- 1Department Ecology and Ecosystem Modelling, University of Potsdam, Potsdam, Germany
- 2Department of Aquatic Environmental Engineering, Karlsruhe Institute of Technology, Karlsruhe, Germany
- 3Leibniz Institute for Baltic Sea Research, Rostock, Germany
- 4Berlin-Brandenburg Institute of Advanced Biodiversity Research (BBIB), Berlin, Germany
Despite the increasing number of species invasions, the factors driving invasiveness are still under debate. This is particularly the case for “invisible” invasions by aquatic microbial species. Since in many cases only a few individuals or propagules enter a new habitat, their genetic variation is low and might limit their invasion success, known as the genetic bottleneck. Thus, a key question is, how genetic identity and diversity of invading species influences their invasion success and, subsequently, affect the resident community. We conducted invader-addition experiments using genetically different strains of the globally invasive, aquatic cyanobacterium Raphidiopsis raciborskii (formerly: Cylindrospermopsis raciborskii) to determine the role of invader identity and genetic diversity (strain richness) at four levels of herbivory. We tested the invasion success of solitary single strain invasions against the invader genetic diversity, which was experimentally increased up to ten strains (multi-strain populations). By using amplicon sequencing we determined the strain-specific invasion success in the multi-strain treatments and compared those with the success of these strains in the single-strain treatments. Furthermore, we tested for the invasion success under different herbivore pressures. We showed that high grazing pressure by a generalist herbivore prevented invasion, whereas a specialist herbivore enabled coexistence of consumer and invader. We found a weak effect of diversity on invasion success only under highly competitive conditions. When invasions were successful, the magnitude of this success was strain-specific and consistent among invasions performed with single-strain or multi-strain populations. A strain-specific effect was also observed on the resident phytoplankton community composition, highlighting the strong role of invader genetic identity. Our results point to a strong effect of the genetic identity on the invasion success under low predation pressure. The genetic diversity of the invader population, however, had little effect on invasion success in our study, in contrast to most previous findings. Instead, it is the interaction between the consumer abundance and type together with the strain identity of the invader that defined invasion success. This study underlines the importance of strain choice in invasion research and in ecological studies in general.
Introduction
In recent years, globalization, along with climate and land use changes, has facilitated the spread and establishment of invasive species, which can have severe ecological impacts and are one of the biggest threats to biodiversity (Sala, 2000). Besides animals and terrestrial plants, high invasion rates are increasingly observed in (aquatic) microorganisms, supported by their high dispersal potential (Ricciardi and MacIsaac, 2010; Strayer, 2010). A prominent example for invasive aquatic microorganism are potentially toxic nostocalean cyanobacteria, an increasingly invasive order in plankton communities (Markensten et al., 2010; Sukenik et al., 2012). Although, microbes have a high potential for invasions and may have severe impacts on local communities (Sala, 2000), they are generally underrepresented in invasion research (Pyšek et al., 2008; Horňák and Corno, 2012; Strayer et al., 2017) and are rather “invisible” invaders (Litchman, 2010).
Generally, the invasiveness of a species is mainly related to three factors: (1) the characteristics of the new habitat, (2) the characteristics of the resident community, and (3) the invader itself (Lonsdale, 1999; Litchman, 2010). However, which of these factors is the main driver of the invasion process is still under debate. (1) As a prerequisite for any invasion, the abiotic conditions of the new habitat must be suitable for the invading species. If this necessity is fulfilled, (2) native communities can counteract invasions by either competitive or consumptive resistance (Alofs and Jackson, 2014). For example, effective consumers can decrease the invasibility of a community by a high consumption leading to high mortality rates of invaders (Petrie and Knapton, 1999; DeRivera et al., 2005; Juliano et al., 2010; Sperfeld et al., 2010), which has been described as the dominant type of resistance in aquatic ecosystems (Alofs and Jackson, 2014). The competitive resistance of a community relies mainly on the resource availability of the habitat, which is often low when native species-richness and/or genetic diversity is high, hampering invasions (Stachowicz et al., 1999, 2002; Fargione and Tilman, 2005; Romanuk and Kolasa, 2005). Besides the impact of the habitat and the resident community, and (3) the specific characteristics of the invader itself drives its invasion success (Crawley et al., 1999). A number of traits are linked to successful invasions, for example: short generation times, efficient resource use, tolerance to a broad range of environmental factors (Sakai et al., 2001; Litchman, 2010; Mächler and Altermatt, 2012; Mallon et al., 2015) and a high plasticity (Richards et al., 2006; Davidson et al., 2011). After establishment, invasive species directly interact with their new environment and actively modify their new habitat through, e.g., consumption or competition (Sakai et al., 2001). Thus they impair the resident community by reducing their fitness and may promote species extinctions within the community (Vilà et al., 2011). This effect on the resident community is often assumed to be species-specific (Hejda et al., 2009; Weithoff et al., 2017). Moreover, successful invasive species can change abiotic and biotic conditions to the point that it promotes the invasion of otherwise inferior invaders, leading to an invasional meltdown (Simberloff and Von Holle, 1999). Using similar reasoning, simultaneous invasions of several strains from a genetically diverse population might promote weaker strains and allows for further invasions, via changes in conditions induced by the more successful strain(s). Since the invader traits might vary among individual strains within a species, strain-specific invasions and interactions with the resident community are expected. The important role of strain identity has been shown for several biological processes, e.g., host–parasite (Barrett et al., 2009) or prey–predator interactions, for example, between Daphnia and Microcystis strains (Van Gremberghe et al., 2009; Lemaire et al., 2012). Only recently, few studies showed that strain identity has a crucial impact on the invasiveness, especially in highly variable and phenotypically plastic species such as in Daphnia (Engel et al., 2011) or in mammals (González-Suárez et al., 2015). Although strain identity can play a role for invasiveness, the genetic diversity of invaders is also positively associated with invasion success (Ahlroth et al., 2003; Crawford and Whitney, 2010; Forsman, 2014). Genetic diversity thereby increases the chance that one “appropriate” strain will succeed in the new habitat.
According to theory, invasive species frequently undergo a genetic bottleneck (Allendorf and Lundquist, 2003; Roman and Darling, 2007), thus decreasing genetic diversity within a species in a new habitat. However, high propagule pressure, several invasion events or invasions from different geographical regions through, e.g., human trade (Kolbe et al., 2004; Therriault et al., 2005; Roman and Darling, 2007; Lockwood et al., 2009) can counteract low genetic diversity of the invader. To date, few studies in invasion ecology have experimentally examined and followed the initial invasion process and subsequent establishment with respect to the genetic diversity and intraspecific variation of the invader (Jeschke and Strayer, 2005; Forsman, 2014; Jeschke and Heger, 2018). In addition, different strains might affect invaded communities differently, due, for example, differences in resource use efficiency.
We conducted laboratory invader-addition experiments to study the underlying mechanisms in the invasion success of the globally invasive and genetically diverse (Abreu et al., 2018) cyanobacterium Raphidiopsis raciborskii (formerly: Cylindrospermopsis raciborskii). Raphidiopsis raciborskii (Aguilera et al., 2018) is a bloom-forming tropical cyanobacterium, which is increasing its global distribution north- and southwards toward the temperate zones (Padisák, 1997; Antunes et al., 2015). Due to its advantageous traits, such as the capability of nitrogen-fixation, a high phosphorus affinity and storage capability and the formation of akinetes (Padisák, 1997; Wiedner et al., 2007a), it has been found to dominate also temperate phytoplankton communities (Hamilton et al., 2005; Budzyńska and Gołdyn, 2017). (i) We studied the role of consumptive and competitive resistance by varying herbivore abundances and types (specialist vs generalist). (ii) We tested the hypothesis that a high genetic (strain) diversity promotes invasions by adding different strains at varying diversity levels. Using amplicon sequencing for the multi-strain invading populations, we could determine the individual strain invasion success. In the case of successful invasions, we investigated (iii) how successful invasions affected the resident species and their community composition.
Materials and Methods
Model Organism
For this study, 10 genetically different R. raciborskii strains from Northeast German lakes were selected, which differ in their growth rates and ingestibility by the generalist rotifer Brachionus calyciflorus (Bolius et al., 2017). The 10 strains used here were chosen from a set of 12 strains (see Bolius et al., 2017) from established laboratory cultures. All strains were isolated between 2004 and 2010 (Wiedner et al., 2007b) and kept in batch stock culture since then.
Experimental Design
We analyzed the invasion success of R. raciborskii into near-native temperate phytoplankton communities as a function of strain traits, the genetic diversity (= strain richness), and herbivory, using invader addition experiments. The resident community consisted of phyto- and zooplankton based on Sperfeld et al. (2010) and was composed of species with different functional traits such as size, growth rate, and edibility (Supplementary Tables 1, 2). To mimic complex natural conditions, competition and predation, the experimental communities consisted of phytoplankton and a herbivore. In particular in aquatic systems, consumers play a stronger role in preventing invasions than competitors (Alofs and Jackson, 2014).
We conducted four experiments, in two differing set-ups (Supplementary Table 1). The first experimental set-up (one experiment) was conducted with a resident community consisting of ten phytoplankton species and a high density of the generalist herbivore B. calyciflorus sensu stricto (Paraskevopoulou et al., 2018; strain from Fussmann et al., 2005). In this set-up, the herbivore went extinct soon after the start and before the invader addition, likely due to an increase of the large and less ingestible resident chlorophyte Pandorina morum. Therefore, this experiment is considered as a no-herbivory experiment. For the logic of an herbivory gradient, this experiment is named experiment IV (Figure 1 and Supplementary Table 1).
