- 1Beijing Key Laboratory of Genetic Engineering Drug and Biotechnology, Institute of Biochemistry and Biotechnology, College of Life Sciences, Beijing Normal University, Beijing, China
- 2Zhejiang Medicine Co., Ltd., Zhejiang, China
- 3Department of Microbiology, College of Life Sciences, Nankai University, Tianjin, China
A conserved open reading frame, dps, is described in Pestalotiopsis microspora, sharing a remarkable similarity with fungal diterpene synthases whose function is less studied. Loss-of-function approach manifested that dps was necessary for the growth and the development of the fungus. A deletion strain, dpsΔ, showed a fundamental retardation in growth, which could deliberately be restored by the addition of exogenous sterols to the media. Gas chromatography–mass spectrometry analysis confirmed the loss of the ability to produce certain sterols. Thus, the tolerance and the resistance of dpsΔ to several stress conditions were impaired. Secondary metabolites, such as the polyketide derivative dibenzodioxocinones, were significantly diminished. At the molecular level, the deletion of dps even affected the expression of genes in the mevalonate pathway. This report adds knowledge about fungal diterpene synthases in Pestalitiopsis microspora.
Introduction
Terpenoids are known as one of the most abundant natural products (Ruan et al., 2019). In fungi, terpenoids are usually synthesized from two C5 units, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) (Li et al., 2015). These two units were condensed via the mevalonate (MVA) pathway and undergo a cyclization step to form terpenoid precursors by various terpene synthases, e.g., monoterpene synthase (Lange, 2015), sesquiterpene synthase (Wang et al., 2016), diterpene synthase, triterpene synthase (Dhingra and Cramer, 2017), and even taxadiene synthase (Bian et al., 2017). Due to bioactivities, many of them have been used in diverse fields, such as medicinal chemistry (Austin et al., 1988), food additives (Ravichandran et al., 2018), and biofuels (Wu et al., 2017). Acquiring terpenoids at the industrial level suffers difficulties, e.g., low content in natural materials. Fermentation by microorganisms is an attractive strategy to obtain these natural products, owing to its large scale and comparatively lower cost if compared to other methods, i.e., extraction from raw material or plant cell culture (Gauvin et al., 2000; Kobayashi and Shigemori, 2002). Many fungi have been reportedly used in the production of terpenoids, i.e., pinene, limonene, camphor, and likely artemisinin (Wu et al., 2017).
P. microspora strain NK17 was isolated by our laboratory and characterized as a producer of a number of dibenzodioxocinones (Niu et al., 2015), with promising application in drug development. A previous study showed that in NK17 there was a diterpene synthase, dps, and it is the only enzyme of this kind in the genome (Chen et al., 2015). Intriguingly, this diterpene synthase shows similarity, to some extent, to the taxadiene synthase which catalyzes the first committed step in the biosynthesis of the antitumor drug paclitaxel in Pacific yew and to the bacterial ent-kaurene synthase in cladogram (Figure 1A). To characterize the role of this ubiquitous enzyme in fungi, we employed gene expression information and metabolite profiling comparatively in the wild type and the mutant strain dpsΔ. We demonstrated that dps played a critical role in the biosynthesis of fungal sterols and was indispensable for the growth of P. microspora NK17.
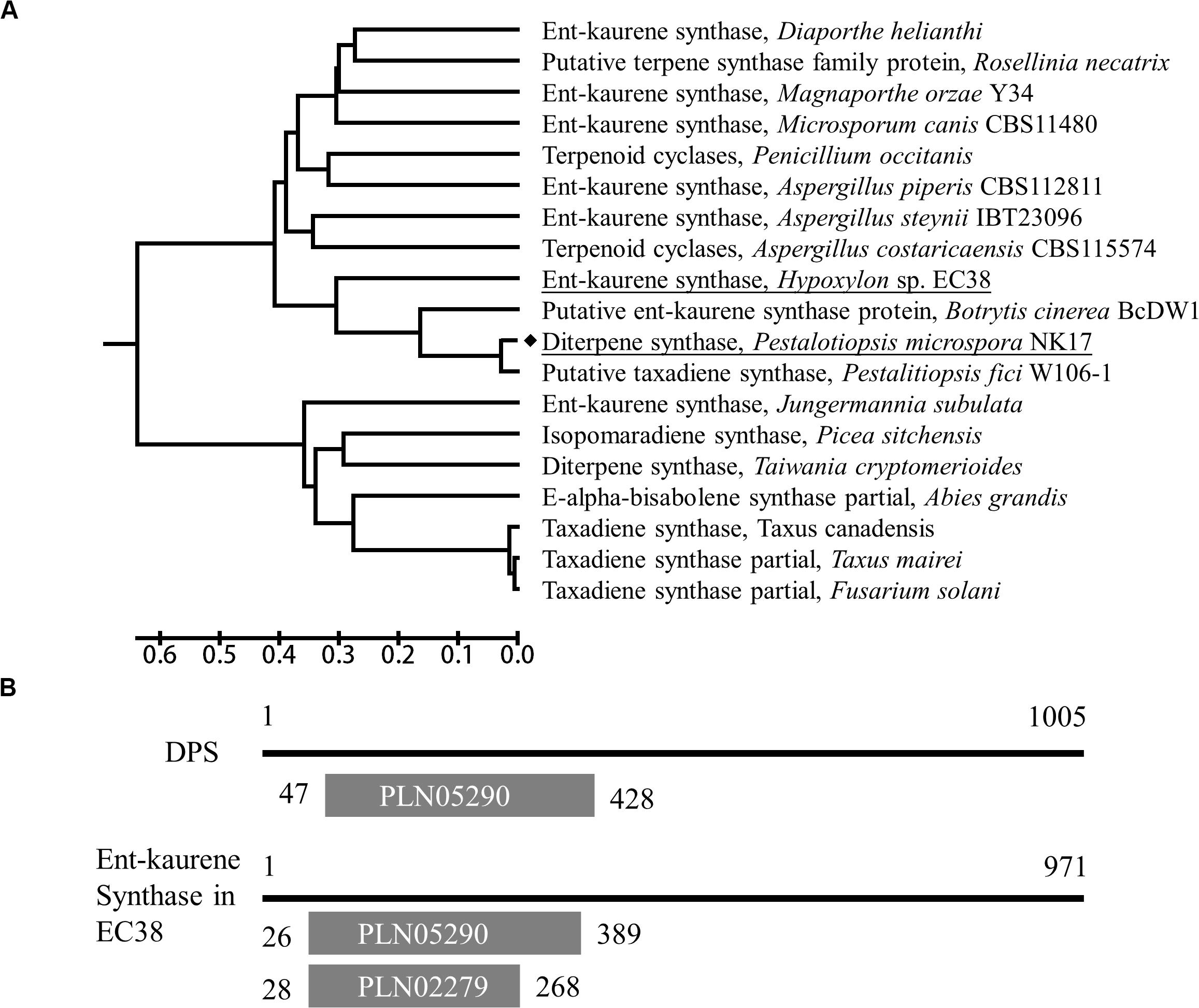
Figure 1. Characterization of the dps gene in NK17. (A) A UPGMA tree based on the full-length protein sequences of dps was built by MEGA7. The dps in NK17 is underlined. Diterpene synthases from several other fungi or taxadiene synthase from Taxus were used for comparison. (B) Protein domain analysis of dps compared to Ent-kaurene synthase of Hypoxylon sp. EC38. PLN05290, ent-copalyl diphosphate synthetase domain; PLN02279, Ent-kaur-16-ene synthase.
Materials and Methods
Strains and Plasmids
The fungus P. microspora NK17 was isolated by our laboratory and was maintained in potato lactose broth (PLB) at 28°C and 200 rpm or on PLA [PLB with 2% (w/w) agar] at 28°C. The orotidine 5’-phosphate decarboxylase-deficient strain ura3Δ was constructed by Chen et al. (2017).
