- 1Departamento de Micro y Nanotecnología, Instituto de Nanociencia y Nanotecnología – Comisión Nacional de Energía Atómica (CNEA), Buenos Aires, Argentina
- 2Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Buenos Aires, Argentina
- 3Departamento de Microbiología, Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario, Rosario, Argentina
Alternative sigma factors have led the core RNA polymerase (RNAP) to recognize different sets of promoters to those recognized by the housekeeping sigma A-directed RNAP. This change in RNAP promoter selectivity allows a rapid and flexible reformulation of the genetic program to face environmental and metabolic stimuli that could compromise bacterial fitness. The model bacterium Bacillus subtilis constitutes a matchless living system in the study of the role of alternative sigma factors in gene regulation and physiology. SigB from B. subtilis was the first alternative sigma factor described in bacteria. Studies of SigB during the last 40 years have shown that it controls a genetic universe of more than 150 genes playing crucial roles in stress response, adaption, and survival. Activation of SigB relies on three separate pathways that specifically respond to energy, environmental, and low temperature stresses. SigB homologs, present in other Gram-positive bacteria, also play important roles in virulence against mammals. Interestingly, during recent years, other unexpected B. subtilis responses were found to be controlled by SigB. In particular, SigB controls the efficiencies of spore and biofilm formation, two important features that play critical roles in adaptation and survival in planktonic and sessile B. subtilis communities. In B. subtilis, SigB induces the expression of the Spo0E aspartyl-phosphatase, which is responsible for the blockage of sporulation initiation. The upregulated activity of Spo0E connects the two predominant adaptive pathways (i.e., sporulation and stress response) present in B. subtilis. In addition, the RsbP serine-phosphatase, belonging to the energy stress arm of the SigB regulatory cascade, controls the expression of the key transcription factor SinR to decide whether cells residing in the biofilm remain in and maintain biofilm growth or scape to colonize new niches through biofilm dispersal. SigB also intervenes in the recognition of and response to surrounding microorganisms, a new SigB role that could have an agronomic impact. SigB is induced when B. subtilis is confronted with phytopathogenic fungi (e.g., Fusarium verticillioides) and halts fungal growth to the benefit of plant growth. In this article, we update and review literature on the different regulatory networks that control the activation of SigB and the new roles that have been described the recent years.
SigB as a Model of Alternative Sigma Factor Present in Bacteria
Gene expression is a fundamental process that is conserved from bacteria to humans. The first step in gene expression is transcription, which is performed by structurally conserved DNA-dependent RNA polymerases (RNAPs), resulting in the synthesis of an RNA molecule from a DNA template. In bacteria, a single species of RNAP is responsible for transcribing both stable RNA (i.e., t- and rRNA), small RNA (i.e., sRNA), and protein-encoding genes RNA (i.e., mRNA). By contrast, eukaryotic systems use three distinct RNAP species to transcribe the different gene classes (RNAP I transcribes most rRNA, RNAP II transcribes mRNA, and RNAP III transcribes tRNA and 5S rRNA) (Lewis et al., 2008). The prokaryotic core RNA polymerase (RNAP) is a large (∼400 kDa) multi-subunit enzyme comprising five (α2ββ’ω) subunits in a crab-claw-like structure.
Although little sequence homology exists between eubacterial RNAP, archaeal RNAP, and eukaryotic RNAPII, the crab-claw structure is remarkably conserved (Zhang et al., 1999; Cramer et al., 2001; Hirata et al., 2008). The two α subunits act as a scaffold to hold the catalytic β and β’ subunits together (Zhang et al., 1999). The exact role of the ω subunit is unclear, but it is related in both structure and sequence to the eukaryotic polymerase subunit Rpb6 (Minakhin et al., 2001). It appears to be responsible for controlling transcription in response to nutrient changes, correct folding of the β’ subunit, and its assembly into the core multi-subunit enzyme (Mukherjee et al., 1999; Vrentas et al., 2005; Chatterji et al., 2007).
The channel formed by β and β’ is referred to as the primary channel, which contains a deep positively charged cleft housing the enzyme’s active site. During transcription, downstream double-stranded DNA separates into a single-stranded DNA template, which enters the primary channel and contacts the active site to allow polymerization of RNA (Borukhov and Nudler, 2008). Due to the crowding of the primary channel by the DNA:RNA hybrid, nucleotide triphosphates (NTPs) must access the active site through an alternative route. They do this through a pore on RNAP called the secondary channel, which allows access to the active site not only for NTPs but also for other regulatory proteins and molecules. The elongating RNA molecule is separated from the DNA template by a wedge-like domain on RNAP to redirect the nascent RNA molecule through a third channel called the RNA exit channel, which then allows upstream DNA to reanneal as it exits RNAP (for a review see Borukhov and Nudler, 2008). Many of the regulatory roles of transcription factors are exerted through interaction with these structural elements (Borukhov et al., 2005). For initiation of transcription to occur, RNAP must first associate with a sixth component, a sigma factor, to form what is termed the holoenzyme (α2ββ’ωσ) RNAP, which allows it to recognize and bind promoter DNA sequences (Murakami, 2015).
In the model bacteria Escherichia coli and Bacillus subtilis, the most important housekeeping sigma factors are Sig70 and SigA, respectively. They are present during planktonic and sessile growth (Losick and Pero, 1981; Gitt et al., 1985) and are responsible for initiating transcription from most promoters under optimal physiological conditions. Promoters whose recognition is mediated by Sig70 and SigA exhibit a canonical sequence centered at positions −35 and −10 base pairs from the start point of transcription (Losick and Pero, 1981). In addition, bacteria also have alternative sigma factors (discovered 40 years ago) to redirect the core RNAP to transcribe a minor set of genes required for specific adaptive responses. The availability of different sets of sigma factors in each bacterial species allows a rapid and reversible adaptation to changes in ecological scenarios. The number of alternative sigma factors present in different bacterial genera is variable and may reflect the lifestyle of the bacterium. It can go from only one sigma factor (e.g., in the intracellular pathogen Mycoplasma genitalium) to more than 60 sigma factors in the soil and antibiotic producing bacterium Streptomyces coelicolor (Gruber and Gross, 2003).
The roles covered by alternative sigma factors are large and diverse. They go from the well characterized roles in stress response, biogenesis of extracellular appendages (e.g., flagella), and developmental programs such as sporulation and biofilm formation; to less characterized processes such as the production of aerial hyphae and photosynthesis regulation in S. coelicolor and cyanobacteria, respectively (Helmann, 2002; Gruber and Gross, 2003). For B. subtilis, the core RNAP can interact with at least 10 different alternative sigma factors to recognize different promoters under diverse environmental and physiologic conditions (Losick and Pero, 1981; Helmann, 2019). Table 1 shows an updated list of the alternative sigma factors present in B. subtilis and their functions. Within this group, the ones involved in spore formation (SigH, SigF, SigE, SigG, and SigK) (Nicholson et al., 1989; Sun et al., 1991; Predich et al., 1992; Roels et al., 1992; Zheng et al., 1992; Piggot and Hilbert, 2004; Aguilar et al., 2007; Kovács, 2016) and stress adaption (SigB and SigH) (Boylan et al., 1991; Predich et al., 1992; Méndez et al., 2004; Bartolini et al., 2019a,b; Nadezhdin et al., 2020) are the best characterized.
Bacillus subtilis offers an excellent platform to learn how alternative sigma factors can be controlled. Both SigF are essential to express the early forespore developmental program (Losick and Stragier, 1992; Piggot and Hilbert, 2004) and SigB regulon (Price, 2000, 2002; Hecker et al., 2007). They are governed by analogous phosphorylation-dependent partner switching mechanisms that involve anti-sigma factors with kinase activities, serine-threonine developmental phosphatases, and anti-anti-sigma factors. SigF is regulated by the anti-sigma factor with serine kinase activity SpoIIAB and the anti-anti-sigma factor SpoIIAA, which are cotranscribed in the same operon with the spoIIAC gene-coding for SigF (i.e., the tricistronic spoIIA operon: spoIIAAspoIIABspoIIAC). Before the formation of the asymmetric septum of the developing sporangium (i.e., forespore plus mother-cell compartments), SpoIIAA remains inactive because it is phosphorylated by the anti-sigma factor SpoIIAB that also captures SigF in an inactive complex. Soon after polar septum formation, SpoIIAA is activated by dephosphorylation in the forespore compartment of the developing sporangium. The compartmentalized activation of SpoIIAA is mediated by the developmental phosphatase SpoIIE that is specifically activated in the forespore compartment (Losick and Stragier, 1992; Piggot and Hilbert, 2004; Pedrido et al., 2013). Once activated in the forespore compartment, SigF directs the expression of spoIIR that codes for a signaling protein (i.e., SpoIIR), which is responsible for the mother-cell restricted activation of SigE, the first sigma factor of the mother-cell linage of sporulation genes. SpoIIR acts as a vectorial signal that activates, in the mother cell compartment, the SpoIIGA protease that is responsible for the proteolytic processing of inactive pro-SigE to active SigE (Piggot and Hilbert, 2004).
This series of early developmental events trigger the compartmentalized expression of forespore and mother-cell genes that will end in the formation of a latent and robust spore cell (Losick and Stragier, 1992; Piggot and Hilbert, 2004). Interestingly, SigB, the first bacterial alternative sigma factor found on the basis of its biochemical properties (Haldenwang and Losick, 1979) is also regulated, like SigF, by proteins with anti-sigma factor, anti-anti-sigma factor and phosphatase activities (see below).
Bacillus subtilis General Stress Response Controlled by SigB Factor
To be able to endure in their natural ecosystems (e.g., soil and host gut) (Hong et al., 2009), B. subtilis has developed complex and interconnected molecular pathways to survive starvation and stress conditions. A clear example of this, apart from sporulation (see above), is the biofilm formation (Branda et al., 2001; Lombardia et al., 2006; Pedrido et al., 2013; Vlamakis et al., 2013; Grau et al., 2015; Hölscher et al., 2015; Kovács and Dragoš, 2019). These responses (i.e., sporulation and biofilm formation) are tightly regulated, time-consuming (e.g., 8–10 h under optimal laboratory conditions), and might be inappropriate to allow a rapid and efficient adaption of B. subtilis cells under unfavorable conditions (Piggot and Hilbert, 2004; Branda et al., 2006; Grau et al., 2015; Kovács and Dragoš, 2019). An almost immediate, but no less sophisticated and efficient, cellular response (i.e., general stress response, GSR) of B. subtilis to a wide range of different stresses is the rapid (e.g., 5–15 min after the imposition of the stress) and short-lived induction of more than 150 general stress proteins (GSP), dependent on the transcription factor SigB (Haldenwang and Losick, 1979; Binnie et al., 1986; Price, 2002; Hecker et al., 2007; Losick and Pero, 2018).
The pioneering work of W. Haldenwang and R. Losick discovered SigB and the first gene (ctc) coding for a member of the GSR (Haldenwang and Losick, 1979), whilst trying to discover the genes and proteins responsible for a cellular escape from vegetative growth and the start of spore formation. Later, but still in the pre-genome-sequencing era, about fifty genes were individualized as members of the SigB regulon through insertional mutagenesis (using suicide vectors and transposons) and proteomic analysis of wild-type and isogenic sigB minus (i.e., ΔsigB) strains under different stress conditions (reviewed in Price, 2002; Hecker et al., 2007). The complete genome sequencing of the B. subtilis strain 168 (Kunst et al., 1997) opened the use of postgenome and “omics” strategies that rapidly expanded the SigB regulon to almost 200 genes (Price, 2000, 2002; Hecker et al., 2007; Nicolas et al., 2012).
Comparative proteomic and genetic analysis of wild-type and ΔsigB B. subtilis strains under different culture conditions (i.e., unstressed and stressed) enabled the identification and assignment of a large set of GSP to the SigB regulon, many of which have proven biochemical activities. SigB is transiently induced after the imposition of a particular stress that halts or slows down the rate of growth, and at the end of the logarithmic phase of growth (in the absence of external stresses), before cells stop active growth and enter into a non-growing state. The presence of GSP in resting or non-growing cells might protect them against stresses that would appear in the future and compromise cell survival. Examples of GSP under SigB control are catalases (KatB, KatX), DNA-protecting enzymes (Dps), proteins repairing oxidative damage (OhrB), and disulfide stress (TxrA), proteins involved in osmotic resistance (OpuD, OpuE, YerD), heat stress resistance (ClpC, ClpP), antibiotic resistance (BmrU, BmrR), cold stress resistance (GsiB), cell envelope protection (GtaA, GtaB), accurate protein synthesis under stress (Ctc), sporulation response (Spo0E), etc. (Price, 2000, 2002; Hecker et al., 2007; Nicolas et al., 2012). Many of the SigB-induced genes are also under dual transcriptional control by other regulatory proteins (e.g., PhoP-PhoR, CtsR, Spx, SigH, and ECF sigma factors SigX and SigW) (Price, 2000, 2002; Hecker et al., 2007; Nicolas et al., 2012).
General stress response is conserved among certain Gram-positive bacteria such as B. subtilis, Bacillus licheniformis, Bacillus halodurans, Bacillus clausii, Bacillus cereus, Clostridium difficile, Oceanobacillus iheyensis, Listeria monocytogenes, Listeria innocua, and Listeria welshimeri, Staphylococcus aureus, Mycobacterium tuberculosis, S. coelicolor, and Ralstonia eutropha. However, some facultative anaerobic Gram-positive bacteria have not developed this conserved developmental program (reviewed in Hecker et al., 2007). In Gram-negative bacteria, SigB orthologs are absent but they express other alternative sigma factors, for example, SigS (RpoS) present in E. coli and SigE (RpoE) present in V. cholera, which shares stress management proficiency with SigB (Kumar et al., 1994; Singh et al., 2017). The alternative transcription factor of SigB and its structural or functional orthologs, are not only the master regulators of the GSR but also control bacterial virulence in pathogens such as L. monocytogenes (Chaturongakul et al., 2008), S. aureus (Shaw et al., 2006), B. cereus (de Been et al., 2010), Bacillus anthracis (Fouet et al., 2000), and Vibrio cholerae (Singh et al., 2017). Table 2 shows an updated list of the SigB structural orthologs present in Gram-positive bacteria.

Table 2. List of alternative sigma factors with structural similarity to B. subtilis SigB present in different Gram-positive bacteria.
