- 1Shandong Provincial Key Laboratory for Biology of Vegetable Diseases and Insect Pests, Department of Entomology, College of Plant Protection, Shandong Agricultural University, Tai’an, China
- 2College of Life Sciences, Shandong Agricultural University, Tai’an, China
Parasitism is a special interspecific relationship in insects. Unlike most other ectoparasites, Nasonia vitripennis spend most of its life cycle (egg, larvae, pupae, and early adult stage) inside the pupae of flies, which is covered with hard puparium. Microbes play important roles in host development and help insect hosts to adapt to various environments. How the microbes of parasitic wasp respond to different fly hosts living in such close relationships motivated this investigation. In this study, we used N. vitripennis and three different fly pupa hosts (Lucilia sericata, Sarcophaga marshalli, and Musca domestica) to address this question, as well as to illustrate the potential transfer of bacteria through the trophic food chains. We found that N. vitripennis from different fly pupa hosts showed distinct microbiota, which means that the different fly hosts could affect the bacterial communities of their parasitic wasps. Some bacteria showed potential horizontal transfer through the trophic food chains, from the food through the fly to the parasitic wasp. We also found that the heritable endosymbiont Wolbachia could transferred from the fly host to the parasite and correlated with the bacterial communities of the corresponding parasitic wasps. Our findings provide new insight to the microbial interactions between parasite and host.
Introduction
As the most abundant animals worldwide, insects benefit greatly from their symbionts, as they help the insects in developing different strategies to adapt to various environments. With the rapid development of high-throughput sequencing technology, the composition and roles of microorganisms associated with insects have aroused extensive interest in the scientific community.
Insect-associated microorganisms participate in many life processes of their hosts, such as nutrition and digestion (Baumann et al., 1995; Wernegreen, 2002; Zientz et al., 2004; Toh, 2005; Geiger et al., 2011), immune defense (Hedges et al., 2008; Mahadav et al., 2008; Teixeira et al., 2008), reproductive modifications (Stouthamer et al., 1999; Lawson et al., 2001), and pesticide resistance (Kikuchi et al., 2012). Some microbes also play vital roles in the coevolution and speciation of their host insects (Hosokawa et al., 2006; Gosalbes et al., 2010; Werren et al., 2010; Brucker and Bordenstein, 2013). As an example, the endosymbiont Wolbachia infects almost 65% of insects (Hilgenboecker et al., 2008) and helps in evolution of host reproductive strategies, such as cytoplasmic incompatibility, male killing, feminization, parthenogenesis, and even speciation (Bordenstein, 2003; Werren et al., 2008; Brucker and Bordenstein, 2012a; Gebiola et al., 2016; Shropshire and Bordenstein, 2016).
Many factors can affect the bacterial communities of insects: firstly, the species of insect hosts, which is known to shape their bacterial communities. For example, the microbial communities of honey bees and bumble bees exhibit similar patterns as the clustering of the host (Martinson et al., 2011; Koch et al., 2013). Secondly, diet can affect the hosts’ bacterial diversity. Previous studies have found that the composition of intestinal microorganisms in larvae of Helicoverpa armigera (Lepidoptera, Noctuidae) is similar to that of the plant community, indicating that food is an important source of intestinal microorganisms (Priya et al., 2012). According to a study on Monochamus alternatus (Coleoptera, Cerambycidae), Enterococcus is the dominant bacterium in the intestinal tract of larvae feeding on natural food, while Lactococcus is the dominant bacterium in the intestinal tract of larvae feeding on artificial feed (Kim et al., 2017). Distantly related hosts that live in close symbiotic relationship can also maintain similar microbial communities. With varying degrees of nest sharing between them, Megalomyrmex social parasites (Solenopsidini) and their fungus-growing ant hosts from the genera Cyphomyrmex, Trachymyrmex, and Sericomyrmex share symbiont bacteria (Liberti et al., 2015). Thirdly, spatial and temporal distributions of the host could influence their bacterial compositions (Zhang and Blackwell, 2002; Falush et al., 2003; Hedlund and Staley, 2004; Papke and Ward, 2004; Martiny et al., 2006). The bacterial structure of termites shows significant differences between diverse sampling sites (Hongoh et al., 2005).
Nasonia vitripennis (Hymenoptera, Pteromalidae) is an important parasitoid whose female wasp stings, injects venom, and lays eggs in many different fly pupae, where their eggs, larvae, pupae, and early-stage adults are developed. N. vitripennis usually lives on species of the family Calliphoridae, Muscidae, and Sarcophagidae. Because of its short generation time, large offspring production, and easy rearing, Nasonia has emerged as a model organism for developmental and evolutionary genetics (Werren and Loehlin, 2009), as well as a research model for host–microbial community interaction studies (Dittmer et al., 2016).
Parasitism is a special symbiotic relationship in insects. Unlike other ectoparasitic wasps that usually live on the surface of their hosts, N. vitripennis spends most of its life cycle inside fly species, which are covered with puparium; thus, it lives in an enclosed environment. The microbiota from both fly species and the wasps may have more opportunities to communicate due to such an enclosed environment (Lynch, 2015). The microbial communities of N. vitripennis have been previously reported (Brooks et al., 2016; Jessica et al., 2016). However, the response of the microbial community composition of the wasps to different hosts in such an enclosed environment is still unknown. In this study, we used N. vitripennis and three different fly hosts (Lucilia sericata, Sarcophaga marshalli, and Musca domestica) with different food resources to address the following issues: (1) how the different hosts affect the bacterial communities of N. vitripennis, (2) whether there is bacterial transfer through food chains, and (3) whether the existence of the endosymbiont Wolbachia in fly species could affect the microbiota of the parasitic wasp.
Materials and Methods
Experimental Insects
Three different fly hosts, L. sericata (Ls, Diptera: Calliphoridae), S. marshalli (Sm, Diptera: Sarcophagidae), and M. domestica (Md, Diptera: Muscidae), were trapped at the campus of Shandong Agricultural University, China, and then have been kept in the laboratory since 2012. Fly traps were baited with household pork to obtain eggs of the carnivorous L. sericata and S. marshalli, as well as wheat bran mixed with water to obtain eggs of M. domestica. Laboratory rearings were performed with pork carrion (Ca) for the carnivorous flies (L. sericata and S. marshalli), whereas M. domestica was reared on wheat bran (Wb). For larval development rearing was done at 28°C, 50% relative humidity (RH) and 16:8 h (L:D) photoperiod. All fly adults were provided with a 10% honey solution (in water) provided ad libitum (Meister, 1962). N. vitripennis was routinely reared on M. domestica, at the same rearing conditions as descried above. Microbial diversity assays were performed starting from this lab strain, as indicated in Figure 1. Briefly, coetaneous N. vitripennis females were split into three batches. Each batch was further reared on L. sericata (NvLs), S. marshalli (NvSm), or M. domestica (NvMd) pupae for 10 generations. After 10 generations, parasitized hosts (1-day-old pupae) were isolated in 50-ml tubes to allow parasitoid development, and unparasitized hosts were used as controls, as indicated in Figure 1.
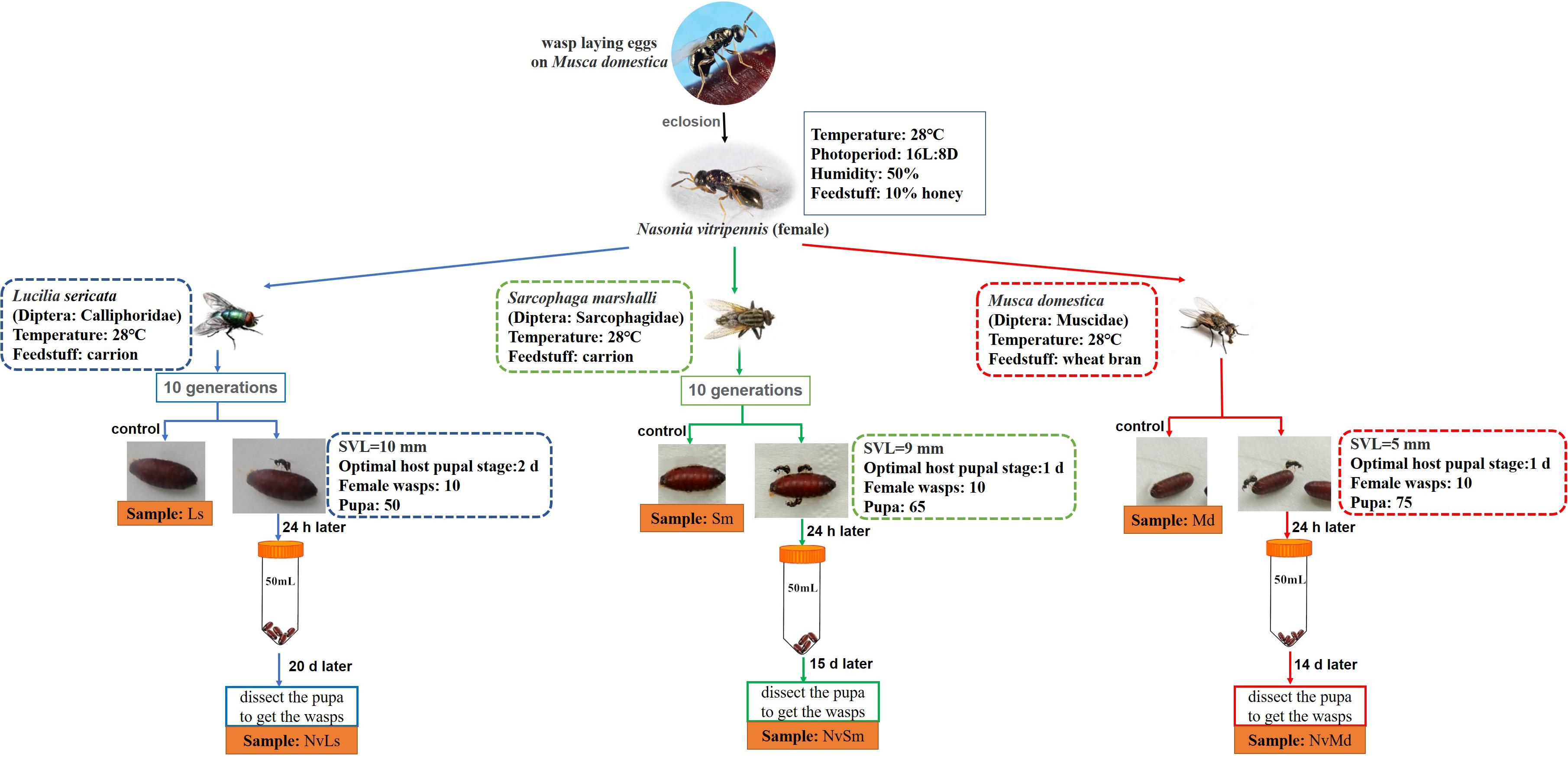
Figure 1. Feeding conditions of experimental insects in laboratory and acquisition of experimental samples. Snout–vent length (SVL) represents the body length of the fly species.