Escherichia coli DB3.1 (Invitrogen, Carlsbad, CA, United States) was used to propagate the pOSCAR plasmid (Chen et al., 2017), while the other plasmids used in this study were generally propagated in E. coli DH5α. Agrobacterium tumefaciens LBA4404 was used for A. tumefaciens-mediated transformation (ATMT) of NK17 (Yu et al., 2015). All of the bacterial strains were cultured in Luria–Bertani medium with appropriate antibiotics at 37°C (for E. coli) or 28°C (for A. tumefaciens) and 200 rpm when needed. An inducing medium (IM) with 200 mg/ml uracil and 20 mg/ml acetosyringone (AS, Sigma, St. Louis, MO, United States) was used as the minimal medium for the selection of NK17 transformants.
Construction of the Deletion Vector pOSCAR-dpsΔ
The deletion vector of dps was constructed by the in-fusion method. A pOSCAR plasmid containing the ura3 gene was used as the marker (Chen et al., 2017). First, the 5’-flank and the 3’-flank were amplified from the NK17 genome via PCR and purified by a DNA Gel Extraction Kit (Axygen, Union City, CA, United States). Then, the plasmid was cut by the enzyme EcoRI, and an in-fusion reaction was performed to add the 5’-flank to pOSCAR, resulting in the intermediate plasmid pOSCAR-dpsΔ-up. The primer pair dpsΔ-up-F/ura3-R (Table 1) was used to verify the plasmid. This intermediate plasmid was next cut by enzyme HindIII, and the 3’-flank was added to the plasmid via an in-fusion reaction. The primer pair ura3-F/dpsΔ-down-R (Table 1) was applied for PCR to verify the final plasmid by the size of the bonds. Thus, the dps deletion vector pOSCAR-dpsΔ was constructed completely.
Construction of dps Deletion Strain dpsΔ
Ten micrograms of the pOSCAR-dpsΔ vector was transformed into A. tumefaciens LBA4404 by ATMT following the protocol described by Chen et al. (2017) and Wang et al. (2017). Then, A. tumefaciens LBA4404 transformants harboring pOSCAR-dpsΔ were co-cultured with 106 conidia of ura3Δ at 28°C on a nitrocellulose filter, which was spread on an IM minimal medium plate. After 2 days, the filter was transferred to a selection medium PLA plate supplemented with kanamycin (100 mg/ml). Another 2 days later, three transformants were transferred to a new PLA plate. Single-spore isolation was applied for every transformant after it had grown for 5 days. Then, the total DNA from each transformant was extracted from mycelium after growing in 100 ml of PLB for 4 days. The primer pair dpsΔ-up/ura3-R was used to verify the transformants. In addition, the strains were further verified by Southern blot following the protocol of Zhang et al. (2016).
RNA Preparation, Reverse-Transcription PCR, and qRT-PCR
TRIzol Kit was used to prepare total RNA from lyophilized mycelia after they had grown for 4 days. Then, reverse-transcription PCR (RT-PCR) was performed as described previously by Zhang et al. (2016).
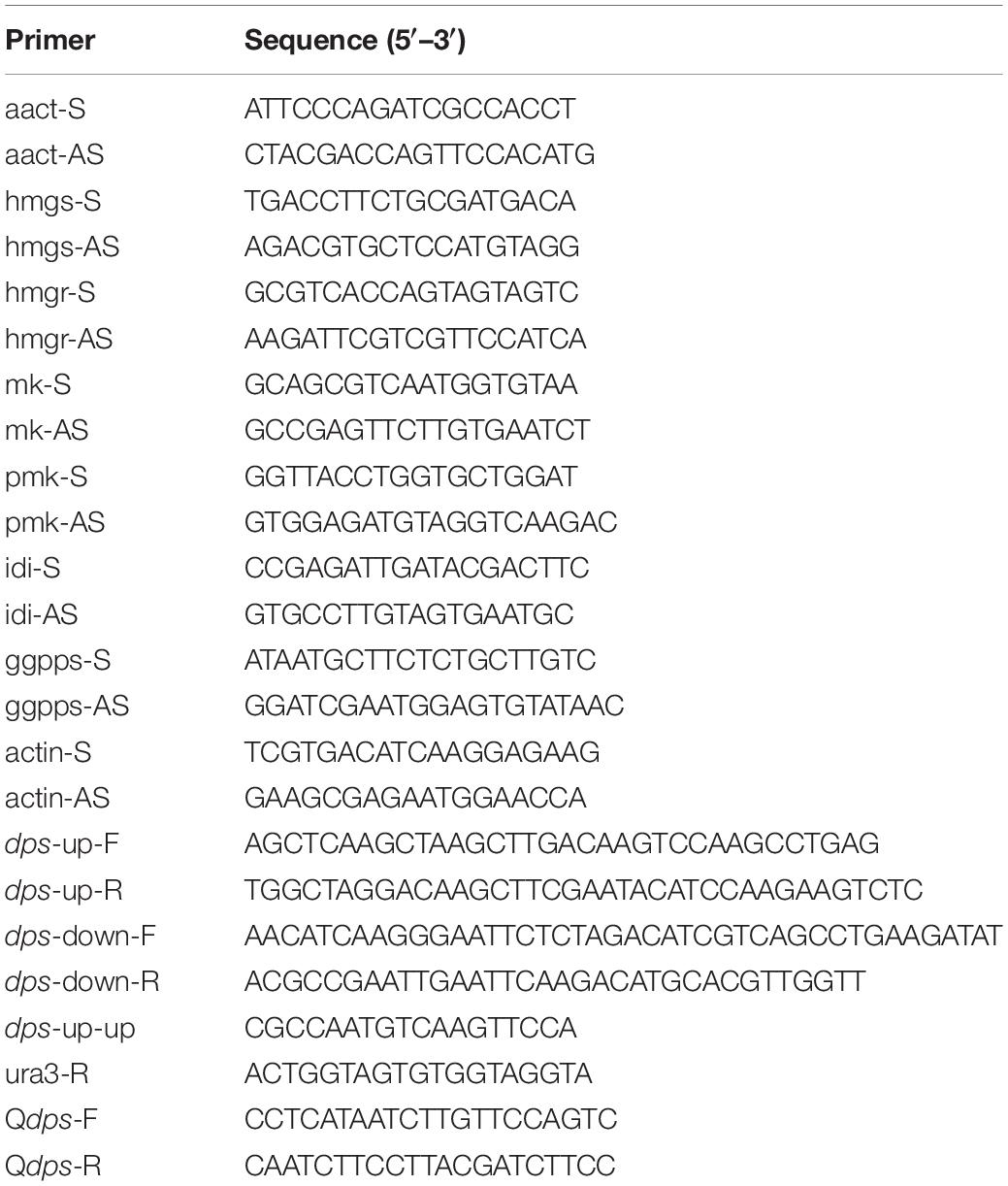
To examine the expression of dps in dpsΔ, two pairs of primers, actin-F/actin-R for the actin-encoding gene ACT and Qdps-F/Qdps-R for dps, were designed to amplify the cDNA of NK17 and dpsΔ by RT-PCR and qRT-PCR. The assay was also performed to examine the expression of MVA genes. The primers used in this assay are listed in Table 1.
Analysis of Secondary Metabolites
The particular metabolites, dibenzodioxocinones, and some other secondary metabolites of NK17 and dpsΔ were analyzed by high-performance liquid chromatography (HPLC) and gas chromatography–mass spectrometry (GC–MS), respectively. First, the two strains, NK17 and dpsΔ, were cultured in 100 ml of PLA medium for 7 days. Then, 1 ml of spore suspension was added to 500 g of sweet potato waste medium for solid fermentation. After 15 days, 500 ml of methyl alcohol was added into the fermentation medium to extract the metabolites. Next, the extracts were collected by filtration and concentrated to approximately 50 ml. Finally, the extracts of NK17 and dpsΔ were analyzed by HPLC or GC–MS. The HPLC analysis (Agilent 1100, Agilent Technologies, Santa Clara, CA, United States) was conducted at 40°C, using A Kromasil C18 ODS column (4.6 mm × 250 mm, AKZO Nobel, Gland, Switzerland). The mobile phase of HPLC was MeOH/H2O (70/30, v/v) at a flow rate of 1 ml/min, and 20 μl of the sample was injected and analyzed at 227 nm. In the GC–MS condition, 1 μl of sample was injected into GC–MS (Bruker 320), using a DB-3 MS (0.25 μm × 0.25 μm × 30 m) column at analyzing temperature of 250°C, scan range, 45–8,000 Da. In this experiment, the sweet potato waste medium without inoculation was analyzed as the negative control.
