
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 June 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01436
Adult females of stag beetles (Coleoptera: Lucanidae) possess an ovipositor-associated mycangium for conveying symbiotic microorganisms. In most lucanid species, their mycangium contains yeast symbionts of the genus Scheffersomyces Kurtzman and Suzuki that are known for their xylose-fermenting capability. The lucanid genus Platycerus Geoffroy, 1762 is a group of small blue stag beetles, in which ten Japanese species constitute a monophyletic clade. Here we examined the evolutionary relationships of these Japanese Platycerus species and their yeast symbionts, together with a Korean Platycerus species and other lucanid species as outgroup taxa. Based on the internal transcribed spacer (ITS) and the intergenic spacer (IGS) sequences, the yeast symbionts of all Platycerus species were closely related to each other and formed a monophyletic clade. There is no variation in ITS sequences of the yeast symbionts of the Japanese Platycerus species. Based on IGS sequences, the yeast symbionts formed clusters that largely reflected the geographic distribution of the host insects, being shared by sympatric Platycerus species except for P. delicatulus Lewis, 1883 and P. viridicuprus Kubota & Otobe, The symbiont phylogeny was globally not congruent with the host COI-based phylogeny, although some local congruences were observed. Statistically significant correlations were detected between the genetic distances of COI sequences of the host insects and those of IGS sequences of the yeast symbionts in Japan. These results suggest that, at least to some extent, the host insects and the yeast symbionts may have experienced co-evolutionary associations. While the Japanese Platycerus species formed a monophyletic clade in the COI phylogeny, the yeast symbionts of Japanese P. viridicuprus were very closely related to those of Korean P. hongwonpyoi Imura & Choe, 1989, suggesting the possibility that a recent secondary contact of the two beetle species during a marine withdrawal, e.g., in the last glacial period, might have resulted in an inter-specific horizontal transmission of the yeast symbiont.
Symbiotic relationships with fungi are known in diverse insect taxa (Vega and Blackwell, 2005; Biedermann and Vega, 2020). Particularly those between wood-inhabiting insects and fungi have attracted much attention, because such associations often enable utilization of woody materials that are abundant in the environment but difficult to digest as food. Since wood mainly consists of polymers such as cellulose, hemicellulose and lignin that are indigestible for most animals, microbial assistance is pivotally important for wood-feeding insects (Parkin, 1940; Haack and Slansky, 1987; Stokland, 2012). Although bacterial or protozoan symbionts of termites and wood-feeding cockroaches are anaerobic gut symbionts (Cleveland, 1924; Slaytor, 1992; Breznak and Brune, 1994), most fungal symbionts of xylophagous insects are aerobic and either gut or external symbionts (Batra, 1963; Suh et al., 2003, 2006; Nguyen et al., 2007; Grünwald et al., 2010). In ambrosia beetles, wood wasps and many other insects, fungal symbionts are carried and dispersed by external pouch-like organs called mycangia or mycetangia (Grebennikov and Leschen, 2010; Biedermann and Vega, 2020).
Stag beetles of the family Lucanidae, which embrace over 1,000 species in the world, mainly feed on decaying wood at their larval stages (Tanahashi et al., 2010; Kim and Farrell, 2015). Adult females of the Lucanidae possess a mycangium, constituted by an invaginated pouch of intersegmental membrane on the dorsal side of the abdominal tip, for conveying symbiotic microorganisms, which have been found in all the lucanid species representing 13 genera so far examined (Aegus Macleay, 1819; Dorcus Macleay, 1819; Prosopocoilus Hope & Westwood, 1845; Lucanus Scopoli, 1763; Neolucanus Thomson, 1862; Prismognathus Motschulsky, 1860; Figulus MacLeay, 1819; Nigidius MacLeay, 1819; Platycerus Geoffroy, 1762; Aesalus Fabricius, 1801; Nicagus LeConte, 1862; Ceruchus Macleay, 1819; Sinodendron Hellwig, 1792) (Tanahashi et al., 2010, 2017; Tanahashi and Hawes, 2016). They commonly harbor yeast symbionts in their mycangia (Tanahashi et al., 2010, 2017; Hawes, 2013) most of which belong to the genus Scheffersomyces Kurtzman & M. Suzuki known as a xylose-fermenting yeast group (Du Preez and Prior, 1985; Jeffries and Kurtzman, 1994; Olsson and Hahn-Hägerdal, 1996). Phylogenetically, the yeast symbionts are closely related (or belong) to S. stipitis (Pignal) Kurtzman & M. Suzuki or S. segobiensis (Santa María & C. García) Kurtzman & M. Suzuki (Tanahashi et al., 2010, 2017). Xylose is a main component of hemicelluloses in broad-leaved trees (Sjöström, 1993). Considering that hemicelluloses are difficult to utilize for most animals, the yeast symbionts of the stag beetles have been suspected to help lucanid larvae digest hemicelluloses (Tanahashi et al., 2010). The presence of mycangium in all lucanid species suggests an intimate relationship between the stag beetles and their yeast symbionts over evolutionary time. The yeast symbionts seem likely to be mutualistic to the host stag beetles, but so far, no proof for this has been found.
The genus Platycerus is a group of small blue stag beetles, widely distributed in the northern hemisphere and comprising more than 50 species, of which over a half are found in East Asia (29 species in China; 1 species in Korea; and 10 species in Japan based on Huang et al., 2010; Imura, 2010a, b, 2011, 2012; Huang and Chen, 2017; Kubota et al., 2008). Kubota et al. (2011) and Zhu et al. (2019), Zhu et al. (unpublished) inferred that all Platycerus species distributed in Japan are monophyletic and have been speciated since several hundred thousand years to some six million years ago. At most, three Platycerus species may be sympatrically found in Japan (Imura, 2010b; Kubota et al., 2011). Since many closely related species are distributed in the Japanese archipelago, the Platycerus stag beetles represent an interesting research target to investigate the evolutionary relationship between the host insects and the yeast symbionts in their speciation process.
Previously, it was shown that the yeast symbionts of each one female of Platycerus delicatulus Lewis, 1883 and P. acuticollis K. Kurosawa, 1969 from Japan and seven females of P. hongwonpyoi Imura & Choe 1989 from South Korea exhibited no genetic variation based on the sequences of the internal transcribed spacer (ITS) region and 26S ribosomal RNA gene (26S rRNA), which have been often used for fungal identification (Tanahashi et al., 2017). These sequences were the most similar to homologous sequences from the fungal symbionts of Prismognathus angularis Waterhouse, 1874 from Japan. In an attempt to detect more genetic variation among the yeast symbionts of the stag beetles, Tanahashi et al. (2017) adopted another molecular marker, intergeneic spacer (IGS) region including 26S rRNA, IGS1, 5S ribosomal RNA gene (5S rRNA), IGS2, and 18S ribosomal RNA gene (18S rRNA) (>2,000 bp). The yeast symbionts showed some IGS sequence differences among collection sites and host species (Tanahashi et al., 2017).
In this study, we analyzed the evolutionary relationship between the stag beetles and their yeast symbionts. By focusing on the Platycerus species in Japan, (1) we investigated the genetic variation of the Platycerus-associated yeast symbionts based on ITS and IGS regions, and (2) we analyzed the presence or absence of co-evolutionary and geographic relationships between the host insects and the yeast symbionts.
In total, ten Platycerus species and five subspecies were collected at the sampling sites listed in Table 1. Most samples were obtained as adults in the field, and some were collected as larvae from decaying wood and reared individually to adulthood with the same wood pieces in which they were sampled in the field. Finally, we obtained in total 61 adult females representing all the Platycerus species and subspecies collected at 30 sites in Japan. We also collected 11 females of eight other lucanid species in Japan as outgroup taxa (Supplementary Tables 1, 2 and Figure 1A).