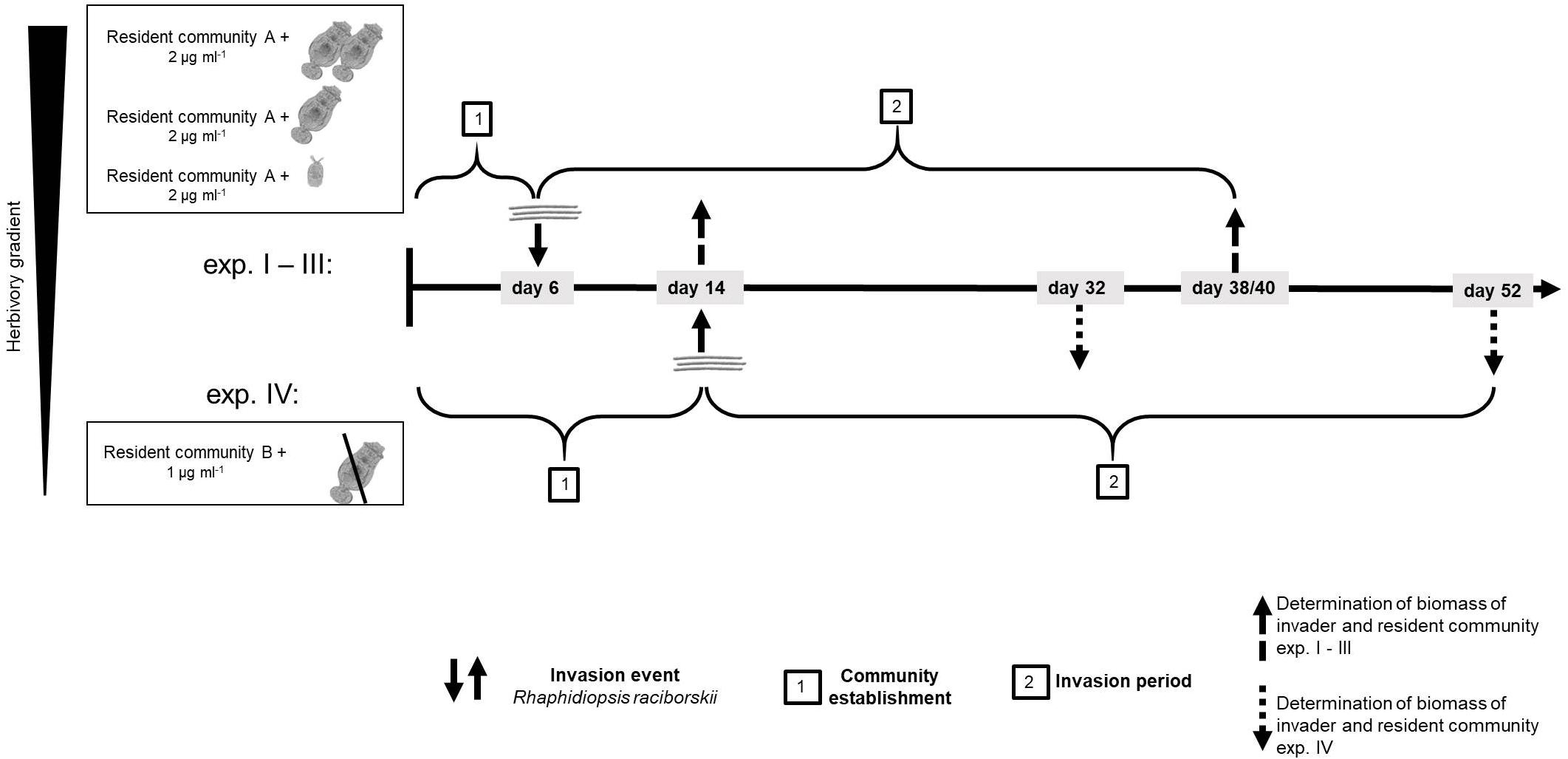
Figure 1. Time line of the experiments showing the four different experiments and how experiment IV differed from experiments I–III. For resident community A and B, see Supplementary Table 1.
To investigate the impact of the herbivory on the invasion of R. raciborskii and to mimic a more natural community, a second experiment was conducted with modifications (set-up 2, three experiments, Figure 1): the composition of the resident phytoplankton community had to be changed, allowing B. calyciflorus to persist. However, this altered resident community enabled a high reproduction and abundance of B. calyciflorus. Thus, in a third experiment, we reduced the initial abundance of the herbivore, while in a fourth experiment, a smaller and more specialist consumer Cephalodella sp. replaced Brachionus (Figure 1). In addition, the sampling interval was modified to include an earlier sampling to detect short-term invasion success and its impacts, because the two (later) samplings of the first set-up resulted in similar high biomasses of the invader (see section “Results”). Thus, experiments IV (set-up 1) differed from the other experiments with respect to the species composition of the resident community, the timing of the invasion and the subsequent sampling regime (Figure 1 and Supplementary Table 1). The detailed design of the four experiments is explained and the initial parameters of the first set-up (experiment IV) are added in parentheses in the text that follows.
At the start of the experiments, the resident community was set up in 300 ml Erlenmeyer flasks (microcosms) with 150 (100) ml of modified sterile freshwater Woods Hole medium (after Nichols, 1973; 2 mM HEPES buffer, pH 8, 80 μg phosphorous L–1, nitrogen:phosphorous = 20:1) and was inoculated with in total 2 (1) μg ml–1 of a resident phytoplankton community (Supplementary Table 1) in equal parts of 0.25 (0.1) μg ml–1 for each species. In addition to the phytoplankton species, the resident communities included an herbivorous consumer: either the generalist rotifer B. calyciflorus or the smaller, more specialist rotifer Cephalodella sp. (Altermatt et al., 2011; Seifert et al., 2015). The four different herbivory levels were generated as follows (the experiments are numbered by decreasing herbivory): for a high level of herbivory, the initial density of B. calyciflorus was 1.5 animals ml–1 (experiment I, set up 2) and for a lower level of herbivory, 0.2 animals ml–1 (experiment II, set up 2). A low and more specific herbivory was generated adding one Cephalodella sp. ml–1 to the microcosms (experiment III, set up 2). Experiment IV started with 2.2 animals ml–1 of B. calyciflorus, but the herbivore declined drastically soon after the start and eventually died out (see above, first experimental set-up), resulting in no grazing pressure during the invasion period. In experiments II and III, three flasks were set-up without herbivory as no-herbivory control. Before adding the animals, food algae from stock cultures were removed by filtering the animals through gauze (for B. calyciflorus 50 μm and for Cephalodella sp. 20 μm). All phytoplankton and rotifer species were pre-cultured at 20°C, 18:6 h light-dark cycle as batch stock cultures. These conditions for temperature and light-dark cycle were chosen as they reflect the typical conditions in summer in NE Germany where the strains were isolated from. After an establishment period for the resident community (experiments I–III: 6 days; experiment IV: 14 days), the invader R. raciborskii was added, mimicking one single invasion event. The invader was added either as a single strain or in multi-strain populations (genetic diversity). In particular, R. raciborskii was added in an increasing genetic diversity gradient from one strain to a genetic mixture of up to 10 strains, with up to 15 replicates per treatment (number of replicates experiments I–III/experiment IV): one (2/3 replicates per strain), three (15/9 replicates, but different strain populations – see Supplementary Figure 1), six (15/9 replicates, but different strain populations), nine (two replicates), all 10 (three replicates), and none (as a control: three replicates without invader addition). In experiment IV the maximum genetic diversity treatment was nine strains (Supplementary Table 1). For all experiments and diversity treatments, the strains were randomly selected from the 10 strain-pool (see above and Supplementary Figure 1) and each time the invader was added with the same total biomass of 0.1 μg ml–1 (5% of initial phytoplankton resident community), resulting in a lower absolute biomass of each strain in the genetic mixtures (0.033 to 0.01 μg ml–1 per strain).
The addition of the invader comprises different forms of replicates: for the single strain treatments and nine respectively 10 strains, the same strain/strains was/were added three times, for the treatments with three and six strains all replicates consisted of different strain combinations (technical replicate, Supplementary Figure 1). In addition to the cyanobacteria, 1 ml of their filtrate (mixture of all strains; 0.8 μm, cellulose nitrate filters, Sartorius, Göttingen, Germany) was added to each flask to exclude a possible effect of potential strain-specific cyanobacteria-associated heterotrophic bacteria on the invasion success.
All experiments were run at 20°C at a light-dark cycle of 16:8 h and a light intensity of 130 μmol photons m–1 s–1 (measured in water with a spherical light sensor, Li-Cor, SQSA 0107, WALZ Mess- und Regeltechnik, Effeltrich, Germany). To reduce sedimentation, all flasks were shaken gently 15 min h–1. Every second day, 20% of the experimental volume was exchanged with fresh medium (dilution rate = 0.16 day–1). A total of 5 ml were used for regular chlorophyll-a fluorescence measurement (Fluorometer TD 700, Turner Designs, Sunnyvale, CA, United States) as an estimate of total phytoplankton density, the remaining volume was fixed with Lugol’s iodine, for subsequent analysis. Halfway through the experiments, the flasks were exchanged to avoid phytoplankton wall growth. The experiments lasted for 40/40/38/52 days (experiments I, II, III, and IV), respectively. Experiment III was two days shorter due to laboratory logistics.
Non-genetic Sample Analyses
The phytoplankton species composition, including the invader R. raciborskii, was determined twice: either 8 days after the invasion (day 14; experiments I–III) for short-term changes, or 22 days after invasion for intermediate term changes (day 36; experiment IV) and in all experiments 34/34/32/38 days after invasion (experimental day 40/40/38/52) for long-term changes. Rotifer abundance was determined for every sampling day. All zoo- and phytoplankton samples were analyzed using inverted light microscopy (AxioVision A1, Carl Zeiss, Jena, Germany; AxiovertS100, Carl Zeiss; ALTRA20, Olympus Soft Imaging Solutions GmbH, Münster, Germany), except for Synechococcus elongatus, which was quantified after acridine orange staining on black 0.2 μm membrane filters (Whatman Nuclepore, GE Healthcare, Maidstone, United Kingdom) using epifluorescence microscopy (Axioscop2, Carl Zeiss). Phytoplankton cell dimensions were measured (30–50 per species) with a computer-aided image system (Cell B, Olympus Soft Imaging Solutions GmbH) and the biovolume was calculated by multiplying the abundances with the cell volume assuming appropriate geometrical shapes (Hillebrand et al., 1999). Since the strains of R. raciborskii cannot be unambiguously morphologically distinguished, biomass data were not separated by strain.