Drug and Stress Sensitivity Assay
To test the effect of dps on the growth and the drug sensitivity of NK17, 2 and 4 μg/ml of amphotericin (AmB) and 300 and 600 μg/ml of hydroxyurea (HU) were added to the PLA plates. In addition, 0.5 and 1 M NaCl were added to the PLA plate to test the osmotic sensitivity of these mutants. The temperature susceptibility of the dpsΔ mutant strains was tested at 15, 22, and 28°C. The wild-type NK17 was used as control. Each set of conditions was applied in triplicates, and the growth of the strains in each assay was recorded daily.
Results
Characterization of dps in NK17
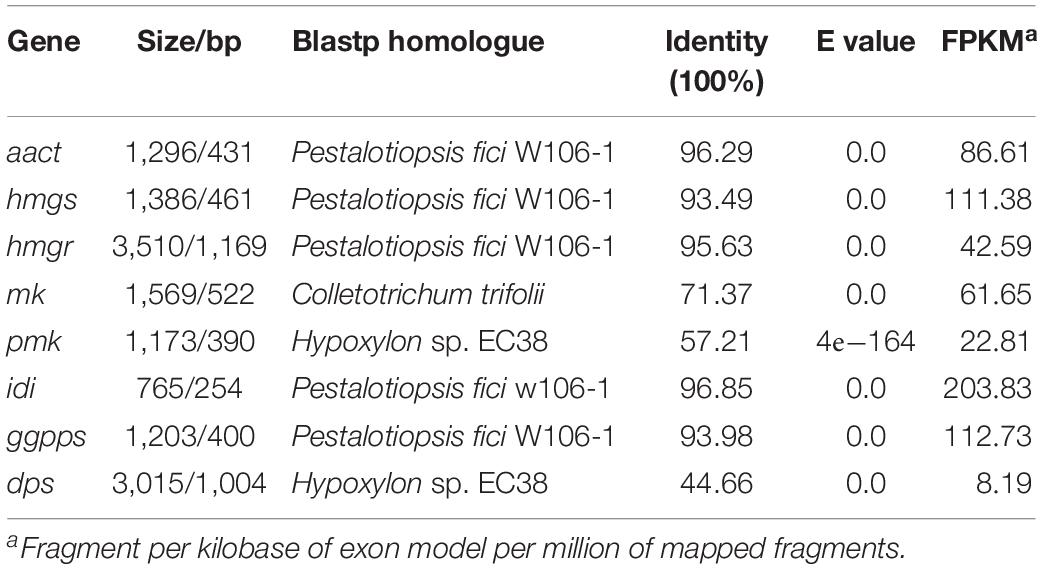
Via functional domain alignment, a diterpene synthase gene in NK17, which shared a remarkable similarity (44.66%) to ent-kaurene synthase in Hypoxylon sp. EC38 (Wu et al., 2017; Figure 1), was found and thus designated as dps (dps in P. microspora NK17). A phylogenetic tree was built by the software MEGA7 package based on the deduced protein sequence of dps and terpene synthase in other fungi (Figure 1A). There is a conserved ent-copalyl diphosphate synthase domain, PLN05290, in the protein (Figure 1B). The sequence similarity suggests that dps was potentially responsible for the biosynthesis of diterpenes in NK17.
To test the function of dps, we created a disruption of dps by homologous targeting via ATMT in NK17 ura3Δ using the selection marker ura3 complementation (Figure 2A). One of the mutation strains, named dpsΔ, was selected for Southern blotting. The 5’ flanking region was used as the probe to hybridize to a fragment of 988 bp (in wild type) and a fragment of 2,452 bp in dpsΔ (Figure 2B). The blotting confirmed the disruption of dps. The result of real-time PCR also showed the loss of expression of dps in dpsΔ (Figure 2C).
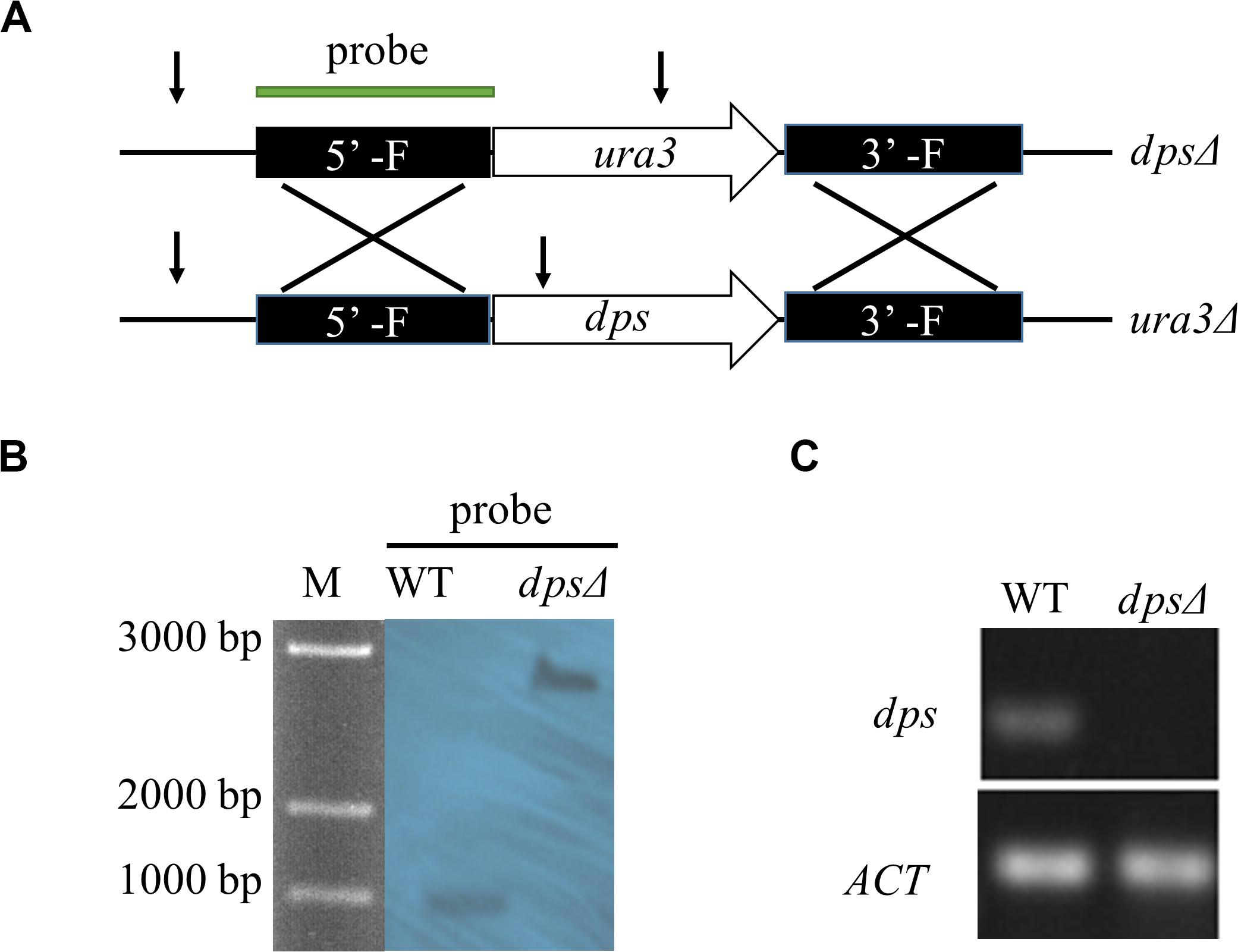
Figure 2. Construction of dps-deleted strain dpsΔ. (A) Schematic of the recipient strain ura3Δ and dps-deleted strain dpsΔ. In the dpsΔ strain, dps was replaced by a NK17 ura3 marker through homologous recombination. (B) Southern blotting to verify the mutations in dpsΔ. Genomic DNA from NK17 and dpsΔ were digested with BglII [cutting site is shown in panel (A) by arrows]. The probe used in Southern blotting is shown in panel (A). When the probe was used, one band of 988 bp was observed in wild-type NK17, and one band of 2,452 bp was observed in both dpsΔ. (C) Transcription of dps in wild type and dpsΔ was detected. No transcription of dps was identified in dpsΔ.
