- Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
(p)ppGpp is at the core of global bacterial regulation as it controls growth, the most important aspect of life. It would therefore be expected that at least across a species the intrinsic (basal) levels of (p)ppGpp would be reasonably constant. On the other hand, the historical contingency driven by the selective pressures on bacterial populations vary widely resulting in broad genetic polymorphism. Given that (p)ppGpp controls the expression of many genes including those involved in the bacterial response to environmental challenges, it is not surprising that the intrinsic levels of (p)ppGpp would also vary considerably. In fact, null mutations or less severe genetic polymorphisms in genes associated with (p)ppGpp synthesis and hydrolysis are common. Such variation can be observed in laboratory strains, in natural isolates as well as in evolution experiments. High (p)ppGpp levels result in low growth rate and high tolerance to environmental stresses. Other aspects such as virulence and antimicrobial resistance are also influenced by the intrinsic levels of (p)ppGpp. A case in point is the production of Shiga toxin by certain E. coli strains which is inversely correlated to (p)ppGpp basal level. Conversely, (p)ppGpp concentration is positively correlated to increased tolerance to different antibiotics such as β-lactams, vancomycin, and others. Here we review the variations in intrinsic (p)ppGpp levels and its consequences across the E. coli species.
Diversity of (p)ppGpp Concentrations–Impact on Growth Rate and Beyond
“The study of bacterial growth is the essence of microbiology” (Jacques Monod).
The success of an organism in evolutionary terms resides in its ability to reproduce and perpetuate its genes. It would thus be expected that bacterial resources would be devoted most of the time to maximizing growth rate. This may be true under some circumstances, especially, under optimal laboratory growth conditions. However, bacteria actually keep growth rate under very tight control. At the core of growth regulation is a small nucleotide that appears in two different forms—guanosine tetra- and penta-phosphate—ppGpp and pppGpp, collectively known as (p)ppGpp. The grip of (p)ppGpp on growth rate is achieved mainly through an effective inhibition of stable RNA (rRNA and tRNA) synthesis during amino acid starvation and other nutritional stresses in a process that became known as the stringent control (Cashel and Gallant, 1968; Potrykus and Cashel, 2008; Potrykus et al., 2011). Nevertheless, the impact of (p)ppGpp on cell physiology goes far beyond stable RNA control. (p)ppGpp also inhibits DNA replication, lipid and protein synthesis and ultimately cell division (Potrykus and Cashel, 2008; Traxler et al., 2008). Whenever the growth conditions deteriorate, (p)ppGpp concentration increases, severely repressing the expression of growth-related genes. This repression is necessary in order to promote the reallocation of resources, which are then shifted from growth promotion to the maintenance of amino acid as well as energy pools and to cell protection and survival. In fact, (p)ppGpp concentration increases stepwise according to the severity of nutrient depletion (Traxler et al., 2011).
In E. coli and related bacterial species, (p)ppGpp is synthesized by two different proteins—RelA and SpoT. These proteins evolved by duplication from a bifunctional ancestral RelA/SpoT Homolog (RSH) possessing both (p)ppGpp synthetic and hydrolytic capabilities, resulting in two proteins with overlapping functionalities (Mittenhuber, 2001; Atkinson et al., 2011). The RelA and SpoT proteins contain 744 and 702 amino acids, respectively. Both proteins can be divided in two parts of similar size (Figure 1). The NTD half of the protein harbors the catalytic HD (hydrolytic) and Synth (Synthetic) domains. In RelA, the HD domain is not active. The CTD portion of the protein contains four regulatory domains: TGS (ThrRS, GTPase, SpoT/RelA domain), AH (α-helical domain), RIS (Ribosome-InterSubunit domain) and ACT (Aspartate kinase-Chorismate mutase-TyrA domain) (Atkinson et al., 2011; Loveland et al., 2016). RelA responds to intracellular amino acid imbalancies, such as amino acid starvation, by synthesizing large amounts of (p)ppGpp (Cashel, 1969). RelA carries an inactive (p)ppGpp-hydrolytic domain and does not hydrolyze the alarmone under any conditions. SpoT is a bifunctional enzyme that contains functional (p)ppGpp-synthetic and hydrolytic domains, but displays weak (p)ppGpp-synthetic activity and strong ppGpp hydrolytic activity. The relA knockout accumulates ppGpp in response to several environmental stresses, such as carbon and nitrogen (Edlin and Donini, 1971), phosphate (Spira et al., 1995), iron (Vinella et al., 2005), and fatty acid (Battesti and Bouveret, 2006) starvation.
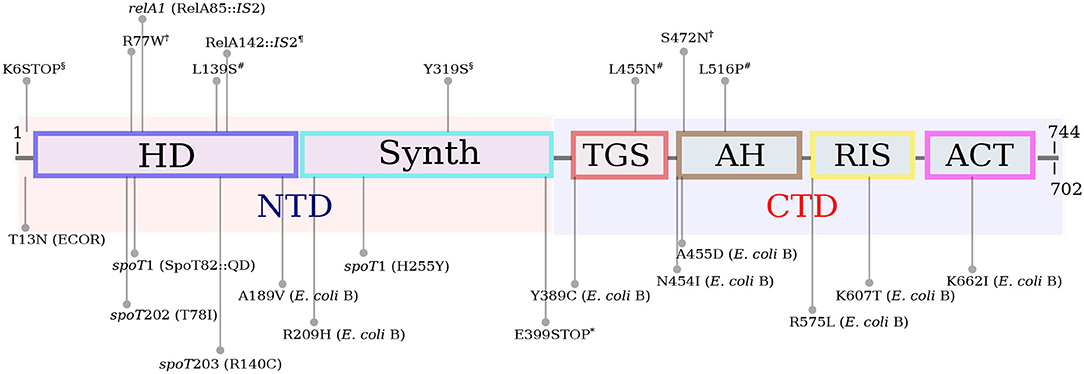
Figure 1. Schematic representation of the long RSH (RelA/SpoT) architecture, as per Atkinson et al. (2011) and Loveland et al. (2016). The RelA and SpoT proteins contain 744 and 702 amino acids, respectively. The NTD half of RSH proteins harbors the catalytic HD (hydrolytic) and Synth (Synthetic) domains. In RelA, the HD domain is not active. The CTD portion of the protein contains four regulatory domains: TGS (ThrRS, GTPase, SpoT/RelA domain), AH (α-helical domain), RIS (Ribosome-InterSubunit domain) and ACT (Aspartate kinase-Chorismate mutase-TyrA domain). The mutations in RelA and SpoT mentioned in the main text are shown above and below the protein diagram, respectively. relA1, spoT1, spoT202 and spoT203 are known mutations present in many E. coli K-12 derivatives. The spoT1 allele consists of two mutations. All mutations, with the exception of those that are followed by E. coli B or ECOR (in parentheses) were found in K-12 strains. The T13N substitution is common in strains of the ECOR collection (Ferenci et al., 2011). The SpoT mutations in E. coli B were selected in an evolution experiment in glucose-limited minimal medium (Cooper et al., 2003). Mutations ending with “*” were selected during adaptation to high temperature (Kishimoto et al., 2010); “¶” labels indicate RelA mutations selected for n-butanol tolerance (Reyes et al., 2012); “#” labels point to mutations in RelA selected under high ethanol concentration (Horinouchi et al., 2015); RelA mutations ending with “†” were selected for isopropanol tolerance (Horinouchi et al., 2017) and “§” labels indicate RelA mutations selected under growth with lactate (Conrad et al., 2009).