As a measurement of a resource use trait, at the end of the experiments I–III, particulate carbon (C), nitrogen (N), and phosphorous (P) were measured and the molar C:N:P ratios of the phytoplankton communities calculated. For particulate C and N, samples were vacuum filtered on GF/C filters (precombusted for 4 h at 450°C; Whatman), dried and measured using an elementary analyzer (EA 3000, EuroVector S.p.A, Milan, Italy; software: Callidus Software, EuroVector S.p.A). Particulate P was measured after filtration on 0.45 μm membrane filters (PALL Corporation, Port Washington, NY, United States). Determination followed the blue molybdate method by Murphy and Riley (1962), after digestion with H2SO4, K2S2O8, and heat (121°C; 1 h autoclaving) at a photometer (880 nm; UV Mini 1240 UV-VIS spectrophotometer, Shimadzu, Kyoto, Japan). For in-depth analysis, including strain-specific invasion success in genetic mixtures and strain-specific effects on the resident community experiment III was analyzed further. In this experiment the invader and the herbivore persistently coexisted.
Genetic Analyses
In experiment III, the composition of the genetic mixtures was analyzed, to identify the individual strain-specific invasion success in the populations.
Amplicon Amplifying and Sequencing
Prior to DNA extraction, the phytoplankton samples of each mixture at the day of the invasion event and at the end (see above) were centrifuged and stored at −20°C. DNA was extracted using Metagenomic DNA Isolation Kit for Water (Epicentre, Madison, WI, United States). The R. raciborskii strains used here did not differ in common marker genes, e.g., cbcBA or ITS (unpublished data) and therefore, we designed primers for unique genetic markers/amplicons to distinguish each strain from all others (see Supplementary Material “Primer Design”). Following DNA extraction, from all mixtures, strain-specific markers (GenBank MT531416–MT531536) were amplified – using unique primers (Supplementary Table 3). After determining the DNA concentration (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA, United States), the amplified products of each mixture were pooled and purified (innuPREP PCRpure Kit, Analytik Jena, Jena, Germany). All PCRs were run in 20 μl aqueous reaction volume containing 1× PCR buffer, 200 μM dNTPs, 0.5 M primer, 0.02 U/μl Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific) and approximately 10 ng of DNA. The initial denaturation was at 98°C for 30 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 64°C for 30 s, extension at 72°C for 20 s, and a final extension at 72°C for 10 min. To determine the relative share of each strain next generation sequencing was used. Libraries were constructed from amplicons and sequenced on an Illumina NextSeq to generate 150 bp paired-end reads. SeqPrep (John, 2011) was used for adapter trimming, quality filtering (q 30), and read merging. This resulted in 1,700 to 908,000 merged read pairs per sample (median 533,000).
To determine which strains were present in each of the 69 samples (33 mixtures of the invasion event and 36 of the end), the merged read pairs from each sample were compared to each of the determined unique sequences that make up the expected PCR products (see Supplementary Material “Primer Design”). To exclude the possibility of cross-contamination of PCR primers/products, this included the comparison of amplicons with sequences of PCR products for which no primer pair had been used in the given experiment. For this analysis, we used the merged read pairs as queries and the determined unique PCR products as database for BLAST searches (Altschul et al., 1990). Only alignments with 100% sequence identity and the exact length of an expected PCR product sequence were scored.
The number of each PCR product was counted as “abundance” and the relative shares of each strain were determined. As the invader was added into the experiment by biomass, we used the relative data of the invasion event (day 6) as the “DNA amount base line” and the relative gain or loss of each strain in the genetic mixtures until the end was calculated from that as a measurement for strain-specific invasion success. When the strains MEL07 and Peter07_149, as well as ZIE05 and ZIE11 were in one mixture, their individual share could not be determined due to the absence of an appropriate discriminant sequence (see Supplementary Material “Primer Design”). Therefore, in genetic mixtures with both strains present, the data were combined (named as “MEL07 and Peter07_149”; “ZIE05 and ZIE11”). One genetic mixture with three strains (sample 47) was excluded from further analysis, as the included strain 27F11 was not detected in the start sample.
Statistical Analysis
To test for an effect of strain richness on the invasion success we calculated the invasion yield as the natural logarithm of the ratio between the observed yield of the invader and the expected yield estimated from single-strain treatments. For each diversity level, the mean and the 95% confidence interval (CI) were calculated. A positive effect of strain richness on the invasion success occurred, when the mean was positive and the 95% CI did not include zero. We included diversity levels of three and six strains. To compare two data-sets, a t-test was applied. Either an ANOVA for normal distributed data, or Kruskal–Wallis test as a non-parametric test was performed for comparing more than two groups using the data analysis software SigmaPlot (13.0). When significant differences were found, a pairwise multiple comparison test was applied (Tukey Post hoc test). Normality of data was tested using the Shapiro–Wilk test. Linear regression or Spearman rank correlation was applied to test for relationships or correlations between variables. To compare the strain-specific invasion differences, we calculated the coefficient of variation (CV) as the standard deviation divided by the mean, calculated from the relative share in the whole community. To evaluate a potential strain-specific effect on the resident communities, we performed a PERMANOVA (in R, R Development Core Team 2018, RStudio 1.0.136, function “adonis” of the package “vegan” with Bray–Curtis distance; with 10,000 permutations) for the one strain treatments. For the PERMANOVA, the composition of the resident communities (in biomass; without invader) was used. The composition of the resident communities was ordinated using a principal component analysis (PCA; in R, package “stats”) to illustrate the similarity of the resident community compositions for experiment III, day 38.
Results
We examined the strain-specific invasion success of the cyanobacterium R. raciborskii at different levels of genetic diversity and four levels of herbivory.
Impact of the Herbivore Pressure on the Invasion Successes
Grazing pressure strongly drove the invasion success of R. raciborskii. On the one hand, when grazing pressure was high due to high abundance of the consumer B. calyciflorus, the invasion of R. raciborskii completely failed (experiment I, not shown). At the first sampling on day 14 (8 days after the invasion), no R. raciborskii was detected using the applied method. On the other hand, without herbivory, due to consumer extinction (experiment IV, Figure 2C), and in the intended rotifer-free treatments (in experiments II and III, Figures 2A,B), R. raciborskii successfully invaded all plankton communities, reaching on average more than 80% of total community biomass (Figure 2). When B. calyciflorus abundance was at an intermediate level on the invasion date (experiment II), an invasion of the invader was possible (Figure 2A). However, the successful invasion was mainly the result of an “either or” pattern: when the consumer eventually went extinct shortly after the invader addition, successful invasions at a low level were observed in six out of 63 cases (day 40), and when the consumer reached high abundances, the invader failed or remained at very low levels (negative significant Spearman rank correlation between abundance of invader and consumer, rho = 0.650, significant at p < 0.0001). Only with Cephalodella sp. as the consumer, was coexistence possible with high abundances of both consumer and invader: 8 days after invasion, R. raciborskii contributed to 1.9 ± 0.9% of the phytoplankton community (Figure 2B). Until day 38 (32 days after invasion), R. raciborskii strongly increased its relative biomass share up to 72% (mean ± SD, 44 ± 17%), dominating most of the communities (67% of all microcosms; Figure 2B). The abundance of Cephalodella sp. was not correlated with the invasion success of R. raciborskii (linear regression, experiment III day 14: R2 = 0.021, p = 0.235; day 38: R2 = 0.001, p = 0.774).
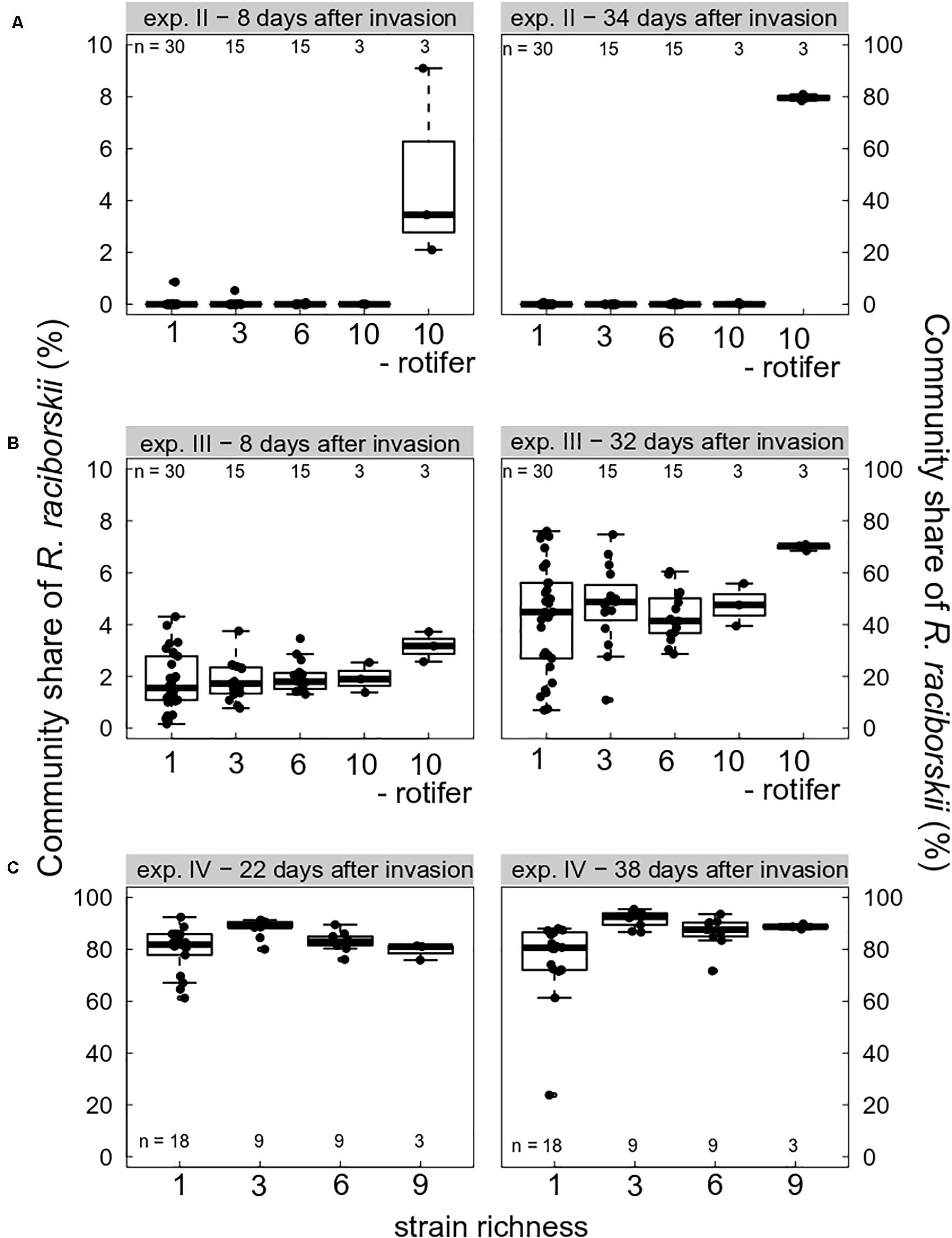
Figure 2. Boxplot (with data points) showing invasion success of Raphidiopsis raciborskii, measured as the biomass percent contribution of the resident community composition and as a function of strain richness (genetic diversity) at days 8, 22, 32/34, and 38 after invasion in experiments (A) II (intermediate grazing pressure, Brachionus calyciflorus), (B) III (low grazing pressure, Cephalodella sp.), and (C) IV (no grazing pressure, extinction of B. calyciflorus). Microcosms without consumers are indicated with “- rotifer.” Note different scales on the y-axes. n indicates the total number of microcosms. For one-strain treatments there were three (A,B) or two (C) replicates per strain; for the diversity levels three and six there were n mixtures with random strains and for levels nine and 10, there was only one combination possible replicated three times.
