
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 July 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01674
Sulfated fucans are important marine polysaccharides widely distributed in brown algae and echinoderms, which gained increasing research interest for their various biological and biomedical activities. Fucanases could serve as tools in the bioconversion and structural investigation of sulfated fucans. A few gene-defined endo-1,4-fucanases have been characterized, while the sequence of endo-1,3-fucanase remain unstudied. Here, an endo-1,3-fucanase gene funA was screened from the genome of marine bacterium Wenyingzhuangia fucanilytica CZ1127T using transcriptomics. None of the previously reported glycoside hydrolase domains were predicted in the enzyme FunA, which hydrolyzed sulfated fucans in a random endo-acting manner. Ultrahigh performance size exclusion chromatography-mass spectrometry and nuclear magnetic resonance analyses revealed that FunA specifically cleaves α-1,3 glycosidic linkage between 2-O-sulfated and non-sulfated fucose residues. FunA exhibited transglycosylating activity with glycerin, methanol, and L-fucose as acceptors. D206 and E264 were critical for the functioning of FunA as identified by the site-directed mutagenesis. Another five homologs of FunA were confirmed to possess endo-1,3-fucanase activities. This is the first report on the sequence of endo-1,3-fucanase. The novelty of FunA and its homologs in sequences and activity shed light on a novel glycoside hydrolase family, GH168.
Sulfated fucans, also known as fucoidans, are sulfated polysaccharides mainly composed of L-fucose and sulfate groups usually extracted from brown algae and echinoderms. Because of their various biological and biomedical characteristics such as being anticoagulant (Colliec et al., 1991; Cumashi et al., 2007), antithrombotic (Min et al., 2012), anti-inflammatory (Cumashi et al., 2007), anticancer (Nagamine et al., 2020), and immunomodulatory (Amin et al., 2020), research interest in sulfated fucans have increased. Additionally, they had been utilized for dermal burn healing (Sezer et al., 2007), wound healing (Murakami et al., 2010) and bone tissue regeneration (Jin and Kim, 2011) due to their excellent biocompatibility. According to the linkage pattern of their backbone, sulfated fucans could be generally classified into two groups (Cumashi et al., 2007): (i) type I sulfated fucans composed of homogeneous 1,3-α-L-fucopyranose residues. They distributed in most of Laminariales and Ectocarpales algae (Schultz-Johansen et al., 2018), and in echinoderms including sea cucumbers (Chen et al., 2012; Yu et al., 2014a, b, 2015; Chang et al., 2016) and sea urchins (Mulloy et al., 1994; Vilela-Silva et al., 1999); (ii) type II sulfated fucans consisting of alternating 1,3- and 1,4-linked α-L-fucopyranose residues. Most of the sulfated fucans from Fucales algae adopted this linkage pattern (Jiao et al., 2011; Schultz-Johansen et al., 2018).
Fucanase is a class of enzyme that specifically catalyzes the hydrolysis of glycoside bonds within sulfated fucans in an endo-acting manner. Fucanases are desirable tools to identify the structure of sulfated fucans (Berteau and Mulloy, 2003) and establish structure-activity relationships (Kusaykin et al., 2016). To date, a few genes of endo-1,4-fucanases (i.e., enzymes cleaving α-1,4 glycosidic linkages in type II sulfated fucan backbones) have been characterized, including FcnA, FFA1, FFA2, Fp273, Fp277, Fp279, P5AFcnA, and P19DFcnA (Colin et al., 2006; Silchenko et al., 2017, 2018; Schultz-Johansen et al., 2018; Vickers et al., 2018). All of the reported endo-1,4-fucanases belong to the glycoside hydrolase (GH) 107 family in the CAZy database1. However, the gene for endo-1,3-fucanase, which cleaves α-1,3 glycosidic linkage in type I or type II sulfated fucans in an endo-acting manner are yet to be identified (Schultz-Johansen et al., 2018).
Marine microbes play a crucial role in biogeochemical cycling (Falkowski et al., 2008). The bacteria of family Flavobacteriaceae are generally considered as specialists for the polysaccharide degradation through the various activities of carbohydrate active enzymes (Zhang H. et al., 2019). In our previous work, we isolated a marine bacterium Wenyingzhuangia fucanilytica CZ1127T (= CCTCC AB 2015089T = KCTC 42864T) (Chang et al., 2010; Chen et al., 2016), which belongs to family Flavobacteriaceae. Catabolic profiling confirmed this bacterium was versatile in polysaccharide bioconversions, and the polysaccharides degradation was usually related to dedicated genes (Barbeyron et al., 2016). Hence, the sequencing of entire genome was completed recently (GenBank accession no. CP014224). Initial studies indicated that W. fucanilytica secreted fucanase using sulfated fucan as the sole carbon source. Utilizing the intracellular enzymes produced by W. fucanilytica, we prepared a series of sulfated fucan oligosaccharides composed of 1,3-α-L-fucopyranose, and clarified the structure of several type I sulfated fucans from sea cucumbers including Acaudina molpadioides (Yu et al., 2014a), Apostichopus japonicus (Yu et al., 2015), Holothuria tubulosa (Chang et al., 2016), and Thelenota ananas (Yu et al., 2014b). Results of the said study showed that W. fucanilytica was capable of producing endo-1,3-fucanase.
For this study then, we aim to determine the sequence coding for endo-1,3-fucanase from W. fucanilytica CZ1127T. Since there was no endo-1,3-fucanase gene described yet, we screened candidate genes from those that were up-regulated when the W. fucanilytica was exposed to sulfated fucans. The candidate genes were cloned and heterologously expressed in Escherichia coli, and the enzyme activity, biochemical characteristics, and hydrolysis patterns were subsequently investigated. The novel enzyme described in this study may promote the application of endo-1,3-fucanase in bio-industry.
Dried sea cucumber Isostichopus badionotus was purchased from a local market in Qingdao (China). The sulfated fucan was extracted following a previously reported method (Chang et al., 2010). Extracts were further purified using an ÄKTATM Prime Plus system (GE healthcare, Uppsala, Sweden) equipped with a HiPrepTM 26/60 Sephacryl S-400 column (GE Healthcare, Uppsala, Sweden). The purified polysaccharide was dialyzed, lyophilized and used for other downstream processing.
Sulfated fucans from the brown algae Fucus vesiculosus and Macrocystis pyrifera were purchased from Sigma-Aldrich (St. Louis, MO, United States); sulfated fucan from Ascophyllum nodosum was purchased from Carbosynth (Suzhou, China). Reagents used in ultrahigh performance size exclusion chromatography (UPSEC) - mass spectrometry (MS) (UPSEC-MS) were LC-MS grade. D2O (99.8%, Sigma-Aldrich, St. Louis, MO, United States) was used in the nuclear magnetic resonance (NMR) experiment. The other reagents were all analytical grade.
For transcriptome profiling, the bacterium was cultivated in the minimal medium (Ensor et al., 1999) containing 0.2% (w:v) D-glucose (Sigma-Aldrich, St. Louis, MO, United States) or sulfated fucan from Isostichopus badionotus, and incubated overnight at 25°C until it reached midlogarithmic growth phase. Total RNA of W. fucanilytica CZ1127T was extracted using RNAprep pure Cell/Bacteria Kit (TIANGEN, Beijing, China). The concentration and quality of RNA were measured using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, United States). The RNA integrity was validated via Agilent 2100 Bioanalyzer (Agilent Technologies, United States). The mRNA was enriched using the Illumina Ribo-Zero rRNA Removal Kit (Bacteria) and split into small-fragment after adding the fragmentation buffer. Di-tagged cDNAs were synthesized by successive random priming with terminal-tagging oligos and purified with the AMPure XP beads (Beckman Coulter, Beverly, MA, United States). The enrichment of cDNAs was amplified and then the final mRNA-seq library was generated. Sequencing was then carried out on the Illumina X-Ten platform in Novogene Co., Ltd. (Beijing, China).
Read mapping was performed on the W. fucanilytica CZ1127T reference genome (RefSeq NZ_CP014224.1) by Bowtie v2.0.6 and TopHat v2.0.7 software. The number of reads mapping to each predicted coding sequence was counted by HTSeq v0.6.1. Normalization for differential expression analysis and statistical tests were performed using DEGSeq R package v1.12.0 software, and a Bonferroni p-value adjustment was used to correct for multiple testing. Genes with an adjusted p-value < 0.05 were considered as differentially expressed.
