- 1Department of Clinical Laboratory, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China
- 2Department of Clinical Laboratory, Hunan Provincial People’s Hospital, The First Affiliated Hospital of Hunan Normal University, Changsha, China
- 3Beijing Key Laboratory of Emerging Infectious Diseases, Institute of Infectious Diseases, Beijing Ditan Hospital, Capital Medical University, Beijing, China
- 4Graduate School, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China
- 5School of Biomedical Sciences, Charles Sturt University, Orange, NSW, Australia
This study was to investigate the prevalence of mcr-1-positive Enterobacteriaceae (MPE) in intra-abdominal infections (IAIs), urinary tract infections (UTIs), and lower respiratory tract infections (LRTIs) in China. A total of 6,401 Enterobacteriaceae isolates were collected consecutively from IAI, UTI, and LRTI patients in 19 hospitals across mainland China during 2014–2016. MPE isolates were screened by PCR detection for the mcr gene. The resistance profiles were tested by antimicrobial susceptibility test. All MPE isolates were characterized by pulsed-field gel electrophoresis (PFGE), multi-locus-sequence typing, O and H serotyping, and whole-genome sequencing. Among the 6,401 Enterobacteriaceae isolates, 17 Escherichia coli strains (0.27%) were positive for the mcr-1 gene. The MPE prevalence rates in IAI, UTI, and LRTI patients were 0.34% (12/3502), 0.23% (5/2154), and 0% (0/745), respectively. The minimum inhibition concentrations (MICs) of colistin against 3 isolates were of 0.5–2 mg/L, and 4–8 mg/L against other 14 isolates. All the 17 isolates were susceptible to meropenem, imipenem, tigecycline, and ceftazidime/avibactam. The 17 MPE isolates belonged to 14 different ST types, and those that belonged to the same STs were not clonal by PFGE. The mcr-1-harboring plasmid of ten MPE isolates could transfer to the recipients by conjugation and the colistin MICs of the transconjugants ranged from 0.5 to 8 mg/L. Mcr-1-carrying plasmids from the 17 MPE isolates could be grouped into four clusters, including 8 IncX4 type, 4 IncI2 type, 4 IncHI2A type, and 1 p0111 type. Multiple-drug resistance genes and virulence genes were detected. In conclusion, the prevalence of MPE in IAI, UTI, and LRTI were low in China, and no clonal transmission was identified in our study. Most MPE isolates exhibited low-level colistin resistance. However, our study indicated that MPE isolates always carried a variety of drug resistance and virulence genes, which should be paid more attention.
Introduction
In the past few decades, multidrug-resistant (MDR) Gram-negative bacilli (mainly Escherichia coli and Klebsiella pneumoniae) increasingly caused infections and spread rapidly, which posed a major threat to global health (Mathers et al., 2015; Potter et al., 2016; Miao et al., 2018). Therefore, colistin is inevitably used in the clinic setting and has been regarded as the last-resort agent against MDR Enterobacteriaceae (Li et al., 2017). The mcr-1 gene was firstly discovered in animals as early as 1980, but few researches have been performed to study its functions and characteristics (Shen et al., 2016). Liu et al. (2016) firstly reported that the mcr-1 gene was related to the colistin resistance in Enterobacteriaceae isolates from animals and humans. They found that not only is the resistance mechanism of polymyxin caused by the chromosomal mutations but also the mcr-1 gene is commonly carried by plasmids. The plasmid-harboring mcr-1 could be horizontally transferred among Enterobacteriaceae. The mcr-1-positive Enterobacteriaceae (MPE) became a high threat to break the last line of defense against the MDR Enterobacteriaceae.
This novel finding caused widespread concern around the world (Liu et al., 2016). Subsequently, South America, Asia, Europe, Africa, and other countries have described mcr-1 in Enterobacteriaceae strains isolated from animals, foods, water, humans, and other ecosystems (Li B. et al., 2018). The gene may be transmitted to human beings through direct contact via the food chain or between humans and animals (Chen et al., 2017). A report from Bolivia described a very high rate (38.3%) of mcr-1 gene carriage in isolates from healthy children (7–11 years old) in rural areas with no sewage treatment systems and where the economy is dominated by subsistence agriculture and animal husbandry (Giani et al., 2018). This illustrates the wide spread of the mcr-1 gene through food, animals, and the environment, to the population. It is understood that the mcr-1 gene has been detected in humans, animals, food, and the environment in more than 30 countries on five continents (Soo et al., 2019). In addition, multiple subtypes of mcr genes, including mcr-1 to mcr-10, have been identified recently (Anyanwu et al., 2020; Wang et al., 2020). The prevalent subtypes in China were mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, and mcr-7 (Nang et al., 2019). Although multiple studies have been performed to investigate the prevalence of MPE in the breeding industry, few were conducted in hospital and human community.
In this study, we performed a multicenter study in China to describe the epidemiology of MPE isolates from patients of intra-abdominal infection (IAI), urinary tract infection (UTI), and lower respiratory tract infection (LRTI). We screened the mcr gene among over six thousand non-repetitive Enterobacteriaceae isolates from the three types of patients to uncover the prevalence and performed whole-genome sequencing to study the genomic characteristics of mcr-1-positive isolates.
Materials and Methods
Bacterial Isolates
We enrolled a total of 6,401 non-repetitive, consecutively collected Enterobacteriaceae isolates from patients with IAIs, UTIs, and LRTIs, in 19 tertiary hospitals in China from the years 2014 to 2016. The study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (Number: S-K238). The majority of the intra-abdominal specimens were obtained during surgery, including paracentesis specimens and peritoneal fluid, as well as sample of gall bladder, appendix, abscesses, liver, and pancreas. UTI isolates were mainly obtained from culture of midstream urine, and LRTI isolates were cultured from qualified lower respiratory tract samples, such as sputum and bronchoalveolar lavage fluid. All duplicate isolates (same species from the same patient) were excluded. Isolates were first identified at the local hospitals by Vitek 2 and then shipped to the central clinical microbiology laboratory of the Peking Union Medical College Hospital for further identification using MALDI-TOF MS (Vitek MS, BioMe ìrieux, France).
Multi-Locus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE)
Multi-locus sequence typing was conducted according to the protocol and primers on the MLST website1. PFGE was carried out by using XbaI. DNA fragments were electrophoresed by the CHEF-MAPPER XA PFGE system (Bio-Rad Laboratories, United States) for 19 h at 14°C, 6 V/cm, 120°, and switch from 2.16 to 63.8 s. PFGE images were analyzed by BioNumerics software (Version 7.6, Applied Maths, Belgium).