Roles of dps in the Biosynthesis of Secondary Metabolites
Diterpene synthase is a key enzyme in the biosynthesis of diterpenes (Koksal et al., 2011; Pelot et al., 2019). To investigate whether dps was required for the group of secondary metabolites, we first conducted HPLC profiling for the deletion strain dpsΔ. Extracts were prepared from sweet potato waste cultures statically grown for 13 days at 28°C. In HPLC profiling, we found that in dpsΔ strains the production of dibenzodioxocinones was reduced to 37% of the wild-type level (Figure 3A). In a previous study, we have reported that dibenzodioxocinones are the number of polyketides and a dominant group of secondary metabolites in NK17 (Figure 3B; Niu et al., 2015; Liu Y. et al., 2019). In this analysis, dibenzodioxocinone 1’,2’-epoxy-3’,4’-didehydropenicillide (Liu Y. et al., 2019) was monitored at a retention time of 17.6 min. In addition, the HPLC analysis found no new compounds after the deletion of the dps gene (Figure 3A).
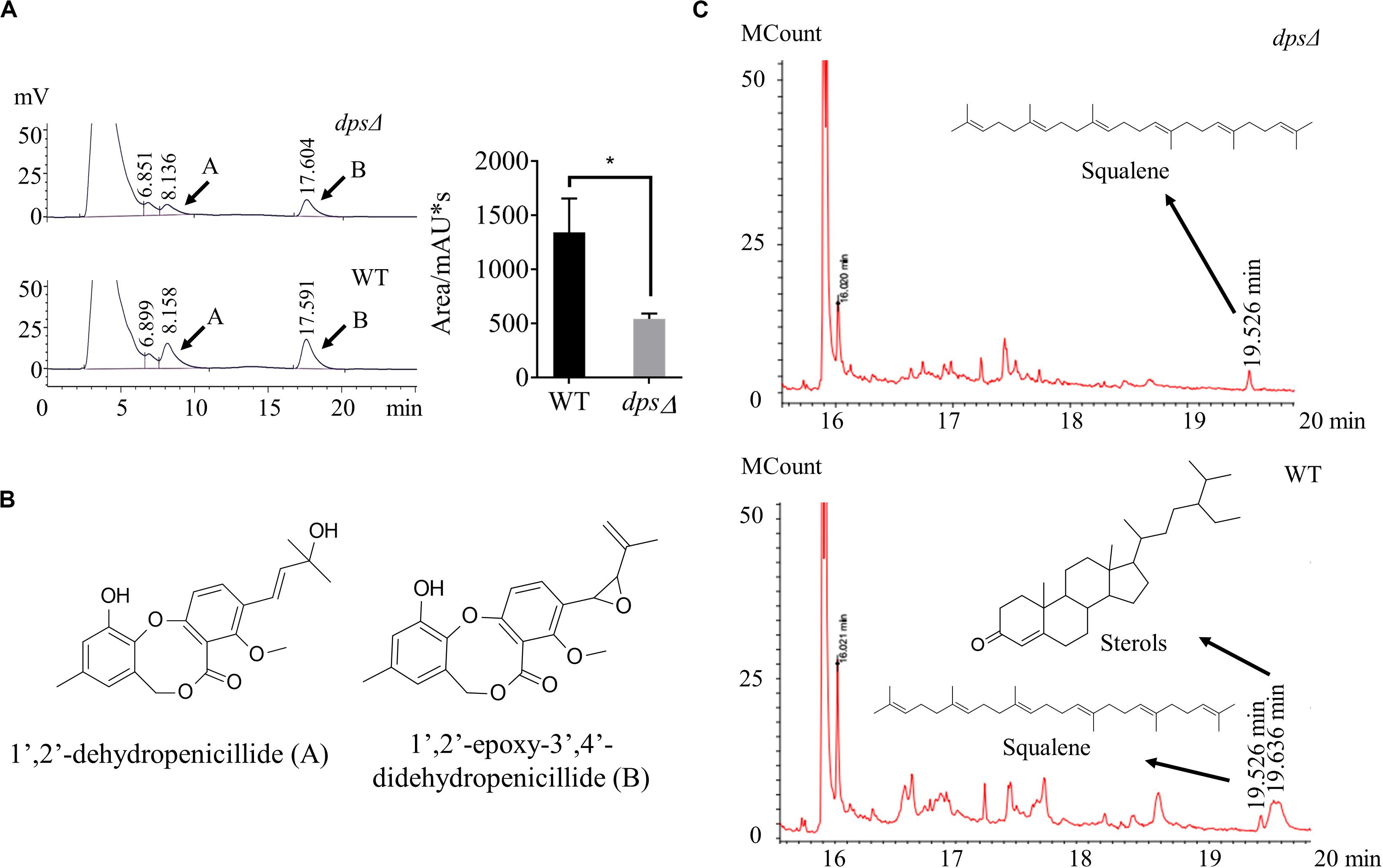
Figure 3. High-performance liquid chromatography (HPLC) and gas chromatography–mass spectrometry (GC–MS) analysis of the secondary metabolites in the mutant strains, dpsΔ and wild type. (A,B) HPLC profiling of the secondary metabolites. The samples were prepared from sweet potato waste fermentation extracts. Compounds (A,B) were two dibenzodioxocinones, the main metabolites of NK17, that had been previously identified (Liu Y. et al., 2019). Each condition was done in three independent assays. The peak area of compound (B) is shown in the right part of panel (A). *p < 0.05. (C) GC–MS profiling of the secondary metabolites in dpsΔ and NK17. The compound at the retention time of 19.526 min in both dpsΔ and NK17 was squalene. However, the peak of sterols at 19.636 min was present in NK17 and absent in dpsΔ.
To better understand the roles of dps in secondary metabolism, GC–MS was applied to compare the production of terpenes in the extracts of dpsΔ and the wild type. As a result, we found that the peak corresponding to sterols, at a retention time of 19.636 min in the total ion current, was absent in the metabolites of dpsΔ (Figure 3C). Sterols are triterpenoids that are usually cyclized by sterol synthase, such as lanosterol synthase and cycloartenol synthase (Nes, 2011), instead of diterpene synthase. Additionally, in the GC–MS spectrum, the peak corresponding to squalene, the key precursor of sterols (Nes, 2011), was observed at a retention time of 19.526 min in both dpsΔ and the wild type (Figure 3C and Supplementary Figure S1). These data suggest that dps participates in the biosynthesis of precursors of triterpenoids, e.g., sterols, in NK17 and we were unable to detect any new diterpenes in the metabolites of the wild-type NK17.
dps Is Required for the Growth of NK17 and Resistance to External Stress
As components in cellular membranes, sterols play a critical role in the physiology of eukaryotic cells, such as in oxidative stress and in membrane structure (Gold et al., 2016; Liu Z. et al., 2019). We observed, as discussed above, that dps could affect the biosynthesis of sterols in NK17 (Figure 3C). To test the role of dps in the growth of NK17, we further analyzed the colony diameters of dpsΔ and the wild type on the PLA plates at 28°C (Figure 4A) and lower temperature, i.e., 15 and 22°C (Figure 4B). The colony diameter of dpsΔ was nearly 50% smaller than that of the wild type (Figure 4A). These results clearly demonstrate that dps plays an important role in the growth of NK17.
