- Microbiology Lab, Biotechnology Programme, M. S. Swaminathan Research Foundation, Chennai, India
Biodegradation of phenol using bacteria is recognized as an efficient, environmentally friendly and cost-effective approach for reducing phenol pollutants compared to the current conventional physicochemical processes adopted. A potential phenol degrading bacterial strain Glutamicibacter nicotianae MSSRFPD35 was isolated and identified from Canna indica rhizosphere grown in distillery effluent contaminated sites. It showed high phenol degrading efficiency up to 1117 mg L–1 within 60 h by the secretion of catechol 1,2-dioxygenase via ortho intradial pathway. The strain MSSRFPD35 possess both the catechol 1,2 dioxygenase and catechol 2,3 dioxygenase coding genes that drive the ortho and meta pathways, but the enzymatic assay revealed that the strain cleaves catechol via ortho pathway. Haldane’s kinetic method was well fit to exponential growth data and the following kinetic parameter was obtained: μ∗ = 0.574 h–1, Ki = 268.1, Ks = 20.29 mg L–1. The true μmax and Sm were calculated as 0.37 h–1 and 73.76 mg L–1, respectively. The Haldane’s constant values were similar to earlier studies and healthy fitness depicted in correlation coefficient value R2 of 0.98. Phenol degrading kinetic’s was predicted using Haldane’s model as qmax 0.983, Ki′ 517.5 and Ks′ 9.152. Further, MSSRFPD35 was capable of utilizing different monocyclic and polycyclic aromatic hydrocarbons and to degrade phenol in the presence of different heavy metals. This study for the first time reports high phenol degrading efficiency of G. nicotianae MSSRFPD35 in the presence of toxic heavy metals. Thus, the strain G. nicotianae MSSRFPD35 can be exploited for the bioremediation of phenol and its derivatives polluted environments, co-contaminated with heavy metals.
Introduction
Phenol and its derivatives namely nitrophenol, halogenated phenol, alkylphenol, etc. are widely used in several industrial plants like petrochemical, phenol resin, pharmaceuticals, paint, textile, leather, pulp mills, coal conversion, and leather processing units (Haddadi and Shavandi, 2013; Jiang et al., 2013; Villegas et al., 2016; Prasse et al., 2018). Gallons of unprocessed effluents polluted with phenol and its derivatives discharged by these industries are reported to contaminate soil, groundwater table and agriculture lands; and to harm the soil and plant health and productivity (Shi et al., 2014; Wu et al., 2018) and also to affect terrestrial and aquatic animals, and humans at very low concentrations (Prasse et al., 2018).
Phenolic compounds have a recalcitrant structure, which consists of an aromatic ring with a hydroxyl group attached to the benzene ring; making it resistant to natural biodegradation and decomposition (Reardon et al., 2000; Ma et al., 2013; Chen et al., 2017). Phenol is highly soluble in water up to the concentration of 10 g L–1 (Bajaj et al., 2009) and thus the effluents discharged from industries contain high concentrations of phenol and its derivatives in the range of 50–2000 mg L–1 (Jusoh and Razali, 2008) concentrations much higher than the permissible limits leading to high risk of polluting the environment. The permissible limits of phenol in industrial effluents to be discharged in the domestic surface water is only 1 mg L–1 (IS: 2490-1974) and 5 mg L–1 in public sewers (IS: 3306-1974) (Hussain et al., 2015) and concentration range of 5–2000 mg L–1 phenol is reported to be carcinogenic for human and toxic to all life forms (Comte et al., 2013).
In addition to phenol, the effluent released from industries contain heavy metals like copper, lead, cadmium, chromium, etc., as co-contaminants in different composition and concentration which are highly toxic, persistent, and non-degradable in nature (Thavamani et al., 2012; Wong et al., 2015). The continued discharge of phenol and heavy metal contaminated industrial effluents leads to its accumulation in the environment including water bodies that reach a threshold harmful to living systems, and acute phenol exposures causes lung and digestive tract carcinoma, liver, kidney, heart, and nervous system disorders in human, enters the food chain and leads to severe socio-environmental problems (Wang et al., 2011; Fan et al., 2017). Therefore, it is imperative to reduce the concentration of phenol and heavy metals in the industrial effluents and maintain defined standards before releasing these pollutants into the environment.
Although numerous physio-chemical methods such as water chlorination, flocculation, photocatalysis, Fenton’s reaction, ozonisation, chemical oxidation, activated carbon adsorption, reverse osmosis, ion exchange with resin, etc., are widely adopted for the removal of phenol and heavy metal contaminants from industrial waste-waters (Mohammadi et al., 2014; Villegas et al., 2016) these approaches are less efficient with high operation cost and produce intermediate compounds as secondary pollutants (Rajasulochana and Preethy, 2016). Therefore biological treatment especially the use of microbial cultures is reported to significantly degrade phenol and its derivatives from different industrial wastewaters, particularly bacterial embedded treatments showed a significant reduction at low cost without secondary pollutants, and is eco-friendly (Banerjee and Ghoshal, 2010b; Wang et al., 2015; Chen et al., 2017; Tiwari et al., 2017).
Several bacterial groups are described to degrade phenol either by anaerobic or aerobic metabolic activity and utilize it as a sole energy source (Thavamani et al., 2012; Wang et al., 2015; Gu, 2016). Phenol degrading bacteria follow either the ortho cleavage pathway which converts catechol into intermediate cis, cis muconic acid or the meta cleavage pathway that converts catechol to 2-hydroxymuconic semialdehyde (2-HMSA) (Hamzah and Al-Baharna, 1994; Banerjee and Ghoshal, 2010b; Hasan and Jabeen, 2015; Wu et al., 2018) during phenol degradation. Different phenolic compounds at varying concentrations are reported to trigger either ortho or meta or both the metabolic cleavage pathways involved in phenol degradation (Wu et al., 2018). Pseudomonas cepacia ATCC 29351, when grown on salicylate, activates only the ortho-pathway, while benzoate activates both ortho and meta pathways (Hamzah and Al-Baharna, 1994; Mahiudddin et al., 2012). Pseudomonas putida ATCC 49451 degrades benzoate at 200–300 mg L–1 involving only the ortho pathway, but at higher concentrations of benzoate, the degradation involves both the pathways (Cao et al., 2008). Bacterial degradation of phenol and its derivatives like benzene and toluene had been inferred and extensively studied in different groups such as Pseudomonas sp. (Reardon et al., 2000; Mahiudddin et al., 2012; Hasan and Jabeen, 2015; Chen et al., 2017; Iqbal et al., 2018), Burkholderia sp. (Arora and Jain, 2012), Kocuria sp. (Wu et al., 2018), Acinetobacter sp. (Jiang et al., 2013; Iqbal et al., 2018), Arthrobacter sp. (Wong et al., 2015), Bacillus sp. (Banerjee and Ghoshal, 2010b; Hasan and Jabeen, 2015; Iqbal et al., 2018), Halomonas sp. (Haddadi and Shavandi, 2013), Arthrobacter sp. W1, etc. (Ma et al., 2013; Shi et al., 2014; Wong et al., 2015).
Degradation kinetics of microbial bioremediation in bioreactor offers evidence for optimum design and efficient bioremediation of phenol contaminated effluents (Banerjee and Ghoshal, 2010b). Generally, the derivatives of phenol and other co-contaminants such as heavy metals discharged from industries pose great challenge for efficient biodegradation and also depends on the tolerance level of the degrading bacterial isolates as these toxic substances negatively correlate with the degradation efficacy (Thavamani et al., 2012; Mohammadi et al., 2014; Satchanska et al., 2015; Wong et al., 2015; Chen et al., 2017; Iqbal et al., 2018). However, only few studies have reported microbial degradation of phenol particularly at high concentrations (i.e.) up to 1100 mg L–1, and limited studies have reported the biological degradation efficiency of phenol under the influence of different heavy metals such as chromium, etc., till date (Lima et al., 2008; Ontanon et al., 2015; Wong et al., 2015). Moreover, few studies have been carried out to assess the phenol degrading efficiency of bacteria isolated from distillery effluent contaminated Canna indica rhizosphere associated soil and to estimate the impact of heavy metals on the rate of phenol degradation. Hence exploration of strains with the capability to degrade phenol with high efficiency and with tolerance to multiple heavy metals can be promising candidates for the bioremediation of phenol contaminated effluents co contaminated with heavy metals which have inhibitory effects on bacterial degradation. So, this study aims (i) to isolate and characterize potential phenol degrading bacterial isolates, (ii) to evaluate kinetic models to determine the degradation efficiency of potential isolates, (iii) to identify molecular and metabolic pathways involved in the degradation of phenol, and (iv) to determine the impact of heavy metals pollutants in phenol biodegradation.
Materials and Methods
Materials and Chemicals
Davis minimal medium (DMM), 4-aminoantipyrine, potassium ferrocyanide, catechol, sodium hydroxide pellets used in this study were purchased from HiMedia Laboratories Pvt. Ltd. Mumbai, phenol, 2-mercaptoethanol, ammonium hydroxide, glycerol, disodium EDTA, primers, acids and solvents were procured from Sigma, Aldrich, Tris Base, Agarose from Bio Basic Inc. All the following heavy metals CoCl2.2H2O, ZnSO4.H2O, (CH3COO)2Pb.3H2O, HgCl2, CdCl2, CuSO4.5H2O, NiCl2.6H2O, MnCl2.4H2O, and K2Cr2O7 were purchased from Merck Millipore, Taq DNA polymerase Master Mix from Ampliqon and PCR purification Kit from Favorgen Biotech Corp. were used.
Isolation of Bacteria From Rhizosphere Soil
Phenol degrading bacterial strains were isolated from rhizosphere soils of Canna indica grown in sites contaminated by effluents from Distillery plant located in Vuyyuru, Krishna District, Andhra Pradesh, India (16°21′52.7″N 80°51′52.2″E). Three individual C. indica plants were uprooted and the rhizosphere soils from each sample were collected aseptically in sterile polythene bags and pooled. About 10 g of rhizosphere soil was used for the isolation of phenol degrading bacterial isolates by aseptically dispensing it into 100 ml of DMM containing dipotassium phosphate (7 g L–1), monopotassium phosphate (2 g L–1), ammonium sulfate (1 g L–1), sodium citrate (0.5 g L–1) and magnesium sulfate (0.1 g L–1) with final pH 7.0 supplemented with 200 mg L–1 of phenol and enriched by incubating in the shaker at 160 rpm at 30°C for 24 h. Then 1 ml of the enriched soil suspension sample was serially diluted and spread on DMM plate amended with 200 mg L–1 of phenol as the sole source of carbon with 1.8% agar and incubated for 48 h at 30°C and the colony-forming units (CFU) were counted. Diverse individual colonies were selected and streaked in 200 mg L–1 phenol amended DMM medium and stored in 25% glycerol (v/v) at −80°C for further analysis.
Screening for Potential Phenol Degrading Strains
The bacteria isolates obtained from the DMM medium amended with 200 mg L–1 of phenol were screened to determine the phenol degrading efficiency by inoculating in DMM agar plates amended with different phenol concentration of 600, 700, 800, 900, 1000, and 1200 mg L–1. The inoculated plates were incubated at 30°C for 72 h and the isolates with efficiency to grow in phenol concentration of 1200 mg L–1 were selected and maintained as pure culture (Sandhu et al., 2009).
DNA Fingerprinting Analysis
Genomic DNA was isolated from efficient bacterial isolates capable of growing in 1000 mg L–1 of phenol and the diversity was analyzed using BOXA1R primer (5′-ACG GCA AGG CGA CGC TGA CG-3′) (Viswanath et al., 2015). Each 20 μl reaction containing 6 μl of 2X ampliqon red mix, 2 μl of 2.5 pmol primer, 2 μl of 50 ng template DNA was made up to 20 μl using double sterilized HPLC water. The PCR was carried by, initial denaturation at 94°C for 5 min, 35 cycles of 94°C for 3 s, 92°C for 30 s, annealing at 50°C for 1 min, extension at 68°C for 8 min, followed by final extension for 10 min then hold at 4°C. Around 20 μl of PCR products were electrophoresed for 6 h on 2% agarose gel prepared in 1X TAE buffer. The BOX-PCR DNA profiles were visualized under UV illumination and documented using a Gel DocTM XR+ Gel Documentation System (Bio-Rad, United States). The fingerprinting profiles were analyzed using GelJv.2 DNA tool by normalization, recognition, and assignment of bands on the gel by the Dice coefficient (Heras et al., 2015). The cluster analysis was performed by unweighted pair group method with arithmetic mean (UPGMA) algorithm and the dendrogram was constructed with similarity matrices.
