
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 July 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01670
 Yanping Li1,2†
Yanping Li1,2† Tengfei Xie2†
Tengfei Xie2† Rui Pang2†
Rui Pang2† Qingping Wu2*
Qingping Wu2* Jumei Zhang2
Jumei Zhang2 Tao Lei2
Tao Lei2 Liang Xue2
Liang Xue2 Haoming Wu2
Haoming Wu2 Juan Wang1
Juan Wang1 Yu Ding3
Yu Ding3 Moutong Chen2
Moutong Chen2 Shi Wu2
Shi Wu2 Haiyan Zeng2
Haiyan Zeng2 Youxiong Zhang2
Youxiong Zhang2 Xianhu Wei2
Xianhu Wei2Vibrio parahaemolyticus is a marine and estuarine bacterium that leads to damage of aquatic industry by foodborne outbreaks and possesses an enormous threat to food safety as well as human health worldwide. In the current study, we investigated 905 food samples (ready-to-eat foods, fish, and shrimp) from 15 provinces in China, and aimed to determine prevalence, biological characteristics and genetic diversity of presumptive V. parahaemolyticus isolates. Firstly, 14.17% of 240 fish samples, 15.34% of 365 shrimp samples and 3.67% of 300 RTE food samples were positive for potential V. parahaemolyticus. Secondly, 69 food samples (14.87%) collected in summer were positive for target isolates, while the rate of positive sample of 441 food samples in winter reached 7.26%. Thirdly, we purified 202 V. parahaemolyticus strains for further research. And antimicrobial susceptibility results of strains tested revealed that the highest resistance rate was observed for ampicillin (79.20%). At the same time, 148 (73.27%) of all isolates were classified and defined as multi-drug resistant foodborne bacteria. The results of PCR assay showed that the isolates being positive for the tdh, trh or both genes, were up to 9.90%, 19.80% or 3.96%. Besides, multiplex PCR test showed that the isolates carrying O2 serogroup were the most prevalent. Furthermore, sequence types (STs) of 108 isolates were obtained via multi-locus sequence typing. Not only 82 STs were detected, but also 41 of which were updated in the MLST database. Thus, our findings significantly demonstrated the high contamination rates of V. parahaemolyticus in fish and shrimp and it may possess potential threat for consumer health. We also provided up-to-date dissemination of antibiotic-resistant V. parahaemolyticus which is important to ensure the high efficacy in the treatment of human and aquatic products infections. Lastly, with the identification of 82 STs including 41 novel STs, this study significantly revealed the high genetic diversity among V. parahaemolyticus. All of our research improved our understanding on microbiological risk assessment in ready-to-eat foods, fish, and shrimp.
Vibrio parahaemolyticus, a food-borne gastroenteritis-causing bacterium, is identified as a gram-negative bacterial cell that is common in seawater, seafood, and aquatic products. V. parahaemolyticus gastroenteritis outbreaks have been reported worldwide, including those in Bangladesh (Akther et al., 2016), Europe (Baker-Austin et al., 2010; Caburlotto et al., 2016; Lopatek et al., 2018), Japan (Arakawa et al., 1999), United States (Shaw et al., 2015) and South America (Raszl et al., 2016). In China, this bacterium is also a common causative agent of food poisoning (Xie et al., 2015; Dong et al., 2016) associated with consumption of fish, shellfish, and shrimp. Recently, not only has it been isolated from samples of a variety of aquatic products, but it has also been found in ready-to-eat (RTE) food (Xie et al., 2015; Cho et al., 2016; Pang et al., 2019). However, to date, the limited risk assessment of V. parahaemolyticus on its prevalence and contamination levels of RTE food leaded less information about monitoring and treatment strategies in China. Therefore, we presented information and provided insights indicating the importance of developing microbiological risk assessment and control strategies for V. parahaemolyticus strains closely related to food safety into the future.
Since the discovery of penicillin in the 1920s, the application of antibiotics makes great contribution to human and animal medical treatment (Aarestrup and Wegener, 1999). Nowadays, increasing researches have obviously indicated that prevalence of antibiotic-resistant V. parahaemolyticus may pose a huge threat to public health and economic development for humans worldwide (Ghenem and Elhadi, 2018; Lopatek et al., 2018). During the past few decades, due to the excessive application of antibiotics in medical treatment and aquaculture industry, antibiotic resistance has emerged and evolved in V. parahaemolyticus (Mazel and Davies, 1999; Cabello, 2006; Elmahdi et al., 2016). Currently, a large number of reports indicate that V. parahaemolyticus isolated from various sources has presented high resistance to single or multiple antibiotics, especially ampicillin (Lee et al., 2018; Fattel et al., 2019; Jiang et al., 2019; Mohamad et al., 2019; Ryu et al., 2019). The frequently occurring phenomenon of multi-drug resistance in V. parahaemolyticus directly affects application of antibiotics and prevention as well as treatment of bacterial infectious diseases (Xu et al., 2016). Therefore, it is urgent and necessary to establish a monitoring system and efficient treatment strategies of the V. parahaemolyticus antimicrobial-resistance profile.
Food poisoning associated with pathogenic V. parahaemolyticus is often caused by consumption of seafood and aquatic products contaminated with pathogen as well as bacterial toxins (Food and Drug Administration [FDA], 2018). V. parahaemolyticus strains carry the tox R gene that encodes an important membrane-localized regulatory protein, and it commonly gets involved in the bacterial regulation of a variety of expression products (Lin et al., 1993; Zhang et al., 2018). For example, tox R expression is able to regulate the production of thermostable direct hemolysin (TDH), TDH related hemolysin (TRH), T3SS1, and T3SS2 (Whitaker et al., 2012; Hubbard et al., 2016; Mala et al., 2016). TDH with hemolytic activity, enterotoxicity and cytotoxicity, is defined as a type of membrane pore protein. TRH, a heat labile toxin, is thought to have a similar hemolytic activity and pathogenic mechanism to TDH (Matsuda et al., 2010; Leoni et al., 2016). However, recent researches even showed that some clinical V. parahaemolyticus strains do not carry both major virulence factors but remains pathogenic indicating putative virulence factors exist, and pathogenicity might be achieved with different strategies employed by different strains (Mahoney et al., 2010; Jones et al., 2012; Cai and Zhang, 2018). To date, tox R, tdh, and trh gene sequences have been identified in V. parahaemolyticus isolates using PCR-based methods (Shirai et al., 1990; Xie et al., 2017).