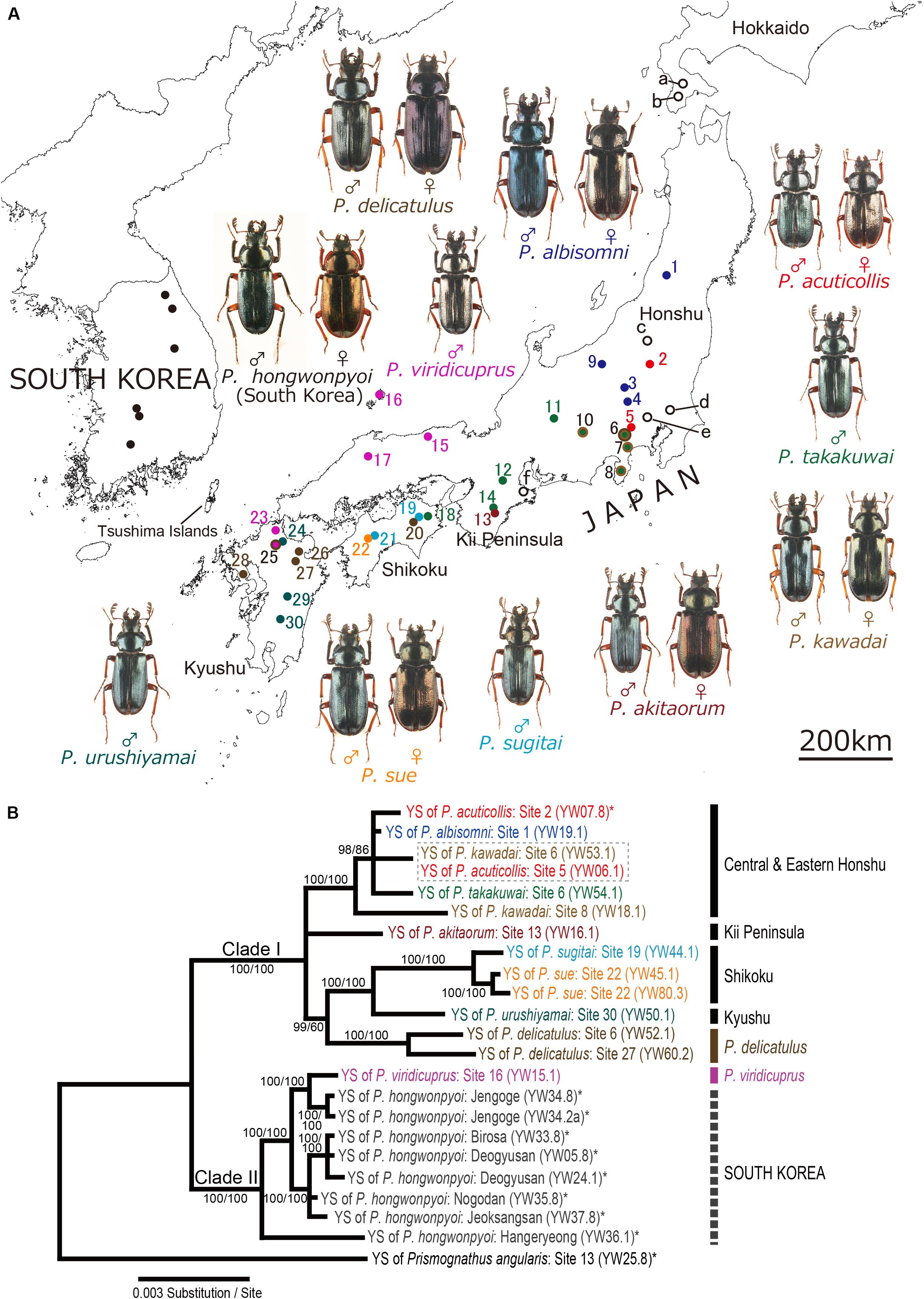
Figure 1. (A) Sample collection sites. Colored circles 1–30, collection sites of Japanese Platycerus species; open circles a-f, collection sites of outgroup lucanid species; filled circles in South Korea, collection sites of P. hongwonpyoi (Tanahashi et al., 2017). Note that sites a–c include several collection points. Also see Supplementary Table 1. (B) Bayesian inference (BI) phylogeny of the yeast symbionts of Platycerus stag beetles based on combined data of ITS and IGS sequences. Numbers at the nodes indicate posterior probability for BI phylogeny (>50%)/that recoding the gaps (50%). YS means yeast symbionts. Numbers following YW indicate the female code number and the strain number (see Supplementary Table 2). Dotted boxes indicate the combined haplotypes shared by more than one species of the host stag beetles. Asterisks indicate the sequences reported in previous studies. F81 model (ITS) and HKY+I model (IGS) were selected as the best-fit substitution model by jModelTest ver. 2.1.7. The BI phylogenies are identical regardless of the gap treatment.
A mycangium, which is located under the eighth abdominal tergite, was dissected from each adult female in sterilized phosphate-buffered saline (PBS) as described in Tanahashi et al. (2017) (Figure 2A). After removing surrounding exoskeletal membranes, the dissected mycangium was homogenized in PBS using a pellet pestle in a plastic tube, and each of 5-fold dilution series of the homogenate (equivalent to 1/51, 1/52, …, 1/55 of the dissected mycangium) was spread onto a potato dextrose agar (PDA: Sigma Aldrich, St. Louis, MO, United States) plate containing 50 μg/ml rifampicin. The plates were incubated at 20°C for 4 days, which had been established as suitable temperature and duration for growth of the Platycerus yeast symbionts (Tanahashi et al., 2017). The number of colony-forming units (CFU) per organ was calculated based on a plate on which an adequate number (usually 30∼300) of colonies appeared (Figure 2A).
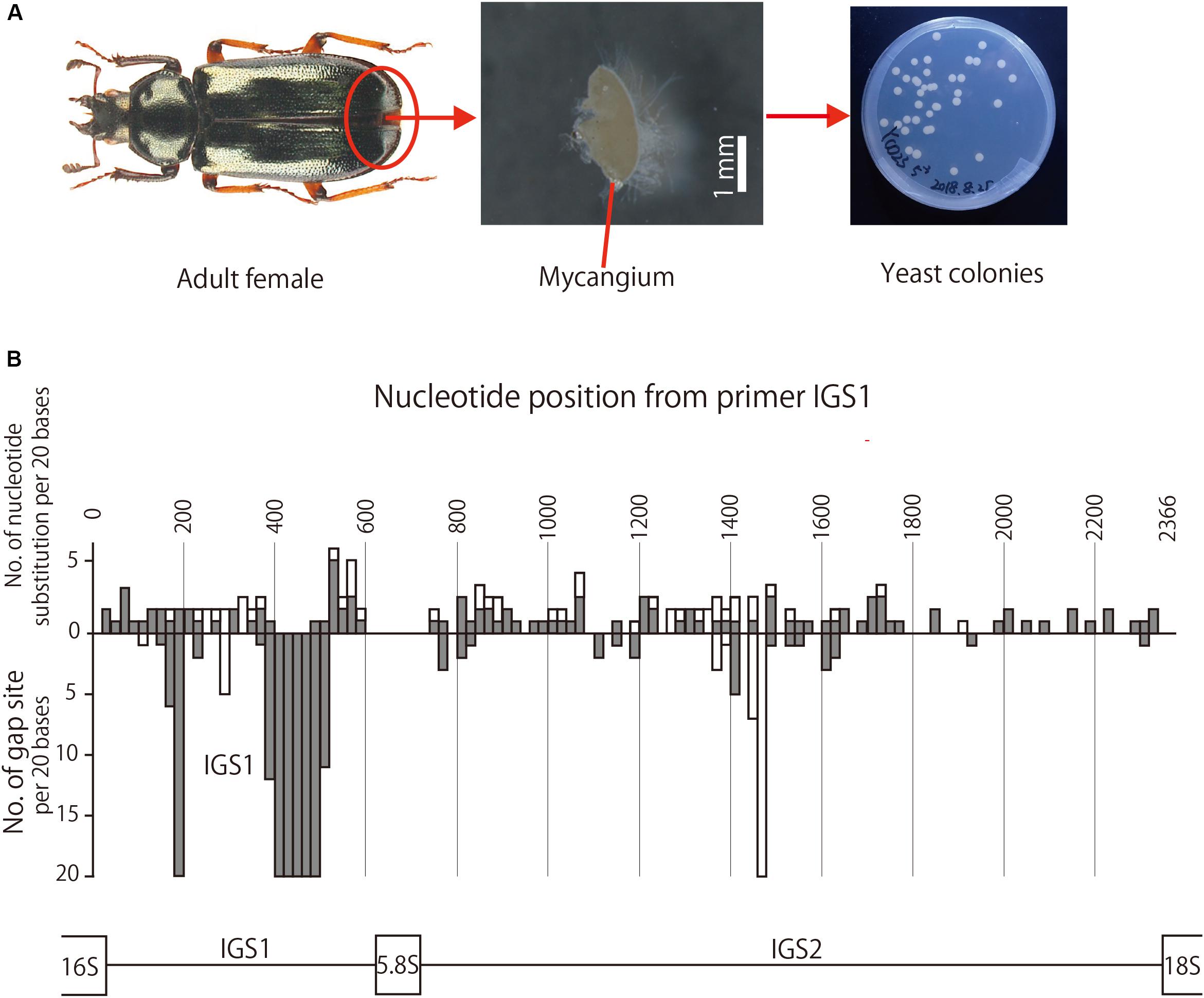
Figure 2. (A) Adult female, dissected mycangium, and isolated yeast colonies of P. kawadai. (B) Site-dependent sequence polymorphisms in IGS region among the yeast symbionts of Japanese Platycerus species (n = 60), P. hongwonpyoi from South Korea (n = 6), and Prismognathus angularis (n = 1). Bars above the horizontal line represent the numbers of nucleotide substitution, and those bellow the horizontal line represent the numbers of gap sites for every 20 bases of sequences. White and gray bars indicate the numbers within all data sets and those within Platycerus species, respectively.