Strain-Specific Invasion Success
We found consistent strain-specific differences in the invasion success in experiments III and IV (Figure 3). Some strains were highly invasive, while others invaded the communities with lower densities, consistently between both experiments (positive Spearman rank correlation between strain invasion success of experiments III and IV, rho = 0.762, significant at p = 0.028), although on different absolute biomass and community composition. In experiment IV the overall invasion success was higher than in experiment III (Figure 2). In experiment III, strain-specific differences were high, expressed as a CV of 0.48, calculated from the relative share in the whole community. In experiment IV, the strains were on a more similar level (CV = 0.11), except for one outlier (MEL07, Figure 3B).
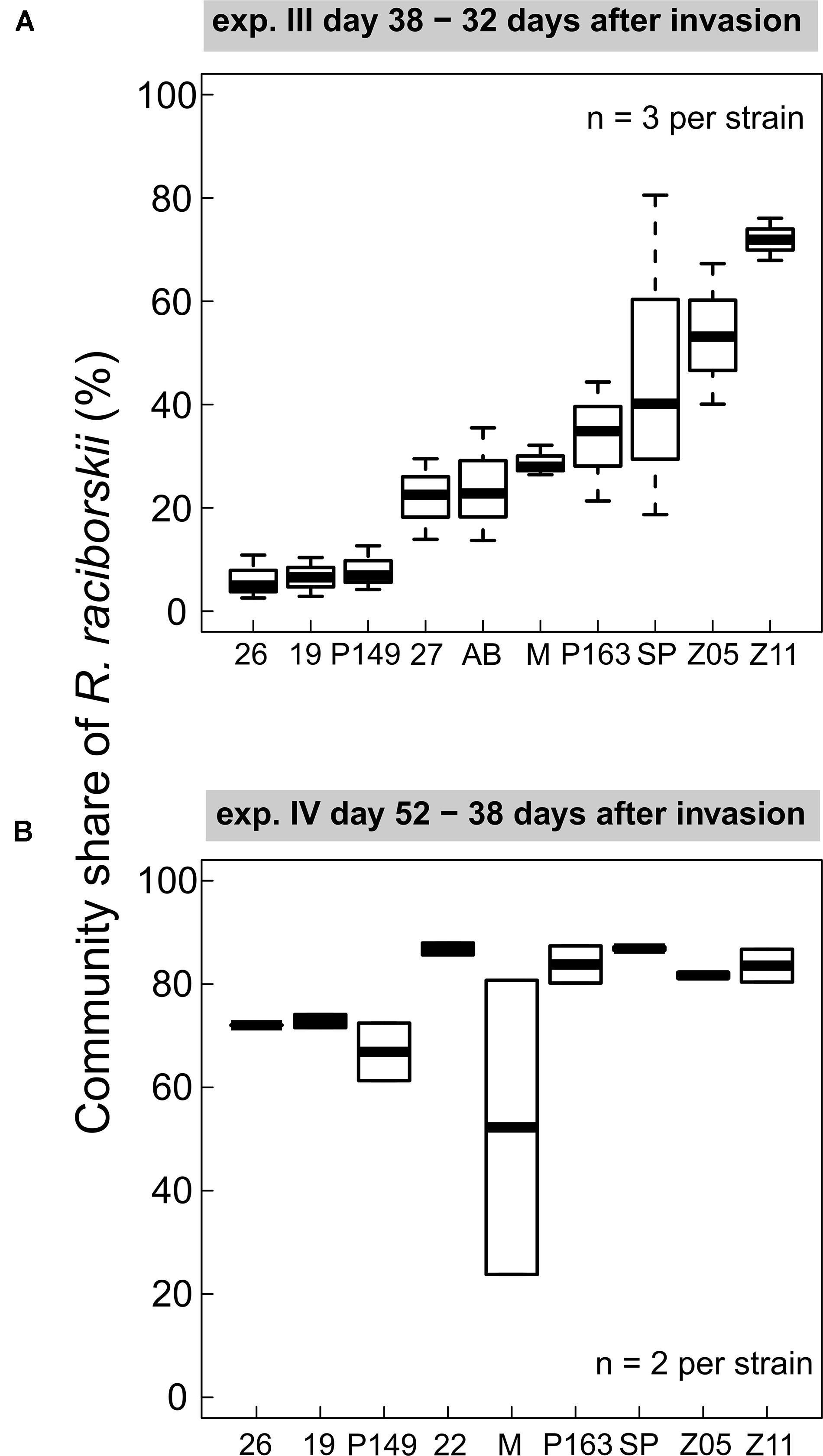
Figure 3. Individual strain invasion success of Raphidiopsis raciborskii (boxplots; in % of total community share) in experiments with (A) Cephalodella sp. (experiment III), 32 days after invasion, n = 3; (B) experiment IV with extinct herbivore, 38 days after invasion, n = 2. Abbreviations of the strains: 26, 26D9; 19, 19F6; P149, Peter07_149; 22, 22F8; 27, 27F11; AB, AB2008/71; M, MEL07; P163, Peter07_163; SP, SP08_4; Z5, ZIE05; Z11, ZIE11.
Low Impact of the Genetic Diversity
The effect of the genetic diversity on the invasion success could only be analyzed for experiments III and IV, with successful invasions. In the experiment with a low grazing pressure imposed by Cephalodella sp., the invasion success of R. raciborskii was not affected by the genetic diversity (experiment III, the invasion yield was not significantly different from zero; Figure 4A). The average R. raciborskii community share was very similar in all treatments, but the variance decreased with increasing diversity on both sampling days (see Figure 2). With no grazing pressure (experiment IV), the relative invasion success of R. raciborskii was only positively affected by the genetic diversity of the three-strain treatments (day 36 and 52) and for the six-strain treatments at day 52 (Figure 4B); comparison of invasion success: day 36 and 52, ANOVA, p < 0.001, Tukey Post hoc test, p < 0.006).
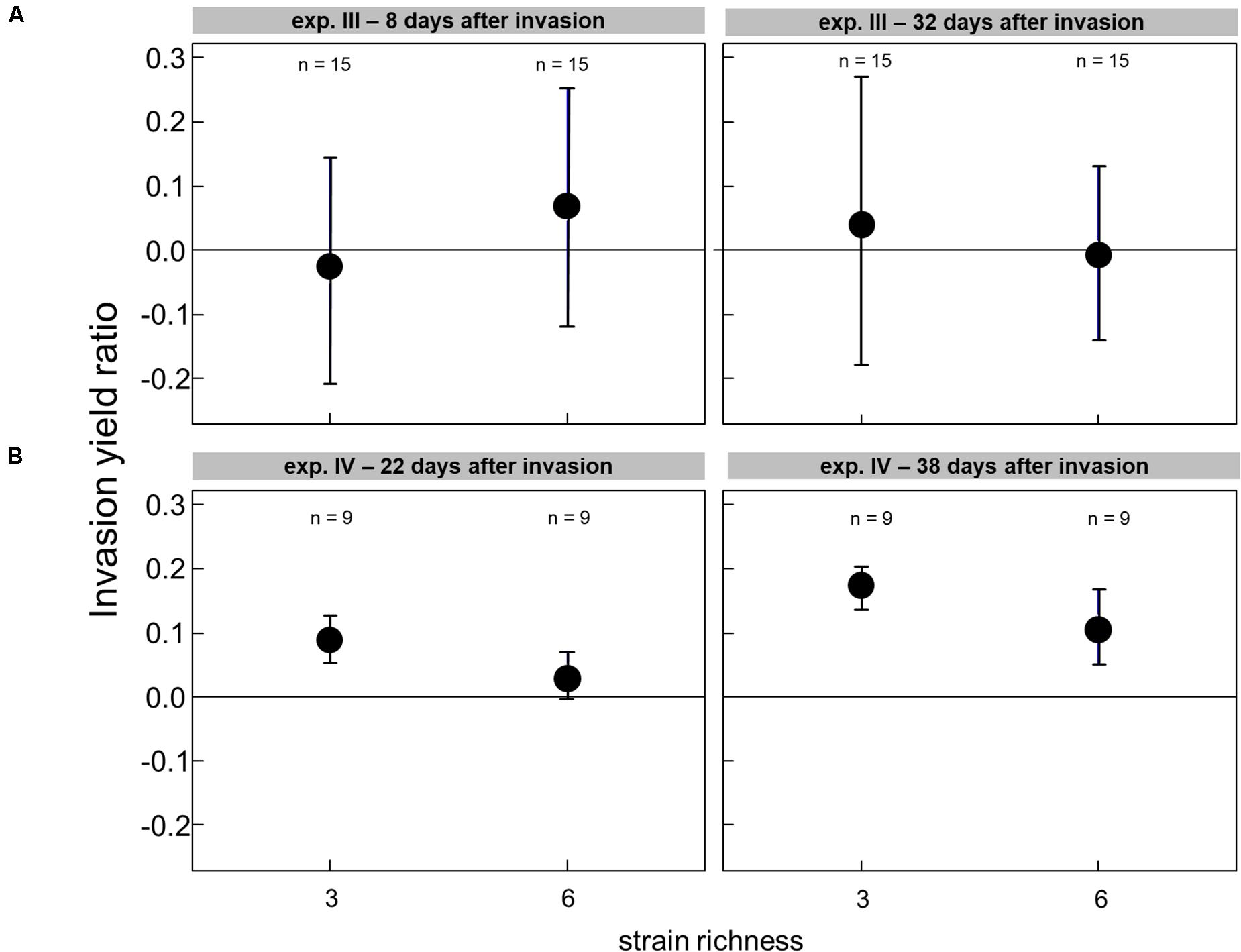
Figure 4. The invasion yield of Raphidiopsis raciborskii in experiments with Cephalodella sp. (experiment III) (A) and extinction of Brachionus calyciflorus (experiment IV) (B) for the diversity levels of three and six strains.