DNA Extraction, PCR Amplification, and Sequencing
Regardless of gender, fresh fly species at the pupal stage (1-day-old) were collected for DNA extraction, although sex could have an impact on microbial communities (Han et al., 2017). Twenty M. domestica pupae, 10 S. marshalli pupae, and 10 L. sericata pupae were selected (members of each fly type were considered as one sample), while more than 100 N. vitripennis individuals were collected before emerging from each of the fly hosts. The fresh fly pupa hosts and parasitic wasps were sterilized in 75% ethanol for 1 min and washed in sterile water for 30 s three times. After sterilization, the samples were collected in 1.5-ml centrifuge tubes and stored in liquid nitrogen. In order to avoid bacterial contamination outside of the puparium, which could affect the bacterial composition of the wasps, after 12 days of the parasite inside the pupa, the puparium was broken with sterilized tweezers and knives. Further, N. vitripennis was transferred to sterilized 1.5-ml centrifuge tubes with small holes to maintain air circulation for the wasps to develop. After 2 days, the wasps were sterilized in 75% ethanol for 1 min and washed in sterile water three times. About 100 individuals were collected in a 1.5-ml centrifuge tube and stored in liquid nitrogen for DNA extraction. Total DNA was extracted from the fly species at the pupal stage and parasitic wasps from different fly hosts, carrion meat, and wheat bran using the OMEGA Soil DNA Extraction Kit. The extracted DNA was amplified by PCR using the bacterial 338F (5′-ACTCCTACGGGAGGCAGCAG-3′)-806R (5′-GGACTACHVGGGTWTCTAAT-3′) primer set targeting the 16S rRNA gene. PCR conditions were as follows: pre-denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 40 s, annealing at 45–59°C (adjusted according to different primers) for 30–60 s, and an extension at 72°C for 1 min; and a final extension for 10 min at 72°C. The qualified DNA samples were selected and sequenced on the Illumina MiSeq platform of Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China.
The gene coding for Wolbachia surface protein (wsp gene) was detected using the Wolbachia-specific primer set of 81F (5′-TGGTCCAATAAGTGATGAAGAAAC-3′) and 691R (5′-AAAAATTAAACGCTACTCCA-3′), and the amplification profiles all followed Xiao et al. (2012). It was observed that L. sericata and M. domestica were not infected with Wolbachia, while S. marshalli was divided into two groups: one group was infected with Wolbachia, while the other group was Wolbachia free.
Bioinformatics and Statistical Analyses
MiSeq sequencing provided double-ended sequence data. First, according to the overlap between paired-end (PE) sequencing reads, pairs of reads were spliced (merge) into a sequence, the reads were filtered based on their quality, and the results were merged by using QIIME2 (Bolyen et al., 2019). The quality filtering standards for sequential reads are as follows. First, the reads with a mass value of less than 20 were filtered, and a 50-bp window was set. If the average mass value in the window is less than 20, the back-end bases were truncated from the window. Reads below 50 bp after quality control were filtered to remove the reads containing N bases. Then, according to the overlap relationship between PE reads, paired reads were spliced (merged) into a sequence, and the minimum overlap length was 10 bp. Then, the maximum error ratio allowed in the overlap area of the mosaicing sequence is 0.2, and the nonconforming sequence is screened. Finally, samples were differentiated according to the barcode and primer at both ends of the sequence, and the sequence direction was adjusted. The allowable mismatches of the barcode were 0, and the maximum primer mismatches were 2. According to the sequence of the two ends of the barcode and the primer sequence, the valid sequences were distinguished from the sample. Then the sequence orientation was corrected, and the optimized data were acquired. The non-repetitive sequences were extracted from the optimized sequences, which could reduce the computational complexity of the intermediate process, the single sequences without duplication were removed, and operational taxonomic units (OTUs) based on the non-repetitive sequences (excluding the single sequence) were clustered according to 97% similarity by using UCHIME. During the clustering process, chimera sequences were removed, and the representative sequences of OTUs were obtained. All the optimized sequences were mapped to the OTU representative sequence, and the sequences with a similarity of more than 97% were selected to generate the OTU table. In order to obtain the information of the species corresponding to each OTU, the RDP classifier Bayesian algorithm was used to analyze 97% similarity of the OTU representative sequence against the SILVA ribosomal RNA gene database using a confidence threshold of 70%.
Based on 97% similarity of the OTUs or other taxonomic levels, the diversity of random sampling in the form of diversity index was calculated using mothur. Alpha diversity and beta diversity were calculated by GmT (Galaxy mothur Toolset) and QIIME2, respectively (Hiltemann et al., 2018; Bolyen et al., 2019). In order to study the similarity or differences in the bacterial composition of the samples, the number of common and unique OTUs in the samples was counted. Based on UniFrac coupled with principal coordinate analysis (PCoA) for all the samples, PCoA and Venn graphs were plotted using the R package. A heatmap plot for abundance analysis was made by using the R package. According to the classification of the composition of the samples and different grouping criteria for linear discriminant analysis (LDA), LDA effect size (LEfSe) found a significant difference in the sample division of the community or species. Differences between populations were analyzed by using one-way ANOVA. Excel was used to show the horizontal transfer of bacteria through the tertiary food chain food–host–parasitic wasp. Information about the species within or between groups was obtained by using the NetworkX. The Network Analysis Toolkit was used to calculate the node degree distribution, the diameter and the average shortest path of the network, the degree of connectivity, and closeness centrality (Hagberg et al., 2008). In Cytoscape 2.8.3, the layout was set as spring embedded, and nodes were used as connection points to reflect the connection between the sample and OTUs and to make a network diagram (Shannon et al., 2003). In order to explore the influence of Wolbachia on other bacteria, SPSS 22.0 was used to calculate the Pearson rank correlation coefficient, and Gephi 0.9.2 (Bickel, 2003) was used for figures. In addition, we took “fly species–parasitic wasp” and “carrion/wheat bran–fly species–parasitic wasp” as the whole, and the bacteria that were shared in host, parasitic wasp, and food in the food chain were figured out using Excel and iTOL software (Letunic and Bork, 2019). FastTree was used to construct the phylogenetic tree based on the maximum likelihood method by selecting the sequences corresponding to OTUs or a class of classified information at different levels. According to the cluster of orthologous groups of proteins (COG) database, the descriptions of each COG and its function were analyzed based on the eggNOG database (Huerta-Cepas et al., 2019). In addition, PICRUSt was used to obtain different levels of metabolic pathway information, as well as the abundance table at each level (Langille et al., 2013).
Results
Nasonia vitripennis Microbiota Profiles in Different Host Species
A total of 754,311 16S rRNA gene sequences were obtained (accession number PRJNA479943). There were 651 OTUs, which were clustered based on 97% 16S rRNA similarity and belonged to 23 prokaryotic phyla. Rarefaction curves showed that all the samples showed enough sequencing depth to represent their bacterial diversity (Supplementary Figure S1).
Interestingly, the network showed a significant microbiota clustering of N. vitripennis and their corresponding fly hosts rather than the bacterial clustering based on fly species (Figure 2). M. domestica and Wolbachia-infected S. marshalli (SmIn) and Wolbachia-free S. marshalli (Sm) showed a closer relationship and more intersections in terms of bacterial community composition with their corresponding parasitic wasps than did L. sericata and its parasitic N. vitripennis; both showed the maximum number of unique OTUs.