We obtained yeast symbiont colonies from mycangial homogenates prepared from 60 of 61 Platycerus females (98.4 %) collected in Japan. We also obtained yeast symbiont colonies from the other lucanid species (Supplementary Table 2). The colonies on PDA plates were uniform, white, circular, and protuberant, having a smooth edge and matte surface. The yeast CFU values ranged from 9.2 × 102 to 5.3 × 104 (n = 59) for the Platycerus species, and from 2.1 × 104 to 4.2 × 105 (n = 11) for the other lucanid species.
Since no morphological differences were observed among the yeast colonies that appeared on the plate, we randomly selected 2–8 colonies for each host individual and transferred them to new PDA plates for subsequent DNA analyses. A small pellet of each yeast colony was suspended in 50 μL of lyticase solution (0.4 U/μL lyticase, 50 mM ethylenediaminetetraacetic acid [EDTA]) and incubated at 37°C for 2 h. The cell suspension was centrifuged at 9,000 rpm for 5 min, and the supernatant containing cell wall saccharides was discarded. The yeast pellet was then re-suspended in 80 μl of the cell lysis solution (1% SDS, 4 M urea, 1 mM EDTA, 150 mM NaCl and 50 mM Tris-HCl; pH 8.0) and incubated at 55°C for 1 h. After cooling, 27 μl (one-third volume of the lysate) of 8 M ammonium acetate was added, mixed vigorously and incubated on ice for 10 min. The solution was centrifuged at 9,000 rpm for 10 min. Crude nucleic acids were obtained from the supernatant (approximately, 80 μl) by isopropanol precipitation followed by 70% ethanol wash. The yeast DNA was dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) as described (Tanahashi et al., 2017).
The entire nucleotide sequences of ITS and IGS regions were amplified by polymerase chain reaction (PCR) at 94°C for 5 min followed by 35 cycles of 95°C for 45 s, 54°C for 45 s, and 72°C for 1 min 30 s (for ITS) or 2 min 15 s (for IGS), by using the primer sets NS7-NL4 (for ITS) and IGS1-IGS4 (for IGS) (Table 1). The PCR products were purified using the Illustra ExoStar clean-up kit (GE Healthcare, Buckinghamshire, United Kingdom). Dye terminator cycle sequencing reactions were performed using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster, CA, United States), and the reaction products were analyzed by an ABI 3130xl genetic analyzer (Applied Biosystems). The primers used for the sequencing analyses were listed in Table 1.
To detect fine genetic variation of the symbiotic yeasts, IGS PCR products from all isolates (i.e., 2–8 colonies per host female) were first sequenced with the primer IGS1. This short sequence (∼700 bp) covered the entire IGS1 region, which was subsequently cut out from the raw sequence data. We used this entire IGS1 sequence (namely, IGS1 haplotype) for a discrimination marker of Scheffersomyces symbiotic yeast strains on the ground that IGS1 is known for the largest genetic variety among IGS regions (Tanahashi et al., 2017). When more than one IGS1 haplotypes were detected from a single host female, we chose one representative isolate for each different IGS1 haplotype for further phylogenetic analyses. This process is the analytical strategy practically adopted by Tanahashi et al. (2017). After that, the whole IGS sequence (approximately 2.2 kb) was determined for each representative isolate for each host female (in total 60 sequences) (Supplementary Table 2) using the sequencing primers IGS2, IGS3, IGS4, IGS7i and IGS8i.
Although IGS regions are suitable for detecting strain-level genetic differences, they are not commonly used for species identification or phylogenetic analysis at higher taxonomic levels, because of too much genetic variation as well as insufficient taxonomic coverage in the databases. Therefore, for the purposes of species identification and phylogenetic analysis of the symbiotic yeasts, we chose 23 symbiotic yeast isolates from 12 females representing all the Japanese Platycerus species (Supplementary Table 2). The ITS PCR products of the representative yeast isolates were sequenced with the primer NS7 and ITS5 (Table 1), which were around 0.7 kb in size and usually contained whole ITS regions (ITS1, 5.8S rRNA, and ITS2) of the Scheffersomyces yeasts. We also determined ITS sequences of 11 symbiotic yeast strains that were isolated from eight outgroup lucanid species (Supplementary Table 2).
For DNA sequencing analysis of host Platycerus beetles, Kubota et al. (2011) reported that cytochrome oxidase subunit I (COI) gene exhibited suitable levels of variation for detecting genetic divergence among species and populations, whereas some nuclear genes did not. Introgressive hybridization has been recognized in some Japanese Platycerus species (e.g., P. takakuwai Fujita, 1987) based on the 28S ribosomal RNA (28S rRNA) and mitochondrial cytochrome oxidase subunit I (COI) genes. It should be noted, however, that both the mitochondria and the symbiotic yeasts are essentially transmitted from mother to offspring (Tanahashi and Hawes, 2016). Since it seems to be reasonable to compare the evolutionary patterns between COI gene of the host beetles and IGS gene of the yeast symbionts, we sequenced COI gene of the host beetles. It should be noted that we were not always able to use the same beetle samples for the yeast isolation and the host DNA analysis. In such cases, we used alternative beetle samples collected at the same field sites. A considerable part of those COI sequences have already been published in previous studies (Kubota and Kubota, 2011; Tanahashi et al., 2017). Therefore, we newly analyzed 18 Japanese Platycerus samples that were needed in this study.
Genomic DNA was extracted from the testes or muscle tissues of adult Platycerus beetles preserved in ethanol using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, United States). The COI gene (primers C1-J-2183 and L2-N-3014, Simon et al., 1994) was amplified by PCR at 94°C for 3 min followed by 30 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 1 min, with a final 7 min extension at 72°C. The PCR products were purified using the Illustra ExoStar clean-up kit (GE Healthcare, Buckinghamshire, United Kingdom). The Dye terminator cycle sequencing reactions were performed using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster, CA, United States), and electrophoresed using an ABI 3130xl genetic analyzer (Applied Biosystems). For the sequencing analysis, the same primer sets as PCR were used.
Besides the 23 ITS sequences, 60 IGS sequences, and 23 COI sequences determined in this study, we also used the following sequences reported in previous studies: 18 yeast ITS sequences, of which 13 were symbionts of lucanid beetles (two from Japanese Platycerus species, eight from Platycerus hongwonpyoi of South Korea, and three from other lucanid species); 11 yeast IGS sequences, of which two were from Japanese Platycerus species, eight were from Platycerus hongwonpyoi of South Korea, and one was from Japanese Prismognathus angularis; 27 Japanese Platycerus COI sequences (Supplementary Table 2).