O and H Serotypes
We used serum agglutination reaction to conduct O antiserum typing. The cooled bacterial suspension was mixed with each of the available O (O1-O180) antisera and observed for agglutination. A bacterial isolate that did not react with all O antisera was classified as non-typeable. H antiserum typing was carried out by PCR and sequencing. The fliC gene was amplified by PCR (primer sequence: fliC-F 5′-ATGGCACAAGTCATTAATACCCAAC-3′; fliC-R: 5′-CTAACCCTGCAGCAG AGACA-3′). The sequences of PCR products were compared with Serotype Finder public database2.
Antimicrobial Susceptibility Test
The antimicrobial susceptibility testing included nine antibiotics (colistin, meropenem, ertapenem, amikacin, levofloxacin, ceftazidime, tigecycline, aztreonam, ceftazidime/avibactam). The MICs of Enterobacteriaceae isolates in this study were determined by standard broth microdilution method. The results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) standard M100-S29 (CLSI, 2019), except for colistin which was interpreted by European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (EUCAST, 2016) and tigecycline by FDA-approved breakpoints. The quality control strains were E. coli ATCC25922 and Pseudomonas aeruginosa ATCC27853.
Screening of the Isolates for mcr-1 and Other Antimicrobial Resistance Genes
All strains were screened for the presence of the mcr-1 gene by PCR as described previously (Liu et al., 2016). The resulting amplicons were verified by Sanger sequencing. We also screened all MPE isolates for the presence of ESBL genes (blaCTX–M, blaSHV, blaTEM), ampC (blaMOX, blaCMY, blaLAT, blaBIL, blaDHA, blaACC, blaMIR, blaFOX, blaACT) and carbapenemase genes (blaIMP, blaVIM, blaNDM, blaAIM, blaSPM, blaKPC, blaDIM, blaBIC, blaGIM, blaSIM, blaOXA–48) by PCR using primer and reaction conditions as previously described (Pérez-Pérez and Hanson, 2002; Queenan and Bush, 2007; Poirel et al., 2011; Hamprecht et al., 2016). The obtained gene sequences were compared with those in the nt database, using the NCBI blast server3.
Conjugation Experiments
Conjugation experiments were carried out to investigate the transferability of the mcr-1 gene in the MPE isolates. The 17 MPE isolates were used as the donors, and the rifampicin-resistant E. coli C600 strain was used as the recipient. The donor and recipient bacteria were incubated to TSB broth and cultured at 35°C for 10–12 h together. The transconjugants were screened on containing colistin (2 μg/ml) and rifampicin (600 μg/ml).
Whole-Genome Sequencing of MPE Isolates and Complete Sequencing of the mcr-1-Carrying Plasmids
The genomic DNA of the MPE isolates was extracted using the Qiagen DNA Mini Kit (Qiagen, Hilden, Germany) and sequenced on the Illumina HiSeq 2500 platform. The genomes were assembled using SPAdes v3.13 by the default parameters (Bankevich et al., 2012). The complete genome sequence of E. coli strain EDL933 was used as the reference to perform phylogenetic analysis. The single-nucleotide polymorphisms (SNPs) were identified using MUMmer (Delcher et al., 2003). The adjacent mutations within 5-bp in an isolate were filtered to avoid recombination. Finally, the concatenated sequences of variant sites (core genome SNPs) conserved in all strains were used to perform phylogenetic analysis using FastTree (Price et al., 2009). The sequences were also determined by the Oxford Nanopore Technologies (ONT) MinION platform and then combined with the Illumina HiSeq 2500 platform-determined sequence. The Illumina read and ONT read were assembled with a hybrid strategy using Unicycler (Wick et al., 2017). Bioinformatic tools were used to analyze these genomes, including ResFinder for resistance genes (Kleinheinz et al., 2014), ISfinder for ISs (Siguier et al., 2006), and PlasmidFinder for plasmid replicon types (Carattoli et al., 2014). Sequence comparisons were performed using BLAST software. The sequences of the plasmids were annotated with Prokka (Seemann, 2014) and edited manually.
Virulence Factor (VF) Determination
The presence of 29 virulence genes associated with extraintestinal pathogenic E. coli was detected by multiplex PCR as described previously (Johnson and Stell, 2000). The tested virulence genes included adhesin VF genes (fimH, papG I, papG II, III, papG allele I, papG allele II, papG allele III, papAH, papC, papEF, sfaS, focG, bmaE, gafD, nfaE, sfa/focDE, afa/draBC), toxin genes (hlyA, cnf1, cdtB), siderophore genes (fyuA, iutA), capsule synthesis-associated VF genes (kpsMT II, kpsMT III, kpsMT K1, kpsMT K5), and miscellaneous VF genes (rfc, ibeA, cvaC, traT, PAI). Pathogenicity island (PAI) from strain CFT073 was used as a PAI marker.
Results
Bacterial Isolates
In total, we collected 3,502 Enterobacteriaceae isolates from IAI patients, 2,154 from UTI patients, and 745 from LRTI patients. The total 6,401 non-repetitive isolates included the following species: E. coli (3598), K. pneumoniae (1840), Enterobacter cloacae (359), Proteus mirabilis (162), Enterobacter aerogenes (124), Klebsiella oxytoca (106), Citrobacter freundii (103), Serratia marcescens (100), and Proteus vulgaris (9). By PCR screening of the mcr-1 gene, we found that 17 isolates (17/6,401, 0.27%) were positive for mcr-1, all of which were E. coli, accounting for 0.47% in this species (17/3,598). Among the 17 MPE isolates, 12 were from IAI patients, 5 from UTI patients, and none from LRTI patients, accounting for the prevalence rates of 0.34% (12/3501), 0.23% (2/2154), and 0% (0/745) among the three types of infection, respectively.