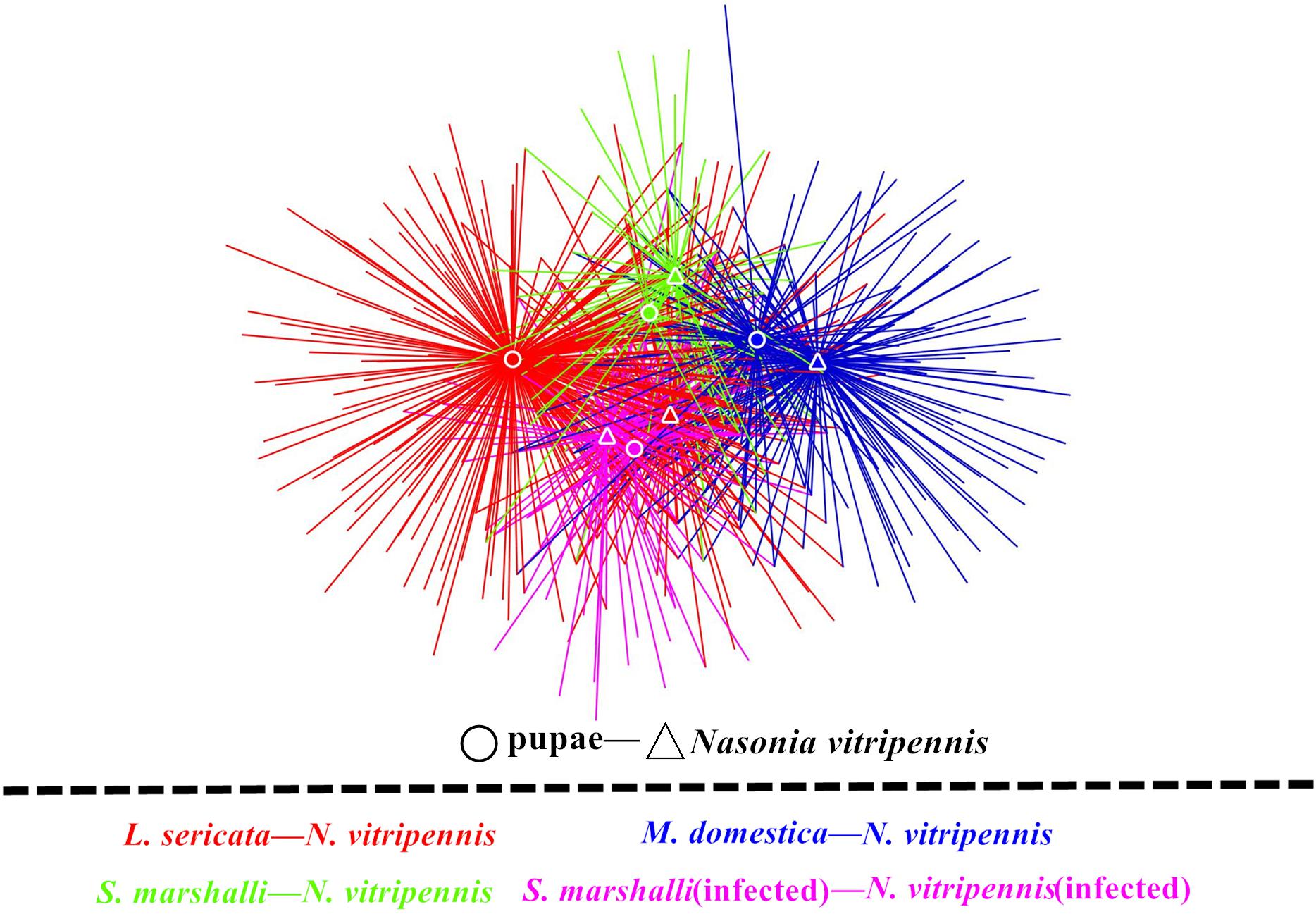
Figure 2. Microbiota network of different fly hosts and their corresponding N. vitripennis. Fly hosts and N. vitripennis are shown as circles and triangles, respectively. Different colors represent the four categories of specifically associated host and parasitic species. Colors red, yellow, green, and purple represent L. sericata, M. domestica, Wolbachia-free S. marshalli, and Wolbachia-infected S. marshalli and their corresponding N. vitripennis, respectively.
Effect of Different Fly Hosts on Microbial Community of Parasitic Wasps
Wasps and their fly hosts showed different bacterial communities. At the phylum level, Proteobacteria was predominant in both L. sericata and S. marshalli whose adults were fed with carrion, while Bacteroidetes and Proteobacteria shared the predominant position in M. domestica whose adults were fed with bran. For N. vitripennis, Proteobacteria was predominant in wasps with L. sericata (NvLs) and S. marshalli (NvSm) as hosts, while Firmicutes was dominant in wasps with M. domestica (NvMd) as the host (Figure 3A). At the genus level, the differences were more defined (Figure 3B). Providencia (41.28%) and Myroides (37.61%) were the predominant genera in L. sericata and M. domestica, respectively. For Wolbachia-free S. marshalli (Sm), Massilia (16.19%), and Sphingobium (12.85%) were the predominant genera, while for Wolbachia-infected S. marshalli (SmIn), Providencia (27.27%) and Vagococcus (16.13%) were the predominant genera, although there was no evident difference at the phylum level due to the presence or absence of Wolbachia. The bacterial communities of the parasitic wasps from different hosts were quite interesting. Gluconobacter (31.24%) and Staphylococcus (81.47%) were the dominant bacterial genera in wasps whose hosts were L. sericata (NvLs) and M. domestica (NvMd), respectively. Proteus (56.03%) and Wolbachia (59.17%) were the dominant genera in N. vitripennis whose hosts were Wolbachia-free (Sm) and Wolbachia-infected S. marshalli (SmIn), respectively (Figure 3B).
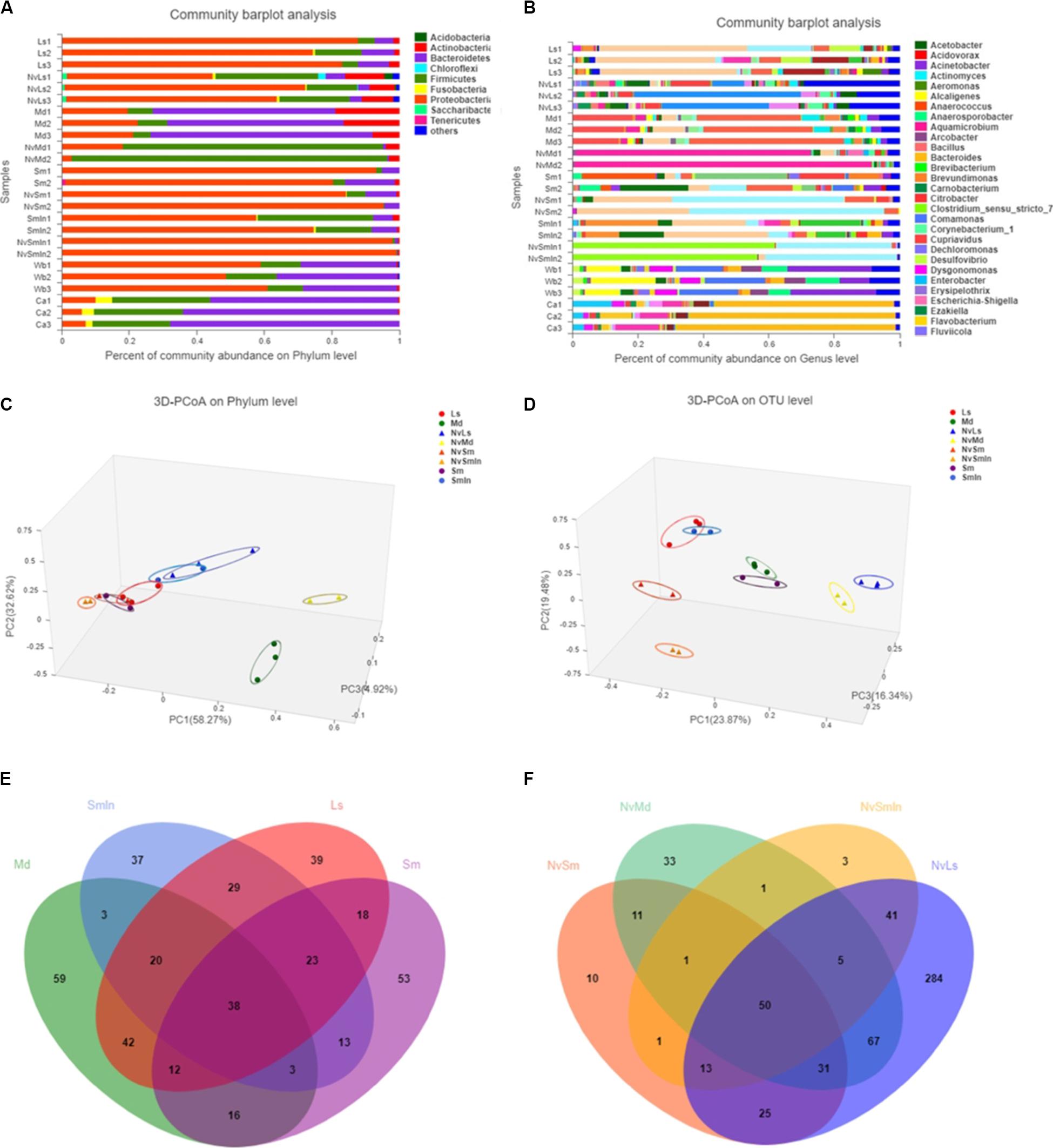
Figure 3. Bacterial composition analysis. Column plots of communities of different samples at phylum level (A) and genus level (B). The length of the columns represents the proportion of species. Bray–Curtis principal coordinate analysis (PCoA) of microbial communities at phylum level (C) and OTU level (D). PC1 and PC2 are two principal coordinate components. Venn diagram at OTU level of fly hosts (L. sericata, M. domestica, Wolbachia-free S. marshalli, and Wolbachia-infected S. marshalli) (E) and corresponding N. vitripennis (F). Different colors represent different samples; the numbers of overlaps and folds represent the shared and the unique species numbers, respectively.
