
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 24 July 2020
Sec. Virology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01795
Interest in coronaviruses because of the 2019 novel coronavirus (SARS-CoV-2) pandemic has generated concern about their occurrence and persistence in aquatic habitats. Coronaviruses are not quantitatively significant constituents of marine virioplankton. Members of the Nidovirales (to which human coronaviruses belong) infect marine mammals, teleosts and possibly invertebrates, and human coronaviruses may persist in marine plankton receiving wastewater effluent. However, virions likely experience significant particle and infectivity decay rates in surface seawater, similar to other enveloped RNA viruses.
The current 2019 novel coronavirus (SARS-CoV-2) pandemic has generated interest and concern about the occurrence and persistence of coronaviruses in aquatic habitats. While the distal origin of SARS-CoV-2 is still undetermined, but likely terrestrial (bats or other terrestrial animals; Andersen et al., 2020) coronaviruses also occur in aquatic mammals and other metazoa as pathogens. Given reports that SARS-CoV-2 can be shed from patients in feces for prolonged periods (Gu et al., 2020; Hindson, 2020; Wu et al., 2020; Yeo et al., 2020; Zhang et al., 2020) and like other human coronaviruses (Bibby and Peccia, 2013) can be detected in wastewater management facilities (Lodder and De Roda Husman, 2020) it is possible that SARS-CoV-2 may be introduced to aquatic habitats through sewage outfall and contact with infected recreational users. The purpose of this mini-review is to summarize currently known marine (and more generally aquatic) coronaviruses, their presence in plankton, and their potential persistence in aquatic habitats.
Coronaviruses belong to the order Nidovirales, a group of viruses rapidly expanding in number mainly as a result of a surge in metatranscriptomic sequencing studies (Shi et al., 2016). Coronavirus virions are large (120–160 nm), and characterized by club-shaped projections on their surface. They bear a ssRNA genome of 25–32 kb, on which (from 5′ to 3′) there are typically two open reading frames encoding non-structural genes, followed by structural genes. Replication of coronaviruses occurs by receptor mediated endocytosis, followed by cytoplasmic replication and assembly of mature virions at the endoplastic reticulum surface, and release by exocytosis (Brian and Baric, 2005). The Nidovirales is currently made up of eight suborders (Abnidovirineae, Arnidovirineae, Cornidovirineae, Mesnidovirineae, Monidovirineae, Nanidovirineae, Ronidovirineae and Tornidovirineae) (King et al., 2018) and virus classification is verified by concatenation and phylogenetic analysis of five protein encoding domains. SARS-CoV-2 belongs to the family Coronaviridae, subfamily Orthocoronavirinae and genus Betacoronavirus.
To date there have been no Betacoronaviruses recovered from any marine animal. However, Alphacoronaviruses and Gammacoronaviruses are described in marine mammals (reviewed in Schütze, 2016; Bossart and Duignan, 2018). These include the Harbor Seal Alphacoronavirus (Bossart and Schwartz, 1990), Pacific Harbor Seal Gammacoronavirus (Nollens et al., 2010), Beluga Whale Gammacoronavirus (Mihindukulasuriya et al., 2008) and the Bottlenose Dolphin Gammacoronavirus (Woo et al., 2014). These viruses are distantly related to SARS-CoV-2. Gammacoronaviruses and Alphacoronaviruses share little homology with SARS-CoV-2 (Figure 1) but are associated with respiratory diseases in pinnipeds and cetaceans. Gammacoronaviruses and Alphacoronaviruses are associated with respiratory secretions and possibly associated with pneumonia in seals (Nollens et al., 2010; Ng et al., 2011) and respiratory disease in cetaceans (Mihindukulasuriya et al., 2008) however, firm pathology has not been established. Wild birds are known viral reservoirs, and birds which live mainly in the marine environment are also known to harbor coronaviruses. For instance, a novel coronavirus within the Gammacoronaviruses was identified in American herring and great black backed gulls (Canuti et al., 2019). Interestingly, these sea bird coronaviruses are within the same clade as marine mammal coronaviruses (Figure 1), suggesting that in the past, transmission between these animals has occurred. Additionally, and perhaps suggesting that the diversity in the marine environment is not yet fully understood, a Nidovirus (PsNV) recovered from Pacific Salmon shares greater similarity with coronaviruses than to other fish or invertebrate nidoviruses. Interestingly, PsNV was localized via in situ hybridization to gill tissue, suggesting a similar tissue tropism and infection strategy to other respiratory coronaviruses (Mordecai et al., 2019, 2020). The morphology, genome organization, and replication of marine gammacoronaviruses is highly similar to human coronaviruses (Mihindukulasuriya et al., 2008; Woo et al., 2014). While no marine gammacoronavirus has been cultivated, the avian gammacoronavirus Infectious Bronchitis Virus (IBV) follows similar entry, replication and shedding as human coronaviruses. While SARS-CoV-2 bind to angiotensin converting enzyme 2 (ACE2) (Yan et al., 2020) IBV binds to sialic acid, and while both infect primarily respiratory tissues, they may have wide tropism and infect multiple organ systems (Winter et al., 2006; Promkuntod et al., 2014; Bande et al., 2016).