In recent years, independent reports have shown that SigB induction produces a noticeable inhibition of the onset of spore formation (Reder et al., 2012a,b; Rothstein et al., 2017). The blockage of sporulation by specific dephosphotylation and inactivation of Spo0A∼Pi (the master regulator of the onset of spore formation) (Piggot and Hilbert, 2004), is driven by the aspartyl phostatase Spo0E encoded by the gene spo0E that possess an active SigB-dependent promoter (Reder et al., 2012b). The proficiency in spore formation is considered a last resort resource because spore formation is a highly energy consuming process and spore germination and outgrowth are also tightly regulated to recover the planktonic growth (Losick and Stragier, 1992; Piggot and Hilbert, 2004; Aguilar et al., 2007; Kovács, 2016). Therefore, it would be beneficial for B. subtilis, either living as single planktonic cells or in social biofilm communities, to explore other less extreme and reversible adaptive responses (i.e., GSR) before it selects the last resort choice (i.e., sporulation) to cope with very unfavorable scenarios. The integration of the SigB activity into the decision-making process of sporulation provides a link to interconnect the two dominant and probably mutually exclusive adaptive responses (sporulation and GSR) in the regulatory network that influences the cell fate of B. subtilis and its relatives (Figure 1; Reder et al., 2012a,b; Rothstein et al., 2017).
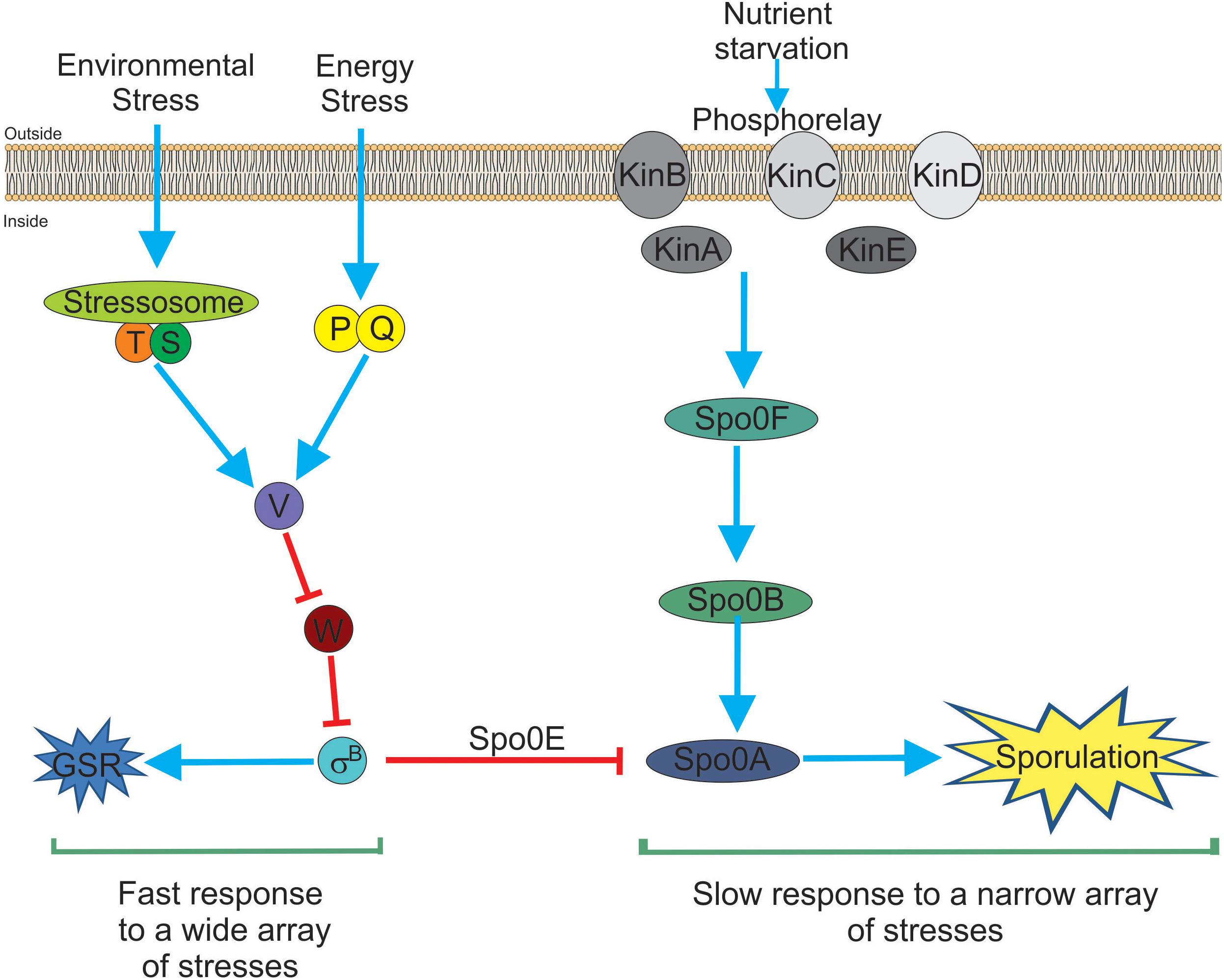
Figure 1. Bacillus subtilis responses to stress. There are two main and interconnected responses that B. subtilis uses to face stress. One response is the general stress response (GSR) triggered by many different types of stresses (for example environmental and energy-depleting stresses). The GSR is rapidly induced after 5–15 min, under laboratory-controlled conditions, it is reversible and depends on the activity of SigB (left part of the figure). This SigB-controlled response would be interpreted as a sort of “panic” and fast response that allows cells (either planktonic or sessile) to cope with multiple stresses. The second stress response (right part of the figure) is sporulation. This response ends with the formation of a mature, highly resistant, and long-lasting spore. Sporulation is time-consuming (at least 8 h to make a mature spore under laboratory-controlled conditions), it is tightly regulated at transcriptional and post-translational levels and responds to fewer stresses (mainly nutrient starvation). Sporulation is under the control of the phosphorelay that activates Spo0A by phosphorylation (i.e., Spo0A∼Pi formation). It is considered the last resort response that B. subtilis (and other bacilli) use to cope and survive under extreme, adverse conditions. Both responses, SigB- and Spo0A-controlled, are interconnected by the aspartyl phosphatase Spo0E induced by SigB. Spo0E inhibits sporulation because of the dephosphorylation of Spo0A∼Pi. In this sense, B. subtilis would be able to first explore less extreme alternatives (i.e., GSR induction) before to trigger the last resort strategy of survival that will end up in the formation of resistant and long-lasting spores. See the text for details.
The SigB-controlled GSR is activated by diverse stressors including high and low temperature, high salt concentrations, ethanol, antibiotics, starvation for glucose, phosphate, and oxygen; inhibitors that decrease the ATP reservoir as well as blue light (Benson and Haldenwang, 1993b; Boylan et al., 1993; Völker et al., 1995; Price, 2000, 2002; Hecker and Völker, 2001; Hecker et al., 2007; Brigulla et al., 2003; Helmann et al., 2003; Mascher et al., 2003; Méndez et al., 2004; Moore et al., 2004; Zhang and Haldenwang, 2005; Gaidenko et al., 2006; Bonilla, 2020). Each of these stimuli induces one of the three SigB regulatory pathways known today (see below). Once activated, SigB binds to the core RNAP and recognizes a particular promoter structure (Haldenwang, 1995). Table 3 shows conservation of the SigB promoter sequences from B. subtilis, L. monocytogenes, S. aureus, and other Gram-positive bacteria.
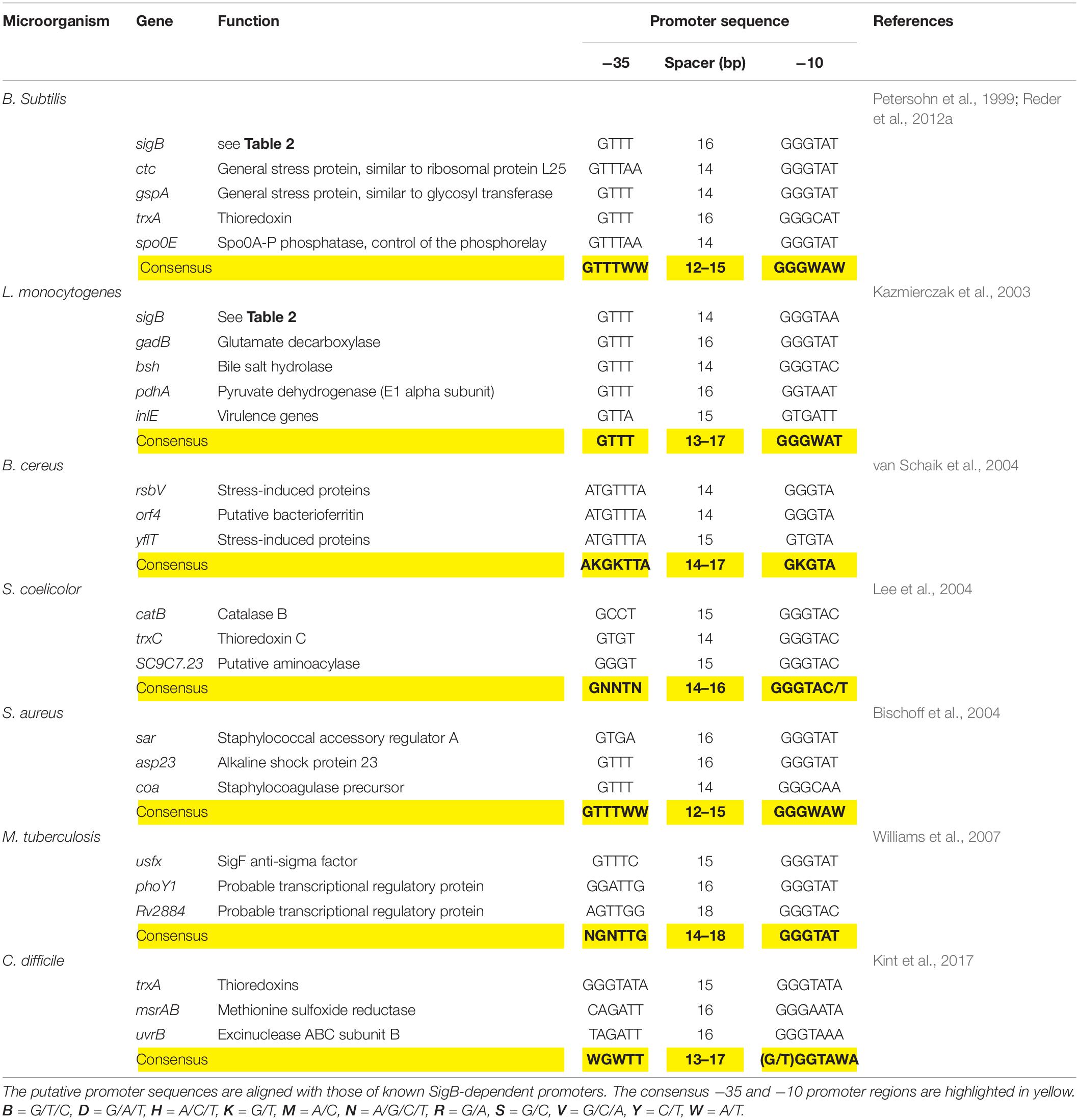
Table 3. Genetic organization of the sigB promoter region of representative genes present in B. subtilis and selected Gram-positive bacteria.
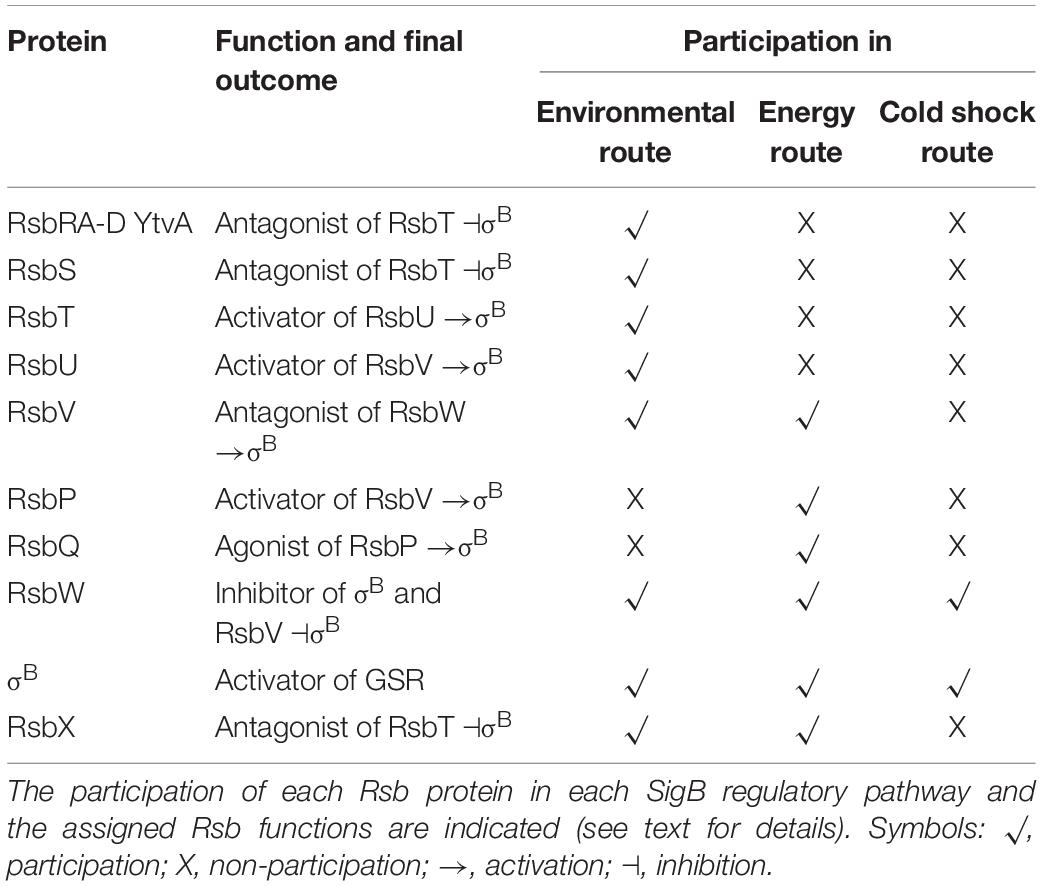
Activation of SigB in response to physical (i.e., environmental) and nutritional (i.e., energy) stresses is separately controlled by two overlapping partner switching mechanisms (Alper et al., 1996; Price, 2002). These partner switching mechanisms are composed of four proteins: an input phosphatase (i.e., RsbP or RsbU, for SigB activation under energy- or environmental-stresses, respectively); a switch kinase with anti-sigma factor activity (RsbW); an antagonist protein or anti-anti-sigma factor (RsbV); and the target protein, the sigma factor (Alper et al., 1996). Under non-stress conditions, the two SigB activating pathways (i.e., energy- and environmental-stress pathways, see below) do not receive stressor inputs, SigB is inactive and the GSR is not induced. This is because the switch protein RsbW (a kinase with anti-sigma factor activity) has two roles: it phosphorylates and inactivates the anti-anti-sigma factor RsbV and sequesters the target SigB forming an inactive complex, and thus preventing its binding to the core RNAP (Benson and Haldenwang, 1993a; Alper et al., 1996; Figure 2).