16S rRNA Based Identification of the Phenol Degrading Bacterial Isolates
The 16S rRNA gene was amplified from the representative isolates of the BOX-PCR cluster groups using universal primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-TAC GGH TAC CTT GTT ACG ACT T-3′) (Sekar et al., 2018). Each 20 μl reaction containing 6 μl of 2X ampliqon red mix, 2 μl of 2.5 pmol 27F and 1492R primers, 2 μl of 50 ng template DNA was made up to 20 μl using double sterilized HPLC water. The PCR conditions were as follows, initial denaturation 94°C for 5 min, 35 cycles of 94°C for 1 min, 55°C for 1 min and 68°C for 8 min. followed by final extension for 10 min then hold at 4°C. PCR products were electrophoresed on 1% agarose gel in 1X TAE buffer at 100 V for 30 min and amplified PCR products were purified using the FavorPrep GEL/PCR Purification Kit (Taiwan) and were sequenced. The taxonomic position of the isolates were identified by sequence similarity blast search against EzTaxon sequence database (Kim et al., 2012). The 16S rRNA phylogenetic tree was constructed using a neighbor-joining algorithm and confidence level in nodes were determined using 1000 bootstrap resampling conducted using the MEGA v6 (Kumar et al., 2016).
Determination of Growth, Phenol Degradation, and Quantification of Substrate
The growth and phenol degrading efficiency of representative isolates from each cluster groups of BOX profiles were determined in DMM broth with 1000 mg L–1 phenol. The culture inoculated broths were incubated in 30°C at 160 rpm for 96 h, the sample were withdrawn at every 24 h and analyzed for phenol concentration. A 1 ml of culture was centrifuged at 8000 rpm for 10 min to remove the cell debris and 50 μl of supernatant was made up to 1 ml and used to determine the phenol residual content through modified 4-amino antipyrine method at absorbance 510 nm by adding 25 μl of 0.5 N NH4OH to sample, followed by 15 μl of PBS (pH-6.9), 11 μl of 4-aminoantipyrine, and 11 μl of potassium ferrocyanide and incubated at 30°C room temperature for 15 min. The appearance of dark red color indicated positive reaction and intensity was measured using MultiskanTM GO Microplate Spectrophotometer at 500 nm (APHA, 2017). Phenol standard was prepared with the concentration range of 100 mg L–1–1200 mg L–1, linear equation Y = 0.002447∗X + 0.1423 was obtained and used to determine phenol concentration of unknown samples (Supplementary Figure S1).
Assessment of Growth Rate and Phenol Degradation Kinetics of MSSRFPD35
The biodegradation of phenol by a potential isolate MSSRFPD35 was determined in batch mode in 250 ml Erlenmeyer flask containing 95 ml of DMM with initial phenol concentration ranging from 0 to 1117 mg L–1. A 5% inoculum of MSSRFPD35 (8 log CFU ml–1) grown in DMM broth with phenol (200 mg L–1) was inoculated in DMM medium amended with phenol concentrations of 0 to 1117 mg L–1, flask without inoculation served as controls and the assay was performed in triplicates. The inoculated flasks were incubated on a rotary shaker at 160 rpm for 120 h at 30°C. At every 12 h time interval samples were withdrawn from each conical flask and the growth rate was measured as absorbance at 600 nm using MultiskanTM GO Microplate Spectrophotometer (APHA, 2017).
Cell Growth Rate Kinetics
The growth absorbance of MSSRFPD35 was converted into dry biomass using linear coefficient derived from growth absorbance vs. dry biomass (X), the cell growth was follows first-order kinetics.
when integrating Eq. (2) with limit t0 (initial time) to t (a time when maximum biomass reached), it becomes
where X0 – initial biomass, X – biomass at time t, and Eq. (3) rewritten as
The experimental specific growth rate (μ) was calculated from the slope of a semi-logarithmic plot of dry biomass ln(X/X0) vs. time.
The growth rate of microbes on inhibitory substrates such as phenol is often described using the substrate inhibition model especially the Haldane’s kinetic model Eq. (5)
Experimental μ was calculated for each initial phenol concentration and this μ value against different initial phenol concentration (S0) was used to predict various kinetic parameters of Haldane’s kinetic model using non-linear regression analysis.
Here μ∗ is one of the fitting parameters of the Haldane model, the true μmax occurs when dμ/dS = 0;
Replacing Eq. (6) in Eq. (5), (Christen et al., 2012).
Where μmax is true maximum growth rate, calculated from Eq. (7).
Degradation Kinetics
The phenol degradation rate from each sample was estimated by using 4-aminoantipyrine method as described above (APHA, 2017). Reduction of phenol content is non-linear concerning time, first-order kinetics was adopted for degradation.
Integrate Eq. (9) with time t0 to t (when complete depletion of phenol)
where S0 – initial phenol concentration, S – phenol concentration at time t.
To determine the degradation rate (q) initial phenol concentration (S0) was acquired from the slope of the linear curve by plotting between −ln(S/S0) and time. The phenol degradation kinetics was acquired by experimental degradation rate of individual phenol concentration (Banerjee and Ghoshal, 2010b; Satchanska et al., 2015). To predict the coefficient values, Haldane’s inhibitory model was used (Eq. 12),
Where Ks′ saturation constant, Ki′ Inhibition constant, S substrate conc. at time t.
Molecular Metabolic Pathway Identification
The degradative enzymes involved in phenol degradation was assessed by targeting the genes coding for catechol 1,2-dioxygenase (CBT77506) and catechol 2,3-dioxygenase (PJJ43550). The nucleotide sequences of these enzymes from Glutamicibacter spp. were extracted from the European Nucleotide Archives (ENA). Primers were designed for catechol 1,2-dioxygenase gene between 321 bp and 730 bp AC12O-F (5′-ATC GAA GGC CCT TAC TAC-3′); AC12O-R (5′-AAG TAC AGC TGG GCG GTG A-3′) and for catechol 2,3-dioxygenase between 3 bp to 819 bp AC23O-F (5′-GAG CAA AGA GAT CGC AAA CC-3′); AC23O-R (5′-GTA GAT CTC GAT GCG GTG GT-3′) using Primer3Plus program (Untergasser et al., 2012). PCR was carried out in a Bio-Rad thermal cycler with 20 μl of reaction mixture containing 2.5 pmol of each forward and reverse primer, ∼50 ng of DNA template, 1X Ampliqon Taq DNA Polymerase Master Mix RED, and the amplification conditions were as follows, an initial denaturation for 5 min at 94°C, followed by 35 cycles at 94°C for 1 min, 56°C for 1 min and 72°C for 1 min, with a final extension at 72°C for 10 min. 5 μl of the PCR product from each sample was electrophoresed using 1% (w/v) agarose gel with 100 bp marker (Thermo, India) and later purified using FavorPrep GEL/PCR Purification Kit (Taiwan). The purified products were sequenced using capillary electrophoresis on an ABI 310 Genetic Analyzer (Applied Biosystems). The identities of the sequenced fragments were determined through BLASTN analysis and CLUSTALW alignment was performed using similarity sequences and the phylogenetic relationship was determined using the neighbor-joining method, with bootstrap analysis (1000 data sets) through Molecular Evolutionary Genetics Analysis (MEGA 6) (Kumar et al., 2016).
Detection of Enzymatic Cleavage Pathway Involved in Phenol Degradation
The strain MSSRFPD35 was inoculated in DMM medium containing 1000 mg L–1 of phenol and harvested at late exponential phase (48 h) by centrifugation at 8000 rpm at 4°C for 10 min. The bacterial cells were washed and resuspended in Tris-HCl buffer (pH 7.6) and kept in ice to avoid heat generation during sonication for 4 min (1 or 2 min off) and to achieve complete lysis. The intracellular crude enzyme present in supernatant were separated from cell debris by centrifugation at 8000 rpm 4°C for 10 min and the cell-free extracts were stored at −20°C. Catechol 1,2-dioxygenase (Ortho enzymatic cleavage assay) and catechol 2,3-dioxygenase (Meta enzymatic cleavage assay) enzyme activities were spectrophotometrically determined using 3.5 ml of quartz cuvette as described by Feist and Hegeman (1969).
The enzymatic assay for Ortho cleavage pathway was performed using 0.7 ml of distilled water, 2 ml of 50 mM Tris-HCL, 0.1 ml of 100 mM β-mercaptoethanol, 0.1 ml of 1 mM catechol and 0.1 ml of cell-free extract. The formation of aromatic ring cleavage product cis, cis muconic acid was measured spectrophotometrically at an absorbance of 260 nm. Similarly, the enzymatic assay for meta cleavage pathway was carried out using 0.6 ml of distilled water, 2 ml of 50 mM Tris-HCl, 0.2 ml of 100 mM catechol and 0.2 ml of cell-free extract mixed well and the increase in absorbance was measured at 375 nm for 5 min. The formation of 2-HMSA from catechol indicates the presence of catechol 2,3-dioxygenase activity at an absorbance of 375 nm. Enzyme activity and specific activities were calculated by equation described by Bhardwaj et al. (2015).
Heavy Metal Tolerance Assay
Heavy metal tolerance and phenol degradation potential of MSSRFPD35 were determined using manganese (II) MnCl2.4H2O – 800 mg L–1; iron (II) FeSO4 – 400 mg L–1; Zinc (II) ZnSO4.H2O – 600 mg L–1; lead (II) (CH3COO)2 Pb.3H2O – 200 mg L–1; cadmium (II) CdCl2 – 50 mg L–1; chromium (VI) K2Cr2O7 – 50 mg L–1; nickel (II) NiCl2.6H2O – 20 mg L–1; copper (II) CuSO4.5H2O – 20 mg L–1; cobalt (II) CoCl2.2H2O – 10 mg L–1; and mercury (II) HgCl2 – 10 mg L–1 in DMM medium with 1000 mg L–1 of phenol. The flasks were inoculated with 5% inoculum of G. nicotianae MSSRFPD35 (8 log CFU ml–1) and incubated on a rotary shaker at 160 rpm for 72 h at 30°C. DMM medium without heavy metals and inoculation were maintained as control and all the treatments were performed in triplicates. The samples from flasks were collected at every 24 h intervals and phenol concentration was measured.
Growth on Different Aromatic Substrates
Growth of MSSRFPD35 in different phenol derivatives like tannic acid 50 mg L–1; cinnamic acid 100 mg L–1; 1-chloro-2,4-dinitro benzene 50 mg L–1; 4-nitrophenol 50 mg L–1; catechol 100 mg L–1; 1-naphthol 50 mg L–1; naphthylamine 50 mg L–1; and gallic acid 200 mg L–1 was screened by inoculating 5% inoculum (8 log CFU/ml) of MSSRFPD35 in DMM broth containing phenol derivatives at the above-mentioned concentration and incubated at 30°C on a rotary shaker at 160 rpm. Bacterial growth was measured at 600 nm absorbance (MultiskanTM GO, Thermo Scientific) after 72 h of incubation. Individual derivatives in DMM without inoculum served as negative control and were performed in triplicates.
Microcosm Study
Microcosms were set up by adding 50 g of soil in 250 ml conical flask and sterilized with three cycles of autoclaving. A uniform volume of 17.5 ml at different concentrations (65 mg L–1, 110 mg L–1 and 240 mg L–1) of phenol in sterile distilled water and 2.5 ml inoculum with 8 log10 CFU ml–1 of G. nicotianae MSSRFPD35 were added into the respective flask under aseptic condition and mixed thoroughly to get a uniform suspension of soil, phenol, and the inoculum. The treatments without microbial inoculum served as control and were performed in triplicates. The flasks were kept in 30°C for 10 days with gentle mixing at every 24 h. About 500 mg of soil samples were collected at the time of inoculation and on the 10th day of incubation from each treatment and suspended in 500 μl of distilled water, vigorously mixed and phenol was extracted by collecting the aqueous phase of the mixture and the phenol concentration was determined from the extracted samples as described above (APHA, 2017).
Statistical Analysis
All experiments were performed in triplicates, regression analysis and kinetics methodology were done using GraphPad Prism 6. The analytical results were compared by applying a one-way ANOVA and significant differences among treatments were determined by Student’s t-test.
Strains and Gene Sequences Submission
The isolated strains from this study were submitted to M. S. Swaminathan Research Foundation Culture Collection [WDCM Registered Number (1220) and accession are provided, MSSRFPD35 = MSSRFCC1542]. The gene sequences from this study were submitted to GenBank/EMBL and the accession numbers for the 16S rRNA gene of the phenol degrading bacterial are KY849351, KY849352, KX901882 – KX901885, catechol 1,2-dioxygenase gene – MK656957 and catechol 2,3-dioxygenase gene – MK656958.