The genes from the up-regulated genes of W. fucanilytica induced by sulfated fucan were selected to be cloned and heterologously expressed in E. coli. Genomic DNA of W. fucanilytica CZ1127T was extracted using the TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China). The presence of signal peptide was predicted using the SignalP 5.0 server (Armenteros et al., 2019). The genes without predicted signal sequences were amplified by PCR using the forward and reverse primer pair (listed in Supplementary Table S1). Purified PCR fragments were subsequently digested using BamHI/XhoI and inserted into the pET 28a(+) vector (Novagen, San Diego, CA, United States) containing a (His)6-tag at the N-terminus. The obtained expression vectors were transformed into E. coli BL21 (DE3) cells. The transformed cells were then selected by using the Luria-Bertani (LB) agar plates containing 30 μg/mL kanamycin, and cultured in the LB broth at 37°C until OD600 reached 0.4. After incubation with 0.5 mM isopropyl β-D-1-thiogalactopyranoside at 17°C for 12 h, cells were collected and disrupted by sonication in 20 mM NaH2PO4-Na2HPO4 buffer (pH 8.0).
To purify the expressed protein, the supernatants of the cell lysate were loaded onto the HisTrap HP columns (GE Healthcare, Uppsala, Sweden) and eluted in a linear gradient of 0-0.5 M imidazole in 20 mM NaH2PO4-Na2HPO4 buffer (pH 8.0) with 0.3 M NaCl at 4°C. The active fractions were combined and desalted using HiPrepTM 26/10 Desalting column (GE Healthcare, Uppsala, Sweden) against 20 mM NaH2PO4-Na2HPO4 buffer (pH 8.0). The purity of the obtained protein was evaluated by SDS-PAGE with a 5% stacking gel and a 12% running gel. Protein bands were visualized by Coomassie brilliant blue (Sigma-Aldrich, St. Louis, MO, United States). The Mw of the enzyme was estimated by comparing with the standard markers (Page Rulerprestained protein ladder, Fermentas, Waltham, MA, United States). The purified enzyme was used in the succeeding experiments.
The enzymatic activity was determined by incubating the purified enzyme with 1 mg/mL substrate in 20 mM NaH2PO4-Na2HPO4 buffer (pH 8.0) at 40°C for 10 min. The released reduced sugar was quantified by a p-hydroxybenzoic acid hydrazide method (Lever, 1972). One unit of activity was defined as the amount of enzyme that released 1.0 μmol reduced sugar (equivalent to L-fucose) per minute. The protein concentration was determined using a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China) with bovine serum albumin as the standard.
To investigate the hydrolysis patterns of the recombinant enzyme, 1 U enzyme solution was incubated with 50 mL sulfated fucan from Isostichopus badionotus (Ib-FUC) solution (2 mg/mL, pH 8.0) at 25°C for 12 h. Aliquots of the resulting reaction products were withdrawn at intervals and heated at 100°C for 10 min to deactivate the enzyme. After 12 h reaction, 1 U enzyme was supplemented, and the incubation was continued for another 12 h to obtain the end product. The products were analyzed using high-performance size exclusion chromatography coupled with a refractive index detector (HPSEC-RID) (Agilent 1260, Agilent Technologies, Santa Cruz, CA, United States). A TSKgel SuperAW4000 column (Tosoh Corporation, Kanagawa, Japan) (0.2 M NaCl as the eluent, flow rate 0.6 mL/min) and a Superdex peptide 10/300 GL column (GE healthcare, Uppsala, Sweden) (50 mM ammonium formate as the eluent, flow rate 0.5 mL/min) were, respectively, utilized to examine the global profile and oligosaccharide pattern.
The effect of temperature on the activity was tested by incubating the enzyme with Ib-FUC at 15–60°C, and thermal stability was evaluated by measuring the residual activity of enzyme after pre-treatment at 4, 25, 30, or 40°C. The optimal pH was determined by incubating the enzyme at pH 4.0-11.0. pH stability was determined by pretreating the enzyme in buffers with varying pH (20 mM citrate-phosphate buffer for pH 4.0–7.0, 20 mM NaH2PO4-Na2HPO4 buffer for pH 6.5–9.0, and 20 mM Na2CO3-NaHCO3 buffer for pH 9.0–11.0) at 30°C for 1 h. The residual activity was finally measured after adjusting the pH back to 8.0. Impacts of metal ions and chemicals were also investigated by, respectively, adding them to the reaction mixture at a final concentration of 1 mM. Particularly, the effect of NaCl on enzyme activity was examined by carrying out the reaction at different salt concentrations of 0–0.5 M.
The kinetic parameters of the enzyme were determined in the presence of 0.1–2.0 mg/mL Ib-FUC. Substrate concentrations and enzyme activities were plotted according to the Michaelis-Menten equation using the software GraphPad Prism (GraphPad Software, San Diego, CA, United States), where Km, Vmax, Kcat, and Kcat/Km were subsequently calculated.
To identify the composition of the reaction products, the products were analyzed using UPSEC-MS. The system consisted of an ultrahigh performance liquid chromatography (UPLC) unit (Agilent 1290, Agilent Technologies, Santa Cruz, CA, United States), an ultra-performance size exclusion column (ACQUITY UPLC Protein BEH SEC 125Å Column, 4.6 mm × 300 mm, Waters, Milford, MA, United States), and a quadrupole/time of flight mass spectrometer (Agilent 6545 LC/Q-TOF, Agilent Technologies, Santa Cruz, CA, United States) with the electrospray ionization source. The eluent was 20% (v/v) methanol containing 10 mM ammonium acetate with a flow rate of 0.2 mL/min. Parameters for the mass spectrometer were the gas temperature of 325°C, gas flow at 8 L/min, nebulizer at 40 psi, sheath gas temperature of 250°C, sheath gas flow at 11 L/min, capillary voltage set at 3500 V, the fragmentor at 180 V; skimmer at 45 V and the scanned mass was 200–1200 Da with a negative ion polarity.
The end product was separated on a HiLoad 26/600 Superdex 30 pg column (GE healthcare, Uppsala, Sweden) by using ÄKTATM Prime Plus system (GE healthcare, Uppsala, Sweden), and eluted with 5 mM ammonium formate (pH 7.0) at a flow rate of 2.6 mL/min. The purified fractions were co-evaporated with D2O twice by lyophilization and consequently dissolved in 500 μL D2O. The external standard was calibrated according to the eternal 4, 4-dimethyl-4-silapentane-1-sulfonic acid (DSS) (0.00 ppm). The 1-dimensional 1H NMR spectrum and 2-dimensional 1H−1H NMR spectra, including the correlation spectrum (COSY), total correlation spectrum (TOCSY) and nuclear overhauser effect spectroscopy (NOESY) were determined by a Bruker AVANCE III 600 HD spectrometer (Bruker, German) at 600 MHz under 25°C with sufficient acquisition time.
Transglycosylation related changes were verified using glycerin, methanol,L-fucose, D-glucose, D-galactose, D-fructose, D-mannose, D-glucosamine or N-acetyl-D-glucosamine as acceptors. Different acceptors (2 mg for solid or 0.20 mL for liquid) with 2 mg Ib-FUC were incubated with 0.02 U enzyme in 1 mL 20 mM NaH2PO4-Na2HPO4 buffer (pH 8.0) at 25°C for 24 h, which were subsequently analyzed in a UPSEC-MS/MS to detect the generation of glycosylated products. The collision energy to dissociate oligosaccharides in the tandem mass spectrum was set at 45 V, and other parameters were as those described in section “UPSEC-MS Analysis.”
The homologs of putative endo-1,3-fucanase sequences in the NCBI non-redundant protein sequence database were searched by BLASTP with a cut-off expected threshold value of 1 × 10–10 (Altschul et al., 1997). Previously defined glycoside hydrolase domains were predicted by dbCAN (Yin et al., 2012). Multiple sequence alignments were performed using ClustalX (Thompson et al., 1997), and the amino acid sequence alignment was produced using the online application WebLogo server (Crooks et al., 2004). A phylogenetic tree was constructed in MEGA6 (Tamura et al., 2013) using the neighbor-joining algorithm, and tree visualization end editing was performed using Figtree version 1.4.4 (FigTree, 2018).
