- State Key Laboratory of Marine Geology, Tongji University, Shanghai, China
Methane seeps are widespread seafloor ecosystems shaped by complex physicochemical-biological interactions over geological timescales, and seep microbiomes play a vital role in global biogeochemical cycling of key elements on Earth. However, the mechanisms underlying the coexistence of methane-cycling microbial communities remain largely elusive. Here, high-resolution sediment incubation experiments revealed a cryptic methane cycle in the South China Sea (SCS) methane seep ecosystem, showing the coexistence of sulfate (SO42–)- or iron (Fe)-dependent anaerobic oxidation of methane (AOM) and methylotrophic methanogenesis. This previously unrecognized methane cycling is not discernible from geochemical profiles due to high net methane consumption. High-throughput sequencing and Catalyzed Reporter Deposition-Fluorescence in situ Hybridization (CARD-FISH) results suggested that anaerobic methane-oxidizing archaea (ANME)-2 and -3 coupled to sulfate-reducing bacteria (SRB) carried out SO42–-AOM, and alternative ANME-2 and -3 solely or coupled to iron-reducing bacteria (IRB) might participate in Fe-AOM in sulfate-depleted environments. This finding suggested that ANME could alter AOM metabolic pathways according to geochemical changes. Furthermore, the majority of methylotrophic methanogens belonged to Methanimicrococcus, and hydrogenotrophic and acetoclastic methanogens were likely inhibited by sulfate or iron respiration. Fe-AOM and methylotrophic methanogenesis are overlooked potential sources and sinks of methane in methane seep ecosystems, thus influencing methane budgets and even the global carbon budget in the ocean.
Introduction
Methane seeps are methane-dependent chemosynthetic ecosystems (Paull et al., 1984) that occur widely in the marine environment, and are considered some of the richest benthic ecosystems on the seabed (Valentine, 2011). The microbially mediated methane cycle dominates methane seeps (Boetius et al., 2000; Knittel and Boetius, 2009) and has an important impact on the global carbon cycle (Boetius and Wenzhöfer, 2013; Offre et al., 2013). Methane is produced from the degradation of organic matter by methanogens in deep sediment (Reeburgh, 2007). The produced methane continuously seeps or erupts from the sedimentary subsurface to the seabed in the form of methane-rich fluids, and more than 90% of the methane is consumed by anaerobic methane-oxidizing archaea (Valentine, 2011), forming an efficient methane biofilter that prevents its diffusion into the seawater.
Previous studies believed that in the ocean, the majority of methane is oxidized anaerobically by anaerobic methane-oxidizing archaea (ANME) coupled with sulfate reduction (Boetius et al., 2000). However, various chemical compounds are thermodynamically more favorable electron acceptors than sulfate for catalyzing anaerobic methane oxidation, such as nitrite (Raghoebarsing et al., 2006), nitrate (Haroon et al., 2013), ferric iron, and manganese (Ettwig et al., 2016; Cai et al., 2018). For instance, biogeochemical profiling evidences indicated the widespread presence of Fe-anaerobic oxidation of methane (AOM) in the ocean, such as in Argentine Basin (Riedinger et al., 2014), Alaskan Beaufort Sea (Treude et al., 2014), North Sea Helgoland mud (Oni et al., 2015), Baltic Sea (Egger et al., 2017), and Mediterranean Sea (Vigderovich et al., 2019). Targeted enrichment with ferrihydrite provides strong evidence for AOM coupled with iron reduction, and ANME-1, Methanococcoides/ANME-3 (Beal et al., 2009), ANME-2a and -2c (Scheller et al., 2016), ANME-2d (Methanoperedens nitroreducens) (Ettwig et al., 2016; Shen et al., 2019), Candidatus Methanoperedens ferrireducens (Cai et al., 2018), and Methanosarcina acetivorans (Yan et al., 2018) might be involved in Fe-AOM. However, the Fe-AOM microorganism physiology and its contribution to the methane consumption remain poorly understood in deep sea (Scheller et al., 2016; He et al., 2018). It is still possible that other unknown microorganisms can perform metal-AOM. For instance, Bar-Or et al. (2017) highlighted the essential role of methanogens and methanotrophic bacteria in the process of Fe-AOM.
In addition, in methane seep ecosystems, acetoclastic and hydrogenotrophic methanogenesis are often considered the dominant methanogenic processes below the Sulfate-methane transition zone (SMTZ) (Zhe et al., 2018). Thermodynamic laws indicate that sulfate-reducing bacteria (SRB) and iron-reducing bacteria (IRB) outcompete methanogens for hydrogen and acetate substrates, thus inhibiting the acetoclastic/hydrogenotrophic methanogenesis pathways (Reeburgh, 2007; Reiche et al., 2010; Zhou et al., 2014; Zhuang et al., 2018). However, the co-occurrence of sulfate reduction, AOM, and methanogenesis has been demonstrated in marine sediments (Sivan et al., 2014). This is also partly supported by the coexistence of ANME and methanogenic archaea in Sonora Margin shallow sediments (0–20 cm) (Vigneron et al., 2015), Peruvian Margin sulfate-reducing zone (0–25 cm) (Maltby et al., 2016), Mediterranean Sea shallow sediment (0–20 cm) (Sela-Adler et al., 2017), and Aarhus Bay surface sediment (Xiao et al., 2017, 2018).
Then, it is becoming clear that methylotrophic methanogenesis plays a major role in marine sediments, especially surficial sediments (Zhuang et al., 2016, 2018; Xiao et al., 2017). Methylotrophic methanogenesis using non-competitive substrates such as (methanol, methylamines, or methyl sulfide and so on) could co-occur with AOM and sulfate reduction (Vigneron et al., 2015; Zhuang et al., 2016). In the presence of sulfate and iron oxides, methanogens may circumvent competition by utilizing ‘non-competitive’ methylated substrates. These non-competitive substrates are ubiquitous in the marine environment, and originate from the degradation of substances such as betaine, choline, lignins, pectin, and creatine, or from the bacterial reduction of trimethylamine oxide (Oremland et al., 1982). Hence, methylotrophic methanogenesis has been suggested to occur in all major oceans (Valentine, 2011; Chronopoulou et al., 2017). However, in methane seep ecosystems, this process is not easily discernible from geochemical profiles due to the overall high net methane consumption in methane seeps. Thus, research on methylotrophic methanogenesis in methane seep is relatively rare, and the contribution of methylated substrates to methanogenesis processes remains elusive (Chronopoulou et al., 2017).
The northern continental slope of the South China Sea (SCS) has various sites of methane seepage, covering a wide range of water depths (200–3000 m). The Jiaolong and Haima methane seeps are the two still-active seep sites in SCS. Previous studies on the seeps in SCS have led to many significant advancements including new insight into the methane seep structure (Liu et al., 2018), mineral carbon/oxygen/sulfur isotopes (Feng et al., 2018), macrobenthos (Dong and Li, 2015; Gong et al., 2015), and microbial lipid biomarkers (Guan et al., 2016, 2018). In addition, the microbial distribution and diversity in SCS methane seeps were also investigated (Jiang et al., 2007; Zhang et al., 2012), and a 16S rRNA gene-based survey indicated the presence of SRB and ANME. However, the biogeochemical evidences and potential activity of methane-cycling microorganisms and their niche differentiation patterns are largely unknown.
In this study, the Jiaolong methane seep was chosen as our research object. In 2018, a remotely operated underwater vehicle, The Remotely Operated Platform for Ocean Science (ROPOS), found a new methane seepage site in this area. Combining high-throughput sequencing, CARD-FISH, and enrichment culture methods with pore water biogeochemistry, we investigated the microbially driven metabolic processes of methane production and consumption. The coexistence of sulfate- or iron-dependent AOM and methylotrophic methanogenesis was found, and the potential rates of these processes were assessed. These results are of great importance to the understanding of the biogeochemical processes in global methane seep ecosystems.
Materials and Methods
Collection of Sediment Samples and Geochemical Analysis
During the R/V Tan Kah Kee 1083 expedition (April–May 2018) in the northern SCS, a remotely operated underwater vehicle (ROPOS, Canadian Scientific Submersible Facility) was used to search for new undisturbed active methane seeps at a water depth of 900–1,200 m. A new active methane seep was found and named Jiaolong F3 site (position 22°6.9678′N, 119°17.0841′E, 1162.53 m). Two push cores with a length of 32 cm were retrieved beneath the black microbial mats by ROPOS (Figure 1). One push core was used for shipboard biogeochemical analyses, and the other push core was stored at 4°C until it was taken back to the laboratory for enrichment experiments.
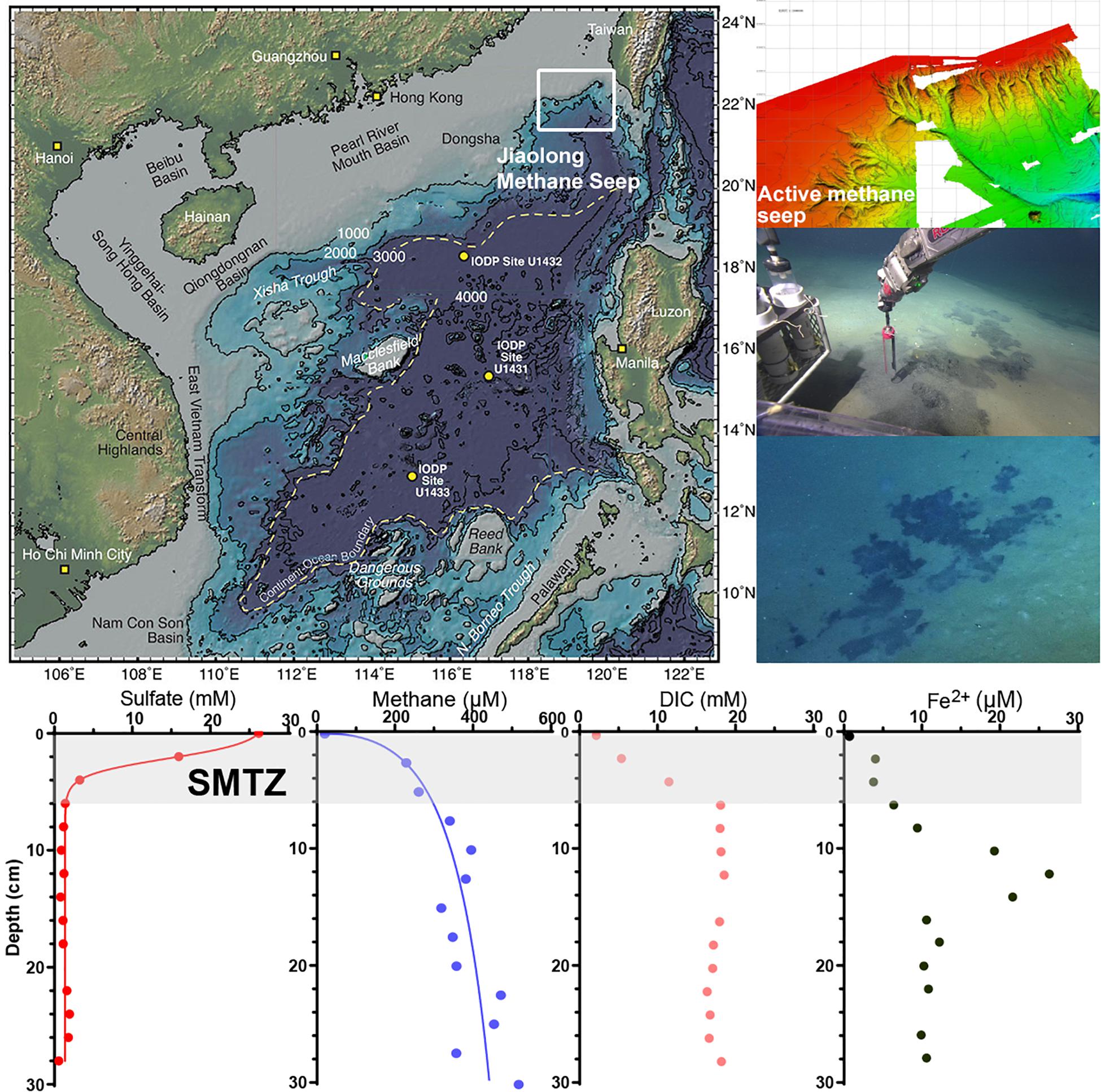
Figure 1. The location of Jiaolong active methane seep and geochemistry profiles. The map is modified from Expedition 349 Scientists (2014).