Results
Isolation and DNA Fingerprinting of Phenol Degrading Bacteria
The enriched Canna indica rhizosphere soil suspension spread plated on DMM agar medium amended with 200 mg L–1 of phenol yielded 7.3 log10 CFU ml–1 after 48 h of incubation at 30°C. Around 128 different morphotype bacterial colonies were isolated and the phenol tolerance assay revealed that 11 isolates which were the most dominant colonies could grow in 1000 mg L–1 of phenol concentration and were coded as MSSRFPD27, MSSRFPD28, MSSRFPD29, MSSRFPD30, MSSRFPD35, MSSRFPD36, MSSRFPH100, MSSRFPH124, MSSRFPH134, MSSRFPH139, and MSSRFPH145. The BOX-PCR fingerprinting analysis showed the existence of polymorphism by amplifying three distinct patterns among the isolates. Dendrogram generated based on BOX-PCR profile with Dice coefficient and the UPGMA clustering method with 90% tolerance level revealed the existence of three diverse cluster groups (Cluster I, II, and III) at 85% similarity coefficient (Supplementary Figure S2).
Taxonomical Identification
Two isolates from each BOX-PCR cluster groups were chosen and the 16S rRNA was amplified and sequenced for taxonomical identification. The EzTaxon analysis revealed that representative isolates of cluster groups I (MSSRFPD27 and MSSRFPD30), II (MSSRFPD28 and MSSRFPD36), and III (MSSRFPD29 and MSSRFPD35), showed 99.9% identity to G. nicotianae DSM 20123T, followed by 99.45% similarity index with G. mysorens LMG 16219T and 99.24% with G. arilaitensis Re117T. Phylogenetic analysis of 16S rRNA of the phenol degrading isolates and closest type strains sequence revealed the isolates from cluster groups I, II, and III formed a monophyletic clade with G. nicotianae DSM 20123T which indicates that MSSRFPD36 and other isolates belong to the group of G. nicotianae with high phenol degrading efficiency (Figure 1).
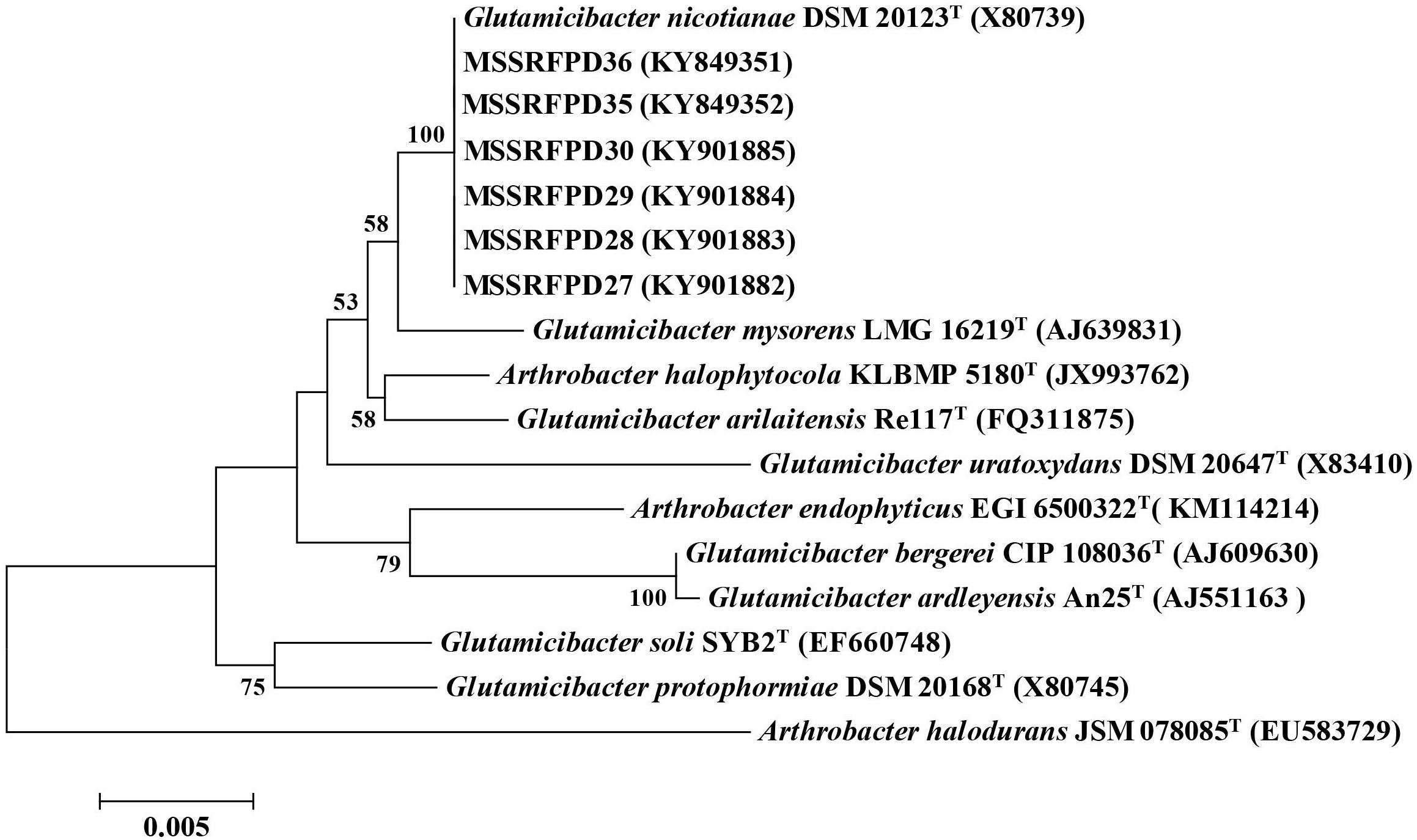
Figure 1. Neighbor-joining tree based on partial 16S rRNA gene sequences of phenol degrading strains and other closely related strains.
Biodegradation of Phenol
Based on the BOX-PCR and 16S rRNA analysis 3 polymorphic isolates MSSRFPD30, MSSRFPD35, and MSSRFPD36 were used to determine the phenol degrading efficiency and rate of degradation at 1000 mg L–1. Among the three strains, MSSRFPD35 rapidly degrade the 1000 mg L–1 of phenol completely within 48 h of incubation, followed by MSSRFPD30 and MSSRFPD36 in 60 h and 96 h, respectively (Figure 2). This indicated strain MSSRFPD35 is highly potential and an efficient phenol degrading isolate obtained from this study.
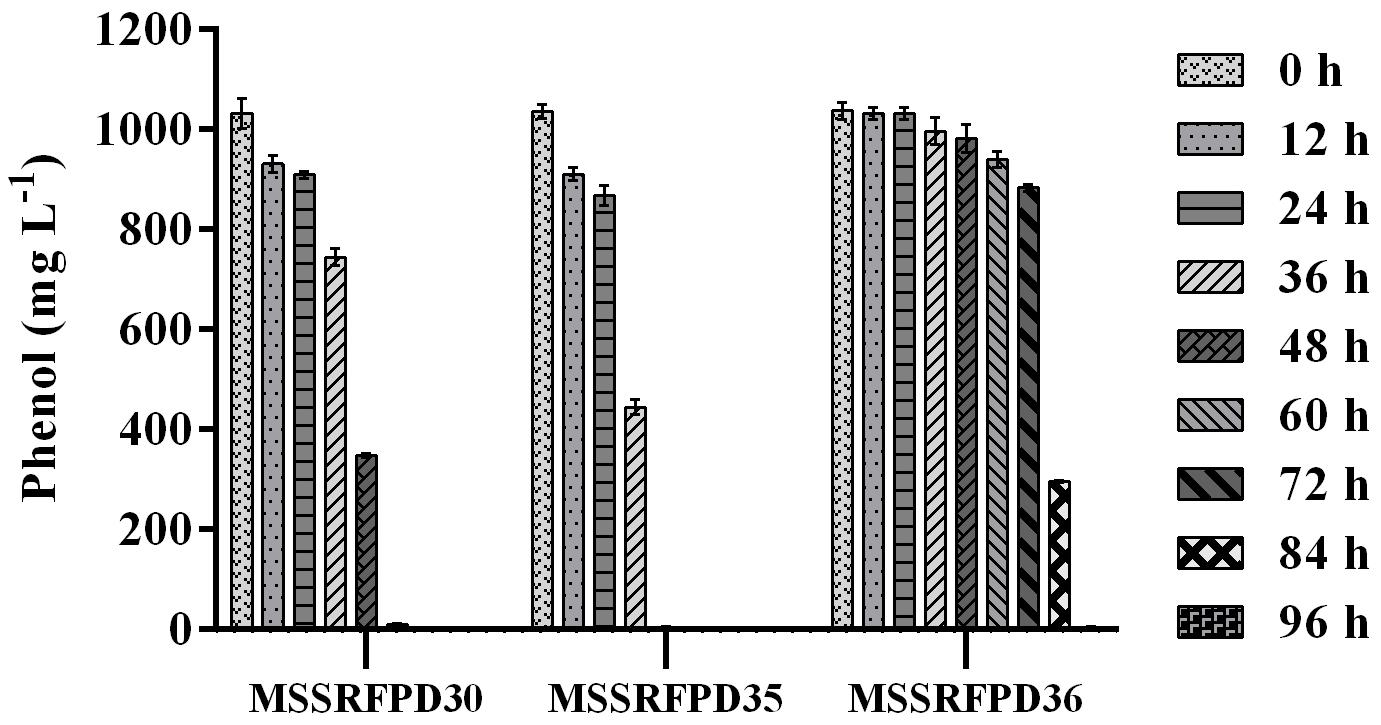
Figure 2. Determination of phenol degradation efficiency of representative isolates from each BOX PCR cluster groups at 1000 mg L–1 phenol.