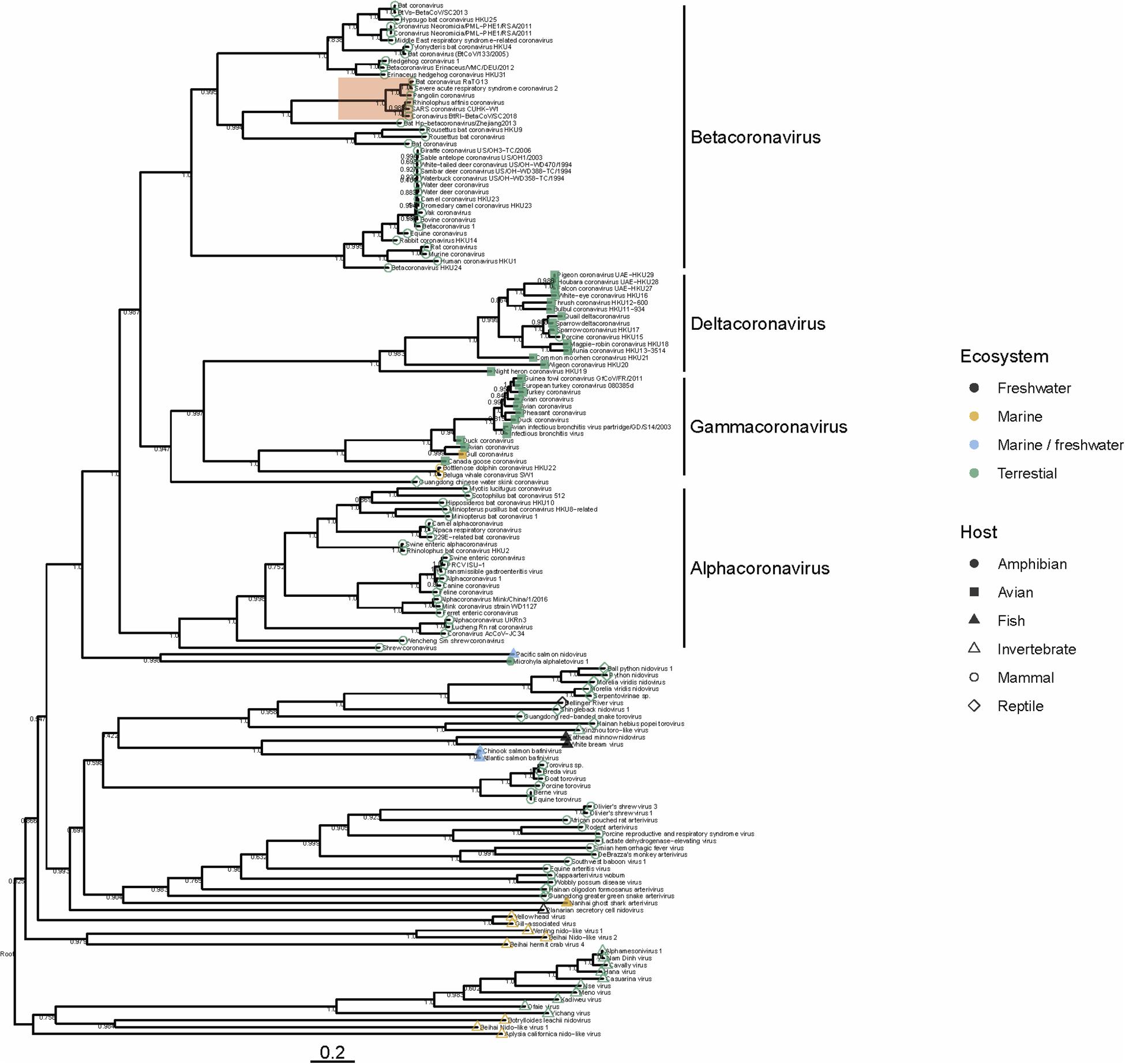
Figure 1. Phylogeny of representative Nidovirales based on ORF1a (replicase) amino acid sequences. Closely related sequences were removed. Tip points are colored by ecosystem and shaped by host. The clade highlighted in red shows the phylogenetic placement of SARS-CoV-2 and closely related viruses. Branches are scaled to the number of amino acid substitutions per site and the tree is mid-point rooted. Node values show FastTree support values (ranging from 0 to 1). Amino acid alignments were carried out using MAFFT (Katoh and Standley, 2013) the tree was built using FastTree (Price et al., 2010) and visualized using ggtree (Yu et al., 2017).
Several nido-like viruses have been detected in invertebrates in the marine environment (Shi et al., 2016; Bukhari et al., 2018). Screening of RNA-specific viral metagenomes prepared from asteroids (NCBI Accessions PRJNA253121 and SAMN08012637 – SAMN08012651; Hewson et al., 2014, 2018b) yielded no definitive coronavirus-like sequences, though weak matches based on amino acid homology were found (Hewson et al., 2018b). Analysis of representative Nidovirales diversity shows that the majority of viral discovery studies are focused on terrestrial mammalian and avian hosts, but the few aquatic representatives in the Nidovirales are represented across the tree (Figure 1), suggesting that their absence might be due to inadequate sampling, rather than a restricted host range. It is becoming clear that despite many newly discovered viruses remain