We constructed a haplotype matrix for each of the ITS and IGS regions of yeasts, and the COI gene of beetles using the DnaSP ver. 5.10.01 software package (Librado and Rozas, 2009). To align the sequences, we used ClustalW implemented in BioEdit ver. 7.2.5 (Hall, 1999). Gaps were treated as missing data. Aligned haplotype sequences were used to construct phylogenetic trees based on maximum likelihood (ML) and Bayesian inference (BI) methods. In addition, 23 combined sequences of ITS and IGS were used to construct phylogenetic trees based on BI methods. The sequence alignments used for the phylogenetic analyses are available as Supplementary Appendices 1–4.
ML trees were constructed using PhyML ver. 3.0 (Guindon and Gascuel, 2003) under the best-fit substitution model selected using jModelTest ver. 2.1.7 (Darriba et al., 2012) based on the Bayesian information criterion (BIC). Confidence at each node was assessed by 100 bootstrap replications.
BI trees were constructed using three runs in MrBayes ver. 3.2.6 (Ronquist and Huelsenbeck, 2003) under the best-fit substitution model selected by jModelTest ver. 2.1.7, for 200 million generations (samplefreq = 20,000) and 2,500 samples burn-in, using Tracer ver. 1.5.0 (Rambaut and Drummond, 2016) to examine convergence toward high effective sample sizes.
For the ITS, IGS, and their combined sequences, we also constructed BI trees based on gap-recoded sequences using FastGap 1.2 (Borchsenius, 2009) to minimize the effect of gaps.
For analyzing co-evolutionary relationships based on genetic sequence data, several methods, e.g., SH-test, AU-test, and likelihood-ratio test (Kawakita, 2008) are available. In these methods, the null hypothesis is that the phylogenies of the hosts and parasites are the same. In this study, since the host insect phylogeny and the yeast symbiont phylogeny seemed to be not at all the same, we adopted the Parafit test (Legendre et al., 2002) using p-distances between IGS sequences of the yeast symbionts and those between COI sequences of the host insects, where null hypothesis is that their phylogenies are not correlated. In addition, Platycerus species and populations are genetically differentiated in Japan, and their genetic divergence seems to be related to their geographic distribution (Kubota et al., 2011). Therefore, we also used the Mantel test to remove the effects of geographical distance.
We randomly chose a symbiont IGS sequence and a host COI sequence for each host beetle population to avoid duplication of beetle populations. Geographic distances between the collection sites of Japanese Platycerus species were estimated using a distance calculation tool available at https://vldb.gsi.go.jp/sokuchi/surveycalc/surveycalc/bl2stf.html (cited 3 May 2018). Genetic p-distances (pairwise nucleotide diversity) of IGS sequences and COI sequences between the insect populations were calculated using Arlequin ver. 3.1 (Excoffier et al., 2005).
We examined the relationship between the geographic distance of the collection sites and the IGS p-distance of the yeast symbionts, the relationship between the geographical distance of the collection sites and the COI p-distance of the host beetles, and the relationship between the IGS p-distance of the yeast symbionts and the COI p-distance of the host beetles by the Mantel test. The Parafit test and the Mantel test were conducted based on 10,000 pseudoreplications of randomization.
All the ITS sequences of the yeast symbionts of the Platycerus species collected in Japan exhibited no sequence variation (containing partial 18S rRNA, ITS1, 5.8S rRNA, ITS2, and partial 26S rRNA; 652 bp; n = 23 from 12 females). These sequences were completely identical to the sequences previously reported from two Japanese Platycerus species and seven Korean individuals of P. hongwonpyoi (Tanahashi et al., 2017) contained only two nucleotide substitutions compared with the sequence of the yeast symbiont from Prosmognathus angularis (Tanahashi et al., 2017) and were the most similar to the sequence of Scheffersomyces segobiensis (Tanahashi et al., 2017) among formally described yeast species. Note that S. segobiensis was isolated from a jewel beetle Chalcophora mariana massiliensis (Villers, 1789) (Coleoptera: Buprestidae) (Santa-María and García-Aser, 1977). Phylogenetic analysis based on the ITS sequences revealed that the yeast symbionts of the Platycerus species constitute a distinct lineage in the clade of the yeast symbionts of the other lucanid species including environmental Scheffersomyces isolates (Supplementary Figure 1).
We first tested genetic variation of the yeast symbionts based on 2–8 colonies within a host female by using the primers IGS1 and IGS2 (covering partial 26S rRNA, IGS1, 5S rRNA, and partial IGS2; >700 bp in size). The IGS1 sequences of multiple yeast colonies isolated from a host female were identical for all the Platycerus females examined, suggesting that there is little genetic variation among the yeast symbionts in the mycangium of a single female. Therefore, a representative isolate was randomly selected for each of the all female samples, and the entire sequences of IGS regions (partial 26S rRNA, IGS1, 5S rRNA, IGS2, and partial 18S rRNA; 2177–2311 bp; n = 59) were sequenced and subjected to the phylogenetic analysis.
For the IGS region, the nucleotide substitution rate was higher in the order of IGS1, the former half of IGS2, and the latter half of IGS2 (Figure 2B). Gap sites were concentrated much in IGS1, and a little in the second quarter of IGS2 (Figure 2B).
In the IGS phylogeny, two major clades were recognized among the yeast symbionts of Platycerus stag beetles: Clade I contained most of the Japanese Platycerus populations, whereas Clade II embraced most populations of P. viridicuprus Kubota et al. (2008) in Japan and all populations of P. hongwonpyoi in South Korea. Most of the Japanese Platycerus species (except for P. delicatulus and most of P. viridicuprus populations) distributed in the same areas tended to share the yeast symbionts belonging to the same subclade: Clade Ia, central and eastern Honshu; Clade Ib, the Kii Peninsula; and Clade Ic, Shikoku and Kyushu (Supplementary Figure 2; also see Figure 1A). The yeast symbionts of P. delicatulus populations formed a monophyletic group of Clade Id. Clade II consisted of the yeast symbionts of most P. viridicuprus populations (western Honshu and northern Kyushu) and all P. hongwonpyoi populations in South Korea. Only one female of P. viridicuprus (Site 25; northern Kyushu) possessed the yeast symbiont that represented the same IGS sequence to P. urushiyamai Imura, 2007 (Supplementary Figure 2). The ITS + IGS combined phylogeny based on 23 samples exhibited the same topology as the IGS phylogeny (Figure 1B).
We constructed COI phylogeny of the host insects (Supplementary Figure 3), and compared it with IGS phylogeny of the yeast symbionts (Figure 3). Globally, the host phylogeny and the symbiont phylogeny were not congruent, indicating that no strict host-symbiont co-speciation has been established in the evolutionary course of the Platycerus stag beetles. On the other hand, it was evident that the same Platycerus species tended to be associated with closely related yeast symbionts (Figure 3) suggesting that stable host-symbiont associations may be locally observed, at least to some extent, in the Platycerus stag beetles.
