- College of Animal Husbandry and Veterinary Science, Henan Agricultural University, Zhengzhou, China
The objective of this study was to explore the genetic and biological features of the tet(M)-harboring plasmid pTS14 in Salmonella enterica strain S14 isolated from a chicken fecal sample. Plasmid pTS14 was identified by conjugation, S1-pulsed-field gel electrophoresis (PFGE), Southern hybridization, and plasmid sequencing. The biological characteristics of pTS14 were assessed via stability, growth kinetics, and starvation survival experiments. Strain S14, belonging to ST3007, harbored a 119-kb tet(M)-bearing IncF2:A1:B1 conjugative plasmid pTS14. The plasmid pTS14 contained a novel transposon Tn6709 with the genetic structure IS26-tnpA1-tnpA2-Δorf13-LP-tet(M)-tnpX-ΔtnpR-IS26, and the resistance genes tet(B), tet(D), strAB, sul2, and blaTEM–1b. In addition, pTS14 was found to be highly stable in the recipient strain E. coli J53. The transconjugant TS14 exhibited a higher survival ratio than E. coli J53 under permanent starvation-induced stress. The tet(M)-bearing IncF2 epidemic plasmid lineage may accelerate the dissemination of tet(M) and other genes by coselection, which could constitute a potentially serious threat to clinical treatment regimens.
Introduction
The tetracycline resistance gene tet(M) encodes a ribosomal protection protein that confers tetracycline resistance to a variety of bacterial species (Franke and Clewell, 1981; Roberts et al., 1986; Donhofer et al., 2012; Roberts and Schwarz, 2016), including 38 genera of Gram-positive bacteria and 39 genera of Gram-negative bacteria, with most associated with other tet genes1, likely through the association with integrative and conjugative transposons located on the chromosome or conjugative plasmids, which facilitate horizontal transfer (Bryan et al., 2004; Jones et al., 2006; Tuckman et al., 2007; de Vries et al., 2009; Hu et al., 2013). In Gram-negative bacteria, the tet(M) gene was first reported in Escherichia coli in 2006 (Jones et al., 2006) and later described in Salmonella enterica isolates from chicken and pig feces in China in 2017 (Ma et al., 2017). To date, several plasmids harboring the tet(M) gene have been reported in E. coli, which include the IncHI2-type plasmids p1106 (MG825373), pECAZ147_2 (CP018993), and pTW4-IncHI2 (MK293945), as well as the hybrid IncN1-IncHI2-type plasmid pHN6DS (MH459020) and IncX1-FI:A:B plasmid pYPE12 (CP041443). The incompatibility group IncF is a main vehicle for the dissemination of the rmtB and/or blaCTX–MS genes in Enterobacteriaceae (Deng et al., 2011; Hou et al., 2012). However, there are relatively few reports of tet(M)-harboring IncF plasmids from Salmonella. Here, we report the complete sequence of the tet(M)-harboring IncF2:A1:B1 plasmid pTS14 isolated from S. enterica. A novel transposon, Tn6709 harboring the tet(M) gene, as well as three other resistance modules, were located on the same plasmid, pTS14. Moreover, the biological characteristics of plasmid pTS14 in Salmonella were further investigated.
Materials and Methods
Bacterial Strains
During a survey of the tet(M) gene in Henan Province, China, conducted in December 2017, one tet(M)-positive Salmonella strain, named S14, was isolated from the feces of a chicken. Strain identification was confirmed by PCR analysis and 16S rRNA sequencing along with MALDI-TOF MS detection (AXIMA Performance; Shimadzu Corporation, Kyoto, Japan) as described previously (Weisburg et al., 1991; Pavlovic et al., 2013). Subsequently, strain S14 was serotyped according to the Kauffmann–White scheme with the use of commercial antiserum.
Susceptibility Testing and Detection of Tetracycline Resistance Genes
The susceptibility of Salmonella strain S14 to 13 antibiotics (Supplementary Table S1) was determined via the broth microdilution method and interpreted in accordance with the guidelines of the Clinical and Laboratory Standards Institute (2017). Minimum inhibitory concentrations were calculated on three independent occasions. E. coli ATCC 25922 was used as a quality control strain. The tet(A), tet(B), tet(C), and tet(D) genes were screened by PCR as described previously (Sun et al., 2018).
Multilocus Sequence Typing (MLST)
To investigate the genetic typing of the isolate, MLST of seven housekeeping genes (thrA, purE, sucA, hisD, aroC, hemD, and dnaN) was performed as described previously (Yap et al., 2016). The sequences were subsequently submitted to the MLST database2 and assigned existing or novel allele type identification numbers. The corresponding sequence types were derived from the set of allelic profiles of each of the seven loci.
Conjugation Experiments
Mating experiments were conducted to evaluate the transferability of the tet(M) gene with the tet(M)-positive strain S14 as the donor and rifampicin-resistant Salmonella strain JS-500 and sodium azide-resistant E. coli strain J53 as the recipients. The transconjugants were selected on MacConkey agar plates supplemented with doxycycline (16 mg/L) and rifampicin (400 mg/L) for Salmonella JS-500 or doxycycline (16 mg/L) and sodium azide (200 mg/L) for E. coli J53. The conjugation frequency was calculated as the ratio of the number of transconjugants per recipient. All transconjugants were confirmed by pulsed-field gel electrophoresis (PFGE). The presence of the tet(M) and other tet genes in all transconjugants was confirmed by PCR analysis and sequencing.