unclassified, members of the family Coronaviridae are no longer limited to viruses of birds and mammals, but also invertebrates (Shi et al., 2018) reptiles (Shi et al., 2018) amphibia (Bukhari et al., 2018) and perhaps also fish (Mordecai et al., 2019; Figure 1). Our analysis suggests that nidoviruses are likely found in all animals, and that much broader sampling and metatranscriptomic sequencing of marine organisms is required to more fully understand the diversity of viruses found in this environment.
To assess the occurrence of coronaviruses as free virions in plankton (i.e., virioplankton), we screened plankton (>0.2 μm) metatranscriptomes of the New York Finger Lakes (NCBI accessions SRR6281416 – SRR6281423), viral metagenomes the Anacostia River in Washington DC (NCBI accession PRJNA637530), along with published RNA viromes from marine and freshwater virioplankton (Culley et al., 2003, 2006, 2014; Djikeng et al., 2009; Lang et al., 2009; Lopez-Bueno et al., 2015; Hewson et al., 2018a) by BLAST, but these yielded no definitive Orthocoronavirus-like sequences. Zeigler et al. (2017) reported coronaviruses-like sequences in plankton recovered from the Baltic Sea through combined metatranscriptome and virome sampling, but concluded these were likely a result of contamination from human sources. While these libraries represent a tiny fraction of the total diversity of aquatic habitats and aquatic animal diversity, it can be inferred that coronaviruses do not represent numerically significant constituents of virioplankton communities.