The molecular identification and classification of Vibrio parahaemolyticus usually used PCR technology and Sanger sequencing to analyze the core genes. For example, based on the variations in somatic O and capsular K antigens, V. parahaemolyticus can be classified into 13 O and 71 K serogroups (Iguchi et al., 1995). It’s believed that various serogroups, associated with high virulence, have been identified in environmental isolates and clinical samples (Mala et al., 2016; Li et al., 2018; Guin et al., 2019). Additionally, the multilocus sequence typing (MLST) scheme, was originally proposed for the identification of closely related bacterial genotypes. However, the genealogical information derived from the DNA sequences also allowed one to address questions about species boundaries and evolutionary relationships (Harismendy et al., 2009; Perez-Losada et al., 2013; Chen et al., 2015; Chen and Perfect, 2017). Both O-serogroup typing and MLST scheme is easy to operate, quick, and facilitates the exchange of data between laboratories via public databases, so that it can be used to investigate the source of infection and route of transmission.
Vibrio parahaemolyticus strains were frequently detected and isolated from seafood and aquatic products. Recently, this bacterium with antibiotic resistant phenotype has also been present in RTE food (Cho et al., 2016; Mala et al., 2016). As a result of an increase in the quality of life in China, RTE food has become a product of mass consumption. This study mainly aims to investigate the seasonal prevalence of V. parahaemolyticus from fish, shrimp, and RTE food in China. We also characterized the prevalence of each isolate and combined phenotyping (antibiotic resistance patterns) and genotyping (MLST) methods to determine and analyze the genetic relatedness and diversity among the tested bacterial isolates. These findings may facilitate the evaluation of the microbiological profiles of edible products and contribute to the effort to ensure food safety in China.
In this study, 905 food samples, including 300 RTE food samples, 240 fish samples and 365 shrimp samples, were collected from retail stores in 15 cities of 15 provinces of China (Figure 1). The climate of sample collection was cold from September 2015 to March 2016 (winter), and hot from March 2016 to September 2016 (summer). All of food samples were collected and placed in sterile sealed plastic bags from environmental microbial contamination, and stored in a cold box at <4°C during transportation. Sample processing and quantification of V. parahaemolyticus were performed immediately. Two V. parahaemolyticus reference strains, ATCC 33847 (O4, tox R+, tdh+, trh–) and ATCC 17802 (O1, tox R+, tdh–, trh+), were obtained from the American Type Culture Collection (ATCC; Manassas, VA, United States).
Sample processing, bacterial load, and qualitative detection of V. parahaemolyticus in the samples were performed, according to the National Food Safety Standards of China Document GB4789.7-2013 with minor modifications. In brief, 25 g of each sample were placed into 225 mL of alkaline peptone water (APW) containing 3% NaCl (Huankai, Guangzhou, China). It was suggested that fish and shrimp samples were taken from surface tissues, intestines or gills. Then, 1 mL suspension was collected from the top 1 cm of each tube, a serial dilution was prepared up to 103, and 1 mL of each dilution was transferred into a new tube with 9 mL of APW (3% NaCl)., The enumeration of presumptive V. parahaemolyticus was determined by the most probable number (MPN) method of culture tubes positive for V. parahaemolyticus to MPN/g using an MPN table.
Purification and identification of suspected V. parahaemolyticus was performed after 18 h of incubation at 37°C in APW (3% NaCl). To form V. parahaemolyticus colonies appeared green or blue green, a loopful from culture medium was streaked on thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates (Huankai, Guangzhou, China) and all plates were incubated at 37°C under aerobic conditions for 18–24 h. One to three forming colonies were purified by streaking onto Chromogenic Vibrio Medium (Huankai, Guangzhou, China) and incubated at 37°C under aerobic conditions for 24 h. Each mauve colony from each Chromogenic Vibrio Medium plate was picked for identification tests including halophilism tests, oxidase activity assessment, Gram staining, the 3.5% NaCl triple-sugar-iron (TSI) test, and API 20E diagnostic strips testing (BioMerieux Company, Marcyl’Étoile, France).
A total of 202 V. parahaemolyticus isolates were subjected for antimicrobial susceptibility testing by disk-diffusion method, according to the detailed guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2018). Nutrient Agar (Huankai, Guangzhou, China), Muller-Hinton agar (Huankai, Guangzhou, China) and a panel of 12 antibiotic disks (Oxoid, Hampshire, United Kingdom) were used for antibiotic susceptibility tests. V. parahaemolyticus isolates were inoculated on Nutrient Agar and cultivated overnight at 37°C. Each bacterial culture was adjusted to the turbidity of a 0.5 McFarland standard and spread onto Muller-Hinton agar plates. After that, the antibiotic disks were placed on the plates that were then incubated for 18 h at 37°C. The following 12 antimicrobial disks were used in this study (with concentrations per disk given in parentheses): ampicillin (10 μg), azithromycin (15 μg), cefazolin (30 μg), cephalothin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), streptomycin (10 μg), trimethoprim-sulfamethoxazole (25 μg), or tetracycline (30 μg). The results of diameter of the inhibition zone around each disk were recorded and expressed as sensitive (S), intermediate (I), and resistant (R), following the methods of the CLSI. Escherichia coli ATCC 25922 and V. parahaemolyticus ATCC 17802 were used as quality control organisms.