S1-PFGE and Southern Hybridization
Prior to PFGE, DNA from the donor strain S14 and the corresponding E. coli J53 transconjugant (named TS14) were treated with S1 nuclease. The location of tet(M) gene was identified by Southern blot hybridization with the use of a probe for the tet(M) gene.
Plasmid Sequencing and Annotation
The plasmid from the transconjugant was extracted using the QIAGEN Plasmid Midi Kit (Qiagen, Hilden, Germany) and sequenced with Illumina Hiseq technology (Illumina, Inc., San Diego, CA, United States). Assembly of the generated sequences with Newbler software v2.6 (Roche Diagnostics Corporation, Indianapolis, IN, United States) generated nine contigs. Gaps between the contigs were closed by PCR and sequencing. The plasmid sequences were initially predicted and annotated using the Subsystem Technology (RAST v2.0) server (Aziz et al., 2008), and corrected manually using the BLASTn and BLASTp algorithms3. The plasmid replicon genotype was identified using the PlasmidFinder database4. Comparative analysis and generation of plasmid maps were performed using the Python application Easyfig and the BLAST Ring Image Generator (Alikhan et al., 2011; Sullivan et al., 2011).
Biological Characteristics of the tet(M)-Harboring Plasmid pTS14
To assess bacterial growth kinetics, the transconjugant TS14 and recipient E. coli J53 were incubated overnight at 37°C in lysogeny broth (LB; 5 mL) and then diluted to an optical density at 600 nm (OD600) of 0.004 in 20 mL of fresh LB with or without doxycycline (16 mg/L). Over a 15-h inoculation period, bacterial growth was measured and recorded every hour. To evaluate plasmid stability, transconjugant E. coli TS14 was maintained for 14 days in daily refreshed LB (100-fold dilution) without antibiotic selection, as previously described (Wu et al., 2018). Approximately 100 colonies were randomly chosen and replica plated onto LB agar plates with doxycycline. All colonies grown on doxycycline-supplemented agar were subjected to PCR analysis to confirm the presence of the tet(M) gene. M9-glycerin (0.2%) minimum medium was used for starvation survival experiments of E. coli J53, TS14, and a mixture of both. The percent survival was calculated as the ratio of the mean number of CFUs divided by the number of CFUs after overnight incubation for each of the triplicate suspensions on days 1, 2, 3, 5, 7, 10, and 15. Experiments were repeated in three separate assays.
Nucleotide Sequence Accession Number
The complete sequence of plasmid pTS14 was submitted to the GenBank under the accession number MN328348.
Results and Discussion
Characterization of Salmonella Strain S14
The tet(M)-positive strain S14 was serotyped as S. enterica, belonging to ST3007. The results of the conjugation assay indicated that the tet(M) gene can be successfully transferred to Salmonella JS-500 and E. coli J53 at frequencies of 1.638 × 10–4 and 1.397 × 10–4, respectively, which were assigned as TS14-JS500 and TS14. Susceptibility testing showed that S14, TS14-JS500, and TS14 were all resistant to doxycycline, tetracycline, gentamicin, sulfamethoxazole-trimethoprim, and amoxicillin (Supplementary Table S1). The transconjugant TS14 was selected for further study. S1-PFGE and Southern hybridization indicated that the tet(M) gene was located on a plasmid of ∼119 kb in size, designated as pTS14 (Supplementary Figure S1). The results of the mating experiments demonstrated that plasmid pTS14 harboring tet(M) can be self-transmissible and spread between and across genera.
Characterization of Plasmid pTS14
Sequencing of the plasmid pTS14 from transconjugant TS14 revealed a 119,493-bp circular episome with a mean G + C content of 50.81% with 155 putative open reading frames (ORFs). The plasmid pTS14, belonging to IncF2:A1:B1, has a typical FII-type backbone and three conserved modules (Figure 1): (i) regions encoding the plasmid replication gene repA; (ii) regions responsible for plasmid transfer and two conserved pilus-encoding loci (tra and trb); (iii) plasmid maintenance region, plasmid segregation system (including ParAB and PsiAB) and a multiple toxin-antitoxin (TA)-based addiction system (including PemI/PemK, CcdA/CcdB, and Hok); and two accessory regions (regions encoding antimicrobial resistance and virulence).