Before sampling, holes were drilled at both sides of the biogeochemistry push cores to collect porewater and methane gas. Holes in one side were drilled at 2 cm intervals, in the other at 2.5 cm, and sealed with diffusion-tight tape. Porewater was extracted using Rhizons (Rhizosphere Research Products, Wageningen, Netherlands) at 2 cm intervals (Seeberg-Elverfeldt et al., 2005). Cut-off syringes were used to sample sediment at a depth interval of 2.5 cm for methane analysis. The push core was then sectioned into 2 cm thick slices and frozen at −80°C for nucleic acid extraction and CARD-FISH analysis. The collected pore water was divided into aliquots for analysis of sulfate, dissolved inorganic carbon (DIC), and dissolved iron (Fe2+).
The pore water sample used for analysis of DIC was quickly transferred into glass vials (2 ml), leaving no headspace for concentration measurements and stored at 4°C. In the laboratory, pore water was measured using a CN analyzer multi N\C 3100 (Analytik Jena AG, Jena, Germany) (Oda et al., 2019). For analysis of the sulfate concentration, 1 ml of pore water was transferred into a vial containing 0.5 ml of ZnAc (5%), which was shaken well and stored at −20°C. In the laboratory, sulfate was measured using a Dionex ICS-1500 Ion Chromatograph and an IonPac AS23 column (eluent: 4.5 mM Na2CO3/0.8 mM NaHCO3, flow: 1 ml/min) (Thermo Fisher Scientific, Sunnyvale, CA, United States). For measurement of dissolved iron, 1 ml of pore water was acidified with ultrapure HCl and assessed using a 797 VA Computrace as previously described (Metrohm, Herisau, Switzerland).
For ex situ measurement of the methane concentration, 3 ml of wet sediment sample was removed from the push core at an interval of 2.5 cm with a cut-off syringe and extruded into a vial (20 ml) containing 6 ml of sodium hydroxide (2.5% w/w). The vial was closed immediately with a butyl rubber stopper, sealed with an aluminum crimp, and stored upside down at 4°C until measurement by gas chromatography (Treude et al., 2005). Before measurement, the sealed bottle was vigorously shaken and headspace methane was determined using gas chromatography-flame ionization detector (Agilent, Santa Clara, CA, United States).
High-Throughput Sequencing
Environmental DNA was extracted using a modified SDS-based extraction method (Zhou et al., 1996). The final DNA concentration was determined by a NanoDrop 2000 (Thermo Scientific, Waltham, MA, United States), and DNA quality was checked by 1% agarose gel electrophoresis. The sequencing steps were conducted by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The bacterial primers were 338F and 806R (Xu et al., 2016), targeting the 16S rDNA V3-V4 region. The archaeal primers were 524F10extF and Arch958R modR (Liu C. et al., 2016), targeting the 16S rDNA V4-V5 region. The PCR protocol was performed according to previously described methods (Liu C. et al., 2016; Xu et al., 2016). The PCR products were extracted from a 2% agarose gel and further purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, United States) and quantified using Quanti FluorTM-ST (Promega, United States). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CA, United States). Raw fastq files were demultiplexed, quality filtered by Trimmomatic, and merged by FLASH. Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version7.1), and chimeric sequences were identified and removed using UCHIME. Taxonomy assignment was performed using the SILVA 16S rRNA database (version 132). The distance-based maximum likelihood was used for phylogenetic analysis. Bootstrap analysis was performed using 1000 replications. Chao1 and Shannon–Weaver diversity indices and rarefaction curves were calculated by MOTHUR. Principal coordinates analysis (PCoA) was computed using PAST. Sequencing data are stored on the NCBI Sequence Read Archive (PRJNA574743, PRJNA574745).
Catalyzed Reporter Deposition-Fluorescence in situ Hybridization (CARD-FISH)
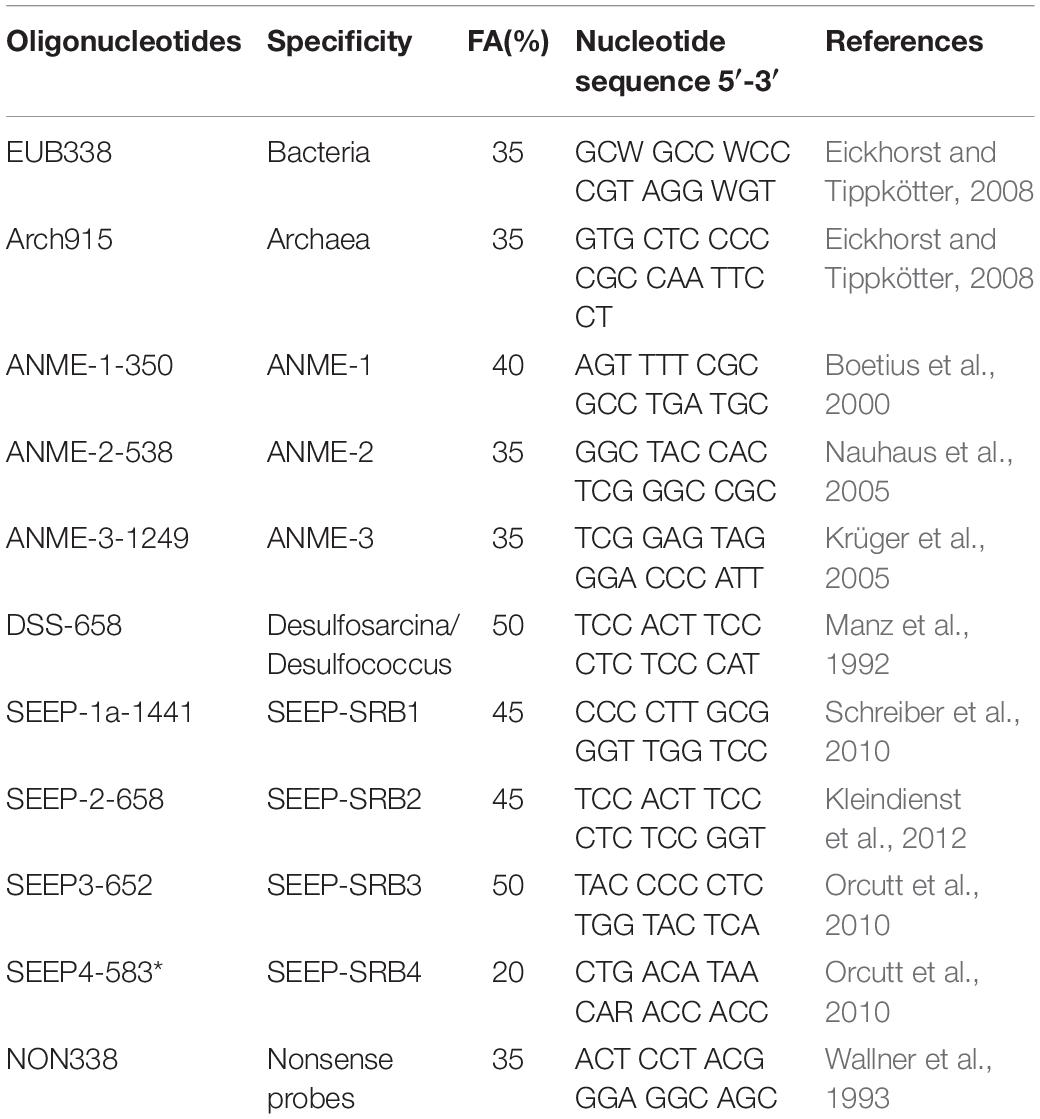
The CARD-FISH protocol was based on a previous study (Pernthaler et al., 2002). The probes-label peroxidase is shown in Table 1. Sediment (0.5 g) was fixed with 4% paraformaldehyde for 24 h at room temperature. The fixed sediments were washed three times by centrifugation (8,000 × g for 10 min) using PBS at 4°C and stored in ethanol/PBS buffer (1:1) at −20°C for further processing. After that, 100 μl of fixed sediments were diluted with 900 μl ethanol/PBS buffer (1:1) and dispersed using ultrasound. Then, 20 μl of dispersed sediments were diluted in 20 ml of Milli Q filtered water. The suspension sediments were filtered on polycarbonate filters, and 0.1% low melting point agarose was dripped onto the filters and dried at 46°C in an incubator. The microbes were permeabilized using 15 μg/ml proteinase K. Then, 3% H2O2 was used to inactivate the endogenous peroxidases.
For hybridization, filters were placed in a tube and mixed with 500 μl hybridization solution [10% dextran sulfate, 2% blocking reagent (Roche, Germany), 0.1% (w/v) sodium dodecyl sulfate, 20 mM Tris–HCl [pH 8.0], 0.9 M NaCl and formamide], and 1 μl of probe working solution (final concentration, 0.028 μM) (Eickhorst and Tippkötter, 2008). Microorganisms were hybridized for at least 60 min on a rotor at 46°C; then, the filters were washed twice using washing solution (Eickhorst and Tippkötter, 2008) (0.01% SDS, 5 mM EDTA [pH 8.0], 20 mM Tris–HCl [pH 8.0] and 3 mM NaCl) at 48°C for 20 min. After washing, filters were mixed with 1000 μl of amplification solution (0.0015% H2O2, 1 × PBS [pH7.4], 0.1% (w/v) blocking reagent) and 1 μl of Alexa488 labeled tyramides (Life TechnologiesTM, Thermo Fisher, United States). The probes were incubated at 46°C in amplification solution for at least 30 min in the dark.
For second hybridizations, the first probe-label peroxidase was inactivated by incubating the filter sections in 0.01 M HCl for 10 min at room temperature and washing the sections with 50 ml of Milli Q water. Then, the CARD-FISH protocol was repeated two times with the same filter sections by using different probes. The second hybridization used Alexa 647 labeled tyramides (Life TechnologiesTM, Thermo Fisher, United States).
Finally, all microorganisms were stained using 4′,6-diamidino-2-phenylindole (DAPI) and mounted with ProLong Gold Antifade reagent (Life Technologies, Carlsbad, CA, United States). Cell counting was performed using ImageJ.
Incubation Experiments for Methane Metabolic Activity
The push core (30 cm length) stored at 4°C was processed for microbial AOM and methanogenesis activity. We determined which substrates were available for AOM and methanogenesis in the Jiaolong methane seep. Push cores were positioned vertically, and the sediment was pushed out of the liner using a plastic plunger. From the center of the push core, samples were taken and incubated for determination of AOM and methanogenesis activity.