Bacterial whole genome was extracted from cell pellet using a commercial Universal DNA Extraction Kit (Sangon, Shanghai, China), according to the manufacturer’s formal instructions. Following DNA extraction, the detection of three virulence genes (tox R, tdh, and trh) in all 202 V. parahaemolyticus isolates was determined through PCR. The primer sequences of tox R were tox R-F: GTCTTCTGACGCAATCGTTG, tox R-R: ATACGAGTGGTTGCTGTCATG (Sangon, Shanghai, China) (Cho et al., 2016; Mala et al., 2016). Detection of the tdh and trh genes was performed as described previously (West et al., 2013), using the primers tdh-F: CTGTCCCTTTTCCTGCCCCCG, tdh-R: AGCCAGACACCGCTGCCATTG; trh-F: ACCTTTTCCTT CTCCWGGKTCSG, and trh-R: CCGCTCTCATATGCYTCG ACAKT (Sangon, Shanghai, China). PCR reagents (total volume, 25 μL) were prepared by mixing 12.5 μL of 2 × PCR Mix (Qiagen), 0.5 μL forward primer, 0.5 μL reverse primer, 0.5 μL genomic DNA, and 11 μL ddH2O. All genes were amplified in a Bio-Rad PTC-200 Thermal Cycler (Bio-Rad, Hercules, CA, United States) using the following PCR protocol: pre-denaturation at 95 °C for 5 min; 40 cycles of 94 °C for 1 min (denaturation), 62 °C for 1 min (annealing), 72 °C for 1 min (extension), and a final extension of 72 °C for 2 min. Before Images were performed in a Gel Image system (Bio-Rad, Hercules, CA, United States), all amplified products were visualized by 2% agarose gel containing GoldView. To validate the PCR performance, whole genome from V. parahaemolyticus strains ATCC33847 (tdh+) and ATCC17802 (trh+) were used as positive control templates, and sterile purified water was used as the negative control.
The serogroups of V. parahaemolyticus isolates were identified using the multiplex PCR technique. The specific primer sequences and PCR protocol were set as described previously (Chen et al., 2012).
Multilocus sequence typing analysis was performed using seven conserved housekeeping genes (i.e., dnaE, gyrB, recA, dtdS, pntA, pyrC, and tnaA) to characterize diversity and epidemiology (Xu et al., 2017). We randomly chosed 108 isolates of V. parahaemolyticus from 202 food-borne isolates. PCR reagents (total volume, 50 μL) were prepared by mixing 5.0 μL of 10× PCR buffer (Takara, Dalian, China), 1 μL forward primer, 1 μL reverse primer, 1 μL genomic DNA, and 42 μL ddH2O. The PCR amplification conditions in a Bio-Rad PTC-200 Thermal Cycler were as follows: a cycle of pre-denaturation at 94 °C for 5 min; 35 cycles at 94 °C for 30 s (denaturation), 55 °C for 30 s (annealing), 72 °C for 2 min (extension), and a final extension at 72 °C for 10 min. Before sequenced on an ABI 3730 sequencer (Applied Biosystems), the target amplification products were purified using a PCR purification kit (Qiagen, Germany). To obtain allele numbers and define sequence types (STs), the sequences were analyzed on the MLST database1. Genotyping analysis and phylogenetic analysis were based on MLST sequences. Both phylogenetic tree and minimum spanning tree were generated by BioNumerics 7.6.
Of the 905 food samples, 101 (11.16%) were positive for V. parahaemolyticus. This included 34 (14.17%) of the 240 fish samples, 56 (15.34%) of the 365 shrimp samples, and 11 (3.67%) of the 300 RTE food samples. Overall, the density of V. parahaemolyticus varied from 1.50 to 100 MPN/g. Of the positive samples, 64.36% contained a bacterial load of 3 to 10 MPN/g, 18.81% contained a load >3 MPN/g, and 16.83% samples exceeded 10 MPN/g (Table 1).
Analysis to determine the seasonal prevalence of the pathogen revealed a higher number of positive samples in the summer compared to winter (Table 2). The rate of V. parahaemolyticus detection in summer reached 14.87%, while it only reached 7.26% in winter. Besides, the prevalence of V. parahaemolyticus in summer were 20.83% (fish samples), 20.11% (shrimp samples), and 4.38% (RTE food samples). On the other hand, the prevalence in winter was 6.67% (fish samples), 12.15% (shrimp samples), and 1.40% (RTE food samples). It meant the contamination level of all samples in summer was higher than that observed in the winter samples Moreover, the mean level of V. parahaemolyticus in samples collected during summer and winter were 8.08 and 5.3 MPN/g, respectively. Therefore, the population of V. parahaemolyticus was certainly abnormal for samples collected in different seasons. In total, 202 potential V. parahaemolyticus isolates from 101 samples were detected and purified for further bacterial classification and epidemiology.
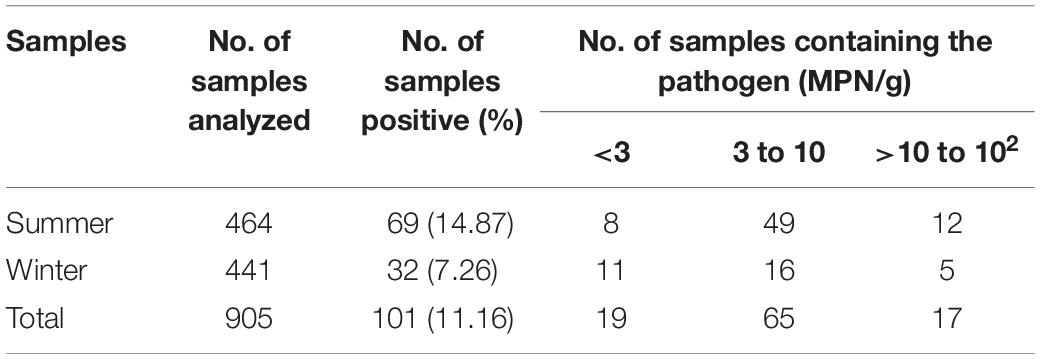
Table 2. Prevalence and levels of Vibrio parahaemolyticus in Chinese samples during different seasons.
The extent of antibiotic resistance was examined in 202 isolates of V. parahaemolyticus. And bacteria exhibited three or more resistant phenotypes was called multidrug-resistant V. parahaemolyticus. The antimicrobial resistance profiles of the isolates are shown in Table 3 and antibiotic-resistant phenotypes of all isolates are summarized in Supplementary Table S1. The isolates were mostly resistant to ampicillin, with 79.20% R ratings and 14.36% I ratings. Additionally, the isolates exhibited relatively high resistance rates of 74.75%, 65.84%, 58.91%, and 44.55%, to cephalothin, streptomycin, cefazolin, and kanamycin, respectively. However, most of the examined isolates were susceptible to nalidixic acid (97.52%), ciprofloxacin (96.04%), and chloramphenicol (90.59%). Among the remaining tested antibiotics, the next highest susceptibility rates were observed for tetracycline (78.71%), trimethoprim-sulfamethoxazole (78.22%), and azithromycin (77.23%). In addition, among all isolates, three were multidrug-resistant (843, 860, and 2928A2), showing resistance to nine antibiotics, and seven isolates showing resistance to eight antibiotics. Of the isolates, 73.27% were resistance to more than three antibiotics.