All experiments were performed at least three times. The SPSS v11.5 (SPSS Inc., Chicago, IL, United States) was utilized to perform Tukey’s post hoc test (ANOVA). A probability value of p-value < 0.05 was considered statistically significant.
There were significant differences in gene expression profiles when W. fucanilytica was cultured in different media (Supplementary Figure S1). Transcriptomics and differential expression analysis revealed 441 genes up-regulated when grown on the minimal medium containing Ib-FUC compared with the minimal medium containing D-glucose. Among these, 25 genes had expressions higher than 26-fold. Since no endo-1,3-fucanase gene has been identified, we focused on 11 genes that had no prior defined GH domains. These sequences were cloned and heterologously expressed in E. coli, five sequences were successfully expressed in the supernatant of cell lysate, and one showed apparent activity on sulfated fucan. This active protein, designated as FunA, was encoded by an open reading frame AXE80_RS10075 (located from 2492018 to 2493244 in the complementary strand, GenBank accession number WP_068826898.1), and composed of 408 amino acid residues containing a putative N-terminal signal peptide (residues M1 to S19). FunA was effectively purified using nickel affinity chromatography (an activity yield of 40.1%). It showed as a single band on SDS-PAGE (Supplementary Figure S3) and an apparent molecular weight (Mw) of 48.0 kDa which is consistent with the expected value (48.2 kDa). Type I sulfated fucan Ib-FUC was used to assay the activity due to its regular and clear primary structure (Supplementary Figure S2). The activity of the purified enzyme on Ib-FUC was 13.7 U/mg.
In the initial hydrolysis stage, the Mw of Ib-FUC sharply decreased (0–20 min, insert of Figure 1). Meanwhile, the polydispersity of the products increased. Several oligosaccharides were also observed on the HPSEC-RID chromatogram (0–10 min, Figure 1), confirming that FunA was a random endo-acting enzyme. Based on the structure of Ib-FUC (Supplementary Figure S2), it could be inferred that FunA was capable of cleaving the α-1,3 glycosidic linkages within the sulfated fucan in an endo-acting manner. FunA was then further examined as a candidate endo-1,3-fucanase.
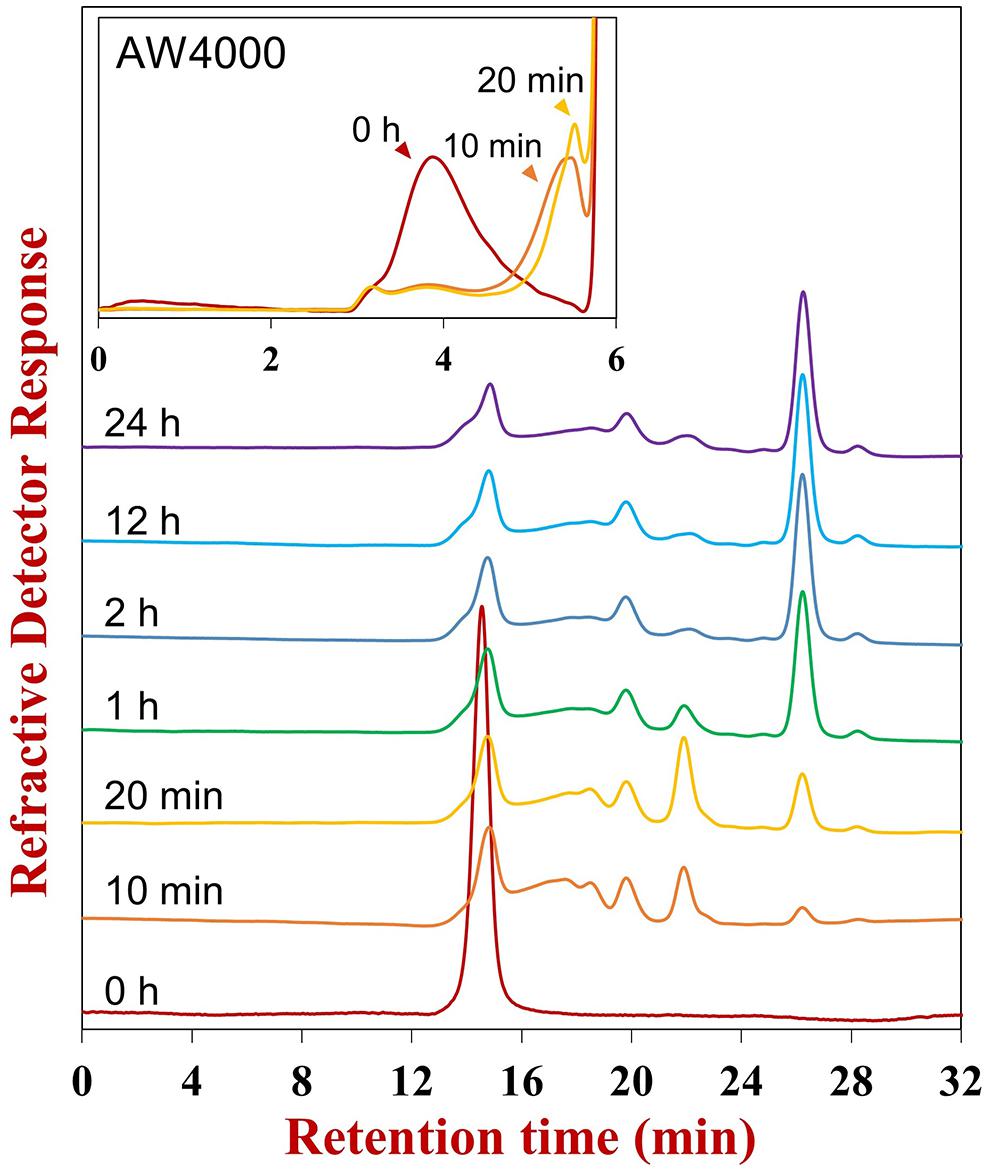
Figure 1. HPSEC-RID chromatograms of hydrolysis products obtained at different reaction times. The reaction time was labeled above the corresponding chromatogram. After 12 h reaction, the fresh enzyme was supplemented into the reaction mixture and incubated for a further 12 h to prepare the end product (24 h). The Superdex peptide 10/300 GL column was employed for examining the oligosaccharide pattern, and the TSKgel SuperAW4000 column was utilized for investigating the global profile (the insert).
FunA was incompetent to hydrolyze sulfated fucans from F. vesiculosus, A. nodosum and M. pyrifera (Supplementary Table S2), which were the effective substrates of GH107 enzymes. Currently, the GH107 family is the only reported fucanase GH family, and its enzymes all exhibited endo-1,4-fucanase activity: MfFcnA was specifically active on α-1,4 glycoside bonds in type II sulfated fucans from F. vesiculosus and A. nodosum (the primary structures were shown in Supplementary Figure S2; Schultz-Johansen et al., 2018); Fp277 and Fp279 could cleave α-1,4 glycoside bonds in A. nodosum (Schultz-Johansen et al., 2018); P5AFcnA and P19DFcnA showed activity on sulfated fucan from M. pyrifera (Vickers et al., 2018), though the primary structure of M. pyrifera was poorly defined. It has not been reported yet whether the current GH107 enzymes could act on α-1,3 glycosidic linkages. These results demonstrate that FunA was distinct from GH107 enzymes.
FunA was most active at 40°C and pH 8.0 (Supplementary Figure S4). It could keep stable at 4 and 25°C after 24 h storage, and retain more than 80% of its original activity after exposure to pH 6.5 to 10.0 for 1 h (Supplementary Figure S4). Further, 94.12% of residual activity could be retained after 90 days storage at 4°C without any protective additives, supporting that it was stable. The effects of metal ions on enzyme activity were summarized in Supplementary Table S3. FunA activity was not significantly influenced by NaCl (Supplementary Figure S4E). The activities of marine polysaccharide enzymes were widely reported to depend on the concentration of NaCl, including Cgi82A (Shen et al., 2017) and Por16A_Wf (Zhang Y. et al., 2019) from W. fucanilytica. The salt-independent property of FunA would be beneficial to its practical applications. The Km, Vmax, Kcat, and Kcat/Km values of FunA were determined as 3.26 ± 0.30 μM (1.05 ± 0.10 mg/mL), 25.45 ± 0.97 U/mg, 20.46 s–1 and 6.28 mM–1 s–1, respectively.