Subtyping of the MPE Isolates and Whole-Genome Sequencing
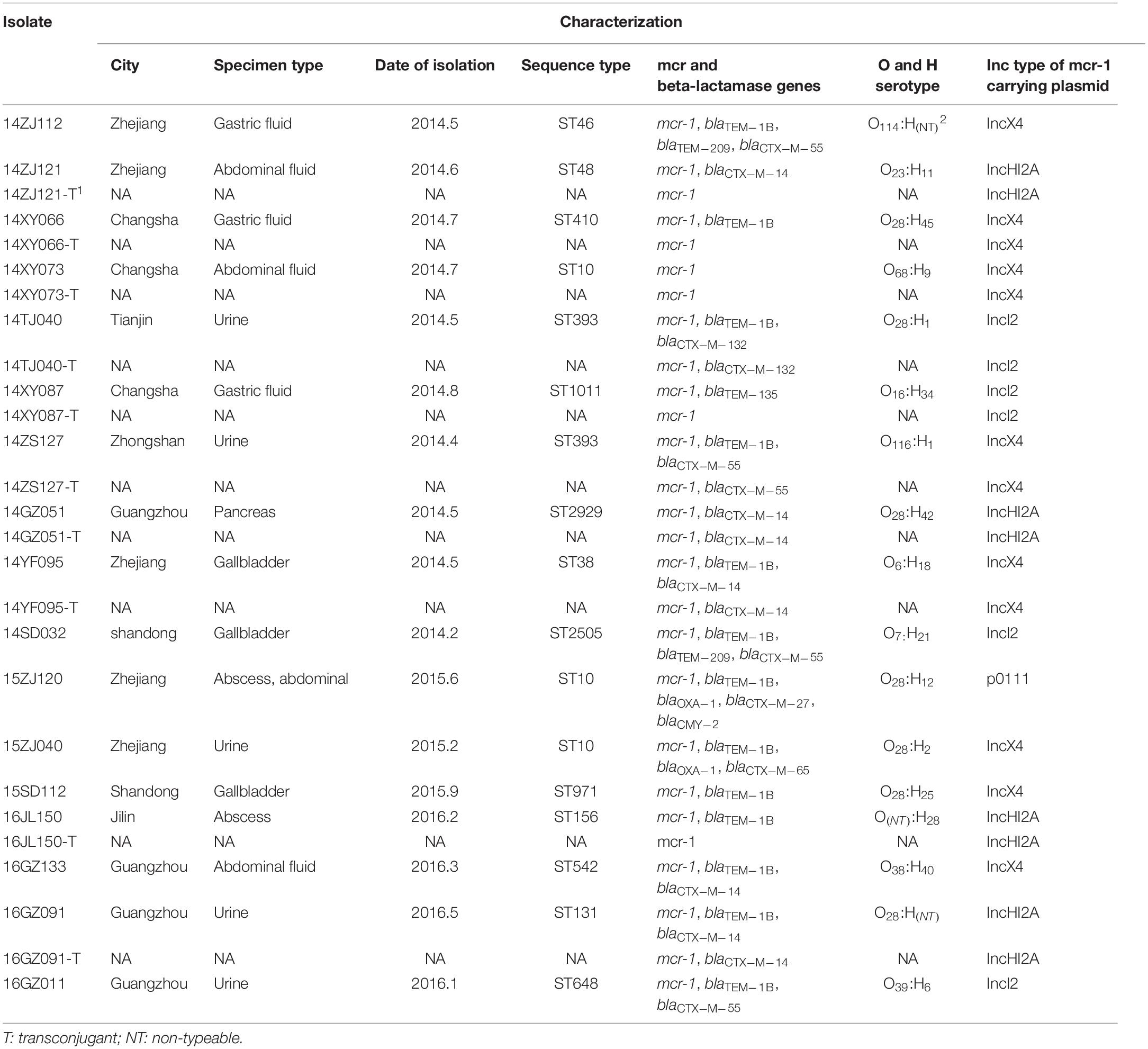
The subtypes of the MPE isolates were analyzed by the PFGE, MLST, and O, H typing. According to the MLST analysis, 17 MPE isolates were grouped into 14 distinct sequence types (STs). Three strains belonged to ST10, two strains belonged to ST393, and the other 12 strains belonged to different types. In the PFGE test, 4 of the 17 MPE strains had no complete bands despite several repeats, and the other 13 strains were divided into 13 different PFGE types (Figure 1). The O and H serotype of 17 isolates included 14 different types and 3 untypable strains.
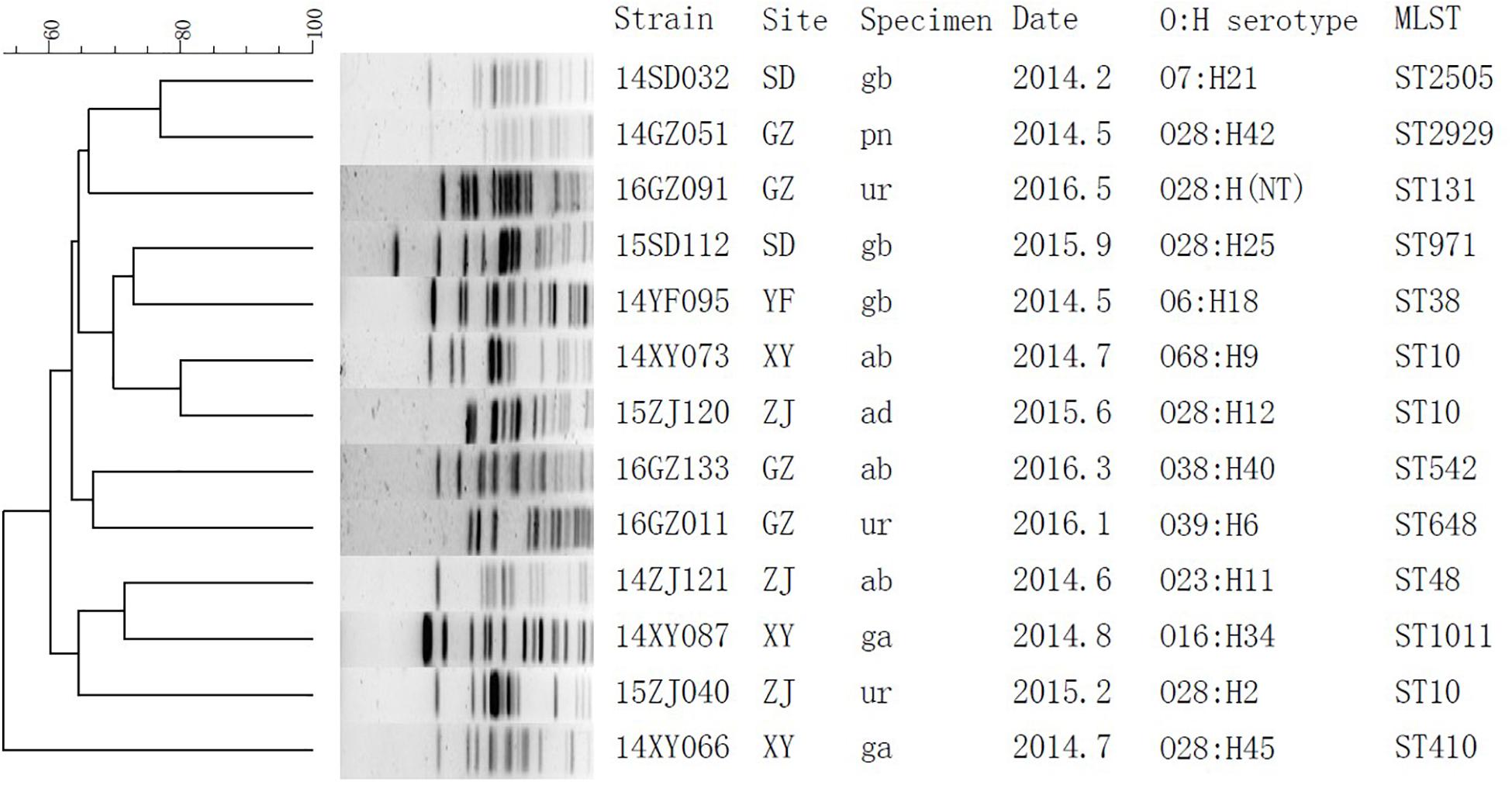
Figure 1. Phylogenetic tree of mcr-1-positive Enterobacteriaceae by PFGE analysis. SD, Shandong; GZ, Guangzhou; YF, Zhejiang; XY, Changsha; ZJ, Zhejiang; gb, gallbladder; pn, pancreas; ur, urine; ab, abdominal fluid; ad, abscess, abdominal; ga, gastric fluid.