All of the 202 V. parahaemolyticus isolates were tested for the presence of tox R, trh and tdh, and the results are shown in Supplementary Table S1. All of the isolates were positive for the tox R gene. Among these, 9.90 and 19.80% of the V. parahaemolyticus strains carried the tdh or the trh genes, respectively, whereas eight isolates harbored both the tdh and trh genes. Among eight isolates (tdh+, trh+), 566, 654, 690, 3154B2, and 709B1 were form shrimp samples whereas the other three were (3331A1, 3478B2, and 3481B3) from fish samples. Lastly, the rates of tdh-positive strains among fish, shrimp, and RTE foods were 15.63, 7.32, and 6.67%, respectively. The rates of trh gene positivity were 18.75, 21.95, and 6.67%, respectively.
The analysis of multiplex PCR-ebased O serogroups is performed to obtain information of DNA fragment size on the distribution of bacterial classification among the 202 V. parahaemolyticus isolates. The results are shown in Table 5. With the exception of serogroups O3, O8, and O9, all other serogroups were detected among the isolates. O2 serogroup was the most prevalent (49.01%), followed by O1 serogroup (22.77%). Besides, the occurrence of V. parahaemolytiucs (O2) were 50.00% (32 of 64 strains from fish samples), 51.22% (63 of 123 strains from fish samples) and 26.67% (4 of 15 strains from RTE food samples). Moreover, there were seven cities (Guiyang, Hangzhou, Hohhot, Hongkong, Macao, Urumchi, and Zhengzhou) whose main prevalent of serogroup was O2 and the occurrence was 52.27, 42.85, 57.89, 67.58, 50.00, 62.50, and 100.00%. The serogroups of V. parahaemolyticus ATCC17802 and ATCC33847, were O1 and O4, respectively.
The results of MLST of the 108 V. parahaemolyticus isolates tested are shown in Table 6, Figures 2, 3. Our research revealed that 81 different STs were identified in 108 isolates derived from fish, shrimp and RTE-food samples (Table 6). In addition, 40 of them were searched in pubMLST Database. Moreover, ST411 (6.5%), ST992 (4.6%), ST423 (3.7%), ST693 (3.7%), and ST1352 (2.8%) were the most common STs detected in this study. More importantly, 41 new STs, including 46 strains, were identified in tested isolates. Lastly, the minimum spanning tree based on allele numbers and O-serogroup typing of the isolates did not reveal a clear clustering pattern connected with the serogroup emergence.
In our study, samples collection, processing and quantification were based on China Document GB4789.7-2013. Meanwhile, V. parahaemolyticus isolates were purified by TCBS agar plate and Chromogenic Vibrio Medium. Our results revealed that these methods were fit to isolation and purification of V. parahaemolyticus specifically. According to epidemiological data, fish and shrimp may suffer from a severe microbial contamination associated with V. parahaemolyticus. Here, we analyzed 300 RTE food, 240 fish and 365 shrimp samples collected in China. From 101 positive samples collected in summer and winter, 202 V. parahaemolyticus isolates were detected, purified and identified. As is showed in Table 1, the most severely contaminated edible was shrimp (15.34%). However, the contamination detection rate was lower than that conveyed in other studies that report a prevalence of 28.0% (Caburlotto et al., 2016). The reason may be that the cities were sample collection was conducted were far away from the ocean, so these types of aquatic products were more scarce. Even though the contamination rate was low, a report indicated that the prevalence of V. parahaemolyticus in aquatic products is an important cause of food poisoning in Shanghai, China (Zhang et al., 2017). On the other hand, the V. parahaemolyticus contamination rate has remained stable at a relatively high level in Shandong and Wenzhou in China (Zhang et al., 2016; Guo et al., 2018). Therefore, to prevent food-borne diseases, it is necessary to highlight the need for microbiological risk assessments of food safety and enhance active monitoring efforts, especially the sanitary management of seafood, aquatic products and RTE food.
Notably, the prevalence of V. parahaemolyticus in summer (14.87%) was higher than that in winter (7.26%), and the mean level as well as population of V. parahaemolyticus was certainly different for samples collected during the winter and summer. The contamination level in the summer was higher than in the winter. This distinction may be related to the differences in the average temperature change between the seasons, as it was in agreement with the results reported in previous studies that showed a seasonal variation in the occurrence of this pathogen (Parveen et al., 2008; Caburlotto et al., 2016; Zhang et al., 2017). Thus, considering temperature as a factor may enhance the efforts of food quality control, in terms of V. parahaemolyticus decontamination, as it is a confirmed pathogen in the WHO risk assessment2. As these results were obtained from a large number of variable samples and from most regions in China, the data presented is more representative of China as a whole.