As SARS-CoV-2 is likely to be released to the marine environment via human effluent, it is important to understand the impact this might have on marine life (if any). Wastewater surveillance strategies to detect SARS-CoV-2 are currently formulated to track human epidemiology, since these precede case loads in the human population (Daughton, 2020; Randazzo et al., 2020). Given the detection of SARS-CoV-2 nucleic acids in wastewater (Ahmed et al., 2020; Lodder and De Roda Husman, 2020; Medema et al., 2020; Orive et al., 2020) coronaviruses may be introduced into aquatic habitats through urban or agricultural runoff or in wastewater effluent. Indeed, emerging reports at the time of publication indicate the presence of SARS-CoV-2 in river water receiving untreated human sewage (Guerrero-Latorre et al., 2020; Haramoto et al., 2020; Rimoldi et al., 2020). Human coronaviruses (HCoV) experience a 99% loss in infectivity in primary and secondary treated wastewater effluent after 1.9–2.4 days (Gundy et al., 2009). A study by Ye et al. (2016) found that murine hepatitis virus (MHV; coronavirus) experienced 90% reduction in infectivity after 13 h in raw wastewater at 25oC, but infectivity was maintained longer (36 h) at 10oC. Taken together, these studies illustrate that intact coronavirus particles may survive in wastewater after excretion, and may be present in coastal waters after discharge. However, coronavirus particles are likely to experience considerable particle decay and loss of infectivity after arriving in aquatic habitats. Virus-like particles, which include both viruses of eukaryotes and phage, experience particle decay in seawater at rates of 2–4 % h–1 (Heldal and Bratbak, 1991) which is generally considered to be a result of sunlight (UV-C radiation) (Wommack et al., 1996; Wilhelm et al., 1998) and through interaction with heat-labile organic matter which may include nucleases and proteases present in marine microorganisms and free in the environment (Noble and Fuhrman, 1997). Different viral groups may have different particle decay rates (Wilhelm et al., 1998) and decay of virus particles is a different process to loss of viral infectivity (Wommack et al., 1996). The decay of coronaviruses in natural waters has not been studied. Several fish pathogens, including viral haemorrhagic septicemia virus (VHSV; Rhabdovirus), infectious salmon anemia virus (ISAV; Orthomyxovirus), Salmon Alphavirus (SA; Togavirus), and Infectious Hematopoeitic Necrosis Virus (IHNV: Rhabdovirus) are enveloped RNA viruses (Crane and Hyatt, 2011) which may be detected in fish pens or holding tanks (Pinto et al., 1993) and natural waters (Lovdal and Enger, 2002). Oye and Rimstad (2001) reported a 3 log reduction in ISAV and VHSV titers after approximately 50 and 9 h, respectively, in sterilized seawater and freshwater. They also found that fish pen water enhanced viral survival compared to sterilized water. Skjold (2014) reported 3 log reductions in SA titer in 12 h. Garver et al. (2013) found that IHNV experienced threefold declines in viral titer <24 h, and that viral decay was much less in open aquaculture compared to closed aquaculture systems or sterile water. Kocan et al. (2001) reported a 50% loss of viral activity in VHSV when inoculated into natural seawater. Hence, viral infectivity decay rates of enveloped RNA viruses may be similar to those of virioplankton in general. The high decay rates present in seawater (detailed above) and high dilution rates suggest coronaviruses may not persist for long periods in natural waters, which would help to minimize the risk of infecting any potential susceptible hosts in the marine environment that could act as animal reservoirs of the virus. However, it is important to note that surviving virions may potentially infect marine mammals, since cetaceans and terrestrial mammals share similar receptor binding domains on ACE2 (Luan et al., 2020; Nabi and Khan, 2020) which may be especially pronounced when such species occur near urban wastewater outfalls. Marine aerosols may also represent another mechanism of re-introduction to terrestrial habitats downstream of wastewater outfalls (Baylor et al., 1977).
In summary, coronaviruses occur uncommonly in marine and freshwater ecosystems as free virions, but this could be because the true diversity in aquatic reservoirs is not well explored. There may be unrecognized coronaviruses infecting marine metazoa that are currently under-sampled relative to terrestrial counterparts. Introduced coronaviruses, such as SARS-CoV-2, may be present in coastal marine waters which are affected by sewage effluent, where they are subject to physical decay and loss of infectivity at rates similar to other aquatic viruses. Monitoring of SARS-CoV-2 in sewage outfalls into these habitats is recommended, since it may provide guidance to recreational users and fisheries to assess risk, especially when such viruses may be concentrated, e.g., by filter feeding organisms or by onshore winds. More study is needed to understand the natural diversity of coronaviruses in marine metazoa through broad viral surveys.
GM and IH wrote the manuscript. Both authors contributed to the article and approved the submitted version.
This work was supported by NSF Grants OCE- 1737127 and OCE- 1537111 to IH. GM is supported by the Liber Ero Fellowship Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Curtis Suttle for comments on an early manuscript draft.
