06.009
Prentice, E., Jerome, W. G., Yoshimori, T., Mizushima, N., and Denison, M. R. (2004). Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279, 10136–10141. doi: 10.1074/jbc.M306124200
Razani, B., Rubin, C. S., and Lisanti, M. P. (1999). Regulation of cAMP-mediated signal transduction via interaction of caveolins with the catalytic subunit of protein kinase A. J. Biol. Chem. 274, 26353–26360. doi: 10.1110/ps.051911706
Riethmüller, J., Riehle, A., Grassmé, H., and Gulbins, E. (2006). Membrane rafts in host-pathogen interactions. Biochim. Biophys. Acta 1758, 2139–2147. doi: 10.1016/j.bbamem.2006.07.017
Rodrigues-Diez, R. R., Tejera-Muñoz, A., Marquez-Exposito, L., Rayego-Mateos, S., Santos Sanchez, L., Marchant, V., et al. (2020). Statins: could an old friend help the fight against COVID-19? Br. J. Pharmacol. doi: 10.1111/bph.15166 [Epub ahead of print].
Romanelli, F., Smith, K. M., and Hoven, A. D. (2004). Chloroquine and hydroxychloroquine as inhibitors of human immunodeficiency virus (HIV-1) activity. Curr. Pharm. Des. 10, 2643–2648. doi: 10.2174/1381612043383791
Rosenberg, E. S., Dufort, E. M., Udo, T., Wilberschied, L. A., Kumar, J., Tesoriero, J., et al. (2020). Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. JAMA 323, 2493–2492. doi: 10.1001/jama.2020.8630
Rosenberger, C. M., Brumell, J. H., and Finlay, B. B. (2000). Microbial pathogenesis: lipid rafts as pathogen portals. Curr. Biol. 10, R823–R825. doi: 10.1016/s0960-9822(00)00788-0
Rubinsztein, D. C., Cuervo, A. M., Ravikumar, B., Sarkar, S., Korolchuk, V., Kaushik, S., et al. (2009). In search of an autophagomometer. Autophagy 5, 585–589. doi: 10.4161/auto.5.5.8823
Sánchez, E. G., Pérez-Núñez, D., and Revilla, Y. (2017). Mechanisms of entry and endosomal pathway of african swine fever virus. Vaccines 5:42. doi: 10.3390/vaccines5040042
Sargiacomo, M., Sudol, M., Tang, Z. L., and Lisanti, M. P. (1993). Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J. Cell Biol. 122, 789–807. doi: 10.1083/jcb.122.4.789
Shakya, A., Bhat, H. R., and Ghosh, S. K. (2018). Update on nitazoxanide: a multifunctional chemotherapeutic agent. Curr. Drug Discov. Technol. 15, 201–213. doi: 10.2174/1570163814666170727130003
Shi, Y., Tan, S. H., Ng, S., Zhou, J., Yang, N. D., Koo, G. B., et al. (2015). Critical role of CAV1/caveolin-1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy 11, 769–784. doi: 10.1080/15548627.2015.1034411
Simmons, G., Zmora, P., Gierer, S., Heurich, A., and Pöhlmann, S. (2013). Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 100, 605–614. doi: 10.1016/j.antiviral.2013.09.028
Simons, K., and Ehehalt, R. (2002). Cholesterol, lipid rafts, and disease. J. Clin. Invest. 110, 597–603. doi: 10.1172/JCI16390
Simons, K., and Toomre, D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39. doi: 10.1038/35036052
Sobo, K., Chevallier, J., Parton, R. G., Gruenberg, J., and Van Der Goot, F. G. (2007). Diversity of raft-like domains in late endosomes. PLoS ONE 2:391. doi: 10.1371/journal.pone.0000391
Song, K. S., Sargiacomo, M., Galbiati, F., Parenti, M., and Lisanti, M. P. (1997). Targeting of a G alpha subunit (Gi1 alpha) and c-Src tyrosine kinase to caveolae membranes: clarifying the role of N-myristoylation. Cell. Mol. Biol. (Noisy-le-grand). 43, 293–303.