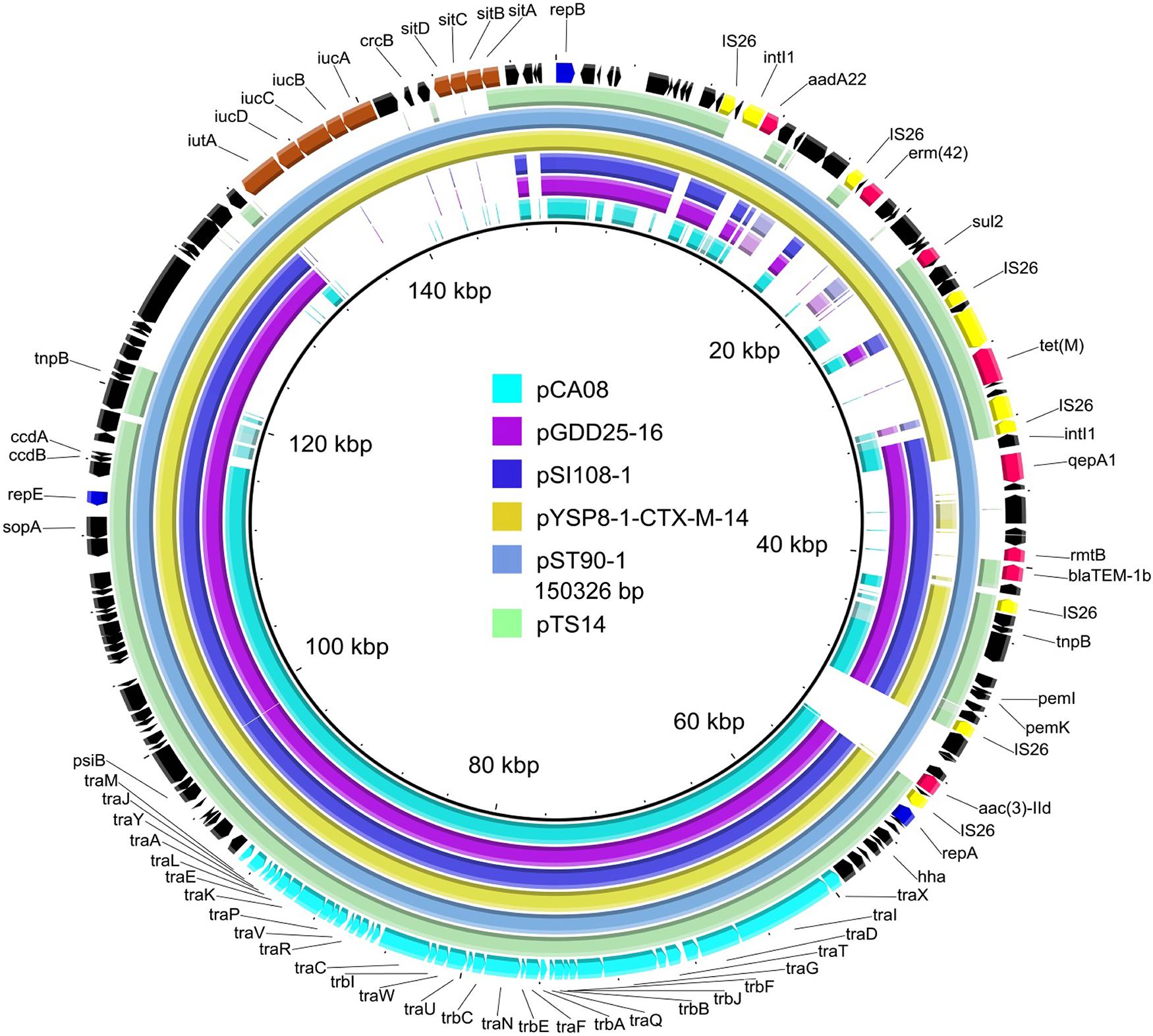
Figure 1. Whole-plasmid sequence of pTS14 and comparison of similar IncF2 plasmids. The 150326-bp pST90-1 was used as a reference plasmid at the highest coverage (79%). Key features of pST90-1 are highlighted in different colors. Replicon genes are in blue; transfer-associated genes in cyan; resistance genes in red; mobile elements in yellow; the loci of stability-associated genes (iutA-iucABCD and sitABCD) in maroon; and hypothetical proteins in black. The outer ring comprises the CoDing sequence of pST90-1. The plasmids in this study included pTS14 (IncF2:A1:B1, MN328348), pST90-1 (IncF2:A1:B58, CP050735), pYSP8-1-CTX-M-14 (IncF2:A1:B1, CP037912), pSI108-1 (IncF2:A1:B58, CP050770), pGDD25-16 (IncF2:A1:B1, MH316135), and pCA08 (IncF2:A16:B20, CP009233).
As shown in Figure 1, BLASTn comparisons demonstrated that plasmid pTS14 (IncF2:A1:B1) shared high homology to S. enterica serovar Typhimurium plasmid pST90-1 (IncF2:A1:B58, CP050735) with 99.97% identity at 81% coverage and to E. coli plasmid pYSP8-1-CTX-M-14 (IncF2:A1:B1, CP037912) with 100% identity at 80% coverage. In general, the incompatibility group IncFII is an epidemic plasmid lineage responsible for the dissemination of the rmtB and/or blaCTX–MS genes in Enterobacteriaceae (Deng et al., 2011; Hou et al., 2012). However, reports of tet(M)-harboring IncF plasmids are very scarce. Interesting, pTS14, pST90-1, and pYSP8-1-CTX-M-14 are the only three IncF2 plasmids known to carry the tet(M) gene (Figure 1). Several other plasmids harboring the tet(M) gene have been reported, such as the IncHI2-type plasmids p1106 (MG825373) and pECAZ147_2 (CP018993), pTW4-IncHI2 (MK293945), and the hybrids IncN1-IncHI2 pHN6DS (MH459020), and IncX1-FI:A:B pYPE12 (CP041443). In addition, pTS14 also exhibited high homology to other IncF2-type plasmids: i.e., the blaCTX–M–14-bearing E. coli plasmid pCA08 (CP009233) with 99.76% identity at 75% coverage, the blaCTX–M–27- and the rmtB-bearing Salmonella plasmid pSI108-1 (CP050770) with 99.97% identity at 73% coverage, and the blaCTX–M–27- and rmtB-bearing Salmonella plasmid pGDD25-16 (MH316135) with 99.97% identity at 71% coverage (Figure 1).