Early in (p)ppGpp research different spontaneous alleles of relA and spoT have been isolated. For instance, the spoT1 allele (Laffler and Gallant, 1974), that confers a spotless phenotype (absence of pppGpp under amino acid starvation), was isolated from the old 58-161 strain and is now common in many K-12 derivatives (Alföldi et al., 1962). Bacteria that carry the spoT1 allele overproduce (p)ppGpp both under nutrient starvation and under normal growth conditions. The spoT1 allele contains two different mutations - a H255Y substitution in the synthetase domain (Synth) and a two-amino acid insertion between residues 82 and 83 (+QD) in the hydrolytic domain (HD), both at the NTD portion of SpoT (Figure 1). The two amino acid insertions in the HD domain are likely to negatively affect the ppGpp-hydrolytic activity of SpoT resulting in high (p)ppGpp basal levels, while the H255Y substitution hits a conserved residue (Atkinson et al., 2011), but its effect on the (p)ppGpp-synthetic activity of SpoT is hard to predict. Interestingly, spoT1 is usually accompanied in many strains by the defective relA1 allele, consisting of an IS2 insertion in the HD domain that is likely to disrupt RelA (p)ppGpp-synthetic activity (Metzger et al., 1989). The relA1 mutant displays lower ppGpp basal level than the relA+ strain (Lagosky and Chang, 1980) and does not accumulate (p)ppGpp in response to amino acid starvation. Apparently, the high ppGpp basal level caused by the spoT1 allele is compensated by the defect in (p)ppGpp synthesis caused by the presence of relA1. It is therefore no wonder that both alleles often appear together in the same genome.
Later on, other spoT alleles, such as spoT201, spoT202 an spoT203 were isolated by selection on amino-triazole plates (Sarubbi et al., 1988). Amino-triazole is a herbicide that inhibits the synthesis of histidine. Bacteria that synthesize high levels of (p)ppGpp overcome histidine starvation by inducing the expression of the his operon (Rudd et al., 1985). A critical difference between spoT201 and the other three alleles was that the former confers an almost normal growth rate. The other alleles (spoT202-203) considerably reduced growth rate and for that reason could be transferred only to a relA1 background, but not to a bacterium that carries a wild-type relA allele. The spoT alleles spoT202 and spoT203 consist, respectively, of T78I and R140C substitutions, both in the HD domain (Potrykus et al., 2011). The molecular nature of the spoT201 mutation has not been published. Given the high (p)ppGpp level in strains bearing these alleles, the spoT201-203 mutations have probably compromised the ppGppase activity of SpoT.
It became evident that an inverse linear correlation exists between the intrinsic level of (p)ppGpp in a bacterium (basal level under unrestricted growth conditions) and the bacterial growth rate (Sarubbi et al., 1988). This negative correlation was confirmed when spoT mutant alleles were transferred to other genetic backgrounds (Spira et al., 2008). The recombinant strains displayed all the hallmarks of the previously analyzed spoT mutations, namely slower growth rate, high levels of the sigma factor RpoS (coordinator of the general stress response) and high resistance to environmental stresses (see below).
The above mentioned relA and spoT alleles and most data on (p)ppGpp physiology and homeostasis were obtained by studying laboratory strains derived from the ancestral K-12 strain. To date very few attempts have been made to analyze (p)ppGpp homeostasis in natural isolates of E. coli. In two of these studies, the basal level and starvation-induced levels of (p)ppGpp were assessed in a set of strains derived from the ECOR collection (Ferenci et al., 2011) and in a collection of Shiga toxin-producing E. coli (STEC) strains (Stella et al., 2017). The ECOR collection contains 72 strains from various locations and environments and from five phylogenetic groups (A, B1, B2, D, and E) that supposedly represents the variability in the E. coli species (Ochman and Selander, 1984). Most ECOR isolates are commensal, but some are pathogenic. The levels of (p)ppGpp in non-limited minimal medium, in response to amino acid starvation or carbon starvation were reported for 33 strains of the ECOR collection. ppGpp concentrations in the ECOR strains treated with serine hydroxamate, an inhibitor of seryl-tRNA synthetase that induces amino acid starvation, were quite similar in all tested strains. However, (p)ppGpp response to carbon starvation was less homogeneous, consistent with the variation in SpoT observed in those strains. A T13N amino acid substitution was common in strains that showed low (p)ppGpp accumulation in response to carbon starvation and was absent in strains presenting high levels of ppGpp (Ferenci et al., 2011). These data suggested that spoT is being subjected to microevolutionary pressures.
It is well-established that the intrinsic concentration of (p)ppGpp is inversely correlated with growth rate (Ryals et al., 1982; Sarubbi et al., 1988; Potrykus et al., 2011; Jin et al., 2012). In fact, gratuitous induction of (p)ppGpp synthesis mediated by relA overexpression causes an almost instantaneous growth arrest (Schreiber et al., 1991; Svitil et al., 1993; Cruvinel et al., 2019). However, the vast majority of studies that analyzed this correlation used isogenic E. coli laboratory strains harboring different relA or spoT alleles. If growth rate is mainly regulated by (p)ppGpp a good correlation between (p)ppGpp levels and growth rate, even in a set of non-isogenic strains, would be expected. Indeed, when the intrinsic ppGpp concentrations of the ECOR isolates growing under non-limited growth conditions are plotted against their respective growth rates, an inverse correlation is observed (Figure 2A), with a Pearson's correlation coefficient = −0.58. Though not perfect, the inverse correlation between ppGpp concentration and growth rate in these strains validates the central role of (p)ppGpp in governing growth rates across the E. coli species, even in strains that come from very different genetic backgrounds as is the case of the ECOR collection. It is worth mentioning that in this as well as in other studies that analyzed (p)ppGpp in exponentially growing bacteria or in response to stresses other than amino acid starvation, pppGpp was below the detection level (Varik et al., 2017; Cruvinel et al., 2019).
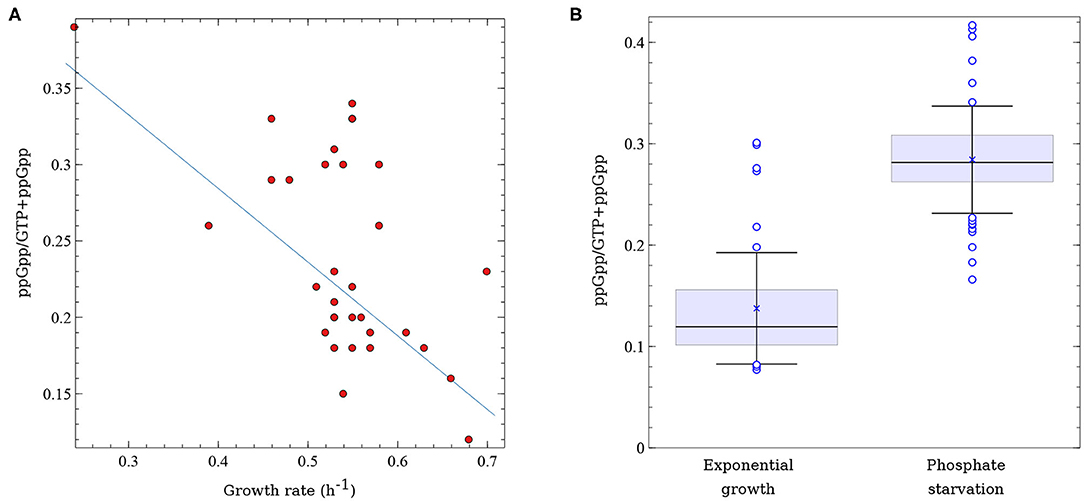
Figure 2. Variability in (p)ppGpp levels across E. coli natural isolates. (A) Correlation between (p)ppGpp basal levels and growth rate (h−1) in 33 strains of the ECOR collection growing exponentially in glucose minimal medium. ppGpp levels correspond to the ratio between ppGpp and GTP + ppGpp. Data was extracted from Ferenci et al. (2011). (B) Distribution of ppGpp levels (ppGpp/GTP+ppGpp) in 50 STEC isolates growing exponentially in non-limited (glucose) minimal medium or under phosphate starvation. x corresponds to the mean values, horizontal lines represent the median and the error bars correspond to 1 standard deviation; represent outliers. Data was extracted from Stella et al. (2017).