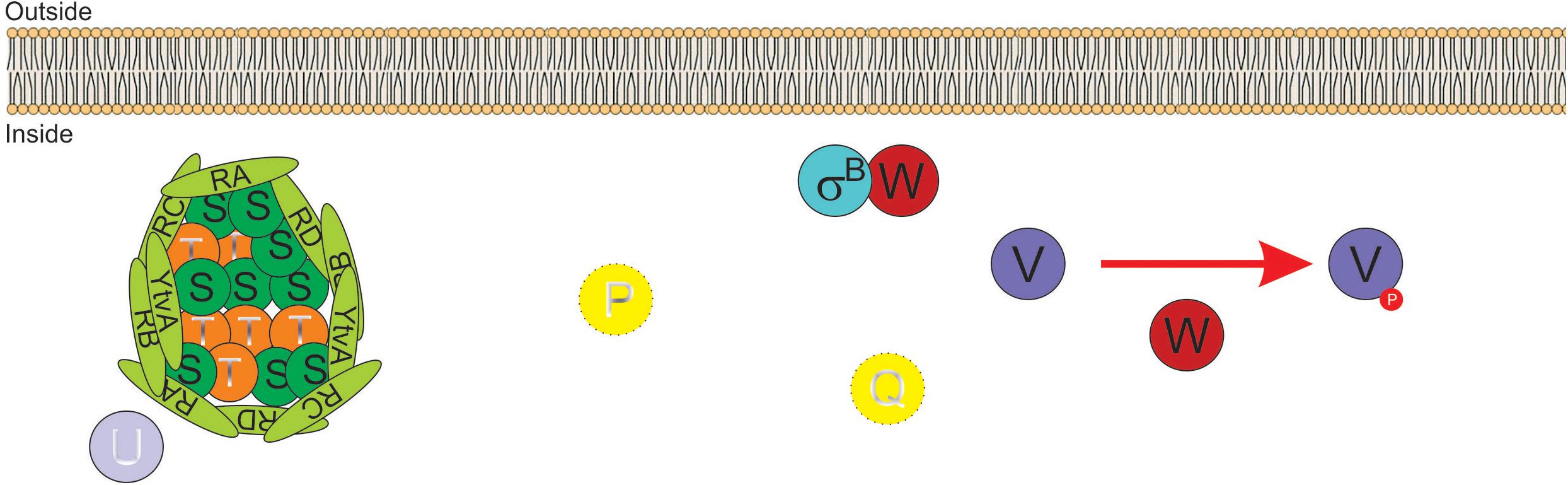
Figure 2. Diagram of SigB regulatory pathways of general stress response under non-stress conditions in Bacillus subtilis. Under non-stress conditions the anti-anti-sigma factor RsbV (V, for simplicity) is phosphorylated (V∼P) by the kinase/anti-sigma factor RsbW (W, for simplicity). W captures SigB (σB) in a stable complex (W:σB), thereby preventing its binding to the RNA polymerase (RNAP). The PP2C-type phosphatase RsbU (U) is inactive. The serine threonine kinase RsbT (T) responsible for the activation of U is inactive and captured in the stressosome, also composed of the antagonist RsbS (S) and the putative sensor proteins RsbRA (RA), RsbRB (RB), RsbRC (RC), RsbRD (RD), and YtvA. Similarly, in the absence of energy stress, the PP2C-type phosphatase RsbP (P) and its activating protein RsbQ (Q) are inactive.
The energy-stress arm of the SigB regulatory cascade is composed by two members: the protein phosphatase 2C (PP2C)-type RsbP (which dephosphorylates RsbV∼Pi); and the agonist RsbQ, which is cotranscribed with RsbP (PArsbPrsbQ) and forms an active complex with it (Vijay et al., 2000; Brody et al., 2001). In the absence of energy-stress, the RsbQ:RsbP complex is not formed, as RsbP is inactive, RsbV remains phosphorylated, and SigB remains captured in the complex with RsbW (Figure 2). The environmental-stress arm of the SigB activation pathway is structurally more complex than the energetic pathway. Similarly, to the requirement of the agonist protein RsbQ for RsbP phosphatase activity, the environmental-stress responding PP2C-type phosphatase RsbU requires the interaction with the serine-threonine kinase RsbT for its activation (Wise and Price, 1995; Yang et al., 1996). In the absence of environmental stress, RsbT is unable to interact with RsbU because it is bound to the antagonist protein RsbS in a large multiproteic complex: the stressosome (Figure 2). The B. subtilis stressosome is composed of a family of homologous or paralog proteins (i.e., RsbRs) thought to be sensors and modulators of environmental stimuli. RsbRA (the best characterized RsbR paralog) is transcribed by rsbRA, the first gene of an eight-gene operon that harbors sigB and other key gene regulators of SigB activity (i.e., the sigB operon, see below). The other RsbR paralogs (i.e., RsbRB, RsbRC, RsbRD, and YtvA) are expressed from diverse genes along the B. subtilis chromosome (Akbar et al., 2001; Chen et al., 2003; Kim et al., 2004a; Delumeau et al., 2006; Gaidenko et al., 2006, 2012).
The stressosome complex adopts a pseudo-icosahedron conformation made of 40 copies of RsbRs, 20 copies of RsbS, and 20 copies of RsbT (Chen et al., 2003; Kwon et al., 2019). Each RsbR paralog contains a variable N-terminal non-heme globin domain (structurally related to globins but lacking conserved histidine residue essential for the incorporation of heme iron), a 13-aminoacid conserved linker domain, and a conserved C-terminal STAS (sulfate transporter antisigma factor antagonist) domain, except for YtvA, which harbors a LOV (light-oxygen-voltage) sensing domain (Gaidenko et al., 2012). Structural analysis of the stressosome structure by cryo-electron microscopy suggests that the non-conserved N-terminal domain of each RsbR paralog protrudes outward the stressosome and the C-terminal domains face and reside inside the complex bound to RsbS and RsbT (Marles-Wright et al., 2008).
The non-conserved amino acid sequence of the N-terminal domains of RsbR paralogs suggests the existence of a different affinity of each paralog, to perceive stress signals and/or different abilities to interact with RsbT. The second and third genes in the sigB operon encode for RsbT and RsbS. This genetic organization opens the possibility that the three proteins (RsbRA, RsbT, and RsbS) could interact with each other soon after their synthesis and enter into the stressosome as a preformed complex (Reeves and Haldenwang, 2006). Once within the stressosome, the RsbRs proteins (RsbRA and its paralogs) seem to be redundant because when many of them are lost, RsbS is unable to retain RsbT in the stressosome. Less characterized stressosome-like complexes are present in other Gram-positive and Gram-negative bacteria, and the B. subtilis stressosome represents a model to better understand their functions and molecular organization (Pané-Farré et al., 2005).
Signal Transduction Pathways of SigB Activation
Activation of SigB by Environmental Stress
Table 4 shows an updated list of all known SigB regulatory proteins and their functions. In the presence of environmental insults (e.g., acid, heat, alcohol stresses), B. subtilis activates the kinase activity of RsbT on RsbRA and RsbS at conserved T171 and S59 residues, respectively (Yang et al., 1996; Kim et al., 2004b; Reeves and Haldenwang, 2006). Diverse genetic, biochemical and modeling studies suggest a progression of events that seem to start when environmental stress increases the kinase activity of RsbT and/or makes RsbS and the RsbR paralogs better suited to become phosphorylated by RsbT (Chen et al., 2003; Kim et al., 2004b; Reeves and Haldenwang, 2006). The phosphorylation of RsbRA at T171 is a prerequisite and facilitates the subsequent phosphorylation of RsbS at S59 by RsbT, and the onset of SigB signaling (Chen et al., 2003; Kim et al., 2004b). RsbS∼Pi is unable to retain RsbT, which is released from the stressosome, and binds to and activates RsbU (Yang et al., 1996). The environmentally activated PP2C-type phosphatase RsbU dephosphorylates the anti-anti-sigma factor, RsbV∼Pi at S56 (Hecker et al., 1996; Yang et al., 1996; Vijay et al., 2000). The RsbV is now free to interact with RsbW, releasing active SigB (Figure 3 and Table 4). Interestingly, additional phosphorylation of RsbRA by RsbT at T205 prevents stressosome hyperactivation and thereby limits SigB activation (Eymann et al., 2011; Liebal et al., 2013). This (first) negative feedback loop would be partially responsible for the transient SigB response after stress imposition, reaching the maximum, under laboratory conditions (growth in nutritional rich broth, with shaking at 37°C), 10–40 min after the shift, but thereafter SigB activity rapidly decreases to a level slightly higher than the pre-shift level (Völker et al., 1995).
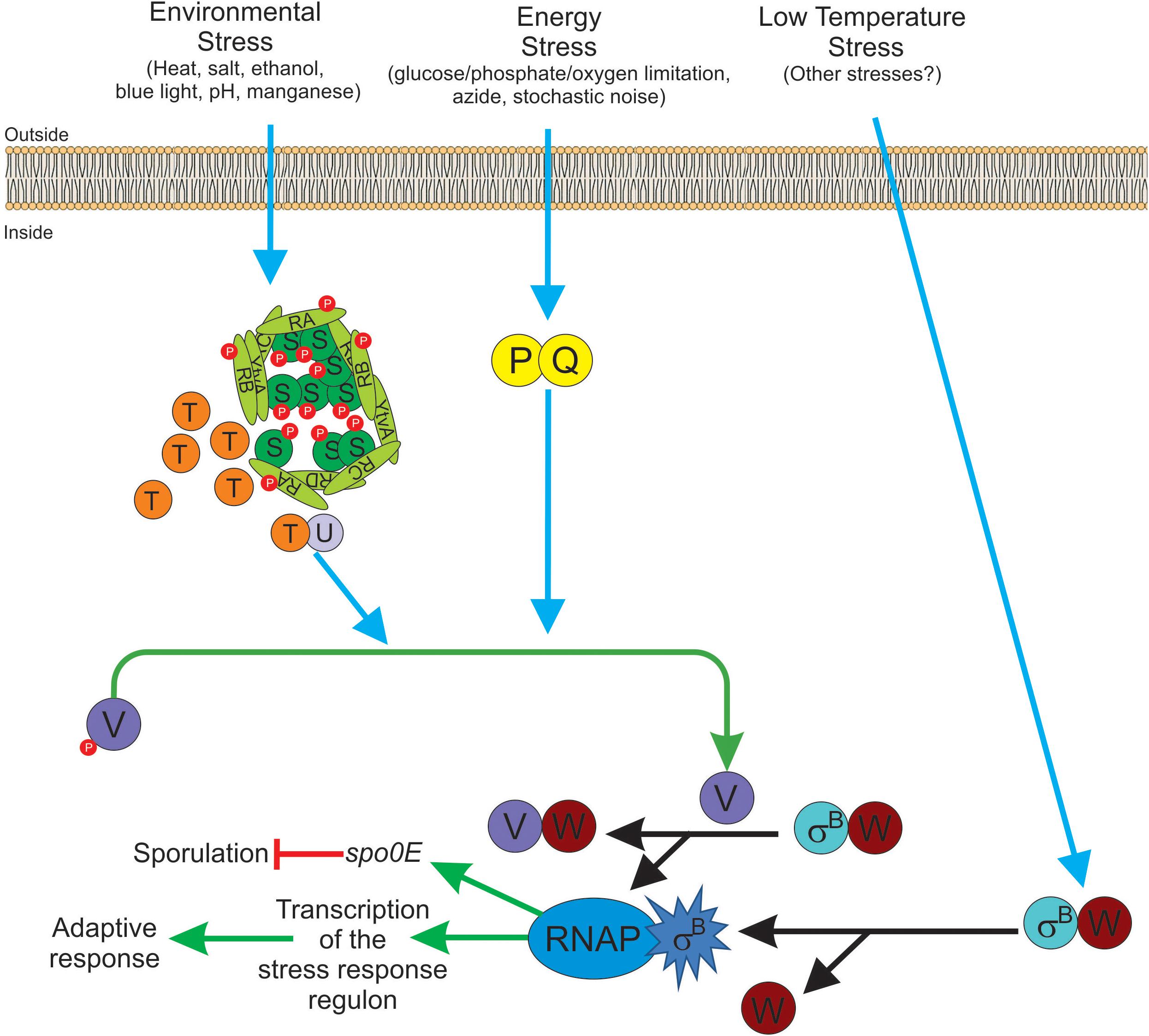
Figure 3. Diagram of SigB regulatory pathways of general stress response under stress conditions in Bacillus subtilis. Energy stress activates the PP2C-type phosphatase RsbP (P for simplicity) and the agonist protein RsbQ (Q for simplicity) which form a complex RsbQP (QP for simplicity) to dephosphorylate RsbV∼Pi (V∼P). Environmental stress induces the kinase activity of RsbT (T for simplicity) to phosphorylate RsbRA (RA∼P), RbsRB (RB∼P), and RsbS (S∼P). Released T from the stressosome binds and activates the PP2C-type phosphatase RsbU (U for simplicity) to dephosphorylate RsbV∼Pi (V∼P). Dephosphorylated RsbV (V), formed by the phosphatases P and/or U, binds to RsbW (W for simplicity), releasing SigB (σB) which in turn binds to RNAP and activates its target genes (i.e., GSR and spo0E). A third SigB activating pathway by low temperature stress operates independently of U, P, and V activities at the level of complex stability between RsbW and SigB (WσB). The RbsX phosphatase is responsible for the dephosphorylation of RsbRA∼Pi, RsbRB∼Pi, and RsbS∼Pi, to restore the levels of SigB activity to the ones present before stress (not shown for simplicity). See the text for further details.