The continuous and extensive abuse of antibiotics in humans as well as animals has led to the urgent state of the outbreak of multidrug-resistant V. parahaemolyticus strains worldwide. In our study, high resistance rate of 202 V. parahaemolyticus isolates was observed for some antibiotics, such as ampicillin (79.20%), cephalothin (74.75%), and streptomycin (65.84%). Similarly, the occurrence and outbreak of streptomycin- and ampicillin-resistant V. parahaemolyticus isolates were also reported in the past 5 years (Hu and Chen, 2016; Jiang et al., 2019). Besides, antibiotic susceptibility tests displayed the highest level of resistance to ampicillin, signifying that ampicillin may be ineffective and invalid for the treatment of Vibrio sp. infections (Hu and Chen, 2016; Jiang et al., 2019). More importantly, ampicillin resistance has been reported to be 100% in other studies (Fattel et al., 2019). Such ampicillin-resistant pattern was closely related to human behavior, such as the application of first generation antibiotics including ampicillin in aquaculture of Vibrio infection. More seriously, some isolates even revealed antibiotic resistant phenotype to gentamicin, tetracycline, or ciprofloxacin, which are first-line antibiotics widely used in clinical treatment for bacterial infection (Elmahdi et al., 2016; Tan et al., 2017). At the same time, 73.27% of 202 strains with multi-drug resistant phenotype were detected and three of them showed resistance to nine antibiotics. The rate was higher than in previous reports (Letchumanan et al., 2015a; Xu et al., 2016). Such observation may be closely related to the abuse of various antibiotics to prevent and control pathogenic bacterial infections in aquatic environments (Cabello et al., 2013). Generally, it is important to evaluate mutation and evolution of resistance determinants in V. parahaemolyticus, as infection caused by emerging of multi-drug antimicrobial-resistant strains plays an essential role in clinical treatment. Recently, the Food and Agriculture Organization (FAO) has designed action plans to increase awareness regarding the urgent state of drug-resistant food-borne pathogens and promote prudent use of antimicrobials3. Moreover, research focusing on providing alternatives to antibiotics is urgently needed, not only for disease control, but also for the sustainable development of the aquaculture industry.
PCR assay for detection of bacterial virulence genes is useful, rapid and efficient. The tox R gene, which mainly gets involved in the regulation of many other genes, such as bacterial persistence, biofilm formation and virulence, has been detected ubiquitously in V. parahaemolyticus. Letchumanan et al. (2015b) discovered that 57.8% (185/320) V. parahaemolyticus isolates carried tox R gene. Kang et al. (2017) detected 31 of the 44 isolates were positive V. parahaemolyticus strains for tox R gene. Thus, it is necessary and urgent to obtain a deeper understanding of the regulatory mechanism of tox R serving as activator of lethality and enterotoxicity. It is well-known that V. parahaemolyticus, especially clinical strains, expressed TDH and TRH encoded by hemolysin genes (tdh and trh) are believed to induce inflammatory gastroenteritis rapidly (Mahoney et al., 2010; Ceccarelli et al., 2013; Raghunath, 2015; Chen et al., 2018). Thus, detecting the hemolysin genes through PCR assay could be primary but efficient method to infer the virulence potential of food derived isolates. In our study, 9.90 and 19.80% of the strains were positive for tdh and trh gene indicating the expression of TDH and TRH exist. Our findings were more serious than those reported previously on V. parahaemolyticus isolates from fish, shrimp and RTE food (Xie et al., 2016). The evolution of tdh or trh isolates may be affected by environmental factors, including interaction with other hosts (Wilson and Salyers, 2003). The evolution of tdh or trh isolates, affected by environmental factors and interacted with other hosts, represents a possible risk to public health. Therefore, monitoring of V. parahaemolyticus pathogenic factors are important to protect aquatic products and RTE food in sales chain and improve food safety in the industry.
O antigen, one of three distinct regions, is an important component of lipopolysaccharide, such as Escherichia coli and Salmonella enterica (Samuel et al., 2004). Depending on external factors, V. parahaemolyticus can produce a capsule with the variability of the O antigen and it was believed as a primary information of bacterial isolates classification. Our data indicated that the serovar O2 (49.01%) was the predominant serogroup in fish (50.00%), shrimp (51.22%), and RTE food (26.67%). Such finding was not in accordance with a previous study that identified the O3 serogroup as the predominant serogroup contaminating shellfish in the eastern coast of China (Zhao et al., 2011). The next highest prevalent was O1 serogroup (22.77%), which may become an epidemic strain. Monitoring serogroup variation could be an effective in improving our understanding of V. parahaemolyticus isolates that cause food poisoning. Besides, The O antigen of Vibrio sp., as a receptor, located on the bacterial surface and made host a major target of specific phages (Seed et al., 2012; Xu et al., 2013). The prevention and control of multi-drug resistant bacteria by phages targeting Vibrio parahaemolyticus O serogroup may be a novel and efficient strategy.
Recently, molecular subtyping has been increasingly used for the analysis of genetic diversity. The MLST method, commonly considered to be molecular typing, can be classified in two basic strategies. In this study, one of these was employed, that relied on allele and ST determination to estimate relatedness among 108 V. parahaemolyticus isolates. On one hand, MLST data can contribute to species discrimination, as it provides both genealogical information and information on recombination, which is critical to V. parahaemolyticus identification. The results showed 81 STs, of which 41 (50.62%) STs, composed of 46 strains, were newly identified and revealed a high degree of diversity among the strains tested. A number of reports have shown similar results. Urmersbach et al. (2014) reported that 130 V. parahaemolyticus strains obtained from a marine environment were classified into 82 STs, and 82.9% of them had not been recorded in the pubMLST database previously. Besides, Lopatek et al. (2018) identified 51 novel STs in the isolates from different species of seafood available on the Polish market that originating from various countries. Moreover, a total of 68 STs were identified in the 90 V. parahaemolyticus isolates, and 41 (60.3%) of them were considered novel (Jiang et al., 2019). Both our study and the research conducted previously have revealed how poorly the current pubMLST data set represents the diversity within V. parahaemolyticus. Therefore, efficient strain identification and accurate information presented in the public database are essential to understand the processes of transmission, perform epidemiological surveillance and subsequently the design of public health control strategies (Comas et al., 2009).
Diarrhea caused by the food-borne pathogen, V. parahaemolyticus, has been a long-standing problem. In summary, this is a comprehensive study that describes the prevalence, serogroup, virulence genes, antibiotic resistant phenotype, molecular classification, and genetic diversity of V. parahaemolyticus detected in aquatic products and RTE foods in China in summer and winter. Significantly, the prevalence of V. parahaemolyticus was inconsistent between summer (14.87%) and winter (7.26%). This study shows the percentage of the isolates that possess the tdh and trh genes, being 9.90 and 19.80%, while there were eight of 202 isolates carried both genes, respectively. More seriously, all isolates were tested positive for tox R gene. In addition, O2 serogroup was found to be the most prevalent of V. parahaemolyticus isolated mainly from seven cities. Moreover, V. parahaemolyticus isolates exhibited resistance to ampicillin, cephalothin and streptomycin were widespread. Lastly, both MLST phylogenetic analysis and MST, basing on 41 novel STs and 40 reported STs, showed large genetic diversity of V. parahaemolyticus tested. As aquatic products and RTE foods consist of popular food choices in China, our findings may be useful in guiding appropriate monitoring strategy, improving the understanding of antimicrobial susceptibility patterns, and providing information for the assessment of exposure to V. parahaemolyticus during food consumption, which are vital to ensure the safety of such products and safeguard human health.