In another analysis of (p)ppGpp fluctuation in natural isolates, ppGpp concentration was measured in 50 STEC strains growing under two different culture conditions—non-limited growth medium and phosphate starvation (Stella et al., 2017). A significant variability in ppGpp levels was observed among the STEC isolates (Figure 2B). On average, ppGpp values were twice as high in bacteria submitted to Pi starvation than in the same bacteria growing exponentially in minimal medium. ppGpp values in this set of strains went from 0.08 to 0.30 units for bacteria growing exponentially and from 0.17 to 0.42 units for phosphate-starved bacteria (units correspond to the ratio of ppGpp over GTP+ppGpp). Though this study did not evaluate the variability of ppGpp with growth rate, it correlated the levels of this alarmone with STEC cytotoxicity, as described below.
Altogether, the data presented here highlight the existence of variability in intrinsic ppGpp concentrations across the E. coli species and that this variability has a substantial impact on growth rate. However, in addition to growth rate control (p)ppGpp directly and indirectly affects many important bacterial characteristics, such as stress responses, virulence, antibiotic resistance and persistence, biofilm formation, genome stability, and more (Potrykus and Cashel, 2008; Dalebroux et al., 2010; Martin-Rodriguez and Romling, 2017; Rasouly et al., 2017; Hobbs and Boraston, 2019). Variability in (p)ppGpp basal levels is thus likely to affect these traits as well.
It is important to notice that in the studies mentioned above that compared (p)ppGpp values in isogenic and non-isogenic strains, (p)ppGpp was assessed using the classical method of formic acid extraction of 32P-labeled bacterial nucleotide pools. These studies did not provide absolute values of (p)ppGpp concentration, but instead presented the level of ppGpp relative to that of GTP+ppGpp as detailed in Cashel (1994). The most relevant limitations of this method is the lack of absolute numerical estimates of (p)ppGpp concentrations and that it leaves out GDP, which constitutes 7.7–15% of the total pool of guanosine nucleotides (Varik et al., 2017), as the resolution of the 32P-labeled nucleotides on the TLC plate is not usually good enough to identify GDP spots on the autoradiogram. Because of these limitations, the ppGpp values obtained in those studies cannot be easily compared to the ones found in other reports. However, the relative values of (p)ppGpp obtained by the classical method are reproducible and give a reasonable estimate of (p)ppGpp status in a particular set of strains. More recent techniques for evaluating (p)ppGpp, based on Ion Chromatography-High-Resolution MS (Patacq et al., 2018), HPLC (Varik et al., 2017), or UPLC (Ihara et al., 2015) largely overcome the disadvantages of the 32P-classical method.
Role of (p)ppGpp in Stress Resistance and Nutritional Competence
(p)ppGpp supports survival by either directly or indirectly stimulating the expression of genes involved in stress protection. The cell response to environmental stresses such as extreme pH and osmolarity, dehydration or oxidative stress is coordinated by the sigma factor RpoS (Landini et al., 2014; Schellhorn, 2014), whose synthesis and stability is enhanced by (p)ppGpp (Gentry et al., 1993; Battesti et al., 2011). The culture history of a bacterial population determines its overall physiology, and more specifically, the strength of its response to environmental challenges (Ryall et al., 2012). The specific hurdles that a bacterial lineage experiences throughout its existence would eventually leave their imprints in its genome. For instance, alleles that maintain high levels of RpoS and other stress-related genes would be selected in a population that is being often exposed to environmental stresses Conversely, bacteria growing in a stress-free environment accumulates mutations in genes that downregulate RpoS synthesis, promotes its proteolysis or even acquire null mutations in the rpoS gene itself (King et al., 2004; Spira and Ferenci, 2008; Wang et al., 2010). Likewise, genes involved in (p)ppGpp metabolism are under selective pressures driven by culture conditions (Spira et al., 2008; Ferenci et al., 2011). ppGpp pleiotropy indicates that variations in intrinsic (p)ppGpp levels might have broad consequences on bacterial physiology and genotypic characteristics of bacterial populations. Bacteria that display intrinsic high levels of (p)ppGpp are more resistant to environmental stresses either because they express high levels of RpoS or because (p)ppGpp directly stimulates the transcription of other genes related to stress protection. However, the correlation between (p)ppGpp and RpoS is not as straightforward as would be expected from extrapolating data on K-12 strains (Gentry et al., 1993; Spira et al., 2008; Battesti et al., 2011). Analysis of E. coli natural isolates does not give a simple relationship in which RpoS concentration is proportional to (p)ppGpp concentration. While some strains exhibit a proportionality between the two measured entities, others display mediocre levels of RpoS but high (p)ppGpp levels (Ferenci et al., 2011). Surely, there are other inputs, other than (p)ppGpp that modulate the levels of RpoS.
Both (p)ppGpp and RpoS directly affect the transcription of dozens of genes and indirectly the transcription of many others (Peano et al., 2015; Wong et al., 2017). RpoS competes with other sigma factors, particularly with σ70 for binding to the core RNA polymerase. The outcome of this competition is that under nutrient limitation or in the stationary phase (circumstances that cause the accumulation of RpoS), the transcription of σ70-dependent genes, i.e., the majority of bacterial genes, is considerably diminished. Hence, the stimulatory effect of (p)ppGpp on RpoS adds another layer of growth control in addition to the already discussed inhibition of stable RNA. Bacterial strains that accumulate high levels of (p)ppGpp or RpoS are less fit for growing on poor carbon sources or under nutrient limitation (King et al., 2004). A trade-off is thus characterized in which a certain bacterial strain cannot simultaneously be nutritionally competent and highly stress resistant (Ferenci, 2016). Figure 3 shows how bacteria with high or low intrinsic (p)ppGpp concentrations deal with environmental challenges.

Figure 3. Diversity in (p)ppGpp levels and its consequences. (p)ppGpp intrinsic concentrations are not constant across E. coli strains. E. coli isolates displaying high (p)ppGpp are benefited under environmental challenges. For instance, these bacteria tolerate higher concentrations of antibiotics and display a higher survival rate when challenged with environmental stresses. Most high-(p)ppGpp pathotypes also show increased ability to colonize host tissues (higher virulence). On the other hand, these strains display lower growth rates and less nutritional competence (ability to utilize alternative carbon sources) than other strains with intrinsically lower (p)ppGpp values. Strains displaying high (p)ppGpp usually carry mutations in the HD domain of SpoT. Mutations in the Synth domain of both RelA or SpoT or in the regulatory domains often cause a reduction in (p)ppGpp levels. The resulting strains display an increased growth rate and higher nutritional competence, but are more sensitive to environmental stresses and antibiotics. STEC is the only known E. coli pathotype whose virulence capability benefits from low (p)ppGpp levels.