To reinforce the transient and controlled SigB response, there is a serine-threonine phosphatase, RsbX, responsible for the dephosphorylation of RsbS∼Pi and RsbRA∼Pi at their S59 and T171 residues, respectively. These dephosphorylations are part of a fine tuning device that allows RsbT to be again sequestered in the stressosome and ensures the transient and controlled nature (second negative feedback regulatory loop) of SigB activation (Yang et al., 1996; Price, 2002). Supporting this model, it has been shown that the phosphatase activity of RsbX is low under non-stress conditions (Chen et al., 2004). Under these conditions, the predominant components of the stressosome are unphosphorylated RsbS and RsbRA. After the imposition of initial stress, the kinase activity of RsbT is activated, and RsbRA and RsbS become phosphorylated by RsbT at T171 and S59 positions, respectively. These environmentally controlled phosphorylations trigger a cascade of biochemical reactions that ends with the liberation of SigB from RsbW and its activation (Figure 3). Because of the positive feedback loop of SigB on its coding operon, the levels of RsbX will increase to compensate for the activation level of RsbT, leading to dephosphorylation of RsbS∼Pi and RsbRA∼Pi at T171 residue, and downregulation of the SigB-dependent response. As expected, in the absence of RsbX, the activity of SigB increases to very high and uncontrolled levels (Benson and Haldenwang, 1992; Boylan et al., 1992; Völker et al., 1997).
How the environmental-related signals are transduced to control the kinase and phosphatase activities of RsbT and RsbX is unknown, and opens the possibility that the stressosome might not constitute the most upstream component of the environmental signaling cascade controlling SigB activation (Kuo et al., 2008). It has been indicated that the ribosomal protein L11 and the essential GTP-binding protein Obg, encoded by the same operon as the phosphorelay gene spo0B, were required for stress activation of SigB (Trach and Hoch, 1989; Scott and Haldenwang, 1999; Zhang et al., 2001). In vitro analysis showed that Obg co-fractionated with ribosomal subunits and the stressosome components RsbR, RsbT, and RsbS (Scott and Haldenwang, 1999). Even though the physiological role of ribosomal subunits and Obg on the environmental-stress arm of the SigB regulatory cascade is unknown, it allows the possibility of a link between the protein synthesis machinery (i.e., the ribosome) and cell-cycle signals (putative mediated by Obg) with the stressosome. Other clues related to the unnoticed existence of upstream participants of the stressosome are found in the Gram-positive pathogen L. monocytogenes where an integral membrane protein required for SigB activation might be the environmental sensor interacting with the stressosome in vivo (Impens et al., 2017).
Activation of SigB by Energy Stress
The input phosphatase RsbP is another PP2C-type specific phosphatase responsible for sensing a depletion of energy (ATP levels). The PP2C-type domain of RsbP is located on its C-terminal region, and the N-terminal region of RsbP contains a Per-Arnt-Sim (PAS) domain that would be involved in the sensing of redox potential, oxygen levels, and protein-protein interactions (Price, 2000, 2002; Hecker et al., 2007). RsbP is cotranscribed and works together with the RsbQ protein (Table 4; Brigulla et al., 2003; Kaneko et al., 2005). RsbPQ dephosphorylates RsbV∼Pi and RsbV activate SigB as described above (Figure 3). Interestingly, in the absence of energy stress, RsbP and RsbQ were responsible for the formation of discrete stochastic pulses of SigB activation (Locke et al., 2011). The significance of the RsbPQ-dependent noise activation of SigB is unclear but it might be advantageous for the survival and persistence of the bacterial population to always have (also in the absence of apparent stress) a minor proportion of cells with active SigB to face sudden and threatening conditions of population survival (i.e., bet-hedging) (Acar et al., 2005; Kussell and Leibler, 2005). Since many bacteria share transcription factors similar to SigB and its regulatory partner switching pathways (Figure 3 and Tables 2, 4), it is tempting to think that the stochastic pulse modulation of the GSR as an important trait that “always” guarantees bacterial survival and persistence.
Soon after the liberation of active SigB and the formation of the RsbW-RsbV complex induced by the environmental and/or energy stresses, RsbW phosphorylates and inactivates RsbV, and released RsbW is now able to be captured and inactivates SigB again (Dufour and Haldenwang, 1994). This event represents a third negative feedback loop responsible for the controlled and transient nature of the SigB response that operates together with the double phosphorylation of RsbRA, and the dephosphorylation of RsbS∼Pi by RsbT kinase and RsbX phosphatase, respectively.
Activation of SigB by Cold Shock Stress
An alternative pathway of SigB activation has been described, which operates during growth at low temperatures (between 17 and 20°C) or after a growth temperature downshift from 37 to 20°C (Brigulla et al., 2003; Méndez et al., 2004). One feature of this pathway is the high and persistent levels of SigB induction (Brigulla et al., 2003; Méndez et al., 2004), compared to the less dramatic and transient nature of the SigB induction observed after bacterial exposition to environmental or energy stresses (Benson and Haldenwang, 1993b; Boylan et al., 1993; Völker et al., 1995; Price, 2000, 2002; Hecker and Völker, 2001; Helmann et al., 2003; Mascher et al., 2003; Moore et al., 2004; Zhang and Haldenwang, 2005; Gaidenko et al., 2006).
Another feature of this induction pathway at low temperature, compared to the environmental and energy pathways, is that SigB cold-shock induction does not need for RsbP, RsbU, and RsbV (Figure 3 and Table 4). The nature of this RsbP-, RsbU-, and RsbV-independent activation pathway is not known but there are some hypotheses proposed by Brigulla et al. (2003). Among these is the idea that at low temperatures an unknown protein or metabolite disrupts the inhibitory RsbW-SigB complex, or it might be the case that the physical interaction between SigB and RsbW could be decreased or impaired at low temperatures (Brigulla et al., 2003). In any of these two hypothetical situations, SigB activity should become high, persistent, and independent of RsbX to improve general stress resistance at low temperature (Brigulla et al., 2003; Méndez et al., 2004).
Interestingly, a recent report showed that B. subtilis survival after oxidative stress (e.g., by addition of sodium nitroprusside) required SigB activation independent from RsbP but also surprisingly, independent of RsbT (Tran et al., 2019). This finding opens the possibility that the RsbT- and RsbP-independent, cold-shock-dependent pathway of SigB activation could recognize a wider spectrum of stresses (Figure 3).
Despite the high SigB activity of B. subtilis growing at low temperature, the sporulation proficiency is extremely low, less than 1% for cultures grown at 17–20°C, compared to a nearly 100% of sporulation efficiency for cultures grown at 37°C (Brigulla et al., 2003; Méndez et al., 2004). Even though an explanation for the low sporulation efficiency at low growth temperatures has not yet been established, we hypothesize that it could result from the high and persistent levels of SigB activity found at low growth temperature, which could trigger the upregulation of the SigB-controlled aspartyl phosphatase Spo0E, responsible for the dephosphorylation of Spo0A∼Pi, which in turn blocks the onset of sporulation (Reder et al., 2012a,b). In any case, why B. subtilis chooses activation of SigB instead Spo0A activation at low temperatures remains a mystery (Figure 3).
Besides the posttranslational regulation imposed by environmental, energy and low temperature stresses, SigB upregulation is also modulated at the transcriptional level (Wise and Price, 1995). SigB is encoded by the seventh genetic unit of the eight-gene sigB operon, and the components of the environmental stress pathway are encoded by the first four genes of this operon. The expression of the sigB operon is under the control of a SigA-dependent promoter (PA) that is responsible for the constitutive and low expression of the operon. There is an extra promoter (PB) under SigB control, which induces the expression of the last four genes of the sigB operon (i.e., PArsbR-rsbS-rsbT-rsbU-PBrsbV-rsbW-sigB-rsbX) (Boylan et al., 1992; Völker et al., 1997; Price, 2002; Hecker et al., 2007). The SigA-dependent expression of the entire sigB operon (and the stochastic noise, see above) ensures that even under non-stress conditions, SigB will always be present in B. subtilis cells to allow a rapid response after sudden and unexpected stress. Thus, any single stress (energy, environmental or low temperature insult) will release SigB from the inhibitory complex formed with RsbW (Figure 3) and upregulate its expression from the PB-dependent promoter. In this sense, active SigB is now able to specifically activate genes expression needed to cope with the inducing stress and also expression of the GSR, thus providing a multiple, unspecific and preventive cell stress adaption (Hecker and Völker, 2001; Price, 2002; Hecker et al., 2007). However, although most of the genes involved in the GSR are induced after specific or unique stress, the activation level of each of the more than 150 genes that integrate the GSR, will depend on the specific imposed stress (i.e., the transcription of not all the SigB-controlled genes respond in the same magnitude to different types of stresses).
The lack of RsbP and RsbQ orthologs does not preclude the ability to induce SigB in response to energy stress. In L. monocytogenes, the stressosome is activated not only after environmental stress but also after energy stress (Martinez et al., 2010). For the opportunistic pathogen S. aureus, and other cocci, it has been observed that there is an absence of RsbP and RsbQ orthologs, but also that stressosome components (i.e., RsbRs, RsbS, and RsbT) are missing (Pané-Farré et al., 2009). Therefore, the components of the SigB regulon in S. aureus are restricted to four members (i.e., RsbU, RsbV, RsbW, and SigB). It seems that only the transcriptional regulation of the S. aureus sigB operon (PArsbUPBrsbVrsbWrsigB) would be responsible for SigB regulation. Transcriptional control of rsbU is sufficient to activate the SigB-dependent stress-response, but surprisingly, the ratio of RsbV to RsbV∼Pi does not increase (as it is the case in B. subtilis) after an environmental insult.
Furthermore, in B. cereus, B. anthracis, and Bacillus thuringiensis the energy- and environmental-stress dependent routes of SigB activation are absent, and the control of the phosphorylation level and kinase activity of the RsbV and RsbW orthologs, respectively, might be performed using other protein modules. In B. cereus, the hybrid and membrane-bound histidine kinase RsbK autophosphorylates under diverse stress conditions at a conserved histidine residue within the H-box domain. RsbK∼Pi is now able to phosphorylate the RsbY protein at its N-terminal receptor domain at a conserved aspartyl residue. Phosphorylation of RsbY activates its C-terminal PP2C-type phosphatase domain to dephosphorylate RsbV∼Pi and so allows SigB activation (de Been et al., 2011).
These observations in L. monocytogenes, S. aureus, and other bacilli, strongly suggest the existence of new and uncharacterized SigB control mechanisms that are absent in B. subtilis but present in other Gram-positive bacteria that expand the significance and complexity of the SigB regulatory network on bacterial GSR for adaption and survival (van Schaik and Abee, 2005; Hecker et al., 2007; O’Byrne and Karatzas, 2008; Pané-Farré et al., 2009; Martinez et al., 2010; de Been et al., 2011).
New Role for the Transcription Factor SigB in Biofilm Growth, Aging, and Dispersal
Biofilms are well organized bacteria ecosystems, in which sessile cells form multicellular aggregations in a self-secreted extracellular matrix with protective and adhesive functions (Vlamakis et al., 2013). Biofilms have the property to adhere to living or non-living surfaces and can be prevalent in natural, industrial, and hospital settings. The proficiency in biofilm formation represents a key feature that many bacterial pathogens share that enables them to resist the action of microbicides and antibiotics, giving rise to the failure of medical therapy and the persistence and dissemination of the pathogen infection (Flemming et al., 2016). Wild-type B. subtilis isolates are predominantly known to form architecturally complex colonies and wrinkled pellicles that serve as models of solid (colony) and liquid (pellicle) bacterial biofilms (Branda et al., 2001; Lombardia et al., 2006; Pedrido et al., 2013; Vlamakis et al., 2013; Grau et al., 2015; Hölscher et al., 2015; Kovács and Dragoš, 2019).
Although most biofilms are studied in pathogenic bacteria and therefore are associated with chronic and persistent infections (such are the cases for S. aureus and Pseudomonas aeruginosa biofilms), we now know that some bacteria produce beneficial biofilms. The clearest example of this is the B. subtilis biofilm formed either on the roots or leaves of plants where they promote plant growth (Beauregard et al., 2013), and biofilms formed in the intestinal mucosa of the eukaryotic host, where they produce an improvement in the host immunity (Donato et al., 2017; Smolentseva et al., 2017). It has recently been shown that B. subtilis can prevent the formation of S. aureus biofilm in both mice and human intestines through a molecular mechanism of quorum-sensing interference that produces the competitive exclusion of the pathogen from the host gut colonized by B. subtilis (Piewngam et al., 2018). By contrast, the co-culture of B. subtilis under biofilm-supporting conditions favored the growth and survival of probiotic lactic acid bacteria (LAB), protected in the extracellular matrix of the biofilm formed by B. subtilis. Otherwise, LABs would perish upon exposure to the acidic pH of the stomach and the microbicide action of intestinal bile (Yahav et al., 2018). More recently, it has been shown in animal models that the B. subtilis biofilm is important in delaying neurodegenerative diseases (i.e., Alzheimer’s and Parkinson’s diseases) (Cogliati et al., 2019; Goya et al., 2020). Therefore, the importance of B. subtilis biofilm in basic and applied research cannot be questioned.
Interestingly, two recent publications (Bartolini et al., 2019a; Nadezhdin et al., 2020) have demonstrated that SigB was induced during B. subtilis biofilm development. These findings expanded the significance of the expression of the SigB-dependent GSR from the self-sufficient and planktonic life style of individual bacteria to the collective and social life style found in biofilm communities (Branda et al., 2001). Bartolini et al. (2019a) reported that the activity of SigB in B. subtilis pellicles (liquid biofilm) during the first hours of cultivation (when the biofilm was juvenile) was not detected. However, the induction of SigB began at the moment that the biofilm decreased its rate of growth, with the highest levels of SigB activity observed when the biofilm grasped its plateau of growth (mature biofilm; Bartolini et al., 2019a). The analysis of biofilm formation at different temperatures in B. subtilis strains lacking key components for SigB activation during environmental (ΔrsbU), metabolic (ΔrsbP), or both types of stresses (ΔrsbUP) showed interesting results (Bartolini et al., 2019a). During biofilm development, neither the environmentally related (RsbU) nor the low temperature-related route of SigB activation was expressed, and only the energy stress related (RsbPQ) route was responsible for activation of SigB inside the biofilm (Figure 4A).