Growth Kinetics and Degradation Kinetics
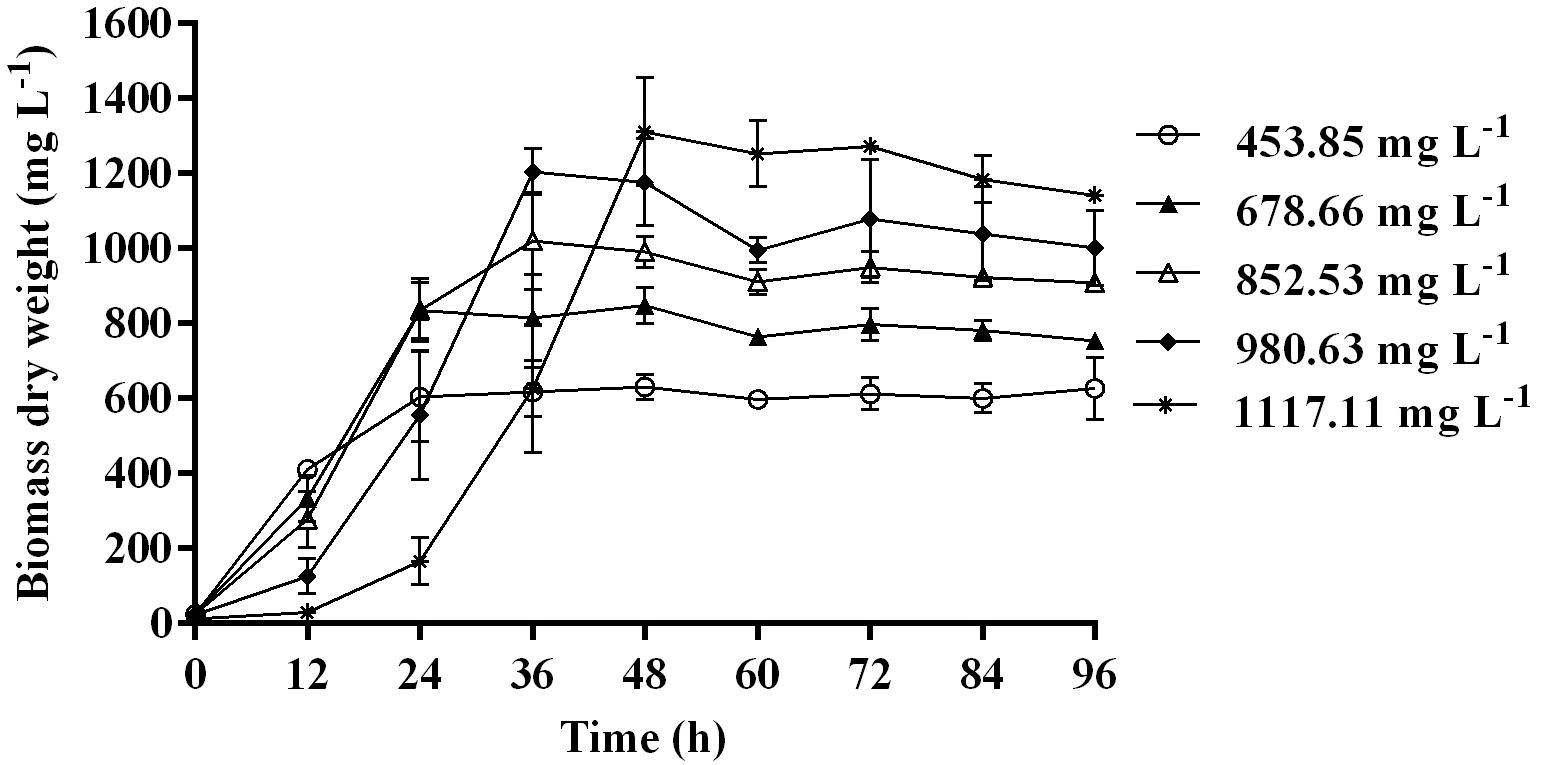
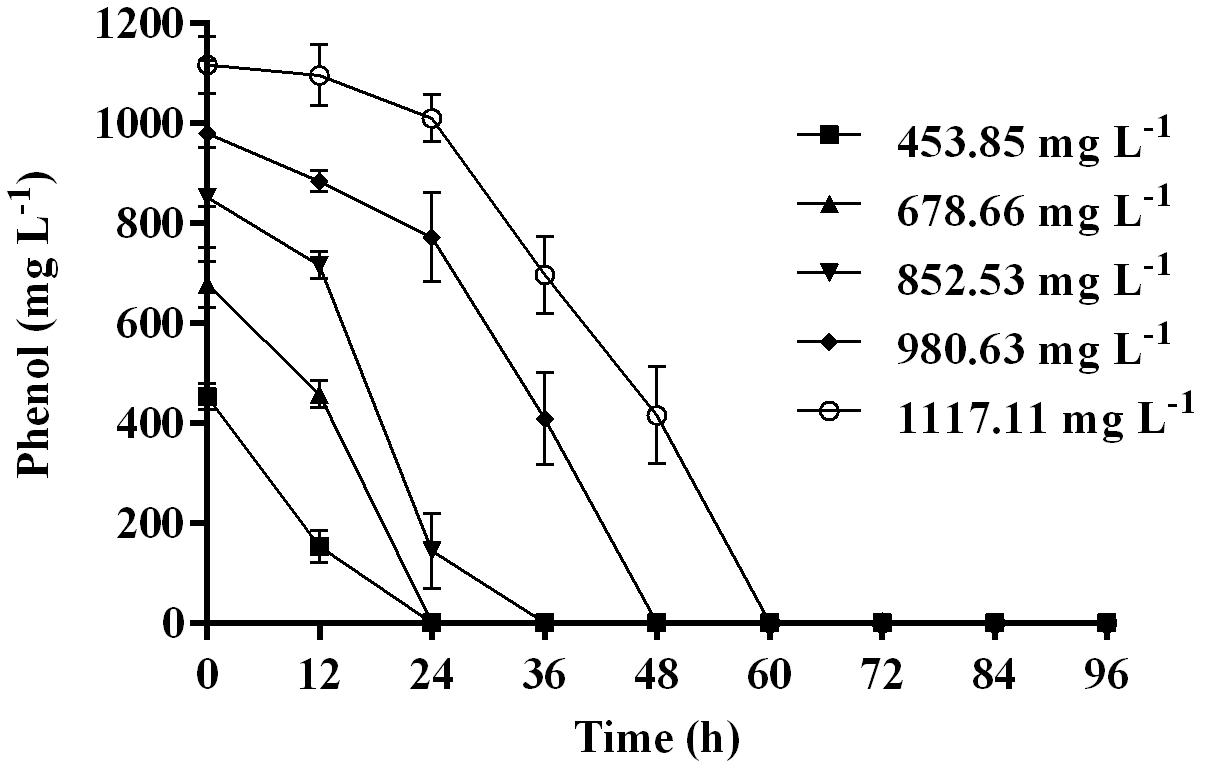
Time course assay of G. nicotianae MSSRFPD35 revealed that degradation of phenol at conc. range of 41.23 to 1117.11 mg L–1 was reached within 6 and 60 h, respectively, with maximum biomass of 1312 mg L–1 (Figure 3A). The biomass at different initial phenol concentration measured every 12 h at OD 600 nm was converted into dry biomass (mg L–1) to calculate growth rate (Figure 4). Haldane model showed a correlation coefficient R2 value of 0.98 when inhibiting phenol concentrations were used as substrate and is well fitted with experimental data (Figure 3B). The predicted kinetic parameters were μ∗ 0.574 h–1, Ks 20.29 mg L–1 and Ki 268.1 mg L–1. The values of kinetic parameters μ∗ predicted maximum growth rate, Ki inhibition constant and Ks half-saturation constant help in identification of substrate’s inhibition character. True maximum growth rate (μmax) and substrate (Sm) concentration at which it occurred were calculated by Eqs. (7) and (6) as 0.37 h–1 and 73.76 mg L–1, respectively. Here graphically predicted growth rate μ∗ (0.574) and true growth rate μmax (0.37) which were overestimated by 55% are shown in Table 1. Depletion of phenol concentration calculated for every 12 h up to 96 h was used to calculate the degradation rate of phenol (Figure 5). Degradation rate for each Si was calculated from the slope of the plot between negative logarithmic substrate concentration, −ln(S/Si) and time, t. The qs values clearly showed an inhibition effect which was reduced when Si was increased. The kinetic parameters were q∗ 1.244, Ks′ 9.152, Ki′ 517.5, Sm′ 68.820 and true maximum degradation rate qmax 0.983 obtained (Table 2) with correlation coefficient value of 0.75 (Figure 3C).
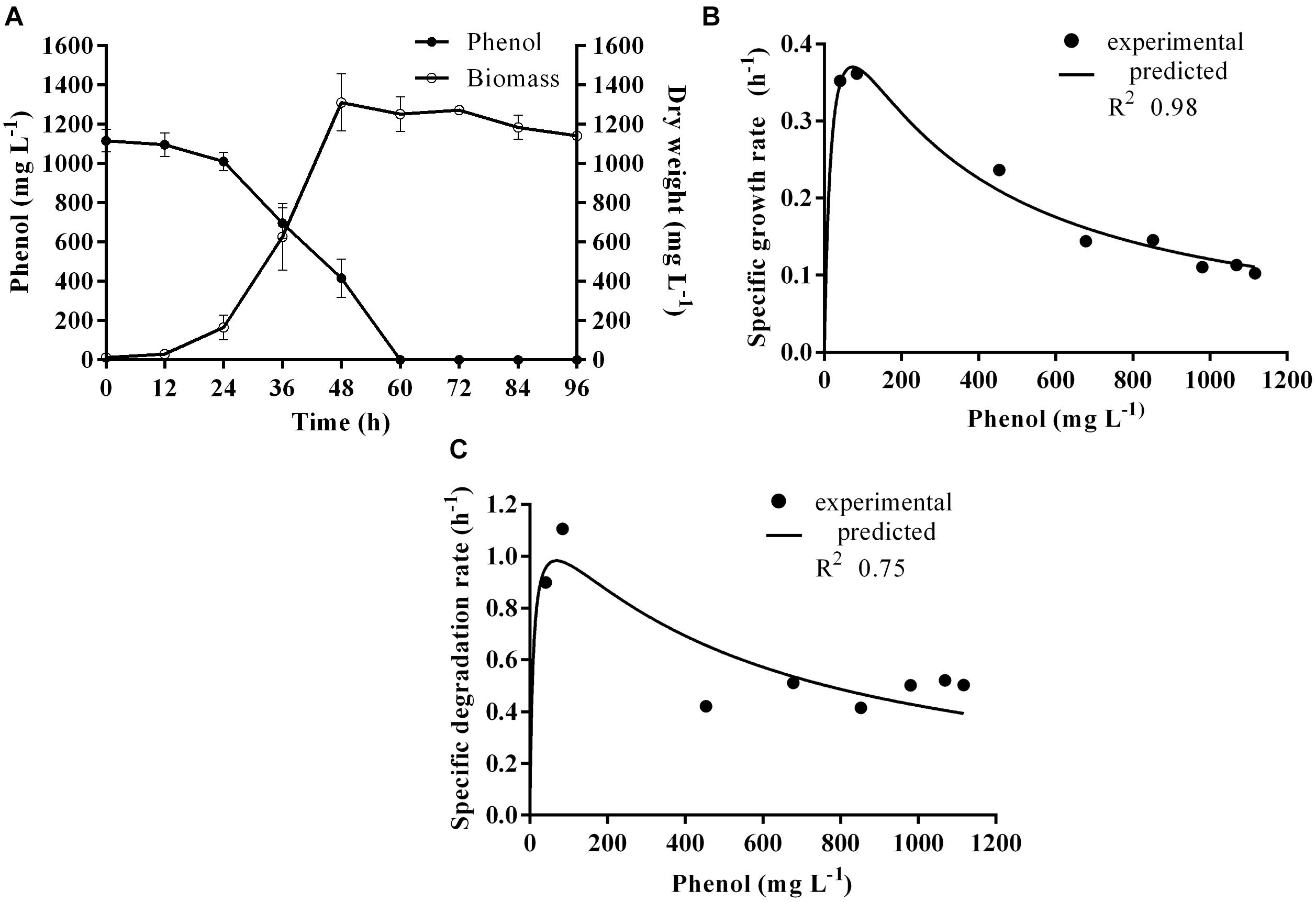
Figure 3. (A) Time course of growth and phenol degradation by G. nicotianae MSSRFPD35 at initial phenol (B) relationship between specific growth rate (μ), and (C) specific degradation rate (q) and initial substrate concentration (Si). Haldane’s model simulation was fitted to the experimental values of μ and q.
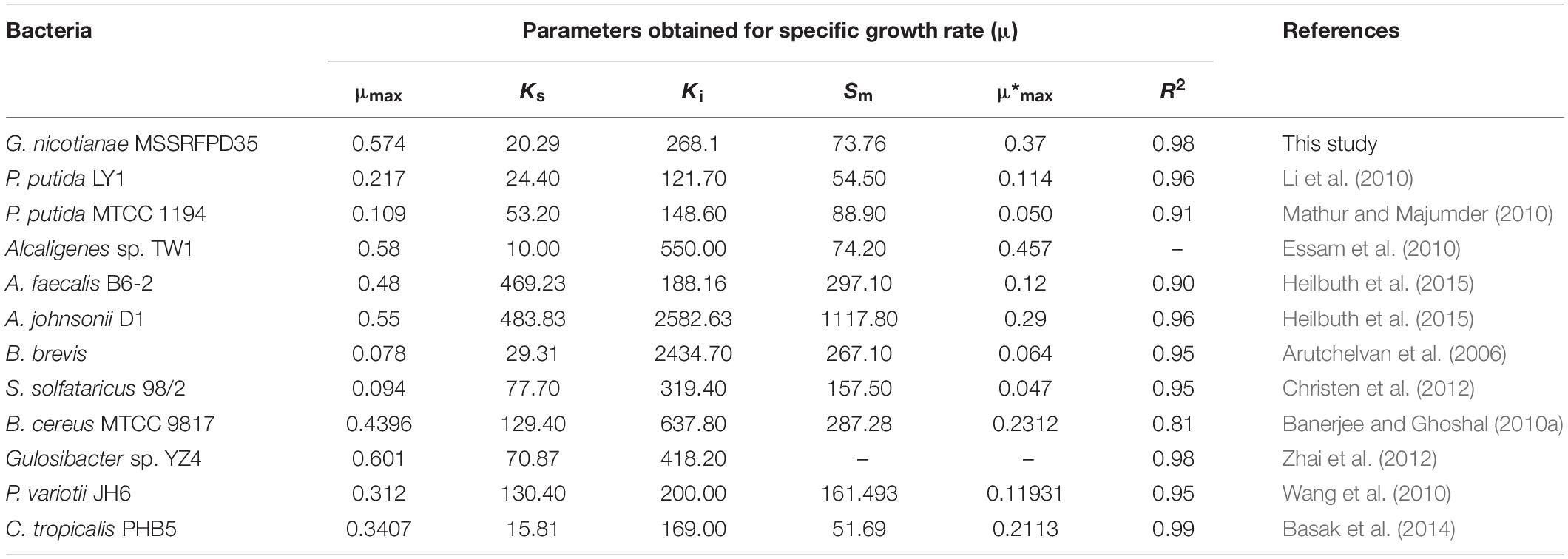
Table 1. Growth kinetic fitting parameter of MSSRFPD35 and calculated parameters of different microbes grown in phenol.