Temporal Dynamics of the Community
Over the course of the experiments, the resident community composition changed and was partly affected by the invasion of R. raciborskii. When R. raciborskii invaded the community without consumers (experiment IV), Chlorella vulgaris, Monoraphidium minutum, Navicula pelliculosa, and Oocystis marsonii had significant different biomasses among invader diversity treatments (ANOVA or Kruskal–Wallis test, p < 0.009): C. vulgaris increased, whereas the other species decreased.
In all four experiments, some species eventually went extinct: Aphanizomen gracile (experiment IV), Stephanodiscus hantzschii (experiments I–III), Peridinium sp. (experiment I–III), and Planktothrix aghardii (experiment I–III). Among the persisting species, the cell size (cell volume for algae; filament width for filamentous cyanobacteria) changed from the experiment start to the end: R. raciborskii and P. aghardii decreased their filaments width (R. raciborskii: 1.8 to 0.9 μm; t-test, p = 0.018; P. aghardii: 5.8 to 4.8 μm; t-test, p = 0.329), the chlorophytes Acutodesmus obliquus (67 to 105 μm3; t-test, p = 0.123) and Chlamydomonas reinhardtii (235 to 416 μm3; t-test, p = 0.014) increased their cell size (see Supplementary Table 2).
Strain-Specific Changes of the Resident Composition
We found an effect of the strain identity on the community compositions at the end of experiment III (Table 1), where R. raciborskii successfully invaded. This resulted in a higher similarity of the resident communities invaded by the same strain (one strain treatments replicates) than for those invaded by different strains, as observed in the PCA of the community composition data in experiment III (in absolute biomass) excluding the invader, day 38 (Figure 5). In addition, the uninvaded control was consistently different from all other invaded treatments (with one exception), indicating a directed change in the resident community after invasion.
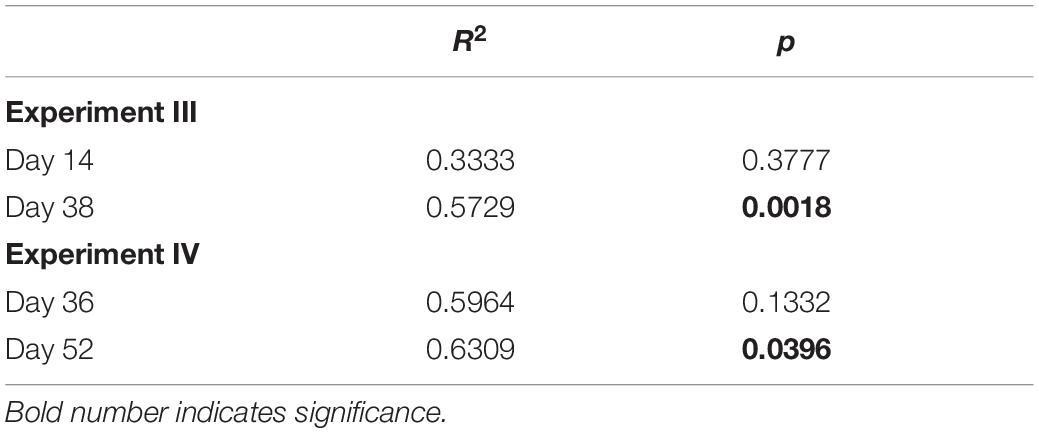
Table 1. Results of PERMANOVA on the effect of strain identity on resident community composition for experiments III and IV.
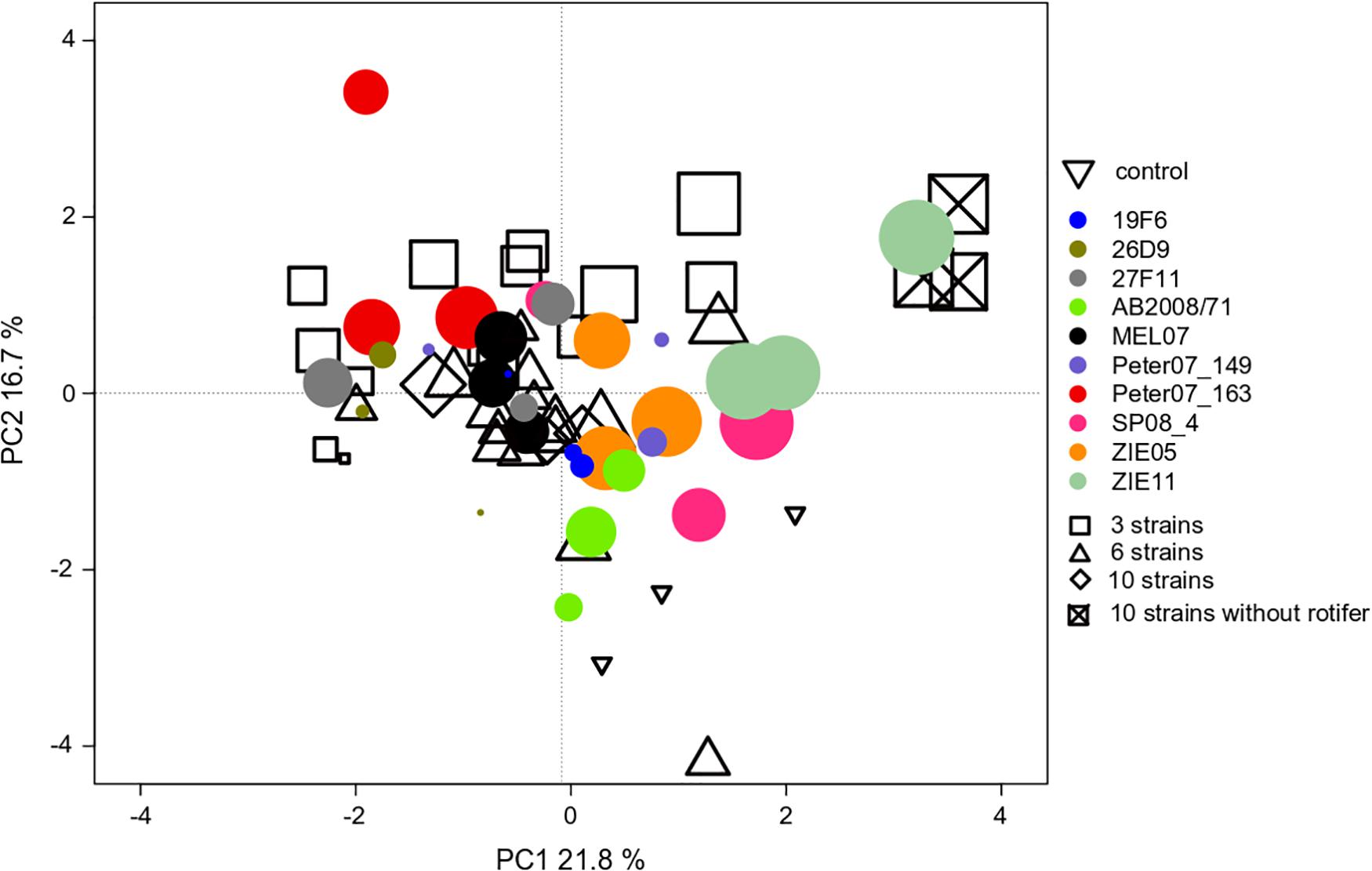
Figure 5. PCA ordination of the individual microcosm community compositions in experiment III with Cephalodella sp. on day 38 (69 microcosms = points in total). The size of the filled circles reflects the biomass contribution of Raphidiopsis raciborskii to the whole phytoplankton community (but not implemented in the ordination). The colors of the filled circles indicate different strains, open symbols indicate the strain richness treatments (3, 6, and 10 strains of genetic mixtures). The open triangles represent the uninvaded control (no invasion of R. raciborskii).
For experiment III, the individual strain share in the invading populations at the time point of the invasion event and at the end of the experiment was determined. The comparison of the relative community share of each strain between start and end revealed that some strains consistently gained and others lost in their relative share (Supplementary Figure 2). In eight mixtures (6 × 3 strains, 2 × 6 strains), one strain was dominant at the end, with a share of more than 66%, including the strains MEL07, Peter07_149, ZIE05, and ZIE11 (Supplementary Figure 3). Apart from a few exceptions, the success of specific strains in mixtures agreed with the success in single strain experiments (Figure 3) with a positive Spearman rank correlation (rho = 0.504, significant at p < 0.01; Figure 6). One extreme outlier (see Figure 6 marked with an asterisk) was result of a low relative share of the strain ZIE11 at the day of the invasion event. Nevertheless, the positive Spearman rank correlation remains significant (rho = 0.496, significant at p < 0.01) even when that outlier is included.