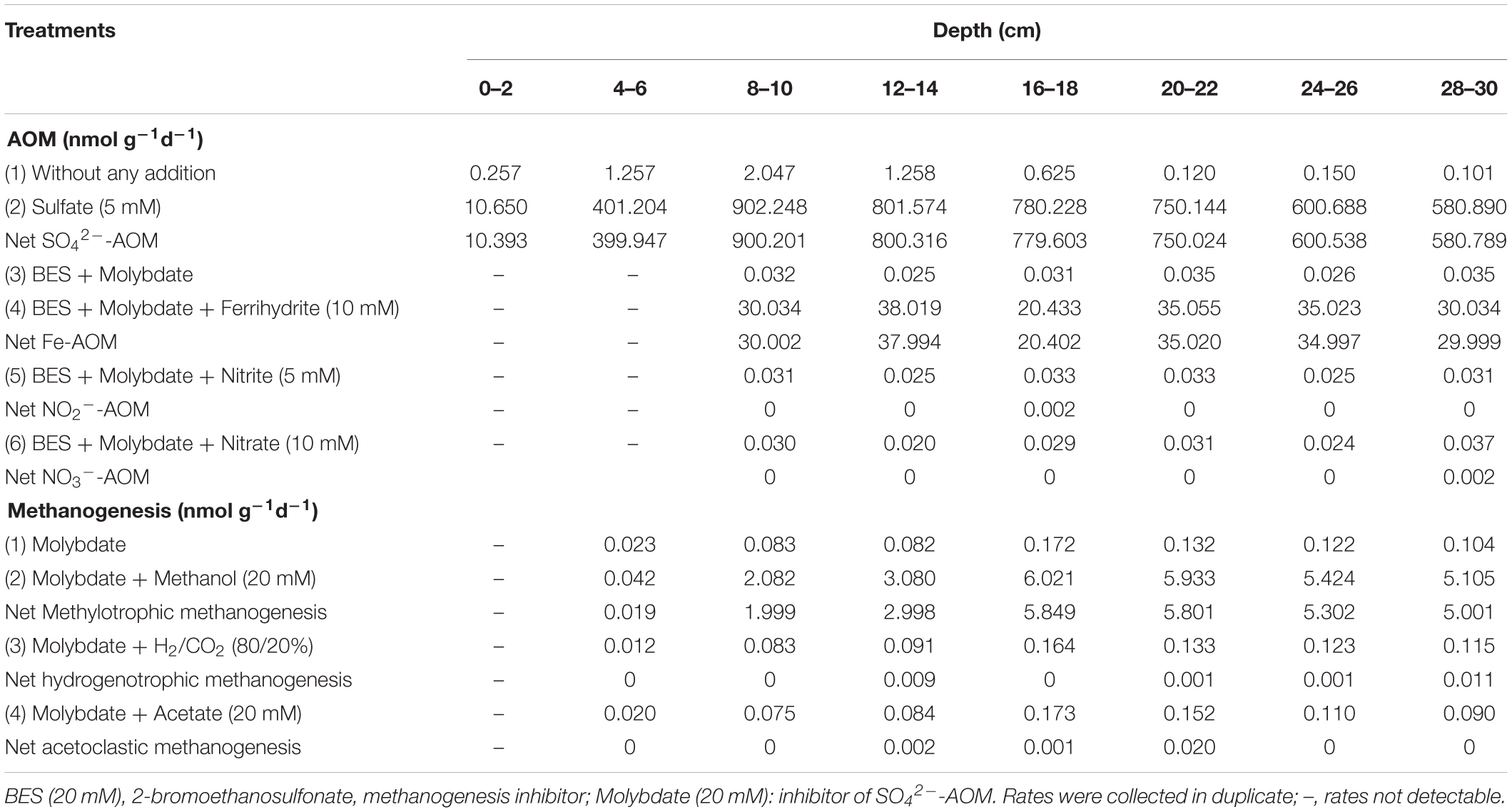
Methods for determining potential rates of AOM have been published previously (Segarra et al., 2015). For measurements of the potential anaerobic methane oxidation rate and mechanism, 5 g of sediment from a different depth was added to 120 ml serum bottles containing 50 ml of artificial mineral medium, with different treatments (see Table 2): (1) 5 mM sulfate, (2) 10 mM Ferrihydrite + 20 mM molybdate, (3) 5 mM Nitrite + 20 mM molybdate + 20 mM 2-bromoethanosulfonate (BES), (4) 10 mM Nitrite + 20 mM molybdate + 20 mM BES, (5) 20 mM molybdate + 20 mM BES, (6) Without any additions. Molybdate as inhibitor for sulfate reduction and BES as inhibitor for methanogenesis (Chidthaisong and Conrad, 2000; Nauhaus et al., 2005) were added. Another reason for BES not inhibiting the ANME archaea (or only partially inhibiting them) could be the formation of syntrophic clusters, especially by ANME-2 archaea, that are not fully permeated by BES (Dekas et al., 2009; Haroon et al., 2013). Negative controls were incubated in parallel for each process within each depth interval so that live samples could be corrected for this ‘background’ activity. Ferrihydrite was synthesized according to protocols described previously (Cai et al., 2018). The serum bottles were sealed with butyl rubber stoppers, crimp-capped, degassed with N2, and reduced with Na2S ⋅ 9H2O (0.5 g/L) and L-cysteine (0.5 g/L). The bottles were filled with 10% methane and incubated for 6 months at 4°C in the dark (Beal et al., 2009). Then, the accumulation of DIC was measured by a C/N elemental analyzer (N/C® 3100, Analytik Jena AG, Germany) as a response to AOM activity. Initial DIC concentrations in the medium and those at time-points DIC were determined.
Furthermore, the potential methane production rate and mechanism were assessed (Buckley et al., 2008; Xiao et al., 2018). Five grams of different depth sediments were added to 120 ml serum bottles containing 50 ml of artificial mineral medium with different treatments: (1) molybdate (20 mM), (2) hydrogen under H2/CO2 80/20 atmosphere + molybdate (20 mM), (3) acetate (20 mM) + molybdate (20 mM), and (4) methanol (20 mM) + molybdate (20 mM). Molybdate was used as an enzymatic inhibitor for sulfate reduction (Oremland and Capone, 1988). Samples with molybdate additions served as negative controls for sulfate-dependent methane oxidation. NaHCO3 was added to a concentration of 10 mM to make a buffer of CO2-HCO3–CO32– to maintain the pH of the medium around neutral. Measurements indicated that the pH was in the range of 7.0–7.2 at the start of the experiment. The serum bottles were sealed with butyl rubber stoppers, crimp-capped, degassed with N2, and reduced with Na2S ⋅ 9H20 (0.5 g/L) and L-cysteine (0.5 g/L). Then, the sediments were incubated for 6 months at 4°C in the dark. Methane production was measured by sampling the headspace of the serum vials by syringe and analyzing with GC-FID (Agilent, United States), using manual injection of 100 μl of headspace gas (Buckley et al., 2008). The needles used for sampling the serum vial headspace were flushed with N2 prior to sampling to prevent oxygen intrusion into the serum vials.
The artificial mineral medium contained (per 1 L): 23 g NaCl, 0.0136 g KH2PO4, 0.0535 g NH4Cl, 0.0147 g CaCl2 ⋅ 2H2O, 0.0204 g MgCl2 ⋅ 6H2O, 1 ml trace element solution (Könneke et al., 2005), and 1 ml vitamin solution (Könneke et al., 2005).
Results
Geochemistry Profiles
The geochemistry results showed that the concentrations of sulfate, methane, and DIC changed rapidly in the depth range of 0–6 cm (Figure 2). The concentration of sulfate decreased from nearly 26.2 mM to 0.43 μM, that of methane increased from 30 μM to 320 μM, and that of DIC increased from 2.20 to 18.34 mM. The rapid decrease in sulfate concentration as well as increase in DIC and methane concentrations indicate that the SMTZ in Jiaolong methane seep area is shallow, within 6 cm below the sediment-water interface. The concentration of pore water ferrous iron was the lowest in the surface sediment (2.6 μM), peaked at a depth of 12 cm (26.6 μM), and then decreased to 10 μM. The highest concentration of ferrous iron appeared below the SMTZ, where sulfate has been exhausted.
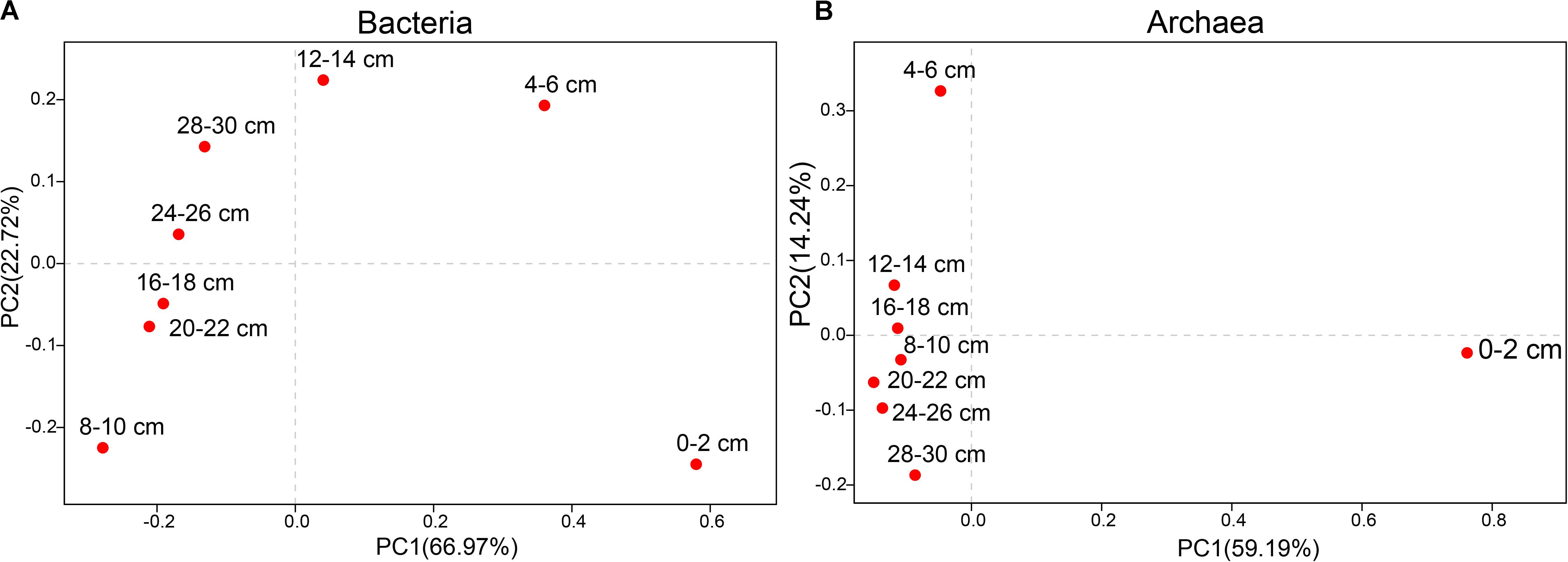
Figure 2. Principal coordinates analysis (PCoA) of different sediments at the genus level. (A) Bacteria. (B) Archaea.