By whole-genome sequencing, we obtained 1.77 ± 0.17 Gb Illumina read data for the MPE isolates. We included 329 published genomes of mcr-1-positive E. coli and obtained 525,318 core genome SNPs by the SNP analysis. By the phylogenetic analysis (Supplementary Figure S1), the MPE isolates obtained in this study were clustered with the genomes of the same STs. The SNP numbers between ST393 strain 14ZS127 and 14TJ040 were 340, and those among ST10 strains were 1376 between 15ZJ120 and 15ZJ040, 2872 between 15ZJ120 and 14XY073, and 2914 between 15ZJ040 and 14XY073. Other 12 MPE strains all belonged to other 12 different STs and spread dispersedly in the tree. The results revealed no clonal transmission in the MPE isolates we obtained.
Antimicrobial Susceptibility Test and Resistance Genes Detection
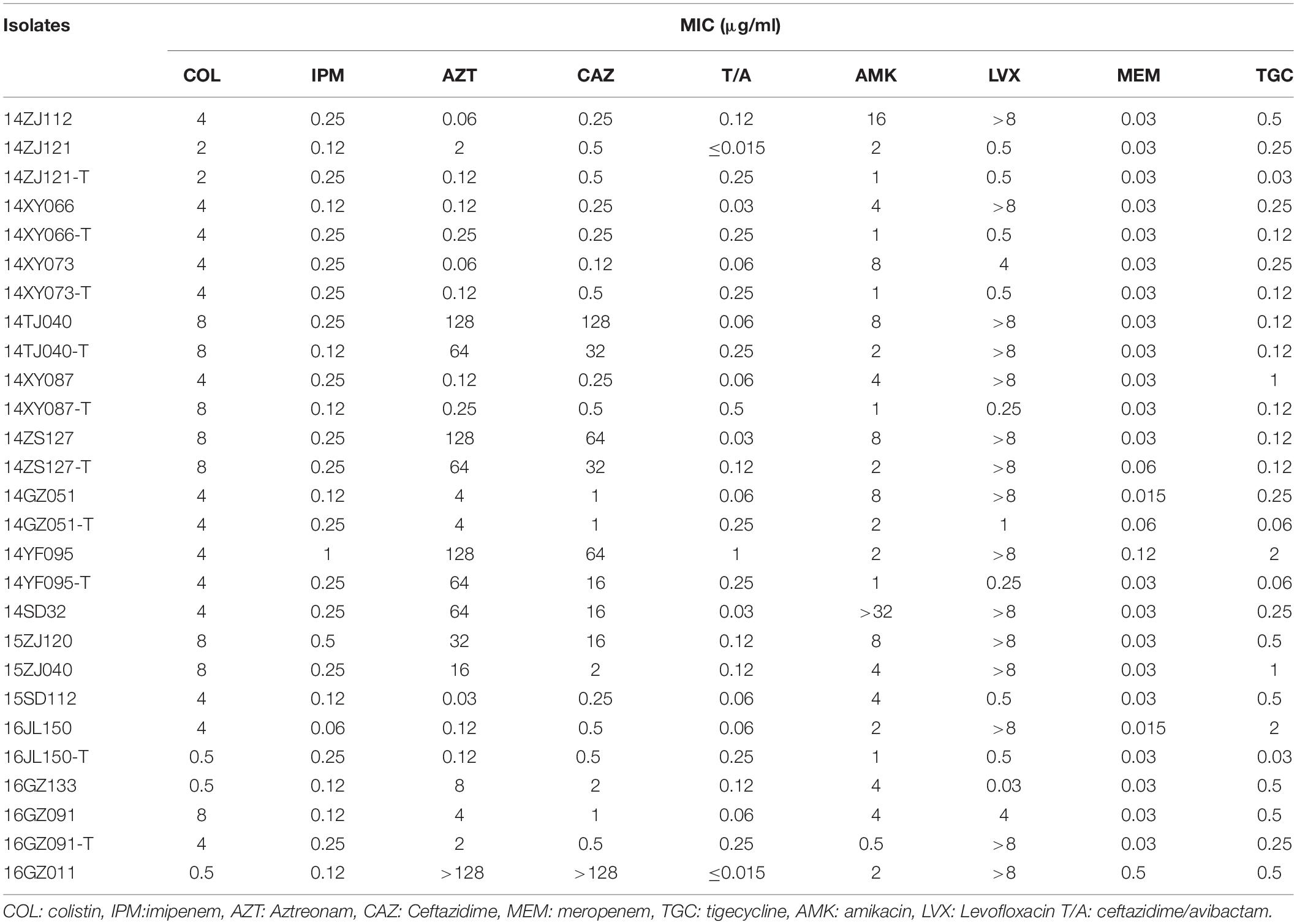
Among the 17 MPE isolates, 14 strains display low-level resistance to colistin (MIC: 4–8 mg/L) and the other 3 strains showed colistin MIC of 0.5–2 mg/L. All MPE isolates were susceptible to meropenem, imipenem, tigecycline, and ceftazidime/avibactam, and only one isolate was non-susceptible to amikacin. Six isolates (35.3%) were resistant to ceftazidime. Seven isolates (41.1%) were resistant to aztreonam. Most of the MPE isolates (82.4%) were resistant to levofloxacin. The detection of resistance genes by PCR revealed that all of the 17 MPE isolates carried multiple antimicrobial resistance genes, including blaTEM–1B (13/17, 76.5%), blaCTX–M–14 (5/17, 29.4%), blaCTX–M–55 (4/17, 23.5%), blaTEM–209 (2/17, 11.8%), blaOXA–1 (2/17, 11.8%), blaCTX–M–132 (1/17, 5.9%), blaCTX–M–65 (1/17, 5.9%), blaTEM–135 (1/17, 5.9%), blaCTX–M–27 (1/17, 5.9%), and blaCMY–2 (1/17, 5.9%) (Table 1).