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O’brien, J. W., et al. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728:138764. doi: 10.1016/j.scitotenv.2020.138764
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., and Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452. doi: 10.1038/s41591-020-0820-9
Bande, F., Arshad, S. S., Omar, A. R., Bejo, M. H., Abubakar, M. S., and Abba, Y. (2016). Pathogenesis and diagnostic approaches of avian infectious bronchitis. Adv. Virol. 2016:4621659.
Baylor, E., Baylor, M., Blanchard, D., Syzdek, L., and Appel, C. (1977). Virus transfer from surf to wind. Science 198, 575–580. doi: 10.1126/science.918656
Bibby, K., and Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 47, 1945–1951. doi: 10.1021/es305181x
Bossart, G. D., and Schwartz, J. C. (1990). Acute necrotizing enteritis associated with suspected coronavirus infection in three harbor seals (Phoca vitulina). J. Zoo Wildlife Med. 21, 84–87.
Brian, D. A., and Baric, R. S. (2005). “Coronavirus genome structure and replication,” in Coronavirus Replication and Reverse Genetics, ed. L. Enjuanes (Berlin: Springer), 1–30. doi: 10.1007/3-540-26765-4_1
Bukhari, K., Mulley, G., Gulyaeva, A. A., Zhao, L., Jiang, J., and Neuman, B. W. (2018). Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology 524, 160–171. doi: 10.1016/j.virol.2018.08.010
Canuti, M., Kroyer, A. N. K., Ojkic, D., Whitney, H. G., Robertson, G. J., and Lang, A. S. (2019). Discovery and characterization of novel RNA viruses in aquatic North American wild birds. Viruses 11:768. doi: 10.3390/v11090768
Crane, M., and Hyatt, A. (2011). Viruses of fish: an overview of significant pathogens. Viruses 3, 2025–2046. doi: 10.3390/v3112025
Culley, A., Lang, A. S., and Suttle, C. A. (2003). High diversity of unknown picorna-like viruses in the sea. Nature 424, 1054–1057. doi: 10.1038/nature01886
Culley, A. I., Lang, A. S., and Suttle, C. A. (2006). Metagenomic analysis of coastal RNA virus communities. Science 312, 1795–1798. doi: 10.1126/science.1127404
Culley, A. I., Mueller, J. A., Belcaid, M., Wood-Charlson, E. M., Poisson, G., and Steward, G. F. (2014). The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. mBio 5:e01210-14.
Daughton, C. G. (2020). Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 736:139631. doi: 10.1016/j.scitotenv.2020.139631
Djikeng, A., Kuzmickas, R., Anderson, N. G., and Spiro, D. J. (2009). Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One 4:e7264. doi: 10.1371/journal.pone.0007264
Garver, K. A., Mahony, A. A., Stucchi, D., Richard, J., Van Woensel, C., and Foreman, M. (2013). Estimation of parameters influencing waterborne transmission of infectious hematopoietic necrosis virus (IHNV) in Atlantic salmon (Salmo salar). PLoS ONE 8:e82296. doi: 10.1371/journal.pone.0082296
Gu, J., Han, B., and Wang, J. (2020). COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology 158, 1518–1519. doi: 10.1053/j.gastro.2020.02.054
Guerrero-Latorre, L., Ballesteros, I., Villacres, I., Granda-Albuja, M. G., Freire, B., and Rios-Touma, B. (2020). First SARS-CoV-2 detection in river water: implications in low sanitation countries. medRxiv [Preprint]. doi: 10.1101/2020.06.14.20131201
Gundy, P. M., Gerba, C. P., and Pepper, I. L. (2009). Survival of coronaviruses in water and wastewater. Food Environ. Virol. 1, 10–14.