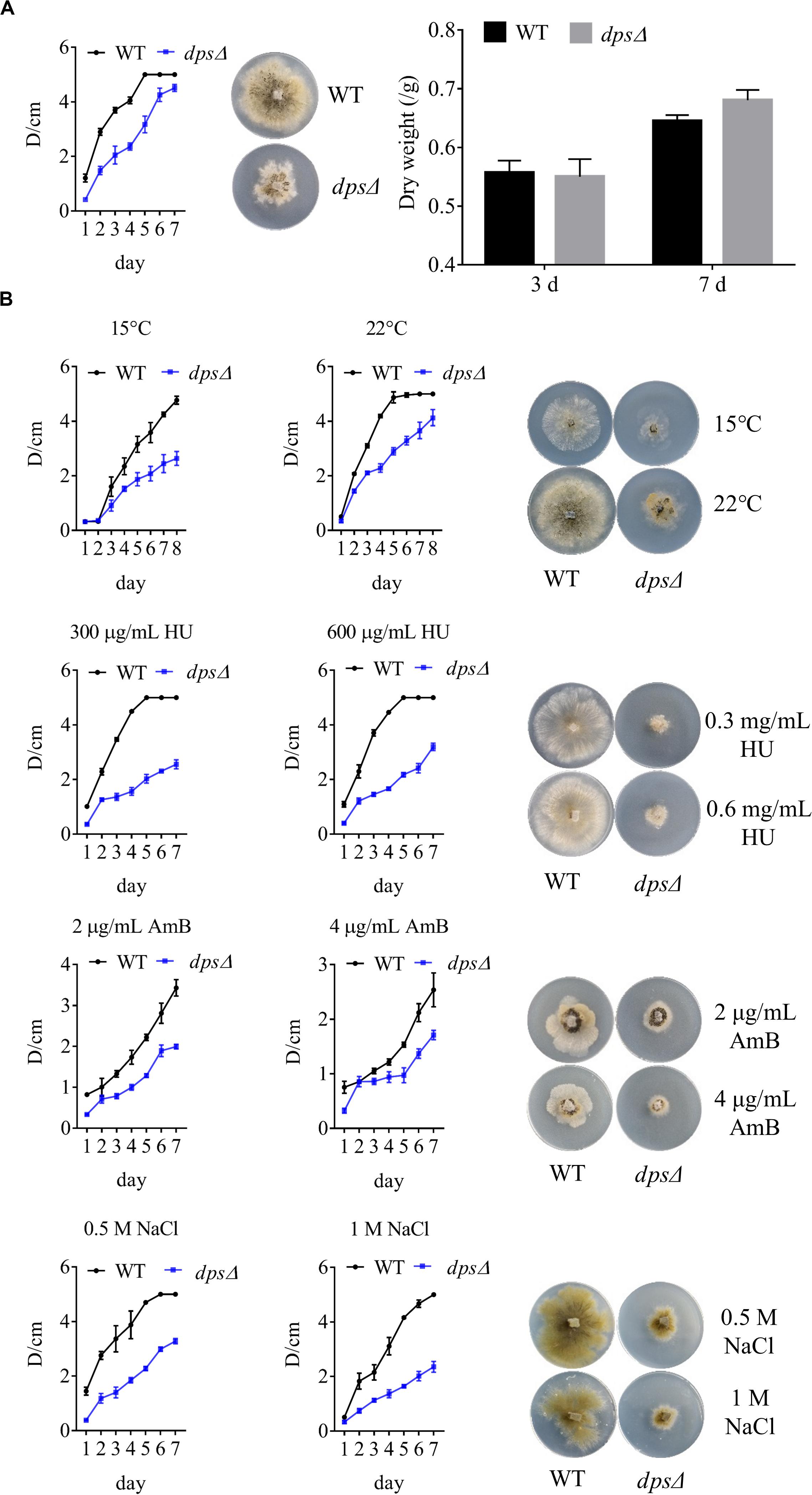
Figure 4. Phenotypic characterization and stress tests of NK17 and dpsΔ. (A) Growth curve and dry weight of mycelia of NK17 and dpsΔ at 28°C. (B) Stress tolerance to different concentrations of HU, NaCl, and AmB and to low temperature on PLB with 2% (w/w) agar (PLA). PLA without any supplement at 28°C (A) served as the control. The growth and the development of dpsΔ were greatly slowed, and the mutant was more sensitive to HU, NaCl, and AmB. In addition, dpsΔ rarely grew when 2 mg/ml AmB was added, although the growth of the wild type was also slightly slowed under these conditions. Each condition was done in three independent assays. The growing status in the plate on the fourth day is shown.
Furthermore, to observe the function of dps against external stress, different concentrations of sodium chloride (NaCl), AmB, and HU were added to the culture (Figure 4B). NaCl is an osmotic reagent that can regulate the osmotic pressure in the cell (Wang et al., 2017), and AmB and HU were added to detect resistance to antifungal fungicides. Treatment with 1 M NaCl or 600 μg/ml HU led to colonies with the diameters of dpsΔ only at 35% that of the wild type (Figure 4B). In addition, dpsΔ was more sensitive than the wild type to AmB and rarely grew in the presence of 2 μg/ml AmB (Figure 4B). The data demonstrated that the growth status of dpsΔ worsened under external stress, which may be due to a disruption of the sterol biosynthesis pathway (Kim and London, 2015; Gold et al., 2016).
Phenotypic Defects of dpsΔ Are Restorable by the Addition of Exogenous Sterols
Because deleting dps affected the synthesis of sterols, which may further inhibit the growth of NK17, we added three sterols, β-sitosterol, stigmasterol, and ergosterol, to a final concentration of 5 mg/L on the PLA plates. As expected, all the sterols restored the growth of dpsΔ (Figure 5A), declaring the direct relation between the biosynthesis of sterols and fungal growth. Besides that, we noticed that the colony diameter of dpsΔ was a little larger than that of the wild type after the addition of stigmasterol after culturing for 3 days (Figures 5A,B). Therefore, it was very likely that the exogenous stigmasterol generated an inhibitory effect in the wild-type NK17 strain.