The mcr-1-Carrying Plasmids and Their Mobility
We obtained the complete sequences of the 17 mcr-1-carrying plasmids from the 17 MPE isolates. Based on the types of origin of replication and sequence similarity of the plasmids, these plasmids could be grouped into four clusters (Figures 2, 3). Eight plasmids belonged to the IncX4 type. Seven of the IncX4 plasmids were highly similar, of which the sizes were 33–34 kb (Figure 2A). In p14YF095-mcr, a 12-kb insertion fragment encoding two copies of blaCTX–M–14-ISEcp1 elements were identified. Four plasmids belonged to the IncI2 type, of which the sizes were 65–67 kb (Figure 2B). Multiple types of origin of replication were identified among other four plasmids, p14GZ051-mcr, p14ZJ121-mcr, p16JL150-mcr, and p16GZ091-mcr, and IncHI2A was the common type of origin of replication in the four plasmids (Figure 3A). The plasmid p15ZJ120-mcr belonged to the p0111 type (Figure 3B).
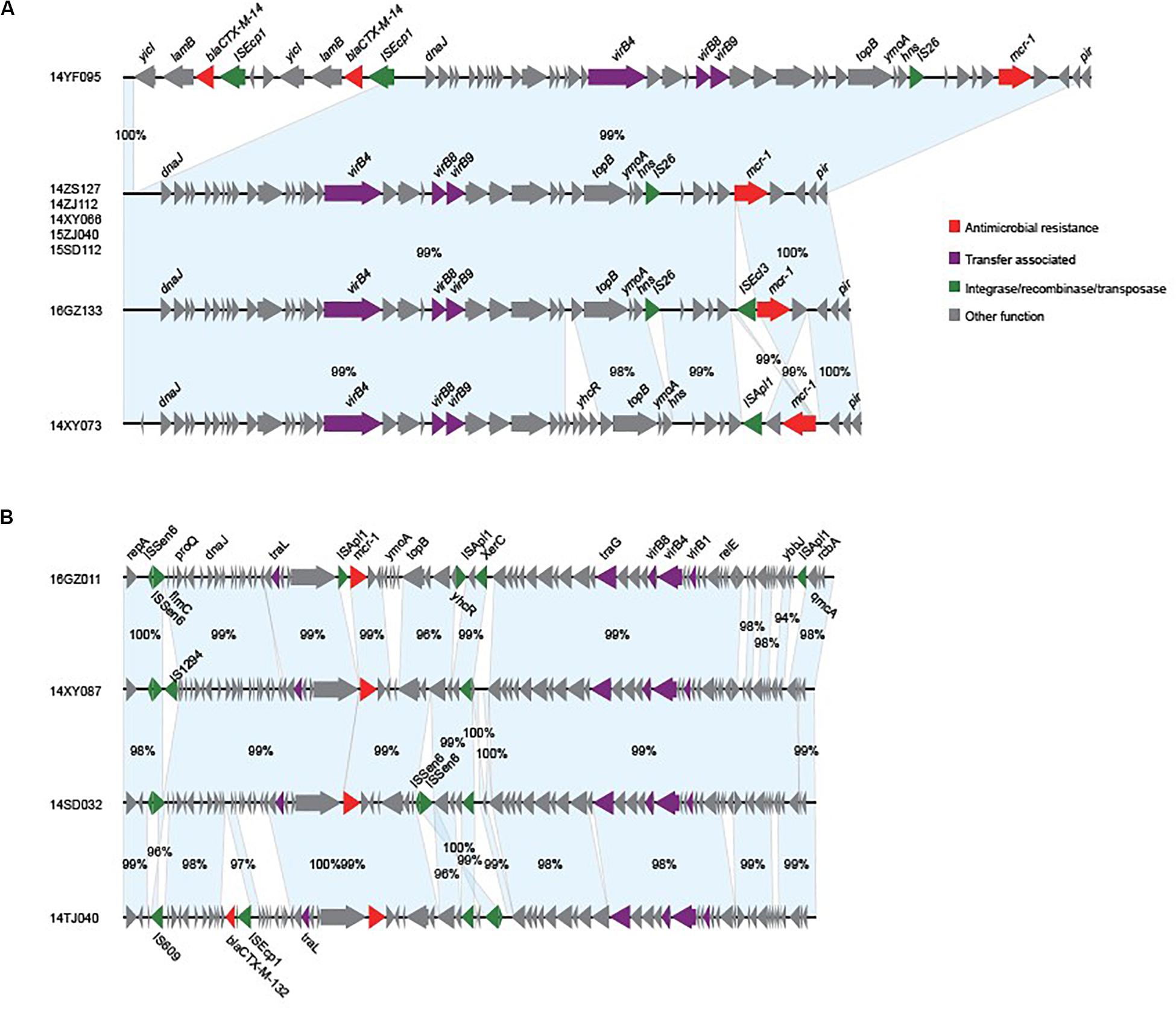
Figure 2. Sequence annotation and alignment of the mcr-1-carrying IncX4 plasmids (A) and IncI2 plasmids (B) obtained in this study. The arrows and triangles represent genes of different functional categories (red: antimicrobial resistance; green: integrase, recombinase, and transposase; purple: transfer associated; gray: other functions). The alignments between two plasmids are represented by light blue blocks, and the sequence identities are marked on the blocks.