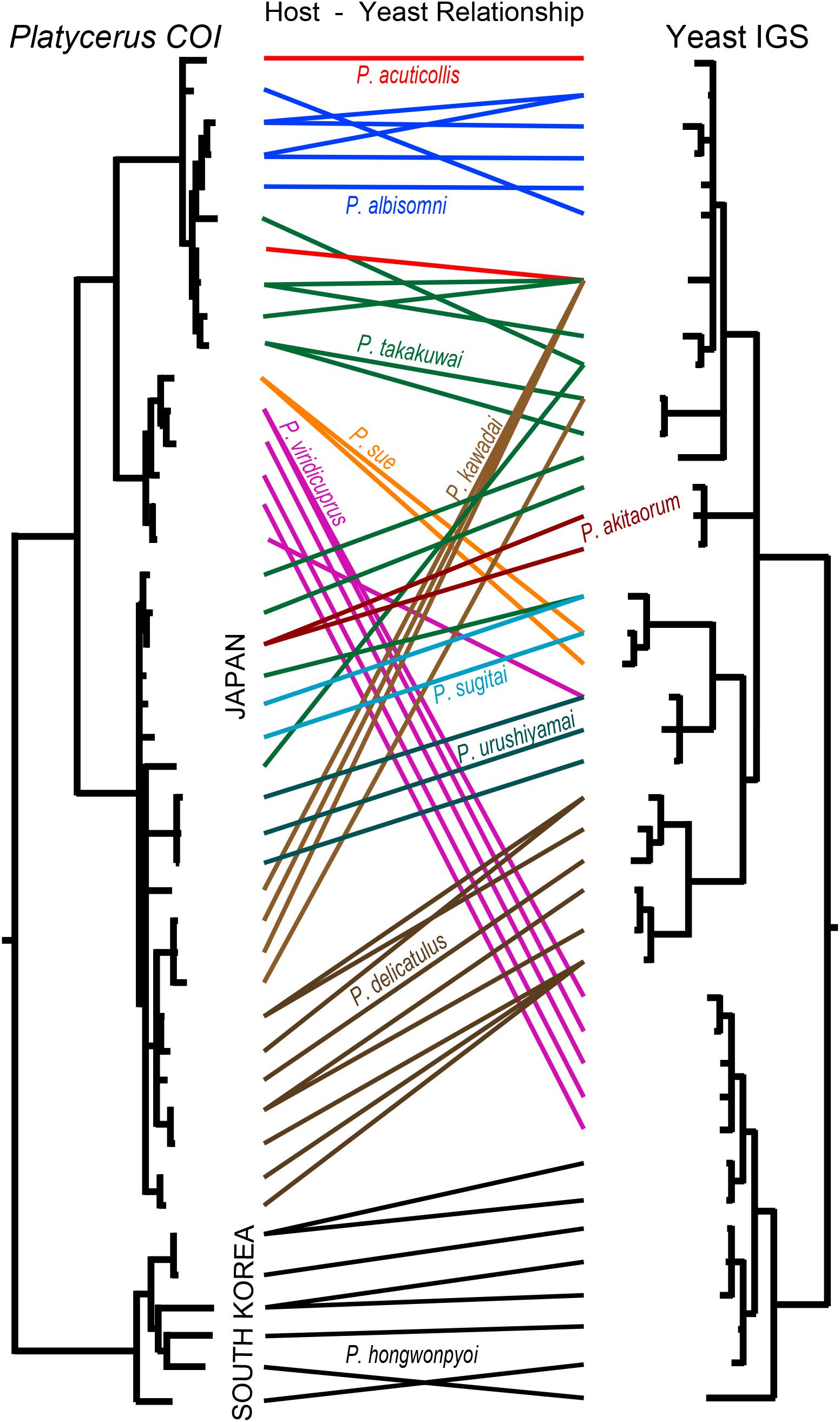
Figure 3. Comparison between IGS phylogeny of the yeast symbionts and COI phylogeny of their host Platycerus stag beetles. The phylogenies are taken and modified from Supplementary Figures 2, 3. The colored lines show the correspondence of the yeast symbionts and their insect hosts.
In each phylogenetic analysis, number of OTUs, length of aligned sequence, numbers of informative sites, gap sites, and recoded gap sites using FastGap 1.2 are shown in Table 2.
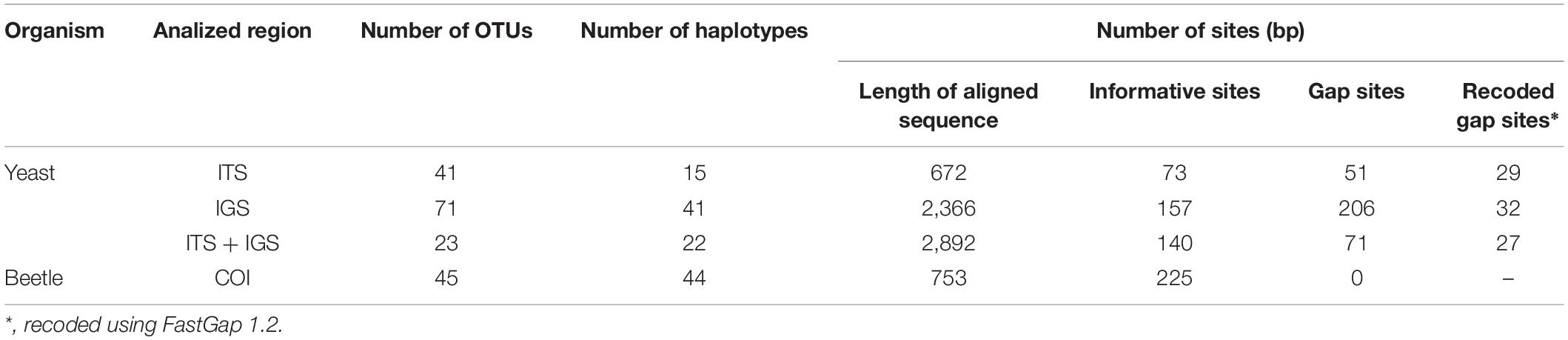
Table 2. Numbers of OTUs and haplotypes, length of aligned sequence, numbers of informative sites, gap sites, and recoded gap sites in each phylogenetic analysis.
Finally, in an attempt to quantitatively evaluate the phylogenetic and geographic aspects of the host-symbiont association in the Platycerus stag beetles, correlations between the IGS p-distances among the yeast symbionts, the COI p-distances among the host insects, and the geographic distances between the collection sites were analyzed statistically. The IGS p-distances between the yeast symbionts were significantly correlated with the COI p-distances between the host insects (P < 0.0001, Parafit test with 10,000 replicates). The IGS p-distances between the yeast symbionts were significantly correlated with the geographical distances between the collection sites (P < 0.0001, Mantel test with 10,000 replicates) (Figure 4A). Similarly, the COI p-distances between the host insects were significantly correlated with the geographical distances between the collection sites (P < 0.0001, Mantel test with 10,000 replicates) (Figure 4B). Reflecting that the Japanese Platycerus species and populations were phylogenetically divided into two large clades (Supplementary Figure 3), their genetic distances form two clusters in Figure 4B.
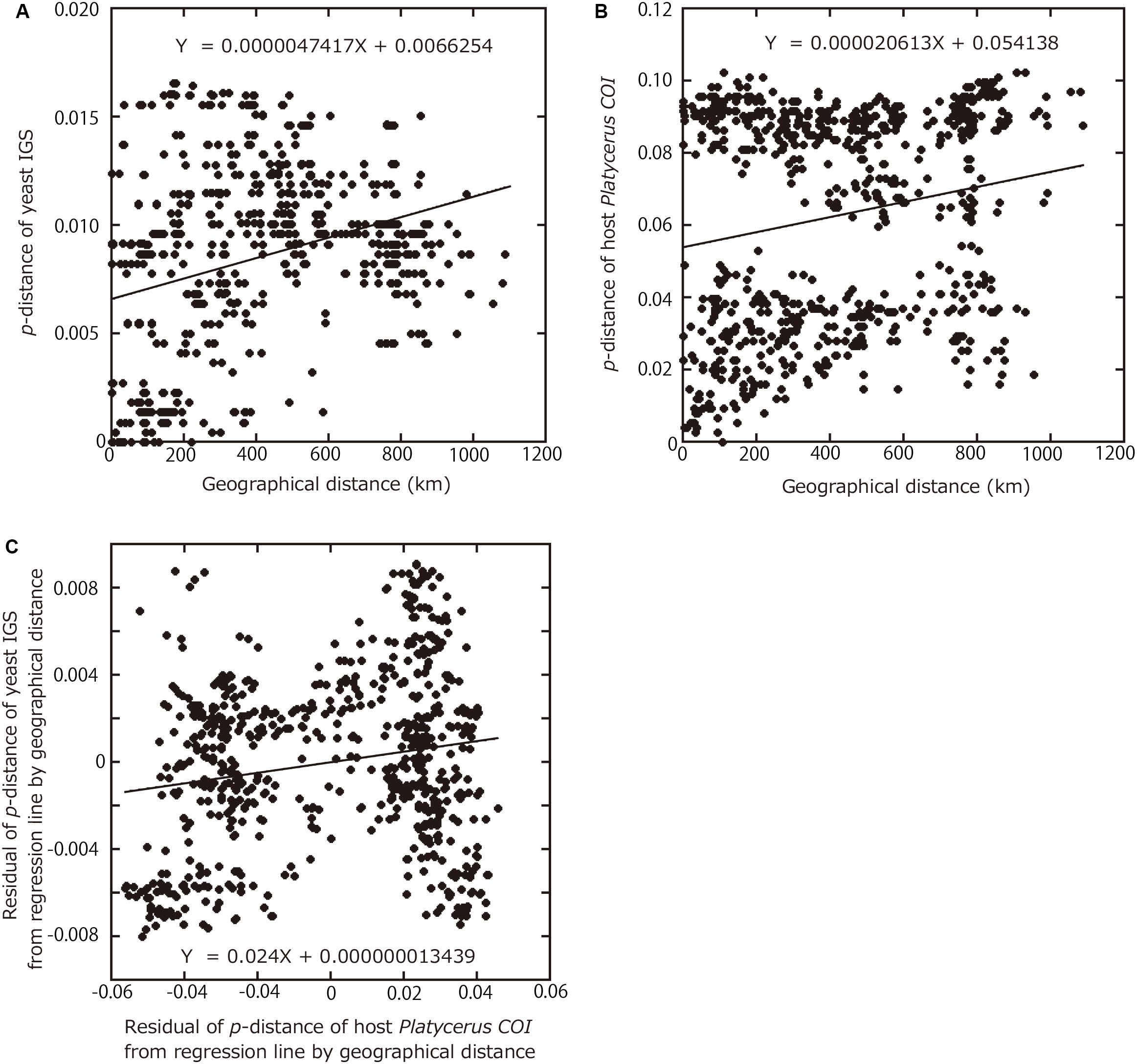
Figure 4. Relationships among geographic distances between collection sites, genetic distances between yeast symbionts, and genetic distances between host Platycerus stag beetles. (A) Relationship of geographical distances between collection sites versus p-distances between the symbiont IGS sequences. (B) Relationship of geographical distances between collection sites versus p-distances of the host COI sequences. (C) Relationship of residual p-distances between the host COI sequences regressed to geographical distances versus residual p-distances between the symbiont IGS sequences regressed to geographical distances.