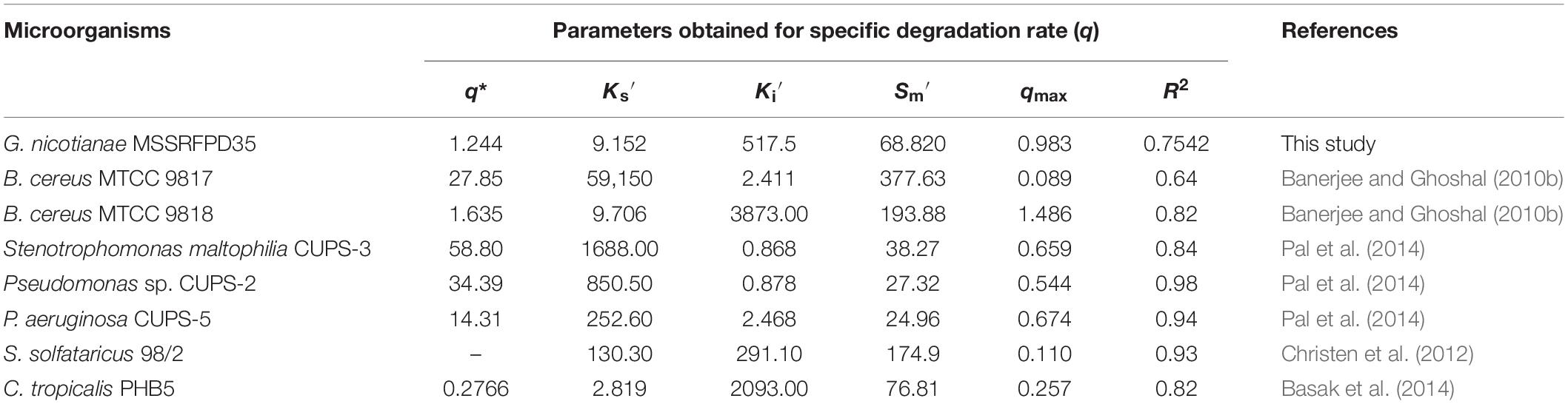
Table 2. Degradation kinetic fitting parameter and calculated parameters of different microbes grown in phenol.
Molecular and Metabolic Pathways Involved in Phenol Degradation
The genes coding for catechol 1,2-dioxygenase and catechol 2,3-dioxygenase that are involved in the degradation of phenol in MSSRFPD35 showed amplification of 409 bp and 816 bp, respectively. The BLASTX analysis of the amplified PCR product sequence showed similarity to catechol 1,2-dioxygenase with 98.25% identity to Glutamicibacter arilaitensis Re117T and Arthrobacter sp. W1, followed by 97.3%, to Arthrobacter sp. MYb213 and Glutamicibacter sp. BW77 (Supplementary Figure S3). The sequence analysis of the amplified product of catechol 2,3-dioxygenase gene showed 96% identity with catechol 2,3-dioxygenase of G. mysorens, followed by 96.43% identity with 3,4-dihydroxyphenylacetate 2,3-dioxygenase of G. arilaitensis Re117T (Supplementary Figure S4). The enzymatic assay of MSSRFPD35 culture extract confirmed that it follows the ortho cleavage pathway by increasing absorbance at 260 nm from 0.035 to 0.079 by the formation of aromatic ring cleavage product cis, cis muconic acid. Enzyme activity of MSSRFPD35 toward catechol 1,2-dioxygenase calculated was 0.046 μmol min–1 and specific activity 0.008 μmol min–1 μg–1, whereas compounds denoting meta cleavage were not detected which was confirmed by no increase in absorbance at 375 nm and no specific enzyme activity was observed. It indicates that isolate MSSRFPD35 follows ortho cleavage pathway for the degradation of phenol though the genome information for meta pathway was also detected.
Phenol Degrading Efficiency of MSSRFPD35 in the Presence of Different Heavy Metals
The phenol degrading efficiency of MSSRFPD35 was not affected in the presence of heavy metal ions such as Mn (800 mg L–1), Zn (600 mg L–1), Pb (200 mg L–1), Ni (20 mg L–1). The phenol degradation was 99% in the presence of heavy metals Pb and Ni, 83% and 93% in the presence of Mn and Zn, respectively, with the time frame of 72 h (Figure 6). Heavy metal Cu (20 mg L–1) and Co (10 mg L–1) amended medium had an inhibitory effect on phenol degradation, were in only 24% and 43% of phenol were degraded, respectively, in 72 h of incubation. While Cd (50 mg L–1), Cr (50 mg L–1), and Hg (200 mg L–1) completely inhibited the growth of MSSRFPD35 and no phenol degradation was observed. The strain MSSRFPD35 grew in the presence of aromatic compounds like 4-nitrophenol (50 mg L–1), gallic acid (200 mg L–1), cinnamic acid (100 mg L–1), naphthol (50 mg L–1), tannic acid (50 mg L–1), naphthylamine (50 mg L–1), and catechol (100 mg L–1) by utilizing all of these as sole carbon source, but was not able to utilize 1-chloro-2,4-dinitrobenzene and failed to grow in the amended medium.
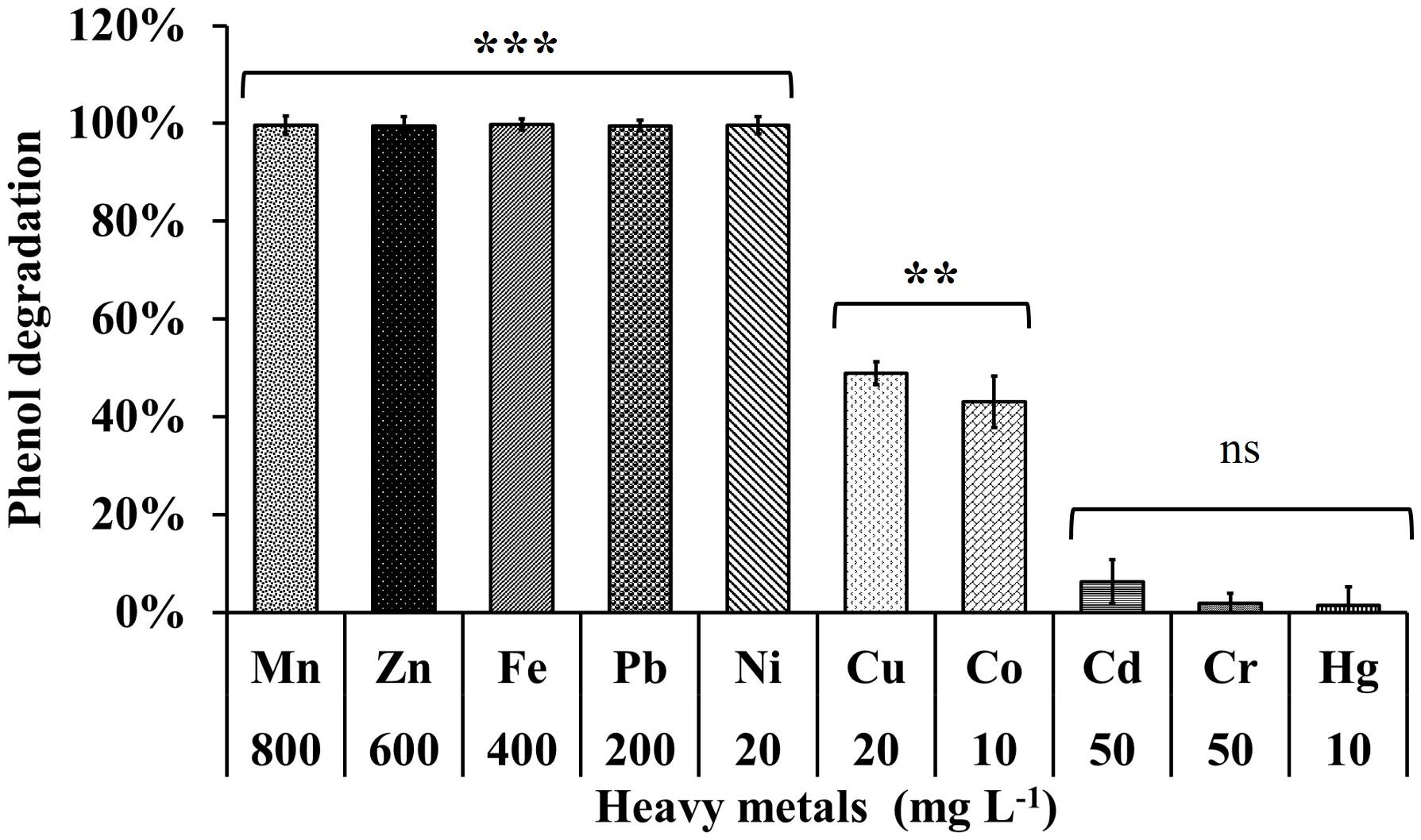
Figure 6. Phenol degradation profiles of MSSRFPD35 under the influence of different heavy metals. Values are mean ± SD of triplicate sets, **P-value = 0.01–0.001, ***P-value = less than 0.001 and nsP-value greater than 0.05 represent the significant difference according to Duncan multiple range test compared to control.
Soil Microcosm
The microcosm experiment conducted with soil slurry spiked with 65 mg L–1, 110 mg L–1 and 240 mg L–1 of phenol showed ∼84% degradation when inoculated with MSSRFPD35 compared to uninoculated soil samples at 10th day of incubation. In the soil slurry amended with 240 mg L–1 of phenol showed 94.9% degradation, followed by 84% of degradation of 65 mg L–1 and 91% of 110 mg L–1 degraded in 10 days of incubation (Figure 7). Control soils without bacterial inoculum showed no significant change in phenol concentration indicating no physiological degradation. Hence, MSSRFPD35 had the potential to degrade phenol not only under laboratory conditions but also in contaminated soils.
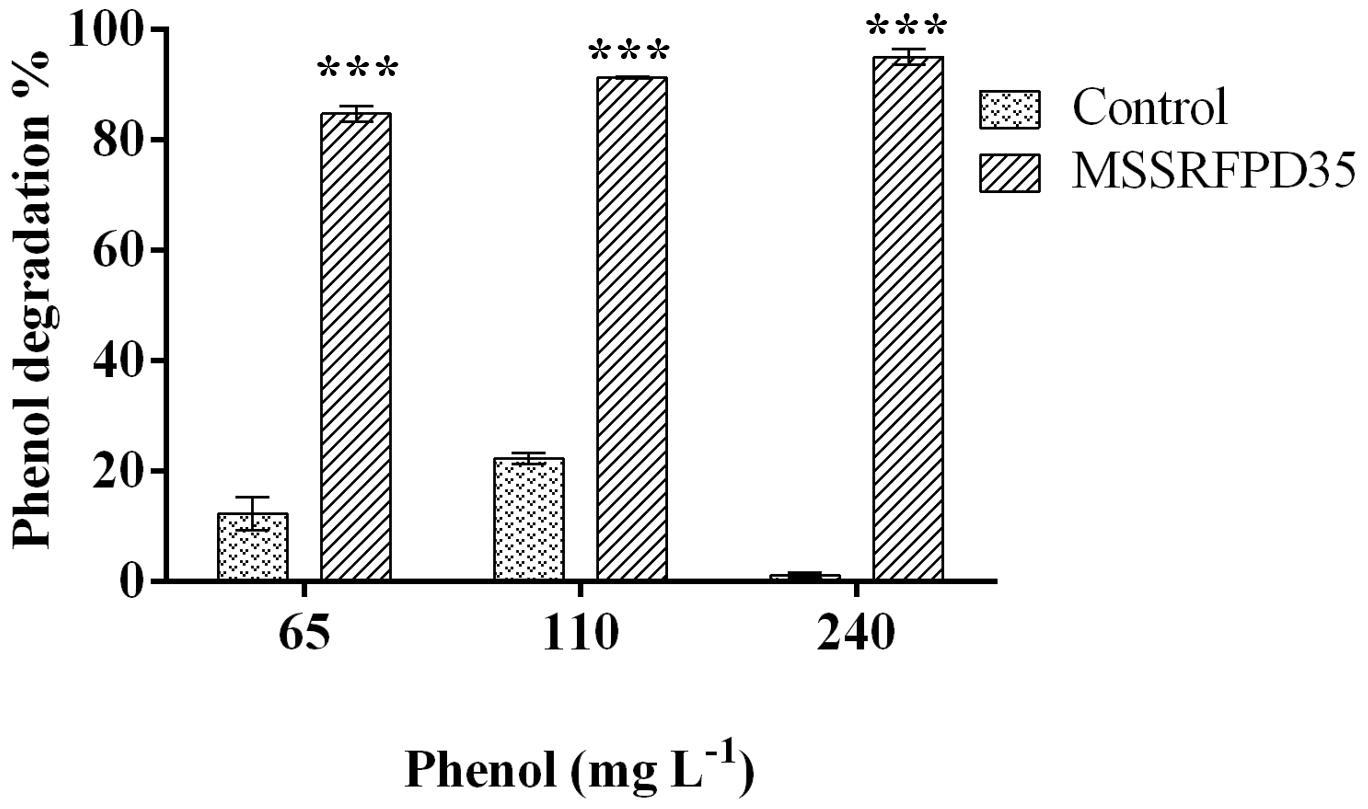
Figure 7. Degradation level of phenol by MSSRFPD35 in sterile soil microcosm. Values are mean ± SD of triplicate sets, ∗∗∗P-value = less than 0.001 represent the significant difference according to Duncan multiple range test compared to control.