Intrinsic (p)ppGpp Concentration as a Target in Evolution Experiments
Given that (p)ppGpp is the most important source of growth rate control (Potrykus et al., 2011), polymorphism in relA and spoT are likely to occur throughout the course of bacterial evolution and adaptation to different environments, especially in those limited in one or more nutrients, a situation that suppresses normal growth. Several evolution experiments, which resulted in the emergence of mutants related to (p)ppGpp both in batch and in continuous cultures, have been conducted to date. The mutations observed in these studies are summarized in Figure 1.
In one of them, 12 E. coli populations have been daily diluted in glucose limited minimal medium and grown for 20,000 generations. Different non-synonymous mutations in spoT have been observed in 8 out of 12 evolved populations (Cooper et al., 2003). The first one, A189V is located at the very end of the HD domain; R209H is at the ~45-residues region between the HD and Synth domains; Y389C is at the regulatory TGS domain; N454I and A455D are located at the beginning of the α-helical domain; the mutations R575L and R607L are in the RIS domain and K662I is at the ACT domain. Although (p)ppGpp levels were not measured in this study, the expression of aminoacyl-tRNA synthetases and ribosomal proteins were shown to be upregulated in one of these spoT mutants (K662I), suggesting that the mutation caused a reduction in (p)ppGpp intrinsic concentration that led to an increase in growth rate. The ACT domain interacts with the ribosome A site in order to activate the (p)ppGpp synthetic activity (Loveland et al., 2016), thus the K662I substitution is likely to interfere with Synth activation resulting in low (p)ppGpp. A non-sense mutation in the TGS domain of spoT (E399*) was observed in another case of adaptive evolution of E. coli growing at 43.2°(Kishimoto et al., 2010). This mutant displayed high growth rate at the high temperature, possibly due to a reduction in intrinsic (p)ppGpp levels. This finding is puzzling, once it has been shown that the truncation of the CTD leads to an upshift in (p)ppGpp synthesis (Mechold et al., 2002; Battesti and Bouveret, 2006). However, this particular evolved strain carried additional mutations in lrp and rho that might have strengthened the observed phenotype.
In another experiment, E. coli subjected to adaptive evolution under high ethanol concentrations acquired different mutations in relA (L139S, L455N, and L519P) that contributed to an increased tolerance in the presence of 5% ethanol (Horinouchi et al., 2015). According to these authors, the relA mutations enabled a relaxed response to ethanol, by diminishing (p)ppGpp concentration, thereby increasing growth rate. The L139S mutation occurred in the pseudo-hydrolytic domain of RelA and is therefore unlikely to affect (p)ppGpp synthesis. The other two mutations—L455N and L516P, were in the TGS and AH domains, respectively. These mutations might have affected the regulation of (p)ppGpp synthesis by RelA as both TGS and AH subunits form the elbow of the boomerang-shaped RelA that interacts with the 30S ribosome and with the deacyl-tRNA (Loveland et al., 2016). A similar study with bacteria growing with increasing concentrations of isopropanol (up to 450 mM) for 210 generations showed that the evolved isolates acquired mutations in relA (Horinouchi et al., 2017). Again, the suggested mechanism was that the relA mutants expressed RelA proteins that synthesized reduced levels of (p)ppGpp in response to isopropanol, resulting in higher growth rates. The mutations—R77W and S472N, were, as before, in the pseudo-HD and AH domains, respectively.
In another experiment of guided evolution, bacteria grown in a chemostat with increasing butanol concentrations (up to 1.3%) for 144 generations acquired mutations in several genes (Reyes et al., 2012). One of the evolved isolates presented an IS2 insertion at the end of the HD domain of RelA, which has probably compromised the integrity of the entire protein, resulting in a RelA-negative phenotype. Mutations in relA also appeared in 2 out of 11 populations growing in lactate minimal medium (Conrad et al., 2009). One mutation—K6*, caused a frameshift at the very beginning of the gene, while the other mutation, Y319S, occurred in the Synth domain of RelA.
In addition to the direct effect of (p)ppGpp on growth, low concentrations of this alarmone also results in reduced levels of RpoS (Gentry et al., 1993; Battesti et al., 2011). Due to the competition between σS and σ70, the former negatively affects the expression of growth-related genes, especially those involved in the uptake and assimilation of alternative carbon sources with a consequent reduction in growth rate (Gentry et al., 1993; King et al., 2004; Magnusson et al., 2005; Spira et al., 2008; Ferenci et al., 2011). Thus, mutations in relA would also improve growth by diminishing RpoS concentration in the cell. Figure 3 summarizes the outcomes of bacterial evolution experiments in which mutations in (p)ppGpp-related genes have been observed.
In conclusion, selection of different relA and spoT alleles in evolution experiments is not uncommon. In fact, in most of these experiments regulatory genes are the primary targets of adaptive selection (Maharjan et al., 2006; Wang et al., 2010). Given the central role that (p)ppGpp plays in the regulation of gene transcription, protein synthesis and growth, it is not surprising that modulation of (p)ppGpp is a primary target for evolution.
Variability in (p)ppGpp Levels and Its Influence on Antibiotic Susceptibility
The stringent response has been linked to bacterial tolerance to β-lactam antibiotics in E. coli. Tolerance to antibiotics is defined as the ability of microorganisms to survive transient exposure to high concentrations of an antibiotic without a change in the minimum inhibitory concentration (MIC) (Brauner et al., 2016). When both the wild-typestrain and relA null mutants were exposed to penicillin under amino acid starvation, only the former was able to avoid cell lysis triggered by the presence of the antibiotic (Goodell and Tomasz, 1980; Kusser and Ishiguro, 1985). Moreover, the protective effect of the stringent response against β-lactam antibiotics was reverted by the addition of chloramphenicol (Kusser and Ishiguro, 1985), a well-known inhibitor of the stringent response (Cortay and Cozzone, 1983). In the aforementioned studies (p)ppGpp levels were not directly measured, however, it has been subsequently shown that mecillinam-tolerant mutants accumulated more (p)ppGpp than mecillinam-sensitive strains (Vinella et al., 1992). It became thus evident that high concentrations of (p)ppGpp increase the level of mecillinam tolerance (Joseleau-Petit et al., 1994). The mechanism by which (p)ppGpp confers tolerance to β-lactams was not entirely elucidated. One possibility is that (p)ppGpp acts by inhibiting the biosynthesis of phospholipids. In fact, treatment with cerulenin, an inhibitor of fatty acid biosynthesis, induced β-lactam resistance in the ΔrelA mutant (Rodionov et al., 1995). In addition, the gratuitous induction of (p)ppGpp accumulation by overexpression of relA resulted in the inhibition of phospholipid and peptidoglycan synthesis and in penicillin tolerance (Rodionov and Ishiguro, 1995) supporting the idea that (p)ppGpp mediates penicillin tolerance through the inhibition of phospholipid synthesis (Rodionov and Ishiguro, 1996). However, a more recent study has demonstrated that antibiotic tolerance to β-lactams occurs even in the absence of RelA (Kudrin et al., 2017). E. coli cells treated with mupirocin, an isoleucyl-tRNA syntethase inhibitor, displayed increased ampicillin tolerance in the wild-type but not in the relaxed strain. In contrast, the combination of trimethoprim with mupirocin, tetracycline or chloramphenicol significantly increased tolerance to ampicillin in both strains. These data indicate that growth arrest/protein synthesis inhibition can, at least in some cases, increase bacterial tolerance to antibiotics in a (p)ppGpp-independent fashion.