Bacteroidetes was the predominant bacteria in carrion, which was used as food for carnivorous insects L. sericata and S. marshalli, while Proteobacteria was the predominant bacteria in wheat bran, which was used as food for the phytophagous fly M. domestica (Figure 3A). The bacterial community of N. vitripennis was quite different from its fly hosts based on PCoA (Figures 3C,D and Supplementary Figure S2). Chao and Ace indices showed that there were significant differences in the bacterial diversity index between different samples (Supplementary Table S1). The bacterial diversity of N. vitripennis with L. sericata (NvLs) as host was significantly higher than that of the other samples. A rank–abundance graph showed that the curve length and span of N. vitripennis with L. sericata (NvLs) as host were higher than those of the other samples, indicating that the species richness and species uniformity of this sample were the highest (Supplementary Figure S3).
The bacterial hierarchical clustering of the samples did not occur based on species. M. domestica and its corresponding N. vitripennis (NvMd) were close to wheat bran in a distance similar to L. sericata and its corresponding N. vitripennis (NvLs). The parasitic wasps hosted by Wolbachia-infected (SmIn) and Wolbachia-free S. marshalli (Sm) were clustered together (Figure 4).
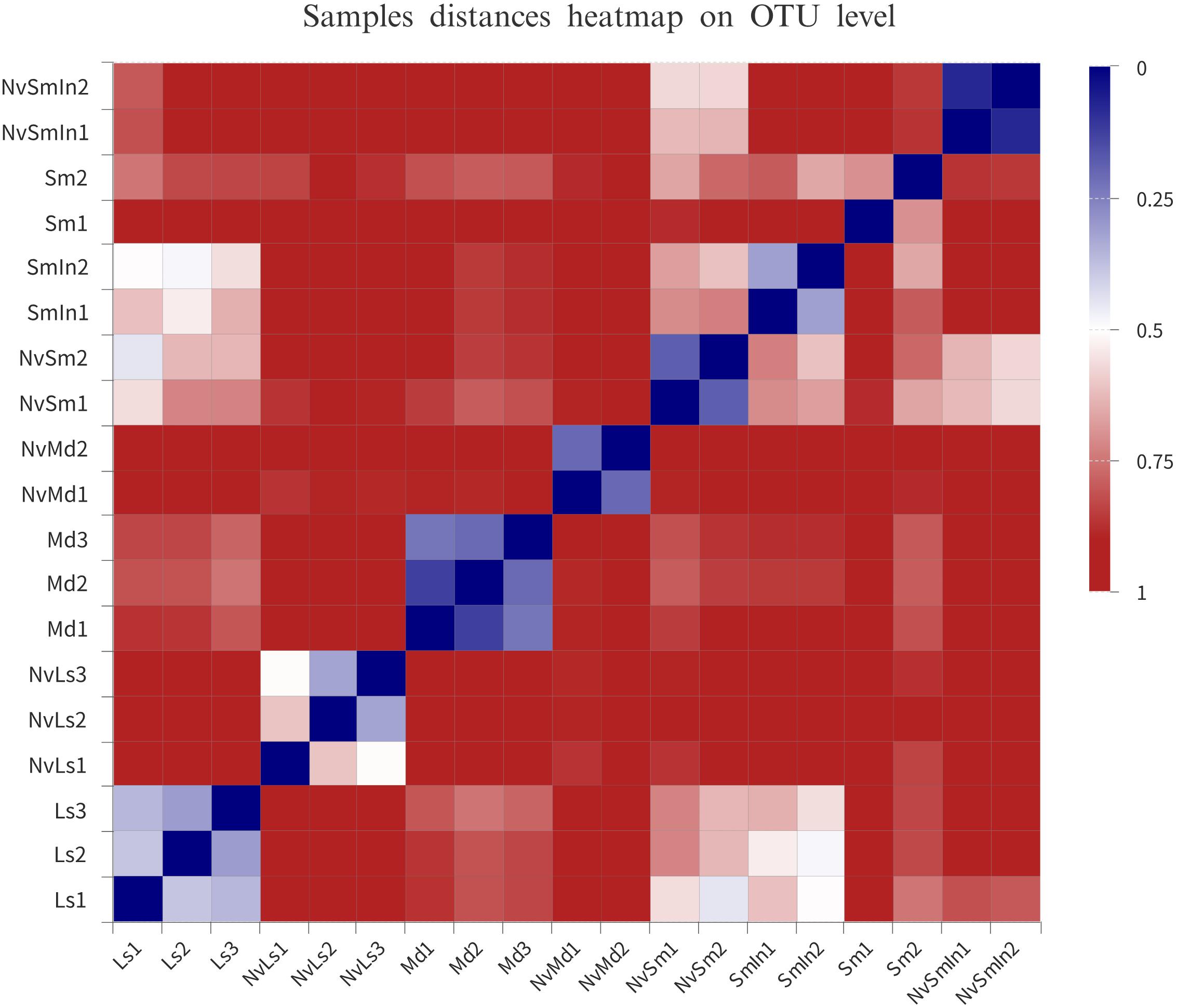
Figure 4. Distance heatmap based on OTU level of the samples. The distance between samples is represented by different color gradients (the value represented by the color gradient on the right of the figure).
There were 221 and 193 OTUs detected in L. sericata and M. domestica, respectively, and there were 166 and 176 OTUs in Wolbachia-infected (SmIn) and Wolbachia-free S. marshalli (Sm), respectively (Figure 3E). Acinetobacter, Bacillus, Citrobacter, Lactococcus, Morganella, and Providencia were common in all types of fly species (Figure 3E). Massilia, Bacillus, Serratia, Providencia, Lactococcus, Staphylococcus, Morganella, Proteus, Bacteroides, Enterobacter, and Myroides were common OTUs in N. vitripennis hosted by all the fly species (Figure 3F). The bacterial diversity of N. vitripennis with L. sericata (NvLs) as host was also higher than that of N. vitripennis inhabiting other hosts. A total of 516 and 199 OTUs were detected in N. vitripennis with L. sericata (NvLs) and M. domestica (NvMd) as hosts, respectively, while there were 142 and 115 OTUs in N. vitripennis with Wolbachia-free (NvSm) and Wolbachia-infected S. marshalli (NvSmIn) as hosts, respectively (Figure 3F). The results showed that bacterial diversity of the same N. vitripennis with a different fly host was significantly different.
Potential Bacterial Transfer via Food–Fly–Wasp Tertiary Food Chain
In this study, the parasitic wasp N. vitripennis was grown in three different fly hosts. S. marshalli and L. sericata were fed with carrion, while M. domestica was fed with wheat bran; thus, the food–pupae–wasp tertiary food chain and the pupae–wasp food chain system were good models to study the potential bacterial transmission (Figure 5 and Supplementary Table S2).
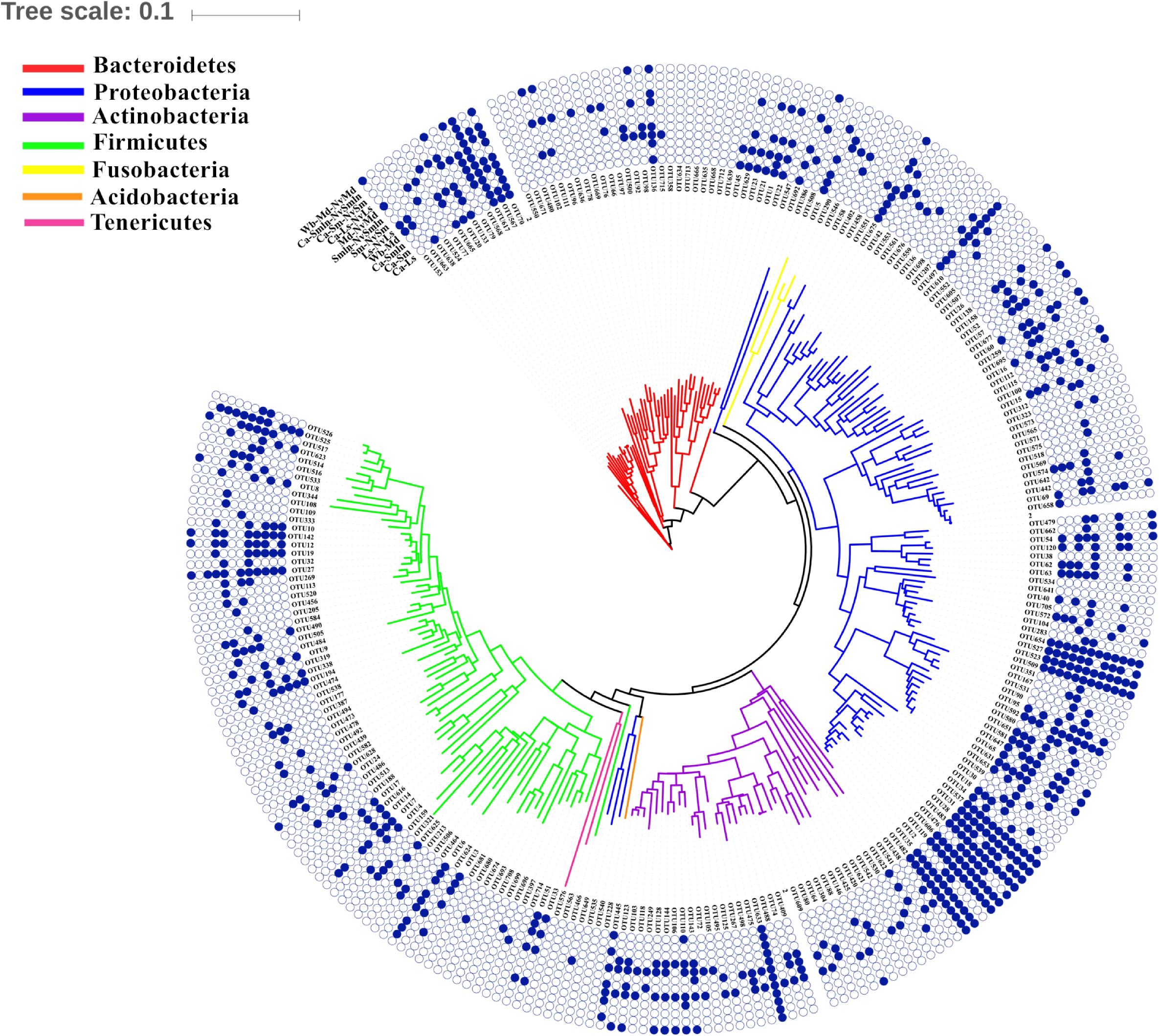
Figure 5. Shared bacteria of different food chains (food–pupae–wasps or pupae–wasps). The phylogenetic tree was constructed by the top 50 OTUs in each sample (internal). The blue solid circle represents the presence of the bacterial OTUs in the food chain, and the hollow circle represents the absence of bacterial OTUs in the food chain. For example, OTU153 showed that it was able to pass through the food chain of the wheat bran–M. domestica pupae–its corresponding N. vitripennis. In addition, it was also able to pass from L. sericata pupae to N. vitripennis.