Comparative analysis revealed that the most noticeable difference among these IncF2 plasmids lay in the region encoding multidrug resistance (MDR) and virulence factors. The MDR region of pTS14 is composed of several segments, successively including a truncated transposon Tn10-like segment carrying the tet(B) and tet(D) genes, a Feo system for ferrous iron utilization, a tet(M)-containing segment, a resistance cluster of sul2-strA-strB, and an incomplete Tn3-harboring blaTEM–1b transposon (Figure 2). The truncated Tn10-like transposon, the first segment, has undergone deletion of IS10L and jemA, which are located upstream of the truncated jemB, and another IS10R that was divided into three segments, which were disrupted by the insertion of IS26 and IS1P with 8-bp direct repeats. The similar structure was observed in plasmid pJSWP006-1 (AP018939), and differed by deleting the IS10R in RCS89-p (LT985304) (Figure 2). This segment along with the Feo system exhibited high (99.99%) identity with those of plasmid pJSWP006-1 (IncF-:A1:B1, AP018939) with the exception of a three-nucleotide substitution (Figure 2). The sul2-strA-strB module, followed by an incomplete Tn3 transposon carrying blaTEM–1b, as the last segment, was located downstream of the tet(M)-containing segment. However, an incomplete Tn3 transposon carrying blaTEM–1b and rmtB was located upstream of the qepA1 in plasmid pST90-1 (Supplementary Figure S2).
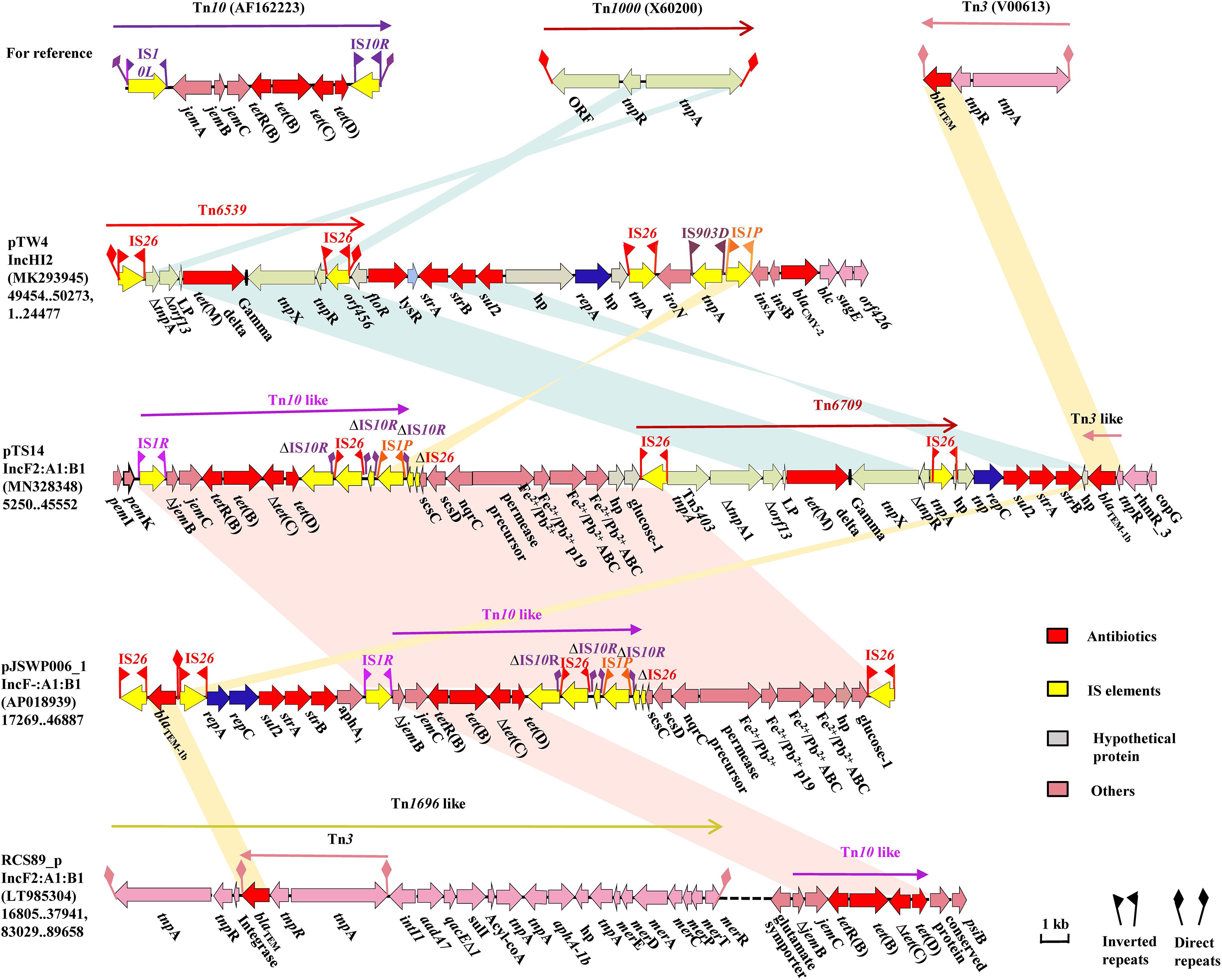
Figure 2. The MDR region of plasmid pTS14 and comparisons with related regions. Genes, mobile elements and other features are colored based on functional classifications. Numbers in parentheses show the nucleotide positions within the corresponding plasmids. Shaded regions denote homologous DNA regions (>97% nucleotide identity). Resistance genes are in red; mobile elements are in yellow; hypothetical proteins are in gray, and others in pink.