Microbial Diversity
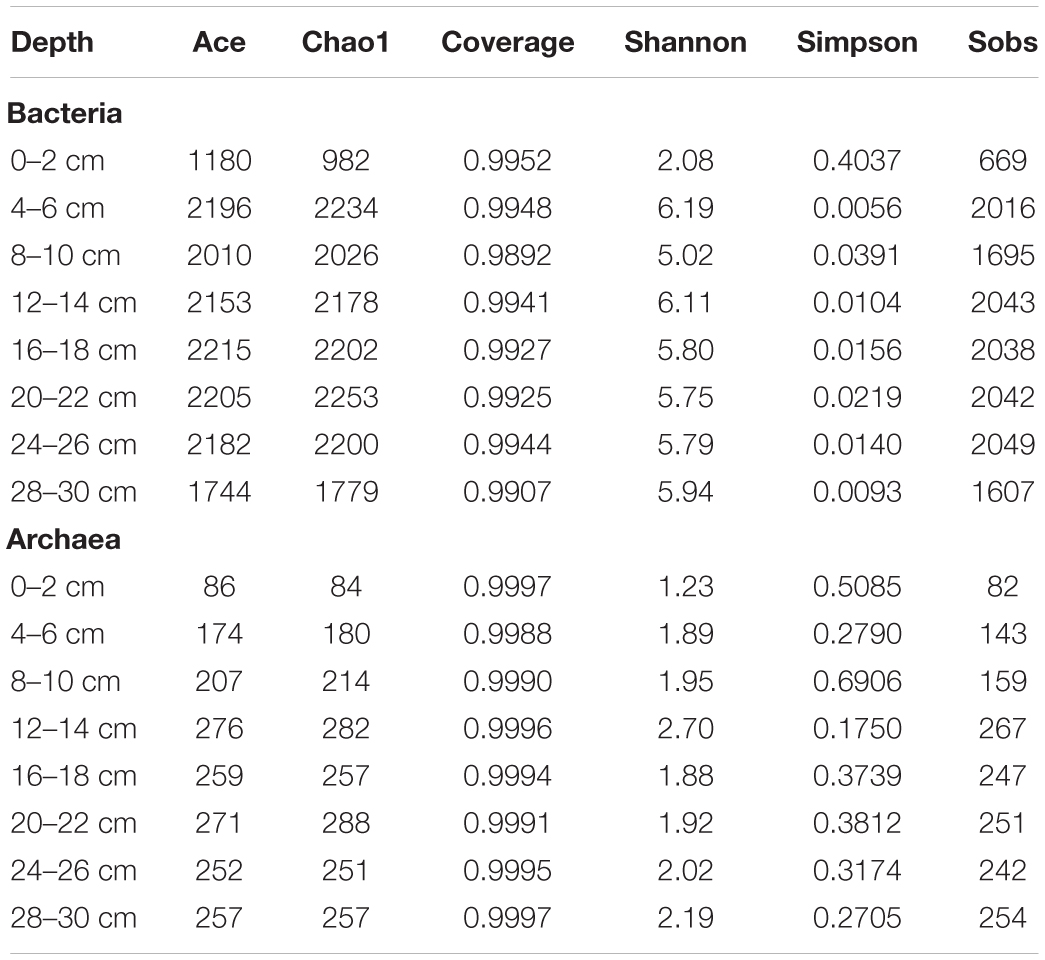
The vertical distribution of the microbial community was investigated in sediments from different depths (Table 3 and Supplementary Figure S1). In total, 848,371 high-quality sequences were generated from the samples (bacteria = 404662 sequences, archaea = 443709 sequences). At the 97% sequence identity level, 114480 OTUs were identified (bacteria = 57767 OTUs, archaea = 56713 OTUs). We found obvious differences in microbial structures among different sediment depths, and archaeal diversity was lower than bacterial diversity.
The PCoA of bacteria from all samples showed differences between surface sediments and deeper sediments (Figure 2). The species diversity was the lowest in surface sediments (Shannon: 2.08), and the highest species diversity was found at the depths of 4–6 cm and 12–14 cm (Shannon: 6.19 and 6.11, respectively). In surface sediments, most of the bacterial sequences were clustered in Sulfurovum (60.0179%), Methyloprofundus (11.1006%), and Sulfurimonas (3%). In the sediments at depths of 4–28 cm, the dominant groups belonged to SRB, including SEEP-SRB1 (12–28%), and Unclassified Desulfobulbaceae (8–20%) (Figures 3, 4). The relative abundance of SEEP-SRB4 was relatively low (0.07–0.4%). In addition, different kinds of IRB were detected including Shewanella (0.01–0.07%), Pseudomonas (0.01–0.1%), Desulfuromonas (0.11-0.92%), Geobacter (0.004–0.02%), and so on (Figure 4).
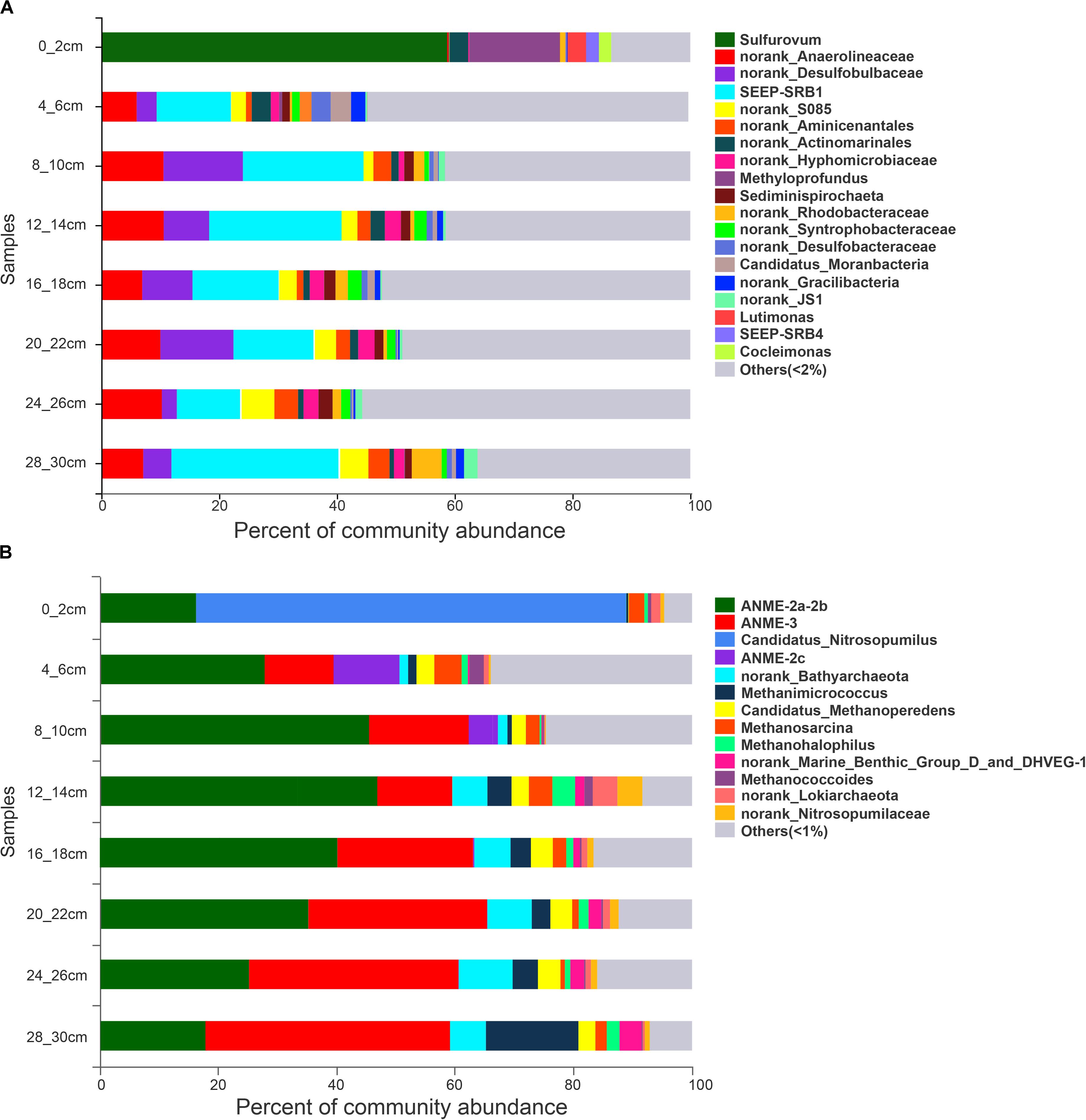
Figure 3. 16S rRNA gene sequence abundance at the genus level obtained from sediments at different depths. (A) bacteria. (B) archaea.
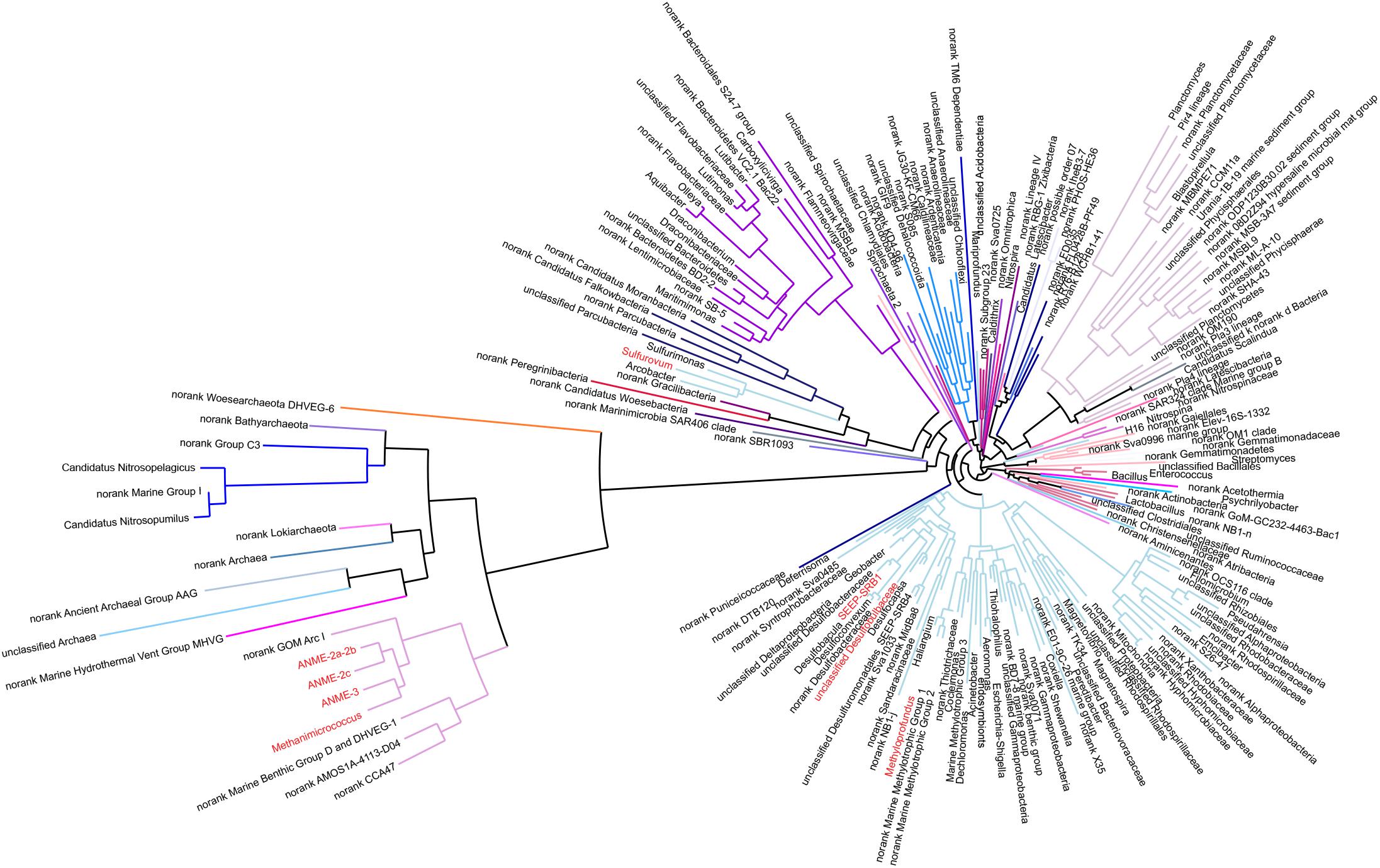
Figure 4. Phylogenetic relationships of 200 representative microbial sequences. Colored lines represent each phylum. The red-colored genus indicates dominant microbes, such as ANME-2a-2b, ANME-2c, ANME-3, Methanimicrococcus, Sulfurovum, SEEP-SRB1, Methyloprofundus etc.
The PCoA of archaea from all samples showed differences between sediments at depths of 0–6 cm and deeper sediments (Figure 2). The species diversity was the lowest in surface sediments (Shannon: 1.23), and the highest species diversity was found at a depth of 12–14 cm (Shannon: 2.70). In surface sediments, the dominant sequence cluster included mainly Nitrosopumilus (70.65%) and ANME-2a/b (18.84%). As the depth of sediment increased, Nitrosopumilus rapidly disappeared, while ANME-2a/b became more abundant, peaking (44.5%) at the depth of 8-10 cm, and then decreased. ANME-3 appeared at the depth of 4–6 cm, and its abundance increased with depth. ANME-2a/b was replaced by ANME-3 at the depth of 20–22 cm. ANME-2c appeared mainly in sediments at depths of 4–6 cm (Figure 3B). Methanogenic archaea were dominated by Methanimicrococcus at depths of 4–30 cm, whose abundance gradually increased from 0.20% to 18.00% with depth, with the highest abundance in the deepest sediments (28–30 cm) (Figures 3, 4). Furthermore, low abundance of methanogens below 4 cm depth was also detected, such as that of norank_Bathyarchaeota (0.01–7%), Methanosphaera (0.0014–0.0024%), Methanomethylovorans (0.0014–0.0076%), Methanosalsum (0.0014–0.0024%), Methanolobus (0.0178–0.0855%), Methanohalophilus (0.31–3.83%), Methanococcoides (0.09–2.34%), Methermicoccaceae (0.004–0.05%), and Methanomassiliicoccus (0.0051–0.0136%) (Figures 3, 4).