All datasets generated for this study are included in the article/Supplementary Material.
QW, YL, TX, RP, and JZ conceived and designed the experiments. YL, YZ, and TX performed the experiments. RP, TL, HZ, HW, and LX analyzed the data. JW, YD, MC, and SW contributed reagents, materials, and analysis tools. YL, TX, QW, and XW contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
We would like to acknowledge the financing support of National Natural Science Foundation of China (No. 3173070), Natural Science Foundation of Guangdong Province (2019A1515010810), and GDAS’ Project of Science and Technology Development (2019GDSYL-020101).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01670/full#supplementary-material
Aarestrup, F. M., and Wegener, H. C. (1999). The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1, 639–644. doi: 10.1016/s1286-4579(99)80064-1
Akther, F., Neogi, S. B., Chowdhury, W. B., Sadique, A., Islam, A., Akhter, M. Z., et al. (2016). Major tdh(+)Vibrio parahaemolyticus serotype changes temporally in the Bay of Bengal estuary of Bangladesh. Infect. Genet. Evol. 41, 153–159. doi: 10.1016/j.meegid.2016.04.003
Arakawa, E., Murase, T., Shimada, T., Yamai, S., and Watanabe, H. (1999). Emergence and prevalence of a novel Vibrio parahaemolyticus O3:K6 clone in Japan. Jpn. J. Infect. Dis. 52, 246–247.
Baker-Austin, C., Stockley, L., Rangdale, R., and Martinez-Urtaza, J. (2010). Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2, 7–18. doi: 10.1111/j.1758-2229.2009.00096.x
Cabello, F. C. (2006). Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8, 1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dolz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Caburlotto, G., Suffredini, E., Toson, M., Fasolato, L., Antonetti, P., Zambon, M., et al. (2016). Occurrence and molecular characterisation of Vibrio parahaemolyticus in crustaceans commercialised in Venice area, Italy. Int. J. Food Microbiol. 220, 39–49. doi: 10.1016/j.ijfoodmicro.2015.12.007
Cai, Q., and Zhang, Y. (2018). Structure, function and regulation of the thermostable direct hemolysin (TDH) in pandemic Vibrio parahaemolyticus. Microb. Pathog. 123, 242–245. doi: 10.1016/j.micpath.2018.07.021
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Chen, M., Guo, D., Wong, H. C., Zhang, X., Liu, F. X., Chen, H. Y., et al. (2012). Development of O-serogroup specific PCR assay for detection and identification of Vibrio parahaemolyticus. Int. J. Food Microbiol. 159, 122–129. doi: 10.1016/j.ijfoodmicro.2012.08.012
Chen, X., Zhu, Q. Y., Yu, F., Zhang, W., Wang, R. N., Ye, X. F., et al. (2018). Serology, virulence and molecular characteristics of Vibrio parahaemolyticus isolated from seafood in Zhejiang province. PLoS One 13:e0204892. doi: 10.1371/journal.pone.0204892
Chen, Y., Frazzitta, A. E., Litvintseva, A. P., Fang, C., Mitchell, T. G., Springer, D. J., et al. (2015). Next generation multilocus sequence typing (NGMLST) and the analytical software program MLSTEZ enable efficient, cost-effective, high-throughput, multilocus sequencing typing. Fungal Genet. Biol. 75, 64–71. doi: 10.1016/j.fgb.2015.01.005
Chen, Y., and Perfect, J. R. (2017). Efficient, cost-effective, high-throughput, multilocus sequencing typing (MLST) method, NGMLST, and the analytical software program MLSTEZ. Methods Mol. Biol. 1492, 197–202. doi: 10.1007/978-1-4939-6442-0_14
Cho, T. J., Kim, N. H., Kim, S. A., Song, J. H., and Rhee, M. S. (2016). Survival of foodborne pathogens (Escherichia coli O157:H7, Salmonella Typhimurium, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus) in raw ready-to-eat crab marinated in soy sauce. Int. J. Food Microbiol. 238, 50–55. doi: 10.1016/j.ijfoodmicro.2016.08.041
Comas, I., Homolka, S., Niemann, S., and Gagneux, S. (2009). Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. doi: 10.1371/journal.pone.0007815
Dong, X., Li, Z., Wang, X., Zhou, M., Lin, L., Zhou, Y., et al. (2016). Characteristics of Vibrio parahaemolyticus isolates obtained from crayfish (Procambarus clarkii) in freshwater. Int. J. Food Microbiol. 238, 132–138. doi: 10.1016/j.ijfoodmicro.2016.09.004
Elmahdi, S., Dasilva, L. V., and Parveen, S. (2016). Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol. 57, 128–134. doi: 10.1016/j.fm.2016.02.008
Fattel, L., Panossian, B., Salloum, T., Abboud, E., and Tokajian, S. (2019). Genomic features of Vibrio parahaemolyticus from Lebanon and comparison to globally diverse strains by whole-genome sequencing. Foodborne Pathog. Dis. 16, 778–787. doi: 10.1089/fpd.2018.2618
Food and Drug Administration [FDA] (2018). FDA Investigated Multistate Outbreak of Vibrio parahaemolyticus Linked to Fresh Crab Meat Imported from Venezuela. Available online at: https://www.fda.gov/food/outbreaks-foodborne-illness/fda-investigated-multistate-outbreak-vibrio-parahaemolyticus-linked-fresh-crab-meat-imported
Ghenem, L., and Elhadi, N. (2018). Isolation, molecular characterization, and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from coastal water in the Eastern Province of Saudi Arabia. J. Water Health 16, 57–69. doi: 10.2166/wh.2017.361
Guin, S., Saravanan, M., Anjay, Chowdhury, G., Pazhani, G. P., Ramamurthy, T., et al. (2019). Pathogenic Vibrio parahaemolyticus indiarrhoeal patients, fish and aquatic environments and their potential for inter-source transmission. Heliyon 5:e01743. doi: 10.1016/j.heliyon.2019.e01743
Guo, S., Lin, D., Wang, L. L., and Hu, H. (2018). Monitoring the results of foodborne diseases in sentinel hospitals in Wenzhou City, China from 2014 to 2015. Iran. J. Public Health 47, 674–681.