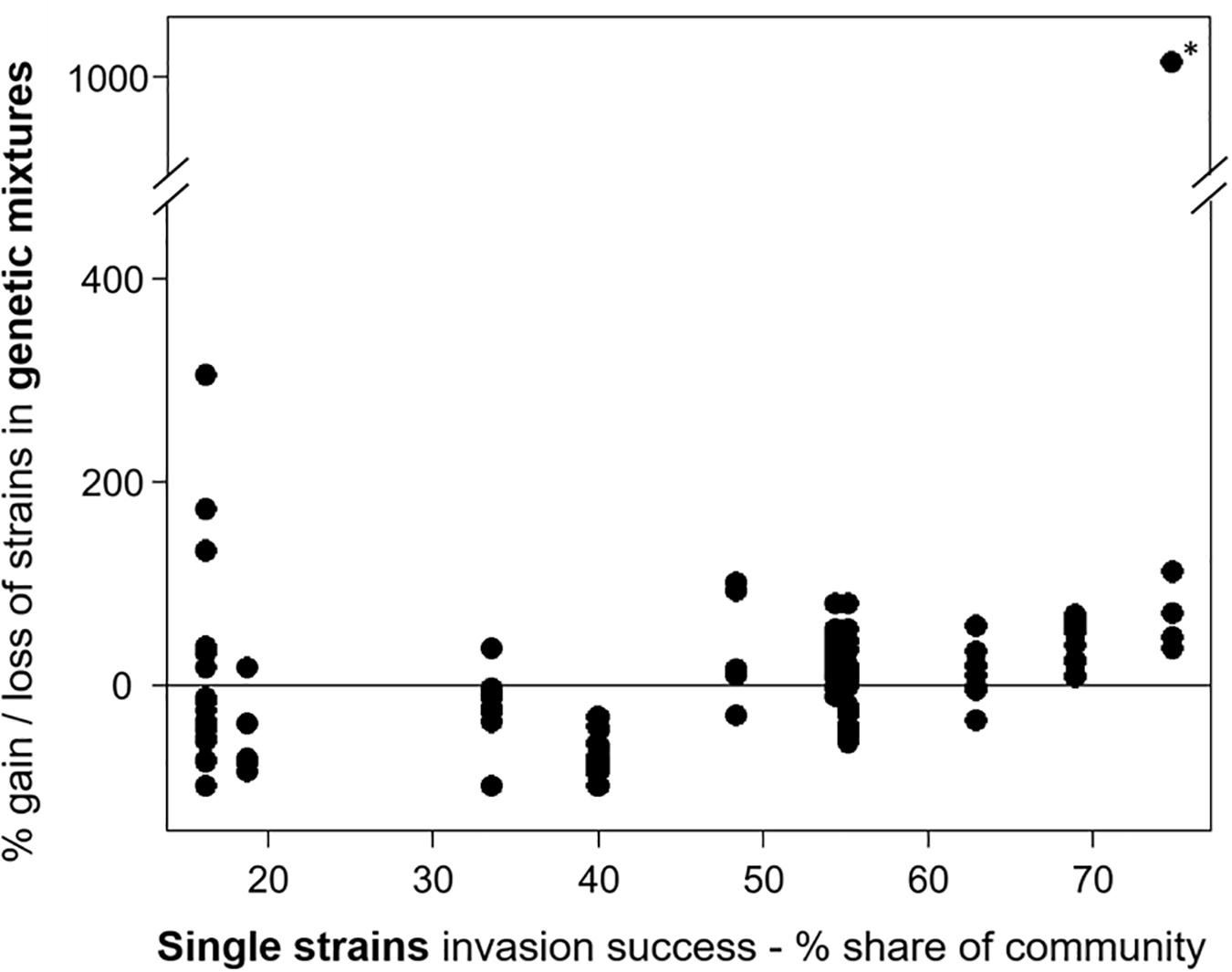
Figure 6. The invasion success of the Raphidiopsis raciborskii strains as their gain or loss in the genetic mixtures (start–end, measurement of invasion success) versus single strains. Asterisk marks the extreme outlier.
Nutrients
At the end of experiment III all microcosms with R. raciborskii exceeded the N-concentration of the medium (51 μmol), by, on average, 78 μmol and the biomass of R. raciborskii was positively correlated with the particulate N (linear regression: R2 = 0.633, p < 0.001), indicating N2-fixation of R. raciborskii. Both, the molar C:N (8:1 to 17:1) and C:P (393:1 to 1,223:1) ratios were considerably higher than the Redfield ratio of 6.6:1 (C:N) 106:1 (C:P), indicating substantial nutrient limitation. In contrast, the controls without the N-fixing R. raciborskii had a nearly two-fold higher C:N ratio, likely because of the lack of N-fixation.
Discussion
High Grazing Pressure Can Prevent Aquatic Invasions
Our results have demonstrated an invasion resistance of resident communities driven by consumption, which is consistent with previous studies (Sperfeld et al., 2010; Alofs and Jackson, 2014; Weithoff et al., 2017). The observed consumptive resistance was somewhat surprising given the highly variable susceptibility to herbivory of the strains we used (Bolius et al., 2017). The comparison of different top-down pressures and types (high vs low and generalist vs specialist) clearly showed that one suitable consumer in the system hampers invasions, as here, most often the generalist (Maron and Vilà, 2001). The high grazing pressure by the generalist rotifer B. calyciflorus utterly prevented the invasion, even though R. raciborskii is a poor single food source for zooplankton (de Bernardi and Giussani, 1990; Panosso and Lürling, 2010). However, a mixed diet with highly edible green algae (Soares et al., 2010) and a low density of R. raciborskii at the invasion event (Panosso and Lürling, 2010; Soares et al., 2010; Sperfeld et al., 2010; Rangel et al., 2016) allowed for persistent herbivory. In contrast, the smaller, specialist consumer Cephalodella sp. could not prevent the invasion and R. raciborskii invaded successfully the communities (experiment III), with no obvious impact of the consumer. Due to its smaller size and likely specific grazing on C. reinhardtii, Cephalodella sp. potentially fed much less on R. raciborskii than the large, generalist B. calyciflorus.
Strain-Specific Invasion Success Can Be the Result of a Higher Resource-Use Efficiency
The overall invasion success of R. raciborskii varied among strains (Figure 3). This strain-specific invasion effect was illustrated by the consistency between the two independent experiments III and IV, even though the resident communities in the two experiments differed in their phytoplankton species composition and the herbivore type and pressure. Such a consistent behavior among the strain’s invasion success was also found between their invasion success as single strains and within genetic mixtures (experiment III): the highly successful single strains also reached a higher gain in the relative share in the invading populations. This high variability in the invasion success is a reflection of their high trait variability. In a previous study, Bolius et al. (2017) found for these strains a high variability in morphology, resource use efficiently and susceptibility to herbivory (see above).
Our data indicate that the most relevant trait for a successful invasion was the intracellular N-content resulting from N-fixation (see Supplementary Table 4) and their differences among the strains. The strain-specific N-fixation correlated positively with the strains’ C:N ratio under nutrient-depleted conditions (Bolius et al., 2017), suggesting a more efficient resource use efficiency and high biomass production of some strains. Another possible supporting mechanism might have been the decrease in filament width over the time course of the experiment. Thinner filaments result in a higher surface to volume ratio, allowing for a better nutrient uptake per unit volume, increasing their competitive abilities (Friebele et al., 1978).
Weak Effect of the Genetic Diversity on the Invasion
In most studies, a positive effect of the genetic diversity on the invasion of species is documented (see the meta-analysis in Forsman, 2014). In our study, we found a strain-specific invasion success, and only a weak effect of genetic diversity (strain richness). This resulted in a relatively constant average invasion success at all diversity levels (experiments III and IV; Figures 2B,C). However, a higher genetic diversity lowered the probability of a very low invasion success, because the presence of one or more successful strains is very likely and, thus, at a strain richness of 10 strains all invading populations had an intermediate invasion success (32 days after invasion, Figure 2B).
In general, positive effects of diversity during invasion are expected, because a high diversity results in a greater chance of including a highly invasive strain, known as portfolio or sampling effect. A low variability among invading strains would result in a low sampling effect due to a limited chance of including a strong competitor. Forsman (2014) showed in a meta-analysis (including 18 studies) a larger effect of the genetic diversity under natural conditions in the field/wild compared to laboratory experiments. He explained this effect by a higher environmental complexity. In our experiments, complexity was reflected by generating a multi-species resident community and by adding a second trophic level to mimic natural conditions. However, we used microcosms with a reduced spatial complexity typically favoring direct interactions over indirect ones (Sarnelle, 1997). Thus, the result of our study might be different from the dominant outcome in the meta-analysis from Forsman (2014).
Alternatively, an increase or a shift in resource use efficiency might occur with increasing strain richness (complementary effect; Fargione and Tilman, 2005). In our study, we found highly invasive strains in both single-strain treatments and in multi-strain populations. To maintain the total invader inoculum constant across all treatments, the individual invader/strain inoculum was ten times higher in the single-strain treatments than in the 10-strain mixtures (0.1 vs 0.01 μg ml–1). As a consequence, competitively successful strains started from a lower population density in mixtures and were accompanied by competitively inferior strains. Since maximal population growth rates under comparable laboratory conditions are at a low to intermediate level ranging from 0.2 to 0.4 d–1, it is likely that superior strains might outcompete others in the long run. At purely competitive conditions (= no consumers), Weis et al. (2007) found that the effect of species richness on biomass production can vary over time and depends on the growth rate-biomass production relationship of the competing species and also on the experimental design: additive versus substitutive (as in the present study). However, in our experiments, in experiment III in particular, the strain-specific effect is most likely much larger than the diversity effect.
In experiment IV, with a different resident community (Supplementary Table 1), the variability among the single-strain-specific invasion success was much lower than in experiment III (Figure 2). Since the consumers went extinct, no nutrient regeneration through feeding took place. Under these competitive conditions, a slightly higher observed invasion success was found than expected from single-strain experiments pointing to a potential complementarity effect (Figure 4).
The establishment of one strong species potentially can lead to an alteration of the resident community allowing otherwise less competitive species also to invade, known as invasional meltdown (Simberloff and Von Holle, 1999), and often observed on the species-level (Jeschke et al., 2012). Transferring this mechanism to our system, we found that weak invaders identified from the single-strain experiments also lost in their relative share over the course of the experiment (Supplementary Figures 2, 3), thus no invasional meltdown was observed in our study. A high invasion success was correlated with high intra-cellular nitrogen content and thus associated with nitrogen-fixation. Another factor for the strain-specific success might be allelopathy of R. raciborskii (Figueredo et al., 2007; Antunes et al., 2012). However, for the strains used in this study, allelopathy was not examined.
Individual Strains Differentially Affect Resident Communities
We found a strain-specific invasion success and additionally a strain-specific differentiation of the resident community compositions (Figure 5 and Table 1), which was independent of the quantity of the invasion success (e.g., Hejda et al., 2009). A similar result was found in a study on the cord-grass Spartina sp., showing strain-specific responses of the neighboring plants (Zerebecki et al., 2017). Since the strains used in our study have a high variability in their traits (Bolius et al., 2017), differential effects on the resident community and their individual species could be expected. The invading strains differed, e.g., in their resource use efficiency, which might have differentially affected the resident communities. In general, this interaction between the specific invading strain and the resident species might then vary in a community context. One important consequence would be that different results are obtained from different experiments with different strains regarding the invasion process and success of species, especially in plastic species like R. raciborskii. This might lead to contrasting results on the invasion potential of species.