The tet(M)-bearing segment, an IS26-bracked composite transposon, was designated as Tn6709 using the Transposon Nomenclature Database5. Tn6709 consists of the insertion sequence IS26, an incomplete transposase tnpA from Tn5403, the incomplete transposase ΔtnpA from the Tn3 family, truncated orf13, the tet(M) leader peptide (LP) gene, the tetracycline resistance gene tet(M), the tnpX and ΔtnpR genes from the Tn1000 transposon, and the IS26 element (Figure 3). Comparative sequence analysis showed that fragment Δorf13-lp-tet(M) was relatively conserved with 100% sequence identity to the other six sequences displayed in Figure 3. Interestingly, the 3′-terminal end of the partial or complete sequence encoding a Tn3-family transposase was located upstream of Δorf13 in six similar sequences and Tn6709, indicating that the transfer of tet(M) was closely related to that of the Tn3 family. Besides, for Tn6709, only two sequences (p41-3 and pTW4) carried the IS26 element upstream of the tnpA or tnpA1 gene. Furthermore, the IS26 element was located downstream of the incomplete tnpR in plasmids pTW4, pYSP8-1-CTX-M-14, and pTS14. The IS26 element plays a key role in the reorganization of the MDR region of plasmids (Harmer et al., 2014). In this study, reverse PCR was performed to explore the existence of a circular intermediate of the IS26-bracked composite transposon Tn6709. However, no circular intermediate could be obtained from strain S14. In addition, sequence analysis revealed that there were no direct repeats flanking the IS26 element in transposon Tn6709, indicating that the IS26-bracked composite transposon Tn6709 acquired by pTS14 may have occurred by recombination rather than transposition.
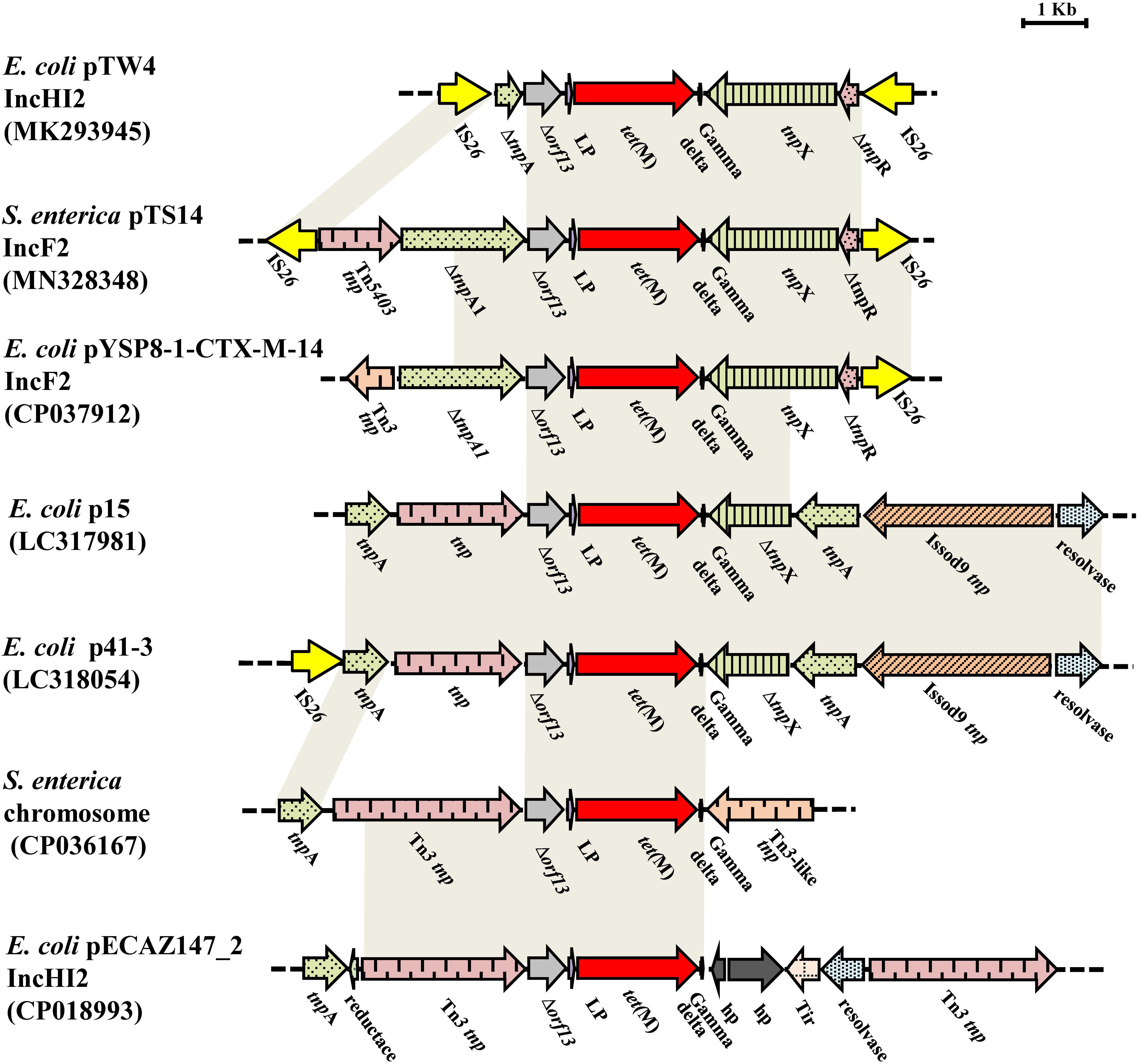
Figure 3. Comparisons of the genetic environment of Tn6709 harboring tet(M) in MN328348, with other sequences carrying tet(M) retrieved from the GenBank database. Similar regions are indicated by dotted lines (hp, gene encoding hypothetical protein; lp, gene encoding tet(M) leader peptide).