There were some unique OTUs found in different food–pupae–wasp tertiary food chains. OTU20 (Myroides) was found only in the food chain beginning with carrion (carrion–L. sericata/S. marshalli–N. vitripennis), while OTU102 (Sphingobacterium), OTU111 (Sphingobacterium), OTU92 (Sphingobacterium), OTU104 (Pseudomonas), OTU106 (Microbacterium), OTU146 (Actinomyces), and OTU662 (Stenotrophomonas) were unique to food chains that began with wheat bran (wheat bran–M. domestica–N. vitripennis); hence, these bacteria were probably transferred from food to fly species to N. vitripennis.
Many unique bacteria only existed in the fly species at the pupal stage to parasitic wasp food chains and were not found in food to fly food chains. Most unique OTUs were found in the L. sericata–N. vitripennis food chain and confirmed the possibility of being transmitted from fly species to N. vitripennis (Figure 5 and Supplementary Table S2). Some of the unique bacteria only existed in one of the two food chain systems, from food to fly species or from fly to wasps; these were rare bacteria appearing at a low abundance.
Effects of Wolbachia on Host Microbiota
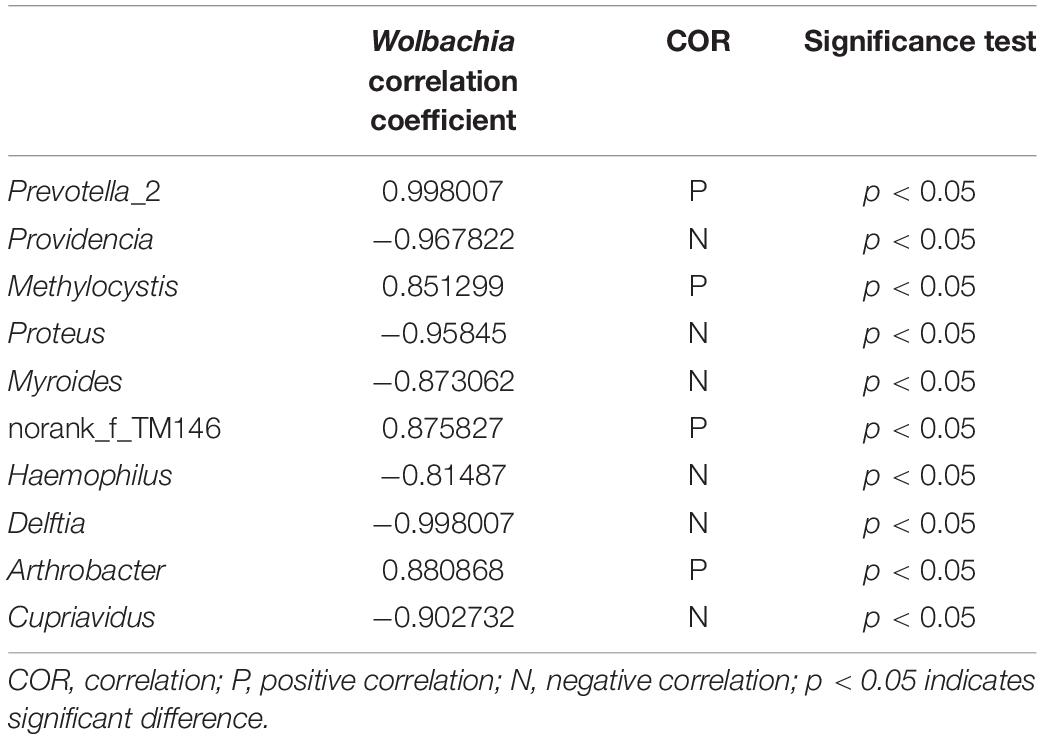
Taking all the bacteria from Wolbachia-infected (NvSmIn) and Wolbachia-free N. vitripennis (NvSm) into account, certain large coenobium was observed in this study. The coenobium, including Proteus, Myroides, Haemophilus, Delftia, Arthrobacter, Cupriavidus, Providencia, Methylocystis, Prevotella_2, and norank_f_TM146 showed a direct relationship with Wolbachia (Figure 6). Except in the cases of Prevotella_2, Methylocystis, Arthrobacter, and norank_f_TM146, the Spearman correlation coefficient between other genera and Wolbachia showed negative correlations, suggesting that Wolbachia and these genera may be in a competitive relationship (Figure 6 and Table 1).
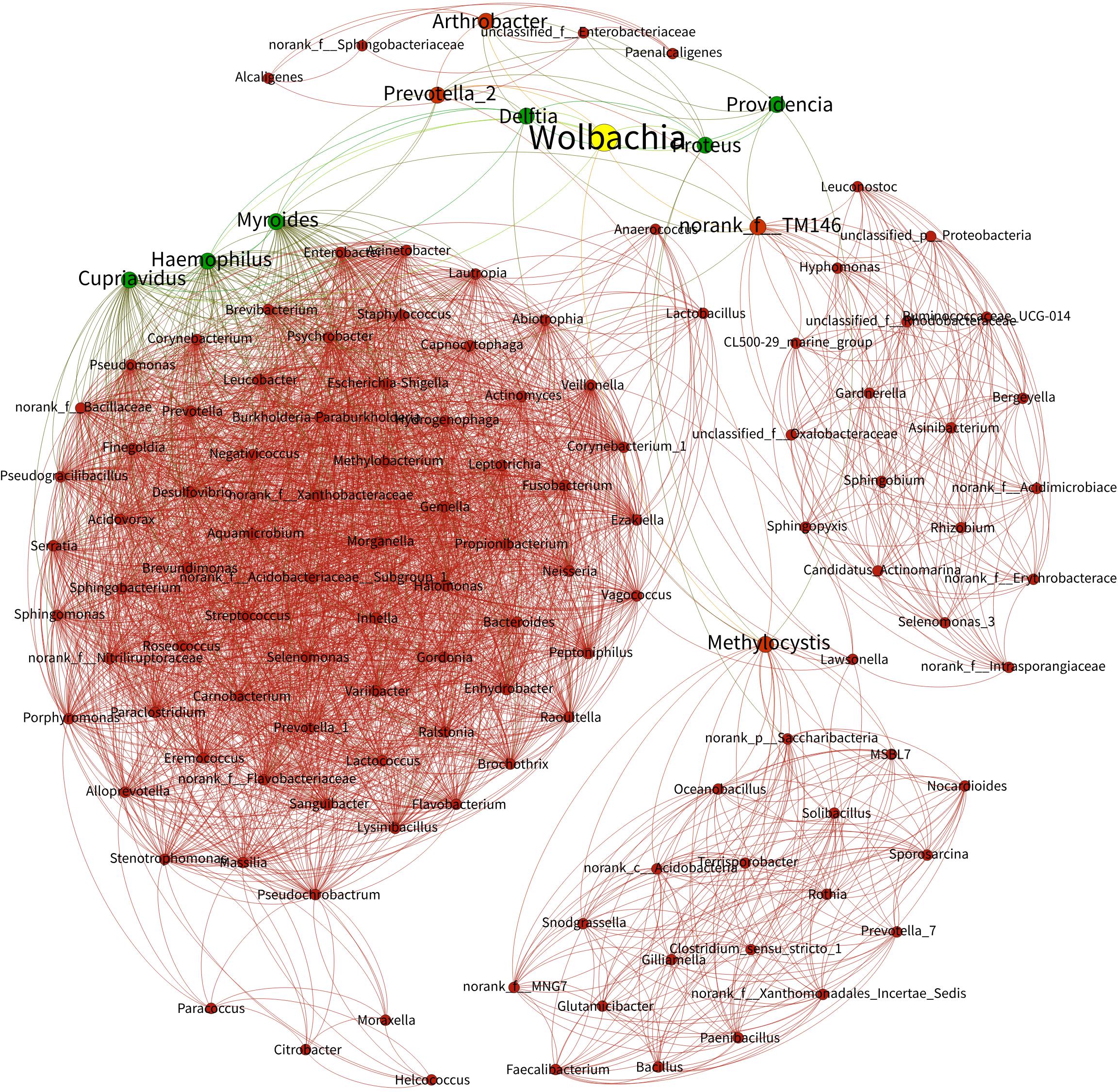
Figure 6. Bacterial networks from Wolbachia-infected and Wolbachia-free N. vitripennis. The Spearman rank correlation coefficient was calculated to reflect the correlation between bacterial species. Red and green lines represent positive and negative correlations, respectively.