For both IGS and COI, regression lines were obtained with geographical distances as the explanatory variables and p-distances as the dependent variables. To remove the effects of the geographic distances, the residuals of p-distance from regression lines were calculated. Even after removing the effects of the geographic distances, the residuals of IGS p-distances between the yeast symbionts were significantly correlated with the residuals of COI p-distances between the host insects (P = 0.0004, Mantel test with 10,000 replicates) (Figure 4C).
In this study, we demonstrated that almost all the adult females of Japanese Platycerus stag beetles are associated with the specific Scheffersomyces yeast symbiont within their mycangium (Figure 2A). Based on ITS sequences, all the yeast symbionts isolated from Japanese and Korean Platycerus species and populations exhibit no sequence variation, constituting a distinct lineage within the Scheffersomyces yeast symbionts of diverse stag beetles (Supplementary Figure 1). These patterns suggest the possibility that the Japanese and Korean Platycerus species are associated with a clade of yeast symbionts that shares the common fungal ancestor with S. segobiensis isolated from a jewel beetle (Supplementary Figure 1).
Based on IGS sequences, we uncovered that the yeast symbionts of Japanese Platycerus stag beetles exhibit considerable genetic and geographic diversity, in which sympatric Platycerus species tend to be associated with the same or closely related yeast symbionts (Supplementary Figure 2). Notable examples are P. kawadai Fujita & Ichikawa, 1982 and P. takakuwai at sites 6, 7 and 8 in Central Honshu; P. takakuwai and P. sugitai Okuda & Fujita, 1987 at sites 18 and 19 in Shikoku; P. sue Imura, 2007 and P. sugitai at sites 21 and 22 in Shikoku; P. urushiyamai and P. viridicuprus at sites 24 and 25 in Kyushu (Supplementary Figure 2). These patterns strongly suggest that the yeast symbionts must have been occasionally transferred between the sympatric host species, plausibly via sharing of the same decaying wood as the larval habitat.
By contrast, we found that, across P. delicatulus populations covering a wide distribution range, their yeast symbionts form a distinct monophyletic clade (Supplementary Figure 2). Considering that P. delicatulus is sympatrically found with many other Platycerus species (Figure 1A) the host-symbiont relationship in P. delicatulus must have been robust without lateral transfer events. It is currently obscure why the host-symbiont connection is stable in P. delicatulus, but we suggest the possibility that ecological aspects of P. delicatulus might be relevant. While larvae of most Platycerus species feed on thin, wet and soft decaying wood on the ground, larvae of P. delicatulus live on large and dry decaying wood, typically standing dead trees (Imura, 2010b). It is conceivable, although speculative, that the distinct ecological niche of P. delicatulus prevents lateral symbiont transfers from other Platycerus species, thereby underpinning the host-symbiont fidelity.
Previous molecular phylogenetic studies reported that Japanese Platycerus species form a monophyletic clade and are genetically distinct from Korean P. hongwonpyoi (Imura and Nagahata, 2009; Kubota et al., 2011) (see Supplementary Figure 3). Notwithstanding this, based on IGS sequences, the yeast symbionts of P. viridicuprus form a well-supported monophyletic clade with the yeast symbionts of P. hongwonpyoi (Figure 1B and Supplementary Figure 2). How can this host-symbiont discrepancy be interpreted? A possible evolutionary scenario is that P. viridicuprus has been maintaining an ancestral type of the yeast symbiont since the ancestral Platycerus species dispersed from the Korean Peninsula to Japan. However, although the Japanese Platycerus clade including P. viridicuprus and the Asian continental Platycerus clade including P. hongwonpyoi diverged about 10 million years ago based on COI sequences (Zhu et al., unpublished) the yeast symbionts of P. viridicuprus are genetically very close to the yeast symbionts of P. hongwonpyoi (Figure 1B and Supplementary Figure 2). These facts do not favor the above idea. An alternative, and more likely evolutionary scenario is that a recent secondary contact of P. viridicuprus and P. hongwonpyoi upon a marine withdrawal around the Tsushima Islands (see Figure 1A) resulted in the lateral symbiont transfer between the two species. Note that one female of P. viridicuprus collected at the western distribution edge (Figure 1A, site 25 in northern Kyushu) exceptionally possessed the yeast symbiont belonging to the Kyushu clade of Clade Ic, which can be explained by a lateral symbiont transfer from P. urushiyamai to P. viridicuprus.
The results of Parafit and Mantel tests showed that the genetic divergence of the Platycerus stag beetles and the genetic divergence of the yeast symbionts are significantly correlated to each other, with or without considering the effects of the geographic distance (Figure 4). These results suggest that the divergence of the host insects has constrained the divergence of the yeast symbionts, and/or vice versa. In other words, the Platycerus stag beetles and the yeast symbionts have co-evolved to some extent but only incompletely, which reflects the following phylogenetic patterns. Globally, the symbiont phylogeny is not at all congruent with the host phylogeny (Figure 3) which strongly suggests that losses, gains, lateral transfers and/or replacements of the Scheffersomyces yeast symbionts must have occurred repeatedly in the evolutionary course of the Platycerus stag beetles. Plausibly, these dynamic evolutionary aspects are relevant to the following factors: (i) the Scheffersomyces yeast symbionts are cultivable and thus capable of surviving outside the symbiotic organ of the insect hosts (Figure 2A), (ii) allied Scheffersomyces yeasts are commonly found in and isolated from the galleries of decaying woods where lucanid larvae inhabit (Kubota et al., 2010) (iii) the Scheffersomyces yeasts are capable of xylose fermentation and thus likely to adapt to proliferation and survival on woody materials (Jeffries et al., 2007; Tanahashi et al., 2010). On the other hand, focusing on local relationships, the same host species tend to be associated with closely related yeast symbionts (Figure 3), suggesting that stable host-symbiont associations may be observed in the Platycerus stag beetles to some extent. In addition, Platycerus species and Prismognathus angularis often coexist in the same wood pieces, each of them is associated with its own yeast symbiont lineage, and these two genera never share the same yeasts (Figure 1B and Supplementary Figure 2; Zhu et al., unpublished). These local host-symbiont affinities are likely to be underpinned by the mycangium-mediated vertical transmission of the yeast symbionts generally found in stag beetles (Tanahashi et al., 2010) and also by presumable low dispersal ability of the insects. Overall, these complex phylogenetic and genetic patterns highlight complex evolutionary trajectories of the host-symbiont relationships in the Platycerus stag beetles.