Discussion
Globally, contamination of phenol and its derivatives in soil and water from industrial effluents are increasing which have major toxic effects to all living organisms (Prasse et al., 2018; Wu et al., 2018). Bioremediation of toxic pollutants and inorganic components by microorganisms is a cost-effective and environmentally safe approach (Satchanska et al., 2015; Tiwari et al., 2017). Gu (2016) discussed the key components for efficient biodegradation of environmental pollutants, and hence understanding of the microbial system with the knowledge on metabolism, nutrient utilization, growth rate and subsequently kinetics are essential. This study for the first time attempted to isolate potential phenol degrading bacteria from distillery effluent contaminated C. indica rhizospheric soils. Among the microorganisms, bacterial species are often dominantly involved in the degradation of phenol and its derivatives (Whiteley et al., 2001; Banerjee and Ghoshal, 2010a; Pal et al., 2014; Shahryari et al., 2018; Wu et al., 2018). Our study also revealed the presence and abundance of diverse potential phenol degrading Arthrobacter strains associated with rhizosphere soils of C. indica. The 16S rRNA analysis of all the cluster groups showed similarity to the strain G. nicotianae, reclassification of Arthrobacter nicotianae (Busse, 2016) but the BOX-DNA fingerprinting analysis significantly showed the existence of an intra-species population. Ellegaard and Engel (2016) reported that intra-species variation cannot be resolved using 16S rRNA based analysis because of high sequence conservation in the 16S rRNA gene. The strains showed different degrees of phenol degradation indicating the intra-species genetic variation in the BOX PCR fingerprinting pattern. The genetic variation among the clustered identical isolates might exhibit significant variation in the phenol degradation efficiency. This indicates that the genotypic and phenotypic plasticity of the strains is due to the lateral gene transfer which was observed in several other genera associated in different niches (Whiteley et al., 2001; Bedhomme et al., 2019).
Among the bacterial genera, Arthrobacter sp. class 1 microorganism are reported as key candidates involved in the degradation of phenolic compounds habited in soil and rhizosphere regions (Arora and Sharma, 2015; TRBA, 2015; Wang et al., 2015; Busse, 2016). Studies using Arthrobacter nicotianae sp. W1 which preferentially grew on ethylbenzene, toluene, catechol, benzene, xylene, and cresol involved a synergistic mechanism in the degradation process (Ma et al., 2013; Wong et al., 2015) of pentachloronitrobenzene (Wang et al., 2015). In this study, we are reporting G. nicotianae MSSRFPD35 with potential to degrade phenol up to 1113 mg L–1 within 60 h under minimal nutrient conditions, comparatively this is one of the efficient phenol degrading strain reported so far. Among the G. nicotianae isolates from this study, the degree of phenol degradation varied significantly, which indicates that degradation potential is not strongly associated with the genus (or) species. The potential of phenol degradation is a specific trait of the individual strain. Several studies reported that degree of phenol degradation and other functional traits varies among the same group, due to its habitat, presence of respective gene and its level of expression (Wang et al., 2015; Tian et al., 2017; Bedhomme et al., 2019). Strain Arthrobacter citreus was reported to degrade 470 mg L–1 phenol in 24 h (Karigar et al., 2006); Arthrobacter chlorophenolicus A6 has potential to degrade high concentrations of 4-CP (up to 347 mg L–1), 4-nitrophenol and 4-bromophenol phenol derivatives (Westerberg et al., 2000). Phenol degradation by Acinetobacter sp. SA01 isolated from farmland contaminated with pesticides and oil refinery pollutants was observed at 1000 mg L–1 of phenol after 60 h under the optimum condition of pH 7, 30°C and 180 rpm (Shahryari et al., 2018). P. putida MTCC 1194 degrade phenol at concentration of 1000 mg L–1 in 162 h (Kumar et al., 2005). Strain Kocuria sp. TIBETAN4 isolated from hyper-saline and alkaline soda lake soil could degrade 470.5 mg L–1 phenol within 3 days, 705 mg L–1 phenol within 4 days, but the degradation of 941 mg L–1 phenol was lengthened to 10 days for complete degradation (Wu et al., 2018).
In the time course assay lag phase of G. nicotianae MSSRFPD35 extended with increasing phenol concentration in the medium, which prolonged the time of biodegradation. Shahryari et al. (2018) reported 18 h lag phase stage of Acinetobacter sp. SA01 and Pseudomonas sp. NCCP-407 to degrade 1000 mg L–1 and 750 mg L–1 of phenol, respectively. Christen et al. (2012) stated that the bacterial cultures that were acclimatized in appropriate substrate concentration had less or no lag phase. However, MSSRFPD35 had no lag phase up to 980 mg L–1 of phenol, but increasing concentration up to 1117 mg L–1 of phenol had 12 h of lag phase which depict the efficiency of MSSRFPD35 to acclimatize to high phenol concentrations. Increasing concentration of phenol can influence changes in the cell membrane and protect the cell from toxic effects of phenol (Murinova and Dercova, 2014) also the cell takes a longer time to adapt to phenol toxicity. The specific growth rate of MSSRFPD35 strain was higher than P. putida LY1 (Li et al., 2010), Bacillus brevis (Arutchelvan et al., 2006) and Sulfolobus solfataricus 98/2 (Christen et al., 2012). It was almost equivalent to earlier reported potential phenol degrading strains like Alcaligenes sp. TW1 (Essam et al., 2010) and B. cereus MTCC 9817 (Banerjee and Ghoshal, 2010b). Gulosibacter sp. YZ4 has a higher specific growth rate of 0.6 mg L–1 which can degrade 2000 mg L–1 of phenol within 72 h (Zhai et al., 2012). Similarly, MSSRFPD35 can degrade 1100 mg L–1 of phenol within 60 h.
The Ks values which depict the affinity toward the phenol substrate was on the lower side for MSSRFPD35 and is equivalent to P. putida LY1, Candida tropicalis PHB5 and Bacillus brevis. Inhibition constant Ki for G. nicotianae MSSRFPD35 was in medium-range among reported strains and it is similar to Paecilomyces variotii JH6 (Wang et al., 2010) and S. solfataricus 98/2 (Christen et al., 2012). The Ki value in this study indicates the higher tolerance of MSSRFPD35 toward phenol when compared to other bacteria reported. Higher values of Ki denote lower inhibition which leads to Monod kinetic model and also indicates the concentration up to which the bacterial strain can tolerate shock loads (Bajaj et al., 2009). Inconsistency among the predicted (μ∗) and true (μmax) maximum growth rate were demonstrated in earlier studies conducted using substrate phenol (Christen et al., 2012; Basak et al., 2014). In this study, graphically predicted μ∗ (0.574) and true growth rate μmax (0.37) were overestimated. However, compared to other bacteria the μmax of MSSRFPD35 was second higher after Alcaligenes sp. TW1 (Essam et al., 2010). Though Sm of MSSRFPD35 was lower among few previous studies, it was higher than C. tropicalis PHB5 and P. putida LY1 but equal to Alcaligenes sp. TW1. This indicates that phenol concentration of 73.76 mg L–1 is the optimal substrate concentration for bacterial growth, while increment in substrate concentration above this inhibited the bacterial growth and biomass formation. Higher values of specific growth rate denoted that MSSRFPD35 can utilize the phenol in a comparatively lesser duration of 60 h. Though specific degradation rate is usually independent of Si, phenol as a substrate showed inhibitory effect on specific degradation rate qs in different studies (Yan et al., 2005; Bai et al., 2007).
Molecular analysis showed the MSSRFPD35 catechol 1,2-dioxygenase gene sequence confirms the similarity to Glutamicibacter spp., while catechol 2,3-dioxygenase enzyme-coding gene sequence showed similarity only with catechol 2,3-dioxygenase of G. mysorens. The sequence results did match with 3,4-dihydroxyphenylacetate and 2,3-dioxygenase gene of Glutamicibacter sp. Even though primers were designed to target a highly specific region of catechol 2,3-dioxygenase enzyme-coding gene, it was not depicted in blast analysis. Its due to the diversity in the enzyme gene sequence regions which is highly variable with less similarity to the target enzyme sequence, whereas both enzymes, homoprotocatechuate 2,3-dioxygenase and catechol 2,3-dioxygenase belongs to the same family of oxidoreductases and is involved in decyclizing of aromatic ring (Sandhu et al., 2009; Tian et al., 2017). The results confirmed that MSSRFPD35 and other Glutamicibacter sp. in this study might possess coding regions for both catechol 1,2-dioxygenase and catechol 2,3-dioxygenase enzymes. Though the gene coding for catechol 2,3-dioxygenase was amplified from the genomic DNA of MSSRFPD35, only catechol 1,2-dioxygenase enzymatic activity was detected indicating that MSSRFPD35 adopted ortho pathway to breakdown catechol intermediate and not meta pathway. Similarly in S. solfataricus 98/2 phenol degradation adopted the meta pathway with catechol 2,3-dioxygenase enzyme, though it possesses both catechol 1,2-dioxygenase and catechol 2,3-dioxygenase genes (Comte et al., 2013). Alkylphenol degrading Pseudomonas sp. TX1, and P. putida TX2 were reported to possess coding genes for both the enzymes (Tuan et al., 2011).
Glutamicibacter spp. were reported to degrade aromatic derivatives like pentachlorobenzene, p-cresol, benzene and toluene, even though the toxicity of aromatic compounds depends on other functional groups attached to the derivatives, most of them formed intermediate in the form of catechol derivatives (Vikram et al., 2013). Wang et al. (2009) reported G. nicotianae W1 adopted ortho pathway in the degradation of phenol p-cresol and mixed phenolic compounds. Presence of catechol dioxygenase enzyme in MSSRFPD35 evidenced that the catechol derivatives could be cleaved and can be converted to simpler forms. For example, distillery effluent contains phenolic acids and its derivatives like gallic acid, cinnamic acids and also other aromatic derivatives that are abundantly used in pesticide, plastic, explosives petrochemical and organic synthesis industries (Arora and Jain, 2012).