The products at different reaction times were identified by liquid chromatograph-mass spectrometry (LC-MS) (Figure 2). The UPSEC was chosen due to its higher separation resolution and better compatibility with mass spectrometry compared with HPSEC. Moreover, UPSEC-MS can achieve the high-throughput detection of the entire glycome and provide abundant structural information. Along with the progress of the enzymatic hydrolysis, two components significantly changed based on total ion chromatograms, which were noted as components I and II (Figure 2A). Component I exhibited an ion peak at m/z 912.0762 (Figure 2B) as [M-H]2– indicating a composition of octasaccharide decorated with eight sulfate groups (Fuc8S8, 1826.1284 Da). Meanwhile, component II exhibited an ion peak at m/z 921.0803 (Figure 2C) as [M-H]–, which was a tetrasaccharide decorated with four sulfate groups (Fuc4S4, 922.0695 Da).
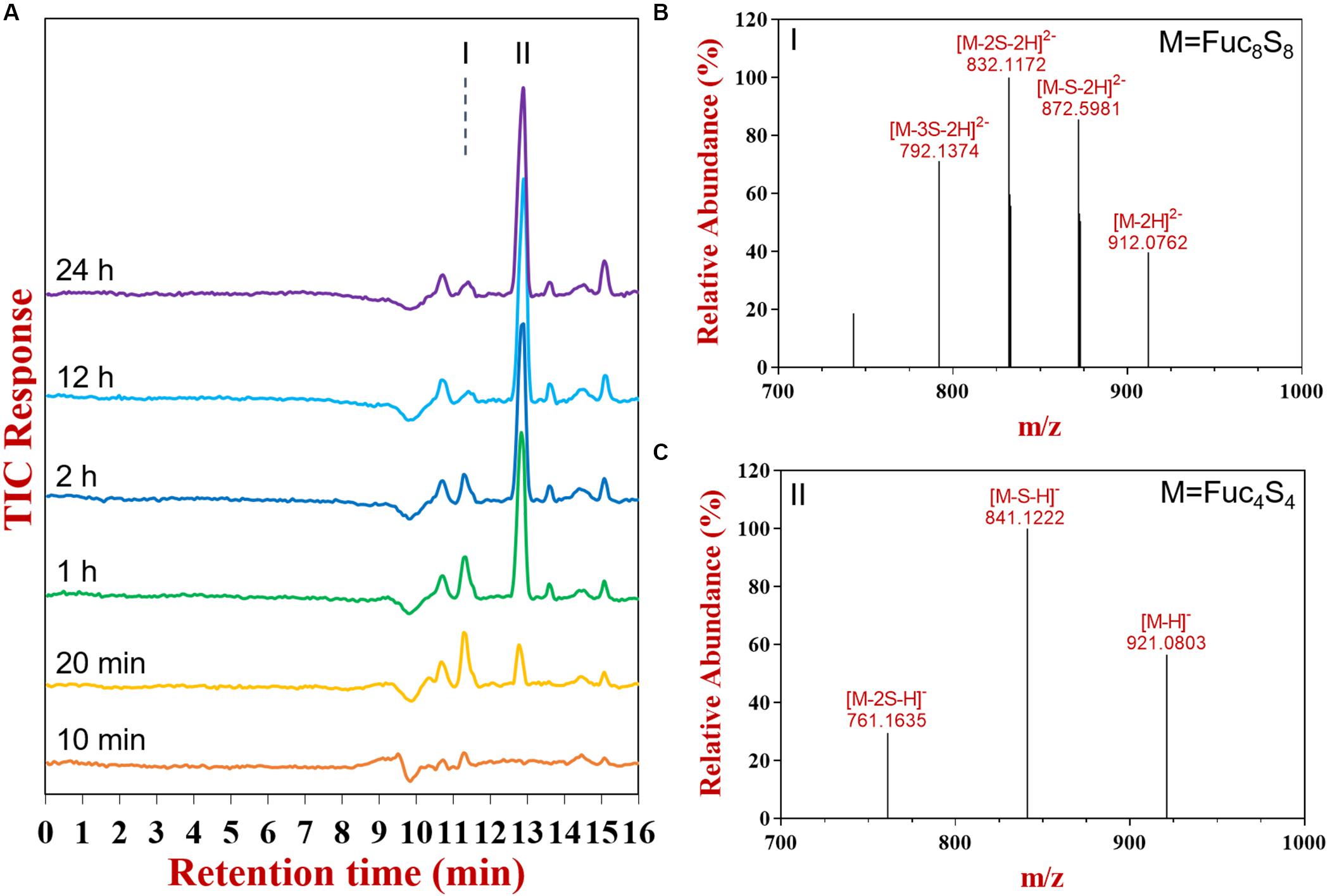
Figure 2. The total ion chromatogram (A), mass spectrums of component I (B) and II (C) as determined by UPSEC-MS.
UPSEC-MS further showed that the octasaccharide (Fuc8S8) increased in the initial stage of the hydrolysis (0–20 min, Supplementary Figure 2A) followed by a decrease, demonstrating the capability of FunA to cleave it. The tetrasaccharide (Fuc4S4) consistently accumulated during the reaction. The end product was obtained after 12 h reaction since the oligosaccharide pattern remained unchanged even after further hydrolysis using the excess enzyme and prolonged incubation time (Figures 1, 2A). Quantitative estimation based on the HPSEC-RID spectrum peak area revealed that the yield of tetrasaccharide against all oligosaccharides products was 87.2%, indicating that the major end product of FunA hydrolysis was tetrasaccharide (Fuc4S4).
The major component of the end product was purified and analyzed by NMR. Four spin systems were found in the COSY spectrum (Figure 3B), and their initiative signals were at δ 5.50, δ 5.39, δ 5.34, and δ 5.10 ppm, which could be attributed to the α-anomeric resonance of four fucopyranoside residues designated as A, B, C, and D (Figure 3A). The other proton chemical shifts (Table 1) were assigned according to their correlation peaks in COSY (Figure 3B) and TOCSY (Supplementary Figure S5) spectra. The positions of sulfated substitution could be elucidated by comparing each proton chemical shift with the corresponding proton chemical shift of the non-sulfated fucose residue (Yu et al., 2013). The O-sulfation would cause the chemical shift of oxymethine protons to downfield by 0.4–0.8 ppm (Yamada et al., 1992). Compared with the proton chemical shifts of non-sulfated fucose residue, residues A, B, C, and D were deduced as 2-O-desulfated, 2,4-O-desulfated, 2-O-desulfated, and non-sulfated α-L-fucose residues, respectively. The glycosidic linkage and the sequence of residues were then validated according to the correlation peaks in NOESY spectrum (insert of Figure 3B). Specifically, H-1 of residue D correlated with H-3 of residue B, H-1 of residue B with H-3 of residue C, and H-1 of residue C to H-3 of residue A. The sequence of residues in the tetrasaccharide was consequently confirmed as D1→3B1→3C1→3A. In result, the major end product was determined to be α-L-Fucp-1→3-α-L-Fucp(2,4OSO3–)-1→3-α-L-Fucp(2OSO3–)-1→3-α-L-Fucp(2OSO3–) (Figure 3C).