Diverse taxa of yeasts are known to be associated with a variety of coleopteran families such as Platypodidae, Scolytidae, Rhynchophoridae, Buprestidae, Bostrychidae, Chrysomelidae, Nitidulidae, Erotylidae, Mycetophagidae, Ciidae, and Lucanidae (Kurtzman et al., 2011; Endoh, 2012). They are isolated from various parts of insect bodies (e.g., whole body, gut, ovary, etc.), larval galleries, and feces. However, only a small fraction of the yeast associates are known to provide mutual benefits to the host beetles. For example, the yeast symbionts of Scolytidae improve the nutritional and environmental conditions of the host beetles (Hunt and Borden, 1990; Rivera et al., 2009). Also the yeast symbionts of Platypodidae are thought to nutritionally contribute to the host beetles (Endoh, 2012). In the beetle bodies, especially within their gut, specific fungal floras including diverse yeasts have been commonly recognized, which often exhibit inter- and intra-specific variations (Shifrine and Phaff, 1956; Leufvén and Nehls, 1986; Suh et al., 2004, 2006; Suh and Blackwell, 2005; Endoh et al., 2011).
Scheffersomyces yeasts are not only found from Platycerus and other groups of the Lucanidae (Van der Walt et al., 1971; Tanahashi et al., 2010, 2017; Hawes, 2013; Tanahashi and Hawes, 2016) but also detected from the larval gut of wood-feeding beetles belonging to the Passalidae, the Cerambycidae and the Scarabaeidae (Suh et al., 2003, 2006; Kubota et al., 2011) although no mycangium-like symbiotic organs have been identified in these non-lucanid beetles (Tanahashi et al., 2010). Considering that Scheffersomyces is the almost only fungus found in the female-specific mycangium of Platycerus, Dorcus, Lucanus and other lucanid beetles (though some fungal flora may exist in their larval gut), it seems likely that the Scheffersomyces yeast symbionts play some important roles for the host stag beetles. Thus far, however, the exact symbiont function has been elusive. In Platycerus species, the yeast symbionts are constantly found in the larval gut and galleries but not in the adult gut (Kubota et al., 2010). On account of the xylose-fermenting capability of Scheffersomyces yeasts (Du Preez and Prior, 1985; Jeffries and Kurtzman, 1994; Olsson and Hahn-Hägerdal, 1996) it is conceivable, although speculative, that the yeast symbionts may contribute to the survival or development of the host insect larvae by helping hemicellulose digestion in the decaying wood (Tanahashi et al., 2010, 2017). In fact, we confirmed that most yeast symbionts of stag beetles examined in this study can utilize xylose, while Saccharomyces cerevisiae cannot (Watanabe et al., unpublished data). How and why specific Scheffersomyces yeast symbionts are selected and colonize the mycangium is of great interest and deserves future studies. To address these questions, we should investigate the horizontal and vertical transmission routes and mechanisms of the yeast symbionts between the host beetles, natural ex-host habitats of the yeast symbionts in the field, the effects of the yeast symbionts on host growth and survival, and other biological aspects of the lucanid-yeast associations.
The datasets generated for this study can be found in the DDBJ: LC438646–LC438745.
KW, KaK, and MT contributed to the data generation of this study. KôK and X-JZ performed genetic analyses. KôK contributed to the study design with the help of MT. KôK and TF wrote the manuscript. All authors approved the final version of the manuscript.
This study was partly supported by the Japan Society for the Promotion of Science KAKENHI Grant Numbers 20248015 and 25292082 to KôK, JP17H06388 to TF, and the Oversea Study Program of Guangzhou Elite Project (No. SUIJING [2015]4) to X-JZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to S. Kobayashi, M. Ono, and N. Kubota for collecting materials, and S.-N. Zhang for drawing the figures.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01436/full#supplementary-material
Batra, L. R. (1963). Ecology of ambrosia fungi and their dissemination by beetles. Trans. Kans. Acad. Sci. 66, 213–236. doi: 10.2307/3626562
Biedermann, P. H. W., and Vega, F. E. (2020). Ecology and evolution of insect-fungus mutualisms. Annu. Rev. Entomol. 65, 431–455. doi: 10.1146/annurev-ento-011019-024910
Breznak, J. A., and Brune, A. (1994). Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 39, 453–487. doi: 10.1146/annurev.en.39.010194
Cleveland, L. R. (1924). The physiological and symbiotic relationships between the intestinal protozoa of termites and their host, with special reference to Reticulitermes flavipes Koller. Biol. Bull. 46, 178–227.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Du Preez, J. C., and Prior, B. A. (1985). A quantitative screening of some xylose-fermenting yeast isolates. Biotechnol. Lett. 7, 241–246. doi: 10.1007/bf01042370
Endoh, R. (2012). Yeasts associated with coleopteran beetles in forest. J. Jpn. For. Soc. 94, 326–334. doi: 10.4005/jjfs.94.326
Endoh, R., Suzuki, M., Okada, G., Takeuchi, Y., and Futai, K. (2011). Fungus symbionts colonizing the galleries of the ambrosia beetle Platypus quercivorus. Microb. Ecol. 62, 106–120. doi: 10.1007/s00248-011-9838-3
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 1–10.
Grebennikov, V. V., and Leschen, R. A. B. (2010). External exoskeletal cavities in Coleoptera and their possible mycangial functions. Entomol. Sci. 13, 81–98. doi: 10.1111/j.1479-8298.2009.00351.x
Grünwald, S., Pilhofer, M., and Holl, W. (2010). Microbial associations in gut system of wood- and bark-inhabiting longhorned beetles [Coleoptera: Cerambycidae]. Syst. Appl. Microbiol. 33, 25–34. doi: 10.1016/j.syapm.2009.10.002
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Haack, R. A., and Slansky, F. (1987). “Nutritional ecology of wood-feeding Coleoptera, lepidoptera, and hymenoptera,” in Nutritional Ecology Of Insects, Mites, And Spiders, eds F. Slansky and J. G. Rodriguez (New York NY: John Wiley), 449–486.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.4236/sgre.2015.64007
Hawes, C. J. (2013). Discovery of a mycangium and associated yeasts in the stag beetle Lucanus cervus (Coleoptera: Lucanidae). White Adm. 85, 22–23.
Huang, H., and Chen, C. C. (2017). Stag Beetles of China III. New Taipei City: Formosa Ecological Company.
Huang, H., Chen, C. C., and Imura, Y. (2010). A new species of the genus Platycerus Geoffroy (Coleoptera, Lucanidae) from Gansu, China. Elytra 38, 233–238.
Hunt, D. W. A., and Borden, J. H. (1990). Conversion of verbenols to verbenone by yeasts isolated from Dendroctonus ponderosae (Coleoptra: Scolytidae). J. Chem. Ecol. 16, 1385–1397. doi: 10.1007/BF01021034
Imura, Y. (2010a). A new Platycerus (Coleoptera, Lucanidae) from the Baotianman Nature Reserve in western Henan, Central China. Elytra 38, 227–232.
Imura, Y. (2011). Two new taxa of the genus Platycerus (Coleoptra, Lucanidae) from China. Spec. Public. Jpn. Soc. Scarabaeoidol. 1, 131–141.
Imura, Y. (2012). Description of a new Platycerus (Coleoptera, Lucanidae) from Sichuan, southwest China, with records of two other species belonging of the same genus. Elytra New Ser. 2, 61–68.
Imura, Y., and Nagahata, Y. (2009). Phylogeny and evolution of the tribe Platycerini (Coleoptera, Lucanidae) of the world inferred from mitochondrial 16S rRNA and COI gene sequences. Elytra 37, 261–272.