Conclusion
To conclude, our experiments revealed that (i) a high level of herbivory hampered invasion pointing to consumptive resistance; (ii) a strong strain-specific impact on the invasion success of the invader, with only a minor effect of genetic diversity of the invading population; and (iii) a strain-specific impact on resident community composition. For R. raciborskii, but also for many other invasive species, genotype-specific pre-adaptation might play an important role for the establishment in a new environment. Since individual strains affect resident communities differently, their long-term persistence might also be different due differences in the competitive arena. In the long run, genetic bottlenecks might increase in importance, if the environment changes and genetic diversity is a key for long-term persistence. Strain-specificity in the invasion process and its potential consequences for long-term persistence under changing environments is a promising research avenue for future studies.
Data Availability Statement
The datasets for this study can be found at https://doi.org/10.5061/dryad.d2547d811.
Author Contributions
GW, CW, and SB conceived the study. SB and KM performed the experiments. SB, KM, and GW analyzed the data. SB and GW wrote the manuscript supported by comments from KM and CW. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Stefanie Hartmann for the bioinformatic analysis and Alexandra Trinks und Michaela Preick for assistance with library preparation and sequencing. We thank Katrin Hinrichs for her assistance with DNA extraction and PCRs. We thank Victoriia Radchuk and Beatrix Beisner for constructive comments on the manuscript. Furthermore, we thank Christina Schirmer, Sabine Donath, Silvia Heim, and Anne Taube for great technical assistance with the experiments. We acknowledge the support of Deutsche Forschungsgemeinschaft (German Science Foundation, WE2556/9-1) and Open Access Publication Fund of University of Potsdam.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01598/full#supplementary-material
References
Abreu, V. A. C., Popin, R. V., Alvarenga, D. O., Schaker, P. D. C., Hoff-Risseti, C., Varani, A. M., et al. (2018). Genomic and genotypic characterization of Cylindrospermopsis raciborskii: toward an intraspecific phylogenetic evaluation by comparative genomics. Front. Microbiol. 9:306. doi: 10.3389/fmicb.2018.00306
Aguilera, A., Gómez, E. B., Kaštovskı, J., Echenique, R. O., and Salerno, G. L. (2018). The polyphasic analysis of two native Raphidiopsis isolates supports the unification of the genera Raphidiopsis and Cylindrospermopsis (Nostocales, Cyanobacteria). Phycologia 57, 130–146. doi: 10.2216/17-2.1
Ahlroth, P., Alatalo, R. V., Holopainen, A., Kumpulainen, T., and Suhonen, J. (2003). Founder population size and number of source populations enhance colonization success in waterstriders. Oecologia 137, 617–620. doi: 10.1007/s00442-003-1344-y
Allendorf, F. W., and Lundquist, L. L. (2003). Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30. doi: 10.1046/j.1523-1739.2003.02365.x
Alofs, K. M., and Jackson, D. A. (2014). Meta-analysis suggests biotic resistance in freshwater environments is driven by consumption rather than competition. Ecology 95, 3259–3270. doi: 10.1890/14-0060.1.sm
Altermatt, F., Schreiber, S., and Holyoak, M. (2011). Interactive effects of disturbance and dispersal directionality on species richness and composition in metacommunities. Ecology 92, 859–870. doi: 10.1890/10-1095.1
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Antunes, J. T., Leão, P. N., and Vasconcelos, V. M. (2012). Influence of biotic and abiotic factors on the allelopathic activity of the cyanobacterium Cylindrospermopsis raciborskii strain LEGE 99043. Microb. Ecol. 64, 584–592. doi: 10.1007/s00248-012-0061-7
Antunes, J. T., Leão, P. N., and Vasconcelos, V. M. (2015). Cylindrospermopsis raciborskii: review of the distribution, phylogeography, and ecophysiology of a global invasive species. Front. Microbiol. 6:473. doi: 10.3389/fmicb.2015.00473
Barrett, L. G., Kniskern, J. M., Bodenhausen, N., Zhang, W., and Bergelson, J. (2009). Continua of specificity and virulence in plant host-pathogen interactions: causes and consequences. New Phytol. 183, 513–529. doi: 10.1111/j.1469-8137.2009.02927.x
Bolius, S., Wiedner, C., and Weithoff, G. (2017). High local trait variability in a globally invasive cyanobacterium. Freshw. Biol. 62, 1879–1890. doi: 10.1111/fwb.13028
Budzyńska, A., and Gołdyn, R. (2017). Domination of invasive Nostocales (Cyanoprokaryota) at 52°N latitude. Phycol. Res. 65, 322–332. doi: 10.1111/pre.12188
Crawford, K. M., and Whitney, K. D. (2010). Population genetic diversity influences colonization success. Mol. Ecol. 19, 1253–1263. doi: 10.1111/j.1365-294X.2010.04550.x
Crawley, M. J., Brown, S. L., Heard, M. S., and Edwards, G. R. (1999). Invasion-resistance in experimental grassland communities: species richness or species identity? Ecol. Lett. 2, 140–148. doi: 10.1046/j.1461-0248.1999.00056.x
Davidson, A. M., Jennions, M., and Nicotra, A. B. (2011). Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecol. Lett. 14, 419–431. doi: 10.1111/j.1461-0248.2011.01596.x
de Bernardi, R., and Giussani, G. (1990). Are blue-green algae a suitable food for zooplankton? An overview. Hydrobiologia 200, 29–41. doi: 10.1007/BF02530326
DeRivera, C. E., Ruiz, G. M., Hines, A. H., and Jivoff, P. (2005). Biotic resistance to invasion: native predator limits abundance and distribution of an introduced crab. Ecology 86, 3364–3376. doi: 10.1890/05-0479
Engel, K., Tollrian, R., and Jeschke, J. M. (2011). Integrating biological invasions, climate change, and phenotypic plasticity. Commun. Integr. Biol. 4, 247–250. doi: 10.4161/cib.4.3.14885
Fargione, J. E., and Tilman, D. (2005). Diversity decreases invasion via both sampling and complementarity effects. Ecol. Lett. 8, 604–611. doi: 10.1111/j.1461-0248.2005.00753.x
Figueredo, C. C., Giani, A., and Bird, D. F. (2007). Does allelopathy contribute to Cylindrospermopsis raciborskii (cyanobacteria) bloom occurrence and geographic expansion? J. Phycol. 43, 256–265. doi: 10.1111/j.1529-8817.2007.00333.x
Forsman, A. (2014). Effects of genotypic and phenotypic variation on establishment are important for conservation, invasion, and infection biology. Proc. Natl. Acad. Sci. U.S.A. 111, 302–307. doi: 10.1073/pnas.1317745111
Friebele, E. S., Correll, D. L., and Faust, M. A. (1978). Relationship between phytoplankton cell size and the rate of orthophosphate uptake: in situ observations of an estuarine population. Mar. Biol. 45, 39–52. doi: 10.1007/BF00388976
Fussmann, G. F., Weithoff, G., and Yoshida, T. (2005). A direct, experimental test of resource vs. consumer dependence. Ecology 86, 2924–2930. doi: 10.1890/06-1692
González-Suárez, M., Bacher, S., and Jeschke, J. M. (2015). Intraspecific trait variation is correlated with establishment success of alien mammals. Am. Nat. 185, 737–746. doi: 10.1086/681105
Hamilton, P. B., Ley, L. M., Dean, S., and Pick, F. R. (2005). The occurrence of the cyanobacterium Cylindrospermopsis raciborskii in constance lake: an exotic cyanoprokaryote new to Canada. Phycologia 44, 17–25. doi: 10.2216/0031-8884(2005)44[17:tootcc]2.0.co;2
Hejda, M., Pyšek, P., and Jarošík, V. (2009). Impact of invasive plants on the species richness, diversity and composition of invaded communities. J. Ecol. 97, 393–403. doi: 10.1111/j.1365-2745.2009.01480.x
Hillebrand, H., Dürselen, C.-D., Kirschtel, D., Pollingher, U., and Zohary, T. (1999). Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 35, 403–424. doi: 10.1046/j.1529-8817.1999.3520403.x
Horňák, K., and Corno, G. (2012). Every coin has a back side: invasion by Limnohabitans planktonicus promotes the maintenance of species diversity in bacterial communities. PLoS One 7:e51576. doi: 10.1371/journal.pone.0051576
Jeschke, J., Gómez Aparicio, L., Haider, S., Heger, T., Lortie, C., Pyšek, P., et al. (2012). Support for major hypotheses in invasion biology is uneven and declining. NeoBiota 14, 1–20. doi: 10.3897/neobiota.14.3435
Jeschke, J. M., and Heger, T eds (2018). Invasion Biology: Hypotheses and Evidence. Wallingford: CABI.
Jeschke, J. M., and Strayer, D. L. (2005). Invasion success of vertebrates in Europe and North America. Proc. Natl. Acad. Sci. U.S.A. 102, 7198–7202. doi: 10.1073/pnas.0501271102
John, J. (2011). SeqPrep: Tool for Stripping Adaptors and/or Merging Paired Reads with Overlap into Single Reads. Available online at: https://github.com/jstjohn/SeqPrep (accessed October 5, 2016).