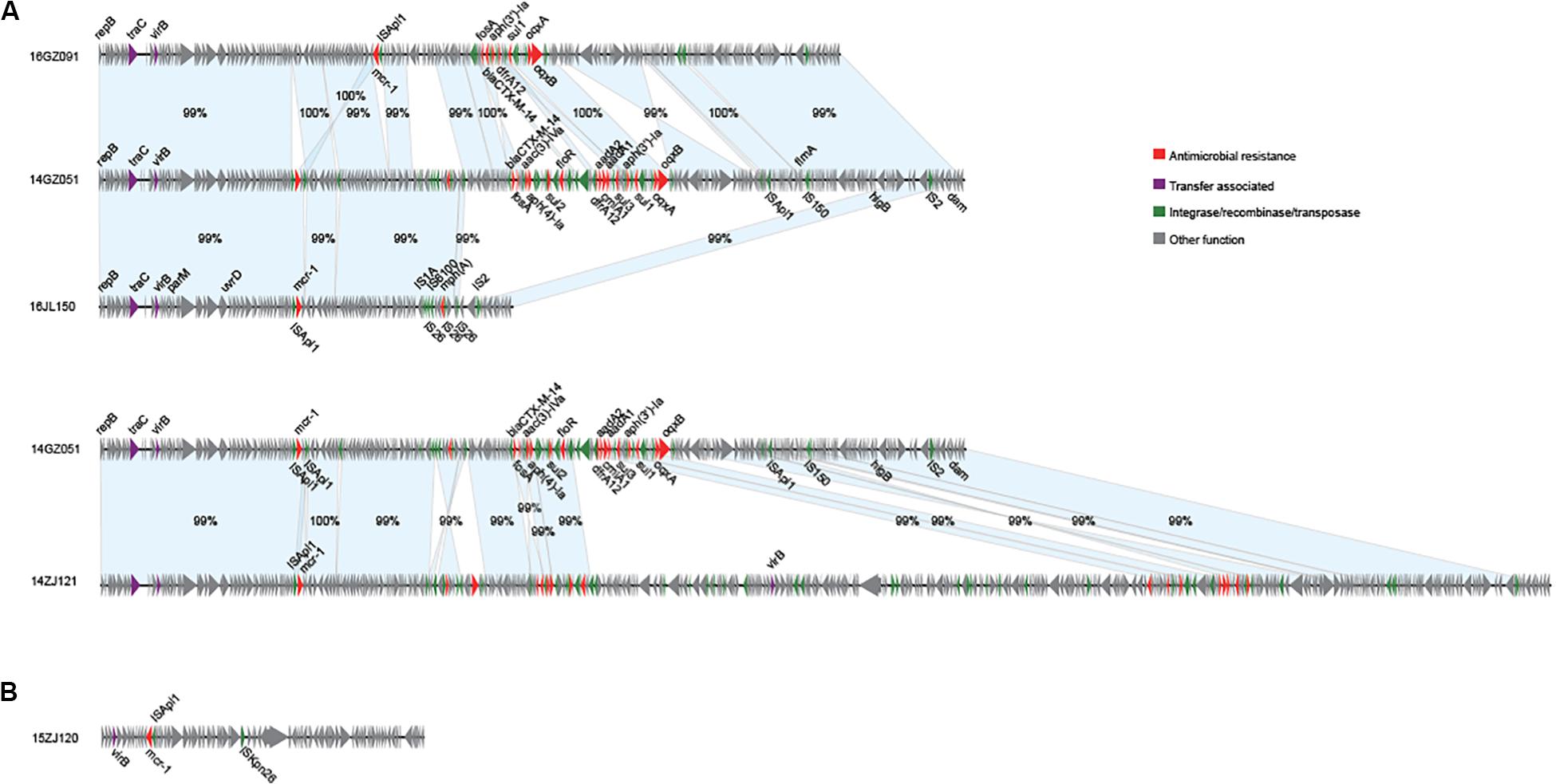
Figure 3. Sequence annotation and alignment of the mcr-1-carrying IncHI2A plasmids (A) and p0111 type plasmid (B) obtained in this study. The arrows and triangles represent genes of different functional categories (red: antimicrobial resistance; green: integrase, recombinase, and transposase; purple: transfer associated; gray: other functions). The alignments between two plasmids are represented by light blue blocks, and the sequence identities are marked on the blocks.
The mcr-1 genes in eight plasmids were identified to be adjacent to IS elements. ISApl1 was the most common element identified in seven plasmids, including an IncX4 plasmid (p14XY073-mcr), an IncI2 plasmid (p16GZ011-mcr), all of the four IncHI2A plasmids, and the p0111 plasmid p15ZJ120-mcr. In plasmid p16GZ133-mcr, the mcr-1 gene was adjacent to ISEcl3. In other plasmids, no IS element was identified adjacent to mcr-1.
The mcr-1 genes of 10 MPE strains were successfully transferred to recipient strain E. coli EC600. The transfer frequencies of the 10 mcr-1-harboring plasmids varied from 10–2 to 10–4. The colistin MIC range of the transconjugants was 0.5–8 mg/L (Table 2).
Virulence Gene Detection
Seventeen MPE isolates carried diverse virulence genes. The most prevalent gene was fimH (16/17, 94.1%), followed by papA (11/17, 64.7%), kpsMT III (10/17, 58.8%), papG allele III (8/17,47.7%), traT (7/17, 41.2%), papEF (6/17, 35.3%), kpsMT K1 (5/17, 29.4%), kpsMT II (4/17, 23.5%), and bmaE (4/17, 23.5%). The remaining 20 virulence genes had a positive rate of less than 20% (Table 3).
Discussion
The mcr-1 gene has been mainly detected in Enterobacteriaceae including E. coli, K. pneumoniae, E. cloacae, E. aerogenes, and Salmonella enterica (Gao et al., 2016). E. coli is the predominant species among MPE (Chen et al., 2017). At present, several countries have detected the mcr-1 gene in Enterobacteriaceae, mainly in food, animals, healthy people, and the environment (Hasman et al., 2015; Fernandes et al., 2016; Kieffer et al., 2017). However, the isolation rate is low in hospitalized patients (0.1–1.0%) (Prim et al., 2016; He et al., 2017; Quan et al., 2017; Wang et al., 2017; Lu et al., 2018). This study investigated the prevalence of the mcr-1 gene in patients with intra-abdominal, urinary tract, and respiratory infections in China from 19 hospitals. Our findings point to a low prevalence (17/6401, 0.27%) of mcr-1-positive isolates among Enterobacteriaceae strains. IAI, UTI, and LRTI were the common infections in the hospital. Our results might reveal a low prevalence of infections caused by MPE in China.
Multi-locus sequence typing analysis showed that 17 MPE isolates belonged to 14 different types, and PFGE and serotyping by O and H antigens both showed high diversity. Among the three ST10 strains, two were isolated from different departments of the same hospital in Zhejiang province 4 months apart (15ZJ120 and 15ZJ040), the genomes of which were of 1376 SNPs. In addition, their PFGE patterns were also of obvious difference; therefore, they were not regarded to be clonal. The two ST393 strains, 14ZS127 and 14TJ040, were from different hospitals. One MPE strain isolated from a urinary tract infection belonged to ST131 which is one of the most important types of extraintestinal infection and has long been recognized as a global threat to human health, often associated with human urinary tract and blood infections (Hasman et al., 2015; Mathers et al., 2015; Shen et al., 2018).
Although most of the MPE strains described to date are resistant to colistin, the present isolates exhibited low-level colistin resistance (MIC: 2–8 μg/ml), and three of them were sensitive to colistin. This is consistent with previous studies showing that mcr-1-carrying E. coli are not always resistant to colistin (Quan et al., 2017; Li B. et al., 2018). This might be caused by the different mcr-1 expression level in these isolates. Among these phylogenetically diverse strains, we identified IncX4 (8 strains) and IncI2 (four strains) types of mcr-1-carrying plasmids with high similarities, respectively, which were the prevalent mcr-1-carrying plasmids (Li et al., 2017; Matamoros et al., 2017; Quan et al., 2017). These indicated that these strains might obtain the plasmids by horizontal gene transfer recently. Therefore, the costs of plasmid fitness might cause different expression levels of the genes in the plasmid (Carroll and Wong, 2018; San Millan and Maclean, 2019), as well as mcr-1, leading to the varied colistin resistance although the strains carried the similar mcr-1-harboring plasmids.