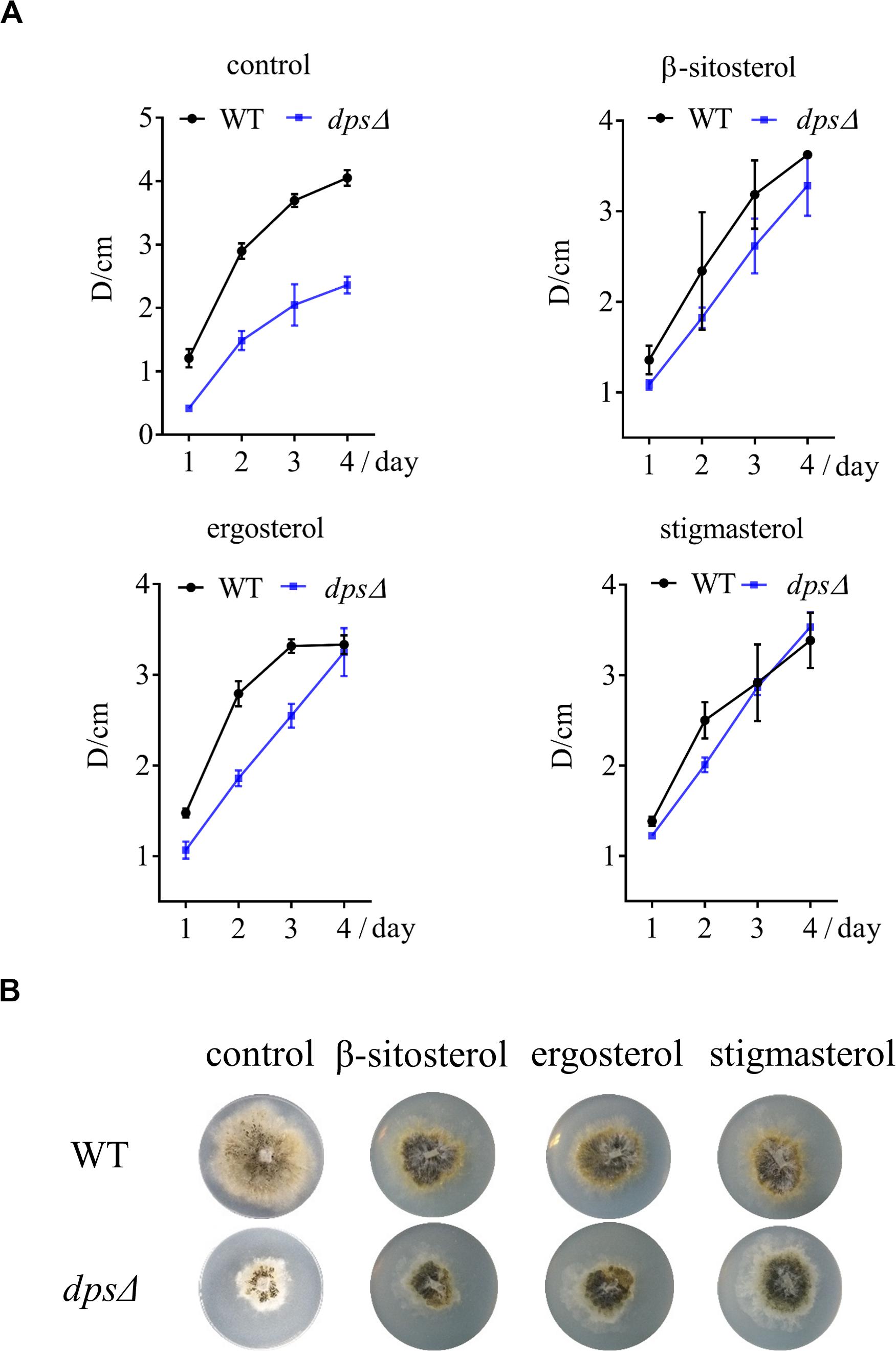
Figure 5. The growth of dpsΔ was recovered by the exogenously supplied sterols to the culture. (A) Growth curve of NK17 and dpsΔ stigmasterol, β-sitosterol, and ergosterol were added separately at 5 mg/L to recover the growth of dpsΔ. (B) After 4 days of culturing, the growth and the development of dpsΔ were recovered. In addition, the growth of NK17 was slightly slowed when sterol was added. Each condition was done in three independent assays.
Disruption of dps Affects the Expression of MVA Pathway Genes
A previous study demonstrated that the biosynthesis of sterols started with IPP and DMAPP, two C5 isoprene units synthesized by the MVA pathway (Engels et al., 2008; Cha et al., 2012; Bian et al., 2017). Total genome sequencing and mRNA sequencing of NK17 were performed to identify the MVA pathway genes in NK17 via protein sequence BLASTP on NCBI. All seven genes in the MVA pathway could be found and the transcriptome sequencing (mRNA-seq) of NK17 suggested that these genes (aact, hmgs, hmgr, mk, pmk, idi, and ggpps) in the MVA pathway could be constitutively expressed (Table 2). Real-time PCR amplification was set to confirm the RNA sequencing data (Figure 6). To detect whether disrupting dps could affect the expression of the MVA pathway, qRT-PCR was applied to analyze the expression of the seven genes in the pathway. Five genes of the seven, aact, hmgs, hmgr, mk, and idi, had a significant lower expression in dpsΔ compared to the wild type (Figure 6), and among them, hmgs (8.3% of the wild-type level in dpsΔ) and hmgr (4.9% in dpsΔ) had the largest decrease in expression (Figure 6). Interestingly, the expression of the other two genes, pmk and ggpps, turned out to be activated instead by more than 60 and 30%, respectively (Figure 6).
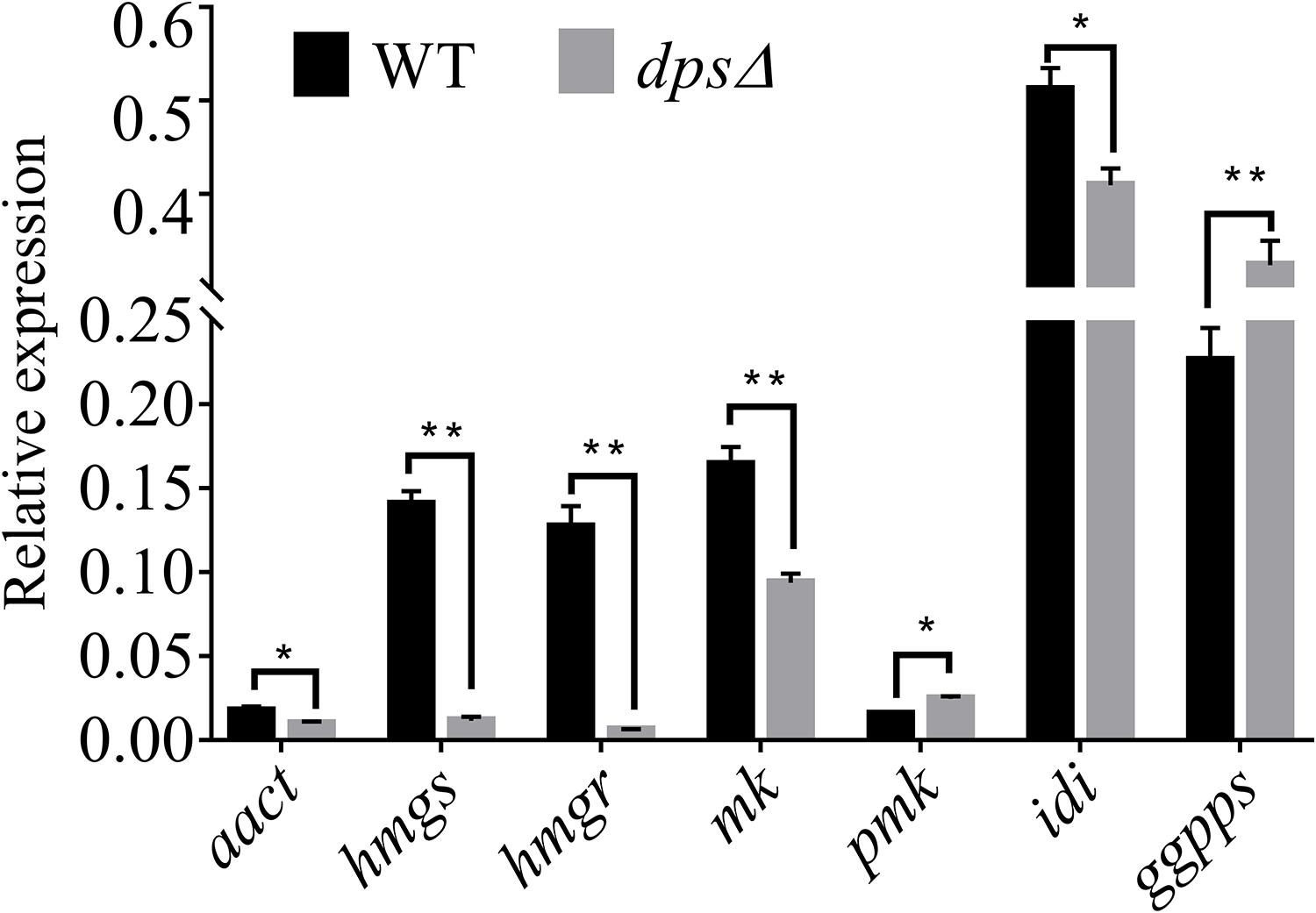
Figure 6. Expression of genes in the mevalonate (MVA) pathway in wild type and dpsΔ. The expression of the MVA pathway genes in NK17 and dpsΔ were detected by qRT-PCR. The values are expressed as a relative expression with respect to the gene for actin as an endogenous control. aact, acetyl-CoA C-acetyltransferase gene; hmgs, 3-hydroxy-3-methylglutaryl CoA synthase gene; hmgr, 3-hydroxy-3-methylglutaryl CoA reductase gene; mk, mevalonate kinase gene; pmk, phosphomevalonate kinase gene; idi, isopentenyl diphosphate isomerase gene; ggpps, geranylgeranyl diphosphate synthase gene. *p < 0.05, **p < 0.01.