Function Prediction Based on 16S rRNA Gene Sequence
All the bacterial KEGG metabolic pathways were mainly classified into amino acid metabolism, carbohydrate metabolism, replication and repair, energy metabolism, glycan biosynthesis and metabolism, enzyme families, membrane transport, etc. (Figure 7). Gastric triacylglycerol lipase helps insects in digestion of fat. Comparison of the differences in gastric triacylglycerol lipase expression in different fly pupae and their corresponding N. vitripennis showed that the lipase expression in carnivorous N. vitripennis was significantly higher than that in the phytophagous M. domestica, which was fed with wheat bran. Moreover, the lipase expression in S. marshalli was significantly higher than that in M. domestica (Figure 8). Although PICRUSt was deemed not suitable for nonhuman microbiomes (Langille et al., 2013), we thought it is believable in our results.
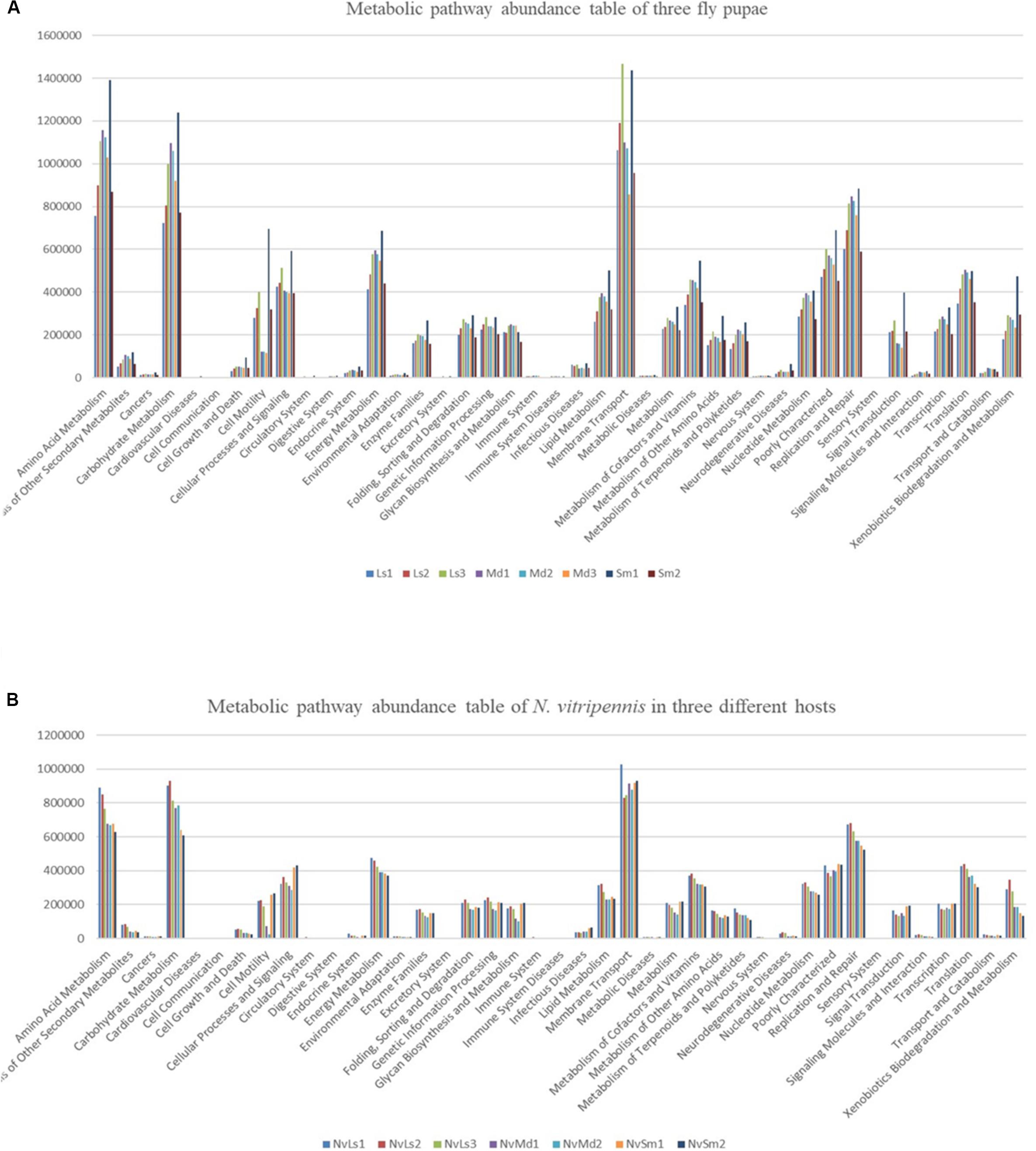
Figure 7. KEGG functional secondary classification of the three different fly pupa hosts (A) and their corresponding N. vitripennis (B).
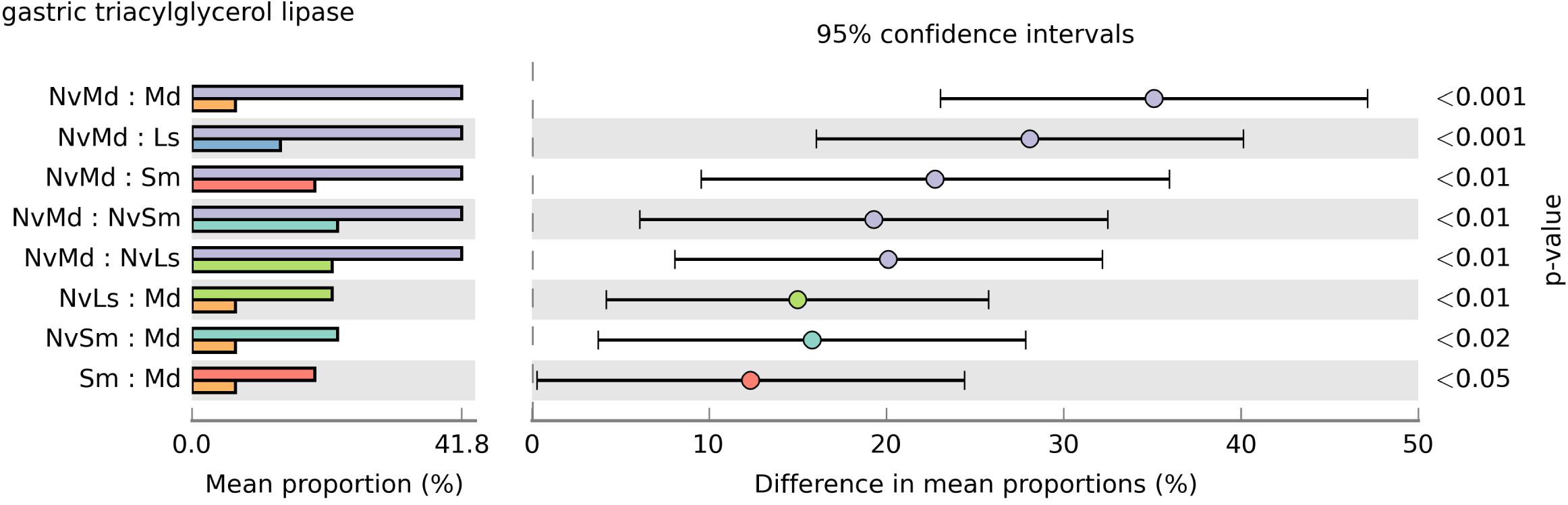
Figure 8. The significance test of gastric triacylglycerol lipase expression abundance based on Welch’s t-test and a two-sided test. The dot color is shown the same as the group color with large expression abundance. Only results with a p-value < 0.05 are shown.
Discussion
Parasitism is a symbiotic insect relationship wherein the parasite consumes its host as nutrition for development, unlike commensalism and mutualism. Nasonia is a special ectoparasite with development of more than one wasp in a single fly pupa (brood size in Nasonia ranges from 3 to 46 per host pupa); they inject their eggs through the puparium of the fly host, where the eggs hatch and then develop into larvae, pupae, and adults. Unlike other ectoparasitoids that usually live on the surface of their hosts, living under the cover of puparium makes Nasonia and its fly hosts good systems to study the interactions between insect parasites and their hosts from different perspectives.