The positive relation between antibiotic tolerance and intrinsic (p)ppGpp concentrations is not restricted to β-lactam antibiotics. The wild-type strain of E. coli displayed higher MIC values for trimethoprim, gentamicin and polymixin when compared to the ΔrelA or ΔrelA ΔspoT mutants (Greenway and England, 1999). The increase in MIC values characterizes an augment in bacterial resistance to these antibiotics (Brauner et al., 2016). Likewise, it has been shown that mutations in the aminoacyl-tRNA synthetase genes leuS and aspS reduced susceptibility to ciprofloxacin, chloramphenicol, rifampicin, mecillinam, ampicillin, and trimethoprim. Deletion of the relA gene in these mutants restored the original MIC values of these antibiotics (Garoff et al., 2018). In another instance bacteria expressing high levels of (p)ppGpp displayed resistance to microcin J25, while strains unable to produce (p)ppGpp were completely sensitive to this antibiotic. In addition, overexpression of relA in a strain naturally susceptible to microcin J25 resulted in high MIC values and higher survival rates in killing curves (Pomares et al., 2008).
Several studies have shown a positive correlation between the expression of hipA, that encodes a serine/threonine-protein kinase that belongs to a type-II toxin/anti-toxin module, (p)ppGpp production and the formation of persisters (Korch et al., 2003; Bokinsky et al., 2013; Germain et al., 2013; Kaspy et al., 2013). Persistence is the ability of a subpopulation of an antibiotic-sensitive strain to survive for longer periods of time in the presence of high concentrations of an antibiotic than the majority of the population (Brauner et al., 2016). Some strains are able to form a higher percentage of persisters than others. For instance, strains carrying the hipA7 allele formed 100-fold more persistent cells than the wild-type strain when exposed to ampicillin. In the absence of (p)ppGpp (ΔrelA ΔspoT double mutant) the hipA7 allele did not confer any advantage regarding antibiotic persistence, suggesting that the high-persistence phenotype elicited by hipA7 is (p)ppGpp-dependent. Accordingly, overexpression of relA in the hipA7 strain increased the frequency of persisters (Korch et al., 2003). On the other hand, overexpression of hipA granted resistance to ampicillin, but only in relA+ bacteria, as bacteria overexpressing hipA but lacking relA were considerably more sensitive to ampicillin. Interestingly, the level of (p)ppGpp in the relA+ strain overexpressing hipA was as high as under amino acid starvation (Bokinsky et al., 2013). Two other studies confirmed the findings of Bokinsky et al. and extended their observations to fluoroquinolone antibiotics (Germain et al., 2013; Kaspy et al., 2013). In addition, these studies suggested a mechanism for hipA stimulation of persistence via (p)ppGpp. In their model hipA inactivates the glutamyl-tRNA synthetase GltX resulting in the accumulation of uncharged tRNAs which ultimately leads to the activation of RelA and (p)ppGpp synthesis.
Formation of persister cells in bacteria exposed to ofloxacin and ampicillin was also observed upon carbon source transitions, a situation that causes the accumulation of (p)ppGpp (Amato et al., 2013; Amato and Brynildsen, 2015). Deletion of relA abolished the formation of ampicillin, but not of ofloxacin persistence, which required the deletion of both relA and spoT. Furthermore, by controlling the level of (p)ppGpp it has been shown that formation of ampicillin persisters required higher concentrations of (p)ppGpp than formation of ofloxacin persisters. It has also been shown that under conditions of nitrogen starvation E. coli accumulates high levels of (p)ppGpp and forms high percentages of persisters when treated with ciprofloxacin, but only in a relA+ strain (Brown, 2019).
Integrons are important elements in the dissemination of antibiotic resistance genes. It has been shown that (p)ppGpp plays a role in the regulation of intI1, which encodes an integrase protein found in class 1 integrons (Strugeon et al., 2016). Accumulation of (p)ppGpp causes the stalling of RNA-polymerase and the formation of R-loops, which in turn activates the SOS response. The autoproteolysis of the intI1 repressor, LexA, ensues resulting in the transcription of intI1. In trans expression of this gene in the ΔrelA ΔspoT double mutant resulted in reduced intI1 promoter activity when compared to the parental strains. Overall, these data indicate that (p)ppGpp helps propagating antibiotic resistance genes through activation of integrase in class 1 integrons.
Variability in (p)ppGpp–Effect on Bacterial Pathogenicity
The expression of virulence-related genes in pathogenic E. coli is very well-integrated with (p)ppGpp homeostasis Dalebroux et al. (2010). For instance, (p)ppGpp influences the ability of enterohemorrhagic E. coli (EHEC) to colonize the host intestine (Nakanishi et al., 2006). This E. coli pathotype secretes a potent cytotoxin—Shiga toxin, that causes serious diseases in humans—bloody diarrhea and HUS (hemolytic uremic syndrome). In addition, bacteria of this pathotype harbor a 35 Kb pathogenicity island known as the Locus of Enterocyte Effacement (LEE), which carries most genes implicated in EHEC intimate adherence (Nguyen and Sperandio, 2012). The passage from the nutrient-rich higher intestine to the nutrient-limited lower intestine triggers the accumulation of (p)ppGpp, which in turn stimulates the transcription of the LEE operons. The EHEC ΔrelA mutant was unable to induce bacterial adherence or expression of the LEE (Nakanishi et al., 2006). Overexpression of relA greatly stimulated the expression of EspB and Tir, two proteins encoded by the LEE and increased the transcription of several LEE genes, implying a positive correlation between (p)ppGpp concentration and EHEC virulence. EPEC (Enteropathogenic E. coli) is another diarrheogenic pathotype that carries the LEE, but unlike EHEC it does not produce Shiga toxin. EPEC strains harbor a plasmid (EAF) that encodes both the BFP fimbria associated with bacterial adherence to the intestine cells and the perABC operon whose products control the transcription of the chromosomal LEE region (Pearson et al., 2016; Serapio-Palacios and Finlay, 2020). Deletion of relA partially impaired EPEC adherence to epithelial cells by diminishing the transcription of the perABC operon that controls the expression of the adhesins BFP and intimin (Spira et al., 2014). However, gratuitous overproduction of (p)ppGpp slightly inhibited the expression of perABC. The antagonistic effects of (p)ppGpp on perABC expression suggests that a fine-tuned concentration of (p)ppGpp is required to maximize EPEC adherence. Even though (p)ppGpp concentrations were not assessed in different EHEC and EPEC isolates the data presented in these studies suggest that the expression of virulence genes and virulence traits are modulated by this alarmone.
Shiga toxin-producing E. coli (STEC) is another diarrheogenic pathotype that secretes Shiga toxin, but unlike EHEC, does not harbors a LEE and, consequently, does not display intimate adherence to intestinal cells (Bryan et al., 2015; Joseph et al., 2020). The role of (p)ppGpp in STEC virulence and particularly in toxin production and secretion has been examined in detail. The stx genes that encode Shiga toxin were introduced in the STEC genome by means of lambdoid bacteriophages, a phenomenon known as phage lysogenic conversion (Harrison and Brockhurst, 2017). The synthesis and release of Shiga toxin is preceded by the induction of the bacteriophage, a development that eventually results in cell lysis (Waldor and Friedman, 2005; Nowicki et al., 2013). Therefore, the level of Shiga-toxin production and release is directly related to the number of STEC bacteria in a population undergoing phage induction. On the other hand, (p)ppGpp has been shown to inhibit stx phage replication, as the ΔrelA ΔspoT double mutant displayed a higher degree of phage DNA replication and formed larger plaques on ΔrelA ΔspoT lawns (Nowicki et al., 2013). A subsequent report has shown that intrinsic (p)ppGpp concentration is indeed inversely correlated with Stx toxin production, as STEC strains showing higher cytotoxicity toward Vero cells (the golden standard method for measuring toxin production and STEC virulence) usually contained lower levels of (p)ppGpp (Stella et al., 2017).