Juliano, S. A., Lounibos, L. P., Nishimura, N., and Greene, K. (2010). Your worst enemy could be your best friend: predator contributions to invasion resistance and persistence of natives. Oecologia 162, 709–718. doi: 10.1007/s00442-009-1475-x
Kolbe, J. J., Glor, R. E., Rodríguez Schettino, L., Lara, A. C., Larson, A., and Losos, J. B. (2004). Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181. doi: 10.1038/nature02807
Lemaire, V., Brusciotti, S., van Gremberghe, I., Vyverman, W., Vanoverbeke, J., and De Meester, L. (2012). Genotype x genotype interactions between the toxic cyanobacterium Microcystis and its grazer, the waterflea Daphnia. Evol. Appl. 5, 168–182. doi: 10.1111/j.1752-4571.2011.00225.x
Litchman, E. (2010). Invisible invaders: non-pathogenic invasive microbes in aquatic and terrestrial ecosystems. Ecol. Lett. 13, 1560–1572. doi: 10.1111/j.1461-0248.2010.01544.x
Lockwood, J. L., Cassey, P., and Blackburn, T. M. (2009). The more you introduce the more you get: the role of colonization pressure and propagule pressure in invasion ecology. Divers. Distrib. 15, 904–910. doi: 10.1111/j.1472-4642.2009.00594.x
Lonsdale, W. M. (1999). Global patterns of plant invasions and the concept of invasibility. Ecology 80, 1522–1536. doi: 10.1890/0012-9658(1999)080[1522:gpopia]2.0.co;2
Mächler, E., and Altermatt, F. (2012). Interaction of species traits and environmental disturbance predicts invasion success of aquatic microorganisms. PLoS One 7:e45400. doi: 10.1371/journal.pone.0045400
Mallon, C. A., Van Elsas, J. D., and Salles, J. F. (2015). Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23, 719–729. doi: 10.1016/j.tim.2015.07.013
Markensten, H., Moore, K., and Persson, I. (2010). Simulated lake phytoplankton composition shifts toward cyanobacteria dominance in a future warmer climate. Ecol. Appl. 20, 752–767. doi: 10.1890/08-2109.1
Maron, J. L., and Vilà, M. (2001). When do herbivores affect plant invasion? Evidence for the natural enemies and biotic resistance hypotheses. Oikos 95, 361–373. doi: 10.1034/j.1600-0706.2001.950301.x
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Nichols, W. H. (1973). Growth Media – Freshwater. Handbook of Phycological Methods. Culture Methods Growth Measurements. Available online at: http://ci.nii.ac.jp/naid/10004055104/ (accessed August 2, 2017).
Padisák, J. (1997). Cylindrospermopsis raciborskii (Woloszynska) Seenayya et Subba Raju, an expanding, highly adaptive cyanobacterium: worldwide distribution and review of its ecology. Arch. Für Hydrobiol. Suppl. Monogr. Beitrage 107, 563–593.
Panosso, R., and Lürling, M. (2010). Daphnia magna feeding on Cylindrospermopsis raciborskii: the role of food composition, filament length and body size. J. Plankton Res. 32, 1393–1404. doi: 10.1093/plankt/fbq057
Paraskevopoulou, S., Tiedemann, R., and Weithoff, G. (2018). Differential response to heat stress among evolutionary lineages of an aquatic invertebrate species complex. Biol. Lett. 14:20180498. doi: 10.1098/rsbl.2018.0498
Petrie, S. A., and Knapton, R. W. (1999). Rapid increase and subsequent decline of Zebra and Quagga mussels in Long Point Bay, Lake Erie: possible influence of waterfowl predation. J. Great Lakes Res. 25, 772–782. doi: 10.1016/S0380-1330(99)70776-8
Pyšek, P., Richardson, D. M., Pergl, J., Jarošík, V., Sixtová, Z., and Weber, E. (2008). Geographical and taxonomic biases in invasion ecology. Trends Ecol. Evol. 23, 237–244. doi: 10.1016/j.tree.2008.02.002
Rangel, L. M., Ger, K. A., Silva, L. H. S., Soares, M. C. S., Faassen, E. J., and Lürling, M. (2016). Toxicity overrides morphology on Cylindrospermopsis raciborskii grazing resistance to the calanoid copepod Eudiaptomus gracilis. Microb. Ecol. 71, 835–844. doi: 10.1007/s00248-016-0734-8
Ricciardi, A., and MacIsaac, H. J. (2010). “Impacts of biological invasions on freshwater ecosystems,” in Fifty Years of Invasion Ecology: The Legacy of Charles Elton, ed. D. M. Richardson (Oxford: Wiley-Blackwell), 211–224. doi: 10.1002/9781444329988.ch16
Richards, C. L., Bossdorf, O., Muth, N. Z., Gurevitch, J., and Pigliucci, M. (2006). Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9, 981–993. doi: 10.1111/j.1461-0248.2006.00950.x
Roman, J., and Darling, J. A. (2007). Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464. doi: 10.1016/j.tree.2007.07.002
Romanuk, T. N., and Kolasa, J. (2005). Resource limitation, biodiversity, and competitive effects interact to determine the invasibility of rock pool microcosms. Biol. Invasions 7, 711–722. doi: 10.1007/s10530-004-0997-8
Sakai, A. K., Allendorf, F. W., Holt, J. S., Lodge, D. M., Molofsky, J., With, K. A., et al. (2001). The population biology of invasive species. Annu. Rev. Ecol. Syst. 32, 305–332. doi: 10.1146/annurev.ecolsys.32.081501.114037
Sala, O. E. (2000). Global biodiversity scenarios for the year 2100. Science 287, 1770–1774. doi: 10.1126/science.287.5459.1770
Sarnelle, O. (1997). Daphnia effects on microzooplankton: comparisons of enclosure and whole-lake responses. Ecology 78, 913–928. doi: 10.1890/0012-9658(1997)078[0913:deomco]2.0.co;2
Seifert, L. I., Weithoff, G., and Vos, M. (2015). Extreme heat changes post-heat wave community reassembly. Ecol. Evol. 5, 2140–2148. doi: 10.1002/ece3.1490
Simberloff, D., and Von Holle, B. (1999). Positive interactions of nonindigenous species: invasional meltdown? Biol. Invasions 1, 21–32. doi: 10.1023/A:1010086329619
Soares, M. C. S., Lürling, M., and Huszar, V. L. M. (2010). Responses of the rotifer Brachionus calyciflorus to two tropical toxic cyanobacteria (Cylindrospermopsis raciborskii and Microcystis aeruginosa) in pure and mixed diets with green algae. J. Plankton Res. 32, 999–1008. doi: 10.1093/plankt/fbq042
Sperfeld, E., Schmidtke, A., Gaedke, U., and Weithoff, G. (2010). Productivity, herbivory, and species traits rather than diversity influence invasibility of experimental phytoplankton communities. Oecologia 163, 997–1010. doi: 10.1007/s00442-010-1594-4
Stachowicz, J. J., Fried, H., Osman, R. W., and Whitlatch, R. B. (2002). Biodiversity, invasion resistance, and marine ecosystem function: reconciling pattern and process. Ecology 83, 2575–2590. doi: 10.2307/3071816
Stachowicz, J. J., Whitlatch, R. B., and Osman, R. W. (1999). Species diversity and invasion resistance in a marine ecosystem. Source Sci. New Ser. 286, 1577–1579. doi: 10.1126/science.286.5444.1577
Strayer, D. L. (2010). Alien species in fresh waters: ecological effects, interactions with other stressors, and prospects for the future. Freshw. Biol. 55, 152–174. doi: 10.1111/j.1365-2427.2009.02380.x
Strayer, D. L., D’Antonio, C. M., Essl, F., Fowler, M. S., Geist, J., Hilt, S., et al. (2017). Boom-bust dynamics in biological invasions: towards an improved application of the concept. Ecol. Lett. 20, 1337–1350. doi: 10.1111/ele.12822
Sukenik, A., Hadas, O., Kaplan, A., and Quesada, A. (2012). Invasion of Nostocales (cyanobacteria) to subtropical and temperate freshwater lakes – physiological, regional, and global driving forces. Front. Microbiol. 3:86. doi: 10.3389/fmicb.2012.00086
Therriault, T. W., Orlova, M. I., Docker, M. F., MacIsaac, H. J., and Heath, D. D. (2005). Invasion genetics of a freshwater mussel (Dreissena rostriformis bugensis) in eastern Europe: high gene flow and multiple introductions. Heredity (Edinb) 95, 16–23. doi: 10.1038/sj.hdy.6800691
Van Gremberghe, I., Vanormelingen, P., Vanelslander, B., Van Der Gucht, K., D’Hondt, S., De Meester, L., et al. (2009). Genotype-dependent interactions among sympatric Microcystis strains mediated by Daphnia grazing. Oikos 118, 1647–1658. doi: 10.1111/j.1600-0706.2009.17538.x
Vilà, M., Espinar, J. L., Hejda, M., Hulme, P. E., Jarošík, V., Maron, J. L., et al. (2011). Ecological impacts of invasive alien plants: a meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 14, 702–708. doi: 10.1111/j.1461-0248.2011.01628.x
Weis, J. J., Cardinale, B. J., Forshay, K. J., and Ives, A. R. (2007). Effects of species diversity on community biomass production change over the course of succession. Ecology 88, 929–939. doi: 10.1890/06-0943
Weithoff, G., Taube, A., and Bolius, S. (2017). The invasion success of the cyanobacterium Cylindrospermopsis raciborskii in experimental mesocosms: genetic identity, grazing loss, competition and biotic resistance. Aquat. Invasions 12, 1879–1890. doi: 10.3391/ai.2017.12.3.07
Wiedner, C., Rücker, J., Brüggemann, R., and Nixdorf, B. (2007a). Climate change affects timing and size of populations of an invasive cyanobacterium in temperate regions. Oecologia 152, 473–484. doi: 10.1007/s00442-007-0683-5
Wiedner, C., Rücker, J., Stüken, A., Preußel, K., Fastner, J., Chorus, I., et al. (2007b). Cylindrospermopsis raciborskii and Cylindrospermopsin in the Lakes of the Berlin Area, ed. C. Wiedner (Berlin: Kompetenzzentrum Wasser Berlin).
Keywords: alien species, genetic diversity, genotype, invasibility, cyanobacteria, consumptive resistance, phytoplankton, Raphidiopsis
Citation: Bolius S, Morling K, Wiedner C and Weithoff G (2020) Genetic Identity and Herbivory Drive the Invasion of a Common Aquatic Microbial Invader. Front. Microbiol. 11:1598. doi: 10.3389/fmicb.2020.01598
Received: 31 December 2019; Accepted: 18 June 2020;
Published: 13 July 2020.
Edited by:
Marcelino T. Suzuki, Sorbonne Universités, FranceReviewed by:
Jorge T. Antunes, University of Porto, PortugalMaria Stockenreiter, Ludwig Maximilian University of Munich, Germany
Copyright © 2020 Bolius, Morling, Wiedner and Weithoff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah Bolius, Ym9saXVzQHVuaS1wb3RzZGFtLmRl; Guntram Weithoff, d2VpdGhvZmZAdW5pLXBvdHNkYW0uZGU=
 Sarah Bolius
Sarah Bolius Karoline Morling2
Karoline Morling2 Guntram Weithoff
Guntram Weithoff