Discussion
P. microspora is an endophytic fungus that is well known for producing a great number of secondary chemicals, including paclitaxel (Strobel et al., 1996). However, it seems that the fungus in our lab lost its capability to produce paclitaxel after extended subculture in the laboratory, although some terpenoids and their derivatives were identified from solid fermentation metabolites by GC–MS, i.e., squalene and sterols (Figure 3C). The biosynthesis of sterols and paclitaxel relied on the common precursors, IPP and DMAPP, the two C5 units (Dhingra and Cramer, 2017). By far, the best-studied biosynthesis pathways of these two C5 units were the methyl-D-erythritol 4-phosphate pathway and the MVA pathway (Nes, 2011). Isotope labeling method in plants and microorganisms demonstrated that both of these two routes existed in higher plants, while in fungi, the biosynthesis of IPP and DMAPP mainly depended on the MVA pathway (Lichtenthaler, 1999; Masse et al., 2004). In this report, we defined the upstream pathway that putatively leads to the biosynthesis of diterpenoids, i.e., the MVA pathway for terpenoid biosynthesis (Engels et al., 2008). As a result, all seven genes in this pathway were determined to be constitutively expressed (Figure 6). The identification of the sterols, which were biosynthesized from squalene, another polymer of IPP and DMAPP (Dhingra and Cramer, 2017), by GC–MS verified this conclusion.
The diterpene synthase in fungi has been well studied to increase the production of diterpenoids or to synthesize new diterpenoids via heterologous expression (Driller et al., 2019). However, there was no report about the function of diterpene synthase on the biosynthesis of triterpenes. By genome sequencing, only one copy of the gene encoding a diterpene synthase was found in NK17, which was initially speculated to be involved in the biosynthesis of paclitaxel. Via ATMT, the diterpene synthase, dps, was disrupted for the purpose of defining its function in NK17 in our study (Figure 2). Interestingly, the peaks of sterols in the GC–MS spectrum were absent in the deletion mutant (Figure 3C). A wide peak, a rather sharp peak, at 19.636 min in the GC–MS spectrum demonstrated that NK17 could produce more than one sterol. Additionally, many sterols could be matched with high scores of R. Match and F. Match by comparing the molecular fragments (Supplementary Figure S2). These findings, together with the changes of MVA pathway gene expression in dpsΔ (Figure 6) and the appearance of squalene in the GC–MS spectrum (Supplementary Figure S1), suggest that dps participates in the biosynthesis of sterols.
Sterols have been extensively studied because of their important roles in drug resistance, developmental regulation, cell signaling, and cell growth. Therefore, a large number of sterols isolated from bacteria, fungi, and plants have been reported and researched (Kim and London, 2015; Lamberson et al., 2017; Rivas-Marin et al., 2019; Ruan et al., 2019). Some sterols have even been implicated in human diseases, including Alzheimer’s disease and cancers (Liu Z. et al., 2019). The disruption of dps in NK17 resulted in increased sensitivity to high osmotic stress and antifungals and decreased growth (Figures 4A,B), which could be improved by the addition of sterols to the medium (Figure 5). In addition, we also noticed that the colony diameters of NK17 with exogenous sterols were smaller than those of the wild type without sterols (Figure 5), demonstrating that the oversupplied sterols in the media could significantly suppress the growth of NK17, in consistence with the result that sterols were particularly important for several cellular processes in fungi and would become a risk factor regardless of whether there was an excess or a shortage (Liu Z. et al., 2019). These data also reinforce the point that the dps of NK17 participated in the biosynthesis of sterols, although the synthesis of squalene was rarely affected (Liu Z. et al., 2019). In addition, the growth of NK17 was apparently slower when dps was knocked out (Figure 4A), which likely is the result from the impaired biosynthesis of sterol production (Gold et al., 2016; Liu Z. et al., 2019). However, when we measured the biomass of the wild type and dpsΔ in PLB, there was hardly a measurable difference between these two strains (Figure 4A). This result demonstrates the fact that there was a great difference between solid and liquid cultures of filamentous fungi in many aspects, for instance, the growth rate, gene expression, colonial morphology, and secondary metabolite biosynthesis (Bigelis et al., 2006). Thus, our data showed that DPS was only indispensable when the fungus was grown on solid plate such as PLA.
In conclusion, by disrupting the diterpene synthase (dps) in NK17, we revealed for the first time that this protein has two functions. One is that dps affects the biosynthesis of triterpenes, such as sterols, although the role, if any, of dps in the biosynthesis of paclitaxel is still uncertain. The other role of the enzyme is that dps is required for the growth and the development of fungi by affecting the biosynthesis of sterols.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XZ and YLiu conceived and designed the study. YLiu, AD, LC, DW, QX, BX, and YLin performed the experiment. LC and XH provided the plasmids. YLiu and XZ wrote the manuscript. QX and XZ reviewed and edited the manuscript. All the authors read and approved the manuscript.
Funding
This work is supported in part by grants from the National Science Foundation of China (#81871629) and the by the National High Technology Research and Development Program (#2012AA023405).
Conflict of Interest
LC was employed by Zhejiang Medicine Co., Ltd., Zhejiang, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Changqi Zhao and Mengxia Xie (Beijing Normal University) for helping us with the characterization of the secondary metabolites.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01426/full#supplementary-material
References
Austin, C. A., Shephard, E. A., Pike, S. F., Rabin, B. R., and Phillips, I. R. (1988). The effect of terpenoid compounds on cytochrome P-450 levels in rat liver. Biochem. Pharmacol. 37, 2223–2229. doi: 10.1016/0006-2952(88)90585-0
Bian, G., Yuan, Y., Tao, H., Shi, X., Zhong, X., Han, Y., et al. (2017). Production of taxadiene by engineering of mevalonate pathway in Escherichia coli and endophytic fungus Alternaria alternata TPF6. Biotechnol. J. 12:1600697. doi: 10.1002/biot.201600697
Bigelis, R., He, H., Yang, H. Y., Chang, L. P., and Greenstein, M. (2006). Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J. Ind. Microbiol. Biotechnol. 33, 815–826. doi: 10.1007/s10295-006-0126-z
Cha, M.-J., Shim, S.-H., Kim, S.-H., Kim, O.-T., Lee, S.-W., Kwon, S.-Y., et al. (2012). Production of taxadiene from cultured ginseng roots transformed with taxadiene synthase gene. BMB Rep. 45, 589–594. doi: 10.5483/BMBRep.2012.45.10.085
Chen, L., Li, Y., Zhang, Q., Wang, D., Akhberdi, O., Wei, D., et al. (2017). Improved pestalotiollide B production by deleting competing polyketide synthase genes in Pestalotiopsis microspora. J. Ind. Microbiol. Biotechnol. 44, 237–246. doi: 10.1007/s10295-016-1882-z