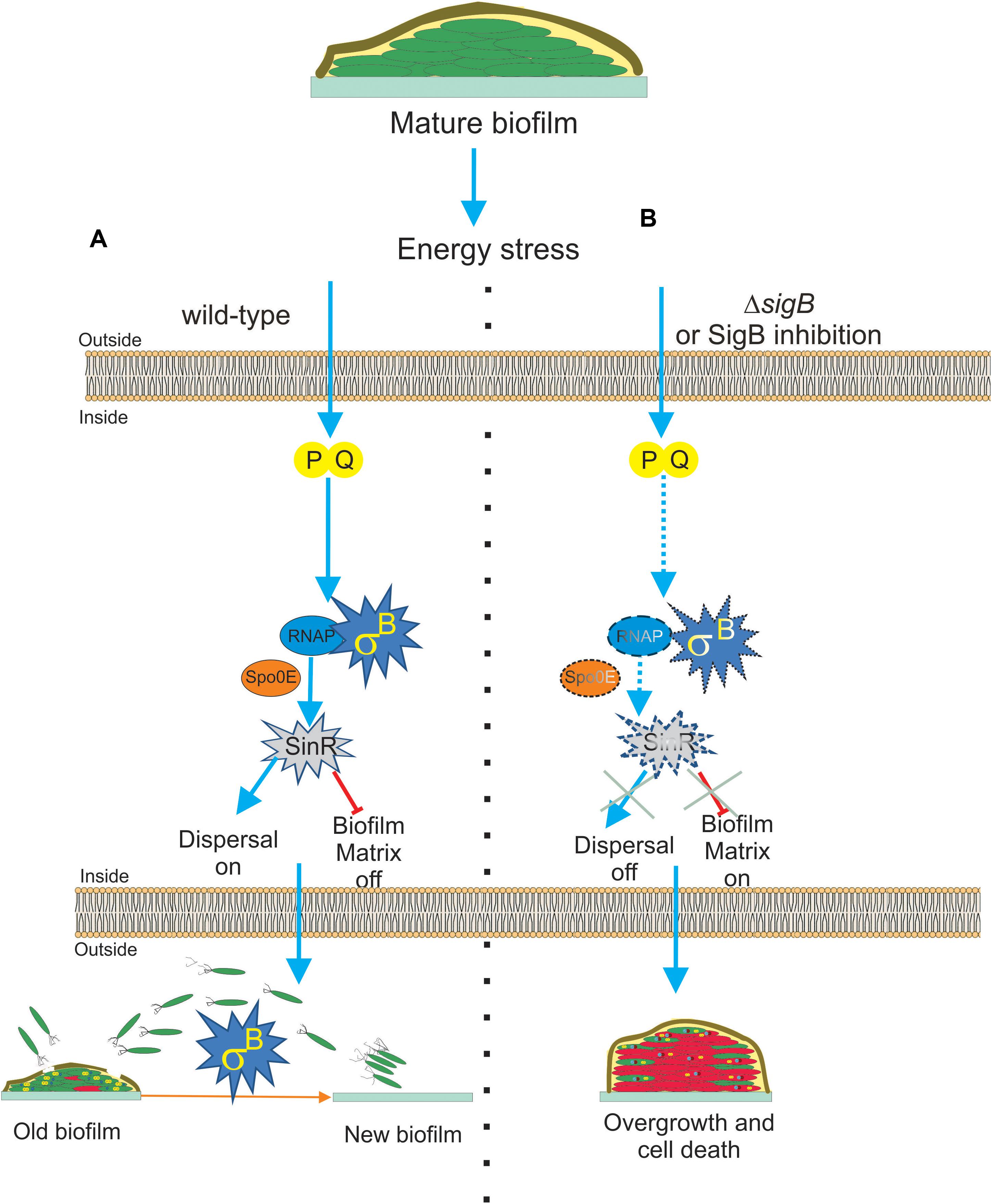
Figure 4. Proposed model for the role of SigB on biofilm growth and fitness. (A) The general stress factor SigB is essential to stop the detrimental overgrowth of the mature biofilm and trigger dispersal before nutrient within the biofilm run out. The signal detected by SigB to accomplish these tasks is transduced through the RsbP/RsbQ energy-related route of the SigB regulatory cascade, which in turn positively upregulates the levels of SinR, the master negative and positive regulator of biofilm formation and motility in B. subtilis, respectively. The positive effect of SigB on spo0E expression would be responsible for the increased levels of SinR activity inside the mature biofilm. (B) Cells in SigB-deficient (ΔsigB) biofilms are unable to sense stress and maintain upregulated levels of SinR (spo0E is not induced) as the biofilm ages. As a consequence of such an energetic imbalance, the biofilm continued to grow and became larger but less resistant to aging and diverse stresses. Dispersal is also downregulated. Development of new drugs that negatively target SigB activity (SigB inhibition in dash lines) in bacterial pathogens of clinical relevance sharing with B. subtilis structural or functional homologs to SigB is an interesting line of research. See the text for details.
Nadezhdin et al. (2020) also reported SigB activation in B. subtilis solid biofilm (wrinkle colonies). Using different state of the art techniques, such as time-lapse imaging and quantitative microscopy, they observed that stochastic pulses of SigB activation in the individual cells of the developing biofilm depend on the functionality of the energy stress pathway of SigB activation, as previously observed by Locke et al. (2011) in B. subtilis planktonic cultures. Therefore, stochastic RsbPQ-mediated SigB activation is a common phenomenon that occurs in planktonic and sessile B. subtilis communities (Locke et al., 2011; Nadezhdin et al., 2020). The stochastic activation of SigB was orchestrated together with the activation of the other main survival pathways present in B. subtilis, the sporulation program (Piggot and Hilbert, 2004; Aguilar et al., 2007; Kovács, 2016). The activities of both adaptive and survival pathways (SigB regulon and sporulation) were expressed as a gradient inside the biofilm, with the peak of expression of SigB located at the top of the biofilm and the sporulation pathway activity restricted to the middle of the biofilm because of the Spo0E-dependent inhibitory effect of SigB on spore formation (see Figure 1 and Nadezhdin et al., 2020). Under non-stress conditions, ΔsigB B. subtilis produced more biofilm compared to the wild-type cells, but these cells died more rapidly because of their increased vulnerability to different stress conditions (Figure 4B). According to these results, it is reasonable to conclude that SigB could prevent undesirable biofilm overgrowth and maintain the fitness of old biofilms (Figure 4A).
SigB might regulate biofilm growth and fitness, as it has been shown that SigB activity was required to maintain expression of sinR in the mature biofilm (Bartolini et al., 2019a, Figure 4A). SinR is an inhibitor of biofilm formation because it represses the operons epsA–O and tapA-sigW-tasA in charge of the synthesis of the exopolysaccharide and the amyloid fiber TasA of the extracellular matrix of the biofilm (Kearns et al., 2005; Chu et al., 2006). Therefore, the biofilm overgrowth observed in ΔsigB sessile cultures of B. subtilis was due to the downregulation of the biofilm inhibitor SinR (Figure 4B). The activity of SinR is also essential for flagellum-mediated B. subtilis motility (swimming and swarming), and sinR mutant strains are non-motile (Fredrick and Helmann, 1996; Kearns and Losick, 2003; Gottig et al., 2005; Grau et al., 2015). Swimming and swarming were reduced in ΔsigB cells, supporting the positive role of SigB on SinR function (Bartolini et al., 2019a). During the biofilm cycle, a phenomenon known as dispersal takes place, where some motile cells detach from the aged biofilm and escape from it to avoid famine (Jefferson, 2004; Liu et al., 2015). According to the requirement of cellular motility for dispersal proficiency (Kaplan, 2010; Dalton et al., 2011; McDougald et al., 2011; Stacy et al., 2014; Barraud et al., 2015; Guilhen et al., 2017), and the motility control by SinR in B. subtilis, it was shown that the biofilm formed by ΔsigB cells had reduced dispersal (Bartolini et al., 2019a), Figure 4B. These findings open the possibility to explore novel strategies for blocking the expression or activity of the SigB homologs present in bacterial pathogenesis as a way to downregulate dispersal and make disease-related biofilms more susceptible to traditional microbicides. However, the mechanism by which SigB controls sinR expression is still unknown, because the sinR promoter lacks a SigB consensus sequence (Nicolas et al., 2012). One possible mechanism is that SigB activates sinR expression indirectly. In line with this, the aspartyl phosphatase Spo0E is a possible candidate because SigB induces expression of spo0E (Reder et al., 2012b). The Spo0E aspartyl phosphatase inactivates Spo0A∼Pi by dephosphorylation and the decreased levels of Spo0A∼Pi releases sinR from its repression by Spo0A∼Pi. Therefore, SigB could be able to increase sinR expression indirectly through Spo0E activity (Figure 4A).
A Novel Positive Role for the Transcription Factor SigB in the Antifungal Proficiency of Bacillus subtilis
Pesticides are extensively used to control plant diseases (Pingali, 2012). However, a negative impact on human health and the environment has been observed (Tago et al., 2014; Kim et al., 2017). Numerous species of Bacilli have been identified as eco-friendly plant-growth promoting bacteria (PGPB) and/or biocontrol agents. PGPB employ direct and indirect mechanisms to enhance plant growth. These direct mechanisms involve phytohormone production, the acquisition of nutrients, and the control of pathogens through the synthesis of hydrolytic enzymes and biopesticides (Ongena et al., 2005; Raja, 2013; Luo et al., 2015). The indirect mechanisms include the triggering of specific defense-related pathways, particularly the induction of systemic resistance (ISR) (Zeriouh et al., 2014; Debois et al., 2015; Aleti et al., 2016), and the release of volatile organic compounds (VOCs) against pathogens (Ongena and Jacques, 2008; Raaijmakers et al., 2010; Cawoy et al., 2014; Cawoy et al., 2015). Most of the characterized plant-beneficial B. subtilis strains display direct and indirect mechanisms to benefit plant growth and can form robust root-associated biofilms.
In a recent study by Bartolini et al. (2019b), it has been shown that SigB and its GSR were largely activated when B. subtilis interacts antagonistically with the phytopathogen Fusarium verticillioides (Figure 5). Specifically, the RsbPQ-route sensing energy stress was required for SigB activation and the antifungal control (Figure 5) when B. subtilis cells were cultured in the presence of live or dead F. verticillioides micelia or cell-free supernatants of the fungi (Bartolini et al., 2019b). In the light of these results, it is assumed that an unknown F. verticillioides metabolite is the primary cause of the energy depletion in B. subtilis. Another interesting finding of Bartolini et al. (2019b), is that biocontrol of F. verticillioides by B. subtilis depended on the production of the antifungal lipopeptide surfactin given that ΔsigB cultures produced fewer quantities of surfactin compared to wild-type B. subtilis, and ΔsrfA B. subtilis cultures showed poor antifungal activity (Figure 6).
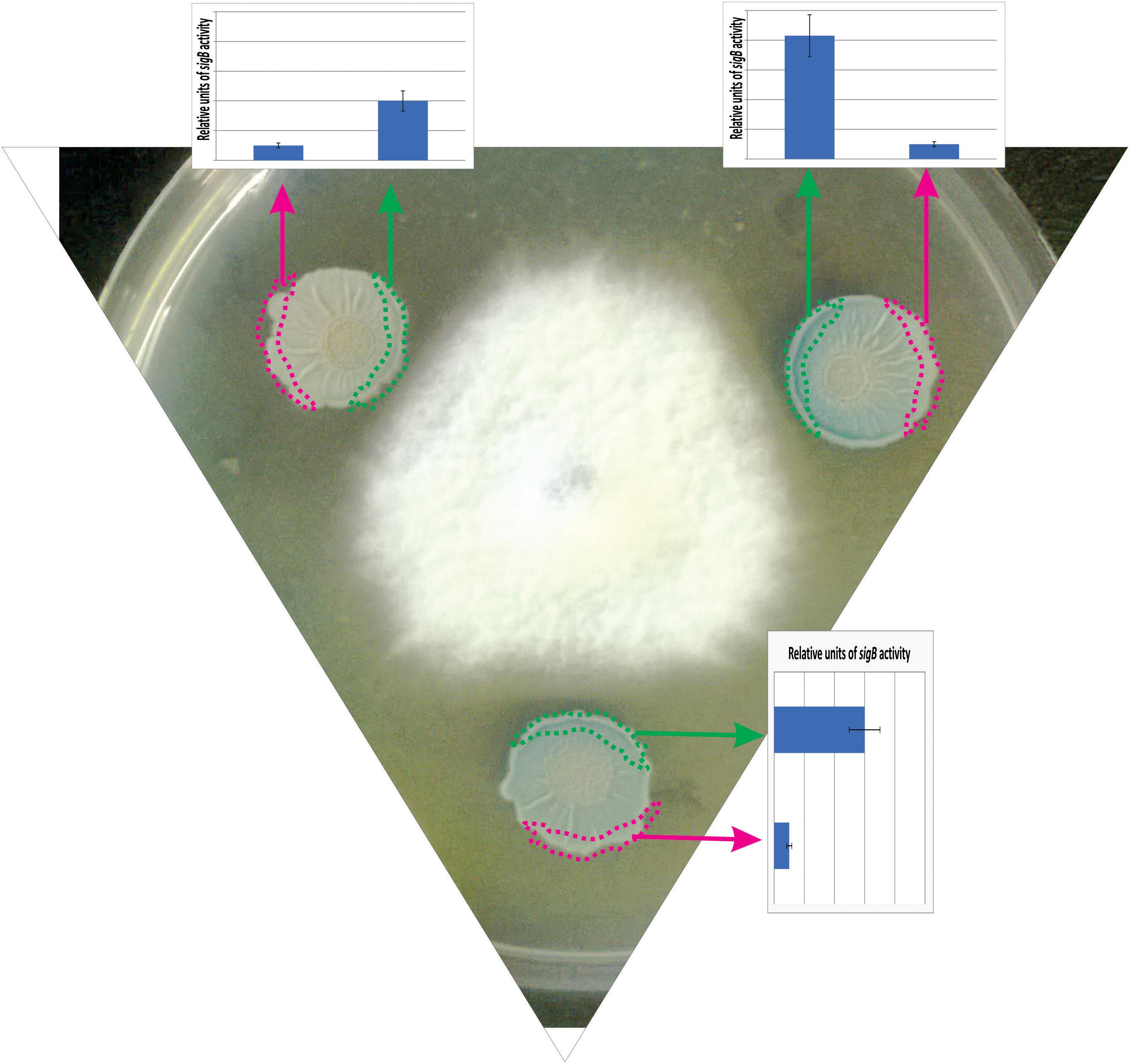
Figure 5. Antagonist response of B. subtilis confronted with the phytopathogen Fusarium verticillioides. The co-culture of a wild-type B. subtilis strain harboring a SigB-reporter gene fusion (ctc-lacZ) with F. verticillioides allows the observation of the antagonistic fungus-bacterium interaction. At the start of the co-incubation, 5 × 105 colony forming units (CFU) of fungal mycelia were poured at the center of the Petri dish, and 1 × 105 colony forming units (CFU) of a stationary phase culture of B. subtilis was placed at three different positions surrounding the fungus to better observe the antagonistic interaction. F. verticillioides-induced SigB activation is evidenced by blue color (derived from the expression of the ctc-lacZ fusion) in the borders of the B. subtilis biofilms (colonies) closer to the fungus. The areas represented by the green and pink rectangles correspond to the biofilm areas used to quantify the β-galactosidase activity (derived from the expressed ctc-lacZ fusion, blue color, inside the biofilm). The relative quantification of the β-galactosidase activity is shown as bar graphs. Microorganisms (B. subtilis and fungi) were co-incubated in PDA (Potato Dextrose Agar) plates at 28°C during 96 h before the photograph is taken.