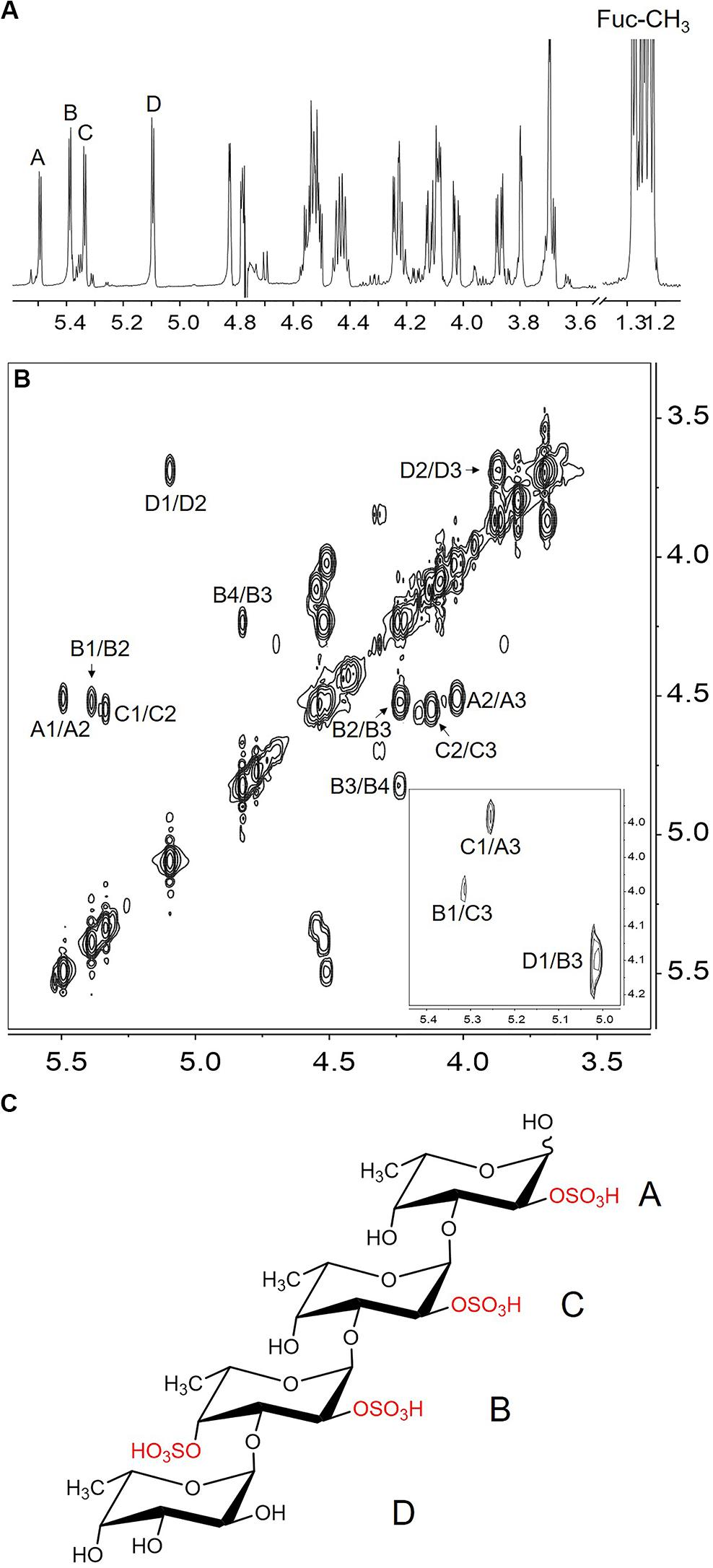
Figure 3. The 1H NMR (A), COSY (B), NOESY (insert of B) spectrum and the structure (C) of the major component in the end product of FunA. A1/A2 indicated the cross-peak between H-1 and H-2 of residue A, etc.
Compared to the structure of Ib-FUC polysaccharide (Supplementary Figure S2), it could be concluded that FunA cleaved the α-1,3-glycoside bonds between 2-O-sulfated and non-sulfated fucose residues in Ib-FUC (Supplementary Figure S6). Besides, FunA was incompetent to hydrolyze sulfated fucans containing α-1,4-glycoside bonds based on the result mentioned above (Supplementary Table S2). Those results further confirmed that FunA is an endo-1,3-fucanase. It should be noted that FunA could not recognize other α-1,3-glycoside bonds in Ib-FUC such as the glycosidic linkages between non-sulfated and 2,4-O-sulfated, between 4-O-sulfated and 2-O-sulfated, or between 2-O-sulfated and 2-O-sulfated fucose residues. It is well known that glycoside hydrolase is a class of enzyme with high specificity to glycosidic linkage and structural modifications. A good example is carrageenase. The κ- and ι-carrageenan share the identical structure except for the additional sulfation on the O-2 position of the anhydro-galactose residue of ι-carrageenan. Nevertheless, their hydrolysis should be implemented by enzymes belonging to two distinctive glycoside hydrolase families, viz., GH16 κ-carrageenase for κ-carrageenan and GH82 ι-carrageenase for ι-carrageenan, demonstrating strong specificity of the enzymes to the sulfated pattern (Matard-Mann et al., 2017). Similarly, the specificity of fucanases might also involve the specific recognition for the sulfated pattern of the substrate. Besides, as shown in section “Confirmation of Endo-1,3-Fucanase Gene Sequence”, FunA was inactive to type II sulfated fucans, confirming that it was incapable of cleaving the α-1,3 glycosidic linkages within the backbone composed of alternating 1,3- and 1,4-linked α-L-fucopyranose residues. FunA has strict specificity, implying that this enzyme could be utilized as a biotechnological tool in the structural investigation of type I sulfated fucans. Besides, FunA could also promote the application of endo-1,3-fucanase in bio-industry such as specific production of α-1,3-linked sulfated fucan oligosaccharides.
The glycoside hydrolases with transglycosylating activity were regarded as the potential biocatalysts for the large-scale synthesis of oligosaccharides or complex carbohydrates (Raich et al., 2016). To confirm the transglycosylating activity for FunA, different acceptors were tested in this experiment. The extracted ion chromatograms of all expected glycosylated products with or without desulfation were thoroughly inspected. No transglycosylating products were observed in sample absented acceptors and the reactions with D-glucose, D-galactose, D-fructose, D-mannose, D-glucosamine, and N-acetyl-D-glucosamine as the acceptor under the conditions of the present study, while the expected ions were detected in the reactions with glycerin, methanol, and L-fucose (Figure 4A and Supplementary Figure S7). To further confirm transglycosylation, the generated products with glycerin as acceptor were detected by UPSEC-MS/MS. The ion [Fuc4S4Gl-2S-2H]2– (m/z 417.0885, Figure 4A) was selected and analyzed in MS/MS due to its high intensity in the MS spectrum. As shown in Figure 4B, critical fragments (Y1 and Y2) containing glycerin aglycone were confirmatively detected in MS/MS, indicating that glycerin was linked to Ib-FUC tetrasaccharide. The results further confirmed FunA is capable to catalyze transglycosylation.
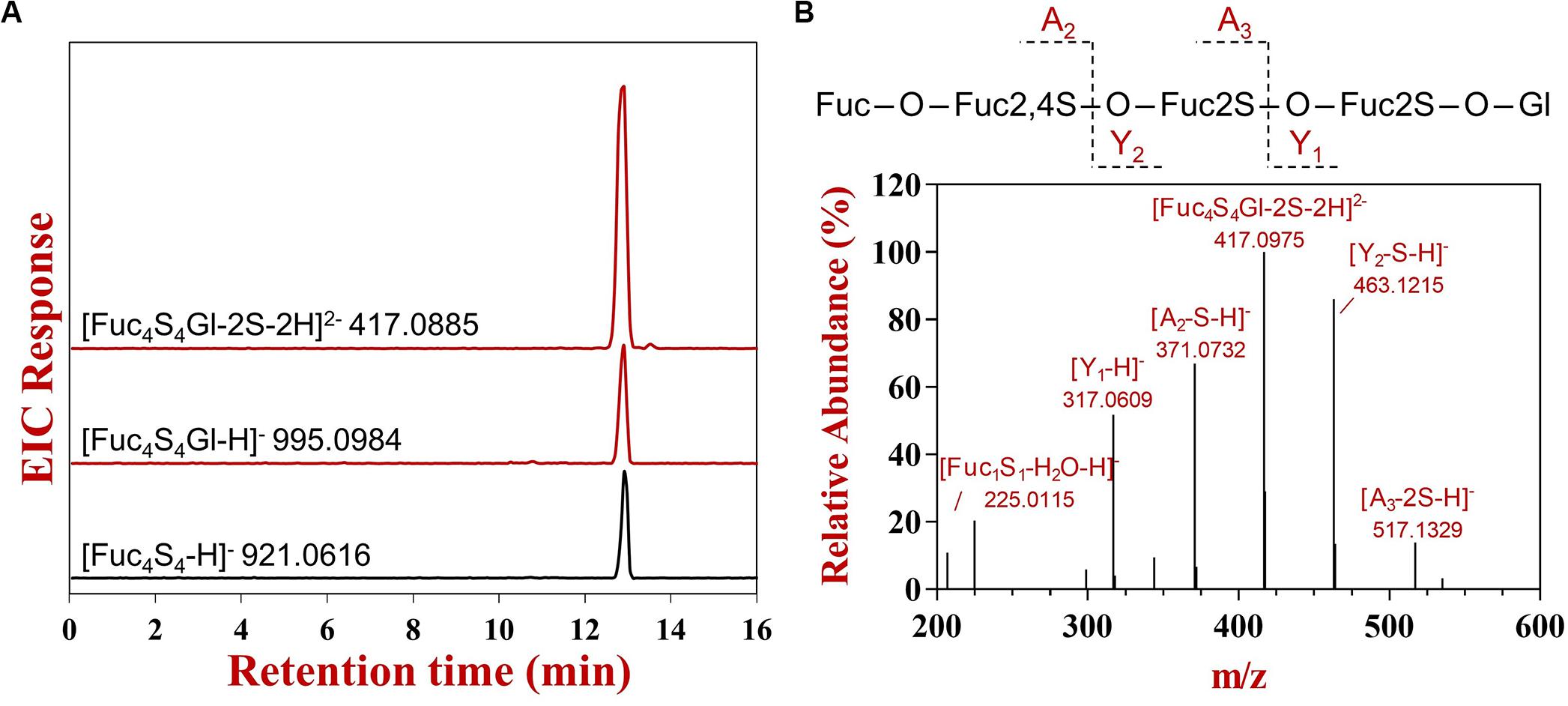
Figure 4. The UPSEC-MS analysis of hydrolysis and transglycosylating products produced by FunA in the reaction system with glycerin as the acceptor. (A) Extraction ion chromatograms; (B) MS/MS spectrum of [Fuc4S4Gl-2S-2H]2–. “Fuc” represented fucose residue, “Gl” represented glycerin residue, “S” represented sulfate group, “O” represented oxygen from the glycosidic bond. The charge number of fragment was provided by the instrumental software (Agilent Masshunter Qualitative Analysis Navigator B.08.00).