Relative Cell Distribution and Abundance of Microorganisms
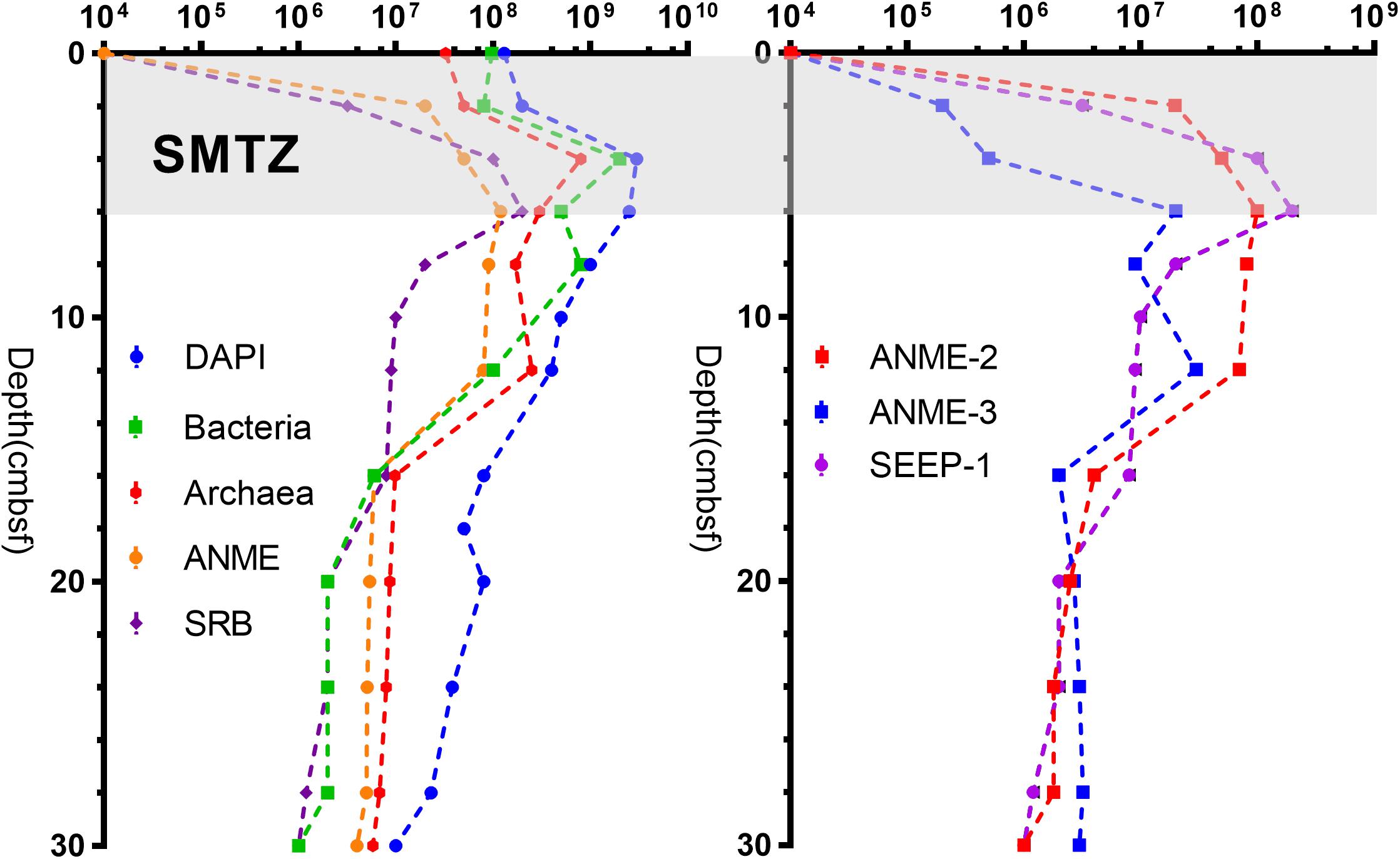
As shown in Figure 5, the distribution of bacteria, archaea, ANME, and SRB were determined by CARD-FISH. In surface sediments, the total number of cells was 1.3 × 108 cells g–1 (3.25 × 107 cells g–1 for archaea and 9.7 × 107 cells g–1 for bacteria) with bacteria as the dominant type. The numbers of SRB and ANME were below the detection limit. At the depth of 2–6 cm, the number of cells increased sharply, reaching 3.0 × 109 cells g–1 at a depth of 4 cm (2.0 × 109 cells g–1 for bacteria and 8.0 × 108 cells g–1 for archaea). Below 6 cm, the total number of cells decreased with increasing depth, reaching 1.0 × 107 cells g–1 at the bottom layer (1.1 × 106 cells g–1 for bacteria and 5.8 × 106 cells g–1 for archaea).
SEEP-SRB1 was the dominant group among SRB, and there was a relatively low concentration of SEEP-SRB4. ANME was dominated by ANME-2 and ANME-3. ANME-1, SEEP-SRB2, and SEEP-SRB3 were not detected by CARD-FISH. The numbers of SRB and ANME were positively correlated at the depths of 0–6 cm, both increasing with depth and peaking at a depth of 6 cm (2.0 × 108 cells g–1 for SRB and 1.4 × 108 cells g–1 for ANME). As the depth increased, the number of SRB gradually decreased to 1.0 × 106 cells g–1. The number of ANMEs was maintained at 9.0 × 107 cells g–1 at depths of 6–12 cm and gradually decreased to 4.1 × 106 cells g–1 at the bottom. Notably, there was no significant change in the abundance of ANME at depths of 6–12 cm, whereas the number of SRB decreased rapidly (Figure 5). In the surface and middle sediments, ANME-2 was dominant, while in the bottom sediments, ANME-3 was dominant (Figure 5).
Morphological studies showed that most of the SEEP-SRB and ANME formed conspicuous AOM consortia (Figure 6). The archaeal core consisted of ANME-2 and ANME-3, and the envelope consisted of SEEP-SRB1 or SEEP-SRB4. SEEP-SRB1 coupled most of ANME-2 and ANME-3, and only a small quantity of ANME-2 and ANME-3 were coupled by SEEP-SRB4. Moreover, we found SEEP-SRB and ANME single cells at different depths. Especially at depths of 10–14 cm, the numbers of ANME-2 and ANME-3 single cells were higher than those in other sediments.
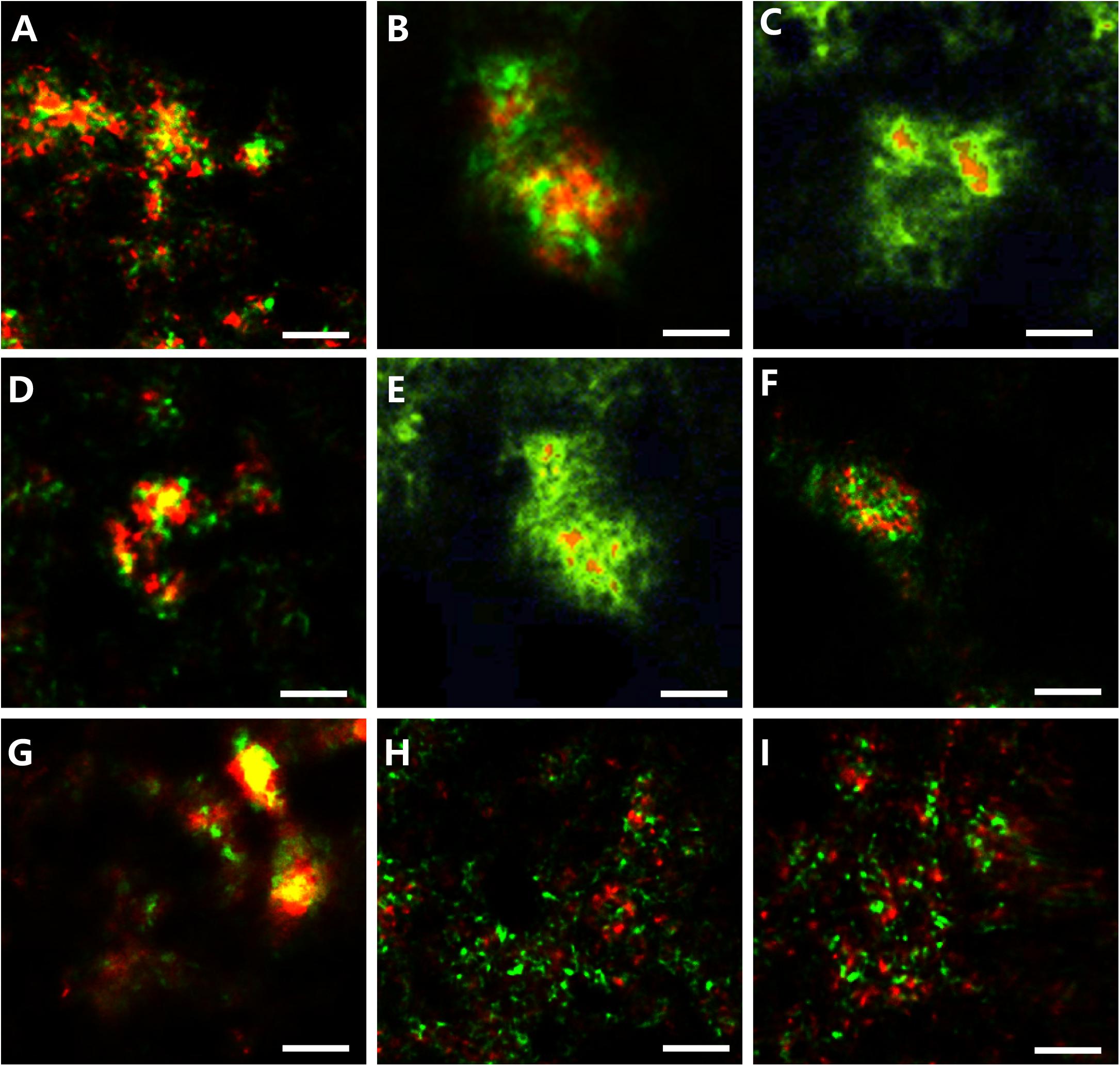
Figure 6. Photomicrographs of CARD-FISH stained samples. (A) Archaeal/bacterial aggregate labeled with ARC915 (red) and EUB338 (green) probes. (B,C) ANME-2 (red, ANME-2-538) and SRB1 (green, probe: SEEP-1a-1441). (D,E) ANME-3 (red, probe: ANME-3-1249) and SEEP-SRB1 (green, probe: SEEP1a-1441). (F) ANME-2 (red, probe: ANME-3-1249) and SEEP-SRB4 (green, probe: SEEP4-583). (G) ANME-3 (red, probe: ANME-3-1249) and SEEP-SRB4 (green, probe: SEEP4-583). (H) Monospecific aggregate of ANME-2 (red, probe: ANME2–538, green probe: EUB338). (I) Monospecific aggregate of ANME-3 (red, probe: ANME3-1249, green probe: EUB338).
Incubation Conditions of Methane-Metabolizing Microorganisms
AOM activities was assessed as shown in Table 2. With the addition of sulfate, the net rate of SO42–-AOM was 10.39 nmol cm–3 day–1 in surface sediments (0–2 cm), and the net rate of SO42–-AOM increased to its maximum of 900.20 nmol cm–3 day–1 at a depth of 8–10 cm, below which the rate decreased, reaching 580.79 nmol cm–3 day–1 at a depth of 28–30 cm.