Harismendy, O., Ng, P. C., Strausberg, R. L., Wang, X. Y., Stockwell, T. B., Beeson, K. Y., et al. (2009). Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 10:R32.
Hu, Q. X., and Chen, L. M. (2016). Virulence and antibiotic and heavy metal resistance of Vibrio parahaemolyticus isolated from crustaceans and shellfish in Shanghai, China. J. Food Protect. 79, 1371–1377. doi: 10.4315/0362-028x.jfp-16-031
Hubbard, T. P., Chao, M. C., Abel, S., Blondel, C. J., Wiesch, P. A., Zhou, X., et al. (2016). Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc. Natl. Acad. Sci. U.S.A. 113, 6283–6288. doi: 10.1073/pnas.1601718113
Iguchi, T., Kondo, S., and Hisatsune, K. (1995). Vibrio parahaemolyticus O-serotypes from O1 to O13 all produce R-type Lipopolysaccharide – Sds-page and compositional sugar analysis. FEMS Microbiol. Lett. 130, 287–292. doi: 10.1111/j.1574-6968.1995.tb07733.x
Jiang, Y., Chu, Y., Xie, G., Li, F., Wang, L., Huang, J., et al. (2019). Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 290, 116–124. doi: 10.1016/j.ijfoodmicro.2018.10.005
Jones, J. L., Ludeke, C. H., Bowers, J. C., Garrett, N., Fischer, M., Parsons, M. B., et al. (2012). Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50, 2343–2352. doi: 10.1128/jcm.00196-12
Kang, C. H., Shin, Y., Jang, S., Yu, H., Kim, S., An, S., et al. (2017). Characterization of Vibrio parahaemolyticus isolated from oysters in Korea: resistance to various antibiotics and prevalence of virulence genes. Mar. Pollut. Bull. 118, 261–266. doi: 10.1016/j.marpolbul.2017.02.070
Lee, L. H., Ab Mutalib, N. S., Law, J. W., Wong, S. H., and Letchumanan, V. (2018). Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front. Microbiol. 9:2513. doi: 10.3389/fmicb.2018.02513
Leoni, F., Talevi, G., Masini, L., Ottaviani, D., and Rocchegiani, E. (2016). Trh (tdh-/trh+) gene analysis of clinical, environmental and food isolates of Vibrio parahaemolyticus as a tool for investigating pathogenicity. Int. J. Food Microbiol. 225, 43–53. doi: 10.1016/j.ijfoodmicro.2016.02.016
Letchumanan, V., Pusparajah, P., Tan, L. T. H., Yin, W. F., Lee, L. H., and Chan, K. G. (2015a). Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015b). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Li, P., Xin, W. W., Xia, S. S., Luo, Y., Chen, Z. W., Jin, D. Z., et al. (2018). MALDI-TOF mass spectrometry-based serotyping of V-parahaemolyticus isolated from the Zhejiang province of China. BMC Microbiol. 18:185. doi: 10.1186/s12866-018-1328-z
Lin, Z., Kumagai, K., Baba, K., Mekalanos, J. J., and Nishibuchi, M. (1993). Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J. Bacteriol. 175, 3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993
Lopatek, M., Wieczorek, K., and Osek, J. (2018). Antimicrobial resistance, virulence factors, and genetic profiles of Vibrio parahaemolyticus from Seafood. Appl. Environ. Microbiol. 84:e00537-18.
Mahoney, J. C., Gerding, M. J., Jones, S. H., and Whistler, C. A. (2010). Comparison of the pathogenic potentials of environmental and clinical Vibrio parahaemolyticus strains indicates a role for temperature regulation in virulence. Appl. Environ. Microbiol. 76, 7459–7465. doi: 10.1128/aem.01450-10
Mala, W., Alam, M., Angkititrakul, S., Wongwajana, S., Lulitanond, V., Huttayananont, S., et al. (2016). Serogroup, virulence, and molecular traits of Vibrio parahaemolyticus isolated from clinical and cockle sources in northeastern Thailand. Infect. Genet. Evol. 39, 212–218. doi: 10.1016/j.meegid.2016.01.006
Matsuda, S., Kodama, T., Okada, N., Okayama, K., Honda, T., and Iida, T. (2010). Association of Vibrio parahaemolyticus thermostable direct hemolysin with lipid rafts is essential for cytotoxicity but not hemolytic activity. Infect. Immun. 78, 603–610. doi: 10.1128/iai.00946-09
Mazel, D., and Davies, J. (1999). Antibiotic resistance in microbes. Cell Mol. Life Sci. 56, 742–754.