Our results demonstrate that different hosts can affect the bacterial communities of parasitic wasps. Species is the most contributing determinant in bacterial composition of different organisms, which was confirmed by our results. Although fly food, parasitic wasps, and fly hosts had some common bacteria, such as Raoultella, Pseudomonas, Providencia, Myroides, Lactococcus, Acinetobacter, and Corynebacterium, they also showed their unique microbiota composition pattern. However, the microbiota of the parasitic wasp N. vitripennis was shaped by its corresponding host doubtlessly. From the microbiota networks of the three fly pupae and their corresponding parasitic wasps, M. domestica and Wolbachia-infected and Wolbachia-free S. marshalli showed a closer relationship and more intersections with their corresponding parasitic wasps than L. sericata and its parasitic wasps; both showed many unique OTUs, individually (Figure 2). This phenomenon needs to be investigated in our next study. Besides organism species, diet and spatial environment are also important factors that influence microbiota of an organism. It is reported that dietary specialization in mutualistic acacia–ants affects the relative abundance of the host-associated bacteria (Rubin et al., 2019). For our parasitic wasps, fly species were not only the sole diet source from egg to adult but also their closed living environment. The unique life history of the ectoparasitic wasp Nasonia also makes it a good model to study the microecology of parasite and hosts.
Different fly hosts and their corresponding N. vitripennis showed different core microbiota. Providencia, Myroides, Massilia, and Sphingobium were predominant in L. sericata, M. domestica, Wolbachia-free S. marshalli, and Wolbachia-infected S. marshalli pupae, respectively. On the other hand, bacterial communities of the same parasitic wasps from different hosts were quite interesting. Gluconobacter, Staphylococcus, Proteus, and Wolbachia were the dominant bacteria genera in wasps whose hosts were L. sericata, M. domestica, Wolbachia-free S. marshalli, and Wolbachia-infected S. marshalli, respectively. Thus, the same parasitic wasp with different hosts showed different dominant bacteria.
Moreover, some of the bacteria mentioned above were also the dominant bacteria in one or more samples. For example, Pseudomonas was one of the most abundant bacteria in Wolbachia-free S. marshalli, and Myroides was one of the most abundant bacteria in M. domestica, which are in accordance with results from a previous study by Zurek and Nayduch (2016). However, Providencia abundantly existed not only in L. sericata but also in Wolbachia-infected S. marshalli. These bacteria were the core bacteria in these samples and had important regulating effects on the physiological activity of the host. Even though there was a close relationship between the parasitic wasps and their fly pupa hosts, as N. vitripennis spends almost its whole life in the fly species, the bacterial composition of the pupae hosts and N. vitripennis showed significant differences.
Different fly hosts showed significant effects on the bacterial diversity of the parasitic wasp N. vitripennis. For this special relationship in animals, parasitism means one can get benefit from the other, such as a developmental environment and nutrition. In case of N. vitripennis, its eggs, larvae, pupae, and young adults all developed inside the fly species at the pupal stage. Three different host sources of N. vitripennis showed different bacterial compositions. Gluconobacter and Staphylococcus were the dominant bacteria in N. vitripennis from L. sericata and M. domestica, respectively. Proteus and Providencia were the dominant bacteria in N. vitripennis with Wolbachia-free S. marshalli as host, while Wolbachia and Proteus were the dominant bacteria in N. vitripennis with Wolbachia-infected S. marshalli as host (Supplementary Figure S7 and Supplementary Table S3). Thus, different fly hosts shaped the bacterial diversity of their parasitic wasps. Brucker and Bordenstein (2012b) found that three species of Nasonia wasps, whose host was Sarcophaga bullata, were dominated by Providencia. Two possible reasons could explain this difference when compared to our results. Firstly, according to our findings, the hosts could affect the microbiota of parasitic wasps, and because different fly hosts were chosen, N. vitripennis from different hosts showed varied bacterial compositions. Secondly, different sequencing methods were used in both studies: Brucker used the first-generation sequencing method, while we used the high-through sequencing method, which can find trace bacteria; hence, our results should be more comprehensive. It is worth noting that the rearing temperature of N. vitripennis was different. Temperature can affect the microbiome of the host. Brucker used 25°C to rear the wasp, while we used 28°C in this study, so temperature is another possible reason for the microbial difference.
We also tried to predict a potential horizontal transfer of bacteria through secondary or tertiary food chains. N. vitripennis inhabiting S. marshalli were infected with Wolbachia, and their corresponding N. vitripennis were infected with Wolbachia too, while when the hosts were not infected with Wolbachia, the parasitic wasps were also Wolbachia free; this phenomenon is substantial evidence for horizontal transfer of bacteria, which was validated by Brucker and Bordenstein (2012b). Taking the wheat bran-M. domestica-N. vitripennis food chain as an example, OTU102 (Sphingobacterium), OTU111 (Sphingobacterium), OTU92 (Sphingobacterium), OTU104 (Pseudomonas), OTU106 (Microbacterium), OTU146 (Actinomyces), and OTU662 (Stenotrophomonas) were found in all three, wheat bran, M. domestica, and N. vitripennis, which suggested the potential horizontal transfer from food to fly and to the parasitic wasp. These food chains are a good model to investigate horizontal transfer of bacteria through a special parasitic relationship. Some trace bacteria were also observed in this study that may play important metabolic functions in their hosts, which were shared by N. vitripennis and their corresponding fly species. While providing substantial insights on the identity, structure, and taxonomic overlap of the microbiota in different food chains, our comparative results could not provide conclusive evidence for the underlying transmission mechanisms of every bacterial symbiont, because some may be shared in nature. To illuminate and confirm the transmission mechanisms, experiments are required in the future.
Endosymbiont Wolbachia is famous for various reproductive manipulations and fitness effects on their hosts. Wolbachia can not only influence the hosts’ mitochondrial DNA (Xiao et al., 2012) but also can alter hosts’ microbiota composition, for example, in the terrestrial isopod Armadillidium vulgare (Isopoda, Oniscoidea) (Dittmer and Bouchon, 2018) and in adult Aedes aegypti (Diptera, Culicidae) mosquitoes (Audsley et al., 2018). In our study, we found both competitive and cooperative relationships between the microbiota. The existence of Wolbachia influenced the presence and abundance of many bacterial taxa within each host population, possibly due to competitive interactions. Proteus, Myroides, Haemophilus, Delftia, Arthrobacter, Cupriavidus, Providencia, Methylocystis, Prevotella_2, and norank_f_TM146 showed symbiotic or competitive relationships with Wolbachia. The microbiota was shaped by interactions not only between the insect hosts and its symbionts but also between the different members of the symbiotic communities (Mouton et al., 2004; Oliver et al., 2006). Competition between Wolbachia and other microorganisms for resources and space in the shared host environments decreased the abundance of some bacteria.
The parasitic wasps and fly hosts are good models to study parasitic interactions from different perspectives. Detailed research on this issues, like the role of bacteria behind successful survival of wasps in new hosts, needs our attention.
Data Availability Statement
The datasets generated for this study have been uploaded to GenBank by the accession number PRJNA479943.
Author Contributions
NW conceived the idea of study and designed the methodology. HX prepared all the experimental insect samples. RD, HX, SG, ZG, and NW participated in data analysis and wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Shandong Province Postdoctoral Innovation Project (201702044), the China Postdoctoral Science Foundation (2017M612312), a Project of Shandong Province Higher Educational Science and Technology Program (J17KA149), and the Provincial Key Research and Development Program of Shandong (2017CXGC0207).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to professor Zhong Zhang in Shandong First Medical University & Shandong Academy of Medical Sciences for assistance in the laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01435/full#supplementary-material
FIGURE S1 | Sparse curve analysis of the samples.
FIGURE S2 | Shannon index at OUT level of the samples.
FIGURE S3 | Rank-abundance curves at OTU level of the samples.
FIGURE S4 | Relationship between Circos samples and species which can reflect the distribution ratio of dominant species in each sample through a visual circle diagram.
FIGURE S5 | Heatmap analysis of the top 50 species abundance in all the samples at the genus level.
FIGURE S6 | Cyclic phylogenetic tree of the top 50 species abundance in all samples at the genus level.
FIGURE S7 | Student’s t-test bar plot on Genus level between two samples.
TABLE S1 | Alpha diversity index table of all the samples.
TABLE S2 | Species abundance of partial OTUs in all samples.
TABLE S3 | Statistical table of difference test between two groups of samples.
References
Audsley, M. D., Seleznev, A., Joubert, D. A., Woolfit, M., O’Neill, S. L., and McGraw, E. A. (2018). Wolbachia infection alters the relative abundance of resident bacteria in adult Aedes aegypti mosquitoes, but not larvae. Mol. Ecol. 27, 297–309.
Baumann, P., Baumann, L., Lai, C. Y., Rouhbakhsh, D., Moran, N. A., and Clark, M. A. (1995). Genetics, physilogy and evolutionary relationship of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 49, 55–94. doi: 10.1146/annurev.mi.49.100195.000415
Bickel, D. R. (2003). Robust cluster analysis of microarray gene expression data with the number of clusters determined biologically. Bioinformatics 19, 818–824. doi: 10.1093/bioinformatics/btg092
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Bokulich, N. A., Abnet, C. C., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotech. 37, 852–857.
Bordenstein, S. R. (2003). “Symbiosis and the origin of species,” in Insect Symbiosis, eds B. Kostas and A. M. Thomas (Boca Raton, FL: CRC Press), 283–304. doi: 10.1201/9780203009918.ch17
Brooks, A. W., Kohl, K. D., Brucker, R. M., van Opstal, E. J., and Bordenstein, S. R. (2016). Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14:e2000225. doi: 10.1371/journal.pbio.2000225
Brucker, R. M., and Bordenstein, S. R. (2012a). Speciation by symbiosis. Trends Ecol. Evol. 27, 443–451. doi: 10.1016/j.tree.2012.03.011
Brucker, R. M., and Bordenstein, S. R. (2012b). The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 66, 349–362. doi: 10.1111/j.1558-5646.2011.01454.x
Brucker, R. M., and Bordenstein, S. R. (2013). The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. doi: 10.1126/science.1240659
Dittmer, J., and Bouchon, D. (2018). Feminizing Wolbachia influence microbiota composition in the terrestrial isopod Armadillidium vulgare. Sci. Rep. 8:6998.