The tet(M) gene was originally designated by Burdett et al. (1982), and was subsequently found to be frequently connected with transposons in Gram-positive bacteria, such as the Tn916 transposon of Streptococcus faecalis (Roberts and Kenny, 1987), the Tn5801-like transposons of Staphylococcus aureus and Enterococcus faecalis (de Vries et al., 2009), and the Tn5397-like transposons of Clostridioides difficile and Enterococcus faecium (Agerso et al., 2006). In Gram-negative bacteria, the tet(M) gene was first reported in E. coli in 2006 as being flanked by IS26 and ISVs1 (Jones et al., 2006), and was first described in S. enterica isolated from chicken and pig feces in China in 2017 (Ma et al., 2017). The tet(M) gene was found to be associated with the Tn6539 transposon (Sun et al., 2018). In the present study, the tet(M) gene was located on the novel composite transposon Tn6709 in IncF2:A1:B1, related to the epidemic plasmid lineage IncF2. Taken together, these results suggest that the tet(M) gene can transfer from Gram-positive bacteria to Gram-negative bacteria and disseminate between different plasmid groups.
Comparative analysis demonstrated that the tet(M)-bearing plasmid pTS14 harbored a Feo system responsible for ferrous iron transportation (Figure 2), while the tet(M)-bearing plasmids pST90-1 and pYSP8-1-CTX-M-14 were absent. Interesting, plasmids pST90-1 and pYSP8-1-CTX-M-14 both contained aerobactin (iutA-iucABCD) and Sit (sitABCD) loci (Figure 1 and Supplementary Figure S2). Iron acquisition systems play important roles in colonization and pathogenicity of many bacteria, particularly the aerobactin (iutA-iucABCD), Sit (sitABCD) and Feo system loci (Sabri et al., 2008; Lau et al., 2016; Khajanchi et al., 2019). In Salmonella, the iron acquisition system can be encoded by chromosomal pathogenicity islands (Janakiraman and Slauch, 2000) or plasmid genes (Khajanchi et al., 2017). In S. enterica, the Sit (sitABCD) and aerobactin iron acquisition (iucABCD-iutA) systems encoded by the IncFIB plasmid have been well characterized, and are reported to enhance virulence potential of intestinal epithelial cells (Khajanchi et al., 2017). In the present study, the Feo system was encoded by the IncF2:A1:B1 plasmid, which may be associated with other biological traits. Thus, the stability of the tet(M)-harboring plasmid was further assessed in E. coli strain TS14.
The Biological Characteristics of the Plasmid pTS14
In the bacterial growth experiment, the growth rate of the transconjugant TS14 was similar to that of the recipient E. coli J53 in LB broth without doxycycline, while that with doxycycline was slightly lower than the recipient in LB broth without doxycycline (Figure 4A). The plasmid pTS14 existed stably in transconjugant TS14 for at least 14 days of passage in an antibiotic-free environment. The survival rate of TS14 was highest, followed by the mixture of TS14 and E. coli J53, and lowest for E. coli J53, which demonstrates that the growth of the transconjugant TS14 harboring pTS14 was not lower than that of E. coli J53. After 15 days, the bacterial numbers of the three cultures all decreased to less than 10% (Figure 4B). Overall, in this initial study, plasmid pTS14 carrying the Feo system did not affect bacterial growth, although there was no benefit to the transconjugant harboring pTS14. Further investigations are warranted to elucidate the role of the Feo acquisition system of plasmids in Salmonella.
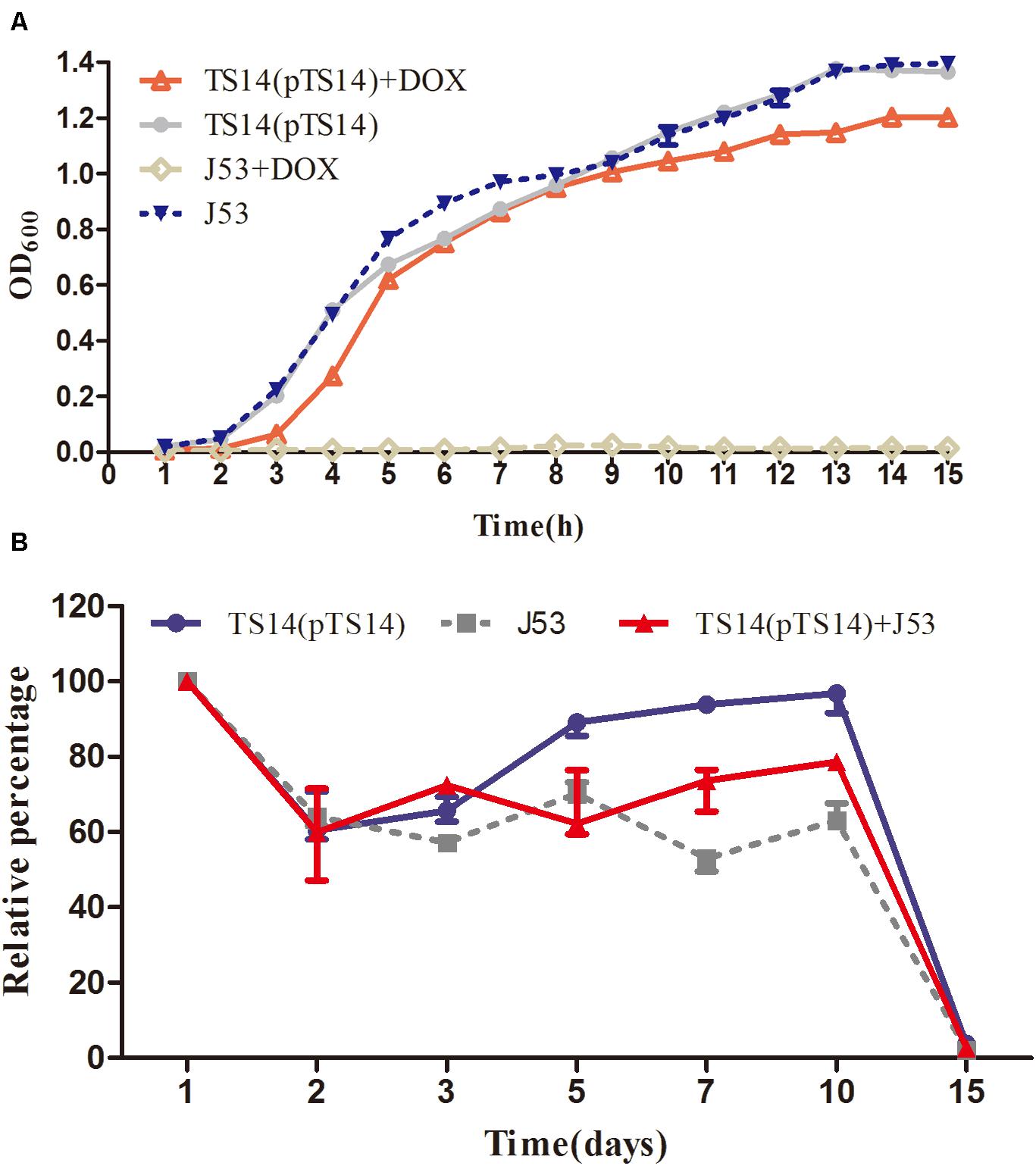
Figure 4. Growth kinetics of the transconjugant TS14 and recipient E. coli J53. (A) The growth kinetics of transconjugant TS14 harboring pTS14 and recipient E. coli J53 over a 15-h inoculation period in the presence and absence of doxycycline (16 μg/mL). (B) The starvation survival of J53, TS14, and their mixture. The values are presented as the mean ± standard deviation.
Conclusion