The glycoside hydrolases generally act via retaining or inverting mechanisms (Mccarter and Withers, 1994). Previous studies showed that glycoside hydrolases with transglycosylating activity were all adopted retaining mechanism, which ascribed to the two steps catalytic mechanism in retaining glycoside hydrolases: glycosylation and deglycosylation. The transglycosylated product is generated when the acceptor was a molecule with hydroxyl groups instead of water in the deglycosylation reaction (Mccarter and Withers, 1994). To the best of our knowledge, no inverting GHs were reported to possess transglycosylating activities. The transglycosylating activity of FunA implied its retaining mechanism in catalysis. This reaction mechanism is in general determined by NMR measurements (Eneyskaya et al., 2001), while the attempt to monitor the changes of anomeric configurations in the reaction by using 1H NMR failed since chemical shifts for β-α-L-Fucp(2OSO3–) could not be figured out.
Using the BLASTP algorithm, 97 potential homologs of FunA were found in the NCBI database using FunA as the query sequence. They were originated from seven phyla of Bacteria (Bacteroidetes, Proteobacteria, Planctomycetes, Kiritimatiellaeota, Lentisphaerae, Verrucomicrobia, and Sphingobacteriia) and three phylum of Eukaryota (Stramenopiles, Choanoflagellida, and Metazoa). None of the previously reported glycoside hydrolase domains were predicted in all of the sequences analyzed. And those potential homologs of FunA were no significant similarity with enzymes of GH107 family according to BLASTP results. The 16 representative sequences, which had distinct clustering in the phylogenetic tree (Figure 5) were further synthesized, cloned, expressed and characterized. The synthesis of candidate α-1,3-fucanase genes was entrusted to Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China), where 13 out of the 16 sequences were successfully expressed in the supernatants of cell lysate (Figure 5). Five proteins showed apparent activity to Ib-FUC, namely WP_081987558.1, WP_068826447.1, WP_068826442.1, OHE80969.1, and WP_083194720.1 (activities were listed in Supplementary Table S4). The sharp decreases of substrate Mw in the initial hydrolysis stage indicated that these five sequences all hydrolyzed Ib-FUC in an endo-acting manner (Supplementary Figure S8). The novelty of FunA and its homologs in sequence and in activity suggests the existence of a novel fucanase family (assigned as the family GH168 by CAZy).
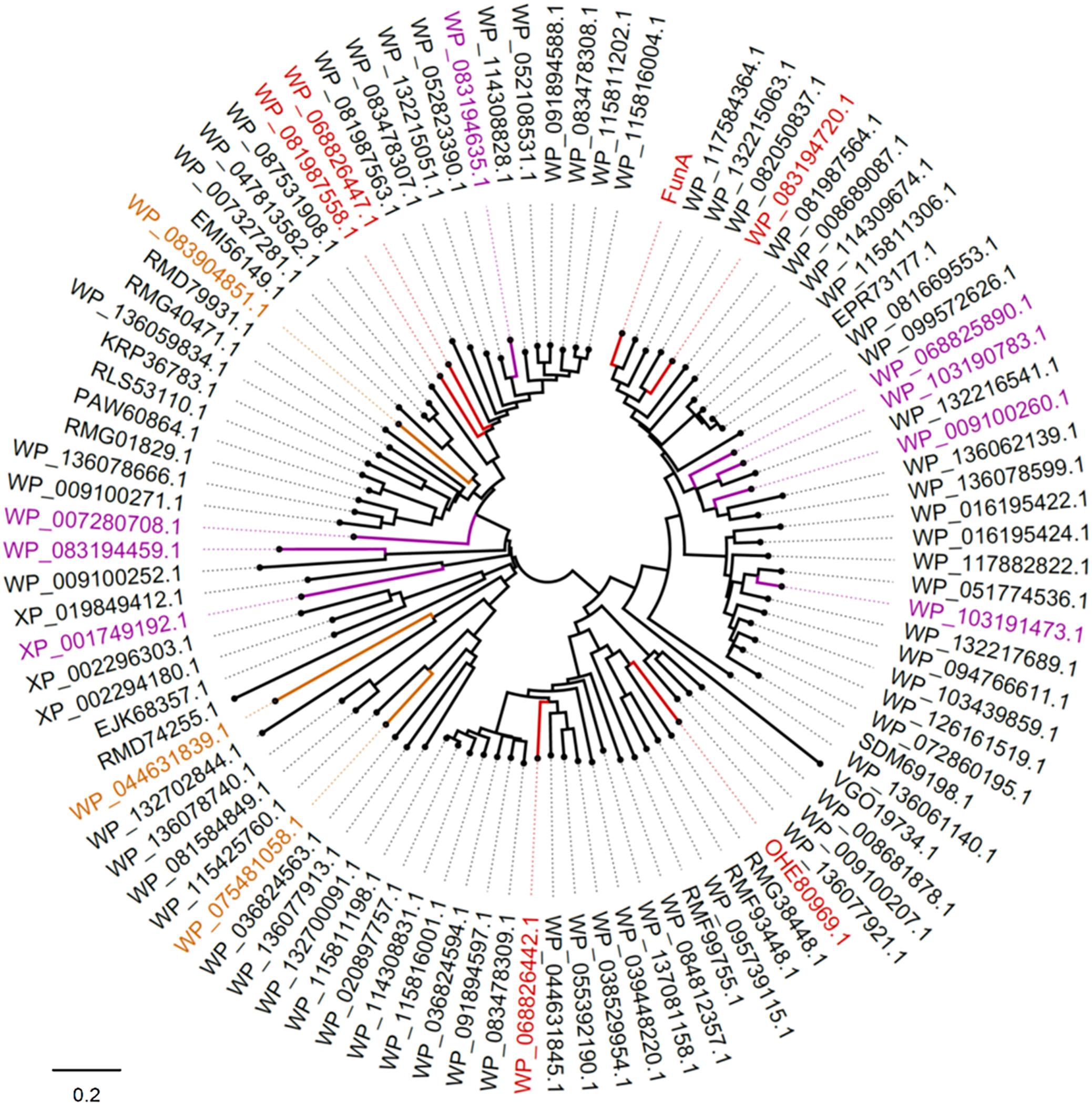
Figure 5. Phylogenetic analysis of FunA and its homologs. Sequences successfully expressed in the supernatant of E. coli cell lysate were highlighted: sequences exhibited activity on Ib-FUC were in red; sequences with no activity on Ib-FUC were in purple; sequences expressed in inclusion body were in yellow.
It should also be noticed that eight sequences (Figure 5, highlighted in purple; Supplementary Figure S9) were successfully expressed while inactive to sulfated fucan substrates employed in the current study. It is well known that many glycoside hydrolase families comprise multiple subfamilies with various substrate specificities, such as GH5, GH16, GH29, and GH95. We supposed that these eight sequences might be specific to other forms of the substrates, warranting further research on the diversity of enzymes in the newly proposed GH family.