The extraintestinal uropathogenic E. coli (UPEC) causes recurrent infections in the urinary tract. A critical mechanism of UPEC infection is the ability to invade the bladder cells by means of Type-I fimbriae. The expression of fimbrial genes is controlled by (p)ppGpp and DksA (Aberg et al., 2008). DksA is a transcription factor that binds to RNA polymerase and greatly enhances the effect of (p)ppGpp on transcription regulation (Gourse et al., 2018). (p)ppGpp activates the promoter of fimB that encodes a recombinase that specifically inverts the promoter of the fimAICDFGH operon. This operon codes for the structural components of the type-I fimbria. By inverting the promoter orientation FimB allows the transcription of the fimAICDFGH operon switching the promoter from “off” to “on” state (Eisenstein, 1981). Amino acid starvation or growth arrest caused by bacteria entering the stationary phase increase (p)ppGpp which activates the fimB and fimA promoters (Aberg et al., 2006). Likewise, relA overexpression also induces the transcription from these promoters resulting in the synthesis of Type-I fimbria and the invasion and colonization of bladder cells. Altogether, the data suggest that UPEC strains with high (p)ppGpp intrinsic levels present higher levels of virulence toward the host.
Lastly, (p)ppGpp is directly associated with the pathogenicity of many bacterial species and is required for the full expression of virulence genes (Dalebroux et al., 2010; Kalia et al., 2013). Interestingly, STEC, the only E. coli pathotype in which a populational study correlating (p)ppGpp and pathogenicity has been performed stands out as an outlier. STEC toxin production is coupled to phage induction, which is inhibited by (p)ppGpp. By inhibiting phage replication (p)ppGpp acts as a legitimate promoter of bacterial survival.
Conclusions
The intrinsic concentration of (p)ppGpp in strains of the species Escherichia coli is not constant. Rather, the level of (p)ppGpp is been shaped by the historical contingency of bacterial populations. There are two types of evidence that support this assertion: direct assessment of (p)ppGpp in E. coli natural isolates and the selection of relA and spoT mutant alleles in evolution experiments. These data indicate that the genes that govern (p)ppGpp synthesis and degradation are subjected to frequent microevolutionary pressures that will eventually determine the optimal concentration of (p)ppGpp in a population. Given the pleiotropic effects of (p)ppGpp in the cell, adjustments of (p)ppGpp intrinsic concentration should have broad implications on bacterial physiology (Figure 3). In fact, intrinsic variations in (p)ppGpp levels differentially affect growth, stress response, virulence and antibiotic resistance. However, the intrinsic levels of (p)ppGpp in E. coli natural isolates do not perfectly correlate with the expected phenotypes. For instance, growth rate and (p)ppGpp inverse correlation across the ECOR strains was significant but not perfect, which suggests that the role of this alarmone in growth is intertwined with other regulatory circuits and that bacterial physiology is always more complex than firstly assumed.
Author Contributions
BS and KO drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by FAPESP grant number 2018/1629-5. BS is a recipient of a CNPq productivity scholarship. KO is a recipient of a CAPES scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aberg, A., Shingler, V., and Balsalobre, C. (2006). (p)ppGpp regulates type 1 fimBriation of Escherichia coli by modulating the expression of the site-specific recombinase fimB. Mol. Microbiol. 60, 1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x
Aberg, A., Shingler, V., and Balsalobre, C. (2008). Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67, 1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x
Alföldi, L., Stent, G. S., and Clowes, R. C. (1962). The chromosomal site of the RNA control (RC) locus in Escherichia coli. J. Mol. Biol. 5, 348–355. doi: 10.1016/S0022-2836(62)80077-1
Amato, S. M., and Brynildsen, M. P. (2015). Persister heterogeneity arising from a single metabolic stress. Curr. Biol. 25, 2090–2098. doi: 10.1016/j.cub.2015.06.034
Amato, S. M., Orman, M. A., and Brynildsen, M. P. (2013). Metabolic control of persister formation in Escherichia coli. Mol. Cell 50, 475–487. doi: 10.1016/j.molcel.2013.04.002
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/spot homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6:e23479. doi: 10.1371/journal.pone.0023479
Battesti, A., and Bouveret, E. (2006). Acyl carrier protein/spot interaction, the switch linking spot-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Bokinsky, G., Baidoo, E. E., Akella, S., Burd, H., Weaver, D., Alonso-Gutierrez, J., et al. (2013). HipA-triggered growth arrest and β-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195, 3173–3182. doi: 10.1128/JB.02210-12
Brauner, A., Fridman, O., Gefen, O., and Balaban, N. Q. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330. doi: 10.1038/nrmicro.2016.34
Brown, D. R. (2019). Nitrogen starvation induces persister cell formation in Escherichia coli. J. Bacteriol. 201:e00622-18. doi: 10.1128/JB.00622-18
Bryan, A., Youngster, I., and McAdam, A. J. (2015). Shiga toxin producing Escherichia coli. Clin. Lab. Med. 35, 247–272. doi: 10.1016/j.cll.2015.02.004
Cashel, M. (1969). The control of ribonucleic acid synthesis in Escherichia coli IV. relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244, 3133–3141.
Cashel, M. (1994). Detection of (p)ppGpp accumulation patterns in Escherichia coli mutants. Methods Mol. Genet. 3, 341–356.
Cashel, M., and Gallant, J. (1968). Control of RNA synthesis in Escherichia coli. I. amino acid dependence of the synthesis of the substrates of RNA polymerase. J. Mol. Biol. 34, 317–330. doi: 10.1016/0022-2836(68)90256-8
Conrad, T. M., Joyce, A. R., Applebee, M. K., Barrett, C. L., Xie, B., Gao, Y., et al. (2009). Whole-genome resequencing of Escherichia coli K-12 mg1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 10:R118. doi: 10.1186/gb-2009-10-10-r118
Cooper, T. F., Rozen, D. E., and Lenski, R. E. (2003). Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 100, 1072–1077. doi: 10.1073/pnas.0334340100
Cortay, J., and Cozzone, A. (1983). Effects of aminoglycoside antibiotics on the coupling of protein and rna syntheses in Escherichia coli. Biochem. Biophys. Res. Commun. 112, 801–808. doi: 10.1016/0006-291X(83)91688-1
Cruvinel, G. T., Neves, H. I., and Spira, B. (2019). Glyphosate induces the synthesis of ppGpp. Mol. Genet. Genome 294, 191–198. doi: 10.1007/s00438-018-1499-1
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C., and Swanson, M. S. (2010). ppGpp conjures bacterial virulence. Micro Mol. Biol. Rev. 74, 171–199. doi: 10.1128/MMBR.00046-09
Edlin, G., and Donini, P. (1971). Synthesis of guanosine 5'-diphosphate, 2'-(or 3'-) diphosphate and related nucleotides in a variety of physiological conditions. J. Biol. Chem. 246, 4371–4373.