Haramoto, E., Malla, B., Thakali, O., and Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 737:140405. doi: 10.1016/j.scitotenv.2020.140405
Heldal, M., and Bratbak, G. (1991). Production and decay of viruses in aquatic environments. Mar. Ecol. Prog. Ser. 72, 205–212. doi: 10.3354/meps072205
Hewson, I., Bistolas, K. S. I., Button, J. B., and Jackson, E. W. (2018a). Occurrence and seasonal dynamics of RNA viral genotypes in three contrasting temperate lakes. PLoS One 13:e0194419. doi: 10.1371/journal.pone.0194419
Hewson, I., Bistolas, K. S. I., Carde, E. M. Q., Button, J. B., Foster, P. J., Flanzenbaum, J. M., et al. (2018b). Investigating the complex association between viral ecology, environment, and Northeast Pacific sea star wasting. Front. Mar. Sci. 5:77. doi: 10.3389/fmars.2018.00077
Hewson, I., Button, J. B., Gudenkauf, B. M., Miner, B., Newton, A. L., Gaydos, J. K., et al. (2014). Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. U.S.A. 111, 17276–17283.
Hindson, J. (2020). COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 17:259. doi: 10.1038/s41575-020-0295-7
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
King, A. M. Q., Lefkowitz, E. J., Mushegian, A. R., Adams, M. J., Dutilh, B. E., Gorbalenya, A. E., et al. (2018). Changes to taxonomy and the International code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2018). Arch. Virol. 163, 2601–2631.
Kocan, R. M., Hershberger, P. K., and Elder, N. E. (2001). Survival of the North American strain of viral hemorrhagic septicemia virus (VHSV) in filtered seawater and seawater containing ovarian fluid, crude oil and serum-enriched culture medium. Dis. Aquat. Organ. 44, 75–78. doi: 10.3354/dao044075
Lang, A. S., Rise, M. L., Culley, A. I., and Steward, G. F. (2009). RNA viruses in the sea. FEMS Microbiol. Rev. 33, 295–323.
Lodder, W., and De Roda Husman, A. M. (2020). SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 5, 533–534. doi: 10.1016/s2468-1253(20)30087-x
Lopez-Bueno, A., Rastrojo, A., Peiro, R., Arenas, M., and Alcami, A. (2015). Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Mol. Ecol. 24, 4812–4825. doi: 10.1111/mec.13321
Lovdal, T., and Enger, O. (2002). Detection of infectious salmon anemia virus in sea water by nested RT-PCR. Dis. Aquat. Organ. 49, 123–128. doi: 10.3354/dao049123
Luan, J., Jin, X., Lu, Y., and Zhang, L. (2020). SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. doi: 10.1002/jmv.25817 [Epub ahead of print].
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., and Brouwer, A. (2020). Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 7, 511–516.
Mihindukulasuriya, K. A., Wu, G., St Leger, J., Nordhausen, R. W., and Wang, D. (2008). Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82, 5084–5088. doi: 10.1128/jvi.02722-07
Mordecai, G. J., Di Cicco, E., Günther, O. P., Schulze, A. D., Kaukinen, K. H., Li, S., et al. (2020). Emerging viruses in British Columbia salmon discovered via a viral immune response biomarker panel and metatranscriptomic sequencing. bioRxiv [Preprint]. doi: 10.1101/2020.02.13.948026
Mordecai, G. J., Miller, K. M., Di Cicco, E., Schulze, A. D., Kaukinen, K. H., Ming, T. J., et al. (2019). Endangered wild salmon infected by newly discovered viruses. eLife 8:e47615.
Nabi, G., and Khan, S. (2020). Risk of COVID-19 pneumonia in aquatic mammals. Environ. Res. 188, 109732–109732.