Spriggs, C. C., Harwood, M. C., and Tsai, B. (2019). How non-enveloped viruses hijack host machineries to cause infection. Adv. Virus Res. 104, 97–122. doi: 10.1016/bs.aivir.2019.05.002
Szczepanski, A., Owczarek, K., Milewska, A., Baster, Z., Rajfur, Z., Mitchell, J. A., et al. (2018). Canine respiratory coronavirus employs caveolin-1-mediated pathway for internalization to HRT-18G cells. Vet Res. 49, 55. doi: 10.1186/s13567-018-0551-9
Takahashi, T., and Suzuki, T. (2011). Function of membrane rafts in viral lifecycles and host cellular response. Biochem. Res. Int. 2011:245090. doi: 10.1155/2011/245090
Thorp, E. B., and Gallagher, T. M. (2004). Requirements for CEACAMs and cholesterol during murine coronavirus cell entry. J. Virol. 78, 2682–2692. doi: 10.1128/jvi.78.6.2682-2692.2004
Wang, H., Sun, Y., Mao, X., Meng, C., Tan, L., Song, C., et al. (2018). Infectious bronchitis virus attaches to lipid rafts and enters cells via clathrin mediated endocytosis. bioRxiv [Preprint] doi: 10.1101/352898
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9. doi: 10.1016/j.cell.2020.03.045
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/JVI.06540-11
Wu, C. H., Yeh, S. H., Tsay, Y. G., Shieh, Y. H., Kao, C. L., Chen, Y. S., et al. (2009). Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 284, 5229–5239. doi: 10.1074/jbc.M805747200
Wudiri, G. A., Schneider, S. M., and Nicola, A. V. (2017). Herpes simplex virus 1 envelope cholesterol facilitates membrane fusion. Front. Microbiol. 8:2383. doi: 10.3389/fmicb.2017.02383
Xu, J., Shi, P. Y., Li, H., and Zhou, J. (2020). Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 6, 909–915. doi: 10.1021/acsinfecdis.0c00052
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762
Yang, C. W., Peng, T. T., and Hsu, H. Y. (2020). Repurposing old drugs as antiviral agents for coronaviruses. Biomed. J. doi: 10.1016/j.bj.2020.05.003 [Epub ahead of print].
Ye, Z. W., Yuan, S., Yuen, K. S., Fung, S. Y., Chan, C. P., and Jin, D. Y. (2020). Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 16, 1686–1697. doi: 10.7150/ijbs.45472
Yu, B., Wang, D. W., and Li, C. (2020). Hydroxychloroquine application is associated with a decreased mortality in critically ill patients with COVID-19. medRxiv [Preprint] doi: 10.1101/2020.04.27.20073379
Yu, L., Chen, Y., and Tooze, S. A. (2018). Autophagy pathway: cellular and molecular mechanisms. Autophagy 14, 207–215. doi: 10.1080/15548627.2017.1378838
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., and Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. doi: 10.1056/NEJMoa1211721
Zhang, J., Xie, B., and Hashimoto, K. (2020). Current status of potential therapeutic candidates for the COVID-19 crisis. Brain Behav. Immun. 87, 59–73. doi: 10.1016/j.bbi.2020.04.046
Zhao, L., Jha, B. K., Wu, A., Elliott, R., Ziebuhr, J., Gorbalenya, A. E., et al. (2012). Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11, 607–616. doi: 10.1016/j.chom.2012.04.011
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Zidovetzki, R., and Levitan, I. (2007). Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim. Biophys. Acta 1768, 1311–1324. doi: 10.1016/j.bbamem.2007.03.026
Zou, D., Xu, J., Duan, X., Xu, X., Li, P., Cheng, L., et al. (2019). Porcine epidemicdiarrhea virus ORF3 proteincausesendoplasmicreticulum stress to facilitate autophagy. Vet. Microbiol. 235, 209–219. doi: 10.1016/j.vetmic.2019.07.005
Keywords: coronavirus, lipid rafts, autophagy, SARS-CoV-2, drugs
Citation: Fecchi K, Anticoli S, Peruzzu D, Iessi E, Gagliardi MC, Matarrese P and Ruggieri A (2020) Coronavirus Interplay With Lipid Rafts and Autophagy Unveils Promising Therapeutic Targets. Front. Microbiol. 11:1821. doi: 10.3389/fmicb.2020.01821
Received: 30 April 2020; Accepted: 10 July 2020;
Published: 11 August 2020.
Edited by:
Joseph Alex Duncan, The University of North Carolina at Chapel Hill, United StatesReviewed by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranRoberta Olmo Pinheiro, Oswaldo Cruz Foundation, Brazil
Copyright © 2020 Fecchi, Anticoli, Peruzzu, Iessi, Gagliardi, Matarrese and Ruggieri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola Matarrese, cGFvbGEubWF0YXJyZXNlQGlzcy5pdA==
†These authors have contributed equally to this work
 Katia Fecchi
Katia Fecchi Simona Anticoli
Simona Anticoli Daniela Peruzzu
Daniela Peruzzu Elisabetta Iessi
Elisabetta Iessi Maria Cristina Gagliardi
Maria Cristina Gagliardi Paola Matarrese
Paola Matarrese Anna Ruggieri
Anna Ruggieri