We found that MPE isolates carried other antimicrobial resistance genes, especially ESBLs, ampC, and carbapenemase genes; particularly in the IncHI2A-type mcr-1-harboring plasmids, multiple resistance genes were identified. This was in agreement with previous studies (Mediavilla et al., 2016; Bi et al., 2017; Rhouma and Letellier, 2017; Li Q. et al., 2018; Zheng et al., 2019). A study in China investigated the coexistence of ESBLs and mcr-1 genes in E. coli isolated from chickens between 2008 and 2014 and concluded that mcr-1-positive strains carry at least one ESBL (Wu et al., 2018). The 16 MPE isolates in the present study carried at least one ESBL resistance gene or carbapenemase gene, and one isolate only carried mcr-1. The most commonly encountered ESBL gene in the present study was blaTEM (16/17, 94.1%). The coexistence of the mcr-1 gene with other drug resistance genes increases the possibility of the emergence of pan-drug-resistant bacteria, leading to less choice of therapeutic drugs.
The 17 MPE isolates carried a variety of virulence genes, mainly adhesion-associated and capsular-associated virulence factors. The adhesion VF genes were mainly fimH (16/17, 94.1%), followed by papA (11/17, 64.7%), papG allele III (8/17, 47.7%), and papEF (6/17, 35.3%), while the capsular-associated virulence genes were mainly kpsMT III (10/17, 58.8%), followed by kpsMT K1 (5/17, 29.4%), and kpsMT II (4/17, 23.5%). Uropathogenic Escherichia coli (UPEC) is the main cause of urinary tract infection, and its virulence factors include adhesion factor, cytotoxic substances, iron carrier, capsule, lipopolysaccharides (Jacobsen et al., 2008). These virulence factors help bacteria break through the host’s immune defense system, colonize the urinary tract, and even invade the epithelial cells of the urethra, leading to urinary tract infection. The adhesion factor is considered to be the most important virulence factor of UPEC. Under the mediation of adhesion factors, UPEC can invade host cells and continue to reproduce to form intracellular colonies, thus avoiding the attack of the host immune system (Lüthje and Brauner, 2014; Subashchandrabose and Mobley, 2015). The main adhesion factors of UPEC are fimbriae (FimABCDEFGH) and pyelonephritis-associated pili (pap). Type I fimbriae mostly in urinary tract infection plays an important role. P pilus mainly plays an important role in upper urinary tract infection and onset of acute pyelonephritis (Snyder et al., 2005; Jacobsen et al., 2008; Lüthje and Brauner, 2014; Subashchandrabose and Mobley, 2015). In the present study, the 17 MPE isolates carried a variety of virulence genes, mainly the pili gene fimH (94.1%) and the P-pilin gene papA (64.7%), followed by the capsular-related gene kpsMT III (58.8%).
Conclusion
Our findings indicate that the prevalence of MPE in IAI, UTI, and LRTI patients was low in China. However, the sequence profile and mobility analyses of the mcr-1-carrying plasmids revealed a frequently horizontal transfer of the plasmids, like the IncX4 and IncI2 plasmids observed in this study, as well as the IncHI2A type MDR plasmids carrying a number of resistance genes. The characteristics of these strains, including high transferability of different resistance genes and multiple virulence factors, can contribute to the rapid spread and increasing mortality of these strains. Controlling of mcr-1-positive stains should be an urgent issue.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (Number: S-K238). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
QY, LX, and YX conceived and designed the work. BJ, QY, PJ, EL, HZ, and DL performed the survey. BJ, PD, and TK analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFE0101800, 2018YFC1200100, and 2018YFC1200105) and CAMS Initiative for Innovative Medicine (Grant No. 2016-I2M-3-014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01611/full#supplementary-material
Footnotes
- ^ http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search
- ^ https://cge.cbs.dtu.dk/services/SerotypeFinder/
- ^ http://blast.ncbi.nlm.nih.gov
References
Anyanwu, M. U., Jaja, I. F., and Nwobi, O. C. (2020). Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: a review. Int. J. Environ. Res. Publ. Health 17:1028. doi: 10.3390/ijerph17031028
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bi, Z. W., Berglund, B., Sun, Q., Nilsson, M., Chen, B., Tarnberg, M., et al. (2017). Prevalence of the mcr-1 colistin resistance gene in extended-spectrum beta-lactamase-producing Escherichia coli from human faecal samples collected in 2012 in rural villages in Shandong Province, China. Int. J. Antimicrob. Agents 49, 493–497. doi: 10.1016/j.ijantimicag.2016.12.018
Carattoli, A., Zankari, E., Garcìa-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carroll, A. C., and Wong, A. (2018). Plasmid persistence: costs, benefits, and the plasmid paradox. Can. J. Microbiol. 64, 293–304. doi: 10.1139/cjm-2017-0609
Chen, K., Chan, E. W., Xie, M., Ye, L., Dong, N., and Chen, S. (2017). Widespread distribution of mcr-1-bearing bacteria in the ecosystem, 2015 to 2016. Euro Surveill. 22, 17–00206. doi: 10.2807/1560-7917.ES.2017.22.39.17-00206
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Ninth Informational Supplement. M100-S29. Wayne, PA: Clinical and Laboratory Standards Institute.