The strain MSSRFPD35 could degrade high levels of synthetic phenol in the presence of different heavy metals; present in industrial effluents that are predominantly contaminated with phenolic and other aromatic derivatives. It is an added functional property of the strain and the application may be expanded in the bioremediation of effluents discharged from the industries. Phenol degrading efficiency in the presence of heavy metals is an important criteria for a strain as these heavy metals are released as co-contaminates along with phenol that would inhibit the bacterial growth and decrease the biodegradation efficiency (Thavamani et al., 2012; Wong et al., 2015). Majority of the heavy metals are reported to be toxic to many bacterial groups by altering the cell morphology, disrupting the cell membrane, directly inhibiting the electron transport enzyme activity, decreasing the biomass, inhibiting growth and damaging the nucleic acid structure (Nies, 1999; Bruins et al., 2000; Sandrin and Maier, 2003; Janicka-Russak et al., 2008). But certain bacterial groups exhibit tolerance to multiple heavy metals at varied concentrations. The major mechanism involved in tolerance is through intracellular and extracellular metal sequestration, metal oxidation, methylation, demethylation, metal-organic complexion, metal-ligand degradation, exclusion by permeability barrier, and production of metal chelators like metallothioneins and exopolysaccharide (Ramasamy and Banu, 2007; Igiri et al., 2018). Silva et al. (2012) reported that the presence of heavy metals Fe3+ and Mn2+ stimulated the catechol dioxygenase activity of Gordonia polyisoprenivorans. Copper metal has a negative reaction toward Fe-S enzymes like ring-hydroxylating dioxygenases and intradiol cleavage dioxygenase which are involved in aromatic metabolic pathway (Grass et al., 2011). Yeom and Yoo (1997) reported that benzene and toluene degradation by Alcaligenes xylosoxidans Y234 were highly inhibited by Co2+ and Ag+ and Cu2+ which affects the catechol 1,2 dioxygenase enzymatic reaction. Lima et al. (2008) reported that Burkholderia cepacia utilized phenol (100 μg ml–1) in the presence of K2Cr2O7 at the concentration of 100 to 200 μg ml–1. Similarly, El-Naas et al. (2009) reported biodegradation of phenol by P. putida at the concentration of 150 mg L–1 and reported heavy metal ions such as iron, aluminum and zinc had no effect on the phenol biodegradation rate. Ontanon et al. (2015) reported degradation of phenol by Bacillus at maximum concentrations of 1000 mg L–1 and reduced Cr (VI) to Cr (III). This study revealed the potential of MSSRFPD35 in degrading phenol in the presence of different heavy metals. Satchanska et al. (2015) also reported that bacterial strains B. subtilis KCMRG5 and P. rhodesiae KCMR5 were able to degrade phenol in the presence of heavy metals. P. fluorescens (BBN1) and P. corrugata (BBB2) were reported to degrade PAH which were able to tolerate 993.6 mg L–1-Pb (NO3)2, 717.83 mg L–1-ZnSO4 and 499.36 mg L–1-CuSO4, respectively (Máthé et al., 2012). However, no evaluation of heavy metal effect on phenol biodegradation by G. nicotianae has previously been performed. The present study indicates that G. nicotianae MSSRFPD35 strain can degrade phenol in the presence of Pb, Ni, Cu, Co, Mn, and Zn metals which might occur as co-pollutants in industrial effluent. Soil microcosm studies also confirmed the in vivo bioremediation potential of MSSRFPD35 and to degrade phenol in contaminated soils. Though there are no studies on soil microcosm with phenol as a substrate, some studies reported 90% degradation of chloro-nitro phenol by Burkholderia sp. RKJ 800 (Arora and Jain, 2012) and Cupriavidus sp. a3 (Tiwari et al., 2017) and 2 nitrobenzene by Arthrobacter sp. SPG in soil (Arora and Sharma, 2015). Strain MSSRFPD35 holding a capacity of tolerating heavy metals, degrading relatively high concentrations of phenol in a shorter time and other phenol derivatives showed promising prospect for application in the remediation of phenol contaminated sites.
Conclusion
The strain G. nicotianae MSSRFPD35 proved to degrade phenol at relatively high concentration via catechol 1,2-dioxygenase directed ortho pathway, specifically in the presence of different heavy metals. The growth and degradation kinetics of the strain MSSRFPD35 utilizing phenol as a sole carbon and nutrient source was well characterized by Haldane model where the μmax, Ki and Ks were described. The strain was capable of growing in various monocyclic and polycyclic aromatic hydrocarbons, tolerate and degrade phenol in the presence of heavy metals like lead, zinc, manganese, iron, and nickel. The versatility of this strain can be exploited for promoting it as a renewable resource for biodegradation of phenol and its derivatives co-contaminated with heavy metals discharged from various industries.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
PD collected the samples, designed and performed the experiments, data analysis and drafted the manuscript. JS and AA supported sample collection, contributed to in vitro experiments, molecular analysis and manuscript correction. PR supervised and supported the study, revised the manuscript and approved for publishing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. M. S. Swaminathan, Founder Chairman, and Executive Director, M. S. Swaminathan Research Foundation, for their support and encouragement. We acknowledge the financial support from the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi, India.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01573/full#supplementary-material
References
APHA (2017). Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association.
Arora, P. K., and Jain, R. K. (2012). Metabolism of 2-chloro-4-nitrophenol in a Gram negative bacterium, Burkholderia sp. RKJ 800. PLoS One 7:e38676. doi: 10.1371/journal.pone.0038676
Arora, P. K., and Sharma, A. (2015). New metabolic pathway for degradation of 2-nitrobenzoate by Arthrobacter sp. SPG. Front. Microbiol. 6:551. doi: 10.3389/fmicb.2015.00551
Arutchelvan, V., Kanakasabai, V., Elangovan, R., Nagarajan, S., and Muralikrishnan, V. (2006). Kinetics of high strength phenol degradation using Bacillus brevis. J. Hazard. Mater. 129, 216–222. doi: 10.1016/j.jhazmat.2005.08.040
Bai, J., Wen, J.-P., Li, H.-M., and Jiang, Y. (2007). Kinetic modeling of growth and biodegradation of phenol and m-cresol using Alcaligenes faecalis. Process. Biochem. 42, 510–517. doi: 10.1016/j.procbio.2006.10.004
Bajaj, M., Gallert, C., and Winter, J. (2009). Phenol degradation kinetics of an aerobic mixed culture. Biochem. Eng. J. 46, 205–209. doi: 10.1016/j.bej.2009.05.021
Banerjee, A., and Ghoshal, A. K. (2010a). Isolation and characterization of hyper phenol tolerant Bacillus sp. from oil refinery and exploration sites. J. Hazard. Mater. 176, 85–91. doi: 10.1016/j.jhazmat.2009.11.002
Banerjee, A., and Ghoshal, A. K. (2010b). Phenol degradation by Bacillus cereus: pathway and kinetic modeling. Bioresour. Technol. 101, 5501–5507. doi: 10.1016/j.biortech.2010.02.018
Basak, B., Bhunia, B., Dutta, S., Chakraborty, S., and Dey, A. (2014). Kinetics of phenol biodegradation at high concentration by a metabolically versatile isolated yeast Candida tropicalis PHB5. Environ. Sci. Pollut. Res. Int. 21, 1444–1454. doi: 10.1007/s11356-013-2040-z
Bedhomme, S., Amoros-Moya, D., Valero, L. M., Bonifaci, N., Pujana, M. A., and Bravo, I. G. (2019). Evolutionary changes after translational challenges imposed by horizontal gene transfer. Genome Biol. Evol. 11, 814–831. doi: 10.1093/gbe/evz031
Bhardwaj, P., Sharma, A., Sagarkar, S., and Kapley, A. (2015). Mapping atrazine and phenol degradation genes in Pseudomonas sp. EGD-AKN5. Biochem. Eng. J. 102, 125–134. doi: 10.1016/j.bej.2015.02.029
Bruins, M. R., Kapil, S., and Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 45, 198–207. doi: 10.1006/eesa.1999.1860
Busse, H. J. (2016). Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 66, 9–37. doi: 10.1099/ijsem.0.000702
Cao, B., Geng, A., and Loh, K. C. (2008). Induction of ortho- and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl. Microbiol. Biotechnol. 81, 99–107. doi: 10.1007/s00253-008-1728-3
Chen, J., Zhang, L., Jin, Q., Su, C., Zhao, L., Liu, X., et al. (2017). Bioremediation of phenol in soil through using a mobile plant-endophyte system. Chemosphere 182, 194–202. doi: 10.1016/j.chemosphere.2017.05.017
Christen, P., Vega, A., Casalot, L., Simon, G., and Auria, R. (2012). Kinetics of aerobic phenol biodegradation by the acidophilic and hyperthermophilic archaeon Sulfolobus solfataricus 98/2. Biochem. Eng. J. 62, 56–61. doi: 10.1016/j.bej.2011.12.012
Comte, A., Christen, P., Davidson, S., Pophillat, M., Lorquin, J., Auria, R., et al. (2013). Biochemical, transcriptional and translational evidences of the phenol-meta-degradation pathway by the hyperthermophilic Sulfolobus solfataricus 98/2. PLoS One 8:e82397. doi: 10.1371/journal.pone.0082397
Ellegaard, K. M., and Engel, P. (2016). Beyond 16S rRNA community profiling: intra-species diversity in the gut microbiota. Front. Microbiol. 7:1475. doi: 10.3389/fmicb.2016.01475
El-Naas, M. H., Al-Muhtaseb, S. A., and Makhlouf, S. (2009). Biodegradation of phenol by Pseudomonas putida immobilized in polyvinyl alcohol (PVA) gel. J. Hazard. Mater. 164, 720–725. doi: 10.1016/j.jhazmat.2008.08.059
Essam, T., Amin, M. A., El Tayeb, O., Mattiasson, B., and Guieysse, B. (2010). Kinetics and metabolic versatility of highly tolerant phenol degrading Alcaligenes strain TW1. J. Hazard. Mater. 173, 783–788. doi: 10.1016/j.jhazmat.2009.09.006
Fan, Y., Zhu, T., Li, M., He, J., and Huang, R. (2017). Heavy metal contamination in soil and brown rice and human health risk assessment near three mining areas in central China. J. Healthc. Eng. 2017:4124302. doi: 10.1155/2017/4124302
Feist, C. F., and Hegeman, G. D. (1969). Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J. Bacteriol. 100, 869–877. doi: 10.1128/jb.100.2.869-877.1969
Grass, G., Rensing, L., and Rensing, C. (2011). Metal toxicity. Metallomics 3, 1095–1097. doi: 10.1039/c1mt90048j
Gu, J.-D. (2016). Biodegradation testing: so many tests but very little new innovation. Appl. Environ. Biotechnol. 1, 92–95.
Haddadi, A., and Shavandi, M. (2013). Biodegradation of phenol in hypersaline conditions by Halomonas sp. strain PH2-2 isolated from saline soil. Int. Biodeterior. Biodegradation. 85, 29–34. doi: 10.1016/j.ibiod.2013.06.005
Hamzah, R. Y., and Al-Baharna, B. S. (1994). Catechol ring-cleavage in Pseudomonas cepacia: the simultaneous induction of ortho and meta pathways. Appl. Microbiol. Biotechnol. 41, 250–256. doi: 10.1007/bf00186968
Hasan, S. A., and Jabeen, S. (2015). Degradation kinetics and pathway of phenol by Pseudomonas and Bacillus species. Biotechnol. Biotechnol. Equip. 29, 45–53. doi: 10.1080/13102818.2014.991638
Heilbuth, N. M., Linardi, V. R., Monteiro, A. S., da Rocha, R. A., Mimim, L. A., and Santos, V. L. (2015). Estimation of kinetic parameters of phenol degradation by bacteria isolated from activated sludge using a genetic algorithm. J. Chem. Technol. Biotechnol. 90, 2066–2075. doi: 10.1002/jctb.4518
Heras, J., Dominguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., et al. (2015). GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 16:270. doi: 10.1186/s12859-015-0703-0
Hussain, A., Dubey, S. K., and Kumar, V. (2015). Kinetic study for aerobic treatment of phenolic wastewater. Water Resour. Ind. 11, 81–90. doi: 10.1016/j.wri.2015.05.002
Igiri, B. E., Okoduwa, S. I. R., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J. Toxicol. 2018:2568038. doi: 10.1155/2018/2568038
Iqbal, A., Arshad, M., Hashmi, I., Karthikeyan, R., Gentry, T. J., and Schwab, A. P. (2018). Biodegradation of phenol and benzene by endophytic bacterial strains isolated from refinery wastewater-fed Cannabis sativa. Environ. Technol. 39, 1705–1714. doi: 10.1080/09593330.2017.1337232
Janicka-Russak, M., Kabała, K., Burzyński, M., and Kłobus, G. (2008). Response of plasma membrane H+-ATPase to heavy metal stress in Cucumis sativu s roots. J. Exp. Bot. 59, 3721–3728. doi: 10.1093/jxb/ern219
Jiang, L., Ruan, Q., Li, R., and Li, T. (2013). Biodegradation of phenol by using free and immobilized cells of Acinetobacter sp. BS8Y. J. Basic Microbiol. 53, 224–230. doi: 10.1002/jobm.201100460
Jusoh, N., and Razali, F. (2008). Microbial consortia from residential wastewater for bioremediation of phenol in a chemostat. J. Teknol. 48, 51–60. doi: 10.11113/jt.v48.233
Karigar, C., Mahesh, A., Nagenahalli, M., and Yun, D. J. (2006). Phenol degradation by immobilized cells of Arthrobacter citreus. Biodegradation 17, 47–55. doi: 10.1007/s10532-005-3048-y
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62(Pt 3), 716–721. doi: 10.1099/ijs.0.038075-0
Kumar, A., Kumar, S., and Kumar, S. (2005). Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem. Eng. J. 22, 151–159. doi: 10.1016/j.bej.2004.09.006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, Y., Li, J., Wang, C., and Wang, P. (2010). Growth kinetics and phenol biodegradation of psychrotrophic Pseudomonas putida LY1. Bioresour. Technol. 101, 6740–6744. doi: 10.1016/j.biortech.2010.03.083
Lima, A., Pereira, M., Geraldo, R., Silva Filho, R., and Hofer, E. (2008). Utilization of phenol in the presence of heavy metals by metal-tolerant nonfermentative gram-negative ba-cteria isolated from wastewater. Rev. Latinoam. Microbiol. 49, 68–73.