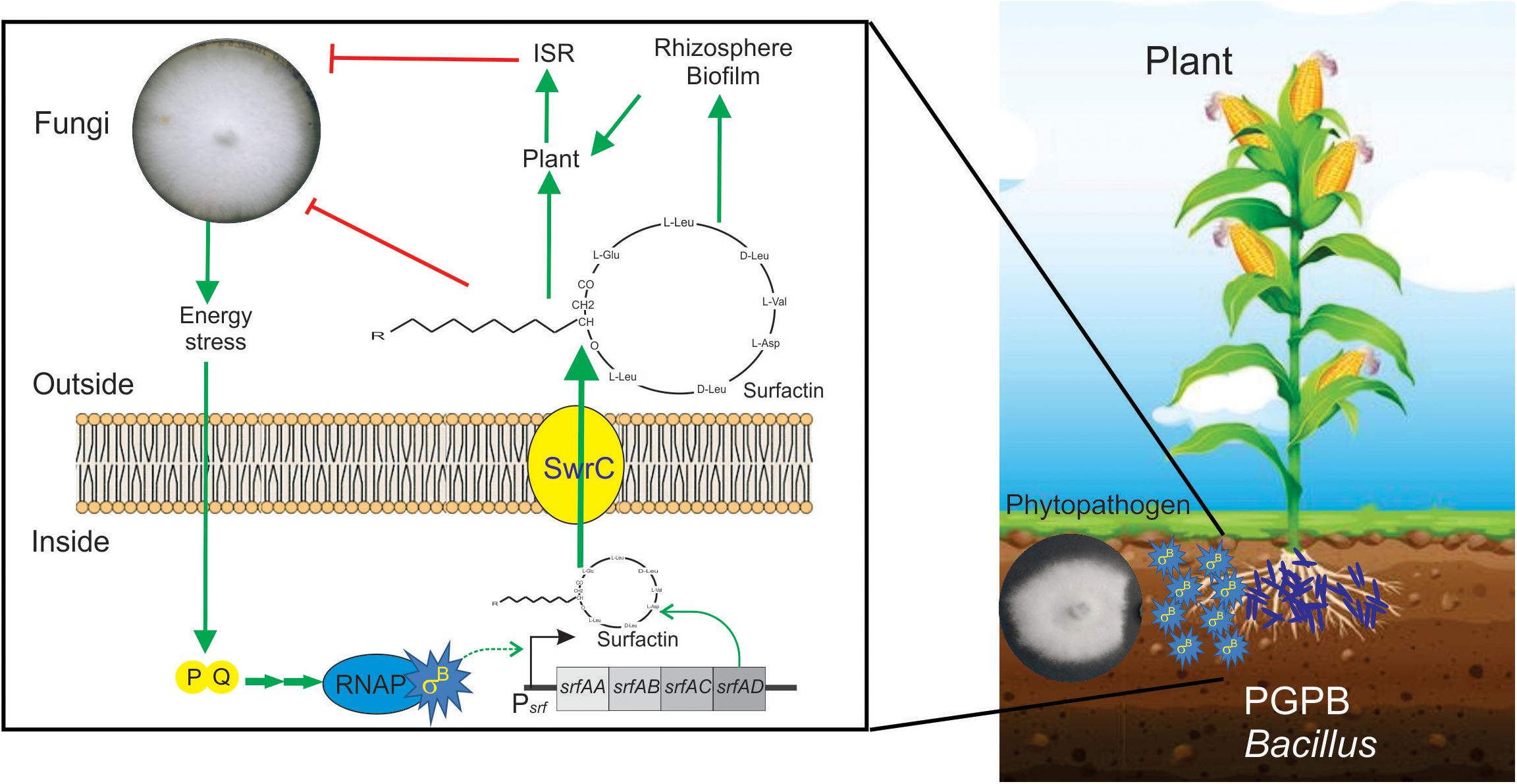
Figure 6. Positive role of SigB in the biocontrol proficiency of PGPB B. subtilis. This cartoon summarizes the beneficial interaction between surfactin-producing B. subtilis cells and plants to resist phytopathogenic fungi. Biofilm- and surfactin-proficient B. subtilis cells can establish a beneficial biofilm at the plant rhizosphere and detect the fungal presence around them. The signal of fungal presence is transduced to the bacterium by an unknown signal that activates the energy stress pathway (PQ in the cartoon) of the SigB regulatory network. Active SigB increases the synthesis of the antifungal lipopeptide surfactin which is exported from the cell through the channel SwrC and/or free cellular membrane diffusion (not shown for simplicity). Outside the bacterial cells, surfactin exerts its fungal growth inhibitory effect. Additionally, surfactins have other two important properties. First, they are important molecules for the proficiency of bacilli at establishing robust and persistent beneficial biofilms at the plant rhizosphere, and second, they can induce plant systemic resistance (ISR) against pathogens. It has been reported that there is specific positive feedback (not shown in the figure for simplicity) from the plant to the bacterium, in which plant polysaccharides stimulate B. subtilis biofilm formation. See the text for details.
A recent report, using fluorescent reporter gene fusions to sigB promoter (Psig-YFP), conclusively demonstrated the existence of stochastic pulsing of SigB activation, absent in ΔsigB cells, during biofilm formation at the interface with the roots of the model plant Arabidopsis thaliana (Nadezhdin et al., 2020). Interestingly, the sigB expression pattern was not affected when the environmental-related stress pathway was inactivated (ΔrsbRU strain) (Nadezhdin et al., 2020). In contrast, sigB expression was severely downregulated when the energy-related pathway (ΔrbsPQ) was inactivated. These observations (Nadezhdin et al., 2020) agree with the results of Bartolini et al. (2019b), showing that only B. subtilis strains proficient in surfactin production and RsbPQ-dependent SigB activation formed beneficial biofilms at the roots of maize plants to protect them from the fungi assault. Overall these results (Bartolini et al., 2019b; Nadezhdin et al., 2020) point to the importance of the energy stress route of SigB activation in the environmentally friendly biofilm established at the plane of B. subtilis-plant interaction against phytopathogenic fungi of agronomic impact (Figure 6).
Concluding Remarks
SigB was the first alternative sigma factor characterized in bacteria, initially hypothesized 40 years ago as a specific transcription factor responsible for the onset of sporulation. Over the years, we have established that SigB plays much broader roles that go beyond spore formation to general stress response, adaption, and survival. Even though early studies involved bacteria living a planktonic lifestyle, recent advances have shown the clear participation of SigB in the regulation of bacterial multicellular lifestyles, such as social biofilms and interaction with other microorganisms. SigB activation is now reported to be casuistically or stochastically established by environmental and energy stresses or noise modulation, respectively. Much work is still needed to establish all the functions regulated by SigB, which is conserved in B. subtilis and many other bacteria.
Author Contributions
FR and RG wrote the main text. FR and MB designed the tables and figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Universidad Nacional de Rosario, CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas) and FONCyT (Fondo para la Investigación Científica y Tecnológica) with the aid of the Pew Latin American Program in Biomedical Sciences (Philadelphia, PA, United States), the Fulbright Committee (Washington, DC, United States), and former Fundación Antorchas (Buenos Aires, Argentina).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acar, M., Becskei, A., and van Oudenaarden, A. (2005). Enhancement of cellular memory by reducing stochastic transitions. Nature 435, 228–232. doi: 10.1038/nature03524
Aguilar, C., Vlamakis, H., Losick, R., and Kolter, R. (2007). Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 10, 638–643. doi: 10.1016/j.mib.2007.09.006
Akbar, S., Gaidenko, T. A., Kang, C. M., O’Reilly, M., Devine, K. M., and Price, C. W. (2001). New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma (B) of Bacillus subtilis. J Bacteriol. 183, 1329–1338. doi: 10.1128/jb.183.4.1329-1338.2001
Aleti, G., Lehner, S., Bacher, M., Compant, S., Nikolic, B., Plesko, M., et al. (2016). Surfactin variants mediate species, specific biofilm formation and root colonization in Bacillus: surfactins mediate species-specific signaling. Environ. Microbiol. 18, 2634–2645. doi: 10.1111/1462-2920.13405
Alper, S., Dufour, A., Garsin, D. A., Duncan, L., and Losick, R. (1996). Role of adenosine nucleotidesin the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260, 165–177. doi: 10.1006/jmbi.1996.0390
Barraud, N., Kjelleberg, S., and Rice, S. A. (2015). Dispersal from microbial biofilms. Microbiol. Spectrum 3:MB-0015-2014. doi: 10.1128/microbiolspec.MB-0015-2014
Bartolini, M., Cogliati, S., Vileta, D., Bauman, C., Rateni, L., Leñini, C., et al. (2019a). Regulation of biofilm aging and dispersal in Bacillus subtilis by the alternative sigma factor SigB. J. Bacteriol. 201:e00473-18. doi: 10.1128/JB.00473-18
Bartolini, M., Cogliati, S., Vileta, D., Bauman, C., Ramirez, W., and Grau, R. (2019b). Stress-responsive alternative sigma factor sigb plays a poe antifungal proficiency of Bacillus subtilis. Appl. Environ. Microbiol. 85:e00178-19. doi: 10.1128/AEM.00178-19
Beauregard, P. B., Chaib, Y., Vlamakis, H., Losick, R., and Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 110, 1621–1630.
Benson, A. K., and Haldenwang, W. G. (1992). Characterization of a regulatory network that controls sigma B expression in Bacillus subtilis. J. Bacteriol. 174, 749–757. doi: 10.1128/jb.174.3.749-757.1992
Benson, A. K., and Haldenwang, W. G. (1993a). Bacillus subtilisσB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 90, 2330–2334. doi: 10.1073/pnas.90.6.2330
Benson, A. K., and Haldenwang, W. G. (1993b). The σB dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J. Bacteriol. 175, 1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147.
Bhaya, D., Watanabe, N., Ogawa, T., and Grossman, A. R. (1999). The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci. U.S.A. 96, 3188–3193. doi: 10.1073/pnas.96.6.3188
Binnie, C., Lampe, M., and Losick, R. (1986). Gene encoding the s37 species of RNA polymerase s factor from Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 83, 5943–5947. doi: 10.1073/pnas.83.16.5943
Bischoff, M., Dunman, P., Kormanec, J., Macapagal, D., Murphy, E., Mounts, W., et al. (2004). Microarray-based analysis of the Staphylococcus aureus sigma B regulon. J. Bacteriol. 186, 4085–4099. doi: 10.1128/jb.186.13.4085-4099.2004
Bonilla, C. Y. (2020). Generally stressed out bacteria: environmental stress response mechanisms in Gram-positive bacteria [published online ahead of print, 2020 Feb 11]. Integr. Comp. Biol. 2020:icaa002.
Borukhov, S., Lee, J., and Laptenko, O. (2005). Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55, 1315–1324. doi: 10.1111/j.1365-2958.2004.04481.x
Borukhov, S., and Nudler, E. (2008). RNA polymerase: the vehicle of transcription. Trends Microbiol. 16, 126–134. doi: 10.1016/j.tim.2007.12.006
Boylan, S. A., Redfield, A. R., Brody, M. S., and Price, C. W. (1993). Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175, 7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993
Boylan, S. A., Rutherford, A., Thomas, S. M., and Price, C. W. (1992). Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174, 3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992
Boylan, S. A., Thomas, M. D., and Price, C. W. (1991). Genetic method to identify regulons controlled by nonessential elements: isolation of a gene dependent on alternate transcription factor sB of Bacillus subtilis. J. Bacteriol. 173, 7856–7866. doi: 10.1128/jb.173.24.7856-7866.1991
Branda, S. S., Chu, F., Kearns, D. B., Losick, R., and Kolter, R. (2006). A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x
Branda, S. S., González-Pastor, J. E., Ben-Yehuda, S., Losick, R., and Kolter, R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 98, 11621–11626. doi: 10.1073/pnas.191384198
Brigulla, M., Hoffmann, T., Krisp, A., Volker, A., Bremer, E., and Volker, U. (2003). Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185, 4305–4314. doi: 10.1128/jb.185.15.4305-4314.2003
Brody, M. S., Stewart, V., and Price, C. W. (2009). Bypass suppression analysis maps the signalling pathway within a multidomain protein: the RsbP energy stress phosphatase 2C from Bacillus subtilis. Mol. Microbiol. 72, 1221–1234. doi: 10.1111/j.1365-2958.2009.06722.x
Brody, M. S., Vijay, K., and Price, C. W. (2001). Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma (B) transcription factor in Bacillus subtilis. J. Bacteriol. 183, 6422–6428. doi: 10.1128/jb.183.21.6422-6428.2001
Burton, A. T., DeLoughery, A., Li, G. W., and Kearns, D. B. (2019). Transcriptional regulation and mechanism of SigN (ZpdN), a pBS32-encoded sigma factor in Bacillus subtilis. mBio 10:e01899-19. doi: 10.1128/mBio.01899-19
Cao, M., and Helmann, J. D. (2004). The Bacillus subtilis extracytoplasmic-function sigma X factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186, 1136–1146. doi: 10.1128/jb.186.4.1136-1146.2004
Cao, M., Wang, T., Ye, R., and Helmann, J. D. (2002). Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma W and sigma M regulons. Mol. Microbiol. 45, 1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x
Cawoy, H., Debois, D., Franzil, L., De Pauw, E., Thonart, P., and Ongena, M. (2015). Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens: lipopeptides as inhibitors of phytopathogens. Mol. Microbiol. 8, 281–295. doi: 10.1111/1751-7915.12238
Cawoy, H., Mariutto, M., Henry, G., Fisher, C., Vasilyeva, N., Thonart, P., et al. (2014). Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant Microbe. Interact. 27, 87–100. doi: 10.1094/mpmi-09-13-0262-r
Chatterji, D., Ogawa, Y., Shimada, T., and Ishihama, A. (2007). The role of the omega subunit of RNA polymerase in expression of the relA gene in Escherichia coli. FEMS Microbiol. Lett. 267, 51–55. doi: 10.1111/j.1574-6968.2006.00532.x
Chaturongakul, S., Raengpradub, S., Wiedmann, M., and Boor, K. J. (2008). Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 16, 388–396. doi: 10.1016/j.tim.2008.05.006
Chen, C. C., Lewis, R. J., Harris, R., Yudkin, M. D., and Delumeau, O. (2003). A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49, 1657–1669. doi: 10.1046/j.1365-2958.2003.03663.x
Chen, F., Di, H., Wang, Y., Cao, Q., Xu, B., Zhang, X., et al. (2016). Small-molecule targeting of a diapophytoene desaturase inhibits S. aureus virulence. Nat. Chem. Biol. 12, 174–179. doi: 10.1038/nchembio.2003
Chen, C. C., Yudkin, M. D., and Delumeau, O. (2004). Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J. Bacteriol. 186, 6830–6836. doi: 10.1128/JB.186.20.6830-6836.2004
Cho, Y. H., Lee, E. J., Ahn, B. E., and Roe, J. H. (2001). SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42, 205–214. doi: 10.1046/j.1365-2958.2001.02622.x
Chu, F., Kearns, D. B., Branda, S. S., Kolter, R., and Losick, R. (2006). Targets the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 59, 1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x
Cogliati, S., Clementi, V., Francisco, M., Crespo, C., Argañaraz, F., and Grau, R. (2019). Bacillus subtilis delays neurodegeneration and behavioral impairment in the Alzheimer’s disease model Caenorhabditis elegans. J. Alzheimer’s Dis. 73, 1055–1072.