Delcher, A. L., Salzberg, S. L., and Phillippy, A. M. (2003). Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinform. 1, 10.3.1–10.3.18. doi: 10.1002/0471250953.bi1003s00103110318
Fernandes, M. R., Moura, Q., Sartori, L., Silva, K. C., Cunha, M. P., Esposito, F., et al. (2016). Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill. 21:30214. doi: 10.2807/1560-7917.ES.2016.21.17.30214
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 12:e1005957. doi: 10.1371/journal.ppat.1005957
Giani, T., Sennati, S., Antonelli, A., Pilato, V. D., Maggio, T. D., Mantella, A., et al. (2018). High prevalence of carriage of mcr-1-positive enteric bacteria among healthy children from rural communities in the Chaco region, Bolivia, September to October 2016. Euro Surveill. 23:1800115. doi: 10.2807/1560-7917.ES.2018.23.45.1800115
Hamprecht, A., Rohde, A. M., Behnke, M., Feihl, S., Gastmeier, P., Gebhardt, F., et al. (2016). Colonization with third-generation cephalosporin-resistant Enterobacteriaceae on hospital admission: prevalence and risk factors. J. Antimicrob. Chemoth. 71, 2957–2963. doi: 10.1093/jac/dkw216
Hasman, H., Hammerum, A. M., Hansen, F., Hendriksen, R. S., Olesen, B., Agerso, Y., et al. (2015). Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 20, 1–5. doi: 10.2807/1560-7917.ES.2015.20.49.30085
He, Q. W., Xu, X. H., Lan, F. J., Zhao, Z. C., Wu, Z. Y., Cao, Y. P., et al. (2017). Molecular characteristic of mcr-1 producing Escherichia coli in a Chinese university hospital. Ann. Clin. Microbiol. Antimicrob. 16;32. doi: 10.1186/s12941-017-0207-z
Jacobsen, S. M., Stickler, D. J., Mobley, H. L., and Shirtliff, M. E. (2008). Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 21, 26–59. doi: 10.1128/cmr.00019-07
Johnson, J. R., and Stell, A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181, 261–272. doi: 10.1086/315217
Kieffer, N., Aires-de-Sousa, M., Nordmann, P., and Poirel, L. (2017). High rate of MCR-1-producing Escherichia coli and Klebsiella pneumoniae among Pigs, Portugal. Emerg. Infect. Dis. 23, 2023–2029. doi: 10.3201/eid2312.170883
Kleinheinz, K. A., Joensen, K. G., and Larsen, M. V. (2014). Applying the resfinder and virulencefinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. doi: 10.4161/bact.27943
Li, B., Ke, B., Zhao, X., Guo, Y., Wang, W., Wang, X., et al. (2018a). Antimicrobial resistance profile of mcr-1 positive clinical isolates of Escherichia coli in China from 2013 to 2016. Front. Microbiol. 9:2514. doi: 10.3389/fmicb.2018.02514
Li, Q., Wang, H., Xu, Y., Bai, X., Wang, J., Zhang, Z., et al. (2018b). Multidrug-Resistant Escherichia albertii: co-occurrence of beta-Lactamase and MCR-1 encoding genes. Front. Microbiol. 9:258. doi: 10.3389/fmicb.2018.00258
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemoth. 72, 393–401. doi: 10.1016/j.ijantimicag.2016.11.026
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Lu, H., Wang, C., Dong, G., Xu, C., Zhang, X., Liu, H., et al. (2018). Prevalence and molecular characterization of Escherichia coli clinical isolates carrying mcr-1 in a Chinese teaching hospital from 2002 to 2016. Antimicrob. Agents Chem. 62:e2623-17. doi: 10.1128/AAC.02623-17
Lüthje, P., and Brauner, A. (2014). Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Microb. Physiol. 65, 337–372. doi: 10.1016/bs.ampbs.2014.08.006
Matamoros, S., Van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:2963. doi: 10.1038/s41598-017-15539-7
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Mediavilla, J. R., Patrawalla, A., Chen, L., Chavda, K. D., Mathema, B., Vinnard, C., et al. (2016). Colistin- and carbapenem-resistant Escherichia coli Harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio 7:3341. doi: 10.3389/fmicb.2018.03341
Miao, M., Wen, H., Xu, P., Niu, S., Lv, J., Xie, X., et al. (2018). Genetic diversity of carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in Eastern China. Front. Microbiol. 9:3341. doi: 10.3389/fmicb.2018.03341
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Pérez-Pérez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/jcm.40.6.2153-2162.2002
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Potter, R. F., D’Souza, A. W., and Dantas, G. (2016). The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Update 29, 30–46. doi: 10.1016/j.drup.2016.09.002
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Prim, N., Rivera, A., Rodriguez-Navarro, J., Espanol, M., Turbau, M., Coll, P., et al. (2016). Detection of mcr-1 colistin resistance gene in polyclonal Escherichia coli isolates in Barcelona, Spain, 2012 to 2015. Euro Surveill. 21:30183. doi: 10.2807/1560-7917.ES.2016.21.13.30183
Quan, J. J., Li, X., Chen, Y., Jiang, Y., Zhou, Z. H., Zhang, H. C., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/s1473-3099(16)30528-x
Queenan, A. M., and Bush, K. (2007). Carbapenemases : the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/cmr.00001-07
Rhouma, M., and Letellier, A. (2017). Extended-spectrum β-lactamases, carbapenemases and the mcr-1 gene: is there a historical link? Int. J. Antimicrob. Agents 49, 269–271.
San Millan, A., and Maclean, R. C. (2019). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5, 65–79. doi: 10.1128/microbiolspec.MTBP-0016-2017
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, Y., Wu, Z., Wang, Y., Zhang, R., Zhou, H. W., Wang, S., et al. (2018). Heterogeneous and flexible transmission of mcr-1 in hospital-associated Escherichia coli. mBio 9:e943-18. doi: 10.1128/mBio.00943-18
Shen, Z., Wang, Y., Shen, Y., Shen, J., and Wu, C. (2016). Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/s1473-3099(16)00061-x
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Snyder, J. A., Haugen, B. J., Lockatell, C. V., Maroncle, N., Hagan, E. C., Johnson, D. E., et al. (2005). Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73, 7588–7596. doi: 10.1128/iai.73.11.7588-7596.2005
Soo, J. S., Huang, R. J., Squire, C. J., and Leung, I. K. H. (2019). MCR-1: a promising target for structure-based design of inhibitors to tackle polymyxin resistance. Crit. Rev. Microbiol. 24, 206–216. doi: 10.1016/j.drudis.2018.07.004
Subashchandrabose, S., and Mobley, H. L. T. (2015). Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol. Spectr. 3, 235–261. doi: 10.1128/microbiolspec.UTI-0015-2012
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, Y., Tian, G. B., Zhang, R., Shen, Y. B., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/s1473-3099(16)30527-8
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, C., Wang, Y., Shi, X., Wang, S., Ren, H., Shen, Z., et al. (2018). Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008-2014. Emerg. Microbes Infect. 7:30. doi: 10.1038/s41426-018-0033-1
Keywords: mcr-1, intra-abdominal infections, urinary tract infections, resistance, virulence
Citation: Jiang B, Du P, Jia P, Liu E, Kudinha T, Zhang H, Li D, Xu Y, Xie L and Yang Q (2020) Antimicrobial Susceptibility and Virulence of mcr-1-Positive Enterobacteriaceae in China, a Multicenter Longitudinal Epidemiological Study. Front. Microbiol. 11:1611. doi: 10.3389/fmicb.2020.01611
Received: 09 April 2020; Accepted: 19 June 2020;
Published: 28 July 2020.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Fupin Hu, Fudan University, ChinaRuichao Li, Yangzhou University, China
Shangshang Qin, Zhengzhou University, China
Copyright © 2020 Jiang, Du, Jia, Liu, Kudinha, Zhang, Li, Xu, Xie and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwen Yang, yangqiwen81@vip.163.com; Liangyi Xie, lyxie78@hunnu.edu.cn
†These authors have contributed equally to this work
 Bin Jiang
Bin Jiang Pengcheng Du3†
Pengcheng Du3† Peiyao Jia
Peiyao Jia Enbo Liu
Enbo Liu Timothy Kudinha
Timothy Kudinha Yingchun Xu
Yingchun Xu Liangyi Xie
Liangyi Xie Qiwen Yang
Qiwen Yang