Jeffries, T. W., Grigoriev, I. V., Grimwood, J., Laplaza, J. M., Aerts, A., Salamov, A., et al. (2007). Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 25, 319–326. doi: 10.1038/nbt1290
Jeffries, T. W., and Kurtzman, C. B. (1994). Strain selection, taxonomy, and genetics of 220 xylose-fermenting yeasts. Enzyme Microb. Technol. 16, 922–932. doi: 10.1016/0141-0229(94)90001-9
Kawakita, A. (2008). “Co-speciation analysis methods for the co-evolution study,” in Ecology of Co-Evolution, The Society of the Study of the Species Biology, eds J. Yokoyama and I. Dozono (Tokyo: Bun-ichi Co., Ltd), 341–354.
Kim, S. I., and Farrell, B. D. (2015). Phylogeny of world stag beetles (Coleoptera: Lucanidae) reveals a Gondwanan origin of Darwin’s stag beetle. Mol. Phylogenet. Evol. 86, 35–48. doi: 10.1016/j.ympev.2015.02.015
Kubota, K., and Kubota, N. (2011). Gene flow between two closely related species in the acuticollis species group (Coleoptera, Lucanidae, genus Platycerus) near their distribution border based on the morphology and mitochondrial gene. Entomol. Sci. 14, 198–202. doi: 10.1111/j.1479-8298.2010.00424.x
Kubota, K., Kubota, N., and Otobe, H. (2010). Mitochondrial gene diversity of Platycerus sue (Coleoptera, Lucanidae), a temporarily designated endangered species based on the law for the conservation of endangered species of wild fauna and flora. Bull. Biogeogr. Soc. Jpn. 65, 151–158.
Kubota, K., Nagahata, Y., Ikeda, H., Kubota, N., Otobe, H., and Umetsu, K. (2011). Diversification process of stag beetles belonging to the genus Platycerus Geoffroy (Coleoptera: Lucanidae) in Japan based on nuclear and mitochondrial genes. Entomol. Sci. 14, 411–427. doi: 10.1111/j.1479-8298.2011.00466.x
Kubota, K., Zhu, X.-J., Ma, T., and Wen, X.-J. (2008). A new species of the genus Platycerus Geoffroy (Coleoptera: Lucanidae) from the Qinling Mountains, Shaanxi Province, China. Elytra New Ser. 8, 175–178.
Kurtzman, C. P., Fell, J. W., and Boekhout, T. (2011). The Yeasts—A Taxonomic Study, 5th Edn, Amsterdam: Elsevier.
Legendre, P., Desdevises, Y., and Bazin, E. (2002). A statistical test of host-parasite coevolution. Syst. Biol. 51, 217–234. doi: 10.1080/10635150252899734
Leufvén, A., and Nehls, L. (1986). Quantification of different yeats associated with the bark beetle, Ips typographus, during its attack on a spruce tree. Microb. Ecol. 12, 237–243. doi: 10.1007/bf02011208
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Nguyen, N. H., Suh, S. O., and Blackwell, M. (2007). Five novel Candida species in insect-associated yeast clades isolated from Neuroptera and other insects. Mycologia 99, 842–858. doi: 10.1080/15572536.2007.11832516
Olsson, L., and Hahn-Hägerdal, B. (1996). Fermentation of lignocellulosic hydrolysates for ethanol production. Enzyme Microb. Technol. 18, 312–331. doi: 10.1016/0141-0229(95)00157-3
Rambaut, A., and Drummond, A. J. (2016). Tracer Version 1.5. MCMC Trace Analysis Tool. Anales del Instituto Nacional de Investigaciones Agrarias. Serie General. Available online at: http://beast.bio.ed.ac.uk/software/tracer (accessed September 12, 2018).
Rivera, F. N., González, E., Gómez, Z., López, N., Hernández-Rodríguez, C., Berkov, A., et al. (2009). Gut-associated yeast in bark beetles of the genus Dendroctonus Erichson (Coleoptera: Curculionidae: Scolitynae). Biol. J. Linn. Soc. 98, 325–342. doi: 10.1111/j.1095-8312.2009.01289.x
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Santa-María, J., and García-Aser, C. (1977). Pichia segobiensis, sp. nov. anales del instituto nacional de investigaciones agrarias. Ser. Gen. 5, 45–50.
Shifrine, M., and Phaff, H. J. (1956). The association of yeasts with certain bark beetles. Mycologia 48, 41–55. doi: 10.2307/3755778
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651–701. doi: 10.1093/aesa/87.6.651
Sjöström, E. (1993). Wood Chemistry: Fundamentals And Applications, 2nd Edn. San Diego CA: Academic Press.
Slaytor, M. (1992). Cellulose digestion in termites and cockroaches: what role do symbionts play? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 103, 775–784. doi: 10.1016/0305-0491(92)90194-v
Stokland, J. N. (2012). “Wood decomposition,” in Biodiversity in Decaying Wood, eds J. N. Stokland, J. Siitonen, and B. G. Jonsson (New York NY: Cambridge University Press), 10–28. doi: 10.1017/CBO9781139025843
Suh, S. O., and Blackwell, M. (2005). Four new yeasts in the Candida mesenterica clade associated with basidiocarp-feeding beetles. Mycologia 97, 167–177. doi: 10.1080/15572536.2006.11832850
Suh, S. O., Marshall, C. J., McHugh, J. V., and Blackwell, M. (2003). Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol. Ecol. 12, 3137–3145. doi: 10.1046/j.1365-294X.2003.01973.x
Suh, S. O., McHugh, J. V., and Blackwell, M. (2004). Expansion of the Candida tanzawaensis yeast clade: 16 novel Candida species from basidiocarp-feeding beetles. Int. J. Syst. Evol. Microbiol. 54, 2409–2429. doi: 10.1099/ijs.0.63246-0
Suh, S. O., McHugh, J. V., Pollock, D. D., and Blackwell, M. (2006). The beetle gut: a hyperdiverse source of novel yeasts. Mycol. Res. 109, 261–265. doi: 10.1017/S0953756205002388
Tanahashi, M., and Hawes, C. J. (2016). The presence of a mycangium in the horned stag beetle Sinodendron cylindricum (Coleoptera: Lucanidae) and the associate yeast symbionts. J. Insect Sci. 16, 76. doi: 10.1093/jisesa/iew054
Tanahashi, M., Kim, J.-K., Watanabe, K., Fukatsu, T., and Kubota, K. (2017). Specificity and genetic diversity of xylose-fermenting Scheffersomyces yeasts associated with small blue stag beetles of the genus Platycerus in East Asia. Mycologia 109, 630–642. doi: 10.1080/00275514.1382648
Tanahashi, M., Kubota, K., Matsushita, N., and Togashi, K. (2010). Discovery of mycangia and associated xylose-fermenting yeasts in stag beetles (Coleoptera: Lucanidae). Naturwissenschaften 97, 311–317. doi: 10.1007/s00114-009-0643-5
Van der Walt, J. P., Scott, D. B., and van der Klift, W. C. (1971). Five new Torulopsis species from South Africa insect sources. Antonie Leeuwenhoek 37, 461–471. doi: 10.1007/bf02218516
Vega, F. E., and Blackwell, M. (eds) (2005). Insect-Fungal Associations: Ecology and Evolution. New York, NY: Oxford University Press.
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide To Methods And Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (London: Academic Press), 315–321.
Keywords: co-evolutionary association, Scheffersomyces, mycangium, ITS, IGS, Japan
Citation: Kubota K, Watanabe K, Zhu X-J, Kawakami K, Tanahashi M and Fukatsu T (2020) Evolutionary Relationship Between Platycerus Stag Beetles and Their Mycangium-Associated Yeast Symbionts. Front. Microbiol. 11:1436. doi: 10.3389/fmicb.2020.01436
Received: 07 March 2020; Accepted: 03 June 2020;
Published: 30 June 2020.
Edited by:
Amparo Latorre, University of Valencia, SpainReviewed by:
Pepijn Wilhelmus Kooij, São Paulo State University, BrazilCopyright © 2020 Kubota, Watanabe, Zhu, Kawakami, Tanahashi and Fukatsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kôhei Kubota, a29oZWlAZnIuYS51LXRva3lvLmFjLmpw; Takema Fukatsu, dC1mdWthdHN1QGFpc3QuZ28uanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.