In other experiments, sulfate reduction was inhibited with the addition of molybdate (20 mM), and methanogenesis was also inhibited by 20 mM BES. When ferrihydrite was supplied, the net Fe-AOM activity appeared below the SMTZ at a depth of 8–10 cm with its rate reaching 30.00 nmol cm–3 day–1. The highest rate of Fe-AOM was 37.99 nmol cm–3 day–1 at a depth of 12–14 cm where the concentration of ferrous iron was the highest (26.60 μM). Furthermore, no obvious increase in nitrite/nitrate-dependent AOM activity had been detected. These results suggested the potential existence of Fe-AOM below the SMTZ in the Jiaolong methane seep.
Sequencing data showed the coexistence of methanogenic archaea and ANME in the sediments, suggesting the cooccurrence of methanogenesis and AOM. Hence, an enrichment experiment was carried out to test our hypothesis. All cultures were incubated at 4°C for 6 months. The data showed that there was no methanogenic potential in the surface sediments. The efflux of methane appeared at the depth of 4–6 cm, with its rate reaching 0.02 nmol cm–3 day–1. The rate peaked at 0.17 nmol cm–3 day–1 at a depth of 16–18 cm, and then decreased to 0.10 nmol cm–3 day–1 at a depth of 28–30 cm (Table 2). Furthermore, methanol, H2/CO2, and acetate were added to the sediments. The results showed that there was no obvious increase in methane production with the addition of H2/CO2 and acetate. However, stimulation of methane production was detected after methanol was added. In the sediments with a depth of 16–18 cm, the net rate of methylotrophic methanogenesis reached its maximum (5.85 nmol cm–3 day–1).
Discussion
Niches and Diversity of Microbes in the Jiaolong Methane Seep
We compared the archaeal and bacterial diversity of the Jiaolong methane seep with those of 23 globally distributed methane seeps, and found that the microbial richness of the Jiaolong methane seep was at a medium level (Supplementary Figure S3). However, microbial abundance data showed that the Jiaolong methane seep hosted higher biomass (5.8 × 106–2.0 × 108 cells g–1 for archaea,1.1 × 106–2.0 × 109 cells g–1 for bacteria) than other methane seeps in the SCS, such as the Haima methane seep (2.8 × 104–3.4 × 106 cells g–1 for archaea, 4.5 × 105–7.4 × 106 cells g–1 for bacteria) (Niu et al., 2017), the GMGS2 gas hydrate station (1.3 × 104–2.7 × 106 cells g–1 for archaea, 3.8 × 104–1.0 × 107 cells g–1 for bacteria) (Cui et al., 2019), and the Haiyang 4 hydrate station (with total cells of 105–106 cells g–1) (the microbial abundance was converted from 16S rDNA qPCR data) (Zhang et al., 2012).
It was found that in the Jiaolong methane seep, the main microbes were SEEP-SRB1 coupled to ANME-2 and ANME-3, and a few SEEP-SRB4 coupled to ANME-2 and ANME-3. No ANME-1, SEEP-SRB2, or SEEP-SRB3 were observed in the sediments. In the surface layer, the dominant sequence cluster belonged mainly to Sulfurovum, Methyloprofundus, Sulfurimonas, Nitrosopumilus, and ANME-2a/b. In contrast, in the deepest sediments, the dominant sequence cluster belonged mainly to ANME-2, ANME-3, and Methanimicrococcus. The microbial diversity obtained in this study obviously differed from those in previous studies at methane seeps in the northern SCS. In June 2013, researchers found that in the Jiaolong methane seep, the majority of the microbial inhabitants at the surface layers (0–6 cm) were Sulfurimonas, Sulfurovum, and ANME-1, while SEEP-SRB1, ANME-1, and ANME-2 dominated the deepest layers (8–14 cm). The percentage of ANME-3 was the lowest in all layers, and SEEP-SRB3 and SEEP-SRB4 were not detected (Wu et al., 2018). At the Haiyang 4 hydrate station, only ANME-1 was detected, and the bacterial groups Chloroflexi and JS1 (Atribacteria) were dominant (Zhang et al., 2012). In the GMGS2 gas hydrate station, the majority groups were ANME-1b, ANME-2c, and bacterial group Desulfobacteraceae (Cui et al., 2019). In Haima active methane seep ecosystems in the southwestern SCS, ANME-2a/b was predominant in the upper and middle layers of the SMTZ, whereas ANME-1b outnumbered ANME-2 below the SMTZ, and ANME-3 was absent (Niu et al., 2017).
Methane seeps are island-like habitats, harboring distinct microbial communities (Ruff et al., 2015). The seep communities comprise bacteria and archaea that occur worldwide but are locally selected by the environment. For example, in situ temperature, methane concentration (Knittel et al., 2005), oxygen concentration (Meulepas et al., 2009), and sulfate concentration (Yanagawa et al., 2011) can significantly affect the ANME species. A total of >1800 ANME sequences have been reported by far including three types of ANME (ANME-1, -2, and -3) across physiochemically contrasting ecological niches. Among them, ANME-1 and ANME-2 are the most widely distributed in the world and tend to be coupled with syntrophic SEEP-SRB1 bacteria. ANME-1 preferentially grows in hydrogen sulfide-rich and sulfate-depleted environments, while ANME-2 is closely associated with sulfate concentration (Yanagawa et al., 2011) and preferentially grows in sulfate-rich areas. ANME-3 is distributed mainly in methane-seeping mud volcanoes and in some methane seeps (Niemann et al., 2006). Furthermore, in marine sediments, an ecological niche separation occurs where ANME-2a/b dominates the upper layers and ANME-2c and/or ANME-1 outcompetes in deeper zones (Timmers et al., 2017). Our results showed that geochemistry could be the primary force shaping the niche differentiation of functional microbial populations associated with methane-cycling in marine environment.
Iron-Mediated Anaerobic Oxidation of Methane
The results of incubation experiments (Table 2) showed both SO42–-AOM and Fe-AOM occurred in Jiaolong methane seep. Fe-AOM activity appeared below SMTZ at a depth of 8–10 cm, and potential net rates ranged from 20.40 to 37.99 nmol cm–3 day–1. Compared with net SO42–-AOM rates ranging from 10.39 to 900.20 nmol cm –3 day –1, net Fe-AOM rates were 10 times lower.
Our study is the first to discover Fe-AOM in the deep sea methane seeps of the SCS, and potential net rates are higher than those in freshwater and coastal sediments. The potential rate of Fe-AOM was 16.44 nmol cm–3 day–1 in Eel River Basin (Beal et al., 2009), 3.61 nmol cm–3 day–1 in brackish coastal sediments (Egger et al., 2015), 3.89 nmol cm–3 day–1 in coastal Georgia (Segarra et al., 2013), and 3.45 nmol cm–3 day–1 in deep lake sediment cores (Sivan et al., 2011). The higher rate of Fe-AOM in this study may result from the sufficient supply of iron oxide and massive methane flux. The slope of the northern SCS is one of the world’s most active areas of modern marine sedimentary processes (Huang and Wang, 2007; Luan et al., 2019). A large amount of river-borne terrigenous sediment input leads to exceptionally high amount of iron oxides in the sediments of the northern SCS (Zhang et al., 2007; Liu Z. F. et al., 2016; Liu et al., 2018). Previous research had suggested that different kinds of iron oxides could serve as electron acceptors for Fe-AOM (Bar-Or et al., 2017). In addition, a large amount of unconsumed methane was released into the bottom seawater (22.23 μM, unpublished data), implying a high methane flux in this area. Thus, the combination of these factors probably stimulated the enhanced rate of Fe-AOM processes in the Jiaolong methane seep.
To the best of our knowledge, there is no representative pure culture of Fe-AOM microorganisms from the marine sediment (Liang et al., 2019). But some microorganisms were suspected of being related to metal-AOM in various earlier studies, which suggested that ANME-1, ANME-3 (Beal et al., 2009), ANME-2a, 2c (Scheller et al., 2016), ANME-2d (Methanoperedens nitroreducens) (Ettwig et al., 2016; Shen et al., 2019), Candidatus Methanoperedens ferrireducens (Cai et al., 2018), or Methanosarcina acetivorans (Yan et al., 2018) might be involved in Fe-AOM. Furthermore, it is still possible that other unknown microorganisms perform metal-AOM. Bar-Or et al. (2017) findings highlight the essential role and participation of methanogens archaea and methanotrophic bacteria in the process of Fe-AOM.
In our study, sequencing data showed that the methanotrophic bacteria (such as Candidatus Methylomirabilis oxyfera, Methylobacter, Methylosarcina, Methylomonas, and Methylococcus) were not detected. Under the condition that 20 mM BES was added to inhibit methanogens archaea in the incubation experiments, the Fe-AOM activity appeared below SMTZ. These evidences excluded the potential participation of methanogenic archaea and methanotrophic bacteria in the process of Fe-AOM. CARD-FISH data showed that there was no significant change in the number of ANME at the depth of 6–12 cm, while the number of SRB decreased rapidly, and some ANME-2 and ANME-3 were not coupled with SRB (Figures 6H,I). These results were similar to those from methane seep enrichment samples of the Eel River Basin and Santa Monica Basin, which contained high abundances of ANME-2a and ANME-3 and could decouple the AOM process from SRB activities when metal compounds were added (Beal et al., 2009; Scheller et al., 2016). So we speculated that ANME-2 and ANME-3 were probably involved in Fe-AOM.
Different kinds of IRB were also detected in the Jiaolong seep sediments, such as Shewanella (0.01–0.07%), Geobacter (0.004–0.02%), Pseudomonas (0.01–0.1%), and Desulfuromonas (0.11–0.92%). Interestingly, the abundance of IRB was relatively higher in the 12–14 cm depth. To date, two potential ways of Fe-AOM were described in previous studies. (1) ANME oxidizes methane and transfer electrons directly to soluble metal ions or complexes, or solid metal oxides (Ettwig et al., 2016; Scheller et al., 2016); (2) ANME should be partnered with metal-reducing microorganisms to perform metal-AOM, in a way similar to the ANME-SRB consortia (Fu et al., 2016; He et al., 2018). It is worth further exploring whether ANME-2 or ANME-3 should be alone or coupled with IRB to perform Fe-AOM process in the Jiaolong methane seep. As the next step, we would like to use 14C-CH4 to enrich and cultivate methane-oxidizing microbial populations from the samples. Stable-isotope probing of active AOM would likely provide more hints on AOM metabolisms.
Furthermore, nitrite/nitrate-dependent AOM activity had not been detected in Jiaolong methane seep. Generally, nitrite-dependent AOM is performed by the NC10 bacteria related to Candidatus Methylomirabilis oxyfera, and nitrate-dependent AOM is performed by the ANME-2d (Ettwig et al., 2016). In our study, no sequences of Candidatus Methylomirabilis oxyfera and ANME-2d were detected. These results suggested that Fe-AOM was probably the dominant non-sulfate AOM pathway in the Jiaolong methane seep.
Methylotrophic Methanogenesis
Generally, there are three major methanogenic pathways: the acetoclastic, hydrogenotrophic, and methylotrophic pathways (Zhuang et al., 2018). Hydrogenotrophic and acetoclastic methanogenesis are often considered the primary pathways in marine deep sediment (Zhe et al., 2018). Based on thermodynamic laws, SRB and IRB outcompete methanogens for both acetate and hydrogen. Therefore, these methanogens are inhibited during active sulfate reduction and iron reduction (Reeburgh, 2007; Reiche et al., 2010; Zhou et al., 2014; Zhuang et al., 2018). In our study, hydrogenotrophic and acetoclastic methanogenesis were not detected due to active sulfate or iron reducing respiration in the Jiaolong methane seep. However, methylotrophic methanogenesis using non-competitive substrates (methanol) appeared below the 4–6 cm sediment layer, and coexisted with SO42–-AOM and Fe-AOM. Most methanogens clustered with the genus Methanimicrococcus. It is an obligatory methylotrophic methanogen, that is, it utilizes only non-competitive substrates, such as methanol or methylated compounds (Sprenger et al., 2000; Zeng et al., 2007; Niemann et al., 2009; Wang et al., 2019). Non-competitive substrates, such as methanol, trimethylamine, methylamines, dimethylsulfide, and dimethylsulfoniopropionate are ubiquitous in the marine environment (Oremland et al., 1982).