Chen, L., Wei, D., Qian, Z., Xi, Y., Yu, W., and Zhu, X. (2015). Orotidine 5’-phosphate decarboxylase-based reusable in situ genetic editing system: development and application in taxol-producing Pestalotiopsis microspora. Eng. Life Sci. 15, 542–549. doi: 10.1002/elsc.201400220
Dhingra, S., and Cramer, R. A. (2017). Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: opportunities for therapeutic development. Front. Microbiol. 8:92. doi: 10.3389/fmicb.2017.00092
Driller, R., Garbe, D., Mehlmer, N., Fuchs, M., Raz, K., Major, D. T., et al. (2019). Current understanding and biotechnological application of the bacterial diterpene synthase CotB2. Beilstein J. Org. Chem. 15, 2355–2368. doi: 10.3762/bjoc.15.228
Engels, B., Dahm, P., and Jennewein, S. (2008). Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 10, 201–206. doi: 10.1016/j.ymben.2008.03.001
Gauvin, A., Smadja, J., Aknin, M., Faure, R., and Gaydou, E. (2000). Isolation of bioactive 5α,8α-epidioxy sterols from the marine sponge L. Can. J. Chem. 78, 986–992. doi: 10.1139/v00-083
Gold, D. A., Grabenstatter, J., Mendoza, A. D., Riesgo, A., Ruiztrillo, I., and Summons, R. E. (2016). Sterol and genomic analyses validate the sponge biomarker hypothesis. PNAS 113, 2684–2689. doi: 10.1073/pnas.1512614113
Kim, J., and London, E. (2015). Using sterol substitution to probe the role of membrane domains in membrane functions. Lipids 50, 721–734. doi: 10.1007/s11745-015-4007-y
Kobayashi, J., and Shigemori, H. (2002). Bioactive taxoids from the Japanese yew Taxus cuspidata. Med. Res. Rev. 22, 305–328. doi: 10.1002/med.10005
Koksal, M., Jin, Y., Coates, R. M., Croteau, R., and Christianson, D. W. (2011). Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469, 116–120. doi: 10.1038/nature09628
Lamberson, C. R., Muchalski, H., McDuffee, K. B., Tallman, K. A., Xu, L., and Porter, N. A. (2017). Propagation rate constants for the peroxidation of sterols on the biosynthetic pathway to cholesterol. Chem. Phys. Lipids 207(Pt B), 51–58. doi: 10.1016/j.chemphyslip.2017.01.006
Lange, B. M. (2015). Biosynthesis and biotechnology of high-Value p-Menthane monoterpenes, including menthol, carvone, and limonene. Adv. Biochem. Eng. Biotechnol. 148, 319–353. doi: 10.1007/10_2014_289
Li, M., Jiang, F., Yu, X., and Miao, Z. (2015). Engineering isoprenoid biosynthesis in Artemisia annua L. for the production of taxadiene: a key intermediate of taxol. Biomed. Res. Int. 2015:504932. doi: 10.1155/2015/504932
Lichtenthaler, H. K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 47–65. doi: 10.1146/annurev.arplant.50.1.47
Liu, Y., Chen, L., Xie, Q., Yu, X., Duan, A., Lin, Y., et al. (2019). A gene cluster for the biosynthesis of dibenzodioxocinons in the endophyte Pestalotiopsis microspora, a Taxol Producer. J. Microbiol. Biotechnol. 29, 1570–1579. doi: 10.4014/jmb.1905.05051
Liu, Z., Jian, Y., Chen, Y., Kistler, H. C., He, P., Ma, Z., et al. (2019). A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum. Nat. Commun. 10:1228. doi: 10.1038/s41467-019-09145-6
Masse, G., Belt, S. T., Rowland, S. J., and Rohmer, M. (2004). Isoprenoid biosynthesis in the diatoms Rhizosolenia setigera (Brightwell) and Haslea ostrearia (Simonsen). Proc. Natl. Acad. Sci. U.S.A. 101, 4413–4418. doi: 10.1073/pnas.0400902101
Nes, W. D. (2011). Biosynthesis of cholesterol and other sterols. Chem. Rev. 111, 6423–6451. doi: 10.1021/cr200021m
Niu, X., Hao, X., Hong, Z., Chen, L., Yu, X., and Zhu, X. (2015). A putative histone deacetylase modulates the biosynthesis of Pestalotiollide B and conidiation in Pestalotiopsis microspora. J. Microbiol. Biotechnol. 25, 579–588. doi: 10.4014/jmb.1409.09067
Pelot, K. A., Hagelthorn, D. M., Hong, Y. J., Tantillo, D. J., and Zerbe, P. (2019). Diterpene synthase-catalyzed biosynthesis of distinct clerodane stereoisomers. Chembiochem 20, 111–117. doi: 10.1002/cbic.201800580
Ravichandran, C., Badgujar, P. C., Gundev, P., and Upadhyay, A. (2018). Review of toxicological assessment of d-limonene, a food and cosmetics additive. Food Chem. Toxicol. 120, 668–680. doi: 10.1016/j.fct.2018.07.052
Rivas-Marin, E., Stettner, S., Gottshall, E. Y., Santana-Molina, C., Helling, M., Basile, F., et al. (2019). Essentiality of sterol synthesis genes in the planctomycete bacterium Gemmata obscuriglobus. Nat. Commun. 10:2916. doi: 10.1038/s41467-019-10983-7
Ruan, R., Chen, Y., Li, H., and Wang, M. (2019). Functional diversification of sterol regulatory element binding proteins following gene duplication in a fungal species. Fungal Genet. Biol. 131:103239. doi: 10.1016/j.fgb.2019.103239
Strobel, G., Yang, X., Sears, J., Kramer, R., Sidhu, R. S., and Hess, W. M. (1996). Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallachiana. Microbiology 142(Pt 2), 435–440. doi: 10.1099/13500872-142-2-5
Wang, C., Park, J. E., Choi, E. S., and Kim, S. W. (2016). Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway - application of PgpB and YbjG phosphatases. Biotechnol. J. 11, 1291–1297. doi: 10.1002/biot.201600250
Wang, D., Akhberdi, O., Hao, X., Yu, X., Chen, L., Liu, Y., et al. (2017). Amino acid sensor kinase Gcn2 is required for conidiation, secondary metabolism, and cell wall integrity in the Taxol-producer Pestalotiopsis microspora. Front. Microbiol. 8:1879. doi: 10.3389/fmicb.2017.01879
Wu, W., Davis, R. W., Tran-Gyamfi, M. B., Kuo, A., LaButti, K., Mihaltcheva, S., et al. (2017). Characterization of four endophytic fungi as potential consolidated bioprocessing hosts for conversion of lignocellulose into advanced biofuels. Appl. Microbiol. Biotechnol. 101, 2603–2618. doi: 10.1007/s00253-017-8091-1
Yu, X., Wang, Y., Pan, J., Wei, D., and Zhu, X. (2015). High frequency of homologous gene disruption by single-stranded DNA in the taxol-producing fungus Pestalotiopsis microspora. Ann. Microbiol. 65, 2151–2160. doi: 10.1007/s13213-015-1055-8
Zhang, Q., Chen, L., Yu, X., Liu, H., Akhberdi, O., Pan, J., et al. (2016). A β-type histone acetyltransferase Hat1 regulates secondary metabolism, conidiation, and cell wall integrity in the taxol-producing fungus Pestalotiopsis microspora. J. Basic. Microbiol. 56, 1380–1391. doi: 10.1002/jobm.201600131
Keywords: Pestalotiopsis microspora, diterpene synthase, MVA genes, sterol biosynthesis, polyketide
Citation: Liu Y, Duan A, Chen L, Wang D, Xie Q, Xiang B, Lin Y, Hao X and Zhu X (2020) A Fungal Diterpene Synthase Is Responsible for Sterol Biosynthesis for Growth. Front. Microbiol. 11:1426. doi: 10.3389/fmicb.2020.01426
Received: 23 February 2020; Accepted: 02 June 2020;
Published: 10 July 2020.
Edited by:
Anindya Chanda, University of South Carolina, United StatesReviewed by:
Christoph Müller, Ludwig Maximilian University of Munich, GermanyAshutosh Singh, University of Lucknow, India
Copyright © 2020 Liu, Duan, Chen, Wang, Xie, Xiang, Lin, Hao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xudong Zhu, emh1MTExODdAYm51LmVkdS5jbg==
 Yanjie Liu
Yanjie Liu Anqing Duan
Anqing Duan Longfei Chen2,3
Longfei Chen2,3 Dan Wang
Dan Wang Biyun Xiang
Biyun Xiang Xiaoran Hao
Xiaoran Hao Xudong Zhu
Xudong Zhu