Eisenstein, B. I. (1981). Phase variation of type 1 Fimbriae in Escherichia coli is under transcriptional control. Science 214, 337–339. doi: 10.1126/science.6116279
Ferenci, T. (2016). Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 24, 209–223. doi: 10.1016/j.tim.2015.11.009
Ferenci, T., Galbiati, H. F., Betteridge, T., Phan, K., and Spira, B. (2011). The constancy of global regulation across a species: the concentrations of ppGpp and rpos are strain-specific in Escherichia coli. BMC Microbiol. 11:62. doi: 10.1186/1471-2180-11-62
Garoff, L., Huseby, D. L., Praski Alzrigat, L., and Hughes, D. (2018). Effect of aminoacyl-trna synthetase mutations on susceptibility to ciprofloxacin in Escherichia coli. J. Antimicrob. Chemother. 73, 3285–3292. doi: 10.1093/jac/dky356
Gentry, D. R., HeRNAndez, V. J., Nguyen, L. H., Jensen, D. B., and Cashel, M. (1993). Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 175, 7982–7989. doi: 10.1128/JB.175.24.7982-7989.1993
Germain, E., Castro-Roa, D., Zenkin, N., and Gerdes, K. (2013). Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254. doi: 10.1016/j.molcel.2013.08.045
Goodell, W., and Tomasz, A. (1980). Alteration of Escherichia coli murein during amino acid starvation. J. Bacteriol. 144, 1009–1016. doi: 10.1128/JB.144.3.1009-1016.1980
Gourse, R. L., Chen, A. Y., Gopalkrishnan, S., Sanchez-Vazquez, P., Myers, A., and Ross, W. (2018). Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184. doi: 10.1146/annurev-micro-090817-062444
Greenway, D. L., and England, R. R. (1999). The intrinsic resistance of Escherichia coli to various antimicrobial agents requires ppGpp and sigma s. Lett. Appl. Microbiol. 29, 323–326. doi: 10.1046/j.1472-765X.1999.00642.x
Harrison, E., and Brockhurst, M. A. (2017). Ecological and evolutionary benefits of temperate phage: what does or doesn't kill you makes you stronger. Bioessays 39:1700112. doi: 10.1002/bies.201700112
Hobbs, J. K., and Boraston, A. B. (2019). (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect. Dis. 5, 1505–1517. doi: 10.1021/acsinfecdis.9b00204
Horinouchi, T., Sakai, A., Kotani, H., Tanabe, K., and Furusawa, C. (2017). Improvement of isopropanol tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. J. Biotechnol. 255, 47–56. doi: 10.1016/j.jbiotec.2017.06.408
Horinouchi, T., Suzuki, S., Hirasawa, T., Ono, N., Yomo, T., Shimizu, H., et al. (2015). Phenotypic convergence in bacterial adaptive evolution to ethanol stress. BMC Evol. Biol. 15:180. doi: 10.1186/s12862-015-0454-6
Ihara, Y., Ohta, H., and Masuda, S. (2015). A highly sensitive quantification method for the accumulation of alarmone ppGpp in Arabidopsis thaliana using UPLC-ESI-qMS/MS. J. Plant Res. 128, 511–518. doi: 10.1007/s10265-015-0711-1
Jin, D. J., Cagliero, C., and Zhou, Y. N. (2012). Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 36, 269–287. doi: 10.1111/j.1574-6976.2011.00279.x
Joseleau-Petit, D., Thévenet, D., and D'Arl, R. (1994). ppGpp concentration, growth without pbp2 activity, and growth-rate control in Escherichia coli. Mol. Microbiol. 13, 911–917. doi: 10.1111/j.1365-2958.1994.tb00482.x
Joseph, A., Cointe, A., Mariani Kurkdjian, P., Rafat, C., and Hertig, A. (2020). Shiga toxin-associated hemolytic uremic syndrome: a narrative review. Toxins 12:67. doi: 10.3390/toxins12020067
Kalia, D., Merey, G., Nakayama, S., Zheng, Y., Zhou, J., Luo, Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. doi: 10.1039/C2CS35206K
Kaspy, I., Rotem, E., Weiss, N., Ronin, I., Balaban, N. Q., and Glaser, G. (2013). HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4, 1–7. doi: 10.1038/ncomms4001
King, T., Ishihama, A., Kori, A., and Ferenci, T. (2004). A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186, 5614–5620. doi: 10.1128/JB.186.17.5614-5620.2004
Kishimoto, T., Iijima, L., Tatsumi, M., Ono, N., Oyake, A., Hashimoto, T., et al. (2010). Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 6:e1001164. doi: 10.1371/journal.pgen.1001164
Korch, S. B., Henderson, T. A., and Hill, T. M. (2003). Characterization of the HipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x
Kudrin, P., Varik, V., Oliveira, S. R. A., Beljantseva, J., Del Peso Santos, T., Dzhygyr, I., et al. (2017). Subinhibitory concentrations of bacteriostatic antibiotics induce RelA-dependent and RelA-independent tolerance to β-lactams. Antimicrob. Agents Chemother. 61:e02173–16. doi: 10.1128/AAC.02173-16
Kusser, W., and Ishiguro, E. E. (1985). Involvement of the RelA gene in the autolysis of Escherichia coli induced by inhibitors of peptidoglycan biosynthesis. J. Bacteriol. 164, 861–865. doi: 10.1128/JB.164.2.861-865.1985
Laffler, T., and Gallant, J. (1974). spoT, a new genetic locus involved in the stringent response in E. coli. Cell 1, 27–30. doi: 10.1016/0092-8674(74)90151-2
Lagosky, P. A., and Chang, F. N. (1980). Influence of amino acid starvation on guanosine 5'-diphosphate 3'-diphosphate basal-level synthesis in Escherichia coli. J. Bacteriol. 144, 499–508. doi: 10.1128/JB.144.2.499-508.1980
Landini, P., Egli, T., Wolf, J., and Lacour, S. (2014). sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ. Microbiol. Rep. 6, 1–13. doi: 10.1111/1758-2229.12112
Loveland, A. B., Bah, E., Madireddy, R., Zhang, Y., Brilot, A. F., Grigorieff, N., et al. (2016). Ribosome∙RelA structures reveal the mechanism of stringent response activation. eLife 5:e17029. doi: 10.7554/eLife.17029
Magnusson, L. U., Farewell, A., and Nyström, T. (2005). ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242. doi: 10.1016/j.tim.2005.03.008
Maharjan, R., Seeto, S., Notley-McRobb, L., and Ferenci, T. (2006). Clonal adaptive radiation in a constant environment. Science. 313, 514–517. doi: 10.1126/science.1129865
Martin-Rodriguez, A. J., and Romling, U. (2017). Nucleotide second messenger signaling as a target for the control of bacterial biofilm formation. Curr. Top. Med. Chem. 17:1928–1944 doi: 10.2174/1568026617666170105144424
Mechold, U., Murphy, H., Brown, L., and Cashel, M. (2002). Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002
Metzger, S., Schreiber, G., Aizenman, E., Cashel, M., and Glaser, G. (1989). Characterization of the RelA1 mutation and a comparison of RelA1 with new RelA null alleles in Escherichia coli. J. Biol. Chem. 264, 21146–21152.
Mittenhuber, G. (2001). Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3, 585–600.