Mohamad, N., Amal, M. N. A., Saad, M. Z., Yasin, I. S. M., Zulkiply, N. A., Mustafa, M., et al. (2019). Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet. Res. 15:176. doi: 10.1186/s12917-019-1907-8
Pang, R., Xie, T., Wu, Q., Li, Y., Lei, T., Zhang, J., et al. (2019). Comparative genomic analysis reveals the potential risk of Vibrio parahaemolyticus isolated from ready-to-eat foods in China. Front. Microbiol. 10:186. doi: 10.3389/fmicb.2019.00186
Parveen, S., Hettiarachchi, K. A., Bowers, J. C., Jones, J. L., Tamplin, M. L., Mckay, R., et al. (2008). Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int. J. Food Microbiol. 128, 354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019
Perez-Losada, M., Cabezas, P., Castro-Nallar, E., and Crandall, K. A. (2013). Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect. Genet. Evol. 16, 38–53. doi: 10.1016/j.meegid.2013.01.009
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Raszl, S. M., Froelich, B. A., Vieira, C. R., Blackwood, A. D., and Noble, R. T. (2016). Vibrio parahaemolyticus and Vibrio vulnificus in South America: water, seafood and human infections. J. Appl. Microbiol. 121, 1201–1222. doi: 10.1111/jam.13246
Ryu, A. R., Mok, J. S., Lee, D. E., Kwon, J. Y., and Park, K. (2019). Occurrence, virulence, and antimicrobial resistance of Vibrio parahaemolyticus isolated from bivalve shellfish farms along the southern coast of Korea. Environ. Sci. Pollut. Res. Int. 26, 21034–21043. doi: 10.1007/s11356-019-05426-1
Samuel, G., Hogbin, J. P., Wang, L., and Reeves, P. R. (2004). Relationships of the Escherichia coli O157, O111, and O55 O-antigen gene clusters with those of Salmonella enterica and Citrobacter freundii, which express identical O antigens. J. Bacteriol. 186, 6536–6543. doi: 10.1128/jb.186.19.6536-6543.2004
Seed, K. D., Faruque, S. M., Mekalanos, J. J., Calderwood, S. B., and Camilli, A. (2012). Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog. 8:e1002917. doi: 10.1371/journal.ppat.1002917
Shaw, K. S., Sapkota, A. R., Jacobs, J. M., He, X., and Crump, B. C. (2015). Recreational swimmers’ exposure to Vibrio vulnificus and Vibrio parahaemolyticus in the Chesapeake Bay, Maryland, USA. Environ. Int. 74, 99–105. doi: 10.1016/j.envint.2014.09.016
Shirai, H., Ito, H., Hirayama, T., Nakamoto, Y., Nakabayashi, N., Kumagai, K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990
Tan, C. W., Malcolm, T. T. H., Kuan, C. H., Thung, T. Y., Chang, W. S., Loo, Y. Y., et al. (2017). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from short mackerels (Rastrelliger brachysoma) in Malaysia. Front. Microbiol. 8:1087. doi: 10.3389/fmicb.2017.01087
Urmersbach, S., Alter, T., Koralage, M. S. G., Sperling, L., Gerdts, G., Messelhäusser, U., et al. (2014). Population analysis of Vibrio parahaemolyticus originating from different geographical regions demonstrates a high genetic diversity. BMC Microbiol. 14:59. doi: 10.1186/1471-2180-14-59
West, C. K. G., Klein, S. L., and Lovell, C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79, 2247–2252. doi: 10.1128/aem.03792-12
Whitaker, W. B., Parent, M. A., Boyd, A., Richards, G. P., and Boyd, E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80, 1834–1845. doi: 10.1128/iai.06284-11
Wilson, B. A., and Salyers, A. A. (2003). Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 11, 347–350. doi: 10.1016/s0966-842x(03)00179-3
Xie, T., Wu, Q., Xu, X., Zhang, J., and Guo, W. (2015). Prevalence and population analysis of Vibrio parahaemolyticus in aquatic products from South China markets. FEMS Microbiol. Lett. 362:fnv178. doi: 10.1093/femsle/fnv178
Xie, T., Xu, X., Wu, Q., Zhang, J., and Cheng, J. (2016). Prevalence, molecular characterization, and antibiotic susceptibility of Vibrio parahaemolyticus from ready-to-eat foods in China. Front. Microbiol. 7:549. doi: 10.3389/fmicb.2016.00549
Xie, T. F., Wu, Q. P., Zhang, J. M., Xu, X. K., and Cheng, J. H. (2017). Comparison of Vibrio parahaemolyticus isolates from aquatic products and clinical by antibiotic susceptibility, virulence, and molecular characterisation. Food Control 71, 315–321. doi: 10.1016/j.foodcont.2016.06.046
Xu, F., Gonzalez-Escalona, N., Haendiges, J., Myers, R. A., Ferguson, J., Stiles, T., et al. (2017). Sequence type 631 Vibrio parahaemolyticus, an emerging foodborne pathogen in North America. J. Clin. Microbiol. 55, 645–648. doi: 10.1128/jcm.02162-16
Xu, J., Zhang, J., Lu, X., Liang, W., Zhang, L., Kan, B., et al. (2013). O antigen is the receptor of Vibrio cholerae serogroup O1 El Tor typing phage VP4. J. Bacteriol. 195, 798–806. doi: 10.1128/jb.01770-12
Xu, X., Cheng, J., Wu, Q., Zhang, J., and Xie, T. (2016). Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol. 16:32. doi: 10.1186/s12866-016-0650-6
Zhang, H. N., Hou, P. B., Chen, Y. Z., Ma, Y., Li, X. P., Lv, H., et al. (2016). Prevalence of foodborne pathogens in cooked meat and seafood from 2010 to 2013 in Shandong Province, China. Iran. J. Public Health 45, 1577–1585.
Zhang, Y., Hu, L., Osei-Adjei, G., Zhang, Y., Yang, W., Yin, Z., et al. (2018). Autoregulation of ToxR and its regulatory actions on major virulence gene Loci in Vibrio parahaemolyticus. Front. Cell Infect. Microbiol. 8:291. doi: 10.3389/fcimb.2018.00291
Zhang, Z. H., Lou, Y., Du, S. P., Xiao, L. L., Niu, B., Pan, Y. J., et al. (2017). Prevalence of Vibrio parahaemolyticus in seafood products from hypermarkets in Shanghai. J. Sci. Food Agric. 97, 705–710. doi: 10.1002/jsfa.7715
Keywords: Vibrio parahaemolyticus, prevalent, antibiotic resistance, serogroup, virulence gene, MLST
Citation: Li Y, Xie T, Pang R, Wu Q, Zhang J, Lei T, Xue L, Wu H, Wang J, Ding Y, Chen M, Wu S, Zeng H, Zhang Y and Wei X (2020) Food-Borne Vibrio parahaemolyticus in China: Prevalence, Antibiotic Susceptibility, and Genetic Characterization. Front. Microbiol. 11:1670. doi: 10.3389/fmicb.2020.01670
Received: 11 February 2020; Accepted: 25 June 2020;
Published: 16 July 2020.
Edited by:
Daniela Ceccarelli, European Commission, BelgiumReviewed by:
Etinosa Igbinosa, University of Benin, NigeriaCopyright © 2020 Li, Xie, Pang, Wu, Zhang, Lei, Xue, Wu, Wang, Ding, Chen, Wu, Zeng, Zhang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t
†These authors have contributed equally to this work as co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.