In summary, the self-transmissible IncF2:A1:B1 plasmid pTS14 from Salmonella carried the tet(M), tet(B) and tet(D) genes, along with the sul2-strA-strB module and the incomplete blaTEM–1b-bearing transposon Tn3. The tet(M) gene was located on the novel composite transposon Tn6709 and flanked by two IS26 elements oriented in the opposite direction. In addition, the plasmid pTS14 harbored a Feo system responsible for ferrous iron transportation.
The epidemic IncF2 plasmids have the potential to carry both virulence and antimicrobial resistance determinants, which might contribute to the further dissemination of the tet(M) gene via co-selection by other antimicrobials. Therefore, there is a need to monitor the dissemination of this MDR- and virulence-associated plasmid among Enterobacteriaceae.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
GH and YP conceived and designed the experiments. YL, XL, and XC performed the experiments. MC, JL, and YL analyzed the data. SL, DH, and LY contributed to reagents, materials, and analysis tools. YL and GH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Key Research and Development Program of China (2016YFD05101304).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01523/full#supplementary-material
FIGURE S1 | S1-PFGE (A) and Southern hybridization (B) of Salmonella strain S14 and the corresponding transconjugant TS14 with the tet(M) gene as a probe. Marker, Salmonella Braenderup H9812.
FIGURE S2 | Comparative analysis of pTS14 with other IncF-type plasmids. Homologous segments generated by a BLASTn comparison (≥97% identity) are shown as gray boxes. Genes are represented by thick arrows. The color code equates to that described in this figure legend.
TABLE S1 | The MICs of S14 and transconjugant in this study.
Footnotes
- ^ http://faculty.washington.edu/marilynr/
- ^ http://mlst.warwick.ac.uk
- ^ http://blast.ncbi.nlm.nih.gov/blast
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://transposon.lstmed.ac.uk/
References
Agerso, Y., Pedersen, A. G., and Aarestrup, F. M. (2006). Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in Enterococci from humans, pigs and poultry. J. Antimicrob. Chemother. 57, 832–839. doi: 10.1093/jac/dkl069
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The rast server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bryan, A., Shapir, N., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507. doi: 10.1128/aem.70.4.2503-2507.2004
Burdett, V., Inamine, J., and Rajagopalan, S. (1982). Heterogeneity of tetracycline resistance determinants in Streptococcus. J. Bacteriol. 149, 995–1004. doi: 10.1128/jb.149.3.995-1004.1982
Clinical and Laboratory Standards Institute (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
de Vries, L. E., Christensen, H., Skov, R. L., Aarestrup, F. M., and Agerso, Y. (2009). Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 64, 490–500. doi: 10.1093/jac/dkp214
Deng, Y., He, L., Chen, S., Zheng, H., Zeng, Z., Liu, Y., et al. (2011). F33:A-:B- and F2:A-:B- plasmids mediate dissemination of rmtB-blaCTX-M-9 group genes and rmtB-qepA in Enterobacteriaceae isolates from pets in China. Antimicrob. Agents Chemother. 55, 4926–4929. doi: 10.1128/AAC.00133-11
Donhofer, A., Franckenberg, S., Wickles, S., Berninghausen, O., Beckmann, R., and Wilson, D. N. (2012). Structural basis for tet(M)-mediated tetracycline resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 16900–16905. doi: 10.1073/pnas.1208037109
Franke, A. E., and Clewell, D. B. (1981). Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145, 494–502. doi: 10.1128/jb.145.1.494-502.1981
Harmer, C. J., Moran, R. A., and Hall, R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14
Hou, J., Huang, X., Deng, Y., He, L., Yang, T., Zeng, Z., et al. (2012). Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 56, 2135–2138. doi: 10.1128/aac.05104-11
Hu, G. Z., Pan, Y. S., Wu, H., Hu, H., Xu, R., Yuan, L., et al. (2013). Prevalence of tetracycline resistance genes and identification of tet(M) in clinical isolates of Escherichia coli from sick ducks in China. J. Med. Microbiol. 62, 851–858. doi: 10.1099/jmm.0.051896-0
Janakiraman, A., and Slauch, J. M. (2000). The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35, 1146–1155. doi: 10.1046/j.1365-2958.2000.01783.x