Nakanishi, N., Abe, H., Ogura, Y., Hayashi, T., Tashiro, K., Kuhara, S., et al. (2006). ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61, 194–205. doi: 10.1111/j.1365-2958.2006.05217.x
Nguyen, Y., and Sperandio, V. (2012). Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2:90. doi: 10.3389/fcimb.2012.00090
Nowicki, D., Kobiela, W., Wȩgrzyn, A., Wegrzyn, G., and Szalewska-Pałasz, A. (2013). ppGpp-dependent negative control of DNA replication of Shiga toxin-converting bacteriophages in Escherichia coli. J. Bacteriol. 195, 5007–5015. doi: 10.1128/JB.00592-13
Ochman, H., and Selander, R. K. (1984). Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157, 690–693. doi: 10.1128/JB.157.2.690-693.1984
Patacq, C., Chaudet, N., and Létisse, F. (2018). Absolute quantification of ppGpp and pppGpp by double-spike isotope dilution ion chromatography-high-resolution mass spectrometry. Anal. Chem. 90, 10715–10723. doi: 10.1021/acs.analchem.8b00829
Peano, C., Wolf, J., Demol, J., Rossi, E., Petiti, L., De Bellis, G., et al. (2015). Characterization of the Escherichia coli σ(s) core regulon by chromatin immunoprecipitation-sequencing (ChiP-seq) analysis. Sci. Rep. 5:10469. doi: 10.1038/srep10469
Pearson, J. S., Giogha, C., Wong Fok Lung, T., and Hartland, E. L. (2016). The genetics of enteropathogenic Escherichia coli virulence. Annu. Rev. Genet. 50, 493–513. doi: 10.1146/annurev-genet-120215-035138
Pomares, M. F., Vincent, P. A., Farías, R. N., and Salomón, R. A. (2008). Protective action of ppGpp in microcin j25-sensitive strains. J. Bacteriol. 190, 4328–4334. doi: 10.1128/JB.00183-08
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Potrykus, K., Murphy, H., Philippe, N., and Cashel, M. (2011). ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575. doi: 10.1111/j.1462-2920.2010.02357.x
Rasouly, A., Pani, B., and Nudler, E. (2017). A magic spot in genome maintenance. Trends Genet. 33, 58–67. doi: 10.1016/j.tig.2016.11.002
Reyes, L. H., Almario, M. P., Winkler, J., Orozco, M. M., and Kao, K. C. (2012). Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab. Eng. 14, 579–590. doi: 10.1016/j.ymben.2012.05.002
Rodionov, D. G., and Ishiguro, E. E. (1995). Direct correlation between overproduction of guanosine 3', 5'-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J. Bacteriol. 177, 4224–4229. doi: 10.1128/JB.177.15.4224-4229.1995
Rodionov, D. G., and Ishiguro, E. E. (1996). Dependence of peptidoglycan metabolism on phospholipid synthesis during growth of Escherichia coli. Microbiology 142, 2871–2877. doi: 10.1099/13500872-142-10-2871
Rodionov, D. G., Pisabarro, A. G., de Pedro, M. A., Kusser, W., and Ishiguro, E. E. (1995). Beta-lactam-induced bacteriolysis of amino acid-deprived Escherichia coli is dependent on phospholipid synthesis. J. Bacteriol. 177, 992–997. doi: 10.1128/JB.177.4.992-997.1995
Rudd, K. E., Bochner, B. R., Cashel, M., and Roth, J. R. (1985). Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J. Bacteriol. 163, 534–542. doi: 10.1128/JB.163.2.534-542.1985
Ryall, B., Eydallin, G., and Ferenci, T. (2012). Culture history and population heterogeneity as determinants of bacterial adaptation: the adaptomics of a single environmental transition. Microbiol. Mol. Biol. Rev. 76, 597–625. doi: 10.1128/MMBR.05028-11
Ryals, J., Little, R., and Bremer, H. (1982). Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 151, 1261–1268. doi: 10.1128/JB.151.3.1261-1268.1982
Sarubbi, E., Rudd, K. E., and Cashel, M. (1988). Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rRNA ribosomal promoter regulation in Escherichia coli. Mol. Genet. Genome 213, 214–222. doi: 10.1007/BF00339584
Schellhorn, H. E. (2014). Elucidating the function of the RpoS regulon. Future Microbiol. 9, 497–507. doi: 10.2217/fmb.14.9
Schreiber, G., Metzger, S., Aizenman, E., Roza, S., Cashel, M., and Glaser, G. (1991). Overexpression of the RelA gene in Escherichia coli. J. Biol. Chem. 266, 3760–3767.
Serapio-Palacios, A., and Finlay, B. B. (2020). Dynamics of expression, secretion and translocation of type III effectors during enteropathogenic Escherichia coli infection. Curr. Opin. Microbiol. 54, 67–76. doi: 10.1016/j.mib.2019.12.001
Spira, B., and Ferenci, T. (2008). Alkaline phosphatase as a reporter of sigma(s) levels and RpoS polymorphisms in different E. coli strains. Arch. Microbiol. 189, 43–47. doi: 10.1007/s00203-007-0291-0
Spira, B., Ferreira, G. M., and de Almeida, L. G. (2014). RelA enhances the adherence of enteropathogenic Escherichia coli. PLoS ONE 9:e91703. doi: 10.1371/journal.pone.0091703
Spira, B., Hu, X., and Ferenci, T. (2008). Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology 154, 2887–2895. doi: 10.1099/mic.0.2008/018457-0
Spira, B., Silberstein, N., and Yagil, E. (1995). Guanosine 3',5'-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for pi. J. Bacteriol. 177, 4053–4058. doi: 10.1128/JB.177.14.4053-4058.1995
Stella, A., Luz, D., Piazza, R., and Spira, B. (2017). ppGpp and cytotoxicity diversity in Shiga toxin-producing Escherichia coli (STEC) isolates. Epidemiol. Infect. 145, 2204–2211. doi: 10.1017/S0950268817001091
Strugeon, E., Tilloy, V., Ploy, M.-C., and Da Re, S. (2016). The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7:e00868-16. doi: 10.1128/mBio.00868-16
Svitil, A. L., Cashel, M., and Zyskind, J. W. (1993). Guanosine tetraphosphate inhibits protein synthesis in vivo. a possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 268, 2307–2311.
Traxler, M. F., Summers, S. M., Nguyen, H.-T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Traxler, M. F., Zacharia, V. M., Marquardt, S., Summers, S. M., Nguyen, H.-T., Stark, S. E., et al. (2011). Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine' gradient in Escherichia coli. Mol. Microbiol. 79, 830–845. doi: 10.1111/j.1365-2958.2010.07498.x
Varik, V., Oliveira, S. R. A., Hauryliuk, V., and Tenson, T. (2017). HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and (p)ppGpp. Sci. Rep. 7:11022. doi: 10.1038/s41598-017-10988-6
Vinella, D., Albrecht, C., Cashel, M., and d'Ari, R. (2005). Iron limitation induces spot-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56, 958–970. doi: 10.1111/j.1365-2958.2005.04601.x
Vinella, D., D'Ari, R., Jaffe, A., and Bouloc, P. (1992). Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 11, 1493–1501. doi: 10.1002/j.1460-2075.1992.tb05194.x
Waldor, M. K., and Friedman, D. I. (2005). Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8, 459–465. doi: 10.1016/j.mib.2005.06.001
Wang, L., Spira, B., Zhou, Z., Feng, L., Maharjan, R. P., Li, X., et al. (2010). Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol. Evol. 2, 478–487. doi: 10.1093/gbe/evq035
Keywords: (p)ppGpp, polymorphism, growth rate, evolution, stress resistance, antibiotic resistance, virulence
Citation: Spira B and Ospino K (2020) Diversity in E. coli (p)ppGpp Levels and Its Consequences. Front. Microbiol. 11:1759. doi: 10.3389/fmicb.2020.01759
Received: 20 May 2020; Accepted: 06 July 2020;
Published: 12 August 2020.
Edited by:
Katarzyna Potrykus, University of Gdansk, PolandCopyright © 2020 Spira and Ospino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beny Spira, YmVueXNAdXNwLmJy
 Beny Spira
Beny Spira Katia Ospino
Katia Ospino