Ng, F. F. T., Wheeler, E., Greig, D., Waltzek, T. B., Gulland, F., and Breitbart, M. (2011). Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J. Gen. Virol. 92, 1318–1323. doi: 10.1099/vir.0.029678-0
Noble, R. T., and Fuhrman, J. A. (1997). Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 63, 77–83. doi: 10.1128/aem.63.1.77-83.1997
Nollens, H. H., Wellehan, J. F., Archer, L., Lowenstine, L. J., and Gulland, F. M. (2010). Detection of a respiratory coronavirus from tissues archived during a pneumonia epizootic in free-ranging Pacific harbor seals Phoca vitulina richardsii. Dis. Aquat. Organ. 90, 113–120. doi: 10.3354/dao02190
Orive, G., Lertxundi, U., and Barcelo, D. (2020). Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 732:139298. doi: 10.1016/j.scitotenv.2020.139298
Oye, A. K., and Rimstad, E. (2001). Inactivation of infectious salmon anaemia virus, viral haemorrhagic septicaemia virus and infectious pancreatic necrosis virus in water using UVC irradiation. Dis. Aquat. Organ. 48, 1–5. doi: 10.3354/dao048001
Pinto, R. M., Jofre, J., Abad, F. X., Gonzalez-Dankaart, J. F., and Bosch, A. (1993). Concentration of fish enveloped viruses from large volumes of water. J. Virol. Methods 43, 31–40. doi: 10.1016/0166-0934(93)90087-8
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Promkuntod, N., Van Eijndhoven, R. E., De Vrieze, G., Gröne, A., and Verheije, M. H. (2014). Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology 448, 26–32. doi: 10.1016/j.virol.2013.09.018
Randazzo, W., Truchado, P., Cuevas-Ferrando, E., Simón, P., Allende, A., and Sánchez, G. (2020). SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 181:115942. doi: 10.1016/j.watres.2020.115942
Rimoldi, S. G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., et al. (2020). Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. medRxiv [Preprint]. doi: 10.1101/2020.05.01.20086009
Schütze, H. (2016). “Chapter 20 - coronaviruses in aquatic organisms,” in Aquaculture Virology, eds F. S. B. Kibenge and M. G. Godoy (San Diego, CA: Academic Press), 327–335. doi: 10.1016/b978-0-12-801573-5.00020-6
Shi, M., Lin, X.-D., Chen, X., Tian, J.-H., Chen, L.-J., Li, K., et al. (2018). The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202. doi: 10.1038/s41586-018-0012-7
Shi, M., Lin, X. D., Tian, J. H., Chen, L. J., Chen, X., Li, C. X., et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. doi: 10.1038/nature20167
Skjold, P. (2014). Survival of Salmonid Alphavirus in Seawater Under Different Physical Conditions. Ph.D. thesis, University of Bergen, Bergen.
Wilhelm, S. W., Weinbauer, M. G., Suttle, C. A., and Jeffrey, W. H. (1998). The role of sunlight in the removal and repair of viruses in the sea. Limnol. Oceanogr. 43, 586–592. doi: 10.4319/lo.1998.43.4.0586
Winter, C., Schwegmann-Wessels, C., Cavanagh, D., Neumann, U., and Herrler, G. (2006). Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J. Gen. Virol. 87, 1209–1216. doi: 10.1099/vir.0.81651-0
Wommack, K. E., Hill, R. T., Muller, T. A., and Colwell, R. R. (1996). Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 62, 1336–1341. doi: 10.1128/aem.62.4.1336-1341.1996
Woo, P. C., Lau, S. K., Lam, C. S., Tsang, A. K., Hui, S. W., Fan, R. Y., et al. (2014). Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J. Virol. 88, 1318–1331. doi: 10.1128/jvi.02351-13
Wu, Y., Guo, C., Tang, L., Hong, Z., Zhou, J., Dong, X., et al. (2020). Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5, 434–435. doi: 10.1016/s2468-1253(20)30083-2
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762
Ye, Y., Ellenberg, R. M., Graham, K. E., and Wigginton, K. R. (2016). Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 50, 5077–5085. doi: 10.1021/acs.est.6b00876
Yeo, C., Kaushal, S., and Yeo, D. (2020). Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 5, 335–337. doi: 10.1016/s2468-1253(20)30048-0
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T.-Y. (2017). ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210x.12628
Zeigler, A. L., Mccrow, J. P., Ininbergs, K., Dupont, C. L., Badger, J. H., Hoffman, J. M., et al. (2017). The Baltic Sea Virome: diversity and transcriptional activity of DNA and RNA viruses. mSystems 2:e00125-16.
Keywords: coronavirus, Nidovirales, virome, virioplankton, disease
Citation: Mordecai GJ and Hewson I (2020) Coronaviruses in the Sea. Front. Microbiol. 11:1795. doi: 10.3389/fmicb.2020.01795
Received: 22 May 2020; Accepted: 09 July 2020;
Published: 24 July 2020.
Edited by:
Andrew S. Lang, Memorial University of Newfoundland, CanadaReviewed by:
Emily S. Bailey, Texas Tech University Health Sciences Center, United StatesCopyright © 2020 Mordecai and Hewson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian Hewson, aGV3c29uQGNvcm5lbGwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.