Multiple sequence alignment of FunA and its homologs showed that D206 and E264 in FunA were strictly conserved in all sequences (Supplementary Figure S10). Generally, the catalytic residues in GH families are acidic amino acids Asp or Glu. To verify the functions of D206 and E264 in FunA, two single-site mutants D206E and E264Q were established and successfully expressed in E. coli (Supplementary Figure S11), and then purified by HisTrap HP columns (Supplementary Figure S11). The purified D206E and E264Q were all inactive on Ib-FUC, indicating that D206 and E264 were critical for the activity of FunA.
The first endo-1,3-fucanase gene funA was found via transcriptomics from marine bacterium W. fucanilytica CZ1127T. The enzymatic properties and hydrolysis patterns of FunA were further characterized. FunA specifically catalyzed α-1,3 glycosidic linkage between 2-O-sulfated and non-sulfated fucose residues in a random endo-acting manner. It exhibited transglycosylating activities with glycerine, methanol, and L-fucose as acceptors, implying a possible retaining hydrolysis mechanism of FunA. D206 and E264 were critical for the activity of FunA. The endo-1,3-fucanase activity of the homologs of FunA was also verified. The discovery and characterization of FunA proposed a novel GH family (GH168) containing endo-1,3-fucanase.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, WP_068826898.1.
JS and YC conceptualized and designed the studies. JS, YZ, and XM performed the experimental operations. JS performed the data analyses. JS and YC wrote the manuscript. YC and CX revised the manuscript. All the authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31671883), the Fundamental Research Funds for the Central Universities (201941005), and Fok Ying-Tong Education Foundation (171024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01674/full#supplementary-material
FIGURE S1 | The volcano plot of differentially expressed genes. The red dots represent the up-regulated genes of W. funcanilytica induced by growth on the sulfated fucans culture medium, and the green dots represent the down-regulated genes.
FIGURE S2 | Structures of sulfated fucans from sea cucumber I. badionotus (Ib), algae F. vesiculosus (Fv) and algae A. nodosum (An).
FIGURE S3 | SDS-PAGE of the purified recombinant FunA.
FIGURE S4 | Biochemical properties of FunA: (A) optimal temperature; (B) thermal stability; (C) optimal pH; (D) pH stability; (E) influence of NaCl concentration.
FIGURE S5 | The TOCSY spectrum of the major component in the end product of FunA. A1/A2 indicated the cross-peak between H-1 and H-2 of residue A, etc.
FIGURE S6 | The cleavage point of FunA acting on Ib-FUC. Arrows represented the cleavage point.
FIGURE S7 | Extracted ion chromatograms of the transglycosylating products with methanol (A) and fucose (B) as acceptors. “Me” represented the methyl group. The ions of corresponding canonical hydrolysis products were shown as references for a comparison of abundance.
FIGURE S8 | Global profiles of the reaction progress in the initial hydrolysis stage of WP_081987558.1 (A), WP_068826447.1 (B), WP_068826442.1 (C), OHE80969.1 (D) and WP_083194720.1 (E). Ib-FUC was used as the substrate. The TSKgel SuperAW4000 column was employed to investigate the global profile.
FIGURE S9 | SDS-PAGE analysis of recombinant potential homologues of FunA. M: Mw standard markers; Line 1: control group; Line 2: cell lysate supernatant of WP_103190783.1; Line 3: cell lysate supernatant of WP_103191473.1; Line 4: cell lysate supernatant of WP_007280708.1; Line 5: cell lysate supernatant of XP_001749192.1; Line 6: control group; Line 7: cell lysate supernatant of WP_083194459.1; Line 8: cell lysate supernatant of WP_009100260.1; Line 9: cell lysate supernatant of WP_068825890.1; Line 10: cell lysate supernatant of WP_083194635.1; Line 11: control group. The over-expressed proteins were indicated with red rectangles.
FIGURE S10 | Multiple sequence alignment of residues in FunA and its homologues. The strictly conserved residues in all sequences were indicated with black triangles.
FIGURE S11 | SDS-PAGE analysis of the mutants. M: Mw standard markers; Line 1: control group; Line 2: cell lysate supernatant of mutant D206E; Line 3: cell lysate supernatant of mutant E264Q; Line 4: the purified D206E; Line 5: the purified E264Q. The over-expressed proteins in cell lysate supernatants were indicated with red rectangles.
TABLE S1 | The forward and reverse primers of FunA and its mutants.
TABLE S2 | The enzymatic activities of FunA hydrolyzed sulfated fucans from brown algae.
TABLE S3 | Effect of metal ions and chemicals on the activity of FunA.
TABLE S4 | Activities of endo-1,3-fucanases acted on sulfated fucan from I. badionotus.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J. H., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Amin, M. L., Mawad, D., Dokos, S., Koshy, P., Martens, P. J., and Sorrell, C. C. (2020). Immunomodulatory properties of photopolymerizable fucoidan and carrageenans. Carbohyd. Polym. 230:115691. doi: 10.1016/j.carbpol.2019.115691
Armenteros, J. J. A., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Barbeyron, T., Thomas, F., Barbe, V., Teeling, H., Schenowitz, C., Dossat, C., et al. (2016). Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans Dsij(T). Environ. Microbiol. 18, 4610–4627. doi: 10.1111/1462-2920.13584
Berteau, O., and Mulloy, B. (2003). Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide. Glycobiology 13, 29R–40R. doi: 10.1093/glycob/cwg058
Chang, Y., Hu, Y., Yu, L., McClements, D. J., Xu, X., Liu, G., et al. (2016). Primary structure and chain conformation of fucoidan extracted from sea cucumber Holothuria tubulosa. Carbohyd. Polym. 136, 1091–1097. doi: 10.1016/j.carbpol.2015.10.016
Chang, Y., Xue, C., Tang, Q., Li, D., Wu, X., and Wang, J. (2010). Isolation and characterization of a sea cucumber fucoidan-utilizing marine bacterium. Lett. Appl. Microbiol. 50, 301–307. doi: 10.1111/j.1472-765X.2009.02792.x
Chen, F., Chang, Y., Dong, S., and Xue, C. (2016). Wenyingzhuangia fucanilytica sp nov., a sulfated fucan utilizing bacterium isolated from shallow coastal seawater. Int. J. Syst. Evol. Micro. 66, 3270–3275. doi: 10.1099/ijsem.0.001184
Chen, S., Hu, Y., Ye, X., Li, G., Yu, G., Xue, C., et al. (2012). Sequence determination and anticoagulant and antithrombotic activities of a novel sulfated fucan isolated from the sea cucumber Isostichopus badionotus. BBA Gen. Subj. 1820, 989–1000. doi: 10.1016/j.bbagen.2012.03.002
Colin, S., Deniaud, E., Jam, M., Descamps, V., Chevolot, Y., Kervarec, N., et al. (2006). Cloning and biochemical characterization of the fucanase FcnA: definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology 16, 1021–1032. doi: 10.1093/glycob/cwl029
Colliec, S., Fischer, A. M., Taponbretaudiere, J., Boisson, C., Durand, P., and Jozefonvicz, J. C. (1991). Anticoagulant properties of a fucoidan fraction. Thromb. Res. 64, 143–154. doi: 10.1016/0049-3848(91)90114-C
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cumashi, A., Ushakova, N. A., Preobrazhenskaya, M. E., D’Incecco, A., Piccoli, A., Totani, L., et al. (2007). A comparative study of the anti-inflammatory, anticoagulant, antiangiogenic, and antiadhesive activities of nine different fucoidans from brown seaweeds. Glycobiology 17, 541–552. doi: 10.1093/glycob/cwm014
Eneyskaya, E. V., Kulminskaya, A. A., Kalkkinen, N., Nifantiev, N. E., Arbatskii, N. P., Saenko, A. I., et al. (2001). An α-l-fucosidase from Thermus sp with unusually broad specificity. Glycoconjug. J. 18, 827–834. doi: 10.1023/A:1021163720282
Ensor, L. A., Stosz, S. K., and Weiner, R. M. (1999). Expression of multiple complex polysaccharide-degrading enzyme systems by marine bacterium strain 2-40. J. Ind. Microbiol. Biot. 23, 123–126. doi: 10.1038/sj.jim.2900696
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
FigTree (2018). Molecular Evolution, Phylogenetics And Epidemiology. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed March 21, 2020).