Cramer, P., Bushnell, D. A., and Kornberg, R. D. (2001). Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876. doi: 10.1126/science.1059493
Dalton, T., Dowd, S. E., Wolcott, R., Sun, Y., Watters, C., Griswold, J., et al. (2011). An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. doi: 10.1371/journal.pone.0027317
de Been, M., Francke, C., Siezen, R. J., and Abee, T. (2011). Novel sigmaB regulation modules of Gram-positive bacteria involve the use of complex hybrid histidine kinases. Microbiology 157(Pt 1), 3–12. doi: 10.1099/mic.0.045740-0
de Been, M., Tempelaars, M. H., van Schaik, W., Moezelaar, R., Siezen, R. J., and Abee, T. (2010). A novel hybrid kinase is essential for regulating the sigma(B)-mediated stress response of Bacillus cereus. Environ. Microbiol. 12, 730–745. doi: 10.1111/j.1462-2920.2009.02116.x
Debarbouille, M., Martin-Verstraete, I., Klier, A., and Rapaport, G. (1991). The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both s54 and phosphotransferase system-dependent regulators. Proc. Natl. Acad. Sci. U.S.A. 88, 2212–2216. doi: 10.1073/pnas.88.6.2212
Debois, D., Fernandez, O., Franzil, L., Jourdan, E., de Brogniez, A., Willems, L., et al. (2015). Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin: Bacillus lipopeptides induced by plant polymers. Environ. Microbiol. Rep. 7, 570–582. doi: 10.1111/1758-2229.12286
Delumeau, O., Chen, C. C., Murray, J. W., Yudkin, M. D., and Lewis, R. J. (2006). High molecular weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 188, 7885–7892. doi: 10.1128/jb.00892-06
DeMaio, J., Zhang, Y., Ko, C., and Bishai, W. R. (1997). Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber. Lung Dis. 78, 3–12. doi: 10.1016/s0962-8479(97)90010-1
DeMaio, J., Zhang, Y., Ko, C., Young, D. B., and Bishai, W. R. (1996). A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci U.S.A. 93, 2790–2794. doi: 10.1073/pnas.93.7.2790
Donato, V., Rodriguez Ayala, F., Cogliati, S., Bauman, C., Costa, J. G., Leñini, C., et al. (2017). Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signaling pathway. Nat. Commun. 8:14332.
Dufour, A., and Haldenwang, W. G. (1994). Interactions between a Bacillus subtilis antisigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176, 1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994
Eymann, C., Schulz, S., Gronau, K., Becher, D., Hecker, M., and Price, C. W. (2011). In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB. Mol Microbiol. 80, 798–810. doi: 10.1111/j.1365-2958.2011.07609.x
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Fouet, A., Namy, O., and Lambert, G. (2000). Characterization of the operon encoding the alternative sigma B factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182, 5036–5045. doi: 10.1128/jb.182.18.5036-5045.2000
Fredrick, K., and Helmann, J. D. (1996). FlgM is the primary regulator of sigD activity, and its absence restores motility to a sinR mutant. J. Bacteriol. 178, 7010–7013. doi: 10.1128/jb.178.23.7010-7013.1996
Gaidenko, T. A., Bie, X., Baldwin, E. P., and Price, C. W. (2012). Two surfaces of a conserved interdomain linker differentially affect output from the RST sensing module of the Bacillus subtilis stressosome. J Bacteriol. 194, 3913–3921. doi: 10.1128/jb.00583-12
Gaidenko, T. A., Kim, T. J., Weigel, A. L., Brody, M. S., and Price, C. W. (2006). The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188, 6387–6395. doi: 10.1128/jb.00691-06
Geiman, D. E., Kaushal, D., Ko, C., Tyagi, S., Manabe, Y. C., Schroeder, B. G., et al. (2004). Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72, 1733–1745. doi: 10.1128/iai.72.3.1733-1745.2004
Giachino, P., Engelmann, S., and Bischoff, M. (2001). Sigma B activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183, 1843–1852. doi: 10.1128/jb.183.6.1843-1852.2001
Gitt, M. A., Wang, L. F., and Doi, R. H. (1985). A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J. Biol. Chem. 260, 7178–7185.
Gottig, N., Pedrido, M. E., Méndez, M., Lombardia, E., Rovetto, A., Philippe, V., et al. (2005). The Bacillus subtilis SinR and RapA developmental regulators are responsible for the inhibition of spore development by alcohol. J. Bacteriol. 187, 2662–2672. doi: 10.1128/jb.187.8.2662-2672.2005
Goya, F., Xue, C., Sampedro-Torres-Quevedo, S., Arnaouteli, L., Riquelme-Dominguez, A., Romanowski, J., et al. (2020). Probiotic Bacillus subtilis prorotects against α-Synuclein aggregation in C. elegans. Cell Rep. 14, 367–380. doi: 10.1016/j.celrep.2019.12.078
Grau, R., de Oña, P., Kunert, M., Leñini, C., Gallegos-Monterrosa, R., Mhatre, E., et al. (2015). A duo of potassium-responsive histidine kinases governs the multicellular destiny of Bacillus subtilis. mBio 6:e00581-15. doi: 10.1128/mBio.00581-15
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Guilhen, C., Forestier, C., and Balestrino, D. (2017). Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol. Microbiol. 105, 188–210. doi: 10.1111/mmi.13698
Haldenwang, W. G. (1995). The sigma factors of Bacillus subtilis. Microbiol. Rev. 59, 1–30. doi: 10.1128/mmbr.59.1.1-30.1995
Haldenwang, W. G., and Losick, R. (1979). A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature 282, 256–260. doi: 10.1038/282256a0
Hecker, M., Pané-Farré, J., and Völker, U. (2007). SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236. doi: 10.1146/annurev.micro.61.080706.093445
Hecker, M., Schumann, W., and Völker, U. (1996). Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19, 417–428. doi: 10.1046/j.1365-2958.1996.396932.x
Hecker, M., and Völker, U. (2001). General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44, 35–91. doi: 10.1016/s0065-2911(01)44011-2
Helmann, J. D. (1991). Alternate sigma factors and the regulation of flagellar gene expression. Mol. Microbiol. 5, 2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x
Helmann, J. D. (2002). The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 46, 47–110. doi: 10.1016/s0065-2911(02)46002-x
Helmann, J. D. (2019). Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol. Microbiol. 112, 335–347. doi: 10.1111/mmi.14309
Helmann, J. D., Wu, M. F., Gaballa, A., Kobel, P. A., Morshedi, M. M., Fawcett, P., et al. (2003). The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185, 243–253. doi: 10.1128/jb.185.1.243-253.2003
Hirata, A., Klein, B. J., and Murakami, K. S. (2008). The X-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854. doi: 10.1038/nature06530
Hölscher, T., Bartels, B., Lin, Y. C., Gallegos-Monterrosa, R., Price-Whelan, A., Kolter, R., et al. (2015). Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J. Mol. Biol. 427, 3695–3708. doi: 10.1016/j.jmb.2015.06.014
Hong, H., Khaneja, R., Tam, N., Cazzato, A., Tan, S., Urdaci, M., et al. (2009). Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 160, 134–143. doi: 10.1016/j.resmic.2008.11.002
Huckauf, J., Nomura, C., Forchhammer, K., and Hagemann, M. (2000). Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors. Microbiology 146(Pt 11), 2877–2889. doi: 10.1099/00221287-146-11-2877
Impens, F., Rolhion, N., Radoshevich, L., Bécavin, C., Duval, M., Mellin, J., et al. (2017). N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat Microbiol. 2:e17005.
Jefferson, K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x
Johnson, W. C., Moran, C. P. Jr., and Losick, R. (1983). Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature 302, 800–804. doi: 10.1038/302800a0
Kaneko, T., Tanaka, N., and Kumasaka, T. (2005). Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis. Protein Sci. 14, 558–565. doi: 10.1110/ps.041170005
Kaplan, J. B. (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218. doi: 10.1177/0022034509359403
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734. doi: 10.1128/jb.185.19.5722-5734.2003
Kearns, D. B., Chu, F., Branda, S. S., Kolter, R., and Losick, R. (2005). A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55, 739–749. doi: 10.1111/j.1365-2958.2004.04440.x
Kearns, D. B., and Losick, R. (2003). Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49, 581–590. doi: 10.1046/j.1365-2958.2003.03584.x
Kim, K. H., Kabir, E., and Jahan, S. A. (2017). Exposure to pesticides and the associated human health effects. Sci. Total Environ. 575, 525–535. doi: 10.1016/j.scitotenv.2016.09.009
Kim, T. J., Gaidenko, T. A., and Price, C. W. (2004a). A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341, 135–150. doi: 10.1016/j.jmb.2004.05.043
Kim, T. J., Gaidenko, T. A., and Price, C. W. (2004b). In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 186, 6124–6132. doi: 10.1128/jb.186.18.6124-6132.2004
Kint, N., Alves Feliciano, C., Hamiot, A., Denic, M., Dupuy, B., and Martin-Verstraete, I. (2019). The σB signalling activation pathway in the enteropathogen Clostridioides difficile. Environ. Microbiol. 21, 2852–2870. doi: 10.1111/1462-2920.14642
Kint, N., Janoir, C., Monot, M., Hoys, S., Soutourina, O., Dupuy, B., et al. (2017). The alternative sigma factor σB plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environ. Microbiol. 19, 1933–1958. doi: 10.1111/1462-2920.13696
Kovács, ÁT. (2016). Bacterial differentiation via gradual activation of global regulators. Curr. Genet 62, 125–128. doi: 10.1007/s00294-015-0524-8
Kovács, ÁT., and Dragoš, A. (2019). Evolved biofilm: review on the experimental evolution studies of Bacillus subtilis Pellicles. J. Mol. Biol. 431, 4749–4759. doi: 10.1016/j.jmb.2019.02.005
Kumar, A., Grimes, B., Fujita, N., Makino, K., Malloch, R. A., Hayward, R. S., et al. (1994). Role of the sigma 70 subunit of Escherichia coli RNA polymerase in transcription activation. J. Mol. Biol. 14, 405–413.
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., et al. (1997). The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256.
Kuo, S., Demeler, B., and Haldenwang, W. G. (2008). The growth-promoting and stress response activities of the Bacillus subtilis GTP binding protein Obg are separable by mutation. J. Bacteriol. 190, 6625–6635. doi: 10.1128/jb.00799-08
Kussell, E., and Leibler, S. (2005). Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078. doi: 10.1126/science.1114383
Kwon, E., Pathak, D., Kim, H. U., Dahal, P., Ha, S. C., Lee, S. S., et al. (2019). Structural insights into stressosome assembly. I.U.Cr.J. 6(Pt 5), 938–947. doi: 10.1107/s205225251900945x
Lee, E. J., Cho, Y. H., Kim, H. S., Ahn, B. E., and Roe, J. H. (2004). Regulation of sigmaB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 186, 8490–8498. doi: 10.1128/jb.186.24.8490-8498.2004
Lewis, P. J., Doherty, G. P., and Clarke, J. (2008). Transcription factor dynamics. Microbiology 154(Pt 7), 1837–1844.