To date, research on the methylotrophic methanogenesis process has been intensively investigated on non-seep sediments, such as those in the Peruvian Margin (with methanol as the substrate, 0.6–1.95 nmol cm–3 d–1) (Maltby et al., 2016), Aarhus Bay (with methanol or trimethylamine as the substrate, 0.83–1.11 nmol cm–3 d–1) (Xiao et al., 2017, 2018), and the Western Mediterranean Sea (with methanol as the substrate, 0.03 nmol cm–3 d–1) (Zhuang et al., 2018). This process is poorly understood in deep sea methane seep ecosystems due to the technical challenges to discern methane production against the overall high background of net methane consumption in methane seeps ecosystem. To the best of our knowledge, there is only one report in the Sonora Margin cold seep showing methane production by Methanococcoides burtonii on non-competitive substrates (with trimethylamine as the substrate, 180∼560 pmol cm–3 day–1) (Vigneron et al., 2015) in shallow sediments above the SMTZ. The rate of methylotrophic methanogenesis in the Jiaolong methane seep reached a maximum of 5.85 nmol cm–3 day–1, which was higher than those observed in other areas. Future study is warranted to elucidate the thermal kinetics underlying this microbially mediated process.
In summary, the incubation experiments revealed the coexistence of sulfate-driven AOM, iron-driven AOM, and methylotrophic methanogenesis in Jiaolong methane seep sediments of the northern SCS where terrigenous sediments rich in iron oxide are imported in large quantities. Fe-AOM and methylotrophic methanogenesis are overlooked potential sources and sinks of methane in SCS methane cycle. Globally, large amounts of iron [∼730 Tg/year] from rivers are transported to ocean continental margins (Jickells et al., 2005), and methane seeps are common along continental margins in areas of high primary productivity and tectonic activity. How this methane cycle, which is affected by the large input of iron oxides, will influence the global carbon cycle is worthwhile to study further.
Data Availability Statement
The datasets generated for this study can be found in the NCBI, PRJNA574743 and PRJNA574745.
Author Contributions
HL, QY, and HZ designed the research and wrote the manuscript. HL performed the experiments and analyzed the data. All authors commented on the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC, No. 91428207).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly appreciate the efforts and assistance of the R/V Tan Kah Kee and ROPOS team. We also greatly appreciate the efforts and assistance of Dr. Zhongjun Jia (Chinese Academy of Sciences) and Xin Dai (Chinese Academy of Sciences) in revising our manuscript. We thank Dr. Hu Wang and Dr. Fuwu Ji (Tongji University) for collecting samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01409/full#supplementary-material
References
Bar-Or, I., Elvert, M., Ecker, W., Kushmaro, A., Vigderovich, H., Zhu, Q. Z., et al. (2017). Iron-coupled anaerobic oxidation of methane performed by a mixed bacterial-archaeal community based on poorly reactive minerals. Environ. Sci. Technol. 51, 12293–12301. doi: 10.1021/acs.est.7b03126
Beal, E. J., House, C. H., and Orphan, V. J. (2009). Manganese- and iron-dependent marine methane oxidation. Science 325, 184–187. doi: 10.1126/science.1169984
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Boetius, A., and Wenzhöfer, F. (2013). Seafloor oxygen consumption fuelled by methane from cold seeps. Nat. Geosci. 6, 725–734. doi: 10.1038/ngeo1926
Buckley, D. H., Baumgartner, L. K., and Visscher, P. T. (2008). Vertical distribution of methane metabolism in microbial mats of the great sippewissett salt marsh. Environ. Microbiol. 10, 967–977. doi: 10.1111/j.1462-2920.2007.01517.x
Cai, C., Leu, A. O., Xie, G. J., Guo, J., Feng, Y., Zhao, J. X., et al. (2018). A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME Journal 12, 1929–1939. doi: 10.1038/s41396-018-0109-x
Chidthaisong, A., and Conrad, R. (2000). Specificity of chloroform, 2-bromoethanesulfonate and fluoroacetate to inhibit methanogenesis and other anaerobic processes in anoxic rice field soil. Soil Biology & Biochemistry 32, 977–988. doi: 10.1016/s0038-0717(00)00006-7
Chronopoulou, P. M., Shelley, F., Pritchard, W. J., Maanoja, S. T., and Trimmer, M. (2017). Origin and fate of methane in the eastern tropical north pacific oxygen minimum zone. ISME Journal 11, 1386–1399. doi: 10.1038/ismej.2017.6
Cui, H., Su, X., Chen, F., Holland, M., Yang, S., Liang, J., et al. (2019). Microbial diversity of two cold seep systems in gas hydrate-bearing sediments in the South China Sea. Marine Environmental Ressearch 144, 230–239. doi: 10.1016/j.marenvres.2019.01.009
Dekas, A. E., Poretsky, R. S., and Orphan, V. J. (2009). Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426. doi: 10.1126/science.1178223
Dong, D., and Li, X. (2015). Galatheid and chirostylid crustaceans (Decapoda: Anomura) from a cold seep environment in the northeastern South China Sea. Zootaxa 4057, 91–105.
Egger, M., Hagens, M., Sapart, C. J., Dijkstra, N., Van Helmond, N. A. G. M., Mogollon, J. M., et al. (2017). Iron oxide reduction in methane-rich deep baltic sea sediments. Geochim. Cosmochim. Acta 207, 256–276. doi: 10.1016/j.gca.2017.03.019
Egger, M., Rasigraf, O., Sapart, C. J., Jilbert, T., Jetten, M. S. M., Rockmann, T., et al. (2015). Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ. Sci. Technol. 49, 277–283. doi: 10.1021/es503663z
Eickhorst, T., and Tippkötter, R. (2008). Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH). Soil Biol. Biochem. 40, 1883–1891. doi: 10.1016/j.soilbio.2008.03.024
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S., and Kartal, B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. U.S.A. 113, 12792–12796. doi: 10.1073/pnas.1609534113
Expedition 349 Scientists (2014). South China Sea tectonics: opening of the South China Sea and its implications for southeast Asian tectonics, climates, and deep mantle processes since the late Mesozoic. Int. Ocean Discov. Progr. Preliminary Rep. 349, 1–109.
Feng, D., Qiu, J.-W., Hu, Y., Peckmann, J., Guan, H., Tong, H., et al. (2018). Cold seep systems in the South China Sea: an overview. J. Asian Earth Sci. 168, 3–16. doi: 10.1016/j.jseaes.2018.09.021
Fu, L., Li, S. W., Ding, Z. W., Ding, J., Lu, Y. Z., and Zeng, R. J. (2016). Iron reduction in the DAMO/Shewanella oneidensis MR-1 coculture system and the fate of Fe(II). Water Res. 88, 808–815. doi: 10.1016/j.watres.2015.11.011
Gong, L., Li, X., and Qiu, J. W. (2015). Two new species of Hexactinellida (Porifera) from the South China Sea. Zootaxa 4034, 182–192.
Guan, H. X., Birgel, D., Peckmann, J., Liang, Q. Y., Feng, D., Yang, S. X., et al. (2018). Lipid biomarker patterns of authigenic carbonates reveal fluid composition and seepage intensity at Haima cold seeps. South China Sea. J. Asian Earth Sci. 168, 163–172. doi: 10.1016/j.jseaes.2018.04.035
Guan, H. X., Feng, D., Wu, N. Y., and Chen, D. F. (2016). Methane seepage intensities traced by biomarker patterns in authigenic carbonates from the South China Sea. Organic Geochem. 91, 109–119. doi: 10.1016/j.orggeochem.2015.11.007
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
He, Z., Zhang, Q., Feng, Y., Luo, H., Pan, X., and Gadd, G. M. (2018). Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci. Total Environ. 61, 759–768. doi: 10.1016/j.scitotenv.2017.08.140
Huang, W., and Wang, P. (2007). Statistics of sediment mass in the South China Sea: method and results. Front. Earth Sci. China 1:88–96. doi: 10.1007/s11707-007-0012-7
Jiang, H., Dong, H., Ji, S., Ye, Y., and Wu, N. (2007). Microbial diversity in the deep marine sediments from the Qiongdongnan Basin in South China Sea. Geomicrobiology Journal 24, 505–517. doi: 10.1080/01490450701572473
Jickells, T. D., An, Z. S., Andersen, K. K., Baker, A. R., Bergametti, G., Brooks, N., et al. (2005). Global iron connections between desert dust, ocean biogeochemistry, and climate. Science 308, 67–71. doi: 10.1126/science.1105959
Kleindienst, S., Ramette, A., Amann, R., and Knittel, K. (2012). Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 14, 2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Ann. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Knittel, K., Losekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/aem.71.1.467-479.2005
Könneke, M., Bernhard, A. E., José, R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Krüger, M., Treude, T., Wolters, H., Nauhaus, K., and Boetius, A. (2005). Microbial methane turnover in different marine habitats. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 6–17. doi: 10.1016/j.palaeo.2005.04.031
Liang, L., Wang, Y., Sivan, O., and Wang, F. (2019). Metal-dependent anaerobic methane oxidation in marine sediment: insights from marine settings and other systems. Sci. China Life Sci. 62, 1287–1295. doi: 10.1007/s11427-018-9554-5
Liu, C., Li, H., Zhang, Y., Si, D., and Chen, Q. (2016). Evolution of microbial community along with increasing solid concentration during high-solids anaerobic digestion of sewage sludge. Bioresour. Technolol. 216, 87–94. doi: 10.1016/j.biortech.2016.05.048
Liu, Z. F., Zhao, Y. L., Colin, C., Stattegger, K., Wiesner, M. G., Huh, C. A., et al. (2016). Source-to-sink transport processes of fluvial sediments in the South China Sea. Earth Sci. Rev. 153, 238–273. doi: 10.1016/j.earscirev.2015.08.005
Liu, J. R., Izon, G., Wang, J. S., Antler, G., Wang, Z., Zhao, J., et al. (2018). Vivianite formation in methane-rich deep-sea sediments from the South China Sea. Biogeosciences 15, 6329–6348. doi: 10.5194/bg-15-6329-2018
Luan, X., Ran, W., Wang, K., Wei, X., Shi, Y., and Zhang, H. (2019). New interpretation for the main sediment source of the rapidly deposited sediment drifts on the northern slope of the South China Sea. J. Asian Earth Sci. 171, 118–133. doi: 10.1016/j.jseaes.2018.11.004
Maltby, J., Sommer, S., Dale, A. W., and Treude, T. (2016). Microbial methanogenesis in the sulfate-reducing zone of surface sediments traversing the Peruvian margin. Biogeosciences 13, 283–299. doi: 10.5194/bg-13-283-2016
Manz, W., Amann, R., Ludwig, W., Wagner, M., and Schleifer, K. H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria - problems and solutions. Syst. Appl. Microbiol. 15, 593–600. doi: 10.1016/s0723-2020(11)80121-9
Meulepas, R. J. W., Jagersma, C. G., Gieteling, J., Buisman, C. J. N., Stams, A. J. M., and Lens, P. N. L. (2009). Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol. Bioeng. 104, 458–470. doi: 10.1002/bit.22412
Nauhaus, K., Treude, T., Boetius, A., and Kruger, M. (2005). Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7, 98–106. doi: 10.1111/j.1462-2920.2004.00669.x
Niemann, H., Fischer, D., Graffe, D., Knittel, K., Montiel, A., Heilmeyer, O., et al. (2009). Biogeochemistry of a low-activity cold seep in the Larsen B area, western Weddell Sea, Antarctica. Biogeosciences 6, 2383–2395. doi: 10.5194/bg-6-2383-2009
Niemann, H., Lösekann, T., De Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Niu, M., Fan, X., Zhuang, G., Liang, Q., and Wang, F. (2017). Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea. FEMS Microbiol. Ecol. 93:fix101.