Jiao, G., Yu, G., Zhang, J., and Ewart, H. S. (2011). Chemical structures and bioactivities of sulfated polysaccharides from marine algae. Mar. Drugs 9, 196–223. doi: 10.3390/md9020196
Jin, G., and Kim, G. H. (2011). Rapid-prototyped PCL/fucoidan composite scaffolds for bone tissue regeneration: design, fabrication, and physical/biological properties. J. Mater. Chem. A. 21, 17710–17718. doi: 10.1039/c1jm12915e
Kusaykin, M. I., Silchenko, A. S., Zakharenko, A. M., and Zvyagintseva, T. N. (2016). Fucoidanases. Glycobiology 26, 3–12. doi: 10.1093/glycob/cwv072
Lever, M. (1972). A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47, 273–279. doi: 10.1016/0003-2697(72)90301-6
Matard-Mann, M., Bernard, T., Leroux, C., Barbeyron, T., Larocque, R., Prechoux, A., et al. (2017). Structural insights into marine carbohydrate degradation by family GH16-carrageenases. J. Biol. Chem. 292, 19919–19934. doi: 10.1074/jbc.M117.808279
Mccarter, J. D., and Withers, S. G. (1994). Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struc. Biol. 4, 885–892. doi: 10.1016/0959-440X(94)90271-2
Min, S., Han, S., Kim, H., Kwon, O., Lee, S., and Kim, J. (2012). Algal fucoidan, unlike heparin, has thrombolytic activity in a murine arterial thrombosis model. Blood Coagul. Fibrin. 23, 359–366. doi: 10.1097/MBC.0b013e3283518815
Mulloy, B., Ribeiro, A. C., Alves, A. P., Vieira, R. P., and Mourao, P. (1994). Sulfated fucans from echinoderms have a regular tetrasaccharide repeating unit defined by specific patterns of sulfation at the O-2 and O-4 positions. J. Biol. Chem. 269, 22113–22123.
Murakami, K., Ishihara, M., Aoki, H., Nakamura, S., Nakamura, S., Yanagibayashi, S., et al. (2010). Enhanced healing of mitomycin C-treated healing-impaired wounds in rats with hydrosheets composed of chitin/chitosan, fucoidan, and alginate as wound dressings. Wound Repair Regen. 18, 478–485. doi: 10.1111/j.1524-475X.2010.00606.x
Nagamine, T., Kadena, K., Tomori, M., Nakajima, K., and Iha, M. (2020). Activation of NK cells in male cancer survivors by fucoidan extracted from Cladosiphon okamuranus. Molecular and clinical oncology 12, 81–88. doi: 10.3892/mco.2019.1943
Raich, L., Borodkin, V., Fang, W., Castro-Lopez, J., van Aalten, D. M. F., Hurtado-Guerrero, R., et al. (2016). A trapped covalent intermediate of a glycoside hydrolase on the pathway to transglycosylation. insights from experiments and quantum mechanics/molecular mechanics simulations. J. Am. Chem. Soc. 138, 3325–3332. doi: 10.1021/jacs.5b10092
Schultz-Johansen, M., Cueff, M., Hardouin, K., Jam, M., Larocque, R., Glaring, M. A., et al. (2018). Discovery and screening of novel metagenome-derived GH107 enzymes targeting sulfated fucans from brown algae. The FEBS journal. 22, 4281–4295. doi: 10.1111/febs.14662
Sezer, A. D., Hatipoglu, F., Cevher, E., Ogurtan, Z., Bas, A. L., and Akbuga, J. (2007). Chitosan film containing fucoidan as a wound dressing for dermal burn healing: preparation and in vitro/in vivo evaluation. AAPS Pharmscitech 8:39.
Shen, J., Chang, Y., Dong, S., and Chen, F. (2017). Cloning, expression and characterization of a ι-carrageenase from marine bacterium Wenyingzhuangia fucanilytica: a biocatalyst for producing ι-carrageenan oligosaccharides. J. Biotechnol. 259, 103–109. doi: 10.1016/j.jbiotec.2017.07.034
Silchenko, A. S., Rasin, A. B., Kusaykin, M. I., Malyarenko, O. S., Shevchenko, N. M., Zueva, A. O., et al. (2018). Modification of native fucoidan from Fucus evanescens by recombinant fucoidanase from marine bacteria Formosa algae. Carbohyd. Polym. 193, 189–195. doi: 10.1016/j.carbpol.2018.03.094
Silchenko, A. S., Ustyuzhanina, N. E., Kusaykin, M. I., Krylov, V. B., Shashkov, A. S., Dmitrenok, A. S., et al. (2017). Expression and biochemical characterization and substrate specificity of the fucoidanase from Formosa algae. Glycobiology 27, 254–263. doi: 10.1093/glycob/cww138
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 12, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. doi: 10.1093/nar/25.24.4876
Vickers, C., Liu, F., Abe, K., Salama-Alber, O., Jenkins, M. L., Springate, C. M., et al. (2018). Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to α-l-fucosidases from GH29. J. Biol. Chem. 47, 18296–18308. doi: 10.1074/jbc.RA118.005134
Vilela-Silva, A., Alves, A. P., Valente, A. P., Vacquier, V. D., and Mourao, P. (1999). Structure of the sulfated alpha-l-fucan from the egg jelly coat of the sea urchin Strongylocentrotus franciscanus: patterns of preferential 2-O and 4-O-sulfation determine sperm cell recognition. Glycobiology 9, 927–933. doi: 10.1093/glycob/9.9.927
Yamada, S., Yoshida, K., Sugiura, M., and Sugahara, K. (1992). One-dimensional and 2-dimensional H-1-NMR characterization of 2 series of sulfated disaccharides prepared from chondroitin sulfate and heparan-sulfate heparin by bacterial eliminase digestion. J. Biochem. 112, 440–447. doi: 10.1093/oxfordjournals.jbchem.a123919
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., and Xu, Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res., W 1, W445–W451. doi: 10.1093/nar/gks479
Yu, L., Ge, L., Xue, C., Chang, Y., Zhang, C., Xu, X., et al. (2014a). Structural study of fucoidan from sea cucumber Acaudina molpadioides: a fucoidan containing novel tetrafucose repeating unit. Food Chem. 142, 197–200. doi: 10.1016/j.foodchem.2013.06.079
Yu, L., Xue, C., Chang, Y., Xu, X., Ge, L., Liu, G., et al. (2014b). Structure elucidation of fucoidan composed of a novel tetrafucose repeating unit from sea cucumber Thelenota ananas. Food Chem. 146, 113–119. doi: 10.1016/j.foodchem.2013.09.033
Yu, L., Xu, X., Xue, C., Chang, Y., Ge, L., Wang, Y., et al. (2013). Enzymatic preparation and structural determination of oligosaccharides derived from sea cucumber (Acaudina molpadioides) fucoidan. Food Chem. 139, 702–709. doi: 10.1016/j.foodchem.2013.01.055
Yu, L., Xue, C., Chang, Y., Hu, Y., Xu, X., Ge, L., et al. (2015). Structure and rheological characteristics of fucoidan from sea cucumber Apostichopus japonicus. Food Chem. 180, 71–76. doi: 10.1016/j.foodchem.2015.02.034
Zhang, H., Yoshizawa, S., Sun, Y., Huang, Y., Chu, X., Gonzalez, J. M., et al. (2019). Repeated evolutionary transitions of flavobacteria from marine to non-marine habitats. Environ. Microbiol. 21, 648–666. doi: 10.1111/1462-2920.14509
Keywords: marine bacterium, sulfated fucan, endo-1, 3-fucanase, transcriptomics, UPSEC-MS, oligosaccharide
Citation: Shen J, Chang Y, Zhang Y, Mei X and Xue C (2020) Discovery and Characterization of an Endo-1,3-Fucanase From Marine Bacterium Wenyingzhuangia fucanilytica: A Novel Glycoside Hydrolase Family. Front. Microbiol. 11:1674. doi: 10.3389/fmicb.2020.01674
Received: 28 April 2020; Accepted: 26 June 2020;
Published: 28 July 2020.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Fu-Li Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaCopyright © 2020 Shen, Chang, Zhang, Mei and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoguang Chang, Y2hhbmd5Z0BvdWMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.