Ma, F., Shi, S. N., Sun, T. H., Li, A., Zhou, J. T., and Qu, Y. Y. (2013). Biotransformation of benzene and toluene to catechols by phenol hydroxylase from Arthrobacter sp. W1. Appl. Microbiol. Biotechnol. 97, 5097–5103. doi: 10.1007/s00253-012-4301-z
Mahiudddin, M., Fakhruddin, A. N., and Abdullah Al, M. (2012). Degradation of phenol via meta cleavage pathway by Pseudomonas fluorescens PU1. ISRN Microbiol. 2012:741820. doi: 10.5402/2012/741820
Máthé, I., Benedek, T., Táncsics, A., Palatinszky, M., Lányi, S., and Márialigeti, K. (2012). Diversity, activity, antibiotic and heavy metal resistance of bacteria from petroleum hydrocarbon contaminated soils located in Harghita County (Romania). Int. Biodeterior. Biodegradation 73, 41–49. doi: 10.1016/j.ibiod.2012.05.018
Mathur, A., and Majumder, C. (2010). Kinetics modelling of the biodegradation of benzene, toluene and phenol as single substrate and mixed substrate by using Pseudomonas putida. Chem. Biochem. Eng. Q. 24, 101–109. doi: 10.15255/cabeq
Mohammadi, S., Kargari, A., Sanaeepur, H., Abbassian, K., Najafi, A., and Mofarrah, E. (2014). Phenol removal from industrial wastewaters: a short review. Desalin. Water Treat. 53, 2215–2234. doi: 10.1080/19443994.2014.883327
Murinova, S., and Dercova, K. (2014). Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014:873081. doi: 10.1155/2014/873081
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. doi: 10.1007/s002530051457
Ontanon, O. M., Gonzalez, P. S., and Agostini, E. (2015). Optimization of simultaneous removal of Cr (VI) and phenol by a native bacterial consortium: its use for bioaugmentation of co-polluted effluents. J. Appl. Microbiol. 119, 1011–1022. doi: 10.1111/jam.12913
Pal, B., Sarkar, P., and Pal, P. (2014). Isolation and characterization of phenol utilizing bacteria from industrial effluent-contaminated soil and kinetic evaluation of their biodegradation potential. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 49, 67–77. doi: 10.1080/10934529.2013.824304
Prasse, C., Ford, B., Nomura, D. K., and Sedlak, D. L. (2018). Unexpected transformation of dissolved phenols to toxic dicarbonyls by hydroxyl radicals and UV light. Proc. Natl. Acad. Sci. U.S.A. 115, 2311–2316. doi: 10.1073/pnas.1715821115
Rajasulochana, P., and Preethy, V. (2016). Comparison on efficiency of various techniques in treatment of waste and sewage water – A comprehensive review. Resour. Efficient Technol. 2, 175–184. doi: 10.1016/j.reffit.2016.09.004
Ramasamy, K., and Banu, S. P. (2007). “Bioremediation of metals: microbial processes and techniques,” in Environmental Bioremediation Technologies, eds S. N. Singh and R. D. Tripathi (Berlin: Springer), 173–187. doi: 10.1007/978-3-540-34793-4_7
Reardon, K. F., Mosteller, D. C., and Bull Rogers, J. D. (2000). Biodegradation kinetics of benzene, toluene, and phenol as single and mixed substrates for Pseudomonas putida F1. Biotechnol. Bioeng. 69, 385–400. doi: 10.1002/1097-0290(20000820)69:4<385::aid-bit5>3.0.co;2-q
Sandhu, A., Halverson, L. J., and Beattie, G. A. (2009). Identification and genetic characterization of phenol-degrading bacteria from leaf microbial communities. Microb. Ecol. 57, 276–285. doi: 10.1007/s00248-008-9473-9
Sandrin, T. R., and Maier, R. M. (2003). Impact of metals on the biodegradation of organic pollutants. Environ. Health Perspect. 111, 1093–1101. doi: 10.1289/ehp.5840
Satchanska, G., Topalova, Y., Dimkov, R., Groudeva, V., Petrov, P., Tsvetanov, C., et al. (2015). Phenol degradation by environmental bacteria entrapped in cryogels. Biotechnol. Biotechnol. Equip. 29, 514–521. doi: 10.1080/13102818.2015.1009167
Sekar, J., Raju, K., Duraisamy, P., and Ramalingam Vaiyapuri, P. (2018). Potential of finger millet indigenous rhizobacterium Pseudomonas sp. MSSRFD41 in blast disease management—growth promotion and compatibility with the resident rhizomicrobiome. Front. Microbiol. 9:1029. doi: 10.3389/fmicb.2018.01029
Shahryari, S., Zahiri, H. S., Haghbeen, K., Adrian, L., and Noghabi, K. A. (2018). High phenol degradation capacity of a newly characterized Acinetobacter sp. SA01: bacterial cell viability and membrane impairment in respect to the phenol toxicity. Ecotoxicol. Environ. Saf. 164, 455–466. doi: 10.1016/j.ecoenv.2018.08.051
Shi, S., Qu, Y., Ma, F., and Zhou, J. (2014). Bioremediation of coking wastewater containing carbazole, dibenzofuran and dibenzothiophene by immobilized naphthalene-cultivated Arthrobacter sp. W1 in magnetic gellan gum. Bioresour. Technol. 166, 79–86. doi: 10.1016/j.biortech.2014.05.036
Silva, A. S., Camargo, F. A. D. O., Andreazza, R., Jacques, R. J. S., Baldoni, D. B., and Bento, F. M. (2012). Enzymatic activity of catechol 1, 2-dioxygenase and catechol 2, 3-dioxygenase produced by Gordonia polyisoprenivorans. Quim. Nova 35, 1587–1592. doi: 10.1590/s0100-40422012000800018
Thavamani, P., Megharaj, M., and Naidu, R. (2012). Bioremediation of high molecular weight polyaromatic hydrocarbons co-contaminated with metals in liquid and soil slurries by metal tolerant PAHs degrading bacterial consortium. Biodegradation 23, 823–835. doi: 10.1007/s10532-012-9572-7
Tian, M., Du, D., Zhou, W., Zeng, X., and Cheng, G. (2017). Phenol degradation and genotypic analysis of dioxygenase genes in bacteria isolated from sediments. Braz. J. Microbiol. 48, 305–313. doi: 10.1016/j.bjm.2016.12.002
Tiwari, J., Naoghare, P., Sivanesan, S., and Bafana, A. (2017). Biodegradation and detoxification of chloronitroaromatic pollutant by Cupriavidus. Bioresour. Technol. 223, 184–191. doi: 10.1016/j.biortech.2016.10.043
TRBA (2015). Classification of Prokaryotes (Bacteria and Archaea) into Risk Groups Committee on Biological Agents (ABAS). Available online at: https://www.baua.de/EN/Tasks/Committee-administration/ABAS/ABAS_node.html (accessed June 6, 2017).
Tuan, N. N., Hsieh, H. C., Lin, Y. W., and Huang, S. L. (2011). Analysis of bacterial degradation pathways for long-chain alkylphenols involving phenol hydroxylase, alkylphenol monooxygenase and catechol dioxygenase genes. Bioresour. Technol. 102, 4232–4240. doi: 10.1016/j.biortech.2010.12.067
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Vikram, S., Pandey, J., Kumar, S., and Raghava, G. P. (2013). Genes involved in degradation of para-nitrophenol are differentially arranged in form of non-contiguous gene clusters in Burkholderia sp. strain SJ98. PLoS One 8:e84766. doi: 10.1371/journal.pone.0084766
Villegas, L. G. C., Mashhadi, N., Chen, M., Mukherjee, D., Taylor, K. E., and Biswas, N. A. (2016). short review of techniques for phenol removal from wastewater. Curr. Pollut. Rep. 2, 157–167. doi: 10.1007/s40726-016-0035-3
Viswanath, G., Jegan, S., Baskaran, V., Kathiravan, R., and Prabavathy, V. R. (2015). Diversity and N-acyl-homoserine lactone production by Gammaproteobacteria associated with Avicennia marina rhizosphere of South Indian mangroves. Syst. Appl. Microbiol. 38, 340–345. doi: 10.1016/j.syapm.2015.03.008
Wang, L., Li, Y., Yu, P., Xie, Z., Luo, Y., and Lin, Y. (2010). Biodegradation of phenol at high concentration by a novel fungal strain Paecilomyces variotii JH6. J. Hazard. Mater. 183, 366–371. doi: 10.1016/j.jhazmat.2010.07.033
Wang, P., Qu, Y., and Zhou, J. (2009). Biodegradation of mixed phenolic compounds under high salt conditions and salinity fluctuations by Arthrobacter sp. W1. Appl. Biochem. Biotechnol. 159, 623–633. doi: 10.1007/s12010-008-8494-7
Wang, Y., Song, J., Zhao, W., He, X., Chen, J., and Xiao, M. (2011). In situ degradation of phenol and promotion of plant growth in contaminated environments by a single Pseudomonas aeruginosa strain. J. Hazard. Mater. 192, 354–360. doi: 10.1016/j.jhazmat.2011.05.031
Wang, Y., Wang, C., Li, A., and Gao, J. (2015). Biodegradation of pentachloronitrobenzene by Arthrobacter nicotianae DH19. Lett. Appl. Microbiol. 61, 403–410. doi: 10.1111/lam.12476
Westerberg, K., Elvang, A. M., Stackebrandt, E., and Jansson, J. K. (2000). Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 50(Pt. 6), 2083–2092. doi: 10.1099/00207713-50-6-2083
Whiteley, A. S., Wiles, S., Lilley, A. K., Philp, J., and Bailey, M. J. (2001). Ecological and physiological analyses of Pseudomonad species within a phenol remediation system. J. Microbiol. Methods 44, 79–88. doi: 10.1016/s0167-7012(00)00231-1
Wong, K. K., Quilty, B., Hamzah, A., and Surif, S. (2015). Phenol biodegradation and metal removal by a mixed bacterial consortium. Bioremediat. J. 19, 104–112. doi: 10.1080/10889868.2014.995368
Wu, L., Ali, D. C., Liu, P., Peng, C., Zhai, J., Wang, Y., et al. (2018). Degradation of phenol via ortho-pathway by Kocuria sp. strain TIBETAN4 isolated from the soils around Qinghai Lake in China. PLoS One 13:e0199572. doi: 10.1371/journal.pone.0199572
Yan, J., Jianping, W., Hongmei, L., Suliang, Y., and Zongding, H. (2005). The biodegradation of phenol at high initial concentration by the yeast Candida tropicalis. Biochem. Eng. J. 24, 243–247. doi: 10.1016/j.bej.2005.02.016
Yeom, S. H., and Yoo, Y. J. (1997). Overcoming the inhibition effects of metal ions in the degradation of benzene and toluene by Alcaligenes xylosoxidans y234. Korean J. Chem. Eng. 14, 204–208. doi: 10.1007/bf02706096
Keywords: phenol biodegradation, distillery effluent, Glutamicibacter sp., Canna indica, Haldane’s kinetics, heavy metal tolerance, soil microcosm
Citation: Purushothaman D, Jegan S, Anu DA and Prabavathy VR (2020) Kinetics of Phenol Biodegradation by Heavy Metal Tolerant Rhizobacteria Glutamicibacter nicotianae MSSRFPD35 From Distillery Effluent Contaminated Soils. Front. Microbiol. 11:1573. doi: 10.3389/fmicb.2020.01573
Received: 27 March 2020; Accepted: 17 June 2020;
Published: 15 July 2020.
Edited by:
Jaco Vangronsveld, University of Hasselt, BelgiumReviewed by:
Zofia Piotrowska-Seget, University of Silesia of Katowice, PolandAng Li, Harbin Institute of Technology, China
Copyright © 2020 Duraisamy, Sekar, Arunkumar and Ramalingam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prabavathy V. Ramalingam, cHJhYmF2YXRoeXZyQG1zc3JmLnJlcy5pbg==
 Purushothaman Duraisamy
Purushothaman Duraisamy Jegan Sekar
Jegan Sekar Anu D. Arunkumar
Anu D. Arunkumar Prabavathy V. Ramalingam
Prabavathy V. Ramalingam