Dittmer, J., van Opstal, E. J., Shropshire, J. D., Bordenstein, S. R., Hurst, G. D., and Brucker, R. M. (2016). Disentangling a holobiont – recent advances and perspectives in Nasonia wasps. Front. Microbiol 7:1478. doi: 10.3389/fmicb.2016.01478
Falush, D., Wirth, T., Linz, B., Pritchard, J. K., Stephens, M., Kidd, M., et al. (2003). Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585. doi: 10.1126/science.1080857
Gebiola, M., Kelly, S. E., Hammerstein, P., Giorgini, M., and Hunter, M. S. (2016). “Darwin’s corollary” and cytoplasmic incompatibility induced by cardinium may contribute to speciation in encarsia wasps (Hymenoptera: Aphelinidae). Evolution 70, 2447–2458. doi: 10.1111/evo.13037
Geiger, A., Fardeau, M. L., Njiokou, F., Joseph, M., Asonganyi, T., Ollivier, B., et al. (2011). Bacterial diversity associated with populations of Glossina spp. from cameroon and distribution within the campo sleeping sickness focus. Microb. Ecol. 62, 632–643. doi: 10.1007/s00248-011-9830-y
Gosalbes, M. J., Latorre, A., Lamelas, A., and Moya, A. (2010). Genomics of intracellular symbionts in insects. Int. J. Med. Microbiol. 300, 271–278. doi: 10.1016/j.ijmm.2009.12.001
Hagberg, A. A., Schult, D. A., and Swart, P. J. (2008). “Exploring network structure, dynamics, and function using NetworkX,” in Proceedings of the 7th Python in Science Conference, eds G. Varoquaux, T. Vaught, and J. Millman (Austin, TX: EuroSciPy), 11–15.
Han, G., Lee, H. J., Jeong, S. E., Jeon, C. O., and Hyun, S. (2017). Comparative analysis of Drosophila melanogaster gut microbiota with respect to host strain, sex, and age. Microb. Ecol. 74, 207–216. doi: 10.1007/s00248-016-0925-3
Hedges, L. M., Brownlie, J. C., O’Neill, S. L., and Johnson, K. N. (2008). Wolbachia and virus protection in insects. Science 322:702. doi: 10.1126/science.1162418
Hedlund, B. P., and Staley, J. T. (2004). “Microbial endemism and biogeography,” in Microbial Diversity and Bioprospecting, ed. A. T. Bull (Washington, DC: Bull ASM Press), 225–231. doi: 10.1128/9781555817770.ch22
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia?–A statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Hiltemann, S. D., Boers, S. A., van der Spek, P. J., Jansen, R., Hays, J. P., and Stubbs, A. P. (2018). Galaxy mothur Toolset (GmT): a user-friendly application for 16S rRNA gene sequencing analysis using mothur. GigaScience 8, 1–5.
Hongoh, Y., Deevong, P., Inoue, T., Moriya, S., Trakulnaleamsai, S., Ohkuma, M., et al. (2005). Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microb. 71, 6590–6599. doi: 10.1128/aem.71.11.6590-6599.2005
Hosokawa, T., Kikuchi, Y., Nikoh, N., Shimada, M., and Fukatsu, T. (2006). Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. doi: 10.1371/journal.pbio.0040337
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314.
Jessica, D., Van, O. E. J., Dylan, S. J., Bordenstein, S. R., Hurst, G. D. D., and Brucker, R. M. (2016). Disentangling a holobiont – recent advances and perspectives in Nasonia wasps. Front. Microbiol. 7:1478.
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 8618–8622. doi: 10.1073/pnas.1200231109
Kim, J. M., Choi, M. Y., Kim, J. W., Lee, S. A., Ahn, J. H., Song, J., et al. (2017). Effects of diet type, developmental stage, and gut compartment in the gut bacterial communities of two Cerambycidae species(coleoptera). J. Microbiol. 55, 21–30. doi: 10.1007/s12275-017-6561-x
Koch, H., Abrol, D. P., Li, J., and Schmid-Hempel, P. (2013). Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 22, 2028–2044. doi: 10.1111/mec.12209
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lawson, E. T., Mousseau, T. A., Klaper, R., Hunter, M. D., and Werren, J. H. (2001). Rickettsia associated with male-killing in a buprestid beetle. Heredity 86(Pt 4), 497–505. doi: 10.1046/j.1365-2540.2001.00848.x
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259.
Liberti, J., Sapountzis, P., and Hansen, L. H. (2015). Bacterial symbiont sharing in Megalomyrmex social parasites and their fungus-growing ant hosts. Mol. Ecol. 24, 3151–3169. doi: 10.1111/mec.13216
Lynch, J. A. (2015). The expanding genetic toolbox of the wasp Nasonia vitripennis and its relatives. Genetics 199, 897–904. doi: 10.1534/genetics.112.147512
Mahadav, A., Gerling, D., Gottlieb, Y., Czosnek, H., and Ghanim, M. (2008). Parasitization by the wasp Eretmocerus mundus induces transcription of genes related to immune response and symbiotic bacteria proliferation in the whitefly Bemisia tabaci. BMC Genomics 9:342. doi: 10.1186/1471-2164-9-342
Martinson, V. G., Danforth, B. N., Minckley, R. L., Rueppell, O., Tingek, S., and Moran, N. A. (2011). A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 20, 619–628. doi: 10.1111/j.1365-294x.2010.04959.x
Martiny, J. B., Bohannan, B. J., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
Meister, G. (1962). Biological observations in the laboratory cultivation of Musca domestica L. (A contribution to the problem of the standardization of cultivation methods). Zeitschrift Fuür Tropenmedizin Und Parasitologie 13:102.
Mouton, L., Dedeine, F., Henri, H., Boulétreau, M., Profizi, N., and Vavre, F. (2004). Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168, 181–189. doi: 10.1534/genetics.104.026716
Oliver, K. M., Moran, N. A., and Hunter, M. S. (2006). Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. B Biol. Sci. 273, 1273–1280. doi: 10.1098/rspb.2005.3436
Papke, R. T., and Ward, D. M. (2004). The importance of physical isolation to microbial diversification. FEMS Microbiol. Ecol. 48, 293–303. doi: 10.1016/j.femsec.2004.03.013
Priya, N. G., Ojha, A., Kajla, M. K., Raj, A., and Rajagopal, R. (2012). Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS One 7:e30768. doi: 10.1371/journal.pone.0030768
Rubin, B. E. R., Kautz, S., Wray, B. D., and Moreau, C. S. (2019). Dietary specialization in mutualistic acacia-ants affects relative abundance but not identity of host-associated bacteria. Mol. Ecol. 28, 900–916. doi: 10.1111/mec.14834
Shannon, P., Markiel, A., and Ozier, O. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shropshire, J. D., and Bordenstein, S. R. (2016). Speciation by symbiosis: the microbiome and behavior. mBio 7:e01785.
Stouthamer, R., Breeuwer, J. A., and Hurst, G. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Teixeira, L., Ferreira, A., and Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6:e2. doi: 10.1371/journal.pbio.1000002
Toh, H. (2005). Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156. doi: 10.1101/gr.4106106
Wernegreen, J. J. (2002). Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3, 850–861. doi: 10.1038/nrg931
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Werren, J. H., and Loehlin, D. W. (2009). Curing Wolbachia infections in Nasonia (parasitoid wasp). Cold Spring Harb. Protoc. 2009:pdb. prot5312.
Werren, J. H., Richards, S., Desjardins, C. A., Niehuis, O., Gadau, J., and Colbourne, J. K. (2010). Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343–348.
Xiao, J. H., Wang, N. X., Murphy, R. W., Cook, J., Jia, L.-Y., and Huang, D.-W. (2012). Wolbachia infection and dramatic intraspecific mitochondrial DNA divergence in a fig wasp. Evolution 66, 1907–1916. doi: 10.1111/j.1558-5646.2011.01561.x
Zhang, N., and Blackwell, M. (2002). Population structure of dogwood anthracnose fungus. Phytopathology 92, 1276–1283. doi: 10.1094/phyto.2002.92.12.1276
Zientz, E., Dandekar, T., and Gross, R. (2004). Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68, 745–770. doi: 10.1128/mmbr.68.4.745-770.2004
Keywords: bacterial community, fly, horizontal transfer, Nasonia vitripennis, parasitism, Wolbachia
Citation: Duan R, Xu H, Gao S, Gao Z and Wang N (2020) Effects of Different Hosts on Bacterial Communities of Parasitic Wasp Nasonia vitripennis. Front. Microbiol. 11:1435. doi: 10.3389/fmicb.2020.01435
Received: 24 December 2019; Accepted: 03 June 2020;
Published: 07 July 2020.
Edited by:
Nicole Marie Gerardo, Emory University, United StatesReviewed by:
Beatriz Sabater-Munoz, Polytechnic University of Valencia, SpainSeth Bordenstein, Vanderbilt University, United States
Copyright © 2020 Duan, Xu, Gao, Gao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ningxin Wang, bnh3YW5nQHNkYXUuZWR1LmNu
†These authors have contributed equally to this work
 Ruxin Duan1†
Ruxin Duan1† Zheng Gao
Zheng Gao Ningxin Wang
Ningxin Wang