Jones, C. H., Tuckman, M., Murphy, E., and Bradford, P. A. (2006). Identification and sequence of a tet(M) tetracycline resistance determinant homologue in clinical isolates of Escherichia coli. J. Bacteriol. 188, 7151–7164. doi: 10.1128/jb.00705-06
Khajanchi, B. K., Hasan, N. A., Choi, S. Y., Han, J., Zhao, S., Colwell, R. R., et al. (2017). Comparative genomic analysis and characterization of incompatibility group FIB plasmid encoded virulence factors of Salmonella enterica isolated from food sources. BMC Genomics 18:570. doi: 10.1186/1471-2164-12-570
Khajanchi, B. K., Xu, J., Grim, C. J., Ottesen, A. R., Ramachandran, P., and Foley, S. L. (2019). Global transcriptomic analyses of Salmonella enterica in Iron-depleted and Iron-rich growth conditions. BMC Genomics 20:490. doi: 10.1186/1471-2164-12-490
Lau, C. K., Krewulak, K. D., and Vogel, H. J. (2016). Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev. 40, 273–298. doi: 10.1093/femsre/fuv049
Ma, S., Lei, C., Kong, L., Jiang, W., Liu, B., Men, S., et al. (2017). Prevalence, antimicrobial resistance, and relatedness of Salmonella isolated from chickens and pigs on farms, Abattoirs, and Markets in Sichuan Province, China. Foodborne Pathog. Dis. 14, 667–677. doi: 10.1089/fpd.2016.2264
Pavlovic, M., Huber, I., Konrad, R., and Busch, U. (2013). Application of MALDI-TOF MS for the identification of food borne bacteria. Open Microbiol. J. 7, 135–141. doi: 10.2174/1874285801307010135
Roberts, M. C., Hillier, S. L., Hale, J., Holmes, K. K., and Kenny, G. E. (1986). Tetracycline resistance and tet(M) in pathogenic urogenital bacteria. Antimicrob. Agents Chemother. 30, 810–812. doi: 10.1128/aac.30.5.810
Roberts, M. C., and Kenny, G. E. (1987). Conjugal transfer of transposon Tn916 from Streptococcus faecalis to Mycoplasma hominis. J. Bacteriol. 169, 3836–3839. doi: 10.1128/jb.169.8.3836-3839.1987
Roberts, M. C., and Schwarz, S. (2016). Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J. Environ. Q. 45, 576–592. doi: 10.2134/jeq2015.04.0207
Sabri, M., Caza, M., Proulx, J., Lymberopoulos, M. H., Bree, A., Moulin-Schouleur, M., et al. (2008). Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain chi7122. Infect. Immun. 76, 601–611. doi: 10.1128/IAI.00789-07
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, Y. W., Liu, Y. Y., Wu, H., Wang, L. F., Liu, J. H., Yuan, L., et al. (2018). IS26-flanked composite transposon Tn6539 carrying the tet(M) gene in IncHI2-type conjugative plasmids from Escherichia coli isolated from ducks in China. Front. Microbiol. 9:3168. doi: 10.3389/fmicb.2018.03168
Tuckman, M., Petersen, P. J., Howe, A. Y., Orlowski, M., Mullen, S., Chan, K., et al. (2007). Occurrence of tetracycline resistance genes among Escherichia coli isolates from the phase 3 clinical trials for tigecycline. Antimicrob. Agents Chemother. 51, 3205–3211. doi: 10.1128/AAC.00625-07
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/JB.173.2.697-703.1991
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness advantage of mcr-1-Bearing IncI2 and IncX4 plasmids in Vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.0331
Keywords: Salmonella enterica, tet(M), Tn6709, biological features, IncF2:A1:B1 plasmid
Citation: Liu Y, Liu X, Cui X, Chen M, Li S, He D, Liu J, Yuan L, Hu G and Pan Y (2020) Characterization of pTS14, an IncF2:A1:B1 Plasmid Carrying tet(M) in a Salmonella enterica Isolate. Front. Microbiol. 11:1523. doi: 10.3389/fmicb.2020.01523
Received: 29 March 2020; Accepted: 12 June 2020;
Published: 03 July 2020.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Séamus Fanning, University College Dublin, IrelandJian-Hua Liu, South China Agricultural University, China
Copyright © 2020 Liu, Liu, Cui, Chen, Li, He, Liu, Yuan, Hu and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gong-zheng Hu, eWFvbGlsYWJAMTYzLmNvbQ==; Yu-shan Pan, cHlsZWFybjIxQDE2My5jb20=
†These authors have contributed equally to this work
 Ying-ying Liu
Ying-ying Liu Xiao-kang Liu†
Xiao-kang Liu† Gong-zheng Hu
Gong-zheng Hu