Liebal, U. W., Millat, T., Marles-Wright, J., Lewis, R. J., and Wolkenhauer, O. (2013). Simulations of stressosome activation emphasize allosteric interactions between RsbR and RsbT. BMC Syst Biol. 7:3. doi: 10.1186/1752-0509-7-3
Liu, J., Prindle, A., Humphries, J., Gabalda-Sagarra, M., Asally, M., Lee, D. Y., et al. (2015). Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554. doi: 10.1038/nature14660
Locke, J. C., Young, J. W., Fontes, M., Hernández Jiménez, M. J., and Elowitz, M. B. (2011). Stochastic pulse regulation in bacterial stress response. Science 334, 366–369. doi: 10.1126/science.1208144
Lombardia, E., Rovetto, A. J., Arabolaza, A. L., and Grau, R. (2006). A LuxS dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J. Bacteriol. 188, 4442–4452. doi: 10.1128/jb.00165-06
Losick, R., and Pero, J. (2018). For want of a template. Cell 172, 1146–1152. doi: 10.1016/j.cell.2018.02.041
Losick, R., and Stragier, P. (1992). Crisscross regulation of cell-type-specific gene expression during development in Bacillus subtilis. Nature 355, 601–604. doi: 10.1038/355601a0
Luo, C., Zhou, H., Zou, J., Wang, X., Zhang, R., Xiang, Y., et al. (2015). Bacillomycin L and surfactin contribute synergistically to the phenotypic features of Bacillus subtilis 916 and the biocontrol of rice sheath blight induced by Rhizoctonia solani. Appl. Microbiol. Biotechnol. 99, 1897–1910. doi: 10.1007/s00253-014-6195-4
Luo, Y., and Helmann, J. D. (2012). Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 83, 623–639. doi: 10.1111/j.1365-2958.2011.07953.x
Marles-Wright, J., Grant, T., Delumeau, O., van Duinen, G., Firbank, S. J., Lewis, P. J., et al. (2008). Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322, 92–96. doi: 10.1126/science.1159572
Martinez, L., Reeves, A., and Haldenwang, W. (2010). Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow σ(B) activation following exposure to either physical or nutritional stress. J. Bacteriol. 192, 6279–6286. doi: 10.1128/jb.00467-10
Mascher, T., Margulis, N. G., Wang, T., Ye, R. W., and Helmann, J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50, 1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P., and Kjelleberg, S. (2011). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695
Méndez, M., Orsaria, L., Philippe, V., Pedrido, M. E., and Grau, R. (2004). Novel Roles of the master transcription factors Spo0A and σB for survival and sporulation of Bacillus subtilis at low growth temperature. J. Bacteriol. 186, 989–1000. doi: 10.1128/jb.186.4.989-1000.2004
Mendez, R., Gutierrez, A., Reyes, J., and Márquez-Magaña, L. (2012). The extracytoplasmic function sigma factor SigY is important for efficient maintenance of the Spβ prophage that encodes sublancin in Bacillus subtilis. DNA Cell Biol. 31, 946–955. doi: 10.1089/dna.2011.1513
Michele, T. M., Ko, C., and Bishai, W. R. (1999). Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43, 218–225. doi: 10.1128/aac.43.2.218
Minakhin, L., Bhagat, S., Brunning, A., Campbell, E. A., Darst, S. A., Ebright, R. H., et al. (2001). Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. U.S.A. 98, 892–897. doi: 10.1073/pnas.98.3.892
Moore, C. M., Nakano, M. M., Wang, T., Ye, R. W., and Helmann, J. D. (2004). Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 186, 4655–4664. doi: 10.1128/jb.186.14.4655-4664.2004
Moran, C. P. Jr., Lang, N., LeGrice, S. F. J., Lee, G., Stevens, M., Sonenshein, A. L., et al. (1982). Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186, 339–346. doi: 10.1007/bf00729452
Mukherjee, K., Nagai, H., Shimamoto, N., and Chatterji, D. (1999). GroEL is involved in activation of Escherichia coli RNA polymerase devoid of the omega subunit in vivo. Eur. J. Biochem. 266, 228–235. doi: 10.1046/j.1432-1327.1999.00848.x
Murakami, K. (2015). Structural biology of bacterial RNA polymerase. Biomolecules 5, 848–864. doi: 10.3390/biom5020848
Murray, E. J., Strauch, M. A., and Stanley-Wall, N. R. (2009). Sigma X is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh. J. Bacteriol. 191, 6822–6832. doi: 10.1128/jb.00618-09
Nadezhdin, E., Murphy, N., Dalchau, N., Phillips, A., and Locke, J. C. W. (2020). Stochastic pulsing of gene expression enables the generation of spatial patterns in Bacillus subtilis biofilms. Nat Commun. 19:950.
Nicholson, W. L., Sun, D. X., Setlow, B., and Setlow, P. (1989). Promoter specificity of sigma G-containing RNA polymerase from sporulating cells of Bacillus subtilis: identification of a group of forespore-specific promoters. J. Bacteriol. 171, 2708–2718. doi: 10.1128/jb.171.5.2708-2718.1989
Nicolas, P., Mader, U., Dervyn, E., Rochat, T., Leduc, A., Pigeonneau, N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106.
O’Byrne, C. P., and Karatzas, K. A. (2008). The role of sigma B (sigma B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140. doi: 10.1016/s0065-2164(08)00605-9
Ondrusch, N., and Kreft, J. (2011). Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS One 6:e16151. doi: 10.1371/journal.pone.0016151
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ongena, M., Jacques, P., Toure, Y., Destain, J., Jabrane, A., and Thonart, P. (2005). Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl. Microbiol. Biotechnol. 69, 29–38. doi: 10.1007/s00253-005-1940-3
Pané-Farré, J., Jonas, B., Hardwick, S., Gronau, K., Lewis, R., Hecker, M., et al. (2009). Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J. Bacteriol. 191, 2561–2575. doi: 10.1128/jb.01514-08
Pané-Farré, J., Lewis, R. J., and Stülke, J. (2005). The RsbRST stress module in bacteria: a signalling system that may interact with different output modules. J. Mol. Microbiol. Biotechnol. 9, 65–76. doi: 10.1159/000088837
Pedrido, M. E., de Oña, P., Ramirez, W., Leñini, C., Goñi, A., and Grau, R. (2013). Spo0A links de novo fatty acid synthesis to sporulation and biofilm development in Bacillus subtilis. Mol. Microbiol. 87, 348–367. doi: 10.1111/mmi.12102
Petersohn, A., Bernhardt, J., Gerth, U., Höper, D., Koburger, T., Völker, U., et al. (1999). Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181, 5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999
Pietiaeinen, M., Gardemeister, M., Mecklin, M., Leskelae, S., Sarvas, M., and Kontinen, V. P. (2005). Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151, 1577–1592. doi: 10.1099/mic.0.27761-0
Piewngam, P., Yue, Z. P., Nguyen, T. H., Dickey, S. W., Joo, H. S., Villaruz, A. E., et al. (2018). Pathogen elimination by probiotic Bacillus via signaling interference. Nature 562, 532–537. doi: 10.1038/s41586-018-0616-y
Piggot, P. J., and Hilbert, D. W. (2004). Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7, 579–586.
Pingali, P. L. (2012). Green revolution: impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. U.S.A. 109, 12302–12308. doi: 10.1073/pnas.0912953109
Predich, M., Nair, G., and Smith, I. (1992). Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sH. J. Bacteriol. 174, 2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992
Price, C. W. (2000). “Protective function and regulation of the general stress response in Bacillus subtilis and related Gram-positive bacteria,” in Bacterial Stress Responses, eds G. Storz and R. Hengge-Aronis (Washington, DC: ASM Press), 179–197.
Price, C. W. (2002). “General stress response,” in Bacillus subtilis and its Closest Relatives. From Genes to Cells, eds A. L. Sonenshein, J. A. Hoch, and R. Losick (Washington, DC: ASM Press), 369–384.
Raaijmakers, J. M., De Bruijn, I., Nybroe, O., and Ongena, M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x
Raja, N. (2013). Biopesticides and biofertilizers: ecofriendly sources for sustainable agriculture. J. Biofertil. Biopestici. 4, 1–2.
Reder, A., Albrecht, D., Gerth, U., and Hecker, M. (2012a). Cross-talk between the general stress response and sporulation initiation in Bacillus subtilis - the σ(B) promoter of spo0E represents an AND-gate. Environ. Microbiol. 14, 2741–2756. doi: 10.1111/j.1462-2920.2012.02755.x
Reder, A., Gerth, U., and Hecker, M. (2012b). Integration of σB activity into the decision-making process of sporulation initiation in Bacillus subtilis. J. Bacteriol. 194, 1065–1074. doi: 10.1128/jb.06490-11
Reeves, A., and Haldenwang, W. G. (2006). Isolation and characterization of dominant mutations in the Bacillus subtilis stressosome components RsbR and RsbS. J. Bacteriol. 189, 1531–1541. doi: 10.1128/jb.01649-06
Roels, S., Driks, A., and Losick, R. (1992). Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174, 575–585. doi: 10.1128/jb.174.2.575-585.1992
Rothstein, D. M., Lazinski, D., Osburne, M. S., and Sonenshein, A. L. (2017). A Mutation in the Bacillus subtilis rsbU gene that limits RNA synthesis during sporulation. J. Bacteriol. 199, e212–e217.
Scott, J. M., and Haldenwang, W. G. (1999). Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor sigma(B). J. Bacteriol. 181, 4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999
Shaw, L., Aish, J., Davenport, J., Brown, M., Lithgow, J., Simmonite, K., et al. (2006). Investigations into σB-modulated regulatory pathways governing extracellular virulence determinant production in Staphylococcus aureus. J. Bacteriol. 188, 6070–6080. doi: 10.1128/jb.00551-06
Singh, P. K., Bartalomej, S., Hartmann, R., Jeckel, H., Vidakovic, L., Nadell, C., et al. (2017). Vibrio cholerae combines individual and collective sensing to trigger biofilm dispersal. Curr. Biol. 27, 3359–3366. doi: 10.1016/j.cub.2017.09.041
Smolentseva, O., Gusarov, I., Gautier, L., Shamovsky, I., De Francesco, A., Losick, R., et al. (2017). Mechanism of biofilm-mediated stress resistance and lifespan extension in Caenorhabditis elegans. Sci. Rep. 7:7137.
Sorokin, A., Bolotin, A., Purnelle, B., Hilbert, H., Lauber, J., Düsterhöft, A., et al. (1997). Sequence of the Bacillus subtilis genome region in the vicinity of the lev operon reveals two new extracytoplasmic function RNA polymerase sigma factors SigV and SigZ. Microbiology 143(Pt 9), 2939–2943. doi: 10.1099/00221287-143-9-2939
Stacy, A., Everett, J., Jorth, P., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2014). Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 111, 7819–7824. doi: 10.1073/pnas.1400586111
Sun, D., Fajardo-Cavazos, P., Sussman, M. D., Tovar-Rojo, F., Cabrera- Martinez, R.-M., and Setlow, P. (1991). Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E-sF: identification of features of good E-sF-dependent promoters. J. Bacteriol. 173, 7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991
Tago, D., Andersson, H., and Treich, N. (2014). “Pesticides and health: a review of evidence on health effects, valuation of risks, and benefit-cost analysis,” in Advances in Health Economics and Health Services Research, eds G. C. Blomquist and K. Bolin (Bingley: Emerald Group Publishing Ltd), 203–295. doi: 10.1108/s0731-2199_2014_0000024006
Thackray, P. D., and Moir, A. (2003). SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185, 3491–3498. doi: 10.1128/jb.185.12.3491-3498.2003
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Trach, K., and Hoch, J. A. (1989). The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171, 1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989
Tran, V., Geraci, K., Midili, G., Satterwhite, W., Wright, R., and Bonilla, C. Y. (2019). Resilience to oxidative and nitrosative stress is mediated by the stressosome, RsbP and SigB in Bacillus subtilis. J, Basic Microbiol. 59, 834–845. doi: 10.1002/jobm.201900076
van der Veen, S., and Abee, T. (2010). Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 76, 7854–7860. doi: 10.1128/aem.01519-10
van Schaik, W., and Abee, T. (2005). The role of sigmaB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr. Opin. Biotechnol. 16, 218–224. doi: 10.1016/j.copbio.2005.01.008
van Schaik, W., Tempelaars, M. H., Wouters, J. A., de Vos, W. M., and Abee, T. (2004). The alternative sigma factor sigmaB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186, 316–325. doi: 10.1128/jb.186.2.316-325.2004
Vijay, K., Brody, M. S., Fredlund, E., and Price, C. W. (2000). A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35, 180–188. doi: 10.1046/j.1365-2958.2000.01697.x
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R., and Kolter, R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Völker, U., Luo, T., Smirnova, N., and Haldenwang, W. (1997). Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J. Bacteriol. 179, 1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997
Völker, U., Völker, A., Maul, B., Hecker, M., Dufour, A., and Haldenwang, W. G. (1995). Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177, 3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995
Vrentas, C. E., Gaal, T., Ross, W., Ebright, R. H., and Gourse, R. L. (2005). Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 19, 2378–2387. doi: 10.1101/gad.1340305
Wiedmann, M., Arvik, T. J., Hurley, R. J., and Boor, K. J. (1998). General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180, 3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998
Wiegert, T., Homuth, G., Versteeg, S., and Schumann, W. (2001). Alkaline shock induces the Bacillus subtilis sigma W regulon. Mol. Microbiol. 41, 59–71. doi: 10.1046/j.1365-2958.2001.02489.x
Williams, E. P., Lee, J. H., Bishai, W. R., Colantuoni, C., and Karakousis, P. C. (2007). Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 189, 4234–4242. doi: 10.1128/jb.00201-07
Wise, A. A., and Price, C. W. (1995). Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals. J. Bacteriol. 177, 123–133. doi: 10.1128/jb.177.1.123-133.1995
Wu, S., de Lencastre, H., and Tomasz, A. (1996). Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178, 6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996
Yahav, S., Berkovich, Z., Ostrov, I., Reifen, R., and Shemesh, M. (2018). Encapsulation of beneficial probiotic bacteria in extracellular matrix from biofilm-forming Bacillus subtilis. Artif. Cells Nanomed. Biotechnol. 46, 974–982. doi: 10.1080/21691401.2018.1476373
Yang, X., Kang, C. M., Brody, M. S., and Price, C. W. (1996). Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10, 2265–2275. doi: 10.1101/gad.10.18.2265
Zellmeier, S., Hofmann, C., Thomas, S., Wiegert, T., and Schumann, W. (2005). Identification of sigma V-dependent genes of Bacillus subtilis. FEMS Microbiol. Lett. 253, 221–229. doi: 10.1111/j.1365-2958.2006.05131.x
Zeriouh, H., de Vicente, A., Perez-Garcia, A., and Romero, D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16, 2196–2211. doi: 10.1111/1462-2920.12271
Zhang, G., Campbell, E. A., Minakhin, L., Richter, C., Severinov, K., and Darst, S. A. (1999). Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A° resolution. Cell 98, 811–824. doi: 10.1016/s0092-8674(00)81515-9
Zhang, S., and Haldenwang, W. G. (2005). Contributions of ATP, GTP, and redox state to nutritional stress activation of the Bacillus subtilis sigmaB transcription factor. J. Bacteriol. 187, 7554–7560. doi: 10.1128/JB.187.22.7554-7560.2005
Zhang, S., Scott, J. M., and Haldenwang, W. G. (2001). Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B). J. Bacteriol. 183, 2316–2321. doi: 10.1128/JB.183.7.2316-2321.2001
Zheng, L., Halberg, R., Roels, S., Ichikawa, H., Kroos, L., and Losick, R. (1992). Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226, 1037–1050. doi: 10.1016/0022-2836(92)91051-p
Keywords: Bacillus subtilis, alternative sigma factors, SigB, general stress response, biofilm fitness, biocontrol, sporulation
Citation: Rodriguez Ayala F, Bartolini M and Grau R (2020) The Stress-Responsive Alternative Sigma Factor SigB of Bacillus subtilis and Its Relatives: An Old Friend With New Functions. Front. Microbiol. 11:1761. doi: 10.3389/fmicb.2020.01761
Received: 25 April 2020; Accepted: 06 July 2020;
Published: 15 September 2020.
Edited by:
Jörg Stülke, University of Göttingen, GermanyReviewed by:
Erhard Bremer, University of Marburg, GermanyRichard A. Daniel, Newcastle University, United Kingdom
Copyright © 2020 Rodriguez Ayala, Bartolini and Grau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Grau, cm9iZXJ0b2dyYXVAZnVsYnJpZ2h0bWFpbC5vcmc=
†These authors have contributed equally to this work
 Facundo Rodriguez Ayala
Facundo Rodriguez Ayala Marco Bartolini
Marco Bartolini Roberto Grau
Roberto Grau