Oda, Y., Sakamoto, M., Iwasaki, Y., Nagasaka, S., Ha, J.-Y., Chang, K.-H., et al. (2019). Inter-clonal variation in copper sensitivity in Bosmina longirostris with different exposure histories. Water Air Soil Pollut. 230:109.
Offre, P., Spang, A., and Schleper, C. (2013). Archaea in biogeochemical cycles. Ann. Rev. Microbiol. 67, 437–457. doi: 10.1146/annurev-micro-092412-155614
Oni, O., Miyatake, T., Kasten, S., Richter-Heitmann, T., Fischer, D., Wagenknecht, L., et al. (2015). Distinct microbial populations are tightly linked to the profile of dissolved iron in the methanic sediments of the Helgoland mud area. North Sea. Front. Microbiol. 6:365. doi: 10.3389/fmicb.2015.00365
Orcutt, B. N., Joye, S. B., Kleindienst, S., Knittel, K., Ramette, A., Reitz, A., et al. (2010). Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep Sea Res. Part II Top. Stud Oceanogr. 57, 2008–2021. doi: 10.1016/j.dsr2.2010.05.014
Oremland, R. S., and Capone, D. G. (1988). Use of “Specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10, 285–383. doi: 10.1007/978-1-4684-5409-3_8
Oremland, R. S., Marsh, L. M., and Polcin, S. (1982). Methane production and simultaneous sulfate reduction in anoxic, salt-marsh sediments. Nature 296, 143–145. doi: 10.1038/296143a0
Paull, C. K., Hecker, B., Commeau, R., Freeman-Lynde, R. P., Neumann, C., Corso, W. P., et al. (1984). Biological communities at the Florida escarpment resemble hydrothermal vent taxa. Science 226, 965–967. doi: 10.1126/science.226.4677.965
Pernthaler, A., Pernthaler, J., and Amann, R. (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101. doi: 10.1128/aem.68.6.3094-3101.2002
Raghoebarsing, A. A., Pol, A., Van De Pas-Schoonen, K. T., Smolders, A. J., Ettwig, K. F., Rijpstra, W. I., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921. doi: 10.1038/nature04617
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 38, 486–513. doi: 10.1021/cr050362v
Reiche, M., Torburg, G., and Küsel, K. (2010). Competition of Fe(III) reduction and methanogenesis in an acidic fen. FEMS Microbiol. Ecol. 65, 88–101. doi: 10.1111/j.1574-6941.2008.00523.x
Riedinger, N., Formolo, M. J., Lyons, T. W., Henkel, S., Beck, A., and Kasten, S. (2014). An inorganic geochemical argument for coupled anaerobic oxidation of methane and iron reduction in marine sediments. Geobiology 12, 172–181. doi: 10.1111/gbi.12077
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. U.S.A. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Scheller, S., Yu, H., Chadwick, G. L., Mcglynn, S. E., and Orphan, V. J. (2016). Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707.
Schreiber, L., Holler, T., Knittel, K., Meyerdierks, A., and Amann, R. (2010). Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ. Microbiol. 12, 2327–2340.
Seeberg-Elverfeldt, J., Schluter, M., Feseker, T., and Kolling, M. (2005). Rhizon sampling of porewaters near the sediment-water interface of aquatic systems. Limnol. Oceanogr. Methods 3, 361–371. doi: 10.4319/lom.2005.3.361
Segarra, K. E., Schubotz, F., Samarkin, V., Yoshinaga, M. Y., Hinrichs, K. U., and Joye, S. B. (2015). High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat. Commun. 6:7477.
Segarra, K. E. A., Comerford, C., Slaughter, J., and Joye, S. B. (2013). Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochim. Cosmochim. Acta 115, 15–30. doi: 10.1016/j.gca.2013.03.029
Sela-Adler, M., Ronen, Z., Herut, B., Antler, G., Vigderovich, H., Eckert, W., et al. (2017). Co-existence of methanogenesis and sulfate reduction with common substrates in sulfate-rich estuarine sediments. Front. Microbiol. 8:766. doi: 10.3389/fmicb.2017.00766
Shen, L. D., Ouyang, L., Zhu, Y., and Trimmer, M. (2019). Active pathways of anaerobic methane oxidation across contrasting riverbeds. ISME J. 13, 752–766. doi: 10.1038/s41396-018-0302-y
Sivan, O., Adler, M., Pearson, A., Gelman, F., Bar-Or, I., John, S. G., et al. (2011). Geochemical evidence for iron-mediated anaerobic oxidation of methane. Limnol. Oceanogr. 56, 1536–1544. doi: 10.4319/lo.2011.56.4.1536
Sivan, O., Antler, G., Turchyn, A. V., Marlow, J. J., and Orphan, V. J. (2014). Iron oxides stimulate sulfate-driven anaerobic methane oxidation in seeps. Proc. Natl. Acad. Sci. U.S.A. 111, 4139–4147.
Sprenger, W. W., Van Belzen, M. C., Rosenberg, J., Hackstein, J. H., and Keltjens, J. T. (2000). Methanomicrococcus blatticola gen. nov., sp. nov., a methanol- and methylamine-reducing methanogen from the hindgut of the cockroach Periplaneta americana. Int. J. Syst. Evol. Microbiol. 6, 1989–1999. doi: 10.1099/00207713-50-6-1989
Timmers, P. H., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S., and Stams, A. J. (2017). Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237.
Treude, T., Krause, S., Maltby, J., Dale, A. W., Coffin, R., and Hamdan, L. J. (2014). Sulfate reduction and methane oxidation activity below the sulfate-methane transition zone in Alaskan Beaufort Sea continental margin sediments: implications for deep sulfur cycling. Geochim.Cosmochim. Acta 144, 217–237. doi: 10.1016/j.gca.2014.08.018
Treude, T., Niggemann, J., Kallmeyer, J., Wintersteller, P., Schubert, C. J., Boetius, A., et al. (2005). Anaerobic oxidation of methane and sulfate reduction along the Chilean continental margin. Geochim.Cosmochim. Acta 69, 2767–2779. doi: 10.1016/j.gca.2005.01.002
Valentine, D. L. (2011). Emerging topics in marine methane biogeochemistry. Ann. Rev. Mar. Sci. 3, 147–171. doi: 10.1146/annurev-marine-120709-142734
Vigderovich, H., Liang, L., Herut, B., Wang, F., Wurgaft, E., Rubin-Blum, M., et al. (2019). Evidence for microbial iron reduction in the methanic sediments of the oligotrophic southeastern Mediterranean continental shelf. Biogeosciences 16, 3165–3181. doi: 10.5194/bg-16-3165-2019
Vigneron, A., L’haridon, S., Godfroy, A., Roussel, E. G., Cragg, B. A., Parkes, R. J., et al. (2015). Evidence of active methanogen communities in shallow sediments of the Sonora Margin cold seeps. Appl. and Environ. Microbiol. 81, 3451–3459. doi: 10.1128/aem.00147-15
Wallner, G., Amann, R., and Beisker, W. (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143. doi: 10.1002/cyto.990140205
Wang, B., Liu, F., Zheng, S., and Hao, Q. (2019). Trophic strategy of diverse methanogens across a river-to-sea gradient. J. Microbiol. 57, 470–478. doi: 10.1007/s12275-019-8482-3
Wu, Y., Qiu, J. W., Qian, P. Y., and Wang, Y. (2018). The vertical distribution of prokaryotes in the surface sediment of Jiaolong cold seep at the northern South China Sea. Extremophiles 22, 1–12.
Xiao, K. Q., Beulig, F., Kjeldsen, K. U., Jørgensen, B. B., and Risgaard-Petersen, N. (2017). Concurrent methane production and oxidation in surface sediment from Aarhus Bay, Denmark. Front. Microbiol. 8:1198. doi: 10.3389/fmicb.2017.01198
Xiao, K. Q., Beulig, F., Røy, H., Jørgensen, B. B., and Risgaard-Petersen, N. (2018). Methylotrophic methanogenesis fuels cryptic methane cycling in marine surface sediment. Limno. Oceanogr. 63, 1519–1527. doi: 10.1002/lno.10788
Xu, N., Tan, G. C., Wang, H. Y., and Gai, X. P. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Yan, Z., Joshi, P., Gorski, C. A., and Ferry, J. G. (2018). A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 9:1642.
Yanagawa, K., Sunamura, M., Lever, M. A., Morono, Y., Hiruta, A., Ishizaki, O., et al. (2011). Niche separation of methanotrophic archaea (ANME-1 and-2) in methane-seep sediments of the eastern japan sea offshore joetsu. Geomicrobiol. J. 28, 118–129. doi: 10.1080/01490451003709334
Zeng, Y., Li, H., and Jiao, N. (2007). Phylogenetic diversity of planktonic archaea in the estuarine region of East China Sea. Microbiol. Res. 162, 26–36. doi: 10.1016/j.micres.2006.03.007
Zhang, Y., Su, X., Chen, F., Wang, Y. Y., Jiao, L., Dong, H. L., et al. (2012). Microbial diversity in cold seep sediments from the northern South China Sea. Geosci. Front. 3, 301–316. doi: 10.1016/j.gsf.2011.11.014
Zhang, Y. G., Ji, J., Balsam, W. L., Liu, L., and Chen, J. (2007). High resolution hematite and goethite records from ODP 1143, South China Sea: co-evolution of monsoonal precipitation and El Niño over the past 600,000 years. Earth Planet. Sci. Lett. 264, 136–150. doi: 10.1016/j.epsl.2007.09.022
Zhe, L., Shao, N., Akinyemi, T., and Whitman, W. B. (2018). Methanogenesis. Curr. Biol. 28, 727–732.
Zhou, J., Bruns, M. A., and Tiedje, J. M. (1996). DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322. doi: 10.1128/aem.62.2.316-322.1996
Zhou, S., Xu, J., Yang, G., and Zhuang, L. (2014). Methanogenesis affected by the co-occurrence of iron(III) oxides and humic substances. FEMS Microbiol. Ecol. 88, 107–120. doi: 10.1111/1574-6941.12274
Zhuang, G. C., Elling, F. J., Nigro, L. M., Samarkin, V., Joye, S. B., Teske, A., et al. (2016). Multiple evidence for methylotrophic methanogenesis as the dominant methanogenic pathway in hypersaline sediments from the Orca Basin. Gulf of Mexico. Geochim. Cosmochim. Acta 187, 1–20. doi: 10.1016/j.gca.2016.05.005
Keywords: methane seeps, South China Sea, anaerobic oxidation of methane, methylotrophic methanogenesis, sulfate reduction, iron reduction
Citation: Li H, Yang Q and Zhou H (2020) Niche Differentiation of Sulfate- and Iron-Dependent Anaerobic Methane Oxidation and Methylotrophic Methanogenesis in Deep Sea Methane Seeps. Front. Microbiol. 11:1409. doi: 10.3389/fmicb.2020.01409
Received: 01 January 2020; Accepted: 29 May 2020;
Published: 08 July 2020.
Edited by:
Ricardo Amils, Autonomous University of Madrid, SpainReviewed by:
Aharon Oren, The Hebrew University of Jerusalem, IsraelAndreas Teske, The University of North Carolina at Chapel Hill, United States
Copyright © 2020 Li, Yang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qunhui Yang, eWFuZ3FoQHRvbmdqaS5lZHUuY24=; Huaiyang Zhou, emhvdWh5QHRvbmdqaS5lZHUuY24=
 Haizhou Li
Haizhou Li Qunhui Yang
Qunhui Yang Huaiyang Zhou
Huaiyang Zhou